Abstract
Microsatellite repeat DNA is best known for its length mutability, which is implicated in several neurological diseases and cancers, and often exploited as a genetic marker. Less well-known is the body of work exploring the widespread and surprisingly diverse functional roles of microsatellites. Recently, emerging evidence includes the finding that normal microsatellite polymorphism contributes substantially to the heritability of human gene expression on a genome-wide scale, calling attention to the task of elucidating the mechanisms involved. At present, these are underexplored, but several themes have emerged. I review evidence demonstrating roles for microsatellites in modulation of transcription factor binding, spacing between promoter elements, enhancers, cytosine methylation, alternative splicing, mRNA stability, selection of transcription start and termination sites, unusual structural conformations, nucleosome positioning and modification, higher order chromatin structure, noncoding RNA, and meiotic recombination hot spots.
Keywords: short, tandem, repeat, transcription, eQTL, review
Introduction
Microsatellites, or short tandem repeats (STRs), also often called short sequence repeats (SSRs), consist of tandem duplications of 1–6 bp motifs. They are highly abundant in the noncoding DNA of all eukaryotic genomes studied, covering 1–3% of the human genome, depending on how they are defined (Lander etal. 2001; Subramanian etal. 2003b; fig. 1A). Their repetitive structure allows strand misalignment, which can result in frequent change of length mutations, at rates as high as 10−4–10−3 per generation (reviewed in Ellegren 2004). Repeats shorter than a threshold length of around five copies, in the case of dinucleotide motifs, or four where the repeated motif is longer, are less mutable and polymorphic than longer STRs (Ananda etal. 2013), and are traditionally not referred to as microsatellites. However, no such threshold length has been found by comparative studies of mutation (Leclercq etal. 2010), and no firm definition is established. The common length polymorphism of microsatellites has been utilized very widely for many years as a marker of genetic difference in diverse fields including gene mapping, population genetics and forensics (reviewed in Hodel etal. 2016), and is probably the attribute for which they are best known among geneticists.
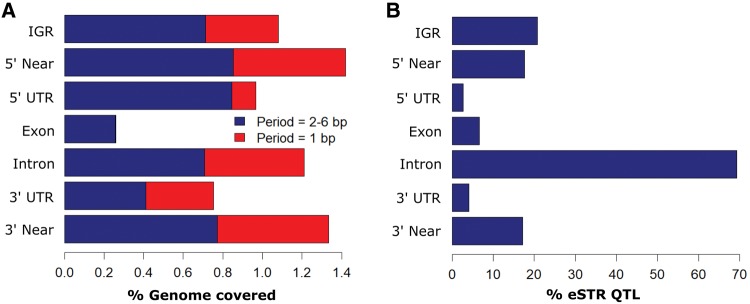
Fig. 1.
—(A) Distribution among human intergenic (IGR), near-genic (within 2 kb of transcription start or end sites), exonic, intronic and untranslated regions of ∼1.4 million microsatellites identified by Willems etal. (2014). Minimum length thresholds were 12 repeat copies for mononucleotide runs, 6 for dinucleotides and 4 for 3–6 bp repeat periods. (B) Distribution of microsatellites (periodicity 2–6 bp) with length variants showing significant effects on transcription (eSTRs) in lymphoblastoid cell lines (Gymrek etal. 2016).
Microsatellites are also well-known for their causative roles in as many as 40 neurological diseases including Huntington’s Disease, Friedreich's Ataxia (FRDA), several of the Spinocelebellar Ataxias (SCA), Fragile X syndrome (FRAXA), and Myotonic Dystrophy types 1 and 2 (DM1 and 2) (reviewed in Pearson etal. 2005; Gatchel and Zoghbi 2005; Groh etal. 2014). In many of these diseases, radical expansions of trinucleotide microsatellites are pathogenic. These mutations begin at threshold levels of around 35–40 repeats and can reach hundreds of copies in affected cells (Pearson etal. 2005). Toxicity often results from hyper-expanded polyglutamine tracts translated from exonic microsatellites, but repeats do not have to encode protein to exert pathogenic effects (Gatchel and Zoghbi 2005). In FRAXA, transcription of the FMR1 gene is silenced in alleles with 200+ copies of its 5′ untranslated region (UTR) CGG repeat due to DNA–RNA hybridization between the repeat in mRNA and the gene itself (Colak etal. 2014). Interestingly, individuals with “preexpansion” 55–200 copy alleles show increased transcription of the gene (Tassone etal. 2007). Reduction of gene expression occurs by a different mechanism in FRDA, in which progression of transcription is inhibited due to a secondary structure formed by the microsatellite, in conjunction with epigenetic modifications (Punga and Buhler 2010; Sakamoto etal. 1999). Another major pathogenic mechanism in microsatellite disease is disruption of splicing (Groh etal. 2014). Several diseases including DM1 and at least two SCAs involve global splicing misregulation due to sequestration of RNA binding proteins by expanded repeats (Echeverria and Cooper 2012; Galka-Marciniak etal. 2012). These effects have also been seen locally, for example, the toxic truncated N-terminal fragment of mutant HTT protein in Huntington’s Disease is generated by CAG repeat length-dependent missplicing (Sathasivam etal. 2013), and the expanded GAA microsatellite associated with FRDA has been shown to affect the splicing efficiency of its gene in model systems (Baralle etal. 2008; Shishkin etal. 2009).
A substantial body of evidence now indicates that many of the transcriptional and RNA-level effects of disease-causing microsatellites are not unique to disease, but instead represent aberrant manifestations of normal microsatellite function. At present, the best known aspect of this is the potential of microsatellites in upstream promoter regions to modulate gene expression levels (reviewed in Sawaya etal. 2012; Press etal. 2014). Scattered examples have been known for many years, and several are now well-replicated. One of the most studied is an (AC)17–39 repeat in the promoter region of the HO-1 gene, polymorphisms of which are associated with cardio-vascular disease, cancer, preeclampsia and Parkinson’s disease, reflecting the antioxidant, anti-inflammatory activities of the HO-1 enzyme (Daenen etal. 2016; Chen etal. 2002; Zhang etal. 2014; Ayuso etal. 2014; Kaartokallio etal. 2014). Others include a (CCTTT)8–17 polymorphism in the promoter of the NOS2 gene, which modifies risk of hypertension and several other conditions including psoriasis (Baloira Villar etal. 2014; Chang etal. 2015; Ryk etal. 2014), and a series of repeats in the AVPR1A gene’s promoter region, which have been associated with social behavior in voles, mice and humans (Donaldson and Young 2013; Hammock and Young 2005; Wang etal. 2016; Walum etal. 2008). One of the most notable examples from a medical standpoint is an A(TA)6–7 TAA polymorphism in the promoter (TATAA box) of the bilirubin UDP-glucuronosyltransferase 1 gene. Individuals with Gilbert’s syndrome are homozygous for the longer allele, which is associated with reduced gene expression (Bosma etal. 1995). It also has major effects on metabolism of the anticancer drug irinotecan (Hoskins etal. 2007). Other well-studied examples of promoter-associated microsatellites are reviewed elsewhere (Sawaya etal. 2012). While promoter loci have been given the most attention to date, single-gene studies have also identified expression-altering microsatellite variants in introns (Zhang etal. 2009; Zakieh etal. 2013; Li etal. 2013; Agarwal etal. 2000; Gebhardt etal. 1999), and UTRs (Chen etal. 2007; Gau etal. 2011; Nagalingam etal. 2014; Galindo etal. 2011; Balasubramaniam etal. 2013; Kumar and Bhatia 2016).
Demonstrated examples of gene expression modulation by microsatellite polymorphism remain isolated at present, but evidence has recently emerged that the phenomenon is widespread in the human genome. Studies of expression quantitative trait loci (eQTL) have shown that a substantial proportion of the heritability of human gene expression levels attributable to common variants in cis is due to STR polymorphism (Gymrek etal. 2016; Quilez etal. 2016). This contribution has likely gone largely unaccounted for in genome-wide association studies (GWAS) because the frequency and diversity of microsatellite polymorphism are much higher than those of single nucleotide polymorphism (SNP) (Willems etal. 2014; Quilez etal. 2016; Gymrek 2017).
Normal microsatellite polymorphism has also been linked to alternative splicing. In 2003, Hui and colleagues showed that splicing efficiency of transcripts from the eNOS gene in minigene constructs depended on the length and sequence of an intronic (CA)19–38 repeat (Hui etal. 2003). The same authors later reported that CA microsatellites in the introns of several other genes could act as splicing enhancers or suppressors (Hui etal. 2005). Other examples include modification of pathogenic splicing in cystic fibrosis by a (CA)9–13 repeat in the CFTR gene (Cuppens etal. 1998).
Adjustment of transcriptional frequency and mRNA splicing are by no means the only aspects of genomic regulation for which the unique properties of microsatellites have been harnessed. Evidence has indicated roles in modulating mRNA stability (Chen etal. 2007), selection of transcription start and termination sites (Kramer etal. 2013; Tseng etal. 2013), enhancer function (Kumar etal. 2013; Gebhardt etal. 1999; Gymrek etal. 2016), nucleosome positioning and modification (Iyer and Struhl 1995; Liu etal. 2006; Zhao etal. 2015; Gymrek etal. 2016; Quilez etal. 2016), higher order chromatin structure (Pathak etal. 2013; McNeil etal. 2006; Subramanian etal. 2003a), noncoding RNAs (ncRNAs) (Amiteye etal. 2013; Zheng etal. 2010), and meiotic recombination hot spots (Gendrel etal. 2000; Kirkpatrick etal. 1999; Choi etal. 2013). Also notable is the surprising importance of exonic microsatellites. These are often highly conserved and are more common than expected in view of their potential to disrupt gene function (Schaper etal. 2014; Loire etal. 2013; Gymrek etal. 2017). They mostly consist of trinucleotide repeats, which have functional roles encoding runs of particular amino acids. Variation in these repeats has been associated with diverse phenotypic changes including skeletal morphology in dogs and receptor protein levels in humans (Fondon and Garner 2004; Brockschmidt etal. 2007). Interestingly, some exonic dinucleotide microsatellites are also maintained, which is mysterious given that their length-changes are expected to cause frameshift mutations in downstream coding sequence (Haasl and Payseur 2014). Indeed, it seems reasonable to speculate that this potential for frameshifts may underlie the propensity of human DNA polymerases to avoid causing mutations that remove interruptions to microsatellites, at least in the case of poly-A repeats (Ananda etal. 2014), although regulatory frameshifting has been described (reviewed in Ketteler 2012; Moxon etal. 2006). As these observations suggest, current understanding of microsatellite biology in general remains limited. However, while the number of microsatellites with demonstrated function remains very small relative to their overall abundance, some functional mechanisms have been described in detail, and several themes are evident (table 1).
Table 1.
Biological Processes Influenced by Microsatellites in Healthy Cells
| Process | Gene (Organism) | Repeat Motif | Ref. |
|---|---|---|---|
| Binding of transcription factors to microsatellite DNA | SLC11A1 (human) | GT (imperfect) | Bayele etal. 2007, |
| ECE-1c (human) | CA (imperfect) | Taka etal. 2013 | |
| TH (human) | TACT | Li etal. 2012 | |
| PIG3 (human) | TGYCC | Albanese etal. 2001 | |
| nadA (N. meningitidis) | TAAA | Contente etal. 2002 | |
| Martin etal. 2005 | |||
| Spacing between promoter elements | GP91-PHOX (human) | CA | Uhlemann etal. 2004 |
| IGF1 (human) | CA | Chen etal. 2016 | |
| Long-range interactions | Intergenic (Drosophila & human) | GATA | Kumar etal. 2013 |
| Transcription start site selection | HO-1 (human) | AC | Kramer etal. 2013 |
| ECE-1c (human) | CA (imperfect) | Li etal. 2012 | |
| Transcription end site selection | ASS1 (human) | GT | Tseng etal. 2013 |
| RNA half-life | FGF9 (human) | TG (imperfect) | Chen etal. 2007 |
| Alternative splicing | APOA2 (human) | GT | Cuppens etal. 1998 |
| CFTR (human) | TG | Hefferon etal. 2004 | |
| eNOS (human) | CA | Hui etal. 2003 | |
| Various (human) | CA | Hui etal. 2005 | |
| Nucleosome packaging | HIS3 (S. cerevisiae) | A | Iyer and Struhl 1995 |
| CSF1 (human) | TG | Liu etal. 2001, Liu etal. 2006 | |
| CYC1 (S. cerevsiae) | CG | Wong etal. 2007 | |
| Genomic (human) | BAA | Zhao etal. 2015 | |
| Histone modification | Genomic (human) | Various | Gymrek etal. 2016 |
| Methylation | Genomic (human & chimpanzee) | CG | Fukuda etal. 2013 |
| Genomic (human) | CG | Quilez etal. 2016 | |
| Noncoding RNA function | Genomic (Drosophila) | AAGAG | Pathak etal. 2013 |
| Genomic (mammals) | GAA | Zheng etal. 2010 | |
| Meiotic recombination | ARG4 (S. cerevisiae)HIS4 (S. cerevisiae) | TGCCGNN | Gendrel etal. 2000, Kirkpatrick etal. 1999 |
| Genomic (A. thaliana) | CCT & CCN | Choi etal. 2013, Shilo etal. 2015 |
Note.—Studies Only Considering Low-Copy STRs Are Not Included
Transcription Factor Binding
Modulation of transcription factor binding by microsatellite length changes may seem the most obvious explanation for STR eQTL, but demonstrated examples of this are quite rare. The GAGA factor, which binds to short GA repeats and modulates chromatin structure, is well-known for its involvement in a significant class of promoters (Adkins etal. 2006; Valipour etal. 2013; Fuda etal. 2015). However, while a small number of long GA microsatellites capable of modulating gene expression in reporter plasmids can be found in promoters (Valipour etal. 2013), functional repeats bound by the GAGA factor are mostly shorter than five copies (Omelina etal. 2011; van Steensel etal. 2003). Few experimental studies have demonstrated direct transcription factor binding to microsatellites as normally defined. Some of the earliest evidence were reported in 2001, for a (TACT)5–10 repeat in the first intron of the human TH gene (Albanese etal. 2001). This microsatellite binds the transcription factor HBP1 and the zinc finger protein ZNF191, and exerts a copy number-dependent silencing effect on the gene. In contrast, another early study showed a stimulatory effect on transcription by a (TGYCC)10–17 repeat located between the positions +451 and +517 of the gene PIG3 (Contente etal. 2002). This study used a variety of methods to show that binding of the microsatellite by the tumor suppressor protein p53 was necessary and sufficient for transcription, the frequency of which correlated with repeat copy number. Evidence for the evolutionary conservation of this functional mechanism has been seen in Neisseria meningitidis. Phase-variable expression of the nadA virulence gene of this pathogenic bacterium is regulated at the transcriptional level by a (TAAA)4–12 promoter microsatellite, which is bound by the transcription factor IHF in a copy number-dependent manner (Martin etal. 2005).
The link with transcription factor binding is more complex in the case of a (GT)5AC(GT)5AC(GT)9–10 microsatellite in the proximal promoter of the gene SLC11A1 (also known as NRAMP1), where gain or loss of a single AC repeat copy can cause a several-fold change in transcription level in reporter plasmid assays (Searle and Blackwell 1999). The functional role of the microsatellite is partly due to two dinucleotide insertions interrupting the repeated motif, which create binding sites for the hypoxia-inducible protein HIF-1 (Bayele etal. 2007). Changes in repeat copy number are associated with the promoter’s response to binding of the transcription factor ATF-3 at an adjacent site (Taka etal. 2013). As outlined below, a possible mechanism for this is microsatellite-mediated disruption of local nucleosomes (fig. 2). A [CA]6[CpG]14–24[CA]30–50 compound-repeat in the ECE-1c gene’s promoter, which is associated with Alzheimer’s disease, also has an unusual relationship with transcription factors. Similarly to the SLC11A1 locus, luciferase assays showed that one particular allele of this microsatellite causes substantially higher levels of ECE-1c expression than any of the other alleles (Li etal. 2012). This effect was linked to binding of the transcription factors SFPQ and PARP-1.
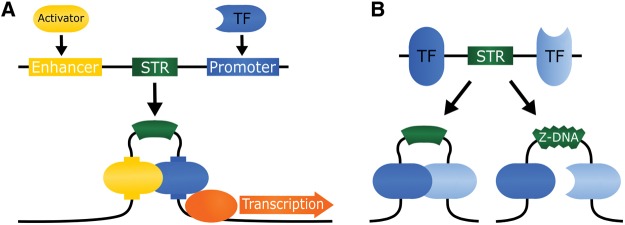
Fig. 2.
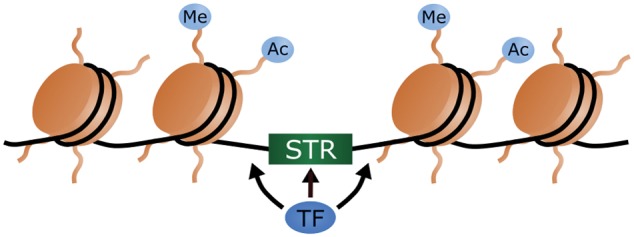
—Several types of microsatellite have been shown to resist packaging into nucleosomes, facilitating the binding of transcription factors (TF) and other proteins to nearby DNA. This sometimes involves induced nonB-DNA structure formation by the microsatellite. TFs also bind directly to some microsatellites. Additionally, STR eQTLs have been associated with epigenetic chemical modifications of nucleosomes, including regulatory methylation and acetylation of histone proteins.
In addition to experimentally verified examples, it has been observed that promoter-associated STR eQTL significantly overlap with known transcription factor binding sites (Quilez etal. 2016). Also consistent with the hypothesis of a common role for transcription factors in mediating microsatellite function are observations of tissue- and cell-type-specific effects of polymorphic microsatellites on target gene expression (Chen etal. 2007; Chiba-Falek and Nussbaum 2001; Albanese etal. 2001; Borrmann etal. 2003). However, it is notable that known examples of transcription factor binding to microsatellites are mostly limited to repeats of longer periodicity and lower uniformity, and because these are less mutable than perfect repeats of short motifs (reviewed in Ellegren 2004), the potential magnitude of their contribution to phenotypic variation is correspondingly lower.
Spacing between Regulatory Elements
The above-mentioned studies show that the direction of correlation between microsatellite length and transcriptional frequency is context-dependent, and maximal activity is sometimes seen for alleles of intermediate length (Morris etal. 2010; Li etal. 2012; Contente etal. 2002). In view of the importance of maintaining particular distances between promoter elements in many contexts (Vardhanabhuti etal. 2007), these observations suggest that the potential of microsatellites to modulate these distances may be underappreciated. At least two studies are found suggestive evidence consistent with the concept. Copy number of a (CA)17–21 microsatellite in the promoter of the IGF1 gene correlates inversely with transcription, but this effect is only seen in the presence of a flanking SNP haplotype (Chen etal. 2016). The haplotype provides a binding site for CCAAT/enhancer-binding-protein δ (C/EBPD), which is essential for the eSTR activity of the microsatellite, and the transcription factor FOXA3 may also be involved (Chen etal. 2016). Other work suggests the possible involvement of DNA looping in at least some distal interactions interposed by microsatellites (fig. 3). The GP91-PHOX gene’s promoter contains a (TA)11–26 repeat, the copy number of which correlates with NADPH-oxidase activity (Uhlemann etal. 2004). The correlation shows regular periodicity, with around five repeat copies between each of three observed maxima. This distance coincides with the approximate length of one helical turn, and similar periodic correlations have been seen at loci where looping is known to occur between two promoter elements either side of a sequence of variable length (Lewis and Adhya 2002; Perez etal. 2000). Also consistent with a function in modulating spacing between functional elements, comparative work has revealed that microsatellites show clear length-specific as well as motif-specific enrichment, with several trends conserved among species (Ramamoorthy etal. 2014).
Fig. 3.
—Microsatellite change of length mutations may act by altering distances between flanking protein binding sites. Observations of high DNA flexibility at microsatellite sequences, and of periodicity approximating helical turn length in a correlation between microsatellite copy number and transcription, suggest the potential for looping to mediate this mechanism where sufficient distance exists between interacting elements, although closer interactions could occur without looping. Two possible scenarios are binding between an enhancer and a transcription factor flanking a microsatellite (TF; A), and between two TFs (B). Adoption of a nonB-DNA structure such as Z-DNA by the microsatellite could also play a role, because such structures absorb supercoiling energy and should thereby reduce the bending potential of surrounding DNA (B).
Links to Enhancer Function
Despite intronic microsatellites being very common among known STR eQTLs (Gymrek etal. 2016; fig. 1B), mechanisms underlying their effects on gene expression (Agarwal etal. 2000; Zakieh etal. 2013) have been studied relatively little in comparison to the promoter-associated loci discussed above. A notable exception is the breast cancer-associated CA14–21 repeat in intron 1 of the EGFR gene, which inhibits transcription by as much as 5-fold at higher copy numbers both invitro and invivo, although other regulatory mechanisms can suppress the effect (Gebhardt etal. 1999, 2000; Buerger etal. 2004). This microsatellite is located between two enhancer elements, one upstream of the promoter and one downstream in intron 1, the activity of which depends on presence of the upstream element (Maekawa etal. 1989). Analysis of the curvature of the repeat DNA and its flanking sequences, based on trinucleotide bending propensity parameters deduced from DNase I digestion data, suggested that the region was highly bendable, and more so at higher repeat copy numbers (Gabrielian etal. 1996). This led to the proposal that the microsatellite could influence interaction between flanking regulatory elements (Gebhardt etal. 1999). The propensity of some microsatellites to form Z-DNA or other structural variants could contribute to such interactions, since while DNA looping may normally require binding of architectural proteins (reviewed in Olson etal. 2013), nonB-DNA structures are expected to modify the process by relieving the torsional tension of nearby DNA, increasing the energy required for it to bend (Benham etal. 2010; Mogil etal. 2016; fig. 3).
Additional evidence consistent with a distal role for some microsatellites in modulating enhancer activity was reported by a study showing that GATA repeats can block interaction between enhancers and promoters invivo (Kumar etal. 2013). A more general role in mediating distal interactions was suggested by a study of long range contacts revealed by 5C experiments, which showed enrichment of low-copy STRs in interacting sequences (Nikumbh and Pfeifer 2017). Low-copy STRs have also been shown to act as functional components of enhancers. A computational analysis in Drosophila identified 2–4 copy repeats of CA, GA, CG, and GATA among the most enriched and discriminative enhancer motifs, and went on to demonstrate that insertion of these elements into nonfunctional sequence could generate enhancer activity (Yanez-Cuna etal. 2014). Supporting a similar role for longer microsatellites, a genome-wide study found enrichment of STR eQTL variants near the enhancer histone mark H3K27ac (Gymrek etal. 2016).
Effects on Alternative Splicing
The best-described examples of functional intronic microsatellites exert their effects at the level of alternative splicing. Perhaps the first reported example of this outside the context of trinucleotide expansion disease was a (GT)16 repeat in the 3′ splice site of the human APOA2 gene’s second intron. This replaces the poly-pyrimidine tract known to be common at 3′ splice sites, and efficient splicing was found to depend on the number of GT repeats present (Shelley and Baralle 1987). Some years later, work on the CFTR gene involved in cystic fibrosis revealed a similar effect for a TG microsatellite located near the exon 9 splice acceptor site. A pathogenic T5 variant at this splice site is associated with exon skipping and disease (Chu etal. 1993), and its effect is modified by the length of an adjacent (TG)9–13 repeat (Cuppens etal. 1998). Several mechanisms have been proposed to explain this (Cuppens etal. 1998; Hefferon etal. 2004; Groman etal. 2004; Zuccato etal. 2004; Buratti etal. 2001). Involvement of the protein TDP-43 was indicated by a study showing that it binds to the microsatellite, and that presence of misspliced mRNA without exon 9 correlates with the expression level of the protein (Buratti etal. 2001). Other evidence has implicated TIA-1 protein in the process (Zuccato etal. 2004). However, based on a study of the effects of replacing the microsatellite with various sequences of similar length, RNA secondary structure may play a role (Hefferon etal. 2004). This study showed greater splicing efficiency in the proximity of sequences with the potential to form RNA hairpins, and two other observations pointed to the importance of this property. Firstly, differences between substituted dinucleotide repeats of various motifs were far greater than differences between (TG)8 and (TG)12, suggesting that the link between microsatellite copy number and splicing efficiency at the locus doesn’t primarily relate to relative positioning of adjacent elements. Second, a similar copy-number dependent suppressive effect to that of poly-TG was shown for a poly-TA substitute, also arguing against binding of sequence-specific splicing effector proteins as the main functional mechanism. Interestingly, splicing was most efficient for sequences predicted to form low-stability hairpins, suggesting transient structure formation (Hefferon etal. 2004). In this context, it is notable that intronic G-qudruplex structures have also been shown to modulate alternative splicing (Ribeiro etal. 2015; Didiot etal. 2008). RNA structures in general may act by causing substantial changes to the distances between elements of the splicing process, or by impeding the progression of RNA polymerase, changing the time-window for splicing regulatory sequences to be recognized (Nieto Moreno etal. 2015).
In contrast, support for protein binding as a primary mediator of microsatellites’ effects on splicing has been seen in the eNOS gene. Copy number of a (CA)19–38 repeat near the 5′ splice site of the gene’s 13th intron correlates with the efficiency with which this intron is excised, and this splicing enhancer activity depends on the RNA-binding protein hnRNP L (Hui etal. 2003). Poly-CA is not structurally equivalent to poly-TG in RNA, and doesn’t form hairpins (Hefferon etal. 2004). Poly-CA microsatellites have also been shown to enhance splicing when inserted at various alternative intronic positions, and generation of cryptic splice sites has been demonstrated in some cases (Hui etal. 2005). Investigating the prevalence of this phenomenon, one study identified several hundred AC microsatellites located close to alternatively spliced exons in the human genome, and performed experimental validation for four of these, demonstrating splice-enhancer effects for two, and suppressive effects by the other two (Hui etal. 2005). Position relative to the splice site was suggested as a potential determinant of positive or negative regulation.
Distinct Functions Observed for UTR Microsatellites
Several UTR microsatellite polymorphisms have been shown to modulate gene expression (Chen etal. 2007; Kumar and Bhatia 2016; Joshi-Saha and Reddy 2015). Like intronic microsatellites the mechanisms underlying their activity haven’t been investigated to the same degree as some promoter-associated loci, but some interesting distinct mechanistic details have emerged. Perhaps the most notable example to date is the complex (TG)3TA(TG)13–16TA(TG)3 microsatellite in the 3′ UTR of the FGF9 gene. One study showed that effects of polymorphism in this repeat on transcription depend on its orientation as well as its position, and are cell-type specific (Chen etal. 2007). The authors of this study noted the microsatellite’s capacity to form hairpin structures and tested its effects on mRNA stability, showing half-life differences of >50% between alleles differing by only one repeat copy. A later study showed binding of the same microsatellite in mRNA by the protein FUBP3, which was associated regulation at the level of translation, though the mechanism for this was not explored (Gau etal. 2011). In contrast, a (TC)8–21 polymorphism in the 5′ UTR of the Tdc gene in Catharanthus roseus was shown to have no effect on translation, but to modulate rate of transcription—an advance on the usually measured parameter transcript abundance, which doesn’t distinguish between effects on transcription and effects on mRNA half-life (Kumar and Bhatia 2016). It seems likely that intronic and exonic microsatellites could also affect mRNA half-life, but to date demonstrated examples of this are lacking.
Microsatellites in UTRs can also influence the location of transcription initiation and termination sites. The human HO-1 gene utilizes several alternative transcription start sites (TSS) downstream of the canonical start codon of its first exon, and the relative abundance of these isoforms correlates with the length of a well-studied poly-AC promoter microsatellite (Kramer etal. 2013). In the ECE-1c gene’s promoter an alternative TSS has been observed within a [CA]6[CpG]14–24[CA]30–50 compound repeat, and this is mediated by binding of the PARP-1 protein to the microsatellite (Kraus and Lis 2003; Li etal. 2012). Observations that STRs are strikingly more common, and also more conserved, close to TSS’s suggest that this may be a common phenomenon (Sawaya etal. 2013). Some evidence also indicates a role for microsatellites in determining the 3′ ends of transcripts. A (GT)14–25 repeat in the 3′ UTR of the human ASS1 gene serves as the poly(A)-downstream GU-rich element to modulate mRNA 3′-end formation, with repeat copy number correlating with the relative abundance of two alternative termination sites (Tseng etal. 2013).
NonB-DNA Structure Formation
In some cases the functional roles of microsatellites have been linked to their potential to adopt nonB-DNA structures. These include several well-described conformations which are energetically less favorable than normal B-form DNA, but inducible by torsional stress (reviewed by Mirkin 2008; fig. 4). The earliest of these to be discovered was Z-DNA. Named for its characteristic zig-zag sugar-phosphate backbone, the left-handed Z-DNA helix is most readily taken up by sequences in which purine and pyrimidine nucleotides alternate, including poly-AC and poly-CG of moderate length (Wang etal. 1979; Wong etal. 2007; Liu etal. 2006). Given that AC is the most commonly repeated motif among mammalian microsatellites, other than mononucleotide arrays (Ellegren 2004), Z-DNA may be the conformational variant most relevant to microsatellite function in mammals. It binds several different proteins (Rich and Zhang 2003; Wang and Vasquez 2007), and can affect gene expression when present in promoter regions (Rothenburg etal. 2001; Wong etal. 2007; Zhang etal. 2006; Liu etal. 2006; Oh etal. 2002).
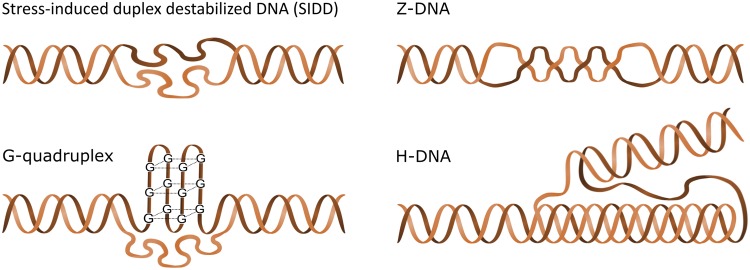
Fig. 4.
—Unusual structural conformations adopted by microsatellites. A/T-rich sequences can form SIDD, Z-DNA can be formed by poly-AC and poly-CG repeats, sequences with four closely spaced stretches of multiple guanine residues can fold into G-quadruplexes, and H-DNA can be adopted by poly-purine sequences with mirror symmetry including poly-AG.
Although they also consist of alternating purines and pyrimidines, Poly-AT microsatellites have much lower Z-DNA forming potential than poly-AC or poly-CG (Ho etal. 1986). However, their low base-pairing stability facilitates formation of cruciform or stress-induced duplex destabilized DNA (SIDD) structures, depending on conditions (Aranda etal. 1997). While cruciforms may be absent from chromosomal DNA invivo, SIDD is quite prevalent (Kouzine etal. 2017), and evidence from Saccharomyces cerevisiae suggests that it may function to relieve the positive supercoiling generated ahead of processing RNA polymerase complexes, potentially also helping to terminate transcription (Benham 1996; Zaret and Sherman 1982).
Duplex melting is also required for formation of G-quadruplexes and intramolecular triplexes/H-DNA. G-quadruplexes are fold-back structures wherein a guanine-rich strand self-associates into square-planar guanine tetrads held together by Hoogsteen hydrogen bonds (Sen and Gilbert 1988). They can be adopted by microsatellites with four or more guanine runs, such as (GGGGTT)4, (GGGT)4, and (GGA)4 (Palumbo etal. 2008; Sundquist and Klug 1989; Ogloblina etal. 2015). In H-DNA, one strand joins adjacent duplex DNA in a triple helix via Hoogsteen bonding (Kohwi and Kohwi-Shigematsu 1988; Dayn etal. 1992). It is most favorable for poly-purine/poly-pyrimidine sequences with mirror symmetry and can be formed by microsatellites including poly-GA and poly-GAA (Potaman etal. 2004; Lu etal. 2003). H-DNA and G-quadruplexes have been linked to regulation of transcription, and also RNA biology (reviewed in Weldon etal. 2016; Murat and Balasubramanian 2014; Jain etal. 2008). However, neither of these two structures is prominent among known STR eQTLs (Gymrek etal. 2016; Quilez etal. 2016).
Notably, the requirement of nonB-DNA structures for supercoiling energy may often be provided invivo by processing polymerases, and it has been suggested that these structures may function to regulate supercoiling (reviewed in Kouzine and Levens 2007; van Holde and Zlatanova 1994). Consistent with this, a recent investigation using permanganate footprinting revealed that 9% of computationally predicted nonB-DNA structures genome-wide were associated with single-stranded DNA in activated mouse B cells, but only in the presence of transcription (Kouzine etal. 2017). H-DNA, G4, Z-DNA, and SIDD were all evident, at genome frequencies of 15,000–23,000 each. The prevalence of G-quadruplex conformations genome-wide has also been demonstrated recently using specific antibodies (Hansel-Hertsch etal. 2016).
Modulation of Chromatin Structure
The unusual structural properties of microsatellites can have functionally relevant influence on chromatin structure. This has been known for many years in trinucleotide repeat disease (Volle and Delaney 2012; Wang 2007; Evans-Galea etal. 2013), though recently it is less commonly addressed by studies of functional microsatellites, which have often been limited to plasmid-based validation work. Some of the most informative experiments connecting normal microsatellites to chromatin structure were done on poly-A runs more than twenty years ago. These extremely common repeats have the potential to resist nucleosome formation due to structural stiffness (Nelson etal. 1987), but are often excluded from definitions of microsatellites, and ignored by functional studies, despite evidence that they can stimulate transcription when present in yeast promoter regions (Iyer and Struhl 1995; Struhl 1985; Schlapp and Rodel 1990). A study of an A15–17 array in the promoter of the HIS3 gene in S. cerevisiae showed that its effect on transcription was not caused by direct protein binding, but was instead due binding of the transcription factor Gcn4 at a distance of 10 bp (Iyer and Struhl 1995). Although poly-G has different structural properties (Panyutin etal. 1989), substituting poly-G tracts of similar length produced similar results (Iyer and Struhl 1995). This study revealed that poly-A tracts perturbed chromatin structure over ∼200 bp, making the Gcn4 binding site more accessible, an effect also associated with increased cytosine methylation at the promoter. Overall similarity of micrococcal nuclease cleavage patterns in the presence or absence of poly-A suggested that altered nucleosome phasing or nucleosome-free DNA was not involved, and it was proposed that nucleosomes covering poly-A may be destabilized and less effective in competition with transcription factors (Iyer and Struhl 1995). However, it is notable that mononucleotide repeats don’t always have the effect of reducing nucleosome stability. A 30 bp poly-A tract in the human HGF promoter region is normally associated with a tightly packaged promoter, inaccessible to DNase I digestion, but truncations of the repeat cause loosening of the chromatin structure, modified protein binding and stimulation of the promoter in breast cancer tissue (Ma etal. 2009).
Several studies have shown that packaging into nucleosomes is disfavored for the Z-DNA-forming sequences poly-CG and poly-AC (Garner and Felsenfeld 1987; Wong etal. 2007; Liu etal. 2001). In the case of the poly-AC microsatellite in the promoter of the CSF1 gene, multiple lines of evidence indicate that Z-DNA formation is stimulated by the BRG1 protein and participates in nucleosome disruption, resulting in transcriptional activation (Liu etal. 2006). The authors of this study proposed that the Z-form may function to relieve negative supercoils induced by nucleosome release, and also to resist replacement of the nucleosome, allowing room to assemble transcriptional machinery. Another example of BRG1 operating in conjunction with a functional poly-AC microsatellite to activate transcription occurs in the HO-1 gene’s promoter, where substituting an alternative Z-DNA-forming sequence produces the same results (Jianyong Zhang etal. 2006). Poly-CG, which has higher Z-DNA forming potential than poly-AC (Ho etal. 1986), can also disrupt nucleosomes in promoter regions and cause position-dependent stimulation of transcription (Wong etal. 2007).
The effects of poly-CG on nucleosomes can also be influenced by CpG methylation (Davey etal. 2004), and the potential evolutionary importance of mutations in these repeats has been shown by a study associating them with divergence in methylation and gene expression between humans and chimpanzees (Fukuda etal. 2013). The link between microsatellites and methylation was further explored by a study of human promoter-associated STRs, which showed that 463 out of 4849 repeat polymorphisms tested correlated significantly with CpG methylation levels within 1 kb, though only 8% of these showed significant effects in the same direction in two populations (Quilez etal. 2016). Interestingly, this study found that 96% of promoter microsatellites significantly associated with gene expression also influenced local cytosine methylation status. Many of these microsatellites overlapped with DNase I hypersensitive sites, indicating open chromatin.
Unsurprisingly, in view of the degree to which they disturb normal B-DNA structure, the conformational variants G-quadruplex, H-DNA, and SIDD are also associated with reduced nucleosome occupancy (Ruan and Wang 2008; Hansel-Hertsch etal. 2016; Kouzine etal. 2017). The significance of these structures to nucleosomes genome-wide was recently demonstrated in activated mouse B cells (Kouzine etal. 2017). This study showed mild to severe nucleosome depletion at sequences shown to form Z-DNA, G4, H-DNA, and SIDD in the presence of transcription. It is notable, however, that potential to form a nonB-DNA structure is not always relevant, even in promoter regions. For example, a poly-AG microsatellite in the Hsp26 gene’s promoter in Drosophila can form H-DNA, but this property cannot substitute for binding of the GAGA transcription factor in creating an open chromatin configuration (Lu etal. 2003).
Some evidence suggests that effects of microsatellites on chromatin are commonly mediated by regulatory chemical modifications of histone proteins. A genome-wide study of STR eQTL showed enrichment in peaks of the histone marks H3K4me1, H3K4me2, H3K4me3, H3K27ac, H3K36me3, and H3K9ac, which are associated with regulatory and transcribed regions, and depleted near the H3K27me3 mark, which is associated with repression of gene expression (Gymrek etal. 2016). This study also showed significant correlations between variation in regulatory chromatin modifications and variation in STR eQTL genotypes.
Suggesting one possible mechanism underlying this epigenetic complexity, blockage of DNA replication, which is known in trinucleotide repeat disease, can result in epigenetic disruption (Svikovic and Sale 2016; Khurana and Oberdoerffer 2015; Gadgil etal. 2016). Long CTG microsatellites have been shown to cause replication stalling by folding into hairpin structures (Liu etal. 2013), and microsatellites of moderate length known to form G-quadruplexes or H-DNA, for example TC20 and TTCC9, can block DNA polymerases invitro (Hile and Eckert 2004). Interestingly, the mononucleotide repeat T11 is also able to stall invitro DNA polymerization, perhaps due to DNA bending, which is known to occur adjacent to sequences consisting entirely of adenine or thymine bases without any TA dinucleotides (Hile and Eckert 2008; Hud and Plavec 2003). Z-DNA has been shown to inhibit RNA polymerase (Ditlevson etal. 2008). Replication stalling at abundant secondary structures can cause epigenetic instability invivo in cells lacking certain factors, though the degree of error-proneness inherent in the systems employed to resolve impediments to replication in normal cells is unclear (Guo etal. 2015; Wu and Spies 2016; Sarkies etal. 2010; Schiavone etal. 2016). However, this and other potential explanations of STR eQTL involving nonB-DNA structures lack currently available evidence that minor changes to microsatellite length are commonly expected to affect these structures substantially. This poses a problem because nearly all microsatellite mutations involve only one or two repeat copies (Sun etal. 2012).
Departure from B-form DNA is not always a necessary component of microsatellites’ effects on chromatin. A human genome-wide survey of GAA microsatellites, known for disrupting nucleosomes when present at extreme lengths corresponding those seen in FRDA (Ruan and Wang 2008), showed that abundant (GAA)6–8 repeats were also associated with substantial nearby nucleosome depletion (Zhao etal. 2015). Milder effects were seen at distances of up to 400 bp from the microsatellites. Intriguingly, poly-A tracts were very frequently found near the 5′ ends of these repeats, where they were associated with further reductions in nucleosome occupancy. Suggesting that the low flexibility of the AA dinucleotide base-step (Fujii etal. 2007) rather than H-DNA formation was responsible for these effects, CAA, TAA, and GAA microsatellites all showed similar patterns of nucleosome depletion. Another study found TGGA repeats to be the most significant feature in random sequence with outstandingly low nucleosome formation invitro, and showed that this was not explained by G-quadruplex formation (Cao etal. 1998).
Evidence also suggests that some microsatellites may have a role in modulating higher order chromatin structure. Repeats of the motif GATA are enriched in the sex chromosomes of some organisms, and their frequency distributions in the human and mouse genomes show a striking peak in frequency at 10–12 copies—a pattern not seen for other tetranucleotide repeats of similar composition (Subramanian etal. 2003a). The distribution of these repeats on sex chromosomes suggests a link to chromatin domain boundaries. They are >10-fold enriched throughout the 10-Mb segment of human Xp22 that escapes inactivation (McNeil etal. 2006), and their flanking sequences contain patterns characteristic of nuclear matrix attachment (Subramanian etal. 2003a). Moreover, work in several organisms has shown that they are bound by Bkm-binding protein, which is predominantly expressed in the germ cells of the heterogametic sex, where sex-determining chromosomes are decondensed and transcriptionally active (Singh etal. 1994).
Roles in Regulatory RNA
Given that 50% or more of mammalian DNA is transcribed, and that many noncoding transcripts are functional (reviewed in Mattick and Makunin 2006; Holoch and Moazed 2015), it is unsurprising that microsatellites have acquired functions in ncRNA as well as in mRNA (fig. 5). An example is the RNA component of the nuclear matrix. One study found that 70% of RNA clones isolated from Drosophila nuclear matrix RNA contained AAGAG microsatellites, transcribed from both strands and likely deriving from pericentromeric regions in which AAGAG repeats are predominantly located (Pathak etal. 2013). Knockdown of poly-AAGAG-containing transcripts by RNA interference resulted in late larval/early pupal lethality. Long ncRNAs primarily consisting of GAA repeats have also been shown to associate with the nuclear matrix (Zheng etal. 2010).
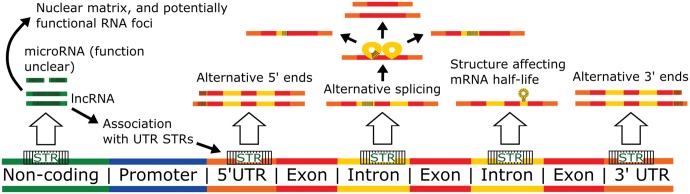
Fig. 5.
—Effects of microsatellites at the level of RNA. Long noncoding RNAs (lncRNAs) predominantly consisting of microsatellites have been observed to function in the nuclear matrix and to aggregate into nuclear foci with indications of functional significance. They have also been shown to associate with DNA microsatellites in UTRs. Microsatellite-dominated microRNAs have been observed, but their function is not yet clear. Intronic microsatellites can modulate splicing efficiency, including exon skipping and splice site selection, and UTR repeats can influence the locations of the sites of transcription initiation and termination. Transcribed microsatellites can also affect mRNA half-life, which may be due to formation of secondary structures such as hairpins.
At present, evidence for other roles in functional ncRNA is sketchy, but some suggestive observations have been made. One study showed that long ncRNAs containing poly-GAA constitute a distinct class of nuclear-retained RNA which forms foci (Zheng etal. 2010). Microsatellite-based RNA foci can be pathogenic in trinucelotide expansion disease (Echeverria and Cooper 2012; Galka-Marciniak etal. 2012), but they may also have regulatory functions. In mouse cell lines GAA-rich lncRNA foci are found in functionally important areas such as the cytokinetic midbody in late-telophase cells, and are redistributed in response to changes in proliferation status. They also associate with genomic GAA repeats, which are enriched near the 5′ and 3′ ends of genes (Zheng etal. 2010).
Another species of ncRNA in which microsatellites may exert some effect is microRNA. The ability of these short 20–24 bp RNAs to regulate transcription has been studied extensively in plants (reviewed in Sunkar and Zhu 2007), and in Boechera it has been observed that microsatellites are prominent features of many of them. Out of 994 microRNAs identified by one study, 673 (67%) predominantly consisted of 2–7 repeats of the trinucleotide motifs GAA, GCA, GGA, GGU, UGA and their compliments, some of which were conserved in Arabidopsis (Amiteye etal. 2013).
Regulation of Meiotic Recombination Hot Spots
In the human and mouse genomes, a proportion of the hot spots in which meiotic recombination events are most frequent are governed by sequence-specific DNA binding proteins such as PRDM9, in concert with epigenetic processes (reviewed in Paigen and Petkov 2010). However, in organisms which lack a functional PRDM9 system, the basis in sequence of hot spot determination is less clear. Hot spots in these species are often found in gene promoters, and in S. cereivisiae it has been demonstrated that they can be affected by the presence of microsatellites including telomeric sequence (G1–3T1)n (White etal. 1993), and poly-AC (Gendrel etal. 2000). The latter study showed inhibition of strand exchange and stimulation of double crossover at the microsatellite. Like transcription, recombination hot spots require an opening of the chromatin structure, and a (CCGNN)12–48 repeat, shown to resist nucleosomes invitro, can also modulate hot spot activity in S. cerevisiae (Kirkpatrick etal. 1999). Hot spots often contain GC-rich and repetitive sequence naturally, an observation which has led to the additional suggestion of replication pausing mediated by epigenetic marks as a determining mechanism, in view of experiments in yeast demonstrating coupling of replication and meiotic double-strand break formation (Borde etal. 2000; Petes 2001; Bagshaw etal. 2008).
More recently, microsatellites have been implicated in plant recombination hot spots (reviewed in Choi and Henderson 2015). One study showed that A-rich and (CTT)2–7 repeat sequences are the most common hot spot-associated motifs in Arabidopsis (Choi etal. 2013). A-rich elements are predominantly found just upstream of hot spot promoter TSSs, overlapping with regions of nucleosome depletion, and CTT repeats are located just downstream of these TSSs, coinciding with crossover peaks. Underlying mechanisms were not explored by this study. As mentioned above, these two repeat types are often found in close proximity in the human genome, and both have been linked with nucleosome depletion (Iyer and Struhl 1995; Zhao etal. 2015). However, hot spot-associated CTT repeats in Arabidopsis are associated with peaks in H2A.Z nucleosome occupancy, and with the H3K4me3 histone modification, which has been linked to recombination and transcription (Choi etal. 2013; Shilo etal. 2015). Arabidopsis hot spots are also enriched for repeats of the motif CCN. These show similar patterns of distribution to CTT repeats with respect to nucleosomes (Shilo etal. 2015).
Prevalence of Functional Microsatellites
Several additional lines of evidence suggest that functional microsatellites are more prevalent than has traditionally been appreciated. Most prominent is recent work associating STR genotypes with transcript abundance genome-wide. One study in lymphoblastoid cell lines identified 2060 significant eQTL STRs, contributing 10–15% to the heritability of human gene expression levels attributable to common variants in cis (Gymrek etal. 2016). As this study was limited to linear correlations between repeat copy number and expression level, it presumably underestimated nonlinear effects, which are likely to be common in view of evidence detailed above that microsatellite alleles of intermediate length often show the most positive associations with transcription. Surprisingly, 69% of the eQTL STRs were in introns, and only 17.7% were in upstream promoters, with 20.8% located >5 kb from any known gene (fig. 1B). A similar study of 4,849 promoter-associated microsatellites found 183 significantly associated with nearby gene expression, but only 5% of these showed significant effects in the same direction in two populations (Quilez etal. 2016). The motif-group most frequently seen was AC, though the most overrepresented motifs in both studies included A-rich tri and tetranucleotide repeats such as AAC and AAAC, which are not known to form nonB-DNA structures.
Promoter microsatellites may be more influential in yeast, where correlations between interstrain divergence in gene expression levels and microsatellite variation showed significant effects at around 25% of promoters (Vinces etal. 2009). Several other comparative studies have indicated widespread microsatellite function. It has been known for many years that some microsatellites, including noncoding repeats, have remained conserved between species across hundreds of millions of years (FitzSimmons etal. 1995; Zhang etal. 2006), and recent studies taking advantage of the wealth of genome sequences made available because the advent of next generation sequencing technology have revealed evolutionary conservation at large numbers of loci, although conservation decays exponentially with phylogenetic distance at many (Buschiazzo and Gemmell 2010; Sawaya etal. 2012). Conservation has been found to be highest for microsatellites in UTR and coding regions, and emergence of new microsatellites happens most often in these areas (Sawaya etal. 2012). Some repeat motifs are more conserved than others, notably AC. The most conserved locations tend to be near TSS, even when 5′ UTR loci are not considered (Sawaya etal. 2012). Supporting the functional significance of conserved microsatellites, a recent comparison of primate genomes revealed that genes with orthologous microsatellites in upstream or transcribed regions consistently show elevated interspecies divergence in gene expression levels across various tissue types (Bilgin Sonay, Carvalho, etal. 2015).
It is notable in this context that microsatellite function is not necessarily limited to highly conserved loci. Evidence suggesting the potential importance of primate-specific repeats includes exceptional expansion or contraction in the primate lineage of core promoter microsatellites in several genes related to neuronal and craniofacial development (Namdar-Aligoodarzi etal. 2015; Ohadi etal. 2015). The potential significance of variation in microsatellites to brain development is also indicated by their enrichment and conservation in genes connected with neurological and other developmental systems, and by the large number of microsatellite-phenotype associations reported for such genes (Bolton etal. 2013; Fondon etal. 2008; Nithianantharajah and Hannan 2007; Sawaya etal. 2012).
Additional support for prevalent microsatellite function has been gathered through interspecies comparisons of their distribution relative to other genomic elements. In maize, for example, microsatellite densities were found to be highest in 5′ UTR, followed by 3′ UTR, promoter, intronic, intergenic, and protein coding regions (Qu and Liu 2013). A study of 29 land plant species also found the highest densities in 5′ UTRs, followed by promoters, while in two algal species, densities were highest in introns and coding regions respectively, with intronic microsatellites concentrated near intron–exon boundaries (Zhao etal. 2014).
Future Perspectives
Genome-wide studies are likely to continue identifying functional microsatellites. Until recently, significant obstacles to the incorporation of large numbers of repeat loci into GWAS in humans included practical difficulties with large scale genotyping, problems with mapping short sequence reads, and statistical hypothesis testing issues generated by multiple alleles per locus—but these are now being alleviated through theoretical and technological developments, including less expensive, longer read sequencing (Press etal. 2014; Li etal. 2017; Shin etal. 2017; Gymrek 2017). Large numbers of microsatellites have already been directly incorporated into GWAS in Drosophila, for example a study involving three traits, 2.5 million SNPs and 78,000 microsatellites found that the representation of microsatellites among significantly phenotype-associated loci at the level of P < 10−6 was 5.6%, even though they only comprised 3% of markers used (Mackay etal. 2012). Relatively cheaper approaches to identifying functional microsatellites include deep sequencing around existing GWAS hits, and investigation of highly conserved loci associated with genes of interest, both of which have found some success (Grunewald etal. 2015; Bagshaw etal. 2017).
Somatic mutation is another aspect of microsatellite biology made visible by high throughput sequencing. Questions of growing tractability include the degree to which this causes ageing and age-related disease (Bavarva etal. 2014; Kurz etal. 2015), and the possibility that it functions in normal brain development (Nithianantharajah and Hannan 2007). One particularly interesting hypothesis is that microsatellite variation provides a mechanism of rapid adaptation for individual developing neurons, mirroring its potential role at the level of whole organisms (Nithianantharajah and Hannan 2007). Also under investigation are the functional effects of global microsatellite instability associated with some colorectal, gastric and other cancers (Kim and Park 2014; Bilgin Sonay, Koletou, etal. 2015; Hogan etal. 2015).
In conjunction with available genome-wide functional data, high throughput technologies will find additional applications in elucidating microsatellites’ functional mechanisms, for example in the identification of molecular networks affected by expression-altering variants. In view of the population specificity of the effects of promoter-associated STR eQTL, exploration of the genetic backgrounds modifying their effects seems particularly relevant (Quilez etal. 2016). Intronic loci are likely to be another immediate focus, given their unexpected enrichment among STR eQTL (Gymrek etal. 2016), and UTR microsatellites are also of increased current interest in view of the above-mentioned study of gene expression divergence between primates, which revealed that 3′ UTR loci have more influence than those in promoter, exonic, or intronic regions (Bilgin Sonay, Carvalho, etal. 2015). As these examples illustrate, it seems likely that microsatellites, once thought of as generally neutral, retain considerable capacity to surprise genomic investigators with their diverse, pervasive functional significance.
Acknowledgments
Funding for this work was provided by the University of Otago, New Zealand. I thank Kateryna Makova and anonymous reviewers for helpful comments on the manuscript.
Literature Cited
- Adkins NL, Hagerman TA, Georgel P.. 2006. GAGA protein: a multi-faceted transcription factor. Biochem Cell Biol. 84(4):559–567. [DOI] [PubMed] [Google Scholar]
- Agarwal AK, et al. 2000. CA-Repeat polymorphism in intron 1 of HSD11B2 : effects on gene expression and salt sensitivity. Hypertens (Dallas Tex 1979) 36(2):187–194. [DOI] [PubMed] [Google Scholar]
- Albanese V, et al. 2001. Quantitative effects on gene silencing by allelic variation at a tetranucleotide microsatellite. Hum Mol Genet. 10(17):1785–1792. [DOI] [PubMed] [Google Scholar]
- Amiteye S, et al. 2013. Novel microRNAs and microsatellite-like small RNAs in sexual and apomictic Boechera species. MicroRNA (Shariqah, United Arab Emirates) 2(1):45–62. [PubMed] [Google Scholar]
- Ananda G, et al. 2013. Distinct mutational behaviors differentiate short tandem repeats from microsatellites in the human genome. Genome Biol Evol. 5(3):606–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananda G, et al. 2014. Microsatellite interruptions stabilize primate genomes and exist as population-specific single nucleotide polymorphisms within individual human genomes. PLoS Genet. 10(7):e1004498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda A, Perez-Ortin JE, Benham CJ, Del Olmo ML.. 1997. Analysis of the structure of a natural alternating d(TA)n sequence in yeast chromatin. Yeast 13(4):313–326. [DOI] [PubMed] [Google Scholar]
- Ayuso P, et al. 2014. An association study between Heme oxygenase-1 genetic variants and Parkinson’s disease. Front Cell Neurosci. 8:298.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagshaw ATM, Horwood LJ, Fergusson DM, Gemmell NJ, Kennedy MA.. 2017. Microsatellite polymorphisms associated with human behavioural and psychological phenotypes including a gene-environment interaction. BMC Med Genet. 18(1):12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagshaw ATM, Pitt JPW, Gemmell NJ.. 2008. High frequency of microsatellites in S. cerevisiae meiotic recombination hotspots. BMC Genomics. 9(1):49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramaniam S, Kumar S, Sharma A, Mitra A.. 2013. Microsatellite (GT)n polymorphism at 3′UTR of SLC11A1 influences the expression of brucella LPS induced MCP1 mRNA in buffalo peripheral blood mononuclear cells. Vet Immunol Immunopathol. 152(3–4):295–302. [DOI] [PubMed] [Google Scholar]
- Baloira Villar A, et al. 2014. CCTTT pentanucleotide repeats in inducible nitric oxide synthase gene expression in patients with pulmonary arterial hypertension. Arch Bronconeumol. 50(4):141–145. [DOI] [PubMed] [Google Scholar]
- Baralle M, Pastor T, Bussani E, Pagani F.. 2008. Influence of Friedreich ataxia GAA noncoding repeat expansions on pre-mRNA processing. Am J Hum Genet. 83(1):77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavarva JH, Tae H, McIver L, Karunasena E, Garner HR.. 2014. The dynamic exome: acquired variants as individuals age. Aging (Albany, NY) 6(6):511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayele HK, et al. 2007. HIF-1 regulates heritable variation and allele expression phenotypes of the macrophage immune response gene SLC11A1 from a Z-DNA forming microsatellite. Blood 110(8):3039–3048. [DOI] [PubMed] [Google Scholar]
- Benham CJ. 1996. Duplex destabilization in superhelical DNA is predicted to occur at specific transcriptional regulatory regions. J Mol Biol. 255(3):425–434. [DOI] [PubMed] [Google Scholar]
- Benham CJ, et al. , editors. 2010. Mathematics of DNA structure, function and interactions. New York: Springer Science & Business Media. [Google Scholar]
- Bilgin Sonay T, Carvalho T, et al. 2015. Tandem repeat variation in human and great ape populations and its impact on gene expression divergence. Genome Res. 25(11):1591–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgin Sonay T, Koletou M, Wagner A.. 2015. A survey of tandem repeat instabilities and associated gene expression changes in 35 colorectal cancers. BMC Genomics. 16(1):702.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton KA, et al. 2013. STaRRRT: a table of short tandem repeats in regulatory regions of the human genome. BMC Genomics. 14:795.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V, Goldman AS, Lichten M.. 2000. Direct coupling between meiotic DNA replication and recombination initiation. Science 290(5492):806–809. [DOI] [PubMed] [Google Scholar]
- Borrmann L, Seebeck B, Rogalla P, Bullerdiek J.. 2003. Human HMGA2 promoter is coregulated by a polymorphic dinucleotide (TC)-repeat. Oncogene 22(5):756–760. [DOI] [PubMed] [Google Scholar]
- Bosma PJ, et al. 1995. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert's syndrome. N Engl J Med. 333(18):1171–1175. [DOI] [PubMed] [Google Scholar]
- Brockschmidt FF, Nöthen MM, Hillmer AM.. 2007. The two most common alleles of the coding GGN repeat in the androgen receptor gene cause differences in protein function. J Mol Endocrinol. 39(1):1–8. [DOI] [PubMed] [Google Scholar]
- Buerger H, et al. 2004. Allelic length of a CA dinucleotide repeat in the egfr gene correlates with the frequency of amplifications of this sequence–first results of an inter-ethnic breast cancer study. J Pathol. 203(1):545–550. [DOI] [PubMed] [Google Scholar]
- Buratti E, et al. 2001. Nuclear factor TDP-43 and SR proteins promote invitro and invivo CFTR exon 9 skipping. EMBO J. 20(7):1774–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschiazzo E, Gemmell NJ.. 2010. Conservation of human microsatellites across 450 million years of evolution. Genome Biol Evol. 2:153–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Widlund HR, Simonsson T, Kubista M.. 1998. TGGA repeats impair nucleosome formation. J Mol Biol. 281(2):253–260. [DOI] [PubMed] [Google Scholar]
- Chang Y-C, et al. 2015. The (CCTTT) n pentanucleotide repeat polymorphism in the inducible nitric oxide synthase gene promoter and the risk of psoriasis in Taiwanese. Arch Dermatol Res. 307(5):425–432. [DOI] [PubMed] [Google Scholar]
- Chen HY, et al. 2016. The mechanism of transactivation regulation due to polymorphic short tandem repeats (STRs) using IGF1 promoter as a model. Sci Rep. 6:38225.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T-M, et al. 2007. Microsatellite in the 3′ untranslated region of human fibroblast growth factor 9 (FGF9) gene exhibits pleiotropic effect on modulating FGF9 protein expression. Hum Mutat. 28(1):98. [DOI] [PubMed] [Google Scholar]
- Chen Y-H, et al. 2002. Microsatellite polymorphism in promoter of heme oxygenase-1 gene is associated with susceptibility to coronary artery disease in type 2 diabetic patients. Hum Genet. 111(1):1–8. [DOI] [PubMed] [Google Scholar]
- Chiba-Falek O, Nussbaum RL.. 2001. Effect of allelic variation at the NACP-Rep1 repeat upstream of the alpha-synuclein gene (SNCA) on transcription in a cell culture luciferase reporter system. Hum Mol Genet. 10(26):3101–3109. [DOI] [PubMed] [Google Scholar]
- Choi K, et al. 2013. Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat Genet. 45(11):1327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Henderson IR.. 2015. Meiotic recombination hotspots – a comparative view. Plant J. 83(1):52–61. [DOI] [PubMed] [Google Scholar]
- Chu CS, Trapnell BC, Curristin S, Cutting GR, Crystal RG.. 1993. Genetic basis of variable exon 9 skipping in cystic fibrosis transmembrane conductance regulator mRNA. Nat Genet. 3(2):151–156.7684646 [Google Scholar]
- Colak D, et al. 2014. Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science 343(6174):1002–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contente A, Dittmer A, Koch MC, Roth J, Dobbelstein M.. 2002. A polymorphic microsatellite that mediates induction of PIG3 by p53. Nat Genet. 30(3):315–320. [DOI] [PubMed] [Google Scholar]
- Cuppens H, et al. 1998. Polyvariant mutant cystic fibrosis transmembrane conductance regulator genes. The polymorphic (Tg)m locus explains the partial penetrance of the T5 polymorphism as a disease mutation. J Clin Invest. 101(2):487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daenen KEL, Martens P, Bammens B.. 2016. Association of HO-1 (GT)n promoter polymorphism and cardiovascular disease: a reanalysis of the literature. Can J Cardiol. 32(2):160–168. [DOI] [PubMed] [Google Scholar]
- Davey CS, Pennings S, Reilly C, Meehan RR, Allan J.. 2004. A determining influence for CpG dinucleotides on nucleosome positioning invitro. Nucleic Acids Res. 32(14):4322–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayn A, Samadashwily GM, Mirkin SM.. 1992. Intramolecular DNA triplexes: unusual sequence requirements and influence on DNA polymerization. Proc Natl Acad Sci U S A. 89(23):11406–11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiot M-C, et al. 2008. The G-quartet containing FMRP binding site in FMR1 mRNA is a potent exonic splicing enhancer. Nucleic Acids Res. 36(15):4902–4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditlevson JV, et al. 2008. Inhibitory effect of a short Z-DNA forming sequence on transcription elongation by T7 RNA polymerase. Nucleic Acids Res. 36(10):3163–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson ZR, Young LJ.. 2013. The relative contribution of proximal 5′ flanking sequence and microsatellite variation on brain vasopressin 1a receptor (Avpr1a) gene expression and behavior. PLoS Genet. 9(8):e1003729.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverria GV, Cooper TA.. 2012. RNA-binding proteins in microsatellite expansion disorders: mediators of RNA toxicity. Brain Res. 1462:100–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. 2004. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 5(6):435–445. [DOI] [PubMed] [Google Scholar]
- Evans-Galea MV, Hannan AJ, Carrodus N, Delatycki MB, Saffery R.. 2013. Epigenetic modifications in trinucleotide repeat diseases. Trends Mol Med. 19(11):655–663. [DOI] [PubMed] [Google Scholar]
- FitzSimmons NN, Moritz C, Moore SS.. 1995. Conservation and dynamics of microsatellite loci over 300 million years of marine turtle evolution. Mol Biol Evol. 12(3):432–440. [DOI] [PubMed] [Google Scholar]
- Fondon JW, Garner HR.. 2004. Molecular origins of rapid and continuous morphological evolution.Proc Natl Acad Sci U S A. 101(52):18058–18063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondon JW 3rd, Hammock EAD, Hannan AJ, King DG.. 2008. Simple sequence repeats: genetic modulators of brain function and behavior. Trends Neurosci. 31(7):328–334. [DOI] [PubMed] [Google Scholar]
- Fuda NJ, et al. 2015. GAGA factor maintains nucleosome-free regions and has a role in RNA polymerase II recruitment to promoters. PLoS Genet. 11(3):e1005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Kono H, Takenaka S, Go N, Sarai A.. 2007. Sequence-dependent DNA deformability studied using molecular dynamics simulations. Nucleic Acids Res. 35(18):6063–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K, et al. 2013. Regional DNA methylation differences between humans and chimpanzees are associated with genetic changes, transcriptional divergence and disease genes. J Hum Genet. 58(7):446–454. [DOI] [PubMed] [Google Scholar]
- Gabrielian A, Simoncsits A, Pongor S.. 1996. Distribution of bending propensity in DNA sequences. FEBS Lett. 393(1):124–130. [DOI] [PubMed] [Google Scholar]
- Gadgil R, Barthelemy J, Lewis T, Leffak M.. 2016. Replication stalling and DNA microsatellite instability. Biophys. Chem. doi:10.1016/j.bpc.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo CL, et al. 2011. A long AAAG repeat allele in the 5′ UTR of the ERR-gamma gene is correlated with breast cancer predisposition and drives promoter activity in MCF-7 breast cancer cells. Breast Cancer Res Treat. 130(1):41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galka-Marciniak P, Urbanek MO, Krzyzosiak WJ.. 2012. Triplet repeats in transcripts: structural insights into RNA toxicity. Biol Chem. 393(11):1299–1315. [DOI] [PubMed] [Google Scholar]
- Garner MM, Felsenfeld G.. 1987. Effect of Z-DNA on nucleosome placement. J Mol Biol. 196(3):581–590. [DOI] [PubMed] [Google Scholar]
- Gatchel JR, Zoghbi HY.. 2005. Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet. 6(10):743–755. [DOI] [PubMed] [Google Scholar]
- Gau B-H, Chen T-M, Shih Y-HJ, Sun HS.. 2011. FUBP3 interacts with FGF9 3′ microsatellite and positively regulates FGF9 translation. Nucleic Acids Res. 39(9):3582–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt F, Burger H, Brandt B.. 2000. Modulation of EGFR gene transcription by secondary structures, a polymorphic repetitive sequence and mutations – a link between genetics and epigenetics. Histol Histopathol. 15(3):929–936. [DOI] [PubMed] [Google Scholar]
- Gebhardt F, Zanker KS, Brandt B.. 1999. Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 1. J Biol Chem. 274(19):13176–13180. [DOI] [PubMed] [Google Scholar]
- Gendrel CG, Boulet A, Dutreix M.. 2000. (CA/GT)(n) microsatellites affect homologous recombination during yeast meiosis. Genes Dev. 14(10):1261–1268. [PMC free article] [PubMed] [Google Scholar]
- Groh M, Silva LM, Gromak N.. 2014. Mechanisms of transcriptional dysregulation in repeat expansion disorders. Biochem Soc Trans. 42(4):1123–1128. [DOI] [PubMed] [Google Scholar]
- Groman JD, et al. 2004. Variation in a repeat sequence determines whether a common variant of the cystic fibrosis transmembrane conductance regulator gene is pathogenic or benign. Am J Hum Genet. 74(1):176–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald TGP, et al. 2015. Chimeric EWSR1-FLI1 regulates the Ewing sarcoma susceptibility gene EGR2 via a GGAA microsatellite. Nat Genet. 47(9):1073–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, et al. 2015. A distinct triplex DNA unwinding activity of ChlR1 helicase. J Biol Chem. 290(8):5174–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymrek M. 2017. A genomic view of short tandem repeats. Curr Opin Genet Dev. 44:9–16. [DOI] [PubMed] [Google Scholar]
- Gymrek M, et al. 2016. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat Genet. 48(1):22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymrek M, Willems T, Erlich Y, Reich D.. 2017. A framework to interpret short tandem repeat variation in humans. bioRxiv. doi:10.1101/092734. [DOI] [PMC free article] [PubMed]
- Haasl RJ, Payseur BA.. 2014. Remarkable selective constraints on exonic dinucleotide repeats. Evolution 68(9):2737–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammock EAD, Young LJ.. 2005. Microsatellite instability generates diversity in brain and sociobehavioral traits. Science 308(5728):1630–1634. [DOI] [PubMed] [Google Scholar]
- Hansel-Hertsch R, et al. 2016. G-quadruplex structures mark human regulatory chromatin. Nat Genet. 48(10):1267–1272. [DOI] [PubMed] [Google Scholar]
- Hefferon TW, Groman JD, Yurk CE, Cutting GR.. 2004. A variable dinucleotide repeat in the CFTR gene contributes to phenotype diversity by forming RNA secondary structures that alter splicing. Proc Natl Acad Sci U S A. 101(10):3504–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hile SE, Eckert KA.. 2008. DNA polymerase kappa produces interrupted mutations and displays polar pausing within mononucleotide microsatellite sequences. Nucleic Acids Res. 36(2):688–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hile SE, Eckert KA.. 2004. Positive correlation between DNA polymerase alpha-primase pausing and mutagenesis within polypyrimidine/polypurine microsatellite sequences. J Mol Biol. 335(3):745–759. [DOI] [PubMed] [Google Scholar]
- Ho PS, Ellison MJ, Quigley GJ, Rich A.. 1986. A computer aided thermodynamic approach for predicting the formation of Z-DNA in naturally occurring sequences. EMBO J. 5(10):2737–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodel RGJ, et al. 2016. The report of my death was an exaggeration: a review for researchers using microsatellites in the 21st century. Appl Plant Sci. 4(6):1600025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan J, DeJulius K, Liu X, Coffey JC, Kalady MF.. 2015. Transcriptional profiles underpin microsatellite status and associated features in colon cancer. Gene 570(1):36–43. [DOI] [PubMed] [Google Scholar]
- Holoch D, Moazed D.. 2015. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 16(2):71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins JM, Goldberg RM, Qu P, Ibrahim JG, McLeod HL.. 2007. UGT1A1*28 genotype and irinotecan-induced neutropenia: dose matters. J Natl Cancer Inst. 99(17):1290–1295. [DOI] [PubMed] [Google Scholar]
- Hud NV, Plavec J.. 2003. A unified model for the origin of DNA sequence-directed curvature. Biopolymers 69(1):144–158. [DOI] [PubMed] [Google Scholar]
- Hui J, et al. 2005. Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing. EMBO J. 24(11):1988–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui J, Stangl K, Lane WS, Bindereif A.. 2003. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nat Struct Biol. 10(1):33–37. [DOI] [PubMed] [Google Scholar]
- Iyer V, Struhl K.. 1995. Poly(dA: dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 14(11):2570–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Wang G, Vasquez KM.. 2008. DNA triple helices: biological consequences and therapeutic potential. Biochimie 90(8):1117–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi-Saha A, Reddy KS.. 2015. Repeat length variation in the 5′UTR of myo-inositol monophosphatase gene is related to phytic acid content and contributes to drought tolerance in chickpea (Cicer arietinum L.). J Exp Bot. 66(19):5683–5690. [DOI] [PubMed] [Google Scholar]
- Kaartokallio T, et al. 2014. Microsatellite polymorphism in the heme oxygenase-1 promoter is associated with nonsevere and late-onset preeclampsia. Hypertens (Dallas Tex 1979) 64(1):172–177. doi: 10.1161/HYPERTENSIONAHA.114.03337. [DOI] [PubMed] [Google Scholar]
- Ketteler R. 2012. On programmed ribosomal frameshifting: the alternative proteomes. Front Genet. 3:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S, Oberdoerffer P.. 2015. Replication stress: a lifetime of epigenetic change. Genes (Basel) 6(3):858–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-M, Park PJ.. 2014. A genome-wide view of microsatellite instability: old stories of cancer mutations revisited with new sequencing technologies. Cancer Res. 74(22):6377–6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DT, Wang YH, Dominska M, Griffith JD, Petes TD.. 1999. Control of meiotic recombination and gene expression in yeast by a simple repetitive DNA sequence that excludes nucleosomes. Mol Cell Biol. 19(11):7661–7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi Y, Kohwi-Shigematsu T.. 1988. Magnesium ion-dependent triple-helix structure formed by homopurine-homopyrimidine sequences in supercoiled plasmid DNA. Proc Natl Acad Sci U S A. 85(11):3781–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzine F, et al. 2017. Permanganate/S1 nuclease footprinting reveals non-B DNA structures with regulatory potential across a mammalian genome. Cell Syst. doi: 10.1016/j.cels.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzine F, Levens D.. 2007. Supercoil-driven DNA structures regulate genetic transactions. Front Biosci. 12:4409–4423. [DOI] [PubMed] [Google Scholar]
- Kramer M, et al. 2013. Alternative 5′ untranslated regions are involved in expression regulation of human heme oxygenase-1. PLoS ONE. 8(10):e77224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL, Lis JT.. 2003. PARP goes transcription. Cell 113(6):677–683. [DOI] [PubMed] [Google Scholar]
- Kumar RP, Krishnan J, Pratap Singh N, Singh L, Mishra RK.. 2013. GATA simple sequence repeats function as enhancer blocker boundaries. Nat Commun. 4:1844.. [DOI] [PubMed] [Google Scholar]
- Kumar S, Bhatia S.. 2016. A polymorphic (GA/CT)n- SSR influences promoter activity of Tryptophan decarboxylase gene in Catharanthus roseus L. Don. Sci Rep. 6:33280.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz C, et al. 2015. Coding microsatellite frameshift mutations accumulate in atherosclerotic carotid artery lesions: evaluation of 26 cases and literature review. Mol Med. doi: 10.2119/molmed.2014.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409(6822):860–921. [DOI] [PubMed] [Google Scholar]
- Leclercq S, Rivals E, Jarne P.. 2010. DNA slippage occurs at microsatellite loci without minimal threshold length in humans: a comparative genomic approach. Genome Biol Evol. 2:325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DEA, Adhya S.. 2002. In vitro repression of the gal promoters by GalR and HU depends on the proper helical phasing of the two operators. J Biol Chem. 277(4):2498–2504. [DOI] [PubMed] [Google Scholar]
- Li J, et al. 2013. A functional fetal HSD11B2[CA]n microsatellite polymorphism is associated with maternal serum cortisol concentrations in pregnant women. Kidney Blood Press Res. 38:132–141. [DOI] [PubMed] [Google Scholar]
- Li Y, et al. 2012. A polymorphic microsatellite repeat within the ECE-1c promoter is involved in transcriptional start site determination, human evolution, and Alzheimer’s disease. J Neurosci. 32(47):16807–16820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, et al. 2017. An accurate and efficient method for large-scale SSR genotyping and applications. Nucleic Acids Res. 45(10):e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Chen X, Leffak M.. 2013. Oligodeoxynucleotide binding to (CTG). (CAG) microsatellite repeats inhibits replication fork stalling, hairpin formation, and genome instability. Mol Cell Biol. 33:571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Mulholland N, Fu H, Zhao K.. 2006. Cooperative activity of BRG1 and Z-DNA formation in chromatin remodeling. Mol Cell Biol. 26:2550–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, et al. 2001. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell 106:309–318. [DOI] [PubMed] [Google Scholar]
- Loire E, Higuet D, Netter P, Achaz G.. 2013. Evolution of coding microsatellites in primate genomes. Genome Biol Evol. 5:283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, et al. 2003. The capacity to form H-DNA cannot substitute for GAGA factor binding to a (CT)n*(GA)n regulatory site. Nucleic Acids Res. 31:2483–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. 2009. Somatic mutation and functional polymorphism of a novel regulatory element in the HGF gene promoter causes its aberrant expression in human breast cancer. J Clin Invest 119(3):478–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC, et al. 2012. The Drosophila melanogaster genetic reference panel. Nature 482:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Imamoto F, Merlino GT, Pastan I, Ishii S.. 1989. Cooperative function of two separate enhancers of the human epidermal growth factor receptor proto-oncogene. J Biol Chem. 264:5488–5494. [PubMed] [Google Scholar]
- Martin P, Makepeace K, Hill SA, Hood DW, Moxon ER.. 2005. Microsatellite instability regulates transcription factor binding and gene expression. Proc Natl Acad Sci U S A. 102:3800–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV.. 2006. Non-coding RNA. Hum Mol Genet. 15(Spec No: R17–R29). [DOI] [PubMed] [Google Scholar]
- McNeil JA, Smith KP, Hall LL, Lawrence JB.. 2006. Word frequency analysis reveals enrichment of dinucleotide repeats on the human X chromosome and [GATA]n in the X escape region. Genome Res. 16(4):477–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin SM. 2008. Discovery of alternative DNA structures: a heroic decade (1979–1989). Front Biosci. 13:1064–1071. [DOI] [PubMed] [Google Scholar]
- Mogil LS, Becker NA, Maher LJ 3rd. 2016. Supercoiling effects on short-range DNA looping in E. coli. PLoS ONE. 11:e0165306.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris EE, et al. 2010. A GA microsatellite in the Fli1 promoter modulates gene expression and is associated with systemic lupus erythematosus patients without nephritis. Arthritis Res Ther. 12:R212.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon R, Bayliss C, Hood D.. 2006. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu Rev Genet. 40:307–333. [DOI] [PubMed] [Google Scholar]
- Murat P, Balasubramanian S.. 2014. Existence and consequences of G-quadruplex structures in DNA. Curr Opin Genet Dev. 25:22–29. [DOI] [PubMed] [Google Scholar]
- Nagalingam S, Uppuluri MV, Gunda P, Ravishanker U, Tirunilai P.. 2014. Evaluation of leptin and leptin receptor gene 3′ UTR polymorphisms in essential hypertension. Clin Exp Hypertens. 36:419–425. [DOI] [PubMed] [Google Scholar]
- Namdar-Aligoodarzi P, et al. 2015. Exceptionally long 5′ UTR short tandem repeats specifically linked to primates. Gene 569:88–94. [DOI] [PubMed] [Google Scholar]
- Nelson HC, Finch JT, Luisi BF, Klug A.. 1987. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature 330(6145):221–226. [DOI] [PubMed] [Google Scholar]
- Nieto Moreno N, Giono LE, Cambindo Botto AE, Munoz MJ, Kornblihtt AR.. 2015. Chromatin, DNA structure and alternative splicing. FEBS Lett. 589:3370–3378. [DOI] [PubMed] [Google Scholar]
- Nikumbh S, Pfeifer N.. 2017. Genetic sequence-based prediction of long-range chromatin interactions suggests a potential role of short tandem repeat sequences in genome organization. BMC Bioinformatics. 18(1):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nithianantharajah J, Hannan AJ.. 2007. Dynamic mutations as digital genetic modulators of brain development, function and dysfunction. Bioessays 29:525–535. [DOI] [PubMed] [Google Scholar]
- Ogloblina AM, et al. 2015. Parallel G-quadruplexes formed by guanine-rich microsatellite repeats inhibit human topoisomerase I. Biochemistry (Mosc) 80:1026–1038. [DOI] [PubMed] [Google Scholar]
- Oh D-B, Kim Y-G, Rich A.. 2002. Z-DNA-binding proteins can act as potent effectors of gene expression invivo. Proc Natl Acad Sci U S A. 99:16666–16671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohadi M, et al. 2015. Core promoter short tandem repeats as evolutionary switch codes for primate speciation. Am J Primatol. 77:34–43. [DOI] [PubMed] [Google Scholar]
- Olson WK, Grosner MA, Czapla L, Swigon D.. 2013. Structural insights into the role of architectural proteins in DNA looping deduced from computer simulations. Biochem Soc Trans. 41:559–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omelina ES, Baricheva EM, Oshchepkov DY, Merkulova TI.. 2011. Analysis and recognition of the GAGA transcription factor binding sites in Drosophila genes. Comput Biol Chem. 35:363–370. [DOI] [PubMed] [Google Scholar]
- Paigen K, Petkov P.. 2010. Mammalian recombination hot spots: properties, control and evolution. Nat Rev Genet. 11:221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo SL, et al. 2008. A novel G-quadruplex-forming GGA repeat region in the c-myb promoter is a critical regulator of promoter activity. Nucleic Acids Res. 36:1755–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panyutin IG, Kovalsky OI, Budowsky EI.. 1989. Magnesium-dependent supercoiling-induced transition in (dG)n.(dC)n stretches and formation of a new G-structure by (dG)n strand. Nucleic Acids Res. 17:8257–8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak RU, et al. 2013. AAGAG repeat RNA is an essential component of nuclear matrix in Drosophila. RNA Biol. 10:564–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CE, Nichol Edamura K, Cleary JD.. 2005. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 6:729–742. [DOI] [PubMed] [Google Scholar]
- Perez N, Rehault M, Amouyal M.. 2000. A functional assay in Escherichia coli to detect non-assisted interaction between galactose repressor dimers. Nucleic Acids Res. 28:3600–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes TD. 2001. Meiotic recombination hot spots and cold spots. Nat Rev Genet. 2:360–369. [DOI] [PubMed] [Google Scholar]
- Potaman VN, et al. 2004. Length-dependent structure formation in Friedreich ataxia (GAA)n*(TTC)n repeats at neutral pH. Nucleic Acids Res. 32:1224–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press MO, Carlson KD, Queitsch C.. 2014. The overdue promise of short tandem repeat variation for heritability. Trends Genet. 30:504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punga T, Buhler M.. 2010. Long intronic GAA repeats causing Friedreich ataxia impede transcription elongation. EMBO Mol Med. 2:120–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J, Liu J.. 2013. A genome-wide analysis of simple sequence repeats in maize and the development of polymorphism markers from next-generation sequence data. BMC Res Notes. 6:403.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilez J, et al. 2016. Polymorphic tandem repeats within gene promoters act as modifiers of gene expression and DNA methylation in humans. Nucleic Acids Res. 44:3750–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy S, Garapati HS, Mishra RK.. 2014. Length and sequence dependent accumulation of simple sequence repeats in vertebrates: potential role in genome organization and regulation. Gene 551:167–175. [DOI] [PubMed] [Google Scholar]
- Ribeiro MM, et al. 2015. G-quadruplex formation enhances splicing efficiency of PAX9 intron 1. Hum Genet. 134:37–44. [DOI] [PubMed] [Google Scholar]
- Rich A, Zhang S.. 2003. Timeline: Z-DNA: the long road to biological function. Nat Rev Genet. 4:566–572. [DOI] [PubMed] [Google Scholar]
- Rothenburg S, Koch-Nolte F, Rich A, Haag F.. 2001. A polymorphic dinucleotide repeat in the rat nucleolin gene forms Z-DNA and inhibits promoter activity. Proc Natl Acad Sci U S A. 98:8985–8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan H, Wang Y-H.. 2008. Friedreich’s ataxia GAA.TTC duplex and GAA.GAA.TTC triplex structures exclude nucleosome assembly. J Mol Biol. 383:292–300. [DOI] [PubMed] [Google Scholar]
- Ryk C, et al. 2014. The (CCTTT)n microsatellite polymorphism in the NOS2 gene may influence lung cancer risk and long-term survival, especially in non-smokers. Tumour Biol. 35:4425–4434. [DOI] [PubMed] [Google Scholar]
- Sakamoto N, et al. 1999. Sticky DNA: self-association properties of long GAA.TTC repeats in R.R.Y triplex structures from Friedreich’s ataxia. Mol Cell. 3:465–475. [DOI] [PubMed] [Google Scholar]
- Sarkies P, Reams C, Simpson LJ, Sale JE.. 2010. Epigenetic instability due to defective replication of structured DNA. Mol Cell. 40:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathasivam K, et al. 2013. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc Natl Acad Sci U S A. 110:2366–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya S, et al. 2013. Microsatellite tandem repeats are abundant in human promoters and are associated with regulatory elements. PLoS ONE. 8:e54710.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya SM, Bagshaw AT, Buschiazzo E, Gemmell NJ.. 2012. Promoter microsatellites as modulators of human gene expression. Adv Exp Med Biol. 769:41–54. [DOI] [PubMed] [Google Scholar]
- Sawaya SM, Lennon D, Buschiazzo E, Gemmell N, Minin VN.. 2012. Measuring microsatellite conservation in mammalian evolution with a phylogenetic birth–death model. Genome Biol Evol. 4:636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaper E, Gascuel O, Anisimova M.. 2014. Deep conservation of human protein tandem repeats within the eukaryotes. Mol Biol Evol. 31:1132–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavone D, et al. 2016. PrimPol is required for replicative tolerance of G quadruplexes in vertebrate cells. Mol Cell. 61:161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlapp T, Rodel G.. 1990. Transcription of two divergently transcribed yeast genes initiates at a common oligo(dA-dT) tract. Mol Gen Genet. 223(3):438–442. [DOI] [PubMed] [Google Scholar]
- Searle S, Blackwell JM.. 1999. Evidence for a functional repeat polymorphism in the promoter of the human NRAMP1 gene that correlates with autoimmune versus infectious disease susceptibility. J Med Genet. 36:295–299. [PMC free article] [PubMed] [Google Scholar]
- Sen D, Gilbert W.. 1988. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 334:364–366. [DOI] [PubMed] [Google Scholar]
- Shelley CS, Baralle FE.. 1987. Deletion analysis of a unique 3′ splice site indicates that alternating guanine and thymine residues represent an efficient splicing signal. Nucleic Acids Res. 15:3787–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilo S, Melamed-Bessudo C, Dorone Y, Barkai N, Levy AA.. 2015. DNA crossover motifs associated with epigenetic modifications delineate open chromatin regions in Arabidopsis. Plant Cell. 27:2427–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin G, et al. 2017. CRISPR-Cas9-targeted fragmentation and selective sequencing enable massively parallel microsatellite analysis. Nat Commun. 8:14291.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishkin AA, et al. 2009. Large-scale expansions of Friedreich’s ataxia GAA repeats in yeast. Mol Cell. 35:82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh L, Wadhwa R, Naidu S, Nagaraj R, Ganesan M.. 1994. Sex- and tissue-specific Bkm(GATA)-binding protein in the germ cells of heterogametic sex. J Biol Chem. 269:25321–25327. [PubMed] [Google Scholar]
- Struhl K. 1985. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 82:8419–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Mishra RK, Singh L.. 2003a. Genome-wide analysis of Bkm sequences (GATA repeats): predominant association with sex chromosomes and potential role in higher order chromatin organization and function. Bioinformatics 19:681–685. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Mishra RK, Singh L.. 2003b. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 4:R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JX, et al. 2012. A direct characterization of human mutation based on microsatellites.Nat Genet.44(10):1161–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist WI, Klug A.. 1989. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature 342:825–829. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Zhu J-K.. 2007. Micro RNAs and short-interfering RNAs in plants. J Integr Plant Biol. 49(6):817–826. [Google Scholar]
- Svikovic S, Sale JE.. 2016. The effects of replication stress on S phase histone management and epigenetic memory. J Mol Biol. doi: 10.1016/j.jmb.2016.11.011. [DOI] [PubMed] [Google Scholar]
- Taka S, Gazouli M, Politis PK, Pappa KI, Anagnou NP.. 2013. Transcription factor ATF-3 regulates allele variation phenotypes of the human SLC11A1 gene. Mol. Biol. Rep. 40:2263–2271. [DOI] [PubMed] [Google Scholar]
- Tassone F, et al. 2007. Elevated FMR1 mRNA in premutation carriers is due to increased transcription. RNA 13:555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng S-H, Cheng C-Y, Huang M-Z, Chung M-Y, Su T-S.. 2013. Modulation of formation of the 3′-end of the human argininosuccinate synthetase mRNA by GT-repeat polymorphism. Int J Biochem Mol Biol. 4:179–190. [PMC free article] [PubMed] [Google Scholar]
- Uhlemann A-C, et al. 2004. DNA phasing by TA dinucleotide microsatellite length determines invitro and invivo expression of the gp91phox subunit of NADPH oxidase and mediates protection against severe malaria. J Infect Dis. 189:2227–2234. [DOI] [PubMed] [Google Scholar]
- Valipour E, et al. 2013. Polymorphic core promoter GA-repeats alter gene expression of the early embryonic developmental genes. Gene 531:175–179. [DOI] [PubMed] [Google Scholar]
- van Holde K, Zlatanova J.. 1994. Unusual DNA structures, chromatin and transcription. Bioessays 16(1):59–68. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Delrow J, Bussemaker HJ.. 2003. Genomewide analysis of Drosophila GAGA factor target genes reveals context-dependent DNA binding. Proc Natl Acad Sci U S A. 100:2580–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardhanabhuti S, Wang J, Hannenhalli S.. 2007. Position and distance specificity are important determinants of cis-regulatory motifs in addition to evolutionary conservation. Nucleic Acids Res. 35:3203–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinces MD, Legendre M, Caldara M, Hagihara M, Verstrepen KJ.. 2009. Unstable tandem repeats in promoters confer transcriptional evolvability. Science 324:1213–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volle CB, Delaney S.. 2012. CAG/CTG repeats alter the affinity for the histone core and the positioning of DNA in the nucleosome. Biochemistry 51:9814–9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walum H, et al. 2008. Genetic variation in the vasopressin receptor 1a gene (AVPR1A) associates with pair-bonding behavior in humans. Proc Natl Acad Sci U S A. 105:14153–14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AH, et al. 1979. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature 282:680–686. [DOI] [PubMed] [Google Scholar]
- Wang G, Vasquez KM.. 2007. Z-DNA, an active element in the genome. Front Biosci. 12:4424–4438. [DOI] [PubMed] [Google Scholar]
- Wang J, et al. 2016. Sex-specific mediation effect of the right fusiform face area volume on the association between variants in repeat length of AVPR1A RS3 and altruistic behavior in healthy adults. Hum Brain Mapp. 37:2700–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-H. 2007. Chromatin structure of repeating CTG/CAG and CGG/CCG sequences in human disease. Front Biosci. 12:4731–4741. [DOI] [PubMed] [Google Scholar]
- Weldon C, Eperon IC, Dominguez C.. 2016. Do we know whether potential G-quadruplexes actually form in long functional RNA molecules? Biochem Soc Trans. 44:1761–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MA, Dominska M, Petes TD.. 1993. Transcription factors are required for the meiotic recombination hotspot at the HIS4 locus in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 90:6621–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems T, Gymrek M, Highnam G, Mittelman D, Erlich Y.. 2014. The landscape of human STR variation. Genome Res. 24:1894–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong B, Chen S, Kwon J-A, Rich A.. 2007. Characterization of Z-DNA as a nucleosome-boundary element in yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 104:2229–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CG, Spies M.. 2016. G-quadruplex recognition and remodeling by the FANCJ helicase. Nucleic Acids Res. 44:8742–8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez-Cuna JO, et al. 2014. Dissection of thousands of cell type-specific enhancers identifies dinucleotide repeat motifs as general enhancer features. Genome Res. 24(7):1147–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakieh A, Simin H, Forousan S, Manoochehr T.. 2013. Polymorphic CT dinucleotide repeat in the GATA3 gene and risk of breast cancer in Iranian women. Med Oncol. 30:504.. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Sherman F.. 1982. DNA sequence required for efficient transcription termination in yeast. Cell 28:563–573. [DOI] [PubMed] [Google Scholar]
- Zhang J, et al. 2006. BRG1 interacts with Nrf2 to selectively mediate HO-1 induction in response to oxidative stress. Mol Cell Biol. 26:7942–7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, et al. 2014. Association between the (GT)n polymorphism of the HO-1 gene promoter region and cancer risk: a meta-analysis. Asian Pac J Cancer Prev. 15:4617–4622. [DOI] [PubMed] [Google Scholar]
- Zhang L, et al. 2006. Conservation of noncoding microsatellites in plants: implication for gene regulation. BMC Genomics. 7:323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, He L, Liu W, Sun C, Ratain MJ.. 2009. Exploring the relationship between polymorphic (TG/CA)n repeats in intron 1 regions and gene expression. Hum Genomics. 3:236–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, et al. 2015. GAA triplet-repeats cause nucleosome depletion in the human genome. Genomics 106:88–95. [DOI] [PubMed] [Google Scholar]
- Zhao Z, et al. 2014. Genome-wide analysis of tandem repeats in plants and green algae. G3 (Bethesda) 4:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng R, et al. 2010. Polypurine-repeat-containing RNAs: a novel class of long non-coding RNA in mammalian cells. J Cell Sci. 123:3734–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato E, Buratti E, Stuani C, Baralle FE, Pagani F.. 2004. An intronic polypyrimidine-rich element downstream of the donor site modulates cystic fibrosis transmembrane conductance regulator exon 9 alternative splicing. J Biol Chem 279:16980–16988. [DOI] [PubMed] [Google Scholar]