Abstract
Each microRNA (miRNA) represses a web of target genes and, through them, controls multiple phenotypes. The difficulties inherent in such controls cast doubt on how effective miRNAs are in driving phenotypic changes. A “simple regulation” model posits “one target–one phenotype” control under which most targeting is nonfunctional. In an alternative “coordinate regulation” model, multiple targets are assumed to control the same phenotypes coherently, and most targeting is functional. Both models have some empirical support but pose different conceptual challenges. Here, we concurrently analyze multiple targets and phenotypes associated with the miRNA-310 family (miR310s) of Drosophila. Phenotypic rescue in the mir310s knockout background is achieved by promoter-directed RNA interference that restores wild-type expression. For one phenotype (eggshell morphology), we observed redundant regulation, hence rejecting “simple regulation” in favor of the “coordinate regulation” model. For other phenotypes (egg-hatching and male fertility), however, one gene shows full rescue, but three other rescues aggravate the phenotype. Overall, phenotypic controls by miR310s do not support either model. Like a thermostat that controls both heating and cooling elements to regulate temperature, redundancy and incoherence in regulation generally suggest some capacity in stability control. Our results therefore support the published view that miRNAs play a role in the canalization of transcriptome and, hence, phenotypes.
MicroRNAs (miRNAs) are a class of small RNAs, 22 nt long, that broadly and weakly repress gene expression in animals (Lai 2003; Bartel 2004, 2009). They are very abundant, are often highly conserved, and, unlike other regulatory genes such as transcription factors (TFs), are exclusive down-regulators of their direct targets (Chen and Rajewsky 2007; Hobert 2008; Martinez and Walhout 2009). The repression by miRNAs is puzzling as each miRNA targets a large number of genes, and naturally, the mean repression is weak. Even highly expressed miRNAs often repress many of their targets by <50% (Baek et al. 2008; Selbach et al. 2008; Eichhorn et al. 2014). The puzzle is further compounded by the existence of many miRNAs expressed at low levels.
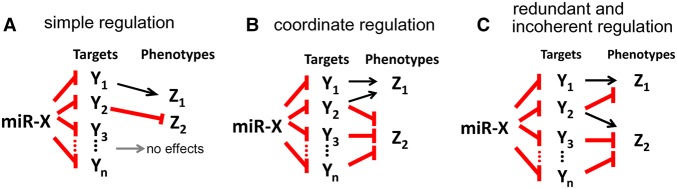
The puzzle of broad and weak target repression suggests to many investigators that miRNA targeting may often be biologically insignificant. An early model invokes “simple regulation,” which posits “one target gene–one phenotype” control. Despite subsequent challenges (e.g., Hunter et al. 2013), this simple model continues to receive recent support (Flynt and Lai 2008; Ecsedi et al. 2015; Pinzon et al. 2017). As illustrated in Figure 1A, a miRNA (miR-X) down-regulates many target genes (Y1, Y2, …, Yn), and the down-regulation leads to changes in a few phenotypes (Z1, Z2, etc). In this view, a major gene Y1 is principally responsible for the phenotype Z1 (Grosshans et al. 2005; Iovino et al. 2009; Bejarano et al. 2010; Smibert and Lai 2010; Ecsedi et al. 2015), and another target gene Y2 may be responsible for a phenotype Z2. Overall, for any miRNA, only a few targets are biologically relevant, and most others are noise (Seitz 2009; Pinzon et al. 2017). The view is tantamount to suggesting that >90% of the total repression capacity of the entire miRNA repertoire is noise (see Discussion).
Figure 1.
Models of phenotypic regulation by miRNA. (A) “Simple regulation”: A miRNA represses one target gene to control a phenotype. Different targets are responsible for different phenotypes. Red (blunt-headed) arrow indicates repression. (B) “Coordinated regulation”: A miRNA represses multiple targets, which in turn govern the same phenotype in the same direction. Pleiotropy, where a single target (e.g., Y2) influences more than one phenotype, is common. (C) In the “redundant and incoherent regulation” model, a miRNA affects each phenotype through multiple targets redundantly and often incoherently. Pleiotropy is also common.
In an alternative “coordinated regulation” hypothesis, multiple targets may be deployed to control the same phenotype in a coordinated manner (Subramanyam et al. 2011; Lai 2015; Cicek et al. 2016; Mahmoudi and Cairns 2017). In Figure 1B, targeting via both Y1 and Y2 enhances phenotype Z1, while Y2, Y3, and Yn targeting reduces phenotype Z2. These actions may be additive but could also be redundant if each target by itself can fully rescue the phenotype. Redundant control has been suggested to be central to miRNA function (Tan et al. 2012; Lee and Hyun 2014) and could account for the large number of target genes. Such a control circuit, however, could be difficult to assemble since all target genes are required to influence the phenotypes in the same direction.
While many studies have supported the “simple regulation” hypothesis by finding a single gene capable of rescuing the phenotype of interest (Karres et al. 2007; Hyun et al. 2009), few have ruled out redundancy, by which other target genes may also regulate the same phenotype to a similar extent. Curiously, in studies that deal with the same miRNA (Let-7) and the same phenotype (vulva rupture), opposite conclusions were reached (Hunter et al. 2013; Ecsedi et al. 2015). To rigorously test the models, it will be necessary to analyze each target gene individually and restore each expression to the wild-type level. Several phenotypes should also be examined simultaneously. Here, we study the miRNA-310 cluster (miR-310/311/312/313, denoted as miR310s here) of Drosophila. Because the four members of miR310s share the same seed (see Fig. 2A; Supplemental Fig. S1; Supplemental Text S4) and are clustered, they are often studied as a single functional unit (Tang et al. 2010; Tsurudome et al. 2010; Pancratov et al. 2013; Cicek et al. 2016). Cross-species rescues using Drosophila pseudoobscura miR310s are also carried out to probe functional conservation.
Figure 2.
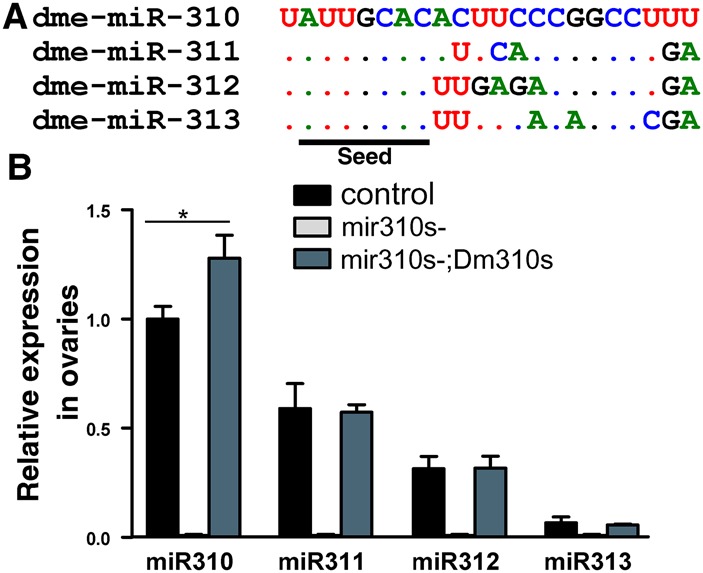
Sequence and expression of miR310s. (A) Mature sequences of the miR310 cluster in Drosophila melanogaster (Dm310s). The four members share a conserved seed (underlined). (B) Relative expression of miR310s in ovaries. mir310s- denotes the knockout line in which no expression is detectable. The miR310s cluster is reintroduced in the mir310s-;UAS-Dm310s line in which miR310s is driven by the UAS element. Assays were taken by qRT-PCR in triplicate using 2S RNA as the endogenous control. Error bars, SEs by Student's t-test. (*) P < 0.05.
Results
The criteria for identifying miRNA targets responsible for a phenotype are as follows (see Fig. 1A,B): First, the phenotype Z is associated with the deletion of miRNA-X. Second, a set of target genes, including gene Y, is shown to be up-regulated in the deletion background. Third, restoring the expression of Y to its wild-type pattern results in the rescue of phenotype Z in the miR-X deletion background. If the phenotypic rescue is complete, gene Y is often declared the major gene for the control of Z. Strictly speaking, it is necessary to rule out other genes having a comparable phenotypic effect, but this is rarely done (see Discussion).
Defects in egg morphology, hatchability, and male fertility
The mir310s deletions were generated in Drosophila melanogaster in our earlier study (Tang et al. 2010). Quantitative real-time PCR (qRT-PCR) measurements show that the knockout lines (mir310s-) lack all miR310s expression (Fig. 2B; Supplemental Fig. S3). Since miR310s are most highly expressed in the ovaries and testes (Bejarano et al. 2012; Marco 2014), eggs and sperm are the tissues we focused on in this study.
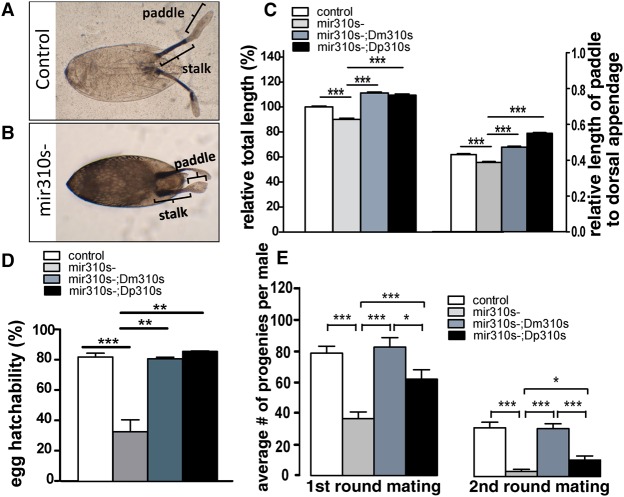
First, we note that the dorsal appendages attached to the eggshell for respiration (Suzanne et al. 2001; Berg 2008) are defective in the mir310s deletion background. The total length of dorsal appendages is reduced by ∼10% in the eggs of mir310s- flies (Fig. 3A,B). The fraction of the total length occupied by the paddle region is 43.2 ± 0.4% in the control and 38.8 ± 0.5% in the mir310s- eggs (Fig. 3C). Second, we measured egg hatchability, which is ∼32% in mir310s- eggs but 81.6% in the control (Fig. 3D). The deformity in the dorsal appendage appears to reflect functional differences inside the eggs.
Figure 3.
Three phenotypic defects in the mir310s- flies rescued by Dm310s or Dp310s. The reintroduced mir310s of either D. melanogaster (Dm310s) or D. pseudoobscura (Dp310s) are used. (A,B) The dorsal appendage of eggshell consists of a stalk and a paddle. Note that the paddle of the mir310s- egg is flattened and shortened. (C) Loss of miR310s significantly shortened the dorsal appendage (left), especially the paddle (right). Either Dm310s or Dp310s rescued the phenotypes (more than 200 eggs examined for each genotype). (D) Loss of miR310s significantly decreased egg hatchability, which could also be fully rescued by Dm310s or Dp310s. (Five replicates, each with more than 100 eggs for each genotype.) (E) Average number of progeny produced by each male in two consecutive matings. Mean progeny count of mir310s- males was reduced by >50% in the first round of mating and 87% in the second round. Dm310s fully restored the fertility, but Dp310s rescue is partial. More than 15 males were individually tested for each genotype; error bars, SEs. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001 by Student's t-test.
Third, for male reproduction, since there is no visible morphological defect, male fertility was measured at two levels of sperm exhaustion by mating males to two batches of females in succession. Since sperm is stored in seminal vesicles, males with a reduced capacity in sperm production would have a lower fertility count only under sperm-depletion conditions (Wu 1983). In mir310s- males, the progeny count was reduced by half in the first round (78 progeny per male vs. 36) and by 87% in the second (30 vs. four) (Fig. 3E).
We then reintroduced mir310s into the mir310s- background to rescue the phenotypes. The construct (Dm310s, for mir310s of D. melanogaster) has been described earlier (Tang et al. 2010). The transgenic line expresses all members of miR310s at a level close to the wild type (Fig. 2B; Supplemental Fig. S3) and exhibits full phenotypic rescue (Fig. 3C–E). These observations confirm that the observed phenotypes are due to the mir310s knockout.
Target identification and the selection of five genes: Dl, E2f2, EcR, Mad, and Mef2
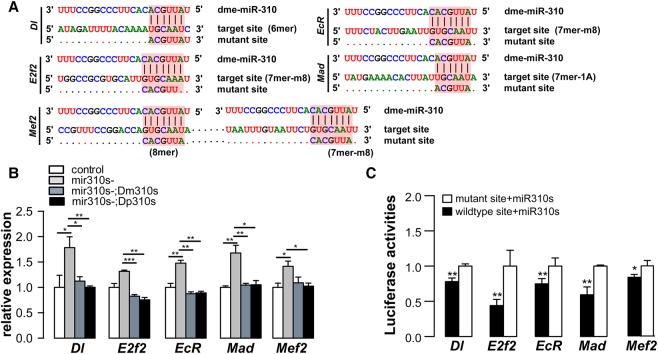
We next aimed to identify target genes through which miR310s exert their phenotypic effects. We predicted miR310s targets using TargetScan, PicTar, and PITA (Grun et al. 2005; Lewis et al. 2005; Kertesz et al. 2007; Kheradpour et al. 2007; Ruby et al. 2007) and selected 47 potential targets for qRT-PCR validation (for details, see Methods and Supplemental Fig. S4). These genes were chosen in consideration of their possible roles in dorsal appendage formation, follicle cell development, and male fertility. In comparison with the control flies, most of the 47 predicted targets are up-regulated during early and late oogenesis in mir310s- females (see Supplemental Fig. S5). We further selected candidate target genes for in vitro validation (for details, see Supplement Text S1). Five genes—Dl, E2f2, EcR, Mad, and Mef2—were chosen for their possible functional involvement (see Methods; Supplemental Text S1), but they merely represent a small sample of the total target pool. All of these genes contain conserved miR310s binding sites in their 3′ UTRs (Fig. 4A). They are overexpressed in mir310s- flies and are repressed in the [mir310s-; Dm310s] and [mir310s-;Dp310s] lines (Fig. 4B).
Figure 4.
Validation of direct targets of miR310s by in vitro mutagenesis. (A) Putative target sites of miR310 on the 3′ UTRs of Dl, E2f2, EcR, Mad, and Mef2. miR310 seed region, target sites, and mutant sites were shaded in red. The mutant site of each gene contained altered base pairs, which disrupt pairing with miR310s. The 3′ UTR of Mef2 has two target sites, both of which were mutated. (B) The expressions of the five target genes are de-repressed in mir310s- flies (in ovaries of 3-d-old females) and repressed again by either Dm310s or Dp310s. (Relative expression were measured by qRT-PCR in triplicate, using rp49 as endogenous control.) (C) The luciferase reporter with wild-type miR310s target sites is significantly inhibited by the cotransfected mir310s vis-à-vis those carrying mutated target sites. Five replicates in Drosophila S2 cells. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001 by Student's t-test.
To stringently confirm the expected repression by miR310s, we cloned sequences with either the wild-type miR310s target site or the mutated site from each of the five genes (see Methods; Fig. 4A). Each of the cloned sequences carrying a luciferase reporter was then transfected into Drosophila S2 cells for the reporter assay (see Methods). The comparisons of luciferase activity in transfected cells are given in Figure 4C. Reporter activity is higher in cells with mutated target sites for each of the five genes, confirming the direct repression by miR310s through the target sites shown in Figure 4A.
Restoring wild-type expression by promoter-driven RNAi
Expression of target genes increases in mir310s knockout lines. As noted before, the restoration of the expression of a major target gene to its wild-type pattern should fully rescue the phenotype. We first established a Gal4 driver under the control of the mir310s promoter. This Gal4 construct drives UAS-RNAi in the same tissues as the native miR310s. We then use the Gal4/UAS-RNAi construct to repress each of the five verified target genes in mir310s- flies. Measured by qRT-PCR, all five genes were significantly repressed by RNAi in ovaries (Fig. 5A).
Figure 5.
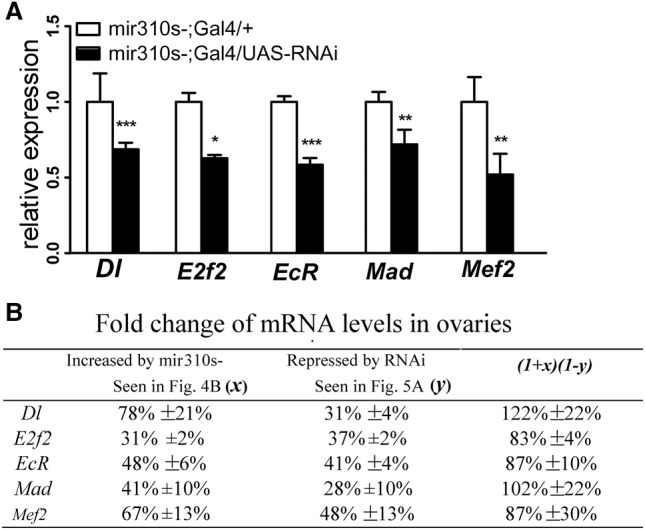
Restoration of target gene expression by RNAi in the mir310s- line. (A) Relative expression of the five targets is measured by qRT-PCR in the ovaries of 3-d-old females. In Gal4/UAS-RNAi flies, genes that are de-repressed in the mir310s- line are repressed by RNAi. Assays were taken in triplicate. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001 by Student's t-test. (B) The restoration of target gene expression in the mir310s- line by RNAi. The second column shows the up-regulation of the five targets due to mir310s knockout, and the third column shows the down-regulation due to RNAi of individual targets. The last column gives the restored level relative to the wild-type expression. Note that the tissue distribution of RNAi is under the control of a Gal4, which is itself driven by the mir310s promoter.
To see whether expression of these target genes has been restored to the wild-type level, we combine expression increase due to mir310s deletion and its decrease due to RNAi in ovaries. As summarized in Figure 5B, each target gene is up-regulated by a percentage in mir310s- ovaries (x; second column), and then down-regulated by a percentage by RNAi (y; third column). The restored value relative to the wild type would be (1 + x) (1 − y), as shown in the last column. The observations fall in the range of 83%–122% for the five genes. The expression rescue is then used for evaluating the phenotypic effect of each target gene.
Phenotypic effects of individual gene expression rescue
With the expression rescue confirmed, we can now address miRNA effects on phenotypes. We follow the common approach, which rescues the phenotypes in the miRNA knockout background (e.g., Hunter et al. 2013). Another approach is to carry out the rescue in the wild-type background (e.g., Ecsedi et al. 2015). The appropriate strategy depends on the genetic interactions underlying the phenotypes. If knocking out a miRNA would result in a phenotype through two target genes independently as shown in Table 1A, then the basis of comparison should be the wild type. On the other hand, if the knockout phenotype requires the epistatic actions of two target genes as shown in Table 1B, the comparison should be done in the knockout background. In reference to Table 1, A and B, it is likely that one approach may reveal more genes for the same phenotype than the other, and the two sets of targets may not overlap. Since strong epistasis requiring mutations at multiple loci to yield a phenotype is common in evolution (Palopoli and Wu 1994; Sun et al. 2004), the approach based on Table 1B is appropriate. The considerations above also dictate the methodology suitable for testing the models of Figure 1. In Supplemental Text S3, a detailed discussion of competing methods is presented.
Table 1.
Modes of target gene interactions

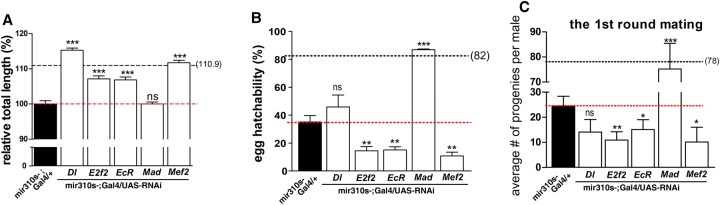
The phenotypic rescues are shown in Figure 6, A through C, where red lines indicate the mir310s- phenotype (0% rescue) and black lines reflect the wild-type values, or 100% rescue. For the dorsal appendage, RNAi experiments show strong rescue by restoring expression of four of the five genes (Fig. 6A). Dorsal appendage length is reduced by 10% in mir310s- lines. Dl and Mef2 RNAis rescue the phenotypes by 15% and 12%, respectively. Similarly, E2f2 and EcR rescue the phenotype by ∼7%. Hence, all four genes can be considered “major genes” with 0.7-, 1.2-, and 1.5-fold rescue (P << 10−3 for all; for detail, see legends), and miR310s regulate the dorsal appendage through several target genes redundantly (Fig. 6A; Supplemental Fig. S7).
Figure 6.
Phenotypic rescue by restoring the expression of individual target genes. The red dotted line, indicating the phenotypic value of mir310s- flies, demarcates positive and negative rescue. The black line indicates the wild-type value, hence denoting full rescue. (A) Dorsal appendages: Four genes with their expression restored by RNAi rescue the phenotypic defect. More than 200 eggs were examined for each genotype. (B) Egg hatchability: Only one gene (Mad) shows full rescue, and a second one (Dl) shows partial but insignificant rescue. The remaining three genes (E2f2, EcR, and Mef2) show negative rescue. (C) Male fertility (progeny count for males in the first round of mating). Only one gene (Mad) shows full rescue. The rest show negative rescue, which is statistically significant for three genes (E2f2, EcR, and Mef2) and insignificant for one (Dl). More than 15 individual males were tested for each genotype. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001 by Student's t-test.
Incoherent control of egg hatchability and male fertility
Mad RNAi could restore the hatchability defect of mir310s knockout to the wild-type level, suggesting that this is the single major gene in the conventional sense. In addition, Dl RNAi increases hatchability by 30%, but the variability is too large for the result to be statistically significant (Fig. 6B). The remaining three genes are intriguing. RNAi experiments not only fail to show rescue but in fact aggravate the phenotype below the red dashed line by 58% (E2f2, P = 0.007), 57% (EcR, P = 0.0082), and 69% (Mef2, P = 0.0032), respectively. We note that the dorsal appendages and hatchability both pertain to the properties of eggs but have divergent genetic interactions.
The control of male fertility by miR310s is similar to that of egg hatchability. Mad RNAi males produced 75.2 progeny (Fig. 6C), very close to the number of wild-type males (78.0) (Fig. 3E). Since another miR310s target, Arm (armadillo), has been shown to be a major gene in previous studies (Pancratov et al. 2013), there is redundancy for male fertility. The reverse rescue observed in Figure 6B is frequently observed in Figure 6C with the exception of Dl RNAi, which goes in the opposite direction for the two phenotypes (although the differences are not statistically significant in either direction). RNAi experiments aggravate the phenotype by 55% (E2f2, P = 0.006), 38% (EcR, P = 0.05), and 58% (Mef2, P = 0.03), respectively.
Discussion
Many previous studies supporting the “simple regulation” model of Figure 1A reach the conclusion by identifying a target gene with near 100% rescue (Karres et al. 2007; Varghese and Cohen 2007; Hyun et al. 2009; Luo and Sehgal 2012). Most did not test other genes as redundancy is assumed to be absent. The issue of redundant control is unsettled even for the same miRNA (let -7) and the same phenotype (vulva rupture) in the same species (C. elegans) (Hunter et al. 2013; Ecsedi et al. 2015; Rausch et al. 2015). Hunter et al. (2013) concluded that a large number of target genes (more than 20) participate in vulva regulation using RNAi rescue for a large panel of target genes. In contrast, Ecsedi et al. (2015) concluded that LIN-41 is the sole target of let-7 driving the vulval rupture phenotype. The stark contradiction is usually explained in technical terms, which are summarized in the Supplement Text S2. Briefly, the RNAi experiments (Hunter et al. 2013) may not have restored wild-type expression, and the “double mutant” assay (Ecsedi et al. 2015) effectively introduced an artificial miRNA for rescue. Either approach might be interpreted to be flawed, but as noted in Table 1, A and B, they may both be correct. Rescue experiments in the wild type (Ecsedi et al. 2015) versus miRNA knockout background (Hunter et al. 2013) may yield complementary, rather than overlapping, results.
To our knowledge, ours may be the first study that restores the expression of each of many target genes to (near) wild-type patterns. Every miRNA phenotype analyzed in this study is indeed governed by multiple target genes, thus supporting the conclusions of previous reports (Grosshans et al. 2005; Subramanyam et al. 2011; Hunter et al. 2013; Lee and Hyun 2014). The claim of the “simple regulation” hypothesis that miRNAs in animals, like those in plants, have few target genes connected to phenotypes is unsupported by these studies. Simple regulation essentially means that the bulk, rather than a small fraction, of miRNA repressions are biologically irrelevant (Pinzon et al. 2017).
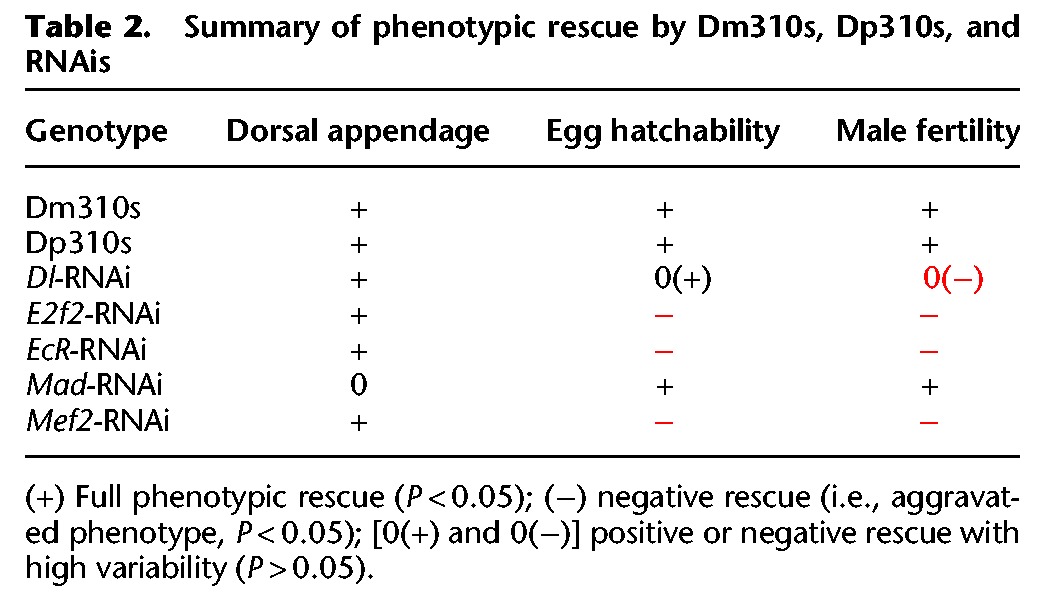
The question is then whether miRNAs evolve gene targeting that coherently governs the phenotypes. In this study, the control is incoherent for two of the three phenotypes (Fig. 6B,C; Supplemental Fig. S8). Since only five targets genes are analyzed, all phenotypes could be incoherently regulated if more target genes were assayed. Such incoherent control is also evident in the results presented by Hunter et al. (2013) (their Fig. 4). The pleiotropic effects of target genes further complicate the task of coherent control. For example, RNAis of E2f2, EcR, and Mef2 rescue dorsal appendages positively but rescue egg hatchability negatively (see Table 2), even though both phenotypes pertain to female fertility.
Table 2.
Summary of phenotypic rescue by Dm310s, Dp310s, and RNAis
With redundancy and, especially, incoherence, miR310s’ actions in driving phenotypic changes seem to be self-canceling (Figs. 1C, 6). Available data from other miRNAs suggest that this conclusion may be general (Subramanyam et al. 2011; Hunter et al. 2013). When a regulator controls the output via two mutually opposing pathways, as in an incoherent feed-forward loop (Hornstein and Shomron 2006; Wu et al. 2009), the control is often indicative of stability and homeostasis (von Dassow et al. 2000; Tsang et al. 2007; Li et al. 2009; Wu et al. 2009). Regulation of the stabilizing kind is further interpreted to manifest stasis in phenotypes. Waddington (1942) first proposed the existence of mechanisms that “canalize” development (Waddington 1942), which are compared with water traveling in defined canals. The molecular basis of canalization has been suggested to be heat shock proteins (Rutherford and Lindquist 1998) and miRNAs, respectively, at the level of proteins and transcripts. The literature on miRNAs’ possible roles in canalization has been quite extensive (Hornstein and Shomron 2006; Martinez et al. 2008; Li et al. 2009; Peterson et al. 2009; Wu et al. 2009; Ebert and Sharp 2012; Gursky et al. 2012; Cassidy et al. 2013; Posadas and Carthew 2014).
The view of miRNAs being the elusive canalizing agent is generally based on the analysis of motifs with two to four nodes (Hornstein and Shomron 2006; Wu et al. 2009). Recently, the analysis of small motifs has been extended to larger gene regulatory networks (GRNs). An interesting and intuitive model is the network of RNA cross-talks (Salmena et al. 2011; Ebert and Sharp 2012; Tay et al. 2014; Thomson and Dinger 2016). In a companion study (Chen et al. 2017), we apply the May-Wigner theory of network stability (May 1972, 2001; Wu et al. 2009; Allesina and Tang 2012; Tang et al. 2014), commonly used in the study of foodweb stability, to GRN stability.
Either at the motif or network level, the canalization view is about miRNAs’ ability to stabilize the output. In this capacity, miRNAs’ function depends on the connectivity and distribution of interaction strength among nodes. For that reason, the edges (i.e., connections between nodes) may often be exchangeable in evolution (Wu et al. 2009; Lyu et al. 2014). Indeed, when Dm310s and Dp310s are inserted into D. melanogaster, 40% of their targets do not overlap (Tang et al. 2010). Nevertheless, the observed phenotypic rescues by Dp310s are comparable with those by Dm310s (Fig. 3C–E), suggesting a level of functional conservation associated with substantial divergence in targeting.
In conclusion, miRNAs are often part of motifs such as incoherent feed-forward loops that stabilize expression output (Tsang et al. 2007; Osella et al. 2011). The observations on hatchability and male fertility (Fig. 6B,C; for summary, see Table 2) support a generalized model of incoherent feed-forward (Fig. 1C). Furthermore, miRNAs as a whole are abundant and stable relative to their target mRNAs (Miska et al. 2007; Alvarez-Saavedra and Horvitz 2010; Song et al. 2014; Sun et al. 2015). All these characteristics suggest a role in maintaining homeostasis (Wu et al. 2009; Herranz and Cohen 2010; Vidigal and Ventura 2015).
Methods
Fly stocks and genetics
The mir310s knockout strain of D. melanogaster (w1118; mir310s-; denoted as mir310s- here) through P-element excision and the knockout control strain of precise P-element excision was obtained as previously described (Tang et al. 2010). To rescue the mir310s knockout, w1118 flies were transformed with pUAST vector expressing mir310s from D. melanogaster (denoted as Dm310s) or D. pseudoobscura (denoted as Dp310s) as previously described (Tang et al. 2010). The resultant flies with genotype w1118; UAS-Dm310s or w1118; UAS-Dp310s and the mir310s- flies were crossed with w1118; Cyo/Sp; TM3,Ser/Sb, following the cross scheme showed in Supplemental Figure S2A, to obtain the rescue strains of w1118; mir310s-; UAS-mir310s (also denoted as mir310s-; Dm310s and mir310s-; Dp310s, respectively).
The GAL4/UAS-RNAi system was used to rescue individual targets in the mir310s- background. To capture the native expression pattern of miR310s in D. melanogaster, we constructed the mir310s-Gal4 strain by transforming the putative promoter region of mir310s into w1118 flies. A ∼2.0-kb fragment upstream of mir310s was amplified from the genomic DNA of w1118 with forward primer 5′-TTAGATCTGAGCAGCAATTGTCGCAGTA-3′ and reverse primer 5′-TGGAATTCGGCTAGAACGATGTCGGGTA-3′, and the amplicon was cloned into BglII and EcoRI sites of pCaSpeR-Gal4 vector after gel purification and restriction digestion. The germline transformation of w1118 flies was conducted using a pTURBO (Flybase. ID:FBmc0002087) helper plasmid, and standard procedures (Rubin and Spradling 1982) finally obtained the flies with genotype w1118; mir310s-Gal4. Five UAS-RNAi strains of the miR310s targets were used, including UAS-Dl-RNAi (Bloomington Stock Center, hereafter, stock no. 28032, genotype: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02867}attP2), UAS-E2f2-RNAi (stock no. 36674, genotype: y1 sc* v1; P{y[+t7.7] v[+t1.8]=TRiP.HMS01562}attP2), UAS-EcR-RNAi (stock no. 29374, genotype: y1 v1; P{y[+t7.7] v[+t1.8]=TRiP.JF02538}attP2), UAS-Mad-RNAi (stock no. 35648, genotype: y1 sc* v1; P{y[+t7.7] v[+t1.8]=TRiP.GLV21013}attP2/TM3, Sb), and UAS-Mef2-RNAi (stock no. 28699, genotype: y1 v1; P{y[+t7.7] v[+t1.8]=TRiP.JF03115}attP2). The UAS-RNAi (y1sc*v1; +; UAS-RNAi), mir310s-Gal4 (w1118; mir310s-Gal4), and mir310s- (w1118; mir310s-) flies were crossed with w1118; Cyo/Sp; TM3,Ser/Sb, following the cross scheme showed in Supplemental Figure S2B to obtain the target rescue strains of mir310s-;mir310s-Gal4/UAS-RNAi and the control strain of mir310s-;mir310s-Gal4/+. All flies were reared on cornmeal-sucrose-yeast medium at 25°C and a 12-h light–dark cycle.
Egg dorsal appendage, hatchability, and male fertility assays
We measured the length of egg dorsal appendages, hatchability, and male fertility for the mir310s knockout, control, and rescue flies. For the length of dorsal appendages, eggs laid within 1-h time frame were collected, aligned on cover slips, and examined under a Leica DMI4000B microscope (Leica Microsystems). Pictures of dorsal appendages were analyzed using the Leica Application Suite (version 3.8.0) to measure the length of dorsal appendages for the paddle and stalk region separately. Dorsal appendages of more than 200 eggs were measured for each genotype. For egg hatchability, seven pairs of virgin females and males were allowed to mate for 24 h in an egg-laying vial, and then the grape juice plate was removed and replaced with a fresh plate. Five vials were set up for each genotype. After 12 h, the eggs laid on the fresh plate were collected and kept in 25°C to allow hatching for 24 h. Hatchability was calculated as the mean proportion of the number of hatched eggs relative to the total number of collected eggs across the five replicates. For male fertility, each 5-d-old male was mated with five 3- to 5-d-old virgin females of w1118 for 48 h and then transferred to a new vial to mate with another five virgin females for 48 h. More than 15 males were examined for each genotype. Male fertility was measured as the average number of progenies per male in each of the two consecutive rounds of mating. A two-tailed t-test was used to determine the statistical significance of phenotypic differences between genotypes.
Quantitative miRNA and mRNA analysis by qRT-PCR
We measured the relative expression levels of individual miR310s members in the strains of mir310s-, the knockout control, and mir310s-;UAS-mir310s using qRT-PCR. Total RNA was extracted from ovaries or testes of 3-d-old flies using TRIzol reagent (Thermo Fisher Scientific, catalog no. 15596026) and subject to the removal of genomic DNA using TURBO DNA-free Kit (Thermo Fisher Scientific, catalog no. AM1907) following the manufacturer's protocols. qRT-PCR of miRNAs was conducted using stem-loop reverse transcription (Chen et al. 2005) followed by TaqMan PCR analysis using the miRNA UPL (Roche Diagnostics) probe assay protocol (Varkonyi-Gasic et al. 2007; He et al. 2016). Three biological replicates were conducted for each genotype, and 2S RNA was used as an endogenous control (primers are listed in Supplemental Table S1).
Total RNA was extracted from ovaries at the early and late stages of oogenesis for qRT-PCR analyses of the 47 predicted target genes or from ovaries of 3-d-old females for the analysis of the five targets involved in UAS-RNAi with TRIzol (Thermo Fisher Scientific, catalog no. 15596026). To collect samples of early oogenesis, young females (<6 h after eclosion) were dissected to collect ovaries, which only contain egg chambers before stage 5. To collect samples of late oogenesis, 3-d-old virgin females were dissected in FBS solution, and egg chambers during stage 9–12 were collected. Total RNA was treated with a TURBO DNA-free kit (Thermo Fisher Scientific, catalog no. AM1907) and reverse-transcribed into cDNA with a PrimeScript first strand cDNA synthesis kit (TAKARA Bio, catalog no. 6110A). qPCR was performed using a SYBR Premix Ex Taq II kit (TAKARA Bio, catalog no. RR82WR) following the manufacturer's instructions. The housekeeping gene rp49 was used as an endogenous control. The experiments were performed in triplicates. Relative expression levels of miRNAs or target genes were obtained by the 2−ΔΔCT method (Livak and Schmittgen 2001) and were statistically analyzed using the two-tailed Student's t-test. All primers used in qRT-PCR were listed in Supplemental Table S3.
Target prediction and identification of functionally relevant targets
To select miR310s candidate targets for experimental validation, targets in D. melanogaster were predicted using three computational tools (Grun et al. 2005; Lewis et al. 2005; Kertesz et al. 2007; Kheradpour et al. 2007; Ruby et al. 2007): (1) TargetScan (TargetScanFly release 6.2, 2012 (the prediction was download from the webpage http://www.targetscan.org/cgi-bin/targetscan/fly_12/targetscan.cgi?gid=&mir_c=miR-92%2F310%2F311%2F312%2F313&mir_nc=&mirg=); (2) PicTar (the miR310/311/312/313 targets were predicted, chosen data set: single miRNA target prediction with the setting S1: high sensitivity; the results were downloaded from http://pictar.mdc-berlin.de/cgi-bin/new_PicTar_fly.cgi?species=fly for each member and then combined); and (3) PITA version 6, which is based on miRbase r11.0 miRNAs and on the Drosophila genome version dm3. The prediction results were downloaded from the webpage https://genie.weizmann.ac.il/pubs/mir07/mir07_data.html, selecting “All” catalogs with no flank, yielding 546 candidate genes. The predicted targets were filtered by requiring they (1) are known to be expressed in the late stage of oogenesis (Yakoby et al. 2008; Tootle et al. 2011); (2) are involved in the KEGG signaling pathways critical for oogenesis or spermatogenesis, such as the BMP, MAPK, Hh, Notch, JAK/STAT, and Wnt signaling pathways (Xie and Spradling 1998; Kiger et al. 2001; Roth 2001; Suzanne et al. 2001; Tulina and Matunis 2001; Chen and McKearin 2003; Kawase et al. 2004; Kirilly et al. 2005; Song et al. 2007; Michel et al. 2012); and (3) produce mutant phenotypes associated with terms “germline stem cells,” “follicle cells,” or “dorsal appendage” as reported in FlyBase (http://flybase.org). This gave a list of 58 predicted targets, which are functionally relevant to the miR310s phenotypes. We further excluded 13 genes expressed at low levels in ovaries according to FlyBase and added another two experimentally validated targets of miR310s, Khc-73 and imd (Robins et al. 2005; Tsurudome et al. 2010), as positive controls. By doing so, we finally obtained a list of 47 genes for further analysis (Supplemental Table S2).
To examine target overlap among the four miR310s members, which share a conserved seed region, we predicted their targets using miRanda and DIANA-microT-CDS (Enright et al. 2003; Betel et al. 2010; Reczko et al. 2012; Paraskevopoulou et al. 2013). Predicted targets were downloaded from http://www.microrna.org/microrna/getDownloads.do (version: August 2010 Release) or searched on http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index (version: 5.0). Venn diagrams were produced by R package “VennDiagram.”
Luciferase reporter assay
A dual-luciferase reporter assay was used to validate the predicted miR310s targets in S2 cells. Full-length 3′ UTRs of five targets, as described in D. melanogaster genome release r6.11, were amplified from w1118 flies. The miR310s target sites at these 3′ UTRs were mutated using fusion PCR (Supplemental Fig. S6). Amplicons of the wild-type or mutant 3′ UTRs were cloned into 5′NotI and 3′Xhol sites of psiCHECK-2 vector (Promega Biotech, catalog no. C8021) and used to transfect S2 cells in 96-well plates with 100 ng of ub-Gal4 vectors, 200 ng of pUAST-mir310s (Tang et al. 2010), and 200 ng of the wild-type or mutant 3′ UTR reporter vectors using Lipofectamine 2000 (Thermo Fisher Scientific, catalog no. 12566014). Luciferase activities were measured 60 h after transfection using the Dual-Glo luciferase assay system (Promega Biotech, catalog no. E2920) following the manufacturer's instruction. Experiments were performed in four replicates for each gene. Primers used are listed in Supplemental Table S4.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation of China (31730046, 91231117, 91331202), the 985 Project (33000-18821105), the National Basic Research Program (973 Program) of China (2014CB542006), the Science Foundation for Outstanding Young Teachers in Higher Education of Guangdong (Yq2013005), and the Fundamental Research Funds for the Central Universities (16lgjc75).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.222505.117.
References
- Allesina S, Tang S. 2012. Stability criteria for complex ecosystems. Nature 483: 205–208. [DOI] [PubMed] [Google Scholar]
- Alvarez-Saavedra E, Horvitz HR. 2010. Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol 20: 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. 2008. The impact of microRNAs on protein output. Nature 455: 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano F, Smibert P, Lai EC. 2010. miR-9a prevents apoptosis during wing development by repressing Drosophila LIM-only. Dev Biol 338: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano F, Bortolamiol-Becet D, Dai Q, Sun K, Saj A, Chou YT, Raleigh DR, Kim K, Ni JQ, Duan H, et al. 2012. A genome-wide transgenic resource for conditional expression of Drosophila microRNAs. Development (Cambridge, England) 139: 2821–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg CA. 2008. Tube formation in Drosophila egg chambers. Tissue Eng Part A 14: 1479–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Koppal A, Agius P, Sander C, Leslie C. 2010. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol 11: R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy JJ, Jha AR, Posadas DM, Giri R, Venken KJ, Ji J, Jiang H, Bellen HJ, White KP, Carthew RW. 2013. miR-9a minimizes the phenotypic impact of genomic diversity by buffering a transcription factor. Cell 155: 1556–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, McKearin D. 2003. Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Curr Biol 13: 1786–1791. [DOI] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. 2007. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet 8: 93–103. [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. 2005. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33: e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Allesina S, Shen Y, Wu C-I. 2017. From foodwebs to gene regulatory networks (GRNs): Weak repressions by microRNAs confer system stability. bioRxiv https://doi.org/10.1101/176701.
- Cicek IO, Karaca S, Brankatschk M, Eaton S, Urlaub H, Shcherbata HR. 2016. Hedgehog signaling strength is orchestrated by the mir-310 cluster of microRNAs in response to diet. Genetics 202: 1167–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. 2012. Roles for microRNAs in conferring robustness to biological processes. Cell 149: 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecsedi M, Rausch M, Grosshans H. 2015. The let-7 microRNA directs vulval development through a single target. Dev Cell 32: 335–344. [DOI] [PubMed] [Google Scholar]
- Eichhorn SW, Guo H, McGeary SE, Rodriguez-Mias RA, Shin C, Baek D, Hsu SH, Ghoshal K, Villen J, Bartel DP. 2014. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol Cell 56: 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. 2003. MicroRNA targets in Drosophila. Genome Biol 5: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Lai EC. 2008. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet 9: 831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. 2005. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev Cell 8: 321–330. [DOI] [PubMed] [Google Scholar]
- Grun D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N. 2005. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput Biol 1: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gursky VV, Surkova SY, Samsonova MG. 2012. Mechanisms of developmental robustness. Bio Syst 109: 329–335. [DOI] [PubMed] [Google Scholar]
- He L, Xie M, Huang J, Zhang T, Shi S, Tang T. 2016. Efficient and specific inhibition of plant microRNA function by anti-microRNA oligonucleotides (AMOs) in vitro and in vivo. Plant Cell Rep 35: 933–945. [DOI] [PubMed] [Google Scholar]
- Herranz H, Cohen SM. 2010. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev 24: 1339–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. 2008. Gene regulation by transcription factors and microRNAs. Science (New York, NY) 319: 1785–1786. [DOI] [PubMed] [Google Scholar]
- Hornstein E, Shomron N. 2006. Canalization of development by microRNAs. Nat Genet 38Suppl: S20–S24. [DOI] [PubMed] [Google Scholar]
- Hunter SE, Finnegan EF, Zisoulis DG, Lovci MT, Melnik-Martinez KV, Yeo GW, Pasquinelli AE. 2013. Functional genomic analysis of the let-7 regulatory network in Caenorhabditis elegans. PLoS Genet 9: e1003353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun S, Lee JH, Jin H, Nam J, Namkoong B, Lee G, Chung J, Kim VN. 2009. Conserved microRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell 139: 1096–1108. [DOI] [PubMed] [Google Scholar]
- Iovino N, Pane A, Gaul U. 2009. miR-184 has multiple roles in Drosophila female germline development. Dev Cell 17: 123–133. [DOI] [PubMed] [Google Scholar]
- Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. 2007. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell 131: 136–145. [DOI] [PubMed] [Google Scholar]
- Kawase E, Wong MD, Ding BC, Xie T. 2004. Gbb/Bmp signaling is essential for maintaining germline stem cells and for repressing bam transcription in the Drosophila testis. Development 131: 1365–1375. [DOI] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. 2007. The role of site accessibility in microRNA target recognition. Nat Genet 39: 1278–1284. [DOI] [PubMed] [Google Scholar]
- Kheradpour P, Stark A, Roy S, Kellis M. 2007. Reliable prediction of regulator targets using 12 Drosophila genomes. Genome Res 17: 1919–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT. 2001. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science 294: 2542–2545. [DOI] [PubMed] [Google Scholar]
- Kirilly D, Spana EP, Perrimon N, Padgett RW, Xie T. 2005. BMP signaling is required for controlling somatic stem cell self-renewal in the Drosophila ovary. Dev Cell 9: 651–662. [DOI] [PubMed] [Google Scholar]
- Lai EC. 2003. microRNAs: Runts of the genome assert themselves. Curr Biol 13: R925–R936. [DOI] [PubMed] [Google Scholar]
- Lai EC. 2015. Two decades of miRNA biology: lessons and challenges. RNA 21: 675–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GJ, Hyun S. 2014. Multiple targets of the microRNA miR-8 contribute to immune homeostasis in Drosophila. Dev Comp Immunol 45: 245–251. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. 2009. A microRNA imparts robustness against environmental fluctuation during development. Cell 137: 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Luo W, Sehgal A. 2012. Regulation of circadian behavioral output via a MicroRNA-JAK/STAT circuit. Cell 148: 765–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu Y, Shen Y, Li H, Chen Y, Guo L, Zhao Y, Hungate E, Shi S, Wu CI, Tang T. 2014. New microRNAs in Drosophila—birth, death and cycles of adaptive evolution. PLoS Genet 10: e1004096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi E, Cairns MJ. 2017. MiR-137: an important player in neural development and neoplastic transformation. Mol Psychiatry 22: 44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco A. 2014. Sex-biased expression of microRNAs in Drosophila melanogaster. Open Biol 4: 140024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Walhout AJ. 2009. The interplay between transcription factors and microRNAs in genome-scale regulatory networks. Bioessays 31: 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Barrasa MI, Hammell M, Sequerra R, Doucette-Stamm L, Roth FP, Ambros VR, Walhout AJ. 2008. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev 22: 2535–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May RM. 1972. Will a large complex system be stable? Nature 238: 413–414. [DOI] [PubMed] [Google Scholar]
- May RM. 2001. Stability and complexity in model ecosystems. Princeton University Press, Princeton, NJ. [Google Scholar]
- Michel M, Kupinski AP, Raabe I, Bokel C. 2012. Hh signalling is essential for somatic stem cell maintenance in the Drosophila testis niche. Development 139: 2663–2669. [DOI] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. 2007. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet 3: e215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osella M, Bosia C, Cora D, Caselle M. 2011. The role of incoherent microRNA-mediated feedforward loops in noise buffering. PLoS Comput Biol 7: e1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palopoli MF, Wu CI. 1994. Genetics of hybrid male sterility between drosophila sibling species: A complex web of epistasis is revealed in interspecific studies. Genetics 138: 329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancratov R, Peng F, Smibert P, Yang S Jr, Olson ER, Guha-Gilford C, Kapoor AJ, Liang FX, Lai EC, Flaherty MS, et al. 2013. The miR-310/13 cluster antagonizes β-catenin function in the regulation of germ and somatic cell differentiation in the Drosophila testis. Development 140: 2904–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T, Hatzigeorgiou AG. 2013. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res 41(Web Server issue): W169–W173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KJ, Dietrich MR, McPeek MA. 2009. MicroRNAs and metazoan macroevolution: insights into canalization, complexity, and the Cambrian explosion. Bioessays 31: 736–747. [DOI] [PubMed] [Google Scholar]
- Pinzon N, Li B, Martinez L, Sergeeva A, Presumey J, Apparailly F, Seitz H. 2017. microRNA target prediction programs predict many false positives. Genome Res 27: 234–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posadas DM, Carthew RW. 2014. MicroRNAs and their roles in developmental canalization. Curr Opin Genet Dev 27: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch M, Ecsedi M, Bartake H, Mullner A, Grosshans H. 2015. A genetic interactome of the let-7 microRNA in C. elegans. Dev Biol 401: 276–286. [DOI] [PubMed] [Google Scholar]
- Reczko M, Maragkakis M, Alexiou P, Grosse I, Hatzigeorgiou AG. 2012. Functional microRNA targets in protein coding sequences. Bioinformatics 28: 771–776. [DOI] [PubMed] [Google Scholar]
- Robins H, Li Y, Padgett RW. 2005. Incorporating structure to predict microRNA targets. Proc Natl Acad Sci 102: 4006–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S. 2001. Drosophila oogenesis: coordinating germ line and soma. Curr Biol 11: R779–R781. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. 2007. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 17: 1850–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S. 1998. Hsp90 as a capacitor for morphological evolution. Nature 396: 336–342. [DOI] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. 2011. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146: 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H. 2009. Redefining microRNA targets. Curr Biol 19: 870–873. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63. [DOI] [PubMed] [Google Scholar]
- Smibert P, Lai EC. 2010. A view from Drosophila: multiple biological functions for individual microRNAs. Semin Cell Dev Biol 21: 745–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Call GB, Kirilly D, Xie T. 2007. Notch signaling controls germline stem cell niche formation in the Drosophila ovary. Development (Cambridge, England) 134: 1071–1080. [DOI] [PubMed] [Google Scholar]
- Song R, Walentek P, Sponer N, Klimke A, Lee JS, Dixon G, Harland R, Wan Y, Lishko P, Lize M, et al. 2014. miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature 510: 115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R. 2011. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol 29: 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Ting CT, Wu CI. 2004. The normal function of a speciation gene, Odysseus, and its hybrid sterility effect. Science (New York, NY) 305: 81–83. [DOI] [PubMed] [Google Scholar]
- Sun K, Jee D, de Navas LF, Duan H, Lai EC. 2015. Multiple in vivo biological processes are mediated by functionally redundant activities of Drosophila mir-279 and mir-996. PLoS Genet 11: e1005245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzanne M, Perrimon N, Noselli S. 2001. The Drosophila JNK pathway controls the morphogenesis of the egg dorsal appendages and micropyle. Dev Biol 237: 282–294. [DOI] [PubMed] [Google Scholar]
- Tan H, Poidevin M, Li H, Chen D, Jin P. 2012. MicroRNA-277 modulates the neurodegeneration caused by Fragile X premutation rCGG repeats. PLoS Genet 8: e1002681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T, Kumar S, Shen Y, Lu J, Wu ML, Shi S, Li WH, Wu CI. 2010. Adverse interactions between micro-RNAs and target genes from different species. Proc Natl Acad Sci 107: 12935–12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Pawar S, Allesina S. 2014. Correlation between interaction strengths drives stability in large ecological networks. Ecol Lett 17: 1094–1100. [DOI] [PubMed] [Google Scholar]
- Tay Y, Rinn J, Pandolfi PP. 2014. The multilayered complexity of ceRNA crosstalk and competition. Nature 505: 344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson DW, Dinger ME. 2016. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 17: 272–283. [DOI] [PubMed] [Google Scholar]
- Tootle TL, Williams D, Hubb A, Frederick R, Spradling A. 2011. Drosophila eggshell production: identification of new genes and coordination by Pxt. PLoS One 6: e19943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A. 2007. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell 26: 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurudome K, Tsang K, Liao EH, Ball R, Penney J, Yang JS, Elazzouzi F, He T, Chishti A, Lnenicka G, et al. 2010. The Drosophila miR-310 cluster negatively regulates synaptic strength at the neuromuscular junction. Neuron 68: 879–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulina N, Matunis E. 2001. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science 294: 2546–2549. [DOI] [PubMed] [Google Scholar]
- Varghese J, Cohen SM. 2007. microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drosophila. Genes Dev 21: 2277–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. 2007. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidigal JA, Ventura A. 2015. The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol 25: 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dassow G, Meir E, Munro EM, Odell GM. 2000. The segment polarity network is a robust developmental module. Nature 406: 188–192. [DOI] [PubMed] [Google Scholar]
- Waddington CH. 1942. Canalization of development and the inheritance of acquired characters. Nature 150: 563–565. [DOI] [PubMed] [Google Scholar]
- Wu CI. 1983. Virility deficiency and the sex-ratio trait in DROSOPHILA PSEUDOOBSCURA. I. Sperm displacement and sexual selection. Genetics 105: 651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI, Shen Y, Tang T. 2009. Evolution under canalization and the dual roles of microRNAs: a hypothesis. Genome Res 19: 734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Spradling AC. 1998. decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell 94: 251–260. [DOI] [PubMed] [Google Scholar]
- Yakoby N, Bristow CA, Gong D, Schafer X, Lembong J, Zartman JJ, Halfon MS, Schupbach T, Shvartsman SY. 2008. A combinatorial code for pattern formation in Drosophila oogenesis. Dev Cell 15: 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.