Abstract
Isolating RNA from insects is becoming increasingly important in molecular entomology. Four methods including three commercial kits RNeasy Mini Kit (Qiagen), SV Total RNA isolation system (Promega), TRIzol reagent (Invitrogen), and a cetyl trimethylammonium bromide (CTAB)-based method were compared regarding their ability to isolate RNA from whole-body larvae of Thaumatotibia leucotreta (Meyrick), Thanatophilus micans (F.), Plutella xylostella (L.), and Tenebrio molitor (L.). A difference was observed among the four methods regarding RNA quality but not quantity. However, RNA quality and quantity obtained was not dependent on the insect species. The CTAB-based method produced low-quality RNA and the Trizol reagent produced partially degraded RNA, whereas the RNeasy Mini Kit and SV Total RNA isolation system produced RNA of consistently high quality. However, after reverse transcription to cDNA, RNA produced using all four extraction methods could be used to successfully amplify a 708 bp fragment of the cytochrome oxidase I gene. Of the four methods, the SV Total RNA isolation system showed the least amount of DNA contamination with the highest RNA integrity number and is thus recommended for stringent applications where high-quality RNA is required. This is the first comparison of RNA isolation methods among different insect species and the first to compare RNA isolation methods in insects in the last 20 years.
Keywords: CTAB, RNA extraction, RNeasy Mini Kit, SV Total RNA isolation system, TRIzol reagent
Isolating RNA from insects is becoming increasingly important as a growing number of molecular biology applications rely on using RNA rather than DNA, demonstrated, e.g., by Baton et al. (2008) , Götz et al. (2012) , Wu et al. (2013) , and Zhang et al. (2013) . Currently, limited research is available on RNA isolation methods from insects. The only published research comparing RNA isolation methods in insects is demonstrated by Noriega and Wells (1993) , where three different RNA isolation protocols were compared. However, as molecular entomology has advanced substantially in the last 20 years, these methods may not be suitable for stringent RNA applications. In addition, isolation methods that have since been developed may produce RNA of a higher quality.
Different RNA isolation methods often produce RNA of varying quantity and quality, which may depend on the type of tissue used. Isolating RNA from tissue requires separation of the nucleotides from secondary metabolites such as polyphenolics, etheric oils, carbohydrates, and lignins; which differ according to tissue composition ( Dong and Dunstan 1996 ). It is therefore vital that the optimal RNA isolation method is selected according to the type of tissue used to provide RNA of a suitably high quantity and quality for the selected downstream application.
Currently, insect gene expression studies such as those performed by Gatehouse et al. (2009) , Xie et al. (2012) , and Ogaugwu and Wimmer (2013) appear to be focussed mainly on agriculturally important insects. Thus, for this study three agriculturally important insect pests were chosen for comparison: the false codling moth Thaumatotibia leucotreta (Meyrick, 1913) (Lepidoptera: Tortricidae) , diamondback moth Plutella xylostella (Linnaeus, 1767) (Lepidoptera: Plutellidae), and mealworm Tenebrio molitor (Linnaeus, 1758) (Coleoptera: Tenebrionidae), as they are particularly damaging to both the commercial and subsistence agriculture sectors in South Africa ( Talekar and Shelton 1993 , Li and Bouwer 2012 , Oppert et al. 2012 ). In addition, the forensically important insect Thanatophilus micans (Castelnau, 1840) (Coleoptera: Silphidae) ( Ridgeway et al. 2014 ) was chosen for comparison. Two different species each of Coleoptera and Lepidoptera were selected to provide a comparison between and within different insect orders. Whole body extractions were analyzed as they cover all potential tissue types for use and are used for gene expression studies ( Gatehouse et al. 2009 ).
A number of criteria determine a good RNA isolation method. The method needs to be highly efficient to limit the chance of RNA degradation ( Tattersall et al. 2005 ), must provide high-quality RNA that contains no contaminants, must produce RNA of a reasonable quantity and overall needs to be consistent and robust. In addition, the safety of the RNA isolation method should also be considered, since many RNA isolation methods include the use of toxic or hazardous chemicals such as phenol, chloroform, or 2-mercaptoethanol. In general, a good RNA isolation method is characterized as being economically viable; where any costs of the method, including isolation time and monetary value, are justified by the above-mentioned criteria. The aim of this study was thus to compare different RNA extraction methods from insect whole-body larvae and to evaluate the RNA produced based on the criteria described earlier. This is the first comparison of RNA isolation methods among different insect species and the first to compare RNA isolation methods in insects in the last 20 years.
Materials and Methods
Insect Material
Late-instar larvae of T. leucotreta, P. xylostella , Te. molitor, and Th. micans were collected from established cultures at Rhodes University. Live individuals were flash frozen in liquid nitrogen and homogenized using a sterile mortar and pestle. In total, 30 mg of homogenized tissue was then placed into a prefrozen microcentrifuge tube. This was the standard sample preparation for all RNA isolation methods. Three replicates were included for every species using each method, resulting in a total of 48 independent RNA isolations.
RNA Isolation
Four RNA isolation methods were compared in this study. Three of these methods are based on commercial kits: TRIzol reagent (Invitrogen, Carlsbad, CA), the SV Total RNA isolation system (Promega, Madison, WI), and the RNeasy Mini Kit (Qiagen, Hilden, Germany). The remaining RNA isolation method is a complex CTAB-based method ( Baiges and Mas 2003 ). Reagents for the CTAB-based method were purchased from the Merck group (Merck, South Africa).
The RNeasy Mini Kit and SV Total RNA isolation system were followed according to manufacturers’ specifications. The CTAB protocol was adjusted by including three ethanol washes instead of two for improved RNA precipitation. In addition, the pellet was suspended in 100 µl of H 2 O instead of the recommended 30 µl before the optional cleanup to better dissolve the RNA pellet. The TRIzol reagent protocol was followed according to the manufacturer’s specifications with the exception that only 30 mg of tissue was used instead of the suggested 50–100 mg to allow direct comparison with the other four RNA isolation methods used in this study. All RNA extracted was stored at −80°C before further analysis.
DNase Digestion
To evaluate the necessity of additional DNase digestion steps, the sample was divided and 10 µl RNA from each method was digested using the RQ1 RNase-Free DNase kit (Promega, Madison, WI) according to manufacturer’s specifications to remove any remaining DNA from the RNA extractions. DNA contamination of RNA isolations before digestion with DNase enzyme was evaluated using polymerase chain reaction (PCR), along with positive and negative controls.
PCR Application
DNA contamination of isolated RNA was evaluated by PCR to amplify a 708 bp fragment of the cytochrome oxidase I (COI) gene using the primers LCO1490 and HCO2198 ( Folmer et al. 1994 ). DNA contamination was assumed to be present if the PCR was successful. PCR tubes contained 12.5 µl Master Mix (Promega, Madison, WI), 1.5 µl of each primer (10 pmol), 1.5 µl MgCl 2 (25 mM), 7 µl H 2 O, and 1 µl RNA, resulting in a total volume of 25 µl. The conditions for each of 40 PCR cycles were as follows: denaturation at 95°C for 30 s, annealing at 54°C for 1 min, and extension at 72°C for 1 min 30 s. The reaction was preceded with an initial denaturation step of 95°C for 5 min and ended with a final extension step of 72°C for 10 min. PCRs were checked for amplification using agarose gel electrophoresis (AGE).
cDNA Conversion
The RNA of T. leucotreta , isolated using the four methods described earlier, was converted to cDNA using the Maxima H Minus Reverse Transcriptase kit with a random hexamer primer (5′-d(NNNNNN) -3 (Thermo Scientific, Wilmington, DL). The sample was incubated at 25°C for 10 min followed by 30 min at 50°C. The reaction was terminated by heating the sample to 85°C for 5 min. cDNA was then tested in a PCR using the conditions described earlier to determine its applicability for a basic downstream application.
Analysis
RNA quality and quantity was analyzed using AGE, a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DL) and an Agilent 2100 Bioanalyser with an RNA 6000 chip kit. Using AGE good quality RNA was considered to be two well-defined fragments, corresponding to the 18S (2,000 nt) and 28S (3,800 nt) ribosomal subunits, with limited smearing. Each gel contained a 100 bp-DNA ladder (Promega, Madison, WI) with 20 and 100 ng Lambda DNA standards to infer concentrations. Gels were loaded with 5 µl RNA sample. The gels were used at a concentration of 1.5% agarose and run at 80 V for 30–40 min. Spectrophotometer 230/260 and 280/260 ratios were used to evaluate RNA quality, with good quality RNA having readings of 1.8–2.0 and 2.0–2.2 for each ratio, respectively. Total RNA analysis pg sensitivity (Eukaryote) was conducted using an Agilent 2100 Bioanalyser. The ladder was used at 1,000 pg/µl concentrations with the minimum and maximum visible range at 17 and 70 s, respectively. RNA integrity number (RIN) and fluorescent graphs were analysed for each method.
Results
RNA Quality
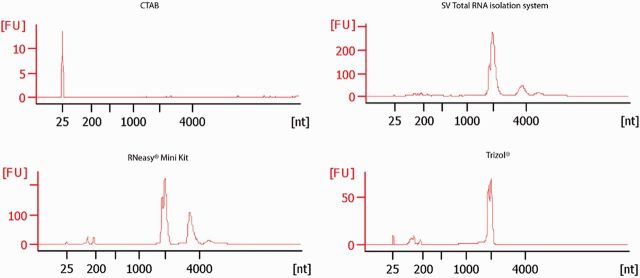
Spectrophotometer readings for RNA isolations conducted using the CTAB isolation protocol were generally poor, with only 70% of the isolations showing acceptable 280/260 and 260/230 ratios. In addition, AGE of RNA extracted using the CTAB isolation protocol only showed faint smearing, with no well-defined bands ( Fig. 1 ). The average RIN for CTAB extractions was 1, with no peak visible for either the18S or 28S fragments on the electropherogram ( Fig. 2 ). The quality of RNA using Trizol reagent was fairly inconsistent as only half of the reactions had acceptable 260/230 ratios with the rest below 1.3. The mean RIN of samples extracted with Trizol reagent was 5.3 (SD ± 0.14), and could thus be considered partially degraded. RNeasy Mini Kit and SV Total RNA isolation system showed acceptable purity ratios, with a 280/260 mean value of 2.01 (SD ± 0.15) and 230/260 ratio of 1.85 (SD ± 0.37). The mean RIN score for RNeasy was 8.2 (SD ± 0.57), which is considered intact RNA. Electropherogram graphs of RNA isolated using the RNeasy Mini Kit showed the highest 28S peak out of the four methods, with a good 5S fragment ( Fig. 2 ) The mean RIN for SV Total RNA extraction kit was 9.25 (SD ± 0.21). These RNA extractions showed well defined peaks with limited contamination ( Fig. 2 ).
Fig. 1.
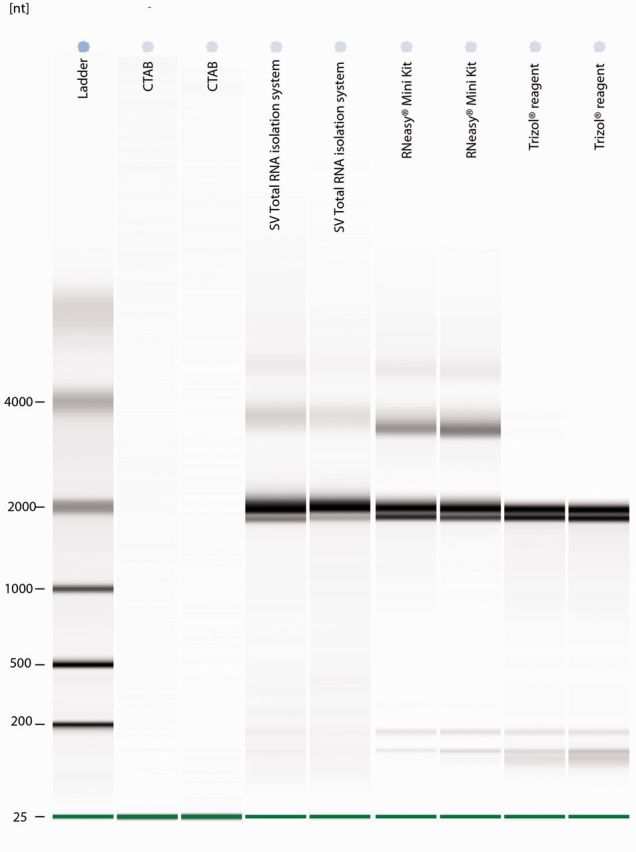
Pseudo-gel image produced using an Agilent 2100 Bioanalyser, showing the results of RNA extracted from T. leucotreta using the RNeasy Mini Kit, a CTAB-based protocol, TRIzol reagent, and the SV Total RNA kit. The 18S RNA subunit is visible at 2,000 nt, the 28S large subunit at 3,800 nt, and a combination of 5S, 5.8S, and tRNAs at ± 180 nt.
Fig. 2.
Agilent 2100 Bioanalyser electropherogram graphs showing RNA extracted using the RNeasy Mini Kit, a CTAB-based protocol, TRIzol reagent and the SV Total RNA kit Markers can be seen at 25 nt, 18S RNA subunit at 2,000 nt, the 28S large subunit at 3,800 nt, and a combination of 5S, 5.8S, and tRNAs at ± 180 nt.
RNA Quantity
According to spectrophotometer results, the mean RNA yield per extraction was 96,319 ng (SD ± 39,463) for CTAB isolations, 52,314 ng (SD ± 2,593) for the SV Total RNA isolation system, 40,127 ng (SD ± 14,407) for RNeasy Mini Kit isolations and 85,079 ng (SD ± 52,154) for TRIzol reagent isolations.
DNA Contamination
RNA isolated using TRIzol reagent, RNeasy Mini Kit, and the CTAB method showed high amounts of DNA contamination, resulting in PCR products of high concentration. Only RNA extracted using the SV Total RNA isolation system did not show high amounts of DNA contamination. Additional DNase digestions were performed for a subsample of RNA extracted using each of the four methods. After DNase digestion, PCR amplification was unsuccessful for RNA extracted using each of the four methods, showing that DNA contamination had been eliminated.
PCR Application
T. leucotreta RNA was reverse transcribed to cDNA and used to amplify a 708 bp fragment of the COI gene in order to compare the success of different RNA isolation methods for a basic downstream application. It was found that RNA isolated using all four methods included in this study could be used to produce well-defined amplification products.
Discussion
RNA Quality
There was no clear difference among the four insect species regarding the quality of RNA extracted. However, there was variation among different RNA isolation methods for both the 280/260, 230/260 quality ratios, and the Agilent Bioanalyser results. The SV Total RNA isolation system and RNeasy Mini Kit extracted the highest quality RNA. RNA extracted using the Trizol reagent showed partially degraded RNA. The low 230/260 quality ratio could indicate a residue of phenol from the Trizol extraction. Although it is preferable for gene expression studies that the RNA is of a high quality, RIN above 5 have been noted as acceptable RNA for microarrays and gene expression studies as long as the product length is <400 bp ( Schoor et al. 2003 , Imbeaud et al. 2005 , Fleige and Pfaffl 2006 ). Based on the electropherogram graphs, it was assumed that high-quality RNA had a 28S peak twice the height of the 18S peak ( Green and Sambrook 2012 ). However, this is rarely achieved as the 28S RNA subunit degrades more rapidly compared with the 18S subunit ( Fleige and Pfaffl 2006 ). The major cause of this denaturing is a result of heating the RNA during the extraction procedure, which may denaturate the 28S ribosomal subunit to produce two fragments similar in size to that of the 18S RNA subunit ( Ishikawa and Newburgh 1972 , Winnebeck et al. 2010 ). SV Total RNA isolation system and the Trizol reagent both had heating steps during the procedure, which may account for the lower 28S peak ( Fig. 2 ). The complete lack of a 28S peak with Trizol extractions is most likely a result of the 15-min heating process at 60°C during the isolation procedure. The CTAB method did not produce RNA of a high quality or quantity. However, this method is costeffective, especially when large sample sizes are used, and can be used for simple downstream applications such as basic PCR for phylogenetic studies. For example, phylogenetics analysis based on RNA may be performed using lower quality RNA, and as such may not require stringent RNA isolation methods ( Kambhampati 1995 , Rokas and Holland 2000 , Kjer 2004 , Gambino et al. 2008 , Regier et al. 2010 , Podolska et al. 2011a , b ).
RNA Quantity
As spectrophotometer results may confuse contamination within the RNA sample as increased RNA concentration, only RNA samples with acceptable 280/260 and 230/260 ratio readings were considered. Thus, no CTAB RNA concentrations readings were analyzed. RNA extractions using Trizol reagent showed the highest concentrations of RNA. The RNeasy Mini Kit and SV Total RNA system extracted similar and consistent concentrations of RNA extracted.
DNA Contamination
As three of the methods showed high levels of DNA contamination, it demonstrated the importance for an additional DNase step following the RNA extraction, with the exception of the SV Total RNA isolation system.
PCR Application
Coposis et al. (2007) found that PCR results are not generally affected by RNA integrity, which would explain why PCR using low-quality RNA, produced using the CTAB-based protocol, were successful. However, the performance of more stringent applications, such as microarray analysis, is affected by RNA integrity. Therefore, depending on the downstream applications the choice of RNA isolation method would vary depending on the quantity and quality of RNA desired the cost of the isolation method, as well as the time taken to perform isolations.
Additional Considerations
Besides the quantity and quality of RNA isolated, an important consideration for selecting an appropriate RNA isolation method is cost. The commercial kits were significantly more expensive per isolation than the CTAB method. The CTAB method costs <$0.2 per isolation if more than 50 reactions were performed, due to the initial start up costs. The RNeasy Mini Kit and SV Total RNA isolation system were the most expensive at $7 and $6.2 per isolation, respectively. The TRIzol method, which cost $4.5 per isolation, was cheaper than these. However, isopropanol and ethanol were required but were not included in the kit, necessitating an additional cost of $20 for 1,000 reactions. In addition to the RNA extraction costs, a further DNase step was considered with a cost of $110 for 1,000 reactions. A further factor to consider when selecting an RNA extraction method is the time taken to perform the isolation. There was an inverse relationship between cost and isolation time for each RNA isolation method included in this study. The RNeasy Mini and SV Total RNA isolation system both averaged bench and total times of 40 min and the CTAB protocol required a bench time of 2.5 h. The safety of the isolation procedure for the user is also an important consideration. The CTAB and TRIzol isolation protocols rely on the use of chloroform and phenol respectively, which are both hazardous to users, thus making the RNeasy Mini Kit and SV Total RNA isolation system the safest isolation methods of the four that were tested.
All four isolation methods evaluated produced RNA of a similar quantity and quality regardless of insect species analyzed, indicating that these results may be applicable to larvae from a range of insect species. RNA isolated using the RNeasy Mini Kit was found to be of high quality and always of a consistent concentration, with the method also being the most robust and safe, since no harmful chemicals were used. The RNeasy Mini Kit does not require any heating steps, allowing for the quantity and quality of the RNA to be determined without any interference due to denaturation of the 28S ribosomal subunit. Even though, TRIzol reagent has been most commonly used for gene expression analysis in insects ( Lawrence et al. 2008 , Gatehouse et al. 2009 , Wu et al. 2011 , Xie et al. 2012 , Rinkevich and Scott 2013 ), which requires very high quality of RNA ( Fleige and Pfaffl 2006 , Copois et al. 2007 , Podolska et al. 2011a , b ), it was found that this method was not consistent and produced a high quantity but lower quality of RNA when compared with RNA isolated using the RNeasy Mini Kit and SV Total RNA isolation system. RNA isolations performed using the SV Total RNA system were of the highest quality and of consistently high quantity. In addition, RNA isolated using this method did not require an additional DNase digestion step, thus saving time and money. The SV Total RNA system is thus recommended for any downstream applications requiring a consistently good quality and quantity of RNA.
Acknowledgments
We would like to thank Tanya Pretorius, Claire Love, and Tamryn Marsberg for invaluable technical assistance, Martin Villet and Sean Moore for advice, Citrus Research International (experiment number 1049) and the Alexander von Humboldt Foundation for funding and Rhodes University for facilities.
References Cited
- Baiges I., Mas A. . 2003. . Good quality Vitis RNA obtained from an adapted DNA isolation protocol . Int. J. Vine Wine Sci. 37 : 59 – 61 . [Google Scholar]
- Baton L., Garver L., Xi Z., Dimopoulos G. . 2008. . Functional genomics studies on the innate immunity of disease vectors . Insect Sci. 15 : 15 – 27 . [Google Scholar]
- Copois V., Bibeau F., Bascoul-Mollevi C., Salvetat N., Chalbos P., Bareil C., Candeil L., Fraslon C., Conseiller E., Granci V., et al. . 2007. . Impact of RNA degradation on gene expression profiles: assessment of different methods to reliably determine RNA quality . J. Biotechnol. 127 : 549 – 59 . [DOI] [PubMed] [Google Scholar]
- Dong J., Dunstan D. I. . 1996. . A reliable method for extraction of RNA from various conifer tissues . PlantCell Rep. 15 : 516 – 521 . [DOI] [PubMed] [Google Scholar]
- Fleige S., Pfaffl M. W. . 2006. . RNA integrity and the effect on the real-time qRT-PCR performance . Mol. Aspects Med. 27 : 126 – 139 . [DOI] [PubMed] [Google Scholar]
- Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. . 1994. . DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates . Mol. Marine Biol. Biotechnol. 3 : 294 – 299 . [PubMed] [Google Scholar]
- Gambino G., Perrone I., Gribaudo I. . 2008. . A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants . Phytochem. Anal. 19 : 520 – 525 . [DOI] [PubMed] [Google Scholar]
- Gatehouse H. S., Poulton J., Markwick N. P., Gatehouse L. N., Ward V. K., Young V. L., Luo Z., Schaffer R., Christeller J. T. . 2009. . Changes in gene expression in the permissive larval host lightbrown apple moth ( Epiphyas postvittana , Tortricidae) in response to EppoNPV (Baculoviridae) infection . Insect Mol. Biol. 18 : 635 – 648 . [DOI] [PubMed] [Google Scholar]
- Götz M., Popovski S., Kollenberg M., Gorovits R., Brown J. K., Cicero J. M., Czosnek H., Winter S., Ghanim M. . 2012. . Implication of Bemisia tabaci heat shock protein 70 in Begomovirus-whitefly interactions . J. Virol. 86 : 13241 – 13252 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R., Sambrook J. . 2012. . Molecular cloning : a laboratory manual, 4th ed. Harbor Laboratory Press . Cold Spring Harbor, N.Y . [Google Scholar]
- Imbeaud S., Graudens E., Boulanger V., Barlet X., Zaborski P., Eveno E., Mueller O., Schroeder A., Auffray C. . 2005. . Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces . Nucleic Acids Res. 33 : e56 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Newburgh R. W. . 1972. . Studies of the thermal conversion of 28 S RNA of Galleria mellonella (L.) to an 18 S product . J. Mol. Biol. 64 : 135 – 144 . [DOI] [PubMed] [Google Scholar]
- Kambhampati S. 1995. . A phylogeny of cockroaches and related insects based on DNA sequence of mitochondrial ribosomal RNA genes . Proc. Natl Acad. Sci. U. S. A. 92 : 2017 – 2020 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjer K. M. 2004. . Aligned 18S and insect phylogeny . Syst. Biol. 53 : 506 – 514 . [DOI] [PubMed] [Google Scholar]
- Lawrence S. D., Novak N. G., Ju C. J-T, Cooke J. E. K. . 2008. . Potato, Solanum tuberosum , defense against Colorado potato beetle, Leptinotarsa decemlineata (Say): microarray gene expression profiling of potato by Colorado potato beetle regurgitant treatment of wounded leaves . J. Chem. Ecol. 34 : 1013 – 1025 . [DOI] [PubMed] [Google Scholar]
- Li H., Bouwer G. . 2012. . The larvicidal activity of Bacillus thuringiensis Cry proteins against Thaumatotibia leucotreta (Lepidoptera: Tortricidae) . Crop Protect. 32 : 47 – 53 . [Google Scholar]
- Noriega F. G., Wells M. A. . 1993. . A comparison of three methods for isolation of RNA from mosquitoes . Insect Mol. Biol. 2 : 21 – 24 . [DOI] [PubMed] [Google Scholar]
- Ogaugwu C. E., Wimmer E. A. . 2013. . Molecular cloning and expression of nanos in the Mediterranean fruit fly, Ceratitis capitata (Diptera: Tephritidae) . Gene Expr. Patterns 13 : 183 – 188 . [DOI] [PubMed] [Google Scholar]
- Oppert B., Martynov A. G., Elpidina E. N. . 2012. . Bacillus thuringiensis Cry3Aa protoxin intoxication of Tenebrio molitor induces widespread changes in the expression of serine peptidase transcripts . Comp. Biochem. Physiol. Part D Genomics Proteomics 7 : 233 – 242 . [DOI] [PubMed] [Google Scholar]
- Podolska A., Kaczkowski B., Kamp Busk P., Søkilde R., Litman T., Fredholm M., Cirera S. . 2011a. . MicroRNA expression profiling of the porcine developing brain . PLoS One. 6 : e14494 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolska A., Kaczkowski B., Litman T., Fredholm M., Cirera S. . 2011b. . How the RNA isolation method can affect microRNA microarray results . Acta Biochim. Pol. 58 : 535 – 540 . [PubMed] [Google Scholar]
- Regier J. C., Shultz J. W., Zwick A., Hussey A., Ball B., Wetzer R., Martin J. W., Cunningham C. W. . 2010. . Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences . Nature 463 : 1079 – 1083 . [DOI] [PubMed] [Google Scholar]
- Ridgeway J. A., Midgley J. M., Collett I. J., Villet M. H. . 2014. . Advantages of using development models of the carrion beetles Thanatophilus micans (Fabricius) and T. mutilatus (Castelneau) (Coleoptera: Silphidae) for estimating minimum post mortem intervals, verified with case data. Int. J . Legal Med. 128 : 207 – 220 . [DOI] [PubMed] [Google Scholar]
- Rinkevich F. D., Scott J. G. . 2013. . Limitations of RNAi of α6 nicotinic acetylcholine receptor subunits for assessing the in vivo sensitivity to spinosad . Insect Sci. 20 : 101 – 108 . [DOI] [PubMed] [Google Scholar]
- Rokas A., Holland P. . 2000. . Rare genomic changes as a tool for phylogenetics . Trends Ecol. Evol. 15 : 454 – 459 . [DOI] [PubMed] [Google Scholar]
- Schoor O., Weinschenk T., Hennenlotter J., Corvin S., Stenzl A., Rammensee H.-G., Stevanović S. . 2003. . Moderate degradation does not preclude microarray analysis of small amounts of RNA . BioTechniques 35 : 1192 – 1196 , 1198–1201 . [DOI] [PubMed] [Google Scholar]
- Talekar N. S., Shelton A. M. . 1993. . Biology, ecology and management of the diamondback moth . Annu. Rev. Entomol. 38 : 275 – 301 . [DOI] [PubMed] [Google Scholar]
- Tattersall E.A.R., Ergul A., Alkayal F., Deluc L., Cushman J. C., Cramer G. R. . 2005. . Comparison of methods for isolating high-quality RNA from leaves of grapevine . J. Enol. 4 : 400 – 407 . [Google Scholar]
- Winnebeck E., Millar C., Warman G. . 2010. . Why does insect RNA look degraded? J. Insect Sci . 10 : 159 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P., Han S., Chen T., Qin G., Li L., Guo X. . 2013. . Involvement of microRNAs in infection of silkworm with Bombyx mori cytoplasmic polyhedrosis virus (BmCPV) . PLoS One 8 : e68209 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P., Wang X., Qin G., Liu T., Jiang Y.-F., Li M.-W., Guo X.-J. . 2011. . Microarray analysis of the gene expression profile in the midgut of silkworm infected with cytoplasmic polyhedrosis virus . Mol. Biol. Rep. 38 : 333 – 341 . [DOI] [PubMed] [Google Scholar]
- Xie W., Lei Y., Fu W., Yang Z., Zhu X., Guo Z., Wu Q., Wang S., Xu B., Zhou X., et al. . 2012. . Tissue-specific transcriptome profiling of Plutella xylostella third instar larval midgut . Int. J. Biol. Sci. 8 : 1142 – 1155 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Li H.-C., Miao X.-X. . 2013. . Feasibility, limitation and possible solutions of RNAi-based technology for insect pest control . Insect Sci. 20 : 15 – 30 . [DOI] [PubMed] [Google Scholar]