Abstract
MEX-3 is a K-homology (KH) domain-containing RNA-binding protein first identified as a translational repressor in Caenorhabditis elegans, and its four orthologs (MEX-3A–D) in human and mouse were subsequently found to have E3 ubiquitin ligase activity mediated by a RING domain and critical for RNA degradation. Current evidence implicates human MEX-3C in many essential biological processes and suggests a strong connection with immune diseases and carcinogenesis. The highly conserved dual KH domains in MEX-3 proteins enable RNA binding and are essential for the recognition of the 3′-UTR and post-transcriptional regulation of MEX-3 target transcripts. However, the molecular mechanisms of translational repression and the consensus RNA sequence recognized by the MEX-3C KH domain are unknown. Here, using X-ray crystallography and isothermal titration calorimetry, we investigated the RNA-binding activity and selectivity of human MEX-3C dual KH domains. Our high-resolution crystal structures of individual KH domains complexed with a noncanonical U-rich and a GA-rich RNA sequence revealed that the KH1/2 domains of human MEX-3C bound MRE10, a 10-mer RNA (5′-CAGAGUUUAG-3′) consisting of an eight-nucleotide MEX-3–recognition element (MRE) motif, with high affinity. Of note, we also identified a consensus RNA motif recognized by human MEX-3C. The potential RNA-binding sites in the 3′-UTR of the human leukocyte antigen serotype (HLA-A2) mRNA were mapped with this RNA-binding motif and further confirmed by fluorescence polarization. The binding motif identified here will provide valuable information for future investigations of the functional pathways controlled by human MEX-3C and for predicting potential mRNAs regulated by this enzyme.
Keywords: crystal structure, E3 ubiquitin ligase, RNA, RNA binding protein, RNA-protein interaction, KH domain, MEX-3C, RNA-protein complex
Introduction
Specific protein–RNA interactions play crucial roles in biological processes, including transcriptional and translational regulation, RNA processing and transport, and mRNA stabilization and degradation (1). RNA-binding proteins often interact with their RNA targets via modular RNA-binding domains (RBDs)2 (2), such as the RNA-recognition motif (3, 4), double-stranded RBD (5–8), zinc-finger domain (9, 10), and heterogeneous nuclear ribonucleoprotein K-homology (KH) domain (11–16). The variable RBD architectures guarantee the proper functioning of RNA-binding proteins when dealing with the sequence and/or structure diversity of their RNA substrates.
The KH domain was originally identified as a conserved triple repeat in human heterogeneous nuclear ribonucleoprotein K that interacts with RNA (12) and is one of the most abundant RNA-binding domains in all three domains of life (14, 17–20). Multiple copies of the KH domain are often observed to enhance protein–nucleic acid–binding affinity and specificity (21–26). The KH domain is comprises ∼70 amino acid residues that form a structure in which a three-stranded β-sheet is packed against three α-helices (13–16, 27). KH domains can be classified into two types (type I and II) on the basis of their topology (18, 28). The type I fold exists typically in eukaryotic proteins, and the type II fold exists in prokaryotic proteins. Both types form a cleft consisting of various structural elements, including the α1 helix, α2 helix, GXXG motif, β2 strand, and the variable loop, that accommodates four single-stranded nucleotides (28, 29). The KH domain-containing proteins perform diverse cellular functions, and the recognition between KH domains and their RNA targets is essential for the establishment of post-transcriptional regulatory networks (28–33). It has been reported previously that, in multiple structures of KH–nucleic acid complexes, adenine and cytosine appear to be the dominant nucleotides in the two central RNA-binding sites of the KH domain, although a G-rich sequence was also reported to specifically interact with the KSRP KH3 domain (15, 27–29, 34). Unlike the other types of single-stranded nucleic acid-binding motifs (1), KH domains recognize the RNA substrates primarily through hydrogen bonding, electrostatic and polar interactions, and hydrophobic interactions, excluding the nucleic acid base-to-protein aromatic side chain stacking interaction.
The Caenorhabditis elegans MEX-3 (ceMEX-3) protein is one member of the KH domain-containing RNA-binding protein family (30). It contains two KH domains and has mainly been regarded as a translational repressor in mRNA post-transcriptional regulation, contributing to early embryonic development and the maintenance of germ line totipotency in adults (30, 31, 35, 36). The primary phenotype caused by a mutated MEX-3 is an abnormal generation of the body-wall muscles from the anterior blastomere. A family of MEX-3–homologous proteins (MEX-3A–D) was subsequently identified in humans and mice with all proteins retaining the highly conserved dual KH domains and acquiring a RING finger domain with ubiquitin E3 ligase activity at the C terminus, which is absent in ceMEX-3 (37). These four homologous mammalian MEX-3 proteins are thought to play similar roles as ceMEX-3. They mediate post-transcriptional regulation in a wide range of biological processes, including hypertension (38), postnatal growth (39), energy metabolism (40), cancer (41), and the immune response (32, 33, 42).
Of these four homologs, MEX-3C is known to be widely expressed in many tissues, especially in the spleen and thymus (43). It has been reported that MEX-3C is associated with several diseases. For instance, polymorphic variations in human MEX-3C (hMEX-3C) have been shown to be responsible for the susceptibility to essential hypertension (38). Other studies demonstrated that MEX-3C promotes postnatal growth through translational regulation of insulin-like growth factor 1 (Igf1) mRNA (39) and has a pivotal role in the regulation of energy balance because its mutation reduces adiposity and increases energy expenditure (40). MEX-3C has been identified as a chromosomal instability suppressor in cancer (41). MEX-3C has also been reported to be involved in the immune response where it directly or indirectly binds to the viral RNA and regulates the retinoic acid-inducible gene-1–mediated antiviral innate immune response (43). Lehner and co-workers (32, 33) revealed that MEX-3C regulates the expression of the HLA-A2 gene by binding to its 3′-UTR, thereby inducing the ubiquitin-dependent degradation of this mRNA. Moreover, RING-deleted MEX-3C maintains the ability to inhibit the translation of mRNA, similar to its C. elegans ancestor, but is no longer capable of degrading the mRNA.
However, the molecular mechanisms of mRNA degradation and translational repression induced by the recognition and binding of MEX-3C KH domains to the 3′-UTR of mRNAs remain to be elucidated. One related challenge would be the identification of the consensus RNA sequence recognized by the MEX-3C KH domains. Using in vitro selection and biochemical analysis, Pagano et al. (36) identified an RNA sequence motif ((A/G/U)(G/U)AGN0–8U(U/A/C)UA) that was strongly bound by the ceMEX-3 protein. In this sequence, two four-nucleotide half-sites separated by 0–8 nucleotides were shown to be recognized preferentially by two different KH domains connected via a 31-amino acid flexible linker. Because of the high sequence identity of the dual KH domains of hMEX-3C and ceMEX-3 (Fig. 1A) (37), this RNA-binding preference could possibly also apply to the hMEX-3C protein.
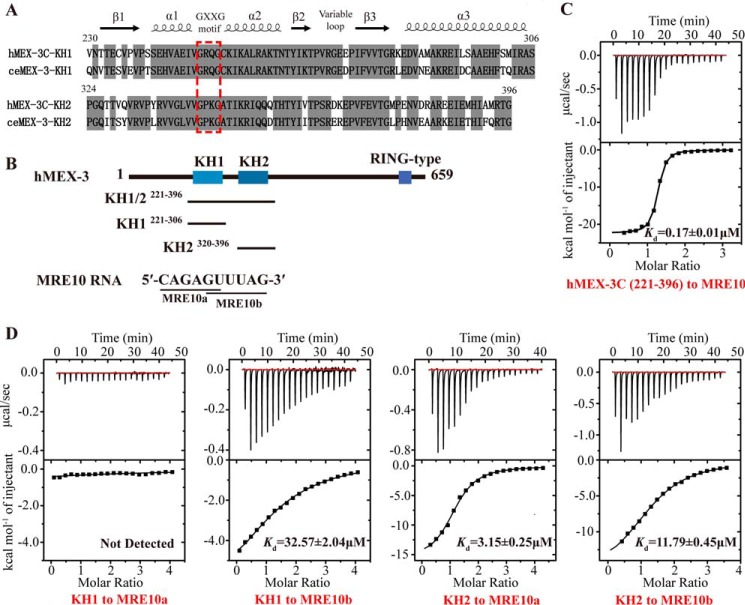
Figure 1.
The hMEX-3C KH domains bind the MRE10 RNA. A, sequence alignment of the KH domains of human MEX-3C and C. elegans MEX-3. Conserved residues are highlighted in gray. The GXXG motif is indicated by a red dashed box. The secondary structure elements of human MEX-3C KH domain assigned on the basis of the X-ray structure are shown at the top. B, the domain architecture of human MEX-3C and the sequence of MRE10 RNA. The black lines represent the hMEX-3C constructs used for ITC and structure determination. C, the ITC fitting result of hMEX-3C KH1/2 domains with MRE10 RNA. D, the ITC fitting results of individual hMEX-3C KH domain with MRE10a or MRE10b.
In this study, we determined the high-resolution crystal structures of hMEX-3C KH1–5′-GUUUAG-3′ and KH2–5′-CAGAGU-3′ complexes and analyzed the crucial interactions stabilizing the protein–RNA–binding interface. Furthermore, isothermal titration calorimetry (ITC) experiments were used to verify the crucial interactions contributing to binding specificity and to determine the consensus RNA motifs for two individual KH domains. Based on these results, we mapped the real RNA targets for the hMEX-3C KH domains at the 3′-UTR of HLA-A2 mRNA, which was further confirmed by fluorescence polarization (FP).
Results
hMEX-3C KH domains bind MRE10 RNA
Using the systematic evolution of ligands by exponential enrichment assay, Pagano et al. (36) previously reported an RNA aptamer, (A/G/U)(G/U)AGN0–8U(U/A/C)UA (MEX-3–recognition element (MRE)), that exhibited strong binding affinities with the KH domains of C. elegans MEX-3. Sequence alignment of hMEX-3C with ceMEX-3 shows that the sequence identity of the dual KH (KH1/2) domains is 73% (Fig. 1A). Because of the high sequence identity, it is reasonable to hypothesize that the KH1/2 domains of hMEX-3C will bind to MRE RNA in a manner similar to that of its C. elegans homolog. Using ITC, we therefore measured the binding affinity of hMEX-3C KH1/2 domains for MRE10, a 10-mer RNA (5′-CAGAGUUUAG-3′) consisting of an eight-nucleotide MRE motif flanked by a cytosine and a guanine at its 5′- and 3′-ends, respectively (Fig. 1B). Our ITC result shows that hMEX-3C KH1/2 domains bind MRE10 with a high affinity (Kd = 0.17 ± 0.01 μm) (Fig. 1C), suggesting that MRE10 could be a potential regulatory target for hMEX-3C.
KH domains can typically accommodate four nucleotides in their binding clefts (28). Pagano et al. (36) suggested that the MRE is a bipartite element that consists of two KH-recognition motifs separated by 0–8 nucleotides, (A/G/U)(G/U)AG and U(U/A/C)UA, and each one can be specifically recognized and bound by an individual KH domain of MEX-3 in C. elegans. To verify this in hMEX-3C, the 10-mer MRE10 was divided into two parts, namely MRE10a comprising the upstream sequence CAGAGU and MRE10b comprising the downstream sequence GUUUAG (Fig. 1B). ITC was used to test their binding affinities for the individual KH domains of hMEX-3C. As summarized in Fig. 1D, hMEX-3C KH1 binds MRE10b with a Kd value of 32.57 ± 1.9 μm, whereas no binding was detected between KH1 and MRE10a. In contrast, KH2 can bind both MRE10a and MRE10b, but the binding affinity for MRE10a was ∼4-fold stronger than that for MRE10b (Kd = 3.15 ± 0.25 μm for MRE10a and 11.79 ± 0.45 μm for MRE10b). Overall, our ITC results indicate that the KH domains in MEX-3 proteins maintain their binding specificities for RNA substrates across different species.
Overall structure of the KH1–MRE10b and KH2–MRE10a complexes
Although efforts have been made previously to determine the interaction preferences in the RNA recognition of KH domains, there remain some uncertainties that need further examination. For instance, although adenine or cytosine was often found to occupy the central position(s) of the nucleic acid-binding sites of KH domains (34), a G-rich RNA sequence has been reported to bind specifically with the KSRP KH3 domain (29). In the case of hMEX-3C, the KH-bound RNA sequences that have been identified (5′-CAGAGU-3′ and 5′-GUUUAG-3′) do not seem to have any apparent similarities with the RNA sequences from those reported KH–RNA complex structures, which inspired us to elucidate the structural details of the interaction between the RNA segments and the KH domains of hMEX-3C. Our initial attempts to crystalize hMEX-3C KH1/2 domains complexed with MRE10 were not successful. NMR experiments showed that the 1H-15N HSQC spectra of individual KH1 and KH2 domains can be aligned well with that of KH1/2 domains (supplemental Fig. S1), suggesting that the KH1 and KH2 domains do not interact with each other in solution. We therefore crystallized the individual KH1 and KH2 domains in the absence and presence of their RNA substrates and eventually solved the apo-form structure of the KH2 domain and the complex structures of KH1–MRE10b and KH2–MRE10a. The final refinement model of the KH1–MRE10b complex had an Rwork value of 17.2% and an Rfree value of 20.9% at 1.8-Å resolution, whereas the structure model of the KH2–MRE10a complex was refined to Rwork and Rfree values of 17.0 and 21.8%, respectively, at 2.0-Å resolution. Additionally, the structure of the KH2 domain in the apo form was determined at 2.5-Å resolution and was refined to a final Rwork value of 22.6% and Rfree value of 27.0% (supplemental Table S1). In the KH1–MRE10b and KH2–MRE10a crystal structures, each asymmetric unit contains only one complex, whereas the crystallographic asymmetric unit in the apo form of the KH2 domain comprises three molecules.
The overall structures of the two complexes and the apo-form KH2 are depicted in Fig. 2 and supplemental Fig. S2A. The structural comparison between the apo and the RNA-bound forms of the KH2 domain indicates no significant conformational changes upon RNA binding (root mean square deviation between Cα atoms of the two structures is 0.37 Å) (supplemental Fig. S2B), which is consistent with previous data about KH–RNA interactions (15, 16). In contrast, because the resolution of the apo-form KH2 is relatively low, the side chains of the residues at the RNA-binding interface in the complex structure were poorly defined and not built in the apo form, which limited further analysis of the structural changes upon RNA binding. A comparison of the KH1 domain in the KH1–MRE10b complex with the KH2 domain from the KH2–MRE10a complex shows high structural similarity with a root mean square deviation for Cα atoms of 0.42 Å (supplemental Fig. S2C). In all these structures, the KH domains form a typical KH fold in which three α-helices pack against a β-sheet composed of three antiparallel β-strands, their topological arrangement being β-α-α-β-β-α, which is common to all type I KH folds. Between the α1 and α2 helices in the KH1 and KH2 domains, there are 310 helices comprising the sequences Gly-Arg-Gln-Gly and Gly-Pro-Lys-Gly, respectively, which refer to the evolutionarily conserved GXXG motif observed in all KH domains. The β2 and β3 strands in both KH domains are linked by an eight-amino acid polypeptide, which represents the variable loop. These secondary structural motifs form a typical RNA-binding cleft where the GXXG motif and the variable loop function together as a “molecular vise” to grip the RNA segment (Fig. 2).
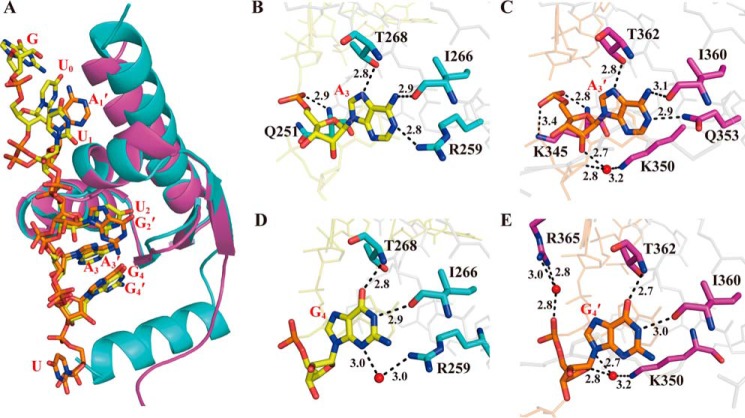
Figure 2.
Structure of the hMEX-3C KH domains in complex with RNA segments. A, schematic representation of the KH1–GUUUAG complex structure. In the left-hand panel, the KH1 domain is shown in cyan, and the MRE10b RNA (GUUUAG) is shown as a stick representation. The right-hand panel shows the electrostatic surface potential of the KH1–GUUUAG complex surface. The N-terminal helix of the KH1 domain is not an original component of KH domain architecture, and its formation may be induced by the addition of the expression tag. B, schematic representation of the KH2–CAGAGU complex structure. The left-hand panel shows the KH2 domain in magenta and the observable nucleotides of MRE10a (AGAGU) as sticks. The right-hand panel shows the electrostatic surface potential of the KH2–CAGAGU complex surface. Regions of electropositive and electronegative potential are shown in blue and red, respectively. The Fo − Fc maps before inclusion of RNAs are shown as green mesh (contoured at 2.0 σ), and the simulated annealing 2Fo − Fc composite omit maps are shown as purple mesh (contoured at 1.0 σ).
The structural alignment of the RNA segments in these two complexes indicates that the KH1 and KH2 domains bind to their RNA ligands in a similar manner (Fig. 4A). All six RNA nucleotides were built in the KH1–GUUUAG complex, whereas only five nucleotides were observed for the KH2–CAGAGU complex because the electron density of the first cytosine was difficult to trace (Fig. 2). In the structure of the KH2–CAGAGU complex (Fig. 2B), the core recognition sequence (AGAG) lies upon the α/β platform and has direct contacts with the protein, resembling the KH–RNA complex structures solved previously (15, 16, 44), at the four canonical RNA-binding positions in the KH domain (referred to as positions 1, 2, 3, and 4 below). Conversely, in the KH1–GUUUAG complex (Fig. 2A), five sequential nucleotides (UUUAG) contact the KH domain. The first uracil interacts specifically at the far side of the binding cleft with the residues from α1, α2, and α3 helices (hereafter called position 0), whereas the core recognition sequence (UUAG) occupies the RNA-binding positions 1–4. The uracil at the 3′-end of the CAGAGU complexed with the KH2 domain and the guanine at the 5′-end of the GUUUAG complexed with the KH1 domain have no obvious interactions with the KH2 and KH1 domains and are most likely stabilized by crystal packing. This is along expected lines for the KH1/2–MRE10 interaction in which the binding modes in both the individual KH–RNA complexes are preserved and the above mentioned uracil and guanine are specifically recognized by the adjacent KH1 and KH2 domains, respectively. Interestingly, the RNA-binding mode in the KH1–GUUUAG complex is somewhat unexpected because it has been suggested previously that UUUA binds the KH1 domain at the four canonical RNA-binding positions (36). Our crystal structure, however, clearly shows that UUAG instead of UUUA binds the KH1 domain at positions 1–4, whereas the first uracil at the 5′-end interacts with this KH domain at position 0 where specific interactions were rarely reported previously.
Figure 4.
Interaction details of the KH domains with the nucleotides at binding positions 3 and 4. A, structural alignment of the KH1–MRE10b and KH2–MRE10a complexes. The KH domains and the RNAs are shown in schematic and stick models, respectively. KH1, KH2, MRE10b, and MRE10a are colored cyan, magenta, yellow, and orange, respectively. B and C, the hydrogen-bonding interactions of A3 and A3′ involved in the KH1–MRE10b and KH2–MRE10a complexes, respectively. D and E, the hydrogen-bonding interactions of G4 and G4′ involved in the KH1–MRE10b and KH2–MRE10a complexes, respectively. The protein and RNA are colored and plotted as in A. Red spheres represent structured water molecules that participate in the hydrogen-bonding network. The hydrogen bonds are indicated as black dashed lines.
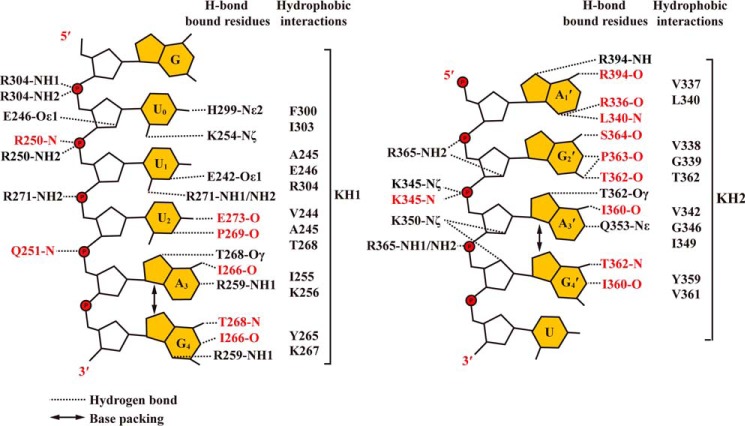
As summarized in Table 1 and Fig. 3, both KH1 and KH2 domains form extensive hydrogen bonds with their bound RNAs. In the KH1–GUUUAG structure, a total of 19 hydrogen bonds are observed between the KH domain and RNA, including seven protein side chain–nucleobase, five side chain–RNA backbone, five main chain–nucleobase, and two main chain–RNA backbone hydrogen-bonding interactions. In the KH2–CAGAGU complex, the total number of H-bonds is 20, and the corresponding numbers of hydrogen bonds of the different types listed above are 3, 6, 10, and 1.
Table 1.
The numbers of hydrogen bonds of different types in the complex structures of KH1–GUUUAG and KH2–CAGAGU
| KH1–GUUUAG |
KH2–CAGAGU |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| U0 | U1 | U2 | A3 | G4 | All | A1′ | G2′ | A3′ | G4′ | All | |
| Side chain–nucleobase | 2 | 2 | 0 | 2 | 1 | 7 | 1 | 0 | 2 | 0 | 3 |
| Side chain–RNA backbone | 3 | 1 | 1 | 0 | 0 | 5 | 0 | 2 | 2 | 2 | 6 |
| Main chain–nucleobase | 0 | 0 | 2 | 1 | 2 | 5 | 3 | 4 | 1 | 2 | 10 |
| Main chain–RNA backbone | 0 | 1 | 0 | 1 | 0 | 2 | 0 | 0 | 1 | 0 | 1 |
| Total | 5 | 4 | 3 | 4 | 3 | 19 | 4 | 6 | 6 | 4 | 20 |
Figure 3.
The schematic representation of protein–RNA interactions in the complex structures of KH1–MRE10b and KH2–MRE10a. Both hydrogen-bonding and hydrophobic interactions are shown. The residues involved in hydrogen-bonding interaction are colored black and red, depending on whether they use the side chain or the main chain to mediate the RNA–protein interaction.
hMEX-3C possesses a highly conserved interface at the RNA-binding positions 3 and 4 in its KH domains
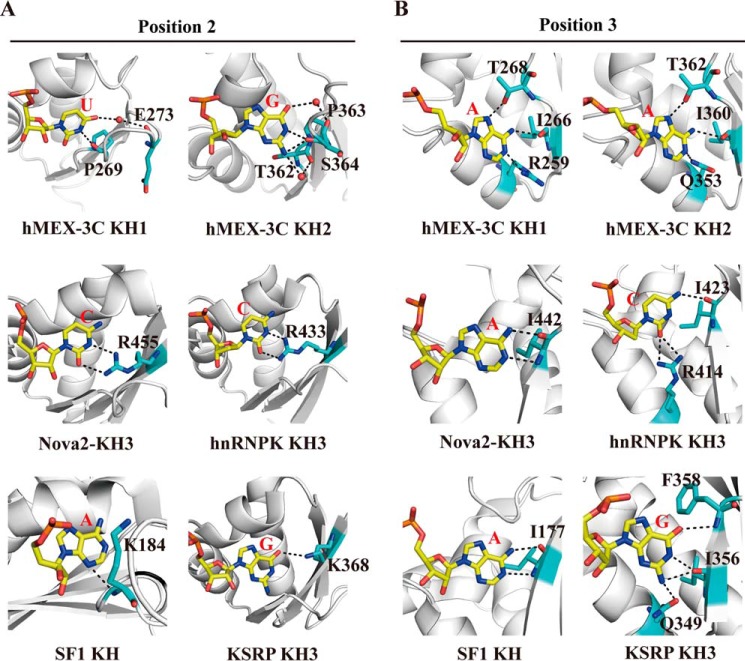
The interactions between the KH domains and RNA segments are highly similar at binding positions 3 and 4 where hydrogen bonds are abundantly observed (Figs. 3 and 4). In both complex structures, the base of the adenine at position 3 (A3 for KH1–GUUUAG or A3′ for KH2–CAGAGU) is anchored by a hydrophobic pocket largely constituting residues from the α2 helix, β2 strand, and variable loop (Ile-255, Arg-259, Ile-266, and Thr-268 for KH1; Val-342, Ile-349, Gln-353, Ile-360, and Thr-362 for KH2). The adenine base is specifically recognized by the surrounding residues via hydrogen bonds (Fig. 4, B and C): the N7 nitrogen of A3 or A3′ with the OH group of the threonine (Thr-268 of KH1 or Thr-362 of KH2), the amino group of A3 or A3′ with the main chain carbonyl of the isoleucine (Ile-266 of KH1 or Ile-360 of KH2), and the N1 nitrogen of A3 or A3′ with the NH2 group of Arg-259 of KH1 or Gln-353 of KH2. In addition to these hydrogen bonds between the base and the protein, the backbone and the ribose of this adenine also contribute to the hydrogen-bonding interactions with the KH domain. The amide group of Gln-251 within the GXXG motif grips the RNA fragment by forming a hydrogen bond with the OP1 oxygen of A3 in the KH1 complex, whereas the main chain NH and the side chain NH2 groups of Lys-345 from the same motif of the KH2 domain form hydrogen bonds with the OP1 and OP2 oxygen of A3′, respectively. In addition, the 2′-hydroxyl group of A3′ forms a water-mediated hydrogen bond with the NH2 group of Lys-350 from the α2 helix in the KH2 complex, further strengthening the interaction.
In the two structures, the guanine base at position 4 (G4 or G4′) is stacked with the A3 or A3′ base, providing a very stable and specific recognition interface on RNA for KH domains (Fig. 4A). Similar to A3 or A3′, G4 or G4′ also interacts with the adjacent threonine (Thr-268 of KH1 or Thr-362 of KH2) and isoleucine (Ile-266 of KH1 or Ile-360 of KH2) via two direct hydrogen bonds: the O6 oxygen of G4 or G4′ with the main chain NH group of the threonine and the imino group of G4 or G4′ with the main chain carbonyl of the isoleucine, respectively (Fig. 4, D and E). Other water-mediated hydrogen bonds are also found in both structures, including the N3 nitrogen of G4 with the side chain NH2 group of Arg-259 in the KH1 complex, the O4′ of the G4′ ribose with the 2′-hydroxyl group of A3′ and the NH2 group of Lys-350, and the OP2 oxygen of G4′ with the NH2 group of Arg-365 in the KH2 complex. Moreover, the side chains of Tyr-265 and Lys-267 in the KH1 complex (or Tyr-359 and Val-361 in the KH2 complex) contribute to the hydrophobic interactions with the G4 base (or G4′) (Fig. 3). Overall, the hydrogen-bonding network as well as the hydrophobic interactions ensures that the two purine nucleotides at positions 3 and 4 can be tightly anchored in the pocket on the surface of the KH domain.
Other parts of the hMEX-3C KH–RNA interfaces are dissimilar
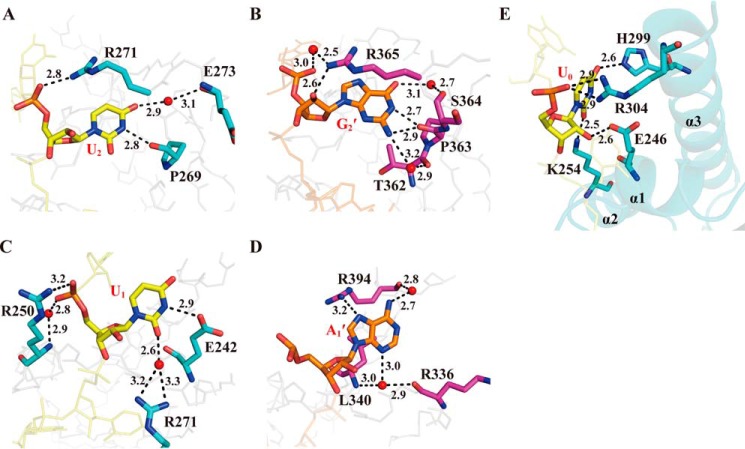
Unlike binding positions 3 and 4, the interactions at other positions on the two KH–RNA interfaces are not similar. In the KH1 complex structure, the uracil base at position 2 (U2) is inserted into a hydrophobic pocket formed by Ser-241, Val-244, and Ala-245 of the α1 helix and Pro-269 and Arg-271 of the variable loop. The U2 base also makes a direct hydrogen bond with the carbonyl group of Pro-269 using its N3H group and a water-mediated hydrogen bond with the main chain carbonyl group of Glu-273 through its O4 oxygen, while its backbone OP2 group is involved in a hydrogen bond with the NH2 group of Arg-271 (Fig. 5A). In contrast, G2′, the nucleotide equivalent to U2 in the KH2 complex, is involved in a more complicated hydrogen-bonding network with the surrounding residues; it is the nucleotide that forms the largest number of hydrogen bonds in both structures (Fig. 5B). Unlike U2, the G2′ base is more likely to be sandwiched by the long side chain of Arg-365 from the variable loop and the hydrophobic surface formed by Val-338 of the α1 helix and Thr-362 and Pro-363 of the variable loop. Both the imino and amino groups of the G2′ base form hydrogen bonds with the main chain carbonyl of Pro-363, while its amino group also makes an alternative water-mediated hydrogen bond with the carbonyl oxygen of Thr-362. Another water-mediated hydrogen bond is formed between the O6 oxygen of the G2′ base and the main chain carbonyl group of Ser-364. In addition, the side chain NH2 group of Arg-365 makes water-mediated and direct hydrogen bonds with the OP1 oxygen and the C2′-ribosyl oxygen of the G2′ backbone, respectively.
Figure 5.
Interaction details of the KH domains with the nucleotides at the binding positions prior to position 3. A and B, the hydrogen-bonding interactions of U2 and G2′ involved in the KH1–MRE10b and KH2–MRE10a complexes, respectively. C and D, the hydrogen-bonding interactions of U1 and A1′ involved in the KH1–MRE10b and KH2–MRE10a complexes, respectively. E, a view highlighting the protein–RNA–binding surface and the hydrogen bonds at position 0 in KH1 domain. Red spheres represent structured water molecules that participate in the hydrogen-bonding network. The black dashed lines indicate the hydrogen bonds. The protein and RNA are represented as in Fig. 4.
The most striking differences between the two KH–RNA–binding interfaces are in the RNA-binding sites prior to position 2. In the KH2 complex structure, there is only one adenine at position 1 (A1′), whereas two uracils are located at positions 0 and 1 (U0 and U1), respectively, in the KH1 complex (Fig. 4A). A1′ is bound to the hydrophobic surface consisting of Val-337 and Leu-340 of the α1 helix as well as Arg-394 and Thr-395 of the α3 helix; its adenine ring is involved in hydrophobic interactions with the side chains of Val-337, Leu-340, and Thr-395. Multiple hydrogen bonds were identified at this interface. The side chain NH2 group of Arg-394 makes a hydrogen bond with the N7 nitrogen of A1′, while the main chain carbonyl of Arg-394 forms a hydrogen bond with the amino group of A1′ via a water molecule (Fig. 5D). In addition, the N3 nitrogen of A1′ forms water-mediated hydrogen bonds with the main chain NH of Leu-340 and the carbonyl oxygen of Arg-336. In contrast, U1 in the KH1 complex exhibits a structural rearrangement with its base ring swinging away from the position of the A1′ base in the KH2 complex toward position 2 (supplemental Fig. S2D); it forms water-mediated hydrogen bonds with the side chain NH2 groups of Arg-271 from the variable loop through its O2 oxygen and a direct hydrogen bond with the side chain carbonyl of Glu-242 through its imino group. Additionally, the phosphate backbone of U1 forms a direct hydrogen bond and a water-mediated hydrogen bond with the side chain of Arg-250 located within the conserved GXXG motif (Fig. 5C). In addition to the above mentioned hydrogen bonds, U1 is also stabilized by hydrophobic interactions with the hydrophobic portions of Glu-246 and Arg-304 side chains (Fig. 3).
Notably, a new specific binding site at position 0 occupied by a uracil was identified in the KH1 complex structure that has rarely been reported in previous studies (Fig. 5E). Recognized specifically by the KH1 domain, U0 interacts with adjacent residues through multiple direct hydrogen bonds, including those between the O2 and O4 oxygen of the U0 base and the NH2 group of Lys-254 and the side chain NH group of His-299, respectively, as well as those between the C2′-ribosyl hydroxyl and backbone OP1 groups and the side chain groups of Glu-246 and Arg-304, respectively. In addition, the side chains of Phe-300 and Ile-303 also contribute to the hydrophobic interactions with the uracil ring of U0 (Fig. 3).
The dual KH1/2 domains maintain the same RNA recognition patterns as those in the individual KH domains
To determine whether the individual complexes reflect real protein–RNA interactions between the KH1/2 domains and the bipartite MRE10, NMR experiments were performed. The assignments of backbone proton and nitrogen of the KH1 and KH2 domains are shown in supplemental Figs. S3 and S4. 1H-15N HSQC spectra of the KH1, KH2, and KH1/2 domains titrated with increasing amounts of MRE10b, MRE10a, and MRE10, respectively, were recorded. The full HSQC spectra and some selected regions are shown in supplemental Figs. S5 and S6. Most likely because the KH1/2–MRE10 interaction falls in the range of intermediate exchange, the RNA titration caused severe line broadening and some peak disappearances in the HSQC spectrum. Despite the poor quality of the 1H-15N HSQC spectrum of KH1/2 with MRE10, some isolated resonances, representing the residues located at the KH–RNA–binding interfaces in our crystal structures, showed similar trends in chemical shift perturbation in the HSQC spectra of the individual KH domains and the KH1/2 domains (supplemental Figs. S5 and S6). This suggests that the recognition patterns in the individual KH–RNA complexes are most likely maintained in the KH1/2–MRE10 complex.
Structure-guided mutagenesis reveals some key residues involved in RNA binding by hMEX-3C KH domains
Based on a careful analysis of the two complex structures we determined, a total of 15 residues were found to form hydrogen-bonding interactions with RNA via their side chain groups, including Glu-242, Glu-246, Arg-250, Lys-254, Arg-259, Thr-268, Arg-271, His-299, and Arg-304 from the KH1 domain and Lys-345, Lys-350, Gln-353, Thr-362, Arg-365, and Arg-395 from the KH2 domain (Fig. 3). To evaluate the individual contributions of these residues to the hMEX-3C KH1/2–MRE10 interaction, we generated alanine mutants of these residues for ITC analysis. CD spectra confirmed that the mutants of hMEX-3C maintain a secondary structure composition similar to that of the wild type (supplemental Fig. S7).
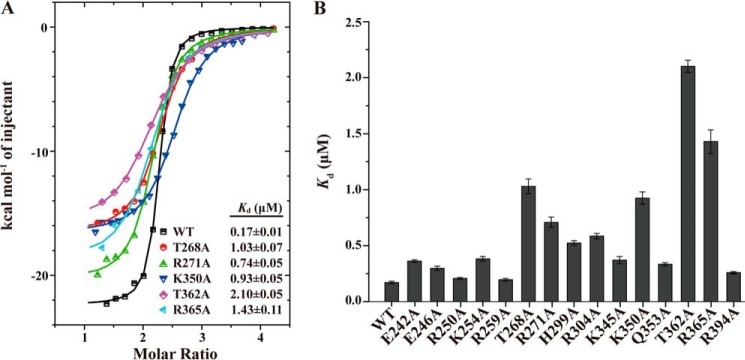
As summarized in Fig. 6 and supplemental Table S2, compared with the wild type, the mutants E242A, E246A, R250A, K254A, and R259A in KH1 and K345A, Q353A, and R394A in KH2 reduced the binding affinities by ∼2-fold or less, implying that the protein–RNA interaction was marginally perturbed by these mutations. A more significant reduction in binding affinity (>3-fold) was detected for mutants H299A and R304A; the His-299 and Arg-304 residue side chains mediate hydrogen-bonding interactions to stabilize U0 in the KH1 complex structure, suggesting that this newly identified RNA-binding site in the hMEX-3C KH domain is specific.
Figure 6.
The key residues involved in RNA binding by MEX-3C KH domains. A, the ITC fitting curves of the wild-type hMEX-3C KH1/2 domains and its mutants to MRE10 RNA. B, the Kd value histogram of the wild-type KH1/2 domains and its mutants to MRE10. The error bars represent S.D.
The conserved threonine (Thr-268 in KH1 and Thr-362 in KH2) in the variable loop stabilizes the hMEX-3C–MRE10 interaction by forming multiple hydrogen bonds with the purine nucleotides at positions 3 and 4 (Fig. 4). Unsurprisingly, mutating these two residues to alanine significantly influenced the protein–RNA interaction. The T268A mutation decreased the binding affinity by ∼6-fold, whereas T362A reduced it by ∼12-fold. The conserved residue Arg-271 in the variable loop of KH1 (or Arg-365 in KH2) is also observed to form multiple hydrogen bonds with the U1 base and U2 backbone in the KH1 complex and with the G2′ and G4′ backbones in the KH2 complex (Figs. 4 and 5). As expected, the corresponding mutants, R271A and R365A, had ∼4- and ∼8-fold lower binding affinities, respectively, compared with the wild type. Additionally, Lys-350 in the α2 helix forms water-mediated hydrogen bonds with the 2′-hydroxyl group of A3′ and the O4′ of G4′ in the KH2 complex structure, and the corresponding alanine mutant K350A weakened the binding affinity by ∼5.4-fold, which is consistent with the results from other mutants where protein–RNA interactions at positions 3 and 4 were disrupted. However, Lys-256, the residue in the KH1 domain that is equivalent to Lys-350, forms no hydrogen bonds with the purine nucleotides at the core positions, which may partially explain the ∼10-fold difference in the binding affinities of the two KH domains for their RNA substrates (Kd = 32.57 ± 2.04 μm for KH1–MRE10b and 3.15 ± 0.25 μm for KH2–MRE10a).
The GXXG motif is highly conserved among all KH domains and is essential for the recognition of the RNA targets. Surprisingly, R250A of KH1 and K345A of KH2 had only marginally lower binding affinities for their RNA-binding partners with ∼1- and ∼2-fold reduction, respectively. We, therefore, propose that the GXXG motif in hMEX-3C KH domains plays a role in defining the RNA phosphate backbone trace rather than contributing to the protein–RNA–binding affinity.
Determination of the hMEX3C consensus RNA sequence
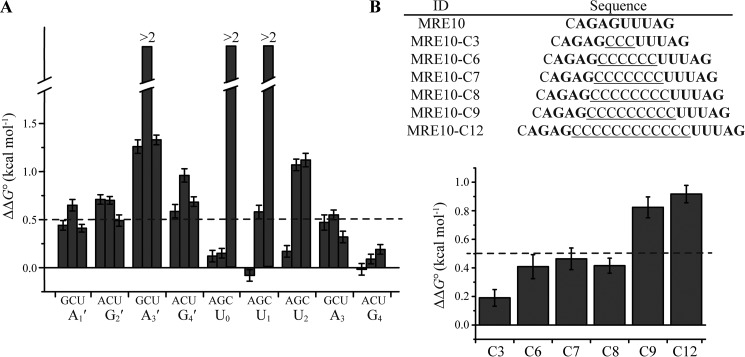
To determine whether a consensus RNA sequence existed for hMEX-3C KH domains, ITC was used to investigate the contribution of each nucleotide of MRE10 to the KH–RNA interaction by measuring the binding affinities of hMEX-3C KH1/2 domains for single-point mutants of each nucleotide (Fig. 7A and supplemental Table S2). ΔΔG0 was determined to evaluate the change in affinity, and ΔΔG0 > 0.5 kcal mol−1 was defined as a significant reduction in the binding affinity. For the KH1-binding sequence UUUAG, cytosine mutants of the three uracil residues and the guanine mutant of the third uracil residue resulted in significant reductions in the binding affinity, whereas all other mutants showed relatively minor effects. For the KH2-recognition sequence AGAG, significant reductions in binding affinity were observed for the cytosine mutants of every nucleotide. The nucleotide at the third position showed a strict preference for adenine. Intriguingly, cytosine was often found in previously determined KH–RNA complexes, especially at positions 2 and 3. However, most positions, if not all, in the case of hMEX-3C KH1/2 domains do not tolerate cytosine at all, which is consistent with previous results obtained for ceMEX-3 (36). This tendency of MEX-3 proteins to disfavor cytosine may contribute to the mechanisms they use to target their specific RNA substrates. Notably, although AG occupies binding positions 3 and 4 in both KH domains, the effects on the binding affinity caused by the mutations of these two nucleotides in KH1–GUUUAG are not comparable with those in KH2–CAGAGU. This is probably because the AG that is bound with KH2 is centrally located in the 10-mer MRE10 RNA, whereas the AG complexed with KH1 is at the 3′-end of the RNA sequence, and the nucleotides may have enough conformational flexibility to adapt to the structural environments in the KH domain, especially for the last guanine. Finally, based on our ITC results, we defined the RNA recognition sequence (A/G/U)(G/U)AG + (A/G/U)(A/U)(A/U)(A/G/U)N for the dual KH domains of hMEX-3C, which is a bipartite element that consists of one four-nucleotide and one five-nucleotide motif recognized by KH2 and KH1, respectively.
Figure 7.
Mutagenesis of MRE10 RNA. A, the MRE10 RNA was mutated systematically, and the Kd value for every single-nucleotide mutation was determined by ITC experiment. The change of standard free energy change is defined as ΔΔG0 = −RTln(1/Kd,mut) − [−RTln(1/Kd,wt)]. The bars represent the ΔΔG0 value, and the single-nucleotide mutants are indicated on the horizontal axis. The broken bars indicate the mutants that can hardly bind to hMEX-3C. B, upper, the RNA sequence of MRE10 and a series of spacing mutants with the background of cytosine are shown. The hMRE motif are shown in bold. Lower, the Kd value for each spacing mutant was determined by ITC experiment. The bars represent the ΔΔG0 value, and the spacing mutants are indicated on the horizontal axis. The error bars represent S.D.
The length of the linker between the two half-sites of MRE has been previously indicated to be important in mediating its binding with MEX-3 (36). To test whether this is also the case for MRE10 with hMEX-3C, a series of mutants with different numbers of cytosines inserted between the two half-sites of MRE10 were generated for ITC experiments. The insertion of cytosine is considered to not cause a change in the binding pattern because this base is not tolerated in MRE10. As summarized in Fig. 7B, hMEX-3C KH1/2 can bind mutants with up to eight cytosine nucleotides between the two half-sites without significantly reducing the protein–RNA–binding affinity. Based on these data, we refined the consensus RNA sequence to (A/G/U)(G/U)AGN0–8(A/G/U)(A/U)(A/U)(A/G/U)N.
hMEX-3C dual KH domains bind specifically to the HLA-A2 transcript
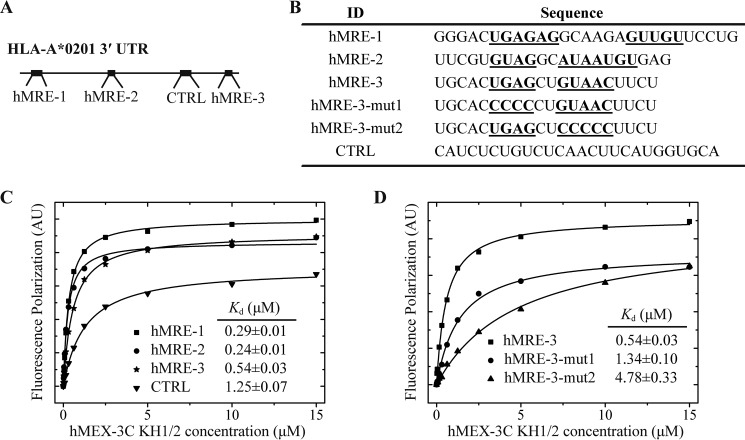
Although it has been found that human MEX-3C can regulate the translation of many mRNAs, HLA-A2, to the best of our knowledge, is the only mRNA that has been experimentally confirmed to interact directly with hMEX-3C KH domains (32, 33). By binding to the HLA-A2 3′-UTR, hMEX-3C can induce the ubiquitin-dependent degradation of this mRNA or inhibit mRNA translation. Therefore, we applied the above defined RNA-recognition motif for hMEX-3C KH domains to the HLA-A2 3′-UTR and screened three potential binding targets for hMEX-3C, named hMRE-1–3 (Fig. 8A). With regard to position, hMRE-1 is located adjacent to the 5′-end of HLA-A2 3′-UTR, whereas hMRE-3 is very close to the 3′-end. To verify their interactions with the KH1/2 domains, FP experiments were used to investigate the binding affinities of hMEX-3C KH1/2 domains with these three hMRE-containing RNA fragments with their lengths varying from 20 to 27 nucleotides and a control region lacking hMRE (Fig. 8B). Our results show that all of the hMRE-containing RNA fragments exhibited high binding affinities (in the nm range) toward the dual KH domains of hMEX-3C, whereas the control region showed relatively low affinity in the μm range (Fig. 8C).
Figure 8.
hMEX-3C binds specifically to hMREs present in HLA-A2 3′-UTR. A, a schematic representation of the hMREs distribution within HLA-A2 3′-UTR. The hMRE motifs and control (CTRL) region are indicated as bold lines. B, the RNA sequences used for FP experiments are shown. The hMRE half-sites are shown in bold and underlined. Note that hMRE-1 comprises two copies of overlapping hMREa elements, and hMRE-2 comprises three copies of overlapping hMREb elements. C, the binding affinities of hMEX-3C for hMRE-1, hMRE-2, hMRE-3, and the control region determined by FP experiments. The FP data were fitted using Equation 1. The hMRE-1–3 sequences exhibited high binding affinities toward the dual KH domains of hMEX-3C, whereas the control region showed relatively low binding affinity. D, the binding affinities of hMEX-3C for hMRE-3, hMRE-3-mut1, and hMRE-3-mut2 determined by FP experiments. The binding affinities of hMRE-3 mutants for hMEX-3C were significantly decreased. AU, absorbance units.
To further verify that hMRE is essential for specific recognition, we generated two variants of hMRE-3 (each half-site of hMRE-3 was mutated to multiple consecutive cytosines) for FP experiments. We chose hMRE-3 because it contains only one copy of hMRE, whereas hMRE-1 and hMRE-2 contain multiple copies of hMREa or hMREb. As expected, the binding affinities of hMRE-3-mut1 and hMRE-3-mut2 for KH1/2 decreased by ∼3- and ∼9-fold, respectively, when compared with hMRE-3 (Fig. 8D). Therefore, our results show that hMRE is essential for the high-affinity interaction between hMEX-3C and HLA-A2 3′-UTR.
Discussion
MEX-3 was first reported as a translational repressor in C. elegans, and its orthologs in vertebrates were subsequently identified to have E3 ubiquitin ligase activity, promoting RING-dependent degradation of certain mRNAs critical to embryonic development, immune responses, metabolism, and even carcinogenesis (32, 33, 38–42). Although efforts have been made recently to elucidate the functions of MEX-3–mediated biological processes (30, 35, 45), the mechanisms MEX-3 proteins use to identify their real RNA targets remain unclear. In this study, we determined two crystal structures of human MEX-3C KH domains with their RNA substrates and identified the amino acids and nucleotides crucial to protein–RNA recognition specificity. The binding motif derived from our results will provide valuable information for future investigations of the functional pathways mediated by hMEX-3C and for predicting potential mRNAs regulated by this enzyme.
In addition to the four classically bound nucleotides, an extra nucleotide at the 5′-end of the single-stranded RNA or DNA is occasionally observed in the reported structures of KH–nucleic acids complexes, whose binding is often stabilized by hydrophobic interactions, van der Waals forces, or packing effects. In the hMEX-3C KH1–GUUUAG complex, however, a uracil located at position 0 interacts with the residues of α1, α2, and α3 helices via multiple hydrogen bonds, exhibiting a high binding specificity. Interestingly, even though the structures of the two hMEX-3C KH domains are highly similar, no extra bound nucleotide is found in the KH2–CAGAGU complex. A comparison of the two structures reveals that the α3 helix in the KH2 domain is three residues shorter, shows a slightly different orientation from that in the KH1 domain, and lacks positively charged residues at positions equivalent to His-299 and Arg-304 in the α3 helix of the KH1 domain (supplemental Figs. S2C and S8). Altogether, these factors could help explain why the KH2 domain does not possess an extra RNA-binding site. To extend our findings, we analyzed the specific binding capacities for an extra nucleotide in other KH domains with reported structures by checking for two criteria, an α3 helix that was suitably long and a charged residue(s) at a position(s) equivalent to His-299 and/or Arg-304 in hMEX-3C KH1 (supplemental Fig. S8). The Nova-2 KH3 and PCBP1 KH1 domains, which satisfy both these conditions, are KH domains that have been reported so far to interact with an extra nucleotide using hydrogen bonds (15, 46). Thus, our findings can be used to predict which KH domains have an extra specific RNA-binding site.
The extra RNA-binding site allows the hMEX-3C KH1 domain to bind its RNA substrate with a higher specificity, which is consistent with the distinct ITC results for the KH1 domain with the MRE10a and MRE10b fragments (Fig. 1D). The KH2 domain, in contrast, exhibits comparable binding affinities for the two RNA fragments (<4-fold difference in binding affinity). Overall, our structural and ITC data suggest that the contributions of the two KH domains to the recognition of RNA targets are different, and the KH2 domain appears to contribute more to binding affinity.
The nucleotides bound at position 2 are dominantly adenine or cytosine in almost all published structures (15, 27, 34, 44). Our structure of hMEX-3C KH1–GUUUAG, for the first time, shows that uracil is bound at this position. Guanine is also rarely found in position 2 of the KH–RNA-DNA–binding interface with the only exception so far being the KSRP KH3–AGGGC complex. In the hMEX-3C KH2–CAGAGU complex, the G2′ base forms as many as four hydrogen bonds with the protein backbone, whereas the corresponding guanine base in the KSRP KH3 complex makes only one hydrogen bond with the side chain of a lysine residue from the β3 strand (Fig. 9A). Notably, both the uracil and guanine at position 2 in our two complex structures make hydrogen bonds with the backbone atoms of two or three different residues in the variable loop adjacent to the β3 strand (Fig. 9A). In contrast, all previously reported complex structures show that the nucleotide bound at this position interacts primarily with the positively charged side chain of a single residue in the β3 strand. Therefore, we have observed a new recognition pattern that has not been seen previously in other KH–nucleic acid complexes. Additionally, in all KH–RNA complex structures, barring the KSRP KH–AGGGC complex, an adenine is bound at position 3. In these structures, this adenine forms hydrogen bonds with the backbone atoms of a highly conserved isoleucine of the KH domain using its Watson-Crick edge, mimicking the classical base-pairing behavior in nucleic acids. The hMEX-3C KH domains also show a strong preference for adenine at binding position 3. However, this adenine establishes multiple hydrogen bonds with three different KH residues using both its Watson-Crick and Hoogsteen edges (Fig. 9B). Moreover, the interactions between the Hoogsteen edge and the side chains of threonine residues (Thr-268/Thr-362) appear to be essential for binding specificity as both the T268A and T362A mutations decreased the binding affinities of the KH1/2 domains for RNA significantly. However, the corresponding residues in other KH domains are nonpolar amino acids, which lack a side chain group capable of forming hydrogen bonds with the Hoogsteen edge of the adenine (supplemental Fig. S8). Another characteristic that distinguishes hMEX-3C KH from other KH domains is that cytosine is not tolerated at most of its RNA-binding positions, a feature that has also been confirmed in ceMEX-3C (36). This preference for non-cytosine nucleotides and the novel “1-to-n” recognition patterns at positions 2 and 3 may represent a unique strategy that the MEX-3 family utilizes to recognize its RNA substrates.
Figure 9.
Comparison of the hydrogen bonds pattern in the central positions of hMEX-3C KH1–GUUUAG/KH2–CAGAGU complexes with other representative KH–nucleic acid complexes. A and B, comparison of the hydrogen bonds pattern in position 2 (A) and in position 3 (B). The residues involved in hydrogen-bonding interactions are labeled and shown as sticks.
Some mechanisms have been proposed to explain how MEX-3C regulates the degradation of mRNAs via their 3′-UTRs, but some of the available evidence shows that without the RING domain MEX-3C can still inhibit the translation of mRNA, probably by sequestering the mRNA. Either way, a specific recognition pattern between the MEX-3C RNA-binding domains and the 3′-UTRs of mRNAs may exist. Interestingly, according to our data, HLA-A2, a previously characterized MEX-3C regulatory target, contains three hMRE-containing sequences in its 3′-UTR. All of these sequences show high binding affinities toward the KH domains of MEX-3C, and the half-site mutants of hMRE decreased the binding affinities significantly, suggesting that MEX-3C may regulate mRNA translation by recognizing and binding the hMRE sites. Further work is needed to verify whether all of these potential binding motifs are functionally relevant and whether hMRE recognition is a ubiquitous regulation mechanism used by MEX-3 family proteins in cells.
Experimental procedures
Protein expression and purification
The gene for hMEX-3C was purchased from Life Technologies. The DNA segments for the human MEX-3C KH1 domain (residues 221–306), KH2 domain (residues 320–396), and dual KH domains (residues 221–396) were amplified by PCR. Subsequently, the KH1 and KH2 domains were cloned into a modified pET28a (Novagen) vector without the thrombin cleavage site (p28a), whereas the dual KH domains were cloned into a modified pET28a vector in which the thrombin cleavage site was substituted with a small ubiquitin-like modifier (SUMO) protease cleavage site. The mutants of the KH1/2 domains were constructed by conventional PCR using the MutanBEST kit (TaKaRa) and verified by DNA sequencing. All recombinant proteins were overexpressed in Escherichia coli BL21 (DE3)-Gold cells (Novagen) cultured in LB medium to an A600 of 1.0 and then induced with 0.03 mm isopropyl 1-thio-β-d-galactopyranoside for 6 h at 37 °C. All proteins were purified on a nickel-chelating column (Qiagen) using 20 mm Tris (pH 7.5), 1 m NaCl and then further purified by size-exclusion chromatography using a Superdex 75 or Superdex 200 column (GE Healthcare) equilibrated with SEC buffer (20 mm Tris (pH 7.5), 2 m NaCl). Finally, the KH1/2 domains and its mutants were treated with ULP1 to remove the His6-SUMO tags. The 15N,13C-labeled MEX-3C KH1, KH2, and KH1/2 domains used for NMR experiments were synthesized by growing the bacteria in LeMaster and Richards medium supplemented with 0.5 g/liter 15N-labeled NH4Cl, 2.5 g/liter labeled glucose and purified by the same procedure as described above.
Isothermal titration calorimetry
RNA oligomers were purchased from Takara Bio, Inc. and dissolved in diethyl pyrocarbonate-treated water to a final concentration of 1 mm. ITC assays were performed using a MicroCal iTC200 calorimeter (GE Healthcare), and all experiments were carried out at 20 °C. A buffer containing 50 mm Tris (pH 7.5), 200 mm NaCl, and 1 mm TCEP was used for sample dilution. The concentrations of the proteins were determined spectrophotometrically. The MRE10 RNA and its mutants were diluted in buffer to a final concentration of 20 μm. The C values of the experiments performed with these RNA oligomers are in the range of 10–133. In contrast, MRE10a and MRE10b were diluted in the same buffer to a final concentration of 30 μm. The C values for their ITC experiments are in the range of 1–10. The ITC experiments involved 20 injections of 2 μl of protein into 200 μl of RNA. In the control experiment, the buffer without protein was injected into RNA to compensate for the heat of protein dilution. Data analysis was done using the ITC data analysis module Origin 7.0 (MicroCal) provided by the manufacturer. The ΔG0 of protein–RNA binding was computed as −RTln(1/Kd) where R, T, and Kd are the gas constant, temperature, and dissociation constant, respectively.
Nuclear magnetic resonance
Backbone resonances of the KH1 and KH2 domains were assigned using 0.5 mm 15N,13C-labeled protein in NMR buffer (50 mm sodium phosphate (pH 6.5), 100 mm NaCl, 5 mm TCEP) with 10% D2O using 3D CBCA(CO)NH and 3D CBCANH on a Bruker DMX 600 MHz at 298 K. The 15N-labeled KH1, KH2, and KH1/2 domains (each at a concentration of 0.1 mm) in NMR buffer containing 10% D2O were titrated with increasing amounts of MRE10b, MRE10a, and MRE10, respectively. After each titration, the 1H-15N HSQC spectra were recorded on a Bruker DMX 600 MHz at 298 K. All NMR data were processed using NMRPipe (47), and the spectra were assigned using Sparky (48).
Crystallization, data collection, structure determination, and refinement
Prior to crystallization, the hMEX-3C KH1 domain was dialyzed against a buffer (pH 7.5) containing 20 mm Tris, 200 mm NaCl, and 2 mm TCEP. The KH2 domain was dialyzed against a buffer (pH 7.5) containing 20 mm Tris and 200 mm NaCl. The RNA oligomers were diluted in diethyl pyrocarbonate-treated water to a final concentration of 4 mm. The crystals of the apo-form KH2 domain were obtained by sitting drop vapor diffusion against 1.6 m ammonium sulfate at 4 °C with a protein concentration of 8 mg/ml. KH1–GUUUAG crystals were obtained by hanging drop vapor diffusion against 5% (v/v) Tacsimate (pH 7.0), 0.1 m HEPES (pH 7.0), and 10% (w/v) polyethylene glycol monomethyl ether 2000 at 20 °C with a protein concentration of 20 mg/ml and a protein:RNA ratio of 2:1. KH2–CAGAGU crystals were obtained by sitting drop vapor diffusion against 0.04 m magnesium acetate tetrahydrate, 0.05 m sodium cacodylate trihydrate (pH 6.0), and 30% (v/v) (±)-2-methyl-2,4-pentanediol at 20 °C with a protein concentration of 4 mg/ml and a protein:RNA ratio of 2:1. The mother liquor supplemented with 20–30% (v/v) glycerol was used as a cryoprotectant for the crystals. Crystals were soaked with the cryoprotectant, harvested using cryoloops, and flash cooled in liquid nitrogen.
X-ray intensity data of the crystals were collected on beamline 18U1 at the Shanghai Synchrotron Radiation Facility at 100 K. The data sets were integrated and scaled using HKL2000 software. For the KH1–GUUUAG complex, the phase problem was solved by molecular replacement with the program MOLREP (49) using the structure of hMEX-3D KH2 (Protein Data Bank code 2DGR) as a search model. The sequence identity between hMEX-3D KH2 and hMEX-3C KH1 is 46%. The model was further improved by manual model building using Coot (50) and refined primarily using PHENIX.refine (51). Final refinement strategies included xyz coordinates, individual B-factors, occupancies, and automated correction of N/Q/H errors. The structures of KH2–CAGAGU and apo-KH2 were processed and refined in a manner similar to the KH1–GUUUAG structure using the latter as a search model for molecular replacement (Protein Data Bank code 5WWW). The refinement strategy for the KH2–CAGAGU complex was the same as that of the KH1–GUUUAG structure. For the structure of apo-form KH2, final refinement strategies included xyz coordinates, individual B-factors, TLS parameters (one TLS group per chain), occupancies, non-crystallographic symmetry restraints, automated correction of N/Q/H errors, and optimization of X-ray/stereochemistry and X-ray/atomic displacement parameter weight. The Fo − Fc map was generated before including RNA in the model. The composite omit 2Fo − Fc map was generated by running the program phenix.composite_omit_map with Cartesian simulated annealing refinement prior to map generation. Crystal diffraction data and refinement statistics are listed in supplemental Table S1. Structure analysis was performed using PyMOL.
CD measurements
Far-UV CD spectra of the hMEX-3C KH1/2 domain and its mutants were determined using an Applied Photophysics Chirascan spectrometer at 298 K. The spectra were recorded at wavelengths between 195 and 260 nm using a 0.05-cm-path length cell. The protein samples were diluted to 0.1 mg/ml with CD buffer (30 mm PBS (pH 7.5), 100 mm NaCl). A buffer-only reference was subtracted from each curve. All samples were tested in triplicate.
hMRE screening
The RNA-recognition element of hMEX-3C KH1/2 domains (hMRE) located within the HLA-A2 3′-UTR was screened using Vector NTI software (InforMax, Inc.). The hMRE RNA is a bipartite element that consists of two half-sites recognized by the KH1 and KH2 domains (hMREa, (A/G/U)(G/U)AG, and hMREb, (A/G/U)(A/U)(A/U)(A/G/U)N). We searched the HLA-A2 3′-UTR using the hMEX-3C recognition element (A/G/U)(G/U)AGN0–8(A/G/U)(A/U)(A/U)(A/G/U)N, and three RNA sequences (named hMRE-1–3) containing hMRE were identified. Note that hMRE-1 comprises two copies of overlapping hMREa elements, and hMRE-2 comprises three copies of overlapping hMREb elements.
Fluorescence polarization assay
Lyophilized 5′-FAM (carboxyfluorescein)-labeled RNA oligomers were purchased from Takara Bio, Inc. and dissolved in FP buffer (20 mm Tris (pH 7.5), 200 mm NaCl) to a final concentration of 50 nm. Equilibrium dissociation constants for different RNAs and hMEX-3C KH1/2 domains were determined by measuring FP as described previously (52). The hMEX-3C KH1/2 domains with the His6-SUMO tag were serially diluted in FP buffer. Before the assay, 190 μl of labeled RNA at 50 nm was mixed with 10 μl of protein at different concentrations and incubated for 10 min. Samples were then excited at 485 nm, and FP was detected at 525 nm using a SpectraMax M5 (Molecular Devices) plate reader at 20 °C. The FP data were fitted to a 1:1 binding model using Equation 1,
| (Eq. 1) |
where FP is the observed total polarization, FPini is the FP value of free RNA, P is the concentration of protein, R is the concentration of RNA, and n is the binding stoichiometry (protein:RNA ratio).
Author contributions
L. Y. performed most of the experiments, analyzed the results, and wrote the paper. L. Y., C. W., and Q. G. designed the experiments. F. L. helped in determining the crystal structure. A. N. helped in revising the manuscript. J. Z. and J. W. helped in performing the NMR experiments. L. Y., C. W., Y. S., and Q. G. analyzed the results. L. Y. and Q. G. revised the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Lijun Wang, Deqiang Yao, Yiyang Jiang, Mengqi Lv, Ling Xu, Bin Wen, Pengzhi Wu, Difei Xu, and Xiaodan Liu for helpful discussions. Huichao Ou is thanked for help with the ITC experiments. We also thank the staff of beamline BL18U1 at Shanghai Synchrotron Radiation Facility for assistance with the data collection.
This work was supported by Strategic Priority Research Program of the Chinese Academy of Science Grants XDB08010101 and XDB08030302 and Chinese National Natural Science Foundation Grants 91540103, 31330018, and 31270782. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S8 and Tables S1 and S2.
The atomic coordinates and structure factors (codes 5WWZ, 5WWW, and 5WWX) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- RBD
- RNA-binding domain
- KH
- K-homology
- MRE
- MEX-3–recognition element
- KSRP
- KH-type splicing-regulatory protein
- ceMEX-3
- C. elegans MEX-3
- hMEX-3C
- human MEX-3C
- ITC
- isothermal titration calorimetry
- FP
- fluorescence polarization
- HSQC
- heteronuclear single quantum coherence
- hMRE
- human MRE
- SUMO
- small ubiquitin-like modifier
- TCEP
- tris(2-carboxyethyl)phosphine.
References
- 1. Nagai K. (1996) RNA-protein complexes. Curr. Opin. Struct. Biol. 6, 53–61 [DOI] [PubMed] [Google Scholar]
- 2. Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B. M., Strein C., Davey N. E., Humphreys D. T., Preiss T., Steinmetz L. M., Krijgsveld J., and Hentze M. W. (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 [DOI] [PubMed] [Google Scholar]
- 3. Nagai K., Oubridge C., Jessen T. H., Li J., and Evans P. R. (1990) Crystal-structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein-A. Nature 348, 515–520 [DOI] [PubMed] [Google Scholar]
- 4. Oubridge C., Ito N., Evans P. R., Teo C. H., and Nagai K. (1994) Crystal structure at 1.92Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 372, 432–438 [DOI] [PubMed] [Google Scholar]
- 5. Green S. R., and Mathews M. B. (1992) Two RNA-binding motifs in the double-stranded RNA-activated protein kinase, DAI. Genes Dev. 6, 2478–2490 [DOI] [PubMed] [Google Scholar]
- 6. St Johnston D., Brown N. H., Gall J. G., and Jantsch M. (1992) A conserved double-stranded RNA-binding domain. Proc. Natl. Acad. Sci. U.S.A. 89, 10979–10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kharrat A., Macias M. J., Gibson T. J., Nilges M., and Pastore A. (1995) Structure of the dsRNA binding domain of E. coli RNase III. EMBO J. 14, 3572–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ramos A., Grünert S., Adams J., Micklem D. R., Proctor M. R., Freund S., Bycroft M., St Johnston D., and Varani G. (2000) RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 19, 997–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu D., Searles M. A., and Klug A. (2003) Crystal structure of a zinc-finger-RNA complex reveals two modes of molecular recognition. Nature 426, 96–100 [DOI] [PubMed] [Google Scholar]
- 10. Hudson B. P., Martinez-Yamout M. A., Dyson H. J., and Wright P. E. (2004) Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat. Struct. Mol. Biol. 11, 257–264 [DOI] [PubMed] [Google Scholar]
- 11. De Boulle K., Verkerk A. J., Reyniers E., Vits L., Hendrickx J., Van Roy B., Van den Bos F., de Graaff E., Oostra B. A., and Willems P. J. (1993) A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat. Genet. 3, 31–35 [DOI] [PubMed] [Google Scholar]
- 12. Siomi H., Matunis M. J., Michael W. M., and Dreyfuss G. (1993) The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 21, 1193–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Castiglone Morelli M. A., Stier G., Gibson T., Joseph C., Musco G., Pastore A., and Travè G. (1995) The KH module has an αβ fold. FEBS Lett. 358, 193–198 [DOI] [PubMed] [Google Scholar]
- 14. Musco G., Stier G., Joseph C., Castiglione Morelli M. A., Nilges M., Gibson T. J., and Pastore A. (1996) Three-dimensional structure and stability of the KH domain: Molecular insights into the fragile X syndrome. Cell 85, 237–245 [DOI] [PubMed] [Google Scholar]
- 15. Lewis H. A., Musunuru K., Jensen K. B., Edo C., Chen H., Darnell R. B., and Burley S. K. (2000) Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell 100, 323–332 [DOI] [PubMed] [Google Scholar]
- 16. Beuth B., Pennell S., Arnvig K. B., Martin S. R., and Taylor I. A. (2005) Structure of a Mycobacterium tuberculosis NusA-RNA complex. EMBO J. 24, 3576–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burd C. G., and Dreyfuss G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science 265, 615–621 [DOI] [PubMed] [Google Scholar]
- 18. Grishin N. V. (2001) KH domain: one motif, two folds. Nucleic Acids Res. 29, 638–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Achsel T., and Bagni C. (2016) Cooperativity in RNA-protein interactions: the complex is more than the sum of its partners. Curr. Opin. Neurobiol. 39, 146–151 [DOI] [PubMed] [Google Scholar]
- 20. Gibson T. J., Thompson J. D., and Heringa J. (1993) The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS Lett. 324, 361–366 [DOI] [PubMed] [Google Scholar]
- 21. McKnight G. L., Reasoner J., Gilbert T., Sundquist K. O., Hokland B., McKernan P. A., Champagne J., Johnson C. J., Bailey M. C., Holly R., O'Hara P. J., and Oram J. F. (1992) Cloning and expression of a cellular high density lipoprotein-binding protein that is up-regulated by cholesterol loading of cells. J. Biol. Chem. 267, 12131–12141 [PubMed] [Google Scholar]
- 22. Ashley C. T. Jr., Wilkinson K. D., Reines D., and Warren S. T. (1993) FMR1 protein: conserved RNP family domains and selective RNA binding. Science 262, 563–566 [DOI] [PubMed] [Google Scholar]
- 23. Siomi H., Siomi M. C., Nussbaum R. L., and Dreyfuss G. (1993) The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell 74, 291–298 [DOI] [PubMed] [Google Scholar]
- 24. Currie J. R., and Brown W. T. (1999) KH domain-containing proteins of yeast: absence of a fragile X gene homologue. Am. J. Med. Genet. 84, 272–276 [PubMed] [Google Scholar]
- 25. O'Donnell W. T., and Warren S. T. (2002) A decade of molecular studies of fragile X syndrome. Annu. Rev. Neurosci. 25, 315–338 [DOI] [PubMed] [Google Scholar]
- 26. Ostareck-Lederer A., and Ostareck D. H. (2004) Control of mRNA translation and stability in haematopoietic cells: the function of hnRNPs K and E1/E2. Biol. Cell 96, 407–411 [DOI] [PubMed] [Google Scholar]
- 27. Backe P. H., Messias A. C., Ravelli R. B., Sattler M., and Cusack S. (2005) X-ray crystallographic and NMR studies of the third KH domain of hnRNP K in complex with single-stranded nucleic acids. Structure 13, 1055–1067 [DOI] [PubMed] [Google Scholar]
- 28. Valverde R., Edwards L., and Regan L. (2008) Structure and function of KH domains. FEBS J. 275, 2712–2726 [DOI] [PubMed] [Google Scholar]
- 29. Nicastro G., García-Mayoral M. F., Hollingworth D., Kelly G., Martin S. R., Briata P., Gherzi R., and Ramos A. (2012) Noncanonical G recognition mediates KSRP regulation of let-7 biogenesis. Nat. Struct. Mol. Biol. 19, 1282–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Draper B. W., Mello C. C., Bowerman B., Hardin J., and Priess J. R. (1996) MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell 87, 205–216 [DOI] [PubMed] [Google Scholar]
- 31. Hollingworth D., Candel A. M., Nicastro G., Martin S. R., Briata P., Gherzi R., and Ramos A. (2012) KH domains with impaired nucleic acid binding as a tool for functional analysis. Nucleic Acids Res. 40, 6873–6886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cano F., Bye H., Duncan L. M., Buchet-Poyau K., Billaud M., Wills M. R., and Lehner P. J. (2012) The RNA-binding E3 ubiquitin ligase MEX-3C links ubiquitination with MHC-I mRNA degradation. EMBO J. 31, 3596–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cano F., Rapiteanu R., Sebastiaan Winkler G., and Lehner P. J. (2015) A non-proteolytic role for ubiquitin in deadenylation of MHC-I mRNA by the RNA-binding E3-ligase MEX-3C. Nat. Commun. 6, 8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nicastro G., Taylor I. A., and Ramos A. (2015) KH-RNA interactions: back in the groove. Curr. Opin. Struct. Biol. 30, 63–70 [DOI] [PubMed] [Google Scholar]
- 35. Hunter C. P., and Kenyon C. (1996) Spatial and temporal controls target pal-1 blastomere-specification activity to a single blastomere lineage in C. elegans embryos. Cell 87, 217–226 [DOI] [PubMed] [Google Scholar]
- 36. Pagano J. M., Farley B. M., Essien K. I., and Ryder S. P. (2009) RNA recognition by the embryonic cell fate determinant and germline totipotency factor MEX-3. Proc. Natl. Acad. Sci. U.S.A. 106, 20252–20257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Buchet-Poyau K., Courchet J., Le Hir H., Séraphin B., Scoazec J. Y., Duret L., Domon-Dell C., Freund J. N., and Billaud M. (2007) Identification and characterization of human Mex-3 proteins, a novel family of evolutionarily conserved RNA-binding proteins differentially localized to processing bodies. Nucleic Acids Res. 35, 1289–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guzmán B., Cormand B., Ribasés M., González-Núñez D., Botey A., and Poch E. (2006) Implication of chromosome 18 in hypertension by sibling pair and association analyses: putative involvement of the RKHD2 gene. Hypertension 48, 883–891 [DOI] [PubMed] [Google Scholar]
- 39. Jiao Y., Bishop C. E., and Lu B. (2012) Mex3c regulates insulin-like growth factor 1 (IGF1) expression and promotes postnatal growth. Mol. Biol. Cell 23, 1404–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jiao Y., George S. K., Zhao Q., Hulver M. W., Hutson S. M., Bishop C. E., and Lu B. (2012) Mex3c mutation reduces adiposity and increases energy expenditure. Mol. Cell. Biol. 32, 4350–4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Burrell R. A., McClelland S. E., Endesfelder D., Groth P., Weller M. C., Shaikh N., Domingo E., Kanu N., Dewhurst S. M., Gronroos E., Chew S. K., Rowan A. J., Schenk A., Sheffer M., Howell M., et al. (2013) Replication stress links structural and numerical cancer chromosomal instability. Nature 494, 492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cano F., and Lehner P. J. (2013) A novel post-transcriptional role for ubiquitin in the differential regulation of MHC class I allotypes. Mol. Immunol. 55, 135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kuniyoshi K., Takeuchi O., Pandey S., Satoh T., Iwasaki H., Akira S., and Kawai T. (2014) Pivotal role of RNA-binding E3 ubiquitin ligase MEX3C in RIG-I-mediated antiviral innate immunity. Proc. Natl. Acad. Sci. U.S.A. 111, 5646–5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu Z., Luyten I., Bottomley M. J., Messias A. C., Houngninou-Molango S., Sprangers R., Zanier K., Krämer A., and Sattler M. (2001) Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science 294, 1098–1102 [DOI] [PubMed] [Google Scholar]
- 45. Ciosk R., DePalma M., and Priess J. R. (2006) Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science 311, 851–853 [DOI] [PubMed] [Google Scholar]
- 46. Yoga Y. M., Traore D. A., Sidiqi M., Szeto C., Pendini N. R., Barker A., Leedman P. J., Wilce J. A., and Wilce M. C. (2012) Contribution of the first K-homology domain of poly(C)-binding protein 1 to its affinity and specificity for C-rich oligonucleotides. Nucleic Acids Res. 40, 5101–5114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 48. Goddard T. D., and Kneller D. G. (2004) SPARKY 3, University of California, San Francisco [Google Scholar]
- 49. Vagin A., and Teplyakov A. (2010) Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 50. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang W., Wang L., Zou Y., Zhang J., Gong Q., Wu J., and Shi Y. (2011) Cooperation of Escherichia coli Hfq hexamers in DsrA binding. Genes Dev. 25, 2106–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.