Abstract
Tyrosine receptor kinase B (TrkB), a receptor for brain-derived neurotrophic factor and neurotrophin-3, includes the alternatively spliced three isoforms specifically in rats. Each isoform, full length TrkB (TrkB FL), truncated TrkB type-1 (TrkB T1) and type-2 (TrkB T2), seems to mediate diverse cellular function. Some studies suggest that TrkB plays a key role in both neural and non-neural systems. In the present study, we examined mRNA and protein expression profile of three TrkB isoforms in normal adult rat multiple tissues. TrkB FL mRNA and protein were both highly expressed exclusively in brain. While TrkB T1 mRNA was highly expressed exclusively in brain, glycosylated TrkB T1 protein was expressed in brain and heart. TrkB T2 mRNA level in brain was the highest. In brain, TrkB FL mRNA expression was higher in cerebral cortex, but lower in brainstem. TrkB T1 mRNA expression was higher in hypothalamus, but lower in cerebellum. TrkB T2 mRNA expression was higher in cerebral cortex and cerebellum, but lower in brainstem. The present study for the first time clarified diverse distribution of three TrkB isoforms in rat multiple tissues and could serve as a useful resource for understanding the physiology and pathophysiology of mammals including human via comparing the expression pattern of TrkB isoforms.
Keywords: brain, gene expression, protein synthesis, rat, receptor
Tyrosine receptor kinase B (TrkB) is a receptor for brain-derived neurotrophic factor (BDNF) and neurotrophin-3 [32]. Three TrkB isoforms, full length TrkB (TrkB FL), truncated TrkB type-1 (TrkB T1) and type-2 (TrkB T2), are alternatively spliced and expressed specifically in rats. All the isoforms possess the same extracellular domain, while they possess each different intracellular domain [24]. TrkB FL mediates the activation of ERK and Akt signaling pathway in a ligand-dependent manner, which affects the neural development, growth, differentiation and survival [13, 16, 29]. TrkB T1 not only exhibits dominant-negative effect for TrkB FL, but also mediates the intracellular Ca2+ mobilization, suppression of Rho-GDPase inhibitor activity and activation of phospholipase C in glial cells [8, 12]. TrkB T2 possibly modifies the intracellular signaling activated through BDNF stimulation [3]. Hence, TrkB isoforms differently regulate the intracellular signal transduction.
Further, TrkB isoforms seem to play roles in various tissues other than neural system. It was reported that TrkB FL mediates the tumor development via regulating metastasis, chemoresistance, angiogenesis and anoikis [4, 11, 30]. In addition, TrkB is expressed in human atherosclerotic lesion [18]. TrkB seems to be necessary for angiogenesis during the development of heart [35]. BDNF-TrkB T1 axis mediates contraction via intracellular Ca2+ increase in mouse hearts [10]. These results suggest that TrkB isoforms could exert various functions in multiple tissues.
Previously, mRNA expression level of TrkB FL and T1 in rats, which are used as a model for understanding the human physiology and pathophysiology, was examined by performing RNA-seq analysis [37]. However, expression level of TrkB T2 mRNA and TrkB isoforms protein in rat multiple tissues still remains to be unknown. In the present study, we aimed to clarify them.
MATERIALS AND METHODS
Animal and tissue harvest
Animal care and procedures were conducted in conformity with the institutional guideline of School of Veterinary Medicine, the Kitasato University. An animal study was approved by the ethical committee of School of Veterinary Medicine, the Kitasato University. Male Wistar rats (5-week-old) (CLEA Japan, Tokyo, Japan) were maintained in a constant temperature and humidity room (22 ± 2°C, 50–60%, 12 hr for lighting). They can freely access the food (CE2, CLEA Japan) and tap water. After the rats (7-week-old) were anesthetized with urethane (1.5 g/kg, i.p.) and euthanized by inferior vena cava exsanguination, tissues were isolated and dissected in approximately 5 mm cubes. The isolated tissues were as follows: testis, coagulating gland, pancreas, spleen, stomach, jejunum, colon, liver, adrenal, kidney, thymus, lung, heart, aorta, thyroid, eye, skeletal muscle and whole brain. In addition, whole brain was separated into 5 regions (cerebral cortex, cerebellum, brainstem, hypothalamus and hippocampus). Then, the tissues were mashed by using Cell Destroyer (Prosense Inc., Tokyo, Japan) for extraction of total RNA and protein.
Materials
The primary antibody sources were as follows: TrkB FL (CSB-PA020174, Flarebio, College Park, MD, U.S.A.) and TrkB T1 (sc-119, Santa Cruz Biotech, Santa Cruz, CA, U.S.A.). Anti-TrkB FL antibody (CSB-PA020174) was produced by immunizing rabbit with a synthetic peptide surrounding Tyr516 in intracellular domain of TrkB FL (NCBI Reference Sequence: NP_036863.1). Thus, the antibody only reacts with TrkB FL (Fig. 2). Anti-TrkB T1 antibody (sc-119) was produced by immunizing rabbit with a synthetic peptide locating at the C-terminus of TrkB T1 (NCBI Reference Sequence: NP_001156640.1). Thus, the antibody only reacts with TrkB T1 (Fig. 4). The secondary antibody was anti-rabbit IgG Horseradish Peroxidase (HRP)-linked Antibody (#7074, Cell Signaling Technology, Beverly, MA, U.S.A.).
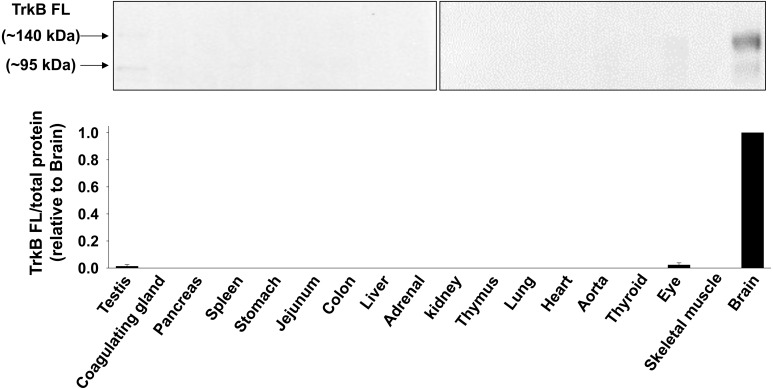
Fig. 2.
Expression level of TrkB FL protein in rat tissues. Total protein was extracted from homogenized tissues of Wistar rats. Expression of TrkB FL protein was determined by Western blotting (n=3−4) using anti-TrkB FL antibody (CSB-PA020174, Flarebio). Equal loading of protein was confirmed by Ponceau-S staining. The data normalized to total protein were shown relative to brain in bar graph. Data were expressed as mean ± SE.
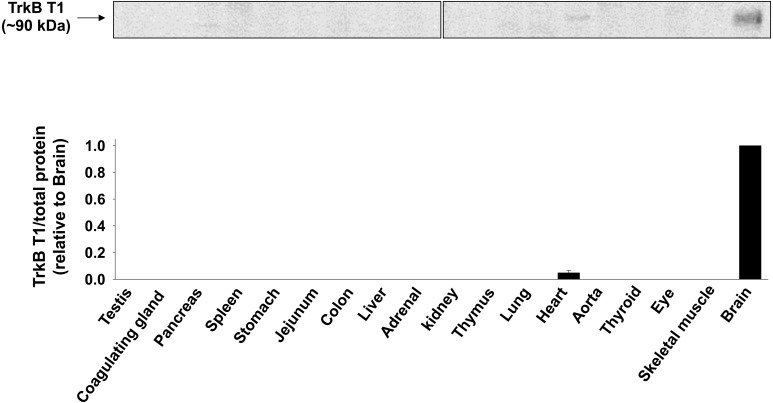
Fig. 4.
Expression level of TrkB T1 protein in rat tissues. Total protein was extracted from homogenized tissues of Wistar rats. Expression of TrkB T1 protein was determined by Western blotting (n=3−4) using anti-TrkB T1 antibody (sc-119, Santa Cruz Biotech). Equal loading of protein was confirmed by Ponceau-S staining. The data normalized to total protein were shown relative to brain in bar graph. Data were expressed as mean ± SE.
RT-PCR
RT-PCR was performed as previously described [26, 28]. Total RNA was extracted from mashed tissues using a TRI Reagent (Molecular Research Center Inc., Cincinnati, OH, U.S.A.). The first strand cDNA was synthesized from 250 ng of total RNA by using the ReverTra Ace qPCR master mix (TOYOBO, Osaka, Japan) at 65°C for 5 min, 37°C for 15 min and 98°C for 5 min. The PCR amplification was performed using Quick Taq HS DyeMix (TOYOBO). After the initial activation at 94°C for 1 min, the 33 (for TrkB FL, T1 and T2)- or 18 (for 18s ribosomal RNA)-cycles of amplifications at 94°C for 30 sec, 58°C for 30 sec and 68°C for 30 sec were done. After the PCR products were electrophoresed in agarose gel containing ethidium bromide (final concentration, 50 µg/ml) at 100 V for 35 min, they were visualized and analyzed by using the ATTO light capture system and CS analyzer 3.0 software (ATTO, Tokyo, Japan). The primer sequences were described in Supplemental Table.
Quantitative RT-PCR (qRT-PCR) analysis
qRT-PCR was performed as previously described [26]. After cDNA was synthesized as described above, PCR amplification was performed using THUNDERBIRD qPCR Mix (TOYOBO) with the pair of gene-specific primers (Supplemental Table). Real-time analysis was performed using PikoReal 96 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, U.S.A.). After initial activation at 95°C for 1 min, 40 cycles of amplifications at 95°C for 1 min and 60°C for 30 sec were done. Melting curve was analyzed from 60 to 90°C. We determined the quantity of cycles at fixed signal intensity (Cq) as 2-ΔCq value relative to 18s ribosomal RNA or glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Western blotting
Western blotting was performed as described previously [27, 28, 33, 34]. Protein lysates were obtained from mashed rat tissues using Cell Lysis Buffer (Cell Signaling Technology) containing 20 mM Tris-HCl (pH7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton-X, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4 and 1 µg/ml leupeptin supplemented with 0.1% protease inhibitor mixture (Nacalai Tesque, Kyoto, Japan) on ice. Protein concentration was determined by using a bicinchoninic acid method (Pierce, Rockford, IL, U.S.A.). After equal amount of protein (10 µg) was separated by SDS-PAGE (7.5%) at 80–120 V for 1.5 hr, they were transferred to a nitrocellulose membrane (Pall Corporation, Ann Arbor, MI, U.S.A.) at 400 mA for 1.5 hr. For confirming the equal loading of protein, the membranes were stained with 0.1% Ponceau-S/5% acetic acid at room temperature for 5 min and washed three times with 1% acetic acid at room temperature for 5 min. The Ponceau-S stained membranes were scanned in visible light by using the ATTO light capture system. Total density of all the visible bands in each lane was measured as the amount of total protein using CS analyzer 3.0 software (Supplemental Fig. 1). After being blocked with 0.5% skim milk, the membranes were incubated with a primary antibody [1:500 dilution in Tris-buffered saline with Tween 20 (TBS-T)] at 4°C overnight. They were detected by using HRP-conjugated secondary antibody (1:10,000 dilution in TBS-T, 1 hr at room temperature) and the EZ-ECL system (Biological Industries, Kibbutz Beit Hesmek, Israel). The results were analyzed using CS analyzer 3.0 software.
RESULTS
Expression level of TrkB FL in rat tissues
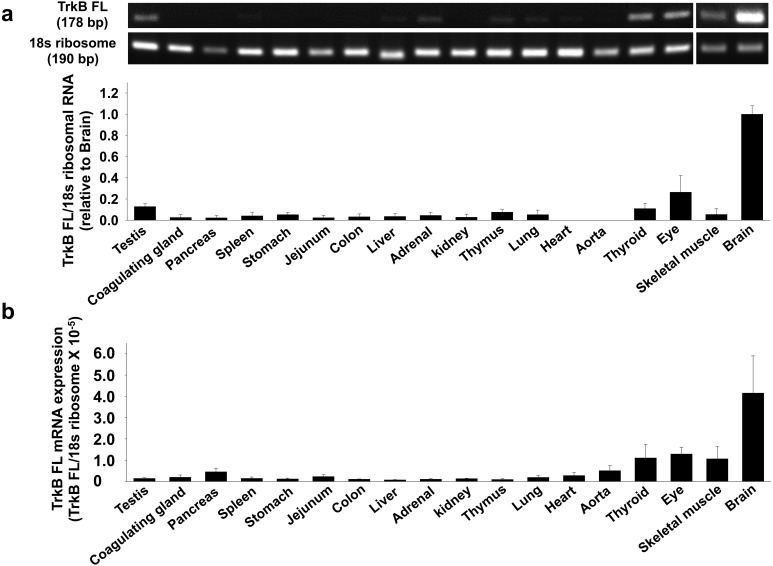
We first examined the expression level of TrkB FL mRNA in rat tissues by RT-PCR and qRT-PCR. TrkB FL mRNA was highly expressed in brain. In other tissues, the expression was much lower compared with brain (Fig. 1a and 1b, n=3–4). We further examined the protein expression level of TrkB FL in rat tissues by Western blotting. The electrophoresed mobility of glycosylated TrkB FL differs from non-glycosylated TrkB FL in SDS-PAGE [15]. Thus, we identified an approximately 95 kDa band as non-glycosylated TrkB FL and a 140 kDa band as glycosylated TrkB FL. The TrkB FL protein was highly expressed in brain, while the TrkB FL protein was barely detected in other tissues, except for testis and eye (Fig. 2, n=3–4), generally reflecting the mRNA expression level of TrkB FL in rat.
Fig. 1.
Expression level of full length tyrosine receptor kinase B (TrkB FL) mRNA in rat tissues. Total RNA was extracted from homogenized tissues of Wistar rats. a) Expression of TrkB FL mRNA was measured by RT-PCR using the gene specific primers to rat TrkB FL (n=3−4). The data normalized to 18s ribosomal RNA were shown as fold increase relative to brain in bar graph. Data were expressed as mean ± SE. b) Expression of TrkB FL mRNA was measured by quantitative RT-PCR (qRT-PCR) using the gene specific primers to rat TrkB FL (n=3). The relative mRNA level to rat 18s ribosomal RNA was calculated using 2-ΔCq values.
Expression level of TrkB T1 in rat tissues
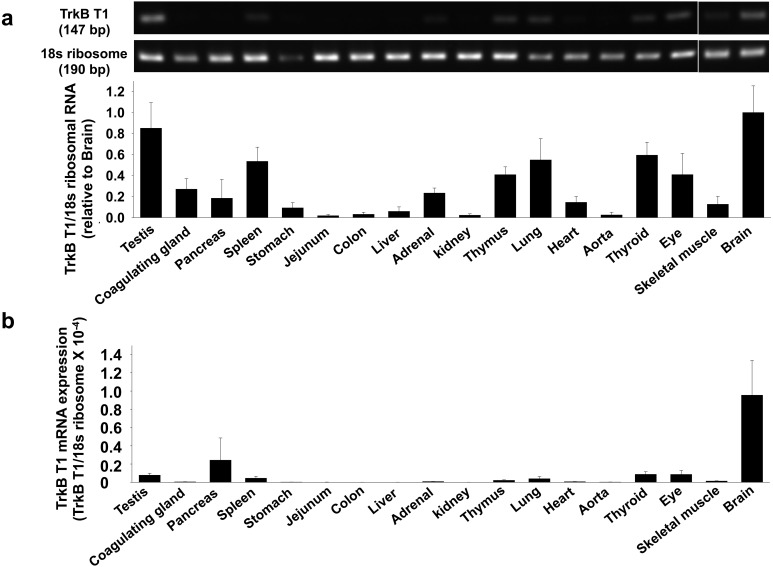
We next examined the expression level of TrkB T1 mRNA in rat tissues by RT-PCR and qRT-PCR. TrkB T1 mRNA was highly expressed in brain (Fig. 3a and 3b, n=3–4). We further measured the protein expression level of TrkB T1 in rat tissues by Western blotting. The molecular weight of rat TrkB T1 protein is approximately 43 kDa predicted from its amino acid sequences, except for signal peptides (NCBI Reference Sequence: NM_001163168.2). Since TrkB T1 has the same extracellular domain as TrkB FL, glycosylation of TrkB T1 could also change its electrophoresed mobility in SDS-PAGE. Thus, we identified an approximately 90 kDa band as glycosylated TrkB T1 [5, 14, 20, 25]. The glycosylated TrkB T1 was highly expressed in brain. While the expression was slightly observed in heart, no visible band was observed in other tissues (Fig. 4, n=3–4).
Fig. 3.
Expression level of truncated TrkB type-1 (TrkB T1) mRNA in rat tissues. Total RNA was extracted from homogenized tissues of Wistar rats. a) Expression of TrkB T1 mRNA was measured by RT-PCR using the gene specific primers to rat TrkB T1 (n=3−4). The data normalized to 18s ribosomal RNA were shown as fold increase relative to brain in bar graph. Data were expressed as mean ± SE. b) Expression of TrkB T1 mRNA was measured by qRT-PCR using the gene specific primers to rat TrkB T1 (n=3). The relative mRNA level to rat 18s ribosomal RNA was calculated using 2-ΔCq values.
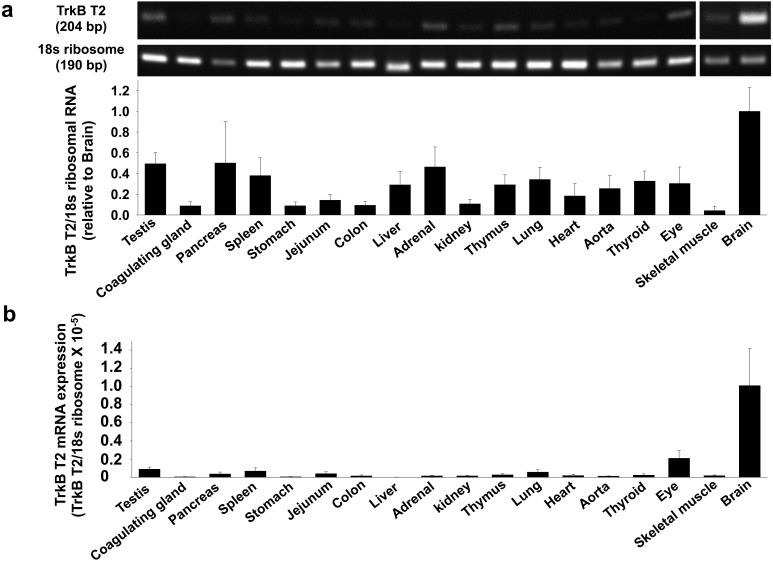
Expression level of TrkB T2 in rat tissues
We next examined the expression level of TrkB T2 mRNA in rat tissues by RT-PCR and qRT-PCR. TrkB T2 was highly expressed in brain. In other tissues, the expression was lower than brain (Fig. 5a and 5b, n=3–4). It should be noted that we cannot examine the TrkB T2 protein expression level because there is no commercially available antibody.
Fig. 5.
Expression level of truncated TrkB type-2 (TrkB T2) mRNA in rat tissues. Total RNA was extracted from homogenized tissues of Wistar rats. a) Expression of TrkB T2 mRNA was measured by RT-PCR using the gene specific primers to rat TrkB T2 (n=3−4). The data normalized to 18s ribosomal RNA were shown as fold increase relative to brain in bar graph. Data were expressed as mean ± SE. b) Expression of TrkB T2 mRNA was measured by qRT-PCR using the gene specific primers to rat TrkB T2 (n=3). The relative mRNA level to rat 18s ribosomal RNA was calculated using 2-ΔCq values.
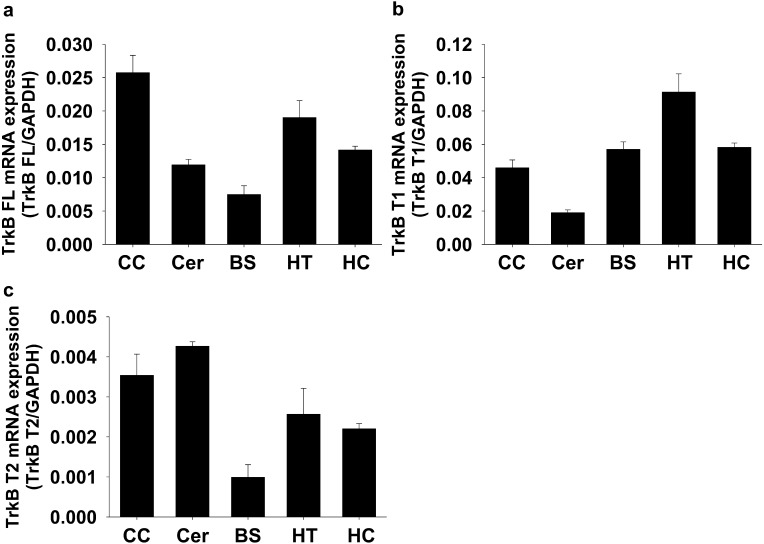
Expression level of TrkB isoforms mRNA in several regions of rat brain
Since all TrkB isoforms mRNA was highly expressed in brain, we finally examined the expression pattern of TrkB isoforms mRNA in several regions of rat brain. TrkB FL mRNA was higher in cerebral cortex, but lower in brainstem (Fig. 6a, n=3). TrkB T1 mRNA was higher in hypothalamus, but lower in cerebellum (Fig. 6b, n=3). TrkB T2 mRNA was higher in cerebral cortex and cerebellum, but lower in brainstem (Fig. 6c, n=3).
Fig. 6.
Expression level of TrkB isoforms mRNA in several regions of rat brain. Total RNA was extracted from homogenized tissues of Wistar rats. Expression level of a) TrkB FL mRNA, b) TrkB T1 mRNA and c) TrkB T2 mRNA was measured by qRT-PCR using the gene specific primers to rat TrkB isoforms (n=3). The relative mRNA level to rat glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was calculated using 2-ΔCq values. CC: Cerebral cortex, Cer: Cerebellum, BS: Brainstem, HT: Hypothalamus, HC: Hippocampus.
DISCUSSION
In the present study, we examined the mRNA and protein expression level of three TrkB isoforms in normal adult rat multiple tissues and obtained the following results; 1) TrkB FL mRNA and protein were both highly expressed exclusively in brain, 2) While TrkB T1 mRNA was highly expressed exclusively in brain, glycosylated TrkB T1 protein was expressed exclusively in brain (higher expression) and heart (lower expression), 3) TrkB T2 mRNA level in brain was the highest, and 4) In brain, TrkB FL mRNA expression was higher in cerebral cortex, but lower in brainstem. TrkB T1 mRNA expression was higher in hypothalamus, but lower in cerebellum. TrkB T2 mRNA expression was higher in cerebral cortex and cerebellum, but lower in brainstem. Collectively, we for the first time determined the diverse distribution of mRNA and protein of three TrkB isoforms in rat multiple tissues.
TrkB FL protein and mRNA were highly expressed exclusively in brain (Figs. 1 and 2). TrkB FL plays a critical role in regulating the neural development, growth, differentiation and survival [13, 16, 29]. The present results in rats well support the concept.
TrkB T1 mRNA and glycosylated TrkB T1 protein were also highly expressed in rat brain similar to the case of TrkB FL, confirming the previous results that TrkB T1 is expressed in rat brain [1, 9]. Interestingly, in heart, glycosylated TrkB T1 protein was detected by Western blotting, while the TrkB T1 mRNA level was lower (Figs. 3 and 4). TrkB T1 protein expression in heart was previously reported [7], which seemed to mediate the ligand-dependent contraction in cardiomyocytes [10]. The results indicate that TrkB T1 protein, even expressed at quite low level, would be expected to have some functions in heart. Likewise, the expression pattern of TrkB T1 mRNA and protein was different between brain and heart. We speculate this is because the regulation of post-transcriptional modifications for TrkB T1, such as proteolytic cleavage [17, 22]and microRNA silencing [23], would be different among these tissues.
It should be noted that we detected approximately 43 kDa bands in rat multiple tissues by Western blotting using anti-TrkB T1 antibody (sc-119) (Supplemental Fig. 2). Since the molecular weight of rat TrkB T1 protein is estimated as approximately 43 kDa (NCBI Reference Sequence: NM_001163168.2), it might be possible that the bands corresponded to non-glycosylated TrkB T1. If so, the non-glycosylated TrkB T1 seems to be ubiquitously expressed in rat multiple tissues. A glycosylation of TrkB potentially mediates the stability of TrkB [19]. Moreover, TrkA, belonging to the Trk family protein, requires a glycosylation for localization at plasma membrane, and non-glycosylated TrkA could not activate cellular signaling [36]. Hence, it is supposed that putative non-glycosylated TrkB T1 expressed in other tissues than brain and heart could not be localized at plasma membrane and do not have ligands-dependent cellular function.
The mRNA expression level of TrkB T2 in brain was the highest (Fig. 5). The expression pattern of TrkB T2 mRNA in rat multiple tissues was similar to that of TrkB FL and T1. While the functions of TrkB T2 remain largely unknown, TrkB T2 could bind to BDNF at least [3]. Additionally, similar to TrkB T1, TrkB T2 has dominant-negative effects for TrkB FL [5, 6]. Thus, TrkB T2 expressed in brain would serve as an antagonistic mediator of BDNF-TrkB FL signal.
In the present study, we further examined the expression pattern of TrkB isoforms mRNA in several regions of rat brain. The mRNA expression pattern of TrkB FL and TrkB T2 in the brain was similar, except for cerebellum (Fig. 6a and 6c). Interestingly, TrkB T2 mRNA in rat cerebellum was the highest compared with other brain regions (Fig. 6c). In cerebellum, it was suggested that TrkB T2 had no direct effects on intracellular function, but may work for the clearance of excess ligand (BDNF) [21]. We also found that mRNA expression of TrkB T1 but not TrkB FL and TrkB T2 was relatively higher in brainstem. It was previously reported that TrkB T1 was highly expressed in astrocyte [31] and non-neural cells abundantly existed in other brain regions than cerebral cortex and cerebellum [2], supporting our present results. Nonetheless, clarification of cell type specific expression pattern (neuron vs. glia) of TrkB isoforms warrants further investigation.
In summary, the present results for the first time clarified several points. First, all the TrkB isoforms were localized in rat brain. Second, while TrkB T1 mRNA was highly expressed specifically in brain, glycosylated TrkB T1 protein was expressed in brain and heart. The present study for the first time determined the diverse distribution of mRNA and protein of three TrkB isoforms in rat multiple tissues. The present findings in rat are hopefully useful for understanding the physiology and pathophysiology of mammals including human via comparing expression pattern of TrkB isoforms.
Supplementary
Acknowledgments
This study was supported by Grant-in-Aid for Japan society for the Promotion of Science Research Fellow Grant (JSPS KAKENHI Grant Number JP17J02328) and School of Veterinary Medicine, The Kitasato University.
REFERENCES
- 1.Allendoerfer K. L., Cabelli R. J., Escandón E., Kaplan D. R., Nikolics K., Shatz C. J.1994. Regulation of neurotrophin receptors during the maturation of the mammalian visual system. J. Neurosci. 14: 1795–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azevedo F. A., Carvalho L. R., Grinberg L. T., Farfel J. M., Ferretti R. E., Leite R. E., Jacob Filho W., Lent R., Herculano-Houzel S.2009. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J. Comp. Neurol. 513: 532–541. doi: 10.1002/cne.21974 [DOI] [PubMed] [Google Scholar]
- 3.Baxter G. T., Radeke M. J., Kuo R. C., Makrides V., Hinkle B., Hoang R., Medina-Selby A., Coit D., Valenzuela P., Feinstein S. C.1997. Signal transduction mediated by the truncated trkB receptor isoforms, trkB.T1 and trkB.T2. J. Neurosci. 17: 2683–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodeur G. M., Minturn J. E., Ho R., Simpson A. M., Iyer R., Varela C. R., Light J. E., Kolla V., Evans A. E.2009. Trk receptor expression and inhibition in neuroblastomas. Clin. Cancer Res. 15: 3244–3250. doi: 10.1158/1078-0432.CCR-08-1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Wit J., Eggers R., Evers R., Castrén E., Verhaagen J.2006. Long-term adeno-associated viral vector-mediated expression of truncated TrkB in the adult rat facial nucleus results in motor neuron degeneration. J. Neurosci. 26: 1516–1530. doi: 10.1523/JNEUROSCI.4543-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eide F. F., Vining E. R., Eide B. L., Zang K., Wang X. Y., Reichardt L. F.1996. Naturally occurring truncated trkB receptors have dominant inhibitory effects on brain-derived neurotrophic factor signaling. J. Neurosci. 16: 3123–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng N., Huke S., Zhu G., Tocchetti C. G., Shi S., Aiba T., Kaludercic N., Hoover D. B., Beck S. E., Mankowski J. L., Tomaselli G. F., Bers D. M., Kass D. A., Paolocci N.2015. Constitutive BDNF/TrkB signaling is required for normal cardiac contraction and relaxation. Proc. Natl. Acad. Sci. U.S.A. 112: 1880–1885. doi: 10.1073/pnas.1417949112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fenner B. M.2012. Truncated TrkB: beyond a dominant negative receptor. Cytokine Growth Factor Rev. 23: 15–24. doi: 10.1016/j.cytogfr.2012.01.002 [DOI] [PubMed] [Google Scholar]
- 9.Fryer R. H., Kaplan D. R., Feinstein S. C., Radeke M. J., Grayson D. R., Kromer L. F.1996. Developmental and mature expression of full-length and truncated TrkB receptors in the rat forebrain. J. Comp. Neurol. 374: 21–40. doi: [DOI] [PubMed] [Google Scholar]
- 10.Fulgenzi G., Tomassoni-Ardori F., Babini L., Becker J., Barrick C., Puverel S., Tessarollo L.2015. BDNF modulates heart contraction force and long-term homeostasis through truncated TrkB.T1 receptor activation. J. Cell Biol. 210: 1003–1012. doi: 10.1083/jcb.201502100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geiger T. R., Peeper D. S.2007. Critical role for TrkB kinase function in anoikis suppression, tumorigenesis, and metastasis. Cancer Res. 67: 6221–6229. doi: 10.1158/0008-5472.CAN-07-0121 [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez A., Moya-Alvarado G., Gonzalez-Billaut C., Bronfman F. C.2016. Cellular and molecular mechanisms regulating neuronal growth by brain-derived neurotrophic factor. Cytoskeleton (Hoboken) 73: 612–628. doi: 10.1002/cm.21312 [DOI] [PubMed] [Google Scholar]
- 13.Gupta V. K., You Y., Gupta V. B., Klistorner A., Graham S. L.2013. TrkB receptor signalling: implications in neurodegenerative, psychiatric and proliferative disorders. Int. J. Mol. Sci. 14: 10122–10142. doi: 10.3390/ijms140510122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haapasalo A., Sipola I., Larsson K., Akerman K. E., Stoilov P., Stamm S., Wong G., Castren E.2002. Regulation of TRKB surface expression by brain-derived neurotrophic factor and truncated TRKB isoforms. J. Biol. Chem. 277: 43160–43167. doi: 10.1074/jbc.M205202200 [DOI] [PubMed] [Google Scholar]
- 15.Haniu M., Talvenheimo J., Le J., Katta V., Welcher A., Rohde M. F.1995. Extracellular domain of neurotrophin receptor trkB: disulfide structure, N-glycosylation sites, and ligand binding. Arch. Biochem. Biophys. 322: 256–264. doi: 10.1006/abbi.1995.1460 [DOI] [PubMed] [Google Scholar]
- 16.Huang E. J., Reichardt L. F.2003. Trk receptors: roles in neuronal signal transduction. Annu. Rev. Biochem. 72: 609–642. doi: 10.1146/annurev.biochem.72.121801.161629 [DOI] [PubMed] [Google Scholar]
- 17.Jadhav T., Geetha T., Jiang J., Wooten M. W.2008. Identification of a consensus site for TRAF6/p62 polyubiquitination. Biochem. Biophys. Res. Commun. 371: 521–524. doi: 10.1016/j.bbrc.2008.04.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang H., Huang S., Li X., Li X., Zhang Y., Chen Z. Y.2015. Tyrosine kinase receptor B protects against coronary artery disease and promotes adult vasculature integrity by regulating Ets1-mediated VE-cadherin expression. Arterioscler. Thromb. Vasc. Biol. 35: 580–588. [DOI] [PubMed] [Google Scholar]
- 19.Kojima S., Nakayama T., Kuwajima G., Suzuki H., Sakata T.1999. TrkB mutant lacking the amino-terminal half of the extracellular portion acts as a functional brain-derived neurotrophic factor receptor. Biochim. Biophys. Acta 1420: 104–110. [DOI] [PubMed] [Google Scholar]
- 20.Kryl D., Barker P. A.2000. TTIP is a novel protein that interacts with the truncated T1 TrkB neurotrophin receptor. Biochem. Biophys. Res. Commun. 279: 925–930. [DOI] [PubMed] [Google Scholar]
- 21.Light K. E., Ge Y., Belcher S. M.2001. Early postnatal ethanol exposure selectively decreases BDNF and truncated TrkB-T2 receptor mRNA expression in the rat cerebellum. Brain Res. Mol. Brain Res. 93: 46–55. [DOI] [PubMed] [Google Scholar]
- 22.Makkerh J. P., Ceni C., Auld D. S., Vaillancourt F., Dorval G., Barker P. A.2005. p75 neurotrophin receptor reduces ligand-induced Trk receptor ubiquitination and delays Trk receptor internalization and degradation. EMBO Rep. 6: 936–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maussion G., Yang J., Yerko V., Barker P., Mechawar N., Ernst C., Turecki G.2012. Regulation of a truncated form of tropomyosin-related kinase B (TrkB) by Hsa-miR-185* in frontal cortex of suicide completers. PLoS ONE 7: e39301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Middlemas D. S., Lindberg R. A., Hunter T.1991. trkB, a neural receptor protein-tyrosine kinase: evidence for a full-length and two truncated receptors. Mol. Cell. Biol. 11: 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohira K., Hayashi M.2003. Expression of TrkB subtypes in the adult monkey cerebellar cortex. J. Chem. Neuroanat. 25: 175–183. [DOI] [PubMed] [Google Scholar]
- 26.Okada M., Suzuki A., Yamawaki H., Hara Y.2013. Levosimendan inhibits interleukin-1β-induced cell migration and MMP-9 secretion in rat cardiac fibroblasts. Eur. J. Pharmacol. 718: 332–339. [DOI] [PubMed] [Google Scholar]
- 27.Okamura Y., Otani K., Sekiguchi A., Kogane T., Kakuda C., Sakamoto Y., Kodama T., Okada M., Yamawaki H.2017. Vasculo-protective effect of BMS-309403 is independent of its specific inhibition of fatty acid-binding protein 4. Pflugers Arch. 469: 1177–1188. [DOI] [PubMed] [Google Scholar]
- 28.Otani K., Okada M., Yamawaki H.2015. Expression pattern and function of tyrosine receptor kinase B isoforms in rat mesenteric arterial smooth muscle cells. Biochem. Biophys. Res. Commun. 467: 683–689. [DOI] [PubMed] [Google Scholar]
- 29.Park H., Poo M. M.2013. Neurotrophin regulation of neural circuit development and function. Nat. Rev. Neurosci. 14: 7–23. [DOI] [PubMed] [Google Scholar]
- 30.Roesler R., de Farias C. B., Abujamra A. L., Brunetto A. L., Schwartsmann G.2011. BDNF/TrkB signaling as an anti-tumor target. Expert Rev. Anticancer Ther. 11: 1473–1475. [DOI] [PubMed] [Google Scholar]
- 31.Rose C. R., Blum R., Pichler B., Lepier A., Kafitz K. W., Konnerth A.2003. Truncated TrkB-T1 mediates neurotrophin-evoked calcium signalling in glia cells. Nature 426: 74–78. [DOI] [PubMed] [Google Scholar]
- 32.Squinto S. P., Stitt T. N., Aldrich T. H., Davis S., Bianco S. M., Radziejewski C., Glass D. J., Masiakowski P., Furth M. E., Valenzuela D. M., et al. 1991. trkB encodes a functional receptor for brain-derived neurotrophic factor and neurotrophin-3 but not nerve growth factor. Cell 65: 885–893. [DOI] [PubMed] [Google Scholar]
- 33.Usui T., Okada M., Hara Y., Yamawaki H.2012. Death-associated protein kinase 3 mediates vascular inflammation and development of hypertension in spontaneously hypertensive rats. Hypertension 60: 1031–1039. [DOI] [PubMed] [Google Scholar]
- 34.Usui T., Nijima R., Sakatsume T., Otani K., Kameshima S., Okada M., Yamawaki H.2015. Eukaryotic elongation factor 2 kinase controls proliferation and migration of vascular smooth muscle cells. Acta Physiol. (Oxf.) 213: 472–480. [DOI] [PubMed] [Google Scholar]
- 35.Wagner N., Wagner K. D., Theres H., Englert C., Schedl A., Scholz H.2005. Coronary vessel development requires activation of the TrkB neurotrophin receptor by the Wilms’ tumor transcription factor Wt1. Genes Dev. 19: 2631–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watson F. L., Porcionatto M. A., Bhattacharyya A., Stiles C. D., Segal R. A.1999. TrkA glycosylation regulates receptor localization and activity. J. Neurobiol. 39: 323–336. [DOI] [PubMed] [Google Scholar]
- 37.Yu Y., Fuscoe J. C., Zhao C., Guo C., Jia M., Qing T., Bannon D. I., Lancashire L., Bao W., Du T., Luo H., Su Z., Jones W. D., Moland C. L., Branham W. S., Qian F., Ning B., Li Y., Hong H., Guo L., Mei N., Shi T., Wang K. Y., Wolfinger R. D., Nikolsky Y., Walker S. J., Duerksen-Hughes P., Mason C. E., Tong W., Thierry-Mieg J., Thierry-Mieg D., Shi L., Wang C.2014. A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat. Commun. 5: 3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.