ABSTRACT
Radioactive strontium (90Sr) leaked into saline environments, including the ocean, from the Fukushima Daiichi Nuclear Power Plant after a nuclear accident. Since the removal of 90Sr using general adsorbents (e.g., zeolite) is not efficient at high salinity, a suitable alternative immobilization method is necessary. Therefore, we incorporated soluble Sr into biogenic carbonate minerals generated by urease-producing microorganisms from a saline solution. An isolate, Bacillus sp. strain TK2d, from marine sediment removed >99% of Sr after contact for 4 days in a saline solution (1.0 × 10−3 mol liter−1 of Sr, 10% marine broth, and 3% [wt/vol] NaCl). Transmission electron microscopy and energy-dispersive X-ray spectroscopy showed that Sr and Ca accumulated as phosphate minerals inside the cells and adsorbed at the cell surface at 2 days of cultivation, and then carbonate minerals containing Sr and Ca developed outside the cells after 2 days. Energy-dispersive spectroscopy revealed that Sr, but not Mg, was present in the carbonate minerals even after 8 days. X-ray absorption fine-structure analyses showed that a portion of the soluble Sr changed its chemical state to strontianite (SrCO3) in biogenic carbonate minerals. These results indicated that soluble Sr was selectively solidified into biogenic carbonate minerals by the TK2d strain in highly saline environments.
IMPORTANCE Radioactive nuclides (134Cs, 137Cs, and 90Sr) leaked into saline environments, including the ocean, from the Fukushima Daiichi Nuclear Power Plant accident. Since the removal of 90Sr using general adsorbents, such as zeolite, is not efficient at high salinity, a suitable alternative immobilization method is necessary. Utilizing the known concept that radioactive 90Sr is incorporated into bones by biomineralization, we got the idea of removing 90Sr via incorporation into biominerals. In this study, we revealed the ability of the isolated ureolytic bacterium to remove Sr under high-salinity conditions and the mechanism of Sr incorporation into biogenic calcium carbonate over a longer duration. These findings indicated the mechanism of the biomineralization by the urease-producing bacterium and the possibility of the biomineralization application for a new purification method for 90Sr in highly saline environments.
KEYWORDS: biomineralization, bioremediation, halophilic, marine environment, radionuclide, urease
INTRODUCTION
At the Fukushima Daiichi Nuclear Power Plant (FDNPP), the nuclear reactors are cooled by the continuous treatment of contaminated water that contains radionuclides. Some of this contaminated water with high saline concentrations has leaked to the port of FDNPP and into groundwater (1). It has been estimated that among the radionuclides, 3.5 PBq of 134Cs and 137Cs and 52 GBq of 90Sr have leaked into the ocean (2). Since 90Sr has a long half-life (28.9 years), humans are potentially exposed to it through bioconcentration from the food chain in the contaminated environments, especially aquatic sites (3). Highly toxic 90Sr has chemical properties similar to those of calcium (Ca); therefore, if ingested, it replaces Ca in the bones and persists in the body, resulting in a long-term negative health impact (3). Therefore, it is morally urgent and socially important to promptly clean leaked radionuclides to minimize their anthropogenic and environmental impacts. Minerals with a capacity for Cs+ and Sr2+ adsorption, such as zeolite, are widely used as removal adsorbents for radionuclides. Nevertheless, under high saline concentrations, the adsorption efficiency of the mineral adsorbents decreases for all radionuclides (4, 5). Hence, it is difficult to use these adsorbent materials directly to immobilize radionuclides from contaminated groundwater with high salinity or even clean the seawater in the port of FDNPP—in the latter case, no effective immobilization method has yet been developed. To solve this problem, a new method to immobilize radionuclides from a highly saline environment should be developed.
Recently, various methods that use biomineralization by microorganisms to remove radionuclides from contaminated solutions have been studied (6–8). Using biogenic minerals, dispersed radionuclides at low concentrations can be removed for a long period at a low cost. Some studies have shown the use of calcium carbonate minerals among the group of biogenic minerals to successfully remove soluble Sr (8, 9). Biogenic calcium carbonate is produced through different metabolic pathways, such as ureolysis, photosynthesis, sulfate reduction, and methane oxidation (10, 11). In addition, metal-binding functional groups, such as carboxyl, hydroxyl, amine, and methyl groups, are present in the extracellular polymeric substance (EPS) and the cell surface and play an important role in Ca2+ and Sr2+ absorption and nucleation (12, 13).
Urease-producing microorganisms generate ammonium and bicarbonate ions through the hydrolysis of urea and, thus, increase pH in the growth environment (equation 1), which promotes calcium carbonate precipitate (equation 2) (8, 9).
| (1) |
| (2) |
Specifically, Sr is removed from solution through incorporation into biogenic minerals during the generation of calcium carbonate or by adsorption onto the surface of the generated calcium carbonate (8, 9). However, to the best of our knowledge, studies of the cleanup of radioactive Sr under high-salinity conditions, such as encountered with seawater, by biomineralization using microorganisms and characterization of its biominerals are still lacking. In the present study, to establish a radionuclide removal method for a highly saline environment, we showed that (i) halotolerant microorganisms that produce carbonate minerals based on the hydrolysis of urea can be isolated from marine sediments and that (ii) soluble Sr is removed effectively from a highly saline solution. In addition, we (iii) examined the morphology of the biogenic carbonate minerals (BCM), together with chemical forms and local conditions of Sr incorporated in BCM using electron microscopy and X-ray absorption fine-structure analyses.
RESULTS
Isolation of a urease-producing microorganism and Sr removal in a highly saline solution.
From the urease-producing candidates isolated from the marine sediment samples, we obtained eight strains that had robust reproducibility of a red halo indicating pH increase on the isolation plate medium. To examine the Sr removal capacity of these eight strains, the concentration of soluble Sr in broth was measured after 12 days of cultivation. Four isolated strains, TK2a, TK2b, TK2d, and TK4c, reduced the soluble Sr from a concentration of 1.0 × 10−3 mol liter−1 at the beginning to 0.7 × 10−3, 0.6 × 10−3, 0.1 × 10−3, and 0.6 × 10−3 mol liter−1, respectively, after 12 days. For the other four strains tested, the soluble Sr barely decreased, similar to the case with the negative control. Although the isolates TK2a, TK2b, and TK2d showed high Ca removal rates, 53, 82, and 93%, respectively, only the TK2d strain was able to remove Sr at a high rate. These results revealed that not all of the urease-producing bacteria eliminated Sr from the medium. Since the TK2d strain removed the largest amount of soluble Sr among four strains, this strain was selected for further study.
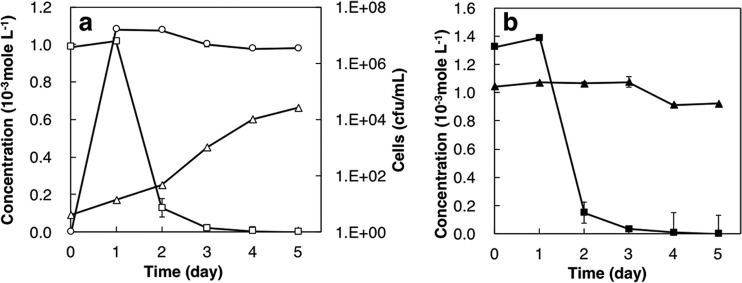
The test for Sr removal by the TK2d strain shows that this strain reduced the concentration of soluble Sr to 0.13 × 10−3 mol liter−1 (i.e., Sr removal of 87%) within 2 days; subsequently, the concentration gradually reduced to below 1.0 × 10−7 mol liter−1 after 4 days (i.e., Sr removal of ca. 100% was attained) (Fig. 1a). In addition, the TK2d strain removed 89% of the initial soluble Ca (1.3 × 10−3 mol liter−1) in 2 days (Fig. 1b), similar to the case with the soluble Sr; however, there was no decrease in soluble Mg (Fig. 1b). The growth of viable cell numbers in the Sr removal test peaked on the first day, to about 1.0 × 107 CFU/ml, but subsequently, there was almost no change through the 5-day experiment (Fig. 1a). The concentration of NH4+ in marine broth 2216 (MB), an indicator of the consumption of urea, gradually increased to 0.66 × 10−3 mol liter−1 at 5 days (Fig. 1a). These results indicated that in a saline solution, the soluble Sr concentration decreases with the concurrent consumption of urea to produce NH4+.
FIG 1.
Time course of soluble Sr, NH4+, and cell growth (a) and soluble Ca and Mg in the Sr removal test (b) using the TK2d strain under 3.0% NaCl. Open squares, soluble Sr concentration; open triangles, NH4+ concentration; open circles, viable cell number; closed squares, soluble Ca concentration; closed triangles, soluble Mg concentration.
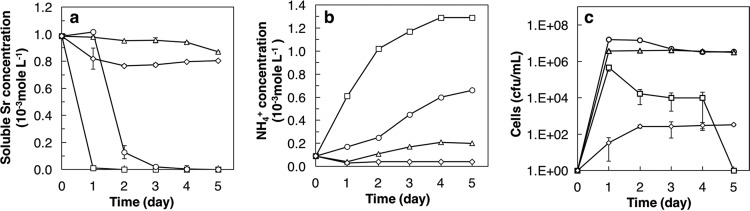
The concentrations of Sr and NH4+ in MB containing 0.3, 3.0, 5.0, or 10% NaCl during the culture of the TK2d strain and the viable cell numbers are shown in Fig. 2a, b, and c, respectively. The concentration of Sr decreased to 0.01 × 10−3 mol liter−1 within 1 day at 0.3% NaCl and 3 days at 3% NaCl and to ca. 0.8 × 10−3 mol liter−1 at 5 days at 5.0 and 10.0% NaCl. The concentrations of NH4+ at 5 days increased to 1.3 × 10−3 mol liter−1, 0.66 × 10−3 mol liter−1, 0.20 × 10−3 mol liter−1, and 0.04 × 10−3 mol liter−1, respectively, at 0.3%, 3.0%, 5.0%, and 10.0% NaCl. The results showed lower concentrations of NH4+ with increasing NaCl concentrations in MB, indicating that the urease activity of the TK2d strain is inhibited by NaCl. The viable cell numbers of the TK2d strain increased to approximately 1.0 × 106 CFU/ml after 1 day under 0.3% NaCl, followed by a rapid decrease until they died out by 5 days. The numbers of viable cells grown in MB depended on the concentration of NaCl and were in the order of 3.0% = 5.0% > 0.3% > 10%; that is, the TK2d strain is a moderate halophile (14). These results indicated that a decrease of the soluble Sr concentration at 3.0% NaCl was preceded by TK2d strain activity and was similar to that at 0.3% NaCl, but the rate of the decrease was lower at 3.0% than at 0.3% NaCl.
FIG 2.
Time courses of soluble Sr (a), soluble NH4+ (b), and viable cell number (c) under various NaCl concentrations. Open squares, 0.3% NaCl; open circles, 3.0% NaCl; open triangles, 5.0% NaCl; open diamonds, 10% NaCl.
The partial 16S rRNA gene sequence analysis showed that the TK2d strain has 96.2% homology with the nonpathogenic bacterium Bacillus humi LMG 22167, 93.9% with Bacillus luteolus YIM 93174, and 93.1% with Bacillus galliciensis BFLP-1. The phylogenetic tree construction based on 16S rRNA revealed that the TK2d strain could be classified into the genus Bacillus (see Fig. S1 in the supplemental material). The morphology of the TK2d strain (Fig. S2) showed that the TK2d strain was rod shaped, with a minor axis of 0.9 to 1.0 μm and a major axis of 1.5 to 2.5 μm with motility. A physiological characterization summarized in Table S1 indicated that the TK2d strain grew well under aerobic conditions and multiplied under conditions of 0.3 to 10% NaCl and 30 to 37°C, forming ellipsoidal spores at the cell ends. Furthermore, the TK2d strain was positive for catalase and oxidase reactions and was Gram positive. Together, the results showed that the TK2d strain is different from B. humi, primarily in terms of urease activity, arbutin fermentation ability, and nitrate reduction ability. Therefore, this single strain was named Bacillus sp. strain TK2d.
Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) analyses of precipitates.
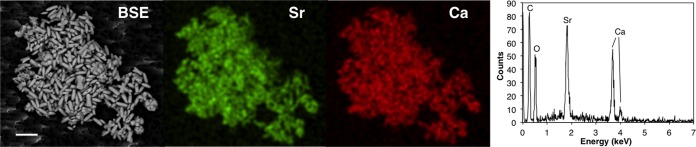
The backscattered image of the precipitates after 3 days of contact in MB containing 1.0 × 10−3 mol liter−1 of Sr and 3.0% NaCl (Fig. 3, BSE) showed that ca. 5.0-μm rod-like shaped solids were aggregated. The energy-dispersive X-ray spectroscopy (EDS) spectrum indicated that the precipitates contained Sr and Ca (Fig. 3, graph). The elemental mapping of Sr and Ca (Fig. 3, Sr and Ca) revealed that the two elements were present at nearly the same position in the precipitates. The presence of C in solids was not identified, because C was used as the base material. These results indicated that the rod-like shaped precipitates with the same shape as the microbes, involving Ca and Sr, were formed during the culture of the TK2d strain in MB. Here, we refer to the precipitates formed by the TK2d strain as BCM.
FIG 3.
SEM-EDX analysis of precipitates in culture of the TK2d strain after 3 days. Back-scattered electron image (BSE), elemental mappings (Sr and Ca), and EDS spectrum are shown. Bar = 10 μm.
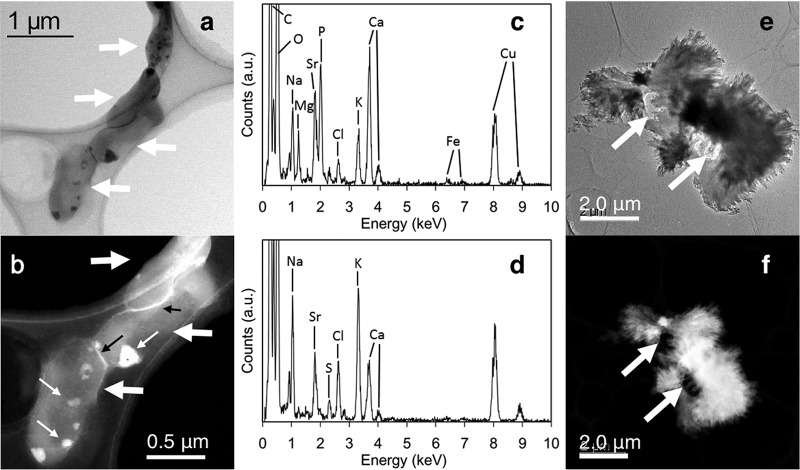
TEM and high-angle annular dark-field scanning TEM (HAADF-STEM) images of the BCM after 2 days (Fig. 4a and b) showed that the cells of the TK2d strain were recognizable (shown by thick white arrows). Figure 4a shows that bright solids formed inside cells (e.g., thin white arrows). The HAADF-STEM and TEM photographs taken from parts of cells (shown in Fig. S3 and S4) support the formation of precipitates inside cells. The HAADF-STEM image (Fig. 4b) shows that the precipitates contained heavier elements than cell components. Portions of the cells were brighter than whole cells, indicating the accumulation of heavy metals on the cell surface (Fig. 4b, black arrows). The EDS spectrum of the precipitates formed inside cells (Fig. 4c) indicated that Mg, Sr, P, and Ca were present. The peak intensity of P was nearly the same as that of Ca, indicating that biogenic calcium-magnesium phosphate minerals (BPM) were formed and involved Sr. The presence of Sr in the BPM indicated intracellular Sr accumulation.
FIG 4.
TEM and HAADF-STEM images of precipitates in culture of the TK2d strain after 2 and 8 days of cultivation. (a and b) Bright-field TEM (BFTEM) (a) and HAADF-STEM (b) images of the cells and the precipitates inside cells after 2 days of cultivation; (c) EDS spectrum of the precipitate inside cells; (d) EDS spectrum of the bright rim (leftmost black arrow) in panel b; (e and f) BFTEM (e) and HAADF-STEM (f) images of the cells and the precipitates after 8 days of cultivation. Thick white arrows in panels a, b, e, and f indicate cells of the TK2d strain. Thin white arrows in panel b indicate the precipitates. a.u., arbitrary units.
The EDS spectrum of the bright rim (Fig. 4d) showed the presence of Na, Sr, S, Cl, K, and Ca, indicating that Sr had accumulated at the rim of the cells. Since the intensity of the Ca peak was high, Ca accumulation occurred at the rim of the cells. Interestingly, a low intensity of the P peak revealed that no phosphate minerals had developed on the rim.
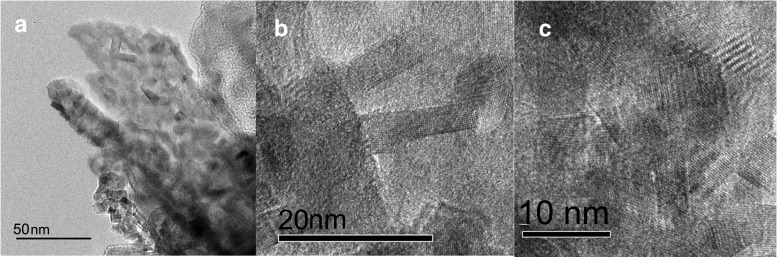
The TEM and HAADF images of the BCM at 8 days (Fig. 4e and f) indicated that the cells (shown by arrows) were surrounded by the BCM containing Sr and Ca, according to EDS analysis (Fig. S5). High-resolution TEM (HRTEM) images (Fig. 5a) and magnified images (Fig. 5b and c) showed that the presence of distinct fringes extended to different directions in the precipitates from outside cells, indicating the formation of polycrystalline minerals. The selected area electron diffraction (SAED) pattern from the BCM in Fig. 5c showed that the interplanar distance values (d-values) of the precipitate were approximately 3.41, 2.81, 2.58, 2.42, 2.03, and 1.81 Å. These d-values were consistent with the structure of carbonate salts as aragonite (15). These results clearly showed that the calcium carbonate formation of aragonite was formed as BCM by a ureolysis process of the TK2d strain.
FIG 5.
TEM analysis of precipitates from the TK2d strain after 10 days of cultivation. (a) HRTEM image; (b and c) magnified HRTEM images of needle-like crystals in panel a. Bars = 50 nm (a), 20 nm (b), and 10 nm (c).
The TEM photograph of the abiotic Sr-CaCO3 (Fig. S6a) shows that at least some of the abiotic calcium carbonate formed euhedral crystals. The size of the crystals was relatively larger than those of the carbonate biominerals produced by the TK2d strain. A five-point analysis of the crystal showed that the concentration of Ca was extremely high, because of the initial concentration ratio between Ca and Sr under synthesizing conditions (Fig. S6b). The addition of Na2CO3 in the MB resulted in the formation of calcium carbonate precipitates. The concentration of Sr, Mg, and Ca decreased in the MB solution, indicating that the abiotically formed calcium carbonate precipitates accumulated not only Sr but also Mg (Fig. S7). In contrast, BCM involved Mg below the detection limit of the EDS analysis. Hence, the incorporation of Sr and Mg by BCM is extremely different from that by abiotically formed calcium carbonate precipitate.
Analysis of the chemical state of Sr solids with XANES.
X-ray absorption near-edge structure (XANES) spectra of Sr in the BCM after 5 days (initial soluble Sr concentration: 1.0 × 10−3 mol liter−1) of cultivation (Fig. 6) along with the spectra of SrCO3 and SrCl2 as standards and abiotic Sr-CaCO3 precipitates show that the first peak appeared at approximately 16,110 eV, whereas the second peak appeared at 16,140 eV. Likewise, in the spectra of the standard Sr sample, the energy of the first peak of SrCO3 was at approximately 16,110 eV, whereas that of the second peak was 16,150 eV. Similarly, for SrCl2 the first and second peaks were at 16,110 and 16,145 eV, respectively. Hence, the energy at which the first peak appeared was nearly identical for all samples, whereas that of the second peak varied slightly depending on the specific sample.
FIG 6.
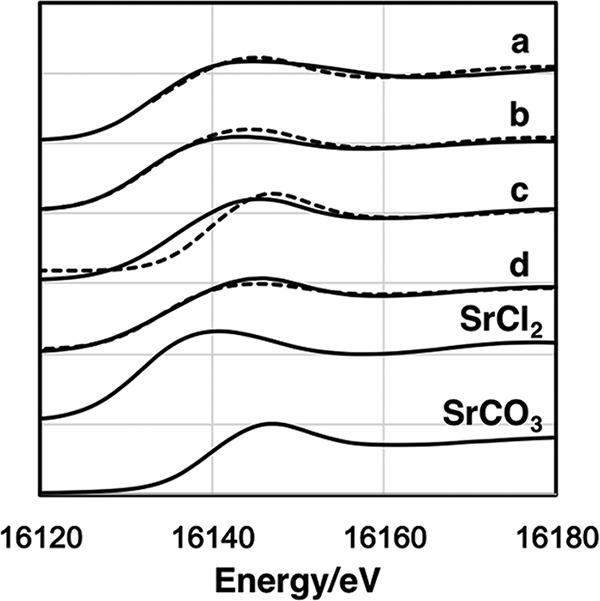
KIII-edge XANES spectra of Sr for abiotic Sr-CaCO3 precipitates (a), BCM of 2 mM Sr for 5 days (b), BCM of 1 mM Sr for 10 days (c), BCM of 1 mM Sr for 5 days (d), SrCl2, and SrCO3. Solid lines and dashed lines show experimental data and a linear combination fitting, respectively.
A linear combination fitting (LCF) of the XANES spectra for Sr in the precipitates formed by the TK2d strain and abiotic Sr-CaCO3 precipitates with the standards is shown in Fig. 6 (dashed lines). The contents of SrCl2 and SrCO3 are listed in Table 1. Larger amounts of the chemical form SrCO3 resulted from longer exposure duration and lower Sr concentrations at the beginning. Interestingly, half of the Sr in the abiotic precipitates was present as SrCl2. These results clearly revealed that Sr changed its chemical form after being incorporated in the BCM.
TABLE 1.
Contents of the chemical forms SrCl2 and SrCO3 in the BCMs and abiotic Sr-CaCO3
| Sample type | Initial Sr concn (10−3 mol liter−1) | Cultivation time (days) | Chemical species (%) |
|
|---|---|---|---|---|
| SrCl2 | SrCO3 | |||
| BCM | 2.0 | 5 | 65 | 35 |
| BCM | 1.0 | 5 | 48 | 52 |
| BCM | 1.0 | 10 | 13 | 87 |
| Sr-CaCO3 precipitate | 1.0 | 0 (no culture) | 55 | 45 |
DISCUSSION
The present study attempted to establish a new biological method in which the concentration of dissolved radionuclides in seawater is reduced and precipitated in ports and bays in the long term. A halotolerant carbonate mineral-producing bacterium was isolated from the marine environment and successfully removed soluble Sr. The results clearly showed not only that the halophilic bacterium Bacillus sp. strain TK2d could be isolated from the sediment in Tokyo Bay (TK) but also that up to 99% of the soluble Sr at NaCl concentrations in the range of 0.3 to 3.0% (Fig. 2a) could be solidified and removed. Furthermore, Sr/Ca molar ratios in the biogenic precipitates after 7 days of cultivation at 0.3% and 3.0% NaCl were 0.77 and 0.75, respectively, higher than that in the initial culture, 0.74. This result suggests that the TK2d strain selectively removed Sr by precipitation. The remarkable ability of this strain to remove Sr indicated that it is suitable for immobilization of radionuclides in highly saline environments, such as marine environments or groundwater with high ionic strength.
The cell surfaces of bacteria contain a range of organic acid functional groups that can adsorb metal cations and protons from solution (12, 13, 16, 17). The pKa values of the functional groups of bacteria were compiled by Borrok and Fein (17). The adsorption of the metal cations of Cd, Pb, Sr, and Zn by different species of bacteria showed similar pH dependences among species (18). Furthermore, bacterial cells were surrounded by EPS with a thin layer and absorbed Ca2+ ions (12, 13). Since Sr is present as Sr2+ ions in neutral pH solutions, the TK2d strain adsorbed Sr2+ ions on the functional groups of the cell wall and/or EPS. The EDS spectrum of the cell surface indicated Sr accumulation (Fig. 4d; see also Fig. S4ii), suggesting that the first step of Sr accumulation by the TK2d strain is adsorption on the cell surface (Fig. S8i). The TK2d strain hydrolyzed urea intracellularly and released HCO3− and NH4+ ions extracellularly, like most ureolytic bacteria (Fig. 2b) (19). After the (chemical) reaction with Sr2+ accumulated by adsorption on the cell surface and HCO3− released from the cells (Fig. S8i and ii), the BCM containing Sr was formed on the cell surface and the bigger one (crystallized products) left the cells (Fig. 3 and 4; see also Fig. S4). These results suggested that Sr carbonate mineralization by the TK2d strain is caused by adsorption on the cell surface as the first step (Fig. S8i), followed by the concurrent incorporation of Sr during the formation of extracellular mineralization (Fig. S8ii). Thus, this stepwise reaction is key for Sr bioaccumulation.
The EDS spectrum at the cell surface showed Ca and Sr, but not Mg, accumulation (Fig. 4d; see also Fig. S4ii). Generally, microbial ureolysis progresses inside cells and then produces bicarbonate and NH3+ ions, which are released outside cells (11, 20); this reaction may have occurred in the TK2d strain. Released bicarbonate ions probably reacted with the accumulated Ca and Sr to form Sr-containing BCM at the cell surface (Fig. 4d; see also Fig. S4ii). Needle-like BCM developed from the cells of the TK2d strain (Fig. 4e and 5), which supports the process of BCM formation at the cell surface. Less containment of Mg in BCM strongly suggests that the formation of BCM progressed not in the bulk solution but at the cell surface. XANES analysis of Sr in BCM showed a tighter association of Sr over a longer duration, indicating that the incorporated Sr in BCM was not soluble in solution until BCM was stable.
Microorganisms adjust the amount of Ca and Mg in their cells through ion channels (21); therefore, in the microspace around microbial cells, the Ca2+/Mg2+ ratio differs from that in the macroenvironment (i.e., the environment “away” from microorganisms). Rivadeneyra et al. reported that a moderate halobacterium, Halobacillus trueperi, preferentially generated calcite in artificial seawater, but when the carbonate species that was inorganically generated under the same condition was calculated using PHREEQC, a geochemical simulation program, it was found that the carbonate containing Mg, dolomite, was the most easily generated (22). Alternatively, as reported by Sánchez-Román et al., calcite and magnesium calcite were formed by Halomonas aquamarina under conditions in which aragonite was inorganically generated (23). In present study, the generation of aragonite BCM containing no Mg by the TK2d strain supported the proposal by Rivadeneyra et al. (22) and Sánchez-Román et al. (23) that a difference in the species of microorganisms leads to a difference in the ratio of Ca2+ to Mg2+ around the cells. Thus, the process of carbonate mineral formation by microorganisms is distinct from the abiotic process; moreover, it will possibly differ based on the type of microorganism involved. Therefore, it is important to select microorganisms that are functionally suited to Sr removal through biomineralization applications. Owing to its Mg ion channel, the TK2d strain that removed Sr with CaCO3 while leaving Mg intact may create an environment in which Ca and Sr are preferentially solidified and precipitated. In this way, the carbonate precipitation method using the TK2d strain shows great potential for immobilization radionuclide 90Sr in highly saline environments.
Homogeneous partitioning coefficients (DSr) described the extent of Sr removal from the liquid phase into the mineral phase and were calculated using the following equation (24):
| (3) |
where X is the solid-phase molar concentration of Sr and Ca and [Sr] and [Ca] are the liquid-phase concentrations. In the study, DSr values were 1.01 to 1.02 at 0.3% NaCl and 0.62 to 1.01 at 3.0% NaCl. Previous studies showed soluble Sr being incorporated into calcite generated by Sporosarcina pasteurii in a nonsaline freshwater environment, and DSr values were 0.6 (9), 0.46 (24), and 1.0 in quasi-two-dimensional (quasi-2D) porous medium reactor (25). The DSr values of the TK2d strain were slightly higher than that of S. pasteurii in previous studies of Sr bioprecipitation, without considering different experimental conditions such as temperature and salinity. In addition, Fujita et al. suggested that DSr values of Sr precipitation by abiotic conditions are lower than for that by microbial ureolysis (9). Furthermore, the potential use of ureolytic microbes has been shown for 90Sr immobilization at the Hanford 100-N area in Washington State (26). These results showed the formation of calcite to incorporate Sr from freshwater. However, our study indicated the elimination of Sr in a highly saline solution by concurrent incorporation into biogenic aragonite and biogenic phosphate minerals by the TK2d strain. To demonstrate the feasibility of 90Sr purification in a highly saline environment by the biomineralization, further work will be focused on the exploration of potential answers for the removal of 90Sr using model and actual contaminated seawater.
MATERIALS AND METHODS
Sediment sampling.
Marine sediments were obtained from seven stations in Tokyo Bay (TK) using Ekman-Barge sediment samplers (Rigo-sha, Tokyo, Japan) on the Hiyodori, a small research and training vessel of the Tokyo University of Marine Science and Technology. The sediment samples were mixed and diluted with a sterilized physiological saline solution at a proportion of 10% (wt/vol). The diluted samples were then used for the isolation of the study microorganisms.
Medium and cultivation conditions.
Marine broth 2216 (MB; 3.74 g/liter) (Becton Dickinson and Company, MD) with NaCl (final concentration, 30 g/liter) was autoclaved and then filtered (0.45-μm pore size; Corning, NY) under aseptic conditions. Filtered MB with urea (final concentration, 20 g/liter) was used as the artificial seawater medium. The medium was solidified with 1.5% (wt/vol) agar for colony formation and isolation. Phenol red (PR; final concentration, 12 mg/liter) was added to the solidified medium at pH 7.6 to detect increases in pH. To test the removal of Sr, Sr was added (final concentration, 1.0 × 10−3 mol liter−1) as SrCl2 to MB. The inoculated medium plates were incubated at 30°C; the liquid culture medium was placed in a rotational shaking incubator at 120 rpm.
Isolation of urease-producing microorganisms.
For the isolation of a urease-producing microbe, ureolysis in the plate medium was evidenced by a red halo that appeared as the pH increased (equation 1) around a colony detected in PR. A diluted sample solution (20 μl) was inoculated onto an MB plate containing PR and then incubated at 30°C to form visible colonies. A colony with a red halo was transferred to a fresh medium plate and incubated. These transfers were repeated twice to isolate a single colony with the red halo indicator.
Sr removal test.
To estimate soluble Sr removal from MB containing Sr, isolates were cultured therein. The isolated colonies were inoculated into 5.0 ml of MB dispensed in a 15-ml tube and incubated at 30°C for 24 h. The 0.5-ml culture broth (optical density at 600 nm [OD600] = 0.1) was inoculated into 50 ml of fresh MB containing 1.0 × 10−3 mol liter−1 of Sr in a 100-ml Erlenmeyer flask and then incubated under the same conditions as described above. The incubation was conducted in triplicate. The culture was sampled (1.0-ml aliquots) every 24 h, and the aliquots were centrifuged at 21,000 × g for 5 min at 4.0°C. The supernatant and precipitate were used for the quantification of the soluble Sr concentration and observation of bacteria and solidified products. The concentration of Sr in the supernatant was measured using inductively coupled plasma-atomic emission spectrometry (ICP-AES; iCAP6300DUO; Thermo Fisher Scientific, Kanagawa, Japan), as described below. The percent removal of soluble Sr from the aqueous solution was determined by the equation
| (4) |
where C0 is the initial concentration of the soluble element and Cx is the concentration of the soluble element after x days of cultivation.
As a negative control, MB containing Sr without any isolates was incubated under the same conditions. Cell growth was quantified during the Sr removal test by recording the CFU count on the MB plate.
To determine the halotolerance of the isolates and their capability to remove soluble Sr, NaCl was added at 3.0 to 10.0% (wt/vol) to MB, and tests for the removal of soluble Sr were performed (by following the same procedure as described earlier).
Measurement of urease activity.
Urease activity was determined by measuring the amount of NH4+ released from urea (equation 1), according to the indophenol blue method (27) and using the reagent set for a Lambda-9000 water analyzer (Kyoritsu Chemical-Check, Tokyo, Japan).
Quantification of soluble elements by ICP-AES.
Concentrations of Sr and other key elements were measured by ICP-AES. Each sample was centrifuged (21,000 × g, 5 min, and 20°C) to separate the supernatant and precipitate (i.e., cell pellet). The supernatant was filtered through a disc filter (0.2-μm pore size; Kurabo, Osaka, Japan) and appropriately diluted with ultrapure water (UPW) (Barnstead NANOpure Diamond; Thermo Fisher Scientific). The concentrations of Sr and other elements were calculated by comparison to a standard curve generated by the mixed standard solution XSTC-622 (SPEX CertiPrep, Metuchen, NJ), and measurements were carried out in triplicate per sample by ICP-AES. The resulting concentrations were within 5% of the sample average.
Cell observations and elemental analyses by SEM-EDS.
After the Sr removal test, the cell pellet was mixed with a glutaraldehyde solution at a final concentration of 2.0% (vol/vol) and incubated at 4.0°C for 1 h. The mixed solution was dropped onto a filter (nanopercolator; JEOL, Tokyo, Japan). The microorganisms on the filter were washed twice with sterile saline and observed using a scanning electron microscope (SEM) (Miniscope TM-3000; Hitachi High-Technologies, Tokyo, Japan) operated at 15 kV. Energy-dispersive X-ray spectroscopy (EDS) (Quantax70; Bruker AXS Microanalysis GmbH, Karlsruhe, Germany) was used to acquire the X-ray spectra and map the elements.
Solid characterization by EM.
The collected precipitates were analyzed by double Cs-corrected scanning transmission electron microscopy (TEM) (ARM200F; JEOL) at 200 kV and at the maximum optical resolution of 0.11 nm. The TEM had two systems: an illumination lens and an imaging lens equipped with an aberration correction function; in addition, the EDS (JED-2200; JEOL) was equipped with an extrasensitive silicon drift detector (solid angle = 0.8 sr). Elemental mapping, spot analysis, and standardless semiquantification were performed using the software JEOL Analysis Station (JEOL). The detection limit in the EDS analysis was approximately 0.1% (wt/wt). Image formations were appropriately carried out using high-angle annular dark-field scanning TEM (HAADF-STEM) and high-resolution TEM (HRTEM). The electron probe diameter is approximately 0.1 nm. Unstained samples were desalinated by washing thrice with UPW and further prepared by drying diluted aliquots of suspension on holey carbon-coated copper (Cu) mesh grids (Okenshoji, Tokyo, Japan).
XANES analysis.
To determine the chemical structure of Sr in the biogenic precipitates, an analysis of the energy of the Sr K-edge X-ray absorption near-edge structure (XANES) was carried out at the Photon Factory Advanced Ring using the BL-27B beam line at the High Energy Accelerator Research Organization (KEK, Tsukuba, Japan). The powders of the reference samples were mixed with boron nitrate (special grade; Wako Pure Chemicals Industries, Osaka, Japan) in a mortar for 30 min and then pressed into a tablet with a diameter of 1.0 cm and thickness of 1.0 mm. The biogenic precipitation samples were captured on a 0.2-μm-pore-size filter and then encapsulated in polyvinyl bags (1.0 cm wide by 1.0 cm high). Analyses of incident (I 0) and transmitted (I) X-rays were monitored within an ionization chamber. A silicon(111) monochromator was used to obtain the incident X-ray beam. The spectra of the solid samples were collected in fluorescence mode. A sample was placed 45° from the incident beam, and a 7-element solid-state detector monitored the fluorescence yield of each sample. Data analyses of spectra were performed using the software REX2000 (Rigaku, Tokyo, Japan).
Standard materials.
SrCl2 (Wako Pure Chemicals Industries), SrCO3 (Wako Pure Chemicals Industries), and abiotic Sr-Ca carbonate (Sr-CaCO3) were used as standard materials for TEM and XANES analyses. Abiotic Sr-CaCO3 was prepared as follows: Na2CO3 (3.3 × 10−1 mol liter−1), CaCl2 (36.6 × 10−3 mol liter−1), and SrCl2 (1.0 × 10−3 mol liter−1) were mixed at 25°C and centrifuged (21,000 × g, 5 min, and 4.0°C), and then collected precipitate was dried at 50°C.
Identification of isolated bacterium.
The morphology of the isolate was determined using a light microscope (BX50F4; Olympus, Tokyo, Japan). Gram staining was performed using a Favor G Nissui kit (Nissui Pharmaceutical, Tokyo, Japan). By following the method of Barrow and Feltham (28), several physiological and biochemical properties were evaluated: catalase and oxidase activities, oxidation/fermentation (O/F) of d-glucose, and acid and gas production from d-glucose. Additional biochemical tests were carried out using an API 50 CHB (bioMérieux, Lyon, France). The nucleotide sequence of the 16S rRNA gene in the isolate was determined as follows: the isolate was cultured for 3 days in MB, the bacterial cells were collected by centrifugation at 8,000 × g at 4.0°C for 5 min, and then they were washed twice with sterilized saline solution. The genomic DNA of the bacterium was extracted by an ISOPLANT DNA extraction kit (Nippon Gene, Tokyo, Japan). The 16S rRNA gene was amplified using a PCR thermal cycler (GeneAmp PCR System 9700; Applied Biosystems, CA), with extracted genomic DNA serving as the template. A primer set composed of 9F (5′-GAGTTTGATCCTGGCTCAG-3′) and 1541R (5′-AAGGAGGTGATCCAGCC-3′) was used for the PCR amplification (29). The nucleotide sequence of the 16S rRNA gene was determined and analyzed with multiple primers (9F, 515F [5′-GTGCCAGCAGCCGCGGT-3′], 1115R [5′-AGGGTTGCGCTCGTTG-3′], and 1541R) (29) and a 3730xl DNA analyzer (Applied Biosystems). The use of the Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi) (30) enabled a homology search on the 16S rRNA gene sequences. Based on the nucleotide sequences of bacterial species with high homology, a phylogenetic tree was constructed by following the neighbor-joining method (31) using the software CLUSTAL W and Molecular Evolutionary Genetics Analysis, version 6.0 (MEGA6).
Accession number(s).
The sequence determined in this study has been deposited in the DNA Data Bank of Japan (DDBJ; http://www.ddbj.nig.ac.jp) under accession number LC034238.
Supplementary Material
ACKNOWLEDGMENTS
This study was partially supported by the JST Initiatives for Atomic Energy Basic and Generic Strategic Research.
We are grateful to Chiaki Imada, Takeshi Terahara, and the captains and officers of the research and training vessel Hiyodori at Tokyo University of Marine Science and Technology for their technical assistance with sediment sampling.
We declare no competing financial interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00855-17.
REFERENCES
- 1.International Atomic Energy Agency. 2015. The Fukushima Daiichi accident. IAEA, Vienna, Austria. [Google Scholar]
- 2.Povinec PP, Hirose K, Aoyama M. 2013. Fukushima radioactivity impact, p 131–275. In Fukushima accident. Elsevier, Waltham, MA. [Google Scholar]
- 3.Agency for Toxic Substances and Disease Registry. 2004. Toxicological profile for strontium. US Department of Health and Human Services, Public Health Service, ATSDR, Atlanta, GA: https://www.atsdr.cdc.gov/toxprofiles/tp159.pdf. [PubMed] [Google Scholar]
- 4.Wajima T. 2013. Ion exchange properties of Japanese natural zeolites in seawater. Anal Sci 29:139–141. doi: 10.2116/analsci.29.139. [DOI] [PubMed] [Google Scholar]
- 5.International Atomic Energy Agency. 2002. Application of ion exchange processes for the treatment of radioactive waste and management of spent ion exchangers. IAEA, Vienna, Austria: http://www-pub.iaea.org/MTCD/publications/PDF/TRS408_scr.pdf. [Google Scholar]
- 6.Newsome L, Morris K, Trivedi D, Bewsher A, Lloyd JR. 2015. Biostimulation by glycerol phosphate to precipitate recalcitrant uranium (IV) phosphate. Environ Sci Technol 49:11070–11078. doi: 10.1021/acs.est.5b02042. [DOI] [PubMed] [Google Scholar]
- 7.Handley-Sidhu S, Hriljac JA, Cuthbert MO, Renshaw JC, Pattrick RAD, Charnock JM, Stolpe B, Lead JR, Baker S, Macaskie LE. 2014. Bacterially produced calcium phosphate nanobiominerals: sorption capacity, site preferences, and stability of captured radionuclides. Environ Sci Technol 48:6891–6898. doi: 10.1021/es500734n. [DOI] [PubMed] [Google Scholar]
- 8.Achal V, Pan X, Zhang D. 2012. Bioremediation of strontium (Sr) contaminated aquifer quartz sand based on carbonate precipitation induced by Sr resistant Halomonas sp. Chemosphere 89:764–768. doi: 10.1016/j.chemosphere.2012.06.064. [DOI] [PubMed] [Google Scholar]
- 9.Fujita Y, Redden GD, Ingram JC, Cortez MM, Ferris FG, Smith RW. 2004. Strontium incorporation into calcite generated by bacterial ureolysis. Geochim Cosmochim Acta 68:3261–3270. doi: 10.1016/j.gca.2003.12.018. [DOI] [Google Scholar]
- 10.Zhu T, Dittrich M. 2016. Carbonate precipitation through microbial activities in natural environment, and their potential in biotechnology: a review. Front Bioeng Biotechnol 4:4. doi: 10.3389/fbioe.2016.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Muynck W, De Belie N, Verstraete W. 2010. Microbial carbonate precipitation in construction materials: a review. Ecol Eng 36:118–136. doi: 10.1016/j.ecoleng.2009.02.006. [DOI] [Google Scholar]
- 12.Dupraz C, Reid RP, Braissant O, Decho AW, Norman RS, Visscher PT. 2009. Processes of carbonate precipitation in modern microbial mats. Earth Sci Rev 96:141–162. doi: 10.1016/j.earscirev.2008.10.005. [DOI] [Google Scholar]
- 13.Obst M, Dynes JJ, Lawrence JR, Swerhone GDW, Benzerara K, Karunakaran C, Kaznatcheev K, Tyliszczak T, Hitchcock AP. 2009. Precipitation of amorphous CaCO3 (aragonite-like) by cyanobacteria: a STXM study of the influence of EPS on the nucleation process. Geochim Cosmochim Acta 73:4180–4198. doi: 10.1016/j.gca.2009.04.013. [DOI] [Google Scholar]
- 14.Kushner DJ. 1978. Life in high salt and solute concentrations: halophilic bacteria, p 317–368. In Kushner DJ. (ed), Microbial life in extreme environments. Academic Press, London, United Kingdom. [Google Scholar]
- 15.Dal Negro A, Ungaretti L. 1971. Refinement of the crystal structure of aragonite. Am Mineral 56:768–772. [Google Scholar]
- 16.Yee N, Fein JB. 2001. Cd adsorption onto bacterial surfaces: a universal adsorption edge? Geochim Cosmochim Acta 65:2037–2042. doi: 10.1016/S0016-7037(01)00587-7. [DOI] [Google Scholar]
- 17.Borrok DM, Fein JB. 2005. The impact of ionic strength on the adsorption of protons, Pb, Cd, and Sr onto the surfaces of Gram negative bacteria: testing non-electro static, diffuse, and triple-layer models. J Colloid Interface Sci 286:110–126. doi: 10.1016/j.jcis.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 18.Ginn BR, Fein JB. 2008. The effect of species diversity on metal adsorption onto bacteria. Geochim Cosmochim Acta 72:3939–3948. doi: 10.1016/j.gca.2008.05.063. [DOI] [Google Scholar]
- 19.Krajewska B. 2009. Ureases I. Functional, catalytic and kinetic properties: a review. J Mol Catal B Enzym 59:9–21. doi: 10.1016/j.molcatb.2009.01.003. [DOI] [Google Scholar]
- 20.Stocks-Fischer S, Galinat JK, Bang SS. 1999. Microbiological precipitation of CaCO3. Soil Biol Biochem 31:1563–1571. doi: 10.1016/S0038-0717(99)00082-6. [DOI] [Google Scholar]
- 21.Rosen B. 1987. Bacterial calcium transport. Biochim Biophys Acta 906:101–110. doi: 10.1016/0304-4157(87)90007-4. [DOI] [PubMed] [Google Scholar]
- 22.Rivadeneyra MA, Párraga J, Delgado R, Ramos-Cormenzana A, Delgado G. 2004. Biomineralization of carbonates by Halobacillus trueperi in solid and liquid media with different salinities. FEMS Microbiol Ecol 48:39–46. doi: 10.1016/j.femsec.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 23.Sánchez-Román M, Rivadeneyra MA, Vasconcelos C, McKenzie JA. 2007. Biomineralization of carbonate and phosphate by moderately halophilic bacteria. FEMS Microbiol Ecol 61:273–284. doi: 10.1111/j.1574-6941.2007.00336.x. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell AC, Ferris FG. 2005. The coprecipitation of Sr into calcite precipitates induced by bacterial ureolysis in artificial groundwater: temperature and kinetic dependence. Geochim Cosmochim Acta 69:4199–4210. doi: 10.1016/j.gca.2005.03.014. [DOI] [Google Scholar]
- 25.Lauchnor EG, Schultz LN, Bugni S, Mitchell AC, Cunningham AB, Gerlach R. 2013. Bacterially induced calcium carbonate precipitation and strontium coprecipitation in a porous media flow system. Environ Sci Technol 47:1557–1564. doi: 10.1021/es304240y. [DOI] [PubMed] [Google Scholar]
- 26.Fujita Y, Taylor JL, Wendt LM, Reed DW, Smith RW. 2010. Evaluating the potential of native ureolytic microbes to remediate a 90Sr contaminated environment. Environ Sci Technol 44:7652–7658. doi: 10.1021/es101752p. [DOI] [PubMed] [Google Scholar]
- 27.McCoy DD, Cetin A, Hausinger RP. 1992. Characterization of urease from Sporosarcina ureae. Arch Microbiol 157:411–416. doi: 10.1007/BF00249097. [DOI] [PubMed] [Google Scholar]
- 28.Barrow GI, Feltham RKA. 1993. Cowan and Steel's manual for the identification of medical bacteria, 3rd ed. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 29.Green MR, Sambrook J. 2012. Molecular cloning: a laboratory manual, 4th ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 30.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 31.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.