Abstract
The ability to identify cancer lesions with endogenous biomarkers is currently limited to tumours ~1 cm in diameter. We recently reported an exogenously administered tumour-penetrating nanosensor that sheds, in response to tumour-specific proteases, peptide fragments that can then be detected in the urine. Here, we report the optimization, informed by a pharmacokinetic mathematical model, of the surface presentation of the peptide substrates to both enhance on-target protease cleavage and minimize off-target cleavage, and of the functionalization of the nanosensors with tumour-penetrating ligands that engage active trafficking pathways to increase activation in the tumour microenvironment. The resulting nanosensor discriminated sub-5 mm lesions in human epithelial tumours and detected nodules with median diameters smaller than 2 mm in an orthotopic model of ovarian cancer. We also demonstrate enhanced receptor-dependent specificity of signal generation in the urine in an immunocompetent model of colorectal liver metastases, and in situ activation of the nanosensors in human tumour microarrays when re-engineered as fluorogenic zymography probes.
Early detection of tumours offers hope for greatly improved outcomes for cancer patients1. For the majority of cancer types, diagnosis when the disease is localized to the organ of origin correlates with significantly greater long-term survival compared to when disease has spread to distant sites, largely because currently available therapeutics are most effective when patients are treated in early stages of disease2–4. Despite the need for technology that can detect early stage disease, the predictive value of existing biomarkers used to diagnose cancers is limited. For example, recent studies have shown that screening with the blood biomarker CA-125 for ovarian cancer diagnosis does not improve patient prognosis5. However, screening with biomarkers that are predictive can significantly improve patient outcomes: colorectal cancer (CRC) mortality at 30-years follow-up was reduced by 32% with annual fecal occult-blood testing6. These studies indicate more research is needed to develop biomarkers that are predictive of disease.
Despite the progress in improving tumour detection tools, clinical detection of tumours is limited to masses ~1 cm in diameter via imaging techniques (e.g. MRI and PET)7,8 and analysis of blood biomarkers shed by the tumour (e.g. proteins and cell free nucleic acids)8,9. It is estimated that it can take up to ten years to establish tumours this size from initial tumourigenesis8, leaving a large window of opportunity for early diagnosis to improve patient outcomes. A minimally invasive tool to screen patients would significantly impact patient prognosis. To this end, we have previously developed an exogenously administered activity-based nanosensor (ABN) that, in response to protease activity in the tumour, sheds peptide fragments that are ultimately concentrated in the urine10,11. Aberrantly expressed proteases are promising candidate enzymes to leverage in both diagnostics and therapeutics as they play critical roles cancer progression12. While this ABN has shown promise for tumour detection in a subcutaneous flank model of CRC, a tool suitable for early detection would require magnitude-fold gains in sensitivity.
Whereas the focus of conventional blood-based biomarkers is to identify specific signatures that are a barometer for disease state and then develop methods to sensitively detect them, our synthetic system has the advantage in that it can be engineered to both maximize the amplitude of signal and specificity of signal generation. A multi-compartment pharmacokinetic model built as a system of ordinary differential equations (ODEs) we have previously built predicted that a 100-fold improvement in parameters related to tumour-specific signal generation would be necessary for the detection sub-5 mm sized tumours13. Inspired and instructed by this model, we chose parameters to optimize our urinary diagnostic with the following design principles: detection of molecular events that are conserved hallmarks of tumour invasiveness, minimal interaction with the non-tumour host biology, and robust sampling of the tumour microenvironment. We pursued two strategies, which we believed would act in synergy: (1) optimal proteolytic substrate presentation on the nanoparticle surface to maximize the relative on- vs. off-target cleavage rates and (2) increased tumour access using tumour-penetrating ligands. Herein, we engineered our ABN platform based on the above design criteria, and achieved sub-5 mm detection of tumour lesions in disease models that recapitulate important features of human disease. Furthermore, we validated that our sensors respond to human cancer biopsy samples.
MMP9 is upregulated across human cancers
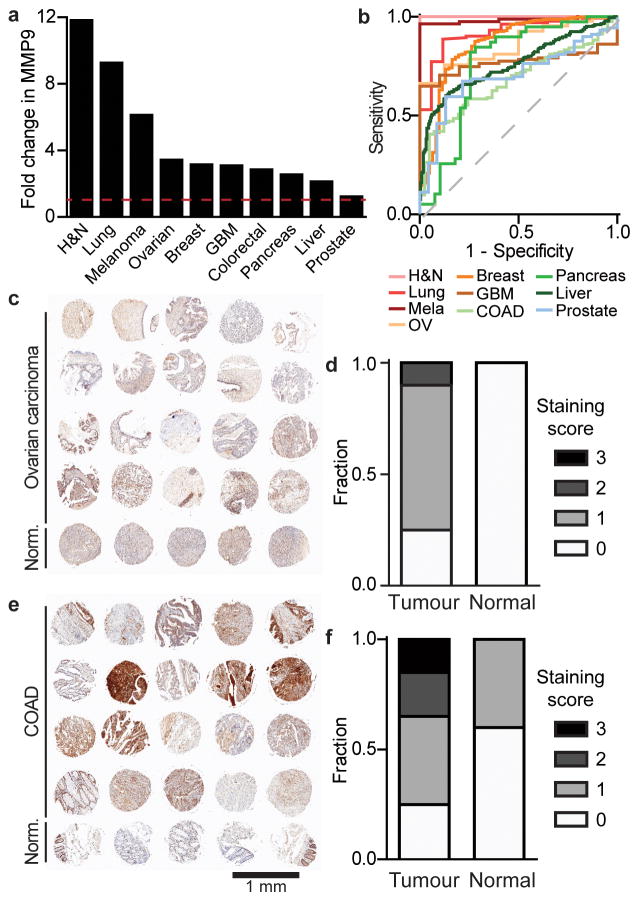
Our first objective was to engineer the ABN based on a proteolytic target that is highly elevated across numerous human cancers and that has a fundamental biological role in tumour progression. Analysis of mRNA expression data from Oncomine and TCGA showed that matrix metalloproteinase 9 (MMP9) levels are significantly upregulated across many human cancer types compared to healthy controls (Fig. 1a), and can classify cancer as distinct from normal tissue, as established by constructing receiver operating characteristic (ROC) curves (median area under the curve (AUC) = 0.81; Fig. 1b). Furthermore, MMP9 mRNA levels are consistent across all stages, indicating it can be used for both early and late stage diagnosis (Fig. S1a–b). Using MMP9 as a disease marker may be valuable in identifying aggressive from indolent cancers as it is known to play a critical role in the angiogenic switch needed for access to host vasculature when tumours reach 1–2 mm in diameter14. Therefore, MMP9 activity measurements may prospectively reflect disease progression, and not merely detect a byproduct of tumour growth, as is the case with many existing blood biomarkers.
Figure 1. MMP9 is upregulated across human cancers.
(a) Analysis of fold change of MMP9 mRNA expression in tumour compared to healthy controls compiled from Oncomine and TCGA data. (b) ROC curves constructed based on MMP9 mRNA expression data to represent how well MMP9 can classify various cancer types compared to healthy controls (median AUC = 0.81). (c) IHC of MMP9 protein in normal and ovarian carcinoma from a human TMA and (d) corresponding staining scores for tumour (n = 20 cores) versus normal (n = 5 cores) tissue. Blinded analysis of staining was done by a pathologist; 3 for the highest level of staining and 0 for no visible staining. (e) IHC of MMP9 protein in normal colon and colon adenocarcinoma (COAD) from a TMA and (f) corresponding expression scores. Full TMA staining shown in Fig. S1d.
We performed MMP9 immunohistochemical (IHC) staining of a tumour tissue microarray (TMA), which was blindly scored by a pathologist and confirmed elevated MMP9 protein levels across many human cancers (Fig. 1c–f, Fig. S1b–c). Thus, based on our analysis of human protein expression and evidence in the literature14,15, we proceeded to engineer our ABN as a probe for MMP9 activity in vivo for platform development, and to establish design principles that could be applied to other tumour-specific proteases found to be upregulated in cancer. Existing point-of-care technologies inadequately access MMP9, as it acts locally at the tumour site14 and thus, cannot be assayed from body fluids with high sensitivity and specificity16. Further exploration into other MMPs in breast cancer samples from TCGA revealed several other biomarker candidates that had elevated mRNA levels (Fig. S1e). These proteases could be explored with multiplexed substrates in future iterations of ABNs.
ABN optimization achieves magnitude-fold improvement
We pursued two strategies to engineer an ultrasensitive nanosensor: (1) presentation of peptide substrates on the nanoparticle surface for maximal on-target and minimal off-target protease cleavage, and (2) modification of ABNs with tumour-penetrating ligands that engage active tumour trafficking pathways initiated by receptor binding. We hypothesized these two strategies would cooperate to increase cancer-specific signal generation.
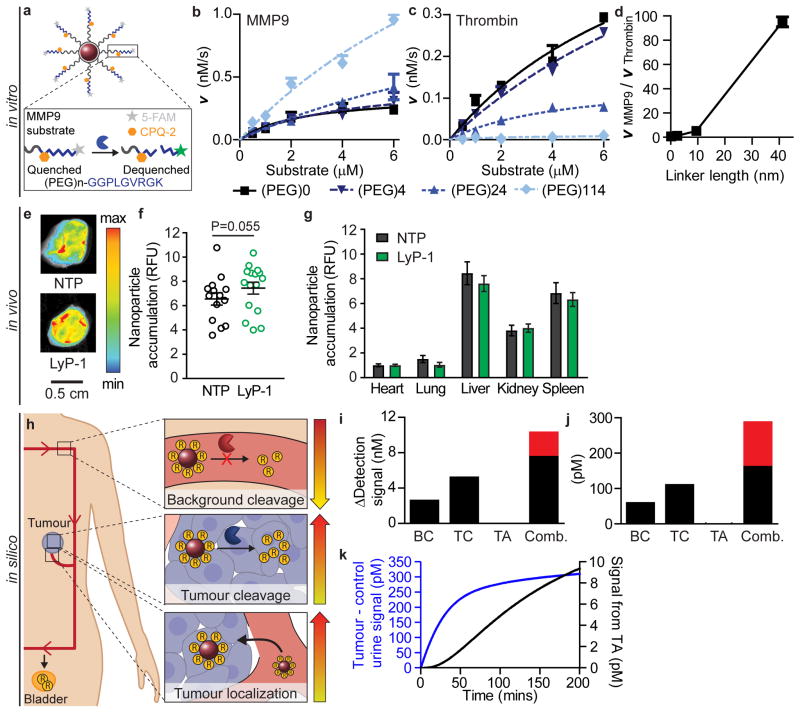
Extrapolating from experimental observations of enzyme-substrate interactions on monolayer surfaces17, we hypothesized surface presentation would allow us to tune specificity between on- and off-target proteolytic cleavage rates of our MMP9 substrate. We tested cleavage kinetics of an MMP9 peptide substrate (PLGVRGK), flanked by a fluorescence resonance energy transfer (FRET) pair, at varying distances from our nanoparticle core (Fig. 2a). MMP9 cleavage rates of this substrate increased with increasing presentation distance for all substrate concentrations tested (0.5, 1, 2, 4, and 6 μM); as an example, cleavage velocity (V) increased from 0.24 to 0.96 nM/s at a 6 μM substrate concentration (Fig. 2b). Unexpectedly, this phenomenon was not generalizable across enzymes; cleavage rates by the serine protease thrombin decreased with increasing presentation length when the same MMP9 substrate was tested, with V (at 6 μM substrate concentration) decreasing from 0.29 to 0.01 nM/s (Fig. 2c). Nor was increasing the tether length generalizable across substrates; thrombin cleavage of a thrombin-selective substrate (fPRSGGG, lower case = D-amino acid) was optimally presented at an intermediate length (Fig. S2). Taken together, these data indicate it is possible to significantly augment the signal-to-noise ratio of ABNs simply by tuning presentation on nanoparticle surfaces (Fig. 2d). Optimizing substrate presentation is a possible solution to the challenge of engineering protease specificity for synthetic substrates8,19 that has been relatively unexplored in the literature and could also be leveraged in other enzyme-activated technologies20,21. We exploited the increased on-target and decreased off-target protease cleavage at long tether lengths to present the MMP9 substrate on our ABN.
Figure 2. In vitro and in vivo experiments and in silico evaluation enables engineering of an optimized ABN.
(a) Schematic showing FRET-based protease substrate displayed on an ABN surface. Cleavage velocities of an MMP9 substrate by (b) MMP9 or (c) thrombin at 0, 4, 24, 114 subunits of PEG molecules. Lines represent fit to Michaelis-Menten equation (mean +SEM, n = 2–3). (d) Ratio of MMP9 and thrombin velocities at 6 μM substrate concentration. Accumulation of LyP-1 ABN and non-penetrating ABN control (NTP) in (e) excised tumours and quantification of ABN signal in (f) tumours and (g) organs (mean ±SEM, n = 7 for NTP and 8 for Lyp-1 ABN). (h) Schematic of parameters varied in a mathematical pharmacokinetic model. Change in detection signal when optimized parameters (BC = background cleavage, TC = tumour cleavage, and TA = tumour accumulation) are applied to (i) 10 mm versus (j) 5 mm tumours (red denotes additional signal gained when parameters are combined compared to BC+TC+TA). (k) Kinetic traces of detected urine signal (blue line) with all three parameters improvements and signal contribution from increasing accumulation term (TA; black line).
Next, we turned to the question of how to localize our MMP9 sensor to sample the tumour microenvironment in a living organism. Passive nanoparticle accumulation via the enhanced permeation and retention (EPR) effect is heterogeneous in human tumours22; however, ligands can be deployed to improve tumour distribution23. Although tumour-targeting ligands may not significantly affect overall bulk accumulation of nanomaterials into the tumour23,24, they can increase cell-specific association and retention in the tumour environment and positively impact clinical outcomes25,26. Furthermore, we hypothesized that the benefit of tissue-level localization is magnified when applied to an activity-based system, due to the synergy of the accumulation and enzymatic amplification effects. To this end, we employed a unique class of peptide ligands discovered by in vivo phage display that mediate active internalization and transport of nanomaterials past tumour stroma and into the tumour tissue, referred to as tumour-penetrating peptides27,28.
We combined the dual strategies of optimized MMP9 substrate presentation and increased tumour tissue access into our engineered urinary diagnostic platform, described previously10. We decorated our ABNs with the cyclic tumour-penetrating peptide, LyP-1 (CGNKRTRGC), previously shown to increase penetration of a variety of nanomaterials deep into the tumour parenchyma by binding its cognate receptor, p32, and engaging a secondary receptor, NRP-1, which triggers an active transport pathway29,30. We employed an MDA-MB-435 subcutaneous flank xenograft model as a model of an epithelial tumour. MDA-MB-435 tumours have elevated p3231 and MMP932 expression. Non-penetrating (NTP) and LyP-1 ABNs were matched for substrate valency (Fig. S3a–b) and were measured to have a ~60 nm hydrodynamic diameter and a slightly negative surface potential (Fig. S3c). Visualization of the fluorescent core of the ABN three hours after administration revealed accumulation in the tumour at similar levels to organs (Fig. 2e–g), with a moderate increase (~20%) in tumour accumulation and concomitant decreases in off-target organ accumulation with the addition of LyP-1 (Fig. 2g).
We employed our previously developed pharmacokinetic mathematical model as a tool to understand how our improvements could affect the performance of the nanosensor13. The model is comprised of a set of five ODEs solved in three separate compartments (blood, tumour, and bladder) derived from transport and biochemical governing equations (e.g., Fick’s Law and Michaelis-Menten kinetics) with variables fit to experimental data (SI Text). Based on in vitro and in vivo experimental ABN data presented above (Fig. 2a–g), we modified three parameters of the model: tumour protease-specific cleavage ( ), off-target cleavage ( ), and tumour accumulation ( ) (Fig. 2h). When applied to tumours of moderate size (10 mm diameter), the model predicted cooperation between the three parameters: increases in detection signal were greater when parameters were input simultaneously compared to the sum of each parameter input individually (Fig. 2i, red bar). In fact, the model ascribed negligible benefit in detection signal with increased tumour accumulation by itself. When we applied the same analysis to small tumours (5 mm diameter), similar trends were observed (Fig. 2j). The model predicted interactions between parameters to be greater for the small tumour size, suggesting that multi-parameter optimizations are important when engineering ABNs for detection of small tumours. Lastly, we looked at the potential benefit from improvements in accumulation when combined with the other two parameters by measuring the difference in detection signal when all three parameters were input versus when just the improved protease cleavage parameters were input (Fig. 2k). The model predicted only modest gains in urine signal. We hypothesized that the contribution from spatial distribution, tissue-level localization, and overall changes in non-tumour biodistribution is greater than estimated. We therefore investigated performance of our optimized ABNs in a series of in vivo models with sub-5 mm tumour sizes.
Tumour-penetrating ABNs detect sub-5 mm diameter tumours
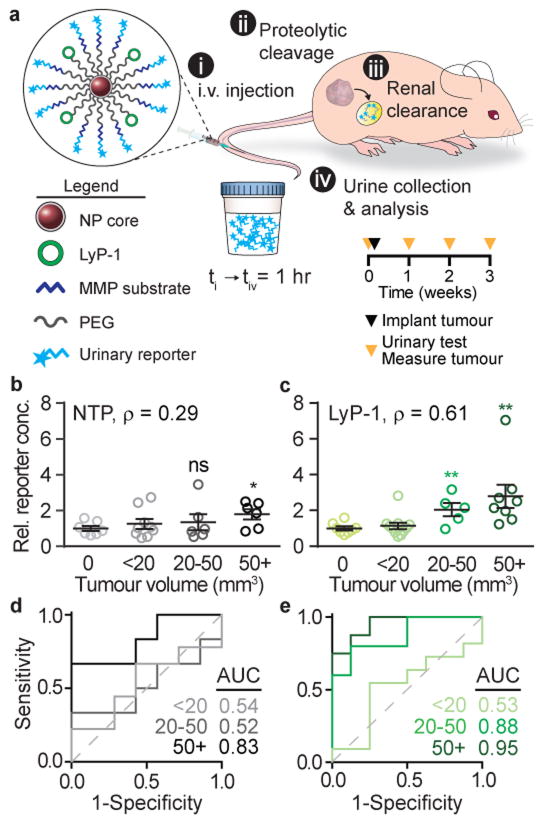
In order to establish the smallest tumour sizes that these tumour-penetrating ABNs could detect, a urinary test (measurement of peptide fragments in the urine 1 hour after nanoparticle administration) was performed at 1, 2, and 3 weeks after initial tumourigenesis of MDA-MB-435 xenografts (Fig. 3a). Measurement of ABN half-life in the blood and the free substrate clearance in the blood and urine show that signal decays rapidly with half-lives far shorter than the 7 day intervals between particle administrations (Fig. S4a–b). At the time of each weekly urine test, we also measured the total tumour volume per mouse as determined by caliper measurements (Fig. S5a–c). Non-penetrating ABNs with optimally presented MMP9 substrate discriminated tumours that were ~150 mm3 in volume (Fig. 3b) – volumes approximately 3-fold smaller than when substrate was directly presented on the nanoparticle surface11. Consistent with the in silico predictions of greater effects from multi-parameter optimization at small tumour sizes (Fig. 2j), the addition of tumour-penetrating ligand improved the detection to tumour volumes between 20–50 mm3, corresponding to 3.4–4.6 mm diameter tumours (Fig. 3c). ROC analysis for tumour volumes of 50 mm3 and greater yielded an AUC value of 0.83 for non-penetrating ABNs (Fig. 3d) compared to 0.95 for LyP-1 ABNs (Fig. 3e), indicating LyP-1 could increase the predictive power of our ABN. Urine signals were also elevated when tracked over time (Fig. S5d–e). The urinary limit of detection markedly improved with tumour-penetrating ligands, from 150 mm3 to 30 mm3, despite the moderate 1.2-fold increase in accumulation. This unanticipated dramatic enhancement may be explained by biological phenomena such as cell- and tissue-level distribution, as observed previously in the literature25,26. Although this level of biological sophistication was not built into our model, because the collective knowledge of nanoparticle-biological interactions is still an active area of research, our result demonstrates the value of molecular-targeting beyond simple measurements of bulk tissue accumulation24 and highlights the need to develop our understanding of tumour localization. Taken together, optimized substrate presentation and tumour-penetration led to over an order of magnitude improvement (~14-fold) in detection size compared with previous technology without these modifications, which detected ~500 mm3 tumour volumes11.
Figure 3. Optimized ABNs detect sub-5 mm diameter tumours.
(a) Top, schematic showing urine testing in a mouse flank model of MDA-MB-435. Bottom, study time course of urine testing. Relative reporter concentrations in the urine normalized to urine signal in healthy mice after (b) non-tumour penetrating (NTP) ABN (avg. tumour size for 50+ bin: 146 mm3) and (c) LyP-1 ABN (avg. tumour size for 50+ bin: 132 mm3) application. Tumours from 7 (NTP ABN) or 8 (LyP-1 ABN) mice measured over three weeks were binned by tumor size. Mean ±SEM, Student’s t-test, two-tailed, *P<0.05, **P<0.01; ρ Spearman correlation of urine signal to tumour size in individual mice. For distribution of sizes see Fig. S4. ROC and calculated AUC values for (d) NTP control and (e) LyP-1 ABN.
ABNs outperform a clinical test in an ovarian cancer model
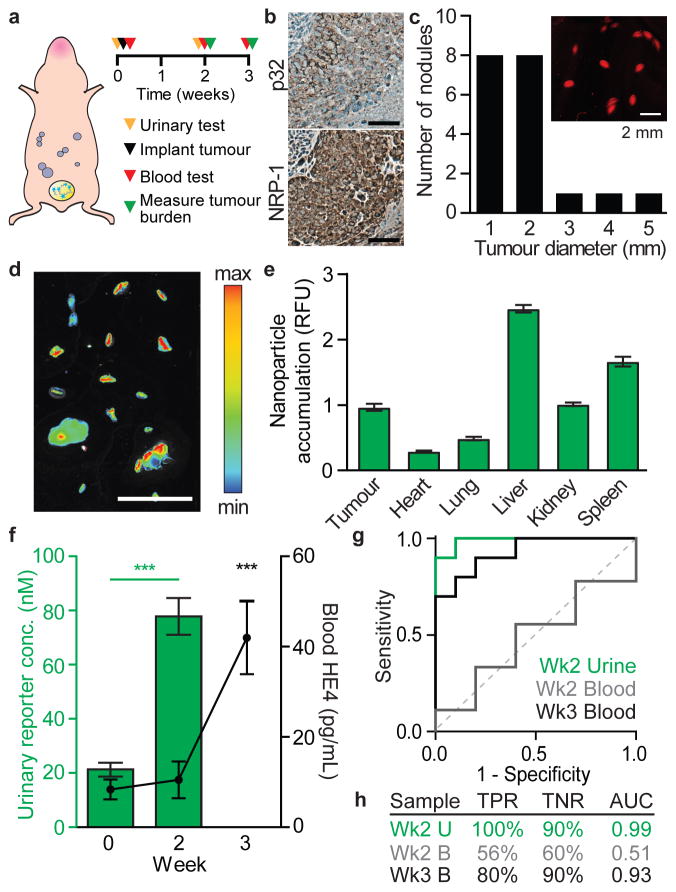
Early detection of ovarian cancer could yield substantial improvements in patient prognosis. 80% of current cases are diagnosed once the tumour has spread past the ovary due to inadequate detection methods33. In addition, 70% of ovarian cancer patients relapse after the standard of care, and therefore would greatly benefit from point-of-care longitudinal monitoring for early indications of relapse. Based on this clinical need, we challenged the tumour-penetrating ABN platform to detect low tumour burden in an orthotopic disseminated xenograft model of high-grade serous ovarian cancer. The mouse model was established by intraperitoneal implantation of the human cell line OVCAR-8 (Fig. 4a), which displays surface p3234 and NRP-1 (Fig. 4b), and secretes HE435 (Fig. S6a–c) and MMP936,37. We confirmed OVCAR-8 conditioned media can cleave the MMP9 substrate, which can be inhibited by the broad spectrum matrix metalloprotease inhibitor, Marimistat (Fig. S6d). Growth of tumours can be monitored by bioluminescence tracking of luciferized cells (Fig. S6e–f). When implanted into the peritoneal space of mice, OVCAR-8 cells form disseminated tumour nodules (timescale of weeks) and mice eventually accumulate ascites (timescale of months) — two disease presentations that also occur in human ovarian cancer patients. Two weeks after initial tumourigenesis, mice showed no overt signs of illness and had an average total tumour burden of 36 mm3 with a median nodule diameter of < 2 mm (Fig. 4c, Fig. S7). After intravenous ABN administration, resected tumours showed significant accumulation of nanoparticles (Fig. 4d, e) and a strong dependence between total tumour accumulation and tumour size (Fig. S7a).
Figure 4. Urinary biomarkers outperform blood biomarkers in detection of millimeter-sized lesions in orthotopic xenograft models of ovarian cancer.
(a) Schematic of LyP-1 ABN testing in an ovarian cancer model. (b) Immunohistochemical staining of excised tumours (scale bar: 100 μm). (c) Size distribution of tumour nodules retrieved from the intraperitoneal space of a representative animal at 2 weeks after tumour initiation. Average total tumour burden at this timepoint was 36 mm3 (Fig. S6b, n = 10). (d) Accumulation of ABNs in tumour nodules in a representative animal (scale bar = 1 cm) and (e) biodistribution of ABNs in organs (n = 10, mean ±SEM). (f) Urinary measurements of ABNs (green bars) and blood measurements of HE4 (black line). (n = 10 per group, mean ±SEM, Student’s t-test, two-tailed, ***P<0.001). (g) ROC of urinary diagnostic and blood biomarker. (h) Calculated sensitivity (true positive rate (TPR)) and specificity (true negative rate (TNR)) based on diagnostic cutoffs and ROC-AUC.
We tested whether our ABNs could: (1) detect low burden ovarian cancer and (2) outperform a clinical blood biomarker. We chose to compare our system to the blood biomarker HE4 as OVCAR-8 cells produce relatively high levels of HE4, on par with four other human ovarian cancer cell lines (Fig. S6b–c). At 36 mm3 total tumour burden, urinary signal from the ABN was significantly elevated compared to mice with no tumours (Fig. 4f). This threshold of sensitivity is critical as clinically approved imaging modalities can typically only resolve individual tumour nodules greater than 5 mm. Additionally, in a retrospective analysis of clinical data, it has been estimated that to decrease serous ovarian cancer mortality by 50% with an annual screen, a test would have to be sensitive to tumours < 5 mm38. In comparison, the blood biomarker HE4 was not elevated to detectable levels at this timepoint and was only significantly elevated three weeks post-tumourigenesis, when the average total burden was 88 mm3 (Fig. 4f, Fig. S7b–c). A comparison of the predictive powers between the two diagnostic systems results in ABN performance at 2-weeks (ROC-AUC = 0.99) exceeding the blood biomarker HE4 at 2-weeks (ROC-AUC = 0.51) and 3-weeks (ROC-AUC = 0.93), with improvement in true positive rate (TPR) and true negative rate (TNR; Fig. 4g–h). We confirmed that our tumour nodule retrieval methodology accounted for greater than 80% of the tumour burden by measuring luminescence pre- and post-resection (Fig. S7d–g). The tumour burden remaining can be attributed to uncollected macroscopic and microscopic lesions.
The difference in tumour volumes between detection via application of HE4 blood serum ELISA and tumour-penetrating ABNs was 2.4-fold. It has previously been estimated that early stage human serous ovarian carcinomas have a doubling time of four months38. Extrapolating the benefit seen in detection compared to HE4 and assuming a simple monoexponential growth model of human ovarian cancer (NT(t) = NT,0e(ln2/[DT])t where NT is the starting tumour size and DT is the doubling time)8, diagnosis could occur 5 months sooner with ABNs than the blood biomarker. Improving diagnosis time by 5 months could significantly impact the prognosis of ovarian cancer patients, especially considering ovarian cancers spend on average 1 year as stage III and IV cancers before they become clinically apparent38.
Ligand-receptor matching improves ABN urinary signal
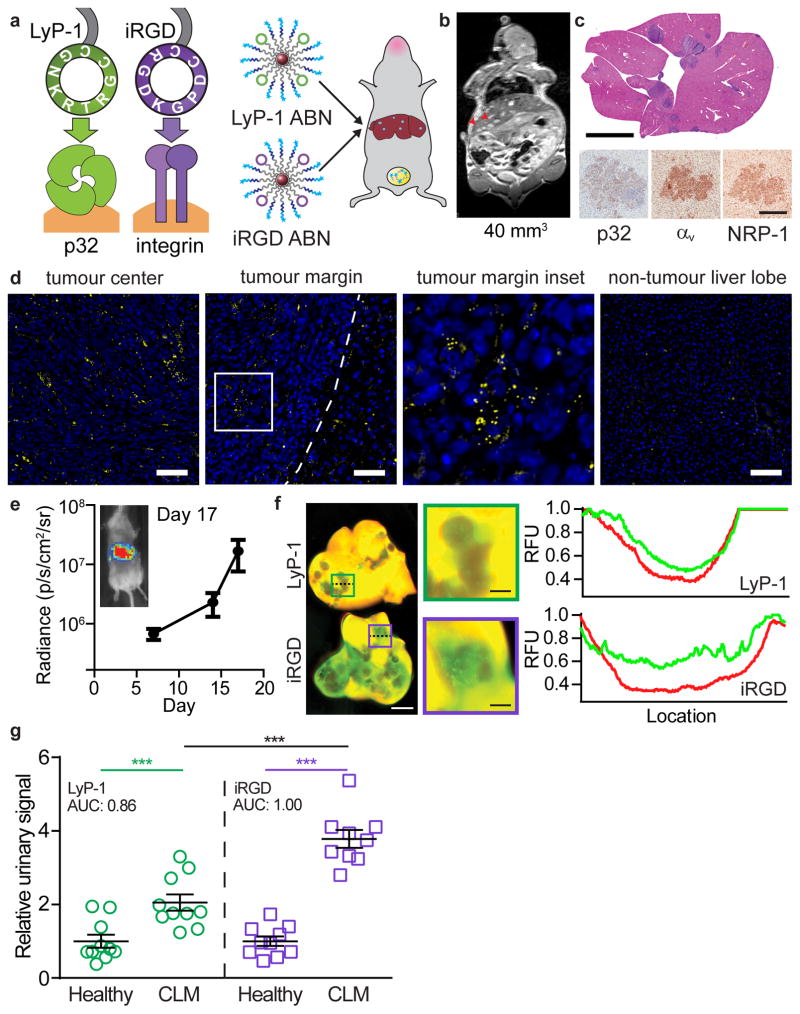
Receptor expression on tumours varies by tumour type and patient. In order to engineer a nanosensor with versatile tumour-penetrating capabilities, we utilized another tumour-penetrating ligand, iRGD (CRGDKGPDC), which engages the same active-internalization pathway as LyP-1, but binds to αvβ3/β5 integrin heterodimers as its primary receptor, which are ectopically overexpressed in a large subset of cancers39,40(Fig. 5a). We applied iRGD and LyP-1 ABNs to an immunocompetent mouse model of colorectal liver metastasis (CLM), induced via an intrasplenic injection of p32-negative, integrin-positive, and NRP-1-positive MC-26 cells (Fig. 5b–c, Fig. S8a–c). MC-26 cells are derived from a mouse colorectal cancer line and secretes MMP941, which can be detected in the tumour nodules as well as in the tumour-adjacent liver but not in non-tumour bearing liver lobes (Fig. 5d). Substrate conjugation on iRGD ABNs was at similar stoichiometries as the LyP-1 ABNs (SI Fig. 9a). Mouse and human MMPs have high homology42, and we confirmed that mouse MMP9 cleaves our substrate (Fig. S9b). A study of ABN clearance in immunocompetent BALB/c mice shows no signal is detectable after 24 hours in the urine, 48 hours in the blood, and 7 days in the tissues (Fig. S10a–c). This was consistent in the model of orthotopic ovarian cancer (Fig. S10d). Furthermore, no signs of toxicity were observed in histopathological assessment of tissues 3 hours, 24 hours, and 7 days after ABN administration compared with PBS injected controls (Fig. S11).
Figure 5. Minimally-invasive receptor classification in syngeneic liver metastasis by targeted ABNs.
(a) LyP-1 and iRGD both engage the same tumour trafficking pathway but rely on different primary receptors. LyP-1 and iRGD ABNs were tested in a liver metastasis model. (b) MRI image of colorectal liver metastasis (CLM) mouse with 40 mm3 of total tumour burden; tumour nodules are indicated by red arrows. (c) Top, H&E staining (scale bar = 5 mm). Bottom, immunohistochemical staining of tumour sections for primary receptors, p32 and αv integrin, and secondary receptor, NRP-1 (scale bar = 200 μm). (d) Imaging of tumour center, margin, and normal liver in sections from CLM mice. Tumour margin is indicated by dotted line and inset is indicated by white box. Sections were stained for nuclei (blue) and MMP9 (yellow), scale bar = 100 μm. (e) Bioluminescence imaging of CLM mice over time (average, ±SEM, n=20). ABNs were administered when luminescence readings reached 107 p/s/cm2/sr. (f) Fluorescent scans of tumours showing ABN accumulation (red: liver autofluorescence, green: nanoparticle, left scale bar = 2 cm, right scale bars = 5 mm). Line traces of ABN and liver autofluorescence corresponding to dashed lines in relative fluorescence units (RFU). (g) Relative reporter concentrations measured in the urine of healthy mice vs. CLM mice after application of LyP-1 or iRGD ABN (n=9 (iRGD) or 10 (LyP-1) mice per group, mean denoted by line, Student’s t-test, two-tailed, ***P<0.001.)
The growth of individual liver metastases was measured by MRI and was well-correlated with bioluminescence signal of luciferized MC-26 cells, allowing us to monitor total tumour burden via bioluminescence imaging (Fig. 5e, Fig. S12). We examined nanoparticle localization after intravenous administration of iRGD or LyP-1 ABNs and observed no changes in overall organ accumulation (Fig. S13a), but increased penetration of iRGD ABNs into the tumour metastases compared to LyP-1 (Fig. 5f, Fig. S13b–c). Application of either LyP-1 or iRGD ABNs in the urinary test resulted in distinct increases in signal when applied to CLM mice compared to healthy mice (Fig. 5g), and compared to mice with sham surgeries (Fig. S14). Consistent with low p32 and high integrin surface expression on MC-26 tumours (Fig. 5c), iRGD ABNs had significantly improved diagnostic performance with relative reporter concentration increases over LyP-1 ABNs (AUC=1.00 vs. 0.86, respectively). Urine signal from iRGD ABNs was also more positively correlated with overall burden than LyP-1 ABNs (Fig. S14b). As might be anticipated from low p32 surface expression in MC-26 tumours, administration of LyP-1 ABNs to CLM mice resulted in relative reporter concentration increases similar to non-penetrating ABNs administered in the hind flank model (Fig. 3b), supporting the interpretation that presence of the primary receptor can generate enhanced urinary signal. ABNs could serve as a minimally invasive modality to monitor patients longitudinally after primary tumour resection in colorectal cancer, where the majority of patients are at-risk to develop CLM3.
ABN zymography tool evaluates substrates in human tissues
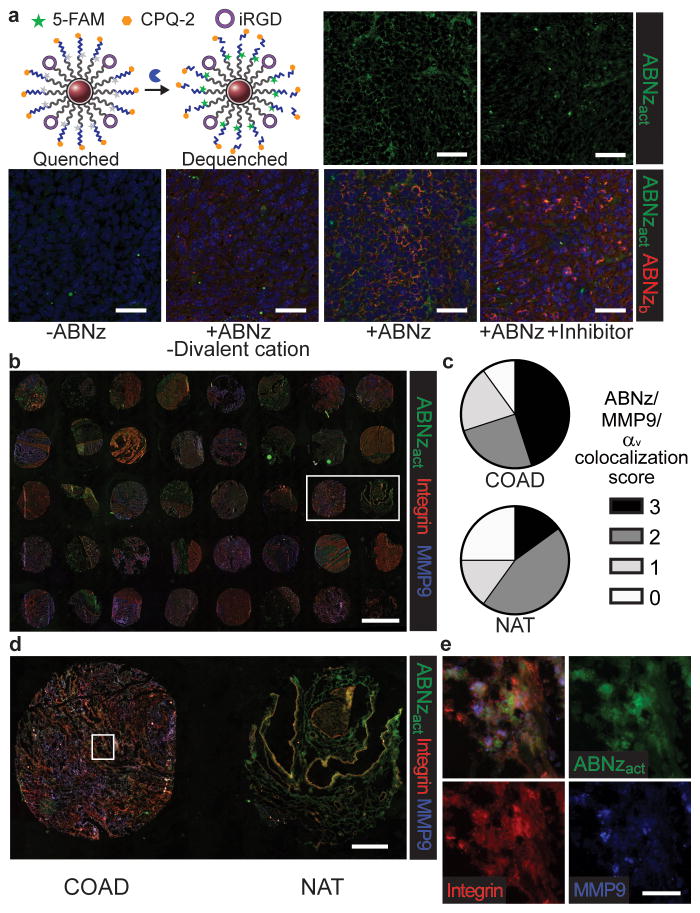
With the long-term goal of translation into patients, we set out to validate that our sensors are responsive in human samples. However, the evaluation and localization of protease activity on synthetic peptide substrates in excised tissues has been challenging because substrates can diffuse away after proteolysis43. To investigate ABN activity in ex vivo tissues, we applied a FRET-pair flanked substrate such that cleavage could be monitored with fluorescence imaging (Fig. 6a). Combining existing zymography techniques44 with our ABN (ABNz) allows the mapping protease activity to tissue localization. Here, the critical step involves binding the nanoparticle to the tissue prior to proteolysis, thereby enabling localization. When iRGD ABNz were applied to frozen liver metastasis sections from CLM mice, signal was dependent on both MMP activity, which was reduced by pharmacological inhibition, and binding, which was reduced when divalent cations necessary for integrin receptor engagement were omitted (Fig. 6a).
Figure 6. ABN zymography is responsive to human tissues.
(a) Top left, schematic for ABN technology re-engineered for zymography (ABNz). Nanoparticles are modified with iRGD targeting ligand and FRET pair flanked substrates that increase in fluorescence upon proteolytic cleavage. Frozen sections of CLM livers collected from mice were bound with PBS only (-ABNz), ABNz without divalent cations necessary for iRGD binding, and ABNz. Tissues were subsequently cleaved in MMP9 buffer with and without MMP9 inhibitor and imaged for the bound ABNz (ABNzb; red) and cleaved ABNz (ABNzact; green) (scale bar = 50 μm). (b) Application of iRGD ABNz to human CRC tumour microarray consisting of 20 colorectal adenocarcinoma (COAD) samples and 20 normal adjacent tissue (NAT) (scale bar = 2 mm). (c) Extent of signal colocalization between COAD and NAT samples scored on a scale of 0 to 3 by a blinded, independent researcher. (d) Higher magnification of one patient sample containing cancer vs normal adjacent tissue from box denoted in (b) (scale bar = 400 μm). (e) Magnified tissue from box denoted in (d) to show cell level colocalization of activated ABNz with integrin and MMP9 staining (scale bar = 40 μm).
Lastly, we applied iRGD ABNz to a frozen human CRC TMA that contained biopsies from tumours and normal adjacent tissue (Fig. 6b). Extent of signal colocalization of activated ABNz, integrin, and MMP9 in cells of each tissue core was blindly scored and revealed that ABNz differentially responded to tumour tissue compared to normal adjacent tissue, with 9/20 tumour cores showing high signal generation compared to only 3/20 normal cores (Fig. 6c). We note that normal adjacent tissues are collected from CRC patients and may have elevated protease levels compared to tissue from healthy patients. A closer examination of the tissue (Fig. 6d–e) shows examples of ABNz activation co-localizing with integrin and MMP9 staining. The finding that ABN-based signal generation correlates with MMP9 and integrin expression supports that this platform may be applicable for detection of human cancers. Beyond this application, our technology could also be used as a tool to profile human tissues for spatial information of protease activity that could support the translation of other protease-sensitive technologies and therapeutics.
Outlook
To extend the ABN technology for application to a wide selection of tumours, a suite of tumour-penetrating ligands specific for a range of primary receptors could be built by incorporation of the C-end Rule motif, K/RXXK/R, which triggers the active internalization pathway28. Recent investigation into the mechanisms of these ligands implicates trafficking pathways distinct from endocytosis30. In this work, we employed two peptides, LyP-1 and iRGD, which bind to different primary receptors, but use the same NRP-1 secondary receptor to initiate tumour-penetration31,45. In particular, the iRGD peptide has been shown to increase the accumulation and activity of the antibody therapy, trastuzumab45. We note that the target of trastuzumab is extracellular HER2, motivating our use of these peptides to sense extracellular MMP9. Furthermore, a corollary to the observation that urinary signal increases when primary receptor is matched with the tumour-penetrating ligand is that this technology can be further developed as a cocktail of tumour-penetrating ABNs to noninvasively and rapidly stratify patients based on receptor expression. This tool could be used as a companion diagnostic to monitor receptor status in tumours treated with precision medicines, such as integrin-targeted therapeutics.
To move this technology towards potential clinical translation, we validated that our ABN technology responds to human tumour biopsy samples. We benchmarked our technology against the blood biomarker HE4, which is shown to perform well in detecting early stage ovarian cancer 46. Our technology was able to outperform HE4, detecting orthotopic tumour volumes that were 2.4-fold lower. Based on simple, monoexponential growth models,8 this would yield a detection advantage of 5 months. We also note that the median ovarian cancer nodule diameter detected by transvaginal ultrasound in humans is 4.7 cm, a tumour volume that is ~1500 larger than the volume we detected in our orthotopic mouse model47. Nonetheless, it is unclear how the limit of detection in mouse models will scale to human tumour detection. While both pre-clinical and clinical characterization and development will be necessary to apply this technology for detection of small tumour lesions in humans, recent translation of protease-activated imaging has shown promise from mouse models to human clinical trials and outlines a path for application in patients26. Looking forward, further improvement in protease-based cancer detection technology can be made as new cancer-specific protease profiles and specific substrates are identified. Our approaches to engineering an ultra-sensitive diagnostic can be coupled with specificity achieved through multiplexing of protease substrates10 and targeting ligands.
Methods
MMP9 expression analysis
MMP9 expression data was queried from Oncomine (https://www.oncomine.org/) and TCGA (http://cancergenome.nih.gov/) where transcriptomic data was available from both tumour and control samples. The list of cancers analyzed can be found in the Supplementary Information. Expression in tumours was normalized to controls from each data set.
Tissue microarray staining and scoring
Multiple Organ Cancer and Normal Tissue Microarray was obtained from US Biomax, Inc. (Catalog number MC5003b). Microarray was stained with anti-MMP9 antibody (Abcam; 1:1000). Blind scoring of cores was done by a pathologist.
Synthesis of peptides and NPs
All peptides were synthesized by CPC Scientific, Inc. For recombinant enzyme studies and ABNz, intramoleculary quenched peptides were used (MMP substrate: 5FAM-GGPLGVRGKK(CPQ2)-PEG2-C; Thrombin Substrate: 5FAM-GGfPRSGGGK(CPQ2)-PEG2-C; FAM: fluorophore, CPQ2: quencher, lower case: D-stereoisomer). In vivo protease sensitive substrates were synthesized to contain a urinary reporter comprised of a protease resistant D-stereoisomer of Glutamate-Fibrinopeptide B with a near-IR dye for urinary detection (Biotin-CGPLGVRGKK(Cy7)eGvndneeGffsar). Targeting peptides were synthesized and cyclized (LyP1: C-K(5FAM)-C6-CGNKRTRGC,; iRGD: C-PEG2-CRGDKGPDC; C6 = 6-aminohexanoic acid linker, Cys2 & Cys3 bridge).
Iron-oxide nanoparticles (IONPs) were formed by reacting iron(III) chloride hexahydrate and iron(II) chloride tetrahydrate with dextran as previously described48. These nanoparticles have been previously shown to be biocompatible and safe, and were cleared from the animal within 5 days10. Nanoparticles were aminated by reacting with ammonium hydroxide. Size measurements were performed by dynamic light scattering (Malvern Instruments Nano ZS90). For conjugation of peptides to NPs, NPs were first reacted with NHS-VivoTag 680 (VT680, PerkinElmer) and MAL-PEG(5k)-SVA (Laysan Bio.) to introduce sulfhydryl reactive handles. Cysteine terminated peptides were then reacted with the NPs and unreacted peptide was filtered using spin filters (Millipore, MWCO = 30 kDa). For experiments to identify optimal PEG lengths, PEG of varying length was purchased (Thermo Scientific) and reacted in the same manner. Nanoparticles were stored in PBS at 4 °C or at -20 °C. Valencies of peptide conjugation and concentrations were quantified by absorbance (Molecular Devices SpectraMax Plus). Typical valencies were: ~60 protease cleavable peptide, ~5 targeting peptide, and ~10 VT680 per nanoparticle.
In vitro recombinant protease assays
Nanoparticles coupled with intramolecularly quenched peptide substrates were reacted with recombinant proteases to identify cleavage velocities. NPs were mixed with 1% (wt/vol) BSA (Sigma) and incubated with recombinant proteases (MMP-9: Enzo Life Sciences; Thrombin: Haematologic Technologies) in a final volume of 50 μL in enzyme specific buffers (MMP9 buffer: 50 mM Tris, 150 mM NaCl, 5 mM CaCl2, 1μM ZnCl2, pH 7.5; Thrombin: PBS) in a 384 well plate for time-lapse fluorimetry to measure dequenching at 37 °C (SpectroMax Gemini EM Microplate Reader). Michaelis-Menten constants were determined by assessing initial cleavage velocities at different substrate concentrations (Prism 5.0). Final MMP-9 concentration was 100 nM and final thrombin concentration was 7 nM.
Cell culture
MDA-MB-435 human cancer cell line (American Type Culture Collection) and MC-26 (Tanabe Laboratory, Massuchusetts General Hospital) mouse colon carcinoma cell line were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S). OVCAR-8 human ovarian cancer cell line (NCI-60) was cultured in Roswell Park Memorial Institute (RPMI) also supplemented with FBS and P/S. All cell lines used in these studies were tested for mycoplasma.
Antibody staining and flow cytometry
To confirm the expression of targetable integrins on the MC-26 cells, cells were harvested with Cell Dissociation Buffer, enzyme-free (ThermoFisher Scientific). Cells were stained for αv integrin (550024 BD Pharmingen; 1:100), NRP-1 (AF566 R&D Biosystems; 1:100), and p32 (AB 2991 Millipore; 1:100) at 4 °C for 1 hour and probed with secondary antibodies conjugated with fluorophores using standard protocols. Cells were analyzed by flow cytometry on a BD LSR II Flow Cytometer.
ELISA for ovarian cancer blood biomarkers
Secreted and cytoplasmic levels of human epididymis protein (HE4) were measured by ELISA, according to manufacturer protocols (R&D Systems). Cultured supernatant and lysed cells from five cell lines were diluted as needed. All measurements were normalized to number of cells and secretion time.
MDA-MB-435 subcutaneous xenograft studies
All animal studies were approved by MIT’s committee on animal care (MIT protocol 0411-036-14). To generate subcutaneous grafts, 3–4 week-old female NCr nude mice (Taconic) were injected bilaterally with 5 × 106 MDA- MB-435 cells per flank. Urine measurements were made prior to tumour inoculation by injecting either 0.5 μM LyP1 or non-penetrating ABNs (by protease cleavable peptide) in 200 μL PBS intravenously at most every week after inoculation. After nanoparticle injection, mice were placed in custom housing with a 96-well plate base for urine collection. After 1 hour, bladders were voided to collect between 100–200 μL of urine. For analysis, urine was diluted between 10- to 25-fold in PBS. Reporter concentration was quantified by fluorescence measurement of Cy7 using a Licor by comparing to a ladder (Odyssey). Tumour sizes were measured using digital electronic calipers (Marathon Management Co.) and volume was calculated as volume = 0.5 × length × width2, where length and width are the larger and smaller dimensions, respectively. Total tumour volume was defined as the sum of the tumour volume in each flank. Mice were binned based on tumour sizes and urine signal was quantified for the bins. Receiver operating characteristic (ROC) curves were generated in GraphPad.
For organ and tumour biodistribution and quantification, mice were sacrificed at 3 hours post-NP injection and organs were removed and scanned on the LI-COR Odyssey Infrared Imaging System. Fluorescence from the nanoparticle scaffold (VT680) and the peptide (Cy7) was quantified using ImageJ software (NIH).
Pharmacokinetic mathematical model
For complete description and derivation of the model, refer to Kwong GA et al, PNAS 201513. We inputted the measured properties of this sensor by modifying the base case values from the previous model. From the data in Figure 2a–g: MMP9 cleavage went up 4-fold at the 6 μM concentration and thrombin cleavage went down 29.1-fold at 6 μM substrate. We used these numbers and inputted them to change the base case values for kcat, tumour and kcat, background by the respective fold changes observed. Here, we assume the change in thrombin cleavage is abstracted to background proteolysis for simplicity. From the data in Figure 2g: tumour accumulation went up 1.2-fold. We updated the permeability term in the ODE, knp_tumour by the observed difference. We further modified the nanoparticle half-life to match that of our nanosensors. For 10-mm tumour, we used the base case model tumour enzyme concentration of 700 nM; for 5-mm tumour, we used a concentration of 7 nM, which is approximately in the range for such a tumour size.
Ovarian cancer orthotopic model studies
To generate an orthotopic model of human ovarian cancer, 3–4 week-old female NCr nude mice were injected intraperitoneally with 2 × 106 OVCAR-8 cells expressing firefly luciferase. Tumour burden was measured using luminescence with an In Vivo Imaging System (IVIS, PerkinElmer). Each mouse was sacrificed and tumours retrieved from all organs in the peritoneal space. Tumours were collected on a glass slide and scanned on a LI-COR Odyssey with 169 μm resolution. Widths and lengths were measured with ImageJ software and tumour volume was calculated as described above for subcutaneous tumours. Reported tumour volumes for each group is the average of 10 mice. The size distribution of nodules recovered from each mouse is shown in Fig. S7c. Prior to tumour induction, urine measurements were performed by injecting mice with 0.5 μM LyP1 ABNs as described above.
Approximately 200 μL of blood was collected and spiked with EDTA to a final concentration of 5 mM and blood cells were pelleted. Plasma was stored at -20 °C prior to HE4 quantification. HE4 levels were measured using the HE4 quantikine ELISA kit according to manufacturer protocols (R&D Systems).
Liver metastasis model studies
Immunocompetent, 6–8 week-old female BALB/c mice were injected with 5 × 104 syngeneic MC-26 cells expressing firefly luciferase in the subsplenic capsule to allow cells to seed the liver. After 90 seconds, the spleen was removed to prevent splenic tumours. Tumour growth was monitored by luminescence and magnetic resonance imaging (MRI). Prior to induction of liver metastases, urine measurements were made by injecting 0.5 μM iRGD or LyP-1 ABNs biomarkers. Post tumour inoculation urine measurements were performed when tumour luminescence reached an average of 1–1.5×107 photons/sec/cm2/sr.
Toxicity and clearance studies
All studies were completed in immunocompetent, 6–8 week-old female BALB/c mice. After administration of ABN or free reporter, 10 μL of blood and urine were collected at the indicated timepoints for fluorescence measurements. For tissue clearance and histopathological examination, animals were sacrificed at the indicated timepoints and organs were fixed in formalin. Organs were imaged for ABN with the LI-COR Odyssey as described above before embedding in paraffin and sectioning.
Histology
All histology sectioning and staining was performed at the KI Histology Core. Organs were fixed using formalin immediately after necropsy for 24 hours and stored at 4 °C prior to embedding into paraffin, sectioning, and staining. OVCAR-8 tumour sections were stained with p32 (Genscript custom antibody; 1:100) and NRP-1 (R&D Biosystems AF3870; 1:100). MC-26 tumour sections were stained with αv integrin (Millipore AB1930; 1:100), p32 (AB 2991 Millipore; 1:100), and NRP-1 (AF566 R&D Biosystems; 1:100).
Application of ABNz
Livers from CLM mice were extracted and immediately embedded and frozen in optimal cutting temperature (OCT) compound. Before application of ABNz, liver sections were air-dried and fixed in cold acetone44. Protease substrates were designed similarly to intramolecular quenched probes for in vitro cleavage experiments except fluorophore/quencher positions were flipped. Fresh frozen acetone fixed human CRC TMA was purchased from US Biomax, Inc (Catalog # FCO405a). After hydration in PBS and blocking in 2% BSA solution for 1 hour at 4 °C, ABNz were applied and incubated for 3 hours at 4 °C with and without divalent cation in the buffer to allow for binding but no cleavage. The slide was washed and the buffer exchanged with MMP9 cleavage buffer (recipe above) and incubated at 37 °C overnight in a humidified box. For inhibited controls, 50 μM marimastat was added to the binding and cleavage buffers. Slides were stained with αv integrin (Biolegend 327902; 1:100) and MMP9 (Abcam ab137867; 1:500) followed by application of the appropriate secondary antibodies (Jackson Immunolabs). Slides were scanned on a Pannoramic 250 Optimum (Perkin Elmer) and co-localization was scored blindly by an independent researcher.
Statistical analyses
All statistical analyses were performed in GraphPad (Prism 5.0) or MATLAB R2013b. Each set of data shown is representative of studies repeated in at least two independent experiments. Sample sizes of n = 7–10 used in animals experiments were estimated using a power test with an expected effect size of 50–100% with a variance of 30–50%. No animals were excluded in analyses. The investigators were not blinded to groups and treatments during experiments. For each animal experiment, groups were established before tumourigenesis and therefore no randomization was used in allocation of groups.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its Supplementary Information.
Code availability
Skeleton MATLAB code of the pharmacokinetic model is available on GitHub at https://github.com/jaidud3/Cancer-activity-based-nanosensor-mathematical-framework.
Supplementary Material
Acknowledgments
We thank Dr. Heather Fleming (MIT) for critical editing of the manuscript, and Dr. Andrew Warren (MIT), Professor Gabe Kwong (Georgia Tech), Dr. Justin Voog (MIT, MGH), Dr. Vyas Ramanan, and Colin Buss (MIT) for helpful discussion. We are grateful to the Koch Institute Swanson Biotechnology Core at MIT, especially Dr. Scott Malstrom, Kathy Cormier, and veterinary pathologist Dr. Roderick T. Bronson. This study was supported in part by a Koch Institute Support Grant P30-CA14051 from the National Cancer Institute (Swanson Biotechnology Center), a Core Center Grant P30-ES002109 from the National Institute of Environmental Health Sciences, the Ludwig Fund for Cancer Research, and the Koch Institute Marble Center for Cancer Nanomedicine. E.J.K. acknowledges support from the Ruth L. Kirschstein National Research Service Award (1F32CA177094-01). J.S.D. thanks the National Science Foundation Graduate Research Fellowship Program for support. S.N.B. is a Howard Hughes Institute Investigator.
Footnotes
Author contributions:
E.J.K. and J.S.D. performed the experiments and analysed the data; E.J.K., J.S.D., and S.N.B. designed experiments and wrote the manuscript.
Competing interests:
The authors declare no competing financial interests.
References
- 1.Etzioni R, et al. The case for early detection. Nat Rev Cancer. 2003;3:243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 2. [Accessed: 23rd June 2016];Cancer Statistics Review, 1975–2013 - SEER Statistics. Available at: http://seer.cancer.gov/csr/1975_2013/
- 3.Kanas GP, et al. Survival after liver resection in metastatic colorectal cancer: review and meta-analysis of prognostic factors. Clin Epidemiol. 2012;4:283–301. doi: 10.2147/CLEP.S34285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bristow RE, Tomacruz RS, Armstrong DK, Trimble EL, Montz FJ. Survival Effect of Maximal Cytoreductive Surgery for Advanced Ovarian Carcinoma During the Platinum Era: A Meta-Analysis. J Clin Oncol. 2002;20:1248–1259. doi: 10.1200/JCO.2002.20.5.1248. [DOI] [PubMed] [Google Scholar]
- 5.Fader AN, et al. The prognostic significance of pre- and post-treatment CA-125 in grade 1 serous ovarian carcinoma: A Gynecologic Oncology Group study. Gynecol Oncol. 2014;132:560–565. doi: 10.1016/j.ygyno.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaukat A, et al. Long-Term Mortality after Screening for Colorectal Cancer. N Engl J Med. 2013;369:1106–1114. doi: 10.1056/NEJMoa1300720. [DOI] [PubMed] [Google Scholar]
- 7.Frangioni JV. New Technologies for Human Cancer Imaging. J Clin Oncol. 2008;26:4012–4021. doi: 10.1200/JCO.2007.14.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hori SS, Gambhir SS. Mathematical Model Identifies Blood Biomarker–Based Early Cancer Detection Strategies and Limitations. Sci Transl Med. 2011;3:109ra116–109ra116. doi: 10.1126/scitranslmed.3003110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henry NL, Hayes DF. Cancer biomarkers. Mol Oncol. 2012;6:140–146. doi: 10.1016/j.molonc.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwong GA, et al. Mass-encoded synthetic biomarkers for multiplexed urinary monitoring of disease. Nat Biotechnol. 2013;31:63–70. doi: 10.1038/nbt.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warren AD, Kwong GA, Wood DK, Lin KY, Bhatia SN. Point-of-care diagnostics for noncommunicable diseases using synthetic urinary biomarkers and paper microfluidics. Proc Natl Acad Sci. 2014;111:3671–3676. [Google Scholar]
- 12.Koblinski JE, Ahram M, Sloane BF. Unraveling the role of proteases in cancer. Clin Chim Acta Int J Clin Chem. 2000;291:113–135. doi: 10.1016/s0009-8981(99)00224-7. [DOI] [PubMed] [Google Scholar]
- 13.Kwong GA, et al. Mathematical framework for activity-based cancer biomarkers. Proc Natl Acad Sci. 2015 doi: 10.1073/pnas.1506925112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kessenbrock K, Plaks V, Werb Z. Matrix Metalloproteinases: Regulators of the Tumor Microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farina AR, Mackay AR. Gelatinase B/MMP-9 in Tumour Pathogenesis and Progression. Cancers. 2014;6:240–296. doi: 10.3390/cancers6010240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu ZS, et al. Prognostic significance of MMP-9 and TIMP-1 serum and tissue expression in breast cancer. Int J Cancer. 2008;122:2050–2056. doi: 10.1002/ijc.23337. [DOI] [PubMed] [Google Scholar]
- 17.Deere J, et al. Kinetics of Enzyme Attack on Substrates Covalently Attached to Solid Surfaces: Influence of Spacer Chain Length, Immobilized Substrate Surface Concentration and Surface Charge. Langmuir. 2008;24:11762–11769. doi: 10.1021/la801932f. [DOI] [PubMed] [Google Scholar]
- 18.Nagase H, Fields GB. Human matrix metalloproteinase specificity studies using collagen sequence-based synthetic peptides. Pept Sci. 1996;40:399–416. doi: 10.1002/(SICI)1097-0282(1996)40:4%3C399::AID-BIP5%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 19.Khersonsky O, Tawfik DS. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu Rev Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 20.Whitley MJ, et al. A mouse-human phase 1 co-clinical trial of a protease-activated fluorescent probe for imaging cancer. Sci Transl Med. 2016;8:320ra4–320ra4. doi: 10.1126/scitranslmed.aad0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desnoyers LR, et al. Tumor-Specific Activation of an EGFR-Targeting Probody Enhances Therapeutic Index. Sci Transl Med. 2013;5:207ra144–207ra144. doi: 10.1126/scitranslmed.3006682. [DOI] [PubMed] [Google Scholar]
- 22.Miller MA, et al. Predicting therapeutic nanomedicine efficacy using a companion magnetic resonance imaging nanoparticle. Sci Transl Med. 2015;7:314ra183–314ra183. doi: 10.1126/scitranslmed.aac6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertrand N, Wu J, Xu X, Kamaly N, Farokhzad OC. Cancer nanotechnology: the impact of passive and active targeting in the era of modern cancer biology. Adv Drug Deliv Rev. 2014;66:2–25. doi: 10.1016/j.addr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilhelm S, et al. Analysis of nanoparticle delivery to tumours. Nat Rev Mater. 2016;1:16014. [Google Scholar]
- 25.Davis ME, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hrkach J, et al. Preclinical Development and Clinical Translation of a PSMA-Targeted Docetaxel Nanoparticle with a Differentiated Pharmacological Profile. Sci Transl Med. 2012;4:128ra39–128ra39. doi: 10.1126/scitranslmed.3003651. [DOI] [PubMed] [Google Scholar]
- 27.Ruoslahti E. SPECIALIZATION OF TUMOUR VASCULATURE. Nat Rev Cancer. 2002;2:83–90. doi: 10.1038/nrc724. [DOI] [PubMed] [Google Scholar]
- 28.Ruoslahti E, Bhatia SN, Sailor MJ. Targeting of drugs and nanoparticles to tumors. J Cell Biol. 2010;188:759–768. doi: 10.1083/jcb.200910104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin KY, Kwon EJ, Lo JH, Bhatia SN. Drug-induced amplification of nanoparticle targeting to tumors. Nano Today. 2014;9:550–559. doi: 10.1016/j.nantod.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pang H-B, et al. An endocytosis pathway initiated through neuropilin-1 and regulated by nutrient availability. Nat Commun. 2014;5 doi: 10.1038/ncomms5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fogal V, Zhang L, Krajewski S, Ruoslahti E. Mitochondrial/Cell-Surface Protein p32/gC1qR as a Molecular Target in Tumor Cells and Tumor Stroma. Cancer Res. 2008;68:7210–7218. doi: 10.1158/0008-5472.CAN-07-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dufour A, et al. Small-Molecule Anticancer Compounds Selectively Target the Hemopexin Domain of Matrix Metalloproteinase-9. Cancer Res. 2011;71:4977–4988. doi: 10.1158/0008-5472.CAN-10-4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zurawski VR, et al. An initial analysis of preoperative serum CA 125 levels in patients with early stage ovarian carcinoma. Gynecol Oncol. 1988;30:7–14. doi: 10.1016/0090-8258(88)90039-x. [DOI] [PubMed] [Google Scholar]
- 34.Ren Y, Hauert S, Lo JH, Bhatia SN. Identification and Characterization of Receptor-Specific Peptides for siRNA Delivery. ACS Nano. 2012;6:8620–8631. doi: 10.1021/nn301975s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drapkin R, et al. Human epididymis protein 4 (HE4) is a secreted glycoprotein that is overexpressed by serous and endometrioid ovarian carcinomas. Cancer Res. 2005;65:2162–2169. doi: 10.1158/0008-5472.CAN-04-3924. [DOI] [PubMed] [Google Scholar]
- 36.Rankin EB, et al. AXL is an Essential Factor and Therapeutic Target for Metastatic Ovarian Cancer. Cancer Res. 2010;70:7570–7579. doi: 10.1158/0008-5472.CAN-10-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alper Ö, et al. Epidermal Growth Factor Receptor Signaling and the Invasive Phenotype of Ovarian Carcinoma Cells. J Natl Cancer Inst. 2001;93:1375–1384. doi: 10.1093/jnci/93.18.1375. [DOI] [PubMed] [Google Scholar]
- 38.Brown PO, Palmer C. The Preclinical Natural History of Serous Ovarian Cancer: Defining the Target for Early Detection. PLOS Med. 2009;6:e1000114. doi: 10.1371/journal.pmed.1000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugahara KN, et al. Tissue-penetrating delivery of compounds and nanoparticles into tumors. Cancer Cell. 2009;16:510–520. doi: 10.1016/j.ccr.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao M, Lam EC, Kelly CR, Zhou W, Wolfe MM. Cyclooxygenase-2 selective inhibition with NS-398 suppresses proliferation and invasiveness and delays liver metastasis in colorectal cancer. Br J Cancer. 2004;90:712–719. doi: 10.1038/sj.bjc.6601489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackson BC, Nebert DW, Vasiliou V. Update of human and mouse matrix metalloproteinase families. Hum Genomics. 2010;4:194–201. doi: 10.1186/1479-7364-4-3-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanman LE, Bogyo M. Activity-Based Profiling of Proteases. Annu Rev Biochem. 2014;83:249–273. doi: 10.1146/annurev-biochem-060713-035352. [DOI] [PubMed] [Google Scholar]
- 44.Withana NP, et al. Labeling of active proteases in fresh-frozen tissues by topical application of quenched activity-based probes. Nat Protoc. 2016;11:184–191. doi: 10.1038/nprot.2016.004. [DOI] [PubMed] [Google Scholar]
- 45.Sugahara KN, et al. Coadministration of a tumor-penetrating peptide enhances the efficacy of cancer drugs. Science. 2010;328:1031–1035. doi: 10.1126/science.1183057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Gorp T, et al. HE4 and CA125 as a diagnostic test in ovarian cancer: prospective validation of the Risk of Ovarian Malignancy Algorithm. Br J Cancer. 2011;104:863–870. doi: 10.1038/sj.bjc.6606092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alcázar JL, Guerriero S, Laparte C, Ajossa S, Jurado M. Contribution of power Doppler blood flow mapping to gray-scale ultrasound for predicting malignancy of adnexal masses in symptomatic and asymptomatic women. Eur J Obstet Gynecol Reprod Biol. 2011;155:99–105. doi: 10.1016/j.ejogrb.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 48.Park JH, et al. Systematic surface engineering of magnetic nanoworms for in vivo tumor targeting. Small Weinh Bergstr Ger. 2009;5:694–700. doi: 10.1002/smll.200801789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its Supplementary Information.