Abstract
Human gut microbiota is an important determinant for health and disease, and recent studies emphasize the numerous factors shaping its diversity. Here we performed a genome-wide association study (GWAS) of the gut microbiota using two cohorts from northern Germany totaling 1,812 individuals. Comprehensively controlling for diet and non-genetic parameters, we identify genome-wide significant associations for overall microbial variation and individual taxa at multiple genetic loci, including the VDR gene (encoding vitamin D receptor). We observe significant shifts in the microbiota of Vdr−/− mice relative to control mice and correlations between the microbiota and serum measurements of selected bile and fatty acids in humans, including known ligands and downstream metabolites of VDR. Genome-wide significant (P < 5 × 10−8) associations at multiple additional loci identify other important points of host–microbe intersection, notably several disease susceptibility genes and sterol metabolism pathway components. Non-genetic and genetic factors each account for approximately 10% of the variation in gut microbiota, whereby individual effects are relatively small.
Microbes inhabiting the human intestine mediate key metabolic, physiological and immune functions1,2, and perturbations of this ecosystem can profoundly influence health and disease3,4. As disease states can also impose secondary changes to the gut microbiota, a fundamental understanding of the forces determining gut micro-bial composition in healthy individuals is essential for deciphering the nature of disease states and developing therapeutic strategies. Assemblage of the gut community begins at birth5,6, and, once established, compositional features are resilient to perturbations7,8. The composition of the gut microbiota is highly variable among adults9,10, although family members tend to harbor more similar communities than unrelated individuals11,12. Both genetic and environmental determinants may underlie this similarity among familial microbiomes. Diet is one of the major environmental drivers for microbial community structure13,14, and other known factors include age and geography11,15 as well as the intake of medication16.
There is increasing support for a host genetic component shaping and/or structuring between-individual variability in the gut microbiota. Using 416 twin pairs, Goodrich et al.12 showed that monozygotic twins display greater overall similarity in their microbial communities than dizygotic twins and identified microbial taxa that were affected by host genetic variation. Influence of single candidate genes on the composition of the microbiome is also suggested by studies of the human gut mucosa (FUT2; ref. 17) or in mouse models (Nod2; ref. 18). A recent study using available Human Microbiome Project (HMP) metagen-omic sequencing data19 assessed associations between genome-wide genetic variation in humans and the microbiome and identified an association between the LCT gene and the abundance of bacteria in the Bifidobacterium genus. However, a small sample size (n = 93) and lack of thorough correction for known confounding factors (such as diet) represent drawbacks of this study. Here we report the results from a well-powered systematic host GWAS of the fecal microbiome in two independent but geographically matched cohorts totaling 1,812 individuals of European ancestry. A dense genomic marker set comprising a total of 6,344,846 genotyped and imputed SNPs and extensive metadata were included in the analyses, which enabled us to study the influence of host genotype, alongside dietary and other environmental factors, on between-individual variability in the gut microbiome.
RESULTS
Establishing covariables for the genetic analysis
Fecal samples were obtained from two independent cohorts of 914 individuals (PopGen20) and 1,115 individuals (Food-Chain Plus; FoCus21), both recruited at the University Hospital Schleswig-Holstein in the city area of Kiel, Germany, through the local Biobank PopGen20. For each of the 2,029 samples, high-quality 16S rRNA gene sequence data (minimum of 10,000 reads/sample) were generated, yielding a total of 38 and 374 identified phyla and genera, respectively. The two cohorts exhibited similar taxon abundance at high (Supplementary Fig. 1) and low (Supplementary Fig. 2) taxo-nomic levels, although small differences in β diversity (Bray–Curtis) were present between the cohorts (r2 = 0.026; P = 1 × 10−3), which were due to differences in age, body mass index (BMI) and sex ratio (Supplementary Table 1). A subset of 1,812 of the 2,029 individuals had available SNP array data in addition to the 16S rRNA data. Unless otherwise noted, results are presented for the combined cohort of 1,812 individuals, that is, PopGen and FoCus (results for individual cohorts are provided in Supplementary Figs. 1–3, Supplementary Table 1 and the Supplementary Note).
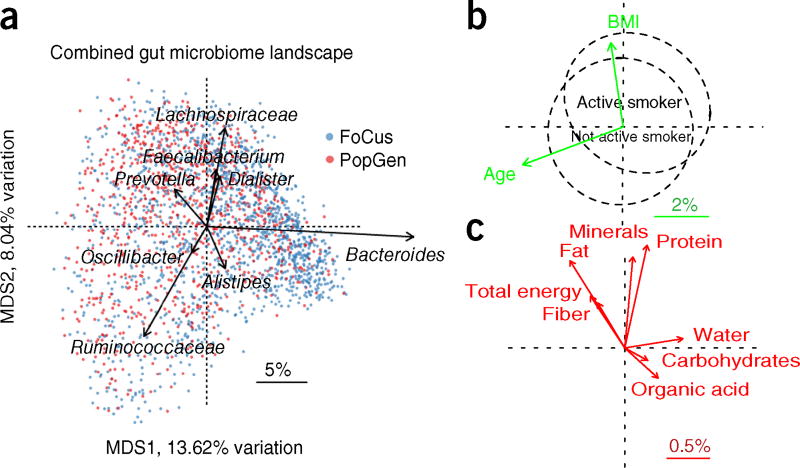
Variables previously reported to influence the gut microbiota, including age, sex, BMI11,12 and smoking status22, all displayed significant correlations with variability in the microbiome (P < 0.05; Fig. 1, Supplementary Fig. 4 and Supplementary Table 1). In terms of the percentage of variation explained (as determined through principal-coordinate analysis (PCoA) applied to Bray–Curtis dissimilarity (BC), a β-diversity measure that reflects between-individual variability), age accounted for the greatest amount (4.74%) in the combined cohort, followed by BMI, smoking and sex (3.79%, 2.14% and 1.79%, respectively; Fig. 1).
Figure 1. Overview of variation in the gut microbiota and significantly associated non-genetic parameters.
(a) PCoA of the combined cohort using BC. Arrows represent increases in the eight most abundant genera (arrow length is proportional to mean abundance; scale bar); n= 1,812. Samples are colored according to cohort. MDS1 and MDS2 are the two major axes from PCoA. (b) Correlation of age, BMI and smoking status with microbiota. For age and BMI, green arrows denote effect size (variation in β diversity explained; scale bar). Differences in smoking status are depicted as two circles with different centroids, with the dashed lines containing 50% of the samples for each group (for visualization). (c) Correlation of major nutrients with microbiota, with red arrows denoting effect size (variation in β diversity explained; scale bar). As most individual nutrients are co-linear with total energy, all arrows, save for the one for total energy, show the increase in standardized nutrient values (calculated by nutrient/total energy).
Moreover, using available food frequency data, we performed a systematic analysis of long-term diet and nutrients with respect to the microbiome. Using either the major food groups (for example, vegetables) or nutrients (for example, dietary protein content; Fig. 1, Supplementary Fig. 4 and Supplementary Table 1), we quantified the contribution to observed variability in the microbiome (PCoA applied to BC). We found that eight of the nine major nutrients and 12 of the 17 food groups displayed significant correlations in at least one cohort (Supplementary Table 1), and overall diet was significantly associated with the landscape of the human gut microbiome and explained 5.79% of the variation in BC (Fig. 1). Addition of the other significant covariates (age, sex, BMI and smoking status; P < 0.05) resulted in a total of 8.87% of the variation in BC explained (Supplementary Fig. 3). Given their detectable influence on between-individual variability, dietary variables, that is, water, alcohol and ‘total energy’ (as the best proxy for other co-linear nutrients with Pearson r > 0.5), were included as covariates in the subsequent host SNP versus microbiome association analyses.
Host genetic loci influence microbial β diversity
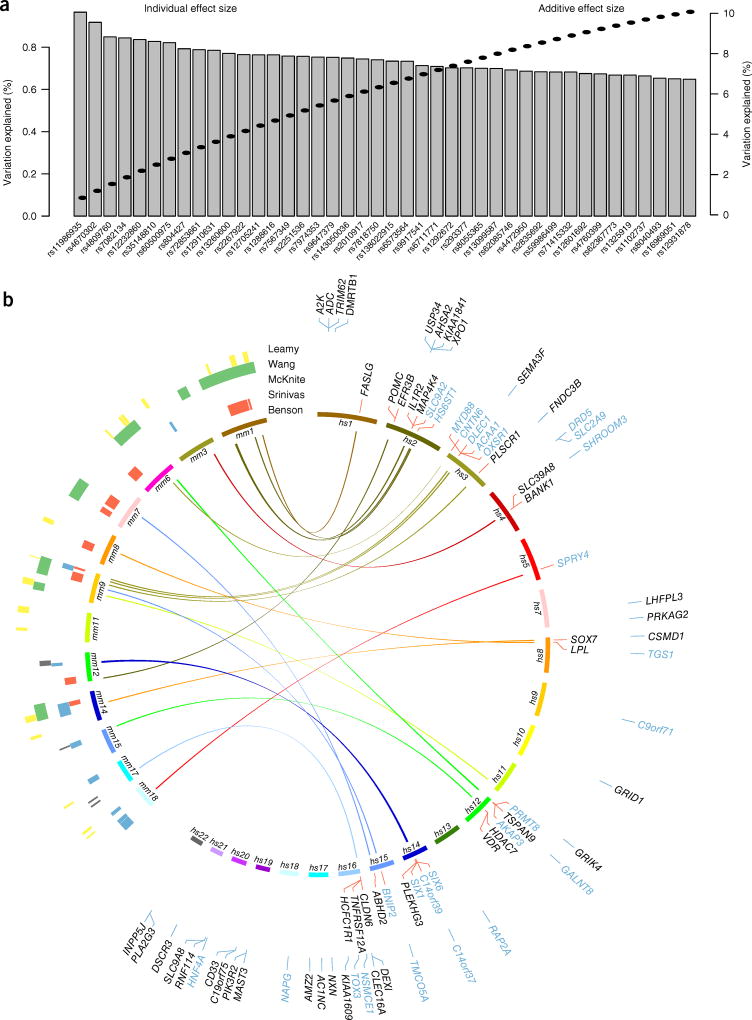
Between-individual variability is measured by β-diversity indices, which represent overall differences between microbial communities in the population and are driven by variation among multiple taxa. To identify individual loci contributing to βdiversity, we employed a multidimensional ANOVA approach, for which significance thresholds were determined for distinct classes of minor allele frequency (MAF) by performing >2 × 107 permutations to simulate the largest possible effect size (percentage variation in β diversity explained) that can occur by chance (for details, see the Online Methods). After stringent filtering based on effect sizes in the cohorts separately as well as in combination (Online Methods), this analysis showed 42 loci to be associated with β diversity (P < 5 × 10−8; Fig. 2 and Table 1), each of which contributed from 0.65 to 0.97% of the variation in community structure (measured by BC) and additively explained 10.43% in the combined cohort (Fig. 2). Of these loci, 21 could be successfully replicated in a smaller, independent cohort composed of obese individuals (FoCus obesity, n = 371), recruited from the same geographic area (Online Methods and Supplementary Table 2).
Figure 2. Individual and combined effects of significant loci and overview of all significant loci identified in this study.
(a) Effect sizes (variation in β diversity explained) for the 42 significant loci (lead SNPs) are shown in decreasing order (left axis), and additive effects (Online Methods) are shown by the dashed line (right axis). (b) Chromosomes on the right side of the plot show the chromosomal position of genes significantly associated with β diversity (black) or an individual taxon (blue). The inner circle includes genes whose mouse homologs were implicated in one or more previously published mouse QTL studies35,47–50(supplementary tables 6 and 7), denoted by a link to the corresponding mouse chromosome and appearing in the same color as the human chromosome on which the gene is located. For genes located in the outer circle, either there is no mouse homolog or the mouse homolog does not fall within a QTL.
Table 1.
Summary of loci significantly associated with β diversity
| Locus | SNP ID | Chr. | A1 | A2 | Locus start | Locus end | Nearest gene | Genes in locus | Effect size (%) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | rs804427 | 1 | A | C | 33,538,964 | 33,623,510 | AK2 | ADC, TRIM62, AK2 | 0.79 |
| 2 | rs1288616 | 1 | G | A | 53,885,577 | 53,965,248 | DMRTB1 | DMRTB1 | 0.76 |
| 3 | rs1102737 | 1 | G | A | 172,700,868 | 172,779,833 | FASLG | 0.66 | |
| 4 | rs72853661 | 2 | T | C | 25,323,083 | 25,453,968 | POMC | POMC, EFR3B | 0.79 |
| 5 | rs7567349 | 2 | A | G | 61,384,324 | 61,853,037 | XPO1 | AHSA2, USP34, XPO1, KIAA1841 | 0.76 |
| 6 | rs2010917 | 2 | T | C | 135,172,338 | 135,197,891 | MGAT 5 | MGAT 5 | 0.74 |
| 7 | rs71415332 | 2 | G | A | 102,309,520 | 102,616,128 | – | IL1R2, MAP4K4 | 0.68 |
| 8 | rs4670302 | 2 | T | G | 33,808,725 | 34,068,392 | FAM98A | FAM98A | 0.92 |
| 9 | rs6711771 | 2 | C | G | 34,339,420 | 34,491,584 | – | – | 0.71 |
| 10 | rs13099587 | 3 | G | A | 146,250,561 | 146,275,555 | PLSCR1 | PLSCR1 | 0.70 |
| 11 | rs9647379 | 3 | G | C | 171,759,410 | 171,833,266 | FNDC3B | FNDC3B | 0.75 |
| 12 | rs143050036 | 3 | C | T | 49,898,318 | 50,208,819 | SEMA3F | RBM5, MST1R, CAMKV, MON1A, RBM6, SEMA3F | 0.75 |
| 13 | rs60500975 | 4 | A | T | 102,769,693 | 102,929,034 | – | BANK1 | 0.82 |
| 14 | rs62367773 | 5 | A | G | 74,171,398 | 74,220,999 | FAM169A | 0.67 | |
| 15 | rs1292672 | 6 | C | T | 87,217,958 | 87,509,434 | HTR1E | 0.70 | |
| 16 | rs35148810 | 7 | C | T | 151,515,842 | 151,530,983 | – | PRKAG2 | 0.83 |
| 17 | rs12705241 | 7 | A | C | 104,219,681 | 104,381,102 | – | LHFPL3 | 0.76 |
| 18 | rs13260600 | 8 | C | T | 3,705,807 | 3,713,004 | CSMD1 | CSMD1 | 0.77 |
| 19 | rs138022915 | 8 | T | C | 19,815,256 | 19,939,049 | LPL | LPL | 0.73 |
| 20 | rs11986935 | 8 | T | A | 10,576,753 | 10,732,050 | SOX7 | SOX7, PINX1 | 0.97 |
| 21 | rs7818750 | 8 | G | A | 135,273,640 | 135,299,611 | ZFAT | 0.74 | |
| 22 | rs1325919 | 9 | C | T | 37,626,956 | 37,650,386 | FRMPD1 | 0.67 | |
| 23 | rs7082134 | 10 | A | G | 87,865,009 | 87,884,110 | GRID1 | GRID1 | 0.84 |
| 24 | rs2251536 | 11 | G | C | 8,852,239 | 8,853,177 | – | ST5 | 0.76 |
| 25 | rs4472950 | 11 | C | T | 120,798,714 | 120,853,675 | – | GRIK4 | 0.69 |
| 26 | rs7974353 | 12 | T | C | 48,256,280 | 48,270,596 | – | VDR | 0.75 |
| 27 | rs4760399 | 12 | T | C | 93,011,759 | 93,081,307 | C12orf74 | 0.67 | |
| 28 | rs6573564 | 14 | T | A | 65,119,676 | 65,157,187 | PLEKHG3 | 0.73 | |
| 29 | rs12910631 | 15 | G | T | 26,603,288 | 26,622,999 | – | 0.79 | |
| 30 | rs8040493 | 15 | T | G | 101,414,167 | 101,418,682 | – | 0.65 | |
| 31 | rs293377 | 15 | G | C | 89,623,490 | 89,635,268 | ABHD2 | ABHD2 | 0.70 |
| 32 | rs8055365 | 16 | T | C | 84,566,729 | 84,581,275 | KIAA1609 | KIAA1609 | 0.70 |
| 33 | rs59986499 | 16 | G | A | 3,065,924 | 3,097,940 | CLDN6 | MMP25, TNFRSF12A, CLDN6, CCDC64B, HCFC1R1, THOC6 | 0.68 |
| 34 | rs12931878 | 16 | A | G | 11,031,741 | 11,207,817 | CLEC16A | DEXI, CLEC16A | 0.65 |
| 35 | rs62085746 | 17 | T | C | 66,166,300 | 66,213,540 | AMZ2 | 0.69 | |
| 36 | rs16969051 | 17 | C | T | 32,248,813 | 32,258,877 | ACCN1 | ACCN1 | 0.65 |
| 37 | rs12601692 | 17 | A | G | 782,416 | 794,333 | – | NXN | 0.68 |
| 38 | rs2267922 | 19 | C | G | 18,217,350 | 18,289,634 | IFI30 | MAST3, IFI30, PIK3R2 | 0.77 |
| 39 | rs273647 | 19 | C | G | 51,739,767 | 51,766,748 | C19orf75 | CD33, C19orf75 | 0.84 |
| 40 | rs4809760 | 20 | A | G | 48,428,863 | 48,591,125 | SLC9A8 | RNF114, SLC9A8, SPATA2 | 0.85 |
| 41 | rs2835692 | 21 | A | G | 38,657,572 | 38,704,886 | DSCR3 | 0.68 | |
| 42 | rs9917541 | 22 | C | A | 31,520,338 | 31,531,133 | PLA2G3 | PLA2G3, INPP5J | 0.71 |
All loci have effect sizes greater than the signifcance threshold calculated from the null distribution (P < 5 × 10−8; Online Methods). The name of the lead SNP, chromosome, position, nearest gene and genes within a locus were determined using DEPICT software. A1 and A2 are the reference/alternative alleles based on the 1000 Genome Project. Chr., chromosome.
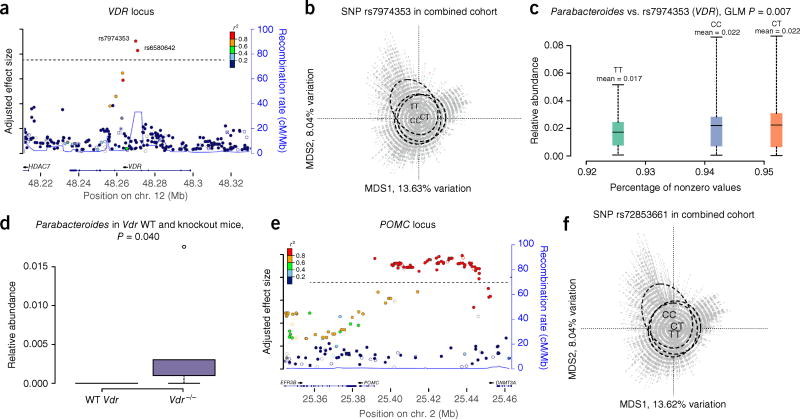
Interestingly, variants in the VDR gene (encoding vitamin D receptor) were among the 42 significant loci and accounted for 0.75% of the variation in the combined cohort (Fig. 3). VDR encodes a nuclear transcription factor, which through heterodimerization with the retinoid X receptor (RXR) exerts a range of physiological effects with many known exogenous and endogenous ligands. Besides vitamin D, both microbial (for example, secondary bile acids) and dietary (for example, fatty acids) metabolites act via the VDR–RXR heterodimer23,24. To further explore this association, we analyzed gut microbiota data from a published Vdr−/− mouse model25, confirming that loss of Vdr in mice substantially affects β diversity (42% variation in BC explained in this controlled setting; Supplementary Fig. 5). Detailed exploration of parallels between human and mouse microbiota also showed that VDR consistently influences individual bacterial taxa such as Parabacteroides (Fig. 3c,d; additional taxa are shown in Supplementary Fig. 6). Incidentally, in another data set, we observed upregulation of VDR in the colonic biopsies of patients with acute inflammation, Crohn’s disease or ulcerative colitis as compared to healthy controls, accompanied by much lower abundance of Parabacteroides, thereby further supporting such interaction (Supplementary Fig. 7 and Supplementary Note). Of note, enrichment analysis of genetic loci significantly associated with individual taxa (Table 2) showed vitamin D response as the fourth most significantly associated gene set (Table 3).
Figure 3. VDR and POMC as examples of genes associated with β diversity.
(a) LocusZoom plot of adjusted effect size (for each SNP, the actual effect size is divided by the significance threshold adjusted according to MAF category, represented by the dashed line; Online Methods) at the VDR locus, where two SNPs passed the significance threshold for association with β diversity (P < 5 × 10−8 for association with overall microbiome variation, measured by BC). (b) Association between genotypes at the lead SNP (rs7974353) and β diversity (BC). Microbiome data are shown in a PCoA plot; the dashed lines contain 50% of the samples for each group (for visualization) and show differences in the centroids for each genotype group; n = 1,812. (c) Meta-analysis in humans shows Parabacteroides to be the most significant taxon correlated with VDR using a GLM (Online Methods). The x axis shows the percentage of nonzero values for each genotype at rs7974353, and boxes and bars summarize 50% and 95% confidence intervals, respectively, for nonzero values; n = 1,812. (d) Knockout of Vdr25 in mice also leads to changes in Parabacteroides abundance. Error bars, 5–95% confidence intervals (n = 3 wild-type (WT) mice and n = 5 knockout mice; supplementary Fig. 6 and supplementary Note). (e) LocusZoom plot for adjusted effect size in the region upstream of POMC, where 78 SNPs passed the significance threshold. (f) Association between the genotypes of the lead SNP at POMC (rs72853661) and β diversity (BC). Microbiome data are shown in a PCoA plot; the dashed lines contain 50% of the samples for each group (for visualization) and show differences in the centroids for each genotype group; n = 1,812.
Table 2.
Loci associated with bacterial abundance
| Locus | Bacteria | SNP | A1 | A2 | Meta P | Meta β | β-div P | Chr. | Locus start | Locus end | Nearest gene | Genes in locus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Unclassifed Enterobacteriaceae | rs938295 | C | T | 2.34 × 10−8 | −0.49 | 0.76 | 1 | 16,087,164 | 16,124,985 | FBLIM1 | FBLIM1 |
| 2 | Unclassifed Acidaminococcaceae | rs75036654 | C | T | 4.94 × 10−10 | −1.39 | 0.06 | 1 | 37,717,219 | 37,780,821 | LINC01137 | |
| 3 | OTU13305 Fecalibacterium Species-level OTU | rs597205 | T | C | 7.68 × 10−9 | −0.62 | 0.85 | 1 | 112,379,026 | 112,415,622 | C1orf183 | C1orf183 |
| 4 | Blautia genus | rs4669413 | T | C | 1.2 × 10−8 | −0.18 | 0.75 | 2 | 9,801,744 | 9,818,596 | RP11–521D12.1 | |
| 5 | Blautia genus | rs79387448 | C | T | 7.68 × 10−11 | −0.31 | 0.66 | 2 | 103,099,953 | 103,239,356 | SLC9A2 | SLC9A2 |
| 6 | Bacilli class | rs10928827 | G | A | 1.02 × 10−8 | −0.22 | 0.19 | 2 | 129,426,740 | 129,473,850 | HS6ST1 | |
| Lactobacillales order | rs10928827 | G | A | 4.19 × 10−9 | −0.23 | 0.19 | ||||||
| 7 | Gammaproteobacteria class | rs4621152 | C | T | 1.4 × 10−8 | −0.29 | 0.79 | 2 | 217,857,450 | 217,924,261 | AC007557.1 | |
| 8 | Unclassifed Acidaminococcaceae | rs56006724 | A | G | 6.35 × 10−10 | −0.88 | 0.93 | 2 | 228,486,044 | 228,523,585 | C2orf83 | C2orf83 |
| 9 | Marinilabiliaceae family | rs11915634 | T | C | 2.99 × 10−10 | −1.30 | 0.14 | 3 | 1,452,602 | 1,517,331 | CNTN6 | |
| Unclassifed Marinilabiliaceae | rs11915634 | T | C | 2.99 × 10−10 | −1.30 | 0.14 | ||||||
| 10 | OTU10032 unclassifed Enterobacteriaceae Species-level OTU | rs3925158 | C | G | 6.29 × 10−9 | −1.00 | 0.78 | 3 | 38,161,078 | 38,313,688 | SLC22A13 | SLC22A13, MYD88, DLEC1, ACAA1, OXSR1 |
| 11 | EscherichiaShigella | rs13096731 | A | G | 2.55 × 10−8 | −0.43 | 0.12 | 3 | 58,014,818 | 58,089,851 | FLNB | FLNB |
| 12 | Lactobacillales order | rs59042687 | T | G | 6.22 × 10−9 | −0.23 | 0.02 | 3 | 95,359,287 | 95,823,523 | LINC00879 | |
| 13 | Unclassifed Marinilabiliaceae | rs9831278 | C | T | 2.53 × 10−8 | −1.16 | 0.49 | 3 | 98,879,786 | 98,942,990 | LINC00973 | |
| Marinilabiliaceae family | rs9831278 | C | T | 2.53 × 10−8 | −1.16 | 0.49 | ||||||
| 14a | Lactobacillales order | rs62295801 | G | T | 5.32 × 10−10 | −0.27 | 0.21 | 3 | 162,444,724 | 163,236,170 | LINC01192 | LINC01192 |
| 15 | Bacilli class | rs7646786 | T | C | 2.29 × 10−8 | −0.22 | 0.5 | 3 | 185,729,634 | 185,742,372 | LOC344887 | |
| 16 | Unclassifed Porphyromonadaceae | rs7656342 | A | G | 2.8 × 10−9 | 0.39 | 0.22 | 4 | 9,721,358 | 9,895,176 | DRD5 | SLC2A9, DRD5 |
| 17b | Marinilabiliaceae family | rs11724031 | G | A | 2.44 × 10−10 | −0.97 | 0.68 | 4 | 77,441,448 | 77,467,405 | SHROOM3 | SHROOM3 |
| Unclassifed Marinilabiliaceae | rs11724031 | G | A | 2.44 × 10−10 | −0.97 | 0.68 | ||||||
| Marinilabiliaceae family | rs9996716 | G | A | 5.58 × 10−9 | −0.69 | 0.2 | ||||||
| Unclassifed Marinila-biliaceae | rs9996716 | G | A | 5.58 × 10−9 | −0.69 | 0.2 | ||||||
| 18 | Erysipelotrichaceae family | rs17421787 | C | G | 3.6 × 10−8 | −0.30 | 0.16 | 4 | 131,293,675 | 131,512,291 | RP11-22J15.1 | |
| Erysipelotrichales order | rs17421787 | C | G | 3.6 × 10−8 | −0.30 | 0.16 | ||||||
| Erysipelotrichia class | rs17421787 | C | G | 3.6 × 10−8 | −0.30 | 0.16 | ||||||
| 19 | Unclassifed Porphyromonadaceae | rs9291879 | C | T | 3.51 × 10−9 | −0.58 | 0.08 | 5 | 66,515,817 | 66,550,855 | CD180 | |
| 20 | OTU10032 unclassifed Enterobacteriaceae | rs249733 | T | C | 4.74 × 10−10 | −0.65 | 0.68 | 5 | 141,877,862 | 141,911,748 | SPRY4 | |
| 21 | Unclassifed Acidaminococcaceae | rs17661843 | T | C | 3.72 × 10−14 | −1.40 | 0.26 | 7 | 48,381,902 | 48,433,594 | ABCA13 | ABCA13 |
| 22 | OTU10032 unclassifed Enterobacteriaceae | rs13276516 | A | G | 5.54 × 10−9 | −0.61 | 0.41 | 8 | 56,589,428 | 56,596,140 | TGS1 | |
| 23 | OTU10032 unclassifed Enterobacteriaceae Species-level OTU | rs2318350 | T | C | 3.65 × 10−9 | −1.15 | 0.95 | 8 | 139,889,972 | 139,942,500 | COL22A1 | COL22A1 |
| 24 | OTU10032 unclassifed Enterobacteriaceae | rs17085775 | C | T | 2.06 × 10−8 | −1.03 | 0.54 | 9 | 71,165,704 | 71,167,878 | C9orf71 | |
| 25 | Lactobacillales order | rs7083345 | T | C | 2.89 × 10−9 | 0.24 | 0.02 | 10 | 7,020,329 | 7,044,987 | RP11-554I8.2 | |
| Bacilli class | rs7083345 | T | C | 3.38 × 10−10 | 0.25 | 0.02 | 10 | 7,020,329 | 7,044,987 | RP11-554I8.2 | ||
| 26 | Lactobacillales order | rs7113056 | C | T | 1.72 × 10−13 | −0.50 | 0.07 | 11 | 122,091,502 | 122,154,110 | RP11-166D19.1 | |
| 27 | Bacilli class | rs479105 | T | C | 1.21 × 10−8 | −0.22 | 0.48 | 12 | 3,357,596 | 3,393,503 | PRMT8 | |
| 28 | OTU10032 unclassifed Enterobacteriaceae Species-level OTU | rs1009634 | G | A | 7.12 × 10−9 | −1.31 | 0.93 | 12 | 4,779,313 | 4,900,344 | AKAP3 | NDUFA9, GALNT8, RP11-234B24.2 |
| 29 | Gammaproteobacteria class | rs9300430 | C | T | 1.3 × 10−9 | −0.61 | 0.12 | 13 | 98,269,478 | 98,306,405 | RAP2A | |
| 30 | Proteobacteria phylum | rs9323326 | A | G | 8.76 × 10−10 | −0.21 | 0.02 | 14 | 58,476,448 | 58,532,709 | SLC35F4 | C14orf37 |
| 31 | Unclassifed Acidaminococcaceae | rs986417 | C | T | 2.63 × 10−9 | −1.40 | 0.47 | 14 | 60,787,269 | 61,122,040 | SIX6 | SIX6, C14orf39, SIX1 |
| 32 | Unclassifed Erysipelotrichaceae | rs11626933 | G | A | 1.83 × 10−8 | −0.24 | 0.55 | 14 | 90,681,816 | 90,810,659 | C14orf102 | C14orf102 |
| 33 | OTU15355 Dialister Species-level OTU | rs12442649 | G | A | 3.72 × 10−8 | −1.49 | 0.85 | 15 | 37,968,393 | 38,035,538 | TMCO5A | |
| 34 | Enterobacteriaceae family | rs35275482 | C | A | 3.72 × 10−11 | −0.54 | 0.06 | 15 | 60,027,987 | 60,128,040 | BNIP2 | |
| Enterobacteriales order | rs35275482 | C | A | 3.72 × 10−11 | −0.54 | 0.06 | ||||||
| 35 | OTU10032 unclassifed Enterobacteriaceae | rs12149695 | A | T | 1.82 × 10−9 | 0.61 | 0.23 | 16 | 27,205,994 | 27,293,886 | FLJ21408 | NSMCE1, FLJ21408, KDM8 |
| 36 | Lactobacillales order | rs1362404 | T | G | 1.56 × 10−8 | 0.23 | 7.5 × 10−5 | 16 | 51,955,443 | 52,017,380 | TOX3 | |
| 37 | Erysipelotrichaceae family | rs11877825 | G | T | 2.82 × 10−11 | −0.27 | 0.34 | 18 | 10,566,345 | 10,595,758 | NAPG | |
| Erysipelotrichia class | rs11877825 | G | T | 2.82 × 10−11 | −0.27 | 0.34 | ||||||
| Erysipelotrichales order | rs11877825 | G | T | 2.82 × 10−11 | −0.27 | 0.34 | ||||||
| 38 | Bacilli class | rs148330122 | C | T | 1.32 × 10−9 | −0.48 | 0.18 | 19 | 38,497,288 | 38,631,252 | SIPA1L3 | SIPA1L3 |
| 39 | Bacilli class | rs2071199 | T | C | 1.24 × 10−8 | −0.32 | 0.58 | 20 | 43,030,809 | 43,037,422 | HNF4A–AS1 | HNF4A |
| 40 | Actinobacteria class | rs34613612 | C | G | 6.34 × 10−10 | 0.25 | 9.87 × 10−3 21 | 32,184,901 | 32,204,347 | KRTAP8-1 | KRTAP8-1 | |
| Actinobacteria phylum | rs34613612 | C | G | 6.34 × 10−10 | 0.25 | 9.87 × 10−3 |
The 54 associations with bacterial abundance are grouped into 40 loci on the basis of LD. “Locus” corresponds to locus number, “Bacteria” corresponds to the trait associated with a locus, “SNP” corresponds to the tag SNP for a locus–trait pair, “A1” is the allele for which association is analyzed, “A2” is the opposite allele, “Meta P” is the meta-analysis P value for A1, “Meta β” is the meta-analysis coeffcient for A1, “β-div P” is the P value for association with β diversity (Online Methods), “Chr.” corresponds to the chromosome, “Locus start” is the genetic position at which the locus starts and “Locus end” is the genetic position at which the locus ends, “Nearest gene” is the nearest gene to the SNP according to DEPICT; “Genes in locus” includes genes found in the locus according to DEPICT.
Locus 14 contains the rs9290183 hit in addition to rs62295801, although PLINK does not clump these SNPs together. bLocus 17 includes rs9996716, which is located 219 bp downstream of the end of the locus according to DEPICT.
Table 3.
Gene set and tissue enrichment results for associations with individual bacterial traits
| Top 20 enriched gene sets
|
Top 20 Enriched Tissues
|
||||||
|---|---|---|---|---|---|---|---|
| Original gene set ID |
Original gene set description | Nominal P | Name | MeSH first-level term |
MeSH second-level term |
Nominal P | MeSH term |
| GO:0007566 | Embryo implantation | 2.29 × 10−5 | Keratinocytes | Cells | Epithelial cells | 2.85 × 10−3 | A11.436.397 |
| MP:0009402 | Decreased skeletal muscle fber diameter | 4.09 × 10−5 | Intestines | Digestive system | Gastrointestinal tract | 5.57 × 10−3 | A03.556.124 |
| GO:0033273 | Response to vitamin | 4.77 × 10−5 | Gastrointestinal tract | Digestive system | Gastrointestinal tract | 6.7 × 10−3 | A03.556 |
| GO:0033280 | Response to vitamin D | 8.8 × 10−5 | Lower gastrointestinal tract |
Digestive system | Gastrointestinal tract | 6.92 × 10−3 | A03.556.249 |
| MP:0006317 | Decreased urine sodium level | 1.19 × 10−4 | Colon | Digestive system | Gastrointestinal tract | 7.07 × 10−3 | A03.556.249.249.356 |
| GO:0071496 | Cellular response to external stimulus | 1.73 × 10−4 | Intestine, large | Digestive system | Gastrointestinal tract | 7.27 × 10−3 | A03.556.249.249 |
| MP:0010027 | Increased liver cholesterol level | 1.73 × 10−4 | Hepatocytes | Cells | Epithelial cells | 9.19 × 10−3 | A11.436.348 |
| MP:0000221 | Digestive system | 1.75 × 10−4 | Ileum | Digestive system | Gastrointestinal tract | 9.96 × 10−3 | A03.556.249.124 |
| GO:0007229 | Integrin-mediated signaling pathway | 1.97 × 10−4 | Rectum | Digestive system | Gastrointestinal tract | 0.01 | A03.556.124.526.767 |
| GO:0031668 | Cellular response to extracellular stimulus | 2.33 × 10−4 | Intestinal mucosa | Digestive system | Gastrointestinal tract | 0.02 | A03.556.124.369 |
| GO:0033189 | Response to vitamin A | 2.5 × 10−4 | Mucous membrane | Tissues | Membranes | 0.02 | A10.615.550 |
| GO:0031669 | Cellular response to nutrient levels | 3.03 × 10−4 | Colon, sigmoid | Digestive system | Gastrointestinal tract | 0.02 | A03.556.249.249.356.668 |
| GO:0055093 | Response to hyperoxia | 3.09 × 10−4 | Epithelial cells | Cells | Epithelial cells | 0.03 | A11.436 |
| ENSG00000215328 | HSPA1A PPI subnetwork | 3.27 × 10−4 | Hypothalamo hypophyseal system | Nervous system | Central nervous system | 0.03 | A08.186.211.730.317.357. 352.435 |
| ENSG00000143393 | PI4KB PPI subnetwork | 3.37 × 10−4 | Neurosecretory systems | Nervous system | Neurosecretory systems | 0.03 | A08.713 |
| MP:0005266 | Abnormal metabolism | 3.51 × 10−4 | Hypothalamus, middle | Nervous system | Central nervous system | 0.03 | A08.186.211.730.317.35 7.352 |
| GO:0048545 | Response to steroid hormone stimulus | 3.54 × 10−4 | Membranes | Tissues | Membranes | 0.04 | A10.615 |
| GO:0009991 | Response to extracellular stimulus | 3.7 × 10−4 | Monocyte macrophage precursor cells | Cells | Myeloid cells | 0.04 | A11.627.624.249 |
| GO:0033143 | Regulation of intracellular steroid hormone receptor signaling pathway | 4.29 × 10−4 | Urinary bladder | Urogenital system | Urinary tract | 0.04 | A05.810.890 |
| GO:0031490 | Chromatin DNA binding | 4.59 × 10−4 | Intestine, small | Digestive system | Gastrointestinal tract | 0.04 | A03.556.124.684 |
Enrichment analysis was performed using DEPICT for the 40 loci associated with individual bacterial traits. The table shows the 20 most enriched gene sets (three left columns) and the 20 most enriched tissues or cell types (fve right columns). For tissue and cell type enrichment, headings are given for minimum two MeSH levels: frst- and second-level terms. The “Name” column contains the name for the lowest level term with enrichment. The MeSH codes for the hierarchical branch are given in the “MeSH term” column. Analysis for enriched genes and analysis for enriched tissues were independent of each other.
The gut microbiome is essential for bile acid metabolism, and bile acids act as both key VDR ligands and regulators of VDR expression23,24,26. In addition, polyunsaturated fatty acids act as ligands for RXR, the heterodimeric partner of VDR, and were shown to compete for ligand binding to VDR23. We therefore performed targeted measurement of bile acids and ω3 and ω6 polyunsaturated fatty acids in human serum in a subset of the PopGen cohort (n = 551). We found significant correlations between several bile acids and β diversity (BC), including taurochenodeoxycholic acid (TCDCA; 2.2% variation explained) and glycochenodeoxycholic acid (GCDCA; 1.4% variation explained; Supplementary Fig. 8 and Supplementary Table 3). Bile acids also significantly associated with individual bacterial taxa, including the secondary bile acids lithocholic acid (LCA; a known VDR ligand) and deoxycholic acid (DCA; Supplementary Table 3), both of which are produced by the gut microbiota24. In addition, genomic analysis showed that Parabacteroides bacteria contain pathways involved in secondary bile acid metabolism (Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway pdi00121) and could thus indeed be associated with bile acid profiles, a hypothesis that is further supported by positive correlations between Parabacteroides abundance and LCA concentration (Supplementary Table 3). Furthermore, functional profiling of the gut microbiome via shotgun metagenomic analysis in a subset of the PopGen cohort (n = 122) also showed differences in bile acid–related gene pathways with respect to VDR genotype (Supplementary Fig. 9). Finally, the above-mentioned data from colonic biopsies also suggested that the interplay between VDR and Parabacteroides involves two genes associated with bile acid metabolism (CYP27A1, encoding cytochrome P450 family 27 subfamily A member 1, and NR5A2, encoding nuclear receptor subfamily 5 group A member 2; Supplementary Fig. 7), with interactions lost in the context of intestinal inflammation (Supplementary Fig. 7). Together, these findings provide evidence that the gut microbiota significantly contributes to human bile acid profiles, as previously reported in mice27. For fatty acids (false discovery rate (FDR) < 0.05), we detected significant correlations between the gut microbiota and 7 of 15 polyunsaturated fatty acids, including arachidonic acid (an ω6 fatty acid that is capable of binding VDR), which correlated with β diversity (1.22% variation explained) and several specific taxa (Supplementary Table 4). Of note, two additional genome-wide significant associations with the gut microbiota are critically involved in bile acid (HNF4A; Table 2) and arachidonic acid (PLA2G3; Table 1) homeostasis28,29. Finally, several loci identified in this study, in addition to VDR, were significantly correlated with bile acid profiles, as shown by regression analysis (Supplementary Table 5).
Many other interesting findings are found among the 42 significant loci (Table 1), in particular the POMC (proopiomelanocortin) gene (rs72853661, P < 5 × 10−8; Fig. 3e). As an extremely functionally diverse protein, POMC participates in multiple physiological processes ranging from antimicrobial activity to appetite regulation (Supplementary Table 6). Furthermore, this locus located upstream of the POMC gene is the largest we discovered (78 SNPs over 54.8 kb; Fig. 3e) and contains multiple SNPs that regulate the expression of POMC in multiple human tissues, as determined by expression quantitative trait locus (eQTL) studies (GTEx database). The associated SNP rs66589178 in particular is predicted to be a VDR binding site (RegulomeDB), and the TRAP analysis tool predicted an almost 200-fold difference in affinity for VDR between the two alleles (Supplementary Fig. 10). Other findings include the HTR1E (serotonin receptor) and GRID1 (glutamate ionotropic receptor) genes, which are potential components of the gut–brain axis30, and genetic variation near CLEC16A (rs12931878, P < 5 × 10−8), a gene associated with multiple autoimmune and inflammatory disorders involving alterations to gut microbiota (Supplementary Table 6). A number of other regions are implicated in disease susceptibility as previously reported by case-control GWAS and can be found in Table 1 and Supplementary Tables 6 and 7 (for example, BANK1 close to SCL39A8).
Finally, a targeted analysis was performed for the human leukocyte antigen (HLA) complex on chromosome 6. The HLA complex shapes the immune repertoire and may influence gut microbiome composition31. Because SNPs do not capture the extreme polymorphism of the classical HLA genes, we imputed HLA alleles using SNP2HLA (Online Methods) and implemented a constrained ordination approach. This approach showed significant association of alleles at HLA-B (HLA-B*52:01) and HLA-C (HLA-C*12:02) in both cohorts (P < 0.05; Supplementary Fig. 11 and Supplementary Table 8). The associated alleles have been implicated in risk for ulcerative colitis in multiple ancestry groups32,33 and in Takayasu arteritis34.
Genetic associations with individual bacterial traits
To detect associations between genetic variants and specific bacterial traits, we first curated the microbiome data and removed rare bacteria by defining a ‘core measurable microbiota’ (ref. 35) (Supplementary Fig. 12 and Supplementary Table 9), which included 40 operational taxonomic units (OTUs) and 58 taxa ranging from the genus to the phylum level, and employed a generalized linear model (GLM) framework incorporating a negative binomial (negbin) distribution. Accordingly, we identified 54 significant associations involving 40 loci and 22 bacterial traits (meta-analysis P < 5 × 10−8 and single-cohort P < 5 × 10−4; Table 2). Of the 22 bacterial traits, the largest number belonged to Firmicutes (n = 10), followed by Proteobacteria (n = 7), Bacteroidetes (n = 3) and Actinobacteria (n = 2), at the phylum level. To identify the nearest and neighboring genes for each locus, we annotated the identified SNPs using DEPICT36 (Table 3).
Among the 54 robust associations, the SLC2A9 gene was associated with unclassified Porphyromonadaceae (rs7656342, meta-analysis P = 2.8 × 10−9) (Supplementary Fig. 13). The SLC2A9 gene encodes a member of the glucose transporter family, which is important for maintaining glucose homeostasis37. Furthermore, a number of long intergenic noncoding RNAs were among the 54 associations, including association of LINC01192 with Lactobacillales (rs62295801, meta-analysis P = 5.32 × 10−10) (Supplementary Fig. 13). Of note, gene set enrichment analysis detected associations for LINC01192 with ‘response to vitamin A’ and for SLC2A9 with both ‘response to vitamin D’ and ‘increased liver cholesterol level’ (Table 3).
Next, we evaluated whether the genetic signal for β diversity is influenced by the abundance of individual bacterial taxa. Indeed, 37 loci that correlated with β diversity also correlated with the abundance of several core measurable microbiota taxa and OTUs (P < 0.01), albeit not at the genome-wide significance level (Supplementary Fig. 14). Conversely, the loci identified in association analyses for individual taxa explained a proportion of the variation in β diversity (six loci with P < 0.05, effect size of 0.29–0.49%) but did not reach our conservative significance threshold of P < 5 × 10−8 (Table 2). Thus, in conclusion, we found that genetic variants correlating with microbiome structure could be either strongly associated with an individual taxon or simultaneously associated with multiple taxa, with each association having a small effect size.
Enrichment analysis of gene sets and tissues
To further assess the functional relevance of the 54 identified associations between genetic variants and specific bacterial traits, we used DEPICT36 to perform both gene set and tissue enrichment analyses (Table 3). DEPICT prioritizes genes in associated regions on the basis of functional relationships and linkage disequilibrium (LD) structure. Of interest, ‘response to vitamin D’ (original gene set ID GO:0033280, P = 8.8 × 10−5) was the fourth most enriched term. Enrichment of response to vitamins in general was also observed, including ‘response to vitamin A’, another fat-soluble vitamin binding to the retinoic acid receptor (RAR) and involved in bile acid homeostasis38,39. The gene set for ‘response to vitamin D’ includes SLC22A13, SLC2A9, COL22A1, ABCA13 and KRTAP8-1 (Table 2). The VDR gene locus itself, however, is not included, as the enrichment analysis was limited to loci associated with single bacterial taxa, and the association with Parabacteroides (Fig. 3c) did not reach the genome-wide significance threshold. Further, the term ‘increased liver cholesterol level’ was among the top enriched gene sets (original gene set ID MP:0010027, P = 1.7 × 10−4) and corresponds to one of the functions of the POMC gene locus identified in the above analysis. Among the bacterial taxa associated with ‘increased liver cholesterol level’ were Gammaproteobacteria, Bacilli, unclassified Porphyromonadaceae and an OTU belonging to Enterobacteriaceae. Furthermore, in a trans-eQTL analysis of the SNPs associated with β diversity or single bacterial taxa (Supplementary Tables 6 and 7), FDFT1, which encodes the first specific enzyme in cholesterol synthesis, was among the top hits, further emphasizing the fact that several hits converge onto the sterol pathway.
In the tissue enrichment analysis, the top 20 results with P < 0.05 (Table 3) showed the Medical Subject Heading (MeSH) terms ‘digestive system’ (10 occurrences), ‘nervous system’ (3 occurrences) and ‘cells’ (3 occurrences) as most significant. The best associated subcategories for ‘digestive system’ were ‘intestinal mucosa’ and ‘mucous membrane’, whereas the subcategories for ‘cells’ included ‘mono-cyte macrophage precursor cells’, ‘epithelial cells’ and ‘hepatocytes’. In sum, the tissue enrichment analysis relates microbial-associated host loci with gastrointestinal and immune-related tissues and cells, thus supporting the functional relevance of the identified loci.
DISCUSSION
We herein present a comprehensive analysis of genome-wide host–microbiota associations. We adhered to rigorous standards by including a large number of samples (1,812 SNP array–16S rRNA microbiome data set pairs) and considering important known and herein identified confounders of variation in the gut microbiome. As geography is a major factor contributing to microbiome composition11,15, we used cohorts recruited from the same country and corrected for population stratification/ancestry in our genetic data set. We discovered genome-wide significant associations between gut microbial characteristics and the VDR gene, in addition to a large number of other host genetic factors, and eventually quantified the total contribution of host genetic loci to β diversity as 10.43%. The non-genetic factors examined (age, sex, BMI, smoking status and dietary patterns) explain 8.87% of the observed variation in the gut microbiome.
As shown in Supplementary Figure 15, the associations at the VDR locus with gut microbial community composition provide compelling follow-up to the finding by Makishima et al.24 that secondary bile acids (bile acids transformed by gut microbial metabolism, that is, LCA, glycine-conjugated LCA and 3-keto-LCA from 7α-dehydroxylated primary CDCA) serve as ligands for VDR. Validation of a relationship between VDR alterations and the gut microbiota in the Vdr−/− mouse model25 substantiates these observations. Results from gene set enrichment analysis and the observation that the bile acid profile in serum associates with variation in the gut microbiome further support this finding. The underlying mechanisms for the observed association between gut microbial profiles and the serum bile acid pool warrant further elaboration. The possibility that VDR-mediated signaling serves as a key mediator in the gut–liver signaling axis and microbial co-metabolism, as previously shown for FXR (farnesoid X receptor27), motivates substantial new research directions. Although the lack of an association at the FXR locus (Supplementary Fig. 16) does not signify the lack of FXR involvement in microbial bile acid co-regulation23 (for example, functional variation may simply not be present in our cohort), the VDR associations detected in the present study add another important player to this relationship.
Insight on interactions between the microbiome and bile acid homeostasis are mostly based on mouse studies27,40,41, for which the transfer of interpretations into the human setting may be considerably biased given the large differences in bile acid profiles between mice and humans. Additional data presented in Supplementary Tables 6 and 7 show cross-validation for a subset of the genes detected in the human analysis, including VDR, whereby differential expression in germ-free and conventionally raised mice further supports the roles of these genes in interacting with and/or maintaining the homeostasis of the gut microbiome. Such overlap between distantly related mammalian hosts provides strong support for our discoveries and, hence, the internal validity of our experiment. Genetic associations at the VDR locus were also detected in human inflammatory bowel disease and liver disease42,43, for which the underlying mechanisms were proposed to be a perturbation of key aspects of host– microbe interactions44. The multidimensional relationship of key factors involved in VDR signaling (bile acids and ω6 fatty acids in particular) and the gut microbiota is even supported by genetic associations at functionally related loci (HNF4A and PLA2G3).
The POMC locus gives rise to a number of proopiomelanocortin-derived peptides involved in various physiological processes, including blood sugar regulation, inflammation and energy intake45, and association of SNP rs66589178, potentially affecting a VDR binding site (Supplementary Fig. 10), is an additional interesting circumstantial observation for the VDR finding. On the basis of their broad influence on bacterial community structure (contribution to β diversity as measured by BC) in our cohorts, VDR and POMC (among other genes) could be major regulators of the gut microbiome. Given that VDR and POMC are further associated with numerous important phenotypes (Supplementary Tables 6 and 7), our results provide a strong indication for genetic associations across phenotypes, including BMI, Crohn’s disease and the intestinal microbiome. However, further dedicated studies are still needed to link these pleiotropic signaling pathways and their associated biology46. Finally, understanding the functional consequences of the genetic variants discovered in this study will also require in-depth exploration, as the functional consequences of the lead SNPs remain unknown (for example, VDR lead SNP rs7974353).
Genome-wide screening for host genetic associations with gut microbiome composition has mostly been performed in mice, for which environmental factors and genetic background are easy to control. Thus, to further validate our findings, we compared our results to previously published QTL studies for the mouse gut microbiome. We found that mouse homologs of numerous GWAS hits in our study are contained in the confidence intervals of mouse QTLs (Fig. 2b). One such overlap even involves association with an identical trait— between the SLC9A2 gene and genus Blautia—in addition to traits at higher taxonomic levels (class or phylum). In addition, among all GWAS performed for human traits as determined by the National Human Genome Research Institute (NHGRI) GWAS Catalog, most loci and genes discovered in our study were previously associated with various traits, including diseases for which there is growing evidence of microbiome involvement in disease etiology (for example, inflammatory bowel disease, obesity and type 2 diabetes; Supplementary Tables 6 and 7). Furthermore, specific associations of genes observed in previous studies (for instance, FUT2, NOD2 and LCT) could be replicated in our data set, but with less contribution in terms of influencing overall microbial variation (Supplementary Figs. 16 and 17).
In summary, we identify several genetic and non-genetic factors that determine the composition of the human gut microbiome. We show that genetic variation at the VDR locus significantly influences micro-bial co-metabolism and the gut–liver axis. Multiple other findings highlight key aspects of the intersections of host physiology with the gut microbiota, including a number of disease susceptibility genes in complex human diseases and the gut–brain axis. Key non-genetic covariable parameters, including diet, cumulatively have a similar magnitude of influence on the microbiome as host genetics, highlighting the importance of controlling for these confounders. Our study also indicates that the effect of individual genes is small and emphasizes the need for adequate statistical power and large sample sizes in future assessments. Following a similar logic to that provided by the outcomes of GWAS, the underlying biology of our observations may far exceed the statistical estimates and is likely to provide a critical framework for future studies of host–microbe interactions in humans.
METHODS
Methods and any associated references are available in the online version of the paper.
ONLINE METHODS
Study subjects and sample collection
Two population-based cohorts from Schleswig-Holstein (Germany) were included in the study. Nine hundred and fourteen individuals from the PopGen cohort and 1,115 individuals from the FoCus (Food Chain Plus) cohort were included. These two study cohorts were recruited independently from each other, and the maximum number of individuals available was included to increase statistical power for various analyses. All samples, as well as corresponding information on phenotype and dietary behavior, were obtained from the PopGen biobank (Schleswig-Holstein, Germany)20. Study participants collected fecal samples at home in standard fecal tubes. Samples were shipped immediately at room temperature or brought to the collection center by the participants. Upon arrival into the study center (within 24 h), samples were stored at −80 °C until processing. Written, informed consent was obtained from all study participants, and all protocols were approved by the institutional ethical review committee in adherence with the Declaration of Helsinki Principles; investigators were blinded to sample identities. Sequence data for the 16S rRNA gene, genotype, nutritional and phenotype data used for the herein described study have been made available to other scientists through PopGen’s biobank general data transfer agreement. A summary of the phenotypes used in this paper is given in Supplementary Table 1.
Genotyping data
Samples of the PopGen and FoCus cohorts were geno-typed on different genotyping arrays. The PopGen samples were typed on the Affymetrix 6.0, Affymetrix Axiom, Illumina 550k, custom Illumina Immunochip and Illumina Metabochip arrays with sample sizes before quality control ranging from 678 to 1,218 and a variant coverage of 196,524 to 934,968 variants. The FoCus samples were typed on the custom Illumina Immunochip and the Omni Express Exome, with 1,024 and 1,713 samples overall before quality control and a variant coverage of 195,732 to 964,193 variants. For each cohort, genotype data for each array were quality controlled separately and then merged and imputed. In total, 17,017,474 single-nucleotide variants (SNVs) were included for the PopGen cohort and 17,340,550 SNVs were included for the FoCus cohort. Consequently, stringent quality filtering was performed for all genotyping data, with details provided in the accompanying Supplementary Note.
Sequencing and processing of bacterial 16S rRNA sequences
Bacterial genomic DNA was extracted using the QIAamp DNA Stool Mini kit from Qiagen on a QIAcube system. For all samples, the V1–V2 region of the 16S rRNA gene was sequenced on the MiSeq platform, using the 27F-338R primer pair and dual MID indexing (8 nt each on the forward and reverse primers) as described by Kozich et al.51. Sequencing was performed with MiSeq Reagent Kit v2. After sequencing, MiSeq fastq files were derived from base calls for read 1 and 2 (R1 and R2), as well as both indices (I1 and I2), using the Bcl2fastq module in CASAVA 1.8.2. Stringent demultiplexing was carried out by allowing no mismatches in either index sequence (instead of the default of one mismatch allowed by MiSeq). Forward and reverse reads were merged with FLASH software (v1.2)52, and quality filtering was subsequently performed with the fastx toolkit, excluding sequences with >5% nucleotides with quality score <30. Chimeras in sequences were removed using UCHIME (v6.0)53. After randomly selecting 10,000 reads for each sample, taxonomical classification and compositional matrices for each taxonomical level were carried out using the RDP classifier54 with the latest reference database (RDP14), where classifications with low confidence at the genus level (<0.8) were organized in an arbitrary taxon of ‘unclassified family’. Species-level OTUs (97% similarity) were created using the UPARSE routine55.
Bile acid and fatty acid measurements on human serum samples
Serum bile acid and polyunsaturated fatty acid composition in plasma was analyzed for 551 PopGen samples by HPLC-MS/MS as recently described56,57. Five bile acids (cholic acid (CA), chenodeoxycholic acid (CDCA), lithocholic acid (LCA), deoxycholic acid (DCA) and ursodeoxycholic acid (UDCA)), including their taurinated (T) and glycinated (G) conjugates, were measured, as well as the following fatty acids: C18:2n-6 (linoleic acid), C18:3n-3 (α-linolenic acid), C18:3n-6 (γ-linolenic acid), C18:4n-3 (stearidonic acid), C20:2n-6 (eicosadienoic acid), C20:3n-6 (dihomo-γ-linolenic acid), C20:4n-3 (eicosatetraenoic acid), C20:4n-6 (arachidonic acid), C20:5n-3 (eicosapentaenoic acid), C21:5n-3 (heneicosapentaenoic acid), C22:2n-6 (docosadienoic acid), C22:4n-6 (adrenic acid), C22:5n-3 (docosapentaenoic acid), C22:5n-6 (docosapentaenoic acid), C22:6n-3 (docosahexaenoic acid).
Statistical analysis
Correlation between microbiome and metadata
In both cohorts, β-diversity measures based on genus-level composition were generated using the ‘vegdist’ function (Bray–Curtis and Jaccard dissimilarities). Community ordination was performed using PCoA based on the calculated dissimilarities using the ‘capscale’ function in ‘vegan’ (v2.3). The ‘envfit’ function in ‘vegan’ was used to correlate either categorical data, for which it performs multidimensional ANOVA on the ordination, or continuous variables, for which the function tests linear correlations between a given variable and the coordinates of microbial communities. This test does not assume a normal distribution, as the significance value is determined by a permutation test.
We considered a range of reported confounding variables that could shape the human gut microbiome: age, sex, BMI, smoking and major nutritional components or food groups derived from diet patterns; similarly, the association analysis was performed for bile acid profiles and fatty acid composition. Dietary patterns were collected via a validated, self-administered, 112-item food frequency questionnaire established for German populations58,59. All participants were given the option of completing the questionnaire preferably as a web-based version and, optionally, on paper. Information on macro-and micronutrient intake was obtained by using the German Food Code and Nutrient Database (vII.3) and provided by the Department of Epidemiology of the German Institute of Human Nutrition Potsdam-Rehbruecke. Before association analysis, all individuals who took antibiotics less than 6 weeks before stool collection were excluded to remove the possible influences of antibiotic medication. The effect size and significance of the mentioned variables were estimated using ‘envfit’, and the variables with significant effects (P < 0.05) were further used in the GWAS analysis as covariates (water, alcohol and all other highly correlated nutritional variables, which were collectively joined under the umbrella ‘total energy’). The combined effect of host metadata was estimated further using the ‘bioenv’ function in the ‘vegan’ package, which calculates the maximum Pearson correlation of microbial variation (Bray–Curtis dissimilarity) and combined dissimilarity in the selected subset of metadata (denoted by Gower distances). To reduce random errors in low-abundance taxa, the analysis focused on the ‘core measurable microbiota’, which was determined using technical replicates according to Benson et al.35. Only taxa with an average of >40 reads per sample (and thus with less error introduced by random processes) were included (Supplementary Fig. 12).
Association of individual bacterial traits with human genetic variation
To identify human genetic variation associated with the abundance of individual gut bacteria, a statistical test for each combination of SNP and taxon was performed. The abundance of bacteria in the human gut is characterized by an increasing number of zeros at lower taxonomic levels, a right-skewed distribution often with a long tail and only positive values. Thus, a model assuming a normal distribution of dependent variables could not be fitted to our data. The GLM with a negative binomial (negbin) distribution and log link was selected for the statistical analysis as the best-fitting model across all bacteria. The hurdle model with a negbin distribution showed increasingly good fit with increasing numbers of zeros. The GLM negbin model was therefore selected as a consistent model across all bacteria, while the analysis of species (97% similarity threshold OTUs) was supported with the hurdle model60.
Our identified ‘core measurable microbiota’ (ref. 35) consists of 64 taxa across five levels (phylum, class, order, family and genus) and 42 species-level OTUs. Taxa with >90% of their counts within the first 5% of the range of counts or with >90% of above-zero counts within the first 5% of the above-zero range were excluded, as they performed poorly with the selected model(s). Forty OTUs and 58 taxa were used for association study with human SNPs. The analyses were preformed on both cohorts separately (986 samples in FoCus and 826 samples in PopGen). In the analyses, outliers defined as 5 s.d. were removed and genetic variants not overlapping in FoCus and PopGen were discarded, while variants with MAF >0.05 and IMPUTE2 INFO criteria >0.8 were included. No population stratification was observed between the two cohorts (λGC = 1.00; Supplementary Fig. 18) b61. The covariates BMI, age, sex, genetic principal components 1–3 and nutritional variables alcohol, water and ‘total energy’ intake were used. The analyses were performed using R Project version 3.2 and the GLM.nb function in ‘MASS’ package version 7.3 for the GLM negbin and the hurdle function in package ‘pscl’ (v1.4).
A meta-analysis of GLM negbin hits across the two cohorts was performed using PLINK (v1.9 64-bit)62, with the command “--meta-analysis +qt”, including information on β coefficients and standard errors. Clumping was performed using PLINK v1.9 with the “--clump” command on SNPs meeting the following filtering criteria: meta-study fixed-effect P value < 5 × 10−8, single-cohort P value < 5 × 10−4, the same β value sign (same direction of association) and AIC (model fit parameter) < 50,000. Clumps with at least two SNPs for which at least one SNP was genotyped were selected. For each selected clump, the SNP with the lowest meta-analysis P value was selected as the tag SNP, and for bacteria containing zero counts the hurdle model was applied as described above. All hits were confirmed to be supported by the count or zero part of the hurdle model with P < 0.05 in both studies.
Genetic variation correlated with overall community differences
We also performed analyses aimed at identifying genetic variation that might not necessarily associate with individual bacterial taxa with genome-wide significance but might rather correlate with overall community differences (β diversity). We performed a simulation and treated genotype at each locus as categorical variables (the distribution of each genotype follows Hardy–Weinberg equilibrium). We measured the genotype association using the ‘envfit’ function in the ‘vegan’ R package (v2.3). This approach calculates the community differences associated with three different genotypes, by comparing the difference in the centroids of each group relative to the total variation, on the basis of the main axes of the PCoA. By shuffling the simulated genotype >2 × 107 times, we effectively obtained a large enough null distribution of effect size. This was performed for six categories of MAF to represent loci with MAFs of 5%, 10%, 20%, 30%, 40% and 50% (whereas in case of a real SNP, it is compared to the category with the closest MAF value; Supplementary Fig. 19), and if a certain locus displays greater effect sizes than the simulated maximum they are extremely unlikely to be observed by chance (P < 5 × 10−8) and can be considered to be genome-wide significant. We have filtered SNPs in a similar fashion as the taxa associations mentioned above.
The additive effect of the significant loci from this analysis was then determined using redundancy analysis based on genus-level composition (‘rda’ in the ‘vegan’ package) and the ‘ordiR2step’ function in the ‘vegan’ package, which optimizes the order of loci in a linear model and sums up the variation of the ordination explained by each additional locus.
HLA analyses were conducted on the respective HLA haplotypes within each locus, coded as carrier or non-carrier for each specific allele. We performed distance-based redundancy analysis after correction for host characteristics (see description of association analysis for factors). These models were then tested using a permutative ANOVA approach (5,000 permutations) as implemented in the ‘vegan’ function ‘anova.cca’, and the coefficients of determination were extracted via ‘RsquareAdj’.
Annotation and enrichment
DEPICT36 was used to annotate and perform tissue and gene set enrichment analyses among the significant single-bacteria associations. DEPICT was used with the following settings: (i) association_pvalue_cutoff: 1 × 10−5, (ii) nr_repititions: 20, and (iii) nr_permutations: 500; all available analysis steps were performed. For genotype data, we used 1000_genomes_project_phase3_CEU/ALL.chr_merged.phase3_sha-peit2_mvncall_integrated_v5.2 0130502.genotypes; for the collection file, we specified ld0.5_collection_depict_150315.txt.gz and for the reconstituted gene sets file we specified GPL570-GPL96-GPL1261-GPL1355TermGeneZScores-MGI_MF_CC_RT_IW_BP_KEGG_z_z.binary.
Analysis of association between bile acids and lead SNPs identified in this study
To identify bile acids associated with lead SNPs identified to be associated with the microbiome in this study, a generalized linear model with an inverse Gaussian distribution and log link was applied. As a supporting model, a two-part model was used comprising a GLM with binomial distribution and logit link for zero versus nonzero values, and a linear regression on log-transformed concentrations plus a constant (c = 1) for nonzero values. For both models, outliers with bile acid levels more than 5 s.d. from the mean were excluded and the covariates age, sex, BMI, vitamin K, alcohol, bile acid batch number and PC1–3 were included. The analysis included 520 samples.
Cis- and trans-eQTL analysis on human data
For SNPs identified as associated with β diversity and/or single bacterial traits, a cis- and trans-eQTL analysis was performed using data on 2,360 individuals. The analysis design and recourse are described in detail in previous studies63,64. In summary, cis-eQTL analysis was performed on SNP–probe pairs for cases where the distance was less than 1 Mb. To consider the effects of SNPs in LD with a disease-associated SNP (trait–SNP), a conditioned analysis was performed by first adjusting the probe expression level for the effect of the strongest associated local SNPs (eSNP) and then repeating the eQTL analysis. Likewise, the P value for the local best SNP was calculated with conditioning on the trait SNP. To control for FDR, sample labels were permuted 100 times to obtain a P-value distribution. Expression probes with a significant association (FDR < 5%, two-way conditional analysis for cis-eQTL analysis) to a trait SNP are given in Supplementary Tables 6 and 7.
Analysis of gut microbiome data from Vdr-knockout mice
Gut microbiome data from Jin et al.25 include fecal samples from three wild-type and five Vdr-knockout mice for which the V4–V6 region of the 16S rRNA gene was sequenced on the 454 GS-FLX platform. Quality filtering, removal of chimeras and classification were performed according to the same procedure described in the previous section. Statistical tests for the effect of Vdr genotype on the microbiome were carried out with the ‘envfit’ function in ‘vegan’ as described for the analysis for human SNPs. Comparison of specific taxa was carried out by the Wilcoxon test. Results are shown in Supplementary Figures 5 and 6.
Analysis of association of bile acids and fatty acids with the microbiome
To identify bacteria associated with the concentration of measured bile acids, including total LCA (the sum of LCA, G.LCA and T.LCA) and total BA (sum of all 15 bile acids), a generalized linear model with an inverse Gaussian distribution and log link was applied, excluding outliers more than 5 s.d. from the mean for bacteria and bile acids, adding a constant (c = 1) to bile acid concentration and including the covariates age, sex, BMI, total energy intake, water, alcohol and bile acid batch number (n = 569). To identify bacteria associated with ω3 and ω6 fatty acids, a linear regression model was applied with a square root transformation of fatty acids, excluding outliers with values more than 5 s.d. from the mean for bacteria and including the covariates age, sex, BMI, total energy intake, water and alcohol. Two samples with negative concentrations found for C22.2n.6 were excluded, leaving 567 samples in the fatty acid analysis. Benjamini–Hochberg corrected P values were calculated for each dependent variable to determine significance (Supplementary Table 4).
Shotgun metagenomic analysis
For a subset of 197 individuals, the same DNA extracts used in 16S rRNA gene sequencing were subjected to shotgun metagenomic sequencing. Samples were prepared following the protocol for the Illumina Nextera DNA Library Preparation kit and sequenced on the HiSeq Platform as 2 × 125 bp paired-end reads. Nextera adaptor sequences were trimmed using Trimmomatic (v0.32)65. Quality control of the sequencing reads was performed with sickle (v1.330), and parameters were set to a sliding-window quality threshold of 20 and a minimum length of 60 after quality trimming. DeconSeq66 was run to identify and remove human reads from the sequencing file, using the hg19 human genome sequence as the reference database. If one of the reads belonging to a read pair was removed at any of the quality control steps, the respective paired read was discarded as well. Samples that passed quality control, with no diagnosed IBD, IBS or diabetes and with genetic data (n = 122), were analyzed using HUMAnN2 with default settings except ‘–bt2_ps sensitive’ for the analysis of pathway and gene family abundance. Tables were normalized to relative abundance using ‘humann2_renorm_table –units relab’. Gene families including the term ‘bile acid’ were selected, and four pathways relevant for bile acid metabolism were selected (bile acid degradation, iso-bile acid biosynthesis I + II, bile acid biosynthesis, neutral pathway and glycocholate metabolism (bacteria)). Association with VDR genotype (rs7974353) was evaluated using GLM with an inverse Gaussian distribution, the covariates BMI, age, sex, alcohol, water and total energy intake and removal of outliers more than 5 s.d. from the mean and a constant (c = 1) added to abundance followed by multiplication by 1 × 106.
Replication in the FoCus obesity cohort
SNPs found to be significantly associated with β diversity in this study were consequently replicated in an additional FoCus obesity cohort (n = 371). The FoCus obesity cohort was recruited from the Obesity Outpatient Centre at the University Hospital in Kiel, which offers both non-surgical and surgical obesity therapies. Similar phenotype and genotyping profiles were obtained for the FoCus control cohort. The recruitment of the FoCus obesity cohort was approved by the local Ethics Committee (A156/03), and each patient gave their informed consent. To replicate associations of lead SNPs with β diversity, the effect size of each SNP was calculated with ‘envfit’, and consequent P values were calculated on the basis of the same empirical null distributions described above; successful replications are defined as having P < 0.05/42 (in total, 42 SNPs were included in the test).
Supplementary Material
Acknowledgments
We thank A.D. Paterson and colleagues for support in selection of models for GWAS. We further thank Der Norddeutsche Verbund für Hoch- und Höchstleistungsrechnen (HLRN) and S. Knief and H. Marten for computational resources and support. This work was supported by German Research Foundation (DFG) Collaborative Research Center 1182, ‘Origin and Function of Metaorganisms’ (J.F.B. and A.F.) and Excellence Cluster 306, ‘Inflammation at Interfaces’ (J.F.B. and A.F.) and by German Federal Ministry of Education and Research (BMBF) project ‘SysINFLAME’ (J.F.B. and A.F.). Project support was also provided by the Norwegian PSC Research Center and the Western Norway Regional Health Authority (grant 911802) (T.H.K.). M.K. is the recipient of a Postdoctoral Research Fellowship from the German Research Foundation (DFG). J.R.H. was funded by the Norwegian Research Council (240787/F20).
Footnotes
URLs. PopGen Biobank, https://www.epidemiologie.uni-kiel.de/biobanking/biobank-popgen; GTEx, http://www.gtexportal.org/; RegulomeDB, http://www.regulomedb.org/; TRAP, http://trap.molgen.mpg.de/cgi-bin/home.cgi; GWAS Catalog, http://www.ebi.ac.uk/gwas; CASAVA, http://support.illumina.com/sequencing/sequencing_software/casava; FastqToolkit, http://hannonlab.cshl.edu/fastx_toolkit/; vegan R package, http://vegan.r-forge.r-project.org/; MASS R package, http://cran.r-project.org/package=MASS; pscl R package, http://cran.r-project.org/package=pscl; HUMAnN2, http://www.bitbucket.org/biobakery/humann2; InnateDB, http://www.innatedb.com/; cisRED, http://www.cisred.org/; sickle, http://github.com/najoshi/sickle; German Food Code and Nutrient Database, www.mri.bund.de/de/service/datenbanken/bundeslebensmittelschluessel/.
Data access. All samples and information on their corresponding phenotypes and dietary behavior were obtained from the PopGen Biobank (Schleswig-Holstein, Germany) and can be accessed through a Material Data Access Form. Information about the Material Data Access Form and how to apply can be found at http://www.uksh.de/p2n/Information+for+Researchers.html.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
A.F., J.F.B. and T.H.K. conceived the project. U.N., W.L., M.L. and K.S. organized recruitment and sample collection for the PopGen and FoCus cohorts. Genotyping data were collected and processed by L.B.T., J. Skiecevicˇienė, J.R.H., F.D. and K.H.; nutritional data were generated and processed by S.S., M.P.-J., M. Koch and U.N.; microbiome data were generated and processed by J.W., P. Rausch, F.-A.H., M.C.R., P. Rosenstiel, K.C.-S., S.K. and J.F.B.; and bile acid and fatty acid data were generated and processed by S.A.-D., P.B., R.K.B., M.D’A. and H.-U.M. T.E., J. Sun, J.B., F.S., D.E., M.H., G.R., P.H., W.-H.P., R.S.-T., R.H. and P. Rosenstiel contributed to additional experiments and data for this study. Statistical analyses were performed by J.W., L.B.T., J. Skiecevicˇienė, P. Rausch and M. Kummen, and J.W., L.B.T., J. Skiecevicˇienė, P. Rausch, M. Kummen, J.R.H., M.D’A., H.-U.M., T.H.K., J.F.B. and A.F. interpreted the results. J.W., L.B.T., J. Skiecevicˇienė, P. Rausch, M. Kummen, J.R.H., T.H.K., J.F.B. and A.F. wrote the manuscript, with input from all other authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 2.Fraune S, Bosch TC. Why bacteria matter in animal development and evolution. BioEssays. 2010;32:571–580. doi: 10.1002/bies.200900192. [DOI] [PubMed] [Google Scholar]
- 3.Sekirov I, Russell SL, Antunes LCBB, Finlay BB. Gut microbiota in health and disease. Physiol. Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 4.Chow J, Mazmanian SK. A pathobiont of the microbiota balances host colonization and intestinal infammation. Cell Host Microbe. 2010;7:265–276. doi: 10.1016/j.chom.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annu. Rev. Microbiol. 2011;65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- 7.Antonopoulos DA, et al. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caporaso JG, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckburg PB, et al. Diversity of the human intestinal microbial fora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodrich JK, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cotillard A, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 14.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rehman A, et al. Geographical patterns of the standing and active human gut microbiome in health and IBD. Gut. 2016;65:238–248. doi: 10.1136/gutjnl-2014-308341. [DOI] [PubMed] [Google Scholar]
- 16.Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rausch P, et al. Colonic mucosa-associated microbiota is infuenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc. Natl. Acad. Sci. USA. 2011;108:19030–19035. doi: 10.1073/pnas.1106408108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rehman A, et al. Nod2 is essential for temporal development of intestinal microbial communities. Gut. 2011;60:1354–1362. doi: 10.1136/gut.2010.216259. [DOI] [PubMed] [Google Scholar]
- 19.Blekhman R, et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015;16:191. doi: 10.1186/s13059-015-0759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krawczak M, et al. PopGen: population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet. 2006;9:55–61. doi: 10.1159/000090694. [DOI] [PubMed] [Google Scholar]
- 21.Müller N, et al. IL-6 blockade by monoclonal antibodies inhibits apolipoprotein (a) expression and lipoprotein (a) synthesis in humans. J. Lipid Res. 2015;56:1034–1042. doi: 10.1194/jlr.P052209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biedermann L, et al. Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS One. 2013;8:e59260. doi: 10.1371/journal.pone.0059260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haussler MR, et al. Vitamin D receptor: molecular signaling and actions of nutritional ligands in disease prevention. Nutr. Rev. 2008;66(Suppl. 2):S98–S112. doi: 10.1111/j.1753-4887.2008.00093.x. [DOI] [PubMed] [Google Scholar]
- 24.Makishima M, et al. Vitamin D receptor as an intestinal bile acid sensor. Science. 2002;296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- 25.Jin D, et al. Lack ofes the functions of the murine intestinal microbiome. Clin. Ther. 2015;37:996–1009. e7. doi: 10.1016/j.clinthera.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 26.D’Aldebert E, et al. Bile salts control the antimicrobial peptide cathelicidin through nuclear receptors in the human biliary epithelium. Gastroenterology. 2009;136:1435–1443. doi: 10.1053/j.gastro.2008.12.040. [DOI] [PubMed] [Google Scholar]
- 27.Sayin SI, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-β-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Inoue Y, Yu AM, Inoue J, Gonzalez FJ. Hepatocyte nuclear factor 4α is a central regulator of bile acid conjugation. J. Biol. Chem. 2004;279:2480–2489. doi: 10.1074/jbc.M311015200. [DOI] [PubMed] [Google Scholar]
- 29.Sato H, et al. Group III secreted phospholipase A2 transgenic mice spontaneously develop infammation. Biochem. J. 2009;421:17–27. doi: 10.1042/BJ20082429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yano JM, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olivares M, et al. The HLA-DQ2 genotype selects for early intestinal microbiota composition in infants at high risk of developing coeliac disease. Gut. 2015;64:406–417. doi: 10.1136/gutjnl-2014-306931. [DOI] [PubMed] [Google Scholar]
- 32.Okada Y, et al. HLA-Cw*1202–B*5201–DRB1*1502 haplotype increases risk for ulcerative colitis but reduces risk for Crohn’s disease. Gastroenterology. 2011;141:864–871. e1. doi: 10.1053/j.gastro.2011.05.048. 5. [DOI] [PubMed] [Google Scholar]
- 33.Arimura Y, et al. Characteristics of Japanese infammatory bowel disease susceptibility loci. J. Gastroenterol. 2014;49:1217–1230. doi: 10.1007/s00535-013-0866-2. [DOI] [PubMed] [Google Scholar]
- 34.Terao C, et al. Two susceptibility loci to Takayasu arteritis reveal a synergistic role of the IL12B and HLA-B regions in a Japanese population. Am. J. Hum. Genet. 2013;93:289–297. doi: 10.1016/j.ajhg.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benson AK, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. USA. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pers TH, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phay JE, Hussain HB, Moley JF. Cloning and expression analysis of a novel member of the facilitative glucose transporter family, SLC2A9 (GLUT9) Genomics. 2000;66:217–220. doi: 10.1006/geno.2000.6195. [DOI] [PubMed] [Google Scholar]
- 38.Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Repa JJ, et al. Regulation of absorption and ABC1-mediated effux of cholesterol by RXR heterodimers. Science. 2000;289:1524–1529. doi: 10.1126/science.289.5484.1524. [DOI] [PubMed] [Google Scholar]
- 40.Wahlström A, Sayin SI, Marschall HU, Bäckhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016;24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 41.Duparc T, et al. Hepatocyte MyD88 affects bile acids, gut microbiota and metabolome contributing to regulate glucose and lipid metabolism. Gut. 2016 doi: 10.1136/gutjnl-2015-310904. http://dx.doi.org/10.1136/gutjnl-2015-310904. [DOI] [PMC free article] [PubMed]
- 42.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of infammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu JZ, et al. Dense genotyping of immune-related disease regions identifes nine new risk loci for primary sclerosing cholangitis. Nat. Genet. 2013;45:670–675. doi: 10.1038/ng.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun J. VDR/vitamin D receptor regulates autophagic activity through ATG16L1. Autophagy. 2016;12:1057–1058. doi: 10.1080/15548627.2015.1072670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krude H, Biebermann H, Gruters A. Mutations in the human proopiomelanocortin gene. Ann. NY Acad. Sci. 2003;994:233–239. doi: 10.1111/j.1749-6632.2003.tb03185.x. [DOI] [PubMed] [Google Scholar]
- 46.Tuoresmäki P, Väisänen S, Neme A, Heikkinen S, Carlberg C. Patterns of genome-wide VDR locations. PLoS One. 2014;9:e96105. doi: 10.1371/journal.pone.0096105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, et al. Analysis of intestinal microbiota in hybrid house mice reveals evolutionary divergence in a vertebrate hologenome. Nat. Commun. 2015;6:6440. doi: 10.1038/ncomms7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srinivas G, et al. Genome-wide mapping of gene-microbiota interactions in susceptibility to autoimmune skin blistering. Nat. Commun. 2013;4:2462. doi: 10.1038/ncomms3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKnite AM, et al. Murine gut microbiota is defned by host genetics and modulates variation of metabolic traits. PLoS One. 2012;7:e39191. doi: 10.1371/journal.pone.0039191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leamy LJ, et al. Host genetics and diet, but not immunoglobulin A expression, converge to shape compositional features of the gut microbiome in an advanced intercross population of mice. Genome Biol. 2014;15:552. doi: 10.1186/s13059-014-0552-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ.Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Magocˇ T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifer for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 56.Abu-Hayyeh S, et al. Prognostic and mechanistic potential of progesterone sulfates in intrahepatic cholestasis of pregnancy and pruritus gravidarum. Hepatology. 2016;63:1287–1298. doi: 10.1002/hep.28265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bjørndal B, et al. Krill powder increases liver lipid catabolism and reduces glucose mobilization in tumor necrosis factor-α transgenic mice fed a high-fat diet. Metabolism. 2012;61:1461–1472. doi: 10.1016/j.metabol.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 58.Nöthlings U, Hoffmann K, Bergmann MM, Boeing H. Fitting portion sizes in a self-administered food frequency questionnaire. J. Nutr. 2007;137:2781–2786. doi: 10.1093/jn/137.12.2781. [DOI] [PubMed] [Google Scholar]
- 59.Dehne LI, Klemm C, Henseler G, Hermann-Kunz E. The German food code and nutrient data base (BLS II.2) Eur. J. Epidemiol. 1999;15:355–359. doi: 10.1023/a:1007534427681. [DOI] [PubMed] [Google Scholar]
- 60.Xu L, Paterson AD, Turpin W, Xu W. Assessment and selection of competing models for zero-infated microbiome data. PLoS One. 2015;10:e0129606. doi: 10.1371/journal.pone.0129606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Degenhardt F, et al. Genome-wide association study of serum coenzyme Q10 levels identifes susceptibility loci linked to neuronal diseases. Hum. Mol. Genet. 2016 doi: 10.1093/hmg/ddw134. http://dx.doi.org/10.1093/hmg/ddw134. [DOI] [PubMed]
- 62.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ellinghaus D, et al. Analysis of fve chronic infammatory diseases identifes 27 new associations and highlights disease-specifc patterns at shared loci. Nat. Genet. 2016;48:510–518. doi: 10.1038/ng.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wood AR, et al. Defning the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bolger AM, Lohse M, Usadel B. Trimmomatic: a fexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmieder R, Edwards R. Fast identifcation and removal of sequence contamination from genomic and metagenomic datasets. PLoS One. 2011;6:e17288. doi: 10.1371/journal.pone.0017288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.