Recycling of internalized membrane proteins back to the cell surface controls diverse cellular processes. MacDonald and Piper genetically dissect a recycling pathway in yeast to reveal a cohort of novel and conserved factors, including the Rag GTPases, which contribute to metabolic control by regulating surface recycling independently of TORC1 signaling.
Abstract
Endocytosed cell surface membrane proteins rely on recycling pathways for their return to the plasma membrane. Although endosome-to-plasma membrane recycling is critical for many cellular processes, much of the required machinery is unknown. We discovered that yeast has a recycling route from endosomes to the cell surface that functions efficiently after inactivation of the sec7-1 allele of Sec7, which controls transit through the Golgi. A genetic screen based on an engineered synthetic reporter that exclusively follows this pathway revealed that recycling was subject to metabolic control through the Rag GTPases Gtr1 and Gtr2, which work downstream of the exchange factor Vam6. Gtr1 and Gtr2 control the recycling pathway independently of TORC1 regulation through the Gtr1 interactor Ltv1. We further show that the early-endosome recycling route and its control though the Vam6>Gtr1/Gtr2>Ltv1 pathway plays a physiological role in regulating the abundance of amino acid transporters at the cell surface.
Introduction
Levels of cell-surface membrane proteins are controlled by the balance between recycling pathways returning them to the plasma membrane and their ubiquitination and endosomal sorting complexes required for transport (ESCRT)–dependent sorting into multivesicular bodies (Maxfield and McGraw, 2004; Grant and Donaldson, 2009; Piper et al., 2014). Recycling can be regulated at the level of individual proteins through specific sorting signals, which are recognized by particular machinery (Hsu et al., 2012). In addition, overall flux through recycling pathways can be regulated globally by signal transduction and metabolic cues, as in the case for growth factor withdrawal, which causes accumulation of a broad set of nutrient transporters in intracellular compartments (Corvera et al., 1986; Tanner and Lienhard, 1987). How global regulation of recycling is orchestrated is unclear, but it is partly controlled through TORC1 signaling, which is constitutively active in some cancer cells, allowing them to sustain an elevated supply of nutrients (Edinger and Thompson, 2002, 2004). Metabolic control over the global trafficking of cell surface proteins is also observed in Saccharomyces cerevisiae (Sc), where a variety of membrane transporters ultimately sort to the vacuole lumen for degradation upon limitation of nitrogen, glucose, or NAD+ (Jones et al., 2012; Becuwe and Léon, 2014; Lang et al., 2014; MacDonald et al., 2015; Müller et al., 2015; O’Donnell et al., 2015). This effect is explained in part by increased ubiquitination of the membrane protein cargoes and more efficient sorting into the multivesicular body (MVB) pathway (Babst and Odorizzi, 2013; Huber and Teis, 2016). TORC1 has been shown to influence this process in part by regulating the activity of particular ubiquitin (Ub)-ligase complexes. However, various interrelated regulatory pathways that connect TORC1 activity to ubiquitination do not adequately explain how global control of their trafficking is mediated by nutrient stress, such as nitrogen limitation (Schmidt et al., 1998; MacGurn et al., 2011; Martín et al., 2011; Merhi and André, 2012; Crapeau et al., 2014; Pfannmüller et al., 2015). One aspect that remains unclear is the extent to which metabolic cues may control flux through the trafficking pathways that convey proteins back to the plasma membrane.
Recycling of membrane proteins back to the surface of mammalian cells can occur along a variety of pathways, including a rapid, direct pathway and a slower route that passes through Rab11-positive recycling endosomes (Maxfield and McGraw, 2004; Grant and Donaldson, 2009; Huotari and Helenius, 2011). Endocytosed recycling proteins, such as the Transferrin receptor, mainly follow these routes and very rarely transit the trans-Golgi network (TGN; Snider and Rogers, 1985; Sheff et al., 1999). However, there is limited understanding of what protein machinery controls these routes and the processes that regulate flux of recycling. Recycling in yeast was first widely recognized from experiments using the styryl endocytic tracer dye, FM4-64, and the fluid phase dye, Lucifer Yellow, both of which are quickly secreted from yeast cells after internalization (Wiederkehr et al., 2000, 2001; Galan et al., 2001). This pathway is distinct from the sorting of late endosomes/MVBs, because FM4-64 efflux is unaltered in vps4Δ mutants (Wiederkehr et al., 2000), which trap MVB cargoes within exaggerated late-endosomal compartments. One critical component of this efflux pathway is Rcy1, an F-box protein whose function defines this process yet whose molecular function remains poorly defined. Loss of Rcy1 and other components involved in this pathway, such as the phospholipid flippase Drs2/Cdc50 complex and the Arf effector, Gcs1, traps endocytosed material in early endosomes (Hua et al., 2002; Chen et al., 2005; Robinson et al., 2006; Furuta et al., 2007; Xu et al., 2013). One well-studied protein that traffics in an Rcy1-dependent route through early endosomes is the SNARE protein Snc1. Unlike the major endocytic recycling pathways in mammalian cells, return of endocytosed Snc1 to the cell surface is thought to occur mainly via transport from early endosomes back to the TGN/Golgi along a retrieval pathway before transit to the cell surface (Tanaka et al., 2011; Sebastian et al., 2012; Feyder et al., 2015; MacDonald and Piper, 2016). This model is rooted in observations showing that a portion of GFP-Snc1 colocalizes with Sec7 (an Arf exchange factor that marks the TGN), and that Snc1 quickly accumulates intracellularly when proteins required for transport through the Golgi are acutely inactivated (Lewis et al., 2000; Chen et al., 2005; Robinson et al., 2006). Indeed, yeast are not currently known to have a direct surface recycling pathway from early endosomes to the cell surface that bypasses a retrieval step to the TGN (MacDonald and Piper, 2016). Here we find that yeast do have such a recycling pathway from early endosomes to the plasma membrane (EE>PM). A genetic screen revealed that recycling requires a signal transduction pathway operating through the Rag GTPases, which in addition to activating TORC1 (Panchaud et al., 2013b), controls recycling through the Gtr1 effector Ltv1 in a manner that is independent of TORC1. Global control over the trafficking of cell-surface proteins upon nitrogen stress is explained by the combined effects of both branches of the bifurcated Gtr1/Gtr2-Rag GTPase pathway, involving retardation of the recycling pathway and inhibition of TORC1.
Results
Cell-surface recycling from early endosomes
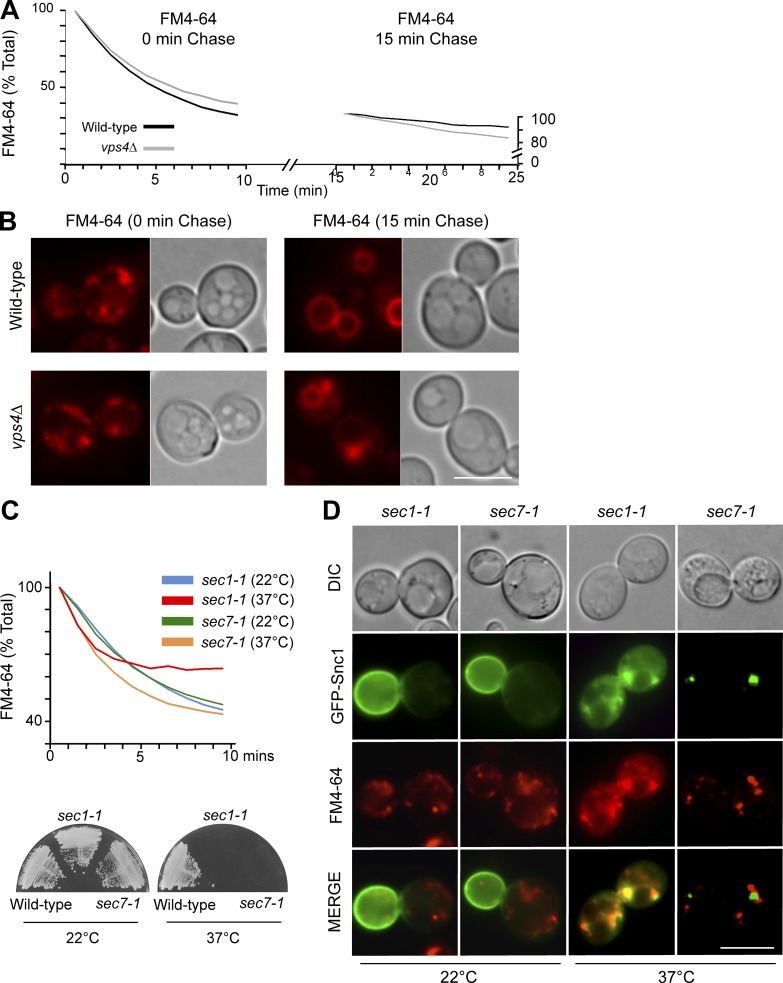
To determine whether an EE>PM recycling route might operate in yeast, we examined the efflux of FM4-64 after a short internalization pulse, which localizes dye to numerous endosomal puncta. Flow cytometry monitoring of the remaining dye showed that ∼70% of the internalized FM4-64 was secreted after 10 min, confirming previous work demonstrating that FM4-64 efflux is rapid and extensive (Wiederkehr et al., 2000). The pool of secreted FM4-64 originated from early endosomes because high efflux rates were observed only when dye was allowed to internalize for short periods of time (Fig. 1 A). When FM4-64 was chased for a further 15 min, under which conditions it reached late endosomes and the limiting membrane of the vacuole (Fig. 1 B), only a low rate of efflux was observed. These data confirm that FM4-64 can readily leave early endosomes and traffic to the plasma membrane, but that efflux from late endosomes is far slower. Similarly, cells lacking the ESCRT-associated Vps4 AAA-ATPase, which dramatically slows flux through late endosomal structures (Babst et al., 1997), had no effect on efflux of FM4-64 from early-endosomal compartments, consistent with previous observations (Wiederkehr et al., 2000). To test how FM4-64 is secreted, we performed dye efflux assays in cells harboring temperature-sensitive (ts) alleles of secretory pathway components: the sec1-1 ts mutant arrests the secretory pathway at the final step of SNARE-mediated vesicle fusion to the plasma membrane, whereas the sec7-1 ts mutant halts Arf1-dependent traffic through the TGN/Golgi apparatus (Novick et al., 1980). At permissive temperature (22°C), the rates of efflux in sec1-1 and sec7-1 cells were similar; at restrictive temperature (37°C), however, sec1-1 cells quickly ceased to secrete FM4-64 (Fig. 1 C). In contrast, efflux was not slowed in sec7-1 cells at 37°C. Previous studies have shown that Snc1, a v-SNARE protein, recycles, colocalizes with the Golgi marker Sec7, and relies on Golgi function to return to the cell surface (Lewis et al., 2000; Robinson et al., 2006), thus following an itinerary from early endosomes to the TGN to join the secretory pathway back to the cell surface. In agreement, we found that when shifted to 37°C, sec7-1 cells rapidly relocated cell-surface GFP-Snc1 to large intracellular puncta similar to the enlarged Golgi compartments that accumulate after Sec7 inactivation (Mioka et al., 2014). However, we found minimal colocalization of FM4-64 with GFP-Snc1 after inactivating Sec7, suggesting that the dye does not travel exclusively through the Golgi during its return to the cell surface (Fig. 1 D). In contrast, Sec1 inactivation caused accumulation of both FM4-64 and GFP-Snc1 in small puncta within the same cellular regions, suggesting that Sec1 is required for the fusion of FM4-64 carrying transport intermediates with the plasma membrane. However, the extent to which GFP-Snc1 and FM4-64 occupied the same vesicular carriers or arrived at the plasma membrane in distinct carriers could not be discerned at this resolution.
Figure 1.
FM4-64 follows a Golgi-independent route from early endosomes to the plasma membrane. FM4-64 efflux measurements (A) and localization of remaining dye (B) from wild-type and vps4Δ cells after 9-min uptake of FM4-64 at 25°C followed by 0- or 15-min chase before the assay. (C) FM4-64 efflux in sec1-1 and sec7-1 ts cells performed at 22°C or 37°C (top), with growth phenotype of ts mutants confirmed (bottom). (D) Colocalization of GFP-Snc1 with FM4-64–labeled endosomes after 10-min uptake assessed in ts mutants at 22°C or 37°C. Bars, 5 µm.
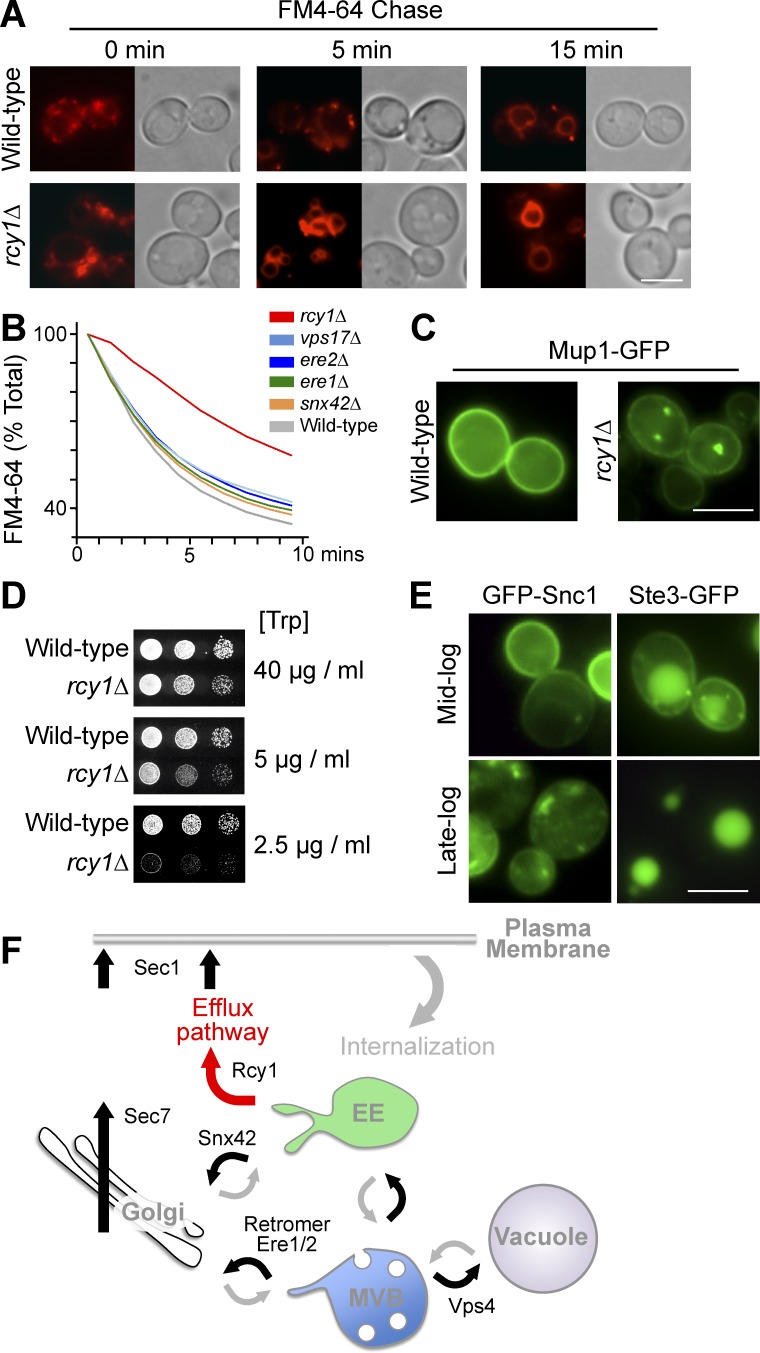
We confirmed previous studies (Wiederkehr et al., 2000) that rcy1Δ cells are defective for FM4-64 recycling, with vacuolar delivery of the remaining dye being accelerated (Fig. 2, A and B). However, no effect on FM4-64 recycling (Fig. 2 B) was found upon loss of components that mediate a retrograde retrieval route back to the TGN from endosomal compartments (e.g., retromer, Ere1, Ere2, and Snx42; Seaman et al., 1998; Hettema et al., 2003; Shi et al., 2011). Together, these data indicate that a portion of internalized FM4-64 can follow a recycling route from early endosomes to the plasma membrane (EE>PM), bypassing the TGN (Fig. 2 F). Additional phenotypes of rcy1Δ cells emphasized the importance of the recycling pathway. The Mup1 methionine transporter, which localizes to the cell surface in the absence of methionine, appears to travel through an Rcy1-dependent recycling route, because it is trapped within intracellular puncta in rcy1Δ mutants (Fig. 2 C). Likewise, we found that growth of trp1 auxotrophic cells in restricted tryptophan (Trp) conditions was dramatically compromised in rcy1Δ cells, indicating their inability to maintain sufficient levels of the Tat2 transporter at the cell surface (Fig. 2 D).
Figure 2.
Various cargoes use an Rcy1-dependent recycling route. (A) Accumulation of FM4-64 at indicated chase times in wild-type and rcy1Δ cells; micrographs were normalized across the series. (B) FM4-64 efflux in indicated mutants. (C) Mup1-GFP localization in wild-type and rcy1Δ cells grown to mid-log phase. (D) Growth assays in wild-type and rcy1Δ cells carrying the trp1 mutation using indicated concentrations of Trp. (E) Wild-type cells expressing either Ste3-GFP or GFP-Snc1 were grown to mid- and late-log phase followed by fluorescence microscopy. (F) Cartography of endocytic pathway: after internalization, cargo enters the Vps4-dependent MVB/degradation pathway or returns from early endosomes or late endosomes to the cell surface via the Golgi/TGN along routes that require Sec7 and Sec1. Cargo such as FM4-64 requires Rcy1 and Sec1, but not Sec7, to return to the plasma membrane. Bars, 5 µm.
An engineered reporter for the EE>PM recycling pathway
We next wanted to identify what cellular machinery was required for operating the EE>PM recycling route. For this, we needed a sensitive and well-defined reporter cargo that could be used in a genetic screen. One problem was that endogenous proteins that follow such a route might also participate in several other trafficking pathways and not be primarily constrained to the EE>PM pathway (Fig. 2 F). Although previous studies have used Snc1 as a reporter to implicate Rcy1, alongside Drs2, Ypt31, and Ypt32, in trafficking out of early endosomes back to the plasma membrane (Galan et al., 2001; Chen et al., 2005; Furuta et al., 2007; Liu et al., 2007), it does not serve as an selective marker of a recycling pathway because Snc1 clearly follows a retrieval pathway back to the TGN/Golgi from early endosomes. Snc1 is also problematic, because its distribution in the cell is highly sensitive to growth conditions (Fig. 2 E) and it is subject to increasing levels of Ub- and ESCRT-dependent sorting into the vacuole during nutrient stress (MacDonald et al., 2015; MacDonald and Piper, 2016). Mup1 might be a candidate, given that it is trapped within endosomes in rcy1Δ cells; but our previous experiments showed that Mup1 undergoes Ub-dependent trafficking to the vacuole when medium lacks a full complement of vitamins or nitrogen, making it unreliable as a phenotypic marker in a high-throughput screen (MacDonald et al., 2015). Previous data suggest that Ste3, the a-factor G protein–coupled receptor, can cycle between endosomes and the cell surface (Davis et al., 1993; Roth and Davis, 1996); however, at steady state, Ste3 is largely localized in the vacuole, owing to its constitutive ubiquitination and MVB sorting (Fig. 2 E).
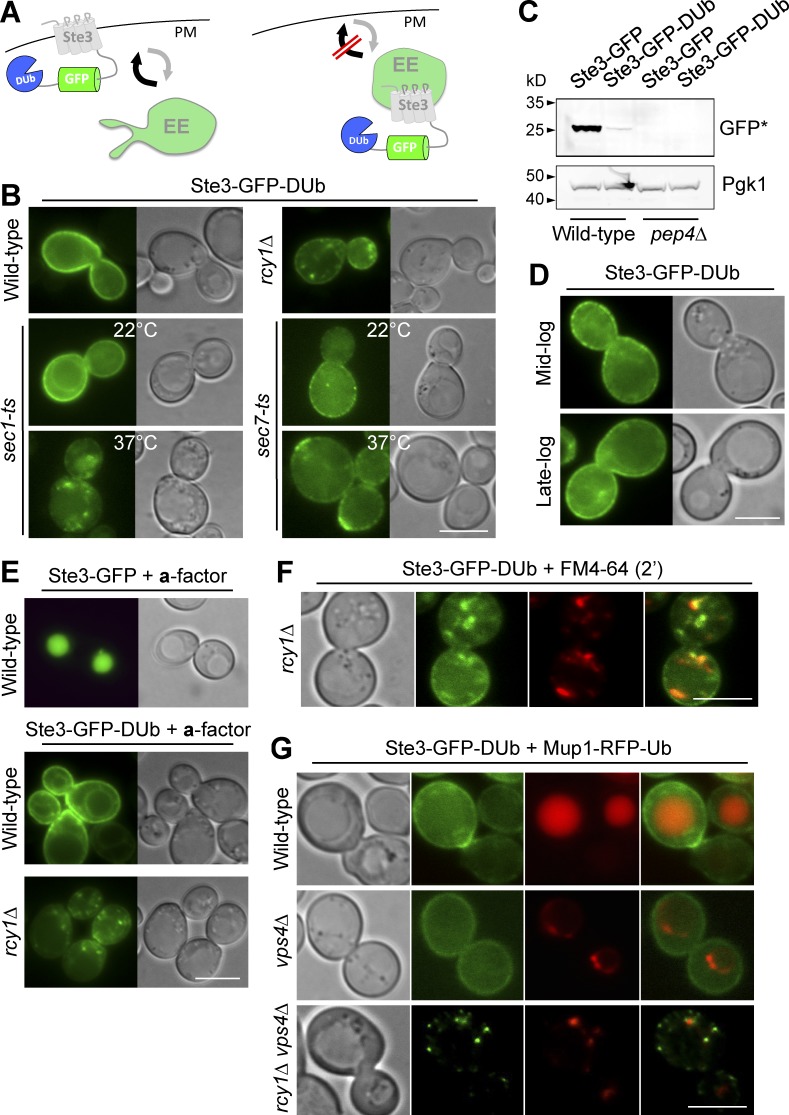
To engineer a better reporter protein of the EE>PM route, we started with Ste3 and blocked its ability to follow the Ub- and ESCRT-dependent route to the vacuole lumen by fusing it to the catalytic domain of the UL36 deubiquitinating peptidase (DUb) and GFP for visualization by microscopy (Stringer and Piper, 2011). We reasoned that fusion of a DUb would prevent trafficking of Ste3 to the vacuole yet still allow it to undergo internalization via its Sla1-binding NPFXD motif (Howard et al., 2002) and recycle via its natural, albeit uncharacterized, route back to the cell surface (Fig. 3 A). In wild-type cells, Ste3-GFP-DUb localized almost exclusively to the plasma membrane (Fig. 3 B), and levels of the vacuolar Pep4-processed form of GFP that is characteristic of delivery to the vacuole were minimal (Fig. 3 C). In addition, Ste3-GFP-DUb retained its cell-surface localization in cells grown to late-log phase (Fig. 3 D), when limiting nutrients cause other cell-surface proteins to shunt to the vacuole and other intracellular compartments (Fig. 2 E). In contrast, Ste3-GFP-DUb accumulated in multiple puncta in rcy1Δ cells, where it colocalized with FM4-64–labeled early endosomes (Fig. 3 F). The addition of the Ste3 ligand a-factor, which stimulates endocytosis and resulted in complete delivery of Ste3-GFP to the vacuole, had only a marginal effect on Ste3-GFP-DUb in wild-type cells. In rcy1Δ cells, however, a-factor drove the residual level of cell surface Ste3-GFP-DUb into endosomal puncta (Fig. 3 E). Like FM4-64, Ste3-GFP-DUb was quickly redistributed to intracellular compartments after inactivation of Sec1 but did not appreciably accumulate in Golgi compartments after inactivation of Sec7 (Fig. 3 B). Ste3-GFP-DUb was exclusively localized to the plasma membrane in vps4Δ cells (Fig. 3 G) and not the exaggerated late-endosomal compartments that accumulate ubiquitinated cargo, such as Mup1-RFP-Ub, in these mutants (Babst et al., 1997). Finally, Ste3-GFP-DUb did not accumulate in late-endosomal compartments of vps4Δ cells when RCY1 was additionally deleted (Fig. 3 G). Collectively, these data support the idea that the Ste3-GFP-DUb reporter is a useful and robust proxy for assessing the function of the EE>PM recycling route, which is dependent on Rcy1 and is followed by a portion of internalized FM4-64.
Figure 3.
A synthetic reporter that follows an Rcy1-dependent EE>PM route. (A) Ste3-GFP-DUb, comprising the Ste3 G protein–coupled receptor fused to GFP and the catalytic domain of DUb, designed to be at the cell surface in wild-type cells and endosomes in recycling mutants. (B) Localization of Ste3-GFP-DUb in wild-type and rcy1Δ cells or sec1-1 (ts) and sec7-1 (ts) cells grown at 22°C and 37°C. (C) Immunoblot of vacuolar-processed GFP cleaved from Ste3-GFP-DUb or Ste3-GFP expressed in wild-type and pep4Δ cells. (D) Ste3-GFP-DUb localization at mid- and late-log phase. (E) Localization of Ste3-GFP and Ste3-GFP-DUb in labeled cells grown in medium enriched with a-factor. (F and G) Ste3-GFP-DUb expressed in indicated mutants colocalized with FM4-64 chased for 2 min (F) or Mup1-RFP-Ub in the presence of 20 µg/nl methionine (G). Bars, 5 µm.
Genetic dissection of the EE>PM recycling pathway
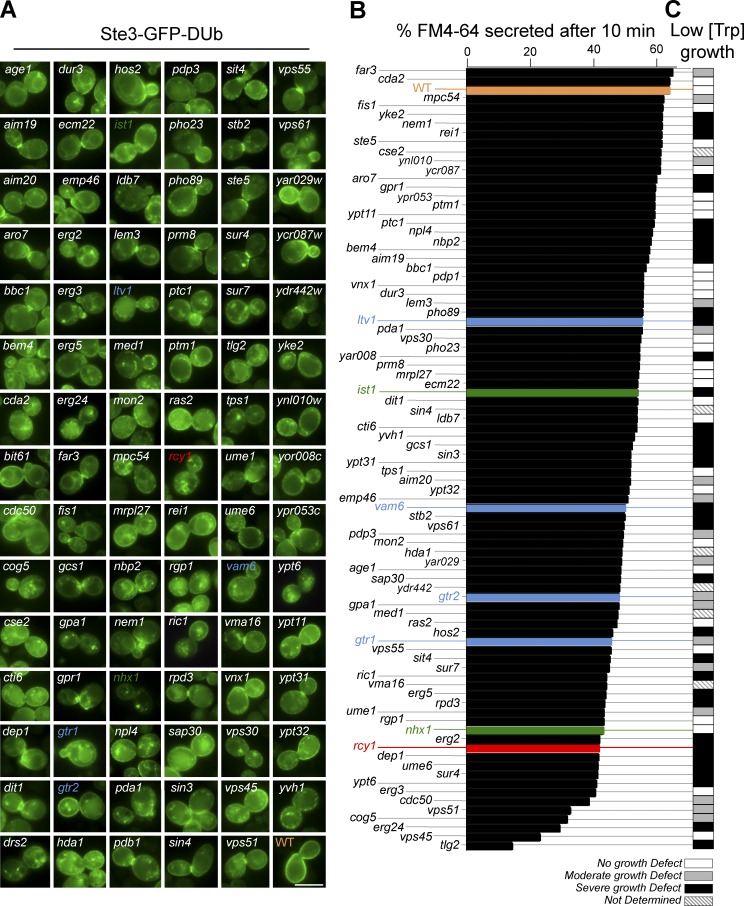
The distribution of Ste3-GFP-DUb was assessed in each of the mutants within the MAT α yeast haploid nonessential gene deletion collection (Winzeler et al., 1999). Initially, 366 mutants were identified using a liberal scoring system of any Ste3-GFP-DUb puncta. These candidates were grown under optimal conditions to reveal 89 mutants clearly defective in localizing Ste3-GFP-DUb to the cell surface. After the identity of the gene deletion was decoded from its position in the mutant array and the genotype of each mutant was confirmed by PCR (Table S1), Ste3-GFP-DUb was localized in cells grown to mid-log phase under identical conditions and imaged for comparison (Fig. 4 A). Mutants were also assessed for defects in FM4-64 efflux using flow cytometry (Fig. 4 B and Fig. S1 b). Finally, each candidate gene was deleted in a Trp− auxotroph parental strain and tested for growth in limiting tryptophan (Figs. 4 C and S1 c). Collectively, the mutants identified in this screen compose the machinery controlling the EE>PM recycling pathway in yeast. Whereas some of the components have clear mechanistic links to a variety of expected functions, such as vesicle tethering, fusion, and tubule formation, others provide unanticipated links to lipid homeostasis, transcriptional programs, and metabolism, while also filling putative functional roles for a handful of uncharacterized proteins (Fig. S1 a). The performance of the screen was validated by its unbiased identification of rcy1Δ, drs2Δ, cdc50Δ, and gcs1Δ mutants, all of which have been previously implicated in transport from early endosomes (Galan et al., 2001; Chen et al., 2005; Robinson et al., 2006; Furuta et al., 2007; Liu et al., 2007). Single mutants of either ypt31Δ or ypt32Δ were defective for Ste3-GFP-DUb localization (Fig. S2), consistent with a specific role in recycling inferred from their physical association with Rcy1, their requirement for trafficking Snc1, and mutations within their TRAPP-II GEF complex that phenocopy the recycling defects of rcy1Δ mutants (Chen et al., 2005; Cai et al., 2007; Furuta et al., 2007). Loss of Nhx1, the endosomal Na/H+ exchanger, caused a profound mislocalization of Ste3-GFP-DUb to large punctate structures that are distinct from late endosomes, accumulation of intracellular Mup1-GFP, and defects in FM4-64 efflux, helping to confirm a major role for Nhx1 in endosomal trafficking, as has been proposed in yeast and mammalian cells (Bowers et al., 2000; Brett et al., 2005; Ohgaki et al., 2010; Kojima et al., 2012; Kondapalli et al., 2015). Overall, these findings validate the screen and its stringency.
Figure 4.
Genetic screen reveals mutants defective in EE>PM recycling. Viable yeast deletion mutants (BY4742 derived) were screened visually for altered localization of plasmid-expressed Ste3-GFP-DUb. (A) Mutants were imaged after growth to mid-log phase under identical conditions. Indicated mutants are color coded: wild-type (orange); rcy1Δ (red); ist1Δ and nhx1Δ (green); gtr1Δ, gtr2Δ, vam6Δ, and ltv1Δ (blue). (B) Summary of FM4-64 efflux assays from mutants plotted as the percentage fluorescence loss after 10-min chase. (C) Summary of growth defects in low tryptophan of mutants made in trp1 auxotrophic (SEY6210 derived) parental strain. Bar, 5 µm.
Metabolic control of cell-surface proteins through the EE>PM recycling pathway
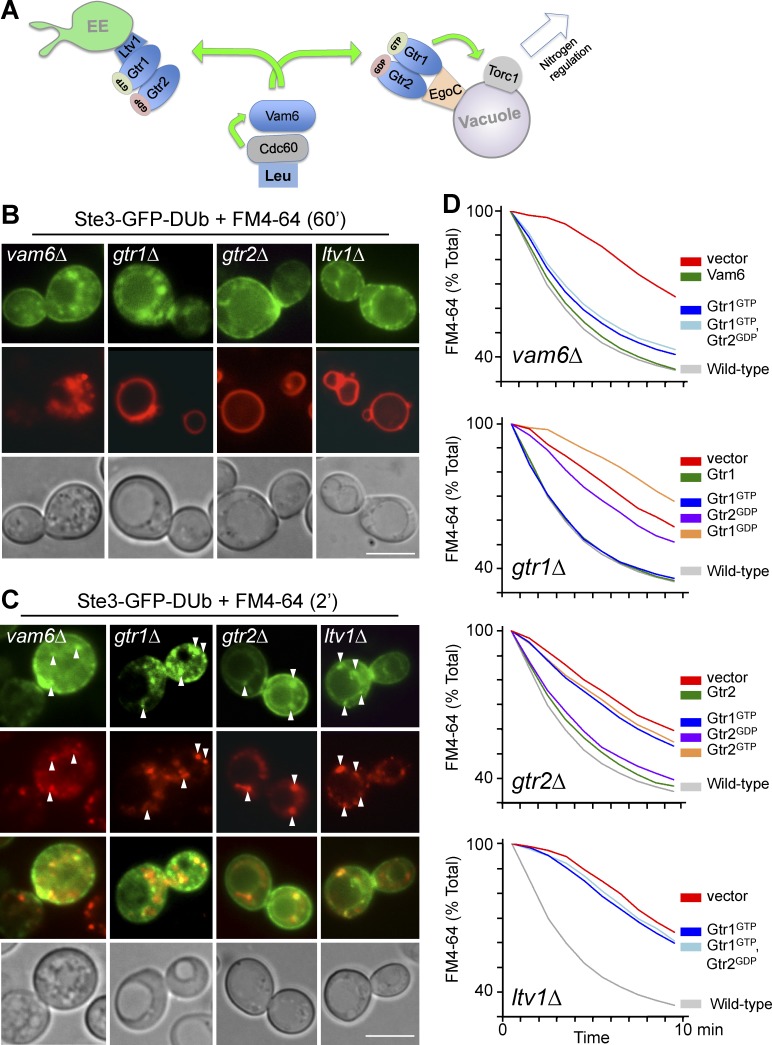
We also identified Gtr1, Gtr2, and Vam6 as defective for recycling, indicating a level of regulation by nitrogen metabolism (Fig. 5 A). Gtr1 and Gtr2, the yeast Rag GTPases, control a conserved mechanism to activate TORC1 (Powis and De Virgilio, 2016). Gtr1 and Gtr2 are localized to the limiting membrane of the vacuole via the Ego complex (Ego1 and Ego3), the yeast homolog of the Ragulator (Dubouloz et al., 2005; Binda et al., 2009). Ego-dependent localization on the vacuole allows GTP-bound Gtr1 and GDP-bound Gtr2 (which are in their activated states) to stimulate TORC1 (Powis et al., 2015; Kira et al., 2016). Upstream of Gtr1 is Vam6, which works as an exchange factor for Gtr1 and senses leucine levels in concert with the leucyl-tRNA synthetase, Cdc60 (Bonfils et al., 2012). Cells lacking Vam6, Gtr1, or Gtr2 mislocalized Ste3-GFP-DUb to early endosomes labeled with FM4-64 (Fig. 5, B and C). Like gtr1Δ and gtr2Δ mutants, loss of Vam6 caused defects in FM4-64 efflux (Fig. 5 D) and slow growth in limited tryptophan (Fig. S1 d). Vam6 is known to have other cellular functions, including roles in SNARE-mediated vacuole fusion in concert with Vam2/Vps41 and the HOPS complex components Vps11, Vps16, and Vps18 (Balderhaar and Ungermann, 2013). However, loss of these other Vam6-interacting components caused no defects in recycling, as measured by localization of Ste3-GFP-DUb (Fig. 6). Epistasis experiments confirmed that Gtr1 functions downstream of Vam6 within their pathway to control recycling, because the defect in FM4-64 efflux of vam6Δ mutants was largely suppressed by expressing a GTP-locked mutant of Gtr1 (Gtr1GTP) or coexpression of Gtr1GTP and Gtr2GDP (Fig. 5 D).
Figure 5.
Gtr1/Gtr2 control of EE>PM recycling is mediated through Vam6. (A) Model for how Gtr1 (GTP-bound)/Gtr2 (GDP-bound) heterodimer localizes to the vacuolar membrane via the Ego complex to activate TORC1. Gtr1 is activated by the GEF Vam6, which senses leucine via the leucyl-tRNA synthetase, Cdc60. Vam6, Gtr1, and Gtr2 are also required for recycling from early endosomes and work through Ltv1. (B and C) Localization of Ste3-GFP-DUb in the indicated mutant cells counter-labeled with FM4-64 for 60 min (B) or 2 min (C). Arrowheads indicate endosomal punta exhibiting colocalization. (D) Efflux of FM4-64 in wild-type cells and gtr1Δ, gtr2Δ, ltv1Δ, and vam6Δ cells carrying vector alone, or plasmids expressing Vam6, Gtr1, Gtr2, or Gtr1 and Gtr2 mutants locked in nucleotide-bound forms, Gtr1GTP, Gtr1GDP, Gtr2GDP, or Gtr2GTP. Bars, 5 µm.
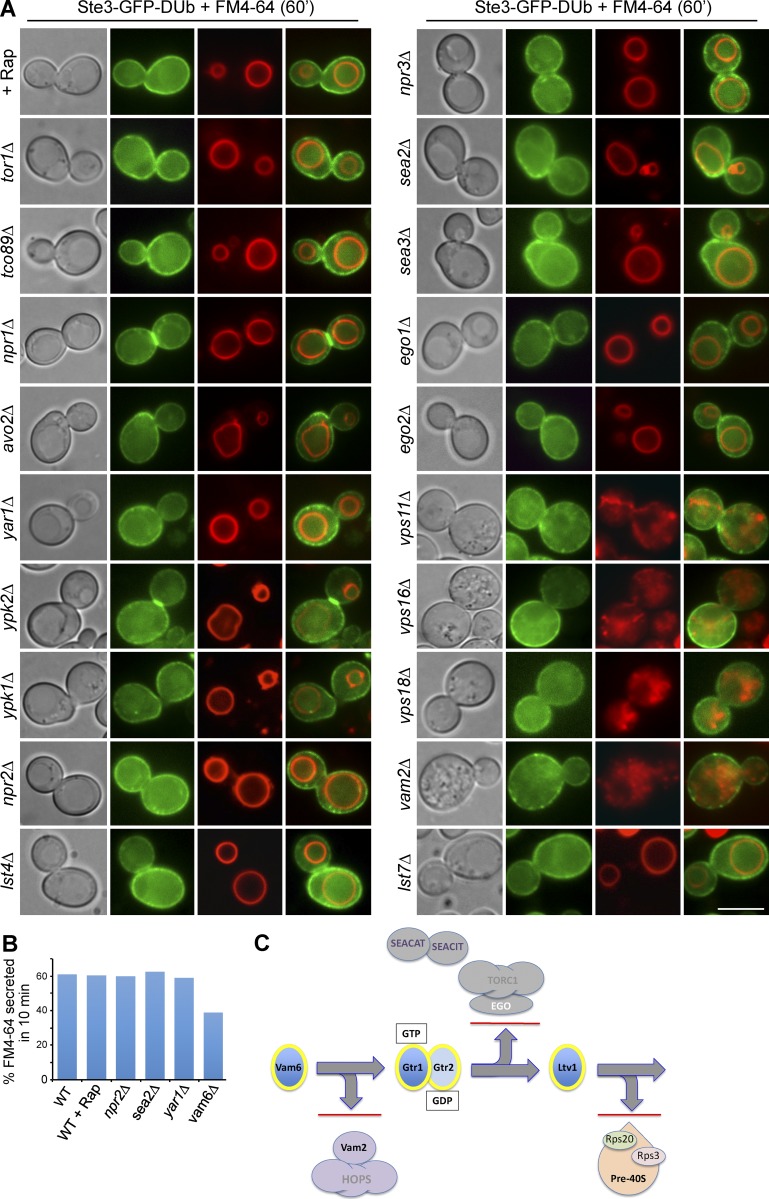
Figure 6.
The Vam6-Gtr1/Gtr2-Ltv1 pathway controls recycling independently of TORC1. (A) Ste3-GFP-DUb localized with FM4-64 after a 60-min chase period in minimal medium lacking dye. (B) FM4-64 efflux of indicated mutants after 10 min. (C) Experimental framework for tracing how the multifunctional proteins Vam6, Gtr1/Gtr2, and Ltv1 work together to control recycling rather than through their other known functions in endosomal fusion (Vam6, as part of the HOPS complex), TORC1 function (Gtr1/Gtr2, stimulators of TORC1 via the Ego complex), and ribosomal biogenesis (Ltv1, as a regulator of pre40S ribosome processing). Bar, 5 µm.
The requirement of Gtr1GTP and Gtr2GDP for the recycling pathway suggested that their downstream effector TORC1 was a regulator of the pathway. However, we found that disrupting TORC1 function or the physical connection between Gtr1/Gtr2 and TORC1 had no effect on recycling. Recycling of Ste3-GFP-DUB was efficient after rapamycin treatment, deletion of the TORC1 subunits Tor1 or Tco89, and loss of the Ego complex (yeast Ragulator) components Ego1 and Ego3, which localize Gtr1/Gtr2 to the vacuole surface near TORC1 (Fig. 6 C). In addition, rapamycin treatment did not inhibit FM4-64 efflux (Fig. 6 A). Previous studies identified several additional proteins that function as GTPase-activating proteins (GAPs) for Gtr1 and Gtr2 that contribute to TORC1 control (Panchaud et al., 2013a,b; Péli-Gulli et al., 2015). However, loss of Npr2, Npr3, Sea2, Sea3, Sea4 (regulators of Gtr1), Lst4, or Lst7 (regulators of Gtr2) did not affect Ste3-GFP-DUb recycling. In addition, loss of Npr2 or Sea2 had no effect on FM4-64 efflux (Fig. 6 B). Instead, our experiments indicated Ltv1, an alternate downstream interactor of Gtr1. Previous studies demonstrated that Ltv1 interacts with Gtr1GTP and disrupts trafficking of Gap1 (Gao and Kaiser, 2006). We found that loss of Ltv1 caused Ste3-GFP-DUb to accumulate within early endosomes (Fig. 5 C), defects in FM4-64 efflux (Fig. 5 D), and slow growth in medium containing low levels of tryptophan (Fig. S1 d). Epistasis experiments indicated that Ltv1 functions downstream of Gtr1/Gtr2, because the defect of FM4-64 efflux of ltv1Δ mutants was not suppressed by expressing Gtr1GTP alone or in combination with Gtr2GDP (Fig. 5 D). Thus, a model emerged wherein a series of multifunctional proteins collaborate within a specific metabolic pathway to execute EE>PM recycling (Figs. 5 A and 6 C). Importantly, although Vam6, Gtr1, and Gtr2 have alternative cellular roles besides controlling EE>PM recycling, we could eliminate the possibility that these additional functions were in control of the recycling pathway (Fig. 6 C).
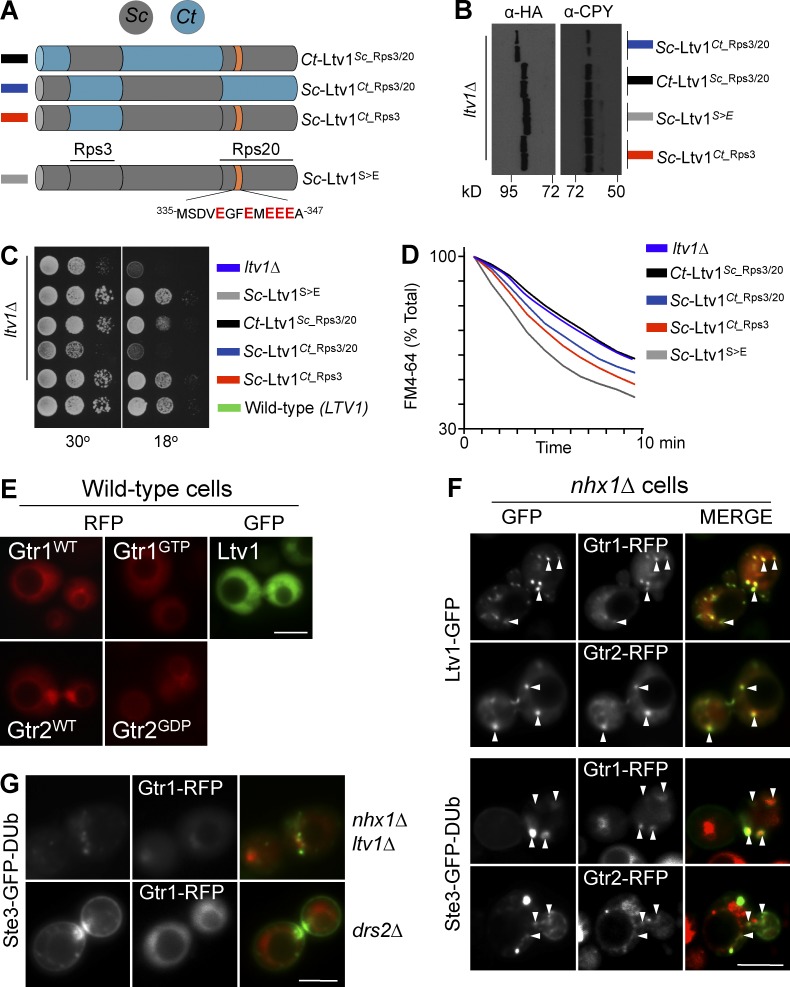
We next focused on the possibility that Ltv1 had an additional direct role in the EE>PM recycling pathway distinct from its known role in ribosome biogenesis. Ltv1 is required for biogenesis of the pre40S ribosome, and loss of Ltv1 causes severe growth defects at low temperature (Seiser et al., 2006; Merwin et al., 2014; de la Cruz et al., 2015). Ltv1 binds Rps3 and Rps20 to help configure the pre40S ribosome and is then ejected before assembly with the 60S subunit, in a process controlled by the phosphorylation of Ltv1 by the yeast casein kinase, Hrr25 (Schäfer et al., 2006; Ghalei et al., 2015). However, the role of Ltv1 in recycling appeared to be independent of pre40S ribosome assembly, because loss of Yar1, which works at a similar step upstream of Ltv1 (de la Cruz et al., 2015), had no effect on Ste3-GFP-DUb recycling or FM4-64 efflux (Fig. 6, A and B). To better distinguish a specific role for Ltv1 in recycling, we sought to genetically separate its potential dual roles in the cell. For this, we made a series of chimeras between Ltv1 from Sc and several distant species including the tree mushroom, Cylindrobasidium torrendii (Ct; Fig. 7 A), which was expressed at comparable levels (Fig. 7 B). To preserve Ltv1 function for pre40S ribosome assembly, we made a Ct chimera (Ct-Ltv1Sc_Rps3/20) containing the known Rsp3- and Rps20-interacting regions from Sc while also changing the Hrr25 phosphorylation sites to glutamate residues, to circumvent the need for Hrr25-dependent phosphorylation (Ghalei et al., 2015). We expressed these chimeras from a low-copy plasmid in ltv1Δ cells and compared these cells to either wild-type cells or ltv1Δ cells expressing the Sc-Ltv1S>E allele comprising Sc LTV1, also carrying the glutamate residue substitutions. Whereas Sc-Ltv1S>E fully restored FM4-64 efflux and normal growth to ltv1Δ mutants, Ct-Ltv1Sc_Rps3/20 was defective for FM4-64 efflux but still restored growth ltv1Δ mutants (Fig. 7, C and D). These data show that the role of Ltv1 in recycling can be clearly dissected from its general role in ribosome assembly and growth. We also found alleles that could partially operate in an opposite manner. Here we found that an Sc chimera (Sc-Ltv1Ct_Rps3/20) in which the Rsp3 and Rps20 binding regions were replaced with the corresponding regions from Ct had a severe growth defect at low temperature yet could partly suppress the FM4-64 recycling defect of ltv1Δ mutants (Fig. 7, C and D).
Figure 7.
The recycling-specific Rag GTPase interactor Ltv1 controls recycling. (A) Ltv1 chimeras from Sc (gray) and Ct (blue) with Rps3 and Rps20 binding sites and phosphomimetic Hrr1 phosphorylation sites shown. (B) FM4-64 efflux of ltv1Δ cells expressing the indicated Ltv1 chimeras. (C) Growth at 30°C and 18°C of the ltv1Δ cells expressing chimeras. (D) Expression levels of HA-tagged chimeras estimated by Western blotting using anti-CPY as a loading control. (E) Wild-type and active nucleotide-bound mutant Rag GTPases tagged with mCherry (RFP) and Ltv1-GFP localized in wild-type cells. (F) Rag GTPases colocalized (arrowheads) with Ltv1-GFP (top) and Ste3-GFP-DUb (bottom) in nhx1Δ cells. (G) Ste3-GFP-Dub and Gtr1-RFP were localized in nhx1Δ ltv1Δ and drs2Δ mutants. Bars, 5 µm.
Localization experiments buttress the model wherein Gtr1/Gtr2 and Ltv1 work directly on the recycling pathway. In wild-type cells (Fig. 7 E), mCherry-tagged Gtr1 and Gtr2, as well as their activated mutant forms (Gtr1GTP and Gtr2GDP), were localized diffusely in the cytosol as well as on the limiting membrane of the vacuole, consistent with previous observations (Powis et al., 2015; Kira et al., 2016). We found that GFP-tagged Ltv1 was mostly localized to the cytosol, with moderate accumulation in the nucleus, consistent with previous studies (Seiser et al., 2006; Merwin et al., 2014). If Gtr1/Gtr2 and Ltv1 transiently associate with endosomes, we reasoned that they might be captured there in mutants that accumulate intermediates along this pathway. Therefore, we performed a localization experiment in nhx1Δ mutants that accumulate large early endosomal structures that capture cargoes such as Ste3-GFP-Ub, FM4-64, and Mup1-GFP (Fig. S2). Loss of Nhx1 caused a distinct relocalization pattern for Gtr1, Gtr2, and Ltv1 in which Ltv1-GFP localizes to Gtr1- and Gtr2-containing puncta (Fig. 7 F). Moreover, Gtr1 and Gtr2 also colocalized with intracellular Ste3-GFP-DUb. Colocalization of Gtr1-RFP with Ste3-GFP-DUb to intracellular puncta in nhx1Δ cells was compromised upon further loss of Ltv1, suggesting that Ltv1 may have a role in recruiting Gtr1 to endosomal structures. Other recycling pathway mutants, such as drs2Δ (Fig. 7 G), did not accumulate Gtr1 in endosomal structures, suggesting that the intermediates accumulated in nhx1Δ mutants are qualitatively different from those in drs2Δ mutants or that Drs2 is part of the protein recruitment machinery for Gtr1 and its associated cohort.
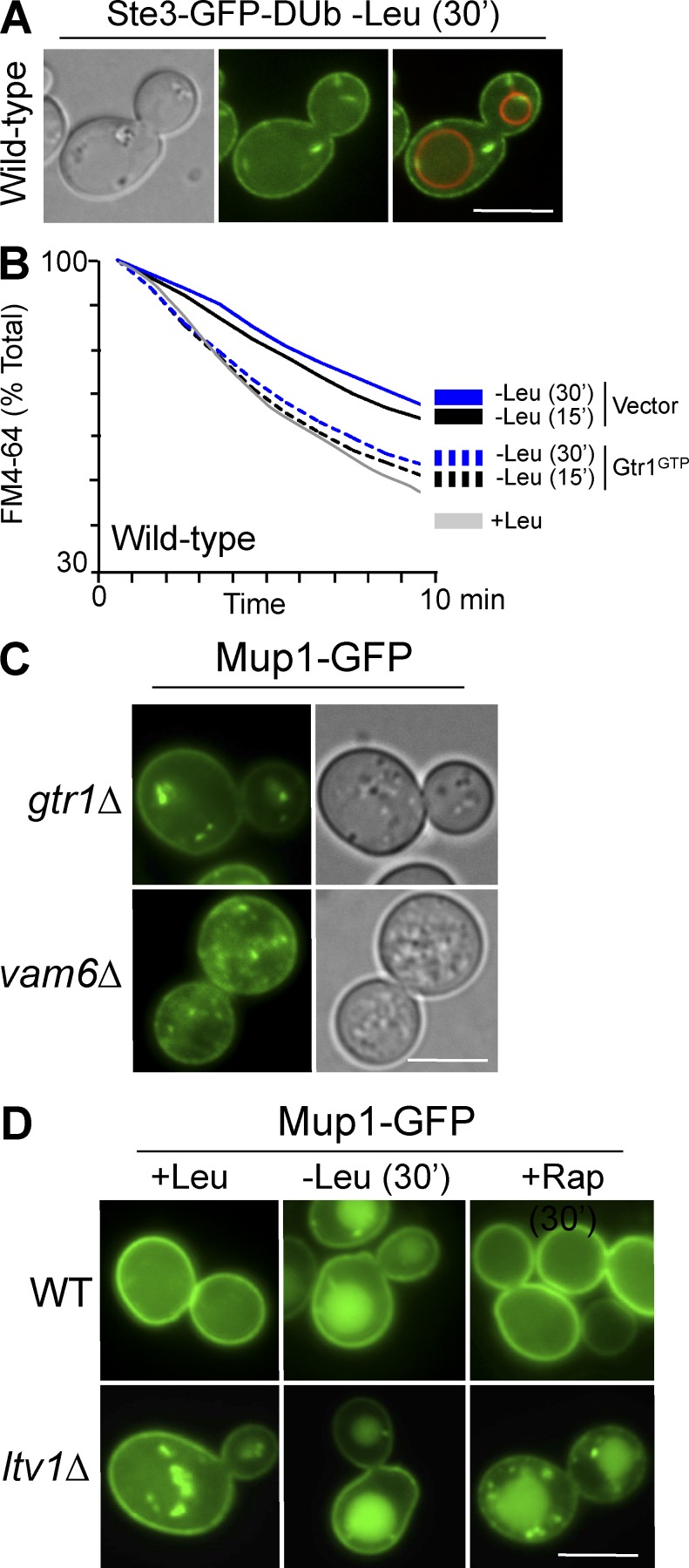
We next assessed how the Vam6>Gtr1/Gtr2>Ltv1 pathway might control the recycling pathway in a physiological context. Vam6 is part of the mechanism that senses leucine, conveying that signal to Gtr1 to regulate TORC1 (Binda et al., 2009; Valbuena et al., 2012). We found that leucine starvation also blocked recycling, as observed by Ste3-GFP-DUb mislocalization (Fig. 8 A) and retardation of FM4-64 efflux (Fig. 8 B). The defect in FM4-64 efflux could be suppressed by expressing the activated Gtr1GTP protein (Fig. 8 B), supporting the model that leucine signals through the Vam6>Gtr1/Gtr2>Ltv1 pathway to control recycling. We next determined how regulation of the recycling pathway could play a role in the overall trafficking of cell surface proteins. Mup1-GFP recycling is defective when the Vam6>Gtr1/Gtr2 pathway is disrupted (Fig. 8 C), and previous studies have shown that nutrient depletion, specifically starvation for leucine, can cause proteins such as Mup1 to traffic to the vacuole (Jones et al., 2012). This effect partly relies on Ub- and ESCRT-mediated trafficking through the MVB pathway, which is responsible for packaging membrane proteins into endosomal lumenal vesicles. Whereas TORC1 inhibition causes global up-regulation of Rsp5 activity (Iesmantavicius et al., 2014) and TORC1 activity controls trafficking of nutrient transporters, opposing modes of TORC1 control over individual transporters have been described. Some stimuli that activate TORC1 cause amino acid transporters such as Can1, Mup1, and Gap1 to traffic to the vacuole through activation of particular arrestin-related substrate-adaptors of the Rsp5 ligase (Schmidt et al., 1998; MacGurn et al., 2011; Martín et al., 2011; Merhi and André, 2012; Crapeau et al., 2014; Pfannmüller et al., 2015). This mechanism does not explain how leucine starvation, which would inactivate TORC1, results in rapid withdrawal of transporters from the cell surface. An alternative explanation is that the effect of leucine starvation reflects a compound phenotype of both TORC1 inactivation and regulation of the recycling pathway via Ltv1 because both would be controlled by the Leucine>Cdc60>Vam6>Gtr1 signaling pathway. Fig. 8 D supports this model in that acute inhibition of TORC1 with rapamycin had no effect on the cell-surface localization of Mup1. Yet in ltv1Δ mutants, where the other branch of the Gtr1 pathway is blocked, rapamycin had a dramatic effect on sorting Mup1-GFP to the vacuole that mimicked the full effect of leucine starvation.
Figure 8.
The Vam6-Gtr1/Gtr2-Ltv1 pathway controls recycling in response to leucine starvation. (A) Localization of Ste3-GFP-Ub after acute leucine starvation. (B) FM4-64 efflux after acute leucine starvation in wild-type cells alone or cells expressing Gtr1GTP. (C) Mup1-GFP was localized in indicated mutants grown to mid-log phase. (D) Localization of Mup1-GFP in wild-type and ltv1Δ cells in replete medium (+Leu), after 30-min leucine starvation (–Leu), or after 30-min rapamycin (200 ng/ml) treatment. Bars, 5 µm.
Discussion
Discovery of an EE>PM pathway in yeast and identification of its molecular machinery using a dedicated synthetic reporter helps clarify functions for several proteins that have clear mammalian homologs (Table S2). These studies also reveal a mechanistic framework for how EE>PM recycling can be regulated by signal transduction pathways that integrate the overall metabolic activity of the cell. Both Snc1 and Ste3-GFP-DUb are internalized from the cell surface and travel out of early endosomes before reaching late endosomes. Both of these cargoes are trapped within early endosomes in cells lacking some of the known protein machinery required for transport out of early endosomes, including Rcy1, Drs2, Cdc50, and the Arf-GAP Gcs1 (Wiederkehr et al., 2000; Hua et al., 2002; Chen et al., 2006; Robinson et al., 2006). We found that the Ste3-GFP-DUb reporter stayed confined to an early endosome/PM itinerary in that it did not travel through late endosomes that accumulate class E/ESCRT vps mutants, and it did not undergo Ub-dependent sorting into multivesicular bodies. Ste3-GFP-DUb also did not accumulate in Golgi structures in sec7 ts cells at the nonpermissive temperature under conditions where GFP-Snc1 did. This suggests that although flux of internalized GFP-Snc1 through the sec7-1–sensitive step through the Golgi is substantial, that of Ste3-GFP-DUb is much less, together illustrating that the route Snc1 takes from endosomes is demonstrably distinct from that of Ste3-GFP-DUb. Importantly, Ste3-GFP-DUb did accumulate intracellularly upon acute loss of Sec1 function (via a sec1 ts allele), showing that the bulk of Ste3-GFP-DUb is internalized and recycled within the time frame of our experiments. These data imply that both Golgi-derived and endosome-derived transport intermediates use a Sec1-sensitive step for fusion with the plasma membrane. Such a model is consistent with data from animal cells, where a cohort of highly homologous SNAREs and associated proteins function in both secretory and endosome-derived transport intermediates to mediate fusion with the plasma membrane (Huynh et al., 2007; Hong and Lev, 2014). Here, such a model obliges that Snc1/2 would populate both intermediates. Retrieval of Snc1 from early endosomes back to the TGN/Golgi has been clearly established (Lewis et al., 2000; Galan et al., 2001; Chen et al., 2005; Robinson et al., 2006). Whereas these data clearly show that a major flux of endosomal Snc1 back to the plasma membrane can travel through TGN/Golgi compartments, they do not exclude the possibility that Snc1 also travels through other pathways, such as the EE>PM pathway proposed here, which bypasses the TGN/Golgi.
In contrast to Snc1, the Ste3-GFP-DUb reporter behaved more similarly to FM4-64, as it did not readily accumulate in Golgi-like compartments upon inactivation of Sec7 under our experimental conditions. We note that previous experiments have shown localization of FM4-64 to Sec7-positive compartments and concluded that FM4-64 traveled back to the Golgi apparatus during its return to the cell surface (Lewis et al., 2000; Bhave et al., 2014). In our experiments, inactivation of Sec7 accumulated Snc1 in compartments markedly distinct from endosomal populations of FM4-64, and dye efflux remained efficient when transit through the Golgi was blocked. One of the difficulties in following FM4-64 is that it travels along multiple pathways, and the basis of its trafficking (as a styryl dye, it binds to a complex of lipids that remain unknown) is unclear. After internalization, some FM4-64 travels to the limiting membrane of the vacuole, and a different portion is effluxed from the cell. Even the mutants defective in FM4-64 recycling exhibit a sizable rate of residual FM4-64 efflux (Figs. 4 and S1). Furthermore, we found that some of these defects are additive, whereby FM4-64 efflux is far worse in an ltv1Δ nhx1Δ double mutant than it is in either single mutant (Fig. S3 g). This could indicate either that loss of each gene results in a partial block of the same pathway or that multiple pathways for FM4-64 efflux exist. It may be that compromise of one pathway would shunt FM4-64 through a different one, including a route via the Golgi apparatus. As Ste-GFP-DUb and the bulk of FM4-64 appeared to follow a recycling route that bypassed the Golgi, and most of the mutants defective for Ste3-GFP-DUb localization had defects in FM4-64 efflux, we conclude that both cargoes mainly report on the direct EE>PM pathway.
For those proteins with less defined roles in the endocytic pathway, discovering a role for them in EE>PM recycling potentially clarifies their primary function. One such protein is Nhx1, an Na+/H+ exchanger localized to early endosomal compartments (Kojima et al., 2012). Loss of Nhx1 in yeast causes changes in late endosome function reminiscent of ESCRT mutants, which have defects in MVB biogenesis and accumulate cargo proteins within enlarged late endosomes (Bowers et al., 2000). We find that nhx1Δ mutants accumulate Ste3-GFP-DUb in endosomal compartments that are distinct from the late endosomal structures that accumulate in ESCRT-deficient cells (Fig. S3), consistent with data from both yeast and animal cells indicating a direct role for Nhx1 in trafficking through early endosomes. Also defective in recycling were mutants lacking Ist1, a protein that is structurally related to ESCRT-III subunits and that binds to the Vps4 AAA-ATPase. Mutant ist1Δ cells are defective in Ste3-GFP-DUb and Mup1 localization, FM4-64 efflux, and growth in low tryptophan (Figs. S1 and S3). Ist1 is not strictly required for ESCRT-dependent sorting into the MVB pathway (Dimaano et al., 2008; Rue et al., 2008; Jones et al., 2012). Recent experiments implicate Ist1 in the scission of tubules emanating from early endosomes (Allison et al., 2013), and cryo-electron microscopy studies indicate that Ist1 can bend membranes in a manner that is topologically distinct from scission of lumenal vesicles mediated by ESCRT-III (McCullough et al., 2015). A distinct role for Ist1 in promoting recycling also fits with previous observations that show delivery of cargo into the MVB pathway being accelerated in the absence of Ist1, which would otherwise rescue cell surface proteins from degradation. We further found that recycling is supported by mutants of Ist1 that lack their MIT-interaction motif, which is required for its functional interaction with Vps4 (Shestakova et al., 2010), underscoring a role for Ist1 that is distinct from ESCRT-III– and Vps4-related functions on late endosomes (Fig. S2). It has been reported that Ist1 is a heavily ubiquitinated and unstable protein whose levels may be affected by various stresses and metabolic conditions (Jones et al., 2012), raising the possibility that the recycling defects of some of the mutants we uncovered were caused by decreased levels of Ist1. However, Ist1 levels appear unchanged in a wide variety of recycling mutants, suggesting that their recycling defects are not caused by simple loss of Ist1 function (Fig. S2).
Nutritional control over the recycling pathway may serve as a way to synchronize surface protein activity (e.g., amino acid transport) with metabolic activity of the cell. As shown previously, starvation for leucine shunts a wide variety of cell surface proteins to the vacuole (Jones et al., 2012). We propose that leucine starvation regulates two coordinated, yet distinct, pathways through the Rag GTPases, Gtr1 and Gtr2. First, surface protein retention in endosomes is achieved by inhibition of the Gtr1/Gtr2>Ltv1 recycling pathway. Second, the sorting of endosomally localized cargo into the MVB pathway is elevated through inhibition of TORC1. It is not yet clear what molecular function Ltv1 provides to promote recycling or how it localizes Gtr1/Gtr2 to endosomes. Ltv1 functions in the assembly or ribosomes and is required for robust growth at lower temperature, yet this alternative function could be genetically separated from its role in the recycling pathway. The effect of Gtr1 and Gtr2 specifically on the recycling pathway is independent of TORC1, because their localization and communication with TORC1 on the vacuole is dependent on the Ego complex, loss of which has no effect on recycling. In addition, acute inhibition of TORC1 with rapamycin or compromise of TORC1 activity by knocking out the nonessential subunits TOR1 and TCO89 also had no effect. TORC1 does have an effect on the trafficking of cell-surface proteins, which is ultimately determined by increased flux through the MVB pathway by increased ubiquitination of cargo. These combined effects speak to complex layers of regulation for nutrient transporters, which we can mimic by the combined inhibition of the recycling pathway (via ltv1Δ mutation) and acute inhibition of TORC1 with a short treatment with rapamycin (Fig. 8 D). This model complements previous work implicating the Rag GTPases and EGO complex in trafficking of the amino acid transporter Gap1 (Gao and Kaiser, 2006). Those studies showed that loss of Gtr1, the Ego complex, or Ltv1 diminishes both total cellular levels and cell-surface levels of Gap1. The effect of the Ego complex mutants would be explained by compromise of TORC1 function and is consistent with the effects of longer-term rapamycin treatment on the ubiquitination and MVB sorting of Gap1, as well as additional transporters shown in other studies (Crapeau et al., 2014). In contrast, loss of another Gtr1 downstream target Ltv1 inhibits return of endocytosed transporters along the EE>PM pathway, sequestering them from the cell surface and increasing their exposure the ubiquitination and MVB trafficking machinery. Such a model emphasizes the intricacies in following endogenous cell surface proteins such as Gap1, that are subject to a variety of trafficking steps at the cell surface, TGN, early endosomes, and MVBs, where many are controlled by ubiquitination (Babst and Odorizzi, 2013; Huber and Teis, 2016). By following Ste3-GFP-DUb and FM4-64, the effects on cargo ubiquitination and specific sorting sequences can be uncoupled, allowing the EE>PM recycling pathway to be revealed.
Materials and methods
Reagents
Yeast strains and plasmids used in this study are listed in Tables S3 and S4. Both Sec1 and Sec7 are essential for growth, and the efficacy of the ts cells used for experiments was verified by absence of growth at 37°C (Fig. 1 D). Plasmids expressing nucleotide-bound locked mutants of Gtr1 and Gtr2 were Gtr1GTP, Gtr1-Q65L(active); Gtr2GTP, Gtr2-Q66L(inactive); Gtr1GDP, Gtr1-S20L(inactive); and Gtr2GDP, Gtr2-S23L(active), as described previously (Binda et al., 2009).
Cell culture conditions
Standard yeast extract peptone dextrose (YPD)-rich medium (2% glucose, 2% peptone, and 1% yeast extract) and synthetic complete (SC) minimal medium (2% glucose, yeast nitrogen base; Research Products International) lacking appropriate amino acids and bases for plasmid selection were used. Geneticin (G418), used at a concentration of 250 µg/ml, and methotrexate (Sigma-Aldrich), used as described (MacDonald and Piper, 2015), were used for selection of yeast integrant strains. For genetic screening, the MAT α haploid deletion library (Winzeler et al., 1999) was purchased from Dharmacon. Expression of plasmids containing the CUP1 promoter was generally induced by the addition of 10–50 µM copper chloride to the medium. Medium enriched in a-factor was achieved by culturing BY4741 MAT A cells transformed with overexpression plasmid pKK16 to increase expression of the mating pheromone a-factor MFA1 and the peptide export transporter STE6 genes (Kuchler et al., 1989), to provide robust production and secretion, respectively, of a-factor. Yeast cells were then removed by centrifugation, and the medium was used directly to induce Ste3 internalization. Cells were routinely harvested from cultures grown to early/mid-log phase (OD600 = 0.5–1.0) or late-log phase (OD600 = 2.0), where labeled, before fluorescence microscopy. For leucine starvation, leu2 mutant cells (BY4742 background) were grown in minimal medium containing leucine, pelleted, and resuspended in minimal medium lacking leucine for 15 or 30 min before assay.
Transformation of yeast deletion library
Transformation of the yeast gene deletion library was performed according to a high-throughput transformation protocol (Gietz and Schiestl, 2007). In brief, yeast strains were grown in 96-well plates, pelleted by centrifugation, and incubated for 12 h at 30°C in 100 μl of 50 mM Tris, pH 8.0, 100 mM LiAc, 0.5 mM EDTA, 50% PEG, and 50 ng/µl plasmid expressing Ste3-GFP-Dub from a low copy (CEN-based pRS316) plasmid under the control of the STE3 promoter. Cells were pelleted, grown, and maintained in minimal medium lacking uracil upon dilution in several new 96-well plates. Cells were grown overnight in minimal medium, pelleted, and resuspended in batches of 24 in minimal medium before imaging by fluorescence microscopy.
Immunoblotting
Yeast cells harvested at mid-log phase were subjected to alkali treatment (0.2 N NaOH) for 3 min followed by resuspension in 50 mM Tris HCl, pH 6.8, 5% SDS, 10% glycerol, and 8 M urea, to prepare whole cell lysates. Proteins resolved by SDS-PAGE were immunodetected with monoclonal antibodies raised against the HA epitope (HA.11; BioLegend), carboxypeptidase Y (10A5B5; Thermo Fisher Scientific), Pgk1 (22C5D8; Thermo Fisher Scientific), and polyclonal antibodies that recognize GFP (Urbanowski and Piper, 1999).
Fluorescence microscopy
Yeast cells were concentrated and resuspended in minimal medium or kill buffer (100 mM Tris HCl, pH 8.0, 0.2% [wt/vol] NaN3, and NaF3), before fluorescence microscopy. GFP, mCherry, mStrawberry, and FM4-64 signals were imaged using a BX60 epifluorescence microscope (Olympus) with a 100× objective lens, NA 1.4. Images were captured at room temperature with a cooled charged-coupled camera (Orca R2; Hamamatsu Photonics) using iVision-Mac software (Biovision Technology). Mup1-mCherry-Ub localization was recorded after 1-h incubation in minimal medium containing 20 µg/ml methionine and 5 µM copper chloride. For localization of early endosomes labeled with FM4-64 (Molecular Probes), cells grown in minimal medium were pelleted and resuspended in 40 µM FM4-64 in YPD medium and incubated for 2 min at 22°C. FM4-64 was added from a 400-µM stock containing 10% DMSO to yield a final labeling concentration of 1% DMSO. Cells were centrifuged and resuspended three times in 0°C kill buffer, resuspended in kill buffer, and stored on ice before microscopy. Labeling of ts mutants (sec1-1 and sec7-1) was accomplished by labeling cells in 40 µM FM4-64 for 2 min at 22°C in YPD. Cells were centrifuged and resuspended in minimal medium prewarmed to either 22°C or 37°C and incubated for an additional 15 min before three washes in 0° kill buffer, storage on ice, and visualization microscopy. For localization with FM4-64–labeled vacuoles, rich medium containing 40 µM FM4-64 was incubated for 10 min at room temperature, followed by three washes with minimal medium and further incubation with minimal medium for 1 h. For localization of Mup1-GFP, Mup1-GFP was expressed from the low-copy LEU2-based plasmid (pPL4070), unless in leucine starvation experiments from the URA3-based (pPL4023) plasmid. Cells were grown to mid-log phase in minimal medium lacking methionine.
FM4-64 efflux assay
To measure FM4-64 efflux under standard conditions, cells were grown in YPD to mid-log phase (OD600 = 1), pelleted, resuspended in 200 µl YPD medium containing 40 µM FM4-64, and incubated at 22°C for 10 min. Cells were transferred to a 0°C water bath for initial chilling and then subjected to three washes (5 min each) in ice-cold buffer followed by pelleting in a cold centrifuge. Cells were stored in a 0°C ice bath at a concentrated density before addition (2,000×) of prewarmed medium at 22°C. FM4-64 fluorescence was measured by flow cytometry using a Becton Dickinson LSR II. Binned averages of ∼100,000 cells in 1-min increments were used to plot results. To measure FM4-64 efflux in sec1-1 and sec7-1 ts mutants, cells were grown in YPD at 22°C, pelleted, resuspended in 200 µl YPD medium containing 40 µM FM4-64, and incubated for 9 min at 22°C. Cells were washed three times in cold minimal medium as in the standard protocol, resuspended in 0°C minimal medium to a concentration of 400 million cells/100 µl of cold minimal medium, and stored in glass tubes immersed in a 0°C water bath. 15 µl cell suspension was then diluted in prewarmed tubes containing 2 ml prewarmed medium at 22°C or 37°C. Temperature was maintained in an insulated chamber comprising a metal cylinder immersed in a beaker containing water at 37°C or 22°C during efflux measurements taken by flow cytometry.
Limited tryptophan growth assay
Cells were grown to mid-log phase, and equivalent volumes were harvested and used to create a serial dilution (1:9) before plating on SC medium plates containing replete (40 µg/ml), moderate (5 µg/ml), or low (2.5 µg/ml) tryptophan concentrations. Wild-type parental strain (SEY6210) control cultures were included on each plate. Plates were then incubated for 48 h at 30°C, and growth was documented.
Online supplemental material
Fig. S1 shows secondary screens used to validate mutants in the recycling pathway. Fig. S2 shows the endosomal proteins that control EE>PM recycling. Fig. S3 shows that TORC1-indepedent recycling relies on specific features of Ltv1. Table S1 is a list of mutants and genotyping information. Table S2 lists mammalian orthologues of recycling mutants. Table S3 lists the strains used in this study. Table S4 lists the plasmids used in this study.
Supplementary Material
Acknowledgments
We thank members of the laboratory for constructs, advice, and helpful discussions. We thank Stanley Winistorfer for help with Ltv1 chimeras and Justin Fishbaugh and the CCOM flow cytometry facility for help with FM4-64 efflux assays. We are grateful to Claudio De Virgilio, Deborah Lycan, Dave Katzmann, and Jeremy Thorner for sharing reagents. We thank Phyllis Hanson and Markus Babst for insightful conversations regarding Ist1 function.
This work was supported by American Heart Association postdoctoral fellowship award 13POST 14710042 to C. MacDonald and National Institutes of Health RO1GM058202 to R.C. Piper.
The authors declare no competing financial interests.
Author contributions: C. MacDonald and R.C. Piper conceived and designed experiments. C. MacDonald performed experiments. C. MacDonald and R.C. Piper wrote the manuscript.
Footnotes
Abbreviations used:
- ts
- temperature-sensitive allele
- Ct
- Cylindrobasidium torrendii
- DUb
- UL36 deubiquitinating peptidase
- EE
- early endosome
- ESCRT
- endosomal sorting complexes required for transport
- MVB
- multivesicular body
- PM
- plasma membrane
- Sc
- Saccharomyces cerevisiae
- Ub
- ubiquitin
References
- Allison R., Lumb J.H., Fassier C., Connell J.W., Ten Martin D., Seaman M.N.J., Hazan J., and Reid E.. 2013. An ESCRT-spastin interaction promotes fission of recycling tubules from the endosome. J. Cell Biol. 202:527–543. 10.1083/jcb.201211045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., and Odorizzi G.. 2013. The balance of protein expression and degradation: An ESCRTs point of view. Curr. Opin. Cell Biol. 25:489–494. 10.1016/j.ceb.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Sato T.K., Banta L.M., and Emr S.D.. 1997. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16:1820–1831. 10.1093/emboj/16.8.1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderhaar H.J.K., and Ungermann C.. 2013. CORVET and HOPS tethering complexes—Coordinators of endosome and lysosome fusion. J. Cell Sci. 126:1307–1316. 10.1242/jcs.107805 [DOI] [PubMed] [Google Scholar]
- Becuwe M., and Léon S.. 2014. Integrated control of transporter endocytosis and recycling by the arrestin-related protein Rod1 and the ubiquitin ligase Rsp5. eLife. 3 10.7554/eLife.03307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhave M., Papanikou E., Iyer P., Pandya K., Jain B.K., Ganguly A., Sharma C., Pawar K., Austin J. II, Day K.J., et al. . 2014. Golgi enlargement in Arf-depleted yeast cells is due to altered dynamics of cisternal maturation. J. Cell Sci. 127:250–257. 10.1242/jcs.140996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda M., Péli-Gulli M.-P., Bonfils G., Panchaud N., Urban J., Sturgill T.W., Loewith R., and De Virgilio C.. 2009. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell. 35:563–573. 10.1016/j.molcel.2009.06.033 [DOI] [PubMed] [Google Scholar]
- Bonfils G., Jaquenoud M., Bontron S., Ostrowicz C., Ungermann C., and De Virgilio C.. 2012. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol. Cell. 46:105–110. 10.1016/j.molcel.2012.02.009 [DOI] [PubMed] [Google Scholar]
- Bowers K., Levi B.P., Patel F.I., and Stevens T.H.. 2000. The sodium/proton exchanger Nhx1p is required for endosomal protein trafficking in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 11:4277–4294. 10.1091/mbc.11.12.4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett C.L., Tukaye D.N., Mukherjee S., and Rao R.. 2005. The yeast endosomal Na+K+/H+ exchanger Nhx1 regulates cellular pH to control vesicle trafficking. Mol. Biol. Cell. 16:1396–1405. 10.1091/mbc.E04-11-0999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Reinisch K., and Ferro-Novick S.. 2007. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell. 12:671–682. 10.1016/j.devcel.2007.04.005 [DOI] [PubMed] [Google Scholar]
- Chen S.H., Chen S., Tokarev A.A., Liu F., Jedd G., and Segev N.. 2005. Ypt31/32 GTPases and their novel F-box effector protein Rcy1 regulate protein recycling. Mol. Biol. Cell. 16:178–192. 10.1091/mbc.E04-03-0258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Wang J., Muthusamy B.-P., Liu K., Zare S., Andersen R.J., and Graham T.R.. 2006. Roles for the Drs2p-Cdc50p complex in protein transport and phosphatidylserine asymmetry of the yeast plasma membrane. Traffic. 7:1503–1517. 10.1111/j.1600-0854.2006.00485.x [DOI] [PubMed] [Google Scholar]
- Corvera S., Davis R.J., Roach P.J., DePaoli-Roach A., and Czech M.P.. 1986. Mechanism of receptor kinase action on membrane protein recycling. Ann. N. Y. Acad. Sci. 488(1 Membrane Path):419–429. 10.1111/j.1749-6632.1986.tb46575.x [DOI] [PubMed] [Google Scholar]
- Crapeau M., Merhi A., and André B.. 2014. Stress conditions promote yeast Gap1 permease ubiquitylation and down-regulation via the arrestin-like Bul and Aly proteins. J. Biol. Chem. 289:22103–22116. 10.1074/jbc.M114.582320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis N.G., Horecka J.L., and Sprague G.F. Jr. 1993. Cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J. Cell Biol. 122:53–65. 10.1083/jcb.122.1.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J., Karbstein K., and Woolford J.L. Jr. 2015. Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu. Rev. Biochem. 84:93–129. 10.1146/annurev-biochem-060614-033917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimaano C., Jones C.B., Hanono A., Curtiss M., and Babst M.. 2008. Ist1 regulates Vps4 localization and assembly. Mol. Biol. Cell. 19:465–474. 10.1091/mbc.E07-08-0747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouloz F., Deloche O., Wanke V., Cameroni E., and De Virgilio C.. 2005. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol. Cell. 19:15–26. 10.1016/j.molcel.2005.05.020 [DOI] [PubMed] [Google Scholar]
- Edinger A.L., and Thompson C.B.. 2002. Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol. Biol. Cell. 13:2276–2288. 10.1091/mbc.01-12-0584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edinger A.L., and Thompson C.B.. 2004. An activated mTOR mutant supports growth factor-independent, nutrient-dependent cell survival. Oncogene. 23:5654–5663. 10.1038/sj.onc.1207738 [DOI] [PubMed] [Google Scholar]
- Feyder S., De Craene J.-O., Bär S., Bertazzi D.L., and Friant S.. 2015. Membrane trafficking in the yeast Saccharomyces cerevisiae model. Int. J. Mol. Sci. 16:1509–1525. 10.3390/ijms16011509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta N., Fujimura-Kamada K., Saito K., Yamamoto T., and Tanaka K.. 2007. Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway. Mol. Biol. Cell. 18:295–312. 10.1091/mbc.E06-05-0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan J.-M., Wiederkehr A., Seol J.H., Haguenauer-Tsapis R., Deshaies R.J., Riezman H., and Peter M.. 2001. Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol. Cell. Biol. 21:3105–3117. 10.1128/MCB.21.9.3105-3117.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., and Kaiser C.A.. 2006. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat. Cell Biol. 8:657–667. 10.1038/ncb1419 [DOI] [PubMed] [Google Scholar]
- Ghalei H., Schaub F.X., Doherty J.R., Noguchi Y., Roush W.R., Cleveland J.L., Stroupe M.E., and Karbstein K.. 2015. Hrr25/CK1δ-directed release of Ltv1 from pre-40S ribosomes is necessary for ribosome assembly and cell growth. J. Cell Biol. 208:745–759. 10.1083/jcb.201409056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D., and Schiestl R.H.. 2007. Microtiter plate transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2:5–8. 10.1038/nprot.2007.16 [DOI] [PubMed] [Google Scholar]
- Grant B.D., and Donaldson J.G.. 2009. Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 10:597–608. 10.1038/nrm2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema E.H., Lewis M.J., Black M.W., and Pelham H.R.. 2003. Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 22:548–557. 10.1093/emboj/cdg062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W., and Lev S.. 2014. Tethering the assembly of SNARE complexes. Trends Cell Biol. 24:35–43. 10.1016/j.tcb.2013.09.006 [DOI] [PubMed] [Google Scholar]
- Howard J.P., Hutton J.L., Olson J.M., and Payne G.S.. 2002. Sla1p serves as the targeting signal recognition factor for NPFX(1,2)D-mediated endocytosis. J. Cell Biol. 157:315–326. 10.1083/jcb.200110027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu V.W., Bai M., and Li J.. 2012. Getting active: Protein sorting in endocytic recycling. Nat. Rev. Mol. Cell Biol. 13:323–328. [DOI] [PubMed] [Google Scholar]
- Hua Z., Fatheddin P., and Graham T.R.. 2002. An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system. Mol. Biol. Cell. 13:3162–3177. 10.1091/mbc.E02-03-0172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber L.A., and Teis D.. 2016. Lysosomal signaling in control of degradation pathways. Curr. Opin. Cell Biol. 39:8–14. 10.1016/j.ceb.2016.01.006 [DOI] [PubMed] [Google Scholar]
- Huotari J., and Helenius A.. 2011. Endosome maturation. EMBO J. 30:3481–3500. 10.1038/emboj.2011.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh K.K., Kay J.G., Stow J.L., and Grinstein S.. 2007. Fusion, fission, and secretion during phagocytosis. Physiology (Bethesda). 22:366–372. 10.1152/physiol.00028.2007 [DOI] [PubMed] [Google Scholar]
- Iesmantavicius V., Weinert B.T., and Choudhary C.. 2014. Convergence of ubiquitylation and phosphorylation signaling in rapamycin-treated yeast cells. Mol. Cell. Proteomics. 13:1979–1992. 10.1074/mcp.O113.035683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C.B., Ott E.M., Keener J.M., Curtiss M., Sandrin V., and Babst M.. 2012. Regulation of membrane protein degradation by starvation-response pathways. Traffic. 13:468–482. 10.1111/j.1600-0854.2011.01314.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kira S., Kumano Y., Ukai H., Takeda E., Matsuura A., and Noda T.. 2016. Dynamic relocation of the TORC1-Gtr1/2-Ego1/2/3 complex is regulated by Gtr1 and Gtr2. Mol. Biol. Cell. 27:382–396. 10.1091/mbc.E15-07-0470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima A., Toshima J.Y., Kanno C., Kawata C., and Toshima J.. 2012. Localization and functional requirement of yeast Na+/H+ exchanger, Nhx1p, in the endocytic and protein recycling pathway. Biochim. Biophys. Acta. 1823:534–543. 10.1016/j.bbamcr.2011.12.004 [DOI] [PubMed] [Google Scholar]
- Kondapalli K.C., Llongueras J.P., Capilla-González V., Prasad H., Hack A., Smith C., Guerrero-Cázares H., Quiñones-Hinojosa A., and Rao R.. 2015. A leak pathway for luminal protons in endosomes drives oncogenic signalling in glioblastoma. Nat. Commun. 6:6289 10.1038/ncomms7289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchler K., Sterne R.E., and Thorner J.. 1989. Saccharomyces cerevisiae STE6 gene product: A novel pathway for protein export in eukaryotic cells. EMBO J. 8:3973–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang M.J., Martinez-Marquez J.Y., Prosser D.C., Ganser L.R., Buelto D., Wendland B., and Duncan M.C.. 2014. Glucose starvation inhibits autophagy via vacuolar hydrolysis and induces plasma membrane internalization by down-regulating recycling. J. Biol. Chem. 289:16736–16747. 10.1074/jbc.M113.525782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M.J., Nichols B.J., Prescianotto-Baschong C., Riezman H., and Pelham H.R.. 2000. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol. Biol. Cell. 11:23–38. 10.1091/mbc.11.1.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Hua Z., Nepute J.A., and Graham T.R.. 2007. Yeast P4-ATPases Drs2p and Dnf1p are essential cargos of the NPFXD/Sla1p endocytic pathway. Mol. Biol. Cell. 18:487–500. 10.1091/mbc.E06-07-0592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald C., and Piper R.C.. 2015. Puromycin- and methotrexate-resistance cassettes and optimized Cre-recombinase expression plasmids for use in yeast. Yeast. 32:423–438. 10.1002/yea.3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald C., and Piper R.C.. 2016. Cell surface recycling in yeast: Mechanisms and machineries. Biochem. Soc. Trans. 44:474–478. 10.1042/BST20150263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald C., Payne J.A., Aboian M., Smith W., Katzmann D.J., and Piper R.C.. 2015. A family of tetraspans organizes cargo for sorting into multivesicular bodies. Dev. Cell. 33:328–342. 10.1016/j.devcel.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGurn J.A., Hsu P.-C., Smolka M.B., and Emr S.D.. 2011. TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell. 147:1104–1117. 10.1016/j.cell.2011.09.054 [DOI] [PubMed] [Google Scholar]
- Martín Y., González Y.V., Cabrera E., Rodríguez C., and Siverio J.M.. 2011. Npr1 Ser/Thr protein kinase links nitrogen source quality and carbon availability with the yeast nitrate transporter (Ynt1) levels. J. Biol. Chem. 286:27225–27235. 10.1074/jbc.M111.265116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield F.R., and McGraw T.E.. 2004. Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5:121–132. 10.1038/nrm1315 [DOI] [PubMed] [Google Scholar]
- McCullough J., Clippinger A.K., Talledge N., Skowyra M.L., Saunders M.G., Naismith T.V., Colf L.A., Afonine P., Arthur C., Sundquist W.I., et al. . 2015. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science. 350:1548–1551. 10.1126/science.aad8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merhi A., and André B.. 2012. Internal amino acids promote Gap1 permease ubiquitylation via TORC1/Npr1/14-3-3-dependent control of the Bul arrestin-like adaptors. Mol. Cell. Biol. 32:4510–4522. 10.1128/MCB.00463-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merwin J.R., Bogar L.B., Poggi S.B., Fitch R.M., Johnson A.W., and Lycan D.E.. 2014. Genetic analysis of the ribosome biogenesis factor Ltv1 of Saccharomyces cerevisiae. Genetics. 198:1071–1085. 10.1534/genetics.114.168294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mioka T., Fujimura-Kamada K., and Tanaka K.. 2014. Asymmetric distribution of phosphatidylserine is generated in the absence of phospholipid flippases in Saccharomyces cerevisiae. MicrobiologyOpen. 3:803–821. 10.1002/mbo3.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Schmidt O., Angelova M., Faserl K., Weys S., Kremser L., Pfaffenwimmer T., Dalik T., Kraft C., Trajanoski Z., et al. . 2015. The coordinated action of the MVB pathway and autophagy ensures cell survival during starvation. eLife. 4:e07736 10.7554/eLife.07736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P., Field C., and Schekman R.. 1980. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 21:205–215. 10.1016/0092-8674(80)90128-2 [DOI] [PubMed] [Google Scholar]
- O’Donnell A.F., McCartney R.R., Chandrashekarappa D.G., Zhang B.B., Thorner J., and Schmidt M.C.. 2015. 2-Deoxyglucose impairs Saccharomyces cerevisiae growth by stimulating Snf1-regulated and α-arrestin-mediated trafficking of hexose transporters 1 and 3. Mol. Cell. Biol. 35:939–955. 10.1128/MCB.01183-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgaki R., Matsushita M., Kanazawa H., Ogihara S., Hoekstra D., and van Ijzendoorn S.C.D.. 2010. The Na+/H+ exchanger NHE6 in the endosomal recycling system is involved in the development of apical bile canalicular surface domains in HepG2 cells. Mol. Biol. Cell. 21:1293–1304. 10.1091/mbc.E09-09-0767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchaud N., Péli-Gulli M.-P., and De Virgilio C.. 2013a Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci. Signal. 6:ra42–ra42. 10.1126/scisignal.2004112 [DOI] [PubMed] [Google Scholar]
- Panchaud N., Péli-Gulli M.-P., and De Virgilio C.. 2013b SEACing the GAP that nEGOCiates TORC1 activation: Evolutionary conservation of Rag GTPase regulation. Cell Cycle. 12:2948–2952. 10.4161/cc.26000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péli-Gulli M.-P., Sardu A., Panchaud N., Raucci S., and De Virgilio C.. 2015. Amino acids stimulate TORC1 through Lst4-Lst7, a GTPase-activating protein complex for the Rag family GTPase Gtr2. Cell Reports. 13:1–7. 10.1016/j.celrep.2015.08.059 [DOI] [PubMed] [Google Scholar]
- Pfannmüller A., Wagner D., Sieber C., Schönig B., Boeckstaens M., Marini A.M., and Tudzynski B.. 2015. The general amino acid permease FfGap1 of Fusarium fujikuroi is sorted to the vacuole in a nitrogen-dependent, but Npr1 kinase-independent manner. PLoS One. 10:e0125487 10.1371/journal.pone.0125487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper R.C., Dikic I., and Lukacs G.L.. 2014. Ubiquitin-dependent sorting in endocytosis. Cold Spring Harb. Perspect. Biol. 6:a016808 10.1101/cshperspect.a016808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powis K., and De Virgilio C.. 2016. Conserved regulators of Rag GTPases orchestrate amino acid-dependent TORC1 signaling. Cell Discov. 2:15049 10.1038/celldisc.2015.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powis K., Zhang T., Panchaud N., Wang R., De Virgilio C., and Ding J.. 2015. Crystal structure of the Ego1-Ego2-Ego3 complex and its role in promoting Rag GTPase-dependent TORC1 signaling. Cell Res. 25:1043–1059. 10.1038/cr.2015.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M., Poon P.P., Schindler C., Murray L.E., Kama R., Gabriely G., Singer R.A., Spang A., Johnston G.C., and Gerst J.E.. 2006. The Gcs1 Arf-GAP mediates Snc1,2 v-SNARE retrieval to the Golgi in yeast. Mol. Biol. Cell. 17:1845–1858. 10.1091/mbc.E05-09-0832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A.F., and Davis N.G.. 1996. Ubiquitination of the yeast a-factor receptor. J. Cell Biol. 134:661–674. 10.1083/jcb.134.3.661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rue S.M., Mattei S., Saksena S., and Emr S.D.. 2008. Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol. Biol. Cell. 19:475–484. 10.1091/mbc.E07-07-0694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer T., Maco B., Petfalski E., Tollervey D., Böttcher B., Aebi U., and Hurt E.. 2006. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 441:651–655. 10.1038/nature04840 [DOI] [PubMed] [Google Scholar]
- Schmidt A., Beck T., Koller A., Kunz J., and Hall M.N.. 1998. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 17:6924–6931. 10.1093/emboj/17.23.6924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.N., McCaffery J.M., and Emr S.D.. 1998. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 142:665–681. 10.1083/jcb.142.3.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian T.T., Baldridge R.D., Xu P., and Graham T.R.. 2012. Phospholipid flippases: Building asymmetric membranes and transport vesicles. Biochim. Biophys. Acta. 1821:1068–1077. 10.1016/j.bbalip.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiser R.M., Sundberg A.E., Wollam B.J., Zobel-Thropp P., Baldwin K., Spector M.D., and Lycan D.E.. 2006. Ltv1 is required for efficient nuclear export of the ribosomal small subunit in Saccharomyces cerevisiae. Genetics. 174:679–691. 10.1534/genetics.106.062117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff D.R., Daro E.A., Hull M., and Mellman I.. 1999. The receptor recycling pathway contains two distinct populations of early endosomes with different sorting functions. J. Cell Biol. 145:123–139. 10.1083/jcb.145.1.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova A., Hanono A., Drosner S., Curtiss M., Davies B.A., Katzmann D.J., and Babst M.. 2010. Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol. Biol. Cell. 21:1059–1071. 10.1091/mbc.E09-07-0572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Stefan C.J., Rue S.M., Teis D., and Emr S.D.. 2011. Two novel WD40 domain-containing proteins, Ere1 and Ere2, function in the retromer-mediated endosomal recycling pathway. Mol. Biol. Cell. 22:4093–4107. 10.1091/mbc.E11-05-0440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider M.D., and Rogers O.C.. 1985. Intracellular movement of cell surface receptors after endocytosis: Resialylation of asialo-transferrin receptor in human erythroleukemia cells. J. Cell Biol. 100:826–834. 10.1083/jcb.100.3.826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer D.K., and Piper R.C.. 2011. A single ubiquitin is sufficient for cargo protein entry into MVBs in the absence of ESCRT ubiquitination. J. Cell Biol. 192:229–242. 10.1083/jcb.201008121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Fujimura-Kamada K., and Yamamoto T.. 2011. Functions of phospholipid flippases. J. Biochem. 149:131–143. 10.1093/jb/mvq140 [DOI] [PubMed] [Google Scholar]
- Tanner L.I., and Lienhard G.E.. 1987. Insulin elicits a redistribution of transferrin receptors in 3T3-L1 adipocytes through an increase in the rate constant for receptor externalization. J. Biol. Chem. 262:8975–8980. [PubMed] [Google Scholar]
- Urbanowski J.L., and Piper R.C.. 1999. The iron transporter Fth1p forms a complex with the Fet5 iron oxidase and resides on the vacuolar membrane. J. Biol. Chem. 274:38061–38070. 10.1074/jbc.274.53.38061 [DOI] [PubMed] [Google Scholar]
- Valbuena N., Guan K.-L., and Moreno S.. 2012. The Vam6 and Gtr1-Gtr2 pathway activates TORC1 in response to amino acids in fission yeast. J. Cell Sci. 125:1920–1928. 10.1242/jcs.094219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr A., Avaro S., Prescianotto-Baschong C., Haguenauer-Tsapis R., and Riezman H.. 2000. The F-box protein Rcy1p is involved in endocytic membrane traffic and recycling out of an early endosome in Saccharomyces cerevisiae. J. Cell Biol. 149:397–410. 10.1083/jcb.149.2.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr A., Meier K.D., and Riezman H.. 2001. Identification and characterization of Saccharomyces cerevisiae mutants defective in fluid-phase endocytosis. Yeast. 18:759–773. 10.1002/yea.726 [DOI] [PubMed] [Google Scholar]
- Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., et al. . 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 285:901–906. 10.1126/science.285.5429.901 [DOI] [PubMed] [Google Scholar]
- Xu P., Baldridge R.D., Chi R.J., Burd C.G., and Graham T.R.. 2013. Phosphatidylserine flipping enhances membrane curvature and negative charge required for vesicular transport. J. Cell Biol. 202:875–886. 10.1083/jcb.201305094 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.