ABSTRACT
The elicitation of broadly and efficiently neutralizing antibodies in humans by active immunization is still a major obstacle in the development of vaccines against pathogens such as the human immunodeficiency virus (HIV), influenza virus, hepatitis C virus or cytomegalovirus. Here, we describe a mammalian cell surface display and monoclonal antibody (mAb)-mediated panning technology that allows affinity-based selection of envelope (Env) variants from libraries. To this end, we established an experimental setup featuring: 1) single and site specific integration of Env to link genotype and phenotype, 2) inducible Env expression to avoid cytotoxicity effects, 3) translational coupling of Env and enhanced green fluorescent protein expression to normalize for Env protein levels, and 4) display on HEK cells to ensure native folding and mammalian glycosylation. For proof of concept, we applied our method to a chimeric HIV-1 Env model library comprising variants with differential binding affinities to the V3-loop-directed mAbs 447–52D and HGN194. Fluorescence-activated cell sorting selectively enriched a high affinity variant up to 56- and 55-fold for 447–52D and HGN194, respectively, after only a single round of panning. Similarly, the low affinity variants for each antibody could be selectively enriched up to 237-fold. The binding profiles of membrane-bound gp145 and soluble gp140 chimeras showed identical affinity ranking, suggesting that the technology can guide the identification of Env variants with optimized antigenic properties for subsequent use as vaccine candidates. Finally, our mAb-based cellular display and selection strategy may also prove useful for the development of prophylactic vaccines against pathogens other than HIV.
KEYWORDS: HIV-1 envelope, neutralizing antibodies, mammalian cell display, panning, library
Abbreviations
- AIDS
acquired immunodeficiency syndrome
- Amp
ampicillin
- APC
allophycocyanin
- BLA
β-lactamase
- bnAb
broadly neutralizing antibody
- CDR3
complementary determining region 3
- CMV
cytomegalovirus
- DMEM
Dul-becco's modified eagle medium
- E. coli
Escherichia coli
- EDTA
Ethylenediaminetetraacetic acid
- eGFP
enhanced green fluorescent protein
- EGTA
ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid
- ELISA
enzyme-linked immunosorbent assay
- Env
envelope protein
- FACS
fluorescence-activated cell sorting
- FBS
fetal bovine serum
- FRT
Flp recombination target
- gDNA
genomic DNA
- gp140
HIV-1 glycoprotein of 140 kDa (terminated before transmembrane-domain)
- gp145
HIV-1 glycoprotein of 145 kDa (terminated after transmembrane-domain)
- gp160
HIV-1 glycoprotein (160 kDa, full-length)
- HEK
human embryonic kidney
- HIV
human immunodeficiency virus
- Hyg
hygromycin
- KD
dissociation constant
- lacZ
β-galactosidase gene
- mAb
monoclonal antibody
- MCS
multiple cloning site
- MFI
mean fluorescence intensity
- NGS
next generation sequencing
- ORF
open reading frame
- ori
origin of replication
- P
Promotor
- PBS
phosphate buffered saline
- pDNA
plasmid DNA
- RT
room temperature
- SV40
simian virus 40
- TaVp2A
Thosea asigna virus peptide 2A
- TERT
telomerase reverse transcriptase
- Tet
tetracycline
- TO
tetracycline operator
- TR
tetracycline repressor
- T-REx
tetracycline-regulated expression
- V1-V5
HIV envelope variable loops 1–5
Introduction
With approximately 35 million infected individuals worldwide and about 39 million AIDS-related deaths so far, the human immunodeficiency virus (HIV)-1 pandemic continues to be a major global public health challenge. According to the World Health Organization, only 49% of HIV-1 positive individuals have access to anti-retroviral therapy, affirming the need for an efficient vaccine.1
Regarding the humoral immune response, the HIV-1 surface protein envelope (Env) is the only virus-encoded determinant present on the virus surface, and thus accessible to antibodies.2 Env is expressed as a gp160 precursor protein that is proteolytically cleaved into gp120 and gp41 by the Golgi-associated furin protease. 3 gp120 and 3 gp41 subunits assemble into the final trimeric (heterohexameric) Env spike, with each gp41 transmembrane subunit being non-covalently associated with the external gp120 subunit, respectively. Whereas gp41 is more conserved, the gp120 subunit has a highly variable and heavily glycosylated surface that includes 5 variable loops (V1–V5).3 The resulting vast number of variants circulating in the human population represents a major challenge for vaccine development, and mainly accounts for the failure of classical vaccine development approaches like chemical inactivation or live attenuation.2
However, after several years of infection, ∼10–50% of patients develop broadly neutralizing antibodies (bnAbs),4,5 which recognize conserved, mostly conformational or quaternary-structure-dependent epitopes on Env.6 Some of these bnAbs can neutralize up to 80–90% of virus strains.5 Prior to 2009, only a few bnAbs were known, and all targeted either the membrane-proximal external region (MPER), the CD4 binding site, or a glycan-dependent epitope in gp120. Recently, however, the development of highly efficient screening methods has resulted in the isolation of a multitude of new bnAbs targeting multiple sites of vulnerability on the trimer.7
Passive immunization of macaques with bnAbs provided complete protection from infection,8,9,10 offering an important starting point for the development of an efficient vaccine. However, due to the high variability of Env,11 an extensive glycan shield,12 conformational masking of target sites,13,14 and conformational instability of Env,3 the elicitation of bnAbs by active immunization is still a major obstacle in vaccine design. To counter these escape strategies, bnAbs often exhibit unique features like high rates of somatic hypermutation and long CDR3-loops, as a result of years of complex co-evolution between virus escape and immune adaptation.15
To properly instruct the development of antibody responses with a broader neutralization profile, several promising, and not mutually exclusive, approaches have been investigated recently, resulting in new generations of envelope immunogens. The approaches include: 1) a directed evolution approach, which identified a chimeric gp120 Env variant (ST-008) eliciting neutralizing antibody responses in rabbits,16 2) the heterologous substitution or deletion of the V1 loop or hyperglycosylation of variable loops to focus antibody responses to more conserved epitopes like the CD4 binding site,17,18 3) chemical cross-linking of Env aiming toward stabilizing the Env trimer in its closed conformation and directing the humoral immune response to neutralizing epitopes,19 and 4) soluble recombinant Env trimers genetically engineered to form stable, closed and well-folded trimers.20,21,22 Notably, such trimers for the first time enabled the induction of neutralizing antibodies against the sequence-matched tier 2 virus in rabbits and macaques.23,24 To mimic the complex co-evolution between Env and the immune system in vivo, sequential immunization with different Env immunogens was applied to first prime naive B cells with a germline B cell receptor, and then enable generation of the mature bnAb over several intermediate stages.25 Recently, sequential immunization strategies have been successfully implemented in knock-in mouse models for germline-reverted versions of the VRC01 and PGT121 families, resulting in high levels of somatic hypermutation and thus guiding the maturation of bnAb responses.26,27,28
It is thus now known that next-generation envelope immunogens need to properly drive the neutralizing antibody response from germline to affinity-matured antibodies. The current set of available broadly neutralizing monoclonal antibodies (mAbs), including their engineered unmutated common ancestors, represents a valuable toolbox to characterize recombinant Env trimers, but possibly even more to select Env variants with the desired binding profiles from libraries comprising either naturally occurring (patient derived), or rationally designed and engineered Env libraries.
We recently reported an evolutionary approach for the identification of such potential HIV-1 vaccine candidates using a first generation retroviral cell display and panning platform, which required 2 rounds of panning for a 20-fold enrichment of a high affinity variant from a small 5-member HIV envelope library.29 However, although low multiplicity of infection was used, multiply transduced cells could not be prevented with this method, overriding an efficient genotype-phenotype linkage in the library.
Here, we describe an alternative selection procedure using a single-integration stable cell line technology for direct mammalian cell display in combination with a fluorescence-activated cell sorting (FACS)-based gating and panning strategy. Using this procedure, we were able to enrich the high affinity variant of a limited model library, which comprises 5 engineered HIV envelope variants with known binding affinities to model antibodies 447–52D and HGN194,30,31 up to 56- and 55-fold, respectively, in a single round of panning. We were also able to selectively enrich the low affinity variants up to 237-fold with the appropriate gating strategy. Hence, using more complex Env libraries, this method could be a valuable tool to select new Env vaccine candidates with enhanced binding to bnAbs and reduced binding to non-neutralizing antibodies for the various approaches mentioned above.
Our mAb-based technology might also prove useful in designing and selecting envelope-based vaccine compounds against other viral pathogens such as influenza virus, hepatitis C virus or even DNA viruses where proper assembly of various envelope subunits to hetero-oligomeric complexes is required to induce high quality quaternary-structure-dependent antibodies, such as the cytomegalovirus (CMV) gH-pentameric complex.32 Lastly, our data provide evidence that the technology can be used to map mAbs against any membrane or envelope protein when, for example, presented as alanine scanning variants on the cell surface.
Results
Cellular display and flow cytometry-based panning procedure
Here, we describe a mammalian cell display and flow cytometry-based panning method that allows strict genotype-phenotype coupling of cell surface-exposed HIV-1 Env variants and enables the identification and selection of variants with modulated antibody affinity (Figure 1). Step 1 comprises the generation of stable HEK293 cell lines, with each cell carrying a single copy of a given Env variant at a defined locus (“linked” cellular library). For this purpose, FlpIn™ T-REx™ 293 cells are transfected with an Env library cloned in a plasmid vector (pQL13), which supports the integration of a single copy Env gene into the predefined Flp recombination target (FRT) site (Figure 1; step 1). After antibiotic selection, each of the resulting HEK293 cells displays, upon induction, one distinct variant on the cell surface, leading to a strict linkage of the Env genotype and phenotype within the mammalian cell library (Figure 1; step 2). Next, Env-expressing HEK293 cells are stained with an antibody of interest (screening antibody, Figure 1; step 3), and high or low affinity variants are sorted by flow cytometry-based cell sorting using the desired gate (Figure 1; step 4). For analytical purposes, the selected Env genes are subsequently amplified from the genomic DNA of the sorted cells and cloned into pQL13 to be further propagated in Escherichia coli. To calculate the enrichment or depletion of the different Env variants, plasmid DNA (pDNA) recovered from E. coli is analyzed either by qPCR or Sanger sequencing of single clones for small libraries, or next generation sequencing (NGS) for larger libraries. Alternatively, genomic DNA (gDNA) of the HEK293 cells isolated before and after FACS can also be directly subjected to NGS. Followed by deconvolution of sequencing data, the level of enrichment or depletion can be calculated for each individual Env variant. If necessary, selected HEK293 cells can be expanded again and used for further panning and selection cycles (not shown in Figure 1).
Figure 1.
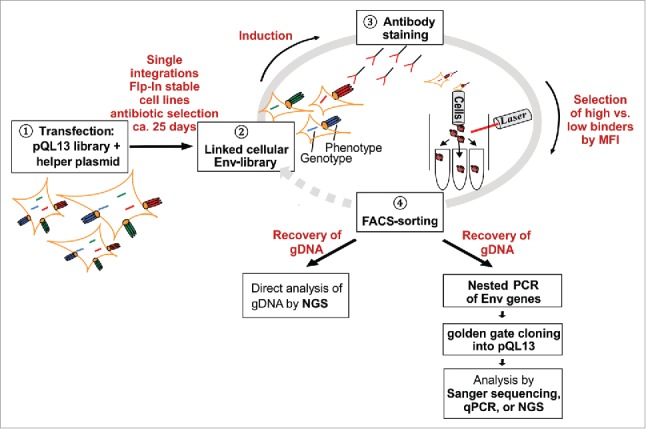
Schematic overview of the flow cytometry-based panning procedure. Stable cell lines are generated by transfection of FlpIn™ T-REx™ 293 cells with pQL13-based Env constructs and the helper plasmid pOG44 carrying the integrase (1). Although multiple Env variants can theoretically enter the human cells and be expressed for a short period of time, a linkage of geno- and phenotype is ultimately achieved because FlpInTM T-RExTM 293 cells possess precisely one FRT site, resulting in single-integration stably transfected HEK293 cells (2). After induction of Env expression, the mammalian cell library is stained with the screening antibody (3), and then envelope-expressing cells yielding the highest or lowest signal to the applied antibody are selected via fluorescence-activated cell sorting (4). For analytical purposes, genomic DNA is recovered from the selected HEK293 cells, envelope genes are amplified by nested PCR, re-cloned into pQL13 and analyzed by qPCR or sequencing of plasmid DNA recovered from single E. coli clones. In the case of larger libraries, genomic DNA from sorted HEK293 cells can be directly subjected to NGS to calculate enrichment or depletion factors for the respective envelope variants. To further enhance enrichment rates, sorted HEK293 cells could be re-expanded to be subjected to a new round of panning.
The HIV-1 chimeric Env/V3 model library
To enable a comparison of the herein described cellular Env display and selection system with a previously described lentiviral Env display system,29 we used the same gp145 Env/V3 model library. It consists of 5 chimeric Env constructs based on a clade C 96ZM651 Env backbone with the third variable domain (V3) seamlessly substituted by the V3 loops derived from isolates MN, RF, CDC42, HXB2 or SF33. This results in a small library with distinct binding profiles to mAbs 447–52D and HGN194 (Figure 5). Using the “golden gate” cloning procedure, we now cloned the Env/V3 model library into a novel vector system, pQL13 (Figure 2) and generated HEK293 stable cell lines with the following features: 1) translational coupling of enhanced green fluorescent protein (eGFP) and Env production via a TaV 2A peptide to enable normalization for Env expression levels according to eGFP fluorescence,29,33 2) single integration of pQL13 into the predefined FRT site, 3) selection for successful integration via hygromycin and 4) Tet-inducible Env and eGFP expression (Figure 2). To facilitate library assembly in E. coli, the introduction of a toxic CcdB cassette in the MCS of the pQL13 target vector allows an efficient selection of envelope positive clones.
Figure 5.
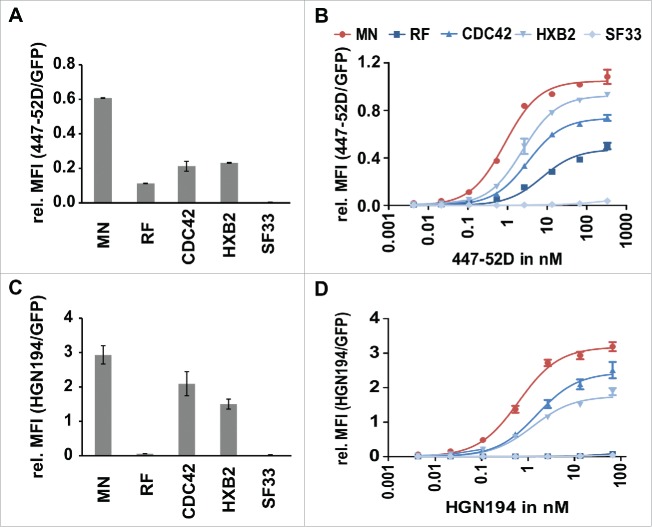
Analysis of the affinities of 447–52D and HGN194 to the individual members of the Env/V3 model library. 24 h post induction of the Env/V3 stable HEK293 cell lines with doxycycline (1 µg/ml), these cells were stained with mAbs 447–52D or HGN194 until equilibrium binding was reached. Relative MFI values of bound antibodies in relation to the eGFP signal (MFI APC/ MFI eGFP) to normalize for Env expression levels are shown (mean values of 3 independent experiments). A and C: Relative MFIs (MFI APC/ MFI eGFP) of HEK293 cell lines stained with 2.7 nM 447–52D (A) and 6.7 nM HGN194 (C) (the same concentrations as used in all sorting experiments, see below) are shown to illustrate the individual binding pattern of the 5 Env/V3 chimeras to the respective antibody. B and D: flow cytometry titration using serial dilutions of 447–52D (B) and HGN194 (D).
Figure 2.
Schematic overview of the pQL13 vector system and the FlpIn™ T-REx™ stable cell lines.In pQL13, Env expression is linked to eGFP expression by a TaV 2A peptide. The eGFP-Env expression cassette is integrated into a distinct Flp Recombination Target (FRT) site within the FlpIn™ T-REx™ HEK293 cells, providing the hygromycin gene with a start-codon and hence resulting in the acquisition of a hygromycin resistance that can be used for selection of stable cell lines. Expression of eGFP and Env is under regulation of the Tet repressor (TR, T-REx™, red triangle) and thus inducible. The locations of the TaV 2A peptide, as well as the ATG leading to the acquisition of the hygromycin resistance, are highlighted in red. Note that the pQL13 vector graphic shows the horizontal map of the circular QL13 plasmid and that the FRT site (52 bp) and the Tet operator sequence (TO, 9 bp, clear diamond) are not in scale in this schematic. P: Promotor (SV40: simian virus 40, CMV: cytomegalovirus, BLA: β-lactamase), lacZ: β-galactosidase ORF, Amp: ampicillin ORF, Hyg: hygromycin ORF, ori: origin of replication, TR: Tet repressor protein, TaVp2A: TaV 2A peptide. Promotor positions are indicated by bent arrows.
Generation of single integration stable cell lines using the pQL13-based chimeric Env/V3 variants
Stable HEK293 cell lines of the 5 different Env/V3 chimeras were generated by targeted transfection of FlpIn™ T-REx™ 293 cells with the 5 different gp145 Env/V3 encoding pQL13 plasmids either individually or collectively. This resulted either in 5 separate clonal cell lines or a bulk HEK293 cell library with each cell expressing one of the 5 Env/V3 variants (Figure 1; step 1). Each FlpIn™ T-REx™ HEK293 cell contains one distinct FRT site, allowing the integration of only one Env variant per cell, even if several different variants may enter the cell after transient transfection of a pQL13-based library. Hence, after successful integration and selection, an efficient linkage of genotype and phenotype within the library is eventually reached. Successful integration events of the eGFP-Env cassette are selected by a hygromycin resistance that is acquired and constitutively expressed only after successful recombination into the FRT site (Figure 2).
After ∼25 d of antibiotic selection, the “linked” cellular Env library is established (Figure 1; step 2). The stable integration of the Tet repressor in the FlpIn™ T-REx™ cells in combination with the doxycycline-inducible CMV promotor of pQL13 thereby results in the regulated expression (T-REx) of the integrated Env gene (Figure 2). Notably, the T-REx system only utilizes regulatory elements from the native Tet operon,34 avoiding the potentially toxic effects of viral transactivation domains used for other doxycycline-regulated systems.35 In sum, the inducible expression of the eGFP-Env cassette allows the cultivation of stable HEK293 cell lines without negative effects attributed to possible Env cytotoxicity.
To reassess the linkage of genotype and phenotype in the cell library, we analyzed relative copy numbers of integrated pQL13 plasmids per HEK293 cell using a copy number assay, which confirmed one integration event per cell for all scenarios tested (Figure 3).
Figure 3.
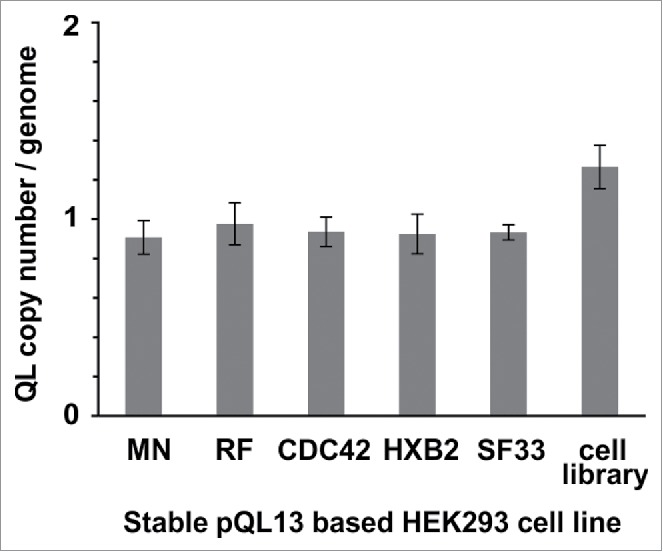
TaqMan® Copy Number assay of stable cell lines. Stable cell lines were generated using either separate transfection of each of the pQL13-based Env/V3 chimeras or a mixture of all 5 Env/V3 chimeras for transfection (HEK293 cell library). Relative copy numbers of integrated pQL13 plasmids were explored using a TaqMan Copy Number Assay on 4 individual samples of genomic DNA of each cell line and probing for eGFP in relation to the human telomerase reverse transcriptase (TERT) genes, resulting in an integration rate of 1 for each human cell line (differences between the 6 stable cell lines statistically n.s., p > 0.05).
Expression of chimeric Env/V3 variants correlates with GFP reporter expression
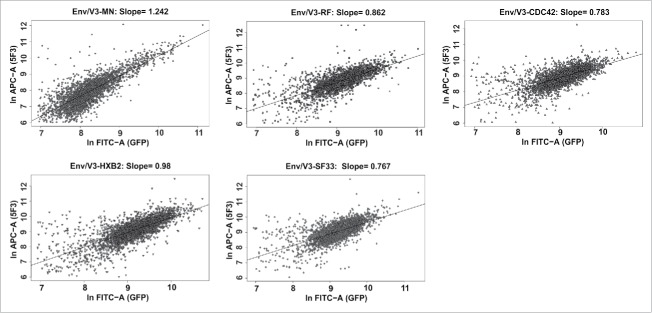
In pQL13, the expression of the marker gene eGFP was genetically coupled to Env expression by a TaV 2A peptide (Figure 2). The insect virus-derived (Thosea asigna virus) 2A peptide impairs peptide bond formation by a ribosomal skip mechanism between the 2A glycine and the 2B proline of the consensus sequence 2A, Asp-Val/Ile-Glu-X-Asn-Pro-Gly; 2B, Pro.33 This should result in an equal expression level for eGFP and Env, while only one residue is N-terminally attached to the envelope protein. To confirm coupled expression of both eGFP and Env, doxycycline-induced HEK293 cells were stained with the antibody 5F3,36 which binds to the extracellular domain of gp41 that is present in all 5 Env/V3 variants. A correlation between eGFP and Env expression was observed for each of the 5 Env/V3 chimeras (Figure 4), qualifying eGFP fluorescence as an indirect signal to normalize for Env expression. Notably, expression rates of the variants differed (with Env/V3-MN displaying the weakest signals), highlighting the importance of expression normalization.
Figure 4.
Correlated expression of V3 envelope variants and eGFP. 3 × 105 cells of each HEK293 stable cell line were induced with doxycycline (1 µg/ml) 24 h before flow cytometry analysis. The cells were harvested and stained with 5F3 and goat anti-human-APC as a secondary antibody. Measurements were performed in duplicates. 2.5 × 104 cells were recorded per sample and the correlation between APC and GFP signal intensities was analyzed for each stable cell line (3000 events shown).
The Env/V3 model library comprises distinct binding profiles to mAbs 447–52D and HGN194
We determined the binding profiles of 2 different V3-specific model-antibodies, 447–52D and HGN194, to the individual variants of the Env/V3 model library by flow cytometry-based titration of the individual stable HEK293 cell lines (Figure 5). The Env/V3 chimera MN displayed the highest affinity toward 447–52D and HGN194, followed by the variants CDC42 and HXB2. Whereas HGN194 bound neither the Env/V3 chimera RF nor SF33 (Figure 5D), 447–52D could still bind Env/V3 chimera RF to some extent (Figure 5B). For these non- or only weakly binding variants, KD values could not be determined. KD values of the variants for which binding affinities to 447–52D and HGN194 could be calculated are summarized in Table 2. In conclusion, the demonstrated distribution of varying affinities was considered useful for the purpose of evaluating the stable cell line-based panning procedure.
Table 2.
Comparison of apparent affinities of membrane-bound gp145 obtained from flow cytometry titration vs. soluble gp140 Env variants obtained from ELISA titration.
| V3 peptides | Membrane-bound gp145 | Soluble gp140 | ||||
|---|---|---|---|---|---|---|
| KD [nM] |
KD [nM] (flow cytometry titration, n = 3) |
KD [nM] (ELISA titration, n = 3) |
||||
| V3 loop substitution | 447–52D | HGN194 | 447–52D | HGN194 | 447–52D | HGN194 |
| MN | 0.6- | NA | 0.9 ± 0.08 (R2 = 0.990) | 0.6 ± 0.04 (R2 = 0.981) | 0.4 ± 0.02 (R2 = 995) | 0.2 ± 0.05 (R2 = 0.926) |
| HXB2 | 24 | 2 ± 0.2 (R2 = 0.991) | 1.3 ± 0.2 (R2 = 0.973) | 0.5 ± 0.002 (R2 = 0.983) | 7 ± 5 (R2 = 0.537) | |
| CDC42 | 5 | 3 ± 0.2 (R2 = 0.996) | 1.5 ± 0.09 (R2 = 0.951) | 1 ± 0.4 (R2 = 0.983) | 0.6 ± 0.2 (R2 = 0.797) | |
| RF | 0.9 | 7 ± 1 (R2 = 0.972) | NA | 74 ± 7 (R2 = 0.992) | NA | |
| SF33 | NA | NA | NA | NA | NA | |
KD values were calculated using nonlinear least squares regression for titration of membrane-bound gp145 Envs and soluble gp140 Envs. Due to low antibody binding, the apparent KD values for RF and SF33 could not be calculated in some cases (NA). Values for V3 peptides were adopted from ref.30
Single round flow cytometry-based panning of the Env/V3 cell library leads to enrichment of high or low affinity variants
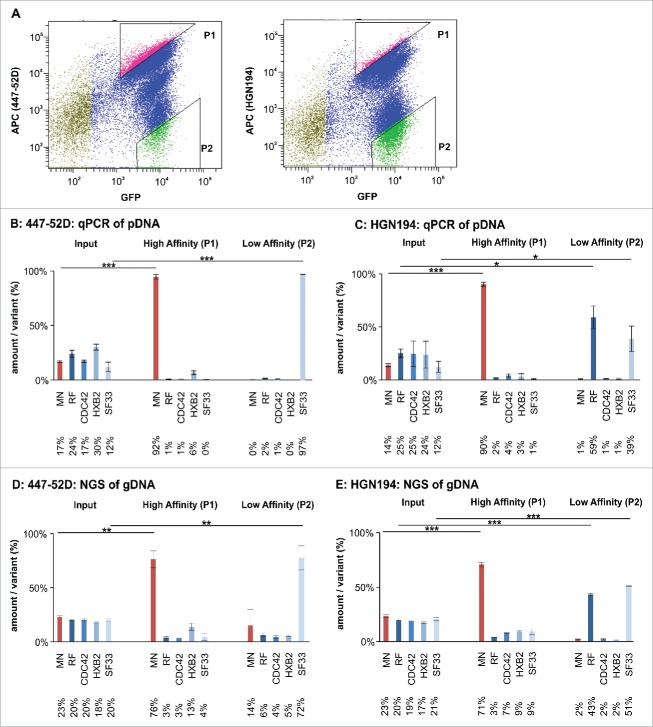
The mammalian cell library displaying the Env/V3 variants was subsequently subjected to flow cytometry-based cell sorting using ∼3 × 107 cells per sort.29 Triangular gates were chosen to sort cells with the highest (P1) or lowest (P2) 447–52D or HGN194 signal normalized by their eGFP signal, respectively (Figure 6A). Genomic DNA (gDNA) from the input sample and the sorted cells (∼30,000 cells/sorting gate) was recovered and Env genes were amplified by nested PCR and re-cloned into pQL13, respectively. The distribution of Env/V3 variants in the input sample and in the sample after one cycle of panning was assessed by qPCR, capillary sequencing of 96 single clones, and NGS of pDNA pools recovered from E. coli. Moreover, gDNA, recovered from the input sample and the sorted HEK293 cells, was subjected directly to NGS to allow for a streamlined analysis approach for larger libraries (Figure 6 and S1).
Figure 6.
Flow cytometry-based panning using the Env/V3 model library stable cell line. A: A representative sorting experiment is shown to illustrate the flow cytometry-based cell sorting strategy (50,000 events). Living, single HEK293 cells were gated according to common hierarchical gating strategies. The remaining cells were then gated for highest APC signals (447–52D or HGN194 with anti-human-APC secondary antibody) in relation to eGFP signals (expression control). Thus, a triangular gate (gate P1) was chosen to select for high affinity binders. Similarly, P2 was chosen to select for low affinity binders to 447–52D and HGN194. B to E: The stable cell line pool containing the complete 5 member Env/V3 model library was used in a flow cytometry-based panning procedure 24 h after induction with doxycycline using 2.7 nM 447–52D (B, D) or 6.7 nM HGN194 (C, E). The envelope genes from the cell line pool prior (Input) and after selection (High Affinity P1 and Low Affinity P2) were PCR-amplified from the genomic DNA, cloned into pQL13, recovered from E. coli and analyzed by qPCR (B and C) or, alternatively, the gDNA of the HEK293 cells was directly subjected to NGS analysis (D and E). The mean values of 3 independent experiments are shown. The percentages of the variants in the input mixture and after one round of panning are given. Statistics were calculated using an unpaired t-test. Asterisks are indicated for *** p < 0.001, ** p < 0.01 and * p < 0.05.
For the 447–52D flow cytometry-based cell sorting experiment, we demonstrate a statistically significant (*** = p < 0.001) enrichment of the high affinity Env/V3 variant MN out of a mixture of all variants (qPCR: 17% MN in input enriched to 92% MN after one round of panning) in gate P1 (Figure 6B). Thus, after only a single round of panning, we achieved an enrichment rate of 56 for the Env/V3 variant MN (for calculation, see Table 1). Although reduced from 30% in the input fraction to 6% after the first round (qPCR), the Env chimera presenting the HXB2-V3 loop scored second best, which is in line with the affinity measurements of surface-exposed envelopes (Figure 5, Table 2).
Table 1.
Specific enrichment rates of the Env/V3 variants selected from the fluorescence-activated cell sorting procedure after one round of panning.
| High Affinity Gate P1 |
Low Affinity Gate P2 |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| |
Env after sorting (%) / Env input (%) |
Fold enrichmenta MN |
Env after sorting (%) / Env input (%) |
Fold enrichmenta RF |
Env after sorting (%) / Env input (%) |
Fold enrichmenta SF33 |
|||
| MN | Non-MN | RF | Non-RF | SF33 | Non-SF33 | ||||
| 447–52D | 5.41 | 0.10 | 56 | 0.08 | 1.29 | 0.06 | 8.08 | 0.03 | 237 |
| HGN194 | 6.43 | 0.12 | 55 | 2.36 | 0.56 | 4 | 3.25 | 0.70 | 5 |
calculations were made according to ref.49 ( , or ) for the samples analyzed by qPCR, #) cycle of panning.
Within the low signal gate P2, we were also able to selectively enrich the low affinity variant SF33, thereby achieving an enrichment factor of 237 for mAb 447–52D (qPCR: input 12%; 1st round 97%) (Table 1). Hence, our panning technology may be applicable to isolate Env variants with particularly low binding to undesired antibodies, like non-neutralizing antibodies toward epitopes presented in the CD4-induced Env conformation.
Thus, the normalized mean fluorescent intensities (MFIs) measured at a given 447–52D antibody concentration (Figure 5A), as well as the binding affinities of the individual Env/V3 chimeras calculated based on the binding curves (Figure 5B and Table 2) chiefly reflected the copy number (qPCR) of the respective sequences quantified after the single round flow cytometry-based cell sorting process.
These findings were confirmed by an independent FACS experiment, where we used an alternative V3-specific mAb, HGN194. Here, we were able to enrich the high affinity variant MN from 14% (input fraction, qPCR) to 90% (Figure 6C) after one round of panning, resulting in an enrichment rate of 55 (Table 1). Similarly, as RF and SF33 both did not show binding to HGN194 (Figure 5C and D, Table 2), the low signal gate P2 selectively enriched those 2 variants from 25% and 12% to 59% and 39%, respectively, which corresponds to enrichment factors of 4 for chimera RF and 5 for chimera SF33 (Figure 6C, Table 1).
However, enrichment analysis by qPCR requires a specific probe for each variant of the library and is therefore only applicable for small libraries with known sequence variations. Hence, the above enrichment data for 447–52D and HGN194 were also confirmed by Sanger sequencing of 96 E. coli single clones that carried the re-cloned plasmid DNA as well as NGS using the Illumina Miseq platform, which allows the analysis of any larger library in the future. NGS was applied to gDNA prepared from the HEK293 cells before and after sorting and, for control, to pDNA of amplified and re-cloned Env genes recovered from E. coli. Importantly, we were able to replicate the above described results with all applied methods, with only minor differences between pDNA and gDNA analyses (Figure 6, Figure S1).
Enrichment factors resulting from the panning procedure for the displayed Env/V3 chimeras are predictive for the affinity ranking determined for the corresponding soluble secreted envelopes
To investigate whether the enrichment factors determined for the cell-surface-displayed gp145 Env/V3 variants are predictive for the affinity-based ranking of the corresponding soluble secreted gp140 envelope proteins, the 5 pQL13 encoded gp140 variants of the Env/V3-model library lacking the cytoplasmic tail as well as the transmembrane domain were purified to trimer homogeneity (Figure S2A). Subsequently, apparent binding affinities of the different variants toward 447–52D and HGN194 were analyzed by solid-phase binding enzyme-linked immunosorbent assay (ELISA) (Figure S2B). This resulted in a very similar ranking according to relative binding affinities compared with the titration of gp145 Env displaying cell lines (Figures 5B and D and S2), and apparent dissociation constants (KD) were found to reflect the same order (Table 2), with MN having the highest affinity, followed by CDC42 and HXB2. Furthermore, in agreement with the membrane-bound variants, 447–52D showed weak binding to RF, while HGN194 bound neither RF nor SF33. Hence, this method is, albeit indirectly, also very valuable for the identification of soluble envelope variants with modified binding characteristics. Noteworthy, whereas the ranking of the Env/V3 proteins according to binding affinities toward 447–52D resulted in the same order for both the membrane displayed and the soluble form of the polypeptide, early experiments using V3 peptides derived from the 5 HIV strains came to a different order,30 suggesting that the overall conformation of the trimeric protein may influence the formation of the 447–52D epitope.
In sum, the data demonstrate a clear relation between: 1) the apparent KD values determined for the individual cell-surface displayed Env/V3 chimeras (Figures 5B and D), 2) the enrichment factors determined for the Env/V3 chimeras after panning in the high affinity gate (Figure 6), and 3) the binding affinities calculated for the soluble secreted gp140/V3 chimeras (Table 2).
Discussion
Here, we describe a mammalian cell surface display-based panning strategy for the selection of Env variants with improved affinity toward a screening antibody. This methodology could in principle select and qualify new HIV-1 Env vaccine candidates based on their affinity to broadly neutralizing antibodies. The proposed strategy is largely driven by the increasing number of these antibodies, which have proven to be helpful in selecting and characterizing envelopes according to their antigenic properties. Our panning technology is based on the display of trimeric envelopes on inducible HEK stable cell lines. Mammalian cells not only allow mammalian glycosylation and folding, but also expression of functional Envs in a membrane context, and are, for example, widely used as a producer cell line for Env pseudoviruses for neutralization assays.37,38,39 In addition, this technology enables the assessment of native full-length Env proteins in their natural membrane environment. Thus, this display technique should fulfill all of the proposed requirements of envelope presentation, including the preservation of promising quaternary structure epitopes of Env and membrane-proximal epitopes that might not be properly represented in truncated Env variants.40
It is worth noting that the integration of our library vector pQL13 at one distinct FRT site allows a highly stringent linkage of genotype and phenotype within a library, while the translational coupling of inducible Env and GFP expression by a TaV 2A peptide sequence represents an accurate means to normalize for Env expression levels. Taken together, these advantages accounted for high enrichment rates in only one single round of panning, as exemplified using our 5-member model library with the mAbs 447–52D and HGN194. While the high affinity chimera 96ZM651-Env/V3-MN could be enriched up to 56- and 55-fold, respectively, the low affinity gate led to the selective enrichment of the sole non-binding variant Env/V3-SF33 for 447–52D, while both non-binding variants displaying the HIV-1 RF and SF33 V3-loop were selected with HGN194. Hence, this stable cell line-based panning procedure represents a substantial improvement to our previously published panning approach, where genotype-phenotype linkage within the library was based on a lentiviral vector system for stable transgene integration, and which resulted in an enrichment of 20-fold for the high affinity chimera MN after 2 rounds of selection.29
If applied in conjunction with a carefully selected panel of bnAbs, our approach also provides opportunities to identify new Env variants with the potential to overcome the various immune escape strategies of Env. We could show that NGS can be directly applied to the genomic DNA recovered from sorted HEK293 cells in comparison to the genomic DNA of the unsorted population, allowing fast analysis of large libraries without the limitations arising from the re-cloning strategy. Hence, this approach would be easily applicable to more comprehensive trimeric Env libraries and highly potent bnAbs like VRC01,41 PG9/16 or members of the PGT family.37,42 Notably, by probing tailor-made envelope libraries using a combination of inferred unmodified germline versions of bnAbs, selected intermediate stages and the mature bnAbs, our panning approach could also be used to inform immunogen design for sequential immunization approaches.43,44
Although not specifically addressed within this study, our method may also be useful for the rapid epitope mapping of newly discovered and yet uncharacterized antibodies. Accordingly, alanine scanning Env libraries substituting surface-exposed residues or glycan scanning libraries with modified N-glycosylation sites or patterns may be screened to establish the footprints of mature bnAbs and their inferred germline versions.
Once identified via the proposed mammalian cell surface display, selected Env variants could be either delivered as a DNA vaccine via nonviral or viral vectors as membrane-bound protein or as purified soluble gp140 Env trimers. Herein, we demonstrate, for the set of analyzed envelope chimeras, that phenotypes of the chimeric membrane-bound Env/V3 library can successfully be transferred to soluble gp140 variants regarding the overall order of binding affinity to 447–52D and HGN194. The apparent magnitudes of KD values varied between the different applied analysis methods (flow cytometry or solid-phase binding ELISA), most probably due to completely different specific inherent conditions of the 2 methods. In sum, this panning approach could also be used for the identification of soluble trimeric vaccine candidates. However, antigenicity profiles might not always be easily transferable from membrane-bound to soluble variants, for example when epitopes are located within the membrane-proximal region of gp41. Therefore, antigenicity profiles for soluble variants would have to be evaluated individually for immunogens identified by this approach.
In conclusion, we provide a methodology for a mammalian cell-based display and selection system that uses antibody-mediated probing of desirable properties of complete trimeric HIV envelopes. The limitations regarding library sizes to be screened in one step and the subsequent resolution need to be explored in greater detail. Based on the results, it is, however, fair to assume that the technology will find a broad range of applications possibly also beyond HIV, for any immunogen design effort where bnAbs are exploited to improve the design of envelopes, like influenza, hepatitis C virus or pathogens such as CMV, where the gH-pentameric complex needs to be properly presented to bind and induce high quality bnAbs.32
Materials and methods
Construction of pQL13
The 2 Esp3I sites contained in the starting construct pcDNA5/FRT/TO (Invitrogen/ThermoFisher Scientific, # V6520–20) were deleted by standard molecular biology methods and the plasmid was linearized with BamHI/XhoI. Ligation with the described previously eGFP (GenBank # U55763) / p2A/ CcdB expression-cassette resulted in the final construct pQL13,45,46,29 which was further propagated in the CcdB resistant E. coli strain DB3.1.
Construction of the chimeric Env/V3 model library
The previously described Env/V3 model library consisted of 5 variants with a 96ZM651 (GenBank, # AF286224) backbone and differing V3 regions (MN, RF, CDC42, HXB2, SF33).29,47 These Env chimeras were seamlessly cloned into pQL13 via golden gate cloning as described previously,29 and verified by Sanger sequencing and restriction analysis.
Construction of gp140 variants
Gp140 constructs were obtained by PCR-amplification from the pQL13-gp145 Env/V3 plasmids. The REKR furin cleavage site of the gp145 variants was mutated to cleavage incompetent REKS via overlap extension PCR and the transmembrane domain was deleted using suitable primers. Gp140 constructs were re-cloned into pQL13 using golden gate cloning (i.e., using type IIS restriction endonucleases) and verified by Sanger sequencing.29
Generation of stable cell lines using Flp-In T-REx 293 cells
Flp-In™ T-Rex™ cells (Invitrogen/ThermoFisher Scientific, # K6500–01) were used to generate stable cell lines with inducible envelope expression regulated under the Tet operator/repressor system. Cells were cultivated in DMEM supplemented with 10% fetal bovine serum (FBS) (tetracycline free, Biochrom, # S0115), 1% Pen/Strep, 100 µg/ml zeocin and 15 µg/ml blasticidin (DMEM+) and seeded to reach 80% confluency on the day of transfection. For this, 5 × 105 Flp-In™ T-Rex™ cells were seeded into 6-well plates the day before transfection. Directly before transfection, the medium was replaced with 1 ml DMEM without supplements (DMEM−). 2 µg of a plasmid mixture containing the pQL13-Env/V3 variants and the helper plasmid pOG44 carrying the integrase (1.6 µg pQL13-Env/V3 variant + 0.4 µg pOG44) were mixed with 8 µl polyethylenimine (Polysciences, # 23966–2, 1 mg/mL) and 100 µl DMEM−. The mixture was incubated for 10 min at room temperature (RT) and then added to the cell culture. After 6 h of incubation, medium was replaced by 2 ml DMEM+. After 48 h, cells were harvested and seeded into a 10 cm dish. After an additional 5 h, the medium was replaced by 10 ml DMEM supplemented with 10% FBS, 1% Pen/Strep, 100 µg/ml hygromycin and 15 µg/ml blasticidin to select cells with stable integration events. Over the following 20–25 d the medium was replaced every 72 h. Then cells were detached and seeded into a 15 cm dish. Established stable cell lines were tested for GFP and Env expression via flow cytometry. Both stable cell lines from an even plasmid mixture of all 5 variants as well as a stable cell line for each variant individually were generated.
TaqMan copy number assay
Genomic DNA was prepared from all established cell lines (Qiagen, Qiaamp DNA Blool Mini Kit, # 51104) and samples were processed with TaqMan® Universal PCR Master Mix (Life technologies/ThermoFisher Scientific, # 4304437). The TaqMan® Copy Number Assays for Markers & Reporters (Life technologies/ThermoFisher Scientific, # 4400291) was used to determine the copy number of integrated pQL13 plasmids in all established stable cell lines. The TaqMan® Copy Number Reference Assay, TERT, Human (Life technologies/ThermoFisher Scientific, # 4403316) was included to compare the number of insertions per cell to a genomic reference (results corrected for 2 copies of human TERT). Analysis was done in a StepOne Plus cycler (Applied Biosystems/ThermoFisher Scientific) according to the manufacturer's instructions. Results were analyzed for statistically significant differences between the 6 cell lines using an unpaired 2-sided t-test.
Flow cytometry analysis of the individual stable Env/V3 cell lines
The stable cell lines were activated to express GFP and Env by supplementing the medium with 1 µg/ml doxycycline. After 24 h, cells were detached with PBE (PBS + 0.5% FBS, 2 mM ethylenediaminetetraacetic acid (EDTA)) and centrifuged at 250 g for 5 min at 4 °C. Then cells were incubated with serial dilutions of 447–52D (Polymun, Klosterneuburg, Austria, # AB014) or HGN194 (kindly provided by Prof. Dr. Antonio Lanzaveccia and Dr. Davide Corti) diluted in PBE for 20 min at 4°C to reach equilibrium. Subsequently, cells were washed 3 times with 1 ml ice cold PBE (centrifugation at 250 g, 5 min, 4°C). The secondary antibody, goat anti-human-IgG- allophycocyanin (APC; Jackson ImmunoResearch, # 109–136–098), diluted in PBE was applied for 1 h at 4°C. After another 3 wash cycles with 1 ml ice cold PBE each (centrifugation at 250 g, 5 min, 4°C), cells were resuspended in PBE and subjected to cytometric analysis using a FACS Canto II (Becton Dickinson). GFP expression (MFI), which was coupled to Env expression via the TaV 2A peptide sequence, was used to calculate normalized Env expression signals (MFI) when determining the affinity of 447–52D and HGN194 to the different Env chimeras. To this end, the quotient of the MFI of APC (Env) and the MFI of GFP (expression control) was calculated, resulting in a relative MFI value.
After subtracting the negative controls (Env-negative 293 cells titrated with the same antibody concentrations) to adjust the starting point of the curves to 0, curves were fitted using non-linear least squares regression (hyberbolar one site binding, GraphPad Prism 5), resulting in apparent KD-values for the membrane-bound envelope variants. To analyze the ratio of Env and GFP expression in each stable cell line, cells were stained with 5F3 antibody (Polymun, Klosterneuburg, Austria, # AB010) as described above,36 and the correlation between GFP and APC was analyzed for all individual FlpIn HEK293 cell lines.
Flow cytometry-based panning: Affinity enrichment and isolation of Env/V3 variants
3 × 107 cells of the Env/V3 cell library generated with an equimolar mixture of all 5 Env/V3 plasmids were seeded into a 15 cm dish and activated with doxycycline (1 µg/ml) the day before the panning procedure. After 24 hours, cells were prepared according to the protocol described in the previous section and were finally resuspended in PBE at a maximal concentration of 6 × 107 cells/ml and filtered with a 30 µm pre-separation filter (Miltenyi Biotech, # 130–041–407). 50 µl of the sample were put aside to serve as an input control. Cells were sorted using a FACS Aria IIu instrument (Becton Dickinson). The instrument was set to “single cell mode” to discard 2-target-events and obtain the most accurate counts for the sorting procedure.48 At least 20,000 cells were collected in each gate. The genomic DNA of both the sorted cells and the input sample were prepared using the Qiaamp DNA Blood Mini kit (Qiagen, # 51104) according to manufacturer's instructions, but without addition of carrier DNA and elution in only 20 µl 10 mM Tris/HCl pH 8.0. The genomic DNA was then used as a template for nested-PCR to amplify the integrated Env genes isolated in the sorting procedure for re-cloning into pQL13 and analysis by qPCR or capillary sequencing or directly subjected to NGS.
Enrichment analysis using Realtime PCR (qPCR)
To determine the distribution of Env variants in a mixture of plasmid DNA, a qPCR reaction was used as described previously.29 Briefly, the reverse-primers were designed to specifically bind one variant of the V3 model library, whereas the probe and forward-primer were designed to bind every variant alike. Furthermore, an additional reverse-primer binding every variant alike was used as a reference primer, determining the total amounts of the applied Env genes. The reactions were performed according to the manufacturer's instructions (Finnzymes, # DyNAmo F-455).
Sequencing of single clones
DNA preparation and Sanger sequencing of single clones was conducted by GATC Biotech.
NGS
NGS was performed on input and first round samples using the Illumina MiSeq platform on both the envelope genes amplified from the gDNA and cloned back into pQL13, as well as directly on the genomic DNA of the selected cells. In both cases, Env/V3 loops were amplified in a 2-step PCR reaction to first add short universal adaptor sequences on the 5´ and 3´ ends, and then add barcoded adaptor sequences according to TruSeq® Small RNA Sample Prep Kits (Illumina). PCR products were purified using Agencourt AMPure XP magnetic beads (Beckman Coulter, # A63881) after each PCR reaction. Quality control and quantification of PCR products was performed with a Bioanalyzer 2100 (Agilent) and High Sensitivity DNA Chips (Agilent, # 5067–4626). After quantification, samples were pooled and analyzed on a 300 cycles MiSeq V2 chip (Illumina, # MS-102–2002).
Purification of soluble gp140 Env variants
His-tagged soluble gp140 proteins were purified from FreeStyle™ 293-F Cells (Life Technologies, # R79007). Cells were cultivated according to manufacturer's instructions in FreeStyle™ 293 Expression Medium (Life Technologies, # 12338018) supplemented with 0.5% Pen/Strep. Before transfection, 9 × 108 cells were harvested at 100 g, 5 min and 4°C and resuspended in 900 ml Freestyle medium without supplements. 900 µg of pQL13-Env/V3 gp140 were mixed with 6.5 ml DMEM without supplements. Simultaneously, 3.6 ml polyethylenimine (Polysciences, # 23966–2, 1 mg/ml) were diluted with 6.5 ml DMEM without supplements. Both mixtures were incubated separately for 5 min at RT and then mixed to be incubated for further 20 min at RT before the transfection mixture was added to the cells. After 6 h of incubation, the medium was changed to 900 ml of Freestyle™ 293 Expression Medium supplemented with 0.5% Pen/Strep. On day 5 after transfection, cells were harvested at 100 g, 5 min at 4°C and the secreted gp140 variants were purified from the supernatant. For this, the supernatant was first loaded onto a lectin column (Galanthus nivalis lectin, agarose bound, Biozol, # VEC-AL 1243–5), using PBS, 1 mM EDTA, 1 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) for column equilibration and washing. Protein was eluted with PBS, 1 mM EDTA, 1 mM EGTA, 1 M methyl-α-D-mannopyranoside (Merck Millipore, # 462711–100GM). Secondly, fractions containing Env protein were pooled and concentrated and trimeric protein was purified to homogeneity using a HiPrep 16/60 Sephacryl S-300 HR size-exclusion chromatography column (GE Healthcare, # GE17–1167–01).
ELISA
ELISA plates (Nunc Maxisorp, # 442404) were coated with 1 µg/ml G. nivalis lectin (Sigma, # L8275–5MG) in PBS overnight at 4°C. All wash cycles were conducted with PBS, 0.05% Tween20 using a Tecan HydroFlex ELISA washer. After coating, plates were washed 3 times and gp140 Env variants were added at a concentration of 1 µg/ml in PBS and incubated for 2 h at RT. Then, plates were blocked for 2 h at RT with 5% (w/v) skimmed milk powder, 5% (v/v) FBS (Gibco), 0.1% Tween20 and then washed 6 times, followed by incubation with a dilution series of 447–52D (Polymun, Klosterneuburg, Austria, # AB014) or HGN194 (kindly provided by Prof. Dr. Antonio Lanzaveccia and Dr. Davide Corti) in PBS, 1% bovine serum albumin (w/v) for 1 h at RT to reach equilibrium binding. Unbound antibody was washed away with 10 wash cycles. Bound antibody was labeled with horseradish peroxidase-coupled rabbit anti-human IgG (Agilent, Dako # P021402–2) for 30 min at RT, followed by 10 wash cycles and incubation with TMB-ultra substrate (ThermoFisher Scientific, # 34028). The reaction was stopped with 1 M H2SO4 and absorption at 450 nm was analyzed. After subtracting the negative controls (no primary antibody) to adjust the starting point of the curves to 0, curves were fitted using non-linear least squares regression (hyberbolar one site binding, GraphPad Prism 5).
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Dr. Susan Zolla-Pazner and Dr. Miroslaw K. Gorny, both New York University School of Medicine, who established and characterized the 447–52D antibody. MAb HGN194 was kindly provided by Prof. Dr. Antonio Lanzaveccia and Dr. Davide Corti.We are grateful to the group of Dr. Petra Hoffmann, especially to Jaqueline Igl for their guidance and support in fluorescent cell sorting experiments.This work was supported by the Bill and Melinda Gates Foundation under Grant 38637 (VDC),by the NIH under Grant 6P01AI066287–02 (HIVRAD Subaward Agreement) and by Grant 031B0036A (BMBF, KMU Innovativ) to R.W. respectively.
References
- 1.UNAIDS. UNAIDS Fact Sheet November 2016. [accessed 2017January08]. http://www.unaids.org/en/resources/fact-sheet.
- 2.Karlsson Hedestam GB, Fouchier RA, Phogat S, Burton DR, Sodroski J, Wyatt RT. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat Rev Microbiol. 2008;6(2):143-55. doi: 10.1038/nrmicro1819. PMID:18197170 [DOI] [PubMed] [Google Scholar]
- 3.Ward AB, Wilson IA. Insights into the trimeric HIV-1 envelope glycoprotein structure. Trends Biochem Sci. 2015;40(2):101-7. doi: 10.1016/j.tibs.2014.12.006. PMID:25600289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS. 2014;28(2):163-9. doi: 10.1097/QAD.0000000000000106. PMID:24361678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore PL, Williamson C, Morris L. Virological features associated with the development of broadly neutralizing antibodies to HIV-1. Trends Microbiol. 2015;23(4):204-11. doi: 10.1016/j.tim.2014.12.007. PMID:25572881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wibmer CK, Moore PL, Morris L. HIV broadly neutralizing antibody targets. Curr Opin HIV AIDS. 2015;10(3):135-43. doi: 10.1097/COH.0000000000000153. PMID:25760932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol. 2015;16(6):571-6. doi: 10.1038/ni.3158. PMID:25988889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parren PW, Marx PA, Hessell AJ, Luckay A, Harouse J, Cheng-Mayer C, Moore JP, Burton DR. Antibody Protects Macaques against Vaginal Challenge with a Pathogenic R5 Simian/Human Immunodeficiency Virus at Serum Levels Giving Complete Neutralization In Vitro. J Virol. 2001;75(17):8340-7. doi: 10.1128/JVI.75.17.8340-8347.2001. PMID:11483779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hessell AJ, Rakasz EG, Poignard P, Hangartner L, Landucci G, Forthal DN, Koff WC, Watkins DI, Burton DR. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 2009;5(5):e1000433. doi: 10.1371/journal.ppat.1000433. PMID:19436712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baba TW, Liska V, Hofmann-Lehmann R, Vlasak J, Xu W, Ayehunie S, Cavacini LA, Posner MR, Katinger H, Stiegler G, et al.. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat Med. 2000;6(2):200-6. doi: 10.1038/72309. PMID:10655110 [DOI] [PubMed] [Google Scholar]
- 11.Sagar M, Wu X, Lee S, Overbaugh J. Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J Virol. 2006;80(19):9586-98. doi: 10.1128/JVI.00141-06. PMID:16973562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al.. Antibody neutralization and escape by HIV-1. Nature. 2003;422(6929):307-12. doi: 10.1038/nature01470. PMID:12646921 [DOI] [PubMed] [Google Scholar]
- 13.Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, et al.. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420(6916):678-82. doi: 10.1038/nature01188. PMID:12478295 [DOI] [PubMed] [Google Scholar]
- 14.Lewis G, Finzi A, DeVico A, Pazgier M. Conformational Masking and Receptor-Dependent Unmasking of Highly Conserved Env Epitopes Recognized by Non-Neutralizing Antibodies That Mediate Potent ADCC against HIV-1. Viruses. 2015;7(9):5115-32. doi: 10.3390/v7092856. PMID:26393642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sather DN, Carbonetti S, Malherbe DC, Pissani F, Stuart AB, Hessell AJ, Gray MD, Mikell I, Kalams SA, Haigwood NL, et al.. Emergence of Broadly Neutralizing Antibodies and Viral Coevolution in Two Subjects during the Early Stages of Infection with Human Immunodeficiency Virus Type 1. J Virol. 2014;88(22):12968-81. doi: 10.1128/JVI.01816-14. PMID:25122781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du SX, Xu L, Zhang W, Tang S, Boenig RI, Chen H, Mariano EB, Zwick MB, Parren PW, Burton DR, et al.. A directed molecular evolution approach to improved immunogenicity of the HIV-1 envelope glycoprotein. PLoS One. 2011;6(6):e20927. doi: 10.1371/journal.pone.0020927. PMID:21738594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ching L, Stamatatos L. Alterations in the immunogenic properties of soluble trimeric human immunodeficiency virus type 1 envelope proteins induced by deletion or heterologous substitutions of the V1 loop. J Virol. 2010;84(19):9932-46. doi: 10.1128/JVI.00868-10. PMID:20660181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selvarajah S, Puffer BA, Lee FH, Zhu P, Li Y, Wyatt R, Roux KH, Doms RW, Burton DR. Focused dampening of antibody response to the immunodominant variable loops by engineered soluble gp140. AIDS Res Hum Retroviruses. 2008;24(2):301-14. doi: 10.1089/aid.2007.0158. PMID:18284327 [DOI] [PubMed] [Google Scholar]
- 19.Schiffner T, Kong L, Duncan CJ, Back JW, Benschop JJ, Shen X, Huang PS, Stewart-Jones GB, DeStefano J, Seaman MS, et al.. Immune focusing and enhanced neutralization induced by HIV-1 gp140 chemical cross-linking. J Virol. 2013;87(18):10163-72. doi: 10.1128/JVI.01161-13. PMID:23843636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Peña AT, Korzun J, et al.. A Next-Generation Cleaved, Soluble HIV-1 Env Trimer, BG505 SOSIP.664 gp140, Expresses Multiple Epitopes for Broadly Neutralizing but Not Non-Neutralizing Antibodies. PLoS Pathog. 2013;9(9):e1003618. doi: 10.1371/journal.ppat.1003618. PMID:24068931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pugach P, Ozorowski G, Cupo A, Ringe R, Yasmeen A, de Val N, Derking R, Kim HJ, Korzun J, Golabek M, et al.. A native-like SOSIP.664 trimer based on an HIV-1 subtype B env gene. J Virol. 2015;89(6):3380-95. doi: 10.1128/JVI.03473-14. PMID:25589637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma SK, deVal N, Bale S, Guenaga J, Tran K, Feng Y, Dubrovskaya V, Ward AB, Wyatt RT. Cleavage-Independent HIV-1 Env Trimers Engineered as Soluble Native Spike Mimetics for Vaccine Design. Cell Rep. 2015;11(4):539-50. doi: 10.1016/j.celrep.2015.03.047. PMID:25892233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, et al.. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349(6244):aac4223. doi: 10.1126/science.aac4223. PMID:26089353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng Y, Tran K, Bale S, Kumar S, Guenaga J, Wilson R, de Val N, Arendt H, DeStefano J, Ward AB, et al.. Thermostability of Well-Ordered HIV Spikes Correlates with the Elicitation of Autologous Tier 2 Neutralizing Antibodies. PLoS Pathog. 2016;12(8):e1005767. doi: 10.1371/journal.ppat.1005767. PMID:27487086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malherbe DC, Doria-Rose NA, Misher L, Beckett T, Puryear W, Schuman JT, Kraft Z, O'Malley J, Mori M, Srivastava I, et al.. Sequential immunization with a subtype B HIV-1 envelope quasispecies partially mimics the in vivo development of neutralizing antibodies. J Virol. 2011;85(11):5262-74. doi: 10.1128/JVI.02419-10. PMID:21430056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dosenovic P, Von Boehmer L, Escolano A, Jardine J, Freund NT, Gitlin AD, McGuire AT, Kulp DW, Oliveira T, Scharf L, et al.. Immunization for HIV-1 Broadly Neutralizing Antibodies in Human Ig Knockin Mice. Cell. 2015;161(7):1505-15. doi: 10.1016/j.cell.2015.06.003. PMID:26091035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Escolano A, Steichen JM, Dosenovic P, Kulp DW, Golijanin J, Sok D, Freund NT, Gitlin AD, Oliveira T, Araki T, et al.. Sequential Immunization Elicits Broadly Neutralizing Anti-HIV-1 Antibodies in Ig Knockin Mice. Cell. 2016;166(6):1445-58. doi: 10.1016/j.cell.2016.07.030. PMID:27610569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Briney B, Sok D, Jardine JG, Nemazee D, Burton DR, Schief WR, Briney B, Sok D, Jardine JG, Kulp DW, et al.. Tailored Immunogens Direct Affinity Maturation toward HIV Neutralizing Antibodies Article Tailored Immunogens Direct Affinity Maturation toward HIV Neutralizing Antibodies. Cell. 2016;166(6):1459-70.e11. doi: 10.1016/j.cell.2016.08.005. PMID:27610570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruun TH, Mühlbauer K, Benen T, Kliche A, Wagner R. A mammalian cell based FACS-panning platform for the selection of HIV-1 envelopes for vaccine development. PLoS One. 2014;9(10):e109196. doi: 10.1371/journal.pone.0109196. PMID:25279768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorny MK, Xu JY, Karwowska S, Buchbinder A, Zolla-Pazner S. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J Immunol. 1993;150(2):635-43. PMID:7678279 [PubMed] [Google Scholar]
- 31.Corti D, Langedijk JP, Hinz A, Seaman MS, Vanzetta F, Fernandez-Rodriguez BM, Silacci C, Pinna D, Jarrossay D, Balla-Jhagjhoorsingh S, et al.. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One. 2010;5(1):e8805. doi: 10.1371/journal.pone.0008805. PMID:20098712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gardner TJ, Stein KR, Duty JA, Schwarz TM, Noriega VM, Kraus T, Moran TM, Tortorella D. Functional screening for anti-CMV biologics identifies a broadly neutralizing epitope of an essential envelope protein. Nat Commun. 2016;7:13627. doi: 10.1038/ncomms13627. PMID:27966523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, Vignali DA. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22(5):589-94. doi: 10.1038/nbt957. PMID:15064769 [DOI] [PubMed] [Google Scholar]
- 34.Yao F, Svensjö T, Winkler T, Lu M, Eriksson C, Eriksson E. Tetracycline repressor, tetR, rather than the tetR-mammalian cell transcription factor fusion derivatives, regulates inducible gene expression in mammalian cells. Hum Gene Ther. 1998;9(13):1939-50. doi: 10.1089/hum.1998.9.13-1939. PMID:9741432 [DOI] [PubMed] [Google Scholar]
- 35.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89(12):5547-51. doi: 10.1073/pnas.89.12.5547. PMID:1319065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, et al.. Generation of Human Monoclonal Antibodies against HIV-1 Proteins; Electrofusion and Epstein-Barr Virus Transformation for Peripheral Blood Lymphocyte Immortalization. AIDS Res Hum Retroviruses. 1994;10(4):359-69. doi: 10.1089/aid.1994.10.359. PMID:7520721 [DOI] [PubMed] [Google Scholar]
- 37.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hu PY, Moyle M, et al.. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477(7365):466-70. doi: 10.1038/nature10373. PMID:21849977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al.. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326(5950):285-9. doi: 10.1126/science.1178746. PMID:19729618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heyndrickx L, Heath A, Sheik-Khalil E, Alcami J, Bongertz V, Jansson M, Malnat M, Montefiori D, Moog C, Morris L, et al.. International Network for Comparison of HIV Neutralization Assays: The NeutNet Report II. PLoS One. 2012;7(5):e36438. doi: 10.1371/journal.pone.0036438. PMID:22590544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moscoso CG, Sun Y, Poon S, Xing L, Kan E, Martin L, Green D, Lin F, Vahlne AG, Barnett S, et al.. Quaternary structures of HIV Env immunogen exhibit conformational vicissitudes and interface diminution elicited by ligand binding. Proc Natl Acad Sci U S A. 2011;108(15):6091-6. doi: 10.1073/pnas.1016113108. PMID:21444771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, O'Dell S, Walker LM, Wu X, Guenaga J, Feng Y, Schmidt SD, McKee K, Louder MK, Ledgerwood JE, et al.. Mechanism of Neutralization by the Broadly Neutralizing HIV-1 Monoclonal Antibody VRC01. J Virol. 2011;85(17):8954-67. doi: 10.1128/JVI.00754-11. PMID:21715490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, de la Peña AT, Cupo A, Julien JP, van Gils M, Lee PS, et al.. Structural Delineation of a Quaternary, Cleavage-Dependent Epitope at the gp41-gp120 Interface on Intact HIV-1 Env Trimers. Immunity. 2014;40(5):669-80. doi: 10.1016/j.immuni.2014.04.008. PMID:24768348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, Cohen KW, Jardine J, Menis S, Scheid JF, West AP, et al.. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J Exp Med. 2013;210(4):655-63. doi: 10.1084/jem.20122824. PMID:23530120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoot S, McGuire AT, Cohen KW, Strong RK, Hangartner L, Klein F, Diskin R, Scheid JF, Sather DN, Burton DR, et al.. Recombinant HIV Envelope Proteins Fail to Engage Germline Versions of Anti-CD4bs bNAbs. PLoS Pathog. 2013;9(1):e1003106. doi: 10.1371/journal.ppat.1003106. PMID:23300456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, Vignali DA. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22(5):589-94. doi: 10.1038/nbt957. PMID:15064769 [DOI] [PubMed] [Google Scholar]
- 46.Bernard P, Gabant P, Bahassi EM, Couturier M. Positive-selection vectors using the F plasmid ccdB killer gene. Gene. 1994;148(1):71-4. doi: 10.1016/0378-1119(94)90235-6. PMID:7926841 [DOI] [PubMed] [Google Scholar]
- 47.Rodenburg CM, Li Y, Trask SA, Chen Y, Decker J, Robertson DL, Kalish ML, Shaw GM, Allen S, Hahn BH, et al.. Near full-length clones and reference sequences for subtype C isolates of HIV type 1 from three different continents. AIDS Res Hum Retroviruses. 2001;17(2):161-8. doi: 10.1089/08892220150217247. PMID:11177395 [DOI] [PubMed] [Google Scholar]
- 48.Shapiro HM. Practical Flow Cytometry. Hoboken: (NJ: ), Wiley:2005. [Google Scholar]
- 49.Khare PD, Rosales AG, Bailey KR, Russell SJ, Federspiel MJ. Epitope selection from an uncensored peptide library displayed on avian leukosis virus. Virology. 2003;315(2):313-21. doi: 10.1016/S0042-6822(03)00530-0. PMID:14585334 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.