Abstract
Single-molecule and single-nanoparticle biosensors are a growing frontier in diagnostics. Digital biosensors are those which enumerate all specifically immobilized biomolecules or biological nanoparticles, and thereby achieve limits of detection usually beyond the reach of ensemble measurements. Here we review modern optical techniques for single nanoparticle detection and describe the single-particle interferometric reflectance imaging sensor (SP-IRIS). We present challenges associated with reliably detecting faint nanoparticles with SP-IRIS, and describe image acquisition processes and software modifications to address them. Specifically, we describe a image acquisition processing method for the discrimination and accurate counting of nanoparticles that greatly reduces both the number of false positives and false negatives. These engineering improvements are critical steps in the translation of SP-IRIS towards applications in medical diagnostics.
Index Terms: Optical biosensing, digital detection, nanoparticle imaging, interferometry, single particle detection
I. Introduction
The development of optical tools for detection and characterization of nanoparticles will impact a broad range of disciplines in biological research from nanomedicine to nanotoxicology [1]. Single-molecule counting or digital detection provides resolution and sensitivity beyond the reach of ensemble measurements. The impressive capabilities of digital detection schemes have led to desire for the translation of these techniques into clinically useful applications [2]. In order to realize the diagnostics potential, future research efforts should focus on the development of practical systems with infrastructural requirements better aligned with the functional realities of a clinical environment.
In vitro detection of nucleic acid and protein biomarkers are an indispensable component of modern clinical practice. The sub-wavelength size scale of these biomolecules makes direct detection through traditional microscopy methods extremely challenging. As such, modern gold standards rely upon amplification methods to generate a detectable signal that scales with analyte concentration. Recent advancements in automated diagnostic platforms based on polymerase chain reaction (PCR) and enzyme-linked immunosorbent assays (ELISA) have enabled the routine detection of trace levels of nucleic acid and protein biomarker. Additionally, parallel research efforts in sample pre-concentration techniques have shown further potential for enhancement of traditional assays [3]. While these techniques achieve impressive results, all rely heavily upon a complicated sequences of sample preparation and amplification processes that limits their effectiveness outside of well-equipped laboratory environments [4]. On the other hand, rapid and point-of-care (POC) testing is commonly performed with lateral-flow style Rapid Diagnostic Tests (RDTs), which achieve qualitative biomarker detection in a robust and easy to use format [5].
Digital detectors offer the potential to fill the diagnostic gap between ultrasensitive molecular amplification tests and qualitative POC tests, by promising both direct and sensitive measurement of health biomarkers. The single-particle interferometric reflectance imaging sensor (SP-IRIS) is one such technique, which can enumerate individual nanoparticles immobilized onto a very flat thin film reflecting substrate [6]. The SP-IRIS instrument is a simple reflectance microscope (Figure 1), in which partially-coherent light shines down onto substrate and is strongly reflected. The faint light scattered by a nanoparticle on the substrate is observed as a small, diffraction-limited perturbation of the reflected light, due to interference. SP-IRIS has been used to directly detect Ebola- and Marburg-pseudotyped vesicular stomatitis virus in serum and whole blood [7] [8] without sample preparation. It has also been used to perform highly-sensitive detection of single molecules via functionalized gold nanoparticle labels which are also individually counted [9].
Figure 1.
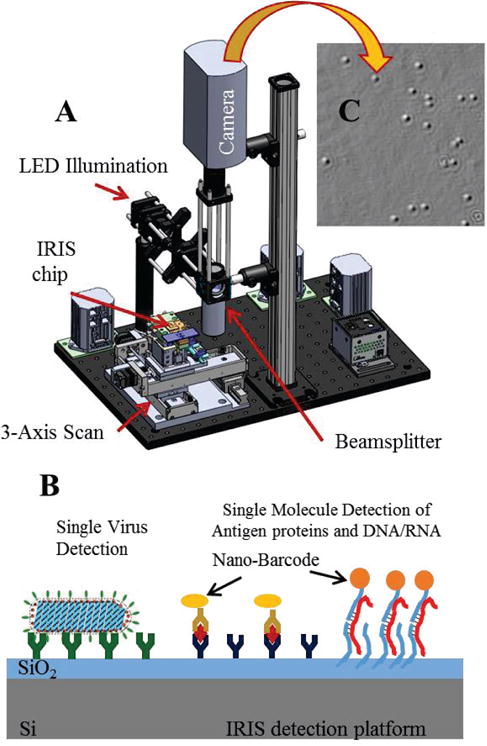
SP-IRIS detection platform. a) optical setup which consist of LED lighting module, imaging objective (50× .8NA) and CCD imaging camera. b) Illustration of SP-IRIS sensor utilized for protein and nucleic acid detection. c) A sample image showing response from individual nanoparticles.
Despite the potential for very high sensitivity demonstrated by SP-IRIS, the robustness of the image acquisition and particle-counting software has been a major challenge. In this paper, we first describe the problem of robustly detecting dim nanoparticles in SP-IRIS images. We then present software and process improvements to SP-IRIS that have improved the sensor’s overall performance thru a significant reduction in the rate of false positives. Many of these improvements have broader relevance to signal processing in imaging biosensors and single-particle detectors.
II. Optical Imaging Techniques for non-Fluorescent Nano-Particle Characterization
Conventional light scattering microscopy cannot detect features that are significantly smaller than the wavelength of illumination. Very small scatterers are blurred by the characteristic point spread function (PSF) of the microscope lenses and illumination, the intensity of which is a very strong function of particle size: quasi-static scattering theory relates the strength of the induced dipole to the polarizability of the particle α as
where r is the particle radius, εp is the particle permittivity, and εm is the surrounding medium permittivity. The observed intensity at the detector scales with the square of the scattered field, resulting in an r6 signal scaling that rapidly drops below the background noise for small nanoparticles. While fluorescence labeling techniques have been successfully employed to increase both the sensitivity and the resolution of the optical microscope [9, 10], persistent issues with photobleaching and nonspecific binding to complex media components present significant obstacles. Furthermore, variability of fluorescence signal masks the information related to the physical size of the biological nanoparticles.
There are several approaches to overcome the difficulties in detection and visualization of non-fluorescent nanoparticles. We focus on methods for optical detection of nanoparticles captured on a solid surface. Here, we briefly review three of the most common methods: (i) optical resonators (such as whispering gallery mode devices), (ii) dark-field or evanescent excitation imaging, and (iii) interferometric enhancement. Resonance is a very powerful technique to enhance weak optical interactions. Since the optical scattering from a nanoparticle is vanishingly small, a promising way to increase the effective interaction is using resonant microcavity structures in which the light samples a nanoparticle many times before being detected [12], [13]. In this case, light circulates in the optical sensors formed by guided-wave devices coupled to resonant structures (such as rings, spheres or disks) and establishes whispering gallery modes (WGM). Nanoparticles captured on the surface of the resonator interact with the evanescent tail of the optical wave and perturbs the resonance behavior of WGMs in the cavity. Using high-Q (quality factor) WGM devices detection of nanoparticles such as viruses and single molecules has been demonstrated [13], [14]. Despite significant advances and exquisite sensitivity, these devices have limited impact on biological detection of nanoparticles. One of the critical limitations is low throughput capacity. More importantly, these type of sensor devices work very well in pure solutions but their sensitivity is significantly hampered in complex biological solutions such as serum or whole blood. Finally, the small active sensor area that is characteristic of such resonators means that their concentration detection limit is strongly limited by diffusion [15].
An interesting single nanoparticle detection method combines resonant enhancement utilizing a photonic crystal sensor surface with imaging. A one-dimensional photonic crystal (PC) surface was utilized to detect surface attachment of individual dielectric and metal nanoparticles without fluorescence through measurement of localized shifts in the resonant wavelength and resonant reflection magnitude [16].
We review the (ii) dark field and (iii) interferometric detection methods in more detail below. These methods have the potential of large area imaging and high throughput as well as applicability in detection of nanoparticles in complex solutions allowing for applications in in vitro diagnostics.
A. Nanoparticle Detection with Evanescent Illumination
Dark-field illumination methods enable detection of directly scattered light through suppression of background signal. Most methods used total internal reflection style illumination to generate scattering with an evanescent field interacting with nanoparticles captured on the surface. Because this field decays exponentially, only the immediate vicinity of the sensor surface is interrogated, and the illuminating field does not propagate to the detector. Compared to standard epi-illumination microscopy utilizing dark-field objectives, total-internal reflection and evanescent excitation provide better suppression of the background.
Total-internal reflection (TIR) illumination has been studied extensively in the context of TIR fluorescence (TIRF) microscopy that images fluorescently labeled biological entities close to the interface and it can observe binding of single chromophores [17]. Utilizing similar excitation schemes and imaging scattered intensity from surface bound nanoparticles allows for direct imaging of ~100nm for dielectric particles and viruses [18] as well as single ~40nm Au nanoparticles [19]. Since scattering intensity scales with approximately r6 these techniques are very sensitive to particle size.
An alternative to prism- and objective-based TIR illumination is guided-wave illumination that provides a highly-controlled evanescent field interacting with nanoparticles on the surface [20]. Guided-wave evanescent excitation has several advantages over conventional TIR illumination including tighter confinement and uniformity of the excitation field over large areas. Waveguide-based dark-field illumination has been utilized for simultaneous observation of scattered and fluorescent light from multiple surface-associated nanoscopic objects using standard microscopes [21].
A contrasting example for evanescent excitation and imaging is Surface Plasmon Resonance Microscopy (SPRM) which operates in bright-field. The resonant excitation of the surface plasmon waves is sensitive to the local refractive index in the vicinity of the metal layer, thus binding of a nanoparticle on the metal surface provides a discernible signature on the spatial image obtained on the array detector. Imaging and detection of sub-100nm silica nanoparticles and single H1N1 viruses have been demonstrated by SPRM [22].
B. Nanoparticle Visualization with Interferometric Imaging
In contrast to dark-field microscopy relying on suppression of the background light, interferometric methods utilize a strong reference field that is interfered with scattered light to enhance the visibility of nanoparticles. In a simplistic manner, the observed intensity can be expressed as:
where θ represents the relative phase angle between the scattered and reference fields. The first term, |Er|2 is the observed intensity of the reference field. The second term, |Es|2 has a very strong size dependence (r6) and is negligibly small for particles significantly smaller than the illumination wavelength. The third cross term has a weaker size dependence (r3) and can be much greater than the purely scattered light for small nanoparticles. This basic concept applies to heterodyne and homodyne interferometric detection techniques. Interferometric microscopy method has demonstrated detection of gold nanoparticles as small as 5nm [23], viruses [24], [25], and even individual unlabeled proteins [26] in laboratory environments and pure sample solutions.
Our optical imaging technique, known as the single-particle interferometric reflectance imaging sensor (SP-IRIS) visualizes single nanoscale particles by utilizing a common-path interferometric enhancement [6]. Optical scattering from individual nanoscale particles are enhanced by the layered dielectric surface acting as an optical antenna. The interference of reference light reflected from the sensor surface with the scattered field produces a distinct signal that reveals the size of the particle. As we describe below in detail, SP-IRIS is capable of detecting dielectric/biological nanoparticles with diameters down to 60nm diameter and gold nanoparticles down to 40nm diameter. These gold nanoparticles are conjugated to secondary recognition probes, and used as labels, allowing protein or DNA assays with single-molecule readout [9], [27]. Below, we describe the evolution of the SP-IRIS technology, review the difficulties in accurate sizing and discrimination in real-life applications, and present a practical and robust image acquisition and processing method.
III. Single-Particle Interferometric Reflectance Imaging Sensor (SP-IRIS)
The Interferometric Reflectance Imaging Sensor (IRIS) is a low-cost, compact and simple to use biosensing platform developed at Boston University. IRIS has demonstrated highthroughput detection and quantification of protein-protein binding, DNA-protein binding and DNA-DNA hybridization in real-time with high sensitivity and reproducibility [6], [28]. Recent significant advancements in IRIS technology have allowed us to identify individual captured nanoparticles through correlation of the observed interference patterns intensity with analyte size and shape. This new modality of IRIS is termed single-particle IRIS (SP-IRIS). SP-IRIS, as illustrated in Fig. 1, shines light from an LED source on nanoparticles bound to the sensor surface, which consists of a silicon dioxide layer on top of a silicon substrate (Fig. 1b). Interference of light reflected from the sensor surface is modified by the presence of particles producing a distinct signal that is captured by a conventional CCD camera. This appears as a dot on the image (Fig. 1c), the peak intensity of with is correlated to the size of the particle using a forward model. Size discrimination reduces the noise from nonspecifically bound particles. In an SP-IRIS image, as many as a million distinct nanoparticles can be simultaneously detected. SP-IRIS relies on efficient collection of scattered light from nanoparticles and thus requires high magnification (50×) and high numerical aperture (0.8), which limits the field of view to less than 0.3 mm × 0.3 mm using conventional CCD cameras. For DNA arrays with a 100 μm pitch, as many as about 10 spots can be imaged at once. To interrogate larger arrays, consecutive images are taken to cover the entire IRIS sensor using an automated scanning stage. SP-IRIS can operate in either a labeled or label-free modality, as some analytes (viruses) can be directly visualized (Figure 1b).
For smaller analytes (such as individual nucleic acid or protein biomarkers), single molecule sensitivity is achieved by secondary functionalization of targets captured on the surface with a small gold or other metallic nanoparticle. Labeling secondary probes with a particle commonly referred to as “mass-tagging” is a prevalent method to enhance sensitivity. Mass-tagging approaches employed with conventional microscopy require at particles on a size scale of hundreds of microns, resulting in limitations due to diffusion and steric hindrance of the secondary tag [15], [29]. In contrast, SP-IRIS can measure the shape and size of individual nanoparticles as small as 20 nm, which is only about twice the hydrodynamic diameter of an antibody, allowing this information to serve as an identifier of the biomolecule attached to the nanoparticle. We term this identifying feature as a “nano-barcode”. Nano-barcode based detection also improves assay specificity, because the nano-barcode has to match the probe it is binding over to be considered a positive binding event.
A. Advancement of SP-IRIS technology
In reviewing the evolution of the technology, we illustrate various difficulties in building a robust system for accurate sizing and discrimination of nanoparticles. SP-IRIS was first introduced in 2010 for high-throughput detection and sizing of individual low-index nanoparticles and viruses for pathogen identification [7]. Size discrimination of nanoparticles with diameters of 70, 100, 150, and 200 nm using an oxide on silicon substrate in a wide-field, reflected-mode microscope was demonstrated. For a particular oxide thickness and illumination wavelength, the observed intensity of a nanoparticle on the surface has a specific size dependence. The images acquired and supporting numerical simulations were conducted for a single focal plane coinciding with the oxide-silicon interface. The successful demonstration of size discrimination was enabled by two factors: (i) the measurements were done in dry conditions and (ii) the samples were prepared by directly depositing the polystyrene and viral nanoparticles on the surface rather than a biologically relevant capture using immobilized probes. Even though these factors allowed for reasonable assumptions concerning the axial location of nanoparticles with respect to the surface, the authors noted that the varying axial position of nanosphere centroids with increasing radius resulted in a single-wavelength sizing curve that became double-valued for larger particles. The difficulty of focusing on a layered reflecting surface was noted and future corrections were suggested utilizing axial scans and fitting the oscillation in phase to the forward model at peak response illustrated by a numerical study [30] and shown in Fig. 2.
Figure 2.
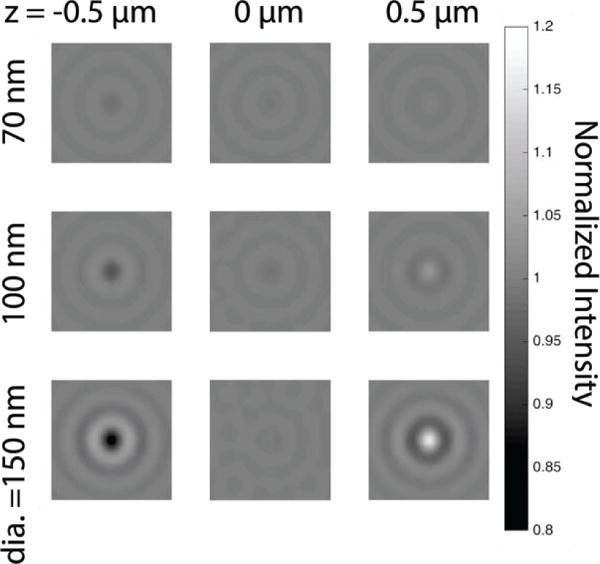
Simulated images of 70 nm (top), 100 nm (middle) and 150 nm (bottom) diameter polystyrene (n=1.60) nanoparticles resting on a 30nm SiO2 IRIS substrate, at three different focus positions with respect to the water-film interface. NA=0.9, water immersion, λ = 525 nm.)
As the SP-IRIS technology evolved, it was applied to direct label-free capture and characterization of viruses from complex media such as blood or serum. Affinity-based capture, size discrimination, and a “digital” detection scheme to count single viruses, yielded a multiplexed and sensitive virus sensing assay [7]. These experiments were conducted on dry samples after viruses were captured on SP-IRIS chips from serum or whole blood contaminated with high levels of bacteria. Size discrimination proved very valuable to reduce the background noise since the antibody surfaces have inherent roughness, and non-specific binding of biological particles in complex solutions can be significant. By combining the advantages of SP-IRIS, with microfluidics, led to real-time digital detection of individual viruses as they bind to an antibody microarray [8]. In liquid, the index ratio between the particle to the surrounding medium is reduced, resulting in a 3-fold reduction compared to dry measurements. Furthermore, the captured viral particles may have an axial position distribution, especially when elevated using immobilized probes with flexible tethers for improved capture efficiency [31].
B. Rigorous modeling of the interferometric signal
SP-IRIS uses Köhler illumination, in which the Fourier plane of a partially coherent LED source is imaged onto the sample plane. Each spatial position (x,y) on the emitter produces a plane wave incident on the sample at corresponding angle (kx, ky). Since each of these components corresponds to an independent photon emission event, these illumination components do not interfere. Each plane wave component of the incident light interacts with the sample independently, resulting in a scattered field from the particle and reflected field from the surface, which have a path length difference less than the temporal coherent length (usu. 10–30 microns) and interfere at the detector. The resulting intensity from this illumination component is added incoherently to the others, resulting in the final observed image. Note that the nanoparticle can be considered as a dipole scatterer since its size is much smaller than the illumination wavelength. The theoretical foundations of SP-IRIS with an emphasis on its key parameters that influence the signal have been studied by our group in [32], and a physical model utilizing the angular spectrum representation [33] was realized based on this study. The calculated intensities for all the driving field components are summed to get the overall interferometric signal.
Many parameters effect the optical visibility of nanoparticles on an IRIS substrate, including the particle size and elevation above the interface as well as immersion medium, collection NA and illumination wavelength. When illuminated by a single plane wave of normal incidence, light scattered by the nanoparticle in the forwards direction (into the substrate) is reflected and interferes with back-scattered light. This self-interference of the scattered fields is stronger in particular directions, depending on the optical path length difference between forward-and back-scattering. Increasing the particle size, or elevation from the Si-SiO2 interface, increases the height of the effective dipole, and shifts back-scattering towards higher angles. Furthermore, unlike a Michaelson interferometer for example, the path length of the reference reflection from the Si-SiO2 interface cannot be dynamically adjusted. Nanoparticles for which the substrate is optimal manifest a single strong positive (or negative) peak in their defocus curves (e.g. Figure 3). Larger, smaller or elevated particles will deviate from this however: at the extreme, they will be in quadrature with the reference field, and therefore exhibit smaller negative and positive peaks of approximately equal magnitude (e.g. Figures 4–5). In summary, changing the particle position not only shifts the scattering radiation pattern towards higher angles but also processes the phase of the scattered fields with respect to the reflected field in a predictable manner. This understanding has motivated the design of improved acquisition and image processing filters, discussed in later sections.
Figure 3.
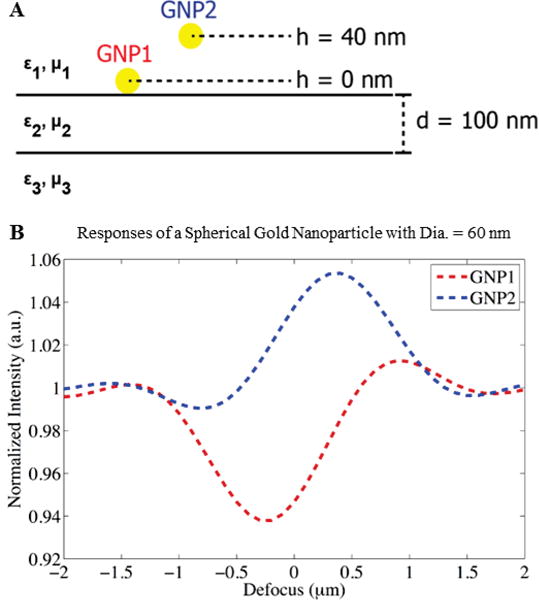
Spherical gold nanoparticles (Dia. = 60 nm) at (a) h = 0 nm (GNP1) and h = 40 nm (GNP2), and (b) their interferometric responses (GNP1 shown in red, and GNP2 shown in blue). Adapted from [32].
Figure 4.
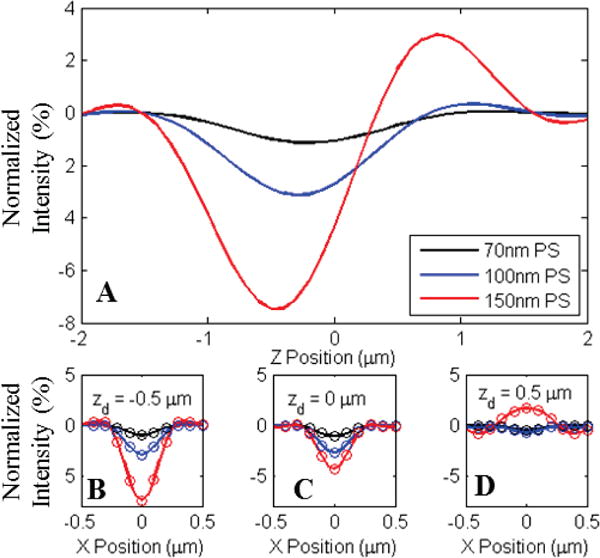
A) Normalized intensity of center pixel for three sizes of polystyrene nanospheres bound to a 30nm oxide on silicon substrate. Significant changes in appearance and defocus behavior are observed due to variation in z-axis position of the radiating dipole with respect to the reference field generated by the reflective surface. B–D) Line profiles of the observed appearance for the three nanospheres simulated for three focal plane offsets (−.5, 0, and +.5 μm) with respect to the oxide surface. Simulations were generated for an .9NA in-water imaging system. Circles in plots B–D represent pixel sampling locations for a 2 μm pixel pitch with 40× magnification.
Figure 5.
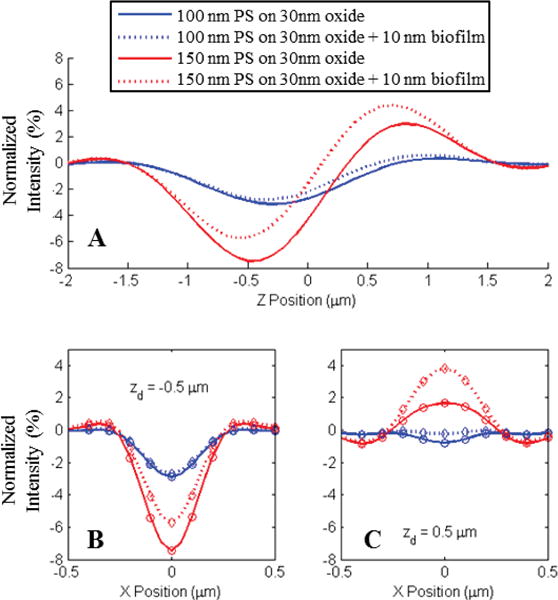
A) Shift in the normalized intensity of center pixels for two sizes of polystyrene spheres (100nm and 150nm) due to the presence of a 10 nm biofilm of capture probes. B–C) Line profiles of the observed appearance for the three nanospheres simulated for two focal plane offsets (−.5 and +.5 μm) with respect to the oxide surface. Simulations were generated for an .9NA in-water imaging system. Circles in plots B and C represent pixel sampling locations for a 2 μm pixel pitch with 40× magnification.
IV. Robust Nanoparticle Identification and Discrimination
Although interferometric imaging methods have been well established as a tool for nanoparticle characterization, the translation of these techniques to a diagnostic context will induce a significant paradigm shift in the validity of many assumptions upon which preliminary studies were based. Clinical contexts require that these techniques function as a concentration measurement instead of as a characterization tool. From this perspective, accurate characterization of nanoparticle properties is only relevant to the extent that it enables accurate discrimination of chemically specific binding events from spurious signals resulting from nonspecifically bound scattering objects and morphological variation in immobilized capture probes. While the spatial multiplexing of conditions through micro-arrayed capture probes on solid-phase substrates enables higher level of assay parallelism than is achievable in solution-based assays, variations in spot morphology and immobilization density increase both the prevalence of nonspecific background signal and the expected variation in the axial height of captured nanoparticles with respect to the reflective surface. Although early studies demonstrated the use of SP-IRIs for concentration measurements of unlabeled viral pathogens, these efforts were heavily dependent well optimized probe morphology, manual focal control by a skilled operator, and the ability to assume of homogenous nanoparticle properties and axial locations [7], [8].
Because diagnostically relevant nanoparticles are typically much smaller than the wavelength of illuminating light, their appearance in wide-field images takes the form of radially symmetric regions of alternating positive and negative normalized intensity. The rotationally invariant nature of these interference patterns makes it possible to identify the centroids of these nanoparticle signatures through simple template matching algorithms, provided that the analysis algorithm is capable of creating a sufficiently accurate simulated template. While this process is widely employed to identify point spread functions (PSFs) in conventional fluorescence and bright-field microscopy images, where PSF appearance is determined solely by optical system parameters, the interferometric nature of SP-IRIS measurements produces a much larger parameter space of possible template appearances. Even without the variability induced by changes in nanoparticle axial location, small errors in substrate alignment (on the order of tenths of a degree) can result in heterogeneous defocus behaviors for identical nanoparticles in different regions of a single field of view. Figure 7 shows a raw image of 100 nm PS nanospheres physisorbed onto a 30 nm oxide substrate in water, displaying signature with varying appearance for different regions. External measurements performed during system calibration indicate a misalignment of no more than one quarter of a degree, demonstrating the extremely high calibration diligence required for accurate nanoparticle characterization over a wide field of view using only single-plane images. While preliminary studies were able to minimize this variability through careful alignment and sample quality control, such strict tolerance requirements will not be compatible with widespread use of these techniques by unskilled operators.
Figure 7.
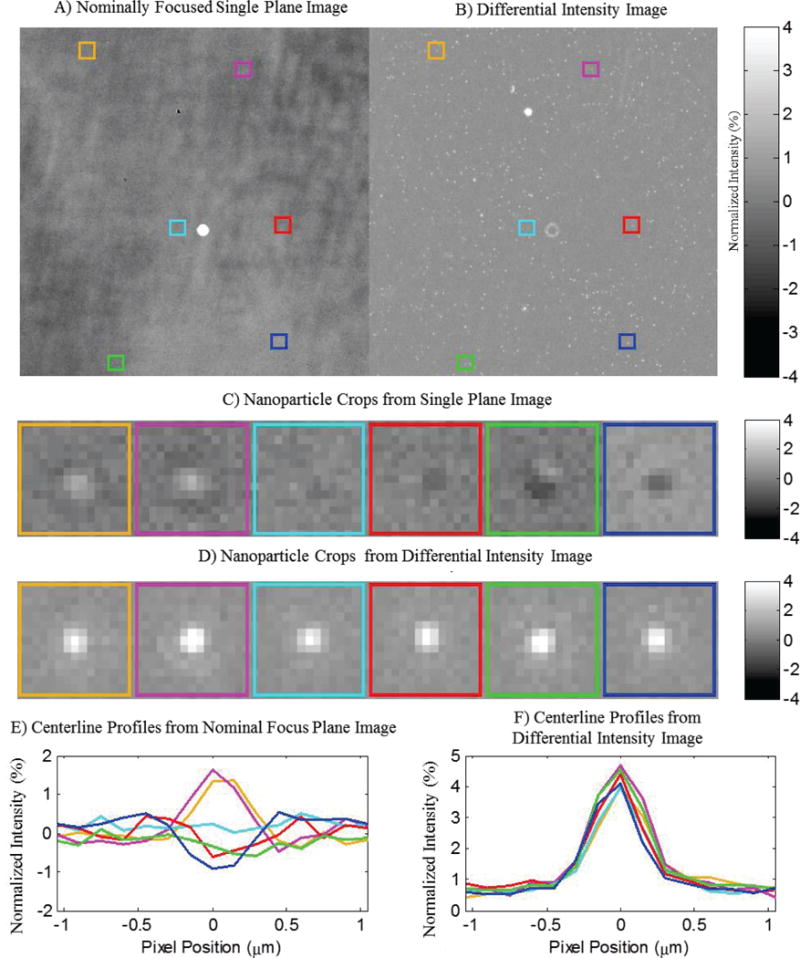
A) 100 nm Polystyrene nanospheres exhibit varying signal levels in a single plane image stemming from morphological variations on the substrate surface. The variations among the nanoparticle signals diminish in the differential normalized intensity image, enabling their identification by template matching. Colored Boxes identify the locations of representative nanoparticles taken from different regions of the field of view. B) Differential intensity image generated from the total peak to peak change in intensity over a 6 μm Z-stack sampled at 200nm increments. C) Crops of the observed appearance of representative nanoparticles from the colored regions in A. D) Differential intensity crops of identical regions from B. E) Centerline profiles of the regions shown in C, displaying varying appearance due to slight sample misalignment. F) Centerline profiles of regions in D, demonstrating consistent signal via differential intensity measurements.
In an effort to overcome the obstacles posed by variable defocus behavior, recent advancements in automated imaging and analysis have shown that the change in intensity experienced by a nanoparticle signature over the extent of a given range of focal planes, or differential intensity, is considerably more predictable than its specific appearance in any single image, as shown through simulations in Figure 6. The nanoparticle response generated by the calculation of differential intensity collapses into a consistent profile regardless of amplitude or defocus behavior, enabling the straightforward identification of nanoparticle locations within an image using simple template matching methods. Furthermore, the amplitude of the differential intensity signature provides a consistent metric for size-based discrimination. Using this concept, we have developed an algorithm for the robust measurement of the concentration of surface-bound nanoparticle populations regardless of heterogeneity in size and axial offset.
Figure 6.
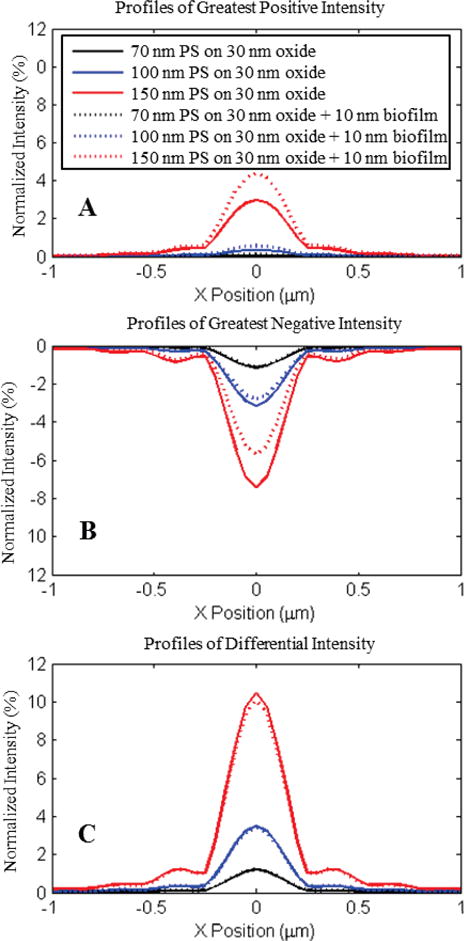
Compressed images for three diameters of Polystyrene nanospheres (70, 100, and 150 nm) imaged in water are generated by collecting the maximum and minimum normalized intensities present over a sequence of sequentially defocused images on a per-pixel basis. Solid lines denote compressed signatures from nanospheres on 30nm oxide, and dashed lines denote signatures from a 40nm oxide. The underlying Z-stacks are composed of 61 images simulated for a .9 NA imaging system in water at 200 nm increments over a 6 μm range centered on the oxide surface.
A) Largest positive normalized intensity values per pixel. B) Largest negative normalized intensity per pixel. C) Total differential intensity generated from the subtraction of plots in B from A. Differential intensity measurements demonstrate strong agreement between conditions despite significant variation in single – plane appearance and defocus behavior.
Initially, a nominal focal plane is identified via an autofocus algorithm utilizing a finite impulse response filter optimized for the critical spatial frequency of the optical system [34], after which a z-stack is acquired at 200nm increments over a 6 micron range centered at the starting point. Each slice of the z-stack is normalized into units of local normalized intensity by dividing the raw image by a low-pass filtered background image. This 3D data structure is used to calculate the maximum peak to peak intensity observed at each pixel over the total range of defocus positions within the z-stack, resulting in a two dimensional image differential intensity in which signatures from a heterogeneous population of nanoparticles are collapsed into a single consistent profile. A simulated template is generated by using the above method to compress simulated images generated by the SP-IRIS physical model. Cross-correlating this template with the differential intensity image results in a 2D correlogram, in which each pixel represents the probability that said location is the center point of a diffraction limited scattering object. The (x,y) locations of probable nanoparticle locations is then generated through simple morphological peak detection after thresholding this 2D correlogram by a high probability integer (typically 90%). These (x,y) locations are then used to extract intensity traces of nanoparticle centroids from the original z-stack, from which nanoparticles sizes are discriminated via their total change in intensities. Finally, the concentration of bound nanoparticles is determined by taking the ratio of the total number of confirmed nanoparticles with desired characteristics to the interrogated area.
V. Conclusions
In this paper, we have reviewed an array of optical imaging techniques capable of direct detection of nanoparticle biomarkers without fluorescent labels. We consider wide-field imaging as the most promising technique due to its simplicity and high throughput. We have focused on interferometric imaging and discussed the evolution of Single-Particle Interferometric Reflectance Imaging Sensor (SP-IRIS) from a laboratory instrument requiring manual operation by a skilled operator to an automated tool for diagnostic applications.
While the performance capabilities of interferometric imaging techniques have been widely demonstrated, their stringent calibration and characterization requirements have often been identified as significant obstacles to their translation to clinical environment. As we demonstrated in this paper, it is crucial to establish a rigorous model for the optical signatures of nanoparticles and develop a robust image acquisition and analysis technique. We have identified challenges associated with defocus, alignment, and nanoparticle orientation, and introduced a concept for robust nanoparticle detection and discrimination despite the presence of these phenomena.
Figure 8.
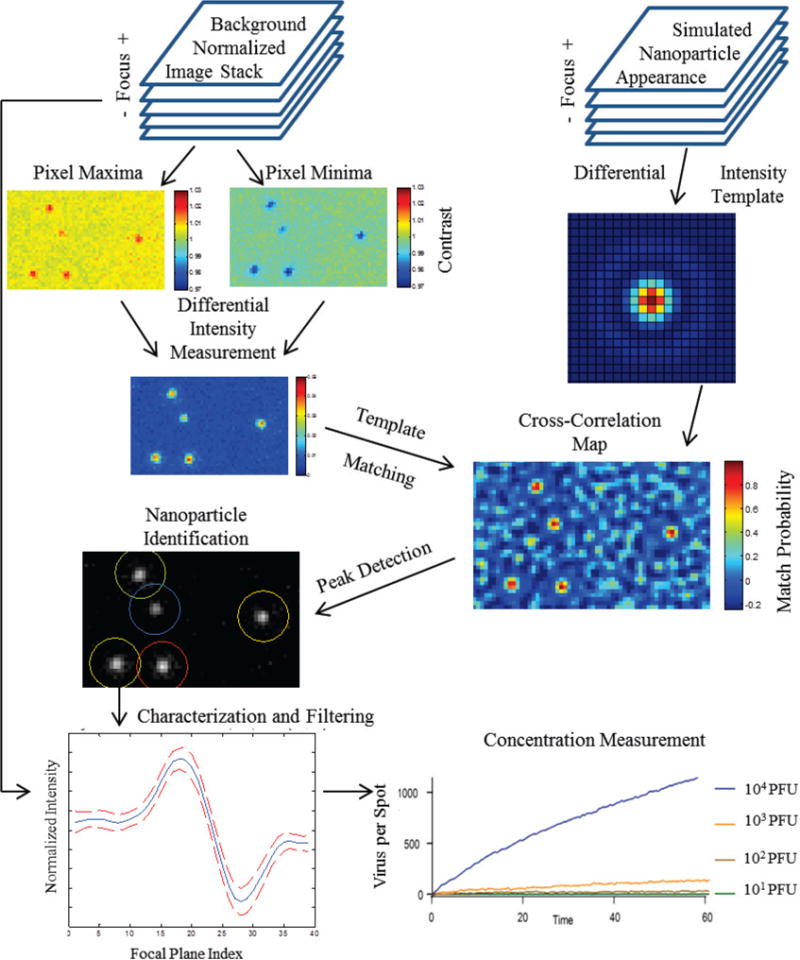
Block diagram of algorithm for nanoparticle detection and counting using z-stacks of incrementally defocused images.
Acknowledgments
We would like to thank Dr. David Freedman and Dr. George Daaboul of NexGen Arrays, LLCfor their invaluable expertise and contributions.
This work was supported in part by the National Institutes of Health (R01AI1096159) and ASELSAN Research Center, Ankara, Turkey. O. Avci acknowledges support from BU-BAU Fellowship.
Biographies

Jacob T. Traeb received the B.S. degree in mechanical engineering from Tufts LTniversity in 2008 and the M.S. degree in mechanical engineering from Boston LTniversity, Boston, MA, LISA, in 2011. He is currently pursuing the Ph.D. degree in mechanical engineering from Boston LTniversity, Boston, MA, USA with Professor M. Selim Ünlü at the Optical Characterization and Nanophotonics (OCN) Laboratory. His research interests lie in the design and control of automated microscopy systems and microfluidic lab-on-chip diagnostic platforms.

Oguzhan Avci received the B.S. degree in electrical and electronics engineering from Bilkent LTniversity, Ankara, Turkey, in 2012, and the M.S. degree in electrical engineering from Boston LTniversity, Boston, MA, USA, in 2014. He is currently pursuing the Ph.D. degree in electrical engineering from Boston LTniversity, Boston, MA, USA at the Optical Characterization and Nanophotonics (OCN) Laboratory. His current research focuses on physical modeling and development of highly sensitive and multiplexed interferometric biosensors for nanoparticle detection and characterization.

Derin Sevenler received a B.S. in Mechanical and Aerospace Engineering from Cornell LTniversity in 2011 and an M.S. in Biomedical Engineering from Boston LTniversity in 2014. He is currently a PhD candidate in Biomedical Engineering at Boston LTniversity. His research focus is the development of novel biosensors and molecular diagnostics to improve understanding and treatment of adaptive cancers and infectious diseases.

John H. Connor received the B.A. degree in chemistry from Swarthmore College, Swarthmore, PA, and the Ph.D. degree in pharmacology from Duke University, Durham, NC, in 1994 and 1999, respectively.
He was a Postdoctoral Fellow at Wake Forest University, Winston-Salem, NC. In 2006, he joined the faculty of the Boston University School of Medicine, Boston, MA, where he is currently an Assistant Professor of microbiology. His research interests include understanding viral pathogenesis and the virus/host interface.

M. Selim Ünlü (M’90-SM’95-F’07) received the B.S. degree from the Middle East Technical University, Ankara, Turkey, in 1986, and the M.S.E.E. (1988) and Ph.D. (1992) degrees from the University of Illinois at Urbana-Champaign, all in electrical engineering. Since 1992, he has been on the faculty of Boston University, currently appointed as Distinguished Professor of Engineering. He is appointed in electrical and computer engineering and affiliated with biomedical engineering, physics, material science and engineering, and graduate medical sciences. He has served as the Associate Dean for Research and Graduate Programs in engineering as well as the Associate Director of Center for Nanoscience and Nanobiotechnology. His research interests are in the areas of nanophotonics and biophotonics focusing on high-resolution solid immersion lens microscopy of integrated circuits and development of biological detection and imaging techniques, particularly in multiplexed detection of single viral pathogens and protein and nucleic acid microarrays.
Dr. Ünlü was the recipient of the National Science Foundation CAREER and Office of Naval Research Young Investigator Awards in 1996. He has been selected as a Photonics Society Distinguished Lecturer for 2005–2007 and Australian Research Council Nanotechnology Network (ARCNN) Distinguished Lecturer for 2007. He has been elevated to IEEE Fellow rank in 2007 for his contributions to optoelectronic devices. In 2008, he was awarded the Science Award by the Turkish Scientific Foundation. His professional service includes the former chair of photodetectors and imaging, and Biophotonics, and founding chair of Nanophotonics technical committees for IEEE Photonics Society, and Editor-in-Chief for IEEE Journal of Quantum Electronics.
Contributor Information
Jacob Trueb, Department of Mechanical Engineering Boston University, 8 St. Mary’s Street, Boston, MA 02215.
Oguzhan Avci, Department of Electrical and Computer Engineering, Boston University, 8 St. Mary’s Street, Boston, MA 02215, USA (617-353-5067).
Derin Sevenler, Department of Biomedical Engineering at Boston University, 44 Cummington Way, Boston MA 02215.
John H. Connor, Microbiology Department, Boston University School of Medicine, Boston, MA 02118, USA
M. Selim Ünlü, Department of Electrical and Computer Engineering, Boston University, 8 St. Mary’s Street, Boston, MA 02215, USA (617-353-5067).
References
- 1.Yurt A, Daaboul GG, Connor JH, Goldberg BB, Ünlü MS. Single nanoparticle detectors for biological applications. Nanoscale. 2012 Feb;4(3):715–26. doi: 10.1039/c2nr11562j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walt DR. Optical methods for single molecule detection and analysis. Anal Chem. 2013;85(3):1258–1263. doi: 10.1021/ac3027178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grilli S, Miccio L, Gennari O, Coppola S, Vespini V, Battista L, Orlando P, Ferraro P. Active accumulation of very diluted biomolecules by nano-dispensing for easy detection below the femtomolar range. Nat Commun. 2014;5:5314. doi: 10.1038/ncomms6314. [DOI] [PubMed] [Google Scholar]
- 4.Hoorfar J, Malorny B, Abdulmawjood A, Cook N, Fach P, Wagner M. Practical Considerations in Design of Internal Amplification Controls for Diagnostic PCR Assays. J Clin Microbiol. 2004;42(5):1863–8. doi: 10.1128/JCM.42.5.1863-1868.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Farrell B. Evolution in Lateral Flow–Based Immunoassay Systems. In: Wong R, Tse H, editors. Lateral Flow Immunoassay. Totowa, NJ: Humana Press; 2009. pp. 1–33. [Google Scholar]
- 6.Avci O, Ünlü N, Özkumur A, Ünlü M. Interferometric Reflectance Imaging Sensor (IRIS)—A Platform Technology for Multiplexed Diagnostics and Digital Detection. Sensors. 2015;15(7):17649–17665. doi: 10.3390/s150717649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daaboul GG, Lopez CA, Chinnala J, Goldberg BB, Connor JH, Selim Ünlü M. Digital sensing and sizing of vesicular stomatitis virus pseudotypes in complex media: A model for ebola and marburg detection. ACS Nano. 2014;8(6):6047–6055. doi: 10.1021/nn501312q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scherr SM, Daaboul GG, Trueb J, Sevenler D, Fawcett H, Connor JH, Ünlü MS. Real-Time Capture and Visualization of Individual Viruses in Com- plex Media. nanoLetters. 2016 doi: 10.1021/acsnano.5b07948. Vol. in print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monroe MR, Daaboul GG, Tuysuzoglu A, Lopez CA, Little FF, Ünlü MS. Single nanoparticle detection for multiplexed protein diagnostics with attomolar sensitivity in serum and unprocessed whole blood. Anal Chem. 2013 Apr;85(7):3698–706. doi: 10.1021/ac4000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lichtman JW, Conchello J. Fluorescence microscopy. Nat Methods. 2005;2(12):910–919. doi: 10.1038/nmeth817. [DOI] [PubMed] [Google Scholar]
- 11.Hell SW. Toward fluorescence nanoscopy. Nat Biotechnol. 2003;21(11):1347–55. doi: 10.1038/nbt895. [DOI] [PubMed] [Google Scholar]
- 12.Armani AM, Kulkarni RP, Fraser SE, Flagan RC, Vahala KJ. Detection with Optical Microcavities. Science. 2007 Aug;317:783–787. doi: 10.1126/science.1145002. 80- [DOI] [PubMed] [Google Scholar]
- 13.Vollmer F, Arnold S. Whispering-gallery-mode biosensing: label-free detection down to single molecules. Nat Methods. 2008;5(7):591–596. doi: 10.1038/nmeth.1221. [DOI] [PubMed] [Google Scholar]
- 14.Shao L, Jiang XF, Yu XC, Li BB, Clements WR, Vollmer F, Wang W, Xiao YF, Gong Q. Detection of single nanoparticles and lentiviruses using microcavity resonance broadening. Adv Mater. 2013;25(39):5616–5620. doi: 10.1002/adma201302572. [DOI] [PubMed] [Google Scholar]
- 15.Sheehan PE, Whitman LJ. Detection limits for nanoscale biosensors. Nano Lett. 2005 Apr;5(4):803–7. doi: 10.1021/nl050298x. [DOI] [PubMed] [Google Scholar]
- 16.Zhuo Y, Hu H, Chen W, Lu M, Tian L, Yu H, Long KD, Chow E, King WP, Singamaneni S, Cunningham BT. Single nanoparticle detection using photonic crystal enhanced microscopy. Analyst. 2014;139:1007–15. doi: 10.1039/c3an02295a. [DOI] [PubMed] [Google Scholar]
- 17.Schneckenburger H. Total internal reflection fluorescence microscopy: Technical innovations and novel applications. Curr Opin Biotechnol. 2005;16(1):13–18. doi: 10.1016/j.copbio.2004.12.004. SPEC. ISS. [DOI] [PubMed] [Google Scholar]
- 18.Enoki S, Iino R, Morone N, Kaihatsu K, Sakakihara S, Kato N, Noji H. Label-Free Single-Particle Imaging of the Influenza Virus by Objective-Type Total Internal Reflection Dark-Field Microscopy. PLoS One. 2012 Nov;7(11):e49208. doi: 10.1371/journal.pone.0049208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He H, Ren J. A novel evanescent wave scattering imaging method for single gold particle tracking in solution and on cell membrane. Talanta. 2008;77(1):166–171. doi: 10.1016/j.talanta.2008.05.059. [DOI] [PubMed] [Google Scholar]
- 20.Agnarsson B, Ingthorsson S, Gudjonsson T, Leosson K. Evanescent-wave fluorescence microscopy using symmetric planar waveguides. Opt Express. 2009;17(7):5075. doi: 10.1364/oe.17.005075. [DOI] [PubMed] [Google Scholar]
- 21.Agnarsson B, Lundgren A, Gunnarsson A, Rabe M, Kunze A, Mapar M, Simonsson L, Bally M, Zhdanov VP, Hook F. Evanescent Light-Scattering Microscopy for Label-Free Interfacial Imaging: From Single Sub-100 nm Vesicles to Live Cells. ACS Nano. 2015;9(12):11849–11862. doi: 10.1021/acsnano.5b04168. [DOI] [PubMed] [Google Scholar]
- 22.Wang S, Shan X, Patel U, Huang X, Lu J, Li J, Tao N. Label-free imaging, detection, and mass measurement of single viruses by surface plasmon resonance. Proc Natl Acad Sci U S A. 2010;107(37):16028–32. doi: 10.1073/pnas.1005264107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobsen V, Stoller P, Brunner C, Vogel V, Sandoghdar V. Interferometric optical detection and tracking of very small gold nanoparticles at a water-glass interface. Opt Express. 2006;14(1):405–14. doi: 10.1364/opex.14.000405. [DOI] [PubMed] [Google Scholar]
- 24.Ewers H, Jacobsen V, Klotzsch E, Smith AE, Helenius A, Sandoghdar V. Label-free optical detection and tracking of single virions bound to their receptors in supported membrane bilayers. Nano Lett. 2007;7(8):2263–2266. doi: 10.1021/nl070766y. [DOI] [PubMed] [Google Scholar]
- 25.Kukura P, Ewers H, Muller C, Renn A, Helenius A, Sandoghdar V. High-speed nanoscopic tracking of the position and orientation of a single virus. Nat Methods. 2009;6(12):923–927. doi: 10.1038/nmeth.1395. [DOI] [PubMed] [Google Scholar]
- 26.Ortega Arroyo J, Andrecka J, Spillane KM, Billington N, Takagi Y, Sellers JR, Kukura P. Label-free, all-optical detection, imaging, and tracking of a single protein. Nano Lett. 2014;14(4):2065–2070. doi: 10.1021/nl500234t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daaboul GG, Gagni P, Benussi L, Bettotti P, Ciani M, Cretich M, Freedman DS, Ghidoni R, Ozkumur AY, Piotto C, Prosperi D, Santini B, Ünlü MS, Chiari M. Digital Detection of Exosomes by Interferometric Imaging. Sci Rep. 2016;6:37246. doi: 10.1038/srep37246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozkumur E, Needham JW, Bergstein DA, Gonzalez R, Cabodi M, Gershoni JM, Goldberg BB, Ünlü MS. Label-free and dynamic detection of biomolecular interactions for high-throughput microarray applications. Proc Natl Acad Sci U S A. 2008 Jun;105(23):7988–92. doi: 10.1073/pnas.0711421105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luchansky MS, Washburn AL, McClellan MS, Bailey RC. Sensitive on-chip detection of a protein biomarker in human serum and plasma over an extended dynamic range using silicon photonic microring resonators and sub-micron beads. Lab Chip. 2011;11(12):2042–2044. doi: 10.1039/c1lc20231f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yurt A, Daaboul GG, Zhang X, Hwang GM, Goldberg BB, Ünlü MS. Widefield interferometric detection and size determination of dielectric nanoparticles. 2010 23rd Annu Meet IEEE Photonics Soc. 2010:197–198. [Google Scholar]
- 31.Seymour E, Daaboul GG, Zhang X, Scherr SM, Ünlü NL, Connor JH, Ünlü MS. DNA-Directed Antibody Immobilization for Enhanced Detection of Single Viral Pathogens. Anal Chem. 2015;87(20):10505–10512. doi: 10.1021/acs.analchem.5b02702. [DOI] [PubMed] [Google Scholar]
- 32.Avci O, Adato R, Ozkumur AY, Ünlü MS. Physical modeling of interference enhanced imaging and characterization of single nanoparticles. Opt Express. 2016;24(6):6094. doi: 10.1364/OE.24.006094. [DOI] [PubMed] [Google Scholar]
- 33.Novotny L, Hecht B. Principles of nano-optics. Cambridge university press; 2012. [Google Scholar]
- 34.Boddeke FR, Van Vliet LJ, Netten H, Young IT. Autofocusing in microscopy based on the OTF and sampling. Bioimaging. 1994;2(4):193–203. [Google Scholar]


