Abstract
RUNX3, runt-domain transcription factor, is a master regulator of gene expression in major developmental pathways. It acts as a tumor suppressor in many cancers but is oncogenic in certain tumors. We observed upregulation of RUNX3 mRNA and protein expression in nasal-type extranodal natural killer (NK)/T-cell lymphoma (NKTL) patient samples and NKTL cell lines compared to normal NK cells. RUNX3 silenced NKTL cells showed increased apoptosis and reduced cell proliferation. Potential binding sites for MYC were identified in the RUNX3 enhancer region. Chromatin immunoprecipitation–quantitative PCR revealed binding activity between MYC and RUNX3. Co-transfection of the MYC expression vector with RUNX3 enhancer reporter plasmid resulted in activation of RUNX3 enhancer indicating that MYC positively regulates RUNX3 transcription in NKTL cell lines. Treatment with a small-molecule MYC inhibitor (JQ1) caused significant downregulation of MYC and RUNX3, leading to apoptosis in NKTL cells. The growth inhibition resulting from depletion of MYC by JQ1 was rescued by ectopic MYC expression. In summary, our study identified RUNX3 overexpression in NKTL with functional oncogenic properties. We further delineate that MYC may be an important upstream driver of RUNX3 upregulation and since MYC is upregulated in NKTL, further study on the employment of MYC inhibition as a therapeutic strategy is warranted.
Introduction
Human runt-related transcription factor (RUNX) family is composed of three members including RUNX1, RUNX2 and RUNX3, are known as the developmental regulators and have been shown to be important in human cancers.1 RUNX family is highly conserved in their runt homology domain, which is involved in the sequence-specific DNA binding and heterodimerization with the common co-factor CBFβ.2 RUNX1 is essential for generation of hematopoietic stem cells and is involved in human leukemia.2, 3 RUNX2 is essential for skeletal development and has an oncogenic potential.1, 4 RUNX3 is expressed in wider ranges of tissues and has multiple roles. Among others, RUNX3 is a major tumor suppressor of gastric, colon and many other solid tumors.2, 5, 6 Inactivation of RUNX3 by hemizygous deletion, promoter hypermethylation, histone modification and protein mislocalization is frequently observed, suggesting a tumor suppressive role for RUNX3.5, 6, 7
In addition to its well-known tumor suppressor role in human cancers, RUNX3 has also recently been reported to play an oncogenic role in a certain subset of cancers. Oncogenic properties of RUNX were first identified by retroviral activation screens in which all three murine RUNX genes were found to cooperate with MYC oncogene to promote leukemogenesis.8 In basal cell carcinomas, RUNX3 was overexpressed in cancer cells compared to normal epidermis.9 RUNX3 is also oncogenic in head and neck squamous cell carcinoma, ovarian cancer and Ewing sarcoma where overexpression of RUNX3 promoted proliferation and tumorigenesis.10, 11, 12 Collectively, these findings suggest that RUNX3 can function as an oncogene and tumor suppressor in a cellular context-dependent manner.
Extranodal NK/T-cell lymphoma nasal-type (NKTL) is a rare and aggressive disease more frequent in Asia and South America than in Europe and North America and is characterized by a neoplastic proliferation of Epstein–Barr virus (EBV)-infected cytotoxic T and NK cells.13 Although several recent studies have explored new treatment modalities for NKTL, the optimal therapy has still not been found. Interestingly, there have been several recent reports implicating the role of RUNX3 in the maturation pathway of NK cells and cytotoxic T-lymphocytes.14 RUNX3 mediates transcriptional activation in cytotoxic T- and NK cells. Functional annotation of shared CD8+ T and NK RUNX3-regulated genes revealed enrichment for those involved in lymphocyte activation, proliferation, cytotoxicity, migration and cytokine production.15 RUNX3 also regulates expression of Eomes and three cardinal markers of the effector cytotoxic T lymphocyte program including IFN-γ, perforin, and granzyme B, indicating a critical role of RUNX3 in the differentiation of NK and cytotoxic T-lymphocytes.16 Interestingly, gene expression data from our previous study on NKTL revealed RUNX3 to be upregulated in tumor compared to normal NK cells.17
In this study, we investigated the role of RUNX3 and demonstrated that RUNX3 is overexpressed in NKTL compared to normal NK cells and downregulation of RUNX3 results in increased apoptosis and reduced proliferation, supporting the role of RUNX3 as an oncogene in NKTL. We further deciphered the mechanisms of RUNX3 upregulation in NKTL and our data revealed that MYC plays a role in the transcriptional regulation of RUNX3 in NKTL. Targeted inhibition of MYC using small-molecule inhibitor, JQ1, resulted in downregulation of MYC and RUNX3 with reduction in cell proliferation and increased cell death.
Materials and methods
Cell culture and NK cell isolation
A panel of NKTL cells (KHYG-1, HANK-1, NK-YS, SNK-1 and SNK-6) were used in this study. Cell lines were incubated at 37 °C in a humidified atmosphere of 5% CO2. The characteristics and the culture conditions are listed in the Supplementary Table S1.
Highly pure untouched normal NK cells were isolated using an indirect magnetic labeling system by depletion of magnetically labeled cells from human peripheral blood mononuclear cells. (STEMCELL Technologies Inc., Vancouver, BC, Canada).
Immunohistochemistry and immunofluorescence
Four-μm sections from tissue microarray (TMA) blocks containing 38 samples of NKTL were stained for RUNX3 (1:500, 5G4 clone from Professor Yoshiaki Ito). The clinical and pathological data of the 38 NKTL samples were previously published and included in Supplementary Table S2.18
The immunohistochemical expression for RUNX3 was scored as a percentage of the total tumor cell population per 1-mm core diameter (400 ×). Positive expression for RUNX3 is defined as positive nuclear expression of RUNX3 in at least 50% of tumor cell population. We obtained the expression values of MYC from our previous study17 and correlated it with RUNX3.
Double immunofluorescence (DIF) to demonstrate co-localization of RUNX3 and MYC was performed on selected NKTL paraffin tissue sections using Opal 7-color Flourophore TSA plus Fluorescence Kit (NEL797001KT, PerkinElmer Inc., Waltham, MA, USA). Appropriate controls were used. Image acquisition and analysis was done with the Vectra 2 multispectral automated imaging system and inForm 2.0 image analysis software (PerkinElmer Inc.).
See Supplementary Methods for details of IHC, scoring and DIF.
RNA extraction, real-time quantitative PCR analysis and western blot analysis
See Supplementary Methods for details.
Knockdown of RUNX3 and c-MYC
Knockdown was achieved via electroporation with optimized pulse conditions in NKTL cells utilizing the NEON Transfection System (Life Technologies, Carlsbad, CA, USA). Non-targeting siRNA pool was used as controls. NKTL cells were transiently transfected with a pool of siRNAs (Thermo Scientific, Waltham, MA, USA) selectively targeting human RUNX3 and c-MYC. NKTL cells were treated to the optimized transfection conditions. Treated cell lines were assessed for protein and mRNA expressions.
Flow cytometric analysis for apoptosis
The apoptotic cell death analyses were carried out using Annexin-V-APC and propidium iodide (PI) detection systems.
Cell proliferation analysis
Cell proliferation was assayed using BrdU Cell Proliferation Assay kit (Cell Signaling Technology, Danvers, MA, USA) according to the manufacturer’s instructions.
Luciferase reporter assay
The RUNX3 enhancer sequence (Supplementary Methods) that contains essential elements was cloned into pGL3-Basic luciferase reporter vector (Promega, Madison, WI, USA) via specific restriction sites. Luciferase assay was analyzed in Hela and NK-YS cells. Cells were lysed, and the activities of firefly luciferase and renilla luciferase in the transfected cells were measured using a Dual-Luciferase Assay System (Promega).
Chromatin immunoprecipitation
Chromatin immunoprecipitation assay was performed in KHYG-1 and SNK-1 cells according to the manufacturer’s protocol (Cell Signaling Technology) with anti-MYC antibody (Cell Signaling Technology). Immunoprecipitation with isotype matched anti-IgG antibody was used as control. The immunoprecipitated DNA was purified as per the manufacturer’s instructions (Cell Signaling Technology). Primers used for RUNX3 enhancer, and control detection were described in detail in the Supplementary Methods.
Cell viability analysis
Cell viability was determined using the MTS assay (Promega). The cells were incubated for 72 h and MTS reagent was then added into each well and incubated for 2 h at 37 °C, followed by the absorbance reading at 490 nm using a microplate reader (TECAN Infinite 200 Pro, Zurich, Switzerland).
MYC Inhibition with JQ1 and Rescue in NKTL cells
The thieno-triazolo-1,4-diazepine (JQ1) compound used in assays was a kind gift from James Bradner (Dana-Farber Cancer Institute, MA, USA), and suspended in DMSO to a stock concentration (10 mM) and subsequently diluted to working concentrations as indicated for treatment assays. In order to assess if the cell death from JQ1 treatment can be rescued by overexpression of MYC, NKTL cells were transfected with pcDNA3-MYC or empty vector, respectively, and incubated overnight. The transfection efficiency was validated using NKTL cells (Supplementary Figure S1). Transfected NKTL cells were treated with JQ1 for 48 h and processed for cell viability assays. Gene expression and western blotting was performed to assess MYC levels in the treated cells.
See Supplementary Information for details of all methods and materials.
Results
RUNX3 is overexpressed in NKTL
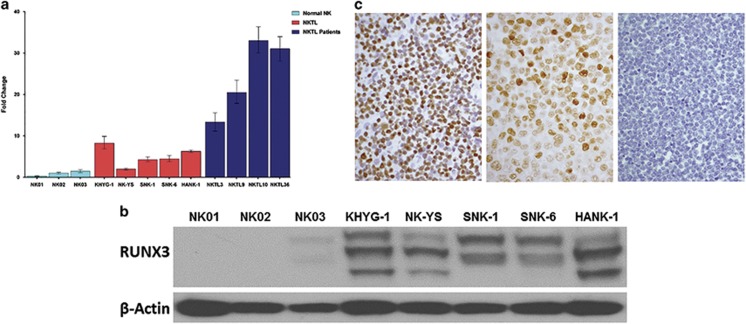
Quantitative real-time RT-PCR revealed RUNX3 mRNA to be overexpressed in NKTL cells and NKTL patient samples compared to normal NK cells (Figure 1a), which supports our previously published gene expression data that showed overexpression of RUNX3 in NKTL samples compared to normal NK cells (Supplementary Figure S2a).17 Similarly, there is a consistent higher protein expression in the NKTL cell lines (KHYG1, HANK1, SNK-1 and SNK-6) compared to normal NK cells (Figure 1b), which show low levels of RUNX3 expression (Supplementary Figure S2b). The protein expression of RUNX3 in the NK cell lines were distinctly higher compared to other known RUNX3 positive cell lines such as SNU-5 (gastric cancer) and THP-1 (monocytic leukemia) (Supplementary Figure S2c). Additional bands (isoforms) of RUNX3 protein are observed in the NKTL cell lines compared to SNU-5 and THP-1 (Figures 1b and 2b, Supplementary Figure S2) and this may be due to alternatively spliced isoforms arising from differential usage of the two alternative RUNX3 promoters (P1 and P2) in tumor cells and/or mobility shift secondary to post-translational modifications, such as phosphorylation and ubiquitination.19, 20
Figure 1.
Endogenous RUNX3 expression in NKTL cells (KHYG1, NK-YS, SNK-1, SNK-6 and HANK-1), NKTL patient samples and normal NK cells (NK01-NK03). (a) mRNA expression profiles of RUNX3 in NKTL cells, normal NK cells and NKTL patient samples. cDNA was converted from total RNA of normal NK, NKTL cells and NKTL patient samples, subsequently assayed by quantitative real-time PCR (RT-PCR). RUNX3 mRNA is upregulated in NKTL cells and NKTL patient samples compared to normal NK cells. (b) Protein profiles of RUNX3 across normal and NKTL cells. NKTL cell lysates were probed with RUNX3 (5G4) antibody. There is overexpression of RUNX3 protein in NKTL cells compared to normal NK cells. (c) Immunohistochemistry showing overexpression of RUNX3 in NKTL patient sample (left) and NKTL cell line (SNK-6, middle) and negative expression in normal NK cells (right).
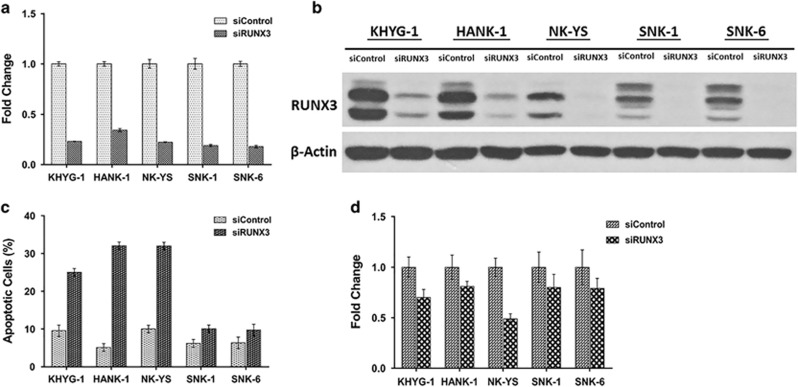
Figure 2.
RUNX3 inhibition in NKTL cells. Silenced RUNX3 NKTL cells (siRUNX3) showed deregulation in cell proliferation. (a) NKTL cells were transiently transfected with control siRNA (siControl) and with siRNA against RUNX3 (siRUNX3). Seventy two hours post transfection, total RNA was prepared and subjected to quantitative RT-PCR. A significant reduction of RUNX3 mRNA was observed across all NKTL cells. (b) Whole-cell lysates were prepared and processed for immunoblotting, probed with anti-RUNX3 antibody. β-Actin was used as a loading control. RUNX3 expression was efficiently silenced after 72 h of transfection using siRUNX3. (c) Transfected NKTL cells were stained with Annexin-V and propidium iodide and analyzed for the induction of apoptosis by flow cytometry. Silenced RUNX3 NKTL cells showed an induction of apoptosis compared to control siRNA (siControl). (d) Transfected NKTL cells were stained with BrdU and analyzed for the effects on the cell proliferation rate. The cell proliferation rate was disrupted in RUNX3 silenced NKTL cells (siRUNX3).
We further analyzed RUNX3 protein expression using immunohistochemistry on tissue microarray sections containing 38 patient samples of NKTL. Using 50% positive expression as cutoff, 27 out of 35 cases showed positive RUNX3 expression (Figure 1c, Supplementary Table S3). Our results confirmed that RUNX3 gene and protein is overexpressed in NKTL cells and patient samples compared to normal NK cells.
RUNX3 inhibition reduced proliferation and increased apoptosis of NKTL cell lines
In order to assess if RUNX3 overexpression is of functional importance in NKTL, we attempted RUNX3 knockdown using siRNA targeting RUNX3 and investigated the effects of RUNX3 inhibition on apoptosis induction and cell proliferation using Annexin-V and Propidium iodide for apoptosis and BrdU incorporation, respectively. Successful knockdown of RUNX3 was achieved in 5 NKTL cells including KHYG-1, NK-YS, HANK1, SNK1 and SNK6, with mRNA expressions showing a distinct RUNX3 reduction and a corresponding reduction in the protein expressions (Figures 2a and b).
Apoptosis assay revealed that RUNX3 depletion resulted in increased levels of apoptosis in all five NKTL cells with the effect most prominently seen in KHYG-1, HANK-1 and NK-YS. (Figure 2c and Supplementary Figure S3). In agreement with the apoptotic assay, the knockdown phenotype of the NKTL cells also displayed a substantial reduction in their proliferation rate, as illustrated by a decrease in their BrdU incorporation (Figure 2d).
Recently, it has been shown in lung cancer mouse model that RUNX3 is necessary for the normal function of the p14ARF–p53 pathway, which induces apoptosis and protects cells from oncogenic stimuli such as activated Ras and MYC.21, 22 In this mouse model, KrasG12D induces RUNX3 expression causing p53 stabilization and tumor suppression, and RUNX3 inactivation by the mutation contributes to tumor development.19 To clarify if the overexpression of RUNX3 is a consequence of deregulation of the p14ARF–p53 pathway instead of RUNX3 acting as an oncogene, we sequenced the entire open reading frame of the p53 gene for mutation in five NKTL cell lines. Four of the five NKTL cell lines showed wild-type p53, indicating that the p14ARF–p53 pathway inactivation by p53 mutation is an unlikely mechanism in NKTL (Supplementary Methods and Supplementary Table S4). We also demonstrated that the p53 signaling pathway is functionally intact. NKTL cells displayed normal functional p53 drug-induced responses as evidenced by the upregulation of total and phosphorylated p53 together with its downstream targets MDM2, p21, PUMA and NOXA following drug-induced genotoxic stress (Supplementary Methods and Supplementary Figures S4a and b). Furthermore, RUNX3 expression in NKTL is in the nucleus rather than cytoplasm where the protein undergoes degradation.23 Together with our knockdown data, our results suggest that the overexpressed RUNX3 is active and functional in NKTL, thereby strengthening the postulation that RUNX3 overexpression is indeed oncogenic in NKTL.
Mechanism of RUNX3 overexpression in NKTL
In order to understand the mechanism of RUNX3 deregulation in NKTL, we looked for mutation and recurrent copy-number changes as possible mechanisms for RUNX3 upregulation. However, amplification and gains involving genomic loci containing RUNX3 have not been detected in NKTL.24, 25 Whole-exome sequencing on NKTL performed also did not detect any mutation of RUNX3.26, 27 Hence, we hypothesize that RUNX3 overexpression is transcriptionally modulated in NKTL.
MYC is involved in the transcriptional regulation of RUNX3
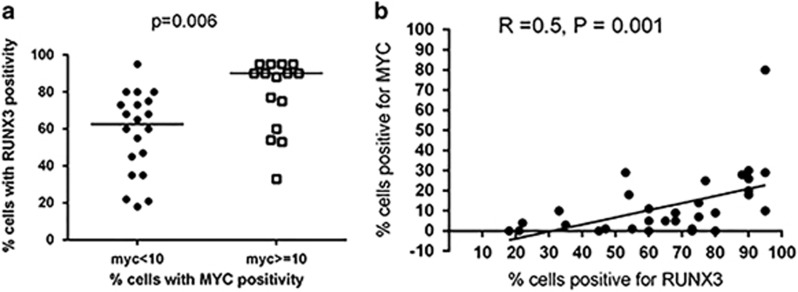
Data from murine retroviral insertional work had demonstrated that RUNX family of genes, including RUNX3, can act as a MYC-collaborating gene in the pathogenesis of thymic T-cell lymphoma in CD2-MYC transgenic mice. These proviral insertions induced overexpression of structurally intact RUNX gene products.28, 29 In addition, we have found MYC to be activated in NKTL in our previous study.17 When we compared the immunohistochemical protein expression of RUNX3 and MYC derived from our previous study, we found that cases of NKTL with high MYC protein expression also showed a significantly higher median expression of RUNX3 (P=0.006; Figure 3a), and there was a moderate correlation between RUNX3 and MYC protein expression using Spearman correlation analysis (r=0.5, P=0.001; Figure 3b and Supplementary Figure S5a). Double immunofluorescence revealed co-localization of MYC and RUNX3 protein within the same tumor nuclei, providing further support for the cooperation between RUNX3 and MYC (Supplementary Methods and Supplementary Figure S5b). Based on these findings, we investigated whether the upregulation of RUNX3 may be transcriptionally modulated by MYC.
Figure 3.
Immunohistochemical expression of MYC and RUNX3 protein in NKTL patient samples. (a) Cases with high MYC protein expression (MYC⩾10%) also showed a significantly higher median expression of RUNX3 using Student’s t-test (P=0.006). (b) MYC and RUNX3 protein expression showed moderate correlation using Spearman correlation analysis (r=0.5, P=0.001).
To this end, we examined for potential binding activity between MYC and RUNX3 using MatInspector, which is a software tool that utilizes a large library of matrix descriptions for transcription factor binding sites to locate matches in DNA sequences.30 We identified 3 potential binding sites for MYC in the RUNX3 enhancer (eR3) region (Supplementary Figure S6). In addition, using Clustal Omega software, which is a sequence alignment tool used to identify regions of similarity that may indicate functional, structural and/or evolutionary relationship between two biological sequences, we found that the MYC binding sites in the eR3 sequences of human, mouse, dog, horse and chimpanzee show a high degree of similarity (Supplementary Figure S6), suggesting that the MYC binding sites are evolutionarily conserved across different species and likely to play an in vivo regulatory role.
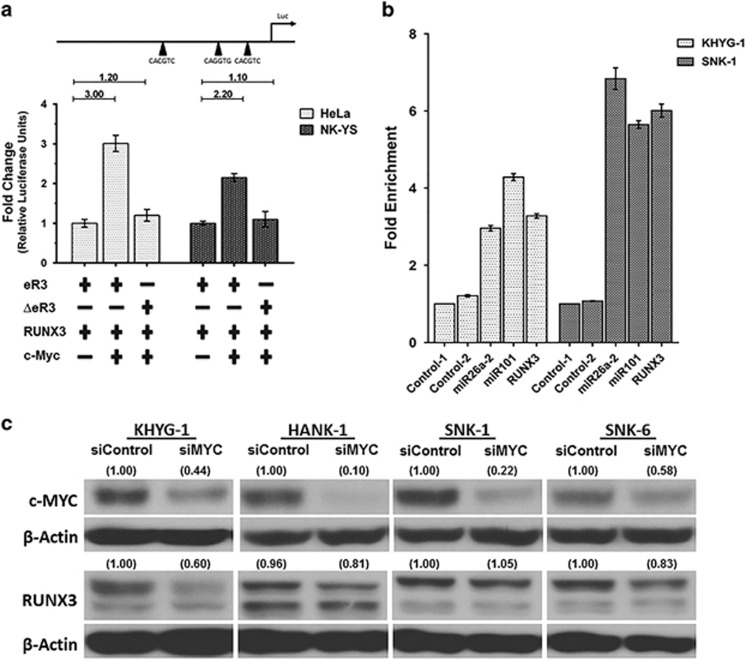
Next we evaluated whether MYC can induce the activity of RUNX3 enhancer in HeLa and NKYS cells. Co-transfection of the MYC expression vector with the RUNX3 enhancer reporter plasmid resulted in considerable activation of RUNX3 enhancer activity than the reference construct in HeLa (3-fold increase) and NK-YS (2.2-fold increase) cells (Figure 4a). An increase in RUNX3 enhancer activity was not seen when the co-transfection was performed with the RUNX3 reporter construct where all three potential MYC binding sites were mutated, indicating that MYC transactivates RUNX3 enhancer in a DNA-binding-dependent manner (see Supplementary Methods for details).
Figure 4.
RUNX3 is transcriptionally activated by MYC. (a) The enhancer activity in cells expressing the RUNX3 enhancer (eR3) and the construct with the mutated MYC binding sites (ΔeR3) co-transfected with c-MYC and RUNX3 was measured 48 h post transfection. RUNX3 was co-transfected due to the presence of RUNX binding sites in the enhancer. The co-transfected c-MYC construct showed substantially higher luciferase activity than the reference (eR3). This activity was reduced back to baseline by mutating the MYC binding sites in the enhancer region, suggesting that the transcriptional activity is due to specific binding by MYC. Black arrowheads on the luciferase constructs indicate the position of MYC binding sites. (b) Chromatin immunoprecipitation (ChIP)–qPCR for endogenous MYC binding to RUNX3 enhancer (eR3) in KHYG-1 and SNK-1 cells. Fold enrichment in the ChIP experiment represents the signal obtained after MYC immunoprecipitation followed by qPCR amplified by primer pairs that spanned the enhancer region. Fold enrichments were calculated by determining the IP efficiency (ratios of the amount of immunoprecipitated DNA to that of the input sample, i.e. percentage recovery) and normalized to the level observed at a control region, which was defined as 1. Control-1 and Control-2 denote the non-targeting negative controls while miR26a-2 and miR101 act as positive controls for the ChIP assay. There is a 3.3-fold (KHYG-1) and 6-fold (SNK-1) enrichment of genomic DNA fragments at the MYC binding sites compared to control pull-down sample, indicating binding of MYC to eR3. (c) Protein expressions of MYC and RUNX3 after MYC knockdown. NKTL cells were treated for 72 h and cell lysates were prepared for immunoblots for the MYC protein levels. RUNX3 expression profiles were analyzed post 96 h after MYC knockdown. Silencing of MYC sequentially deregulated RUNX3 expression in NKTL cells.
We further examined whether MYC binds to the RUNX3 enhancer (eR3) using ChIP-qPCR assays in KHYG-1 and SNK-1 cells. Using an antibody directed against MYC, the immunoprecipitated chromatin was amplified using primers flanking the MYC binding sites in the RUNX3 enhancer. We observed a specific enrichment of genomic DNA fragments at the MYC binding sites (3.3-fold: KHYG-1 and 6-fold: SNK-1) in reference to control pull-down sample. (Figure 4b and Supplementary Methods for details). Taken together, this reaffirms the MYC-RUNX3 interaction and the involvement of MYC in the regulation of RUNX3.
Further validation that MYC is an upstream regulator of RUNX3 was achieved by selective knockdown of MYC and RUNX3 using siRNAs specific to MYC and RUNX3. Suppression of c-MYC resulted in reduction of RUNX3 mRNA in NKTL cell lines. (Supplementary Figure S7a). There was a corresponding reduction in RUNX3 protein expression following knockdown of MYC in KHYG-1, HANK-1 and SNK-6 (Figure 4c). However, when we knockdown RUNX3, MYC mRNA levels remained unaffected (Supplementary Figure S7b). Collectively, these results indicate that MYC is upstream of RUNX3 and positively regulates RUNX3 transcription. However, we noticed that a large proportion of cases show marked RUNX3 overexpression without a corresponding upregulation of MYC (Figure 3b), suggesting that MYC may not be a direct upstream regulator of RUNX3 in NKTL and other factors may be involved in the overexpression of RUNX3.
Therapeutic implications of MYC Inhibition in NKTL
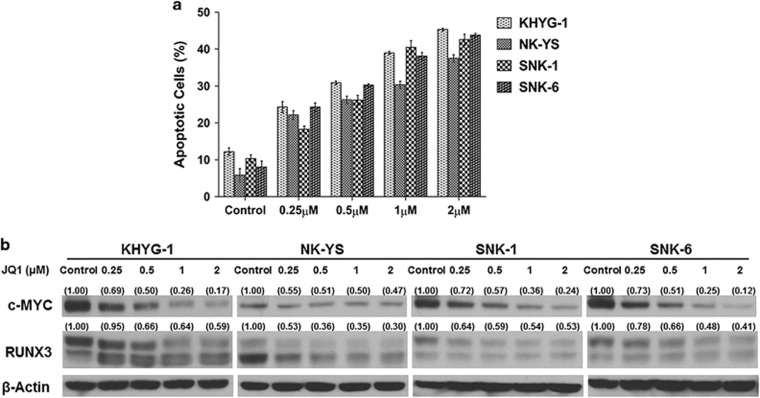
Since MYC is activated in NKTL and downregulation of MYC results in RUNX3 suppression with consequent increase in apoptosis and reduction in proliferation, we explored the use of JQ1, a novel small-molecule inhibitor capable of suppressing MYC transcription by binding selectively to the conserved bromodomain and extra-terminal domain (BET) protein family and selectively targeting malignant cells by disrupting chromatin-mediated signal transduction and reducing transcription at oncogene loci, most notably MYC.31, 32 Treatment with JQ1 resulted in a robust downregulation of c-MYC mRNA and protein with accompanying reduction of RUNX3 mRNA and protein in KHYG-1, NK-YS, SNK-1 and SNK-6 (Figure 5b and Supplementary Figure S8a). JQ1 treatment suppressed NKTL cell viability with increased apoptosis in a dose-dependent manner in all 4 NKTL cell lines tested (KHYG-1, NK-YS, SNK-1 and SNK-6; Figure 5a). All cell lines were sensitive to the treatment as indicated by the inhibitory concentration (IC50) values below 10 μM (Supplementary Figure S8b). Based on the IC50 values, NKTL cells were treated with varying concentrations with a 2-fold difference between each dose. In line with the cell viability results, the treated cells also showed a dose-dependent induction of apoptosis, opening the possibility of MYC inhibition as a potential therapeutic strategy in NKTL but requires further evaluation.
Figure 5.
Effects of MYC inhibition in NKTL cells using BET bromodomain inhibitor (JQ1). RUNX3 downregulation was observed in a dose-dependent manner upon treatment with a small-molecule inhibitor (JQ1) that downregulates MYC. Downregulation of MYC and RUNX3 by JQ1 lead to apoptosis in NKTL cells. (a) Protein expression of MYC and RUNX3 in JQ1 treated NKTL cells at the respective concentrations. Increasing doses of JQ1 effectively reduced MYC and RUNX3 expression levels in NKTL cells. (b) JQ1-treated NKTL cells were stained with Annexin-V and propidium iodide and analyzed for the induction of apoptosis by flow cytometry. The effect of JQ1 treatment on the NKTL cells was a distinct increase in apoptosis in a dose-dependent manner in all NKTL cells.
Growth inhibition upon depletion of MYC by JQ1 treatment can be rescued by exogenous expression of MYC
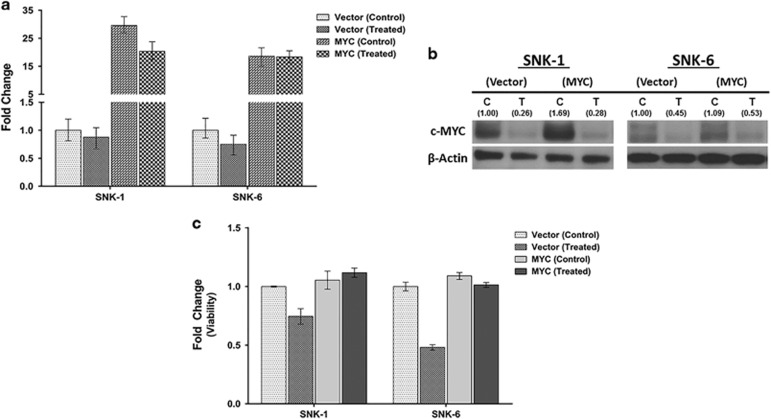
We next explored whether the depletion of MYC is responsible for the apoptosis induced by JQ1 treatment in malignant NKTL cells. To achieve this, MYC was transfected into SNK-1 and SNK-6 using an expression vector to bring about overexpression of MYC in the cell lines. This was followed by JQ1 treatment to assess if exogenous expression of MYC can prevent the growth reduction mediated by JQ1 treatment. Indeed, exogenous expression of MYC mRNA and protein (Figure 6a and b, left) in both control and JQ1-treated SNK-1 cell line was able to abort the decrease in cell viability and rescue the tumor cells from cell death compared to cells transfected with empty vector, with and without JQ-1 treatment (Figure 6c, left). Similar results were observed in another NKTL cell line, SNK-6 (Figure 6a–c, right). The rescue experiment results indicate that the JQ1-induced cell death in NKTL cells is in large part due to the decrease in MYC levels rather than an off-target effect. Unlike MYC mRNA levels, which was not inhibited by JQ1 treatment following ectopic MYC expression (Figure 6a), we noticed only minimal rescue effect on MYC protein levels (Figure 6b). It is uncertain if this is due to the short half-life of MYC protein and variability in MYC degradation.33
Figure 6.
Effects of MYC ectopic expression in NKTL cells using BET bromodomain inhibitor (JQ1). Overexpression of c-MYC in NKTL cells (SNK-1 and SNK-6) by transfection rescues the cell viability observed with JQ1 treatment, arguing that MYC downregulation by JQ1 contributes functionally to cell growth in NKTL. (a) mRNA were prepared after treating with JQ1 (0.25 μm, 48 h) or DMSO control. MYC expression was rescued (MYC treated) in NKTL cells. (b) Protein profiles from cell lysates of empty and MYC overexpression vector after treating with JQ1 (0.25 μm, 48 h). Immunoblotting showed MYC expression was upregulated in part upon MYC overexpression (MYC-T) compared to empty vector (Vector-T). (c) Cell viability analysis of empty and MYC-overexpressing NKTL cells treated with JQ1 (0.25 μm, 48 h). Cell viability increased in MYC transfected cells (MYC-treated) compared to Vector (treated).
Discussion
It appears that both loss and gain of RUNX3 function can contribute to cancer development. The dual role of RUNX3 as a tumor suppressor and oncogene remains unclear but appears to be tissue dependent or organ specific rather than the histologic type of cancer34 and indicates a complex mode of molecular activation and partnering.
In the recent years, there is increasing recognition that RUNX3 can also act as an oncogene in skin cancers including basal cell carcinomas (BCC), ovarian cancers, squamous cell carcinomas (SCC) of the head and neck, and Ewing sarcoma.9, 10, 11, 12, 34, 35, 36 Moreover, high level expression of RUNX3 has been correlated with poor prognosis in a subset of acute myeloid leukemia carrying FLT3 mutations.37 In BCC and SCC of head and neck, the overexpressed RUNX3 proteins are full-length and intact without mutation and fully functional.9, 34 In gastric and other tumors, RUNX3 can be overexpressed due to protein mislocalization,5, 38 which may be caused by MDM2-mediated ubiquitination and degradation of RUNX3 via p14ARF-MDM2 surveillance pathway.21 However, when RUNX3 transcription factor is mislocalized to the cytoplasm, it is in an inactive state and unlikely to function as an oncogene.5, 38 Importantly, our data demonstrate that RUNX3 overexpression in NKTL is functionally active and shows oncogenic phenotype. First, RUNX3 is overexpressed in the tumor nuclei of NKTL, and not mislocalized to the cytoplasm where it may be inactivated. Second, knockdown of RUNX3 resulted in an increase in apoptosis and reduction in proliferation. Third, the lack of p53 mutation and an intact p53 pathway in the NKTL cell lines excludes the possibility that the overexpression of RUNX3 is as a consequence of deregulation of RUNX3-p14ARF–p53 pathway.39 Thus, our findings have uncovered a crucial and novel role of RUNX3 in promoting viability and proliferation. We have also demonstrated the upregulation of RUNX3 in the majority of high-grade B-cell lymphomas and peripheral T-cell lymphomas (Supplementary Table S5 and Supplementary Figure S9), suggesting that the oncogenic role of RUNX3 may also be operational in other lymphoid malignancies and warrants further investigation.
While there has been a profusion of publications describing the role of RUNX3 as a tumor suppressor in a large variety of human cancers, the role of RUNX3 as an oncogene and the mechanisms of RUNX3 upregulation are poorly understood.9, 10, 11, 12, 34, 36 Notably, we have demonstrated in this study that the oncogenic role of RUNX3 is transcriptionally regulated by MYC in NKTL. This is in line with data implicating the RUNX genes as MYC collaborating genes whereby RUNX targeting by retroviruses in T- or B-lymphoid cells have been identified in transgenic models harboring MYC.28, 29, 40 Furthermore, reciprocal experiment in which CD2-RUNX2 mice were infected with murine leukemia virus revealed a major bias towards targeting of MYC family genes in the resultant T-cell lymphomas, again suggesting a preferential relationship in oncogenic collaboration between RUNX and MYC.41 Although the precise mechanism of this collaboration remains unclear, it has been postulated that the resultant ectopic expression of RUNX isoforms contribute to lymphomagenesis by causing upregulation of target genes, such as T-cell receptor (TCR) complex genes and CD3ε.29 Activation of TCR/CD3 expression is a crucial event in T-cell development42 and transcriptional stimulation of the component genes may promote cell transit through growth or differentiation checkpoints or enabling survival of cells carrying TCRs with aberrant affinity for major histocompatibility complex.29
MYC can be upregulated in cancers due to multiple mechanisms, including chromosomal translocation, gene amplification, mutation of upstream signaling pathways, and mutation that enhance the stability of the protein. Although MYC is often overexpressed in NKTL, neither amplification nor mutation of MYC was found in NKTL.43 MYC is a transcriptional target of the EBV proteins EBNA244 and LMP1.45 Since NKTL shows EBV latency pattern II characterized by absence of EBNA-2 and presence of LMP-1,46 it is interesting to postulate that the activation of MYC in NKTL could be through the activity of an EBV-related protein, such as LMP1.17 This would be consistent with the importance of EBV infection in the pathogenesis of NKTL. Another mechanism of MYC overexpression in NKTL may be via the phosphorylation activation of STAT3 since MYC is a known target of STAT3 transcriptional activity.24, 47, 48
Besides regulating the transcriptional activation of RUNX3, data from our previous study demonstrated that MYC also upregulates EZH2 by inducing repression of its regulatory microRNAs in NKTL.49 These results are consistent with MYC being a master regulator of diverse cellular functions and, hence, the proposition that MYC functions as a universal amplifier of gene expression rather than an on-off specifier of distinct transcriptional program(s).50 Nevertheless, this proposal remains a subject of contention as regulation of distinct subsets of genes by MYC have also been demonstrated.51, 52 In this study, we have demonstrated that MYC is involved in the transcriptional regulation of RUNX3 in NKTL. However, whether this is a result of MYC directly and specifically upregulating RUNX3 or a phenomenon of indirect amplification of global transcripts via downstream effects of MYC-induced subsets of genes require further characterization.
MYC is long considered a compelling therapeutic target because of its diverse role in human malignancies. However, pharmacologic inhibition of MYC function has proven challenging because of both the diverse mechanisms driving its aberrant expression and the challenge of disrupting protein–DNA interactions. Since MYC is activated in NKTL from our previous study,17 we explored a novel compound, JQ1, which is able to reduce MYC transcription and reported to be therapeutically effective in pre-clinical animal models.31, 32, 53 BET bromodomain inhibition by JQ1 confers a selective repression of transcriptional networks induced by c-MYC and the anti-tumor effect of JQ1 is not accompanied by a nonspecific, toxic effect on all hematopoietic cells.32 Our findings with JQ1 treatment demonstrate that pharmacologic inhibition of MYC is achievable in NKTL cell lines through targeting BET bromodomains, resulting in downregulation of RUNX3, induction of apoptosis and reduced cell viability. This opens the possibility of targeting MYC as a potential therapeutic strategy in NKTL. Furthermore, the development of novel therapeutic strategies geared towards the modulation of MYC expression, either alone or in combination with other therapies, may be of potential therapeutic significance for aggressive and difficult to treat tumors, such as NKTL.
Acknowledgments
S-BN was supported by the Singapore Ministry of Health’s National Medical Research Council Transition Award (NMRC/TA/0020/2013). This work is supported in part by Singapore Ministry of Education Academic Research Fund Tier 1 (WBS No: R-179-000-046-112). W-JC was supported by the Singapore Ministry of Health’s National Medical Research Council Clinician Scientist Investigator Award. Ethics approval was obtained from IRB, National University of Singapore, ID: 10‐107.
Author contributions
W-JC conceived and designed study, and analyzed the data; S-BN conceived and designed the study, analyzed the data and wrote the paper. VS performed experiments and wrote the paper; GSSN, MO, JY, DC-CV and YI provided vital reagents and interpreted findings; T-HC performed bioinformatics analysis; MFH, MS-T, S-NC and SF constructed TMA, performed IHC and DIF; NS maintained and contributed cell lines.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
The authors declare no conflict of interest.
Supplementary Material
References
- Ito Y. Oncogenic potential of the RUNX gene family: 'Overview'. Oncogene 2004; 23: 4198–4208. [DOI] [PubMed] [Google Scholar]
- Ito Y. RUNX genes in development and cancer: regulation of viral gene expression and the discovery of RUNX family genes. Adv Cancer Res 2008; 99: 33–76. [DOI] [PubMed] [Google Scholar]
- Kundu M, Compton S, Garrett-Beal L, Stacy T, Starost MF, Eckhaus M et al. Runx1 deficiency predisposes mice to T-lymphoblastic lymphoma. Blood 2005; 106: 3621–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong DT, Lim J, Goh X, Pratap J, Pereira BP, Kwok HS et al. Cancer-related ectopic expression of the bone-related transcription factor RUNX2 in non-osseous metastatic tumor cells is linked to cell proliferation and motility. Breast Cancer Res 2010; 12: R89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Liu Q, Salto-Tellez M, Yano T, Tada K, Ida H et al. RUNX3, a novel tumor suppressor, is frequently inactivated in gastric cancer by protein mislocalization. Cancer Res 2005; 65: 7743–7750. [DOI] [PubMed] [Google Scholar]
- Lee CW, Ito K, Ito Y. Role of RUNX3 in bone morphogenetic protein signaling in colorectal cancer. Cancer Res 2010; 70: 4243–4252. [DOI] [PubMed] [Google Scholar]
- Fujii S, Ito K, Ito Y, Ochiai A. Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by increasing histone H3 methylation. J Biol Chem 2008; 283: 17324–17332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron ER, Neil JC. The Runx genes: lineage-specific oncogenes and tumor suppressors. Oncogene 2004; 23: 4308–4314. [DOI] [PubMed] [Google Scholar]
- Salto-Tellez M, Peh BK, Ito K, Tan SH, Chong PY, Han HC et al. RUNX3 protein is overexpressed in human basal cell carcinomas. Oncogene 2006; 25: 7646–7649. [DOI] [PubMed] [Google Scholar]
- Tsunematsu T, Kudo Y, Iizuka S, Ogawa I, Fujita T, Kurihara H et al. RUNX3 has an oncogenic role in head and neck cancer. PLoS ONE 2009; 4: e5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CWL, Chuang LSH, Kimura S, Lai SK, Ong CW, Yan B et al. RUNX3 functions as an oncogene in ovarian cancer. Gynecol Oncol 2011; 122: 410–417. [DOI] [PubMed] [Google Scholar]
- Bledsoe KL, McGee-Lawrence ME, Camilleri ET, Wang X, Riester SM, van Wijnen AJ et al. RUNX3 facilitates growth of Ewing sarcoma cells. J Cell Physiol 2014; 229: 2049–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JKC, Quintanilla-Martinez L, Ferry JA, Peh SC. Extranodal NK/T-cell lymphoma, nasal type. In: Swerdlow SH et alWHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th edn. IRAC Press: Lyon, France, 2008, pp 285–288. [Google Scholar]
- Ohno S, Sato T, Kohu K, Takeda K, Okumura K, Satake M et al. Runx proteins are involved in regulation of CD122, Ly49 family and IFN-gamma expression during NK cell differentiation. Int Immunol 2008; 20: 71–79. [DOI] [PubMed] [Google Scholar]
- Lotem J, Levanon D, Negreanu V, Leshkowitz D, Friedlander G, Groner Y. Runx3-mediated transcriptional program in cytotoxic lymphocytes. PLoS ONE 2013; 8: e80467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG et al. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med 2009; 206: 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Selvarajan V, Huang G, Zhou J, Feldman AL, Law M et al. Activated oncogenic pathways and therapeutic targets in extranodal nasal-type NK/T cell lymphoma revealed by gene expression profiling. J Pathol 2011; 223: 496–510. [DOI] [PubMed] [Google Scholar]
- Ng SB, Yan J, Huang G, Selvarajan V, Tay JL, Lin B et al. Dysregulated microRNAs affect pathways and targets of biologic relevance in nasal-type natural killer/T-cell lymphoma. Blood 2011; 118: 4919–4929. [DOI] [PubMed] [Google Scholar]
- Ito Y, Bae SC, Chuang LS. The RUNX family: developmental regulators in cancer. Nat Rev Cancer 2015; 15: 81–95. [DOI] [PubMed] [Google Scholar]
- Puig-Kröger A, Aguilera-Montilla N, Martínez-Nuñez R, Domínguez-Soto A, Sánchez-Cabo F, Martín-Gayo E et al. The novel RUNX3/p33 isoform is induced upon monocyte-derived dendritic cell maturation and downregulates IL-8 expression. Immunobiology 2010; 215: 812–820. [DOI] [PubMed] [Google Scholar]
- Chi XZ, Kim J, Lee YH, Lee JW, Lee KS, Wee H et al. Runt-related transcription factor RUNX3 is a target of MDM2-mediated ubiquitination. Cancer Res 2009; 69: 8111–8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS. Runx3 inactivation is a crucial early event in the development of lung adenocarcinoma. Cancer cell 2013; 24: 603–616. [DOI] [PubMed] [Google Scholar]
- Yamada C, Ozaki T, Ando K, Suenaga Y, Inoue K-i, Ito Y et al. RUNX3 modulates DNA damage-mediated phosphorylation of tumor suppressor p53 at Ser-15 and acts as a co-activator for p53. J Biol Chem 2010; 285: 16693–16703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, de Reynies A, de Leval L, Ghazi B, Martin-Garcia N, Travert M et al. Gene expression profiling identifies emerging oncogenic pathways operating in extranodal NK/T-cell lymphoma, nasal type. Blood 2010; 115: 1226–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal J, Kucuk C, Deleeuw RJ, Srivastava G, Tam W, Geng H et al. Genomic analyses reveal global functional alterations that promote tumor growth and novel tumor suppressor genes in natural killer-cell malignancies. Leukemia 2009; 23: 1139–1151. [DOI] [PubMed] [Google Scholar]
- Jiang L, Gu ZH, Yan ZX, Zhao X, Xie YY, Zhang ZG et al. Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat Genet 2015; 47: 1061–1066. [DOI] [PubMed] [Google Scholar]
- Koo GC, Tan SY, Tang T, Poon SL, Allen GE, Tan L et al. Janus kinase 3-activating mutations identified in natural killer/T-cell lymphoma. Cancer Discov 2012; 2: 591–597. [DOI] [PubMed] [Google Scholar]
- Stewart M, MacKay N, Cameron ER, Neil JC. The common retroviral insertion locus Dsi1 maps 30 kilobases upstream of the P1 promoter of the murine Runx3/Cbfa3/Aml2 gene. J Virol 2002; 76: 4364–4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. Proviral insertions induce the expression of bone-specific isoforms of PEBP2[alpha]A (CBFA1): evidence for a new myc collaborating oncogene. Proc Natl Acad Sci USA 1997; 94: 8646–8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 2005; 21: 2933–2942. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O et al. Selective inhibition of BET bromodomains. Nature 2010; 468: 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011; 146: 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory MA, Hann SR. c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt's lymphoma cells. Mol Cell Biol 2000; 20: 2423–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo Y, Tsunematsu T, Takata T. Oncogenic role of RUNX3 in head and neck cancer. J Cell Biochem 2011; 112: 387–393. [DOI] [PubMed] [Google Scholar]
- Nevadunsky NS, Barbieri JS, Kwong J, Merritt MA, Welch WR, Berkowitz RS et al. RUNX3 protein is overexpressed in human epithelial ovarian cancer. Gynecol Oncol 2009; 112: 325–330. [DOI] [PubMed] [Google Scholar]
- Lee JH, Pyon JK, Kim DW, Lee SH, Nam HS, Kang SG et al. Expression of RUNX3 in skin cancers. Clin Exp Dermatol 2011; 36: 769–774. [DOI] [PubMed] [Google Scholar]
- Lacayo NJ, Meshinchi S, Kinnunen P, Yu R, Wang Y, Stuber CM et al. Gene expression profiles at diagnosis in de novo childhood AML patients identify FLT3 mutations with good clinical outcomes. Blood 2004; 104: 2646–2654. [DOI] [PubMed] [Google Scholar]
- Lau QC, Raja E, Salto-Tellez M, Liu Q, Ito K, Inoue M et al. RUNX3 is frequently inactivated by dual mechanisms of protein mislocalization and promoter hypermethylation in breast cancer. Cancer Res 2006; 66: 6512–6520. [DOI] [PubMed] [Google Scholar]
- Lee YS, Lee JW, Jang JW, Chi XZ, Kim JH, Li YH et al. Runx3 inactivation is a crucial early event in the development of lung adenocarcinoma. Cancer Cell 2013; 24: 603–616. [DOI] [PubMed] [Google Scholar]
- Blyth K, Vaillant F, Hanlon L, Mackay N, Bell M, Jenkins A et al. Runx2 and MYC collaborate in lymphoma development by suppressing apoptotic and growth arrest pathways in vivo. Cancer Res 2006; 66: 2195–2201. [DOI] [PubMed] [Google Scholar]
- Blyth K, Terry A, Mackay N, Vaillant F, Bell M, Cameron ER et al. Runx2: a novel oncogenic effector revealed by in vivo complementation and retroviral tagging. Oncogene 2001; 20: 295–302. [DOI] [PubMed] [Google Scholar]
- Boyd RL, Hugo P. Towards an integrated view of thymopoiesis. Immunol Today 1991; 12: 71–79. [DOI] [PubMed] [Google Scholar]
- Sugimoto KJ, Kawamata N, Sakajiri S, Oshimi K. Molecular analysis of oncogenes, ras family genes (N-ras, K-ras, H-ras), myc family genes (c-myc, N-myc) and mdm2 in natural killer cell neoplasms. Jpn J Cancer Res 2002; 93: 1270–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C, Laux G, Eick D, Jochner N, Bornkamm GW, Kempkes B. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J Virol 1999; 73: 4481–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirmeier U, Hoffmann R, Kilger E, Schultheiss U, Briseno C, Gires O et al. Latent membrane protein 1 of Epstein-Barr virus coordinately regulates proliferation with control of apoptosis. Oncogene 2005; 24: 1711–1717. [DOI] [PubMed] [Google Scholar]
- Ng SB, Khoury JD. Epstein-Barr virus in lymphoproliferative processes: an update for the diagnostic pathologist. Adv Anat Pathol 2009; 16: 40–55. [DOI] [PubMed] [Google Scholar]
- Coppo P, Gouilleux-Gruart V, Huang Y, Bouhlal H, Bouamar H, Bouchet S et al. STAT3 transcription factor is constitutively activated and is oncogenic in nasal-type NK/T-cell lymphoma. Leukemia 2009; 23: 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol 2007; 7: 41–51. [DOI] [PubMed] [Google Scholar]
- Yan J, Ng SB, Tay JL, Lin B, Koh TL, Tan J et al. EZH2 overexpression in natural killer/T-cell lymphoma confers growth advantage independently of histone methyltransferase activity. Blood 2013; 121: 4512–4520. [DOI] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 2012; 151: 68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 2014; 511: 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz S, Lorenzin F, Morton J, Wiese KE, von Eyss B, Herold S et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature 2014; 511: 483–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA 2011; 108: 16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.