Abstract
T regulatory cells (Tregs) play a key role in modulating T cell responses. Clinical trials showed that Tregs modulate graft-versus-host disease (GvHD) after allogeneic hematopoietic stem cell transplantation (allo-HSCT). However, their ability to mediate anti-leukemic activity (graft-versus-leukemia [GvL]) is largely unknown. Enforced interleukin-10 (IL-10) expression converts human CD4+ T cells into T regulatory type 1 (Tr1)-like (CD4IL-10) cells that suppress effector T cells in vitro and xenoGvHD in humanized mouse models. In the present study, we show that CD4IL-10 cells mediate anti-leukemic effects in vitro and in vivo in a human leukocyte antigen (HLA) class I-dependent but antigen-independent manner. The cytotoxicity mediated by CD4IL-10 cells is granzyme B (GzB) dependent, is specific for CD13+ target cells, and requires CD54 and CD112 expression on primary leukemic target blasts. CD4IL-10 cells adoptively transferred in humanized mouse models directly mediate anti-tumor and anti-leukemic effects. In addition, when co-transferred with peripheral blood mononuclear cells (PBMCs), CD4IL-10 cells contribute to the GvL activity but suppress xenoGvHD mediated by the PBMCs. These findings provide for the first time a strong rationale for CD4IL-10 cell immunotherapy to prevent GvHD and promote GvL in allo-HSCT for myeloid malignancies.
Keywords: tolerance, immunotherapy, gene transfer
Graphical Abstract
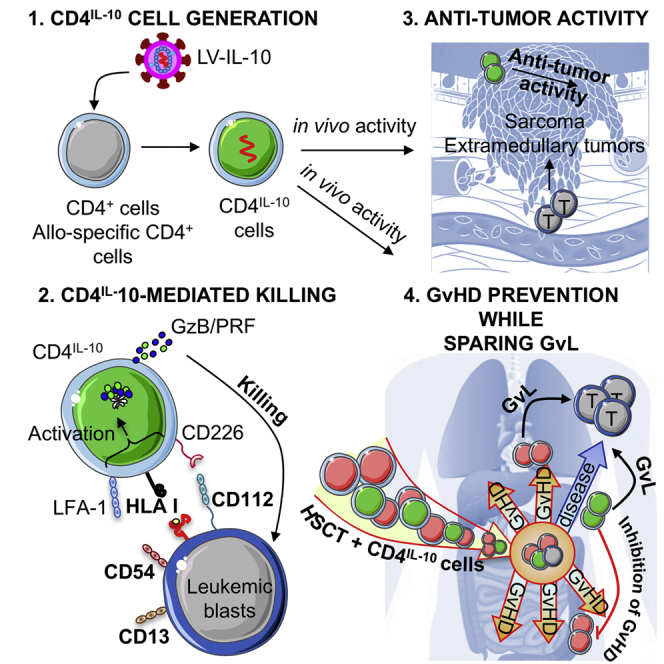
Tr1 cells are generated by overexpressing IL-10 in CD4+ T cells (CD4IL-10). In this issue of Molecular Therapy, Locafaro et al. (2017) show that CD4IL-10 cells kill myeloid leukemia in an HLA class I-dependent mechanism, mediate anti-tumor and anti-leukemic effects, and contribute to GvL while preventing GvHD in humanized mice.
Introduction
T regulatory cells (Tregs) promote and maintain immunological tolerance. Among Tregs, FOXP3+ Tregs1 and the T regulatory type 1 (Tr1) cells have been extensively investigated.2 Tr1 cells are induced in the periphery upon chronic antigen (Ag) stimulation in the presence of interleukin 10 (IL-10);2 co-express CD49b and LAG-3;3 and secrete IL-10, transforming growth factor β (TGF-β), variable amounts of interferon (IFN)-γ, and low IL-2, IL-4, and IL-17.2, 3, 4 Tr1 cells suppress T cell responses via the secretion of IL-10 and TGF-β,2, 5 and the specific killing of myeloid cells via granzyme B (GzB).6 Ag-specific Tr1 cells can be generated in vitro in the presence of IL-10 or tolerogenic DC-10.7
In the past decade, much effort has been devoted to develop methods for the in vitro induction and/or expansion of Tregs suitable for cell therapy.8 Treg-based cell therapy has been extensively tested in pre-clinical models of graft-versus-host disease (GvHD),9, 10, 11, 12 and proof-of-principle clinical trials in allogeneic hematopoietic stem cell transplantation (allo-HSCT) have demonstrated the safety of Treg-based cell therapy.13, 14, 15, 16, 17 Freshly isolated14, 15, 17 or in-vitro-expanded polyclonal CD25+ Tregs13, 16 were infused after allo-HSCT for hematological malignancies to prevent GvHD. In these studies, a reduction in GvHD severity was observed as compared with historical controls. Furthermore, it has been reported that in CD25+ Treg-treated patients, the cumulative incidence of relapse was significantly lower than in controls.17 In a clinical trial aimed at promoting immune reconstitution in the absence of severe GvHD, we demonstrated the safety and feasibility of Tr1 cell infusion in hematological cancer patients undergoing haploidentical HSCT.18 Host-specific IL-10-anergized donor T cells (IL-10 donor lymphocyte infusion [DLI]), containing Tr1 cells, were infused in the absence of immunosuppression in a small cohort of patients. Results demonstrated that after IL-10 DLI infusion, only moderate GvHD was observed and a tolerance signature was achieved. Furthermore, the treatment accelerated immune reconstitution and correlated with long-lasting disease remission.18 A major difference between the CD25+ Treg-based trials and the IL-10 DLI trial is that, in the former, a pool of polyclonal non-Ag-specific cells was administered, whereas in the latter, anergized donor-derived T cells, which contain host-specific Tr1 cells, were infused.
Despite the significant progress in establishing in vitro protocols to induce alloantigen-specific IL-10-anergized T cells for Tr1-based immunotherapy, the resulting population still contains a large proportion of effector T cells that could potentially limit the in vivo efficacy of Tr1 cells.19 To overcome this limitation, we developed a method to generate a selected population of IL-10-producing Tr1 cells by lentiviral vector (LV)-mediated human IL-10 gene transfer. IL-10-engineered CD4+ (CD4IL-10) cells display a cytokine profile and phenotype super-imposable to bona fide Tr1 cells, suppress T cell responses, lyse myeloid cell lines in vitro, and prevent xenoGvHD in vivo.20
In the present study, we investigate the anti-leukemic and anti-tumor activity of polyclonal and alloantigen-specific CD4IL-10 cells in vitro and in vivo. We demonstrate that CD4IL-10 cells generated with an LV encoding for human IL-10 and ΔNGFR (LV-IL-10) specifically kill myeloid leukemic cell lines in a human leukocyte Ag (HLA) class I-dependent, but Ag-independent, manner. CD4IL-10 cells kill also primary myeloid blasts in vitro, and this anti-leukemic activity is dependent on CD13, CD54, and CD112 expression on target cells. Furthermore, CD4IL-10 cells have a direct anti-tumor and anti-leukemic effect, and collaborate with allogeneic peripheral blood mononuclear cells (PBMCs) to mediate graft-versus-leukemia (GvL), while inhibiting xenoGvHD in vivo. These data strongly support the use of CD4IL-10 Tr1 cells as immunotherapy to prevent GvHD and promote GvL in allo-HSCT for myeloid malignancies.
Results
Polyclonal CD4IL-10 Cells Have a Tr1 Phenotype and Kill Myeloid Leukemic Cell Lines In Vitro
CD4IL-10 cells were generated by transducing CD4+ T cells with a bidirectional LV co-encoding for human IL-10 and ΔNGFR, as a clinical-grade marker gene (LV-IL-10/Δ nerve growth factor receptor [NGFR]; Figure S1A). Transduction efficiency of CD4+CD45RO− T cells was on average 51.3%, and the percentage of cell recovery after selection of ΔNGFR-expressing cells reflected the percentage of transduced cells (57% ± 19%, mean ± SD, n = 8; Table S1). After selection, in vitro expansion of polyclonal CD4IL-10 cells cultured with feeder mixture was lower compared with control cells transduced with LV-GFP/ΔNGFR (CD4ΔNGFR) (Table S1). CD4IL-10 cells generated with LV-IL-10/ΔNGFR recapitulated the Tr1 cell phenotype, as previously described for LV-IL-10/GFP.20 Expanded CD4IL-10 cells are memory (CD45RO+CD45RA−) T cells that express CD25, but not FOXP3 (Figure S1B). Moreover, CD4IL-10 cells, in contrast with control LV-transduced CD4ΔNGFR cells, but similar to naturally occurring Tr1 cells,3 secreted significantly higher levels of IL-10 and IFN-γ, and low amounts of IL-4 and IL-17 (Figure S1C). CD4IL-10 cells, but not CD4ΔNGFR cells, suppressed polyclonal T cell responses in vitro (Figure S1D). Importantly, similar to Tr1 cells,6 CD4IL-10 cells expressed significantly higher levels of CD18, CD2, and CD226 as compared with freshly isolated CD4+ memory T cells and CD4ΔNGFR cells, which have been previously demonstrated to be critical markers for Tr1-mediated cytotoxicity (Figure S2).
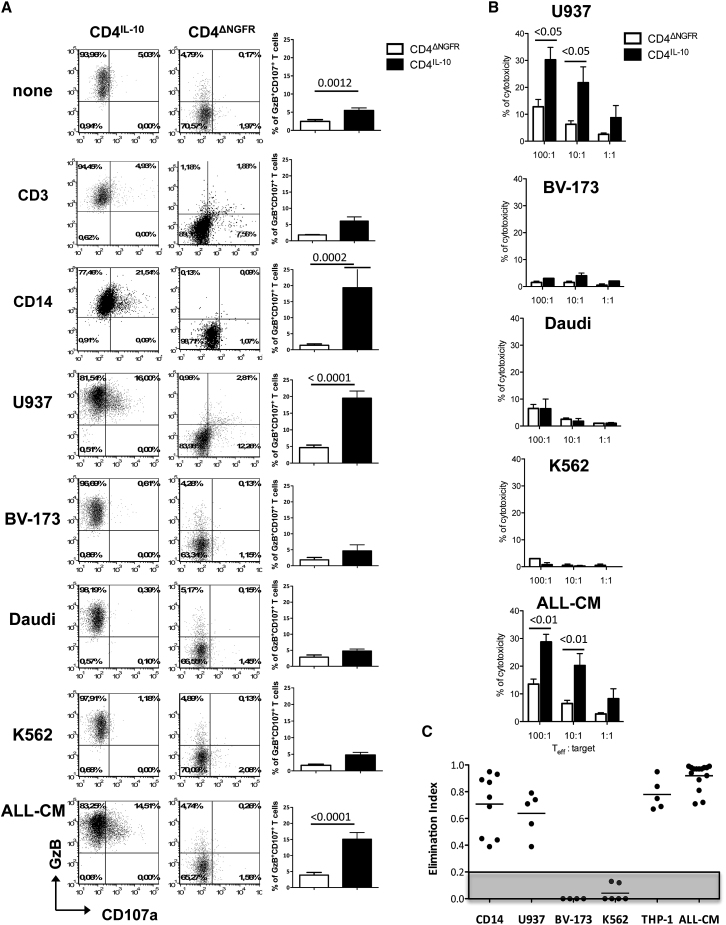
Next, we evaluated the ability of CD4IL-10 cells to kill a panel of immortalized leukemic cell lines. Freshly isolated T lymphocytes (CD3) and monocytes (CD14) were used as negative and positive control target cells, respectively. A significantly higher proportion of GzB+CD107a+ cells was observed in co-cultures of CD4IL-10 cells with CD14 cells, U937, a monocytic cell line, or ALL-CM, a cell line derived from a patient suffering from a lymphoblastic crisis of chronic myelogenous leukemia21, 22 as compared with CD4ΔNGFR cells (Figure 1A). Conversely, CD4IL-10 cells did not degranulate when cultured with CD3 cells, BV-173,23 Daudi, or K562, an erythroleukemic cell line (Figure 1A), and the percentage of GzB+CD107a+ cells in the later conditions was similar to that observed with unstimulated CD4IL-10 cells alone. CD4IL-10 cells also killed U937 and ALL-CM, but not BV-173, Daudi, or K562, cell lines at a 100:1 (effector:target) ratio at significantly higher levels than CD4ΔNGFR cells (Figure 1B). Consistently, in 4-day co-culture experiments, CD4IL-10 cells eliminated CD14 cells, U932, ALL-CM, and THP-1, but not BV-173 or K562 cell lines (Figures 1C and S3).
Figure 1.
CD4IL-10 Cells Kill Myeloid Cell Lines In Vitro
(A) Degranulation of CD4IL-10 cells. CD4IL-10 and CD4ΔNGFR cells were co-cultured with CD3, CD14, U937, BV-173, Daudi, K562, and ALL-CM target cells at a 5:1 (E:T) ratio for 6 hr, and the frequency of CD107a+GzB+ cells was measured by FACS. One representative donor (left panels) and mean ± SEM of n = 20 for CD4IL-10 and CD4ΔNGFR cells cultured alone or with ALL-CM or U937 cell lines, n = 4 for CD4IL-10 cells and n = 2 for CD4ΔNGFR cells co-cultured with CD3+ cells, n = 8 for CD4IL-10 and CD4ΔNGFR cells co-cultured with CD14+ cells, n = 5 for CD4IL-10 and CD4ΔNGFR cells co-cultured with the BV-173 cell line, n = 12 for CD4IL-10 and CD4ΔNGFR cells co-cultured with the Daudi cell line, and n = 17 for CD4IL-10 cells and CD4ΔNGFR cells co-cultured with the K562 cell line (right panels) are reported. p, Mann-Whitney test. (B) Cytotoxicity of CD4IL-10 cells. Cytotoxicity against U937, BV-173, Daudi, K562, and ALL-CM cell lines was determined by 51Cr-release assay. Mean ± SEM of cytotoxicity performed in duplicated is reported; n = 4 independent donors for CD4IL-10 and CD4ΔNGFR cells against ALL-CM and U937 cell lines, tested in two independent experiments, and n = 2 for CD4IL-10 and CD4ΔNGFR cells against BV-173, Daudi, and K562 cell lines tested in one experiment. p, two-sided Mann-Whitney test. (C) CD4IL-10 cells killed myeloid cell lines. CD4IL-10 and CD4ΔNGFR cells were co-cultured with CD14, U937, BV-173, K562, THP-1, and ALL-CM cells at a 1:1 ratio for 3 days. Residual leukemic cell lines (CD45lowCD3−) were counted by FACS, and cytolysis mediated by CD4IL-10 cells was measured as elimination index (see Materials and Methods) for each target cells. Analysis was performed in five independent experiments. Dots represent the elimination index of CD4IL-10 cells generated from different healthy donors, and lines represent mean values of the elimination index. Gray area indicates the threshold of cytotoxicity.
Overall, these data demonstrate that CD4IL-10 cells kill specifically myeloid cells and myeloid leukemic cell lines.
Alloantigen-Specific CD4IL-10 Cells Kill Myeloid Leukemic Cell Lines In Vitro in an Ag-Independent Manner
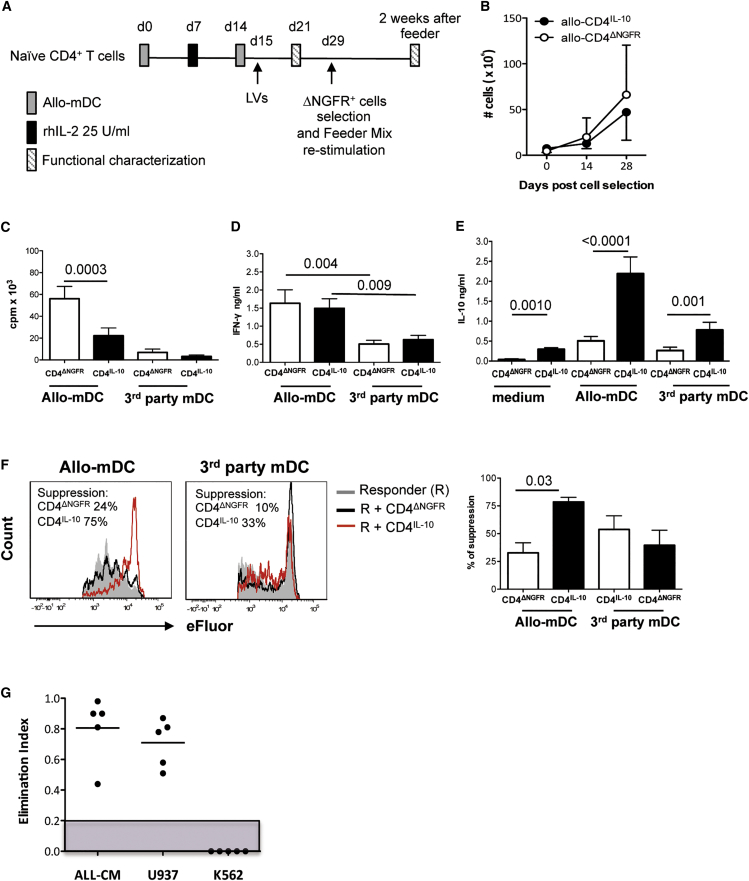
Alloantigen-specific LV-10-transduced T (allo-CD4IL-10) cells were generated by stimulation of naive CD4+ T cells with allo-mDCs and transduced upon secondary stimulation (Figure 2A). Transduction efficiency was on average 52.5%, and after selection of ΔNGFR-expressing cells, the percentage of cell recovery (45% ± 9.9%, mean ± SD, n = 10) reflected that of transduced cells (Table S2). Allo-CD4IL-10 and allo-CD4ΔNGFR cells equally expanded in vitro when cultured with feeder mixture (Figure 2B; Table S2). In contrast, allo-CD4IL-10 cells re-stimulated with allo-mDCs proliferated significantly less compared with allo-CD4ΔNGFR cells (Figure 2C), despite similar levels of IFN-γ production (Figure 2D). As expected, allo-CD4IL-10 cells spontaneously released significantly higher IL-10 amounts compared with allo-CD4ΔNGFR cells. Importantly, upon allo-mDC stimulation, IL-10 production by allo-specific CD4IL-10 cells increased and was significantly higher compared with that of control cells (Figure 2E). Allo-CD4IL-10 and allo-CD4ΔNGFR cells had similar low proliferative capacity and IFN-γ production in response to third-party Ags (third-party mDCs) (Figures 2C and 2D), but allo-CD4IL-10 cells produced higher levels of IL-10 (Figure 2E). Moreover, allo-CD4IL-10, but not allo-CD4ΔNGFR, cells efficiently suppressed the proliferation of allo-specific T cell lines activated with allo-mDCs, but not with third-party mDC (Figure 2F). Finally, allo-CD4IL-10 cells killed ALL-CM and U937, but not K562 leukemic, cell lines, similar to what we observed with polyclonal CD4IL-10 cells (Figure 2G).
Figure 2.
Enforced IL-10 Expression in Alloantigen-Specific CD4+ T Cells Promotes Their Conversion into Tr1-like Cells with the Ability to Kill Myeloid Cells
(A) Protocol to generate allo-CD4IL-10 cells. Enriched alloantigen-specific CD4+ T cells were transduced with LV-IL-10 or LV-GFP at an MOI of 20 during a secondary stimulation with the same allo-mDCs used for priming (see also Materials and Methods). (B) Allo-CD4IL-10 and allo-CD4ΔNGFR cells equally expanded in vitro. Purified allo-CD4IL-10 and allo-CD4ΔNGFR cells were expanded with feeder mixture (see Materials and Methods), and numbers of recovered cells at the indicated time points are presented. Mean ± SD of n = 6 independent experiments are presented. (C) The proliferative responses of allo-CD4IL-10 and allo-CD4ΔNGFR cells. Allo-CD4IL-10 and allo-CD4ΔNGFR cells were stimulated with allogeneic (allo-mDCs) or 3rd party mDCs (ratio 10:1), and proliferation was evaluated by [3H]thymidine incorporation on days 3 and 5, respectively. Mean ± SEM of n = 19 for allogeneic stimulation and n = 16 for 3rd-party stimulation tested in more than five independent experiments. All samples were tested in duplicate-triplicate. p, Wilcoxon matched-pairs signed rank test. (D and E) Cytokine production of allo-CD4IL-10 and allo-CD4ΔNGFR cells. (D and E) Allo-CD4IL-10 or allo-CD4ΔNGFR cells were stimulated with allo-mDCs or a third-party mDC (ratio 10:1), and IFN-γ (D) and IL-10 (E) production were determined in culture supernatants 48 hr after activation by ELISA. Mean ± SEM of n = 12–26 donors tested in five independent experiments. All samples were tested in duplicate-triplicate. p, Wilcoxon matched-pairs signed rank test. (F) Suppressive activity of allo-CD4IL-10 cells. Autologous allo-specific CD4+ T cell lines (responders) were labeled with eFluor dye and stimulated with allo-mDCs (ratio 10:1) alone or in the presence of allo-CD4IL-10 or allo-CD4ΔNGFR cells at a 1:1 ratio. After 3 days of culture the suppressive ability was determined by CFSE/eFluor dilution of CD4+ΔNGFR− cells. One representative donor (left panel) and mean ± SEM of n = 6 for allogeneic stimulation and 3rd-party stimulation tested in three independent experiments (right panel) are shown. The suppression mediated by allo-CD4IL-10 cells or allo-CD4ΔNGFR cells was calculated as follows: ([proliferation of responders in the presence of transduced cells/proliferation of responders alone] × 100). p, Wilcoxon matched-pairs signed rank test. (G) Allo-CD4IL-10 cells killed myeloid cell lines. CD4IL-10 and CD4ΔNGFR cells were co-cultured with ALL-CM, U937, and K562 cell lines at 1:1 ratio for 3 days. Residual leukemic cell lines (CD45lowCD3−) were counted by FACS. Cytolysis mediated by CD4IL-10 cells was measured as elimination index (see Materials and Methods) for each target cell. Analysis was performed in two independent experiments. Dots represent the elimination index of CD4IL-10 cells generated from different healthy donors, and lines represent mean values of the elimination index. Gray area indicates the threshold of cytotoxicity.
These results show that enforced IL-10 expression in allo-specific CD4+ T cells promotes their conversion into Tr1-like suppressor cells able to kill myeloid cell lines.
CD4IL-10 Cells Kill Myeloid Cell Lines in a GzB-Dependent and HLA Class I-Mediated Recognition
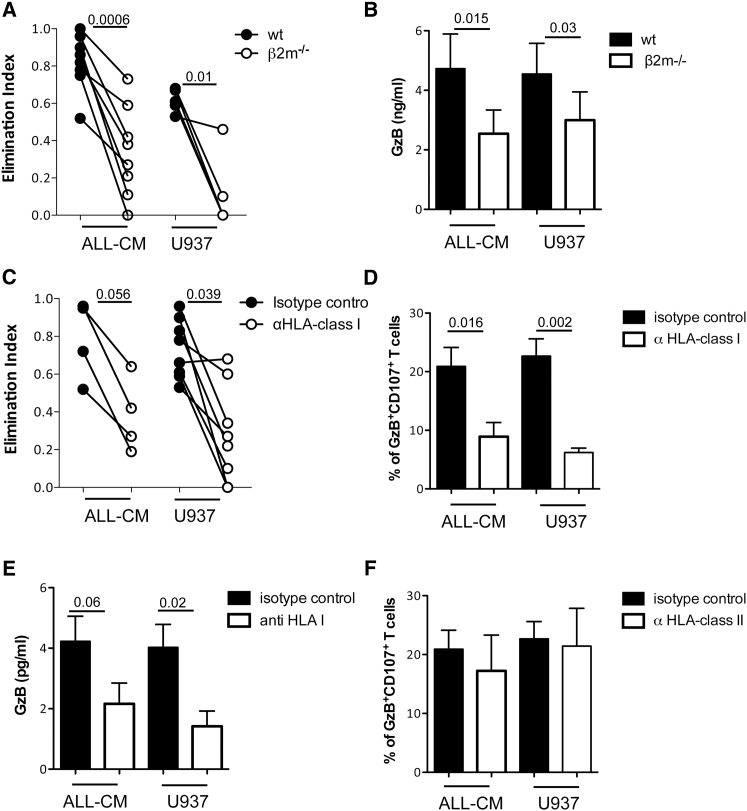
In non-activating conditions, >95% of CD4IL-10 cells expressed GzB, whereas <5% of CD4ΔNGFR cells were GzB+ (Figure 1A). Addition of the GzB inhibitor Z-AAD-CMK inhibited the killing of ALL-CM and U937 cell lines by CD4IL-10 cells in a dose-dependent manner (Figure 3), indicating that GzB is required for CD4IL-10-mediated killing.
Figure 3.
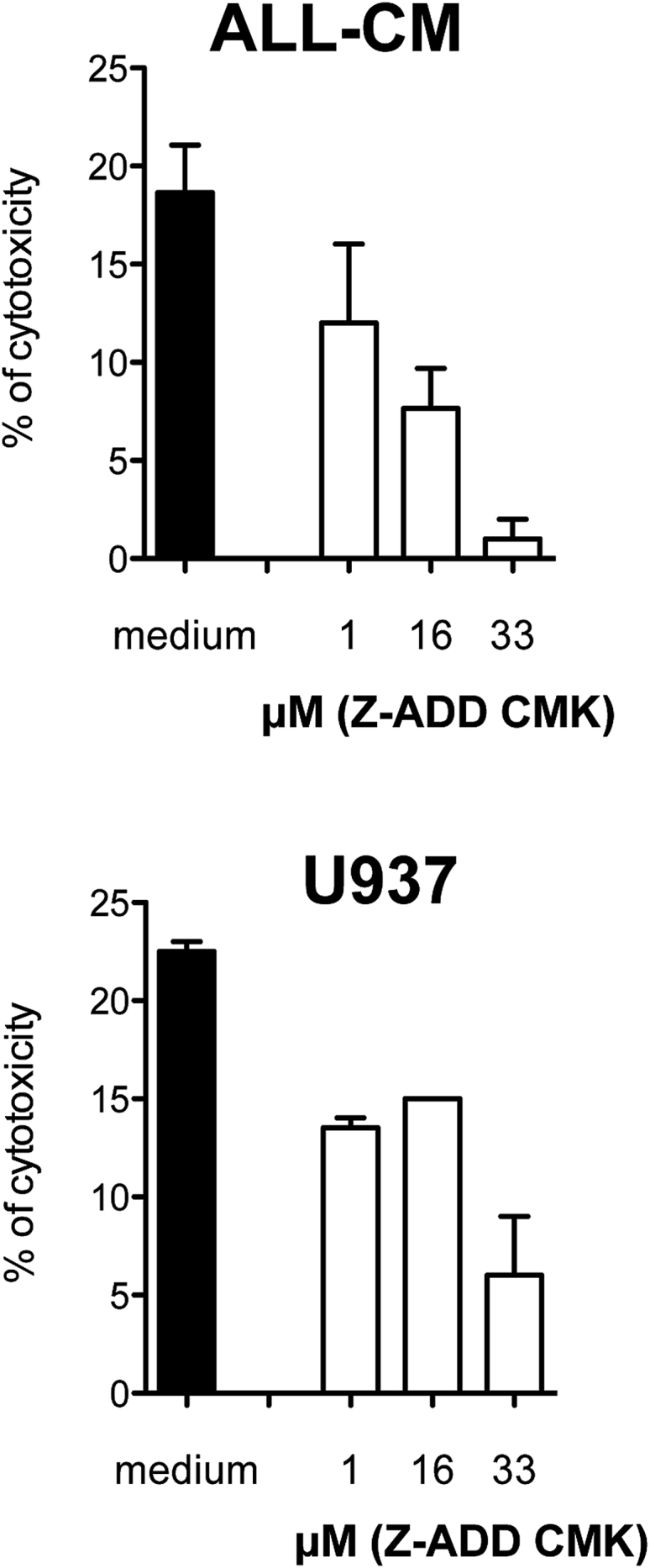
CD4IL-10 Cell-Mediated Killing of Myeloid Cell Lines Is GzB-Dependent
CD4IL-10 cells were pre-incubated with the GzB-inhibitor Z-AAD-CMK at the indicated concentrations and cultured with ALL-CM and U937 target cell lines at a 50:1 (E:T) ratio. Cytotoxicity was determined by 51Cr release. Mean ± SEM of n = 3 independent donors performed in triplicates is reported. Analysis was performed in one experiment.
The U937, THP-1, and ALL-CM cell lines that were killed by CD4IL-10 cells expressed CD54, HLA class I, CD58, CD155, and CD112, whereas K562 and Daudi cell lines that were not killed by CD4IL-10 cells were HLA class I-negative and expressed CD54 and variable levels of CD58, CD155, or CD112 (Figure S4). The BV173 cell line, which was not killed by CD4IL-10 cells, expressed HLA class I and CD58, but not CD54 and CD155, and it was CD112 dim (Figure S4). These findings suggest that CD4IL-10-mediated killing requires HLA class I expression, stable CD54/LFA-1-mediated adhesion, and CD2/CD58- and CD155-CD112/CD226-mediated activation. To determine the need of HLA class I expression on target cells for an efficient CD4IL-10-mediated killing, we selectively deleted HLA class I expression on ALL-CM and U937 cell lines by disrupting the β2-microglobulin (β2 m) encoding gene. β2 m-deficient (β2 m−/−). ALL-CM and U937 cell lines were killed by CD4IL-10 cells at significantly lower levels compared with wild-type ALL-CM and U937 cell lines (Figure 4A). CD4IL-10 cells activated by β2 m−/− ALL-CM and U937 cell lines also released significantly lower levels of GzB compared with CD4IL-10 cells activated by wild-type ALL-CM and U937 cell lines (Figure 4B). Further, the addition of a pan anti-HLA class I monoclonal antibody (mAb) inhibited CD4IL-10-mediated killing of ALL-CM and U937 cell lines (Figure 4C) and prevented the degranulation of CD4IL-10 cells, as shown by the significantly lower frequency of GzB+CD107a+ cells (Figure 4D). The lower degranulation observed in the presence of an anti-HLA class I mAb paralleled the significantly lower levels of GzB released by CD4IL-10 cells (Figure 4E). Interestingly, addition of a pan anti-HLA class II mAb during co-culture of CD4IL-10 cells with ALL-CM or U937 cell lines did not prevent degranulation and release of GzB (Figure 4F; data not shown).
Figure 4.
CD4IL-10 Cell-Mediated Killing of Myeloid Cell Lines Is HLA Class I Dependent
(A) HLA class I-deficient cell lines are resistant to CD4IL-10-mediated killing. CD4IL-10 and CD4ΔNGFR cells were co-cultured with ALL-CM, β2 m−/− ALL-CM, U937, or β2 m−/− U937 target cell lines at a 1:1 (E:T) ratio for 3 days. Residual leukemic cell lines (CD45lowCD3−) were counted by FACS. Cytolytic effect mediated by CD4IL-10 cells was measured as elimination index for each target cell. Dots represent CD4IL-10 generated from eight different healthy donors co-cultured with ALL-CM and β2 m−/− ALL-CM and five different healthy donors co-cultured with U937 and β2 m−/− U937 tested in two independent experiments. p, two-sided Wilcoxon matched-pairs signed rank test. (B) CD4IL-10 cells activated with HLA class I-deficient cell lines released low GzB. CD4IL-10 and CD4ΔNGFR cells were co-cultured with ALL-CM, β2 m−/− ALL-CM, U937, or β2 m−/− U937 target cell lines at a 1:1 (E:T) ratio, and the levels of GzB in culture supernatants were analyzed by ELISA 24 hr after activation. Mean ± SEM of GzB released by seven different healthy donors tested in two independent experiments is reported. p, two-sided Wilcoxon matched-pairs signed rank test. (C) Addition of neutralizing anti-HLA class I mAb prevents killing of ALL-CM and U937 cell lines. CD4IL-10 and CD4ΔNGFR cells were co-cultured with ALL-CM or U937 target cell lines at a 1:1 (E:T) ratio in the presence of 10 μg/mL anti-HLA class I or isotype control mAbs for 3 days. Residual leukemic cell lines (CD45lowCD3−) were counted by FACS. Cytolytic effect by CD4IL-10 cells was measured as elimination index for each target cell. Dots represent CD4IL-10 cells generated from four different healthy donors co-cultured with ALL-CM and eight different healthy donors co-cultured with U937 tested in two independent experiments. p, two-sided Wilcoxon matched-pairs signed rank test. (D) Degranulation of CD4IL-10 cells is prevented by the addition of neutralizing anti-HLA class I mAb. CD4IL-10 and CD4ΔNGFR cells were co-cultured with ALL-CM or U937 target cell lines at 5:1 (E:T) ratio in the presence of 10 μg/mL anti-HLA class I or isotype control mAbs for 6 hr. The percentage of CD107a+GzB+ cells was measured by FACS. Mean ± SEM of GzB+CD107a+ cells generated from eight different healthy donors tested in two independent experiments is reported. P, two-sided Wilcoxon matched-pairs signed rank test. (E) CD4IL-10 cells activated in the presence of neutralizing anti-HLA class I mAb released low GzB. CD4IL-10 cells were co-cultured with ALL-CM or U937 target cell lines at 1:1 (E:T) ratio in the presence of 10 μg/mL anti-HLA class I or isotype control mAbs, and the levels of GzB in culture supernatants were analyzed by ELISA 24 hr after activation. Mean ± SEM of GzB released by 11 different healthy donors co-cultured with ALL-CM and U937 in the presence of isotype control Abs and 4 different healthy donors co-cultured with ALL-CM and U937 in the presence of anti-HLA class I Abs tested in two independent experiments are reported. p, two-sided Wilcoxon matched-pairs signed rank test. (F) Degranulation of CD4IL-10 cells is not prevented by the addition of neutralizing anti-HLA class II mAb. CD4IL-10 and CD4ΔNGFR cells were co-cultured with ALL-CM or U937 target cell lines at 5:1 (E:T) ratio in the presence of 10 μg/mL anti-HLA class II or isotype control mAbs for 6 hr. The percentage of CD107a+GzB+ cells was measured by FACS. Mean ± SEM of GzB+CD107a+ cells generated from four different healthy donors tested in two independent experiments is reported.
These findings indicate that HLA class I expression on myeloid target cells is required for efficient CD4IL-10-mediated lysis, and that the interaction of CD4IL-10 cells with HLA class I-expressing target cells mediates their activation and the consequent release of GzB.
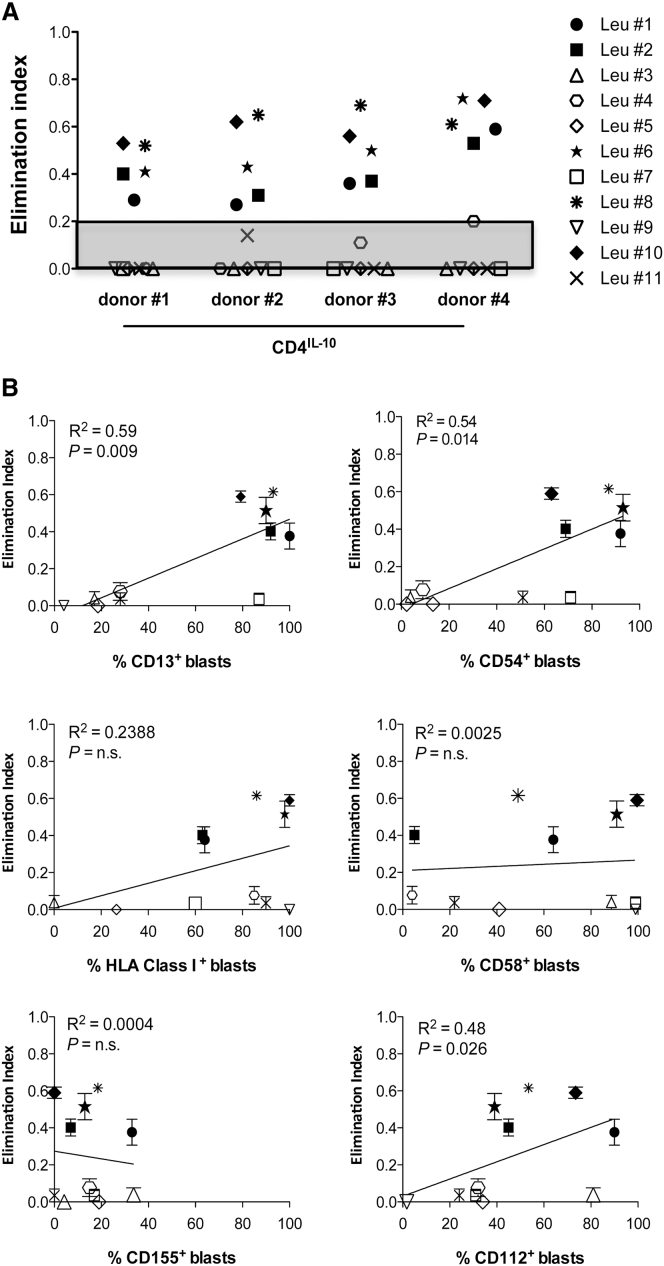
CD4IL-10 Cells Specifically Kill Myeloid Leukemic Primary Blasts
We next tested the ability of CD4IL-10 cells to kill primary blasts isolated from patients with acute myeloid leukemia (AML blasts) at diagnosis and classified according to the French-American-British (FAB) subtypes and World Health Organization (WHO) categories (Table 1). As negative controls, we used primary non-myeloid blasts isolated from patients with acute lymphoblastic leukemia (ALL blasts) at diagnosis (Table 1). CD4IL-10 cells killed four out of eight primary AML blasts (Figure 5A). CD4IL-10-mediated lysis correlated with the expression of CD13, CD54, and CD112 on AML blasts, but not with CD58 or CD155 (Figure 5B). CD4IL-10 cells eliminated CD13+CD54+CD112+ AML blasts from patients Leu #1, Leu #2, Leu #6, and Leu #8, but they did not kill AML blasts from patients Leu 3, Leu 4, and Leu #5, which were CD13− and CD54−. Moreover, CD4IL-10 cells did not eliminate CD13+CD54+CD112− AML blasts from patient Leu #7 (Figure 5B). Interestingly, CD4IL-10 cells eliminated blasts expressing CD13, CD54, and CD112 from one of the three ALL patients (Leu #10) (Figure 5B).
Table 1.
Patient Characteristics
| Leu # | Patients | Sex/Age (year) | FABa | WHOb | Cytogenetics | Molecular Markers | Source | Blasts (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | AML 61 | F/71 | M0 | AML | NA | Flt3 ITD+/NPMI+ | PB | 92 |
| 2 | AML 63 | F/83 | M0 | AML with minimal maturation | del7, t(1;7), t(4;12) | Flt3 ITD+/NPMI+ | PB | 98 |
| 3 | AML 39 | F/47 | M1 | AML without maturation | 46, XX | Flt3 ITD−/NPMI− | BM | 79 |
| 4 | AML 60 | F/64 | M1 | AML | 46, XX | Flt3 ITD+/NPMI− | BM | 78 |
| 5 | AML 1 | F/50 | M1 | AML without maturation | NA | Flt3 ITD−/NPMI+ | PB | 98 |
| 6 | AML 64 | M/60 | M2 | AML with t(8;21)RUNX1-RUNKX1T1 | del9, t(8;21) | Flt3 ITD+/NPMI+ | PB | 26 |
| 7 | AML 37 | F/59 | M2 | AML with maturation | 46, XX | Flt3 ITD−/NPMI+ | BM | 54 |
| 8 | AML 5 | F/66 | M2 | AML with maturation | dup8 | Flt3 ITD+/NPMI+ | PB | 65 |
| 9 | ALL 57 | M/38 | ALL | ALL-T | NA | NA | PB | 46 |
| 10 | ALL 48 | F/22 | ALL | ALL | NA | NA | BM | 85 |
| 11 | ALL 62 | F/76 | ALL | ALL-B L2 | NA | Flt3 ITD+/NPMI+ | PB | 91 |
BM, bone marrow; F, female; Flt3 ITD, Flt3 internal-tandem duplication; M, male; NA, not applicable; NPM1, nucleophosmin; PB, peripheral blood.
AML and ALL subtypes according to the French-American-British (FAB) classification.
AML and ALL categories according to the World Health Organization (WHO) classification.
Figure 5.
CD4IL-10 Cells Specifically Kill Primary Blasts Expressing CD13, CD54, and CD112 In Vitro
(A) CD4IL-10 and CD4ΔNGFR cells were co-cultured with primary blasts at a 1:1 (E:T) ratio for 3 days. Residual leukemic blasts (CD45lowCD3−) were counted by FACS. Cytolytic effect of CD4IL-10 cells was measured as elimination index (see Materials and Methods) for each target cell. Dots represent each primary blast co-cultured with CD4IL-10 cells. Analysis was performed in two independent experiments. Gray area indicates the threshold of cytotoxicity. (B) Correlation between cytotoxicity and expression of specific markers on primary blasts. Plots represent percentages of CD13, CD54, HLA class I, CD58, CD155, and CD112-positive primary blasts versus elimination index of CD4IL-10 cells for each primary blast tested in a co-culture assay (mean ± SEM). The line represents the linear regression. The p value of the correlation and the coefficient of determination (R2) are reported (two-tailed test).
Although a small cohort of patients was tested, these data suggest that CD4IL-10 cells selectively eliminate CD13+ leukemic cells and that optimal CD4IL-10-mediated killing requires stable CD54/LFA-1-mediated adhesion and CD112/CD226-mediated activation.
CD4IL-10 Cells Mediate Anti-tumor Effects In Vivo
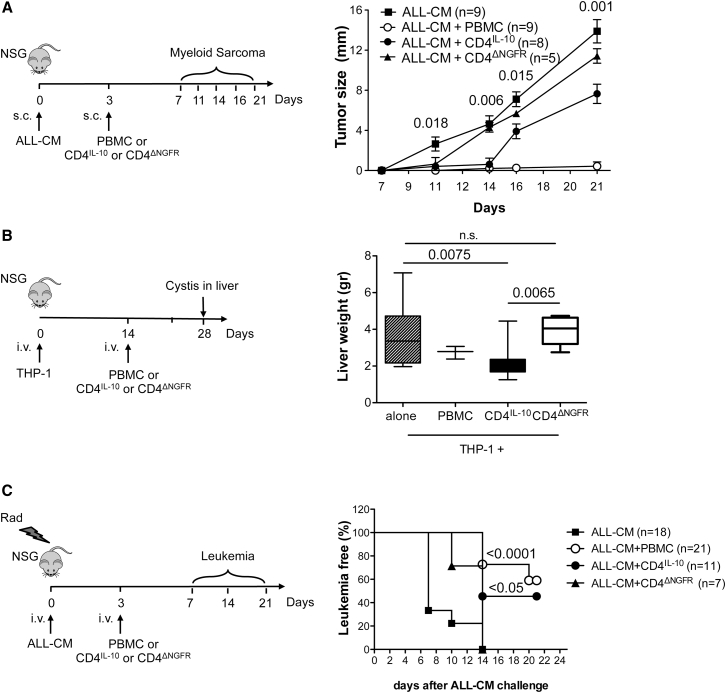
We tested the anti-leukemic activity of CD4IL-10 cells in vivo in four different humanized models: the subcutaneous myeloid sarcoma, the extra-medullary THP-1 myeloid tumor,24 and two ALL-CM leukemia models. Non-obese diabetic scid gamma (NSG) mice subcutaneously injected with ALL-CM cells developed a myeloid sarcoma 3 weeks later. Subcutaneous injection of CD4IL-10 cells day 3 after ALL-CM infusion significantly delayed myeloid sarcoma growth (Figure 6A). No tumor development was detected within the first 14 days in CD4IL-10-treated mice, and on day 21 the tumor size was significantly smaller in CD4IL-10-treated mice as compared with both ALL-CM control mice and CD4ΔNGFR-injected cells (p = 0001 and p = 0.029, respectively; Figure 6A). CD4ΔNGFR-injected mice developed myeloid sarcomas similar to mice injected with ALL-CM alone. Injection of allogeneic PBMCs, used as positive controls, completely prevented tumor growth (Figure 6A). CD4IL-10 cells did not induce a significant reduction in the tumor size in NSG mice subcutaneously injected with β2 m−/− ALL-CM cell lines (Figure S5), indicating that HLA class I expression on tumor cells is required for CD4IL-10 cell in vivo anti-tumor activity.
Figure 6.
CD4IL-10 Cells Mediate Anti-tumor Activity In Vivo
(A) CD4IL-10 cells prevent sarcoma growth. NSG mice were subcutaneously injected with ALL-CM (2 × 106 cells/mouse) and on day 3, mice received allogeneic PBMCs (2 × 106 cells/mouse), CD4IL-10 cells, or CD4ΔNGFR cells (1 × 106 cells/mouse). Tumor growth was measured at the indicated time points. Data obtained from three independent experiments are presented. Statistical analysis was performed by comparing CD4IL-10 cell-treated mice (ALL-CM + CD4IL-10) versus un-treated control mice (ALL-CM): p, two-sided Mann-Whitney test. (B) CD4IL-10 cells mediate anti-tumor activity in a model of extra-medullary tumor. NSG mice were intravenously injected with THP-1 leukemia cells (2 × 106 cells/mouse), and 14 days later mice received allogeneic PBMCs (2 × 106 cells/mouse), CD4IL-10 cells, or CD4ΔNGFR cells (1 × 106 cells/mouse). Tumor growth was analyzed by measuring the weight of THP-1-infiltrated livers. Mean ± SEM of n = 12 mice for THP-1, n = 3 mice for THP-1+ PBMCs, n = 8 mice for THP-1 + CD4IL-10 cells, and n = 7 mice for THP-1 + CD4ΔNGFR cells are presented. Data obtained from two independent experiments are presented; p, two-sided Mann Whitney test. (C) CD4IL-10 cells delay leukemia development. NSG mice were sub-lethally irradiated and intravenously injected with ALL-CM (5 × 106 cells/mouse). On day 3, mice received allogeneic PBMCs (5 × 106 cells/mouse), CD4IL-10 cells, or CD4ΔNGFR cells (2.5 × 106 cells/mouse). Percentages (%) of leukemia-free mice, presence of less than 50% of circulating human blast hCD45+hCD3− in peripheral blood, are shown. Data obtained from three independent experiments are presented. Statistical analyses were performed by comparing treated mice versus un-treated control mice (ALL-CM): p, one-way ANOVA plus Bonferroni posttest.
Next, we assessed the anti-leukemic activity of CD4IL-10 cells on a THP-1 myeloid tumor.24 In this model, mice injected with THP-1 cell lines developed cysts in the liver starting from week 2, and at week 4 the liver weight was 3.7 ± 1.7 g (mean ± SD, n = 12; Figure 6B). Injection of PBMCs 2 weeks after THP-1 cell infusion prevented tumor growth as demonstrated by the reduced liver weight (2.7 ± 0.3 g, mean ± SD, n = 3; Figure 6B) and the reduced number of liver cysts and liver-infiltrating THP-1 cells (Figure S6). Adoptive transfer of CD4IL-10 cells 2 weeks after THP-1 injection significantly inhibited tumor growth (liver weight: 2.1 ± 0.9 g, mean ± SD, n = 8; Figure 6B). In addition, CD4IL-10-treated mice had a lower number of liver cyst and liver-infiltrating THP-1 cells (Figure S6). In contrast, no differences were observed in tumor development in CD4ΔNGFR-treated versus untreated THP-1-injected mice (liver weight: 3.9 ± 0.8 g, mean ± SD, n = 7, and 3.7 ± 1.7 g, mean ± SD, n = 12, respectively; Figures 6B and S6).
We next evaluated whether CD4IL-10 cells mediate anti-leukemic effects in an ALL-CM leukemia model. NSG mice were sub-lethally irradiated, injected with ALL-CM cell lines, and after 3 days with allogeneic PBMCs, CD4IL-10 cells, or CD4ΔNGFR cells (Figure 6C). Adoptive transfer of CD4IL-10 cells significantly delayed leukemia progression, whereas treatment with CD4ΔNGFR cells did not. Importantly, the leukemia progression in CD4IL-10-injected mice was not significantly different from that observed with PBMC-injected mice (Figure 6C).
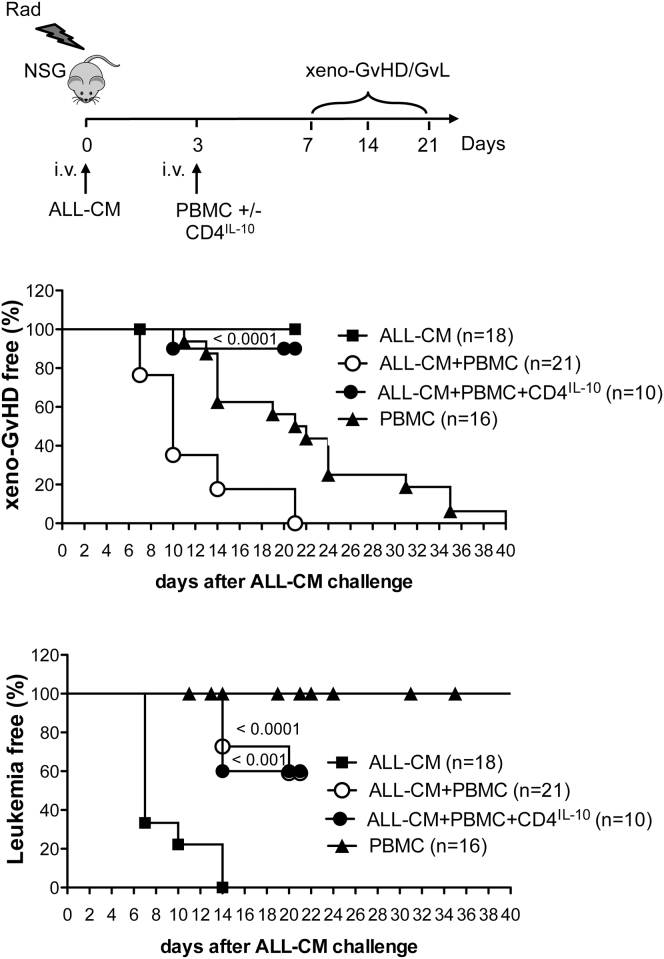
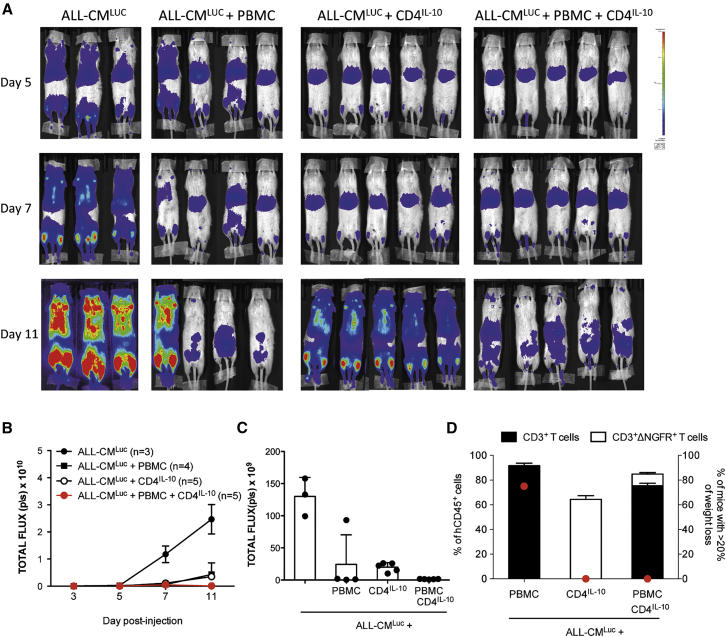
We previously showed that adoptive transfer of CD4IL-10 cells prevents xenoGvHD mediated by human CD4+ T cells in non-obese diabetic-severe combined immunodeficiency (NOD-SCID) mice.20 Here, we tested the effects of CD4IL-10 cells in irradiated NSG mice injected with ALL-CM cell lines and PBMCs. In this setting, PBMCs protected mice from leukemia, but induced a lethal xenoGvHD (Figure 7). Mice injected with PBMCs in the absence of ALL-CM developed xenoGvHD with a slower kinetic compared with mice injected with ALL-CM+PBMCs, and as expected, they did not develop leukemia (Figure 7). This different kinetic could be because of the presence of allogeneic ALL-CM, which may provide an additional signal for PBMC activation in vivo. Interestingly, adoptive transfer of CD4IL-10 cells together with PBMCs mediated protection from leukemia in 60% of the mice and prevented development of xenoGvHD in the mice (Figure 7). Using a bioluminescence analysis, we observed that CD4IL-10 cells infused in combination with PBMCs contributed in preventing leukemia development (Figures 8A–8C) and at the same time completely blocked lethal xenoGvHD mediated by PBMCs (Figure 8D).
Figure 7.
Adoptive Transfer of CD4IL-10 Cells Inhibits Leukemia Development and Prevents XenoGvHD Mediated by Human Allogeneic PBMCs
NSG mice were sub-lethally irradiated and intravenously injected with ALL-CM (5 × 106 cells/mouse). On day 3, mice received allogeneic PBMCs (5 × 106 cells/mouse) alone or in combination with CD4IL-10 cells (2.5 × 106 cells/mouse). As control, irradiated mice were intravenously injected on day 3 with PBMCs (5 × 106 cells/mouse) alone. Percentages (%) of xenoGvHD-free mice, presence of less than 50% of circulating human T lymphocytes hCD45+hCD3+ in peripheral blood with a loss of weight lower than 20% (middle panel), and % of leukemia-free mice, presence of less than 50% of circulating human blast hCD45+hCD3− in peripheral blood (lower panel), are shown. Data obtained from nine independent experiments are presented. Statistical analyses were performed by comparing ALL-CM + PBMCs + CD4IL-10 versus ALL-CM + PBMCs (middle panel) and ALL-CM + PBMCs + CD4IL-10 or ALL-CM + PBMCs versus un-treated control mice (ALL-CM) (lower panel): P, one-way ANOVA plus Bonferroni posttest.
Figure 8.
CD4IL-10 Cells Contribute with Allogeneic PBMCs in Mediating GvL while Inhibiting XenoGvHD
NSG mice were sub-lethally irradiated and intravenously injected with ALL-CMLuc cells (5 × 106 cells/mouse), and received allogeneic PBMCs (5 × 106 cells/mouse) or CD4IL-10 cells (2.5 × 106 cells/mouse) alone or in combination. (A and B) Leukemia development was monitored by BLI at the indicated time points. (A and B) Single mice scanning (A) and mean ± SEM of the total flux (photons/seconds) of single mice over time (B) are shown. (C) Analysis of the total flux (photons/seconds) at day 11. Bars represent mean ± SEM of the total flux of mice analyzed at day 11 after ALL-CMLuc cell injection; dots represent a single mouse. (D) Analysis of xenoGvHD occurrence. Bars represent the mean ± SEM of the % of human CD3+ lymphocytes and CD3+ΔNGFR+ cells within hCD45+ cells in peripheral blood of mice analyzed at day 11 post-treatment, and dots represent the mean % of treated mice with a loss of weight >20%. Data obtained from one representative experiment out of two performed are presented.
Overall, these findings demonstrate that CD4IL-10 cells have direct anti-tumor and anti-leukemic effects in vivo in humanized mouse models. In addition, CD4IL-10 cells and PBMCs have a cumulative GvL effect, but CD4IL-10 cells prevent xenoGvHD induced by allogeneic PBMCs.
Discussion
We previously reported that enforced expression of IL-10 confers a Tr1-like phenotype and function to human CD4+ T cells, including killing of myeloid cells.20 In this study, we show that Tr1 (CD4IL-10) cells generated with a novel bidirectional LV encoding for human IL-10 and ΔNGFR, as a clinical-grade marker gene, upregulate the expression of CD18, CD2, and CD226, and secrete GzB. CD4IL-10 cells acquire cytotoxic activity restricted to myeloid cells, including myeloid leukemic cell lines and, of note, primary myeloid leukemic blasts. We demonstrate that alloantigen-specific Tr1 cells obtained by LV-10 transduction of allogeneic T cells (allo-CD4IL-10 cells) are also able to kill myeloid cells in an Ag-independent manner. The myeloid-specific killing mediated by CD4IL-10 is HLA class I dependent; however, HLA-class I expression on target CD13+ primary blasts is not sufficient to promote the CD4IL-10-mediated cytotoxicity, which also requires stable CD54-mediated adhesion and activation via CD226. In humanized mouse models, CD4IL-10 cells mediate potent direct anti-tumor and anti-leukemic effects, prevent xenoGvHD mediated by allogeneic effector cells, and contribute to the GvL effect.
Previous studies indicated that Tregs can kill effector T cells through a granzyme-dependent mechanism and that this cytotoxicity contributes to their suppressive function.25, 26 In addition, we showed that Tr1 cells eliminate myeloid, but not lymphoid, cells.6 In the present study, we demonstrate that the cytotoxic activity of CD4IL-10 cells is specifically directed against myeloid leukemic cells and myeloid blasts. Furthermore, we show that CD4IL-10 cells have cytotoxic activity also in vivo. We prove that the lack of HLA class I on target cells, or the inhibition of HLA class I recognition by neutralizing mAbs, abrogates the CD4IL-10- mediated killing in vitro and in vivo, suggesting that activation of CD4IL-10 cells through receptor-HLA class I interaction is necessary for GzB release and killing of target cells. Conversely, inhibition of HLA class II does not impair CD4IL-10 cell activation and the elimination of target myeloid cells. This mechanism of target recognition and killing mediated by CD4IL-10 cells resembles the recognition and activation of NK cells; however, CD4IL-10 cells do not express specific killer-cell immunoglobulin-like receptors (KIRs) or CD94/NKG2D (data not shown). Future studies are required to identify the specific activating receptor that recognizes HLA class I molecules on target cells triggering the activation of CD4IL-10 and Tr1 cells.
The correlation between the expression of the myeloid marker CD13 on target cells and the ability of CD4IL-10 cells to eliminate primary leukemic blasts suggests that CD13 expression determines the myeloid specificity of CD4IL-10 cell-mediated cytotoxicity. Killing of primary blasts by CD4IL-10 cells also correlates with CD54 expression, showing the importance of adhesion between target cells and CD4IL-10 cells for effective cytolysis. CD54 on myeloid blasts allows stable and prolonged interaction with LFA-1 on CD4IL-10 cells, which enables the target-directed secretion of lytic granules containing GzB. In addition, similar to what was described for NK-mediated lysis of AML blasts,27 the interaction between CD112 on myeloid leukemic target blasts and CD226 on CD4IL-10 cells is required for CD4IL-10 cell activation. Because CD226 is involved in the downstream signaling cascade of LFA-1,28 we speculate that CD112/CD226 interaction contributes to the stabilization of CD4IL-10-target cell conjugates that triggers CD4IL-10 cell degranulation. CD226-mediated activation of T cells is inhibited by T cell immunoreceptor with Ig and ITIM domains (TIGIT),29 another receptor for CD112 and CD155.30 TIGIT binds to CD155 with higher affinity than CD226 and inhibits the cytotoxic activity and IFN-γ production by NK cells.31, 32 However, primary blasts express CD155 at lower levels than CD112,27, 33 and CD4IL-10-mediated lysis does not correlate with CD155 expression. Thus, although CD4IL-10 cells express both TIGIT (data not shown) and CD226, activation via CD226/CD112 is dominant over the inhibitory signal mediated by TIGIT/CD155. Overall, our findings indicate that CD13, CD54, and CD112 expression on leukemic blasts may be used as biomarkers predictive of the anti-leukemic effect mediated by CD4IL-10 cells.
CD4IL-10 cells mediate anti-tumor and anti-leukemic effect in vivo in humanized mouse models of solid myeloid tumors and leukemia. The anti-tumor effect was strictly related to the co-localization of CD4IL-10 and the tumor cells. CD4IL-10 cells displayed anti-tumor activity when locally injected within the myeloid sarcoma, or when administered systemically in mice with liver-bearing myeloid tumors, or in mice with leukemia where CD4IL-10 cells home to the target tissue and co-localize with the tumor cells (data not shown). The finding that CD4IL-10 cells eliminate myeloid leukemia in a TCR-independent, but HLA class I-dependent, manner suggests their possible use to limit, and possibly to overcome, leukemia relapse caused by the loss of not-shared HLA alleles after allo-HSCT.34, 35, 36
Recognition of host alloantigens by donor T lymphocytes is responsible for GvHD after allo-HSCT, a major cause of transplant-related morbidity and mortality. Donor T cells also recognize alloantigens on leukemic cells, thus mediating a beneficial GvL.37 Prevention and treatment of GvHD currently relies on depletion of T cells from the graft or on general immunosuppression, which, however, abrogates the GvL effect, increasing the incidence of leukemia relapse.38 Establishing a regimen that prevents GvHD without affecting GvL represents a key challenge in allo-HSCT treatment of hematological malignancies. Pre-clinical studies showed that immunotherapy with Tregs12 or CD4IL-10 cells20 inhibits xenoGvHD. In addition, clinical studies with Tr1 cell-containing DLI18 or CD25+ Tregs15, 16, 17 suggest that these cells are able to inhibit GvHD. Here, we demonstrate that adoptive transfer of CD4IL-10 cells prevents xenoGvHD mediated by allogeneic PBMCs and delays leukemia development. This unique property of CD4IL-10 cells is due to their intrinsic anti-leukemic function and to their ability to suppress the peripheral T cells, mediating GvHD. In vivo bioluminescence analyses confirm that CD4IL-10 cells collaborate with PBMCs to promote a GvL effect, but inhibit the xenoGvHD mediated by PBMCs. We envision that CD4IL-10 cells, upon HLA class I- and CD226-mediated activation, exert a direct anti-leukemic effect eliminating CD13+ myeloid leukemic cells, whereas after TCR-mediated activation, they secrete IL-10 and suppress allo(xeno)-reactive human T cells. Moreover, transgenic IL-10 expression favors the expansion of CD8+ T cells with anti-tumor activity;39 thus, it is possible that CD4IL-10 cells preferentially suppress the effector CD4+ T cells, which mediate GvHD, but spare effector CD8+ T cells, which mediate GvL.
In conclusion, we have established reproducible and robust methods to generate a large and homogeneous pool of polyclonal and alloantigen-specific human Tr1-like cells. We have demonstrated that CD4IL-10 cells (1) specifically kill CD13+CD54+CD112+ myeloid leukemic cells; (2) kill myeloid cells in a HLA class I-dependent but Ag-independent manner; (3) mediate anti-tumor and anti-leukemic activities in vivo; and (4) contribute to the GvL activity, but suppress the xenoGvHD mediated by the PBMCs in vivo. These findings pave the way for the use of CD4IL-10 cell immunotherapy to prevent GvHD and promote GvL in allo-HSCT for myeloid malignancies.
Materials and Methods
Subjects
All protocols were approved by the Institutional Review Board, and samples were collected under written informed consent according to the Declaration of Helsinki. AML and ALL patient characteristics referred to the San Raffaele Hematology and Bone Marrow Transplantation Unit at diagnosis are listed in Table 1.
Cell Preparation and Cell Lines
PBMCs were prepared by centrifugation over Ficoll-Hypaque gradients. CD4+ T cells were purified with the CD4 T cell isolation kit (Miltenyi Biotec), resulting in purity of >95%. CD4+ T cells were then depleted of CD45RO+ cells using anti-CD45RO-coupled magnetic beads and LD-negative selection columns (Miltenyi Biotech). The proportion of CD4+CD45RA+ in the selected population was consistently >90%. CD14+ and CD3+ T cells were purified by positive selection with CD14+ and CD3+ Microbeads (Miltenyi Biotec), respectively, with a resulting purity of >95%. U937 (monocytic cell line), K562 (erythroleukemic cell line), BV-173 (a pre-B lymphoblastic leukemia),23 Daudi (B lymphoblastic cell line), and THP-1 (myelomonocytic leukemia) cell lines were obtained from the ATCC. The ALL-CM cell line derived from a CML patient suffering from a Philadelphia chromosome-positive lymphoid blast crisis was kindly provided by Dr. A. Bondanza. To generate β2 m-deficient ALL-CM and U937 cell lines, cells were nucleofected by Amaxa 4D Nucleofector System with X-unit (Lonza) using the EP100 program. In brief, 3 × 105 cells were re-suspended in a solution containing 20 μL of SF solution (Lonza) and 3 μL of pre-mixed Cas9 plasmid (500 ng) and the specific B2M guide #18 CRISPR plasmid (5′-GAGTAGCGCGAGCACAGCTA-3′ cut in B2M exon 1, 250 ng). After nucleofection, cells were expanded in culture. To generate ALL-CM-expressing luciferase (ALL-CMLuc), the ALL-CM cell line was transduced with an LV encoding for phosphoglycerate kinase (PGK)-luciferase (kindly provided by Prof. Ferrari G.) at an MOI of 50 and expanded in X-VIVO15 medium with 5% human serum (BioWhittaker-Lonza), 100 U/mL penicillin-streptomycin (BioWhittaker). All cell lines were routinely tested for mycoplasma contamination.
Plasmid Construction
The coding sequence of human IL-10 was excised from pH15C (ATCC 68192), and the 549 bp fragment was cloned into the multiple cloning site of pBluKSM (Invitrogen) to obtain pBluKSM-human interleukin-10 (hIL-10). A fragment of 555 bp was obtained by excision of hIL-10 from pBluKSM-hIL-10 and ligation to 1074.1071.hPGK.GFP.WPRE.mhCMV.dNGFR.SV40PA (here named LV-ΔNGFR), to obtain LV-IL-10/ΔNGFR. The presence of the bidirectional promoter (human PGK promoter plus minimal core element of the cytomegalovirus (CMV) promoter in the opposite direction) allows co-expression of the two transgenes. The sequence of LV-IL-10/ΔNGFR was verified by pyrosequencing (Primm).
Vector Production and Titration
VSV-G-pseudotyped third generation bidirectional lentiviral vectors (bdLVs) were produced by Ca3PO4 transient four-plasmid co-transfection into 293T cells and concentrated by ultracentrifugation as described previously.20 Titer was estimated by limiting dilution, vector particles were measured by HIV-1 Gag p24 Ag immune capture (NEN Life Science Products), and vector infectivity was calculated as the ratio between titer and particle. Titers ranged from 5 × 108 to 6 × 109 transducing units/mL, and infectivity from 5 × 104 to 105 transducing units/ng p24.
Dendritic Cell Differentiation
Peripheral blood CD14+ monocytes were positively selected using CD14+ Microbeads (Miltenyi Biotec) according to the manufacturer’s instructions. Cells were cultured in RPMI 1640 (Lonza) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin/streptomycin (Lonza), 2 mM l-glutamine (Lonza), at 37°C in the presence of 10 ng/mL recombinant human (rh) IL-4 (R&D Systems) and 100 ng/mL recombinant human granulocyte-macrophage colony-stimulating factor (rhGM-CSF) (Genzyme) for 5 days. To generate mature dendritic cells (mDCs), on day 5 we matured cells with 1 mg/mL lipopolysaccharide (LPS; Sigma) for an additional 2 days. At day 7, DCs were collected, phenotypically analyzed, and used to stimulate T cells. The purity and maturation state of DCs were checked by flow cytometry to determine expression of CD1a, CD14, CD86, CD83, and HLA-DR.
Transduction of Human CD4+ T Cells
Polyclonal CD4-transduced cells were obtained as previously described.20 CD4+ΔNGFR+ cells were beads-sorted using CD271+ Microbeads (Miltenyi Biotec) and expanded in X-VIVO15 medium with 5% human serum (BioWhittaker-Lonza), 100 U/mL penicillin-streptomycin (BioWhittaker), and 50 U/mL rhIL-2 (PROLEUKIN; Novartis). To generate alloantigen-specific transduced cells, we stimulated naive CD4+ T cells (106/well) with allogeneic mDCs (105/well). At days 7 and 10, medium was replaced by fresh medium supplemented with 25 U/mL rhIL-2. At day 14, cells were collected, washed, and 24 hr after the secondary stimulation with the same allo-mDCs used for priming, cells were transduced with LV-GFP/ΔNGFR (allo-CD4ΔNGFR) or LV-IL-10/ΔNGFR (allo-CD4IL-10) at an MOI of 20. Transduced CD4+ΔNGFR+ T cells were purified and stimulated every 2 weeks with allogeneic feeder mixture as previously described.20 Purity of selected cells was consistently greater than 95%. Allo-CD4ΔNGFR and allo-CD4IL-10 cells after two to three feeder mixture expansions were used to perform in vitro and in vivo experiments.
Cytokine Determination
In vitro experiments were performed with CD4ΔNGFR and CD4IL-10 cells at least 12 days after feeders. To measure cytokine production, we stimulated CD4ΔNGFR and CD4IL-10 cells with immobilized anti-CD3 (10 μg/mL) and soluble anti-CD28 (1 μg/mL) mAbs in a final volume of 200 μL of medium (96-well round-bottom plates, 2 × 105/well), whereas allo-CD4ΔNGFR and allo-CD4IL-10 cells were stimulated with allo- or third-party mDCs at a 10:1 ratio (105/well CD4ΔNGFR or CD4IL-10 cells with 104/well mDCs in a final volume of 200 μL of medium, 96-well round-bottom plates). Supernatants were harvested after 48 hr of culture, and levels of IL-4, IL-10, IFN-γ, and IL-17 were determined by ELISA according to the manufacturer’s instructions (BD Biosciences). To measure GzB production, we co-cultured CD4IL-10 or CD4DNGFR cells with ALL-CM or U937 target cell lines at 1:1 (effector: target [E:T]) ratio in the presence of 10 μg/mL anti-HLA class I or isotype control mAbs or with wild-type (WT) or β2 m−/− target cell lines. Supernatants were harvested after 24 hr of culture, and levels of GzB were determined by ELISA according to the manufacturer’s instructions (R&D).
Proliferation and Suppression Assays
To evaluate alloantigen-specific proliferative responses, we stimulated allo-CD4ΔNGFR and allo-CD4IL-10 cells with allo- or third-party mDCs at a 10:1 ratio (105/well CD4ΔNGFR or CD4IL-10 cells with 104/well mDCs in a final volume of 200 μL of medium, 96-well round-bottom plates). After the indicated time, cells were pulsed for 16 hr with 1 μCi/well [3H]thymidine. To test the suppressive capacity of CD4ΔNGFR and CD4IL-10 cells, we labeled Responder cells with carboxyfluorescein succinimydl ester (CFSE) (Molecular Probes) or eFluor670 (Invitrogen) before stimulation with immobilized anti-CD3 (10 μg/mL) and soluble anti-CD28 (1 μg/mL) mAbs or with allo- or third-party mDCs at a 10:1 ratio. Suppressor cells were added at a 1:1 ratio. After 4–6 days of culture, proliferation of responder cells was determined by analyzing the CFSE/eFluor670 dilution of CD4+ ΔNGFR− T cells.
Flow Cytometry Analysis
Fluorescence-activated cell sorting (FACS) analyses were performed on CD4ΔNGFR and CD4IL-10 cells at least 12 days after feeders. For the detection of cell surface Ags, CD4IL-10 and CD4ΔNGFR cells were stained with anti-CD4 (clone SK3; BD Pharmingen), anti-CD25 (clone 2A3; BD Pharmingen), anti-CD45RO (clone UCHC1; BioLegend), anti-CD45RA (clone HI100; BioLegend), anti-CD226 (clone DX11; BioLegend or Miltenyi Biotec), anti-CD2 (clone RPA-2.10; BD Pharmingen), and anti-CD18 (clone 6.7; BD Pharmingen) mAbs after a 2.4G2 blocking step. For the detection of cell surface Ags on target cells, leukemic cell lines and primary blasts were stained with anti-CD45 (clone HI30; BioLegend), anti-HLA class I (clone W6/32; BioLegend), anti-CD112 (clone TX31; BioLegend), anti-CD155 (clone SKII4; BioLegend), anti-CD13 (clone WM15; BD Pharmingen), anti-CD54 (clone HA58; BD Pharmingen), and anti-CD58 (clone 1C3; BD Pharmingen). Cells were incubated with the aforementioned mAbs for 30 min at 4°C in PBS 2% FBS, washed twice, and fixed with 0.25% formaldehyde. For the detection of FOXP3 (clone 259D; BioLegend) after surface staining, cells were fixed, permeabilized, and stained with the Foxp3 staining Buffer Set according to the manufacturer’s instructions (eBioscience). For the expression of GzB (clone MHGB04; Invitrogen) after surface staining, cells were fixed, permeabilized, and stained with the BD Cytofix/Cytoperm Kit according to the manufacturer’s instructions (catalog no. 554714; BioLegend). To evaluate human chimerism in peripheral blood of treated NSG mice, we co-stained cells with anti-human CD45 (clone HI30; BioLegend), anti-human CD3 (clone SK7; BD Biosciences), anti-human CD33 (clone AC104.3E3; Miltenyi Biotec), anti-CD271 (clone ME20.4; BioLegend), and anti-murine CD45 (clone 30F11; BD Biosciences) mAbs. Samples were acquired using a FACS Canto II flow cytometer (Becton Dickinson), and data were analyzed with FCS express (De Novo Software). Quadrant markers were set to unstained controls.
Cytotoxicity Assays
CD4ΔNGFR and CD4IL-10 cell degranulation was evaluated in CD107a flow cytometric assay, and their cytotoxic activity was analyzed in a standard 51Cr-release assay, as described by Magnani et al.6 Cytotoxicity of transduced cells was also analyzed in co-culture experiments. In brief, target and effector cells were plated in a 1:1 ratio for 3 days. At the end of co-culture, cells were harvested and surviving cells were counted and analyzed by FACS. In some experiments, anti-HLA class I (clone W6/32; BioLegend), anti-HLA class II (anti-HLA-DR, -DQ, -DR; BioLegend), and isotype control (IgG2a,k; BD Pharmingen) mAbs, or the GzB inhibitor (Benzyloxycarbonyl-Ala-Ala-Asp-chlormethylkethone) (Z-AAD-CMK) (Sigma) were added at the indicated concentrations. Elimination index (EI) was calculated as 1 − (number of targets that remained in the co-culture with CD4IL-10 per number of targets that remained in the co-culture with CD4ΔNGFR).
Mice
NSG female mice were purchased from Charles-River Italia. All mice were fed standard laboratory diet and maintained under standard laboratory conditions free of specific pathogens. All animal care procedures were performed according to protocols approved by the OSR Institutional Animal Care and Use Committee (IACUC protocol 488), following the 3R principles (replacement, reduction and refinement) and the Decreto Legislativo #116 dated January 27th, 1992, from the Italian Parliament.
Humanized Mouse Models
In all experiments 6- to 8-week-old female NSG mice were used.
Subcutaneous ALL-CM Tumor Model
On day 0 mice were subcutaneously injected with ALL-CM cells (2 × 106) and on day 3 with PBMCs (2 × 106), CD4IL-10 cells, or CD4ΔNGFR (1 × 106) cells. Sarcoma growth was monitored at least three times per week, and moribund mice were euthanized for ethical reasons.
THP-1 Tumor Model
On day 0 mice were intravenously injected with THP-1 cells (2 × 106) and on day 14 with PBMCs (2 × 106), CD4IL-10 cells, or CD4ΔNGFR (1 × 106) cells. Survival and weight loss were monitored at least three times per week as previously described.21 At week 4, mice were euthanized to analyze human cells in the liver.
Graft-versus-Leukemia Model, ALL-CM Leukemia Model
On day 0 mice received total body irradiation with a single dose of 175–200 cGy from a linear accelerator according to the weight of the mice and were intravenously injected with ALL-CM cells (5 × 106) and on day 3 with PBMCs (5 × 106), CD4IL-10cells, or CD4ΔNGFR (2.5 × 106) cells. Survival and weight loss were monitored at least three times per week as previously described21 and were euthanized for ethical reasons. At weekly intervals mice were bled and human chimerism was determined by calculating the frequency of human CD45+ cells within the total lymphocyte population.
Graft-versus-Leukemia and GvHD Model
On day 0 mice received total body irradiation as above and were intravenously injected with ALL-CM cells (5 × 106). On day 3 mice were injected with PBMCs (5 × 106) alone or in combination with CD4IL-10 or CD4ΔNGFR (2.5 × 106) cells. As control, irradiated mice were intravenously injected on day 3 with PBMCs (5 × 106 cells/mouse). Survival, weight loss, and human chimerism were monitored as above.
Bioluminescence Image Acquisition
Small-animal bioluminescence imaging (BLI) was performed using the IVIS Spectrum CT System (Perkin Elmer). The system is composed of a low-noise, back-thinned, back-illuminated charge-coupled device (CCD) camera cooled at −90°C with a quantum efficiency in the visible range above 85%. Each mouse received an intraperitoneal injection of 150 mg luciferin/kg body weight 10 min before BLI. During image acquisition, the animals were kept at 37°C and under gaseous anesthesia (2%–3% isoflurane and 1 L/min oxygen). Dynamic BLI was performed by acquiring a set of images every 2 min from 10 to 20 min after luciferin injection to detect the highest BLI signal. The images were obtained using the following settings: exposure time = auto, binning = 8, f = 1, and field of view = 13 cm (field C). Dark images were acquired before and then subtracted to bioluminescence images; no emission filters were used during BLI acquisitions.
Bioluminescence Image Analysis
BLI image analysis was performed by placing a region of interest (ROI) over the mouse and by measuring the total flux (photons/seconds) within the ROI. Images were acquired and analyzed using Living Image 4.5 (Perkin Elmer).
Statistical Analysis
The number of replicates per mouse to be used for each experiment has been estimated in order to get a power in identifying the variables considered >90% alpha error = 0 and on the basis of previous experience with the animal strains and animal models. No randomization was applied to animal studies, and investigators were not blinded. All data produced were analyzed by statistical methods chosen according to number and distribution of the determinations and variance and variation of the experimental groups. All the results were tested for normal distribution of the variables analyzed, and parametric or non-parametric test will be used accordingly. All these statistical analyses were performed with the consulting assistance of the University Centre for Statistics in the Biomedical Sciences (CUSSB) at the San Raffaele University, Milan. Statistical analyses on the functional data were performed using a Mann-Whitney U test for non-parametric data and a two-way analysis of variance test. One-way ANOVA tests and Bonferroni’s multiple comparisons were used to analyze the data from the in vivo experiments. The p values less than 0.05 were considered significant. Statistic calculations were performed with the Prism program 5.0 (GraphPad Software). CD4IL-10 cells that did not display a cytokine profile (IL-10/IL-4 ratio > 4) and suppressive activity were not included in the analysis.
Author Contributions
G.L. and G.A. performed experiments; collected, analyzed, and interpreted data; performed statistical analysis; and contributed to the writing of the manuscript. F.R., L.C., and B.C. performed the in vivo experiments. A.S. performed the bioluminescence assays and analyzed the data. F.C. provided patients’ samples. A.L. contributed vital new reagents. A.B. contributed to the supervision of the in vivo experiments and provided scientific advice. M.G.R. supervised the experiments, analyzed and interpreted data, provided scientific advice, and wrote the manuscript. S.G. designed the study, analyzed and interpreted data, coordinated and supervised the project, and wrote the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
We thank Prof. Luigi Naldini for the kind gift of the bidirectional LV, Prof. Giuliana Ferrari for the gift of LV encoding for luciferase, and Dr. Cristina Tresoldi for providing clinical samples. This work was supported by a grant from the Italian Telethon Foundation “Comitato Telethon Fondazione Onlus” Core grant TIGET TGT11E02, the Italian Association for Cancer Research project IG 2013 N. 14555 (Associazione Italiana per la Ricerca sul Cancro [AIRC]), and a European grant for European Cooperation in Science and Technology (Action BM1305: Action to Focus and Accelerate Cell-based Tolerance-inducing Therapies; http://www.afactt.eu).
Footnotes
Supplemental Information includes six figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2017.06.029.
Contributor Information
Maria Grazia Roncarolo, Email: mg1@stanford.edu.
Silvia Gregori, Email: gregori.silvia@hsr.it.
Supplemental Information
References
- 1.Sakaguchi S., Sakaguchi N., Asano M., Itoh M., Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol. 1995;155:1151–1164. [PubMed] [Google Scholar]
- 2.Groux H., O’Garra A., Bigler M., Rouleau M., Antonenko S., de Vries J.E., Roncarolo M.G. A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature. 1997;389:737–742. doi: 10.1038/39614. [DOI] [PubMed] [Google Scholar]
- 3.Gagliani N., Magnani C.F., Huber S., Gianolini M.E., Pala M., Licona-Limon P., Guo B., Herbert D.R., Bulfone A., Trentini F. Coexpression of CD49b and LAG-3 identifies human and mouse T regulatory type 1 cells. Nat. Med. 2013;19:739–746. doi: 10.1038/nm.3179. [DOI] [PubMed] [Google Scholar]
- 4.Bacchetta R., Bigler M., Touraine J.L., Parkman R., Tovo P.A., Abrams J., de Waal Malefyt R., de Vries J.E., Roncarolo M.G. High levels of interleukin 10 production in vivo are associated with tolerance in SCID patients transplanted with HLA mismatched hematopoietic stem cells. J. Exp. Med. 1994;179:493–502. doi: 10.1084/jem.179.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roncarolo M.G., Gregori S., Battaglia M., Bacchetta R., Fleischhauer K., Levings M.K. Interleukin-10-secreting type 1 regulatory T cells in rodents and humans. Immunol. Rev. 2006;212:28–50. doi: 10.1111/j.0105-2896.2006.00420.x. [DOI] [PubMed] [Google Scholar]
- 6.Magnani C.F., Alberigo G., Bacchetta R., Serafini G., Andreani M., Roncarolo M.G., Gregori S. Killing of myeloid APCs via HLA class I, CD2 and CD226 defines a novel mechanism of suppression by human Tr1 cells. Eur. J. Immunol. 2011;41:1652–1662. doi: 10.1002/eji.201041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bacchetta R., Gregori S., Serafini G., Sartirana C., Schulz U., Zino E., Tomiuk S., Jansen U., Ponzoni M., Paties C.T. Molecular and functional characterization of allogantigen-specific anergic T cells suitable for cell therapy. Haematologica. 2010;95:2134–2143. doi: 10.3324/haematol.2010.025825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregori S., Passerini L., Roncarolo M.G. Clinical outlook for type-1 and FOXP3(+) T regulatory cell-based therapy. Front. Immunol. 2015;6:593. doi: 10.3389/fimmu.2015.00593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann P., Ermann J., Edinger M., Fathman C.G., Strober S. Donor-type CD4(+)CD25(+) regulatory T cells suppress lethal acute graft-versus-host disease after allogeneic bone marrow transplantation. J. Exp. Med. 2002;196:389–399. doi: 10.1084/jem.20020399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edinger M., Hoffmann P., Ermann J., Drago K., Fathman C.G., Strober S., Negrin R.S. CD4+CD25+ regulatory T cells preserve graft-versus-tumor activity while inhibiting graft-versus-host disease after bone marrow transplantation. Nat. Med. 2003;9:1144–1150. doi: 10.1038/nm915. [DOI] [PubMed] [Google Scholar]
- 11.Cohen J.L., Trenado A., Vasey D., Klatzmann D., Salomon B.L. CD4(+)CD25(+) immunoregulatory T cells: new therapeutics for graft-versus-host disease. J. Exp. Med. 2002;196:401–406. doi: 10.1084/jem.20020090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gregori S., Goudy K.S., Roncarolo M.G. The cellular and molecular mechanisms of immuno-suppression by human type 1 regulatory T cells. Front. Immunol. 2012;3:30. doi: 10.3389/fimmu.2012.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trzonkowski P., Bieniaszewska M., Juścińska J., Dobyszuk A., Krzystyniak A., Marek N., Myśliwska J., Hellmann A. First-in-man clinical results of the treatment of patients with graft versus host disease with human ex vivo expanded CD4+CD25+CD127- T regulatory cells. Clin. Immunol. 2009;133:22–26. doi: 10.1016/j.clim.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Edinger M., Hoffmann P. Regulatory T cells in stem cell transplantation: strategies and first clinical experiences. Curr. Opin. Immunol. 2011;23:679–684. doi: 10.1016/j.coi.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Di Ianni M., Falzetti F., Carotti A., Terenzi A., Castellino F., Bonifacio E., Del Papa B., Zei T., Ostini R.I., Cecchini D. Tregs prevent GVHD and promote immune reconstitution in HLA-haploidentical transplantation. Blood. 2011;117:3921–3928. doi: 10.1182/blood-2010-10-311894. [DOI] [PubMed] [Google Scholar]
- 16.Brunstein C.G., Miller J.S., Cao Q., McKenna D.H., Hippen K.L., Curtsinger J., Defor T., Levine B.L., June C.H., Rubinstein P. Infusion of ex vivo expanded T regulatory cells in adults transplanted with umbilical cord blood: safety profile and detection kinetics. Blood. 2010;117:1061–1070. doi: 10.1182/blood-2010-07-293795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martelli M.F., Di Ianni M., Ruggeri L., Falzetti F., Carotti A., Terenzi A., Pierini A., Massei M.S., Amico L., Urbani E. HLA-haploidentical transplantation with regulatory and conventional T-cell adoptive immunotherapy prevents acute leukemia relapse. Blood. 2014;124:638–644. doi: 10.1182/blood-2014-03-564401. [DOI] [PubMed] [Google Scholar]
- 18.Bacchetta R., Lucarelli B., Sartirana C., Gregori S., Lupo Stanghellini M.T., Miqueu P., Tomiuk S., Hernandez-Fuentes M., Gianolini M.E., Greco R. Immunological outcome in haploidentical-HSC transplanted patients treated with IL-10-anergized donor T cells. Front. Immunol. 2014;5:16. doi: 10.3389/fimmu.2014.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gregori S., Tomasoni D., Pacciani V., Scirpoli M., Battaglia M., Magnani C.F., Hauben E., Roncarolo M.G. Differentiation of type 1 T regulatory cells (Tr1) by tolerogenic DC-10 requires the IL-10-dependent ILT4/HLA-G pathway. Blood. 2010;116:935–944. doi: 10.1182/blood-2009-07-234872. [DOI] [PubMed] [Google Scholar]
- 20.Andolfi G., Fousteri G., Rossetti M., Magnani C.F., Jofra T., Locafaro G., Bondanza A., Gregori S., Roncarolo M.G. Enforced IL-10 expression confers type 1 regulatory T cell (Tr1) phenotype and function to human CD4(+) T cells. Mol. Ther. 2012;20:1778–1790. doi: 10.1038/mt.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bondanza A., Valtolina V., Magnani Z., Ponzoni M., Fleischhauer K., Bonyhadi M., Traversari C., Sanvito F., Toma S., Radrizzani M. Suicide gene therapy of graft-versus-host disease induced by central memory human T lymphocytes. Blood. 2006;107:1828–1836. doi: 10.1182/blood-2005-09-3716. [DOI] [PubMed] [Google Scholar]
- 22.Bondanza A., Hambach L., Aghai Z., Nijmeijer B., Kaneko S., Mastaglio S., Radrizzani M., Fleischhauer K., Ciceri F., Bordignon C. IL-7 receptor expression identifies suicide gene-modified allospecific CD8+ T cells capable of self-renewal and differentiation into antileukemia effectors. Blood. 2011;117:6469–6478. doi: 10.1182/blood-2010-11-320366. [DOI] [PubMed] [Google Scholar]
- 23.Pegoraro L., Matera L., Ritz J., Levis A., Palumbo A., Biagini G. Establishment of a Ph1-positive human cell line (BV173) J. Natl. Cancer Inst. 1983;70:447–453. [PubMed] [Google Scholar]
- 24.Casucci M., Nicolis di Robilant B., Falcone L., Camisa B., Norelli M., Genovese P., Gentner B., Gullotta F., Ponzoni M., Bernardi M. CD44v6-targeted T cells mediate potent antitumor effects against acute myeloid leukemia and multiple myeloma. Blood. 2013;122:3461–3472. doi: 10.1182/blood-2013-04-493361. [DOI] [PubMed] [Google Scholar]
- 25.Grossman W.J., Verbsky J.W., Barchet W., Colonna M., Atkinson J.P., Ley T.J. Human T regulatory cells can use the perforin pathway to cause autologous target cell death. Immunity. 2004;21:589–601. doi: 10.1016/j.immuni.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 26.Grossman W.J., Verbsky J.W., Tollefsen B.L., Kemper C., Atkinson J.P., Ley T.J. Differential expression of granzymes A and B in human cytotoxic lymphocyte subsets and T regulatory cells. Blood. 2004;104:2840–2848. doi: 10.1182/blood-2004-03-0859. [DOI] [PubMed] [Google Scholar]
- 27.Pende D., Spaggiari G.M., Marcenaro S., Martini S., Rivera P., Capobianco A., Falco M., Lanino E., Pierri I., Zambello R. Analysis of the receptor-ligand interactions in the natural killer-mediated lysis of freshly isolated myeloid or lymphoblastic leukemias: evidence for the involvement of the Poliovirus receptor (CD155) and Nectin-2 (CD112) Blood. 2005;105:2066–2073. doi: 10.1182/blood-2004-09-3548. [DOI] [PubMed] [Google Scholar]
- 28.Shibuya K., Lanier L.L., Phillips J.H., Ochs H.D., Shimizu K., Nakayama E., Nakauchi H., Shibuya A. Physical and functional association of LFA-1 with DNAM-1 adhesion molecule. Immunity. 1999;11:615–623. doi: 10.1016/s1074-7613(00)80136-3. [DOI] [PubMed] [Google Scholar]
- 29.Lozano E., Dominguez-Villar M., Kuchroo V., Hafler D.A. The TIGIT/CD226 axis regulates human T cell function. J. Immunol. 2012;188:3869–3875. doi: 10.4049/jimmunol.1103627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu X., Harden K., Gonzalez L.C., Francesco M., Chiang E., Irving B., Tom I., Ivelja S., Refino C.J., Clark H. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat. Immunol. 2009;10:48–57. doi: 10.1038/ni.1674. [DOI] [PubMed] [Google Scholar]
- 31.Georgiev H., Danisch S., Chambers B.J., Shibuya A., Förster R., Bernhardt G. To the editor: TIGIT versus CD226: hegemony or coexistence? Eur. J. Immunol. 2014;44:307–308. doi: 10.1002/eji.201343925. [DOI] [PubMed] [Google Scholar]
- 32.Stanietsky N., Rovis T.L., Glasner A., Seidel E., Tsukerman P., Yamin R., Enk J., Jonjic S., Mandelboim O. Mouse TIGIT inhibits NK-cell cytotoxicity upon interaction with PVR. Eur. J. Immunol. 2013;43:2138–2150. doi: 10.1002/eji.201243072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez-Correa B., Gayoso I., Bergua J.M., Casado J.G., Morgado S., Solana R., Tarazona R. Decreased expression of DNAM-1 on NK cells from acute myeloid leukemia patients. Immunol. Cell Biol. 2012;90:109–115. doi: 10.1038/icb.2011.15. [DOI] [PubMed] [Google Scholar]
- 34.Vago L., Perna S.K., Zanussi M., Mazzi B., Barlassina C., Stanghellini M.T., Perrelli N.F., Cosentino C., Torri F., Angius A. Loss of mismatched HLA in leukemia after stem-cell transplantation. N. Engl. J. Med. 2009;361:478–488. doi: 10.1056/NEJMoa0811036. [DOI] [PubMed] [Google Scholar]
- 35.Villalobos I.B., Takahashi Y., Akatsuka Y., Muramatsu H., Nishio N., Hama A., Yagasaki H., Saji H., Kato M., Ogawa S., Kojima S. Relapse of leukemia with loss of mismatched HLA resulting from uniparental disomy after haploidentical hematopoietic stem cell transplantation. Blood. 2010;115:3158–3161. doi: 10.1182/blood-2009-11-254284. [DOI] [PubMed] [Google Scholar]
- 36.Toffalori C., Cavattoni I., Deola S., Mastaglio S., Giglio F., Mazzi B., Assanelli A., Peccatori J., Bordignon C., Bonini C. Genomic loss of patient-specific HLA in acute myeloid leukemia relapse after well-matched unrelated donor HSCT. Blood. 2012;119:4813–4815. doi: 10.1182/blood-2012-02-411686. [DOI] [PubMed] [Google Scholar]
- 37.Appelbaum F.R. Haematopoietic cell transplantation as immunotherapy. Nature. 2001;411:385–389. doi: 10.1038/35077251. [DOI] [PubMed] [Google Scholar]
- 38.Warren E.H., Deeg H.J. Dissecting graft-versus-leukemia from graft-versus-host-disease using novel strategies. Tissue Antigens. 2013;81:183–193. doi: 10.1111/tan.12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Groux H., Bigler M., de Vries J.E., Roncarolo M.G. Inhibitory and stimulatory effects of IL-10 on human CD8+ T cells. J. Immunol. 1998;160:3188–3193. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.