Abstract
Infertility is a relatively common disorder of the reproductive system and remains unexplained in many cases. In vitro fertilization techniques have uncovered previously unrecognized infertility phenotypes, including oocyte maturation arrest, the molecular etiology of which remains largely unknown. We report two families affected by female-limited infertility caused by oocyte maturation failure. Positional mapping and whole-exome sequencing revealed two homozygous, likely deleterious variants in PATL2, each of which fully segregates with the phenotype within the respective family. PATL2 encodes a highly conserved oocyte-specific mRNP repressor of translation. Previous data have shown the strict requirement for PATL2 in oocyte-maturation in model organisms. Data gathered from the families in this study suggest that the role of PATL2 is conserved in humans and expand our knowledge of the factors that are necessary for female meiosis.
Keywords: maturation arrest, meiosis, IVF, P100, PAT1A, GVBD
Main Text
Infertility, a reproductive-system disorder defined by the failure to achieve a clinical pregnancy after 12 months or more of regular unprotected sexual intercourse, affects 10.7%–15.5% of couples.1 Although infertility is highly heterogeneous in etiology, investigating the cause is necessary for guiding treatment options. Additionally, molecular understanding of infertility has the potential to reveal fundamental insight into human reproduction. This is particularly true when mechanical and hormonal causes are excluded and abnormal cellular phenotypes are observed, which is now possible with the advent of in vitro fertilization (IVF) techniques. One example is the discovery of TLE6 and PADI6 (MIM: 612399 and 610363, respectively) mutations causing failure of zygote cleavage, which revealed a critical role of the extracellular maternal complex and zygotic gene activation in humans and their conserved role in other species.2, 3
The syndrome of oocyte maturation failure is an extremely rare cause of primary female infertility: only a few cases have been reported to date. This maturation arrest can represent failure to complete any of the various stages of meiosis I or II.4, 5 The resulting incompetence of the oocyte to be fertilized even with intracytoplasmic sperm injection poses a significant management challenge. Several mutants (typically mice) have been described as having a maturation-arrest phenotype.6, 7, 8, 9, 10, 11, 12, 13 These include Cdc25b and Pde3a knockouts (KOs) that arrest at the germinal vesicle (GV) stage, Mei1, Cks2, Mlh1, and Lfng KOs that arrest at meiosis I, and Smc1b and Mos KOs that arrest at meiosis II.6, 7, 8, 9, 10, 11, 12, 13 Interestingly, although some of these mutants are associated with female-limited sterility, others (e.g., Mei, Cks2, Mlh1, and Smc1b mutants) display a sexually dimorphic infertility phenotype. No mutations, however, have been reported in the human orthologs of these genes in the context of infertility. Thus, it remains unknown what causes maturation arrest in human female individuals with infertility. In this study, we suggest that PATL2 (MIM: 614661) mutations are one such etiology on the basis of human genetics data and the gene’s established role in oocyte maturation in model organisms.
Affected individuals were recruited after providing informed consent under a research protocol approved by King Faisal Specialist Hospital and Research Center (KFSHRC) research advisory council 2121053. Meiosis I maturation arrest was defined as the failure of resumption of meiosis after controlled ovarian stimulation and follicular maturation triggering by luteinizing hormone and human chorionic gonadotropin. Venous blood samples were obtained from the index individuals and available relatives in each family for DNA extraction. For positional mapping, the Axiom SNP Chip (Affymetrix) platform was used for genome-wide genotyping. Runs of homozygosity (ROHs) > 1 Mb were considered surrogates of autozygosity by AutoSNPa.14 Linkage analysis was performed with the easyLINKAGE package under a fully penetrant autosomal-recessive female-limited model.15 For exome analysis, we selected the index individual from each family. The samples were prepared according to the preparation guide of the Agilent SureSelect Target Enrichment Kit, and the resulting libraries were sequenced with the Illumina HiSeq 2000 sequencer. The Genome Analysis Toolkit was used for variant calling. We considered only variants that are novel or very rare (minor allele frequency [MAF] < 0.001) in the ExAC Browser and 2,379 Saudi exomes. We also performed computational structural analysis of candidate variants. Sequences were retrieved from UniProt. We used SwissModel to produce homology models.16 RaptorX was used for prediction of secondary structures and protein disorders.17 Models were manually inspected, and mutations were evaluated with PyMOL.
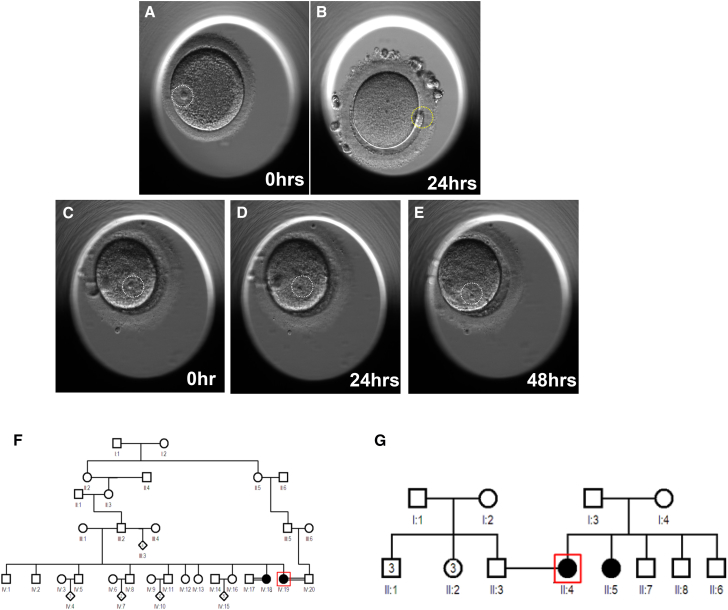
Family 1 consists of two Arab sisters (IV:18 and IV:19, currently 35 and 27 years of age, respectively) who presented with primary infertility but regular menstrual cycles to our IVF center at KFSHRC. The index individual (IV:19) had a 7-year history of primary infertility. She had undergone seven failed IVF cycles, each with an adequate number of retrieved eggs (5–23 oocytes), but all arrested in meiosis I, as indicated by the presence of a GV (Figures 1C–1E, Movie S1, and Supplemental Note; Figures 1A and 1B and Movie S2 show normal meiosis I for comparison). Hormone levels were measured on the third day of the cycle, and her follicle-stimulating hormone (FSH) level was 6.6 IU/L. There was no indication of polycystic ovary disorder on hormonal or imaging studies. Oocytes were cultured in the in vitro maturation media, but there was no progress on meiotic maturation for 48 hr. Her sister (IV:18) also had a 6-year history of primary infertility. She had undergone one IVF cycle, and four retrieved eggs were arrested in meiosis I (Figures S1A–S1C). Like her sister, she lacked features of polycystic ovary disorder, and her FSH level was 5.6 IU/L on the third day of the cycle. Although consanguinity was denied, both parents belong to the same tribe. Please refer to the Supplemental Note for detailed clinical information.
Figure 1.
Identification of Individuals with a Phenotype Involving Oocyte Maturation Arrest
(A and B) Images of an immature oocyte with a germinal vesicle (A, white circle) and a mature oocyte with a polar body (B, yellow circle) from a normal woman.
(C–E) Images of immature oocytes from the individual with a maturation arrest phenotype from family 1. (C) An oocyte retrieved from a stimulated ovary. (D and E) Immature oocytes after 24 (D) and 48 (E) hr culture in in vitro maturation media indicate failure in maturation. Pedigrees of family 1 (F) and family 2 (G) are shown. WES was performed on the index individual (indicated by a red box) in each family.
Family 2 also consists of two Arab sisters with primary infertility. The index individual (II:4), 33 years old, presented to the IVF center at Habib Medical Group Hospital and underwent five trials of failed intrauterine insemination followed by an unsuccessful IVF cycle. A total of 20 oocytes were retrieved, but none were mature (Figure S1D). She was noted to have a failure of resumption of meiotic arrest. Her sister, 26 years old, has a 3-year history of primary infertility but has not yet had any IVF cycles. Both sisters had regular menstrual cycles, and their workups ruled out polycystic ovary disorder. Similar to the parents in family 1, the sisters’ parents are not consanguineous but have the same tribal origin.
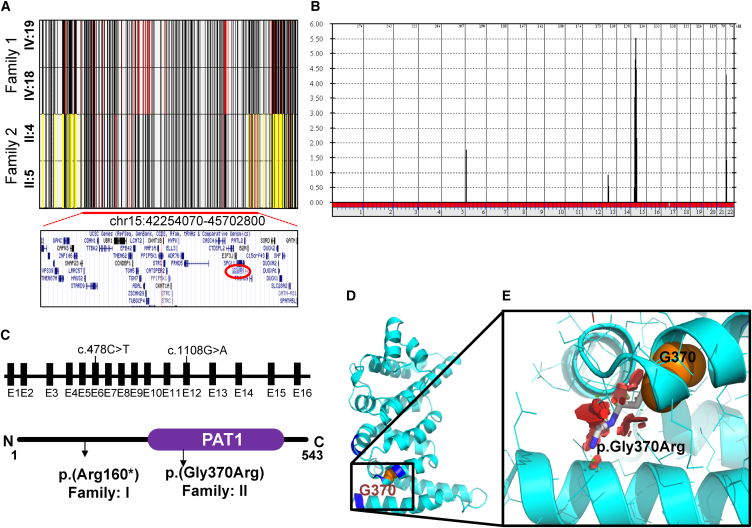
Given the extreme rarity of their phenotype, we hypothesized that both families are affected by a similar underlying molecular defect. Although neither family is apparently consanguineous, the shared tribal origin of the parents (endogamy) makes it still possible that the phenotype is caused by autozygosity for ancestral recessive mutations, which can be traced by positional mapping. Examination of the ROHs revealed only one overlapping autozygous region of 3.4 Mb (chr15: 42,254,070–45,702,800) (Figure 2A). Linkage analysis confirmed that both families map to this candidate locus (Figure 2B). However, the haplotype of the affected members was clearly different between the two families, which suggests that each family has a different mutation in the candidate gene, consistent with the difference in their tribal affiliation (Figure 2A and Table S1). Exome sequencing revealed 63,890 variants in the index individual of family 1 and 72,370 variants in the index individual of family 2. However, only one rare homozygous variant was identified within the candidate locus in each family, and both variants involved PATL2: c.478C>T (p.Arg160∗) (GenBank: NM_001145112.1) in family 1 and c.1108G>A (p.Gly370Arg) (GenBank: NM_001145112.1) in family 2 (Table S2). No other candidate homozygous variants outside the linkage interval or compound-heterozygous variants were identified. Segregation analysis using Sanger sequencing confirmed that the variant is homozygous in the affected members of each respective family (Figure S2). Interestingly, homozygosity for c.478C>T (p.Arg160∗) was compatible with fertility in males, as demonstrated by the father and two brothers in family 1 (Figure S2A). Both variants were completely absent in 2,379 Saudi exomes and very rare or absent in the ExAC Browser (MAF = 0.00003362 and 0 for p.Arg160∗ and p.Gly370Arg, respectively).
Figure 2.
Identification of PATL2 Variants in Individuals with a Phenotype Involving Oocyte Maturation Arrest
(A) Shared haplotypes (chr15: 42,254,070–45,702,800; UCSC Genome Browser hg19) of the affected members from family 1 (IV:19 and IV:18) and family 2 (II:4 and II:5). Yellow lines indicate heterozygous SNPs. Red lines indicate rare homozygous SNPs. The lower panel shows genes that are within the shared ROH region. The position of PATL2 (chr15: 44,966,961–45,003,514; UCSC Genome Browser hg19) is highlighted with a red circle.
(B) Genome-wide linkage analysis showing a chr15 peak with a pLOD score of 5.5 (16 individuals are included; see Figure S2 for details).
(C) Genomic context and structural presentation of PTAL2, including the two variants.
(D and E) The PAT1 domain contains the residues (Arg301, Leu302, Arg368, Ala327, Arg402, and Arg403) corresponding to the PATL1 residues involved in RNA binding (shown in dark blue). (D) Based on the structure of the corresponding region of PATL1 (PDB: 2XEQ; 34% sequence identity), the secondary-structure representation of the homology model of the C-terminal domain of PATL2 (residues 299–540) is shown. Gly370 is shown as orange spheres. (E) Magnified view of the outlined region in (D). In addition to Gly370 (orange), the putative p.Gly370Arg rotamer side chain (gray) with the least possible clashes (red discs) is shown, illustrating that p.Gly370Arg compromises the folding and function of PATL2.
PATL2 encodes an RNA-binding protein that acts as a translational repressor. PAT proteins contain a conserved N-terminal sequence, a proline-rich region, a Mid domain, and a C-terminal domain. Prediction of secondary structure and disorder indicated that the N-terminal 290 residues of the 541-residue protein PATL2 are highly mobile and unstructured. The PATL2 residues 299–540 are predicted to adopt a stable three-dimensional (3D) fold. A 3D structure of this C-terminal domain can be modeled with good confidence on the basis of the 34% identical C-terminal domain of PAT1 (PDB: 2XES and 2XEQ; modeling QMEAN score of −2.79). Gly370 localizes in helix 3 of the C-terminal domain. It is oriented toward the hydrophobic core of the superhelical fold of this domain (Figure 2D). An arginine in this position would result in severe steric clashes (Figure 2E) and lead to the unfavorable introduction of a charge within the hydrophobic core. The p.Gly370Arg variant is therefore expected to strongly destabilize this protein region (PolyPhen-2: 0.915; SIFT: 0.03; and CADD: 33). In PAT1, this region is involved in RNA binding.18 This region is highly conserved, and stereochemical features are preserved in PATL2 (Figures 2D and 2E). It is therefore expected that p.Gly370Arg will lead to loss of RNA and protein ligands that are central to the function of PAT proteins. The glycine residue at the 370th position is highly conserved from humans to yeast (Figure S3). The premature stop codon at position 160, on the other hand, leads to complete loss of the RNA-binding domain, so this truncation is expected to result in severe loss of function of PATL2 (we found no evidence of nonsense-mediated decay).
In humans, oogenesis commences in utero and stalls in meiosis I until ovulation (when meiosis I is completed), whereas meiosis II is completed only after fertilization. During this process, the chromatin condensation does not permit active transcription; as a result, intracellular signaling is primarily controlled at the translational level. A pivotal factor in this process is a large mRNP complex that sequesters RNA in the oocyte.19 This complex is functionally very similar to the P bodies in the somatic cells.20, 21 Completion of meiosis is marked by GV breakdown (GVBD), a critical event that is triggered by a marked decline in CPEB and consequent translational activation of MOS (MIM: 190060) and CCNB1 (MIM: 123836). 22, 23, 24 Interestingly, Mos deficiency has been shown to result in meiosis arrest in mice.25
PATL2 and PATL1 are vertebrate paralogs of Pat1p in yeast, PATR-1 in roundworm, and HPat in fruit fly.26 The ancestral ortholog served a dual function (RNA decapping and translational repression), and its deficiency has been shown to result in translational repression with deleterious consequences.27 PATL2 was first identified in 1992 as a Xenopus oocyte-specific protein that binds single-stranded DNA, and it was labeled P100.28 It was later found that PATL2 is a component of a complex involving the cytoplasmic polyadenylation element and its binding factor CPEB, and this complex is a critical temporal regulator of translation during oocyte maturation.26, 29 PATL2 is highly abundant in early stages of meiosis but declines precipitously, similarly to CPEB (with which it physically interacts), with the onset of GVBD. Indeed, it has been shown that this decline in the amount of PATL2 is necessary for GVBD and that ectopic expression completely blocks meiosis I progression.26, 30 Importantly, PATL1 does not compensate for the role of PATL2 in meiosis given that it is only detectable in later stages. PATL2 has been shown to harbor RNA binding activity and resulting translational repression, but decapping activity has not been shown.26 On PATL1, the region necessary for RNA binding has been shown to involve a basic patch on the C-terminal domain (residues Arg519, Arg520, Arg591, Arg595, Lys625, and Lys626, corresponding to Arg301, Leu302, Arg368, Ala327, Arg402, and Arg403, respectively, in PATL2);18 this patch is completely lost as a result of the truncating variant in family 1 and is expected to be severely impaired by the missense variant in family 2.
The function of PATL2 in humans is unknown. Despite the inbred nature of the local population, the complete lack of homozygous deleterious variants in our local exome database suggests that it is highly constrained in the recessive sense. Indeed, the data we present in this study suggest a role in human oogenesis that is only lost in the setting of biallelic mutations. Although data from Xenopus indicate that the decline in the amount of PATL2 ortholog coincides with and probably triggers GVBD, we show that its deficiency in humans results in a highly similar meiosis I arrest. This suggests a strict requirement for temporal control of the PATL2 expression level for normal oocyte maturation. Interestingly, we show that the effect of PATL2 is limited to females, given that at least three males who are homozygous for a severe truncating variant are fertile.
In conclusion, we suggest that PATL1 mutations arrest meiosis I and thus lead to female-limited infertility in humans. This rare etiology of infertility expands our knowledge of factors required for normal human oogenesis and suggests a highly conserved network that controls this process across species. We hope that this and future discoveries of the molecular underpinning of human infertility will inform and advance new therapeutic strategies.
Acknowledgments
We thank the study participants for their enthusiastic participation. We also thank the Sequencing and Genotyping Core Facilities at the King Faisal Specialist Hospital and Research Center for their technical help. This work was funded in part by the King Abdulaziz City for Science and Technology (13-BIO1113-20), and we acknowledge the support of the Saudi Human Genome Project. The research reported by S.T.A. in this publication was supported by funding from the King Abdullah University of Science and Technology.
Published: September 28, 2017
Footnotes
Supplemental Data include a Supplemental Note, three figures, two tables, and two movies and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.08.009.
Web Resources
ClustalW2, http://www.ebi.ac.uk/Tools/msa/clustalw2/
Exome Aggregation Consortium (ExAC) Browser, http://exac.broadinstitute.org/
OMIM, http://www.omim.org
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
PyMOL, http://pymol.org/
RaptorX, http://raptorx.uchicago.edu/
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
UCSC Genome Browser, http://genome.ucsc.edu
UniProt, http://www.uniprot.org/
Supplemental Data
References
- 1.Thoma M.E., McLain A.C., Louis J.F., King R.B., Trumble A.C., Sundaram R., Buck Louis G.M. Prevalence of infertility in the United States as estimated by the current duration approach and a traditional constructed approach. Fertil. Steril. 2013;99:1324–1331.e1. doi: 10.1016/j.fertnstert.2012.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alazami A.M., Awad S.M., Coskun S., Al-Hassan S., Hijazi H., Abdulwahab F.M., Poizat C., Alkuraya F.S. TLE6 mutation causes the earliest known human embryonic lethality. Genome Biol. 2015;16:240. doi: 10.1186/s13059-015-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maddirevula S., Coskun S., Awartani K., Alsaif H., Abdulwahab F.M., Alkuraya F.S. The human knockout phenotype of PADI6 is female sterility caused by cleavage failure of their fertilized eggs. Clin. Genet. 2017;91:344–345. doi: 10.1111/cge.12866. [DOI] [PubMed] [Google Scholar]
- 4.Beall S., Brenner C., Segars J. Oocyte maturation failure: a syndrome of bad eggs. Fertil. Steril. 2010;94:2507–2513. doi: 10.1016/j.fertnstert.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mrazek M., Fulka Jr J., Jr. Failure of oocyte maturation: possible mechanisms for oocyte maturation arrest. Hum. Reprod. 2003;18:2249–2252. doi: 10.1093/humrep/deg434. [DOI] [PubMed] [Google Scholar]
- 6.Lincoln A.J., Wickramasinghe D., Stein P., Schultz R.M., Palko M.E., De Miguel M.P., Tessarollo L., Donovan P.J. Cdc25b phosphatase is required for resumption of meiosis during oocyte maturation. Nat. Genet. 2002;30:446–449. doi: 10.1038/ng856. [DOI] [PubMed] [Google Scholar]
- 7.Masciarelli S., Horner K., Liu C., Park S.H., Hinckley M., Hockman S., Nedachi T., Jin C., Conti M., Manganiello V. Cyclic nucleotide phosphodiesterase 3A-deficient mice as a model of female infertility. J. Clin. Invest. 2004;114:196–205. doi: 10.1172/JCI21804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Libby B.J., De La Fuente R., O’Brien M.J., Wigglesworth K., Cobb J., Inselman A., Eaker S., Handel M.A., Eppig J.J., Schimenti J.C. The mouse meiotic mutation mei1 disrupts chromosome synapsis with sexually dimorphic consequences for meiotic progression. Dev. Biol. 2002;242:174–187. doi: 10.1006/dbio.2001.0535. [DOI] [PubMed] [Google Scholar]
- 9.Spruck C.H., de Miguel M.P., Smith A.P., Ryan A., Stein P., Schultz R.M., Lincoln A.J., Donovan P.J., Reed S.I. Requirement of Cks2 for the first metaphase/anaphase transition of mammalian meiosis. Science. 2003;300:647–650. doi: 10.1126/science.1084149. [DOI] [PubMed] [Google Scholar]
- 10.Edelmann W., Cohen P.E., Kane M., Lau K., Morrow B., Bennett S., Umar A., Kunkel T., Cattoretti G., Chaganti R. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996;85:1125–1134. doi: 10.1016/s0092-8674(00)81312-4. [DOI] [PubMed] [Google Scholar]
- 11.Hahn K.L., Johnson J., Beres B.J., Howard S., Wilson-Rawls J. Lunatic fringe null female mice are infertile due to defects in meiotic maturation. Development. 2005;132:817–828. doi: 10.1242/dev.01601. [DOI] [PubMed] [Google Scholar]
- 12.Revenkova E., Eijpe M., Heyting C., Hodges C.A., Hunt P.A., Liebe B., Scherthan H., Jessberger R. Cohesin SMC1 β is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat. Cell Biol. 2004;6:555–562. doi: 10.1038/ncb1135. [DOI] [PubMed] [Google Scholar]
- 13.Araki K., Naito K., Haraguchi S., Suzuki R., Yokoyama M., Inoue M., Aizawa S., Toyoda Y., Sato E. Meiotic abnormalities of c-mos knockout mouse oocytes: activation after first meiosis or entrance into third meiotic metaphase. Biol. Reprod. 1996;55:1315–1324. doi: 10.1095/biolreprod55.6.1315. [DOI] [PubMed] [Google Scholar]
- 14.Carr I.M., Flintoff K.J., Taylor G.R., Markham A.F., Bonthron D.T. Interactive visual analysis of SNP data for rapid autozygosity mapping in consanguineous families. Hum. Mutat. 2006;27:1041–1046. doi: 10.1002/humu.20383. [DOI] [PubMed] [Google Scholar]
- 15.Lindner T.H., Hoffmann K. easyLINKAGE: a PERL script for easy and automated two-/multi-point linkage analyses. Bioinformatics. 2005;21:405–407. doi: 10.1093/bioinformatics/bti009. [DOI] [PubMed] [Google Scholar]
- 16.Arnold K., Bordoli L., Kopp J., Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 17.Källberg M., Margaryan G., Wang S., Ma J., Xu J. RaptorX server: a resource for template-based protein structure modeling. Methods Mol Biol. 2014;1137:17–27. doi: 10.1007/978-1-4939-0366-5_2. [DOI] [PubMed] [Google Scholar]
- 18.Braun J.E., Tritschler F., Haas G., Igreja C., Truffault V., Weichenrieder O., Izaurralde E. The C-terminal α-α superhelix of Pat is required for mRNA decapping in metazoa. EMBO J. 2010;29:2368–2380. doi: 10.1038/emboj.2010.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouvet P., Wolffe A.P. A role for transcription and FRGY2 in masking maternal mRNA within Xenopus oocytes. Cell. 1994;77:931–941. doi: 10.1016/0092-8674(94)90141-4. [DOI] [PubMed] [Google Scholar]
- 20.Parker R., Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 21.Kotaja N., Sassone-Corsi P. The chromatoid body: a germ-cell-specific RNA-processing centre. Nat. Rev. Mol. Cell Biol. 2007;8:85–90. doi: 10.1038/nrm2081. [DOI] [PubMed] [Google Scholar]
- 22.Ballantyne S., Daniel D.L., Jr., Wickens M. A dependent pathway of cytoplasmic polyadenylation reactions linked to cell cycle control by c-mos and CDK1 activation. Mol. Biol. Cell. 1997;8:1633–1648. doi: 10.1091/mbc.8.8.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Moor C.H., Richter J.D. The Mos pathway regulates cytoplasmic polyadenylation in Xenopus oocytes. Mol. Cell. Biol. 1997;17:6419–6426. doi: 10.1128/mcb.17.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radford H.E., Meijer H.A., de Moor C.H. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochimica et Biophysica Acta. 2008:217–2229. doi: 10.1016/j.bbagrm.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colledge W.H., Carlton M.B., Udy G.B., Evans M.J. Disruption of c-mos causes parthenogenetic development of unfertilized mouse eggs. Nature. 1994;370:65–68. doi: 10.1038/370065a0. [DOI] [PubMed] [Google Scholar]
- 26.Marnef A., Maldonado M., Bugaut A., Balasubramanian S., Kress M., Weil D., Standart N. Distinct functions of maternal and somatic Pat1 protein paralogs. RNA. 2010;16:2094–2107. doi: 10.1261/rna.2295410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haas G., Braun J.E., Igreja C., Tritschler F., Nishihara T., Izaurralde E. HPat provides a link between deadenylation and decapping in metazoa. J. Cell Biol. 2010;189:289–302. doi: 10.1083/jcb.200910141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rother R.P., Frank M.B., Thomas P.S. Purification, primary structure, bacterial expression and subcellular distribution of an oocyte-specific protein in Xenopus. Eur. J. Biochem. 1992;206:673–683. doi: 10.1111/j.1432-1033.1992.tb16973.x. [DOI] [PubMed] [Google Scholar]
- 29.Hake L.E., Richter J.D. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell. 1994;79:617–627. doi: 10.1016/0092-8674(94)90547-9. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura Y., Tanaka K.J., Miyauchi M., Huang L., Tsujimoto M., Matsumoto K. Translational repression by the oocyte-specific protein P100 in Xenopus. Dev. Biol. 2010;344:272–283. doi: 10.1016/j.ydbio.2010.05.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.