Abstract
Colon cancer is a multifactorial disease associated with a variety of lifestyle factors. Alterations in the gut microbiota and the intestinal metabolome are noted during colon carcinogenesis, implicating them as critical contributors or results of the disease process. Diet is a known determinant of health, and as a modifier of the gut microbiota and its metabolism, a critical element in maintenance of intestinal health. This review summarizes recent evidence demonstrating the role and responses of the intestinal microbiota during colon tumorigenesis and the ability of dietary bioactive compounds and probiotics to impact colon health from the intestinal lumen to the epithelium and systemically. We first describe changes to the intestinal microbiome, metabolome, and epithelium associated with colon carcinogenesis. This is followed by a discussion of recent evidence indicating how specific classes of dietary bioactives, prebiotics, or probiotics affect colon carcinogenesis. Lastly, we briefly address the prospects of using multiple ‘omics’ techniques to integrate the effects of diet, host, and microbiota on colon tumorigenesis with the goal of more fully appreciating the interconnectedness of these systems and thus, how these approaches can be used to advance personalized nutrition strategies and nutrition research.
Keywords: Bioactives, colon cancer, microbiome, metabolome
1. Introduction
Colon cancer is associated with various risk factors including abdominal obesity, chronic inflammation, and mutagen exposure [1–5]. The classically-defined hallmarks of cancer are: 1) self-sufficiency in growth signals, 2) insensitivity to growth-inhibitory signals, 3) evasion of apoptosis, 4) limitless replicative potential, 5) sustained angiogenesis, and 6) tissue invasion and metastasis [6, 7]. More recently, two emerging hallmarks of cancer have been proposed: 1) deregulated cellular energetics, and 2) avoidance of immune destruction [8]. Indeed, cancer cells are known to exhibit altered metabolism (i.e., the Warburg effect) [9–11]. In addition, tumor-promoting inflammation, and genome instability and mutation have been proposed as cancer enabling characteristics [8]. Alterations in the epigenome, such as global hypomethylation and site-specific hypermethylation of tumor suppressor genes, regulate gene expression patterns and have been associated with colon carcinogenesis [12].
The fundamental model of sporadic cancer development involves three steps: initiation, promotion, and progression [13]. Initiation refers to an irreversible mutation event in the DNA sequence following exposure to mutagens, such as ionizing radiation, aromatic amines, polycyclic hydrocarbons, and oxygen free radicals [13, 14]. Promotion is characterized by alterations in cell apoptosis and proliferation that occur intrinsically in response to initiation (e.g., overexpression of mutant p53) and/or to promoters that drive proliferation and/or inhibit apoptosis of initiated cells [13]. Progression is characterized by structural changes in cell karyotype, further accumulation of genetic mutations, anaplasia, development of malignant neoplasms, as well as tumor invasion and metastatic growth [13, 15]. Interestingly, cells within the primary tumor are known to harbor heterogeneous mutation profiles making targeted treatment strategies more difficult [15]. Therefore, significant efforts are made to inhibit carcinogenesis at the initiation and promotion stages.
Colon epithelial cells exist in a mutualistic relationship with bacteria residing in the lumen. The gut microbiota is estimated to contain over 1,000 different ‘species-level’ phylotypes [16]. One recent metagenomics study reported a microbial gene catalog of 3.3 million genes, which is ~150 times larger than the human gene complement [16–18]. The microbial metagenome permits a repertoire of metabolic functions that supplement host metabolism, and the gut microbiota has been implicated in a number of physiological processes, including modulation of intestinal motility, regulation of luminal pH, stimulation of immune function, and metabolism of undigested food [19]. Dysbiosis refers to perturbations in microbial populations, and has been linked to obesity, cancer, and promotion of colonic inflammation (e.g., inflammatory bowel disease (IBD) and colitis) [2, 20–25]. Inflammation can occur in response to a variety of biological insults, including infection by pathogens, immune sensitivity, autoimmunity, and exposure to ionizing radiation, and has been implicated in both sporadic colorectal cancer (CRC) and colitis-associated colon cancer (CAC) [2, 5, 26–29].
Diet is a major contributor to human health and metabolism, and research has demonstrated that diet also contributes to the composition and metabolism of the gut microbiota [30, 31]. Epidemiological studies have revealed that consumption of fruits, vegetables, and whole grains reduces CRC risk [32–34]. The chemoprotective activity afforded by fruit, vegetable, and whole grain consumption is believed to be due to the fiber content, antioxidant compounds, and other bioactive compounds (i.e., extra-nutritional elements of food that can affect living tissue) [32, 33, 35]. Diets containing bioactive compounds that directly inhibit proliferation and promote apoptosis of transformed cells are promising strategies for the prevention of colon cancer. Additionally, certain food constituents can undergo metabolism by the microbiota to produce secondary bioactive compounds that then affect host physiology, such as the fermentation of dietary fiber into short-chain fatty acids (SCFA) [36–40]. In this review, we give an overview of the current understanding of dietary bioactives in modulating the molecular, microbial, and metabolome changes that occur during colon carcinogenesis.
2. Host and microbe-associated alterations as contributors to colon carcinogenesis
2.1 Epithelial perturbations and their outcomes
Colonocytes serve several important functions including nutrient absorption, mucin secretion, and endocrine activity. The differentiated cell types performing these functions are derived from stem cell precursors that reside at the base of the intestinal crypt [41]. To date, there has been extensive debate regarding the identity of stem cells within the colon crypt. However, the leucine-rich-repeat-containing G-protein-coupled receptor 5 positive (Lgr5+) crypt base columnar cells are a strong candidate, as these cells undergo cell division daily and their daughter cells constitute the transit-amplifying crypt compartment [41]. Furthermore, lineage tracing studies using an inducible Cre knock-in allele and the Rosa26-lacZ reporter strain have demonstrated restricted expression of Lgr5+ cells to the base of the crypt that can generate all epithelial lineages over a 60-day period [41, 42]. Daughter cells derived from the stem cell population will continue to divide and eventually differentiate into mature colonocytes as they migrate away from the base of the intestinal crypt toward the intestinal lumen. Mature, senescent cells, which are lost at the luminal surface via sloughing or spontaneous apoptosis, are normally replaced at an equal rate by proliferating cells [43]. Coordinated proliferation and migration of colonic epithelial cells are necessary for the maintenance of barrier function and epithelial restitution following an injury to the epithelial cells [43, 44].
Adult colon stem cells sustain self-renewal and are target cells for cancer-initiating mutations [42, 45]. Tissue-specific mutation accumulation in colon stem cells resulting in alterations in differentiation/plasticity, and stem cell location/proliferation are generally believed to represent the earliest step towards colon tumorigenesis [46, 47]. The onset of colon tumorigenesis is often driven by mutations in the Wnt signaling pathway [48–50]. Specifically, adenomatous polyposis coli (APC) loss-of-function or β-catenin gain-of-function mutations result in stabilization of free β-catenin, which leads to aberrant Wnt signaling that can drive tumorigenesis [48–50]. Importantly, mutational activation of the Wnt pathway in Lgr5+ cells gives rise to intestinal tumors with greater efficiency than other intestinal cell types [42]. According to the cancer stem cell hypothesis, the population of cells that propagate tumor formation is self-renewing and multipotent [51]. Thus, intestinal stem cells are considered the cell-of-origin of cancer because these cells already exhibit self-renewing capacity and multipotency [41, 42, 52]. In addition to the concept that cancer stem cells are the direct products of neoplastically transformed normal adult stem cells, there is also evidence that transit amplifying cells with mutant genomes can dedifferentiate and enter the stem cell state [53]. Adding to the complexity, recent evidence indicates that multiple stem cell hierarchies exist in the intestine and that plasticity within stem cell hierarchies mediates cellular fate in response to extrinsic factors such as nutrition, inflammation, and physical stress signals [54–58]. Interestingly, a role for diet in the regulation of intestinal stem cells has been reported [59–62].
Although the expansion of initiated cells is required for tumor formation, previous work has shown cell proliferation to be a poorer predictor of tumor development than markers of differentiation and apoptosis [63]. Furthermore, tissues with relatively higher rates of proliferation do not appear to exhibit a higher incidence of spontaneous tumor development [64]. Apoptosis is a key regulator of physiological development and tissue homeostasis [65]. To date, two major mechanisms of apoptosis induction in mammalian cells have been elucidated in detail. The extrinsic (i.e., death receptor) and intrinsic (i.e., mitochondrial) apoptosis pathways involve caspase activation cascades leading to controlled cell death [65]. The extrinsic apoptosis pathway requires activation of membrane receptors of the tumor necrosis factor (TNF) receptor superfamily such as CD95 (APO-1/Fas) or TNF-related apoptosis-inducing ligand (TRAIL) receptors [65, 66]. Activation of the intrinsic pathway, which is often dysregulated in colon cancer, involves mitochondrial outer membrane permeabilization (MOMP) and subsequent release of cytochrome c and other proteins from the mitochondria, triggering caspase-3 activation through the formation of an apoptosome complex [67, 68]. B-cell lymphoma-2 (Bcl-2) family proteins, among other factors, regulate the intrinsic mitochondrial apoptosis pathway and appear to dictate the sensitivity or resistance of the cell to apoptosis induction [68–70]. Activation of the intrinsic pathway in response to DNA damage provides a mechanism whereby DNA damaged cells can be removed from the tissue, thus preventing the propagation of transformed cells or retention of mutated progenitor cells [68]. Inhibition of apoptosis is an important mechanism of tumor promotion that can arise from genetic and epigenetic alterations (e.g., p53 loss-of-function mutations or bcl-2 promoter hypomethylation, respectively) and exposure to chemical promoters [71–74]. Additional evidence of epigenetic alterations contributing to colon carcinogenesis is the hypermethylation of the DNA repair gene, human MutL homolog 1 (hMLH1), which has been linked to microsatellite instability (MSI) associated colon carcinogenesis [75]. Therefore, dietary bioactives targeting the colonocyte epigenome are promising strategies for reprogramming aberrant processes associated with colon cancer, such as inhibition of apoptosis in transformed cells [13, 72, 76].
The unique molecular and genetic features observed in normal and diseased tissues of the proximal and distal colon reveal differing susceptibilities and mechanisms for disease initiation. One study examining gene expression profiles carried out on healthy adult human biopsy samples revealed more than 1000 differentially expressed genes between the ascending and descending colon [77]. The Cancer Genome Anatomy Project (CGAP) functional grouping of the differentially expressed genes revealed major pathways implicated in colon cancer including epidermal growth factor (EGF), transforming growth factor beta (TGF-β), Wnt, Ras, insulin, and integrin signaling [77]. Differences in gene expression between the proximal and distal colon may result from the observed differential DNA methylation patterns in colonocytes [78]. Studies comparing cancers of the proximal and distal colon have revealed numerous differences in molecular and clinicopathological features, which reflect different susceptibilities to neoplastic transformation, such as the frequency of V-Ki-Ras2 Kirsten Rat Sarcoma 2 Viral Oncogene Homolog (KRAS) and B-Raf Proto-Oncogene, Serine/Threonine Kinase (BRAF) mutations, and the prevalence of MSI [79–81]. In response to dietary fiber and carcinogen treatment, differences in colonocyte proliferation and differentiation between the proximal and distal rat colon have been observed [82]. Future research should take into consideration the unique characteristics of these sites during experimental design.
As mentioned previously, inflammation is a known promoter of sporadic colon cancer and CAC. Although the cause of onset remains unknown, CAC is characterized by elevated reactive oxygen species (ROS), increases in pro-inflammatory cytokines and elevated expression of transcription factors involved in inflammation signaling [2]. This chronic inflammation can lead to genome instability and mutations in critical regulatory genes, such as the tumor suppressor gene, p53, that can drive carcinogenesis [26, 71, 83]. For example, we have recently demonstrated that loss of p53 function in stem cells enables colonic tumor formation only when combined with DNA damage and chronic inflammation [52]. In sporadic CRC, inflammation is often a result of immune cell migration to the tumor site where activated immune cells produce ROS, reactive nitrogen intermediates, and pro-inflammatory cytokines, which can further damage DNA and drive cell proliferation [2, 29, 84]. Additionally, there is evidence linking inflammation to carcinogenesis via epigenetic modifications at critical regulatory gene loci [85–88]. It is apparent that inflammation can impact all stages of colon cancer development, and nutritional strategies aimed at modulating inflammation and ROS production may help mitigate disease risk.
2.2 Changes to microbiome/metabolome and their role in tumorigenesis
Currently, investigations into the physiological mechanisms by which the microbiota can influence colon cancer risk are limited due to the diversity and complexity of the microbiota. Despite the ongoing discussion concerning whether changes in the microbiota occur prior to or as a result of colon carcinogenesis, some significant observations have been made that suggest a causal role of the microbiota in the disease process. For example, studies employing rodent models of spontaneous, chemically-induced (e.g., azoxymethane, AOM), or genetically predisposed (e.g., APCmin/+) colon cancer demonstrate enhanced tumorigenesis in conventionally raised animals versus germ-free or antibiotic-treated animals, suggesting a promotive effect of the microbiota on tumor formation [89–95]. Often, the enhanced tumorigenicity in conventionally raised mice is attributed to microbially-sustained levels of chronic low-grade inflammation that acts as a tumor promoter. However, activation of the innate immune system through toll-like receptor (TLR) and NOD-like receptor (NLR) agonists has been shown to enhance anti-tumor activity of the innate immune system [96, 97]. Taken together, these observations suggest a double-edged role of the microbiota in colon carcinogenesis that may be dependent on the degree and mechanism of the innate immune response activation.
Because inflammation is a known driver of CRC, work has been conducted to better understand how the microbiota are altered by, or contribute to, pro-inflammatory states. A recent metagenomics study revealed that the fecal microbiome of patients with IBD has, on average, 25% fewer microbial genes than individuals not suffering from IBD [18]. These findings are in agreement with other studies documenting a reduction in gut microbe diversity in association with IBD [98–100]. An experimental animal model of IBD has shown that increases in colonic injury are negatively associated with Firmicutes, Actinobacteria, Lactobacillales and Lactobacillus [101]. Dysbiosis has also been observed following spaceflight and radiation exposure in rodent models demonstrating the susceptibility of the microbiota to environmental factors other than the host diet [102]. These observations suggest that microbial dysbiosis may act as a driver and/or a consequence of colon disease development.
Fecal profiling experiments of CRC patients and healthy subjects have revealed numerous taxonomic differences in the microbiota and these findings should be useful in directing mechanistic studies. The 454 pyrosequencing of fecal samples from 46 CRC patients and 56 healthy volunteers identified 48 operational taxonomic units (OTUs) associated with the segregation of normal and CRC samples by UniFrac analysis [103]. OTUs belonging to the genera Enterococcus, Escherichia/Shigella, Klebsiella, Streptococcus, and Peptostreptococcus were significantly more abundant in CRC patients. Conversely, OTUs related to the genus Roseburia and other butyrate-producing members of Lachnospiraceae were less abundant in CRC patients. In addition, quantitative polymerase chain reaction (qPCR) analysis of microbial butyrate synthesis genes confirmed lower levels of butyrate-producing bacteria in CRC patients, supporting the identity of butyrate-producing microbes as contributors to intestinal health. These observations were similarly replicated in a more recent study that found nine OTUs represented by the butyrate-producing genera Faecalibacterium and Roseburia to be significantly less abundant in CRC patients versus healthy control subjects [104]. Another study revealed a greater presence of Fusobacterium nucleatum and Enterobacteriaceae in CRC patients versus healthy control subjects [105].
In addition to overall changes in the gut microbiota of CRC patients versus healthy controls, there is evidence demonstrating microbial differences between normal and diseased tissue sites within a given individual. One study characterizing the microbial structure of the intestinal lumen, cancerous tissue, and matched noncancerous normal tissue, revealed numerous differences between these sampling sites [106]. Microbial diversity was significantly lower in tumor tissue than noncancerous tissue, suggesting a more selective microenvironment exists in proximity to diseased tissue. In cancerous tissue, Lactobacillales was enriched, whereas Faecalibacterium was reduced. With respect to mucosa-adherent bacteria, Bifidobacterium, Faecalibacterium, and Blautia were reduced in CRC patients whereas Fusobacterium, Porphyromonas, Peptostreptococcus, and Mogibacterium were increased. Lastly, in the lumen, bacteria associated with metabolic disorders or metabolic exchange with the host, such as Erysipelotrichaceae, Prevotellaceae, and Coriobacteriaceae, were increased in CRC patient samples. In another study, lower abundances of Staphylococcus and Bacillus and higher abundances of Escherichia-Shigella and Prevotella were observed in mucosal samples derived from polyp biopsies compared to adjacent healthy tissues. Altogether, these findings suggest certain bacteria may better compete in the transformed niche and that characterizing these microenvironments could reveal novel mechanisms by which the microbiota influence disease progression. Future studies attempting to define causal relationships between specific microbiota and disease processes would greatly benefit from more precise sampling techniques, such as laser capture microdissection (LCM). Application of site-specific sampling techniques in combination with more sensitive analytical approaches in future experiments will improve the ability to detect diet-induced modifications of host-microbe interactions in colon cancer models.
Metabolomics analysis of CRC patients versus healthy controls is a useful tool for investigating alterations in co-metabolism or metabolic exchange between host cells and the gut microbiota and permits identification of novel CRC metabolite biomarkers. Studies of the fecal metabolome have revealed alterations in SCFA, amino acid (protein), and unsaturated fatty acid metabolism in CRC patients [107]. Another study using serum samples from colorectal adenoma, CRC, or healthy control subjects, revealed pathways for urea, caffeine, and galactose metabolism were related to CRC progression [108]. Similar to microbial observations between normal and diseased tissue, metabolomics analyses have revealed different metabolite signatures between stool, cancers, and healthy adjacent mucosa [109]. In that study, metabolic pathway analysis of significantly different metabolites revealed aberrant SCFA metabolism, fructose, mannose, and galactose metabolism, and glycolytic, gluconeogenic, and pyruvate metabolism in CRC tissue of 17 CRC patients, versus healthy adjacent tissue. Furthermore, whereas nearly all of the metabolites detected in CRC tissue were found in adjacent mucosa, less than 50% of metabolites (213 out of 500 total metabolites) detected in the stool were shared with either tissue site. These findings reinforce the need to consider metabolomic profiles derived from multiple adjacent sampling sites to enhance biomarker discovery and personalized treatment and prevention strategies.
To define a beneficial microbiota composition that is capable of suppressing CRC, it will be important to first understand the mechanisms by which the microbiota can predispose host cells to carcinogenesis. Some bacteria have been shown to directly initiate carcinogenesis via the production of toxins that can damage DNA [110]. For example, colibactin toxin produced by E. coli belonging to the B2 phylogroup causes DNA crosslinks and double strand breaks (DSB) that can lead to mutations [111]. Bacteroides fragilis toxin is known to activate Wnt and NF-κB signaling pathways, increase cell proliferation, enhance epithelial release of pro-inflammatory molecules, and induce DNA damage in vitro [112–114]. More recently, the gene encoding Bacteroides fragilis toxin was found to be more abundant in tumor mucosa samples than control biopsies, especially in late-stage CRC, suggesting that the bacteria capable of synthesizing the toxin play a persistent role in promoting tumor growth through sustained activation of tumor-promoting pathways [115]. The Salmonella protein AvrA has recently been shown to activate the STAT3 pathway in a CAC mouse model (AOM plus dextran sulfate sodium, DSS) [116]. The role of the STAT3 pathway and signaling in colon cancer has been well established [117–120]. Indirect effects of specific bacteria on carcinogenesis have been characterized that are largely dependent on the immune system for manifestation. In addition to inducing DNA modifications, the E. coli strain that produces colibactin has recently been shown to encourage pro-tumoral activities of tumor-associated macrophages by infecting and persisting within the immune cells. As a result, these bacteria induce sustained cyclooxygenase-2 (COX-2) expression by macrophages, which is a hallmark of the inflammation associated with colon cancer [121]. A recently proposed mechanism for bacterial-induced chromosomal instability (CIN) was reported by Wang and Huycke [122] in which macrophage polarization to an M1 phenotype by commensal bacteria leads to a trans-4-hydroxy-2-nonenol (4-HNE) mediated bystander effect in colon epithelial cells. The 4-HNE released by M1 macrophages in response to commensal invasion during periods of intestinal barrier dysfunction causes mutations, DSB, and spindle dysfunction leading to CIN in epithelial cells. Taken together, these experiments demonstrate the potential of direct and indirect mechanisms of gut microbiota for initiating and promoting colon carcinogenesis.
3. Dietary mediators of colon tumorigenesis
3.1 Overview
A recent review points to the importance of many different diet-derived biologically active compounds in the inhibition of carcinogenesis [7]. Dietary compounds may reach the colon because they are: 1) too large to be absorbed in the small intestine, 2) bioavailable compounds that escape deglycosylation and absorption in the small intestine, or 3) aren’t accessible to the host due to their intercalation in the food matrix [39]. Those compounds absorbed in the small intestine, conjugated by the liver, and returned to the intestine via enterohepatic circulation also may reach the large intestine [39]. Additionally, there is research to suggest that processing bioactive-containing foodstuffs can affect the bioaccessibility of bioactives, which may result in some molecules reaching the colon that otherwise might have been absorbed in the small intestine [123, 124].
Dietary bioactives have the potential to directly modify and/or mitigate tumorigenic processes via multiple pathways. These pathways may be microbe-independent (i.e., direct), microbe-dependent (i.e., indirect), or both. The microbe-independent pathway represents a direct action of bioactive compounds on the intestinal epithelium that may be additive, synergistic, or antagonistic. Microbe-dependent pathways include diet-induced modifications in the substrates reaching the colon that alter the total number and/or population characteristics of the colonic microbiota, or changes in the production of microbial metabolites [125]. Some dietary components exert bactericidal functions and are, therefore, direct modifiers of the gut microbiota [126–128]. Similar to the observed differences in physiology and cancer phenotype/genotype between the proximal and distal colon mentioned previously, the composition of the microbiota is also known to differ spatially along the colorectal axis and between mucosal and luminal sites [17, 129, 130]. The observed differences between lumen- and mucosa-associated microbial communities likely reflect the nature of the host-microbe interaction for these sub-populations and importantly, the availability of their preferred substrates.
The most often ascribed function of dietary bioactives, particularly for polyphenolics, is their antioxidant capacity, and studies have estimated that 90–95% of dietary polyphenols escape absorption in the small intestine due to their size [131]. The gut microbiota are capable of a variety of metabolic reactions that affect the structure and functional groupings of bioactives including ring-C cleavage, dihydroxylation, decarboxylation, and demethylation [39]. Indeed, there is substantial evidence that much of the biological response to dietary bioactives is due to microbial derivatives rather than the native compounds [132–135]. A large consortium of authors reviewing the potential of bioactive compounds in cancer inhibition stressed the importance of exposure to these molecules during the early stages of cell transformation [7]. They noted their efficacy is greatest during this time when most of the cellular changes are likely to be epigenetic in nature.
3.2 Fiber and prebiotics
Nondigestible carbohydrates (i.e., dietary fiber) pass through the small intestine into the cecum and large intestine where they undergo metabolism by the colonic microbiota [136]. Many bacteria preferentially metabolize carbohydrates over other energy sources such as protein [137]. Predominantly saccharolytic fermentation occurs in the cecum and proximal colon of humans and rodents, where most SCFA production takes place [138–140]. The metabolic fate of fiber depends largely on the solubility/fermentability of the fiber itself and the microbiota present in the colon during digestion [141]. Readily fermentable fibers such as oat bran, pectin, and guar are easily fermented by the microbiota to produce methane, carbon dioxide, and SCFA, whereas cellulose and wheat bran are poorly fermented [141, 142]. Acetate, propionate, and butyrate are the most abundant SCFA in the colon, and concentrations typically decrease from the proximal to the distal colon [17, 79]. Benefits of consuming both readily fermentable and poorly fermentable fibers have been established, with the latter being due to its hydrophilic bulking activity, which serves to dilute carcinogens, pro-carcinogens, and other potential tumor promoters in the intestinal lumen [143]. Furthermore, stool-bulking agents have also been shown to increase the rate of digesta passage through the intestine, which can both minimize exposure to toxic compounds in the lumen and increase the levels of butyrate in the distal colon [144]. In rats exposed to AOM, consumption of a wheat bran diet significantly lowered tumor incidence compared to animals consuming an oat bran diet [145]. In addition, consumption of the wheat bran diet was associated with lower body weight, more normalized ratios of SCFA in the intestinal lumen, and increased fecal mass and bulk, which suggests increased bulking ability and dilution potential of wheat bran versus the more readily fermentable oat bran diet.
If dietary fiber and resistant carbohydrates are limiting, the gut microbiota in the distal colon will need to rely on proteolytic fermentation to meet their energy needs [137]. Proteolytic fermentation results in SCFA, along with a number of putatively toxic metabolites such as ammonia and sulphur-containing compounds [137]. Dietary protein levels also may influence butyrate utilization as a high protein diet has been found to decrease expression of the proton coupled monocarboxylate transporter 1 (MCT1, a butyrate transporter) in the colon of piglets [146]. In contrast, Liu et al., found no effect of dietary protein on MCT1 expression or luminal butyrate levels in rats [147].
Butyrate is a four carbon SCFA and the preferred energy substrate of colonocytes [148]. In healthy humans, absolute concentrations of butyrate range from 11 to 25 mM in the feces [140]. Bacteria synthesize butyrate from butyryl-coenzyme A (CoA) derived from acetyl-CoA using one of two enzymes, butyrate kinase or butyryl-CoA:acetate-CoA transferase [149]. Butyrate is absorbed by colonocytes via passive diffusion and by active transport with various ion exchange transporters, such as the sodium-coupled monocarboxylate transporter 1 (Slc5a8) and MCT1 (also known as Slc16a1) [140]. Butyrate has been intensively studied for its ability to inhibit inflammation and carcinogenesis, reduce oxidative stress, and promote colonic barrier function [140]. Butyrate has also been shown to affect gene expression via epigenetic modification of chromatin. In cancer cells exhibiting the Warburg effect, butyrate accumulates in the cell and inhibits histone deacetylase (HDAC) activity and stimulates histone acetyltransferase (HAT) activity by acting as an acetyl-CoA donor, thereby regulating genes that enhance apoptosis and reduce cell proliferation [148, 150]. Our lab has demonstrated increased Bcl-2 promoter methylation and levels of apoptosis in rats consuming a fish oil and pectin diet versus animals consuming a diet containing corn oil and cellulose [72]. Additional in vitro experiments demonstrate butyrate’s ability to reduce global and Bcl-2-like protein 11 (BCL2L11) promoter methylation in HCT-116 colon cancer cells [76]. These data support the identity of butyrate as a modifier of the epigenome and promoter of apoptosis in colon cancer cells. Colonic mucosa from slc5a8−/− knockout mice exhibits dramatically increased expression of inducible nitric oxide synthase (iNOS), a marker of inflammation, in addition to genes of the interferon gamma (IFN-γ) and TGF-β signaling pathways compared to wild-type mice [151]. Indeed, Slc5a8 plays a critical role in the ability of butyrate to suppress colon inflammation and cancer [152–154]. Additionally, butyrate was shown to suppress colonic inflammation by two mechanisms: 1) inhibition of STAT1 phosphorylation by IFN-γ in colonic epithelial cells leading to a reduction in iNOS expression, and 2) inhibition of Fas promoter-bound HDAC1 leading to increased T cell expression of Fas and, thus, enhanced sensitivity to apoptosis of activated T cells thereby reducing T cell production of IFN-γ [151]. Butyrate has also been shown to induce apoptosis via a non-mitochondrial, Fas-mediated, extrinsic pathway in colonocytes [155, 156].
Research has also demonstrated the ability of SCFA to affect immune function. Kim et al. recently demonstrated that SCFA impact the metabolism of B lymphocytes by increasing acetyl-CoA levels, oxidative phosphorylation, glycolysis, and fatty acid synthesis, and that these changes in metabolism support the production of antibodies necessary for preventing infection by pathogens [157]. SCFA were also found to regulate gene expression programs needed for plasma B cell differentiation. Importantly, dietary fiber and SCFA both increased intestinal (Immunoglobulin A, IgA) and systemic (IgA and IgG) antibody levels supporting the notion that the gut microbiota, by way of metabolism of dietary fiber, promotes systemic immunity. The gut microbiota and its metabolites (SCFA, particularly butyrate) have also been shown to affect the differentiation of naïve T cells in the gut epithelium [158]. The mechanisms responsible for the differentiation of naïve T cells into Treg cells include direct actions of butyrate on naïve T cells (i.e., histone H3 acetylation at the Foxp3 locus and Gpr109a activation) and indirect actions via changes in epithelial cell cytokine production (e.g., TGF-β).
Taken together, these observations support a pleiotropic role of butyrate on the colon epithelium as a modulator of inflammation and colonocyte physiology. Interestingly, there is some evidence that butyrate may promote colon tumorigenesis [159–161] or that some dietary interventions reduced colon carcinogenesis in a manner not related to fecal butyrate levels [145]. These findings suggest that microbial metabolites such as short chain fatty acids contribute to colon cancer risk in a complex, context-dependent manner. Additional work needs to be conducted to better understand how gut microbial metabolites impact gut health. Specifically, understanding how other dietary components may interact with microbial metabolite production and their effects on host tissue should be explored if we are to identify dietary patterns capable of suppressing colon carcinogenesis.
3.3 Lipids
The n-3 polyunsaturated fatty acids (PUFAs), eicosapentaenoic (EPA; 20:5n-3) and docosahexaenoic acid (DHA; 22:6n-3) have been extensively studied for their bioactive and chemoprotective properties in the colon, which include immunomodulatory functions, regulation of apoptosis, and epigenome modification [70, 76, 162–171]. For example, rats consuming fish oil and exposed to AOM have significantly fewer O6-methylguanine DNA adducts during the initiation period (6–12 hours post-injection) than corn oil fed rats [169]. In another study, rats exposed to AOM and consuming dietary fish oil had a lower adenocarcinoma incidence and increased levels of apoptosis and cellular differentiation compared to corn oil rats [172]. Recent work has demonstrated that n-3 PUFA in combination with dietary fibers that result in high colon butyrate concentrations (e.g., pectin), or with direct butyrate administration, beneficially alters whole genome and gene-specific DNA methylation and histone acetylation. As mentioned previously our lab has demonstrated in vivo that dietary fish oil plus pectin increases bcl-2 promoter methylation and apoptosis in carcinogen-induced colon tumors, compared to animals consuming corn oil plus cellulose [72]. Further investigation using HCT-116 colon cancer cells has demonstrated DHA and butyrate enhance apoptosis induction in part by demethylation of proapoptotic gene promoters [76]. In addition to the effects of n-3 PUFA and butyrate on the epigenetic state of cells, it has been demonstrated that DHA in combination with butyrate enhances mitochondrial lipid oxidation and reduces mitochondrial membrane potential, which contributes to the induction of apoptosis [70]. Thus, n-3 PUFA and butyrate appear to protect against colon carcinogenesis by sensitizing cells to apoptosis through epigenetic-dependent and independent mechanisms. More recent work has demonstrated that fish oil is especially effective in inducing apoptosis in colon adult stem cells exhibiting DNA damage [173]. Although the clinical relevance of these cogent preclinical data remains to be determined, a large landmark cohort study (n = 96,354) of Seventh Day Adventists recently demonstrated that the risk of CRC was reduced by 22% among all vegetarians combined compared to non-vegetarians, but protection was greatest among pescovegetarians who consume high amounts of both fiber and n-3 PUFA-containing fish [174].
3.4 Phenolic bioactives and other small bioactive compounds
Most fruits and vegetables contain bioactive compounds. The phytochemical profile can vary between different varieties of the same food and can be influenced by how the foods are processed [175–177]. The concentration of phytochemicals also varies throughout the fruit (e.g. seed, peel, pulp) and is typically highest in the fruit peel where the compounds function as antioxidants to prevent damage from ultraviolet radiation [178, 179]. Many phytochemicals are known to have anti-microbial and anti-inflammatory properties [180–182]. The mechanisms by which the compounds exert these effects in vivo and to what extent they are absorbed and metabolized by the host and gut microbiota remains to be fully characterized. For example, polyphenols are a structural class of phytochemicals with multiple phenolic units found naturally in fruits, vegetables, and whole grains [183–186]. Numerous studies have revealed chemoprotective effects of polyphenolic compounds in vivo and in vitro [187–190]. Additionally, there is evidence that polyphenols may exert their chemoprotective effects through epigenetic mechanisms [191–193]. The bioavailability of polyphenols varies depending on the individual compound; however, it has been shown that human colonic bacteria can metabolize polyphenols into lower-molecular-weight bioactive compounds, which may be more bioavailable to the host [133, 190, 194, 195]. Further, consumption of dietary polyphenols is known to alter the composition of the gut microbiota, which can affect host health [196–198]. Understanding how foods containing dietary polyphenols can shape the microbiota composition and metabolism will be important for defining bioactive profiles. Likewise, characterizing the metabolites generated from microbial metabolism of polyphenols, and their presence in the intestinal lumen and systemic circulation will be important for future studies seeking to elucidate biomolecular mechanisms.
Because a whole food (e.g., fruit or vegetable) and combinations of foods contain a mixture of bioactive compounds in combination with the micro and macronutrients of the food, it will be important to characterize how compounds synergize in vivo. For example, combining the dietary polyphenol curcumin with fish oil, rich in n-3 polyunsaturated fatty acids, was recently shown to reduce nuclear β-catenin in aberrant crypt foci in mice exposed to AOM [62]. Furthermore, this combination of dietary bioactives was shown to synergistically increase targeted apoptosis in Lgr5+ stem cells. Mechanisms associated with the anti-cancer effects of n-3 PUFA and curcumin involve regulation of apoptotic proteins (e.g., bcl-2) and priming or activation of the intrinsic mitochondrial-dependent apoptosis pathway [73, 167, 168, 199]. Within a whole food, such as prunes (dried plums), there are a variety of bioactive compounds including phenolic acids (e.g., quercetin, chlorogenic acid, and neochlorogenic acid), sugar alcohols (e.g., sorbitol and mannitol), and hydroxycinnamic acids (e.g., sinapinic acid), as well as a mixture of soluble and insoluble fiber [200, 201]. Prune consumption is commonly associated with a reduction in constipation and improved intestinal motility [200]. These effects can be attributed in part to the dietary fiber found in prunes; however, they are also noted when consuming prune juice, which lacks the insoluble fiber found in the whole fruit. Hence, not all of the bioactive properties ascribed to prune consumption are due to its fiber content. Our lab has shown that quercetin and chlorogenic acid influence fecal SCFA concentrations, suggesting alterations in the gut microbiota or in the absorption of SCFA by the host [202]. Furthermore, our lab has demonstrated the ability of quercetin to reduce the numbers of high multiplicity aberrant crypt foci (HMACF), which are thought to be most indicative of eventual tumor formation [203, 204]. A reduction in proliferation and an increase in apoptosis were also observed in rats fed quercetin, and these effects are thought to be related to observed reductions in pro-inflammatory mediators, cyclooxygenase 1 and 2 (COX-1 and COX-2) [204], as well as through impacts on other signaling pathways including NF-κB, PI3K/Akt/mTOR, and JNK/JUN [204–207]. Quercetin has also been shown to reduce the incidence, multiplicity, and size of colorectal tumors in male F344 rats treated with AOM, compared to control diet animals [208]. Sorbitol is a sugar alcohol found in high concentrations in dried plums and contributes to the laxative effect associated with prune consumption [209]. Sorbitol has been shown to induce apoptosis in HCT-116 cancer cells via activation of the p38 mitogen-activated protein kinase (MAPK) pathway, which activates the mitochondrial death cascade [210]. Additionally, in rats consuming a dried plum powder, analysis of the microbial metagenome revealed a reduction in the relative abundance of microbial genes responsible for the synthesis of secondary bile acids, which are known to induce DNA damage and promote colon cancer [211–213]. Recent evidence documents the critical nature of farnesoid X receptor mediated bile acid metabolism to colon inflammation and carcinogenesis [214].
Another major source of dietary phytochemicals is whole grains [184, 215]. For example, sorghum is an ancient grain predominantly grown in dry, arid zones. Sorghum varieties contain different levels and types of phenolic and polyphenolic compounds [184]. Sumac sorghum contains high levels of condensed tannins (i.e., polymerized flavonols), which contribute to its high phenolic content and oxygen radical absorbance capacity [184]. Indeed, sumac sorghum has been characterized as having greater antioxidant capacity than many other grains, blueberries, and pomegranates [216, 217]. Decortication of the sorghum grain to produce sorghum bran increases the concentration (3–6 times higher than the whole grain) of the phenolic compounds [217]. Our lab has previously shown that polyphenol-rich sorghum brans alter the microbial composition and beneficially affect microbial diversity and richness in a rat model of colitis [101]. In addition, we have shown that less soluble fibers with lower fermentability, such as sorghum bran and cellulose, are more protective against colonic injury during bouts of colitis than highly fermentable fibers, such as pectin [218]. Although these findings are in contrast to the previously discussed protective effects of soluble fibers during carcinogenesis, it is evident that fibers of varying fermentability/solubility exert their effects on the colonic mucosa in a disease-dependent context that includes dysbiosis. Further, our lab has demonstrated that diets containing sumac bran alter fecal SCFA levels, suggesting these polyphenol-rich fiber sources may alter microbial metabolism or host SCFA absorption [219].
As previously mentioned, it will be important to characterize the mechanisms by which dietary bioactives exert their chemoprotective activities in order to better understand how specific phytochemicals can prevent or inhibit specific diseases. This includes identifying and characterizing receptors for native phytochemicals and their microbial metabolites. For example, the aryl hydrocarbon receptor (AhR) is a ligand-activated transcription factor that is widely expressed in various cell types [220]. Several studies have demonstrated the important role of AhR and ligands for this receptor in mediating gastrointestinal function [221–223]. For example, there are reports showing that aromatic hydrocarbons such as 2,3,7,8-tetrachloro-dibenzo-p-dioxin (TCDD; an AhR agonist) decrease inflammation associated with Crohn’s disease and other AhR agonists protect against IBD [222]. It has also been reported that AhR silencing or lack of AhR ligands compromises the maintenance of intraepithelial lymphocytes in the skin and intestine [224].
AhR also binds other ligands unrelated to TCDD, including dietary botanical-derived compounds such as flavonoids, indole-3-carbinol (I3C) and diindolylmethane (DIM), and several tryptophan metabolites including formylindolo-[3,2-b]-carbazole (FICZ) [225–227]. Dietary tryptophan can be metabolized by the gut microbiota into indole-3-acetate, indole-3-aldehyde, indole, and tryptamine, which are selective AhR modulators [134]. We recently demonstrated that four compounds produced from tryptophan by the intestinal microbiota – indole, indole-3-acetate, indole-3-aldehyde, and tryptamine – are ligands for the AhR [228]. Consistent with our findings, Zelante et al. recently reported that indole-3-aldehyde is also a microbiota-derived AhR ligand that contributes to a microbiota-AhR signaling axis in the intestine [229]. Thus, AhR agonists/antagonists produced by the intestinal microbiota may have a profound effect on gastrointestinal biology. In support of this rationale, it has been demonstrated that APCMin/+ mice that develop intestinal tumors are remarkably impacted by AhR. The loss of AhR by crossing APCMin/+ with AhR−/− mice enhances intestinal tumor formation and treatment of APCMin/+ mice with AhR ligands [e.g., cruciferous vegetable-derived I3C and DIM] inhibits tumor formation [221]. Similar results indicating that AhR is a repressor of inflammation-associated colon cancer have also been recently described [230]. Collectively, these findings clearly demonstrate that the AhR and its dietary and microbial-derived ligands have a profound impact on gastrointestinal homeostasis and tumorigenesis.
3.5 Probiotics and synbiotics
The potential role of probiotics, “live microorganisms which, when administered in adequate amounts, confer a health benefit on the host” [231], in CRC and CRC prevention has been reviewed across decades of scientific research. Early reports emphasized an indirect role for probiotics in colon cancer prevention and treatment of cancer therapy-induced diarrhea with a number of publications reporting either beneficial [232, 233] or detrimental [234] effects. On the role of probiotics in cancer prevention, studies showed that selected strains of the Lactic Acid Bacteria (LAB) group were capable of binding to mutagenic amines generated by cooking a protein-rich food [235], degrade nitrosamines [236], reduce specific activities of bacterial β-glucuronidases, nitroreductases and azoreductases (enzymes potentially involved in transforming pro-carcinogens into active carcinogens [237]), and inhibit 2-amino-3-methylimidazol[4,5-f]quinoline (IQ)-induced incidence of colon tumors in rats experiments [238]. Hence, there is a general agreement that individual probiotic strains can beneficially affect metabolic activities that occur in the gastrointestinal tract and enhance the host’s immune response. Newer research has implicated a more complex role for probiotics in CRC considering novel evidence showing that composition of the gut microbiome could influence, for example, effectiveness of cancer immunotherapies [239]. In fact, in a recent study, direct administration of Bifidobacterium to mice with established tumors from either Jackson Laboratory or Taconic Farms showed improved tumor-specific immunity and response to alpha programmed death-ligand 1 (αPD-L1) monoclonal antibody therapy only in Taconic Farms mice [240], clearly providing enlightenment to the responders versus non-responders conundrum often reported in prebiotic, probiotic and synbiotic interventional studies.
A beneficial effect of probiotics is clearly associated with particular bacterial taxa, and more precisely, bacterial strains. For example, species of Bifidobacterium and Lactobacillus, specifically B. lactis [241], B. longum [242–245], L. acidophilus [243, 246], L. casei [243, 247], and L. rhamnosus [246, 248] prevented or reduced the number of ACFs in animal models, while Enterococcus [249] and Lactococcus [250] were mostly ineffective. However, although Enterococcus faecium CRL 183 indeed failed to inhibit the formation of ACF in 1,2-dimethylhydrazine (DMH)-induced rats [249], strains of Enterococcus durans were capable of producing butyrate [251], an anti-proliferative and counter carcinogenic metabolite [252].
Comprehensive mechanistic studies of probiotics’ beneficial effects on tumor suppression and tumor progression are limited (reviewed in Azcarate-Peril et al. [253]). A tumor-suppressive molecule has been identified from culture supernatants of Lactobacillus gasseri ATCC334, a well-characterized probiotic [254]. The molecule ferrichrome (a hydrophilic metal chelating agent or siderophore generated by ATCC334) exhibited its tumor-suppressive effect through the induction of apoptosis in colon cancer cells via activation of the c-jun N-terminal kinase (JNK) pathway [255]. Another bacterial metabolite produced by specific strains of LAB is conjugated linoleic acid (CLA), capable of inhibiting cell proliferation through a tumor suppressor p-53 dependent mechanism in breast and colon cancer cells [256]. CLA generated by Pediococcus pentosaceus GS4 restricted proliferation of human colon cancer cells, which triggered apoptosis during biohydrogenation of free linoleic acid. Moreover, administration of GS4 to a mouse model of colon cancer resulted in enhancement of the gut microbiota biohydrogenation capability and subsequent stimulation of colonocyte apoptosis [257].
Microbiome studies are providing a better understanding of diet-microbiota and microbe-microbe interactions in the gastrointestinal tract although even more large-scale studies are needed to evaluate the efficacy of probiotic interventions to prevent or treat CRC. Undoubtedly the enormous amount of data generated by microbiome association studies will lead to the identification of benefits provided by novel strains, and possibly to a new definition of probiotics. Not only do we know now that bacteria not currently acknowledged as probiotics provide benefits to their host, like Bacteroides fragilis and Clostridium clusters IV and XIVa, which protect mice against trinitrobenzene sulfonic acid (TNBS)- or DSS-induced colitis [258, 259], but also studies have demonstrated microbial interactive networks that condition the host’s responses to interventions.
4. Novel/emerging ‘omics’ techniques and other approaches to address identified knowledge gaps
Early investigations to characterize and profile the microbiome utilized culture methods and microscopy, but these efforts were limited by cultivability, nonspecific substrate utilization, and the inability to distinguish morphologically similar bacteria from one another [260]. Early next-generation-sequencing techniques enabled the classification of bacteria from a sample at the taxonomic level by sequencing variable regions of the 16S ribosomal RNA (rRNA) gene. Taxonomic microbial profiling from 16S rRNA sequencing yields relative abundances of bacteria classified into OTUs, based on sequence similarity of sample reads to known bacterial sequences [261]. These data can be used to estimate diversity within and between samples based on the number of species in a sample (richness) and their relative abundance (evenness) [262]. Taxonomic marker gene survey data (e.g., 16S rRNA) can also be used to predict the metagenome of a sample, thus, providing some insight into the functional gene content of the microbiota present [263]. High-throughput, next-generation metagenome sequencing removes the need for prediction but is computationally intensive and yields much larger data sets requiring more data storage. In metagenome sequencing analysis, sequences are aligned to a gene catalog and the functional potential assessed by mapping genes to a KEGG database or through gene ontology (GO) enrichment analysis [264, 265].
The ultimate goal for microbial community profiling is to accurately assess which microbes are present in a sample and their collective genomes (metagenomics), which gene programs and protein networks are active at a given moment in time (metatranscriptomics and metaproteomics, respectively), and the metabolites present (metabolomics). Metabolomics enables the relative assessment of thousands of metabolites including microbe-, diet-, and host-derived molecules and represents an instantaneous manifestation of the combined cellular processes taking place in the sample. A recent study employing systematic comparison of metagenomic and metatranscriptomics data from healthy human subjects demonstrated that 41% of microbial gene transcripts were not differentially regulated relative to their genomic abundance and that the microbial transcriptome profile was significantly more individualized than the metagenome profile [266]. Therefore, understanding how changes in the microbial transcriptome relate to their abundance in the metagenome and how diet impacts this relationship will be important. Furthermore, there is a clear necessity to go beyond metagenomics and metatranscriptomics towards a complete ‘systems biology’ approach that includes the metabolome. Integrated omics approaches will be necessary to elucidate the complex network of interactions between the diet, microbiota, and host. This will enable identification of specific genes and microorganisms involved in metabolism and conversion of dietary bioactives. This information can then be used to construct intra- and inter-organismal metabolic pathways that will provide a framework for monitoring the microbiome response to specific bioactive exposures. For example, research conducted by Sridharan et al. used in silico microbial metabolism reaction networks coupled with two independent mass spectrometry metabolomics analyses to identify and characterize microbial metabolites which were then used in vitro to determine mechanisms of action [267]. ‘Omics’ data from the host can also be employed to predict and monitor the host response to the microbiome and its regulation thereof, particularly in individualized clinical settings where host genetics is already a primary consideration.
The appropriateness of targeted versus untargeted ‘omics’ approaches is debated but ultimately depends on the question being asked. For example, although untargeted metabolomics by multiplatform mass spectrometry permits relative quantification of a greater number of metabolites, it inherently leads to more noise and artifacts than a targeted metabolite assay coupled with appropriate standards [268]. For the discovery of novel disease biomarkers and construction of metabolic pathways, untargeted metabolomic approaches appear to be the most suitable due to the wealth of information gathered. Conversely, for hypothesis-driven experiments, targeted metabolite assays afford greater sensitivity and robustness. In addition, as evidenced by significant differences in metabolome and microbiome profiles collected from adjacent sampling sites (e.g., lumen versus mucosa or diseased versus normal adjacent tissue), targeted sampling procedures should be utilized for untargeted ‘omics’ analyses thereby improving the confidence of the interpretation and explanation of the data generated.
Despite the overall reduction in colon cancer incidence, the increase in distal cancer incidence in individuals younger than the recommended initial screening age (50 years old) supports the need for earlier screening [269]. Earlier colon screening coupled with fecal microbial profiling to identify cancer-associated microbes would enhance colon cancer prevention. It is estimated that the transition from precancerous cell to malignant colorectal tumor takes ~10–15 years implicating a substantial window for pre-cancer screening and implementation of prevention techniques [270]. As critical bioinformatics approaches become available for the integration of multi-omics analyses and as the required sample input continues to decrease it will be extremely beneficial to adopt more precise sampling procedures for characterizing the microbiota and interactions within a given microenvironment. The resolution afforded by passive fecal collection is useful as a clinical diagnostic test for cancer associated microbes; however, patients already diagnosed with colon cancer or found to have polyps following colonoscopy could benefit from more precise diagnostic protocols. Similar to the procedures used for genotyping a tumor biopsy to inform targeted therapeutic strategies, targeted tumor- or polyp-associated microbial profiling could reveal additional host-microbe signaling networks that can be targeted for therapy. Distinguishing between host- and microbial-derived growth signals could help explain the discrepancy in treatment responses of CRC patients. Although challenging, identifying critical microbial and metabolic biomarkers should improve screening and early disease detection [160]. Understanding these interactions will also aid in prevention because dietary bioactives that select against specific bacteria (versus broad-spectrum antibiotics) or that can inhibit signaling pathways downstream of microbial activities will improve the usefulness of diagnostic information gathered from fecal microbiome profiling.
Nutrigenomics and nutrigenetics are ‘multi-omics’ manifestations aimed at understanding nutrient-gene interactions. Indeed, diet can function at multiple levels to regulate the flow of genetic information [271–274]. These sciences and the knowledge acquired from them will be necessary for the development of personalized nutrition and disease prevention/treatment strategies, many of which are already used to some degree for the treatment of diet-related diseases, such as inflammatory bowel disease, obesity, diabetes, and cancer [275]. Understanding how and to what extent specific nutrients affect the expression of microbial genes adds an additional layer of complexity due to the considerable microbial variation between individuals and the ostensibly infinite number of combinations between host and microbial genomes. Presumably, however, better health outcomes from nutritional interventions would be achieved by taking into consideration an individual’s inherited and acquired genetic characteristics, age, dietary and lifestyle preferences, and gut microbiome profile.
5. Conclusions
Our understanding of the capacity of the gut microbiota to affect host physiology continues to increase and reinforces the necessity to study modifiers of the gut microbiome. It is irrefutable that the gut microbiota plays a critical role in regulating the intestinal epithelium and that the consequences of alterations in the gut microbiome can extend beyond the intestines. The concept that the gut microbiota can impact the development and activity of the host immune system, that the host immune system is a governor of the gut microbiota, and that microbial profiles are far more individualized than originally perceived, underscores the necessity to understand how individual genetic polymorphisms can ‘personalize’ the gut microbiota and, consequently, the ability of diet to influence these processes. The fact that the diet can shape the composition and function of the gut microbiota and that the microbiota can influence the health of the host beyond the intestinal epithelium implies a third mechanism by which diet can promote intestinal health; physiological changes in response to microbially-derived metabolites or ligands in other tissue sites (e.g., liver, muscle, etc.) may provide feedback to the intestinal epithelium that affect colon cancer risk and the risk of other associated diseases. Recent evidence suggesting a role of the gut microbiota in modulating host satiety and brain function (i.e., gut-brain axis) is remarkably fascinating and further emphasizes the need for understanding the capacity for diet to shape gut microbiota function and metabolism [278–282].
Despite the increasing acceptance of this paradigm, causative roles for specific bacteria are only suggested by current research. To effectively define the direct contribution of specific bacterium (or bacteria) to colon health/disease will require tightly controlled animal studies utilizing well-characterized dietary, environmental and disease conditions. Data produced by these studies may be more generalizable to human populations. Although fecal bacteria profiling has not yet been employed as a clinical tool for CRC diagnosis, research into this area of noninvasive diagnosis shows promise [276]. Similarly, despite changes in the microbiota being associated with disease, there are no clear bacterial biomarkers of colorectal cancer; however, elucidation of microbial pathways involved in the synthesis or detoxification of genotoxic agents (e.g., colonic sulfur metabolism, secondary bile acid production, etc.) will enable identification of novel compounds and biomarkers for this purpose. Although the sale and use of probiotics continues to increase, it is important to reinforce the concept that the mode of action cannot be generalized to all strains and will depend on other factors such as prebiotic use and current microbiota composition and health status [277]. With respect to colon health, the gut microbiota influences cellular processes by internal and external mechanisms, which include receptor binding and signal transduction/inhibition, modulation of gene and protein expression, epigenetic modifications, changes in metabolism, and cell differentiation/fate. Furthermore, dietary phytochemicals that affect the composition and metabolism of the microbiota can synergize with their direct effects, or those of other bioactives, to enhance or alter their bioactivity (Fig. 1). Importantly, for compounds with unknown mechanisms but observed phenotypic responses, it is entirely possible that the function is completely microbial-dependent and the efficacy, therefore, contingent on the presence of the microbe(s) and their metabolic response to the dietary stimulus. Studies employing germ-free animals and animals with discrete microbial populations, in addition to traditional cell culture and co-culture experimentation, will be necessary to dissect the mechanisms and co-dependencies of specific diet-host-microbe interactions. The correlation, or often lack of correlation, between data from stool versus healthy or diseased tissue samples adds enormous complexity to the microbe-host relationship. Therefore, to improve our understanding of these relationships, “omics” studies should be used to interrogate concomitant changes in the diseased mucosa and the stool in order to maximize the information gathered from passive fecal sampling. Experiments should use multiple “omics” approaches to identify those biomarkers that most accurately and robustly characterize the disease state. Furthermore, it will be necessary to assess these relationships throughout the disease process in order to improve the sensitivity and specificity of diagnostic screening approaches for CRC prevention and treatment.
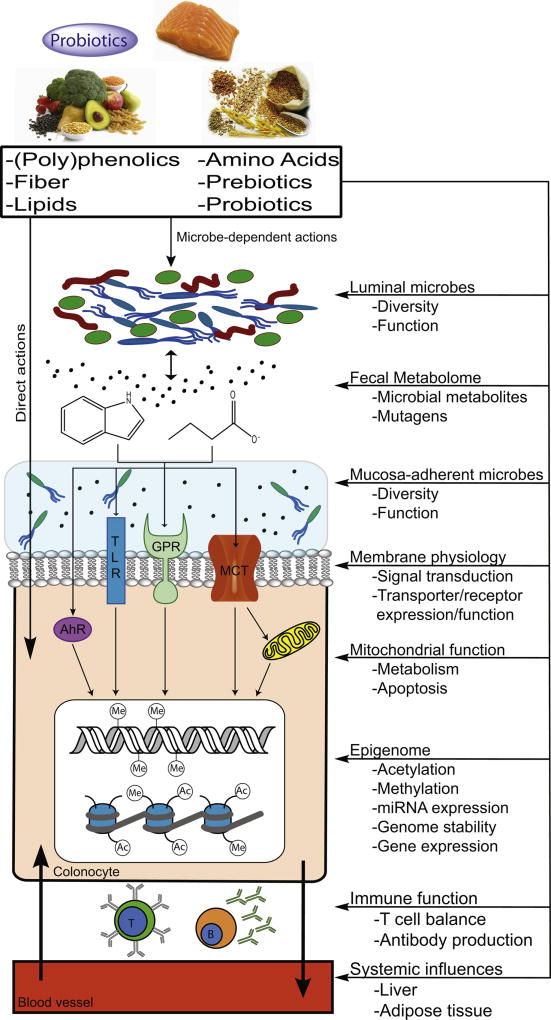
Fig 1. Graphical depiction of the diet-host-microbe interaction.
This overview figure represents example mechanisms whereby diet-microbe-host interactions are associated with colon carcinogenesis. In addition to direct effects of diet on host health and metabolism, diet can be a modifier of the gut microbiome (e.g., its composition and functional attributes). Gut microbiota are capable of furtherer metabolizing dietary bioactives to generate secondary bioactive compounds, which can affect host physiology. Exchange of metabolites between different microbes in the gut is possible and adds complexity to the diet-microbe-host interaction network. Native dietary bioactives and microbially-derived metabolites can act on epithelial cells in the colon by extrinsic and intrinsic mechanisms. Dietary bioactives and their derivatives can act as ligands for receptors or alter receptor function, serve as metabolic fuel for cells, function as modifiers of the epigenome, etc. These compounds can also influence immune function and host health beyond the epithelium. Our understanding of the capacity of diet to direct a functionally beneficial gut microbiome for both localized (i.e., colon) and systemic health will continue to increase with improved ‘omics’ techniques and advances in ‘omics’ data integration.
Acknowledgments
Work originating the from the authors laboratories described in this review was supported by the United Sorghum Checkoff Program (HVM006-12), the California Dried Plum Board (PN12-20), NASA (NNX15AD64G), the National Institutes of Health (R01 CA168312, R35CA197707, RO1CA202697, RO1ES025713, P30ES02351), the NIH/National Institute of Diabetes and Digestive and Kidney Diseases grant (P30DK34987), and the Cancer Prevention Research Institute of Texas (RP160589IH). Derek Seidel also was supported by the Mentored Research Program in Space Life Sciences funded by the National Space Biomedical Research Institute through NCC 9-58 (EO02001).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors have no competing interests.
References
- 1.Ahmed RL, Schmitz KH, Anderson KE, Rosamond WD, Folsom AR. The metabolic syndrome and risk of incident colorectal cancer. Cancer. 2006;107:28–36. doi: 10.1002/cncr.21950. [DOI] [PubMed] [Google Scholar]
- 2.Terzic J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–2114. e2105. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 3.Datta K, Suman S, Kallakury BV, Fornace AJ., Jr Heavy ion radiation exposure triggered higher intestinal tumor frequency and greater beta-catenin activation than gamma radiation in APC(Min/+) mice. PloS One. 2013;8:e59295. doi: 10.1371/journal.pone.0059295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim JH, Lim YJ, Kim YH, Sung IK, Shim SG, Oh SO, Park SS, Yang S, Son HJ, Rhee PL, et al. Is metabolic syndrome a risk factor for colorectal adenoma? Cancer Epidemiology, Biomarkers and Prevention. 2007;16:1543–1546. doi: 10.1158/1055-9965.EPI-07-0199. [DOI] [PubMed] [Google Scholar]
- 5.Kim SB, Bozeman RG, Kaisani A, Kim W, Zhang L, Richardson JA, Wright WE, Shay JW. Radiation promotes colorectal cancer initiation and progression by inducing senescence-associated inflammatory responses. Oncogene. 2016;35:3365–3375. doi: 10.1038/onc.2015.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 7.Block KI, Gyllenhaal C, Lowe L, Amedei A, Amin AR, Amin A, Aquilano K, Arbiser J, Arreola A, Arzumanyan A. Seminars in Cancer Biology. Elsevier; 2015. Designing a broad-spectrum integrative approach for cancer prevention and treatment; pp. S276–S304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feron O. Pyruvate into lactate and back: from the Warburg effect to symbiotic energy fuel exchange in cancer cells. Radiotherapy and Oncology. 2009;92:329–333. doi: 10.1016/j.radonc.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 11.Xie J, Wu H, Dai C, Pan Q, Ding Z, Hu D, Ji B, Luo Y, Hu X. Beyond Warburg effect--dual metabolic nature of cancer cells. Scientific Reports. 2014;4:4927. doi: 10.1038/srep04927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammoud SS, Cairns BR, Jones DA. Epigenetic regulation of colon cancer and intestinal stem cells. Current Opinion in Cell Biology. 2013;25:177–183. doi: 10.1016/j.ceb.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pitot HC. The molecular biology of carcinogenesis. Cancer. 1993;72:962–970. doi: 10.1002/1097-0142(19930801)72:3+<962::aid-cncr2820721303>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 14.Moody CS, Hassan HM. Mutagenicity of oxygen free radicals. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:2855–2859. doi: 10.1073/pnas.79.9.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yokota J. Tumor progression and metastasis. Carcinogenesis. 2000;21:497–503. doi: 10.1093/carcin/21.3.497. [DOI] [PubMed] [Google Scholar]
- 16.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanauchi O, Matsumoto Y, Matsumura M, Fukuoka M, Bamba T. The Beneficial Effects of Microflora, Especially Obligate Anaerobes, and Their Products on the Colonic Environment in Inflammatory Bowel Disease. Current Pharmaceutical Design. 2005;11:1047–1053. doi: 10.2174/1381612053381675. [DOI] [PubMed] [Google Scholar]
- 20.Marchesi JR, Dutilh BE, Hall N, Peters WH, Roelofs R, Boleij A, Tjalsma H. Towards the human colorectal cancer microbiome. PloS One. 2011;6:e20447. doi: 10.1371/journal.pone.0020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 22.Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, Corthier G, Tran Van Nhieu J, Furet JP. Microbial dysbiosis in colorectal cancer (CRC) patients. PloS One. 2011;6:e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamboli CP, Neut C, Desreumaux P, Colombel JF. Dysbiosis in inflammatory bowel disease. Gut. 2004;53:1–4. doi: 10.1136/gut.53.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turner ND, Ritchie LE, Bresalier RS, Chapkin RS. The microbiome and colorectal neoplasia: environmental modifiers of dysbiosis. Curr Gastroenterol Rep. 2013;15:346. doi: 10.1007/s11894-013-0346-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irrazábal T, Belcheva A, Girardin SE, Martin A, Philpott DJ. The multifaceted role of the intestinal microbiota in colon cancer. Molecular Cell. 2014;54:309–320. doi: 10.1016/j.molcel.2014.03.039. [DOI] [PubMed] [Google Scholar]
- 26.Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: how are they linked? Free Radical Biology and Medicine. 2010;49:1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greer JB, O'Keefe SJ. Microbial induction of immunity, inflammation, and cancer. Frontiers in Physiology. 2011;1:168. doi: 10.3389/fphys.2010.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karin M, Lawrence T, Nizet V. Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell. 2006;124:823–835. doi: 10.1016/j.cell.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 29.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nature Reviews: Gastroenterology & Hepatology. 2016;13:691–706. doi: 10.1038/nrgastro.2016.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salonen A, de Vos WM. Impact of diet on human intestinal microbiota and health. Annual Review of Food Science and Technology. 2014;5:239–262. doi: 10.1146/annurev-food-030212-182554. [DOI] [PubMed] [Google Scholar]
- 32.Riboli E, Norat T. Epidemiologic evidence of the protective effect of fruit and vegetables on cancer risk. American Journal of Clinical Nutrition. 2003;78:559S–569S. doi: 10.1093/ajcn/78.3.559S. [DOI] [PubMed] [Google Scholar]
- 33.Terry P, Giovannucci E, Michels KB, Bergkvist L, Hansen H, Holmberg L, Wolk A. Fruit, vegetables, dietary fiber, and risk of colorectal cancer. Journal of the National Cancer Institute. 2001;93:525–533. doi: 10.1093/jnci/93.7.525. [DOI] [PubMed] [Google Scholar]
- 34.Trock B, Lanza E, Greenwald P. Dietary fiber, vegetables, and colon cancer: critical review and meta-analyses of the epidemiologic evidence. Journal of the National Cancer Institute. 1990;82:650–661. doi: 10.1093/jnci/82.8.650. [DOI] [PubMed] [Google Scholar]
- 35.Kris-Etherton PM, Hecker KD, Bonanome A, Coval SM, Binkoski AE, Hilpert KF, Griel AE, Etherton TD. Bioactive compounds in foods: their role in the prevention of cardiovascular disease and cancer. The American Journal of Medicine. 2002;113:71–88. doi: 10.1016/s0002-9343(01)00995-0. [DOI] [PubMed] [Google Scholar]
- 36.Papandreou D, Noor ZT, Rashed M. The role of soluble, insoluble fibers and their bioactive compounds in cancer: a mini review. Food and Nutrition Sciences. 2015;6:1. [Google Scholar]
- 37.Monagas M, Urpi-Sarda M, Sanchez-Patan F, Llorach R, Garrido I, Gomez-Cordoves C, Andres-Lacueva C, Bartolome B. Insights into the metabolism and microbial biotransformation of dietary flavan-3-ols and the bioactivity of their metabolites. Food & Function. 2010;1:233–253. doi: 10.1039/c0fo00132e. [DOI] [PubMed] [Google Scholar]
- 38.Aura AM, Martin-Lopez P, O'Leary KA, Williamson G, Oksman-Caldentey KM, Poutanen K, Santos-Buelga C. In vitro metabolism of anthocyanins by human gut microflora. European Journal of Nutrition. 2005;44:133–142. doi: 10.1007/s00394-004-0502-2. [DOI] [PubMed] [Google Scholar]
- 39.Aura A-M. Microbial metabolism of dietary phenolic compounds in the colon. Phytochemistry Reviews. 2008;7:407–429. [Google Scholar]
- 40.Clavel T, Henderson G, Alpert CA, Philippe C, Rigottier-Gois L, Dore J, Blaut M. Intestinal bacterial communities that produce active estrogen-like compounds enterodiol and enterolactone in humans. Applied and Environmental Microbiology. 2005;71:6077–6085. doi: 10.1128/AEM.71.10.6077-6085.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 42.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 43.Potten CS, Wilson JW, Booth C. Regulation and significance of apoptosis in the stem cells of the gastrointestinal epithelium. Stem Cells. 1997;15:82–93. doi: 10.1002/stem.150082. [DOI] [PubMed] [Google Scholar]
- 44.Ruthig DJ, Meckling-Gill KA. Both (n-3) and (n-6) fatty acids stimulate wound healing in the rat intestinal epithelial cell line, IEC-6. Journal of Nutrition. 1999;129:1791–1798. doi: 10.1093/jn/129.10.1791. [DOI] [PubMed] [Google Scholar]
- 45.Clevers H. The cancer stem cell: premises, promises and challenges. Nature Medicine. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 46.Blokzijl F, de Ligt J, Jager M, Sasselli V, Roerink S, Sasaki N, Huch M, Boymans S, Kuijk E, Prins P, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016;538:260–264. doi: 10.1038/nature19768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huels DJ, Sansom OJ. Stem vs non-stem cell origin of colorectal cancer. British Journal of Cancer. 2015;113:1–5. doi: 10.1038/bjc.2015.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–320. doi: 10.1016/s0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- 49.Suraweera N, Robinson J, Volikos E, Guenther T, Talbot I, Tomlinson I, Silver A. Mutations within Wnt pathway genes in sporadic colorectal cancers and cell lines. International Journal of Cancer. 2006;119:1837–1842. doi: 10.1002/ijc.22046. [DOI] [PubMed] [Google Scholar]
- 50.Polakis P. Wnt signaling in cancer. Cold Spring Harbor Perspectives in Biology. 2012;4:a008052. doi: 10.1101/cshperspect.a008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rycaj K, Tang DG. Cell-of-Origin of Cancer versus Cancer Stem Cells: Assays and Interpretations. Cancer Research. 2015;75:4003–4011. doi: 10.1158/0008-5472.CAN-15-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davidson LA, Callaway ES, Kim E, Weeks BR, Fan YY, Allred CD, Chapkin RS. Targeted Deletion of p53 in Lgr5-Expressing Intestinal Stem Cells Promotes Colon Tumorigenesis in a Preclinical Model of Colitis-Associated Cancer. Cancer Research. 2015;75:5392–5397. doi: 10.1158/0008-5472.CAN-15-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chaffer CL, Weinberg RA. How does multistep tumorigenesis really proceed? Cancer Discovery. 2015;5:22–24. doi: 10.1158/2159-8290.CD-14-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Visvader JE, Clevers H. Tissue-specific designs of stem cell hierarchies. Nature Cell Biology. 2016;18:349–355. doi: 10.1038/ncb3332. [DOI] [PubMed] [Google Scholar]
- 55.Wu S, Powers S, Zhu W, Hannun YA. Substantial contribution of extrinsic risk factors to cancer development. Nature. 2016;529:43–47. doi: 10.1038/nature16166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tetteh PW, Basak O, Farin HF, Wiebrands K, Kretzschmar K, Begthel H, van den Born M, Korving J, de Sauvage F, van Es JH, et al. Replacement of Lost Lgr5-Positive Stem Cells through Plasticity of Their Enterocyte-Lineage Daughters. Cell Stem Cell. 2016;18:203–213. doi: 10.1016/j.stem.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 58.Tetteh PW, Farin HF, Clevers H. Plasticity within stem cell hierarchies in mammalian epithelia. Trends in Cell Biology. 2015;25:100–108. doi: 10.1016/j.tcb.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 59.Beyaz S, Mana MD, Roper J, Kedrin D, Saadatpour A, Hong SJ, Bauer-Rowe KE, Xifaras ME, Akkad A, Arias E, et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature. 2016;531:53–58. doi: 10.1038/nature17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DeClercq V, McMurray DN, Chapkin RS. Obesity promotes colonic stem cell expansion during cancer initiation. Cancer Letters. 2015;369:336–343. doi: 10.1016/j.canlet.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Igarashi M, Guarente L. mTORC1 and SIRT1 Cooperate to Foster Expansion of Gut Adult Stem Cells during Calorie Restriction. Cell. 2016;166:436–450. doi: 10.1016/j.cell.2016.05.044. [DOI] [PubMed] [Google Scholar]
- 62.Kim E, Davidson LA, Zoh RS, Hensel ME, Salinas ML, Patil BS, Jayaprakasha GK, Callaway ES, Allred CD, Turner ND, et al. Rapidly cycling Lgr5+ stem cells are exquisitely sensitive to extrinsic dietary factors that modulate colon cancer risk. Cell Death & Disease. 2016;7:e2460. doi: 10.1038/cddis.2016.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang WCL, Chapkin RS, Lupton JR. Predictive value of proliferation, differentiation and apoptosis as intermediate markers for colon tumorigenesis. Carcinogenesis. 1997;18:721–730. doi: 10.1093/carcin/18.4.721. [DOI] [PubMed] [Google Scholar]
- 64.Ward JM, Uno H, Kurata Y, Weghorst CM, Jang JJ. Cell proliferation not associated with carcinogenesis in rodents and humans. Environmental Health Perspectives. 1993;101:125–135. doi: 10.1289/ehp.93101s5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reed JC. Mechanisms of apoptosis. American Journal of Pathology. 2000;157:1415–1430. doi: 10.1016/S0002-9440(10)64779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fulda S, Debatin KM. Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene. 2006;25:4798–4811. doi: 10.1038/sj.onc.1209608. [DOI] [PubMed] [Google Scholar]
- 67.Saelens X, Festjens N, Vande Walle L, van Gurp M, van Loo G, Vandenabeele P. Toxic proteins released from mitochondria in cell death. Oncogene. 2004;23:2861–2874. doi: 10.1038/sj.onc.1207523. [DOI] [PubMed] [Google Scholar]
- 68.Ichim G, Tait SW. A fate worse than death: apoptosis as an oncogenic process. Nature Reviews Cancer. 2016 doi: 10.1038/nrc.2016.58. [DOI] [PubMed] [Google Scholar]
- 69.Green DR, Kroemer G. The pathophysiology of mitochondrial cell death. Science. 2004;305:626–629. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- 70.Ng Y, Barhoumi R, Tjalkens RB, Fan YY, Kolar S, Wang N, Lupton JR, Chapkin RS. The role of docosahexaenoic acid in mediating mitochondrial membrane lipid oxidation and apoptosis in colonocytes. Carcinogenesis. 2005;26:1914–1921. doi: 10.1093/carcin/bgi163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rodrigues NR, Rowan A, Smith ME, Kerr IB, Bodmer WF, Gannon JV, Lane DP. p53 mutations in colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:7555–7559. doi: 10.1073/pnas.87.19.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cho Y, Turner ND, Davidson LA, Chapkin RS, Carroll RJ, Lupton JR. A chemoprotective fish oil/pectin diet enhances apoptosis via Bcl-2 promoter methylation in rat azoxymethane-induced carcinomas. Experimental Biology and Medicine (Maywood, NJ) 2012;237:1387–1393. doi: 10.1258/ebm.2012.012244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hong MY, Chapkin RS, Davidson LA, Turner ND, Morris JS, Carroll RJ, Lupton JR. Fish oil enhances targeted apoptosis during colon tumor initiation in part by downregulating Bcl-2. Nutrition and Cancer. 2003;46:44–51. doi: 10.1207/S15327914NC4601_06. [DOI] [PubMed] [Google Scholar]
- 74.Wright SC, Zhong J, Larrick JW. Inhibition of Apoptosis as a Mechanism of Tumor Promotion. FASEB Journal. 1994;8:654–660. doi: 10.1096/fasebj.8.9.8005393. [DOI] [PubMed] [Google Scholar]
- 75.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cho Y, Turner ND, Davidson LA, Chapkin RS, Carroll RJ, Lupton JR. Colon cancer cell apoptosis is induced by combined exposure to the n-3 fatty acid docosahexaenoic acid and butyrate through promoter methylation. Experimental Biology and Medicine (Maywood, NJ) 2014;239:302–310. doi: 10.1177/1535370213514927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Glebov OK, Rodriguez LM, Nakahara K, Jenkins J, Cliatt J, Humbyrd CJ, DeNobile J, Soballe P, Simon R, Wright G, et al. Distinguishing right from left colon by the pattern of gene expression. Cancer Epidemiology, Biomarkers and Prevention. 2003;12:755–762. [PubMed] [Google Scholar]
- 78.Barnicle A, Seoighe C, Golden A, Greally JM, Egan LJ. Differential DNA methylation patterns of homeobox genes in proximal and distal colon epithelial cells. Physiological Genomics. 2016;48:257–273. doi: 10.1152/physiolgenomics.00046.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Minoo P, Zlobec I, Peterson M, Terracciano L, Lugli A. Characterization of rectal, proximal and distal colon cancers based on clinicopathological, molecular and protein profiles. International Journal of Oncology. 2010;37:707–718. doi: 10.3892/ijo_00000720. [DOI] [PubMed] [Google Scholar]
- 80.Yamauchi M, Morikawa T, Kuchiba A, Imamura Y, Qian ZR, Nishihara R, Liao X, Waldron L, Hoshida Y, Huttenhower C, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61:847–854. doi: 10.1136/gutjnl-2011-300865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Watanabe T, Kobunai T, Toda E, Yamamoto Y, Kanazawa T, Kazama Y, Tanaka J, Tanaka T, Konishi T, Okayama Y, et al. Distal colorectal cancers with microsatellite instability (MSI) display distinct gene expression profiles that are different from proximal MSI cancers. Cancer Research. 2006;66:9804–9808. doi: 10.1158/0008-5472.CAN-06-1163. [DOI] [PubMed] [Google Scholar]
- 82.Hong MY, Chang WCL, Chapkin RS, Lupton JR. Relationship among colonocyte proliferation, differentiation, and apoptosis as a function of diet and carcinogen. Nutrition and Cancer-an International Journal. 1997;28:20–29. doi: 10.1080/01635589709514548. [DOI] [PubMed] [Google Scholar]
- 83.Wiseman H, Halliwell B. Damage to DNA by reactive oxygen and nitrogen species: role in inflammatory disease and progression to cancer. Biochemical Journal. 1996;313(Pt 1):17–29. doi: 10.1042/bj3130017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meira LB, Bugni JM, Green SL, Lee CW, Pang B, Borenshtein D, Rickman BH, Rogers AB, Moroski-Erkul CA, McFaline JL, et al. DNA damage induced by chronic inflammation contributes to colon carcinogenesis in mice. Journal of Clinical Investigation. 2008;118:2516–2525. doi: 10.1172/JCI35073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hahn MA, Hahn T, Lee DH, Esworthy RS, Kim BW, Riggs AD, Chu FF, Pfeifer GP. Methylation of polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Research. 2008;68:10280–10289. doi: 10.1158/0008-5472.CAN-08-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Molecular Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30:1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 88.Edwards RA, Witherspoon M, Wang K, Afrasiabi K, Pham T, Birnbaumer L, Lipkin SM. Epigenetic repression of DNA mismatch repair by inflammation and hypoxia in inflammatory bowel disease-associated colorectal cancer. Cancer Research. 2009;69:6423–6429. doi: 10.1158/0008-5472.CAN-09-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vannucci L, Stepankova R, Kozakova H, Fiserova A, Rossmann P, Tlaskalova-Hogenova H. Colorectal carcinogenesis in germ-free and conventionally reared rats: different intestinal environments affect the systemic immunity. International Journal of Oncology. 2008;32:609–617. [PubMed] [Google Scholar]
- 90.Sacksteder MR. Occurrence of spontaneous tumors in the germfree F344 rat. Journal of the National Cancer Institute. 1976;57:1371–1373. doi: 10.1093/jnci/57.6.1371. [DOI] [PubMed] [Google Scholar]
- 91.Reddy BS, Weisburger JH, Narisawa T, Wynder EL. Colon carcinogenesis in germ-free rats with 1,2-dimethylhydrazine and N-methyl-n'-nitro-N-nitrosoguanidine. Cancer Research. 1974;34:2368–2372. [PubMed] [Google Scholar]
- 92.Reddy BS, Narisawa T, Wright P, Vukusich D, Weisburger JH, Wynder EL. Colon carcinogenesis with azoxymethane and dimethylhydrazine in germ-free rats. Cancer Research. 1975;35:287–290. [PubMed] [Google Scholar]
- 93.Uronis JM, Muhlbauer M, Herfarth HH, Rubinas TC, Jones GS, Jobin C. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PloS One. 2009;4:e6026. doi: 10.1371/journal.pone.0006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, Taniguchi K, Yu GY, Osterreicher CH, Hung KE, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17- mediated tumour growth. Nature. 2012;491:254–258. doi: 10.1038/nature11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Klimesova K, Kverka M, Zakostelska Z, Hudcovic T, Hrncir T, Stepankova R, Rossmann P, Ridl J, Kostovcik M, Mrazek J, et al. Altered gut microbiota promotes colitis-associated cancer in IL-1 receptor-associated kinase M-deficient mice. Inflammatory Bowel Diseases. 2013;19:1266–1277. doi: 10.1097/MIB.0b013e318281330a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Garaude J, Kent A, van Rooijen N, Blander JM. Simultaneous targeting of toll- and nod-like receptors induces effective tumor-specific immune responses. Science Translational Medicine. 2012;4:120ra116. doi: 10.1126/scitranslmed.3002868. [DOI] [PubMed] [Google Scholar]
- 97.Pradere JP, Dapito DH, Schwabe RF. The Yin and Yang of Toll-like receptors in cancer. Oncogene. 2014;33:3485–3495. doi: 10.1038/onc.2013.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ott S, Musfeldt M, Wenderoth D, Hampe J, Brant O, Fölsch U, Timmis K, Schreiber S. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sepehri S, Kotlowski R, Bernstein CN, Krause DO. Microbial diversity of inflamed and noninflamed gut biopsy tissues in inflammatory bowel disease. Inflammatory Bowel Diseases. 2007;13:675–683. doi: 10.1002/ibd.20101. [DOI] [PubMed] [Google Scholar]
- 101.Ritchie LE, Sturino JM, Carroll RJ, Rooney LW, Azcarate-Peril MA, Turner ND. Polyphenol-rich sorghum brans alter colon microbiota and impact species diversity and species richness after multiple bouts of dextran sodium sulfate-induced colitis. FEMS Microbiology Ecology. 2015;91:fiv008. doi: 10.1093/femsec/fiv008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ritchie LE, Taddeo SS, Weeks BR, Lima F, Bloomfield SA, Azcarate-Peril MA, Zwart SR, Smith SM, Turner ND. Space Environmental Factor Impacts upon Murine Colon Microbiota and Mucosal Homeostasis. PloS One. 2015;10:e0125792. doi: 10.1371/journal.pone.0125792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, Jia W, Cai S, Zhao L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. The ISME journal. 2012;6:320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, Chen Y, Yang F, Lu N, Wang Z, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microbial Ecology. 2013;66:462–470. doi: 10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- 105.Mira-Pascual L, Cabrera-Rubio R, Ocon S, Costales P, Parra A, Suarez A, Moris F, Rodrigo L, Mira A, Collado MC. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. Journal of Gastroenterology. 2015;50:167–179. doi: 10.1007/s00535-014-0963-x. [DOI] [PubMed] [Google Scholar]
- 106.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PloS One. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PloS One. 2013;8:e70803. doi: 10.1371/journal.pone.0070803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sanchez-Espiridion B, White L, Mishra L, Raju GS, Kopetz S, Gu J, Ye Y, Wu X, Liang D. Abstract 8: Global and targeted metabolomic profiling of colorectal cancer progression. Cancer Research. 2016;76:8–8. [Google Scholar]
- 109.Brown DG, Rao S, Weir TL, O'Malia J, Bazan M, Brown RJ, Ryan EP. Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016;4:11. doi: 10.1186/s40170-016-0151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gagnaire A, Nadel B, Raoult D, Neefjes J, Gorvel JP. Collateral damage: insights into bacterial mechanisms that predispose host cells to cancer. Nature Reviews: Microbiology. 2017;15:109–128. doi: 10.1038/nrmicro.2016.171. [DOI] [PubMed] [Google Scholar]
- 111.Vizcaino MI, Crawford JM. The colibactin warhead crosslinks DNA. Nature Chemistry. 2015;7:411–417. doi: 10.1038/nchem.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wu S, Powell J, Mathioudakis N, Kane S, Fernandez E, Sears CL. Bacteroides fragilis enterotoxin induces intestinal epithelial cell secretion of interleukin-8 through mitogen-activated protein kinases and a tyrosine kinase-regulated nuclear factor-κB pathway. Infection and Immunity. 2004;72:5832–5839. doi: 10.1128/IAI.72.10.5832-5839.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu S, Shin J, Zhang G, Cohen M, Franco A, Sears CL. The Bacteroides fragilis toxin binds to a specific intestinal epithelial cell receptor. Infection and Immunity. 2006;74:5382–5390. doi: 10.1128/IAI.00060-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Goodwin AC, Destefano Shields CE, Wu S, Huso DL, Wu X, Murray-Stewart TR, Hacker-Prietz A, Rabizadeh S, Woster PM, Sears CL, Casero RA., Jr Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15354–15359. doi: 10.1073/pnas.1010203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Boleij A, Hechenbleikner EM, Goodwin AC, Badani R, Stein EM, Lazarev MG, Ellis B, Carroll KC, Albesiano E, Wick EC, et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clinical Infectious Diseases. 2015;60:208–215. doi: 10.1093/cid/ciu787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lu R, Wu S, Zhang YG, Xia Y, Zhou Z, Kato I, Dong H, Bissonnette M, Sun J. Salmonella Protein AvrA Activates the STAT3 Signaling Pathway in Colon Cancer. Neoplasia. 2016;18:307–316. doi: 10.1016/j.neo.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Corvinus FM, Orth C, Moriggl R, Tsareva SA, Wagner S, Pfitzner EB, Baus D, Kaufman R, Huber LA, Zatloukal K, et al. Persistent STAT3 Activation in Colon Cancer Is Associated with Enhanced Cell Proliferation and Tumor Growth. Neoplasia. 2005;7:545–555. doi: 10.1593/neo.04571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lin L, Liu A, Peng Z, Lin HJ, Li PK, Li C, Lin J. STAT3 is necessary for proliferation and survival in colon cancer-initiating cells. Cancer Research. 2011;71:7226–7237. doi: 10.1158/0008-5472.CAN-10-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H, Eckmann L, Karin M. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bromberg J, Wang TC. Inflammation and cancer: IL-6 and STAT3 complete the link. Cancer Cell. 2009;15:79–80. doi: 10.1016/j.ccr.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Raisch J, Rolhion N, Dubois A, Darfeuille-Michaud A, Bringer MA. Intracellular colon cancerassociated Escherichia coli promote protumoral activities of human macrophages by inducing sustained COX-2 expression. Laboratory Investigation. 2015;95:296–307. doi: 10.1038/labinvest.2014.161. [DOI] [PubMed] [Google Scholar]
- 122.Wang X, Huycke MM. Colorectal cancer: role of commensal bacteria and bystander effects. Gut Microbes. 2015;6:370–376. doi: 10.1080/19490976.2015.1103426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Anson NM, Selinheimo E, Havenaar R, Aura AM, Mattila I, Lehtinen P, Bast A, Poutanen K, Haenen GR. Bioprocessing of wheat bran improves in vitro bioaccessibility and colonic metabolism of phenolic compounds. Journal of Agricultural and Food Chemistry. 2009;57:6148–6155. doi: 10.1021/jf900492h. [DOI] [PubMed] [Google Scholar]
- 124.Hemery YM, Anson NM, Havenaar R, Haenen GR, Noort MW, Rouau X. Dry-fractionation of wheat bran increases the bioaccessibility of phenolic acids in breads made from processed bran fractions. Food Research International. 2010;43:1429–1438. [Google Scholar]
- 125.Maciorowski KG, Turner ND, Lupton JR, Chapkin RS, Shermer CL, Ha SD, Ricke SC. Diet and carcinogen alter the fecal microbial populations of rats. Journal of Nutrition. 1997;127:449–457. doi: 10.1093/jn/127.3.449. [DOI] [PubMed] [Google Scholar]
- 126.Taguri T, Tanaka T, Kouno I. Antimicrobial activity of 10 different plant polyphenols against bacteria causing food-borne disease. Biological and Pharmaceutical Bulletin. 2004;27:1965–1969. doi: 10.1248/bpb.27.1965. [DOI] [PubMed] [Google Scholar]
- 127.Sójka M, Kołodziejczyk K, Milala J, Abadias M, Viñas I, Guyot S, Baron A. Composition and properties of the polyphenolic extracts obtained from industrial plum pomaces. Journal of Functional Foods. 2015;12:168–178. [Google Scholar]
- 128.O’Keefe SJ, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, Posma JM, Kinross J, Wahl E, Ruder E. Fat fibre and cancer risk in African Americans and rural Africans. Nature Communications. 2015;6 doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang Z, Geng J, Tang X, Fan H, Xu J, Wen X, Ma ZS, Shi P. Spatial heterogeneity and co-occurrence patterns of human mucosal-associated intestinal microbiota. ISME J. 2014;8:881–893. doi: 10.1038/ismej.2013.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Consortium HMP. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Clifford M. Diet-derived phenols in plasma and tissues and their implications for health. Planta Medica. 2004;70:1103–1114. doi: 10.1055/s-2004-835835. [DOI] [PubMed] [Google Scholar]
- 132.Williamson G, Clifford MN. Colonic metabolites of berry polyphenols: the missing link to biological activity? British Journal of Nutrition. 2010;104(Suppl 3):S48–66. doi: 10.1017/S0007114510003946. [DOI] [PubMed] [Google Scholar]
- 133.Zhang LS, Davies SS. Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions. Genome Medicine. 2016;8:46. doi: 10.1186/s13073-016-0296-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cheng Y, Jin UH, Allred CD, Jayaraman A, Chapkin RS, Safe S. Aryl Hydrocarbon Receptor Activity of Tryptophan Metabolites in Young Adult Mouse Colonocytes. Drug Metabolism and Disposition: The Biological Fate of Chemicals. 2015;43:1536–1543. doi: 10.1124/dmd.115.063677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nature Reviews: Microbiology. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 136.Turner ND, Lupton JR. Dietary fiber. Advances in Nutrition: An International Review Journal. 2011;2:151–152. doi: 10.3945/an.110.000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Macfarlane GT, Macfarlane S. Human colonic microbiota: ecology, physiology and metabolic potential of intestinal bacteria. Scandinavian Journal of Gastroenterology Supplement. 1997;222:3–9. doi: 10.1080/00365521.1997.11720708. [DOI] [PubMed] [Google Scholar]
- 138.Lupton JR, Kurtz PP. Relationship of colonic luminal short-chain fatty acids and pH to in vivo cell proliferation in rats. Journal of Nutrition. 1993;123:1522–1530. doi: 10.1093/jn/123.9.1522. [DOI] [PubMed] [Google Scholar]
- 139.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Alimentary Pharmacology and Therapeutics. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 141.Lupton JR. Microbial degradation products influence colon cancer risk: the butyrate controversy. Journal of Nutrition. 2004;134:479–482. doi: 10.1093/jn/134.2.479. [DOI] [PubMed] [Google Scholar]
- 142.Lupton JR, Turner ND. Potential protective mechanisms of wheat bran fiber. American Journal of Medicine. 1999;106:24S–27S. doi: 10.1016/s0002-9343(98)00343-x. [DOI] [PubMed] [Google Scholar]
- 143.Gazzaniga JM, Lupton JR. Dilution effect of dietary fiber sources: an in vivo study in the rat. Nutrition Research. 1987;7:1261–1268. [Google Scholar]
- 144.Lewis SJ, Heaton KW. Increasing butyrate concentration in the distal colon by accelerating intestinal transit. Gut. 1997;41:245–251. doi: 10.1136/gut.41.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zoran DL, Turner ND, Taddeo SS, Chapkin RS, Lupton JR. Wheat bran diet reduces tumor incidence in a rat model of colon cancer independent of effects on distal luminal butyrate concentrations. Journal of Nutrition. 1997;127:2217–2225. doi: 10.1093/jn/127.11.2217. [DOI] [PubMed] [Google Scholar]
- 146.Tudela CV, Boudry C, Stumpff F, Aschenbach JR, Vahjen W, Zentek J, Pieper R. Down-regulation of monocarboxylate transporter 1 (MCT1) gene expression in the colon of piglets is linked to bacterial protein fermentation and pro-inflammatory cytokine-mediated signalling. British Journal of Nutrition. 2015;113:610–617. doi: 10.1017/S0007114514004231. [DOI] [PubMed] [Google Scholar]
- 147.Liu X, Blouin J-M, Santacruz A, Lan A, Andriamihaja M, Wilkanowicz S, Benetti P-H, Tomé D, Sanz Y, Blachier F. High-protein diet modifies colonic microbiota and luminal environment but not colonocyte metabolism in the rat model: the increased luminal bulk connection. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2014;307:G459–G470. doi: 10.1152/ajpgi.00400.2013. [DOI] [PubMed] [Google Scholar]
- 148.Donohoe DR, Curry KP, Bultman SJ. Microbial oncotarget: bacterial-produced butyrate, chemoprevention and Warburg effect. Oncotarget. 2013;4:182–183. doi: 10.18632/oncotarget.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio. 2014;5:e00889. doi: 10.1128/mBio.00889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Donohoe DR, Collins LB, Wali A, Bigler R, Sun W, Bultman SJ. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Molecular Cell. 2012;48:612–626. doi: 10.1016/j.molcel.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zimmerman MA, Singh N, Martin PM, Thangaraju M, Ganapathy V, Waller JL, Shi H, Robertson KD, Munn DH, Liu K. Butyrate suppresses colonic inflammation through HDAC1-dependent Fas upregulation and Fas-mediated apoptosis of T cells. American Journal of Physiology: Gastrointestinal and Liver Physiology. 2012;302:G1405–1415. doi: 10.1152/ajpgi.00543.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ganapathy V, Gurav A, Singh N. The butyrate transporter SLC5A8 is a tumor suppressor in colon linked to dietary fiber content. AACR. 2015 [Google Scholar]
- 153.Gurav A, Sivaprakasam S, Bhutia YD, Boettger T, Singh N, Ganapathy V. Slc5a8, a Na+-coupled high-affinity transporter for short-chain fatty acids, is a conditional tumour suppressor in colon that protects against colitis and colon cancer under low-fibre dietary conditions. Biochemical Journal. 2015;469:267–278. doi: 10.1042/BJ20150242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gurav A, Singh N, Ganapathy V. A critical role for Slc5a8 in the suppression of colonic inflammation by commensal bacteria-derived metabolites (MUC9P. 821) The Journal of Immunology. 2014;192:199.198–199.198. [Google Scholar]
- 155.Chapkin RS, Fan Y, Lupton JR. Effect of diet on colonic-programmed cell death: molecular mechanism of action. Toxicology Letters. 2000;112–113:411–414. doi: 10.1016/s0378-4274(99)00263-5. [DOI] [PubMed] [Google Scholar]
- 156.Fan YY, Zhang J, Barhoumi R, Burghardt RC, Turner ND, Lupton JR, Chapkin RS. Antagonism of CD95 signaling blocks butyrate induction of apoptosis in young adult mouse colonic cells. American Journal of Physiology. 1999;277:C310–319. doi: 10.1152/ajpcell.1999.277.2.C310. [DOI] [PubMed] [Google Scholar]
- 157.Kim M, Qie Y, Park J, Kim CH. Gut Microbial Metabolites Fuel Host Antibody Responses. Cell Host Microbe. 2016;20:202–214. doi: 10.1016/j.chom.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Furusawa Y, Obata Y, Hase K. Seminars in Immunopathology. Springer; 2015. Commensal microbiota regulates T cell fate decision in the gut; pp. 17–25. [DOI] [PubMed] [Google Scholar]
- 159.Crim KC, Sanders LM, Hong MY, Taddeo SS, Turner ND, Chapkin RS, Lupton JR. Upregulation of p21Waf1/Cip1 expression in vivo by butyrate administration can be chemoprotective or chemopromotive depending on the lipid component of the diet. Carcinogenesis. 2008;29:1415–1420. doi: 10.1093/carcin/bgn144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Belcheva A, Irrazabal T, Martin A. Gut microbial metabolism and colon cancer: can manipulations of the microbiota be useful in the management of gastrointestinal health? Bioessays. 2015;37:403–412. doi: 10.1002/bies.201400204. [DOI] [PubMed] [Google Scholar]
- 161.Belcheva A, Irrazabal T, Robertson SJ, Streutker C, Maughan H, Rubino S, Moriyama EH, Copeland JK, Kumar S, Green B, et al. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell. 2014;158:288–299. doi: 10.1016/j.cell.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 162.Triff K, Kim E, Chapkin RS. Chemoprotective epigenetic mechanisms in a colorectal cancer model: Modulation by n-3 PUFA in combination with fermentable fiber. Curr Pharmacol Rep. 2015;1:11–20. doi: 10.1007/s40495-014-0005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chapkin RS, DeClercq V, Kim E, Fuentes NR, Fan YY. Mechanisms by Which Pleiotropic Amphiphilic n-3 PUFA Reduce Colon Cancer Risk. Current Colorectal Cancer Reports. 2014;10:442–452. doi: 10.1007/s11888-014-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kolar SS, Barhoumi R, Lupton JR, Chapkin RS. Docosahexaenoic acid and butyrate synergistically induce colonocyte apoptosis by enhancing mitochondrial Ca2+ accumulation. Cancer Research. 2007;67:5561–5568. doi: 10.1158/0008-5472.CAN-06-4716. [DOI] [PubMed] [Google Scholar]
- 165.Chapkin RS, Davidson LA, Ly L, Weeks BR, Lupton JR, McMurray DN. Immunomodulatory effects of (n-3) fatty acids: putative link to inflammation and colon cancer. Journal of Nutrition. 2007;137:200S–204S. doi: 10.1093/jn/137.1.200S. [DOI] [PubMed] [Google Scholar]
- 166.Davidson LA, Nguyen DV, Hokanson RM, Callaway ES, Isett RB, Turner ND, Dougherty ER, Wang N, Lupton JR, Carroll RJ, Chapkin RS. Chemopreventive n-3 polyunsaturated fatty acids reprogram genetic signatures during colon cancer initiation and progression in the rat. Cancer Research. 2004;64:6797–6804. doi: 10.1158/0008-5472.CAN-04-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hong MY, Chapkin RS, Barhoumi R, Burghardt RC, Turner ND, Henderson CE, Sanders LM, Fan YY, Davidson LA, Murphy ME, et al. Fish oil increases mitochondrial phospholipid unsaturation, upregulating reactive oxygen species and apoptosis in rat colonocytes. Carcinogenesis. 2002;23:1919–1925. doi: 10.1093/carcin/23.11.1919. [DOI] [PubMed] [Google Scholar]
- 168.Chapkin RS, Hong MY, Fan YY, Davidson LA, Sanders LM, Henderson CE, Barhoumi R, Burghardt RC, Turner ND, Lupton JR. Dietary n-3 PUFA alter colonocyte mitochondrial membrane composition and function. Lipids. 2002;37:193–199. doi: 10.1007/s11745-002-0880-8. [DOI] [PubMed] [Google Scholar]
- 169.Hong MY, Lupton JR, Morris JS, Wang N, Carroll RJ, Davidson LA, Elder RH, Chapkin RS. Dietary fish oil reduces O6-methylguanine DNA adduct levels in rat colon in part by increasing apoptosis during tumor initiation. Cancer Epidemiology, Biomarkers and Prevention. 2000;9:819–826. [PubMed] [Google Scholar]
- 170.Hong MY, Turner ND, Murphy ME, Carroll RJ, Chapkin RS, Lupton JR. In vivo regulation of colonic cell proliferation, differentiation, apoptosis, and P27Kip1 by dietary fish oil and butyrate in rats. Cancer Prevention Research (Philadelphia, Pa) 2015;8:1076–1083. doi: 10.1158/1940-6207.CAPR-15-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Vanamala J, Glagolenko A, Yang P, Carroll R, Murphy M, Newman R, Ford J, Braby L, Chapkin R, Turner N. Dietary fish oil and pectin enhance colonocyte apoptosis in part through suppression of PPARδ/PGE2 and elevation of PGE3. Carcinogenesis. 2008;29:790–796. doi: 10.1093/carcin/bgm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chang WCL, Chapkin RS, Lupton JR. Fish oil blocks azoxymethane-induced rat colon tumorigenesis by increasing cell differentiation and apoptosis rather than decreasing cell proliferation. Journal of Nutrition. 1998;128:491–497. doi: 10.1093/jn/128.3.491. [DOI] [PubMed] [Google Scholar]
- 173.Kim E, Davidson LA, Patil BS, Jayaprakasha GK, Callaway ES, Turner ND, Chapkin RS. Effects of chemoprotective diets on crypt adult stem cells: the cells of origin of colon cancer (819.1) The FASEB Journal. 2014;28 [Google Scholar]
- 174.Orlich MJ, Singh PN, Sabate J, Fan J, Sveen L, Bennett H, Knutsen SF, Beeson WL, Jaceldo-Siegl K, Butler TL, et al. Vegetarian dietary patterns and the risk of colorectal cancers. JAMA Intern Med. 2015;175:767–776. doi: 10.1001/jamainternmed.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Liu RH. Health benefits of fruit and vegetables are from additive and synergistic combinations of phytochemicals. American Journal of Clinical Nutrition. 2003;78:517S–520S. doi: 10.1093/ajcn/78.3.517S. [DOI] [PubMed] [Google Scholar]
- 176.Tsao R, Yang R, Young JC, Zhu H. Polyphenolic profiles in eight apple cultivars using highperformance liquid chromatography (HPLC) Journal of Agricultural and Food Chemistry. 2003;51:6347–6353. doi: 10.1021/jf0346298. [DOI] [PubMed] [Google Scholar]
- 177.Vanamala J, Cobb G, Turner ND, Lupton JR, Yoo KS, Pike LM, Patil BS. Bioactive compounds of grapefruit (Citrus paradisi Cv. Rio Red) respond differently to postharvest irradiation, storage, and freeze drying. Journal of Agricultural and Food Chemistry. 2005;53:3980–3985. doi: 10.1021/jf048167p. [DOI] [PubMed] [Google Scholar]
- 178.Dinkova-Kostova AT. Phytochemicals as protectors against ultraviolet radiation: versatility of effects and mechanisms. Planta Medica. 2008;74:1548–1559. doi: 10.1055/s-2008-1081296. [DOI] [PubMed] [Google Scholar]
- 179.Alothman M, Bhat R, Karim A. Effects of radiation processing on phytochemicals and antioxidants in plant produce. Trends in Food Science & Technology. 2009;20:201–212. [Google Scholar]
- 180.Cowan MM. Plant products as antimicrobial agents. Clinical Microbiology Reviews. 1999;12 doi: 10.1128/cmr.12.4.564. 564-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Holt EM, Steffen LM, Moran A, Basu S, Steinberger J, Ross JA, Hong CP, Sinaiko AR. Fruit and vegetable consumption and its relation to markers of inflammation and oxidative stress in adolescents. Journal of the American Dietetic Association. 2009;109:414–421. doi: 10.1016/j.jada.2008.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Siriwardhana N, Kalupahana NS, Cekanova M, LeMieux M, Greer B, Moustaid-Moussa N. Modulation of adipose tissue inflammation by bioactive food compounds. The Journal of Nutritional Biochemistry. 2013;24:613–623. doi: 10.1016/j.jnutbio.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 183.Scalbert A, Williamson G. Dietary intake and bioavailability of polyphenols. Journal of Nutrition. 2000;130:2073S–2085S. doi: 10.1093/jn/130.8.2073S. [DOI] [PubMed] [Google Scholar]
- 184.Dykes Phenolic Compounds in Cereal Grains and Their Health Benefits. Cereal Foods World. 2007;52:105–111. [Google Scholar]
- 185.Vanamala J, Leonardi T, Patil BS, Taddeo SS, Murphy ME, Pike LM, Chapkin RS, Lupton JR, Turner ND. Suppression of colon carcinogenesis by bioactive compounds in grapefruit. Carcinogenesis. 2006;27:1257–1265. doi: 10.1093/carcin/bgi318. [DOI] [PubMed] [Google Scholar]
- 186.Leonardi T, Vanamala J, Taddeo SS, Davidson LA, Murphy ME, Patil BS, Wang N, Carroll RJ, Chapkin RS, Lupton JR, Turner ND. Apigenin and naringenin suppress colon carcinogenesis through the aberrant crypt stage in azoxymethane-treated rats. Experimental Biology and Medicine (Maywood, NJ) 2010;235:710–717. doi: 10.1258/ebm.2010.009359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yang CS, Landau JM, Huang MT, Newmark HL. Inhibition of carcinogenesis by dietary polyphenolic compounds. Annual Review of Nutrition. 2001;21:381–406. doi: 10.1146/annurev.nutr.21.1.381. [DOI] [PubMed] [Google Scholar]
- 188.Lea MA, Ibeh C, desBordes C, Vizzotto M, Cisneros-Zevallos L, Byrne DH, Okie WR, Moyer MP. Inhibition of growth and induction of differentiation of colon cancer cells by peach and plum phenolic compounds. Anticancer Research. 2008;28:2067–2076. [PubMed] [Google Scholar]
- 189.Lea MA, Ibeh C, Han L, Desbordes C. Inhibition of growth and induction of differentiation markers by polyphenolic molecules and histone deacetylase inhibitors in colon cancer cells. Anticancer Research. 2010;30:311–318. [PubMed] [Google Scholar]
- 190.Miene C, Weise A, Glei M. Impact of polyphenol metabolites produced by colonic microbiota on expression of COX-2 and GSTT2 in human colon cells (LT97) Nutrition and Cancer. 2011;63:653–662. doi: 10.1080/01635581.2011.552157. [DOI] [PubMed] [Google Scholar]
- 191.Link A, Balaguer F, Goel A. Cancer chemoprevention by dietary polyphenols: promising role for epigenetics. Biochemical Pharmacology. 2010;80:1771–1792. doi: 10.1016/j.bcp.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Banerjee N, Kim H, Talcott ST, Turner ND, Byrne DH, Mertens-Talcott SU. Plum polyphenols inhibit colorectal aberrant crypt foci formation in rats: potential role of the miR-143/protein kinase B/mammalian target of rapamycin axis. Nutrition Research. 2016;36:1105–1113. doi: 10.1016/j.nutres.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 193.Shankar E, Kanwal R, Candamo M, Gupta S. Seminars in Cancer Biology. Elsevier; 2016. Dietary phytochemicals as epigenetic modifiers in cancer: promise and challenges; pp. 82–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Rechner AR, Smith MA, Kuhnle G, Gibson GR, Debnam ES, Srai SK, Moore KP, Rice-Evans CA. Colonic metabolism of dietary polyphenols: influence of structure on microbial fermentation products. Free Radical Biology and Medicine. 2004;36:212–225. doi: 10.1016/j.freeradbiomed.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 195.Déprez S, Brezillon C, Rabot S, Philippe C, Mila I, Lapierre C, Scalbert A. Polymeric Proanthocyanidins Are Catabolized by Human Colonic Microflora into Low-Molecular-Weight Phenolic Acids. The Journal of nutrition. 2000;130:2733–2738. doi: 10.1093/jn/130.11.2733. [DOI] [PubMed] [Google Scholar]
- 196.Cardona F, Andres-Lacueva C, Tulipani S, Tinahones FJ, Queipo-Ortuno MI. Benefits of polyphenols on gut microbiota and implications in human health. The Journal of Nutritional Biochemistry. 2013;24:1415–1422. doi: 10.1016/j.jnutbio.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 197.Duenas M, Munoz-Gonzalez I, Cueva C, Jimenez-Giron A, Sanchez-Patan F, Santos-Buelga C, Moreno-Arribas MV, Bartolome B. A survey of modulation of gut microbiota by dietary polyphenols. Biomed Res Int. 2015;2015:850902. doi: 10.1155/2015/850902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Etxeberria U, Fernandez-Quintela A, Milagro FI, Aguirre L, Martinez JA, Portillo MP. Impact of polyphenols and polyphenol-rich dietary sources on gut microbiota composition. Journal of Agricultural and Food Chemistry. 2013;61:9517–9533. doi: 10.1021/jf402506c. [DOI] [PubMed] [Google Scholar]
- 199.Guo LD, Chen XJ, Hu YH, Yu ZJ, Wang D, Liu JZ. Curcumin inhibits proliferation and induces apoptosis of human colorectal cancer cells by activating the mitochondria apoptotic pathway. Phytotherapy Research. 2013;27:422–430. doi: 10.1002/ptr.4731. [DOI] [PubMed] [Google Scholar]
- 200.Stacewicz-Sapuntzakis M, Bowen PE, Hussain EA, Damayanti-Wood BI, Farnsworth NR. Chemical composition and potential health effects of prunes: a functional food? Critical Reviews in Food Science and Nutrition. 2001;41:251–286. doi: 10.1080/20014091091814. [DOI] [PubMed] [Google Scholar]
- 201.Fang N, Yu S, Prior RL. LC/MS/MS characterization of phenolic constituents in dried plums. Journal of Agricultural and Food Chemistry. 2002;50:3579–3585. doi: 10.1021/jf0201327. [DOI] [PubMed] [Google Scholar]
- 202.Piefer LA, Weeks B, Carroll R, Byrne D, Ambrus A, Turner N. Quercetin and chlorogenic acid affect butyrate excretion, NF-κB activity, and cell proliferation in DSS treated rats. The FASEB Journal. 2012;26:263.266–263.266. [Google Scholar]
- 203.Turner N, Paulhill K, Warren C, Davidson L, Chapkin R, Lupton J, Carroll R, Wang N. Quercetin suppresses early colon carcinogenesis partly through inhibition of inflammatory mediators; II International Symposium on Human Health Effects of Fruits and Vegetables: FAVHEALTH 2007 841; 2007. pp. 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Warren CA, Paulhill KJ, Davidson LA, Lupton JR, Taddeo SS, Hong MY, Carroll RJ, Chapkin RS, Turner ND. Quercetin may suppress rat aberrant crypt foci formation by suppressing inflammatory mediators that influence proliferation and apoptosis. The Journal of nutrition. 2009;139:101–105. doi: 10.3945/jn.108.096271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zhang XA, Zhang S, Yin Q, Zhang J. Quercetin induces human colon cancer cells apoptosis by inhibiting the nuclear factor-kappa B Pathway. Pharmacognosy Magazine. 2015;11:404–409. doi: 10.4103/0973-1296.153096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Refolo MG, D'Alessandro R, Malerba N, Laezza C, Bifulco M, Messa C, Caruso MG, Notarnicola M, Tutino V. Anti Proliferative and Pro Apoptotic Effects of Flavonoid Quercetin Are Mediated by CB1 Receptor in Human Colon Cancer Cell Lines. Journal of Cellular Physiology. 2015;230:2973–2980. doi: 10.1002/jcp.25026. [DOI] [PubMed] [Google Scholar]
- 207.Han M, Song Y, Zhang X. Quercetin Suppresses the Migration and Invasion in Human Colon Cancer Caco-2 Cells Through Regulating Toll-like Receptor 4/Nuclear Factor-kappa B Pathway. Pharmacognosy Magazine. 2016;12:S237–244. doi: 10.4103/0973-1296.182154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Dihal AA, de Boer VC, van der Woude H, Tilburgs C, Bruijntjes JP, Alink GM, Rietjens IM, Woutersen RA, Stierum RH. Quercetin, but not its glycosidated conjugate rutin, inhibits azoxymethane-induced colorectal carcinogenesis in F344 rats. The Journal of nutrition. 2006;136:2862–2867. doi: 10.1093/jn/136.11.2862. [DOI] [PubMed] [Google Scholar]
- 209.Peters R, Lock RH. Laxative effect of sorbitol. British Medical Journal. 1958;2:677–678. doi: 10.1136/bmj.2.5097.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Lu X, Li C, Wang YK, Jiang K, Gai XD. Sorbitol induces apoptosis of human colorectal cancer cells via p38 MAPK signal transduction. Oncology Letters. 2014;7:1992–1996. doi: 10.3892/ol.2014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Yang Y, Gallaher DD. Effect of dried plums on colon cancer risk factors in rats. Nutrition and Cancer. 2005;53:117–125. doi: 10.1207/s15327914nc5301_14. [DOI] [PubMed] [Google Scholar]
- 212.Glinghammar B, Inoue H, Rafter JJ. Deoxycholic acid causes DNA damage in colonic cells with subsequent induction of caspases, COX-2 promoter activity and the transcription factors NF-kB and AP-1. Carcinogenesis. 2002;23:839–845. doi: 10.1093/carcin/23.5.839. [DOI] [PubMed] [Google Scholar]
- 213.Baijal PK, Clow EP, Fitzpatrick DW, Bird RP. Tumor-enhancing effects of cholic acid are exerted on the early stages of colon carcinogenesis via induction of aberrant crypt foci with an enhanced growth phenotype. Canadian Journal of Physiology and Pharmacology. 1998;76:1095–1102. doi: 10.1139/cjpp-76-12-1095. [DOI] [PubMed] [Google Scholar]
- 214.Selmin OI, Fang C, Lyon AM, Doetschman TC, Thompson PA, Martinez JD, Smith JW, Lance PM, Romagnolo DF. Inactivation of Adenomatous Polyposis Coli Reduces Bile Acid/Farnesoid X Receptor Expression through Fxr gene CpG Methylation in Mouse Colon Tumors and Human Colon Cancer Cells. The Journal of nutrition. 2016;146:236–242. doi: 10.3945/jn.115.216580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Liu RH. Whole grain phytochemicals and health. Journal of Cereal Science. 2007;46:207–219. [Google Scholar]
- 216.Dykes L, Rooney LW. Sorghum and millet phenols and antioxidants. Journal of Cereal Science. 2006;44:236–251. [Google Scholar]
- 217.Awika JM, McDonough CM, Rooney LW. Decorticating sorghum to concentrate healthy phytochemicals. Journal of Agricultural and Food Chemistry. 2005;53:6230–6234. doi: 10.1021/jf0510384. [DOI] [PubMed] [Google Scholar]
- 218.Ritchie L, Taddeo S, Weeks B, Carroll R, Dykes L, Rooney L, Turner N. Impact of Novel Sorghum Bran Diets on DSS-Induced Colitis. Nutrients. 2017;9:330. doi: 10.3390/nu9040330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Ritchie LE, Sturino JM, Azcarate-Peril MA, Turner ND. Novel sorghum brans containing bioactive compounds alter colon microbiota in response to a DSS-induced chronic inflammatory state. FASEB Journal. 2013;27:247.242. [Google Scholar]
- 220.Walisser JA, Glover E, Pande K, Liss AL, Bradfield CA. Aryl hydrocarbon receptor-dependent liver development and hepatotoxicity are mediated by different cell types. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17858–17863. doi: 10.1073/pnas.0504757102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kawajiri K, Kobayashi Y, Ohtake F, Ikuta T, Matsushima Y, Mimura J, Pettersson S, Pollenz RS, Sakaki T, Hirokawa T. Aryl hydrocarbon receptor suppresses intestinal carcinogenesis in ApcMin/+ mice with natural ligands. Proceedings of the National Academy of Sciences. 2009;106:13481–13486. doi: 10.1073/pnas.0902132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Benson JM, Shepherd DM. Aryl hydrocarbon receptor activation by TCDD reduces inflammation associated with Crohn's disease. Toxicological Sciences. 2011;120:68–78. doi: 10.1093/toxsci/kfq360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Furumatsu K, Nishiumi S, Kawano Y, Ooi M, Yoshie T, Shiomi Y, Kutsumi H, Ashida H, Fujii-Kuriyama Y, Azuma T, Yoshida M. A role of the aryl hydrocarbon receptor in attenuation of colitis. Digestive Diseases and Sciences. 2011;56:2532–2544. doi: 10.1007/s10620-011-1643-9. [DOI] [PubMed] [Google Scholar]
- 224.Li Y, Innocentin S, Withers DR, Roberts NA, Gallagher AR, Grigorieva EF, Wilhelm C, Veldhoen M. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147:629–640. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 225.Wincent E, Bengtsson J, Mohammadi Bardbori A, Alsberg T, Luecke S, Rannug U, Rannug A. Inhibition of cytochrome P4501-dependent clearance of the endogenous agonist FICZ as a mechanism for activation of the aryl hydrocarbon receptor. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:4479–4484. doi: 10.1073/pnas.1118467109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Mohammadi-Bardbori A, Bengtsson J, Rannug U, Rannug A, Wincent E. Quercetin, resveratrol, and curcumin are indirect activators of the aryl hydrocarbon receptor (AHR) Chemical Research in Toxicology. 2012;25:1878–1884. doi: 10.1021/tx300169e. [DOI] [PubMed] [Google Scholar]
- 227.DiNatale BC, Murray IA, Schroeder JC, Flaveny CA, Lahoti TS, Laurenzana EM, Omiecinski CJ, Perdew GH. Kynurenic acid is a potent endogenous aryl hydrocarbon receptor ligand that synergistically induces interleukin-6 in the presence of inflammatory signaling. Toxicological Sciences. 2010;115:89–97. doi: 10.1093/toxsci/kfq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Jin UH, Lee SO, Sridharan G, Lee K, Davidson LA, Jayaraman A, Chapkin RS, Alaniz R, Safe S. Microbiome-derived tryptophan metabolites and their aryl hydrocarbon receptor-dependent agonist and antagonist activities. Molecular Pharmacology. 2014;85:777–788. doi: 10.1124/mol.113.091165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D'Angelo C, Massi-Benedetti C, Fallarino F, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 230.Diaz-Diaz CJ, Ronnekleiv-Kelly SM, Nukaya M, Geiger PG, Balbo S, Dator R, Megna BW, Carney PR, Bradfield CA, Kennedy GD. The Aryl Hydrocarbon Receptor is a Repressor of Inflammationassociated Colorectal Tumorigenesis in Mouse. Annals of Surgery. 2016;264:429–436. doi: 10.1097/SLA.0000000000001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Reid G, Sanders ME, Gaskins HR, Gibson GR, Mercenier A, Rastall R, Roberfroid M, Rowland I, Cherbut C, Klaenhammer TR. New scientific paradigms for probiotics and prebiotics. J Clin Gastroenterol. 2003;37:105–118. doi: 10.1097/00004836-200308000-00004. [DOI] [PubMed] [Google Scholar]
- 232.Delia P, Sansotta G, Donato V, Messina G, Frosina P, Pergolizzi S, De Renzis C, Famularo G. Prevention of radiation-induced diarrhea with the use of VSL# 3, a new high-potency probiotic preparation. The American journal of gastroenterology. 2002;97:2150. doi: 10.1111/j.1572-0241.2002.05946.x. [DOI] [PubMed] [Google Scholar]
- 233.Osterlund P, Ruotsalainen T, Korpela R, Saxelin M, Ollus A, Valta P, Kouri M, Elomaa I, Joensuu H. Lactobacillus supplementation for diarrhoea related to chemotherapy of colorectal cancer: a randomised study. British Journal of Cancer. 2007;97:1028–1034. doi: 10.1038/sj.bjc.6603990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Giralt J, Regadera JP, Verges R, Romero J, de la Fuente I, Biete A, Villoria J, Cobo JM, Guarner F. Effects of probiotic Lactobacillus casei DN-114 001 in prevention of radiation-induced diarrhea: results from multicenter, randomized, placebo-controlled nutritional trial. International Journal of Radiation Oncology, Biology, Physics. 2008;71:1213–1219. doi: 10.1016/j.ijrobp.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 235.Orrhage K, Sillerstrom E, Gustafsson JA, Nord CE, Rafter J. Binding of mutagenic heterocyclic amines by intestinal and lactic acid bacteria. Mutation Research. 1994;311:239–248. doi: 10.1016/0027-5107(94)90182-1. [DOI] [PubMed] [Google Scholar]
- 236.Rowland IR, Grasso P. Degradation of N-nitrosamines by intestinal bacteria. Applied Microbiology. 1975;29:7–12. doi: 10.1128/am.29.1.7-12.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Goldin B, Gorbach SL. Alterations in fecal microflora enzymes related to diet, age, lactobacillus supplements, and dimethylhydrazine. Cancer. 1977;40:2421–2426. doi: 10.1002/1097-0142(197711)40:5+<2421::aid-cncr2820400905>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 238.Reddy BS, Rivenson A. Inhibitory effect of Bifidobacterium longum on colon, mammary, and liver carcinogenesis induced by 2-amino-3-methylimidazo[4,5-f]quinoline, a food mutagen. Cancer Research. 1993;53:3914–3918. [PubMed] [Google Scholar]
- 239.Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Le Leu RK, Hu Y, Brown IL, Woodman RJ, Young GP. Synbiotic intervention of Bifidobacterium lactis and resistant starch protects against colorectal cancer development in rats. Carcinogenesis. 2010;31:246–251. doi: 10.1093/carcin/bgp197. [DOI] [PubMed] [Google Scholar]
- 242.Challa A, Rao DR, Chawan CB, Shackelford L. Bifidobacterium longum and lactulose suppress azoxymethane-induced colonic aberrant crypt foci in rats. Carcinogenesis. 1997;18:517–521. doi: 10.1093/carcin/18.3.517. [DOI] [PubMed] [Google Scholar]
- 243.Bolognani F, Rumney CJ, Pool-Zobel BL, Rowland IR. Effect of lactobacilli, bifidobacteria and inulin on the formation of aberrant crypt foci in rats. European Journal of Nutrition. 2001;40:293–300. doi: 10.1007/s394-001-8359-7. [DOI] [PubMed] [Google Scholar]
- 244.Rowland IR, Rumney CJ, Coutts JT, Lievense LC. Effect of Bifidobacterium longum and inulin on gut bacterial metabolism and carcinogen-induced aberrant crypt foci in rats. Carcinogenesis. 1998;19:281–285. doi: 10.1093/carcin/19.2.281. [DOI] [PubMed] [Google Scholar]
- 245.Kulkarni N, Reddy BS. Inhibitory Effect of Bifidobacterium Iongum Cultures on the Azoxymethane-Induced Aberrant Crypt Foci Formation and Fecal Bacterial β-Glucuronidase. Experimental Biology and Medicine. 1994;207:278–283. doi: 10.3181/00379727-207-43817. [DOI] [PubMed] [Google Scholar]
- 246.McIntosh GH, Royle PJ, Playne MJ. A probiotic strain of L. acidophilus reduces DMH-induced large intestinal tumors in male Sprague-Dawley rats. Nutrition and Cancer. 1999;35:153–159. doi: 10.1207/S15327914NC352_9. [DOI] [PubMed] [Google Scholar]
- 247.Yamazaki K, Tsunoda A, Sibusawa M, Tsunoda Y, Kusano M, Fukuchi K, Yamanaka M, Kushima M, Nomoto K, Morotomi M. The effect of an oral administration of Lactobacillus casei strain shirota on azoxymethane-induced colonic aberrant crypt foci and colon cancer in the rat. Oncology Reports. 2000;7:977–982. doi: 10.3892/or.7.5.977. [DOI] [PubMed] [Google Scholar]
- 248.Gamallat Y, Meyiah A, Kuugbee ED, Hago AM, Chiwala G, Awadasseid A, Bamba D, Zhang X, Shang X, Luo F, Xin Y. Lactobacillus rhamnosus induced epithelial cell apoptosis, ameliorates inflammation and prevents colon cancer development in an animal model. Biomedicine and Pharmacotherapy. 2016;83:536–541. doi: 10.1016/j.biopha.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 249.Silva MF, Sivieri K, Rossi EA. Effects of a probiotic soy product and physical exercise on formation of pre-neoplastic lesions in rat colons in a short-term model of carcinogenic. Journal of the International Society of Sports Nutrition. 2009;6:17. doi: 10.1186/1550-2783-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Li W, Li CB. Lack of inhibitory effects of lactic acid bacteria on 1,2-dimethylhydrazine-induced colon tumors in rats. World Journal of Gastroenterology. 2003;9:2469–2473. doi: 10.3748/wjg.v9.i11.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Raz I, Gollop N, Polak-Charcon S, Schwartz B. Isolation and characterisation of new putative probiotic bacteria from human colonic flora. British Journal of Nutrition. 2007;97:725–734. doi: 10.1017/S000711450747249X. [DOI] [PubMed] [Google Scholar]
- 252.Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. Journal of Clinical Gastroenterology. 2006;40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 253.Azcarate-Peril MA, Sikes M, Bruno-Barcena JM. The intestinal microbiota, gastrointestinal environment and colorectal cancer: a putative role for probiotics in prevention of colorectal cancer? American Journal of Physiology: Gastrointestinal and Liver Physiology. 2011;301:G401–424. doi: 10.1152/ajpgi.00110.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Azcarate-Peril MA, Altermann E, Goh YJ, Tallon R, Sanozky-Dawes RB, Pfeiler EA, O'Flaherty S, Buck BL, Dobson A, Duong T, et al. Analysis of the genome sequence of Lactobacillus gasseri ATCC 33323 reveals the molecular basis of an autochthonous intestinal organism. Applied and Environmental Microbiology. 2008;74:4610–4625. doi: 10.1128/AEM.00054-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Konishi H, Fujiya M, Tanaka H, Ueno N, Moriichi K, Sasajima J, Ikuta K, Akutsu H, Tanabe H, Kohgo Y. Probiotic-derived ferrichrome inhibits colon cancer progression via JNK-mediated apoptosis. Nature communications. 2016;7:12365. doi: 10.1038/ncomms12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Kemp MQ, Jeffy BD, Romagnolo DF. Conjugated linoleic acid inhibits cell proliferation through a p53-dependent mechanism: effects on the expression of G1-restriction points in breast and colon cancer cells. The Journal of nutrition. 2003;133:3670–3677. doi: 10.1093/jn/133.11.3670. [DOI] [PubMed] [Google Scholar]
- 257.Dubey V, Gosh AR, Bishayee K, Khuda-Bukhsh AR. Appraisal of the anti-cancer potential of probiotic Pediococcus pentosaceus GS4 against colon cancer: in vitro and in vivo approaches. Journal of Functional Foods. 2016;23:66–79. [Google Scholar]
- 258.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 259.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.O'toole PW, Flemer B. Seminars in Liver Disease. Thieme Medical Publishers; 2016. Studying the Microbiome:“Omics” Made Accessible; pp. 306–311. [DOI] [PubMed] [Google Scholar]
- 261.Weinstock GM. Genomic approaches to studying the human microbiota. Nature. 2012;489:250–256. doi: 10.1038/nature11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Kuczynski J, Stombaugh J, Walters WA, González A, Caporaso JG, Knight R. Current Protocols in Microbiology. John Wiley & Sons, Inc.; 2005. Using QIIME to Analyze 16S rRNA Gene Sequences from Microbial Communities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Franzosa EA, Morgan XC, Segata N, Waldron L, Reyes J, Earl AM, Giannoukos G, Boylan MR, Ciulla D, Gevers D, et al. Relating the metatranscriptome and metagenome of the human gut. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E2329–2338. doi: 10.1073/pnas.1319284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Sridharan GV, Choi K, Klemashevich C, Wu C, Prabakaran D, Pan LB, Steinmeyer S, Mueller C, Yousofshahi M, Alaniz RC, et al. Prediction and quantification of bioactive microbiota metabolites in the mouse gut. Nat Commun. 2014;5:5492. doi: 10.1038/ncomms6492. [DOI] [PubMed] [Google Scholar]
- 268.Gelman SJ, Patti GJ. Profiling cancer metabolism at the 'omic' level: a last resort or the next frontier? Cancer Metab. 2016;4:2. doi: 10.1186/s40170-016-0144-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA: A Cancer Journal for Clinicians. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 270.Boursi B, Arber N. Current and future clinical strategies in colon cancer prevention and the emerging role of chemoprevention. Current Pharmaceutical Design. 2007;13:2274–2282. doi: 10.2174/138161207781368783. [DOI] [PubMed] [Google Scholar]
- 271.Phillips CM. Nutrigenetics and metabolic disease: current status and implications for personalised nutrition. Nutrients. 2013;5:32–57. doi: 10.3390/nu5010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Ordovas JM, Corella D. Nutritional genomics. Annu Rev Genomics Hum Genet. 2004;5:71–118. doi: 10.1146/annurev.genom.5.061903.180008. [DOI] [PubMed] [Google Scholar]
- 273.Stover PJ. Nutritional genomics. Physiological Genomics. 2004;16:161–165. doi: 10.1152/physiolgenomics.00204.2003. [DOI] [PubMed] [Google Scholar]
- 274.Elliott R, Ong TJ. Nutritional genomics. BMJ: British Medical Journal. 2002;324:1438. doi: 10.1136/bmj.324.7351.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Trujillo E, Davis C, Milner J. Nutrigenomics, Proteomics, Metabolomics, and the Practice of Dietetics. Journal of the American Dietetic Association. 2006;106:403–413. doi: 10.1016/j.jada.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 276.Liang Q, Chiu J, Chen Y, Huang Y, Higashimori A, Fang J, Brim H, Ashktorab H, Ng SC, Ng SSM, et al. Fecal Bacteria Act as Novel Biomarkers for Noninvasive Diagnosis of Colorectal Cancer. Clinical Cancer Research. 2017;23:2061–2070. doi: 10.1158/1078-0432.CCR-16-1599. [DOI] [PubMed] [Google Scholar]
- 277.Venugopalan V, Shriner KA, Wong-Beringer A. Regulatory Oversight and Safety of Probiotic Use. Emerging Infectious Diseases. 2010;16:1661–1665. doi: 10.3201/eid1611.100574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Breton J, Tennoune N, Lucas N, Francois M, Legrand R, Jacquemot J, Goichon A, Guérin C, Peltier J, Pestel-Caron M. Gut commensal E. coli proteins activate host satiety pathways following nutrientinduced bacterial growth. Cell Metabolism. 2016;23:324–334. doi: 10.1016/j.cmet.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 279.Khan MJ, Gerasimidis K, Edwards CA, Shaikh MG. Role of gut microbiota in the aetiology of obesity: proposed mechanisms and review of the literature. Journal of Obesity. 2016;2016 doi: 10.1155/2016/7353642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Fuentes CT, Schellekens H, Hoevenaars N, Ross P, Roy B, Stanton C, Dinan TG, Cryan JF. Identification of Novel Probiotics to Modify Appetite and Satiety Directly Targeting the Ghrelin Receptor. The FASEB Journal. 2016;30:717.712–717.712. [Google Scholar]
- 281.Burcelin R. When gut fermentation controls satiety: A PYY story. Molecular Metabolism. 2017;6:10–11. doi: 10.1016/j.molmet.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Fetissov SO. Role of the gut microbiota in host appetite control: bacterial growth to animal feeding behaviour. Nature Reviews Endocrinology. 2017;13:11–25. doi: 10.1038/nrendo.2016.150. [DOI] [PubMed] [Google Scholar]