Abstract
Salmonella reduces flagella biogenesis to avoid detection within host cells by a largely unknown mechanism. We identified an EAL-like protein STM1697 as required and sufficient for this process. STM1697 surges to a high level after Salmonella enters host cells and restrains the expression of flagellar genes by regulating the function of flagellar switch protein FlhD4C2, the transcription activator of all other flagellar genes. Unlike other anti-FlhD4C2 factors, STM1697 does not prevent FlhD4C2 from binding to target DNA. A 2.0 Å resolution STM1697–FlhD structure reveals that STM1697 binds the same region of FlhD as STM1344, but with weaker affinity. Further experiments show that STM1697 regulates flagella biogenesis by restricting FlhD4C2 from recruiting RNA polymerase and the regulatory effect of STM1697 on flagellar biogenesis and virulence are all achieved by interaction with FlhD. Finally, we describe a novel mechanism mediated by STM1697 in which Salmonella can inhibit the production of flagella antigen and escape from the host immune system.
INTRODUCTION
Salmonella is an important facultative pathogen causing food-borne illness and typhoid fever in humans, with a high rate of morbidity and mortality (1). Flagella in Salmonella allow locomotion and are also involved in the invasion process to infect host cells and trigger the host immune system (2,3). The formation and assembly of the flagellum is a well-organized, hierarchical process in Salmonella (4). Flagellar genes in Salmonella are divided into three classes based on the order of transcription (5). The FlhD4C2 complex, the only product of the flagellar class I operon, is essential for the expression of other flagellar genes (6–8). Four FlhD subunits and two FlhC subunits form the FlhD4C2 heterohexamer to bind to the DNA upstream of flagellar class II promoters, recruit RNA polymerase, and promote flagellar class II gene transcription (8,9). Our previous study showed that FlhD4C2 binds to its target DNA through the FlhC subunit, and FlhD is essential for DNA binding by maintaining the ring-like structure of FlhD4C2 (10).
The translation and expression of the flhDC operon is tightly regulated by a series of regulators. The AMP-catabolite gene activator protein complex activates flhDC transcription when there is a high intracellular concentration of cAMP (11). RcsB, H-NS and RtsB also regulate cell motility by interacting with the flhDC promoter (11–13). CsrA, an RNA-binding protein, regulates flagellar gene expression by enhancing translation of flhDC mRNA, and DnaK, a chaperone protein, converts the native FlhDC to a functional protein form (14–16). The intracellular FlhD4C2 complex can be degraded by the ClpXP protease (7). Additionally, three proteins, STM1344 (also named YdiV or RflP), STM1697 and FliT, were recently identified as negative regulators of flagellum in Salmonella by direct protein–protein interactions: FliT binds to the FlhC subunit, and STM1344 and STM1697 interact with the FlhD subunit (17–22).
STM1344 and STM1697 belong to the EAL-like domain family, whose members are often related to hydrolysis of the second signaling messenger bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP) but STM1344 and STM1697 do not exhibit catalytic activity to c-di-GMP or the ability to bind c-di-GMP (17,23,24). Our previous studies showed that YdiV (Escherichia coli homologue of STM1344) formed multiple types of complexes with FlhD4C2 (10). YdiV1–FlhD4C2 and YdiV2–FlhD4C2 could bind DNA, but YdiV3–FlhD4C2 and YdiV4–FlhD4C2 did not. The third and fourth YdiV molecules bound to the FlhD4C2 complex by squeezing into the ring-like structure of FlhD4C2 between the two internal D subunits. This opened the ring-like structure, causing the complex to lose its DNA-binding ability. Thus, a reasonable model is that YdiV inhibits FlhD4C2 only at relatively high concentrations (10).
Interestingly, although both belong to the EAL-like domain family, a variety of clues show that the molecular mechanism by which STM1697 interacts with FlhD and regulates flagellar gene expression is different from that of YdiV. STM1697 shares only low-sequence identity with STM1344 and E. coli YdiV (22 and 29%, respectively) (17). Additionally, key residues (Phe155, Phe168, Phe181 and Ala184) of YdiV in the YdiV–FlhD interface are missing or substituted in the sequence of STM1697, suggesting a novel interactional mode (Supplementary Figure S1). More importantly, results from recent studies have shown that the FlhDL22H mutant disrupts the interaction with YdiV but is still able to interact with STM1697, further indicating distinct interactions of FlhD with STM1697 and YdiV (17).
Previous findings have demonstrated that STM1697 not only functions as a flagellar regulator but also is involved in virulence phenotypes during Salmonella infection (17). The deletion of STM1697 promoted Salmonella internalization into epithelial cells and the secretion of the type three secretion system 1 effector protein. However, the molecular mechanisms of those regulatory processes remain largely undefined.
Here, we examined the role of STM1697 in the suppression of flagellin production to escape the immune system that occurs when Salmonella invades host cells. STM1697 remains a high level after Salmonella enters host cells and restrains the expression of flagellar genes by regulating the function of the flagellar switch protein FlhD4C2. Unlike other anti-FlhD4C2 factors, STM1697 could not promote the separation of the FlhD4C2–DNA complex even at a very high concentration. To better understand the detailed molecular mechanisms for the STM1697-mediated FlhDC regulation, we determined the crystal structure of STM1697–FlhD at 2.0 Å resolution. Structural analysis showed that STM1697 and YdiV bound to the same region of FlhD protein, though the interface of the STM1697–FlhD complex is not as tight as that of YdiV–FlhD. Most importantly, our studies suggested that STM1697 regulates flagellar formation by inhibiting FlhD4C2 from recruiting RNA polymerase to the promoters of class II flagellar genes rather than by disturbing the interaction between FlhD4C2 and DNA. Gene knockout and complementation experiments demonstrated that STM1697 regulates flagellar formation and virulence phenotypes through its interaction with FlhD. Interface mutants of STM1697 that specifically perturb the interaction between STM1697 and FlhD eliminated the flagellar inhibition and virulence regulation activities. Our studies provide new insights into the complex regulation of bacterial flagellum and revise our understanding of the inhibition mechanism of the anti-FlhD4C2 factors.
MATERIALS AND METHODS
Construction of Stm1697 knock-out strain
The gene disruption mutant of STM1697 was constructed using the lambda Red recombinase system as described previously (25).
Plasmid construction
For in vivo study, STM1697 was cloned into the pBAD24 vector. For expression of STM1344, STM1697, stFlhD and stFlhD4C2 proteins using E. coli, the STM1344, STM1697 and stFlhD genes were cloned into the expression vector pGL01, and the entire flhDC operon was cloned into pET21b. The stFlhD gene was also cloned into pET29b without a his-tag for STM1697–stFlhD production. Mutational derivative plasmids of STM1697 were amplified from the above corresponding plasmids using a Site-Directed Mutagenesis system (TransGen biotech, Beijing, China).
Protein expression and purification
STM1697 and mutants were expressed in E. coli BL21 (DE3) using NZM medium, and then purified by Ni2+-NTA affinity column and Superdex 200 successively.
The STM1697–stFlhD complex was obtained by co-expression in E. coli BL21 (DE3) in Luria-Bertani (LB) medium as described (10). The STM1697–stFlhD complex was lysed by trypsin and purified using a Source Q ion exchange column and Superdex 200 chromatography.
The expression and purification of stFlhD4C2 was executed as described (10).
Crystallization and structure determination
Crystals of the STM1697–stFlhD complex were grown by hanging drop vapour diffusion at 20°C. The crystallization buffer contained 0.2 M KCl, 0.01 M MgCl2, 0.05 M MES pH5.6 and 5% PEG 8000. Diffraction data were collected at the Shanghai Synchrotron Radiation facility (SSRF) beamline BL17u1. The datasets were processed using the HKL2000 software suite. The atomic model was built using Coot and refined using PHENIX. Data collection and structure refinement statistics are summarized in Supplementary Table S1.
Protein pull-down assay
STM1697 and mutants with the his-tag were used as bait protein. FlhD without the his-tag was used as the prey protein.
EMSA experiment
For the electrophoretic mobility shift assay (EMSA) experiment presented in Figure 4, a 49-bp flhB promoter in Salmonella was synthesized as the target DNA. Next, 10 pmol DNA was pre-incubated with different ratios of proteins for 10 min. Then samples were separated by a native 5% polyacrylamide gel and stained with Ethidium Bromide and Coomassie brilliant blue.
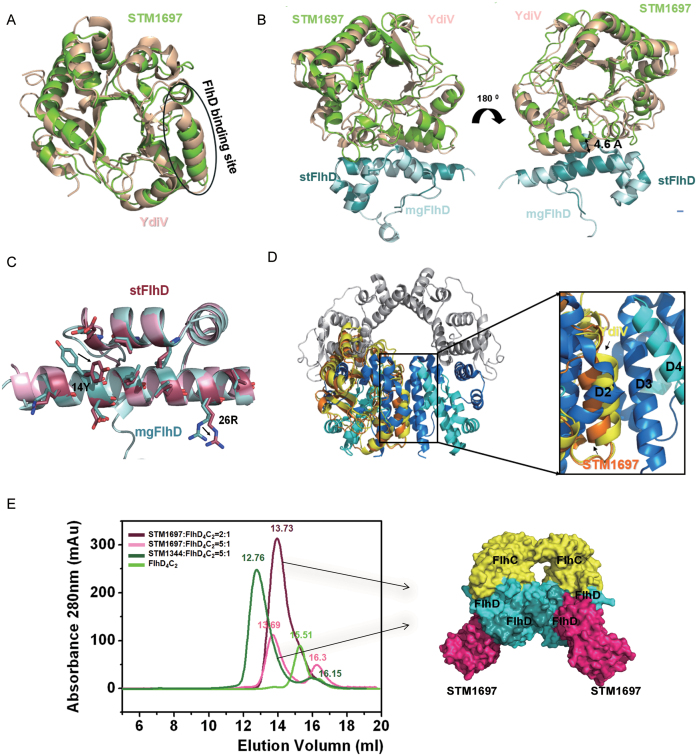
Figure 4.
Structural comparison of STM1697–stFlhD and YdiV–mgFlhD. (A) Structural superposition of STM1697 and YdiV. The two structures are shown in cartoon mode (YdiV: light yellow, STM1697: green). The FlhD binding sites for STM1697 and YdiV are indicated by circles. (B) Structural superposition of STM1697–stFlhD and YdiV–FlhD. The stFlhD in the STM1697–stFlhD structure is used as the reference structure and YdiV-FlhD is superimposed onto the STM1697–stFlhD complex (stFlhD: blue, mgFlhD: light blue, YdiV: light yellow, STM1697: green). (C) Details of changes in the stFlhD and mgFlhD (stFlhD from Salmonella typhimurium, mgFlhD from Escherichia coli MG1655) in the complex structure. Residues located in the interface are shown in stick mode with different colors (stFlhD: hotpink, mgFlhD: blue). The positional changes of the side-chains of 14Y and 26R are indicated by arrows. (D) The structure of FlhD4C2 is used as a model, and STM1697–stFlhD and YdiV–FlhD are separately superimposed onto the third FlhD molecule of FlhD4C2 (STM1697: orange, YdiV: yellow, FlhD: blue). Whole structures are shown in cartoon mode. The right figure presents a close-up view of the interface between the second and the third FlhD molecules of FlhD4C2. All interfaces are overlapped, and the interface of YdiV–FlhD shows more interactions than STM1697–stFlhD and D2–D3. (E) STM1697 and FlhD4C2 cannot form the (STM1697)4 FlhD4C2 hetero-decamer. Size-exclusion chromatography (SEC) results of the 2:1 and 5:1 mixtures of STM1697 and FlhD4C2. The STM1344 and FlhD4C2 mixture is used as a positive control because it forms the (STM1344)4 FlhD4C2 hetero-decamer in vitro. FlhD4C2 alone is used as another control. The elution volume of every peak value is marked in the corresponding colors. STM16972FlhD4C2 complex appeared in solution, but no hetero-decamer formed when the ratio of STM1697 and FlhD4C2 reached 5:1. The right figure presents the model of the STM16972FlhD4C2 hetero-hexamer.
Size-exclusion chromatography
For the size-exclusion chromatography (SEC) experiment presented in Figure 4, proteins were mixed at the corresponding ratio with concentrations varying between 50 and 200 μM for 10 min at 4°C and then injected for SEC using a superdex 200 column.
For the SEC experiment presented in Figure 6, a 145-bp DNA fragment containing the fliA promoter was amplified from the Salmonella genome. Polymerase chain reaction (PCR) products were purified using the E.Z.N.A. Cycle Pure Kit (OMEGA). Proteins and DNA were mixed for 10 min at 4°C and then injected for SEC using a Superdex 200 increase column.
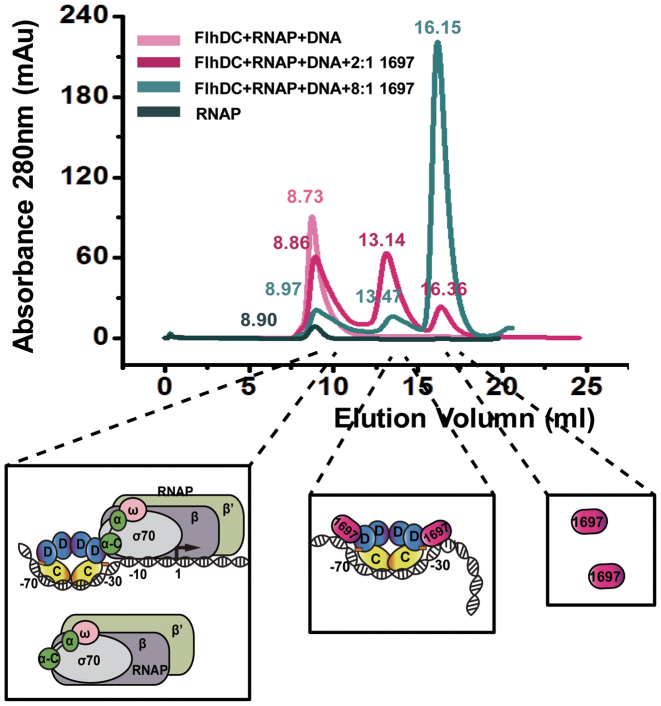
Figure 6.
STM1697 promotes dissociation of FlhDC–RNAP–DNA complex. SEC of the mixtures of FlhD4C2, RNAP and DNA with different concentration of STM1697 were performed. RNAP alone is used as a control. The elution volume of every peak value is marked in the corresponding colors. The cartoon models of protein complexes eluting in corresponding volume were represented below.
Native gel experiment
For the native gel experiment shown in Figure 5, a 269-bp DNA fragment containing the fliA promoter was amplified from the Salmonella genome. PCR products were purified using the E.Z.N.A. Cycle Pure Kit (OMEGA). About 10 pmol DNA was pre-incubated with different ratios of protein. Then, the samples were separated by native 4% polyacrylamide gel and stained with Gel Red and Coomassie brilliant blue G250.
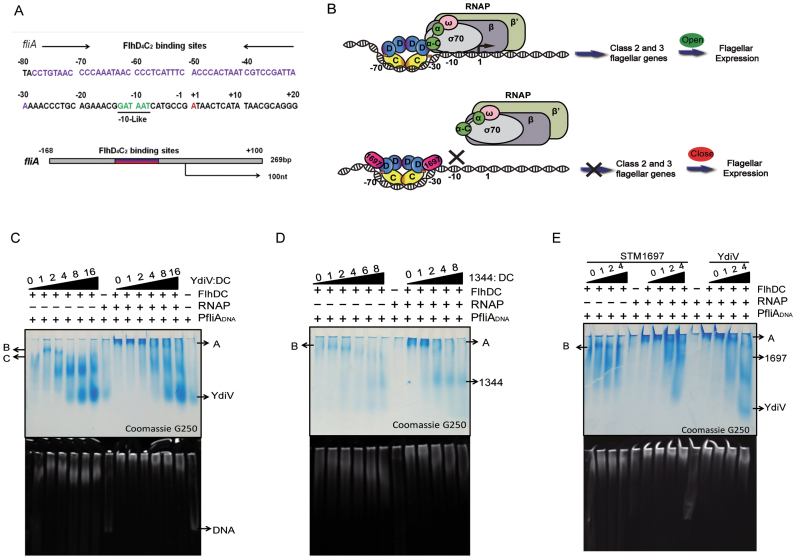
Figure 5.
The binding of STM1697 to FlhD4C2 restrains RNAP recruitment. (A) Sequence analysis of the upstream region of the class II flagellar gene fliA. The FlhD4C2 binding region is highlighted in purple, and the −10 like region of the promoter that serves as a σ70 RNA polymerase binding site is colored green. The transcriptional start site is marked in red. The endpoints of the 269 bp DNA used for the following experiments is also shown. (B) Upper: FlhD4C2 complex interacts with target DNA through the Zn-cys cluster and the positive-charge-enriched region of the FlhC dimer. After binding to the −30 to −70 region of the target gene, the FlhD4C2 complex recruits RNA polymerase to form a transcription initiation complex, then the transcription of class 2 flagellar genes is turned on. Lower: the STM1697 molecules bind to two peripheral FlhD subunits to form three-protein complexes with FlhD4C2. The incorporation of STM1697 prevents RNA polymerase from binding the promoter. As a result, the subsequent expression of flagellar genes is repressed. (C–E) Native gel results preformed with different ratios of protein mixture. Twenty picomoles of FlhD4C2, 10 pmol of DNA and 40 pmol of RNAP were used in the reaction systems. Different amounts of YdiV, STM1344 or STM1697 were added according to the indicated ratio to FlhD4C2. The upper and lower pictures show the same gel stained with Coomassie brilliant blue and Gel red, respectively. Band A is the complex of RNAP, FlhD4C2 and DNA, and band B contains FlhD4C2 and DNA. When YdiV, STM1344 or STM1697 were added, band A disappeared.
Preparation of strains used in functional studies
Salmonella typhimurium ATCC14028 and mutant strains were grown overnight in 10 ml LB medium with 100 μg/ml Ampicillin, and then subcultured into 5 ml LB medium with 100 μg/ml Ampicillin and 0.1% L-arabinose to induce protein expression. To induce invasion phenotype, LB + 0.3M NaCl were used to culture bacteria. When the OD600 reached 0.4–0.6, all strains were adjusted to the same OD600 by the addition of 0.9% NaCl. Bacterial cells were then harvested for functional study. The detailed information of strains used in this study are listed in Supplementary Table S2.
Cell culture, bacterial infection model and RNA extraction
The human colon adenocarcinoma cell line HT-29 was maintained at 37°C in a humidified atmosphere with 5% CO2 in complete RPMI-1640 medium (Gibco) containing 10% fetal bovine serum (Gibco). Cells were seeded at 1 × 107 in 100-mm-wide tissue culture dishes. Salmonella typhimurium ATCC14028 and the STM1697 knock-out strain were added to the HT-29 cell monolayer at an Multiplicity Of Infection (MOI) = 10. After 1 h at 37°C, 100 μg ml−1 gentamicin was added to the cells to kill the extracellular bacteria. At each time point, 2, 4, 6 and 8 h after infection, the HT-29 monolayers were washed, lysed in TRIzol reagent (TIANGEN) and then stored at −70°C.
RNA isolation and real-time quantitative PCR
Total RNA was isolated using the RNAprep Pure Cell/Bacteria Kit (TIANGEN). The reverse transcription reactions were performed using the RevertAid cDNA Synthesis Kit (Thermo) according to the manufacturer's instructions. The quantitative real-time-PCR (qRT-PCR) reactions were performed on an Applied Biosystems 7500 Sequence Detection system (Applied Biosystems, Foster, CA, USA) using iTaq Univerial SYBR Green Supermix (BIO-RAD).
Western blot
Bacterial cells were lysed and examined by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). Polyclonal antibody of FliC (abcam) and HRP-Conjugated Goat anti Rabbit IgG (h+l) (abcam) were used in this study.
Swimming motility assay
Bacterial cells were diluted to OD600 = 10 and inoculated on 0.3% LB agar plates with 100 μg/ml Ampicillin and 0.1% L-arabinose. Swimming motility was observed after 5 h at 30°C.
Invasion assay
For the experiments in Figure 7, Bacteria were diluted using RPMI-1640 medium and then seeded on confluent HT-29 cells grown in 24-well plates at a multiplicity of infection of 20. One hour post-infection, gentamicin (100 μg ml−1) was added to the cells for 1 h to kill the remaining extracellular bacteria. Cells were washed gently with phosphate-buffered saline and then disrupted with 1% Triton X-100 (Sigma Chemical). The number of intracellular bacteria was determining the colony-forming units (CFU) counts of viable colonies.
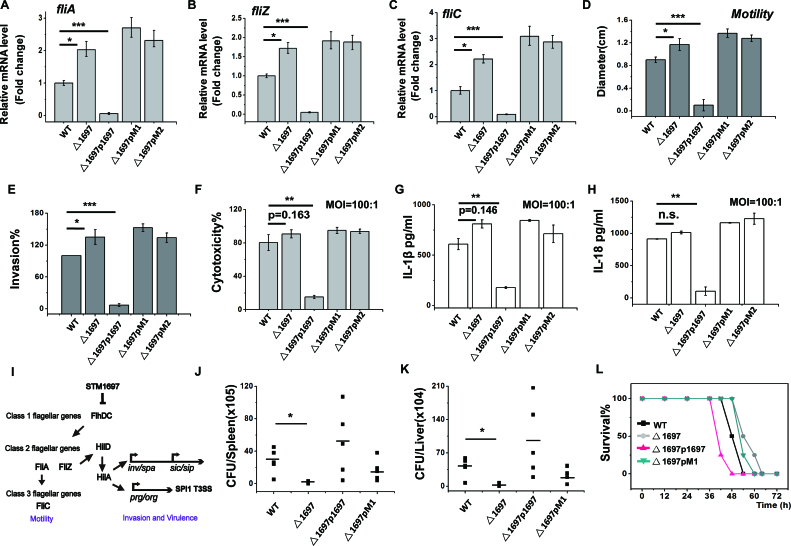
Figure 7.
The suppression of STM1697 of cell motility and virulence phenotypes is mediated by its interaction with FlhD. (A–C) The expression levels of flagellar gene fliA, fliZ and fliC in the wild-type strain, the STM1697 knockout strain and the STM1697 or mutant over-expressed strains were separately tested by qRT-PCR. GADPH was used as an internal reference. (D) The motility of the wild-type strain, the STM1697 knockout strain and STM1697 or mutant over-expressed strains were measured on a 0.3% soft agar plate. (E) Invasion capability of those strains for HT-29 cells. (F) Bone marrow-derived macrophages (BMDM) from BALB/c mice were infected by the strains. Cytotoxicity was determined by LDH release 4 h post-infection. (G and H) IL-1βor IL-18 secretion of BMDM infected with MOI 100:1 were determined by ELISA after 4 h post-infection. (I) Model illustrating the mechanism of STM1697-mediated invasion and virulence regulation. (J and K) BALB/c mice were infected with equal numbers of those strains by intraperitoneal injection. CFU in the spleen and liver were determined after 6 h. (L) BALB/c mice were infected with equal numbers of the wild-type strain, the STM1697 knockout strain, and over-expressed STM1697 or mutant strains by intraperitoneal injection. Survival was monitored. Bars represent data from three independent experiments. Statistical significance is indicated by *P < 0.05, **P < 0.01 and ***P < 0.001 as compared with wild-type Salmonella typhimurium ATCC14028 using a t-test. WT: S. typhimurium ATCC14028 VC; Δ1697: Δstm1697 VC; Δ1697p1697: Δstm1697p1697; Δ1697pM1: Δstm1697pM1; Δ1697pM2: Δstm1697pM2. VC = pBAD24, p1697 = STM1697 cloned into pBAD24, pM1 = STM1697 F160A, L173A cloned into pBAD24 and pM2 = STM1697 L172A, L173A, H176A cloned into pBAD24. In all experiments, protein expression was monitored by qPCR and sodium dodecyl sulphate-polyacrylamide gel electrophoresis (see Supplementary Figure S3).
Macrophage cytotoxicity assay
Bone marrow-derived macrophages were isolated as described. Macrophages were infected at the indicated MOI and then incubated for 4 h. Cytotoxicity was determined by LDH assay (CytoTox 96, Promega). IL-1β and IL-18 secretion was determined by ELISA kit (R&D Systems and MBL, respectively).
In vivo infection of BALB/c mice
Six- to eight-week-old female BALB/c mice were used in this study. For bacterial infections, mice were infected intraperitoneally (IP) with 1 × 106 of each S. typhimurium strain. The mice were sacrificed 6 h post-infection. The number of bacteria located in the spleen or liver was detected by CFU counts of viable colonies. For survival, each cohort of mice (n = 5) was infected intraperitoneally with 1 × 106 bacteria in 100 μl, and the death of mice were observed every 2–3 h.
RESULTS
Salmonella upregulates STM1697 expression during invasion
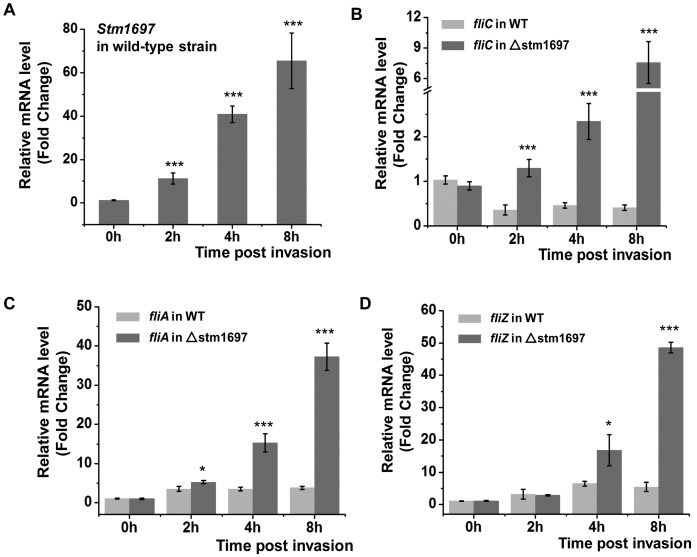
The intracellular expression of STM1697 before and after Salmonella entering into host cells was monitored by qRT-PCR (Figure 1A). Compared with the one before invasion, the expression of STM1697 increased 11.2-, 40.8- and 65.5-times when detected respectively in 2, 4 and 8 h after invasion. These data suggest that STM1697 may play an important regulatory role when Salmonella survive in the host cells.
Figure 1.
STM1697 effectively restrains flagellin expression within host cells. (A) Salmonella upregulates STM1697 expression during invasion. The intracellular concentrations of STM1697 mRNA before and after Salmonella invasion were detected by qRT-PCR. ***P < 0.001 as compared with the one before invasion (0 h) using a t-test. (B–D) The expression of fliC, fliA and fliZ genes in wild-type Salmonella and △stm1697 strain before and after they enter into host cells was monitored by qRT-PCR. Statistical significance is indicated by *P < 0.05 and ***P < 0.001 as compared with wild-type strain at the same time using a t-test.
STM1697 is essential for flagellar inhibition during invasion
To investigate the role of STM1697 on expression of flagellar genes during invasion, the expression of fliC, fliA, fliZ and flhD genes in wild-type Salmonella and △stm1697 strain before and after entering to host cells was monitored by qRT-PCR (Figure 1B–D; Supplementary Figure S2). Interestingly, in wild-type Salmonella, the expression of flagellin (FliC) decreased 2- to 3-fold after invasion while the expression of Class II flagellar genes (FliA and FliZ) increased 3- to 5-fold. In contrast, the expression of FliC, FliA and FliZ in △stm1697 strain was drastically increased (∼8× for FliC and 40–50× for FliA and FliZ) 8 h post-invasion suggesting some signals of host cells may trigger the high expression of flagellin, and STM1697 effectively restrains flagellin expression to a low level. The mRNA level of the flagellar master gene flhD was not significantly different in the wild-type Salmonella and the △stm1697 strain implying that the inhibition of flagella mediated by STM1697 is realized after transcription of FlhDC.
STM1697 formed stable complexes with FlhD4C2 but did not prevent FlhDC from binding to DNA
STM1697 has been reported to interact with FlhD4C2 (17). We found that STM1697 did form stable complexes with FlhD or FlhDC and STM1697–FlhD and STM1697–FlhDC complexes represent good properties in SEC. However, the molecular mechanism of STM1697 mediated flagellar inhibition remained unclear. As described before, interactions of FlhD with STM1697 and YdiV seem to be different. Therefore, one of the goals of our research was to determine the details of the interaction of STM1697 and FlhD4C2 to better understand their activity.
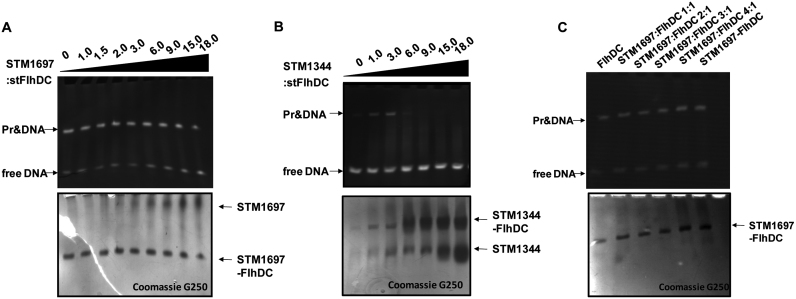
To determine whether STM1697 inhibits the DNA-binding activity of FlhD4C2, we performed a series of EMSAs. STM1344 was used as a positive control of a protein that inhibits FlhD4C2 binding to DNA. Interestingly, the purified STM1697 protein did not effectively inhibit the DNA binding activity of FlhD4C2 even if the molar ratio of STM1697:FlhDC was as high as 18:1. In contrast, purified STM1344 protein prevented FlhDC from binding to DNA at a ratio of STM1344:FlhDC of 3:1 or higher (Figure 2A and B).
Figure 2.
STM1697 did not prevent FlhD4C2 from binding to DNA. (A) EMSA with stFlhD4C2 and its target DNA at different concentrations of STM1697. Sixty picomoles of FlhD4C2 was mixed with different ratios of STM1697 and then the DNA-binding ability was examined by EMSA using 10 pmol DNA. The upper and lower pictures show the same gel stained with ethidium bromide and Coomassie brilliant blue, respectively. (B) EMSA with stFlhD4C2 and its target DNA at different concentration of STM1344. Sixty picomoles of FlhD4C2 was mixed with different ratios of STM1344 and then the DNA-binding ability was examined by EMSA using 30 pmol DNA. (C) EMSA of the stFlhD4C2-STM1697 mixture and purified STM1697–FlhDC complex. Hundred picomoles of purified STM1697–FlhD4C2 or STM1697 and FlhD4C2 mixture (premix for 10 min) were mixed for 10 pmol DNA and examined by EMSA.
In Figure 2A, STM1697, FlhD4C2 and DNA were added to the reaction system simultaneously. It is possible that the formation of FlhDC and DNA complex in reaction system might be prior to the formation of STM1697 and FlhDC which might be the reason for the negative result. So another EMSA experiment was performed in which STM1697 and FlhDC was mixed at first and then target DNA was added. Purified STM1697–FlhDC complex was also test to further verify that STM1697–FlhDC complex can still bind to DNA (Figure 2C). Results showed that premixing of STM1697 and FlhDC and purified STM1697–FlhD4C2 complex all exhibited DNA binding activity demonstrating that STM1697 does not prevent FlhD4C2 from binding to DNA, even at high concentration, unlike its homologue YdiV.
Structure of the STM1697–FlhD complex
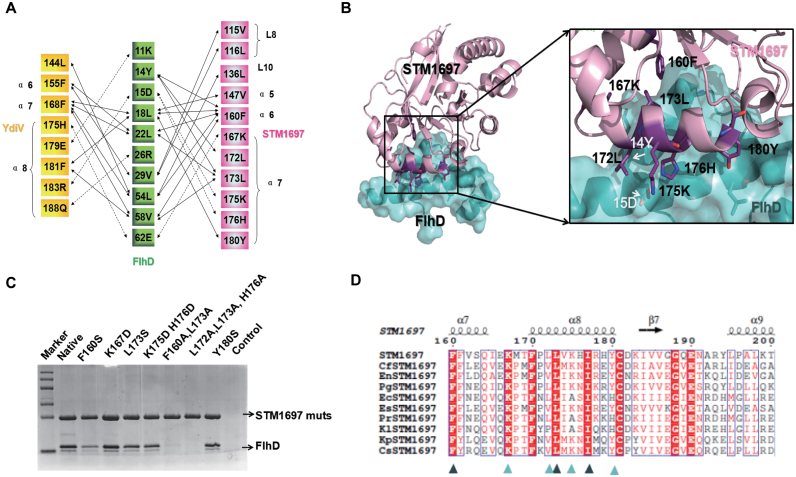
In order to elucidate the interactional mechanism between STM1697 and FlhD4C2, we crystallized the STM1697–FlhD complex. The crystals diffracted to about 2.0 Å resolution and belonged to space group P31 with the unit-cell parameters of a = b = 61.01Å, c = 166.89 Å, α = β = 90° and γ = 120° (Supplementary Table S1). Surprisingly, structural analysis showed that STM1697 bound to the same region of FlhD (the first and fourth α helix) that YdiV binds (Figure 3A). Eleven residues of STM1697 (Val115, Leu116, Leu136, Val147, Phe160, Lys167, Leu172, Leu173, Lys175, His176 and Tyr180) directly participate in the interaction with FlhD, forming a close interface through hydrophobic and electrostatic interactions. Remarkably, Phe160 and Leu173 of STM1697 are located in the center of the molecular interface penetrating into hydrophobic pockets of FlhD with their hydrophobic side chains. Additionally, three pairs of polar contacts (Lys167S/Glu62F, Lys175S/Asp15F and His176S/Asp15F) further strengthen this interface (Figure 3B). Mutagenesis was next employed to validate the importance of residues of STM1697 located in the protein-protein interface in the structural model. Eight mutants of STM1697 were constructed and their ability to bind FlhD was investigated using pull-down assays (Figure 3C). The binding affinity of the STM1697 F160S mutant to FlhD was significantly decreased, and two mutants (F160A/L173A and L172A/L173A/H176A) lost their affinity to FlhD, confirming the significance of these residues in FlhD-binding.
Figure 3.
Structure of the STM1697–stFlhD complex. (A) Schematic diagram of essential residues denoting molecular interactions between STM1697 or YdiV and FlhD. Van der Waals interactions (<4 Å) and polar interactions (hydrogen bonds or salt bridges) are indicated using solid arrows and dashed arrows, respectively. (B) The interface between STM1697 and stFlhD. Residues in the interface are shown in stick model and labeled in relevant color (purple: STM1697, white: stFlhD). (C) Pull down of stFlhD by native/mutants of STM1697 with the His-tag. stFlhD without the his-tag was used as the negative control. The two mutants of STM1697 lose interaction with stFlhD. (D) CLUSTALW alignment between Salmonella STM1697 and STM1697 from other bacteria. Forty residues (160–200 aa) of STM1697 are used in this alignment and the most conserved residues of STM1697 are marked by arrowheads. Cf: Citrobacter freundii, En: Enterobacter sp. GN02600, Pg:Pluralibacter gergoviae, Ec: Enterobacter cloacae, Es: Escherichia coli, Pr: Proteobacteria, Kl: Klebsiella oxytoca, Kp: Klebsiella pneumoniae and Cr: Cronobacter sakazakii.
By sequence alignment, we found that the key residues (Phe160 and Leu173) of STM1697 in the interactional surface are highly conserved across its orthologs, suggesting this binding interaction of STM1697–FlhD may be employed widely (Figure 3D).
Structural comparison between STM1697–FlhD and YdiV–FlhD
Structural analysis suggested a topological structure of STM1697 of a modified TIM-like barrel fold similar to that of a conventional EAL domain protein, but with low-sequence identities. Structural comparison of STM1697 with all other PDB entries using the Dali server (http://ekhidna.biocenter.helsinki.fi/dali_server) indicated that, as expected, YdiV was the most similar homolog with a root mean square deviation (RMSD) of ∼2.0 Å for 235 Cα atoms. Additionally, we superimposed the STM1697 molecule from the STM1697–FlhD structure and the YdiV molecule from the YdiV–FlhD structure and found that most of regions from STM1697 fit well with those of YdiV except in the region involved in FlhD-binding (Figure 4A). When the superposition was performed using FlhD as a template, surprisingly, STM1697 did not match YdiV at all due to position deviation of the entire molecule (Figure 4B). The square deviation of the two FlhD-binding helixes (α8 in YdiV and α7 in STM1697) was about 4.6 Å, suggesting that the bond strength of YdiV–FlhD and STM1697–FlhD might be discrepant. Detailed analysis showed that the conformation of FlhD from STM1697–FlhD was almost the same as the one from YdiV–FlhD except the side chain deflection of essential residues (Tyr14 and Lys26) located in the interface of STM1697–FlhD (Figure 4C).
Previous study showed that YdiV forms four kinds of complexes with FlhD4C2: YdiV1FlhD4C2, YdiV2FlhD4C2, YdiV3FlhD4C2 and YdiV4FlhD4C2 (10). In order to form YdiV3FlhD4C2 and YdiV4FlhD4C2 complexes, YdiV must enter the ring-like structure of FlhD4C2, disrupting the interface between the central FlhD subunits. As a result, the ring-like structure of FlhD4C2 is disrupt and the structure of complex rearrange into an opened one that can not bind DNA. To analyze the interaction of those complexes, STM1697–FlhD and YdiV–FlhD were separately superimposed with the central FlhD subunits (D3) of FlhD4C2. It is evident that the distance between STM1697 and FlhD molecule was furthest, compared with that of YdiV–FlhD and FlhD2–FlhD3 complexes, suggesting the weakest interaction between STM1697 and FlhD (Figure 4D). According to above analysis, whether STM1697 can squeeze into the ring-like structure of FlhD4C2, as its homologue YdiV, remains uncertain.
STM1697 cannot form STM16973FlhD4C2 or STM16974FlhD4C2 complexes with FlhD4C2in vitro
We next performed experiments using SEC to determine if STM1697 can bind FlhD4C2 to form STM16973FlhD4C2 or STM16974FlhD4C2 complexes. Purified STM1697 protein (26.5 kD) was mixed with FlhD4C2 protein (96.4 kD) at molar ratios of 5:1 or 2:1 and analyzed by a size-exclusion column. As a control, a 5:1 mixture of STM1344 (26.4 kD) and FlhD4C2 was used, and should form a stable hetero-decamer (STM13444FlhD4C2) in solution. FlhD4C2 alone was also examined as a control. As shown in Figure 4E, the STM1697–FlhD4C2 mixture with the molar ratio of 2:1 showed a single elution peak at a position (13.73 ml) consistent with a larger complex than the FlhD4C2 hexamer (15.51 ml), suggesting the STM1697–FlhD4C2 complex formed in solution. When the ratio of STM1697 and FlhD4C2 was increased to 5:1, two peaks (13.69 and 16.30 ml) appeared. The first peak was the newly-formed STM1697–FlhD4C2 complex and the second one was the excessive amount of free STM1697. The 5:1 mixture of STM1344 and FlhD4C2 eluted at two positions, 12.76 and 16.15 ml. The elution volume measured in the 2:1 STM1697–FlhD4C2 mixture (13.69 ml) was similar to that for the 5:1 mixture (13.73 ml), and were both larger than that of STM13444FlhD4C2 (12.76 ml), suggesting that STM1697 cannot form the STM16973FlhD4C2 or STM16974FlhD4C2 complex with FlhD4C2 under these conditions. Given the above-mentioned data, we propose that unlike STM1344, STM1697 interacts with FlhD4C2 to form STM16971–FlhD4C2 or STM16972–FlhD4C2 complex, but does not prevent FlhD4C2 from binding to target DNA.
The binding of STM1697 to FlhD4C2 restrains RNA polymerase recruitment
Previous studies showed that class II flagellar genes are only transcribed by σ70-containing RNA polymerase in the presence of the FlhD4C2 complex (26,27). The FlhD4C2 complex binding sites are located ∼30–80 bp upstream of the transcription start site of class II flagellar genes (28,29). Here, the upstream sequence of fliA operon was used to study the transcription initiation of class II flagellar genes. There is no obvious −35 promoter element (TTGACA) in the upstream sequence of the fliA operon. At this promoter, the −30 to −79 bp contain the FlhD4C2 binding site (28,30). This positioning and the lack of a strong −35 promoter element suggest that FlhD4C2 may promote σ70 RNA polymerase recruitment (Figure 5A). The C-terminal domain of the RNA polymerase α subunit was reported to be required for FlhD4C2-mediated transcription activation and structural simulation showed that FlhD4C2 was able to directly interact with the α subunit and δ subunit (27). Therefore, we hypothesize that, when STM1697 binds to the FlhD subunits of the FlhD4C2 complex, steric exclusion prevents σ70 RNA polymerase from binding to template DNA, thus blocking transcription(Figure 5B). To test our hypothesis, we examined the ability of RNA polymerase to bind a 269-bp fragment of promoter DNA by native gel electrophoresis (Figure 5A, C–E). The results showed that σ70 RNA polymerase could not form a complex with target DNA in the absence of the FlhD4C2 complex. Once FlhD4C2 was added, FlhD4C2, RNA polymerase and target DNA formed a large complex (band A in Figure 5C–E). Interestingly, upon addition of the anti-factors of FlhD4C2 (YdiV, STM1344 or STM1697), the large DNA–proteins complex disappeared (Figure 5C–E). These data support our hypothesis that the binding of anti-factors (YdiV or STM1697) to the periphery of the FlhDC complex prevents RNA polymerase from binding to the promoter.
SEC was used to further determine that STM1697 promotes dissociation of FlhDC–RNAP–DNA complex. The mixtures of FlhD4C2, RNAP and DNA in absence and presence of STM1697 were detected. RNAP alone is used as a control (Figure 6). FlhDC–RNAP–DNA mixture (∼600 kD) and RNAP (∼500 kD) alone were eluted at the similar position (8.6–8.9 ml). When STM1697 was added to the complex, a second peak appeared at 13.14 ml which might be the complex of STM1697–flhDC–DNA (∼180 kD). Excessive STM1697 was eluted at 16.1–16.4 ml.
Those results well support our hypothesis that STM1697 prevents σ70 RNA polymerase from binding to template DNA.
The STM1697 suppression of cell motility is mediated by its interaction with FlhD
Five Salmonella strains containing wild-type, stm1697 knockout strain, or that strain with over-expression of STM1697 or two mutants of STM1697 shown above to be defective in FlhD binding (FlhD-free, Mut-1: F160A/L173A and Mut-2: L172A/L173A/H176A) were constructed for functional study. The STM1697 mutants expressed by Mut-1 and Mut-2 were unable to bind to FlhD in vitro, as shown above. High-level expression of native STM1697 or mutant proteins was detected at the RNA level by qRT-PCR and at the protein level by SDS-PAGE (Supplementary Figure S3). To evaluate the transcription levels of flagellar genes in those strains, qRT-PCR was performed of three flagellar genes (fliA, flhB and fliC). The transcription of all three genes was significantly decreased in the STM1697-expression strain and upregulated in △stm1697, compared with the WT strain. Differently, the two STM1697 mutant strains did not show obvious variation compared with the △stm1697 strain. Those data indicated that STM1697 directly represses class II and class III flagellar genes and this inhibitory effect depends on its interaction with FlhD (Figure 7A–C). Subsequently, the expression of FliC protein in those strains was detected by western blotting, and the protein levels were consistent with the qRT-PCR results (Supplementary Figure S4). Next, the motility behavior of those strains was determined using soft agar plates. In accordance with the report from Ahmad et al., the strain deleted for STM1697 exhibited significantly enhanced swimming capability compared to the wild-type strain, and the strain in which STM1697 was over-expressed did not swim at all (17). The expression of the FlhD-free mutants of STM1697 showed similar motility phenotypes as the △stm1697 strain suggesting that the motility inhibition of STM1697 is directly mediated by interaction with FlhD (Figure 7D). These data supported a model in which the suppression of cell motility by STM1697 was mediated by its interaction with FlhD.
The effect of STM1697 on virulence phenotypes is also mediated by its interaction with FlhD
The STM1697 protein was reported to be involved in virulence phenotypes during Salmonella infection (17). To better understand the mechanism by which STM1697 regulates virulence phenotypes of Salmonella, the effect of STM1697 and mutants on virulence phenotypes was investigated. First, the ability to invade HT-29 cells was measured for the five strains (Figure 7E). Consistent with previous studies, the invasion ability of the △stm1697 strain was enhanced compared with the wild-type strain (17). Remarkably, the STM1697 over-expression strain showed a defect in invasion of HT-29, as did the two FlhD-free STM1697 mutants, indicating that the ability of STM1697 to regulate invasion phenotypes requires FlhD.
Next, macrophage cytotoxicity of these strains was determined through measurement of lactate dehydrogenase (LDH) release, indicating cell membrane damage. The release of the IL-1β and IL-18 cytokines was also measured using an ELISA kit (Figure 7F–H). The release of LDH, IL-1β and IL-18 in the △stm1697 strain was weakly increased compared with the wild-type strain. In contrast, when STM1697 was over-expressed, there was a significant reduction in the levels of LDH, IL-1β and IL-18. The two FlhD-free STM1697 mutants over-expressed strains show similar phenotype as the △stm1697 strain. The above data revealed that the expression of STM1697 can significantly reduce Salmonella-caused pyroptosis of macrophages and result in immunosuppression, and these activities require interaction with FlhD. Previous study has shown that the class II flagellar gene FliZ regulates the Salmonella pathogenicity island 1 by the activation of HilD and HilA (31,32). Therefore, the mechanism by which STM1697 regulates invasion and macrophage cytotoxicity in Salmonella may be achieved by the inhibition of the FliZ gene (Figure 7I).
To investigate the affect of STM1697 on Salmonella virulence in vivo, animal experiments were conducted (Figure 7J–L). The Salmonella strain lacking STM1697 showed significantly attenuated ability to colonize the spleen and in the liver when infected intraperitoneally. Interestingly, the multiplication of the STM1697 over-expressed strain in the liver and the spleen were irregular for unknown reasons, and the FlhD-free STM1697 mutant over-expressed strain behaved similarly. Consistent with this observation, the survival curve of mice infected with equal amount of bacteria revealed that the pathogenicity of the △stm1697 strain and the FlhD-free STM1697 mutant over-expressed strain were weaker than wild-type, and the STM1697 over-expressed strain exhibited enhanced virulence. These results indicated that the effect of STM1697 on virulence phenotypes was also mediated by its interaction with FlhD.
DISCUSSION
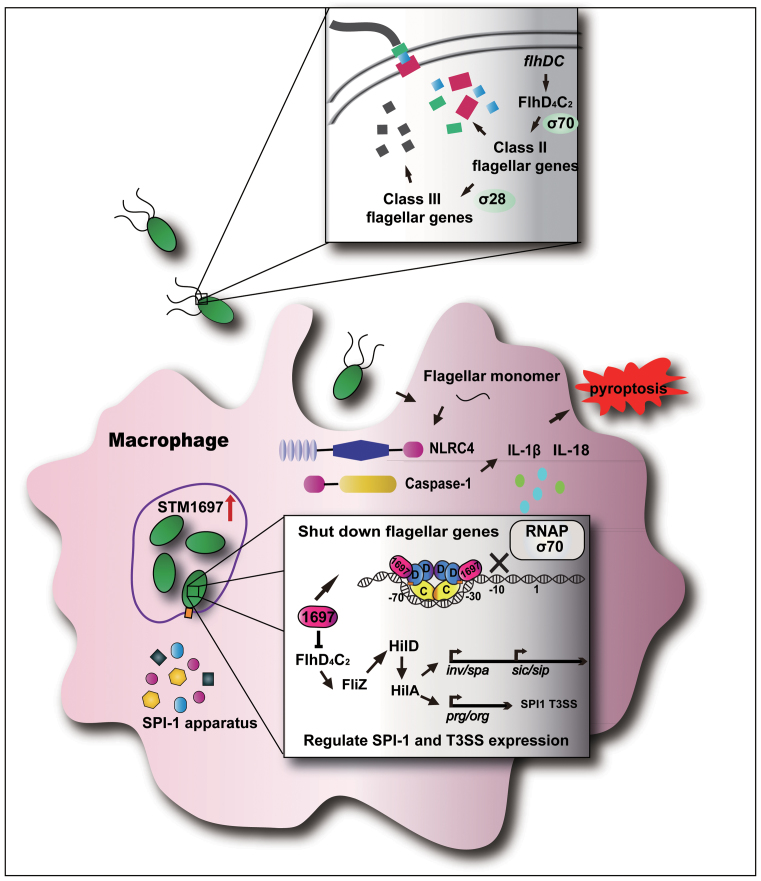
The flagellum is indispensable when Salmonella is invading host cells, however, after entering host cells, the situation changes. Flagellin can be detected by the host pattern recognition receptor TLR5, which activates the host immune system and triggers pyroptosis of macrophages (33,34). Interestingly, maybe due to long-term evolution, Salmonella reduced the expression of flagellin to escape from the host immune system (35,36). However, the mechanism involved in this process remains ill-defined. Here, we revealed that the EAL-like protein STM1697 suppressed the formation of flagellum after Salmonella invasion into host cells. Significantly, the intracellular concentration of STM1697 increased dozens of times after entering host cells. Very surprisingly, we found that the expression of flagellin increased ∼10× in the △stm1697 strain after entering into host cells, indicating that some signals in host cells may trigger the high expression of flagellin. STM1697 effectively restrains flagellin expression to a low level and avoids detection of Salmonella by the host immune system. Thus, a new model by which STM1697 regulates flagellar biogenesis during invasion and infection was established (Figure 8).
Figure 8.
The role of STM1697 in the Salmonella infection process. Before entry into the host cell, the expression of STM1697 remains at a low level. FlhD4C2 complex binds to the upstream region of flagellar genes and recruits RNA polymerase to express flagellar genes. When Salmonella enters the host cell, the expression of STM1697 is stimulated by some unknown mechanism. STM1697 then binds to the peripheral FlhD of the FlhD4C2 complex, which restrains the recruitment of RNA polymerase by steric exclusion. As a result, the expression of flagellar genes is repressed, and then the expression of FliZ-regulated genes is also downregulated. Salmonella benefits from this process to escape the host immune system.
Model for STM1697-dependent flagellar regulation during infection
Before entry of host cells, the expression of STM1697 is low. FlhD4C2 complex binds to the upstream region of flagellar genes and recruits RNA polymerase to express flagellar genes. When Salmonella enters the host cell, the expression of STM1697 is stimulated by some unknown mechanism. STM1697 then binds to the peripheral FlhD of the FlhD4C2 complex, which restrains the recruitment of RNA polymerase, repressing the expression of flagellar genes. At the same time, the expression of FliZ-regulated genes is also downregulated.
This is an effective strategy for Salmonella. Flagellin is a major activator of NF-κB signaling pathways and pro-inflammatory gene activation in intestinal epithelial cells (37,38). The STM1697-mediated repression of flagellin enables Salmonella to evade TLR5-mediated cytokine production in epithelial cells. Additionally, the reduction of flagellin allows Salmonella to escape induction of macrophage pyroptosis and replicate successfully within macrophages.
New insight into the mechanism of anti-FlhD4C2 factor-mediated motility control
It is thought that anti-FlhD4C2 factors inhibit flagellar formation by disrupting the interaction between FlhD4C2 and its target DNA. In this study, a novel mechanism was discovered: STM1697 does not disrupt the interaction between FlhD4C2 and DNA but prevents FlhD4C2 from recruiting RNA polymerase. This research provides new insight into the mechanism of anti-FlhD4C2 factors and demonstrates how anti-FlhD4C2 can function even at low concentration.
Two EAL-like proteins in Salmonella, STM1344 and STM1697, act as anti-FlhD4C2 factors by direct interaction with FlhD. Those two proteins share low-sequence identity (<22%). Previously, we have reported that the E. coli homolog of STM1344 regulates motility in a concentration-dependent manner (10). Our previous studies showed that E. coli YdiV formed multiple types of complexes with FlhD4C2. YdiV1–FlhD4C2 and YdiV2–FlhD4C2 can bind DNA, but YdiV3–FlhD4C2 and YdiV4–FlhD4C2 are unable to bind DNA. The third and fourth YdiV molecule altered the FlhD4C2 complex by entering the ring-like structure of FlhD4C2 between the two internal D subunits. This caused opening of the ring-like structure, eliminating DNA-binding ability. Thus, it was thought that YdiV-inhibited FlhD4C2 only at relatively high concentrations. However, based on the data in this paper, the model in which YdiV regulates flagellar production can be improved. When the intracellular concentration of YdiV is low, YdiV binds the exterior FlhD subunits to prevent FlhDC from recruiting RNA polymerase in a dynamical and reversible regulatory process. When the intracellular concentration of YdiV slowly accumulates to a certain level, YdiV then strips FlhD4C2 from DNA and facilitates ClpXP protease-mediated FlhD4C2 degradation, which is lasting and irreversible.
Why does Salmonella employ two similar factors to regulate FlhD4C2?
The activation of stm1344 and stm1697 genes may be mediated by different factors sensing different signals. The AI-2 binding protein SdiA can activate the expression of stm1344 but not stm1697, although the RNA binding protein CsrA (Carbon Storage Regulator A) is reported to reduce the expression of both stm1344 and stm1697 in Salmonella (39–42). We found the production of STM1344 was upregulated more than 10-fold when Salmonella was cultured in M9 restrictive medium, but no such changes were observed in the expression of STM1697 (unpublished data). Overall, the explanation of the activation of the stm1697 gene after Salmonella enters host cells requires further investigation.
STM1344 and STM1697 function in different ways. It has reported that some anti-FlhD4C2 factors, like YdiV and FliT, promote hydrolysis by the ClpXP protease of the FlhD4C2 complex (21,22). However, our unpublished data showed that the degradation of FlhD4C2 by the ClpXP protease was promoted by STM1344 but not by STM1697. Furthermore, we found that FlhD4C2 can preferentially form a complex with STM1344 and, most importantly, STM1344 could separate the STM1697–FlhD complex to form an STM1344–FlhD complex, but STM1697 cannot disrupt the STM1344–FlhD complex. All these phenomena suggest differences in the interactions of these proteins that could potentially explain differences in the regulatory mechanisms mediated by STM1344 or STM1697. More studies need to be done to determine the complete pathways to activate the expression of STM1344 and STM1697 and then inhibit flagellar genesis when Salmonella moves back and forth in different environments.
ACCESSION NUMBER
The accession number of STM1697–FlhD structure in this paper is PDB ID: 5HXG.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff at beamline BL17U at the SSRF for supporting with data collection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31500050, 31270786]; Innovation Project of Shandong Academy of Medical Sciences; Department of Health and Family-plan Bureau, Shandong Province [2016WS0525, 2015WS0188]; Shandong Academy of Medical Sciences Grant [2015–27]; Shandong Provincial College of Traditional Chinese Medicine Antiviral Collaborative Innovation Center [XTCX2014B01–05]; National Science and Technology Major Special Project of China [2014ZX09509–001001008]. Funding for open access charge: Innovation Project of Shandong Academy of Medical Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1. Martin L.J., Fyfe M., Dore K., Buxton J.A., Pollari F., Henry B., Middleton D., Ahmed R., Jamieson F., Ciebin B. et al. Increased burden of illness associated with antimicrobial-resistant Salmonella enterica serotype typhimurium infections. J. Infect. Dis. 2004; 189:377–384. [DOI] [PubMed] [Google Scholar]
- 2. Hayashi F., Smith K.D., Ozinsky A., Hawn T.R., Yi E.C., Goodlett D.R., Eng J.K., Akira S., Underhill D.M., Aderem A.. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001; 410:1099–1103. [DOI] [PubMed] [Google Scholar]
- 3. Ramos H.C., Rumbo M., Sirard J.C.. Bacterial flagellins: mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol. 2004; 12:509–517. [DOI] [PubMed] [Google Scholar]
- 4. Chilcott G.S., Hughes K.T.. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol. Mol Biol. Rev. 2000; 64:694–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kutsukake K., Ohya Y., Iino T.. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J. Bacteriol. 1990; 172:741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kutsukake K. Autogenous and global control of the flagellar master operon, flhD, in Salmonella typhimurium. Mol. Gen. Genet. 1997; 254:440–448. [DOI] [PubMed] [Google Scholar]
- 7. Tomoyasu T., Takaya A., Isogai E., Yamamoto T.. Turnover of FlhD and FlhC, master regulator proteins for Salmonella flagellum biogenesis, by the ATP-dependent ClpXP protease. Mol. Microbiol. 2003; 48:443–452. [DOI] [PubMed] [Google Scholar]
- 8. Liu X., Matsumura P.. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 1994; 176:7345–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang S., Fleming R.T., Westbrook E.M., Matsumura P., McKay D.B.. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J. Mol. Biol. 2006; 355:798–808. [DOI] [PubMed] [Google Scholar]
- 10. Li B., Li N., Wang F., Guo L., Huang Y., Liu X., Wei T., Zhu D., Liu C., Pan H. et al. Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility. Nucleic Acids Res. 2012; 40:11073–11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Soutourina O., Kolb A., Krin E., Laurent-Winter C., Rimsky S., Danchin A., Bertin P.. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J. Bacteriol. 1999; 181:7500–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Francez-Charlot A., Laugel B., Van Gemert A., Dubarry N., Wiorowski F., Castanie-Cornet M.P., Gutierrez C., Cam K.. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 2003; 49:823–832. [DOI] [PubMed] [Google Scholar]
- 13. Ellermeier C.D., Slauch J.M.. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 2003; 185:5096–5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wei B.L., Brun-Zinkernagel A.M., Simecka J.W., Pruss B.M., Babitzke P., Romeo T.. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 2001; 40:245–256. [DOI] [PubMed] [Google Scholar]
- 15. Yakhnin A.V., Baker C.S., Vakulskas C.A., Yakhnin H., Berezin I., Romeo T., Babitzke P.. CsrA activates flhDC expression by protecting flhDC mRNA from RNase E-mediated cleavage. Mol. Microbiol. 2013; 87:851–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Takaya A., Matsui M., Tomoyasu T., Kaya M., Yamamoto T.. The DnaK chaperone machinery converts the native FlhD2C2 hetero-tetramer into a functional transcriptional regulator of flagellar regulon expression in Salmonella. Mol. Microbiol. 2006; 59:1327–1340. [DOI] [PubMed] [Google Scholar]
- 17. Ahmad I., Wigren E., Le Guyon S., Vekkeli S., Blanka A., El Mouali Y., Anwar N., Chuah M.L., Lunsdorf H., Frank R. et al. The EAL-like protein STM1697 regulates virulence phenotypes, motility and biofilm formation in Salmonella typhimurium. Mol. Microbiol. 2013; 90:1216–1232. [DOI] [PubMed] [Google Scholar]
- 18. Hung C.C., Haines L., Altier C.. The flagellar regulator fliT represses Salmonella pathogenicity island 1 through flhDC and fliZ. PLoS One. 2012; 7:e34220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wada T., Morizane T., Abo T., Tominaga A., Inoue-Tanaka K., Kutsukake K.. EAL domain protein YdiV acts as an anti-FlhD4C2 factor responsible for nutritional control of the flagellar regulon in Salmonella enterica Serovar Typhimurium. J. Bacteriol. 2011; 193:1600–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hengge R., Galperin M.Y., Ghigo J.M., Gomelsky M., Green J., Hughes K.T., Jenal U., Landini P.. Systematic nomenclature for GGDEF and EAL domain-containing cyclic Di-GMP turnover proteins of Escherichia coli. J. Bacteriol. 2015; 198:7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sato Y., Takaya A., Mouslim C., Hughes K.T., Yamamoto T.. FliT selectively enhances proteolysis of FlhC subunit in FlhD4C2 complex by an ATP-dependent protease, ClpXP. J. Biol. Chem. 2014; 289:33001–33011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takaya A., Erhardt M., Karata K., Winterberg K., Yamamoto T., Hughes K.T.. YdiV: a dual function protein that targets FlhDC for ClpXP-dependent degradation by promoting release of DNA-bound FlhDC complex. Mol. Microbiol. 2012; 83:1268–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chou S.H., Galperin M.Y.. Diversity of cyclic Di-GMP-binding proteins and mechanisms. J. Bacteriol. 2016; 198:32–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Romling U., Liang Z.X., Dow J.M.. Progress in understanding the molecular basis underlying functional diversification of cyclic dinucleotide turnover proteins. J. Bacteriol. 2017; 199, doi:10.1128/JB.00790-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Datsenko K.A., Wanner B.L.. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Arnosti D.N. Regulation of Escherichia coli sigma F RNA polymerase by flhD and flhC flagellar regulatory genes. J. Bacteriol. 1990; 172:4106–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu X., Fujita N., Ishihama A., Matsumura P.. The C-terminal region of the alpha subunit of Escherichia coli RNA polymerase is required for transcriptional activation of the flagellar level II operons by the FlhD/FlhC complex. J. Bacteriol. 1995; 177:5186–5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ikebe T., Iyoda S., Kutsukake K.. Promoter analysis of the class 2 flagellar operons of Salmonella. Genes Genet. Syst. 1999; 74:179–183. [DOI] [PubMed] [Google Scholar]
- 29. Tanabe Y., Wada T., Ono K., Abo T., Kutsukake K.. The transcript from the sigma (28)-dependent promoter is translationally inert in the expression of the sigma(28)-encoding gene fliA in the fliAZ operon of Salmonella enterica serovar Typhimurium. J. Bacteriol. 2011; 193:6132–6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee Y.Y., Barker C.S., Matsumura P., Belas R.. Refining the binding of the Escherichia coli flagellar master regulator, FlhD4C2, on a base-specific level. J. Bacteriol. 2011; 193:4057–4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chubiz J.E., Golubeva Y.A., Lin D., Miller L.D., Slauch J.M.. FliZ regulates expression of the Salmonella pathogenicity island 1 invasion locus by controlling HilD protein activity in Salmonella enterica serovar typhimurium. J. Bacteriol. 2010; 192:6261–6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jubelin G., Lanois A., Severac D., Rialle S., Longin C., Gaudriault S., Givaudan A.. FliZ is a global regulatory protein affecting the expression of flagellar and virulence genes in individual Xenorhabdus nematophila bacterial cells. PLoS Genet. 2013; 9:e1003915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fink S.L., Bergsbaken T., Cookson B.T.. Anthrax lethal toxin and Salmonella elicit the common cell death pathway of caspase-1-dependent pyroptosis via distinct mechanisms. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:4312–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miao E.A., Leaf I.A., Treuting P.M., Mao D.P., Dors M., Sarkar A., Warren S.E., Wewers M.D., Aderem A.. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat. Immunol. 2010; 11:1136–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stewart M.K., Cummings L.A., Johnson M.L., Berezow A.B., Cookson B.T.. Regulation of phenotypic heterogeneity permits Salmonella evasion of the host caspase-1 inflammatory response. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:20742–20747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stewart M.K., Cookson B.T.. Mutually repressing repressor functions and multi-layered cellular heterogeneity regulate the bistable Salmonella fliC census. Mol. Microbiol. 2014; 94:1272–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tallant T., Deb A., Kar N., Lupica J., de Veer M.J., DiDonato J.A.. Flagellin acting via TLR5 is the major activator of key signaling pathways leading to NF-kappa B and proinflammatory gene program activation in intestinal epithelial cells. BMC Microbiol. 2004; 4:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fink S.L., Cookson B.T.. Pyroptosis and host cell death responses during Salmonella infection. Cell. Microbiol. 2007; 9:2562–2570. [DOI] [PubMed] [Google Scholar]
- 39. Zhou X., Meng X., Sun B.. An EAL domain protein and cyclic AMP contribute to the interaction between the two quorum sensing systems in Escherichia coli. Cell Res. 2008; 18:937–948. [DOI] [PubMed] [Google Scholar]
- 40. Jonas K., Edwards A.N., Ahmad I., Romeo T., Romling U., Melefors O.. Complex regulatory network encompassing the Csr, c-di-GMP and motility systems of Salmonella Typhimurium. Environ. Microbiol. 2010; 12:524–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Leng Y., Vakulskas C.A., Zere T.R., Pickering B.S., Watnick P.I., Babitzke P., Romeo T.. Regulation of CsrB/C sRNA decay by EIIA(Glc) of the phosphoenolpyruvate: carbohydrate phosphotransferase system. Mol. Microbiol. 2016; 99:627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perez-Morales D., Bustamante V.H.. The global regulatory system Csr senses glucose through the phosphoenolpyruvate: carbohydrate phosphotransferase system. Mol. Microbiol. 2016; 99:623–626. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.