ABSTRACT
In biotechnological workhorses like Streptococcus thermophilus and Bacillus subtilis, natural competence can be induced, which facilitates genetic manipulation of these microbes. However, in strains of the important dairy starter Lactococcus lactis, natural competence has not been established to date. However, in silico analysis of the complete genome sequences of 43 L. lactis strains revealed complete late competence gene sets in 2 L. lactis subsp. cremoris strains (KW2 and KW10) and at least 10 L. lactis subsp. lactis strains, including the model strain IL1403 and the plant-derived strain KF147. The remainder of the strains, including all dairy isolates, displayed genomic decay in one or more of the late competence genes. Nisin-controlled expression of the competence regulator comX in L. lactis subsp. lactis KF147 resulted in the induction of expression of the canonical competence regulon and elicited a state of natural competence in this strain. In contrast, comX expression in L. lactis NZ9000, which was predicted to encode an incomplete competence gene set, failed to induce natural competence. Moreover, mutagenesis of the comEA-EC operon in strain KF147 abolished the comX-driven natural competence, underlining the involvement of the competence machinery. Finally, introduction of nisin-inducible comX expression into nisRK-harboring derivatives of strains IL1403 and KW2 allowed the induction of natural competence in these strains also, expanding this phenotype to other L. lactis strains of both subspecies.
IMPORTANCE Specific bacterial species are able to enter a state of natural competence in which DNA is taken up from the environment, allowing the introduction of novel traits. Strains of the species Lactococcus lactis are very important starter cultures for the fermentation of milk in the cheese production process, where these bacteria contribute to the flavor and texture of the end product. The activation of natural competence in this industrially relevant organism can accelerate research aiming to understand industrially relevant traits of these bacteria and can facilitate engineering strategies to harness the natural biodiversity of the species in optimized starter strains.
KEYWORDS: Lactococcus lactis, natural competence, comparative genomics
INTRODUCTION
Horizontal gene transfer (HGT) plays an important role in the evolution of bacteria (1–4). In several species, an important mechanism for HGT is natural competence. This phenomenon is defined as a cellular state that enables internalization of exogenous DNA, followed by autonomous replication as a plasmid or incorporation into the chromosome via homologous recombination. Among Gram-positive bacteria, natural competence was first described in Streptococcus pneumoniae (5, 6). More recently, it was found that among lactic acid bacteria (LAB), the important yogurt bacterium Streptococcus thermophilus can enter a state of natural competence upon culturing in a chemically defined medium (7). When Gram-positive bacteria enter a state of natural competence, exogenous DNA translocates through the DNA uptake machinery, a multiprotein complex comprising the proteins ComEA, ComEC, ComFA, and ComFC and a nuclease (EndA in S. pneumoniae) encoded by the late competence (com) genes (8, 9). Other late competence genes encode proteins that compose pilus-like structures (ComGA-GG) or protect internalized DNA against degradation (SsbA, SsbB, DprA, and RecA) (8–10). Expression of these genes is positively regulated by the competence master regulator ComX, which acts as an alternative sigma factor (11–13). In S. thermophilus, expression of comX is initiated upon formation of the quorum-sensing ComRS complex, comprising the pheromone-like peptide ComS and transcriptional regulator ComR and encoded by the comRS operon (14, 15). Addition of a synthetic peptide that resembles the active competence pheromone has proven to be a successful strategy to induce natural competence in several bacterial species. For example, addition of a synthetic ComS peptide to S. thermophilus cultures in the early logarithmic phase of growth enabled the activation of natural competence and highly efficient DNA transformation (14, 16). Analogously, other streptococci, including S. pneumoniae, utilize the comCDE regulatory module to control natural competence, involving the competence-stimulating peptide (CSP) (encoded by comC) and a two-component system (encoded by comD and comE [17, 18]), and the addition of synthetic CSP leads to development of natural competence in this species.
Strains of Lactococcus lactis are of great importance in the dairy industry, primarily in the production of cheese, butter, and buttermilk (19). So far, a comRS- or comCDE-like system has not been identified in L. lactis. Nevertheless, complete sets of late competence genes appear to be present in several L. lactis genomes (20–22; this study). In addition, increased expression of competence genes has been observed in L. lactis subsp. lactis IL1403 and KF147 under specific conditions that included carbon starvation (23, 24). Unfortunately, in neither of these strains, or any other L. lactis strain, could natural competence development be experimentally established (20, 24). As an alternative route to establish natural competence, overexpression of comX has been employed, aiming to enhance expression of the complete late competence regulon. Such an approach has been successful in S. thermophilus (25) but failed in L. lactis IL1403 (20). Nevertheless, the observations that complete sets of late competence genes are apparently present in some of the L. lactis genomes (26, 27) and that their expression can be induced under specific conditions (23, 24) deserve a more dedicated bioinformatic and experimental effort.
Here, we present a comparative genomics analysis of 43 L. lactis genomes to assess their potential to enter a state of natural competence. Moreover, by employment of controlled expression of ComX, we demonstrate enhanced expression of the late competence regulon and concomitant induction of natural competence, which was successful only in strains predicted to encode a complete late competence machinery. The discovery of natural competence in L. lactis will enable transfer of genetic information without the use of genetically modified organisms (GMO), resulting in the improvement of the industrial performance of strains of this species and the enhancement of the quality of fermented products.
RESULTS
Genomic analyses show complete sets of competence genes in several L. lactis strains.
To evaluate whether L. lactis strains possess the genetic capacity to enter a state of natural competence, late-competence-associated genes were initially identified in the L. lactis KF147 genome by using the known competence genes of the naturally competent Streptococcus thermophilus LMD-9 (7, 14). This strain was selected for this primary analysis based on previous work that reported that many of the late competence genes were induced in this strain under starvation, nongrowth conditions (24). Similar proteins (similar both in length and sequence) were identified to be encoded within the KF147 genome for all selected late competence genes of S. thermophilus LMD-9, corroborating that a complete competence gene set is present in this L. lactis strain (see Table S1 and Fig. S1 in the supplemental material).
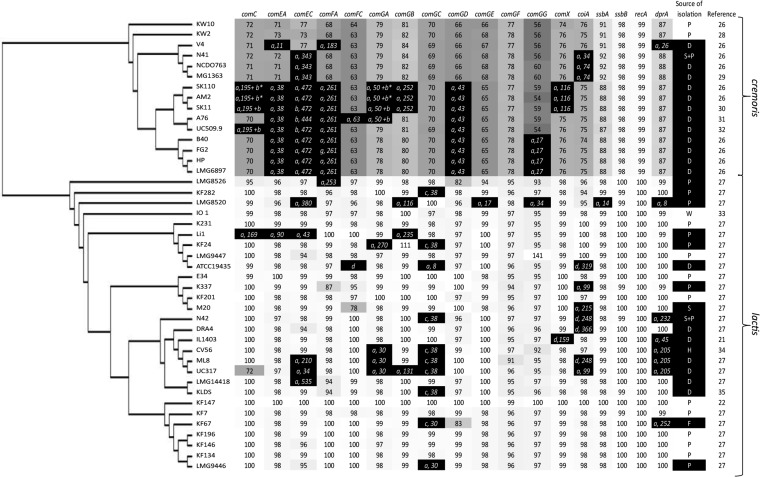
Subsequently, the identified KF147 competence protein sequences were used as a reference set for identification and comparison to the orthologous groups (OGs) of genes carried by 42 other L. lactis strains (21, 22, 26–35). Full-length protein sequence identity to the KF147 OGs was calculated for all 42 L. lactis strains (Fig. 1). This analysis revealed considerable genomic decay in several of the strains of both L. lactis subsp. lactis and L. lactis subsp. cremoris. Moreover, for the OGs that were intact, there was a clear distinction between the levels of identity observed for strains belonging to L. lactis subsp. lactis (which includes strain KF147) and L. lactis subsp. cremoris (Fig. 1), exemplifying the genetic distinction between these two subspecies (36). Among the strains belonging to L. lactis subsp. cremoris, only strains KW2 and KW10 appeared to encode full-length homologues of all the late competence proteins selected in KF147, albeit with identity scores ranging from 56 to 99% (Fig. 1). Notably, when the late competence gene set of the KW2 strain instead of that of strain KF147 was used to determine full-length protein sequence identity levels, it was apparent that late competence proteins displayed a high degree of conservation within L. lactis subsp. cremoris but were distinct from their orthologues in L. lactis subsp. lactis (see Fig. S2 in the supplemental material). Among the 28 strains belonging to L. lactis subsp. lactis, at least 10 appeared to encode a full set of late competence proteins.
FIG 1.
Genomic analysis of 43 L. lactis strains to assess genetic capacity to develop natural competence. A concatenated core genome single-nucleotide polymorphism (SNP) tree of 43 L. lactis strain was combined with full-length protein identity scores (%) for the selected subset of late-competence-associated proteins in comparison to their homologues in L. lactis strain KF147, which was used as a reference. Protein identity scores are depicted within each cell and reflected by gray scales based on the L. lactis KF147 query protein sequences, in which at least 90% full-length alignment is considered indicative of gene presence. Genetic events leading to competence gene decay (black cells in the figure) are specified as a premature stop codon within the first 90% of the gene (a), transposon insertion (b), prophage insertion (c), or absence of gene, a mutated/alternative start, or lengthened/fused protein at least more than 25% of its total length (d), followed by the position within the protein sequence where the event is detected relative to its N terminus. Source of isolation: P, plant; D, dairy; S, soil; W, water; H, human body; F, fruit (72). References for the genome sequences are given when available.
The genomic decay within these late competence genes in the L. lactis subsp. cremoris strains displayed several conserved disruptive mutations in specific genes, including IS982 insertions in comEC (strains SK11, A76, and UC509.9) and comGA (strains SK11 and A76), although with some variation with respect to the precise position of insertion (Fig. 1; see Fig. S3 in the supplemental material). Various strains of L. lactis subsp. cremoris contained conserved premature stop codons within one or more of their late competence genes, suggesting that these strains derive from a common ancestor, in which conserved and strain-specific mutations have shaped the decay pattern of the late competence genes. For example, strains SK110, AM2, SK11, A76, UC5099, B40, FG2, HP, and LMG6897 share similar mutational events in comEA, comEC, comFA, and comGD, whereas strains N41, NCDO763, and MG1363 harbor common mutations in comEC (see Fig. S4 in the supplemental material). In contrast, the disruptive mutations observed in the late competence genes of strains of L. lactis subsp. lactis appeared to be more scattered (Fig. 1), suggesting that degenerative mutations accumulated more recently in this subspecies. Nevertheless, several strains (KF282, KF24, N42, CV56, ML8, KLDS, UC317, and KF67) contain a (remnant of a) prophage insertion within the comGC gene (see Fig. S5 in the supplemental material). Remarkably, these phage sequences are always inserted at the same position within the comGC sequence, suggesting site-specific integration at a conserved sequence element within the comGC gene.
In summary, these findings indicate that in the majority of L. lactis strains, one or more late competence functions are compromised, suggesting that these strains are not able to develop a state of natural competence. The analysis also implies that in some strains, including L. lactis KF147, the genetic capacity to enter a state of naturally competence appears to be intact. Finally, it is noteworthy that within the present panel of strains, there are no dairy isolates that appear to encode a complete set of intact late competence proteins, which may reflect the high level of genome decay that has been reported for strains in the milk environment before (37–39).
Moderate overexpression of the late competence regulon regulator ComX results in a state of natural competence in L. lactis KF147.
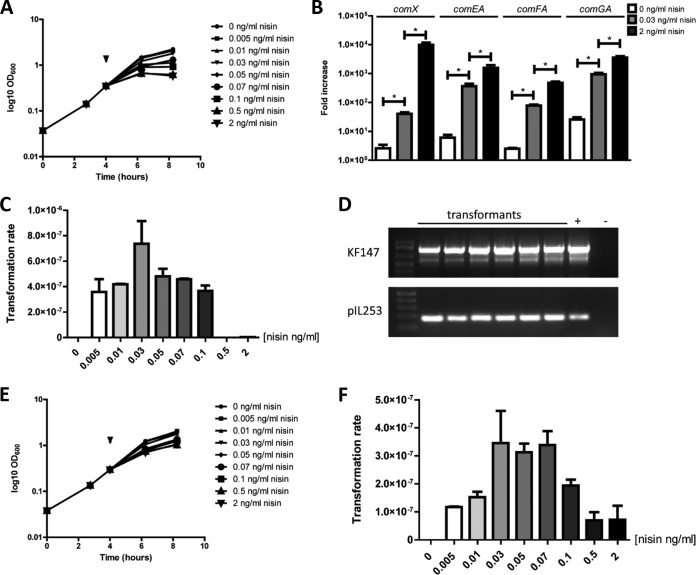
In order to test whether the identified competence machinery can be activated and is functional, we set out to overexpress the predicted competence regulator ComX. From the subset of strains predicted to harbor a complete set of competence genes, L. lactis KF147 harbors a chromosomal copy of nisRK but does not produce nisin (40), allowing nisin-inducible comX expression by cloning of this gene under the control of PnisA in pNZ8150 (41). This comX expression strategy led to a dose-dependent inhibition of growth (Fig. 2A), which was not observed in the control strains harboring pNZ8150 (see Fig. S6 in the supplemental material) (41) or pNZ8040, a vector enabling nisin-inducible expression of pepN (Fig. S6) (42). Hence, the observed growth retardation is not caused by the addition of nisin or the overexpression of proteins as such but is specifically caused by the presence of ComX.
FIG 2.
Impact of ComX expression on growth, late competence gene expression, and natural competence phenotype in L. lactis KF147. (A) Dose-dependent growth inhibition upon nisin induction of L. lactis KF147 harboring pNZ6200. The arrowhead indicates the time point of nisin induction. (B) comX, comEA, comFA, and comGA expression levels after nisin induction. *, significant differences (P < 0.05). (C and D) Number of colonies obtained (C) and confirmation of their genetic identity (D). (E and F) Same analysis for strain L. lactis KF147 harboring pNZ6201.
To investigate the impact of elevated ComX levels on the expression levels of the late competence genes, their transcript levels were determined by reverse transcription-quantitative PCR (RT-qPCR) on RNA derived from L. lactis KF147 harboring pNZ8150 or pNZ6200, either uninduced or moderately or fully induced with nisin. Under uninduced conditions, comX expression levels were 2.5- to 6-fold increased in L. lactis KF147 harboring pNZ6200 compared to the pNZ8150-harboring cells, which likely reflects low-level “promoter leakage” due to the presence of PnisA on a high-copy-number plasmid (Fig. 2B). Induction of comX expression in L. lactis KF147 harboring pNZ6200 with either 0.03 or 2 ng/ml nisin for 2 h led to 15- to 20-fold and 1,500- to 4,000-fold induction of comX expression relative to that in the uninduced control of the same strain, respectively (Fig. 2B). Similarly, expression of the late competence genes comEA, comFA, and comGA was induced, illustrating the strongly enhanced expression of the late competence regulon as a consequence of the elevated levels of its regulator, ComX (Fig. 2B). These induction conditions for the activation of late competence genes were employed to test whether the corresponding phenotype could also be observed, by adding pIL253 (43) to the culture medium at the same time point that comX induction was initiated using a range of nisin concentrations. As expected, no pIL253 transformants were obtained for L. lactis KF147 harboring pNZ8150 under any of the conditions tested (data not shown). In contrast, pIL253 transformants were obtained for L. lactis KF147 harboring pNZ6200 following induction with nisin concentrations ranging from 0.005 to 0.1 ng/ml nisin, with an approximate transformation rate of 10−7 to 10−6 (transformants/total cell number/μg plasmid DNA). The highest transformation rates were obtained after induction with 0.03 ng/ml nisin (Fig. 2C). Both strain identity and pIL253 presence was confirmed by PCR in all transformants tested (Fig. 2D). Notably, full nisin induction (2.0 ng/ml nisin) of comX expression in pNZ6200-harboring L. lactis KF147 did not result in any transformants. To check whether comX of the L. lactis subsp. cremoris strain MG1363 is still functional, transformation of L. lactis KF147 harboring pNZ6201 upon nisin induction was also tested. Similar results were obtained when comX of L. lactis subsp. cremoris MG1363 was expressed in L. lactis subsp. lactis KF147 (Fig. 2E and F), indicating that comX derived from an L. lactis subsp. cremoris strain is also fully functional. Taken together, these results demonstrate that activation of moderate expression, but not high-level expression, of endogenous comX in L. lactis KF147 elicits the natural competence phenotype in this strain. The observation that this does not occur at a high level of comX expression may be a consequence of the observed growth defect under these conditions, which may interfere with completion of the competence machinery assembly and/or recovery of potential transformants after plating. Such a notion is supported by the observation that expression of a heterologous copy of comX (derived from L. lactis subsp. cremoris) induced less severe growth defects upon maximal nisin induction and still led to detectable natural competence development, albeit with reduced efficiency compared to that with moderate nisin induction levels.
ComX-induced transformation in L. lactis depends on the late competence operon comEA-EC.
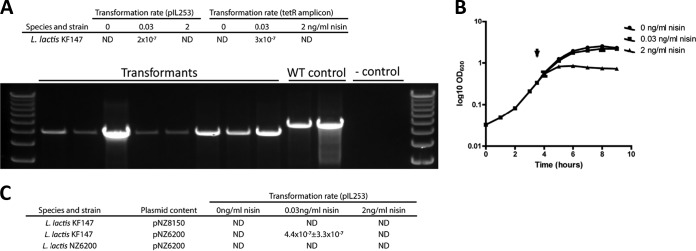
The experiments described above do not provide direct proof for a functional dependency of the observed transformation phenotype on the expression of the late competence genes, although this is likely, considering the fact that these genes encode the DNA uptake machinery. Therefore, we constructed a comEA-EC-negative derivative of L. lactis KF147 through the integration of a linear fragment harboring a tetracycline resistance-encoding tetR gene flanked by regions homologous to the 5′ and 3′ regions surrounding the comEA-EC operon. The procedure for moderate comX induction was applied to transform this linear mutagenesis fragment into strain KF147 harboring pNZ6200. Integrants with the anticipated genotype (ΔcomEA-EC::tetR) (Fig. 3A) were obtained with an efficiency similar to that observed for pIL253 transformation. Subsequent comX expression induction experiments in the ΔcomEA-EC::tetR derivative of strain KF147 (NZ6200) harboring pNZ6200 showed that full induction of comX led to a growth rate reduction in this strain (Fig. 3B) similar to that observed for the parental KF147 strain. Importantly, nisin concentration-dependent comX overexpression and corresponding upregulation of expression of the comFA and comGA, but not comEA, genes was similar to what was established in L. lactis KF147 harboring pNZ6200 (see Fig. S7 in the supplemental material). However, in contrast to the case for the parental strain KF147, transformation of NZ6200 harboring pNZ6200 with pIL253 did not yield any transformants (Fig. 3C), establishing the involvement of the comEA-EC operon in comX-induced competence in L. lactis KF147.
FIG 3.
Natural competence of L. lactis KF147 depends on the late competence operon comEA-EC. (A) By use of a linear mutagenesis fragment, a comEA-EC-negative derivative of KF147 harboring pNZ6200 (NZ6200) could be obtained with high efficiency. (B and C) NZ6200 displayed a similar growth defect upon full nisin induction (B), but transformation with pIL253 is not feasible (C).
Expansion of the natural competence phenotype to a broader set of L. lactis strains.
In order to employ the comparative genomics analysis performed in this study as a predictor for competence potential, L. lactis subsp. cremoris strains KW2 (28), and NZ9000 (41, 44) and L. lactis subsp. lactis IL-9000 (45) were tested for transformability upon comX induction. Strains NZ9000 and IL-9000 are derivatives of MG1363 and IL1403, respectively, which contain the nisRK genes integrated in their chromosome, thereby allowing the use of the nisin-controlled expression system. Strain KW2 does not harbor nisRK in its genome, and to facilitate the use of the nisin induction system in this strain, pNZ9531 was first introduced into this strain, thereby expressing nisRK in this background from a low-copy-number plasmid vector that is compatible with the pNZ8150 backbone of the pNZ6200 and pNZ6201 vectors used for comX expression (46). The plasmid pNZ8150, pNZ6200, or pNZ6201 was transformed into electrocompetent cells of IL-9000, NZ9000, and pNZ9531-harboring L. lactis KW2. These transformants were induced with 0, 0.03 or 2 ng/ml nisin, and the induction of the competence phenotype was evaluated in these strains by transformation with pIL253 (for NZ9000 and IL-9000) or pNZ6202 (a tetracycline-selectable pIL253 derivative). As anticipated, none of the conditions employed allowed the activation of natural competence in strain NZ9000 (Table 1), which is in good agreement with the comEC and coiA mutations observed in its parental strain MG1363 (Fig. 1). In contrast, transformants were obtained for pNZ6200 and pNZ6201 harboring derivatives of IL-9000, despite its incomplete dprA gene, and when these plasmids were transformed to KW2 harboring pNZ9531 (Table 1). Notably, although the efficiency of transformation appeared to be the highest for the cells in which moderate comX expression was induced (i.e., with 0.03 ng/ml nisin), transformants also were obtained under uninduced conditions and upon high-level induction of comX expression (i.e., with 2 ng/ml nisin). The observed transformation under uninduced conditions might be caused by the previously reported higher levels of “leakage” of the nisA promoter activity in L. lactis strains harboring the nisRK expression vector pNZ9531 (46), whereas the dprA mutation in the IL-9000 parental strain (IL1403) (Fig. 1) may require lower levels of comX expression to activate competence, as the DprA function has been associated with competence shutoff (47, 48). The observation that high-level comX expression still allowed competence development in KW2 and IL-9000 despite the growth-inhibitory consequences of this level of induction, which is in contrast with the results obtained for strain KF147, remains to be determined. Finally, in pNZ9531-harboring KW2, induced expression of the comX derived from L. lactis subsp. cremoris (i.e., as expressed from pNZ6201) allowed competence development, confirming the bidirectional functional exchangeability of the comX genes from these two L. lactis subspecies. These results confirm the predictions made by comparative genomics (Fig. 1) with respect to the capacity to develop a natural competence phenotype in L. lactis strains, and they establish that strains of both subspecies have the capacity of natural competence which can be induced by controlled expression of the comX-encoded regulator from either of the subspecies.
TABLE 1.
Assessing transformation with plasmid pIL253/pNZ6202 by controlled expression of comX in L. lactis IL-9000, NZ9000, and pNZ9531-harboring KW2
| L. lactis strain | Plasmid content | Transformation rate (pIL253/pNZ6202)a with the following [nisin] (ng/ml): |
||
|---|---|---|---|---|
| 0 | 0.03 | 2 | ||
| IL-9000 | pNZ6200 | 1.5 × 10−7 ± 8.2 × 10−8 | 1.7 × 10−7 ± 1.5 × 10−7 | 3.4 × 10−7 ± 1.7 × 10−7 |
| pNZ8150 | ND | ND | ND | |
| KW2 | pNZ9531 + pNZ6200 | 1.5 × 10−6 ± 1.1 × 10−6 | 1.0 × 10−5 ± 9.2 × 10−7 | 5.2 × 10−6 ± 3.1 × 10−6 |
| pNZ9531 + pNZ6201 | 1.1 × 10−6 ± 1.1 × 10−6 | 6.2 × 10−6 ± 5.7 × 10−6 | 3.1 × 10−7 ± 2.2 × 10−7 | |
| pNZ9531 + pNZ8150 | ND | ND | ND | |
| NZ9000 | pNZ6200 | ND | ND | ND |
| pNZ8150 | ND | ND | ND | |
The transformation rate was calculated as number of pIL253 or pN6202 transformants/total number of cells/μg DNA. Values are means ± standard deviations. ND, not detected.
DISCUSSION
This study demonstrates that the L. lactis strains KF147, KW2, and IL1403 possess a functional DNA uptake machinery, which can be activated by the ComX regulator. This implies that identification of a complete set of late competence genes through comparative genomics represents an appropriate approach to predict the capacity of a strain to enter a state of natural competence, and it seems likely that most, if not all, of the other strains identified here as carrying complete gene sets can be made naturally competent via the same strategy of comX overexpression. It should be noted that the expression of a much larger set of over 100 genes is regulated upon addition of the competence pheromone in streptococci (12, 49, 50). For instance, development of natural competence usually occurs in concert with increased expression of proteins involved in DNA recombination, thereby facilitating integration of acquired DNA (51), a feature that has been observed in this study for L. lactis KF147, as well suggesting expression of such proteins in L. lactis KF147 upon competence development. Nevertheless, we show that the dedicated assessment of only the canonical late competence genes is a valid predictor for competence potential in L. lactis strains.
It is commonly assumed that the L. lactis ancestor strain prior to subspeciation into L. lactis subsp. lactis and L. lactis subsp. cremoris originated from a plant-associated niche and that strains adapted to increase their fitness in the nutritionally rich dairy environment (40, 52, 53). Remarkably, none of the dairy isolates of L. lactis that were analyzed here appear to carry a complete set of late competence genes, suggesting that during the adaptation to the dairy niche, there was no significant environmental fitness benefit associated with the capacity to become naturally competent. This may relate to a real lack of fitness benefit of this phenotype within the dairy environment or may be due to highly consistent suppression of the phenotype during growth in milk, thereby preventing the possible fitness benefit to become apparent, which may allow the decay of encoding genes without an apparent fitness cost for the bacteria. The latter scenario appears to be in agreement with the observed activation of the expression of late competence genes in L. lactis during carbon starvation conditions (23, 24), which are not likely to occur within the dairy niche, as it is very rich in lactose. The genomic decay events associated with dairy-derived L. lactis strains include prophage disruption of the comGC locus in strains of L. lactis subsp. lactis (28, 54) and insertion of IS982 into several com genes in strains of L. lactis subsp. cremoris (40, 55–57). Notably, the phylogenetic relatedness of L. lactis subsp. cremoris strains predicted on the basis of competence gene decay events displayed a topology that was remarkably similar to that observed for the core genome relatedness of these strains (see Fig. S4 in the supplemental material). Importantly, typical dairy environment-associated lactic acid bacteria quite commonly display genomic decay as a consequence of the adaptation to this nutritionally rich environment (37–39). For example, loss-of-function events have been observed in S. thermophilus, Lactobacillus helveticus, and Lactobacillus bulgaricus upon prolonged culturing in milk, with mutations accumulating in genes encoding transport-, energy metabolism-, and virulence-associated functions, implying that these functions do not contribute to fitness in the dairy niche (37, 39, 58, 59). Analogously, experimental evolution of L. lactis KF147 to enhanced fitness and growth in milk was shown to be associated with suppression of gene repertoires associated with the import and utilization of a variety of typically plant environment-associated carbon sources, as well as mutations leading to functional reconstitution and elevated transcription of the peptide import system (opp) of this strain (52). Paradoxically, dairy strains of S. thermophilus still possess the genetic and phenotypic capacity to develop natural competence (14, 25, 39, 60), suggesting that competence development in this species contributes to fitness in this habitat. In contrast to the case for L. lactis, where carbon starvation has been associated with induction of late competence expression (23, 24), similar conditions have not been implied in competence regulation in S. thermophilus. This may suggest that S. thermophilus actively expresses the competence phenotype in the dairy environment, which may contribute to this species' fitness in the milk environment.
In nature, natural competence in bacteria is commonly a transient phenotypic state with a small window of opportunity to take up DNA (61), the activation and shutdown of which are subject to subtle regulation (47, 48) to prevent futile activation of the costly process and to sustain genomic stability. Analogously, optimal induction in L. lactis was achieved with a moderate level of ComX induction, whereas high-level induction of this regulator failed to lead to competence development (strain KF147) or led to significantly reduced levels of transformation (strains KW2, and IL-9000). Analogously, full induction of the late competence gene expression in Lactobacillus sakei was achieved by moderate levels of induction of its central regulator sigH (62). Moreover, high-level comX expression was consistently associated with reduced growth efficiency of the strains used in this study, which is illustrative of the tight connection between competence and growth (63). Previous comX expression studies using L. lactis IL1403, the parental strain of IL-9000, failed to elicit natural competence (20), which may have been due to inappropriate expression levels of comX or might have been caused by the fact that the endogenous comX gene of IL1403 was used, which contains an alternative start codon and appears to be truncated.
Taken together, the results of this study show that in L. lactis strains that carry complete late competence gene sets, a state of competence can be induced by controlled expression of comX; in particular, moderate expression of this regulator appears to be effective in activation of this phenotype. Naturally competent L. lactis strains could internalize plasmid and linear DNA from their environment with similar efficiencies. The conditions that naturally activate comX expression and contribute to the regulation of competence development in L. lactis remain to be established. Unraveling the in situ control mechanisms of natural competence in L. lactis would offer opportunities to exploit this phenotype for strain improvement purposes in this industrially important species.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strains used in this study are listed in Table 2. The publicly available draft genome sequences of 43 L. lactis strains (21, 22, 26–35) were used for comparative genomics of late competence genes by employing OrthoMCL to obtain orthologous group (OG) sequences in order to construct an orthologous gene matrix (64; M. Wels et al., unpublished data). L. lactis strains were routinely cultivated in M17 (Tritium, Eindhoven, The Netherlands) supplemented with 1% (wt/vol) glucose (Tritium, Eindhoven, The Netherlands) at 30°C without agitation. For competence experiments, L. lactis strains were cultivated in chemically defined medium (65, 66) supplemented with 1% (wt/vol) glucose (GCDM) (Tritium, Eindhoven, The Netherlands). Upon recovery after electrotransformation or natural transformation, L. lactis cells were cultivated in recovery medium (M17 supplemented with 1% glucose, 200 mM MgCl2, and 20 mM CaCl2). Escherichia coli TOP10 (Invitrogen, Breda, The Netherlands) was routinely cultivated in TY (Tritium, Eindhoven, The Netherlands) at 30°C with agitation. Antibiotics were added when appropriate: 5.0 μg/ml chloramphenicol, 10 μg/ml erythromycin, and 12.5 μg/ml tetracycline.
TABLE 2.
Strains, plasmids, and primers used for the experiments in this study
| Strain, plasmid, or primer | Relevant features or sequencea | Reference(s) |
|---|---|---|
| Strains | ||
| Lactococcus lactis | ||
| KF147 | Plant-derived strain belonging to L. lactis subsp. lactis | 22 |
| NZ6200 | ΔcomEA-EC::tetR derivative of strain KF147 | This study |
| IL-9000 | Dairy-derived strain belonging to L. lactis subsp. lactis harboring nisRK integrated into its genome | 21, 45 |
| NZ9000 | Dairy-derived strain belonging to L. lactis subsp. cremoris harboring nisRK integrated into its genome | 29, 44 |
| KW2 | Plant-derived strain belonging to L. lactis subsp. cremoris | 28 |
| Escherichia coli TOP10 | Cloning host | |
| Plasmids | ||
| pIL253 | Emr; high-copy-number plasmid replicative in L. lactis | 43 |
| pNZ8040 | Cmr; pNZ123 derivative containing pepN downstream of the nisin promoter | 42 |
| pNZ8150 | Cmr; pNZ123 derivative with ScaI site downstream of the nisin promoter for translational fusion | 41 |
| pNZ6200 | Cmr; pNZ8150 derivative containing comX from L. lactis KF147 downstream of the nisin promoter | This study |
| pNZ6201 | Cmr; pNZ8150 derivative containing comX from L. lactis MG1363 downstream of the nisin promoter | This study |
| pNZ6202 | Tetr; pIL253 derivative with eryR replacement by tetR from pGhost8 | This study |
| pGhost8 | Tetr; vector with thermosensitive replicon, tetR | 69 |
| Primers | ||
| C1 | ATGACATATTACTTGGAAGAAGAGG | |
| C2 | CCTTGGTACCTCACTCTTCGTCTTCTGAAAATAAGATG | |
| C3 | ATGACATATTACCTGGAAGAAAATGAATTCG | |
| C4 | CCTTGGTACCTTAATCATCATCTCGAGAAAATAGTATATTTTTG | |
| C5 | AGATCTAGTCTTATAACTATACTGAC | |
| C6 | GCCTTGGTTTTCTAATTTTGGTTC | |
| C7 | GGAATGAAACGAGCAGATGCCC | |
| C8 | GTAATCATGGTCATAGCTGTTTCCACTTTTATATACGAAAAAACTCTTGGA | |
| C9 | TCCAAGAGTTTTTTCGTATATAAAAGTGGAAACAGCTATGACCATGATTAC | |
| C10 | GTCTTTTGCTCACTTTTTCCTTTCATGTTGTAAAACGACGGCCAGTG | |
| C11 | GGCACTGGCCGTCGTTTTACAACATGAAAGGAAAAAGTGAGCAAAAGAC | |
| C12 | ATTCATTGGAAGAAGACCTTTCGG | |
| C13 | CCCATAAAGCCGTAAACCAAGTGAAAG | |
| C14 | GAAGACCAAATTCTTTATTTTGCGG | |
| C15 | GATATCGAATTCCTGCAGCCCG | |
| C16 | CCTTAGTACTCTACAGAATATTACTATACACTCCAGAAG | |
| SS1 | CAGCGGAAGAGAACCGTATT | |
| SS2 | CTCAGTTCCTTGGATGCCAT | |
| PS1 | AGCAGCATAATAGATTTATTGAATAGG | |
| PS2 | GCATCTAATTTAACTTCAATTCCTATTATAC | |
| Q1 | CCTGGCGTACGTGAAGATGTC | |
| Q2 | TTTCGTCAGCCGGAACATAGC | |
| Q3 | TCTATTAGAAGAGCAGAGCGATGGTC | |
| Q4 | CTTGATAATGTGCGCTCAAGCCTTC | |
| Q5 | GTCAGCAGGCAAAGCTCTGTC | |
| Q6 | ACTTGACTAGTGACCGAATTAGCAGAG | |
| Q7 | ATGGCGACAACTATTTCCGAGCTCC | |
| Q8 | CAAGTTTGTCAGTAGAAGTTGCGGTC | |
| Q9 | CGCAGACGAGTTCAATTGGGAG | |
| Q10 | CGAGCCTACTGGATCAGCAAAGAG |
Underlining indicates restriction sites used in subsequent cloning procedures. Emr, erythromycin resistance; Cmr, chloramphenicol resistance; Tetr, tetracycline resistance.
DNA manipulations.
Plasmid DNA from E. coli and L. lactis was isolated using a Jetstar 2.0 maxiprep kit (ITK Diagnostics bv, Uithoorn, The Netherlands). Notably, phenol-chloroform extraction was performed prior to loading on the Jetstar column for plasmid isolation from L. lactis cultures (67). Primers were synthesized by Sigma-Aldrich (Zwijndrecht, The Netherlands). PCR was performed using KOD polymerase according to the manufacturer's instructions (Merck Millipore, Amsterdam, The Netherlands). PCR products and DNA fragments in agarose gel were purified using the Wizard SV gel and PCR clean-up system (Promega, Leiden, The Netherlands). PCR-grade chromosomal DNA was isolated by using InstaGene Matrix (Bio-Rad, Veenendaal, The Netherlands). Ligations were performed using T4 ligase, and, when applicable, the products were transformed into either electrocompetent E. coli TOP10 (Invitrogen, Breda, The Netherlands) or L. lactis NZ9000, IL-9000, KW2, or KF147 (68).
Plasmid and mutant construction.
To enable controlled expression of comX in L. lactis, the comX gene was amplified by PCR using the primer pair C1-C2 or C3-C4 and L. lactis KF147 or MG1363 chromosomal DNA as a template, respectively. The resulting 502-bp comX amplicons were digested with KpnI (introduced in primers C2 and C4) and ligated into KpnI-ScaI-digested pNZ8150 (41), yielding pNZ6200 and pNZ6201, respectively. These comX overexpression vectors were transformed into electrocompetent L. lactis KF147, NZ9000, IL-9000, and KW2 (68). The natural competence potential in L. lactis strains was evaluated using pIL253 (43) when possible, but because of incompatibility of antibiotic resistance markers in strain KW2, an alternative plasmid in which the erythromycin resistance (eryR) gene was replaced by a tetracycline resistance (tetR) gene was constructed. To this end, a 1,644-bp tetR amplicon was generated using primers C15 and C16 with pGhost8 (69) as a template and cloned as a PstI-SacI fragment into similarly digested pIL253, yielding pNZ6202.
A comEA-EC deletion derivative of the L. lactis KF147 mutant was constructed using double-crossover recombination. To construct the mutagenesis fragment, the 5′ and 3′ flanking regions of the comEA-EC operon were amplified using chromosomal DNA of strain KF147 as a template and primer pairs C7-C8 and C11-C12, respectively. The tetracycline resistance-encoding gene tetR was amplified from pNZ7103 (70) using primers C9 and C10. Splicing by overhang extension PCR (SOE PCR) (71) was employed to join the three amplicons using the compatible sequence overhangs introduced by the primers in the individual PCRs (Table 2) and primers C7 and C12 for amplification in this PCR. The 6-kb SOE amplicon was purified from 1% agarose gels and transformed to naturally competent L. lactis KF147 (see Results). The anticipated comEA-EC deletion in the resulting derivatives of L. lactis KF147, yielding L. lactis NZ6200, was confirmed by PCR using the C13 and C14 primers.
Induction of competence in L. lactis.
Cells harboring pNZ6200, pNZ6201, or pNZ8150 were grown overnight in GCDM with appropriate antibiotics, followed by subculturing (1:65) in the same medium to an optical density at 600 nm (OD600) of 0.3, at which point Ultrapure nisin A (Handary, Brussels, Belgium) was added to the medium at a final concentration of 0.005, 0.01, 0.03, 0.05, 0.07, 0.1, 0.5, or 2 ng/ml. In parallel, 1 μg of plasmid DNA was added. Samples were incubated for 2 h at 30°C, after which 5 ml recovery medium was added and incubation was continued for another 2 h. Bacteria were pelleted by centrifugation at 4,000 × g for 8 min, and transformants were enumerated by plating of serial dilutions on GM17 plates. KF147 transformants were subjected to PCR analysis to assess the presence of the transformed plasmid with primers PS1 and PS2, whereas the strain-specific primers SS1 and -2 were used to confirm strain identity.
Analysis of competence gene expression.
RNA was isolated from L. lactis cultures using the High Pure RNA isolation kit (Roche Diagnostics Nederland B.V., Almere, The Netherlands), including an on-column DNase treatment. Eluted RNA was again treated with DNase (1 U; Thermo Fisher Scientific, Waltham, MA, USA) for 45 min at room temperature to remove remaining DNA, followed by DNase inactivation by the addition of EDTA to a final concentration of 25 mM and then heating at 75°C for 15 min. cDNA was prepared using 10 ng total RNA and random hexamer primers in the reverse transcription reaction (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's protocol. Control RNA samples that were not reverse transcribed were included as negative controls to ensure the absence of DNA contamination. Transcripts of competence genes were quantified using 2 μl cDNA and locus-specific primers for each competence-associated target gene (primers Q1 to Q10 [Table 2]) in SYBR green-quantified PCR (Bio-Rad, Veenendaal, The Netherlands). Transcript copy numbers were calculated using a template standard curve and normalized to the housekeeping control transcript of rpoA. These RT-qPCR analyses were performed in triplicate for each sample using the Freedom EVO 100 robot system (Tecan, Männedorf, Switzerland), and amplicon identities were verified using melting curve analysis. The nonparametric Mann-Whitney U test (one-tailed) was used to determine whether gene expression levels were significantly different between uninduced and induced conditions (P < 0.05).
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Jan Kok for providing L lactis strain IL-9000. We thank Sabri Cebeci and Koen Giesbers of NIZO for technical assistance.
This work was carried out within the BE-Basic R&D Program, which was granted an FES subsidy from the Dutch Ministry of Economic Affairs.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01320-17.
REFERENCES
- 1.Lerat E, Daubin V, Ochman H, Moran NA. 2005. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol 3:807–814. doi: 10.1371/journal.pbio.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dagan T, Artzy-Randrup Y, Martin W. 2008. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc Natl Acad Sci U S A 105:10039–10044. doi: 10.1073/pnas.0800679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Treangen TJ, Rocha EPC. 2011. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet 7:e1001284. doi: 10.1371/journal.pgen.1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeuchi N, Kaneko K, Koonin EV. 2014. Horizontal gene transfer can rescue prokaryotes from Muller's Ratchet: benefit of DNA from dead cells and population subdivision. G3 4:325–339. doi: 10.1534/g3.113.009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffith F. 1928. The significance of pneumococcal types. J Hyg (Lond) 27:113–159. doi: 10.1017/S0022172400031879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blokesch M. 2016. Natural competence for transformation. Curr Biol 26:R1126–R1130. doi: 10.1016/j.cub.2016.08.058. [DOI] [PubMed] [Google Scholar]
- 7.Gardan R, Besset C, Guillot A, Gitton C, Monnet V. 2009. The oligopeptide transport system is essential for the development of natural competence in Streptococcus thermophilus strain LMD-9. J Bacteriol 191:4647–4655. doi: 10.1128/JB.00257-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muschiol S, Balaban M, Normark S, Henriques-Normark B. 2015. Uptake of extracellular DNA: competence induced pili in natural transformation of Streptococcus pneumoniae. Bioessays 37:426–435. doi: 10.1002/bies.201400125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnston C, Martin B, Fichant G, Polard P, Claverys JP. 2014. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol 12:181–196. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 10.Mann JM, Carabetta VJ, Cristea IM, Dubnau D. 2013. Complex formation and processing of the minor transformation pilins of Bacillus subtilis. Mol Microbiol 90:1201–1215. doi: 10.1111/mmi.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo P, Morrison DA. 2003. Transient association of an alternative sigma factor, ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus pneumoniae. J Bacteriol 185:349–358. doi: 10.1128/JB.185.1.349-358.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterson S, Cline RT, Tettelin H, Sharov V, Morrison DA. 2000. Expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J Bacteriol 182:6192–6202. doi: 10.1128/JB.182.21.6192-6202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee MS, Morrison DA. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J Bacteriol 181:5004–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontaine L, Boutry C, de Frahan MH, Delplace B, Fremaux C, Horvath P, Boyaval P, Hols P. 2010. A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J Bacteriol 192:1444–1454. doi: 10.1128/JB.01251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fontaine L, Wahl A, Flechard M, Mignolet J, Hols P. 2015. Regulation of competence for natural transformation in streptococci. Infect Genet Evol 33:343–360. doi: 10.1016/j.meegid.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 16.Fontaine L, Dandoy D, Boutry C, Delplace B, de Frahan MH, Fremaux C, Horvath P, Boyaval P, Hols P. 2010. Development of a versatile procedure based on natural transformation for marker-free targeted genetic modification in Streptococcus thermophilus. Appl Environ Microbiol 76:7870–7877. doi: 10.1128/AEM.01671-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pestova EV, Havarstein LS, Morrison DA. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol 21:853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- 18.Håvarstein LS, Coomaraswamy G, Morrison DA. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci U S A 92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leroy F, De Vuyst L. 2004. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci Technol 15:67–78. doi: 10.1016/j.tifs.2003.09.004. [DOI] [Google Scholar]
- 20.Wydau S, Dervyn R, Anba J, Dusko Ehrlich S, Maguin E. 2006. Conservation of key elements of natural competence in Lactococcus lactis ssp. FEMS Microbiol Lett 257:32–42. doi: 10.1111/j.1574-6968.2006.00141.x. [DOI] [PubMed] [Google Scholar]
- 21.Bolotin A, Wincker P, Mauger S, Jaillon O, Malarme K, Weissenbach J, Ehrlich SD, Sorokin A. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res 11:731–753. doi: 10.1101/gr.GR-1697R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siezen RJ, Bayjanov J, Renckens B, Wels M, van Hijum SA, Molenaar D, van Hylckama Vlieg JE. 2010. Complete genome sequence of Lactococcus lactis subsp. lactis KF147, a plant-associated lactic acid bacterium. J Bacteriol 192:2649–2650. doi: 10.1128/JB.00276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Redon E, Loubiere P, Cocaign-Bousquet M. 2005. Transcriptome analysis of the progressive adaptation of Lactococcus lactis to carbon starvation. J Bacteriol 187:3589–3592. doi: 10.1128/JB.187.10.3589-3592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ercan O, Wels M, Smid EJ, Kleerebezem M. 2015. Genome-wide transcriptional responses to carbon starvation in nongrowing Lactococcus lactis. Appl Environ Microbiol 81:2554–2561. doi: 10.1128/AEM.03748-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blomqvist T, Steinmoen H, Havarstein LS. 2006. Natural genetic transformation: a novel tool for efficient genetic engineering of the dairy bacterium Streptococcus thermophilus. Appl Environ Microbiol 72:6751–6756. doi: 10.1128/AEM.01156-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wels M, Backus L, Boekhorst J, Dijkstra AR, Beerthuyzen M, Siezen RJ, Bachmann H, van Hijum SAFT. 2017. Draft genome sequences of 11 Lactococcus lactis subsp. cremoris strains. Genome Announc 5:e01739-16. doi: 10.1128/genomeA.01739-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Backus L, Wels M, Boekhorst J, Dijkstra AR, Beerthuyzen M, Kelly WJ, Siezen RJ, van Hijum SAFT, Bachmann H. 2017. Draft genome sequences of 24 Lactococcus lactis strains. Genome Announc 5:e01737-16. doi: 10.1128/genomeA.01737-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelly WJ, Altermann E, Lambie SC, Leahy SC. 2013. Interaction between the genomes of Lactococcus lactis and phages of the P335 species. Front Microbiol 4:257. doi: 10.3389/fmicb.2013.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wegmann U, O'Connell-Motherway M, Zomer A, Buist G, Shearman C, Canchaya C, Ventura M, Goesmann A, Gasson MJ, Kuipers OP, van Sinderen D, Kok J. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J Bacteriol 189:3256–3270. doi: 10.1128/JB.01768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, Shakhova V, Grigoriev I, Lou Y, Rohksar D, Lucas S, Huang K, Goodstein DM, Hawkins T, Plengvidhya V, Welker D, Hughes J, Goh Y, Benson A, Baldwin K, Lee JH, Diaz-Muniz I, Dosti B, Smeianov V, Wechter W, Barabote R, Lorca G, Altermann E, Barrangou R, Ganesan B, Xie Y, Rawsthorne H, Tamir D, Parker C, Breidt F, Broadbent J, Hutkins R, O'Sullivan D, Steele J, Unlu G, Saier M, Klaenhammer T, Richardson P, Kozyavkin S, Weimer B, Mills D. 2006. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci U S A 103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolotin A, Quinquis B, Ehrlich SD, Sorokin A. 2012. Complete genome sequence of Lactococcus lactis subsp. cremoris A76. J Bacteriol 194:1241–1242. doi: 10.1128/JB.06629-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ainsworth S, Zomer A, de Jager V, Bottacini F, van Hijum SA, Mahony J, van Sinderen D. 2013. Complete genome of Lactococcus lactis subsp. cremoris UC5099, host for a model lactococcal P335 bacteriophage. Genome Announc 1:e00119-12. doi: 10.1128/genomeA.00119-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato H, Shiwa Y, Oshima K, Machii M, Araya-Kojima T, Zendo T, Shimizu-Kadota M, Hattori M, Sonomoto K, Yoshikawa H. 2012. Complete genome sequence of Lactococcus lactis IO-1, a lactic acid bacterium that utilizes xylose and produces high levels of l-lactic acid. J Bacteriol 194:2102–2103. doi: 10.1128/JB.00074-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao Y, Lu Y, Teng KL, Chen ML, Zheng HJ, Zhu YQ, Zhong J. 2011. Complete genome sequence of Lactococcus lactis subsp. lactis CV56, a probiotic strain isolated from the vaginas of healthy women. J Bacteriol 193:2886–2887. doi: 10.1128/JB.00358-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X, Wang Y, Huo G. 2013. Complete genome sequence of Lactococcus lactis subsp. lactis KLDS 40325. PLoS One 1:e00962-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Godon J, Delorme C, Ehrlich SD, Renault P. 1992. Divergence of genomic sequences between Lactococcus lactis subsp. lactis and Lactococcus lactis subsp. cremoris. Appl Environ Microbiol 58:4045–4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayo B, van Sinderen D, Ventura M. 2008. Genome analysis of food grade lactic acid producting bacteria from basics to applications. Curr Genomics 9:169–183. doi: 10.2174/138920208784340731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altermann E, Russell WM, Azcarate-Peril MA, Barrangou R, Buck BL, McAuliffe O, Souther N, Dobson A, Duong T, Callanan M, Lick S, Hamrick A, Cano R, Klaenhammer TR. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc Natl Acad Sci U S A 102:3906–3912. doi: 10.1073/pnas.0409188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolotin A, Quinquis B, Renault P, Sorokin A, Ehrlich SD, Kulakauskas S, Lapidus A, Goltsman E, Mazur M, Pusch GD, Fonstein M, Overbeek R, Kyprides N, Purnelle B, Prozzi D, Ngui K, Masuy D, Hancy F, Burteau S, Boutry M, Delcour J, Goffeau A, Hols P. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat Biotechnol 22:1554–1558. doi: 10.1038/nbt1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siezen RJ, Bayjanov JR, Felis GE, van der Sijde MR, Starrenburg M, Molenaar D, Wels M, van Hijum SA, van Hylckama Vlieg JE. 2011. Genome-scale diversity and niche adaptation analysis of Lactococcus lactis by comparative genome hybridization using multi-strain arrays. Microb Biotechnol 4:383–402. doi: 10.1111/j.1751-7915.2011.00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mierau I, Kleerebezem M. 2005. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl Microbiol Biotechnol 68:705–717. doi: 10.1007/s00253-005-0107-6. [DOI] [PubMed] [Google Scholar]
- 42.de Ruyter PGGA, Kuipers OP, de Vos WM. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol 62:3662–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon D, Chopin A. 1988. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie 70:559–566. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
- 44.Kuipers OP, de Ruyter PGGA, Kleerebezem M, de Vos WM. 1998. Quorum sensing controlled gene expression in lactic acid bacteria. J Bacteriol 64:15–21. [Google Scholar]
- 45.Pinto JP, Zeyniyev A, Karsens H, Trip H, Lolkema JS, Kuipers OP, Kok J. 2011. pSEUDO, a genetic integration standard for Lactococcus lactis. Appl Environ Microbiol 77:6687–6690. doi: 10.1128/AEM.05196-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kleerebezem M, Beerthuyzen MM, Vaughan EE, de Vos WM, Kuipers OP. 1997. Controlled gene expression systems for lactic acid bacteria transferable nisin inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl Environ Microbiol 63:4581–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mirouze N, Berge MA, Soulet AL, Mortier-Barriere I, Quentin Y, Fichant G, Granadel C, Noirot-Gros MF, Noirot P, Polard P, Martin B, Claverys JP. 2013. Direct involvement of DprA, the transformation-dedicated RecA loader, in the shut-off of pneumococcal competence. Proc Natl Acad Sci U S A 110:E1035–E1044. doi: 10.1073/pnas.1219868110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weng L, Piotrowski A, Morrison DA. 2013. Exit from competence for genetic transformation in Streptococcus pneumoniae is regulated at multiple levels. PLoS One 8:e64197. doi: 10.1371/journal.pone.0064197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peterson SN, Sung CK, Cline R, Desai BV, Snesrud EC, Luo P, Walling J, Li H, Mintz M, Tsegaye G, Burr PC, Do Y, Ahn S, Gilbert J, Fleischmann RD, Morrison DA. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol Microbiol 51:1051–1070. doi: 10.1046/j.1365-2958.2003.03907.x. [DOI] [PubMed] [Google Scholar]
- 50.Zaccaria E, Wells JM, van Baarlen P. 2016. Metabolic context of the competence-induced checkpoint for cell replication in Streptococcus suis. PLoS One 11:e0153571. doi: 10.1371/journal.pone.0153571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mell JC, Redfield RJ. 2014. Natural competence and the evolution of DNA uptake specificity. J Bacteriol 196:1471–1483. doi: 10.1128/JB.01293-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quiberoni A, Rezaïki L, El Karoui M, Biswas I, Tailliez P, Gruss A. 2001. Distinctive features of homologous recombination in an ‘old’ microorganism, Lactococcus lactis. Res Microbiol 152:131–139. doi: 10.1016/S0923-2508(01)01183-4. [DOI] [PubMed] [Google Scholar]
- 53.Bachmann H, Starrenburg MJ, Molenaar D, Kleerebezem M, van Hylckama Vlieg JE. 2012. Microbial domestication signatures of Lactococcus lactis can be reproduced by experimental evolution. Genome Res 22:115–124. doi: 10.1101/gr.121285.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feiner R, Argov T, Rabinovich L, Sigal N, Borovok I, Herskovits AA. 2015. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat Rev Microbiol 13:641–650. doi: 10.1038/nrmicro3527. [DOI] [PubMed] [Google Scholar]
- 55.van Hylckama Vlieg JE, Rademaker JL, Bachmann H, Molenaar D, Kelly WJ, Siezen RJ. 2006. Natural diversity and adaptive responses of Lactococcus lactis. Curr Opin Biotechnol 17:183–190. doi: 10.1016/j.copbio.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 56.Siezen RJ, Renckens B, van Swam I, Peters S, van Kranenburg R, Kleerebezem M, de Vos WM. 2005. Complete sequences of four plasmids of Lactococcus lactis subsp. cremoris SK11 reveal extensive adaptation to the dairy environment. Appl Environ Microbiol 71:8371–8382. doi: 10.1128/AEM.71.12.8371-8382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu W, Mierau I, Mars A, Johnson E, Dunny G, McKay LL. 1995. Novel insertion sequence like element IS982 in lactococci. Plasmid 33:218–225. doi: 10.1006/plas.1995.1023. [DOI] [PubMed] [Google Scholar]
- 58.Callanan M, Kaleta P, O'Callaghan J, O'Sullivan O, Jordan K, McAuliffe O, Sangrador-Vegas A, Slattery L, Fitzgerald GF, Beresford T, Ross RP. 2008. Genome sequence of Lactobacillus helveticus, an organism distinguished by selective gene loss and insertion sequence element expansion. J Bacteriol 190:727–735. doi: 10.1128/JB.01295-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van de Guchte M, Penaud S, Grimaldi C, Barbe V, Bryson K, Nicolas P, Robert C, Oztas S, Mangenot S, Couloux A, Loux V, Dervyn R, Bossy R, Bolotin A, Batto JM, Walunas T, Gibrat JF, Bessieres P, Weissenbach J, Ehrlich SD, Maguin E. 2006. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc Natl Acad Sci U S A 103:9274–9279. doi: 10.1073/pnas.0603024103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goh YJ, Goin C, O'Flaherty S, Altermann E, Hutkins R. 2011. Specialized adaptation of a lactic acid bacterium to the milk environment: the comparative genomics of Streptococcus thermophilus LMD-9. Microb Cell Fact 10:S22. doi: 10.1186/1475-2859-10-S1-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zaccaria E, van Baarlen P, de Greeff A, Morrison DA, Smith H, Wells JM. 2014. Control of competence for DNA transformation in Streptococcus suis by genetically transferable pherotypes. PLoS One 9:e99394. doi: 10.1371/journal.pone.0099394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmid S, Bevilacqua C, Crutz-Le Coq A-M. 2012. Alternative sigma factor σH activates competence gene expression in Lactobacillus sakei. BMC Microbiol 12:32. doi: 10.1186/1471-2180-12-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Desai K, Mashburn-Warren L, Federle MJ, Morrison DA. 2012. Development of competence for genetic transformation of Streptococcus mutans in a chemically defined medium. J Bacteriol 194:3774–3780. doi: 10.1128/JB.00337-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fischer S, Brunk BP, Chen F, Gao X, Harb OS, Iodice JB, Shanmugam D, Roos DS, Stoeckert CJ Jr. 2011. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinformatics Chapter 6:Unit 6.12.11.19. doi: 10.1002/0471250953.bi0612s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Otto R, ten Brink BV, Konings HWN. 1983. The relation between growth rate and electrochemical proton gradient of Streptococcus cremoris. FEMS Microbiol Lett 16:69–74. doi: 10.1111/j.1574-6968.1983.tb00261.x. [DOI] [Google Scholar]
- 66.Poolman B, Konings WN. 1988. Relation of growth of Streptococcus lactis and Streptococcus cremoris to amino acid transport. J Bacteriol 170:700–707. doi: 10.1128/jb.170.2.700-707.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 68.Wells JM, Wilson PW, Le Page RWF. 1993. Improved cloning vectors and transformation procedure for Lactococcus lactis. J Appl Bacteriol 74:629–636. doi: 10.1111/j.1365-2672.1993.tb05195.x. [DOI] [PubMed] [Google Scholar]
- 69.Maguin E, Prevost H, Ehrlich SD, Gruss A. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol 178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bron PA, Grangette C, Mercenier A, de Vos WM, Kleerebezem M. 2004. Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J Bacteriol 186:5721–5729. doi: 10.1128/JB.186.17.5721-5729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Horton RM, Cai Z, Ho SN, Pease LR. 1990. Gene splicing by overlap extension tailor-made genes using the polymerase chain reaction. Biotechniques 8:528–535. [PubMed] [Google Scholar]
- 72.Dijkstra AR, Setyawati MC, Bayjanov JR, Alkema W, van Hijum SA, Bron PA, Hugenholtz J. 2014. Diversity in robustness of Lactococcus lactis strains during heat stress, oxidative stress, and spray drying stress. Appl Environ Microbiol 80:603–611. doi: 10.1128/AEM.03434-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.