Summary
Inositol pyrophosphates are novel signaling molecules possessing high-energy pyrophosphate bonds and involved in a number of biological functions. Here, we report the correct identification and characterization of the kinases involved in the inositol pyrophosphate biosynthetic pathway in Trypanosoma brucei: inositol polyphosphate multikinase (TbIPMK), inositol pentakisphosphate 2-kinase (TbIP5K) and inositol hexakisphosphate kinase (TbIP6K). TbIP5K and TbIP6K were not identifiable by sequence alone and their activities were validated by enzymatic assays with the recombinant proteins or by their complementation of yeast mutants. We also analyzed T. brucei extracts for the presence of inositol phosphates using polyacrylamide gel electrophoresis and high performance liquid chromatography. Interestingly, we could detect inositol phosphate (IP), inositol 4,5-bisphosphate (IP2), inositol 1,4,5-trisphosphate (IP3) and inositol hexakisphosphate (IP6) in T. brucei different stages. Bloodstream forms unable to produce inositol pyrophosphates, due to downregulation of TbIPMK expression by conditional knockout, have reduced levels of polyphosphate and altered acidocalcisomes. Our study links the inositol pyrophosphate pathway to the synthesis of polyphosphate in acidocalcisomes, and may lead to better understanding of these organisms and provide new targets for drug discovery.
Keywords: Acidocalcisome, polyphosphate, inositol pyrophosphate, calcium, Trypanosoma brucei
Graphical Abstract
We report the correct identification and characterization of the kinases involved in the inositol pyrophosphate biosynthetic pathway in Trypanosoma brucei and identified the substrates and products of these reactions. We provide functional evidence of their activity by complementation of yeast mutants, and we establish the relevance of this pathway for acidocalcisome polyphosphate synthesis in trypanosomes.
Introduction
Myo-inositol is an essential precursor for the synthesis of soluble inositol phosphates (IPs) and lipid-bound inositols called phosphoinositides (PIPs) (Irvine et al., 2001). After inositol incorporation into the lipid phosphatidylinositol (PI), the inositol ring is phosphorylated to PIPs such as phosphatidylinositol 4,5-bisphosphate (PIP2) through the action of a phosphatidylinositol phosphate (PIP) kinase. PIP2 is cleaved by a phosphoinositide phospholipase C (PI-PLC) (Cocco et al., 2015) to inositol 1,4,5-trisphosphate (IP3) (Fig. 1) and 1,2-diacylglycerol (DAG), which are important second messengers. While DAG stimulates a protein kinase C (Nishizuka, 1986), IP3 stimulates an IP3 receptor to release Ca2+ from intracellular stores (Berridge, 2009) and can be further metabolized to other soluble IPs by several kinases and phosphatases.
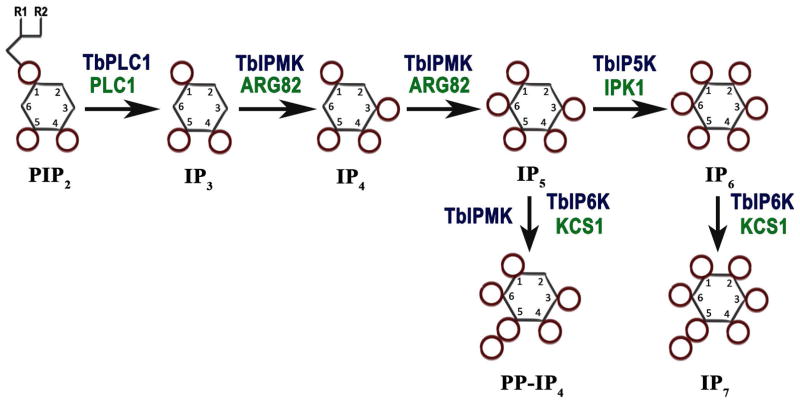
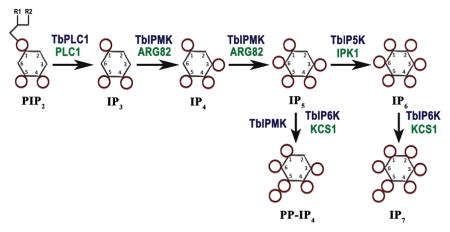
Fig. 1.
Inositol phosphate pathway in Trypanosoma brucei. The soluble IP pathway starts with hydrolysis of PIP2 by TbPI-PLC1, releasing IP3 that is phosphorylated by TbIPMK to generate IP4 and IP5. IP5 is phosphorylated by TbIP5K to generate IP6. IP5 and IP6 can be further phosphorylated by TbIPMK or TbIP6K to generate inositol pyrophosphates PP-IP4 and IP7. Names of the equivalent yeast enzymes are in green.
The inositol phosphate multikinase (IPMK) has dual 3-kinase/6-kinase activity and catalyzes the conversion of IP3 into inositol tetrakisphosphate (IP4) and inositol pentakisphosphate (IP5). IP5 is converted into inositol hexakisphosphate (IP6), the fully phosphorylated myo-inositol also known as phytic acid, by the 2-kinase activity of inositol pentakisphosphate kinase (IP5K, or IPPK). Further phosphorylation of IP6 by the inositol hexakisphosphate kinase (IP6 kinase or IP6K) results in the production of diphosphoinositol polyphosphates (PP-IPs), also known as inositol pyrophosphates. These are IPs characterized by containing one or more high-energy pyrophosphate moiety. PP-IPs were discovered in the early 1990’s, in Dictyostelium discoideum (Europe-Finner et al., 1991, Mayr GW, 1992, Stephens et al., 1993), Entamoeba histolytica (Martin et al., 1993), and in mammalian cells (Menniti et al., 1993). The best-characterized member of this class is 5-diphosphoinositol pentakisphosphate (5-PP-P5 or IP7), which has five of the myo-inositol hydroxyls monophosphorylated, while the sixth, at the 5-position, contains a pyrophosphate group (Albert et al., 1997). The IP6K can also metabolize IP5 to disphosphoinositol tetrakisphosphate (PP-IP4) (Saiardi et al., 2000, Losito et al., 2009). Another isomer of IP7, containing a pyrophosphate at the 1-position, can also be formed by a more recently identified enzyme termed diphosphoinositol pentakisphosphate kinase (PP-IP5K), though this enzyme appears to be predominantly associated physiologically with the formation of diphosphoinositol hexakisphosphate (PP2-IP4 or IP8) (Choi et al., 2007).
Among the many roles attributed to PP-IPs are the regulation of telomere length (Saiardi et al., 2005, York et al., 2005), DNA repair by homologous recombination (Luo et al., 2002, Jadav et al., 2013), response to hyperosmotic stress (Pesesse et al., 2004, Choi et al., 2007), vesicle trafficking (Saiardi et al., 2000, Saiardi et al., 2002), apoptosis (Morrison et al., 2001, Nagata et al., 2005), autophagy (Nagata et al., 2010), binding of pleckstrin homology domains to phospholipids and proteins (Luo et al., 2003, Gokhale et al., 2013), transcription of glycolytic enzymes (Szijgyarto et al., 2011), hemostasis (Ghosh et al., 2013), phagocytic and bactericidal activities of neutrophils (Prasad et al., 2011), epigenetic modifications to chromatin (Burton et al., 2013) and exocytic insulin secretion (Illies et al., 2007). PP-IPs may signal through allosteric interaction with proteins (i.e. binding to pleckstrin homology (PH) or other domains of proteins) or by phosphotransfer reactions (Saiardi, 2012, Shears, 2015, Wild et al., 2016). The phosphotransfer reaction is non-enzymatic and requires a phospho-serine residue within an acidic region and consists in adding a second phosphate to the phosphor-serine resulting in pyrophosphorylation (Saiardi, 2012).
Trypanosoma brucei, which belongs to the group of parasites that causes African trypanosomiasis (sleeping sickness), possesses a PI-PLC that is stimulated by very low Ca2+ concentrations (King-Keller et al., 2015) and an IP3 receptor that localizes to the acidocalcisomes instead of the endoplasmic reticulum (Huang et al., 2013). We now found that they also possess orthologs to IPMK, IP5K and IP6K, but do not have recognizable orthologs to PP-IP5K, inositol 1,4,5-trisphosphate 3-kinases (ITPKs) and inositol tetrakisphosphate 3-kinase 1 (ITPK1) (Table S1). The ortholog to IPMK (TbIPMK) was recently reported as essential for the bloodstream forms of the parasites (Cestari et al., 2015), suggesting that the soluble inositol phosphate pathway is essential for the parasite. The orthologs to IP5K and IP6K were not recognizable by sequence only and were wrongly annotated as a putative hypothetical protein and as inositol polyphosphate-like protein, respectively. In the present study, we thoroughly characterized the soluble inositol phosphate pathway of T. brucei. We cloned, expressed and biochemically characterized the recombinant enzymes from T. brucei, complemented yeast mutants to demonstrate their function, analyzed their products, studied the inositol phosphate metabolism of T. brucei cells, and revealed the link of this pathway to the synthesis of polyphosphate in acidocalcisomes.
Results
Sequence analysis of T. brucei inositol phosphate kinases
Gene homology searches followed by validation of their activity (see below) have allowed to identify in the T. brucei genome (http://www.tritrypdb.org/tritrypdb/) the presumably gene orthologs to the inositol phosphate kinases encoding inositol polyphosphate multikinase (IPMK in mammals, and Arg82p or Ipk2p in yeast) (Tb427tmp.211.3460); the IP5 kinase (IPPK or IP5K in mammals, and Ipk1p in yeast) (Tb427.04.1050); and the IP6 kinase (IP6K in mammals, and Kcs1p in yeast) (Tb427.07.4400), (Fig. 1), and named TbIPMK, TbIP5K, and TbIP6K, respectively (Table S1). No orthologs to diphosphoinositol pentakisphosphate kinase (PP-IP5K in mammals, or Vip1 in yeasts) were found, although orthologs to this gene are present in Apicomplexan (Laha et al., 2015) and Giardia (EuPathDB). The orthologs to TbIPMK, TbIP5K, and TbIP6K identified in T. cruzi (TcCLB.510741.110, TcCLB.506405.90, TcCLB.504213.90) and Leishmania major (LmjF.35.3140, LmjF.34.3700, LmjF.14.0340) shared 45%, 36%, 35%, and 29%, 28%, 24% amino acid identity, respectively. Those of T. brucei share 15%, 16%, and 15% identity with the human enzymes, respectively. Structural analyses (ELM and TMHMM servers) predicted no transmembrane domains. A signal peptide was predicted for TbIP5K, but not for TbIPMK or TbIP6K. Mature proteins of 342, 461, and 756 amino acids with predicted molecular weights of 38.8, 51, and 82.6 kDa, for TbIPMK, TbIP5K, and TbIP6K, respectively, were also predicted. Amino acids 138–147 of TbIPMK, and 588–596 of TbIP6K contained the conserved sequence PCVLDL(I)KL(M)G demonstrated previously as the putative inositol phosphate binding site that catalyzes the transfer of phosphate from ATP to inositol phosphates (Bertsch et al., 2000). TbIP5K possesses the sequence PVLDIELL (amino acids 269–276) instead. Both TbIPMK and TbIP6K have a SASLL or TSSLL domain present in most members of this family of enzymes and required for enzymatic activity (Saiardi et al., 2001b, Nalaskowski et al., 2002).
We utilized homologous recombination to add a hemagglutinin (HA) or c-Myc tag to the endogenous loci (Oberholzer et al., 2006) of TbIPMK, TbIP5K and TbIP6K. All three inositol phosphate kinases are expressed in procyclic forms (PCF) of T. brucei (Fig. 2A). Although the predicted MW of TbIP6K is 82.6 kDa the enzyme has multiple phosphorylations (Urbaniak et al., 2013) and these post-translational modifications (in addition to the HA tag) could result in a higher apparent MW. Interestingly, TbIP5K revealed no expression when using the HA-tag, but a protein with the expected size was detected when using a c-Myc tag (Fig. 2A). In addition, we tagged the three IP kinases in T. brucei bloodstream forms (BSF) but no clear bands were detected by western blot analyses although the tagged genes were expressed at the mRNA level (data not shown), suggesting that protein expression is lower in BSF than in PCF.
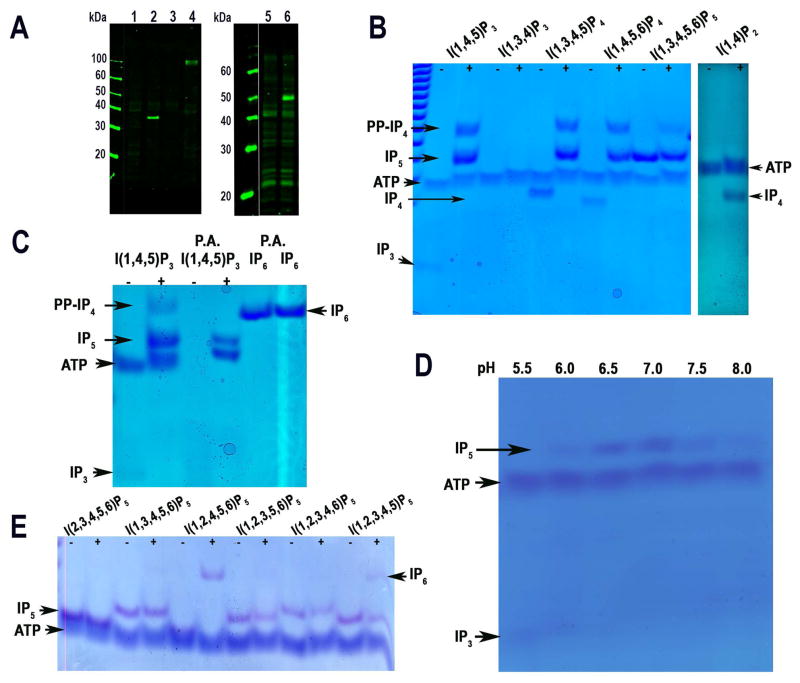
Fig. 2. Western blot analyses and enzymatic activity of TbIPMK.
A. Western blot analyses of T. brucei PCF expressing epitope-tagged TbIPMK, TbIP5K and TbIP6K. Left panel are HA tagged cell lines: 1, wild-type; 2, TbIPMK-HA; 3, wild-type; 4, TbIP6K-HA. Right panel is a c-Myc tagged line: 5, wild-type; 6, TbIP5K-cMyc.
B. Kinase reactions performed with recombinant TbIPMK (2 μg) using the indicated substrates at 250 μM for 1 hour at 37°C. TbIPMK can phosphorylate I(1,4,5)P3 but not I(1,3,4)P3 to produce I(1,3,4)P5 and PP-IP4, and can phosphorylate I(1,3,4,5)P4, and I(1,4,5,6)P4 to produce IP5 and PP-IP4. It can also phosphorylate I(1,3,4,5,6)P5 to PP-IP4. Other arrows show bands corresponding to ATP, IP4, and IP3. TbIPMK can phosphorylate I(1,4)P2 to produce IP4, and I(1,4,5)P3 to produce IP5 and PP-IP4.
C. Treatment of the sample with perchloric acid (PA) eliminates the band corresponding to PP-IP4 but has no effect on IP6. Other arrows indicate bands corresponding to ATP and IP3.
D. Optimum pH for TbIPMK activity is within the physiological range.
E. TbIPMK can only phosphorylate positions 3 and 6 of different IP5 derivatives to generate IP6. Note the lower synthesis of PP-IP4 using I(1,3,4,5,6)P5 as substrate compared to results obtained in (B) and (C). We observed that shorter enzymatic reaction time resulted in less PP-IP4 synthesis.
All results are representative of three or more independent experiments.
Characterization of the inositol phosphate multikinase (TbIPMK)
To characterize the enzymatic activity of TbIPMK we expressed it as fusion protein with an N-terminal polyhistidine tag, purified and tested its activity in vitro. We found that it catalyzes the formation of IP5 from IP3 or IP4, as detected by polyacrylamide gel electrophoresis (Fig. 2B). Inositol-1,4,5-trisphosphate (I(1,4,5)P3) but not inositol-1,3,4-trisphosphate (I(1,3,4)P3) could be used as substrate while both inositol-1,3,4,5-tetraphosphate (I(1,3,4,5)P4) and inositol-1,4,5,6-tetraphosphate (I(1,4,5,6)P4) could be used for the generation of inositol-1,3,4,5,6-pentakisphosphate (I(1,3,4,5,6)P5) (Fig. 2B), indicating that TbIPMK has a dual 3-kinase/6-kinase activity. An additional product, which runs closely but not identically to IP6, was also detected when IP3, IP4, or IP5 was used as substrate (Figs. 2B and 2C). The ability of IPMK to form PP-IP4, an inositol pyrophosphate containing 6 phosphates and thus migrating closely to IP6, has been demonstrated for the mammalian and yeast ortholog (Saiardi et al., 2001a, Zhang et al., 2001), and we therefore suspected that TbIPMK could have the same activity. A treatment with perchloric acid (PA), which degrades high-energy phosphoanhydride bonds (pyrophosphates) and is inactive against the phosphoester bond of IP6 (Fig. 2C) (Pisani et al., 2014), demonstrated that the highly phosphorylated product of TbIPMK is a pyrophosphate containing species, therefore PP-IP4. The pH optimum of rTbIPMK was determined. TbIPMK has the maximum activity for IP3 at the pH range of 6.5–7.0 (Fig. 2D). We also tested the ability of TbIPMK to phosphorylate different isomers of IP5. Recombinant TbIPMK was able to phosphorylate I(1,2,4,5,6)P5 and I(1,2,3,4,5)P5 to IP6 after short incubation times, but it was not able to use I(2,3,4,5,6)P5, I(1,3,4,5,6)P5, (I(1,2,3,5,6)P5, or (I(1,2,3,4,6)P5 as substrate (Fig. 2E). Although I(1,2,4,5,6)P5 and I(1,2,3,4,5)P5 would not be physiological substrates, the results again confirms a 3/6-kinase activity. Interestingly, TbIPMK could also phosphorylate I(1,4)P2 to IP4 (Fig. 2B). The mammalian IPMK has been reported to have PI3-kinase activity that produces PIP3 from PIP2 (Resnick et al., 2005). However, our in vitro activity tests using PIP2 as substrate revealed no such activity (data not shown) in agreement with the results of a previous report (Cestari et al., 2016).
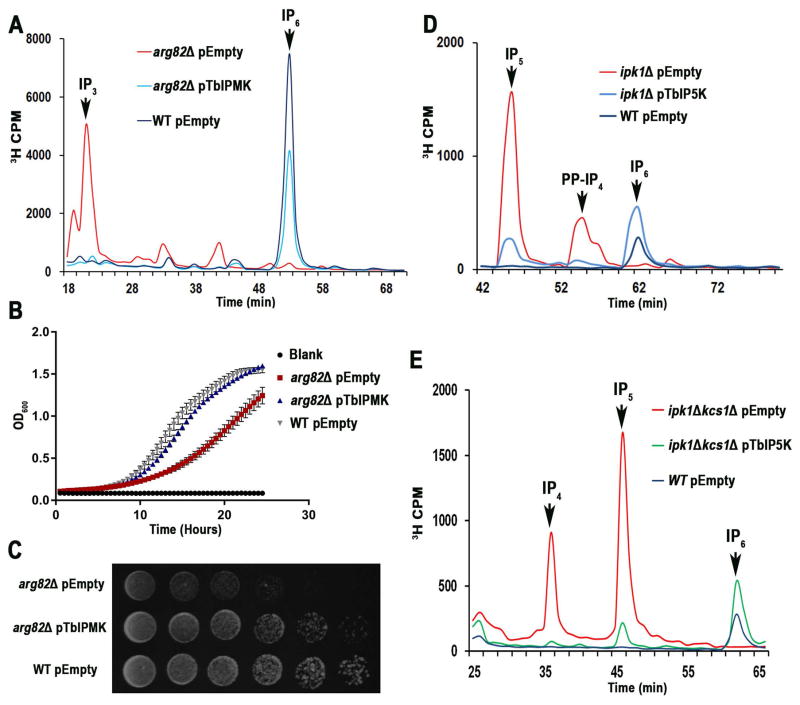
The ability of TbIPMK to act on IP3 in vivo was tested by complementation of a null mutant for its ortholog ARG82 (arg82Δ in Saccharomyces cerevisiae. Fig. 3A shows the HPLC analysis of soluble inositol phosphates isolated from yeast labeled with [3H]inositol. Arg82p phosphorylates IP3 to produce IP4 and IP5, and in its absence there is accumulation of IP3, instead of the accumulation of IP6 that occurs in wild type yeast (Fig. 3A). The metabolic pathway from IP3 to IP6 was restored by complementation with TbIPMK (Fig. 3A). These results indicate that TbIPMK function as part of the IP6 biosynthetic pathway established in yeast (York et al., 1999). We also examined the ability of TbIPMK to rescue the growth defect of arg82Δ yeast. Complementation of arg82Δ with TbIPMK rescued their growth defect (Fig. 3B, and 3C). Therefore, TbIPMK was able to complement yeast deficient in its ortholog Arg82p, providing molecular evidence of its function. The results also suggest that the pathway for IP5 synthesis is similar to that present in yeast with conversion of I(1,4,5)P3 into I(1,4,5,6)P4 and I(1,3,4,5,6)P5, TbIPMK acting as a 3/6-kinase. This is different from the pathway for synthesis of I(1,3,4,5,6)P5 present in humans, where the major activity of IP4 kinase is phosphorylation at the D-5 position (Chang et al., 2002).
Fig. 3. TbIPMK, and TbIP5K complementation of yeast mutants.
A. HPLC analysis of soluble inositol phosphates of S. cerevisiae arg82Δ mutants transformed with an empty vector (red) or a vector containing the entire open reading frame of TbIPMK (blue), and compared to those of wild-type (WT) yeast transformed with empty vector (black).
B. Growth of the same cells in liquid medium as estimated by measuring optical density at 660 nm. arg82Δ mutants had reduced growth, which was restored by expression of TbIPMK. Mean ± s.d. for three independent experiments, each one with 6 duplicates.
C. WT, and arg82Δ transformed with empty vector or arg82Δ transformed with TbIPMK (serially diluted 10-fold, 106–10 cells/spot from left to right) were spotted on YPD plates and incubated at 30°C for 2 days.
D. HPLC analysis of soluble inositol phosphates of Scipk1Δ mutants transformed with an empty vector (red) or a vector containing the entire open reading frame of TbIPMK (blue) and compared with wild type transformed with an empty vector (black).
E. HPLC analysis of Scipk1ΔKcs1Δ complemented with empty vector (red) or TbIP5K (green) shows reconstitution of IP6 synthesis. In black, wild type transformed with empty vector.
All results are representative of three or more independent experiments.
Characterization of the inositol pentakisphosphate kinase (TbIP5K)
Although expression of polyhistidine-tagged TbIP5K was obtained in bacteria and the recombinant protein had the expected molecular mass, we were not able to detect its activity in vitro, even in the presence of different isomers of IP5 (data not shown) suggesting that additional post-translational modifications are needed. In this regard, activity of human IP5K could only be obtained when expressed in insect cells (Verbsky et al., 2002). However, TbIP5K was able to complement null mutant yeast deficient in its ortholog IPK1 (Ipk1Δ) (Fig. 3D). Ipk1p phosphorylates IP5 to produce IP6, and in its absence there is accumulation of IP5, instead of the accumulation of IP6 that occurs in wild type yeast. The metabolic pathway from IP5 to IP6 was restored by complementation with TbIP5K (Fig. 3D). The presence of a shoulder close to the PP-IP4 eluting peak in the mutant yeast suggests the existence of two isomeric PP-IP4 species. We also complemented yeast mutants for both ipk1Δ (IP5K) and kcs1Δ (IP6K). These mutants accumulate IP2, IP3, IP4, and IP5 but no PP-IPs. While complementation with either TbIPMK or TbIP6K (not shown) alone did not change appreciably the inositol polyphosphate profile, synthesis of IP6 was restored by complementation with TbIP5K alone (Fig. 3E), demonstrating that TbIP5K is the only inositol phosphate kinase identified in T. brucei genome that can produce IP6.
Characterization of the inositol hexakisphosphate kinase (TbIP6K)
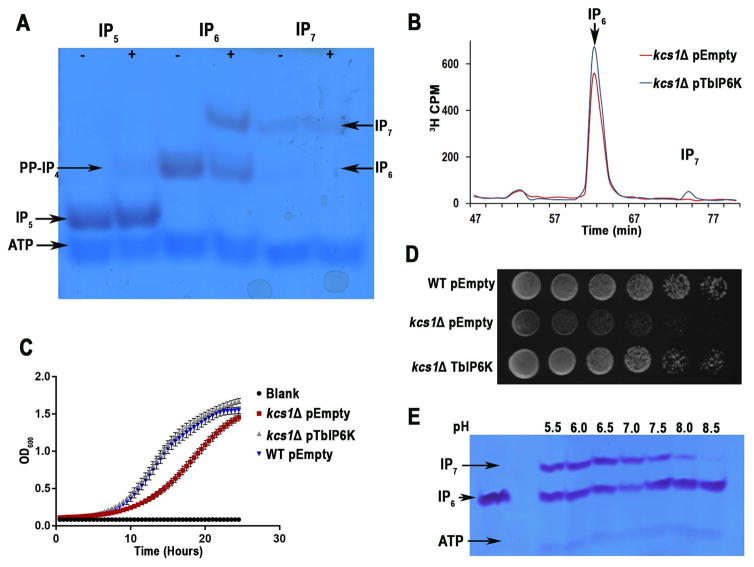
TbIP6K catalyzes the formation of IP7 from IP6. TbIP6K was also tagged with an HA tag using homologous recombination with the endogenous gene loci (Oberholzer et al., 2006). We detected expression of the enzyme in T. brucei procyclic forms (PCF) by western blot analysis (Fig. 2A). Recombinant TbIP6K was found to generate PP-IP4 from IP5 and IP7 from IP6 (Fig. 4A). Interestingly TbIP6K was not able to generate IP8 using a 5PP-IP7 as substrate, which suggests that, as IP6K from yeast and mammals, TbIP6K phosphorylates phosphate position D-5. Therefore, TbIP6K is able to generate two PP-IPs in vitro: PP-IP4, and IP7. The activity of TbIP6K was tested in vivo by complementation of a null mutant for its IP6K ortholog (KCS1) in S. cerevisiae. In the absence of KCS1 there is no accumulation of IP7, but the metabolic pathway from IP6 to IP7 is restored by complementation with TbIP6K (Fig. 4B). Complementation of Kcs1Δ TbIP6K also rescued the growth defect of these mutants (Fig. 4C and 4D). The TbIP6K enzymatic activity has optimum pH 6.0–7.0 (Fig. 4E).
Fig. 4. TbIP6K activity and complementation of yeast mutants.
A. Kinase reactions performed with recombinant TbIP6K (2 μg) using the indicated substrates at 150 μM for 1 hour at 37°C. TbIP6K can phosphorylate I(1,3,4,5,6)P5 to PP-IP4 and IP6 to produce IP7 (5PP-IP5) but cannot phosphorylate IP7 to produce IP8. Other arrows show bands corresponding to ATP, and IP5.
B. HPLC analysis of soluble inositol phosphates of S. cerevisiae kcs1Δ mutants transformed with an empty vector (red) or a vector containing the entire open reading frame of TbIP6K (blue).
C. Growth of the same cells in liquid medium as estimated by measuring optical density at 660 nm. kcs1Δ mutants had reduced growth, which was restored by expression of TbIP6K. Mean ± s.d. for three independent experiments, each one with 6 duplicates.
D. WT, and kcs1Δ transformed with empty vector or kcs1Δ transformed with TbIP6K (serially diluted 10-fold, 106–10 cells/spot from left to right) were spotted on YPD plates and incubated at 30°C for 2 days.
E. Optimum pH for TbIP6K activity is under acidic conditions. We detected a higher activity at pH 6.0 and 6.5.
All results are representative of three or more independent experiments.
Characterization of inositol phosphates from T. brucei cells
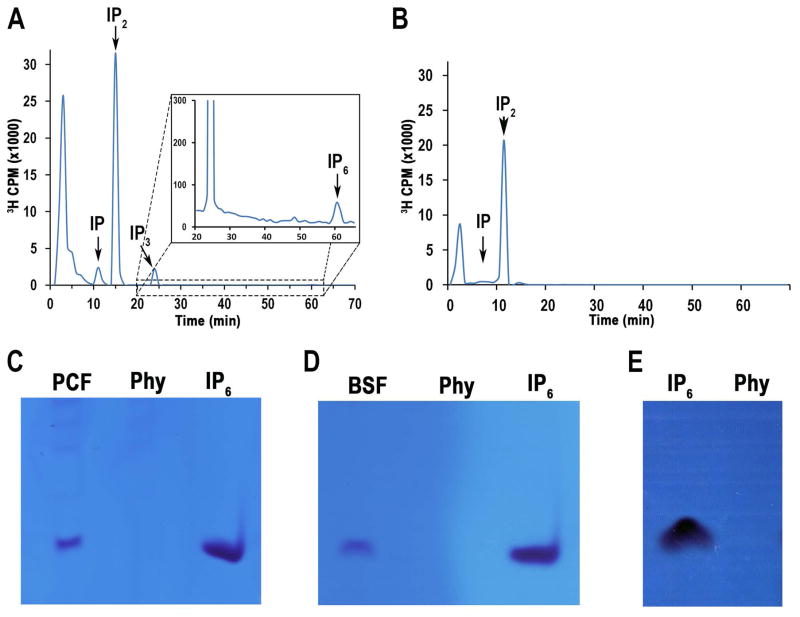
Previous attempts to characterize soluble inositol phosphates from T. brucei (Moreno et al., 1992) and T. cruzi (Docampo et al., 1991) only detected IP, IP2 and IP3. We used increased labeling time to 40 hours (BSF) and 75 hours (PCF) with [3H]inositol and used an improved protocol for purifying and analyzing inositol phosphates (see Materials and methods). Using these conditions, we were able to detect a small peak of IP6 in PCF but not in BSF of the parasite (Figs. 5A, and 5B). The inability to detect radiolabeled IP6 in the BSF might simply reflect the lower number of cells that can be obtained in culture. To improve the detection of IP6 we used a different approach that does not require metabolic labeling with [3H]inositol. We extracted IPs from large amounts of cells (see Materials and methods) and assayed extracts by 35% polyacrylamide gel electrophoresis (PAGE). A band that runs like the IP6 standard and that disappears after treatment of the extracts with phytase (Phy) was observed in both PCF and BSF (Figs. 5C, and 5D). Other highly phosphorylated inositol phosphates were not detected. These results confirm that both PCF and BSF TbIPMK and TbIP5K can sequentially synthesize IP6 in T. brucei.
Fig. 5. HPLC and PAGE analyses of soluble inositol phosphates from T. brucei PCF and BSF.
A. PCF showed the presence of IP, IP2, IP3 and IP6.
B. BSF showed the presence of IP, and IP2. Cells were labeled with [3H]inositol as described under Experimental Procedures.
C–E. PAGE analyses of extracts from PCF (C) or BSF (D) or standard IP6 (E). Samples in (C) and (D) (5 × 109 cells) were treated with phytase (Phy) (0.1 mg/ml, pH 5.0, at 37°C for 1 hour) to confirm that the bands correspond to IP6.
E. Phytase control activity with IP6 standard.
All results are representative of three or more independent experiments.
Biological relevance of the TbIPMK pathway
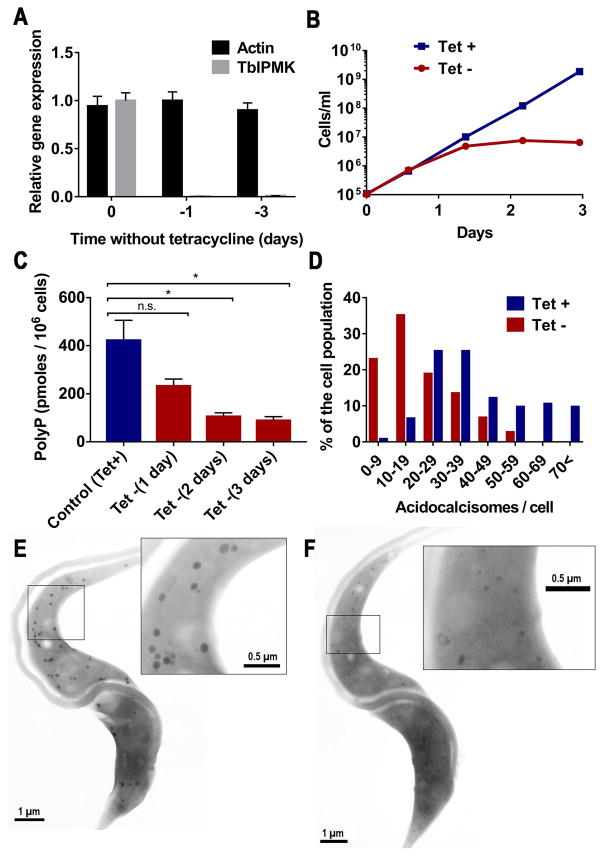
Yeast lacking Arg82p have no observable inorganic polyphosphate accumulation (Lonetti et al., 2011). As polyphosphate has important roles in trypanosomes, including growth, response to osmotic stress, and maintenance of persistent infections (Lander et al., 2016), we investigated whether deletion of soluble inositol polyphosphates affected the levels of polyphosphate in T. brucei. We used the TbIPMK conditional knockout BSF cell line previously described (Cestari et al., 2015). Removal of tetracycline to induce the knockdown of TbIPMK dramatically reduced its expression more than 100-fold (Fig. 6A). Growth stalled after the first day without tetracycline (Fig. 6B). A resulting progressive reduction in polyphosphate levels was detected (Fig. 6C). Acidocalcisomes are the main cellular storage compartment for polyphosphate in trypanosomes (Lander et al., 2016). However, examination of the cells by super-resolution microscopy with antibodies against the vacuolar proton pyrophosphatase (TbVP1) showed no apparent difference in labeling or distribution of acidocalcisomes between control and TbIPMK mutant cells (Figure S1). In previous work we demonstrated that a knockdown of the TbVtc4, which catalyzes the synthesis and translocation of polyphosphate into acidocalcisomes, results in less electron-dense organelles, as examined by electron microscopy (Ulrich et al., 2014). We hypothesized that if the polyphosphate reduction observed (Fig. 6C) was primarily within acidocalcisomes, we should observe similar changes in the TbIPMK mutant cells. Indeed, electron microscopy of the TbIPMK mutants showed a reduction in the number (Fig. 6D), size, and electron density (compare Fig. 6E and 6F) of electron-dense organelles identifiable as acidocalcisomes. This result indicates that acidocalcisome polyphosphate synthesis is disrupted by ablation of the inositol phosphate signaling pathway.
Fig. 6. Phenotypic changes of mutant BSF deficient in TbIPMK.
A. qRT-PCR analysis of gene expression of TbIPMK at time 0 and after 1 and 3 days in the absence of tetracycline as compared to expression of control actin. Values are means ± s.e.m., n = 3. P < 0.001 at days 1 and 3 without tetracycline. Student’s t test.
B. In vitro growth of BSF in the presence (+Tet) or absence (-Tet) of 1 μM tetracycline. Values are means ± s.e.m., n = 3 (bars are smaller than symbols).
C. Quantification of short-chain polyphosphate in control (+Tet) and induced (-Tet) TbIPMK conditional knockout BSF. Values are means ± s.e.m, n = 3, *P < 0.05. Student’s t test.
D. Numeric distribution of acidocalcisomes in BSF. Whole unfixed parasites were observed by transmission electron microscopy and the number of acidocalcisomes per cell in ~100 cells of control (+Tet) and conditional TbIPMK mutants (-Tet) were counted (the results from 3 independent experiments were combined).
E, F. Scanning transmission electron microscopy (STEM) images from control (E) or TbIPMK conditional mutant BSF showing acidocalcisomes. Bar = 1 μm. Insets show acidocalcisomes highlighted in (E) and (F) at higher magnification. Bars = 0.5 μm.
Discussion
Our work establishes the presence of an inositol pyrophosphate (PP-IPs) synthesis pathway in T. brucei. We demonstrated that genes encoding proteins with homology to kinases involved in the generation of IP5 from IP4 and IP3 (TbIPMK), of IP6 from IP5 (TbIP5K), and of IP7 from IP6 (TbIP6K) are present in the T. brucei genome (TbIPMK, TbIP5K, and TbIP6K). To demonstrate that these genes encode for functional enzymes we complemented yeast strains deficient in their corresponding orthologs and compared their products with those produced in the wild type strain providing in vivo genetic evidence of their function. We did not compare them with the knockout strains overexpressing the endogenous genes because the heterologous gene expression is often hampered by a diverse genetic code usage and by the lack of yeast specific post-translational processing. Thus, the heterologous genes are regularly expressed from a stronger promoter. The overexpressing of the endogenous gene from a stronger promoter might generate, to the contrary, ‘hyper’ phenotype and not a normal WT phenotype and our aim was to demonstrate their function and not to compare the activities to those of the overexpressed endogenous genes. Suppression of this pathway in T. brucei BSF resulted in a significant decrease in polyphosphate levels and in morphological alterations of the acidocalcisomes. The results suggest that this pathway is important for polyphosphate synthesis in acidocalcisomes.
Examination of the protein sequences of TbIPMK, TbIP5K, and TbIP6K indicated low identity with the mammalian enzymes but conservation of the putative binding site that catalyzes the transfer of phosphate from ATP to IPs, as well as of other domains required for enzymatic activity. The expression of these three kinases is very low in BSF since no clear bands were detected by western blot analyses of endogenous tagged lines, although gene expression is detectable at the mRNA level. Conversely, all three kinases can be easily identified by western blot analysis of PCF. Our results suggest, in agreement with the presence of these enzymes in other unicellular organisms such as D. discoideum (Europe-Finner et al., 1991, Mayr GW, 1992, Stephens et al., 1993), and E. histolytica (Martin et al., 1993), an early emergence of this pathway preceding the origin of multicellularity.
The application of polyacrylamide gel electrophoresis (PAGE) and toluidine blue staining (Losito et al., 2009, Pisani et al., 2014) allowed the characterization of the IPs synthesizing kinases of T. brucei and the identification of the products of each reaction bypassing the need for extraction under the strong acidic conditions required for HPLC analysis that has been shown to degrade some of the most highly phosphorylated species (Losito et al., 2009).
Previous work has indicated that TbIPMK is essential for growth (Cestari et al., 2015) and infectivity (Cestari et al., 2016) of T. brucei BSF, and partially characterized the recombinant enzyme (Cestari et al., 2016). We confirmed that TbIPMK prefers I(1,4,5)P3 and I(1,3,4,5)P4 as substrates (Cestari et al., 2016) and found that it does not phosphorylate I(1,3,4)P3. We also confirmed that TbIPMK cannot phosphorylate the lipid PIP2 to PIP3 (Cestari et al., 2016), as the human enzyme does (Resnick et al., 2005). In addition, we found that the enzyme can use I(1,3,4,5)P4 and I(1,4,5,6)P4 for the generation of I(1,3,4,5,6)P5 indicating that TbIPMK has a dual 3-kinase/6-kinase activity. This is in contrast to the human enzyme, where the major activity of IP4 kinase is phosphorylation at the D-5 position (Chang et al., 2002). Moreover, we demonstrated that TbIPMK is able to generate PP-IP4 in vitro, using either I(1,4,5)P3, I(1,3,4,5)P4 or I(1,3,4,5,6)P5, as well as IP6 from I(1,2,4,5,6)P5, or (I(1,2,3,4,5)P5 as substrate, again indicating a 3/6-kinase activity. TbIPMK has a neutral pH optimum for phosphorylation of both IP3 and IP4. Previous work (Cestari et al., 2016) described inhibitors of this enzyme that inhibited T. brucei BSF growth. However, their IC50s against the enzymes were higher (3.4–5.33 μM) than the EC50s for their growth inhibition (0.51–0.83 μM), suggesting that either the drugs are accumulated or other targets might be involved in the sensitivity of T. brucei BSF to those inhibitors. The search for more specific inhibitors is warranted to demonstrate the relevance of this pathway to human disease and drug therapy. Interestingly, a recombinant multi-domain protein from Plasmodium knowlesi termed PkIPK1 was shown to have IPMK-like activity and was able to generate I(1,3,4,5)P4 from I(1,4,5)P3 and I(1,2,4,5,6)P5 from either I(1,2,5,6)P4 or I(1,3,4,6)P4, showing 3/5-kinase activity (Stritzke et al., 2012).
We were not able to detect activity of the recombinant IP5K in the presence of different isomers of IP5 suggesting that, as proposed for the mammalian enzyme, post-translational modifications are needed for its activity (Verbsky et al., 2002). However, TbIP5K was able to complement null mutant yeast deficient in its ortholog IPK1 (Ipk1Δ), providing genetic evidence of its function.
Recombinant TbIP6K was able to generate PP-IP4 from IP5 and IP7 from IP6, but was not able to generate IP8 using a 5-PP-IP5 as substrate suggesting that, as IP6K from mammalian cells (Draskovic et al., 2008), TbIP6K phosphorylates phosphate at position D-5. Therefore, TbIP6K is able to generate two PP-IPs in vitro: PP-IP4, and IP7. Complementation of yeast deficient in its ortholog confirmed the function of this enzyme.
T. brucei incorporates poorly the radioactive tracer [3H]inositol a feature previously observed in Dictyostelium discoideum (Losito et al., 2009). Nevertheless, improved metabolic labeling with [3H]inositol resulted in detection of IP, IP2, IP3 and IP6 by HPLC analysis of PCF extracts. In contrast to the results obtained using similar methods in yeasts (Azevedo et al., 2006), plants (Phillippy et al., 2015) or animal cells (Guse et al., 1993), only very low levels of IP6 were detected and no labeled IP6 was detected by HPLC using BSF extracts. However, IP6 was clearly detected by PAGE and toluidine blue staining when large numbers of parasites were used. No inositol pyrophosphates were detected since to purify and visualize IPs we removed the abundant inorganic polyphosphate (polyP) by acidic treatment, procedure that would degrade IP7 to IP6. However, the absence of IP7 could be also attributed to the high turnover of these important signaling molecules (Glennon et al., 1993, Burton et al., 2009). Some cells accumulate IP6 and produce IP7 upon signaling events. For instance, Cryptococcus neoformans requires synthesis of IP7 for successful establishment of infection (Li et al., 2016). A recent study demonstrated that IP7 binds the SPX domain of proteins involved in phosphate homeostasis in plants, yeast and humans with high affinity and specificity and postulated the role of this domain as a polyphosphate sensor domain (Wild et al., 2016, Azevedo et al., 2017). Two proteins in T. brucei possess SPX domains, TbVtc4 (Lander et al., 2013), which is involved in polyphosphate synthesis and translocation, and TbPho91 (Huang et al., 2014), a phosphate transporter. Both proteins localize to acidocalcisomes (Huang et al., 2014), the main polyphosphate storage of these cells. Our results, showing lower levels of polyphosphate and altered acidocalcisomes in TbIPMK BSF mutants, support the link between PP-IPs and polyphosphate metabolism.
In summary, both recombinant enzymes, TbIPMK and TbIP6K, are able to generate inositol pyrophosphates. The essentiality of the first enzyme of this pathway, TbIPMK, for growth and infectivity of T. brucei BSF (Cestari et al., 2015, Cestari et al., 2016) suggests that the study of the PP-IPs pathway in trypanosomes could lead to the elucidation of potentially multiple important roles of these compounds, possibly linked to the synthesis of polyphosphate. Differences between mammalian and trypanosome metabolism of these compounds could provide potential targets for drug development.
Experimental procedures
Chemicals and reagents
Mouse antibodies against HA were from Covance (Hollywood, FL). Inositol, myo-[1,2-3H(N)] (60 Ci/mmol, ART 0261A) was from American Radiolabeled Chemicals, Inc. Goat anti-mouse antibodies were from LI-COR Biosciences (Lincoln, NE). Laemmli sample buffer was from Bio-Rad Laboratories (Hercules, CA). The bicinchoninic (BCA) protein assay kit was from Pierce (Thermo Fisher Scientific, USA). Titanium dioxide (TiO2) beads (Titansphere ToO 5 μm) were from GL Sciences (USA). PrimeSTAR HS DNA polymerase was from Clontech Laboratories Inc. (Takara, Mountain View, CA). Vector pET32 Ek/LIC was from Novagen (Merck KGaA, Darmstadt, Germany). Acrylamide mix was from National Diagnostics (Chapel Hill, NC). CelLytic M cell lysis reagent, P8340 protease inhibitor, protease inhibitors, Benzonase Nuclease, antibody against c-Myc, inositol phosphates, and other analytical reagents were from Sigma-Aldrich (St. Louis, MO).
Cell cultures
T. brucei Lister strain 427 BSF and PCF were used. The BSF were cultivated at 37°C in HMI-9 medium (Hirumi et al., 1989) supplemented with 10% heat inactivated fetal bovine serum (FBS, Sigma). The PCF were cultivated at 28°C in SDM-79 medium (Cunningham, 1977) supplemented with 10% heat-inactivated FBS and hemin (7.5 μg/ml). To determine the presence of IP6 by PAGE analysis T. brucei BSF were also isolated from infected mice (Balb/c, female, 6–8 weeks old) and rats (Wistar, male retired breeders), as described previously (Cross, 1975). T. brucei IPMK conditional knockout cell line was obtained and grown as described previously (Cestari et al., 2015).
Yeast strains
The yeast strains used in this study are isogenic to DDY1810 (MATa leu2-3,112 trp1-Δ901 ura3-52 prb1-1122 pep4-3 prc1-407), except for the ipk1Δkcs1Δ strain that is isogenic to BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and was previously described (Saiardi et al., 2002). The DDY1810 protease deficient strain is often used to increase the expression of exogenous proteins upon overexpression due to a deletion on the Pep4 protease. The generation of DDY1810 kcs1Δ strain was previously described (Onnebo et al., 2009). The arg82Δ, ipk1Δ yeast strains in the DDY1810 genetic background were generated following standard homologous recombination techniques (Gueldener et al., 2002) using oligonucleotides listed in Table S2. Initially diagnostic PCR was performed to confirm the correct integration of the deletion constructs. Subsequently, the soluble inositol polyphosphate profile of these new strains was used to phenotypically validate the correct homologous recombination event.
Epitope tagging, cloning expression and biochemical characterization of inositol phosphate kinases
We followed a one-step epitope-tagging method (Oberholzer et al., 2006) to produce the C-terminal HA- or cMyc-tagging cassettes for transfection of T. brucei PCF (Table S2). Briefly, the tagging cassettes containing selection markers were generated for cell transfection by PCR using pMOTag4H and pMOTag33M as templates with the corresponding PCR primers of the genes (Table S2). Transfection was performed using 2.5 × 107 PCF parasites from log phase. Cells were harvested at 1,000 × g for 10 min, washed with 10 ml of ice-cold sterile Cytomix buffer (2 mM EGTA, 3 mM MgCl2, 120 mM KCl, 0.5% glucose, 0.15 mM CaCl2, 0.1 mg/ml bovine serum albumin, 10 mM K2HPO4/KH2PO4, 1 mM hypoxanthine, 25 mM Hepes, pH 7.6), centrifuged at 1,000 × g for 7 min, suspended in 0.5 ml Cytomix and transferred to an ice-cold 4 mm gap cuvette (Bio-Rad) containing 15 μg of PCR amplicon. Cuvettes were incubated 5 min on ice and immediately electroporated twice in Bio-Rad GenePulser Xcell™ Electroporation System at 1.5 kV, 25 μF. Cuvettes were kept on ice for one minute between electroporation pulses. Cell mixture was transferred to SDM-79 medium with 15% FBS. After 6 h appropriate antibiotics were added. The sequences of the three kinases TbIPMK, TbIP5K and TbIP6K were amplified from genomic DNA by PCR (Table S2) using PrimeSTAR HS DNA polymerase and cloned into ligation independent expression vector pET32 Ek/LIC, as recommended by the manufacturer. Constructs were cloned into Escherichia coli BL21-CodonPlus(DE3) and protein expression was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) in Luria Bertani broth for 3 h. Protein purification was performed using affinity chromatography HIS-Select® Cartridge, according to the manufacturer’s instructions. We tested activity of the kinases on commercially available substrates. Enzyme assays were performed at 37°C using approximately 50 ng of recombinant protein, 20 mM Hepes buffer, pH 7.0, 0.2–0.5 mM substrate, 6 mM MgCl2, 100 mM NaCl, 1 mM dithiotreitol (DTT), 0.5 mM ATP, 10 mM phosphocreatine, and 40 U creatine kinase. Enzymatic reactions were stopped with 3 μl of 100 mM EDTA and kept on ice or frozen until further use. Reaction products were resolved by PAGE using 35% acrylamide/bis-acrylamide 19:1 gels in Tris/Borate/EDTA (TBE) buffer as described by (Losito et al., 2009). Gels were stained with toluidine blue (Losito et al., 2009).
RNA quantification
The TbIPMK conditional knockout cell line was grown with or without 1 μg/ml tetracycline and harvested at room temperature. RNA was extracted with TRI reagent (Sigma) and used as template for cDNA synthesis with SuperScript III RNA Polymerase (ThermoFisher) and oligo-dT as recommended by the manufacturer. We then performed qRT-PCR analysis using specific primers (Table S2) and SYBR Green Supermix (Bio-Rad). Relative TbIPMK gene expression relative to actin was calculated using CFX Manager™ Software (Bio-Rad).
Western blot analyses
Cells were harvested, washed twice in PBS, and lysed with CelLytic M cell lysis reagent containing protease inhibitor cocktail (Sigma P8340) diluted 1:250, 1 mM EDTA, 1 mM phenylmethanesulfonyl fluoride (PMSF), 20 μM trans-epoxysuccinyl-L-leucylamido(4-guanidino)butane (E64) and 50 U/ml Benzonase Nuclease (Millipore). The protein concentration was determined by using a BCA protein assay kit. The total cell lysates were mixed with 2X Laemmli sample buffer at 1:1 ratio (vol/vol) and directly loaded in 10% SDS-PAGE. The separated proteins were transferred onto nitrocellulose membranes using a Bio-Rad transblot apparatus. The membranes were blocked with 5% (wt/vol) nonfat milk in PBS containing 0.5% Tween-20 (PBS-T) at 4°C overnight. The blots were incubated for 1 hour with mouse antibodies against HA (1:1000) or mouse antibodies against c-Myc (1:1000). After five washings with PBS-T the blots were incubated with goat anti-mouse antibodies at a dilution of 1:15000 and developed using an Odyssey CLx Infrared Imaging System (LI-COR) according to the manufacturer instructions.
Yeast complementation
S. cerevisiae strains generated from DDY1810 were used: arg82Δ, ipk1Δ, kcs1Δ, ipk1Δkcs1Δ. TbIPMK, TbIP5K and TbIP6K were amplified from T. brucei Lister 427, cloned into plasmid pADH:GST (pYES-ADH1-GST) (Azevedo et al., 2009). Yeast cells were grown for 48 h in CSM plates. One colony was collected and suspended in 0.2 M lithium acetate with 25% polyethylene glycol solution and 0.1 M DTT. Cells were homogenized in 100 μl of solution with 100 ng of plasmid DNA and 5 μl of salmon sperm (Sigma D76560). Cells were incubated at 42°C for 30 min and immediately plated in CSM -URA plates. Colonies were used for further experiments.
Titanium dioxide bead extraction
We adapted the method of Wilson et al. (Wilson et al., 2015) for cell extraction of inositol polyphosphates. Cells (5 × 109) were harvested and washed twice in washing buffer A with glucose (BAG, 116 mM NaCl, 5.4 mM KCl, 0.8 mM MgSO4, 50 mM Hepes, ph 7.3, and 5.5 mM glucose). The pellet was then mixed with 1 M perchloric acid, resuspended by sonication (40% amplitude) for 10 s and kept at room temperature for 15 min. The sample was centrifuged art 18,000 × g for 5 min and the supernatant was transferred to a new tube and boiled for 30 min to remove the large amount of polyphosphate present in T. brucei. Seven mg of TiO2 beads were washed with water and 1 M perchloric acid, and added to the sample and left rotating for 30 min. Beads were centrifuged at 3,500 × g and inositol phosphates eluted with 1 M KOH, 10 mM EDTA. The sample was neutralized with perchloric acid and split into two. One half was digested with phytase (0.1 mg/ml) in the same medium at pH 5.0 and 37°C for 1 h. Extracts were resolved by 35% PAGE analysis as described above.
HPLC analysis
Inositol phosphate analysis was performed according to (Azevedo et al., 2006). Briefly, yeast liquid cultures were diluted to OD600 0.005 in inositol free media supplemented with 5 μCi/ml [3H] inositol and grown overnight at 30°C with shaking. Cells were washed twice with water and immediately incubated with ice-cold 1 M perchloric acid and 3 mM EDTA. Glass beads were added and cells lysed by vortexing at 4°C for 2 min, 3 times. Lysates were centrifuged and supernatants neutralized with 1 M K2CO3 and 3 mM EDTA. Samples were analyzed by strong anion exchange HPLC using SAX 4.6125 mm column (Whatman cat. no. 4621-0505). The column was eluted with two slightly different gradients generated by mixing buffer A (1 mM Na2EDTA) and buffer B [buffer A plus 1.3 M (NH4)2HPO4 (pH 3.8 with H3PO4)] as follows: 0–5 min, 0% B; 5–10 min, 0–30% B; 10–60 min, 30–100% B; 60–80 min, 100% B; or as follow: 0–5 min, 0% B; 5–10 min, 0–10% B; 10–85 min, 20–100% B; 85–100 min 100% B. Four mL of Ultima-Flo AP liquid scintillation cocktail (Perkin-Elmer cat. no. 6013599) was added to each fraction, mixed and radioactivity quantified in a scintillation counter.
T. brucei labeling for HPLC analysis
T. brucei PCF (~3×106 cells) were labeled with 5 μCi/ml of 1,2-[3H]-inositol in SDM-79 medium (with 10% FBS) and grown for approximately 72 h. T. brucei BSF (~2×105 cells) were labeled with 5 μCi/ml of 1,2-[3H]-inositol in HMI-9 medium (with 10% FBS) and grown for approximately 40 h. Cells were washed with PBS or BAG twice and frozen immediately. Soluble inositol phosphates were extracted and analyzed as described before (Azevedo et al., 2006), with minor modifications. Briefly, cells were suspended in ice-cold perchloric acid and broken by vortexing for 2 min. All steps were performed at 4°C. Lysates were centrifuged for 5 min at 18,000 × g and supernatants transferred to new tubes, where the pH was neutralized with 1 M K2CO3 and 3 mM EDTA. Samples were stored at 4°C and resolved by HPLC.
Polyphosphate extraction and measurement
Short chain polyphosphate was extracted from BSF T. brucei and quantified as described previously (Ulrich et al., 2014).
Immunofluorescence Assay
T. brucei BSF were washed with BAG and fixed with 2% paraformaldehyde in BAG for 1 h at room temperature. Then they were adhered to poly-L-lysine coated coverslips and permeabilized with 0.1% Triton X-100 in PBS for 5 min. Blocking was performed overnight at 4°C in PBS containing 100 mM NH4Cl, 3% BSA, 1% fish gelatin and 5% goat serum. Cells were then incubated with anti-TbVP1 polyclonal Guinea pig antibody (1:100) for 1 h and subsequently with Alexa 488-conjugated goat anti-Guinea pig antibody (1:1000) for 1h. Microscopy images were taken with a 100X oil immersion objective, a high-power solid-state 405 nm laser and EM-CCD camera (Andor iXon) under nonsaturating conditions in a Zeiss ELYRA S1 (SR-SIM) super resolution microscope. Images were acquired and processed with ZEN 2011 software with SIM analysis module.
Electron microscopy
Imaging of whole T. brucei BSF and determination of morphometric parameters were done as described previously (Ulrich et al., 2014).
Statistical analysis
All experiments were repeated at least three times (biological replicates) with several technical replicates as indicated in the figure legends, and where indicated results are expressed as means ± s.d. or s.e.m. of n experiments. Statistical analyses were performed using the Student’s t-test. Results are considered significant when P < 0.05.
Supplementary Material
Acknowledgments
We thank Noelia Lander for help with pET32 cloning, Thomas Seebeck for the pMOTag vectors, Igor Cestari and Ken Stuart for the TbIPMK conditional knockout cell line, Muthugapatti Kandasamy and the Biomedical Microscopy Core of the University of Georgia for help with the super-resolution microscope, and John Shields and Mary Ard from the Georgia Electron Microscopy for help with electron microscopy. This work was funded by U.S. National Institutes of Health (grant AI077538) and supported by the Medical Research Council (MRC) core support to the MRC/UCL Laboratory for Molecular Cell Biology University Unit (MC_UU_1201814). C.C was partially supported by an European Molecular Biology Organization (EMBO) Short Term Fellowship and Center for Tropical and Emerging Global Diseases (CTEGD) travel fellowship.
Footnotes
Competing interests
The authors declare no competing or financial interests
Author contributions
C.C., A.S. and R.D. designed the experiments and analyzed the data. C.C. and A.S. conducted the experiments. R.D. wrote the majority of the manuscript, with specific sections contributed by C.C., and A.S. R.D. and A.S. supervised the work and contributed to the analysis of experiments.
Supplementary information available online at:
References
- Albert C, Safrany ST, Bembenek ME, Reddy KM, Reddy K, Falck J, et al. Biological variability in the structures of diphosphoinositol polyphosphates in Dictyostelium discoideum and mammalian cells. Biochem J. 1997;327:553–560. doi: 10.1042/bj3270553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C, Burton A, Ruiz-Mateos E, Marsh M, Saiardi A. Inositol pyrophosphate mediated pyrophosphorylation of AP3B1 regulates HIV-1 Gag release. Proc Nat Acad Sci USA. 2009;106:21161–21166. doi: 10.1073/pnas.0909176106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C, Saiardi A. Extraction and analysis of soluble inositol polyphosphates from yeast. Nat Prot. 2006;1:2416–2422. doi: 10.1038/nprot.2006.337. [DOI] [PubMed] [Google Scholar]
- Azevedo C, Saiardi A. Eukaryotic Phosphate Homeostasis: The Inositol Pyrophosphate Perspective. Trends Biochem Sci. 2017;42:219–231. doi: 10.1016/j.tibs.2016.10.008. [DOI] [PubMed] [Google Scholar]
- Berridge MJ. Inositol trisphosphate and calcium signalling mechanisms. Biochim Biophys Acta. 2009;1793:933–940. doi: 10.1016/j.bbamcr.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Bertsch U, Deschermeier C, Fanick W, Girkontaite I, Hillemeier K, Johnen H, et al. The second messenger binding site of inositol 1,4,5-trisphosphate 3-kinase is centered in the catalytic domain and related to the inositol trisphosphate receptor site. j Biol Chem. 2000;275:1557–1564. doi: 10.1074/jbc.275.3.1557. [DOI] [PubMed] [Google Scholar]
- Burton A, Azevedo C, Andreassi C, Riccio A, Saiardi A. Inositol pyrophosphates regulate JMJD2C-dependent histone demethylation. Proc Nat Acad Sci USA. 2013;110:18970–18975. doi: 10.1073/pnas.1309699110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton A, Hu X, Saiardi A. Are inositol pyrophosphates signalling molecules? J Cell Physiol. 2009;220:8–15. doi: 10.1002/jcp.21763. [DOI] [PubMed] [Google Scholar]
- Cestari I, Haas P, Moretti NS, Schenkman S, Stuart K. Chemogenetic characterization of inositol ophosphate metabolic pathway reveals druggable enzymes for targeting kinetoplastid parasites. Cell Chem Biol. 2016;23:608–617. doi: 10.1016/j.chembiol.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cestari I, Stuart K. Inositol phosphate pathway controls transcription of telomeric expression sites in trypanosomes. Proc Nat Acad Sci USA. 2015;112:E2803–2812. doi: 10.1073/pnas.1501206112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SC, Miller AL, Feng Y, Wente SR, Majerus PW. The human homolog of the rat inositol phosphate multikinase is an inositol 1,3,4,6-tetrakisphosphate 5-kinase. J Biol Chem. 2002;277:43836–43843. doi: 10.1074/jbc.M206134200. [DOI] [PubMed] [Google Scholar]
- Choi JH, Williams J, Cho J, Falck JR, Shears SB. Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J Biol Chem. 2007;282:30763–30775. doi: 10.1074/jbc.M704655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocco L, Follo MY, Manzoli L, Suh PG. Phosphoinositide-specific phospholipase C in health and disease. J Lipid Res. 2015;56:1853–1860. doi: 10.1194/jlr.R057984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross GA. Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology. 1975;71:393–417. doi: 10.1017/s003118200004717x. [DOI] [PubMed] [Google Scholar]
- Cunningham I. New culture medium for maintenance of tsetse tissues and growth of trypanosomatids. J Protozool. 1977;24:325–329. doi: 10.1111/j.1550-7408.1977.tb00987.x. [DOI] [PubMed] [Google Scholar]
- Docampo R, Pignataro OP. The inositol phosphate/diacylglycerol signalling pathway in Trypanosoma cruzi. Biochem J. 1991;275:407–411. doi: 10.1042/bj2750407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draskovic P, Saiardi A, Bhandari R, Burton A, Ilc G, Kovacevic M, et al. Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem Biol. 2008;15:274–286. doi: 10.1016/j.chembiol.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Europe-Finner GN, Gammon B, Newell PC. Accumulation of [3H]-inositol into inositol polyphosphates during development of Dictyostelium. Biochem Biophys Res Commun. 1991;181:191–196. doi: 10.1016/s0006-291x(05)81400-7. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Shukla D, Suman K, Lakshmi BJ, Manorama R, Kumar S, Bhandari R. Inositol hexakisphosphate kinase 1 maintains hemostasis in mice by regulating platelet polyphosphate levels. Blood. 2013;122:1478–1486. doi: 10.1182/blood-2013-01-481549. [DOI] [PubMed] [Google Scholar]
- Glennon MC, Shears SB. Turnover of inositol pentakisphosphates, inositol hexakisphosphate and diphosphoinositol polyphosphates in primary cultured hepatocytes. Biochem J. 1993;293:583–590. doi: 10.1042/bj2930583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale NA, Zaremba A, Janoshazi AK, Weaver JD, Shears SB. PPIP5K1 modulates ligand competition between diphosphoinositol polyphosphates and PtdIns(3,4,5)P3 for polyphosphoinositide-binding domains. Biochem J. 2013;453:413–426. doi: 10.1042/BJ20121528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse AH, Greiner E, Emmrich F, Brand K. Mass changes of inositol 1,3,4,5,6-pentakisphosphate and inositol hexakisphosphate during cell cycle progression in rat thymocytes. J Biol Chem. 1993;268:7129–7133. [PubMed] [Google Scholar]
- Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- Huang G, Bartlett PJ, Thomas AP, Moreno SN, Docampo R. Acidocalcisomes of Trypanosoma brucei have an inositol 1,4,5-trisphosphate receptor that is required for growth and infectivity. Proc Nat Acad Sci USA. 2013;110:1887–1892. doi: 10.1073/pnas.1216955110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Ulrich PN, Storey M, Johnson D, Tischer J, Tovar JA, et al. Proteomic analysis of the acidocalcisome, an organelle conserved from bacteria to human cells. PLoS Pathog. 2014;10:e1004555. doi: 10.1371/journal.ppat.1004555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illies C, Gromada J, Fiume R, Leibiger B, Yu J, Juhl K, et al. Requirement of inositol pyrophosphates for full exocytotic capacity in pancreatic beta cells. Science. 2007;318:1299–1302. doi: 10.1126/science.1146824. [DOI] [PubMed] [Google Scholar]
- Irvine RF, Schell MJ. Back in the water: the return of the inositol phosphates. Nature reviews Mol Cell Biol. 2001;2:327–338. doi: 10.1038/35073015. [DOI] [PubMed] [Google Scholar]
- Jadav RS, Chanduri MV, Sengupta S, Bhandari R. Inositol pyrophosphate synthesis by inositol hexakisphosphate kinase 1 is required for homologous recombination repair. J Biol Chem. 2013;288:3312–3321. doi: 10.1074/jbc.M112.396556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Keller S, Moore CA, Docampo R, Moreno SN. Ca2+ regulation of Trypanosoma brucei phosphoinositide phospholipase C. Eukaryot Cell. 2015;14:486–494. doi: 10.1128/EC.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laha D, Johnen P, Azevedo C, Dynowski M, Weiss M, Capolicchio S, et al. VIH2 regulates the synthesis of inositol pyrophosphate InsP8 and jasmonate-dependent defenses in Arabidopsis. Plant Cell. 2015;27:1082–1097. doi: 10.1105/tpc.114.135160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander N, Cordeiro C, Huang G, Docampo R. Polyphosphate and acidocalcisomes. Biochem Soc Trans. 2016;44:1–6. doi: 10.1042/BST20150193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander N, Ulrich PN, Docampo R. Trypanosoma brucei vacuolar transporter chaperone 4 (TbVtc4) is an acidocalcisome polyphosphate kinase required for in vivo infection. J Biol Chem. 2013;288:34205–34216. doi: 10.1074/jbc.M113.518993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Lev S, Saiardi A, Desmarini D, Sorrell TC, Djordjevic JT. Identification of a major IP5 kinase in Cryptococcus neoformans confirms that PP-IP5/IP7, not IP6, is essential for virulence. Sci Rep. 2016;6:23927. doi: 10.1038/srep23927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonetti A, Szijgyarto Z, Bosch D, Loss O, Azevedo C, Saiardi A. Identification of an evolutionarily conserved family of inorganic polyphosphate endopolyphosphatases. J Biol Chem. 2011;286:31966–31974. doi: 10.1074/jbc.M111.266320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losito O, Szijgyarto Z, Resnick AC, Saiardi A. Inositol pyrophosphates and their unique metabolic complexity: analysis by gel electrophoresis. PLoS One. 2009;4:e5580. doi: 10.1371/journal.pone.0005580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo HR, Huang YE, Chen JC, Saiardi A, Iijima M, Ye K, et al. Inositol pyrophosphates mediate chemotaxis in Dictyostelium via pleckstrin homology domain-PtdIns(3,4,5)P3 interactions. Cell. 2003;114:559–572. doi: 10.1016/s0092-8674(03)00640-8. [DOI] [PubMed] [Google Scholar]
- Luo HR, Saiardi A, Yu H, Nagata E, Ye K, Snyder SH. Inositol pyrophosphates are required for DNA hyperrecombination in protein kinase c1 mutant yeast. Biochemistry. 2002;41:2509–2515. doi: 10.1021/bi0118153. [DOI] [PubMed] [Google Scholar]
- Martin JB, Bakker-Grunwald T, Klein G. 31P-NMR analysis of Entamoeba histolytica. Occurrence of high amounts of two inositol phosphates. Eur J Biochem. 1993;214:711–718. doi: 10.1111/j.1432-1033.1993.tb17972.x. [DOI] [PubMed] [Google Scholar]
- Mayr GWRT, THiel U, Vogel G, Stephens LR. Phosphoinositol diphosphates: non-enzymic formation in vitro and occurrence in vivo in the cellular slime mold Dictysotelium. Carbohyd Res. 1992;234:247–262. [Google Scholar]
- Menniti FS, Miller RN, Putney JW, Jr, Shears SB. Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J Biol Chem. 1993;268:3850–3856. [PubMed] [Google Scholar]
- Moreno SN, Docampo R, Vercesi AE. Calcium homeostasis in procyclic and bloodstream forms of Trypanosoma brucei. Lack of inositol 1,4,5-trisphosphate-sensitive Ca2+ release. J Biol Chem. 1992;267:6020–6026. [PubMed] [Google Scholar]
- Morrison BH, Bauer JA, Kalvakolanu DV, Lindner DJ. Inositol hexakisphosphate kinase 2 mediates growth suppressive and apoptotic effects of interferon-beta in ovarian carcinoma cells. J Biol Chem. 2001;276:24965–24970. doi: 10.1074/jbc.M101161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata E, Luo HR, Saiardi A, Bae BI, Suzuki N, Snyder SH. Inositol hexakisphosphate kinase-2, a physiologic mediator of cell death. J Biol Chem. 2005;280:1634–1640. doi: 10.1074/jbc.M409416200. [DOI] [PubMed] [Google Scholar]
- Nagata E, Saiardi A, Tsukamoto H, Satoh T, Itoh Y, Itoh J, et al. Inositol hexakisphosphate kinases promote autophagy. Int J Biochem & Cell Biol. 2010;42:2065–2071. doi: 10.1016/j.biocel.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Nalaskowski MM, Deschermeier C, Fanick W, Mayr GW. The human homologue of yeast ArgRIII protein is an inositol phosphate multikinase with predominantly nuclear localization. Biochem J. 2002;366:549–556. doi: 10.1042/BJ20020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizuka Y. Studies and perspectives of protein kinase C. Science. 1986;233:305–312. doi: 10.1126/science.3014651. [DOI] [PubMed] [Google Scholar]
- Oberholzer M, Morand S, Kunz S, Seebeck T. A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol biochem Parasitol. 2006;145:117–120. doi: 10.1016/j.molbiopara.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Onnebo SM, Saiardi A. Inositol pyrophosphates modulate hydrogen peroxide signalling. Biochem J. 2009;423:109–118. doi: 10.1042/BJ20090241. [DOI] [PubMed] [Google Scholar]
- Pesesse X, Choi K, Zhang T, Shears SB. Signaling by higher inositol polyphosphates. Synthesis of bisdiphosphoinositol tetrakisphosphate (“InsP8”) is selectively activated by hyperosmotic stress. J Biol Chem. 2004;279:43378–43381. doi: 10.1074/jbc.C400286200. [DOI] [PubMed] [Google Scholar]
- Phillippy BQ, Perera IY, Donahue JL, Gillaspy GE. Certain malvaceae plants have a unique accumulation of myo-Inositol 1,2,4,5,6-pentakisphosphate. Plants (Basel) 2015;4:267–283. doi: 10.3390/plants4020267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisani F, Livermore T, Rose G, Chubb JR, Gaspari M, Saiardi A. Analysis of Dictyostelium discoideum inositol pyrophosphate metabolism by gel electrophoresis. PLoS One. 2014;9:e85533. doi: 10.1371/journal.pone.0085533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A, Jia Y, Chakraborty A, Li Y, Jain SK, Zhong J, et al. Inositol hexakisphosphate kinase 1 regulates neutrophil function in innate immunity by inhibiting phosphatidylinositol-(3,4,5)-trisphosphate signaling. Nat Immunol. 2011;12:752–760. doi: 10.1038/ni.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick AC, Snowman AM, Kang BN, Hurt KJ, Snyder SH, Saiardi A. Inositol polyphosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity. Proc Nat Acad Sci USA. 2005;102:12783–12788. doi: 10.1073/pnas.0506184102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A. Cell signalling by inositol pyrophosphates. Subcell Biochem. 2012;59:413–443. doi: 10.1007/978-94-007-3015-1_14. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Caffrey JJ, Snyder SH, Shears SB. The inositol hexakisphosphate kinase family. Catalytic flexibility and function in yeast vacuole biogenesis. J Biol Chem. 2000;275:24686–24692. doi: 10.1074/jbc.M002750200. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Nagata E, Luo HR, Sawa A, Luo X, Snowman AM, Snyder SH. Mammalian inositol polyphosphate multikinase synthesizes inositol 1,4,5-trisphosphate and an inositol pyrophosphate. Proc Nat Acad Sci USA. 2001a;98:2306–2311. doi: 10.1073/pnas.041614598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A, Resnick AC, Snowman AM, Wendland B, Snyder SH. Inositol pyrophosphates regulate cell death and telomere length through phosphoinositide 3-kinase-related protein kinases. Proc Nat Acad Sci USA. 2005;102:1911–1914. doi: 10.1073/pnas.0409322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A, Sciambi C, McCaffery JM, Wendland B, Snyder SH. Inositol pyrophosphates regulate endocytic trafficking. Proc Nat Acad Sci USA. 2002;99:14206–14211. doi: 10.1073/pnas.212527899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears SB. Inositol pyrophosphates: why so many phosphates? Adv Biol Reg. 2015;57:203–216. doi: 10.1016/j.jbior.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens L, Radenberg T, Thiel U, Vogel G, Khoo KH, Dell A, et al. The detection, purification, structural characterization, and metabolism of diphosphoinositol pentakisphosphate(s) and bisdiphosphoinositol tetrakisphosphate(s) J Biol Chem. 1993;268:4009–4015. [PubMed] [Google Scholar]
- Stritzke C, Nalaskowski MM, Fanick W, Lin H, Mayr GW. A Plasmodium multi-domain protein possesses multiple inositol phosphate kinase activities. Mol Biochem Parasitol. 2012;186:134–138. doi: 10.1016/j.molbiopara.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Szijgyarto Z, Garedew A, Azevedo C, Saiardi A. Influence of inositol pyrophosphates on cellular energy dynamics. Science. 2011;334:802–805. doi: 10.1126/science.1211908. [DOI] [PubMed] [Google Scholar]
- Ulrich PN, Lander N, Kurup SP, Reiss L, Brewer J, Soares Medeiros LC, et al. The acidocalcisome vacuolar transporter chaperone 4 catalyzes the synthesis of polyphosphate in insect-stages of Trypanosoma brucei and T. cruzi. J Eukaryot Microbiol. 2014;61:155–165. doi: 10.1111/jeu.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak MD, Martin DM, Ferguson MA. Global quantitative SILAC phosphoproteomics reveals differential phosphorylation is widespread between the procyclic and bloodstream form lifecycle stages of Trypanosoma brucei. J Proteome Res. 2013;12:2233–2244. doi: 10.1021/pr400086y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbsky JW, Wilson MP, Kisseleva MV, Majerus PW, Wente SR. The synthesis of inositol hexakisphosphate. Characterization of human inositol 1,3,4,5,6-pentakisphosphate 2-kinase. J Biol Chem. 2002;277:31857–31862. doi: 10.1074/jbc.M205682200. [DOI] [PubMed] [Google Scholar]
- Wild R, Gerasimaite R, Jung JY, Truffault V, Pavlovic I, Schmidt A, et al. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science. 2016;352:986–990. doi: 10.1126/science.aad9858. [DOI] [PubMed] [Google Scholar]
- Wilson MS, Bulley SJ, Pisani F, Irvine RF, Saiardi A. A novel method for the purification of inositol phosphates from biological samples reveals that no phytate is present in human plasma or urine. Open Biol. 2015;5:150014. doi: 10.1098/rsob.150014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York JD, Odom AR, Murphy R, Ives EB, Wente SR. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science. 1999;285:96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]
- York SJ, Armbruster BN, Greenwell P, Petes TD, York JD. Inositol diphosphate signaling regulates telomere length. J Biol Chem. 2005;280:4264–4269. doi: 10.1074/jbc.M412070200. [DOI] [PubMed] [Google Scholar]
- Zhang T, Caffrey JJ, Shears SB. The transcriptional regulator, Arg82, is a hybrid kinase with both monophosphoinositol and diphosphoinositol polyphosphate synthase activity. FEBS Lett. 2001;494:208–212. doi: 10.1016/s0014-5793(01)02351-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.