Abstract
Tris(1,3-dichloro-2-propyl)phosphate (TDCIPP) is an organophosphate flame retardant that impacts zebrafish epiboly – an effect that may be associated with genome-wide hypomethylation. Using zebrafish as a model, the objectives of this study were to (1) quantify concentration-dependent impacts of TDCIPP on epiboly; (2) determine whether co-exposure with folic acid (FA) – a methyl donor – mitigates TDCIPP-induced impacts; and (3) using ten previously identified TDCIPP-susceptible loci, rely on bisulfite amplicon sequencing (BSAS) to monitor CpG methylation dynamics across multiple TDCIPP concentrations in the presence or absence of FA. Embryos were exposed to TDCIPP from 0.75 h post-fertilization (hpf) to 2, 4, 6, or 24 hpf in the presence or absence of 1 mM FA. Although TDCIPP delayed epiboly up to 3 h by 6 hpf and induced malformations by 24 hpf, FA was unable to mitigate TDCIPP-induced effects at all stages evaluated. Moreover, while no differences in global methylation were detected using a 5-methylcytosine (5-mC) DNA ELISA, BSAS revealed that TDCIPP-induced effects on CpG methylation were dependent on concentration and developmental stage, and that early effects on methylation do not persist despite continuous exposure. Our findings demonstrate that TDCIPP delays zebrafish epiboly, a phenotype that is preceded by complex, dynamic alterations in DNA methylation.
TOC image
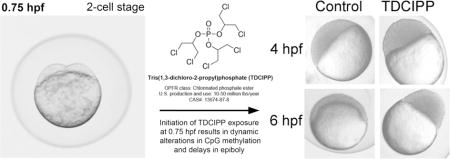
Introduction
Early zebrafish development is characterized by a series of coordinated cell divisions and movements that prepare the embryo for proper differentiation and patterning. Beginning at the end of the blastula period, cells amassed on the animal pole of the yolk spread over the surface in a process called epiboly. This represents the onset of gastrulation and cell fate determination. Proper cell migration during epiboly is critical for normal development, and epiboly mutants induce embryo mortality and severe abnormalities1. Similarly, chemicals that delay or arrest epiboly may lead to downstream abnormalities later in development.
Key early developmental events such as epiboly are preceded by reprogramming of the DNA methylome. Cytosine methylation of DNA is an epigenetic mark that regulates transcription and differentiation. During cleavage within zebrafish embryos, parentally inherited 5-methylcytosines (5-mCs) are passively erased and, beginning at 2 hpf, patterns are reestablished to reach paternal methylation levels by gastrulation2,3. Treatment of early zebrafish embryos with 5-azacytidine (5-azaC) – a DNA methyltransferase inhibitor – decreased DNA methylation and induced abnormal somite patterning4, demonstrating that DNA methylation is critical for patterning and differentiation.
Tris(1,3-dichloro-2-propyl)phosphate (TDCIPP) is a high-production volume flame retardant found within indoor environments, including house dust, sofas, and baby furniture5,6. Metabolites of TDCIPP have been detected in urine from mother:toddler pairs7 and human exposure has increased over the last 10 years8. TDCIPP has been linked to impacts on behavior9,10,11, reproduction12,13, receptor signaling14, and gene expression15. Furthermore, previous studies demonstrated that TDCIPP exposure within the first two hours of zebrafish embryogenesis leads to enhanced toxicity later in development16, and others have reported that early exposure to TDCIPP delays epiboly and impacts segmentation15. However, the mechanisms of these effects during early embryonic development within zebrafish remain unknown. Therefore, as chronic human exposure to TDCIPP following migration from treated products is likely common within the United States, additional studies within animal models are needed to (1) provide a foundation for understanding the impacts of TDCIPP exposure on early embryonic development and (2) help formulate mechanistic-based hypotheses for prenatal developmental toxicity studies within rodents and epidemiological studies within human populations.
Using a restriction enzyme-based approach, our earlier studies demonstrated that the cleavage period (0.75–2.25 hpf) during embryogenesis is susceptible to TDCIPP-induced delays in zygotic genome methylation16, suggesting that downstream effects on embryonic development may be associated with impacts on methylation during cleavage. Therefore, we conducted whole-genome bisulfite sequencing (WGBS) to identify the magnitude and extent, if any, of 5-azaC- and TDCPP-induced effects on the entire DNA methylome at 2 hpf (following exposure from 0.75–2 hpf) at single base-pair resolution17. Within this study, we found that rapid embryonic uptake of 250 μM 5-azaC or 2 μM TDCIPP from 0.75 to 2 hpf resulted in chemical- and chromosome-specific alterations in cytosine methylation at 2 hpf, and that TDCIPP-induced impacts to the zebrafish DNA methylome at 2 hpf are complex and primarily localized to regions outside of classical CpG islands17. Importantly, this study yielded positional information about altered CpG methylation within TDCIPP-susceptible loci – data that may be useful for identifying future biomarkers of TDCIPP exposure within zebrafish and humans.
Interestingly, TDCIPP did not inhibit DNA methyltransferase activity in vitro up to 500 μM17, suggesting that TDCIPP may induce CpG hypomethylation via depletion of methyl donors (e.g., S-adenosyl-L-methionine). Methyl supplementation has the potential to reverse chemically-induced DNA methylation18,19 and maternal supplementation with methyl donors (such as folic acid) has been shown to alter methylation profiles in offspring20,21,22. Therefore, the objectives of the current study were to: (1) quantify TDCIPP-induced impacts on epiboly as a function of concentration; (2) determine whether co-exposure with folic acid mitigates TDCIPP-induced impacts on epiboly; and (3) based on TDCIPP-susceptible loci identified from our previous WGBS study, rely on bisulfite amplicon sequencing (BSAS) – a cost-effective, targeted sequencing-based strategy23 – to monitor CpG methylation dynamics across multiple TDCIPP concentrations and developmental stages in the presence or absence of folic acid.
Materials and Methods
Animals
Adult 5D zebrafish (Danio rerio) were maintained and bred on a recirculating system as previously described by Vliet et al.24. All embryos were collected immediately after spawning and staged according to previously described methods25. Adult breeders were handled and treated in accordance with an Institutional Animal Care and Use Committee (IACUC)-approved animal use protocol (#20150035) at the University of California, Riverside.
Chemicals
TDCIPP (98% purity) and folic acid (FA) (≥97%) were purchased from ChemService (West Chester, PA) and Sigma-Aldrich (Saint Louis, MO), respectively. TDCIPP stocks and working solutions were prepared in embryo media (EM) as previously described16. FA treatments were prepared in EM immediately prior to each experiment to minimize degradation.
Epiboly and Deformity Assessments
Viable 5D embryos (30 initial embryos per replicate beaker) were staged within 30 min after spawning, randomly distributed to clean 50-mL glass beakers (three beakers per treatment for 4- and 6-hpf time-points; nine beakers per treatment for 24-hpf time-point), and then exposed to 10 mL of vehicle (0.1% DMSO) or TDCIPP (0.78, 1.56, or 3.12 μM) in the presence or absence of 1 mM FA. Thirty initial embryos per replicate beaker were exposed to ensure that a sufficient number of embryos were remaining for imaging procedures described below. TDCIPP concentrations were selected based on our previous work16, a study published by Fu et al.15, and additional range-finding exposures using epiboly progression as a read-out. The FA concentration selected for this study was based on a previous study that successfully mitigated selenite-induced cardiac and neural abnormalities using 1 mM FA supplementation19. In addition, as 1 mM FA (~0.44 mg/ml FA) approached the aqueous solubility at room temperature and pH 7 (all FA treatment solutions in this study exhibited a yellowish-orange color), this concentration represented the maximum allowable concentration for determining whether FA mitigates TDCIPP-induced effects.
All exposures were conducted under static conditions in the dark (to prevent photolysis of FA) at 28°C from 0.75 hpf to 4, 6, or 24 hpf. At 4 or 6 hpf, embryos were chilled for 30 min at 4°C to cease development for imaging, oriented in lateral recumbency within 4% methyl cellulose (10 embryos per beaker), and imaged using a Leica MZ10 F stereomicroscope equipped with a DMC2900 camera. Epiboly progression was assessed by quantifying cell height above the yolk at 4 and 6 hpf, and as percent epiboly at 6 hpf; all measurements were performed within Image J (https://imagej.nih.gov/ij/). At 24 hpf, embryos were imaged directly in EM (20 embryos per beaker) and scored as normal, deformed, or dead.
Genomic DNA Extractions
Embryos were exposed to vehicle (0.1% DMSO) or TDCIPP (0.78 or 3.12 μM) in the presence or absence of 1 mM FA as described above (60 initial embryos per replicate beaker; 12 beakers per treatment for 2 hpf; four beakers per treatment for 4 and 6 hpf). Sixty initial embryos per replicate beaker were exposed to ensure that 50 embryos per replicate were remaining for genomic DNA extractions. At 2 hpf, three beakers containing 50 embryos each were pooled into a single 2-mL cryovial and snap-frozen in liquid nitrogen, resulting in four replicate pools containing 150 embryos per pool; at 4 and 6 hpf, 50 embryos from each beaker were transferred to four 2-mL cryovials and snap-frozen in liquid nitrogen, resulting in four replicate pools containing 50 embryos per pool. Genomic DNA was extracted from 2-, 4-, or 6-hpf embryos using a slightly modified version of the Wizard Genomic DNA Purification Kit (Promega, Madison, WI). Following homogenization, each homogenate was spiked with 10 μL proteinase K (>600 mAU/ml) and 1 μL 100 mg/ml RNase A, incubated at 55°C while shaking at 300 rpm, and then incubated with an additional 1 μL RNase A for 4 h at 37°C. Samples were cleaned and concentrated using a Genomic DNA Clean & Concentrator-10 Kit (Zymo Research Corp., Irvine, CA) per the manufacturer’s instructions. DNA was quantified using a Qubit 2.0 Fluorometer, and DNA quality was confirmed on a 1% agarose gel.
Bisulfite Amplicon Sequencing (BSAS)
BSAS was adapted based on previously published protocols23. Genomic DNA samples (50 ng for 2 hpf; 150 ng for 4 hpf; 150 ng for 6 hpf) were bisulfite-treated using an EZ DNA Methylation-Lightning Kit (Zymo). Ten regions of interest (ROIs) were identified based on TDCIPP-susceptible loci identified from our previous WGBS study (SI Table S1)17. These 10 ROIs were selected based on five CpGs that were also impacted by FA within humans21, while the remaining five ROIs were selected to represent exonic and intergenic regions with a range of methylation differences relative to vehicle controls. Primers were designed with Zymo’s Bisulfite Primer Seeker (http://www.zymoresearch.com/tools/bisulfite-primer-seeker) to amplify 300- to 400-bp ROIs (SI Table S1). ROIs were amplified within a 25-μL reaction containing ZymoTaq PreMix, 5 ng bisulfite-treated DNA, and 2.5 μM of each primer using an Eppendorf Mastercycler Nexus Thermocycler. PCR conditions were: 95°C for 10 min, followed by 35 cycles of 95°C for 30 s, 51–65°C (depending on the ROI) for 30 s, and 72°C for 30 s, with a final extension step at 72°C for 7 min. PCR products were cleaned and purified using a QIAquick 96 PCR Purification Kit (Qiagen), and amplicons were quantified using a Qubit 2.0 Fluorometer. Amplicon quality was confirmed using an Agilent 2100 Bioanalyzer system and high-sensitivity DNA kit. Amplicons were pooled prior to library preparation at a concentration of 0.1 ng per amplicon. Library preparation was performed with the Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA) and indexed by treatment replicate. Library quality and quantity were confirmed using a Qubit 2.0 Fluorometer and 2100 Bioanalyzer system. Libraries were then pooled by stage (2, 4, or 6 hpf), diluted to a concentration of 1.3 pM (with 40% PhiX control), and paired-end (2×150) sequenced on our Illumina MiniSeq Sequencing System using a 300-cycle Mid Output Reagent Kit.
All sequencing data were uploaded to Illumina’s BaseSpace in real-time for downstream analysis of quality control and potential differences in methylation. All three sequencing runs resulted in >92% of reads with ≥Q30. Reads were aligned and duplicate reads were removed using a ROI-based enrichment manifest within MethylSeq. MethylKit was used on positions with >10X coverage to assess significant methylation differences of >1% relative to stage-matched vehicle controls; q≤0.01 was used to minimize false positives associated with multiple comparisons. To identify potential interactions between TDCIPP and FA, a quasi-binomial model was used to run logistic regression analysis on all CpG sites with >10X coverage using RStudio (https://www.rstudio.com).
DNA Methylation Quantitation
Global 5-methylcytosine (5-mC) was measured using 10 ng of genomic DNA (three samples per treatment) and a 5-mC DNA ELISA kit (Zymo) per the manufacturer’s instructions. Only two replicates were used for 2-hpf samples, as there was <10 ng of genomic DNA within two out of four replicate samples following BSAS.
Statistical Analyses
All statistical analyses were performed within RStudio (https://www.rstudio.com). Two-way analysis of variance (ANOVA) was performed on 5-mC ELISA data, log-transformed cell height data, and arcsin-transformed percent epiboly data to meet assumptions for normality and equal variance. Pair-wise comparisons were analyzed using Tukey-based post-hoc tests. Percent mortality and deformity data at 24 hpf were analyzed using binomial logistic regressions. Significance was determined at p≤0.05.
Accession Number
Raw Illumina (fastq.gz) sequencing files (144 files totaling 2.59 GB) are available via NCBI’s BioProject database under BioProject ID PRJNA395080.
Results and Discussion
During early zebrafish embryogenesis, cells proliferate and aggregate on the animal pole until 3.25 hpf prior to epiboly initiation and progression. As TDCIPP was previously shown to delay epiboly at a single concentration (3 μM)15, we quantified the effects of TDCIPP on epiboly as a function of TDCIPP concentration. Moreover, we investigated whether co-exposure with a methyl donor (folic acid, FA) mitigated the effects of TDCIPP on epiboly. Initiation of TDCIPP exposure at 0.75 hpf resulted in an increase in average cell height in a concentration-dependent manner at 4 and 6 hpf (Figure 1A and 1B), and embryos exposed to 3.12 μM TDCIPP were delayed by as much as 3 h at 6 hpf relative to vehicle controls (Figure 1C). Embryo mortality at 24 hpf was significantly increased following exposure to 1.56 and 3.12 μM TDCIPP (Figure 1D), and TDCIPP exposure resulted in an increase in abnormalities at all test concentrations following exposure from 0.75–24 hpf (Figure 1E and 1F). Interestingly, while exposure to FA alone from 0.75–24 hpf did not affect epiboly progression at 4 or 6 hpf nor result in deformities or mortality at 24 hpf, FA was unable to mitigate TDCIPP-induced effects at all three developmental stages evaluated (Figure 1A–1E).
Figure 1.
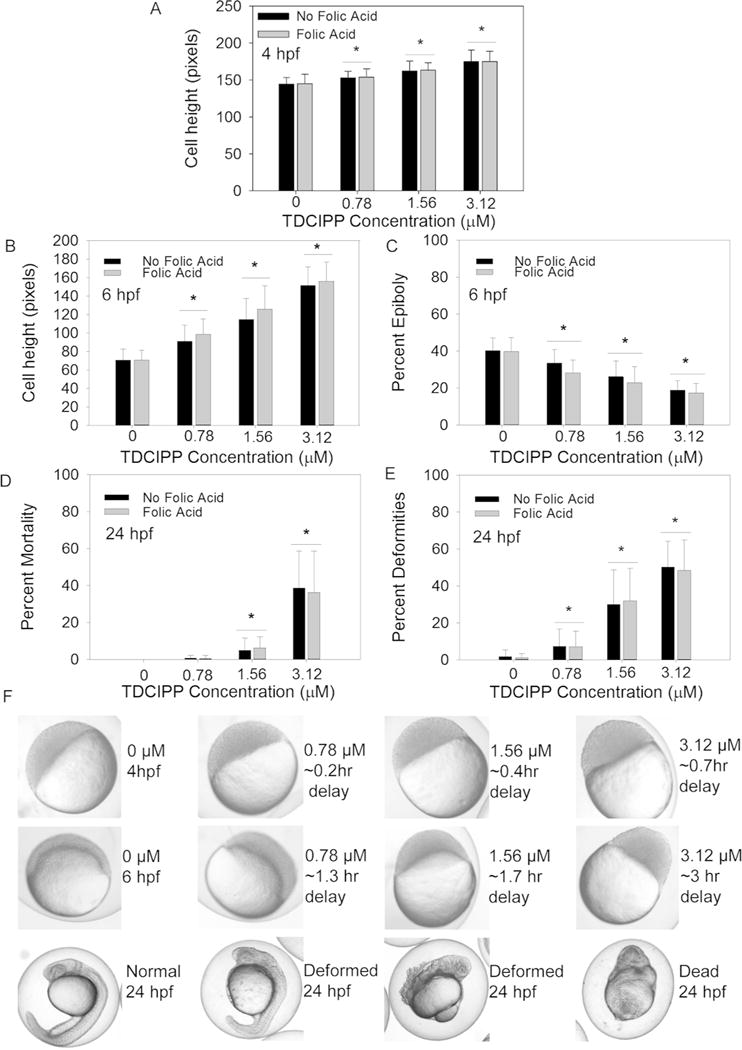
Developmental impacts of vehicle (0.1% DMSO) or TDCIPP (0.78, 1.56, or 3.12 μM) in the presence or absence of 1 mM FA on (A) cell height at 4 hpf; (B) cell height at 6 hpf; (C) percent epiboly at 6 hpf; (D) percent mortality at 24 hpf; and (E) percent deformities at 24 hpf. (F) Representative images of epiboly delays at 4 and 6 hpf and deformities at 24 hpf (embryos are approximately 0.7 mm in diameter). Data are presented as mean ± standard deviation. Black bars represent TDCIPP-only treatments in EM, while gray bars represent TDCIPP treatments containing 1 mM FA. Asterisks denote significant differences (p≤0.05) within TDCIPP treatments (with or without FA) relative to respective vehicle controls. For 4- and 6-hpf data, a total of 30 total embryos were imaged across three replicate beakers (10 embryos per beaker; for 24-hpf data, mean percentages are based on nine beakers containing 20 imaged embryos per beaker.
Folate is a cofactor in the formation of S-adenosyl-L-methionine – a substrate required for cytosine methylation. FA supplementation is recommended for women of reproductive age through 12 weeks of gestation to decrease the incidence of neural tube defects26, and these impacts are thought to be mediated by DNA methylation. Within animal models, FA has also been used as a methyl donor to rescue chemically-induced adverse developmental phenotypes. For example, a FA-containing diet fed to female agouti mice restored Avy locus methylation levels and offspring coat color altered by bisphenol A exposure18. Similarly, co-exposure with 1 mM FA significantly decreased the effect of selenite on zebrafish embryo hatch and survival19. However, in our study, co-exposure to 1 mM FA was unable to block or mitigate TDCIPP-induced effects on zebrafish epiboly, suggesting that TDCIPP-induced impacts on embryonic development may not be due to methyl donor depletion resulting from TDCIPP exposure.
To determine whether FA mitigated TDCIPP-induced effects on DNA methylation, we examined global methylation levels in genomic DNA from embryos exposed to TDCIPP in the presence and absence of FA. Consistent with previously published data2, global methylated cytosine ranged between 4–6% for all treatments (Figure S1). However, similar to our previous study that relied on an LC/MS-MS-based method17, no significant differences in global methylation were detected across all treatments using a 5-mC DNA ELISA, suggesting that global methylation assays are not sensitive enough to detect base resolution changes induced by TDCIPP in zebrafish embryos. Therefore, using our existing WGBS dataset for TDCIPP17, we relied on BSAS as a targeted sequencing strategy to monitor differentially methylated CpGs within TDCIPP-susceptible loci.
Out of 10 amplicons analyzed, nine amplicons were aligned and 94 total CpG sites were detected, resulting in 12–13M reads passing filter (Table S2) with an average of 143,000 targeted unique aligned reads and a mean target coverage of 4,038 reads (Table S3). Overall, consistent with previous studies2, CpG methylation averaged between 81–88% (Figure S2) while non-CG methylation ranged from 0.6–1.09% (Table S3), indicating that bisulfite conversion was successful across all samples. Interestingly, clustered heat maps revealed variable impacts of TDCIPP and FA at different stages (Figure 2A; Table S4); raw percent methylation data for all treatments (including vehicle controls) and stages are available within Tables S5 (2 hpf), S6 (4 hpf), and S7 (6 hpf). Relative to vehicle controls, the magnitude of the effect on methylation decreased from 2 to 6 hpf despite being continuously immersed in TDCIPP, suggesting that TDCIPP-effects on methylation are reversible within a 4-h exposure period even though internal TDCIPP doses increase from 2 to 6 hpf17. In addition, the direction of response (hyper- vs. hypo-methylation) was not consistent from 2 to 6 hpf within each treatment. Logistic regression analysis was used to detect the potential interaction of FA and TDCIPP on CpG methylation at each stage (Figure 2B; Figure S3). While a significant interaction between TDCIPP and FA (p≤0.05) was detected for nine CpG positions at 2, 4, or 6 hpf (Figure 2B), none of these positions were shared among stages. Finally, based on pair-wise comparisons relative to stage-matched vehicle controls, the number of differentially methylated bases decreased from 2 to 6 hpf (Figure 2C) and included both hypo- and hyper-methylated positions. Overall, our BSAS data suggest that methylation dynamics are strongly dependent on TDCIPP concentration and developmental stage, and that the majority of TDCIPP-induced effects on methylation at 2 hpf do not persist at 4 and 6 hpf despite continuous exposure.
Figure 2.
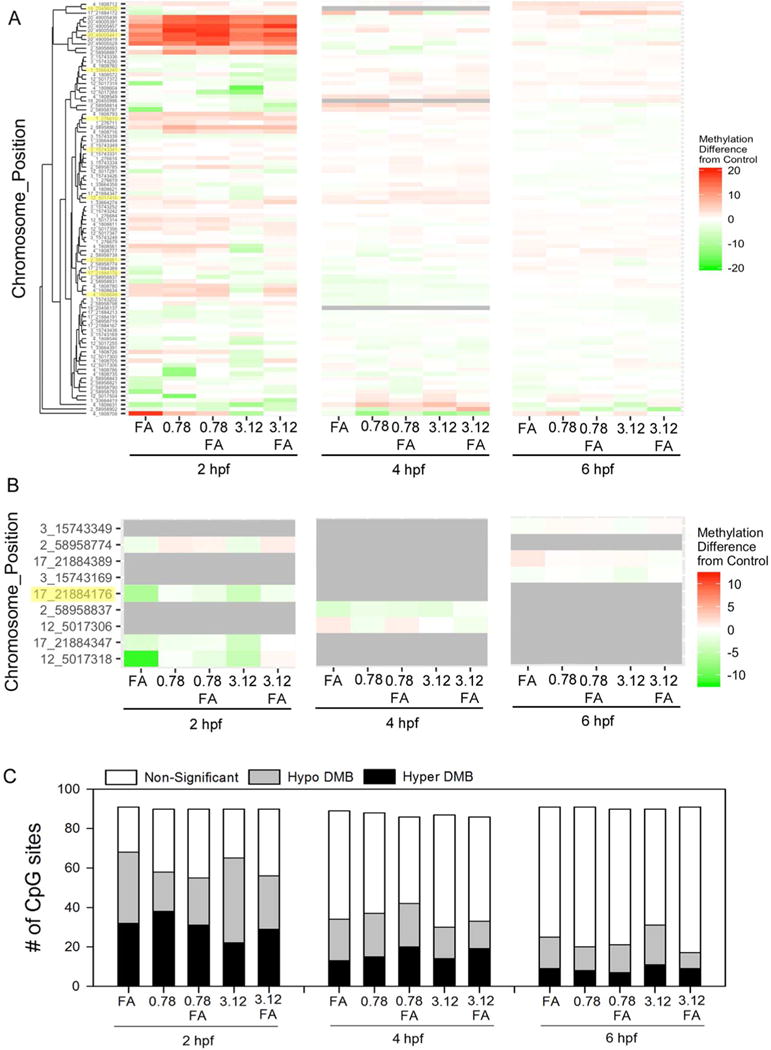
Bisulfite amplicon sequencing of ten amplicons from embryos exposed to vehicle (0.1% DMSO) or TDCIPP (0.78 or 3.12 μM) in the presence or absence of 1 mM FA from 0.75 hpf to 2, 4, or 6 hpf. Complete linkage-based clustering of methylation differences (relative to stage-matched vehicle controls) by position for (A) all CpG sites or (B) CpG sites with a significant interaction (p≤0.05) between TDCIPP and FA at 2, 4, or 6 hpf. Within Panels A and B, positions highlighted yellow represent CpG sites that were previously identified based on whole-genome bisulfite sequencing. (C) Summary of MethylKit-based pairwise comparisons between treatments and stage-matched vehicle controls. White, gray, and black bars represent the number of non-significant, hypo-methylated, and hyper-methylated CpG sites, respectively (q≤0.01). N = four genomic DNA samples per treatment.
In conclusion, our findings demonstrate that TDCIPP significantly delays zebrafish epiboly following initiation of exposure at 0.75 hpf, a phenotype that is preceded by complex, dynamic alterations in DNA methylation. In the current study, TDCIPP impacted DNA methylation in a highly stochastic manner where the magnitude and direction of alterations in DNA methylation were not consistent throughout epiboly, suggesting that (1) these epigenetic marks are not stable during this brief window of embryonic development within zebrafish and (2) if these experiments were repeated multiple times, the magnitude and direction of response for TDCIPP-induced effects on these same CpG sites at 2, 4, and 6 hpf may vary from experiment-to-experiment. Thus, although a subset of CpG sites were methylated in a TDCIPP-dependent manner, we were unable to identify positions that may serve as biomarkers of TDCIPP exposure, raising questions about the utility and reliability of CpG methylation as biomarkers of exposure within animal models and human populations. Nevertheless, this study demonstrates the importance of follow-up, targeted approaches such as BSAS for monitoring DNA methylation dynamics following identification and prioritization of susceptible loci using WGBS. In addition, this study further provides insight into chemically-induced impacts on zebrafish methylation during embryogenesis that, given the strong level of variability within a highly controlled laboratory study, may help explain potential sources of uncertainty within human population-based environmental epigenetic studies.
Supplementary Material
Acknowledgments
Funding was provided by the National Institutes of Health grants (R21ES022797 and R01ES027576) and USDA National Institute of Food and Agriculture Hatch Project 1009609 to DCV. The authors greatly appreciate Michael Park for his assistance with programming in R.
Footnotes
Supporting Information
Supporting Information Files 1 and 2 contain supporting information regarding BSAS amplicons, sequencing quality, and sequencing results. 5-mC DNA ELISA data and logistic regression results are provided in Supporting Information File 2. This information is available free of charge via the Internet at http://pubs.acs.org/journal/estlcu.
Conflict of Interest Statement
The authors declare no conflict of interest.
References
- 1.Kane DA, Hammerschmidt M, Mullins MC, Maischein HM, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Haffter P, Heisenberg CP, Jiang YJ, Kelsh RN, Odenthal J, Warga RM, Nusslein-Volhard C. The zebrafish epiboly mutants. Development. 1996;123:47–55. doi: 10.1242/dev.123.1.47. [DOI] [PubMed] [Google Scholar]
- 2.Potok ME, Nix DA, Parnell TJ, Cairns BR. Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell. 2013;153:759–772. doi: 10.1016/j.cell.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang L, Zhang J, Wang JJ, Wang L, Zhang L, Li G, Yang X, Ma X, Sun X, Cai J, et al. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell. 2013;153:773–84. doi: 10.1016/j.cell.2013.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin CC, Laforest L, Akimenko MA, Ekker M. A role for DNA methylation in gastrulation and somite patterning. Dev Biol. 1999;206:189–205. doi: 10.1006/dbio.1998.9105. [DOI] [PubMed] [Google Scholar]
- 5.Stapleton HM, Klosterhaus S, Eagle S, Fuh J, Meeker JD, Blum A, Webster TF. Detection of organophosphate flame retardants in furniture foam and U.S. house dust. Environ Sci Technol. 2009;43:7490–7495. doi: 10.1021/es9014019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stapleton HM, Klosterhaus S, Keller A, Ferguson PL, van Bergen S, Cooper E, Webster TF, Blum A. Identification of flame retardants in polyurethane foam collected from baby products. Environ Sci Technol. 2011;45:5323–31. doi: 10.1021/es2007462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butt CM, Congleton J, Hoffman K, Fang M, Stapleton HM. Metabolites of Organophosphate Flame Retardants and 2-Ethylhexyl Tetrabromobenzoate (EH-TBB) in Urine from Paired Mothers and Toddlers. Environ Sci Technol. 2014;48:10432–8. doi: 10.1021/es5025299. [DOI] [PubMed] [Google Scholar]
- 8.Hoffman K, Butt CM, Webster TF, Preston EV, Hammel SC, Makey C, Lorenzo A, Cooper E, Carignan C, Meeker J, Hauser R, Soubry A, Murphy SK, Price T, Hoyo C, Mendelsohn E, Congleton J, Daniels JL, Stapleton HM. Temporal Trends in Exposure to Organophosphate Flame Retardants in the United States. Environ Sci Technol Lett. 2017;4:112–118. doi: 10.1021/acs.estlett.6b00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarema KA, Hunter DL, Shaffer RM, Behl M, Padilla S. Acute and developmental behavioral effects of flame retardants and related chemicals in zebrafish. Neurotoxicol Teratol. 2015;52(Pt B):194–209. doi: 10.1016/j.ntt.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dishaw LV, Hunter DL, Padnos B, Padilla S, Stapleton HM. Developmental exposure to organophosphate flame retardants elicits overt toxicity and alters behavior in early life stage zebrafish (Danio rerio) Toxicol Sci. 2014;142:445–54. doi: 10.1093/toxsci/kfu194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noyes PD, Haggard DE, Gonnerman GD, Tanguay RL. Advanced morphological - behavioral test platform reveals neurodevelopmental defects in embryonic zebrafish exposed to comprehensive suite of halogenated and organophosphate flame retardants. Toxicol Sci. 2015;145:177–95. doi: 10.1093/toxsci/kfv044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, Ji K, Jo A, Moon HB, Choi K. Effects of TDCPP or TPP on gene transcriptions and hormones of HPG axis, and their consequences on reproduction in adult zebrafish (Danio rerio) Aquat Toxicol. 2013:134–135. 104–11. doi: 10.1016/j.aquatox.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Wang Q, Lam JC, Han J, Wang X, Guo Y, Lam PK, Zhou B. Developmental exposure to the organophosphorus flame retardant tris(1,3-dichloro-2-propyl) phosphate: estrogenic activity, endocrine disruption and reproductive effects on zebrafish. Aquat Toxicol. 2015;160:163–71. doi: 10.1016/j.aquatox.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Liu C, Wang Q, Liang K, Liu J, Zhou B, Zhang X, Liu H, Giesy JP, Yu H. Effects of tris(1,3-dichloro-2-propyl) phosphate and triphenyl phosphate on receptor-associated mRNA expression in zebrafish embryos/larvae. Aquat Toxicol. 2013:128–129. 147–57. doi: 10.1016/j.aquatox.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 15.Fu J, Han J, Zhou B, Gong Z, Santos EM, Huo X, Zheng W, Liu H, Yu H, Liu C. Toxicogenomic responses of zebrafish embryos/larvae to tris(1,3-dichloro-2-propyl) phosphate (TDCPP) reveal possible molecular mechanisms of developmental toxicity. Environ Sci Technol. 2013;47:10574–10582. doi: 10.1021/es401265q. [DOI] [PubMed] [Google Scholar]
- 16.McGee SP, Cooper EM, Stapleton HM, Volz DC. Early zebrafish embryogenesis is susceptible to developmental TDCPP exposure. Environ Health Perspect. 2012;120:1585–91. doi: 10.1289/ehp.1205316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Volz DC, Leet JK, Chen A, Stapleton HM, Katiyar N, Kaundal R, Yu Y, Wang Y. Tris(1,3-dichloro-2-propyl) phosphate induces genome-wide hypomethylation within early zebrafish embryos. Environ Sci Technol. 2016;50:10255–10263. doi: 10.1021/acs.est.6b03656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 2007;104:13056–61. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Y, Wu M, Li D, Li X, Li P, Zhao J, Luo M, Guo C, Gao X, Lu C, Ma X. Embryonic developmental toxicity of selenite in zebrafish (Danio rerio) and prevention with folic acid. Food Chem Toxicol. 2012;50:2854–2863. doi: 10.1016/j.fct.2012.04.037. [DOI] [PubMed] [Google Scholar]
- 20.Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr. 2002;132:2393S–2400S. doi: 10.1093/jn/132.8.2393S. [DOI] [PubMed] [Google Scholar]
- 21.Joubert BR, den Dekker HT, Felix JF, Bohlin J, Ligthart S, Beckett E, Tiemeier H, van Meurs JB, Uitterlinden AG, Hofman A, et al. Maternal plasma folate impacts differential DNA methylation in an epigenome-wide meta-analysis of newborns. Nat Commun. 2016;7:10577. doi: 10.1038/ncomms10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barua S, Kuizon S, Brown WT, Junaid MA. DNA methylation profiling at single-base resolution reveals gestational folic acid supplementation influences the epigenome of mouse offspring cerebellum. Front Neurosci. 2016;10:1–14. doi: 10.3389/fnins.2016.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masser DR, Stanford DR, Freeman WM. Targeted DNA methylation analysis by next-generation sequencing. J Visualized Exp. 2015;96 doi: 10.3791/52488. e5248810.3791/52488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vliet SM, Ho TC, Volz DC. Behavioral screening of the LOPAC1280 library in zebrafish embryos. Toxicol App Pharmacol. 2017;329:241–248. doi: 10.1016/j.taap.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 26.De-Regil LM, Peña-Rosas JP, Fernández-Gaxiola AC, Rayco-Solon P. Effects and safety of periconceptional oral folate supplementation for preventing birth defects. Cochrane Database Syst Rev. 2015;12:CD007950. doi: 10.1002/14651858.CD007950.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


