Abstract
Heme (iron protoporphyrin IX) is an essential protein prosthetic group and signaling molecule required for most life on Earth. All heme-dependent processes require the dynamic and rapid mobilization of heme from sites of synthesis or uptake to hemoproteins present in virtually every subcellular compartment. The cytotoxicity and hydrophobicity of heme necessitate that heme mobilization be carefully controlled to mitigate the deleterious effects of this essential toxin. Indeed, a number of disorders, including certain cancers, cardiovascular diseases, and aging and age-related neurodegenerative diseases, are tied to defects in heme homeostasis. However, the molecules and mechanisms that mediate heme transport and trafficking, and the dynamics of these processes, are poorly understood. This is in large part due to the lack of physical tools for probing cellular heme. Herein, we discuss the recent development of fluorescent probes that can monitor and image kinetically labile heme with respect to its mobilization and role in signaling. In particular, we will highlight how heme gazing with these tools can uncover new heme trafficking factors upon being integrated with genetic screens and illuminate the concentration, subcellular distribution, and dynamics of labile heme in various physiological contexts. Altogether, the monitoring of labile heme, along with recent biochemical and cell biological studies demonstrating the reversible regulation of certain cellular processes by heme, is challenging us to reconceptualize heme from being a static cofactor buried in protein active sites to a dynamic and mobile signaling molecule.
Graphical Abstract
Heme (iron protoporphyrin IX) is an essential cofactor and potential toxin that is required by virtually every organism across all of the kingdoms of life. As a protein prosthetic group, heme performs a diverse array of critical life-enabling functions that span chemical catalysis, electron transfer, and gas binding and transport.1 Rather ironically, heme is also cytotoxic because of its hydrophobicity and pro-oxidant redox activity, challenging cells to tightly regulate its concentration and availability.2–4 This has contributed to the long-held view that there is no “free” or “labile” heme and that it is tightly bound and buried in the active sites of kinetically inert hemoproteins or degraded if not utilized by heme enzymes.5 However, this notion is incompatible with the fact that heme must be rapidly mobilized to cognate hemoproteins residing in virtually every subcellular compartment and emerging evidence indicating heme can act as a reversible and dynamic signaling molecule.1, 3, 6–13 How then do cells mobilize heme for insertion into hemoproteins and signaling while taming its inherent hydrophobicity and toxicity? In this review, we describe new fluorescent probes for heme and how they can be used to reveal the molecules and mechanisms that mediate the transport and trafficking of heme and their dynamics.14, 15 We place these latest fluorescence-based approaches to characterizing heme homeostasis in the context of other modalities for monitoring heme and more traditional biochemical and genetic approaches. While our focus will be on eukaryotic heme homeostasis, the principles and approaches discussed herein are applicable to prokaryotic systems, as well. As a number of human disorders are associated with defects in heme homeostasis, including certain cancers,8 cardiovascular disease,16 and aging and age-related neurodegenerative diseases,17–19 a detailed knowledge of the thermodynamics and kinetics governing heme mobilization and availability will undoubtedly lead to new heme-based therapeutics and diagnostics.
The concentration and bioavailability of cellular heme pools are the primary factors that contribute to the safe assimilation of heme into physiological processes. The concentration of heme is dictated by its biosynthesis and degradation, which are both very well understood.1 With few exceptions, all eukaryotes synthesize heme through a highly conserved eight-step pathway that is partitioned between the mitochondria and the cytosol; the first and the last three steps occur in the mitochondria, and the other reactions occur in the cytosol. Heme synthesis begins with the condensation of glycine with succinylcoenzyme A to form 5-aminolevulinic acid (5-ALA), the first committed precursor, and ends with the insertion of iron into protophorphyrin IX to form heme on the matrix side of the mitochondrial inner membrane. Heme degradation is performed by heme oxygenase, a resident of the endoplasmic reticulum membrane that catalyzes the oxidative cleavage of the heme ring at the α-methylene bridge to form biliverdin, carbon monoxide (CO), and ferrous iron (Fe2+). All of the genes involved in heme synthesis and degradation have been cloned, and there are atomic-resolution structures and mechanistic insight into all of the proteins involved.1
In striking contrast to heme synthesis and degradation, there is comparatively little understanding of the molecules and mechanisms that regulate heme bioavailability. Once heme is synthesized on the matrix side of the mitochondrial inner membrane, it is not known what factors regulate the movement of heme to hemoproteins present in the matrix lumen, inner membrane, or intermembrane space, or the transit of heme out of the mitochondria to the cytosol and beyond.20 It is also unclear what binds and buffers heme in a manner that would mitigate its aggregation and deleterious redox activity. As heme has properties of both a metal and a lipid, its transport and trafficking likely mirror those of transition metals like iron or copper and mitochondrially derived lipids like cardiolipin (CL) and phosphatidylethanolamine (PE).20 These properties of heme invoke the existence of heme transporters, chaperones, and carrier proteins, as well as mechanisms involving the mobilization of heme through membrane contact points between different organelles and vesicular transport.20 The recent development of fluorescent probes for imaging and monitoring bioavailable heme has facilitated the discovery of key molecules and mechanisms that mediate heme mobilization and its dynamics, especially upon being integrated with genetic and biochemical approaches.
BIOLOGICAL HEME MONITORING AND IMAGING
Total cellular heme can be conceptualized as a sum of the contributions from kinetically inert and labile heme pools. Kinetically inert heme, which constitutes most cellular heme, is unavailable for new heme-dependent functions and corresponds to heme buried in the active sites of high-affinity hemoproteins like globins and cytochromes. 14, 15, 20, 21 Kinetically labile heme (LH), on the other hand, is available for new heme-dependent functions and corresponds to heme that can readily exchange between biomolecules on physiologically relevant time scales that support heme-dependent functions. In other words, the labile heme pool defines bioavailable heme. While LH is critical for understanding heme transport, trafficking, and signaling, its nature—concentration, speciation, oxidation state, and localization—and dynamics are not well understood. This is in large part due to the lack of tools available to monitor and image LH.
There are a number of approaches that have been developed to probe cellular heme, but until recently, none to monitor LH or bioavailable heme relevant to its trafficking and role in signaling in intact living cells. The most common analytical methods for monitoring heme involve disruption of cells or tissues, acid, and/or heat denaturation to remove heme from proteinaceous material, heme extraction using organic solvents (e.g., acetone and butanol), and finally quantification by (a) UV/visible (UV/vis) spectroscopy of porphyrin π—π* transitions, (b) fluorescence spectroscopy of the demetalated porphyrin, or (c) reversed-phase high-pressure liquid chromatography (HPLC) coupled to UV/vis or fluorescence detectors. 22, 23 However, these methods probe only total heme and do not differentiate between kinetically inert and labile heme.
To exact information about bioavailable heme, the activities of various heme-dependent enzymes in cell or tissue extracts are often profiled, including cytochrome P450 enzymes (ER), tryptophan 2,3-dioxygenase (cytosol), and peroxidases that can be genetically encoded and targeted to different subcellular compartments.21, 24–26 Collectively, while these methods have served as powerful tools for identifying changes in steady-state heme concentration and bioavailability in a number of contexts, including between healthy and diseased cells, they suffer from a number of drawbacks that arise from the fact that cells or tissues are homogenized and time-intensive heme analyses and enzyme assays are employed. These traditional methods (a) cannot probe subcellular heterogeneity in heme pools, (b) may artifactually alter bioavailable heme pools due to the repartitioning of heme upon cell lysis, (c) may be unsuitable for probing the spatial and temporal dynamics of cellular heme due to the challenges associated with quickly purifying organelles and/or isolating them in sufficient quantities, and (d) may be blind to cell-to-cell variation in heme pools because large populations of cells are required for these traditional methods of heme analysis.
More recently, innovative nondisruptive methods for imaging heme have been developed that circumvent all the drawbacks associated with the traditional methods of heme quantification just described. These new approaches include Raman27 and photothermal lens microscopies,28 as well as the development of fluorescent sensors for heme.14, 15 While Raman and photothermal lens microscopies are powerful label-free methods for probing heme in cells, they report on only the most abundant high-affinity hemoproteins like globins and cytochromes because of the poorly defined spectral characteristics of labile heme and insufficient sensitivity to resolve the low concentration of this heme pool.27, 28 On the other hand, recently developed ratiometric fluorescent reporters for heme have offered unparalleled insight into subcellular LH pools relevant for heme trafficking and signaling.
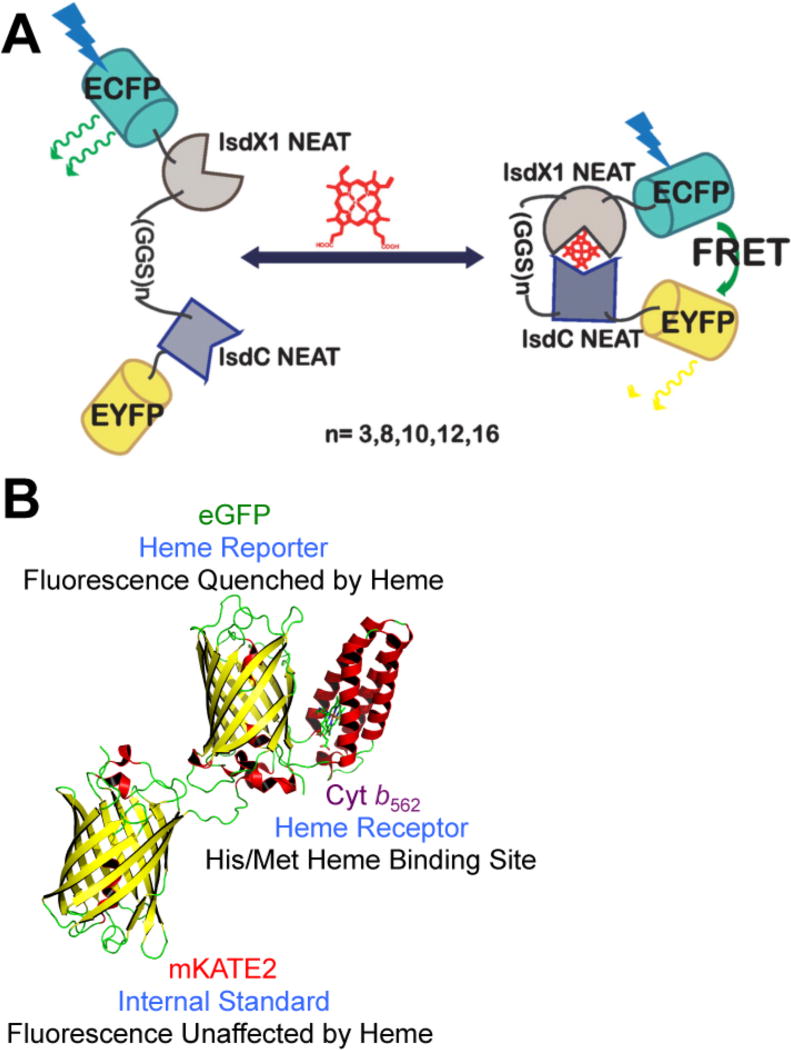
Inspired by a large number of Forster resonance energy transfer (FRET)-based probes for a wide range of biological analytes, including transition metals like copper29 and zinc,30–32 we and others have developed FRET sensors for heme built around hemoprotein scaffolds found in Nature. These new heme sensors allow for the dynamic ratiometric imaging of heme in intact living cells and subcellular compartments, circumventing the need for cell disruption and time-intensive enzyme assays, and have collectively illuminated fundamental aspects of heme trafficking, signaling, and cellular dynamics. Song et al. reported the first FRET sensor for cellular heme imaging. The sensor, CISDY, consists of a heme sensory module containing NEAT domains of two heme transfer chaperones, IsdX1 and IsdC, tethered by a linker, and flanked by ECFP and EYFP at the N- and C-termini, respectively (Figure 1A). Heme binding induces the heterodimerization of IsdX1 and IsdC, resulting in an increase in the FRET efficiency of the ECFP–EYFP pair. Expression of CISDY across various human cell lines and in locations spanning the cytosol nucleus, mitochondria, and ER not only revealed labile heme levels to be ~20 nM across these compartments but also imaged the spatiotemporal dynamics of incorporation of heme into heme deficient cells.
Figure 1.
Genetically encoded fluorescent heme sensors (A) CISDY and (B) HS1. Panel A was adapted with permission from ref 14. Copyright 2015 American Chemical Society. The molecular model of HS1 was generated using PyMol and is derived from the X-ray structures of mKATE [Protein Data Bank (PDB) entry 3BXB] and CG6 (PDB entry 3U8P).
Contemporaneously, Hanna et al. reported the FRET-based heme sensor 1 (HS1), a fusion of cytochrome b562 (Cyt b562), enhanced green fluorescent protein (EGFP), and a red fluorescent protein, Katushka 2 (mKATE2)14 (Figure 1B). Binding of heme to Cyt b562, a four-helix bundle His/Met heme binding protein from Escherichia coli,33 results in >90% quenching of EGFP fluorescence via FRET, whereas fluorescence emission of mKATE2 is relatively insensitive to heme binding because it is a poor FRET donor for heme.14 Thus, the ratio of heme-sensitive EGFP fluorescence to hemeinsensitive mKATE2 fluorescence provides a readout of cellular heme that is independent of sensor concentration. The development and application of HS1 to probe heme homeostasis not only serve to highlight key principles in sensor development but also have uncovered fundamental insight into heme trafficking, dynamics, and signaling.
HEME SENSOR DEVELOPMENT: SUCCESSES AND CHALLENGES
There are a number of key considerations when conceptualizing fluorescent probes for heme, including heme selectivity, appropriately tuned heme dissociation constants, and minimizing perturbations to heme homeostasis upon sensor expression. The development of heme probes CISDY and HS1 highlights both successes and challenges in realizing these considerations. In terms of heme selectivity over other potentially interfering metabolites, including heme biosynthetic intermediates, e.g., protoporphyrin IX, heme degradation products, e.g., bilirubin or biliverdin, and other metals, e.g., iron, copper, or zinc, both CISDY and HS1 are selective for heme over protoporphyrin IX, bilirubin, biliverdin, and a range of first row transition metals, including manganese, iron, copper, and zinc. The success in achieving heme selectivity is likely due to the fact that the heme sensory modules of CISDY and HS1 are natural hemoproteins that evolved to utilize heme over other cellular metabolites.
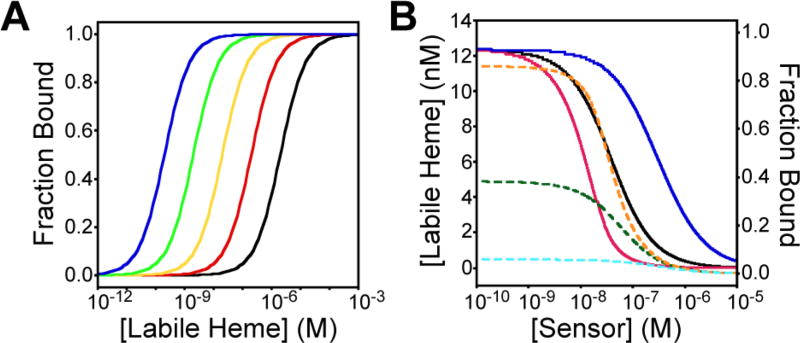
In terms of heme binding affinities, the heme dissociation constant, KD, of the probe must be tuned such that it is not over- or undersaturated with heme for optimal heme imaging. On the basis of the single-site binding model depicted in eqs 1–3, the relationship among labile heme concentration, heme sensor KD, and the fractional saturation of the sensor is depicted in Figure 2A. It can be seen that the KD of the heme sensor must be similar to the concentration of labile heme to ensure that the sensor is 50% bound with analyte so that both increases and decreases in heme concentration can be easily monitored.
| (1) |
| (2) |
| (3) |
Figure 2.
Simulations of the dependence of the (A) fractional heme saturation of the sensor on labile heme concentrations and (B) observed labile heme concentration and fractional saturation of the heme sensor on sensor expression level. (A) The relationship between labile heme concentration and fractional heme saturation of the sensor is shown for sensors with heme dissociation constants of 2000 nM (black), 200 nM (red), 20 nM (yellow), 2 nM (green), and 0.2 nM (blue). Optimal heme sensing is achieved when the Kd of the heme sensor is similar to the concentration of labile heme. The simulation is based on the model depicted in eqs 1–3. (B) Dependence of the observed sensor fractional saturation (dashed lines) and the concentration of labile heme (solid lines) on sensor expression. The simulations were conducted using Hyperquad Simulation and Speciation HySS 2009, assuming a hypothetical competition between the heme sensor and a cellular heme buffer, present at 20 nM and having a heme KD value of 20 nM, for 20 nM heme. Simulations are shown for three sensors with KD values of 2 nM (orange and red), 20 nM (green and black), and 200 nM (light blue and dark blue).
The challenge of designing a heme sensor with an appropriate heme binding affinity for heme sensing is made even more complicated by the fact that heme can be present in two oxidation states, oxidized ferric Fe3+-heme and reduced ferrous Fe2+-heme. The relatively reducing cellular environment [Emcytosol ~ – 320 mV vs the normal hydrogen electrode (NHE)], which is governed by the ratio of oxidized to reduced glutathione, coupled with the reduction potential of aqueous heme, estimated to be between −50 and −220 mV versus the NHE, suggests that LH is biased toward the reduced state.14 However, the actual fraction of LH that is reduced is dependent on its speciation and the degree to which it equilibrates with the glutathione redox buffer, both of which are unknown.
The prototype heme sensor, HS1, exhibited ferric and ferrous heme KD values of 3 and <1 nM at pH 7.0, respectively. However, HS1 was quantitatively saturated in cells across multiple locales, e.g., cytosol, mitochondria, and nucleus, indicating that its heme affinities were too tight for cellular heme sensing. Mutation of the heme-coordinating methionine residue to alanine in the Cyt b562 domain of HS1 generated the sensor variant HS1-M7A, which exhibited ferric and ferrous heme KD values of 2 µM and 25 nM at pH 7.0, respectively. In the yeast cytosol, HS1-M7A was found to be 20–50% saturated, making it well suited for heme sensing experiments. However, in the nucleus and mitochondria, HS1-M7A was found not to bind heme. If LH is largely reduced, these data would suggest that the LH concentrations in the cytosol are ~20 and <2.5 nM in the mitochondria and nucleus, respectively. However, these estimates of LH would change depending on the degree to which LH is oxidized in cells.14 Prima facie evidence for LH being largely reduced comes from the observation that the heme sensor CISDY had similar amounts of heme bound as HS1-M7A in cells despite having an ~30-fold difference in ferric heme dissociation constants, 2 µM for HS1-M7A14 versus 63 nM for CISDY15 at pH 7. If LH were largely ferric, the fraction of heme bound to CISDY would be very different from that of HS1-M7A (Figure 2A). On the other hand, the similar fraction of heme bound to CISDY and HS1-M7A in cells would suggest these sensors have similar ferrous heme affinities, but, unfortunately, the affinity of ferrous heme for CISDY was never determined.15 Future work should be focused on expanding the range of sensor heme binding affinities and achieving heme oxidation-state selectivity to better elucidate the role of LH in heme trafficking, signaling, and dynamics in different compartments and cell types.
Another important consideration is the expression of the heme probe. The expression of the sensor can alter the observed labile heme pool, and sensor expression can also interfere with heme homeostasis or other aspects of metabolism and physiology. Figure 2B depicts the relationship among sensor heme binding affinity, sensor expression level, fractional saturation of the sensor, and the observed concentration of labile heme. The simulation in Figure 2B is based on a hypothetical competition model between heme sensors exhibiting varying heme affinities, KD values of 2, 20, and 200 nM, and a cellular heme buffer, present at 20 nM and having a heme KD value of 20 nM, for 20 nM heme. At a low level of sensor expression relative to heme, <20 nM, the observed labile heme pool and the heme saturation of the sensor are independent of sensor concentration, which reflects the ideal scenario in which the sensor is not present at sufficient concentrations to perturb the heme binding equilibria of heme buffering factors. As the level of sensor expression exceeds the concentration of heme, >20 nM, one observes that the fractional saturation of the sensor decreases because of the presence of excess sensor relative to cellular heme. This results in a decrease in the concentration of the observed labile heme pool (see eq 3). Lower-affinity heme sensors are less disruptive to the measurement of labile heme at higher levels of sensor expression, but the trade-off is that they are less saturated with heme (Figure 2B). In practice, one can determine the dependence of the observed LH concentration on sensor expression by driving sensor expression using regulatable promoters or promoters of different strengths,14 or using flow cytometry to correlate cell–cell variations in sensor expression on observed LH.31
Another important consideration is that heme sensor expression can impact heme homeostasis or other aspects of physiology. To account for this, one should ensure that cells expressing the sensor do not exhibit defects in cell viability or the activity of various heme-dependent functions, e.g., the activities of heme-regulated transcription factors, respiration, or catalase activity. Titration of HS1-M7A in yeast cells using strong, medium, and weak promoters did not result in changes in the fractional heme saturation of the sensors or defects in heme-dependent processes, indicating that heme sensor expression did not itself impact heme availability. These types of experiments described for HS1, which are rarely performed with most metal sensors, including CISDY,15 are vital for using the sensors to dissect heme cell biology in an unbiased manner.
IDENTIFICATION OF HEME TRAFFICKING FACTORS USING HEME SENSORS
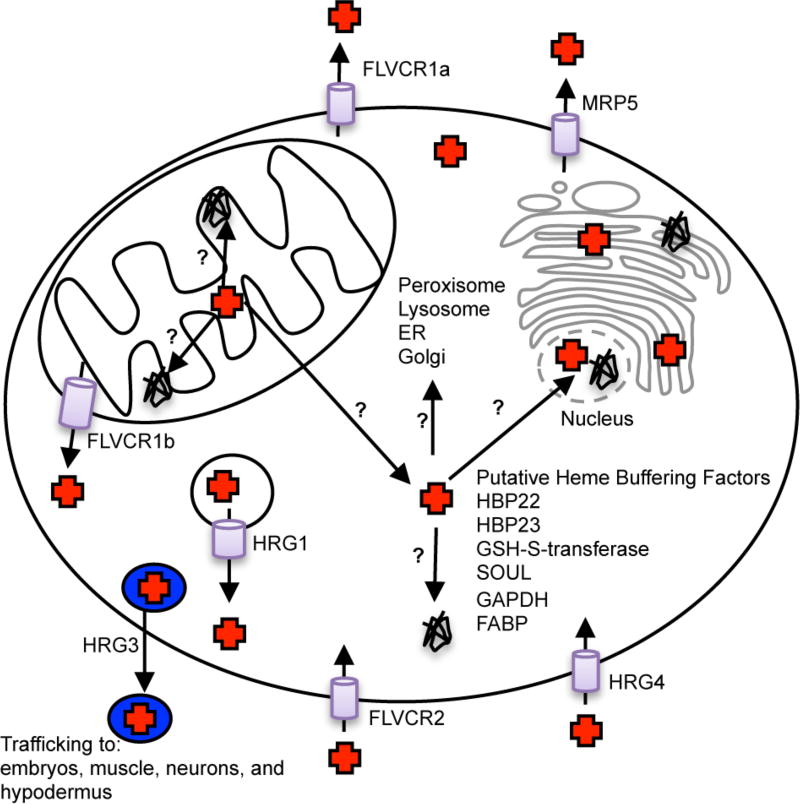
The development of fluorescence-based heme imaging agents offers an unparalleled set of new tools for elucidating heme homeostatic mechanisms and can complement traditional molecular genetics and biochemical approaches to identifying heme transport and trafficking factors. The ability to utilize genetic approaches to easily dissect heme transport and trafficking pathways is complicated by contributions from heme biosynthetic factors.1, 20 The establishment of Caenorhabditis elegans, a heme auxotroph lacking the heme biosynthetic machinery, as a simple animal model for studying heme homeostatic pathways in the absence of confounding contributions from heme synthesis has revolutionized our ability to study heme transport and trafficking.20 Indeed, several proteins that facilitate the intra- and intercellular distribution of heme were identified in C. elegans using heme-dependent gene expression profiling coupled with gene silencing/overexpression studies and phenotypic analyses.20 These heme-regulated genes include cytosolic heme importers HRG-1 and HRG-4,34 the intercellular heme chaperone, HRG-3,35 a putative intracellular heme chaperone, HRG-2,36 and ABC-type heme exporter MRP-537 (Figure 3). HRG-4, which lacks mammalian homologues, localizes to the plasma membrane, whereas HRG-1, which is conserved from arthopods to vertebrates, including mammals, is present in endosomal compartments and mobilizes heme into the cytosol. The intercellular heme chaperone, HRG-3, is an 8 kDa protein that binds and delivers maternal heme to developing oocytes. Heme is exported from cells via the ABC transporter MRP-5/ABCC5.37 In the C. elegans hypodermis, HRG-2, a single-pass type I transmembrane protein containing a thioredoxin-like fold, facilitates heme utilization, albeit its mechanism of action is not well-understood.
Figure 3.
Model of eukaryotic heme transport and trafficking. The final step of heme synthesis occurs in the mitochondrial matrix, and heme must be transported out of the mitochondria and incorporated into a multitude of hemoproteins found in different compartments. This process is likely mediated by heme chaperones and transporters. Proteins previously implicated in heme transport, trafficking, or buffering are identified, and trafficking pathways that are currently unknown are marked with question marks.
In addition to the development of tractable genetic model systems like C. elegans for the dissection of heme homeostatic pathways, heme trafficking factors have also been serendipitously discovered when certain pathological conditions have been studied. For instance, cats infected with feline leukemia virus (FeLV) develop aplastic anemia because of a loss of erythroid progenitors. It was subsequently determined that the cell surface receptor for FeLV, FLVCR1a, a 12-transmembrane domain (TMD) plasma membrane protein of the major facilitator superfamily of transporter proteins, was a cytoplasmic heme exporter that protects developing erythroid cells from heme toxicity and susceptibility to anemia11, 38 (Figure 3). Additionally, it was later determined that an alternative transcriptional start site located within the first intron of FLVCR1a results in a shorter transcript that produces a truncated six-TMD isoform, FLVCR1b, which transports heme out of the mitochondria39 (Figure 3). In addition, the homologous cell surface protein, FLVCR2, which is also a receptor for FeLV and implicated in disorders of the brain vasculature, e.g., Fowler’s syndrome, was demonstrated to serve as a heme importer40 (Figure 3).
Biochemically driven approaches to probing heme homeostatic mechanisms have also been instrumental. For instance, a number of cellular heme binding proteins (HBPs) have been identified on the basis of their interaction with heme-agarose or blue-sepharose columns and submicromolar heme dissociation constants. These HBPs include 22 kDa HBP, 23 kDa HBP, SOUL, glutathione S-transferase, fatty acid binding protein (FABP), and glyceraldehyde phosphate dehydrogenase (GAPDH)1, 20 (Figure 3). However, with the exception of GAPDH (vide infra), it is unknown if these proteins play a role in heme trafficking beyond simply buffering excess heme.
While genetic and biochemical approaches to characterizing heme homeostasis have been incredibly fruitful, the application of fluorescent heme sensors to probe heme trafficking pathways promises to greatly expedite the discovery process for uncovering heme homeostatic mechanisms across multiple cell types, organisms, and (patho)-physiological contexts. Given the widespread availability of instrumentation capable of high-throughput fluorescence measurements, including plate readers and flow cytometers, heme sensors like CISDY and HS1 can be integrated with genome-wide overexpression or deletion screens to rapidly identify new heme homeostatic factors in a deliberate and methodical manner. Indeed, this was recently demonstrated when heme sensor HS1-M7A was used to screen the Saccharomyces cerevisiae gene deletion collection for genes that impact heme bioavailability.14 After a partial screen of the yeast knockout libary, TDH3, encoding GAPDH, was found to control cytosolic heme availability; tdh3Δ cells exhibited a 2–4-fold increase in HS1-M7A–detected cytosolic labile heme concentration, but not total heme concentration. Further, it was found that heme availability to the nuclear heme-dependent transcription factor, heme activated protein 1, Hap1p, was diminished in tdh3Δ cells. Taken together, the data are consistent with a model in which GAPDH buffers cytosolic heme and regulates its availability to Hap1p. Given that GAPDH is present at concentrations approaching ~1 mM41 and it exhibits nanomolar heme dissociation constants,42 it is likely a primary constituent of the cytosolic heme buffer. However, it is unclear at this time if GAPDH binds and delivers heme directly to Hap1p in the nucleus or if there are other intermediary factors involved. It is worth noting that the identification of GAPDH as a heme homeostatic factor using the heme sensor was supported by prior biochemical and cell biological studies that demonstrated that GAPDH acts as a heme chaperone in mammalian cells and can bind and deliver heme to nitric oxide synthase in a NO-dependent manner (vide infra).43
ELUCIDATION OF THE SPATIOTEMPORAL DYNAMICS OF HEME USING HEME SENSORS
Heme trafficking and signaling necessitate that there is a pool of kinetically labile heme (LH) and molecules and mechanisms in place that can safely and rapidly distribute heme for a multitude of heme-dependent or regulated processes.20 The subcellular distribution, mobilizing factors, and the spatiotemporal dynamics of LH are not well understood. The recent development and application of fluorescent heme sensors have illuminated a number of critical aspects of cellular heme distribution and dynamics relevant to heme trafficking and signaling and promise to continue shedding light on heme mobilization dynamics.
The concentration and distribution of LH define the heme pool that can be utilized for heme trafficking and signaling. Heme imaging experiments with heme sensor HS1-M7A revealed that the cytosol has the most LH, ~20–40 nM, with the nucleus and mitochondria having considerably less LH, <2.5 nM. Given this subcellular distribution of heme, we proposed that the cytosol serves as a heme reservoir for trafficking and signaling, with GAPDH constituting the bulk of the cytosolic heme buffer (vide supra).14 However, the physiological significance of cytosolic LH has yet to be established.
Rather surprisingly, mitochondria, which are sites of heme biosynthesis and have a very high demand for heme due to the heme requirements of the respiratory complexes, have a low concentration of LH, <2.5 nM or fewer than one molecule.14 For reference, total ferrous heme in the yeast mitochondria has been estimated to be ~30 µM, or ~9000 molecules.44 This low concentration of mitochondrial LH suggests that, once synthesized, matrix heme is trafficked in a manner that limits its availability and circumvents the LH pool. This notion is consistent with the recent identification of mitochondrial heme metabolism complexes that traffic heme via transient protein–protein interactions.45
The nucleus also has a limited LH pool, <2.5 nM or fewer than six molecules. On one hand, this small amount of nuclear LH is not surprising given the toxicity of heme and the need to limit its exposure to genetic material.2–4 On the other hand, this observation raises the intriguing question of how nuclear heme-regulated transcription factors acquire heme given that the heme regulatory motifs (HRMs) of many of these factors exhibit micromolar heme dissociation constants (vide infra).46
How dynamic are labile heme pools, and what are the factors that mobilize them for trafficking and signaling? Experiments with HS1-M7A probing cellular LH have revealed considerable insight into cellular heme dynamics. For instance, flow cytometry of a population of exponential-phase yeast cells revealed that there is a dynamic trimodal distribution of cells exhibiting high (~40 nM), medium (~20 nM), and low (<1 nM) concentrations of LH, suggesting LH may be dynamically regulated.14 It is tempting to speculate that this trimodal distribution may be due to a mixed population of cells in different phases of the cell cycle, e.g., G1, S, and G2/M phases, or metabolic cycle, e.g., oxidative (Ox), reductive/building (RB), or reductive/charging (RC) phases.47 Indeed, with respect to the latter, the expression of genes involved in heme biosynthesis and degradation, as well as the concentration of heme biosynthetic intermediates, exhibits periodic oscillations corresponding to different metabolic states.47 However, further experiments are required to establish the origin and physiological consequences of the cell-to-cell heterogeneity in LH.
The signaling circuits that couple heme mobilization to heme utilization are poorly understood. One possibility is that there are transient and/or periodic changes in heme synthesis and degradation for controlling heme-regulated factors, as was demonstrated during metabolic cycling47 (vide supra). Another possibility is that there are physiological stimuli that can mobilize or redistribute LH for use in signaling or trafficking. Two notable examples of this stem from prior work demonstrating the ability of signaling molecules like hydrogen peroxide (H2O2) and nitric oxide (NO) to mobilize subcellular heme pools. For instance, in yeast, H2O2 triggers heme transfer between cytochrome c peroxidase (Ccp1p) and catalase (Cta1p) in the mitochondria.48 Respiratory chain-derived H2O2 labilizes heme from Ccp1 by oxidizing heme-coordinating His175 to 2-oxo-His, leading to heme labilization and transfer to Cta1p.
NO also profoundly impacts LH dynamics and heme bioavailability. For instance, Stuehr and colleagues found that NO blocked insertion of heme into a number of hemoproteins, including inducible nitric oxide synthase (iNOS), hemoglobin, catalase, and cytochrome p450 enzymes.43, 49 Using iNOS as a model hemoprotein to elucidate the mechanism of NO-regulated heme insertion, it was found that GAPDH is a heme chaperone that regulates insertion of heme into iNOS in a NO-dependent manner. S-Nitrosation of GAPDH at Cys152 weakened heme–GAPDH interactions and prevented GAPDH from transferring heme to iNOS.
In addition, NO mobilizes cell-wide heme pools. On the basis of experiments performed 20 years ago in endothelial cells demonstrating that NO increases the size of cellular iron pools due to the enhanced activity of the heme-degrading enzyme, heme oxygenase, it was proposed that NO induced the release of heme from a number of hemoproteins.50 Application of heme sensor HS1-M7A in yeast cells provided a direct demonstration of NO-mediated heme mobilization for the first time.14 Treatment of yeast cells with the small molecule NO source, NOC-7, resulted in a rapid increase in the LH concentration in the cytosol (from 17 to 40 nM) and nucleus (from <2.5 to 218 nM) within 20 min, followed by a slower re-establishment of the initial steady-state heme levels over the course of ~80 min. However, at this time, a proteome-wide description of proteins that bind and release heme in an NO-dependent manner is not known, and the physiological consequences of NO-mediated heme mobilization, and in particular the reason for the exquisite sensitivity of nuclear LH to NO, are not understood.
Mechanistically, there are a number of pathways that may lead to NO-dependent heme release and mobilization from cellular factors. For instance, nitrosylation of ferrous hemoproteins can labilize the trans ligand, e.g., His, and weaken the heme affinity, causing heme release. 51, 52 Alter-natively, S-nitrosation of certain heme-coordinating cysteine thiols53 or allosteric Cys sites that may impact heme binding43 can also result in heme dissociation. Indeed, the thiol-specific alkylating agent, iodoacetimide (IAM), impacted NO-dependent changes in LH as measured by HS1-M7A, suggesting that the sites of heme mobilization may in fact be thiol-containing factors.14 Importantly, hydrogen sulfide (H2S), another physiologically relevant signaling molecule, may labilize heme and regulate heme mobilization in a manner analogous to that of NO.54 For instance, H2S may bind and reduce the heme iron center, releasing ferrous heme in proteins that bind it more weakly than ferric heme. Alternatively, H2S-dependent persulfidation may result in allosteric changes in certain hemoproteins that result in heme dissociation.
NATURE OF HEME SIGNALING
Long conceptualized to act solely as a protein cofactor, heme serves as a dynamic signaling molecule, as strongly suggested by more recent studies. Here we discuss the meaning of heme signaling and how the distribution and spatiotemporal dynamics of LH may impact heme signaling pathways. We define heme-based signal transduction as any pathway in which binding of heme to or dissociation of heme from a target protein alters its activity, leading to changes in metabolism and physiology. A defining feature of heme signaling is that there is a distribution of heme-bound and unbound states of the target protein during active signaling. By this definition, heme signaling does not include the large number of heme-dependent proteins like respiratory complexes and enzymes like cytochrome P450s that are found almost exclusively in their heme-bound states and not regulated at the level of heme binding or dissociation. Therefore, a thermodynamic signature of heme signaling is that the heme dissociation constant of the target protein is similar in magnitude to the local LH pool, i.e., KD ~ [LH]. If KD ≫ [LH] or KD ≪ [LH], large fluctuations from the steady-state LH concentration are required to support heme-based signaling, which would pose significant energetic barriers.
There are a number of illustrative examples of heme signaling in Nature on which fluorescent heme probes have shed light. For instance, binding of heme to a number of nuclear transcription factors, including Hap1 in yeast and p53, Bach1, Rev-erb-α, and Rev-erb-β in mammals, alters their activity and collectively impacts diverse functions spanning oxygen sensing, iron homeostasis, the antioxidant stress response, mitochondrial respiration and biogenesis, mitophagy, apoptosis, circadian rhythms, cell cycle progression, and proliferation. 7–9, 46, 55–58 As noted earlier, many heme-regulated nuclear transcription factors exhibit micromolar heme dissociation constants,8, 46 but that of nuclear labile heme was found to be <2.5 nM.14 This would suggest that these transcription factors are not populated with enough heme to alter their transcriptional activity. One explanation is that the affinities of many heme-regulated factors are mischaracterized. Indeed, a re-evaluation of the heme dissociation constant of HRM-containing transcription factor Rev-erbβ found that heme binds 100-fold tighter than previously estimated, ~20 nM versus 2–6 µM.59 A number of factors likely contribute to the mischaracterization of heme binding affinities, including the propensity of heme to aggregate in aqueous buffers60 and the characterization of peptide fragments of the heme binding domains rather than the full length native proteins.46 With regard to the latter, as heme binding is a combination of heme iron axial ligand coordination, protein–porphyrin hydrophobic interactions, and ionic interactions between the heme propionates and charged protein residues, peptide fragments of native proteins are likely devoid of many key structural determinants of the energetics of heme binding.61
Another intriguing possibility to account for the population of low-affinity heme regulatory sites of transcription factors with heme is that there may be transient increases in LH concentration caused by active signaling processes. Indeed, this concept is supported by the fact that signaling molecules like NO can result in a >10-fold increase, from <2.5 nM to as much as ~200 nM, in nuclear LH concentration as determined using heme sensor HS1-M7A.14
Heme can also regulate certain ion channels.62, 63 For instance, binding of heme to His and Cys residues on a cytoplasmic CXXHX16H motif of cardiac KATP channels results in increased currents.63 Heme increases KATP currents in a dose-dependent manner with a maximal response achieved with 500 nM heme, and the half-maximal increase in KATP channel open probability was achieved with ~100 nM heme. These regulatory concentrations of heme are in the physiological range of cytosolic LH, found to be ~20–40 nM using heme sensors HS1-M7A and CISDY.14, 15 Thus, these KATP channels are heme sensors, and their activity can be dramatically altered with even small perturbations to LH.
Another very interesting case of heme signaling is the ability of heme to regulate micro RNA (miRNA) processing.64 Heme activates DGCR8, a RNA binding protein that cooperates with the RNase III enzyme Drosha to recognize and cleave long primary transcript pri-miRNA into precursor miRNAs (pre-miRNAs) in the nucleus. Ferric and ferrous heme dissociation constants for DGCR8 were estimated to be <200 fM and >1 µM, respectively,64 leading to the conclusion that ferric heme is the active effector for DGCR8-mediated miRNA processing. However, the current lack of oxidation-state specific heme sensors makes it difficult to evaluate the relative contributions of ferric versus ferrous LH toward heme signaling processes.
CONCLUSION
The past decade has witnessed an explosion in our understanding of heme cell biology. The development of new and innovative genetic, biochemical, and biophysical approaches, and in particular fluorescent heme imaging platforms, has revealed unprecedented insight into heme transporters and chaperones, and the spatiotemporal dynamics of labile heme relevant to its trafficking and role in signaling. The continued development of next-generation heme sensors with an expanded range of heme binding affinities, oxidation-state selectivities, and fluorescence emission wavelengths for simultaneous imaging of different subcellular compartments and tissues will greatly expand the tools available to deepen our understanding of heme trafficking and signaling in multiple organisms and physiological contexts.
Acknowledgments
We thank Dr. Rebecca Donegan for critical reading of the manuscript and helpful suggestions.
Funding
We are grateful for research funding from the U.S. National Institutes of Health (ES025661 and GM118744 to A.R.R.), the U.S. National Science Foundation (MCB-1552791 to A.R.R.), the Blanchard Professorship and start-up funding from the Georgia Institute of Technology (to A.R.R.), and the U.S. Department of Education GAANN Program (Grant P200A120081 to O.M.-G.).
Footnotes
Author Contributions
D.A.H. and O.M.-G. contributed equally to this work.
Notes
The authors declare no competing financial interest.
References
- 1.Severance S, Hamza I. Trafficking of Heme and Porphyrins in Metazoa. Chem. Rev. 2009;109:4596–4616. doi: 10.1021/cr9001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar S, Bandyopadhyay U. Free Heme Toxicity and Its Detoxification Systems in Human. Toxicol. Lett. 2005;157:175–188. doi: 10.1016/j.toxlet.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Chiabrando D, Vinchi F, Fiorito V, Mercurio S, Tolosano E. Heme in Pathophysiology: A Matter of Scavenging, Metabolism and Trafficking across Cell Membranes. Front. Pharmacol. 2014;5:61. doi: 10.3389/fphar.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sassa S. Why Heme Needs to Be Degraded to Iron, Biliverdin Ixalpha, and Carbon Monoxide? Antioxid. Redox Signaling. 2004;6:819–824. doi: 10.1089/ars.2004.6.819. [DOI] [PubMed] [Google Scholar]
- 5.Ponka P, Sheftel AD, English AM, Scott Bohle D, Garcia-Santos D. Do Mammalian Cells Really Need to Export and Import Heme? Trends Biochem. Sci. 2017 doi: 10.1016/j.tibs.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Mense SM, Zhang L. Heme: A Versatile Signaling Molecule Controlling the Activities of Diverse Regulators Ranging from Transcription Factors to Map Kinases. Cell Res. 2006;16:681–692. doi: 10.1038/sj.cr.7310086. [DOI] [PubMed] [Google Scholar]
- 7.Igarashi K, Sun J. The Heme-Bach1 Pathway in the Regulation of Oxidative Stress Response and Erythroid Differentiation. Antioxid. Redox Signaling. 2006;8:107–118. doi: 10.1089/ars.2006.8.107. [DOI] [PubMed] [Google Scholar]
- 8.Shen J, Sheng X, Chang Z, Wu Q, Wang S, Xuan Z, Li D, Wu Y, Shang Y, Kong X, Yu L, Li L, Ruan K, Hu H, Huang Y, Hui L, Xie D, Wang F, Hu R. Iron Metabolism Regulates P53 Signaling through Direct Heme-P53 Interaction and Modulation of p53 Localization, Stability, and Function. Cell Rep. 2014;7:180–193. doi: 10.1016/j.celrep.2014.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Identification of Heme as the Ligand for the Orphan Nuclear Receptors Rev-Erb-alpha and Rev-Erb-beta. Nat. Struct. Mol. Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamza I, Dailey HA. One Ring to Rule Them All: Trafficking of Heme and Heme Synthesis Intermediates in the Metazoans. Biochim. Biophys. Acta, Mol. Cell Res. 2012;1823:1617–1632. doi: 10.1016/j.bbamcr.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keel SB, Doty RT, Yang Z, Quigley JG, Chen J, Knoblaugh S, Kingsley PD, De Domenico I, Vaughn MB, Kaplan J, Palis J, Abkowitz JL. A Heme Export Protein Is Required for Red Blood Cell Differentiation and Iron Homeostasis. Science. 2008;319:825–828. doi: 10.1126/science.1151133. [DOI] [PubMed] [Google Scholar]
- 12.Haldar M, Kohyama M, So AY, Kc W, Wu X, Briseno CG, Satpathy AT, Kretzer NM, Arase H, Rajasekaran NS, Wang L, Egawa T, Igarashi K, Baltimore D, Murphy TL, Murphy KM. Heme-Mediated Spi-C Induction Promotes Monocyte Differentiation into Iron-Recycling Macrophages. Cell. 2014;156:1223–1234. doi: 10.1016/j.cell.2014.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dutra FF, Bozza MT. Heme on Innate Immunity and Inflammation. Front. Pharmacol. 2014;5:115. doi: 10.3389/fphar.2014.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanna DA, Harvey RM, Martinez-Guzman O, Yuan X, Chandrasekharan B, Raju G, Outten FW, Hamza I, Reddi AR. Heme Dynamics and Trafficking Factors Revealed by Genetically Encoded Fluorescent Heme Sensors. Proc. Natl. Acad. Sci. U. S. A. 2016;113:7539–7544. doi: 10.1073/pnas.1523802113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song Y, Yang M, Wegner SV, Zhao J, Zhu R, Wu Y, He C, Chen PR. A Genetically Encoded Fret Sensor for Intracellular Heme. ACS Chem. Biol. 2015;10:1610–1615. doi: 10.1021/cb5009734. [DOI] [PubMed] [Google Scholar]
- 16.Wu ML, Ho YC, Lin CY, Yet SF. Heme Oxygenase-1 in Inflammation and Cardiovascular Disease. Am. J. Cardiovasc. Dis. 2011;1:150–158. [PMC free article] [PubMed] [Google Scholar]
- 17.Schipper HM, Song W, Zukor H, Hascalovici JR, Zeligman D. Heme Oxygenase-1 and Neurodegeneration: Expanding Frontiers of Engagement. J. Neurochem. 2009;110:469–485. doi: 10.1111/j.1471-4159.2009.06160.x. [DOI] [PubMed] [Google Scholar]
- 18.Atamna H, Killilea DW, Killilea AN, Ames BN. Heme Deficiency May Be a Factor in the Mitochondrial and Neuronal Decay of Aging. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14807–14812. doi: 10.1073/pnas.192585799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atamna H, Frey WH., 2nd A Role for Heme in Alzheimer’s Disease: Heme Binds Amyloid Beta and Has Altered Metabolism. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11153–11158. doi: 10.1073/pnas.0404349101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddi AR, Hamza I. Heme Mobilization in Animals: A Metallolipid’s Journey. Acc. Chem. Res. 2016;49:1104–1110. doi: 10.1021/acs.accounts.5b00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan X, Rietzschel N, Kwon H, Walter Nuno AB, Hanna DA, Phillips JD, Raven EL, Reddi AR, Hamza I. Regulation of Intracellular Heme Trafficking Revealed by Subcellular Reporters. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E5144–5152. doi: 10.1073/pnas.1609865113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinclair PR, Gorman N, Jacobs JM. Current Protocols in Toxicology. Unit 8.3. Chapter 8. Wiley; New York: 2001. Measurement of Heme Concentration. [DOI] [PubMed] [Google Scholar]
- 23.Oh JY, Hamm J, Xu X, Genschmer K, Zhong M, Lebensburger J, Marques MB, Kerby JD, Pittet JF, Gaggar A, Patel RP. Absorbance and Redox Based Approaches for Measuring Free Heme and Free Hemoglobin in Biological Matrices. Redox Biol. 2016;9:167–177. doi: 10.1016/j.redox.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonkovsky HL, Healey JF, Lourie AN, Gerron GG. Intravenous Heme-Albumin in Acute Intermittent Porphyria: Evidence for Repletion of Hepatic Hemoproteins and Regulatory Heme Pools. Am. J. Gastroenterol. 1991;86:1050–1056. [PubMed] [Google Scholar]
- 25.Puy H, Gouya L, Deybach JC. Porphyrias. Lancet. 2010;375:924–937. doi: 10.1016/S0140-6736(09)61925-5. [DOI] [PubMed] [Google Scholar]
- 26.Atamna H, Brahmbhatt M, Atamna W, Shanower GA, Dhahbi JM. Apohrp-Based Assay to Measure Intracellular Regulatory Heme. Metallomics. 2015;7:309–321. doi: 10.1039/c4mt00246f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Manen HJ, Uzunbajakava N, van Bruggen R, Roos D, Otto C. Resonance Raman Imaging of the Nadph Oxidase Subunit Cytochrome B558 in Single Neutrophilic Granulocytes. J. Am. Chem. Soc. 2003;125:12112–12113. doi: 10.1021/ja036973r. [DOI] [PubMed] [Google Scholar]
- 28.Lu S, Min W, Chong S, Holtom GR, Xie XS. Label-Free Imaging of Heme Proteins with Two-Photon Excited Photothermal Lens Microscopy. Appl. Phys. Lett. 2010;96:113701. [Google Scholar]
- 29.Wegner SV, Arslan H, Sunbul M, Yin J, He C. Dynamic Copper(I) Imaging in Mammalian Cells with a Genetically Encoded Fluorescent Copper(I) Sensor. J. Am. Chem. Soc. 2010;132:2567–2569. doi: 10.1021/ja9097324. [DOI] [PubMed] [Google Scholar]
- 30.Carter KP, Young AM, Palmer AE. Fluorescent Sensors for Measuring Metal Ions in Living Systems. Chem. Rev. 2014;114:4564–4601. doi: 10.1021/cr400546e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dittmer PJ, Miranda JG, Gorski JA, Palmer AE. Genetically Encoded Sensors to Elucidate Spatial Distribution of Cellular Zinc. J. Biol. Chem. 2009;284:16289–16297. doi: 10.1074/jbc.M900501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aper SJ, Dierickx P, Merkx M. Dual Readout BRET/FRET Sensors for Measuring Intracellular Zinc. ACS Chem. Biol. 2016;11:2854–2864. doi: 10.1021/acschembio.6b00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng Y, Sligar SG, Wand AJ. Solution Structure of Apocytochrome b562. Nat. Struct. Biol. 1994;1:30–35. doi: 10.1038/nsb0194-30. [DOI] [PubMed] [Google Scholar]
- 34.Rajagopal A, Rao AU, Amigo J, Tian M, Upadhyay SK, Hall C, Uhm S, Mathew MK, Fleming MD, Paw BH, Krause M, Hamza I. Haem Homeostasis Is Regulated by the Conserved and Concerted Functions of HRG-1 Proteins. Nature. 2008;453:1127–1131. doi: 10.1038/nature06934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C, Samuel TK, Sinclair J, Dailey HA, Hamza I. An Intercellular Heme-Trafficking Protein Delivers Maternal Heme to the Embryo During Development in C. Elegans. Cell. 2011;145:720–731. doi: 10.1016/j.cell.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen C, Samuel TK, Krause M, Dailey HA, Hamza I. Heme Utilization in the Caenorhabditis Elegans Hypodermal Cells Is Facilitated by Heme-Responsive Gene-2. J. Biol. Chem. 2012;287:9601–9612. doi: 10.1074/jbc.M111.307694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korolnek T, Zhang J, Beardsley S, Scheffer GL, Hamza I. Control of Metazoan Heme Homeostasis by a Conserved Multidrug Resistance Protein. Cell Metab. 2014;19:1008–1019. doi: 10.1016/j.cmet.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quigley JG, Yang Z, Worthington MT, Phillips JD, Sabo KM, Sabath DE, Berg CL, Sassa S, Wood BL, Abkowitz JL. Identification of a Human Heme Exporter That Is Essential for Erythropoiesis. Cell. 2004;118:757–766. doi: 10.1016/j.cell.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 39.Chiabrando D, Marro S, Mercurio S, Giorgi C, Petrillo S, Vinchi F, Fiorito V, Fagoonee S, Camporeale A, Turco E, Merlo GR, Silengo L, Altruda F, Pinton P, Tolosano E. The Mitochondrial Heme Exporter Flvcr1b Mediates Erythroid Differentiation. J. Clin. Invest. 2012;122:4569–4579. doi: 10.1172/JCI62422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duffy SP, Shing J, Saraon P, Berger LC, Eiden MV, Wilde A, Tailor CS. The Fowler Syndrome-Associated Protein Flvcr2 Is an Importer of Heme. Mol. Cell. Biol. 2010;30:5318–5324. doi: 10.1128/MCB.00690-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peralta D, Bronowska AK, Morgan B, Doka E, Van Laer K, Nagy P, Grater F, Dick TP. A Proton Relay Enhances H2o2 Sensitivity of Gapdh to Facilitate Metabolic Adaptation. Nat. Chem. Biol. 2015;11:156–163. doi: 10.1038/nchembio.1720. [DOI] [PubMed] [Google Scholar]
- 42.Hannibal L, Collins D, Brassard J, Chakravarti R, Vempati R, Dorlet P, Santolini J, Dawson JH, Stuehr DJ. Heme Binding Properties of Glyceraldehyde-3-Phosphate Dehydrogenase. Biochemistry. 2012;51:8514–8529. doi: 10.1021/bi300863a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chakravarti R, Aulak KS, Fox PL, Stuehr DJ. Gapdh Regulates Cellular Heme Insertion into Inducible Nitric Oxide Synthase. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18004–18009. doi: 10.1073/pnas.1008133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garber Morales J, Holmes-Hampton GP, Miao R, Guo Y, Munck E, Lindahl PA. Biophysical Characterization of Iron in Mitochondria Isolated from Respiring and Fermenting Yeast. Biochemistry. 2010;49:5436–5444. doi: 10.1021/bi100558z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Medlock AE, Shiferaw MT, Marcero JR, Vashisht AA, Wohlschlegel JA, Phillips JD, Dailey HA. Identification of the Mitochondrial Heme Metabolism Complex. PLoS One. 2015;10:e0135896. doi: 10.1371/journal.pone.0135896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang L, Guarente L. Heme Binds to a Short Sequence That Serves a Regulatory Function in Diverse Proteins. EMBO J. 1995;14:313–320. doi: 10.1002/j.1460-2075.1995.tb07005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. Cyclic Changes in Metabolic State During the Life of a Yeast Cell. Proc. Natl. Acad. Sci. U. S. A. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kathiresan M, Martins D, English AM. Respiration Triggers Heme Transfer from Cytochrome c Peroxidase to Catalase in Yeast Mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2014;111:17468–17473. doi: 10.1073/pnas.1409692111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waheed SM, Ghosh A, Chakravarti R, Biswas A, Haque MM, Panda K, Stuehr DJ. Nitric Oxide Blocks Cellular Heme Insertion into a Broad Range of Heme Proteins. Free Radical Biol. Med. 2010;48:1548–1558. doi: 10.1016/j.freeradbiomed.2010.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yee EL, Pitt BR, Billiar TR, Kim YM. Effect of Nitric Oxide on Heme Metabolism in Pulmonary Artery Endothelial Cells. Am. J. Physiol. 1996;271:L512–L518. doi: 10.1152/ajplung.1996.271.4.L512. [DOI] [PubMed] [Google Scholar]
- 51.Cooper CE. Nitric Oxide and Iron Proteins. Biochim. Biophys. Acta, Bioenerg. 1999;1411:290–309. doi: 10.1016/s0005-2728(99)00021-3. [DOI] [PubMed] [Google Scholar]
- 52.Herzik MA, Jonnalagadda R, Kuriyan J, Marletta MA. Structural Insights into the Role of Iron-Histidine Bond Cleavage in Nitric Oxide-Induced Activation of H-Nox Gas Sensor Proteins. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E4156–E4164. doi: 10.1073/pnas.1416936111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weichsel A, Maes EM, Andersen JF, Valenzuela JG, Shokhireva T, Walker FA, Montfort WR. Heme-Assisted S-Nitrosation of a Proximal Thiolate in a Nitric Oxide Transport Protein. Proc. Natl. Acad. Sci. U. S. A. 2005;102:594–599. doi: 10.1073/pnas.0406549102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mishanina TV, Libiad M, Banerjee R. Biogenesis of Reactive Sulfur Species for Signaling by Hydrogen Sulfide Oxidation Pathways. Nat. Chem. Biol. 2015;11:457–464. doi: 10.1038/nchembio.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pfeifer K, Kim KS, Kogan S, Guarente L. Functional Dissection and Sequence of Yeast Hap1 Activator. Cell. 1989;56:291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- 56.Ogawa K, Sun J, Taketani S, Nakajima O, Nishitani C, Sassa S, Hayashi N, Yamamoto M, Shibahara S, Fujita H, Igarashi K. Heme Mediates Derepression of Maf Recognition Element through Direct Binding to Transcription Repressor Bach1. EMBO J. 2001;20:2835–2843. doi: 10.1093/emboj/20.11.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, Lazar MA. Rev-Erbalpha, a Heme Sensor That Coordinates Metabolic and Circadian Pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 58.Woldt E, Sebti Y, Solt LA, Duhem C, Lancel S, Eeckhoute J, Hesselink MK, Paquet C, Delhaye S, Shin Y, Kamenecka TM, Schaart G, Lefebvre P, Neviere R, Burris TP, Schrauwen P, Staels B, Duez H. Rev-Erb-Alpha Modulates Skeletal Muscle Oxidative Capacity by Regulating Mitochondrial Biogenesis and Autophagy. Nat. Med. 2013;19:1039–1046. doi: 10.1038/nm.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gupta N, Ragsdale SW. Thiol-Disulfide Redox Dependence of Heme Binding and Heme Ligand Switching in Nuclear Hormone Receptor Rev-Erb-Beta. J. Biol. Chem. 2011;286:4392–4403. doi: 10.1074/jbc.M110.193466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Villiers KA, Kaschula CH, Egan TJ, Marques HM. Speciation and Structure of Ferriprotoporphyrin IX in Aqueous Solution: Spectroscopic and Diffusion Measurements Demonstrate Dimerization, but Not Mu-Oxo Dimer Formation. JBIC, J. Biol. Inorg. Chem. 2007;12:101–117. doi: 10.1007/s00775-006-0170-1. [DOI] [PubMed] [Google Scholar]
- 61.Reddi AR, Reedy CJ, Mui S, Gibney BR. Thermodynamic Investigation into the Mechanisms of Proton-Coupled Electron Transfer Events in Heme Protein Maquettes. Biochemistry. 2007;46:291–305. doi: 10.1021/bi061607g. [DOI] [PubMed] [Google Scholar]
- 62.Hou S, Reynolds MF, Horrigan FT, Heinemann SH, Hoshi T. Reversible Binding of Heme to Proteins in Cellular Signal Transduction. Acc. Chem. Res. 2006;39:918–924. doi: 10.1021/ar040020w. [DOI] [PubMed] [Google Scholar]
- 63.Burton MJ, Kapetanaki SM, Chernova T, Jamieson AG, Dorlet P, Santolini J, Moody PC, Mitcheson JS, Davies NW, Schmid R, Raven EL, Storey NM. A Heme-Binding Domain Controls Regulation of ATP-Dependent Potassium Channels. Proc. Natl. Acad. Sci. U. S. A. 2016;113:3785–3790. doi: 10.1073/pnas.1600211113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barr I, Smith AT, Chen Y, Senturia R, Burstyn JN, Guo F. Ferric, Not Ferrous, Heme Activates Rna-Binding Protein DGCR8 for Primary Microrna Processing. Proc. Natl. Acad. Sci. U. S. A. 2012;109:1919–1924. doi: 10.1073/pnas.1114514109. [DOI] [PMC free article] [PubMed] [Google Scholar]