Abstract
The switch of pyruvate kinase (PK) M1 to PKM2 is pivotal for glucose metabolism in cancers. The PKM1/M2 shift is controlled by the alternative splicing of two mutually exclusive exons in the PKM gene. PKM1 is expressed in differentiated tissues, whereas PKM2 is expressed in cancer tissues. This study revealed that the haplodeficiency of FUSE-binding protein (FBP)-interacting repressor (FIR), a transcriptional repressor of the c-myc gene, contributed to the splicing of PKM1 to PKM2 in mice thymic lymphoma and/or T-cell type acute lymphoblastic leukemia (T-ALL) using six-plex tandem mass tag (TMT) quantitative proteomic analysis. TMT revealed 648 proteins that were up- or downregulated in mice thymic lymphoma tissues compared with wild type mouse. These proteins included transcription factors and proteins involved in DNA damage repair, DNA replication, T-cell activation/proliferation, apoptosis, etc. Among them, PKM2 protein, but not PKM1, was upregulated in the thymic lymphoma as well as T-ALL. Using qRT-PCR, we revealed that the activation of PKM2 mRNA was higher in thymic lymphoma cells of FIR+/−TP53−/− mice than that in control lymphocytes of FIR+/+TP53−/− sorted by flow cytometry. FIR knockdown by siRNA suppressed hnRNPA1 expression in HeLa cells. These results indicated that FIR haplodeficiency contributes the alternative splicing of PKM1 to PKM2 by partly inhibiting hnRNPA1 expression in the thymic lymphoma cells prior to T-ALL. Taken together, our findings suggest that FIR and its related spliceosomes are potential therapeutic targets for cancers, including T-ALL.
Keywords: FUSE-binding protein-interacting repressor, LC-MS/MS, pyruvate kinase M2, six-plex tandem mass tags labeling, thymic lymphoma
INTRODUCTION
Pyruvate kinase (PK) M2 is activated in cancers through alternative splicing. PKM2 activation supports aerobic glycolysis: the so-called “Warburg effect” [1]. The PKM1/M2 splicing switch is affected by many factors, including c-Myc [2]; however, the mechanism of the PKM1 switch to PKM2 in carcinogenesis is largely unclear. This study revealed that the c-myc regulator FBP-interacting receptor (FIR) affects the splicing of PKM1 to PKM2, at least in part, in mice, as shown by six-plex tandem mass tag (TMT) quantitative proteomic analysis. c-Myc is a transcription factor that has several functions, including cell proliferation, differentiation, tumorigenesis, apoptosis, and cell cycle control [3]. The activation of c-Myc is frequently observed in many malignancies and places a burden on RNA and protein synthesis as well as affects metabolism even during pro-tumor stages [4]. Myc-stress in particular may affect alternative splicing through spliceosome complexes in cancers [4, 5]; therefore, core spliceosomes are a promising target for the treatment of c-Myc-driven cancers [4, 5, 6]. FIR is a c-myc gene transcriptional suppressor and binds to FUSE-binding protein (FBP), which is a transcription factor essential for the c-myc gene. FIR represses c-myc gene transcription by suppressing TFIIH/p89/XPB helicase [7]. FIR⊿exon2 is an alternatively spliced variant of FIR that lacks exon2, which includes the transcriptional repression domain [8]. FIR⊿exon2 was recently reported to be activated in different cancers and functions as a dominant negative form of FIR, potentially inducing c-Myc in colorectal cancer tissues [8]. In addition, disturbed alternative splicing of FIR increased c-myc gene transcription [9] in hepatocellular carcinoma [10] and non-small cell lung cancer [11]. FIR has also been co-immunoprecipitated with spliceosome complex proteins, including SAP155 (SF3B1) and hnRNPA1 [12], whereas SAP155 is required for pre-mRNA splicing of FIR [12, 13]. Mutations of SAP155 are associated with various cancers, including hematopoietic malignancies [14], whereas FIR+/−TP53−/− mice generated thymic lymphoma/T-call type acute lymphoblastic leukemia (T-ALL) [15]. In this study, comparative protein profiles were examined in thymic lymphomas of FIR+/−TP53−/− mice by six-plex TMT analysis to investigate the molecular mechanisms of tumor development. TMT is a useful gel-free tool for comparative quantitative proteomics and biomarker identification in body fluids such as sera and gingival fluid [17–19]. Notably, FIR haplodeficiency promoted the splicing of PKM1 to PKM2 in mice thymic lymphoma without circulating tumor cells/bone marrow invasion revealed by flow cytometry analysis through potentially inhibiting hnRNPA1 expression.
RESULTS
Proteins preparation for TMT analysis of FIR+/−TP53−/− mouse thymic lymphoma tissues
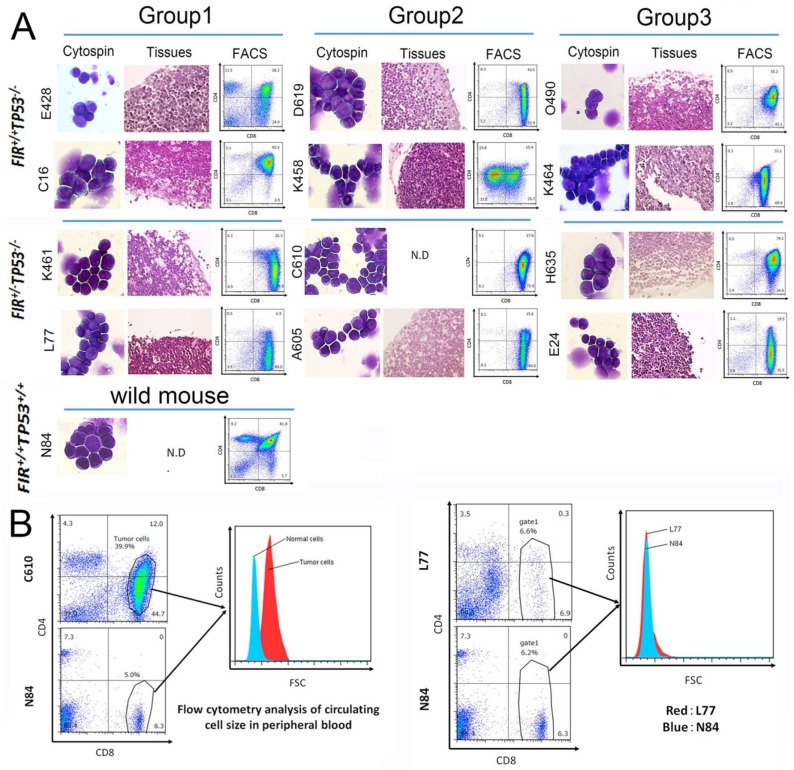
Thirteen thymic lymphoma tissues, (one FIR+/+TP53+/+, six FIR+/+TP53−/−, and six FIR+/−TP53−/−) were used for six-plex TMT analysis (Table 1). Histopathologic and flow cytometry of tumor cells were indicate (Figure 1A). Cytospin preparation and flow cytometry analysis revealed that more than 10% of T-ALL cells were observed in bone marrows of two FIR+/−TP53−/− mice and one FIR+/+TP53−/− mouse (D619, C610 and H635 in Table 1, Figure 1A). Mice were diagnosed as thymic lymphoma in case no circulating tumor cells were detected whereas T-ALL with circulating tumor cells revealed by flow cytometry analysis (Figure 1B, Supplementary Figure 1). Six different samples can be compared in a single experiment using TMT to identify and quantity proteins by MS/MS. Samples were divided into three groups for TMT analysis. Six samples can be labeled simultaneously in our TMT protocol, therefore six FIR+/+TP53−/− and FIR+/−TP53−/−samples were divided into three groups of two. Normal thymus tissue samples from FIR+/+TP53+/+ mice were analyzed as internal controls (Table 1, Figure 2A).
Table 1. Characteristics of the mice used in this study.
| Mouse no. | Genotype | Phenotype | Surface marker | TMT label reagent | Thymus weight (g) | Bone marrow invasion (%) | |
|---|---|---|---|---|---|---|---|
| Wild | N84 | FIR+/+TP53+/+ | Wild | — | TMT6-126 | 0.07 | N.D. |
| Group 1 | E428 | FIR+/+ TP53−/− | Thymic lymphoma | CD4low+/CD8+ | TMT6-128 | 0.96 | 1.1 |
| C16 | Leukemia/thymic lymphoma | CD4+/CD8+ | TMT6-129 | 0.71 | N.D. | ||
| K461 | FIR+/− TP53−/− | Thymic lymphoma | CD4low+/CD8+ | TMT6-130 | 0.80 | N.D. | |
| L77 | Thymic lymphoma | CD4low+/CD8+ | TMT6-131 | 1.17 | N.D. | ||
| Group 2 | D619 | FIR+/+ TP53−/− | Leukemia/thymic lymphoma | CD4low+/CD8+ | TMT6-128 | 1.12 | 61.2 |
| K458 | Leukemia/thymic lymphoma | CD4+/CD8low+ | TMT6-129 | 0.29 | 5.0 | ||
| C610 | FIR+/− TP53−/− | Leukemia/thymic lymphoma | CD4low+/CD8+ | TMT6-130 | 1.20 | 42.6 | |
| A605 | Leukemia/thymic lymphoma | CD4low+/CD8+ | TMT6-131 | 0.90 | N.D. | ||
| Group 3 | O490 | FIR+/+ TP53−/− | Leukemia/thymic lymphoma | CD4+/CD8+ | TMT6-128 | 0.26 | N.D. |
| K464 | Thymic lymphoma | CD4low+/CD8+ | TMT6-129 | 0.88 | 6.7 | ||
| H635 | FIR+/− TP53−/− | Leukemia/thymic lymphoma | CD4+/CD8+ | TMT6-130 | 1.01 | 22.6 | |
| E24 | Thymic lymphoma | CD4low+/CD8+ | TMT6-131 | 1.57 | N.D. |
Figure 1. Features of thymic lymphoma in FIR+/+ TP53−/− and FIR+/− TP53−/− groups using TMT analysis.
(A) Histologic features of thymic lymphoma in FIR+/+ TP53−/− (E428, C16, D619, K458, O490, and K464) and FIR+/− TP53−/− (K461, L77, C610, A605, H635 and E24). Cytospin preparation and flow cytometry analysis of thymic lymphoma cells and hematoxylin and eosin (H&E) staining of paraffin-embedded thymic lymphoma tissues. TMT analyses were performed in three groups. (B) Flow cytometry analysis indicated the average circulating cell size by FSC (Forward scatter) at x-axis. The average circulating cell size of T-ALL mouse (C610) was larger but that of thymic lymoma mouse (L77) was as same as that of normal mouse (N84), indicated by gated area.
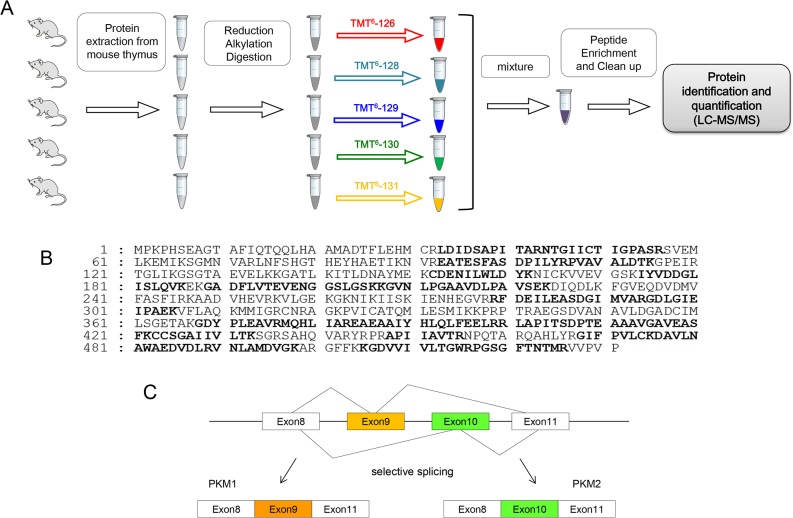
Figure 2.
(A) Schematic diagram of experimental protocol. (B) Identification of PKM2 by LC-MS/MS. The amino acid sequence of PKM2 is shown. Matching peptide sequences are printed in bold. (C) Schematic diagram of PKM splicing.
One FIR+/+TP53+/+ sample was labeled with reporter ions at m/z = 126, two FIR+/+TP53−/− samples were labeled with m/z = 127, 128, and two FIR+/−TP53−/− samples were labeled with m/z = 129, 130, respectively (Figure 2A). Proteomic analysis of these three groups revealed 1098, 1074, 1052 proteins, respectively, 648 of which were common (Supplementary Table 3). Of these, 45 proteins were quantified 1.5 times higher/lower in FIR+/−TP53−/− samples compared with normal FIR+/+TP53+/+ (Table 2, Supplementary Table 4). Identified proteins were involved in several functions, including transcriptional control, DNA repair, DNA replication, T-cell activation, T-cell proliferation, and apoptosis induction. Furthermore, hemoglobin, serum albumin, and keratin samples from the mice were included (Table 2). Five proteins were up/downregulated differently, and they included hemopexin, 60S ribosomal protein L29, fibronectin, spermidine synthase, and keratin, type II cytoskeletal 1.
Table 2. Up- or downregulated proteins identified by LC-MS/MS in T-ALL mice (FIR+/−TP53−/− and FIR+/+TP53−/−) compared to wild type mice (FIR+/+TP53+/+).
| Accession | Description | # Amino acids | MW [kDa] | T-ALL mice / wild type | Number of samples examined by TMT | |||
|---|---|---|---|---|---|---|---|---|
| More than 1.5 times-upregulated | Less than 0.67 times-downregulated | |||||||
| 1 | ↑ | P12265 | Beta-glucuronidase | 648 | 74.2 | 2.19 | 12 | 0 |
| 2 | ↑↓ | Q91×72 | Hemopexin | 460 | 51.3 | 2.07 | 10 | 1 |
| 3 | ↑ | Q9DCA5 | Ribosome biogenesis protein BRX1 homolog | 353 | 41.2 | 1.92 | 9 | 0 |
| 4 | ↑ | P47911 | 60S ribosomal protein L6 | 296 | 33.5 | 1.86 | 10 | 0 |
| 5 | ↑↓ | P47915 | 60S ribosomal protein L29 | 160 | 17.6 | 1.77 | 8 | 1 |
| 6 | ↑ | P52480 | Pyruvate kinase isozymes M1/M2 | 531 | 57.8 | 1.75 | 10 | 0 |
| 7 | ↑↓ | P11276 | Fibronectin | 2477 | 272.3 | 1.53 | 5 | 1 |
| 8 | ↑ | Q6ZQL4 | WD repeat-containing protein 43 | 677 | 75.3 | 1.52 | 5 | 0 |
| 9 | ↑↓ | Q64674 | Spermidine synthase | 302 | 34.0 | 1.34 | 5 | 3 |
| 10 | ↓ | P07724 | Serum albumin | 608 | 68.6 | 1.05 | 2 | 2 |
| 11 | ↓ | P16045 | Galectin-1 | 135 | 14.9 | 0.74 | 0 | 6 |
| 12 | ↓ | Q00896 | Alpha-1-antitrypsin 1-3 | 412 | 45.8 | 0.73 | 1 | 6 |
| 13 | ↓ | Q05816 | Fatty acid-binding protein, epidermal | 135 | 15.1 | 0.69 | 0 | 7 |
| 14 | ↓ | Q61233 | Plastin-2 | 627 | 70.1 | 0.68 | 0 | 7 |
| 15 | ↓ | P51885 | Lumican | 338 | 38.2 | 0.68 | 0 | 5 |
| 16 | ↑↓ | P04104 | Keratin, type II cytoskeletal 1 | 637 | 65.6 | 0.68 | 1 | 7 |
| 17 | ↓ | Q64105 | Sepiapterin reductase | 261 | 27.9 | 0.62 | 0 | 9 |
| 18 | ↓ | O88531 | Palmitoyl-protein thioesterase 1 | 306 | 34.5 | 0.62 | 0 | 7 |
| 19 | ↓ | Q8CGP6 | Histone H2A type 1-H | 128 | 13.9 | 0.61 | 0 | 7 |
| 20 | ↓ | Q9EPB4 | Apoptosis-associated speck-like protein containing a CARD | 193 | 21.4 | 0.57 | 0 | 9 |
| 21 | ↓ | Q60611 | DNA-binding protein SATB1 | 764 | 85.8 | 0.54 | 0 | 11 |
| 22 | ↓ | P30681 | High mobility group protein B2 | 210 | 24.1 | 0.54 | 0 | 10 |
| 23 | ↓ | P01896 | H-2 class I histocompatibility antigen, alpha chain (Fragment) | 185 | 20.4 | 0.52 | 0 | 11 |
| 24 | ↓ | Q61599 | Rho GDP-dissociation inhibitor 2 | 200 | 22.8 | 0.52 | 0 | 10 |
| 25 | ↓ | P99029 | Peroxiredoxin-5, mitochondrial | 210 | 21.9 | 0.51 | 0 | 11 |
| 26 | ↓ | P07356 | Annexin A2 | 339 | 38.7 | 0.51 | 0 | 10 |
| 27 | ↓ | P01942 | Hemoglobin subunit alpha | 142 | 15.1 | 0.51 | 0 | 9 |
| 28 | ↓ | P01831 | Thy-1 membrane glycoprotein | 162 | 18.1 | 0.50 | 0 | 9 |
| 29 | ↓ | P02088 | Hemoglobin subunit beta-1 | 147 | 15.8 | 0.50 | 0 | 8 |
| 30 | ↓ | P48036 | Annexin A5 | 319 | 35.7 | 0.50 | 0 | 12 |
| 31 | ↓ | P68033 | Actin, alpha cardiac muscle 1 | 377 | 42.0 | 0.50 | 0 | 10 |
| 32 | ↓ | Q922U2 | Keratin, type II cytoskeletal 5 | 580 | 61.7 | 0.49 | 0 | 10 |
| 33 | ↓ | P28654 | Decorin | 354 | 39.8 | 0.44 | 0 | 10 |
| 34 | ↓ | O89053 | Coronin-1A | 461 | 51.0 | 0.41 | 0 | 12 |
| 35 | ↓ | P16125 | L-lactate dehydrogenase B chain | 334 | 36.5 | 0.38 | 0 | 12 |
| 36 | ↓ | P15864 | Histone H1.2 | 212 | 21.3 | 0.38 | 0 | 12 |
| 37 | ↓ | P02089 | Hemoglobin subunit beta-2 | 147 | 15.9 | 0.27 | 0 | 12 |
| 38 | ↓ | P02301 | Histone H3.3C | 136 | 15.3 | 0.27 | 0 | 12 |
| 39 | ↓ | P62806 | Histone H4 | 103 | 11.4 | 0.26 | 0 | 12 |
| 40 | ↓ | Q62426 | Cystatin-B | 98 | 11.0 | 0.22 | 0 | 12 |
| 41 | ↓ | P08074 | Carbonyl reductase [NADPH] 2 | 244 | 25.9 | 0.18 | 0 | 12 |
| 42 | ↓ | Q9QWL7 | Keratin, type I cytoskeletal 17 | 433 | 48.1 | 0.18 | 0 | 12 |
| 43 | ↓ | Q9DB60 | Prostamide/prostaglandin F synthase | 201 | 21.7 | 0.15 | 0 | 12 |
| 44 | ↓ | P11679 | Keratin, type II cytoskeletal 8 | 490 | 54.5 | 0.11 | 0 | 12 |
| 45 | ↓ | P05784 | Keratin, type I cytoskeletal 18 | 423 | 47.5 | 0.08 | 0 | 12 |
Details of each sample are listed in Supplementary Table 4
PKM2 was activated at protein and mRNA levels in thymic lymphoma of FIR+/−TP53−/− mice
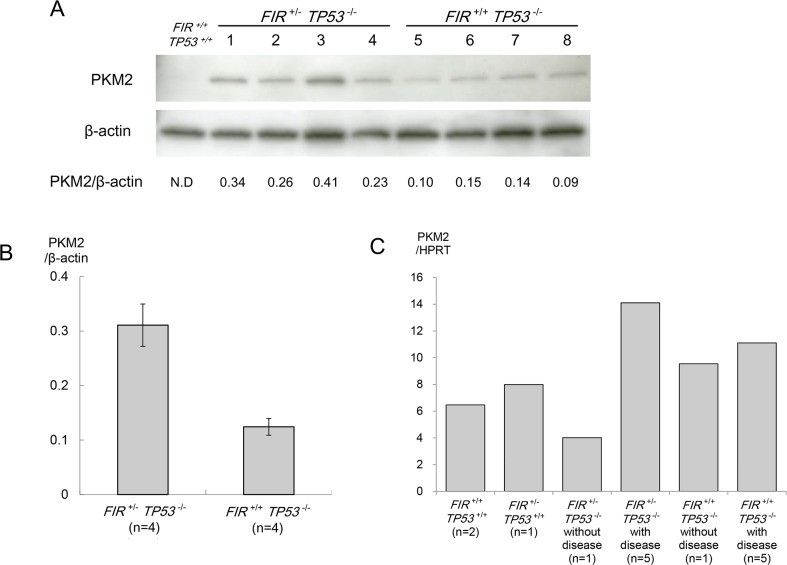
PKM2 protein was upregulated in the thymic lymphoma of FIR+/−TP53−/− mice compared with FIR+/+TP53−/− mice (Figure 2B, 2C). Western blotting confirmed that PKM2 was overexpressed 2.5 fold in the thymic lymphoma of FIR+/−TP53−/− mice compared with FIR+/+TP53−/− mice (p = 0.002) (Figure 3A, 3B), while PKM2 mRNA was elevated approximately twice more in the thymic lymphoma of FIR+/−TP53−/− mice than the FIR+/+TP53−/− mice (Figure 3C). These results indicated that FIR haplodeficiency promotes splicing of PKM1 to PKM2 in benign thymic lymphoma at the transcriptional level.
Figure 3. Western blot analysis of pyruvate kinase (PK) M2.
(A) Proteins extracted from each mouse thymus were separated by 10.0–12.0% SDS-PAGE and probed with anti-PKM2. Expression levels are relatively higher in FIR+/− TP53−/− samples than FIR+/+ TP53−/−. (B) Differences in expression were analyzed using the Student's t-test. The expression levels of these proteins were upregulated significantly in FIR+/− TP53−/− group (p = 0.002). Band intensities were quantified by image analysis software. The line indicates the range and the rectangle indicates the mean values. (C) PKM2 mRNA expression was significantly upregulated in thymic lymphoma of FIR+/−TP53−/− mice.
Knockdown of FIR suppressed hnRNPA1 expression
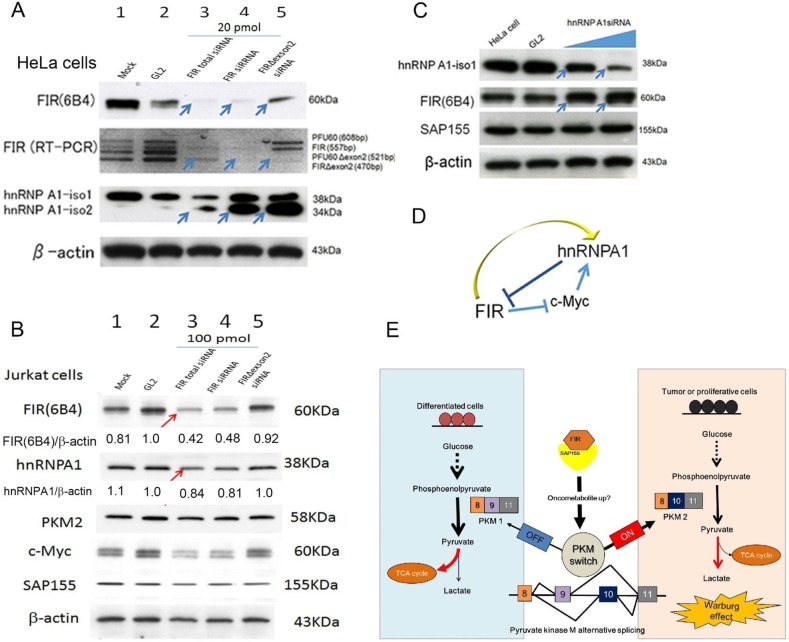
Haplodeficiency of FIR upregulated PKM2 at protein and mRNA levels (Figure 2B, 2C), and HnRNPA1 promotes splicing of PKM1 to PKM2 [21, 22]. Notably, FIR was co-immunoprecipitated with hnRNPA1 in nuclear extracts of HeLa cells [12], suggesting that FIR affects the ratio of PKM1:PKM2 via hnRNPA1. Based on these results, we investigated the relationship between FIR and hnRNPA1. Knockdown of FIR by siRNA suppressed hnRNPA1 expression in HeLa cells (Figure 4A, arrows) and Jurkat cells (Figure 4B, arrows), indicating that altered FIR expression affected PKM1/PKM2 splicing through hnRNPA1 expression. FIR haplodeficiency activates c-Myc in mice [15], which itself directly induces hnRNPA1 [23]; therefore FIR-mediated hnRNPA1 suppression requires c-Myc. Further study is required to elucidate this feedback mechanism (Figure 4C, 4D). In conclusion, FIR haplodeficiency switches PKM1 to PKM2 in thymic lymphoma, possibly by affecting hnRNPA1 expression (Figure 4E).
Figure 4. Knockdown of FIR suppressed hnRNPA1 expression.
FIR and hnRNPA1 expression was examined after siRNA treatment of HeLa cells. GL2 siRNA was transfected as the negative control. After 48 h and 72 h of incubation, whole-cell extracts were analyzed by western blotting. (A) Three different types of FIR siRNAs were transfected into HeLa cells. Lane 2 is the GL2 siRNA control transfection, lane 3 is 20 pmol of FIR total (FIR and FIRΔexon2) siRNA, lane 4 is 20 pmol FIR-specific siRNA transfection, lane 5 is 20 pmol FIRΔexon2-specific siRNA. HnRNPA1 expression was down-regulated by total FIR siRNAs (Lane 3 as compared to Lanes 1, 2, arrows). (B) Three different types of FIR siRNAs were transfected into Jurkat cells. Lane 2 is the GL2 siRNA control transfection, lane 3 is 100 pmol of FIR total (FIR and FIRΔexon2) siRNA, lane 4 is 100 pmol FIR-specific siRNA transfection, lane 5 is 100 pmol FIRΔexon2-specific siRNA. HnRNPA1 expression was down-regulated by total FIR siRNAs (Lane 3 as compared to Lanes 1, 2, arrows). (C) HnRNPA1 siRNAs were transfected into HeLa cells in dose-dependent manner. Lane 2 is GL2 siRNA, lane 3 is 20 pmol type V1 hnRNPA1, hnRNPA1 siRNA, and lane 4 is 20 pmol type G5 hnRNPA1 siRNA. FIR expression was upregulated by hnRNPA1 siRNA (arrows). (D) Schematic view of the relationship among FIR, hnRNPA1 and c-Myc. (E) FIR haplodeficiency promotes splicing to PKM2 through affecting alternative splicing switch in mice thymic lymphoma tissues using TMT analysis.
DISCUSSION
FIR+/−TP53−/− and FIR+/+TP53−/− mice that develop thymic lymphoma and T-ALL were generated previously [15]. We performed proteomic and qRT-PCR analyses in thymic lymphoma tissues and revealed that PKM2 was significantly higher at both mRNA and protein levels in thymic lymphoma of FIR+/−TP53−/− mice than in FIR+/+TP53−/− mice. This indicates that FIR haplodeficiency switches PKM1 to PKM2 at the transcriptional level, FIR knockdown suppressed hnRNPA1 expression, and hnRNPA1 promotes splicing of PKM1 to PKM2 and was reported to be coimmunoprecipitaed with FIR [12]. This study performed the requisite statistical analysis to make an argument that FIR and its related spliceosomes are potential therapeutic targets for cancers, including T-ALL.
TMT quantitative analysis is a gel-free proteomic technique and can be used to analyze various samples, including sera/plasma, body fluids, or gingival crevicular fluids [17–19]. Using this approach, we examined the altered protein expression profile of thymic lymphoma tissues under FIR haplodeficient conditions. In addition, we revealed that FIR/FIRΔexon2 was co-immunoprecipitated with PTB, hnRNP L/M, and PCBP1, which are known splicing factors of CD44 [12]. This suggested that altered FIR expression affects PKM1/PKM2 splicing by modifying hnRNPA1 expression. Moreover, FIR haplodeficiency activates c-Myc in thymic lymphoma cells in mice [15]. c-Myc directly induces hnRNPA1 [23], indicating that hnRNPA1 suppression by FIR siRNA depends partly on the c-Myc pathway. Further study is required to elucidate this feedback mechanism.
FIR links c-myc transcriptional regulation and alternative splicing [13]. FIR variants are activated in colorectal cancers [8], hepatoma [10], non-small cell lung cancer [11], and leukemia [15]. FIR is also called as PUF60, which is a member of U2AF splicing factor family [24]. An alternatively spliced dominant negative form of FIR is activated in cancer tissues. Notably, FIR forms a complex with SAP155 (SF3B1), which is frequently mutated in hematopoietic malignancies [25] and affects alternative splicing [14]. The FIR-SAP155 (SF3B1) interaction affects both hnRNPA1 and c-Myc expression and may represent a therapeutic target in c-Myc-driven cancer.
Normal differentiated cells depend on mitochondrial oxidative phosphorylation to generate energy. In cancer cells, energy is produced by aerobic glycolysis, called “the Warburg effect” [1]. Switching from PKM1 to PKM2 promotes aerobic glycolysis and tumor formation [23]. This study revealed that the switch from PKM1 to PKM2 occurs in lymphoma cells prior to T-ALL, indicating that PKM2 activation is required but not sufficient for T-ALL progression in this mouse model. Moreover, FIR haplodeficiency potentially affects PKM1/PKM2 splicing by modifying hnRNPA1 expression.
In the case of tumors originating from differentiated cells, high levels of PKM2 in tumor or cancer initiating cells are attributed to the switch of PKM gene splicing from PKM1 to PKM2. In contrast, PKM2 expression is directly inherited from the precursor/stem cells [26]. Our results support the notion above that PKM2 is activated in non-malignant thymic lymphoma. Therefore, FIR is a potential target for cancer therapy. Because the function of PKM2 in tumor progression seems to be regulated in multiple ways [26], further studies are required to fully elucidate this complicated mechanism.
MATERIALS AND METHODS
Preparation of genetically engineered mice tissues
Genetically manipulated mice were generated as previously described [15]. Thymus lymphoma and/or T-ALL was generated in FIR++TP53+/+ and FIR+/−TP53−/− mice as previously described [15] and thymus tissues were extracted. Details of all samples are described in Table 1. Mice with similar phenotypes were divided into groups for the quantitative protein assay and FIR++TP53+/+ mice were used as controls.
Cancer cell lines
HeLa and Jurkat cell lines (acute T-cell leukemia cells) were cultured in IMDM and RPMI 1640 (ThermoFisher Scientific Diagnostics CO., LTD, Tokyo, Japan), respectively, supplemented with 10% FBS and 1% penicillin–streptomycin. Cells were grown at 37°C in a 5% CO2 incubator.
Western blotting and antibodies for cancer cell lines
Culture medium was removed, and cells were washed twice with cold (4°C) PBS. Then, cells were lysed with 1:20 β-mercaptoethanol in 2X sample buffer and incubated at 100°C for 5 min. Whole-cell lysates were assayed for protein content (Bio-Rad, Hercules, CA, USA), and 10 μg of proteins were separated by SDS-PAGE on 7.5% or 10–20% XV PANTERA gels before being transferred onto polyvinylidene fluoride membranes using a tank transfer apparatus. The membranes were blocked with 0.5% skimmed milk in PBS overnight at 4°C. Antigens were detected with enhanced chemiluminescence detection reagents (GE Healthcare UK Ltd., Buckinghamshire, UK). Membranes were incubated with primary antibodies for 1 h at room temperature, followed by three 10-min washes with 1XPBS/0.01% Tween 20. Membranes were then incubated with commercial secondary antibodies, followed by three 15-min washes with 1XPBS/0.01% Tween 20. The primary mouse monoclonal antibody against the FIR C-terminus (Total FIRs 6B4) was prepared by Dr. Nozaki [16].
Flow cytometry and cell sorting
Flow cytometry and cell sorting were performed as described previously [15]. Thymic lymphoma cells were stained with APC-conjugated CD4, PE-conjugated CD8α and Pacific Blue-conjugated CD45.2 (BioLegend). Dead cells were detected by staining with 1 μg/mL propidium iodide (Sigma-Aldrich), and flow cytometry and cell sorting were performed on a FACSCanto II or FACSAria II flow cytometer (BD Biosciences).
Protein extraction from mice thymus
Thymi from FIR+/+TP53−/− and FIR+/−TP53−/− mice were frozen in liquid nitrogen and then floated in 20X 4 M urea/100 mM ammonium bicarbonate. Floating pieces of thymus tissue were homogenized and centrifuged at 24,000 rpm at room temperature using a POLYTRON® homogenizer (KINEMATICA, Schweiz). After centrifugation, thymus cells were completely disrupted by sonication three times for 10 sec each, and after homogenization and sonication, samples were centrifuged at 50,000 rpm at 4°C for 60 min before the supernatant was collected. Extracted protein supernatant samples were stored at −80°C before protein digestion.
Protein in-solution digestion
Extracted samples of 100 μg protein were mixed with 100 μL of 4 M urea/100 mM ammonium bicarbonate after measuring protein concentrations. Then, 100 μL of 4 M urea/100 mM ammonium bicarbonate, including 2% 200 mM DTT, was added and incubated at 57°C for 30 min. After incubation, 10 μL of 600 mM iodacetoamide was added and incubated at room temperature for 30 min in the dark. Twenty microliters of lysyl endopeptidase were incubated in 200 μL of distilled water at 37°C for 3 h, and then, 25 μL of trypsin were incubated with 200 μL of distilled water overnight at 37°C. The resulting peptides were added to 50 μL of 10% TFA.
Protein extraction, reduction and TMT labeling
After protein extraction and reduction using Stage Tip (Thermo Fisher Scientific Inc, USA), samples were digested and lyophilized before being resuspended in 40 μL of 0.1% TFA. After vortexing at room temperature for 5 min, 40 μL of triethyl ammonium bicarbonate were added and mixed. An Isobaric Label Reagent set (Pierce, Idaho, ID, USA) was used for six-plex TMT according to the manufacturer's instructions. Each mass-tagging reagent tube contained 0.8 mg of different isobaric chemical tags with 42 μL of anhydrous acetonitrile, dissolved for 5 minutes with occasional vortexing and brief centrifugation to gather the solution. Samples were then incubated in 42 μL of the unique reagent for 1 h at room temperature. The reaction was quenched by incubation in 8 μL of 5% hydroxylamine for 30 min. Samples were cleaned using C18 spin tips, and stored at −80°C until LC-MS/MS analysis (Figure 2A).
LC-MS/MS
Enriched digested peptides were injected into a trap column (C18, 0.3×5 mm, DIONEX, CA, USA) and analytical column (monolithic silica-C18, 0.1×2000 mm, GL sciences, Tokyo, Japan), which was attached to the Ultimate 3000 (DIONEX, CA, USA).
The flow rate of the mobile phase was 500 nL/min, with a solvent composition programmed to change in 480 min cycles with varying ratios of solvent A (2% v/v CH3CN and 0.1% v/v HCOOH) to solvent B (90% v/v CH3CN and 0.1% v/v HCOOH) as follows; 5–10% B 60 min, 10–30% B 280 min, 30–40 % B 60 min, 40–100% B 2 min, 100% B 5 min, 100–5% B 2 min, and 5% B 71 min. Purified peptides from HPLC were introduced to a LTQ-Orbitrap XL hybridion-trap Fourier transform mass spectrometer (Thermo Scientific, San Jose, CA, USA) using a Pico Tip (New Objective, MA, USA). A cycle scan event included one full scan and three data-dependent, CID, high-energy, collision-activated, dissociation dual MS/MS scans for TMT reporter ions [19]. The database search engine (Proteome discoverer; version 1.3.0, Thermo Scientific, San Jose, CA, USA) was used to identify proteins based on mass, tandem mass spectra, and reporter ion spectra of peptides. Peptide mass data were matched by searching the UniProtKB Mouse database (SwissProt 2014, 16951 entries), using the following search parameters: peptide mass tolerance, 2 ppm; fragment tolerance, 0.6 Da; enzyme set to trypsin, allowing up to two missed cleavages; dynamic modifications, methionine oxidation; static modifications, cysteine carbamidomethylation, and; TMT tagged N-terminus and lysine. The minimum criteria of protein identification were filtered with Xcorr versus charge state and set as false discovery rate (FDR) < 1%. The FDR was estimated by comparing with a randomized decoy database created by the Proteome Discoverer 1.3.0 program supplied by Thermo Scientific.
Quantitative real-time PCR (qRT-PCR)
Quantitative real-time PCR to measure PKM2 was performed as previously described [9, 12] (Supplementary Table 1).
Western blotting
Details of primary and secondary antibodies are given in Supplementary Table 2A. Briefly, 20 μg mouse thymus protein extracts were separated using SDS polyacrylamide gel electrophoresis and transferred to a PVDF membrane (Millipore, Bedford, MA) as described previously [20]. The membranes were incubated with anti-PKM2 antibody (ab154816; rabbit monoclonal, Abcam, Cambridge, MA, USA) at 1:200 dilutions for 60 min at room temperature. After primary antibody incubation, membranes were washed three times for 30 min in PBS-Tween (0.1%), prior to incubation in the appropriate horseradish peroxidase-linked secondary antibody (goat anti-rabbit IgG horseradish peroxidase-linked antiserum) diluted 1:2500 for 60 min at room temperature. Finally, membranes were washed three times as previously described, and immunoreactive proteins were revealed with an enhanced chemiluminescence substrate reaction using Pierce® ECL Plus western blotting substrate (Thermo Scientific, San Jose, CA, USA) according to the manufacturer's instructions. Total-Lab TL12 imaging analysis software (Shimadzu Co., Ltd. Kyoto, Japan) was used for quantification of bands intensities and statistical analysis.
FIR and hnRNPA1 siRNA
Three FIR siRNAs and two hnRNPA1 siRNAs were purchased from Sigma-Aldrich (Supplementary Table 2B). siRNAs were transiently transfected using Lipofectamine 2000 (Invitrogen) for HeLa cells and HiPerFect Transfection Reagent (QIAGEN) for Jurkat cells according to the manufacturer's instructions. After transfection with FIR siRNA, cells were cultured for 48 h and after transfection with hnRNPA1 siRNAs, cells were cultured for 72 h, at 37°C in a CO2 incubator.
Statistical analysis
Numerical data were presented as mean ± standard deviation (SD). Statistical significance was assessed using the Student's t-test and P values less than 0.05 were considered statistically significant.
SUPPLEMENTARY MATERIALS FIGURE AND TABLES
Abbreviations
- PK
pyruvate kinase
- FBP
FUSE-binding protein
- FIR
FBP-interacting repressor
- T-ALL
T-cell type acute lymphoblastic leukemia
- TMT
six-plex tandem mass tag
Footnotes
CONFLICTS OF INTEREST
We have no conflicts of interest to declare.
GRANT SUPPORT
This study was supported by JSPS Grant-in-Aid for Scientific Research(C) from “the Ministry of Education, Science, Sports and Culture of Japan” (KAKENHI), Grant Numbers 26460667, 16K08972 and was partly supported by Extramural Collaborative Research Grant of Cancer Research Institute, Kanazawa University to KM.
REFERENCES
- 1.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avigan MI, Strober B, Levens D. A far upstream element stimulates c-myc expression in undifferentiated leukemia cells. J Biol Chem. 1990;265:18538–18545. [PubMed] [Google Scholar]
- 4.Hsu TY, Simon LM, Neill NJ, Marcotte R, Sayad A, Bland CS, Echeverria GV, Sun T, Kurley SJ, Tyagi S, Karlin KL, Dominguez-Vidaña R, Westbrook TF, et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015;525:384–388. doi: 10.1038/nature14985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stine ZE, Dang CV. Splicing and diceing MYC-mediated synthetic lethality. Cancer Cell. 2015;28:405–406. doi: 10.1016/j.ccell.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 6.Alderton GK. MYC: splicing up your survival. Nat Rev Cancer. 2015;15:574–575. doi: 10.1038/nrc4013. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, He L, Collins I, Ge H, Libutti D, Li J, Egly JM, Levens D. The FBP interacting repressor targets TFIIH to inhibit activated transcription. Mol Cell. 2000;5:331–341. doi: 10.1016/s1097-2765(00)80428-1. [DOI] [PubMed] [Google Scholar]
- 8.Matsushita K, Tomonaga T, Shimada H, Shioya A, Higashi M, Matsubara H, Harigaya K, Nomura F, Libutti D, Levens D, Ochiai T. An essential role of alternative splicing of c-myc suppressor FUSE-binding protein-interacting repressor in carcinogenesis. Cancer Res. 2006;66:1409–1417. doi: 10.1158/0008-5472.CAN-04-4459. [DOI] [PubMed] [Google Scholar]
- 9.Kajiwara T, Matsushita K, Itoga S, Tamura M, Tanaka N, Tomonaga T, Matsubara H, Shimada H, Habara Y, Matsuo M, Nomura F. SAP155-mediated c-myc suppressor far-upstream element-binding protein-interacting repressor splicing variants are activated in colon cancer tissues. Cancer Sci. 2013;104:149–156. doi: 10.1111/cas.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malz M, Bovet M, Samarin J, Rabenhorst U, Sticht C, Bissinger M, Roessler S, Bermejo JL, Renner M, Calvisi DF, Singer S, Ganzinger M, Breuhahn K, et al. Overexpression of far upstream element (FUSE) binding protein (FBP)-interacting repressor (FIR) supports growth of hepatocellular carcinoma. Hepatology. 2014;60:1241–1250. doi: 10.1002/hep.27218. [DOI] [PubMed] [Google Scholar]
- 11.Müller B, Bovet M, Yin Y, Stichel D, Malz M, González-Vallinas M, Middleton A, Ehemann V, Schmitt J, Muley T, Meister M, Herpel E, Breuhahn K, et al. Concomitant expression of far upstream element (FUSE) binding protein (FBP) interacting repressor (FIR) and its splice variants induce migration and invasion of non-small cell lung cancer (NSCLC) cells. J Pathol. 2015;237:390–401. doi: 10.1002/path.4588. [DOI] [PubMed] [Google Scholar]
- 12.Matsushita K, Kajiwara T, Tamura M, Satoh M, Tanaka N, Tomonaga T, Matsubara H, Shimada H, Yoshimoto R, Ito A, Kubo S, Natsume T, Nomura F, et al. SAP155-mediated splicing of FUSE-binding protein-interacting repressor serves as a molecular switch for c-myc gene expression. Mol Cancer Res. 2012;10:787–799. doi: 10.1158/1541-7786.MCR-11-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsushita K, Tamura M, Tanaka N, Tomonaga T, Matsubara H, Shimada H, Levens D, He L, Liu J, Yoshida M, Nomura F. Interactions between SAP155 and FUSE-binding protein-interacting repressor bridges c-Myc and P27Kip1 expression. Mol Cancer Res. 2013;11:689–698. doi: 10.1158/1541-7786.MCR-12-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alsafadi S, Houy A, Battistella A, Popova T, Wassef M, Henry E, Tirode F, Constantinou A, Piperno-Neumann S, Roman-Roman S, Dutertre M, Stern MH. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branch-point usage. Nat Commun. 2016;7:10615. doi: 10.1038/ncomms10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsushita K, Kitamura K, Rahmutulla B, Tanaka N, Ishige T, Satoh M, Hoshino T, Miyagi S, Mori T, Itoga S, Shimada H, Tomonaga T, Nomura F, et al. Haploinsufficiency of the c-myc transcriptional repressor FIR, as a dominant negative-alternative splicing model, promoted p53-dependent T-cell acute lymphoblastic leukemia progression by activating Notch1. Oncotarget. 2015;6:5102–5117. doi: 10.18632/oncotarget.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimura K, Nozaki N, Enomoto T, Tanaka M, Kikuchi A. Analysis of M phase-specific phosphorylation of DNA topoisomerase II. J Biol Chem. 1996;271:21439–21445. doi: 10.1074/jbc.271.35.21439. [DOI] [PubMed] [Google Scholar]
- 17.Sinclair J, Timms JF. Quantitative profiling of serum samples using TMT protein labelling, fractionation and LC-MS/MS. Methods. 2011;54:361–369. doi: 10.1016/j.ymeth.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez JC. Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Anal Chem. 2008;80:2921–2931. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- 19.Tsuchida S, Satoh M, Kawashima Y, Sogawa K, Kado S, Sawai S, Nishimura M, Ogita M, Takeuchi Y, Kobyashi H, Aoki A, Kodera Y, Nomura F, et al. Application of quantitative proteomic analysis using tandem mass tags for discovery and identification of novel biomarkers in periodontal disease. Proteomics. 2013;13:2339–2350. doi: 10.1002/pmic.201200510. [DOI] [PubMed] [Google Scholar]
- 20.Kimura A, Sogawa K, Satoh M, Kodera Y, Yokosuka O, Tomonaga T, Nomura F. The application of a three-step serum proteome analysis for the discovery and identification of novel biomarkers of hepatocellular carcinoma. Int J Proteomics. 2012;62:3190. doi: 10.1155/2012/623190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen M, David CJ, Manley JL. Concentration-dependent control of pyruvate kinase M mutually exclusive splicing by hnRNP proteins. Nat Struct Mol Biol. 2012;19:346–354. doi: 10.1038/nsmb.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luan W, Wang Y, Chen X, Shi Y, Wang J, Zhang J, Qian J, Li R, Tao T, Wei W, Hu Q, Liu N, You Y. PKM2 promotes glucose metabolism and cell growth in gliomas through a mechanism involving a let-7a/c-Myc/hnRNPA1 feedback loop. Oncotarget. 2015;6:13006–13018. doi: 10.18632/oncotarget.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen M, Zhang J, Manley JL. Turning on a fuel switch of cancer: hnRNP proteins regulate alternative splicing of pyruvate kinase mRNA. Cancer Res. 2010;70:8977–8980. doi: 10.1158/0008-5472.CAN-10-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corsini L, Hothorn M, Stier G, Rybin V, Scheffzek K, Gibson TJ, Sattler M. Dimerization and protein binding specificity of the U2AF homology motif of the splicing factor Puf60. J Biol Chem. 2009;284:630–639. doi: 10.1074/jbc.M805395200. [DOI] [PubMed] [Google Scholar]
- 25.Hahn CN, Venugopal P, Scott HS, Hiwase DK. Splice factor mutations and alternative splicing as drivers of hematopoietic malignancy. Immunol Rev. 2015;263:257–278. doi: 10.1111/imr.12241. [DOI] [PubMed] [Google Scholar]
- 26.Li Z, Yang P, Li Z. The multifaceted regulation and functions of PKM2 in tumor progression. Biochim Biophys Acta. 2014;1846:285–296. doi: 10.1016/j.bbcan.2014.07.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.