Abstract
Activation of Ras signalling occurs in ~30% of human cancers; however, activated Ras alone is not sufficient for tumourigenesis. In a screen for tumour suppressors that cooperate with oncogenic Ras (RasV12) in Drosophila, we identified genes involved in the autophagy pathway. Bioinformatic analysis of human tumours revealed that several core autophagy genes, including GABARAP, correlate with oncogenic KRAS mutations and poor prognosis in human pancreatic cancer, supporting a potential tumour-suppressive effect of the pathway in Ras-driven human cancers. In Drosophila, we demonstrate that blocking autophagy at any step of the pathway enhances RasV12-driven epithelial tissue overgrowth via the accumulation of reactive oxygen species and activation of the Jun kinase stress response pathway. Blocking autophagy in RasV12 clones also results in non-cell-autonomous effects with autophagy, cell proliferation and caspase activation induced in adjacent wild-type cells. Our study has implications for understanding the interplay between perturbations in Ras signalling and autophagy in tumourigenesis, which might inform the development of novel therapeutics targeting Ras-driven cancers.
Introduction
Mutations that lock the small G-protein RAS in its activated form, such as RASV12, are found in over 30% of human cancers, making it one of the most important oncogenes.1, 2, 3 However, RAS activation alone is not sufficient for the development of cancer because of the induction of senescence.4, 5, 6, 7, 8 Identification of genes or pathways that cooperate with Ras to drive cancer progression is therefore paramount. With its ease of manipulation and expansive repertoire of available genetic tools, the vinegar fly Drosophila melanogaster has served a pioneering role in the study of cancer-causing genes and cooperative tumourigenesis.9, 10, 11 In Ras-driven epithelial overgrowth, studies in Drosophila have revealed that impairment of cell polarity or mitochondrial functions enhance tumour growth and invasion via activation of the Jun kinase (JNK) stress response pathway, faithfully recapitulating some of the features responsible for tumour progression in human cancers.11, 12, 13, 14, 15
Autophagy is a widely conserved catabolic process, and basal autophagy levels are necessary for cell homeostasis, clearing aberrant or unnecessary cytoplasmic material, such as misfolded proteins, supernumerary or defective organelles, the accumulation of which would otherwise generate toxic stress that is detrimental to proper cell metabolism and function.16, 17 In some developmental contexts, autophagy has also been shown to induce programmed cell death.18 Indeed, there is a tight relationship between autophagy and apoptosis, as members of the BCL-2 antiapoptotic family of protein interact directly with the autophagy machinery at the nucleation step.19 Moreover, autophagy is induced in response to external stresses, such as starvation or hypoxia, allowing cells to cope with transient nutrient deprivation or lower levels of oxygen.20
In tumourigenesis, the role of autophagy is yet to be fully resolved and appears context dependent.21, 22, 23 Several lines of evidence point to a role for autophagy in the survival of tumour cells in the hypoxic, nutrient-deprived microenvironment of an early tumour where a blood supply is yet to be established. Indeed, higher autophagy levels are present in most aggressive forms of human cancers to sustain the growth of the tumour, and blocking autophagy in mouse models of cancer restrains growth and progression towards more aggressive types of tumours.24, 25 Of note, tumours with activating Ras mutations are particularly dependent on functional autophagy.24, 26, 27, 28, 29 Conversely, inactivating mutations in several autophagy genes, such as UVRAG, AMBRA1 and BECN1, have been identified in breast and ovarian cancers,30 and Becn1, Atg5 or Atg7 depletion in the mouse are linked to increased tumourigenesis, highlighting the tumour-suppressive function of autophagy in these contexts.31, 32
In this study, following our identification of the core autophagy regulator Atg8a/GABARAP, Atg7 and Atg9 in a large-scale screen for RasV12-cooperating tumour suppressors in Drosophila, we investigate the tumour-suppressive role of autophagy in Ras-activated tissues. We show that knockdown of autophagy at any step of the process synergises with oncogenic Ras to promote growth of Drosophila epithelial tissues. We bioinformatically identify low expression of various autophagy genes in pancreatic adenocarcinoma and correlate it with oncogenic KRAS status and poor prognosis. In Drosophila, we uncover non-cell-autonomous proliferation and apoptosis in wild-type tissue surrounding RasV12-autophagy-impaired mutant clones. We show that autophagy inhibition in a Ras-activated background leads to induction of oxidative stress and JNK pathway activation. Genetic knockdown of oxidative stress or JNK is sufficient to prevent RasV12-autophagy-impaired tissue overgrowth. Our study reveals extensive cross-talk between Ras signalling and the autophagy pathway and suggests that therapeutic inhibition of autophagy could be contraindicated in certain subsets of cancers.
Results
A screen for RasV12 -cooperative tumour suppressors identifies genes involved in the autophagy pathway
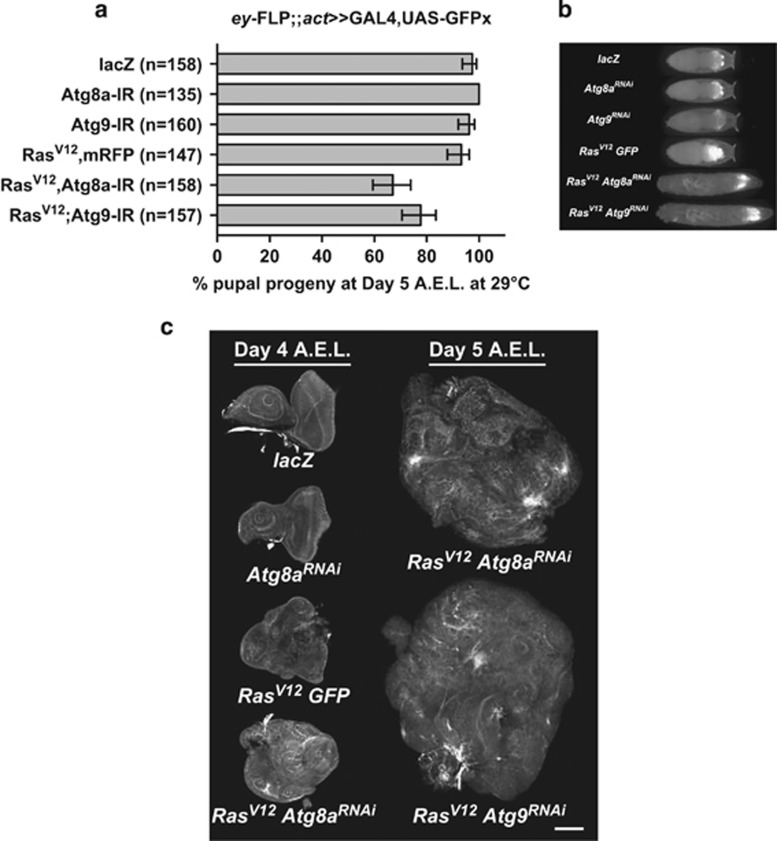
To identify new tumour suppressor genes that cooperate with constitutively active oncogenic Ras (RasV12), a high-throughput genome-wide in vivo RNA interference (RNAi) screen in the Drosophila eye-antennal epithelium was performed (Zoranovic et al., in preparation). Hits were selected based on their ability to delay larval development when knocked down in eye-antennal imaginal discs, a characteristic that correlates with uncontrolled growth of the tissue and subsequent failure to timely induce pupariation.33, 34 Three genes of the autophagy pathway, Atg7, Atg8a and Atg9, were identified among the hits, suggesting that autophagy can suppress the growth of RasV12-expressing epithelial tissue. Indeed, knockdown of Atg8a or Atg9 in RasV12-expressing eye-antennal epithelial tissue using the eyeless-FLP-out, actin-GAL4 system (see Materials and methods) delayed pupariation, with 20% of animals still crawling as giant larvae 5 days after egg lay (AEL) when raised at 29 °C, whereas controls formed pupae at 4 days (Figures 1a and b). Third instar larval RasV12 AtgRNAi eye-antennal epithelial tissues displayed loss of architecture and overgrowth compared with control discs (Figure 1c). Furthermore, RasV12 AtgRNAi eye-antennal epithelial tissue overgrew (Figure 1c—day 5 AEL) and was associated with a giant larvae phenotype, when all control flies had pupariated (Figures 1a and b). Notably, knocking down Atg8a or Atg9 alone did not induce any significant defect in size or shape of the eye-antenna discs nor delayed pupariation (Figures 1a–c, and data not shown), and animals developed into healthy, fertile adults (Supplementary Figure 1a). Thus, knockdown of Atg8a or Atg9 cooperates with RasV12 to result in neoplastic tumours and blocks metamorphosis at the larval–pupal stage, a characteristic observed previously with other Ras-cooperating mutations, for example, in the apicobasal polarity genes, scrib, dlg, lgl and baz,12, 13, 35 or upon overexpression of actin cytoskeletal genes, RhoGEF2, pbl (RhoGEF), Rac1 and bsk (Jun kinase).36, 37
Figure 1.
A screen for Ras-cooperative tumour suppressor genes identifies members of the autophagy pathway. (a) Pupariation count of ey-FLP-out Act»GAL4 larvae expressing the indicated transgenes at 5 days AEL. Knockdown of Atg8a or Atg9 in combination with RasV12 expression delayed pupariation in ~20% of animals, while neither single knockdown of Atg8a or Atg9, nor expression of RasV12 alone, did. (b) At day 5 AEL, giant larvae with enhanced GFP-positive masses can be observed in RasV12 Atg8aRNAi or RasV12 Atg9RNAi crosses by fluorescent microscopy, while all control animals have turned into pupae. (c) F-actin (phalloidin) staining of eye-antennal discs of ey-FLP-out Act»GAL4 wandering L3 larvae at day 4 AEL or giant larvae at day 5 AEL expressing the indicated transgenes. RasV12 AtgRNAi tissues lose morphology and keep growing after all control flies have pupariated. Error bars=95% confidence intervals. Scale bar: 50 μm.
All steps of autophagy are required to limit overgrowth of RasV12-expressing epithelial tissues
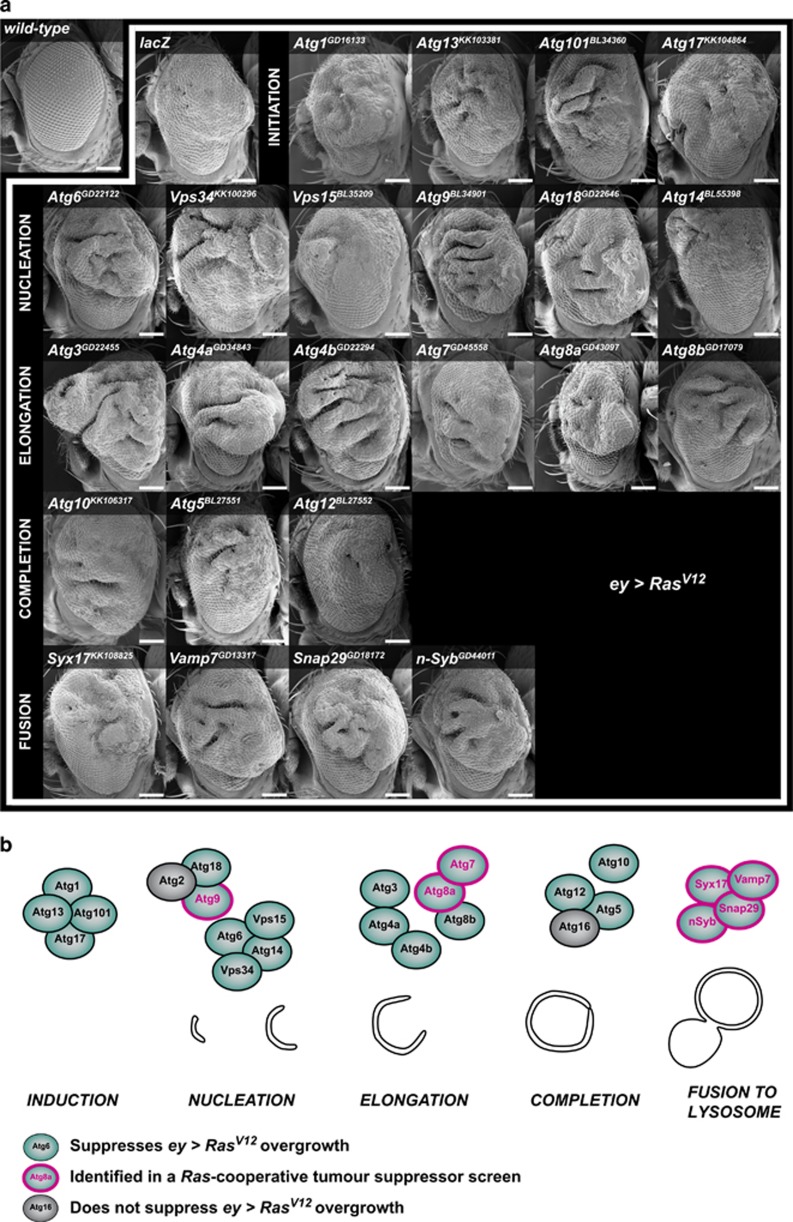
Because the three autophagy-related genes identified in our screen mapped to two distinct steps of the process—formation of the isolation membrane for Atg9, or phagophore elongation for Atg7 and Atg8a, we questioned whether the tumour-suppressive effect was limited to these genes, or whether autophagy in general was necessary to limit RasV12-driven overgrowth of fly epithelial tissue. To test this systematically, we switched to a well-characterised tissue overgrowth model in which the eyeless-GAL4 (ey-GAL4) driver expresses RasV12 and/or Atg-RNAis in the eye-antennal epithelium.36, 38 Knockdown of Atg8a, Atg7 or Atg9 each enhanced the overgrowth and roughness of ey-GAL4, RasV12 (ey>RasV12) eyes, as anticipated (Figure 2a and Supplementary Figure 2a), which was not observed with the controls (Supplementary Figure 2b). We then assessed the growth of adult eyes expressing RasV12 and RNAis for most of the documented genes involved in autophagy in Drosophila (Table 1, Supplementary Table 1 and Supplementary Figure 2a), and confirmed that the RNAi lines could effectively block autophagy in a Ras-activated developing epithelium by measuring accumulation of the autophagic cargo adapter Ref(2)P/p62 (Supplementary Figure 3). This included genes required at the initiation (Atg1, Atg101, Atg13, Atg17), nucleation (Atg6, Vps34, Vps15, Atg14, Atg18a), elongation (Atg3, Atg4a, Atg4b, Atg8b) and completion (Atg5, Atg10, Atg12) steps. Strikingly, knockdown of almost all genes involved in the autophagy pathway enhanced RasV12-induced overgrowth (Figures 2a and b, Supplementary Figure 2a and Supplementary Table 1). The only exceptions were that of Atg2 and Atg16, for which the single RNAi line available for each did not modify ey>RasV12 eye size or roughness (Supplementary Figure 2a), and Atg18b, which we did not assess in this study.
Figure 2.
Impaired autophagy enhances Ras-driven overgrowth of the adult Drosophila eye. (a) Scanning electron micrographs of ey-GAL4 adult female Drosophila eyes crossed to the indicated RNAi lines (superscript) or lacZ control. Inhibition of autophagy at any step of the process results in enhanced overgrowth of RasV12-expressing eyes. (b) Schematic of the autophagy pathway. Genes that knockdown enhances Ras-driven overgrowth are highlighted in green, with those identified in the RasV12-cooperative tumour suppressor primary screen circled in red. Scale bars: 100 μm.
Table 1. Autophagy-related genes in Drosophila and their corresponding human orthologues.
| Autophagy step | Drosophila gene | CG no. | Human orthologue | Molecular function |
|---|---|---|---|---|
| Induction | Atg1 | CG10967 | ULK1, ULK2 | Kinase |
| Atg101 | CG7053 | ATG101 | Protein binding | |
| Atg13 | CG7331 | ATG13 | Protein kinase binding | |
| Atg17/FIP200 | CG1347 | RB1CC1 | Protein kinase binding | |
| Nucleation | Atg6 | CG5429 | BECN1 | Protein binding |
| Atg14 | CG11877 | ATG14 | Protein binding | |
| Vps15/ird1 | CG9746 | PIK3R4 | Kinase | |
| Vps34/Pi3K59F | CG5373 | PIK3C3 | Kinase | |
| Atg18a | CG7986 | WIPI2 | PIP2 binding | |
| Atg9a | CG3615 | ATG9A, ATG9B | Protein binding | |
| Atg2 | CG1241 | ATG2A, ATG2B | Protein binding | |
| Elongation | Atg3/Aut1 | CG6877 | ATG3 | Ubiquitin-like ligase |
| Atg4a | CG4428 | ATG4A, ATG4B | Cysteine-type endopeptidase | |
| Atg4b | CG6194 | ATG4C | Cysteine-type endopeptidase | |
| Atg7a | CG5489 | ATG7 | Ubiquitin-activating enzyme | |
| Atg8aa | CG32672 | GABARAP | Ubiquitin-like | |
| Atg8b | CG12334 | MAP1LC3C, MAP1LC3B2 | Ubiquitin-like modifying enzyme | |
| Completion | Atg10 | CG12821 | ATG10 | Ubiquitin-like ligase |
| Atg12 | CG10861 | ATG12 | Ubiquitin-like | |
| Atg5 | CG1643 | ATG5 | Ubiquitin-like ligase | |
| Atg16 | CG31033 | ATG16L1, ATG16L2 | Ubiquitin-like ligase | |
| Fusion to the lysosome | Syx17a | CG7452 | STX17 | SNAP/SNARE protein binding |
| Vamp7a | CG1599 | VAMP7, VAMP8 | SNARE protein binding | |
| Snap29/usnpa | CG11173 | SNAP29 | SNAP receptor | |
| nSyba | CG17248 | VAMP1, VAMP2, VAMP3 | SNARE protein binding |
Bold Italics indicate genes identified in a screen for Ras-cooperative tumour suppressor genes.
These data suggested that other autophagy-related hits might be present in our screen. Interestingly, analysis of protein interactions within the 947 candidate genes identified in the RasV12-cooperative tumour suppressor screen revealed a cluster of Snap/SNARE proteins, composed of Syx17, Snap29 and Vamp739 (Supplementary Figure 1b), that have recently been shown to regulate autophagy at the last step of the process by promoting fusion of autophagosomes to lysosomes.40 Of interest, the neuronal synaptobrevin, nSyb, predicted by protein interaction studies to be part of this cluster,39 and whose paralogue Syb was recently confirmed to form a functional complex with Syx17, Snap29 and Vamp7,41 was also identified in our screen. Similarly to other components of the autophagy pathway above, we were able to confirm that Syx17, Vamp7, Snap29 or nSyb knockdown could also enhance RasV12-induced overgrowth (Figures 2a and b, Supplementary Figure 2a and Supplementary Table 1).
Taken together, our data show that blocking autophagy at any step of the pathway can potentiate Ras-driven overgrowth (Figure 2b), suggesting that functional basal autophagy is required to limit overgrowth of RasV12-expressing epithelial tissues.
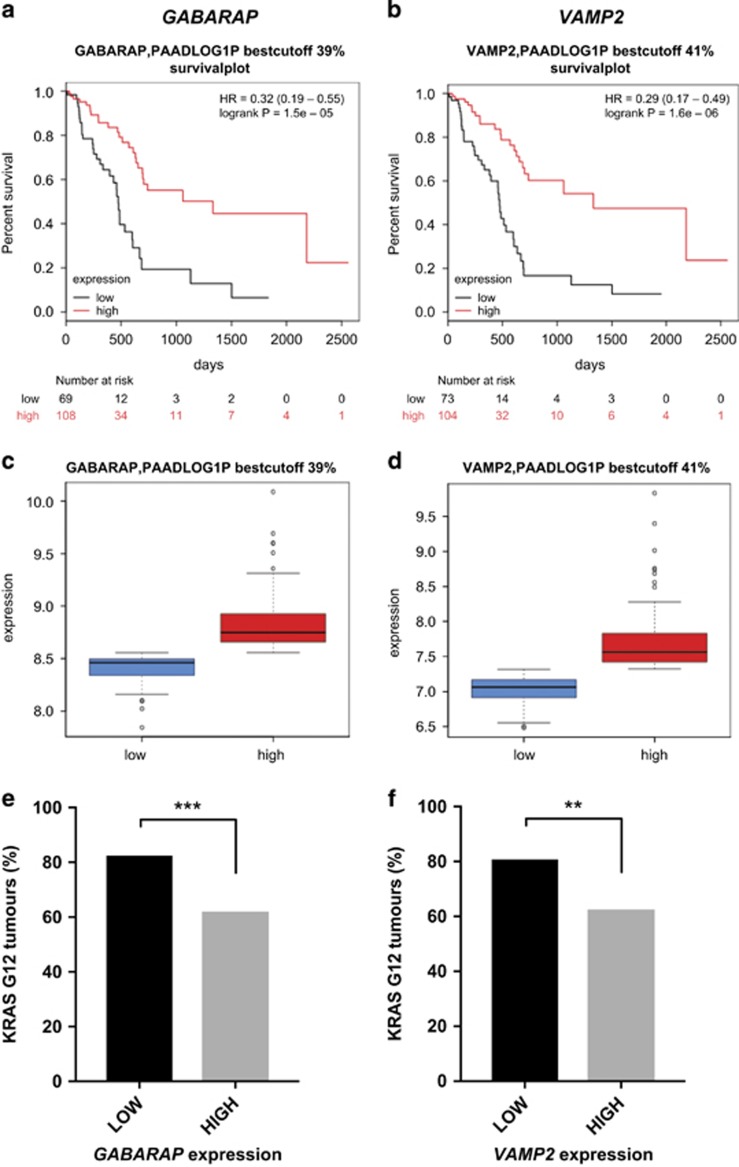
Lower expression of GABARAP and VAMP2 correlates with worse prognosis in KRAS-G12-positive pancreatic adenocarcinoma
The autophagy pathway has ambivalent roles in tumourigenesis (reviewed in Ávalos et al.22). Our Drosophila data argues for a tumour-suppressive function of the pathway in Ras-driven tumourigenesis. To investigate the expression of autophagy-related genes in human tumours with documented KRAS mutations, we queried available RNAseq data from The Cancer Genome Atlas. We focused on pancreatic ductal adenocarcinoma (PAAD), as incidence of KRAS-activating mutations in this tumour type is above 80%. We performed K-means clustering of tumours into low- or high-expressing groups for each of the human orthologues of the autophagy genes identified in the fly screen, and survival distribution was assessed for both groups. Patients with tumours exhibiting low mRNA expression for GABARAP, GABARAPL1, GABARAPL2, GABARAPL3 or VAMP2 showed significant association with worse clinical outcome (hazard ratio <1) and significant enrichment for KRAS-G12-activating mutations (P<0.01) (Figures 3a–d, Supplementary Table 2 and Supplementary Data sets 1 and 2), suggesting that in PAAD with G12 KRAS mutations, these genes individually behave as tumour suppressors. Noteworthy, this correlation between low-expressing autophagy, KRAS-activating mutations and poor clinical outcome was still observed when tumours were ranked for the mean expression of the entire subset of autophagy genes identified in the primary fly screen (Supplementary Figure 4), although this effect was mostly, but not exclusively, driven by the GABARAP family and VAMP2 (Supplementary Table 3).
Figure 3.
Low expression of GABARAP and VAMP2 is prognostic for worse survival in KRAS-G12-positive PAAD. Survival curves of PAAD patients with low (black) or high (red) expression of GABARAP (a) and VAMP2 (b). Low expression of GABARAP (c) or VAMP2 (d) correlates with worse clinical outcome in patients (a and b) and with enrichment for G12-activating mutations in KRAS (e and f), suggesting that these autophagy-related genes behave as tumour suppressors in PAAD with KRAS-activating mutations. Statistics: Fisher’s exact t-test.
On the other hand, poorer clinical outcome was associated with higher expression of other autophagy-related genes such as VAMP7 and VAMP8 (Supplementary Table 2 and Supplementary Data sets 1 and 2). While this suggests that not all genes of the pathway act as tumour suppressors, in tumours where VAMP7 or VAMP8 showed higher expression, other autophagy genes, such as ATG9B or GABARAP, often showed reduced expression (Supplementary Figure 4a and Supplementary Data set 2), and this upregulation could reflect a mechanism to compensate for a blockage of the pathway at another level.
Altogether, this analysis shows that low expression of the GABARAP family and VAMP2 correlates with a higher risk of poorer outcome in PAAD and might serve as a good indicator of poor prognosis in KRAS-G12 bearing tumours.
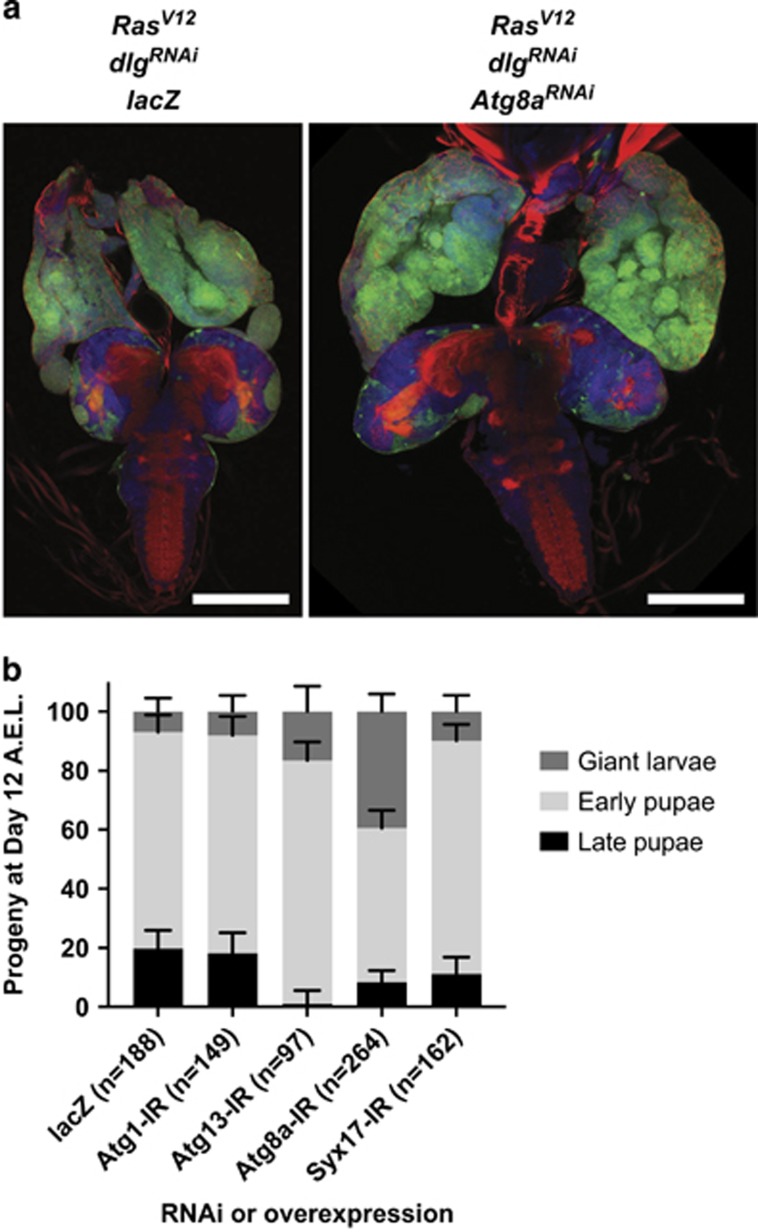
Autophagy limits the growth of RasV12 dlgRNAi epithelial tumours in Drosophila
It has been proposed that the tumour-suppressive role of autophagy in cancer would be limited to early stages of tumour development, while later stages require autophagy to sustain their growth (reviewed in White21 and Galluzzi et al.23). To investigate whether autophagy could also have a tumour-suppressive effect in a more aggressive model of tumourigenesis, we inhibited the pathway in the eye-antennal epithelium in which RasV12 was expressed together with knockdown of the cell polarity gene, dlg. Loss of dlg has been previously shown to cooperate with RasV12 in tumourigenesis,13, 42, 43 and human dlg orthologs are mutated or downregulated in cancer.44, 45, 46, 47 In the ey-FLP-out Act»GAL4 RasV12 dlgRNAi model,43 mutant tissue (marked by green fluorescent protein (GFP)) overgrows, loses architecture, the eye-antennal discs fuse together and invasion occurs into the brain lobes and ventral nerve chord (Figure 4a—control lacZ). As a consequence, normal development is altered: the majority of animals (73%) die early after pupariation, with only 20% of animals reaching a later developmental stage, with adult structures visible through the pupal case, and 7% of animals failing to pupariate and dying as giant larvae (Figure 4b—control lacZ). Inhibiting autophagy at the elongation step, using Atg8aRNAi, increased third instar larval eye-antennal disc size (Figure 4a), and accentuated developmental delay, with nearly 40% of animals dying as giant larvae, and only 8% reaching the late pupal stage (Figure 4b). Inhibiting autophagy at the autophagosome–lysosome fusion step, using Syx17RNAi, also delayed the development of ey-FLP-out Act»GAL4 RasV12 dlgRNAi larvae, although not as strongly as Atg8aRNAi (Figure 4b). Blocking the initiation step, using Atg13RNAi, markedly enhanced the developmental delay (Figure 4b). However, inhibiting the pathway at the same step, using Atg1RNAi, did not have any substantial effect on ey-FLP-out Act»GAL4 RasV12 dlgRNAi development. A difference in the knockdown efficiency in the eye-antennal disc is plausible; however, we have shown that both RNAis efficiently block the autophagic flux in Ras-activated wing epithelium (Supplementary Figure 4). Thus, specific functions of Atg1 and Atg13 are more likely to account for this difference. Indeed, it has recently been shown that Atg1 has autophagy-independent roles in tissue growth.48 Altogether, these data show that blocking autophagy at multiple steps of the pathway can also have a tumour-suppressive role in a more advanced Ras-driven tumoural setting.
Figure 4.
Blocking autophagy enhances polarity-impaired RasV12-driven epithelial tumours. (a) Eye-antennal imaginal discs of ey-FLP-out Act»RasV12 dlgRNAi lacZ (left) or ey-FLP-out Act»RasV12 dlgRNAi Atg8aRNAi (right) are shown attached to the brain and ventral nerve chord. (b) Autophagy inhibition at the induction (Atg13RNAi), elongation (Atg8aRNAi) or fusion to lysosome (Syx17RNAi) step of the pathway enhanced developmental delay in an ey-FLP-out Act»RasV12 dlgRNAi cooperative model of tumourigenesis. Inhibition of autophagy at the elongation step, using Atg8aRNAi, accentuated developmental delay, with nearly 40% of animals dying as giant larvae, and only 8% reaching a later pupal stage. Inhibiting autophagy at the autophagosome–lysosome fusion step, using Syx17RNAi, also delayed development of ey-FLP-out Act»GAL4 RasV12 dlgRNAi larvae, although not as strongly as Atg8aRNAi, with ~10% dying as giant larvae, and only 11% making it to the later pupal stage. Blocking the initiation step, using Atg13RNAi, markedly enhanced developmental delay, with only 1% of animals reaching the late pupae stage, and 16% dying as giant larvae. However, inhibiting the pathway at the same step, using Atg1RNAi, did not have any substantial effect on ey-FLP-out Act»GAL4 RasV12 dlgRNAi development. Error bars=95% confidence intervals.
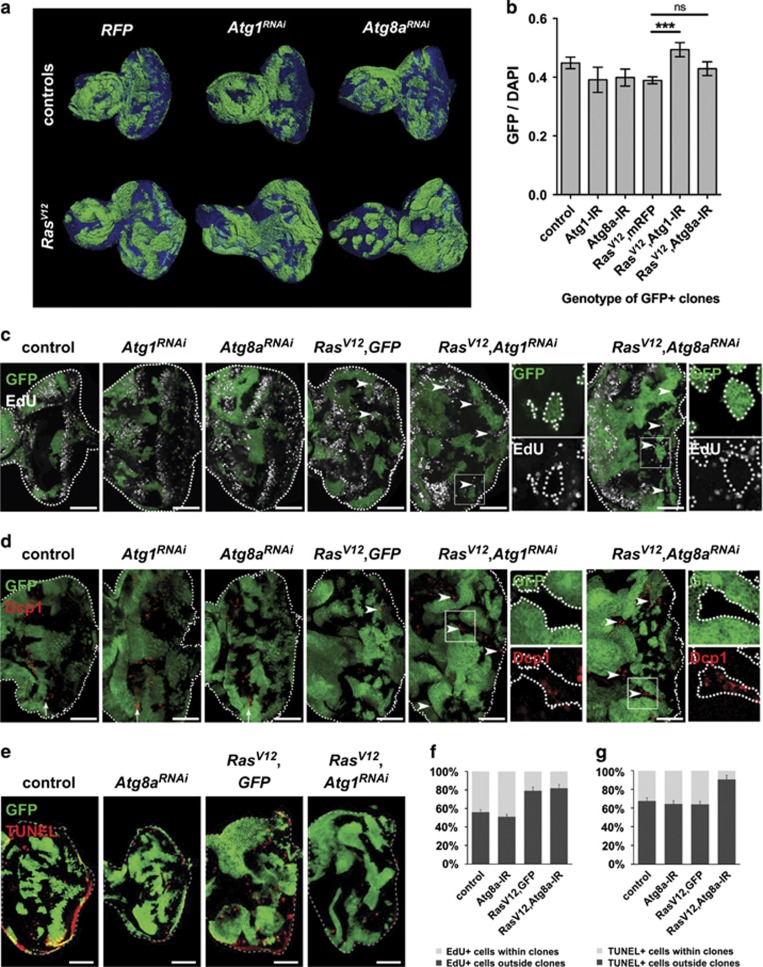
Non-cell-autonomous ectopic proliferation, caspase activation and cell death in RasV12 AtgRNAi mosaic tissues
To further investigate the effect of autophagy inhibition in RasV12-expressing epithelial tissue in a setting that more closely mimics the development of mammalian tumours, we turned to a clonal system using the mosaic analysis with a repressible cell marker (MARCM) technique,49 where UAS-transgene-expressing clones are marked by the expression of GFP from a UAS-GFP construct. We first examined the representation of GFP-positive tissue in eye-antennal discs of third instar larvae using three-dimensional reconstruction of confocal z-sections (Figure 5a). In clones where autophagy was inhibited at the initiation or elongation steps with Atg1RNAi or Atg8aRNAi, respectively, the ratio of GFP/4',6-diamidino-2-phenylindole (DAPI) volume was not significantly different to the control (Figure 5b). In discs expressing both RasV12 and Atg1RNAi in clones, the proportion of GFP tissue was significantly higher relative to control RasV12 mRFP samples (Figure 5b), consistent with the overgrowth we observed in a whole-tissue setting (Figure 2a). This trend was also observed in RasV12 Atg8aRNAi, although failing to meet statistical significance.
Figure 5.
Autophagy inhibition in Ras-activated tissues results in cell- and non-cell-autonomous effects on tissue growth. (a) Three-dimensional (3D) reconstruction of RasV12AtgRNAi and control eye-antennal mosaic discs from L3 wandering larvae. Mutant clones are GFP+ and tissues were counterstained with DAPI (blue). (b) In discs expressing both RasV12 and Atg1RNAi in clones, the proportion of GFP tissue was significantly higher than in the control RasV12 mRFP samples (49.4±2.4%, compared to 38.9±1.2%, P=0.00028). This trend was also observed in RasV12 Atg8aRNAi, although the difference was not significantly different (42.9±2.3%, P=0.068). (c) Non-cell-autonomous proliferation in wild-type tissue (non-GFP) surrounding RasV12GFP, RasV12 Atg1RNAi or RasV12Atg8aRNAi clones (GFP+), as seen by EdU incorporation. (d) Non-cell-autonomous caspase activation in wild-type tissue (non-GFP) surrounding RasV12Atg1RNAi or RasV12Atg8aRNAi clones (GFP+), as seen by Dcp1 staining. Arrows indicate the apoptotic wave before the MF. (e) Apoptosis is confirmed by TUNEL assay around clones expressing RasV12 and Atg8a-RNAi. (f') Quantification of data in (d). (g) Quantification of data in (e). Error bars=s.e.m. Statistics: one-way ANOVA with Tukey's multiple correction test. Scale bars: 50 μm.
We then examined proliferation using 5-ethynyl-2′-deoxyuridine (EdU) incorporation. Ectopic proliferation posterior to the morphogenetic furrow (MF) was observed in RasV12 GFP clones (Figure 5c, arrowheads), consistent with the role of activated Ras in driving cell proliferation.50 Strikingly, in RasV12 GFP, RasV12 Atg1RNAi and RasV12 Atg8aRNAi mosaic discs we observed that most EdU-positive cells surrounded mutant GFP clones, suggesting that mutant tissue induced non-cell-autonomous proliferation of the neighbouring wild-type tissue (Figure 5c, arrowheads and insets, quantified in Figure 5f). Since the overall mutant-to-wild-type tissue ratio was slightly increased in RasV12 AtgRNAi samples compared with RasV12 mRFP (Figures 5a and b), cell proliferation in the wild-type tissue must be overcompensated by apoptosis. Indeed, staining for Drosophila death caspase 1 (Dcp1) in mosaic discs revealed that, posterior to the MF, Dcp1 activation occurred preferentially in the wild-type tissue surrounding RasV12 AtgRNAi mutant clones, and that much less Dcp1-positive cells were detected in RasV12 GFP mosaic discs (Figure 5d). Indeed, terminal deoxynucleotidyl transferase dUTP nick-end labelling (TUNEL) staining further confirmed that apoptosis was preferentially induced in the vicinity of RasV12 Atg8aRNAi tissue, with very little apoptosis detected within clones (Figure 5e, quantified in Figure 5 g). Overall, the reduced apoptosis observed within RasV12 Atg8aRNAi clones compared with RasV12 GFP clones could explain the overgrowth of RasV12 AtgRNAi tissues in mosaic discs (Figures 6a and b).
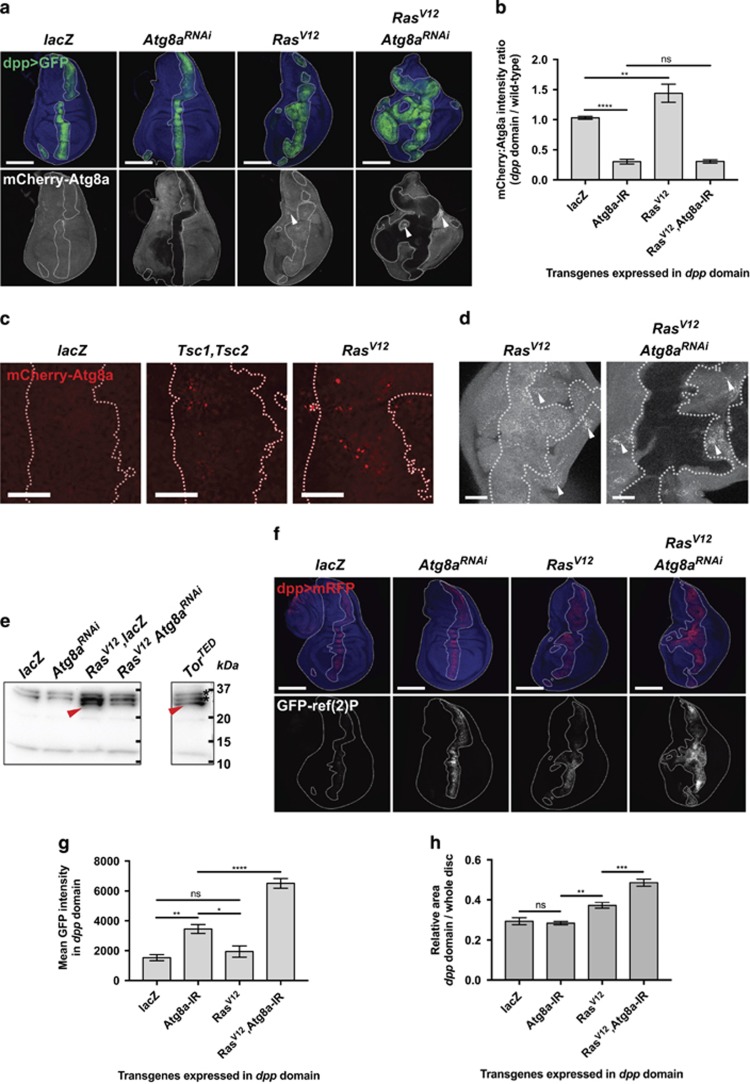
Figure 6.
RasV12 expression induces autophagy. (a–d) Effect of Atg8aRNAi, RasV12 and RasV12 Atg8aRNAi expressed via the dpp-GAL4 driver on pmCherry-Atg8a expression in L3 wing discs. mCherry-Atg8a levels (b) are increased upon Ras activation. (c) mCherry-Atg8a punctae are detected in the Dpp domain (dotted lines) upon expression of Tsc1 and Tsc2 transgenes (positive control) or RasV12, while no puncta is detected upon expression of a control lacZ. (d) Non-cell-autonomous activation of autophagy is also observed in wild-type tissue surrounding RasV12 and RasV12 Atg8aRNAi tissue (arrowheads). (e) Monitoring of autophagy flux induction by detection of free mCherry in mCherry-Atg8a tissues. A 27 kDa band corresponding to free mCherry is detected in wing discs expressing RasV12 and RasV12 Atg8aRNAi in the Dpp domain, as well as in the positive control expressing TorTED. *, unspecified band. (f) Effect of Atg8aRNAi, RasV12 and RasV12 Atg8aRNAi expressed via the dpp-GAL4 driver on GFP-Ref(2)P accumulation in L3 wing discs. Atg8a knockdown in the Dpp domain blocks autophagic flux as seen by accumulation of GFP-Ref(2)P aggregates. Slight accumulation of Ref(2)P aggregates is detected upon Ras activation, and blocking autophagic flux in this context leads to massive accumulation of Ref(2)P aggregates in the Dpp domain, quantified in (g). (h) As in the developing eye epithelium, autophagy inhibition in a Ras-activated background leads to tissue overgrowth, with proportion of GFP+ tissue higher in RasV12 Atga-RNAi compared with Ras-only or Atg8a-RNAi-only controls. Scale bars: (a and e) 100 μm, (c) 20 μm and (d) 50 μm. Error bars: s.e.m. Statistics: one-way ANOVA with Tukey's multiple correction.
High levels of Ras activity has also been shown to induce cell-autonomous differentiation;51 however, autophagy inhibition did not alter this effect as seen by staining for the neuronal marker Elav (Supplementary Figure 4). Interestingly, we saw no alterations in proliferation or apoptosis in RasV12 AtgRNAi clones. Thus altogether, our data suggest that autophagy inhibition in Ras-activated clones induced non-cell-autonomous proliferation and also caspase activation, and cell death, in the surrounding wild-type tissue, resulting in an overall overgrowth of the mutant tissue.
Autophagy is induced upon Ras activation
To test whether our observations on inhibiting autophagy in the eye-antennal epithelium was tissue-specific or a more generalised phenomenon for other epithelial tissues, we expressed RasV12 and AtgRNAis along the anteroposterior boundary of the wing imaginal disc using the dppblk-GAL4 driver (dpp>) (Figure 6). In this setting, we examined the effect of the RasV12 and AtgRNAi transgenes on autophagic flux in vivo by monitoring Atg8a expression (using ubiquitous pmCherry-Atg8a expressed under endogenous promoter control)52 or Ref(2)P levels and aggregation (using UAS-GFP-ref(2)P).53
In controls expressing a lacZ construct, faint but homogeneous expression of mCherry-Atg8a (under endogenous promoter control) was detected throughout the disc (Figure 6a, quantified in Figure 6b), while low levels of GFP-Ref(2)P (under UAS control) accumulated in the dpp expression domain (Dpp strip) (Figure 6f, quantified in Figure 6g). As expected upon Atg8aRNAi expression, we observed reduced levels of Atg8a (Figures 6a and b—Atg8aRNAi) and accumulation of Ref(2)P aggregates in the Dpp strip (Figures 6f and g—Atg8aRNAi), confirming that in Atg8aRNAi tissue, autophagy flux is efficiently impaired. In RasV12-expressing samples, mCherry-Atg8a levels were elevated compared with wild-type surrounding tissue (Figures 6a and b—RasV12). Higher magnification of the Dpp domain showed mCherry-Atg8a puncta accumulating upon RasV12 expression, similarly to that observed with Tsc1,Tsc2 overexpression, which inhibits mTORC1 and induces autophagy54 (Figure 6c). This finding suggests that basal autophagy is increased upon Ras activation. Interestingly, we also noticed increased Atg8a levels and punctae in the wild-type tissue adjacent to the Dpp domain expressing RasV12, and similarly in the wild-type tissue adjacent to the Dpp domain expressing RasV12 Atg8aRNAi (Figure 6d). These data suggest that basal autophagy is also induced in a non-cell-autonomous manner in the vicinity of RasV12 and RasV12 Atg8aRNAi tissue (Figure 6d, arrowheads; see also Figure 6a). To investigate the functional induction of the autophagy flux, we sought to detect the release of free mCherry that occurs after the mCherry-Atg8a reporter is processed by the lysosome. Overexpression of a TorTED construct, a well-known inducer of autophagy flux,55 under the dpp-GAL4 driver used in Figures 6a–d leads to the detection of a ~27kDa protein corresponding to the molecular weight of free mCherry (Figure 6e, right lane, arrowhead). Upon expression of lacZ or Atg8aRNAi, no such band is detected, suggesting that mCherry is not cleaved from Atg8a and that flux is not occurring. However, upon Ras expression, we were able to detect free mCherry, confirming that Ras induces autophagy flux in developing Drosophila epithelium. Moreover, detection of free mCherry in discs expressing a strip of RasV12 Atg8aRNAi tissue also reveals the non-cell-autonomous induction of the pathway, as mCherry is knocked down together with Atg8a in the presence of the RNAi in the strip, and only mCherry-Atg8a expressed in the wild-type portion of the disc is detected (Figures 6a, b and d).
Increased levels of GFP-Ref(2)P were also observed upon RasV12 expression (Figures 6f and g—RasV12). Oncogene-induced upregulation of p62 has been shown to occur consecutive to activation of the Nrf2-dependent unfolded protein response downstream of Myc in Drosophila,56 as well as upon RAS activation in mammalian systems.57, 58 Thus, upregulation of p62 levels is not necessarily the sign of autophagy blockage, but can also reflect upregulation of the pathway from a low baseline (as in developing epithelia). In our case, endogenous p62 upregulation could increase the pool of p62 and explain the difference in clearance of the GFP-p62 fusion protein compared with control (see also Figure 7c). Supporting this hypothesis, blocking autophagic flux in the RasV12-expressing Dpp strip by expressing Atg8a-RNAi led to massive accumulation of Ref(2)P aggregates, much more than upon Atg8a knockdown alone (Figures 6f and g—RasV12 Atg8aRNAi), suggesting that RasV12 expression sensitises the cells to autophagy impairment.
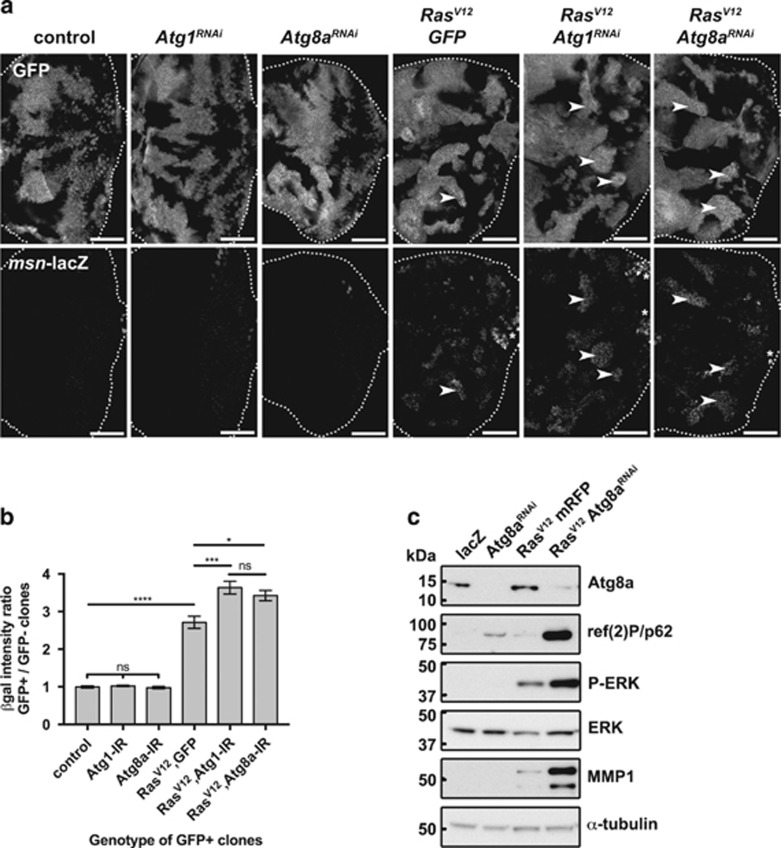
Figure 7.
JNK signalling is induced upon autophagy inhibition in RasV12-expressing tissues. (a) JNK pathway activation in RasV12Atg1RNAi or RasV12Atg8aRNAi clones, as detected by the expression of the msn-lacZ reporter (arrowheads). Stronger induction of msn-lacZ can be seen in RasV12AtgRNAi clones compared with RasV12 GFP clones (quantified in (b)). Asterisks show endogenous msn-lacZ expression in glial cells. (c) Western blot analyses of Atg8a, Ref(2)P, phospho-ERK, total-ERK and MMP1 relative to α-tubulin in ey-FLP-out, Act»GAL4 lacZ, Atg8aRNAi, RasV12 or RasV12Atg8aRNAi lysates. RasV12 signalling induces autophagy in the eye epithelium, as seen by higher level of Atg8a in RasV12-expressing tissue compared with control lacZ tissue. Ref(2)P accumulation confirms inhibition of the autophagic flux in Atg8aRNAi and RasV12Atg8aRNAi discs. Strong accumulation of Ref(2)P is observed in RasV12Atg8aRNAi. Ras-MAPK signalling is increased in RasV12Atg8aRNAi tissue, compared with RasV12 alone, as seen by increased phospho-ERK levels. Strong induction of the JNK target MMP1 in RasV12AtgRNAi discs reveals synergistic activation of the JNK pathway in RasV12 tissue upon autophagy inhibition. Scale bars: 50 μm. Statistics (b): mean±s.e.m., one-way ANOVA with Tukey's multiple correction.
Importantly, dpp>RasV12 expression induced overgrowth of the Dpp strip, which was enhanced by Atg8a knockdown (Figure 6h). Of note, the wing disc outside of the Dpp strip was distorted in RasV12 Atg8aRNAi samples (Figures 6a and f), suggesting that non-cell-autonomous tissue growth effects might also be occurring, as was observed in the eye-antennal disc.
In summary, our data shows that, in developing epithelia, autophagy is induced cell and non-cell autonomously in response to Ras activation. Impairing autophagy (by Atg8a knockdown) in a Ras-activated background leads to massive accumulation of Malumbres and Barbacid2 P aggregates and promotes non-cell-autonomous autophagy (revealed by Atg8a aggregates). Finally, as in the eye epithelium, autophagy inhibition enhanced growth of the RasV12-expressing wing epithelium, confirming the tumour-suppressive role of autophagy in Ras-activated Drosophila epithelia.
ERK and JNK signalling are enhanced upon autophagy blockage in Ras-activated tissues
We and others have previously shown that Ras-cooperative overgrowth often depends on the co-activation of the stress-induced JNK branch of the mitogen-activated protein kinase (MAPK) pathway.35, 36, 37 To test whether blocking autophagy in a Ras-activated background could induce the JNK pathway, we assessed the levels of the well-known JNK target misshapen (msn), using a msn-lacZ enhancer-trap reporter59 in eye-antennal disc clones (Figure 7a). While no expression of the msn-lacZ reporter was detected in control, Atg1RNAi or Atg8aRNAi clones, strong induction of msn-lacZ was detected in RasV12 Atg1RNAi and RasV12 Atg8aRNAi clones, relative to the milder induction that occurred in RasV12 GFP clones (Figure 7a, quantified in Figure 7b).
Cross-talk and feedback mechanisms between autophagy and MAPK signalling have been reported previously in mammals.60, 61, 62, 63 To investigate further the activation of the extracellular signal-regulated kinase (ERK) and JNK branches of the MAPK pathway when autophagy is blocked in a Ras-activated background, we looked at protein levels in lysates of ey-FLP-out, Act»GAL4 eye-antennal discs (Figure 7c). First, we confirmed our previous observation that basal autophagy is induced upon Ras activation, as seen by increased Atg8a levels (Figure 7c—lane 3, quantified in Supplementary Figure 6). Again, blocking autophagic flux with Atg8aRNAi in this context led to a strong accumulation of Ref(2)P (Figure 7c—lane 4 and Supplementary Figure 6).
Then, we monitored the levels of phospho-ERK (active) and total-ERK in eye tissue lysates. Phospho-ERK was below detectable levels in control lacZ and Atg8aRNAi samples, and was elevated as expected in RasV12 mRFP samples (Figure 7c—lanes 1–3). Strikingly, strong upregulation of phospho-ERK levels was detected in RasV12 Atg8aRNAi samples, suggesting that blocking autophagy increased the flux through the Ras-ERK pathway (Figure 7c—lane 4 and Supplementary Figure 6). These observations are consistent with recent mammalian studies showing that autophagy proteins regulate ERK phosphorylation and that autophagosomes can serve as intracellular scaffolds for the MAPK cascade.61
Finally, we monitored the levels of MMP1, a known JNK target. MMP1 was upregulated by RasV12 relative to the lacZ control and Atg8aRNAi alone (Figure 7c—lane 3), and a synergistic increase in MMP1 levels was observed in RasV12 Atg8aRNAi tissue (Figure 7c—lane 4, quantified in Supplementary Figure 6). This observation, along with the high levels of msn-lacZ induced in RasV12 Atg8aRNAi mosaic discs (Figure 7a), confirms that, upon autophagy inhibition, the JNK pathway is robustly induced in Ras-activated tissues.
Autophagy inhibition in Ras-activated tissue induces massive accumulation of reactive oxygen species
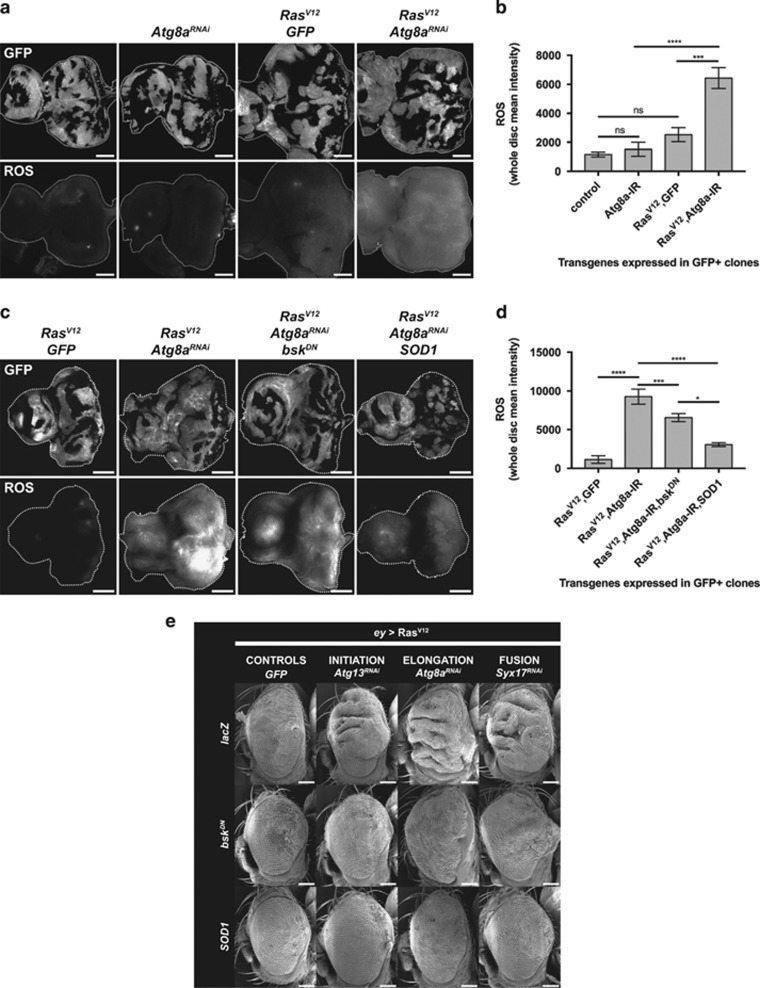
Autophagy is a key homeostasis mechanism by which cells remove defective organelles and protein aggregates to prevent oxidative stress.16, 17 Moreover, it has been documented that the JNK stress response module is activated in response to reactive oxygen species (ROS) accumulation.64 Thus, we sought to investigate whether the enhanced overgrowth observed in RasV12 AtgRNAi tissues is caused by ROS-induced JNK activation.
First, we monitored ROS levels upon autophagy inhibition of Ras-activated tissues by performing CellROX Deep Red staining on mosaic eye-antennal discs (Figure 8a). Expression of lacZ control or Atg8a-RNAi only did not lead to ROS accumulation, and expression of RasV12 alone only slightly increased oxidative stress levels (Figure 8a, quantified in Figure 8b). However, when autophagy was blocked in RasV12-expressing clones, we observed massive accumulation of ROS (Figures 8a and b—RasV12 Atg8aRNAi). Surprisingly, the accumulation of CellROX staining was not entirely cell autonomous, suggesting that either ROS diffused from mutant clones into the microenvironment or that mutant tissue induced ROS production in neighbouring wild-type tissue (Figure 8a and Supplementary Figure 7a—RasV12 Atg8aRNAi).
Figure 8.
Upon autophagy inhibition, ROS accumulate in Ras-activated tissue and lead to cooperative tissue overgrowth via JNK activation. (a) Oxidative stress levels in eye-antennal mosaic discs as detected by CellROX Deep Red staining. Autophagy inhibition using Atg8aRNAi did not augment ROS levels compared with control discs. Ras activation (RasV12 GFP) slightly increased ROS levels. However, massive accumulation of ROS was detected in RasV12Atg8aRNAi mosaic tissue. Interestingly, ROS are not limited to RasV12Atg8aRNAi clones, but spread in the whole disc, suggesting a non-cell-autonomous regulation of oxidative stress levels in RasV12AtgRNAi mosaic tissue. Scale bar: 50 μm. (b) Quantification of ROS levels in whole discs from (a). (c) ROS levels in RasV12 Atg8aRNAi mosaic discs are reduced upon SOD1 overexpression, but not upon JNK blockage, confirming that JNK activation is downstream of ROS accumulation in RasV12 Atg8aRNAi discs. Note that overexpression of SOD1 rescues both cell-autonomous and non-cell-autonomous accumulation of ROS, and restores the size and shape of RasV12 Atg8aRNAi eye discs. (d) Quantification of ROS levels in whole discs from (c). (e) Scanning electron micrographs of adult eyes expressing RasV12 and GFP (first column), Atg13RNAi (second column), Atg8aRNAi (third column) or Syx17RNAi (fourth column), in combination with control lacZ (first row), bskDN (second row) or SOD1 (third row). RasV12AtgRNAi eye overgrowth is rescued by overexpression of bskDN or SOD1, showing that autophagy-impaired RasV12-driven tumourigenesis is due to elevated ROS levels and JNK signalling. Note that overexpression of SOD1 and bskDN also rescues Ras overgrowth alone (first column). Error bars=s.e.m. Statistics: one-way ANOVA with Tukey's multiple correction.
Blocking JNK by overexpressing a dominant-negative form of the JNK orthologue basket (bskDN) in RasV12 Atg8aRNAi clones restored the architecture of the tissue but only partially reduced ROS levels (Figure 8c, quantified in Figure 8d), confirming that oxidative stress cues are mostly upstream of JNK activation. However, reducing oxidative stress by overexpressing superoxide dismutase 1 (SOD1) strikingly rescued disc overgrowth, restored tissue architecture and markedly reduced ROS levels (Figures 8c and d).
Accumulation of ROS and JNK activation are responsible for the cooperation of Atg knockdown and RasV12 in tissue overgrowth
To test whether JNK activation was causal to the overgrowth seen in RasV12 AtgRNAi adult Drosophila eye, we overexpressed bskDN in Ras-activated tissues in which autophagy was blocked at the induction (Atg13RNAi), elongation (Atg8aRNAi) or fusion to the lysosome (Syx17RNAi) steps (Figure 8c). bskDN rescued the overgrowth of RasV12 Atg13RNAi, RasV12 Atg8aRNAi and RasV12 Syx17RNAi eye epithelial tissue (Figure 8c, quantified in Supplementary Figure 7b). Strikingly, reducing ROS levels, by overexpressing SOD1, strongly rescued the overgrowth of RasV12 Atg13RNAi, RasV12 Atg8aRNAi and RasV12 Syx17RNAi eye epithelial tissue to a greater extent than blocking JNK (Figure 8c and Supplementary Figure 7b).
Altogether, these data show that, in Ras-activated tissues, blocking autophagy at several steps of the pathway leads to marked accumulation of ROS and JNK activation, and that these events are responsible for cooperative overgrowth with activated Ras.
Discussion
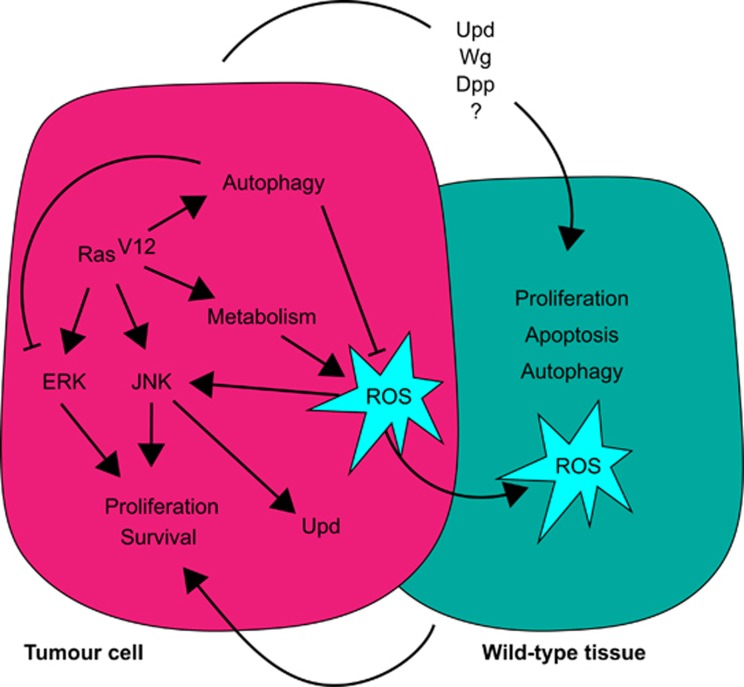
Autophagy’s involvement in cancer initiation and progression has drawn much attention over the past few years. Autophagy can be oncogenic as well as tumour suppressive depending on context and tumour stage.21, 22, 23 In this study, following up on a genome-wide screen for Ras-cooperative tumour suppressors that identified members of the autophagy pathway, we investigate the mechanisms by which autophagy contains the growth of Ras-activated tissues. We show that, in RasV12-expressing epithelial tissues, inhibition of autophagic flux at different steps of the pathway results in the accumulation of ROS and activation of the JNK stress response module, leading to overgrowth of mutant tissue (Figure 9).
Figure 9.
Model of cooperation between Ras and the autophagy pathway. Autophagy induction upon Ras-activation limits the accumulation of reactive oxygen species (ROS) and the activation of the ERK pathway. When autophagy is blocked in RasV12-expressing cells, ERK is further activated, ROS accumulate and activate the JNK pathway, resulting in cooperative overgrowth of RasV12 tissues by cell-autonomous and non-cell autonomous mechanisms. ROS are detected in neighbouring wild-type tissue, and non-cell autonomous activation of autophagy, proliferation, caspase activation and cell death is observed, which might feed back to promote the proliferation and survival of the RasV12 cells.
In our model, functional autophagy is required to restrain the growth of Ras-activated epithelial tissue growth in a clonal or whole-tissue setting, as well as in combination with impaired cell polarity. This is in contrast with a previous report where autophagy depletion was reported to reduce the growth of Ras-activated Drosophila tissues, alone or in combination with scrib loss of function.65 One factor that might explain the difference between the two studies is the different promoters used to drive the Ras oncogene, and the overall number of UAS constructs in the system, raising questions about levels of Ras expression in our study and theirs. With Ras levels possibly determining the differential output of autophagy,26 this observation could be instrumental for further investigating the interplay between the two pathways.
In our analysis, we observed cross-talk between the autophagy and Ras signalling pathways: activating Ras-induced autophagy, and blocking autophagy in this context further increased ERK activity (Figures 7 and 9). By its activation of the PI3K/AKT/mTOR pathway, Ras is predicted to inhibit autophagy.66 However, there is increasing evidence that Ras positively regulates autophagy, and higher levels of autophagy are observed in Ras-expressing tumour cells.24, 58, 67, 68 Furthermore, the proto-oncogene Myc, a downstream activator of Ras signalling, also induces autophagy and tissue growth in the Drosophila wing disc.56 We show here that positive regulation of autophagy by Ras is conserved in Drosophila. Conversely, autophagy regulates the Ras signalling pathway. For instance, it has been suggested that autophagosomes can act as intracellular scaffolds for players of the MAPK cascade, and that members of the LC3 family, including GABARAP, colocalise with phospho-ERK at the surface of autophagosomes.61 This direct regulation of intracellular signalling by autophagy is not unique: it has been shown in a lung cancer cell line that autophagy specifically promotes RHOA degradation and therefore downregulates RHOA signalling.69 Thus, these observations raise the possibility that some of the effects of autophagy inhibition on Ras-driven overgrowth lie in direct regulation of intracellular signalling by the autophagic machinery.
An important insight from our study is our demonstration that ROS accumulation and the induction of the JNK stress response pathway potentiates the tissue growth induced by activated Ras. ROS accumulation upon autophagy inhibition in Ras-driven cancer is usually detrimental to tumour growth, as it leads to DNA damage, chromosomal instability, and apoptosis.70, 71 In line with these observations, increasing ROS levels has been used as a way to trigger apoptosis in pancreatic cancer cells,71 and strong induction of ROS and JNK activity also triggers apoptosis in Drosophila models of chromosomal instability.72 In our model however, the apoptosis-inducing effects of ROS and JNK are prevented by the cell survival properties of the Ras oncogene, thereby repurposing the action of JNK towards blocking differentiation, pupariation and promoting an invasive phenotype.9, 11, 73
The interplay between autophagy, ROS and JNK has been evidenced in several mammalian studies: in response to oncogenic RAS, JNK is critical for Atg5 and Atg7 upregulation and autophagy induction,62 63 and treatment with antioxidants is sufficient to abolish RAS-induced JNK activation.62 We show that lowering oxidative stress levels by overexpressing SOD1 is able to rescue overgrowth of autophagy-impaired, Ras-activated tissues, to a greater extent than blocking JNK. This suggests that there might be JNK-independent pathways that are activated by ROS that contribute to tissue overgrowth. Indeed, other studies have shown that the p38 MAPK stress response pathway is also activated upon oxidative stress,74 and ROS-induced JNK and p38 signalling synergise to promote tissue regeneration in Drosophila.75
One issue that we are yet to resolve is the origin of ROS upon autophagy impairment in Drosophila RasV12-expressing tissues. Interestingly, in mammalian cells, p62 accumulation promotes tumourigenesis via upregulation of oxidative stress, and removal of p62 by autophagy is sufficient to reduce tumour burden.76 Thus, the strong accumulation of Ref(2)P/p62 we see upon autophagy inhibition in the Ras-activated background raises the possibility that a similar p62-dependent mechanism might be at work in our Drosophila model. On the other hand, oncogenic Ras-driven transformation is associated with upregulation of metabolic pathways and increased ROS production due to the accumulation of dysfunctional mitochondria,26, 77 a situation that would be exacerbated by blocking autophagy. Supporting this model, ROS-induced JNK activation and non-cell-autonomous induction of proliferation has been documented in mosaic Drosophila tissues expressing oncogenic Ras and inactivating mutations in mitochondrial genes.14
In our system, we also observed non-cell-autonomous effects on wild-type tissues surrounding RasV12-autophagy-impaired mutant cells, including the induction of autophagy, increased active caspase, cell death and cell proliferation. Since blocking autophagy in RasV12-expressing cells markedly augments ROS levels, ROS could be leading to these effects on the surrounding wild-type tissue by diffusion (Figure 9). In parallel, the upregulation of ROS and JNK could lead to the induction of paracrine factors, such as Dpp (TGFβ), Wingless (WNT) or Upd (IL6), that affect the surrounding tissue75, 78 (Figure 9). Similar non-cell-autonomous effects of autophagy impairment have been observed in mammalian systems, where blocking autophagy results in an inflammatory microenvironment.79, 80 It is possible that this ROS-mediated inflammatory signalling also participates in the non-cell-autonomous growth we observe in our model. In line with this hypothesis, a new study has highlighted the importance of extracellular ROS in inducing apoptosis-induced proliferation via the recruitment of macrophage-like cells (haemocytes) to regenerating Drosophila tissues,81 raising the possibility that similar cross-talk between immune cells and the epithelium drives overgrowth of Ras-activated tissues upon autophagy inhibition.
During the course of this work, a study by Katheder et al.82 highlighted the non-cell-autonomous contribution of autophagy to RasV12, scrib−/− tumour growth. In mosaic tissues, autophagy is induced in the wild-type cells surrounding RasV12 clones, and is further enhanced in the neighbouring cells of RasV12, scrib−/− clones to sustain their proliferation, suggesting that non-cell-autonomous regulation of autophagy and proliferative/apoptotic mechanisms in tissues expressing a constitutively active form of the Ras oncogene, or in Ras-cooperative tumourigenesis settings, is a general mechanism.
In summary, our Drosophila model of Ras-driven cooperative tumourigenesis has elucidated that ROS and JNK activation are critical for synergistic tissue growth. Given the effect of autophagy inhibition on ROS levels upon Ras activation, and the active development of autophagy blockers as cancer therapeutic agents, it is now crucial to identify situations in which inhibiting this pathway could lead to enhanced tumourigenesis rather than block progression of tumours. Indeed, recent studies are revealing that the protective effect of autophagy on ROS levels is sufficient to suppress tumour progression and metastasis.83 Therefore, understanding the complex interplay between Ras, ROS and autophagy is crucial to enable the development of new therapeutic protocols targeting the autophagy pathway in cancer patients. Our data indicate that Drosophila will be a powerful model by which to decipher this interplay.
Materials and methods
Fly genetics
Drosophila stocks used:
y,w,ey-FLP1;act>CD2>GAL4,UAS-GFP (eyeless-FLP-out, actin-GAL4 system)
ey-GAL4,UAS-Ras V12 /CyO
y,w,ey-FLP1,UAS-mCD8-GFP;tub-GAL4,FRT82B,tub-GAL80/TM6B,Tb (MARCM3R)
ey-FLP1;UAS-dlg RNAi ,UAS-Ras V12 ;act>CD2>GAL4,UAS-GFP
ey-GAL4,UAS-Ras V12 /CyO,tub-GAL80;UAS-SOD1
ey-GAL4,UAS-Ras V12 /CyO,tub-GAL80;UAS-bsk DN
ey-GAL4,UAS-Ras V12 /CyO,tub-GAL80;UAS-GFP
pmCherry-Atg8a;dpp blk -GAL4,UAS-GFP/SM6a-TM6B,Tb
UAS-GFP- ref(2 )P;dpp blk -GAL4,UAS-His2Av:mRFP/SM6a-TM6B,Tb
UAS-Ras V12 ,UAS-Atg8a-IR KK109654
UAS-Ras V12 ,UAS-Atg1-IR GD16133
UAS-Ras V12 ;UAS-Atg9-IR GD10045
UAS-Ras V12 ,UAS-GFP
UAS-Ras V12 ,UAS-myr-mRFP
UAS-SOD1 (BL33606, kindly provided by S Gregory)
UAS-bsk DN
UAS-lacZ
msn-lacZ,FRT82B (P(PZ)msn 06948 )
pmCherry-Atg8a (a gift from D Denton)
UAS-GFP- ref(2 )P (a gift from T Neufeld)
In all stocks, UAS-RasV12 refers to UAS-dRas85DV12 on the second chromosome.
RNAi lines
All stocks and experimental crosses were performed on molasses and yeast based food, in 25 °C or 29 °C incubators. In all experiments involving dissection of larvae, flies were left laying for 8 to 16 h, and wandering L3 larvae were collected at day 5 AEL) (when raised at 25 °C), or day 4 AEL (when raised at 29 °C).
Pupariation assays
In ey-FLP-out, Act»GAL4 pupariation assays, late second to early third instar larvae of correct genotypes were collected at day 4 AEL, counted and transferred (50 individuals) into fresh vials of food. Pupae were counted every 24 h after sorting, from day 5 AEL to day 9 AEL. Hatched adults were counted from day 10 AEL to day 14 AEL. Results were plotted in GraphPad Prism (GraphPad Software, La Jolla CA, USA) with 95% confidence intervals. Pupariation assays were performed two times.
Western blot analysis
Twenty to 40 pairs of eye/antennal or wing imaginal discs were dissected in cold phosphate-buffered saline (PBS) and homogenised in RIPA. Six micrograms of proteins were separated by electrophoresis on a 4–12% Bis-Tris gel (Thermo Fisher, Waltham, MA, USA) and blotted onto a PVDF membrane. Blocking and antibody incubations were performed in TBS+0.1% Tween-20 (TBST)+5% skimmed milk. Three biological repeats were performed for the western blot experiment in Figure 7c (quantified in Supplementary Figure 6). Western blot in Figure 6e was performed once.
Immunofluorescence analysis
Wandering third instar larvae were dissected in cold PBS and inverted cuticles were fixed in 4% paraformaldehyde. Blocking and antibody incubations were performed in PBS+0.3% Triton X-100+2% goat serum. Primary incubations were performed overnight at 4 °C, protected from light on a rotating wheel. Secondary incubations were performed for 2 h at room temperature (RT), protected from light on a nutator platform. All washes were performed in PBS+0.3% Triton X-100. Samples were then immersed in 80% glycerol: 20% PBS, and mounted
Antibodies
Primary antibodies used were as follows: rabbit polyclonal anti-ref(2)p (1:10 000 (WB), 1:2000 (IF); a gift from G Juhász), rabbit polyclonal anti-Atg8a (1:5000 (WB); a gift from T Neufeld), mouse anti-MMP1 (1:100 (WB); Developmental Studies Hybridoma Bank, University of Iowa, IA, USA), mouse monoclonal anti-Elav (1:200; Developmental Studies Hybridoma Bank), rabbit polyclonal anti-β-galactosidase (1:200; Molecular Probes, Eugene, OR, USA), rabbit polyclonal anti-Dcp1 (1:200; Cell Signaling, Danvers, MA, USA; a kind gift of L Cheng), mouse monoclonal anti-activated ERK1/2 (phospho-ERK 1:5000; Sigma, St Louis, MO, USA), rabbit polyclonal anti-ERK1/2 (total-ERK 1:5000; Cell Signaling), mouse monoclonal anti-α-tubulin (1:10 000; Sigma), rabbit polyclonal anti-mCherry (1:6000; Kerafast, Boston, MA, USA; EMU109). The secondary antibodies used were as follows: goat anti-rabbit:HRP (1:5000), goat anti-mouse:HRP (1:15 000), goat anti-rabbit:Alexa-647(1:500; Molecular Probes) and goat anti-mouse:Alexa-568 (1:500; Molecular Probes). Dcp1 staining experiments were conducted once, mCherry-Atg8a detection experiments was performed more than five times and GFP-p62 detection experiments were performed three times.
EdU incorporation assay
EdU incorporation assay (Life Technologies, Thermo Fisher) was performed as follows. Inverted L3 larvae were incubated for 20 min in PBS+50 nm EdU (check) at RT. Tissue was fixed in 4% paraformaldehyde for 20 min at RT, washed 3x times with PBS+0.5% Triton X-100, incubated with antibody against GFP and washed and incubated with secondary antibodies, before EdU revelation using Click-iT protocol (Life Technologies, Thermo Fisher). EdU assays were carried out two times.
TUNEL assay
TUNEL assay was performed using the Click-iT Plus TUNEL assay (Molecular Probes; cat. no. C10619) following guidelines for cells with the following differences: permeabilization of tissues was performed overnight in PBS+0.3% Triton X-100, TdT reaction was performed by incubating overnight followed by 1.5 h at 37 °C, and incubation with Click-iT reagent was performed for 45 min at 37 °C. TUNEL experiments were performed two times.
Detection of ROS
Detection of ROS was performed in imaginal discs using CellROX Deep Red (Life Technologies, Thermo Fisher), following the protocol published previously.72 Briefly, eye-antennal imaginal discs were dissected in Shields and Sang M3 medium at RT, and transferred into a 1.5 ml Eppendorf tube. Dissection medium was removed and replaced by a freshly made solution of 5 μm CellROX Deep Red in M3 medium at RT. Samples were incubated for 8 min on a nutator, protected from light. CellROX was removed and samples were washed quickly two times in RT PBS, and briefly fixed with 4% paraformaldehyde for 4 min on a nutator, protected from light. Paraformaldehyde was removed and samples were washed quickly two times in RT PBS. Discs were immersed in 80% glycerol in PBS, mounted on SuperFrost microscope slides with coverslips and imaged within 30 min of mounting. For detection of CellROX, acquisition parameters were set up on RasV12 Atg8aRNAi samples, as they accumulated the most dye, and the same parameters were used on to image other genotypes. ROS assays were carried out three times.
Confocal microscopy
Confocal images were taken on a Nikon C2 confocal microscope (Nikon, Minato, Tokyo, Japan) or Zeiss LSM 780 Laser Scanning Microscope (Zeiss, Oberkochen, Germany).
Quantification of immunofluorescence data
For quantification of GFP clones in Figure 4, image stacks were imported into Imaris 7.4 (BIOT). GFP and DAPI volumes were calculated for at least eight discs of each genotype, using the same parameters across samples and genotypes. The ratio of GFP/DAPI was calculated and averaged within each genotype and statistical significance was assessed by one-way analysis of variance (ANOVA) with multiple comparisons in GraphPad Prism (GraphPad Software), which was also used to generate graphical results. To control for dosage effects of the UAS/GAL4 system, we used a UAS-RasV12,UAS-mRFP stock as a control for UAS-RasV12,UAS-Atg1RNAi and UAS-RasV12,UAS-Atg8aRNAi lines. This allowed us to directly compare GFP levels between RasV12,mRFP, RasV12,Atg1RNAi and RasV12,Atg8aRNAi samples. Summary of UAS constructs in MARCM crosses: control FRT=1xUAS (UAS-CD8-GFP); Atg1RNAi and Atg8aRNAi=2xUAS (UAS-CD8-GFP and UAS-RNAi); RasV12,mRFP, RasV12,Atg1RNAi, RasV12,Atg8aRNAi=3xUAS (UAS-CD8-GFP, UAS-RasV12 and UAS-mRFP or UAS-RNAi).
For quantification of EdU and TUNEL, positive cells anterior to the MF were counted in at least four eye-antennal discs.
For quantification of βgal and Elav staining (Figure 7a and Supplementary Figure 5), fluorescence intensity was measured in Photoshop in a 30px diameter area within a GFP clone, and in a 30px area in the wild-type tissue neighbouring the measured clone. Three to six individual discs, and at least six clones per disc, were measured.
For mCherry-Atg8a quantification (Figure 6b), at least three discs of each genotype were imaged and maximum intensity projections were generated. In Photoshop (Adobe Systems Inc., San Jose, CA, USA), a mask was applied over the GFP channel to highlight the region of the dppblk-GAL4 driver expression (Dpp domain). mCherry intensity was measured in an area of 30x30 px2 within the Dpp domain or within the wild-type domain of the same disc, measurements were exported and intensity ratios were (Dpp/wt) were calculated.
For GPF-ref(2)P quantification at x20 (Figure 6e), mean intensity was measured in single slices throughout the Dpp domain. For quantification of punctae (Supplementary Figures 4b and c), single images were acquired with the x40 objective and a 2x zoom in the middle of the wing pouch. Images were processed in Image J (Bethesda, MD, USA) as follows: first, same threshold was applied on the GFP channel to generate binary images, then a watershed filter was used to separate juxtaposed punctae, and finally, particles were counted in a 80 × 80 px2 box within the Dpp domain using the analyse particles plugin. At least eight independent discs were measured per RNAi line.
All measurements were exported and plotted into GraphPad Prism (GraphPad Software), and statistical significance was calculated by one-way ANOVA with multiple comparisons with Tukey's multiple correction test. All statistical tests were two-sided, and the variance is presented as s.e.m.
Electron microscopy
Adult flies were progressively dehydrated in concentrations of ethanol ranging from 25 to 100% over the course of 4 days on a nutator. Flies were then desiccated by critical point drying in a Leica critical point dryer (Wetzlar, Germany), mounted on steel stubs and coated with 20 nm of gold particles. Representative images of each phenotype were taken on a JEOL JCM-6000 NeoScope scanning electron microscope (Akishima, Tokyo, Japan) at x80 and x300 magnifications. Images were cropped and aligned in Adobe Photoshop.
Assessment of adult eye overgrowth
ey-GAL4, UAS-RasV12/CyO (ey>RasV12) female flies were crossed to UAS-AtgRNAi males or UAS-lacZ males for control. Crosses were raised at 29 °C for 11 days before scoring. Enhancement of Ras-driven overgrowth was assessed by eye in at least 20 F1 ey-GAL4, UAS-RasV12/UAS-AtgRNAi females by comparing with ey-GAL4, UAS-RasV12/UAS-lacZ females.
Human data analysis
Human orthologues of Drosophila autophagy genes (Atg8a, Atg9, Atg7, Syx17, Vamp7, Snap29, nSyb) were defined from the DIOPT v.5.384 reported at FlyBase. We retrieved Cancer Genome Atlas patient samples of RNAseq, KRAS mutation profiles and overall survival data (version 2016_01_28) from the BROAD GDAC database (https://gdac.broadinstitute.org/) for PAAD. The mutational profiles (variant calls) were based on the MutSig algorithm85 for PAAD (TP.MutSigNozzleReport2.0.Level_4.2016012800).
For the survival analysis we considered only patient samples that were present in the MutSig report ‘TP.final_analysis_set.maf’ and had clinical overall survival information (last day follow up and status). We retrieved normalised illumina hiseq rnaseqv2 RSEM Level 3 data, the Clinical Level_1 for PAAD. For the analysis only primary solid tumour samples were considered.
For PAAD 132/177 (74%) RNAseq patient samples with KRAS mutations were available. We categorised the patient samples in ‘G12’, ‘other’ for samples with KRAS mutations assigned and ‘none’ for the remaining RNAseq samples.
For the survival analysis based on individual autophagy genes, we split patient samples for each gene into a low expression and a high expression group based on multiple cutoff of gene expression percentiles ranging from 0.2 to 0.8. For the best split cutoff that was estimated based on the lowest P-value of a log-rank test, we tested for the enrichment and for the underrepresentation of KRAS-G12 mutations in the low group compared with the high group using a one-sided Fisher’s exact test. For the enrichment and underrepresentation test for G12 mutations, the category ‘none’ and ‘other’ were aggregated.
For the survival analysis that considered multiple autophagy genes jointly, we performed a log-rank test between two patient groups that were defined based on a k-means clustering. The patient cohort with a larger average RNAseq expression was denoted as ‘high’ and the patient cohort with lower average expression as ‘low’. The k-means clustering procedure was repeated 100 times on all samples. The patient samples were designated either to the low or high expression cohort based on their most frequently assigned group. For the clustering procedure, we log_e(1+x) transformed the normalised RSEM gene RNAseq samples and scaled the data (z-transformed and centred) for each gene. For the heatmap visualisations, we ranged the data to values between 0 and 1 by a normal probability distribution function.
The processing and analysis of the data was performed in R. For the survival analysis, we used the survival R package84 and survplot function (http://www.cbs.dtu.dk/~eklund/survplot/). The heatmap visualisations were performed with the Complex Heatmap Bioconductor package.
Acknowledgments
We thank Donna Denton, Sharad Kumar, Louise Cheng, Tor Erik Rusten, Lee Jun, Gabor Juhasz, Tom Neufeld, Stephen Gregory and Zeeshan Shaukat for reagents, and Tor Erik Rusten, Andreas Bergmann, Donna Denton, Sharad Kumar and Stephen Gregory for discussions. We thank Edwina McGlinn for critical reading of the manuscript. We are grateful to the Peter MacCallum Microscopy core and the LIMS Bioimaging facility for their help with imaging and analysis. We are grateful to OzDros and to Peter Burke for quarantine, shipment and maintenance of Drosophila stocks. The project was supported by grants from the National Health and Medical Research Council (NHMRC, no. 1020525) and from the PeterMac Foundation to HER, JMP, KJS and POH, and funds from LIMS and La Trobe University. JM was supported by an NHMRC grant (no. 1020525) and LIMS and La Trobe University, SB by a Australian Postgraduate Research Award and TZ by a Marie Curie Excellence grant. HER and POH are supported by fellowships of the (NHMRC) and funds from LIMS and La Trobe University.
Author contributions
JM, HER, POH, TZ and JP designed experiments. JM, SB, RDMS and TZ performed research. JM, POH and HER wrote the manuscript. KJS, POH, CM, JP and HER supervised the project.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer 2011; 11: 761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer 2003; 3: 459–465. [DOI] [PubMed] [Google Scholar]
- Fernández-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes Cancer 2011; 2: 344–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ARJ, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JFJ et al. Autophagy mediates the mitotic senescence transition. Genes Dev 2009; 23: 798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco E, Spinelli M, Vanoni M. Approaches to Ras signaling modulation and treatment of Ras-dependent disorders: a patent review (2007–present). Expert Opin Ther Pat 2012; 22: 1263–1287. [DOI] [PubMed] [Google Scholar]
- Dimauro T, David G. Ras-induced senescence and its physiological relevance in cancer. Curr Cancer Drug Targets 2010; 10: 869–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman ML, Marshall CJ, Olson MF. RAS and RHO GTPases in G1-phase cell-cycle regulation. Nat Rev Mol Cell Biol 2004; 5: 355–366. [DOI] [PubMed] [Google Scholar]
- Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet 2012; 44: 1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumby AM, Richardson HE. Using Drosophila melanogaster to map human cancer pathways. Nat Rev Cancer 2005; 5: 626–639. [DOI] [PubMed] [Google Scholar]
- Gonzalez C. Drosophila melanogaster: a model and a tool to investigate malignancy and identify new therapeutics. Nat Rev Cancer 2013; 13: 172–183. [DOI] [PubMed] [Google Scholar]
- Cheng L, Parsons LM, Richardson HE. Modelling Cancer in Drosophila—The Next Generation (Version 20). Encyclopedia Life Sciences (eLS Wiley): West Sussex, UK, 2013. [Google Scholar]
- Brumby AM, Richardson HE. Scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. EMBO J 2003; 22: 5769–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini RA, Xu T. A genetic screen in Drosophila for metastatic behavior. Science 2003; 302: 1227–1231. [DOI] [PubMed] [Google Scholar]
- Ohsawa S, Sato Y, Enomoto M, Nakamura M, Betsumiya A, Igaki T. Mitochondrial defect drives non-autonomous tumour progression through Hippo signalling in Drosophila. Nature 2012; 490: 547–551. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Ohsawa S, Igaki T. Mitochondrial defects trigger proliferation of neighbouring cells via a senescence-associated secretory phenotype in Drosophila. Nat Commun 2014; 5: 5264. [DOI] [PubMed] [Google Scholar]
- Murrow L, Debnath J. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu Rev Pathol 2013; 8: 105–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, White E. Autophagy in tumorigenesis and energy metabolism: friend by day, foe by night. Curr Opin Genet Dev 2011; 21: 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH et al. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr Biol 2009; 19: 1741–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005; 122: 927–939. [DOI] [PubMed] [Google Scholar]
- Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal 2014; 20: 460–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer 2012; 12: 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ávalos Y, Canales J, Bravo-Sagua R, Criollo A, Lavandero S, Quest AFG. Tumor suppression and promotion by autophagy. Biomed Res Int 2014; 2014: 603980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F et al. Autophagy in malignant transformation and cancer progression. EMBO J 2015; 34: 856–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Chen H-Y, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev 2011; 25: 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Wei S, Gan B, Peng X, Zou W, Guan J-L. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev 2011; 25: 1510–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellot GL, Liu D, Pervaiz S. ROS, autophagy, mitochondria and cancer: Ras, the hidden master? Mitochondrion 2013; 13: 155–162. [DOI] [PubMed] [Google Scholar]
- Schmukler E, Kloog Y, Pinkas-Kramarski R. Ras and autophagy in cancer development and therapy. Oncotarget 2014; 5: 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. Exploiting the bad eating habits of Ras-driven cancers. Genes Dev 2013; 27: 2065–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang XD, Lapi E, Sullivan A, Jia W, He Y-W et al. Autophagic activity dictates the cellular response to oncogenic RAS. Proc Natl Acad Sci USA 2012; 109: 13325–13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfanelli V, Fuoco C, Lorente M, Salazar M, Quondamatteo F, Gherardini PF et al. AMBRA1 links autophagy to cell proliferation and tumorigenesis by promoting c-Myc dephosphorylation and degradation. Nat Cell Biol 2015; 17: 20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Tortola L, Perlot T, Wirnsberger G, Novatchkova M, Nitsch R et al. A dual role for autophagy in a murine model of lung cancer. Nat Commun 2014; 5: 3056. [DOI] [PubMed] [Google Scholar]
- Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev 2011; 25: 795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menut L, Vaccari T, Dionne H, Hill J, Wu G, Bilder D. A mosaic genetic screen for Drosophila neoplastic tumor suppressor genes based on defective pupation. Genetics 2007; 177: 1667–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombani J, Andersen DS, Leopold P. Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science 2012; 336: 582–585. [DOI] [PubMed] [Google Scholar]
- Leong GR, Goulding KR, Amin N, Richardson HE, Brumby AM. Scribble mutants promote aPKC and JNK-dependent epithelial neoplasia independently of Crumbs. BMC Biol 2009; 7: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumby AM, Goulding KR, Schlosser T, Loi S, Galea R, Khoo P et al. Identification of novel Ras-cooperating oncogenes in Drosophila melanogaster: a RhoGEF/Rho-family/JNK pathway is a central driver of tumorigenesis. Genetics 2011; 188: 105–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo P, Allan K, Willoughby L, Brumby AM, Richardson HE. In Drosophila, RhoGEF2 cooperates with activated Ras in tumorigenesis through a pathway involving Rho1-Rok-Myosin-II and JNK signalling. Dis Model Mech 2013; 6: 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim FD, Chang HC, Therrien M, Wassarman DA, Laverty T, Rubin GM. A screen for genes that function downstream of Ras1 during Drosophila eye development. Genetics 1996; 143: 315–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruharsha KG, Rual JF, Zhai B, Mintseris J, Vaidya P, Vaidya N et al. A protein complex network of Drosophila melanogaster. Cell 2011; 147: 690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takats S, Nagy P, Varga A, Pircs K, Karpati M, Varga K et al. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J Cell Biol 2013; 201: 531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli E, Ginefra P, Mastrodonato V, Beznoussenko GV, Rusten TE, Bilder D et al. Multiple functions of the SNARE protein Snap29 in autophagy, endocytic, and exocytic trafficking during epithelial formation in Drosophila. Autophagy 2014; 10: 2251–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doggett K, Grusche FA, Richardson HE, Brumby AM. Loss of the Drosophila cell polarity regulator Scribbled promotes epithelial tissue overgrowth and cooperation with oncogenic Ras-Raf through impaired Hippo pathway signaling. BMC. Dev Biol 2011; 11: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willecke M, Toggweiler J, Basler K. Loss of PI3K blocks cell-cycle progression in a Drosophila tumor model. Oncogene 2011; 30: 4067–4074. [DOI] [PubMed] [Google Scholar]
- Fuja TJ, Lin F, Osann KE, Bryant PJ. Somatic mutations and altered expression of the candidate tumor suppressors CSNK1 epsilon, DLG1, and EDD/hHYD in mammary ductal carcinoma. Cancer Res 2004; 64: 942–951. [DOI] [PubMed] [Google Scholar]
- Liu Z, Niu Y, Xie M, Bu Y, Yao Z, Gao C. Gene expression profiling analysis reveals that DLG3 is down-regulated in glioblastoma. J Neurooncol 2014; 116: 465–476. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Guo Y, Liu Y, Zhang F, Wang Y, Shen B et al. Methylation-mediated silencing of Dlg5 facilitates bladder cancer metastasis. Exp Cell Res 2015; 331: 399–407. [DOI] [PubMed] [Google Scholar]
- Tomiyama L, Sezaki T, Matsuo M, Ueda K, Kioka N. Loss of Dlg5 expression promotes the migration and invasion of prostate cancer cells via Girdin phosphorylation. Oncogene 2015; 34: 1141–1149. [DOI] [PubMed] [Google Scholar]
- Li M, Lindblad JL, Perez E, Bergmann A, Fan Y. Autophagy-independent function of Atg1 for apoptosis-induced compensatory proliferation. BMC Biol 2016; 14: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci 2001; 24: 251–254. [DOI] [PubMed] [Google Scholar]
- Karim FD, Rubin GM. Ectopic expression of activated Ras1 induces hyperplastic growth and increased cell death in Drosophila imaginal tissues. Development 1998; 125: 1–9. [DOI] [PubMed] [Google Scholar]
- Halfar K, Rommel C, Stocker H, Hafen E. Ras controls growth, survival and differentiation in the Drosophila eye by different thresholds of MAP kinase activity. Development 2001; 128: 1687–1696. [DOI] [PubMed] [Google Scholar]
- Denton D, Chang T-K, Nicolson S, Shravage B, Simin R, Baehrecke EH et al. Relationship between growth arrest and autophagy in midgut programmed cell death in Drosophila. Cell Death Differ 2012; 19: 1299–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvezin C, Ayala C, Braden CR, Kim J, Neufeld TP. Assays to monitor autophagy in Drosophila. Methods 2014; 68: 134–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM et al. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol 2008; 181: 655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell F, Wang S, Katheder N, Rusten TE, Samakovlis C. Two-tiered control of epithelial growth and autophagy by the insulin receptor and the ret-like receptor, stitcher. PLoS Biol 2013; 11: e1001612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P, Varga A, Pircs K, Hegedus K, Juhász G. Myc-driven overgrowth requires unfolded protein response-mediated induction of autophagy and antioxidant responses in Drosophila melanogaster. PLoS Genet 2013; 9: e1003664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J, Kang Y, Zhao R, Xia Q, Lee DF, Chang Z et al. KrasG12D-induced IKK2/beta/NF-kappaB activation by IL-1alpha and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell 2012; 21: 105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargini R, Garcia-Escudero V, Izquierdo M, Wandosell F. Oncogene-mediated tumor transformation sensitizes cells to autophagy induction. Oncol Rep 2016; 35: 3689–3695. [DOI] [PubMed] [Google Scholar]
- Galko MJ, Krasnow MA. Cellular and genetic analysis of wound healing in Drosophila larvae. PLoS Biol 2004; 2: E239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MJ, Gamez G, Menke C, Hernandez A, Thorburn J, Gidan F et al. Regulation of autophagy and chloroquine sensitivity by oncogenic RAS in vitro is context-dependent. Autophagy 2014; 10: 1814–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lopez N, Athonvarangkul D, Mishall P, Sahu S, Singh R. Autophagy proteins regulate ERK phosphorylation. Nat Commun 2013; 4: 2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M-J, Woo S-J, Yoon C-H, Lee J-S, An S, Choi Y-H et al. Involvement of autophagy in oncogenic K-Ras-induced malignant cell transformation. J Biol Chem 2011; 286: 12924–12932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun JY, Yoon CH, An S, Park IC, Kang CM, Kim MJ et al. The Rac1/MKK7/JNK pathway signals upregulation of Atg5 and subsequent autophagic cell death in response to oncogenic Ras. Carcinogenesis 2009; 30: 1880–1888. [DOI] [PubMed] [Google Scholar]
- Wu H, Wang MC, Bohmann D. JNK protects Drosophila from oxidative stress by transcriptionally activating autophagy. Mech Dev 2009; 126: 624–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez E, Das G, Bergmann A, Baehrecke EH. Autophagy regulates tissue overgrowth in a context-dependent manner. Oncogene 2015; 34: 3369–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta S, Hidaka E, Ogata A, Yokota S, Kamata T. Ras is involved in the negative control of autophagy through the class I PI3-kinase. Oncogene 2004; 23: 3898–3904. [DOI] [PubMed] [Google Scholar]
- Alves S, Castro L, Fernandes MS, Francisco R, Castro P, Priault M et al. Colorectal cancer-related mutant KRAS alleles function as positive regulators of autophagy. Oncotarget 2015; 6: 30787–30802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell 2011; 22: 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaid A, Cerezo M, Chargui A, Corcelle-Termeau E, Pedeutour F, Giuliano S et al. Autophagy plays a critical role in the degradation of active RHOA, the control of cell cytokinesis, and genomic stability. Cancer Res 2013; 73: 4311–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev 2007; 21: 1367–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev 2011; 25: 717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaukat Z, Liu D, Choo A, Hussain R, O'Keefe L, Richards R et al. Chromosomal instability causes sensitivity to metabolic stress. Oncogene 2015; 34: 4044–4055. [DOI] [PubMed] [Google Scholar]
- Doggett K, Turkel N, Willoughby LF, Ellul J, Murray MJ, Richardson HE et al. BTB-zinc finger oncogenes are required for Ras and Notch-driven tumorigenesis in Drosophila. PLoS One 2015; 10: e0132987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CH, Iskandar KB, Yadav SK, Hirpara JL, Loh T, Pervaiz S. Simultaneous induction of non-canonical autophagy and apoptosis in cancer cells by ROS-dependent ERK and JNK activation. PLoS ONE 2010; 5: e9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santabarbara-Ruiz P, Lopez-Santillan M, Martinez-Rodriguez I, Binagui-Casas A, Perez L, Milan M et al. ROS-induced JNK and p38 signaling is required for unpaired cytokine activation during Drosophila regeneration. PLoS Genet 2015; 11: e1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen H-Y et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009; 137: 1062–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Lu W, Chen G, Wang P, Chen Z, Zhou Y et al. K-ras(G12V) transformation leads to mitochondrial dysfunction and a metabolic switch from oxidative phosphorylation to glycolysis. Cell Res 2012; 22: 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TT. Non-autonomous consequences of cell death and other perks of being metazoan. AIMS Genet 2015; 2: 54–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006; 10: 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 2008; 456: 259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty CE, Diwanji N, Lindblad JL, Tare M, Amcheslavsky A, Makhijani K et al. Extracellular reactive oxygen species drive apoptosis-induced proliferation via Drosophila macrophages. Curr Biol 2016; 26: 575–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katheder NS, Khezri R, O'Farrell F, Schultz SW, Jain A, Rahman MM et al. Microenvironmental autophagy promotes tumour growth. Nature 2017; 541: 417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Li C, Zheng W, Guo Q, Zhang Y, Kang M et al. Inhibition of autophagy promotes metastasis and glycolysis by inducing ROS in gastric cancer cells. Oncotarget 2015; 6: 39839–39854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Flockhart I, Vinayagam A, Bergwitz C, Berger B, Perrimon N et al. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics 2011; 12: 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013; 499: 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.