Abstract
Cryptococcus species cause invasive infections in humans. Lipids play an important role in the progression of these infections. Independent studies done by our group and others provide some detail about the functions of these lipids in Cryptococcus infections. However, the pathways of biosynthesis and the metabolism of these lipids are not completely understood. To thoroughly understand the physiological role of these Cryptococcus lipids, a proper structure and composition analysis of Cryptococcus lipids is demanded. In this study, a detailed spectroscopic analysis of lipid extracts from Cryptococcus gattii and Cryptococcus grubii strains is presented. Sphingolipid profiling by LC-ESI-MS/MS was used to analyze sphingosine, dihydrosphingosine, sphingosine-1-phosphate, dihydrosphingosine-1-phosphate, ceramide, dihydroceramide, glucosylceramide, phytosphingosine, phytosphingosine-1-phosphate, phytoceramide, α-hydroxy phytoceramide, and inositolphosphorylceramide species. A total of 13 sterol species were identified using GC-MS, where ergosterol is the most abundant species. The 31P-NMR-based phospholipid analysis identified phosphatidylcholine, phosphatidylethanolamine, phosphatidylinositol, phosphatidylserine, phosphatidyl-N,N-dimethylethanolamine, phosphatidyl-N-monomethylethanolamine, phosphatidylglycerol, phosphatidic acid, and lysophosphatidylethanolamine. A comparison of lipid profiles among different Cryptococcus strains illustrates a marked change in the metabolic flux of these organisms, especially sphingolipid metabolism. These data improve our understanding of the structure, biosynthesis, and metabolism of common lipid groups of Cryptococcus and should be useful while studying their functional significance and designing therapeutic interventions.
Keywords: lipid profiling, mass spectrometry, nuclear magnetic resonance, phosphoglycerides
Cryptococcus species are common environmental fungi and are well-known to cause invasive pulmonary infections in humans (1). Cryptococcal meningitis, also known as meningoencephalitis, is the most frequent and serious form of infection caused by the Cryptococcus species. According to a global estimate, “approximately 1 million cases of cryptococcal meningitis occur each year, resulting in over 600,000 deaths by 3 months of infection” (Ref. 2; p. 1). Generally, Cryptococcus infection cases are common in immunocompromised patients (2); however, in rare instances, cryptococcal infections have been reported in immunocompetent patients with an underlying liver cirrhosis condition or an end stage renal condition, also known as cryptococcal peritonitis (3), as well as immunocompetent patients with uncommon underlying disease conditions (4). Cryptococcus grubii and Cryptococcus gattii are the two most prevalent species that cause cryptococcosis in humans (5). While C. grubii is more common in immunocompromised patients, C. gattii is quite capable of infecting immunocompetent individuals as well (1).
In the last decade, a serious effort has been made to understand the mechanisms of the virulence and pathogenesis of Cryptococcus. We know that a large capsule on the outer surface, melanin synthesis, and urease activity are some of the unique features of Cryptococcus among other pathogenic fungi like Candida (6). The ability of Cryptococcus species to grow at low pH helps them to survive the killing in phagolysosomes inside the host (7). Fast dissemination of this fungus from lungs to brain allows a narrow time window to kill it; this is considering the fact that the fungal infection more often than not is secondary in nature (8). Hence, limiting the growth of this fungus in a patient with HIV, for example, becomes quite difficult. Lipids play an underlying and important role in these mechanisms regulating fungal pathogenicity (9).
For instance, several lipid biosynthetic pathway genes are now known to directly impact virulence and pathogenesis mechanisms (6, 9–12). This corroborates the fact that deletion of these lipid biosynthetic genes renders fungal strains avirulent. For example, deletion of phosphatidylserine decarboxylase (Psd)1, Psd2, and Cho2 resulted in loss of virulence in Candida albicans (13, 14). Changes in phospholipid levels can result in changes in membrane potential and fluidity, which in turn can affect the functionality of proteins associated with these lipids (15, 16). Sterols and sphingolipids are also an important membrane component, usually associated with membrane rafts (17). More complex sugar or acyl chain-linked sterols and sphingolipids are linked to the vesicular secretion pathway, capsule integrity, and virulence mechanisms in Cryptococcus (6, 10, 12, 17, 18). Loss of ergosterol biosynthetic pathway protein (Erg) genes and sphingolipid biosynthesis genes also results in avirulent fungal strains (19–22). Lately, more specifically for Cryptococcus infections, sphingolipids have been emphasized as key regulators of pathogenesis. Our group has previously shown that: 1) Glucosylceramide (GlcCer; a glycosphingolipid) is critical for growth at alkaline/neutral pH and for dissemination of Cryptococcus. The strain harboring a deletion in the GlcCer synthase 1 gene (Δgcs1) was avirulent and confined to the lung granuloma. GlcCer is an important structural component of the capsule and the biomembranes of Cryptococcus (19). 2) Inositolphosphorylceramide (IPC), a complex sphingolipid, is usually associated with lipid rafts as an important structural component in fungi. In Cryptococcus, the conditional GAL7::IPC1 mutant, in which IPC synthase 1 (IPC1) is controlled by the GAL7 promoter, shows poor growth in acidic pH when Ipc1 is downregulated (23). 3) Phytoceramides (PhytoCers) usually accumulate by the catabolism of IPC derivatives by the inositol phosphosphingolipid phospholipase C (Isc1) enzyme. Interestingly, the Δisc1 mutant shows poor oligomerization, but decent membrane raft localization, of Pma1 (an ATPase responsible for maintaining the H+ gradient across the plasma membrane) (24). Supplementation of PhytoCer reverts the oligomeric capabilities of Pma1 in the Δisc1 mutant (25). 4) The small sphingolipid molecules, sphingosine (Sph), dihydrosphingosine (dhSph), sphingosine-1-phosphate (Sph1P), and dhSph-1-phosphate (dhSph1P), have an important role in modulation of host immune response toward Cryptococcus infection (26, 27). Altogether, these published reports by our group and others have emphasized the significance of the sphingolipid biosynthetic pathway for the development inhibitors as treatment strategies against Cryptococcus (28, 29).
Lipid biosynthetic pathways have been a major resource in the development treatment measures for cryptococcal infections (28). The antifungals, amphotericin B (a polyene) and fluconazole (an azole), are commonly used for targeting fungal ergosterol biosynthesis (30). The cell wall-inhibiting compounds, like echinocandins, are ineffective in managing cryptococcal infections (31). Moreover these drugs show poor killing efficiency and have higher toxicity, demanding the discovery of new and more effective antifungal drugs (28). During the last two decades, several groups, including our group, have reported the identification and characterization of various inhibitors of the fungal sphingolipid pathway (28, 29). Despite this effort, no antifungal targeting this pathway has reached the clinic.
Considering the importance of lipids in aspects like virulence and pathogenesis, and possibly in development of therapeutic strategies, it becomes important to understand the lipid metabolism processes of Cryptococcus in greater detail. A more detailed analysis of the lipid metabolic network in Cryptococcus is necessary in order to determine the lipid structures synthesized in Cryptococcus and their routes of biosynthesis.
In the present study, using different spectroscopic approaches, we have characterized three major lipid groups of Cryptococcus, specifically, phosphoglycerides using 31P-NMR spectroscopy, sphingolipids using LC-ESI-MS/MS, and sterols using GC-MS. Between C. grubii and C. gattii strains, marked changes were revealed in the lipid compositions. Quantification of lipid subclasses among C. grubii and C. gattii strains shows that GlcCer(d19:2/18:0h) and ergosterol are the two most abundant lipids in Cryptococcus. Further, the metabolite fluxes of ceramides (Cers), dihydroceramides (dhCers), PhytoCers, α-hydroxy PhytoCers (αOHPhytoCers), and their corresponding IPCs provide useful insights into the sphingolipid metabolism of C. grubii and C. gattii strains. Analysis of the structures involved in GlcCer biosynthesis point to the key enzymes that are involved in this pathway, many of which are still uncharacterized. Structural analysis of sterol intermediates suggests the presence of three alternate pathways of ergosterol biosynthesis in Cryptococcus. Further, the pathway for phosphoglyceride biosynthesis as well as the phosphoglyceride species composition in Cryptococcus is discussed. Finally, we also discuss the most important lipid changes observed in C. grubii and C. gattii in comparison to other Cryptococcus strains, namely, Cryptococcus albidus and Cryptococcus laurentii, which are mostly nonpathogenic. Overall, these analyses significantly add to our understanding of Cryptococcus lipid structures and their biosynthesis. These findings will contribute toward characterizing the genes involved in the biosynthetic pathways of these lipids and may contribute to the development of new biological targets and treatment strategies.
MATERIALS AND METHODS
Materials
Lipid standards were purchased from Sigma-Aldrich and Avanti Polar Lipids Inc. All solvents and chemicals, unless specifically mentioned, were LC-MS grade purchased from Sigma-Aldrich and Fisher Scientific, UK.
Strains, culture conditions, and lipid extraction
Strains used in this work include: C. grubii (H99), C. gattii (R265, R272), C. albidus, and Cryptococcus laurentii (all strains from the Del Poeta laboratory at Stony Brook University, Stony Brook, NY). Cell cultures were maintained on YPD-agar plates (yeast extract, peptone, and dextrose; Difco) at 30°C. For lipid extraction, cells were grown in YNB (yeast nitrogen base containing 20 g/l glucose and 25 mM HEPES, pH 7.4) at 30°C for 16 h. Cells were centrifuged for 10 min at 2,472 g at room temperature. Supernatant was removed and the cell pellet was suspended in PBS and cells counted. For lipid extraction, a two step extraction procedure was used. The first extraction was performed using the method of Mandala (32). Briefly, 5 × 108 cells were washed twice with distilled sterile water and pelleted and suspended in 1.5 ml of Mandala lipid extraction buffer (ethanol:dH2O:diethylether:pyridine:14.2 N ammonium hydroxide; 15:15:5:1:0.018; v/v/v/v/v). The lipids, C17 Cer and C17 Sph, were added as internal standards prior to lipid extraction. Tubes were vortexed and sonicated twice at intervals of 20 s and incubated in a 60°C water bath for 15 min. The process was repeated. Cell debris was removed by centrifugation (2,472 g, 4°C, 10 min) and the supernatant was dried in a SPD210 SpeedVac system (ThermoFisher Scientific, Waltham, MA). Next, Bligh and Dyer extraction was performed by dissolving the pellet into chloroform:methanol in the ratio 1:2 (v/v) and incubating at 37°C for 1 h (33). Phase separation was performed by adding 1 ml chloroform and 1 ml dH2O with vortexing. The hydrophobic layer was taken, dried in a Speed Vac, and the lipid dry weight measured. At this stage, one-tenth of each sample was aliquoted for the determination of inorganic phosphate (Pi) (21). For Pi estimation, 0.6 ml washing buffer (10 N H2SO4:10% perchloric acid:water; 9:1:40; v:v:v) was added to each tube and incubated overnight at 150°C. Samples were then cooled and 0.9 ml ultrapure water, 0.5 ml of 0.9% ammonium molybdate, and 0.2 ml of 9% ascorbic acid solution were added to each sample in this order with vortexing and incubated at 45°C for 30 min. Readings were noted at OD820 nm and the Pi concentration was determined against the NaH2PO4 standard curve. Pi estimations were done on each sample as an additional control of the lipid extraction method efficacy (data not shown). One-half of the remaining sample, post Bligh and Dyer extraction, was used for phospholipid analysis by 31P-NMR.
The remaining half was used to perform mild alkaline hydrolysis (21, 34). Briefly, 0.5 ml chloroform and 0.5 ml 0.6 M methanolic KOH were added to the dry lipid samples, vortexed, and incubated at room temperature for 1 h. Then 0.325 ml 1 M HCl and 0.125 ml dH2O were added and vortexed. The hydrophobic layer was dried in a SpeedVac, flushed with N2, and stored at −20°C until analyzed. These samples were used for either sterol analysis by GC-MS or sphingolipid analysis by LC-ESI-MS/MS.
Sphingolipid profiling using LC-ESI-MS/MS
Sphingolipid species were detected in base hydrolyzed lipid samples by the multiple reaction monitoring (MRM) approach described previously (34, 35), using a TSQ Quantum Ultra™ triple quadrupole mass spectrometer (Thermo Scientific). Extracted lipids were suspended in a buffer containing 1 mM ammonium formate and 0.2% formic acid in methanol (buffer B). A 10 μl sample was delivered by Accela pump/autosampler (Thermo Finnigan) to the HPLC system fitted with a 3 μm C8SR, 150 × 3.0 mm column (Peeke Scientific). A two buffer mobile system was used: 2 mM ammonium formate and 0.2% formic acid in water (buffer A) and buffer B. C17 Sph (C17 base; D-erythro-Sph) and C17 Cer (d18:1/17:0; N-heptadecanoyl-D-erythro-Sph) were added as an internal standard for these analyses prior to lipid extraction. Identification of sphingolipid species by MRM scanning represents a more targeted approach. MRM depends on recording the m/z of a select parent ion unique to a sphingolipid species (Q1) and its fragmentation production which is the m/z of a select daughter ion (Q3). Molecular ions of each species were detected as [M + H]+. Specifically, dhSph (d18:0), Sph (d18:1), and phytosphingosine (PhytoSph; t18:0) could be using parent ions of m/z 302, 300, and 318, where a loss of one water molecule from the sphingoid backbone results in daughter ions, m/z 284, 282, and 300. Ions of m/z 266, 264, and 282 are a result of loss of two water molecules from the sphingoid backbone. dhSph1P, Sph1P, and PhytoSph-1-phosphate (PhytoSph1P) were detected using parent ions of m/z 382, 380, and 398 and their respective daughter ions of m/z 284, 264, and 282 that result from a loss of the phosphate moiety and one water molecule from their parents. Similarly, the complex sphingolipids, Cer/α-hydroxy Cers (αOHCers), dhCer, and PhytoCer/αOHPhytoCer were detected using characteristic daughter fragments of m/z 266, 264, and 282 (characteristic to t18:0 backbone and results from the loss of two water molecules) or by the daughter ions resulting from the loss of one water molecule from the parent molecular ion (soft fragmentation). GlcCer species were detected using daughter ions of m/z 264 (d18:1), 262 (d18:2), and 276 (d19:2) that resulted from the loss of sphingoid base and two water molecules from the parent ion. Notably, the identity of GlcCer species were confirmed using the GlcCer purified from Candida utilis, which has similar GlcCers to Cryptococcus (19, 36). IPC species were detected using daughter ions of m/z 282 (for t18:0 backbone) and m/z 310 (for t20:0 backbone). IPC species resulting from d18:0 or d18:1 backbones were detected using water loss fragments of the parent ions. IPCs are known to show poor ionization efficiency in positive ion mode. For this reason, we analyzed IPC species in negative ion mode as well (see below) (34). MRM settings for the target sphingolipid species detected in TSQ Quantum Ultra™ triple quadrupole mass spectrometer operating in positive ion mode are listed in supplemental Sheet S1. Mass instrumentation settings were as follows: HESI was used as ion source, spray voltage at 3,500 V, vaporizer temperature at 400°C, sheath gas pressure at 60 (a.u.), ion sweep gas pressure at 2.0 (a.u.), auxiliary gas pressure at 15 (a.u.), and capillary temperature was kept at 300°C. Processing of the collected data was performed using Xcaliber and LCquan software systems. Quantitation of lipid species was based on internal standard normalization using the linear regression model, as described previously (supplemental Fig. S1) (35). The levels of different sphingolipid molecular species in each sample were normalized to lipid dry weight in milligrams.
IPC species are readily ionized in negative ion mode where molecular ions are detected as [M − H]− (34, 37, 38). IPC species were also analyzed using negative precursor ion scanning using m/z 241 as the precursor ion and scanned the mass range between m/z 600 and 1,500. Unfractionated lipid extracts were directly introduced by continuous infusion into the TSQ Quantum Access triple quadrupole mass spectrometer instrument (Thermo Fisher Scientific). Mass instrumentation settings were as follows: ESI was used as ion source, spray voltage at 3,500 V, sheath gas pressure at 15 (a.u.), capillary temperature at 270°C, and collision energy at 35 V. Processing of the collected data was performed using the Xcaliber software system.
Sterol and fatty acid analysis by GC-MS
Extracted and base hydrolyzed lipid samples were derivatized using 100 μl N,O-bis(trimethylsilyl) trifluoroacetamide/trimethylchlorosilane reagent (Sigma-Aldrich) at 85°C for 90 min. Next, 50 μl of n-hexane were added to the derivatized sample and vortexed. The samples were then analyzed using a HP5-MS column (30 m × 250 μm × 0.25 μm) on an Agilent 7890 GC-MS instrument (Agilent Technologies, Santa Clara, CA). Sterol analysis was performed as described previously (39–41). Briefly, the initial column temperature of 100°C was held for 0.5 min and ramped at 35°C/min to 240°C and at 3°C/min to 305°C with a hold of 5 min. All EI-mass spectra were recorded at 70 eV with an ion source temperature of 230°C. The front inlet temperature was kept at 290°C splitless and the MSD transfer line temperature was kept at 280°C. The retention time and mass spectral patterns of cholesterol standard (Sigma) were used as a reference (42). Twenty-five micrograms of cholesterol were added as an internal standard for these analyses prior to lipid extraction. Sterol species contents in each sample were normalized to lipid dry weight in milligrams. The structures of various sterol species were confirmed either by authentic standards or by comparing previously published mass spectral fragmentation patterns or both. Characteristic identifiers of various sterol species are listed in supplemental Table S1. For fatty acid analyses, N,O-bis(trimethylsilyl) trifluoroacetamide/trimethylchlorosilane-derivatized samples were also analyzed on an HP-5MS column (30 m × 250 μm × 0.25 μm). The initial column temperature of 70°C was held for 2 min and ramped at 5°C/min to 200°C and at 20°C/min to 300°C with a hold of 5 min. All EI-mass spectra were recorded at 70 eV with an ion source temperature of 230°C. The front inlet temperature was kept at 280°C with a 10:1 split and the MSD transfer line temperature was kept at 280°C. The retention time and mass spectral patterns of fatty acid methyl ester standard mix (Sigma) were used as a reference to identify respective fatty acids.
Phosphoglyceride analysis using 31P-NMR spectroscopy
31P-NMR analysis was based on the method of Lehnhardt et al. (43) and was performed in triplicate. The phospholipid classes in the total lipid extracts were analyzed by 31P-NMR in a sodium cholate detergent system, as described by MacKenzie, Vyssotski, and Nekrasov (44). The sodium cholate detergent solution was prepared containing: sodium cholate (10% w/w), EDTA (1% w/w), and phosphonomethylglycine as an internal standard for quantification (0.3 g/l); pH was adjusted to 7.1 using sodium hydroxide; the detergent solution was an aqueous solution containing 20% D2O for deuterium field-frequency lock capability. Samples were mixed with the detergent solution, ultrasonicated (60°C, 10 min), and pH adjusted with aqueous NaOH if necessary. The quantitative phosphorus NMR spectra were recorded using a two-channel Bruker Avance300 instrument. A capillary containing 0.4% phosphoric acid was included in the NMR tube for measuring relative chemical shifts. The identities of peaks were confirmed using chemical shifts compared with those of pure standards. Analysis of one sample after deacylation using monomethylamine was also used to aid in the identification of the minor phospholipids [e.g., phosphatidyl-N,N-dimethylethanolamine (PDME) and phosphatidyl-N-monomethylethanolamine (PMME)] (45). Average molecular weights of the lipid classes were calculated based on fatty acid profiles of the lipid extracts (discussed below). 31P-NMR analysis was performed on non-base hydrolyzed lipid extracts. Phospholipid contents in each sample were normalized to lipid dry weight in milligrams and represented as %w/w. Furthermore, lipid species compositions of select lipid classes were determined by a shotgun approach using multiplexed precursor ion and neutral loss scanning experiments in positive ion mode on an AB SCIEX QTRAP® system. Lipid species for each class of phosphoglyceride were selected as: Pre 184.1 [phosphatidylcholine (PC)], NL 141 [phosphatidylethanolamine (PE)], and NL 185 [phosphatidylserine (PS)] (46). Data analysis was performed using LipidView software from SCIEX.
Statistical analyses
The Student’s t-test was employed to determine the statistical significance between data sets. P < 0.05 was considered to be significant. Data represent the mean ± SEM of at least three independent experiments. To highlight the statistical significance among different datasets, principal component analysis (PCA) was performed using the XLSTAT software.
RESULTS
Several environmental and human isolate species of Cryptococcus have been identified to cause invasive infections in humans, leading to high morbidity (1, 5). Among these, C. grubii (H99) and C. gattii strains (R265 and R272) are the most well-studied (1, 5), but little is known about their lipid profiles (19, 20, 47). In the present study, we have aimed to analyze the major groups of lipids, specifically, sphingolipids, sterols, and phosphoglycerides. For lipid analysis, Cryptococcus cells were cultured in YNB complete medium at 30°C and lipids were extracted using Mandala extraction followed by subsequent Bligh and Dyer extraction and base hydrolysis when appropriate, as described in the Materials and Methods. The extracted lipids were analyzed using different MS approaches to determine the levels of lipid intermediates.
Sphingolipid profile of C. grubii (H99) and C. gattii (R265 and R272) strains
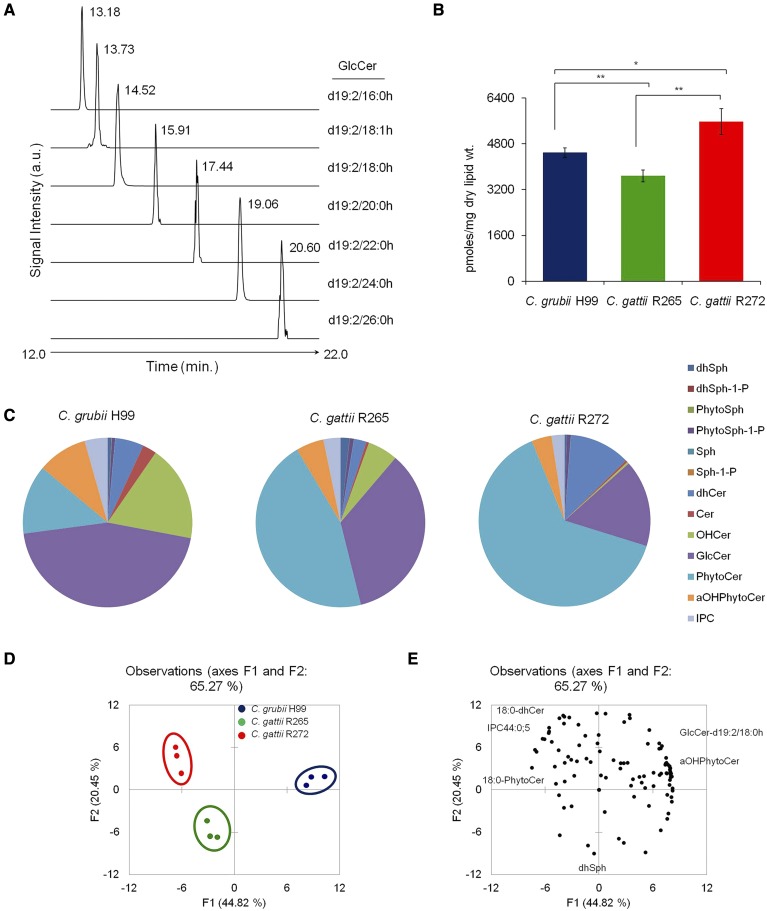
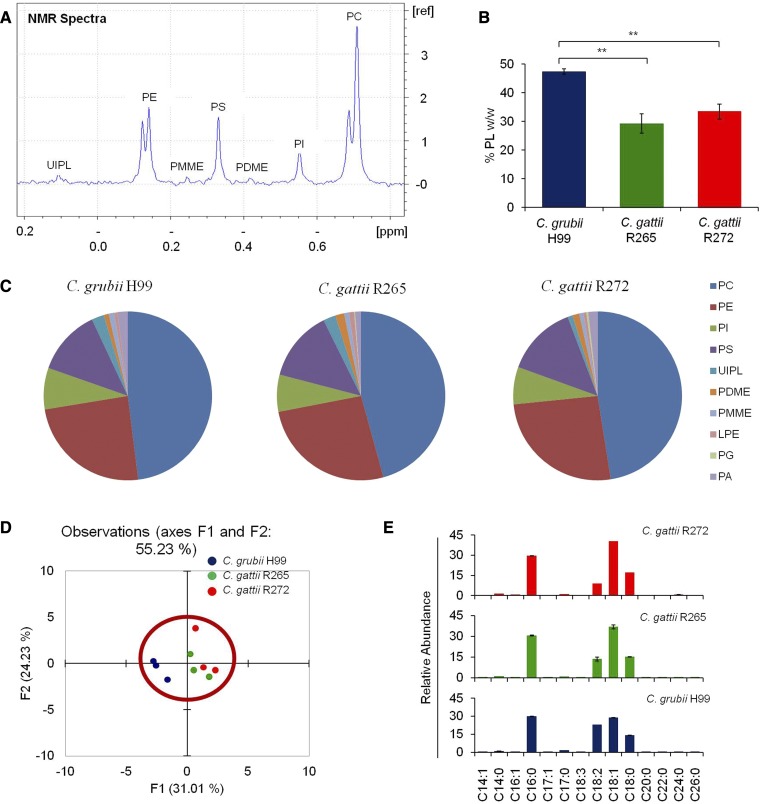
Sphingolipids represent one of the more complex lipid systems in fungi, with differences in their sphingoid backbone or long chain base (LCB), N-linked fatty acyl chain lengths, hydroxylations, and hexose linkages (10, 34). To get an in-depth map of various sphingolipid structures and contents, we employed a MRM-based LC-MS method (34, 35, 46). Lipid analytes were separated on a HPLC instrument and their identity was confirmed by MRM scanning on a mass spectrometer in positive ion mode. In Fig. 1A, it can clearly be seen that all targeted GlcCer species were resolved into individual peaks; where an increase in retention time directly corresponded to the increase in the N-linked fatty acyl chain length. Using this approach, we determined the absolute levels of 13 different classes of sphingolipids. Among these, six classes of less abundant LCBs (which are important signaling molecules) were analyzed, namely, dhSph, dhSph1P, Sph, Sph1P, PhytoSph, and PhytoSph1P, and seven classes of complex sphingolipids were analyzed, namely, dhCer, Cer, αOHCer, PhytoCer, αOHPhytoCer, GlcCer, and IPCs. The total sphingolipid content of H99, R265, and R272 was 4,490, 3,678, and 5,569 pmoles/mg dry lipid weight, respectively (Fig. 1B, supplemental Table S2). Evidently, the sphingolipid content of R272 was as much as 34% higher as compared with the H99 or R265 strains.
Fig. 1.
Sphingolipid profiling of Cryptococcus by MRM scanning using LC-MS. A: MRM monitoring chromatograms representing the identification of d19:2-sphingoid base containing GlcCer. Only GlcCer detection is shown here as a representative of sphingolipid groups analyzed. B: Total sphingolipid content in C. grubii (H99) and C. gattii (R265 and R272) strains. Mean ± SEM is shown (n = 3). *P < 0.05, ** P < 0.01 (Student’s t-test). C: Sphingolipid class composition of C. grubii and C. gattii strains. Data represents percent abundance and the means of three replicates are plotted. D: PCA of sphingolipid species analyzed shows that the three replicates are grouped, C. grubii (blue), C. gattii R265 (green), and C. gattii R272 (red), and that their lipid profiles are different. Factors F1 (x axis) and F2 (y axis) represent the two most variable principal components. E: PCA loading plots representing the contribution of individual species to the sample variability.
Of the total sphingolipid pool, the total LCB (dhSph, dhSph1P, Sph, Sph1P, PhytoSph, and PhytoSph1P) contents were between 1.2% and 2.6% in the analyzed strains. The overall LCB content did not vary much between strains, except that dhSph was 1.7- to 2.7-fold higher in R265 when compared with H99 and R272. The Cer structures (dhCer, Cer, αOHCer, PhytoCer, and αOHPhytoCer) contributed 49–80% of the total sphingolipids (Fig. 1C). However, the composition of these sphingolipids varied significantly between strains. The dhCers were abundant in R272, Cers, αOHCer, and αOHPhytoCer in H99, and PhytoCer in R265 and R272. The GlcCer content ranged between 16% and 45% among strains. GlcCer amounts were significantly higher (>2-fold) in H99 and R265, compared with R272. The IPC content ranged between 2.4% and 4.4% and was found to be highest in the C. grubii H99 strain.
Sphingolipid species profile.
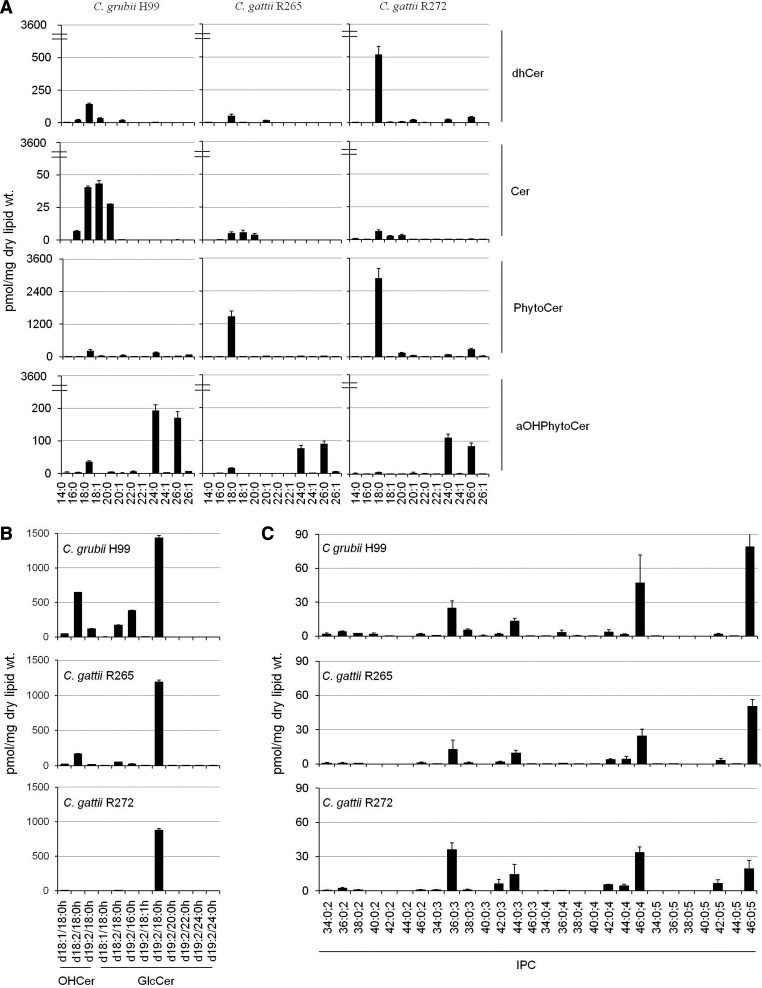
In fungi, the sphingolipid structure is comprised of a LCB backbone amide linked to a fatty acid at the C2 position and an ester linked to a polar head group at the C1 position (34). The LCBs detected in our analysis contained a d18:0 backbone, which is the major sphingoid backbone found in Cryptococcus species (34). We further analyzed various Cer-related structures based on the amide-linked fatty acyl chain on the LCB (Fig. 2A). The de novo biosynthesis of complex sphingolipid structure is marked by amide linkage of a fatty acyl to the LCB by Cer synthases (CerSs) to form dhCer. In our analysis, we scanned for fatty acyls of varying chain lengths ranging from 14 to 26 carbons. The dhCer(d18:0/18:0) species (m/z 568.6) was the major dhCer species in all three strains and most abundant in R272. A Δ4-desaturation of the LCB of dhCer forms Cer structures. Cer(d18:1/18:0) (m/z 566.5), Cer(d18:1/18:1) (m/z 564.4), and Cer(d18:1/20:0) (m/z 594.5) species were the major Cer species. These Cer species were significantly higher in H99, which also showed the presence of the Cer(d18:1/16:0) species.
Fig. 2.
Comparative sphingolipid species analyses of C. grubii and C. gattii strains. A: Cer, dhCer, PhytoCer, and αOHPhytoCer species content in C. grubii and C. gattii strains. Plots depict sphingolipid species from 14 to 26 carbon chain lengths (x axis). B: OHCer and GlcCer species content in C. grubii and C. gattii strains. C: IPC species content in C. grubii and C. gattii strains. IPC species are represented as total number of carbons in the sphingoid base and acyl chains:total number of carbon-carbon double bonds in the sphingoid base and acyl chains;number of hydroxyl groups present in the sphingoid base and acyl chains. Mean ± SEM is shown (n = 3). Breaks in the y axis of (A) have been used for better visualization of datasets.
We have previously shown that Cryptococcus contains αOHCer species, where the fatty acyl linked to the LCB is α-hydroxylated (19, 20). These are detected more prominently in the GlcCer biosynthetic pathway mutants, like Δgcs1 (mutant for GlcCer synthase) and Δsmt1 (mutant for sphingolipid C9-methyl transferase), or upon treatment with inhibitors of the enzymes synthesizing GlcCer (19, 20). These are αOHCer(d18:1/18:0h) (m/z 582.4), αOHCer(d18:2/18:0h) (m/z 580.4), and αOHCer(d19:2/18:0h) (m/z 594.4). The Δ8-desaturation of the LCB of Δ4-Cer [αOHCer(d18:1/18:0h); d18:1 backbone; 4-sphingenine] will result in formation of Δ4,Δ8-Cer [αOHCer(d18:1/18:0h); d18:2 backbone; 4,8-sphingadienine]. Further, the addition of a methyl group at the C9 position of the 4,8-sphingadienine base of Δ4,Δ8-Cer forms 9-methyl-Δ4,Δ8-Cer [αOHCer(d19:2/18:0h); d19:2 backbone; 9-methyl-4,8-sphingadienine]. All three species, αOHCer(d18:1/18:0h), αOHCer(d18:2/18:0h), and αOHCer(d19:2/18:0h), were detected in H99 and R265, but were negligible in R272 (Fig. 2B).
Formation of GlcCer requires the transfer of the glucose moiety from (UDP)-glucose to the C1 hydroxyl group of the αOHCer (19). Earlier, we showed that the 9-methyl-4,8-sphingadienine backbone containing GlcCer(d19:2/18:0h) (m/z 756.4) species marks the end product of the GlcCer biosynthetic pathway (19). This fact is confirmed by the current analysis, as the GlcCer(d19:2/18:0h) species was the most abundant GlcCer structure in three tested strains. In this study, we additionally scanned for six other fatty acyls (16:0h, 18:1h, 20:0h, 22:0h, 24:0h, and 26:0h) in the 9-methyl-4,8-sphingadienine backbone containing the GlcCer pool (Fig. 2B). Indeed, we found a considerable amount of GlcCer(d19:2/16:0h) species (m/z 728.4) in H99 and a detectable level in R265. Almost all of the GlcCer pool in R272 was comprised of GlcCer(d19:2/18:0h) species only. Among the alternate GlcCer species reported in Cryptococcus [namely, GlcCer(d18:1/18:0h) (m/z 744.4) and GlcCer(d18:2/18:0h) (m/z 742.4)], the GlcCer(d18:2/18:0h) species was present in H99. It is noteworthy that we also detected low levels of GlcCer species with nonhydroxylated fatty acids like GlcCer(d19:2/18:0) (m/z 740.4) [data not shown (19)].
The addition of a hydroxyl group at the C4 position of the dhSph backbone results in the production of PhytoSph (t18:0 backbone). Transfer of a nonhydroxylated fatty acyl to the amide group at the C2 position will form PhytoCer structures (34). PhytoCer(d18:0/18:0) (m/z 584.7), PhytoCer(d18:0/24:0) (m/z 668.8), and PhytoCer(d18:0/26:0) (m/z 696.7) species were detected as the major PhytoCer species and were more abundant in R265 and R272, compared with H99 (Fig. 2A). The α-hydroxylation of the fatty acyl moiety of PhytoCers forms αOHPhytoCers. The αOHPhytoCer(d18:0/24:0h) (m/z 684.5) and αOHPhytoCer(d18:0/26:0h) (m/z 712.5) species were detected as the major αOHPhytoCer species and were more abundant in H99 (Fig. 2A).
Further, both PhytoCer and αOHPhytoCer structures can be converted to IPC structures by transfer of a phosphorylinositol group (23). Although the IPC structures and pathway of synthesis have been characterized in several fungi (36, 48–54), they remain poorly studied in Cryptococcus (23). In our MRM analysis (positive ion mode), we scanned for 28 IPC structures (Fig. 2C). IPC species are represented as “total number of carbons in the sphingoid backbone and the fatty acyls:total number of double bonds;total number of hydroxyl groups in the sphingoid backbone and the fatty acyls.” IPC species, IPC36:0;3 (m/z 826.7), IPC42:0;3 (m/z 910.8), IPC44:0;3 (m/z 938.7), IPC42:0;4 (m/z 926.5), IPC44:0;4 (m/z 954.5), IPC46:0;4 (m/z 982.5), IPC42:0;5 (m/z 942.5), and IPC46:0;5 (m/z 998.5), were detectable (IPC derivatives 3B, 4, and 5; based on the total number of hydroxyl groups in the sphingoid backbone and the fatty acyls), along with some other low-level IPC structures (IPC derivatives 2 and 3A). The amounts of these IPC species were quite similar in the H99, R265, and R272 strains. Previous studies have used the negative ion mode to detect IPC structure because better ionization could be achieved. To confirm our MRM data, we analyzed various IPC structures using precursor ion scanning for m/z 241 in negative ion mode (supplemental Fig. S2). These analyses confirmed that the IPC species detected in MRM scanning were true structures. However, some IPC structures did show a much better ionization in the negative ion mode compared with the positive ion MRM scans, for example, IPC(36:0;3) (supplemental Table S3). In both analyses, we could detect low levels of IPC structures with two hydroxyl groups in the structure, for example, IPC(36:0;2), suggestive of the fact that the transfer of a phosphorylinositol group to dhCer species can also contribute to the IPC pool. The caution here remains that there are two major drawbacks in analyzing IPC structures by MS methods: variations in ionization efficiencies and lack of appropriate standards. Notably, the complex IPC derivatives, namely the mannosylinositolphosphorylceramide and mannosyldiinositolphosphorylceramide structures, were below detection in our analysis (data not shown).
To validate the statistical significance of data sets, we performed PCA. PCA plots between the two most variable principal components (contributing to 65% variance) showed that the overall sphingolipid species content was significantly different between H99, R265, and R272 (Fig. 1D). The sphingolipid species contributing most to this strain distribution in the PCA plots are depicted by PCA loading plots (Fig. 1E).
Sphingolipid biosynthetic pathway.
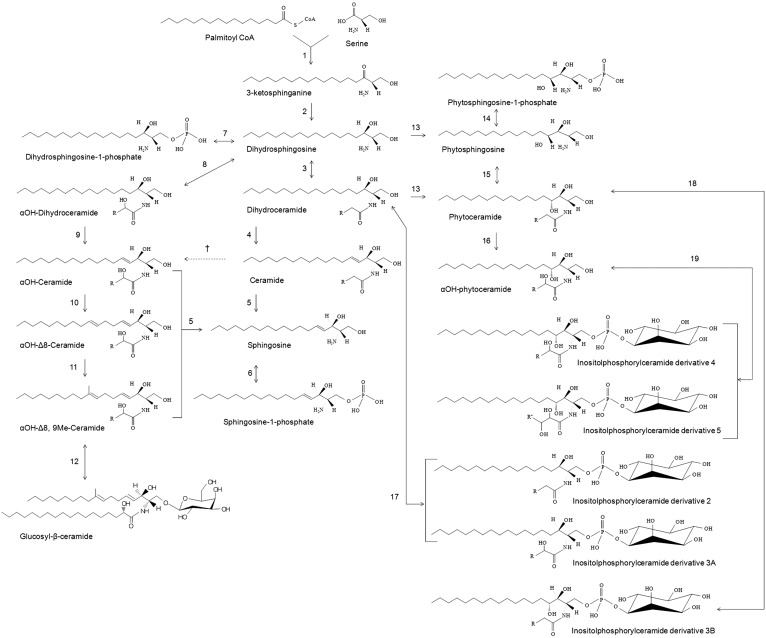
Although the genes involved in the biosynthesis of various sphingolipid structures are well-characterized in other model yeasts, like Saccharomyces cerevisiae, and, in part, also in C. albicans (34, 49, 50), only a few genes have been characterized in Cryptococcus species (19, 21, 23, 24). The primary step in understanding the overall sphingolipid metabolism was to understand the structure and composition of various sphingolipid species present in Cryptococcus. Therefore, in this study we aimed to detect and quantify various sphingolipid species of Cryptococcus. Our results clearly show that similar sphingolipid species are present in both C. grubii (H99) and C. gattii (R265 and R272) strains (Fig. 2), suggesting that the sphingolipid biosynthesis pathway is largely conserved in these strains. However, we did find differences in the amounts of various sphingolipid species in the H99, R265, and R272 strains (Fig. 2). One possible reason for these observed differences in the amounts of sphingolipid metabolites could be that the enzymes involved at the catalytic steps have variable substrate specificities in each strain. This altered sphingolipid metabolic flux, of course, will depend on the overall physiological requirement of each strain and may be linked to the different environment with which they are mostly associated: birds for C. grubii and trees for C. gattii. The structure and composition analyses data obtained in this study allowed us to chart the sphingolipid biosynthetic pathway of Cryptococcus (Fig. 3).
Fig. 3.
Pathway of sphingolipid biosynthesis in Cryptococcus. Based on the profile obtained for H99, R265, and R272, the pathway of sphingolipid biosynthesis in Cryptococcus is composed of the following enzymatic steps (reverse step enzymes are in square brackets): (1) SPT; (2) KDR; (3) dhCer synthases [ceramidases]; (4) dhCer desaturases or Δ4-desturase; (5) ceramidases; (6) Sph kinase [sphingosine-1-phosphate phosphatase]. It is important to note here that several different sphingoid bases (or LCBs) have been reported in pathogenic fungi [reviewed in (34)]; (7) dhSph kinase [dhSph-1-phosphate phosphatase]; (8) dhCer synthases [ceramidases]; (9) dhCer desaturases or Δ4-desturase; (10) Δ8-desturase; (11) sphingolipid C9 methyltransferase; (12) GlcCer synthases [glycoceramidases]; (13) C4-hydroxylase; (14) PhytoSph kinase [PhytoSph1P phosphatase]; (15) CerSs; (16) α-hydroxylase; (17, 18, 19) IPC synthases [inositol phosphosphingolipid phospholipase C]. “R” represents carbons 14 to 24 acyl chains; † α-hydroxylase, putative step.
The de novo biogenesis of sphingolipids in Cryptococcus commences with the condensation of palmitoyl-CoA and L-serine [by a serine palmitoyltransferase (SPT)], and subsequent reduction to dhSph (d18:0 backbone or LCB) by 3-ketodihydrosphingosine reductase (KDR). CerSs then amide link a fatty acyl chain to the dhSph structure forming dhCer (34). Our data show that CerSs majorly utilize 18:0 fatty acyl in Cryptoccocus, but can also use 16:0 or 18:1 fatty acyls (as in H99) and 24:0 or 26:0 fatty acyls (as in R272) to form dhCer structures. A Δ4-desaturation of LCB in the dhCer structure by a Δ4-desaturase forms Cer structures containing the d18:1 backbone.
Notably, if the Cer structures are comprised of nonhydroxylated fatty acyls, then these Cers do not contribute much toward the GlcCer pool of Cryptococcus. This is supported by the fact that the major GlcCer species is GlcCer(d19:2/18:0h) in Cryptococcus and contains a hydroxylated fatty acid [also reported earlier by our group (19, 21)] and that only low levels of GlcCer species with nonhydroxylated fatty acyls, like GlcCer(d19:2/18:0), were observed (data not shown) (19). The GlcCer(d19:2/18:0h) structure is marked by three different characteristics: a double bond at the C8 position of the LCB, methylation at the C9 position of the LCB, and the amide-linked α-hydroxylated fatty acyl (10). It is likely that the GlcCer biogenesis pathway in Cryptococcus runs parallel to the routine Cer pathway, with specific CerSs being able to amide link a hydroxylated fatty acyl chain to the dhSph structure forming αOHdhCer. The αOHdhCers are rapidly desaturated by Δ4-desaturase at the C4 position of the LCB to form αOHCer, the major species being Cer(d18:1/18:0h). Further desaturation by Δ8-desaturase at the C8 position of the LCB should form the Cer(d18:2/18:0h) structure. A methylation step at the C9 position of the LCB by sphingolipid C9-methyl transferase [characterized as Smt1 in Cryptococcus (21)] results in formation of Cer(d19:2/18:0h). Finally, transfer of the glucose moiety to the C1 hydroxyl group by GlcCer synthase [characterized as Gcs1 in Cryptococcus (19)] forms the end product GlcCer(d19:2/18:0h). The fact that we could detect other GlcCer species, like GlcCer(d19:2/16:0h), in our analysis shows that biosynthesis of αOHCer structure is a critical step for GlcCer biosynthesis. It is noteworthy that the activity of Gcs1 is not entirely limited to αOHCer structures. Gcs1 can, to some extent, utilize nonhydroxylated Cer structures as well as synthesize GlcCer (19). Further, based on our data, we are tempted to speculate whether the nonhydroxylated fatty acyl-linked Cer structures could be converted to αOHCer structures by an α-hydroxlase (Fig. 3).
Another group of sphingolipids containing a t18:0 backbone was detected in our analysis. A dhSph-C4-hydroxylase transfers a hydroxyl group at the C4 position of dhSph to form PhytoSph, resulting in a t18:0 backbone. A specific CerS should catalyze the transfer of nonhydroxylated fatty acyls (C18:0, C24:0, or C26:0) to the amide group of PhytoSph at the C2 position to form PhytoCer structures [for example, PhytoCer(t18:0/18:0), PhytoCer(t18:0/24:0), or PhytoCer(t18:0/26:0)]. A sphingolipid α-hydroxylase will transfer a hydroxyl group at the C2 position on PhytoCers to form αOHPhytoCer structures [for example, αOHPhytoCer(t18:0/18:0h), αOHPhytoCer(t18:0/24:0h), or αOHPhytoCer(t18:0/26:0h)]. Transfer of a phosphorylinositol group onto the PhytoCer and αOHPhytoCer structures is catalyzed by IPC synthase (characterized as Ipc1 in Cryptococcus) resulting in the formation of IPC derivatives (derivatives 4, 5, and 3B; see Fig. 3). Formation of IPC derivative 5 is speculated to be due to transfer of an additional hydroxyl group onto the fatty acyl of IPC derivative 4 by an unknown hydroxylase. In our analysis, we could also detect IPC derivatives 2 and 3A, suggestive of the fact that Ipc1 can also utilize dhCer and αOHdhCer as substrates.
Pathways of sphingolipid biosynthesis and genes involved therein are well-characterized in several fungi, such as S. cereviseae, Aspergillus fumigatus, Aspergillus nidulans, C. albicans, Pichia pastoris, and others (34, 37, 47–54). In one example of P. pastoris, the pathway of GlcCer biosynthesis very closely fits with the pathway present in Cryptococcus (19, 21, 54). However, in Cryptococcus, only four genes have been studied in detail (SMT1, GCS1, IPC1, and ISC1) (19, 21, 23, 24). Therefore, it becomes demanding to determine the substrate specificities and kinetics of various uncharacterized enzymes involved in GlcCer biosynthesis, specifically SPT, KDR, CerS, Δ4-desaturase, and Δ8-desaturase, in Cryptococcus. It will also be interesting to study to determine whether Cryptococcus is able to utilize the PhytoCer backbone for GlcCer biosynthesis, as found in GCS overexpressed strains of P. pastoris (54). Similar studies are required for the uncharacterized or unknown enzymes involved in IPC biosynthesis, namely, dhSph-C4-hydroxylase and CerS. For example, the presence of closely related Cer, αOHCer, and PhytoCer structures in the sphingolipid pool suggests that there could be several CerSs in Cryptococcus, each with specific activity. Multiple CerSs have been reported in other systems, namely, Lip1/Lac1/Lag1 (in S. cereviseae) (55), BarA/LagA (in A. nidulans) (56), and Lac1/Lag1 (in C. albicans) (49).
Several intermediates involved in the sphingolipid breakdown pathway were also identified in all three tested strains. These were Sph, Sph1P, dhSph1P, and PhytoSph-1P [LCB-1-phosphates (LCB-1Ps)]. Degradation of Cer by ceramidase enzyme results in formation of Sph, which is then phosphorylated by Sph kinase into Sph1P. Similarly, dhSph kinase and PhytoSph kinase catalyze the conversion of dhSph into dhSph1P and PhytoSph into PhytoSph-1P, respectively. The phosphorylation step is reversible and is catalyzed by specific phosphatases. These LCB-1P structures play an important role in the regulation of phagocytic response in Cryptococcus (26, 27). Therefore, this kinase-phosphatase regulation of LCB-1P is crucial for maintaining LCB-1P levels. In the final degradation step, the LCB-1Ps are catabolized by LCB-1P lyases. The enzymes involved in sphingolipid degradation are known in other fungi (57–59), but require a much needed characterization in Cryptococcus. The majority of these enzymes are fungi specific and, therefore, are potential targets for drug development.
Sterol profile of C. grubii (H99) and C. gattii (R265 and R272) strains
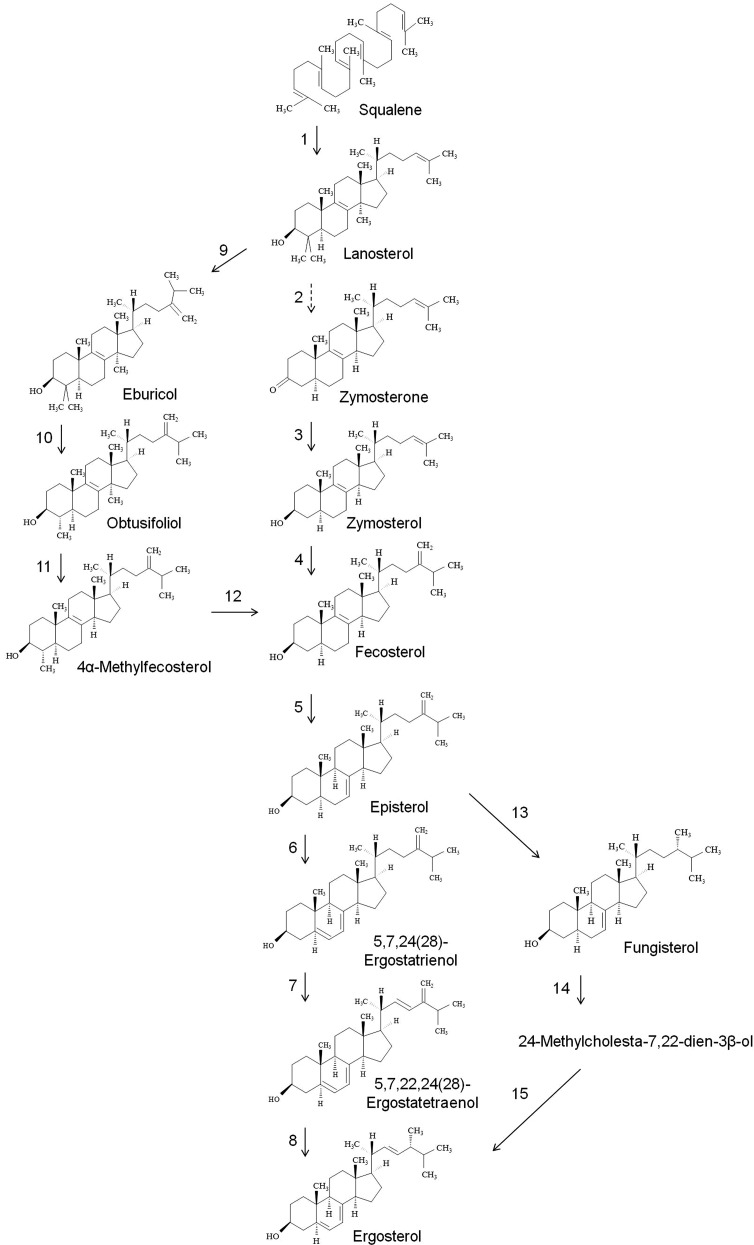
The sterol composition of Cryptococcus is of high significance, as all the major antifungals used currently in clinics target the ergosterol biosynthetic pathway (28). Ergosterol has a key role in the regulation of membrane fluidity, protein folding and localization, and the cell cycle (12, 17). Therefore, an analysis of the ergosterol biosynthetic pathway of Cryptococcus will be useful in designing new antifungals. The ergosterol biosynthetic pathway has been studied in S. cereviease, A. fumigatus, C. albicans, and C. glabrata, and is comprised of over 20 enzymatic steps, with multiple alternative pathways for the ergosterol biosynthesis (60–64). In Cryptococcus, our knowledge of the ergosterol biosynthetic pathway is limited to only a few studies (39, 60, 65, 66). The ergosterol studies performed earlier by both Nes et al. (39) and Ghannoum et al. (60) were performed only on C. grubii strains. Both studies used specific inhibitors to block the ergosterol biosynthetic pathway, but presented a contradictory composition profile for sterol intermediates (39, 60). Therefore, further studies to understand the ergosterol biosynthetic pathway in Cryptococcus are required.
Here, we describe the analysis of various intermediates of the ergosterol biosynthetic pathway of several Cryptococcus strains (both C. grubii and C. gattii). These analyses provide valuable pieces of information about ergosterol biosynthesis in Cryptococcus, including sterol intermediates and possible routes of biosynthesis. Various sterol structures were analyzed by the GC-MS technique, as described in the Materials and Methods.
Ergosterol pathway intermediates.
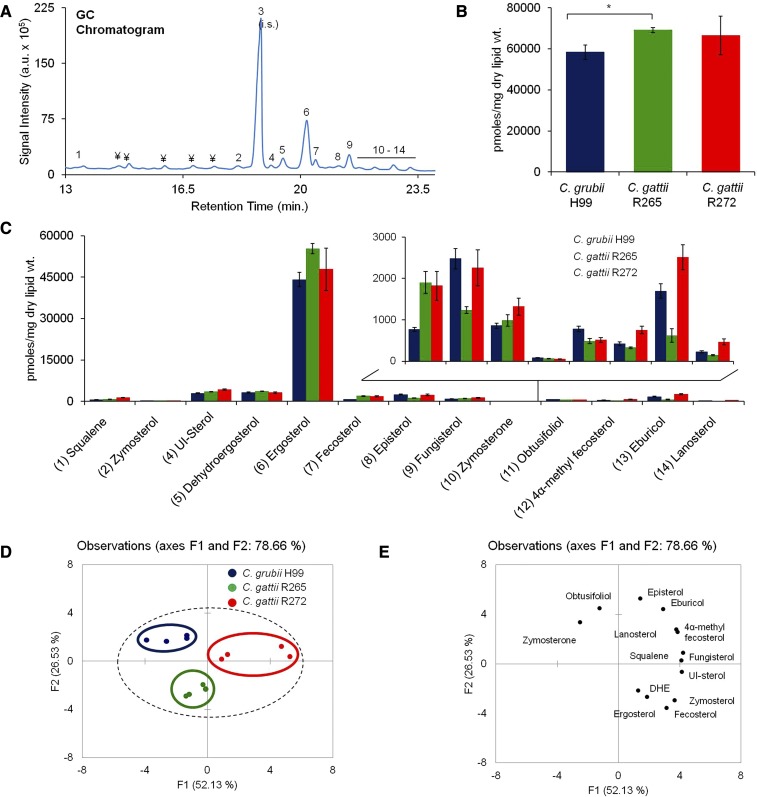
In our GC-MS analysis, we detected 13 intermediate metabolites involved in ergosterol biosynthesis. Various structures were identified by comparison with known sterol spectra and 12 sterol intermediates were identified. Figure 4A represents the order of elution (or the retention time in GC) for various sterol structures. These structures are: 1) squalene: 2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene; 2) zymosterol: (3b,5a)-cholesta-8,24-dien-3-ol; 3) cholesterol: (3β)-cholest-5-en-3-ol (internal standard); 4) unidentified sterol; 5) dehydroergosterol: (3β,22E)-ergosta-5,7,22,24(26)-tetraen-3-ol; 6) ergosterol: 3β,22E)-ergosta-5,7,22-trien-3-ol; 7) fecosterol: (3β,5α)-ergosta-8,24(28)-dien-3-ol; 8) episterol: (3β,5α)-ergosta-7,24(28)-dien-3-ol; 9) fungisterol: (3β,5α)-ergost-7-en-3-ol; 10) zymosterone: (5α)-cholesta-8,24-dien-3-one; 11) obtusifoliol: (3β,4α,5α)-4,14-dimethylergosta-8,24(28)-dien-3-ol; 12) 4α-methyl fecosterol: (3β,4α,5α)-4-methylergosta-8,24(28)-dien-3-ol; 13) eburicol: (3β)-24-methylenelanost-8-en-3-ol; and 14) lanosterol: (3β)-lanosta-8,24-dien-3-ol. The total sterol content was 58,347 pmol/mg dry lipid weight for H99, 69,089 pmol/mg dry lipid weight for R265, and 66,552 pmol/mg dry lipid weight for R272 strain (Fig. 4B). Ergosterol contributed to about 72–80% of the total sterols. Although the ergosterol content of H99 was slightly lower compared with R265 and R272 strains, these changes were not dramatic (Fig. 4C).
Fig. 4.
Sterol analysis of Cryptococcus by using GC-MS. A: A representative total ion chromatogram showing detection of sterols in the C. grubii strain. (1) squalene; (2) zymosterol; (3) cholesterol, internal standard; (4) unidentified sterol (UI-sterol); (5) dehydroergosterol; (6) ergosterol; (7) fecosterol; (8) episterol; (9) fungisterol; (10) zymosterone; (11) obtusifoliol; (12) 4α-methyl fecosterol; (13) eburicol; (14) lanosterol. Peaks marked with “¥” are nonsterol metabolites. B: Total sterol content in C. grubii and C. gattii strains. Mean ± SEM is shown (n = 3). *P < 0.05 (Student’s t-test). C: Sterol class composition of C. grubii and C. gattii strains. D: PCA of sterol species analyzed shows that the three replicates are grouped, C. grubii (blue), C. gattii R265 (green), C. gattii R272 (red), and that their lipid profiles are marginally different. Factors F1 (x axis) and F2 (y axis) represent the two most variable principal components. E: PCA loading plots representing the contribution of individual species to the sample variability.
While ergosterol was detected as the major peak in the Cryptococcus lipid extracts, other sterol intermediates ranged between <1% and 6.4% between the different tested strains (Fig. 4C). We observed significant changes in the levels of the less abundant sterol species, as compared with the most abundant ergosterol and dehydroergosterol species. Among the minor sterol species, zymosterol, fecosterol, and unidentified sterol were present in higher amounts (as much as 2.5-fold) in R265 and R272 compared with the H99 strain. Also, zymosterone and obtusifoliol were present in lower amounts (as much as 1.6-fold) in R265 and R272 compared with the H99 strain. The levels of other intermediates, namely, squalene, fungisterol, 4α-methyl fecosterol, eburicol, and lanosterol, were high (as much as 2.2-fold) in R272 compared with H99 or R265. These data were statistically validated by PCA, which showed that, indeed, the differences in minor sterol intermediates were significant enough to effectively separate H99, R265, and R272 strains on PCA plots (Fig. 4D, E). The overall PCA plots show that the sterol profiles of H99, R265, and R272 strains are quite close, mainly because a significant contribution to the sterol pool comes from ergosterol, the levels of which do not change much between strains.
Sterol biosynthesis.
Sterol biosynthesis in fungi, depending on the species and strain, is known to occur via several different routes (60–65). The steps involved in ergosterol biosynthesis are catalyzed by ERG genes. More than 20 Erg proteins are known to be involved in ergosterol biosynthesis. While most have been characterized in different fungal strains, only a few, like Erg11 (lanosterol 14α-demethylase), are characterized in Cryptococcus species (65, 66). To date, only a few studies have attempted to explain the biosynthesis of sterols in Cryptococcus (39, 60, 65, 66). Based on the structures detected in our sterol analysis, the ergosterol biosynthetic pathway can be divided into three categories (Fig. 5). First is the classical pathway of ergosterol biosynthesis that has been established in fungi. The beginning of ergosterol biosynthesis is marked by cyclization of squalene into lanosterol by squalene monooxygenase (Erg1) and 2,3-oxidosqualene-lanosterol cyclase (Erg7). Subsequent conversion of lanosterol into fecosterol in several enzymatic steps is catalyzed by lanosterol 14-α-demethylase (Erg11), C14 sterol reductase (Erg24), C4 methyl sterol oxidase (Erg25), C-3 sterol dehydrogenase (Erg26), 3-keto sterol reductase (Erg27), and C24-sterol methyltransferase (Erg6). The fact that very low levels of the intermediates, zymosterol and zymosterone (<0.5% of the total sterols), were detected in our analysis, suggests that the conversion steps of lanosterol to fecosterol occur at very rapid rates. Further, fecosterol is converted into ergosterol in the several steps catalyzed by C8-sterol isomerase (Erg2), C5-sterol desaturase (Erg3), C22-sterol desaturase (Erg5), and C24-sterol reductase (Erg4). Running parallel to this pathway is the first alternative pathway of sterol biosynthesis involving the formation of eburicol, obtusifoliol, and 4α-methyl fecosterol (Fig. 5). These sterol intermediates have been known to accumulate in cells deficient in 14α-sterol demethylase enzymes (Cyp51A or Cyp51B) or in the cells treated with fluconazole or itraconazole drugs. In our analysis, the total level of eburicol, obtusifoliol, and 4α-methyl fecosterol was 2.1–5.6% (percent of total sterols), suggesting that this pathway could also be directly involved in the biosynthesis of ergosterol. This pathway has been shown to be active in several pathogenic fungi and involves Erg6, Erg25, Erg26, Erg27, and Erg11 proteins (62–68). Briefly, methylation of C24 in lanosterol structure results in formation of eburicol. Two subsequent demethylations at C4 and C14 result in formation of 4α-methyl fecosterol, which can be further demethylated by Erg11-like proteins [Cyp51A or Cyp51B (66)] to form fecosterol.
Fig. 5.
The pathway of sterol biosynthesis in Cryptococcus. Based on the profile obtained for H99, R265, and R272, the pathway of sterol biosynthesis in Cryptococcus is composed of the following enzymatic steps: (1) squalene monooxygenase (Erg1), 2,3-oxidosqualene-lanosterol cyclase (Erg7); (2) lanosterol 14-α-demethylase (Erg11), C14 sterol reductase (Erg24), C4 methyl sterol oxidase (Erg25), C-3 sterol dehydrogenase (Erg26); (3) 3-keto sterol reductase (Erg27); (4) C24-sterol methyltransferase (Erg6); (5) C8-sterol isomerase (Erg2); (6) C5-sterol desaturase (Erg3); (7) C22-sterol desaturase (Erg5); (8) C24-sterol reductase (Erg4); (9) Erg6; (10, 11, 12)£ Erg25, Erg26, Erg27 and Erg11; (13) Erg4; (14) Erg5, and (15) Erg3. “£” represents putative enzymes involved in the step.
The second alternative pathway involves the biosynthesis of (3β,5α)-ergost-7-en-3-ol (or fungisterol) intermediate (Fig. 5). Fungisterol, detected earlier in the sterol extracts of pathogenic fungi as well as in several Erg mutants (68, 69), was also detected in Cryptococcus lipid extracts in this study (as much as ∼2% of the total sterol). Erg4 can use episterol as substrate and catalyze a reduction step at C24 to form fungisterol. Fungisterol can then be converted to ergosterol in two subsequent steps involving a desaturation at C22 (which forms 24-methylcholesta-7,22-dien-3β-ol) by Erg5 and a reduction at C24 by Erg3. Ergosterol biosynthesis remains a key drug target for treating pathogenic fungal infections. However, the most commonly used antifungal drug, fluconazole, has been largely ineffective to treat Cryptococcus infections (22, 60). This could be because there are multiple pathways of ergosterol biosynthesis in Cryptococcus. Although we did find differences in sterol composition of various Cryptococcus strains, these differences do contribute much toward fluconazole susceptibility (60).
Phosphoglyceride profile of C. grubii (H99) and C. gattii (R265 and R272) strains
Phosphoglycerides are critical components of all fungal membranes and to the physiological processes happening therein (11, 12). For example, gene mutants defective in amino-phosphoglycerides, like PE and PS, are unable to grow on nonfermentable carbon sources, have abnormal mitochondria, are temperature sensitive, and are avirulent in some cases (13, 70). The biophysical properties of different phosphoglyceride structures have been explored using in vitro vesicle studies (71–73). The majority of these works have been done on fungi like S. cereviseae (74), Schizosaccharomyces pombe (75) and C. albicans (13, 14). Although the phosphoglyceride biosynthetic pathway is quite conserved in fungi (74), the studies focusing on characterizing the phosphoglyceride structures and compositions of Cryptococcus are very limited. In order to understand the phosphoglyceride metabolism of Cryptococcus in detail, we assessed the composition of phosphoglycerides by 31P-NMR in lipid extracts of H99, R265, and R272 strains (Fig. 6).
Fig. 6.
Phosphoglyceride profiling of Cryptococcus using 31P-NMR. A: A representative spectra of 31P-NMR showing detection of phospholipids in the C. grubii H99 strain. UIPL, unidentified phospholipid. B: Total phospholipid content in the C. grubii and C. gattii strains. Data represents %w/w. Mean ± SEM is shown (n = 3). **P < 0.01 (Student’s t-test). C: Phospholipid class composition of C. grubii and C. gattii strains. The percent abundance and means of three replicates are plotted. D: PCA of phospholipids shows that the three replicates are grouped, C. grubii (blue), C. gattii R265 (green), C. gattii R272 (red), and that these lipid profiles are similar. Factors F1 (x axis) and F2 (y axis) represent the two most variable principal components. E: Fatty acid composition of C. grubii and C. gattii strains as detected using GC-MS. Relative abundances of fatty acyls with 14–26 carbons are plotted as mean ± SEM (n = 3).
Phosphoglyceride classes.
31P-NMR is a reliable method for determining the phospholipid class profile from lipid extracts. In our 31P-NMR analysis (43–45), 10 subclasses of phosphoglycerides were detected in all three tested strains: PC, PE, phosphatidylinositol (PI), PS, PDME, PMME, lysophosphatidylethanolamine (LPE), phosphatidylglycerol (PG), phosphatidic acid (PA), and an unidentified phospholipid. The total phosphoglyceride levels measured by 31P-NMR for H99 (47.4% w/w) were significantly lower than the R265 (29.3% w/w) or the R272 strain (33.5% w/w) (Fig. 6B).
The 31P-NMR spectrum of lipid extracts of Cryptococcus strains showed baseline peak resolution of major phosphoglycerides, namely, PC, PE, PI, and PS (Fig. 6A). Quantitative measurements showed that these major phosphoglycerides constituted 92.7–94.2 mol% of the total phosphoglycerides in tested Cryptococcus strains. While the minor phosphoglyceride classes, namely, PDME, PMME, LPE, PG, PA, and unidentified phospholipid, constituted 5.8–7.3 mol% of the total phosphoglycerides in tested Cryptococcus strains. Evidently there was no significant difference in levels of various phosphoglyceride classes between H99, R265, and R272 strains (Fig. 6C). Other lysophosphoglycerides, namely, lyso-PC, LPE, lyso-PI, lyso-PS, and lyso-PG, were below detection limits in our 31P-NMR analysis. The statistical significance of these data was validated by PCA, which showed no apparent differences in the phosphoglyceride profiles of H99, R265, and R272 strains (Fig. 6D).
Fatty acyl composition.
In a phosphoglyceride structure, the carbonyl groups of two fatty acyls are esterified to the hydroxyl groups at the C1 and C2 position of a glycerol backbone, while a phosphoric acid group is esterified to the hydroxyl group at the C3 position (74). Based on their position, chain lengths, and degree of unsaturation, these fatty acyls can easily modulate the biophysical properties of the phosphoglycerides in the membrane (71, 72). Our GC-MS analysis showed that four major fatty acyls, namely, C16:0, C18:0, C18:1, and C18:2, contributed to 96 mol% of the total fatty acyl pool in H99, R265, and R272 strains (Fig. 6E). We used these fatty acyl profiles to calculate the average molecular weights of the lipid classes in the 31P-NMR analysis. We observed a high level of C18:2 fatty acyl in H99 (23 mol% of total fatty acyls) compared with R265 (13.6 mol% of total fatty acyls) or R272 (8.7 mol% of total fatty acyls) strains. Concurrently, a low level of C18:1 fatty acyl in H99 (28.7 mol% of total fatty acyls) compared with R265 (36.9 mol% of total fatty acyls) or R272 (40.3 mol% of total fatty acyls) strains was observed. It is possible that the activity of fatty acyl desaturases [like Ole1 and Ole2 in C. albicans (76)] could contribute to a high C18:2/C18:1 ratio (of 0.8) in the H99 strain > C18:2/C18:1 ratio (of 0.4) in the R265 strain > low C18:2/C18:1 ratio (of 0.2) in the R272 strain. It is important to note that we used a HP5-MS column to separate fatty acyls, which shows poor resolution between C18:2 and C18:3 fatty acyls. Therefore, some part of the C18:2 mass spectral signal could be contributed by the C18:3 mass spectral signal.
Phosphoglyceride species composition.
To determine the individual phosphoglyceride species, we used multiple precursor ion and neutral loss scans, described previously in (46). The analysis was performed on H99 lipid extract and lipid species belonging to three major phosphoglycerides, namely, PC, PE, and PS, were determined (supplemental Fig. S3). The phosphoglyceride species are represented as the total number of carbons in the two fatty acyl chains:total number of double bonds in the two fatty acyl chains. In Cryptococcus, PC species, namely, 34:1, 34:2, 34:3, 36:1, 36:2, 36:3, 36:4, and 36:5, contribute to 84% of the total PC pool. PE species, namely, 34:1, 34:2, 36:1, 36:2, 36:3, 36:4, 46:0, and 46:1, contribute to 74% of the total PE pool. PS species, namely, 34:0, 34:1, 34:2, 36:1, 36:2, 36:3, 36:4, and 36:5, contribute to 98% of the total PS pool. These data suggest that phosphoglyceride species in Cryptococcus are mainly composed of nonhydroxylated 16C and 18C fatty acyls, and are mostly mono- or polyunsaturated. Contrastingly, complex sphingolipid structures, like the IPCs, in Cryptococcus contain very long chain hydroxylated fatty acyls (24C or 26C). These results are consistent with a previous study on Δcap67 (an acapsular mutant of Cryptococcus neoformans), which showed similar phosphoglyceride composition (47). Nonetheless, a more targeted study is required to confirm the nonexistence of phosphoglyceride species with hydroxylated fatty acyls.
Phosphoglyceride biosynthesis.
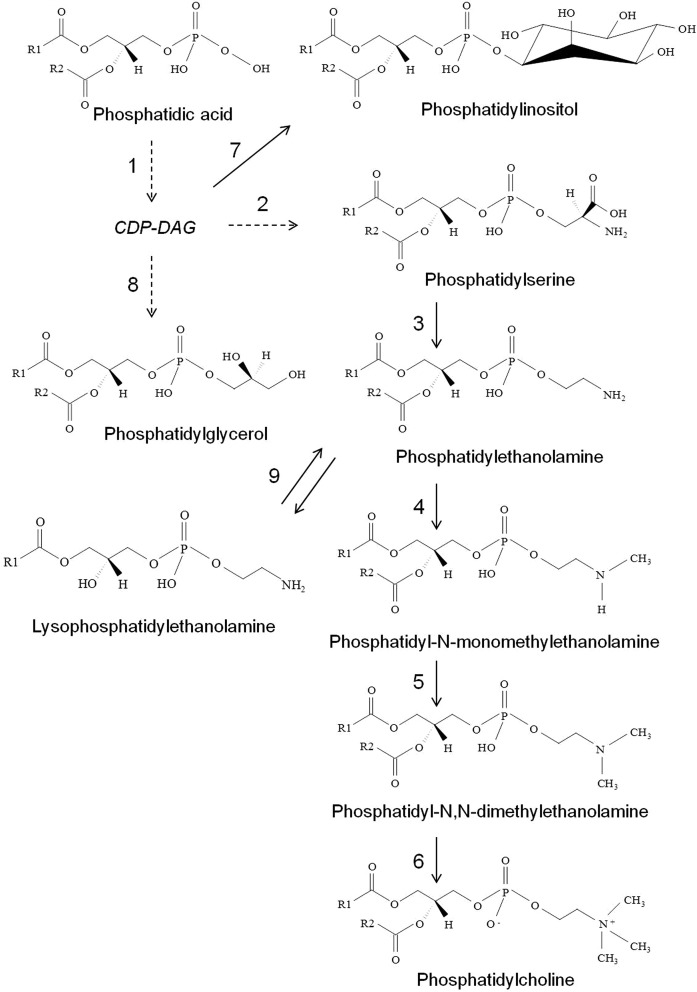
The steps involved in the phosphoglyceride biosynthetic pathway are largely conserved in fungi and share many commonalities to mammals (13, 74, 75). Our data suggest that the phosphoglyceride biosynthesis in Cryptococcus is quite similar to that reported for S. cereviseae (74) and C. albicans (13) (Fig. 7). The de novo biosynthesis of phosphoglyceride primarily begins by conversion of PA into cytidyldiphosphate-diacylglycerol (CDP-DAG) using a phosphatidate cytidylyltransferase enzyme (Cds1 in S. cereviseae) (74). CDP-DAG is the precursor for all major phosphoglycerides (PC, PE, PI, PS, and PG) (74). In the initial step, an enzyme, PS decarboxylase (Cho1 in S. cereviseae and C. albicans), catalyzes the conversion of CDP-DAG and serine into PS by releasing cytidylmonophosphate (CMP). Further, PS decarboxylases (Psd1/Psd2 in S. cereviseae and C. albicans) convert PS into PE. Three subsequent methylations by specific methyltransferases [Cho2 in S. cereviseae (77) and C. albicans (13) and Opi3 in S. cereviseae (78)] on PE result in the formation of PC. CDP-DAG is converted into PI by the activity of three enzymes: L-myo-inositol-1-phosphate synthase (Ino1 in S. cereviseae), inositol monophosphatase (Inm1 in S. cereviseae), PI synthase (Pis1 in S. cereviseae) (74, 79). CDP-DAG is converted into PG by phosphatidylglycerolphosphate synthase [Pgs1 in S. cereviseae (80) and C. glabrata (81)] and a phosphatidylglycerophosphatase [Gep4 in S. cereviseae (82)]. PE is converted to LPE by phospholipase B [Plb1 in C. gattii (83)]. The majority of enzymes involved in phosphoglyceride metabolism are well-characterized in S. cereviseae and C. albicans (13, 74), but none (to the best our knowledge) have been characterized in Cryptococcus species. Based on these analyses, it is apparent that similar enzymes will be involved in the phosphoglyceride biosynthesis in Cryptococcus as well (as described in KEGG pathway database). Also, as reported for eukaryotic systems, the Kennedy pathway of phosphoglyceride biosynthesis involving CDP-choline and CDP-ethanolamine branches is most likely to be involved in PE and PC biosynthesis in Cryptococcus as well (13, 74). Considering that deletion of PSD1, PSD2, CHO1, and CHO2 had compromised phospholipid levels, drastic effects on virulence and pathogenicity traits of C. albicans (13, 14), and that these genes are nonidentical to any human gene, this presents a strong opportunity to exploit these genes as therapeutic targets in Cryptococcus as well.
Fig. 7.
Pathway of phospholipid biosynthesis in Cryptococcus. Based on the profile obtained for H99, R265, and R272, the pathway of phospholipid biosynthesis in Cryptococcus is composed of the following enzymatic steps: (1) phosphatidate cytidylyltransferase; (2) PS synthase; (3) PS decarboxylase; (4) PE methyltransferase; (5, 6) methylene-fatty-acyl-phospholipid synthase or phospholipid methyltransferase; (7) L-myo-inositol-1-phosphate synthase, inositol monophosphatase, PI synthase; (8) phosphatidylglycerolphosphate synthase, phosphatidylglycerophosphatase; (9) phospholipase B (1-acyl-sn-gylcerol-3-phosphate acyl transferase). Dashed lines represent that multiple enzymatic steps are involved. CDP-ethanolamine and CDP-choline branches of the Kennedy pathway and the biosynthesis of lysophospholipids (72), except LPE, are not shown in this pathway.
Comparative analysis of GlcCer and sterols of other Cryptococcus strains
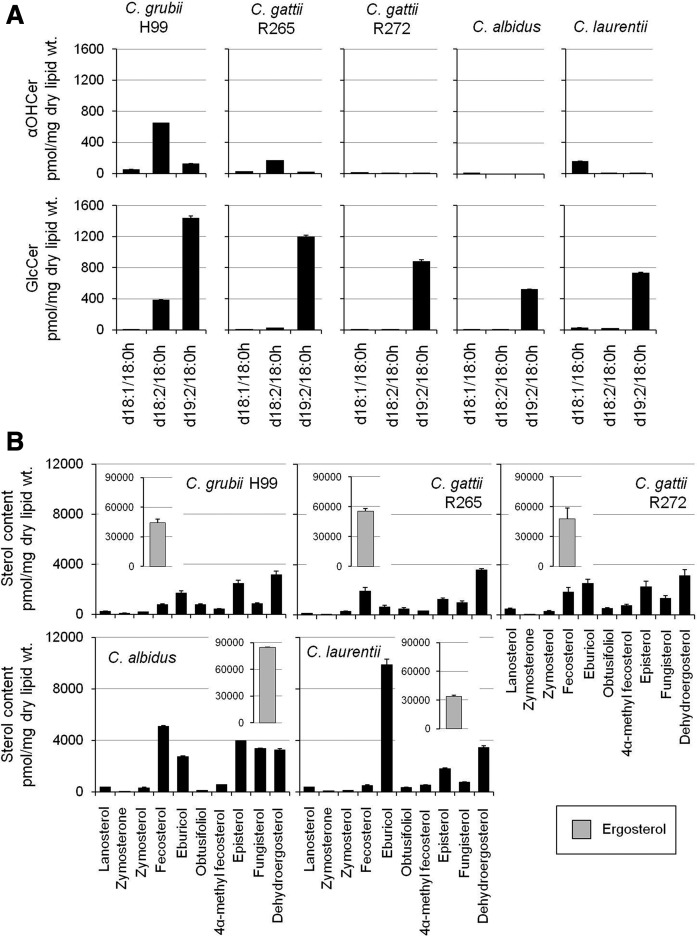
The biosynthetic pathways of sphingolipids and ergosterol represent the two most fungal-specific pathways that are currently being targeted for antifungal drug development studies (28). Mutants of both sphingolipid and sterol pathway genes show a wide range of phenotypic defects in fungi, the most common being the loss of virulence traits and the loss of fungal growth/cellular replication (12, 19–25). This is most likely because deletion of these genes disrupts the structural component(s) of plasma membranes, such as the formation of ordered domains (10, 17). For instance, the end product of the GlcCer biosynthetic pathway, GlcCer(d19:2/18:0h), is a unique complex sphingolipid synthesized by fungi. The biosynthetic process involves activity of Δ8-desaturase (uncharacterized), Smt1, and Gcs1 in Cryptococcus (19, 21). In the absence of Smt1, Gcs1 is able to recognize Cer(d18:1/18:0h) and Cer(d18:2/18:0h) as substrates and converts them to respective GlcCers, GlcCer(d18:1/18:0h) and GlcCer(d18:2/18:0h). To evaluate the status of the GlcCer biosynthetic pathway in several other Cryptococcus strains, we compared their levels of αOHCer and respective GlcCer structures (Fig. 8A). The analysis was performed in C. grubii H99, C. gattii R265, C.gattii R272, C. albidus, and C. laurentii strains. The analyses show that GlcCer(d19:2/18:0h) is the most abundant GlcCer synthesized in all tested strains. The C. grubii H99 showed the highest levels of Cer(d18:2/18:0h) structure. The levels of GlcCer biosynthetic intermediates were least in C.gattii R272, C. albidus, and C. laurentii strains.
Fig. 8.
Comparative lipid profile of major human pathogenic Cryptococcus species. A: αOHCer and GlcCer compositions of various Cryptococcus species. B: Sterol compositions of various Cryptococcus species. Ergosterol content is shown in the inset. Mean ± SEM is shown (n = 3). Different scales have been used for better visualization of datasets.
Ergosterol is the most abundant sterol in all Cryptococcus strains (Fig. 8B) (29, 39). The C. albidus strain showed high ergosterol content and the C. laurentii strain showed the lowest ergosterol levels. The presence of structures like eburicol, obtusifoliol, 4α-methyl fecosterol, and fungisterol strongly suggests that the alternate pathways of ergosterol biosynthesis (apart from the classical pathway) are also active in Cryptococcus. For example, high levels of eburicol and low levels of zymosterone and zymosterol were detected in the C. laurentii strain. This suggests that the activity of Erg11 is possibly altered in C. laurentii and that ergosterol biosynthesis is occurring via the alternate pathway 1 (eburicol →→ 4α-methyl fecosterol → fecosterol). Similarly noticeable amounts of fungisterol were present in all Cryptococcus strains, most in C. albidus. This suggests that alternate pathway 2 (fungisterol →→ ergosterol) is active in Cryptococcus strains (Fig. 5).
Our data suggest: 1) the biosynthetic pathways of GlcCer and ergosterol are conserved; and 2) the levels of various metabolites of the biosynthetic pathway are significantly variable between different Cryptococcus strains. Further, the four key properties of GlcCer(d19:2/18:0h) structure, specifically, the Δ8 double bond, the methyl group at the C9 position, the α-hydroxyl group of the fatty acyl, and the glucosyl moiety, can all affect the membrane biophysical properties or membrane order (10). Changes in their level will eventually change the biophysical properties with the consequent effect on virulence traits. Similarly, structural differences in various sterol structures, such as the 3-hydroxyl group, the location of double bonds, aliphatic side chain, and flatness, can all affect the membrane biophysical properties and membrane order (17). Although ergosterol is the major membrane sterol in fungi, recruitment of altered sterol intermediates (minor sterols) into the membrane could significantly alter membrane order (84). Considering the fact that GlcCer(d19:2/18:0h) and several sterol structures are the most argued lipids for their role in virulence, in vitro studies using synthetic vesicles may shed light on their role in membrane physical properties (84–87).
DISCUSSION
In the last decade, we have seen a steep rise in invasive C. grubii and C. gattii infections in both immunocompromised and immunocompetent patients (1, 2, 4, 5). Currently used antifungals are proving ineffective because of their toxicity, narrow spectrum, and the development of tolerance (28, 30, 31). Therefore, we urgently need to work in the direction of developing new antifungals. In this regard, the lipid biosynthetic pathways of Cryptococcus have emerged as potential targets (10, 28). This is primarily because the structure and biosynthesis of several fungal lipids is different from mammalian lipids (10). In Cryptococcus, several studies have shown the role of lipids in its growth and virulence, but not many studies have been done to characterize their compositions or the genetic makeup of the biosynthetic pathways (19, 20, 47, 60). In this study, we have determined the compositions of the most common lipids of Cryptococcus, using various spectroscopic approaches. These are sphingolipids, sterols, and phosphoglycerides.
To characterize the sphingolipids, we have used a MRM-based LC-ESI-MS/MS approach. This technique is well-suited for targeted lipidomics and gives picomole level quantification (35, 46). In our analysis, we were able to detect 100 species belonging to 12 classes of sphingolipids. The experiments showed that the sphingolipid biosynthetic pathway is conserved in C. grubii (H99) and C. gattii (R265 and R272) strains. This is similar to the pathway that has been proposed in other fungi, such as C. albicans, A. fumigatus, and A. nidulans (34, 37, 48–54). Sphingolipid biosynthesis can be divided into two parts with a common precursor, dhSph (d18:0). The first part is the GlcCer biosynthesis pathway and the second is the IPC biosynthesis pathway (10). GlcCer biosynthesis is mainly dependent on the activities of five enzymes: CerS, Δ4-desaturase, Δ8-desaturase, Smt1 (21), and Gcs1 (19). The end product of this pathway is the GlcCer(d19:2/18:0h) species (19). Our data indicate that a specific CerS will recognize α-hydroxylated fatty acyls (mostly C18:0h and, to some extent, C16:0h) and sphingoid base (d18:0) as substrates, leading to downstream GlcCer synthesis. Low levels of Cer structures with nonhydroxylated fatty acyls (C14:0 to C26:0) were also detected in both C. gattii and C. grubii strains. Some of these Cer structures with nonhydroxylated fatty acyls are also converted into the respective GlcCer structure (19). Upon incorporation into the membranes, these Cer and GlcCer structures play an important role in fungal growth at alkaline or neutral pH, virulence, and pathogenesis [reviewed in (10)]. IPC biosynthesis is dependent on CerS that can utilize PhytoSph as one of the substrates (possibly different from the CerS involved in Cer biosynthesis) and Ipc1 enzymes (23). The IPC species composition data indicates that Ipc1 of C. gattii and C. grubii strains can utilize four different Cer-related structures as substrates to synthesize the complex IPC derivatives: dhCer (C18:0 fatty acyl), αOHdhCer (C18:0h fatty acyl), PhytoCer (C18:0, C24:0 and C26:0 fatty acyls), and αOHPhytoCer (C24:0h and C26:0h fatty acyls). We did find some interesting differences when we compared the sphingolipid profiles of highly virulent Cryptococcus strains (H99 and R265) with a less virulent strain (R272). For example, the R272 strain was abundant in dhCer(d18:0/18:0) and PhytoCer(d18:0/18:0) compared with the H99 or R265 strain. Considering the fact that the downstream complex sphingolipids (GlcCers or IPCs) of the dhCer or PhytoCer did not change significantly between H99, R265, and R272, it is possible that these Cer structures may play a greater role in virulence than what is currently attributed to them (10, 88). Thus, studies on CerSs of C. neoformans will address how these Cers are synthesized. In Cryptococcus, deletion of Isc1 results in depletion of PhytoCers, which alters the plasma membrane homeostasis, in turn inhibiting oligomerization of Pma1 proton pump (a plasma membrane ATPase) resulting in hypersensitivity to acidic pH (intracellular survival). This phenotype could be reversed by supplementing the growth medium with PhytoCers (24, 25). This is strong evidence suggesting that sphingolipids, like PhytoCers and perhaps Cer(s), may play a key role in the regulation of membrane stability, which will translate into the regulation of fungal virulence and pathogenesis.
Further, we characterized the sterols using a simple GC-MS-based method and could detect 13 sterol structures. These 13 sterol intermediates are detectable in all tested Cryptococcus strains. Our data indicate that the sterol biosynthesis in Cryptococcus has three alternate routes: 1) the classical pathway (Erg11 dependent) via zymosterol → fecosterol →→→→ ergosterol; 2) the eburicol pathway (Erg11 independent) via eburicol → obtusifoliol → 4α-methylfecosterol → fecosterol →→→→ ergosterol; and 3) the fungisterol pathway via episterol → fungisterol →→ ergosterol. It is important to note that in an earlier study by Ghannoum et al. (60), obtusifoliol was reported as the major sterol (21.1–68.2% of the total sterol) in Cryptococcus. The high obtusifoliol content detected could be due to either strain-specific sterol differences or sterol analysis methodology-related differences, like variations in ionization efficiencies or some kind of stress that pushed cells toward the eburicol pathway leading to higher obtusifoliol content. In contrast, in our analysis, we report that ergosterol is the major sterol structure in Cryptococcus strains (64.3–87.4% of the total sterol) and our analysis is similar to the one reported by Nes et al. (39). We did find, however, some strain-specific differences in sterol species (39), which may explain the difference with the Ghannoum analysis (60). For example, compared with H99 or R265, the R272 strain has a high amount (as much as 2.2-fold) of methylated sterol structures, namely, lanosterol, eburicol, and 4α-methyl fecosterol, which could result from minor alteration of Erg11 activity (39, 64, 65, 67). In summary, Cryptococcus can synthesize ergosterol via multiple routes, even when the key enzymes might be inhibited (39, 64, 65, 67).
Finally, to characterize the phosphoglycerides, we used a 31P-NMR approach and could detect 10 classes of phosphoglycerides. Analysis of the phosphoglyceride species showed that the major phosphoglyceride classes, namely, PC, PE, and PS, contain 16C or 18C fatty acyls (nonhydroxylated) in their structure that may be mono- or polyunsaturated. The amounts of various phosphoglyceride classes were similar in the H99, R265, and R272 strains. The only major difference found was that the fatty acyl ratio of C18:2/C18:1 was low in R272 compared with the H99 or R265 strain. Possibly certain fatty acyl desaturases are less active in R272 (76). Nonetheless, our 31P-NMR data clearly showed: 1) the phosphoglyceride compositions of Cryptococcus strains; and 2) the CDP-DAG pathway of phosphoglyceride biosynthesis is functional in Cryptococcus (74).
In summary, we have provided the compositions and the models for composition-based biosynthetic pathways of sphingolipid, sterol, and phosphoglyceride lipid groups of Cryptococcus strains. We observed certain similarities, few strain-specific differences, and some virulence-related changes between C. gattii and C. grubii strains. This analysis was performed through a spectroscopy-based measurement of substrates and products of the three biosynthetic pathways, which are the direct indicators of the expression or the activity of the enzyme (50, 89). Except for a few, the majority of enzymes involved in these biosynthetic processes are uncharacterized. The structural data, methods of analysis, and biosynthetic pathways discussed in this study will provide a reference point for characterizing the unknown genes involved in this lipid biosynthesis. Altogether, the understanding of these complex lipid pathways could help in developing new and highly selective antifungals.
Supplementary Material
Acknowledgments
The authors appreciate the constructive discussions from John, Rob, Toni, Izolda, Nadia, and Justin of the proteomics/lipidomics core facility at HSC, Stony Brook University, during the course of this study and Paul Baker at SCIEX for QTRAP analysis.
Footnotes
Abbreviations:
- CDP-DAG
- cytidyldiphosphate-diacylglycerol
- Cer
- ceramide
- CerS
- ceramide synthase
- dhCer
- dihydroceramide
- dhSph
- dihydrosphingosine
- dhSph1P
- dihydrosphingosine-1-phosphate
- Erg
- ergosterol biosynthetic pathway protein
- Gcs1
- glucosylceramide synthase 1
- GlcCer
- glucosylceramide
- IPC
- inositolphosphorylceramide
- Ipc1
- inositolphosphorylceramide synthase 1
- Isc1
- inositol phosphosphingolipid phospholipase C
- KDR
- 3-ketodihydrosphingosine reductase
- LCB
- long chain base
- LCB-1P
- long chain base-1-phosphate
- LPE
- lysophosphatidylethanolamine
- MRM
- multiple reaction monitoring
- αOHCer
- α-hydroxy ceramide
- αOHPhytoCer
- α-hydroxy phytoceramide
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PCA
- principal component analysis
- PDME
- phosphatidyl-N,N-dimethylethanolamine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PhytoCer
- phytoceramide
- PhytoSph
- phytosphingosine
- PhytoSph1P
- phytosphingosine-1-phosphate
- Pi
- inorganic phosphate
- PI
- phosphatidylinositol
- PMME
- phosphatidyl-N-monomethylethanolamine
- PS
- phosphatidylserine
- Psd
- phosphatidylserine decarboxylase
- Smt1
- sphingolipid C9-methyl transferase
- Sph
- sphingosine
- Sph1P
- sphingosine-1-phosphate
- SPT
- serine palmitoyltransferase
This work was supported by National Institutes of Health Grants AI125770 and AI116420 and by Merit Review Grant I01BX002624 from the Veterans Affairs Program in Biomedical Laboratory Research and Development to M.D.P. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. M.D.P. is a co-founder and Chief Scientific Officer of MicroRid Technologies Inc. All other authors have no financial and competing interests with the subject matter or materials discussed in the manuscript.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Gibson J. F., and Johnston S. A.. 2015. Immunity to Cryptococcus neoformans and C. gattii during cryptococcosis. Fungal Genet. Biol. 78: 76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park B. J., Wannemuehler K. A., Marston B. J., Govender N., Pappas P. G., and Chiller T. M.. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 23: 525–530. [DOI] [PubMed] [Google Scholar]
- 3.El-Kersh K., Rawasia W. F., Chaddha U., and Guardiola J.. 2013. Rarity revisited: cryptococcal peritonitis. BMJ Case Rep. 2013: bcr2013009099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chau T. T., Mai N. H., Phu N. H., Nghia H. D., Chuong L. V., Sinh D. X., Duong V. A., Diep P. T., Campbell J. I., Baker S., et al. . 2010. A prospective descriptive study of cryptococcal meningitis in HIV uninfected patients in Vietnam - high prevalence of Cryptococcus neoformans var grubii in the absence of underlying disease. BMC Infect. Dis. 10: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwon-Chung K. J., Fraser J. A., Doering T. L., Wang Z., Janbon G., Idnurm A., and Bahn Y.. 2014. Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb. Perspect. Med. 4: a019760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Almeida F., Wolf J. M., and Casadevall A.. 2015. Virulence-associated enzymes of Cryptococcus neoformans. Eukaryot. Cell. 14: 1173–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levitz S. M., Nong S. H., Seetoo K. F., Harrison T. S., Speizer R. A., and Simons E. R.. 1999. Cryptococcus neoformans resides in an acidic phagolysosome of human macrophages. Infect. Immun. 67: 885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vu K., Tham R., Uhrig J. P., Thompson G. R. III, Na Pombejra S., Jamklang M., Bautos J. M., and Gelli A.. 2014. Invasion of the central nervous system by Cryptococcus neoformans requires a secreted fungal metalloprotease. MBio. 5: e01101–e01114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shea J. M., Henry J. H., and Del Poeta M.. 2006. Lipid metabolism in Cryptococcus neoformans. FEMS Yeast Res. 6: 469–479. [DOI] [PubMed] [Google Scholar]
- 10.Del Poeta M., Nimrichter L., Rodrigues M. L., and Luberto C.. 2014. Synthesis and biological properties of fungal glucosylceramide. PLoS Pathog. 10: e1003832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prasad R., and Singh A.. 2013. Lipids of Candida albicans and their role in multidrug resistance. Curr. Genet. 59: 243–250. [DOI] [PubMed] [Google Scholar]
- 12.Rella A., Farnoud A. F., and Del Poeta M.. 2016. Plasma membrane lipids and their role in fungal virulence. Prog. Lipid Res. 61: 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y. L., Montedonico A. E., Kauffman S., Dunlap J. R., Menn F. M., and Reynolds T. B.. 2010. Phosphatidylserine synthase and phosphatidylserine decarboxylase are essential for cell wall integrity and virulence in Candida albicans. Mol. Microbiol. 75: 1112–1132. [DOI] [PubMed] [Google Scholar]
- 14.Cassilly C. D., Farmer A. T., Montedonico A. E., Smith T. K., Campagna S. R., and Reynolds T. B.. 2017. Role of phosphatidylserine synthase in shaping the phospholipidome of Candida albicans. FEMS Yeast Res. 17: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohli A., Smriti, Mukhopadhyay K., Rattan A., and Prasad R.. 2002. In vitro low-level resistance to azoles in Candida albicans is associated with changes in membrane lipid fluidity and asymmetry. Antimicrob. Agents Chemother. 46: 1046–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh A., Rella A., Schwacke J., Vacchi-Suzzi C., Luberto C., and Del Poeta M.. 2015. Transmembrane transporter expression regulated by the glucosylceramide pathway in Cryptococcus neoformans. BMC Res. Notes. 8: 681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farnoud A. M., Toledo A. M., Konopka J. B., Del Poeta M., and London E.. 2015. Raft-like membrane domains in pathogenic microorganisms. Curr. Top. Membr. 75: 233–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodrigues M. L., Nakayasu E. S., Oliveira D. L., Nimrichter L., Nosanchuk J. D., Almeida I. C., and Casadevall A.. 2008. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot. Cell. 7: 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rittershaus P. C., Kechichian T. B., Allegood J. C., Merrill A. H. Jr., Hennig M., Luberto C., and Del Poeta M.. 2006. Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans. J. Clin. Invest. 116: 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pasrija R., Panwar S. L., and Prasad R.. 2008. Multidrug transporters CaCdr1p and CaMdr1p of Candida albicans display different lipid specificities: both ergosterol and sphingolipids are essential for targeting of CaCdr1p to membrane rafts. Antimicrob. Agents Chemother. 52: 694–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh A., Wang H., Silva L. C., Na C., Prieto M., Futerman A. H., Luberto C., and Del Poeta M.. 2012. Methylation of glycosylated sphingolipid modulates membrane lipid topography and pathogenicity of Cryptococcus neoformans. Cell. Microbiol. 14: 500–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sionov E., Chang Y. C., Garraffo H. M., Dolan M. A., Ghannoum M. A., and Kwon-Chung K. J.. 2012. Identification of a Cryptococcus neoformans cytochrome P450 lanosterol 14α-demethylase (Erg11) residue critical for differential susceptibility between fluconazole/voriconazole and itraconazole/posaconazole. Antimicrob. Agents Chemother. 56: 1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mare L., Iatta R., Montagna M. T., Luberto C., and Del Poeta M.. 2005. APP1 transcription is regulated by inositol-phosphorylceramide synthase 1-diacylglycerol pathway and is controlled by ATF2 transcription factor in Cryptococcus neoformans. J. Biol. Chem. 280: 36055–36064. [DOI] [PubMed] [Google Scholar]
- 24.Henry J., Guillotte A., Luberto C., and Del Poeta M.. 2011. Characterization of inositol phospho-sphingolipid-phospholipase C 1 (Isc1) in Cryptococcus neoformans reveals unique biochemical features. FEBS Lett. 585: 635–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farnoud A. M., Mor V., Singh A., and Del Poeta M.. 2014. Inositol phosphosphingolipid phospholipase C1 regulates plasma membrane ATPase (Pma1) stability in Cryptococcus neoformans. FEBS Lett. 588: 3932–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McQuiston T., Luberto C., and Del Poeta M.. 2011. Role of sphingosine-1-phosphate (S1P) and S1P receptor 2 in the phagocytosis of Cryptococcus neoformans by alveolar macrophages. Microbiology. 157: 1416–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farnoud A. M., Bryan A. M., Kechichian T., Luberto C., and Del Poeta M.. 2015. The granuloma response controlling Cryptococcosis in mice depends on the sphingosine kinase 1-sphingosine 1-phosphate pathway. Infect. Immun. 83: 2705–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rollin-Pinheiro R., Singh A., Barreto-Bergter E., and Del Poeta M.. 2016. Sphingolipids as targets for treatment of fungal infections. Future Med. Chem. 8: 1469–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mor V., Rella A., Farnoud A. M., Singh A., Munshi M., Bryan A., Naseem S., Konopka J. B., Ojima I., Bullesbach E., et al. . 2015. Identification of a new class of antifungals targeting the synthesis of fungal sphingolipids. MBio. 6: e00647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Archibald L. K., Tuohy M. J., Wilson D. A., Nwanyanwu O., Kazembe P. N., Tansuphasawadikul S., Eampokalap B., Chaovavanich A., Reller L. B., Jarvis W. R., et al. . 2004. Antifungal susceptibilities of Cryptococcus neoformans. Emerg. Infect. Dis. 10: 143–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker L. A., Gow N. A. R., and Munro C. A.. 2010. Fungal echinocandin resistance. Fungal Genet. Biol. 47: 117–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mandala S. M., Thornton R. A., Frommer B. R., Curotto J. E., Rozdilsky W., Kurtz M. B., Giacobbe R. A., Bills G. F., Cabello M. A., Martín I., et al. . 1995. The discovery of australifungin, a novel inhibitor of sphinganine N-acyltransferase from Sporormiella australis. Producing organism, fermentation, isolation, and biological activity. J Antibiot. (Tokyo). 48: 349–356. [DOI] [PubMed] [Google Scholar]
- 33.Bligh E. G., and Dyer W. J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 34.Singh A., and Del Poeta M.. 2016. Sphingolipidomics: an important mechanistic tool for studying fungal pathogens. Front. Microbiol. 7: 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bielawski J., Szulc Z. M., Hannun Y. A., and Bielawska A.. 2006. Simultaneous quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods. 39: 82–91. [DOI] [PubMed] [Google Scholar]
- 36.Mor V., Farnoud A. M., Singh A., Rella A., Tanno H., Ishii K., Kawakami K., Sato T., and Del Poeta M.. 2016. Glucosylceramide administration as a vaccination strategy in mouse models of cryptococcosis. PLoS One. 11: e0153853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh A., Prasad T., Kapoor K., Mandal A., Roth M., Welti R., and Prasad R.. 2010. Phospholipidome of Candida: each species of Candida has distinctive phospholipid molecular species. OMICS. 14: 665–677. [DOI] [PubMed] [Google Scholar]
- 38.Shui G., Guan X. L., Low C. P., Chua G. H., Goh J. S., Yang H., and Wenk M. R.. 2010. Toward one step analysis of cellular lipidomes using liquid chromatography coupled with mass spectrometry: application to Saccharomyces cerevisiae and Schizosaccharomyces pombe lipidomics. Mol. Biosyst. 6: 1008–1017. [DOI] [PubMed] [Google Scholar]
- 39.Nes W. D., Zhou W., Ganapathy K., Liu J., Vatsyayan R., Chamala S., Hernandez K., and Miranda M.. 2009. Sterol 24-C-methyltransferase: an enzymatic target for the disruption of ergosterol biosynthesis and homeostasis in Cryptococcus neoformans. Arch. Biochem. Biophys. 481: 210–218. [DOI] [PubMed] [Google Scholar]
- 40.Singh A., Mahto K. K., and Prasad R.. 2013. Lipidomics and in vitro azole resistance in Candida albicans. OMICS. 17: 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang Y. C., Lamichhane A. K., Garraffo H. M., Walter P. J., Leerkes M., and Kwon-Chung K. J.. 2014. Molecular mechanisms of hypoxic responses via unique roles of Ras1, Cdc24 and Ptp3 in a human fungal pathogen Cryptococcus neoformans. PLoS Genet. 10: e1004292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guan X. L., Riezman I., Wenk M. R., and Riezman H.. 2010. Yeast lipid analysis and quantification by mass spectrometry. Methods Enzymol. 470: 369–391. [DOI] [PubMed] [Google Scholar]
- 43.Lehnhardt F. G., Röhn G., Ernestus R., Grüne M., and Hoehn M.. 2001. 1H- and 31P-NMR spectroscopy of primary and recurrent human brain tumors in vitro: malignancy-characteristic profiles of water soluble and lipophilic spectral components. NMR Biomed. 14: 307–317. [DOI] [PubMed] [Google Scholar]
- 44.MacKenzie A., Vyssotski M., and Nekrasov E.. 2009. Quantitative analysis of dairy phospholipids by 31P NMR. J. Am. Oil Chem. Soc. 86: 757–763. [Google Scholar]
- 45.Clarke N. G., and Dawson R. M.. 1981. Alkaline O→N-transacylation. A new method for the quantitative deacylation of phospholipids. Biochem. J. 195: 301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brügger B., Erben G., Sandhoff R., Wieland F. T., and Lehmann W. D.. 1997. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc. Natl. Acad. Sci. USA. 94: 2339–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Longo L. V., Nakayasu E. S., Pires J. H., Gazos-Lopes F., Vallejo M. C., Sobreira T. J., Almeida I. C., and Puccia R.. 2015. Characterization of lipids and proteins associated to the cell wall of the acapsular mutant Cryptococcus neoformans Cap 67. J. Eukaryot. Microbiol. 62: 591–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bennion B., Park C., Fuller M., Lindsey R., Momany M., Jennemann R., and Levery S. B.. 2003. Glycosphingolipids of the model fungus Aspergillus nidulans: characterization of GIPCs with oligo-alpha-mannose-type glycans. J. Lipid Res. 44: 2073–2088. [DOI] [PubMed] [Google Scholar]
- 49.Cheon S. A., Bal J., Song Y., Hwang H. M., Kim A. R., Kang W. K., Kang H. A., Hannibal-Bach H. K., Knudsen J., Ejsing C. S., et al. . 2012. Distinct roles of two ceramide synthases, CaLag1p and CaLac1p, in the morphogenesis of Candida albicans. Mol. Microbiol. 83: 728–745. [DOI] [PubMed] [Google Scholar]
- 50.Ejsing C. S., Sampaio J. L., Surendranath V., Duchoslav E., Ekroos K., Klemm R. W., Simons K., and Shevchenko A.. 2009. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA. 106: 2136–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuroda M., Hashida-Okado T., Yasumoto R., Gomi K., Kato I., and Takesako K.. 1999. An aureobasidin A resistance gene isolated from Aspergillus is a homolog of yeast AUR1, a gene responsible for inositol phosphorylceramide (IPC) synthase activity. Mol. Gen. Genet. 261: 290–296. [DOI] [PubMed] [Google Scholar]
- 52.Voynova N. S., Mallela S. K., Vazquez H. M., Cerantola V., Sonderegger M., Knudsen J., Ejsing C. S., and Conzelmann A.. 2014. Characterization of yeast mutants lacking alkaline ceramidases YPC1 and YDC1. FEMS Yeast Res. 14: 776–788. [DOI] [PubMed] [Google Scholar]
- 53.Alves de Castro P., Dos Reis T. F., Dolan S. K., Oliveira Manfiolli A., Brown N. A., Jones G. W., Doyle S., Riaño-Pachón D. M., Squina F. M., Caldana C., et al. . 2016. The Aspergillus fumigatus SchASCH9 kinase modulates SakAHOG1 MAP kinase activity and it is essential for virulence. Mol. Microbiol. 102: 642–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ternes P., Wobbe T., Schwarz M., Albrecht S., Feussner K., Riezman I., Cregg J. M., Heinz E., Riezman H., Feussner I., et al. . 2011. Two pathways of sphingolipid biosynthesis are separated in the yeast Pichia pastoris. J. Biol. Chem. 286: 11401–11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guillas I., Kirchman P. A., Chuard R., Pfefferli M., Jiang J. C., Jazwinski S. M., and Conzelmann A.. 2001. C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. EMBO J. 20: 2655–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li S., Du L., Yuen G., and Harris S. D.. 2006. Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans. Mol. Biol. Cell. 17: 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim S., Fyrst H., and Saba J.. 2000. Accumulation of phosphorylated sphingoid long chain bases results in cell growth inhibition in Saccharomyces cerevisiae. Genetics. 156: 1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Serra M., and Saba J. D.. 2010. Sphingosine 1-phosphate lyase, a key regulator of sphingosine 1-phosphate signaling and function. Adv. Enzyme Regul. 50: 349–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kondo N., Ohno Y., Yamagata M., Obara T., Seki N., Kitamura T., Naganuma T., and Kihara A.. 2014. Identification of the phytosphingosine metabolic pathway leading to odd-numbered fatty acids. Nat. Commun. 5: 5338. [DOI] [PubMed] [Google Scholar]
- 60.Ghannoum M. A., Spellberg B. J., Ibrahim A. S., Ritchie J. A., Currie B., Spitzer E. D., Edwards J. E. Jr., and Casadevall A.. 1994. Sterol composition of Cryptococcus neoformans in the presence and absence of fluconazole. Antimicrob. Agents Chemother. 38: 2029–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shobayashi M., Mitsueda S., Ago M., Fujii T., Iwashita K., and Iefuji H.. 2005. Effects of culture conditions on ergosterol biosynthesis by Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 69: 2381–2388. [DOI] [PubMed] [Google Scholar]
- 62.Alcazar-Fuoli L., and Mellado E.. 2013. Ergosterol biosynthesis in Aspergillus fumigatus: its relevance as an antifungal target and role in antifungal drug resistance. Front. Microbiol. 3: 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanglard D., Ischer F., Parkinson T., Falconer D., and Bille J.. 2003. Candida albicans mutations in the ergosterol biosynthetic pathway and resistance to several antifungal agents. Antimicrob. Agents Chemother. 47: 2404–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hull C. M., Parker J. E., Bader O., Weig M., Gross U., Warrilow A. G., Kelly D. E., and Kelly S. L.. 2012. Facultative sterol uptake in an ergosterol-deficient clinical isolate of Candida glabrata harboring a missense mutation in ERG11 and exhibiting cross-resistance to azoles and amphotericin B. Antimicrob. Agents Chemother. 56: 4223–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Revankar S. G., Fu J., Rinaldi M. G., Kelly S. L., Kelly D. E., Lamb D. C., Keller S. M., and Wickes B. L.. 2004. Cloning and characterization of the lanosterol 14alpha-demethylase (ERG11) gene in Cryptococcus neoformans. Biochem. Biophys. Res. Commun. 324: 719–728. [DOI] [PubMed] [Google Scholar]
- 66.Sheng C., Miao Z., Ji H., Yao J., Wang W., Che X., Dong G., Lü J., Gu W., and Zhang W.. 2009. Three-dimensional model of lanosterol 14 alpha-demethylase from Cryptococcus neoformans: active-site characterization and insights into azole binding. Antimicrob. Agents Chemother. 53: 3487–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alcazar-Fuoli L., Mellado E., Garcia-Effron G., Lopez J. F., Grimalt G. O., Cuenca-Estrella J. M., and Rodriguez-Tudela J. L.. 2008. Ergosterol biosynthesis pathway in Aspergillus fumigatus. Steroids. 73: 339–347. [DOI] [PubMed] [Google Scholar]
- 68.Osumi T., Taketani S., Katsuki H., Kuhara T., and Matsumoto I.. 1978. Ergosterol biosynthesis in yeast. Pathways in the late stages and their variation under various conditions. J. Biochem. 83: 681–691. [DOI] [PubMed] [Google Scholar]
- 69.Kaneshiro E. S., Amit Z., Swonger M. M., Kreishman G. P., Brooks E. E., Kreishman M., Jayasimhulu K., Parish E. J., Sun H., Kizito S. A., et al. . 1999. Pneumocysterol [(24Z)-ethylidenelanost-8-en-3beta-ol], a rare sterol detected in the opportunistic pathogen Pneumocystis carinii hominis: structural identity and chemical synthesis. Proc. Natl. Acad. Sci. USA. 96: 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Singh A., Yadav V., and Prasad R.. 2012. Comparative lipidomics in clinical isolates of Candida albicans reveal crosstalk between mitochondria, cell wall integrity and azole resistance. PLoS One. 7: e39812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Janmey P. A., and Kinnunen P. K.. 2006. Biophysical properties of lipids and dynamic membranes. Trends Cell Biol. 16: 538–546. [DOI] [PubMed] [Google Scholar]
- 72.van Meer G., Voelker D. R., and Feigenson G. W.. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9: 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Spanova M., Zweytick D., Lohner K., Klug L., Leitner E., Hermetter A., and Daum G.. 2012. Influence of squalene on lipid particle/droplet and membrane organization in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1821: 647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carman G. M., and Han G. S.. 2011. Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu. Rev. Biochem. 80: 859–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kanipes M. I., Hill J. E., and Henry S. A.. 1998. The Schizosaccharomyces pombe cho1+ gene encodes a phospholipid methyltransferase. Genetics. 150: 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oh C. S., and Martin C. E.. 2006. Candida albicans Spt23p controls the expression of the Ole1p Delta9 fatty acid desaturase and regulates unsaturated fatty acid biosynthesis. J. Biol. Chem. 281: 7030–7039. [DOI] [PubMed] [Google Scholar]
- 77.Summers E. F., Letts V. A., McGraw P., and Henry S. A.. 1988. Saccharomyces cerevisiae cho2 mutants are deficient in phospholipid methylation and cross-pathway regulation of inositol synthesis. Genetics. 120: 909–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McGraw P., and Henry S. A.. 1989. Mutations in the Saccharomyces cerevisiae opi3 gene: effects on phospholipid methylation, growth and cross-pathway regulation of inositol synthesis. Genetics. 122: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shetty A., and Lopes J. M.. 2010. Derepression of INO1 transcription requires cooperation between the Ino2p-Ino4p heterodimer and Cbf1p and recruitment of the ISW2 chromatin-remodeling complex. Eukaryot. Cell. 9: 1845–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chang S. C., Heacock P. N., Clancey C. J., and Dowhan W.. 1998. The PEL1 gene (renamed PGS1) encodes the phosphatidylglycero-phosphate synthase of Saccharomyces cerevisiae. J. Biol. Chem. 273: 9829–9836. [DOI] [PubMed] [Google Scholar]
- 81.Batová M., Dzugasová V., Borecká S., Goffa E., Oblasová Z., and Subík J.. 2009. Molecular and phenotypic analysis of mutations causing anionic phospholipid deficiency in closely related yeast species. Folia Microbiol. (Praha). 54: 30–36. [DOI] [PubMed] [Google Scholar]
- 82.Osman C., Haag M., Wieland F. T., Brügger B., and Langer T.. 2010. A mitochondrial phosphatase required for cardiolipin biosynthesis: the PGP phosphatase Gep4. EMBO J. 29: 1976–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Latouche G. N., Sorrell T. C., and Meyer W.. 2002. Isolation and characterisation of the phospholipase B gene of Cryptococcus neoformans var. gattii. FEMS Yeast Res. 2: 551–561. [DOI] [PubMed] [Google Scholar]
- 84.Kim J., and London E.. 2015. Using sterol substitution to probe the role of membrane domains in membrane functions. Lipids. 50: 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li G., Kim J., Huang Z., St Clair J. R., Brown D. A., and London E.. 2016. Efficient replacement of plasma membrane outer leaflet phospholipids and sphingolipids in cells with exogenous lipids. Proc. Natl. Acad. Sci. USA. 113: 14025–14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Raj S., Nazemidashtarjandi S., Kim J. H., Joffe L., Zhang X, Singh A., Mor V., Desmarini D., Djordjevic J., Raleigh D. P., et al. . 2017. Changes in glucosylceramide structure affect virulence and membrane biophysical properties of Cryptococcus neoformans. Biochim. Biophys. Acta - Biomembranes. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim J. H., Singh A., Del Poeta M., Brown D., and London E.. 2017. Effects of sterol structure and sterol ability to form ordered membrane domains upon cellular endocytosis. J. Cell Sci. 130: 2682–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heung L. J., Luberto C., and Del Poeta M.. 2006. Role of sphingolipids in microbial pathogenesis. Infect. Immun. 74: 28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., and Mann M.. 2008. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 455: 1251–1254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.