Abstract
Background
Our previous studies have demonstrated that ginsenoside-Rg1 can promote angiogenesis in vitro and in vivo through activation of the glucocorticoid receptor (GR). Furthermore, microRNA (miRNA) expression profiling has shown that Rg1 can modulate the expression of a subset of miRNAs to induce angiogenesis. Moreover, Rb1 was shown to be antiangiogenic through activation of a different pathway. These studies highlight the important functions of miRNAs on ginseng-regulated physiological processes. The aim of this study was to determine the angiogenic properties of Korean Red Ginseng extract (KGE).
Methods and Results
Combining in vitro and in vivo data, KGE at 500 μg/mL was found to induce angiogenesis. According to the miRNA sequencing, 484 differentially expressed miRNAs were found to be affected by KGE. Among them, angiogenic-related miRNAs; miR-15b, -23a, -214, and -377 were suppressed by KGE. Meanwhile, their corresponding angiogenic proteins were stimulated, including vascular endothelial growth factor, vascular endothelial growth factor receptor-2, endothelial nitric oxide synthase, and MET transmembrane tyrosine kinase. The miRNAs-regulated signaling pathways of KGE were then found by Cignal 45-Pathway Reporter Array, proving that KGE could activate GR.
Conclusion
KGE was found capable of inducing angiogenesis both in vivo and in vitro models through activating GR. This study provides a valuable insight into the angiogenic mechanisms depicted by KGE in relation to specific miRNAs.
Keywords: angiogenesis, endothelial cells, Korean Red Ginseng extract, microRNAs
1. Introduction
Angiogenesis is the formation of new blood vessels from pre-existing blood vessels. It is involved in both physiological and pathological conditions such as embryo development [1], wound healing [2], atherosclerosis [3], and tumor growth [4]. During angiogenesis, complex cell–cell interactions and various ligand–receptor activations are involved; endothelial cells play a central role in this process [5]. Once activated by angiogenic factors, endothelial cells release proteolytic enzymes, migrate, and invade surrounding extracellular matrix, where they assemble into new blood vessels. Besides, endothelial cells are also important in regulating vascular functions, including vasodilation and blood vessel integrity. Endothelial dysfunction is associated with diverse vascular diseases, such as atherosclerosis, stroke, and hypertension [6].
Significant progress has been made in elucidating the molecular basis of endothelial functions. Recent studies have highlighted the importance of microRNAs (miRNAs) [7]. miRNAs are a group of small RNAs of approximately 18–24 bp. Although miRNAs are noncoding RNAs, they are important in regulating over 30% of gene expression at the post-transcriptional level [8]. Mature miRNAs in the cytoplasm recognize the 3′-untranslated region of target mRNAs, and their partial complementary binding to the 3′-untranslated region may lead to translational repression of the mRNA.
Ginseng (Panax ginseng Meyer), a traditional Chinese medicine, has been used for thousands of years. It is a slow-growing perennial herb, with large fleshy roots. Among the 11 species of ginseng, the two major species are the Asian (Chinese and Korean: P. ginseng) ginseng and American ginseng, Panax quinquefolius. It is well-known for its diverse benefits, includes immunomodulation, anti-inflammation, -allergy, -atherosclerosis, -hypertension, -diabetes, -stress, and -carcinogenesis, as well as wound healing [9], [10].
Ginsenosides, the major bioactive ingredient in ginseng extracts, are a class of steroid glycosides and triterpene saponins. More than 100 ginsenosides have been identified from the ginseng extracts, and they are classified into protopanaxadiol and protopanaxatriol types according to their structure [11]. Among the several types of protopanaxatriol (e.g., Rf, Rh1, and Rg1) and protopanaxadiol (e.g., Rb1, Rd, and Rh2), Rg1, and Rb1 were found to be in the highest content. Our previous studies have demonstrated that ginsenoside-Rg1 can promote angiogenesis in vitro [12] and in vivo [13] through activation of the glucocorticoid receptor (GR) [14]. Furthermore, miRNA expression profiling has shown that Rg1 can modulate the expression of a subset of miRNAs to induce angiogenesis [16], [17]. Moreover, Rb1 was shown to induce type I collagen expression in human dermal fibroblast by reducing the miR-25 expression [17].
These studies throw light on the important functions of miRNAs on ginseng-regulated physiological processes. By contrast, the role of miRNAs in Korean Red Ginseng extract (KGE) affecting physiological responses has not been studied so far. In this project, we aim to study the functional role of miRNAs and the underlying mechanism in KGE-regulated angiogenesis. Our results show that KGE stimulates angiogenesis in vitro and in vivo through activation of the GR.
2. Materials and methods
2.1. Reagents and chemicals
KGE was provided by Korea Ginseng Corporation (Seoul, Korea). Stock solution of KGE (50mM) was prepared in sterile water. Chemicals not specified were obtained from USB Chemicals (Cleveland, OH, USA). KGE was prepared from the roots of a 6-yr-old fresh P. ginseng Meyer. KGE was yielded from red ginseng water extract and the water content of the pooled extract was 36% of total weight, contained major ginsenoside-Rb1: 33.05%, Rg1: 7.95%, Re: 8.26%, Rc: 13.51%, Rb2: 11.51%, Rd: 4.04%, Rf: 5.51%, Rh1: 4.49%, Rg2S: 5.51%, and Rg3S: 6.18%.
2.2. Cell culture
Human umbilical vein endothelial cells (HUVECs; Lonza, Walkersville, MD, USA), were maintained in medium M199 supplemented with heparin (90 mg/L), heat-inactivated fetal bovine serum (20%, v/v), endothelial cell growth supplement (20 μg/mL), and penicillin and streptomycin (1%, v/v). They were kept at 37°C in humidified air with 5% CO2 and were used within passages 2–8. The cells were seeded overnight and treated with KGE in M199 containing fetal bovine serum (1%, v/v) and endothelial cell growth supplement (10 μg/mL).
2.3. Cell proliferation assay
Cell proliferation was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide salt (MTT) kit (USB). Equal numbers of HUVECs (1 × 104 cells/well) were seeded onto 96-well plates and incubated overnight. After the indicated time, cells were incubated with MTT solution (0.5 mg/mL) in assay medium for 4 h. Then the residual MTT was removed and the crystals were dissolved by incubation with DMSO solution for color development. The absorbance at wavelengths 450 nm and 690 nm (reference) were measured using a microplate reader (ELx800; Biotek, Winooski, VT, USA).
2.4. Cell migration assay
To evaluate the migration ability of the cells, HUVECs (3 × 104 cells/well) were seeded onto 96-well plates and incubated overnight. A denuded cell area was created by scratching the 100% confluent cell monolayer using a mechanical wounder [18]. After scratching, culture medium was replaced with fresh medium with or without KGE, and images of each well at the beginning (At0) and after 16 h (At16) were captured. The scratched area was measured using the Image J software (http://rsb.onfo.nih.gov). The migration of cells toward the denuded area was expressed as the percentage of recovery:
| Percentage of recovery = (At0 − At16 / At0) × 100%. | (1) |
2.5. Endothelial tube formation assay
A 96-well plate pre-coated with growth factor-reduced Matrigel (BD Bioscience, San Jose, CA, USA) was allowed to solidify at 37°C for 1 h. HUVECs (3 × 104 cells/well) were then plated on the Matrigel substratum and cultured in medium with or without KGE. Tube network in each well were captured after 8 h, the angiogenic activities were determined by counting the number of branch points of the formed tubes in each well.
2.6. Zebrafish endogenous alkaline phosphatase-based vascular staining
Zebrafish embryos (24 h postfertilization) were dechorionated by pronase (2 mg/mL) for 15 min. The embryos were then incubated with various concentrations of KGE in water containing 1-phenyl-2-thiourea at 28.5°C for another 48 h. Embryos (72 h postfertilization) were euthanized, and alkaline phosphatase activity were assayed after fixation for 30 min at 4°C in 4% paraformaldehyde. Then, fish embryos were treated with ethanol (50% and 100%) for 5 min, respectively. Dehydrated embryos were then incubated in pre-chilled acetone for 30 mins at −20°C and rinsed with phosphate-buffered saline with 0.1% Tween-20. For staining, embryos were equilibrated with alkaline phosphatase buffer at room temperature for 15 min and subsequently stained with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl-phospate (AMRESCO, Solon, OH, USA) at room temperature for 30 min in dark. The subintestinal vessels (SIV) basket of the stained zebrafish was examined under stereomicroscope (Olympus SZX16) with attached digital camera (Olympus DP71; Olympus America, San Jose, CA, USA). Areas of SIVs were quantified by Image J software (http://rsb.onfo.nih.gov).
2.7. Western blot analysis
After treatment, cells were washed twice with ice-cold phosphate-buffered saline and lysed in lysis buffer (Novagen, Madison, WI, USA) containing protease (0.5%, v/v) and phosphatase inhibitor cocktails (0.5%, v/v; Calbiochem, Billerica, MA, USA). The cells were harvested by scraping, and the cell lysate was collected after centrifugation. The protein concentration of the cell lysates was determined by the DC protein assay (Bio-Rad, Hercules, CA, USA). Equal amounts of protein were separated by 10% SDS-PAGE followed by electroblotting onto nitrocellulose membrane. The membrane was soaked in blocking buffer (1% nonfat milk) and then incubated with primary antibody overnight at 4°C. The washed membrane was then further incubated with horseradish peroxidase-conjugated goat-anti-rabbit or -mouse Immunoglobulin G (IgG) secondary antibody (Invitrogen, Carlsbad, CA, USA), and the signal was visualized using the Chemiluminescent Western Detection kit (Bio-Rad).
2.8. Small RNA transcriptomic analysis
HUVECs were treated with KGE (500 μg/mL) and RNA was extracted. Small RNA transcriptomic analysis was performed using Next Generation Sequencing for identification of KGE-regulated miRNAs through a contract service provided by Beijing Genomics Institute (BGI; Shenzhen, China). Total miRNA was separated into different sizes using polyacrylamide gel, and strips of the gel between 18 nucleotides and 30 nucleotides were cut out, followed by blending and centrifugation. Reverse transcription was performed to convert the miRNA into double-stranded form for amplification. Afterwards, the PCR products were purified for transcriptome quantification and structural analysis using HiSeq4000 (Illumina, San Diego, CA, USA), the differential miRNA expression between control and KGE- treated group were analyzed.
2.9. TaqMan microRNA assay
To confirm miRNA expression, total RNA of HUVECs was extracted using TRIzol (Invitrogen). Quantitative analysis of miRNA expression was performed using real-time PCR with the TaqMan microRNA assay (Applied Biosystems, Foster City, CA, USA) and was detected by a StepOnePlus real-time PCR system (Applied Biosystems). The level of U6 small nuclear RNA 2 (U6B) was used to normalize the relative expression of mature hsa-miR-15b, -23a, and -214.
2.10. Cignal 45-Pathway Reporter Array
To perform screening on the signaling pathways that are affected by KGE, Cignal 45-Pathway Reporter Array (SA Biosciences, Frederick, MD, USA) was used. COS-7 cells were transfected with various luciferase reporters provided in the kit and further incubated with KGE. By examining the activity of the reporters, the receptor-regulated signaling pathways were delineated in this model.
2.11. Cignal GRE Reporter Assay
To confirm that KGE can activate the activity of GR-induced signal transduction pathways, Cignal GRE Reporter Assay (QIAGEN, Hilden, Germany) was used. COS-7 cells were transfected with glucocorticoid transcriptional response element (GRE) reporter provided in the kit and further incubated with KGE. The change in the activity of the GRE reporter was determined by comparing the normalized luciferase activities of the reporter in KGE-treated versus untreated transfectants.
2.12. Statistical analysis
All results are expressed as the mean ± standard derivation of at least three independent experiments. All data were analyzed by Student t test and or one-way analysis of variance (SigmaPlot version 12.5, Systat Software Inc). Values of p < 0.05 were considered as statistically significant.
3. Results
3.1. KGE induces proliferation, migration, and tube formation of HUVECs
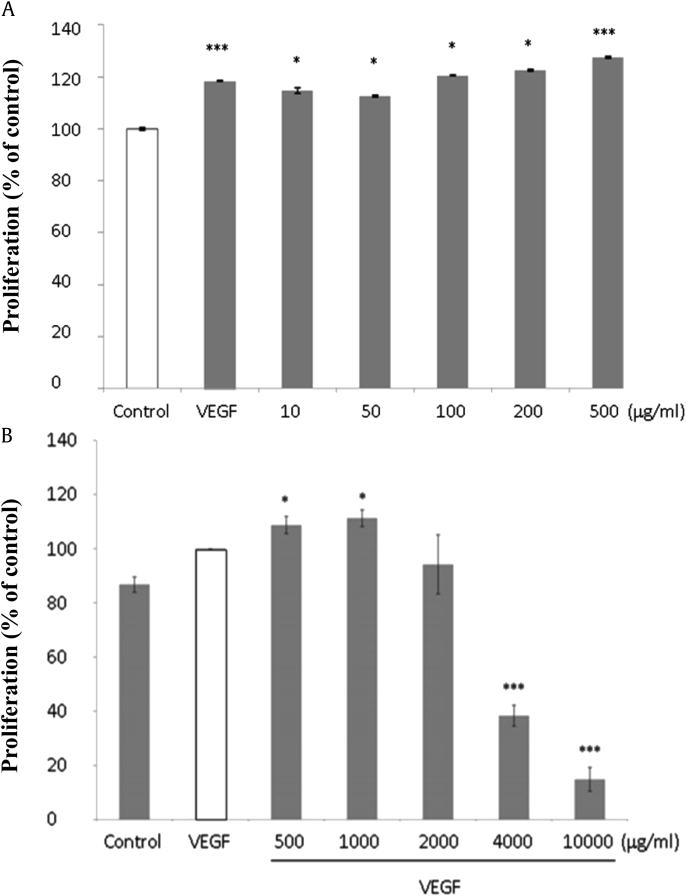
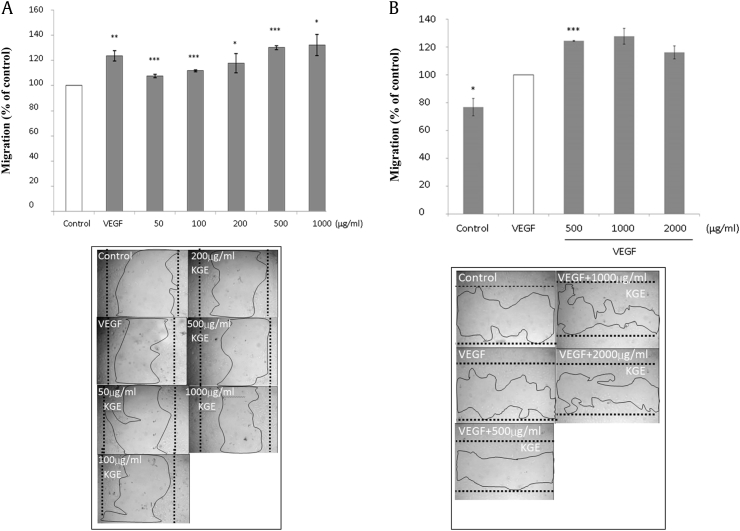
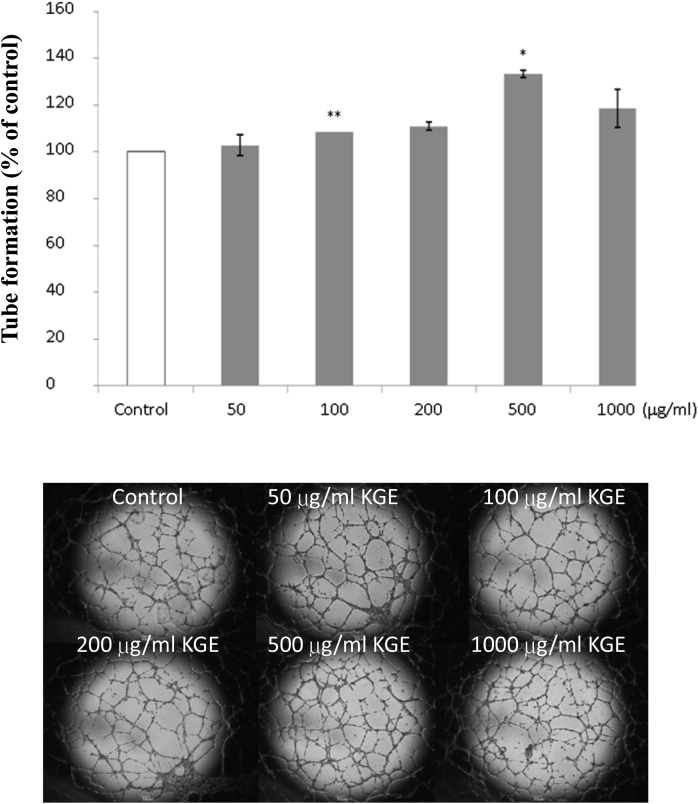
We first examined whether KGE would regulate endothelial cell angiogenesis, which was indicated by cell proliferation, migration, and tube formation in this study in vitro. HUVECs were treated with various concentrations of KGE ranging from 10 μg/mL to 10,000 μg/mL in 1% M199 for preliminary purpose, and KGE was found capable of inducing cell proliferation (Fig. 1A), migration (Fig. 2A), and tube formation (Fig. 3) in a dose-dependent manner, with 25%, 30%, and 33% increase at 500 μg/mL, respectively. The angiogenic effects of KGE on vascular endothelial growth factor (VEGF)-treated HUVECs were also examined. Increasing concentrations of KGE from 2,000 μg/mL to 10,000 μg/mL were found to suppress HUVECs proliferation (Fig. 1B) and migration (Fig. 2B).
Fig. 1.
Effects of KGE on HUVECs proliferation. (A) HUVECs were incubated with various concentrations of KGE (10–500 µg/ml) for 48 h. VEGF (10 µg/ml) was added as positive control. (B) HUVECs were incubated with various concentrations of KGE (500–10,000 µg/ml) with VEGF for 48 h. VEGF (10 µg/ml) was added as positive control. Cell proliferation was examined by MTT assay. Values are presented as the means ± SD of three independent experiments. All data were analyzed by ANOVA with multiple post hoc testing. * p < 0.05, *** p < 0.001 vs control or VEGF control.
Fig. 2.
(A) Effects of KGE on HUVECs migration. HUVECs were incubated with various concentrations of KGE (50–1000 µg/ml) for 24 h. VEGF (10 ng/ml) was added as positive control. The upper panel indicated the quantification of the cell migration. The lower panel showed the migration of endothelial cells. Values are presented as the means ± SD of three independent experiments. All data were analysed by ANOVA with multiple post hoc testing. * p < 0.05, ** p < 0.01, *** p < 0.001 vs control or VEGF control. (B) Effects of KGE on HUVECs migration. HUVECs were incubated with various concentrations of KGE (500–2000 μg/ml) with VEGF for 24 h. VEGF (10 ng/ml) was added as positive control. The upper panel indicated the quantification of the cell migration. The lower panel showed the migration of endothelial cells. Values are presented as the means ± SD of three independent experiments. All data were analysed by ANOVA with multiple post hoc testing. * p < 0.05, ** p < 0.01, *** p < 0.001 vs control or VEGF control.
Fig. 3.
Effects of KGE on HUVECs endothelial tube formation. HUVECs were incubated with various concentrations of KGE (50–1000 µg/ml) for 24 h. The upper panel indicated the quantification of tube formation and the lower one showed the tube network formed by HUVECs. Values are presented as the means ± SD of three independent experiments. All data were analysed by ANOVA with multiple post hoc testing. * p < 0.05, ** p < 0.01 vs control.
3.2. KGE induces angiogenesis in vivo
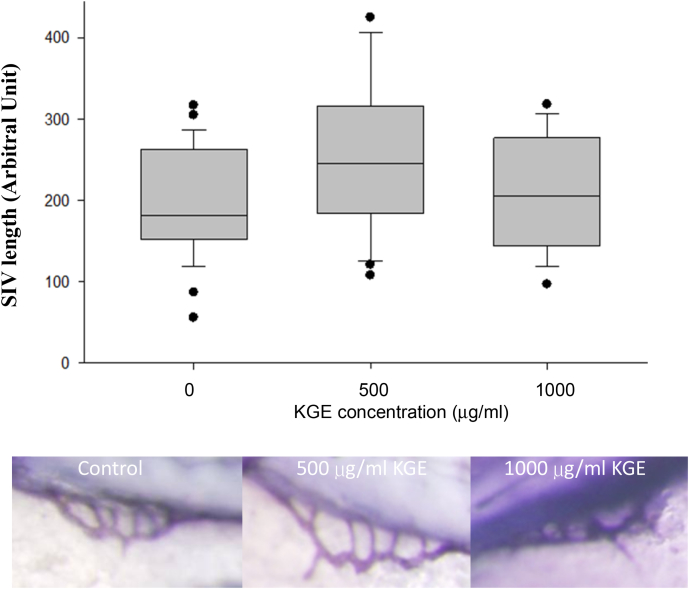
To further investigate whether KGE regulates in vivo angiogenesis, we examined the effect of KGE on-angiogenesis in zebrafish embryos. Zebrafish embryos serve as an excellent in vivo model for monitoring angiogenesis. Concomitant result was found in zebrafish model (Fig. 4), KGE at 500 μg/mL significantly increased SIV growth. However, increasing concentration of KGE did not further stimulate the SIV growth. These results provide clear evidence that KGE can promote angiogenesis in vitro and in vivo.
Fig. 4.
Effects of KGE on zebrafish embryos SIV length. Zebrafish embryos at 24 h.p.f. were incubated with KGE (500 or 1000 µg/ml) for 48 h. The embryos were stained and images were captured under microscope. The upper panel indicated the quantification of the length of SIV by Image J software, the lower panel showed the SIV of the zebrafish. Values are presented as the means ± SD. At least 30 embryos were assessed in each condition. All data were analysed by ANOVA with multiple post hoc testing. * p < 0.05 vs control.
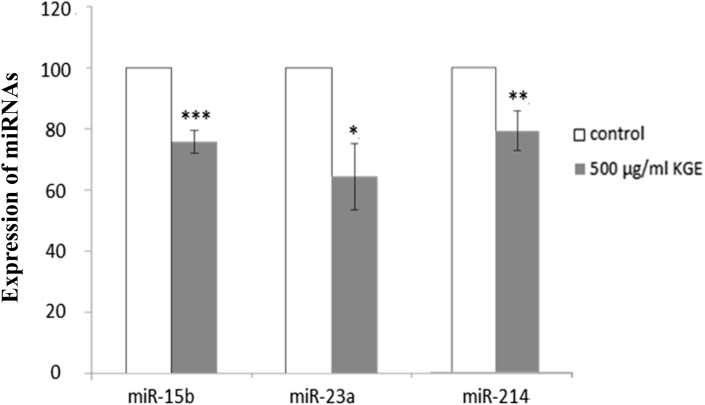
3.3. KGE suppresses miR-15b, -23a, -214, and -377 expression in HUVECs
Small RNA transcriptomic analysis was performed using next generation sequencing for the identification of KGE-regulated miRNAs. The differential expression of miRNA candidates between the control and KGE-treated (500 μg/mL) groups were compared and 484 differentially expressed miRNAs were found (Fig. S1). Among these miRNAs, those that are related to angiogenesis were discussed. Our previous finding showed that the expressions of miR-15b [16], -23a [19], and -214 [15] were closely correlated to the angiogenic activity. Interestingly, the trend of the miRNA sequencing data was similar to the previous finding. KGE (500 μg/mL) was also found to suppress the expressions of miR-15b, -23a, and -214 by TaqMan microRNA assay (Fig. 5). Our previous study discovered that miR-377 was correlated to Rg1-induced angiogenesis [20]; the expression of miR-377 was downregulated after treatment with KGE.
Fig. 5.
Effects of KGE on miRNA expressions as determined by TaqMan microRNA assay. HUVECs were incubated with KGE (500 µg/ml) for 24 h. The graph indicated the quantification of miR-15b, -23a and -214 expressions. Values are presented as the means ± SD of three independent experiments. All data were analysed by ANOVA with multiple post hoc testing. * p < 0.05, ** p < 0.01 and *** p < 0.001 vs control.
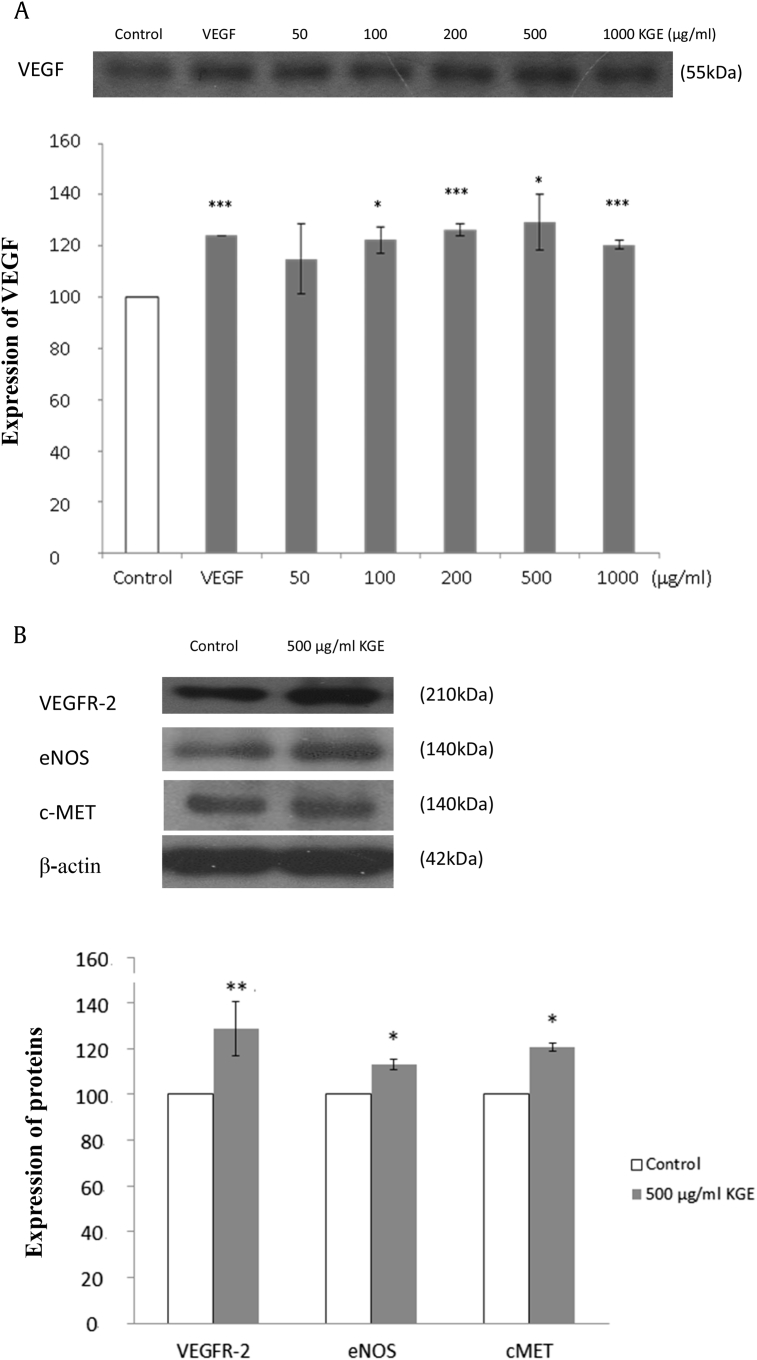
3.4. KGE induces VEGF, VEGF receptor-2, MET transmembrane tyrosine kinase, and endothelial nitric oxide synthase expression in HUVECs
According to the results of miRNA sequencing and TaqMan microRNA assay, decreases in miR-15b, -23a, -214, and -377 expression were found, which suggests an increase in the expression of angiogenic proteins VEGF, VEGF receptor-2 (VEGFR-2), endothelial nitric oxide synthase (eNOS), and MET transmembrane tyrosine kinase (c-MET), respectively. To validate the expression of proteins, western blot analysis was performed. VEGF, the potent angiogenic protein and essential growth factor for vascular endothelial cells [20], was examined with respect to miR-377 downregulation [20]. Results indicate that KGE at 100 μg/mL, 200 μg/mL, 500 μg/mL, and 1,000 μg/mL was found to increase VEGF expression significantly, and KGE at 500 μg/mL is the optimal concentration to induce VEGF expression (Fig. 6A). In response to the suppression of miR-15b, -23a, and -214 expression, the expression of the corresponding proteins, VEGFR-2 [21], c-MET [22], and eNOS [23], respectively, were investigated. KGE at 500 μg/mL was found to increase VEGFR-2, eNOS, and c-MET expression significantly (Fig. 6B), thereby inducing angiogenesis, which correlated very well to the result of downregulation of the corresponding miR-15b, -23a, and -214 expression.
Fig. 6.
(A) Effects of KGE on angiogenic protein expressions. Effect of KGE on VEGF expression. HUVECs were incubated with various concentrations of KGE (50–1000 µg/ml) for 24 h. VEGF (10 ng/ml) was added as positive control. HUVECs were incubated with KGE (500 µg/ml) for 24 h. The bottom graphs showed the quantification of VEGF expression and the upper chamber showed the expressions of VEGF by western blot analysis. Values are presented as the means ± SD of three independent experiments. All data were analysed by ANOVA with multiple post hoc testing. * p < 0.05, ** p < 0.01 and *** p < 0.001 vs control. (B) Effects of KGE on angiogenic protein expressions. Effect of KGE on VEGFR-2, eNOS and c-MET expressions. HUVECs were incubated with KGE (500 μg/ml) for 24 h. The bottom graphs showed the quantification of protein expression normalized with β-actin and the upper chamber showed the expressions of VEGFR-2, eNOS and c-MET by western blot analysis. Values are presented as the means ± SD of three independent experiments. All data were analysed by ANOVA with multiple post hoc testing. * p < 0.05, ** p < 0.01 and *** p < 0.001 vs control.
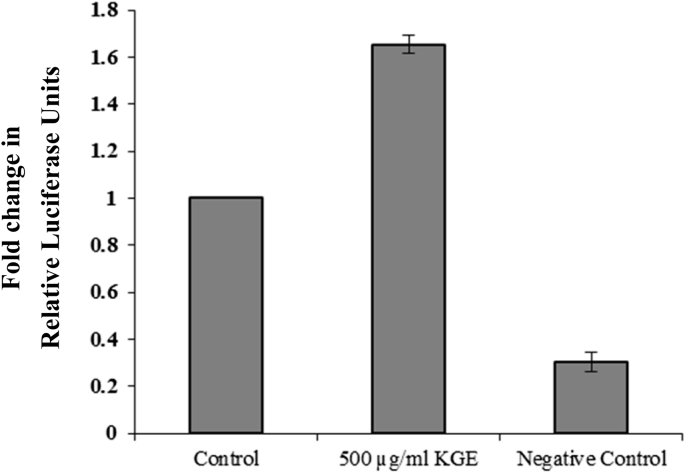
3.5. KGE induces angiogenesis via activation of GR
The receptor-regulated signaling pathways of KGE (500 μg/mL) were found by Cignal 45-Pathway Reporter Array, using Renilla reporter for internal normalization. KGE was found to upregulate the GRE reporter in this study (Fig. S2), suggesting that KGE may stimulate angiogenesis through GR. Cignal GRE Reporter Assay was used to validate the upregulation of GR pathway activity by KGE (500 μg/mL). By normalizing the activity of GRE-responsive firefly luciferase affected by KGE with Renilla luciferase activity, a change in the activity of GRE reporter was found. KGE stimulated the GRE reporter activity by 65%, supporting that KGE can stimulate the GR pathway (Fig. 7).
Fig. 7.
Effect of KGE on glucocorticoid receptor. COS-7 cells were incubated with KGE (500 µg/ml) for 24 h. The graph indicated the quantification of the GRE reporter expression. Values are presented as the means ± SD of three independent experiments. A non-inducible firefly luciferase construct served as a negative control. All data were analysed by ANOVA with multiple post hoc testing. * p < 0.05, ** p < 0.01 and *** p < 0.001 vs control.
4. Discussion
It is well-accepted that P. ginseng is implicated as a panacea to have diverse pharmacological benefits. The major bioactive ginsenosides have been demonstrated to exert beneficial effects on the cardiovascular system, in which ginsenoside-Rg1 [13] and -Re [24], [25] have been shown to induce angiogenesis that contributes to wound healing promotion, while others, such as ginsenosides-Rb1 [20], -Rb2, and -Rg3 [26], [27] are reported to suppress VEGF-induced in vitro and in vivo angiogenesis, leading to tumor growth inhibition. Despite the understanding of the single ginsenosides, the angiogenic properties of the total ginseng extract are still unclear.
Our results show that KGE (10–1,000 μg/mL), containing a wide spectrum of ginsenosides, stimulates angiogenesis in a dose-dependent manner in vivo and in vitro. KGE at 500 μg/mL was found to be optimal for induction of cell proliferation, migration, and tube formation of HUVECs as well as SIV formation in zebrafish model. Thus, this concentration was selected for further investigation. By contrast, increasing concentrations of KGE (2,000–10,000 μg/mL) caused cell death of HUVECs and therefore suppressed angiogenesis.
To further examine the pharmacological mechanism of the ginseng extract on angiomodulation, small RNA transcriptomic analysis was performed using next generation sequencing. The differential expression of miRNA candidates between the control and KGE-treated (500 μg/mL) groups were compared and 484 differentially expressed miRNAs were found. According to the microRNA microarray profiling performed in our previous studies, Rg1 was shown to modulate the expression of a subset of miRNAs to induce angiogenesis [16], [17]. Based on these microarray data, angiogenic-mediated miRNAs expressions have been investigated. Interestingly, the trend of miR-15b, -23a, -214, and -377 expression in miRNA sequencing data was similar to our previous findings. KGE (500 μg/mL) was able to suppress the expression of miR-15b, -23a, and -214 as confirmed by real-time PCR.
With this background information, the expression levels of the respective angiogenic proteins corresponding to the downregulated miRNAs were investigated. According to the miRNA sequencing result, the expression of miR-377 was downregulated after treating with KGE indicating the possible enhanced expression of the angiogenic protein VEGF [20]. VEGF is the potent angiogenic protein that is involved in the induction of endothelial cell proliferation, cell migration, and neovascularization [28]. From the western blotting, VEGF expression was induced by increasing concentration of KGE. From our previous study, miR-214 was found to modulate the angiogenic activity of HUVECs through suppressing eNOS expression, to inhibit cell migration and tube formation [15]. Later, we found that diminishing miR-15b could increase VEGFR-2 expression on HUVECs, as reflected by promotion of cell migration and tubulogenesis [15]. Recently, we have also demonstrated the role of miR-23a on mediating angiogenesis by downregulating c-MET expression, resulting in reducing recruitment of different cytoplasmic adaptor proteins and HUVECs proliferation [20], [29]. In response to the suppression of miR-15b, -23a, and -214 expression, the expression of the corresponding proteins, VEGFR-2, c-MET, and eNOS were investigated. KGE at 500 μg/mL was found to increase expression of those proteins significantly, thereby inducing angiogenesis.
The receptor-regulated signaling pathways of KGE (500 μg/mL) were screened by Cignal 45-Pathway Reporter Array. Similar to our previous study, KGE can activate GRE reporter expression in GRE reporter assay. These data clearly indicate that KGE-stimulated angiogenesis is correlated with the activation of GR. Thus, many GR-mediated signaling pathways would be activated by KGE. The activated GR complex can downregulate the inflammatory effect [30] and the atherosclerotic effect [31]. These findings provide new insights into the angiogenic mechanism of the ginseng extract.
5. Further perspective
We hypothesized that the angiogenic property of KGE may be similar to the single ginsenoside-Rg1 activity with the suppression of targeted miRNAs, induction of angiogenic proteins, and activation of the GR, which could further be developed into wound healing or diabetic ulcer skin care products. During wound healing, KGE helps in stimulating the angiogenic proteins (VEGF, VEGFR-2, c-MET, and eNOS) through suppressing the expression of respective miRNAs. Consequently, it boosts the proliferation, migration, and microvascular angiogenic capillaries sprout to invade the fibronectin-rich wound clot, and then form a mesh of blood vessels [32]. KGE plays a role in stimulating the angiogenesis process, thereby allowing scar formation in the wound. In context with the extraordinary quality of KGE in wound healing through angiogenesis, we speculate that KGE could be introduced in the herbal treatment of wound anomalies with huge success.
Conflicts of interest
All authors declare no conflicts of interest.
Acknowledgments
The grant and materials were supported from Korean Society of Ginseng and Korea Ginseng Corporation.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jgr.2016.08.011.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Heinke J., Patterson C., Moser M. Life is a pattern: vascular assembly within the embryo. Front Biosci (Elite Ed) 2012;4:2269–2288. doi: 10.2741/541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li W.W., Talcott K.E., Zhai A.W., Kruger E.A., Li V.W. The role of therapeutic angiogenesis in tissue repair and regeneration. Adv Skin Wound Care. 2005;18:491–500. doi: 10.1097/00129334-200511000-00013. [DOI] [PubMed] [Google Scholar]
- 3.Bochkov V.N., Philippova M., Oskolkova O., Kadl A., Furnkranz A., Karabeg E., Afonyushkin T., Gruber F., Breuss J., Minchenko A. Oxidized phospholipids stimulate angiogenesis via autocrine mechanisms, implicating a novel role for lipid oxidation in the evolution of atherosclerotic lesions. Circ Res. 2006;99:900–908. doi: 10.1161/01.RES.0000245485.04489.ee. [DOI] [PubMed] [Google Scholar]
- 4.Carmeliet P., Jain R.K. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 5.Augustin H.G., Kozian D.H., Johnson R.C. Differentiation of endothelial cells: analysis of the constitutive and activated endothelial cell phenotypes. Bioessays. 1994;16:901–906. doi: 10.1002/bies.950161208. [DOI] [PubMed] [Google Scholar]
- 6.Yildiz O. Vascular smooth muscle and endothelial functions in aging. Ann N Y Acad Sci. 2007;1100:353–360. doi: 10.1196/annals.1395.038. [DOI] [PubMed] [Google Scholar]
- 7.Condorelli G., Latronico M.V., Cavarretta E. MicroRNAs in cardiovascular diseases: current knowledge and the road ahead. J Am Coll Cardiol. 2014;63:2177–2187. doi: 10.1016/j.jacc.2014.01.050. [DOI] [PubMed] [Google Scholar]
- 8.Carthew R.W., Sontheimer E.J. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen L.P. Ginsenosides chemistry, biosynthesis, analysis, and potential health effects. Adv Food Nutr Res. 2009;55:1–99. doi: 10.1016/S1043-4526(08)00401-4. [DOI] [PubMed] [Google Scholar]
- 10.Kwak Y.S., Park J.D., Yang J.W. Present and its prospect of red ginseng efficacy research. Food Ind Nutr. 2003;8:30–37. [Google Scholar]
- 11.Sengupta S., Toh S.A., Sellers L.A., Skepper J.N., Koolwijk P., Leung H.W., Yeung H.W., Wong R.N., Sasisekharan R., Fan T.P. Modulating angiogenesis: the yin and the yang in ginseng. Circulation. 2004;110:1219–1225. doi: 10.1161/01.CIR.0000140676.88412.CF. [DOI] [PubMed] [Google Scholar]
- 12.Yue P.Y., Wong D.Y., Ha W.Y., Fung M.C., Mak N.K., Yeung H.W., Leung H.W., Chan K., Liu L., Fan T.P. Elucidation of the mechanisms underlying the angiogenic effects of ginsenoside Rg(1) in vivo and in vitro. Angiogenesis. 2005;8:205–216. doi: 10.1007/s10456-005-9000-2. [DOI] [PubMed] [Google Scholar]
- 13.Leung K.W., Cheng Y.K., Mak N.K., Chan K.K., Fan T.P., Wong R.N. Signaling pathway of ginsenoside-Rg1 leading to nitric oxide production in endothelial cells. FEBS Lett. 2006;580:3211–3216. doi: 10.1016/j.febslet.2006.04.080. [DOI] [PubMed] [Google Scholar]
- 14.Leung K.W., Pon Y.L., Wong R.N., Wong A.S. Ginsenoside-Rg1 induces vascular endothelial growth factor expression through the glucocorticoid receptor- related phosphatidylinositol 3-kinase/Akt and beta-catenin/ T-cell factor-dependent pathway in human endothelial cells. J Biol Chem. 2006;281:36280–36288. doi: 10.1074/jbc.M606698200. [DOI] [PubMed] [Google Scholar]
- 15.Chan L.S., Yue P.Y., Mak N.K., Wong R.N. Role of microRNA-214 in ginsenoside-Rg1-induced angiogenesis. Eur J Pharm Sci. 2009;38:370–377. doi: 10.1016/j.ejps.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Chan L.S., Yue P.Y., Wong Y.Y., Wong R.N. MicroRNA-15b contributes to ginsenoside-Rg1-induced angiogenesis through increased expression of VEGFR-2. Biochem Pharmacol. 2013;86:392–400. doi: 10.1016/j.bcp.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 17.Kwok H.H., Yue P.Y., Mak N.K., Wong R.N. Ginsenoside Rb(1) induces type I collagen expression through peroxisome proliferator-activated receptor-delta. Biochem Pharmacol. 2012;84:532–539. doi: 10.1016/j.bcp.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 18.Yue P.Y., Leung E.P., Mak N.K., Wong R.N. A simplified method for quantifying cell migration/wound healing in 96-well plates. J Biomol Screen. 2010;15:427–433. doi: 10.1177/1087057110361772. [DOI] [PubMed] [Google Scholar]
- 19.Kwok H.H., Chan L.S., Poon P.Y., Yue P.Y., Wong R.N. Ginsenoside-Rg1 induces angiogenesis by the inverse regulation of MET tyrosine kinase receptor expression through miR-23a. Toxicol Appl Pharmacol. 2015;287:276–283. doi: 10.1016/j.taap.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 20.Wen Z., Huang W., Feng Y., Cai W., Wang Y., Wang X., Liang J., Wani M., Chen J., Zhu P. MicroRNA-377 regulates mesenchymal stem cell-induced angiogenesis in ischemic hearts by targeting VEGF. PLoS One. 2014;9:e104666. doi: 10.1371/journal.pone.0104666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahimi N. (2006) VEGFR-1 and VEGFR-2: two non- identical twins with a unique physiognomy. Front Biosci. 2006;11:818–829. doi: 10.2741/1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venepalli N.K., Goff L. Targeting the HGF-cMET axis in hepatocellular carcinoma. Int J Hepatol. 2013;2013:341636. doi: 10.1155/2013/341636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukumura D., Gohongi T., Kadambi A., Izumi Y., Ang J., Yun C.O., Buerk D.G., Huang P.L., Jain R.K. Predominant role of endothelial nitric oxide synthase in vascular endothelial growth factor-induced angiogenesis and vascular permeability. Proc Natl Acad Sci U S A. 2001;98:2604–2609. doi: 10.1073/pnas.041359198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung K.W., Leung F.P., Huang Y., Mak N.K., Wong R.N. Non-genomic effects of ginsenoside-Re in endothelial cells via glucocorticoid receptor. FEBS Lett. 2007;581:2423–2428. doi: 10.1016/j.febslet.2007.04.055. [DOI] [PubMed] [Google Scholar]
- 25.Huang Y.C., Chen C.T., Chen S.C., Lai P.H., Liang H.C., Chang Y., Yu L.C., Sung H.W. A natural compound (ginsenoside Re) isolated from Panax ginseng as a novel angiogenic agent for tissue regeneration. Pharm Res. 2005;22:636–646. doi: 10.1007/s11095-005-2500-3. [DOI] [PubMed] [Google Scholar]
- 26.Mochizuki M., Yoo Y.C., Matsuzawa K., Sato K., Saiki I., Tono-oka S., Samukawa K., Azuma I. Inhibitory effect of tumor metastasis in mice by saponins, ginsenoside-Rb2, 20(R)- and 20(S)- ginsenoside-Rg3, of Red ginseng. Biol Pharm Bull. 1995;18:1197–1202. doi: 10.1248/bpb.18.1197. [DOI] [PubMed] [Google Scholar]
- 27.Yue P.Y., Wong D.Y., Wu P.K., Leung P.Y., Mak N.K., Yeung H.W., Liu L., Cai Z., Jiang Z.H., Fan T.P. The angiosuppressive effects of 20(R)- ginsenoside Rg3. Biochem Pharmacol. 2006;72:437–445. doi: 10.1016/j.bcp.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 28.Duffy A.M., Bouchier-Hayes D.J., Harmey J.H. Vascular endothelial growth factor (VEGF) and its role in non-endothelial cells: autocrine signalling by VEGF. Madame Curie Bioscience Database. 2000 [Google Scholar]
- 29.You W.K., McDonald D.M. The hepatocyte growth factor/c-Met signaling pathway as a therapeutic target to inhibit angiogenesis. BMB Rep. 2008;41:833–839. doi: 10.5483/bmbrep.2008.41.12.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vandevyver S., Dejager L., Tuckermann J., Libert C. New insights into the anti-inflammatory mechanisms of glucocorticoids: an emerging role for glucocorticoid-receptor-mediated transactivation. Endocrinology. 2013;154:993–1007. doi: 10.1210/en.2012-2045. [DOI] [PubMed] [Google Scholar]
- 31.Asai K., Funaki C., Hayashi T., Yamada K., Naito M., Kuzuya M., Yoshida F., Yoshimine N., Kuzuya F. Dexamethasone- induced suppression of aortic atherosclerosis in cholesterol-fed rabbits. Arterioscler Thromb. 1993;13:892–899. doi: 10.1161/01.atv.13.6.892. [DOI] [PubMed] [Google Scholar]
- 32.Tonnesen M.G., Feng X., Clark R.A. Angiogenesis in wound healing. J Investig Dermatol Symp Proc. 2000;5:40–46. doi: 10.1046/j.1087-0024.2000.00014.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.