Abstract
The application of global “-omics” technologies to exercise has introduced new opportunities to map the complexity and interconnectedness of biological networks underlying the tissue-specific responses and systemic health benefits of exercise. This review will introduce major research tracks and recent advancements in this emerging field, as well as critical gaps in understanding the orchestration of molecular exercise dynamics that will benefit from unbiased omics investigations. Furthermore, significant research hurdles that need to be overcome to effectively fill these gaps related to data collection, computation, interpretation, and integration across omics applications will be discussed. Collectively, a cross-disciplinary physiological and omics-based systems approach will lead to discovery of a wealth of novel exercise-regulated targets for future mechanistic validation. This frontier in exercise biology will aid the development of personalized therapeutic strategies to improve athletic performance and human health through precision exercise medicine.
Integrated omics efforts focused on genes, proteins, peptides, posttranslational modifications, lipids, and metabolites in a range of samples will help us better understand human physiological responses to exercise.
The dynamic human physiological responses to exercise have been widely studied in the context of metabolism and mechanical stress. Health benefits of exercise in prevention, delay, and/or treatment of pathophysiology associated with metabolic disorders and aging are widely appreciated. However, the intricacies of molecular networks and biological mechanisms underlying how humans adapt to exercise and acquire health benefits are not fully understood. In response to perturbations in whole-body homeostasis induced by physical activity and exercise, biological networks are stimulated in various cell types and organs to manage systemic metabolic and mechanical demands (Hawley et al. 2014). Targeted, reductionist-based approaches have laid the foundation for our understanding of distinct biological mechanisms regulating acute versus repeated exercise adaptations using a range of experimental models from cells to animals and humans. However, the complexity and integrated nature of the whole-body exercise response warrants global unbiased systems approaches to unravel the interwoven networks underlying the benefits of exercise in health and disease.
In this early stage of the exercise and omics revolution, there is a wealth of molecular information now within reach to help build on our understanding of human metabolism and exercise physiology. There is tremendous potential for omics approaches to fill critical gaps in our understanding of the integrative networks underlying the health benefits of exercise (Zierath and Wallberg-Henriksson 2015). Advancements in omics-based technologies have facilitated recent applications to more comprehensively map these networks. The establishment of centers dedicated to this omics research scope was the focus of a recent National Institutes of Health (NIH) workshop including contributions from leading exercise researchers. This workshop led to the creation of an NIH Common Fund program titled “Molecular Transducers of Physical Activity in Humans.” The overall aim of this initiative is to define the “exercise responsome” and establish the biological function of molecules eliciting the systemic effects of exercise (Neufer et al. 2015).
To achieve these aims, single omics applications alone will not be sufficient to map the complexity of genetic and environmental contributions to the exercise response. Integrated omics efforts applied to human exercise will not come without significant challenges. A multidisciplinary approach across international centers providing physiological, omics, and bioinformatics expertise will be required to complete such arduous sample collection and omics data analysis efforts to catalog exercise-regulated networks. Rather than focusing on specific molecular targets, this review will broadly introduce omics research capabilities to exercise biologists to foster training initiatives and collaborative research. To further set the stage for future omics studies applied to exercise biology, the major omics research tracks and recent advances from human studies in the past 5 years will be reviewed. Following this background, potential aspects of exercise biology that can benefit from omics approaches will be discussed along with the potential applications, hurdles, solutions and short-term gains associated with achieving these global discovery efforts.
OMICS AND EXERCISE RESEARCH TRACKS
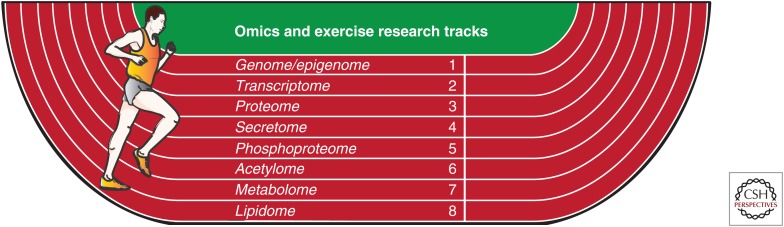
Mapping and interrogating the dynamic molecular networks underpinning exercise will involve omics approaches focused on genes, proteins, peptides, posttranslational modifications, lipids, and metabolites in a range of tissue samples and bodily fluids. In no particular order, eight major omics and exercise research tracks are depicted in Figure 1. It is important to note that these omics applications should not be considered as completely distinct tracks, because integration across these tracks will be crucial for delineating molecular networks and linkages. In addition, each track can be further subdivided into distinct fields involving various omics technological approaches. The research tracks most relevant to human exercise will be briefly outlined below to provide a broad introduction of omics approaches to date and discussion of short-term gains for researchers interested in this exciting renaissance in exercise biological research.
Figure 1.
Omics and exercise research tracks. Eight major global omics-based research themes, or “tracks,” are depicted that can be applied to map various aspects of exercise biological networks and discover novel targets underlying exercise responses and training adaptations in health and disease (i.e., genome/epigenome, transcriptome, proteome, secretome, phosphoproteome, acetylome, metabolome, and lipidome). Although these research themes are depicted as distinct tracks and in no particular order, multidisciplinary omics data integration across these tracks will be critical to the identification and validation of novel exercise-regulated orchestration of targets within these networks.
GENOME AND EPIGENOME
Together, the genomics and epigenomics research tracks have extended our understanding of the interactions of genetic background and environment in human athletic performance and exercise responsiveness. Genomics technological advances now provide platforms for cheaper and faster deep sequencing of the human genome. Previous observational genetic studies, primarily based on smaller, heterogeneous cohorts (Wang et al. 2016) have assessed candidate genes and variants. Genome-wide association studies (GWAS) have pinpointed gene variants (e.g., ACTN3 and ACE) and single-nucleotide polymorphisms (SNPs) potentially impacting human exercise performance, health benefits of exercise (Tanaka et al. 2016), and predictors of responsiveness to exercise training (Sarzynski et al. 2016). The molecular bases of gene interactions underlying the human exercise response are complex; however, it is well appreciated that genetic variation impacts gene–exercise interactions (Bouchard et al. 2011). Unbiased hypothesis-free whole-genome mapping efforts in larger cohorts will help unravel this level of intricacy. Multicenter research efforts, such as the Athlome Project Consortium, have recently been established (Pitsiladis et al. 2016) to help achieve this global strategy. These efforts will substantially expand the catalog of gene–exercise interactions and lead to future mechanistic studies of how these genes are linked to changes in an individual’s environment to determine functional bases of exercise training adaptations and responsiveness.
The growing field of epigenomics and exercise is aimed at further establishing these genetic–environmental links by mapping global methylation patterns on DNA and posttranslational modifications on regulatory chromosomal proteins (i.e., histone methylation, phosphorylation, and acetylation). These environmentally regulated modifications of DNA and histones impact transcriptional control by altering spatial accessibility of DNA and chromatin structure, as well as recruitment of histone-modifying factors. Epigenetic programming is impacted by both acute and chronic exercise and central to skeletal muscle metabolic regulatory control (Howlett and McGee 2016) and maintenance of skeletal muscle mass with aging (Sharples et al. 2016). DNA methylation is dramatically altered by exercise training in both human skeletal muscle (Nitert et al. 2012) and adipose tissue (Ronn et al. 2013). Hypomethylation impacts the promoter regions of key exercise-regulated metabolic genes (e.g., PGC-1α, PDK4, and PPAR-δ), and reduced methylation status occurs in parallel with respective exercise-triggered increases in gene expression (Barres et al. 2012). Recent efforts have also applied epigenomics to unravel how blood metabolic gene-specific methylation patterns such as AMP-activated protein kinase (AMPK) are altered by exercise (King-Himmelreich et al. 2016). Future hypothesis-free studies, integration across omics platforms such as a recent human muscle integrated transcriptomics study (Lindholm et al. 2014), along with more mechanistic epigenomic studies dissecting target genes and variants will be paramount for moving this exercise genomic/epigenomic field forward.
TRANSCRIPTOME
Complementary to epigenomic regulation, exercise induces dynamic changes in gene transcription by stimulating a range of protein kinases and cellular signaling pathways. Repeated exercise bouts reinforce tissue transcriptional adaptations in response to exercise mechanical and metabolic stimuli. Together, these acute and chronic effects of exercise reprogram the transcriptome, which encompasses biomarkers of training adaptations and exercise health benefits. Transcriptomics technologies such as microarray and RNA sequencing (RNA-seq) platforms help decipher how the array of genomic and epigenomic environmental regulatory mechanisms cumulatively affect exercise-regulated gene transcription networks, such as the plasticity of human skeletal muscle. In extensions of prevalent literature in this area, recent studies have uncovered novel insights into endurance exercise-regulated muscle transcriptomic network remodeling during recovery (Neubauer et al. 2014), protein supplementation (Rowlands et al. 2011, 2016), and impairments in endurance-trained subjects lacking metabolic responsiveness (i.e., training improvements in insulin sensitivity) (Bohm et al. 2016). The human muscle resistance exercise training–responsive transcriptional networks in the settings of aging (Raue et al. 2012), aging-associated muscle frailty (Hangelbroek et al. 2016), stress-induced immune activation (Gordon et al. 2012), and combined aerobic training modes (Lundberg et al. 2016) are also continuing to be unraveled using transcriptomics. New studies assessing other tissues and biological fluids have uncovered adipose tissue networks (Ronn et al. 2014) and transcriptional landscapes involving human plasma biomarkers of triglyceride responses (Sarzynski et al. 2015) and peripheral blood cell biomarkers of oxygen uptake responsiveness to endurance training (Dias et al. 2015). Although not discussed in detail, small noncoding RNA molecules such as microRNA are also important contributors to exercise-regulated posttranscriptional modulation and represent an exciting area for future investigation in epigenetic control of the exercise response (as systematically reviewed in Flowers et al. 2015). Given the dynamic nature of acute and chronic exercise transcriptional responses and number of tissues involved in systemic exercise responses, exercise transcriptomics is addressing large-scale computational challenges to unravel how various training regimens integrate the genetic and environmental nature of exercise-induced tissue adaptations.
PROTEOME
These exercise-induced transcriptome changes are intimately linked to the downstream alterations in protein translation and posttranslational regulation that occur in the hours and days following exercise. These changes in protein activity and/or abundance are central to tissues’ molecular functionality. Transcriptional changes are not always accurate markers of protein status because of discordant gene–protein expression level timing and proportionality, as well as the array of mechanisms regulating both messenger RNA (mRNA) and protein abundance, localization, and turnover. However, the transcriptome, proteome, and integrated profiles provide a global snapshot of how tissues are reprogrammed by exercise. Mass spectrometry (MS)-based proteomics has recently been applied to human tissues to accurately identify and quantify global exercise-regulated remodeling of thousands of proteins with exercise training, tissue damage, aging, and disease. Exercise-related network blueprints to date are primarily focused on the skeletal muscle proteome (as reviewed in Petriz et al. 2016) to map adaptations following acute and endurance exercise. Moreover, protein remodeling in the settings of varied training status (Schild et al. 2015), myositis (Munters et al. 2016), and type 2 diabetes (T2D) (Hussey et al. 2013) relevant to muscle metabolic health has been uncovered using proteomics. Bioinformatics analysis of network adaptations has revealed important exercise variables underlying skeletal muscle (Padrao et al. 2016) and heart (Ferreira et al. 2015) plasticity. Sophisticated proteomics applications have even narrowed down analyses to the single muscle fiber level (Murgia et al. 2015). Future proteomic analyses of other accessible biopsies (e.g., adipose tissue) and plasma (Leggate et al. 2012), which is capable of being profiled in samples from thousands of subjects (Cominetti et al. 2016), will help unravel tissue-specific and systemic proteome dynamics occurring with exercise.
SECRETOME
In addition to resident proteins, an emerging subfield of proteomics involves global characterization of factors secreted from cells and tissues (e.g., skeletal muscle, adipose tissue). These secreted factors function via autocrine, paracrine, and/or endocrine mechanisms to communicate exercise-regulated signals. Global MS-based analysis of the exercise-regulated secretome addresses tissue cross-talk mechanisms involved in the systemic exercise response. Two rapidly expanding secretome subfields involve identification of panels of released peptides and cytokines (e.g., myokines, adipokines) and profiling cargo from cellular-derived exosome vesicles in response to exercise. Skeletal muscle secretes myokines that communicate with several other tissues (e.g., adipose tissue, liver, pancreas, brain, and bone) to program exercise-induced metabolic and mechanical adaptations (Pedersen and Febbraio 2012). Early myokinome profiling efforts in human muscle involved translating secreted proteins in cultured human muscle cells to expression of candidate myokines in human muscle biopsies following strength training (Norheim et al. 2011). More recently, global microarray (Catoire et al. 2014) and MS-based (Hartwig et al. 2014) approaches have uncovered novel components of the human muscle secretome in response to acute exercise and exercise training. Proteomics-based discovery of the adipokinome has expanded our understanding of secreted proteins from human adipocytes (Lehr et al. 2012). Future efforts to apply these techniques to other secretory organs and blood samples in the context of exercise intervention are critical for mapping novel exercise cross-talk mechanisms. For example, the secretome isolated from human cells such as lymphocytes (Al-Dabbagh et al. 2015) can be used to determine how secreted factors impact other tissues such as skeletal muscle in a cellular experimental setting. Ongoing efforts to uncover the global exercise-regulated secretome will also involve identification of secreted proteins and nucleotides transported between cells and tissues via exosomes and microvesicles, as recently reviewed (Safdar et al. 2016). Together, this secreteome research track represents exciting discovery potential for solving the puzzle of how secreted, circulating candidate factors elicit systemic exercise effects and modulate metabolism in health and disease (Weigert et al. 2014).
PHOSPHOPROTEOME AND ACETYLOME
Although exercise is well appreciated to trigger diverse protein posttranslational modifications (PTMs), the application of omics technologies to globally map and quantify PTM transducers of exercise is at a very early stage. MS-based phosphoproteomics and acetylomics will be specifically discussed; however, it must be appreciated that other PTMs also underlie subtleties in human exercise responses. Reversible protein phosphorylation is regulated by a balance of kinase and phosphatase activity that dynamically impacts molecular substrates. This dynamic switch mechanism underlies many biological functions including key regulatory aspects of metabolism (Humphrey et al. 2015). Most previous research of exercise-induced phosphorylation has focused on a small subset of protein kinases and pathways. The first application of global MS-based phosphoproteomics to acute exercise-induced regulation of human skeletal muscle revealed extensive complexity and interconnectedness in exercise signal transduction (Hoffman et al. 2015). This landmark study revealed >1000 phosphorylation sites regulated by a single bout of high-intensity endurance exercise, including a large proportion of regulated sites and kinases never previously associated with the exercise response. Future global phosphoproteomics efforts will determine how this intricate network of kinases and substrates are impacted by a range of exercise and human subject variables. More targeted MS approaches will help profile subsets of exercise-regulated proteins and PTMs using interrogation of MS libraries in samples from larger subject cohorts to complement the deep coverage provided by global phosphoproteomics.
In addition to phosphorylation, exercise fine-tunes metabolism by inducing protein acetylation on lysine residues. Acetylomics is aimed at enriching this PTM from complex samples to map how acetylation alters protein function and metabolism. Protein acetylation in mitochondria serves as a reversible regulatory metabolic control mechanism that is dynamically regulated by deacetylases, including sirtuins such as SIRT3 (Baeza et al. 2016). Lysine acetylation regulates key exercise-responsive aspects of mitochondrial function in skeletal muscle (Philp et al. 2014). Acetylation status correlates with human insulin sensitivity (Mielke et al. 2014) and age-related adaptive responses to exercise training that help maintain quality and protect against oxidative damage of mitochondrial proteins (Johnson et al. 2015). Together, these findings warrant further global acetylome investigations in human tissues. Adding to the level of complexity of mapping single exercise-regulated PTMs is the subfield of PTM cross talk. Regulation of one PTM may program protein conformational changes that promote substrate accessibility and/or additional PTMs that underlie downstream biological functions. Multiple proteomics-based applications for sample enrichment and PTM profile comparisons, in addition to other protein regulatory mechanisms relevant to exercise (e.g., redox proteomics) (McDonagh et al. 2014), will be essential to map and further understand mechanisms of exercise cross talk that translate to protein functional responses.
METABOLOME AND LIPIDOME
In addition to mapping gene, protein, and PTM networks regulated by exercise, omics technologies can also be applied to identify and quantify metabolite and lipid biomarkers underlying tissue-specific and systemic metabolic adaptations of exercise. Exercise-induced adaptations in expression and/or activity of metabolic enzymes affect an abundance of small-molecule metabolites serving a wide range of biological functions. Metabolomics applications to human exercise have become more prevalent with improved global and targeted detection capabilities using MS and nuclear magnetic resonance (NMR) spectroscopy. Recent applications of metabolomics have uncovered metabolic networks impacted by high-intensity interval training programs (Kuehnbaum et al. 2014; Peake et al. 2014). Insights into branched-chain amino acid metabolism in response to resistance training have also been revealed from metabolomics analysis of skeletal muscle biopsies from healthy and frail older subjects (Fazelzadeh et al. 2016), as well as plasma from overweight subjects following resistance and endurance training (Glynn et al. 2015). Plasma metabolomics profiling of healthy versus T2D subjects, along with integrated global transcriptomics, has mapped disease alterations in acute exercise responses (Hansen et al. 2015). Moreover, metabolomics studies have uncovered biomarkers of human cardiometabolic fitness in skeletal muscle (Huffman et al. 2014) and aerobic capacity in serum (Lustgarten et al. 2013). Current and future directions in this growing field include globally profiling the metabolome signature of plasma and other biological fluids (e.g., urine [Wu and Gao 2015], saliva [Wallner-Liebmann et al. 2016], and sweat [Hooton et al. 2016]) from healthy, athletic, sedentary, and diseased subjects exposed to a range of exercise programs. A subfield of exercise metabolomics, termed “sportomics,” will translate metabolite markers associated with the metabolic challenges of exercise training and competition to similar stresses occurring in disease settings (Bassini and Cameron 2014). Moreover, systems-based approaches studying extreme environments (e.g., high altitude [Edwards and Thiele 2013]) will reveal important aspects of physiological adaptations relevant to health and athletic performance.
Mapping the intricacies in exercise-induced remodeling of thousands of lipid species involved in metabolic pathways has benefited from global MS-based lipidomics approaches, and therefore the latest translational studies in human tissues and serum will be highlighted. Lipid remodeling in skeletal muscle has been the focus of several recent studies, as intramyocellular lipids play important roles in human health, athletic training, and the progression of metabolic derangements such as insulin resistance and T2D (Coen and Goodpaster 2012). Lipidomics analysis of human muscle biopsies following endurance exercise has explored exercise-induced alterations in phospholipid species and their relation to subject metabolic health status (Newsom et al. 2016). Complementary lipidomics-based studies have also focused on specific subspecies of ceramides and sphingolipids modified by acute exercise in human muscle (Bergman et al. 2016) and serum (Bergman et al. 2015). Furthermore, human blood analyses have uncovered how the lipidome composition is associated with subjects’ aerobic fitness levels and responses to lipid-based metabolic challenges (Morris et al. 2015). Roles for lipid mediators underlying inflammatory responses to resistance exercise in blood (Markworth et al. 2013) and skeletal muscle (Markworth et al. 2016) have also emerged from lipidomics approaches. Together, these recent human lipidomics efforts have characterized dynamic changes in tissue and blood lipid profiles and their potential impacts on exercise-regulated cellular permeability, stability of cell surface receptors, and inflammatory responses.
It will take decades to fully understand and integrate the large datasets generated from these distinct omics research tracks. However, there are several short-term gains in the next 5 years that will significantly advance exercise and medical science with complementary efforts from both large-scale consortia and individual specialized investigators. Short-term gains will be maximized by establishing specific standardized acute and repeated exercise protocols that are known to elicit robust physiological responses and health benefits to facilitate data harmonization and increase the size of datasets available for future integration. Importantly, the first short-term gain will be establishment of an exercise responsome signature for these standardized exercise interventions. Second, compiling and comparing interactions between epigenomic and transcriptomic signatures with known protein, PTM, metabolite, and lipid networks for each standardized protocol will begin to connect acute exercise modifications with downstream targets. This will be especially beneficial in building on existing databases of gene and protein interactions to map blueprints of how both known and novel exercise-induced responses program functional training and health/disease adaptations. Pinpointing and prioritizing key nodes in these blueprints that can be mechanistically validated in the years ahead will be extremely useful as in vitro exercise models and target screening platforms are developed.
The establishment and growth of multicenter consortia such as those resulting from the Athlome Project Consortium and NIH Common Fund program will be critical to accessing large subject populations and streamlining multiple-omics-based approaches from international partners focusing on specific exercise interventions and research questions. For example, these consortia will facilitate integration of cross-omics data obtained from identical subjects’ samples and improve data harmonization and accessibility from multiple centers. Individual investigators will also provide invaluable contributions to global omics efforts by establishing key partnerships that will align subject access with critical human exercise physiology, omics-based technology, and bioinformatics expertise. Importantly, individual investigators will initiate new cross-disciplinary research collaborations and provide training opportunities for the current and next generation of scientists to continue to grow these omics and exercise research tracks and provide longer-term scientific and medical gains.
CRITICAL GAPS AND VARIABLES UNDERLYING EXERCISE BIOLOGICAL NETWORK DYNAMICS
Collectively, recent omics applications have started to catalog the intricate networks underlying various aspects of exercise in health, sport, aging, and disease. In future cross-disciplinary collaborative studies, it will be crucial for omics-based researchers to appreciate that one type of exercise does not fit all. Embracing the distinct responses of exercise interventions in a range of populations and environments will facilitate personalized exercise prescription and more effective data comparisons. In the following section, important exercise and subject variables will be discussed that can benefit from omics-based global approaches and may lead to more personalized exercise interventions in health and disease settings.
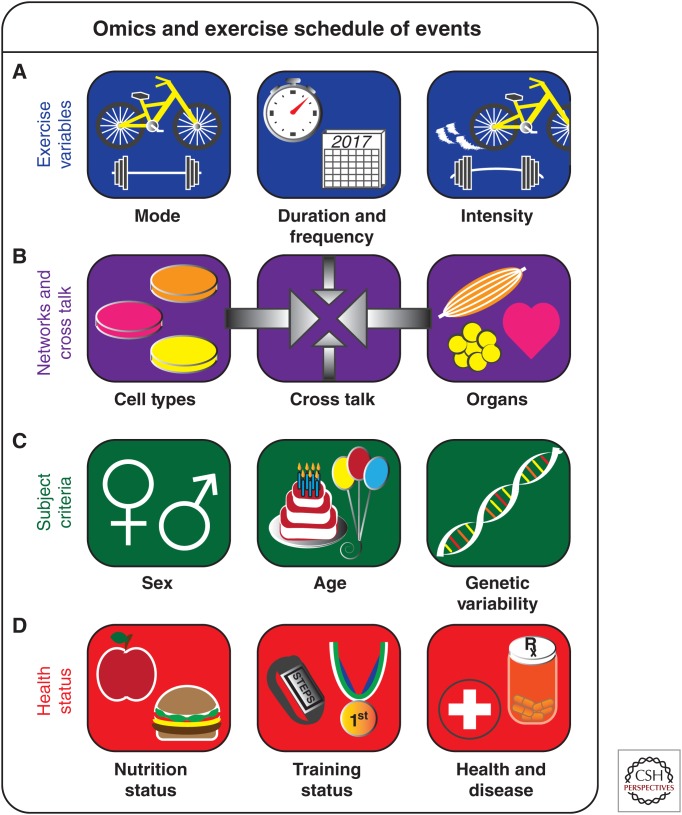
First, diversities in biological networks underlying exercise variables (Fig. 2A), such as mode, frequency, duration, and intensity, will need to be considered from study conception and design to post hoc study comparisons. Although several pathways are known to be distinct between endurance and resistance-based exercise networks, for example, reductionist studies have uncovered only the surface of these differences. A global omics approach is warranted to map how different gene, protein, metabolite, and lipid responses are regulated by aerobic, anaerobic, and concurrent exercise programs to maximize downstream responses and subsequent health benefits. Furthermore, deciphering how acute, transient molecular responses program downstream training adaptations is highly dependent on the duration and frequency of exercise intervention. Variations in exercise program timing, intensities, and training schedules are therefore required to capture how divergent biological networks are recruited and regulated.
Figure 2.
Omics and exercise schedule of events. The critical gaps in our understanding and range of variables associated with studying exercise in a global unbiased fashion are illustrated, which set the stage for the future exercise and omics field “schedule of events.” These events involve (A) exercise variables associated with establishing and comparing acute and chronic studies; (B) cellular and organ (e.g., skeletal muscle, adipose tissue, and heart) biological networks and the exercise-mediated cross talk between these pathways; (C) subject criteria considerations for studying human populations; and (D) subjects’ health, nutrition, training, and disease status that impact exercise biological networks. A detailed understanding of how each variable and network impacts systemic exercise responses will reveal how exercise interventions can be used to improve training adaptations and human health.
Another major variable associated with mapping the exercise responsome is the quantity of cellular/tissue networks and complicated cross-talk mechanisms (Fig. 2B) involved in systemic effects and health benefits. Tissues such as skeletal muscle, adipose, liver, pancreas, brain, heart, and the vascular system have both distinctive and redundant exercise-regulated pathways. Given the invasive nature of obtaining some human tissue samples, complementary mechanistic studies in cellular and animal models will be required to unravel these intricacies. The study of homeostatic regulatory control involving tissue cross talk is facilitated by the relative ease of obtaining periodic human blood samples across study time points. This secretome research area has tremendous discovery potential for omics approaches to identify novel secreted circulating factors that stimulate aspects of the exercise response.
In addition to these cellular/tissue networks and cross talk, it is important to note that exercise can induce dynamic remodeling in the cellular composition of tissues and vasculature that will affect data interpretation across multiple omics applications. For example, inflammation and tissue injury resulting from exercise can lead to dramatic changes in cellular composition involving infiltration of immune cells (e.g., monocytes/macrophages) to facilitate the regeneration and repair of injured skeletal muscle. Cell-type remodeling, therefore, represents a potential confounding factor of the exercise-induced responsome that must be considered when interpreting how tissues such as skeletal muscle adapt following exercise intervention. Subject criteria such as sex, age, and genetic variability (Fig. 2C) will also need to be carefully considered in omics study design and comparison. The majority of human studies obtaining skeletal muscle biopsies have enrolled only male subjects, and sex differences in muscle exercise-regulated pathways are poorly understood. The influence of sex on exercise and hormonal responses is an important variable understudied to date, as highlighted by sex-specific variations in resistance exercise training transcriptional network responses observed in skeletal muscle (Liu et al. 2010). More studies in female subjects, from mechanistic animal-based to global human-based studies, will be required to unravel sex differences in exercise networks. Moreover, exercise is beneficial in the prevention, delay, and/or treatment of disorders that become prevalent with aging. Omics studies addressing how molecular networks adapt with age to identify biomarkers associated with early-life intrauterine environmental exposures (Vrijheid et al. 2014; Smith and Ryckman 2015), growth and development (Radom-Aizik and Cooper 2016), and later-life frailty (Erusalimsky et al. 2016) have provided invaluable insights into critical targets contributing to the pathological consequences of environmental and aging processes. Genetic variability also affects how an individual of each sex and population will participate in and respond to exercise. Omics approaches will aid future analysis of complicated research questions such as the molecular bases for willingness to participate in voluntary exercise (Kelly et al. 2015). Large global omics efforts to characterize biological networks of both heterogeneous and specific populations around the world, such as those applied in the genomics of athletic performance, are warranted. Collectively, multicenter clinical trials addressing key variables of racial diversity, sex, and age will begin to tailor personalized exercise medicine programs and reap exercise’s full range of health benefits.
Subject nutritional, training, and health status (Fig. 2D) represent important hereditary and environmentally acquired exercise variables in human omics-based research projects. Several aspects of nutrigenomics and nutritional environment (e.g., diet composition and tissue nutrient availability) can affect molecular networks, exercise performance, and training adaptations. Microbiome composition may also emerge as an important biomarker of tissue exercise–environment network interactions. Given the potential of nutritional variables to modulate the exercise networks from several research tracks depicted in Figure 1, this important future area of omics approaches will pinpoint key nutrient-regulated molecular nodes within networks. These may represent feasible nutritional avenues for amplifying exercise-regulated signals, training adaptations, and health benefits in individuals across the spectrum of sedentary, intermittent, recreationally trained, athletic, and elite athlete status. Efforts to control diet and/or provide detailed study subject dietary monitoring, facilitated by cross-disciplinary collaborations with nutrition and dietetics experts, will better integrate omics data from multiple studies and trials. Furthermore, detailed reporting of exercise protocols and subjects’ fitness will help benchmark training adaptations across omics studies.
Central to the overall goal of linking exercise molecular networks to improved health outcomes is the consideration of how exercise networks and key molecular components are regulated and dysregulated in health and disease. Exercise is a promising therapy for metabolic diseases and disorders of aging such as sarcopenia. To effectively compare biological networks and identify therapeutically relevant targets in healthy versus diseased states, randomized controlled trials should use more omics-based sample analyses to uncover novel therapeutic opportunities for exercise stimuli to prevent, delay onset, and/or treat obesity and a range of associated chronic diseases. A catalog of epigenomic alterations in T2D, for example, will be useful in determining how genetic–environmental interactions contribute to disease and promote the therapeutic potential of exercise.
Taken together, global unbiased approaches have tremendous potential uses and applications in the area of personalized exercise prescription, especially as these omics datasets continue to accrue and become integrated. Compared with targeted reductionist approaches, omics-based discovery approaches can be used to map and identify not only known but also novel and low-abundance biological molecules. These may represent critical nodes of exercise biological networks that can be engaged by various modes, durations, and intensities. For example, a global signature of endurance versus resistance exercise adaptations in health and disease can be used to pinpoint biomarkers of training status and disease progression. Differences between individual signatures can then be integrated with genomic information and whole body physiological data to help provide biological explanations for variability in responsiveness to training, suitability for an individual to be performing a specific type of exercise, and the health benefits induced by a particular exercise regimen. Ultimately, mapping and interrogating exercise biological networks will help explain biology underlying targets that become dysregulated with disease and can be targeted with exercise intervention based on subject variables such as sex, age, and genetic makeup. The overall goal and application of these findings to personalized exercise medicine will be to identify the exercise program(s) that can elicit the maximal health benefits in the setting of each individual or subset of a population. In addition to just exercise interventions, mining these cellular/tissue biological network signatures may provide value in predicting predisposition and development of diseases, associated morbidities/complications, and responsiveness of biological network maps to potential future exercise-mimetic therapies.
Although these numerous exercise and subject variables should be taken into consideration from initial study design to data interpretation, an overall feasible solution is crucial to overcoming many of these critical gaps. Nutrition is one key variable of health status that is well appreciated to impact multiple layers of the exercise responsome from gene and protein expression/activity to metabolite and lipid levels. Nutrition status can be feasibly studied and intervened in subjects across age groups, sexes, and diverse populations. Interventions such as modulation of diet composition and tissue substrate availability are promising solutions to enhance performance, training adaptations, and overall health status in a variety of exercise modes and intensities. Therefore, well-controlled omics-based studies in the next 5 years and beyond should be dedicated to studying the impacts of nutrition status on multiple layers of exercise-regulated biological networks from the epigenome and transcriptome to the proteome/phosphoproteome, metabolome, and lipidome. These nutrition-focused omics efforts will help determine key biological nodes that can be targeted or amplified by diet to feasibly and realistically enhance the health-promoting effects of exercise.
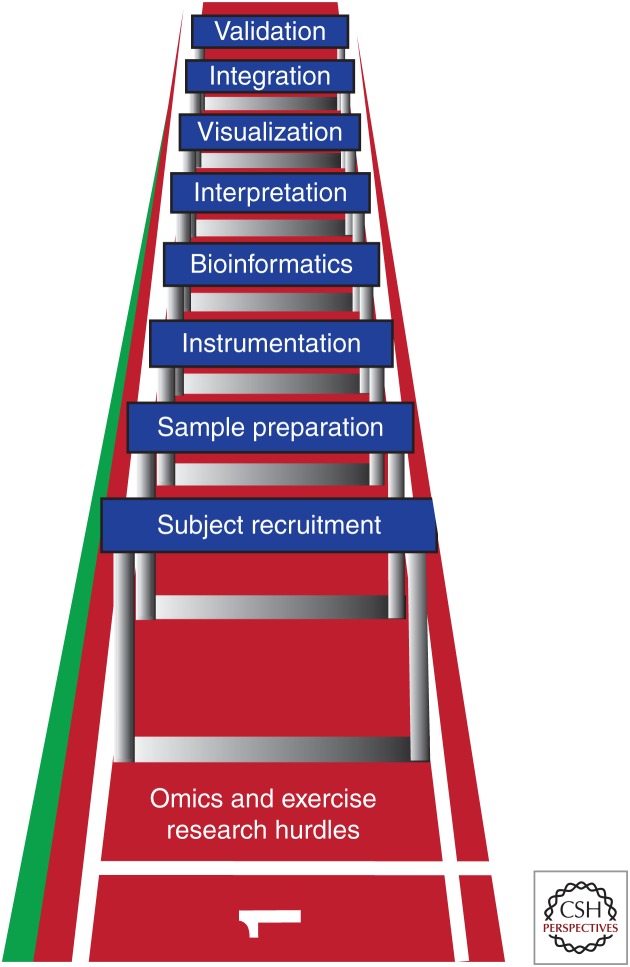
OMICS AND EXERCISE RESEARCH HURDLES
The renaissance of omics-based discovery approaches and integrating data across studies to unravel complex exercise networks will not be easily achieved with efforts from a single discipline. It will require cross-disciplinary efforts from exercise physiologists, geneticists, biochemists, clinical trial managers, computational biologists, bioinformaticists, and data visualization specialists to overcome significant hurdles associated with this emerging field. As depicted in Figure 3, several distinct hurdles associated with data collection, computation, and interpretation need to be overcome to develop the omics and exercise research tracks. Encouragingly, the establishment of key collaborations and consortia, as well as advancements in both technology and data analysis over the past 5 years highlighted below, has shown the feasibility of these global omics-based discovery approaches. These recent efforts have helped make significant progress at overcoming these hurdles and setting the stage for an exciting era of the omics and exercise field in the next 5 years and beyond.
Figure 3.
Omics and exercise research hurdles. The challenges, or “hurdles,” associated with the development of multidisciplinary omics and exercise research projects are illustrated. These hurdles are associated with project design and establishment (i.e., subject recruitment, sample preparation, and instrumentation), data analysis and presentation (i.e., bioinformatics, interpretation, and visualization), and advancement of these data in the context of previous findings and experimental interrogation of novel exercise-regulated biological targets (i.e., integration and validation).
First, subject recruitment and associated ethics approvals for large multicenter collaborative human studies will represent a major challenge to catalog variable exercise responses in healthy and diseased subjects from a spectrum of genetic backgrounds, ages, and training statuses. Expert consultation in sample preparation is another critical hurdle for optimal tissue- and fluid-collection timing, storage, and processing before omics analyses. Given the transient, intermittent nature of exercise responses, timing of sample collection is critical to ensure the molecular networks impacted by exercise are most efficiently preserved for downstream mapping. As discussed in Neufer et al. (2015), multicenter omics studies will benefit from standardized and accessible biological specimen-collection procedures to minimize study variability. Moreover, protocol access and consistency will facilitate collaborative applications of multiple omics platforms and dataset validation across centers.
Technological considerations regarding instrumentation and data-collection protocols from each research track depicted in Figure 1 are constantly evolving. Each omics methodology has a broad range of instruments, as well as sample preparation reagent and methodological considerations that can impact quality and robustness of datasets. Not every instrument or platform will be ideal for a given application or research question. Therefore, to overcome hurdles related to instrumentation, strategic cross-disciplinary collaborations will need to combine access to human subjects and samples with optimal omics instrumentation.
A detailed discussion of all the advantages and available technologies for each global omics application is out of the scope of this review. In summary, there are several major advantages of omics platforms (i.e., deep sequencing of the genome, epigenome, and transcriptome, and global MS-based analysis of proteins, PTMs, metabolites, and lipids) over more targeted approaches (i.e., polymerase chain reaction [PCR], microarray, immunoblotting, metabolite, and lipid extraction measurements). First, the unbiased discovery nature of omics approaches allows identification and quantification of both known and novel targets from a complex biological sample (e.g., both coding and noncoding RNAs using RNA sequencing). Second, the depth of omics coverage allows a significantly larger number of biological targets to be quantified. Compared with PCR, microarray, and immunoblotting procedures that measure single or relatively smaller subsets of biological targets, unbiased discovery approaches such as RNA sequencing and MS-based proteomics allow accurate identification and quantitative measures of thousands of RNAs and proteins/PTMs. The increasing capabilities of integrating large multi-omics datasets have tremendous discovery potential over linking only a few known biological targets. Finally, the dynamic range and sensitivity of omics approaches, which are now becoming cheaper and faster to use than ever before, also allow detection of lower abundance biological targets (e.g., rare transcripts and proteins). Together, the constantly improving unbiased, sensitive detection capabilities of omics approaches can help uncover novel intricacies and biomarkers underlying health and disease that were never capable before using only reductionist approaches. Research hurdles and current disadvantages associated with global omics approaches will be discussed further below and include limitations in accessing state-of-the-art instrumentation, increased cost of enlisting some omics platforms, and computational expertise required including trained bioinformatics personnel and software access for comprehensive data analysis.
One of the largest omics hurdles to overcome is access to computational and bioinformatics expertise for data interpretation and extraction of important biological targets. The interpretation of a dataset comprising thousands of molecular changes can be overwhelming for a reductionist biologist to distil and interpret. Establishment of interdisciplinary teams and collaborations will need to provide increased exposure to tools required for data computation, interpretation, and integration across omics platforms. Exposure to even basic principles of bioinformatics approaches used will ensure that appropriate statistical considerations are made from study design to completion. Moreover, visualizing large datasets in a clear interpretable fashion also represents a major challenge and has sparked a renaissance in how omics data and complex biological networks are portrayed. Another level of complexity is integration of datasets across multiple omics research tracks and physiological interventions to pinpoint key nodes of network regulatory control (Yugi et al. 2016). This is an especially fruitful area for determining how epigenomic, transcriptomic, and PTM program downstream tissue adaptations and major strides in cross-omics data integration have been made over the past 5 years. For example, integrated global analyses of DNA methylation and transcriptomics in skeletal muscle have permitted assessment of epigenomic contributions (Lindholm et al. 2014) and miRNA signatures (Rowlands et al. 2014) to exercise gene expression. Omics data integration will require sophisticated database curation, sharing, and searching capacities across institutions, such as recent efforts to integrate human muscle transcriptomics data across various exercise interventions (Vissing and Schjerling 2014). Given the breadth of experimental models required to validate targets, cross-species comparisons will also be required to compare orthologous omics networks. Although some genes and protein sequences are well-conserved among mammals, others will be more difficult to match and compare among large-scale datasets. Such cross-species comparisons of tissue gene expression and secretome data have led to the identification of a panel of novel candidate adipomyokines (Schering et al. 2015) and transcriptional signatures of endurance exercise training responsiveness (Keller et al. 2011). Development of accessible tools for cross-species database extraction is central to overcoming this hurdle. For example, the creation of PhosphOrtholog to align and compare protein sequences has helped integrate PTM-based omics data across species (Chaudhuri et al. 2015) and aided discovery of novel AMPK substrates from global phosphoproteomic datasets (Hoffman et al. 2015). More automated tools that mine current molecular interaction databases will also help link changes in exercise-regulated genes, proteins, PTMs, lipids, and metabolites.
Unbiased discovery approaches have potential to identify a wealth of novel exercise-regulated biological targets and network interactions. Although this review has stressed the power of such global approaches, complementary reductionist approaches are also required to provide mechanistic biological validation of novel targets. This final hurdle depicted in Figure 3 represents a major challenge based on the limited capacity and experimental models available to screen a large number of exercise targets in a physiologically relevant manner. Cellular and animal knockout and knockin models will be useful in pinpointing roles for novel molecules within exercise networks. However, being able to effectively “exercise” cells is challenging because of the absence of physiological components central to the exercise response (e.g., hormones, secreted factors, neural input) (Neufer et al. 2015). The development of standardized validated in vitro exercise models, such as electronically stimulated human muscle cell models to detect secreted proteins (Scheler et al. 2015) as well as experimental animal models and training protocols, will need to be standardized to foster target screening efforts. Given the multifaceted nature of the exercise response, this will be a major challenge to translate and target a subset of molecular “hits” found in exercise network omics screens to meaningful biological outcomes related to the health benefits of exercise.
CONCLUDING REMARKS
In conclusion, the complexity and interconnectedness of exercise biological networks will not be fully understood by studying single tissues or molecular targets alone. The advancing field of omics and exercise will map these networks in a holistic, unbiased, and integrated manner to identify how various cells and organs contribute to systemic adaptations in a range of exercise environments in health and disease. This promising era of exercise biology will continue to evolve with technological and computational advances to overcome significant hurdles associated with establishing and mining large datasets. As the field of omics and exercise continues to grow, new opportunities arise for exercise scientists to collaborate and unravel the intricate nature of exercise that have not been achievable without these global approaches. These cross-disciplinary research projects set the stage for future schedules of events to biologically validate candidate exercise-regulated targets and develop personalized strategies to mimic exercise and achieve its full potential of health benefits.
ACKNOWLEDGMENTS
This work is supported by an Australian University Catholic Research Funding (ACURF) Early Career Researcher Grant awarded to N.J.H. The author acknowledges that all publications from the rapidly advancing exercise and omics field could not be discussed because of space limitations and the primary focus on human studies.
Footnotes
Editors: Juleen R. Zierath, Michael J. Joyner, and John A. Hawley
Additional Perspectives on The Biology of Exercise available at www.perspectivesinmedicine.org
REFERENCES
- Al-Dabbagh S, McPhee JS, Murgatroyd C, Butler-Browne G, Stewart CE, Al-Shanti N. 2015. The lymphocyte secretome from young adults enhances skeletal muscle proliferation and migration, but effects are attenuated in the secretome of older adults. Physiol Rep 3: e12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeza J, Smallegan MJ, Denu JM. 2016. Mechanisms and dynamics of protein acetylation in mitochondria. Trends Biochem Sci 41: 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres R, Yan J, Egan B, Treebak JT, Rasmussen M, Fritz T, Caidahl K, Krook A, O’Gorman DJ, Zierath JR. 2012. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab 15: 405–411. [DOI] [PubMed] [Google Scholar]
- Bassini A, Cameron LC. 2014. Sportomics: Building a new concept in metabolic studies and exercise science. Biochem Biophys Res Commun 445: 708–716. [DOI] [PubMed] [Google Scholar]
- Bergman BC, Brozinick JT, Strauss A, Bacon S, Kerege A, Bui HH, Sanders P, Siddall P, Kuo MS, Perreault L. 2015. Serum sphingolipids: Relationships to insulin sensitivity and changes with exercise in humans. Am J Physiol Endocrinol Metab 309: E398–E408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman BC, Brozinick JT, Strauss A, Bacon S, Kerege A, Bui HH, Sanders P, Siddall P, Wei T, Thomas MK, et al. 2016. Muscle sphingolipids during rest and exercise: A C18:0 signature for insulin resistance in humans. Diabetologia 59: 785–798. [DOI] [PubMed] [Google Scholar]
- Bohm A, Hoffmann C, Irmler M, Schneeweiss P, Schnauder G, Sailer C, Schmid V, Hudemann J, Machann J, Schick F, et al. 2016. TGFβ contributes to impaired exercise response by suppression of mitochondrial key regulators in skeletal muscle. Diabetes 58: 2849–2861. [DOI] [PubMed] [Google Scholar]
- Bouchard C, Rankinen T, Timmons JA. 2011. Genomics and genetics in the biology of adaptation to exercise. Compr Physiol 1: 1603–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoire M, Mensink M, Kalkhoven E, Schrauwen P, Kersten S. 2014. Identification of human exercise-induced myokines using secretome analysis. Physiol Genomics 46: 256–267. [DOI] [PubMed] [Google Scholar]
- Chaudhuri R, Sadrieh A, Hoffman NJ, Parker BL, Humphrey SJ, Stockli J, Hill AP, James DE, Yang JY. 2015. PhosphOrtholog: A web-based tool for cross-species mapping of orthologous protein post-translational modifications. BMC Genomics 16: 617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen PM, Goodpaster BH. 2012. Role of intramyocellular lipids in human health. Trends Endocrinol Metab 23: 391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominetti O, Nunez Galindo A, Corthesy J, Oller Moreno S, Irincheeva I, Valsesia A, Astrup A, Saris WH, Hager J, Kussmann M, et al. 2016. Proteomic biomarker discovery in 1000 human plasma samples with mass spectrometry. J Proteome Res 15: 389–399. [DOI] [PubMed] [Google Scholar]
- Dias RG, Silva MS, Duarte NE, Bolani W, Alves CR, Junior JR, da Silva JL, de Oliveira PA, Alves GB, de Oliveira EM, et al. 2015. PBMCs express a transcriptome signature predictor of oxygen uptake responsiveness to endurance exercise training in men. Physiol Genomics 47: 13–23. [DOI] [PubMed] [Google Scholar]
- Edwards LM, Thiele I. 2013. Applying systems biology methods to the study of human physiology in extreme environments. Extrem Physiol Med 2: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erusalimsky JD, Grillari J, Grune T, Jansen-Duerr P, Lippi G, Sinclair AJ, Tegner J, Vina J, Durrance-Bagale A, Minambres R, et al. 2016. In search of “omics”-based biomarkers to predict risk of frailty and its consequences in older individuals: The FRAILOMIC Initiative. Gerontology 62: 182–190. [DOI] [PubMed] [Google Scholar]
- Fazelzadeh P, Hangelbroek RW, Tieland M, de Groot LC, Verdijk LB, van Loon LJ, Smilde AK, Alves RD, Vervoort J, Muller M, et al. 2016. The muscle metabolome differs between healthy and frail older adults. J Proteome Res 15: 499–509. [DOI] [PubMed] [Google Scholar]
- Ferreira R, Moreira-Goncalves D, Azevedo AL, Duarte JA, Amado F, Vitorino R. 2015. Unraveling the exercise-related proteome signature in heart. Basic Res Cardiol 110: 454. [DOI] [PubMed] [Google Scholar]
- Flowers E, Won GY, Fukuoka Y. 2015. MicroRNAs associated with exercise and diet: A systematic review. Physiol Genomics 47: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn EL, Piner LW, Huffman KM, Slentz CA, Elliot-Penry L, AbouAssi H, White PJ, Bain JR, Muehlbauer MJ, Ilkayeva OR, et al. 2015. Impact of combined resistance and aerobic exercise training on branched-chain amino acid turnover, glycine metabolism and insulin sensitivity in overweight humans. Diabetologia 58: 2324–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon PM, Liu D, Sartor MA, IglayReger HB, Pistilli EE, Gutmann L, Nader GA, Hoffman EP. 2012. Resistance exercise training influences skeletal muscle immune activation: A microarray analysis. J Appl Physiol (1985) 112: 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangelbroek RW, Fazelzadeh P, Tieland M, Boekschoten MV, Hooiveld GJ, van Duynhoven JP, Timmons JA, Verdijk LB, de Groot LC, van Loon LJ, et al. 2016. Expression of protocadherin γ in skeletal muscle tissue is associated with age and muscle weakness. J Cachexia Sarcopenia Muscle 7: 604–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JS, Zhao X, Irmler M, Liu X, Hoene M, Scheler M, Li Y, Beckers J, Hrabe de Angelis M, Haring HU, et al. 2015. Type 2 diabetes alters metabolic and transcriptional signatures of glucose and amino acid metabolism during exercise and recovery. Diabetologia 58: 1845–1854. [DOI] [PubMed] [Google Scholar]
- Hartwig S, Raschke S, Knebel B, Scheler M, Irmler M, Passlack W, Muller S, Hanisch FG, Franz T, Li X, et al. 2014. Secretome profiling of primary human skeletal muscle cells. Biochim Biophys Acta 1844: 1011–1017. [DOI] [PubMed] [Google Scholar]
- Hawley JA, Hargreaves M, Joyner MJ, Zierath JR. 2014. Integrative biology of exercise. Cell 159: 738–749. [DOI] [PubMed] [Google Scholar]
- Hoffman NJ, Parker BL, Chaudhuri R, Fisher-Wellman KH, Kleinert M, Humphrey SJ, Yang P, Holliday M, Trefely S, Fazakerley DJ, et al. 2015. Global phosphoproteomic analysis of human skeletal muscle reveals a network of exercise-regulated kinases and AMPK substrates. Cell Metab 22: 922–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooton K, Han W, Li L. 2016. Comprehensive and quantitative profiling of the human sweat submetabolome using high-performance chemical isotope labeling LC-MS. Anal Chem 88: 7378–7386. [DOI] [PubMed] [Google Scholar]
- Howlett KF, McGee SL. 2016. Epigenetic regulation of skeletal muscle metabolism. Clin Sci (Lond) 130: 1051–1063. [DOI] [PubMed] [Google Scholar]
- Huffman KM, Koves TR, Hubal MJ, Abouassi H, Beri N, Bateman LA, Stevens RD, Ilkayeva OR, Hoffman EP, Muoio DM, et al. 2014. Metabolite signatures of exercise training in human skeletal muscle relate to mitochondrial remodelling and cardiometabolic fitness. Diabetologia 57: 2282–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey SJ, James DE, Mann M. 2015. Protein phosphorylation: A major switch mechanism for metabolic regulation. Trends Endocrinol Metab 26: 676–687. [DOI] [PubMed] [Google Scholar]
- Hussey SE, Sharoff CG, Garnham A, Yi Z, Bowen BP, Mandarino LJ, Hargreaves M. 2013. Effect of exercise on the skeletal muscle proteome in patients with type 2 diabetes. Med Sci Sports Exerc 45: 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ML, Irving BA, Lanza IR, Vendelbo MH, Konopka AR, Robinson MM, Henderson GC, Klaus KA, Morse DM, Heppelmann C, et al. 2015. Differential effect of endurance training on mitochondrial protein damage, degradation, and acetylation in the context of aging. J Gerontol A Biol Sci Med Sci 70: 1386–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller P, Vollaard NB, Gustafsson T, Gallagher IJ, Sundberg CJ, Rankinen T, Britton SL, Bouchard C, Koch LG, Timmons JA. 2011. A transcriptional map of the impact of endurance exercise training on skeletal muscle phenotype. J Appl Physiol (1985) 110: 46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SA, Villena FP, Pomp D. 2015. The “omics” of voluntary exercise: Systems approaches to a complex phenotype. Trends Endocrinol Metab 26: 673–675. [DOI] [PubMed] [Google Scholar]
- King-Himmelreich TS, Schramm S, Wolters MC, Schmetzer J, Moser CV, Knothe C, Resch E, Peil J, Geisslinger G, Niederberger E. 2016. The impact of endurance exercise on global and AMPK gene-specific DNA methylation. Biochem Biophys Res Commun 474: 284–290. [DOI] [PubMed] [Google Scholar]
- Kuehnbaum NL, Gillen JB, Gibala MJ, Britz-McKibbin P. 2014. Personalized metabolomics for predicting glucose tolerance changes in sedentary women after high-intensity interval training. Sci Rep 4: 6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggate M, Carter WG, Evans MJ, Vennard RA, Sribala-Sundaram S, Nimmo MA. 2012. Determination of inflammatory and prominent proteomic changes in plasma and adipose tissue after high-intensity intermittent training in overweight and obese males. J Appl Physiol (1985) 112: 1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehr S, Hartwig S, Lamers D, Famulla S, Muller S, Hanisch FG, Cuvelier C, Ruige J, Eckardt K, Ouwens DM, et al. 2012. Identification and validation of novel adipokines released from primary human adipocytes. Mol Cell Proteomics 11: M111.010504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm ME, Marabita F, Gomez-Cabrero D, Rundqvist H, Ekstrom TJ, Tegner J, Sundberg CJ. 2014. An integrative analysis reveals coordinated reprogramming of the epigenome and the transcriptome in human skeletal muscle after training. Epigenetics 9: 1557–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Sartor MA, Nader GA, Gutmann L, Treutelaar MK, Pistilli EE, Iglayreger HB, Burant CF, Hoffman EP, Gordon PM. 2010. Skeletal muscle gene expression in response to resistance exercise: Sex specific regulation. BMC Genomics 11: 659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg TR, Fernandez-Gonzalo R, Tesch PA, Rullman E, Gustafsson T. 2016. Aerobic exercise augments muscle transcriptome profile of resistance exercise. Am J Physiol Regul Integr Comp Physiol 310: R1279–R1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustgarten MS, Price LL, Logvinenko T, Hatzis C, Padukone N, Reo NV, Phillips EM, Kirn D, Mills J, Fielding RA. 2013. Identification of serum analytes and metabolites associated with aerobic capacity. Eur J Appl Physiol 113: 1311–1320. [DOI] [PubMed] [Google Scholar]
- Markworth JF, Vella L, Lingard BS, Tull DL, Rupasinghe TW, Sinclair AJ, Maddipati KR, Cameron-Smith D. 2013. Human inflammatory and resolving lipid mediator responses to resistance exercise and ibuprofen treatment. Am J Physiol Regul Integr Comp Physiol 305: R1281–R1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markworth JF, Maddipati KR, Cameron-Smith D. 2016. Emerging roles of pro-resolving lipid mediators in immunological and adaptive responses to exercise-induced muscle injury. Exerc Immunol Rev 22: 110–134. [PubMed] [Google Scholar]
- McDonagh B, Sakellariou GK, Jackson MJ. 2014. Application of redox proteomics to skeletal muscle aging and exercise. Biochem Soc Trans 42: 965–970. [DOI] [PubMed] [Google Scholar]
- Mielke C, Lefort N, McLean CG, Cordova JM, Langlais PR, Bordner AJ, Te JA, Ozkan SB, Willis WT, Mandarino LJ. 2014. Adenine nucleotide translocase is acetylated in vivo in human muscle: Modeling predicts a decreased ADP affinity and altered control of oxidative phosphorylation. Biochemistry 53: 3817–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris C, O’Grada CM, Ryan MF, Gibney MJ, Roche HM, Gibney ER, Brennan L. 2015. Modulation of the lipidomic profile due to a lipid challenge and fitness level: A postprandial study. Lipids Health Dis 14: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munters LA, Loell I, Ossipova E, Raouf J, Dastmalchi M, Lindroos E, Chen YW, Esbjornsson M, Korotkova M, Alexanderson H, et al. 2016. Endurance exercise improves molecular pathways of aerobic metabolism in patients with myositis. Arthritis Rheumatol 68: 1738–1750. [DOI] [PubMed] [Google Scholar]
- Murgia M, Nagaraj N, Deshmukh AS, Zeiler M, Cancellara P, Moretti I, Reggiani C, Schiaffino S, Mann M. 2015. Single muscle fiber proteomics reveals unexpected mitochondrial specialization. EMBO Rep 16: 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer O, Sabapathy S, Ashton KJ, Desbrow B, Peake JM, Lazarus R, Wessner B, Cameron-Smith D, Wagner KH, Haseler LJ, et al. 2014. Time course-dependent changes in the transcriptome of human skeletal muscle during recovery from endurance exercise: from inflammation to adaptive remodeling. J Appl Physiol (1985) 116: 274–287. [DOI] [PubMed] [Google Scholar]
- Neufer PD, Bamman MM, Muoio DM, Bouchard C, Cooper DM, Goodpaster BH, Booth FW, Kohrt WM, Gerszten RE, Mattson MP, et al. 2015. Understanding the cellular and molecular mechanisms of physical activity-induced health benefits. Cell Metab 22: 4–11. [DOI] [PubMed] [Google Scholar]
- Newsom SA, Brozinick JT, Kiseljak-Vassiliades K, Strauss AN, Bacon SD, Kerege AA, Bui HH, Sanders P, Siddall P, Wei T, et al. 2016. Skeletal muscle phosphatidylcholine and phosphatidylethanolamine are related to insulin sensitivity and respond to acute exercise in humans. J Appl Physiol (1985) 120: 1355–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitert MD, Dayeh T, Volkov P, Elgzyri T, Hall E, Nilsson E, Yang BT, Lang S, Parikh H, Wessman Y, et al. 2012. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes 61: 3322–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norheim F, Raastad T, Thiede B, Rustan AC, Drevon CA, Haugen F. 2011. Proteomic identification of secreted proteins from human skeletal muscle cells and expression in response to strength training. Am J Physiol Endocrinol Metab 301: E1013–E1021. [DOI] [PubMed] [Google Scholar]
- Padrao AI, Ferreira R, Amado F, Vitorino R, Duarte JA. 2016. Uncovering the exercise-related proteome signature in skeletal muscle. Proteomics 16: 816–830. [DOI] [PubMed] [Google Scholar]
- Peake JM, Tan SJ, Markworth JF, Broadbent JA, Skinner TL, Cameron-Smith D. 2014. Metabolic and hormonal responses to isoenergetic high-intensity interval exercise and continuous moderate-intensity exercise. Am J Physiol Endocrinol Metab 307: E539–E552. [DOI] [PubMed] [Google Scholar]
- Pedersen BK, Febbraio MA. 2012. Muscles, exercise and obesity: Skeletal muscle as a secretory organ. Nat Rev Endocrinol 8: 457–465. [DOI] [PubMed] [Google Scholar]
- Petriz BA, Gomes CP, Almeida JA, de Oliveira GP Jr, Ribeiro FM, Pereira RW, Franco OL. 2016. The effects of acute and chronic exercise on skeletal muscle proteome. J Cell Physiol 232: 257–269. [DOI] [PubMed] [Google Scholar]
- Philp A, Rowland T, Perez-Schindler J, Schenk S. 2014. Understanding the acetylome: Translating targeted proteomics into meaningful physiology. Am J Physiol Cell Physiol 307: C763–C773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitsiladis YP, Tanaka M, Eynon N, Bouchard C, North KN, Williams AG, Collins M, Moran CN, Britton SL, Fuku N, et al. 2016. Athlome Project Consortium: A concerted effort to discover genomic and other “omic” markers of athletic performance. Physiol Genomics 48: 183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radom-Aizik S, Cooper DM. 2016. Bridging the gaps: The promise of omics studies in pediatric exercise research. Pediatr Exerc Sci 28: 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raue U, Trappe TA, Estrem ST, Qian HR, Helvering LM, Smith RC, Trappe S. 2012. Transcriptome signature of resistance exercise adaptations: Mixed muscle and fiber type specific profiles in young and old adults. J Appl Physiol (1985) 112: 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronn T, Volkov P, Davegardh C, Dayeh T, Hall E, Olsson AH, Nilsson E, Tornberg A, Dekker Nitert M, Eriksson KF, et al. 2013. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet 9: e1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronn T, Volkov P, Tornberg A, Elgzyri T, Hansson O, Eriksson KF, Groop L, Ling C. 2014. Extensive changes in the transcriptional profile of human adipose tissue including genes involved in oxidative phosphorylation after a 6-month exercise intervention. Acta Physiol (Oxf) 211: 188–200. [DOI] [PubMed] [Google Scholar]
- Rowlands DS, Thomson JS, Timmons BW, Raymond F, Fuerholz A, Mansourian R, Zwahlen MC, Metairon S, Glover E, Stellingwerff T, et al. 2011. Transcriptome and translational signaling following endurance exercise in trained skeletal muscle: Impact of dietary protein. Physiol Genomics 43: 1004–1020. [DOI] [PubMed] [Google Scholar]
- Rowlands DS, Page RA, Sukala WR, Giri M, Ghimbovschi SD, Hayat I, Cheema BS, Lys I, Leikis M, Sheard PW, et al. 2014. Multi-omic integrated networks connect DNA methylation and miRNA with skeletal muscle plasticity to chronic exercise in type 2 diabetic obesity. Physiol Genomics 46: 747–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlands DS, Nelson AR, Raymond F, Metairon S, Mansourian R, Clarke J, Stellingwerff T, Phillips SM. 2016. Protein-leucine ingestion activates a regenerative inflammo-myogenic transcriptome in skeletal muscle following intense endurance exercise. Physiol Genomics 48: 21–32. [DOI] [PubMed] [Google Scholar]
- Safdar A, Saleem A, Tarnopolsky MA. 2016. The potential of endurance exercise-derived exosomes to treat metabolic diseases. Nat Rev Endocrinol 12: 504–517. [DOI] [PubMed] [Google Scholar]
- Sarzynski MA, Davidsen PK, Sung YJ, Hesselink MK, Schrauwen P, Rice TK, Rao DC, Falciani F, Bouchard C. 2015. Genomic and transcriptomic predictors of triglyceride response to regular exercise. Br J Sports Med 49: 1524–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarzynski MA, Ghosh S, Bouchard C. 2016. Genomic and transcriptomic predictors of response levels to endurance exercise training. J Physiol 10.1113/JP272559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheler M, de Angelis MH, Al-Hasani H, Haring HU, Weigert C, Lehr S. 2015. Methods for proteomics-based analysis of the human muscle secretome using an in vitro exercise model. Methods Mol Biol 1295: 55–64. [DOI] [PubMed] [Google Scholar]
- Schering L, Hoene M, Kanzleiter T, Jahnert M, Wimmers K, Klaus S, Eckel J, Weigert C, Schurmann A, Maak S, et al. 2015. Identification of novel putative adipomyokines by a cross-species annotation of secretomes and expression profiles. Arch Physiol Biochem 121: 194–205. [DOI] [PubMed] [Google Scholar]
- Schild M, Ruhs A, Beiter T, Zugel M, Hudemann J, Reimer A, Krumholz-Wagner I, Wagner C, Keller J, Eder K, et al. 2015. Basal and exercise induced label-free quantitative protein profiling of m. vastus lateralis in trained and untrained individuals. J Proteomics 122: 119–132. [DOI] [PubMed] [Google Scholar]
- Sharples AP, Stewart CE, Seaborne RA. 2016. Does skeletal muscle have an “epi”-memory? The role of epigenetics in nutritional programming, metabolic disease, aging and exercise. Aging Cell 15: 603–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, Ryckman KK. 2015. Epigenetic and developmental influences on the risk of obesity, diabetes, and metabolic syndrome. Diabetes Metab Syndr Obes 8: 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Wang G, Pitsiladis YP. 2016. Advancing sports and exercise genomics: moving from hypothesis-driven single study approaches to large multi-omics collaborative science. Physiol Genomics 48: 173–174. [DOI] [PubMed] [Google Scholar]
- Vissing K, Schjerling P. 2014. Simplified data access on human skeletal muscle transcriptome responses to differentiated exercise. Sci Data 1: 140041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrijheid M, Slama R, Robinson O, Chatzi L, Coen M, van den Hazel P, Thomsen C, Wright J, Athersuch TJ, Avellana N, et al. 2014. The human early-life exposome (HELIX): Project rationale and design. Environ Health Perspect 122: 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner-Liebmann S, Tenori L, Mazzoleni A, Dieber-Rotheneder M, Konrad M, Hofmann P, Luchinat C, Turano P, Zatloukal K. 2016. Individual human metabolic phenotype analyzed by 1H NMR of saliva samples. J Proteome Res 15: 1787–1793. [DOI] [PubMed] [Google Scholar]
- Wang G, Tanaka M, Eynon N, North KN, Williams AG, Collins M, Moran CN, Britton SL, Fuku N, Ashley EA, et al. 2016. The future of genomic research in athletic performance and adaptation to training. Med Sport Sci 61: 55–67. [DOI] [PubMed] [Google Scholar]
- Weigert C, Lehmann R, Hartwig S, Lehr S. 2014. The secretome of the working human skeletal muscle—A promising opportunity to combat the metabolic disaster? Proteomics Clin Appl 8: 5–18. [DOI] [PubMed] [Google Scholar]
- Wu J, Gao Y. 2015. Physiological conditions can be reflected in human urine proteome and metabolome. Expert Rev Proteomics 12: 623–636. [DOI] [PubMed] [Google Scholar]
- Yugi K, Kubota H, Hatano A, Kuroda S. 2016. Trans-omics: How to reconstruct biochemical networks across multiple “omic” layers. Trends Biotechnol 34: 276–290. [DOI] [PubMed] [Google Scholar]
- Zierath JR, Wallberg-Henriksson H. 2015. Looking ahead perspective: Where will the future of exercise biology take us? Cell Metab 22: 25–30. [DOI] [PubMed] [Google Scholar]