Abstract
Sex determining region Y-box protein 12 (SOX12) plays an important role in the tumorigenesis of hepatocellular carcinoma. The involvement of SOX12 in human lung cancer is not well-understood. The aim of the current study was to explore the expression pattern and function of SOX12 in lung cancer. SOX12 expression in lung cancer tissues was elevated as assessed by analyzing The Cancer Genome Atlas (TCGA) lung cancer cohort and real-time PCR data of our own cohort. We found that SOX12 mRNA expression was up-regulated in 83.3% (75/90) of the lung cancer tissues in comparison with paired normal tissues. Moreover, high SOX12 expression predicted reduced overall survival. We also found that knockdown of SOX12 in SPC-A-1 and A549 cells impaired lung cancer cell proliferation, migration and invasion in vitro, but promoted lung cancer cell apoptosis. In vivo tumorigenesis experiments showed that inhibition of SOX12 expression significantly suppressed the growth of xenograft tumors. Finally, the mRNA and protein levels of cell growth (PCNA and Cyclin E), apoptosis (Bcl-2 and Bax), invasion (matrix metalloproteinase-9) and epithelial-mesenchymal transition (EMT; Twist1 and E-cadherin) related moderators were affected by SOX12 knockdown. Chromatin immunoprecipitation assays showed that Cyclin E and Twist1 were direct transcriptional targets of SOX12. Taken together, our study suggests that SOX12 functions as an oncogenic molecule during the development of human lung cancer.
Keywords: SOX12, lung cancer, cell growth, migration, invasion
Introduction
Lung cancer is one of the most common diagnosed cancer in the world. Despite the advances in surgical resection, radiotherapy and anti-cancer drugs, lung cancer still remains the most common cause of cancer-related mortality with the 5-year survival rate of less than 15% [1]. At the time of diagnosis, the stage is often advanced. The main reason for the high mortality rate is the sustained proliferation and high metastatic potential of cancer cells [2,3]. Therefore, a better understanding of the molecular mechanisms associated with the proliferation and metastasis in lung cancer is crucial to develop more effective therapies for lung cancer.
Sex determining region Y-box (SOX) genes encode a family of transcription factors, which belong to the high mobility group (HMG) superfamily. SOX12, together with SOX4 and SOX11 belongs to group C of SOX genes (SOXC) [4]. SOX proteins recognize and interact with the common SOX target sequence A/TAACAA/T and activate the transcription of target genes [5,6]. SOX genes are important for diverse developmental processes, including the formation of germ layers, organ development and cell differentiation [7]. Therefore, deletion or mutation of SOX gene often cause developmental defects and congenital diseases [8]. The expression of SOX genes is altered in a wide range of tumors [8,9], and work as oncogenes or tumor suppressor genes. SOX4 [10-13] and SOX11 [14-17], the other two members of SOXC, are overexpressed in various cancers including lung cancers, and their expression may be closely related with poor prognosis and advanced clinical stage of some tumors. Recent data also demonstrated the overexpression of SOX12 and the potential prognostic value in hepatocellular carcinoma (HCC) [18] and breast cancer [19]. However, the potential roles and biological mechanisms of SOX12 in lung cancer have not been studied.
In the current study, we showed that SOX12 was overexpressed in lung cancer samples based on analyses on a public available dataset and our own real-time PCR results. SOX12 expression was strongly associated with the prognosis of lung cancer patients. Silencing SOX12 in lung cancer cell lines was sufficient to inhibit cell growth, migration and invasion in vitro, as well as tumor xenograft growth in vivo. Further, abrogation of its expression in lung cancer cell lines affected the mRNA and protein expression of cell cycle-, cell apoptosis- and metastasis-related proteins.
Materials and methods
Human lung cancer cohort
SOX12 expression was re-analyzed with a RNA-sequencing dataset downloaded from The Cancer Genome Atlas (TCGA, https://tcga-data.nci.nih.gov/tcga/, Version: 7/29/2014). The human lung cancer dataset includes 488 lung cancer samples and 58 control (non-tumorous) samples.
Clinical specimen and cell lines
This study was approved by the Institute Research Medical Ethics Committee of Xin Hua Hospital Affiliated to Shanghai Jiao Tong University (Shanghai, China). Written informed consent was obtained from all patients. A total of 90 patients with lung cancer undergoing surgical resection at Department of Cardiothoracic Surgery, Xin Hua Hospital from January 2013 to December 2014 was enrolled in this study. Complete clinical and pathological follow-up data were obtained from all patients. Fresh tissues (paired tumor samples and matched adjacent normal tissue samples) were resected for SOX12 expression detection.
All cell lines were obtained from Cellular Institute of Chinese Academy of Science (Shanghai, China) and maintained at 37°C under a 5% CO2 atmosphere in appropriate medium supplemented with 10% fetal bovine serum (FBS; Life Technologies, Carlsbad, CA, USA), 100 U/ml penicillin and 100 μg/ml streptomycin. HEK293T, NCI-H446, A549 and SPC-A-1 were grown in Dulbecco’s minimum essential medium (DMEM; HyClone, UT, USA). BEAS-2B, NCI-H358, NCI-H1975 and NCI-H292 were grown in RPMI-1640 medium (Hyclone).
RNA isolation and real-time PCR
Total RNA from the tissue samples or lung cancer cell lines was extracted by using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and treated with DNase to remove genomic DNA. A total of 2 μg RNA was reverse-transcribed into cDNA with the M-MLV reverse transcriptase (Promega, Madison, WI, USA) according to the manufacturer’s protocol. Quantitative real-time PCR was performed by mixing the template and primers with the SYBR Green mix (Thermo Fisher Scientific, Rockford, IL, USA) and detected on the ABI Prism 7300 sequence detection system (Applied Biosystems, Foster City, CA, USA). Primer sequences were listed in Table 1. The relative gene expression was normalized to GAPDH.
Table 1.
Primers sequences for real-time PCR
| Primer | Primer sequence | Size (bp) |
|---|---|---|
| SOX12 (NM_006943.3) | F: 5’-AGCACCCGTGTGACTCTTTCC-3’ | 209 |
| R: 5’-AGCAGAACCAAGCCCTGTCTC-3’ | ||
| PCNA (NM_002592.2) | F: 5’-GGTGTTGGAGGCACTCAAGG-3’ | 115 |
| R: 5’-CAGGGTGAGCTGCACCAAAG-3’ | ||
| Cyclin E (NM_001238.2) | F: 5’-TGCGTATGTGACAGATGGAG-3’ | 111 |
| R: 5’-AGCCAGGACACAATAGTCAG-3’ | ||
| Bax (NM_004324.3) | F: 5’-AGCTGAGCGAGTGTCTCAAG-3’ | 248 |
| R: 5’-TGTCCAGCCCATGATGGTTC-3’ | ||
| Bcl-2 (NM_000633.2) | F: 5’-CCACCAGCACCATAGAAG-3’ | 131 |
| R: 5’-GAGCAGGCACAGAGAAAG-3’ | ||
| MMP-9 (NM_004994.2) | F: 5’-AAGGGCGTCGTGGTTCCAACTC-3’ | 210 |
| R: 5’-AGCATTGCCGTCCTGGGTGTAG-3’ | ||
| Twist (NM_000474.3) | F: 5’-AGTCCGCAGTCTTACGAG-3’ | 228 |
| R: 5’-GCTTGCCATCTTGGAGTC-3’ | ||
| E-cadherin (NM_004360.3) | F: 5’-GAGAACGCATTGCCACATACAC-3’ | 164 |
| R: 5’-AAGAGCACCTTCCATGACAGAC-3’ | ||
| GADPH (NM_001256799.1) | F: 5’-CACCCACTCCTCCACCTTTG-3’ | 110 |
| R: 5’-CCACCACCCTGTTGCTGTAG-3’ |
Immunohistochemical (IHC) staining
Tissues were fixed in 4% paraformaldehyde, paraffin embedded and sectioned into 5-mm-thick slices. After deparaffinization, IHC staining was conducted using anti-SOX12, anti-Ki67 or anti-Twist1 (Abcam, Cambridge, MA, USA; Ab175430). The images were captured under a microscope (200 × magnification).
Western blotting and quantification
Western blotting was performed as previously described [20]. Briefly, cells were lysed with RIPA lysis buffer containing protease inhibitor mixture (Sigma). Total protein lysates containing 30 µg protein were separated by SDS-PAGE and electrotransferred to NC membrane. After blocking with 5% skim milk, the membranes were incubated with primary antibodies overnight at 4°C and then with secondary antibodies labeled with horseradish peroxidase for 1 h at room temperature. GAPDH was served as a loading control. Protein bands were visualized with Clarity Western ECL kit (Bio-Rad Laboratories Inc., Richmond, CA, USA) followed by image analysis using ImageJ software (Bethesda, MD, USA; http://rsb.info.nih.gov/ij/). The following primary antibodies were used: SOX12 (Abcam, Cambridge, MA, USA; Ab167614), PCNA (Cell Signaling Technology, Danvers, MA, USA; #13110), Cyclin E (Abcam, Ab3927), Bcl-2 (Santa Cruz, Santa Cruz, CA, USA; Sc-492), Bax (Santa Cruz, Sc-493), MMP-9 (Abcam, Ab119906), Twist1 (Abcam, Ab175430), E-cadherin (Cell Signaling Technology, #14472) and GAPDH (Cell Signaling Technology, #5174).
shRNA constructs and establishment of stable SOX12 knockdown cell lines
shRNA against human SOX12 (shSOX12) and a non-specific control shRNA (shNC) was cloned into the pLKO-1 lentiviral vector. The SOX12-targeting sequence was as follows: CATGGCGGATTACCCGGACTA [18]. pLKO-shRNA-containing plasmids were co-transfected into HEK293T cells with psPAX2 and PMD2G (Addgene, Cambridge, MA, USA) by using lipofectamine 2000 (Life Technologies) following the manufacture’s instruction. At 48 h after transfection, lentivirus particles were collected to infected SPC-A-1 and A549 cells. Clones stably expressing shSOX12 were selected by puromycin resistance (Sigma, St. Louis, MO, USA). Total RNA and cell lysates were prepared for real time RT-PCR or Western blotting analysis to validate the knockdown effect of shSOX12.
Expression constructs
Full length human SOX12 and shRNA-resistant SOX12 were synthesized and cloned into pCDNA3.1 vector (Invitrogen). The shRNA-resistant SOX12 construct encodes the same amino acid as wild-type SOX12, but contains a different nucleotide sequence in SOX12 shRNA target region: Wild-type SOX12, 5’-CATGGCGGATTACCCGGACTA-3’; rescue SOX12, 5’-CATGGCCGACTATCCAGATTA-3’. Mutated nucleotides are shown in bold.
Cell proliferation assays
To determine cell growth, cells were plated onto 96-well plates at a density of 2 × 103 per well. At 0, 24, 48 and 72 h, Cell Count Kit-8 (CCK-8, Dojindo Laboratories) solution was added to each well and incubated at 37°C for 1 h. Optical density (OD) values at 450 nm were detected by a microplate reader (BioTek, VT, USA).
Evaluation of apoptosis by flow cytometry
The rate of cells undergoing early apoptosis was assessed by double labeling with Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI). Cells were harvested, washed twice with PBS and then double-labeled with Annexin V-FITC and PI (eBioscience) as described by the manufacturer. Cells were analyzed using a flow cytometer (Accuri C6; BD Biosciences, San Jose, CA, USA). Early apoptosis was defined by Annexin V-positive and PI-negative staining.
Transwell assay
The migration assays were performed in 24-well Transwell chambers (Corning, NY, USA). Stable cells were trypsinized, re-suspended in medium without FBS and plated in the upper chamber (4 × 104 cells in 100 μl of medium). Medium containing 20% FBS (600 μl) was added to the lower chamber as an attractant. After incubating at 37°C under 5% CO2 for 24 h, and non-migrating cells were completely removed from the upper surface of the membrane with a cotton swab. Migrated cells remained in the lower surface of the membrane were fixed with 4% paraformaldehyde, stained with 0.5% crystal violet and then washed twice with PBS. Stained cells were viewed under a microscope (200 × magnification), and the number of migrated cells was counted in 10 random fields.
For cell invasion assay, the filters were pre-coated with Matrigel matrix (1 μg/μL; BD Biosciences) at 4°C overnight. The rest of the assay was conducted in the same manner as migration assay.
In vivo tumorigenesis experiments
For the xenograft tumor growth assay, A549 cells stably infected with shNC or shSOX12 (2 × 106 cells per mouse) suspended in PBS were injected subcutaneously into the right flank of 5-week-old BALB/c nude mice (Shanghai Laboratory Animal Company, Shanghai, China). Tumor size was measured every 3 days with a caliper, and tumor volume (v) was calculated with this formula: v = 0.5 × (length × width2). After 24 days, xenograft tumor tissues were isolated, photographed, and stored in liquid nitrogen for real-time PCR and Western blotting analyses. All experiments were approved by the Committee on the Ethics of Animal Experiments of Shanghai Jiao Tong University.
Chromatin immunoprecipitation (ChIP) assays
ChIP assay was performed on A549 cells stably infected with shNC or shSOX12. Briefly, cells were fixed with 1% formaldehyde and lysed with RIPA Buffer. Following sonication, the chromatin solutions were incubated with Protein A agarose and anti-SOX12 (Abcam) or control IgG antibody overnight at 4°C. Binding was detected by PCR with the primers for Cyclin E or Twist1 promoter. The primers were listed in Table 2.
Table 2.
Primers sequences for ChIP
| Twist | Primer 1 | F: 5’-TCAGCTTGAGATATCTGC-3’ |
| R: 5’-GGAGTCAGTACACAGTGT-3’ | ||
| Primer 2 | F: 5’-CAGGACAGTCTCCTCCGA-3’ | |
| R: 5’-TTGGCCTGACGTGAGGAG-3’ | ||
| Cyclin E | Primer 1 | F: 5’-TCTCATTAGCCGGTAAGCCC-3’ |
| R: 5’-CACAGTCTTGGTGCCTCTCT-3’ |
Statistical analysis
All statistics were performed using GraphPad Prism, version 6.0 (GraphPad, San Diego, CA, USA). The RNA-seq data of lung adenocarcinoma were obtained from the TCGA project (http://cancergenome.nih.gov). Survival analysis was carried out using the log-rank test and Kaplan-Meier curves. Statistical analyses were performed using student’s t test. All in vitro experiments assays were performed in triplicate and the results were presented as the mean value ± standard deviation (SD). For all analyses, a P value of <0.05 was set as statistically significant.
Results
SOX12 expression is elevated in lung cancer patients
We re-analyzed lung cancer dataset from TCGA and found that SOX12 expression was significantly higher in lung cancer tissues than normal tissues (Figure 1A, P<0.0001). The elevated expression of SOX12 in lung cancer was validated by real-time PCR analysis on 90 pairs of lung cancer and normal tissues obtained from Shanghai Xinhua Hospital (Shanghai, China) (P<0.0001). SOX12 mRNA expression was presented as the log2 ratio between tumor samples and normal samples. Positive and negative log2 indicated the increased and decreased expression of SOX12 in tumor sample in comparison with the paired normal samples, respectively. As depicted in Figure 1B, SOX12 mRNA was elevated in 83.3% (75/90) of the tested tumor tissues.
Figure 1.
SOX12 is overexpressed in lung cancer. A. Analysis of SOX12 expression in lung cancer and normal tissues based on TCGA dataset. B. SOX12 mRNA expression was detected in 90 tumor tissues and matched normal tissues by real-time PCR. SOX12 mRNA was normalized with GAPDH expression. SOX12 mRNA levels in 90 pairs of lung cancer and normal tissues were determined by real-time PCR analysis. The positive and negative log2 (Tumor/Normal) on the y-axis indicated the increased and decreased expression of SOX12 in tumor tissue, respectively. C. Overall survival analysis by log-rank test and Kaplan-Meier curves. D. IHC staining of SOX12 in tumor and matched normal tissues. E. SOX12 protein expression was determined by Western blotting. N1-8, normal tissues; T1-8, tumor tissues.
Further, according to the relative SOX12 mRNA expression in tumor tissues, the 90 patients were classified into two groups: SOX12 high group (n = 45) and SOX12 low group (n = 45). Kaplan-Meier analysis and log-rank test showed that the overall survival time in lung cancer patients with high expression of SOX12 was shorter than that with low expression (Figure 1C).
SOX12 protein level was examined in lung cancer clinical specimens and normal lung tissues by IHC staining and Western blotting analyses. A higher level of SOX12 protein expression was observed in tumor tissues in comparison with the paired normal tissues (Figure 1D and 1E). Our data demonstrated that SOX12 expression was significantly elevated in tumor tissues both at transcription and translation levels.
Knockdown of SOX12 in lung cancer cells
Real-time PCR and Western blot analysis detected SOX12 mRNA and protein expression in 6 lung cancer cell lines (Figure 2A and 2B), respectively. SPC-A-1 and A549 cells expressed high levels of SOX12. Thus, to explore the loss-of-function phenotype of SOX12, we stably knocked down its expression in these two cell lines. As shown in Figure 2C and 2D, SOX12 protein and mRNA were efficiently suppressed in two selected clones (shSOX12-1 and shSOX12-2) compared with control shRNA (shNC) stable cells.
Figure 2.

Knockdown of SOX12 in lung cancer cells. (A) SOX12 mRNA levels were detected in 6 human lung cancer cells by real-time PCR and normalized with GAPDH. (B) SOX12 protein levels were determined in human lung cancer cells by Western blot. GAPDH was served as the loading control. (C, D) Validation of SOX12 knockdown in SPC-A-1 and A549 cells by real-time PCR (C) and Western blot (D). shNC: cells stably expressing control shRNA; shSOX12-1 and shSOX12-2, cell clones stably expressing SOX12 shRNA. ***P<0.001 VS shNC.
SOX12 knockdown suppressed cell proliferation, but induced cell apoptosis
We determined the effect of SOX12 knockdown on cell proliferation by CCK-8 assay. The results showed that knockdown of endogenous SOX12 (shSOX12-1 and shSOX12-2) significantly inhibited the proliferation of SPC-A-1 and A549 cells compared to the negative controls (shNC) (Figure 3A). On the contrary, SOX12 overexpression increased the proliferation of NCI-H292, which had lower-SOX12 expression (Figure S2A and S2B).
Figure 3.
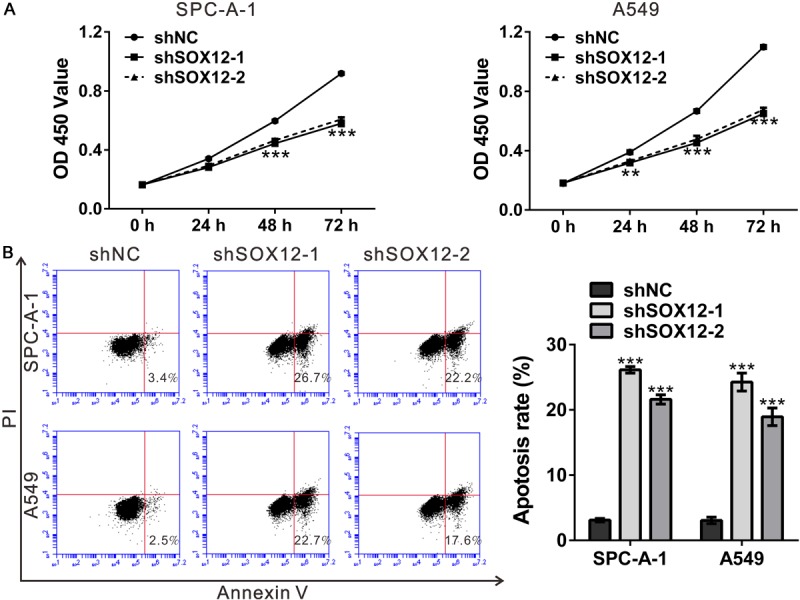
Knockdown of SOX12 suppressed cell proliferation, but induced cell apoptosis. A. Knockdown of SOX12 inhibited the growth of SPC-A-1 and A549 cells as indicated by CCK-8 assay. B. Cell apoptosis of SPC-A-1 and A549 cells analyzed by Annexin V-FITC/PI staining and flow cytometry. shNC: cells stably expressing control shRNA; shSOX12-1 and shSOX12-2, cell clones stably expressing SOX12 shRNA. **P<0.01 and ***P<0.001 VS shNC.
To explore whether SOX12 affected early apoptosis, we performed flow cytometry analysis of lung cancer cells double labeled with Annexin V/PI. Early apoptosis is defined by Annexin V-positive and PI-negative staining. As illustrated in Figure 3B, early apoptosis rate was notably induced by SOX12 knockdown in both cell lines.
SOX12 knockdown inhibited migration and invasion of lung cancer cells
The promoted effects of SOX12 have been observed on the invasion and metastasis of HCC [18]. To examine whether SOX12 plays a role in facilitating the migration and invasion of lung cancer cells, we performed Transwell assays. As shown in Figure 4, migration and invasion capacities of SPC-A-1 and A549 cells infected with shSOX12 virus were decreased compared to the control group. Complementary results were observed in NCI-H292 cells stably overexpressed SOX12 (Figure S2C). These data indicate that SOX12 could promote the migratory and invasive phenotype of lung cancer cells.
Figure 4.
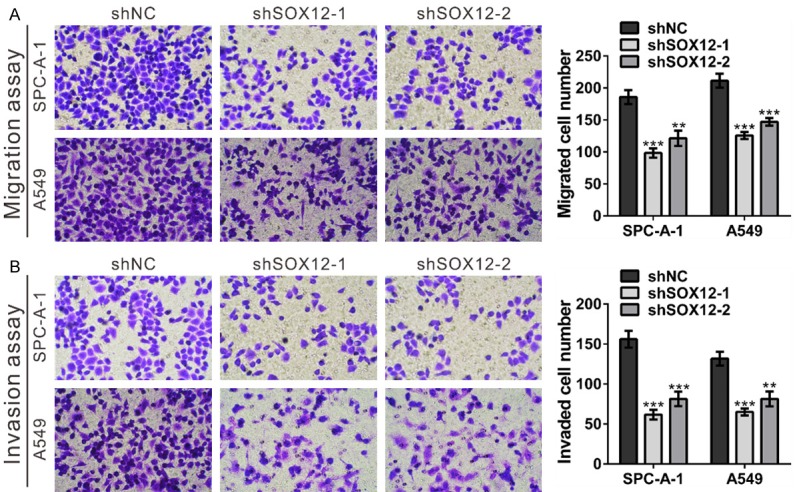
SOX12 knockdown inhibited the migration and invasion of lung cancer cells. A. Migration capacities of SPC-A-1 and A549 cells stably expressing the control shRNA (shNC) or SOX12 shRNA (shSOX12). Left panels: representative images; Right panels: quantifications of average migrated cell number. B. Invasion capacities of SPC-A-1 and A549 cells stably expressing the control shRNA (shNC) or SOX12 shRNA (shSOX12). Left panels: representative images; Right panels: quantifications of average migrated cell number. shNC: cells stably expressing control shRNA; shSOX12-1 and shSOX12-2, cell clones stably expressing SOX12 shRNA. **P<0.01 and ***P<0.001 VS shNC.
Effects of re-expression of shRNA-resistant SOX12
To further confirm the effects of SOX12 knockdown, shRNA-resistant SOX12 expression plasmid was constructed. SPC-A-1 and A549 cells with SOX12 silenced (shSOX12-1) were transfected with Vector or shRNA-resistant SOX12 (Re-expression). As shown in Figure 5, the defect in cell proliferation, migration and invasion could be rescued by expressing shRNA-resistant SOX12 in the shSOX12-1 cells, but not by Vector alone.
Figure 5.
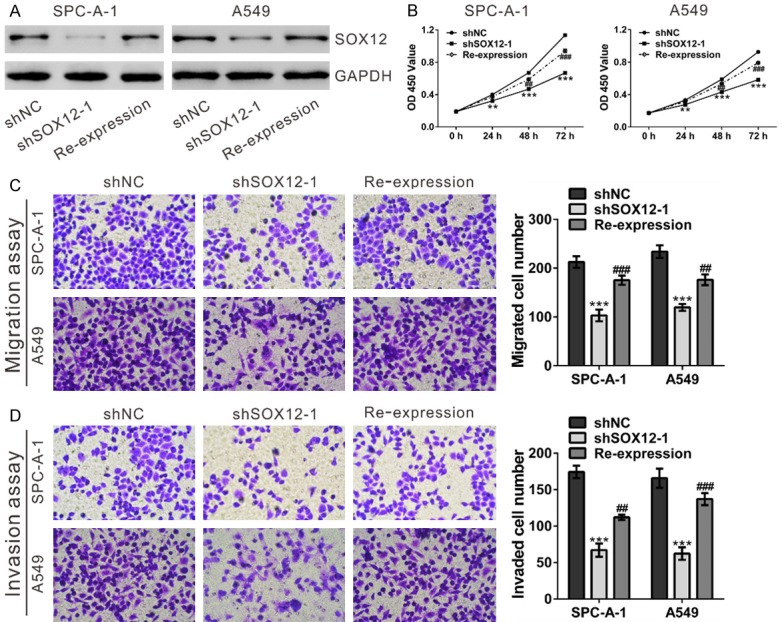
Re-expression of shRNA-resistant SOX12 rescued the inhibitory effects of shSOX12 on cell proliferation, migration and invasion. SPC-A-1 and A549 cells stably expressing SOX12 shRNA (shSOX12-1) were transfected with shRNA-resistant SOX12 or Vector using Lipofectamine (Invitrogen). (A) SOX12 protein expression was determined at 48 after transfection. (B) Cell proliferation was assessed by CCK-8 assay. (C, D) Cell migration (C) and invasion (D) were evaluated by Transwell assays. shNC: cells stably expressing control shRNA; shSOX12-1, cell clone stably expressing SOX12 shRNA were transfected with Vector; Re-expression, shSOX12-1 cells were transfected with shRNA-resistant SOX12. **P<0.01 and ***P<0.001 VS shNC; ##P<0.01 and ###P<0.001 VS shSOX12-1.
Knockdown of SOX12 suppressed the growth of xenograft tumors
We then investigated whether knockdown of SOX12 in lung cancer cells could reduce tumor growth in vivo. A549 cells stably transducted with shSOX12 or shNC were established and subcutaneously injected to nude mice (n = 6/group). Tumor size was measured every 3 days. As shown in Figure 6A, the tumors formed by shSOX12-1 cells grew significantly slower than that from shNC cells. After 24 days, the xenograft tumors were resected (Figure 6B) and SOX12 expression was detected. SOX12 mRNA expression decreased in xenograft formed by shSOX12-1 cells as compared to xenograft formed by shNC cells (Figure 6C) (n = 4/group). Similar results were obtained in SOX12 protein expression (Figure 6D). Further, IHC staining revealed that the Ki67 and Twist1 positive cells were significantly decreased in tumors formed from shSOX12-1 cells compared with that in shNC cells (Figure 6E). These data suggested that suppression of SOX12 in lung cancer cells repressed cell proliferation in vivo.
Figure 6.
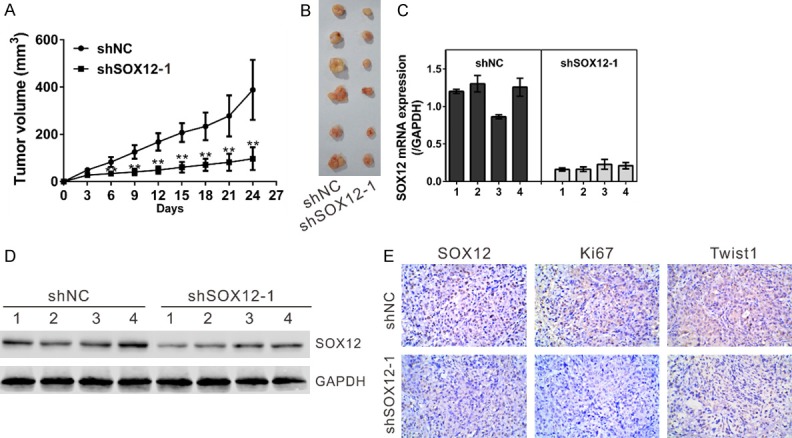
SOX12 knockdown reduced the growth of xenograft tumors. BALB/c nude mice were inoculated with A549 cells stably infected with shNC or shSOX12 virus (n = 6). The tumor size was monitored every three days. (A) Growth curve of xenograft tumors derived from A549 cells stably expressing the control shRNA (shNC) or SOX12 shRNA (shSOX12). (B) After 24 days, xenograft tumors were dissected and photographed. (C, D) The expression of SOX12 in xenograft was determined by real-time PCR (C) and Western blot (D). (E) The expression of SOX12, Ki67 and Twist1 in xenograft was determined by IHC staining. **P<0.01 and ***P<0.001 VS shNC.
Mechanisms of SOX12 exerts its function
Real-time PCR and Western blot assays were performed to analyze the expression of possible genes downstream of SOX12 (Figure 7A-C). In accordance with in vitro functional experiments, knockdown of SOX12 significantly down-regulated cell growth (PCNA and Cyclin E), anti-apoptosis (Bcl-2), metastasis (MMP-9) and epithelial-mesenchymal transition (EMT) (Twist1) related factors in both SPC-A-1 and A549 cell lines compared with the control cells, while remarkably up-regulated the apoptosis factor (Bax) and the main factor of EMT (E-cadherin).
Figure 7.
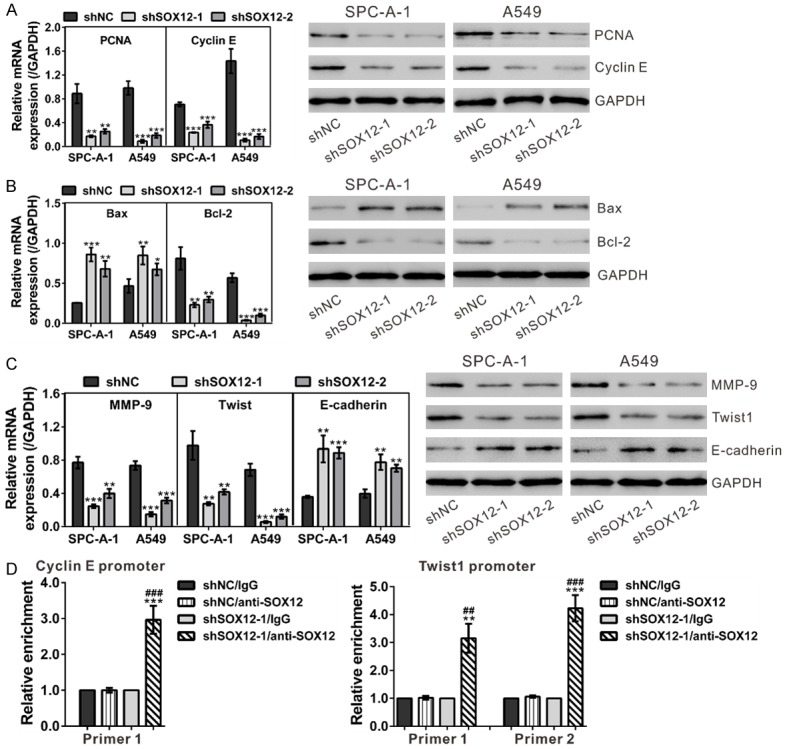
Mechanisms of SOX12 exerts its function. A. mRNA and protein expression of cell growth-related factors (PCNA and Cyclin E) was evaluated by real-time PCR (left panel) and Western blot (right panel), respectively. B. mRNA (left panel) and protein (right panel) expression of apoptotic factors (Bcl-2 and Bax) was assessed. C. mRNA (left panel) and protein (right panel) expression of metastasis (MMP-9) and EMT (Twist1 and E-cadherin) related factors was evaluated. shNC: cells stably expressing control shRNA; shSOX12-1 and shSOX12-2, cell clones stably expressing SOX12 shRNA. **P<0.01 and ***P<0.001 VS shNC. D. SOX12 knockdown affected the binding of SOX12 to Cyclin E promoter and Twist1 promoter in A549 cells as demonstrated by ChIP analysis. **P<0.01 and ***P<0.001 VS shNC/anti-SOX12; ##P<0.01 and ###P<0.001 VS shSOX12-1/IgG.
SOX proteins can recognize a similar motif (A/TAACAA/T) and then activate the transcription of target genes [5,6]. We found that there are one and two candidate motifs in Cyclin E and Twist1 promoter, respectively. Chromatin immunoprecipitation (ChIP) assay confirmed the direct binding of SOX12 to the promoters of Cycline E and Twist1 in A549 cells (Figure 7D). These data suggested that SOX12 transactivated the expression of both genes.
Discussion
SOX12 belongs to SOXC family. It is suggested that SOXC genes may act in redundancy to control cell differentiation and development [21]. Members of SOXC family, SOX4, SOX11 and SOX12, have been found to be up-regulated in human cancer tissues [10-18]. However, the expression of SOX12 in lung cancer have not been clarified. In this study, we found that SOX12 expression was elevated in lung cancer tissues based on analyses on a public available dataset, our own real-time PCR results and Western blot results (Figure 1). SOX12 expression in lung cancer cells were also increased as compared to BEAS-2B, a human normal bronchial epithelial cell line (Figure 2B and Figure S1). Moreover, lung cancer patients with higher SOX12 expression had shorter overall survival time, whereas patients with lower SOX12 expression had better survival (Figure 1C). Thus, we revealed the potential clinical value of SOX12 for patients with lung cancer. The sustained proliferation and high metastatic potential are main hallmarks of cancer cells [22]. Then we examined the roles of SOX12 on the cellular behaviors of lung cancer cells by knocking down its expression. Our data revealed that inhibition of SOX12 in SPC-A-1 and A549 cells could impede cell growth (Figure 3A), migration and invasion (Figure 4), which could be rescued by re-expression of SOX12 (Figure 5). On the contrary, ectopic expression of SOX12 in NCI-H292 cells with lower expression of SOX12 caused an inverse effect (Figure S2). Moreover, SOX12 knockdown could suppressed in vivo tumorigenicity (Figure 6). These data suggested for the first time that SOX12 is a potential oncogene in lung cancer.
We then tried to investigate the molecular mechanisms by which SOX12 promote tumor development and metastasis. SOX proteins harbor HMG box domain, which can interact with the promoter of target genes and then activate their transcription. In HCC cells, overexpression of SOX12 can induce EMT by transactivating the expression of Twist1 [18]. We supposed that SOX12 promotes the growth and metastasis of lung cancer by regulating the expression of key moderators in tumor progression. As expected, knockdown of SOX12 in both SPC-A-1 and A549 cells led to a significant decrease in the mRNA and protein expression of cell growth (PCNA and Cyclin E), anti-apoptosis (Bcl-2), invasion (MMP-9) and EMT (Twist1) related factors. These data suggested that the regulatory role of SOX12 on these key moderators is at the transcriptional level. Moreover, it is known that SOX proteins can recognize a similar DNA motif (A/TAACAA/T) [5,6]. One and two candidate motifs were observed in Cyclin E and Twist1 promoter, respectively. ChIP assay confirmed that SOX12 can directly binding to the promoters of Cycline E and Twist1 in A549 cells (Figure 7D). SOX12 may exert its function through its transcription factor activity. Furthermore, forkhead box Q1 (FOXQ1) is reported to interact with the promoter of SOX12 and transactivate its expression in HCC [18]. The overexpression of FOXQ1 is closely related with poor prognosis and EMT phenomenon of non-small cell lung cancer [23]. Further studies will be necessary to explore the association of SOX12 and FOXQ1.
In summary, this study provides evidence that SOX12 functions as an oncogenic molecule during the development of human lung cancer. High levels of SOX12 seems as an unfavorable prognosis factor for patients with lung cancer, and may serve as a therapeutic target in the future.
Acknowledgements
This study was supported by Shanghai Yangfan Program for Young Scientific and Technological Talents (15YF1407800) and National Natural Science Foundation of China (81602418 and 81572248).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, Alteri R, Robbins AS, Jemal A. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64:252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 3.Malvezzi M, Bertuccio P, Rosso T, Rota M, Levi F, La Vecchia C, Negri E. European cancer mortality predictions for the year 2015: does lung cancer have the highest death rate in EU women. Ann Oncol. 2015;26:779–786. doi: 10.1093/annonc/mdv001. [DOI] [PubMed] [Google Scholar]
- 4.Dy P, Penzo-Mendez A, Wang H, Pedraza CE, Macklin WB, Lefebvre V. The three SoxC proteins--Sox4, Sox11 and Sox12--exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008;36:3101–3117. doi: 10.1093/nar/gkn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hosking BM, Muscat GE, Koopman PA, Dowhan DH, Dunn TL. Trans-activation and DNAbinding properties of the transcription factor, Sox-18. Nucleic Acids Res. 1995;23:2626–2628. doi: 10.1093/nar/23.14.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosking BM, Wyeth JR, Pennisi DJ, Wang SC, Koopman P, Muscat GE. Cloning and functional analysis of the Sry-related HMG box gene, Sox18. Gene. 2001;262:239–247. doi: 10.1016/s0378-1119(00)00525-4. [DOI] [PubMed] [Google Scholar]
- 7.Lefebvre V, Dumitriu B, Penzo-Mendez A, Han Y, Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int J Biochem Cell Biol. 2007;39:2195–2214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong C, Wilhelm D, Koopman P. Sox genes and cancer. Cytogenet Genome Res. 2004;105:442–447. doi: 10.1159/000078217. [DOI] [PubMed] [Google Scholar]
- 9.Castillo SD, Sanchez-Cespedes M. The SOX family of genes in cancer development: biological relevance and opportunities for therapy. Expert Opin Ther Targets. 2012;16:903–919. doi: 10.1517/14728222.2012.709239. [DOI] [PubMed] [Google Scholar]
- 10.Aaboe M, Birkenkamp-Demtroder K, Wiuf C, Sorensen FB, Thykjaer T, Sauter G, Jensen KM, Dyrskjot L, Orntoft T. SOX4 expression in bladder carcinoma: clinical aspects and in vitro functional characterization. Cancer Res. 2006;66:3434–3442. doi: 10.1158/0008-5472.CAN-05-3456. [DOI] [PubMed] [Google Scholar]
- 11.Liao YL, Sun YM, Chau GY, Chau YP, Lai TC, Wang JL, Horng JT, Hsiao M, Tsou AP. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene. 2008;27:5578–5589. doi: 10.1038/onc.2008.168. [DOI] [PubMed] [Google Scholar]
- 12.Medina PP, Castillo SD, Blanco S, Sanz-Garcia M, Largo C, Alvarez S, Yokota J, Gonzalez-Neira A, Benitez J, Clevers HC, Cigudosa JC, Lazo PA, Sanchez-Cespedes M. The SRY-HMG box gene, SOX4, is a target of gene amplification at chromosome 6p in lung cancer. Hum Mol Genet. 2009;18:1343–1352. doi: 10.1093/hmg/ddp034. [DOI] [PubMed] [Google Scholar]
- 13.Liu P, Ramachandran S, Ali Seyed M, Scharer CD, Laycock N, Dalton WB, Williams H, Karanam S, Datta MW, Jaye DL, Moreno CS. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res. 2006;66:4011–4019. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- 14.Brennan DJ, Ek S, Doyle E, Drew T, Foley M, Flannelly G, O’Connor DP, Gallagher WM, Kilpinen S, Kallioniemi OP, Jirstrom K, O’Herlihy C, Borrebaeck CA. The transcription factor Sox11 is a prognostic factor for improved recurrence-free survival in epithelial ovarian cancer. Eur J Cancer. 2009;45:1510–1517. doi: 10.1016/j.ejca.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 15.Ek S, Dictor M, Jerkeman M, Jirstrom K, Borrebaeck CA. Nuclear expression of the non B-cell lineage Sox11 transcription factor identifies mantle cell lymphoma. Blood. 2008;111:800–805. doi: 10.1182/blood-2007-06-093401. [DOI] [PubMed] [Google Scholar]
- 16.Dictor M, Ek S, Sundberg M, Warenholt J, Gyorgy C, Sernbo S, Gustavsson E, Abu-Alsoud W, Wadstrom T, Borrebaeck C. Strong lymphoid nuclear expression of SOX11 transcription factor defines lymphoblastic neoplasms, mantle cell lymphoma and Burkitt’s lymphoma. Haematologica. 2009;94:1563–1568. doi: 10.3324/haematol.2009.008474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weigle B, Ebner R, Temme A, Schwind S, Schmitz M, Kiessling A, Rieger MA, Schackert G, Schackert HK, Rieber EP. Highly specific overexpression of the transcription factor SOX11 in human malignant gliomas. Oncol Rep. 2005;13:139–144. [PubMed] [Google Scholar]
- 18.Huang W, Chen Z, Shang X, Tian D, Wang D, Wu K, Fan D, Xia L. Sox12, a direct target of FoxQ1, promotes hepatocellular carcinoma metastasis through up-regulating Twist1 and FGFBP1. Hepatology. 2015;61:1920–1933. doi: 10.1002/hep.27756. [DOI] [PubMed] [Google Scholar]
- 19.Ding H, Quan H, Yan W, Han J. Silencing of SOX12 by shRNA suppresses migration, invasion and proliferation of breast cancer cells. Biosci Rep. 2016;36:e00389. doi: 10.1042/BSR20160053. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Wang LN, Wang Y, Lu Y, Yin ZF, Zhang YH, Aslanidi GV, Srivastava A, Ling CQ, Ling C. Pristimerin enhances recombinant adeno-associated virus vector-mediated transgene expression in human cell lines in vitro and murine hepatocytes in vivo. J Integr Med. 2014;12:20–34. doi: 10.1016/S2095-4964(14)60003-0. [DOI] [PubMed] [Google Scholar]
- 21.Penzo-Mendez AI. Critical roles for SoxC transcription factors in development and cancer. Int J Biochem Cell Biol. 2010;42:425–428. doi: 10.1016/j.biocel.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 23.Feng J, Zhang X, Zhu H, Wang X, Ni S, Huang J. FoxQ1 overexpression influences poor prognosis in non-small cell lung cancer, associates with the phenomenon of EMT. PLoS One. 2012;7:e39937. doi: 10.1371/journal.pone.0039937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



