Abstract
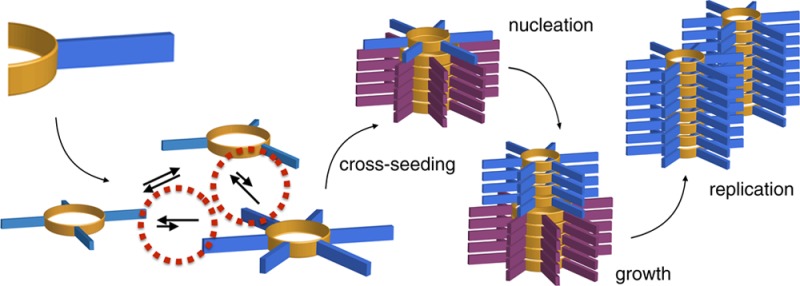
Our knowledge regarding the early steps in the formation of evolvable life and what constitutes the minimal molecular basis of life remains far from complete. The recent emergence of systems chemistry reinvigorated the investigation of systems of self-replicating molecules to address these questions. Most of these studies focus on single replicators and the effects of replicators on the emergence of other replicators remains under-investigated. Here we show the cross-catalyzed emergence of a novel self-replicator from a dynamic combinatorial library made from a threonine containing peptide building block, which, by itself, only forms trimers and tetramers that do not replicate. Upon seeding of this library with different replicators of different macrocycle size (hexamers and octamers), we observed the emergence of hexamer replicator consisting of six units of the threonine peptide only when it is seeded with an octamer replicator containing eight units of a serine building block. These results reveal for the first time how a new replicator can emerge in a process that relies critically on the assistance by another replicator through cross-catalysis and that replicator composition is history dependent.
How life originated and how life may be synthesized de novo are among the grand challenges in contemporary science. Research in these areas has focused on the biomolecules essential to current life (proteins, RNA and DNA) or on the bottom-up construction of chemical systems that mimic the essential characteristics of life. Over the last decades, the fields of systems chemistry,1 and dynamic combinatorial chemistry2 in particular, have developed synthetic systems that capture some of the essential characteristics of life: compartmentalization,3 reaction networks (addressing the issue of metabolism)4 and systems featuring self- and cross-replicating molecules.5
In this study, we will focus on self-replication. Until now, relatively few replicators have been reported. Following pioneering work by von Kiedrowski,6 completely synthetic replicators were developed by Rebek7 and Philp.8 Joyce, Lehman and Szostak developed systems of replicating RNAs9 whereas Chmielewski,10 Ghadiri,11 Ashkenasy12 and us13 focused on peptide-based replicators. Though in the majority of these studies replicators were designed in full structural detail, we explored how replicators emerged from complex mixtures where the structure of the emerging replicator was not predetermined. For this purpose, we developed dynamic combinatorial libraries (DCLs) made from building-blocks featuring two thiol groups for reversible disulfide chemistry13b and a peptide that is predisposed to β-sheet formation by virtue of alternating hydrophobic and hydrophilic amino-acid residues. In solution, reaction between these building blocks and oxygen from the air yields a DCL that consists of a mixture of disulfide macrocycles of different ring sizes (Scheme 1). If one of the macrocycles can stabilize itself through self-assembly, the product distribution shifts toward this compound at the expense of the other compounds in the library. Assembly occurs through a nucleation–growth mechanism which allows exponential replication to be achieved: growing fibers break into fragments by mechanical agitation (i.e., stirring or shaking), which increases the number of ends from where the fibers grow.13b
Scheme 1. (a) A dynamic Combinatorial Library of Differently Sized Macrocyclic Disulfides Is Formed upon Oxidation of a Threonine Containing Peptide Functionalized Dithiol; (b) Selective Formation of Replicator 16 upon Cross-Seeding; (c) Schematic Representation of the Tentative Mechanism through Which Replicator 48 Gives Rise to Replicator 16.

In most of our previous studies,13d the emergence of replicators occurred spontaneously. In an intriguing recent study,13a we showed that assembly driven self-replication could also be triggered by a template that raises the concentration of the potential replicator above its critical aggregation concentration. However, until now, the role of existing replicators on the emergence of new ones has received little attention. Yet, cross-catalysis may be a powerful mechanism for the diversification and evolution of replicators and the development of replicator “ecosystems”.14
We now report a system in which autonomous replicator emergence is not observed, but where replicator emergence requires cross-catalysis by another pre-existing replicator. Cross-catalyzed emergence was mediated only by a replicator with a specific ring size and peptide sequence. We also show that replication is strongly dependent on the sample history marking an important step in the direction of self-replicator evolution.
In the course of a systematic investigation of the effect of the structure of the peptide building blocks with the general architecture shown in Scheme 1 on the formation of replicators, we encountered unexpected behavior upon using building block 1. In agitated DCLs made from 2–5, individually, self-replicators emerged spontaneously in all cases. Yet in similar experiments starting from building block 1, we did not observe any replicators. Using our standard protocol, we rapidly oxidized a solution of 1 (3.8 mM in 50 mM borate buffer pH 8.1) to 80% (conversion of thiols into disulfides) using sodium perborate solution (80 mM), followed by slower further oxidation mediated by oxygen present in the air. Solutions were stirred at 1200 rpm or left nonagitated. The kinetic profiles of these libraries, monitored by UPLC-MS, show the formation trimers (13) and tetramers (14), neither of which self-assembles or self-replicates (Figure 1a; see SI Figure S1 for the nonagitated sample). The absence of any replicator might be explained with the library reaching complete oxidation before any replicator had the chance to emerge (disulfide exchange requires a catalytic amount of thiol). In order to prevent freezing the library by complete oxidation, we repeated the experiment at a constant oxidation level of 80% and monitored the composition of the small DCL over the course of one month under an inert atmosphere. Also in this experiment, 13 and 14 dominated the library and no replicator emergence was observed (Figure 1b).
Figure 1.
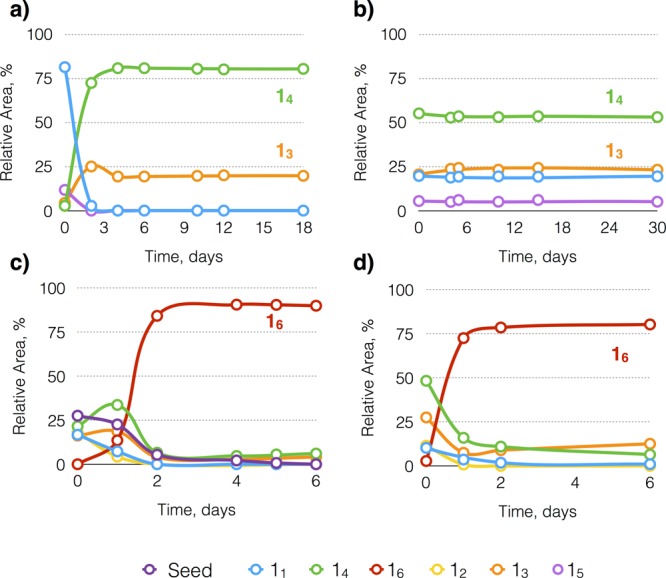
Kinetic profile of a dynamic combinatorial library made from building block 1 (3.8 mM in 50 mM borate buffer, pH 8.1) stirred at 1200 rpm and (a) kept under ambient conditions, (b) 80% oxidized and kept under an inert atmosphere; kinetic profile of a dynamic combinatorial library made from an 80% oxidized solution of building block 1 (3.8 mM in 50 mM borate buffer, pH 8.1) seeded with (c) 48 (cross-seeding), (d) 16 (self-seeding), stirred at 1200 rpm.
We then investigated whether the formation of replicators made from 1 could be induced by cross-seeding with preformed replicators made from building blocks 2, 3, 4 or 5. Thus, 10 mol % of 26, 36, 46, 48, 58, prepared following published protocols,13c,13d was added to stirred DCLs made from 1 and the compositions of the mixtures were monitored over time. These experiments failed to induce the formation of new macrocycles (see SI Figure S2) with one notable exception: only cross-seeding with 48 induced the formation of a new macrocycle (16, Figure 1c). The sigmoidal growth of 16 is consistent with self-replication.
To confirm that 16 is a self-replicator, we added it as a seed to a DCL made from 1. Figure 1d shows that 16 grows rapidly upon seeding and accounts for 72% of the library material within 1 day (see SI Figure S5 for results of seeding with smaller amounts of seed).15 Note that the lag phase, that was observed in the library to which 48 was added, diminished when 16 was used as a seed instead of 48. The presence of this lag phase suggests that growth of 16 occurs on the ends of fibers of 48, but it is a relatively rare event and that fragmentation of these nuclei of 16 (induced by 48) into secondary nuclei is required before replication of 16 becomes efficient (see Scheme 1c). This interpretation was further supported by the fact that seeding a library made from 1 with 48 in the absence of mechanical agitation dramatically slowed down the rate of replication of 16 (Figure S3). Detailed analysis of the early stage of the growth of 16 seeded by 48 (Figure S4) did not reveal any mixed macrocycles containing both building blocks, lending further support for the proposed mechanism.
The structure of the assemblies formed by 16 was characterized by transmission electron microscopy (TEM), circular dichroism (CD) spectroscopy and thioflavin T fluorescence assays (Figure 2). Negative staining TEM micrographs of a sample dominated by 16 revealed laterally associated fibers that were approximately 100 nm long (Figure 2c). In contrast, libraries made from 1 containing mostly cyclic trimers and tetramers did not show any ordered or disordered aggregates. The CD spectrum of samples dominated by 16 showed positive helicity at 196 nm and negative helicity at 218 nm, indicative of a β-sheet structure (Figure 2a).16 We observed only random coil secondary structure for all other libraries made from 1 that were dominated by trimers and tetramers. Thioflavin T fluorescence measurements17 also confirmed a β-sheet amyloid-fibril-like18 structure of 16, evident from a more than 50-fold increase in emission intensity, whereas all libraries dominated by 13 and 14 showed a 3-fold increase at most (Figure 2b).
Figure 2.
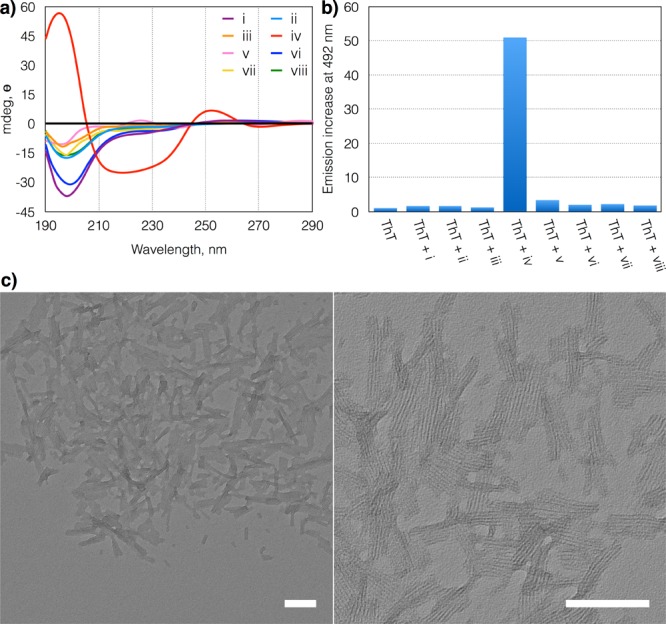
(a) CD spectra and (b) maximum thioflavin T fluorescence emission intensity (at 492 nm) of nonseeded and seeded (10 mol %) libraries made from building block 1 (3.8 mM in 50 mM borate buffer, pH 8.1): i, stirred; ii, inert atm.; iii, nonagitated; iv, seeded with 48; v, seeded with 58; vi, seeded with 26; vii, seeded with 36; viii, seeded with 46. (c) TEM micrographs of the library corresponding to Figure 1c. Scale bars are 100 nm.
In order to investigate whether cross-catalysis between the 16 and 48 replicators is reciprocal, libraries were made from building block 4 and the effect of seeding by 16 was probed. Mechanical agitation was not applied, as agitation induces the autonomous formation of the 48 replicator. We observed the emergence of 48 and 46 alongside some mixed hexamers containing both building blocks 1 and 4 in the library seeded with 16, whereas the nonseeded control only led to the formation of nonassembling 43 and 44 macrocycles (Figure 3a,b). Thus, cross-catalysis appears to be reciprocal, albeit not completely symmetrical: 48 induces the formation of 16 but not 18 whereas 46 does not give rise to any replicators based on 1, within the time frame of our experiments. Conversely, 16 induces the formation of both 46 and 48. These perhaps nonintuitive cross-catalytic effects prompted us to explore their origin. The fact that replicators based on the more hydrophobic building block 1 are hexamers, whereas those based on the more hydrophilic 4 are predominantly octamers fits with the general trend that we reported previously:13d more hydrophobic building blocks allow for self-assembly and concomitant replication already at a smaller ring size. What remains puzzling is why the replication by 16 is only triggered by 48 and not by any other of our established replicators. We suspect that this could be due to specific interactions involving the OH-groups of the serine residue in 4. This hypothesis is supported by the observation that octamers of alanine containing building block 5, thus lacking these OH groups, do not induce the formation of any replicators based on 1.
Figure 3.

Kinetic profile of a nonagitated dynamic combinatorial library made from building block 4 (3.8 mM in 50 mM borate buffer, pH 8.1) 80% oxidized (a) nonseeded (b) after addition of 10 mol percent of preformed 16 as seed. (c) Kinetic profile of a library made by mixing peptide 1 and peptide 4 (3.42 mM in 1 and 0.38 mM in 4) in borate buffer (pH 8.1, 50 mM) to form a 3.8 mM library. The library was then oxidized to 80% with freshly prepared perborate solution (80 mM) and stirred at 1200 rpm under an inert atmosphere.
Finally, we investigated to what extent the replicator distribution is dependent on sample history. We prepared a DCL with the same overall building block composition as the one shown in Figure 3b (3.42 mM in 1 and 0.38 mM in 4) but now mixed these building blocks at the start of the experiment. We monitored the sample over a period of 40 days but did not detect the emergence of replicators 16 or 48 (Figure 3c). This experiment shows that the history of the sample is a decisive factor in determining replicator presence or absence, just like the evolutionary history dictates the species composition in current life. Though history-dependence is a widespread phenomenon in materials science19 and protein folding,20 it had not yet been reported to dictate the nature of self-replicating molecules. Moreover, the history dependence observed here involves the interaction history between molecules in the mixture, and not merely the history in terms of physical properties, such as pH or temperature.
In conclusion, we have shown how the emergence of a new replicator based on threonine-containing building block 1 relies on the presence of a specific pre-existing replicator containing serine residues. Cross-catalysis between replicators (and therefore replicator mutation) in this system was found to be remarkably specific, as structurally closely related replicators failed to show the same effect. Our results also constitute an important first step in the development of abiotic systems of replicators in the direction of primitive life. While in previous work the presence of the building blocks of a replicator was typically sufficient for replicators to emerge (after some lag phase), in contemporary life, new species only derive from pre-existing ones (for as far as we know, no new life forms emerge solely from abiotic materials). Thus, in current life, the species present at any given time point reflect not only the available resources but also the evolutionary history of the various biological species. Our results represent a first step in the transition from a regime where replicator abundance is governed by building block availability to one where pre-existing replicators control the new replicator population. So, instead of replicator distributions being defined solely by the present conditions, the systems (evolutionary) history is now also becoming an essential controlling factor. Such history-dependent systems of self-replicators represent an important strategic direction in research on the origin of life and the development of de novo life.
Acknowledgments
We are grateful for support from the ERC, the NWO, COST Action CM1304 and the Ministry of Education, Culture and Science (Gravitation program 024.001.035). Check marks in the Scheme 1 are designed by Freepik.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.7b07346.
UPLC and LC-MS methods and data, kinetic data for all libraries with and without seeding, CD spectra, ThT fluorescence experiments for the DCLs made from peptides 1–5, and TEM micrographs for DCLs made from peptides 1–5 (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- a Mattia E.; Otto S. Nat. Nanotechnol. 2015, 10, 111–119. 10.1038/nnano.2014.337. [DOI] [PubMed] [Google Scholar]; b Ruiz-Mirazo K.; Briones C.; de la Escosura A. Chem. Rev. 2014, 114, 285–366. 10.1021/cr2004844. [DOI] [PubMed] [Google Scholar]; c Ludlow R. F.; Otto S. Chem. Soc. Rev. 2008, 37, 101–108. 10.1039/B611921M. [DOI] [PubMed] [Google Scholar]
- a Cougnon F. B. L.; Sanders J. K. M. Acc. Chem. Res. 2012, 45, 2211–2221. 10.1021/ar200240m. [DOI] [PubMed] [Google Scholar]; b Moulin E.; Cormos G.; Giuseppone N. Chem. Soc. Rev. 2012, 41, 1031–1049. 10.1039/C1CS15185A. [DOI] [PubMed] [Google Scholar]; c Ladame S. Org. Biomol. Chem. 2008, 6, 219–226. 10.1039/B714599C. [DOI] [PubMed] [Google Scholar]; d Corbett P. T.; Leclaire J.; Vial L.; West K. R.; Wietor J.-L.; Sanders J. K. M.; Otto S. Chem. Rev. 2006, 106, 3652–3711. 10.1021/cr020452p. [DOI] [PubMed] [Google Scholar]; e Miljanic O. S. Chem. 2017, 2, 502–524. 10.1016/j.chempr.2017.03.002. [DOI] [Google Scholar]; f Jin Y.; Yu C.; Denman R. J.; Zhang W. Chem. Soc. Rev. 2013, 42, 6634–6654. 10.1039/c3cs60044k. [DOI] [PubMed] [Google Scholar]
- a Schrum J. P.; Zhu T. F.; Szostak J. W. Cold Spring Harbor Perspect. Biol. 2010, 2, a002212. 10.1101/cshperspect.a002212. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hanczyc M. M. Science 2003, 302, 618–622. 10.1126/science.1089904. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Brea R. J.; Hardy M. D.; Devaraj N. K. Chem. - Eur. J. 2015, 21, 12564–12570. 10.1002/chem.201501229. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Walde P.; Umakoshi H.; Stano P.; Mavelli F. Chem. Commun. 2014, 50, 10177–10197. 10.1039/C4CC02812K. [DOI] [PubMed] [Google Scholar]
- a Nijemeisland M.; Abdelmohsen L. K. E. A.; Huck W. T. S.; Wilson D. A.; van Hest J. C. M. ACS Cent. Sci. 2016, 2, 843–849. 10.1021/acscentsci.6b00254. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Le Saux T.; Plasson R.; Jullien L. Chem. Commun. 2014, 50, 6189–6195. 10.1039/C4CC00392F. [DOI] [PubMed] [Google Scholar]; c Hordijk W.; Kauffman S. A.; Steel M. Int. J. Mol. Sci. 2011, 12, 3085–3101. 10.3390/ijms12053085. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Keller M.; Kampjut D.; Harrison S.; Ralser M. Nat. Ecol. Evol. 2017, 1, 0083. 10.1038/s41559-017-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Duim H.; Otto S. Beilstein J. Org. Chem. 2017, 13, 1189–1203. 10.3762/bjoc.13.118. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Vidonne A.; Philp D. Eur. J. Org. Chem. 2009, 2009, 593–610. 10.1002/ejoc.200800827. [DOI] [Google Scholar]; c Bissette A. J.; Fletcher S. P. Angew. Chem., Int. Ed. 2013, 52, 12800–12826. 10.1002/anie.201303822. [DOI] [PubMed] [Google Scholar]; d Clixby G.; Twyman L. Org. Biomol. Chem. 2016, 14, 4170–4184. 10.1039/C6OB00280C. [DOI] [PubMed] [Google Scholar]
- a von Kiedrowski G. Angew. Chem., Int. Ed. Engl. 1986, 25, 932–935. 10.1002/anie.198609322. [DOI] [Google Scholar]; b Ploeger T. A.; von Kiedrowski G. Org. Biomol. Chem. 2014, 12, 6908–6914. 10.1039/C4OB01168F. [DOI] [PubMed] [Google Scholar]; c Luther A.; Brandsch R.; von Kiedrowski G. Nature 1998, 396, 245–248. 10.1038/24343. [DOI] [PubMed] [Google Scholar]; d Kindermann M.; Stahl I.; Reimold M.; Pankau W.; von Kiedrowski G. Angew. Chem., Int. Ed. 2005, 44, 6750–6755. 10.1002/anie.200501527. [DOI] [PubMed] [Google Scholar]; e Dieckmann A.; Beniken S.; Lorenz C. D.; Doltsinis N. L.; von Kiedrowski G. Chem. - Eur. J. 2011, 17, 468–480. 10.1002/chem.201002325. [DOI] [PubMed] [Google Scholar]
- a Tjivikua T.; Ballester P.; Rebek J. J. Am. Chem. Soc. 1990, 112, 1249–1250. 10.1021/ja00159a057. [DOI] [Google Scholar]; b Pieters R.; Huc I.; Rebek J. Angew. Chem., Int. Ed. Engl. 1994, 33, 1579–1581. 10.1002/anie.199415791. [DOI] [Google Scholar]
- a Kosikova T.; Mackenzie H.; Philp D. Chem. - Eur. J. 2016, 22, 1831–1839. 10.1002/chem.201503740. [DOI] [PubMed] [Google Scholar]; b del Amo V.; Philp D. Chem. - Eur. J. 2010, 16, 13304–13318. 10.1002/chem.201000423. [DOI] [PubMed] [Google Scholar]; c Sadownik J. W.; Philp D. Angew. Chem., Int. Ed. 2008, 47, 9965–9970. 10.1002/anie.200804223. [DOI] [PubMed] [Google Scholar]; d Vidonne A.; Philp D. Eur. J. Org. Chem. 2009, 2009, 593–610. 10.1002/ejoc.200800827. [DOI] [Google Scholar]; e Bottero I.; Huck J.; Kosikova T.; Philp D. J. Am. Chem. Soc. 2016, 138, 6723–6726. 10.1021/jacs.6b03372. [DOI] [PubMed] [Google Scholar]
- a Paul N.; Joyce G. Curr. Opin. Chem. Biol. 2004, 8, 634–639. 10.1016/j.cbpa.2004.09.005. [DOI] [PubMed] [Google Scholar]; b Hayden E. J.; von Kiedrowski G.; Lehman N. Angew. Chem., Int. Ed. 2008, 47, 8424–8428. 10.1002/anie.200802177. [DOI] [PubMed] [Google Scholar]; c Szostak J. W. J. Biomol. Struct. Dyn. 2013, 31, 2–3. 10.1080/07391102.2013.786330. [DOI] [Google Scholar]; d Jia T. Z.; Fahrenbach A. C.; Kamat N. P.; Adamala K. P.; Szostak J. W. Nat. Chem. 2016, 8, 915–921. 10.1038/nchem.2551. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yao S.; Ghosh I.; Zutshi R.; Chmielewski J. Nature 1998, 396, 447–450. 10.1038/24814. [DOI] [PubMed] [Google Scholar]
- a Ura Y.; Beierle J. M.; Leman L. J.; Orgel L. E.; Ghadiri M. R. Science 2009, 325, 73–77. 10.1126/science.1174577. [DOI] [PubMed] [Google Scholar]; b Lee D.; Severin K.; Yokobayashi Y.; Ghadiri M. Nature 1997, 390, 591–594. 10.1038/37569. [DOI] [PubMed] [Google Scholar]; c Lee D.; Granja J.; Martinez J.; Severin K.; Ghadiri M. Nature 1996, 382, 525–528. 10.1038/382525a0. [DOI] [PubMed] [Google Scholar]
- a Rubinov B.; Wagner N.; Matmor M.; Regev O.; Ashkenasy N.; Ashkenasy G. ACS Nano 2012, 6, 7893–7901. 10.1021/nn302223v. [DOI] [PubMed] [Google Scholar]; b Bourbo V.; Matmor M.; Shtelman E.; Rubinov B.; Ashkenasy N.; Ashkenasy G. Origins Life Evol. Biospheres 2011, 41, 563–567. 10.1007/s11084-011-9257-y. [DOI] [PubMed] [Google Scholar]; c Tena-Solsona M.; Nanda J.; Diaz-Oltra S.; Chotera A.; Ashkenasy G.; Escuder B. Chem. - Eur. J. 2016, 22, 6687–6694. 10.1002/chem.201600344. [DOI] [PubMed] [Google Scholar]; d Dadon Z.; Wagner N.; Alasibi S.; Samiappan M.; Mukherjee R.; Ashkenasy G. Chem. - Eur. J. 2015, 21, 648–654. 10.1002/chem.201405195. [DOI] [PubMed] [Google Scholar]; e Mukherjee R.; Cohen-Luria R.; Wagner N.; Ashkenasy G. Angew. Chem., Int. Ed. 2015, 54, 12452–12456. 10.1002/anie.201503898. [DOI] [PubMed] [Google Scholar]
- a Nowak P.; Colomb-Delsuc M.; Otto S.; Li J. J. Am. Chem. Soc. 2015, 137, 10965–10969. 10.1021/jacs.5b04380. [DOI] [PubMed] [Google Scholar]; b Colomb-Delsuc M.; Mattia E.; Sadownik J. W.; Otto S. Nat. Commun. 2015, 6, 7427. 10.1038/ncomms8427. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Leonetti G.; Otto S. J. Am. Chem. Soc. 2015, 137, 2067–2072. 10.1021/ja512644f. [DOI] [PubMed] [Google Scholar]; d Malakoutikhah M.; Peyralans J. J.-P.; Colomb-Delsuc M.; Fanlo-Virgos H.; Stuart M. C. A.; Otto S. J. Am. Chem. Soc. 2013, 135, 18406–18417. 10.1021/ja4067805. [DOI] [PubMed] [Google Scholar]; e Carnall J. M. A.; Waudby C. A.; Belenguer A. M.; Stuart M. C. A.; Peyralans J. J. P.; Otto S. Science 2010, 327, 1502–1506. 10.1126/science.1182767. [DOI] [PubMed] [Google Scholar]
- Lee D.; Severin K.; Ghadiri M. Curr. Opin. Chem. Biol. 1997, 1, 491–496. 10.1016/S1367-5931(97)80043-9. [DOI] [PubMed] [Google Scholar]
- The total peak area in DCLs made from 1 dominated by trimer and tetramer was comparable to that of a DCL dominated by hexamer, indicating that the molar absorptivity of the building block is independent of the macrocycle in which it resides. Therefore, peak areas are representative of concentrations of individual library members (Figure S42).
- Greenfield N. J. Nat. Protoc. 2007, 1, 2876–2890. 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar P.; Culling C. Arch. Pathology 1959, 68, 487–498. [PubMed] [Google Scholar]
- Levine H. Protein Sci. 1993, 2, 404–410. 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge J. J. Appl. Polym. Sci. 1967, 11, 1199–1207. 10.1002/app.1967.070110717. [DOI] [Google Scholar]
- a Luft J. R.; Newman J.; Snell E. H. Acta Crystallogr., Sect. F: Struct. Biol. Commun. 2014, 70, 835–853. 10.1107/S2053230X1401262X. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Calonder C.; Tie Y.; Van Tassel P. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 10664–10669. 10.1073/pnas.181337298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


