Abstract.
Early stage diagnosis of laryngeal squamous cell carcinoma (SCC) is of primary importance for lowering patient mortality or after treatment morbidity. Despite the challenges in diagnosis reported in the clinical literature, few efforts have been invested in computer-assisted diagnosis. The objective of this paper is to investigate the use of texture-based machine-learning algorithms for early stage cancerous laryngeal tissue classification. To estimate the classification reliability, a measure of confidence is also exploited. From the endoscopic videos of 33 patients affected by SCC, a well-balanced dataset of 1320 patches, relative to four laryngeal tissue classes, was extracted. With the best performing feature, the achieved median classification recall was 93% [interquartile range ]. When excluding low-confidence patches, the achieved median recall was increased to 98% (), proving the high reliability of the proposed approach. This research represents an important advancement in the state-of-the-art computer-assisted laryngeal diagnosis, and the results are a promising step toward a helpful endoscope-integrated processing system to support early stage diagnosis.
Keywords: laryngeal cancer, tissue classification, texture analysis, surgical data science
1. Introduction
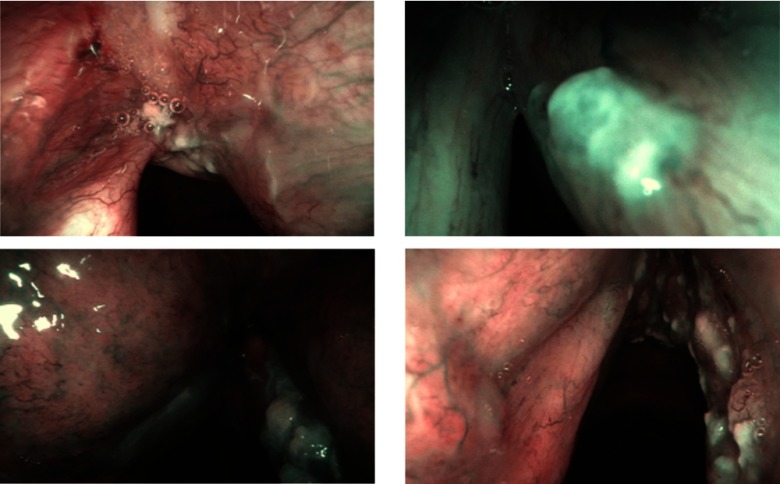
Squamous cell carcinoma (SCC) is the most common cancer of the laryngeal tract, arising from 95% to 98% of all cases of laryngeal cancer.1 It is well known from medical literature that early stage SCC diagnosis can lower mortality rate and preserve both laryngeal anatomy and vocal fold function.2 Histopathological examination of tissue samples extracted with biopsy is currently the gold-standard for diagnosis. However, the relevance of tissue visual analysis for screening purposes has led, in the past few years, to the development of optical-biopsy techniques such as narrow-band imaging (NBI) endoscopy,3 which has become state of the art for laryngeal tract inspection. The identification of suspicious tissues during the endoscopic examination is, however, challenging due to the late onset of symptoms and to the small modifications of the mucosa, which can pass unnoticed to the human eye.4 Main modifications occur to the mucosa vascular tree, with the presence of longitudinal hypertrophic vessels and dot-like vessels, known as intraepithelial papillary capillary loops (IPCL).3 Changes in the epithelium aspect not related to the vascular tree, such as thickening and whitening of the epithelial layer (leukoplakia), are associated with increased risk of developing SCC, too.5 Visual samples of laryngeal endoscopic video frames of patients affected by SCC are given in Fig. 1.
Fig. 1.
Visual samples of NBI laryngeal endoscopic frames of patients affected by SCC.
Considering the clinical challenges in diagnosis, some preliminary attempts of computer-assisted diagnosis have been presented,6,7 despite only Barbalata and Mattos6 specifically focus on early stage diagnosis. The study proposes an algorithm for the classification of early stage vocal fold cancer based on the segmentation and analysis of blood vessels. Vessel segmentation is performed with matched filtering coupled with first-order derivative of Gaussian. Vessel tortuosity, thickness, and density are used as features to discriminate between malignant and benign tissue by means of linear discriminant analysis (LDA). Despite the good results (), the classification proposed in Ref. 6 is strongly sensitive to a priori set parameters, e.g., vessel width and orientation. Moreover, focusing on vessels alone does not allow one to take into account epithelial modifications that do not affect the vascular tree (i.e., in case of leukoplakia).
The emerging and rich literature on surgical data science for tissue classification outside the field of laryngoscopy has recently focused on more sophisticated techniques, which mainly exploit machine learning algorithms to classify tissues according to texture-based information.8 In Ref. 9, the histogram of local binary patterns (LBP) is exploited to classify ulcer and healthy regions in capsule endoscopy images using multilayer perceptron. In Ref. 10, the LBP histogram is combined with intensity-based features to classify abdominal tissues in laparoscopic images by means of support vector machines (SVM). Similarly, in Ref. 11, intensity-based features and LBP histogram are used to characterize lesions in gastric images. In Ref. 12, the LBP histogram is combined with gray-level co-occurrence matrix (GLCM)-based features to classify gastroscopy images. AdaBoost is used to perform the classification. In Ref. 13, Gabor filter-based features are used to classify healthy and cancerous tissue in gastroscopy images by means of SVM. A recent work14 exploits NBI data for colorectal image analysis. Colorectal tissues are classified as neoplastic or healthy by means of GLCM-based features and SVM.
Inspired by these recent and promising studies, in this paper, we aim at investigating if texture-based approaches applied to laryngeal tissue classification in NBI images can provide reliable results, to be used as support for early stage diagnosis. Specifically, we investigate the following two hypotheses:
-
•
Hypothesis 1 (H1): Machine-learning techniques can classify laryngeal tissues in NBI images by exploiting textural information.
-
•
Hypothesis 2 (H2): By estimating the level of classification confidence and discarding low-confidence samples, the number of incorrectly classified cases can be lowered.
The importance of estimating the level of classification confidence with a view to improving system performance has been widely highlighted in several research fields, such as face recognition,15 spam-filtering,16 and glioma and colon cancer recognition.17 In particular, it has been reported that allowing a system to produce “do not know” results can potentially reduce the number of incorrectly classified cases.18 In the analyzed scenario, estimating the classification confidence would be beneficial since tissue biopsy would be required only for low-confidence regions in the image.
To the best of our knowledge, we are the first to investigate the use of texture-based classification algorithms for laryngeal tissue analysis.
2. Materials and Methods
This section explains the proposed approach to automatic laryngeal tissue classification (Sec. 2.1), as well as the evaluation protocol (Sec. 2.2) used to investigate the two hypotheses introduced in Sec. 1.
2.1. Automatic Laryngeal Tissue Classification
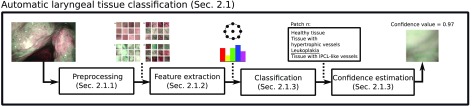
The proposed method consists of the following steps: (i) preprocessing (Sec. 2.1.1), (ii) feature extraction (Sec. 2.1.2), (iii) classification (Sec. 2.1.3), and (iv) confidence estimation (Sec. 2.1.4). The workflow of the approach is shown in Fig. 2.
Fig. 2.
Workflow of the proposed approach to laryngeal tissue classification in NBI endoscopic video frames.
2.1.1. Preprocessing
Anisotropic diffusion filtering19 is used to lower noise while preserving sharp edges in NBI images. Specular reflections (SR), usually present due to the wet and smooth laryngeal surface, are automatically identified exploiting their low saturation and high brightness and then masked.6 After denoising, squared patches are selected from the image, as described in Sec. 2.2.
SR masking is necessary because it may not always be possible selecting patches without SR. This is due to the small extension of early stage cancerous tissues in the image, which may overlap with SR (especially for the case of intrapapillary capillary loop-like vessels).
2.1.2. Feature extraction
As laryngeal endoscopic images are captured under various illumination conditions and from different viewpoints, the features that encode the tissue texture information should be robust to the pose of the endoscope as well as to the lighting conditions. Furthermore, with a view of a real-time computer-aided application, they should be computationally cheap. In this paper, we investigate the use of the following descriptors to characterize the texture of laryngeal tissues:
Texture-based global descriptors. Among classic texture-based global descriptors, LBP are widely considered as state of the art for medical image texture analysis.20 LBP are gray-scale invariant and provide low-complexity, well matching the requisite of this application. The first formulation of LBP () introduced in the literature requires defining, for a pixel , a spatial circular neighborhood of radius with equally spaced neighbor points ():
| (1) |
where and denote the gray values of the pixel and of its ’th neighbor , respectively, and is defined as
| (2) |
The most often adopted LBP formulation is the uniform rotation-invariant one ().21 Rotation invariance is suitable for the purpose of this paper since the endoscope pose during the larynx inspection is constantly changing. From , the L2-normalized histogram of () is computed and used as feature vector.
For comparison, the GLCM, a second widely used descriptor, is tested. GLCM calculates how often pair of pixels with specific values and in a specified spatial relationship occur in an image. The spatial relationship is defined by and , which are the angle and distance between and . The GLCM width (), equal to the GLCM height (), corresponds to the number of quantized image intensity gray-levels. For the intensity gray-level, the GLCM computed with and is defined as
| (3) |
From the normalized , as suggested in Ref. 22, a feature set () is extracted, which consists of GLCM contrast, correlation, energy, and homogeneity. The normalized , which expresses the probability of gray-level occurrences, is obtained by dividing each entry by the sum of all entries.
First-order statistics. Intensity mean, variance, and entropy [Eq. (4)] in each patch are computed and concatenated to form a single-intensity-based feature set (). The entropy is defined as
| (4) |
where refers to the image histogram counts of the bin. As recommended in Ref. 23, such features are adopted to integrate the texture-based information encoded in .
In addition to these descriptors, we tested two feature combinations (, ), as suggested in Ref. 23 for applications in colorectal image analysis.
2.1.3. Classification
To perform tissue classification, SVM are used.24 SVM are chosen since they allow overcoming the curse-of-dimensionality that arises when analyzing our high-dimensional feature space.25,26 The kernel trick prevents parameter proliferation, lowering computational complexity, and limiting over-fitting. Moreover, the SVM decisions are only determined by the support vectors, which make SVM robust to noise in training data. Here, SVM with the Gaussian kernel () are used. For a binary classification problem, given a training set of data , where is the ’th input feature vector and is the ’th output label, the SVM decision function takes the form of
| (5) |
where
| (6) |
is a real constant and is retrieved as follows:
| (7) |
with
| (8) |
In this paper, and are retrieved with grid search, as explained in Sec. 2.2. To implement multiclass SVM classification, the one-versus-one scheme is used.
For the sake of completeness, the performance of other classifiers, such as -nearest neighbors (kNN),27 naive Bayes (NB),28 and random forest (RF),29 are also investigated.
Prior to classification, the feature matrices are normalized within each feature dimension. Specifically, the feature matrices are preprocessed by removing the mean (centering) and scaling to unit variance.
2.1.4. Confidence estimation
As a prerequisite for our confidence estimation, we compute the probability [] of the ’th patch to belong to the ’th class, with and the number of considered tissue classes. For the probability computation, the Platt scaling method revised for multiclass classification problems is used.30 The Platt scaling method consists of training the parameters of an additional sigmoid function to map SVM outputs to probabilities.
To estimate the reliability of the SVM classification of the ’th patch, inspired by the work in Ref. 31 for abdominal tissue classification applications, we evaluate the dispersion of among the classes using the Gini coefficient (GC)32
| (9) |
where is the Lorentz curve, which is the cumulative probability among laryngeal classes rank-ordered by decreasing values of their individual probabilities. The GC has value 0 if all the probabilities are equally distributed (maximum uncertainty) and 1 for maximum inequality (the classifier is 100% confident in assigning the label). The classification of a patch is considered to be confident if GC is higher than a threshold ()
2.2. Evaluation
In this study, four tissue classes, which are typically evaluated during early stage diagnosis with NBI laryngoscopy, are considered: (i) tissue with IPCL-like vessels, (ii) leukoplakia, (iii) tissue with hypertrophic vessels, and (iv) healthy tissue. We retrospectively analyzed 33 NBI videos, which refer to 33 different patients affected by SCC. SCC was diagnosed with histopathological examination. Videos were acquired with a NBI endoscopic system (Olympus Visera Elite S190 video processor and an ENF-VH rhino-laryngo videoscope) with frame rate of 25 fps and image size of .
A total number of 330 in-focus images (10 per video) was manually selected from the videos, in such a way that the distance between the endoscope and the tissue could be considered constant and approximatively equal to 1 mm for all the images. This distance is suggested in clinics for correct evaluation of tissues during NBI endoscopy examination.33
The NBI images were preprocessed as in Sec. 2.1.1. The parameters used for anisotropic diffusion filtering were set as in Ref. 34. The saturation and brightness thresholding values used to mask SR were set as in Ref. 6.
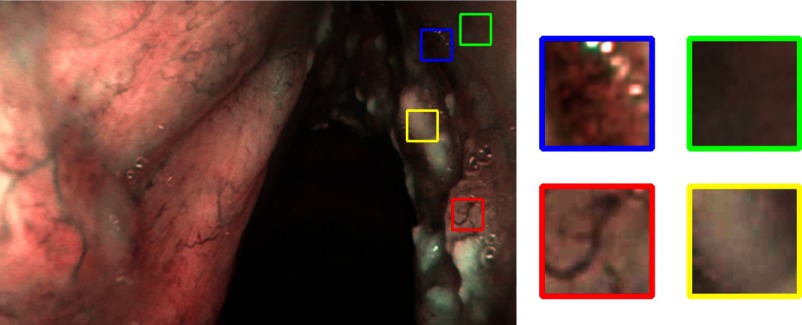
For each of the 330 images, 4 patches were manually cropped with a size of , for a total of 1320 patches, equally distributed among the 4 classes (Table 1). Each patch was cropped from a portion of tissue relative to only one of the four considered classes (tissue with IPCL-like vessels, leukoplakia, hypertrophic vessels, and healthy tissue), thus avoiding tissue overlap in one patch. The selection was performed under the supervision of an expert clinician (otolaryngologist specialized in head and neck oncology). A visual example of four patches cropped from a NBI frame is shown in Fig. 3. We decided to select only one patch per tissue class because, in most of the images, we were not able to select more than a single patch for the IPCL-like class. This is due to the small extension of this vascular alteration in early stage cancer.
Table 1.
Evaluation dataset. For each of the 33 patients’ video, 10 images are used for a total of 330 images. From each image, four tissue patches are extracted for a total of 1320 patches relative to the four considered tissue classes: healthy tissue, tissue with hypertrophic vessels, leukoplakia, and tissue with IPCL-like vessels. For a robust evaluation, the dataset is split at patient level to perform threefold cross validation. In each fold, 11 patients are included, for a total of 110 images per fold. Each fold contains 440 patches equally distributed among the laryngeal tissue classes.
| Fold 1 | Fold 2 | Fold 3 | Total | |
|---|---|---|---|---|
| Patient ID | 1–11 | 12–22 | 23–33 | 33 |
| No. of images | 110 (10 per patient) | 110 (10 per patient) | 110 (10 per patient) | 330 |
| No. of patches | 440 patches (4 per image) | 440 patches (4 per images) | 440 patches (4 per image) | 1320 |
Fig. 3.
Four patches, relative to the four analyzed laryngeal tissue classes, are manually cropped from the image. Blue: tissue with IPCL-like vessels; yellow: tissue with leukoplakia; green: healthy tissue; and red: tissue with hypertrophic vessels.
For the feature extraction described in Sec. 2.1.2, the were computed with the following combinations: (1; 8), (2; 16), (3; 24), and the corresponding were concatenated. Such choice allows a multiscale, and therefore, a more accurate description of the texture, as suggested in Ref. 10. Twelve were computed using all the possible combinations of , with and , and the corresponding sets were concatenated. The chosen interval of allows one to approximate rotation invariance, as suggested in Ref. 22. The values of were chosen to be consistent with the scale used to compute . , , and were computed for each channel in the NBI image. All the tested feature vectors and their length are reported in Table 2.
Table 2.
Tested feature vectors and corresponding number of features. , intensity mean, variance, entropy; , GLCM-based descriptors; and , normalized histogram of rotation-invariant uniform LBPs.
| Feature vector | |||||
|---|---|---|---|---|---|
| Number of features | 9 | 144 | 153 | 162 | 171 |
As for performing the classification presented in Sec. 2.1.3, the SVM hyperparameters were retrieved via grid-search and cross validation on the training set. The grid-search space for and was set to and , respectively, with six values spaced evenly on scale in both cases. Similarly, we retrieved the number of neighbors for kNN with a grid-search space set to [2,10] with nine values spaced evenly, and the number of trees in the forest for RF with a grid-search space set to [40,100] with six values spaced evenly.
The computation of , , and was implemented using OpenCv.35 The classification was implemented with scikit-learn.36
Investigation of H1. In order to assess our hypothesis that machine-learning techniques can characterize laryngeal tissues in NBI images by exploiting textural information, we first evaluated the classification performance of the texture descriptors without confidence estimation (base case).
To obtain a robust estimation of the classification performance, threefold cross validation was performed, separating data at patient level to prevent data leakage. The 1320-patch dataset was split to obtain well-balanced folds both at patient-level and tissue-level, as shown in Table 1. Each time, two folds were used for training and the remaining one for testing purpose only. This evaluation does not lead to biased results since our dataset is balanced over the three folds.
Inspired by Ref. 10, we computed the class-specific recall () to evaluate the classification performance
| (10) |
where is the number of elements of the ’th class correctly classified (true positive of the ’th class) and is the number of elements of the ’th class wrongly assigned to one of the three left classes (false negative of the ’th class). We further evaluated the class-specific precision ()
| (11) |
where the number of false positive of the ’th class, and the score (), where
| (12) |
For a comprehensive analysis, we computed the area under the receiver operating characteristic (ROC) curve (AUC). Since our task is a multiclass classification problem and our dataset is balanced, we computed the macroaverage ROC curve. The gold-standard classification was obtained by labeling the patches under the supervision of an expert clinician.
We used the Wilcoxon signed-rank test (significance level ) for paired sample to assess whether the classification achieved with our best performing (highest median value) feature vector significantly differs from the ones achieved with the other feature sets in Table 2. Similarly, we evaluated whether the classification achieved with SVM differs (Wilcoxon signed-rank test with ) from the ones achieved with the other tested classifiers (kNN, NB, RF).
For the sake of completeness, we compared the performance of our best-performing feature set with those of the most recent—and so far, the only one—method6 published on the topic of laryngeal tissue classification in NBI endoscopy, applying the latter to our dataset. As introduced in Sec. 1, the method requires setting the vessel segmentation parameters, which were here set as in Ref. 6. The feature classification was performed with SVM, instead of LDA, for fair comparison. The comparison was repeated excluding the leukoplakia class, to avoid privileging the proposed method. Indeed, the method in Ref. 6 focuses on the analysis of vessels, which, however, are not visible in case of leukoplakia due to the thickening of the epithelial layer.
Investigation of H2. To investigate the hypothesis that, by estimating the level of classification confidence and discarding low-confidence samples, the number of incorrectly classified cases can be lowered, we evaluated how , , and obtained with our best performing feature vector change considering different thresholds () on the GC value. Since, once the low-confidence patches are excluded, the balance between classes did not hold, we computed the ROC curves for each of the four laryngeal classes (and not the macroaverage ones as for H1).
3. Results
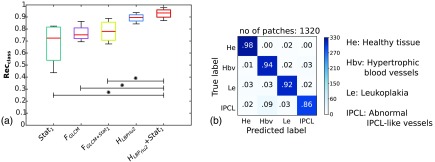
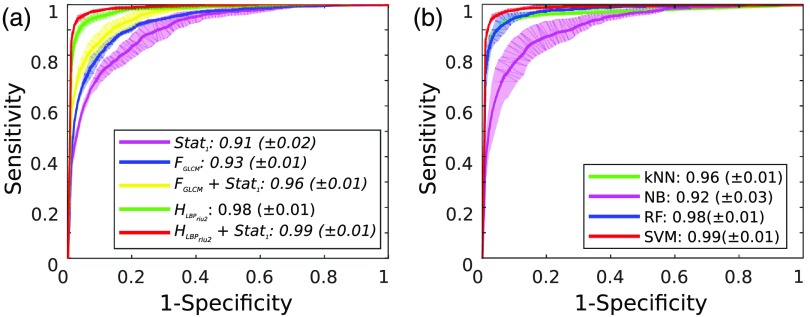
For the base case, the best performance [median , interquartile range ] was obtained with and SVM classification, as shown in Table 3. The same was observed also when considering (, ) and (, ). The classification statistics relative to all the analyzed features are reported in Fig. 4(a). Significant differences () were found when comparing with , , and . The normalized confusion matrix for is shown in Fig. 4(b). In Fig. 5(a), the macroaverage ROC curves are reported for all tested features and SVM classification. The mean AUC across the three folds was 0.99 for .
Table 3.
Median (first quartile to third quartile) class-specific recall (), precision (), and score () obtained testing different feature vectors for the base case (i.e., without the inclusion of confidence on classification estimation). Classification is obtained with SVM. , intensity mean, variance, entropy; , GLCM-based descriptors; , normalized histogram of rotation-invariant uniform LBPs.
| 72 (54–82) | 75 (72–81) | 78 (71–86) | 90 (87–92) | 93 (90–96) | |
| 67 (57–80) | 75 (71–80) | 78 (72–84) | 90 (88–92) | 94 (91–95) | |
| 70 (56–81) | 74 (71–80) | 79 (72–85) | 90 (89–91) | 92 (91–95) |
Fig. 4.
Comparison of different features without including the classification confidence estimation. Classification is obtained with SVM. (a) Boxplots of class-specific recall () for different features. , intensity mean, variance, entropy; , GLCM-based descriptors; and , histogram of rotation-invariant uniform LBPs. The stars indicate significant differences (Wilcoxon test, ). (b) Normalized confusion matrix for . The colorbar indicates the number of patches. The total number of patches (no. of patches) is reported.
Fig. 5.
Macroaverage ROC curves. The mean ( standard deviation) curves obtained from the three cross-validation folds are reported in bold (transparent area). The mean ( standard deviation) area under the ROC curve is reported in the legend. (a) ROC curves for the tested features. Classification is obtained using SVM. , intensity mean, variance, entropy; , GLCM-based descriptors; and , histogram of rotation-invariant uniform LBPs. (b) ROC curves for the tested classifiers. Classification is obtained using the histogram of LBP and first-order statistics. kNN, -nearest neighbors; NB, naive Bayes; RF, random forest; and SVM, support vector machines.
As shown in Table 4, SVM has shown comparable performance with respect to kNN and RF in terms of , , and , whereas SVM outperformed () NB. The same can be noticed from the ROC curve analysis in Fig. 5(b).
Table 4.
Comparison of different classifiers. Median (first quartile to third quartile) class-specific recall (), precision (), and score () are reported for the four different tissue classes. Classification is obtained using the histogram of LBPs and first-order statistics. kNN, -nearest neighbors; NB, naive Bayes; RF, random forest; and SVM, support vector machines.
| NB | RF | SVM | ||
|---|---|---|---|---|
| 90 (84–93) | 78 (74–82) | 89 (84–91) | 93 (90–96) | |
| 89 (86–91) | 81 (73–84) | 87 (86–89) | 94 (91–95) | |
| 89 (86–91) | 79 (74–83) | 89 (86–90) | 92 (91–95) |
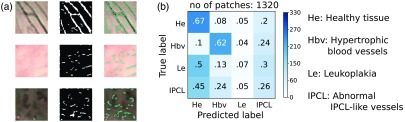
When applying the algorithm proposed by Barbalata and Mattos6 to our dataset, a median value of 42% was obtained, with IQR of 48%. Significant differences () were found when comparing the algorithm results with those obtained exploiting . Visual examples of the vessel segmentation obtained with the method proposed in Ref. 6 are reported in Fig. 6(a) for patches with hypertrophic vessels, healthy tissue, and IPCL-like vessels. The confusion matrix for the classification obtained with the method in Ref. 6 is reported in Fig. 6(b). The Barbalata and Mattos algorithm correctly labeled leukoplakias and abnormal IPCL only in the 7% and 26% of all cases, respectively. Almost half of leukoplakias and abnormal IPCL were misclassified as healthy tissues. When excluding the leukoplakia class, the was: 62% (healthy tissue), 70% (tissue with hypertrophic vessels), and 28% (tissue with ICPL-like vessels).
Fig. 6.
Performance of the state of art. (a) Visual samples of the vessel segmentation obtained applying Barbalata and Mattos6 algorithm to patches with hypertrophic vessels (first row), healthy tissue (second row), and IPCL-like vessels (third row). From left to right, original patch, vessel mask, and vessel mask superimposed on the original patch. (b) Normalized confusion matrix obtained applying Barbalata and Mattos6 algorithm to our dataset. Colorbar indicates the number of patches. The total number of patches (no. of patches) is reported.
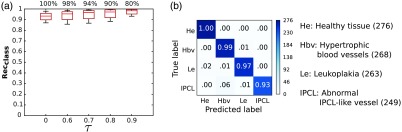
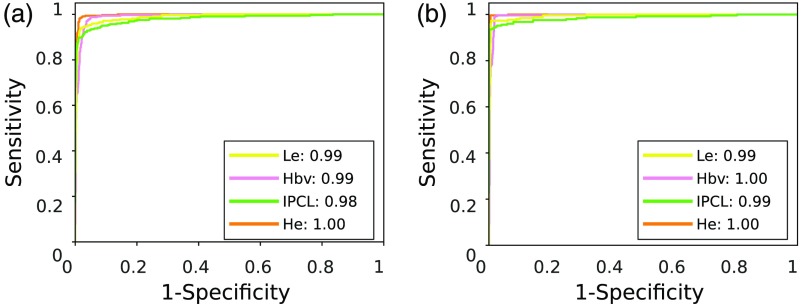
As shown in Table 5, when varying in , the median for monotonically increased from 93% (base case) to 98% (). The corresponding statistics are shown in Fig. 7(a). In particular, as can be seen by comparing the confusion matrices in Figs. 4(b) and 7(b), the classification recall increased from 98% (healthy tissue), 94% (hypertrophic vessels), 92% (leukoplakia), 86% (IPCL-like vessels) to 100% (healthy tissue), 99% (hypertrophic vessels), 97% (leukoplakia), and 93% (IPCL-like vessels). Despite the fact that a slightly lower improvement was observed for the IPCL class with respect to the other classes, it is worth noting that, with the proposed approach, no misclassification occurred between the IPCL class and the healthy tissue one. The same happened also for the hypertrophic vessel class while only the 2% of samples with leukoplakia was misclassified as healthy. The same trend was observed for and . The ROC curves for and are shown in Fig. 8. The AUC is reported for each of the analyzed classes [ ()]. The increment came at the cost of a reduction of the percentage of confident patches to 80% () of all the patches in the testing set, which corresponds to patches. However, as shown in Fig. 6(b), with the exclusion of low-confidence patches, even in the worst case (classification of tissue with IPCL-like vessels), the accuracy still reached 93%.
Table 5.
Median (first quartile to third quartile) class-specific recall (), precision (), and score () are reported at different level of confidence () on SVM classification.
| 93 (90–96) | 95 (91–97) | 96 (92–99) | 98 (93–99) | 98 (95–100) | |
| 94 (91–95) | 95 (92–96) | 97 (93–98) | 97 (95–98) | 99 (96–100) | |
| 92 (91–95) | 94 (93–96) | 95 (94–97) | 96 (95–97) | 98 (97–99) |
Fig. 7.
Effect of varying the threshold () on the classification confidence level. Classification is obtained using LBP and first-order statistics with SVM. (a) Boxplot of the class-specific accuracy-rate () for different . The percentage of confident patches for each is reported above each boxplot. refers to classification without confidence estimation. (b) Normalized confusion matrix for . The number of patches for each class is reported in parenthesis.
Fig. 8.
ROC curves at different level of confidence on classification. Each curve refers to one of the laryngeal tissue classes. He, healthy tissue; Hbv, tissue with hypertrophic vessels; Le, leukoplakia; and IPCL, tissue with IPCL-like vessels. The area under the ROC curve, for each curve, is reported in the legend. Classification is obtained using LBP and first-order statistics with SVM. (a) ROC curves for . (b) ROC curves for .
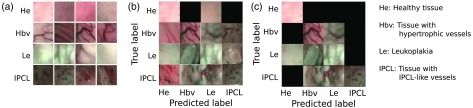
Figure 9 shows visual samples of patches in our dataset [Fig. 9(a)], as well as samples of patch classification results at the base case [Fig. 9(b)] and after the introduction of the confidence measure () [Fig. 9(c)].
Fig. 9.
Visual samples of classification results. Classification is obtained using LBP and first-order statistics with SVM. (a) Examples of patches for the four tissue classes in our dataset. (b) Visual confusion matrices for the base case, i.e., without the inclusion of confidence estimation and (c) after including the confidence estimation with . Black squares indicate the absence of misclassification between the true and predicted label.
4. Discussion
In this paper, we presented and fully evaluated an innovative approach to the computer-aided classification of laryngeal tissues in NBI laryngoscopy. Different textural features were tested to investigate the best feature set to characterize malignant and healthy laryngeal tissues: texture-based global descriptors ( and ) and first-order statistics (). A confidence measure on the SVM-based classification was used to estimate the reliability of the classification results.
When comparing noncombined features (, , ), the highest classification performance was obtained with . In general, performed worse with respect to . This is probably due to the GLCM lack of robustness to illumination condition changes, which are typically encountered during endoscopic examination.
SVM has shown comparable performance with respect to RF and kNN, while significant differences () were found with respect to NB. This is probably due to NB not being able to handle high-dimensional feature spaces such as ours. This is in accord with previous findings in the literature.25,37,38 Accordingly, for the tested dataset, we could not conclude that SVM performance was better than the one obtained with RF and kNN.
When comparing the proposed method with the state of the art, the classification based on significantly outperformed () the one proposed by Barbalata and Mattos,6 also when excluding the leukoplakia class. Since the method in Ref. 6 relies on accurate vessel segmentation (to extract vascular shape-based features), a possible reason of such result could be related to the challenging nature of our validation dataset, which, however, summarizes the diagnostic scenario well. Indeed, vessel segmentation was not trivial [Fig. 6(a)] due to (i) the noisy nature of NBI data, (ii) the low contrast of vessels in patches with healthy tissue and leukoplakia, and (iii) the irregular shape of IPCL-like vessels. With texture-based features, higher classification performance was achieved with respect to shape-based features since texture-based feature computation does not require vessel segmentation. Moreover, the texture-based features here used are invariant to illumination changes and endoscope pose, which makes them suitable for the analyzed scenario.
The classification performance obtained with was further increased by estimating the confidence of the SVM classification, with few misclassifications of confident patches that mainly occurred with high-challenging vascular patterns, whose classification is not trivial also for the human eye [Fig. 9(c)]. Such results support our hypothesis that the proposed approach is suitable for classifying laryngeal tissues with high reliability, since it automatically estimates its own confidence level and provides high classification accuracy for confident patches.
A limitation of the proposed study could be seen in its patch-based nature. Note, however, that the choice of focusing on patches manually extracted under the supervision of an expert clinician was driven by the necessity of having a controlled and representative dataset to fairly evaluate different features. As future work, instead of manually selecting squared patches, we plan to implement more automatic strategies, such as superpixel segmentation.39 The features could be directly extracted from superpixels, as to classify each superpixel as belonging to one of the analyzed laryngeal classes. Moreover, considering that recent researches on gastrointestinal image classification40,41 are focusing more and more on convolutional neural networks (CNN), it would be interesting to exploit also CNN as feature extractor for comparison.
Our expectation is that research on the classification of laryngeal tissues will be empowered by the proposed work, becoming a topic of interest for the scientific community, which until now has mainly focused on other anatomical sites, such as the gastrointestinal tract. Moreover, we hope this study will motivate a more structured and widespread data collection in clinics and the sharing of such data through public databases. Despite the dimension of the analyzed dataset (330 images) is comparable with that of similar researches (e.g., Barbalata and Mattos6 with 120 images, and Turkmen et al.7 with 70 images), larger amounts of data would bring the possibility of further exploring machine-learning classification algorithms, e.g., to classify a larger number of laryngeal malignant tissues.
In conclusion, the most significant contribution of this work is showing that LBP-based features and SVM can differentiate laryngeal tissues accurately. This is highly beneficial for practical uses. Comparing with other state-of-the-art methods in the area, the proposed method is simpler and the result is more accurate. It is acknowledged that further research is required to further ameliorate the algorithm as to offer all possible support for diagnosis, but the results presented here are surely a promising step toward a helpful endoscope-integrated processing system to support the diagnosis of early stage SCC.
Biographies
Sara Moccia received her BSc degree in 2012 and her MSc degree in 2014. She is a PhD student in the Department of Advanced Robotics at the Istituto Italiano di Tecnologia (IIT) and in the Department of Information, Electronics and Bioengineering at Politecnico di Milano. Her research interests include computer vision, image processing, and machine learning applied to the medical field.
Elena De Momi received her MSc degree in 2002 and her PhD in 2006. She is an assistant professor in the Electronic, Information, and Bioengineering Department (DEIB) of Politecnico di Milano. She was the cofounder of the Neuroengineering and Medical Robotics Laboratory, in 2008, being responsible of the medical robotics section. She has been an associate editor of the Journal of Medical Robotics Research, the International Journal of Advanced Robotic Systems, and Frontiers in Robotics and AI. Since 2016, she has been an associated editor of the IEEE International Conference on Robotics and Automation.
Marco Guarnaschelli received his BSc degree in 2014 and his MSc degree in 2017. He graduated from the Politecnico di Milano in biomedical engineering. His research interests include computer vision, image processing, and machine learning.
Matteo Savazzi received his BSc degree in 2014 and his MSc degree in 2017. He graduated from the Politecnico di Milano in biomedical engineering. His research interests include computer vision, image processing, and machine learning.
Andrea Laborai received his MSc degree in 2014. He has been working as a resident in otorhinolaryngology at the Ospedale Policlinico San Martino University of Genova since 2014. He spent six months of the residency program at the Spedali Civili of Brescia.
Luca Guastini has been working since 2012 both as a clinician of Genoa’s IRCCS San Martino-IST University Hospital and as an assistant professor of otorhinolaryngology at the University of Genoa’s Medical and Postgraduate School. So far, he has published 38 papers for a selection of prestigious national and international journals, 12 book chapters, and as many as 110 congress and symposium-related publications. He is a member of the European Laryngological Society and Italian Otorhinolaryngology Society.
Giorgio Peretti received his MSc degree in 1983. He is a full professor at Genoa University and the director of the Department of Otorhinolaryngology, University of Genoa, Italy. So far, he has authored 92 papers for a selection of national and international journals, 45 book chapters, and as many as 355 congress and symposium-related publications. His main fields of research include angiogenesis in early stage laryngeal carcinoma, narrow-band imaging in diagnosis and surveillance of head and neck cancer, combination of endoscopy and imaging in staging, and surveillance of head and neck cancer.
Leonardo S. Mattos received his BSc degree in 1998, his MSc degree in 2003, and his PhD in 2007. He is a team leader at the Istituto Italiano di Tecnologia (IIT). His research interests include robotic microsurgery, user interfaces and systems for safe and efficient teleoperation, computer vision, and automation. He has been a researcher at IIT’s Advanced Robotics Department since 2007. He was the PI and coordinator of the European project and is currently the PI and coordinator of the project TEEP-SLA.
Disclosures
The authors have no conflict of interest to disclose.
References
- 1.Markou K., et al. , “Laryngeal cancer: epidemiological data from Northern Greece and review of the literature,” Hippokratia 17(4), 313–318 (2013). [PMC free article] [PubMed] [Google Scholar]
- 2.Unger J., et al. , “A noninvasive procedure for early-stage discrimination of malignant and precancerous vocal fold lesions based on laryngeal dynamics analysis,” Cancer Res. 75(1), 31–39 (2015). 10.1158/0008-5472.CAN-14-1458 [DOI] [PubMed] [Google Scholar]
- 3.Piazza C., et al. , “Narrow band imaging in endoscopic evaluation of the larynx,” Curr. Opin. Otolaryngol. Head Neck Surg. 20(6), 472–476 (2012). 10.1097/MOO.0b013e32835908ac [DOI] [PubMed] [Google Scholar]
- 4.Poels P. J., de Jong F. I., Schutte H. K., “Consistency of the preoperative and intraoperative diagnosis of benign vocal fold lesions,” J. Voice 17(3), 425–433 (2003). 10.1067/S0892-1997(03)00010-9 [DOI] [PubMed] [Google Scholar]
- 5.Isenberg J. S., Crozier D. L., Dailey S. H., “Institutional and comprehensive review of laryngeal leukoplakia,” Ann. Otol. Rhinol. Laryngol. 117(1), 74–79 (2008). 10.1177/000348940811700114 [DOI] [PubMed] [Google Scholar]
- 6.Barbalata C., Mattos L. S., “Laryngeal tumor detection and classification in endoscopic video,” IEEE J. Biomed. Health Inf. 20(1), 322–332 (2016). 10.1109/JBHI.2014.2374975 [DOI] [PubMed] [Google Scholar]
- 7.Turkmen H. I., Karsligil M. E., Kocak I., “Classification of laryngeal disorders based on shape and vascular defects of vocal folds,” Comput. Biol. Med. 62, 76–85 (2015). 10.1016/j.compbiomed.2015.02.001 [DOI] [PubMed] [Google Scholar]
- 8.Maier-Hein L., et al. , “Surgical data science: enabling next-generation surgery,” arXiv preprint, arXiv:1701.06482 (2017).
- 9.Li B., Meng M. Q.-H., “Texture analysis for ulcer detection in capsule endoscopy images,” Image Vision Comput. 27(9), 1336–1342 (2009). 10.1016/j.imavis.2008.12.003 [DOI] [Google Scholar]
- 10.Zhang Y., et al. , “Tissue classification for laparoscopic image understanding based on multispectral texture analysis,” Proc. SPIE 9786, 978619 (2016). 10.1117/12.2216090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang P., Cong Y., Guan M., “A computer-aided lesion diagnose method based on gastroscopeimage,” in Int. Conf. on Information and Automation (ICIA), pp. 871–875, IEEE; (2012). 10.1109/ICInfA.2012.6246904 [DOI] [Google Scholar]
- 12.Shen X., et al. , “Lesion detection of electronic gastroscope images based on multiscale texture feature,” in IEEE Int. Conf. on Signal Processing, Communication and Computing (ICSPCC), pp. 756–759, IEEE; (2012). 10.1109/ICSPCC.2012.6335638 [DOI] [Google Scholar]
- 13.Van Der Sommen F., et al. , “Computer-aided detection of early cancer in the esophagus using HD endoscopy images,” Proc. SPIE 8670, 86700V (2013). 10.1117/12.2001068 [DOI] [Google Scholar]
- 14.Misawa M., et al. , “Accuracy of computer-aided diagnosis based on narrow-band imaging endocytoscopy for diagnosing colorectal lesions: comparison with experts,” Int. J. Comput. Assisted Radiol. Surg. 12(5), 757–766 (2017). 10.1007/s11548-017-1542-4 [DOI] [PubMed] [Google Scholar]
- 15.Orozco J., et al. , “Confidence assessment on eyelid and eyebrow expression recognition,” in 8th IEEE Int. Conf. on Automatic Face and Gesture Recognition, pp. 1–8, IEEE; (2008). 10.1109/AFGR.2008.4813454 [DOI] [Google Scholar]
- 16.Delany S. J., et al. , “Generating estimates of classification confidence for a case-based spam filter,” in Int. Conf. on Case-Based Reasoning (ICCBR), Vol. 3620, pp. 177–190, Springer; (2005). [Google Scholar]
- 17.Zhang C., Kodell R. L., “Subpopulation-specific confidence designation for more informative biomedical classification,” Artif. Intell. Med. 58(3), 155–163 (2013). 10.1016/j.artmed.2013.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLaren B., Ashley K., “Helping a CBR program know what it knows,” in Int. Conf. on Case-Based Reasoning Research and Development, pp. 377–391 (2001). [Google Scholar]
- 19.Mendrik A. M., et al. , “Noise reduction in computed tomography scans using 3-D anisotropic hybrid diffusion with continuous switch,” IEEE Trans. Med. Imaging 28(10), 1585–1594 (2009). 10.1109/TMI.2009.2022368 [DOI] [PubMed] [Google Scholar]
- 20.Nanni L., Lumini A., Brahnam S., “Local binary patterns variants as texture descriptors for medical image analysis,” Artif. Intell. Med. 49(2), 117–125 (2010). 10.1016/j.artmed.2010.02.006 [DOI] [PubMed] [Google Scholar]
- 21.Ojala T., Pietikainen M., Maenpaa T., “Multiresolution gray-scale and rotation invariant texture classification with local binary patterns,” IEEE Trans. Pattern Anal. Mach. Intell. 24(7), 971–987 (2002). 10.1109/TPAMI.2002.1017623 [DOI] [Google Scholar]
- 22.Haralick R. M., et al. , “Textural features for image classification,” IEEE Trans. Syst. Man Cybern. SMC-3(6), 610–621 (1973). 10.1109/TSMC.1973.4309314 [DOI] [Google Scholar]
- 23.Onder D., Sarioglu S., Karacali B., “Automated labelling of cancer textures in colorectal histopathology slides using quasi-supervised learning,” Micron 47, 33–42 (2013). 10.1016/j.micron.2013.01.003 [DOI] [PubMed] [Google Scholar]
- 24.Burges C. J., “A tutorial on support vector machines for pattern recognition,” Data Min. Knowl. Discovery 2(2), 121–167 (1998). 10.1023/A:1009715923555 [DOI] [Google Scholar]
- 25.Csurka G., et al. , “Visual categorization with bags of keypoints,” in Workshop on Statistical Learning in Computer Vision, ECCV, Vol. 1, Prague, pp. 1–22 (2004). [Google Scholar]
- 26.Lin Y., et al. , “Large-scale image classification: fast feature extraction and SVM training,” in IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), pp. 1689–1696, IEEE; (2011). 10.1109/CVPR.2011.5995477 [DOI] [Google Scholar]
- 27.Keller J. M., Gray M. R., Givens J. A., “A fuzzy K-nearest neighbor algorithm,” IEEE Trans. Syst. Man Cybern. SMC-15(4), 580–585 (1985). 10.1109/TSMC.1985.6313426 [DOI] [Google Scholar]
- 28.Lewis D. D., “Naive (Bayes) at forty: the independence assumption in information retrieval,” in European Conf. on Machine Learning, pp. 4–15, Springer; (1998). [Google Scholar]
- 29.Breiman L., “Random forests,” Mach. Learn. 45(1), 5–32 (2001). 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 30.Wu T.-F., Lin C.-J., Weng R. C., “Probability estimates for multi-class classification by pairwise coupling,” J. Mach. Learn. Res. 5, 975–1005 (2004). [Google Scholar]
- 31.Moccia S., et al. , “Uncertainty-aware organ classification for surgical data science applications in laparoscopy,” arXiv preprint arXiv:1706.07002 (2017). [DOI] [PubMed]
- 32.Marcot B. G., “Metrics for evaluating performance and uncertainty of Bayesian network models,” Ecol. Modell. 230, 50–62 (2012). 10.1016/j.ecolmodel.2012.01.013 [DOI] [Google Scholar]
- 33.Lukes P., et al. , “The role of NBI HDTV magnifying endoscopy in the prehistologic diagnosis of laryngeal papillomatosis and spinocellular cancer,” BioMed Res. Int. 2014, 1–7 (2014). 10.1155/2014/285486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moccia S., et al. , “Automatic workflow for narrow-band laryngeal video stitching,” in IEEE 38th Annual Int. Conf. of the Engineering in Medicine and Biology Society (EMBC), pp. 1188–1191, IEEE; (2016). 10.1109/EMBC.2016.7590917 [DOI] [PubMed] [Google Scholar]
- 35.http://docs.opencv.org/3.1.0/index.html.
- 36.http://scikit-learn.org/stable/index.html.
- 37.Duro D. C., Franklin S. E., Dubé M. G., “A comparison of pixel-based and object-based image analysis with selected machine learning algorithms for the classification of agricultural landscapes using SPOT-5 HRG imagery,” Remote Sens. Environ. 118, 259–272 (2012). 10.1016/j.rse.2011.11.020 [DOI] [Google Scholar]
- 38.Bosch A., Zisserman A., Munoz X., “Image classification using random forests and ferns,” in IEEE 11th Int. Conf. on Computer Vision (ICCV), pp. 1–8, IEEE; (2007). 10.1109/ICCV.2007.4409066 [DOI] [Google Scholar]
- 39.Li Z., Chen J., “Superpixel segmentation using linear spectral clustering,” in IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), pp. 1356–1363, IEEE; (2015). 10.1109/CVPR.2015.7298741 [DOI] [Google Scholar]
- 40.Zhu R., Zhang R., Xue D., “Lesion detection of endoscopy images based on convolutional neural network features,” in 8th Int. Congress on Image and Signal Processing (CISP), pp. 372–376, IEEE; (2015). 10.1109/CISP.2015.7407907 [DOI] [Google Scholar]
- 41.Park S. Y., Sargent D., “Colonoscopic polyp detection using convolutional neural networks,” Proc. SPIE 9785, 978528 (2016). 10.1117/12.2217148 [DOI] [Google Scholar]