Abstract
Numerous aquatic invertebrates remain dormant for decades in a hydrated state as encysted embryos. In search for functional pathways associated with this form of dormancy, we used label-free quantitative proteomics to compare the proteomes of hydrated encysted dormant embryos (resting eggs; RE) with nondormant embryos (amictic eggs; AM) of the rotifer Brachionus plicatilis.
A total of 2631 proteins were identified in rotifer eggs. About 62% proteins showed higher abundance in AM relative to RE (Fold Change>3; p = 0.05). Proteins belonging to numerous putative functional pathways showed dramatic changes during dormancy. Most striking were changes in the mitochondria indicating an impeded metabolism. A comparison between the abundance of proteins and their corresponding transcript levels, revealed higher concordance for RE than for AM. Surprisingly, numerous highly abundant dormancy related proteins show corresponding high mRNA levels in metabolically inactive RE. As these mRNAs and proteins degrade at the time of exit from dormancy they may serve as a source of nucleotides and amino acids during the exit from dormancy. Because proteome analyses point to a similarity in functional pathways of hydrated RE and desiccated life forms, REs were dried. Similar hatching and reproductive rates were found for wet and dried REs, suggesting analogous pathways for long-term survival in wet or dry forms. Analysis by KEGG pathways revealed a few general strategies for dormancy, proposing an explanation for the low transcriptional similarity among dormancies across species, despite the resemblance in physiological phenotypes.
Many organisms have evolved the capacity to enter dormancy as a way of surviving environmental conditions that are incompatible with maintenance of their regular life activities. With resumption of favorable conditions, they recommence activities, normal development and reproduction. The term dormancy refers here to a long-term phenomenon and differs from an annual or periodic event, which characterizes insect diapause. In insects, diapause is a genetically programmed response and is regulated by neuro-hormones (1). This puzzling phenomenon whereby organisms survive long periods in a state of suspended animation or latent life (2) has captured the imagination of many scientists. It occurs in many evolutionary unrelated organisms—from prokaryotes to mammals (3, 4). Important questions that are asked regarding dormancy include: how organisms enter dormancy, how they maintain their viability for long periods, and how they revive and exit dormancy. In general, the entrance into dormancy occurs in conditions conducive to normal development (5, 6). The signal varies between species, ranging from changes in photoperiod, crowding, temperature, salinity and others (1, 7). Surprisingly, similar phenotypes and common functional pathways have been identified, regardless of the diversity and complexity in the survival strategies of organisms displaying dormancy. These include depression of metabolic pathways, suspension of the cell cycle, changes in carbohydrate and lipid metabolism, resistance to stress and protection of cellular structures (1, 3, 4, 6, 8, 9). Conversely, there is little transcriptional similarity among dormancies across species (10).
One of the best ways of preserving long-term viability during dormancy is by desiccation. Spectacular examples include the revival of plant seeds after centuries of storage (11, 12). A few adult invertebrate species survive desiccation for numerous years, including nematodes, bdelloid rotifers and tardigrades (13, 14). Other known examples of organisms surviving desiccation or dehydration include Artemia cysts (8, 9), insect springtails (15), and the Sleeping chironomid (16, 17). In organisms where dormancy is associated with desiccation, there is a formation of a glassy state, which entails an increase in viscosity, leading to a dramatic reduction in chemical reactions and consequently in metabolism and other functional pathways (18). There are, however, several examples of survival for several decades or even centuries in a nondesiccated state in aquatic organisms such as rotifers, cladocerans, or copepods, in the form of resting eggs or ephippia (19–24). Most amazing is the survival of Daphnia ephippia in a lake sediment cores for over six centuries (25). Dormancy in these species is developmentally programmed at embryonic stages. Developmental arrest at embryonic developmental stages is also well known in insects (1), fish (26), and mammals (27–30). One of the most famous examples of diapause occurs in developing embryos of the silkworm, Bombyx mori (31), where a diapause hormone is excreted by the subesophageal ganglion of the mother and acts on the developing eggs, which later exhibit diapause during embryonic development. Recently, a diapause hormone receptor-like gene was identified in Artemia with a function in inducing and maintenance of diapause in embryos (32). The brine shrimp, Artemia franciscana embryos enter diapause or dormancy in the gastrula stage of embryogenesis and is one of the most studied species as it endures desiccation (5, 9). Significant advances were made in revealing functional changes during dormancy in model and nonmodel organisms (1, 6). Nevertheless, diverse experimental organisms offer additional unique advantages for investigating dormancy by contributing important insight into the wide scope of the dormancy phenomenon. Here, we aim at expanding our knowledge on dormancy in embryos, by studying resting eggs of rotifers, a nonmodel aquatic invertebrate species.
The resting eggs (RE) or encysted embryos formed by the rotifer B. plicatilis are an excellent model for investigating long-term survival in the hydrated state. Rotifers are minute metazoans (∼50–2000 μm in length; ∼1000 cells), and one of the major groups of zooplankton inhabiting streams, rivers, lakes, and estuaries, functioning as secondary producers in the aquatic ecosystem (33, 34). Numerous rotifer species populate temporary ponds that may evaporate, ponds or lakes with unfavorable low or high temperatures, appearance and vanishing of suitable sources of food, and advent of predators (35). Consequently, rotifers appear and die out rapidly, with large fluctuations in the live population, but the strategy of forming dormant RE by different species or genetic populations facilitates the survival of genetic pools beyond these unfavorable conditions, in the form of a genetic bank, mostly in the hydrated form but on occasions also in a dry form (23). The ecological significance of dormancy in rotifers has been extensively reviewed (36–41) and molecular information was published in recent years (42–51). Including, the expression patterns in amictic and resting eggs of genes encoding proteins with putative functions in dormancy (52, 53; e.g. small heat-shock proteins, ferritin, glutathione-S-transferase, and trehalose-6-phosphate synthase). Two mRNAs and proteins were identified in rotifers for group 3 Late embryogenesis abundant (LEA) proteins (54).
The production of RE requires a switch from asexual to sexual reproduction (see Fig. 1 in (52)). The nondormant or amictic egg (AM)1 develops within hours into an amictic diploid female, through ameiotic parthenogenesis. The developing embryo within the amictic egg is carried by the mother until the embryo completes its development and hatches. The diploid RE is formed, however, after internal fertilization and the developing RE is carried by the mother for about 2 days. It is then released and sinks to the bottom of the culture container, pond or lake. The RE is an encased or encysted embryo consisting of 26–160 nuclei in a syncytial mass at early or late stages of gastrulation, probably before zygotic transcription (55). Interestingly, transport of maternal factors (RNA, protein and lipid droplets), from the vitellarium into the cytoplasm of maturing amictic eggs was demonstrated (56). Production of different rotifer life stages, including RE or AM eggs, is easily achieved for experimental purposes in the laboratory. RE stored in the dark after their formation, will hatch after an obligatory dormant period (∼ one month), if exposed to light. A batch of eggs exposed to light will hatch over a period of a few days (57). Specific genes were reported to be expressed during early stages of hatching (0.5–4 h of illumination) in the rotifer Brachiounus manjavacas (44).
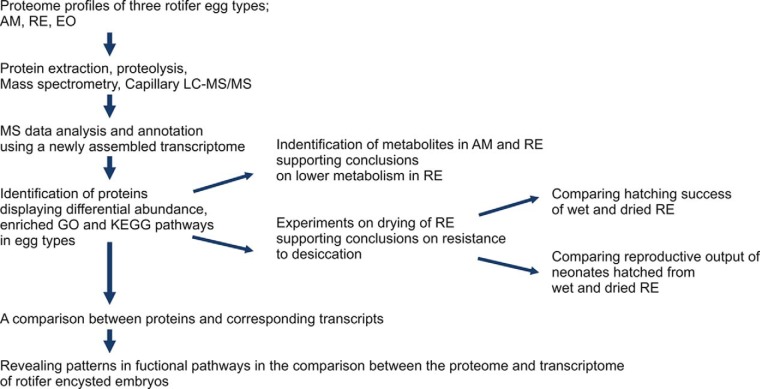
Fig. 1.
An overview of the experimental procedure, including proteomics and data analyses in dormant resting eggs (RE, EO) and nondormant amictic eggs (AM), metabolome profiling and desiccation experiments.
Studies on Artemia cysts for the past six decades (reviewed in 5, 6, 8, 9, 58–60, and others), have provided a wealth of information on the arrest of development, cell cycle, transcription and translation, bioenergetic transition, downregulation of metabolism and the protection of biological macromolecules during embryonic diapause. Most notably are studies on resistance to desiccation, including the role of tehalose in preserving membranes and proteins during desiccation, the function of multiple LEA proteins, safeguarding cell organelles and proteins during desiccation, small heat-shock proteins functioning in stress tolerance and the contribution of trehalose and LEA proteins to cellular vitrification at ambient temperatures (reviewed in 9).
Molecular insight into cell cycle revealed a role for Polo-like kinase (plk1) and p90 ribosomal S6 kinase 1 during cell division and cyst formation (61) and a role for ribosomal S6 kinase in the early events of pre-emergence (62). In addition, miR-100 and miR-34, were found to regulate cell cycle arrest during entry into dormancy in Artemia (63). Alternative metabolic pathways were also reported for the dauer stage of Caenorhabditis elegans (64) and insects during diapause (65). Moreover, a general lack of synchronization was observed between the arrest of cell cycle and metabolic down-regulation in dormant forms such as Artemia and killifish embryos (26). In addition, it has been demonstrated that the transcription factor FOXO is a prime candidate for activating many physiological pathways that generate the diapause phenotype in insects, including regulation of metabolism and the cell cycle (6, 26).
Gene expression profiles linked with diapause have been studied in numerous insects (e.g. 15, 66–72 and others), but almost all of them were not performed on embryos, except for Tu et al. 2015 (73). A few studies were performed on Brachionid rotifers forming resting eggs (42, 51–53, 74). Gene expression profiles, however, are insufficient for explaining gene function, as mRNAs do not fully correlate with protein levels (75, 76). This lack of correlation was also demonstrated for prediapausing and nondiapausing locust eggs (73). Only a limited number of dormancy related proteins were revealed by proteome profiling studies of dormant or diapausing invertebrate embryos. Proteome profiling of Artemia encysted and postdiapause embryos revealed 75 proteins with differential abundance in Artemia franciscana cysts (77) and 33 unique proteins in cysts of Artemia sinica (78). Shotgun proteome analyses identified 192 diapause unique proteins in the silkworm (79) and 212 differential abundant proteins between prediapause and nondiapause locust eggs (73).
To reveal the wide scope of functional changes taking place during dormancy of encysted embryos we used a shotgun, label free proteomics approach and identified 2631 protein groups and grouped a large number of them into putative functional pathways. Because proteome profiling showed that numerous pathways in hydrated encysted embryos of rotifers are similar with those of Artemia cysts and life forms showing resistance to desiccation (6, 9), we dried REs. The viability of desiccated rotifer RE was tested by comparing the hatching percent and the reproductive rates of neonates hatching from wet and dried RE. In this study we also aim at addressing two additional topics: (1) How similar are the functional pathways in encysted dormant rotifer eggs that survive in a hydrated form for long periods of time, with those described so far for other forms of dormancy or diapause in embryos? (2) Whether proteome profiling can assist in explaining the small transcriptional similarity among dormancies across species, despite the resemblance in physiological phenotypes.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
This study consists (Fig. 1) of: (a) proteome analyses of three rotifer egg types; amictic eggs (AM), resting eggs (RE), and resting eggs ready for hatching (E0) to discover the proteins with functions in dormancy. Triplicate samples were examined for each egg type. LC-MS/MS data was analyzed by comparing the amino acid sequences with an in silico translated newly assembled reference transcriptome. Principle Component Analysis (PCA) was used to reveal the similarities between the nine analyzed samples based on their global proteomic profiles. To discover proteins showing differential abundance in AM relative to RE or E0, 1-way ANOVA test was carried out with the contrasts of AM versus RE and AM versus E0., and in addition, proteins which were present above a certain background level in one egg type and absent in the other were identified. Gene ontology (GO) and KEGG pathway enrichment analyses were performed to identify overrepresented biological themes within the differentially abundant proteins. To ascertain the relationship between the proteome and transcriptome, the abundance of proteins was compared with gene expression levels; (b) Metabolome profiling was used to support the conclusions on differences in the metabolism of AM and RE, using t test on results of metabolites, from triplicate samples of each egg type; and (c) Analysis of functional pathways suggested a resistance of RE to desiccation. Viability of dried RE was determined by comparing (five replicates for each time point) the hatching percent of dried RE after 1–56 days of dry storage, by Tukey Post-Hoc tests, after a Chi-square test, showing a normal distribution of the percent of hatching. In addition, reproductive output by neonates hatching from 56-day dry stored RE (n = 110) was compared with wet stored RE (n = 113), by 1-way ANOVA, after a Chi-square test, showing a normal distribution of the results. More detailed information is provided in the following sections.
Rotifer Proteome Samples
Rotifers were cultured as described previously (51, 53). Resting eggs (REs) were manually collected from the bottom of culture vessels of 14–25-day-old rotifer cultures, cleaned of the debris and placed in a microfuge vial and flash frozen by liquid nitrogen and stored at −80 °C. Three biological replicate samples, each consisting of ∼5000 RE, were collected from three different cultures. In addition, three replicate samples of RE were collected from the same cultures, except that they were placed in vials with seawater (10 ppt) and incubated at 25 °C in the dark, in a temperature controlled room (treatment E0). After 3 months of incubation, these eggs (named E0) were flash frozen in liquid nitrogen and stored at −80 °C. For collection of amictic eggs (AM), females from the same cultures were concentrated on a 60 μm mesh plankton sieve. They were re-suspended in 10–15 ml of sterile sea water (10 ppt) and the AM eggs were removed from the females by intensive up and down aspiration with a 5-ml pipette tip. Following this treatment, AM eggs were transferred from the bottom of the vial into Petri dishes. The AM eggs were manually collected and three replicate samples from the three different cultures were placed in a microfuge vial (∼5000 AM eggs per vial) and flash frozen by liquid nitrogen and stored at −80 °C.
Metabolome Samples
Rotifers were cultured in artificial sea water (Red Sea, Israel; 10.8 gm L−1) and fed with Nannochloropsis sp. (Galil Algae, Israel). RE were collected after 8–10 days of culture from three different cultures. Amictic eggs (AM) were collected from three cultures after 3–4 culture days. AM and RE were collected into microfuge vials, washed twice with Dulbecco's phosphate buffered saline (Sigma-Aldrich, UK), frozen in LN2 and stored at −80 °C.
Extraction of proteins, proteolysis, and Mass Spectrometry
Proteins were extracted from samples that were also used for RNA extraction (51) with the Guanidinium thiocyanate-phenol-chloroform extract (TRI Reagent; Sigma, St. Louis, MO) following the manufacturer instructions. It was assumed that similar efficiencies in protein extraction were obtained with the TRI Reagent for the three egg types. The proteins in 8 m Urea, 100 mm ammonium bicarbonate were reduced with 2.8 mm DTT for 30 min at 60 °C, modified with 8.8 mm iodoacetamide in 100 mm ammonium bicarbonate in the dark for 30 min at room temperature, and digested in 2 m Urea, 25 mm ammonium bicarbonate with modified trypsin (Promega, Madison, WI) at a 1:50 enzyme-to-substrate ratio, overnight at 37 °C. An additional second trypsinization was done for 4 h to ensure complete cleavage. The tryptic peptides were desalted using C18 tips (Harvard-Apparatus) dried and re-suspended in 0.1% formic acid. The resulting tryptic peptides from the supernatant were analyzed by LC-MS/MS using a Q-Exactive mass spectrometer (Thermo Scientific, Bremen, Germany) fitted with a capillary HPLC (Easy nLC 1000, Thermo Scientific). The peptides were loaded onto a C18 homemade capillary column (20 cm, 75 micron ID) packed with Reprosil C18-Aqua (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany) in solvent A (0.1% formic acid in water). The peptides mixture was resolved with a (5 to 28%) linear gradient of solvent B (95% acetonitrile with 0.1% formic acid) for 180 min, followed by gradient of 5 min gradient of 28 to 95% and 25 min at 95% acetonitrile with 0.1% formic acid in water at flow rates of 0.15 μl/min. Mass spectrometry was performed in a positive ion mode (at mass range of m/z 350–1800 AMU and resolution of 70,000) using repetitively full MS scan followed by collision induces dissociation (HCD, at 35 normalized collision energy) of the 10 most dominant ions (>1 charges) selected from the first full MS scan. The AGC settings were set to 3 × 106 for the full MS and 1 × 105 for the MS/MS scans. The intensity threshold for triggering MS/MS analysis was 1 × 104. A dynamic exclusion list was enabled with exclusion duration of 20 s.
Mass Spectrometry Data Analysis
The mass spectrometry data was analyzed using the MaxQuant software 1.4.1.2. (www.coxdocs.org, 80) for peak picking identification and quantitation, using the Andromeda search engine (81) and searching for tryptic peptides against the translated rotifer transcriptome described below (15,102 proteins; supplemental Table S1) with mass tolerance of 20 ppm and 4.5 ppm after recalibration for the precursor masses and 20 ppm for the fragment ions. Methionine oxidation, and protein N terminus acetylation were accepted as variable modifications and carbamidomethyl on cysteine was accepted as static modifications. Minimal peptide length was set to six amino acids and a maximum of two miscleavages was allowed. Peptide- and protein-level false discovery rates (FDRs) were filtered to 1% using the target-decoy strategy. The protein table was filtered to eliminate the identifications from the reverse database, and common contaminants. Identification parameters also included minimum PEP score of 1 and at least two unique peptides for identification. The term “protein” was used in this manuscript for the identified peptides, instead of the more appropriate term of “protein groups.” The data was quantified by label free analysis using the MaxQuant software, based on extracted ion currents (XICs) of peptides enabling quantitation from each LC/MS run for each peptide identified in any of experiments (supplemental Table S1). The mass spectrometry proteomics data were deposited at the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD003395.
Transcriptome Assembly
A reference Brachionus plicatilis transcriptome was constructed from several sequence collections: (1) 52,143 Expressed Sequence Tag (EST) sequences from dbEST (NCBI), after trimming of adapter sequences, low complexity sequences and terminal poly-A's, (2) 84 mRNA sequences from RefSeq, and (3) 18,751 contigs from previous RNA-Seq analysis (51). All sequences were assembled using CAP3 with the following parameters: -c 12 -o 30 -p 75 -s 500 -y 250 -z 2. The resulting transcriptome, containing the contigs and “singlets” produced by CAP3 comprised a total of 16,680 putative transcripts. The assembled transcriptome is available at http://bioinfo.bgu.ac.il/genomes/Esther_Lubzens/public/(Username: ester; Password: retse).
Transcriptome Conceptual Translation and Annotation
Transcripts longer than 60 bp were compared with the RefSeq protein database using BLASTX and were also submitted to OrfPredictor to identify the frame with the longest open reading frame. Transcripts were subsequently translated through their entire length at the frame detected by BlastX, if they had a hit with e-value < 10−5, or otherwise, they were translated at the frame detected by OrfPredictor. The resulting amino acid sequences were annotated using Blast2GO through BLASTX versus RefSeq (considering up to top 20 hits, e-value cutoff 10−3), and were assigned KEGG Orthology (KO) IDs using the KEGG KAAS server. Transcript and protein-level annotations are provided in supplemental Table S2. The in-silico translated reference transcriptome was used as the reference database in the MaxQuant analysis described above.
Proteins in Rotifer Eggs
A total of 2928 proteins were identified for the three egg types of RE, AM, and E0 and the list of identified proteins is shown in supplemental Table S3A. Peptide counts were taken from the “razor + unique peptides” column in MaxQuant file. Protein intensity values from MaxQuant were Log2 transformed. A protein with zero intensity in a certain sample was regarded as “not detected” (ND) in that sample and indicated by a blank cell in supplemental Table S3A and S3B. Intensity distribution plots and Principal Component Analysis (PCA), carried out in Partek® Genomics Suite®, showed that sample E0_3 was exceptional (supplemental Fig. S1A, S1B). Therefore, this sample was excluded from further analyses. As shown in supplemental Fig. S1A, the intensity values in all three replicates of AM were generally higher than in RE and E0 (the distribution plot is shifted to the right). Because we could not know whether this is caused by a biological effect or by a technical bias, we thought it would be more appropriate to stay on the safe side and avoid normalization. Normalization may introduce its own bias, distort the data, and hide real changes in protein abundance among the samples. the use of unnormalized data, a stringent fold of change (FC) cutoff of 3 was used for differential abundance. As shown in supplemental Table S3B, the obtained results were like those achieved with LFQ-normalized data which underwent the same statistical analysis described below except that a more traditional FC cutoff of 1.5 was used.
For a given protein in a given egg type, the following terms were defined: (a) the protein was considered as “present” in that egg type if it was detected in at least 2 replicates with 2 or more peptides in each replicate; (b) “present above background level” if it was “present” and its average intensity value across replicates was higher than 24 (which is the 10% percentile value of all intensity values in the data set), (c) “absent” if it was not detected (ND) in all of the replicates and (d) “uncertain” if it had an intensity value in only one replicate. A total of 2342, 1751, and 1482 proteins were found to be present in AM, RE and E0 egg types, respectively (supplemental Table S3B). Altogether, 2631 proteins were found to be present in at least one of the egg types, and these proteins were submitted to subsequent analyses (supplemental Table S3B).
Identification of Proteins Showing Differential Abundance Between Dormant and Nondormant Eggs
As indicated above, prior to statistical testing, all zero intensity values were replaced by missing values. This was applied, because when one of the egg types had zero intensities and the other did not, the zeros created artificially inflated fold of change values and extremely low p values in the statistical tests. Such a phenomenon is especially severe in proteomics data, where the lowest nonzero intensities are usually around 2 to the power of 18–20. Therefore, in the current study, the statistical test neglected the zeros, and separate criteria were applied to identify proteins with apparently no expression in one egg type and apparent expression in another egg type.
A 1-Way ANOVA Test was Carried Out in Partek with the Contrasts
AM versus RE, AM versus E0, RE versus E0. Significance p values were adjusted for multiple testing using the False Discovery Rate (FDR) method. Fold of change (FC) in each contrast was expressed in linear scale, where positive and negative values indicate higher or lower abundance, respectively. A protein was regarded as showing differentially abundance between two egg types, if it had a fold change larger than 3 and FDR-adjusted p value <0.05. This resulted in 1088 proteins in the comparison between AM and RE and 1042 proteins in the comparison of AM and E0. In addition, proteins fulfilling any of the following criteria were added to the list of differentially abundant proteins: (a) It was absent in one group and present above background level (see above) in the other; (b) It was “uncertain” (see above) in one group and present above background level in the other group and the fold change was larger than 10. As shown in supplemental Table S3B, the final list of “differentially abundant proteins” included 1538 proteins for the AM versus RE contrast, and 1570 proteins for the AM versus E0 contrast. Uniprot (http://www.uniprot.org/) was used for retrieving functional information on proteins.
Analysis of RNA-Seq Data and Comparison Between Proteins and Corresponding Transcripts
RNA-Seq data from Clark et al., 2012 (51) was used for AM and RE (one replicate sample per egg type, based on the extraction of ∼5000 eggs for each egg type) was reanalyzed using the current reference transcriptome. Single-end 32bp reads of AM and RE samples were aligned to the assembled transcriptome using STAR 2.3 and the number of reads per transcript were counted using BEDTools.
Normalization of the raw counts to account for library size was performed using the DESeq R package. Normalized count values were log2 transformed, after values < 1 were floored to 1. Transcripts with normalized count value < 6 in both samples were filtered out.
Protein intensity values and their corresponding RNA-Seq normalized counts (both in log2 scale) were loaded into R. For each egg type (AM, RE) a simple linear regression and a residual analysis were carried out to compare RNA and protein expression. Genes having studentized residual >2 or <−2 were considered discordant. They comprised 4.2% of the proteins. Genes having studentized residual between −0.7 and 0.7, comprising 50% of the proteins, were considered concordant.
Gene Ontology (GO) Annotation and Enrichment Analysis
Gene Ontology (GO) annotation and enrichment analyses were carried out using Blast2GO. Enrichment was done using Fischer exact test, and the entire transcriptome was used as the background set. The list of enriched GO terms was further reduced using Blast2GO pro's “Reduce to most specific terms” algorithm. This algorithm filters the results by removing parent terms of already existing, statistically significant, child GO terms. GO terms having enrichment FDR p value < 0.05 were reported. Further processing of the enrichment results was done using a Perl script (supplemental Tables S4, S6, and S9).
KEGG Pathway Enrichment Analysis
Enrichment analyses of KEGG pathways in selected protein sets, was carried out using Expander (82). The sets included (supplemental Table S5A). Three sets for proteins in AM, RE or E0, (b) Two sets of differentially abundant proteins in the comparison of AM versus RE and AM versus E0, and c) Two sets of proteins in concordance with RNA for AM and RE. All KO IDs identified in the reference transcriptome served as the background set. Drawing and coloring of KEGG pathways was done using “KEGG Mapper - Search & Color Pathway” tool.
Hierarchical Clustering
Hierarchical clustering was carried out in Partek. Prior to clustering, intensity values were z-scored so that for each protein the mean and standard deviation were 0 and 1, respectively. In the heat maps, missing values were marked in white.
Sequence Analyses of Selected Proteins
LEA proteins from selected organisms were searched against the reference transcriptome using blast and hmmsearch (supplementary File Text S1).
Lists of Proteins of Specific Interest
Proteins of specific interest were prepared by manual curation and are shown in Table I and supplemental Table S3B.
Table I. Differential abundance of hallmark proteins known for their association with dormancy. The list contains proteins that significantly differed between AM and RE or E0 (FDR p < 0.05; FC<3), or were present only in RE or E0 but not in AM. The minus sign indicates lower abundance in the comparison between AM vs. RE or AM vs. E0 (or higher abundance in RE or E0). Numerous proteins were not detected in AM (ND AM).
| Protein ID | Majority protein IDs | Majority protein titles | Fold-change (AM vs. RE) | Fold-change (AM vs. E0) |
|---|---|---|---|---|
| Ferritin | ||||
| 364 | Contig 7007 | ferritin heavy chain | −2443.33 | −1743.58 |
| 66 | Contig 2851 | ferritin heavy chain a-like | −28.04 | −21.49 |
| 116 | Contig 9535 | soma ferritin-like | −1238.12 | −1359.40 |
| 183 | Contig 11553; Contig 717 | soma ferritin-like; soma ferritin-like | −53.07 | −43.94 |
| Glutathione s-transferase | ||||
| 1899 | Contig 7113 | glutathione s-transferase | −23.26 | −32.67 |
| 269 | Contig 9208 | glutathione s-transferase a-like | −2.81 | −2.97 |
| 1863 | Contig 570 | glutathione s-transferase-like | −10.38 | −3.22 |
| 127 | Contig 533 | glutathione s-transferase-like | −3243.34 | −3295.31 |
| 347 | Contig 276; Contig 7601 | hematopoietic prostaglandin d synthase; glutathione s-transferase-like | −5.77 | −6.96 |
| Glutathione peroxidase | ||||
| 570 | Contig 3114 | glutathione peroxidase | −3.58 | −4.62 |
| Peroxiredoxin | ||||
| 214 | Contig 9623 | peroxiredoxin prdx5 | −285.68 | −293.61 |
| 106 | Contig 9620 | peroxiredoxin prdx5 | −1020.36 | −850.76 |
| Superoxide dismutase | ||||
| 145 | Contig 10586 | superoxide dismutase | −4.88 | −4.13 |
| 426 | Contig 5597 | superoxide dismutase | −4.41 | −3.36 |
| 1496 | Contig 1507 | superoxide dismutase | −8.922 | −3.66 |
| 1963 | Singlet 16655 | superoxide dismutase | ND AM | |
| 2452 | Contig 11562 | superoxide dismutase | ND AM | |
| 536 | Contig 7232 | copper chaperone for superoxide dismutase | ND AM | |
| Thioredoxin | ||||
| 532 | Contig 665 | thioredoxin | −5.00 | −4.96 |
| 1453 | Contig 11560 | thioredoxin h-type | −3.06 | −3.64 |
| Catalase | ||||
| 61 | Contig 3897 | catalase | −21.84 | −18.15 |
| Oxidoreductase | ||||
| 4 | Contig 411 | uncharacterized oxidoreductase -like | −2223.68 | −1861.37 |
| 11 | Contig 448 | uncharacterized oxidoreductase -like | −811.40 | −813.88 |
| 175 | Contig 8056 | dehydrogenase reductase sdr family member 1 | −5.52 | −3.24 |
| 566 | Contig 2703 | dehydrogenase reductase sdr family member 1 | −10.37 | −5.51 |
| 226 | Contig 12126 | dehydrogenase reductase sdr family member 12 | −2.56 | −2.37 |
| Selenium | ||||
| 25 | Contig 11382 | selenium-binding protein | −517.75 | −506.53 |
| 35 | Contig 5327 | selenium-binding protein | −518.03 | −711.96 |
| 1805 | Contig 3315 | selenoprotein o | −6.39 | −5.95 |
| 508 | Contig 169 | selenoprotein o-like | −6.69 | −7.67 |
| LEA proteins | ||||
| 1 | Contig 9631 | protein lea- isoform k | −994.73 | −1161.00 |
| 2 | Contig 227;Contig 3165 | late embryogenesis abundant protein family protein; late embryogenesis abundant protein family protein | −1399.92 | −1111.06 |
| Alpha crystallin | ||||
| 9 | Contig 7561 | alpha-crystallin a chain-like | −537.82 | −523.10 |
| Trehalose | ||||
| 1088 | Contig 14054 | trehalose-phosphate synthase | −15.88 | −23.99 |
| Heat-shock proteins | ||||
| 46 | Contig 11379 | heat shock hsp20 | −1260.69 | −1261.17 |
| 147 | Contig 11391 | heat shock protein | −217.35 | −186.24 |
| 349 | Contig 2953 | heat shock protein beta-1-like | −3.49 | −4.16 |
| 16 | Contig 11373; Singlet 16498 | heat shock protein hsp-like; heat shock protein beta-1 | −4077.68 | −4275.32 |
| 521 | Contig 404 | heat shock protein hsp20 | ND AM | |
Metabolome Profiling Samples
All steps were carried out in metaSysX GmbH, Potsdam Germany. Frozen samples (from −80 °C) of AM and RE were extracted according to a modified protocol from Giavalisco et al., 2009 (83). After extractions volumes collected for further analysis were adjusted to same number of eggs (∼24,000 eggs per sample). LC-MS, MS/MS and GC-MS measurements, data processing, data annotation and data normalization, are provided in supplementary File Text S2.
Desiccation Experiments of RE
The experiments consisted of three stages: (a) preparation, collections, storage and drying of RE, (b) comparing the viability of dried and wet RE in hatching experiments, and (c) comparing the reproductive rates of neonates hatched from wet and dried RE.
(a) Preparation, Collection and Storage of RE
Resting eggs (RE) were produced by rotifers cultured in artificial sea water (Red Sea Ltd., Israel) at a salinity of 10.8 gm L−1 (equivalent to 10 ppt sea water) and fed with thawed concentrated Nannochloropsis sp. (Galil Algae, Israel). RE were collected after 9–11 days of culture, cleaned from debris and stored at room temperature (24–26 °C) in 20 ml scintillation glass vials filled with 10 ml sea water (10 ppt; 300–1000 RE per vial). Each vial was wrapped with aluminum foil and several vials were prepared from each culture. RE were stored for 6–15 months from three different cultures (Culture 1 for 14 months, Culture 2 for 6 months, and Culture 3 for 15 months).
(b) Comparing the Viability of Dried and Wet RE in Hatching Experiments
On the first day of the experiment (Day 0), RE were removed from one vial and distributed into five 35 mm glass Petri dishes containing 10 ml sea water (10ppt) and 50–70 RE per dish. This procedure was repeated for RE of vials from three different cultures. The Petri dishes were placed under a cold light fluorescent lamp (200–400 μEinstein m−2sec−1), at 24–26 °C. The number of hatched neonates was counted after 48 and 72 h. The number of unhatched RE was counted after 96 h. The percent of hatching (H) was calculated as %H = (H/H + UH)×100, where H is the number of hatched neonates and UH is the number of unhatched REs. In parallel, 50–70 RE from each one of three cultures were distributed to 20 Petri dishes with a small volume of seawater (50–100 μl; 10 ppt) and were dried by evaporation. The RE seemed desiccated after 15–20 min as displayed by the appearance of salt crystals. They were further dried for 24 h at room temperature (24–26 °C). After 24 h (Day 1), 10 ml of sea water (10 ppt) were added to five Petri dishes from each of three cultures (a total of 15 Petri dishes for the three replicate cultures) and the Petri dishes were subjected to hatching conditions as described above. The remaining 15 Petri dishes for each one of the three cultures (a total of 45 dishes) containing the dried RE, were wrapped in aluminum foil and stored at 24–26 °C. Hatching experiments were performed after 14, 28, and 56 days of dry storage, using five Petri dishes on each day, from each culture. The hatching percent was compared between dried RE at the different time points and wet RE of day 0.
(c) Comparing the Reproductive Rates of Neonates Hatched from Wet and Dried RE
Neonates hatching from dried RE (stored for 56 days) and neonates that hatched from the corresponding wet stored RE, were incubated in 24-well culture plates (Nunc, Denmark). Each well contained one neonate in 1 ml of seawater (10 ppt) and algae (Nannochloropsis sp. at 12–18 × 106 cells). The number of rotifers and eggs were counted after 72 h of incubation, at 24–26 °C.
Statistical analyses of the hatching experiments, was performed by Tukey Post-Hoc tests (after a Chi-square test, showing a normal distribution of the percent of hatching). One-way ANOVA and t test were used to test the differences in reproductive performance of neonates from wet and dried RE (after a Chi-square test showing normal distribution of results). Main Effects ANOVA, Tukey Post-Hoc and t- tests (after a Chi-square test, showing a normal distribution) were performed by using the DellTM StatisticaTM 13.2 package for Windows software. Statistical significant differences were considered at p < 0.05.
RESULTS
General Information on the Proteome Profiles of Resting Eggs and Amictic Eggs
In this study we carried out a comparison between the proteomes of encysted dormant embryos (“resting eggs”; RE or E0) with nondormant embryos (amictic eggs; AM). Morphological differences between AM and RE are shown in Fig. 2.
Fig. 2.
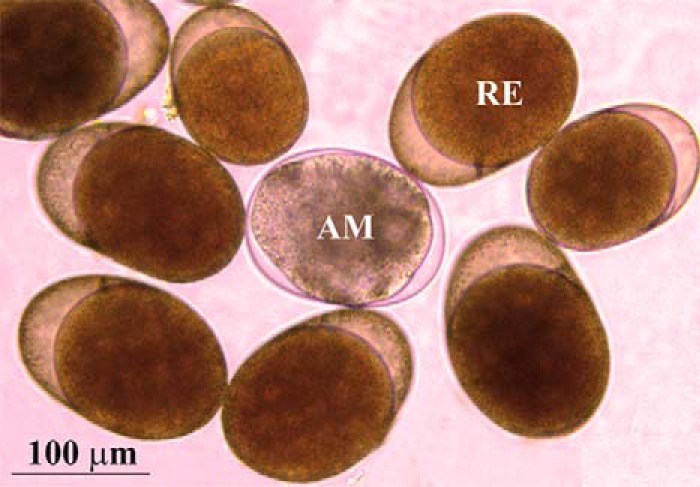
A microscopic view of amictic (AM) and resting eggs (RE). The picture was taken by a Leica DMi8 inverted microscope equipped with a Leica DFC 310 color camera.
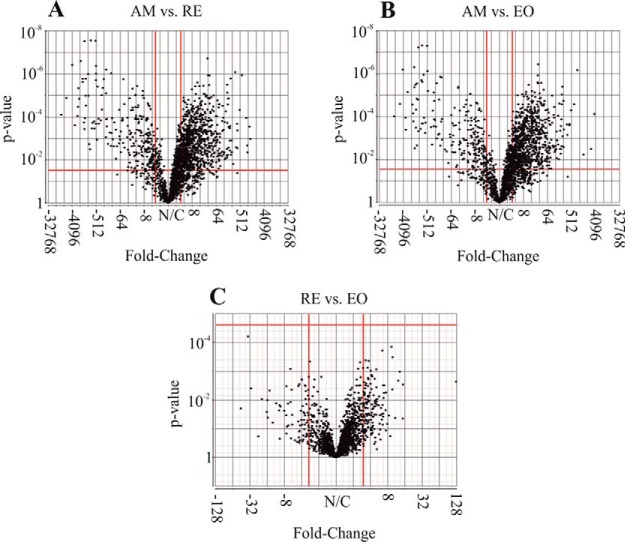
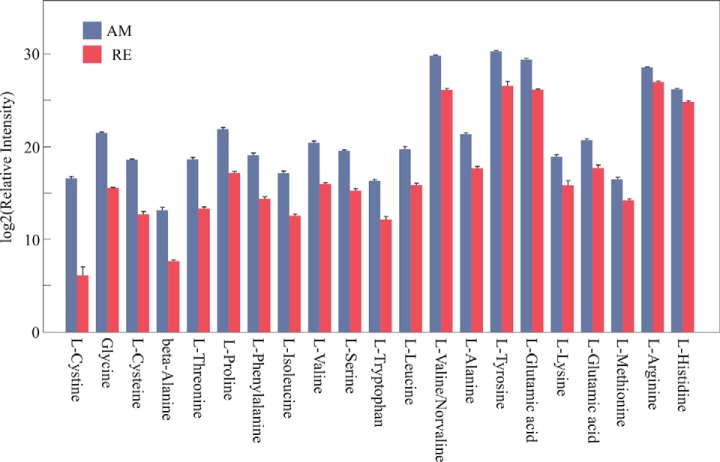
A total of 2928 proteins were identified in the nine examined samples (supplemental Table S3A), using the translated and annotated B. plicatilis transcriptome as a reference (supplemental Tables S2A and S2B). As described in the Experimental Procedures, 2631 proteins displayed at least two peptides in two replicate egg samples (“Present proteins,” supplemental Table S3B). Of these proteins, 2351 proteins were annotated (89.4%) and 1492 (64%) were assigned with KEGG KO numbers. In general, most of the proteins were shared by all egg types (Fig. 3) and relatively a small number of proteins were specific to one egg type (supplemental Table S3B). Quantitative analysis showed larger differences in the proteins' abundance between AM and RE and smaller differences between RE and E0 groups (supplemental Fig. S1A, S1B). Although E0 consists only of two biological replicates, the detailed analyses of their proteome profiles were retained as they underscore the differences between RE versus AM. In the comparison of RE versus E0, no proteins were found to have a significantly different abundance (Fig. 4). An additional selection for proteins, which occur in one egg type and not the other or have significantly different abundance among the egg types (FDR p value<0.05, Fold change>±3, Fig. 3), revealed 1538 proteins differing between AM and RE (AM versus RE; supplemental Table S3B). Pairwise comparisons show that ∼62% of the proteins showed higher abundance (FC>3; FDR p value < 0.05) in AM versus RE, with numerous proteins displaying FC>100 (supplemental Table S3B). For comparison, LFQ-normalized data showed 1498 proteins differing between AM and RE, of these 1068 proteins were identical with those of the unnormalized data mentioned before (supplemental Table S3B; see explanation for unnormalized versus LFQ normalization in the Experimental Procedures). The most abundant proteins differ greatly between AM and resting eggs as seen in Fig. 5A and 5B, and these results were similar to the LFQ-normalized data (results not shown in detail but can be retrieved from data in supplemental Table S3B). AM eggs show an abundance of cytoskeleton proteins (tubulin, actin), ATP synthase, fatty acid binding protein, α-crystalline b chain and the regulatory protein 14-3-3. Most of them were also found in RE. Resting eggs show high abundance of LEA proteins, ferritin, peroxiredoxin, selenium binding protein, a mammalian ependymin-related protein 1 and α-crystalline a chain. Elongation factor α-2 is abundant in all egg types.
Fig. 3.
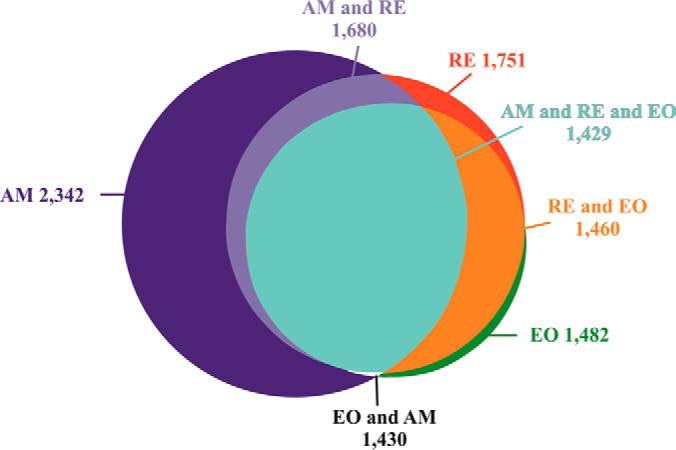
A Venn diagram showing the relative distribution and overlap of proteins of three egg types; AM (2342 proteins), RE (1,751 proteins), and E0 (1482 proteins). The number of shared proteins by AM, RE or E0 and proteins differing between AM and RE or E0 is also shown. Proteins are considered ”present“ in an egg type if they have at least 2 replicates with 2 or more peptides each in that egg type.
Fig. 4.
Volcano plots showing fold-change (FC) and p value per protein, in a 1-Way ANOVA of protein intensity values. Each panel shows a certain contrast included in the statistical model (AM versus RE, AM versus E0, RE versus E0). Each dot indicates a protein. The y-axis shows nominal p values. Cutoff lines (in red) indicate FDR-adjusted p value of 0.05 and FC of 3 and −3 (in linear scale). As shown, multiple proteins show significant difference in abundance in the comparison of AM versus RE and AM versus E0. However, none of the proteins passed the cutoff lines in the comparison RE versus E0.
Fig. 5.
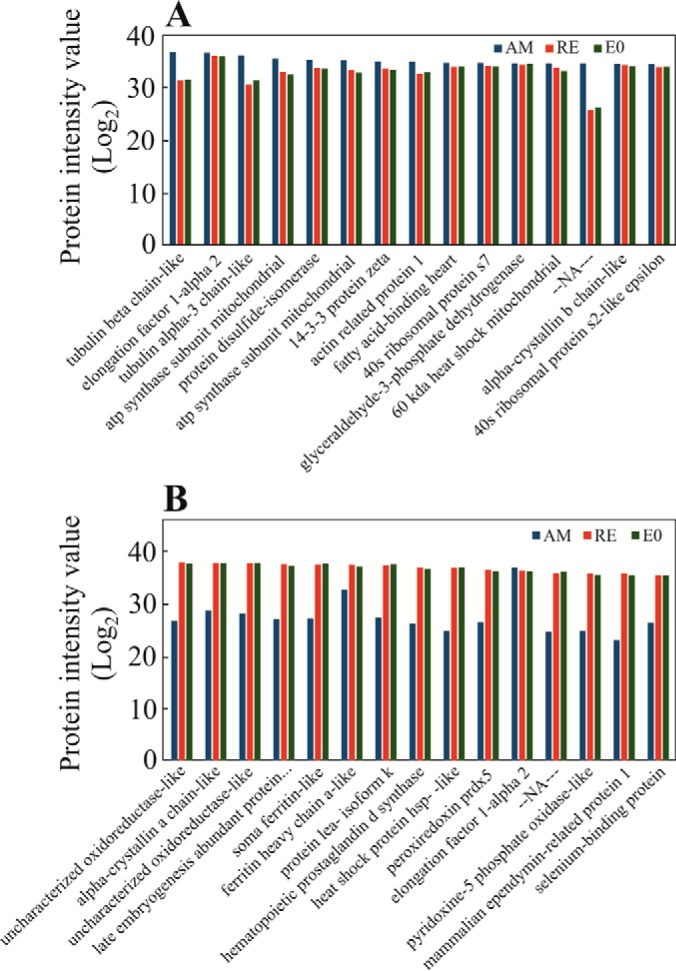
The most abundant proteins in A, amictic eggs (AM); B, resting eggs (RE) and resting eggs ready for hatching (E0).
Differences Between Resting Eggs and Nondormant Eggs in Functional Pathways
Gene Ontology and KEGG Pathways
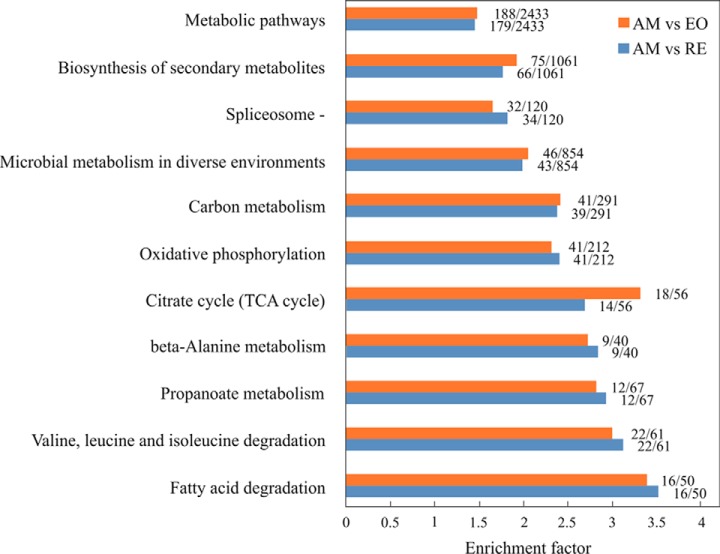
Functional differences between the three egg types (AM, RE and E0), were highlighted by a search for enriched pathways using Gene Ontology (GO; supplemental Table S4; supplemental Fig. S2) and KEGG pathways for each egg type (supplemental Table S5; supplemental Fig. S3). In addition, we carried out enrichment analyses for KEGG pathways and GO terms of proteins showing differential abundance in the comparisons of AM versus RE and AM versus E0. (Fig. 6 and supplemental Table S5 for KEGG pathways; supplemental Fig. S4 and supplemental Table S6 for GO terms). KEGG pathways and GO terms highlight differences in energy metabolism, especially in processes with an association of the mitochondria (e.g. tricarboxylic acid (TCA or citric cycle), electron transport, oxidation phosphorylation), lipid and fatty acid metabolism, amino acid metabolic processes, transcription, translation, splicing, protein processing and remodeling, developmental processes, and proteins with a function in response to oxidative stress and environmental contaminants.
Fig. 6.
Enriched KEGG pathways of the proteins with significant differences in their abundance in the comparisons of AM versus RE (blue) and AM versus E0 (red). For each pathway, the nominator shows the number of proteins in the comparison of AM versus RE or AM versus E0. The denominator shows the total number of proteins in the translated reference transcriptome that were associated with each pathway.
Six general patterns were identified, by comparing the protein abundance in functional pathways, between dormant and nondormant embryos. For simplicity, we present the KEGG maps for the comparison between proteins of AM and RE as there were no significant differences in the abundance between RE and E0 (supplemental Table S3B; Fig. 4).
(a) Lower Abundance of a Large Number of Functional Proteins (or Enzymes) in a Pathway
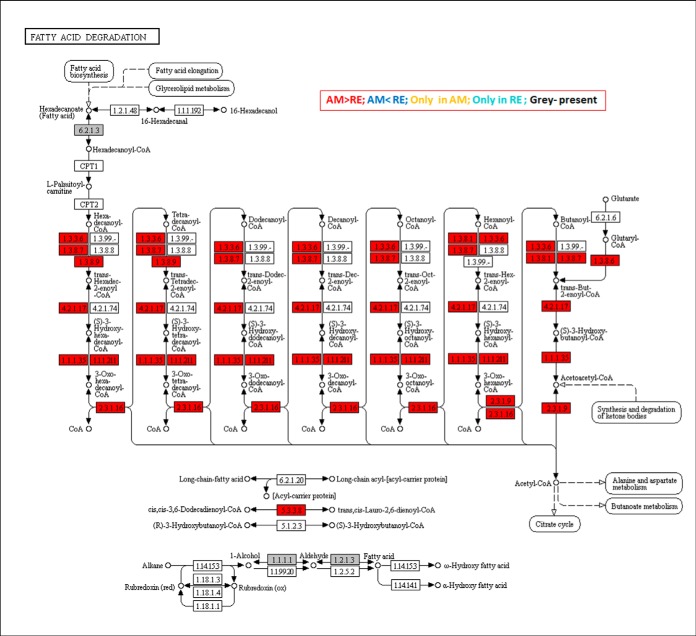
Lower abundance was observed for a large number of proteins in fatty acid degradation (Fig. 7 and supplemental Fig. S5) and in valine, leucine and isoleucine degradation pathways (supplemental Fig. S6). In these pathways, the proteins remain available but at a lower abundance and can be recruited during exit from dormancy.
Fig. 7.
A KEGG map displaying the proteins in association with the Fatty acid degradation pathway. Boxes in gray show the proteins that were identified in rotifer eggs. Boxes in red show proteins that were significantly more abundant in AM versus RE. Statistically significant differences were indicated by 1-Way ANOVA test (FDR p value 0.05, FC>3).
(b) Lower Abundance of Proteins With a Key Function in a Pathway
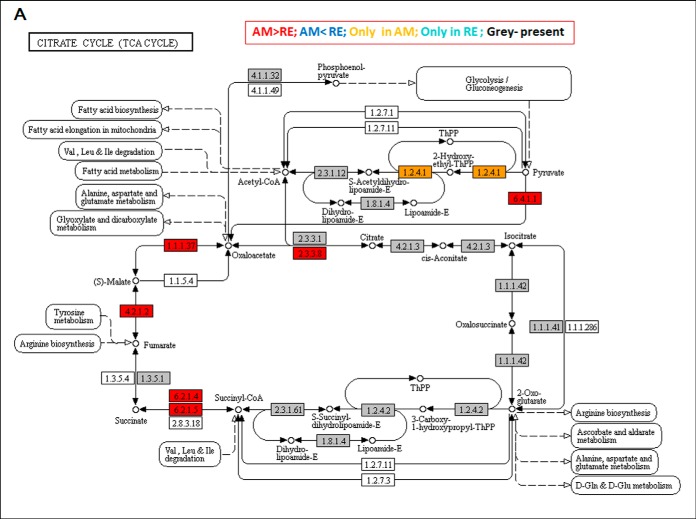
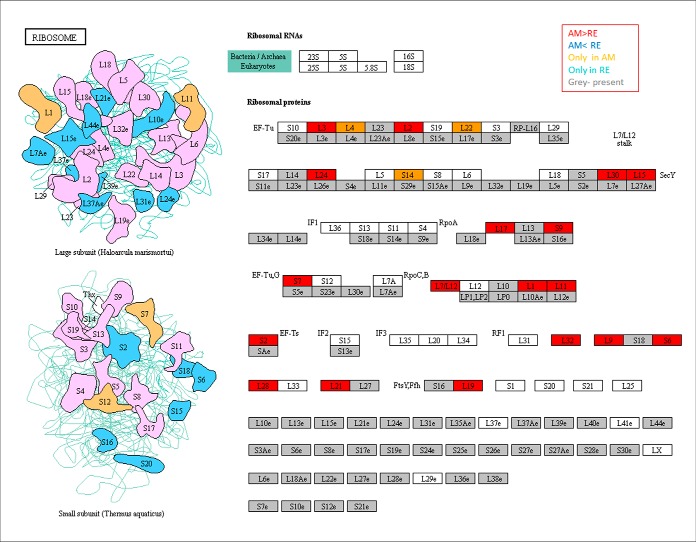
Lower abundance of proteins composing the pyruvate dehydrogenase (PDH) complex. PDH is one of the central enzymes in aerobic metabolism (see role in Artemia; 83) serving as a gateway between glycolysis (supplemental Figs. S7 and S8) and the citrate acid cycle (TCA cycle Fig. 8A and supplemental Fig. S9). It catalyzes the overall conversion of pyruvate to acetyl-CoA and is composed of multiple copies of three enzymatic components: pyruvate dehydrogenase (E1), dihydrolipoamide acetyltransferase (E2) and lipoamide dehydrogenase (E3). All three are also found in the 2-oxoglutarate dehydrogenase complex (ID1503), which is neither detected in RE nor E0 (supplemental Table S3B). The lower abundance of PDH and the putative absence of 2-oxoglutarate dehydrogenase in RE suggest a functional impairment of the mitochondria. The lower abundance or absence of PDH is also relevant to the pyruvate pathway (supplemental Figs. S10 and S11). Another example showing lower abundance or absence of one or more proteins in a pathway includes the ribosome, where low abundance of numerous ribosomal proteins was found (Fig. 9). Notably, mitochondrial ribosomal proteins were more abundant in AM than RE and three ribosomal proteins were not detected in RE. Low abundance of proteins was also found in the cell cycle, spliceosome and mRNA surveillance pathways (supplemental Figs. S12, S13, and S14, respectively).
Fig. 8.
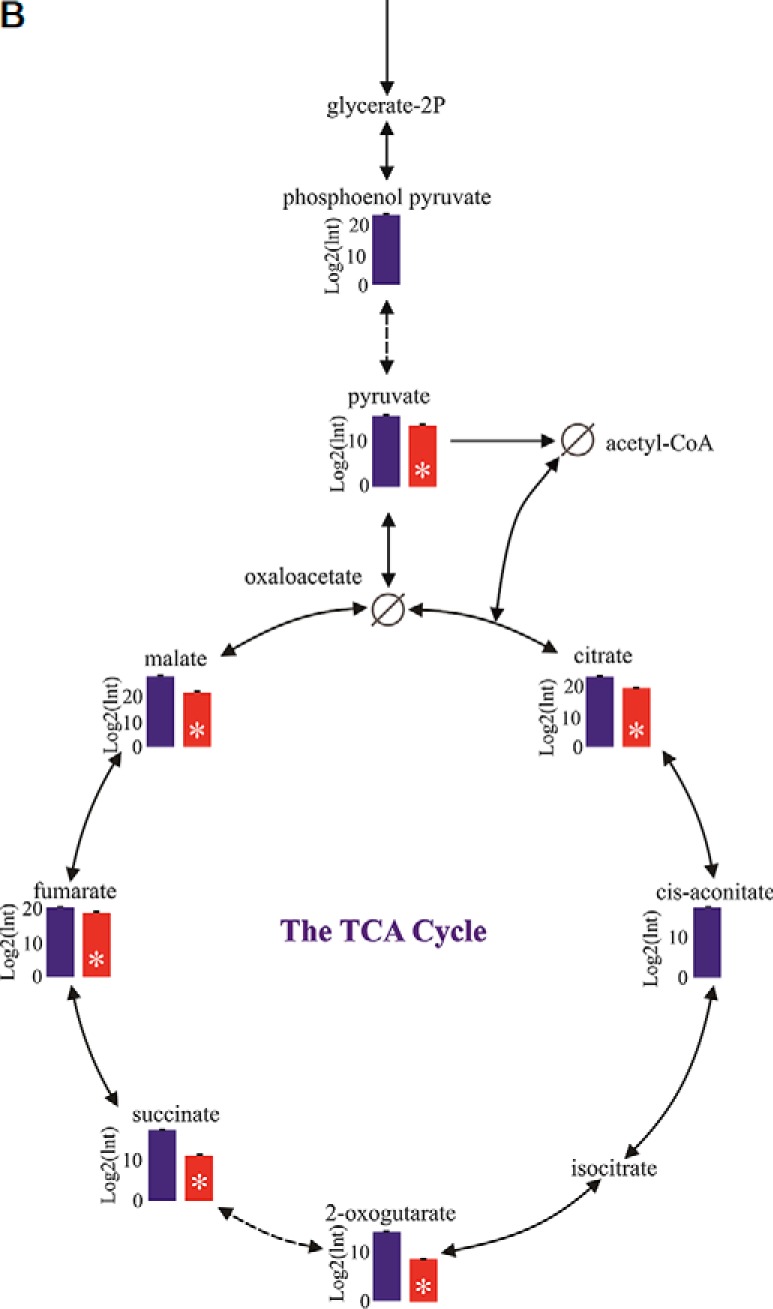
The Citrate (TCA) Cycle. A, A KEGG map showing the proteins in association with the Citrate cycle (TCA) pathway. Boxes in gray show the proteins that were identified in rotifer eggs. Boxes in red show proteins that were significantly more abundant in AM versus RE. Yellow boxes show the proteins that were detected only in AM eggs. Statistically significant differences were indicated by 1-Way ANOVA test (FDR p value 0.05, FC>3), B, A comparison of the log2 relative intensities of compounds between AM and RE, as determined by metabolome analyses. Blue columns - AM eggs, Red columns- RE. A star indicates statistical significant differences (t test; p < 0.05).
Fig. 9.
A KEGG map showing the proteins in association with the Ribosome. Boxes in gray show the proteins that were identified in rotifer eggs. Boxes in red show proteins that were significantly more abundant in AM versus RE. Yellow boxes show the proteins that were detected only in AM eggs.
(c) Lower Abundance of Multitask Proteins Such as Kinases
About 65 kinases were identified and 28 showed differential abundance in the comparison of dormant and nondormant embryos, and 11 were not detected in dormant embryos (supplemental Table S3B). Most notably, MAPK (Mitogen-activated protein kinase; ID785), with a crucial regulatory function, was not detected in RE and E0. Other examples include MAP2K1 (dual specificity mitogen-activated protein kinase kinase; ID2315) - a component of the MAP kinase signal transduction pathway and CDK1 (Cyclin-dependent kinase 1-like; ID806), a key player in cell cycle regulation, showed lower abundance in RE versus AM (FC<14.9). Also, CDK2AP2 (Cyclin-dependent kinase 2-associated protein 2; ID2362), with a role in regulating the self-renewal of embryonic stem cells (ESCs) and regulation of microtubule organization of metaphase II oocytes (see http://www.uniprot.org/uniprot/O75956), showed lower abundance in RE versus AM (FC<13.4). Interesting putative pathways with proteins showing differential abundance displayed in dormant versus nondormant embryos include the Neurotrophin signaling pathway, MAPK signaling pathway, GnRH signaling pathway, Toll-like receptor signaling pathway, Wnt signaling pathway, FOXO signaling pathway, Insulin signaling pathway, Oocyte meiosis, Progesterone-mediated and oocyte maturation (See http://www.kegg.jp; supplemental Table S7). The kinases with significantly differing abundance between AM and RE or E0, include the MAPK, JNK, and ERK 1/2.
(d) Higher Abundance of Proteins with a Putative Inhibitory Function in a Pathway
The protein PI13 (ID110) showed higher abundance in the proteasome of RE (supplemental Fig. S15). This protein was described as a proteasome inhibitor of mammalian cells and a modulator of proteasome formation. Recent studies indicate that in Drosophila melanogaster the overexpression of DmPI13 increased proteasome activity in vivo. However, in vitro studies with purified DmPI13, revealed that the protein activates 26S proteasomes but inhibits the activity of the proteasome core particle (85).
(e) Other Patterns
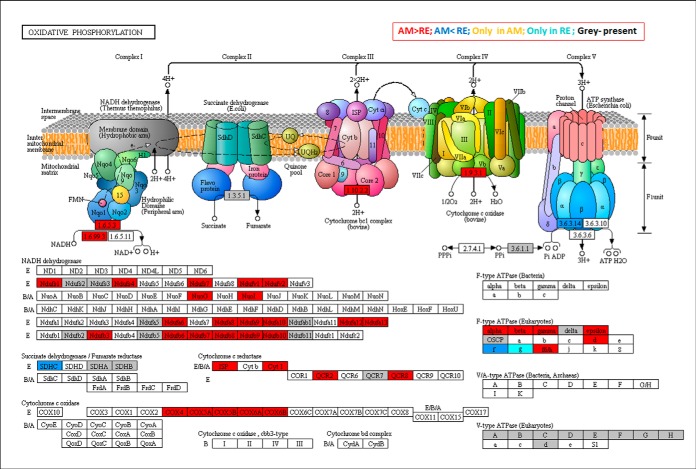
Mixed patterns of higher and lower abundance of proteins or occurrence of proteins only in RE in a functional pathway. This pattern was found in oxidative phosphorylation (Fig. 10 and supplemental Fig. S16) and the pentose phosphate pathway (supplemental Figs. S17 and S18). Additional examples include protein processing and endoplasmic reticulum pathways (supplemental Fig. S19).
Fig. 10.
A KEGG map showing the proteins in association with Oxidative phosphorylation pathway. Boxes in gray show the proteins that were identified in rotifer eggs. Boxes in red and blue show proteins that were significantly more abundant in AM or RE, respectively. Turquoise boxes show the proteins that were detected only in RE. Statistically significant differences were indicated by 1-Way ANOVA test (FDR p value 0.05, FC>3).
(f) Higher Abundance of Hallmark Proteins with a Specific Protective Function
Numerous proteins with specific functions in dormant forms showed differential abundance in the comparison of AM and RE (Table I). This list includes LEA proteins, ferritin, proteins associated with cell stress such as glutathione s-transferase like proteins, peroxidredoxin, thioredoxin, superoxide dismustase, catalase, oxidoreductase, heat shock proteins, α-crystalline proteins, selenium proteins, and proteins associated with synthesis of trehalose (52, 53). Most noticeable are the high abundance of proteins in the comparison of RE versus AM to the extent of FC>500–1,000. Additional information on these proteins is found in the supplementary File Text S1.
Detailed Description of Pathways with a Known Association in Dormancy or Diapause
(a) Energy Metabolism and the Mitochondria
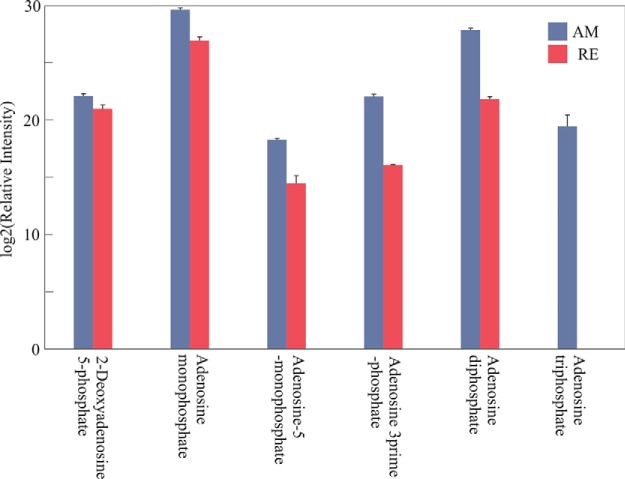
Dormancy has been associated with a depression in metabolism (84) or alternative metabolic pathways (64, 86, 87), and results shown above support the contention of downregulation of metabolism in RE in association with oxidative phosphorylation, the citrate cycle, and pyruvate metabolism (Fig. 10 and supplemental Fig. S16; Fig 8A and supplemental Fig. S9; supplemental Fig. S10 and supplemental Fig. S11). Proteins associated with NADH dehydrogenase, cytochrome c oxidase, cytochrome c reductase and F-type ATPase, showed lower abundance in RE as displayed in the oxidative phosphorylation pathway (Fig 10). The putative significant down-regulation of metabolism is supported by metabolome analyses that showed the low abundance/absence of ATP in RE (Fig 11) and significantly lower abundance of metabolites in the citrate cycle (Fig 8B). Because the pentose phosphate pathway was shown to play a role in embryonic diapause of mouse blastocysts (30) we searched for the abundance of proteins in the pentose phosphate pathway (supplemental Figs. S17 and S18). Two proteins showed higher abundance in RE; glucokinase/gluconate (ID1707) and transaldolase (ID67), which are associated with phosphorylation of d-gluconate and conversion of d-ribose-5 phosphate to d-glyceradehyde-3-phosphate, respectively. d-gluconate or β-d- glucose-6-phosphate could lead in RE to the synthesis of d-glyceraldehyde-3P and a participation in glycolysis. Ribose-phosphate pyrophosphokinase (ID1431), which is the gateway to nucleotide metabolism from d-ribose-5-phosphate, showed lower abundance in RE, suggesting a possible diversion from metabolism of purine, pyrimidine or histidine to the nonoxidative metabolic pathway of the Pentose Phosphate Pathway.
Fig. 11.
A comparison of the log2 relative intensities of adenosine compounds between AM and RE, as determined by metabolome analyses. Blue columns, AM eggs; Red columns, RE. All differences between AM and RE were statistically significant (t test; p < 0.05).
Another source of energy involves the degradation or metabolism of amino acids. In general, proteins of pathways associated with amino acid metabolism were more abundant in AM versus RE. One example includes proteins that were associated with valine, leucine and isoleucine degradation mentioned above (supplemental Fig. S6), with connections to the citrate cycle (through the synthesis of acetyl-CoA from amino acids). Statistically significant lower amounts of amino acids were identified in RE (Fig 12).
Fig. 12.
A comparison of the log2 relative intensities of amino acids between AM and RE, as determined by metabolome analyses. Blue columns, AM eggs; Red columns, RE. All differences between AM and RE were statistically significant (t test; p < 0.05).
(b) Lipid, Fatty Acid Metabolism and Lipoproteins
Changes in the lipid metabolism were often associated with dormancy in several organisms (65, 88). In rotifers, lipid droplets were identified in resting eggs (89) and therefore these pathways were examined here in more detail. Numerous proteins with functions in lipid and fatty acid metabolism were identified in eggs but only a few showed differences in abundance between RE and AM (supplemental Table S3B). As mentioned before, proteins functioning in fatty acid degradation, which are a potential source for generating energy, were less abundant in RE (Fig. 7 and supplemental Fig. S5). Of the fatty acid binding proteins, only fatty acid-binding brain protein (ID 1335) showed higher abundance in RE. Interestingly, lipid storage droplets surface-binding protein 2-like (ID49), a perilipin homolog, was significantly more abundant in RE versus AM. Similarly, lipid phosphate phosphohydrolase 1 isoform (ID1738) with a broad function in lipid metabolic processes, was found in RE but not in AM. A monoacylglycerol lipase (ID1993; abhd12-like protein) that hydrolyzes glycerol monoesters of long-chain fatty acids was not detected in RE (90). Nonspecific lipid transfer protein (ID776, ID1027) with a function in mediating the transfer of all common phospholipids, cholesterol, and gangliosides between membranes and a role in regulating steroidogenesis (91), was highly abundant in AM versus RE (FC>70., FC>108.2, respectively). Also, vigilin (ID297; supplemental Table S3B) was highly abundant in AM in comparison with RE.
(c) DNA Synthesis, Replication and the Cell Cycle
Suspension of the cell cycle is one of the characteristics of dormancy (see the Introduction; 1, 6). A relatively small number of proteins participating in DNA replication and cell cycle were identified in rotifer eggs (supplemental Fig. S12). These include the Mcm2–7 complex, DNA ligase1-like (ID1639), DNA polymerase delta subunit 3 (ID2467), were not detected in RE. DNA polymerase delta subunit 2 (ID2874) was more abundant in AM versus RE (FC>24.6; supplemental Table S3B). In contrast DNA polymerase beta (POLB; ID75) performing base excision repair and required for DNA maintenance, replication and recombination, was higher in RE. A few regulatory proteins with an association of the cell cycle, were less abundant or were not detected in RE. PCNA (Proliferating cell nuclear antigen; ID1400) showed lower abundance by 84.7 fold in RE (versus AM). Zinc finger homeobox protein 2-like (ID2338) and homeobox-containing protein 1 isoform x2 (ID2638) were not detected in RE. PCNA is an auxiliary protein of DNA polymerase delta and is involved in the control of eukaryotic DNA replication. GO annotations for the homeobox proteins include sequence-specific DNA binding transcription factor activity and sequence-specific DNA binding. In addition, mitotic checkpoint protein (bub3-like protein; ID1923), with a putative function in regulation of chromosome segregation during oocyte meiosis in mouse (92), was not detected in RE.
(d) Transcription, Translation, Degradation and Proteome Remodeling
In general, numerous proteins associated with these functions showed lower abundance in RE, indicating an impairment of these functions. DNA-directed RNA polymerases I and III subunit rpac1 (ID667) and DNA-directed RNA polymerase II subunit rpb3 (ID1089), which catalyze the transcription of DNA to RNA, showed lower abundance in RE in comparison with AM. RNA polymerase II-associated factor 1 homolog (ID1030), mediator of RNA polymerase II transcription subunit 8 isoform x1 (ID1692) and RNA polymerase II subunit a c-terminal domain phosphatase ssu72 (ID1976), with multiple functions during transcription, were also with lower abundance in RE (supplemental Table S3B). As shown before, a relatively large number of cytoplasmic ribosomal proteins were identified in rotifer eggs (Fig. 9) and numerous with higher abundance of proteins in AM. Of interest is the elongator complex proteins 6 (ID2003) which was highly elevated in RE (FC>24). This protein is a histone acetyltransferase component of the RNA polymerase II (Pol II) holoenzyme and involved in transcriptional elongation (93). The elongator may play a role in chromatin remodeling and acetylation of histones H3 and probably H4 and functions in cell migration (see below). The proteins of the spliceosome showed differential abundance in the comparison of AM and RE (supplemental Fig. S13). In the mRNA Surveillance pathways (supplemental Fig. S14), many proteins were significantly higher in AM and numerous proteins were not detected in RE, suggesting lower translation and processing of mRNAs in RE. One of the most abundant proteins in the comparison of AM versus RE (FC>88) was a 116 kDa u5 small nuclear ribonucleoprotein component-like protein and part of the U4/U6-U5 tri-snRNP complex (ID2264; supplemental Table S3B), which is required for pre-mRNA splicing (94).
In contrast to the pathways discussed above, most of the proteins that were associated with protein degradation by the proteasome, showed similar abundance in AM and RE (supplemental Fig. S15). The proteasome activator complex subunit 3 (ID960) which may be involved in regulation of the cell cycle was higher in AM. Two proteins that were higher in RE include the Rpn7 (ID137), a member of the regulatory particle and the PI31, which was discussed previously. A protein annotated as proteasome activator subunit 3 (PA28 gamma; ID960) was higher in AM. Proteins associated protein processing in the endoplasmic reticulum were identified in AM and RE (supplemental Fig. S19), with differential display of a numerous proteins.
(d) Histones and Chromatin
Histones play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. We identified proteins annotated as histone proteins (histone h1, histone h2a, histone h4, several histone-like proteins) and histone deacetylases (Supplemental Table S3B) which promote the deacetylation of lysine residues on the N-terminal part of the core histones. A histone chaperone asf1-like protein (ID1693), with a function in association with the chromatin assembly factor 1 (CAF-1) promotes replication-dependent chromatin assembly and plays a role in the formation of silent heterochromatin (95). Most of the proteins showed higher abundance in AM in comparison with RE or E0, or were not detected in RE or E0. However, one protein with significantly higher abundance in RE versus AM (FC>64), is the core histone macro-like isoform x2 (ID486). This is a variant of histone H2A which replaces conventional H2A in a subset of nucleosomes, where it represses transcription, nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Replication protein A (RPA) is a heterotrimeric protein complex of three polypeptides of 70, 34, and 13 kDa and is required for many DNA metabolic processes, including DNA replication, repair and DNA damage checkpoint activation. This protein (ID2067, ID1068, and ID267), showed higher abundance in AM versus AM with FC ranging 3.7 to 4.5. It is also required for the recruitment of the DNA double-strand break repair factors RAD52 and RAD51 to chromatin in response to DNA damage. Additional proteins with a lower abundance in RE and a function in modeling chromatin, are the chromobox proteins (ID1407, ID599), which are components of heterochromatin.
(e) Cytoskeleton
A relatively large number of cytoskeleton associated proteins with a diversity of functions, showed highly differentially abundance between AM and RE (supplemental Table S3B). This includes highly abundant proteins of AM versus RE, like microtubule-associated protein futsch-like (ID584; FC>110) with a neuromuscular function during embryogenesis and troponin I (ID663) that was not detected in RE and troponin t (ID796) that showed lower abundance in RE (FC<244). In contrast, a higher abundance in RE (with a FC ranging from −9 to −17 in AM versus RE) was found for tubulin alpha (ID1417, 1833), which is the major constituent of microtubules and functions in tight and gap junctions.
(f) Oocyte, Spermatozoa and Cilia Proteins
Proteins associated with oocytes and spermatozoa were generally more abundant in AM (supplemental Table S3B). Two proteins with high differences in abundance between AM and RE include the nuclear auto-antigenic sperm protein (ID48), with a FC >334 and the oocyst wall protein (96), with a FC >179 (ID251). As proteins associated with sperm may involve proteins associated with cilia (as mentioned above), additional proteins that were annotated as cilia proteins are given in supplemental Table S3B. Proteins identified as radial spoke head proteins (ID1022, ID1267, ID1409, ID2058, and ID 2472) showed lower abundance (AM versus RE, FC ranging from 34.6 to 186.2) or were not detected in RE. These proteins form a probable component of the axonemal radial spoke head. Radial spokes (97) are regularly spaced along cilia, sperm and flagella axonemes. Similarly, dynein proteins (ID1033, ID1381, ID1791, ID2168, ID2533), except for two (ID929, ID2694), showed differences in their abundance between AM and RE (FC ranging from 12 to 31) or were not detected in RE. The hu-li tai shao-like protein (adducing; ID597) with a function in the structure of egg chamber proteins (in Drosophila, 98), was more abundant in AM versus RE. Conversely, Piwi-like protein 1 ID315, ID1045, ID1138, ID2880) with a function in germline stem cell division (99) showed higher abundance in RE (FC ranging from 2.6 to 7.2).
(g) Ras Proteins
The Ras superfamily of small guanosine triphosphatases (GTPAses) function as GDP/GTP-regulated molecular switches (100). Twenty-one out of the 29 identified proteins in this family (supplemental Table S3B) showed differential abundance between AM and RE and three were not detected in RE. On the other hand, ras-related protein rab-5a (ID2445) with a putative function in fusion of plasma membranes and early endosomes, was not detected in AM.
(h) Regulation
Transcription Factors, Hormones, and Signaling Pathways—In search for pathways that could be involved in regulation of dormancy in RE, we identified putative proteins that were associated with transcription factors, hormones with known functions in rotifers and other organisms and proteins associated with signaling pathways (supplemental Table S3B). All of the proteins were more abundant in AM and some of them were not detected in RE or E0. In this list, we find two members of the 14-3-3 proteins, 14-3-3 protein epsilon (ID616; AM versus RE, FC>71.7) and 14-3-3 protein zeta D64; versus RE, FC>2.64). Two proteins were annotated as insulin-like growth factor 2 mRNA-binding protein1 (ID1263, ID1511), with higher abundance in AM (FC>29.1 and FC>35.6). One of them (ID1263), with a putative function as an RNA-binding factor, recruiting target transcripts to cytoplasmic protein-RNA complexes (mRNPs), allowing mRNA transport and transient storage and shields from endonuclease attacks or microRNA-mediated degradation (http://www.uniprot.org/uniprot/Q9Y6M1).
A juvenile hormone esterase binding protein (ID1562), suggesting a role for JH in rotifers, was identified in rotifer eggs. Juvenile hormone esterase degrades juvenile hormone, which acts in conjunction with ecdysteroids to control gene expression in insects (101). Proteins associated with steroids include two proteins annotated as estradiol 17-beta-dehydrogenase 8 or 12, a membrane-associated progesterone receptor component 2-like and a steroid receptor RNA activator 1. An enzyme with a function in processing of protein hormones and of neuropeptides from their precursors (ID966; neuroendocrine convertase 2-like; 102) showed highly differentially abundance in the comparison of AM and RE. A protein associated with the activation of Notch-1 (delta and notch-like epidermal growth factor-related receptor; ID2657), was identified in AM but was not detected in RE or E0. Insulin signaling pathway has been implicated in diapause regulation in insects (103) and two proteins associated with insulin were mentioned before. Many proteins could be associated with downstream functions of signaling pathways (e.g. kinases; Ras family proteins; calmodulin and others).
(i) Calcium
Proteins annotates as calmodulin, calreticulin, calcineurin, calnexin with an association to calcium storage or binding of calcium, were more abundant in AM versus RE, or were absent in RE (supplemental Table S3B). Interestingly, one protein of the neural calcium sensor 2 (ID265), was highly more abundant in RE (FC>408 for RE versus AM) but others showed variable patterns. Moreover, mammalian ependymin-related protein 1 (ID219) with a putative role in calcium-dependent cell adhesion, was one of the most abundant proteins in RE and E0 (FC>6742 in RE versus AM). Similarly, recoverin (ID54), was highly abundant in RE in comparison with AM (FC>523.8 in RE versus AM). This is a neuronal calcium-binding protein with a key role in the inhibition of rhodopsin kinase, which regulates the phosphorylation of rhodopsin and is involved in the recovery phase of visual excitation and in adaptation to background light (104). This protein might play a role in light sensing, vital to hatching from RE.
Within the list of putative enzymes, a few enzymes (showing differential abundance in the comparison of AM and RE), with interesting functions were identified, such as chitinase (three proteins; ID 1770, chitinase domain-containing protein 1; ID2398 acidic mammalian chitinase-like; ID2466 probable chitinase 3-like), aurora kinase a (ID2279) with a function during meiosis and mitosis (105); where its activity peaks during the G2 phase to M phase transition in the cell cycle. Another enzyme of interest is the chorion peroxidase-like (ID543, ID1521; identified only in RE or E0) that catalyzes protein cross-linking through dityrosine formation and phenol oxidase-catalyzed chorion melanization in insects (106). A protein associated with cleavage of β-carotene was identified (carotene-9 oxygenase, ID1789; beta,beta-carotene 9′,10′-dioxygenase, BCDO2), with small differences in abundance in the two egg types.
Correlation Between Proteome and Transcriptome Expression Profiles
The relationship between protein abundance and RNA expression levels show a negligible positive correlation (107) for AM (Pearson's correlation coefficient, R2 = 0.2112) and a low positive correlation (Pearson's correlation coefficient, R2 = 0.4357) for RE (Fig 13A, 13B), with p value < 2.2e-16 for both egg types (see also supplemental Table S8). Three categories were defined in comparing the abundance levels of the proteins with the corresponding expression levels of mRNAs: (a) A group with an apparent concordance between proteins and the corresponding mRNAs (Fig 13A,13B; in blue),(b) A group with an apparent discordance between proteins and the corresponding mRNAs (Fig 13A, 13B; in red) and (c) A middle group for which neither concordance nor disconcordance can be shown (Fig 13A, 13B; black). The proteins in apparent concordance with the corresponding RNAs of RE show the following enriched KEGG pathways (supplemental Table S5G): ribosome, oxidative phosphorylation, valine, leucine and isoleucine degradation, RNA transport, carbon metabolism and citrate cycle. On the other hand, proteins of the AM group display an apparent concordance with mRNAs for only two KEGG enriched pathways, oxidative phosphorylation and spliceosome (supplemental Table S5F). The GO terms for the comparison of proteins and the corresponding mRNAs of AM and RE, are shown in supplemental Table S9. Interestingly, the hallmark proteins (protein lea- isoform k, LEA protein family protein, alpha-crystallin a chain-like, heat shock hsp20, soma ferritin-like; ferritin heavy chain) with high abundance in RE and/or E0 also show relatively high levels of the corresponding mRNAs (Fig 13B).
Fig. 13.
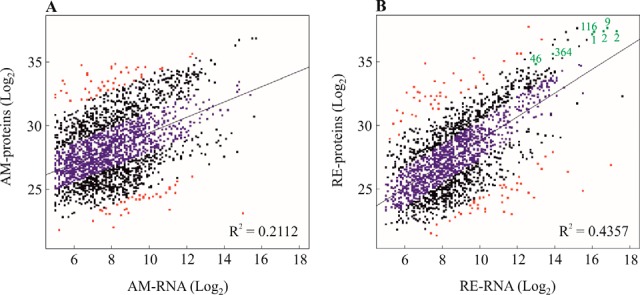
The scatter plot showing the mRNA expression levels and the abundance of proteins in amictic eggs (AM, on the left; y = 0. 60197x + 23.44552, p value < 2.2e-16) and resting eggs (RE, on the right; y = 0.9342x + 19.4816, p value < 2.2e-16). The Pearson correlation coefficients (R2) are shown on the figure. Apparent concordant proteins and RNAs are shown in blue and apparent disconcordant proteins are shown in red. The hallmark proteins of RE (in B) displaying high mRNA levels and the corresponding proteins are shown in green; 1 - protein lea- isoform k; 2- late embryogenesis abundant protein family protein; 9- alpha-crystallin a chain-like; 46- heat shock hsp20; 116- soma ferritin-like; 364- ferritin heavy chain.
Effect of Drying on RE Hatching and Reproductive Rates of Hatched Neonates
Drying of RE had no adverse effect on the percent of hatching after 1, 14, and 56 days (Fig 14). Differences in hatching of RE from the three cultures were not statistically different, although they were kept for different times at room temperature before the drying experiments (Univariate tests of significance; p = 0.77). Statistically significant higher hatching was observed between wet and dried RE after 28 days of drying. Each one of the neonates that hatched from the 56-day dried RE, produced a total of 3.54 ± 1.48 (mean ± S.D.) rotifers and eggs (n = 110), after 72 h of incubation, like the number (3.37 ± 0.98, mean ± S.D., n = 113; p = 0.32, t test) produced by neonates hatching from the control wet RE.
Fig. 14.
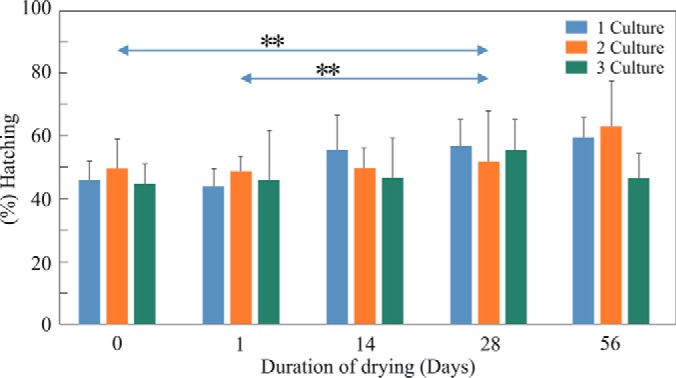
The percent of hatching of neonates from wet and dried RE. RE were obtained from three different cultures and stored in the dark, in three separate glass vials with sea water (10 ppt) for 6–15 months. RE from each vial were distributed to 25 Petri plated (50 RE per plate). Five plates were placed in the light (time 0) and 20 other plates were dried for 1 day and the hatching of neonates was tested after 1, 14, 28, and 56 days. Plates that were tested on days 14, 28, and 56 were stored in the dark, at 24–26 °C. Bars on the top show statistically significant differences (p < 0.01) between hatching of wet RE (day 0) or RE dried for 1 day and RE after 28 days of dried storage. The differences between the RE from the three cultures were not statistically significant (p > 0.05).
DISCUSSION
The phenomenon of dormancy is of wide interest as it occurs in a large number of organisms from prokaryotes to mammals (3, 4). However, proteomic information on dormancy is limited, particularly in nonmodel organism with dormancy exhibited during embryonic development. The intention of the current study is to reveal mechanisms of importance for maintaining dormancy in encysted rotifer embryos by quantitative proteomics, as most wide scale studies in rotifers and other organisms exhibiting dormancy, focused mostly on transcriptome profiling. The overall relatively low correlation between expression levels of mRNAs and abundance of proteins (75,76; in the comparison of pre-diapause and nondiapause locust eggs (73) and Fig 13), renders proteome profiling an important tool for providing an additional valuable view on putative functional changes during dormancy.
A total of 2631 proteins were identified with at least two peptides in two replicate egg samples, comprising ∼15% of the current available transcriptome. Assuming conservation in transcript and protein sequences, it provides information on many putative conserved functional, cellular and molecular pathways, some of which were supported by metabolome profiling. Remarkably, a relative large amount of information was obtained by quantitative proteome profiling while using an assembled transcriptome, as the genome of this species has not yet been sequenced. Proteome profiling provides here a one-time snapshot on the abundance of many proteins in three egg types. It should be noted that protein levels may not fully reflect the functional levels of a specific protein within a pathway, as this may also depend on protein turnover rates (108). However, low or high abundance, or detection in one group and absence (below detection level) in another group, provides insight into putative functional directions. Taking these restrictions into account, the proteome profiles were analyzed with an attempt to provide a coherent view of the functional roles of the identified proteins in supporting dormancy.
About 62% of the proteins were significantly more abundant in AM compared with RE and E0, and 47 and 42 proteins showed FC>100 in these respective comparisons (supplemental Fig. S3). The difference between RE and E0 versus AM is also demonstrated in the list of the most highly abundant proteins of each group (Fig 5A, 5B). Differences in the abundance of proteins between resting eggs before (RE) and after (E0) the obligatory dormant period (Fig. 4; supplemental Table S3B), were not significant but RE displayed 22 proteins that were not determined for E0. Although functional differences were suggested during the obligatory dormant period by E0 enriched KEGG pathways (supplemental Fig. S5), these were not statistically significant (Fig. 4).
About 16 and 28 proteins were similar to those described for cysts of Artemia sinica and Artemia franciscana, respectively (77, 78). Only one of the proteins (calcineurin), unique to the dormant silkworm, was also found as specific to RE (79). In the locust proteome (73), ten of the annotated proteins showing differential abundance between prediapause and nondiapause locust eggs, were found in rotifers. Five showed higher abundance in pre-diapause eggs versus nondiapause eggs and in RE versus AM, including glutathione S-transferase, hydroxysteroid dehydrogenase-like protein 2-like, alcohol dehydrogenase class-3, glucosamine-6-phosphate isomerase 2, isocitrate dehydrogenase. Dolichyl-Diphosphooligosaccharide—protein glycosyltransferase (which is involved in the pathway of protein glycosylation), showed lower differential abundance in the comparison in prediapause eggs versus, nondiapause eggs and in RE versus AM.
Proteome profiling revealed the large number of functional pathways that were putatively compromised in the dormant encysted RE, especially those in mitochondria (Fig 15 and Fig 16). It is usually assumed that metabolism is reduced during dormancy to conserve resources for long-term survival and resumption of life activities. We show here that metabolism is highly reduced in rotifer encysted embryos, as ATP levels were below detection levels (Fig 11). These lower ATP levels can be attributed to a significantly lower activity of the TCA cycle as demonstrated by proteome and metabolome profiling (Fig 8A, 8B; supplemental Fig. S11). In addition, proteome profiling suggests compromised function of the pyruvate metabolic pathway, the oxidative phosphorylation pathway and mitochondrial ribosomes. However, differences were not observed in the abundance of most proteins of the glycolysis or pentose phosphate pathways, suggesting that the proteins of these pathways are a source for low levels of energy during the dormant period or can be quickly recruited to provide energy at the time of resumption of metabolism, during the exit from dormancy. A diversion from the citrate cycle to the pentose phosphate pathway is characteristic of mammalian preimplantation embryos, which may exhibit a form of dormancy (73), as this would lead to synthesis of macromolecules for creating new biomass (109).
Fig. 15.
An overview of the differentially expressed cellular pathways during dormancy.
Fig. 16.
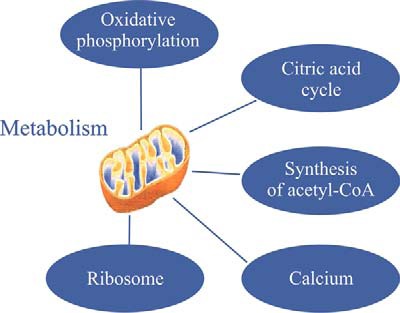
An overview of the differentially expressed pathways in the mitochondria during dormancy.
Long-term survival of hydrated Artemia encysted embryos was attributed to a role of V-ATPases in intracellular acidification leading to metabolic depression (6). As there were no significant differences in the levels of the proteins between AM and RE (Fig 10) like the observation in Artemia (6), the role of V-type ATPases in dormant rotifer eggs remains unclear. Interestingly, F-type ATPase that transforms the energy of ATP hydrolysis to electrochemical potential differences was more abundant in RE versus AM (Fig 10). Moreover, a transcript encoding a V-type H(+)-translocating pyrophosphatase was differentially expressed in 30 min illuminated REs (44), possibly indicating the up-regulation of the oxidative phosphorylation pathway during initial steps of revival from dormancy.
Lipids were described as an energy source during dormancy in many organisms, ranging from C. elegans to hibernating bears (110, 111). In the Asian tiger mosquito, however, downregulation of genes involved in lipid catabolism was observed during diapause (87). The picture for utilization of lipids in rotifer REs is far from being clear. Proteins involved in Fatty acid degradation and fatty acid metabolism showed significantly lower abundance in RE and E0 (Fig. 7 and supplemental Fig. S5). Proteins associated with beta-oxidation, oxidation of long chain fatty acids, and biosynthesis of saturated and unsaturated fatty acids, were higher in AM than RE (supplemental Table S3B), possibly indicating lower utilization of lipids in encysted embryos because of an overall reduction in metabolism. Vigilin, a high-density lipoprotein, with a function in cell sterol metabolism was highly abundant in AM. On the other hand, a few proteins with an association to formation of lipid droplets, lipase activity and lipid transport, were higher in RE. The occurrence of the yolk protein vitellogenin was reported for rotifers (112), but we could not find any matches for this protein in our analyses (results not shown).
Protein and amino acid degradation is another option for generating energy. The lower abundance of amino acids in RE (Fig 12) and the lower abundance of proteins in the Valine, leucine and isoleucine metabolism pathway (supplemental Fig. S6) indicates lower availability of amino acids for metabolism and translation. In general, there were no differences between AM and RE in the level of most proteins of the proteasome but the differential abundance of three proteins, especially of PI131 may suggest regulation of the proteasome function in REs (supplemental Fig. S15). PI13 is an inhibitor of the proteasome in mammals but in Drosophila melanogaster it stimulates proteasome activity while being regulated by the F-box protein Nutcracker during normal development (84).
Additional functions suggested as being compromised in REs include the cell cycle progression, cytoskeleton proteins, replication, transcription, translation, amino acid metabolism, proteome remodeling among others (supplemental Table S3B). Similar general trends were reported for proteome studies in insect diapause (73, 79, 113). Most noticeable is the large number of enzymes and protein kinases that showed differentially abundance between dormant and nondormant eggs (supplemental Table S3B). Most interestingly, mitogen-activated protein kinase 1-like (MAP), regulating many functions, including cell cycle and with a role in adaptation to desiccation in nematodes (114) was not detected in RE or E0. In addition, numerous Ras family proteins showed significantly lower abundance in AM versus RE or AM versus E0. The Ras superfamily of small GTPases, act as binary molecular switches and function as modulators of a remarkably complex and diverse range of cellular processes and transcription factors (supplemental Table S3B; 100).
Additional interesting differences include lower protein abundance in RE and E0 of proteins associates with histones and chromatin, playing a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. In Artemia, acetylation of chromatin-associated histone H3 lysine 56 inhibits the development of cysts (115). Hanson et al., 2013 (74), suggest a role for histones during sexual reproduction in B. calyciflorus. Information on the involvement of key signaling molecules, transcription factors, regulatory peptides or hormones is scarce, probably because of their lower abundance (supplemental Table S3B) but some showed higher abundance in AM versus RE or E0. They include most notably the 14-3-3 proteins with a wide array of regulatory functions (116). The 14-3-3 proteins constitute a highly conserved protein family, with more than 100 signaling proteins serving as ligands for other proteins (117). They play an important role in a wide range of cellular processes, including signal transduction, apoptosis, cell cycle progression, regulation of the Ras/Raf/MAPK cascade, the anterior-posterior axis during embryonic development and serve as checkpoint activators within eukaryotic cells. 14-3-3 proteins participate in light signaling and bind to phytochrome interacting factors in plants (118) and melatonin synthesis in mammals (119). As mentioned before, light promotes hatching of rotifer REs (44, 57). In addition, 14-3-3 showed differential abundance in the comparison of dormant and nondormant silkworm eggs (79). Moreover, 14-3-3 proteins are also associated with the forkhead box O (FOXO) transcription factors (120). It was demonstrated recently that numerous distinct groups of genes that are targets of FOXO promote the diapause phenotype in insects. FOXO is a transcription factor functioning downstream of insulin and juvenile hormone signaling in insects and the targets include genes that are associated with metabolism, development/cell cycle, intra- and extracellular signaling, transcription, translation, protein modification, stress response, cell death and apoptosis (6, 121). The results presented here suggest a role for FOXO in dormancy of encysted rotifer eggs as a few proteins with an association to insulin and juvenile hormone metabolism were identified here, but these results require additional verification. Additional hormones may be involved in regulation as proteins with an association to estradiol synthesis and progesterone with known functions in rotifers, were also identified in the current study (43, 122).
Like other dormant organisms, a unique set of genes is evoked and highly abundant in rotifer REs (51–53). The abundance of stress tolerance related proteins was significantly altered in dormant eggs (Table I) like the changes observed in other organisms displaying dormancy (3, 8, 9). These include glutathione S- transferase (GST) with antioxidant activity, protecting cellular components from oxidative damage. Ferritin is one of the most abundant proteins in dormant RE and E0 and shows higher abundance in diapausing insects (123, 124) and copepods (125). It has been shown that the concentration of ferritin increases in response to stresses such as anoxia (126). Artemia cysts accumulate artemin, a diapause-specific ferritin homologue with chaperon activity (127). Ferritin serves as an iron storage protein and is the main component of yolk proteins in snail eggs (128). LEA proteins with an important role in facilitating desiccation (129), show high abundance in RE (Fig. 5; Table I and supplementary Text S1).
There is a low correlation between the lists of the most abundant proteins in AM and resting eggs (RE or E0; Fig. 5A, 5B) and the list of most abundant proteins does not match fully with the list of most abundant transcripts (51). A detailed comparison in the level of transcripts and the corresponding proteins indicates higher concordance in RE in comparison with AM (Fig 13A, 13B), possibly because of a lower protein turnover in RE (108). Furthermore, the apparent concordant proteins of RE display a larger number of enriched KEGG pathways than those of AM proteins (supplemental Table S5). Interestingly, hallmark proteins with a specific function in RE (e.g. LEA proteins; ferritin, alpha crystalline, superoxide dismutase) also display high levels of mRNAs (Fig 13B). These proteins degrade during revival from dormancy (52–54), raising the question whether these mRNAs function in translation during the dormant period or do they serve as a reservoir for nucleic acids.
In general, rotifer dormant embryos display pathways like dormant stages that survive desiccation, such as Artemia cysts and the sleeping chironomid (8, 9, 16). We did not find differences in the hatchability of dried and wet stored RE up to 56 days of storage. Moreover, there were no differences in the reproductive output of neonates hatching from these dry or wet stored RE. Similar hatching percent of dried and wet RE after 3 months of storage were demonstrated for the freshwater rotifer species Brachionus calyciflorus (130) but most other studies (e.g. 131, 132) do not provide quantitative information on the comparative success of hatching from wet and dried field samples or quantitative information on subsequent viability. RE withstand desiccation and this trait is probably embedded already at the time of their formation, as they survive after a short desiccation period, in contrast to a slow dehydration regime required for high recovery, by bdelloid rotifers (133) and some tardigrade species that do not survive quick drying (134). Moreover, metabolism is probably reduced shortly after their formation, explaining the long-term survival in the hydrated or desiccated form. The functional events taking place during the obligatory dormant period require additional studies.
In conclusion, the current study expands our knowledge and provides insight on the wide scope of functions that are regulated in dormant encysted embryos. The interpretations from proteome profiling on a reduced metabolism and translation in dormant embryos were supported by metabolome analyses, strengthening to some extent, the conclusions, on other putative functional pathways that were identified in our analyses.
The functional activity of a pathway depends on numerous enzymes or proteins involved in this pathway. In some pathways, one or more of the proteins were not detected, suggesting a malfunction of the pathway. In different organisms, the malfunction of a pathway may depend on changes in different proteins within the pathway, while leading to a similar functional effect (phenotypic function). Consequently, it can be suggested that a divergence between organisms (10) may be because of differences in the regulated genes and/or proteins within a functional pathway, while leading to the same result. Moreover, these differences may also reflect a divergence in the gene regulatory pathways (e.g. hormones, epigenetic changes etc.) between organisms. The relatively low concordance between transcriptome and proteome profiling provides another explanation for the low transcriptional similarity among dormancies across species, despite the resemblance in physiological phenotypes.
DATA AVAILABILITY
Mass spectrometry proteomics data were deposited at the ProeomeXchange Consortium via Pride partner repository with the data set identifier PXD003395 (https://www.ebi.ac.uk/pride/archive/simpleSearch?q=PXD003395&submit=Search). The assembled transcriptome is available at http://bioinfo.bgu.ac.il/genomes/Esther_Lubzens/public/ (Username: ester; Password: retse).
Supplementary Material
Acknowledgments
We thank Reini Malka Hamo for technical assistance. The proteomics analyses were performed at the Smoler Proteomics Center Technion and the Technical assistance of Ilana Navon in operating the mass spectrometers is much appreciated. The purchase of the Q-Exactive-Plus mass spectrometer was Supported by the Israel Science Foundation (ISF), the I-CORE Program of the Planning and Budgeting Committee and the ISF (Grant 1775/12).
Footnotes
Author contributions: T.Z., N.D., A.A., and E.L. performed research; T.Z., V.C.-C., I.P., S.S.K., I.B., A.A., and E.L. analyzed data; T.Z., V.C.-C., A.A., and E.L. wrote the paper; N.D., A.A., and E.L. designed research; T.Z. equal contribution with Vered Chalifa-Caspi (below); V.C.-C. supervised and participated in forming the transcriptome assembly; A.A. co-Corresponding author.
* This study was supported by the Israel Science Foundation grant #1524/13.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- AM
- Amictic eggs
- ANOVA
- analysis of variance
- E0
- Resting eggs before emergence
- FC
- fold change
- FDR
- false discovery rate
- GO
- Gene ontology
- LEA
- Late embryogenesis abundant protein
- PCA
- principal component analysis
- PDH
- pyruvate dehydrogenase
- RE
- resting eggs.
REFERENCES
- 1. Denlinger D. L., Yocum G. D., and Rinehart J. P. (2012) Hormonal control of diapause. In: Gilbert L. I. ed. Insect Endocrinology, pp. 430–463, Elsevier, Amsterdam [Google Scholar]
- 2. Keilin D. (1959) The problem of anabioisis or latent life: history and current concept. Proc. R. Soc. Lond. B 150, 149–191 [DOI] [PubMed] [Google Scholar]
- 3. Lubzens E., Cerdà J., and Clark M. S. (2010) Topics in Current Genetics 21, pp. 1–283, Dormancy and Resistance in Harsh Environments, Springer, Heidelberg, Dordrecht, London, NY [Google Scholar]
- 4. Lubzens E. (2015) Frére Jacques/Dormez vous? Dormancy, an intriguing phenomenon shared by many forms of life. Mol. Reprod. Dev. 82, DOI: 10.1002/mrd.22480 (Editorial) [DOI] [PubMed] [Google Scholar]
- 5. Hand S. C., Menze M. A., Borcar A., Patil Y., Covi J. A., Reynolds J. A., and Toner M. (2011) Metabolic restructuring during energy-limited states: Insights from Artemia franciscana embryos and other animals. J. Insect. Physiol. 57, 584–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hand S. C., Denlinger D. L., Podrabsky J. E., and Roy R. (2016) Mechanisms of animal diapause: recent developments from nematodes, crustaceans, insects, and fish. Am. J. Physiol. Regul. Integr. Comp. Physiol. 310, R1193–R211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kostál V. (2006) Eco-physiological phases of insect diapause. J. Insect. Physiol. 52, 113–127 [DOI] [PubMed] [Google Scholar]
- 8. MacRae T. H. (2010) Gene expression, metabolic regulation and stress tolerance during diapause. Cell Mol. Life Sci. 67, 2405–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. MacRae T. H. (2016) Stress tolerance during diapause and quiescence of the brine shrimp, Artemia. Cell Stress Chaperones 21, 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ragland G. J., Denlinger D. L., and Hahn D. A. (2010) Mechanisms of suspended animation are revealed by transcript profiling of diapause in the flesh fly. Proc. Natl. Acad. Sci. U.S.A. 107, 14909–14914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shen-Miller J., Mudgett M. B., Schopf J. W., Clarke S., and Berger R. (1995) Exceptional seed longevity and robust growth: ancient sacred lotus from China. Am. J. Bot. 82, 1367–1380 [Google Scholar]
- 12. Sallon S., Solowey E., Cohen Y., Korchinsky R., Egli M., Woodhatch I., Simchoni O., and Kislev M. (2008) Germination, genetics, and growth of an ancient date seed. Science 320, 1464. [DOI] [PubMed] [Google Scholar]
- 13. Fontenato D., Bunnefeld N., and Westberg M. (2012) Long-term survival of microscopic animals under desiccation is not so long. Astrobiology 12, 863–869 [DOI] [PubMed] [Google Scholar]
- 14. Hashimoto T., Horikawa D. D., Saito Y., Kuwahara H., Kozuka-Hata H., Shin-I T., Minakuchi Y., Ohishi K., Motoyama A., Aizu T., Enomoto A., Kondo K., Tanaka S., Hara Y., Koshikawa S., Sagara H., Miura T., Yokobori S., Miyagawa K., Suzuki Y., Kubo T., Oyama M., Kohara Y., Fujiyama A., Arakawa K., Katayama T., Toyoda A., and Kunieda T. (2015) Extremotolerant tardigrade genome and improved radiotolerance of human cultured cells by tardigrade-unique protein. Nat. Commun. 7, 12808. doi: 10.1038/ncomms12808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clark M. S., and Worland M. R. (2008) How insects survive the cold: molecular mechanisms- a review. J. Comp. Physiol. B. 178, 917–933 [DOI] [PubMed] [Google Scholar]
- 16. Cornette R., and Kikawada T. (2011) The induction of anhydrobiosis in the sleeping chironomid: current status of our knowledge. IUBMB Life 63, 419–429 [DOI] [PubMed] [Google Scholar]
- 17. Gusev O., Suetsugu Y., Cornette R., Kawashima T., Logacheva M. D., Kondrashov A. S., Penin A. A., Hatanaka R., Kikuta S., Shimura S., Kanamori H., Katayose Y., Matsumoto T., Shagimardanova E., Alexeev D., Govorun V., Wisecaver J., Mikheyev A., Koyanagi R., Fujie M., Nishiyama T., Shigenobu S., Shibata T. F., Golygina V., Hasebe M., Okuda T., Satoh N., and Kikawada T. (2014) Comparative genome sequencing reveals genomic signature of extreme desiccation tolerance in the anhydrobiotic midge. Nat. Commun. 5, 4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Berjak B. (2006) Unifying perspectives of some mechanisms basic to desiccation tolerance across life forms. Seed Sci. Res. 16, 1–15 [Google Scholar]
- 19. Carvalho G. R., and Wolf H. G. (1989) Resting eggs of lake- Daphnia I. Distribution, abundance and hatching of eggs collected from various depths in lake sediments. Freshwater Biol. 22, 459–470 [Google Scholar]
- 20. Hairston N. G., Vanbrunt R. A., Kearns C. M., and Engstrom D. R. (1995) Age and survivorship of diapausing eggs in a sediment egg bank. Ecology 76, 1706–1711 [Google Scholar]
- 21. Marcus N. H., and Boero F. (1998) Minireview: The importance of benthic-pelagic coupling and the forgotten role of life cycles in coastal systems. Limnology Oceanogr. 43, 763–768 [Google Scholar]
- 22. Kotani T., Ozaki M., Matsuoka K., Snell T. W., and Hagiwara A. (2001) Reproductive isolation among geographically and temporally isolated marine Brachionus strains. Hydrobiologia 446, 283–290 [Google Scholar]
- 23. Garcia-Roger E. M., Carmona M. J., and Serra M. (2006) Patterns in rotifer diapausing egg banks: Density and viability. J. Exp. Mar. Biol. Ecol. 336, 198–210 [Google Scholar]
- 24. Radzikowski J. (2013) Resistance of dormant stages of planktonic invertebrates to adverse environmental conditions. J. Plankton Res. 35, 707–723 [Google Scholar]
- 25. Frisch D., Morton P. K., Chowdhury P. R., Culver B. W., Colbourne J. K., Weider L. J., and Jeyasingh P. D. (2014) A millennial-scale chronicle of evolutionary responses to cultural eutrophication in Daphnia. Ecol. Lett. 17, 360–368 [DOI] [PubMed] [Google Scholar]
- 26. Podrabsky J. E., and Hand S. C. (2015) Physiological strategies during animal diapause: lessons from brine shrimp and annual killifish. J. Exp. Biol. 218, 1897–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lopes F. L., Desmarais J. A., and Murphy B. D. (2004) Embryonic diapause and its regulation. Reproduction 128, 669–678 [DOI] [PubMed] [Google Scholar]
- 28. Ptak G. E., Tacconi E., Czernik M., Toschi P., Modlinski J. A., and Loi P. (2012) Embryonic diapause is conserved across mammals. PLoS ONE 7, e33027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fenelon J. C., Banerjee A., and Murphy B. D. (2014) Embryonic diapause: development on hold. Int. J. Dev. Biol. 58, 163–174 [DOI] [PubMed] [Google Scholar]
- 30. Fu Z., Wang B., Wang S., Wu W., Wang Q., Chen Y., Kong S., Lu J., Tang Z., Ran H., Tu Z., He B., Zhang S., Chen Q., Jin W., Duan E., Wang H., Wang Y. L., Li L., Wang F., Gao S., and Wang H. (2014) Integral proteomic analysis of blastocysts reveals key molecular machinery governing embryonic diapause and reactivation for implantation in mice. Biol. Reprod. 90, 52:1–11 [DOI] [PubMed] [Google Scholar]
- 31. Yamashita O. (1996) Diapause Hormone of the Silkworm, Bombyx mori: Structure, Gene Expression and Function. J. Insect. Phvsiol. 42, 669–679 [Google Scholar]
- 32. Ye H. L., Li D. R., Yang J. S., Chen D. F., De Vos S., Vuylsteke M., Sorgeloos P., Van Stappen G., Bossier P., Nagasaw H., and Yang W. J. (2017) Molecular characterization and functional analyses of a diapause hormone receptor-like gene in parthenogenetic Artemia. Peptides (in press) [DOI] [PubMed] [Google Scholar]
- 33. Wallace R. L. (2002) Rotifers: Exquisite metazoan. Integ. Comp. Biol. 42, 660–667 [DOI] [PubMed] [Google Scholar]
- 34. Wallace R. L., Snell T. R., Ricci C., and Nogrady T. (2006) Rotifera. In: Segers H., ed. Biology, Ecology and Systematics Guides to the Identification of the Microinvertebrates of the Continental Waters of the World 23 Backhuys Publishers, Leiden [Google Scholar]
- 35. Walsh E. J., Smith H. A., and Wallace R. L. (2014) Rotifers of temporary waters. Int. Rev. Hydrobiol. 99, 3–19 [Google Scholar]
- 36. Ricci C. (2001) Dormancy patterns in rotifers. Hydrobiologia 446/447, 1–11 [Google Scholar]
- 37. Gilbert J. J. (2004) Population density, sexual reproduction and diapause in monogonont rotifers: new data for Brachionus and a review. J. Limnol. 63, 32–36 [Google Scholar]
- 38. Gilbert J. J. (2007) Timing of diapauses in monogonont rotifers: mechanisms and strategies, In: Alekseev V. R., De Stasio B., Gilbert J. J., eds. Diapause in aquatic invertebrates: Theory and human use Monographiae Biologicae, 84, pp. 11–27. Springer, Dordrecht, The Netherlands [Google Scholar]
- 39. Gilbert J. J., and Schröder T. (2004) Rotifers from diapausing, fertilized eggs: unique features and emergence. Limnology Oceanog. 49, 1341–1354 [Google Scholar]
- 40. Gilbert J. J., and Schröder T. (2007) Intraclonal variation in propensity for mixis in several rotifers: variation among females and maternal age. Hydrobiologia 593, 121–128 [Google Scholar]
- 41. Schröder T. (2005) Diapause in monogonont rotifers. Hydrobiologia 564, 291–306 [Google Scholar]
- 42. Denekamp N. Y., Suga T., Hagiwara A., Reinhardt R., and Lubzens E. (2010) A role for molecular studies in unveiling the pathways for formation of rotifer resting eggs and their survival during dormancy, In: Lubzens E., Cerdà J., Clark M. S. eds., Topics in Current Genetics 21, pp. 109–132. Dormancy and Resistance in Harsh Environments, Springer, Heidelberg, Dordrecht, London, NY [Google Scholar]
- 43. Stout E. P., La Clair J. J., Snell T. W., Shearer T. L., and Kubanek J. (2010) Conservation of progesterone hormone function in invertebrate reproduction. Proc. Natl. Acad. Sci. 107, 11859–11864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim H.-J., Suga K., Kim B.-M., Rhee J.-S., Lee J.-S., and Hagiwara A. (2015) Light-dependent transcriptional events during resting egg hatching of the rotifer Brachionus manjavacas. Marine Genomics 20, 25–31 [DOI] [PubMed] [Google Scholar]
- 45. Kim R. O., Rhee J. S., Won E. J., Lee K. W., Kang C. M., Lee Y. M., and Lee J. S. (2011) Ultraviolet B retards growth, induces oxidative stress, and modulates DNA repair-related gene and heat shock protein gene expression in the monogonont rotifer, Brachionus sp. Aquat. Toxicol. 101, 529–539 [DOI] [PubMed] [Google Scholar]
- 46. Lee B. Y., Kim H. S., Hwang D. S., Won E. J., Choi B. S., Choi I. Y., Park H. G., Rhee J. S., Lee J. S. (2015) Whole transcriptome analysis of the monogonont rotifer Brachionus koreanus provides molecular resources for developing biomarkers of carbohydrate metabolism. Comp. Biochem. Physiol. D Genomics Proteomics 14, 33–41 [DOI] [PubMed] [Google Scholar]
- 47. Lee J. S., Kim R.-O., Rhee J. S., Han J., Hwang D. S., Choi B. S., Lee C. J., Yoon Y. D., Lim J. S., Lee Y. M., Park G. S., Hagiwara A., and Choi I. Y. (2011) Sequence analysis of genomic DNA (680Mb) by GS-FLX-Titanium sequencer in the monogonont rotifer, Brachionus ibericus. Hydrobiologia 662, 65–75 [Google Scholar]
- 48. Rhee J. S., Kim R. O., Choi H. G., Lee J., Lee Y. M., and Lee J. S. (2011) Molecular and biochemical modulation of heat shock protein 20 (Hsp20) gene by temperature stress and hydrogen peroxide (H2O2) in the monogonont rotifer, Brachionus sp. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 154, 19–27 [DOI] [PubMed] [Google Scholar]
- 49. Rhee J. S., Kim R. O., Kim B. M., Dahms H. U., and Lee J. S. (2012) Genomic organization of selected genes in the small monogonont rotifer, Brachionus koreanus. Gene 505, 108–113 [DOI] [PubMed] [Google Scholar]
- 50. Snell T. W. (2011) A review of the molecular mechanisms of monogonont rotifer reproduction. Hydrobiologia 662, 89–97 [Google Scholar]
- 51. Clark M. S., Denekamp N. Y., Thorne M. A. S., Reinhardt R., Drugowski M., Albrecht M. W., Klags S., Beck A., Kube M., and Lubzens E. (2012) Long-term survival of hydrated resting eggs from Brachionus plicatils. PLoS ONE 7, e29365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Denekamp N. Y., Thorne M. A. S., Clark M. S., Kube M., Reinhardt R., and Lubzens E. (2009) Discovering genes associated with dormancy in the monogonont rotifer Brachionus plicatilis. BMC Genomics 10, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Denekamp N. Y., Reinhardt R., Albrecht M. W., Drungowski M., Kube M., and Lubzens E. (2011) The expression pattern of dormancy-associated genes in multiple life-history stages in the rotifer Brachionus plicatilis. Hydrobiologia 662, 51–63 [Google Scholar]
- 54. Denekamp N. Y., Reinhardt R., Kube M., and Lubzens E. (2010) Late Embryogenesis Abundant (LEA) proteins in nondesiccated, encysted, and diapausing embryos of rotifers. Biol. Repro. 82, 714–724 [DOI] [PubMed] [Google Scholar]
- 55. Boschetti C., Leasi F., and Ricci C. (2011) Developmental stages in diapausing eggs: an investigation across monogonont rotifer species. Hydrobiologia 662, 149–155 [Google Scholar]
- 56. Smith J. M., Cridge A. G., and Dearden P. K. (2010) Germ cell specification and ovary structure in the rotifer Brachionus plicatilis. EvoDevo 1, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Minkoff G., Lubzens E., and Kahan D. (1983) Environmental factors affecting hatching of rotifer (Brachionus plicatilis) resting eggs. Hydrobiologia 104, 61–69 [Google Scholar]
- 58. Clegg J. S. (2001) Cryptobiosis - a peculiar state of biological organization. Comp. Biochem. Physiol. B 128, 613–624 [DOI] [PubMed] [Google Scholar]
- 59. Warner A. H., MacRae T. H., and Bagshaw J. C. (1989) Cell and molecular biology of Artemia development. Plenum Press, New York and London, 453pp [Google Scholar]
- 60. Abatzopoulos Th J., Beardmore J. A., Clegg J. S., and Sorgeloos P. (2002) Artemia: Basic and applied biology. Springer Science+Business Media Dodrecht, 286pp [Google Scholar]
- 61. Li R., Chen D. F., Zhou R., Jia S. N., Yang J.-S., Clegg J. S., and Yang W. J. (2012) Involvement of polo-like kinase 1(Plk1) in mitotic arrest by inhibition of Mitogen-activated proteinkinase-extracellular signal-regulated kinase-ribosomal S6 kinase 1 (MER-ERK-RSK1) cascade. J. Biol. Chem. 287, 15923–15934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Malarkey K., Coker K. J., and Stuggill T. W. (1998) Ribosomal S6 kinase is activated as an early event in preemergence development of encysted embryos of Artemia salina. Eur. J. Biochem. 251, 269–274 [DOI] [PubMed] [Google Scholar]
- 63. Zhao L. L., Jin F., Ye X., Zhu L., Yang J. S., and Yang W. J. (2015) Expression profiles of miRNAs and involvement of miR-100 and miR-34 in regulation of cell cycle arrest in Artemia. Biochem. J. 470, 223–231 [DOI] [PubMed] [Google Scholar]
- 64. Burnell A. M., Houthoofd K., O'Hanlon K., and Vanfleteren J. R. (2005) Alternate metabolism during the dauer stage of the nematode Caenorhabditis elegans. Exp. Gerontol. 40, 850–856 [DOI] [PubMed] [Google Scholar]
- 65. Hahn D. A., and Denliner D. L. (2011) Energetics of insect diapause. Ann. Rev. Entomol. 56, 103–121 [DOI] [PubMed] [Google Scholar]
- 66. Baker D. A., and Russell S. (2009) Gene expression during Drosophila melanogaster egg development before and after reproductive diapause. BMC Genomics 10, 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kankare M., Salminen T., Laiho A., Vesala L., and Hoikkala A. (2010) Changes in gene expression linked with adult reproductive diapause in a northern malt fly species: a candidate gene microarray study. BMC Ecol. 10, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rinehart J. P., Robich R. M., and Denlinger D. L. (2010) Isolation of diapause-regulated genes from the flesh fly, Sarcophaga crassipalpis by suppressive subtractive hybridization. J. Insect. Physiol. 56, 603–609 [DOI] [PubMed] [Google Scholar]
- 69. Yocum G. D., Rinehart J. P., Chirumamilla-Chapara A., and Larson M. L. (2009) Characterization of gene expression patterns during the initiation and maintenance phases of diapause in the Colorado potato beetle, Leptinotarsa decemlineata. J. Insect. Physiol. 55, 32–39 [DOI] [PubMed] [Google Scholar]
- 70. Clark M. S., Thorne M. A., Purać J., Burns G., Hillyard G., Popović Z. D., Grubor-Lajsić G., and Worland M. R. (2009) Surviving the cold: molecular analyses of insect cryoprotective dehydration in the Arctic springtail Megaphorura arctica (Tullberg). BMC Genomics 10, 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Burns G., Thorne M. A., Hillyard G., Clark M. S., Convey P., and Worland M. R. (2010) Gene expression associated with changes in cold tolerance levels of the Antarctic springtail, Cryptopygus antarcticus. Insect Mol. Biol. 19, 113–120 [DOI] [PubMed] [Google Scholar]
- 72. Poupardin R., Schöttner K., Korbelová J., Provazník J., Doležel D., Pavlinic D., Beneŝ V., and Koŝtál V. (2015) Early transcriptional events linked to induction of diapause revealed by RNAseq in larvae of drosophilid fly, Chymomyza costata. BMC Genomics 16, 720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tu T., Wang J., Hao K., Whitman D. W., Fan Y., Cao G., and Zhang Z. (2015) Transcriptomic and proteomic analysis of pre-diapause and nondiapause eggs of migratory locust, Locusta migratoria L. (Orthoptera: Acridoidea). Scient. Rep. 5, 11402 10.1038/srep11402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hanson S. J., Stelzer C. P., Welch D. B., and Logsdon J. M. (2013) Comparative transcriptome analysis of obligately asexual and cyclically sexual rotifers reveals genes with putative functions in sexual reproduction, dormancy, and asexual egg production. BMC Genomics 14, 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Griffin T. J., Gygi S. P., Ideker T., Rist B., Eng J., Hood L., and Aebersold R. (2002) Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol. Cell Proteomics 1, 323–333 [DOI] [PubMed] [Google Scholar]
- 76. Ghazalpour A., Bennett B., Petyuk V. A., Orozco L., Hagopian R., Mungrue I. N., Farber C. R., Sinsheimer J., Kang H. M., Furlotte N., Park C. C., Wen P. Z., Brewer H., Weitz K., Camp D. G. 2nd, Pan C., Yordanova R., Neuhaus I., Tilford C., Siemers N., Gargalovic P., Eskin E., Kirchgessner T., Smith D. J., Smith R. D., and Lusis A. J. (2011) Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 7, e1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wang W., Meng B., Chen W., Ge X., Liu S., and Yu J. (2007) A proteome study on postdiapused embryonic development of brine shrimp (Artemia franciscana). Proteomics 7, 3580–3591 [DOI] [PubMed] [Google Scholar]
- 78. Zhou Q., Wu C., D B., Liu F., and Xiang J. (2008) The encysted dormant embryo proteome of Artemia sinica. Mar. Biotechnol. 10, 438–446 [DOI] [PubMed] [Google Scholar]
- 79. Fan L., Lin J., Zhong Y., and Liu J. (2013) Shotgun proteomic analysis on the diapause and nondiapause eggs of domesticated silkworm Bombyx mori. PLoS ONE 8, e60386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda – a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 82. Ulitsky I., Maron-Katz A., Shavit S., Sagir D., Linhart C., Elkon R., Tanay A., Sharan R., Shiloh Y., and Shamir R. (2010) Expander: from expression microarrays to networks and functions. Nat. Prot. 5, 303–322 [DOI] [PubMed] [Google Scholar]
- 83. Giavalisco P., Köhl K., Hummel J., Seiwert B., and Willmitzer L. (2009) 13C isotope-labeled metabolomes allowing for improved compound annotation and relative quantification in liquid chromatography-mass spectrometry-based metabolomic research. Anal. Chem. 81, 6546–6551 [DOI] [PubMed] [Google Scholar]
- 84. Patil Y. N., Marden B., Brand M. D., and Hand S. C. (2013) Metabolic downregulation and inhibition of carbohydrate catabolism during diapause in embryos of Artemia franciscana. Physiol. Biochem. Zool. 86, 106–118 [DOI] [PubMed] [Google Scholar]
- 85. Schmidt M., and Finley D. (2014) Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta 1843, 13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Holt S. J., and Riddle D. L. (2003) SAGE surveys C. elegans carbohydrate metabolism: evidence for an anaerobic shift in the long-lived dauer larva. Mech. Ageing Dev. 124, 779–800 [DOI] [PubMed] [Google Scholar]
- 87. Mádi A., Mikkat S., Koy C., Ringel B., Thiesen H. J., and Glocker M. O. (2008) Mass spectrometric proteome analysis suggests anaerobic shift in metabolism of Dauer larvae of Caenorhabditis elegans. Biochim. Biophys. Acta 1784, 1763–1770 [DOI] [PubMed] [Google Scholar]
- 88. Reynolds J. A., Poelchau M. F., Rahman Z., Armbruster P. A., and Denlinger D. L. (2012) Transcript profiling reveals mechanisms for lipid conservation during diapause in the mosquito, Aedes albopictus. J. Insect Physiol. 58, 966–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wurdak E. S., Gilbert J. J., and Jagels R. (1978) Fine structure of the resting eggs of the rotifers Brachionus calyciflorus and Asplanchna sieboldi. Trans. Am. Microsc. Soc. 97, 49–72 [PubMed] [Google Scholar]
- 90. Labar G., Wouters J., and Lambert D. M. (2010) A review on the monoacylglycerol lipase: at the interface between fat and endocannabinoid signalling. Curr. Med. Chem. 17, 2588–2607 [DOI] [PubMed] [Google Scholar]
- 91. Westerman J., Wirtz K. W., Westerman J., Wirtz K. W., Berkhout T., van Deenen L. L., Radhakrishnan R., and Khorana H. G. (1985) The primary structure of the nonspecific lipid transfer protein (sterol carrier protein 2) from bovine liver. Eur. J. Biochem. 132, 441–449 [Google Scholar]
- 92. Li M., Li S., Yuan J., Wang Z. B., Sun S. C., Schatten H., and Sun Q. Y. (2009) Bub3 is a spindle assembly checkpoint protein regulating chromosome segregation during mouse oocyte meiosis. PLoS ONE 4, e7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Svejstrup J. Q. (2007) Elongator complex: how many roles does it play? Curr. Opin. Cell Biol. 19, 331–336 [DOI] [PubMed] [Google Scholar]
- 94. Fabrizio P., Laggerbauer B., Lauber J., Lane W. S., Lührmann R. (1997) An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 16, 4092–4106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Moshkin Y. M., Armstrong J. A., Maeda R. K., Tamkun J. W., Verrijzer P., Kennison J. A., and Karch F. (2002) Histone chaperone ASF1 cooperates with the Brahma chromatin-remodelling machinery. Genes Dev. 16, 2621–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Templeton T. J., Lancto C. A., Vigdorovich V., Liu C., London N. R., Hadsall K. Z., and Abrahamsen M. S. (2004) The Cryptosporidium oocyst wall protein is a member of a multigene family and has a homolog in Toxoplasma. Infect. Immun. 72, 980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zhu X., Liu Y., and Yan P. (2016) Radial Spokes—A Snapshot of the motility regulation, assembly, and evolution of cilia and flagella. Cold Spring Harb. Perspect Biol. December a028126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zaccai M., and Lipshitz H. D. (1996) Role of Adducin-like (hu-li tai shao) mRNA and protein localization in regulating cytoskeletal structure and function during Drosophila oogenesis and early embryogenesis. Dev. Genet. 19, 249–257 [DOI] [PubMed] [Google Scholar]
- 99. Cox D. N., Chao A., Baker J., Chang L., Qiao D., and Lin H. (1998) A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12, 3715–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wennerberg K., Rossman K. L., and Der C. J. (2005) The Ras superfamily at a glance. J. Cell Sci. 118, 843–846 [DOI] [PubMed] [Google Scholar]
- 101. Shanmugavelu M., Baytan A. R., Chesnut J. D., and Bonning B. C. (2000) A novel protein that binds juvenile hormone esterase in fat body tissue and pericardial cells of the tobacco hornworm Manduca sexta L. J. Biol. Chem. 275, 1802–1806 [DOI] [PubMed] [Google Scholar]
- 102. Husson S. J., Clynen E., Baggerman G., Janssen T., and Schoofs L. (2006) Defective processing of neuropeptide precursors in Caenorhabditis elegans lacking proprotein convertase 2 (KPC-2/EGL-3): mutant analysis by mass spectrometry. J. Neurochem. 98, 1999–2012 [DOI] [PubMed] [Google Scholar]
- 103. Sim C., and Denlinger D. L. (2013) Insulin signaling and the regulation of insect diapause. Front. Physiol. 4, 189:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kawamura S., Hisatomi O., Kayada S., Tokunaga F., and Kuo C. H. (1993) Recoverin has S-modulin activity in frog rods. J. Biol. Chem. 268, 14579–14582 [PubMed] [Google Scholar]
- 105. Crane R., Gadea B., Littlepage L., Wu H., and Ruderman J. V. (2004) Aurora, meiosis and mitosis. Biol. Cell 96, 215–229 [DOI] [PubMed] [Google Scholar]
- 106. Li J., Hodgeman B. A., and Christensen B. M. (1996) Involvement of peroxidase in chorion hardening in Aedes aegypti. Insect Biochem. Mol. Biol. 26, 309–317 [DOI] [PubMed] [Google Scholar]
- 107. Mukata M. M. (2012) Statistics Corner: A guide to appropriate use of Correlation coefficient in medical research. Malawi Medical J. 24, 69–71 [PMC free article] [PubMed] [Google Scholar]
- 108. Galland M., Huguet R., Arc E., Cueff G., Job D., and Rajjou L. (2014) Dynamic proteomics emphasizes the importance of selective mRNA translation and protein turnover during Arabidopsis seed germination. Mol. Cell Proteomics 13, 252–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Krisher R. L., and Prather R. S. (2012) A role for the Warburg effect in preimplantation embryo development: metabolic modification to support rapid cell proliferation. Mol. Reprod. Dev. 79, 311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Carey H. V., Andrews M. T., and Marin S. L. (2003) Mammalian hibernation: cellular and molecular responses. Physiol. Rev. 83, 1153–1181 [DOI] [PubMed] [Google Scholar]
- 111. Narbonne P., and Roy R. (2009) Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature 45, 210–214 [DOI] [PubMed] [Google Scholar]
- 112. Jones B. L., Schneider D. M., and Snell T. W. (2012) Thermostable proteins in the diapausing eggs of Brachionus manjavacas (Rotifera). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 162, 193–199 [DOI] [PubMed] [Google Scholar]
- 113. Pavlides S. C., Pavlides S. A., and Tammariello S. P. (2011) Proteomic and phosphoproteomic profiling during diapause entrance in the flesh fly, Sarcophaga crassipalpis. J. Insect Physiol. 57, 635–644 [DOI] [PubMed] [Google Scholar]
- 114. Banton M. C., and Tunnacliffe A. (2012) MAPK phosphorylation is implicated in the adaptation to desiccation stress in nematodes. J. Exp. Biol. 215, 4288–4298 [DOI] [PubMed] [Google Scholar]
- 115. Zhou R., Yang F., Chen D. F., Sun X. Y., Yang J. S., and Yang W. J. (2013) Acetylation of chromatin-associated histone H3 Lysine 56 inhibits the development of encysted Artemia embryos. PLoS ONE 8, e68374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Darling D. L., Yingling J., and Wynshaw-Boris A. (2005) Role of 14-3-3 proteins in eukaryotic signaling and development. Curr. Top. Dev. Biol. 68, 281–315 [DOI] [PubMed] [Google Scholar]
- 117. Morrison D. K. (2009) The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 19, 16–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Adams E., Diaz C., Hong J. P., and Shin R. (2014) 14-3-3 proteins participate in light signaling through association with phytochrome interacting factors. Int. J. Mol. Sci. 15, 22801–22814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Ganguly S., Weller J. L., Ho A., Chemineau P., Malpaux B., and Klein D. C. (2005) Melatonin synthesis: 14-3-3-dependent activation and inhibition of arylalkylamine N-acetyltransferase mediated by phosphoserine-205. Proc. Natl. Acad. Sci. U.S.A. 102, 1222–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Hay N. (2011) Interplay between FOXO, TOR, and Akt. Biochim. Biophys. Acta 1813, 1965–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Sim C., Kang D. S., Kim S., Bai X., and Denlinger D. L. (2015) Identification of FOXO targets that generate diverse features of the diapause phenotype in the mosquito Culex pipiens. Proc. Natl. Acad. Sci U.S.A. 112, 3811–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Gallardo W. G., Hagiwara A., and Snell T. W. (2000) Effect of juvenile hormone and serotonin (5-HT) on mixis induction of the rotifer Brachionus plicatilis Muller. J. Exp. Mar. Biol. Ecol. 252, 97–107 [DOI] [PubMed] [Google Scholar]
- 123. Wolschin F., and Gadau J. (2009) Deciphering proteomic signatures of early diapause in Nasonia. PLoS ONE 4, e6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Zhang Q., Lu Y. X., and Xu W. H. (2013) Proteomic and metabolomic profiles of larval hemolymph associated with diapause in the cotton bollworm, Helicoverpa armigera. BMC Genomics 14, 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Tarrant A. M., Baumgartner M. F., Verslycke T., and Johnson C. L. (2008) Differential gene expression in diapausing and active Calanus finmarchicus (Copepoda). Mar. Ecol. Prog. Ser. 355, 193–207 [Google Scholar]
- 126. Larade K., and Storey K. B. (2004) Accumulation and translation of ferritin heavy chain transcripts following anoxia exposure in a marine invertebrate. J. Exp. Biol 207, 1353–1360 [DOI] [PubMed] [Google Scholar]
- 127. Chen T., Villeneuve T. S., Garant K. A., Amons R., and MacRae T. H. (2007) Functional characterization of artemin, a ferritin homolog synthesized in Artemia embryos during encystment and diapause. FEBS J. 274, 1093–1101 [DOI] [PubMed] [Google Scholar]
- 128. Bottke W. (1982) Isolation and properties of vitellogenic ferritin from snails. J. Cell Sci. 58, 225–240 [DOI] [PubMed] [Google Scholar]
- 129. Tunnacliffe A., Hincha D. K., Leprince O., and Macherel D. (2010) LEA proteins: versatility of form and function, In: Lubzens E., Cerdà J., Clark M. S. eds., Topics in Current Genetics 21, pp. 91–108. Dormancy and Resistance in Harsh Environments, Springer, Heidelberg, Dordrecht, London, NY [Google Scholar]
- 130. Kanagasabapathi V., and Munuswamy N. (2011) Preservation, development, and hatching of resting eggs in the freshwater rotifer Brachionus calyciflorus Pallas. Microsc. Res. Tech. 74, 744–748 [DOI] [PubMed] [Google Scholar]
- 131. Gilbert J. J. (2001) Spine development in Brachionus quadridentatus from an Australian billabong“ genetic variation and induction by Asplanchna. Hydrobiologia 446/447, 19–28 [Google Scholar]
- 132. Garcia-Roger E. M., Carmona M. J., and Serra M. (2006) Hatching and viability of rotifer diapausing eggs collected from pond sediments. Freshwater Biol. 51, 1351–1358 [Google Scholar]
- 133. Ricci C., Melone G., Santo N., and Caprioli M. (2003) Morphological response of a bdelloid rotifer to desiccation. J. Morphol. 257, 246–253 [DOI] [PubMed] [Google Scholar]
- 134. Boothby T. C., Tapia H., Brozena A. H., Piszkiewicz S., Smith A. E., Giovannini I., Rebecchi L., Pielak G. J., Koshland D., and Goldstein B. (2017) Tardigrades Use Intrinsically Disordered Proteins to Survive Desiccation. Mol. Cell 65, 975–984.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry proteomics data were deposited at the ProeomeXchange Consortium via Pride partner repository with the data set identifier PXD003395 (https://www.ebi.ac.uk/pride/archive/simpleSearch?q=PXD003395&submit=Search). The assembled transcriptome is available at http://bioinfo.bgu.ac.il/genomes/Esther_Lubzens/public/ (Username: ester; Password: retse).