Mitosis involves complex interactions of multiple pathways, some yet uncharacterized. Tipton et al. use bioinformatics to implicate GTSE1 in mitosis. Depletion of GTSE1 disrupts chromosome alignment and spindle pole integrity, in part because of hyperstabilization of spindle microtubules and depletion of Aurora B kinase and Kif4A chromokinesin on chromosome arms.
Abstract
In mitosis, the dynamic assembly and disassembly of microtubules are critical for normal chromosome movement and segregation. Microtubule turnover varies among different mitotic spindle microtubules, dictated by their spatial distribution within the spindle. How turnover among the various classes of spindle microtubules is differentially regulated and the resulting significance of differential turnover for chromosome movement remains a mystery. As a new tactic, we used global microarray meta-analysis (GAMMA), a bioinformatic method, to identify novel regulators of mitosis, and in this study, we describe G2- and S phase–expressed protein 1 (GTSE1). GTSE1 is expressed exclusively in late G2 and M phase. From nuclear envelope breakdown until anaphase onset, GTSE1 binds preferentially to the most stable mitotic spindle microtubules and promotes their turnover. Cells depleted of GTSE1 show defects in chromosome alignment at the metaphase plate and in spindle pole integrity. These defects are coupled with an increase in the proportion of stable mitotic spindle microtubules. A consequence of this reduced microtubule turnover is diminished recruitment and activity of Aurora B kinase on chromosome arms. This decrease in Aurora B results in diminished binding of the chromokinesin Kif4A to chromosome arms.
Introduction
The alignment of chromosomes at metaphase is a well-conserved feature of mitosis in higher eukaryotes. This feature of mitosis promotes the equal distribution of sister chromatids into the two daughter cells at anaphase. Stepwise mitotic progression requires proteolytic degradation of key mitotic substrates that fosters chromatid separation followed by cytokinesis and mitotic exit (Musacchio and Salmon, 2007). The force-producing machines, including microtubule dynamics, microtubule-associated proteins, and microtubule motors, are under intense investigation, but we still lack complete understanding of how bipolar metaphase alignment is achieved and maintained (Walczak et al., 2010). Therefore, it is essential to unveil novel mitotic regulators and understand how they interact with and influence known pathways that drive and maintain metaphase.
Factors important in chromosome movement and spindle assembly are microtubule-binding proteins, microtubule-dependent motors, and microtubule depolymerases (Rieder and Salmon, 1994; Kosco et al., 2001; Kapoor and Compton, 2002; Kline-Smith et al., 2004; Schneider et al., 2007; Bakhoum et al., 2009a; Verhey and Hammond, 2009; Fu et al., 2010; Barisic et al., 2014). Balance between poleward forces primarily acting at kinetochores and polar ejection forces acting upon chromosome arms is likely driven by regulated microtubule assembly and disassembly and by microtubule-dependent motor proteins, which play important roles in mediating chromosome alignment and spindle stability during mitosis. In early mitosis, dynein, a minus end–directed motor protein found on kinetochores, moves chromosomes toward the spindle poles (Li et al., 2007; Yang et al., 2007; Vorozhko et al., 2008). The plus end–directed motor protein CENP-E (kinesin 7) at kinetochores functions to transport chromosomes trapped near spindle poles along microtubules toward the cell equator (Kapoor et al., 2006; Cai et al., 2009; Kim et al., 2010). Additionally, the activities of the plus end–directed chromokinesins Kid (kinesin 10) and Kif4A (kinesin 4) found on chromosome arms generate ejection forces pushing the arms away from the poles (Rieder et al., 1986; Antonio et al., 2000; Funabiki and Murray, 2000; Brouhard and Hunt, 2005; Stumpff et al., 2012; Wandke et al., 2012). Although much has been revealed regarding the regulation of kinetochores in moving chromosomes, the roles of arm-based ejection force pathways remain relatively unexplored and controversial.
G2- and S phase–expressed protein 1 (GTSE1) is a microtubule-associated protein originally identified as a p53-inducible gene that was previously described to function in controlling DNA damage–induced apoptosis by down-regulating p53 function during interphase (Utrera et al., 1998; Collavin et al., 2000; Monte et al., 2000, 2003, 2004). Additionally, GTSE1 has been shown to function as an EB1-dependent plus end–tracking protein that is required for cell migration during interphase (Scolz et al., 2012). After nuclear envelope breakdown (NEB) in mitosis, GTSE1 microtubule plus end tip tracking is inhibited until anaphase onset, and instead, the protein decorates the microtubule lattice. Additionally, GTSE1 becomes hyperphosphorylated upon entry into mitosis (Collavin et al., 2000; Scolz et al., 2012). Most recently, GTSE1 was reported to inhibit mitotic centromere-associated kinesin (MCAK) microtubule depolymerase activity during mitosis and thereby promote microtubule stability in mitosis (Bendre et al., 2016). In this study, in contrast, we provide evidence that GTSE1 fosters turnover of the most stable microtubules within the mitotic spindle from prometaphase to anaphase onset. At anaphase onset, GTSE1 redistributes to the astral microtubules, concomitant with its return to tip tracking. We speculate that this redistribution aids in stabilizing midzone microtubules during anaphase and telophase. Cells depleted of GTSE1 display hyperstabilized spindle microtubules, which in turn affects the activity of the mitotic kinase Aurora B—specifically on chromosome arms. The loss of Aurora B activity on chromosome arms diminishes accumulation of the chromokinesin Kif4A. Depletion of Kif4A induces multipolar spindles. In sum, we have identified a novel pathway in which GTSE1 is an upstream regulator of microtubule stability, chromosome alignment, spindle pole integrity, and timely mitotic progression.
Results
Bioinformatic identification of a novel mitotic effector, GTSE1
A bioinformatic algorithm called global microarray meta-analysis (GAMMA) was used to identify potential novel mitotic regulators among gene products with little or no previously established function in mitosis (see the Bioinformatic characterization of GTSE1 section of Materials and methods; Wren, 2009; Dozmorov et al., 2011). GAMMA uses a “guilt by association” approach to infer the function of genes based on what is known about their most correlated transcripts, and it can be used to predict functions even if no literature exists on the gene being analyzed. Among the candidate mitotic regulators, we selected GTSE1 for further analysis because previous work had shown GTSE1 to interact with microtubules (Utrera et al., 1998; Monte et al., 2003, 2004; Bublik et al., 2010; Liu et al., 2010; Scolz et al., 2012). GAMMA was then used to predict specific functions and interactions of GTSE1. The relevant associations confirmed in this study as well as those found in previous work are summarized in Table 1. The complete list of GAMMA-predicted functional, phenotypic, and genetic associations is provided in Table S1.
Table 1. Summary of highest-scoring new and previously confirmed associations of GTSE1 as predicted by GAMMA analysis.
| Predicted Association | GAMMA Score | References |
|---|---|---|
| Kif4A | 405 | This study |
| Cyclin B2 | 358 | Wang and Adjaye, 2011; Barlin et al., 2015; Gridley et al., 2015 |
| Spindle assembly | 341 | This study |
| Aurora B | 336 | This study |
| Spindle microtubules | 304 | This study |
| Mitotic spindle | 281 | This study |
| Chromosome congression | 209 | This study |
| Misaligned chromosomes | 164 | This study |
| Chromosome biorientation | 152 | This study |
| Nek2 | 142 | Pfleger and Kirschner, 2000 |
| PLK1 | 131 | Liu et al., 2010; Wang and Adjaye, 2011 |
| Cyclin B1 | 121 | Taylor and Stark, 2001 |
| APC/C | 120 | Pfleger and Kirschner, 2000 |
A higher GAMMA score indicates a stronger predicted association. A complete table of predictions is shown in Table S1.
Endogenous GTSE1 protein expression is highly restricted to M phase and localizes to spindle microtubules during mitosis
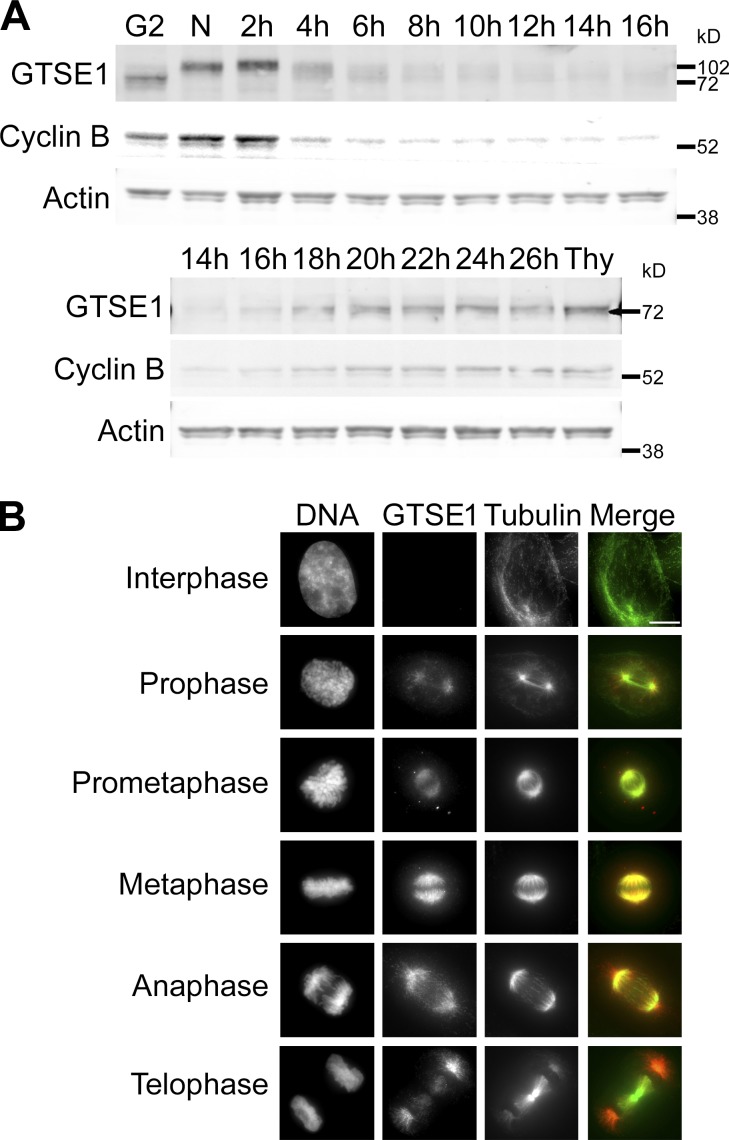
To map the expression and distribution of endogenous GTSE1, we generated a polyclonal antibody targeting the N terminus of the protein. In agreement with previous work (Collavin et al., 2000), we found that GTSE1 protein levels became detectable in an enriched G2 phase population, peaked during mitosis (N lane), and fell to very low levels in G1 (Fig. 1 A, top), consistent with its identification as a target for the mitotic ubiquitin ligase, the anaphase-promoting complex/cyclosome (APC/C; Pfleger and Kirschner, 2000). GTSE1 protein levels remained very low during G1 and S phase until ∼20 h after release from nocodazole, consistent with reentry into the G2 phase of the cell cycle (Fig. 1 A, bottom). Importantly, the GTSE1 protein expression profile mimicked that of the known mitotic regulator cyclin B. GTSE1 also undergoes posttranslational modification during mitosis, as demonstrated by the increase in its electrophoretic mobility during exit from mitosis (Fig. 1 A, top). This mitotic shift had been shown previously to be a result of M phase–specific phosphorylation (Collavin et al., 2000; Scolz et al., 2012). Of note, GTSE1 protein levels accumulated in cells arrested in S phase by thymidine treatment (Thy lane; Fig. 1 A, bottom). This finding provides a plausible explanation for past work describing high levels of GTSE1 expression in S phase because thymidine was used in synchronization (Liu et al., 2010). Upon tracking endogenous GTSE1 by immunofluorescence, we found that GTSE1 was largely undetectable in most interphase cells. However, GTSE1 was readily detectable in mitotic cells and found to decorate the mitotic spindle throughout mitosis (Fig. 1 B). The localization of GTSE1 to spindle microtubules during mitosis is consistent with it being a known microtubule-binding protein (Scolz et al., 2012). Interestingly, GTSE1 appears to predominantly decorate astral microtubules in anaphase and telophase cells. Collectively, these data indicate that GTSE1 protein expression is strongly cell cycle–regulated, with protein levels peaking during mitosis. GTSE1 binds mitotic spindle microtubules from prometaphase to anaphase onset, where it then concentrates on astral microtubules.
Figure 1.
GTSE1 protein expression peaks during G2 and M phase and localizes to mitotic spindle microtubules. (A) Western blot showing GTSE1 protein expression from HeLa cells after release from nocodazole for the indicated times. The N lane indicates cells in nocodazole and the 0 time point. The G2 lane indicates the adherent cell population that was collected after mitotic shakeoff from the 0 time point (N lane). The Thy lane indicates cells that were released from nocodazole after 16 h directly into thymidine for 24 h. Cyclin B protein levels were also probed to show cell cycle specificity. Actin is shown as a control for loading levels. Cells exit mitosis by ∼4 h after release from nocodazole as denoted by cyclin B degradation. GTSE1 levels remained very low after exit from mitosis until ∼20 h after release from nocodazole, which is a time consistent with reentry into G2 phase. During thymidine-induced S phase arrest (Thy lane), GTSE1 protein accumulated. (B) Immunofluorescence images of endogenous GTSE1 in unsynchronized HeLa cells during interphase and different stages of mitosis. GTSE1 was largely undetectable in most interphase cells. During late G2/prophase, it began to accumulate near centrosomes and became concentrated on mitotic spindle microtubules during mitosis. Cells were coimmunostained for DNA (DAPI) and α-tubulin. Images represent maximum-intensity projections. GTSE1 images are scaled equivalently. Bar, 10 µm.
GTSE1 is required for chromosome alignment and a timely progression through mitosis
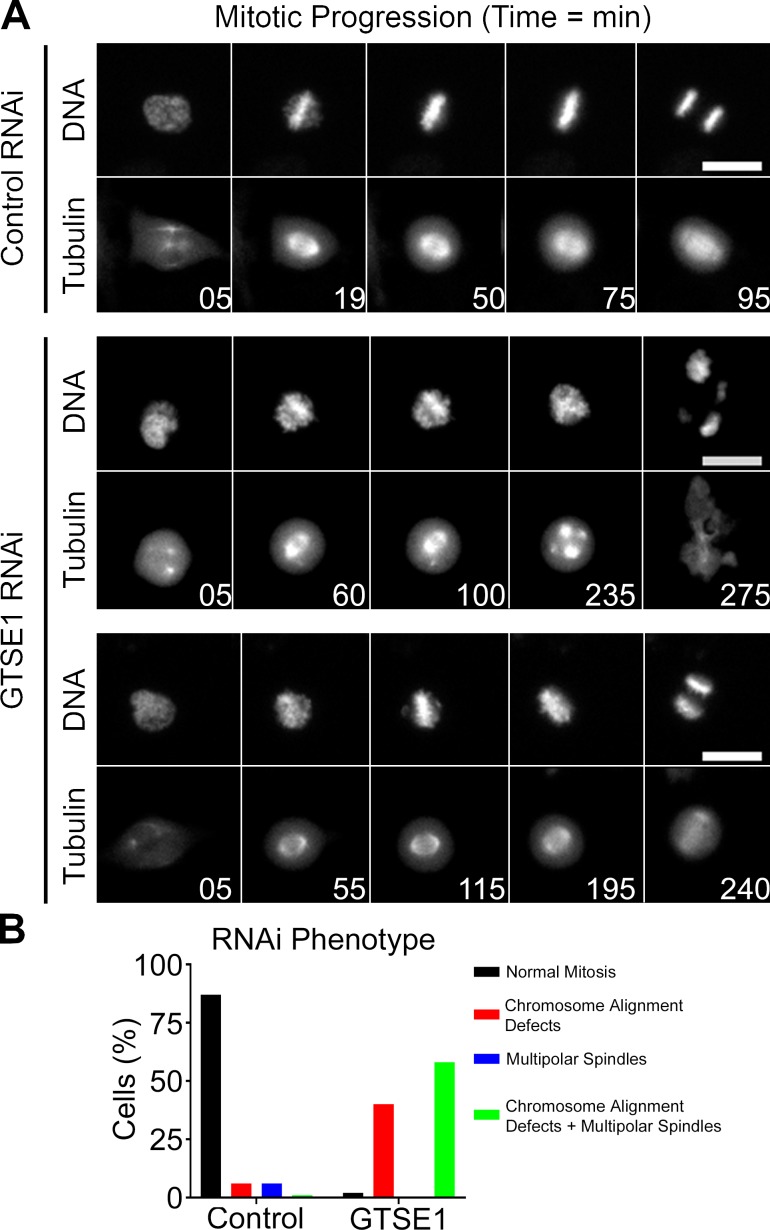
The localization data were consistent with a potential role for GTSE1 as a novel mitotic regulator. To test this idea, we performed RNAi knockdown combined with time-lapse microscopy experiments in HeLa cells stably expressing H2B-RFP and GFP-tubulin (Fig. 2 A and Videos 1, 2, and 3). siRNA transfection resulted in a reduction of ≥97% of the respective protein levels as determined by immunoblotting after 48 h (Fig. S4 B; GTSE1 [GT] lane). 36 h after depletion of GTSE1, the number of cells undergoing a successful mitosis (see Fig. 2 legend) was dramatically reduced from 87% in control siRNA–transfected cells to 2% in GTSE1 siRNA–transfected cells (Fig. 2 B). The majority of GTSE1-depleted cells displayed both defects in chromosome alignment and formed multipolar spindles that prevented cells from achieving a bipolar metaphase (Fig. 2 A, middle; and Fig. 2 B). In some instances, GTSE1-depleted cells showed chromosome alignment defects without becoming multipolar (Fig. 2 A, bottom; and Fig. 2 B). When mitotic duration was monitored, i.e., the time from NEB to anaphase onset, we found that the mean duration of mitosis in cells depleted of GTSE1 was 285 min compared with 92 min in control cells (Fig. S1 A). This increase in mitotic duration was caused by difficulty achieving metaphase after NEB (43 min in control cells and 215 min in GTSE1-depleted cells; Fig. S1 B). The duration of metaphase was also slightly longer in GTSE1-depleted cells (70 min in GTSE1-depleted cells vs. 49 min in control cells; Fig. S1 C). Similar results were obtained when GTSE1 depletion combined with time-lapse microscopy experiments were performed in U2OS cells (Fig. S1, D–G; Bendre et al., 2016). Thus, GTSE1 is a critical regulator of normal chromosome movement and spindle integrity during mitosis.
Figure 2.
GTSE1 depletion causes defects in mitotic progression. (A) Select frames from live-cell imaging of HeLa cells stably expressing H2B-RFP and GFP-tubulin after transfection of either control or GTSE1 siRNA (see Videos 1, 2, and 3). For cells transfected with GTSE1 siRNA, examples of both chromosome alignment defects and multipolar spindles (middle) and chromosome alignment defects alone (bottom) are shown. Bars, 10 µm. (B) Quantification of mitotic phenotypes as assessed from live-cell imaging in A. Note that in GTSE1-depleted cells, only a small population (2%) underwent normal mitosis, whereas the majority of cells displayed both chromosome alignment defects and multipolar spindles (58% of cells) or chromosome alignment defects alone (40% of cells). As shown in the middle, most cells exhibited multipolar spindles after prolonged prometaphase arrest. Cells completing a bipolar anaphase within 3 h of NEB were classified as undergoing normal mitosis. Chromosome alignment defects and multipolar spindles were scored using DNA and microtubule morphology. Control n = 140 cells, and GTSE1 n = 100 cells, from two independent experiments.
GTSE1 regulates microtubule dynamics during mitosis
Our immunolabeling results and a previous study of exogenously expressed GFP-tagged GTSE1 (Scolz et al., 2012) showed that GTSE1 bound the walls of spindle microtubules from prometaphase to anaphase onset. This localization suggested a role for GTSE1 in regulating spindle microtubule dynamics. To test this, we measured fluorescence dissipation after photoactivation of microtubules in U2OS cells stably expressing photoactivatable GFP (PAGFP)-tubulin. Cells transfected with either control or GTSE1 siRNA were analyzed. Only cells entering mitosis during the time frame of imaging were photoactivated to ensure tubulin turnover was not affected by prolonged mitotic arrest. A bar-shaped region within the spindle was photoactivated perpendicular to the long axis of the mitotic spindle and adjacent to the mass of chromosomes as determined by phase-contrast microscopy. Time-lapse images were acquired before and after photoactivation by spinning-disk confocal microscopy. Representative images depict fluorescence in prephotoactivated and postphotoactivated cells (Fig. 3 A, top). In cells depleted of GTSE1, the photoactivated population of tubulin persisted longer as compared with control cells. Measurements of fluorescence intensity minus background were collected after photoactivation at each time interval and were corrected for photobleaching using measurements of photoactivated spindles in cells treated with 10 µM taxol. Plots were generated for control cells and GTSE1-depleted cells by averaging corrected data at each time point and fit with a double exponential curve F = A1 × exp(−k1 × t) + A2 × exp(−k2 × t), where A1 and A2 represent the percent total fluorescence contribution of the fast turnover (less stable) and slow turnover (more stable) microtubule populations, and k1 and k2 represent the respective decay rate constants (Fig. 3 A, bottom). Although the total fluorescence contribution of the fast and slow microtubule populations in control cells was measured to be 56.29% (53.32 and 59.26%; 95% CI) and 43.98% (41.77 and 46.19%; 95% CI), respectively, cells depleted of GTSE1 had both a reduction in the fast microtubule population to 41.53% (38.33 and 44.73%; 95% CI) and an increase in the slow microtubule population to 58.61% (56.21 and 61.02%; 95% CI) when compared with controls. This suggests that in the absence of GTSE1, the stability of microtubule populations is altered, favoring an increase in the slow microtubule population. When microtubule half-lives were examined, little change was observed between control and GTSE1-depleted cells in the fast microtubule population (control t1/2 = 10 s; GTSE1 t1/2 = 12 s). In contrast, the half-life of the slow population significantly increased after GTSE1 depletion (t1/2 = 587 s) when compared with controls (t1/2 = 289 s). A modeling approach was used to elucidate the effects GTSE1 depletion has on the turnover rate, the amount, or both the turnover rate and amount of the slow microtubule population. The GTSE1 fluorescence after dissipation curve was plotted along with control curves varying either the rate (k2), the amount (A2), or both the rate and amount (k2 and A2 from the double exponential curve above; Fig. S2, A–C). Varying the rate of turnover (k2) alone of the slow microtubule population in the control curve (Fig. S2 A) did not provide close modeling for the data obtained after GTSE1 depletion. Varying the amount (A2) alone of the slow microtubule population (Fig. S2 B) generated a better fit. However, varying both the turnover rate and amount (k2 and A2) of the slow population of microtubules (Fig. S2 C) provided the best fit to the GTSE1 fluorescence after dissipation data. These findings are consistent with GTSE1 depletion affecting the amount and, to a lesser extent, the rate of the slow population.
Figure 3.
Spindle microtubule stability is enhanced by depletion of GTSE1. (A, top) Select frames from live-cell imaging of tubulin photoactivation in metaphase U2OS cells stably expressing PAGFP-tubulin after transfection with either control or GTSE1 siRNA. Only cells entering mitosis during imaging were photoactivated to ensure tubulin turnover was not affected by prolonged mitotic arrest. Time is in s. (Bottom) Fluorescence dissipation after photoactivation. The filled and unfilled circles represent the mean values recorded at each time point after photoactivation. The bars represent SEM. Control n = 22 cells; GTSE1 n = 18 cells from four independent experiments. Lines indicate fitted curves (control R2 = 0.998; GTSE1 R2 = 0.995). F = A1 × exp(−k1 × t) + A2 × exp(−k2 × t) where A1 and A2 represent the percent total fluorescence, and k1 and k2 represent the respective decay rate constants. (B, left) Immunofluorescence of α-tubulin levels in methanol-fixed HeLa cells transfected with either control or GTSE1 siRNA. Images represent maximum-intensity projections. α-Tubulin levels are scaled equivalently. (Right) Inner spindle microtubule (MT) fluorescence intensity quantification of α-tubulin after control or GTSE1 siRNA transfection. The fluorescence intensity from a region encompassing only inner spindle microtubules was analyzed (the dashed circle in the top left image marks region of interest). GTSE1 depletion enhanced the levels of α-tubulin found in the inner spindle when compared with control cells. SD is plotted for the mean from each condition. n = 24 cells from two independent experiments for each condition. **, P < 0.01. (C) Immunofluorescence of GTSE1 in HeLa cells that were untreated or cold treated to examine localization to the more stable microtubule population, or transfected with Hec1 siRNA to examine localization in cells unable to make kinetochore–microtubule interactions. GTSE1 labeling of spindle microtubules was largely unaffected by cold treatment, but it failed to accumulate on microtubules after depletion of Hec1. Images represent maximum-intensity projections. Bars, 10 µm.
To further examine the changes in microtubule stability after GTSE1 depletion, we measured the microtubule density within the inner spindle by quantitative immunofluorescence (Fig. 3 B). Consistent with the increase of slow turnover microtubules observed in Fig. 3 A, cells depleted of GTSE1 showed an increase in α-tubulin fluorescence intensity in the inner spindle when compared with control cells. We next sought to determine whether GTSE1 preferentially localized to a subset of microtubules in the mitotic spindle. To test this, we examined GTSE1 localization in cells that had been cold-treated to remove highly dynamic microtubules. We compared these with cells depleted of the essential microtubule-binding component of kinetochores Hec1/Ndc80 that were preventing formation of stable kinetochore–microtubule interactions and depleting the more stable spindle microtubules (Fig. 3 C). Interestingly, although GTSE1 remained strongly distributed on the cold-resistant microtubules, little GTSE1 remained associated with the less stable spindle microtubules in cells depleted of Hec1/Ndc80 that lacked kinetochore-attached microtubules (see Discussion). Collectively with the photoactivation data, these findings suggest that GTSE1 preferentially localizes to and promotes the turnover of more stable spindle microtubules.
Aurora B kinase activity is sensitive to microtubule dynamics
Aurora B kinase is among the strongest mitotic regulators predicted by GAMMA to interact with GTSE1. Roles for Aurora B in regulating chromosome alignment during mitosis are well established (Adams et al., 2001; Kallio et al., 2002; Ditchfield et al., 2003; Jelluma et al., 2008). Previous work has also shown that Aurora B kinase activity is promoted by microtubules both in vitro and in vivo (Rosasco-Nitcher et al., 2008; Tseng et al., 2010; Banerjee et al., 2014). We examined the ability of Aurora B to phosphorylate one of its major mitotic targets on chromatin, serine 10 on histone H3 in mitotic HeLa cells in control cells, or in cells treated with nocodazole or taxol (Fig. 4). Although little change in histone H3 S10 phosphorylation was observed when comparing prometaphase and metaphase cells, a significant reduction in phosphorylation was observed in cells treated with either spindle poison (Fig. 4, bottom). These results indicate that full endogenous Aurora B kinase activity requires intact microtubules with normal dynamics.
Figure 4.
Full Aurora B kinase activity on chromosome arms requires intact and dynamic microtubules. (Top) Immunofluorescence of histone H3 pS10 levels in HeLa cells treated with DMSO, nocodazole (Noc), or taxol for 2 h. Images represent maximum-intensity projections. Histone H3 pS10 levels are scaled equivalently. Bars, 10 µm. (Bottom) Fluorescence intensity quantification of chromosome-bound histone H3 pS10 immunofluorescence in mitotic HeLa cells treated as above. Prometaphase (Prometa) and metaphase (Meta) cells were quantified separately to determine whether histone H3 pS10 levels change during mitosis. Treatment with either nocodazole or taxol for 2 h to depolymerize or stabilize microtubules, respectively, greatly reduced the levels of histone H3 pS10 on chromosomes. SD is plotted for the mean from each condition. n = 15 cells from two independent experiments. Asterisks indicate statistical significance of mean value difference when compared with prometaphase as assessed by the Kruskal-Wallis test. **, P < 0.01; ****, P < 0.0001. FI, fluorescence intensity.
Aurora B kinase activity is regulated by GTSE1 during mitosis
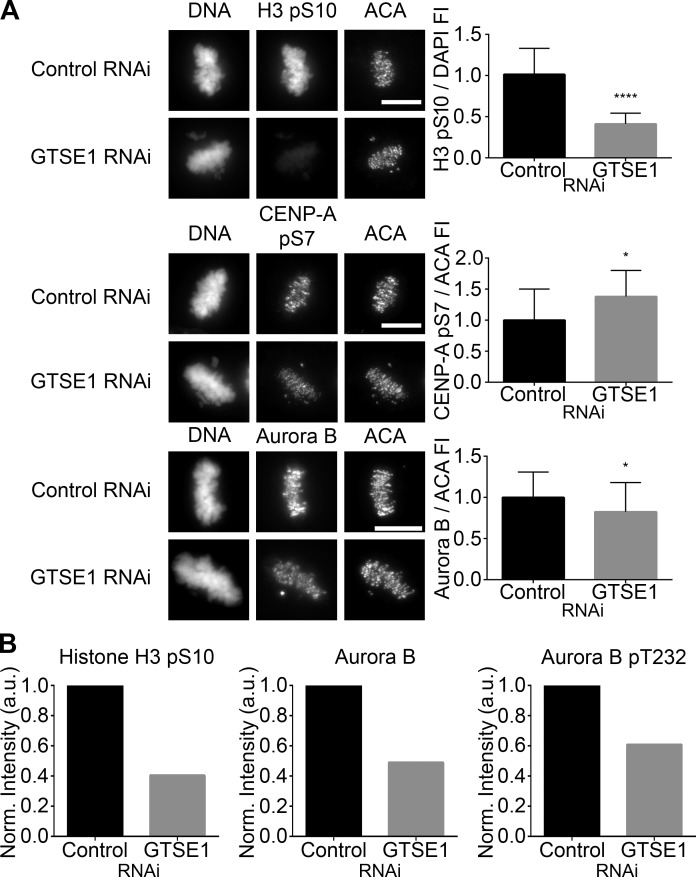
The data obtained in Figs. 3 and 4 implicate GTSE1 as a novel regulator of Aurora B activity via its ability to accelerate microtubule dynamicity during mitosis. To assess the potential link between GTSE1 and Aurora B activity, mitotic HeLa cells were transfected with control or GTSE1 siRNA, and the levels of two Aurora B phosphorylation events, histone H3 pS10 found along chromosome arms and CENP-A pS7 found at the centromere, along with the levels of kinetochore-localized Aurora B, were examined (Figs. 5 A and S3 A). The levels of histone H3 pS10 localized on chromosome arms were greatly reduced (Fig. 5 A, top). In contrast, although the amount of Aurora B detected at centromeres was slightly reduced (Fig. 5 A, bottom), phosphorylation of CENP-A on S7 was modestly increased. Similarly, strong reduction in histone H3 pS10 and slight increase in CENP-A pS7 were detected in prometaphase cells, although the change in CENP-A pS7 did not reach statistical significance (Fig. S3 A). The strong disparity of the Aurora B target on chromosome arms versus centromeres suggests that these activities are subject to differential regulation. To test these results with another method of analysis, cytosolic and chromosome fractions were isolated from mitotic HeLa cells transfected with either control or GTSE1 siRNA and analyzed by Western blot (Fig. S3 B). HeLa cells were arrested with nocodazole for 16 h to accumulate mitotic cells and then released for 25 min into drug-free medium to allow spindles to form. Quantification of the normalized protein levels revealed that although the reduction of histone H3 pS10 levels in chromosome fractions were similar to the results obtained by immunofluorescence in Fig. 5 A, there was a significant reduction, ∼51%, in the total amount of Aurora B found in the chromosome fraction from cells depleted of GTSE1 (Fig. 5 B). Collectively, this suggests that GTSE1 depletion results in loss of approximately half of the Aurora B from chromosomes. Blotting of chromosome fractions of GTSE1-depleted cells with an anti-phosphoepitope antibody recognizing the activating phosphorylation of Aurora B at T232 also showed ∼39% reduction in labeling compared to chromosome fractions from control cells. This reduction in Aurora B autophosphorylation after GTSE1 depletion and subsequent increased microtubule stability is consistent with the observations that full Aurora B kinase activity requires both intact and dynamic microtubules (Fig. 4; Rosasco-Nitcher et al., 2008; Tseng et al., 2010; Banerjee et al., 2014). Altogether, these observations suggest that GTSE1 promotes binding of Aurora B to chromosome arms. In contrast, GTSE1 depletion has more limited effects on Aurora B accumulation and activity at centromeres. However, it is currently unclear mechanistically how GTSE1 mediation of microtubule dynamics promotes the localization and activity of Aurora B preferentially on chromosome arms.
Figure 5.
GTSE1 modulates Aurora B kinase activity during mitosis. (A, left) Immunofluorescence of histone H3 pS10, CENP-A pS7, or Aurora B levels in metaphase HeLa cells transfected with either control or GTSE1 siRNA. Cells were coimmunostained for DNA and kinetochores. Images represent maximum-intensity projections. Histone H3 pS10, CENP-A pS7, and Aurora B levels are scaled equivalently in each panel. Bars, 10 µm. (Right) Fluorescence intensity quantification of chromosome-bound histone H3 pS10 and kinetochore-localized CENP-A pS7 and Aurora B immunofluorescence after control or GTSE1 siRNA transfection. Depletion of GTSE1 greatly reduced the levels of histone H3 pS10 on chromosome arms. In contrast, the level of pS7 on kinetochore-localized CENP-A was slightly increased, whereas the total amount of kinetochore-associated Aurora B was slightly decreased. Thus, GTSE1 depletion diminishes Aurora B activity on chromosome arms but increases Aurora B activity at kinetochores. SD is plotted for the mean from each condition. Histone H3 pS10 n = 20 cells, CENP-A pS7 n = 10 cells, and Aurora B n = 30 cells from two independent experiments for each condition. *, P < 0.05; ****, P < 0.0001. FI, fluorescence intensity. (B) Quantification of the chromosome-bound fractions from the immunoblot shown in Fig. S3 B. Histone H3 pS10, total Aurora B, and Aurora B pT232 levels were normalized to histone H3. Although the reduction in histone H3 pS10 levels by immunoblot were comparable to data obtained by immunofluorescence (A, top), in the absence of GTSE1, total chromosome-bound Aurora B levels were further reduced when compared with kinetochore-localized Aurora B (A, bottom), suggesting that in cells depleted of GTSE1, there is a greater loss of Aurora B from chromosome arms compared with Aurora B at kinetochores. Similarly, a fraction of Aurora B autophosphorylation activity was reduced in the absence of GTSE1.
Aurora B kinase activity regulates Kif4A localization
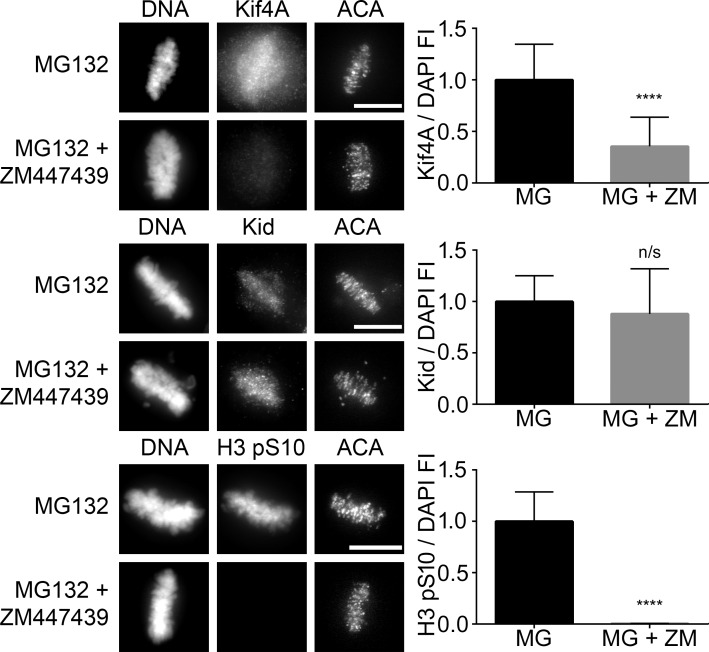
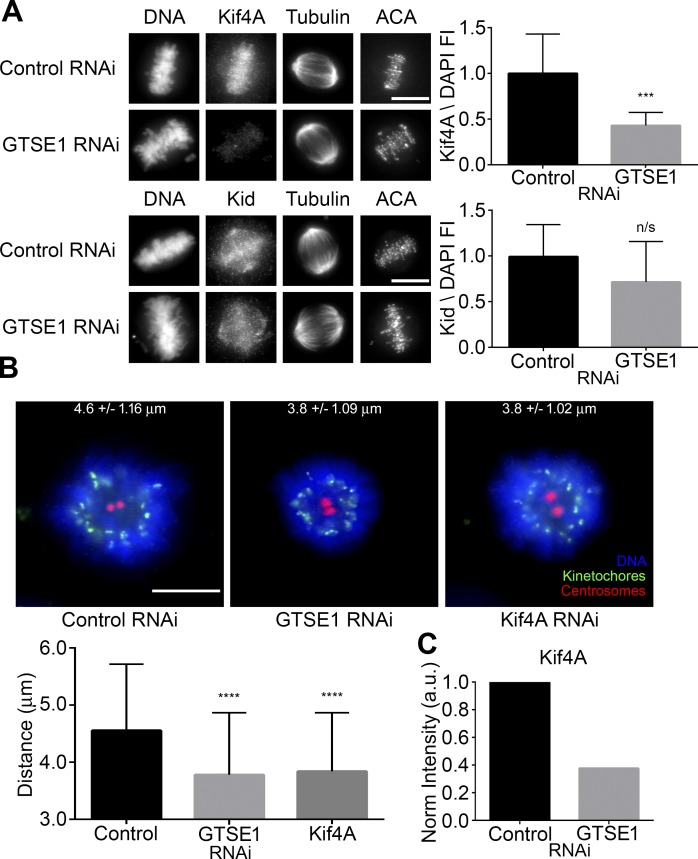
Although the roles of Aurora B at centromeres and kinetochores have been well characterized (Adams et al., 2001; Kallio et al., 2002; Ditchfield et al., 2003; DeLuca et al., 2006, 2011; Jelluma et al., 2008; Liu et al., 2009; van der Horst and Lens, 2014), it remains unclear whether and how Aurora B on chromosome arms might impact chromosome movement and spindle maintenance. The chromokinesins Kif4A and Kid localize to chromosome arms, and past studies implicate them in chromosome alignment during mitosis (Wandke et al., 2012; Barisic et al., 2014). GAMMA predicted a very high association of GTSE1 for Kif4A but not Kid (Tables 1 and S1). Kif4A was identified by proteomic and phosphoproteomic analysis to be specifically phosphorylated during mitosis, requiring the activity of both CDK1 and Aurora B (Ohta et al., 2016; Takahashi et al., 2016). In anaphase cells, it has been shown that maximal microtubule-dependent ATPase activity of Kif4A and its localization to the central spindle was dependent on its phosphorylation by Aurora B (Nunes Bastos et al., 2013). We examined both Kif4A and Kid localization during early mitosis in the presence or absence of ZM447439, a chemical inhibitor of Aurora B kinase. These experiments were performed in the presence of the proteasome inhibitor MG132 to prevent mitotic exit induced by global Aurora B inhibition. Although the levels of Kid remained relatively unaffected after inhibition of Aurora B kinase activity, the levels of Kif4A were reduced both on chromosomes and microtubules (Fig. 6, top and middle). Because Aurora B kinase activity requires both intact and dynamic microtubules, we examined Kif4A localization in mitotic HeLa cells treated with nocodazole or taxol (Fig. S3 C). A significant reduction in Kif4A was observed in cells treated with either spindle poison (Fig. S3 C, right). Collectively, these results indicate that Aurora B functions in the recruitment of Kif4A to chromosome arms in prometaphase.
Figure 6.
Aurora B inhibition reduces Kif4A levels on mitotic chromosomes. (Left) Immunofluorescence of Kif4A, Kid, or histone H3 pS10 levels in metaphase HeLa cells treated with MG132 (MG) alone or in combination with ZM447439 (ZM) for 2 h to inhibit Aurora B activity. MG132 was included to prevent mitotic exit. Cells were labeled for DNA and kinetochores. Images represent maximum-intensity projections. Kif4A, Kid, and histone H3 pS10 levels are scaled equivalently in each panel. Bars, 10 µm. (Right) Fluorescence intensity quantification of chromosome-bound Kif4A, Kid, or histone H3 pS10 immunofluorescence after treatments. Loss of Aurora B kinase activity dramatically reduced Kif4A levels without affecting the levels of Kid. Loss of histone H3 pS10 levels was used to demonstrate inhibition of Aurora B kinase activity. SD is plotted for the mean from each condition. n = 20 cells from two independent experiments for each condition. ****, P < 0.0001. FI, fluorescence intensity.
GTSE1 regulates Kif4A localization and polar ejection forces
We reasoned that GTSE1-affecting microtubule dynamics may be functioning upstream of a pathway involving chromosome arm Aurora B–mediated recruitment of Kif4A to mitotic chromosomes. We examined both Kif4A and Kid levels in control and GTSE1-depleted cells by immunofluorescence. Although Kid bound to chromatin remained relatively unaffected, Kif4A bound to chromatin and microtubules was reduced after GTSE1 depletion (Figs. 7 A and S3 D). The result was further confirmed by Western blotting of cytosolic and chromosome-bound fractions from mitotic HeLa cells transfected with either control or GTSE1 siRNA. Although the cytosolic levels of Kif4A remained similar, the chromosome-bound fraction of Kif4A was reduced (Figs. 7 C and S3 B). To determine whether global Kif4A levels are affected by GTSE1 depletion, whole-cell extracts from mitotic HeLa cells were examined by Western blot for Kif4A. Consistent with these data, cells depleted of GTSE1 showed lower global levels of Kif4A when compared with controls (Fig. S5, A and B).
Figure 7.
GTSE1 depletion reduces Kif4A levels on mitotic chromosomes and polar ejection forces in monastrol-treated cells. (A, left) Immunofluorescence of Kif4A or Kid levels in metaphase HeLa cells transfected with either control or GTSE1 siRNA. Cells were coimmunostained for DNA, tubulin, and kinetochores. Images represent maximum-intensity projections. Both Kif4A and Kid levels are scaled equivalently in each panel. (Right) Fluorescence intensity quantification of chromosome-bound Kif4A or Kid after control or GTSE1 siRNA transfection. Depletion of GTSE1 greatly reduced the levels of Kif4A bound to chromatin during metaphase, whereas Kid levels were slightly reduced but not to a significant degree. n = 15 cells from two independent experiments for each condition. FI, fluorescence intensity. (B, top) HeLa cells transfected with either control, GTSE1, or Kif4A siRNA (Wandke et al., 2012; Barisic et al., 2014) were treated with monastrol for 2 h and immunostained for DNA, kinetochores, and centrosomes (γ-tubulin). Numbers indicate mean kinetochore-to-pole distances ± SD of the pooled data from four independent experiments. Control n = 50 cells, GTSE1 n = 40 cells, and Kif4A n = 38. (Bottom) Depletion of either GTSE1 or Kif4A resulted in a significant decrease in the distance between kinetochores and centrosomes, suggesting a reduction in polar ejection forces. SD is plotted for the mean from each condition. Asterisks indicate statistical significance of mean value difference when compared with control as assessed by Kruskal-Wallis test. Bars, 10 µm. ***, P < 0.001; ****, P < 0.0001. (C) Quantification of the chromosome-bound Kif4A fraction from the immunoblot shown in Fig. S3 B. Kif4A levels were normalized to histone H3. Note that the reduction in KIf4A levels by immunoblot was comparable to data obtained by immunofluorescence (A, top) in the absence of GTSE1.
A customary test of polar ejection forces is measuring the distributions of chromosomes around monopolar spindles induced by treatment of cells with the Eg5 inhibitor monastrol (Stumpff et al., 2012; Wandke et al., 2012; Barisic et al., 2014). We found that depletion of GTSE1 diminished the distance between the spindle poles and kinetochores, suggesting that polar ejection forces were compromised in the GTSE1-depleted cells. A similar decrease was induced by depletion of Kif4A (Figs. 7 B and S4 C), which is consistent with certain previously published studies (Wandke et al., 2012; Barisic et al., 2014) but is in contrast with another study (Stumpff et al., 2012). To further verify the roles of both GTSE1 and Kif4A in polar ejection forces, an additional pool of four siRNAs to GTSE1 and six other siRNA sequences targeting Kif4A taken from previously published studies (Stumpff et al., 2012; Voets et al., 2015) were tested for knockdown efficiency by Western blotting (Fig. S4, A and B) and then examined for differences in polar ejection forces in the monastrol assay described in this section (Fig. S4 C). All siRNAs tested achieved >93% reduction in GTSE1 or Kif4A levels except for one Kif4A siRNA that reduced levels by only 20%. Consistent with the data described above, the additional siRNAs tested resulted in diminished distances between spindle poles and kinetochores in the monastrol assay, with the siRNA that reduced Kif4A by only 20% having the most modest effect. These data indicate that depletion of GTSE1 or Kif4A reduces polar ejection forces and that GTSE1 functions upstream of Kif4A localization to chromosome arms.
Additionally, we analyzed the phenotypes of RNAi knockdown of Kif4A using time-lapse microscopy experiments in HeLa cells stably expressing H2B-RFP and GFP-tubulin (Fig. S5, C and D; and Videos 4 and 5). The percentage of the respective protein levels after siRNA transfection was determined in Fig. S4 A. 40 h after depletion of Kif4A, the number of cells undergoing a successful mitosis was dramatically reduced from 92% in control siRNA–transfected cells to 29% in Kif4A siRNA (1), 41% in Kif4A siRNA (2), and 62% in Kif4A siRNA (SP) transfected cells (Fig. S5 C). The majority of Kif4A-depleted cells formed multipolar spindles that resulted in erroneous chromosome segregation (Videos 4 and 5). This observation is consistent with Mazumdar et al. (2004). When mitotic duration was monitored, i.e., the time from NEB to anaphase onset, we found that no significant difference between control and Kif4A-depleted cells existed when comparing cells undergoing a normal mitosis (Fig. S5 D). When comparing Kif4A depletion phenotypes with GTSE1 depletion phenotypes, chromosome alignment defects were not apparent after the loss of Kif4A. However, the increase in multipolar spindle formation after the loss of Kif4A was consistent with GTSE1 depletion. In GTSE1-depleted cells, we only detected multipolar spindle formation after chromosome alignment defects. The differences in phenotypes observed may reflect the fact that GTSE1 depletion only diminishes, but does not eliminate, Kif4A on chromosome arms (Figs. 7 A and S3, B and D). Alternatively, depletion of GTSE1 and its consequences for Aurora B on chromosome arms may affect other mitotic pathways beyond Kif4A depletion.
Discussion
Although research of yeast and other model systems has been instrumental in identifying key basic components of cell division, vertebrates have elaborated additional regulatory mechanisms, at least some of which remain to be identified and characterized. In this study, we used bioinformatics to identify GTSE1 as a novel mitotic regulator. Depletion of GTSE1 from cells greatly enhances the incidence of chromosome alignment defects and formation of multipolar spindles. One molecular consequence of GTSE1 depletion is reduced recruitment of the chromokinesin Kif4A to chromosomes. Previous studies have reported that depletion of Kif4A results in chromosome alignment defects (Mazumdar et al., 2004; Wandke et al., 2012; Barisic et al., 2014), and a previous study revealed regulation of Kif4A by Aurora B phosphorylation in the spindle midzone of anaphase cells (Nunes Bastos et al., 2013). However, we found the strongest effect of Kif4A depletion on spindle pole integrity, consistent with Mazumdar et al. (2004). Mechanistically, GTSE1 depletion increases the stability of the slow turnover population of microtubules in the mitotic spindle, and this in turn reduces Aurora B recruitment and activity on chromosome arms. Parenthetically, we note that many mitosis researchers using tubulin photoactivation to study microtubule dynamics equate the slow turnover population with kinetochore microtubules and the fast population with nonkinetochore microtubules. However, in our opinion, this categorization has never been rigorously tested, is most likely simplistic, and more likely reflects the limitations of our analysis. We believe it possible and in fact likely that spindle microtubules exhibit a broad range of dynamics and that our current methods are too crude to detect all. For this reason, we prefer to discuss the range of mitotic microtubule turnover rates from slow to fast, the two patterns distinguishable by photoactivation.
During mid-mitosis, GTSE1 stops tip tracking and associates with the lattice of spindle microtubules, specifically with those undergoing slow turnover (Fig. 3 C). Depletion of GTSE1 increases the proportion of slow turnover microtubules and also decreases their rate of turnover (Figs. 3 A and S2). The differences in half-life observed in the more stable microtubules after GTSE1 depletion may be in part a consequence of higher stability, leading to a decrease in tubulin dimer availability. Overall, our data indicate that GTSE1 accumulates on and primarily regulates the more stable microtubules within the mitotic spindle by promoting their turnover. The precise molecular mechanisms by which GTSE1 regulates turnover of microtubules will require in vitro analysis to be pursued in future studies.
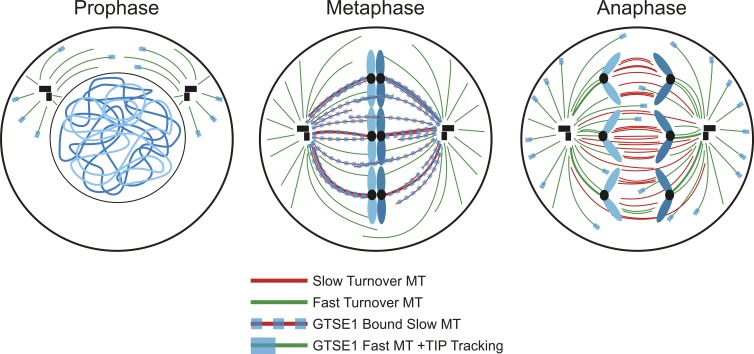
Of interest is the observation that GTSE1 plus end tip tracking via binding to EB1 is inhibited from NEB to anaphase onset during mitosis. Instead, during this time, GTSE1 is found to bind the lattice of the slow-turnover microtubules of the mitotic spindle (Fig. 3 C). This observation may provide a key link between GTSE1 localization throughout mitosis and regulation of its plus end microtubule tip tracking activity. After NEB, GTSE1 plus end tip tracking is suppressed, allowing GTSE1 to bind and promote the turnover of the slow population of microtubules. This redistribution of GTSE1 and the increased turnover of the slow microtubule population may enhance the ability of EB1 at microtubule plus ends to activate Aurora B on chromosome arms. Upon anaphase onset, GTSE1 resumes plus end tip tracking, eliminating any potential destabilizing influence of GTSE1 on midzone microtubules (Fig. 8).
Figure 8.
GTSE1 undergoes dynamic relocalization during mitosis. Before NEB, GTSE1 plus end tip tracks on microtubules (MTs) in an EB1-dependent fashion. After NEB, GTSE1 microtubule plus end tip tracking is suppressed, allowing the relocalization of GTSE1 to the lattice of slow-turnover microtubules in the mitotic spindle. GTSE1 relocalization allows it to promote turnover of the slow-turnover microtubule population. Upon anaphase onset, GTSE1 microtubule plus end tip tracking is reinstated, allowing GTSE1 to primarily decorate the fast-turnover astral microtubule population.
It is noteworthy that both destabilization and hyperstabilization of spindle microtubules reduce Aurora B activity on chromosome arms (Fig. 4). This finding suggests that the proper balance of dynamic microtubules is essential to maintain optimal Aurora B activity on arms. The plus end tracking protein EB1 interacts with Aurora B and enhances its activity by blocking its dephosphorylation (Sun et al., 2008). Further work has shown that EB1 activates Aurora B kinase at centromeres/kinetochores and on chromatin (Banerjee et al., 2014). We speculate that EB1 targeted by dynamic fast turnover microtubules may stimulate Aurora B kinase activity to recruit and possibly activate Kif4A on chromosome arms. We also noted that the total levels of both Aurora B and Kif4A are diminished in cells depleted of GTSE1. Both Aurora B and Kif4A are substrates of the E3 ubiquitin ligase APC/C and are subject to proteasome degradation (Stewart and Fang, 2005; Floyd et al., 2008; Singh et al., 2014). Although the APC/C is restrained by the spindle checkpoint before anaphase, many APC/C substrates are partially or fully targeted earlier in mitosis (Geley et al., 2001; Hames et al., 2001; Brito and Rieder, 2006). We speculate that loss of normal association with chromosome arms may result in degradation of these proteins. However, we cannot rule out the possibility that GTSE1 depletion affects the synthesis of Aurora B and Kif4A.
Kif4A is known to be phosphorylated by Aurora B in anaphase (Takahashi et al., 2016). Whether Kif4A is the direct target of phosphorylation by Aurora B in prometaphase or whether the effect of Aurora B for Kif4A association with chromosome arms is indirect is a critical question for future investigation. Importantly, GTSE1 depletion allowed us to strongly manipulate Aurora B activity on chromosome arms. At centromeres, depletion of GTSE1 had a modest effect, revealing a slight decrease in the amount of Aurora B but a slight increase in Aurora B kinase activity. The functional consequence of these slight changes in Aurora B localization and activity at centromeres after GTSE1 depletion remains unknown. Aurora B is recruited to centromere regions by Haspin, Bub1, and Mps1 kinases, which are concentrated there (Kelly et al., 2010; Wang et al., 2010; Yamagishi et al., 2010; van der Waal et al., 2012). The important roles of Aurora B in regulating kinetochore assembly, functions in chromosome movement, and checkpoint signaling are well documented (Adams et al., 2001; Kallio et al., 2002; Ditchfield et al., 2003; DeLuca et al., 2006, 2011; Jelluma et al., 2008; Liu et al., 2009; van der Horst and Lens, 2014). However, it has been difficult to study the roles of Aurora B at chromosome arms because of the difficulty of separating functions there from those at centromeres and kinetochores. Our evidence indicates that GTSE1 promotion of Aurora B on chromosome arms functions to promote normal chromosome movement and spindle integrity. One target is Kif4A, but increased microtubule stability caused by GTSE1 depletion may also affect other mitotic pathways including those that impact kinetochores.
The study of GTSE1 has a relatively brief but highly varied history. It has been proposed to function in several pathways during interphase, including control of apoptosis after DNA damage via regulation of p53 function, mediation of p21CIP1/WAF1 stability, counteracting paclitaxel-induced cytotoxicity, and regulation of cell migration in an EB1-dependent fashion (Monte et al., 2003, 2004; Bublik et al., 2010; Scolz et al., 2012). However, endogenous GTSE1 protein levels are very tightly regulated throughout the cell cycle, becoming detectable only in G2, peaking during mitosis, and being wholly degraded upon mitotic exit and entry into G1 (Fig. 1 A; Collavin et al., 2000; Pfleger and Kirschner, 2000). Endogenous GTSE1 is largely undetectable in most interphase cells (Fig. 1 B). One potential explanation for previous suggestions of interphase roles for GTSE1 is artifactual accumulation of GTSE1 during drug-induced S phase arrest (Fig. 1 A). A second problem has been the reliance on exogenous expression of GTSE1 in cells, which results in exaggerated expression of GTSE1 at points in the cell cycle when endogenous levels are extremely low (Monte et al., 2003; Bublik et al., 2010; Liu et al., 2010; Scolz et al., 2012). A third potential explanation could be differences in cell cycle regulation in different cell types.
Recently, Bendre et al. (2016) suggested that GTSE1 regulates microtubule stability during mitosis via inhibition of MCAK microtubule depolymerase activity. Their data regarding the interaction between GTSE1 and MCAK appear compelling, and consistent with our observations, they report chromosome alignment defects and prolonged mitosis upon siRNA-mediated depletion of GTSE1. However, whereas we report that depletion of GTSE1 increases stability of the slow turnover population of spindle microtubules, they report the opposite, namely that GTSE1 loss leads to higher turnover because of the reduced inhibition of MCAK microtubule depolymerase activity. What might underlie this discrepancy? Both our study and that of Bendre et al. (2016) used tubulin photoactivation in the same U2OS cell line developed by Bakhoum et al. (2009b). However, in control cells, Bendre et al. (2016) reported a t1/2 of 955 s (15.9 min) for the turnover of the slower microtubule population. In contrast, we report a t1/2 of 289 s (4.8 min) for the same population. We note that our turnover half time for the slow population is similar to those previously reported for this cell line (t1/2 of ∼5–7 min; Bakhoum et al., 2009b; Kabeche and Compton, 2013). Interestingly, in the GTSE1-depleted cells, our results, t1/2 of 587 s (9.8 min), are close to those of Bendre et al. (2016), t1/2 of 478 s (8 min), for the slow turnover microtubules. This indicates that the primary difference in observations between the two studies rests in the measurements of microtubule turnover in the controls.
The interaction of chromosome arms with mitotic spindle microtubules generates polar ejection forces that play roles in chromosome movement and spindle assembly. One potential mediator of this interaction is the chromokinesin Kif4A, which distributes along arms in mitosis before anaphase onset. However, the role of Kif4A as a mediator of polar ejection forces is controversial, with previous studies concluding that it is an important component, that it plays only a minor role, that it antagonizes ejection forces, or that it promotes them (Mazumdar et al., 2004; Stumpff et al., 2012; Wandke et al., 2012; Barisic et al., 2014). The reasons for these discrepancies are unclear. A potential explanation could be variability among different cell types. Comparing several published siRNA sequences (Stumpff et al., 2012; Wandke et al., 2012; Barisic et al., 2014; Voets et al., 2015), we found that depletion of Kif4A reduced polar ejection forces in HeLa cells (Figs. 7 B and S4 C), consistent with results published by Barisic et al. (2014). Kif4A has also been shown to play a direct role in regulating microtubule dynamics during mitosis (Bringmann et al., 2004; Stumpff et al., 2012; Wandke et al., 2012; Nunes Bastos et al., 2013; Voets et al., 2015). However, Kif4A depletion is reported to decrease microtubule stability primarily in the fast turnover or astral microtubule populations (Stumpff et al., 2012; Wandke et al., 2012; Voets et al., 2015). Additionally, cells depleted of Kif4A displayed defects in mitotic spindle integrity, suggesting a requirement for Kif4A in mitotic spindle maintenance (Mazumdar et al., 2004). Similar to Mazumdar et al. (2004), we found that depletion of Kif4A was a strong inducer of multipolar spindles, thus perhaps providing an explanation of one of the GTSE1 phenotypes. But unlike GTSE1 depletion, loss of Kif4A did not strongly impact chromosome alignment. There are many potential mechanisms. Perhaps the simplest is that GTSE1 depletion resulting in loss of arm Aurora B has consequences for other important mitotic regulators such as MCAK (Bendre et al., 2016). How Kif4A depletion and the resulting effects on polar ejection forces may impact pole integrity remain important questions for the future.
In summary, we have identified GTSE1 as a novel regulator of chromosome alignment and spindle pole integrity that functions by modulating microtubule dynamics during mitosis. The ability of GTSE1 to increase microtubule dynamics promotes the localization of Aurora B specifically to chromosome arms, resulting in the subsequent recruitment of chromokinesin Kif4A and generation of polar ejection forces. This work not only establishes a role for GTSE1 as a novel mitotic regulator, but it also brings to light new pathways needed for the proper alignment of chromosomes to the metaphase plate and maintenance of spindle pole integrity, both of which contribute to the prevention of chromosome missegregation events and subsequent aneuploidy. Up-regulation of GTSE1 expression correlates with aggressive malignancy in several types of cancer, and in some cases, knockdown of GTSE1 was found to diminish proliferation, migration, and invasion in xenograft models (Bublik et al., 2010; Scolz et al., 2012; Subhash et al., 2015; Zekri et al., 2015; Guo et al., 2016; Tian et al., 2016). Thus, GTSE1 is a potential biomarker and putative therapeutic target for individualized cancer treatment.
Materials and methods
Bioinformatic characterization of GTSE1
An algorithm called GAMMA (Wren, 2009; Dozmorov et al., 2011) was used to identify potential mitotic regulators, of which GTSE1 was chosen for closer investigation. GAMMA was then used to predict function, phenotype, and genetic associations for GTSE1. Full details on how GAMMA works have been published previously (Wren, 2009; Dozmorov et al., 2011), but in brief, GAMMA first identifies highly correlated transcripts using publicly available microarray data from NCBI’s GEO database. For each query gene, GAMMA first identifies a set of the 40 most correlated transcripts. Then, for this transcriptionally correlated gene set, a literature-mining algorithm (Wren et al., 2004) identifies which commonalities (e.g., genes, phenotypes, diseases, chemical compounds, drugs, metabolites, and Gene Ontology categories) it has within published Medline abstracts. This “guilt by association” approach infers the function of genes based upon what is known about their most correlated transcripts and can be used to predict function even if no literature exists on the gene being analyzed. GAMMA has been experimentally validated in several studies to date (Daum et al., 2009; Lupu et al., 2011; Clemmensen et al., 2012; Towner et al., 2013a,b; Fisch et al., 2015; Gandhapudi et al., 2015).
siRNA
The siRNA sequences 5′-GUACAAAGAAGGUCACUUA-3′ and 5′-CUACUAUCUAGACUCGAAU-3′ targeted GTSE1 and siRNA 5′-GCAAGAUCCUGAAAGAGAU-3′ (Wandke et al., 2012; Barisic et al., 2014), 5′-CAAUGUGCUCAGACGUAA-3′ (Stumpff et al., 2012), 5′-CCAAAUCAUUUGCCGAG-3′ (Stumpff et al., 2012), 5′-AGGCGUACAUUCUCCCUUA-3′ (Voets et al., 2015), 5′-GAAGAGGCCCACUGAAGUU-3′ (Voets et al., 2015), 5′-UGAAAGAGAUGUGCGAUGU3′ (Voets et al., 2015), and 5′-UGACUCGACUGCUUCAAGA-3′ (Voets et al., 2015) targeting Kif4A were synthesized by Sigma-Aldrich. The siRNA sequence 5′-GAGUAGAACUAGAAUGUGA-3′ targeting Hec1 was synthesized by QIAGEN. ON-TARGET plus SMARTpool siRNAs targeting GTSE1 (5′-ACACGUGGCUGUAGGAUCU-3′, 5′-GAACUGAACCAACAAGGGA-3′, 5′-UAAAUAAUCCGGUUCCCGA-3′, and 5′-GUACAAAGAAGCUCACUUA-3′) and Kif4A (5′-AGGCGUACAUUCUCCCUUA-3′, 5′-GAAGAGGCCCACUGAAGUU-3′, 5′-UGAAAGAGAUGUGCGAUGU-3′, and 5′-UGACUCGACUGCUUCAAGA-3′) were synthesized by GE Healthcare. Nontargeting control siRNA was synthesized by Bioneer.
Cell culture, transfection, and drug treatments
Parental HeLa cells, HeLa cells stably expressing GFP-tubulin and H2B-RFP, and U2OS cells stably expressing PAGFP-tubulin (gifts from D. Compton, Dartmouth College, Hanover, NH) were cultured in DMEM with 10% FBS supplemented with penicillin and streptomycin, 20 mM Hepes, and 0.1 mM nonessential amino acids at 37°C with 5% CO2. To synchronize cells in mitosis, cultures were treated with 2.5 mM thymidine for 24 h and released into media containing 200 ng/ml nocodazole for 12 h. Alternatively, cultures were treated with 200 ng/ml nocodazole for 16 h. Transient transfection was performed using TransIT-LT1 (Mirus) transfection reagent according to the manufacturer’s instructions. For siRNA transfection, cells were transfected with 20 nM siRNA for 48 h using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s protocol. To inhibit Aurora B activity, cells were treated with 10 µM ZM447439 for 2 h. 20 µM MG132 was also added to prevent mitotic exit. Taxol was used at 10 µM. Monastrol was used at 100 µM for 2 h to induce monopolar spindles.
Production of GTSE1 polyclonal antibody
A fragment of GTSE1 corresponding with amino acids 1–243 was cloned into the pDEST15 vector (Invitrogen) and expressed in Rosetta2 (DE3) pLysS (Novagen). The GST fusion protein was purified using glutathione-sepharose 4B (GE Healthcare) and used to generate rabbit polyclonal antiserum (Cocalico Biologicals).
Western blotting
For whole-cell lysates, cells were lysed in cell lysis buffer (PBS, 0.5% NP-40, 1 mM tris(2-carboxyethyl)phosphine, and 10% glycerol) supplemented with protease inhibitor cocktail (Sigma-Aldrich) and phosphatase inhibitors (100 mM NaF, 1 mM Na3VO4, 60 mM β-glycerophosphate, and 100 nM microcystin-LR). Alternatively, to isolate cytosolic and chromosome-bound fractions or to generate whole-cell extracts, cells were first swollen in swelling buffer (10 mM Hepes, 40 mM KCl, 5 mM EGTA, and 4 mM MgSO4) at 37°C, and then lysed in extraction and lysis buffer (PHEM [60 mM Pipes, pH 6.9, 25 mM Hepes, 10 mM EGTA, and 4 mM MgSO4], 0.5% Triton X-100, 5 mM tris(2-carboxyethyl)phosphine, and 10% glycerol) containing protease and phosphatase inhibitors. Chromosome and cytosolic fractions were separated by centrifugation, and chromosome pellets were suspended in extraction and lysis buffer. The protein concentration of lysates was measured using the BCA Protein Assay kit (Thermo Fisher Scientific). For electrophoresis, sample loading buffer (Invitrogen) and DTT to a final concentration of 50 mM were added. Proteins were separated with a NuPAGE gel electrophoresis system (Invitrogen) and transferred to a 0.45-µm PVDF membrane (Immobilon-FL; EMD Millipore). Membranes were blocked in 10% SEA BLOCK blocking buffer (Thermo Fisher Scientific) and 0.05% Tween-20 in PBS (PBST). The following primary antibodies were used: rabbit anti-GTSE1 (Cocalico Biologicals), rabbit anti–α-tubulin (Abcam), mouse anti-AIM1 (BD), rabbit anti–histone H3 (Abcam), mouse anti-Kif4A (a gift from T. Misteli, National Cancer Institute, Bethesda, MD), rabbit anti–Aurora B pT232 (Rockland), mouse anti–histone H3 pS10 (Cell Signaling Technology), mouse anti–cyclin B (BD), and rabbit anti-actin (Abcam). Membranes were washed in PBST. Secondary goat anti–mouse and goat anti–rabbit antibodies were conjugated to either IR700 or IR800 dyes (Azure Biosystems). Western blots were imaged on an Azure c600 Imaging System. Western blots were quantified by calculating integrated band intensities using MetaMorph software (Molecular Devices) and Excel (Microsoft).
Immunofluorescence
HeLa cells were grown on glass coverslips and treated as detailed in Figs. 1, 3, 4, 5, 6, 7, S3, and S4. Cells were fixed in 2% paraformaldehyde/PHEM solution containing 0.5% Triton X-100 for 15 min. Coverslips were washed in MBST (10 mM MOPS, 150 mM NaCl, and 0.05% Tween-20), blocked in 20% boiled normal goat serum, and incubated overnight with primary antibodies. Samples were then incubated with secondary antibodies for 1 h, stained with DNA dye DAPI, and mounted using Vectashield (Vector Laboratories). Alternatively, to measure inner spindle intensity or polar ejection forces in monastrol-treated cells, cells were fixed in methanol for 20 min at −20°C and were processed for immunofluorescence as described above. Cold-treated cells were incubated for 10 min in 4°C in 1×PBS on ice. The following primary antibodies were used: human anticentromere antigen (ACA)/CREST (Antibody, Inc.), mouse anti–γ-tubulin (Sigma-Aldrich), rabbit anti–α-tubulin (AbD Serotec), rabbit anti-GTSE1 (Cocalico Biologicals), mouse anti-Kif4A (a gift from T. Misteli), rabbit anti-Kif22/Kid (Bethyl Laboratories, Inc.), mouse anti-AIM1 (BD), mouse anti–histone H3 pS10 (Cell Signaling Technology), and rabbit anti–CENP-A pS7 (Upstate Biologicals). Secondary antibodies used were goat anti–rabbit antibodies conjugated to Cy3 or FITC (Jackson ImmunoResearch Laboratories, Inc.), goat anti–rat antibodies conjugated to Cy3 or FITC, goat anti–mouse antibodies conjugated to Cy3 or FITC, and goat anti–human antibody conjugated to Cy3, FITC, or Cy5. The images were acquired using an Axioplan II microscope (ZEISS) equipped with a 100× 1.4 NA objective and an ORCA-ER camera (Hamamatsu Photonics) and processed using MetaMorph software. Distances between kinetochores and spindle poles were measured using MetaMorph. Quantification of immunofluorescence images was performed as previously described (Daum et al., 2009). The graphs depict mean fluorescence value with SD in each condition. The Mann-Whitney two-tailed test or Kruskal-Wallis test (with Dunn’s post hoc comparison) in Prism (GraphPad Software) was used to determine statistical significance among groups.
Live-cell imaging
Cells were grown in Nunc chambered coverslips (Thermo Fisher Scientific). To maintain appropriate pH levels and avoid evaporation during imaging, culture media was exchanged to Leibovitz’s L-15 medium supplemented with 10% FBS, penicillin, and streptomycin, and the medium was overlaid with mineral oil. Cells were treated as detailed in Figs. 2 and 3. Time-lapse fluorescence images were collected using 20× 0.5 NA or 40× 1.3 NA objectives on an Axiovert 200M inverted microscope (ZEISS) equipped with an objective heater, air curtain, ORCA-ER camera, and MetaMorph software. Images were captured every 5 min for 20 h. Time-lapse videos displaying the elapsed time between consecutive frames were assembled using MetaMorph software. The first time frame after NEB, metaphase chromosome alignment, and anaphase onset/mitotic exit each was noted in Excel, and the mitotic interval from NEB to metaphase (alignment time), metaphase to anaphase (metaphase duration), or NEB to anaphase onset/mitotic exit was calculated.
For photoactivation experiments, PAGFP-tubulin expressing U2OS cells were cultured as described in the previous paragraph and imaged using a 100× 1.4 NA objective on an Axio Observer inverted microscope (ZEISS) equipped with an objective heater, air curtain, spinning disk (Yokogawa Electric Corporation), Mosaic (digital mirror device; Photonic Instruments/Andor), an ORCA-Flash4.0LT camera (Hamamatsu Photonics), and Slidebook software (Intelligent Imaging Innovations). Photoactivation was achieved by targeting a selected area with filtered light from the HBO 100 via the Mosaic, and confocal GFP images across z were acquired at 15-s intervals for 5 min. Fluorescence intensity of the activated region was corrected for photobleaching and background using MetaMorph software. Normalized measurements were fitted to a double exponential decay curve F = A1 × exp(−k1 × t) + A2 × exp(−k2 × t) using MatLab (Mathworks), where A1 and A2 represent the less stable and more stable microtubule populations with decay rates of k1 and k2, respectively. t is the time after photoactivation. The turnover half-life for each population of microtubules was calculated as ln2/k.
Photoactivation simulation
Control data were plotted using nonlinear least squares fit to a double-exponential curve of the equation f(x) = a*exp(−b*x) + c*exp(−d*x). This was done in MatLab using the fit function with a custom fitType. This method was verified using the fit function and “exp2” fitType as well as with the curve-fitting toolbox in MatLab by fitting to a double exponential curve from which identical coefficients and goodness of fit values were obtained. This general model was then applied to GTSE1 data, first by varying all parameters to best fit the data and then by varying the parameters individually to test the effect of GTSE1 loss on each parameter. The unvaried parameters were fixed to control values, and the parameter being tested was varied an order of magnitude approximately surrounding the control value. When varying parameters pertaining to the percentage of the population (a and c), both parameters were varied simultaneously such that the total population was always equal to one. When varying parameters pertaining to the percentage of the population (a and c) and the rate, population parameters were varied simultaneously as above, the fast rate (b) was fixed to control values, and the slow rate (d) was determined by a nonlinear least squares fit to find the optimal rate. Simulations were performed using 100 evenly spaced time points (x) within the range of experimental data.
Online supplemental material
Fig. S1 shows the mitotic timing and RNAi phenotype in HeLa and U2OS cells depleted of GTSE1 as assessed by time-lapse microscopy. Fig. S2 shows the curve fittings derived from simulation of the photoactivation data obtained in Fig. 3 A. Fig. S3 shows Aurora B phosphoepitopes and Kif4A levels by immunostaining and Western blotting in the presence and absence of GTSE1 in HeLa cells. Fig. S3 also shows Kif4A levels in the presence and absence of nocodazole or taxol by immunostaining in HeLa cells. Fig. S4 shows GTSE1 and Kif4A protein levels by Western blotting after transfection of various siRNAs as well as the distances between kinetochores and spindle poles in cells depleted of either GTSE1 or Kif4A in monastrol-treated HeLa cells. Fig. S5 shows Aurora B and Kif4A levels in control and GTSE1-depleted HeLa whole-cell extracts by Western blotting along with the mitotic timing and RNAi phenotype in control and Kif4A-depleted HeLa cells as assessed by time-lapse microscopy. Video 1 shows the mitotic progression of a control siRNA-transfected cell. Videos 2 and 3 show the mitotic progression and phenotypes of GTSE1 siRNA-transfected cells. Videos 4 and 5 show the mitotic progression and phenotypes of Kif4A siRNA-transfected cells. Table S1 shows the predicted functions, phenotypes, and genetic associations from GAMMA’s analysis of the GTSE1 transcriptional network.
Supplementary Material
Acknowledgments
We thank Dr. Ted Salmon for assistance in analysis of photoactivation data.
Support for this project was provided in part by the National Institute of General Medical Sciences (grant 5R01GM111731) and the Oklahoma Center for the Advancement of Science and Technology (grant HR12-177) to G.J. Gorbsky. A.R. Tipton was supported by the National Cancer Institute National Research Service Award postdoctoral fellowship (grant 1F32 CA189450). J.D. Wren was supported in part by the National Institute of General Medical Sciences (grant 5P20GM103636). G.J. Gorbsky was also supported by the McCasland Foundation.
The authors declare no competing financial interests.
Author contributions: A.R. Tipton, J.R. Daum, and G.J. Gorbsky designed the experiments. A.R. Tipton and J.R. Daum executed the experiments. J.D. Wren designed and conducted bioinformatics analysis. J.C. Siefert conducted the mathematical simulations. A.R. Tipton and G.J. Gorbsky wrote the manuscript.
Footnotes
Abbreviations used:
- APC/C
- anaphase-promoting complex/cyclosome
- GAMMA
- global microarray meta-analysis
- MCAK
- mitotic centromere-associated kinesin
- NEB
- nuclear envelope breakdown
- PAGFP
- photoactivatable GFP
References
- Adams R.R., Maiato H., Earnshaw W.C., and Carmena M.. 2001. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153:865–880. 10.1083/jcb.153.4.865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonio C., Ferby I., Wilhelm H., Jones M., Karsenti E., Nebreda A.R., and Vernos I.. 2000. Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell. 102:425–435. 10.1016/S0092-8674(00)00048-9 [DOI] [PubMed] [Google Scholar]
- Bakhoum S.F., Genovese G., and Compton D.A.. 2009a Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr. Biol. 19:1937–1942. 10.1016/j.cub.2009.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum S.F., Thompson S.L., Manning A.L., and Compton D.A.. 2009b Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat. Cell Biol. 11:27–35. 10.1038/ncb1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee B., Kestner C.A., and Stukenberg P.T.. 2014. EB1 enables spindle microtubules to regulate centromeric recruitment of Aurora B. J. Cell Biol. 204:947–963. 10.1083/jcb.201307119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barisic M., Aguiar P., Geley S., and Maiato H.. 2014. Kinetochore motors drive congression of peripheral polar chromosomes by overcoming random arm-ejection forces. Nat. Cell Biol. 16:1249–1256. 10.1038/ncb3060 [DOI] [PubMed] [Google Scholar]
- Barlin J.N., Zhou Q.C., Leitao M.M., Bisogna M., Olvera N., Shih K.K., Jacobsen A., Schultz N., Tap W.D., Hensley M.L., et al. . 2015. Molecular subtypes of uterine leiomyosarcoma and correlation with clinical outcome. Neoplasia. 17:183–189. 10.1016/j.neo.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendre S., Rondelet A., Hall C., Schmidt N., Lin Y.C., Brouhard G.J., and Bird A.W.. 2016. GTSE1 tunes microtubule stability for chromosome alignment and segregation by inhibiting the microtubule depolymerase MCAK. J. Cell Biol. 215:631–647. 10.1083/jcb.201606081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann H., Skiniotis G., Spilker A., Kandels-Lewis S., Vernos I., and Surrey T.. 2004. A kinesin-like motor inhibits microtubule dynamic instability. Science. 303:1519–1522. 10.1126/science.1094838 [DOI] [PubMed] [Google Scholar]
- Brito D.A., and Rieder C.L.. 2006. Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr. Biol. 16:1194–1200. 10.1016/j.cub.2006.04.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard G.J., and Hunt A.J.. 2005. Microtubule movements on the arms of mitotic chromosomes: Polar ejection forces quantified in vitro. Proc. Natl. Acad. Sci. USA. 102:13903–13908. 10.1073/pnas.0506017102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bublik D.R., Scolz M., Triolo G., Monte M., and Schneider C.. 2010. Human GTSE-1 regulates p21(CIP1/WAF1) stability conferring resistance to paclitaxel treatment. J. Biol. Chem. 285:5274–5281. 10.1074/jbc.M109.045948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S., O’Connell C.B., Khodjakov A., and Walczak C.E.. 2009. Chromosome congression in the absence of kinetochore fibres. Nat. Cell Biol. 11:832–838. 10.1038/ncb1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemmensen S.N., Bohr C.T., Rørvig S., Glenthøj A., Mora-Jensen H., Cramer E.P., Jacobsen L.C., Larsen M.T., Cowland J.B., Tanassi J.T., et al. . 2012. Olfactomedin 4 defines a subset of human neutrophils. J. Leukoc. Biol. 91:495–500. 10.1189/jlb.0811417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collavin L., Monte M., Verardo R., Pfleger C., and Schneider C.. 2000. Cell-cycle regulation of the p53-inducible gene B99. FEBS Lett. 481:57–62. 10.1016/S0014-5793(00)01969-4 [DOI] [PubMed] [Google Scholar]
- Daum J.R., Wren J.D., Daniel J.J., Sivakumar S., McAvoy J.N., Potapova T.A., and Gorbsky G.J.. 2009. Ska3 is required for spindle checkpoint silencing and the maintenance of chromosome cohesion in mitosis. Curr. Biol. 19:1467–1472. 10.1016/j.cub.2009.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., and Salmon E.D.. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982. 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- DeLuca K.F., Lens S.M., and DeLuca J.G.. 2011. Temporal changes in Hec1 phosphorylation control kinetochore–microtubule attachment stability during mitosis. J. Cell Sci. 124:622–634. 10.1242/jcs.072629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., and Taylor S.S.. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267–280. 10.1083/jcb.200208091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozmorov M.G., Giles C.B., and Wren J.D.. 2011. Predicting gene ontology from a global meta-analysis of 1-color microarray experiments. BMC Bioinformatics. 12(Suppl 10):S14 10.1186/1471-2105-12-S10-S14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisch A.S., Yerges-Armstrong L.M., Backman J.D., Wang H., Donnelly P., Ryan K.A., Parihar A., Pavlovich M.A., Mitchell B.D., O’Connell J.R., et al. . 2015. Genetic variation in the platelet endothelial aggregation receptor 1 gene results in endothelial dysfunction. PLoS One. 10:e0138795 10.1371/journal.pone.0138795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd S., Pines J., and Lindon C.. 2008. APC/C Cdh1 targets aurora kinase to control reorganization of the mitotic spindle at anaphase. Curr. Biol. 18:1649–1658. 10.1016/j.cub.2008.09.058 [DOI] [PubMed] [Google Scholar]
- Fu W., Tao W., Zheng P., Fu J., Bian M., Jiang Q., Clarke P.R., and Zhang C.. 2010. Clathrin recruits phosphorylated TACC3 to spindle poles for bipolar spindle assembly and chromosome alignment. J. Cell Sci. 123:3645–3651. 10.1242/jcs.075911 [DOI] [PubMed] [Google Scholar]
- Funabiki H., and Murray A.W.. 2000. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 102:411–424. 10.1016/S0092-8674(00)00047-7 [DOI] [PubMed] [Google Scholar]
- Gandhapudi S.K., Tan C., Marino J.H., Taylor A.A., Pack C.C., Gaikwad J., Van De Wiele C.J., Wren J.D., and Teague T.K.. 2015. IL-18 acts in synergy with IL-7 to promote ex vivo expansion of T lymphoid progenitor cells. J. Immunol. 194:3820–3828. 10.4049/jimmunol.1301542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geley S., Kramer E., Gieffers C., Gannon J., Peters J.M., and Hunt T.. 2001. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J. Cell Biol. 153:137–148. 10.1083/jcb.153.1.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gridley D.S., Pecaut M.J., Mao X.W., Wroe A.J., and Luo-Owen X.. 2015. Biological effects of passive versus active scanning proton beams on human lung epithelial cells. Technol. Cancer Res. Treat. 14:81–98. 10.7785/tcrt.2012.500392 [DOI] [PubMed] [Google Scholar]
- Guo L., Zhang S., Zhang B., Chen W., Li X., Zhang W., Zhou C., Zhang J., Ren N., and Ye Q.. 2016. Silencing GTSE-1 expression inhibits proliferation and invasion of hepatocellular carcinoma cells. Cell Biol. Toxicol. 32:263–274. 10.1007/s10565-016-9327-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hames R.S., Wattam S.L., Yamano H., Bacchieri R., and Fry A.M.. 2001. APC/C-mediated destruction of the centrosomal kinase Nek2A occurs in early mitosis and depends upon a cyclin A-type D-box. EMBO J. 20:7117–7127. 10.1093/emboj/20.24.7117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelluma N., Brenkman A.B., van den Broek N.J., Cruijsen C.W., van Osch M.H., Lens S.M., Medema R.H., and Kops G.J.. 2008. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell. 132:233–246. 10.1016/j.cell.2007.11.046 [DOI] [PubMed] [Google Scholar]
- Kabeche L., and Compton D.A.. 2013. Cyclin A regulates kinetochore microtubules to promote faithful chromosome segregation. Nature. 502:110–113. 10.1038/nature12507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio M.J., McCleland M.L., Stukenberg P.T., and Gorbsky G.J.. 2002. Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr. Biol. 12:900–905. 10.1016/S0960-9822(02)00887-4 [DOI] [PubMed] [Google Scholar]
- Kapoor T.M., and Compton D.A.. 2002. Searching for the middle ground. J. Cell Biol. 157:551–556. 10.1083/jcb.200202073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor T.M., Lampson M.A., Hergert P., Cameron L., Cimini D., Salmon E.D., McEwen B.F., and Khodjakov A.. 2006. Chromosomes can congress to the metaphase plate before biorientation. Science. 311:388–391. 10.1126/science.1122142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A.E., Ghenoiu C., Xue J.Z., Zierhut C., Kimura H., and Funabiki H.. 2010. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 330:235–239. 10.1126/science.1189505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Holland A.J., Lan W., and Cleveland D.W.. 2010. Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell. 142:444–455. 10.1016/j.cell.2010.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith S.L., Khodjakov A., Hergert P., and Walczak C.E.. 2004. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol. Biol. Cell. 15:1146–1159. 10.1091/mbc.E03-08-0581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosco K.A., Pearson C.G., Maddox P.S., Wang P.J., Adams I.R., Salmon E.D., Bloom K., and Huffaker T.C.. 2001. Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol. Biol. Cell. 12:2870–2880. 10.1091/mbc.12.9.2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Yu W., Liang Y., and Zhu X.. 2007. Kinetochore dynein generates a poleward pulling force to facilitate congression and full chromosome alignment. Cell Res. 17:701–712. 10.1038/cr.2007.65 [DOI] [PubMed] [Google Scholar]
- Liu D., Vader G., Vromans M.J., Lampson M.A., and Lens S.M.. 2009. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 323:1350–1353. 10.1126/science.1167000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.S., Li H., Song B., and Liu X.. 2010. Polo-like kinase 1 phosphorylation of G2 and S-phase-expressed 1 protein is essential for p53 inactivation during G2 checkpoint recovery. EMBO Rep. 11:626–632. 10.1038/embor.2010.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupu C., Zhu H., Popescu N.I., Wren J.D., and Lupu F.. 2011. Novel protein ADTRP regulates TFPI expression and function in human endothelial cells in normal conditions and in response to androgen. Blood. 118:4463–4471. 10.1182/blood-2011-05-355370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar M., Sundareshan S., and Misteli T.. 2004. Human chromokinesin KIF4A functions in chromosome condensation and segregation. J. Cell Biol. 166:613–620. 10.1083/jcb.200401142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte M., Collavin L., Lazarevic D., Utrera R., Dragani T.A., and Schneider C.. 2000. Cloning, chromosome mapping and functional characterization of a human homologue of murine Gtse-1 (B99) gene. Gene. 254:229–236. 10.1016/S0378-1119(00)00260-2 [DOI] [PubMed] [Google Scholar]
- Monte M., Benetti R., Buscemi G., Sandy P., Del Sal G., and Schneider C.. 2003. The cell cycle-regulated protein human GTSE-1 controls DNA damage-induced apoptosis by affecting p53 function. J. Biol. Chem. 278:30356–30364. 10.1074/jbc.M302902200 [DOI] [PubMed] [Google Scholar]
- Monte M., Benetti R., Collavin L., Marchionni L., Del Sal G., and Schneider C.. 2004. hGTSE-1 expression stimulates cytoplasmic localization of p53. J. Biol. Chem. 279:11744–11752. 10.1074/jbc.M311123200 [DOI] [PubMed] [Google Scholar]
- Musacchio A., and Salmon E.D.. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393. 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Nunes Bastos R., Gandhi S.R., Baron R.D., Gruneberg U., Nigg E.A., and Barr F.A.. 2013. Aurora B suppresses microtubule dynamics and limits central spindle size by locally activating KIF4A. J. Cell Biol. 202:605–621. 10.1083/jcb.201301094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S., Montaño-Gutierrez L.F., de Lima Alves F., Ogawa H., Toramoto I., Sato N., Morrison C.G., Takeda S., Hudson D.F., Rappsilber J., and Earnshaw W.C.. 2016. Proteomics analysis with a nano random forest approach reveals novel functional interactions regulated by SMC complexes on mitotic chromosomes. Mol. Cell. Proteomics. 15:2802–2818. 10.1074/mcp.M116.057885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger C.M., and Kirschner M.W.. 2000. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., and Salmon E.D.. 1994. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J. Cell Biol. 124:223–233. 10.1083/jcb.124.3.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Davison E.A., Jensen L.C., Cassimeris L., and Salmon E.D.. 1986. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J. Cell Biol. 103:581–591. 10.1083/jcb.103.2.581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosasco-Nitcher S.E., Lan W., Khorasanizadeh S., and Stukenberg P.T.. 2008. Centromeric Aurora-B activation requires TD-60, microtubules, and substrate priming phosphorylation. Science. 319:469–472. 10.1126/science.1148980 [DOI] [PubMed] [Google Scholar]
- Schneider L., Essmann F., Kletke A., Rio P., Hanenberg H., Wetzel W., Schulze-Osthoff K., Nürnberg B., and Piekorz R.P.. 2007. The transforming acidic coiled coil 3 protein is essential for spindle-dependent chromosome alignment and mitotic survival. J. Biol. Chem. 282:29273–29283. 10.1074/jbc.M704151200 [DOI] [PubMed] [Google Scholar]
- Scolz M., Widlund P.O., Piazza S., Bublik D.R., Reber S., Peche L.Y., Ciani Y., Hubner N., Isokane M., Monte M., et al. . 2012. GTSE1 is a microtubule plus-end tracking protein that regulates EB1-dependent cell migration. PLoS One. 7:e51259 10.1371/journal.pone.0051259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.A., Winter D., Kirchner M., Chauhan R., Ahmed S., Ozlu N., Tzur A., Steen J.A., and Steen H.. 2014. Co-regulation proteomics reveals substrates and mechanisms of APC/C-dependent degradation. EMBO J. 33:385–399. 10.1002/embj.201385876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S., and Fang G.. 2005. Destruction box-dependent degradation of aurora B is mediated by the anaphase-promoting complex/cyclosome and Cdh1. Cancer Res. 65:8730–8735. 10.1158/0008-5472.CAN-05-1500 [DOI] [PubMed] [Google Scholar]
- Stumpff J., Wagenbach M., Franck A., Asbury C.L., and Wordeman L.. 2012. Kif18A and chromokinesins confine centromere movements via microtubule growth suppression and spatial control of kinetochore tension. Dev. Cell. 22:1017–1029. 10.1016/j.devcel.2012.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subhash V.V., Tan S.H., Tan W.L., Yeo M.S., Xie C., Wong F.Y., Kiat Z.Y., Lim R., and Yong W.P.. 2015. GTSE1 expression represses apoptotic signaling and confers cisplatin resistance in gastric cancer cells. BMC Cancer. 15:550 10.1186/s12885-015-1550-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Gao J., Dong X., Liu M., Li D., Shi X., Dong J.T., Lu X., Liu C., and Zhou J.. 2008. EB1 promotes Aurora-B kinase activity through blocking its inactivation by protein phosphatase 2A. Proc. Natl. Acad. Sci. USA. 105:7153–7158. (published erratum appears in Proc. Natl. Acad. Sci. USA 2008. 105:7153) 10.1073/pnas.0710018105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Tanaka K., Wakai T., and Hirota T.. 2016. Phosphoproteomic analysis of human mitotic chromosomes identified a chromokinesin KIF4A. Biomed. Res. 37:161–165. 10.2220/biomedres.37.161 [DOI] [PubMed] [Google Scholar]
- Taylor W.R., and Stark G.R.. 2001. Regulation of the G2/M transition by p53. Oncogene. 20:1803–1815. 10.1038/sj.onc.1204252 [DOI] [PubMed] [Google Scholar]
- Tian T., Wang Y., Li J., Zhou F., Cao J., Wang Z., He X., and Su H.. 2016. Clinical significance of GTSE1 expression in colorectal carcinoma. Int. J. Clin. Exp. Pathol. 9:1810–1818. [Google Scholar]
- Towner R.A., Jensen R.L., Colman H., Vaillant B., Smith N., Casteel R., Saunders D., Gillespie D.L., Silasi-Mansat R., Lupu F., et al. . 2013a ELTD1, a potential new biomarker for gliomas. Neurosurgery. 72:77–91. 10.1227/NEU.0b013e318276b29d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner R.A., Jensen R.L., Vaillant B., Colman H., Saunders D., Giles C.B., and Wren J.D.. 2013b Experimental validation of 5 in-silico predicted glioma biomarkers. Neuro-oncol. 15:1625–1634. 10.1093/neuonc/not124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng B.S., Tan L., Kapoor T.M., and Funabiki H.. 2010. Dual detection of chromosomes and microtubules by the chromosomal passenger complex drives spindle assembly. Dev. Cell. 18:903–912. 10.1016/j.devcel.2010.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utrera R., Collavin L., Lazarević D., Delia D., and Schneider C.. 1998. A novel p53-inducible gene coding for a microtubule-localized protein with G2-phase-specific expression. EMBO J. 17:5015–5025. 10.1093/emboj/17.17.5015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst A., and Lens S.M.. 2014. Cell division: control of the chromosomal passenger complex in time and space. Chromosoma. 123:25–42. 10.1007/s00412-013-0437-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Waal M.S., Saurin A.T., Vromans M.J., Vleugel M., Wurzenberger C., Gerlich D.W., Medema R.H., Kops G.J., and Lens S.M.. 2012. Mps1 promotes rapid centromere accumulation of Aurora B. EMBO Rep. 13:847–854. 10.1038/embor.2012.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey K.J., and Hammond J.W.. 2009. Traffic control: regulation of kinesin motors. Nat. Rev. Mol. Cell Biol. 10:765–777. 10.1038/nrm2782 [DOI] [PubMed] [Google Scholar]
- Voets E., Marsman J., Demmers J., Beijersbergen R., and Wolthuis R.. 2015. The lethal response to Cdk1 inhibition depends on sister chromatid alignment errors generated by KIF4 and isoform 1 of PRC1. Sci. Rep. 5:14798 10.1038/srep14798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorozhko V.V., Emanuele M.J., Kallio M.J., Stukenberg P.T., and Gorbsky G.J.. 2008. Multiple mechanisms of chromosome movement in vertebrate cells mediated through the Ndc80 complex and dynein/dynactin. Chromosoma. 117:169–179. 10.1007/s00412-007-0135-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak C.E., Cai S., and Khodjakov A.. 2010. Mechanisms of chromosome behaviour during mitosis. Nat. Rev. Mol. Cell Biol. 11:91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandke C., Barisic M., Sigl R., Rauch V., Wolf F., Amaro A.C., Tan C.H., Pereira A.J., Kutay U., Maiato H., et al. . 2012. Human chromokinesins promote chromosome congression and spindle microtubule dynamics during mitosis. J. Cell Biol. 198:847–863. 10.1083/jcb.201110060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., and Adjaye J.. 2011. A cyclic AMP analog, 8-Br-cAMP, enhances the induction of pluripotency in human fibroblast cells. Stem Cell Rev. 7:331–341. 10.1007/s12015-010-9209-3 [DOI] [PubMed] [Google Scholar]
- Wang F., Dai J., Daum J.R., Niedzialkowska E., Banerjee B., Stukenberg P.T., Gorbsky G.J., and Higgins J.M.. 2010. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 330:231–235. 10.1126/science.1189435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren J.D. 2009. A global meta-analysis of microarray expression data to predict unknown gene functions and estimate the literature-data divide. Bioinformatics. 25:1694–1701. 10.1093/bioinformatics/btp290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren J.D., Bekeredjian R., Stewart J.A., Shohet R.V., and Garner H.R.. 2004. Knowledge discovery by automated identification and ranking of implicit relationships. Bioinformatics. 20:389–398. 10.1093/bioinformatics/btg421 [DOI] [PubMed] [Google Scholar]
- Yamagishi Y., Honda T., Tanno Y., and Watanabe Y.. 2010. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 330:239–243. 10.1126/science.1194498 [DOI] [PubMed] [Google Scholar]
- Yang Z., Tulu U.S., Wadsworth P., and Rieder C.L.. 2007. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr. Biol. 17:973–980. 10.1016/j.cub.2007.04.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zekri A.R., Hassan Z.K., Bahnassy A.A., Khaled H.M., El-Rouby M.N., Haggag R.M., and Abu-Taleb F.M.. 2015. Differentially expressed genes in metastatic advanced Egyptian bladder cancer. Asian Pac. J. Cancer Prev. 16:3543–3549. 10.7314/APJCP.2015.16.8.3543 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.