Significance
Bacterial protein synthesis requires the assembly of the 30S and 50S ribosomal subunits on mRNA to form the translationally competent 70S complex. Nontranslating 70S ribosomes homodimerize into a translationally inactive 100S ribosome to promote bacterial survival. The conversion between the 70S and 100S complexes is reversible, but the disassembly pathway of the 100S ribosome is unknown. We show that an evolutionarily conserved GTPase (HflX) splits the Staphylococcus aureus 100S ribosome in a GTP-dependent fashion; the abundance of 100S is influenced by the opposing functions of the dimerizing factor (HPF) and HflX, whose actions are coregulated by the heat stress response and (p)ppGpp-mediated stringent control. These findings provide insights into the crosstalk between the 100S ribosome biogenesis and stress response.
Keywords: ribosome, GTPase, HPF, HflX, stress response
Abstract
The bacterial hibernating 100S ribosome is a poorly understood form of the dimeric 70S particle that has been linked to pathogenesis, translational repression, starvation responses, and ribosome turnover. In the opportunistic pathogen Staphylococcus aureus and most other bacteria, hibernation-promoting factor (HPF) homodimerizes the 70S ribosomes to form a translationally silent 100S complex. Conversely, the 100S ribosomes dissociate into subunits and are presumably recycled for new rounds of translation. The regulation and disassembly of the 100S ribosome are largely unknown because the temporal abundance of the 100S ribosome varies considerably among different bacterial phyla. Here, we identify a universally conserved GTPase (HflX) as a bona fide dissociation factor of the S. aureus 100S ribosome. The expression levels hpf and hflX are coregulated by general stress and stringent responses in a temperature-dependent manner. While all tested guanosine analogs stimulate the splitting activity of HflX on the 70S ribosome, only GTP can completely dissociate the 100S ribosome. Our results reveal the antagonistic relationship of HPF and HflX and uncover the key regulators of 70S and 100S ribosome homeostasis that are intimately associated with bacterial survival.
The biogenesis and function of bacterial 30S and 50S ribosomal subunits and the 70S complex have been studied extensively, but the significance of the 100S ribosome (homodimeric 70S) has only begun to emerge in recent years (1). The 100S ribosome is ubiquitously found in all bacterial phyla and is important for bacterial survival during nutrient limitation (2–6), antibiotic stress (7), host colonization (8), dark adaptation (9), and biofilm formation (10, 11). A common feature of these biological processes is that cells generally conserve energy by undergoing metabolic and translational dormancy because protein synthesis accounts for >50% of energy costs (12, 13). The dimerization of 70S ribosomes has been shown to down-regulate translational efficiency in vivo (3) and in vitro (3, 14), and bacteria lacking 100S ribosomes are prone to early cell death concomitant with rapid ribosome degradation (3, 10, 15, 16). These studies lead to a model whereby the formation of the 100S complex sequesters the ribosome pool away from active translation, and 70S self-dimerization prevents ribosome degradation by an unknown pathway (3, 17). During the stationary phase, the 100S ribosomes are presumably dissociated and reused for new cycles of translation, thereby maintaining cell viability (1, 3, 16, 18). The process and dissociation factors involved in the reversible transition of silent 100S to a translationally competent 70S ribosome remain poorly understood.
By contrast, the 70S dimerizing factor has been characterized in many bacterial species (1, 2, 4, 14). In Firmicutes (such as Staphylococcus aureus and Bacillus subtilis), a single long form of hibernation-promoting factor (HPF) provides the binding platform to conjoin the 30S subunits of the two 70S monomers via a direct interaction between the C-terminal domains (CTDs) in the HPF dimer, each of which is tethered on an individual 70S (Fig. 1). In addition, the N-terminal domain (NTD) of HPF inhibits translation by sterically occluding the mRNA and the anti-codon tRNA binding sites (18–20). During HPF-induced dimerization, a 30S head rotation of the S. aureus 100S ribosome stabilizes the dimerization interface consisting of the rRNA h26, and h40 and the ribosomal protein uS2 (19). This 30S swiveling was not observed in the B. subtilis 30S–70S subcomplex (18).
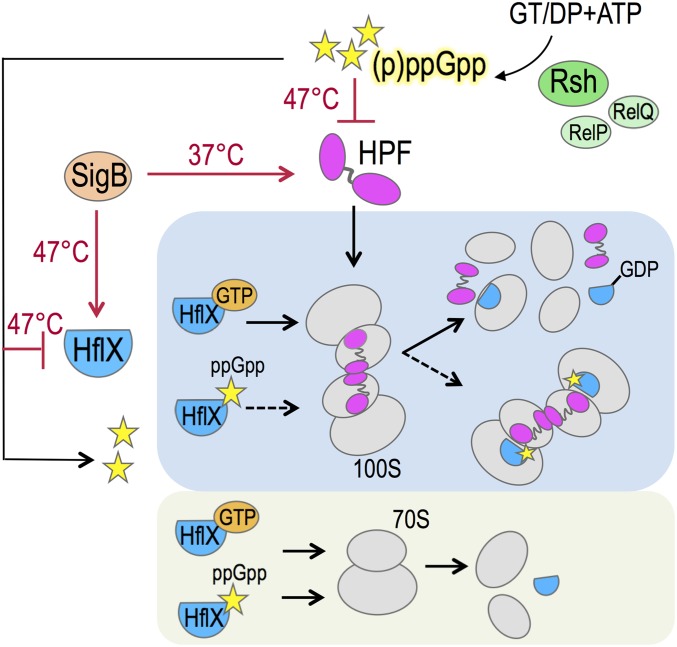
Fig. 1.
A model summarizing the coregulation and opposing roles of HPF and HflX. The stringent response alarmone (p)ppGpp in S. aureus is synthesized from the substrates GT(D)P and ATP primarily by the Rsh (RelA/SpoT homolog) enzyme and, to a lesser extent, by two alternative synthetases, RelP and RelQ (55). The N-terminal domain of HPF binds to the decoding center of the 30S subunit and inhibits translation, whereas the C-terminal domain (CTD) tethers the two 70S monomers via direct interaction of the HPF-CTD dimer to form the 100S complex (19). The production of (p)ppGpp strongly inhibits the synthesis of hpf and hflX under heat stress. ppGpp also binds to HflX. HflX⋅ppGpp is unable to split the 100S complex but is sufficient for 70S dissociation. HflX binds to the peptidyltransferase center in the 50S subunit and stimulates subunit dissociation by disrupting intersubunit bridges (46). The effective stoichiometry of HflX⋅GTP-100S remains to be determined. GTP hydrolysis presumably promotes the release of HPF and HflX simultaneously with 100S breakdown, possibly by way of a 70S intermediate. The general stress response sigma-factor B (SigB) activates the expression of hpf at 37 °C and moderately up-regulates the HflX level at 47 °C. Red arrows indicate a positive regulatory role, bar-headed lines denote repression, and a dashed arrow indicates a loss of action.
The dimerization mechanism of the 100S ribosome in γ-proteobacterial Escherichia coli is distinct from that in S. aureus and B. subtilis. In addition to a short-form HPF analogous to the NTD of long-form HPF, E. coli 70S dimerization requires the cooperative action of the ribosome modulator factor (RMF) (21–24). Rather than a “side-to-side” orientation of the 70S dimer in S. aureus and B. subtlis, E. coli 70S dimerization involves a “head-to-head” configuration (25, 26). The X-ray crystal structure of E. coli HPF and RMF in complex with the heterologous Thermus thermophilus 70S ribosome has shown that the short-form HPF occupies the decoding sites, overlapping with tRNA and mRNA binding, whereas RMF binding stimulates 30S movement to form an expanded interaction pocket comprising the rRNA h39 and the ribosomal proteins uS2, uS3, and uS5 without direct contact between the RMF molecules from each 70S (27). E. coli also possesses an HPF paralog (referred to as YfiA or pY or RaiA) that silences the 70S ribosome (27–29) and antagonizes the formation of the 100S complex (21, 23). YfiA is absent from Firmicutes and the majority of bacteria but is functionally homologous to PSRP1 in plant chloroplasts (30, 31).
The two 70S monomers in the 100S complex are not irreversibly interlocked; instead, the disassembly of the E. coli 100S ribosome can occur within 1 min after transferring the aged culture into fresh medium (16, 32). The 100S ribosomes connected by the long-form HPFs in Firmicutes are more stable by one order of magnitude than the ones generated by the short-form HPF and RMF (2, 14). Furthermore, the parallel beta-sheet interactions between the two 70S ribosomes are not rigid (19), and disrupting the flexible loop linking the CTD and NTD impairs dimerization (18). These findings suggest that the dissociation of 100S ribosome involves an active mechanism to dislodge the dimerizing factors from the ribosome. Although factors such as initiation factor 3 (IF3) and ribosome recycling factor (RRF)/elongation factor G (EF-G) have been implicated in counteracting the formation of hibernating 100S and PSRP1-mediated 70S ribosomes in vitro (30, 33), the proposed activity of these factors on hibernating ribosomes does not corroborate with their canonical roles in ribosome recycling (34, 35) (Discussion, and see Fig. S7). The 100S disassembly factor and the exact order of HPF release and 100S splitting have yet to be identified. Furthermore, the Firmicutes 100S ribosomes are constitutively produced from the lag-logarithmic phase through the stationary phase (3, 14, 36), whereas 100S ribosomes are formed only in cyanobacteria and γ-proteobacteria (including E. coli) during darkness and in the stationary phase (1, 9). The molecular basis underpinning the variations in temporal abundance of the 100S ribosome in different bacterial systems has remained elusive.
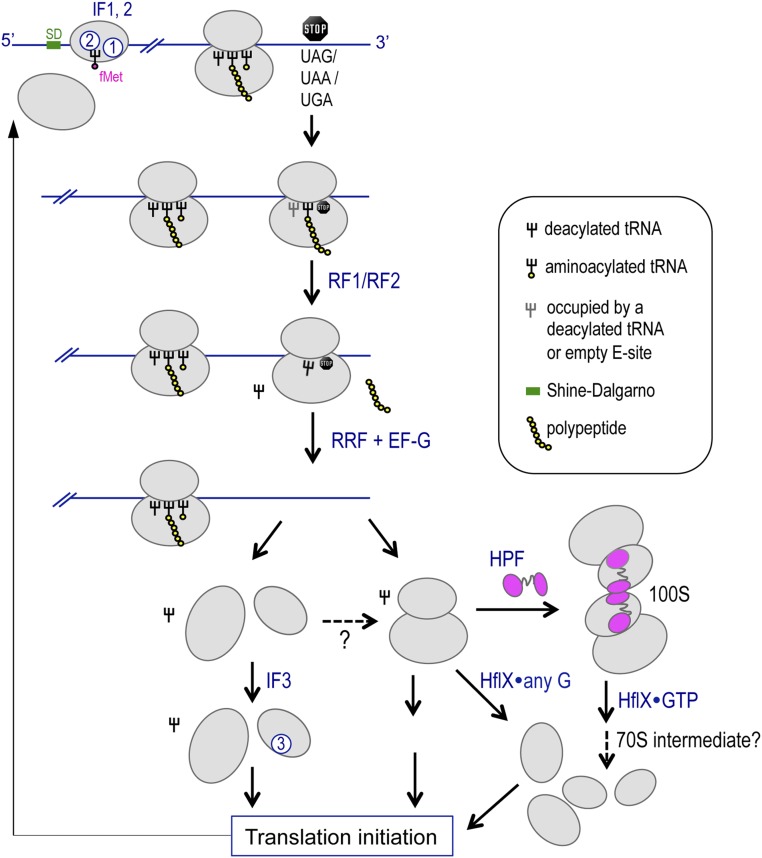
Fig. S7.
A simplified model depicting possible relationships of RRF, IF3, and HflX during ribosome recycling. The initiation of bacterial protein synthesis involves the recruitment of initiation factor 2 (IF2), IF3, mRNA, and the initiator fMet-tRNAfMet to the 30S subunit. IF2 and IF3 eventually dissociate, and the 50S subunit binds to form the initiation complex with the initiator tRNA position at the P site. Subsequent elongation steps are facilitated by the actions of EF-Tu, GTP, and aminoacylated tRNAs with the aid of EF-G to promote translocation. When a termination codon enters the decoding center, it is recognized by the cognate release factor 1 (RF1) or RF2. RF1 and RF2 catalyze the hydrolysis of the peptidyl–tRNA bond, thus liberating the newly synthesized protein. RF3 promotes the dissociation of RF1 and RF2 from the ribosome. During the recycling phase, ribosome recycling factor (RRF) and GTPase EF-G concertedly stimulate the dissociation of the 70S ribosome from the mRNA, followed by tRNA release and in most cases dissociation of the 30S and 50S subunits (35). The binding of IF3 to the 30S subunit prevents 30S–50S rejoining. Without IF3, the subunits may reassociate to form the silent 70S complex. HflX disassembles the 70S complexes released from mRNA and from subunit rejoining in the presence of guanosine binding. HPF dimerizes the 70S complexes into translationally inactive 100S ribosomes. Under specific conditions, GTP-bound HflX dissociates the 100S ribosome into subunits with the possibility of a 70S intermediate during disassembly and with the dislodging of HPF and HflX upon GTP hydrolysis. The free subunits can be recycled for new rounds of translation.
In this study, we demonstrate how the S. aureus 100S ribosome is disassembled by GTPase HflX and how the abundance of the 100S ribosome is regulated via general stress and stringent response pathways that modulate the expression and activity of HflX and HPF (Fig. 1). HflX homologs are conserved from bacteria to humans (37) (Fig. S1); thus, our data may offer general principles of HflX function in ribosome metabolism and translational control.
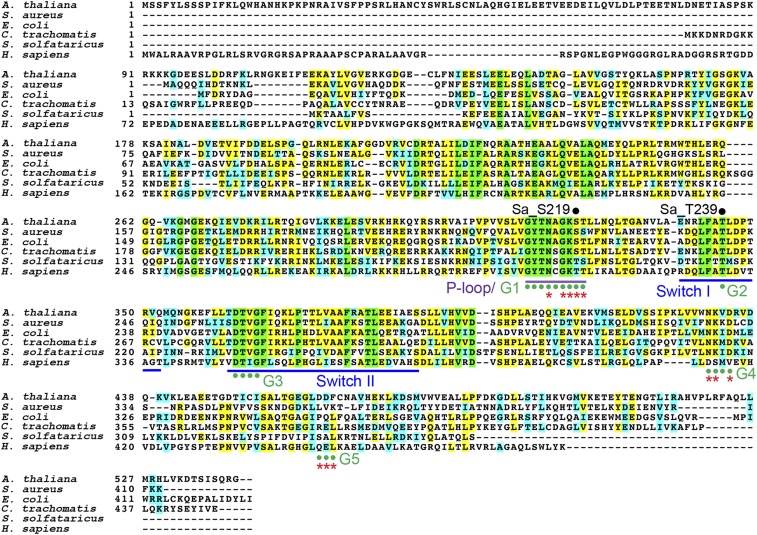
Fig. S1.
Multiple sequence alignment of HflX homologs. HflX homologs are universally conserved from bacteria and plants to humans (33–50% homology). Sequences were extracted from the National Center for Biotechnology Information GenBank with the following accession numbers: Arabidopsis thaliana, AED96978.1; Staphylococcus aureus, ABD21413; Escherichia coli, AAC77130; Chlamydia trachomatis, AAC67975; Sulfolobus solfataricus, AAK40607; and Homo sapiens, O43824 (also known as GTPBP6). Domain structures of P loop, G domains, and Switch I–II regions are color coded. Red asterisks indicate GTP and Mg2+ binding sites. Residues S219 and T239 of S. aureus HflX are labeled.
Results
HflX Dissociates S. aureus 100S Ribosome in a GTP-Dependent Manner.
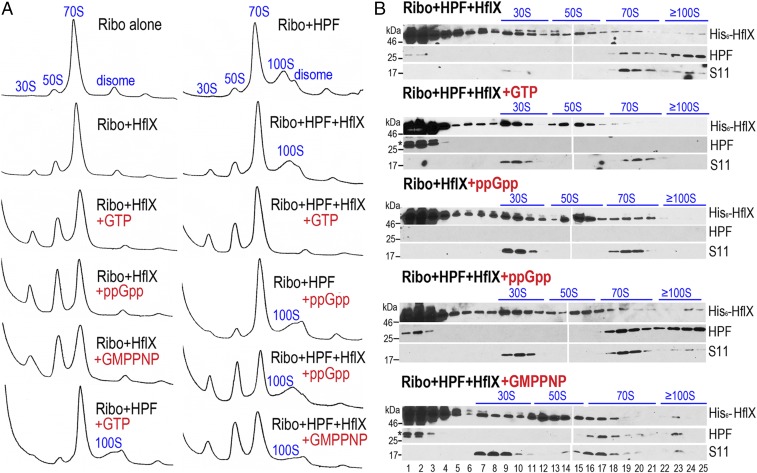
The conversion of the posttermination complex (PoTc) into the vacant 70S complex or free 30S and 50S subunits is required for translational reinitiation (38–41). We hypothesized that the disassembly of the 100S ribosome may involve known inhibitors of 30S–50S subunit joining {IF3, ObgE (42), RatA (43), RsgA (44), RsfS (45), 70S splitter [HflX, IF3 (46)] or PoTc recycling factors [RRF+EF-G (35)]}. With the exception of RatA, homologs of these factors are present in S. aureus. Null mutations of IF3, ObgE, RRF, and EF-G are deleterious. We found that the production of 100S ribosome in the rsfS and rsgA mutants during the log and stationary phase is indistinguishable from 100S ribosome production in wild-type S. aureus (Fig. S2A). The recombinant GTPase ObgE protein also failed to dissociate the 100S ribosome in vitro (Fig. S2B), even at 10-fold molar excess and 2 mM GTP. However, we found that the GTPase HflX could dissociate both S. aureus 70S and 100S ribosomes in vitro (Fig. 2). The in vitro experiments involve the dimerization of salt-washed 70S ribosomes with purified HPF at a molar ratio of 1:1 (3), followed by the addition of HflX and guanosine analogs. The distribution of ribosomal particles was analyzed by sucrose gradient density ultracentrifugation and immunoblotting with antibodies against HPF, His-tag, and ribosomal protein S11. Consistent with previous findings in E. coli (46, 47), S. aureus HflX exhibited very weak or no splitting activity toward the 70S ribosome either by itself (Fig. 2A, Left) or in the presence of ATP or GDP (Fig. S3 B and C), but 70S splitting was significantly enhanced in the presence of GTP, with the maximal activity achieved using nonhydrolyzable guanosine 5′-[β,γ-imido]triphosphate (GMP-PNP). S. aureus HflX is also a target of the stringent response alarmone (p)ppGpp (44). We found that HflX⋅ppGpp dissociates the 70S ribosome as effectively as HflX⋅GMP-PNP. In stark contrast, only HflX⋅GTP could completely dissociate the 100S ribosome (Fig. 2A, Right), whereas HflX⋅ppGpp or HflX⋅GMP-PNP remained 100S bound but were unable to dissociate the complex (Fig. 2B). These results strongly suggest that the disassembly of the 100S ribosome strictly requires GTP hydrolysis, whereas the disassembly of the 70S ribosome is nonselectively stimulated by all of the tested guanosines.
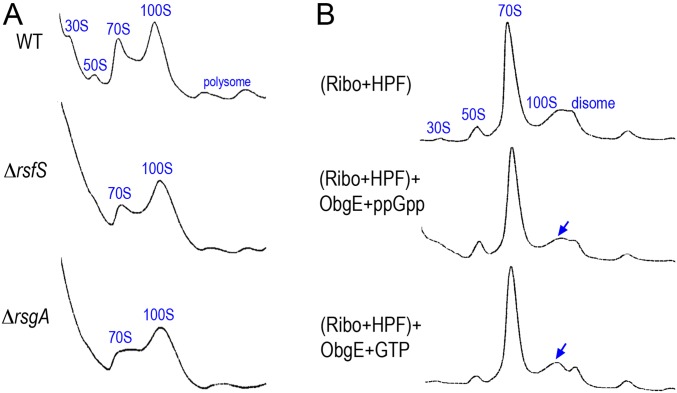
Fig. S2.
The ribosome-binding GTPases RsfS, RsgA, and ObgE do not significantly alter the 100S ribosome abundance. (A) The sucrose density gradient (5–25%, x axis) profiles of S. aureus JE2 (WT) and its rsfS and rsgA null mutants during midlog growth phases (OD600 ∼1.4). Each panel represents 2.5 Abs260 units of RNA input. The y axes show absorbance at 254 nm. (B) In vitro 100S dimerization and dissociation reactions were analyzed by 5–25% sucrose density gradient fractionation (x axis). The 70S dimerization reaction consisted of 0.2 μM salt-washed ribosomes and 2 μM recombinant HPF. Disassembly reactions were programmed with the dimerization reaction supplemented with 2 μM of recombinant ObgE and 2 mM of ppGpp or GTP and incubated at 37 °C for 30 min. The 100S peaks (blue arrows) are not significantly reduced in the presence of ObgE.
Fig. 2.
In vitro disassembly of 70S and 100S ribosomes by HflX. (A) All tested guanosine analogs stimulate 70S dissociation by HflX, whereas only HflX⋅GTP is able to fully dissociate the 100S ribosome. In vitro 70S dimerization was done by incubating equal molar amounts (final 0.2 μM) of salt-washed ribosomes with purified HPF. Dissociation was initiated by the addition of 2 μM His6-HflX and 2 mM nucleotides. The samples were fractionated by 5–25% sucrose density sedimentation (x axis), and ribosome species were monitored by absorbance at 254 nm (y axis). (B) Representative Western blots showing the distribution of HPF and His6-HflX in each ribosomal species separated in A using anti-HPF (1:4,000 dilutions) and monoclonal anti-His5 (1:1,000 dilutions). The 30S ribosomal protein S11 (1:4,000 dilutions) serves as the fractionation marker. HflX⋅ppGpp and HflX⋅GMP-PNP remain bound to the 100S complex without subunit dissociation (lanes 23–25). An asterisk above the HPF band indicates residual His6-HPF from incomplete removal of the affinity tag after thrombin cleavage.
Fig. S3.
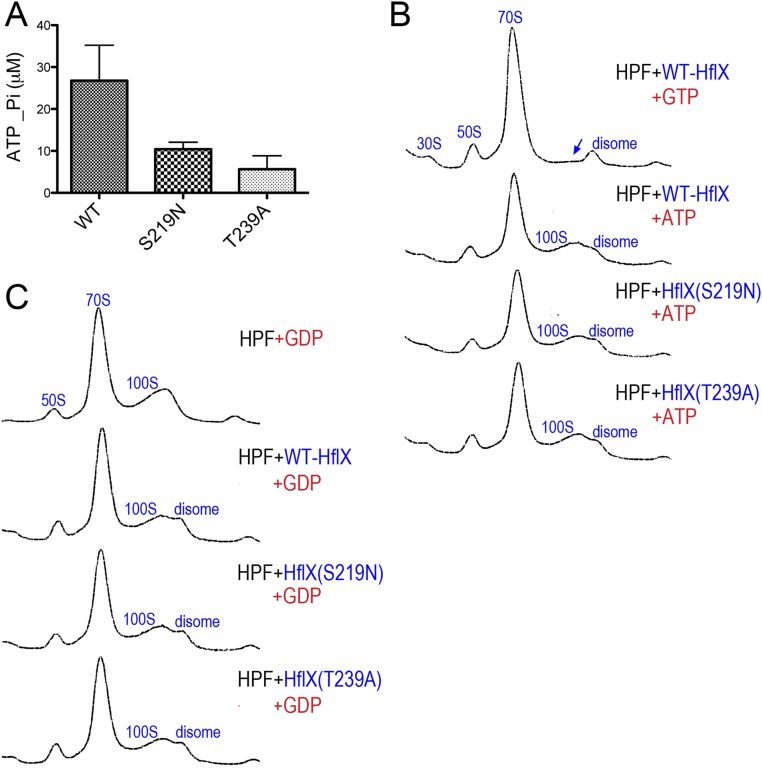
ATP and GDP do not promote the ribosome dissociation activity of HflX. (A) Malachite green ATPase assay showing ∼80% reduction of ATP hydrolysis in the HflXS219N and HflXT239A mutants. Hydrolysis was measured by quantitating the liberated inorganic phosphate (Pi) using a Pi standard curve. Each reaction contains 4 μM HflX proteins and 0.4 mM ATP. Error bars are SEs of three independent replicates. (B) Ribosome profiles of 100S disassembly reactions in the presence of 2 mM GTP or ATP. Ribosome species were analyzed by 5–25% sucrose gradient sedimentation (x axis). Y axes show absorbance at 254 nm. The blue arrow marks the location of diminishing 100S ribosome with the addition of GTP. (C) Same as in B, except that 2 mM GDP was used in place of ATP or GTP.
Importance of HflX GTP Hydrolysis in 100S Ribosome Disassembly.
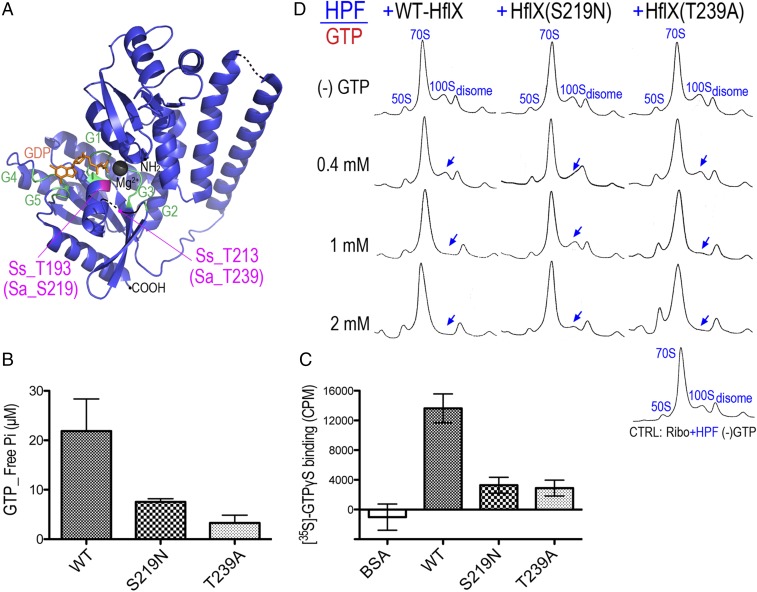
The key regions accounting for the catalytic activity of HflX include the P loop, switches I and II, and the G1–G5 domains (48, 49) (Fig. 3A and Fig. S1). E. coli HflX also binds and hydrolyzes ATP (50, 51). To test how the GTPase activity of HflX influences the dissociation of the 100S ribosome, we introduced S219N and T239A substitutions located in the P-loop/G1 and switch I/G2 regions that correspond to T193 and T213 of the archaeon Sulfolobus solfataricus (hereafter referred as Ss_T193 and Ss_T213). The Ss_T193N mutation perturbs the magnesium site and prevents proper loading of GTP (48), and the equivalent S243N mutation of Chlamydophila pneumoniae loses GTP hydrolysis activity (52). Threonine-239 of S. aureus HflX (Ss_T213, C. pneumoniae T263) is invariant across all bacterial and eukaryotic HflX homologs (Fig. S1). It resides in an unstructured switch I/G2 region, and mutation in this residue impairs GTP binding and hydrolysis (52). We found that the HflXS219N and HflXT239A mutants both had GTP hydrolysis and binding activities reduced by ∼75% (Fig. 3 B and C) and ATP hydrolysis by ∼80% (Fig. S3A). However, the HflXS219N mutant showed a more severe defect in 100S dissociation than the HflXT239A variant regardless of GTP concentration. At 1 mM GTP, the wild-type HflX fully dissociated the 100S ribosome, but under the same conditions, a residual 100S peak was observed in the HflXT239A mutant. A higher GTP concentration could partially offset the mutation, as the HflXT239A mutant efficiently split the 100S complex (Fig. 3D). The GTP concentrations we used are physiologically relevant because bacterial intracellular GTP levels usually range from 0.9 to 1.7 mM (53). The slight differences in the degree of 70S dissociation between Figs. 2A and 3D occurred because the proteins in Fig. 3 were purified from Hepes buffer instead of the routinely used phosphate buffer to avoid a high background of free phosphate in Fig. 3 B and C. The reason for the strong 100S dissociation activity of HflXT239A despite its diminished GTP binding and hydrolysis activities remains to be investigated. Nevertheless, our results confirmed that GTP hydrolysis is absolutely required for 100S disassembly, whereas ATP hydrolysis is dispensable.
Fig. 3.
GTP hydrolysis is required for the HflX-mediated disassembly of the 100S ribosome. (A) Crystal structure of archaeon S. solfataricus HflX in complex with GDP (orange) (Protein Data Bank 3KXI). Residues critical for GTP binding/hydrolysis are colored in magenta, and equivalent positions in the S. aureus HflX are denoted as Sa_S219 and Sa_T239. Magnesium is in gray. G1–G5 domains are colored in green. Dashed lines denote the unstructured regions in the crystal. Overall sequence homology (34%) between SsHflX and SaHflX is shown in Fig. S1. (B) Malachite green GTPase assay showing the reduction of GTPase activity in the S219N and T239A mutants. Each reaction contains 4 μM WT HflX or its mutants and 0.4 mM GTP substrate for a total of 200 μL. The amount of free inorganic phosphate (Pi) released upon GTP hydrolysis was calculated from a Pi standard curve. Error bars are SEs from four independent experiments using three different batches of purified proteins. (C) The S219N and T239A mutants are impaired in GTP binding. Filter binding assays of HflX variants (5 μM) mixed with 0.2 μM of nonhydrolyzable [35S]-GTPγS. The amount of radiolabeled GTP [expressed in counts per million (cpm)] bound to the nitrocellulose membrane-adhered HflX was measured on a liquid scintillation counter. The same amount of non-GTP binder BSA was used in place of HflX to serve as a negative control. The y axis represents the values after background (reaction without any protein) subtraction. Error bars are SEs from three independent experiments. (D) The S219N and T239A mutant proteins do not dissociate the 100S ribosome as efficiently as wild-type (WT) HflX. Shown are 5–25% sucrose gradient density profiles of the in vitro 70S dimerization reactions treated with either WT HflX or its catalytic mutants in the presence of a range of GTP. The entire reaction includes 0.2 μM ribosomes and HPF, 2 μM HflX, and 0–2 mM GTP. Blue arrows mark the diminishing 100S peaks.
Thermoresponsive Phenotypes of the ΔhflX Mutant.
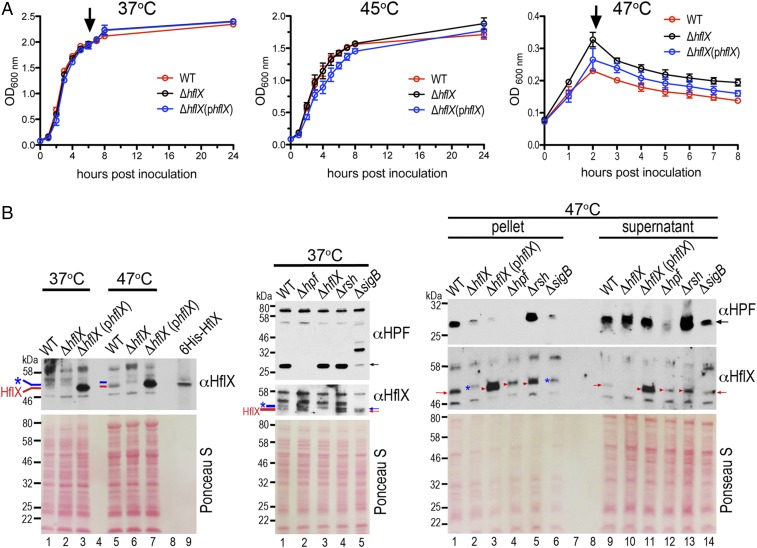
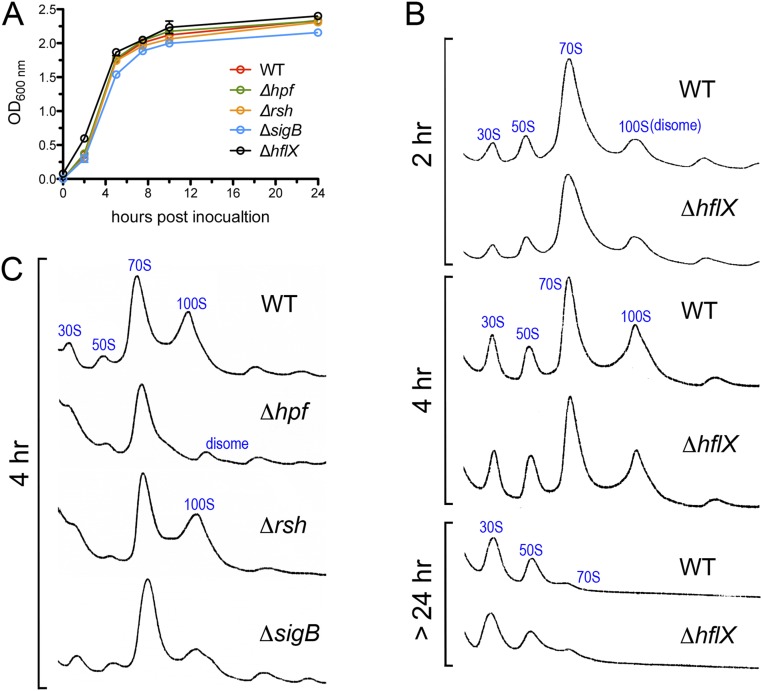
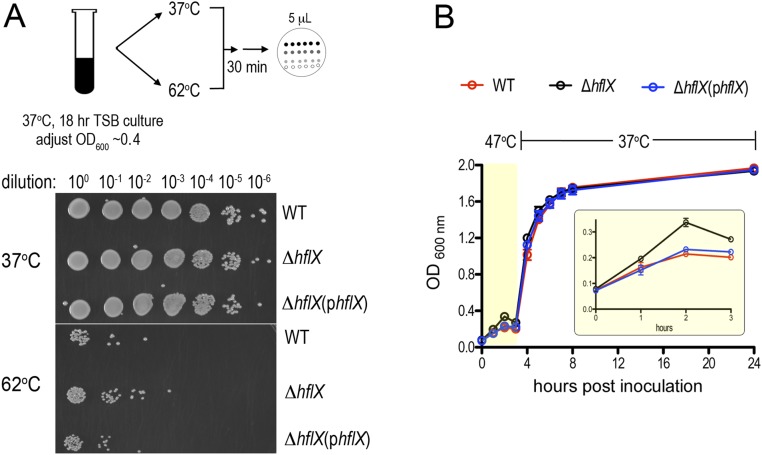
The expression of E. coli HflX is induced by heat through σ32 regulation (54) and is thought to rescue stalled ribosomes under heat stress. E. coli HflX binds to the peptidyltransferase center in the 50S subunit and stimulates subunit dissociation by disrupting intersubunit bridges (46). The hflX knockout strains of E. coli and S. aureus exhibit no detectable growth defects under standard laboratory conditions (30–37 °C in rich medium) (44, 46). Consistent with these results, no growth defect was observed in S. aureus ΔhflX cultured at 37–45 °C (Fig. 4A and Fig. S4A), and no observable difference in ribosome profiles was seen in the ΔhflX mutant relative to the wild-type during various growth phases at 37 °C (Fig. S4B). Paradoxically, and in contrast to the E. coli ΔhflX mutant phenotype, we found that S. aureus ΔhflX was more viable at 47 °C than the wild type (Fig. 4A) and more resistant to acute heat killing (Fig. S5A). The phenotype could be partially rescued by expressing hflX in trans on a plasmid under the control of its native promoter (Fig. 4A). The overproduction of plasmid-borne HflX (>10-fold greater than the wild type, Fig. 4B) may interfere with the proper stoichiometry of the HflX ribosome, which explains the lack of a full rescue. The growth arrest of the WT and the complementing strains grown at 47 °C were reversed to the ΔhflX level upon a temperature downshift to nonstress 37 °C conditions (Fig. S5B). We noted that a transposon insertion mutant of hflX (strain NE1325) also phenocopied the in-frame ΔhflX deletion under the same conditions.
Fig. 4.
Coregulation of HPF and HflX by the general stress and stringent response pathways. (A) Growth curves of S. aureus JE2, ΔhflX null, and ΔhflX (phflX) complementing strain at 37 °C, 45 °C, and 47 °C in tryptic soy broth (TSB) medium, measured by optical density (OD) at 600 nm. Error bars are SEs of three independent experiments. Arrows denote the time points at which cells were collected for immunoblotting, shown in B. (B) Expression of HPF and HflX at 37 °C and 47 °C. (Left) Comparison of HflX levels at 37 °C versus 47 °C. Each lane corresponds to 0.1 Abs280 units of total lysate. Several proteins are up-regulated by heat, thus generating different protein profiles on the Ponceau S stain. The same membrane was stripped and reprobed with the second antibody. (Middle) Expression of HPF and HflX in different mutant backgrounds at 37 °C. Each lane corresponds to 0.1 Abs280 units of total lysate. A ΔsigB mutant produces a distinct protein profile and shows reduced HPF production (lane 5). (Right) Probing the levels of HPF and HflX in the ribosome/complex-bound (pellet) and free protein (supernatant) fractions after spinning through 0.2 Abs280 units of 47 °C-derived lysates through a 1.1-M sucrose cushion. Blue asterisks and lines denote a nonspecific band that exists in the ΔhflX. Red lines and arrows mark the HflX protein. Ponceau S staining of the membranes before immunoblotting serves as the loading control. Anti-HPF and anti-HflX were used at 1:4,000 and 1:1,000 dilutions, respectively.
Fig. S4.
Bacterial growth and the abundance of 100S ribosomes at 37 °C in different genetic backgrounds. (A) Only the ΔsigB mutant was slightly attenuated in growth. Cell growth was measured by monitoring optical density (OD) at 600 nm. Error bars are SEs of three independent biological replicates. (B) Sucrose density gradient (5–25%) profiles of S. aureus JE2 and the ΔhflX mutant during early-log, midlog, and stationary phases. No significant difference in 100S abundance was observed between the wild-type (WT) and the ΔhflX null under these conditions. Each panel represents 2.5 Abs260 units of RNA input. (C) Comparison of 100S abundance in the Δhpf, Δrsh, and ΔsigB null mutants. Ribosome profiles were analyzed as in B. HPF synthesis is greatly reduced in the ΔsigB mutant (Fig. 4B), thereby decreasing 70S dimerization.
Fig. S5.
Better fitness of the ΔhflX mutant under heat stress. (A) Acute thermal killing assay. The ΔhflX mutant is at least sixfold more resistant to heat killing. Stationary phase TSB cultures were diluted with fresh TSB to an OD600 of 0.4, split into two portions and incubated at 37 °C and 62 °C in a thermomixer shaker at 300 rpm. After 30 min of incubation, the cells were subjected to 10-fold serial dilutions with sterile 1× PBS buffer. Five microliters of each dilution was spotted on TSB agar plates and incubated for 18 h at 37 °C. The results are representative of three independent biological replicates. (B) A temperature downshift from 47 °C to a permissive temperature of 37 °C restores the growth of the WT and ΔhflX complementing strains. The cells were grown at 47 °C for 3 h, during which the OD600 of the ΔhflX mutant peaked at 2 h and gradually decreased (yellow box, Inset), whereas the WT and complementing strains were less viable (see also Fig. 4A). Upon heat removal, the WT and complementing strains regrew to levels comparable to the ΔhflX mutant. The error bars are SEs from four independent experiments.
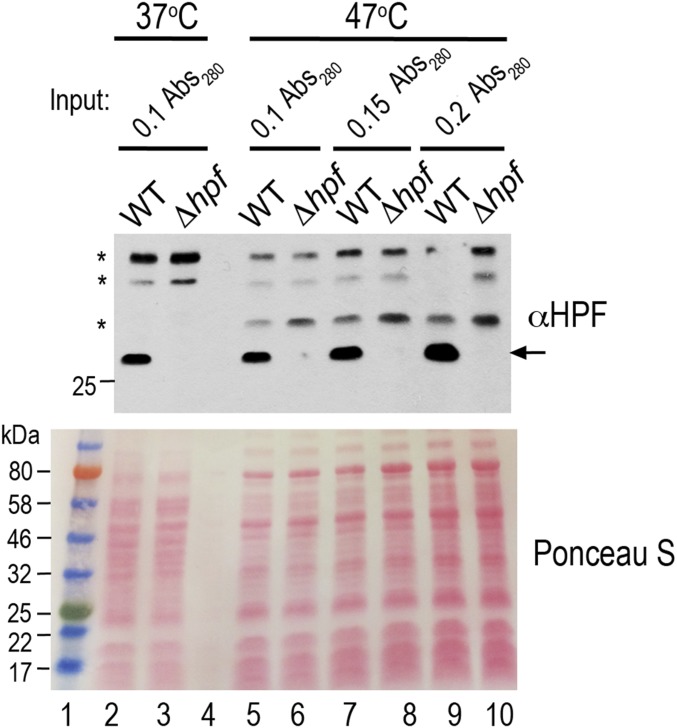
The S. aureus 100S ribosome is constitutively produced at 37 °C throughout all growth phases (3, 36). HflX⋅GTP antagonizes the stability of the 100S ribosome (Fig. 2A). We posit that the abundance of the 100S ribosome is due to a high level of HPF concomitant with low cellular HflX or GTP. We tested the counterbalance between HPF and HflX by examining their expression at different growth temperatures. The HflX antibody that we raised also recognizes a recalcitrant background protein that is 1–2 kDa away from HflX. Despite intensive efforts toward antibody purification and optimizing electrophoresis conditions, we were unable to completely eliminate the background. To reduce the cross-reaction, we preadsorbed antibody with the ΔhflX lysate, and to probe higher levels of HflX, we performed ultracentrifugation to separate the high molecular weight fraction (ribosome-/complex-bound background) from the free proteins in the supernatant (Fig. 4B, Right, compare lanes 2 and 10). Regardless of growth temperatures, the null mutation of hpf did not affect the expression of hflX compared with the wild type, and vice versa.
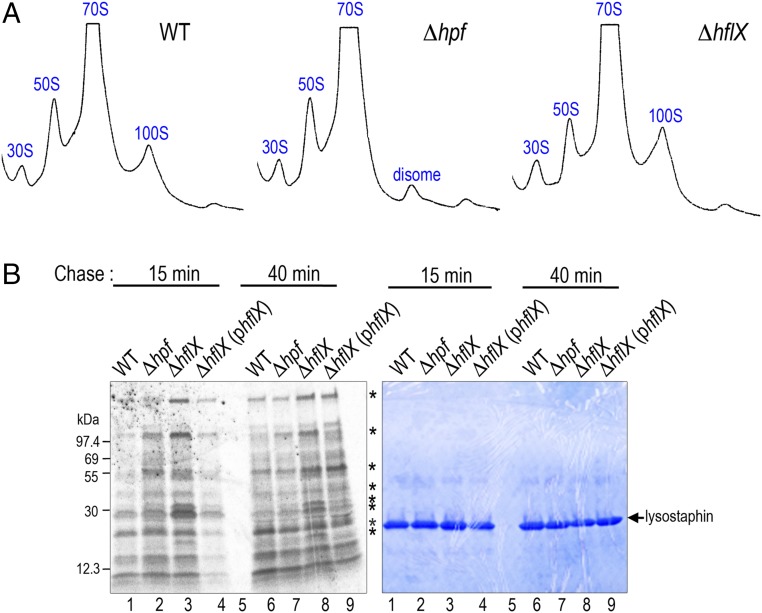
At 37 °C, HflX was barely detectable, and HPF was abundant (Fig. 4B, Left and Fig. S6), explaining the constitutive generation of the 100S ribosome at 37 °C (36) (Fig. S4B). At 47 °C, the HPF level was not significantly altered (Fig. S6), but HflX was heat induced (Fig. 4B, Left), thereby reducing the amount of 100S ribosome (Fig. 5A, WT). Likewise, a ΔhflX mutant generated more 70S and 100S complexes (Fig. 5A, Right). Pulse-chase analysis suggested that the growth advantage of the ΔhflX mutant at 47 °C might be in part due to better translational efficiency (Fig. 5B). Cells were pulse labeled with [35S]-methionine for 3 min, and the synthesis of radiolabeled nascent proteins was monitored by chasing with cold casamino acids after 15 min and 40 min. At both time points, ΔhflX synthesized more proteins (or slower protein turnover) than the wild-type and the complementing strain.
Fig. S6.
Growth temperatures did not significantly affect HPF expression. Asterisks mark the nonspecific cross-reactions of anti-HPF (1:4,000 dilutions). Ponceau S staining shows the equal normalization of protein loading.
Fig. 5.
In vivo contribution of HflX during 47 °C growth. (A) Ribosome profiles of S. aureus JE2 and its hpf, hflX null mutants. A ΔhflX null deficient in ribosome dissociation has more 70S and 100S particles than the wild-type (WT). Ribosome species were analyzed on a 5–25% sucrose density gradient (x axis) and the absorbance was monitored at 254 nm (y axis). Each panel represents 2.5 Abs260 units of RNA input. (B) Pulse-chase analysis showing a reduction of translational capacity in the WT and the ΔhflX (phflX) complementing strain at 47 °C. Cells were pulse labeled with [35S]-methionine for 3 min and subsequently chased with excess cold casamino acids. After 15-min and 40-min incubation, translation was stopped by pouring 1 mL culture over ice. Cell lysis was performed on the ice containing a final concentration of 15 μg/mL lysostaphin. Asterisks mark the major differentially produced proteins. (Right) The same gel as in the phosphorimage, stained with Coomassie Brilliant Blue G, to indicate equal loading.
The General Stress and Stringent Response Pathways Control hpf and hflX Expression.
To further investigate how the expression levels of hpf and hflX affect 100S ribosome production, we turned to potential regulators of hpf and hflX. HflX⋅GTP dissociates the 100S ribosome, but HflX⋅ppGpp does not (Fig. 2A). In S. aureus, (p)ppGpp is produced from GT(D)P and ATP by the major synthetase Rsh (RelA/SpoT homolog) and two alternative proteins RelP and RelQ (55, 56). Close inspection of the 5′-untranslated region (5′-UTR) of comF-hpf operon revealed an alternative sigma factor (SigB or σB) binding motif (57). S. aureus SigB regulates an arsenal of virulence and general stress response genes (57). We analyzed the levels of HPF and HflX by Western blotting using cell lysates harvested at the late log phase (37 °C) and after maximal growth at 47 °C (2–3 h). As expected, we found that a sigB null was attenuated in HPF synthesis and growth at 37 °C; HflX expression was slightly, or not at all, increased (Fig. 4B, Middle and Fig. S4A), which was consistent with the dramatic reduction in the 100S ribosome (Fig. S4C). At 47 °C, no significant changes in HPF level were found in the sigB mutant, but HflX was reduced by approximately twofold (Fig. 4B, Right). At 37 °C, an rsh mutant did not significantly affect both cell growth and 100S abundance (Fig. S4 A and C), presumably due to functional redundancies of RelP and RelQ. At 47 °C, however, both HPF and HflX were derepressed upon rsh knockout (Fig. 4B, Right). It is unclear how (p)ppGpp negatively integrated transcriptional regulation and splitting activity on HflX in response to thermal stress, given that S. aureus HflX binds better to ppGpp (kd = 0.87 ± 0.15 μM) than to GTP (kd = 19.39 ± 5.11 μM) (44). From the expression profiles, we concluded that SigB and (p)ppGpp exert positive and negative regulatory roles on hpf and hflX expression that are strongly influenced by the temperature (Fig. 1).
Discussion
The molecular mechanisms of 70S dimerization and the importance of the 100S ribosome in bacterial resuscitation from nutrient deprivation have been well defined in several bacterial species. Four common themes emerged from these studies. First, the 100S ribosomes are inactive in translation due to the occlusion of ribosomal decoding sites by HPF. The 100S ribosome-mediated down-regulation of translation is one of the survival strategies in slow-growing and dormant cells to reduce energy consumption. Second, the 100S ribosomes in Firmicutes are tethered by direct HPF contact as opposed to allosteric control of dimerization by RMF in E. coli. Third, the formation of the 100S ribosome extends the bacterial life span, which is correlated with the prolonged ribosome integrity. The precise role of dimerization is unclear. It is possible that the “masked” 30S–30S interfaces in the dimer are more resistant to nucleolytic attack, although these regions are not known to be the targets of RNases (58–60). Perhaps the conformational changes in the 30S subunits (19) reduce the accessibility to RNases, and moreover, translational attenuation may alter the expression of specific factors involved in the ribosome decay pathway. Alternatively, dimerization may simply avoid spurious interactions of RNA polymerase (RNAP) with the inactive 70S ribosome because the dimerization interface, e.g., uS2, overlaps with the RNAP–ribosome contact sites (61). Fourth, nutrient replenishment promotes the conversion of 100S ribosomes to 70S concomitantly with an increase in translation and bacterial regrowth. The factor participating in the active dissociation of the 100S ribosome is unclear.
Our biochemical and genetic analyses demonstrate that the GTPase HflX is an antagonizing factor of HPF-induced 70S dimerization in the human pathogen S. aureus (Fig. 1). The counterbalances between HPF and HflX and GTP-ppGpp homeostasis contribute to 100S ribosome abundance. HflX dissociates both 70S and 100S ribosomes. GTP hydrolysis stimulates 100S disassembly, whereas ppGpp binding inhibits it. It is unclear whether a 70S intermediate is necessary for the final conversion into subunits. In contrast, HflX is less selective about guanosines, and the binding of GTP, GMP-PNP, or ppGpp efficiently dissociates the 70S complex. Future kinetic studies are needed to delineate the precise sequence of 100S disassembly and the fates of HflX and HPF upon dissociation as well as the hierarchical order of IF3 and RRF/EF-G with respect to 100S dissociation during ribosome recycling (Fig. S7).
Unlike the γ-proteobacterial 100S ribosome, which is formed only under stress, S. aureus 100S ribosomes are produced from lag-log growth throughout the stationary phase partly because the expression of S. aureus HPF appears to be less tightly controlled (3, 36), and multiple transcriptional pathways may orchestrate hpf expression (see below). Intriguingly, the production of the S. aureus 100S ribosome during the logarithmic phase does not reduce growth and global translation in rich medium and only moderately reduces growth in minimal medium (3). We propose that posttermination vacant 70S complexes are the precursors of 100S ribosomes (Fig. S7). It has been shown that 70S ribosomes do not always undergo subunit dissociation during termination; instead reinitiation could occur in a 70S-scanning mode (38). Moreover, cells exclusively carrying covalently linked 70S ribosomes are viable (40). The 30S and 50S subunits released from mRNA may also reassociate to form 70S ribosomes when the 30S subunit is not bound to the antiassociation factor IF3, given that the in vivo concentration of IF3 is at least 100-fold less than is required to prevent all subunit joining (62). HPF also dimerizes 30S monomers in vivo in chemically defined minimal medium (3). Although unlikely, it is possible that HPF assembles the 100S ribosome by a stepwise 30S:30S → 50S → 100S sequence. The effective stoichiometry of HflX⋅GTP-100S is unclear, but low levels of HflX under nonstress conditions (37 °C, Fig. 4) appear to be sufficient to avoid the buildup of 100S ribosome to a threshold that compromises translation and growth. In fact, one interpretation is that the accumulation of 100S ribosomes over time with the highest-level peaks during the stationary phase may be due to the inhibition of HflX splitting activity by (p)ppGpp, a global regulator induced during the stationary phase when nutrients are limited (55).
A sigB knockout does not completely abolish hpf synthesis (Fig. 4B, Middle), which suggests that the transcription of hpf is regulated by multiple pathways, in addition to SigB and (p)ppGpp. Notably, the comF-hpf 5′-UTR carries a putative binding motif of housekeeping SigA (or σ70). B. subtilis hpf is positively controlled by σB and σH (2), and thus, it is possible that other alternative sigma-factors, e.g., SigH and SigS, whose binding sites are not yet defined (57), also activate the expression of S. aureus hpf upon sensing disparate environmental stimuli. S. aureus hpf is regulated by the global transcriptional repressor CodY (refs. 63 and 64, hpf locus annotated as SA0815 or SACOL0815). Binding to GTP and binding to branched-chain amino acids promote CodY repression. Because intracellular GTP concentration drops as a result of (p)ppGpp synthesis, a potential crosstalk of the CodY–SigB–Rsh pathways in shaping the 100S ribosome output may be envisioned.
The impact of HflX appears to be highly condition dependent. We found that an ΔhflX knockout exhibits no obvious phenotype under standard laboratory conditions. This lack of phenotype may be due to compensation by other functionally redundant factors. For example, E. coli RRF, in conjunction with EF-G, is able to partially complement the growth defect of an E. coli ΔhflX mutant upon heat exposure (46). Since E. coli HflX is thought to rescue translational stalling under heat stress because HflX splits both vacant 70S and elongation-arrested 70S with an empty A site (46), the negative impact that S. aureus HflX exerts on cell growth and translation at 47 °C seems counterintuitive (Figs. 4A and 5B). We speculate that HflX dissociates 100S ribosomes and heat-induced stalled 70S ribosomes, which leads to an increase in translational restart and in turn exacerbates the accumulation of stalled ribosomes. In contrast, an ΔhflX mutant undergoes the translational restart-stalling cycle at a slower rate, which explains why the ΔhflX mutant is more viable than the wild type at 47 °C. Furthermore, by preserving a subpool of 100S ribosomes, the ΔhflX mutant becomes more resistant to acute thermal killing (Fig. S5A), which is consistent with our recent observation that a loss of HPF dimerization function sensitizes cells to heat killing (19). More work is certainly needed to better integrate the opposing actions of HPF and HflX into the complex heat stress and stringent response network.
Heterologous expression of a metagenomics isolated hflX homolog in E. coli confers macrolide resistance through an unknown pathway (65). HflX homologs are widespread in both prokaryotes and eukaryotes. The function of HflX in higher eukaryotes has not been explored. Plant HflX is localized in the chloroplast (37), and 100S dimerization in photosynthetic cyanobacteria is induced by darkness (9); thus, HflX may exert a similar splitting activity on the hibernating 70S chlororibosome (31). The overexpression of the human homolog (GTPBP6) is linked to neuropsychiatric disorders, and many somatic mutations in human cancer have been mapped to GTPBP6 (66–68). Our findings will offer general principles of HflX function in the ribosome metabolism and in translational regulation across all biological systems.
Experimental Procedures
Strains, Plasmids, Chemicals, and Growth Conditions.
Strain JE2 is a community-associated methicillin-resistant S. aureus (CA-MRSA) of USA300 lineage (69). The JE2 mutant derivatives carry a bursa aurealis transposon insertion (ins.) within the coding regions of hpf (ins. 47 nt, strain NE838), rsh (ins. 1691 nt, strain NE1714), sigB (ins. 143 nt, strain NE1109), hflX (ins. 310 nt, strain NE1325), rsfS (ins. 95 nt, strain NE712), and rsgA (ins. 220 nt, strain NE352). Transposon mutants were acquired from Biodefense and Emerging Infections Research Resources Repository (BEI Resources) (National Institute of Allergy and Infectious Diseases, NIH).
The in-frame hflX deletion mutant (strain MNY102) was constructed as follows: A 2-kb flanking region of the hflX was PCR amplified with the primer pairs P1054 (SacI, 5′-GAC AGA GCT CTA GAG AAT TTT AAA GCA AAC CAA-3′)/P1055 (SmaI, 5′-TGC CGC GTA CTC TGC GCC CGG GAT AGC CAT GTT ATA CAT CTC CTT AAT AAA ATC-3′) and P1056 (SmaI, 5′-ATC CCG GGC GCA GAG TAC GCG GCA TAA TAA AAG GAC GAA ATT CAA ATG-3′)/P1057 (SalI, 5′-ATT TAA TCT ATT ATA TAA GTC CTT GTT CTG TCG ACT GAT-3′) via two-step PCR using S. aureus JE2 genomic DNA as the template. The product was digested with SacI and SalI and cloned into the same sites of pBT2 (70). The resulting pBT2ΔhflX was digested with SmaI, dephosphorylated, and ligated to the blunt-ended ∼1.3-kb erythromycin (Erm) resistance cassette that was released from pBTE (70) by KpnI and HindIII digestion. The resulting construct pBT2ΔhflX::Erm was passaged through S. aureus RN4200, and the plasmid was reisolated, electroporated into S. aureus JE2, and selected at 30 °C on tryptic soy broth agar (TSA) plates supplemented with 10 μg/mL chloramphenicol. The integrant was further selected by a 43 °C temperature upshift on chloramphenicol-containing TSA plates. The homologous recombinant was resolved by 30 °C passages and cycloserine enrichment following the published procedures (70). A total of 5 μg/mL of erythromycin was used for recombinant selection. The complementing plasmid of hflX was constructed by cloning the 1.5-kb hflX region into the EcoRI and BamHI sites of pLI50 (71) under the control of its native promoter. The hflX DNA fragment was generated by PCR amplification using primers P1060 (EcoRI, 5′-CAC CTG AAT TCG TTT CTA CTA CAA AAT CAT AAA TT-3′) and P1061 (BamHI, 5′-GAA TTG GAT CCT TTT ATT ATT TTT TAA ATC C-3′).
To overexpress the HflX recombinant protein, an ∼1.3-kb coding region of hflX was PCR amplified with primers P998 (NdeI, 5′-GTA TCA TAT GGC TCA GCA ACA AAT TCA TGA TAC-3′) and P999 (XhoI, 5′-TTC CTC GAG TTA TTA TTT TTT AAA TCC TTT TAT ACG ATA AAC A-3′) and the genomic DNA of JE2 as the template. The PCR product was ligated to an IPTG-inducible pET28a (Novagen), resulting in plasmid pET28::hflX. HflX GTP hydrolysis mutants S219N and T239A were made with the Quik-Change site-direct mutagenesis kit (Agilent Technologies). The cloning and overproduction of the ribosomal protein S11 and ObgE were performed using the same strategy, except that primers P996 (NdeI, 5′-ATG CCA TAT GGC ACG TAA ACA AGT ATC TCG T-3′) and P977 (XhoI, 5′-ATG CCT CGA GAT CCG CTT ATA CAC GAC GAC GTT TTG GTG G-3′) were used to amplify the rpsK region. To amplify obgE, primers P1000 (NcoI, 5′-ATC CAT GG TTG TCG ATC AAG TCA AAA TAT CT-3′) and P1001 (HindIII, 5′-TCA AGC TTC TAA TGA TGG TGA TGG TGA TGA TGA TGT TCA ACG AAT TCA AAT TCT CCG CC-3′) were used and ligated to the NcoI and HindIII sites of pET28a, yielding a C-terminal 8His-tagged ObgE. E. coli BL21(DE3) cells carrying pET28a derivatives were grown in LB medium supplemented with 50 μg/mL kanamycin. S. aureus strains were routinely grown in tryptic soy broth (TSB; BD Difco) or chemically defined minimal medium (72) supplemented with 10 μg/mL chloramphenicol when maintenance of the pLI50 and pBT2 derivatives was necessary. All strains were cultured at 37 °C unless otherwise noted. All chemicals and antibiotics were purchased from Sigma-Aldrich except ppGpp (TriLink Biotechnologies).
Protein Overexpression, Purification, and Antibody Production.
The purification of the HPF recombinant protein had been described previously (73). The N-terminal His6-tag was removed from HPF by the thrombin CleanCleve Kit (Sigma-Aldrich) (3). Polyclonal anti-HPF had been generated previously (73). Hexahistidine-tagged S11 was purified with Ni-NTA affinity chromatography under denaturing conditions according to the manufacturer’s manual [Molecular Cloning Laboratories (MCLAB)]. The protein bands of S11 were excised from SDS/PAGE gels and used to raise antibody in rabbits (Josman). Hexahistidine-tagged HflX proteins were purified under phosphate buffer-based native conditions as previously described (3). The wild-type and mutant HflX recombinant proteins became insoluble during 37 °C growth. To increase solubility, E. coli BL21(DE3) carrying the overexpression plasmid was grown at 37 °C until OD600 0.4–0.5. The cultures were then moved to a 16 °C shaker incubator and induced by 0.5 mM IPTG (IBI Scientific) overnight. In the case when HflX proteins were used for GTPase/ATPase assays, phosphate buffer was substituted with 20 mM Hepes/KOH (pH 7.5) in all purification buffers to eliminate free phosphate contamination. Polyclonal anti-HflX was raised in rabbits (Josman).
In Vitro 100S Formation and Dissociation.
In vitro 70S dimerization was performed as described previously (3). Briefly, a 1:1 molar ratio of salt-washed 70S ribosomes (final 0.2 μM, calculated using 1 Abs260 = 23 pmol/mL 70S) (74) and purified HPF were mixed in a 100 μL reaction containing 1× binding buffer [10×: 50 mM Hepes (pH 7.5)/500 mM KOAc/100 mM NH4Cl/100 mM Mg(OAc)2]. Dimerization was stimulated by 37 °C incubation for 30 min. For 100S dissociation, a total of 30 μL of HflX or one of its mutant proteins was preincubated with various guanosine analogs at different concentrations at room temperature (∼23 °C) for 15 min, followed by mixing with the dimerization reaction in a total of 130 μL containing final concentrations of 2 μM of HflX and 0–2 mM of guanosine analogs. Dissociation reactions were incubated at 37 °C for 30 min, and finally the samples were layered on a 5–25% sucrose density gradient made in buffer C [20 mM Hepes (pH 7.5)/10 mM MgCl2/100 mM NH4Cl] that was equilibrated with a BioComp Gradient Master. The gradients were centrifuged at 210,000 × g at 4 °C in an SW41 rotor for 2.5 h. Fractionation was performed using a Brandel fractionation system equipped with a UA-6 UV/Vis detector. Sucrose fractions (∼250 μL/fraction) were precipitated with a final concentration 10% trichloroacetic acid, and the pellets were washed once with acetone, neutralized with 50 mM Tris base containing Laemmli sample buffer, and resolved by 4–20% TGX SDS/PAGE (Bio-Rad). To probe the distribution of HPF, S11, HflX, or 6His-HflX in each sucrose fraction, monoclonal anti-His5 (Qiagen) and polyclonal anti-HPF, anti-S11, and anti-HflX were used for Western blotting at dilution rates of 1/1,000, 1/4,000, 1/4,000, and 1/1,000, respectively. Anti-HflX was preadsorbed with 1 Abs280 unit of ΔhflX lysate at 4 °C overnight before immunoblotting to reduce a background band that closely comigrates with HflX on SDS/PAGE.
GTP/ATP Hydrolysis and GTP Binding Assays.
The PiColorLock Gold kit (Innova Biosciences) was used to determine the GTPase and ATPase activity of HflX by measuring the amount of free inorganic phosphate (Pi) that binds to the malachite green dye. Reactions were carried out in a 96-well microplate containing final concentrations of 4 μM HflX proteins and 400 μM GTP or ATP substrate in a total volume of 200 μL in 1× binding buffer. The mixtures were incubated on a 37 °C shaker (500 rpm) for 30 min followed by the addition of 1/4× volumes of dye. After 5 min, the reactions were stopped by adding 0.1× volumes of stabilizer. Pi release was measured calorimetrically at 620 nm and quantitated with a phosphate standard curve. The final Pi values correspond to micromolar-free Pi after subtraction of a background that contains the same amount of nucleotide substrate and protein elution buffer in 1× binding buffer (HflX omitted).
A filter binding assay was used to measure GTP binding. Five micromolar HflX proteins were mixed with final 0.2 μM of nonhydrolyzable [35S]-GTPγS (12.5 mCi/mL, 1,250 Ci/mmol; Perkin-Elmer) in a 50-μL reaction containing 1× binding buffer. A background control was prepared with the same components without any protein. Equal amounts of BSA, which does not bind GTP, were used in place of HflX to serve as a negative control. Samples were incubated at 37 °C for 1 h and stopped by the addition of 25× volumes of precipitation buffer [20%(wt/vol) PEG-6000 in 1× binding buffer]. The samples were applied to a Millipore model 1224 vacuum filtration manifold that was preassembled with a prewetted nitrocellulose BA85 disk (25 mm, 0.45 μm; Sigma) and two layers of 25-mm filter paper discs (Sigma). The disk sandwiches were washed three times with 8 mL of 1× binding buffer. Finally, the air-dried nitrocellulose discs were placed in scintillation vials containing 3 mL of Scintisafe Econo F (Fisher Scientific) mixtures, and radioactivity was measured on a Perkin-Elmer Tri-Carb 2910 instrument. The amount of GTP binding was expressed in counts per million (cpm) after background subtraction.
Ribosome Profiles and Cell Lysate Preparation.
Crude ribosomes were isolated from S. aureus Δhpf knockout grown in TSB cultures and prepared by the cryomilling method (3, 73). Two-and-a-half Abs260 units of lysate were loaded on a 5–25% sucrose gradient and fractionated as described above. For Western blotting, S. aureus cell pellets were resuspended in buffer A [20 mM Hepes/KOH (pH 7.5)/14 mM Mg(OAc)2/100 mM KCl/1 mM DTT/0.5 mM PMSF] and homogenized with Lysing Matrix B (100 mg beads/mL cells; MP Biomedicals) on a Retsch MM400 mixer mill at 15 Hz in five 3-min cycles. Clarified lysates were recovered by spinning at 20,817× g at 4 °C for 5 min to remove cell debris. A total of 0.1–0.2 Abs280 units of cell lysate were analyzed on 4–20% TGX SDS/PAGE gels (Bio-Rad). To separate ribosome-bound and unbound fractions, 0.2 Abs280 units of cell lysates were spun through a 1.1 M sucrose cushion made of buffer A in a Beckman TLA-120 rotor at 4 °C, 100,000× g for 2.5 h. The pellet was directly resuspended in Laemmli buffer, whereas the supernatant was precipitated by 10% trichloroacetic acid. Both fractions were resolved on 4–20% TGX SDS/PAGE (Bio-Rad) and subjected to immunoblotting.
Pulse-Chase Labeling.
Pulse-chase analyses were performed as described with minor modifications (75). Briefly, overnight TSB cultures were washed three times with defined minimal medium 4 (M4), diluted into fresh M4, and adjusted to OD600 ∼0.1. After growing in a 47 °C water bath shaker until OD600 ∼0.25 (∼135 min), cells were pulse labeled with 30 μCi/mL of Tran35S-label (MP Biomedicals) for 3 min and chased with an excess amount of unlabeled methionine/casamino acid mix at various time points. At each time point, 1 mL of culture was poured over 0.5 mL of ice in a prechilled microtube containing a final concentration of 15 μg/mL lysostaphin (AMBI). Cell lysis was achieved by incubation on ice for 20 min, followed by 10% trichloroacetic acid precipitation. The total proteins were analyzed by 4–20% TGX SDS/PAGE (Bio-Rad), and radiolabeled proteins were visualized using a Typhoon phosphorimager (GE Healthcare).
Acknowledgments
We thank Anthony Richardson (University of Pittsburgh) for the protocols and plasmids for S. aureus genetic knockouts. Transposon mutants were obtained through the Network on Antimicrobial Resistance in Staphylococcus aureus for distribution by BEI Resources, National Institute of Allergy and Infectious Diseases, NIH. M.-N.F.Y. is supported by the PEW Charitable Trusts, the Edward Mallinckrodt Jr. Foundation, and NIH Grant R01GM121359.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1709588114/-/DCSupplemental.
References
- 1.Yoshida H, Wada A. The 100S ribosome: Ribosomal hibernation induced by stress. Wiley Interdiscip Rev RNA. 2014;5:723–732. doi: 10.1002/wrna.1242. [DOI] [PubMed] [Google Scholar]
- 2.Akanuma G, et al. Ribosome dimerization is essential for the efficient regrowth of Bacillus subtilis. Microbiology. 2016;162:448–458. doi: 10.1099/mic.0.000234. [DOI] [PubMed] [Google Scholar]
- 3.Basu A, Yap MN. Ribosome hibernation factor promotes Staphylococcal survival and differentially represses translation. Nucleic Acids Res. 2016;44:4881–4893. doi: 10.1093/nar/gkw180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puri P, et al. Lactococcus lactis YfiA is necessary and sufficient for ribosome dimerization. Mol Microbiol. 2014;91:394–407. doi: 10.1111/mmi.12468. [DOI] [PubMed] [Google Scholar]
- 5.El-Sharoud WM, Niven GW. The influence of ribosome modulation factor on the survival of stationary-phase Escherichia coli during acid stress. Microbiology. 2007;153:247–253. doi: 10.1099/mic.0.2006/001552-0. [DOI] [PubMed] [Google Scholar]
- 6.Yamagishi M, et al. Regulation of the Escherichia coli rmf gene encoding the ribosome modulation factor: Growth phase- and growth rate-dependent control. EMBO J. 1993;12:625–630. doi: 10.1002/j.1460-2075.1993.tb05695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKay SL, Portnoy DA. Ribosome hibernation facilitates tolerance of stationary-phase bacteria to aminoglycosides. Antimicrob Agents Chemother. 2015;59:6992–6999. doi: 10.1128/AAC.01532-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kline BC, McKay SL, Tang WW, Portnoy DA. The Listeria monocytogenes hibernation-promoting factor is required for the formation of 100S ribosomes, optimal fitness, and pathogenesis. J Bacteriol. 2015;197:581–591. doi: 10.1128/JB.02223-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hood RD, Higgins SA, Flamholz A, Nichols RJ, Savage DF. The stringent response regulates adaptation to darkness in the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci USA. 2016;113:E4867–E4876. doi: 10.1073/pnas.1524915113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akiyama T, et al. Resuscitation of Pseudomonas aeruginosa from dormancy requires hibernation promoting factor (PA4463) for ribosome preservation. Proc Natl Acad Sci USA. 2017;114:3204–3209. doi: 10.1073/pnas.1700695114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williamson KS, et al. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J Bacteriol. 2012;194:2062–2073. doi: 10.1128/JB.00022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell JB, Cook GM. Energetics of bacterial growth: Balance of anabolic and catabolic reactions. Microbiol Rev. 1995;59:48–62. doi: 10.1128/mr.59.1.48-62.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai X, et al. Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nat Microbiol. 2016;2:16231. doi: 10.1038/nmicrobiol.2016.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ueta M, et al. Conservation of two distinct types of 100S ribosome in bacteria. Genes Cells. 2013;18:554–574. doi: 10.1111/gtc.12057. [DOI] [PubMed] [Google Scholar]
- 15.Shcherbakova K, Nakayama H, Shimamoto N. Role of 100S ribosomes in bacterial decay period. Genes Cells. 2015;20:789–801. doi: 10.1111/gtc.12273. [DOI] [PubMed] [Google Scholar]
- 16.Wada A. Growth phase coupled modulation of Escherichia coli ribosomes. Genes Cells. 1998;3:203–208. doi: 10.1046/j.1365-2443.1998.00187.x. [DOI] [PubMed] [Google Scholar]
- 17.Wada A, Mikkola R, Kurland CG, Ishihama A. Growth phase-coupled changes of the ribosome profile in natural isolates and laboratory strains of Escherichia coli. J Bacteriol. 2000;182:2893–2899. doi: 10.1128/jb.182.10.2893-2899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beckert B, et al. Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. EMBO J. 2017;36:2061–2072. doi: 10.15252/embj.201696189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matzov D, et al. The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus. Nat Commun. 2017 doi: 10.1038/s41467-017-00753-8. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khusainov I, et al. Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF. EMBO J. 2017;36:2073–2087. doi: 10.15252/embj.201696105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maki Y, Yoshida H, Wada A. Two proteins, YfiA and YhbH, associated with resting ribosomes in stationary phase Escherichia coli. Genes Cells. 2000;5:965–974. doi: 10.1046/j.1365-2443.2000.00389.x. [DOI] [PubMed] [Google Scholar]
- 22.Ueta M, et al. Role of HPF (hibernation promoting factor) in translational activity in Escherichia coli. J Biochem. 2008;143:425–433. doi: 10.1093/jb/mvm243. [DOI] [PubMed] [Google Scholar]
- 23.Ueta M, et al. Ribosome binding proteins YhbH and YfiA have opposite functions during 100S formation in the stationary phase of Escherichia coli. Genes Cells. 2005;10:1103–1112. doi: 10.1111/j.1365-2443.2005.00903.x. [DOI] [PubMed] [Google Scholar]
- 24.Wada A, Igarashi K, Yoshimura S, Aimoto S, Ishihama A. Ribosome modulation factor: Stationary growth phase-specific inhibitor of ribosome functions from Escherichia coli. Biochem Biophys Res Commun. 1995;214:410–417. doi: 10.1006/bbrc.1995.2302. [DOI] [PubMed] [Google Scholar]
- 25.Kato T, et al. Structure of the 100S ribosome in the hibernation stage revealed by electron cryomicroscopy. Structure. 2010;18:719–724. doi: 10.1016/j.str.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 26.Ortiz JO, et al. Structure of hibernating ribosomes studied by cryoelectron tomography in vitro and in situ. J Cell Biol. 2010;190:613–621. doi: 10.1083/jcb.201005007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polikanov YS, Blaha GM, Steitz TA. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science. 2012;336:915–918. doi: 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agafonov DE, Spirin AS. The ribosome-associated inhibitor A reduces translation errors. Biochem Biophys Res Commun. 2004;320:354–358. doi: 10.1016/j.bbrc.2004.05.171. [DOI] [PubMed] [Google Scholar]
- 29.Vila-Sanjurjo A, Schuwirth BS, Hau CW, Cate JH. Structural basis for the control of translation initiation during stress. Nat Struct Mol Biol. 2004;11:1054–1059. doi: 10.1038/nsmb850. [DOI] [PubMed] [Google Scholar]
- 30.Sharma MR, et al. PSRP1 is not a ribosomal protein, but a ribosome-binding factor that is recycled by the ribosome-recycling factor (RRF) and elongation factor G (EF-G) J Biol Chem. 2010;285:4006–4014. doi: 10.1074/jbc.M109.062299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bieri P, Leibundgut M, Saurer M, Boehringer D, Ban N. The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J. 2017;36:475–486. doi: 10.15252/embj.201695959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aiso T, Yoshida H, Wada A, Ohki R. Modulation of mRNA stability participates in stationary-phase-specific expression of ribosome modulation factor. J Bacteriol. 2005;187:1951–1958. doi: 10.1128/JB.187.6.1951-1958.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida H, Ueta M, Maki Y, Sakai A, Wada A. Activities of Escherichia coli ribosomes in IF3 and RMF change to prepare 100S ribosome formation on entering the stationary growth phase. Genes Cells. 2009;14:271–280. doi: 10.1111/j.1365-2443.2008.01272.x. [DOI] [PubMed] [Google Scholar]
- 34.Hirokawa G, et al. The role of ribosome recycling factor in dissociation of 70S ribosomes into subunits. RNA. 2005;11:1317–1328. doi: 10.1261/rna.2520405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwakura N, et al. Chemical and structural characterization of a model Post-Termination Complex (PoTC) for the ribosome recycling reaction: Evidence for the release of the mRNA by RRF and EF-G. PLoS One. 2017;12:e0177972. doi: 10.1371/journal.pone.0177972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ueta M, Wada C, Wada A. Formation of 100S ribosomes in Staphylococcus aureus by the hibernation promoting factor homolog SaHPF. Genes Cells. 2010;15:43–58. doi: 10.1111/j.1365-2443.2009.01364.x. [DOI] [PubMed] [Google Scholar]
- 37.Suwastika IN, et al. Evidence for lateral gene transfer (LGT) in the evolution of eubacteria-derived small GTPases in plant organelles. Front Plant Sci. 2014;5:678. doi: 10.3389/fpls.2014.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto H, et al. 70S-scanning initiation is a novel and frequent initiation mode of ribosomal translation in bacteria. Proc Natl Acad Sci USA. 2016;113:E1180–E1189. doi: 10.1073/pnas.1524554113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirokawa G, et al. Post-termination complex disassembly by ribosome recycling factor, a functional tRNA mimic. EMBO J. 2002;21:2272–2281. doi: 10.1093/emboj/21.9.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orelle C, et al. Protein synthesis by ribosomes with tethered subunits. Nature. 2015;524:119–124. doi: 10.1038/nature14862. [DOI] [PubMed] [Google Scholar]
- 41.Zavialov AV, Hauryliuk VV, Ehrenberg M. Splitting of the posttermination ribosome into subunits by the concerted action of RRF and EF-G. Mol Cell. 2005;18:675–686. doi: 10.1016/j.molcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 42.Feng B, et al. Structural and functional insights into the mode of action of a universally conserved Obg GTPase. PLoS Biol. 2014;12:e1001866. doi: 10.1371/journal.pbio.1001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Inouye M. RatA (YfjG), an Escherichia coli toxin, inhibits 70S ribosome association to block translation initiation. Mol Microbiol. 2011;79:1418–1429. doi: 10.1111/j.1365-2958.2010.07506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corrigan RM, Bellows LE, Wood A, Gründling A. ppGpp negatively impacts ribosome assembly affecting growth and antimicrobial tolerance in gram-positive bacteria. Proc Natl Acad Sci USA. 2016;113:E1710–E1719. doi: 10.1073/pnas.1522179113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Häuser R, et al. RsfA (YbeB) proteins are conserved ribosomal silencing factors. PLoS Genet. 2012;8:e1002815. doi: 10.1371/journal.pgen.1002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, et al. HflX is a ribosome-splitting factor rescuing stalled ribosomes under stress conditions. Nat Struct Mol Biol. 2015;22:906–913. doi: 10.1038/nsmb.3103. [DOI] [PubMed] [Google Scholar]
- 47.Coatham ML, Brandon HE, Fischer JJ, Schümmer T, Wieden HJ. The conserved GTPase HflX is a ribosome splitting factor that binds to the E-site of the bacterial ribosome. Nucleic Acids Res. 2016;44:1952–1961. doi: 10.1093/nar/gkv1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang B, et al. Functional study on GTP hydrolysis by the GTP-binding protein from Sulfolobus solfataricus, a member of the HflX family. J Biochem. 2010;148:103–113. doi: 10.1093/jb/mvq039. [DOI] [PubMed] [Google Scholar]
- 49.Wu H, et al. Structure of the ribosome associating GTPase HflX. Proteins. 2010;78:705–713. doi: 10.1002/prot.22599. [DOI] [PubMed] [Google Scholar]
- 50.Jain N, Vithani N, Rafay A, Prakash B. Identification and characterization of a hitherto unknown nucleotide-binding domain and an intricate interdomain regulation in HflX-a ribosome binding GTPase. Nucleic Acids Res. 2013;41:9557–9569. doi: 10.1093/nar/gkt705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shields MJ, Fischer JJ, Wieden HJ. Toward understanding the function of the universally conserved GTPase HflX from Escherichia coli: A kinetic approach. Biochemistry. 2009;48:10793–10802. doi: 10.1021/bi901074h. [DOI] [PubMed] [Google Scholar]
- 52.Polkinghorne A, et al. Chlamydophila pneumoniae HflX belongs to an uncharacterized family of conserved GTPases and associates with the Escherichia coli 50S large ribosomal subunit. Microbiology. 2008;154:3537–3546. doi: 10.1099/mic.0.2008/022137-0. [DOI] [PubMed] [Google Scholar]
- 53.Buckstein MH, He J, Rubin H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J Bacteriol. 2008;190:718–726. doi: 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chuang SE, Blattner FR. Characterization of twenty-six new heat shock genes of Escherichia coli. J Bacteriol. 1993;175:5242–5252. doi: 10.1128/jb.175.16.5242-5252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol. 2015;13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geiger T, Kästle B, Gratani FL, Goerke C, Wolz C. Two small (p)ppGpp synthases in Staphylococcus aureus mediate tolerance against cell envelope stress conditions. J Bacteriol. 2014;196:894–902. doi: 10.1128/JB.01201-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mäder U, et al. Staphylococcus aureus transcriptome architecture: From laboratory to infection-mimicking conditions. PLoS Genet. 2016;12:e1005962. doi: 10.1371/journal.pgen.1005962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paier A, Leppik M, Soosaar A, Tenson T, Maiväli Ü. The effects of disruptions in ribosomal active sites and in intersubunit contacts on ribosomal degradation in Escherichia coli. Sci Rep. 2015;5:7712. doi: 10.1038/srep07712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Piir K, Paier A, Liiv A, Tenson T, Maiväli U. Ribosome degradation in growing bacteria. EMBO Rep. 2011;12:458–462. doi: 10.1038/embor.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zundel MA, Basturea GN, Deutscher MP. Initiation of ribosome degradation during starvation in Escherichia coli. RNA. 2009;15:977–983. doi: 10.1261/rna.1381309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kohler R, Mooney RA, Mills DJ, Landick R, Cramer P. Architecture of a transcribing-translating expressome. Science. 2017;356:194–197. doi: 10.1126/science.aal3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gualerzi CO, Pon CL. Initiation of mRNA translation in prokaryotes. Biochemistry. 1990;29:5881–5889. doi: 10.1021/bi00477a001. [DOI] [PubMed] [Google Scholar]
- 63.Majerczyk CD, et al. Direct targets of CodY in Staphylococcus aureus. J Bacteriol. 2010;192:2861–2877. doi: 10.1128/JB.00220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reiss S, et al. Global analysis of the Staphylococcus aureus response to mupirocin. Antimicrob Agents Chemother. 2012;56:787–804. doi: 10.1128/AAC.05363-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lau CH, van Engelen K, Gordon S, Renaud J, Topp E. Novel antibiotic resistance determinants from agricultural soil exposed to antibiotics widely used in human medicine and animal farming. Appl Environ Microbiol. 2017 doi: 10.1128/aem.00989-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hong JJ, et al. Microarray analysis in Tourette syndrome postmortem putamen. J Neurol Sci. 2004;225:57–64. doi: 10.1016/j.jns.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 67.Lee NR, et al. Dosage effects of X and Y chromosomes on language and social functioning in children with supernumerary sex chromosome aneuploidies: Implications for idiopathic language impairment and autism spectrum disorders. J Child Psychol Psychiatry. 2012;53:1072–1081. doi: 10.1111/j.1469-7610.2012.02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lek M, et al. Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fey PD, et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio. 2013;4:e00537–e12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fuller JR, et al. Identification of a lactate-quinone oxidoreductase in Staphylococcus aureus that is essential for virulence. Front Cell Infect Microbiol. 2011;1:19. doi: 10.3389/fcimb.2011.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee CY, Buranen SL, Ye ZH. Construction of single-copy integration vectors for Staphylococcus aureus. Gene. 1991;103:101–105. doi: 10.1016/0378-1119(91)90399-v. [DOI] [PubMed] [Google Scholar]
- 72.Pattee PA, Neveln DS. Transformation analysis of three linkage groups in Staphylococcus aureus. J Bacteriol. 1975;124:201–211. doi: 10.1128/jb.124.1.201-211.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Davis AR, Gohara DW, Yap MN. Sequence selectivity of macrolide-induced translational attenuation. Proc Natl Acad Sci USA. 2014;111:15379–15384. doi: 10.1073/pnas.1410356111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spedding G. Ribosomes and Protein Synthesis: A Practical Approach. Oxford University Press; Oxford: 1990. [Google Scholar]
- 75.Schneewind O, Model P, Fischetti VA. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]