Summary
Regulation is central to the functional versatility of cytoplasmic dynein, a motor involved in intracellular transport, cell division, and neurodevelopment. Previous work established that Lis1, a conserved regulator of dynein, binds to its motor domain and induces a tight microtubule-binding state in dynein. The work we present here—a combination of biochemistry, single-molecule assays, and cryo-electron microscopy—led to the surprising discovery that Lis1 has two opposing modes of regulating dynein, being capable of inducing both low and high affinity for the microtubule. We show that these opposing modes depend on the stoichiometry of Lis1 binding to dynein and that this stoichiometry is regulated by the nucleotide state of dynein’s AAA3 domain. The low affinity state requires Lis1 binding to dynein at a novel conserved site, mutation of which disrupts Lis1’s function in vivo. We propose a new model for the regulation of dynein by Lis1.
ETOC
The binding of dynein with microtubules can be weak or tight depending its mode of regulation by the lissencephaly protein Lis1, providing new insight into Lis1’s conflicting cellular roles.
Introduction
Cytoplasmic dynein-1 (“dynein”) is a microtubule (MT)-based motor that transports cellular cargos towards the minus-ends of MTs. In human and many other eukaryotic cells, dynein distributes and organizes organelles, proteins, RNAs, and viruses, in addition to playing essential roles in cell division. Mutations in the dynein machinery cause a range of neurological diseases (Franker and Hoogenraad, 2013).
Type-1 lissencephaly, a neurodevelopmental disease characterized by a smooth cerebral surface is caused by mutations in one of dynein’s conserved regulators, Lis1 (Reiner et al., 1993). S. cerevisiae, where the dynein pathway has a single, non-essential function in positioning the mitotic spindle (Eshel et al., 1993; Li et al., 1993), also has a Lis1 gene, making it a powerful model system for dissecting how Lis1 regulates dynein (Huang et al., 2012; Roberts et al., 2014; Toropova et al., 2014).
There are conflicting models for the role of Lis1 in dynein regulation. Lis1 has been implicated in many dynein-dependent functions (Cianfrocco et al., 2015), such as (1) localizing and/or maintaining dynein at MT plus ends (Lee et al., 2003; Sheeman et al., 2003; Splinter et al., 2012); (2) initiating dynein transport from MT plus ends (Egan et al., 2012; Lenz et al., 2006; Moughamian et al., 2013); and (3) enabling dynein to move against high loads (McKenney et al., 2010; Reddy et al., 2016; Yi et al., 2011). Other studies have looked at the effects of altering Lis1 expression levels on dynein-based transport. Most of these showed that deletion or depletion of Lis1 reduces transport (Dix et al., 2013; Klinman and Holzbaur, 2015; Moughamian et al., 2013; Pandey and Smith, 2011; Shao et al., 2013; Smith et al., 2000; Yi et al., 2011), while one found that Lis1 depletion increases cargo transport (Vagnoni et al., 2016).
How does Lis1 regulate dynein? Because Lis1 binds directly to dynein’s motor domain (Huang et al., 2012; Toropova et al., 2014), we must consider Lis1 regulation in the context of dynein’s mechanochemical cycle. Dynein is a dimer of two identical “heavy chains”, which can be divided into two functional units: the N-terminal “tail”, required for dimerization, binding the accessory subunits, and cargo interactions; and the “motor”, which is built around an AAA+ (ATPase Associated with diverse cellular Activities) ring containing 6 AAA domains (Figure 1A) (Cianfrocco et al., 2015). Dynein’s motility requires cycles of ATP hydrolysis at AAA1, which are coupled to cycles of MT binding and release at dynein’s MT binding domain (MTBD).
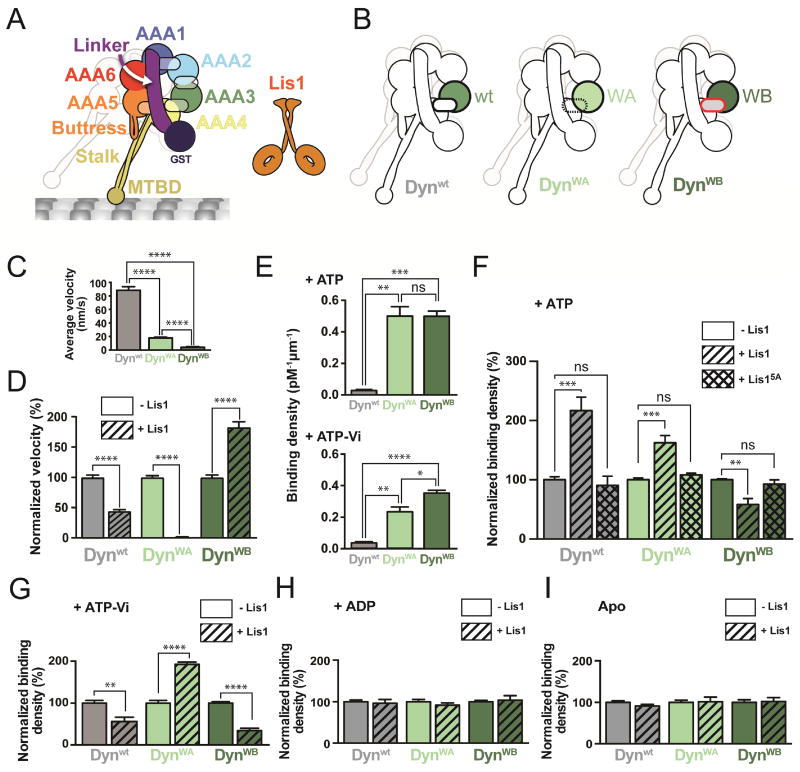
Figure 1. Lis1 has two modes of regulating dynein.
(A) Schematic of the dynein construct used in this study. The semi-transparent ovals represent the nucleotide-binding sites in AAA1-4. The second protomer is faded out for clarity. The β-propellers in the Lis1 dimer (orange) are represented as rings. (B) AAA3 variants used in this study: Dynwt (wild type AAA3), DynWA (Walker A mutation in AAA3), and DynWB (Walker B mutation in AAA3). (C) Average velocities of dynein variants (n>103 events per data point). See Figure S1B for representative kymographs. (D) Normalized average velocities of dynein variants in the absence (solid bars) and presence (hatched bars) of 300 nM Lis1 (n>103 events per data point). Velocities were normalized by setting those in the absence of Lis1 [from (C)] to 100%. (E) Binding densities of dynein variants in the presence of ATP (top) or ATP-Vi (bottom) (n>8 fields of view per data point). See also Table S1. (F) Normalized binding densities of dynein variants alone (solid bars), or in the presence of 300 nM Lis1 (hatched bars), or 300 nM Lis15A (cross-hatched bars) in the presence of ATP. Binding densities were normalized by setting those in the absence of Lis1 [from (E), top] to 100% (n=12 fields of view per data point). (G–I) Normalized binding densities of dynein variants alone (solid bars), or in the presence of 300 nM Lis1 (hatched bars) in the presence of 1 mM ATP-Vi (G), 1 mM ADP (H) and 2.5 units/mL apyrase (“Apo”) (I) (n=8 fields of view per data point). Normalized binding densities in the absence of Lis1 shown in (G) are those in (E, bottom) without normalization. Statistical significance was calculated using an unpaired t-test with Welch’s correction for both velocity (C and D) and binding density (E–I). P-values: ns, not significant; *, <0.05; **, <0.01; ***, <0.001, ****, <0.0001. Data are shown as mean and standard error of mean.
Current evidence suggests that Lis1 acts on dynein by disrupting the coupling between ATP hydrolysis and MT binding and release. Lis1 is a dimer of two β-propellers (Figure 1A) (Tarricone et al., 2004), with a single β-propeller binding to AAA4 on dynein’s ring (Huang et al., 2012; Toropova et al., 2014). In vitro motility assays showed that Lis1 decreases dynein’s velocity (Huang et al., 2012; Yamada et al., 2008) by causing dynein to remain tightly bound to MTs in the presence of ATP, which would normally result in MT release (Huang et al., 2012; McKenney et al., 2010). This model for Lis1 regulation of dynein is at odds with some of the proposed functions of Lis1 discussed above, where depletion of Lis1 leads to a decrease in cargo transport.
The regulation of dynein motility also involves intra-motor control. The nucleotide state at AAA3 regulates dynein’s motility by acting on its mechanochemical cycle (DeWitt et al., 2014; Nicholas et al., 2015): stepping dynein contains ADP at AAA3, while ATP or no nucleotide (“apo”) at this site results in a slower dynein that is tightly bound to MTs (DeWitt et al., 2014). This AAA3-mediated inhibition occurs even when ATP binds to AAA1, which normally triggers MT release.
We were struck by the similarities in the behavior of dynein in the presence of Lis1 or when its AAA3 is in an apo or ATP-bound state. The proximity of Lis1’s binding site to AAA3’s ATP-binding site made us wonder if Lis1 regulates dynein by acting on AAA3. Here we used mutants in yeast dynein to control the nucleotide state of AAA3 and a combination of single-molecule imaging, high-resolution cryo-electron microscopy (cryo-EM), in vitro reconstitutions, and in vivo assays to understand the relationship between Lis1 and the nucleotide state of dynein’s AAA3 domain. We made the surprising discovery that Lis1 can regulate dynein in two distinct and opposing ways, depending on the nucleotide state of AAA3. Based on our data we propose a new model for the regulation of dynein by Lis1 that can explain the multiple and conflicting cellular roles of Lis1.
Results
Lis1 has two modes of regulating dynein
Previous studies led us to hypothesize that the nucleotide state at AAA3 might play a role in the regulation of dynein by Lis1 (DeWitt et al., 2014; Nicholas et al., 2015; Toropova et al., 2014). To test this, we used a minimal S. cerevisiae dynein construct dimerized by GST (Dynwt) (Figure 1A) that is similar to full-length dynein in its motile properties (DeWitt et al., 2012; Reck-Peterson et al., 2006) and regulation by Lis1 (Huang et al., 2012). To mimic different nucleotide states at AAA3, we made well-characterized mutations that disrupt its conserved Walker A and Walker B motifs (Cho et al., 2008; DeWitt et al., 2014; Kon et al., 2004). The Walker A mutant (DynWA) impairs ATP binding, while the Walker B mutant (DynWB) allows ATP to bind, but prevents its hydrolysis (Figure 1B). Lis1 had a similar binding affinity for all three dynein variants (Figure S1A and Table S1).
We first asked if the nucleotide state at AAA3 altered Lis1’s effect on dynein’s velocity using single-molecule motility assays (Figures 1C, 1D, S1B and Table S1). In agreement with previous studies, Lis1 decreased the velocity of Dynwt (Huang et al., 2012; McKenney et al., 2010; Toropova et al., 2014; Yamada et al., 2008) (Figure 1D). While both DynWA and DynWB had slower velocities than Dynwt on their own (Figure 1C and Table S1), the effects of adding Lis1 to them were striking. DynWA was hypersensitive to Lis1, with a velocity reduction of 99% (Figure 1D). Unexpectedly, the Lis1 effect was reversed with DynWB; the velocity of this mutant almost doubled in the presence of Lis1 (Figure 1D). These results suggest that Lis1 can regulate dynein’s velocity in opposite ways depending on the nucleotide state at AAA3.
Next, we asked if the effects of Lis1 on dynein’s velocity were a result of its effect on dynein’s affinity for MTs. To measure MT binding affinities, we used single-molecule total internal reflection fluorescence (TIRF) microscopy to quantify the density of dynein bound to MTs. We first characterized the binding densities of the dynein constructs by themselves in the presence of ATP (Figure 1E and Table S1). Although DynWA and DynWB have significantly higher affinities for MTs than Dynwt (Cho et al., 2008; DeWitt et al., 2014), their affinities are indistinguishable. Given this, we normalized most binding data with the motor alone set to 100% (non-normalized data can be found in Table S1).
As before (Huang et al., 2012), we observed increased binding of Dynwt to MTs with Lis1 and ATP (Figure 1F). In agreement with our velocity data (Figure 1D), the two AAA3 variants were regulated by Lis1 in opposite ways: Lis1 increased the binding density of DynWA by 62% and decreased that of DynWB by 42% with ATP (Figure 1F). Importantly, a Lis1 mutant that cannot bind to dynein (Lis15A) (Toropova et al., 2014) had no effect on any of the variants (Figure 1F). Thus, Lis1 can also regulate dynein’s MT binding in opposing manners depending on the nucleotide state at AAA3.
Given how unexpected Lis1’s effects on DynWB were and the intrinsic high affinity of DynWB for MTs, we wanted to rule out mutant-specific effects. We repeated our MT binding experiments with Dynwt trapped with AAA3 in a nucleotide state as similar to that of DynWB (ATP) as possible. For this we used ATP plus Vanadate (ATP-Vi), which generates ADP-Vi upon ATP hydrolysis (Burgess et al., 2003). Dynwt bound MTs with the same affinity with ATP-Vi or ATP (Figure 1E and Table S1). More importantly, under ATP-Vi conditions, Dynwt and DynWB both showed decreased MT binding with Lis1 (Figure 1G), in striking contrast to the increased MT binding we saw for Dynwt with ATP (Figure 1F), where AAA3 is expected to be mainly in an ADP state (DeWitt et al., 2014). The affinity of DynWA, which cannot bind nucleotide at AAA3, still increased with ATP-Vi and Lis1 (Figure 1G). Performing these experiments with ADP or no nucleotide abolished all relative differences among the three variants (Figure 1H, I), suggesting that dynein’s AAA1 must be in an ATP or ADP-Pi state for Lis1 regulation to be apparent.
We next sought to understand the mechanistic basis for the dual role of Lis1 by determining the structures of dynein:Lis1 complexes using different AAA3 variants.
Structural basis for the tight microtubule binding state of dynein induced by Lis1
Figure 2A summarizes aspects of dynein’s structure relevant to the results presented below. More detailed reviews of dynein’s structure can be found elsewhere (Cianfrocco et al., 2015; Gleave et al., 2014).
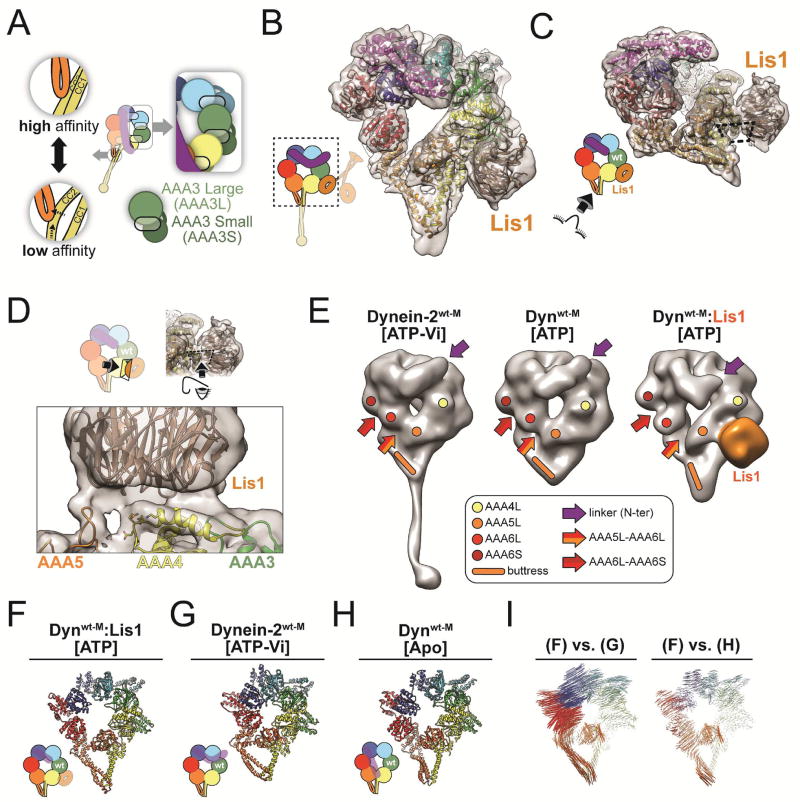
Figure 2. Structural basis for the tight microtubule binding state of dynein induced by Lis1.
(A) Additional structural features of the schematic shown in Figure 1A. The inset on the right shows the architecture of the AAA ring, with each AAA domain composed of both large (AAAL) and small (AAAS) subdomains (exemplified for AAA3). Large and small subdomains are arranged in two separate planes. Semi-transparent ovals represent nucleotide-binding sites. The linker domain forms a third layer, above the AAA ring. Insets on the left illustrate how the buttress couples the conformation of dynein’s ring to dynein’s affinity for MTs by changing the register between the two helices (CC1 and CC2) in the stalk’s coiled coil. (B) Cryo-EM structure of the Dynwt-M:Lis1 complex, solved in the presence of ATP. The cryo-EM map was filtered using local resolution and is shown as a semi-transparent surface, with the atomic model shown as a ribbon diagram. The cartoon (bottom left) indicates the portion of dynein observed in our cryo-EM map. (C) The structure viewed from the stalk, with the Dynwt-M-Lis1 interface indicated by the dashed rectangle. (D) Close-up view of the Dynwt-M-Lis1 interface, seen in a direction perpendicular to the dashed rectangle. Side chains shown on AAA4 are residues that prevent Lis1 binding when mutated (KDEE) (Huang et al., 2012). AAA domains that contribute motifs to the interface are labeled. (E) Ring architecture of human Dynein-2wt-M (ATP-Vi) (Schmidt et al., 2014) (PDB: 4RH7), yeast Dynwt-M (ATP) (Bhabha et al., 2014) (EMDB 6054), and our map of Dynwt-M:Lis1 (ATP). Colored dots and rod highlight equivalent positions in AAA4 (yellow), AAA5 (orange) and AAA6 (red). Colored arrows indicate the N-terminus of the linker (purple), the boundary between AAA5 and AAA6L (orange/red), and the boundary between AAA6L and AAA6S (red/dark red). The structure of human dynein-2 was converted into an EM-like density and both it and our Dynwt-M:Lis1 map were filtered to 20Å. The structure of Dynwt-M (ATP) (EMDB: 6054) has a resolution of 17Å. (F–I) Ring conformations of the Dynwt-M:Lis1 structure (F); the low-affinity, wild-type human Dynein-2wt-M solved in the presence of ATP-Vi (PDB: 4RH7) (G); and the high-affinity, wild-type S. cerevisiae dynein solved in the absence of nucleotide (PDB: 4AKI) (H). We removed the linker and, when present, Lis1 for clarity. (I) Maps of pairwise alpha carbon interatomic distances between the Dynwt-M:Lis1 structure and the low-affinity, wild-type human Dynein-2wt-M (left), and high-affinity, wild-type S. cerevisiae Dynwt-M (right). Structures were aligned using their AAA4L domains. The length and thickness of the vectors are proportional to the calculated interatomic distances. See also Figure S2.
We first used single particle cryo-EM to determine the structure of Dynein:Lis1 in the presence of ATP, a state with high affinity for MTs. We used a monomeric dynein construct lacking GST (Dynwt-M), which is otherwise identical to the dimeric dynein constructs used above. We solved a 7.7Å resolution structure of Dynwt-M bound to Lis1 in the presence of ATP and built an atomic model into the density using Rosetta (Figures 2B, 2C, S2, and Table S2) (Wang et al., 2016).
This map, at higher resolution than our previous structure of the same complex (Toropova et al., 2014), provided a detailed view of the interface between dynein and Lis1 and revealed the conformation of dynein’s ring. As before, a single Lis1 β-propeller was bound to Dynwt-M, even though Lis1 was present as a dimer in our sample. The second β-propeller, as well as the rest of Lis1, are likely disordered and thus averaged out during the reconstruction process. The EM density suggested that Lis1 interacts not only with an alpha helix in AAA4 (Toropova et al., 2014), but also with a loop from AAA5, and possibly another from AAA3 (Figure 2C, D), although the resolution of the map does not establish this last interaction unambiguously.
Next we examined how Lis1 affected the conformation of dynein’s ring by comparing our structure to existing structures of dynein. Our map of Dynwt-M:Lis1 appears to show the linker in its pre-power stroke position; although the N-terminal half of the linker is not visible in the 7.7Å map, an extension pointing towards AAA2 becomes apparent at lower resolution (Figures 2E and S2J). Given the pre-power stroke position of the linker and the fact that we obtained the structure in ATP, AAA1 must contain either ATP or ADP-Pi, as phosphate release results in the power stroke (Mogami et al., 2007). Since this conformation of the linker was observed in the structures of human dynein-2 with ATP-Vi (Schmidt et al., 2014) and yeast Dynwt-M with ATP (Bhabha et al., 2014) we chose them for our comparison (Figure 2E).
Human dynein-2 (ATP-Vi) and yeast Dynwt-M (ATP) both show a closed conformation of the AAA ring, associated with a low-affinity state (Figure 2E). Dynwt-M:Lis1 (ATP), however, shows an open-ring conformation, associated with a high-affinity state (Figure 2E). Given that the only difference between our structure and that of Dynwt-M (ATP) is the presence of Lis1, the open conformation of the ring must be a consequence of Lis1 binding to dynein.
The ring opening and the similarities between the Dynwt-M:Lis1 and nucleotide-free dynein structures are further highlighted by an analysis of interatomic distances between alpha carbons in Dynwt-M:Lis1, human dynein-2 (ATP-Vi) (Schmidt et al., 2014) and nucleotide-free yeast dynein (Schmidt et al., 2012) (Figure 2F–I).
Two Lis1 β-propellers are bound to dynein in the weak microtubule binding state
We then moved on to the most puzzling aspect of our data: why does introducing a Walker B mutation in AAA3 lead to such a dramatic difference in how Lis1 regulates dynein? For this, we solved a 10.5Å-resolution cryo-EM structure of the DynWB-M:Lis1 complex in the presence of ATP-Vi and used Rosetta to build an atomic model (Figures 3A, 3B, S3, and Table S2). As in the Dynwt-M:Lis1 structure, DynWB-M is a monomeric construct lacking GST and Lis1 is a dimer.
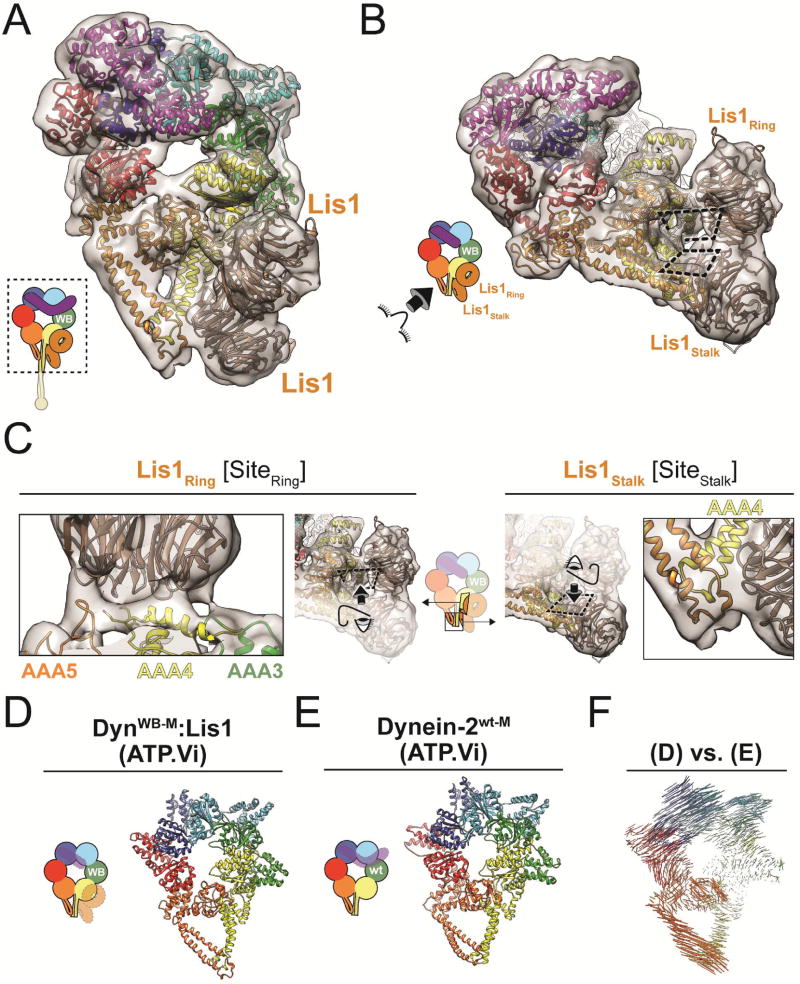
Figure 3. Two Lis1 β-propellers are bound to dynein in the weak microtubule binding state.
(A) Cryo-EM structure of the DynWB-M:Lis1 complex, solved in the presence of ATP-Vi. The cryo-EM map is shown as a semi-transparent surface, with the atomic model generated with Rosetta shown as a ribbon diagram. The cartoon (bottom left) indicates the portion of dynein observed in our cryo-EM map. (B) Structure viewed from the stalk, with the two DynWB-M-Lis1 interfaces indicated by the dashed rectangles. (C) Close-up views of the two DynWB-M-Lis1 interfaces, located on dynein’s ring (SiteRing) and stalk (SiteStalk), viewed perpendicular to the dashed rectangles. AAA domains that contribute motifs to the interfaces are labeled. (D–E) Ring conformations of the DynWB-M:Lis1 structure (D) and the low-affinity, wild-type human dynein-2 solved in the presence of ATP-Vi (PDB: 4RH7) (E). We removed the linker and, when present, Lis1 for clarity. (F) Map of pairwise alpha carbon interatomic distances between the DynWB-M:Lis1 structure and the low-affinity, wild-type human dynein-2. Structures were aligned using their AAA3L domains. The length and thickness of the vectors are proportional to the calculated interatomic distances. See also Figure S3.
Surprisingly, the cryo-EM map of DynWB-M:Lis1 showed not one, but two Lis1 β-propellers bound to dynein (Figure 3A, B). One β-propeller binds to the previously identified site on the ring (AAA4) (“SiteRing”), while the other binds to dynein’s stalk, specifically coiled-coil 1 (CC1) (“SiteStalk”) (Figure 3A–C). Each Lis1 β-propeller uses a different surface to interact with dynein (Figure 3C). The density encompassing the two Lis1 β-propellers is contiguous, suggesting they interact with each other (Figure 3B), although our current resolution does not allow us to determine their rotational orientations and thus the nature of this interface. Analysis of the stoichiometry of our Dynwt-M:Lis1 complex using size-exclusion chromatography suggests that both β-propellers in the structure belong to the same Lis1 dimer (Figure S4).
The closed conformation of dynein’s ring in the DynWB-M:Lis1 structure, similar to that seen in the crystal structure of the low MT affinity state of human dynein-2 (Figure 3D, E) (Schmidt et al., 2014), is consistent with the weak MT binding of the complex. This similarity is further supported by an analysis of interatomic distances between alpha carbons in our DynWB-M:Lis1 structure and those in the structure of human dynein-2 (ATP-Vi) (Figure 3F).
Lis1’s opposite modes of dynein regulation are associated with rigid body motion conformational changes in dynein’s ring
To understand how structural changes in dynein’s ring relate to the two modes of Lis1 regulation and how Lis1 binding at SiteStalk in DynWB-M:Lis1 stabilizes a low affinity state in dynein we calculated inter-atomic distances between Dynwt-M:Lis1 and DynWB-M:Lis1 (Figure 4A). We found that the differences were captured by a rigid motion of elements on one half of dynein’s ring—comprising the stalk, AAA5, AAA6 and part of AAA1— relative to the rest (Figure 4A, B).
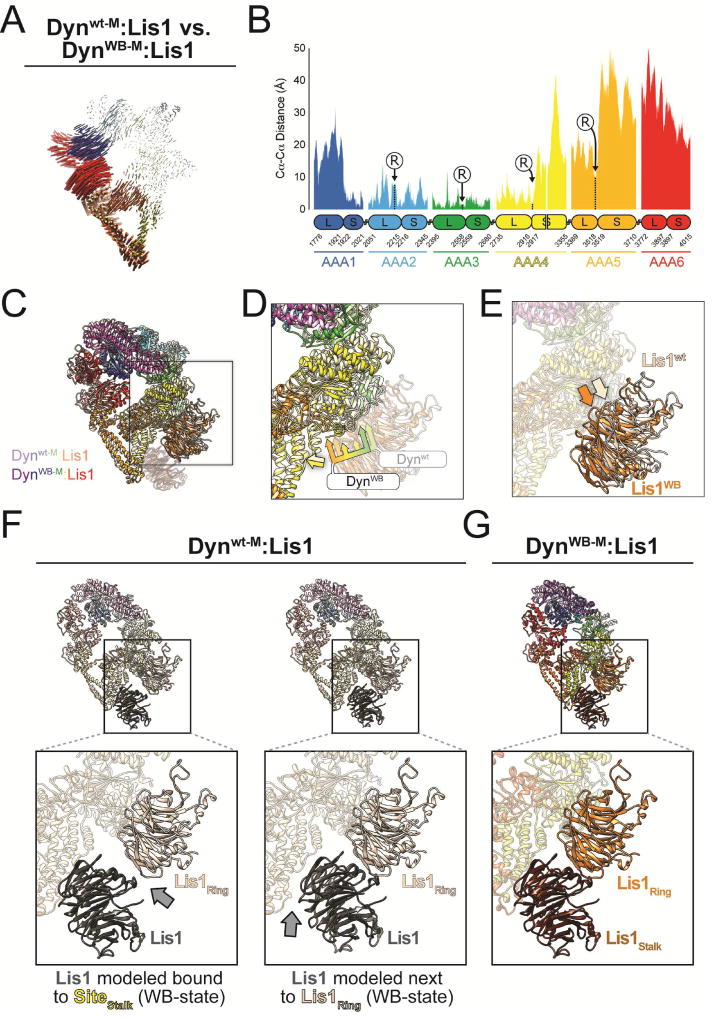
Figure 4. Lis1’s opposite modes of dynein regulation are associated with rigid body motion conformational changes in dynein’s ring.
(A) Map of pairwise alpha carbon interatomic distances between the Dynwt-M:Lis1 and DynWB-M:Lis1 structures. We removed the linker and Lis1 for clarity. Structures were aligned using their AAA3L domains. The length and thickness of the vectors are proportional to the calculated interatomic distances. (B) 1D plot of the interatomic distances shown in panel (A). Large and small AAA subdomains are indicated below the plot, along with amino acid numbers at their boundaries. The positions of the arginine fingers of domains AAA2-5, which act on domains AAA1-4, are labeled (“R”). (C) Superposition of the Dynwt-M:Lis1 and DynWB-M:Lis1 structures, aligned using their AAA4S domains. The Dynwt-M:Lis1 structure is shown in lighter colors and the Lis1 bound to SiteStalk in DynWB-M:Lis1 was faded for clarity. The square highlights SiteRing, and is the area represented in panels (D) and (E). (D) Close-up of the SiteRing in dynein, with Lis1 faded for clarity. The bi-tone yellow arrow indicates good alignment for the base of the stalks. The tri-color multi-headed arrows point to the AAA3, AAA4 and AAA5 elements in SiteRing in both Dynwt-M:Lis1 and DynWB-M:Lis1. (E) Positions of the SiteRing-bound Lis1 in the Dynwt-M:Lis1 and DynWB-M:Lis1 structures. Same view as in (D) but with dynein faded for clarity. Light and dark orange arrows point to the equivalent positions in Lis1 in the Dynwt-M:Lis1 and DynWB-M:Lis1 structures, respectively. (F) Modeling of a second Lis1 into the Dynwt-M:Lis1 structure interacting either with SiteStalk (left) or with the Lis1 bound at SiteRing (right). Grey arrows point to gaps present in the models. (G) For comparison, we show the same view of the experimentally observed DynWB-M:Lis1 structure.
The conformational change illustrated in Figure 4A results in the two Lis1 binding sites (SiteRing and SiteStalk) being closer together in DynWB-M:Lis1 relative to Dynwt-M:Lis1 (Figure 4C–E). This shorter distance means that the interaction between the two Lis1’s cannot be satisfied in the Dynwt-M:Lis1 structure, where we only observed a single Lis1 β-propeller bound (Figure 4).
The putative second Lis1 binding site is conserved
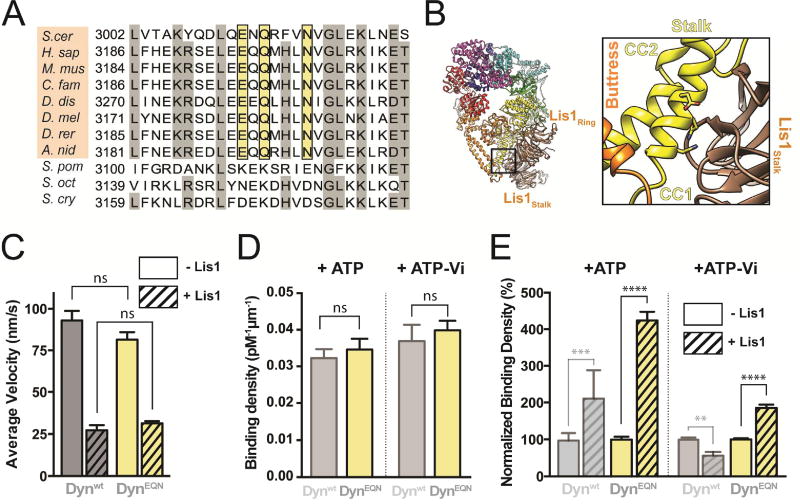
The resolution of our cryo-EM map of the DynWB-M:Lis1 complex is not sufficient to unambiguously dock Lis1 and determine which residues interact with SiteStalk in dynein. However, the segment of CC1 containing SiteStalk is short, limiting the number of candidate binding residues (Figure 3C). We aligned dynein sequences corresponding to this region of the stalk and identified three residues—E3012, Q3014 and N3018 in the yeast sequence (Figure 5A, B)—that are conserved from yeast to humans (Figure 5A). These residues are not conserved in a subset of fission yeasts. Supporting the idea that these residues are central features of SiteStalk, we could not identify a Lis1 orthologue in these fission yeast genomes, while organisms with a conserved EQN triad have clear Lis1 orthologues (Figure 5A).
Figure 5. The second Lis1 binding site is required for the Lis1-induced weak microtubule binding state of dynein.
(A) Sequence conservation around the putative SiteStalk. Sequence in the region of dynein’s stalk identified as the putative second binding site for Lis1, extracted from a full sequence alignment of dynein heavy chain genes. Residues with 70% conservation or higher are shaded grey. The three residues we mutated—E, Q, N, shaded in yellow—are conserved in model organisms that have Lis1 orthologs in their genomes (Saccharomyces cerevisiae; Homo sapiens; Mus musculus; Canis familiaris; Dictyostelium discoideum; Drosophila melanogaster; Danio rerio; Aspergillus nidulans, shaded in orange), but not in a group of fission yeasts (Schizosaccharomyces pombe; Schizosaccharomyces octosporus; Schizosaccharomyces cryophilus) that do not appear to have a Lis1 ortholog in their genome. (B) Atomic model of the DynWB-M:Lis1 complex and close-up view of SiteStalk. The conserved EQN triad is shown in stick representation and nearby dynein motifs are labeled. DynEQN is a construct that carries an EQN to AAA mutation but has a wild-type AAA3. (C) Average velocities of Dynwt (grey) and DynEQN (yellow) in the absence (solid bars) or presence (hatched bars) of 300 nM Lis1 (n>154 events per data point). See Figure S5B for representative kymographs. (D) Binding densities of Dynwt (semi-transparent grey; data reproduced from Figure 1 for comparison) and DynEQN (yellow) in the presence of ATP (left) or ATP-Vi (right). See also Table S1. (E) Normalized binding densities of Dynwt (semi-transparent grey) and DynEQN (yellow) in the absence (solid bars) or presence (hatched bars) of 300 nM Lis1 (n=12 fields of view per data point), in the presence of ATP (left) or ATP-Vi (right). Binding densities were normalized by setting those in the absence of Lis1 to 100%. Data for Dynwt are reproduced from Figure 1 for comparison). Statistical significance was calculated using unpaired t-test with Welch’s correction for both velocity (C) and binding density (D–E). P-values: ns, not significant; **, <0.01; ***, <0.001, ****, <0.0001. Data are shown as mean and standard error of mean.
The second Lis1 binding site is required for Lis1-mediated weak microtubule binding
Our data show that Lis1 induces or stabilizes a weak MT binding state in dynein when AAA3 carries a Walker B mutation, and that in this complex (DynWB-M:Lis1) a second Lis1 β-propeller interacts with dynein’s stalk (SiteStalk). To determine if SiteStalk is required for Lis1 to induce the weak binding state in dynein we mutated the conserved E3012, Q3014, and N3018 residues to alanine (DynEQN). Our prediction was that the Lis1-induced weak binding state would no longer be available in DynEQN.
By itself, DynEQN moved with the same velocity as Dynwt, suggesting that the SiteStalk mutations do not grossly impair dynein’s structure or function (Figures 5C and S5A). DynEQN binds to Lis1 with reduced affinity compared to Dynwt in ATP-Vi (Figure S5B), which is compatible with the loss of one of the two Lis1 binding sites. In addition, the velocities of DynEQN and Dynwt were lowered by similar amounts in the presence of 300 nM Lis1 (Figure 5C), suggesting that mutation of SiteStalk does not impair Lis1’s interaction with dynein at SiteRing.
To test our hypothesis that SiteStalk is required for the Lis1-induced weak MT binding state, we measured DynEQN’s affinity for MTs under conditions that would normally lead to this state (ATP-Vi). Our prediction was that DynEQN would show higher affinity for MTs than Dynwt under those conditions. In the absence of Lis1, DynEQN and Dynwt had a similar affinity for MTs, suggesting that mutating SiteStalk does not alter the ability of DynEQN to interact with MTs (Figure 5D). In the presence of Lis1, as we predicted, DynEQN showed an 85% increase in MT affinity relative to Dynwt (Figure 5E).
With ATP, where AAA3 can sample different nucleotide states, both weak and tight binding states should be available to Dynwt in the presence of Lis1, while the weak binding state would be unavailable to DynEQN. Dynwt’s affinity for MTs in the presence of Lis1 should reflect a mixture of weak and tight binding states, while that of DynEQN should come from tight binding states only. As predicted, in the presence of ATP, Lis1 increased DynEQN’s binding density on MTs two-fold more than it did for Dynwt (Figure 5E).
We also mutated the conserved EQN triad to alanine in dynein with a AAA3 Walker B mutation (DynWB/EQN). While Lis1 increased the velocity of DynWB, it decreased the velocity of DynWB/EQN (Figures 1D, S5C and S5D). Similarly, while Lis1 decreased the MT affinity of DynWB, this decrease was no longer observed in DynWB/EQN with ATP or ATP-Vi (Figure S5E). These results suggest that the conserved second Lis1 binding site (SiteStalk) is required to decrease dynein’s MT affinity when AAA3 contains ATP or ADP-Pi.
The second binding site for Lis1 is required for dynein localization
Yeast dynein positions the mitotic spindle (Eshel et al., 1993; Li et al., 1993) by “pulling” on spindle pole body (SPB)-anchored MTs from the cell cortex (Figure 6A) (Adames and Cooper, 2000). Dynein reaches the cortex by localizing to MT plus ends, either via kinesin-dependent transport or recruitment from the cytosol (Carvalho et al., 2004; Caudron et al., 2008; Markus et al., 2009). Dynein’s plus-end-localization, kinesin-dependent transport, and later “off-loading” to the cell cortex all require Lis1 (Lee et al., 2003; Li et al., 2005; Markus and Lee, 2011; Markus et al., 2009; 2011; Sheeman et al., 2003). The requirement of Lis1 for plus-end-localization and off-loading to the cortex is compatible with a model where Lis1 promotes a tight MT-binding state in dynein. However, this model is at odds with Lis1’s requirement in the kinesin-dependent transport of dynein to MT plus ends, as efficient transport by kinesin would be favored when dynein weakly interacts with MTs (Roberts et al., 2014).
Figure 6. The second binding site for Lis1 is required for dynein’s microtubule plus end localization in vivo and in vitro.
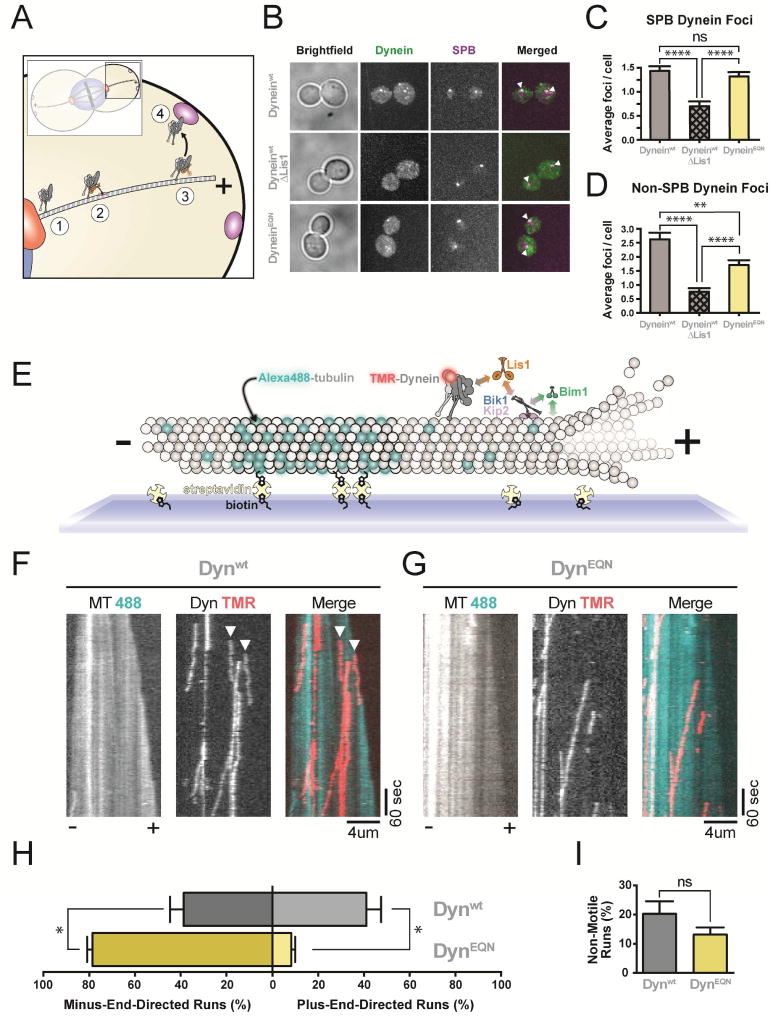
(A) Schematic of dynein and Lis1 function in spindle positioning in S. cerevisiae. Dynein is localized to the SPB (1), transported to the MT plus end by a kinesin in a process that also requires Lis1 (2), maintained at the MT plus end by Lis1 (3), and “off-loaded” to the cell cortex (4), where it pulls on SPB-attached MTs to position the mitotic spindle. (B) Dynein localization in dividing S. cerevisiae. First column: Representative brightfield images. Second column: maximum projections of 3xGFP-labeled dynein (Dyneinwt or DyneinEQN). Third column: maximum projections of tdTomato-labeled SPC110, a SPB marker. Fourth column: merged 3xGFP-dynein and tdTomato-SPB images. White arrowheads: co-localized dynein and SPB signals. Strains imaged: Dyneinwt (top row); Dyneinwt in a ΔLis1 background (middle row); DyneinEQN (bottom row). Both dynein-3xGFP and SPC110-tdTomato are expressed under their endogenous promoters. (C, D) Quantification of the data presented in (B). (C) Average number of dynein foci per cell colocalized with SPBs; and (D) Average number of dynein foci per cell not colocalized with SPBs for Dyneinwt (grey), Dyneinwt/ΔLis1 (hatched grey) and DyneinEQN (yellow) strains (n>50 cells per data point). (E) Schematic representation of our in vitro reconstitution of kinesin-mediated dynein transport to the MT plus end. Brightly-labeled, GMPCPP-stabilized MT seeds are attached to the coverslip via biotin-streptavidin interactions. A dimly-labeled MT extension grows faster at the plus end of the seed, allowing MT polarity to be determined. Addition of dynein (labeled with TMR), Lis1, Bik1, Kip2 and Bim1 results in plus-end-directed transport of dynein by kinesin. Known interactions are shown with double-headed arrows color-coded according to the proteins involved. MT plus and minus ends are labeled. (F, G) Representative kymographs from the assay outlined in (E), with MT (488) and dynein (TMR) channels shown in black and white, and the merged image in pseudocolor, for Dynwt (F) and DynEQN (G). Plus (+) and minus (−) indicate MT polarity. White arrowheads point to the start of plus-end-directed runs. (H) Quantification of the percentage of plus- and minus-end-directed runs for Dynwt (Grey) and DynEQN (Yellow). (I) Quantification of the percentage of non-motile runs. (n = 4 technical replicates). Statistical significance was calculated using Mann-Whitney test for both average number of foci per cell (C, D) and percentage of runs (H, I). P-values: ns, not significant; *, < 0.05; **, <0.01; ****, <0.0001. Data are shown as mean and standard error of mean. See also Figure S6.
To determine the cellular role of SiteStalk, we monitored dynein’s localization in vivo by fusing 3X-GFP to full-length dynein or a version carrying the EQN SiteStalk mutation (dyneinEQN) at the endogenous locus. We also determined dynein localization in cells lacking Lis1 (ΔLis1). Dynein localizes to both the SPB and MT plus ends/ the cortex (Figure 6B) (Lee et al., 2003). We could not distinguish between MT plus ends and the cortex in our experiments, but localization to both sites requires Lis1. Deletion of Lis1 caused mislocalization of dynein (Lee et al., 2003; Sheeman et al., 2003) (Figure 6B), with ~2- and ~3-fold reductions in dynein “foci” (diffraction-limited spots) at the SPB (Figure 6C) and MT plus ends/cortex (Figure 6D), respectively. Interestingly, while yeast carrying the dyneinEQN mutant contained the same number of dynein foci at the SPB as wild type yeast, they showed significantly reduced dynein at the MT plus end/cortex (Figure 6B, C). These data suggest that SiteStalk is required for dynein’s localization in vivo.
The second binding site for Lis1 is required for dynein to be transported by kinesin
Kinesin is required for the plus-end-localization of dynein in yeast (Carvalho et al., 2004; Caudron et al., 2008; Markus et al., 2009), filamentous fungi (Zhang et al., 2010), and mammalian neurons (Twelvetrees et al., 2016). Previously, we reconstituted yeast dynein plus-end-localization in vitro, showing that dynein is transported by a kinesin, Kip2, to MT plus ends in a process that requires Lis1 and two MT plus tip proteins Bik1 (CLIP170) and Bim1 (EB1) (Roberts et al., 2014). During transport, dynein and kinesin engage in a tug-of-war that results in dynein being slowly pulled towards the plus end of MTs by kinesin, with Bik1 and Bim1 enhancing kinesin’s processivity.
We hypothesized that binding of Lis1 to SiteStalk might be required for kinesin to localize dynein to MT plus ends, explaining the in vivo localization defect of DynEQN (Figure 6B, C). To test this, we reconstituted Dynwt and DynEQN trafficking to the plus ends of dynamic MTs (Figure 6E). We monitored the directionality of dynein movement in the presence of Kip2, Lis1, Bik1, and Bim1. Dynwt was transported to the plus end in 41% of events (Figure 6F, H). In contrast, only 8.2% of events were plus-end-directed with DynEQN (Figure 6G, H). The number of non-motile runs and the average velocity of plus-end runs observed with Dynwt and DynEQN were not statistically different, although the average velocity of minus-end runs for DynEQN was slightly faster than those for Dynwt (Figure 6I and S6A). Dynein movement in the plus end direction indicates that the transport complex can form (Roberts et al., 2014). To test if the EQN mutation affected complex formation we monitored the ability of dynein to be recruited to MTs by kinesin (in the presence of Lis1, Bik1 and Bim1). This experiment must be done in a dynein background engineered to have a weak affinity for MTs (Dynweak) (Redwine et al., 2012; Roberts et al., 2014). As expected, Lis1 was required for the recruitment of both Dynweak/wt and Dynweak/EQN to MTs. Dynweak/wt was recruited to MTs ~5 times more efficiently than Dynweak/EQN (Figure S6B), showing that the ability of Lis1 to bind at Sitestalk leads to more efficient kinesin transport complex formation. Thus, Sitestalk is required for dynein to reach MT plus ends and promotes its interaction with the kinesin transport complex.
Discussion
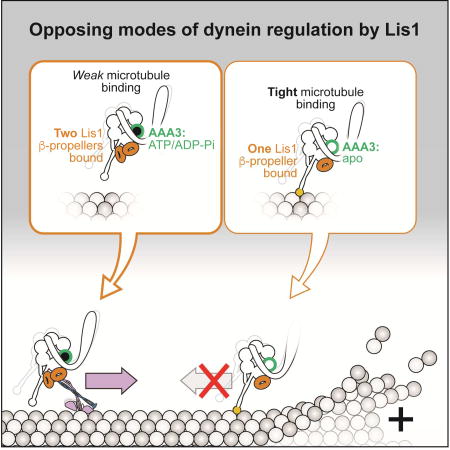
Here we report the surprising discovery that Lis1 has two opposing modes of regulating dynein, being capable of enhancing or inhibiting its affinity for MTs (Figure 7). This is a major revision of the model for how Lis1 regulates dynein with important implications for the biological roles of this ubiquitous and highly conserved dynein regulator.
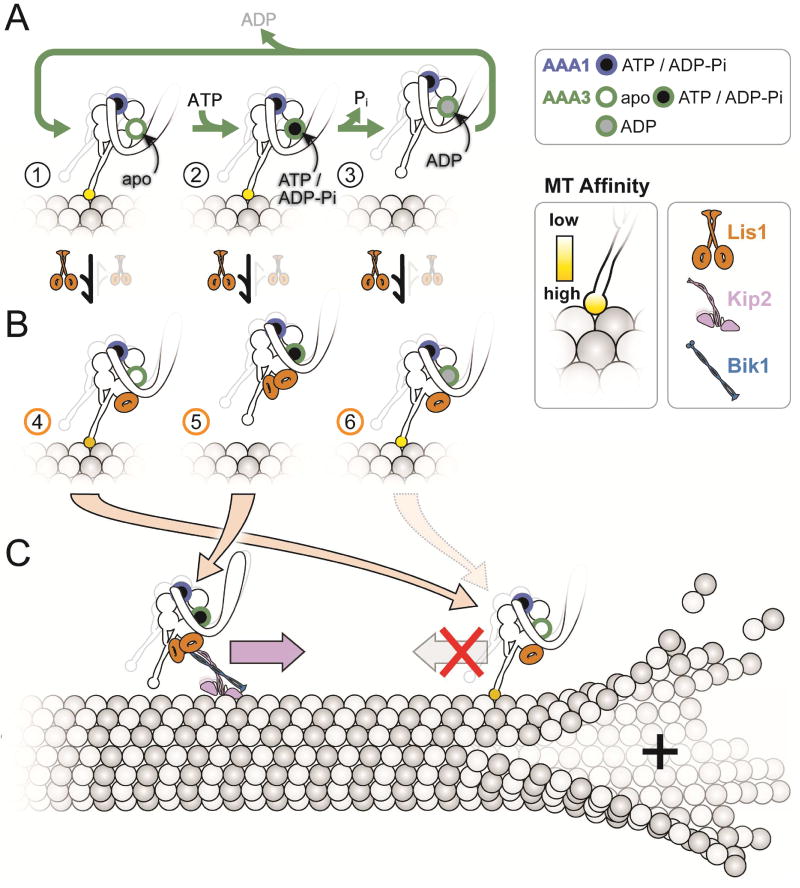
Figure 7. Model for the opposing modes of regulation of dynein by Lis1).
(A, B) The nucleotide state of AAA3 determines which regulatory mode is used by Lis1. The figure illustrates how the ATP hydrolysis cycle at AAA3 affects the affinity of dynein for MTs (A) and how Lis1 acts on these different states (B). We show dynein with ATP/ADP-Pi bound at its AAA1 site to reflect that our data suggest this is the state where Lis1 regulation is apparent. Dynein alone has high affinity for MTs when its AAA3 is either empty (“apo”) (A, state 1), or bound to ATP/ADP-Pi (A, state 2). Phosphate release from AAA3 leads to the AAA3:ADP-bound, low-affinity state of the motor (A, state 3), which is expected to be the predominant state when dynein walks along MTs. Lis1 acts in opposite ways on states 1/3 versus 2: one β-propeller binds when AAA3 is either empty or contains ADP, leading to tight MT binding (B, states 4 and 6); while 2 β-propellers bind when AAA3 has ATP/ADP-Pi, leading to weak MT binding (B, state 5). (C) Proposed biological roles of the Lis1-mediated weak and tight MT-binding states of dynein. In S. cerevisiae, dynein is transported towards the plus-end of MTs by Kip2, a process that requires Lis1. Binding of two Lis1 β-propellers (B, state 5) keeps the motor in a weak affinity state and promotes the formation of the kinesin transport complex. Once at the plus-end of the MT, dynein cycles to a AAA3:ADP or AAA3:apo state (by a process not currently understood); binding of one Lis1 β-propeller (B, states 4 and 6) keeps the motor in a tightly bound to the MT in preparation for cargo loading.
Structural basis for the opposing modes of dynein regulation by Lis1
How does Lis1 stabilize dynein’s ring in a high affinity conformation when AAA3 is nucleotide-free? The higher resolution of the Dynwt-M:Lis1 structure presented here revealed that in addition to the interaction with AAA3 and AAA4 (Huang et al., 2012; Toropova et al., 2014), Lis1 also interacts with AAA5 (Figure 2D). The transition from the high affinity, open conformation of the ring to its low affinity, closed conformation involves the rotation of AAA5 relative to a rigid module comprised of AAA2–4 (Schmidt et al., 2014). This motion leads to the buttress-mediated change in the register of the stalk, and ultimately in changes in the conformation and affinity of the MTBD (Figure 2A) (Cianfrocco et al., 2015). We propose that Lis1 keeps dynein in a high MT affinity state by clamping AAA3/4 and AAA5 together, thus blocking the ring rearrangements necessary for dynein to adopt a low MT affinity state. We were unable to test this hypothesis here because the region of AAA5 where Lis1 interacts with dynein had the lowest local resolution in our structure (Figure S2H, I).
How do the two Lis1 β-propellers prevent dynein from switching to a high-affinity state upon binding to MTs when AAA3 contains ATP or ADP-Pi? Normally, dynein binding to MTs triggers a conformational change in its MTBD that alters the register of its coiled coil stalk and ultimately results in the open, high-affinity conformation of dynein’s ring (Cianfrocco et al., 2015). The sliding of the two helices in the stalk (CC1 and CC2) changes their position with respect to the ring (Schmidt et al., 2014) (Figure 2A) as well as the distance between SiteStalk and SiteRing. Our DynWB-M:Lis1 structure showed that SiteRing is closer to SiteStalk than in Dynwt-M:Lis1 (Figure 4C–E), likely allowing for the interaction between the two Lis1 β-propellers, as suggested by the continuous density we observed (Figure 3A, B). We hypothesize that bridging of CC1 and the ring by the two Lis1 β-propellers bound to dynein when AAA3 contains ATP or ADP-Pi prevents CC1 from sliding towards its high-affinity register. Future experiments could test this idea by disrupting the interaction between the two β-propellers, but this will require a higher resolution map where the rotational orientation of the β-propellers is unambiguous.
We had previously shown that the displacement of the linker from its docking site at AAA5 by Lis1 bound at SiteRing played a role in the formation of the high-affinity state; shortening the linker to a point where it would no longer be obstructed by Lis1 made dynein Lis1-insensitive (Toropova et al., 2014). While it is likely that the position of the linker affects allosteric changes in dynein’s ring, establishing how the short linker circumvents the effect of Lis1 at SiteRing will require a high-resolution structure of Dynwt-M:Lis1 carrying a short linker docked at AAA5.
Both modes of Lis1 regulation are required for dynein function
In S. cerevisiae and other organisms, some of Lis1’s known functions have been difficult to reconcile with its reported molecular function to induce a tight MT binding state in dynein. Our new model for Lis1 regulation of dynein can resolve these apparent contradictions.
As described above, dynein’s function in spindle positioning in yeast is composed of multiple steps (Figure 6A). Our data showed that less dynein was found at MT plus ends/ the cortex in dynein with a mutant SiteStalk (Figure 6B, D). We found that less dynein moved in the plus end direction (Figure 6F–H), but also that a mutant SiteStalk reduced the formation of the kinesin transport complex (Figure S6B). Thus, the defect in plus-end-localization of the dynein SiteStalk mutant could be due to a reduction in the formation of the transport complex and/or dynein’s inability to access the weak MT binding state. Our current experiments do not allow us to differentiate between these two possibilities or the relative contributions of each. However, our results show that dynein’s SiteStalk is required for dynein’s localization to MT plus ends both in vivo and in vitro. Other dynein functions in yeast require high affinity MT binding, such as being retained at MT plus ends. In this context we propose that Lis1 engages dynein only at SiteRing.
The dual functionality of Lis1 we have uncovered (Figure 7) provides a possible explanation for the cellular function of Lis1 in other organisms, some of which have been difficult to reconcile with a model where Lis1 exclusively promotes high-affinity MT binding by dynein. For example, Lis1 overexpression increases the velocity of acidic organelles in mouse neurons (Pandey and Smith, 2011) while its removal decreases the velocity of mRNAs in Drosophila embryos (Dix et al., 2013), pointing to a role for Lis1 in reducing dynein’s affinity for MTs (and thus increasing dynein velocity). We propose that in these cases both Lis1 β-propellers bind to dynein. In fact, binding of Lis1 to an activated human dynein/dynactin complex has recently been shown to increase its velocity in vitro (Baumbach et al., 2017; Gutierrez et al., 2017). Other functions for dynein are more aligned with Lis1 increasing dynein’s affinity for MTs. These include stabilizing dynein at MT plus ends (Li et al., 2005; Splinter et al., 2012), facilitating cargo loading on dynein (Egan et al., 2012; Lenz et al., 2006; Moughamian et al., 2013), aiding in dynein trafficking of high-load cargo (McKenney et al., 2010; Reddy et al., 2016; Yi et al., 2011), and slowing motility of some dynein cargo (Vagnoni et al., 2016). We propose that in these cases Lis1 is bound to dynein by a single β-propeller at SiteRing.
How are the two modes of Lis1 activity regulated?
The underlying basis for the switch between the two modes of Lis1 activity must involve regulating the nucleotide state at AAA3. There are several mechanisms we envision that could achieve this. Local fluctuations in the concentration of ATP or ADP could alter the rate of hydrolysis at AAA3, potentially making Lis1 regulation tunable to different cellular locations or events. Other dynein-binding partners, including Lis1 itself, could play a role in controlling ATP turnover at AAA3. For example, NudE/NudEL binds both dynein and Lis1 and has been implicated in many Lis1-dependent functions (Cianfrocco et al., 2015). Finally, an intriguing possibility is that a backward force exerted on dynein (such as that from a pulled cargo) could influence the nucleotide state of AAA3 (Nicholas et al., 2015). If a backward force on dynein promoted an apo state in AAA3, this would favor a single Lis1 β-propeller binding (at SiteRing) and maintain dynein in a high MT affinity state, which would be advantageous when dynein is transporting a high load cargo. Future work will be required to understand this complex process.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents can be directed to, and will be fulfilled by the Lead Contact, Samara Reck-Peterson (sreckpeterson@ucsd.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Yeast strains
S. cerevisiae strains used in this study are listed in Supplemental Table S3. The endogenous genomic copies of DYN1, PAC1, NDL1, KIP2, TUB1 and SPC110 were modified or deleted using PCR-based methods and transformed using the lithium acetate method. Cultures of S. cerevisiae for protein purification were grown, harvested and frozen as described (Reck-Peterson et al., 2006).
METHODS DETAILS
Protein Purification
Protein purification steps were done at 4 °C unless otherwise indicated. Dynein constructs were purified from S. cerevisiae as described previously (Reck-Peterson et al., 2006). Briefly, liquid nitrogen-frozen yeast cell pellets were lysed by grinding with a chilled coffee grinder and resuspending in dynein lysis buffer (DLB: final concentration 30 mM HEPES [pH 7.4], 50 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 10% glycerol, 1 mM DTT) supplemented with 0.1 mM Mg-ATP, 0.5 mM Pefabloc, 0.05% Triton and cOmplete EDTA-free protease inhibitor cocktail tablet (Roche). The lysate was clarified by centrifuging at 264,900 × g for 1 hr or at 125,100 × g for 2 hr. The clarified supernatant was incubated with IgG sepharose beads (GE Healthcare Life Sciences) for 1.5 hr. The beads were transferred to a gravity flow column, washed with DLB buffer supplemented with 250 mM potassium chloride, 0.1 mM Mg-ATP, 0.5 mM Pefabloc and 0.1% Triton, and with TEV buffer (10 mM Tris–HCl [pH 8.0], 150 mM potassium chloride, 10% glycerol, 1 mM DTT, 0.1 mM Mg-ATP and 0.5 mM Pefabloc). GST-dimerized dynein constructs were labeled with 5 µM Halo-TMR (Promega) in the column for 10 min at room temperature and unbound dyes were washed with TEV buffer at 4°C. Dynein was cleaved f rom IgG beads via incubation with 0.15 mg/mL TEV protease for 1 hr at 16°C. For dynei n monomer constructs, the TEV cleavage step was done overnight at 4 °C and the cle aved proteins were concentrated using 100K MWCO concentrator (EMD Millipore) to 1.5–5 mg/mL. Cleaved proteins were filtered by centrifuging with Ultrafree-MC VV filter (EMD Millipore) in a tabletop centrifuge and flash frozen in liquid nitrogen.
Lis1 and Kip2 were purified from S. cerevisiae as described previously (Huang et al., 2012; Roberts et al., 2014). Lysis and clarification steps were similar to dynein purification except buffer A (final concentration: 50 mM potassium phosphate [pH 8.0], 150 mM potassium acetate, 150 mM sodium chloride, 2mM magnesium acetate, 5mM β-mercaptoethanol, 10% glycerol, 0.2% Triton, 0.5 mM Pefabloc) supplemented with 10 mM imidazole (pH 8.0) and cOmplete EDTA-free protease inhibitor cocktail tablet was used as lysis buffer. The clarified supernatant was incubated with Ni-NTA agarose (Qiagen) for 1 hr. The Ni beads were transferred to the column, washed with buffer A + 20 mM imidazole (pH 8.0) and eluted with buffer A + 250 mM imidazole (pH 8.0). The eluted protein was incubated with IgG sepharose beads for 1 hr. IgG beads were transferred to a gravity flow column, washed with buffer A + 20 mM imidazole (pH 8.0) and with modified TEV buffer (50 mM Tris–HCl [pH 8.0], 150 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 10% glycerol, 1 mM DTT, and 0.5 mM Pefabloc). TEV cleavage was done as described for dynein purification.
Bik1 was purified from Baculovirus as described previously (Roberts et al., 2014). Cell pellets were resuspended in buffer B (final concentration: 50 mM Tris-HCl [pH 8.5], 300 mM potassium chloride, 5 mM β-mercaptoethanol, 5% glycerol, 10 mM imidazole) supplemented with 1% NP-40 and cOmplete EDTA-free protease inhibitor cocktail tablet and lysed using a Dounce homogenizer (15 strokes with loose plunger and 10 strokes with tight plunger). The lysate was clarified by centrifuging at 183,960 × g for 30 min. The clarified supernatant was incubated with Ni-NTA agarose for 1 hr. The Ni beads were transferred to a gravity flow column, washed with buffer C (20 mM Tris-HCl [pH 8.5], 500 mM potassium chloride, 5 mM β-mercaptoethanol, 20 mM imidazole), buffer D (20 mM Tris-HCl [pH 8.5], 1 M potassium chloride, 5 mM β-mercaptoethanol, 20 mM imidazole), buffer C and buffer E (20 mM Tris-HCl [pH 7.5], 200 mM potassium chloride, 5 mM β-mercaptoethanol, 10% glycerol). Bik1 was eluted with buffer E + 300 mM imidazole and flash frozen in liquid nitrogen.
Bim1 was purified from E. Coli as described previously (Roberts et al., 2014). Protein expression was induced in BL-21[DE3] cells (NEB) at OD 0.6 with 0.1 mM IPTG for 16 hr at 18°C. Cell pellets were resuspended in buffer B with 1 mg/mL lysozyme, incubated for 30 min on ice and lysed by sonication. The lysate was clarified by centrifuging at 154,980 × g for 30 min. The clarified supernatant was passed over Strep-Tactin agarose resin (IBA Life Sciences) three times in a gravity flow column. The resin was washed with buffer D and with modified TEV buffer. TEV cleavage was done as described for the dynein purification.
TIRF microscopy
Imaging was performed with an inverted microscope (Nikon, Ti-E Eclipse) equipped with a 100× 1.49 N.A. oil immersion objective (Nikon, Plano Apo). The xy position of the stage was controlled by ProScan linear motor stage controller (Prior). The microscope was equipped with a MLC400B laser launch (Agilent), with 405 nm, 488 nm, 561 nm and 640 nm laser lines. The excitation and emission paths were filtered using appropriate single bandpass filter cubes (Chroma). The emitted signals were detected with an electron multiplying CCD camera (Andor Technology, iXon Ultra 888). Illumination and image acquisition is controlled by NIS Elements Advanced Research software (Nikon).
Single-molecule motility and microtubule binding assay on taxol-stabilized microtubules
Single-molecule motility and MT binding assays were performed in flow chambers made with double stick tape using the TIRF microscopy set up described above. No. 1–1/2 coverslips (Corning) were used for the flow chamber assembly and sonicated in 100% ethanol for 10 min to reduce non-specific binding. Taxol-stabilized MTs with ~10% biotin-tubulin and ~10% Alexa488-tubulin were attached to the flow chamber via biotin-BSA and streptavidin as described previously (Huang et al., 2012). Dynein was labeled with Halo-TMR for visualization. For each frame, Alexa488-tubulin and TMR-dynein were exposed for 100 ms with the 488 nm laser and 561 nm laser, respectively.
For motility assays, 1–25 pM dynein was incubated with 300 nM Lis1 or modified TEV buffer (to buffer match for experiments without Lis1) for 10 min on ice, and flowed into the flow chamber pre-assembled with taxol-stabilized MTs. The final imaging buffer contained DLB supplemented with 50 mM potassium acetate (hence a total of 100 mM potassium acetate), 20 µM taxol, 1 mM Mg-ATP, 1 mg/mL casein, 71.5 mM β-mercaptoethanol and an oxygen scavenger system (0.4% glucose, 45 µg/ml glucose catalase, and 1.15 mg/ml glucose oxidase). MTs were imaged first by taking a single-frame snapshot. Dyneins were imaged every 1 sec (for wild-type dynein) or 2 sec (for mutant dyneins) for 10 min. At the end, MTs were imaged again by taking a snapshot to check for stage drift. Movies showing significant drift were not analyzed. Each sample was imaged no longer than 30 min.
For the single-molecule MT binding assays, 0.5–10 pM dynein was incubated with 300 nM Lis1 or modified TEV buffer (to buffer match for experiments without Lis1) for 10 min on ice. Dynein concentrations were varied to ensure that single dynein molecules could be resolved on each MT. Based on the dissociation constants measured (Figure S1 and Table S1) and using a one-site model of association, we have calculated that in the presence of 300nM Lis1, between 85%-88% of each dynein variant is in complex with Lis1, regardless of the dynein concentration. Given that the Lis1-dynein dissociation constants were determined with monomeric dynein constructs and at a higher salt than is present in the MT binding experiments, it is likely that the complex percentages we report here are underestimations. The final imaging buffer contained DLB supplemented with 20 µM taxol, 1 mg/mL casein, 71.5 mM β-mercaptoethanol, an oxygen scavenger system, and 1 mM nucleotides (Mg-ATP, Mg-ATP/NaVO4, Mg-ADP) or 2.5 units/mL apyrase for the no nucleotide condition. Nucleotides or apyrase were added to the incubated dynein samples immediately before adding to the flow chamber. Dynein was incubated for an additional 10 min in the flow chamber at room temperature to reach steady-state before imaging. For MT binding assays with ATP, dynein was imaged by taking a single-frame snapshot. For MT binding assays with other nucleotide conditions, dynein was imaged every 1 sec for a total of 5 sec. Each sample was imaged at 4 different fields of view and there were between 5 and 10 MTs in each field of view. In single-molecule assays, it is not possible to precisely control the amount of MTs that attach to the coverslip surface on different days due to variations in the surface quality of the coverslips and small variations in the concentration of each taxol-stabilized MT preparation. Therefore, in order to compare and combine replicates taken on different days without bias, the samples with and without Lis1 for each nucleotide condition were imaged in two separate flow chambers made on the same coverslip on the same day with the same stock of polymerized tubulin. The sample without Lis1 served as an internal control for normalization of the binding density (see below).
Size exclusion chromatography to determine the binding stoichiometry
2 µM DynWB-M and 4 µM Lis1 were mixed and incubated for 10 min in DLB supplemented with 200 µM Mg-ATP/NaVO4. Samples were fractionated on a Superdex 200 Increase 3.2/300 using an ÄKTAmicro system (GE Healthcare) that had been equilibrated in DLB buffer supplemented with 200 µM Mg-ATP/NaVO4. Fractions were analyzed by SDS-PAGE on 4–12% Bis-Tris gels (Invitrogen) and visualized with SYPRO Red (Invitrogen). Peak fractions were then diluted so that both DynWB-M and Lis1 were within the 50–500 ng range. Diluted samples were then re-analyzed via SDS-PAGE with an actin standard curve on the same gel to determine the absolute amount (ng) of each protein. Moles of each protein were calculated using 331,000 Da for DynWB-M and 113,800 Da for dimeric Lis1. Molar ratios were then determined. We ensured that each protein binds SYPRO Red in a linear fashion by running titrations of DynWB-M and Lis1 on SDS-PAGE gels and staining with SYPRO Red.
Cryo-EM sample preparation
Protein samples were thawed quickly and kept on ice prior to grid preparation. For Dynwt-M:Lis1, the sample was prepared using the following steps: dynein and Lis1 were incubated on ice for 10 min, after which ATP was added to a final concentration of 5 mM and the sample was incubated on ice for another 10 min. This resulted in both dynein and Lis1 at a final concentration of 0.75 µM in buffer (50 mM Tris–HCl [pH 8.0], 150 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 1 mM DTT, and 5 mM Mg-ATP).
For DynWB-M:Lis1, the sample was prepared using the following method: dynein and Lis1 were incubated on ice for 10 min, after which Mg-ATP/NaVO4 was added to a final concentration of 1.2 mM and the sample was incubated on ice for another 10 min. This resulted in both dynein and Lis1 at a final concentration of 0.75 µM in buffer (50 mM Tris–HCl [pH 8.0], 150 mM potassium acetate, 1 mM EGTA, 1 mM DTT, 1.2 mM Mg- ATP/NaVO4).
After incubation, both samples were treated identically: 4 µl of sample was applied directly to an untreated (no glow discharge or plasma cleaning) UltrAuFoil 1.2/1.3 grid (Quantifoil) in a Vitrobot (FEI Company) kept at 100% humidity and 4°C. After applying the sample, the excess liquid was immediately blotted in the Vitrobot using a blot force of 20 and a blot time of 4 sec prior to plunge-freezing into liquid ethane.
Cryo-EM data collection and image analysis
Both datasets (Dynwt-M:Lis1 and DynWB-M:Lis1) were collected on a Talos Arctica transmission electron microscope (FEI Company) operating at 200 keV with a K2 Summit direct electron detector (Gatan Inc.) (See also Table S2). Images were collected automatically using Leginon (Suloway et al., 2005) in super-resolution mode with a calibrated pixel size of 0.60 Å/pixel. The movies were then processed in the Appion pipeline (Lander et al., 2009) for all subsequent steps. Initial movie alignment and gain reference correction were performed with MotionCor (Li et al., 2013).
For the Dynwt-M:Lis1 dataset (see also Figure S2 and Table 2), 485,102 particles were picked from 5,614 micrographs using FindEM (Roseman, 2004) with templates generated from forward projections at 25 degree angular increments of EMDB 6013 (Toropova et al., 2014) (dynein motor domain without nucleotide). Using these particle coordinates, particles were extracted from micrographs that were aligned using MotionCor2 (Zheng et al., 2017) in Relion-1.4 (Scheres, 2012) using defocus values calculated by CTFFIND4 (Rohou and Grigorieff, 2015) on MotionCor2 micrographs. Micrographs were discarded if CTF confidence fits from CTFFIND4 did not go beyond 10 Å. For initial 2D classification in Relion-1.4, particles were extracted at a box size of 80 × 80 pixels and a pixel size of 4.8 Å/pixel. These particles were classified into 200 classes over 11 iterations with a mask diameter of 210 Å From the resulting averages, 347,462 particles were selected from classes that did not have contaminating ice or gold particulates. These particles were subjected to another round of 2D classification using Relion-1.4, classifying them into 200 classes over 25 iterations with a mask diameter of 190 Å From the resulting class averages (of which a subset is shown in Figure S2B), 151,470 particles (80 × 80 pixels; 4.8 Å/pixel) were selected for 3D refinement to determine a structure at 9.96 Å using gold-standard FSC=0.143, using EMDB6016 filtered to 60 Å as an initial model. A summary of the 3D refinement and classification strategy is shown in Figure S2C. This refinement and all subsequent 3D classification and refinement routines were performed using Relion-2.0beta (Kimanius et al., 2016) on Amazon Web Services using EM-packages-in-the-cloud-v4.0-GPU (Cianfrocco and Leschziner, 2015). Next, particles were re-extracted at a pixel size of 1.2 Å/pixel (324 × 324 pixels) and subjected to 3D classification without alignment, using orientations determined in the previous 3D refinement step. After classifying into 3 groups over 15 iterations, 10 more iterations of classification were performed using a local angular search range of 10 degrees. From this classification, one class was selected for further refinement (45,219 particles) to obtain a 9.48 Å structure using gold-standard FSC=0.143 because the two other classes did not have high resolution features present. With this refined 3D structure, we classified the particles into 2 groups over 15 iterations, followed by 10 iterations of local angular search ranges of 10 degrees. This resulted in one class containing high resolution features that was used for a final round of 3D refinement (25,520 particles). After this last 3D refinement step, the overall resolution was calculated to be 7.7 Å applying a B-factor of −50 Å2 after combining the half-maps and masking during post-processing (Figure S2D). However, due to the presence of flexible regions of the structure, we calculated local resolution and filtered the map using Blocres in Bsoft (Heymann and Belnap, 2007), which displayed a range of resolutions from 6 – 10 Å (Figure S2E).
For the DynWB-M:Lis1 dataset (see also Figure S3 and Table S2), 414,277 particles were picked from 4,826 micrographs using FindEM (Roseman, 2004) with templates generated from forward projections at 25 degree angular increments of EMDB 6013 (Toropova et al., 2014) (dynein motor domain without nucleotide). Using these particle coordinates, particles were extracted from micrographs that were aligned using MotionCor2 (Zheng et al., 2017) in Relion-1.4 (Scheres, 2012) using defocus values calculated by CTFFIND4 (Rohou and Grigorieff, 2015) on MotionCor2 aligned micrographs. Micrographs were discarded if CTF confidence fits from CTFFIND4 did not go beyond 10 Å Particles were extracted using a box size of 64 × 64 pixels and a pixel size of 4.8 Å/pixel. Prior to 2D classification, all particles that had gold particulates (as defined by size and pixel values) were removed from the extracted particle stack. This produced a dataset that had 223,981 particles that were subsequently classified into 250 classes using Relion-1.4 (Figure S3B). After this classification, particles that belong to homogenous classes were selected for further 3D analysis (107,273 particles). Prior to 3D analysis, particles were re-extracted at a box size of 128 × 128 pixels and a pixel size of 2.4 Å/pixel. After performing 3D classification into 3 classes (Figure S3C) using EMDB6016 as a starting model, filtered to 60 Å, we obtained a single class that could be determined to an overall resolution 10.2 Å with a B-factor of −800 Å2 using Relion-1.4 (Figure S3D). Local resolution assessment using Blocres in Bsoft (Heymann and Belnap, 2007) displayed a range of resolutions from 9 – 13 Å (Figure S3E), however the map was filtered to a single value of 10.2 Å with a B-factor of −800 Å2 using Relion-1.4.
All figures were generated using UCSF Chimera.
Model building using Rosetta
The initial model for dynein was generated based on homology detection to the S. cerevisiae dyn1 sequence using Hidden Markov Model as implemented in HHpred (https://toolkit.tuebingen.mpg.de/hhpred), using the top 3 scoring homologous models (PDB: 4RH7 (Schmidt et al., 2014), 3VKG (Kon et al., 2012), and 4AKG (Schmidt et al., 2012)). This model was split into three parts: 1) Linker -> AAA4(Stalk CC1), 2) AAA4(Stalk CC2) -> AAA5 Large, 3) AAA5S -> C-terminus, and each was refined using Rosetta with cryo-EM maps Dynwt-M:Lis1 and DynWB-M:Lis1 (Wang et al., 2016). In each case 200 models were generated, and the RMSD values for the top five models are shown in Figures S2H and S3H, with most RMSD values being <1 Å for Cα backbone atoms. Finally, the top-scoring model from each part was combined for a final refinement to ensure inter-domain contacts were satisfied.
Cα distance calculations
For Cα distance measurements and comparisons, atomic coordinates were used for only large and small AAA domains in the dynein ring. PDB coordinates from the previously published structure of human dynein-2 with ADP.Vi (PDB 4RH7) (Schmidt et al., 2014) were used to create a homology model using the sequence from S. cerevisiae with SWISS-MODEL (Biasini et al., 2014). Using these coordinates, distances between Cα atoms were calculated and displayed using UCSF Chimera (Pettersen et al., 2004). For each distance measurement the lines shown represent the distance between the Cα atoms, and the thickness of the linearly scaled with the distance between atoms. Program is available upon request to the authors.
Single-molecule motility assay on dynamic microtubules
Single-molecule motility assays on dynamic MTs were performed using the TIRF microscopy set up described above. Flow-chambers were prepared as described above using biotin-PEG-functionalized coverslips (Microsurfaces). Brightly-labeled, biotinylated and GMPCPP-stabilized MT seeds were prepared as described (Roberts et al., 2014). Flow chambers were incubated sequentially with the following solutions, interspersed with two washes with assay buffer (BRB80 [80 mM PIPES-KOH pH 6.8, 1 mM magnesium chloride, 1 mM EGTA], 0.5 mg/mL casein and 1 mM DTT): (1) 0.8% pluronic F-127 and 5 mg/mL casein in water (6 min incubation); (2) 0.5 mg/mL streptavidin in BRB80 (3 min incubation); (3) a fresh dilution of MT seeds in assay buffer (3 min incubation); and (4) the final imaging solution containing 2.5–5 pM dynein-TMR, 1 nM Kip2, 5 nM Bim1, 50 nM Bik1, 25 nM Lis1, 15 µM tubulin (~7.5% Alexa488 labeled and ~92.5% unlabeled), 1 mM Mg-ATP, 1 mM Mg-GTP, 0.1% methylcellulose, 71.5 mM β-mercaptoethanol and an oxygen scavenger system in assay buffer. Two-color sequential TIRF movies of MTs and dynein were imaged every 3 sec for a total of 10 min. For each frame, Alexa488-tubulin and TMR-dynein were exposed for 100 ms with the 488 nm laser and for 200 ms with the 561 nm laser, respectively.
Microtubule recruitment assay of weak microtubule binding dynein variants
MT recruitment assay was performed using the TIRF microscopy set up described above. Flow chambers were incubated with the following solutions, with two washes of assay buffer (BRB80, 20µM taxol, 0.5 mg/mL casein and 1 mM DTT) in between: (1) 0.5 mg/mL streptavidin in BRB80 (3 min incubation); (2) a fresh dilution of taxol-stabilized Alexa488-labeled MTs in assay buffer (3 min incubation); and (3) the final imaging solution containing 100pM Dynweak-variant-TMR, 2 nM Kip2, 5 nM Bim1, 50 nM Bik1, 25 nM Lis1, 71.5 mM β-mercaptoethanol and an oxygen scavenger system in assay buffer. The weak MT binding dynein variants contains the following additional mutations: K3166A, K3117A, E3122A, R3124A (Redwine et al., 2012). The final solution containing dynein was incubated for an additional 5 min in the flow chamber at room temperature to reach steady-state before imaging. Dynein was imaged every 1 sec for a total of 5 sec. Each sample was imaged at 4 different fields of view.
Lis1 affinity capture to determine Lis1-dynein binding affinities
Sixteen µL of magnetic SNAP-Capture beads (NEB) were incubated with increasing concentrations of SNAP-Lis1 (0–600 nM) in modified TEV buffer for 1 hour at room temperature with agitation. The supernatant was removed, the beads were washed with 1 ml of modified TEV buffer followed by 1 ml of TEV buffer supplemented with 1 mM DTT, 0.1% NP40, 2 mM MgCl2, 1 mM ATP, and 1 mM NaVO4. 20 nM Dyn(variant)-M was incubated with the beads conjugated to Lis1 for 30 min at room temperature with agitation. The supernatant was removed, ran on a 4–12% Bis Tris gel, and stained with Sypro Red (Thermo Fisher) to visualize the fraction of dynein depleted.
Sequence alignment
Protein sequences of dynein were obtained from UniProt. Sequence alignments were performed with Clustal Omega web services (http://www.ebi.ac.uk/Tools/msa/clustalo/) and annotated using Jalview (http://www.jalview.org/).
Yeast in vivo dynein localization assay
To quantify dynein localization during mitosis, yeast strains containing 3xGFP-labeled dynein and tdTomato-labeled SPB marker, SPC110, were used. Overnight cultures grown from a single colony were diluted to OD600 of 0.1 in 10 mL YPD. Diluted cultures were then grown for 2–3 hours with rotation at 30°C to reach mid-log phase (OD600 0.6–0.8). 100–200 µL of mid-log phase culture were spun down using a table-top centrifuge, the media was discarded and the cells were resuspended in 6 µL of phosphate-saline buffer with calcium and magnesium. The resupended cells were then added to freshly-made synthetic complete media agarose pad on glass slide. No. 1–1/2 coverslips (Corning) were placed on top of the sample and sealed with nail polish. Imaging was performed with a spinning-disk confocal (Yokogawa, CSU10) inverted microscope (Nikon, Eclipse TE2000-E) equipped with a 100× 1.40 N.A. oil-immersion objective (Nikon, Plano Apo), and an electron multiplying CCD (Andor Technology, iXon DV887). The excitation and emission paths were filtered using appropriate single bandpass filter cubes (Chroma). Images were collected for 15 × 500 nm Z-sections (7.5 µm total Z stack). For each Z section, a bright field image, a 3xGFP-labeled dynein image via 488 nm laser excitation, and a tdTomato-labeled SPB image via 568 nm laser excitation were collected. Illumination and image acquisition is controlled by iQ2.6 imaging software (Andor Technology).
QUANTIFICATION AND STATISTICAL ANALYSIS
Single-molecule motility assay on taxol-stabilized or dynamic microtubules
Dynein velocity was calculated from kymographs generated using an ImageJ macro as described (Roberts et al., 2014). In the assay with dynamic MTs, MT plus ends were assigned as the ends with faster growth rate. The directionality of individual dynein runs was subsequently assigned as a plus-end run if it moved toward MT plus end or a minus-end run if it moved in the opposite directions. Each pixel corresponds to 157 nm in our single-molecule assays. Only runs that lasted at least 4 frames were included in the analysis. Bright aggregates, which were less than 5% of the population, were excluded from the analysis. Statistical analyses for velocities were done using unpaired t-test with Welch’s correction in Prism7 (GraphPad). Statistical comparisons of the percentage of plus-end moving events, minus-end moving events or non-motile events were performed using Mann-Whitney test in Prism7. Exact value of n and evaluation of statistical significance are described in the corresponding figure legends.
Single-molecule microtubule binding assay
Dynein binding density on MTs and recruitment to MTs by Kip2 was calculated using ImageJ. A minimum projection of 5 movie frames of dynein was generated to minimize counting non-specific binding events. In the MT decoration assay with ATP, a single-frame snapshot of dynein was used due to dynein motility in the presence of ATP. Intensity profiles of dynein spots were generated over a 5-pixel wide line drawn perpendicular to the long axis of the MT. Intensity peaks at least 2-fold higher than the background intensity were counted as dynein spots bound to MTs. Bright aggregates that were 5-fold brighter than neighboring intensity peaks were not counted as dynein spots. The total number of dynein spots was divided by the total MT length in each field of view to calculate the binding density. Normalized binding density was calculated by dividing by the average binding density of dynein without Lis1 collected on the same coverslip (see above) in each nucleotide condition. Statistical significance was determined using an unpaired t-test with Welch’s correction in Prism7. The exact value of n and evaluation of statistical significance are described in the corresponding figure legends.
Yeast in vivo dynein localization assay
Only yeast cells with large buds and two SPB foci were included in the analysis. Maximum intensity projections of the GFP-Dynein and tdTomato-SPC110 channels were generated. Dynein and SPB foci were identified using the ImageJ plugin, Find Maxima, and a maxima cutoff of at least 1.5-fold higher than the neighboring background pixel intensity value. Dynein foci were separated into two categories: localized at SPB and not localized at SPB. The latter category includes cortical and MT plus-end-localized dynein. We cannot differentiate between these two populations because MTs were not imaged (or labeled in our strains). Statistical comparisons of the average number of dynein foci per cell were done using a Mann-Whitney test in Prism7. The exact value of n and evaluation of statistical significance are described in the corresponding figure legends.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| cOmplete EDTA-free protease inhibitor cocktail tablet | Roche | 11873580001 |
| IgG sepharose 6 fast flow beads | GE healthcare life sciences | 17-0969-01 |
| Ni-NTA agarose | Qiagen | 30210 |
| Strep-Tactin agarose | IBA Life Sciences | 2-1201-010 |
| HaloTag TMR ligand | Promega | G8251 |
| SNAP-Capture magnetic beads | NEB | S9145S |
| NuPage 4–12% Bis Tris Protein gel | Thermo Fisher Scientific | NP0321 |
| Deposited Data | ||
| Coordinates of Dynwt-M:Lis1 | This study | wwPDB: D_1000227341 |
| Coordinates of DynWB-M:Lis1 | This study | wwPDB: D_1000227343 |
| Cryo-EM map of Dynwt-M:Lis1 | This study | wwPDB: D_1000227341 |
| Cryo-EM map of DynWB-M:Lis1 | This study | wwPDB: D_1000227343 |
| Experimental Models: Organisms/Strains | ||
| S. cerevisiae strains used in this study | This study | Supplemental Table S3 |
| Recombinant DNA | ||
| pKL-His8-ZZ-Tev-Bik1 | (Roberts et al., 2014) | N/A |
| pDEST17-StrepII-gsTEV-Bim1 | (Roberts et al., 2014) | N/A |
| Software and Algorithms | ||
| Leginon | (Suloway et al., 2005) | leginon.org |
| Appion | (Lander et al., 2009) | appion.org |
| MotionCorr | (Li et al., 2013) | http://cryoem.ucsf.edu/software/driftcorr.html |
| MotionCor2 | (Zheng et al., 2017) | http://msg.ucsf.edu/em/software/motioncor2.html |
| FindEM | (Roseman, 2004) | N/A |
| CTFFIND4 | (Rohou and Grigorieff, 2015) | http://grigoriefflab.janelia.org/ctffind4 |
| RELION-1.4 | (Scheres, 2012) | http://www2.mrc-lmb.cam.ac.uk/relion |
| RELION-2.0 | (Kimanius et al., 2016) | http://www2.mrc-lmb.cam.ac.uk/relion |
| EM-Packages-in-the-cloud-v3.96 | (Cianfrocco and Leschziner, 2015) | https://sites.google.com/site/emcloudprocessing/ |
| EM-Packages-in-the-cloud-v4.0-GPU | (Cianfrocco and Leschziner, 2015) | https://sites.google.com/site/emcloudprocessing/ |
| Rosetta | (Wang et al., 2016) | https://www.rosettacommons.org/ |
| Bsoft (Blocres) | (Heymann and Belnap, 2007) | https://lsbr.niams.nih.gov/bsoft/programs/blocres.html |
| UCSF Chimera | (Pettersen et al., 2004) | https://www.cgl.ucsf.edu/chimera |
| ImageJ | https://imagej.nih.gov/ij/ | N/A |
| Jalview | http://www.jalview.org/ | N/A |
| GraphPad Prism7 | GraphPad Software | N/A |
| Other | ||
| R1.2/1.3 300 Mesh UltAuFoil holey carbon grids | Quantifoil | N/A |
| Biotin-PEG-functionalized coverslips | Microsurfaces | Bio-02 |
Highlights.
Lis1 has two modes of regulating dynein, set by the nucleotide state of AAA3
Lis1 can induce either tight or weak microtubule-binding states in dynein
The states are set by the number (one or two) of Lis1 β-propellers bound to dynein
The binding site for the second β-propeller is required for Lis1’s function in vivo
Acknowledgments
We thank E. Kellog (UC Berkeley) for the distance comparison script, the UCSD Cryo-EM facility, A. Desai for access to a spinning disk microscope, the UCSD Physics IT group, and W. Lee (U Mass Amherst) for yeast strains. We used the Extreme Science and Engineering Discovery Environment (XSEDE) for computing allocations (MCB160079 to ZMH and MCB140257 to AEL), supported by NSF grant ACI-1548562. MED is supported by the Jane Coffin Childs Memorial Fund, MAC by the Damon Runyon Cancer Research Foundation, ZMH by a NSF graduate research fellowship, SRP and AEL by NIH grant R01GM107214, and SRP by HHMI and the Simons Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
MED, MAC, ZMH, SLRP and AEL designed the experiments; MED, MAC, ZMH, and PTT performed the experiments; MED, MAC, ZMH, SLRP and AEL interpreted the data; and MED, MAC, ZMH, SLRP and AEL wrote the manuscript.
Accession Numbers
Cryo-EM maps and Rosetta models for the structures of Dynwt:Lis1 (PDB ID: 5VH9, EMDB: EMD-8673) and DynWB:Lis1 (PDB ID: 5VLJ, EMDB: EMD-8706) have been deposited in the wwPDB.
References
- Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J. Cell Biol. 2000;149:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbach J, Murthy A, McClintock MA, Dix CI, Zalyte R, Hoang HT, Bullock SL. Lissencephaly-1 is a context-dependent regulator of the human dynein complex. eLife. 2017;6 doi: 10.7554/eLife.21768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhabha G, Cheng H-C, Zhang N, Moeller A, Liao M, Speir JA, Cheng Y, Vale RD. Allosteric communication in the Dynein motor domain. Cell. 2014;159:857–868. doi: 10.1016/j.cell.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Dynein structure and power stroke. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- Carvalho P, Gupta ML, Jr, Hoyt MA, Pellman D. Cell Cycle Control of Kinesin-Mediated Transport of Bik1 (CLIP-170) Regulates Microtubule Stability and Dynein Activation. Dev. Cell. 2004;6:815–829. doi: 10.1016/j.devcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Caudron F, Andrieux A, Job D, Boscheron C. A new role for kinesin-directed transport of Bik1p (CLIP-170) in Saccharomyces cerevisiae. J. Cell. Sci. 2008;121:1506–1513. doi: 10.1242/jcs.023374. [DOI] [PubMed] [Google Scholar]
- Cho C, Reck-Peterson SL, Vale RD. Regulatory ATPase Sites of Cytoplasmic Dynein Affect Processivity and Force Generation. Journal of Biological Chemistry. 2008;283:25839–25845. doi: 10.1074/jbc.M802951200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfrocco MA, Leschziner AE. Low cost, high performance processing of single particle cryo-electron microscopy data in the cloud. eLife. 2015;4 doi: 10.7554/eLife.06664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfrocco MA, Desantis ME, Leschziner AE, Reck-Peterson SL. Mechanism and regulation of cytoplasmic dynein. Annu. Rev. Cell Dev. Biol. 2015;31:83–108. doi: 10.1146/annurev-cellbio-100814-125438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt MA, Chang AY, Combs PA, Yildiz A. Cytoplasmic Dynein Moves Through Uncoordinated Stepping of the AAA+ Ring Domains. Science. 2012;335:221–225. doi: 10.1126/science.1215804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt MA, Cypranowska CA, Cleary FB, Belyy V, Yildiz A. The AAA3 domain of cytoplasmic dynein acts as a switch to facilitate microtubule release. Nature Structural & Molecular Biology. 2014;22:73–80. doi: 10.1038/nsmb.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix CI, Soundararajan HC, Dzhindzhev NS, Begum F, Suter B, Ohkura H, Stephens E, Bullock SL. Lissencephaly-1 promotes the recruitment of dynein and dynactin to transported mRNAs. J. Cell Biol. 2013;202:479–494. doi: 10.1083/jcb.201211052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MJ, Tan K, Reck-Peterson SL. Lis1 is an initiation factor for dynein-driven organelle transport. J. Cell Biol. 2012;197:971–982. doi: 10.1083/jcb.201112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel D, Urrestarazu LA, Vissers S, Jauniaux JC, van Vliet-Reedijk JC, Planta RJ, Gibbons IR. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc. Natl. Acad. Sci. U.S.a. 1993;90:11172–11176. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franker MAM, Hoogenraad CC. Microtubule-based transport - basic mechanisms, traffic rules and role in neurological pathogenesis. J. Cell. Sci. 2013;126:2319–2329. doi: 10.1242/jcs.115030. [DOI] [PubMed] [Google Scholar]
- Gleave ES, Schmidt H, Carter AP. A structural analysis of the AAA+ domains in Saccharomyces cerevisiae cytoplasmic dynein. J. Struct. Biol. 2014;186:367–375. doi: 10.1016/j.jsb.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez PA, Ackermann BE, Vershinin M, McKenney RJ. Differential effects of the dynein-regulatory factor Lissencephaly-1 on processive dynein-dynactin motility. Journal of Biological Chemistry. 2017 doi: 10.1074/jbc.M117.790048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann JB, Belnap DM. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Huang J, Roberts AJ, Leschziner AE, Reck-Peterson SL. Lis1 Acts as a “‘Clutch’” between the ATPase and Microtubule-Binding Domains of the Dynein Motor. Cell. 2012;150:975–986. doi: 10.1016/j.cell.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimanius D, Forsberg BO, Scheres SH, Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife. 2016;5 doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinman E, Holzbaur ELF. Stress-Induced CDK5 Activation Disrupts Axonal Transport via Lis1/Ndel1/Dynein. Cell Reports. 2015;12:462–473. doi: 10.1016/j.celrep.2015.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T, Nishiura M, Ohkura R, Toyoshima YY, Sutoh K. Distinct functions of nucleotide-binding/hydrolysis sites in the four AAA modules of cytoplasmic dynein. Biochemistry. 2004;43:11266–11274. doi: 10.1021/bi048985a. [DOI] [PubMed] [Google Scholar]
- Kon T, Oyama T, Shimo-Kon R, Imamula K, Shima T, Sutoh K, Kurisu G. The 2.8Å crystal structure of the dynein motor domain. Nature. 2012;484:345–350. doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau P-W, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W-L, Oberle JR, Cooper JA. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J. Cell Biol. 2003;160:355–364. doi: 10.1083/jcb.200209022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz JH, Schuchardt I, Straube A, Steinberg G. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. Embo J. 2006;25:2275–2286. doi: 10.1038/sj.emboj.7601119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lee W-L, Cooper JA. NudEL targets dynein to microtubule ends through LIS1. Nat. Cell Biol. 2005;7:686–690. doi: 10.1038/ncb1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc. Natl. Acad. Sci. U.S.a. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus SM, Lee W-L. Regulated offloading of cytoplasmic dynein from microtubule plus ends to the cortex. Dev. Cell. 2011;20:639–651. doi: 10.1016/j.devcel.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus SM, Plevock KM, St Germain BJ, Punch JJ, Meaden CW, Lee W-L. Quantitative analysis of Pac1/LIS1-mediated dynein targeting: Implications for regulation of dynein activity in budding yeast. Cytoskeleton (Hoboken) 2011;68:157–174. doi: 10.1002/cm.20502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus SM, Punch JJ, Lee W-L. Motor- and tail-dependent targeting of dynein to microtubule plus ends and the cell cortex. Curr. Biol. 2009;19:196–205. doi: 10.1016/j.cub.2008.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 2010;141:304–314. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogami T, Kon T, Ito K, Sutoh K. Kinetic characterization of tail swing steps in the ATPase cycle of Dictyostelium cytoplasmic dynein. J. Biol. Chem. 2007;282:21639–21644. doi: 10.1074/jbc.M701914200. [DOI] [PubMed] [Google Scholar]
- Moughamian AJ, Osborn GE, Lazarus JE, Maday S, Holzbaur ELF. Ordered recruitment of dynactin to the microtubule plus-end is required for efficient initiation of retrograde axonal transport. J. Neurosci. 2013;33:13190–13203. doi: 10.1523/JNEUROSCI.0935-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas MP, Berger F, Rao L, Brenner S, Cho C, Gennerich A. Cytoplasmic dynein regulates its attachment to microtubules via nucleotide state-switched mechanosensing at multiple AAA domains. Proc. Natl. Acad. Sci. U.S.a. 2015;112:6371–6376. doi: 10.1073/pnas.1417422112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey JP, Smith DS. A Cdk5-Dependent Switch Regulates Lis1/Ndel1/Dynein-Driven Organelle Transport in Adult Axons. J. Neurosci. 2011;31:17207–17219. doi: 10.1523/JNEUROSCI.4108-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy BJN, Mattson M, Wynne CL, Vadpey O, Durra A, Chapman D, Vallee RB, Gross SP. Load-induced enhancement of Dynein force production by LIS1-NudE in vivo and in vitro. Nat Commun. 2016;7:12259. doi: 10.1038/ncomms12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redwine WB, Hernández-López R, Zou S, Huang J, Reck-Peterson SL, Leschziner AE. Structural basis for microtubule binding and release by dynein. Science. 2012;337:1532–1536. doi: 10.1126/science.1224151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- Roberts AJ, Goodman BS, Reck-Peterson SL. Reconstitution of dynein transport to the microtubule plus end by kinesin. eLife. 2014;3:e02641. doi: 10.7554/eLife.02641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman AM. FindEM--a fast, efficient program for automatic selection of particles from electron micrographs. J. Struct. Biol. 2004;145:91–99. doi: 10.1016/j.jsb.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Gleave ES, Carter AP. Insights into dynein motor domain function from a 3.3-Å crystal structure. Nature Structural & Molecular Biology. 2012;19:492–497. doi: 10.1038/nsmb.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Zalyte R, Urnavicius L, Carter AP. Structure of human cytoplasmic dynein-2 primed for its power stroke. Nature. 2014;518:435–438. doi: 10.1038/nature14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C-Y, Zhu J, Xie Y-J, Wang Z, Wang Y-N, Wang Y, Su L-D, Zhou L, Zhou T-H, Shen Y. Distinct functions of nuclear distribution proteins LIS1, Ndel1 and NudCL in regulating axonal mitochondrial transport. Traffic. 2013;14:785–797. doi: 10.1111/tra.12070. [DOI] [PubMed] [Google Scholar]
- Sheeman B, Carvalho P, Sagot I, Geiser J, Kho D, Hoyt MA, Pellman D. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr. Biol. 2003;13:364–372. doi: 10.1016/s0960-9822(03)00013-7. [DOI] [PubMed] [Google Scholar]
- Smith DS, Niethammer M, Ayala R, Zhou Y, Gambello MJ, Wynshaw-Boris A, Tsai LH. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat. Cell Biol. 2000;2:767–775. doi: 10.1038/35041000. [DOI] [PubMed] [Google Scholar]
- Splinter D, Razafsky DS, Schlager MA, Serra-Marques A, Grigoriev I, Demmers J, Keijzer N, Jiang K, Poser I, Hyman AA, et al. BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Mol. Biol. Cell. 2012;23:4226–4241. doi: 10.1091/mbc.E12-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tarricone C, Perrina F, Monzani S, Massimiliano L, Kim M-H, Derewenda ZS, Knapp S, Tsai L-H, Musacchio A. Coupling PAF signaling to dynein regulation: structure of LIS1 in complex with PAF-acetylhydrolase. Neuron. 2004;44:809–821. doi: 10.1016/j.neuron.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Toropova K, Zou S, Roberts AJ, Redwine WB, Goodman BS, Reck-Peterson SL, Leschziner AE, Kuriyan J. Lis1 regulates dynein by sterically blocking its mechanochemical cycle. eLife. 2014;3:e03372. doi: 10.7554/eLife.03372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twelvetrees AE, Pernigo S, Sanger A, Guedes-Dias P, Schiavo G, Steiner RA, Dodding MP, Holzbaur ELF. The Dynamic Localization of Cytoplasmic Dynein in Neurons Is Driven by Kinesin-1. Neuron. 2016;90:1000–1015. doi: 10.1016/j.neuron.2016.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnoni A, Hoffmann PC, Bullock SL. Reducing Lissencephaly-1 levels augments mitochondrial transport and has a protective effect in adult Drosophila neurons. J. Cell. Sci. 2016;129:178–190. doi: 10.1242/jcs.179184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RY-R, Song Y, Barad BA, Cheng Y, Fraser JS, DiMaio F. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. eLife. 2016;5 doi: 10.7554/eLife.17219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Toba S, Yoshida Y, Haratani K, Mori D, Yano Y, Mimori-Kiyosue Y, Nakamura T, Itoh K, Fushiki S, et al. LIS1 and NDEL1 coordinate the plus-end-directed transport of cytoplasmic dynein. Embo J. 2008;27:2471–2483. doi: 10.1038/emboj.2008.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JY, Ori-McKenney KM, McKenney RJ, Vershinin M, Gross SP, Vallee RB. High-resolution imaging reveals indirect coordination of opposite motors and a role for LIS1 in high-load axonal transport. J. Cell Biol. 2011;195:193–201. doi: 10.1083/jcb.201104076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhuang L, Lee Y, Abenza JF, Peñalva MA, Xiang X. The microtubule plus-end localization of Aspergillus dynein is important for dynein-early-endosome interaction but not for dynein ATPase activation. J. Cell. Sci. 2010;123:3596–3604. doi: 10.1242/jcs.075259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.