Abstract
The vast majority of streptococci colonizing the human upper respiratory tract are commensals, only sporadically implicated in disease. Of these, the most pathogenic is Mitis group member, Streptococcus pneumoniae. Phenotypic and genetic similarities between streptococci can cause difficulties in species identification. Using ribosomal S2-gene sequences extracted from whole-genome sequences published from 501 streptococci, we developed a method to identify streptococcal species. We validated this method on non-pneumococcal isolates cultured from cases of severe streptococcal disease (n = 101) and from carriage (n = 103), and on non-typeable pneumococci from asymptomatic individuals (n = 17) and on whole-genome sequences of 1157 pneumococcal isolates from meningitis in the Netherlands. Following this, we tested 221 streptococcal isolates in molecular assays originally assumed specific for S. pneumoniae, targeting cpsA, lytA, piaB, ply, Spn9802, zmpC and capsule-type-specific genes. Cluster analysis of S2-sequences showed grouping according to species in line with published phylogenies of streptococcal core genomes. S2-typing convincingly distinguished pneumococci from non-pneumococcal species (99.2% sensitivity, 100% specificity). Molecular assays targeting regions of lytA and piaB were 100% specific for S. pneumoniae, whereas assays targeting cpsA, ply, Spn9802, zmpC and selected serotype-specific assays (but not capsular sequence typing) showed a lack of specificity. False positive results were over-represented in species associated with carriage, although no particular confounding signal was unique for carriage isolates.
Keywords: mitis group streptococci, Streptococcus pneumoniae, carriage, invasive disease, ribosomal S2-typing
1. Background
Viridans streptococci are Gram-positive bacteria, many of which have evolved alongside the human host as commensals of the upper airways and oral cavity [1,2]. Interspecies genetic recombination has played a large role in their evolution [1,3] and often makes taxonomical classification a difficult task [4]. Streptococci of the Mitis group [5] present particular challenges as phylogenetic studies report tight associations within the group, reflected by multiple evolutionary lineages with boundaries that are hard to define [2,5–7]. The exception to this, comprising a single evolutionary lineage, is Streptococcus pneumoniae—considered to be the most pathogenic of all Mitis group members [2,5,8].
Asymptomatic colonization of the upper respiratory tract by S. pneumoniae is considered prerequisite for disease, of which the most severe forms are meningitis and bacteraemia with or without pneumonia, collectively described as invasive pneumococcal disease (IPD). The lower pathogenic potential of other Mitis group members is reflected by smaller genomes relative to S. pneumoniae [6]. This is likely the result of a reductive evolutionary process leading to the loss of virulence genes [6], which in turn increases genome stability [1,6]. The most important virulence factor of S. pneumoniae is considered to be its polysaccharide capsule; unencapsulated (non-typeable) strains seldom cause IPD [9,10]. The genomic flexibility of pneumococci has assisted in the great antigenic diversity of the capsular polysaccharides, with evidence of capsular genes imported not only from Mitis group species but also from more distant groups of Anginosus and Salivarius streptococci [2,11]. This has resulted in the classification of over 90 pneumococcal serotypes [12]. To date, the capsule remains the only target for currently marketed pneumococcal vaccines. However, conjugated polysaccharide vaccines (PCVs; protective in all ages) target a maximum of 13 serotypes [13]. Following vaccine introduction, surveillance of pneumococcal disease and carriage have been important measures of direct and indirect effects of vaccination on serotype distribution [14].
The gold standard method for pneumococcal detection is conventional diagnostic culture [15] which relies on colony morphology, sensitivity to optochin and solubility in bile salts. However, some streptococci generate atypical reactions in these assays [1,6,16–20], requiring additional biochemical, serological or genetic tests for species determination [21]. Following the identification of a strain as S. pneumoniae, serotype is usually determined by the capsule swelling (Quellung) [22] or co-agglutination methods [23].
Culture-independent, molecular diagnostic methods of pneumococcal detection are reported to be of higher sensitivity as compared to conventional culture [24–26]. In carriage surveillance, the sensitivity of S. pneumoniae and pneumococcal serotype molecular detection can be further increased by sampling the oral niche in addition to the standard nasopharyngeal swab [15,25,27–30]. However, the high microbial diversity in the oropharynx and saliva [31] is reflected by a greater abundance of other streptococci, carrying homologues of pneumococcal genes [1,3,11,32] and increasing the risk of non-specific results [33,34]. This has become evident throughout the evolution of molecular assays developed for the discrimination and detection of S. pneumoniae, exemplified by assays targeting genes ply (encoding pneumolysin) [35] and lytA (encoding the major autolysin) [36] and DNA fragment Spn9802 (unknown function) [37]. Despite high sensitivity all initial assays proved to be lacking in specificity, subject to confounding by close relatives including Streptococcus pseudopneumoniae and Streptococcus mitis [17,19,35,38–40]. The development of quantitative-PCR (qPCR) overcame this limitation for lytA [35], which is now widely accepted as the molecular determinant of pneumococci, proving both highly sensitive and specific [19,27,29,35,39–41]. Owing to the challenges in achieving both high sensitivity and specificity in molecular assays developed by others, S. pneumoniae gene piaB (encoding the iron acquisition ABC transporter lipoprotein PiaB) has gained our interest [27,42]. With piaB never being detected in oral streptococci, including S. mitis isolates known to possess ply and lytA [16,43], and with the protein being 100% conserved between pneumococcal isolates, it has been suggested that PiaB is unable to evolve through the process of horizontal DNA transfer (HDT) and thus is unlikely to be transferred to species related to S. pneumoniae [16].
In this study, we introduce a new molecular method for the identification of streptococcal species, based on ribosomal multilocus sequence typing (rMLST), developed by Jolley et al. [44] for bacterial strain classification. Of the 53 ribosomal protein (rps) genes analysed in rMLST, we identified a region in rpsB (a single-copy gene encoding the 30S ribosomal protein S2) which could potentially discern species of streptococci. We validated this new method on over 200 streptococcal strains cultured from patients with severe streptococcal disease and from asymptomatic carriage and used this collection to further assess the specificity of molecular assays designed by us [27] and others [26,29,38,45–50] to detect pneumococcal gene sequences (including serotype-specific sequences and sequences encoding potential virulence factors) in clinical samples. We selected these assays primarily based on their previous application in diagnostic settings [26,29,38,45–50]. In the case of assays for which false positivity was already reported [24,28,34,42], we aimed to identify non-pneumococcal species responsible for confounding. We found that only those assays targeting S. pneumoniae-unique sequences within the genes of lytA and piaB were fully specific for pneumococci. We identified species which may confound diagnostic methods of S. pneumoniae and pneumococcal serotype detection. These findings stress the importance of critical interpretation of results from genotypic tests used both in clinical settings and in epidemiological surveillance on carriage and disease.
2. Methods
2.1. Study isolates
Streptococcal strains were isolated from patients with streptococcal disease and asymptomatic carriers (table 1). From disease, we selected 101 non-pneumococcal streptococcal strains received by the Netherlands Reference Laboratory for Bacterial Meningitis (RLBM) between 2000 and 2015. Of these, 70 were isolated from cerebrospinal fluid (CSF), 24 from blood, three from sputa and one isolate each from a wound, joint puncture, bronchoalveolar lavage and an unrecorded sample type. We included all α-haemolytic strains and a maximum of 10 isolates per species of β-haemolytic streptococci received by RLBM in this period. From asymptomatic carriage, one α-haemolytic, catalase-negative colony was selected per culture per individual. In total, 103 strains were isolated from saliva of older adults (n = 51, greater than or equal to 60 years old) and from nasopharyngeal samples from children (n = 52, less than or equal to 2 years old). We also included 17 strains classified upon isolation in previous carriage studies [24,27] as optochin-sensitive yet Quellung non-typeable [15], thus unencapsulated, S. pneumoniae. In addition, nine strains of non-streptococcal species common in the upper respiratory tract, one each of Bacteroides fragilis, Haemophilus influenzae, Neisseria meningitidis, Klebsiella oxytoca, Klebsiella pneumoniae, Pseudomonas aeruginosa, Staphylococcus epidermidis, Staphylococcus aureus and Moraxella catarrhalis, were also included.
Table 1.
Overview of streptococcal strains included in the study (n.a., demographic data not available).
| strain type | source | n | infant | adult | CSFb | blood | sputa | otherc | NPd | OPe/saliva |
|---|---|---|---|---|---|---|---|---|---|---|
| non-pneumococcala | disease | 101 | n.a. | n.a. | 70 | 24 | 3 | 4 | — | — |
| carriage | 103 | 52 | 51 | — | — | — | — | 53 | 50 | |
| NTf pneumococci | carriage | 17 | 12 | 5 | — | — | — | — | 17 | — |
| overall | 221 | 64 | 56 | 70 | 24 | 3 | 4 | 70 | 50 |
aStrains annotated based on sequence analysis of the ribosomal S2 gene.
bCerebrospinal fluid.
cWound, joint puncture, bronchoalveolar lavage fluid, unknown.
dNasopharynx.
eOropharynx.
fNT, non-typeable (presumably acapsular) pneumococci.
2.2. Bacterial DNA extraction
Columbia agar plates supplemented with 5% sheep blood were inoculated from a frozen stock of a single-colony passed culture of a strain and incubated overnight (37°C, 5% CO2). DNA was extracted from plate growth harvests using the DNeasy Tissue kit (QIAGEN, Venlo, the Netherlands). Template DNA concentration was determined by 16S-based real-time qPCR [31].
2.3. Streptococcal species identification (S2-typing)
Streptococcal S2-typing was designed analogous to the method for the identification of Neisseria species [51]. Briefly, genes encoding streptococcal ribosomal proteins were extracted from the NCBI database (https://www.ncbi.nlm.nih.gov). Potentially suitable genes were selected based on their length, variability among streptococci, the ability to discriminate between streptococcal species among a limited set of strains and the presence of conserved sequences with a space of approximately 400 base pairs, appropriate for designing PCR primers. Eventually, rpsB encoding ribosomal protein S2 was selected and primers S2F (5′-ATGGCAGTAATTTCAATG-3′) and S2R (5′-GAATTTTTCAAGACG-3′), targeting an approximately 408 bp variable region (position 2135025–2134618 in the genome of S. pneumoniae TIGR4, GenBank accession no. AE005672.3) were designed to assess streptococcal species identification. This 408 bp sequence was validated using a reference dataset which we created from 501 S2-sequences from streptococcal species, extracted from whole-genome sequences available in the NCBI database (https://www.ncbi.nlm.nih.gov/genome) [2,52]. These reference S2-sequences were aligned using the MUSCLE algorithm in MEGA v6.0 [53] and phylogenetic analysis was performed using the minimum evolution method with nucleotide substitution type and Maximum Composite Likelihood Substitution Model with bootstrap analysis based on 500 replicates. The resulting tree was compared to that of whole-genome sequences published on the online PATRIC database of over 100 000 consistently annotated microbial genomes collected from GenBank and RefSeq [54]. Next, we used the S2 primers (10 µM each) in 25 µl reaction volumes with DreamTaq Master Mix (ThermoFisher Scientific, Landsmeer, the Netherlands) including 2.5 µl of a template (minimum 1 ng, average 70 ng of genomic DNA) to generate a PCR product for all 221 isolates included in the study. PCR conditions were as follows: 95°C for 15 min, then 40 cycles of 94°C for 30 s, 54°C for 1 min and 72°C for 1 min, followed by 60°C for 30 min. S. pneumoniae serotype 19A strain SJD86 was included as a positive control in each run [55]. Amplicons between 400 and 450 bp (approximately, because amplicons generated with the S2F-S2R primer pair can vary in size) were gel purified using the GeneJET PCR Purification kit (ThermoFisher Scientific), then 5 µl was mixed with the S2F (10 µM) or S2R (3 µM) primer and sequenced by Macrogen (Amsterdam, the Netherlands). Sequences generated were assembled using BioNumerics v5.10 (Applied Maths NV, http://www.applied-maths.com) and cross-referenced with the reference dataset for species annotation. Streptococcal strains included in the reference dataset and the S2-sequences (and accession numbers) of the study isolates are detailed in the electronic supplementary material, table S1. Strains within the Mitis group were designated according to the nomenclature proposed by Jensen et al. [5].
The sensitivity and specificity of S2-typing to discriminate streptococcal species was assessed using the 431 non-pneumococcal S2-sequences in the reference dataset and a total of 1227 pneumococcal S2-sequences (70 isolates in the reference dataset and 1157 pneumococcal meningitis isolates retrieved from the collection of the Netherlands Reference Laboratory for Bacterial Meningitis, NRLBM). Read data, assembled and annotated contigs of the 1157 pneumococcal meningitis isolates from NRLBM are deposited in the European Nucleotide Archive (ENA): study accession number PRJEB4909 (http://www.ebi.ac.uk/ena/data/view/PRJEB4909).
2.4. Detection of species-specific DNA sequences
All strains were tested in molecular assays targeting sequences (originally) reported to be unique for S. pneumoniae genes, namely lytA [26], piaB [24,27], Spn9802 [38], cpsA [47] and ply [48]. DNA of S. pneumoniae strain SJD86 was included as a positive control in every molecular assay [55]. In addition, all strains were tested for the presence of the pneumococcal virulence factor zinc metalloproteinase C gene, zmpC.
The presence of sequences matching pneumococcal genes lytA and piaB was assessed using previously described probe-based qPCRs [24,26,27]. The presence of Spn9802 [39] was assessed by qPCR using SYBRgreen chemistry (ThermoFisher Scientific) and primers described by Abdeldaim et al. [38]. Positivity for qPCR-signal was determined when CT values matched 16S DNA concentrations. Conventional PCR (cPCR) was used to detect ply and cpsA (or wzg, a gene within the capsular polysaccharide biosynthesis operon) [47]. When reported, amplicons generated in cPCR were sequenced and analysed in SeqMan Pro (DNASTAR Lasergene v12.2.0, Madison, WI, USA) for homology to published sequences [34].
The presence of zmpC, was detected with cPCR using primers (ShortZmpC-F 5′-CAGCTGGTAACAGCCATGCAA-3′, ShortZmpC-R 5′-CAATGCACCATTTTCTAATCTACCD-3′) targeting a 563 bp fragment corresponding to position 75858–76420 bp in the genome sequence of S. pneumoniae strain TIGR4. One microlitre of DNA template (minimum 0.35 ng bacterial genomic DNA, average 28 ng) was tested with the ShortZmpC F-R primer pair (10 µM each) in a 12.5 µl reaction volume using DreamTaq Master Mix (ThermoFisher Scientific). PCR conditions were as described for the S2-typing PCR except for a 90 s annealing step (54°C). Amplicons (approx. 560 bp) were sequenced as described above. Strains which generated sequences 100% homologous to any published for S. pneumoniae were revisited with primers designed in S. pneumoniae to amplify the approximately 5000 bp [45] and approximately 8000 bp [46] fragments of zmpC. In 50 µl reaction volumes using GoTaq® Long PCR Master Mix (Promega, Madison, USA), 4 µl of DNA template was tested with each primer pair (10 µM each). PCR conditions were as for the 560 bp assay but with longer steps at 72°C (5 min for 5000 bp, 8 min for 8000 bp products, per cycle). DNA of S. pneumoniae serotype 33F strain 2080133 was included as a positive control in all PCRs targeting zmpC.
2.5. Detection of pneumococcal capsule-type-specific DNA sequences
All strains were tested for pneumococcal serotype-specific signal in qPCR assays using primers and probes targeting serotypes/serogroups 1, 3, 6A/B/C/D, 7A/F, 8, 9A/N/V, 10A/B, 12A/B/F, 14, 15A/B/C, 19A, 20, 23F, 33A/F, 35B, 38 [29], 11A/D, 16F, 23A [50], 4, 5, 18B/C, 19F, 22A/F [29,50] and 35F [25]. The pneumococcal serotyping method of capsular sequence typing (CST) was also applied to all DNA templates [49]. For each serotype-specific qPCR, DNA of a clinical pneumococcal strain of the serotype(s) targeted was included as a positive control [34].
2.6. Conventional serotyping
Non-pneumococcal strains generating positive signals in serotype-specific qPCR assays were tested for the expression of capsular polysaccharides by the co-agglutination method of the Pneumotest kit [23] (Staten Serum Institut, SSI Diagnostica, Hillerød, Denmark) and by the Quellung method [22] using type-specific sera (Staten Serum Institut, SSI Diagnostica).
2.7. Statistics
Statistical analyses were conducted using GraphPad Prism v6.02 for Windows (GraphPad Software, CA, USA). Statistical significance was determined using Fisher's Exact test (unless otherwise stated) and defined as p < 0.05.
3. Results
3.1. Phylogenetic tree based on S2-sequences shows clustering according to streptococcal species
Phylogenetic analysis of the streptococcal S2-reference dataset sequences showed grouping according to streptococcal species (figure 1a), comparable to the results from phylogenetic analyses of whole-genome sequences, which can be viewed on the PATRIC website (https://www.patricbrc.org/view/Taxonomy/1301#view_tab=phylogeny) [54].
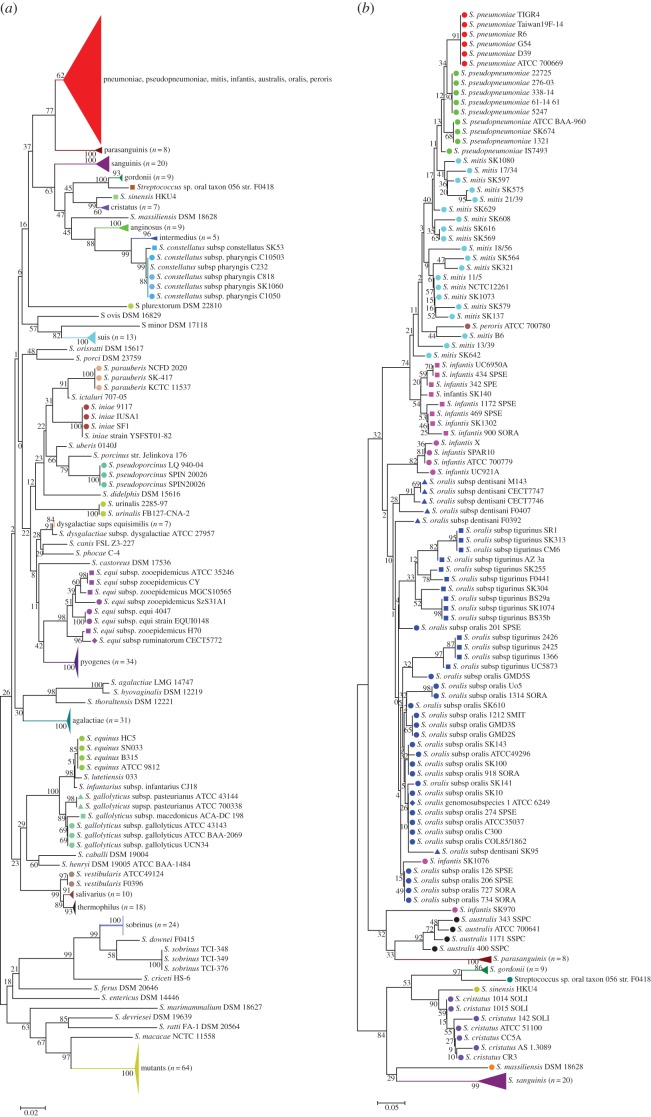
Figure 1.
Cluster analyses of the S2-sequences of (a) the 498 streptococcal strains in the reference dataset (strains of streptococcal species pneumoniae, pseudopneumoniae, mitis, infantis, australis, oralis and peroris are collapsed together, red triangle) and (b) the 360 strains included in the study, belonging to the Mitis group. Phylogenetic analysis was performed using the minimum evolution method with nucleotide substitution type and Maximum Composite Likelihood Substitution Model with bootstrap analysis based on 500 replicates. Strains of the same species are grouped by colour. Different symbols of the same colour indicate subspecies. Magenta squares and circles indicate the two S. infantis clusters identified by Jensen et al. [5] (circles = cluster 1; squares = cluster 2). Different dark blue symbols indicate the different S. oralis subspecies identified by Jensen et al. [5].
Within the Mitis group, the S2-sequences of pneumococcal isolates clustered in a single clade with high reliability (figure 1b). Overall, S2-sequences of S. pneumoniae were very homogeneous. Examination of additional S2-sequences extracted from 1157 invasive S. pneumoniae whole genomes showed that except for one, all S2-sequences grouped together. The exception was an S2-sequence from a pneumococcal isolate, serotype 35B (2060880) from CSF, which was identical to that of S. mitis SK575. The blood isolate of the same patient had the same serotype and S2-type. Among the 1226 pneumococcal S2-sequences, excluding the one of isolate 2060880, only 11 polymorphic sites resulting in the same number of different alleles were found. Analyses of the complete rpsB sequence showed 19 polymorphisms unique to strain 2060880 and S. mitis SK575 dispersed over the entire gene.
Non-pneumococcal streptococci grouped into clusters according to newly proposed species nomenclature [5] but with deep branches. With the exception of SK970 and SK1076, Streptococcus infantis strains resolved into two clusters, according to S. infantis cluster 1 and cluster 2 observed by Jensen et al. [5] after phylogenetic analyses based on whole-genome sequences. Streptococcus oralis were distributed in two groups and did not group into the subspecies cluster observed with whole-genome analyses [5].
3.2. S2-typing of streptococcal strains from disease and carriage for species identification
Amplicons of expected size were generated for all streptococci tested in the study. None of the non-streptococcal strains yielded an S2-cPCR product. By cross-referencing the S2-sequences of the study isolates with the S2-sequences in the reference dataset, streptococcal species were clearly assigned to all 120 isolates from carriers and all 101 isolates cultured from disease. Of the 221 study isolates in total, 146 (66%) were classified as belonging to the Mitis group (32 isolates from disease, 114 isolates from carriage). This included the 17 S. pneumoniae strains (all from carriage and all Quellung non-typeable) which formed a distinct branch within the Mitis group cluster. Altogether, using S2-sequencing to discriminate S. pneumoniae from non-pneumococcal streptococci had a sensitivity of 99.92% (1226/1227) and a specificity of 100% (430/430). S2-sequences and species annotations are published under GenBank accessions MF375925–MF376145.
With differences in the selection criteria for clinical versus carriage isolates included in the study (clinical isolates included both α-haemolytic and β-haemolytic strains, whereas non-pneumococcal isolates from carriage were exclusively randomly selected α-haemolytic strains), we only tested for differences in the distribution of α-haemolytic non-pneumococcal streptococci in disease (n = 53) versus carriage (n = 103). We found Streptococcus salivarius, Streptococcus sanguinis and Streptococcus gallolyticus to be over-represented among strains of α-haemolytic streptococci from disease, whereas S. infantis and S. mitis to be over-represented among carriage isolates (table 2).
Table 2.
Detection of pneumococcal-specific sequences in Streptococcus spp. strains, per S2-type. n.a., not applicable. Strains of this species were not included and tested in the current study. NT denotes all 17 S. pneumoniae strains were non-typeable. Significant difference (third column) in the proportion of α-haemolytic non-pneumococcal streptococci of the species cultured from disease (n = 53 total) versus carriage (n = 103 total), or (columns 4–10) in proportion of individual species strains positive for a particular molecular target when cultured from disease versus carriage.
| streptococcal species according to S2-type | n | disease/carriage (n/n) | lytA | piaB | ply | cps | Spn9802 | zmpC (560 bp) | zmpC (5 kb) | zmpC (8 kb) | qPCR serotype | CST serotype |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β-haemolytic streptococci | 48 | 48/n.a. | ||||||||||
| S. agalactiae | 9 | 9/n.a. | —a | — | — | — | — | — | — | — | — | — |
| S. anginosus | 2 | 2/n.a. | — | — | — | — | — | — | — | — | — | — |
| S. constellatus | 1 | 1/n.a. | — | — | — | — | — | — | — | — | — | — |
| S. dysgalactiae subsp. equisimilis | 10 | 10/n.a. | — | — | 1 | — | — | — | — | — | — | — |
| S. equi subsp. zooepidemicus | 2 | 2/n.a. | — | — | — | — | — | 1 | — | — | — | — |
| S. intermedius | 4 | 4/n.a. | — | — | — | — | — | — | — | — | — | — |
| S. pyogenes | 10 | 10/n.a. | — | — | — | — | — | — | — | — | — | — |
| S. suis | 10 | 10/n.a. | — | — | — | — | — | — | — | — | — | — |
| α-haemolytic streptococci | 174 | 53/103 | ||||||||||
| S. australis | 2 | 0/2 | — | — | — | — | — | — | — | — | — | — |
| S. cristatus | 3 | 2/1 | — | — | — | — | — | — | — | — | — | — |
| S. gallolyticus subsp. Gallolyticusc | 2 | 2/0 | — | — | — | — | — | — | — | — | — | — |
| S. gallolyticus subsp. Pasteurianusc | 2 | 2/0 | — | — | — | — | — | — | — | — | — | — |
| S. gordonii | 1 | 1/0 | — | — | — | — | — | — | — | — | — | — |
| S. infantis | 22 | 3/19* | — | — | — | — | — | 0/3b | 0/1 | — | 0/1 | — |
| S. mitis | 68 | 13/56*** | — | — | 5/13 | — | — | 0/2 | 0/1 | — | 1/3 | — |
| S. oralis | 25 | 10/15 | — | — | — | 1/0 | 1/0 | 0/1 | — | — | — | — |
| S. parasanguinis | 7 | 0/7 | — | — | — | — | — | — | — | — | — | — |
| S. pseudopneumoniae | 6 | 3/3 | — | — | 2/1 | — | 3/0 | — | — | — | — | — |
| S. salivarius | 12 | 11/1# | — | — | — | — | — | — | — | — | — | — |
| S. sanguinis | 7 | 6/1** | — | — | — | — | — | — | — | — | — | — |
| S. pneumoniaeNT | 17 | n.a./17 | 17 | 3 | 15 | 3 | 7 | 10 | 10 | — | 3 | 3 |
| total | 221 | 101/120 | 0/17 | 0/3 | 8/29 | 1/3 | 4/7 | 1/16 | 0/12 | 0/0 | 1/7 | 0/3 |
aNone of the strains tested of the particular species generated an amplicon with 100% sequence homology to sequences published for S. pneumoniae.
bStrains which generated amplicons with 100% sequence homology to sequences published for S. pneumoniae.
cAlso γ-haemolytic.
*p < 0.05, **p < 0.01, ***p < 0.001 #p < 0.0001 (Fisher's exact test).
3.3. Distribution of pneumococcal-specific genes among non-pneumococcal streptococci
All 221 streptococcal and nine non-streptococcal isolates were assessed in molecular assays used to detect S. pneumoniae. Results according to S2-type are detailed in table 2 (summarized for disease versus carriage isolates in the electronic supplementary material, table S2). Individual strains testing positive for any molecular target are detailed further in table 3. All pneumococcus-specific PCRs remained negative when the non-streptococcal strains were tested.
Table 3.
Detailed results of non-pneumococcal streptococcal strains generating false positive signals when tested in molecular assays for common pneumococcal molecular targets.
| lytA | piaB | ply | cpsA | Spn9802 | zmpCb (560 bp) | zmpC (5 kb) | zmpC (8 kb) | qPCR serotype | |
|---|---|---|---|---|---|---|---|---|---|
| strains from invasive diseasea | |||||||||
| S. dysgalactiae subsp. equisimilis | + | ||||||||
| S. equi subsp. zooepidemicus | + | ||||||||
| S. mitis (n = 4) | + | ||||||||
| S. mitis | + | 19F | |||||||
| S. oralis | + | ||||||||
| S. oralis | + | ||||||||
| S. pseudopneumoniae | + | ||||||||
| S. pseudopneumoniae (n = 2) | + | + | |||||||
| strains from asymptomatic carriagea | |||||||||
| S. infantis | 9A/N/V | ||||||||
| S. infantis | + | + | |||||||
| S. infantis (n = 2) | + | ||||||||
| S. mitis | 5 | ||||||||
| S. mitis | 18B/C | ||||||||
| S. mitis | 19F | ||||||||
| S. mitis | + | + | |||||||
| S. mitis | + | + | + | ||||||
| S. mitis (10 strains) | + | ||||||||
| S. oralis | + | ||||||||
| S. pseudopneumoniae | + | ||||||||
aStrains annotated based on sequence analysis of the ribosomal S2 gene.
bStrains which generated PCR amplicons with full homology to sequence published for S. pneumoniae.
All 204 non-pneumococcal isolates were negative in lytA- and piaB-specific qPCRs, yet 4 (2%) were positive for Spn9802. cPCRs targeting cpsA and ply yielded amplicons of correct size in one (0.5%) and 22 (11%) isolates, respectively. None of the cpsA or ply amplicons were of full homology to any sequence published for S. pneumoniae.
Twelve non-pneumococcal streptococci yielded amplicons of expected size in the 560 bp zmpC-specific cPCR. Following amplicon-sequencing, seven isolates (disease: Streptococcus equi subsp. zooepidemicus H70, n = 1; carriage: S. mitis, n = 2; S. infantis, n = 3; S. oralis, n = 1) were 100% homologous to the nucleotide sequences of unencapsulated S. pneumoniae strain NT_110_58 (GenBank accession no. CP007593.1) and encapsulated strains of serotypes 19F (CP001015.1) and 11A (CP001015.1). Of the seven isolates positive for the 563 bp zmpC amplicons with sequence homologous to that reported in S. pneumoniae, one S. mitis and one S. infantis strain also generated product in the 5000 bp cPCR of size reported for some non-typeable S. pneumoniae [45]. None of the 221 study isolates was positive for the 8000 bp cPCR product reported for encapsulated pneumococci, demonstrating better specificity of this cPCR for detecting pneumococcal-specific zmpC sequences [46]. Of note, among α-haemolytic non-pneumococcal streptococcal strains the confounding results for zmpC were observed exclusively among isolates from carriers (n = 6 of 103 strains from carriage versus none of 53 clinical isolates, p = 0.096; table 2).
3.4. Distribution of pneumococcal-specific genes among non-typeable pneumococci
All 17 pneumococci non-typeable by Quellung were lytA-positive, yet only three (18%) were piaB-positive (table 4). This was consistent with previous reports from us [27] and others [43] but also with the piaB gene being absent in published sequences of non-typeable pneumococci (https://www.ncbi.nlm.nih.gov/genome). The three piaB-positive non-typeable pneumococci were also the only unencapsulated strains cpsA-positive by cPCR. When tested in the ply-cPCR however, 15 (88%) non-typeable isolates produced amplicons of expected size. None of the cpsA or ply amplicons were of full homology to any sequence published for S. pneumoniae. In addition, six (35%) non-typeable isolates generated sequence-specific signal in the Spn9802-qPCR.
Table 4.
Streptococcus pneumoniae strains non-typeable (thus, unencapsulated) by the conventional diagnostic method, positive when tested in molecular assays for common pneumococcal molecular targets.
| number of isolates | lytA | piaB | ply | cpsA | Spn9802 | zmpCa (560 bp) | zmpC (5 kb) | zmpC (8 kb) | qPCR serotype | CST serotype |
|---|---|---|---|---|---|---|---|---|---|---|
| 9 | + | + | + | + | ||||||
| 3 | + | + | + | |||||||
| 1 | + | + | + | + | + | |||||
| 1 | + | |||||||||
| 1 | + | + | + | + | + | 22A/F | 22F | |||
| 1 | + | + | + | + | + | 25F | ||||
| 1 | + | + | + | + | 19A | 19A | ||||
| total n (%) | 17 (100) | 3 (18) | 15 (90) | 3 (18) | 7 (40) | 10 (59) | 10 (59) | 0 (0) | 2 (12) | 3 (18) |
aStrains which generated PCR amplicons with full homology to sequences published for S. pneumoniae.
When tested for zmpC, 10 non-typeable strains (59% of 17) produced size-specific amplicon in the 560 bp cPCR which following sequencing were all 100% homologous to the published sequences for the S. pneumoniae strains detailed above. Subsequently, all 10 also produced size-specific amplicons in the 5000 bp cPCR, but none in the 8000 bp cPCR.
3.5. Detection of serotype-specific sequences and capsular polysaccharides
Of 204 non-pneumococcal streptococcal isolates, 42 (21%) yielded amplicons of expected size in CST-cPCR [49]. However, all sequences of these amplicons showed less than 75% sequence homology to those reported for S. pneumoniae and CST results were therefore regarded as negative.
Five non-pneumococcal streptococci (2% of 204 isolates) yielded a serotype-specific signal in pneumococcal serotype/serogroup-specific qPCRs (tables 3 and 4) [28,34]. Of disease strains, a single isolate S2-typed as S. mitis (also ply-positive in cPCR) generated serotype-specific signal in the 19F-qPCR assay published by Carvalho et al. [29]. Among carriage isolates, serotype-specific signals were yielded in qPCR assays published by the same authors and targeting serotypes/serogroups 5, 18B/C, 19F [29] (three isolates S2-typed as S. mitis, each positive in a single serotyping-qPCR assay) and 9A/N/V [29] (single isolate S2-typed as S. infantis). None of the isolates yielded a positive result in any assay published by Pimenta et al. [50]. Of the five non-pneumococcal strains yielding positive signals in serotyping-qPCR assays, two were also positive for the corresponding capsular type (serotype 5 and serogroup 9) in the co-agglutination test. However, none was typeable by the Quellung method. None of the nine non-streptococcal isolates yielded a serotype-specific signal in any of the genotyping assays.
Of the 17 Quellung non-typeable pneumococci from carriage, three (18%) were CST-positive (one strain positive for each of the serotypes 19A, 22F and 25F). All three were also piaB- and cpsA-positive. From these three strains, CST-positivity for serotypes 19A and 22A/F was in agreement with our panel of qPCR serotyping assays (an assay for detection of serotype 25F is not available within this qPCR panel).
4. Discussion
Quicker and more accurate diagnostic methods of pathogen detection advance treatment of infection and contribute to our understanding of disease aetiology [25,35,47,55,56]. Molecular-based diagnostic methods continue to evolve, improving detection of aetiological agents causing streptococcal disease. This also contributes to advances in surveillance of disease and carriage of the clinically most relevant streptococcal species, S. pneumoniae [25,35,57]. This is of particular importance following the introduction of commercial vaccines targeting pneumococcal disease, with molecular method-based surveillance studies already being implemented to monitor vaccine effects in disease and in carriage [58–61]. We demonstrate, however, that this is not without its challenges. Our current study highlights important considerations for the transition from conventional to molecular diagnostic methods. We showed that S2-sequencing discriminated S. pneumoniae from non-pneumococcal streptococci with high sensitivity (greater than 99%) and specificity (100%).
Jolley et al. [44] recently demonstrated that streptococci grouped according to species in phylogenetic analyses using sequences of all ribosomal protein genes. In this study, we classified species of streptococcal strains through sequencing of a variable region of the ribosomal S2 gene. The resulting trees (figure 1a,b) do not completely follow the topology of the trees based on whole-genome sequences, likely due to the much smaller S2-sequences used in our study [5,44]. However, the deviations observed are not of relevance, because S2-sequencing is not intended for studying evolution, but rather for use as a potent and fast tool for streptococcal species identification.
Outside the Mitis group strains grouped accordingly, with the exception of three Streptococcus sobrinus strains which formed a branch separated from the other 24 S. sobrinus strains and Streptococcus agalactiae LMG14747 grouped together with Streptococcus hyovaginalis DSM12219 but apart from the other 32 S. agalactiae. Analyses based on whole-genome sequences are needed to determine whether these strains are genuine S. sobrinus and S. agalactiae, respectively, or have to be reclassified. Within the Mitis group, S. pneumoniae could be clearly and robustly distinguished from non-pneumococcal strains. S. sanguinis, Streptococcus parasanguinis, Streptococcus cristatus, Streptococcus gordonii and the two clusters of S. infantis (with the exception of two strains) also grouped with high fidelity. S. pseudopneumoniae, S. mitis and S. oralis also formed distinct clusters but with low fidelity consistent with the deep branching of the S. mitis and S. oralis strains in whole-genome sequences and the close relatedness between these species and between S. pseudopneumoniae to both S. mitis and S. pneumoniae [1,2,5,7,62]. Ultimately, these observations exemplify the difficulty in streptococcal species annotation through biochemical (immunological) and genetic identification among streptococci colonizing the human upper respiratory tract and oral cavity. This arises from frequent HDT among Mitis group streptococci, which includes genes and their products targeted by diagnostic tests [2,11].
Clear examples of this were evident in our study. Among the 1227 S2-sequences from pneumococci, we observed one with a S. mitis type. This CSF isolate with serotype 35B and ST558 from a meningitis patient was a genuine pneumococcal isolate according to in silico DNA–DNA hybridization values (electronic supplementary material, data) and was lytA and piaB-positive. In addition, the isolate from the blood of the same patient tested positive for the identical S2-sequence and serotype. Close examination of the nucleotide sequence flanking rpsB in the whole-genome sequence of the CSF isolate showed a sequence of approximately 1 kbp upstream of rpsB, comprising an open reading frame (ORF) putatively encoding an amidase, with the highest nucleotide identities to the sequence in S. mitis strain SVGS_061 (95%), while the nucleotide identities with the sequence in S. pneumoniae strains was lower (87%). In addition, we saw evidence of HDT in non-pneumococcal streptococcal species testing positive for sequences of cpsA, ply, zmpC and capsular genes, all previously regarded as unique for S. pneumoniae and for products of these genes, namely pneumococcal capsular polysaccharides detected with the co-agglutination method. The detection of these genes among non-pneumococcal Mitis group streptococci highlights that caution must be taken when interpreting PCR results in assays targeting cpsA, ply, Spn9802 and zmpC when applied to polymicrobial samples and/or samples culture-negative for S. pneumoniae; positive signal may in fact represent confounders which could skew results of disease and carriage surveillance.
Interestingly, non-pneumococcal strains harbouring homologues of genes coding for S. pneumoniae virulence factors have been reported as more commonly associated with disease isolates when compared with carriage isolates of the corresponding species [1]. This does not seem to be the case for the sequences targeted in molecular diagnostic assays in our study. Although none of the confounding results was unique for streptococcal strains cultured from either disease or carriers, ‘false positivity’ was more common among the species of α-haemolytic streptococci that were over-represented in carriage—S. infantis and S. mitis in particular.
There is always a potential that genetic exchange will impede identification based on single targets as compared to identification based on whole-genome sequencing. However, considering the strength of species grouping by the S2-typing method introduced here, we propose the much simpler and more time-efficient S2-typing for use in reference laboratories for the identification of streptococcal species isolated from disease, particularly for the distinction of pneumococci from streptococcal strains confounding pneumococcal diagnostic tests. In addition, with adaptation to a deep-sequencing format, S2-typing could improve the annotation of streptococcal species in microbiome studies currently being based on the less discriminatory 16S gene sequencing, particularly in studies of the respiratory or oral microbiomes [63].
For the analysis of polymicrobial samples, molecular assays targeting specific DNA sequences increase the sensitivity of detection when compared with culture-based methods [25,27–30,34]. For surveillance on pneumococcal carriage, the higher sensitivity of molecular methods for detecting pneumococci can often only be inferred from samples from which live pneumococci cannot be isolated. Therefore, it is essential that assay specificity is carefully assessed. For the molecular detection of pneumococci, the lytA-qPCR assay is fast becoming the standard. However, with lytA homologues in non-pneumococcal streptococci and on prophages [1,17,19,20,35,38,43,64] and with one recent report of a S. pseudopneumoniae strain testing positive in the lytA-specific qPCR tested in this study [56], targeting a second pneumococcal-specific gene in polymicrobial samples reduces the likelihood of misclassification due to false positivity—the chance that confounding bacteria would acquire two genetic markers is low. Given the high specificity observed in the current study and the high concordance between qPCR results and the presence of live pneumococci in samples from children, adults and the elderly [27,28,34], we and others [43] recommend piaB as a suitable countermark to the lytA-qPCR for pneumococcal detection. The piaB distribution in streptococcal strains reported here is in line with results published by us [27] and others [43,65] showing piaB being unique for S. pneumoniae yet absent exclusively from non-typeable pneumococci. Interestingly, because piaB-negative non-typeable pneumococci are absent from IPD but not from carriage, acquired immune responses specific to Pia proteins [66,67] could potentially not only protect against disease by Pia-positive, presumably encapsulated pneumococci, but also increase the fitness costs for such strains competing within the respiratory niche, thus promoting carriage of less virulent non-typeable pneumococci.
Pneumococcal serotyping is also progressively transitioning from a reliance on phenotypic serological methods, such as Quellung and co-agglutination assays, to genotypic methods of serotype determination [25,49,68,69]. Here, we demonstrate the specificity of CST for serotyping pneumococcal isolates [70]. While targeting other serotype-specific genes has demonstrated a lack of specificity due to homologous sequences in non-pneumococcal Mitis group streptococci [11,71], sequencing a wzh gene fragment of the capsular locus was highly specific, supporting its potential as a reliable alternative to culture-dependent pneumococcal serotyping or to molecular methods requiring multiple assays. However, despite the specificity of molecular methods when applied to pure pneumococcal isolates [25,49,68], their reliability when applied to polymicrobial samples must be carefully monitored, due to reports of false positive signals from non-pneumococcal species [24,33,34,42]. In this study, comparatively few non-pneumococcal strains generated signal in serotype-specific assays. This implies that the validation of any serotype-specific assay should not only include testing pneumococcal strains of other serotypes and non-pneumococcal strains, but should also include testing of polymicrobial samples negative for pneumococcus-specific signal.
It should also be stressed that phenotypic methods for pneumococcal serotyping are also not exempt from confounding by (non)pneumococcal strains producing atypical reactions [72,73]. Here, we detected false positivity in the co-agglutination test used to determine type of pneumococcal capsular polysaccharide present in a sample, presumably through the presence of antigenic determinants common to those of S. pneumoniae [11,74,75]. While this is not a new observation [11,20], it has important implications for diagnostic strategies developed on this immunochemistry.
Owing to the importance of capsule for pneumococcal virulence and vaccination strategies, historically, studies of unencapsulated (non-typeable) pneumococci seldom progressed beyond identification as S. pneumoniae [1]. Non-typeable pneumococci are being increasingly detected in carriage surveillance following PCV implementation [76,77], an important consideration due to their higher rates of recombination [78] and greater number of mobile elements [79] than encapsulated strains. However, their prevalence may be skewed when grouped with non-pneumococcal confounders of culture-based methods, or if overlooked due to their atypical phenotype on culture plates [79]. Here, all non-typeable pneumococci were convincingly S2-typed as S. pneumoniae. Despite a lack of capsule, unencapsulated pneumococci have been shown to colonize the nasopharynx of mice as efficiently as encapsulated strains [73,77] and are disproportionately identified as the aetiological agent of highly contagious pneumococcal conjunctivitis [80]. One such gene that might play a role in this is zmpC, suggested to have been only recently acquired by pneumococci [81] and present in only a limited number of strains [46,69]. While studies have previously speculated a role for ZmpC in invasive disease [46,69], it has recently been demonstrated that ZmpC suppresses S. pneumoniae virulence in experimental models of pneumococcal meningitis [82]. Owing to its prevalence in non-typeable pneumococci in the current study (strains which are seldom isolated from invasive disease [9]) and its association with increased adhesion to host mucosal cells [46], our findings further support a role for ZmpC in colonization rather than in pneumococcal disease [81].
In conclusion, in the current study we further demonstrate the potential for misidentification of streptococci, usually carried as harmless respiratory commensals, but with the ability to cause severe disease. While we target the most pathogenic of these—S. pneumoniae—with vaccination programmes, accurate species identification is crucial for the reliable monitoring of pneumococcal disease and effects of vaccination strategies. Conventional diagnostic methods are insensitive in both disease diagnosis [29,56] and in carriage studies [24,28,34]. New methods are required but carry a risk of over-detection or misidentification if subject to confounding from co-occurring species. Here, we employed S2-typing to identify streptococci which may confound both phenotypic and genotypic methods of pneumococcal detection and serotype determination [17,83]. We propose S2-typing for use in reference laboratories to assist in species annotation of streptococcal strains and for the classification of S. pneumoniae reliably from strains confounding pneumococcal diagnostic tests. Furthermore, S2-typing provides a sensitive method to distinguish non-typeable pneumococcal strains from other streptococci generating atypical reactions and to identify individual species contributing genes coding virulence factors to the genetic pool [73]. For enhanced detection of pneumococci in carriage surveillance studies using polymicrobial samples, to increase sensitivity of pneumococcal detection our findings support the use of qPCR assays targeting species-specific regions of the genes lytA and piaB.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank all members of the laboratory staff of Reference Laboratory for Bacterial Meningitis, Academic Medical Center, Amsterdam, the Netherlands, and of the research laboratory at the University Medical Center, Utrecht, the Netherlands. In particular, we thank James Groot, Eline van de Meel and Stijn Wijkstra for laboratory assistance.
Authors' contributions
A.L.W., Y.P., A.vd.E. and K.T. conceived and designed the experiments, A.L.W., S.B. and J.v.E.G. performed the experiments, A.L.W., Y.P., B.F., D.vd.B., A.vd.E. and K.T. analysed the data, A.L.W., Y.P., S.B., B.F., D.vd.B., E.A.M.S., A.vd.E. and K.T. contributed reagents, materials and analysis tools, and A.L.W., E.A.M.S., A.vd.E. and K.T. wrote the paper. All authors read and approved the final manuscript.
Competing interests
E.A.M.S. declares to have received research grants from Pfizer and GSK and fees paid to the institution for advisory boards and participation in independent data monitoring committees for Pfizer and GSK. A.vd.E. declares to have received research grants from Pfizer and fees paid to the institution for consultancy for GSK. K.T. has received consulting fees from Pfizer and grant support for studies on pneumococcal carriage from Pfizer. All other authors report no competing interests.
Funding
The Netherlands Reference Laboratory for Bacterial Meningitis is financially supported by the National Institute of Public Health and the Environment. The study was supported from internal funds from Wilhelmina Children's hospital of UMC Utrecht. Otherwise, this research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
References
- 1.Whatmore AM, Efstratiou A, Pickerill AP, Broughton K, Woodard G, Sturgeon D, George R, Dowson CG. 2000. Genetic relationships between clinical isolates of Streptococcus pneumoniae, Streptococcus oralis, and Streptococcus mitis: characterization of ‘atypical’ pneumococci and organisms allied to S. mitis harboring S. pneumoniae virulence factor-encoding genes. Infect. Immun. 68, 1374–1382. (doi:10.1128/IAI.68.3.1374-1382.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kilian M, Riley DR, Jensen A, Brüggemann H, Tettelin H. 2014. Parallel evolution of Streptococcus pneumoniae and Streptococcus mitis to pathogenic and mutualistic lifestyles. MBio 5, e01490–e01414. (doi:10.1128/mBio.01490-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balsalobre L, Ortega M, de la Campa AG. 2013. Characterization of recombinant fluoroquinolone-resistant pneumococcus-like isolates. Antimicrob. Agents Chemother. 57, 254–260. (doi:10.1128/AAC.01357-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Facklam R. 2002. What happened to the Streptococci: overview of taxonomic and nomenclature changes. Clin. Microbiol. Rev. 15, 613–630. (doi:10.1128/CMR.15.4.613-630.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jensen A, Scholz CFP, Ilian M. 2016. Re-evaluation of the taxonomy of the Mitis group of the genus Streptococcus based on whole genome phylogenetic analyses, and proposed reclassification of Streptococcus dentisani as Streptococcus oralis subsp. dentisani comb. nov., Streptococcus tigurinus. Int. J. Syst. Evol. Microbiol. 66, 4803–4820. (doi:10.1099/ijsem.0.001433) [DOI] [PubMed] [Google Scholar]
- 6.Kilian M, Poulsen K, Blomqvist T, Håvarstein LS, Bek-Thomsen M, Tettelin H, Sørensen UBS, Ahmed N. 2008. Evolution of Streptococcus pneumoniae and its close commensal relatives. PLoS ONE 3, e2683 (doi:10.1371/journal.pone.0002683) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop CJ, Aanensen DM, Jordan GE, Kilian M, Hanage WP, Spratt BG. 2009. Assigning strains to bacterial species via the internet. BMC Biol. 7, 3 (doi:10.1186/1741-7007-7-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dowson CG. 2004. What is a pneumococcus. In The pneumococcus (eds Tuomanen EI, Mitchell TJ, Morrison DA, Spratt BG), pp. 3–14. Washington, DC: ASM Press. [Google Scholar]
- 9.Jansen AGSC, Rodenburg GD, van der Ende A, van Alphen L, Veenhoven RH, Spanjaard L, Sanders EAM, Hak E. 2009. Invasive pneumococcal disease among adults: associations among serotypes, disease characteristics, and outcome. Clin. Infect. Dis. 49, e23–e29. (doi:10.1086/600045) [DOI] [PubMed] [Google Scholar]
- 10.Browall S, et al. 2014. Intraclonal variations among Streptococcus pneumoniae isolates influence the likelihood of invasive disease in children. J. Infect. Dis. 209, 377–388. (doi:10.1093/infdis/jit481) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skov Sørensen UB, Yao K, Yang Y, Tettelin H, Kilian M. 2016. Capsular polysaccharide expression in commensal Streptococcus species: genetic and antigenic similarities to Streptococcus pneumoniae. MBio 7, e01844-16 (doi:10.1128/mBio.01844-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park IH, Geno KA, Yu J, Oliver MB, Kim K-H, Nahm MH. 2015. Genetic, biochemical, and serological characterization of a new pneumococcal serotype, 6H, and generation of a pneumococcal strain producing three different capsular repeat units. Clin. Vaccine Immunol. 22, 313–318. (doi:10.1128/CVI.00647-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moberley S, Holden J, Tatham DP, Andrews RM. 2013. Vaccines for preventing pneumococcal infection in adults. Cochrane Database Syst Rev. 1, CD000422 (doi:10.1002/14651858.CD000422.pub3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinberger DM, Bruden DT, Grant LR, Lipsitch M, O'Brien KL, Pelton SI, Sanders EAM, Feikin DR. 2013. Using pneumococcal carriage data to monitor postvaccination changes in invasive disease. Am. J. Epidemiol. 178, 1488–1495. (doi:10.1093/aje/kwt156) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Satzke C, et al. 2013. Standard method for detecting upper respiratory carriage of Streptococcus pneumoniae: updated recommendations from the World Health Organization Pneumococcal Carriage Working Group. Vaccine 32, 165–179. (doi:10.1016/j.vaccine.2013.08.062) [DOI] [PubMed] [Google Scholar]
- 16.Whalan RH, Funnell SGP, Bowler LD, Hudson MJ, Robinson A, Dowson CG. 2006. Distribution and genetic diversity of the ABC transporter lipoproteins PiuA and PiaA within Streptococcus pneumoniae and related streptococci. J. Bacteriol. 188, 1031–1038. (doi:10.1128/JB.188.3.1031-1038.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki N, Yuyama M, Maeda S, Ogawa H, Mashiko K, Kiyoura Y. 2006. Genotypic identification of presumptive Streptococcus pneumoniae by PCR using four genes highly specific for S. pneumoniae. J. Med. Microbiol. 55, 709–714. (doi:10.1099/jmm.0.46296-0) [DOI] [PubMed] [Google Scholar]
- 18.Pikis A, Campos JM, Rodriguez WJ, Keith JM. 2001. Optochin resistance in Streptococcus pneumoniae: mechanism, significance, and clinical implications. J. Infect. Dis. 184, 582–590. (doi:10.1086/322803) [DOI] [PubMed] [Google Scholar]
- 19.Rolo D, Simões AS, Domenech A, Fenoll A, Liñares J, de Lencastre H, Ardanuy C, Sá-Leão R. 2013. Disease isolates of Streptococcus pseudopneumoniae and non-typeable S. pneumoniae presumptively identified as atypical S. pneumoniae in Spain. PLoS ONE 8, e57047 (doi:10.1371/journal.pone.0057047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arbique JC, et al. 2004. Accuracy of phenotypic and genotypic testing for identification of Streptococcus pneumoniae and description of Streptococcus pseudopneumoniae sp. nov. J. Clin. Microbiol. 42, 4686–4696. (doi:10.1128/JCM.42.10.4686-4696.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kellogg JA, Bankert DA, Elder CJ, Gibbs JL, Smith MC. 2001. Identification of Streptococcus pneumoniae revisited. J. Clin. Microbiol. 39, 3373–3375. (doi:10.1128/JCM.39.9.3373-3375.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Statens Serum Institute. 2013. Streptococcus pneumoniae: textbook in serotyping, virulence factors and enzyme-linked immunosorbent assay (ELISA) for measuring pneumococcal antibodies. Copenhagen, Denmark: Statens Serum Institut. [Google Scholar]
- 23.Lalitha MK, Thomas K, Satish Kumar R, Steinhoff MC. et al. 1999. Serotyping of Streptococcus pneumoniae by coagglutination with 12 pooled antisera. J. Clin. Microbiol. 37, 263–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wyllie AL, et al. 2016. Molecular surveillance of nasopharyngeal carriage of Streptococcus pneumoniae in children vaccinated with conjugated polysaccharide pneumococcal vaccines. Sci. Rep. 6, 23809 (doi:10.1038/srep23809) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Satzke C, Dunne EM, Porter BD, Klugman KP, Mulholland EK. 2015. The PneuCarriage project: a multi-centre comparative study to identify the best serotyping methods for examining pneumococcal carriage in vaccine evaluation studies. PLoS Med. 12, e1001903 (doi:10.1371/journal.pmed.1001903) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carvalho MDG, et al. 2010. Revisiting pneumococcal carriage by use of broth enrichment and PCR techniques for enhanced detection of carriage and serotypes. J. Clin. Microbiol. 48, 1611–1618. (doi:10.1128/JCM.02243-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trzciński K, et al. 2013. Superiority of trans-oral over trans-nasal sampling in detecting Streptococcus pneumoniae colonization in adults. PLoS ONE 8, e60520 (doi:10.1371/journal.pone.0060520) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krone CL, et al. 2015. Carriage of Streptococcus pneumoniae in aged adults with influenza-like-illness. PLoS ONE 10, e0119875 (doi:10.1371/journal.pone.0119875) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Azzari C, et al. 2010. Realtime PCR is more sensitive than multiplex PCR for diagnosis and serotyping in children with culture negative pneumococcal invasive disease. PLoS ONE 5, e9282 (doi:10.1371/journal.pone.0009282) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner P, Hinds J, Turner C, Jankhot A, Gould K, Bentley SD, Nosten F, Goldblatt D. 2011. Improved detection of nasopharyngeal cocolonization by multiple pneumococcal serotypes by use of latex agglutination or molecular serotyping by microarray. J. Clin. Microbiol. 49, 1784–1789. (doi:10.1128/JCM.00157-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biesbroek G, Sanders EAM, Roeselers G, Wang X, Caspers MPM, Trzciński K, Bogaert D, Keijser BJF, Gilbert JA. 2012. Deep sequencing analyses of low density microbial communities: working at the boundary of accurate microbiota detection. PLoS ONE 7, e32942 (doi:10.1371/journal.pone.0032942) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Majewski J, Zawadzki P, Pickerill P, Cohan FM, Dowson CG. 2000. Barriers to genetic exchange between bacterial species: Streptococcus pneumoniae transformation. J. Bacteriol. 182, 1016–1023. (doi:10.1128/JB.182.4.1016-1023.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carvalho MDG, et al. 2013. Non-pneumococcal mitis-group streptococci confound detection of pneumococcal capsular serotype-specific loci in upper respiratory tract. PeerJ. 1, e97 (doi:10.7717/peerj.97) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wyllie AL, et al. 2014. Streptococcus pneumoniae in saliva of Dutch primary school children. PLoS ONE 9, e102045 (doi:10.1371/journal.pone.0102045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carvalho MDG, et al. 2007. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J. Clin. Microbiol. 45, 2460–2466. (doi:10.1128/JCM.02498-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gillespie SH, Ullman C, Smith MD, Emery V. 1994. Detection of Streptococcus pneumoniae in sputum samples by PCR. J. Clin. Microbiol. 32, 1308–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki N, Seki M, Nakano Y, Kiyoura Y, Maeno M. 2005. Discrimination of Streptococcus pneumoniae from viridans group streptococci by genomic subtractive hybridization. Infect. Immun. 43, 4528–4534. (doi:10.1128/JCM.43.9.4528-4534.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abdeldaim GMK, Strålin K, Olcén P, Blomberg J, Herrmann B. 2008. Toward a quantitative DNA-based definition of pneumococcal pneumonia: a comparison of Streptococcus pneumoniae target genes, with special reference to the Spn9802 fragment. Diagn. Microbiol. Infect. Dis. 60, 143–150. (doi:10.1016/j.diagmicrobio.2007.08.010) [DOI] [PubMed] [Google Scholar]
- 39.Stralin K, Herrmann B, Abdeldaim GMK, Olcen P, Holmberg H, Molling P. 2014. Comparison of sputum and nasopharyngeal aspirate samples and of the PCR gene targets lytA and Spn9802 for quantitative PCR for rapid detection of pneumococcal pneumonia. J. Clin. Microbiol. 52, 83–89. (doi:10.1128/JCM.01742-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wessels E, Schelfaut JJG, Bernards AT, Claas ECJ. 2012. Evaluation of several biochemical and molecular techniques for identification of Streptococcus pneumoniae and Streptococcus pseudopneumoniae and their detection in respiratory samples. J. Clin. Microbiol. 50, 1171–1177. (doi:10.1128/JCM.06609-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dhoubhadel BG, et al. 2014. A novel high-throughput method for molecular serotyping and serotype-specific quantification of Streptococcus pneumoniae using a nanofluidic real-time PCR system. J. Med. Microbiol. 63 528–539. (doi:10.1099/jmm.0.071464-0) [DOI] [PubMed] [Google Scholar]
- 42.Wyllie AL, Rümke LW, Arp K, Bosch AATM, Bruin JP, Rots NY, Wijmenga-Monsuur AJ, Sanders EA, Trzciński K. 2016. Molecular surveillance on Streptococcus pneumoniae carriage in non-elderly adults; little evidence for pneumococcal circulation independent from the reservoir in children. Sci. Rep. 6, 34888 (doi:10.1038/srep34888) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simões AS, Tavares DA, Rolo D, Ardanuy C, Goossens H, Henriques-Normark B, Linares J, de Lencastre H, Sá-Leão R. 2016. lytA-based identification methods can misidentify Streptococcus pneumoniae. Diagn. Microbiol. Infect. Dis. 85, 141–148. (doi:10.1016/j.diagmicrobio.2016.03.018) [DOI] [PubMed] [Google Scholar]
- 44.Jolley KA, et al. 2012. Ribosomal multilocus sequence typing: universal characterization of bacteria from domain to strain. Microbiology 158, 1005–1015. (doi:10.1099/mic.0.055459-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joloba ML, et al. 2010. Comparison of transformation frequencies among selected Streptococcus pneumoniae serotypes. Int. J. Antimicrob. Agents 36, 124–128. (doi:10.1016/j.ijantimicag.2010.03.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Surewaard BGJ, et al. 2013. Pneumococcal immune evasion: ZmpC inhibits neutrophil influx. Cell Microbiol. 15, 1753–1765. (doi:10.1111/cmi.12147) [DOI] [PubMed] [Google Scholar]
- 47.Pai R, Gertz RE, Beall B. 2006. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J. Clin. Microbiol. 44, 124–131. (doi:10.1128/JCM.44.1.124-131.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whatmore AM, King SJ, Doherty NC, Sturgeon D, Chanter N, Dowson CG. 1999. Molecular characterization of equine isolates of Streptococcus pneumoniae: natural disruption of genes encoding the virulence factors pneumolysin and autolysin molecular characterization of equine isolates of Streptococcus pneumoniae: natural disruption. Infect. Immun. 67, 2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elberse KEM, van der Heide HGJ, Witteveen S, van de Pol I, Schot CS, van der Ende A, Berbers GAM, Schouls LM. 2012. Changes in the composition of the pneumococcal population and in IPD incidence in The Netherlands after the implementation of the 7-valent pneumococcal conjugate vaccine. Vaccine 30, 7644–7651. (doi:10.1016/j.vaccine.2012.04.021) [DOI] [PubMed] [Google Scholar]
- 50.Pimenta FC, et al. 2013. Sequential triplex real-time pcr assay for detecting 21 pneumococcal capsular serotypes that account for a high global disease burden. J. Clin. Microbiol. 51, 647–652. (doi:10.1128/JCM.02927-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bennett JS, Watkins ER, Jolley KA, Harrison OB, Maiden MCJ. 2014. Identifying Neisseria species by use of the 50S ribosomal protein L6 (rplF) gene. J. Clin. Microbiol. 52, 1375–1381. (doi:10.1128/JCM.03529-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kilian M. 2013. Within the Mitis group, phylogenetic analysis clearly distinguishes Streptococcus mitis and S. pseudopneumoniae from close relatives S. oralis and S. infantis, all having expanded into multiple evolutionary lineages. In The 23rd European Congress of Clinical Microbiology and Infectious Diseases (ECCMID), Berlin, Germany The European Society of Clinical Microbiology and Infectious Diseases. [Google Scholar]
- 53.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. (doi:10.1093/molbev/mst197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wattam AR. et al 2014. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 42(D1), D581–D591. (doi:10.1093/nar/gkt1099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou J, Enright MC, Spratt BG. 2000. Identification of the major Spanish clones of penicillin-resistant pneumococci via the Internet using multilocus sequence typing. J. Clin. Microbiol. 38, 977–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Azzari C, Moriondo M, Indolfi G, Massai C, Becciolini L, de Martino M, Resti M. 2008. Molecular detection methods and serotyping performed directly on clinical samples improve diagnostic sensitivity and reveal increased incidence of invasive disease by Streptococcus pneumoniae in Italian children. J. Med. Microbiol. 57, 1205–1212. (doi:10.1099/jmm.0.2008/000935-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scholz CFP, Poulsen K, Kilian M. 2012. Novel molecular method for identification of Streptococcus pneumoniae applicable to clinical microbiology and 16S rRNA sequence-based microbiome studies. J. Clin. Microbiol. 50, 1968–1973. (doi:10.1128/JCM.00365-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valente C, Hinds J, Pinto F, Brugger SD, Gould K, Mühlemann K, de Lencastre H, Sá-Leão R, Miyaji EN. 2012. Decrease in pneumococcal co-colonization following vaccination with the seven-valent pneumococcal conjugate vaccine. PLoS ONE 7, e30235 (doi:10.1371/journal.pone.0030235) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brugger SD, Frey P, Aebi S, Hinds J, Mühlemann K. 2010. Multiple colonization with S. pneumoniae before and after introduction of the seven-valent conjugated pneumococcal polysaccharide vaccine. PLoS ONE 5, e11638 (doi:10.1371/journal.pone.0011638) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sherwin RL, Gray S, Alexander R, McGovern PC, Graepel J, Pride MW, Purdy J, Paradiso P, File TM. 2013. Distribution of 13-valent pneumococcal conjugate vaccine Streptococcus pneumoniae serotypes in US adults aged 50 years with community-acquired pneumonia. J. Infect. Dis. 208, 1813–1820. (doi:10.1093/infdis/jit506) [DOI] [PubMed] [Google Scholar]
- 61.Bewick T, Sheppard C, Greenwood S, Slack M, Trotter C, George R, Lim WS. 2012. Serotype prevalence in adults hospitalised with pneumococcal non-invasive community-acquired pneumonia. Thorax 67, 540–545. (doi:10.1136/thoraxjnl-2011-201092) [DOI] [PubMed] [Google Scholar]
- 62.Shelburne SA, Sahasrabhojane P, Saldana M, Yao H, Su X, Horstmann N, Thompson E, Flores AR. 2014. Streptococcus mitis strains causing severe clinical disease in cancer patients. Emerg. Infect. Dis. 20, 762–771. (doi:10.3201/eid2005.130953) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Steenhuijsen Piters WAA, et al. 2016. Dysbiosis of upper respiratory tract microbiota in elderly pneumonia patients. ISME J. 10, 97–108. (doi:10.1038/ismej.2015.99) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Severina E, Ramirez M, Tomasz A. 1999. Prophage carriage as a molecular epidemiological marker in Streptococcus pneumoniae. J. Clin. Microbiol. 37, 3308–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tavares DA, Simões AS, Bootsma HJ, Hermans PW, de Lencastre H, Sá-Leão R. 2014. Non-typeable pneumococci circulating in Portugal are of cps type NCC2 and have genomic features typical of encapsulated isolates. BMC Genomics 15, 863 (doi:10.1186/1471-2164-15-863) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jomaa M, Yuste J, Paton JC, Jones C, Dougan G, Brown JS. 2005. Antibodies to the iron uptake ABC transporter lipoproteins PiaA and PiuA promote opsonophagocytosis of Streptococcus pneumoniae. Infect. Immun. 73, 6852–6859. (doi:10.1128/IAI.73.10.6852-6859.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brown JS, Gilliland SM, Ruiz-Albert J, Holden DW. 2002. Characterization of Pit, a Streptococcus pneumoniae iron uptake ABC transporter. Infect. Immun. 70, 4389–4398. (doi:10.1128/IAI.70.8.4389-4398.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Siira L, Kaijalainen T, Lambertsen L, Nahm MH, Toropainen M, Virolainen A. 2012. From Quellung to multiplex PCR, and back when needed, in pneumococcal serotyping. J. Clin. Microbiol. 50, 2727–2731. (doi:10.1128/JCM.00689-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cremers AJH, Kokmeijer I, Groh L, de Jonge MI, Ferwerda G. 2014. The role of ZmpC in the clinical manifestation of invasive pneumococcal disease. Int. J. Med. Microbiol. 8, 984–989. (doi:10.1016/j.ijmm.2014.06.005) [DOI] [PubMed] [Google Scholar]
- 70.Knol MJ, Wagenvoort GHJ, Sanders EAM, Elberse K, Vlaminckx BJ, de Melker HE, van der Ende A. 2015. Invasive pneumococcal disease 3 years after introduction of 10-valent pneumococcal conjugate vaccine, the Netherlands. Emerg. Infect. Dis. 21, 2040–2044. (doi:10.3201/eid2111.140780) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aanensen DM, Mavroidi A, Bentley SD, Reeves PR, Spratt BG. 2007. Predicted functions and linkage specificities of the products of the Streptococcus pneumoniae capsular biosynthetic loci. J. Bacteriol. 189, 7856–7876. (doi:10.1128/JB.00837-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yahiaoui RY, den Heijer CD, Wolfs P, Bruggeman CA, Stobberingh EE. 2016. Evaluation of phenotypic and molecular methods for identification of Streptococcus pneumoniae. Future Microbiol. 11, 43–50. (doi:10.2217/fmb.15.124) [DOI] [PubMed] [Google Scholar]
- 73.Park IH, Kim KH, Andrade AL, Briles DE, Mcdaniel LS, Nahma MH. 2012. Nontypeable pneumococci can be divided into multiple cps types, including one type expressing the novel gene pspK. Am. Soc. Microbiol. 3, e00035-12. (doi:10.1128/mBio.00035-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilson R, et al. 2017. Naturally acquired human immunity to pneumococcus is dependent on antibody to protein antigens. PLoS Pathog. 13, e1006137 (doi:10.1371/journal.ppat.1006137) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rukke HV, et al. 2014. Protective role of the capsule and impact of serotype 4 switching on Streptococcus mitis. Infect. Immun. 82, 3790–3801. (doi:10.1128/IAI.01840-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sá-Leão R, et al. 2009. Changes in pneumococcal serotypes and antibiotypes carried by vaccinated and unvaccinated day-care centre attendees in Portugal, a country with widespread use of the seven-valent pneumococcal conjugate vaccine. Clin. Microbiol. Infect. 15, 1002–1007. (doi:10.1111/j.1469-0691.2009.02775.x) [DOI] [PubMed] [Google Scholar]
- 77.Scott JR, et al. 2012. Nontypeable pneumococcal isolates among Navajo and White Mountain Apache communities: are these really a cause of invasive disease? J. Infect. Dis. 206, 73–80. (doi:10.1093/infdis/jis307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chewapreecha C, et al. 2014. Dense genomic sampling identifies highways of pneumococcal recombination. Nat. Genet. 46, 305–309. (doi:10.1038/ng.2895) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hilty M, et al. 2014. Global phylogenomic analysis of nonencapsulated Streptococcus pneumoniae reveals a deep-branching classic lineage that is distinct from multiple sporadic lineages. Genome. Biol. Evol. 6, 3281–3294. (doi:10.1093/gbe/evu263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martin M, et al. 2003. An outbreak of conjunctivitis due to atypical Streptococcus pneumoniae. N. Engl. J. Med. 348, 1112–1121. (doi:10.1056/NEJMoa022521) [DOI] [PubMed] [Google Scholar]
- 81.Bek-Thomsen M, Poulsen K, Kilian M. 2012. Occurrence and evolution of the paralogous zinc metalloproteases IgA1 Protease, ZmpB, ZmpC, and ZmpD in Streptococcus pneumoniae and related commensal species. MBio. 3, e00303-12 (doi:10.1128/mBio.00303-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yamaguchi M, Nakata M, Sumioka R, Hirose Y, Wada S, Akeda Y, Sumitomo T, Kawabata S. In press Zinc metalloproteinase ZmpC suppresses experimental pneumococcal meningitis by inhibiting bacterial invasion of central nervous systems. Virulence. (doi:10.1080/21505594.2017.1328333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Carvalho MDG, et al. 2012. Potential nonpneumococcal confounding of PCR-based determination of serotype in carriage. J. Clin. Microbiol. 50, 3146–3147. (doi:10.1128/JCM.01505-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.