Abstract
A tribute to Mary Lyon was held in October 2016. Many remarked about Lyon's foresight regarding many intricacies of the X-chromosome inactivation process. One such example is that a year after her original 1961 hypothesis she proposed that genes with Y homologues should escape from X inactivation to achieve dosage compensation between males and females. Fifty-five years later we have learned many details about these escapees that we attempt to summarize in this review, with a particular focus on recent findings. We now know that escapees are not rare, particularly on the human X, and that most lack functionally equivalent Y homologues, leading to their increasingly recognized role in sexually dimorphic traits. Newer sequencing technologies have expanded profiling of primary tissues that will better enable connections to sex-biased disorders as well as provide additional insights into the X-inactivation process. Chromosome organization, nuclear location and chromatin environments distinguish escapees from other X-inactivated genes. Nevertheless, several big questions remain, including what dictates their distinct epigenetic environment, the underlying basis of species differences in escapee regulation, how different classes of escapees are distinguished, and the roles that local sequences and chromosome ultrastructure play in escapee regulation.
This article is part of the themed issue ‘X-chromosome inactivation: a tribute to Mary Lyon’.
Keywords: X-chromosome inactivation, sexual dimorphism, epigenetic regulation, escapee, inactive X chromosome, lupus
1. Introduction
Lyon hypothesized in 1961 that one X chromosome in female mice became inactivated [1]. In 1962, she followed this prescient paper with an extension of the hypothesis to humans, including the suggestion that genes with Y homologues would escape from ‘X-chromosome inactivation’ (XCI) [2]. In the intervening 55 years, we have learned much about the process of inactivation, and, somewhat surprisingly, have found that escape from inactivation is not rare, with over 20% of human genes being escapees that are expressed from both the active and the inactive X chromosome. Many epigenetic factors, including the long non-coding (lnc)RNA expressed only from the inactive X chromosome (and thus named XIST for Xi-specific transcripts), DNA methylation, histone modifications, chromatin remodellers and histone variants, chromosome ultrastructure and nuclear localization act synergistically to silence and then maintain silencing of the chromosome. Yet despite the remarkably stable inactivation maintained by these factors, many genes escape from inactivation, albeit generally with less expression than from the active X allele. Determining which genes escape from inactivation reveals an important source of sexually dimorphic gene expression and, therefore, we start this review by a brief discussion of the role of inactive X expression on human phenotype. We then discuss which genes escape inactivation, in human and mouse, an area that is rapidly progressing as new technologies are able to address multiple primary tissues. Identifying the mechanism by which these exceptions occur will inform our understanding of XCI and broader questions of selective epigenetic repression of genes. We thus discuss the factors and features of the X chromosome that probably contribute mechanistically to escape from inactivation. We conclude this review with a discussion of the potential roles of the X chromosome in systemic lupus erythematosus (lupus) predisposition as an example that underscores the importance of considering the impact of the inactive X in complex diseases.
2. Escape matters: impact of X-chromosome inactivation on human disease
It is generally accepted that XCI was an evolutionary response to achieve dosage equivalence for X-linked genes between males and females following the decay of the Y homologues (see [3] for review). The majority of genes on the X chromosome no longer show recognizable homology with the Y chromosome; however, Y homology is enriched among genes escaping XCI. The potential impact of escape from XCI with respect to dosage equivalency between the sexes depends on the status of the Y homologue (table 1). For some genes, escape from XCI maintains dosage equivalence as expressed homologues are retained on the Y chromosome—these are the genes that Lyon predicted would escape from XCI [2]. The largest cohort of X/Y homologous genes in humans is found in the pseudoautosomal region (PAR1); but intriguingly, the lack of full expression from the inactive X still results in some dosage imbalance, with greater expression in males (table 1) [8]. For genes with expressed Y homologues that map outside the PAR1, the lack of recombination with the X chromosome has allowed divergence of the X and Y versions, so that while both continue to be expressed, they may not be functionally equivalent (e.g. the histone demethylase activity encoded by KDM6A and KDM6C differs due to point substitutions [9]). Many other human escape genes lack expressed Y homologues (see §3), and thus result in dosage imbalance with excess expression in females relative to males. How much impact do these expression differences have? Clearly, in the case of aneuploidy for the X chromosome, altered expression levels from the inactive X will be important, but genes expressed from the inactive X may also protect from effects of or predispose towards somatic mutation, and are also likely to be important contributors to differences between males and females.
Table 1.
Impact of genes on the inactive X. Xa, active X; Xi, inactive X; Xm, maternal X; Xp, paternal X; PAR, pseudoautosomal region.
| males |
females |
impacta |
||
|---|---|---|---|---|
| chromosomes | XY | XX | no male : male transmission of X | |
| subject gene expression | XaY | = | XmaXp i or XmiXpa | Mutation—females are mosaic; males are hemizygous Imprinting—males only have maternal Xb |
| escape gene expression—PAR1 | Xa + Y | > | Xa + Xi | Homology maintained by male recombination Higher expression in males Likely contributors to Turner syndrome |
| escape gene expression—expressed Y homologue (not PAR1) | Xa + Y | ≠ | Xa + Xi | Y version may be functionally distinct Females show higher expression (of X copy) Likely contributors to Turner syndrome |
| escape gene expression—no expressed Y homologue | Xa | < | Xa + Xi | Females show higher expression Potentially protective against Xa mutation Express Xi mutations |
| subject gene expression in aging/cancer | Xa | < | Xa + Xi | Reactivation of genes from Xi reported in some cancers [4] or cells [5] |
| escape gene expression in aging/cancer | XaXaY | < | Xa < XaXi | Loss of X or Y often seen in haematological cancers [6] or with ageing [7] |
aSee text for additional discussion and references.
bImprinted genes are reported for a small number of genes in somatic tissue of mouse X but have not yet been identified on the human X.
(a). Sex chromosome aneuploidies
The viability of 39,X mice (i.e. lacking either an inactive X or Y chromosome) was cited by Lyon as evidence to support her hypothesis of inactivation [1]. However, in humans, over 99% of 45,X conceptuses fail to survive, suggesting a much more important role for the second sex chromosome, and alluding to differences in escapees between humans and mice that is further explored in §3. Hook and Warburton suggested that all viable 45,X (Turner syndrome) females in fact have an undetected cryptic mosaicism, as true 45,X is not viable [10]. The importance of either the inactive X or Y chromosome in normal human development implicates a role for one or more of the 17 human genes that retain X–Y homology, particularly the nine in which Y (and inactive X) expression is not shared with mouse [11]. As the X–Y conserved gene pairs' products are enriched in regulatory functions, they can have widespread impact on the transcriptome. Their continued presence on the Y chromosome, and escape from inactivation on the X chromosome, argues for a strict requirement for continued dosage equivalence, an observation that is reinforced by the number of X-linked intellectual disability syndromes caused by haploinsufficiency (or duplication) of many of these genes (KDM5C, KDM6A, NLGN4X, DDX3X, EIF2S3X, SMC1A and USP9X).
Aneuploidies involving supernumerary X chromosomes (47,XXY and 47,XXX predominantly, although higher numbers of sex chromosomes are viable) are seen at frequencies of approximately 1/500 men and women, respectively. In contrast to the prenatal demise of 45,X, it appears that gain of an X is better tolerated. Yet overall epidemiological surveys suggest an increased mortality, corresponding to a standardized mortality ratio of 2.5 for 47,XXX in Denmark or the UK [12,13] and 1.5 for 47,XXY in the UK [14]. However, these estimates are clearly influenced by ascertainment, which often follows diagnostic evaluation for developmental delay or learning disability. The lack of a distinct and/or severe phenotype for 47,XXX means that many individuals remain undiagnosed, and these are probably females with less severe outcomes, in agreement with the observation that prenatally ascertained 47,XXX females generally had better cognitive and adaptive functioning [15]. While gain of an X chromosome seems detrimental to some cognitive abilities, it has limited impact, or may even be protective for autism spectrum disorders, which generally show a 4 : 1 male predominance [16]. Unbiased ascertainment is essential to better define the phenotypic spectrum associated with supernumerary X chromosomes. Our growing knowledge of the transcriptional regulation of escapees gives hope to the idea of unravelling the contribution of escapees to the profound impact that loss or gain of an inactive X can have on individuals, particularly cognitively and behaviourly.
(b). X-linked mutations
Classically, X-linked mutations manifest in hemizygous males, with mosaicism arising from random XCI providing protection to heterozygous females. Furthermore, for some disorders cellular selection skews the mosaic cell population, further reducing the impact of X-linked mutations in females. There are more than 100 genes identified to be mutated in males with X-linked intellectual disability, often displaying variable severity in females that ranges from unaffected, but rarely equals that of males. More recently, advanced genomic analyses have been applied to females with undiagnosed causes of intellectual disability, uncovering mutations in the escapees USP9X [17] and DDX3X [18]. While missense mutations in these genes have been identified in males, the more severe mutations observed in females may lead to male lethality, resulting in the unique restriction to females who have compensating expression from the other X chromosome. XCI is not the only factor influencing gender-specific prevalence of X-linked mutations. There is generally a higher mutation rate in males (e.g. [19]), and paternal X-linked mutations are not transmitted to sons; however, germ-line mutation frequencies are both gene- and mutation-specific (e.g. [20]).
Cancer often shows a male predominance, and a recent study suggests that a portion of this male excess is attributable to mutations in X-linked tumour suppressor genes that escape inactivation in females: the argument being that females are protected from developing cancer by expression from their second expressed allele [21]. For example, escape from inactivation was previously suggested to protect females from cancers caused by X-linked mutations in KDM6A driving T-cell acute lymphoblastic leukaemia [22]. Nevertheless, the inactive X chromosome (or Y chromosome) is often lost in cancers, which may result in loss of this protective effect [21]. By contrast, somatic mutations have been seen to be enriched on the inactive X chromosome in cancers [23], and in combination with expression of escape genes from the inactive X chromosome may underlie the observation that hypermutation of X-linked genes can be oncogenic drivers in female cancers, including the DDX3X gene that escapes from inactivation [24]. In addition to mutations that occur in cancer, instability of the inactivation of the X has also been reported in female cancers, most notably breast cancer [4], resulting in derepression of a set of previously X-inactivated genes in some cells.
Altogether, the presence of a second X chromosome is generally protective against germ-line X-linked mutations in females through a combination of mosaicism and selection. For somatic mutations, functional hemizygosity is achieved by inactivation, making risk similar for males and females; however, protection may still be afforded by those genes that escape inactivation. On the downside for XX females, instability of inactivation can increase disease risk for females and differences in mutation rate may favour somatic mutations on the heterochromatic inactive X chromosome. In addition to these sex differences from mutations in X-linked genes, the differential expression of escapees may give rise to general male : female differences in disease predisposition.
(c). Sexual dimorphism
Sex differences are common, and generally attributed to the hormonal changes induced by gonadal differentiation; however, there is considerable evidence supporting a role for the sex chromosomes in sexual dimorphism, distinct from sex determination (reviewed in [25]). Mouse breeding schemes that generate XX males and XY females to compare with normal XY males and XX females (known as the ‘Four-Core Genotypes’ model) have revealed a role for the sex chromosomes in sexually dimorphic traits including adiposity, metabolic disease, cardiovascular injury and behaviour [25]. The role is likely to be even more pronounced in humans due to the larger number of escape genes (see §2), but separation of effects from hormonal influences is challenging in the absence of such controlled breeding. As mentioned above, X-chromosome aneuploidy can provide support for a role for the inactive X (or Y chromosome) in disease predispositions. For example, men and women show distinctions in innate and adaptive immune responses (reviewed in [26]). Defects in 45,X individuals suggest a suppressed immune system that is proposed in part to be related to escapee KDM6A, which plays important roles in T-cell differentiation [27]. Additionally, autoimmune disorders consistently show a female excess, with 70–90% of lupus patients being female. Moreover, an over-abundance of 47,XXY males also develop lupus, implicating the presence of the inactive X as a risk factor for this disorder. While a full assessment of the many potential impacts of the X on disease is beyond the scope of this review, we will return to a consideration of the specific case of lupus as an example of the role of the inactive X in complex disease predisposition, after we have discussed what is known about escape from inactivation.
3. The landscape of escape genes on the inactive X chromosome
An understanding of the approaches that have been used to assess X-inactivation status is important for comparing results between both species and studies. New genomic methodologies have not only improved the identification of escapees, but also allowed expansion to multiple tissues and individuals and have yielded insights into their unique chromatin and structural environment relative to other genes on the X. This section summarizes our current understanding of escape gene organization and epigenetic landscape in both human and mouse.
(a). Identification of genes on the human X that escape X inactivation
Multiple approaches have been employed to establish which of the approximately 1100 X chromosome genes escape XCI, in part because direct assessment of inactive X expression is complicated by the random nature of XCI that results in mosaicism within tissues whereby each parental X chromosome is active in only a subset of cells. While each XCI profiling strategy has advantages and disadvantages, results are largely complementary [28] and a clear picture of the human XCI landscape has emerged and continues to be refined.
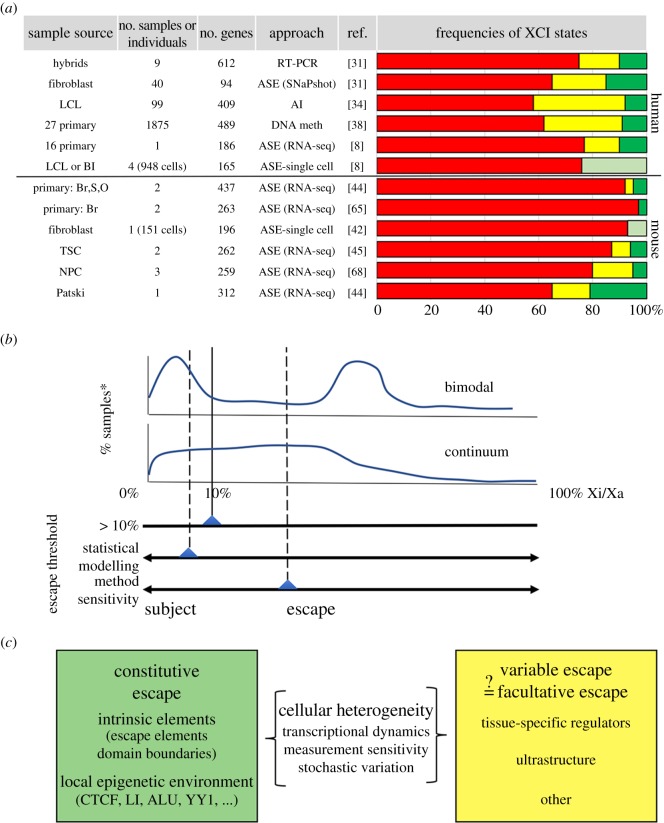
To overcome mosaicism, early studies to evaluate XCI status in humans used cells from female carriers of X-linked disorders and assayed protein variants in clonal cell populations or enzyme activity from human inactive X chromosomes segregated in rodent–human somatic cell hybrids [29,30]. The first human chromosome-wide X-inactivation profile also assessed somatic cell hybrids to directly measure inactive X gene expression, and reported that genes that escape XCI are not rare [31]. While the epigenetic state in somatic cell hybrids is relaxed [32], gene silencing is largely maintained [28]. Indeed, XCI escapees were confirmed by measuring allele-specific expression (ASE) of heterozygous transcribed SNPs in primary cells with preferential inactivation of one of the two Xs [31]. In this latter approach, genes that are X inactivated are monoallelically expressed whereas escapees are bi-allelic. Initial efforts developed quantitative assays to assess each gene individually [31]. Similar allele-specific analysis of RNA-sequencing (seq) data rapidly expands the number of scorable genes [8] (figure 1a). Nevertheless, assessment is still limited to the number of expressed heterozygous SNPs in each individual.
Figure 1.
Frequencies, expression patterns and sources of variable escape. (a) Summary of select XCI surveys. XCI states are coloured red (subject), yellow (variable escape) and green (escape). A lighter green denotes the frequency of bi-allelic genes reported in single-cell RNA-seq surveys, but does not consider cellular heterogeneity as a source of variable escape. The number of genes listed is as reported in the indicated references and may not necessarily correlate with more recent genome annotations (e.g. [31]). Abbreviations include: tissues, blood (Bl), brain (Br), spleen (S) and ovary (O), trophectoderm stem cells (TSC), neuronal precursor cells (NPC); approaches, allele-specific expression (ASE), allelic imbalance (AI). (b) Variable escape between individuals or between tissues can exhibit a bimodal pattern of inactive X expression levels, but more frequently represents a continuum. Inactive X expression for each sample is generally measured relative to active X levels (Xi/Xa) with XCI status depending, in part, on criteria for escape. (c) Classification of escape genes and contributing factors. Cellular heterogeneity probably impacts both constitutive escapees and variable escape genes. Variable escapees may be akin to facultative escape and in addition to tissue-specific regulators and ultrastructure could be impacted by factors influencing epigenetic states or developmental timing. As discussed in text, the constitutive escapees are controlled by intrinsic elements and also factors that influence the local chromatin neighbourhood of which known contributors are cited, although additional factors are probably involved.
While ASE assays allow definitive assignment of inactive X expression, cell samples with non-random inactivation are rare and may be caused by an underlying defect. Therefore, methods to infer inactive X expression using more recently developed genome-wide approaches provide a broader perspective by enabling assessment of karyotypically normal cells and tissues (recently reviewed in [33]). In samples with partial XCI skewing, XCI status can be inferred by allelic imbalance following normalization for XCI skewing [34]. This approach successfully identified escapees by comparing allelic expression ratios that were detected by hybridization to SNP microarrays [34]. Nevertheless, it is most effective for samples with highly skewed XCI, which are rare in the general population.
A strategy that circumvents both issues with XCI mosaicism and the need for informative transcribed SNPs is to infer escape by comparing male and female expression profiles to identify gender differences. Sensitivity from RNA-seq (e.g. [8]) greatly improves previous microarray analyses (e.g. [35]), and many escapees are expressed at higher levels in females. An interesting exception is that the pseudoautosomal genes in PAR1, despite identity to homologues on the Y chromosome, are expressed at higher levels in males [8]. A recent variation of this approach compares male/female cap analysis of gene expression (CAGE) datasets of the 5′ end of transcripts [36] and expands and refines human XCI maps, as alternative transcript start sites can have different XCI status [37]. A clear advantage to female-biased escape gene analyses is the ability to evaluate all expressed X genes in each sample. Nevertheless, female bias is more difficult to detect at genes with partial inactive X expression. Further, gender bias can also reflect hormonal or other sex-specific influences.
Male–female comparisons can be extended to epigenomic features that differentiate active and inactive genes as a means to proxy XCI status. Many escapees are distinguishable from their X-inactivated counterparts by promoter and gene-body DNA methylation signatures [38–40]. X-chromosome-wide comparison of CpG island promoter methylation has particularly high concordance with previous assessments of XCI status [28], although approximately 10% of X genes with CpG islands have intermediate methylation levels precluding XCI status assignment [38]. Open chromatin assessed by the Assay for Transposase-Accessible Chromatin (ATAC)-seq, has also been used to determine XCI status and detects gender differences at escapees with higher power and accuracy using fewer samples than expression microarrays [41].
Most recently, allele-specific analysis of single-cell RNA-seq has emerged as a powerful tool for directly assessing inactive X expression without complications from XCI mosaicism [8]. Current technologies reliably assay the most highly expressed X genes. Challenges such as allele drop out due to fluctuations in gene expression, including transcription bursts, can complicate analyses and cause escapees to appear monoallelic. Initial XCI studies in human, as well as mouse, indicate that adequate cell sampling and accurate haplotype phasing help overcome this hurdle [8,42]. It is encouraging that the escape genes and escape frequencies appear similar to those identified by approaches that assessed mosaic cell samples, yet whether genes that are only partially expressed from the inactive X are accurately identified as escapees has not been fully addressed. This approach offers opportunities to directly examine cells from primary tissues as well as to query unique aspects of escapees, such as intra-individual heterogeneity [42], a question that was previously only addressable at an individual gene level by RNA fluorescence in situ hybridization (FISH). It will be exciting as such studies expand to evaluate inactive X expression in an increasing number of tissues and individuals using single-cell RNA-seq or new advances in FISH [43].
(i). A chromosomal perspective of human inactive X expression
To date, the XCI status for more than 680 genes on the human X chromosome has been catalogued in up to 29 tissues (figure 1a). This includes almost 80% of protein coding genes [28], not considering the multi-copy testis-expressed genes that have accumulated on the X. Importantly, a comparison indicates that discordant XCI classification between studies is relatively rare (estimated at approximately 10%) [28]. Differing XCI assignments can reflect differences in the samples assayed, as escape is probably increased in cultured cells and hybrids compared with primary tissues [28,44], but may also reflect differential assay sensitivity and/or criteria to detect and classify low, but potentially biologically significant, expression from the inactive X (figure 1b).
Integrating results from current XCI profiles confirms that a large number of human genes escape XCI, with estimates ranging from 12 to 20% [8,28] (figure 1a). As discussed above, this list includes PAR1 genes and 12 of 14 genes tested with functional X–Y homologues that map outside of PARs [11]. Yet, many additional genes escape XCI, but lack expressed Y homologues. Notably, escapees are consistently only partially expressed from the inactive X, averaging 33% of levels relative to their active X counterpart [8,34]. Even PAR1 genes are not fully dosage-compensated between sexes, with inactive X expression at most 80% of levels on the active X or Y chromosome. XCI escape was initially defined for genes with inactive X expression at or above 10% of active X levels [31]. More recently, escape definitions have been expanded using statistical methods to validate even lower levels of inactive X expression [44,45]. The biological consequence of very low inactive X expression remains unknown. It is feasible that appreciable levels of inactive X expression could influence traits or dampen carrier phenotypes, as has been seen for autosomal genes (e.g. [46]). With respect to XCI mechanisms, differences between fully inactivated genes and those with very low inactive X expression may help pinpoint altered XCI regulatory features that predispose to reactivation. Additionally, whether genes with very low inactive X expression share XCI regulatory features with other escapees will need to be addressed.
(ii). It is not always black and white; genes that variably escape X inactivation
As XCI profiling has been expanded to larger numbers of cell types and individuals, the prevalence of genes that variably escape XCI has grown. Variable escape is a classification that has been adopted to describe the substantial portion of X genes (greater than 10%) with XCI states that differ between individuals or between cells or tissues within an individual [28,47]. Expression for some variable escapees is bimodal; that is, some genes escape XCI in a subset of samples but are X inactivated in others (figure 1b). More commonly, however, variable escape genes exhibit a continuous range of inactive X expression levels across assigned/calculated thresholds for inactivation or escape [34] (figure 1b). Understanding how, why and which genes variably escape XCI can give unique perspective into cellular, tissue-specific, genetic and/or epigenetic differences that influence expression on the inactive X chromosome, as well as provide insight into the role that these genes play in traits and disorders that show gender bias.
A number of studies have examined tissue differences as a source of variable escape. Very few examples of tissue-specific inactive X variation were detected by DNA methylation analysis, although neuronal tissues may be one exception [38,40]. Epigenetic alterations in the placenta [48] may impact XCI states, but placenta was not included in several recent chromosome-wide studies in human [49], although trophoblast has been well examined in mouse (see below). Female-biased expression analysis of RNA-seq data across 29 tissues confirmed variation between tissues in escape [8]. Serendipitously, Tukiainen et al. [8] identified a single normal female with complete non-random XCI that was included in the genotype tissue expression project (GTEx) [49]. This donor provides a rare opportunity to directly validate XCI tissue variation in humans without concerns about genetic background effects or other inter-individual parameters. Allele-specific analysis of RNA-seq data from 16 tissues from this individual importantly confirmed minimal variation between tissues for almost half of the 43 escape genes identified. Yet inactive X expression levels varied for the other escape genes, with 6% of all X genes tested having bi-allelic expression (i.e. escape) in only a single tissue. For at least some of these genes, overall variation in tissue-specific expression levels may be a limiting factor in adequately discerning variable escape from XCI between tissues, as partial escape at even 10% of active X levels may be below the detection threshold in tissues with already low active X chromosome expression.
Even within a tissue or cell line, cellular heterogeneity is observed for many escapees, with inactive X expression detected only in a subset of cells [50]. Variability may be due to partial XCI escape or lack of sensitivity to detect low inactive X expression in all cells, particularly if active X expression is low. Additionally, heterogeneity can reflect stochastic cellular processes associated with gene expression [51]. Consequently, as sample sizes increase and escape thresholds are impacted not only by approaches that consider very low levels of inactive X expression, but also by the sensitivity of particular methods to detect inactive X expression, an increasing number of genes are seen to escape XCI in at least a subset of samples (figure 1a,b). Recent single-cell RNA-seq experiments provide additional insight into variable XCI escape. For the variable escapee TIMP1, high inactive X expression was detected from a small subset of cells in an individual [8], consistent with previous results in somatic cell hybrids [52] and suggesting cell to cell variation in the propensity to escape, at least for some X genes. These data are complemented by recent single-cell RNA-seq analysis in mouse that showed inactive X variation for one gene, FundC1, was mitotically heritable in cell clones [42], supporting their conclusion that clonal variability can contribute to cellular heterogeneity in allelic expression in females.
An additional source of XCI variation is between individuals, even within the same tissue. While some of this variation may be stochastic, a number of observations support genetic influences. The strongest argument for heritability comes from the analysis of 51 variable escape genes in 31 monozygotic twin pairs [38]. Concordant escape levels between twins were higher than expected by chance, with only 3% of genes differing between twin pairs. These data not only support a substantial genetic component, but also indicate that genetics alone cannot explain all inter-individual variation. Further pointing towards a genetic basis, three studies have documented population differences in XCI escape, with higher escape in Nigerian YRI samples [34,35,53], although it will be important to confirm this lymphoblast cell line-based conclusion in primary cells. Finally, although current single-cell data are limited, XCI escape for at least one gene, ASMTL, differs between haplotypes [8], similar to mouse, where several examples of strain-specific escape have been identified [45,54]. cis-regulated allele-biased expression impacts a high percentage of transcripts throughout the genome and has spurred identification of expression or epigenetic quantitative trait loci (eQTLs or epiQTLs) [55]. Whether similar loci specifically modulate inactive X expression remains unknown, but would be of considerable interest for defining the molecular basis of variable escape and its role in traits. The ability to search for such regulatory loci at variable escape genes should become feasible as datasets allowing allele-specific XCI profiling expand.
(iii). Human escape gene organization
The distribution of escape genes on the human X both reflects sex chromosome evolution and gives insight into regulation. Escape genes are non-randomly organized along the X, with most mapping to the distal short arm. The human X short arm is a recent evolutionary addition and most escapees map within the youngest strata [31,56]. Many of these escape genes retain Y homology, although Y copies have been pseudogenized or have acquired distinct functions or expression patterns (e.g. [57,58]). Acquisition of XCI has been proposed to be a response to Y-gene degradation, suggesting some escapees are at an intermediate stage in this process [59]. Many escape genes, particularly those in the short arm, are clustered in large domains that include at least one gene with Y homology. This could suggest that some adjacent genes simply escape as a bystander effect, lacking strict dosage requirements themselves. Genes at domain boundaries are more frequently discordant between different XCI surveys [28], suggesting that boundaries vary between individuals or that genes at these active/inactive transition regions are more vulnerable to reactivation, particularly in cultured cells. Interestingly, a higher proportion of tissue-specific variable escape genes map to the human X long arm [8], further indicating that chromosome regions respond differently to XCI. That both escapees and variable escape genes without Y homology are under strong purifying selection, similar to their counterparts with Y partners [60,61], may hint that conserved features at escape loci underlie inactive X expression.
(b). Escape in mouse: differences and similarities
Since Lyon first proposed her landmark X-inactivation hypothesis, studies of mouse models have continued to provide insights into the process of XCI. Mice offer numerous experimental advantages, including access to developmental time points throughout XCI establishment, female embryonic stem cells that undergo ex vivo XCI, breeding schemes that produce highly informative offspring, and the ability to generate mutations. Yet differences between human and mouse continue to be revealed, a major one being that in mice there is imprinted inactivation of the paternal X in extra-embryonic tissues, while in humans, expression of the zygotic genes, as well as XCI, initiates later and inactivation is random in both embryonic and extra-embryonic tissues (reviewed in [62]). Furthermore, there are differences in humans and mice in the regulatory sequences controlling XIST (reviewed in [62]), and early human embryos show XIST expression from both Xs [63] as well as evidence for initial ‘dampening’ of X expression [64]. Thus, we turn to mouse to describe the silencing events on the X.
Expression profiles of the mouse inactive X chromosome have been established in both cultured lines and in primary tissues. Evaluation of XCI status in mouse by ASE is facilitated by the use of interspecific crosses to maximize genetic heterogeneity and non-randomly X-inactivated cell samples that result from mutations in XCI regulatory genes, selection [44,65], or the use of extra-embryonic tissues (or cultured lines) that preferentially inactivate the paternal X chromosome [45]. Allelic analysis of single-cell RNA-seq is also aided by interspecific crosses with defined genomes [42]. Studies performed to date agree, in large part, that escape genes are less frequent in mouse than human, with estimates ranging from 3 to 7% of X-linked genes analysed [44] (figure 1a). Nevertheless, orthologues to many mouse escapees also escape XCI on the human X [47], suggesting conserved gene dosage requirements. In contrast to human escapees, most of these mouse escape genes are not clustered, although some lie adjacent to a single lncRNA that also escapes XCI [66,67]. Such juxtaposition could imply a functional role for these lncRNAs, but perhaps more probably highlights the transcriptionally permissive environment at these escape gene loci.
As more tissues and cell samples have been evaluated and, as discussed above, definitions of XCI escape have been expanded to consider lower levels of inactive X expression, the number of variable escapees reported in mouse has increased to greater than 3% (figure 1a,b) [44,45,68,69]. Heterogeneity in inactive X expression levels can be seen within and between tissues types or cells, and even between mouse strains. Nevertheless, many of the F1 crosses used to examine escape in mouse use the same parental lines (Mus musculus strains 129 and C57/BL6, Mus spretus, Mus castaneus), and thus escape has been examined for only a limited number of haplotypes relative to the diversity of X chromosomes profiled in humans. Interestingly, the genes that escape XCI are, in general, the same across strains, although some strain specificity has been reported [45,54]. Therefore, most XCI variation identified to date in mouse cannot be genetic; however, because the variable genes identified are consistently observed across studies, it is also unlikely to be random stochastic instability.
As in human, many mouse genes escape in only a single tissue, although because of recent consortium efforts to comprehensively profile the human transcriptome [49], the number of tissues evaluated in mouse to date is not as extensive. Importantly, the tissue-specific escape seen in primary tissues is mostly confined to genes with tissue-specific functions [44]. Compared with primary tissues, escape is substantially higher in the embryonic kidney-derived Patski cell line, ES-differentiated neuronal precursor cells (NPCs), and extra-embryonic trophoblast stem cells [44,45,68,69], pointing towards epigenetic relaxation in cultured cells and/or cell type–specific differences that favour inactive X transcription. Notably, higher variable escape was also detected or expected in neuronal and extra-embryonic tissues in human [38,40]. Interestingly, in mouse cells [68,69], some variable escapees are clustered in domains, reminiscent of human escape gene organization and perhaps suggesting that more aspects of escape regulation are similar between species than were originally believed.
What distinguishes genes that escape XCI in a tissue or lineage-restricted fashion from other escapees? Most recently, these classes of escape genes have been designated ‘facultative’ and ‘constitutive’ escapees, respectively [69,70] (figure 1c). Data now are emerging that hint at mechanistic differences. Facultative and constitutive escapees are reported to differ not only in chromosome organization as indicated above [68], but also in their expression patterns at the onset of XCI and underlying ultrastructure [68–70]. These topics are discussed in more detail below.
(c). A unique epigenetic escape gene environment on the inactive X
Many groups have profiled the epigenetic landscape of the X in mouse and human to understand how genes that escape inactivation differ from those that are X inactivated. These efforts have not only identified novel escapees, as discussed above, but also aid in pinpointing potential regulatory sequences, as long-range regulatory sequences or escape gene domain boundaries could be epigenetically demarcated. Chromatin profiling indicates that, relative to X-inactivated genes, escape gene promoters are depleted in repressive chromatin marks such as H3K27me3 and are enriched in active modifications, e.g. H3K4me3 (recently reviewed in [33]). Similarly, escape genes can be characterized by promoter mCG hypomethylation and non-canonical promoter mCH hypermethylation and mCH gene-body methylation [38–40]. Additionally, DNAse hypersensitive sites and the related ATAC-seq accessible regions primarily map at human and mouse promoters and human introns of escapees [41,45,69]. It is feasible that chromatin marks, and even accessibility, may simply be a reflection of their transcriptionally active state. Nevertheless, based on these epigenetic signatures, several groups have concluded that escape is often regulated via promoter-proximal sequences [45,69]. Consistent with this idea, YY1 motifs and ChIP-seq peaks are overrepresented at human escapee start sites and are proposed to facilitate escape [36].
Most ATAC-seq accessible sites on the inactive X in mouse identify CTCF sites [69], adding to accumulating evidence that CTCF, a multifunctional protein with broad roles in gene expression and chromosome architecture [71], is an important player in escape gene regulation. CTCF involvement in XCI escape was initially proposed based on inactive X chromosome enrichment at the 5′ end of escapee Kdm5c [72]. Nevertheless, it has been difficult to envision how CTCF could play a role in escape gene regulation, as the vast majority (almost 90%) of the thousands of X-chromosome sites bind equally well to both Xs [73]. However, allele-specific analysis of CTCF binding reveals that some inactive X-specific CTCF sites cluster and colocalize with escape genes [44]. Inactive X CTCF binding was found not only at escape gene promoters, but also at boundaries between escape and X-inactivated genes, pointing to a role in inactive X compartmentalization. Moreover, in the case of a variable escape gene cluster, CTCF binding correlated with the size of the escape domain [44]. Altogether data are consistent with CTCF playing a key role in delimiting escape gene boundaries. It will be important to see if deletion of CTCF binding sites alters inactive X expression at escape gene transitions.
Chromosome three-dimensional structure and nuclear organization also differentiate escape genes and play an essential role in the XCI process. Xist promotes silencing by utilizing three-dimensional proximity to spread to distal sites [74,75]. XIST is excluded from escape genes [76], that are positioned outside of the Xist RNA-enriched silent compartment [77], although such positioning is neither necessary nor sufficient for a gene to escape XCI, at least in trophectoderm stem cells [45]. Allele-specific genome-wide chromosome conformation capture (Hi-C) has recently revealed that the inactive X is largely devoid of topologically associating domain (TAD) structure that characterizes the rest of the genome [78,79]. Instead, the inactive X in both mouse and human is divided into bipartite superdomains [78,79]. Within these domains, CTCF-mediated super loops form between Xist, multiple lncRNA genes, and the DXZ4 locus. Of these, Firre ‘anchors’ the X to the nucleolus [80], while DXZ4 centres the bipartite superdomain structure. Despite overall lack of apparent TAD structure on the inactive X, escape genes appear to be a striking exception, including human escapee clusters [68] and facultative escapee clusters in mouse [68,69]. Disruption of DXZ4 has a remarkable effect on structure of the inactive X, but surprisingly induces only modest changes in escape gene expression; in one of the NPC clones examined by Giorgetti et al. [69] the majority of facultative escape genes were silenced upon deletion (but not constitutive escape genes). Thus, the permissive epigenetic landscape for escape genes is becoming much better defined at a local and global X perspective. Roles for proteins like CTCF in escape gene regulation are being resolved; however, it is also becoming clear that all escapees may not be the same.
4. Beyond epigenetics: factors involved in escape from inactivation
While we have learned much about the landscape of the inactive X and the epigenetic features of the genes that escape inactivation, we still lack an understanding of how escapees avoid the silencing that engulfs the majority of the genes on the inactive X chromosome. The challenge is even greater to explain why some genes escape inactivation in only a subset of tissues, people or cells. Below we incorporate our current knowledge of escapees to evaluate potential contributors to escape from inactivation.
(a). Role for an intrinsic element in escape from inactivation
In aggregate, there is considerable circumstantial evidence that there are sequences that favour escape from inactivation. The constitutive escape genes seem able to avoid heterochromatic recruitment and inactivation, and approximately one-half of the mouse constitutive escapees are shared between mouse and human [28]. Definitive evidence for such an element was provided by Li and Carrel when they analysed random X-linked integrations of BACs (175–186 kb of DNA) containing the mouse escape gene Kdm5c in a female mouse ESC line. Four different single-copy integrations showed escape from inactivation for Kdm5c while flanking genes were silenced, delimiting a 112 kb region containing such an intrinsic element [81]. Further analysis of deletions of additional BAC integrations also showed evidence for a distal boundary that when disrupted allowed spread of euchromatin (escape) into flanking genes [82]. CTCF has long been implicated as a boundary [72], and has clear roles in the structure of the inactive X (discussed above) as well as generally defining TADs; however, CTCF itself is too abundant on the X chromosome to be the sole delineator of escape genes, thus other factors must interact. Interestingly, as mentioned above, Giorgetti et al. [69] saw loss of escape upon removal of DXZ4, suggesting that the topological structure limits the spread of silencing along the chromosome, at least for some escapees; whereas the Horvath et al. [82] study suggests elements limiting the spread of escape along the chromosome. Overall, there is strong evidence supporting a model whereby an intrinsic element prevents inactivation with boundaries limiting both the spread of escape and silencing, resulting in the expression from the inactive X being dependent on both local sequence and the structure of the inactive X.
(b). Distance seems to be a contributor to attenuation of spread
It is striking that human escape genes cluster on the X short arm, with the genes in PAR1 all escaping inactivation. However, the distance is also inter-connected with evolutionary age (as discussed above), complicating assessment of a distance effect. It has been seen that X material translocated beyond the Xp terminus can still be inactivated [83], suggesting that distance alone is not the sole determining factor for escape from XCI. Nevertheless, Marks et al. [68] identified distance from the Xic as correlating with escape from inactivation in mouse, as well as with timing of inactivation, as genes that inactivate latest tend to be located more distally. Further, a recent developmental analysis suggested the earliest silencing correlated not only with distance from the Xist locus, but most highly with proximity to Xist entry sites along the X [54]. While both mouse and human demonstrate a distance effect, more genes escape inactivation in humans, suggesting some additional factor that differs between the species. One considerable difference is the presence of the centromere between the human short and long arm, while the mouse X (like all mouse chromosomes) is acrocentric [84]. The Xist/XIST loci are mid-chromosome in both species, however, the lncRNA FIRRE locus and the macrosatellite DXZ4 that are also important contributors to the inactive X structure, are differentially positioned along the mouse and human X chromosome. The DXZ4 locus is central in mouse but two-thirds of the way along the X in humans, while the Firre/FIRRE locus is one-third of the way along the mouse X but more distally located on the human X. Whether these structural differences contribute to the greater number of human than mouse escape genes will need further exploration.
In unbalanced X;A translocations inactivation can spread into autosomal material in cis; however, this spread is more limited than seen on the X. While cell selection to limit dosage imbalance may bias ascertainment and partially explain this observation, Gartler & Riggs proposed the existence of ‘waystations’ or ‘booster sequences’ that were more abundant on the X chromosome and would amplify the inactivation signal [85]. Mary Lyon suggested LINE-1 (L1) repetitive elements as potential waystations [86]. Sequence comparison confirmed that L1 repeats are depleted at escape genes relative to levels elsewhere on the X chromosome [87–89]. Mouse studies have sought to clarify a role for L1s in XCI. Xist RNA is not targeted to L1 sequences or other repeats [74,90]; however, early silencing of L1 transcripts at the onset of XCI has been suggested to facilitate heterochromatin formation [91] and may explain why escape genes in L1 depleted regions are not recruited into the Xist RNA silent compartment [77]. A second proposed function for L1 elements is to aid silencing of escape prone genes via adjacent full-length L1s that are transcriptionally active during late stages of XCI establishment [91]. Whether novel methylation that has been recently reported at some of these full-length L1s is involved in this process will need to be addressed [92]. Nevertheless, despite abundant data on L1 enrichment and expression at the onset of XCI that is highly correlated with silenced loci, it is difficult to differentiate a functional role from evolutionary influences on sequence abundance. Furthermore, as waystations are defined functionally as elements supporting the spread of XCI, they may be conceptually expanded to include features that impact structure or three-dimensional nuclear ultrastructure.
Evidence for chromatin neighbourhoods that facilitate or attenuate the spread of XCI is also provided by recent genome-wide studies of X;A translocations that used DNA methylation as a proxy for spread of silencing to autosomal genes and again saw an enrichment of L1s in genes that became silenced [93,94]. The most enriched factor/feature correlating with spread of inactivation was EZH2/H3K27me3 [94]. These observations are also consistent with transgenic integrations of XIST where XIST-induced silencing is generally best closest to the integration, but is highly dependent on context including pre-existing H3K27me3 [95]. In addition, the patterns of genes that are subject to, or escape from, inactivation in X;A translocations are similar in independent translocations involving the same autosome [34], supporting the concept that there are sequences favouring silencing or escape from silencing. ALU enrichment was observed at regions that escape from XCI in the X;A translocations [93,94], as well as X escape regions [87–89]. Thus, while distance from the X-inactivation centre may contribute to escape from inactivation, the consistency and clustering of escape genes and ability for inactivation to spread long distances given the appropriate environment suggests that regions that escape from XCI are contextually different from those with X-inactivated genes at the sequence and/or chromosome ultrastructure level.
(c). Role for Xist and associated remodellers in escapee regulation remains poorly defined
The chromatin remodelling and silencing pathways recruited by Xist/XIST appear to function in a synergistic, yet independent, manner (e.g. [95,96]). Somatic deletion of Xist results in the structure of the inactive X becoming more similar to the active X, but only showed a minor elevation in reactivation frequency, with the majority of genes remaining silenced in the absence of Xist [75,97] or XIST [98,99]. Recent surveys [97,100,101] and screens [102–104] have expanded the list of proteins interacting with Xist and, thus, probably involved in the inactivation process (well reviewed in numerous articles, including [105]). Identification of these proteins provides insights into the mechanism of inactivation; however, their roles and pathways are still being dissected. Reduction of multiple factors in somatic cells can elevate the reactivation frequency of X-linked genes, particularly transgenic reporters (e.g. [97,104,106,107]), supporting the concept of co-operative pathways in maintenance of XCI. Many of the Xist-interacting proteins or chromatin remodelling complexes are embryonic lethal or abrogate XCI in mouse ESCs (e.g. [91–93]), and thus germ-line deletions have not been examined for impact on X silencing when absent at the onset of inactivation. The few players that have been examined in detail show distinct subsets of genes that fail to silence. In the absence of SETDB1-mediated H3K9me3, a subset of approximately 150 genes fail to silence [96]; while in the absence of SMCHD1, approximately 10% of genes fail to silence (although a larger group of genes lack DNA methylation) [108]. In neither case are the genes that fail to be silenced substantially enriched for variable escape genes, so to date there is not a clear correlation with escape from inactivation and loss of any particular feature of the inactive X. Studies of the frequency of variable escape in females have not identified females with significantly elevated escape from inactivation, supporting the idea that there is not a simple trans-acting factor limiting escape from inactivation [31,34].
Interestingly, the deletion of the DXZ4 region in mouse ESC resulted not only in structural re-organization in one of the derived NPC clones examined by Giorgetti et al. [69] but also loss of escape from inactivation for many of the facultative escape genes [69]. Deletion of DXZ4 in human fibroblasts resulted in restructuring of the X, but had limited impact on gene expression [109], and similarly, Firre knockdown in mouse fibroblasts does not lead to reactivation (although it disrupts H3K27me3 and perinucleolar targeting) [80]. Further studies will be required to determine if differences between these studies reflect differences between species, developmental timing or sporadic events; however, they point towards an emerging role for chromosomal ultrastructure in escape from XCI.
Overall, Xist interacts with multiple proteins enabling a series of synergistic silencing pathways. Disruption of Xist, or the proteins and lncRNAs interacting with Xist, in somatic cells increases the frequency of stochastic gene reactivation. Disruption of these pathways at the time of inactivation can abrogate silencing of the chromosome, or silencing of a subset of genes, but to date only the ultrastructure of the inactive X has been implicated in enabling certain genes to escape from X inactivation in some contexts.
(d). Escape may include some early developmental reactivation of silent genes
The term ‘escape’ from inactivation suggests that the genes are refractory to the initial silencing; however, most surveys that identify escape genes have assessed adult somatic cells, particularly in humans. Thus, escapees could include both genes that escape from silencing as well as genes that reactivate quite quickly following an initial inactivation. The outcome of rapid reactivation or complete avoidance of silencing is the same—expression from the inactive X and thus potential contribution to the phenotypes reviewed in §2. However, mechanistically it may be useful to differentiate between them in considering how escape occurs. The simplest interpretation of tissue-specific escape from inactivation would be that a gene is silenced and then reactivates in the tissue in question; however, an alternate scenario would involve very late silencing of some genes in some tissue precursors, and there appears to be evidence to support both scenarios. A transgene was seen to inactivate at different timepoints in different lineages; however, the endogenous genes examined seemed to inactivate earlier [110]. The facultative escapees analysed by Giorgetti et al. [69] are reported to be silenced and then reactivate [70], while the tissue-specific facultative escape genes observed by Marks et al. [68] were generally among the latest genes to inactivate, or not seen to be inactivated during the time points analysed during ES cell differentiation in vitro. Furthermore, in that latter study most genes escaping XCI in mouse showed little silencing early, although heterogeneity during XCI made it challenging to preclude some reactivation as had been reported previously for the Kdm5c gene [111]. The issue of heterogeneity during in vitro differentiation is an important one, and can be impacted by the method of differentiation, which often varies between studies. An additional challenge to these studies is that in addition to XCI to achieve dosage equivalence between males and females, there is upregulation of the single active X in order to achieve diploid dosage, which may impact genes differentially and also occurs early in development [112].
While assessing early events of X-linked gene dosage in mouse is challenging, taking those studies to humans presents both logistic and ethical challenges. However, a recent single-cell RNA-seq study of early human embryos managed to generate transcriptomes of 1529 individual cells from 88 human preimplantation embryos [64]. Surprisingly, they report dosage compensation in the presence of bi-allelic XIST expression [64], consistent with a previous report that initial events differ dramatically from mouse, and that XIST-dependent silencing may be a later developmental event [63]. Nevertheless, evaluation of escapees at the onset of human XCI has been hampered, as human ESCs generally retain an inactive X chromosome that erodes upon culturing, which has limited the study of early human XCI in vitro. However, recent work suggests that human naïve ESC can be isolated that model the earliest steps of human XCI [113], and thus it may be possible to determine whether human escapees are ever silenced.
Reactivation may also occur later in development, with potential phenotypic consequences. An important question is thus whether some genes are prone to (or even programmed for) later reactivation. Evidence that some genes may reactivate in humans comes from the observation in somatic cells that a gene (TRAPPC2) that is associated with poised polymerase despite being silenced, is observed to be expressed in additional clones from the same female cell line [114]. Similarly, in some clones, acetylation was detected at the promoter of the variable escapee TIMP1 even when silenced [52]. Drug treatment of mouse cells to induce reactivation shows that some genes seem to be more prone to reactivation [97]. Thus, some genes, in some individuals, may be prone to reactivation, and observed recurrently to reactivate. Early studies of select human genes failed to identify evidence of ageing-related reactivation [115], while in mouse the Otc gene reactivates with age [116]. As chromosome-wide analysis of XCI expands, more genes may be identified that are prone to reactivation with age. This late reactivation of individual genes or regions might contribute to the stochastic cellular heterogeneity observed in single-cell RNA-seq, genes with tissue-specific escape, and also to the increased frequency of escape genes in cultured cells. As mentioned above, reactivation is observed in some cancers [4], and a recent report also suggests a failure to maintain silencing in mature lymphocytes [5]. It is important to note that this discussion considers reactivation of single genes or regions. Full reactivation of the X chromosome is normally only observed developmentally. Overall, while some escape may reflect reactivation, the majority of escape genes seem to avoid the inactivation process rather than fail to maintain silencing; but this is a topic warranting further exploration and extremely challenging to study in humans.
In conclusion, we see multiple factors contributing to the ability of genes to escape from XCI. There is strong evidence for an intrinsic element that directs genes to escape from inactivation; however, the nature of this element remains to be determined. The domains of escape generated by these elements are limited by boundaries, which probably involve CTCF and additional regulators. CTCF is also involved in the formation of TADs across the genome, and the unique superdomain structure of the inactive X chromosome, both of which contribute to the ability of regions to escape from XCI. Whether there is a single element (such as LI repeats) that serves as the hypothesized way station is not yet clear, but additional sequences, perhaps including L1 repeats and PRC2 enrichment, contribute to the ability to spread silencing along the X chromosome. The correlation of escape with distance from the Xist/XIST gene may reflect decreasing density of way station sequences enabling the spread of XCI, but may also reflect ultrastructure of the inactive X or additional features. Expression of XIST/Xist recruits a cascade of chromatin remodellers, which are important for XCI; however, silencing of most X-linked genes is able to be maintained in the absence of many of these, including XIST/Xist itself but may increase the liability to reactivation. Overall, escape reflects an interplay between both genetic and epigenetic features, with the individual actors still shrouded by the challenge of separating causation from correlation.
5. Bringing an understanding of escape genes to human complex disease
As discussed at the beginning of this review, escapees can have impact on human health and disease, including cancer. Expression from the inactive X can offer protection against de novo and inherited X-linked mutations, but has also been proposed to contribute to the over-representation of females for some complex traits, such as the autoimmune disorders. As an example of the contributions of escape genes to disease, we will briefly examine potential roles for the inactive X chromosome in lupus.
(a). Systemic lupus erythematosus
Systemic lupus erythematosus (lupus) occurs with a frequency of greater than 1/1000, with over 90% of patients being female. Lupus has a substantial genetic contribution, with an estimated heritability as high as 66% [117] and a higher incidence among African Americans. Lupus-associated loci have been identified by genome-wide association studies, but explain less than 20% of heritability [118]. Without a better understanding of the aetiology of the disease, a significant proportion of patients, even some who receive optimal medical care, progress to permanent tissue damage including end-stage renal disease. Gender bias in lupus reflects both hormonal and sex chromosome differences between men and women. Multiple studies point to an X-chromosomal role in lupus (e.g. [119]). Using the four-core genotypes with transgenic mice predisposed to lupus reveals accelerated lupus in mice with two X chromosomes relative to females with only a single X chromosome [120]. In humans, a role for genes on the inactive X chromosome is supported by the observation that lupus prevalence is increased in 47,XXY males [119] and decreased in 45,X Turner Syndrome [121]. Even more dramatically, 47,XXX females show a threefold excess in lupus incidence over females in general [122]. Several X-linked genes are overexpressed and/or hypomethylated in women with lupus, including the immune-relevant toll-like receptor genes TLR7, TLR8 and CD40 ligand, CD40LG [123,124]. Strikingly, the CD40LG gene is a variable escape gene, and thus expression levels could vary between females as well as between males and females. The mouse Tlr8 candidate gene is suggested to escape from XCI in bone marrow-derived macrophages demonstrating a potential role for tissue-specific escape from XCI [125]. Furthermore, additional X chromosome genes are implicated, as variants mapping to at least four other loci on the human X are associated with lupus [119,126,127]. Recently, it was reported that female lymphoblast cell lines show some bi-allelic expression of CD40LG, CXCR3 and TLR7 by FISH, and that lymphoblasts from lupus patients often showed higher proportions of these bi-allelic cells, which thus have expression from both the active and inactive X chromosome [5]. In aggregate these studies suggest that variable escape from XCI—whether as a result of genetic variation or relaxed epigenetic control permitting reactivation—may contribute to SLE predisposition in females.
Overexpression of escapees may not be the only mechanism by which an inactive X contributes to autoimmune disorders. The inactive X has been proposed as a sink for heterochromatic silencing proteins [128,129], potentially altering the capacity for females to silence or maintain silencing in the presence of additional sequence variations or cellular stresses. Hypomethylation is a general feature of lupus, and other autoimmune disorders (reviewed in [130]). In addition, lupus-prone mouse models develop autoimmune responses against the envelope (gp70) protein of endogenous retroviruses, and one susceptibility locus maps to the macroH2A locus, a repressive protein strongly enriched on the inactive X [131]. It is also possible that the inactive X is ‘immunogenic’ under some conditions. Autoimmune sera containing antibodies against the Barr body have been identified, although at low frequency [132]. Antibodies to the Lamin B Receptor (LBR), an Xist-interacting protein [133], are seen with the female-biased, autoimmune disorder primary biliary cirrhosis [134], which interestingly often shows increased loss of the inactive X (or Y chromosome in rarer male cases). Mutations in LBR have been identified in Reynolds syndrome, which comprised the related, female-biased, autoimmune disorders scleroderma and primary biliary cirrhosis. Furthermore, females are generally mosaics of cells with the maternal or the paternal X active; however, skewed inactivation has been observed in autoimmune thyroid disorders such as Graves' disease [135]. It has been suggested that such skewing could be the cause, or consequence, of auto-reactivity within a female to cells expressing antigens from the alternate X chromosome.
There are so many different intersections of the inactive X chromosome and autoimmune disorders that it has been challenging to determine whether these are cause or consequence, or even chance. The associations, in particular, the involvement of genes that are variable in their escape from XCI, highlight the need to consider the inactive X chromosome as a potential contributor to disease, rather than a silent evolutionary oddity of sex determination.
6. Concluding thoughts
As outlined above, tremendous strides have been made recently in characterizing the escape landscape on both the human and mouse X chromosomes. The addition of single-cell RNA-seq data, as well as data from multiple tissues, strengthens the evidence for large numbers of genes that escape XCI, particularly on the human X, and has highlighted the profound variation for some escapees in only a subset of tissues, individuals or cells. While there are clear differences between studies (figure 1), overall in humans 65–75% of genes are subject to XCI; 10–15% escape and 10–25% show variable escape. Current data also continue to detect species differences in overall escape levels, as only 6% of mouse genes escape XCI and 4–14% are variable. Yet, as more samples are examined, the number of variable genes in both mouse and humans is on the rise. At a minimum, this suggests that the potential phenotypic impact from escapees is probably broader than once envisioned, but may also raise a cautionary flag that escape classification is in part sensitive to technical and statistical methods (figure 1), which may complicate efforts to identify common escapee features. Indeed, the distinction of constitutive and facultative escapees in mouse is already revealing differences at the ultrastructure level. Whether human escape genes are similarly parsed is unknown, but may become clearer as the underlying basis of variable escapees is better understood.
Mechanistically, the correlations between escape genes and the local epigenetic environment have been strengthened. Repetitive element profiles (low L1, high ALU), CTCF and additional factors such as YY1 associate with escapees. Current data strongly support the presence of local intrinsic elements that allow the constitutive escape genes to continue to be expressed. Yet, the nature of these elements or definitive evidence that any one particular feature is necessary or sufficient for a gene to escape XCI is still elusive. How, or even whether, ultrastructure intersects with these intrinsic elements also remains to be defined. Genetic differences between variable escape genes or refined mapping of escapee-containing transgenes have potential to identify these elements and will probably begin to unravel the complex interactions of sequence, local chromatin structure and three-dimensional chromosome ultrastructure. Identification of these escapee regulatory elements will not only bring key insight into XCI escape mechanisms but should have broader therapeutic potential, to either reactivate an otherwise silent gene copy in females, or to better abrogate silencing of gene therapy vectors. Fifty-five years after Lyon's initial proposition of escape, we still have much to learn about how inactivation silences 80% of X-linked human genes, and how the other 20% continue to make noise from the lyonized X chromosome.
Acknowledgements
C.J.B. thanks members of the lab as well as the Wasserman and Simpson groups for many enlightening discussions on XCI and escape. L.C. acknowledges members of her lab, D. Liu and N. Olsen for their insights on escape genes and their intersection with complex traits.
Data accessibility
This article has no additional data.
Competing interests
We declare we have no competing interests.
Funding
Funding for research in C.J.B.’s lab is provided by CIHR. L.C. acknowledges funding from the PA-DOH and PSU CTSI.
References
- 1.Lyon MF. 1961. Gene action in the X-chromosome of the mouse (Mus musculus L). Nature 190, 372–373. ( 10.1038/190372a0) [DOI] [PubMed] [Google Scholar]
- 2.Lyon MF. 1962. Sex chromatin and gene action in the mammalian X-chromosome. Am. J. Hum. Genet. 14, 135–148. [PMC free article] [PubMed] [Google Scholar]
- 3.Schulz EG, Heard E. 2013. Role and control of X chromosome dosage in mammalian development. Curr. Opin. Genet. Dev. 23, 109–115. ( 10.1016/j.gde.2013.01.008) [DOI] [PubMed] [Google Scholar]
- 4.Chaligne R, et al. 2015. The inactive X chromosome is epigenetically unstable and transcriptionally labile in breast cancer. Genome Res. 25, 488–503. ( 10.1101/gr.185926.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, Syrett CM, Kramer MC, Basu A, Atchison ML, Anguera MC. 2016. Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proc. Natl Acad. Sci. USA 113, E2029–E2038. ( 10.1073/pnas.1520113113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weng S, Stoner SA, Zhang DE. 2016. Sex chromosome loss and the pseudoautosomal region genes in hematological malignancies. Oncotarget 7, 72 356–72 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Machiela MJ, et al. 2016. Female chromosome X mosaicism is age-related and preferentially affects the inactivated X chromosome. Nat. Commun. 7, 11843 ( 10.1038/ncomms11843) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tukiainen T, et al. 2016. Landscape of X chromosome inactivation across human tissues. bioRxiv. ( 10.1101/073957) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walport LJ, Hopkinson RJ, Vollmar M, Madden SK, Gileadi C, Oppermann U. 2014. Human UTY(KDM6C) is a male-specific N-methyl lysyl demethylase. J. Biol. Chem. 289, 18 302–18 313. ( 10.1074/jbc.M114.555052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hook EB, Warburton D. 2014. Turner syndrome revisited: review of new data supports the hypothesis that all viable 45,X cases are cryptic mosaics with a rescue cell line, implying an origin by mitotic loss. Hum. Genet. 133, 417–424. ( 10.1007/s00439-014-1420-x) [DOI] [PubMed] [Google Scholar]
- 11.Bellott DW, et al. 2014. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508, 494–499. ( 10.1038/nature13206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stochholm K, Juul S, Gravholt CH. 2010. Mortality and incidence in women with 47,XXX and variants. Am. J. Med. Genet. A 152A, 367–372. ( 10.1002/ajmg.a.33214) [DOI] [PubMed] [Google Scholar]
- 13.Swerdlow AJ, Schoemaker MJ, Higgins CD, Wright AF, Jacobs PA. 2005. Mortality and cancer incidence in women with extra X chromosomes: a cohort study in Britain. Hum. Genet. 118, 255–260. ( 10.1007/s00439-005-0043-7) [DOI] [PubMed] [Google Scholar]
- 14.Swerdlow AJ, Schoemaker MJ, Higgins CD, Wright AF, Jacobs PA, Group UKCC. 2005. Cancer incidence and mortality in men with Klinefelter syndrome: a cohort study. J. Natl Cancer Inst. 97, 1204–1210. ( 10.1093/jnci/dji240) [DOI] [PubMed] [Google Scholar]
- 15.Wigby K, D'Epagnier C, Howell S, Reicks A, Wilson R, Cordeiro L, Tartaglia N. 2016. Expanding the phenotype of Triple X syndrome: A comparison of prenatal versus postnatal diagnosis. Am. J. Med. Genet. A. 170, 2870–2881. ( 10.1002/ajmg.a.37688) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Printzlau F, Wolstencroft J, Skuse DH. 2017. Cognitive, behavioral, and neural consequences of sex chromosome aneuploidy. J. Neurosci. Res. 95, 311–319. ( 10.1002/jnr.23951) [DOI] [PubMed] [Google Scholar]
- 17.Reijnders MR, et al. 2016. De Novo loss-of-function mutations in USP9X cause a female-specific recognizable syndrome with developmental delay and congenital malformations. Am. J. Hum. Genet. 98, 373–381. ( 10.1016/j.ajhg.2015.12.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snijders BL, et al. 2015. Mutations in DDX3X are a common cause of unexplained intellectual disability with gender-specific effects on Wnt signaling. Am. J. Hum. Genet. 97, 343–352. ( 10.1016/j.ajhg.2015.07.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldmann JM, et al. 2016. Parent-of-origin-specific signatures of de novo mutations. Nat. Genet. 48, 935–939. ( 10.1038/ng.3597) [DOI] [PubMed] [Google Scholar]
- 20.Grimm T, Meng G, Liechti-Gallati S, Bettecken T, Muller CR, Muller B. 1994. On the origin of deletions and point mutations in Duchenne muscular dystrophy: most deletions arise in oogenesis and most point mutations result from events in spermatogenesis. J. Med. Genet. 31, 183–186. ( 10.1136/jmg.31.3.183) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunford A, et al. 2017. Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nat. Genet. 49, 10–16. ( 10.1038/ng.3726) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van der Meulen J, et al. 2015. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood 125, 13–21. ( 10.1182/blood-2014-05-577270) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jager N, et al. 2013. Hypermutation of the inactive X chromosome is a frequent event in cancer. Cell 155, 567–581. ( 10.1016/j.cell.2013.09.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng F, Liu C, Lin CC, Zhao J, Jia P, Li WH, Zhao Z, Zhou XJ. 2015. A gene gravity model for the evolution of cancer genomes: a study of 3,000 cancer genomes across 9 cancer types. PLoS. Comput. Biol. 11, e1004497 ( 10.1371/journal.pcbi.1004497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnold AP. 2017. A general theory of sexual differentiation. J. Neurosci. Res. 95, 291–300. ( 10.1002/jnr.23884) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein SL, Flanagan KL. 2016. Sex differences in immune responses. Nat. Rev. Immunol. 16, 626–638. ( 10.1038/nri.2016.90) [DOI] [PubMed] [Google Scholar]
- 27.Thrasher BJ, Hong LK, Whitmire JK, Su MA. 2016. Epigenetic dysfunction in Turner syndrome immune cells. Curr. Allergy Asthma Rep. 16, 36 ( 10.1007/s11882-016-0612-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balaton BP, Cotton AM, Brown CJ. 2015. Derivation of consensus inactivation status for X-linked genes from genome-wide studies. Biol. Sex Differ. 6, 35 ( 10.1186/s13293-015-0053-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Migeon BR. 1983. Glucose-6-phosphate dehydrogenase as a probe for the study of X-chromosome inactivation in human females. Isozymes Curr. Top. Biol. Med. Res. 9, 189–200. [PubMed] [Google Scholar]
- 30.Mohandas T, Shapiro LJ, Sparkes RS, Sparkes MC. 1979. Regional assignment of the steroid sulfatase-X-linked ichthyosis locus: implications for a noninactivated region on the short arm of human X chromosome. Proc. Natl Acad. Sci. USA 76, 5779–5783. ( 10.1073/pnas.76.11.5779) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carrel L, Willard HF. 2005. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434, 400–404. ( 10.1038/nature03479) [DOI] [PubMed] [Google Scholar]
- 32.Hansen RS, Canfield TK, Stanek AM, Keitges EA, Gartler SM. 1998. Reactivation of XIST in normal fibroblasts and a somatic cell hybrid: abnormal localization of XIST RNA in hybrid cells. Proc. Natl Acad. Sci. USA 95, 5133–5138. ( 10.1073/pnas.95.9.5133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balaton BP, Brown CJ. 2016. Escape artists of the X chromosome. Trends Genet. 32, 348–359. ( 10.1016/j.tig.2016.03.007) [DOI] [PubMed] [Google Scholar]
- 34.Cotton AM, Ge B, Light N, Adoue V, Pastinen T, Brown CJ. 2013. Analysis of expressed SNPs identifies variable extents of expression from the human inactive X chromosome. Genome Biol. 14, R122 ( 10.1186/gb-2013-14-11-r122) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnston CM, Lovell FL, Leongamornlert DA, Stranger BE, Dermitzakis ET, Ross MT. 2008. Large-scale population study of human cell lines indicates that dosage compensation is virtually complete. PLoS Genet. 4, e9 ( 10.1371/journal.pgen.0040009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen CY, et al. 2016. YY1 binding association with sex-biased transcription revealed through X-linked transcript levels and allelic binding analyses. Sci. Rep. 6, 37324 ( 10.1038/srep37324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goto Y, Kimura H. 2009. Inactive X chromosome-specific histone H3 modifications and CpG hypomethylation flank a chromatin boundary between an X-inactivated and an escape gene. Nucleic Acids Res. 37, 7416–7428. ( 10.1093/nar/gkp860) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cotton AM, Price EM, Jones MJ, Balaton BP, Kobor MS, Brown CJ. 2015. Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum. Mol. Genet. 24, 1528–1539. ( 10.1093/hmg/ddu564) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lister R, et al. 2013. Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905 ( 10.1126/science.1237905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultz MD, et al. 2015. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 523, 212–216. ( 10.1038/nature14465) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qu K, Zaba LC, Giresi PG, Li R, Longmire M, Kim YH, Greenleaf WJ, Chang HY. 2015. Individuality and variation of personal regulomes in primary human T cells. Cell. Syst. 1, 51–61. ( 10.1016/j.cels.2015.06.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reinius B, Mold JE, Ramskold D, Deng Q, Johnsson P, Michaelsson J, Frisén J, Sandberg R. 2016. Analysis of allelic expression patterns in clonal somatic cells by single-cell RNA-seq. Nat. Genet. 48, 1430–1435. ( 10.1038/ng.3678) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beliveau BJ, et al. 2015. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat. Commun. 6, 7147 ( 10.1038/ncomms8147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berletch JB, Ma W, Yang F, Shendure J, Noble WS, Disteche CM, Deng X, Bartolomei MS. 2015. Escape from X inactivation varies in mouse tissues. PLoS Genet. 11, e1005079 ( 10.1371/journal.pgen.1005079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calabrese JM, et al. 2012. Site-specific silencing of regulatory elements as a mechanism of X inactivation. Cell 151, 951–963. ( 10.1016/j.cell.2012.10.037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhuiyan ZA, Momenah TS, Amin AS, Al-Khadra AS, Alders M, Wilde AA, Mannens MAM. 2008. An intronic mutation leading to incomplete skipping of exon-2 in KCNQ1 rescues hearing in Jervell and Lange-Nielsen syndrome. Prog. Biophys. Mol. Biol. 98, 319–327. ( 10.1016/j.pbiomolbio.2008.10.004) [DOI] [PubMed] [Google Scholar]
- 47.Peeters SB, Cotton AM, Brown CJ. 2014. Variable escape from X-chromosome inactivation: identifying factors that tip the scales towards expression. BioEssays 36, 746–756. ( 10.1002/bies.201400032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson WP, Price EM. 2015. The human placental methylome. Cold Spring Harb. Perspect. Med. 5, a023044 ( 10.1101/cshperspect.a023044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.The GTEx Consortium. 2015. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660. ( 10.1126/science.1262110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Al Nadaf S, Deakin JE, Gilbert C, Robinson TJ, Graves JA, Waters PD. 2012. A cross-species comparison of escape from X inactivation in Eutheria: implications for evolution of X chromosome inactivation. Chromosoma 121, 71–78. ( 10.1007/s00412-011-0343-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raj A, van Oudenaarden A. 2008. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135, 216–226. ( 10.1016/j.cell.2008.09.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anderson CL, Brown CJ. 2005. Epigenetic predisposition to expression of TIMP1 from the human inactive X chromosome. BMC Genet. 6, 48 ( 10.1186/1471-2156-6-48) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moen EL, Litwin E, Arnovitz S, Zhang X, Zhang W, Dolan ME, Godley LA. 2015. Characterization of CpG sites that escape methylation on the inactive human X-chromosome. Epigenetics 10, 810–818. ( 10.1080/15592294.2015.1069461) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Borensztein M, et al. 2017. Xist-dependent imprinted X inactivation and the early developmental consequences of its failure. Nat. Struct. Mol. Biol. 24, 226–233. ( 10.1038/nsmb.3365) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kilpinen H, Dermitzakis ET. 2012. Genetic and epigenetic contribution to complex traits. Hum. Mol. Genet. 21, R24–R28. ( 10.1093/hmg/dds383) [DOI] [PubMed] [Google Scholar]
- 56.Ross MT, et al. 2005. The DNA sequence of the human X chromosome. Nature 434, 325–337. ( 10.1038/nature03440) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shpargel KB, Sengoku T, Yokoyama S, Magnuson T. 2012. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 8, e1002964 ( 10.1371/journal.pgen.1002964) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vakilian H, et al. 2015. DDX3Y, a male-specific region of Y chromosome gene, may modulate neuronal differentiation. J. Proteome Res. 14, 3474–3483. ( 10.1021/acs.jproteome.5b00512) [DOI] [PubMed] [Google Scholar]
- 59.Jegalian K, Page DC. 1998. A proposed path by which genes common to mammalian X and Y chromosomes evolve to become X inactivated. Nature 394, 776–780. ( 10.1038/29522) [DOI] [PubMed] [Google Scholar]
- 60.Park C, Carrel L, Makova KD. 2010. Strong purifying selection at genes escaping X chromosome inactivation. Mol. Biol. Evol. 27, 2446–2450. ( 10.1093/molbev/msq143) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Slavney A, Arbiza L, Clark AG, Keinan A. 2016. Strong constraint on human genes escaping X-inactivation is modulated by their expression level and breadth in both sexes. Mol. Biol. Evol. 33, 384–393. ( 10.1093/molbev/msv225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Migeon BR. 2016. An overview of X inactivation based on species differences. Semin. Cell. Dev. Biol. 56, 111–116. ( 10.1016/j.semcdb.2016.01.024) [DOI] [PubMed] [Google Scholar]
- 63.Okamoto I, et al. 2011. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 472, 370–374. ( 10.1038/nature09872) [DOI] [PubMed] [Google Scholar]
- 64.Petropoulos S, et al. 2016. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 165, 1012–1026. ( 10.1016/j.cell.2016.03.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu H, et al. 2014. Cellular resolution maps of X chromosome inactivation: implications for neural development, function, and disease. Neuron 81, 103–119. ( 10.1016/j.neuron.2013.10.051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lopes AM, Arnold-Croop SE, Amorim A, Carrel L. 2011. Clustered transcripts that escape X inactivation at mouse XqD. Mamm. Genome 22, 572–582. ( 10.1007/s00335-011-9350-6) [DOI] [PubMed] [Google Scholar]
- 67.Reinius B, et al. 2010. Female-biased expression of long non-coding RNAs in domains that escape X-inactivation in mouse. BMC Genomics 11, 614 ( 10.1186/1471-2164-11-614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marks H, et al. 2015. Dynamics of gene silencing during X inactivation using allele-specific RNA-seq. Genome Biol. 16, 149 ( 10.1186/s13059-015-0698-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Giorgetti L, et al. 2016. Structural organization of the inactive X chromosome in the mouse. Nature 535, 575–579. ( 10.1038/nature18589) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patrat C, Okamoto I, Diabangouaya P, Vialon V, Le Baccon P, Chow J, Heard E. 2009. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc. Natl Acad. Sci. USA 106, 5198–5203. ( 10.1073/pnas.0810683106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ong CT, Corces VG. 2014. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 15, 234–246. ( 10.1038/nrg3663) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Filippova GN, Cheng MK, Moore JM, Truong JP, Hu YJ, Nguyen DK, Tsuchiya KD, Disteche CM. 2005. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev. Cell. 8, 31–42. ( 10.1016/j.devcel.2004.10.018) [DOI] [PubMed] [Google Scholar]
- 73.Ding Z, et al. 2014. Quantitative genetics of CTCF binding reveal local sequence effects and different modes of X-chromosome association. PLoS Genet. 10, e1004798 ( 10.1371/journal.pgen.1004798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Engreitz JM, et al. 2013. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341, 1237973 ( 10.1126/science.1237973) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Splinter E, et al. 2011. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 25, 1371–1383. ( 10.1101/gad.633311) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Murakami K, Ohhira T, Oshiro E, Qi D, Oshimura M, Kugoh H. 2009. Identification of the chromatin regions coated by non-coding Xist RNA. Cytogenet. Genome Res. 125, 19–25. ( 10.1159/000207514) [DOI] [PubMed] [Google Scholar]
- 77.Chaumeil J, Le Baccon P, Wutz A, Heard E. 2006. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 20, 2223–2237. ( 10.1101/gad.380906) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rao SS, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680. ( 10.1016/j.cell.2014.11.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deng X, et al. 2015. Bipartite structure of the inactive mouse X chromosome. Genome Biol. 16, 152 ( 10.1186/s13059-015-0728-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang F, et al. 2015. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 16, 52 ( 10.1186/s13059-015-0618-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li N, Carrel L. 2008. Escape from X chromosome inactivation is an intrinsic property of the Jarid1c locus. Proc. Natl Acad. Sci. USA 105, 17 055–17 060. ( 10.1073/pnas.0807765105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Horvath LM, Li N, Carrel L. 2013. Deletion of an X-inactivation boundary disrupts adjacent gene silencing. PLoS Genet. 9, e1003952 ( 10.1371/journal.pgen.1003952) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Becroft DM, Costello JM, Shaw RL. 1977. 46, X,X-X terminal rearrangement/45, X mosaicism in a child with short stature. Clin. Genet. 11, 122–127. ( 10.1111/j.1399-0004.1977.tb01289.x) [DOI] [PubMed] [Google Scholar]
- 84.Disteche CM. 1999. Escapees on the X chromosome. Proc. Natl Acad. Sci. USA 96, 14 180–14 182. ( 10.1073/pnas.96.25.14180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gartler SM, Riggs AD. 1983. Mammalian X-chromosome inactivation. Annu. Rev. Genet. 17, 155–190. ( 10.1146/annurev.ge.17.120183.001103) [DOI] [PubMed] [Google Scholar]
- 86.Lyon MF. 1998. X-chromosome inactivation: a repeat hypothesis. Cytogenet. Cell. Genet. 80, 133–137. ( 10.1159/000014969) [DOI] [PubMed] [Google Scholar]
- 87.Bailey JA, Carrel L, Chakravarti A, Eichler EE. 2000. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivati. Proc. Natl Acad. Sci. USA 97, 6634–6639. ( 10.1073/pnas.97.12.6634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carrel L, Park C, Tyekucheva S, Dunn J, Chiaromonte F, Makova KD. 2006. Genomic environment predicts expression patterns on the human inactive X chromosome. PLoS Genet. 2, e151 ( 10.1371/journal.pgen.0020151) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang Z, Willard HF, Mukherjee S, Furey TS. 2006. Evidence of influence of genomic DNA sequence on human X chromosome inactivation. PLoS Comput. Biol. 2, e113 ( 10.1371/journal.pcbi.0020113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Simon MD, et al. 2013. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 504, 465–469. ( 10.1038/nature12719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chow JC, et al. 2010. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell 141, 956–969. ( 10.1016/j.cell.2010.04.042) [DOI] [PubMed] [Google Scholar]
- 92.Wu TP, et al. 2016. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature 532, 329–333. ( 10.1038/nature17640) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bala TN, Brahmachary M, Garg P, Borel C, Alnefaie R, Watson CT, Thomas NS, Sharp AJ. 2014. DNA methylation profiling in X;autosome translocations supports a role for L1 repeats in the spread of X chromosome inactivation. Hum. Mol. Genet. 23, 1224–1236. ( 10.1093/hmg/ddt553) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cotton AM, Chen CY, Lam LL, Wasserman WW, Kobor MS, Brown CJ. 2014. Spread of X-chromosome inactivation into autosomal sequences: role for DNA elements, chromatin features and chromosomal domains. Hum. Mol. Genet. 23, 1211–1223. ( 10.1093/hmg/ddt513) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kelsey AD, et al. 2015. Impact of flanking chromosomal sequences on localization and silencing by the human non-coding RNA XIST. Genome Biol. 16, 208 ( 10.1186/s13059-015-0774-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Minkovsky A, Barakat TS, Sellami N, Chin MH, Gunhanlar N, Gribnau J, Plath K. 2013. The pluripotency factor-bound intron 1 of Xist is dispensable for X chromosome inactivation and reactivation in vitro and in vivo. Cell Rep. 3, 905–918. ( 10.1016/j.celrep.2013.02.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Minajigi A, et al. 2015. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 349, aab2276 ( 10.1126/science.aab2276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rack KA, et al. 1994. Absence of the XIST gene from late-replicating isodicentric X chromosomes in leukaemia. Hum. Mol. Genet. 3, 1053–1059. ( 10.1093/hmg/3.7.1053) [DOI] [PubMed] [Google Scholar]
- 99.Brown CJ, Willard HF. 1994. The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature 368, 154–156. ( 10.1038/368154a0) [DOI] [PubMed] [Google Scholar]
- 100.Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. 2015. Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416. ( 10.1016/j.cell.2015.03.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McHugh CA, et al. 2015. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236. ( 10.1038/nature14443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Monfort A, Di Minin G, Postlmayr A, Freimann R, Arieti F, Thore S, Wutz A. 2015. Identification of Spen as a crucial factor for Xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep. 12, 554–561. ( 10.1016/j.celrep.2015.06.067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Moindrot B, Cerase A, Coker H, Masui O, Grijzenhout A, Pintacuda G, Schermelleh L, Nesterova TB, Brockdorff N. 2015. A pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for Xist RNA-mediated silencing. Cell Rep. 12, 562–572. ( 10.1016/j.celrep.2015.06.053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lessing D, et al. 2016. A high-throughput small molecule screen identifies synergism between DNA methylation and Aurora kinase pathways for X reactivation. Proc. Natl Acad. Sci. USA 113, 14 366–14 371. ( 10.1073/pnas.1617597113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mira-Bontenbal H, Gribnau J. 2016. New Xist-interacting proteins in X-chromosome inactivation. Curr. Biol. 26, R338–R342. ( 10.1016/j.cub.2016.03.022) [DOI] [PubMed] [Google Scholar]
- 106.Csankovszki G, Nagy A, Jaenisch R. 2001. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J. Cell. Biol. 153, 773–784. ( 10.1083/jcb.153.4.773) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sripathy S, et al. 2017. Screen for reactivation of MeCP2 on the inactive X chromosome identifies the BMP/TGF-β superfamily as a regulator of XIST expression. Proc. Natl Acad. Sci. USA 114, 1619–1624. ( 10.1073/pnas.1621356114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gendrel AV, Tang YA, Suzuki M, Godwin J, Nesterova TB, Greally JM, Heard E, Brockdorff N. 2013. Epigenetic functions of Smchd1 repress gene clusters on the inactive X chromosome and on autosomes. Mol. Cell. Biol. 33, 3150–3165. ( 10.1128/MCB.00145-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Darrow EM, et al. 2016. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc. Natl Acad. Sci. USA 113, E4504–E4512. ( 10.1073/pnas.1609643113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tan SS, Williams EA, Tam PP. 1993. X-chromosome inactivation occurs at different times in different tissues of the post-implantation mouse embryo. Nat. Genet. 3, 170–174. ( 10.1038/ng0293-170) [DOI] [PubMed] [Google Scholar]
- 111.Lingenfelter PA, Adler DA, Poslinski D, Thomas S, Elliott RW, Chapman VM, Disteche CM. 1998. Escape from X inactivation of Smcx is preceded by silencing during mouse development. Nat. Genet. 18, 212–213. ( 10.1038/ng0398-212) [DOI] [PubMed] [Google Scholar]
- 112.Deng X, Berletch JB, Ma W, Nguyen DK, Hiatt JB, Noble WS, Shendure J, Disteche CM. 2013. Mammalian X upregulation is associated with enhanced transcription initiation, RNA half-life, and MOF-mediated H4K16 acetylation. Dev. Cell 25, 55–68. ( 10.1016/j.devcel.2013.01.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sahakyan A, et al. 2017. Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell. 20, 87–101. ( 10.1016/j.stem.2016.10.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kucera KS, Reddy TE, Pauli F, Gertz J, Logan JE, Myers RM, Willard HF. 2011. Allele-specific distribution of RNA polymerase II on female X chromosomes. Hum. Mol. Genet. 20, 3964–3973. ( 10.1093/hmg/ddr315) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Migeon BR, Axelman J, Beggs AH. 1988. Effect of ageing on reactivation of the human X-linked HPRT locus. Nature 335, 93–96. ( 10.1038/335093a0) [DOI] [PubMed] [Google Scholar]
- 116.Wareham KA, Lyon MF, Glenister PH, Williams ED. 1987. Age related reactivation of an X-linked gene. Nature 327, 725–727. ( 10.1038/327725a0) [DOI] [PubMed] [Google Scholar]
- 117.Guerra SG, Vyse TJ, Cunninghame Graham DS. 2012. The genetics of lupus: a functional perspective. Arthritis Res. Ther. 14, 211 ( 10.1186/ar3844) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sun C, et al. 2016. High-density genotyping of immune-related loci identifies new SLE risk variants in individuals with Asian ancestry. Nat. Genet. 48, 323–330. ( 10.1038/ng.3496) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Webb R, et al. 2009. Variants within MECP2, a key transcription regulator, are associated with increased susceptibility to lupus and differential gene expression in patients with systemic lupus erythematosus. Arthritis Rheum. 60, 1076–1084. ( 10.1002/art.24360) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sasidhar MV, Itoh N, Gold SM, Lawson GW, Voskuhl RR. 2012. The XX sex chromosome complement in mice is associated with increased spontaneous lupus compared with XY. Ann. Rheum Dis. 71, 1418–1422. ( 10.1136/annrheumdis-2011-201246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang L, et al. 2009. Somatic single hits inactivate the X-linked tumor suppressor FOXP3 in the prostate. Cancer Cell. 16, 336–346. ( 10.1016/j.ccr.2009.08.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu K, et al. 2016. X chromosome dose and sex bias in autoimmune diseases: increased prevalence of 47,XXX in systemic lupus erythematosus and Sjogren's syndrome. Arthritis Rheumatol. 68, 1290–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hewagama A, et al. 2013. Overexpression of X-linked genes in T cells from women with lupus. J. Autoimmun. 41, 60–71. ( 10.1016/j.jaut.2012.12.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B. 2007. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J. Immunol. 179, 6352–6358. ( 10.4049/jimmunol.179.9.6352) [DOI] [PubMed] [Google Scholar]
- 125.McDonald G, et al. 2015. Female bias in systemic lupus erythematosus is associated with the differential expression of X-linked toll-like receptor 8. Front. Immunol. 6, 457 ( 10.3389/fimmu.2015.00457) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang Y, et al. 2015. Meta-analysis of GWAS on two Chinese populations followed by replication identifies novel genetic variants on the X chromosome associated with systemic lupus erythematosus. Hum. Mol. Genet. 24, 274–284. ( 10.1093/hmg/ddu429) [DOI] [PubMed] [Google Scholar]
- 127.D'Amico F, et al. 2016. Association between rs2294020 in X-linked CCDC22 and susceptibility to autoimmune diseases with focus on systemic lupus erythematosus. Immunol. Lett. 181, 58–62. ( 10.1016/j.imlet.2016.11.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Blewitt ME, Vickaryous NK, Hemley SJ, Ashe A, Bruxner TJ, Preis JI, Arkell R, Whitelaw E. 2005. An N-ethyl-N-nitrosourea screen for genes involved in variegation in the mouse. Proc. Natl Acad. Sci. USA 102, 7629–7634. ( 10.1073/pnas.0409375102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Juriloff DM, Harris MJ. 2012. Hypothesis: the female excess in cranial neural tube defects reflects an epigenetic drag of the inactivating X chromosome on the molecular mechanisms of neural fold elevation. Birth Defects Res. A Clin. Mol. Teratol. 94, 849–855. ( 10.1002/bdra.23036) [DOI] [PubMed] [Google Scholar]
- 130.Wu H, Zhao M, Tan L, Lu Q. 2016. The key culprit in the pathogenesis of systemic lupus erythematosus: aberrant DNA methylation. Autoimmun. Rev. 15, 684–689. ( 10.1016/j.autrev.2016.03.002) [DOI] [PubMed] [Google Scholar]
- 131.Baudino L, Changolkar LN, Pehrson JR, Izui S. 2010. The Sgp3 locus derived from the 129 strain is responsible for enhanced endogenous retroviral expression in macroH2A1-deficient mice. J. Autoimmun. 35, 398–403. ( 10.1016/j.jaut.2010.08.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hong B, Reeves P, Panning B, Swanson MS, Yang TP. 2001. Identification of an autoimmune serum containing antibodies against the Barr body. Proc. Natl Acad. Sci. USA 98, 8703–8708. ( 10.1073/pnas.151259598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen CK, et al. 2016. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354, 468–472. ( 10.1126/science.aae0047) [DOI] [PubMed] [Google Scholar]
- 134.Worman HJ. 2007. Nuclear envelope protein autoantigens in primary biliary cirrhosis. Hepatol. Res. 37(Suppl. 3), S406–S411. ( 10.1111/j.1872-034X.2007.00227.x) [DOI] [PubMed] [Google Scholar]
- 135.Simmonds MJ, Kavvoura FK, Brand OJ, Newby PR, Jackson LE, Hargreaves CE, Franklyn JA, Gough SCL. 2014. Skewed X chromosome inactivation and female preponderance in autoimmune thyroid disease: an association study and meta-analysis. J. Clin. Endocrinol. Metab. 99, E127–E131. ( 10.1210/jc.2013-2667) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.