Neural crest cells are both highly migratory and significant to vertebrate organogenesis. However, the signals that regulate neural crest cell migration remain unclear. In this study, we identify DAN as a novel factor that inhibits uncontrolled neural crest and metastatic melanoma invasion in a manner consistent with the inhibition of BMP signaling.
Abstract
Neural crest cells are both highly migratory and significant to vertebrate organogenesis. However, the signals that regulate neural crest cell migration remain unclear. In this study, we test the function of differential screening-selected gene aberrant in neuroblastoma (DAN), a bone morphogenetic protein (BMP) antagonist we detected by analysis of the chick cranial mesoderm. Our analysis shows that, before neural crest cell exit from the hindbrain, DAN is expressed in the mesoderm, and then it becomes absent along cell migratory pathways. Cranial neural crest and metastatic melanoma cells avoid DAN protein stripes in vitro. Addition of DAN reduces the speed of migrating cells in vivo and in vitro, respectively. In vivo loss of function of DAN results in enhanced neural crest cell migration by increasing speed and directionality. Computer model simulations support the hypothesis that DAN restrains cell migration by regulating cell speed. Collectively, our results identify DAN as a novel factor that inhibits uncontrolled neural crest and metastatic melanoma invasion and promotes collective migration in a manner consistent with the inhibition of BMP signaling.
Introduction
Neural crest cells are multipotent and highly invasive, yet they follow stereotypical migratory pathways. In the vertebrate head, neural crest cells move collectively in discrete streams to reach precise targets. In the hindbrain region, neural crest cells exit the neural tube and are sculpted into three distinct streams adjacent to rhombomeres 2 (r2), r4, and r6 (Trainor and Krumlauf, 2000) such that neural crest cell exclusion zones form adjacent to r3 and r5 (Farlie et al., 1999). Aberrant cranial neural crest cell migration may result from respecification of rhombomere segment identity (Trainor and Krumlauf, 2000) or disruption of signals within the hindbrain (Kulesa and Gammill, 2010). Thus, the proper anterior-to-posterior formation of neural crest–derived tissues of the face and neck (Le Douarin and Kalcheim, 1999) crucially relies on neural crest cell migration.
A small number of signaling molecules have been identified as expressed in the hindbrain region and shown to control initial neural crest cell trajectories into the paraxial mesoderm. These include members of the Eph/ephrin family (Smith et al., 1997; Mellott and Burke, 2008), ErbB4/neuregulin (Dixon and Lumsden, 1999; Golding et al., 2000, 2002, 2004), versican (Landolt et al., 1995; Perris et al., 1996; Kerr and Newgreen, 1997; Perissinotto et al., 2000; Dutt et al., 2006a,b; Szabó et al., 2016), and neuropilin2/semaphorin3F (Eickholt et al., 1999; Osborne et al., 2005; Yu and Moens, 2005; Gammill et al., 2007). Several of these members are secreted factors that are thought to diffuse from the dorsal hindbrain into the local paraxial mesoderm (for example, ErbB4/neuregulin and neuropilin2/semaphorin3F); however, it is largely unknown what signals exist within the paraxial mesoderm that inhibit uncontrolled neural crest cell invasion. Although some neural crest cell inhibitory signals have been identified in several different model organisms, there has not been a systematic approach to isolate tissue and compare gene expression within cranial neural crest cell–free zones that would identify and test the function of novel inhibitory factors and unify existing hypotheses.
Paralleling the invasive ability of the embryonic neural crest, and ancestrally related to this process, melanoma is among the most aggressive human metastatic cancers (Kulesa et al., 2013). However, efforts to constrain melanoma cell invasion have yielded minimal results, making this disease often fatal (Flaherty et al., 2012; Holderfield et al., 2014; Millet et al., 2017). Interestingly, human metastatic melanoma cells transplanted into the chick embryonic neural crest microenvironment have been shown to replicate aspects of the neural crest migration program to promote invasion and plasticity (Kulesa et al., 2006; Bailey et al., 2012; Bailey and Kulesa, 2014). Transplanted human metastatic melanoma cells have been observed to avoid chick embryonic neural crest cell–free zones (Kulesa et al., 2006; Bailey and Kulesa, 2014), suggesting that inhibitory signals that sculpt and maintain discrete embryonic neural crest cell migratory streams may constrain aggressive melanoma cell invasion (Kulesa et al., 2013; Bailey and Kulesa, 2014). Thus, inhibitory molecules within the embryonic neural crest microenvironment should be functionally tested for their potential to inhibit melanoma cell invasion.
We identified differential screening-selected gene aberrant in neuroblastoma (DAN) in a microarray screen that compared gene expression within the chick paraxial mesoderm isolated from adjacent to r3 to migrating r4 neural crest cells. DAN is a secreted factor that was originally identified in a screen examining genes down-regulated upon cell viral transformations (Ozaki and Sakiyama, 1993). DAN acts as a bone morphogenetic protein (BMP) antagonist by binding to BMPs, thereby preventing interaction with BMP receptors (Hsu et al., 1998; Stanley et al., 1998; Pearce et al., 1999). Studies in chick have revealed that DAN signaling is involved in left–right axis formation and inner ear development (Ogita et al., 2001; Gerlach-Bank et al., 2002, 2004; Yamanishi et al., 2007; Katsu et al., 2012). However, no other roles in early development have been investigated, nor has DAN mRNA expression been reported during neural crest migration. In addition, DAN has been identified as a potential marker of malignancy in pancreatic cancer (Olakowski et al., 2009). Thus, the involvement of DAN signaling in embryonic development and pancreatic cancer and our identification of mRNA expression in the neural crest microenvironment make DAN an excellent candidate to further examine as a potential inhibitor of neural crest and metastatic melanoma cell invasion.
In this study, we combined modeling and experimentation to test the function of DAN signaling. We characterized the 3D spatial pattern of DAN expression with respect to the dynamics of the cranial neural crest cell migratory pattern and tested the in vitro function of DAN to inhibit neural crest and metastatic melanoma cell invasion. To determine the in vivo function of DAN, we used gain or loss of function of DAN in the paraxial mesoderm before cranial neural crest migration and measured changes in the resulting migratory pattern and cell behaviors. Simulations that integrated the experimental data into our previous agent-based cell-induced gradient model (McLennan et al., 2015a) were run on an expanded 2D domain to include neural crest cell–free zones and test a mechanistic hypothesis of DAN function. Collectively, our data demonstrate that DAN plays a critical role in cranial neural crest migration and is a potential inhibitor of human metastatic melanoma invasion.
Results
DAN expression correlates with the neural crest cell migratory pattern
To determine the mRNA expression pattern of DAN during cranial neural crest cell migration, we performed a detailed expression analysis across developmental stages (HH Stage [St] 5–16; Hamburger and Hamilton, 1951). Using multiplexed FISH, we discovered that the pattern of DAN expression in the early embryo is more elaborate and dynamic than had previously been characterized with traditional in situ methods (Gerlach-Bank et al., 2002). By combining optical sectioning of fixed, whole-mounted, and cleared chick embryos over a series of developmental stages, we were able to more fully examine and measure fluorescence levels of DAN and FoxD3 (a marker of migratory neural crest cells) expression, which we describe below.
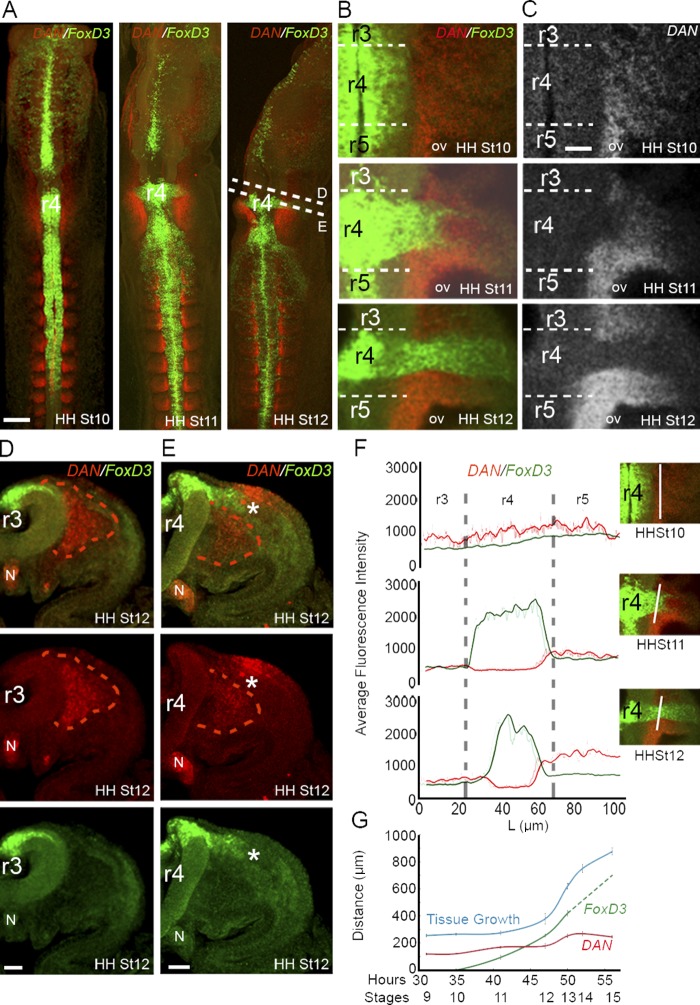
In the hindbrain and before cranial neural crest cell exit from mid-r3 through mid-r5 (HH St 10, 10 somite stage), DAN expression was high in the paraxial mesoderm adjacent to r3–r5 (Fig. 1 A and Video 1). DAN expression extended ventrally in the mesoderm adjacent to the neural tube and lateral along the presumptive dorsal-to-lateral neural crest cell migratory pathway (Fig. 1, A–C; Fig. S1 B; and Video 1). DAN expression was also high in the notochord throughout early development (Fig. 1, D and E; and Videos 1, 2, 3, and 4). As neural crest cells began to exit the dorsal neural tube adjacent to r4 (HH St 11, 13 somite stage), DAN expression remained high in the paraxial mesoderm adjacent to r3 and r5 and directly underneath the dorso-to-lateral neural crest cell migratory pathway (Fig. 1, A–C and E; Fig. S1 B; and Video 2). However, DAN expression was absent in tissue through which neural crest cells have traveled, and FoxD3-positive neural crest cells directly abutted DAN-positive paraxial mesenchymal cells (Fig. 1, B-F; and Video 2). This could be seen clearly through high-resolution consecutive z planes (Video 3). At HH St 12 (16 somite stage), DAN expression remained high in the tissue that surrounds the neural crest cell migratory stream adjacent to r4 (Fig. 1, A–C; and Video 4). DAN was also expressed in the dorsal ectoderm directly overlaying the neural crest cell migratory stream adjacent to r4 but was absent in the ectoderm overlying the paraxial mesoderm adjacent to r3 (Fig. 1, D and E; Fig. S1 B; and Video 4). DAN expression was low in the lateral plate mesoderm (∼200 µm) from the dorsal midline and absent from the forming second branchial arch (Fig. 1 G and Video 4). In summary, neural crest cells initially migrate through a DAN-rich paraxial mesoderm, forming a corridor that is surrounded by DAN-positive regions adjacent to r3, r5, and the ventral mesoderm.
Figure 1.
DAN is expressed by the paraxial mesoderm adjacent to the cranial neural tube. (A) Maximum projections of DAN (red) and FoxD3 (green, neural crest marker) expression in whole embryos at HH St 10, 11, and 12 (n = 10, 12, and 4 embryos, respectively). DAN is expressed in the paraxial mesoderm in the cranial region and segmented in the somites in the trunk regions. (B) Single z planes of DAN and FoxD3 expression in the paraxial mesoderm and the developing otic vesicle (OV) at the axial level of the r4 neural crest cell stream. DAN in expressed by the mesodermal cells immediately bordering the neural crest cell stream. (C) Same images as shown in B, except with DAN expression only. (D) Transverse section of DAN and FoxD3 expression at r3 axial level at HH St 12. DAN is expressed by the paraxial mesoderm. (E) Transverse section of DAN and FoxD3 expression at r4 axial level at HH St 12. DAN is expressed by the paraxial mesoderm and may limit the migrating neural crest cells (asterisks) to the dorsolateral pathway. DAN is also expressed in the thickening ectoderm associated with otic vesicle development. (F) Mean intensity of DAN (red) and FoxD3 (green) expression at HH St 10, 11, and 12 measured adjacent to the neural tube (NT) from mid-r3 to mid-r5 as shown in images on the right (white lines). (G) Width of FoxD3 and DAN expression measured as the distance from the edge of the neural tube to the end of expression over time (hours and HH stages shown). Neural crest cells initially migrate into but eventually migrate past the DAN-rich region (Stage 11–12). The dashed FoxD3 line is predicted neural crest migration at later stages. Bars: (A) 200 µm; (B–E) 50 µm. N, notochord; L, length.
DAN expression in the midbrain is dynamic. We observed low DAN expression in the paraxial mesoderm all along the neural tube from the midbrain to the hindbrain at HH St 8 (4-somite stage; Fig. S1 A). By HH St 9 (7-somite stage), DAN expression was stronger in the ventral paraxial mesoderm, and it persisted during neural crest migration through HH St 15 (24–27-somite stage; Fig. S1 A and Video 5). DAN also appeared highly expressed in the ectoderm overlaying the mesencephalic neural crest cell migratory pathway at HH St 12 and persisted to HH St 15 (Fig. S1 A).
In the trunk, we observed DAN expression restricted to the newly formed caudal somite halves, and this expression pattern expanded to the rostral somite in the anterior presomitic mesoderm (Fig. 1 A; Fig. S1 C; and Videos 1, 2, 3, and 4). As trunk neural crest cells emerged from the dorsal neural tube and moved ventrally in between the neural tube and somite, the expression of DAN was in both rostral and caudal somite halves but was restricted to the medioventral portion of the sclerotome (Fig. S1 C). At these later stages, DAN expression was ventral to the migrating neural crest in the sclerotome (Fig. S1 C). In a similar manner to the head, DAN expression remained within tissue that surrounded the trunk neural crest cell streams as the lead cells reached ventral target sites.
Cranial neural crest and metastatic melanoma cells avoid DAN protein in vitro
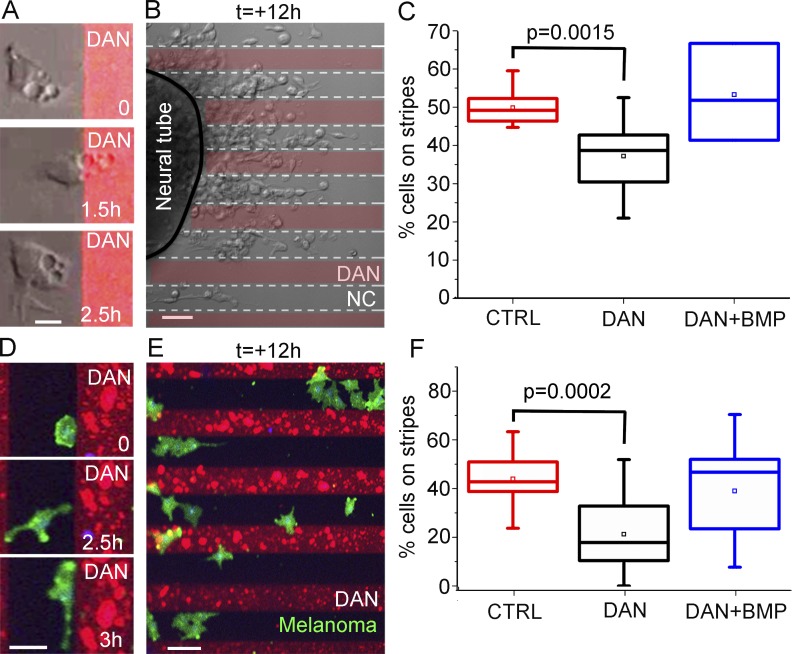
To determine whether DAN affects the invasive ability of cranial neural crest cells in vitro, we explanted cranial neural tubes onto alternating stripes of DAN protein and fibronectin as well as onto fibronectin only (Fig. 2). Measurements showed that significantly more neural crest cells were found on fibronectin-only stripes rather than on DAN protein and fibronectin stripes (Fig. 2, A–C). Time-lapse imaging revealed that neural crest cells migrated out from neural tube explants, encountered the DAN protein stripes, and either rapidly moved across a DAN stripe or retracted protrusions and moved within a fibronectin-only stripe (Fig. 2 A and Video 6). Measurements of cells closer to the neural tube (within 50 µm of the neural tube explant) showed an equal distribution of neural crest cells on/off a DAN stripe (Fig. S4, A and B). This phenomenon appeared to be cell density related; cells that exited the neural tube explanted adjacent to a DAN stripe, requiring space to be made available by other neighboring exiting cells moving off the DAN stripe (Fig. 2 B). When we increased the concentration of DAN protein in the stripes (8–20 µg/ml), we continued to observe cells moving off the DAN stripes wherever space allowed (Fig. 2 B). These data suggest that DAN is inhibitory rather than repulsive to migrating neural crest cells.
Figure 2.
Neural crest and melanoma cells avoid DAN in vitro. (A) Images of a time lapse showing how a single cranial neural crest (NC) cell contacts DAN (red) and then changes the direction of migration. Bar, 15 µm. (B) Image from a time lapse of migrating cranial neural crest cells grown on alternating stripes containing DAN after 12 h of imaging. Bar, 50 µm. (C) Box plot of the percentage of neural crest cells on stripes either containing no factor (red, n = 10 cultures), DAN (black, n = 9 cultures), or DAN and BMP4 (blue, n = 3 cultures). (D) Images of a time lapse of a single melanoma cell (green) contacting and then migrating along the edge of the DAN (red). Bar, 30 µm. (E) Image from a time lapse of migrating melanoma cells grown on alternating stripes containing DAN after 12 h of imaging. Bar, 50 µm. (F) Box plot of the percentage of melanoma cells on stripes either containing no factor (red, n = 12 cultures), DAN (black, n = 18 cultures), or DAN and BMP4 (blue, n = 20 cultures). Two-sided Student’s t test.
To assess whether DAN can inhibit human neural crest–derived cancer cell types, we introduced C8161 metastatic melanoma cells to DAN protein stripes in vitro (Fig. 2, D–F). Measurements showed that the majority of C8161 melanoma cells were found between DAN stripes (Fig. 2 F). When C8161 melanoma cells were observed by time-lapse confocal microscopy, cells actively avoided DAN stripes (Video 7). In a typical time-lapse imaging session, a C8161 melanoma cell moved and extended protrusions within the width of a fibronectin-only stripe (Fig. 2 D). Because DAN is a known BMP antagonist (Hung et al., 2012; Katsu et al., 2012; Nolan et al., 2015), we asked whether the inhibitory response of neural crest and/or C8161 melanoma cells to DAN could be abrogated by the addition of an exogenous BMP source. When we added BMP4 to the DAN stripe assay, both the neural crest and C8161 melanoma cells did not show a preference to avoiding DAN stripes (Fig. 2, C and F). Thus, based on the DAN mRNA expression data and time-lapse analysis of cell behaviors on DAN/fibronectin stripe assays, we decided to examine the in vivo function of DAN.
Addition of exogenous DAN in the in vivo microenvironment reduced neural crest cell migration
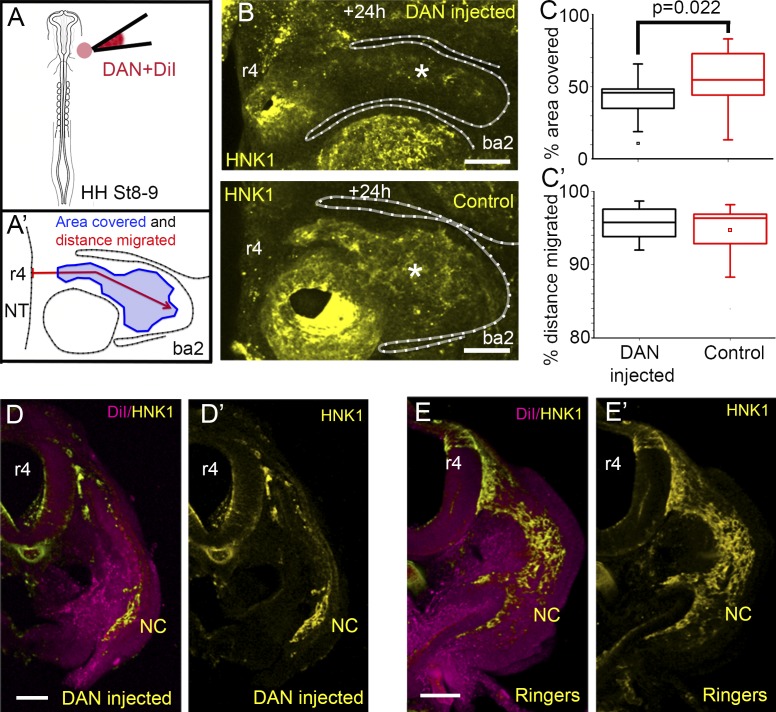
To test the function of DAN in cranial neural crest cell migration, we microinjected soluble human DAN protein into one side of the paraxial mesoderm adjacent to the hindbrain before neural crest cell emigration (Fig. 3 A). After reincubation, we observed a substantial decrease in the number of migrating neural crest cells on the injected versus noninjected control side in the same embryo (Fig. 3 B). Specifically, when we measured the fluorescence intensity of HNK-1–labeled neural crest cells within a subregion of the second branchial arch, the percentage of area covered by HNK-1–positive cells showed a significant decrease after addition of exogenous DAN when compared with control sides in the same embryo (Fig. 3, A′ and C). A similar significant decrease in the percentage of area covered was seen 12 h after addition of exogenous DAN but not 24 h after, suggesting degradation of the exogenous DAN protein (Fig. S2, A–C). Examination of transverse sections showed that migrating neural crest cells formed thinner streams and avoided regions containing exogenous DAN protein (Fig. 3 D; compare with Fig. 3 E). We observed significantly fewer neural crest cells that were able to escape through or around the DAN injection site to reach the distal portion of the second branchial arch (Fig. S2 D), but the total distance migrated was not significantly different from controls (Fig. 3, C′ and D). Furthermore, we observed a reduction in the numbers of migrating neural crest cells when we injected human DAN full-length (FL) lentiviral particles into one side of the paraxial mesoderm adjacent to the hindbrain compared with the control side in the same embryo, consistent with addition of exogenous DAN protein (Fig. S2 E). Collectively, this suggests that enhanced DAN inhibition in vivo reduces the number of invading neural crest cells, but those cells that escape may reach branchial arch targets.
Figure 3.
Neural crest cell migration is decreased when DAN is overexpressed in the paraxial mesoderm. (A) Schematic representation of DAN protein/DiI injections in to the paraxial mesoderm. (A′) Schematic representation of the measurements made for analysis of DAN-related phenotypes. (B) HH St 15 embryo after injection of DAN protein into the paraxial mesoderm showing the injected side (top) and uninjected control side (bottom). Less neural crest (NC) cells migrate toward the branchial arch when DAN is overexpressed (asterisks). (C) Area of the branchial arch region containing neural crest cells 16 h after injection. n = 22 embryos. (C′) Distance of the branchial arch that neural crest cells reached 16 h after injection. n = 22 embryos. Two-sided Student’s t test. (D) Transverse section through an embryo in which DAN and DiI (magenta) have been injected into the paraxial mesoderm. (D′) Same section as in D, neural crest cells only. (E) Transverse section through an embryo in which Ringers and DiI (magenta) have been injected into the paraxial mesoderm. (E’) Same section as in E, neural crest cells only. Bars: (B) 100 µm; (D and E) 50 µm. NT, neural tube; OV, otic vesicle.
Knockdown of DAN function in vivo enhanced neural crest cell migration
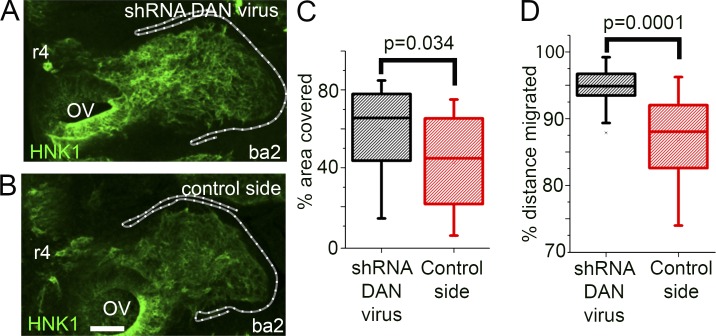
To determine whether knockdown of DAN signaling has an in vivo effect on the cranial neural crest cell migratory pattern, we injected DAN shRNA lentiviral particles into one side of the paraxial mesoderm adjacent to the hindbrain before neural crest cell emigration (HH St 9; at the r4 axial level; Figs. 4 A and S5). After 24 h of egg reincubation, the resulting neural crest migration pattern was compared with the control side in the same embryo (Fig. 4, A and B) as well as those from experiments repeated using scrambled shRNA and empty control lentiviral particles (Fig. S3, A and B; and Fig. S5). Knockdown of DAN expression in the paraxial mesoderm after injection of DAN shRNA lentiviral particles was confirmed by FISH compared in embryos injected with the scrambled shRNA (control) or DAN shRNA lentiviral particles (Fig. S3, E–G).
Figure 4.
Neural crest migration is increased when DAN is knocked down in the paraxial mesoderm. (A) HH St 15 embryo after injection of shRNA DAN lentiviral particles into the paraxial mesoderm. (B) Control side of embryo shown in A. Bar, 50 µm. (C) Area of the branchial arch region containing neural crest cells. (D) Distance of the branchial arch that neural crest cells reached. n = 20 embryos injected with shRNA DAN lentiviral particles. Two-sided Student’s t test. OV, otic vesicle.
We found significantly more migrating neural crest cells on the DAN shRNA-injected side compared with the control side in the same embryo as measured by the percentage of area covered (Fig. 4, A–C). We also found a significant increase in the percent distance migrated of neural crest cells in the DAN shRNA-injected embryos, suggesting that cells traveled faster to reach the second branchial arch (Fig. 4 D). Control empty or scrambled shRNA lentiviral particles injected into the mesoderm did not alter the neural crest cell area covered or total distance migrated (Fig. S3, C and D). We did not observe aberrant neural crest cell invasion into typical neural crest cell–free zones adjacent to r3 and r5 (Fig. 4 A), suggesting that endogenous inhibitory cues in those regions were unaffected. A similar phenotype was observed when DAN was knocked down in the paraxial mesoderm via morpholino (MO) transfection (Fig. S3, H–K). In summary, loss of DAN signaling in the paraxial mesoderm led to increased cell invasion along the neural crest migratory pathway but not uncontrolled invasion into typical neural crest cell–free zones.
The influence of DAN on neural crest cell migration is consistent with inhibition of BMP signaling
DAN is known to be a moderate BMP antagonist (Nolan et al., 2015). To investigate whether the influence of DAN on neural crest cell migration was consistent with BMP modulation, we performed two BMP-specific experiments. First, because DAN is a known antagonist of BMP2 and BMP4 (Hung et al., 2012; Katsu et al., 2012), we knocked down the production of BMP2 in premigratory cranial neural crest cells via MO transfection (Fig. 5 A). BMP4 has been shown to be a key regulator of neural crest delamination in conjunction with noggin (Sela-Donenfeld and Kalcheim, 1999, 2000), so we did not perturb BMP4. Recently, we have discovered that BMP2 is expressed by lead neural crest cells at significantly higher levels than trailing cells during migration (unpublished data). BMP2 has also been implicated in mouse neural crest cell migration (Zhang and Bradley, 1996; Kanzler et al., 2000; Tribulo et al., 2003; Anderson et al., 2006; Correia et al., 2007). Thus, we postulated that knockdown of BMP2 signaling in migrating cranial neural crest cells would result in a phenotype similar to the addition of exogenous DAN. That is, we anticipated fewer migrating neural crest cells in BMP2 MO embryos.
Figure 5.
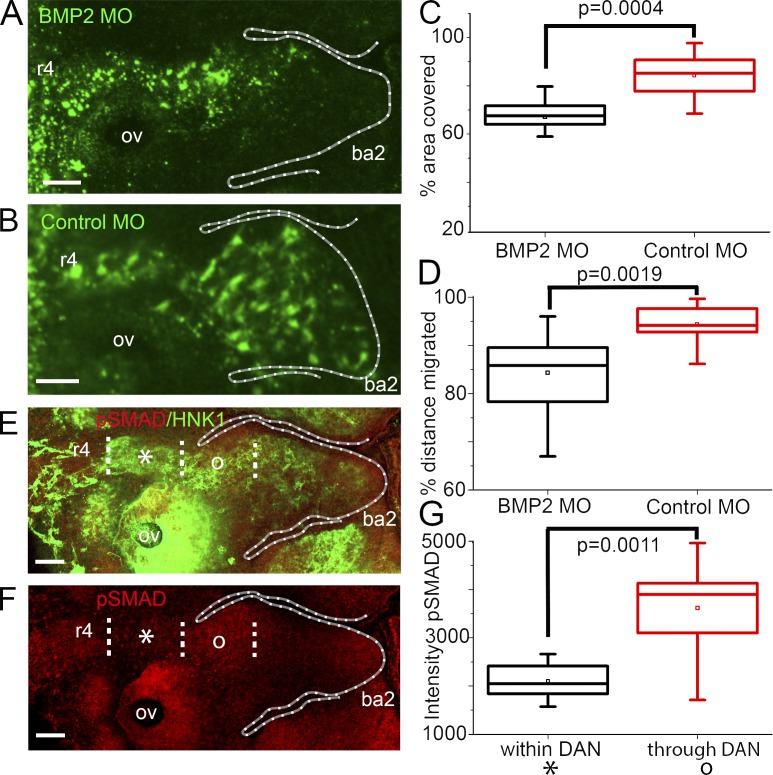
DAN perturbations are consistent with BMP signaling. (A) HH St 15 embryo in which neural crest cells are transfected with BMP2 MO. (B) HH St 15 embryo in which neural crest cells are transfected with control MOs. (C) Area of the branchial arch region containing neural crest cells. n = 21 BMP2 MO embryos; n = 8 control MO embryos. (D) Distance migrated by neural crest into ba2. n = 21 BMP2 MO embryos; n = 8 control MO embryos. (E) HH St 15 embryo labeled with HNK-1 (green) and pSMAD (red). (F) Same embryo as in E but with pSMAD only. Dotted lines represent neural crest cells within (asterisks) and through (o) the DAN-rich region. Bars, 50 µm. (G) Intensity of pSMAD antibody stain within and through DAN. n = 8 embryos. Two-sided Student’s t test. OV, otic vesicle.
We found that BMP2 MO transfection resulted in reduced neural crest migration when compared with control MO transfection (Fig. 5, A and B). Measurements showed that BMP2 MO–transfected neural crest cells did not migrate as far as in control embryos and that there were fewer migrating neural crest cells as measured by the percentage of the area covered by a typical neural crest migratory stream (Fig. 5, C and D).
Because BMPs bind to BMP receptors that consequently lead to downstream activation of SMADs (Reddi, 2001), we next investigated the activation of BMPs by visualizing phosphorylated SMAD signaling. We anticipated that migrating cranial neural crest cells that have passed through the DAN-rich paraxial mesoderm show increased BMP signaling as indicated by an increase in phosphorylated SMAD activity. We found that analysis of the intensity of phosphorylated SMADs is significantly increased in migrating neural crest cells after cells exit the DAN expression region at distances >200 µm from the neural tube when compared with neural crest cells within the DAN expression region (≤75 µm from the dorsal neural tube exit; Fig. 5, E–G). Collectively, these data suggest that our manipulations of DAN signaling and the resulting effects on the neural crest cell migratory pattern are consistent with DAN acting as a BMP antagonist to retard neural crest cell migration.
DAN reduced neural crest cell speed and proliferation in vitro
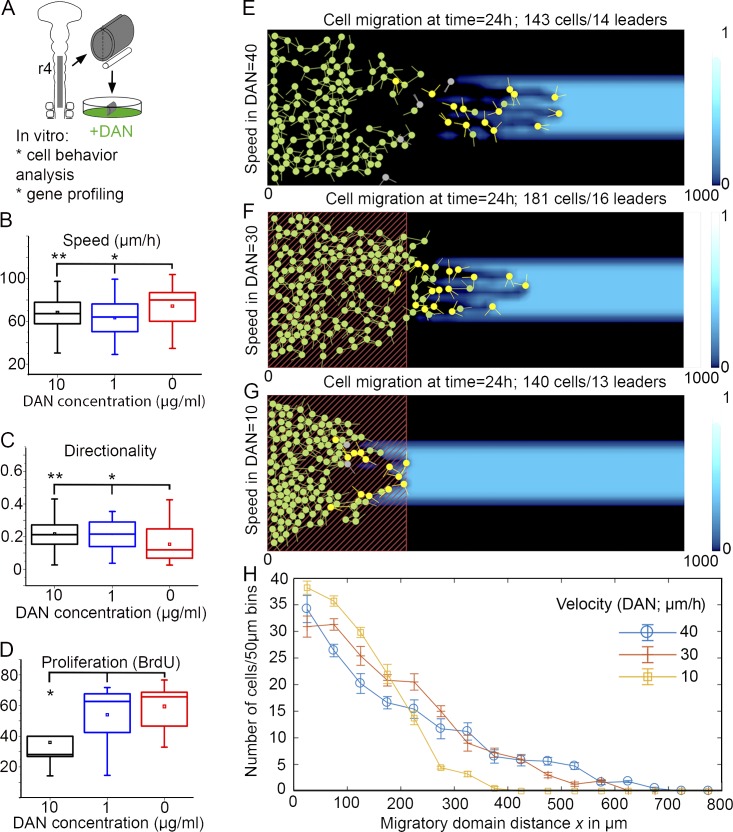
To test the direct effects of DAN on neural crest cell speed without the influence of the embryonic microenvironment, we isolated cranial neural crest cells in vitro and exposed them to 0, 1, and 10 µg/ml of DAN protein (Fig. 6 A). This experimental design was used as opposed to the stripe assays because in the stripe assays, a cell can be outside a DAN-containing stripe and yet still touch and respond to DAN. Therefore, analyzing speed on and off DAN-containing stripes was not informative (Fig. S4 C). Cell trajectories were tracked from time-lapse confocal imaging sessions, and cell proliferation was examined using BrdU (Fig. 6, A–D). We found that the speed of neural crest cells upon exposure to DAN, at all concentrations tested, was significantly reduced (Fig. 6 B). Neural crest cell directionality was slightly increased upon exposure to DAN (Fig. 6 C), and cell proliferation was significantly reduced upon exposure to high concentrations of DAN (Fig. 6 D). DAN protein bound to the culture dish, as opposed to soluble DAN, showed no significant effects upon neural crest cell speed and direction (Fig. S4, D and E). Collectively, these results support our in vivo results that soluble DAN protein reduces neural crest cell speed and proliferation.
Figure 6.
In vitro and in silico experimental data reveal that DAN reduces neural crest cell speed. (A) Schematic representation of experiment. (B) Speed of neural crest cells grown in vitro, exposed to 0 (n = 46 cells), 1 µg/ml (n = 30 cells), and 10 µg/ml (n = 70 cells) of DAN protein. *, P < 0.013; **, P < 0.043. (C) Directionality of the same cells tracked in B. *, P < 0.0079; **, P < 0.0012. (D) BrdU incorporation as an indicator of cell proliferation when exposed to 0 (n = 7 neural tube cultures, 961 cells), 1 µg/ml (n = 8 neural tube cultures, 847 cells), and 10 µg/ml (n = 9 neural tube cultures, 980 cells). *, P = 0.016. Two-sided Student’s t test. (E) No reduction in speed: the model simulated is as in McLennan et al. (2015b) on a widened domain with cell speed 40 µm/h. Neural crest cells are shown in green, cells in a leader state are in yellow, detached trailing cells in green, and the background blue color shows VEGF chemoattractant concentration. (F) Moderate reduction in speed: inside the area representing DAN expression (red dashed rectangle), cell speed is reduced in proportion to DAN concentration (see the Computer modeling supports the hypothesis… section of Results). The minimum cell speed is 30 µm/h. (G) Modeling increased DAN: the simulation is set up as in B, but the cell speed is now reduced down to 10 µm/h (at peak concentration of DAN). (H) Migration profiles: mean cell number versus distance migrated for 10 repeated simulations as in E–G. Error bars show SEM.
When C8161 melanoma cells were exposed to soluble DAN protein, cell trajectories showed an increase in cell speed and a decrease in directionality (Fig. S4, F and G). C8161 melanoma cell proliferation was increased in the presence of DAN in culture as assessed by BrdU labeling (Fig. S4 H). These results suggest that even though both melanoma and neural crest cells avoid DAN in vitro, they respond to DAN by different mechanisms.
Computer modeling supports the hypothesis that DAN restrains cell migration by regulating cell speed
To test the mechanistic features of our hypothesis difficult to probe in vivo, we extended the computational modeling framework of our cell-induced (or self-generated) gradient model (McLennan et al., 2015a) to explore two novel hypotheses. In vivo, there is no physical boundary between the neural crest migratory pathway and neural crest cell–free zones that arise adjacent to r3 and r5. We previously confined neural crest cells to enter a 2D growing migratory domain corresponding with the width of a single rhombomere segment, r4, and respond to a dynamic VEGF chemoattractant on this growing domain (McLennan et al., 2012, 2015a). We included no flux boundary conditions along the anterior and posterior sides of the 2D migratory domain in order to restrict cell movements to a self-imposed corridor. In a typical simulation, parameters were set so that cells responded to the VEGF chemoattractant with a constant sensing accuracy and had constant speed.
In this study, when we performed simulations on an extended 2D migratory domain that expanded the no-flux boundaries beyond the neighboring neural crest cell–free zones, we found that the model reproduced typical neural crest cell stream formation and migration patterns (Fig. 6 E). However, we noted two differences. First, cells within the stream in silico reached a total distance migrated that was significantly increased relative to the typical in vivo stream (Fig. 6 E and Video 8). In addition, the in silico migratory stream often separated, with the majority of the trailing cells remaining near the neural tube because of their loss of direction information from leaders (Fig. 6 E). These simulation results show that our model remains robust both without (McLennan et al., 2015a) and with (Fig. 6 E) an extended neural crest entry domain. Furthermore, it revealed that if small numbers of lead cells escape from the migratory front, collective cell migration is interrupted.
Given our simulation results and our in vitro DAN data, we hypothesized that DAN functions to slow lead neural crest cells to enhance the overall collective migration of the stream. To test this hypothesis, we included the presence of DAN into the model 2D microenvironment by modeling DAN production within a rectangle adjacent to the entry location of the cells. The size and changes in production rate of DAN over time were based on measurements of DAN expression from the embryo (Fig. S1 D). Importantly, we integrated DAN inhibition into the model by decreasing cell speed according to the level of DAN expression. If we assumed that the cell speed dropped to 30 µm/h in the DAN domain (Fig. 6 B), cells were more evenly distributed in the stream, and leaders were increasingly restricted to the migratory front (Fig. 6, F and H; and Video 8). Cells that did migrate completely through the DAN region were able to travel further distally within the migratory domain compared with normal embryos (Fig. 6, E and F). When we overexpressed DAN in silico by reducing the cell speed to 10 µm/h, simulations predicted that very few cells migrated through and out of the DAN region (Fig. 6, G and H; and Video 8). Thus, model simulations that tested the function of DAN to modulate cell speed through a domain near the entrance to the migratory route predicted that collective neural crest cell migration can be facilitated by a spatial pattern of DAN expression through regulation of cell speed within range of the dorsal neural tube.
Knockdown of DAN increased neural crest cell speed, directionality, and proliferation in vivo
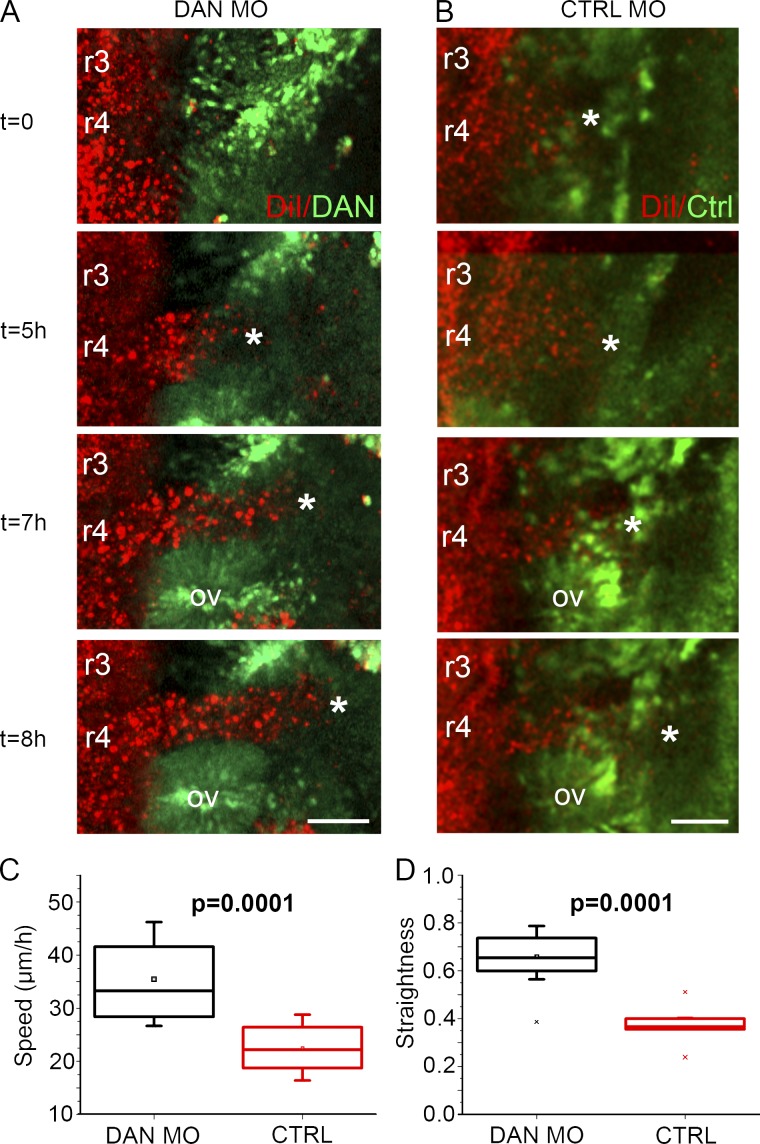
Based on the results of our in vitro analysis and computer simulations, we next wanted to determine whether DAN influences migratory and proliferative properties in vivo. Therefore, we reduced endogenous DAN in the cranial mesoderm via DAN MO transfection and performed time-lapse confocal imaging (Fig. 7). When endogenous DAN was knocked down in vivo, neural crest cells invaded more efficiently than unperturbed neural crest cells (Fig. 7, A and B; and Video 9). Specifically, after DAN knockdown, neural crest cells migrated with a speed of 35.4 µm/h, which was significantly faster than the neural crest cell speed of 22.3 µm/h after electroporation of control MOs into the same area (Fig. 7 C). The control MO neural crest speed determined in this study is consistent with previous cranial neural crest cell tracking experiments (Ridenour et al., 2014). Knockdown of endogenous DAN expression also significantly increased the directionality of neural crest cells (Fig. 7 D). Finally, after DAN knockdown, there was significantly higher proliferation in the cranial neural crest cell stream when compared with the control sides of the same embryos (Fig. S3 L). These in vivo data are consistent with our in vitro data as well as our static observations and further support our hypothesis that DAN influences cell invasion and proliferation.
Figure 7.
In vivo neural crest cell speed, directionality, and proliferation are increased when DAN is knocked down in the paraxial mesoderm. (A) Selected images from a time lapse showing neural crest cells (red) migrating into the paraxial mesoderm transfected with DAN MOs (green). (B) Selected images from a time lapse showing neural crest cells migrating into paraxial mesoderm transfected with control MOs (green). Asterisks indicate the front of the neural crest migratory stream. Bars, 50 µm. (C) Speed of neural crest cells migrating into paraxial mesoderm transfected with DAN MOs (n = 13) and control MOs (n = 9). (D) Directionality of the same cells tracked in C. Two-sided Student’s t test. OV, otic vesicle.
Exposure to exogenous DAN protein alters the molecular profile of in vitro migrating neural crest cells
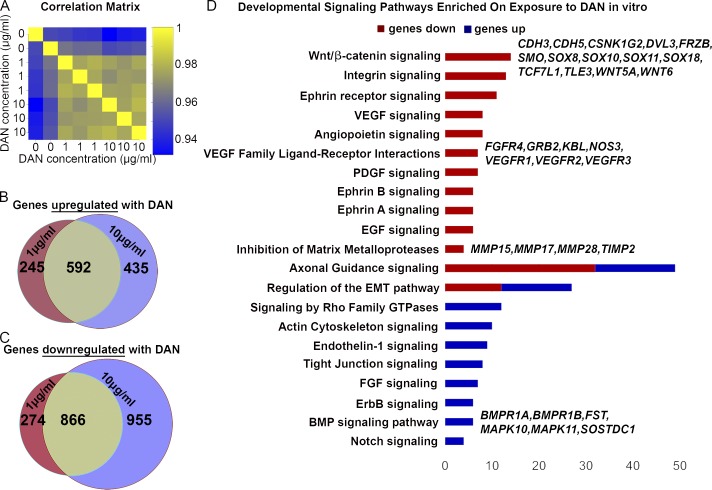
To test the direct effects of DAN on gene expression of migrating neural crest cells, isolated cranial neural crest cells were again exposed to 0, 1, and 10 µg/ml of DAN in vitro, collected, and profiled by RNAseq (Fig. 8 A). Results upon exposure to the two different concentrations of DAN were very similar to each other (Fig. 8 A), with an overlap of 592 up-regulated (80 are receptors and kinases) and 886 down-regulated genes (Fig. 8, B and C; and Table S1). Pathway analysis showed that DAN exposure decreased the expression of many guidance receptors in signaling pathways such as VEGF, Eph/ephrins, PDGF, and EGF (Fig. 8 D). Furthermore, upon exposure to DAN, there was increased expression of actin cytoskeleton and GTPase signaling (Fig. 8 D). Specific examples of receptors, guidance factors, and transcription factors whose expression changed after the addition of DAN included Eph, FGFs, BMP, and VEGF receptors as well as SOX and TFAP transcription factors that are involved in neural crest migration and specification (Table S1). These results are consistent with DAN influencing a cell’s ability to respond to chemotaxis and neighboring cells.
Figure 8.
DAN influences gene expression of neural crest cells. (A) Correlation matrix of the biological replicates exposed to different concentrations of DAN. (B) Venn diagram of genes up-regulated upon exposure to DAN. (C) Venn diagram of genes down-regulated upon exposure to DAN. (D) Integrated pathway analysis displaying up- and down-regulation of genes associated with developmentally relevant pathways. (E) Graph of representative guidance cues and transcription factors that are up- or down-regulated upon exposure to DAN.
Discussion
Our discovery of high DAN expression in the paraxial mesoderm adjacent to r3 (neural crest cell–free zone) by differential screening suggested its potential as an inhibitor of neural crest cell migration. By optimizing a novel multiplexed FISH method in chicks (Choi et al., 2014; McLennan et al., 2015a), we overcame previous hurdles to accurately detect and resolve mRNA expression borders with respect to neural crest migratory streams in whole embryos. This resulted in the characterization of a richer pattern of DAN expression in both the head and trunk. By blending modeling and experiments with in vitro stripe assays and in vivo gain and loss of function, we learned that the role of DAN is more complex and includes four aspects of DAN function.
First, we discovered the DAN expression pattern is dynamic in space and time, initially detected within the paraxial mesoderm adjacent to the hindbrain at a stage before neural crest cell emigration, and expanded laterally commensurate with tissue growth (Figs. 1 and 9). As neural crest cells migrate through the DAN-rich region, a DAN-negative corridor forms (Fig. 1). Whether migrating neural crest cells displace the loosely connected DAN-positive paraxial mesodermal cells or alter DAN expression within these cells is unclear. Although we do not show DAN protein expression, BMP antagonists are thought to rapidly associate with the extracellular matrix and therefore only influence nearby cells (Miyazaki et al., 2008). At the neural crest cell invasive front, it is possible to detect small fingers of DAN-negative subregions that correlate with neural crest cells moving into those subregions (Figs. 1 and S1 B). Further analysis with time-lapse imaging of embryos in which the neural crest and paraxial mesoderm are fluorescently labeled or static imaging of a fluorescently marked paraxial mesoderm could reveal whether there is a physical displacement of mesodermal cells by the invasive neural crest.
Figure 9.
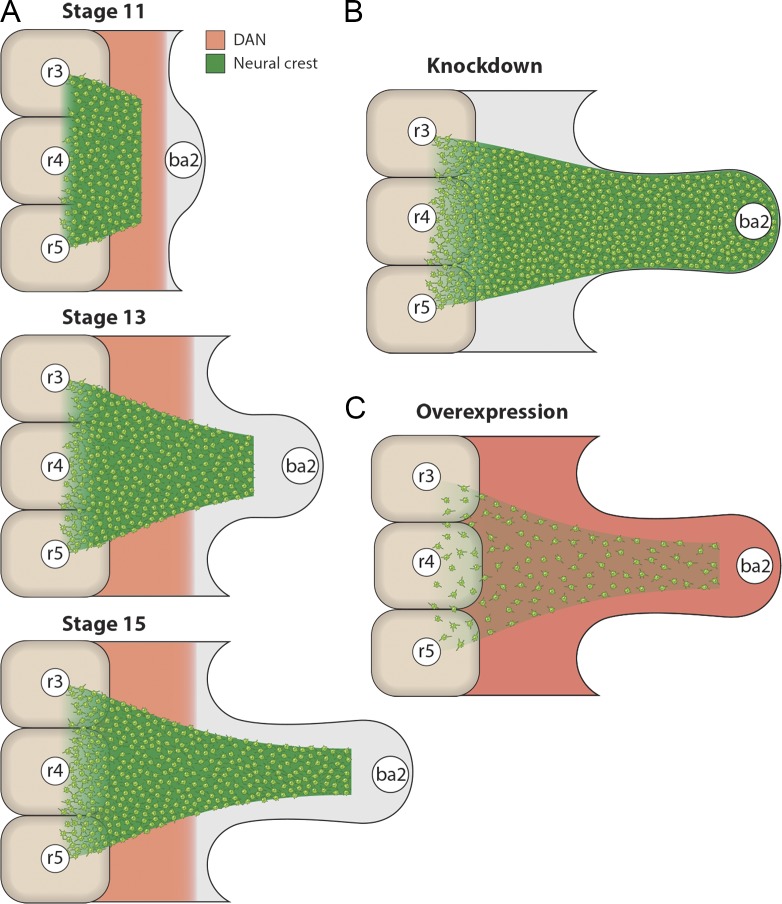
Schematic representations of expression and perturbations of DAN. (A) Expression of DAN in the mesoderm and migrating neural crest cells at HH St 11, 13, and 15. Neural crest cells initially are migrating within the DAN-rich corridor (HH St 11), but then lead neural crest cells migrate through the DAN-rich corridor (HH St 13) and go on to invade the branchial arch target tissue (HH St 15). The width of the DAN-rich corridor increases over time, which is consistent with domain growth. (B) When DAN expression is knocked down, more neural crest cells are present in the stream. (C) When DAN is overexpressed, less neural crest cells are present in the stream.
In the trunk, DAN is expressed throughout the presomitic mesoderm (Fig. S1 C). DAN becomes restricted to the dorsomedial sclerotome within the rostral and caudal somite halves (Fig. S1 C). Thus, by the time trunk neural crest cells emigrate through the rostral somite half, DAN is expressed in a region adjacent to the neural tube in a pattern similar to that reported in the head (Fig. 1). Further studies may elucidate whether DAN plays a similar function in the trunk, because neural crest cells migrate in loosely connected streams (Kasemeier-Kulesa et al., 2005). Expression of other BMP antagonists (e.g., Noggin), including DAN family members (e.g., Gremlin and Cerberus), has not been observed in the cranial paraxial mesoderm. Rather, Noggin and Gremlin are expressed in migrating cranial neural crest cells and have been implicated in the neural crest cell differentiation program to form vertebrate head muscle (Tzahor et al., 2003; Bothe et al., 2011). Chordin is expressed in the notochord (Streit et al., 1998), whereas Cerebus is unilaterally expressed on the left side of the early chick embryo (Zhu et al., 1999) and is suggested to play a role in laterality decisions. Thus, the DAN expression pattern in the trunk mimics that in the head, and future studies of DAN and other DAN family members may provide insights into their potential interaction.
In the mouse, DAN is expressed by the branchial arches, as opposed to adjacent to the neural tube; however, the trunk expression pattern is similar between the two species (Stanley et al., 1998; Pearce et al., 1999; Kim and Pleasure, 2003). DAN knockout mice are viable and have no obvious defects, but any possible defects specific to neural crest migration are yet to be investigated (Dionne et al., 2001). In mice, BMP2 is required for cranial neural crest cell migration, not their delamination (Correia et al., 2007), and therefore it may be that a different combination of BMP antagonists work with BMP2 to control collective neural crest cell migration in mice than in chicks.
Second, we observed a reduction in neural crest cell distance migrated in vivo in DAN gain-of-function embryos and reduced individual cell speed in vitro in the presence of DAN protein (Figs. 3, 6, and 9). We also observed enhanced and faster neural crest cell migration in DAN loss-of-function embryos by both MO and shRNA knockdown in the paraxial mesoderm and static and time-lapse imaging (Figs. 4 and 7). Our computer model simulations that interpreted DAN-rich regions to reduce individual cell speeds supported this proposed functional role for DAN (Fig. 6). Interestingly, knockdown of DAN in vivo not only increased cell speed but also directionality. After taking into account the molecular profiles of neural crest cells grown in vitro in the presence of DAN, we speculate that after knockdown of DAN in vivo, neural crest cells up-regulate important guidance receptors while reducing tight junctions, which would result in cells that are faster and able to migrate in a more directed manner.
Our results are consistent with the function of DAN as a BMP inhibitor during neural crest migration. It is tempting to speculate that DAN is involved in the BMP-regulated delamination of the neural crest, and it indeed may play a minor role in delamination. However, upon addition of exogenous DAN, we did not see larger dorsal neural tubes, indicative of a delamination defect. With other stronger BMP antagonists expressed by delaminating neural crest (Sela-Donenfeld and Kalcheim, 1999, 2000; Tzahor et al., 2003; Bothe et al., 2011) and our data demonstrating that DAN influences speed, directionality, and proliferation, it is clear that the major role of DAN is during the collective migration of the neural crest. We showed that BMP2-MO–transfected neural crest cells do not migrate as far as WT cells (Fig. 5), a phenotype similarly observed when we overexpressed DAN in vivo in the paraxial mesoderm (Fig. 3). Exogenous DAN protein or DAN FL lentiviral particles microinjected into the paraxial mesoderm resulted in fewer migrating neural crest cells and a significant reduction in the number of cells reaching branchial arch 2 (ba2; Fig. 4). A similar reduction was observed when Noggin was added to mouse gut cultures and blocked BMP2/4-expressing enteric neural crest cells (Goldstein et al., 2005). Goldstein et al. (2005) speculated that GDNF-stimulated enteric neural crest cell chemoattraction was reduced in Noggin-treated gut cultures. Interestingly, we found a significant reduction in the expression of VEGF and axonal guidance signaling after in vitro exposure of neural crest cells to DAN protein (Fig. 8). Because we also observed a significant increase in neural crest cell directionality in vivo after the knockdown of DAN, it is attractive to suggest that DAN functions to reduce VEGF- or other guidance factor–stimulated chemotaxis. Future studies that coculture neural tube explants with VEGF-soaked beads or isolated ba2 tissue in the presence and absence of DAN protein should address this hypothesis.
Third, exogenous DAN protein in culture reduced neural crest cell proliferation, whereas endogenous knockdown of DAN in vivo increased neural crest cell proliferation, suggesting an in vivo role to maintain mitotic quiescence in migrating neural crest cells until cells reached peripheral targets. The computer modeling was done independently of the wet laboratory experiments, with only the DAN expression pattern and initial static data from the addition of exogenous DAN experiments to work from. Therefore, proliferation was not a component to the resulting simulations. To further support our wet laboratory observations of DAN’s role in proliferation, we previously showed that migrating chick cranial neural crest cells are mitotically quiescent until reaching the branchial arches (Ridenour et al., 2014). Additionally, DAN has previously been shown to repress cell growth (Ozaki and Sakiyama, 1994; Ozaki et al., 1995; Hanaoka et al., 2000). Thus, DAN inhibition of BMP signaling was in concert with neural crest cell speed, directionality, and proliferation activity.
Fourth, we found that DAN is a candidate inhibitor of human metastatic melanoma, suggesting that the establishment of nonpermissive tissue boundaries that are critical for maintaining discrete embryonic neural crest cell migratory streams may be mined for factors such as DAN to control cancer cell invasion. Our data revealed that aggressive human C8161 melanoma cells avoid DAN protein stripes in stripe assays (Fig. 2), and we have previously shown that these cells follow host embryonic neural crest cell migratory pathways when introduced into the chick dorsal neural tube (Kulesa et al., 2006; Bailey et al., 2012; Bailey and Kulesa, 2014). Other known neural crest cell inhibitors expressed in the hindbrain region and shown to sculpt neural crest cells onto stereotypical migratory pathways, such as semaphorin3F, restrict the growth and metastasis of colorectal cancer (Wu et al., 2011; Gao et al., 2015). Thus, further mechanistic studies of DAN and the identification of other factors expressed within embryonic neural crest cell–free zones may represent a fertile area of research for targets to inhibit cancer cell invasion.
In conclusion, our results demonstrate that collective migration of cranial neural crest cells is driven by a combination of factors that include inhibitory signals within the paraxial mesoderm. Rather than uniquely defining nonpermissive tissue boundaries to establish neural crest cell–free zones, our data support a model wherein the BMP antagonist DAN has a dual role to restrain lead neural crest cell invasion and proliferation that, in turn, promote collective cell migration. That is, by slowing down neural crest cells at the start of migration, the population is able to maintain better cohesion because the dispersal is restrained. The influence of DAN on neural crest cell migration was consistent with BMP signaling. Whether DAN restricts trunk neural crest cells to a dorsolateral migratory pathway and reduces trunk neural crest cell speed and proliferation remain to be explored in silico in an extended 3D model and in experiment. Furthermore, we are now poised to examine up- and downstream signals that regulate DAN expression to help us better understand craniofacial birth defects related to mistakes in neural crest cell migration. Lastly, our discovery of DAN as a candidate inhibitor of metastatic melanoma reveals a molecular inroad to test in a broader range of human metastatic melanoma cell lines/patient tissue samples and in more sophisticated models that are clinically relevant.
Materials and methods
Embryos and in ovo cell perturbations
Fertilized leghorn chickens (Centurion Poultry) were incubated at 38°C in a humidified incubator until the desired stages of development (Hamburger and Hamilton, 1951). Plasmid DNA (H2B mCherry; 2.5 µg/µl) and fluorescein-tagged MOs (0.5 mM; standard control MO, 5′-CCTCTTACCTCAGTT-3′; gallus BMP2 MO, 5′-CGGGTGGCGGCAACCATGATCAACC-3′; Gene Tools) were injected into the neural tube and electroporated at HH St 9 as previously described (McLennan and Kulesa, 2007). In brief, plasmid DNA and/or MOs were mixed with Fast Green FCF (F-7258; Sigma-Aldrich) for visualization and then injected into the lumen of the neural tube at the axial level of the rostral hindbrain using a pulled borosilicate glass needle (BF100-50-10; Suter). Platinum electrodes were then placed 3–5 mm apart parallel to the embryo hindbrain, and five pulses of 20–25 V at 50-ms duration were applied through the electrodes using an Electro Square ECM 830 (BTX). After the addition of sterile Ringer’s solution to the embryo, the egg was resealed with adhesive tape and reincubated until the desired stage.
For DAN protein injections into the paraxial mesoderm, embryos at HH St 10 were injected with human recombinant DAN protein (100–250 µg/ml; 955-DA-050; R&D Systems) only or DAN protein mixed with CellTracker CM-DiI (C-7001; Thermo Fisher Scientific) and Fast Green FCF (for visualization of injected protein) at multiple sites adjacent to the cranial neural tube. For lentiviral injections, live lentiviral particles (human FL DAN, LPP-G0810-LV214-200, DAN shRNA-A, TL316630VA, DAN shRNA-B, TL316630VB, DAN shRNA-C, TL316630VC, DAN shRNA-D, TL316630VD, scrambled, TR30021V, mCherry, LPP-MCHR-LV105-100-C, eYFP, and LPP-EYFP-LV105-100-C; Genecopoeia) at 1.8–2.1 × 107 U/ml were injected into HH St 9 embryos at multiple sites adjacent to the cranial neural tube. For MO electroporations into the paraxial mesoderm, embryos at HH St 9 were injected with fluorescein-labeled MOs (gallus DAN MO, 5′-AGACGGCGGGCACATCCCAT-3′; Gene Tools) mixed with H2B mCherry (1:1) at multiple sites. The positive electrode was then placed above the paraxial mesoderm and the negative electrode below. Although some ectoderm was transfected by this approach, the majority of cells transfected were mesoderm.
Identification of DAN
We performed a microarray screen to identify genes highly enriched in the neural crest cell–free zone adjacent to r3. Tissue was isolated from an area adjacent to r3 at HH St 11, 13, and 15 (in triplicate) for microarray analysis. Microarray analysis was also performed on lead r4 neural crest cells at the same stages. The neural crest microarray analysis was then subtracted from the area adjacent to the r3 microarray analysis, yielding a list of genes highly enriched in the area adjacent to r3. One of the candidate genes from this screen was identified as DAN.
Fluorescence multiplex in situ hybridization (hybridization chain reaction) and immunohistochemistry
Transcripts were visualized in whole embryos and tissue sections by fluorescent multiplex in situ hybridization as previously described (Choi et al., 2014). In brief, chicken embryos were incubated to specific developmental stages, rapidly collected in 0.1% diethyl pyrocarbonate PBS, and fixed in fresh 4% paraformaldehyde at room temperature for 2 h. Embryos were then dehydrated and rehydrated with methanol-based gradients before the hybridization chain reaction was performed according to the manufacturer’s instructions (Molecular Instruments). Embryos were then cleared using FRUIT gradients (Hou et al., 2015). Initially, embryos were placed in 35% fructose, 8 M urea, and 0.5% α-thioglycerol and then gradually transferred up to 100% fructose, 8 M urea, and 0.5% α-thioglycerol, at which stage they were completely cleared (Hou et al., 2015).
Immunostaining with HNK-1 (neural crest marker; TIB-200; ATCC) in whole heads was performed. HNK-1 (1:20) diluted in 10% goat serum, 4% bovine serum albumen, and 0.1% Triton X-100 in PBS were incubated with whole heads overnight at 4°C. After repeated washes, secondary antibody was applied and incubated at 4°C overnight. The secondary antibodies used for HNK-1 were Alexa Fluor goat anti–mouse IgM 488 (A-21042), Alexa Fluor goat anti–mouse IgM 546 (A-21045), or Alexa Fluor goat anti–mouse IgM 647 (A-21238; Thermo Fisher Scientific). Immunostaining with phospho-SMAD1/5/9 rabbit monoclonal antibody (13820S; Cell Signaling Technology) was used at 1:500 following the HNK-1 protocol. Cell death and proliferation were investigated using a cleaved caspase-3 rabbit monoclonal antibody at 1:25 (9579S; Cell Signaling Technology) and a histone H3 sheep polyclonal antibody at 1:100 (NB100-747; Novus Biologicals). BrdU rabbit polyclonal antibody (PA5-32256; Thermo Fisher Scientific) was performed using standard protocol.
In vitro neural crest assays
For neural tube cultures, cranial neural tubes (r3–r5) were isolated as previously described (McLennan et al., 2010). In brief, embryos at HH St 8 were removed from eggs and washed in Ringer’s solution. Then, using a tungsten needle and forceps, transverse cuts were made at the axial levels of the r2/r3 and r5/r6 boundaries. The cranial sections were also cut down the midline before being placed in 1 mg/ml dispase for 7 min on ice. The sections were then manually moved to fresh Ham’s F-12 nutrient mix (11765047; Thermo Fisher Scientific) and agitated to separate neural tubes from ectoderm and other tissues. After a few washes in Ham’s F-12 nutrient mix, the isolated neural tubes were ready for plating onto glass-bottomed dishes (P06G-1.5-20-F; MatTek Corporation) coated with fibronectin (F1141; Sigma-Aldrich) and poly-d-lysine (P7886; Sigma-Aldrich) or stripe assays. To coat glass-bottomed dishes, Ringer’s solution with 20 µg/ml of fibronectin and 20 µg/ml of poly-d-lysine was added to each dish and allowed to sit for at least 30 min at room temperature. The liquid was then removed, and dishes were allowed to air dry for 30 min before use. When the neural tubes were ready for plating, coated dishes were rinsed with Ringer’s solution, and neural tubes were immediately plated with as little liquid as possible. Neural tubes were allowed to adhere for 5–10 min at 37°C (depending on amount of liquid transferred with them) before being covered with Ham’s F-12 nutrient mix.
Stripe assays using cranial neural tubes (r3–r5) were performed as previously described (Krull et al., 1997). In brief, for neural crest stripe assays, 40-µm parallel assay matrices were used (2C; Karlsruhe Institute of Technology). Matrices were cleaned and sterilized before being placed in the middle of a glass-bottomed dish. PE 10 tubing (63018-623; VWR) from the matrices was attached to 30.5 gauge needle (BD305106; VWR) with a 1-ml syringe (53498-486; VWR) containing fibronectin, poly-d-lysine, an Alexa Fluor secondary antibody (1:500, Thermo Fisher Scientific), and a protein of interest. For the experimental stripes, human recombinant DAN protein was used at 8 µg/ml with or without recombinant human BMP4. Recombinant human BMP4 (314-BP-010/CF; R&D Systems) was used at 2:1 molar ratio with DAN, which was 4.8 µg/ml. The Alexa Fluor secondary antibody allowed visualization of the stripes. Using the plunger insert of the 1-ml syringe, each matrix was filled with solution three times, once every 20 min. The matrices were then carefully peeled off of the glass-bottomed dish, which was then rinsed with Ringer’s before fresh Ringer’s with 20 µg/ml of fibronectin and 20 µg/ml of poly-d-lysine was added to each dish and allowed to sit for at least 30 min at room temperature. When ready for use, the solution was removed from each dish and rinsed with Ringer’s, and then neural tubes were plated perpendicular to the stripes. After the neural tubes had adhered to the dish for 5–10 min at 37°C, media containing 5 µg/ml Hoechst (B2261; Sigma-Aldrich) was added for 10 min. This was then replaced with Ham’s F-12 nutrient mix.
For BrdU analysis, DAN protein was added at 0, 1, or 10 µg/ml to the media, and the cultures were incubated for 6 h. BrdU labeling reagent (000103; Thermo Fisher Scientific) was added (1:100), and cultures were reincubated for 2 h. The cultures were then fixed with 4% paraformaldehyde for 30 min at room temperature, and BrdU immunohistochemistry was performed.
For time lapses, the dishes were moved immediately after neural tube plating (and Hoechst staining) to an LSM 800 microscope (ZEISS) with an attached incubator box, set to 37°C, and allowed to equilibrate for 1–2 h. Human recombinant DAN protein, 1 µg/ml or 10 µg/ml, was added to the media overlaying the plated neural tubes 1 h before beginning time lapses or into the fibronectin solution used to coat the plates at 10 µg/ml. Single z stack time lapses were then imaged overnight using a Plan Apochromat 10× 0.45 NA M27 objective with 2.5–5-min intervals.
In vitro melanoma assays
C8161 human metastatic melanoma cells were used (provided by M. Hendrix, Children’s Memorial Research Center, Chicago, IL). C8161 cells were maintained in a culture medium consisting of RPMI + 10% serum. For the stripe assays, 25,000 cells were added to 3 ml culture medium plus pen/strep antibiotic (1:1,000 volume/volume ratio). Stripe assays were prepared as described in the previous section. However, because melanoma cells are larger than the neural crest cells, 90-µm parallel assays were used (2A; Karlsruhe Institute of Technology). For exposure to DAN in the media, c8161-EGFP cells were plated on fibronectin-coated (0.025 µg/ml) glass-bottomed cell culture plates (CLS-1812-024; VWR) at a concentration of 4 × 105 cells/ml. After overnight incubation, 10 µg/ml human recombinant DAN protein was added to the media.
For time lapses, plates containing melanoma cells with and without DAN were immediately moved to the incubated microscope. 1–2 h later, single z stack time lapses were imaged overnight using a Plan Apochromat 10× 0.45 NA M27 objective with 5-min intervals. For BrdU analysis, cultures were exposed to DAN for 6 h, then BrdU labeling reagent was added, and cultures were reincubated for 30 min. Cultures were then fixed with 4% paraformaldehyde for 30 min, and BrdU immunohistochemistry was performed. Initially, melanoma cells were exposed to BrdU labeling reagent for 2 h (same amount of time as the neural crest cells were exposed); however, this lead to BrdU labeling all melanoma cells. Therefore, the time was reduced to 30 min.
Imaging
All embryos were harvested at desired HH stages and fixed in 4% paraformaldehyde for 2 h at room temperature or overnight at 4°C. After three washes in PBS, excess membranes were removed. For any embryos that were HH St 13 (19-somite stage) or older, the trunk portion of the embryo was removed, and a sharpened tungsten needle was used to cut the cranial embryo in half down the midline. By doing this, control and experimental sides of the embryos could be imaged easily. Embryos were then mounted onto glass slides as previously described (Teddy and Kulesa, 2004) and imaged by confocal microscopy on an LSM 780 or 800 microscope using the provided software (ZEISS). In brief, a ring of vacuum grease (05054-AB; SPI Supplies) was placed on a clean glass slide. The embryo was then placed in the ring with a small amount of PBS, and a coverslip was mounted onto the grease, creating a sealed chamber. All images were taken using a Plan Apochromat 10× 0.45 NA M27 or an LD C-Apochromat 40× 1.1 NA W Korr M27 objective.
For time-lapse confocal imaging of neural crest cells in vitro, single z planes of each neural tube were taken every 2.5 or 5 min for 12–18 h. Both control and experimental time lapses were imaged concurrently. For in vivo time-lapse confocal imaging, embryos were transfected with DAN or control MOs immediately followed by injections of DiI in the lumen of the neural tube. 8 h later, embryos were removed from the egg, mounted dorsal side down on Petri dishes containing a mixture of agar and thin albumen (Chapman et al., 2001), and moved to the microscope. After allowing the embryo to settle for 1–2 h, small z stacks of the region of interest were taken using a Plan Apochromat 10× 0.45 NA M27 objective every 5 min (McKinney et al., 2013).
RNAseq
Neural tube cultures were prepared and plated as described in the In vitro neural crest assays section. DAN was added to the media at 0, 1, and 10 µg/ml. After overnight incubation, neural tubes and any ectoderm were removed from in vitro cultures using a tungsten needle and pipette. Media was then removed from neural tube cultures, and the adherent neural crest cells were gently rinsed once with 0.1% diethyl pyrocarbonate PBS. Two or three biological replicates were harvested for each of the three conditions (untreated, 1 µg/ml, and 10 µg/ml). cDNA was synthesized using SMART-seq v4 Ultra Low Input RNA kit (634888; Takara Bio Inc.) with subsequent library preparation by Nextera XT DNA Sample Preparation kit (FC-131-1096; Illumina) and Index kits (FC-131-1002; Illumina). cDNA samples and libraries were both confirmed on a Bioanalyzer 2100 before RNAseq. Libraries were sequenced as 75-bp high output paired reads on a NextSeq (Illumina). Each sample generated in excess of 20 million fastq counts.
Bioinformatics
Fastq files were mapped to the chicken genome galGal4 from the University of California, Santa Cruz, using TopHat (2.0.13) with options -x 1 -g 1. Ensembl 80 annotations were used to define gene coordinates. Quality of the samples was assessed using FastQC (0.10.1). The R (3.2.2) environment was used for the statistical analysis of the data. RPKM values and differentially expressed genes were found using edgeR (3.12.0). Correlations between samples were calculated using Pearson correlations. Genes were considered differentially expressed if they had a p-value <0.05 and a log2 fold change >1 or <−1. Pathway enrichment analysis was performed in the Ingenuity Pathway Analysis software (version 33559992; QIAGEN) from the overlap of 1 µg/ml compared with untreated and 10 µg/ml compared with untreated differential expression gene lists.
Analysis
We calculated the percentage of area covered using the “Surfaces” function of Imaris (Bitplane) to create a surface mask by manually drawing the outline of the whole ba2. Next, we calculated the area of the HNK-1 fluorescence signal using the masked ba2 surface. We set a consistent intensity threshold to the same value for each dataset, a surface grain size of 1 µm was set, the diameter of the largest sphere was set to 1 µm, and then the automatic “Surfaces” function was applied. When the percentage of area was calculated using a signal other than HNK-1, we manually measured the area of the ba2 and separately manually measured the area surrounding the signal within the ba2, then creating a “Surfaces.” We calculated the percentage of the total arch the HNK-1 signal covered by comparing the two values. The percentage of distance the cells migrated was calculated by measuring the total distance of the ba2 and measuring what distance the cells actually migrated. The intensity of pSMAD was calculated by generating a “Spot” (≥60 µm in diameter) in Imaris, which was the same diameter as the width of the r4 stream adjacent to the otic vesicle (50–75 µm from the neural tube). Another spot the same size was placed at least 200 µm from the edge of the neural tube in the middle of the r4 stream. The mean intensity of the channel with the pSMAD signal was recorded for each spot. For the neural tube cultures with different concentrations of DAN protein, cells were manually tracked using “Spot” detection in Imaris for ≥10 h. The spots were set to ≥9 µm in size. The mean speed and directionality of each track was calculated using Imaris. The box plots in each figure were generated by using the values from each dataset indicated. X indicates outliers, and the box plots and whiskers indicate the quartiles and range, respectively, of each dataset. P-values were calculated using a standard Student’s t test. Data distribution was assumed to be normal, but this was not formally tested.
Computer model/simulations
Computational methods and model parameters were as previously described (McLennan et al., 2015a,b). In brief, cell movement was implemented as an off-lattice position-jump process, whereas the VEGF concentration was described by a continuous variable in a reaction-diffusion equation, and both of these are simulated on a growing rectangular domain. Compared with previously published versions of the model, we made the following exceptions: the domain width was increased to 360 µm, and the width of the initial chemoattractant profile and its background production was kept at 120 µm. The DAN-zone extends over the left third of the migratory domain and stretches as the domain grows. Within the DAN-zone, cells move at reduced speed, and the reduction in speed is proportional to the relative concentration of DAN. To model the dynamic expression of DAN, its concentration was modeled to linearly increase from 0 to 1 (relative units) in the first 12 h and then linearly decrease to half of the maximum concentration over the next 6 h. Thus, if the speed in the DAN-zone at peak concentration is 10 µm/h, it increases (as DAN concentration decreases) to 25 µm/h at t = 18 h (half the reduction in speed). Further information is provided in Text S1.
Accession numbers
RNAseq data is located in the NCBI Gene Expression Omnibus under accession GSE101746.
Online supplemental material
Fig. S1 shows further analysis of DAN expression in the head and trunk. Fig. S2 shows further functional testing of increasing DAN in vivo. Fig. S3 shows further functional testing of DAN knockdown in vivo. Fig. S4 shows further neural crest and melanoma in vitro analysis. Fig. S5 shows the phenotypes observed after DAN shRNA lentiviral particle injections. Table S1 shows the RNAseq data of neural crest cells exposed to DAN in vitro. Text S1 further describes the computational methods and model parameters used in the computer simulations.
Supplementary Material
Acknowledgments
We thank members of the Microscopy, Histology, and Molecular Biology core facilities as well as Cathy McKinney for analysis support and Mark Miller, Scientific Illustrator at the Stowers Institute for Medical Research. Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/LIBPB-1133.
P.M. Kulesa would like to acknowledge kind and generous funding from the Stowers Institute for Medical Research. R.E. Baker would like to thank the Royal Society for a Wolfson Research Merit Award.
The authors declare no competing financial interests.
Author contributions: R. McLennan carried out all experiments on the neural crest. C.M. Bailey and J.C. Kasemeier-Kulesa performed the in vitro assays with metastatic melanoma cells. L.J. Schumacher performed the computer model simulations with consultation and interpretation from R.E. Baker and P.K. Maini. J.M. Teddy performed the image analysis and measurements. J.A. Morrison, L.A. Wolfe, M.M. Gogol, and R. McLennan performed the gene expression profiling and analysis. All authors discussed and interpreted the results. R. McLennan and P.M. Kulesa wrote the manuscript and figures with critical comments and suggestions from L.J. Schumacher, R.E. Baker, and P.K. Maini. P.M. Kulesa supervised the overall project. All authors read and approved the final manuscript.
Footnotes
Abbreviations used:
- BMP
- bone morphogenetic protein
- DAN
- differential screening-selected gene aberrant in neuroblastoma
- FL
- full length
- MO
- morpholino
References
- Anderson R.M., Stottmann R.W., Choi M., and Klingensmith J.. 2006. Endogenous bone morphogenetic protein antagonists regulate mammalian neural crest generation and survival. Dev. Dyn. 235:2507–2520. 10.1002/dvdy.20891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey C.M., and Kulesa P.M.. 2014. Dynamic interactions between cancer cells and the embryonic microenvironment regulate cell invasion and reveal EphB6 as a metastasis suppressor. Mol. Cancer Res. 12:1303–1313. 10.1158/1541-7786.MCR-13-0673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey C.M., Morrison J.A., and Kulesa P.M.. 2012. Melanoma revives an embryonic migration program to promote plasticity and invasion. Pigment Cell Melanoma Res. 25:573–583. 10.1111/j.1755-148X.2012.01025.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothe I., Tenin G., Oseni A., and Dietrich S.. 2011. Dynamic control of head mesoderm patterning. Development. 138:2807–2821. 10.1242/dev.062737 [DOI] [PubMed] [Google Scholar]
- Chapman S.C., Collignon J., Schoenwolf G.C., and Lumsden A.. 2001. Improved method for chick whole-embryo culture using a filter paper carrier. Dev. Dyn. 220:284–289. [DOI] [PubMed] [Google Scholar]
- Choi H.M., Beck V.A., and Pierce N.A.. 2014. Next-generation in situ hybridization chain reaction: Higher gain, lower cost, greater durability. ACS Nano. 8:4284–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia A.C., Costa M., Moraes F., Bom J., Nóvoa A., and Mallo M.. 2007. Bmp2 is required for migration but not for induction of neural crest cells in the mouse. Dev. Dyn. 236:2493–2501. 10.1002/dvdy.21256 [DOI] [PubMed] [Google Scholar]
- Dionne M.S., Skarnes W.C., and Harland R.M.. 2001. Mutation and analysis of Dan, the founding member of the Dan family of transforming growth factor β antagonists. Mol. Cell. Biol. 21:636–643. 10.1128/MCB.21.2.636-643.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon M., and Lumsden A.. 1999. Distribution of neuregulin-1 (nrg1) and erbB4 transcripts in embryonic chick hindbrain. Mol. Cell. Neurosci. 13:237–258. 10.1006/mcne.1999.0749 [DOI] [PubMed] [Google Scholar]
- Dutt S., Kléber M., Matasci M., Sommer L., and Zimmermann D.R.. 2006a Versican V0 and V1 guide migratory neural crest cells. J. Biol. Chem. 281:12123–12131. 10.1074/jbc.M510834200 [DOI] [PubMed] [Google Scholar]
- Dutt S., Matasci M., Sommer L., and Zimmermann D.R.. 2006b Guidance of neural crest cell migration: The inhibitory function of the chondroitin sulfate proteoglycan, versican. Sci. World J. 6:1114–1117. 10.1100/tsw.2006.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickholt B.J., Mackenzie S.L., Graham A., Walsh F.S., and Doherty P.. 1999. Evidence for collapsin-1 functioning in the control of neural crest migration in both trunk and hindbrain regions. Development. 126:2181–2189. [DOI] [PubMed] [Google Scholar]
- Farlie P.G., Kerr R., Thomas P., Symes T., Minichiello J., Hearn C.J., and Newgreen D.. 1999. A paraxial exclusion zone creates patterned cranial neural crest cell outgrowth adjacent to rhombomeres 3 and 5. Dev. Biol. 213:70–84. 10.1006/dbio.1999.9332 [DOI] [PubMed] [Google Scholar]
- Flaherty K.T., Hodi F.S., and Fisher D.E.. 2012. From genes to drugs: targeted strategies for melanoma. Nat. Rev. Cancer. 12:349–361. 10.1038/nrc3218 [DOI] [PubMed] [Google Scholar]
- Gammill L.S., Gonzalez C., and Bronner-Fraser M.. 2007. Neuropilin 2/semaphorin 3F signaling is essential for cranial neural crest migration and trigeminal ganglion condensation. Dev. Neurobiol. 67:47–56. 10.1002/dneu.20326 [DOI] [PubMed] [Google Scholar]
- Gao X., Tang C., Shi W., Feng S., Qin W., Jiang T., and Sun Y.. 2015. Semaphorin-3F functions as a tumor suppressor in colorectal cancer due to regulation by DNA methylation. Int. J. Clin. Exp. Pathol. 8:12766–12774. [PMC free article] [PubMed] [Google Scholar]
- Gerlach-Bank L.M., Ellis A.D., Noonen B., and Barald K.F.. 2002. Cloning and expression analysis of the chick DAN gene, an antagonist of the BMP family of growth factors. Dev. Dyn. 224:109–115. 10.1002/dvdy.10079 [DOI] [PubMed] [Google Scholar]
- Gerlach-Bank L.M., Cleveland A.R., and Barald K.F.. 2004. DAN directs endolymphatic sac and duct outgrowth in the avian inner ear. Dev. Dyn. 229:219–230. 10.1002/dvdy.10414 [DOI] [PubMed] [Google Scholar]
- Golding J.P., Trainor P., Krumlauf R., and Gassmann M.. 2000. Defects in pathfinding by cranial neural crest cells in mice lacking the neuregulin receptor ErbB4. Nat. Cell Biol. 2:103–109. 10.1038/35000058 [DOI] [PubMed] [Google Scholar]
- Golding J.P., Dixon M., and Gassmann M.. 2002. Cues from neuroepithelium and surface ectoderm maintain neural crest-free regions within cranial mesenchyme of the developing chick. Development. 129:1095–1105. [DOI] [PubMed] [Google Scholar]
- Golding J.P., Sobieszczuk D., Dixon M., Coles E., Christiansen J., Wilkinson D., and Gassmann M.. 2004. Roles of erbB4, rhombomere-specific, and rhombomere-independent cues in maintaining neural crest-free zones in the embryonic head. Dev. Biol. 266:361–372. 10.1016/j.ydbio.2003.11.003 [DOI] [PubMed] [Google Scholar]
- Goldstein A.M., Brewer K.C., Doyle A.M., Nagy N., and Roberts D.J.. 2005. BMP signaling is necessary for neural crest cell migration and ganglion formation in the enteric nervous system. Mech. Dev. 122:821–833. 10.1016/j.mod.2005.03.003 [DOI] [PubMed] [Google Scholar]
- Hamburger V., and Hamilton H.L.. 1951. A series of normal stages in the development of the chick embryo. J. Morphol. 88:49–92. 10.1002/jmor.1050880104 [DOI] [PubMed] [Google Scholar]
- Hanaoka E., Ozaki T., Nakamura Y., Moriya H., Nakagawara A., and Sakiyama S.. 2000. Overexpression of DAN causes a growth suppression in p53-deficient SAOS-2 cells. Biochem. Biophys. Res. Commun. 278:20–26. 10.1006/bbrc.2000.3758 [DOI] [PubMed] [Google Scholar]
- Holderfield M., Deuker M.M., McCormick F., and McMahon M.. 2014. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat. Rev. Cancer. 14:455–467. 10.1038/nrc3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou B., Zhang D., Zhao S., Wei M., Yang Z., Wang S., Wang J., Zhang X., Liu B., Fan L., et al. . 2015. Scalable and DiI-compatible optical clearance of the mammalian brain. Front. Neuroanat. 9:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu D.R., Economides A.N., Wang X., Eimon P.M., and Harland R.M.. 1998. The Xenopus dorsalizing factor Gremlin identifies a novel family of secreted proteins that antagonize BMP activities. Mol. Cell. 1:673–683. 10.1016/S1097-2765(00)80067-2 [DOI] [PubMed] [Google Scholar]
- Hung W.T., Wu F.J., Wang C.J., and Luo C.W.. 2012. DAN (NBL1) specifically antagonizes BMP2 and BMP4 and modulates the actions of GDF9, BMP2, and BMP4 in the rat ovary. Biol. Reprod. 86:158. [DOI] [PubMed] [Google Scholar]
- Kanzler B., Foreman R.K., Labosky P.A., and Mallo M.. 2000. BMP signaling is essential for development of skeletogenic and neurogenic cranial neural crest. Development. 127:1095–1104. [DOI] [PubMed] [Google Scholar]
- Kasemeier-Kulesa J.C., Kulesa P.M., and Lefcort F.. 2005. Imaging neural crest cell dynamics during formation of dorsal root ganglia and sympathetic ganglia. Development. 132:235–245. 10.1242/dev.01553 [DOI] [PubMed] [Google Scholar]
- Katsu K., Tokumori D., Tatsumi N., Suzuki A., and Yokouchi Y.. 2012. BMP inhibition by DAN in Hensen’s node is a critical step for the establishment of left–right asymmetry in the chick embryo. Dev. Biol. 363:15–26. 10.1016/j.ydbio.2011.12.015 [DOI] [PubMed] [Google Scholar]
- Kerr R.S., and Newgreen D.F.. 1997. Isolation and characterization of chondroitin sulfate proteoglycans from embryonic quail that influence neural crest cell behavior. Dev. Biol. 192:108–124. 10.1006/dbio.1997.8731 [DOI] [PubMed] [Google Scholar]
- Kim A.S., and Pleasure S.J.. 2003. Expression of the BMP antagonist Dan during murine forebrain development. Brain Res. Dev. Brain Res. 145:159–162. 10.1016/S0165-3806(03)00213-X [DOI] [PubMed] [Google Scholar]
- Krull C.E., Lansford R., Gale N.W., Collazo A., Marcelle C., Yancopoulos G.D., Fraser S.E., and Bronner-Fraser M.. 1997. Interactions of Eph-related receptors and ligands confer rostrocaudal pattern to trunk neural crest migration. Curr. Biol. 7:571–580. 10.1016/S0960-9822(06)00256-9 [DOI] [PubMed] [Google Scholar]
- Kulesa P.M., and Gammill L.S.. 2010. Neural crest migration: Patterns, phases and signals. Dev. Biol. 344:566–568. 10.1016/j.ydbio.2010.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesa P.M., Kasemeier-Kulesa J.C., Teddy J.M., Margaryan N.V., Seftor E.A., Seftor R.E., and Hendrix M.J.. 2006. Reprogramming metastatic melanoma cells to assume a neural crest cell-like phenotype in an embryonic microenvironment. Proc. Natl. Acad. Sci. USA. 103:3752–3757. 10.1073/pnas.0506977103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesa P.M., Morrison J.A., and Bailey C.M.. 2013. The neural crest and cancer: A developmental spin on melanoma. Cells Tissues Organs (Print). 198:12–21. 10.1159/000348418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landolt R.M., Vaughan L., Winterhalter K.H., and Zimmermann D.R.. 1995. Versican is selectively expressed in embryonic tissues that act as barriers to neural crest cell migration and axon outgrowth. Development. 121:2303–2312. [DOI] [PubMed] [Google Scholar]
- Le Douarin N., and Kalcheim C.. 1999. The neural crest. Cambridge University Press, Cambridge, UK. 445 pp. 10.1017/CBO9780511897948 [DOI] [Google Scholar]
- McKinney M.C., Fukatsu K., Morrison J., McLennan R., Bronner M.E., and Kulesa P.M.. 2013. Evidence for dynamic rearrangements but lack of fate or position restrictions in premigratory avian trunk neural crest. Development. 140:820–830. 10.1242/dev.083725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan R., and Kulesa P.M.. 2007. In vivo analysis reveals a critical role for neuropilin-1 in cranial neural crest cell migration in chick. Dev. Biol. 301:227–239. 10.1016/j.ydbio.2006.08.019 [DOI] [PubMed] [Google Scholar]
- McLennan R., Teddy J.M., Kasemeier-Kulesa J.C., Romine M.H., and Kulesa P.M.. 2010. Vascular endothelial growth factor (VEGF) regulates cranial neural crest migration in vivo. Dev. Biol. 339:114–125. 10.1016/j.ydbio.2009.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan R., Dyson L., Prather K.W., Morrison J.A., Baker R.E., Maini P.K., and Kulesa P.M.. 2012. Multiscale mechanisms of cell migration during development: theory and experiment. Development. 139:2935–2944. 10.1242/dev.081471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan R., Schumacher L.J., Morrison J.A., Teddy J.M., Ridenour D.A., Box A.C., Semerad C.L., Li H., McDowell W., Kay D., et al. . 2015a Neural crest migration is driven by a few trailblazer cells with a unique molecular signature narrowly confined to the invasive front. Development. 142:2014–2025. 10.1242/dev.117507 [DOI] [PubMed] [Google Scholar]
- McLennan R., Schumacher L.J., Morrison J.A., Teddy J.M., Ridenour D.A., Box A.C., Semerad C.L., Li H., McDowell W., Kay D., et al. . 2015b VEGF signals induce trailblazer cell identity that drives neural crest migration. Dev. Biol. 407:12–25. 10.1016/j.ydbio.2015.08.011 [DOI] [PubMed] [Google Scholar]
- Mellott D.O., and Burke R.D.. 2008. Divergent roles for Eph and ephrin in avian cranial neural crest. BMC Dev. Biol. 8:56 10.1186/1471-213X-8-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet A., Martin A.R., Ronco C., Rocchi S., and Benhida R.. 2017. Metastatic melanoma: Insights into the evolution of the treatments and future challenges. Med. Res. Rev. 37:98–148. 10.1002/med.21404 [DOI] [PubMed] [Google Scholar]
- Miyazaki T., Miyauchi S., Tawada A., Anada T., Matsuzaka S., and Suzuki O.. 2008. Oversulfated chondroitin sulfate-E binds to BMP-4 and enhances osteoblast differentiation. J. Cell. Physiol. 217:769–777. 10.1002/jcp.21557 [DOI] [PubMed] [Google Scholar]
- Nolan K., Kattamuri C., Luedeke D.M., Angerman E.B., Rankin S.A., Stevens M.L., Zorn A.M., and Thompson T.B.. 2015. Structure of neuroblastoma suppressor of tumorigenicity 1 (NBL1): Insights for the functional variability across bone morphogenetic protein (BMP) antagonists. J. Biol. Chem. 290:4759–4771. 10.1074/jbc.M114.628412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogita J., Isogai E., Sudo H., Sakiyama S., Nakagawara A., and Koseki H.. 2001. Expression of the Dan gene during chicken embryonic development. Mech. Dev. 109:363–365. 10.1016/S0925-4773(01)00522-6 [DOI] [PubMed] [Google Scholar]
- Olakowski M., Tyszkiewicz T., Jarzab M., Król R., Oczko-Wojciechowska M., Kowalska M., Kowal M., Gala G.M., Kajor M., Lange D., et al. . 2009. NBL1 and anillin (ANLN) genes over-expression in pancreatic carcinoma. Folia Histochem. Cytobiol. 47:249–255. 10.2478/v10042-009-0031-1 [DOI] [PubMed] [Google Scholar]
- Osborne N.J., Begbie J., Chilton J.K., Schmidt H., and Eickholt B.J.. 2005. Semaphorin/neuropilin signaling influences the positioning of migratory neural crest cells within the hindbrain region of the chick. Dev. Dyn. 232:939–949. 10.1002/dvdy.20258 [DOI] [PubMed] [Google Scholar]
- Ozaki T., and Sakiyama S.. 1993. Molecular cloning and characterization of a cDNA showing negative regulation in v-src-transformed 3Y1 rat fibroblasts. Proc. Natl. Acad. Sci. USA. 90:2593–2597. 10.1073/pnas.90.7.2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki T., and Sakiyama S.. 1994. Tumor-suppressive activity of N03 gene product in v-src-transformed rat 3Y1 fibroblasts. Cancer Res. 54:646–648. [PubMed] [Google Scholar]
- Ozaki T., Nakamura Y., Enomoto H., Hirose M., and Sakiyama S.. 1995. Overexpression of DAN gene product in normal rat fibroblasts causes a retardation of the entry into the S phase. Cancer Res. 55:895–900. [PubMed] [Google Scholar]
- Pearce J.J., Penny G., and Rossant J.. 1999. A mouse cerberus/Dan-related gene family. Dev. Biol. 209:98–110. 10.1006/dbio.1999.9240 [DOI] [PubMed] [Google Scholar]
- Perissinotto D., Iacopetti P., Bellina I., Doliana R., Colombatti A., Pettway Z., Bronner-Fraser M., Shinomura T., Kimata K., Mörgelin M., et al. . 2000. Avian neural crest cell migration is diversely regulated by the two major hyaluronan-binding proteoglycans PG-M/versican and aggrecan. Development. 127:2823–2842. [DOI] [PubMed] [Google Scholar]
- Perris R., Perissinotto D., Pettway Z., Bronner-Fraser M., Mörgelin M., and Kimata K.. 1996. Inhibitory effects of PG-H/aggrecan and PG-M/versican on avian neural crest cell migration. FASEB J. 10:293–301. [DOI] [PubMed] [Google Scholar]
- Reddi A.H. 2001. Interplay between bone morphogenetic proteins and cognate binding proteins in bone and cartilage development: noggin, chordin and DAN. Arthritis Res. 3:1–5. 10.1186/ar133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridenour D.A., McLennan R., Teddy J.M., Semerad C.L., Haug J.S., and Kulesa P.M.. 2014. The neural crest cell cycle is related to phases of migration in the head. Development. 141:1095–1103. 10.1242/dev.098855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela-Donenfeld D., and Kalcheim C.. 1999. Regulation of the onset of neural crest migration by coordinated activity of BMP4 and Noggin in the dorsal neural tube. Development. 126:4749–4762. [DOI] [PubMed] [Google Scholar]
- Sela-Donenfeld D., and Kalcheim C.. 2000. Inhibition of noggin expression in the dorsal neural tube by somitogenesis: a mechanism for coordinating the timing of neural crest emigration. Development. 127:4845–4854. [DOI] [PubMed] [Google Scholar]
- Smith A., Robinson V., Patel K., and Wilkinson D.G.. 1997. The EphA4 and EphB1 receptor tyrosine kinases and ephrin-B2 ligand regulate targeted migration of branchial neural crest cells. Curr. Biol. 7:561–570. 10.1016/S0960-9822(06)00255-7 [DOI] [PubMed] [Google Scholar]
- Stanley E., Biben C., Kotecha S., Fabri L., Tajbakhsh S., Wang C.C., Hatzistavrou T., Roberts B., Drinkwater C., Lah M., et al. . 1998. DAN is a secreted glycoprotein related to Xenopus cerberus. Mech. Dev. 77:173–184. 10.1016/S0925-4773(98)00139-7 [DOI] [PubMed] [Google Scholar]
- Streit A., Lee K.J., Woo I., Roberts C., Jessell T.M., and Stern C.D.. 1998. Chordin regulates primitive streak development and the stability of induced neural cells, but is not sufficient for neural induction in the chick embryo. Development. 125:507–519. [DOI] [PubMed] [Google Scholar]
- Szabó A., Melchionda M., Nastasi G., Woods M.L., Campo S., Perris R., and Mayor R.. 2016. In vivo confinement promotes collective migration of neural crest cells. J. Cell Biol. 213:543–555. 10.1083/jcb.201602083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teddy J.M., and Kulesa P.M.. 2004. In vivo evidence for short- and long-range cell communication in cranial neural crest cells. Development. 131:6141–6151. 10.1242/dev.01534 [DOI] [PubMed] [Google Scholar]
- Trainor P.A., and Krumlauf R.. 2000. Patterning the cranial neural crest: Hindbrain segmentation and Hox gene plasticity. Nat. Rev. Neurosci. 1:116–124. 10.1038/35039056 [DOI] [PubMed] [Google Scholar]
- Tribulo C., Aybar M.J., Nguyen V.H., Mullins M.C., and Mayor R.. 2003. Regulation of Msx genes by a Bmp gradient is essential for neural crest specification. Development. 130:6441–6452. 10.1242/dev.00878 [DOI] [PubMed] [Google Scholar]
- Tzahor E., Kempf H., Mootoosamy R.C., Poon A.C., Abzhanov A., Tabin C.J., Dietrich S., and Lassar A.B.. 2003. Antagonists of Wnt and BMP signaling promote the formation of vertebrate head muscle. Genes Dev. 17:3087–3099. 10.1101/gad.1154103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhou Q., Yang J., Duan G.J., Ou J.J., Zhang R., Pan F., Peng Q.P., Tan H., Ping Y.F., et al. . 2011. Endogenous axon guiding chemorepulsant semaphorin-3F inhibits the growth and metastasis of colorectal carcinoma. Clin. Cancer Res. 17:2702–2711. 10.1158/1078-0432.CCR-10-0839 [DOI] [PubMed] [Google Scholar]
- Yamanishi T., Katsu K., Funahashi J., Yumoto E., and Yokouchi Y.. 2007. Dan is required for normal morphogenesis and patterning in the developing chick inner ear. Dev. Growth Differ. 49:13–26. 10.1111/j.1440-169X.2007.00900.x [DOI] [PubMed] [Google Scholar]
- Yu H.H., and Moens C.B.. 2005. Semaphorin signaling guides cranial neural crest cell migration in zebrafish. Dev. Biol. 280:373–385. 10.1016/j.ydbio.2005.01.029 [DOI] [PubMed] [Google Scholar]
- Zhang H., and Bradley A.. 1996. Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development. 122:2977–2986. [DOI] [PubMed] [Google Scholar]
- Zhu L., Marvin M.J., Gardiner A., Lassar A.B., Mercola M., Stern C.D., and Levin M.. 1999. Cerberus regulates left–right asymmetry of the embryonic head and heart. Curr. Biol. 9:931–938. 10.1016/S0960-9822(99)80419-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.