Abstract
Endogenous retroviruses (ERVs) are involved in placentation; perhaps, the most well-known ERVs are the syncytins, actively transcribed env genes involved in cell–cell fusion and possible morphological variations. However, ERVs other than syncytins that play an important role in placental development have not been well characterized. To identify ERV genes expressed during the onset of placentation in the bovine species, we characterized the expression profiles of bovine conceptus transcripts during the peri-attachment period using RNA-seq analysis, and confirming some candidates through real-time PCR. Using in silico and PCR analyses, we identified a novel ERV proviral sequence derived from a gag region, designated bovine endogenous retroviruses (BERV)-K3, containing Gag_p10 and Gag_p24, zinc finger domain. Initial expression of this ERV in bovine conceptuses was on day 20 (day 0 = day of estrus), soon after conceptus attachment to the endometrial epithelium, and its high placental expression was maintained up to the middle of pregnancy. The BERV-K3 transcript was also found in the uterine luminal and glandular epithelia, liver, kidney, intestine, and skin. BERV-K3 is located on chromosome 7 and integrated within LOC100848658, from which noncoding RNA could be transcribed. Furthermore, the expression of endogenous BERV-K3 in bovine trophoblast cell lines was induced by a WNT agonist, a signaling system common to genes expressed in placentas. These data support the argument that during the evolutionary process, mammals incorporated not only similar ERV sequences, but also ERVs unique to individual species. BERV-K3 is in the latter case, likely providing functions unique to ruminant gestation.
Keywords: bovine, endogenous retrovirus, placentation
Introduction
The endogenous retroviruses (ERVs) are thought to be derived from ancient viral infections of germ cells or their precursors with a retrovirus in mammals and other vertebrates [1]. Indeed, sequences of retroviral origins make up 8 % of the human genome, 10% of the mouse genome, and 18% of the bovine genome [2–4]. Most ERVs have been inactivated by insertions, deletions, and substitutions of nucleotides, and/or epigenetic modifications such as DNA methylation and histone modifications. However, a few open-reading frames (ORFs) of ERV sequences remain intact and are expressed as virus-derived proteins in the host cells [5].
The placenta is remarkably distinct among mammalian species, suggesting a history of rapid evolutionary diversification, resulting from the genes acquired in individual species. It has become apparent that ERV genes play an important role in the development of the placenta and the trophoblast cell lineage in mammalian species, and that during evolution different species may have utilized ERVs of the same as well as different origins. Indeed, different env genes, syncytins, essential for placental morphogenesis have been independently integrated into the genome of humans [6–10], mice [11], rabbits [12], dogs [13], cats [13], sheep [14–18], and cattle [19–23], sheep [21–23], the Rodentia squirrel-related clade [24], Afrotherian tenrecs [25], and marsupials [26]. All identified syncytin env genes exist in different genome sequences and chromosomal locations among species, but the functions such as cell fusion and immune suppression are all shared in mammals. However, the exact evolutionary pathways and the extent to which ERVs function in placental development are still unclear.
It has been determined that the WNT signaling pathway is an important regulator of embryo/conceptus and maternal interaction such as implantation and placental development in mice, sheep, cow, and humans [27–30]. The WNT can induce two downstream signaling cascades, known as the canonical and noncanonical pathways [31]. The canonical WNT pathway is activated when WNT binds to Frizzled (FZD) family receptors and low-density lipoprotein receptor-related protein (LRP) 5/6 coreceptors. This, in turn, leads to cellular accumulation and nuclear translocation of β-catenin (CTNNB), followed by a complex formation with T-cell fate (TCF) to activate transcription of WNT-target genes.
It was shown in mice that one of the Wnt receptors, Fzd5, was necessary for the expression of glial cells missing homolog 1 (Gcm1), an important transcription factor for placental labyrinth development, followed by induction of syncytin-A expression [32]. Matsuura et al. [33] also showed that Wnt/Ctnnb signaling induces Gcm1-syncytin-A, the cell fusion pathway. Although the WNT signal is an important regulator for the expression of ERV during the period of invasive placental formation, whether the WNT signal induces ERV expression in the noninvasive bovine placenta has not been elucidated.
Unlike primate or murine species, conceptus attachment to the uterine endometrial epithelium and subsequent placentation in most ruminants do not occur soon after blastocyst formation [34]. In fact, the conceptus spends a prolonged period within the uterine lumen before developing a definitive attachment to the endometrial epithelium and subsequent formation of placental structures [35]. In the bovine species, ERVs such as BERV-K1 [20], bERVE-A [21], and syncytin-Rum1 [22] have been identified and their potential functions studied. It should be noted, however, that these ERVs are all from env regions; ERVs from other regions such as gag or pol, which could also function in ruminants' placental development, have not been identified or characterized. We hypothesized that ERVs of regions other than env would exist and function in the trophectoderm during the period of placental formation and functioning.
We searched for nucleotide structures of ERV origin, which were expressed in bovine conceptuses during the peri-attachment periods (the criteria are shown in Supplementary Figure S1). Using RNA-seq data, we found that one candidate gene with gag/pol, located on chromosome 7, was minimally expressed on day 20 (day 0 = day of estrus), when conceptus implantation to the uterine epithelium is initiated. Its expression was found to be elevated on day 22, when the placental formation is initiated, and its high expression continued until the middle of pregnancy. Expression of endogenous ERV gene in bovine trophoblast cells was induced by a WNT agonist, a common intracellular signaling for genes expressed in placentas.
Materials and methods
Animals and sampling
All animal procedures in the present study were approved by the Committee for Experimental Animals at Zen-noh Embryo Transfer (ET) Center, Hokkaido and the University of Tokyo, Tokyo, Japan. Estrous synchronization, super-ovulation, artificial insemination and ET processes were performed as previously described [36]. Day 7 blastocysts were collected from super-ovulated and artificially inseminated Japanese black cattle. Sixteen blastocysts derived from the super-ovulation were transferred nonsurgically into the uterine horn of eight estrous synchronized Holstein heifers (n = 2 blastocysts per transfer), ipsilateral to the CL on day 7. Elongated conceptuses were then collected nonsurgically by uterine flushing (500 ml PBS) on day 17 (n = 2), 20 (n = 2), or 22 (n = 4) [37], corresponding to the period before conceptus attachment to the uterine epithelium, when conceptus attachment is initiated, or 2 days into conceptus attachment, respectively. Days 17 and 20 conceptuses (n = 4 each day) in the uterine flushing media were obtained by centrifugation at 400×g for 5 min, snap-frozen and transferred to Animal Resource Science Center at the University of Tokyo. The remaining day 22 pregnant heifers (n = 2) were killed and hysterectomized, from which uteri were excised and frozen immediately using the OCT compound (Sakura Finetek, Tokyo, Japan).
Hysterectomy was performed to collect days 45 and 150 pregnant uteri (n = 3 each) from Japanese black cattle at Azabu University Veterinary Teaching Hospital (AUVTH), Kanagawa, Japan and the National Institute of Agrobiological Sciences (NIAS), Ibaraki, Japan, respectively, from which cotyledons and caruncles were dissected, immediately frozen and stored at −80°C until use. At NIAS, various tissues, heart, liver, kidney, intestine, lung, muscle, skin, lymph node, and spleen were harvested from day 150 pregnant Japanese black cattle, and the uteri were removed from three nonpregnant Japanese black cattle. All tissues dissected were frozen immediately and stored at −80°C until use.
RNA extraction from bovine conceptus and whole-genome sequencing by the RNA-seq (SOLiD3) system
RNA extraction from whole conceptus tissues was performed using Isogen (Nippon Gene, Tokyo, Japan) according to the manufacturer's protocol [38]. For a RNA-seq analysis, total RNA was depleted of rRNAs using the RiboMinus Eukaryote Kit (Life Technologies, Carlsbad, CA, U.S.A.). Details of the sequencing procedure using a SOLiD3 sequencer (Life Technologies), and data analysis have been described previously [37]. In brief, high-throughput sequencing libraries were prepared according to the SOLiD3 whole transcriptome library preparation protocol [39]. ERV-derived sequences in the bovine genome (Btau_4.0.55.dna.toplevel.fa) were identified through the use of RetroTector, which was designed to detect and characterize entire or fragmented ERVs in a given genome [40]. Four types of retroviral-like sequences identified in the genome are gag, pro, pol, and env. Processed nucleotide sequences from the RNA-seq for each of the 3 days examined were aligned against the bovine genome. The Applied Biosystems Whole Transcriptome Analysis Pipeline was used to map short reads. During this mapping phase, up to two mismatches were allowed and short reads that aligned to more than 10 locations were removed. In this analysis, we used the reads whose sequence quality scores were 24 or higher, following the standard parameters of the Applied Biosystems Whole Transcriptome Analysis Pipeline [39]. Matching locations were subsequently used to generate counts for identified ERVs and Ensemble-provided coding sequences (Bos_taurus.Btau_4.0.55.dna.toplevel.fa). To evaluate gene expression level independent of variance in gene lengths and the number of reads among samples, the method of reads per kilobase of exon model per million mapped reads (RPKM), a widely recognized quantification measurement, was applied [41]. The RNA-seq data can be downloaded from the DDBJ Sequence Read Archive [42] with accession number DRA000549 [37].
Cell culture
All cells were maintained at 38.5°C in humidified 5% CO2 and cultured up to confluence. Bovine trophoblast BT-1 [43], CT-1 [44], and F3 [45] cells were kindly provided by Prof. K. Hashizume (Iwate University, Japan), Prof. A. Ealy (Virginia Polytech Institute, U.S.A.), and Prof. C. Pfarrer (University of Veterinary Medicine, Hannover, Germany), respectively. BT-1 cells were cultured on plastic plates coated with type I collagen (Nitta Gelatin, Osaka, Japan) in Dulbecco's modified Eagle medium (DMEM)/Ham's F12 medium (F12) (Invitrogen, Carlsbad, CA, U.S.A.) supplemented with 10% (v/v) fetal bovine serum (FBS; JRH Biosciences, Lenexa, KS, U.S.A.) and antibiotic/antimycotic solution (Invitrogen). CT-1 cells were maintained in DMEM (Invitrogen) containing 10% (v/v) FBS (JRH Biosciences) supplemented with 4.5 g/l d-glucose, nonessential amino acids, 2 mM glutamine, 2 mM sodium pyruvate, 55 mM β-mercaptoethanol, and antibiotic/antimycotic solution (Invitrogen). F3 cells were cultured in DMEM/F12 medium supplemented with 10% FBS (JRH Biosciences) and antibiotic/antimycotic solution (Invitrogen). Bovine primary uterine endometrial epithelial cells (EECs) and stromal cells (STRs) were kindly provided by Prof. K. Okuda (Obihiro University, Japan) and were cultured in DMEM/F12 medium (Invitrogen) supplemented with 5% FBS (JRH Biosciences) and antibiotic/antimycotic solution. Bovine intestinal epithelial cells (Bie) were kindly provided by Prof. H. Aso (Tohoku University, Japan) and were maintained in DMEM/F12 (Invitrogen) supplemented with 10% FBS (JRH Biosciences) and antibiotic/antimycotic solution. Bovine ovarian cumulus-granulosa (oCG) cells were obtained from ovarian follicles that had been collected at a local abattoir, and ear-derived fibroblast (EF) cells were obtained from biopsied ear skin of Japanese black bulls (4 months old) [38], both cells were cultured in DMEM (Invitrogen) containing 5% (v/v) FBS (JRH Biosciences) and antibiotic/antimycotic solution. Bovine kidney cell lines (MDBK and CKT-1) and bovine macrophage cells (BoMAC) were cultured in DMEM (Invitrogen) supplemented with 10% v/v FBS (JRH Biosciences) and antibiotic/antimycotic solution. To study the effects of trophoblast attachment to the uterine endometrial epithelial cells, CT-1 cells were cultured without or with a cell culture insert (Falcon, BD Biosciences, Tokyo, Japan), allowing direct CT-1 cell contact to EECs or indirect cell association with EECs, respectively. To further characterize whether any of the candidate ERV genes could be regulated by Wnt signaling, cultured CT-1 or F3 cells were treated with 1 µM Wnt agonist (sc-222416, Santa Cruz Biotechnology, Dallas, TX, U.S.A.) for 24 h.
RNA isolation from bovine tissues and cultured cells
RNA isolation from bovine tissues and cultured cells was performed using the ISOGEN protocol (Nippon Gene), as described previously [38]. Bovine tissues, heart, liver, kidney, intestine, lung, muscle, skin, lymph node, spleen, and uterus were harvested from three Japanese black cattle at NIAS, Ibaraki, Japan. Excised tissues were submerged in RNAlater (Qiagen, Tokyo, Japan) to prevent RNA degradation, and RNA was then extracted from each tissue. RNA was also isolated from bovine cell lines, including trophoblast cell lines (BT-1, CT-1, and F3), EEC, STR, CKT-1, MDBK, Bie, EF, oCG, and BoMAC. Extracted RNAs were then stored at −30°C until use.
PCR analysis
For PCR and real-time PCR analyses of conceptus RNA, isolated RNA (total 0.5 µg) was reverse-transcribed to cDNA using the ReverTra Ace qPCR RT kit (TOYOBO, Osaka, Japan) in a 10 µl reaction volume, and the resulting cDNA (RT template) was stored at 4°C until use. The cDNA reaction mixture was diluted 1 : 10 using DNase-, RNase-free molecular biology grade water. RT template (cDNA) was subjected to PCR or real-time PCR amplification using specific primers (Table 1). PCR-amplified products were separated on 1.5% (w/v) agarose gels after 32 cycles, from which PCR products were subcloned and verified by DNA sequencing. Quantitative PCRs were performed using the SYBR Green kit (Takara Biomedicals, Tokyo, Japan) and the Applied Biosystems thermal cycle system (7900HT, Applied Biosystems, Tokyo, Japan), as previously described [38]. Real-time PCR was performed under the following thermal cycling conditions: 10 min at 95°C, and 40 cycles of 95°C for 10 s followed by 60°C for 30 s. Average cycle threshold (Ct) values for all mRNAs examined were calculated and normalized to Ct values for ACTB mRNA.
Table 1. Primer sets for BERV-K3 and neighboring gene transcripts.
| BERV-K3 (P1 in Figure 1B) |
| For detection of the transcript and preparation of probes for in situ hybridization |
| P1F: 5′-TCGCCCGAAAGCAGGCTAGTGCTAA-3′ |
| P1R: 5′-CAAGGGCGCAGGCTGTTACCTGTTC-3′ |
| For confirming if BERV-K3 is expressed (in Supplementary Figure S3A) |
| P1F: 5′-TCGCCCGAAAGCAGGCTAGTGCTAA-3′ |
| P1R′: 5′-GCAAGGTTCCGTTTTATGG-3′ |
| For 5′-RACE primer (in Supplementary Figure S3B) |
| P1R: 5′-CAAGGGCGCAGGCTGTTACCTGTTC-3′ |
| For isolated predicted full length (in Supplementary Figure S3C) |
| PFF: 5′-ACGGGTAACAAGGAGTCAAAAG-3′ |
| PFR: 5′-CCCTGATGACAAAGTGACCTCC-3′ |
| The primer set to detect LOC100848658 (P2 in Figure 1B) transcript |
| F: 5′- GCGTCTACCCCAAAACCAGA-3′ |
| R: 5′- ACAGAGAAAGGTGGTCAGGG-3′ |
| The primer set to detect TCF7 (NM_001099186.2) |
| F: 5′- CTGTGAGCTGGTTCACCCAT-3′ |
| R: 5′- TCCGCAATGACTTTGGCTCT-3′ |
| The primer set to detect ACTB (NM_173979.3) |
| F: 5′-CTCTTCCAGCCTTCCTTCCT-3′ |
| R: 5′-GGGCAGTGATCTCTTTCTGC-3′ |
F, forward primer; R, reverse primer.
RNA isolation from bovine conceptus tissues and 5′-RACE for the characterization of the 5′-side of a full-length BERV-K3 transcript
Total RNA was extracted from day 22 bovine conceptuses using the RNeasy Mini Kit together with the RNase-free DNase Set (Qiagen). To identify a full-length BERV-K3 transcript, 5′-RACE with the primer (P1R prime, Table 1 and Supplementary Figure S2) was used to synthesize a first-strand cDNA using the SMARTer RACE 5′/3′ kit (Takara Bio, Inc., Shiga, Japan) according to the manufacturer's instructions. PCR was performed using PrimeSTAR Max (Takara) under the following thermal cycling conditions: 5 cycles of 98°C for 10 s followed by 72°C for 90 s, 5 cycles of 98°C for 10 s followed by 68°C for 90 s, 30 cycles of 98°C for 10 s, 55°C for 10 s followed by 72°C for 90 s, and 1 cycle of 72°C for 10 min. The PCR product was separated by agarose gel electrophoresis and then visualized with a UV trans-illuminator, from which PCR products were subcloned and verified by DNA sequencing.
In situ hybridization
Frozen day 22 uteri embedded in OCT compound were sectioned (10 µm), fixed in 10% neutral buffered formalin, and then subjected to hybridization. Various bovine tissues were fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned (5 µm), and mounted on silane-coated slides (Zyagen, San Diego, CA, U.S.A.). Slide sections were blocked with Block Ace at room temperature for 1 h, incubated with DIG-labeled antisense or sense riboprobe, and then prepared using the BERV-K3 fragment (nucleotide 150 bases) in the pGEM-T Easy Vector with T7 and SP6 promoters (Promega, Madison, WI, U.S.A.) and an RNA transcription kit (Toyobo, Tokyo, Japan) according to the manufacturer's protocol. As described previously [46], hybridization was performed in a humidified chamber at 42°C for 18 h, and bound probes were visualized using alkaline phosphatase-conjugated anti-DIG antibody with nitro blue tetrazolium and 5-brome-4-chloro-3-indolyl phosphate (Promega) as substrates.
Statistical analysis
Quantitative data were subjected to one-way analysis of variance using the Statistical Analyses System (SAS Institute, Cary, NC, U.S.A.); the SEMs shown in the figures were derived from this. When a significant effect of day of pregnancy was detected (P < 0.05), the data were analyzed by Dunnett's test. In these analyses, day of pregnancy was considered an independent source of variation, and replicate was a dependent source.
Results
Searching for ERV-derived elements expressed during the peri-attachment period
We experimentally validated that the expression of ERV-derived sequences that met all of the following criteria: (1) was identified as an ERV-derived sequence from the bovine genome (Btau4.0) by RetroTector, possessing an ORF at least 100 amino acids or has at least one transmembrane domain, (2) was overlapped or located near or in between transcript regions annotated in the Ensembl database (http://www.ensembl.org), allowing two mismatches and annotated more than 10 multiloci, and (3) was detected on at least 2 days (day 17, 20, or 22) by the SOLiD3 sequencing (Supplementary Figure S1). Using these criteria, 10 putative ERVs that are located in between functional genes of the bovine genome were identified (Table 2 and Supplementary Table S1), after which another criterion, up-regulation of transcripts on day 22, was imposed in the analysis. Although the expression of 7 out of 10 putative ERVs declined, the expression of three candidate transcripts was up-regulated in day 20 or 22 pregnant conceptuses. Two transcripts, that both exist on chromosome 7 (ENSBTAT00000052494 and ENSBTAT00000052446, respectively), increased on day 22 (Table 2). In addition, transcript of a third candidate that exists on chromosome 2 (ENSBTAT00000012578) was detected on day 20 when trophoblast attachment to uterine endometrial epithelial cells was initiated, but not on day 22, and was excluded from the subsequent studies.
Table 2. The listing of candidate 10 retroelements detected.
| Transcript ID | Origin | Tag numbers | RPKM | Length | Chr. | Strand | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| D17 | D20 | D22 | D17 | D20 | D22 | |||||
| ENSBTAT00000012578 | Znf | 89 | 105 | 54 | 10.814 | 8.406 | 5.539 | 707 | 2 | − |
| ENSBTAT00000052900 | Gag | 9 | 3 | 0 | 0.312 | 0.069 | 0 | 2472 | 4 | − |
| ENSBTAT00000057217 | Gag | 407 | 7 | 0 | 18.440 | 0.209 | 0 | 1896 | 6 | − |
| ENSBTAT00000053802 | Gag | 435 | 11 | 0 | 19.462 | 0.324 | 0 | 1920 | 6 | − |
| ENSBTAT00000007724 | Env | 8 | 6 | 0 | 0.376 | 0.186 | 0 | 1827 | 6 | − |
| ENSBTAT00000052494 | Gag | 6 | 1 | 44 | 0.318 | 0.035 | 1.973 | 1617 | 7 | + |
| ENSBTAT00000052446 | Gag | 7 | 1 | 56 | 0.244 | 0.030 | 1.649 | 2463 | 7 | + |
| ENSBTAT00000045982 | Env | 2 | 1 | 0 | 0.102 | 0.034 | 0 | 1680 | 11 | − |
| ENSBTAT00000056638 | Gag | 2 | 4 | 0 | 0.070 | 0.092 | 0 | 2469 | 24 | + |
| ENSBTAT00000029917 | Env | 157 | 14 | 0 | 6.801 | 0.400 | 0 | 1983 | X | − |
RPKM, reads per kilo base-pair per million mapped reads; Chr., chromosome.
Identification of candidate ERV sequence and neighboring genes and search for its LTR-like sequences
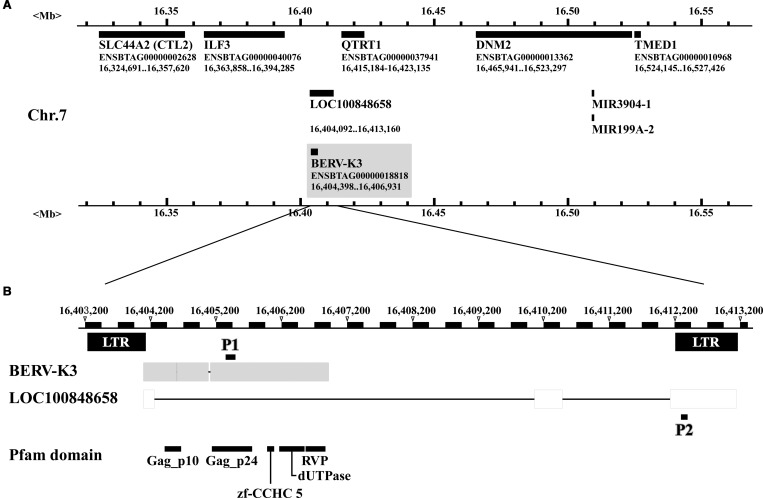
Using in silico analysis, we re-annotated the equivalent region on chromosome 7 with the recent database, Bos Taurus UMD3.1 version; however, the candidate ERVs previously identified were not detected. PCR analysis subsequently carried out using the primers (P1, Figure 1B) identified a portion of the ERV transcript, ENSBTAT00000052446 (Btau4.0) predicted from the original RNA-seq analysis, from which the sequences were extended through the use of RACE method, resulting in the identification of one ERV sequence (Supplementary Figure S2) existed on chromosome 7 as shown in Figure 1A. Subsequently, we identified the ERV mRNA on chromosome 7, containing Gag_p10, Gag_p24, and zinc finger (ZnF_CCHC) motifs, and short introns in between ORF 1 and 2 (2 nt) and between ORF 2 and 3 (36 nt) as shown in Figure 1B and Supplementary Figure S2. The analysis was extended to determine whether actual transcripts existed in the bovine conceptuses, and found that the single ERV transcript was, in fact, amplified by RT-PCR (Supplementary Figure S3A). The amino acid sequence of the ERV transcript was then used as a query against an ERV ORF database called gEVE [47], identifying that the ERV sequence was quite similar to the gag sequence of BERV-K2 [20] (Supplementary Figure S4A). Because the presence of LTRs (long terminal repeats) or LTR-like sequences was expected, attempts were made to identify the presence of LTR or LTR-like sequences on the chromosomal regions between 20 kb upstream of and 20 kb downstream from the ERV sequence. Two 988 and 1036 nt alignable sequences, located −333 nt upstream of and 6097 nt downstream from the ERV, respectively, were found, of which sequences were also similar to those of BERV-K2 (Supplementary Figure S4B). We, therefore, named the putative ERV transcript as BERV-K3.
Figure 1. Identification of BERV-K3 and their neighboring genes on chromosome 7 of the bovine genome.
(A) Genes neighboring BERV-K3. Solid-black bar indicates the location of nucleotide sequences and their identity is given below the solid-black bar. Black bars within the gray box is BERV-K3. (B) Structures of BERV-K3, Pfam domains, and primers set to detect transcripts. P1, indicated by a short-black line, is the primer set 1, which detects BERV-K3. P2, indicated by a short-black line, is the primer to detect LOC100848658. Pfam domains (Gag_p10, Gag_p24, zf-CCHC5, dUTPase and RVP) relative to BERV-K3 structures are given below.
BERV-K3 was located between interleukin enhancer-binding factor 3 (ILF3) and QTRT1 genes, and LOC100848658 was also found as a gene that shares a partial sequence (Figure 1A,B). The BERV-K3 sequence was then expected to be located on equivalent regions of chromosomes in human, murine, and sheep genomes; however, BERV-K3 sequence was not found in these species. These data indicated that the BERV-K3 is unique to the bovine species. In some cases, if these LTRs harbored appropriate binding sites for trophoblast-specific transcription factors, they may function as active promoters or enhancers with the potential to modulate neighboring gene expression [48]. We, therefore, examined the expression of ILF3 and QTRT1 with SOLiD3 data (Table 3); however, these gene expressions were not parallel to that of BERV-K3 expression.
Table 3. The Tag numbers and RPKM values of BERV-K3 neighboring genes.
| Transcript ID | Gene name | Tag numbers | RPKM | ||||
|---|---|---|---|---|---|---|---|
| D17 | D20 | D22 | D17 | D20 | D22 | ||
| ENSBTAT00000003403 | SLC44A2 (CTL2) | 6650 | 11 253 | 2454 | 175.50 | 195.68 | 54.67 |
| ENSBTAT00000040076 | ILF3 | 1360 | 1220 | 1036 | 36.04 | 21.30 | 23.17 |
| ENSBTAT00000056840 | QTRT1 | 71 | 28 | 47 | 11.25 | 2.92 | 6.28 |
| ENSBTAT00000036121 | DNM2 | 984 | 853 | 627 | 24.52 | 14.00 | 13.18 |
| ENSBTAT00000014563 | TMED1 | 268 | 310 | 416 | 15.33 | 11.68 | 20.08 |
Expression of ERV-derived transcripts in bovine conceptuses and other tissues
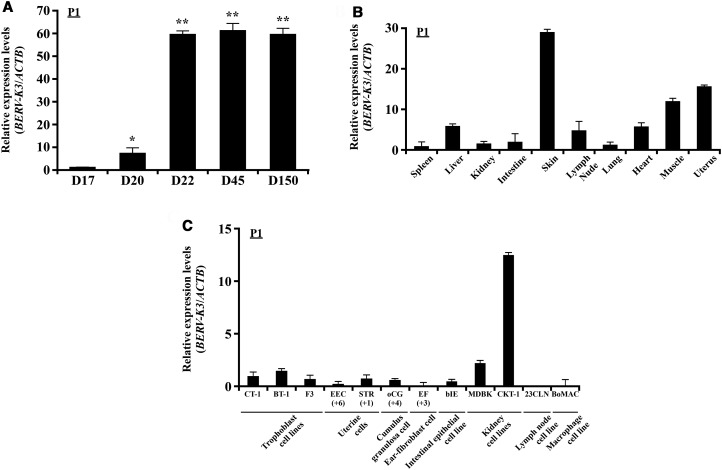
Using real-time PCR analysis, the expression of candidate transcripts was found in days 17, 20, and 22 bovine conceptuses and days 45 and 150 cotyledons. In particular, the expression of BERV-K3 was detected on day 20, when conceptuses begin to attach to uterine epithelial cells, in agreement with RNA-seq and real-time PCR analyses (Figure 2A). LOC100848658 gene was similarly expressed with BERV-K3 in bovine conceptuses, but not in other tissues examined (Figure 2 and Supplementary Figure S5). BERV-K3 was highly expressed in cotyledon on days 45 and 150 pregnancy, and either expression levels were similar to that in day 22 conceptuses. The expression of BERV-K3 in various bovine tissues was then examined (Figure 2B) and found that BERV-K3 was expressed in muscle, uterus, and skin. The expression of BERV-K3 in various bovine cell lines was further examined (Figure 2C) and found that although some expressions were detected in bovine trophoblast cell lines, BERV-K3 was highly expressed in bovine kidney CKT-1 cells.
Figure 2. Expression of BERV-K3 transcript in bovine conceptuses and various tissues and cell lines.
(A) Changes in BERV-K3 expression. Using primer set P1 described in Figure 1B, expression levels of transcript were determined in day 17, 20, and 22 (D17, D20, and D22) conceptuses (n = 4, each day) and day 45 and 150 cotyledons (n = 3, each day). RNAs from four or three of conceptuses or cotyledons, respectively, were used to perform the qPCR analysis. Data are expressed as fold difference relative to its expression on D17. *, **Statistically significant differences in mRNA levels (P < 0.05 or P < 0.01, respectively) when compared with that in D17. (B) BERV-K3 expression in various tissues. BERV-K3 transcripts in various tissues of day 150 pregnant animals, except that uterine sample was harvested from nonpregnant Japanese black cattle. RNAs were extracted from the spleen, liver, kidney, intestine, skin, lymph node, lung, heart, muscle, and uterus (cyclic) (n = 3 each). Muscle and skin were taken from the upper left hind leg area. Data are expressed as fold difference relative to the expression in the spleen. (C) BERV-K3 expression in bovine cell lines. BERV-K3 transcripts in cultured bovine primary cells and cell lines. CT-1, BT-1, and F3 are bovine trophoblast cell lines. Primary uterine cells (EPI and STR; indicated passage times below), primary cumulus-granulosa cell (oCG; indicated passage times below), primary ear-fibroblast cell (EF; indicated passage times below), intestinal epithelial cell line (bIE), kidney cell lines (MDBK and CKT-1), lymph node cell line (23CLN), and macrophage cell line (BoMAC) were subjected to the present study. Three samples each were used and three independent experiments were conducted. Data are expressed as fold difference relative to the expression in CT-1.
In situ localization of BERV-K3 transcript in the uterus and other tissues
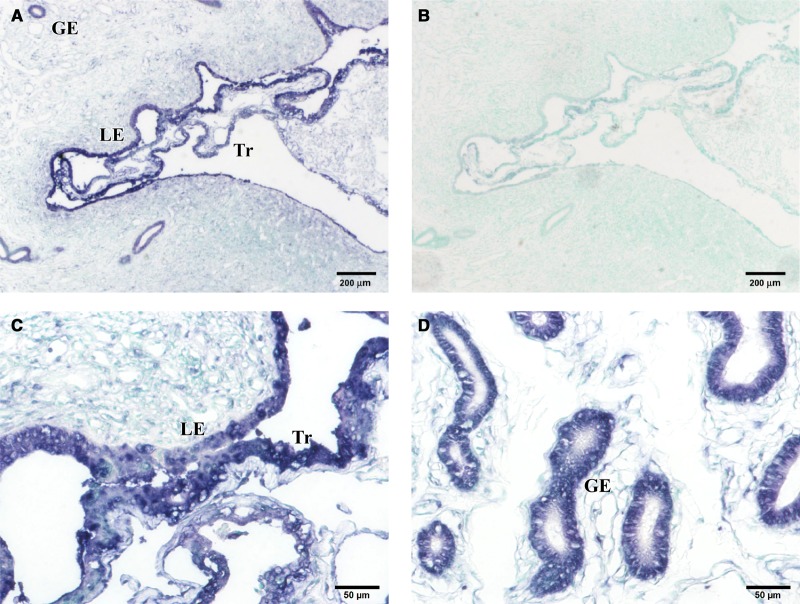
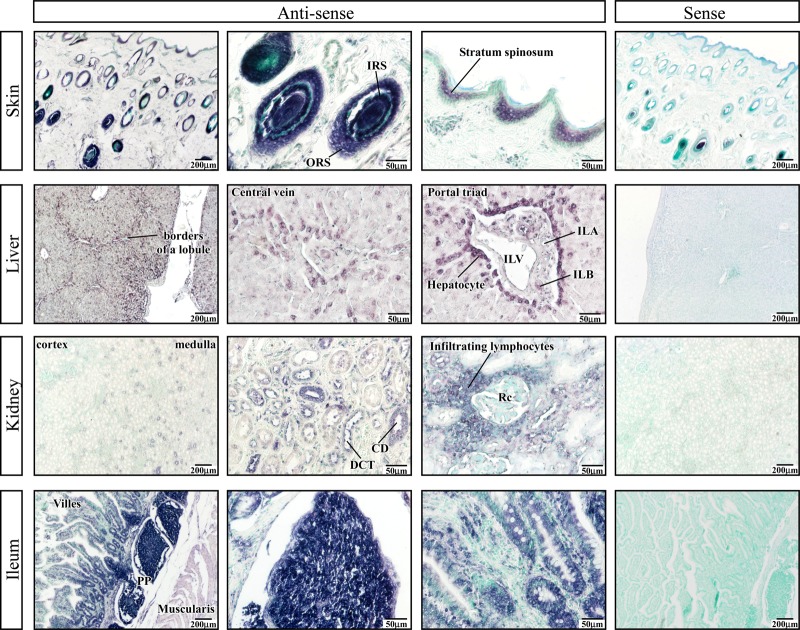
BERV-K3 transcript was found in day 22 uteri, including conceptus, uterine epithelial, and glandular epithelial cells (Figure 3). In addition, BERV-K3 transcript was found in the skin, liver, kidney, and intestine (Figure 4). In the skin, BERV-K3 was localized in outer root and inner root sheath (ORS and IRS, respectively) areas of hair follicles and the stratum spinosum of epidermis. In the liver, BERV-K3 was detected in hepatic laminas of lobule areas, particularly in hepatocytes surrounding central and interlobular veins (ILVs), and in interlobular bile ducts (ILB) of portal canal areas. In the kidney, BERV-K3 transcript was found in epithelial cells of renal-collecting ducts and distal convoluted tubules, and in infiltrating lymphocytes around renal corpuscle; however, BERV-K3 transcript was not found in the cortex region. In the ileum, BERV-K3 was localized in intestinal epithelia and enterocytes in glandules, and in Peyer's patches region. Positive staining was also detected on lymphocytes located in ileum lymphoid follicles and in infiltrating lymphocytes near renal corpuscles. These results indicated that the expression of BERV-K3 was not restricted to the reproductive tissues.
Figure 3. In situ localization of BERV-K3 transcript in day 22 pregnant uteri.
(A) In situ localization of BERV-K3 transcript in day 22 pregnant animals. (B) In situ localization with the sense-strand probe (negative control). (C) Higher magnification showing luminal epithelia and elongated trophoblasts. (D) Higher magnification showing glandular epithelial regions. GE, glandular epithelia; LE, luminal epithelia; Tr, trophoblast.
Figure 4. In situ localization of BERV-K3 transcript in the skin, liver, kidney, and ileum.
Bovine tissue sections were purchased from Zymogen (San Diego, CA, U.S.A.). In the skin, BERV-K3 transcript is found in external root and internal root sheath areas of hair follicles and stratum spinosum of epidermis. ORS, outer root sheath; IRS, inner root sheath. In the liver, BERV-K3 is seen in hepatic laminas of lobule areas, particularly in hepatocytes surrounding central veins and ILV, and in interlobular bile ducts of portal canal areas. ILA, interlobular artery; ILB, interlobular bile duct; ILV, interlobular vein. In the kidney, BERV-K3 transcript is found in epithelial cells of renal-collecting ducts and distal convoluted tubules, and in infiltrating lymphocytes around renal-collecting ducts. CD, collecting duct; DCT, distal convoluted tubule; Rc, renal corpuscle. In the ileum, BERV-K3 is seen in intestinal epithelia and enterocytes in glandules, and in PP regions. PP, Peyer's patch. In each, the far right images represent those with the sense-strand cRNA probe.
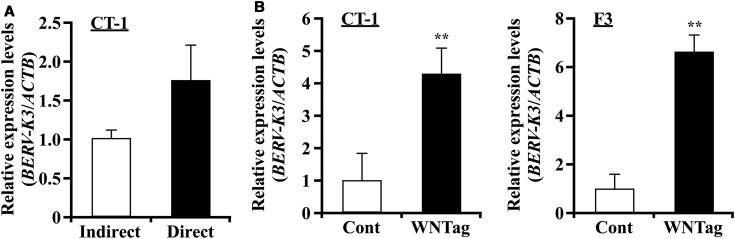
Induction of BERV-K3 transcript by cell-to-cell contact or canonical WNT agonist
When CT-1 cells were cultured with cell culture insert, not allowing direct CT-1 contact to EECs, no increase in BERV-K3 transcripts was found. However, when CT-1 cells were cultured without the cell insert, allowing direct cell-to-cell contact, increase in BERV-K3 was found (Figure 5A). Because WNT signal is known to be important for placentation following conceptus attachment to the uterine epithelium in the bovine species [29] and our previous study [37] showed that WNT2B and its receptor FZDs mRNA were detected during the conceptus attachment period, we then treated trophoblast CT-1 and F3 cells with 1 µM of canonical WNT agonist for 24 h. BERV-K3 and TCF7, but not LOC100848658, were induced by the WNT agonist in both cells (Figure 5B and Supplementary Figure S6).
Figure 5. Induction of BERV-K3 expression in CT-1 by cell-to-cell contact or in CT-1 or F3 cells by treatment with the canonical WNT agonist.
(A) Cell-to-cell contact on BERV-K3 expression. CT-1 cells were co-cultured without or with a cell culture insert on EECs for 24 h. (B) Effect of WNT agonist on BERV-K3 expression. CT-1 (left) or F3 (right) cells were treated with 1 µM of canonical WNT agonist for 24 h. Data are expressed as fold difference relative to the expression in control. **Statistically significant differences in mRNA levels (P < 0.05) when compared with that in control.
Discussion
In the present study, we found ERV, putative gag/pol-derived BERV-K3, transcripts specifically expressed in the bovine placenta from early- to mid-gestation. Trophectoderm and fetal membranes had weak expression of BERV-K3 transcript on day 20 and had greater expression from days 22 to 150. Accordingly, the transcripts were detected in bovine trophoblast CT-1, BT-1 and F3 cells. However, the transcripts were also found in the uterus, skin, liver, kidney, and ileum, indicating that their expression appeared somewhat ubiquitous. Cornelis et al. [22] similarly reported that the Bos-Env2 was expressed in the skin, spleen, and muscle, whereas only limited expression was detected in the bovine placenta. To elucidate molecular mechanisms associated with BERV-K3 transcription, the co-culture system with CT-1 cells and EECs [49] was used to study difference in BERV-K3 expression between days 20 and 22, when bovine conceptus begins its attachment to the uterine epithelium cells on day 20, followed by the tight attachment between two cell types on day 22. When CT-1 cells were cultured without the cell culture insert, increase in BERV-K3 transcripts was observed, suggesting that the cell-to-cell contact of trophoblasts to the uterine epithelial cells is required for BERV-K3 transcription to increase.
In our in silico analysis, LTRs (ERVK-type) were found to be located on both upstream and downstream regions of the BERV-K3 gene. However, while BERV-K3 was expressed in the trophectoderm and uterus, additional expression in the skin, liver, kidney, and ileum indicates that these LTRs are not pregnancy-specific. Transcripts of syncytin-Rum1, a fusogenic protein required for ruminants' placentation, were also detected in maternal (caruncle and intercarunclar endometrium) and fetal membranes (chorioallantois) [50]. Several genes are located in close proximity to BERV-K3 on chromosome 7 of the bovine genome, including ILF3 and CTL2 (also known as SLC44A2), located on the upstream region, and DNM2 and TMED, located on the downstream region of the BERV-K3 locus. Increase in BERV-K3 transcripts in day 22 bovine conceptuses was initially considered trophoblast/placenta-specific; however, the expression of these transcripts other than in the trophectoderm indicates that the insertion of BERV-K3 did not cause the placenta-specific expression of TMED and CTL2. It should be noted that although human and murine genomes lack BERV-K3, TMED, and CTL2 are expressed in placentas of these species. In fact, TMED2/p24b1 protein is required for the morphogenesis of the mouse embryo and placental development [51]. The expression of CTL2 transcripts in bovine conceptuses was high, peaking on day 20 when the conceptus begins its attachment to the uterine endometrial epithelium. Rather than the insertion of BERV-K3 in between TMED and CTL2 genes affected the transcriptional regulation of these genes, it is probable that these genes may play a role in the regulation of BERV-K3 gene expression in bovine trophoblasts. Although viral integration to the host genome could occur in a random manner, the integration would have to be locus-specific, if the integrated gene was to become active in certain cell types and/or physiological conditions.
WNT agonist used in the present study as a WNT signal activator was found to induce endogenous BERV-K3 expression in bovine trophoblast CT-1 and F3 cells. The WNT signaling pathway has been established as an important regulator of embryo/conceptus implantation and placental development in mice, sheep, cow, and humans [27–30]. Our previous study in peri-attachment conceptuses revealed that WNT2B and its receptor 9 FZDs mRNA were detected, while DKK-1, which acts as an antagonist of the canonical WNT signaling pathway, was decreased in day 22 bovine conceptuses [37]. Although an experiment determining the effect and function of DKK-1 is beyond the scope of the present study, it is possible that the decrease in the DKK-1 expression after trophoblast attachment to the uterine epithelial cells could lead to activation of canonical WNT signaling. In response to activation of CTNNB and TCF-dependent transcription by the Wnt agonist, BERV-K3 mRNA was increased in trophoblast CT-1 and F3 cells, suggesting that BERV-K3 could be one of the target genes in the canonical WNT pathway. It was also found that epithelial–mesenchymal transition (EMT) occurs in day 22 bovine conceptuses [52,53]. Because the WNT signal is involved in the EMT process, together with WNT, BERV-K3 may play a role in the bovine conceptus EMT and/or its attachment to the uterine epithelium.
Although envelope proteins of HERVs expressed in human trophoblast cells have fusogenic activity and immunosuppressive properties [9,54], the roles that gag polyproteins play have not been definitively elucidated. Most prominent gag- or gag/pol-derived gene is PEG10, which is thought to have been derived from the Ty3/Gypsy family of retrotransposons and is expressed as two proteins: gag-like protein containing a CCHC-type zinc finger domain and gag/pol-like fusion protein with an additional aspartic protease motif [55,56]. The Peg10 knockout mice showed early embryonic lethality due to defects in placental formation [57]. Although the amino acid sequences differ substantially between PEG10 and BERV-K3 (data not shown), BERV-K3 is also a gag/pol-derived ERV and has similar motifs to those of PEG10. BERV-K3 was found to consist of Gag_p24 and Gag_p10 motifs: Gag_p24 forms inner protein layer of the nucleocapsid, while Gag_p10, a retroviral GAG (core) protein, encompasses the p10 region producing the p10 protein upon proteolytic cleavage of GAG by retroviral protease. The p10 is, in fact, associated with viral envelope glycoproteins in most mammalian retroviruses and may be involved in viral particle assembly, transport, and budding [58]. BERV-K3 also consists of the motifs that potentially encode structural proteins, including zinc finger domain, UTPase, and retroviral aspartyl protease; however, the function of these motifs in BERV-K3 has not been elucidated. Nevertheless, because of its insertion in this specific locus, BERV-K3, unique to the bovine species, could have acquired its capability for expression and/or possibly functioning together with neighboring genes in the bovine placenta.
In the present study, BERV-K3 was also detected in IRS, ORS, and stratum spinosum in skins. It was found that the Wnt/Ctnnb signaling system was detected to function in mouse IRS and ORS [59]. In addition, retroviral-like aspartic protease (SASPase) is expressed in the stratum granulosum of mouse skin and is involved in wrinkle formation and adjustments of the stratum corneum hydration [60,61]. BERV-K3 transcripts found in skin epidermis suggest that BERV-K3 protein could be involved in skin development, although further experimentation is required to characterize its molecular mechanism associated with these events. In the liver, BERV-K3 was detected in hepatocytes surrounding central vein and ILV, in which the Wnt/Ctnnb was detected in pericentral hepatocytes in mice [62]. Involvement of canonical Wnt signaling in intestinal epithelial cell proliferation has also been demonstrated [63]. These and results from the present study suggest that in addition to placental formation and its maintenance, BERV-K3 may play a role in the development and/or maintenance of skin, liver, and intestines.
In conclusion, we found the new gag/pol-derived ERV transcript in RNAs extracted from days 20 to 22 bovine conceptuses, of which ERV was integrated into the chromosome 7. The expression of BERV-K3 gene increased in bovine conceptuses following its attachment to uterine epithelia and its high expression was maintained in the placenta up to 150-day gestation. Although the function of the genes is yet to be elucidated, available data suggest that BERV-K3, unique to the bovine species, could be controlled initially through cell-to-cell contact and then by WNT signaling and plays developmental and/or maintenance roles in placenta, skin, liver, and intestines.
Acknowledgements
We thank Prof. Gaku Ichihara, Department of Occupational and Environment Health, Tokyo University of Science, for generously providing us with his reagents and equipment. Our appreciation is extended to Dr Junyou Li and Mr Masanori Ikeda for taking care of experimental animals at the University of Tokyo farm. We also acknowledge Dr Toru Takahashi, National Institute of Agrobiological Sciences, and Dr Kazuyuki Uchida, Laboratory of Veterinary Pathology, the University of Tokyo, for collection of bovine tissues and their morphological evaluation of various cell/tissue types, respectively. The authors thank Mr Robert Moriarty for his thorough evaluation of the manuscript.
Abbreviations
- BERVs
bovine endogenous retroviruses
- Bie
bovine intestinal epithelial cells
- CTNNB
cellular accumulation and nuclear translocation of β-catenin
- DMEM
Dulbecco's modified Eagle medium
- EECs
endometrial epithelial cells
- EF
ear-derived fibroblast
- EMT
epithelial–mesenchymal transition
- ERVs
endogenous retroviruses
- FBS
fetal bovine serum
- FZD
Frizzled
- Gcm1
glial cells missing homolog 1
- ILB
interlobular bile duct
- ILF3
interleukin enhancer-binding factor 3
- ILV
interlobular vein
- LTRs
long terminal repeats
- oCG
ovarian cumulus-granulosa
- ORFs
open-reading frames
- ORS and IRS
outer root and inner root sheath
- RPKM
reads per kilobase of exon model per million mapped reads
- STRs
stromal cells
- TCF
T-cell fate
Author Contribution
J.Y., T.M., and K.I. co-ordinated the project. S.N. performed analyses related to bioinformatics. T.S., H.B., R.B., K.K., A.I., Y.A., K.K., and K.I. equally performed the experiments. T.S. and K.I. wrote the manuscript, but all authors were involved in discussion throughout the experimentation and manuscript preparation. All authors thoroughly read and approved the manuscript.
Funding
The present study was initially supported by the program for Promotion of Basic Research Activities for Innovative Bioscience (BRAIN) to K.I. This work was also supported by Japan Society for the Promotion of Science (JSPS) KAKENHI grant numbers [15K07697] (to T.S.) and [16K21386] (to S.N.) and by Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry (to K.I.).
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
Supplementary Material
References
- 1.Weiss R.A. (2006) The discovery of endogenous retroviruses. Retrovirology 3, 67 10.1186/1742-4690-3-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.International Human Genome Sequencing Consortium (2004) Finishing the euchromatic sequence of the human genome. Nature 431, 931–945 10.1038/nature03001 [DOI] [PubMed] [Google Scholar]
- 3.Chinwalla A.T., Cook L.L., Delehaunty K.D., Fewell1 G.A., Fulton L.A., Fulton R.S. et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 10.1038/nature01262 [DOI] [PubMed] [Google Scholar]
- 4.Adelson, D.L., Raison, J.M. and Edgar, R.C. (2009) Characterization and distribution of retrotransposons and simple sequence repeats in the bovine genome. Proc. Natl Acad. Sci. U.S.A. 106, 12855–12860 10.1073/pnas.0901282106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeke, J.D. and Stoye, J.P. (1997) Retrotransposons, endogenous retroviruses, and the evolution of retroelements In Retroviruses (Coffin J.M., Hughes S.H. and Varmus H.E., eds), Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 6.Mi, S., Lee, X., Li, X.-p., Veldman, G.M., Finnerty, H., Racie, L. et al. (2000) Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403, 785–789 10.1038/35001608 [DOI] [PubMed] [Google Scholar]
- 7.Blaise, S., de Parseval, N., Bénit, L. and Heidmann, T. (2003) Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl Acad. Sci. U.S.A. 100, 13013–13018 10.1073/pnas.2132646100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kudaka, W., Oda, T., Jinno, Y., Yoshimi, N. and Aoki, Y. (2008) Cellular localization of placenta-specific human endogenous retrovirus (HERV) transcripts and their possible implication in pregnancy-induced hypertension. Placenta 29, 282–289 10.1016/j.placenta.2007.11.009 [DOI] [PubMed] [Google Scholar]
- 9.Esnault, C., Priet, S., Ribet, D., Vernochet, C., Bruls, T., Lavialle, C. et al. (2008) A placenta-specific receptor for the fusogenic, endogenous retrovirus-derived, human syncytin-2. Proc. Natl Acad. Sci. U.S.A. 105, 17532–17537 10.1073/pnas.0807413105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lokossou, A.G., Toudic, C. and Barbeau, B. (2014) Implication of human endogenous retrovirus envelope proteins in placental functions. Viruses 6, 4609–4627 10.3390/v6114609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupressoir, A., Marceau, G., Vernochet, C., Bénit, L., Kanellopoulos, C., Sapin, V. et al. (2005) Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc. Natl Acad. Sci. U.S.A. 102, 725–730 10.1073/pnas.0406509102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heidmann, O., Vernochet, C., Dupressoir, A. and Heidmann, T. (2009) Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: a new ‘syncytin’ in a third order of mammals. Retrovirology 6, 107 10.1186/1742-4690-6-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornelis, G., Heidmann, O., Bernard-Stoecklin, S., Reynaud, K., Véron, G., Mulot, B. et al. (2012) Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proc. Natl Acad. Sci. U.S.A. 109, E432–E441 10.1073/pnas.1115346109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunlap, K.A., Palmarini, M., Varela, M., Burghardt, R.C., Hayashi, K., Farmer, J.L. et al. (2006) Endogenous retroviruses regulate periimplantation placental growth and differentiation. Proc. Natl Acad. Sci. U.S.A. 103, 14390–14395 10.1073/pnas.0603836103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunlap, K.A., Palmarini, M. and Spencer, T.E. (2006) Ovine endogenous betaretroviruses (enJSRVs) and placental morphogenesis. Placenta 27 (suppl.), 135–140 10.1016/j.placenta.2005.12.009 [DOI] [PubMed] [Google Scholar]
- 16.Dunlap, K.A., Palmarini, M., Adelson, D.L. and Spencer, T.E. (2005) Sheep endogenous betaretroviruses (enJSRVs) and the hyaluronidase 2 (HYAL2) receptor in the ovine uterus and conceptus. Biol. Reprod. 73, 271–279 10.1095/biolreprod.105.039776 [DOI] [PubMed] [Google Scholar]
- 17.Armezzani, A., Arnaud, F., Caporale, M., di Meo, G., Iannuzzi, L., Murgia, C. et al. (2011) The signal peptide of a recently integrated endogenous sheep betaretrovirus envelope plays a major role in eluding gag-mediated late restriction. J. Virol. 85, 7118–7128 10.1128/JVI.00407-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murcia, P.R., Arnaud, F. and Palmarini, M. (2007) The transdominant endogenous retrovirus enJS56A1 associates with and blocks intracellular trafficking of Jaagsiekte sheep retrovirus Gag. J. Virol. 81, 1762–1772 10.1128/JVI.01859-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morozov, V.A., Morozov, A.V. and Lagaye, S. (2007) Endogenous JSRV-like proviruses in domestic cattle: analysis of sequences and transcripts. Virology 367, 59–70 10.1016/j.virol.2007.05.018 [DOI] [PubMed] [Google Scholar]
- 20.Baba, K., Nakaya, Y., Shojima, T., Muroi, Y., Kizaki, K., Hashizume, K. et al. (2011) Identification of novel endogenous betaretroviruses which are transcribed in the bovine placenta. J. Virol. 85, 1237–1245 10.1128/JVI.01234-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koshi, K., Suzuki, Y., Nakaya, Y., Imai, K., Hosoe, M., Takahashi, T. et al. (2012) Bovine trophoblastic cell differentiation and binucleation involves enhanced endogenous retrovirus element expression. Reprod. Biol. Endocrinol. 10, 41 10.1186/1477-7827-10-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cornelis, G., Heidmann, O., Degrelle, S.A., Vernochet, C., Lavialle, C., Letzelter, C. et al. (2013) Captured retroviral envelope syncytin gene associated with the unique placental structure of higher ruminants. Proc. Natl Acad. Sci. U.S.A. 110, E828–E837 10.1073/pnas.1215787110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakaya, Y., Koshi, K., Nakagawa, S., Hashizume, K. and Miyazawa, T. (2013) Fematrin-1 is involved in fetomaternal cell-to-cell fusion in Bovinae placenta and has contributed to diversity of ruminant placentation. J. Virol. 87, 10563–10572 10.1128/JVI.01398-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Redelsperger, F., Cornelis, G., Vernochet, C., Tennant, B.C., Catzeflis, F., Mulot, B. et al. (2014) Capture of syncytin-Mar1, a fusogenic endogenous retroviral envelope gene involved in placentation in the Rodentia squirrel-related clade. J. Virol. 88, 7915–7928 10.1128/JVI.00141-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cornelis, G., Vernochet, C., Malicorne, S., Souquere, S., Tzika, A.C., Goodman, S.M. et al. (2014) Retroviral envelope syncytin capture in an ancestrally diverged mammalian clade for placentation in the primitive Afrotherian tenrecs. Proc. Natl Acad. Sci. U.S.A. 111, E4332–E4341 10.1073/pnas.1412268111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cornelis, G., Vernochet, C., Carradec, Q., Souquere, S., Mulot, B., Catzeflis, F. et al. (2015) Retroviral envelope gene captures and syncytin exaptation for placentation in marsupials. Proc. Natl Acad. Sci. U.S.A. 112, E487–E496 10.1073/pnas.1417000112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayashi, K., Burghardt, R.C., Bazer, F.W. and Spencer, T.E. (2007) WNTs in the ovine uterus: potential regulation of periimplantation ovine conceptus development. Endocrinology 148, 3496–3506 10.1210/en.2007-0283 [DOI] [PubMed] [Google Scholar]
- 28.Sonderegger, S., Haslinger, P., Sabri, A., Leisser, C., Otten, J.V., Fiala, C. et al. (2010) Wingless (Wnt)-3A induces trophoblast migration and matrix metalloproteinase-2 secretion through canonical Wnt signaling and protein kinase B/AKT activation. Endocrinology 151, 211–220 10.1210/en.2009-0557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu, W., Tu, Z., Wang, S., Lu, J., Wang, Q., Wang, W. et al. (2013) Spatiotemporal expression of Wnt signaling pathway components during bovine placental development. Theriogenology 80, 893–902 10.1016/j.theriogenology.2013.07.015 [DOI] [PubMed] [Google Scholar]
- 30.Li, Q., Kannan, A., Das, A., Demayo, F.J., Hornsby, P.J., Young, S.L. et al. (2013) WNT4 acts downstream of BMP2 and functions via β-catenin signaling pathway to regulate human endometrial stromal cell differentiation. Endocrinology 154, 446–457 10.1210/en.2012-1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nayeem, S.B., Arfuso, F., Dharmarajan, A. and Keelan, J.A. (2016) Role of Wnt signalling in early pregnancy. Reprod. Fertil. Dev. 28, 525–544 10.1071/RD14079 [DOI] [PubMed] [Google Scholar]
- 32.Lu, J., Zhang, S., Nakano, H., Simmons, D.G., Wang, S., Kong, S. et al. (2013) A positive feedback loop involving Gcm1 and Fzd5 directs chorionic branching morphogenesis in the placenta. PLoS Biol. 11, e1001536 10.1371/journal.pbio.1001536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuura, K., Jigami, T., Taniue, K., Morishita, Y., Adachi, S., Senda, T. et al. (2011) Identification of a link between Wnt/β-catenin signalling and the cell fusion pathway. Nat. Commun. 2, 548 10.1038/ncomms1551 [DOI] [PubMed] [Google Scholar]
- 34.Imakawa, K., Kusama, K. and Yasuda, J. (2014) Early placentation and local immune regulation. Proceedings of the 9th Reproduction in Domestic Ruminants (Juengel, J.L., Miyamoto, A., Price, C., Reynolds, L.P., Smith, M.F., Webb, R., eds), 25–29 August 2014, Obihiro, Japan. Context; 2014, 37K38 [Google Scholar]
- 35.Bazer, F.W., Spencer, T.E. and Johnson, G.A. (2009) Interferons and uterine receptivity. Semin. Reprod. Med. 27, 90–102 10.1055/s-0028-1108013 [DOI] [PubMed] [Google Scholar]
- 36.Ideta, A., Urakawa, M., Aoyagi, Y. and Saeki, K. (2007) Early development in utero of bovine nuclear transfer embryos using early G1 and G0 phase cells. Cloning Stem Cells 9, 571–580 10.1089/clo.2007.0017 [DOI] [PubMed] [Google Scholar]
- 37.Nakagawa, S., Bai, H., Sakurai, T., Nakaya, Y., Konno, T., Miyazawa, T. et al. (2013) Dynamic evolution of endogenous retrovirus-derived genes expressed in bovine conceptuses during the period of placentation. Genome Biol. Evol. 5, 296–306 10.1093/gbe/evt007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakurai, T., Sakamoto, A., Muroi, Y., Bai, H., Nagaoka, K., Tamura, K. et al. (2009) Induction of endogenous interferon tau gene transcription by CDX2 and high acetylation in bovine nontrophoblast cells. Biol. Reprod. 80, 1223–1231 10.1095/biolreprod.108.073916 [DOI] [PubMed] [Google Scholar]
- 39.Ashton-Beaucage, D., Udell, C.M., Lavoie, H., Baril, C., Lefrançois, M., Chagnon, P. et al. (2010) The exon junction complex controls the splicing of MAPK and other long intron-containing transcripts in Drosophila. Cell 143, 251–262 10.1016/j.cell.2010.09.014 [DOI] [PubMed] [Google Scholar]
- 40.Sperber, G.O., Airola, T., Jern, P. and Blomberg, J. (2007) Automated recognition of retroviral sequences in genomic data—RetroTector©. Nucleic Acids Res. 35, 4964–4976 10.1093/nar/gkm515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mortazavi, A., Williams, B.A., McCue, K., Schaeffer, L. and Wold, B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 42.Kodama, Y., Shumway, M., Leinonen, R. and International Nucleotide Sequence Database Collaboration (2012) The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res. 40, D54–D56 10.1093/nar/gkr854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimada, A., Nakano, H., Takahashi, T., Imai, K. and Hashizume, K. (2001) Isolation and characterization of a bovine blastocyst-derived trophoblastic cell line, BT-1: development of a culture system in the absence of feeder cell. Placenta 22, 652–662 10.1053/plac.2001.0702 [DOI] [PubMed] [Google Scholar]
- 44.Talbot, N.C., Caperna, T.J., Edwards, J.L., Garrett, W., Wells, K.D. and Ealy, A.D. (2000) Bovine blastocyst-derived trophectoderm and endoderm cell cultures: interferon tau and transferrin expression as respective in vitro markers. Biol. Reprod. 62, 235–247 10.1095/biolreprod62.2.235 [DOI] [PubMed] [Google Scholar]
- 45.Hambruch, N., Haeger, J.-D., Dilly, M. and Pfarrer, C. (2010) EGF stimulates proliferation in the bovine placental trophoblast cell line F3 via Ras and MAPK. Placenta 31, 67–74 10.1016/j.placenta.2009.10.011 [DOI] [PubMed] [Google Scholar]
- 46.Bai, H., Sakurai, T., Konno, T., Ideta, A., Aoyagi, Y., Godkin, J.D. et al. (2012) Expression of GATA1 in the ovine conceptus and endometrium during the peri-attachment period. Mol. Reprod. Develop. 79, 64–73 10.1002/mrd.21409 [DOI] [PubMed] [Google Scholar]
- 47.Nakagawa, S. and Takahashi, M.U. (2016) gEVE: a genome-based endogenous viral element database provides comprehensive viral protein-coding sequences in mammalian genomes. Database 2016, baw087 10.1093/database/baw087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen, C.J., Lock, W.M. and Mager, D.L. (2009) Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene 448, 105–114 10.1016/j.gene.2009.06.020 [DOI] [PubMed] [Google Scholar]
- 49.Sakurai, T., Bai, H., Bai, R., Arai, M., Iwazawa, M., Zhang, J. et al. (2012) Coculture system that mimics in vivo attachment processes in bovine trophoblast cells. Biol. Reprod. 87, 1–11 10.1095/biolreprod.112.100180 [DOI] [PubMed] [Google Scholar]
- 50.McLean, K.J., Crouse, M.S., Crosswhite, M.R., Black, D.N., Dahlen, C.R., Borowicz, P.P. et al. (2016) Expression of an endogenous retroviral element, during early gestation in beef heifers. J. Anim. Sci. 94, 4452–4456 10.2527/jas.2016-0793 [DOI] [PubMed] [Google Scholar]
- 51.Jerome-Majewska, L.A., Achkar, T., Luo, L., Lupu, F. and Lacy, E. (2010) The trafficking protein Tmed2/p24β1 is required for morphogenesis of the mouse embryo and placenta. Dev. Biol. 341, 154–166 10.1016/j.ydbio.2010.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamakoshi, S., Bai, R., Chaen, T., Ideta, A., Aoyagi, Y., Sakurai, T. et al. (2012) Expression of mesenchymal-related genes by the bovine trophectoderm following conceptus attachment to the endometrial epithelium. Reproduction 143, 377–387 10.1530/REP-11-0364 [DOI] [PubMed] [Google Scholar]
- 53.Kusama, K., Bai, R., Ideta, A., Aoyagi, Y., Okuda, K. and Imakawa, K. (2016) Regulation of epithelial to mesenchymal transition in bovine conceptuses through the interaction between follistatin and activin A. Mol. Cell Endocrinol. 434, 81–92 10.1016/j.mce.2016.06.017 [DOI] [PubMed] [Google Scholar]
- 54.Mangeney, M., Renard, M., Schlecht-Louf, G., Bouallaga, I., Heidmann, O., Letzelter, C. et al. (2007) Placental syncytins: genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc. Natl Acad. Sci. U.S.A. 104, 20534–20539 10.1073/pnas.0707873105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ono, R., Kobayashi, S., Wagatsuma, H., Aisaka, K., Kohda, T., Kaneko-Ishino, T. et al. (2001) A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics 73, 232–237 10.1006/geno.2001.6494 [DOI] [PubMed] [Google Scholar]
- 56.Youngson, N.A., Kocialkowski, S., Peel, N. and Ferguson-Smith, A.C. (2005) A small family of sushi-class retrotransposon-derived genes in mammals and their relation to genomic imprinting. J. Mol. Evol. 61, 481–490 10.1007/s00239-004-0332-0 [DOI] [PubMed] [Google Scholar]
- 57.Ono, R., Nakamura, K., Inoue, K., Naruse, M., Usami, T., Wakisaka-Saito, N. et al. (2006) Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet. 38, 101–106 10.1038/ng1699 [DOI] [PubMed] [Google Scholar]
- 58.van der Kuyl, A.C., Mang, R., Dekker, J.T. and Goudsmit, J. (1997) Complete nucleotide sequence of simian endogenous type D retrovirus with intact genome organization: evidence for ancestry to simian retrovirus and baboon endogenous virus. J. Virol. 71, 3666–3676 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lim, X., Tan, S.H., Koh, W.L.C., Chau, R.M.W., Yan, K.S., Kuo, C.J. et al. (2013) Interfollicular epidermal stem cells self-renew via autocrine Wnt signaling. Science 342, 1226–1230 10.1126/science.1239730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsui, T., Kinoshita-Ida, Y., Hayashi-Kisumi, F., Hata, M., Matsubara, K., Chiba, M. et al. (2006) Mouse homologue of skin-specific retroviral-like aspartic protease involved in wrinkle formation. J. Biol. Chem. 281, 27512–27525 10.1074/jbc.M603559200 [DOI] [PubMed] [Google Scholar]
- 61.Matsui, T., Miyamoto, K., Kubo, A., Kawasaki, H., Ebihara, T., Hata, K. et al. (2011) SASPase regulates stratum corneum hydration through profilaggrin-to-filaggrin processing. EMBO. Mol. Med. 3, 320–333 10.1002/emmm.201100140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang, J., Mowry, L.E., Nejak-Bowen, K.N., Okabe, H., Diegel, C.R., Lang, R.A. et al. (2014) β-Catenin signaling in murine liver zonation and regeneration: a Wnt-Wnt situation! Hepatology 60, 964–976 10.1002/hep.27082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gregorieff, A., Pinto, D., Begthel, H., Destrée, O., Kielman, M. and Clevers, H. (2005) Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology 129, 626–638 10.1016/j.gastro.2005.06.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.