Abstract
Black-and-gold howler monkeys Alouatta caraya, are arboreal primates, inhabitants of Neotropical forests, highly susceptible to the yellow fever virus, considered early 'sentinels' of outbreaks, and thus, of major epidemiological importance. Currently, anthropogenic habitat loss and modifications threatens their survival. Habitat modification can prevent, reduce or change dispersal behavior, which, in turn, may influence patterns of gene flow. We explored past and contemporary levels of genetic diversity, elucidated genetic structure and identified its possible drivers, in ten populations (n = 138) located in the southernmost distribution range of the species in South America, in Argentina and Paraguay. Overall, genetic variability was moderate (ten microsatellites: 3.16 ± 0.18 alleles per locus, allelic richness of 2.93 ± 0.81, 0.443±0.025 unbiased expected heterozygosity; 22 haplotypes of 491-bp mitochondrial Control Region, haplotypic diversity of 0.930 ± 0.11, and nucleotide diversity of0.01± 0.007). Significant evidence of inbreeding was found in a population that was, later, decimated by yellow fever. Population-based gene flow measures (FST = 0.13; θST = 018), hierarchical analysis of molecular variance and Bayesian clustering methods revealed significant genetic structure, grouping individuals into four clusters. Shared haplotypes and lack of mitochondrial differentiation (non-significant θST) among some populations seem to support the hypothesis of historical dispersal via riparian forests. Current resistance analyses revealed a significant role of landscape features in modeling contemporary gene flow: continuous forest and riparian forests could promote genetic exchange, whereas disturbed forests or crop/grassland fields may restrict it. Estimates of effective population size allow anticipating that the studied populations will lose 75% of heterozygosity in less than 50 generations. Our findings suggest that anthropogenic modifications on native forests, increasingly ongoing in Northeastern Argentina, Southern Paraguay and Southeastern Brazil, might prevent the dispersal of howlers, leading to population isolation. To ensure long-term viability and maintain genetic connectivity of A. caraya remnant populations, we recommend preserving and restoring habitat continuity. To conserve the species genetic pool, as well, the four genetic clusters identified here should be considered separate Management Units and given high conservation priority. In light of our findings and considering complementary non-genetic information, we suggest upgrading the international conservation status of A. caraya to “Vulnerable”.
Introduction
Howler monkeys (Primates: Atelidae) are amongst the largest New World monkeys, inhabitants of several Neotropical ecoregions, from central Mexico to northeastern Argentina [1]. Nowadays, these Primates are being increasingly affected by anthropogenic activities, such as deforestation for agriculture and cattle ranching, and flooding of large areas for dam building which derive in loss, modification, reduction or isolation of native forest habitats [2–5]. Such changes trigger secondary processes in primate populations including dispersal restrictions, resource depletion, and pathogen exposure [6–9], which can reduce genetic diversity and effective population size, decreasing the adaptive potential of populations, increasing local extinction risks, and affecting the long-term survival of species [10]. If distance between habitat patches or modification of natural landscape structure prevents individual dispersal, gene flow between populations can be prevented, compromising the adaptation capacity and survival of the species in the long-term [10]. Population genetics can therefore provide insights into how anthropogenic changes affect primate populations, as well as into the historical and contemporary processes that shape the population structure [10], including the influence of landscape features on gene flow [8]. Describing the patterns of distribution of genetic diversity can help define population units important for effective management and conservation [10]. Moreover, population genetic parameters can be used in a holistic framework to support recommendations regarding official international conservation rankings [11]. To date, while some primate groups have been more explored regarding their genetic structure, others remain poorly studied [12]. Relative to other primates, Neotropical species represent an understudied group regarding population genetics [13]. New studies can help deepen our understanding on the factors that influence gene flow in these primates.
Here, we focus on black-and-gold howler monkeys (Alouatta caraya; hereafter denoted as BGHM), arboreal primates which inhabit several ecoregions in South America (Fig 1). Some of these ecorregions, such as the Dry and Humid Chaco forests in Bolivia, Paraguay and Argentina [2, 3], and the Atlantic Forest in Brazil, Paraguay and Argentina [5], are subjected to major anthropogenic modifications, and are entirely fragmented, with native remnants mostly isolated. Population densities and social organization of BGHM differ remarkably along their distribution area [14]. Previous studies indicate that BGHM disperse through riparian forests, which act as biological corridors [15, 16], but habitat fragmentation severely limits their ability to disperse [7]. Demographic records show that both females and males leave their natal groups (social units); therefore, within groups, the adults are expected to be unrelated [17]. Although BGHM can survive in fragmented and impoverished habitats, including those that have undergone selective logging [18], indiscriminate deforestation and destruction of riparian forests could threaten their survival at the southernmost part of the species range [19, 20].
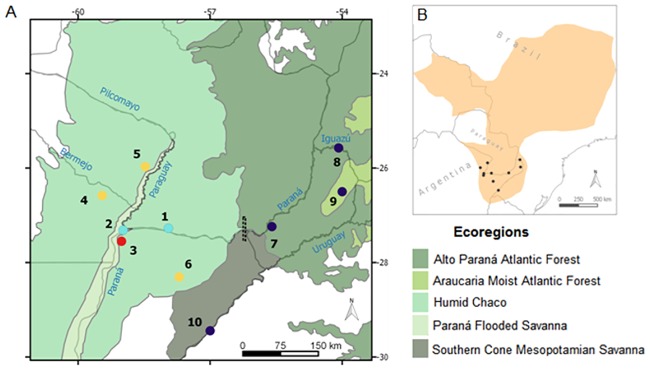
Fig 1. Map of sampling sites.
Maps showing: (A) black-and-gold howler monkeys Alouatta caraya sampling locations in Northeastern Argentina and Southern Paraguay; populations (1 to 10) are represented by circles color-coded to mirror their allocation to the four main genetic clusters identified by Structure analyses. The different background colors indicate different ecoregions [31]. Main rivers are shown in blue font. The dotted black bar next to population 7 indicates the location of the Yacyretá Hydroelectric Dam; (B) Map showing the distribution range of BGHM modified from IUCN database with recent data from [17], with black dots showing location of sampling sites. Full names of sampling sites are given in Table 1.
BGHM have major epidemiological importance because they are sensitive to the yellow fever virus and show high mortality when infected, therefore acting as early sentinels for virus detection [21]. BGHM abundance in the Atlantic Forest of Argentina drastically dropped after the 2008–2009 sylvatic yellow fever outbreak [22] and a recent study reported no evidence of the presence of BGHM in this area [15]. In southern Brazil, as well, the same outbreak decimated many BGHM populations [23], and a recent outbreak in February 2017 caused thousands of monkeys’ deaths [24, 25]. Currently, BGHM are globally classified as “Least Concern” [26]. However, they are being increasingly affected by loss and modification of their native habitat, hunting and trafficking for pet trade, and thus, considered “Near Threatened” in Brazil [27] and Bolivia [28]. Moreover, in the southern limit of the specie’ range (Fig 1), BGHM are classified even under higher risk categories, such as “Vulnerable” in Argentina [1], and “Endangered” in southern Brazilian states [27], highlighting the effects of increasing deforestation and the vulnerability of the monkeys to the yellow fever epidemics. Under such degree of pathogen exposure and habitat degradation, the long-term persistence of BGHM populations is of high concern. In this sense, assessing the levels of genetic diversity and gene flow in small and geographically distant remaining wild BGHM populations is fundamental to support their conservation as well as to clarify the international ranking of the species.
In this study, we used nuclear and mitochondrial markers to investigate ten BGHM populations inhabiting different ecoregions of northeastern Argentina and southern Paraguay (Fig 1), and subject to different degrees of habitat loss, modification and degradation. Our specific objectives were to: (1) describe past- and present-day levels of genetic diversity; (2) assess historical and contemporary patterns of genetic structure; (3) investigate possible drivers of genetic structure, specifically, geographic distance, differences between ecoregions, or variable levels of habitat loss and modification; and (4) provide conservation guidelines and key information for management or reintroduction projects involving these Neotropical primates, by, for example, identifying Management Units (MUs) [29]. Given that historical dispersal routes of BGHM have supposedly gone through unflooded and seasonally flooded riparian forests [16], populations connected by rivers in the past are expected to share mitochondrial similarities. Moreover, given that BGHM disperse through continuous forests, effective dispersal will be likely affected by anthropogenic modifications. Arboreal primates and non-primates are expected to depend on forest continuity to disperse. Previous studies in BGHM occupying a fragmented habitat interrupted by grassland extensions suggest a reduction in the dispersal rate between groups residing at distances greater than 1000 m [7]. Therefore, landscape modification, such as crop monocultures or grasslands used for cattle ranching, may isolate populations because dispersing individuals from their natal groups would have to descend to the ground, being highly susceptible to predation or other sources of mortality before reaching another fragment [7, 30]. Thus, populations of BGHM not connected by continuous forests are expected to show restricted gene flow and to be genetically different from one another.
Material and methods
Ethics statement
This study was carried out in strict accordance with Argentinean laws for research on non-human primates, and following the recommendations of ‘Principles for the Ethical Treatment of Primates’ of the American Society of Primatologists (available at: https://www.asp.org/society/resolutions/EthicalTreatmentOfNonHumanPrimates.cfm). We received specific approval to conduct this study by the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) from Argentina (no. 11420110100322CO). Additional specific sampling permits were obtained from Chaco and Corrientes Provinces, Argentina (Permit Number: 01071), and from Ministry of Ecology, Misiones Province, Argentina (Permit Number: permit no. 04/15); tissue samples were collected as part of an investigation conducted jointly by the Global Health Program, Wildlife Conservation Society and the Ministry of Ecology, Misiones Province, Argentina (Permit Number: 304/09). All methods used for tissue sampling complied with the guidelines recommended by the Protocol for Primate Sample Methods (available from: http://www.vetmed.ucdavis.edu/ohi/local_resources/pdfs/PREDICT_Protocol_Primate_Sampling_29Feb12.pdf). Fecal collection was conducted without capturing the animals and therefore does not cause any harm to the studied species. The specific coordinates for each sampling location are: Pop1: 27,275°S 57,684°W; Pop2: 27,314°S, 58,646°W; Pop3: 27,550°S, 58,679°W; Pop4: 26,791°S, 59,631°W; Pop5: 25,970°S, 58,177°W; Pop6: 28,307°S, 57,457°W; Pop7: 27,467°S, 55,827°W; Pop8: 25,574°S, 54,075°W; Pop9: 26,500°S, 53,833°W; Pop10: 29,445°S, 56,800°W.
Biological sampling and DNA extraction
We sampled 163 BGHM from ten populations inhabiting five ecoregions [31] (Fig 1), in the southernmost edge of the species’ geographic range in northeastern Argentina and southern Paraguay, subjected to different types and degrees of environmental modification. To minimize the sampling of relatives, as BGHM generally disperse during juvenile stages [30], two fecal samples were collected from each adult. In groups 4, 5, 6, 8, 9 and 10, feces were collected immediately after defecation. In group 7, tissue samples were collected from individuals found dead and kept refrigerated at 4°C until necropsy. All samples were preserved at 24°C in 50 ml screw-top tubes containing solid NaCl [32] until DNA extraction (three months to one year later). DNA was extracted from feces using the QIAamp DNA Stool Mini Kit, (QIAGEN, Valencia, USA), following the manufacturer’s protocol with slight modifications, and from tissue samples using standard SDS/Proteinase K digestion followed by phenol–chloroform organic extraction [33]. Appropriate precautions were taken to avoid sample contamination: every step of the experiment was performed in designated laboratory spaces, under laminar flow (with negative pressure) conditions, and using aerosol-resistant filter tips.
Microsatellites amplification
Genotypes from populations 1, 2 and 3 were obtained in previous studies [7, 32]. The remainder samples were amplified at ten autosomal STR markers, previously used in studies in BHGM [34] (AC14, AC17, AC 45, D8S165, D17S804, LL1118, LL157, Tgms1, Tgms2 and AB7), following a two-step multiplex PCR method [35] with minor modifications, in a 25-μL final volume, with 50 ng of DNA (including negative controls with no template DNA),1xGoTaqbuffer (Promega, USA), 1.75 mM MgCl2, 0.2 mM of each dNTP, 1 U GoTaq DNA polymerase (Promega, USA), 4 pmol of each forward primer with an M13 tail, 4 pmol of each reverse primer, and 0.4 mg of Bovine Serum Albumin (BSA, Promega, USA). The second step consisted of a 12.5-μL reaction with 5 μL of a 5:100 dilutions of the first multiplex PCR product as template, 0.875 mM MgCl2, 4 pmol of a forward primer with an M13 tail, 4 pmol reverse primer, and 4 pmol 5´FAM or HEX-labeled M13 primer [34]. Cycling parameters were: initial denaturation (95°C for 4 min), followed by 35 cycles of denaturation (94°C for 45 s), annealing (90 s at 58–60°C), and extension (72°C for 60 s), and a final extension (5 min at 72°C) [34]. Products from the second amplification step of different markers, labeled with different fluorochromes, were combined and separated by electrophoresis on an ABI PRISM 310 Genetic Analyzer. Alleles were manually scored by visual inspection of electropherograms after developing of the bin panel for each locus in GeneMapper ID-X v. 1.2 (Applied Biosystems), using HD400-ROX as internal size standard. To detect possible genotyping errors due to allelic, three independent amplification reactions were performed for each DNA extract (totaling six independent PCRs per marker, per individual). Each homozygous genotype was re-amplified and genotyped three additional times, from the two separate fecal samples per individual.
Mitochondrial DNA amplification
A491-bp fragment of the left domain of the mitochondrial DNA Control Region (mtDNA, CR) was amplified using primers How RA-1 (5’-CTACCATCAACACCCAAAGC-3’) [16] and RC-BugioR (5’-CCAGGTTAAGAGGGTGATAGC-3’, this paper). Amplifications were performed at a final volume of 25 μL, containing 25 ng of single DNA extractions,1x GoTaq buffer (Promega, USA), 1.75 mm MgCl2, 0.2 mM of each dNTP, 1 U GoTaq DNA polymerase (Promega, USA), 4 pmol of each primer, and 0.4 mg BSA. Cycling parameters were: initial denaturation (5 min at 94°C), followed by 35–40 cycles of denaturation (1 min at 94°C), annealing (30 s at 50°C), extension (1 min at 72°C), and a final extension(3 min at 72°C). All products were sequenced bi-directionally in an Applied Biosystems 3500 Genetic Analyzer using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA), and with the samereverse and forward primers used foramplifications. Quality of the sequences was eyed-checked inspecting the electropherograms in Sequencher 5.3 software (LifeCodes, USA).Sequences were edited and aligned using the Musclealgorithm [36] in MEGAv6.0 [37]. As in [38], the possible presence of nuclear mitochondrial insertions (NUMTS) was inspected by performing a BLASTn® search in the National Center for Biotechnology Information (NCBI) website (https://blast.ncbi.nlm.nih.gov/Blast.cgi). For this search, the filters and mask options were clicked off, word size was set to a value of 28, match/mismatch scores were set to 1/-2 and gap creation/extension penalties were set to ´linear´.
Statistical analysis of microsatellites
Genetic diversity, effective population size and demographic parameters
Genotypes were screened for null alleles, stuttering, or scoring errors using Micro-Checker v2.2.3 [39]. Conformation to Hardy-Weinberg equilibrium (HWE) and linkage disequilibrium (LD) was assessed performing exact tests in Genepop v4.2[40] with default settings for Markov chain parameters.The number of alleles per locus, number of private alleles, observed heterozygosity, unbiased expected heterozygosity, inbreeding coefficient, and the probability that two matching genotypes taken at random come from siblings(PID-SIBS)[41] were computed in GenAlEx v6.5 [42]. To account for differing sample sizes, we computed a rarefied measure of allelic diversity (Allelic Richness) in Fstat v2.9.3.2[43], based on a standard sample size of n = 5, the smallest sample with complete genotypes at all loci (Paraguay sample, see Table 1). Statistical differences between groups regarding diversity statistics were evaluated using Kruskal-Wallis rank-sum tests in R statistical environment [44], applying Bonferroni correction to adjust significance levels for multiple comparisons. The pairwise relatedness estimator, R, of [45] was computed to identify first-order relatives (i.e, R ≥ 0.375) [46] that could lead to biased inferences of the population structure. When necessary, we randomly removed one individual from each of the highly related pairs and further analyzed the population genetic structure using the trimmed datasets (see below). The effective population size (Ne) of each group was estimated using a single-sample linkage disequilibrium method with jackknifing, as implemented in LD Ne v3.1 [47], for a minimum allele frequency of 0.05 [48]. The premise of the LD method is that the magnitude of the correlation between allele frequencies is a function of the effective population size and reflects the past finite population history; also, as a function of the sample size (n), the correlation in allelic frequencies arises from sampling a limited number of individuals from the population for estimating gene frequencies and disequilibrium [49]. The LD method for estimating Ne is based on the expectation that small populations accumulate more disequilibrium over time [49]. The method is robust to population size reductions and can be corrected for possible biases when the sample size is lower than the real Ne [47].
Table 1. Sampling information and summary estimates of diversity at ten microsatellites for black-and-gold howler monkeys.
Type of habitat regarding tree-cover, population codes, names and number of sampled social units (groups) are given; n: number of samples analyzed (amplified at a minimum of seven loci); Na: number of different alleles, AR: allelic richness, PA: number of private alleles, Ho: observed heterozygosity ± standard deviation, UHe: unbiased expected heterozygosity ± standard deviation, FIS: inbreeding coefficient, with an asterisk indicating the significant value after Bonferroni correction (adjusted significance level: 0.0005), and PID-Sibs: multilocus probability that two matching genotypes taken at random come from siblings.
| Habitat | Code | Pop. name | Groups | n | Na | AR | PA | Ho | uHe | FIS | PID-Sibs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Modified | Pop 1 | Paraguay | 2 | 5 | 2.8 ± 0.44 | 2.59 | 1 | 0.38 ± 0.08 | 0.42 ± 0.07 | 0.11 | 0.009 |
| Continuous | Pop 2 | Isla Brasilera | 7 | 38 | 5.0 ± 0.91 | 2.80 | 6 | 0.50 ± 0.08 | 0.50 ± 0.08 | -0.08 | 0.003 |
| Modified | Pop 3 | EBCo | 11 | 42 | 4.5 ± 0.93 | 2.60 | 2 | 0.46 ± 0.08 | 0.44 ± 0.07 | -0.06 | 0.006 |
| Modified | Pop 4 | PN Chaco | 4 | 8 | 2.7 ± 0.26 | 2.30 | 1 | 0.39 ± 0.06 | 0.39 ± 0.05 | 0.01 | 0.013 |
| Modified | Pop 5 | Guaycolec | 6 | 10 | 3.5 ± 0.04 | 2.62 | 1 | 0.46 ± 0.06 | 0.46 ± 0.06 | -0.01 | 0.005 |
| Modified | Pop 6 | San Alonso | 5 | 9 | 2.3 ± 0.39 | 1.96 | 0 | 0.41 ± 0.10 | 0.32 ± 0.08 | -0.26 | 0.003 |
| Continuous | Pop 7 | Garupá | 3 | 6 | 2.7 ± 0.39 | 2.38 | 1 | 0.46 ± 0.10 | 0.36 ± 0.07 | -0.24 | 0.002 |
| Continuous | Pop 8 | Yacutinga | 2 | 4 | 2.1 ± 0.34 | 2.10 | 1 | 0.45 ± 0.11 | 0.35 ± 0.07 | -0.30 | 0.002 |
| Continuous | Pop 9 | Piñalito | 4 | 8 | 3.2 ± 0.44 | 2.70 | 2 | 0.34 ± 0.05 | 0.50 ± 0.05 | 0.29* | 0.003 |
| Modified | Pop 10 | Yapeyú | 4 | 8 | 3.0 ± 0.39 | 2.44 | 2 | 0.35 ± 0.10 | 0.37 ± 0.08 | 0.05 | 0.002 |
Genetic structure
We first investigated genetic structure using microsatellites genotypic data by running both individual- and population-based analyses, at fine and regional spatial scales. For example, we evaluated spatial genetic autocorrelation at a fine scale, using GenAlExv6.5. This method is appropriate when there is no a priori way to predict the genetic structure. Under restricted dispersal, we would expect a pattern of positive autocorrelation, where individual-by-individual genetic distances should be more similar at shorter geographical distances [50]. Geographical distance classes were chosen to ensure that the intervals included an even number of pairwise comparisons, ranging from the minimum (0–27 km, the first distance class, including comparison of dyads within the same site) to the maximum distance between sampling sites (578 km). Statistical significance was assessed with 10,000 random permutations and one-tailed probability tests, and 95% confidence intervals around the autocorrelation coefficient, r, were calculated with 10,000 bootstraps. The distance class at which r is no longer significant can be interpreted as an approximation of the extent of detectable positive spatial genetic structure [51]. In addition, to assess contemporary gene flow, we ran non-spatial Bayesian clustering using models in Structure v.2.3.4 [52]. A series of 20 independent runs per K (ranging from 1 to 10) was conducted using the admixture model with correlated allele frequencies, sampling locations as prior (LOCPRIOR), and 1,000,000 MCMC iterations after a burn-in of 50,000 replicates. Given our uneven sampling sizes we applied the correction method proposed by [53] to account for a possible downward bias in the number of genetic clusters recovered by Structure. For this, we analyzed: 1) the full dataset excluding first-order relatives (n = 138 individuals, 10 sites), and three trimmed datasets prepared to achieve more even sampling schemes; 2) a dataset obtained by randomly removing individual genotypes, without replacement, from the two best sampled populations to reach a sample size of 15 individuals in each, plus all other samples (n = 88, 10 sites); 3) the original dataset, removing the populations with n ≤ 6 (n = 123, seven sites); and 4) the sub-sampled even-sized reduced dataset, plus removing the populations with n ≤ 6 (n = 73, seven sites). For each one of these sampling strategies, runs were performed with the above-mentioned settings. We then collected the outputs and computed the corrected estimators MedMeaK, MaxMeaK, MedMedK, and MaxMedK[53] which represent the number of different clusters to which at least one of the populations (e.g., individuals grouped by sampling location) belongs to. These indexes are robust to uneven sampling schemes, and perform equally well or better than other commonly used clustering methods [53].For each sampling strategy, we computed all four estimators with population membership coefficient thresholds varying from 0.50 to 0.80. Then, to determine the number of clusters in our sample, we looked for the K identified by MaxMedK and MedMedK for a 0.80 threshold, as these should be more conservative, less influenced by the presence of migrants, and less affected by an incorrect a priori grouping of some individuals into populations [53]. We used Pophelper v.1 [54] to visualize the results of Structure analyses for the most likely number of K, identified as detailed above.
Landscape analyses
We analyzed the possible influence of landscape features on the genetic structure of BGHM by conducting analyses based on current theory in Circuitscape v 4.0 [55]. For this, we generated raster resistance maps representing the difficulty of BGHM to disperse through different habitats. We drew raster maps with landscape cover of 2009 using data from the European Space Agency portal (http://due.esrin.esa.int/page_globcover.php). The elements of the landscape were classified into three categories: 1) “tree cover” (including both native forests and pine plantations, as these have been shown to be used by BGHM to disperse [7], 2) rivers, and 3) crops/grassland/roads. Accordingly, we assigned resistance values representing the cost of movements for BGHM through these features: a low-resistance value of one for tree-covered pixels, an intermediate-resistance value of 25 for river pixels, and a high-resistance of 50 for crops/grassland/roads pixels. Additional analyses changing these arbitrary values did not change our results significantly. We transformed resistance maps into pairwise resistance distances between sampling sites, using an 8-neighbours correction scheme. Then, using the ‘vegan’ v2.4–3 package [56] in R, we conducted Mantel tests between genetic (Dps = 1 –proportion of shared alleles between populations) and resistance distances, as well as partial Mantel tests between Dps and resistance, controlling for Euclidean distances (in kilometers, computed from geographical coordinates), and between Dps and geographic distance, partialling out the effects of resistance distances [57]. The proportion of shared alleles by pairs of populations was calculated with PopGenReport v2.2.2 package [58] in R. If landscape features influence gene flow, we would expect the Mantel statistic between Dps and resistance and the partial Mantel statistic (controlling for the effect of geographic distance) to be significant, whereas the partial Mantel statistic between Dps and geographic distance (controlling for resistance distances), to be non-significant [57]. Population-based gene flow measures (i.e., pairwise FST values) were also computed between all pairs of populations in Fstat v2.9.3.2.
Statistical analysis of mitochondrial DNA
Haplotype frequencies, nucleotide composition, the number of transitions and transversions, and the number of polymorphic sites were calculated in Arlequin v3.522 [59]. Haplotypic (h) and nucleotidic diversity (ð) were calculated in DnaSP v5.0 [60]. Standard tests of selective neutrality, R2[61], Tajima’s D [62], and Fu's FS [63], and their 95% confidence intervals, were conducted in DnaSP v5 with 1,000 simulations and a neutral infinite-sites model assuming a large constant population size. A constant population size represents the null hypothesis under the neutral model (i.e., the standard coalescent) [64]. Under the constant size hypothesis, when sample sizes are small, as is the case for some of our samples, the statistical tests Fu's Fs and R2 have more power to reject the null hypothesis [64]. Selective neutrality was rejected if small R2 and negative Fu's FS values were significant. A Median-Joining haplotype network [65] was built in PopArt program [66]. To inspect for historical demographic processes undergone by the populations, we carried out an analysis of ‘mismatch distribution’ [67] in Arlequinv3.522with 10,000 bootstraps. Populations at a demographic equilibrium or declining are expected to exhibit a multimodal distribution pattern of pairwise differences between haplotypes, whereas populations that have experienced a sudden demographic expansion are expected to display a unimodal distribution [68]. The smoothness of the mismatch distribution curve was measured using the raggedness (Rg) and the Sums of Squared Deviations (SSD) indexes [68]. The significance of the test was evaluated through 1,000 coalescent simulations, assuming a neutral infinite-sites model and a constant population size. The timing of the demographic processes was estimated by computing Tau [67], using the formula τ = 2ut, where u is the mutation rate of the assayed fragment. We used 0.15 mutations per site per million years, as in previous studies in howler monkeys [16]. To express the time since the expansion in years, we used a generation time of 5 years, which is the average age of first breeding of BGHM [69]. Finally, for mtDNA sequence data, genetic structure was examined in Arlequinv3.522by conducting an analysis of molecular variance (AMOVA) [70] comparing all sampling sites separately, as well as a hierarchical AMOVA grouping the populations within five ecoregions (1. Humid Chaco, HC: Pop 1, 3, 4, 5 and 6; 2. Paraná Flooded Savanna, PFS: Pop 2; 3. Alto Paraná Atlantic Forest, APF: Pop 7 and 8; 4. Araucaria Moist Atlantic Forest, AMAF: Pop 9; and 5. Southern Cone Mesopotamian Savanna, SCMS: Pop 10, see Fig 1 for further reference), and computing θST between populations and population groups.
Results
Genetic diversity, effective population size and demographic parameters
For most samples (n = 98) all ten loci were amplified, whereas in 40 samples a minimum of seven loci were amplified. We found no evidence of linkage between any pair of loci (P< 0.05), nor evidence of significant deviations from Hardy-Weinberg Equilibrium. We found evidence of first-order relatives (R ≥ 0.375) within populations (Pop) 2 and 3, and removed one individual from each of those dyads for further diversity and structuring analyses (final complete dataset n = 138). BGHM populations exhibited moderate levels of microsatellite diversity (Table 1). The mean number of alleles, NA, was3.16 ± 0.18; the mean allelic richness. AR, was2.93 ± 0.81, the lowest value was detected in Pop 6, whereas the highest in Pop 2, the latter also showing the highest number of private alleles. Mean unbiased expected heterozygosity (uHe) was 0.443±0.025 overall populations; uHe of Pop 2 differed significantly from uHe of Pop 6, 7, and 10, and uHe of Pop 9 differed significantly from that of Pop 7 and 8 (Kruskal-Wallis tests, p< 0.05). A significant signal of inbreeding was found only in Pop 9 (Table 1). Ne estimates were 40 (95%CI: 21–112) for Pop 2, and Ne = 26 (95%CI: 15–52) for Pop 3. The remainder populations yielded Ne with infinite 95% CIs.
Thirty-six transitions defined 22 mtDNA CR haplotypes (n = 72), 15 of which were new to this study (Genbank accession numbers MF095740, MF095741, MF095742, MF095743, MF095744, MF095745, MF095746, MF095747, MF095748, MF095749, MF095750, MF095751, MF095752, MF095753 and MF095754), an overall haplotypic diversity of 0.930 ± 0.11, and an overall nucleotidic diversity of 0.01± 0.007. Pop 2 was the most diverse in terms of number of haplotypes, but these were moderately divergent from each other (Fig 2, Table A in S1 File). Pop 3, 6, and 8 showed one haplotype each. Five individuals of Pop 9 showed one haplotype (H_19) separated by a minimum of 20 mutational steps from its ancestral haplotype (Fig 2). The BLAST search of the 491-bpmtDNA fragment of Alouatta caraya as a query sequence retrieved only mitochondrial sequences from Platyrrhini primates from Atelidae family (nine species) and Cebidae (three species) family. Identity of fragment sequences ranged from 77% to 99% with E-values ranging from 0.00 to 0.0057, respectively. Therefore, the presence of NUMTs in our fragment sequences can be considered negligible.
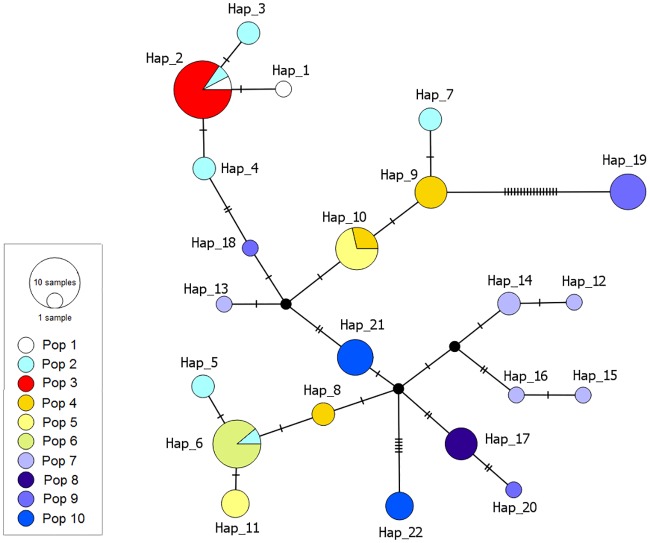
Fig 2. Haplotype network of black-and-gold howler monkeys.
Median-Joining network of 22 haplotypes observed in black-and-gold howler monkeys from northeastern Argentina and southern Paraguay. Circle sizes are proportional to haplotypic frequencies; small black circles indicate median vectors; black lines indicate mutational steps. Full names of populations are given in Table 1, and colors approximate to those defining population clusters.
There was no correlation between mtDNA CR lineages and geographical distribution; some haplotypes were shared between geographically distant populations and different lineages occurred within a population (Fig 2). For the complete BGHM dataset, D (-0.071), Fs (-1.889), and R2(0.099) were non-significant, supporting the neutrality of the assayed fragment. The unimodal pattern of the mismatch distribution (data not shown) was consistent with a demographic expansion scenario estimated to have occurred 9,183 years ago (Tau = 7,053). At the population level, significant SSD and Rg were observed in Pop 5 and Pop 9, which could be interpreted as a demographic expansion (Table A in S1 File).
Genetic structure
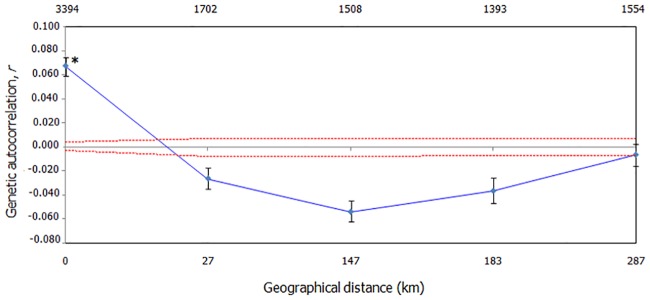
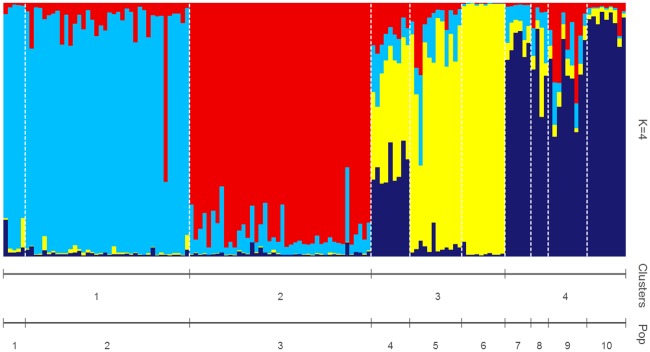
The average inter-individual genetic distance was 7.0 ± 3.0. Results of spatial autocorrelation analyses revealed a significant positive spatial structure (r = 0.067, P< 0.01) only at the first distance class (0–27 km), indicating that BGHM were more closely related to other members of the same population than to BGHM from other populations (Fig 3). The results of the corrected Structure procedure identified four genetically differentiated population clusters as best explaining the nuclear genetic variation observed in BGHM. All datasets analyzed (i.e., the full corrected dataset as well as the sub-sampling strategies) resulted in the same number of clusters (Table B in S1 File). The plots of ancestry membership proportions showed that the four clusters comprise: i) Pop 1 and 2; ii) Pop 3; iii) Pop 4, 5, and 6; and iv) Pop 7, 8, 9, and 10 (Fig 4). In agreement, the maps produced in Circuitscape showed a path of low resistance comprising populations 7, 8, 9, and 10, and another comprising populations 2, 4, and 5, whereas populations 1, 3, and 6 remained more isolated from the others (Fig 5). The Mantel test between Dps and resistance distances was significant (r = 0.528, P = 0.006), while that between Dps and geographic distances was not (r = 0.267, P = 0.081). The partial Mantel test between Dps and resistance (partialling out geographic distance) was significant (r = 0.648, P = 0.001), while that between Dps and geographic distance (partialling out resistance) was not (r = 0.503, P = 0.061).
Fig 3. Spatial autocorrelation in black-and-gold howler monkeys.
Spatial correlogram for howlers (n = 138) showing the genetic correlation coefficient (r) as a function of geographic distance across defined spatial distance classes. Dashed red lines represent upper (U) and lower (L) bounds of the null hypothesis of no spatial structure based on 10,000 random permutations. Error bars represent 95% confidence intervals about r based on 10,000 bootstraps. The asterisk denotes significantly positive r at α = 0.05. The number of pairwise comparisons within each distance class is shown above the plot.
Fig 4. Genetic structure in black-and-gold howler monkeys.
Membership bar-plots of black-and-gold howler monkeys (n = 138) sampled across ten sites in northeastern Argentina and southern Paraguay, resulting from Bayesian clustering analyses in Structure [52] based on genotypic data from 10 microsatellites. Individuals are represented by vertical lines (y-axis) broken into color-segments proportional to their membership coefficients to each cluster (K = 4), and were grouped into populations of sampling, separated with a white dashed line. Equally colored populations share genetic ancestry and are differentiated from the others. Full names of populations are given in Table 1.
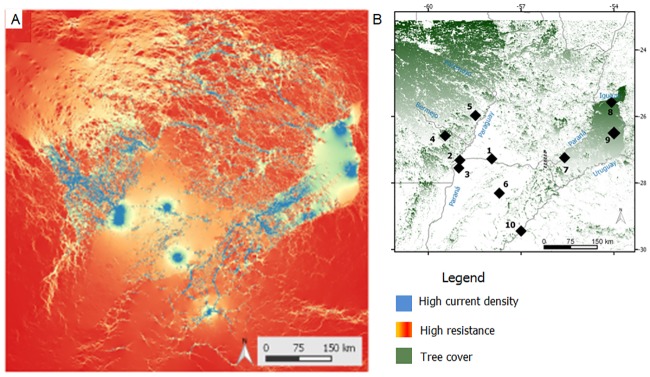
Fig 5. Maps of connectivity between populations of black-and-gold howler monkeys.
Current maps of northeastern Argentina and southern Paraguay showing flow of current. Blue-colored areas represent highest current densities (higher connectivity) whereas light yellow-orange areas represent highest resistance (lowest current densities). Under the hypothesis of dispersal along tree-covered areas and riparian forests, areas in blue will therefore facilitate gene flow whereas areas in light yellow may restrict gene flow. Maps were generated in Circuitscape program, and re-colored in ArcGIS v 10.1 (http://www.esri.com/software/arcgis/arcgis-for-desktop); (B) Map of the studied region showing the 2009 tree cover surface used for Circuitscape analyses. For reference, the sampling sites are indicated with numbers (1–10).
The global FST for microsatellites was 0.13 ± 0.34 and significantly different from zero (p = 0.001). Pairwise FST values between populations indicated substantial genetic differentiation, with 32 significant out of 45 comparisons (Table C in S1 File). Global θST for mtDNA CR fragments was 0.18 ± 0.22 and significantly different from zero (p = 0.001). Pairwise θST values showed that Pop 3 and Pop 6 differed significantly from the others, except for Pop 1 and 2; in turn, Pop 5 differed significantly from 6 and 10, and Pop 10 differed significantly from Pop 9 (Table C in S1 File). The hierarchical AMOVA for microsatellites grouping populations belonging to the same ecoregion showed that 6.20% of variation was significantly distributed among populations within ecoregions (p< 0.0001, d.f. = 5), and that 2.62% variation was significantly partitioned among ecoregions (p< 0.0001,d.f. = 4); likewise, pairwise FST values between ecoregions were significant, except for APF-AMAF and AMAF-SCMS comparisons. In agreement, results of the hierarchical AMOVA for mtDNA showed that 46.14% of variation was significantly distributed among populations within ecoregions (p< 0.0001, d.f. = 5) and that 11.56% of variation was significantly partitioned among ecoregions (p< 0.0001, d.f. = 4). Pairwise θST values showed significant genetic differentiation (p< 0.0001) between AMAF and the other ecoregions, and between SCMS and all ecoregions except for PFS. The Mantel statistic for mtDNA genetic distance and geographic distance between populations was non-significant (null hypothesis: r ≤ 0; Z = -38.90, r = 0.0009, p = 0.476).
Discussion
Loss or alteration of native forest and yellow fever outbreaks represent severe threats to black-and-gold howler monkeys in the southernmost limit of the species range. In light of this scenario, this study assessed their regional genetic diversity, gene flow and connectivity patterns, revealing that they conform four genetically differentiated clusters. Our results may contribute to the upgrade of BGHM conservation status, and provide guidelines for the future management of remnant populations.
Genetic diversity and past demography of BGHM southernmost populations
In natural populations, genetic diversity provides the basis for the maintenance of evolutionary potential and adaptive capacity of individuals to face threats such as environmental change and disease [10]. The populations of black-and-gold howler monkeys studied here occupy sites in the southernmost part of the species’ range in northeastern Argentina and southern Paraguay (Fig 1) and showed mean microsatellites’ diversity (0.420 ± 0.082) similar to or lower than populations of congeneric species studied with some of the same markers employed here (Alouatta pigra: 0.430 [71] and 0.588 [72], Alouatta belzebul:0.640 [73], Alouatta palliata: 0.584 [74], suggesting that they are genetically impoverished, compared to other howlers. The population 2, located in the lower Paraná River, showed the highest genetic diversity estimates and shared haplotypes (Fig 2, Table A in S1 File), suggesting that it historically exchanged migrants with three other populations. Population 6, which inhabits the middle of the Iberá wetlands, showed the lowest genetic diversity estimates, suggesting that it could have been founded by a few individuals which faced dispersal limitations due to strong environmental restrictions. Population 9, located in the Argentinean Araucaria Moist Forest, showed five individuals with a unique, divergent haplotype (Fig 2), suggesting that it may be potentially reproductively isolated from the others. Alternatively, individuals with intermediate mitochondrial variants may have been extirpated by past yellow fever outbreaks in this region [22, 23]. BGHM from Pop 9 (sampled in 2006) showed a significant signal of inbreeding (Table 1), which could have been caused by genetic drift, suggesting that those howlers were already facing the erosion of genetic diversity, common to small endangered populations and, therefore, could have been immunologically depressed when the yellow fever virus attacked in 2007–2008. Our findings gain major importance in the context of recent virus outbreaks, since small populations of BGHM may exhibit increased susceptibility. In the next few decades, the high susceptibility of BGHM to yellow fever [24, 25] could act synergistically with other threats, putting these small and isolated populations at high extinction risk. It is worth noting that, in an epidemiological sense, the BGHM populations of humid Chaco are of utmost importance as genetic reservoirs of the species because yellow fever deaths have not been registered in this area during the last two episodes (2008/9-2017) [24, and Almeida MAB, pers. com.].
Past population expansion estimated with mitochondrial DNA data suggests that the populations located to the west of our sampling area expanded earlier than those located to the east (Fig 1, Table A in S1 File). Mismatch distribution results indicated that all populations expanded in South America during the post-glacial period (Last Glacial Maximum: 20,000–14,000 years ago, [75]). Moreover, the dating of the expansion event of populations 4 and 5 (Table A in S1 File), located in humid Chaco, is consistent with a recent comparative biogeographic study of neotropical primates, which suggests that most species currently inhabiting drier open habitats (such as the humid Chaco) have arrived there in the Pleistocene, from nearby rainforest habitats [76]. The lack of a strong evidence for demographic expansion in populations 7 and 8, located in the Alto Paraná Atlantic Forest, suggests demographic stability for this biome during the Pleistocene, in line with previous studies in other forest-dependent taxa inhabiting this region (e.g., birds, [77]). Conversely, Pop 9, which lies in the extreme west of the total distribution of the AMAF ecoregion (Fig 1), showed significant SSD and Rg indexes, which could be interpreted as a demographic expansion (Table A in S1 File), but more data are needed to explore this hypothesis.
Genetic structure of BGHM southernmost populations
Black-and-gold howler monkeys are arboreal primates still present in some patchy and impoverished forests [19, 30]. The different analytical methods with varying assumptions employed in this study (Bayesian clustering, FST, hierarchical AMOVA, and landscape analyses) were concordant in detecting significant present-day genetic structuring among the examined BGHM populations (Fig 4, Tables B and C in S1 File). Four distinct genetic clusters seem to best explain the nuclear diversity of BGHM inhabiting the southernmost part of the species range. The observed clustering pattern cannot be explained by an Isolation-by-Distance model (non-significant Mantel statistic), but, rather, seems to reflect the concurrent effects of multiple ecological, environmental and contemporary anthropogenic factors acting on the populations inhabiting different ecoregions. For example, populations 1 and 2, which share genetic ancestry and differentiate from others, are located nearby each other and connected by the riparian forest remnants of the Paraguay and Paraná Rivers (Fig 1).This result, as well as the mtDNA shared diversity and lack of historical differentiation among humid Chaco, Paraná Flooded Savanna and Alto Paraná Atlantic Forest (as indicated by θST) seem to support the hypothesis of dispersal via riparian forests, which act as biological corridors enabling the movement of BGHM [16]. Past immigration of BGHM through the riparian forests of the Paraná and Paraguay Rivers could have also contributed to the high genetic diversity observed in Pop 2, compared to other BGHM populations (Table 1) However, more recently, the movements of howlers through these dispersal routes may have been prevented by recent deforestation. In northeastern Argentina and southern Paraguay, the ecosystems were flooded as a consequence of the building of the Yacyretá Hydroelectric Dam in the 1970s [3,4], interrupting the terrestrial and riparian forest corridors that BGHM may have used in the past. In addition, we found that Pop 3, which lies in a remnant protected forest (Estación Biológica de Corrientes), surrounded by grassland, crops, and urbanized areas, clustered separately from all other populations. This significant genetic differentiation could have been caused a long history of forest exploitation and disturbance in this area, promoted by non-native human settlements documented since early 17th century [18, 78]. Populations 4, 5 and 6, which inhabit patches of humid Chaco forest of different size and under variable degrees of protection, comprised another genetically differentiated cluster. Finally, populations 7, 8 and 9, which all lie in Misiones province in Argentina, upstream the Yacyretá Dam on the Paraná River, clustered together with Pop 10, located in Corrientes province, more distant but connected to the others via the Uruguay River riparian forest (Fig 1). Therefore, clusters 1, 2 and 3 involve populations placed upstream the Yacyretá Hydroelectric Dam on the Paraná River, while cluster 4 includes populations located downstream this dam. Misiones populations seem to maintain gene flow, but remain genetically isolated from more distant western populations that are immersed in more disturbed and isolated forest patches. This pattern of connectivity between nearby Misiones BGHM populations could be promoted via dispersal through a relatively well-preserved forest, as consequence of protection policies implemented in this province, such as the “Misiones Green Corridor” [79].
Landscape resistance seems to play a significant role in influencing the patterns of genetic structure observed in BGHM populations, as indicated by Circuistscape analyses (Fig 5), significant partial Mantel statistic between genetic distance and resistance, and non-significant partial Mantel statistic between genetic and geographic distance. The flow of the current showed in the maps (Fig 5) connects populations in a manner that corresponds to the four genetic clusters identified by microsatellites-based Bayesian approaches (Fig 4). Overall, our results suggest that crop/grassland fields could exert resistance to dispersal, and, consequently, to gene flow in BGHM. Populations connected by continuous forest or by relatively well-preserved riparian forests seem to share more similarities at nuclear loci than with populations immersed in more disturbed forests or with populations isolated by anthropogenic modifications, such as deforestation or building of dams. Therefore, our findings suggest that different levels of forest fragmentation that affect the studied populations may have exerted an important impact on the dispersal of the howlers, indicating that connectivity of the monkeys’ habitats is highly relevant for maintaining genetic connectivity across the landscape. These results highlight the importance of preserving continuous native forests, including riparian vegetation, for BGHM dispersal. In line with previous studies in other mammals inhabiting this region, our findings seem to indicate that anthropogenic modifications on native forests and depletion of continuous riparian forests, increasingly ongoing in northeastern Argentina, southern Paraguay and southeastern Brazil, prevent the dispersal of native fauna, and may lead to population isolation [80].
Implications for the conservation and management of BGHM
Based on significant differences in allele frequency distributions and significant divergence in mitochondrial and nuclear loci, BGHM populations inhabiting northeastern Argentina and southern Paraguay comprise four different Management Units. Hence, we recommend that, to preserve the BGHM gene pool in the species’ southernmost range, these four main differentiated population clusters must be given high conservation priority. The pattern of significant genetic differentiation and restricted gene flow between BGHM populations revealed in the present study might result from increasing levels of forest loss and other anthropogenic modifications, such as the flooding of large habitat areas, derived from dam building, which severely limits the howlers’ ability to disperse and cross intermediate habitat regions [7]. In order to maintain or restore the natural gene flow between BGHM populations in the studied regions, the continuous forest patches, as well as the remaining riparian forests, should be protected and preserved. Further genetic studies spanning the entire geographical range of BGHM should help expand our knowledge on the patterns of gene flow and complement these conservation guidelines.
Based on the estimates of the effective population size obtained for the two biggest populations (Ne = 26 and 40), applying the equation for heterozygosity loss (Eqn. 4 in [11]), we can anticipate that the studied BGHM populations will lose heterozygosity below the 25% quantile of the current values in less than 50 generations. We also found that the populations at the southernmost limit of the species range have a reduced effective size and may be genetically depleted to face threatening events such as yellow fever outbreaks, which could rapidly affect all individuals in most of the species’ distribution area. Therefore, following the recently proposed genetic IUCN criterion (see Fig 2a in [11]), the studied BGHM populations should be classified as “Endangered”. We considered that the current IUCN global classification of BGHM as “Least Concern” [26] underestimates the treats to which each of the remaining BGHM populations are subjected; therefore, we propose a re-classification of the global status of the species to “Vulnerable”. This proposed global re-classification category is in line with country-level rankings of Argentina (“Vulnerable”) [81] and Brazil (“Near Threatened”) [27], that comply with the IUCN criteria “A4cd” which refers to a population reduction of 30% in 3 generations (4), where the reduction or its causes may not have ceased, may not be understood, or may not be reversible; mainly due to: (c) a decline in the area of occupancy, extent of occurrence, and/or quality of habitat, and (d) exploitation levels due to hunting or illegal traffic (pet trade). Brazil also adheres to the criterion (e) which refers to the effects of pollutants, introduced taxa, hybridization, competitors, pathogens, or parasites, referring to BGHM vulnerability to yellow fever epidemics. We recommend that, given the high susceptibility of black-and-gold howler monkeys to the yellow fever virus, this criterion should be also adopted by the international IUCN ranking. The current IUCN global status of Alouatta caraya seems to rely heavily on the species’ wide geographic distribution range (Fig 1), which includes large areas of unsuitable habitat and, therefore, does not adequately mirror the actual population density. Thereby, as it occurs with other taxa [82], the actual distribution range of BGHM is overestimated, while their level of risk is underestimated. Based on concurrent genetic and non-genetic evidence mentioned above, we recommend that the IUCN upgrades the global conservation status of Alouatta caraya to “Vulnerable”.
In sum, the present study contributes novel evidence supporting contemporary restricted gene flow between BGHM inhabiting the southernmost portion of the species’ geographic distribution range, and identifies four distinct Management Units for conservation. We also anticipate that most of the studied populations would loss heterozygosity in the mid-term and recommend that the IUCN global conservation status of BGHM is upgraded to “Vulnerable”. Lost habitat connectivity can play a significant role in preventing gene flow between isolated populations and, if not reverted, such a pattern may severely affect the survival capacity of BGHM. Our results have direct implications for the conservation of howlers and should be taken into account by policy makers when taking decisions, drafting management plans or designing reintroduction projects of these vulnerable Neotropical primates.
Supporting information
Table A. Summary estimates of mitochondrial diversity. Genetic diversity estimates, results of neutrality tests and demographic parameters for black-and-gold howler monkeys sampled in ten populations from Northeastern Argentina and Southern Paraguay, based on 491-bp mtDNA Control Region fragment sequences. H: number of haplotypes (n: number of sequences). PS: number of polymorphic sites; h: haplotypic diversity± standard deviation (SD); π: nucleotidic diversity ± standard deviation (SD); Tajima’s (1989) D; Fu’s (1997) Fs; Ramos-Onsins and Rozas’s (2002) R2; SSD: Sum of Squared Deviations, Rg: Raggedness index; Tau: mode of the unimodal mismatch distribution; TSE: time since population expansion (in years before present). Table B. Results of modified Structure procedure. The most likely number of differentiated genetic groups (K = 4, column shaded in grey) found to better explain the variation observed in the genotypic dataset (ten microsatellite loci) of black-and-gold howler monkeys from Northeastern Argentina and Southern Paraguay. The Max-ofMedian (MaxMedK) and Median-of-Median (MedMedK) indexes (Puechmaille, 2016), taken at the 0.80 threshold of membership proportion, were used as conservative estimators of K. The different sampling schemes tested were: 1) the full dataset (n = 138, ten sites), 2) subsampling Pop 2 and 3 to obtain more even sample sizes (n = 88, ten sites), 3) original dataset but removing the least sampled populations (n = 123, seven sites), and 4) the dataset obtained by a combination of the two latter strategies (n = 73, seven sites) (see main text for further details). For each sampling scheme, 20 replicate STRUCTURE runs were performed from K = 2 to K = 10. Table C. Genetic structure observed in populations of black-and-gold howler monkeys from Northeastern Argentina and Southern Paraguay. Pairwise FST values for ten microsatellite loci (below diagonal) and pairwise ΦST values for 512-bp mtDNA Control Region fragment sequences (above diagonal). Significant values after Bonferroni correction are shown in bold (α = 0.0005).
(PDF)
Acknowledgments
The authors are grateful to H. Argibay, M. Uhart, H. Ferreyra and Global Health Program of Wildlife Conservation Society for providing access to tissue samples; E. Fernandez-Duque for facilitating the sampling of population 5; L Jerusalinsky, V Fortes, MAB Almeida, R Leny Cuellar, JL Cartes, and S Saldivar Bellassai for providing updated information on the species conservation status in Brazil, Bolivia, and Paraguay; and C De Angelo for helping with Circuitscape analyses. This study used resources provided by the Servicio de Huellas Digitales Genéticas, Facultad de Farmacia y Bioquímica, Universidad de Buenos Aires. Special thanks are due to Julián, Ivy and Atenea Baigorria, and to Sergio and Indio Quintana for their time and patience. LIO, CIM, MC, and DC are members of Carrera del Investigador Cientfico of CONICET (Consejo Nacional de Investigaciones Científicas y Técnicas, Argentina).
Data Availability
Genotypic data are available from 10.5281/zenodo.846390. Mitochondrial DNA sequences have been uploaded to GenBank (accession numbers: MF095740, MF095741, MF095742, MF095743, MF095744, MF095745, MF095746, MF095747, MF095748, MF095749, MF095750, MF095751, MF095752, MF095753, MF095754).
Funding Statement
This project was funded by a PIP CONICET Research Grant to LIO (no. 114 201101 00322; www.conicet.gov.ar). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Groves CP, Wilson DE, Reeder DM, editors. Mammal Species of the World: A Taxonomic and Geographic Reference, 3rd ed Baltimore, USA: Johns Hopkins UniversityPress; 2005; pp. 148–152. [Google Scholar]
- 2.Boletta PE, Ravelo AC, Planchuelo AM, Grilli M. Assessing deforestation in the Argentine Chaco. Forest Ecol Manag. 2006; 228: 108–114. [Google Scholar]
- 3.Grau HR, Aide TM. Globalization and Land-Use Transitions in Latin America. Ecol. Soc. 2008; 13, 16 Available at: http://www.ecologyandsociety.org/vol13/iss2/art16/. [Google Scholar]
- 4.Bauni V, Schivo F, Capmourteres V, Homberg M. Ecosystem loss assessment following hydroelectric dam flooding: The case of Yacyretá, Argentina. Remote Sens App: Soc Environ. 2015; 1: 50–60. [Google Scholar]
- 5.Ribeiro MC, Metzger JP, Martensen AC, Ponzoni FJ, Hirota MM. The Brazilian Atlantic Forest: How much is left, and how is the remaining forest distributed? Implications for conservation. Biol Cons. 2009; 142: 1141–1153. [Google Scholar]
- 6.Estrada A, Garber PA, Rylands AB, Roos C, Fernandez-Duque E, Di Fiore A et al. Impending extinction crisis of the world’s primates: Why primates matter. Sci Adv. 2017; 3: e1600946 Available from: http://advances.sciencemag.org/content/3/1/e1600946. doi: 10.1126/sciadv.1600946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oklander LI, Kowalewski MM, Corach D. Genetic Consequences of Habitat Fragmentation in Black-and-Gold Howler (Alouatta caraya) Populations from Northern Argentina. Int J Primatol. 2010; 31: 813–832. [Google Scholar]
- 8.Lecompte E, Bouanani M-A, de Thoisy B, Crouau-Roy B. How do rivers, geographic distance, and dispersal behavior influence genetic structure in two sympatric New World monkeys? Am J Primatol. 2017; doi: 10.1002/ajp.22660 [DOI] [PubMed] [Google Scholar]
- 9.Wang W, Yao M. Fine-scale genetic structure analyses reveal dispersal patterns in a critically endangered primate, Trachypithecus leucocephalus. Am J Primatol. 2017; doi: 10.1002/ajp.22635 [DOI] [PubMed] [Google Scholar]
- 10.Allendorf FW, Luikart GH, Aitken SN. Conservation and the Genetics of Populations 2nd ed New Jersey, USA: Wiley-Blackwell; 2010; pp. 587. [Google Scholar]
- 11.Willoughby JR, Sundaram M, Wijayawardena BK, Kimble SJA, Ji Y, Fernandez NB, et al. The reduction of genetic diversity in threatened vertebrates and new recommendations regarding IUCN conservation rankings. Biol Cons. 2015; 191: 495–503. [Google Scholar]
- 12.Vigilant L, Guschanski K. Using genetics to understand the dynamics of wild primate populations. Primates. 2009; 50: 105–120. doi: 10.1007/s10329-008-0124-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruiz-García M, Shostell JM. Phylogeny, Molecular Population Genetics, Evolutionary Biology and Conservation of the Neotropical Primates. Hauppauge NY, USA: Nova Science Pub Inc; 2016; pp. 669. [Google Scholar]
- 14.Zunino GE, González V, Kowalewski M, Bravo S. Alouatta caraya. Relationships among habitat, density and social organization. Primate Report. 2001; 61: 37–46. [Google Scholar]
- 15.Agostini I, Pizzio E, De Angelo C, Di Bitetti MS. Population Status of Primates in the Atlantic Forest of Argentina. Int J Primatol. 2015; 36: 244–258. [Google Scholar]
- 16.Ascunce MS, Hasson E, Mulligan CJ, Mudry MD. Mitochondrial sequence diversity of the southernmost extant New World monkey, Alouatta caraya. Mol Phylogenet Evol. 2007; 43: 202–215. doi: 10.1016/j.ympev.2006.10.004 [DOI] [PubMed] [Google Scholar]
- 17.Rumiz DI. Alouatta caraya: Population density and demography in Northern Argentina. Am J Primatol. 1990; 21: 279–294. [DOI] [PubMed] [Google Scholar]
- 18.Brown A.D. & Zunino GE. Hábitat, densidad y problemas de conservación de los primates de Argentina. Vida Silv Neotrop. 1994; 3: 30–40. [Google Scholar]
- 19.Bicca-Marques JC. How do howler monkeys cope with habitat fragmentation? In:Marsh LK, Chapman CA, editors. Primates in fragments: complexity and resilience. New York, USA:Springer Press; 2013, pp. 283–303. [Google Scholar]
- 20.Silva FE, Bicca-Marques JC. Do Patch Size and Interpatch Distance Influence the Distribution of Brown Howler Monkeys (Alouatta guaribaclamitans) in a Fragmented Landscape in South Brazil? In:Marsh LK, Chapman CA, editors. Primates in fragments: complexity and resilience. New York, USA: Springer Science+Business Media; 2013, pp. 137–145. [Google Scholar]
- 21.Moreno ES, Agostini I, Holzmann I, Di Bitetti MS, Oklander LI, Kowalewski MM, et al. Yellow fever impact on brown howler monkeys (Alouatta guariba clamitans) in Argentina: a metamodelling approach based on population viability analysis and epidemiological dynamics. Mem Inst Oswaldo Cruz. 2015; 110: 865–876. doi: 10.1590/0074-02760150075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holzmann I, Agostini I, Areta JI, Ferreyra H, Beldomenico P, Di Bitetti MS. Impact of yellow fever outbreaks on two howler monkey species (Alouatta guariba clamitans and A. caraya) in Misiones, Argentina. Am J Primatol. 2010; 72: 475–480. doi: 10.1002/ajp.20796 [DOI] [PubMed] [Google Scholar]
- 23.de Almeida MAB, dos Santos E, da Cardoso CJ, da Fonseca DF, Noll CA, Silveira VR, et al. Yellow fever outbreak affecting Alouatta populations in southern Brazil (Rio Grande do Sul State), 2008–2009. Am J Primatol. 2012; 74: 68–76. doi: 10.1002/ajp.21010 [DOI] [PubMed] [Google Scholar]
- 24.Bicca-Marques JC, Calegaro-Marques C, Rylands AB, Strier KB, Mittermeier RA, De Almeida MAB, et al. Yellow fever threatens Atlantic Forest primates. Sci. Adv. e-letter. 2017. Available at: http://advances.sciencemag.org/content/3/1/e1600946/tab-e-letters. [Google Scholar]
- 25.Kean S. Onthetrail of yellowfever. On the trail of yellow fever. Science 357 (6352), 637–64. doi: 10.1126/science.357.6352.637 [DOI] [PubMed] [Google Scholar]
- 26.Fernandez-Duque E, Wallace RB, Rylands AB. Alouatta caraya. The IUCN Red List of Threatened Species 2008; e.T41545A10496784. http://dx.doi.org/10.2305/IUCN.UK.2008.RLTS.T41545A10496784.en
- 27.Ludwig G, Bicca-Marques JC, Rímoli J, Teixeira da Cunha RG, Alves SL, Martins V, et al. Avaliação do Risco de Extinção de Alouatta caraya (Humboldt,1812) no Brasil. 2015; 1. http://www.icmbio.gov.br/portal/faunabrasileira/estado-de-conservacao/7176-mamiferos-alouatta-caraya-bugio-preto.
- 28.Aguirre LF, Aguayo R, Balderrama JA, Cortez C, Tarifa T, Rocha O. Libro Rojo de la fauna silvestre de vertebrados de Bolivia. La Paz, Bolivia: Ministerio de Medio Ambiente y Agua, 2009; pp. 204 http://bolivianamphibianinitiative.org/wp-content/uploads/2015/07/Libro-Rojo-Bolivia_2009_vs1.pdf. [Google Scholar]
- 29.Moritz C. Applications of mitochondrial DNA analysis in conservation: a critical review. Mol Ecol. 1994; 3: 401–411. [Google Scholar]
- 30.Zunino GE, Kowalewski MM, Oklander LI, González V. Habitat fragmentation and population size of the black and gold howler monkey (Alouatta caraya) in a semideciduous forest in Northern Argentina. Am J Primatol. 2007; 69: 966–975. doi: 10.1002/ajp.20389 [DOI] [PubMed] [Google Scholar]
- 31.Olson DM, Dinerstein E, Wikramanayake ED, Burgess ND, Powell GV, Underwood EC, et al. Terrestrial Ecoregions of the World: A New Map of Life on Earth: A new global map of terrestrial ecoregions provides an innovative tool for conserving biodiversity. BioSci 2001; 51: 933–938. [Google Scholar]
- 32.Oklander LI, Marino M, Zunino GE, Corach D. Preservation and extraction of DNA from feces in howler monkeys (Alouatta caraya). Neotrop Primates. 2004; 12: 59–63. [Google Scholar]
- 33.Green MR, Sambrook J. Molecular Cloning: A Laboratory Manual, 4th ed New York, USA: Cold Spring Harbor Laboratory Press; 2012, pp. 47–54. [Google Scholar]
- 34.Oklander LI, Zunino GE, Di Fiore A, Corach D. Isolation, characterization and evaluation of 11 autosomal STRs suitable for population studies in black and gold howler monkeys Alouatta caraya. Mol Ecol Notes. 2007; 7: 117–120. [Google Scholar]
- 35.Arandjelovic M, Guschanski K, Schubert G, Harris TR, Thalmann O, Siedel H, et al. Two-step multiplex polymerase chain reaction improves the speed and accuracy of genotyping using DNA from noninvasive and museum samples. Mol Ecol Res. 2009; 9: 28–36. [DOI] [PubMed] [Google Scholar]
- 36.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32: 1792–1797. doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura K, Stecher G, Peterson D, Filipski A, and Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013; 30: 2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soto-Calderón ID, Clark Nicholas J, Wildschutte Julia VH, DiMattio K, Jensen-Seaman MI, Anthony NM. Identification of species-specific nuclear insertions of mitochondrial DNA (NUMTs) in gorillas and their potential as population genetic markers. Mol Phyl Evol. 2014; 81: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. 2004; MolEcol Notes, 4: 535–538. [Google Scholar]
- 40.Rousset F. genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Res. 2008; 8: 103–106. [DOI] [PubMed] [Google Scholar]
- 41.Waits LP, Luikart G, Taberlet P. Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Mol Ecol. 2001; 10: 249–256. [DOI] [PubMed] [Google Scholar]
- 42.Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics. 2012; 28: 2537–2539. doi: 10.1093/bioinformatics/bts460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goudet J. FSTAT (Version 1.2): A Computer Program to Calculate F-Statistics. J Hered. 1995; 86: 485–486. [Google Scholar]
- 44.R Development Core Team. 2017. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: URL: http://www.R-project.org. [Google Scholar]
- 45.Queller DC, Goodnight KF. Estimating Relatedness Using Genetic Markers. Evolution. 1989; 43: 258–275. doi: 10.1111/j.1558-5646.1989.tb04226.x [DOI] [PubMed] [Google Scholar]
- 46.Blouin MS, Parsons M, Lacaille V, Lotz S. Use of microsatellite loci to classify individuals by relatedness. Mol Ecol. 1996; 5: 393–401. [DOI] [PubMed] [Google Scholar]
- 47.Waples RS, Do C. LDNE: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Resour. 2008; 8: 753–756. doi: 10.1111/j.1755-0998.2007.02061.x [DOI] [PubMed] [Google Scholar]
- 48.Waples RS. A bias correction for estimates of effective population size based on linkage disequilibrium at unlinked gene loci. Conserv Genet. 2006; 7: 167. [Google Scholar]
- 49.Hill WG. Estimation of effectivepopulation size from data on linkage disequilibrium. Genetics Res. 1981; 38: 209–216. [Google Scholar]
- 50.Smouse PE, Peakall R. Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity. 1999; 82: 561–573. [DOI] [PubMed] [Google Scholar]
- 51.Peakall R, Ruibal M, Lindenmayer DB. Spatial autocorrelation analysis offers new insights into gene flow in the Australian bush rat, Rattus fuscipes. Evolution. 2003; 57: 1182–1195. [DOI] [PubMed] [Google Scholar]
- 52.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000; 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Puechmaille SJ. The program structure does not reliably recover the correct population structure when sampling is uneven: subsampling and new estimators alleviate the problem. Mol Ecol Res. 2016; 16: 608–627. [DOI] [PubMed] [Google Scholar]
- 54.Francis RM. pophelper: an R package and web app to analyse and visualize population structure. Mol Ecol Res. 2017; 17: 27–32. [DOI] [PubMed] [Google Scholar]
- 55.McRae BH. Isolation by resistance. Evolution 2006; 60: 1551–1561. [PubMed] [Google Scholar]
- 56.Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003; 14: 927–930. [Google Scholar]
- 57.Chiappero MB, Sommaro LV, Priotto JW, Wiernes MP, Steinmann AR, Gardenal CN. Spatio-temporal genetic structure of the rodent Calomys venustus in linear, fragmented habitats. J Mammal. 2016; 97: 424–435. [Google Scholar]
- 58.Adamack AT, Gruber B. PopGenReport: simplifying basic population genetic analyses in R. Methods Ecol Evol 2014; 5(4): 384–387. [Google Scholar]
- 59.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res. 2010; 10: 564–567. [DOI] [PubMed] [Google Scholar]
- 60.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009; 25: 1451–1452. doi: 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 61.Ramos-Onsins SE, Rozas J. Statistical Properties of New Neutrality Tests Against Population Growth. Mol Biol Evol. 2002; 19: 2092–2100. [DOI] [PubMed] [Google Scholar]
- 62.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989; 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997; 147: 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hudson RR. Gene genealogies and the coalescent process. Oxf Surv Evol Biol. 1990; 7: 1–44. [Google Scholar]
- 65.Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999; 16: 37–48. [DOI] [PubMed] [Google Scholar]
- 66.Leigh JW, Bryant D. popart: full-feature software for haplotype network construction. Methods Ecol Evol. 2015; 6: 1110–1116. [Google Scholar]
- 67.Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol. 1992; 9: 552–569. [DOI] [PubMed] [Google Scholar]
- 68.Harpending HC. Signature of Ancient Population Growth in a Low-Resolution Mitochondrial DNA Mismatch Distribution. Human Biol. 1994; 66: 591–600. [PubMed] [Google Scholar]
- 69.Kowalewski MM, Garber PA. Mating promiscuity and reproductive tactics in female black and gold howler monkeys (Alouatta caraya) inhabiting an island on the Parana river, Argentina. Am J Primatol. 2010; 72: 734–748. doi: 10.1002/ajp.20838 [DOI] [PubMed] [Google Scholar]
- 70.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992; 131: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Winkler LA, Zhang X, Ferrell R, Wagner R, Dahl J, Peter G, Sohn R. Geographic microsatellite variability in Central American howling monkeys. Int J Primatology. 2004;25(1): 197–210. [Google Scholar]
- 72.Van Belle S, Estrada A, Strier KB, Di Fiore A. Genetic structure and kinship patterns in a population of black howler monkeys, Alouatta pigra, at Palenque National Park, Mexico. Am J Primatol. 2012; 74: 948–957. doi: 10.1002/ajp.22047 [DOI] [PubMed] [Google Scholar]
- 73.Gonçalves EC, Silva A, Barbosa MSR, Schneider MPC. Isolation and characterization of microsatellite loci in Amazonian red‐handed howlers Alouatta belzebul (Primates, Plathyrrini). Mol Ecol Res. 2004; 4: 406–408. [Google Scholar]
- 74.Milton K, Lozier JD, Lacey EA. Genetic structure of an isolated population of mantled howler monkeys (Alouatta palliata) on Barro Colorado Island, Panama. Conserv Genet. 2009; 10: 347. [Google Scholar]
- 75.Anderson DE, Goudie AS, Parker AG. Global Environments through the Quaternary, 2nd ed Oxford, UK: Oxford University Press; 2013; pp. 424. [Google Scholar]
- 76.Lynch Alfaro JW, Cortés-Ortiz L, Di Fiore A, Boubli JP. Special issue: Comparative biogeography of Neotropical primates. Mol Phylogenet Evol. 2015; 82: 518–529. doi: 10.1016/j.ympev.2014.09.027 [DOI] [PubMed] [Google Scholar]
- 77.Batalha-Filho H, Cabanne GS, Miyaki CY. Phylogeography of an Atlantic forest passerine reveals demographic stability through the last glacial maximum. Mol Phylogenet Evol. 2012; 65: 892–902. doi: 10.1016/j.ympev.2012.08.010 [DOI] [PubMed] [Google Scholar]
- 78.Gasparri NI, Grau HR Deforestation and fragmentation of Chaco dry forest in NW Argentina (1972–2007). Forest EcolManag. 2009; 258(6): 913–921. [Google Scholar]
- 79.De Angelo C, Paviolo A, Wiegand T, Kanagaraj R, Di Bitetti MS. Understanding species persistence for defining conservation actions: A management landscape for jaguars in the Atlantic Forest. Biol Cons. 2013; 159: 422–433. [Google Scholar]
- 80.Paviolo A, Angelo CD, Ferraz KMPMB, Morato RG, Pardo JM, Srbek-Araujo AC, et al. A biodiversity hotspot losing its top predator: The challenge of jaguar conservation in the Atlantic Forest of South America. Sci Reports. 2016; 6: srep37147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Agostini I, Aprile G, Baldovino MC, Brividoro M, Di Bitetti M, Fantini L, et al. Orden Primates In: Libro rojo de los mamíferos amenazados de la Argentina. Ojeda RA, Chillo V, Díaz Isenrath GV, eds. Mendoza, Argentina: SAREM; 2012, pp. 81–86. [Google Scholar]
- 82.Ramesh V, Gopalakrishna T, Barve S, & Melnick DJ. IUCN greatly underestimates threat levels of endemic birds in the Western Ghats. Biol Cons.2017; 210: 205–221. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table A. Summary estimates of mitochondrial diversity. Genetic diversity estimates, results of neutrality tests and demographic parameters for black-and-gold howler monkeys sampled in ten populations from Northeastern Argentina and Southern Paraguay, based on 491-bp mtDNA Control Region fragment sequences. H: number of haplotypes (n: number of sequences). PS: number of polymorphic sites; h: haplotypic diversity± standard deviation (SD); π: nucleotidic diversity ± standard deviation (SD); Tajima’s (1989) D; Fu’s (1997) Fs; Ramos-Onsins and Rozas’s (2002) R2; SSD: Sum of Squared Deviations, Rg: Raggedness index; Tau: mode of the unimodal mismatch distribution; TSE: time since population expansion (in years before present). Table B. Results of modified Structure procedure. The most likely number of differentiated genetic groups (K = 4, column shaded in grey) found to better explain the variation observed in the genotypic dataset (ten microsatellite loci) of black-and-gold howler monkeys from Northeastern Argentina and Southern Paraguay. The Max-ofMedian (MaxMedK) and Median-of-Median (MedMedK) indexes (Puechmaille, 2016), taken at the 0.80 threshold of membership proportion, were used as conservative estimators of K. The different sampling schemes tested were: 1) the full dataset (n = 138, ten sites), 2) subsampling Pop 2 and 3 to obtain more even sample sizes (n = 88, ten sites), 3) original dataset but removing the least sampled populations (n = 123, seven sites), and 4) the dataset obtained by a combination of the two latter strategies (n = 73, seven sites) (see main text for further details). For each sampling scheme, 20 replicate STRUCTURE runs were performed from K = 2 to K = 10. Table C. Genetic structure observed in populations of black-and-gold howler monkeys from Northeastern Argentina and Southern Paraguay. Pairwise FST values for ten microsatellite loci (below diagonal) and pairwise ΦST values for 512-bp mtDNA Control Region fragment sequences (above diagonal). Significant values after Bonferroni correction are shown in bold (α = 0.0005).
(PDF)
Data Availability Statement
Genotypic data are available from 10.5281/zenodo.846390. Mitochondrial DNA sequences have been uploaded to GenBank (accession numbers: MF095740, MF095741, MF095742, MF095743, MF095744, MF095745, MF095746, MF095747, MF095748, MF095749, MF095750, MF095751, MF095752, MF095753, MF095754).