Abstract
Human prion diseases such as Creutzfeldt-Jakob disease are transmissible brain proteinopathies, characterized by the accumulation of a misfolded isoform of the host cellular prion protein (PrP) in the brain. According to the prion model, prions are defined as proteinaceous infectious particles composed solely of this abnormal isoform of PrP (PrPSc). Even in the absence of genetic material, various prion strains can be propagated in experimental models. They can be distinguished by the pattern of disease they produce and especially by the localization of PrPSc deposits within the brain and the spongiform lesions they induce. The mechanisms involved in this strain-specific targeting of distinct brain regions still are a fundamental, unresolved question in prion research. To address this question, we exploited a prion conversion in vitro assay, protein misfolding cyclic amplification (PMCA), by using experimental scrapie and human prion strains as seeds and specific brain regions from mice and humans as substrates. We show here that region-specific PMCA in part reproduces the specific brain targeting observed in experimental, acquired, and sporadic Creutzfeldt-Jakob diseases. Furthermore, we provide evidence that, in addition to cellular prion protein, other region- and species-specific molecular factors influence the strain-dependent prion conversion process. This important step toward understanding prion strain propagation in the human brain may impact research on the molecular factors involved in protein misfolding and the development of ultrasensitive methods for diagnosing prion disease.
Keywords: neurodegenerative disease, prion, prion disease, protein aggregation, protein misfolding, amyloid, PMCA, regional targeting, strains
Introduction
According to the prion hypothesis, prions, the causative agents of prion diseases, are composed primarily or exclusively of molecules of abnormal prion protein (PrPSc), which replicate via the recruitment and conformational conversion of the host-encoded cellular prion protein (PrPC) (1). Despite the absence of a nucleic acid genome, multiple prion strains can be propagated in experimental models (2) and are characterized by the distinct patterns of disease they produce, and especially by the localization of PrPSc accumulation and strain-induced lesions. Experimental results provided evidence that strain diversity is also present in human prion disorders, including both infectious and sporadic forms of Creutzfeldt-Jakob disease (CJD)2 (3–7). Among patients with sporadic CJD (sCJD), a wide range of clinical and neuropathological phenotypes has been observed. The molecular basis of such phenotypic diversity involves a methionine/valine (Met/Val) polymorphism at codon 129 of the prion protein gene (PRNP), combined with different folding patterns of the PrPSc that accumulates in the brain of affected individuals. The latter is reflected by different sizes of the residual, protease-resistant core fragments of PrPSc (21 kDa: type 1; 19 kDa: type 2a) detected by Western blot analysis following digestion with proteinase-K (PK), suggesting different PK cleavage sites for different conformations of PrPSc (8). Recently, some sCJD subtypes have been associated with distinct human prion strains by transmission to nonhuman primates and transgenic mice (5, 6). A higher proportion of diglycosylated forms characterizes the type 2b PrPSc that is found in variant CJD (vCJD), which results from the cattle to human transmission of the prion strain responsible for the epizootic bovine spongiform encephalopathy (BSE) (9). vCJD is associated with a particular involvement of the posterior thalamus (10). One of the fundamental challenges in prion research is the development and implementation of in vitro models, as protein misfolding cyclic amplification (PMCA) (11), for the investigation of the mechanisms underlying the strain-specific targeting of brain regions. Using PrPSc from CJD-infected brain homogenate as seed and an excess of PrPC from normal brain homogenate as substrate, PMCA conducted in a cyclic manner, conceptually analogous to polymerase chain reaction (PCR), is able to amplify minute quantities of PrPSc present in a sample. This technology has proven very useful to study prion strain behaviors such as genotypic barrier of transmission and strain mutation/adaptation (12–15).
To address this still unresolved issue regarding human prion diseases, we took advantage of PMCA. In this study, we provide the first evidence that a model of specific targeting by human prion strains can be obtained in vitro using region-specific PMCA (rsPMCA). Our results also indicate that (i) strain targeting depends not only on neuroanatomical pathways or cell to cell interactions, which cannot operate in PMCA, but also on local molecular factors and (ii) in addition to cellular prion protein, other region- and species-specific molecular factors influence the conversion process in a strain-dependent-manner.
Results
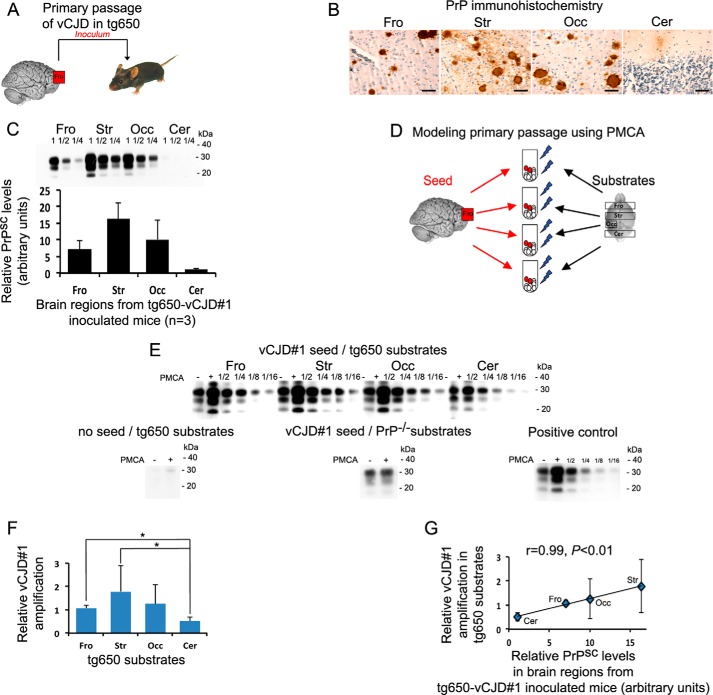
Modeling of region-specific targeting observed in experimental prion diseases
When transmitted to transgenic mice (Fig. 1A) overexpressing human PrPC with a methionine at codon 129 (tg650), as shown by PrP immunohistochemistry (Fig. 1B) and Western blot detection of PrPSc (Fig. 1C), the vCJD strain replicated variously in the different brain regions with high PrPSc load notably in the striatum and the occipital cortex and a low involvement of the cerebellum in agreement with a previous study (16). Using rsPMCA (Fig. 1D), we observed that frontal cortex, striatum, and occipital cortex from tg650 mice amplified vCJD seed more efficiently than cerebellar cortex (Fig. 1E). The values of relative amplification obtained with frontal cortex and striatum were significantly different (p < 0.05) compared with the value obtained with cerebellum (Fig. 1F). Interestingly, when amplification results were plotted as a function of relative PrPSc levels in corresponding brain regions from vCJD-infected tg650 mice (Fig. 1G), we observed a strong and significant correlation (r = 0.99, p < 0.01). This suggests that the strain tropism observed in experimental prion disease could be mimicked in vitro by rsPMCA.
Figure 1.
rsPMCA modeling of region-specific targeting was observed after primary passage of vCJD to transgenic mice expressing human PrPC. A, schematic representation of in vivo experiment. B and C, PrPSc accumulation in different brain regions of vCJD-inoculated tg650 mice that overexpress M129 human PrPC as illustrated by PrP immunochemistry (B) and revealed by Western blotting (C). D, schematic representation of in vitro modeling. E, Western blot illustration of the amplification obtained using vCJD seed and brain substrates from tg650 mice. No amplification was observed when the vCJD seed was omitted and when frontal substrates from PrP knock-out mice were used as substrate. F, relative rsPMCA amplification obtained with vCJD and the different substrates from tg650 mice. G, relative rsPMCA amplification plotted as a function of relative PrPSc level measured in the corresponding brain areas of vCJD-inoculated tg650 mice. Scale bar, 50 μm. Fro, frontal cortex; Str, striatum; Occ, occipital cortex; Cer, cerebellar cortex; PMCA−, no PMCA was applied; PMCA+, one round of PMCA was applied; 1/2, 1/4, 1/8, 1/16, rsPMCA product dilution. Error bars represent S.D.; *, p < 0.05. PMCA results are representative of four independent experiments performed in duplicates.
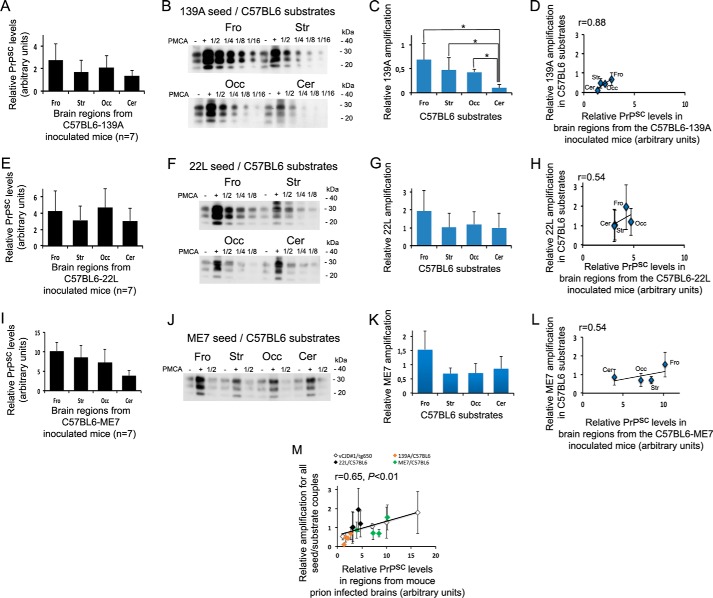
To further explore this phenomenon, we used three well characterized experimental scrapie strains (139A, 22L, and ME7) stabilized in a C57BL/6 background. These strains induce distinct lesion profiles (Fig. 2, A, E, and I) and have shown distinct behaviors when propagated in primary neurons from various brain origins (17). The amplification obtained with rsPMCA partially matched the spread of the scrapie agents observed in vivo (Fig. 2, B and C, F and G, J and K) showing a correlation coefficient of 0.88 for the 139A strain, of 0.54 for the 22L strain, and of 0.54 for the ME7 strain (Fig. 2, D, H, and L). To note, the proportion of diglycosylated forms of PK-resistant PrP was increased in PMCA products as compared with infected brain homogenates (see supplemental Fig. S1). A very similar change in the glycosylation pattern of PrPSc with PMCA conditions has been already observed with the 139A strain and using the same anti-PrP 6D11 antibody (18).
Figure 2.
rsPMCA modeling of region-specific targeting was observed in experimental scrapie. A, E, and I, PrPSc accumulation was observed in different brain regions from terminally ill C57BL/6 mice inoculated with 139A (A), 22L (E), and ME7 (I) strains. B, F, and J, amplifications using 139A (B), 22L (F), and ME7 (J) seeds and brain substrates from C57BL/6 mice are shown on Western blot illustrations. C, G, and K, amplifications obtained with 139A (C), 22L (G), and ME7 (K) strains as seed and C57BL/6 brain regions as substrates are shown and in D, H, and L have been plotted as a function of the PrPSc accumulation measured in the corresponding regions of infected mice. M, amplifications were obtained with all the experimental seed/substrate couples plotted as a function of the PrPSc accumulation measured in the corresponding brain areas of infected mice. Fro, frontal isocortex; Str, striatum; Occ, occipital isocortex; Cer, cerebellar cortex. Error bars represent S.D.; *, p < 0.05. PMCA results are representative of three independent experiments performed in duplicates.
When all the experimental amplification results are plotted as a function of relative PrPSc levels in corresponding C57BL/6-infected brain regions, we observed a positive and significant correlation (r = 0.65; p < 0.01) (Fig. 2M).
Modeling of region-specific targeting observed in Creutzfeldt-Jakob disease
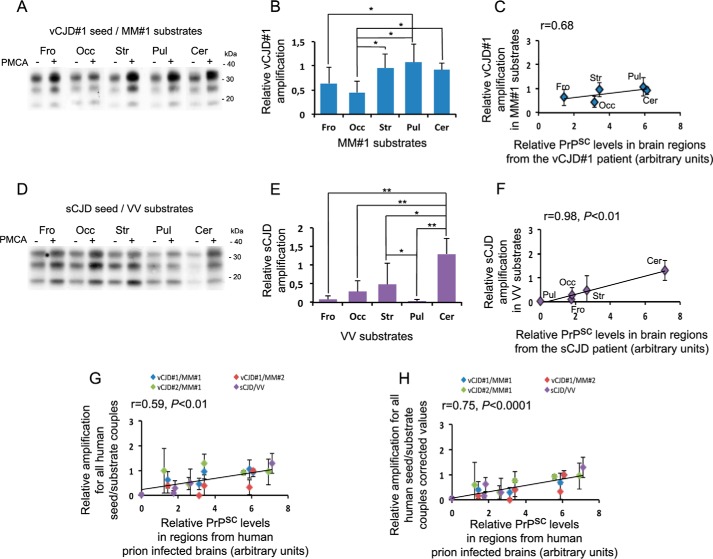
In the context of an interspecies transmission (vCJD to tg650) and despite the expression of the M129 human PrP by mice, the regional brain tropism of the vCJD agent is different from that observed in vCJD patients where the pulvinar nuclei and the cerebellar cortex are strongly involved (10). To assess whether rsPMCA could mimic the brain tropism observed in humans, we next performed rsPMCA using vCJD PrPSc seed and human brain substrates from codon 129 MM individuals.
The same vCJD seed was efficiently but variously amplified using different areas sampled from human MM brains (Fig. 3A). Significant interregional differences in amplification results were observed (p < 0.05, frontal cortex versus pulvinar, occipital cortex versus striatum, pulvinar, and cerebellum) (Fig. 3B). The Pearson correlation coefficient between amplification values and PrPSc levels in the corresponding brain areas of the vCJD patient was 0.68 (Fig. 3C). We validated this result using (i) additional substrates from a distinct MM individual and the same vCJD seed and (ii) an additional seed from another vCJD isolate with the first set of substrates. When using an additional vCJD seed, the correlation coefficient between amplification values and PrPSc levels in corresponding areas was 0.12. When the relative amplification values were corrected for PrPC relative levels of each substrate, the coefficient value reached 0.78 (close to that obtained using the first vCJD case, i.e. 0.84). When we used an additional set of human substrates from a codon 129 MM individual, we obtained a final correlation coefficient of 0.68 (supplemental Fig. S2). To note, despite similar post-mortem delay, the relative level of PrPC in the different human brain areas varied between both MM individuals. Interestingly, the amplifications obtained with human cerebellum were in contrast with those obtained using tg650 cerebellar substrates.
Figure 3.
rsPMCA modeling of region-specific targeting was observed in Creutzfeldt-Jakob disease. A and D, amplification using vCJD#1/MM#1 (A) and sCJD/VV (D) seed/substrate couples are shown on Western blot illustrations. B and E, amplifications obtained with each seed/substrate couple are shown and in C and F have been plotted as a function of the PrPSc accumulation measured in the corresponding regions from the patient who provided the seed. Gand H, amplifications obtained with all human seed/substrate couples are plotted as a function of the PrPSc accumulation measured in the corresponding brain areas of the different patients who provided seeds (G), and corrected values for PrPC relative levels (H). Fro, frontal isocortex; Occ, occipital isocortex; Str, striatum; Pul, pulvinar; Cer, cerebellar cortex. Error bars represent S.D.; *, p < 0.05; **, p < 0.01. PMCA results are representative of three independent experiments performed in duplicates.
Next, we assessed a sCJD isolate characterized by a distinct genotype at codon 129 of PRNP, and, in contrast with vCJD, mild involvement of the pulvinar region and strong targeting of the cerebellar cortex (i.e. the ataxic VV2a subtype of sCJD with early cerebellar ataxia and late dementia). We were able to amplify this seed with various brain regions selected from a human VV individual (Fig. 3D). Interregional variations in amplification results were observed (p < 0.01, cerebellum versus frontal cortex, pulvinar, and occipital cortex; p < 0.05, striatum versus cerebellum and pulvinar) (Fig. 3E). Pulvinar substrate showed poor amplification efficiency, and, as expected, cerebellar substrate efficiently amplified VV2a PrPSc seed. We found a high correlation between amplification values and PrPSc levels in the corresponding brain areas of the VV2a-sCJD patient (r = 0.98, p < 0.01) (Fig. 3F).
When all human amplification results were plotted as a function of relative PrPSc levels in corresponding infected brain regions (Fig. 3G), a significant correlation was observed (r = 0.59, p < 0.01). The correlation was better (r = 0.75, p < 0.001) when the relative amplification values were corrected for PrPC levels of each substrate (Fig. 3H).
Evidence for the role of local brain factors other than PrPC
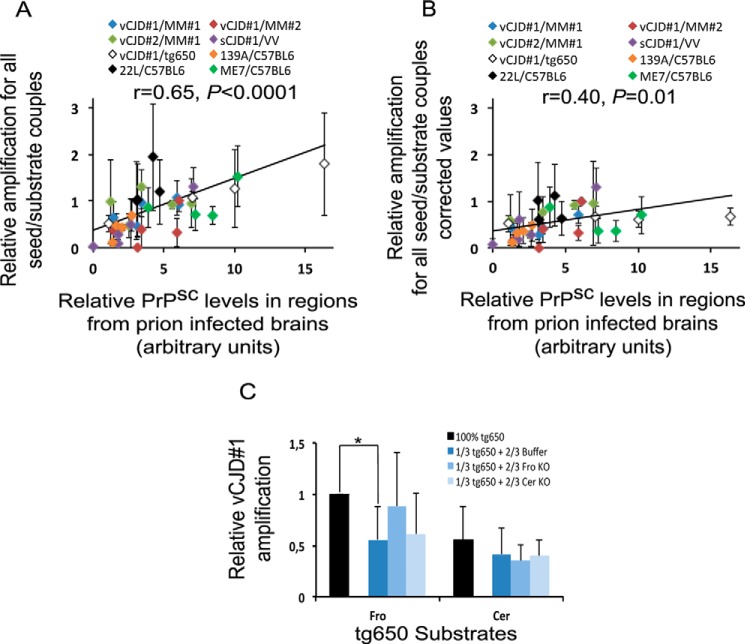
Finally, when all seed/substrate couples (n = 8) were pooled (Fig. 4A), we found a highly significant correlation between the regional tropism of prion strains and the relative amplification values obtained using rsPMCA (r = 0.65, p < 0.0001).
Figure 4.
rsPMCA modeling of region-specific targeting suggests that molecular factors other than PrPc modulate strain tropism. A and B, amplifications obtained with all seed/substrate couples were plotted as a function of the PrPSc accumulation measured in the corresponding brain areas from the affected individuals who provided seeds (A), and corrected values for PrPC relative levels (B). C, PMCA results obtained using vCJD seed and tg650 substrates, diluted or not, by conversion buffer or substrates from PrP knock-out mouse. Fro, frontal cortex; Cer, cerebellar cortex. Error bars represent S.D.; *, p < 0.05. PMCA results are representative of five independent experiments.
When the relative amplification values were corrected for PrPC relative levels of each substrate (Fig. 4B), the correlation between in vivo and rsPMCA results decreased (r = 0.40) consistently with a positive role of PrPC level in PMCA efficacy. Interestingly, a significant correlation was maintained (p = 0.01), however, suggesting that PrPC is not the sole factor involved in the region-specific conversion of PrP. To further investigate this hypothesis, we assessed whether homogenates from distinct brain regions of mice devoid of PrPC could modulate the PMCA efficacy. In vCJD amplification experiment, we observed that the significant decrease of PMCA efficacy induced by the dilution of tg650 frontal cortex substrate with conversion buffer was abolished by replacing the conversion buffer with a homogenate prepared with the frontal cortex of PrP knock-out mouse. Interestingly, no effect was observed when a homogenate prepared with the cerebellum from a PrP knock-out mouse was used. When similar experiments were performed with tg650 substrate prepared with the cerebellum, no effect of the homogenates prepared with the frontal cortex or the cerebellum of a PrP knock-out mouse was noted (Fig. 4C).
Discussion
Our study demonstrates that results obtained using rsPMCA are consistent with in vivo observations in mice and humans. This was observed using seeds from two different forms of human prion diseases associated with distinct brain involvement: An infectious form acquired by peripheral contamination (vCJD) and a sporadic subtype of CJD (VV2a sCJD). Both strains were selected because of their ability to be amplified in our hands without addition of PMCA supplements such as digitonin or heparin (19, 20).
Because it is well known that region-specific targeting of a given strain varies during interspecies transmission and that interspecies transmission does not depend only on PrP sequence homology (1), we assessed whether rsPMCA could also mimic such interspecies variations in prion spread. To this purpose, using the first of the two vCJD seeds amplified by the human MM substrate, we simulated a primary transmission of vCJD in the brain of tg650 mice expressing human PrP with a methionine at codon 129. Amplification levels obtained using rsPMCA matched the spread of the vCJD agent observed in vivo when primary transmission of vCJD in tg650 was performed, showing high involvement of striatum and occipital cortex with important PrPSc deposits, and weak targeting of cerebellar cortex. This result was distinct from that obtained using human MM substrates where a high level of amplification was observed using cerebellar cortex. This implies that the molecular factors involved in vCJD-specific targeting are not only region-specific but also vary between species.
Finally, when all the results were pooled, we found a highly significant correlation between in vivo and in vitro regional brain targeting, suggesting that the mechanisms involved are common to various prion strains. From a fundamental point of view, the ability of rsPMCA to mimic prion tropism supports the idea that PrP conversion plays a central role in strain behavior and that differences in conformation and aggregation properties of PrPSc encipher strain diversity (21, 22).
In contrast with our results, Hu et al. (23) observed that in vitro conversion did not reproduce the pattern of deposition observed in vivo using scrapie-infected hamsters. The discrepancy between the two studies may be explained by the use of distinct prion strains (hamster-adapted scrapie strains versus mouse-adapted scrapie strains and human strains). It is worth noting that the effect of PMCA cofactors may vary with species. Deleault et al. (24) reported that, whereas hamster PrPSc preferentially utilizes RNA as a cofactor, RNA fails to facilitate mouse PrPSc amplification. In addition, a clarification step of the substrate using centrifugation was performed in the hamster study and may have removed some of the components responsible for region-specific modulation of PMCA.
Hu et al. (23) suggested that in vitro conversion in hamsters was dependent on the availability of PrPC. Using mouse-adapted strains, human isolates amplified using human brain areas, and mouse transgenic brains, we confirmed this results. In addition, our data suggest that brain region targeting depends, in part, on local molecular factors distinct from PrPC. Indeed, a significant correlation was preserved when values were corrected for the relative PrPC level of each substrate, suggesting that additional cofactors may be involved during conversion using distinct brain regions. We also observed that the substrate containing the highest PrPC level is not necessarily the one associated with the highest PMCA efficiency. Moreover, supplementation experiments using homogenates prepared with brain areas from PrP knock-out mice suggested the presence of region-specific modulators of PrP conversion, other than PrPC. Altogether, these results suggest that strain-specific tropism not only depends on neuroanatomical pathways or cell-to-cell interactions, but also on both PrPC level and locally expressed non-PrPC cofactors.
It has been suggested that a species-specific cofactor designated protein X, which might function as a molecular chaperone, interferes with the conversion of PrPC into PrPSc (25, 26). Numerous studies evidenced different receptor molecules for prions (27) and protein microarray identified 47 human PrPC–interacting proteins, the great majority of which were annotated as proteins involved in the recognition of nucleic acids (28). Whether molecular factors relevant in PMCA, such as a component of the lipid raft fraction of the cell membrane (20) and ribonucleic acids (29–31), sufficiently vary between brain area to alter prion conversion in various species remains to be established.
PMCA has contributed to our understanding of the nature of the prion agent, and is increasingly used to set up potential sensitive method for early prion diagnosis (19, 32). The ability to mimic prion strain tropism, in an accelerated model and under controlled conditions in vitro, provides a unique and valuable tool for identifying local molecular cofactors that could highly influence the prion protein conversion process and might even constitute therapeutic targets in human prion diseases. Furthermore, rsPMCA may have various applications in the diagnosis field, risk assessment, and from the public health point of view. Exploiting rsPMCA (for example by using the most efficient brain area as a substrate to amplify a specific strain) may be useful to improve the sensitivity of prion detection in early cases, body fluids, or medical material, as well as in studies of interspecies barrier of transmission, especially in those dealing with the zoonotic risk associated with emerging prion diseases in animals.
Experimental procedures
Mouse inoculations
Inoculations were carried out in strict accordance with the recommendations from the Guide for the Care and Use of Laboratory Animals, as provided by the French Ministry of Agriculture and of the European Union (authorization number of the project: 02298.03). Inocula were prepared extemporaneously from vCJD frontal cortex and whole brains from C57BL/6 mice infected with experimental scrapie strains (139A, 22L, or ME7) (17) in a class II microbiological cabinet using disposable equipment as described (16).
Tg650 mice, overexpressing human PrPC with a methionine at codon 129 on a murine PrP knock-out background (16), were inoculated with vCJD frontal cortex and C57BL/6 mice were inoculated with the three experimental mouse-adapted scrapie strains.
Twenty microliters of a 10% (w/v) brain homogenate in 5% glucose were inoculated in 6- to 10-week-old mice by the intracerebral route using disposable syringes as described (16). Mice were monitored daily and were euthanized at the terminal stage of the disease. Brains were removed and either snap frozen in liquid nitrogen for PrPSc biochemistry or fixed in 4% phosphate-buffered formalin for morphological study.
Post-mortem human brain tissues
Brains from terminally ill mice inoculated with prion strains were selected to provide in vivo study or seed material for PMCA experiments. Frozen brain substrates used for PMCA experiments were sampled from healthy tg650 or C57BL/6 mice. In some experiments, frozen brains from PrP knock-out mice (33) were used as negative controls for PMCA.
Human tissues were selected on the basis of the availability of autopsy-obtained frozen brain material and informed consent from patients' relatives for autopsy and research use. The neuropathological examination of various brain regions was realized by standard staining methods, PrP immunohistochemistry (34), and Western blot detection of PrPSc. Three confirmed CJD patients and three non-CJD patients were identified as providers of seed or substrate materials allowing efficient PMCA. Seeds were from two vCJD patients with methionine homozygosity at codon 129 of PRNP and type 2b PrPSc (MM2b), and one sCJD patient with valine homozygosity at codon 129 of PRNP and with type 2a PrPSc (VV2a). Substrates were collected from patients with nonprion and nonneurodegenerative diseases: An MM patient with a post-mortem delay of 24 h and lymphoma, an MM patient with a post-mortem delay of 22 h and status epilepticus, and a VV patient with a post-mortem delay of 33 h and carcinoma.
Genetic analysis
The prion protein gene (PRNP) was analyzed as described previously (35) to obtain the genotype at the codon 129 polymorphism.
Preparation of seeds and substrates
Tissues were carefully dissected to avoid white matter and major blood vessels as much as possible. For extemporaneous use, substrates from frozen cerebral regions were homogenized (10% w/v) in PMCA conversion buffer (PBS with 150 mm NaCl, 1% Triton X-100, 0.005% EDTA pH 8, and cOmplete Protease Inhibitor Mixture 1× (Roche)) using a FastPrep®-24 instrument (MP Biomedical) for 45 s at speed 6.5. Seeds were prepared similarly from CJD frontal isocortex or whole brain from infected mice and stored at −80 °C.
Region-specific protein misfolding cyclic amplification
rsPMCA setup is based on standard PMCA technology (one round only) to investigate the early process of prion strain tropism in the brain of affected individuals. It uses, as substrates, key brain regions that are known to be differentially affected by PrPSc deposition in various human prion disorders and scrapie experimental model.
In humans, a set of five available cerebral regions were studied as PMCA substrates: frontal isocortex (Fro), occipital isocortex (Occ), striatum (Str), pulvinar (Pul), and cerebellar cortex (Cer). In mice, a set of four available cerebral regions were studied as PMCA substrates: frontal isocortex (Fro), occipital isocortex (Occ), striatum (Str), and cerebellar cortex (Cer).
For each experiment, a set of fresh substrates was prepared extemporaneously. Mouse substrates were prepared by pooling similar brain structures from three animals for each experiment. Human substrates were prepared for each experiment by using a set of frozen brain samples from each individual.
Each seed was diluted in the brain region substrates at the appropriate ratio and dispensed into 200-μl PCR tubes that fit with the tube holder of the sonicator. A fraction of each mixture was taken and immediately digested with PK (PMCA−). The remaining volume was subjected to one round of PMCA (PMCA+). Using human substrates, each cycle comprised a 40-s burst of sonication at output 7.5 (∼170 watts; Misonix S4000 sonicator) followed by an incubation period of 29 min and 20 s at 37 °C (40 cycles) (12–14). Using murine substrates, experiments were performed as described previously (15). Each experiment was controlled with the correct amplification of vCJD seed in tg650 brain (internal standard of amplification) and with unseeded substrate as negative control. rsPMCA results are representative of three to four independent experiments performed in duplicates. Amplification levels obtained using these procedures were globally similar to those described in previous studies (15, 36).
Western blotting
Samples were digested with proteinase K at 100 μg/ml for 1 h at 37 °C under continuous agitation. Digested samples were mixed with loading buffer and heated at 95 °C for 5 min. A 10-μl volume of each final mixture (samples of murine infected brains or post-rsPMCA product) was loaded onto precast 4–12% BIS-TRIS gels (Life Technologies) at different serial dilutions (1/2, 1/4, 1/8, 1/16). Proteins were transferred to PVDF membranes. PrP detection was performed using anti-PrP monoclonal antibody anti-CD230 (3F4 clone; BioLegend), epitope 109–112 of human PrP sequence (37) at a dilution of 1/10000, or anti-PrP monoclonal antibody 6D11, epitope 93–109 of human PrP sequence (Covance Inc.) at a dilution of 1/5000. Horseradish peroxidase–conjugated goat anti-mouse secondary antibody (Pierce Biotechnology) was used at a dilution of 1/5000. Signals were visualized using ECL (GE Amersham Biosciences).
Histopathology and immunohistochemistry of tg650 mouse brain
PrP immunohistochemistry was performed as described previously (34) on 7 μm–thick sections from paraffin-embedded samples, using the 12F10 mouse monoclonal antibody (Bertin Pharma) at a dilution of 1/200. A saturation step using 3% BSA for 20 min, hydrated autoclaving, and formic acid pretreatment was applied. The BenchMark XT IHC automated system for immunohistochemistry and the ultraView universal DAB detection kit were used according to the manufacturer's instructions (Ventana Medical Systems).
Densitometric analysis of PrPSc Western blot data
Semi-quantification of PrPSc levels was performed using a GS-800 calibrated densitometer and Quantity One software (Bio-Rad Laboratories). Values for the three PrPSc glycoforms were measured on blots with a nonsaturated signal as described (10, 38). When the signal was saturated, the amount of PrPSc was measured on the last quantifiable dilution (y). The final value (PMCA+) was obtained using the formula: (y) × dilution factor as described (36). For each studied region, we calculated the amplification of PrPSc signal by PMCA as follows: amplificationregion = ((PMCA+) − (PMCA−))/(PMCA−). The relative amplificationregion was calculated as a function of the value of the internal standard of amplification contained in each experiment (amplificationstandard): Relative amplification = amplificationregion/amplificationstandard.
In all experiments, the relative PrPC levels in each normal brain region used in substrate preparation were calculated as a function of the PrPC value in the cerebellar cortex. In a final analysis, we also corrected relative amplification with the results for the relative PrPC level (relative amplification/relative PrPC level) detected in each substrate. Relative PrPSc levels in each brain region from infected subjects were calculated as a function of the value of a PrPSc-positive internal standard present on all blots.
Statistical analysis
For statistical comparisons between regions, and according to the sample size, the nonparametric Mann-Whitney test was used. For correlations between in vivo regional targeting and in vitro PMCA results the Pearson test was used. Analyses were performed using Statview Software version 4.0 (Abacus Concepts) and SigmaStat software version 3.5 (Systat Software). Data are presented as the mean ± S.E., p < 0.05 was considered to be statistically significant.
Author contributions
N. P. was the primary experimentalist. S. H. designed the study. N. P., E. L., Sa. H., and S. Y. performed experiments. J.-L. L. performed PRNP genotyping. V. B. and S. H. performed mouse inoculations. D. S., J.-P. B., and S. H. contributed to the clinicopathological analysis. All authors participated in analyzing and interpreting data and contributed to the manuscript. N. P. and S. H. wrote the paper.
Supplementary Material
Acknowledgments
We are grateful to Dr. V. Sazdovitch for the supervision of the CJD brain collection, M. Bertrand for statistical assistance, and E. Morain and A. Matos for helpful technical assistance. We thank the late Dr. Henry Baron, who carefully read the manuscript and provided useful scientific comments.
This work was supported by the Institut national de Veille Sanitaire. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1 and S2.
- CJD
- Creutzfeldt-Jakob disease
- PMCA
- protein misfolding cyclic amplification
- PK
- proteinase-K
- sCJD
- sporadic CJD
- vCJD
- variant CJD
- rsPMCA
- region-specific PMCA
- MM
- Met/Met genotype at codon 129 of the prion protein gene
- VV
- Val/Val genotype at codon 129 of the prion protein gene.
References
- 1. Prusiner S. B. (1998) Prions. Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aguzzi A. (2008) Staining, straining and restraining prions. Nat. Neurosci. 11, 1239–1240 [DOI] [PubMed] [Google Scholar]
- 3. Bruce M. E., Will R. G., Ironside J. W., McConnell I., Drummond D., Suttie A., McCardle L., Chree A., Hope J., Birkett C., Cousens S., Fraser H., and Bostock C. J. (1997) Transmissions to mice indicate that 'new variant' CJD is caused by the BSE agent. Nature 389, 498–501 [DOI] [PubMed] [Google Scholar]
- 4. Scott M. R., Will R., Ironside J., Nguyen H. O., Tremblay P., DeArmond S. J., and Prusiner S. B. (1999) Compelling transgenetic evidence for transmission of bovine spongiform encephalopathy prions to humans. Proc. Natl. Acad. Sci. U.S.A. 96, 15137–15142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Parchi P., Cescatti M., Notari S., Schulz-Schaeffer W. J., Capellari S., Giese A., Zou W. Q., Kretzschmar H., Ghetti B., and Brown P. (2010) Agent strain variation in human prion disease: Insights from a molecular and pathological review of the National Institutes of Health series of experimentally transmitted disease. Brain 133, 3030–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bishop M. T., Will R. G., and Manson J. C. (2010) Defining sporadic Creutzfeldt-Jakob disease strains and their transmission properties. Proc. Natl. Acad. Sci. U.S.A. 107, 12005–12010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Giles K., Glidden D. V., Patel S., Korth C., Groth D., Lemus A., DeArmond S. J., and Prusiner S. B. (2010) Human prion strain selection in transgenic mice. Ann. Neurol. 68, 151–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parchi P., Giese A., Capellari S., Brown P., Schulz-Schaeffer W., Windl O., Zerr I., Budka H., Kopp N., Piccardo P., Poser S., Rojiani A., Streichemberger N., Julien J., Vital C., Ghetti B., Gambetti P., and Kretzschmar H. (1999) Classification of sporadic Creutzfeldt-Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann. Neurol. 46, 224–233 [PubMed] [Google Scholar]
- 9. Collinge J., and Rossor M. (1996) A new variant of prion disease. Lancet 347, 916–917 [DOI] [PubMed] [Google Scholar]
- 10. Brandel J. P., Heath C. A., Head M. W., Levavasseur E., Knight R., Laplanche J. L., Langeveld J. P., Ironside J. W., Hauw J. J., Mackenzie J., Alpérovitch A., Will R. G., and Haïk S. (2009) Variant Creutzfeldt-Jakob disease in France and the United Kingdom: Evidence for the same agent strain. Ann. Neurol. 65, 249–256 [DOI] [PubMed] [Google Scholar]
- 11. Saborio G. P., Permanne B., and Soto C. (2001) Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 411, 810–813 [DOI] [PubMed] [Google Scholar]
- 12. Jones M., Peden A. H., Prowse C. V., Gröner A., Manson J. C., Turner M. L., Ironside J. W., MacGregor I. R., and Head M. W. (2007) In vitro amplification and detection of variant Creutzfeldt-Jakob disease PrPSc. J. Pathol. 213, 21–26 [DOI] [PubMed] [Google Scholar]
- 13. Jones M., Wight D., Barron R., Jeffrey M., Manson J., Prowse C., Ironside J. W., and Head M. W. (2009) Molecular model of prion transmission to humans. Emerg. Infect. Dis. 15, 2013–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Castilla J., Gonzalez-Romero D., Saá P., Morales R., De Castro J., and Soto C. (2008) Crossing the species barrier by PrPSc replication in vitro generates unique infectious prions. Cell 134, 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Levavasseur E., Privat N., Martin J. C., Simoneau S., Baron T., Flan B., Torres J. M., and Haïk S. (2014) Molecular modeling of prion transmission to humans. Viruses 6, 3766–3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Béringue V., Le Dur A., Tixador P., Reine F., Lepourry L., Perret-Liaudet A., Haïk S., Vilotte J. L., Fontés M., and Laude H. (2008) Prominent and persistent extraneural infection in human PrP transgenic mice infected with variant CJD. PLoS One 3, e1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hannaoui S., Maatouk L., Privat N., Levavasseur E., Faucheux B. A., and Haïk S. (2013) Prion propagation and toxicity occur in vitro with two-phase kinetics specific to strain and neuronal type. J. Virol. 87, 2535–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao C., Han J., Zhang J., Wei J., Zhang B. Y., Tian C., Zhang J., Shi Q., and Dong X. P. (2017) Protein misfolding cyclic amplification cross-species products of mouse-adapted scrapie strain 139A and hamster-adapted scrapie strain 263K with brain and muscle tissues of opposite animals generate infectious prions. Mol. Neurobiol. 54, 3771–3782 [DOI] [PubMed] [Google Scholar]
- 19. Moda F., Gambetti P., Notari S., Concha-Marambio L., Catania M., Park K. W., Maderna E., Suardi S., Haïk S., Brandel J. P., Ironside J., Knight R., Tagliavini F., and Soto C. (2014) Prions in the urine of patients with variant Creutzfeldt-Jakob disease. N. Engl. J. Med. 371, 530–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Abid K., Morales R., and Soto C. (2010) Cellular factors implicated in prion replication. FEBS Lett. 584, 2409–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Prusiner S. B. (2001) Shattuck lecture—neurodegenerative diseases and prions. N. Engl. J. Med. 344, 1516–1526 [DOI] [PubMed] [Google Scholar]
- 22. Legname G., Nguyen H. O., Peretz D., Cohen F. E., DeArmond S. J., and Prusiner S. B. (2006) Continuum of prion protein structures enciphers a multitude of prion isolate-specified phenotypes. Proc. Natl. Acad. Sci. U.S.A. 103, 19105–19110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hu P. P., Morales R., Duran-Aniotz C., Moreno-Gonzalez I., Khan U., and Soto C. (2016) Role of prion replication in the strain-dependent brain regional distribution of prions. J. Biol. Chem. 291, 12880–12887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deleault N. R., Kascsak R., Geoghegan J. C., and Supattapone S. (2010) Species-dependent differences in cofactor utilization for formation of the protease-resistant prion protein in vitro. Biochemistry 49, 3928–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Telling G. C., Scott M., Mastrianni J., Gabizon R., Torchia M., Cohen F. E., DeArmond S. J., and Prusiner S. B. (1995) Prion propagation in mice expressing human and chimeric PrP transgenes implicates the interaction of cellular PrP with another protein. Cell 83, 79–90 [DOI] [PubMed] [Google Scholar]
- 26. Kaneko K., Zulianello L., Scott M., Cooper C. M., Wallace A. C., James T. L., Cohen F. E., and Prusiner S. B. (1997) Evidence for protein X binding to a discontinuous epitope on the cellular prion protein during scrapie prion propagation. Proc. Natl. Acad. Sci. U.S.A. 94, 10069–10074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vana K., Zuber C., Nikles D., and Weiss S. (2007) Novel aspects of prions, their receptor molecules, and innovative approaches for TSE therapy. Cell. Mol. Neurobiol. 27, 107–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Satoh J., Obayashi S., Misawa T., Sumiyoshi K., Oosumi K., and Tabunoki H. (2009) Protein microarray analysis identifies human cellular prion protein interactors. Neuropathol. Appl. Neurobiol. 35, 16–35 [DOI] [PubMed] [Google Scholar]
- 29. Deleault N. R., Lucassen R. W., and Supattapone S. (2003) RNA molecules stimulate prion protein conversion. Nature 425, 717–720 [DOI] [PubMed] [Google Scholar]
- 30. Deleault N. R., Harris B. T., Rees J. R., and Supattapone S. (2007) Formation of native prions from minimal components in vitro. Proc. Natl. Acad. Sci. U.S.A. 104, 9741–9746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Geoghegan J. C., Valdes P. A., Orem N. R., Deleault N. R., Williamson R. A., Harris B. T., and Supattapone S. (2007) Selective incorporation of polyanionic molecules into hamster prions. J. Biol. Chem. 282, 36341–36353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Concha-Marambio L., Pritzkow S., Moda F., Tagliavini F., Ironside J. W., Schulz P. E., and Soto C. (2016) Detection of prions in blood from patients with variant Creutzfeldt-Jakob disease. Sci. Transl. Med. 8, 370ra183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Büeler H., Fischer M., Lang Y., Bluethmann H., Lipp H. P., DeArmond S. J., Prusiner S. B., Aguet M., and Weissmann C. (1992) Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature 356, 577–582 [DOI] [PubMed] [Google Scholar]
- 34. Privat N., Laffont-Proust I., Faucheux B. A., Sazdovitch V., Frobert Y., Laplanche J. L., Grassi J., Hauw J. J., and Haïk S. (2008) Human prion diseases: From antibody screening to a standardized fast immunodiagnosis using automation. Mod. Pathol. 21, 140–149 [DOI] [PubMed] [Google Scholar]
- 35. Laplanche J. L., Delasnerie-Lauprêtre N., Brandel J. P., Chatelain J., Beaudry P., Alpérovitch A., and Launay J. M. (1994) Molecular genetics of prion diseases in France. French Research Group on Epidemiology of Human Spongiform Encephalopathies. Neurology 44, 2347–2351 [DOI] [PubMed] [Google Scholar]
- 36. Bucalossi C., Cosseddu G., D'Agostino C., Di Bari M. A., Chiappini B., Conte M., Rosone F., De Grossi L., Scavia G., Agrimi U., Nonno R., and Vaccari G. (2011) Assessment of the genetic susceptibility of sheep to scrapie by protein misfolding cyclic amplification and comparison with experimental scrapie transmission studies. J. Virol. 85, 8386–8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kascsak R. J., Rubenstein R., Merz P. A., Tonna-DeMasi M., Fersko R., Carp R. I., Wisniewski H. M., and Diringer H. (1987) Mouse polyclonal and monoclonal antibody to SAF (PrP) protein. J. Virol. 61, 3688–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Levavasseur E., Laffont-Proust I., Morain E., Faucheux B. A., Privat N., Peoc'h K., Sazdovitch V., Brandel J. P., Hauw J. J., and Haïk S. (2008) Regulating factors of PrP glycosylation in Creutzfeldt-Jakob disease—implications for the dissemination and the diagnosis of human prion strains. PLoS One 3, e2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.