Summary
CYCLING DOF FACTOR 1 (CDF1) and its homologs play an important role in the floral transition by repressing the expression of floral activator genes such as CONSTANS (CO) and FLOWERING LOCUS T (FT) in Arabidopsis. The day-length specific removal of CDF1-dependent repression is a critical mechanism in photoperiodic flowering. However, the mechanism by which CDF1 represses CO and FT transcription remained elusive. Here we demonstrate that Arabidopsis CDF proteins contain non-EAR motif-like conserved domains required for interaction with the TOPLESS (TPL) co-repressor protein. This TPL interaction confers a repressive function on CDF1, as mutations of the N-terminal TPL binding domain largely impair the ability of CDF1 protein to repress its targets. TPL proteins are present on specific regions of the CO and FT promoters where CDF1 binds during the morning. In addition, TPL binding increases when CDF1 expression is elevated, suggesting that TPL is recruited to these promoters in a time-dependent fashion by CDFs. Moreover, reduction of TPL activity induced by expressing a dominant negative version of TPL (tpl-1) in phloem companion cells results in early flowering and a decreased sensitivity to photoperiod in a manner similar to a cdf loss-of-function mutant. Our results indicate that the mechanism of CDF1 repression is through the formation of a CDF-TPL transcriptional complex, which reduces the expression levels of CO and FT during the morning for seasonal flowering.
Keywords: Arabidopsis thaliana, photoperiodic flowering, transcriptional repression. CONSTANS, FLOWERING LOCUS T
Introduction
The transition from a juvenile to a reproductive stage of development is a critical regulatory process in plants. For annual flowering plants for which a single reproductive event defines the life cycle, timing of this developmental transition is especially important for the fitness of a given individual (Andrés and Coupland, 2012; Johansson and Staiger, 2015; Song et al., 2015). The progression of the axial tilt of Earth’s path throughout the year leads to a predictable yearly flux in temperature and in light duration (photoperiod). This change is especially dramatic at increasing distances from the equator (Thomas and Vince-Prue, 1996). For this reason, many plants measure the photoperiod change to predict the period in which to begin reprogramming from the vegetative to the reproductive phase of the life cycle.
In Arabidopsis thaliana, for which increasing day length (long days) is inductive towards flowering, the signal that initiates this process is the expression of the mobile protein FLOWERING LOCUS T (FT) (Corbesier et al., 2007; Jaeger and Wigge, 2007; Jaeger et al., 2013). FT protein is produced in the phloem companion cells, and transported to the shoot apex (Corbesier et al., 2007; Mathieu et al., 2007; Notaguchi et al., 2008; Liu et al., 2012). There it interacts with downstream regulatory factors to commit the shoot apical meristem to a reproductive cell fate (Abe et al., 2005; Wigge et al., 2005; Taoka et al., 2011; Jaeger et al., 2013). Temporal and developmental regulation of FT transcription is critical for a proper timing of flowering.
For day-length information to be integrated into the expression of FT, several factors are required. The transcription factor CONSTANS (CO) is a primary activator of FT in long days (Suárez-López et al., 2001). The activity of CO results in upregulation of FT toward the dusk of long days. This upregulation is due in part to the stabilization of CO protein by the blue-light photoreceptor FLAVIN BINDING, KELCH REPEAT, F-BOX 1 (FKF1) and GIGANTEA (GI) complex in the long-day afternoon (Song et al., 2012; Song et al., 2014). The stabilization of the CO protein is also accomplished through the action of TIMING OF CAB EXPRESSION 1 (TOC1), and PSEUDO RESPONSE REGULATOR 5 (PRR5), PRR7, and PRR9 proteins, which physically interact with CO and prevent its degradation (Hayama et al., 2017).
The constraint of FT expression to only the afternoon of long days is accomplished through the action of CYCLING DOF FACTOR (CDF) transcription factors. CDF1, CDF2, CDF3, and CDF5 act redundantly to repress the expression of CO and FT (Imaizumi et al., 2005; Fornara et al., 2009). CDF1, CDF2, CDF3 and CDF5 are expressed during the mornings, and their expression is a direct output of the circadian clock (Imaizumi et al., 2005; Fornara et al., 2009). The core clock components, CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), positively act in CDF expression during the late night and morning (Nakamichi et al., 2007; Niwa et al., 2007). CDF transcription is directly inhibited after zeitgieber time (ZT) 4 through the action of PRR5, PRR7, and PRR9 throughout the remainder of the day (Nakamichi et al., 2007). During the afternoon of long days, FKF1-GI complex degrades CDF proteins through ubiquitin mediated degradation (Sawa et al., 2007). This regulation of CDFs, both by the endogenous circadian clock, and through blue light signaling, constrains the activity of these CO and FT repressors to the morning. This results in generating a day-length specific time-window before dusk in which FT can be specifically activated in long days.
CDF homologs in other flowering plant species appear to have largely conserved function. LATE BLOOMER 2 (LATE2) in Pisum sativum and StCDF1 in Solanum tuberosum function as repressors of flowering and tuberization, respectively, in response to photoperiod (Kloosterman et al., 2013; Ridge et al., 2016). CDF proteins belong to the larger family of plant specific DOF (DNA binding with one finger) transcription factors (Yanagisawa, 2002; Yanagisawa, 2004). CDFs compose a distinct subclade of the DOF protein family in Arabidopsis, of which 37 are present in the genome (Yanagisawa, 2002). The DOF DNA binding domain belongs to the C2C2-type zinc finger domain, but is unusual in that only a single zinc coordinating “finger” is present on each protein to assist in DNA binding (Yanagisawa, 2002). This single finger complicates the prediction of DOF cis-elements in target promoters, as the consensus binding sequence of “AAAG” is widely found in most promoters. At least at the CO promoter in Arabidopsis, the presence of a tandemerized DOF binding site upstream of the transcription start site is critical for CDF-dependent repression (Rosas et al., 2014). Potentially, this multimerization could be a mechanism through which DOF target loci are regulated.
TOPLESS (TPL) proteins are common mediators of transcriptional repression in a variety of molecular pathways in Arabidopsis (Liu and Karmarkar, 2008). These include auxin, jasmonic acid, and ethylene hormone signaling, as well as several developmental pathways, including the circadian clock and flowering time regulation (Liu and Karmarkar, 2008; Wang et al., 2013). The originally characterized dominant negative mutant tpl-1 shows a pleiotropic phenotype that strikingly displays a lack of apical cell fate under higher temperatures (Long et al., 2006). TPL and its homologs TOPLESS RELATED 1 (TPR1), TPR2, TPR3, and TPR4 are important co-repressors that assist transcription factors in mediating repression at their target loci (Causier et al., 2012). This mechanism is perhaps most clear for TPL’s role in auxin signaling, where TPL and INDOLE-3-ACETIC ACID INDUCIBLE (IAA) transcription factors repress auxin inducible loci until the IAAs are degraded in response to auxin (Gray et al., 2001). Although the direct mechanism of TPL repression is unclear, TPL interacts with several histone deacetylase (HDAC) enzymes, as well as the CDK8/HUA ENHANCER3 (HEN3) mediator complex module (Wang et al., 2013; Ito et al., 2016). A combination of these interactions likely facilitates the direct blocking and/or accessibility of target promoters to transcriptional activators (Ito et al., 2016). The crystal structure of the N-terminal domain of TPL has been resolved; similar to how the analogous protein Groucho (Gro) functions in Drosophila melanogaster, TPL functions as a tetrameric complex (Ke et al., 2015). The LisH (Lis homology domain) and CtLH (C-terminal to the LisH motif) domains of TPL are important for transcription factor binding. The sites on TPL that EAR (ethylene-responsive element binding factor-associated amphiphilic repression) motif containing factors bind to were identified, but not for other interaction motifs (Ke et al., 2015). Several transcription factors that bind to TPL in a yeast two-hybrid assay are involved in the regulation of FT transcription, including TARGET OF EAT 1 (TOE1) and TOE2, SCHLAFMÜTZE (SMZ), SCHNARCHZAPFEN (SNZ) and TEMPRANILLO 1 (TEM1) and TEM2 (Schmid et al., 2003; Causier et al., 2012; Osnato et al., 2012; Zhang et al., 2015). TOE1 and TOE2 are involved in miRNA172 age-dependent regulation of flowering time (Jung et al., 2007). TEM1, and TEM2 are involved in FT regulation during the afternoon of long days, are antagonistic to CO activity, and are downstream of GI and gibberellin hormones in the photoperiodic flowering pathway (Castillejo and Pelaz, 2008; Osnato et al., 2012). Recently, it was also found that a B-box containing small protein MICROPROTEIN 1A (miP1a), and miP1b, interact with both TPL and CO during the mornings of long days to reduce CO activity (Graeff et al., 2016).
Heretofore, the mechanism of CDF-dependent repression of the CO and FT genes was unknown. We enumerate here that CDF1 recruits TPL to reduce CO and FT transcription during the morning. We show that CDF1 and TPL together make a protein complex that likely forms mainly when CDFs are abundant. Additionally, our analysis of CDF1 and TPL interacting domains shows that the regions of CDF1 that interact with TPL are needed for full repressive activity. In long days, TPL associates with the CO and FT promoters mainly in the morning. Elevating the amount of CDF1 increases TPL binding even in the afternoon. Reduction of TPL activity specifically in phloem companion cells leads to early flowering. We demonstrate that TPL activity through CDFs during the mornings of both long days and short days is critical for constricting CO and FT expression to enable the correct interpretation of seasonal information by the plant.
Results
CDF proteins contain a conserved motif at their N-termini, which is important for binding to TPL
CDF1 and related CDF transcription factors all function as repressors of CO transcription and flowering time in Arabidopsis (Fornara et al., 2009). To explore the possible mechanisms through which CDF transcription factors are able to confer repression on CO and FT transcription, we first looked for regions of high conservation at the amino acid level, as this would imply a retained function. We constructed an amino acid alignment of CDF proteins from Arabidopsis, functionally characterized CDFs from other plant species, as well as CDF-related DOF transcription factors from a variety of green plant lineages (Lijavetzky et al., 2003; Yang et al., 2010; Hernando-Amado et al., 2012; Sugiyama et al., 2012; Kloosterman et al., 2013; Corrales et al., 2014; Lucas-Reina et al., 2015; Kang et al., 2016; Ridge et al., 2016). We found several stretches of amino acid conservation in CDF proteins (Figure S1). In addition to the DOF DNA-binding domain, we found a short conserved sequence (IKLFG) at the N-terminal ends of CDFs and nearly all CDF related DOF transcription factors (Figure S1). This motif exactly overlaps with a previously described (R/K)LFGV motif for binding to the co-repressor protein TPL (Causier et al., 2012). The IKLFG residues are highly conserved among CDF-like DOF proteins found throughout land plants (Figure S1) (Cai et al., 2013; Ma et al., 2015; Kang et al., 2016). While not the canonical EAR motif (the LxLxL amphiphilic domain which composes the majority of TPL interactors), CDFs possess the (R/K)LFGV motif, similar to TEM1 protein and present in 12.3 percent of the established TPL interactome (Causier et al., 2012). Although no DOF domain transcription factors have been reported as being likely TPL interactors, we hypothesized that CDFs bring TPL to target loci to repress transcription.
We next investigated if these residues were critical for interaction with TPL. We first used a yeast two-hybrid (Y2H) approach to see if CDF1 interacted with TPL, and if corresponding mutations in the N-terminal motif of CDF1 impaired complex formation. We found that CDF1 interacted with TPL, but the incorporation of either a deletion of the N-terminal end residues (amino acid positions: 1–19, designated as CDF1-ΔN) including the IKLFG motif, or a replacement of the interior three residues into alanine (A) in the motif (IKLFG to IKAAA; designated as CDF1-mut), was sufficient to prevent the interaction (Figure 1a,b). In either case, the mutated CDF1 proteins could interact with the N-terminal fragment of the GIGANTEA (GI) protein, which binds to the CDF1 C-terminus (Kloosterman et al., 2013). This indicates that these CDF1 variant proteins were expressed in this system, and were still able form complexes with GI. To verify the CDF1-TPL interaction results in planta, we used both Bimolecular Fluorescent Complementation (BiFC) and Co-immunoprecipitation (Co-IP) experiments. CDF1 bound to TPL in BiFC in epidermal cells of Nicotiana benthamiana, but the inclusion of these mutations in the CDF1 N-terminus abolished the interaction with TPL (Figure 1c,S2a). We confirmed that both CDF1-ΔN and CDF1-mut proteins were localized to the nucleus, similar to non-mutated CDF1 (Figure S2b). In addition, reciprocal Co-IP of TPL protein transiently co-expressed in tobacco leaves also showed a recovery of CDF1 protein bound to TPL (Figure 1d). These results together indicate that the N-terminal conserved motif of CDF1 functions as the binding site to TPL.
Figure 1. CDF1 and TPL form a protein complex through a conserved binding motif located at the N-terminus of CDF1.
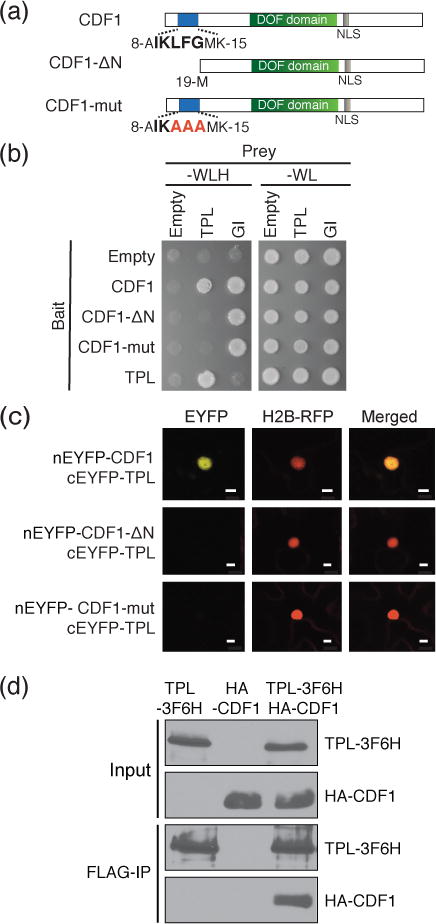
(a) Schematic representation of CDF1 full length protein and the N-terminal CDF variants used in this study. The N-terminal amino acid sequences (IKLFG) conserved in CDF family proteins (Figure S1), which overlap with the TPL binding motif (R/KLFGV), are shown in bold. The mutated amino acids in CDF1-mut protein are shown in red. The relative positions of DOF DNA binding domain (DOF domain) and nuclear localization signal (NLS) are indicated. (b) Y2H analysis of CDF1-TPL protein interaction. The –WLH plate shows the interaction of bait and prey proteins, while the –WL plate shows the presence of both bait and prey constructs. N-terminus of GI protein was used as a positive control for CDF1 variants. (c) BiFC interaction analysis in transiently transfected N. benthamiana leaves between full-length of CDF1 protein, CDF1-ΔN and CDF1-mut variants, and TPL protein. Histone H2B-RFP was used to determine the position of the nucleus in the same cell. Scale bars show 10 μm. (d) TPL-CDF1 interaction using coimmunoprecipitation assay of transiently transfected N. benthamiana leaves.
The TPL interaction motif at the N-termini of CDF family proteins is important for repression of the flowering time genes CO and FT.
To investigate whether the N-terminal motif in CDFs is required for the repressive function of the transcription factor, we generated transgenic plants that overexpress an epitope-tagged CDF1 protein (35S:CDF1-3F6H) or two mutated variants of tagged CDF1 proteins (35S:CDF1-ΔN-3F6H and 35S:CDF1-mut-3F6H) (Figure 1a). We tested the flowering time response of transgenic lines with similar expression levels of CDF1 transcripts (Figure 2a,d,e), and found that 35S:CDF1-3F6H overexpression plants were late-flowering in long days compared to WT as previously shown (Imaizumi et al., 2005). The flowering time of 35S:CDF1-ΔN-3F6H and 35S:CDF1-mut-3F6H lines in which similar or higher levels of CDF1 transcripts was expressed (Figure 2b,c) were not significantly different from that of WT plants, although we noticed that the size of 35S:CDF1-ΔN-3F6H and 35S:CDF1-mut-3F6H rosettes seemed to be slightly smaller than wild-type plants (Figure 2d,e). This result suggests that these mutations at least attenuated the repressive function of the protein for floral induction. Flowering time among all of the lines under short-day photoperiods was similar, although 35S:CDF1-3F6H lines and 35S:CDF1-mut-3F6H #30 were slightly delayed compared to WT plants (Figure S3).
Figure 2. Removal of TPL interacting residues attenuates CDF1 repressor function.
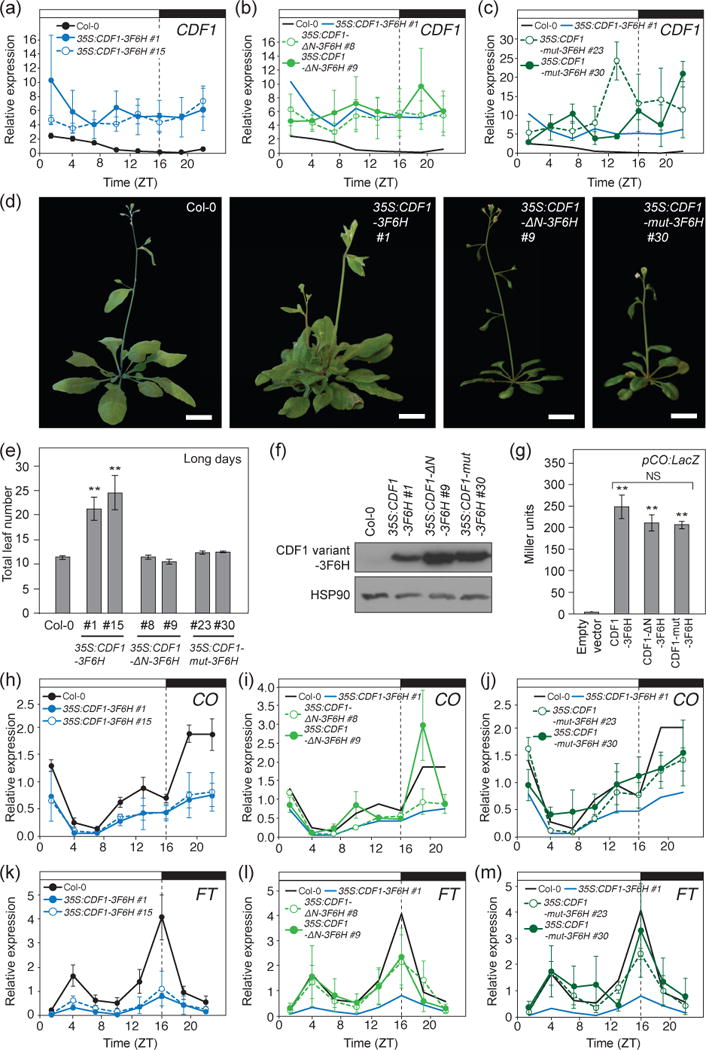
(a, b, and c) CDF1 expression levels in Col-0, 35S:CDF1-3F6H (a), 35S:CDF1-ΔN-3F6H (b), and 35S:CDF1-mut-3F6H plants (c) grown in long days for 14 days. Means +/− SEM were calculated from four independent experiments. The traces of diurnal CDF1 expression changes in Col-0 and 35S:CDF1-3F6H #1 shown in (a) are included in (b) and (c) for comparison. (d) Representative images of WT, 35S:CDF1-3F6H, 35S:CDF1-ΔN-3F6H, and 35S:CDF1-mut-3F6H plants at flowering in long days. Scale bars=2 cm. (e) Quantification of flowering time by total leaf number at bolting from (d) under long days. Means +/- SEM were calculated from N=16 individuals. **P < 0.01 (one-tailed t test). (f) CDF1-3F6H, CDF1-ΔN-3F6H, and CDF1-mut-3F6H protein expression at ZT4 time point in 14-day-old long-day grown transgenic seedlings. HSP90 served as a loading control. (g) Y1H analysis of CDF1-3F6H, CDF1-ΔN-3F6H, CDF1-mut-3F6H proteins binding to a 500 bp region of the CO promoter (pCO:LacZ), which contains a previously characterized DOF binding cis-element repeats. LacZ activity is displayed in Miller units. Means +/− SEM were calculated from three independent experiments. **P < 0.01 (one-tailed t test), NS = non-significant. (h, i, and j) Gene expression analysis of CO in Col-0, 35S:CDF1-3F6H (h), 35S:CDF1-ΔN-3F6H (i), and 35S:CDF1-mut-3F6H plants (j). Plants were grown in long days for 14 days. Experiments were repeated four times independently, and the means +/- SEM derived from four experiments are shown. The traces of diurnal CO expression changes in Col-0 and 35S:CDF1-3F6H #1 shown in (h) are included in (i) and (j) for comparison. (k, l, and m) Gene expression analysis of FT in Col-0, 35S:CDF1-3F6H (k), 35S:CDF1-ΔN-3F6H (l), and 35S:CDF1-mut-3F6H plants (m). Plants were grown in long days for 14 days. Experiments were repeated four times independently, and the means +/− SEM derived from four experiments are shown. The traces of diurnal FT expression changes in Col-0 and 35S:CDF1-3F6H #1 shown in (h) are included in (i) and (j) for comparison.
To eliminate the possibility that the loss of floral repression in the CDF1 variant overexpressors was caused by destabilization of the proteins, we analyzed the expression levels of these CDF1 variants. All CDF1 variant proteins in the transgenic lines with similar expression levels of CDF1 transcripts were expressed at levels slightly higher than non-mutated CDF1 protein during the morning (Figure 2f). This result clearly indicates that the lack of flowering phenotype in 35S:CDF1-ΔN-3F6H and 35S:CDF1-mut-3F6H lines is not due to the lack of the expression of CDF1-ΔN-3F6H and CDF1-mut-3F6H proteins.
We also confirmed that these mutations did not affect the ability to bind to DNA. We used a yeast one-hybrid (Y1H) approach to ascertain if CDF1-ΔN and CDF1-mut could still bind to a 500 bp fragment of the CO promoter, which contains a cluster of DOF binding sites, where CDF1 binds (Imaizumi et al., 2005). We found that the mutated CDF1 proteins fused with the GAL4 activation domain could activate the LacZ reporter similar to the normal CDF1 protein, indicating that these mutations did not interfere with their DNA binding activities (Figure 2g).
We then analyzed whether CO or FT expression was altered in 35S:CDF1-ΔN-3F6H or 35S:CDF1-mut-3F6H plants compared to Wild-type (WT) and 35S:CDF1-3F6H. Under long-day photoperiods, 35S:CDF1-3F6H have a reduction in CO mRNA in the afternoon and night, and FT mRNA levels are lower throughout the day (Figure 2h,k). In 35S:CDF1-ΔN-3F6H and p35S:CDF1-mut-3F6H plants, levels of CO transcript were slightly lower but more similar to that in WT throughout the day, comparing against those in 35S:CDF1-3F6H plants (Figure 2k,j). Overall FT expression patterns in 35S:CDF1-ΔN-3F6H and 35S:CDF1-mut-3F6H lines showed a similar trend throughout the day to that in WT, although a peak FT expression in those lines might be slightly lower (Figure 2l,m). These results suggest that repressive activity of CDF1 is largely ascribed to interaction with the TPL co-repressor through the N-terminal motif of CDF1. Taken together, the results imply that the conserved N-terminal TPL binding site among CDFs is important to confer on CDF1 its repression ability.
TPL associates with the CO and FT promoters during the mornings of long days and in a CDF1 expression dependent manner
If CDF proteins are concerting repressive activity at the CO and FT promoters through recruiting TPL complex during the morning, when CDFs are abundant, we hypothesized that TPL protein should also exist at these loci at the same time as CDF1 (Sawa et al., 2007; Song et al., 2012). We analyzed whether TPL binds to the CO promoter especially in the morning. We performed ChIP-qPCR assays using TPL:TPL-3HA transgenic line in which TPL-3HA cDNA was expressed under its native promoter (Wang et al., 2013). We harvested the samples at ZT4 and ZT16 time points, which correspond to an abundant CDF1 protein time point, and the peak level of FT expression (when CDF1 is low), respectively. Our results showed that TPL protein strongly associated with the CO promoter region at ZT4 (Figure 3a). The binding occurred specifically at amplicons within approximately the first 500 bp upstream of the transcription start site (amplicons 5 and 6) as well as several sites further upstream (amplicons 1 and 3) (Figure 3a). The −500 bp upstream region contains the cluster of DOF binding sites, where CDF1 binds (Imaizumi et al., 2005), and has also been shown to be a highly conserved region among CO homologs in many Arabidopsis-related Brassicaceae species (Simon et al., 2015). Therefore, it likely represents bona fide CDF1 binding sites on the CO promoter. This assertion is also supported by previous CDF1 ChIP experiments (Sawa et al., 2007). Nevertheless, it is also possible that other factors may contribute to recruit TPL to these sites in the CO promoter. We found an insignificant amount of binding of TPL to the CO promoter (except at amplicon 6) during the ZT16 time point (Figure 3a). Similarly at the FT promoter, we found TPL binding to an amplicon upstream of the transcription start site at −800 bp, as well as binding to the 3′-UTR region only in the samples harvested at ZT4 (Figure 3b). CDF1 has been found to bind to near the transcription start site (amplicons 12 and 13), and possibly around amplicon 8 (Song et al., 2012). This indicates that other factors may also recruit TPL to the FT promoter. We found little binding of TPL to the FT promoter at the ZT16 time point (Figure 3b).
Figure 3. TPL associates with the CO and FT promoter regions during the morning in a CDF1-dependent manner in vivo.
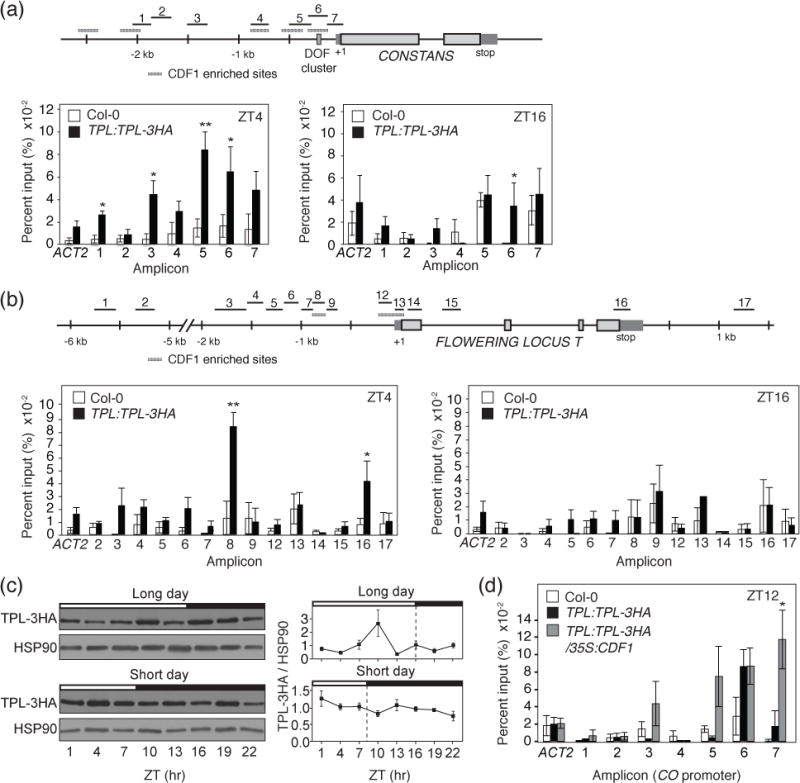
(a and b) ChIP assay-showing TPL binding to regions of the CO and FT genomic locus at ZT4 and ZT16 from 14-day-old TPL:TLP-3HA and Col-0 plants grown under long-day photoperiods. The schematic diagrams of positions of each ChIP amplicon scattered in CO and FT promoter regions are shown. The results are means and +/− SEM calculated from four independent experiments. Col-0 plants were used as negative controls. *P < 0.05,**P < 0.01 (one-tailed t test). Hatched grey boxes indicate regions of CDF1 enrichment from Sawa et al., 2007 and Song et al., 2012, respectively. (c) Daily protein expression profiles of TPL in long days and short days. 14-day-old TPL:TLP-3HA plants were used for experiments. Experiments were performed three times independently. Means +/− SEM were calculated from ratios of signal strength of TPL proteins and HSP90 loading control. (d) ChIP experiment for TPL binding to the CO genomic locus at ZT12, in Col-0, TPL:TPL:3HA, and 35S:CDF1 TPL:TPL:3HA plants. All calculations were performed as in (a) and (b). Plants were 14 days old and grown under long-day photoperiods.
TPL is widely expressed throughout development (Wei et al., 2015; Espinosa-Ruiz et al., 2017); we wondered what mechanism led TPL protein to be differentially recruited to both CO and FT promoter regions between morning and night. A previous study demonstrated that TPL protein levels changed under 12-hour light/12-hour dark conditions (Wang et al., 2013). Therefore, we analyzed the protein expression profiles of TPL in long days and short days using TPL:TPL-3HA plants. TPL was expressed at similar levels throughout the day in both day-length conditions, although we saw a slight increase in TPL levels around ZT10 in long days (Figure 3c). This result shows that the expression levels of TPL protein did not correlate with the degrees of association of TPL to specific regions of CO and FT loci between morning and night.
As CDF1 physically interacts with TPL, and because CDF1 binds to some overlapping DNA regions (such as CO amplicons 1, 5, and 6) during the morning, we hypothesized that the morning enriched binding of TPL to the CO and FT promoters was at least in part due to CDF1 binding to these promoter regions. To assess this possibility, we analyzed the binding of TPL to the CO promoter in plants with higher levels of CDF1 of which CO is repressed throughout the day. We used TPL:TPL-3HA and TPL:TPL-3HA/35S:CDF1 plants with similar levels of TPL-3HA protein (Figure S4a,b). We chose the ZT12 time point, as CDF1 levels in WT should be low due to degradation through the FKF1-GI complex, but in the 35S:CDF1 line CDF1 levels are still sufficiently high enough to repress CO and FT (Imaizumi et al., 2005). We saw increased enrichment of TPL in 35S:CDF1 background around amplicon 7, within the first 500 bp region upstream of the transcription start site (Figure 3d), where CDF1 was found to bind (Sawa et al., 2007). This data suggests that the recruitment of TPL to specific regions of CO and FT promoter is likely conveyed by the binding of CDF protein that widely fluctuates in expression level throughout the day.
Loss of TPL function in phloem companion cells causes early flowering under long-day and short-day conditions
If CDF transcription factors repress target loci through TPL recruitment, we reasoned that the loss of TPL function should mimic the loss of CDFs. Dominant negative tpl-1 mutants have been utilized in a variety of studies to observe the function of TPL and related TPRs (Long et al., 2006). Although the molecular function of the tpl-1 mutation is unknown, its presence seems to interfere with the function of normally expressed TPL and TPRs, resulting in a dominant negative phenotype (Long et al., 2006). However, the pleiotropic nature of the tpl-1 phenotype, and in particular the loss of meristematic function and lateral organ defects make detailed analysis of photoperiodic flowering time challenging. In order to more specifically characterize the role of TPL in flowering time regulation, we aimed to lessen the function of TPL and TPRs only in leaf phloem tissues where CDFs, CO and FT are expressed. We generated transgenic lines that expressed tpl-1 mutant protein from the control of a SUCROSE-PROTON SYMPORTER 2 (SUC2) phloem companion cell-specific promoter (SUC2:HA-tpl-1). To our knowledge, this is this first study that utilizes a tissue-specific promoter driven tpl-1 transgenic line to study the effect of reduction of TPL/TPR function in specific tissues. The cdf1,2,3,5 mutant (hereafter referred to as the cdfq mutant) is very early flowering in both long days and short days, due to an upregulation of CO and FT during the morning (Fornara et al., 2009). We anticipated that in SUC2:HA-tpl-1 plants, CDFs would be unable to recruit functional TPL protein, and would thus be unable to repress their target loci. In other words, we predicted that a flowering time phenotype of SUC2:HA-tpl-1 likely resembles that of cdfq.
We first analyzed the flowering time phenotype of these plants. Similar to cdfq mutants, SUC2:HA-tpl-1 plants flower early in long days and especially short days (Figure 4). This result implies that having proper function of TPL/TPRs in phloem is important for photoperiodic flowering regulation.
Figure 4. Expressing a dominant negative form of TPL in phloem companion cells phenocopies cdfq mutants, and lessens the late flowering phenotype of CDF1 overexpressors.
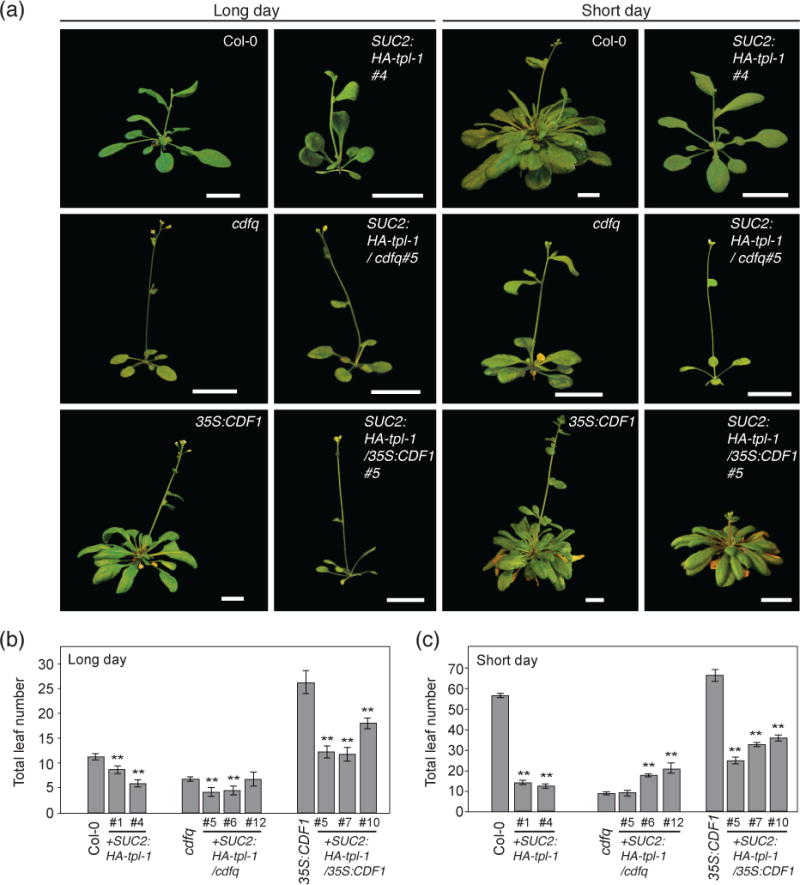
(a) Representative images of SUC2:HA-tpl-1, SUC2:HA-tpl-1/cdfq, and SUC2:HA-tpl-1/35S:CDF1 plants under long-day and short-day photoperiods at flowering. Scale bars=2 cm. (b and c) Quantification of flowering time by total leaf number at bolting from (a) under long days (b) and short days (c). Means +/− SEM were calculated from N=16 individuals. **P < 0.01 (one-tailed t test).
Because TPL can interact with several other repressors of FT transcription, we wanted to check if the cdfq mutant phenotype was synergistic with SUC2:HA-tpl-1. We expected that in cdfq mutant backgrounds, if CDFs were genetically epistatic to TPL, then the flowering time phenotype of SUC2:HA-tpl-1/cdfq would not be additive. We generated SUC2:HA-tpl-1/cdfq lines with similar to or higher levels of TPL (tpl-1) transcripts than that in SUC2:HA-tpl-1 (Figure S4c). We saw a slight hastening of flowering time under long-day conditions in several SUC2:HA-tpl-1/cdfq lines (which have higher amount of TPL (tpl-1) transcripts). Under short-day conditions, SUC2:HA-tpl-1/cdfq lines were the same or later flowering than cdfq (Figure 4). This suggests that any additional contribution from other TPL-interacting factors likely occurs under long-day conditions, however with flowering occurring at four to five total leaf number in long days, these plants may be encroaching on a developmental minimum of leaves before inflorescence initiation can occur. In short days, as several SUC2:HA-tpl-1/cdfq lines showed slightly later flowering than cdfq, TPL may work with other factors that also influence flowering time.
Conversely, we wanted to investigate if the expression of tpl-1 in leaf phloem could mitigate the flowering effect caused by the overexpression of CDF1. If CDF1 represses its target loci mainly through TPL in leaf phloem, then introgression of the SUC2:HA-tpl-1 transgene into a 35S:CDF1 genetic background should severely lessen the late flowering phenotype of the 35S:CDF1 plants. We found that in both long-day and short-day conditions the SUC2:HA-tpl-1/35S:CDF1 plants flowered significantly earlier than the 35S:CDF1 plants (Figure 4). This indicates that CDF1 represses flowering mainly through the function of TPL/TPRs. The SUC2:HA-tpl-1/35S:CDF1 plants, however, flowered later than their corresponding SUC2:HA-tpl-1 plants in wild type background. This suggests that CDFs may also have some repressor activity unrelated to TPL function, or that increased CDF1 concentration can make use of TPL complexes that do not contain the tpl-1 mutant protein.
Taken together, these data suggest that the loss of TPL function impairs the native CDF proteins from being able to repress the photoperiodic flowering pathway.
Loss of TPL function in phloem interferes with CDF-dependent repression of CO and FT
We next investigated whether SUC2:HA-tpl-1 plants had increased expression of CDF target loci, similar to the cdfq mutants. We measured the gene expression of CO and FT in both long days and short days in WT, cdfq, and SUC2:HA-tpl-1 lines (Figure 5,S5). In long days, SUC2:HA-tpl-1 plants showed increased levels of CO mRNA throughout the middle of the day (Figure 5a). Additionally, they showed increased expression of FT throughout the day, similar to cdfq mutants (Figure 5b). We also analyzed the expression levels of FT downstream genes, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), FRUITFUL (FUL), LEAFY (LFY), and APETALA 1 (AP1) (Andrés and Coupland, 2012; Song et al., 2015) (Figure S5). Especially in the SUC2:HA-tpl-1#4 line in which TPL (tpl-1) levels are relatively higher, the expression level of SOC1, FUL, LFY, and AP1 were higher than those in WT plants (Figure S5). In short days, CO mRNA level was upregulated during the daytime in both SUC2:HA-tpl-1 plants and cdfq mutants, and FT expression level was upregulated relative to WT (Figure 5c,d). These data support the notion that SUC2:HA-tpl-1 affects flowering time and expression of CO and FT genes in a manner similar to cdfq.
Figure 5. SUC2:HA-tpl-1 transgenic plants have increased FT and CO expression in long days and short days, as well as over developmental time.
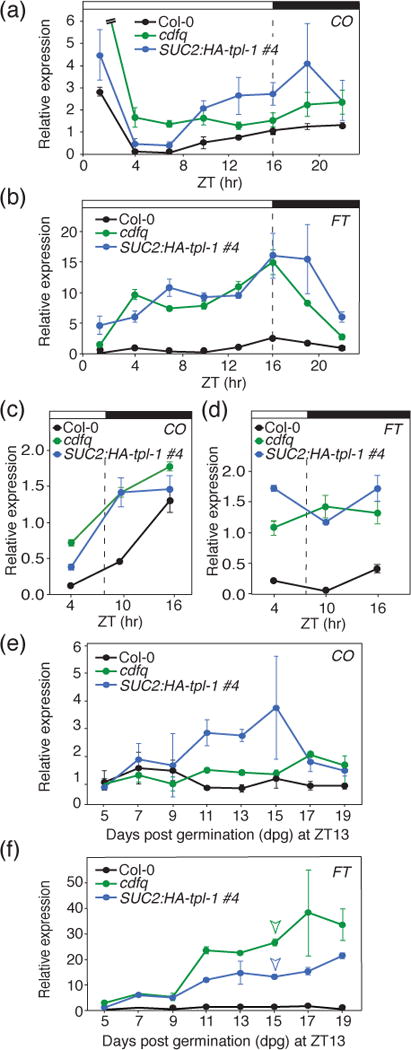
(a) and (b) Diurnal gene expression analysis of CO and FT in Col-0, cdfq, and SUC2:HA-tpl-1 plants. Experiments were performed on 14-day-old seedlings grown in long days, with 3-hour resolution. Means +/− SEM were calculated from four independent experiments. (c and d) Gene expression analysis of CO (c) and FT (d) in Col-0, cdfq, and SUC2:HA-tpl-1 plants in short days. Experiments were performed on 14-day-old seedlings in short days, with 6-hour resolution. Means +/- SEM were calculated from four independent experiments. (e and f) Gene expression analysis of CO (e) and FT (f) in Col-0, cdfq, and SUC2:HA-tpl-1 plants over developmental time. Plants were harvested over time from 5-days to 19-days old at the ZT13 time point of long days. Means +/− SEM were calculated from four independent experiments. Arrowheads indicate the day at which either cdfq or SUC2:HA-tpl-1 plants were first observed having a single plant begin flowering (the wild-type plants did not start flowering during the experiment).
Both FT and CO expression levels increase over developmental time (Kotake et al., 2003). We wanted to ascertain whether the increase of CO and FT expression seen in cdfq mutants on a daily scale were present over developmental time, and whether SUC2:HA-tpl-1-dependent increases in CO and FT expression followed a similar trajectory across early development. We performed a gene expression analysis of WT, cdfq, and SUC2:HA-tpl-1 plants from day 5 after germination until day 19. We harvested the tissues at the ZT13 time point, which is the daytime peak of CO expression, and close to the daily peak of FT expression. During the experiment, both cdfq mutants and SUC2:HA-tpl-1 plants, but not WT, started flowering on day 15 (Figure 5f). We observed that CO expression increased in both cdfq and SUC2:HA-tpl-1 plants from day 11 onward compared to WT, and FT levels increased gradually in both cdfq and SUC2:HA-tpl-1 throughout the experimental period (Figure 5e,f). We saw a similar trend when looking at the expression of downstream targets of FT, SOC1, LFY, and AP1 (Figure S6). These data suggest that CDF and TPL-dependent repression of flowering occurs throughout seedling development.
Both SUC2:HA-tpl-1/gi and SUC2:HA-tpl-1/fkf1 plants flowered earlier than their parental mutants in long days and short days
In addition to CDF1 related genetic backgrounds, we wanted to further validate the potential CDF-dependent nature of the SUC2:HA-tpl-1 phenotype. In fkf1 and gi mutant backgrounds, increased levels of CDF1 protein accumulates in the middle of the day, lengthening the period of CDF dependent repression of CO and FT, which contributes in their later flowering phenotypes (Imaizumi et al., 2005; Sawa et al., 2007; Fornara et al., 2009). We introduced the SUC2:HA-tpl-1 construct into fkf1-2 and gi-2 mutant backgrounds, and analyzed the flowering time phenotype. In long days, fkf1-2 and gi-2 mutants flower significantly later than WT plants, but mutants expressing the SUC2:HA-tpl-1 transgene are comparable or earlier than WT (Figure 6a,b). In short days, we saw a similar phenotype to SUC2:HA-tpl-1/35S:CDF1 transgenic plants, with a significant reduction in flowering time, but later than WT plants under long-day conditions as well as later than SUC2:HA-tpl-1/ Col-0 plants of similar transgene expression level (Figure 6c). Again, this suggests that some CDF activity independent of TPL may also be contributing to a delay in flowering time phenotype under long-day and short-day conditions.
Figure 6. SUC2:HA-tpl-1 plants in fkf1 and gi backgrounds are early flowering in long days and short days.
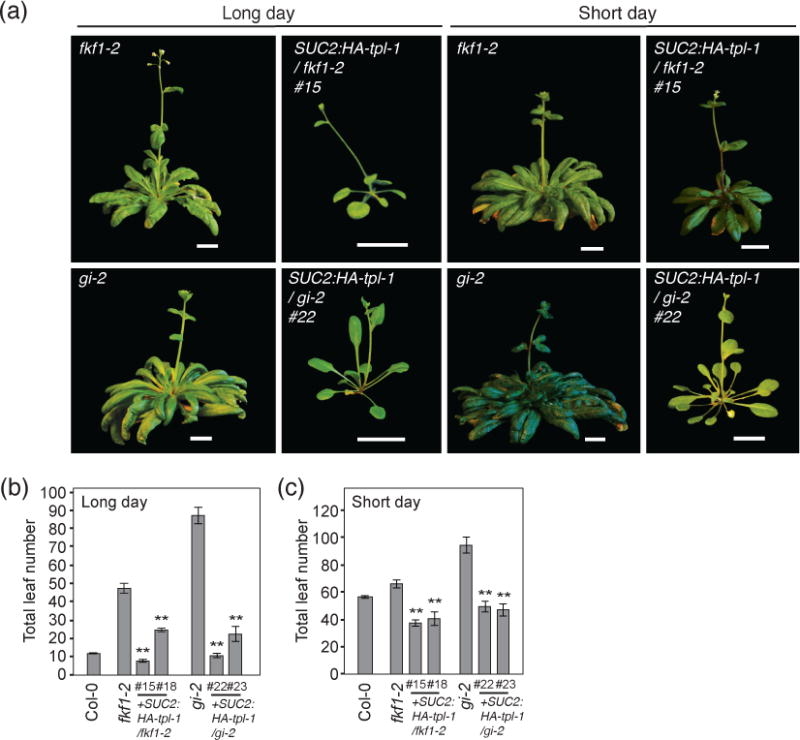
(a) Representative images of SUC2:HA-tpl-1 plants (SUC2:HA-tpl-1/fkf1-2, and SUC2:HA-tpl-1/gi-2) and their respective genetic backgrounds (fkf1-2, and gi-2) under long-day and short-day photoperiods at flowering. Scale bars=2 cm. (b and c) Quantification of flowering time by total leaf number at bolting from (a) under long days (b) and short days (c). Means +/− SEM were calculated from N=16 individuals. **P < 0.01 (one-tailed t test).
TPL regulates TSF levels and overexpression of tpl-1 in leaf phloem accelerates flowering time in co and ft mutants
If the introduction of the SUC2:HA-tpl-1 transgene impairs the ability of CDFs to repress transcription, then we reasoned that the loss of the key activators which CDFs regulate would revert the early flowering phenotype seen in both SUC2:HA-tpl-1 to a late flowering phenotype. To investigate this, we transformed SUC2:HA-tpl-1 into the ft-101 and co-101 mutant genetic background, and analyzed the flowering time phenotype. We found that under long-day conditions, SUC2:HA-tpl-1/ft-101 and SUC2:HA-tpl-1/co-101 plants flowered later than SUC2:HA-tpl-1 plants, however, they flowered earlier than ft-101 or co-101 mutants (Figure 7a,b). In short days, we found this phenotype was attenuated and the SUC2:HA-tpl-1/ft-101 and SUC2:HA-tpl-1/co-101 plants flowered more similar to, but still earlier to that of ft-101 and co-101 plants (Figure 7a,c). TPL has been previously reported to act in repressing downstream components of the flowering pathway such as LFY at the shoot apex (Wu et al., 2015). Due to the tissue specific activity of the SUC2 promoter at these developmental stages however, it is unlikely that the SUC2:HA-tpl-1 transgene is expressed at the shoot apex. We checked the gene expression of several downstream and parallel components of the flowering pathway in WT, cdfq, and SUC2:HA-tpl-1 under long-day conditions to investigate the potential cause of early flowering in ft-101 and co-101 mutants (Figure S5). Upon checking the gene expression of the FT paralog TWIN SISTER OF FT (TSF), which is expressed in vascular tissues (Yamaguchi et al., 2005), we found that TSF expression was highly upregulated in SUC2:HA-tpl-1 plants throughout the day (Figure 7d). In cdfq mutants, we saw an upregulation of TSF transcripts during both the morning and afternoon, but not of the same magnitude as in SUC2:HA-tpl-1 plants (Figure 7d). This suggested that CDFs may partially regulate TPL recruitment to the TSF gene, and another factor may also play an important role to regulate TSF through TPL in the afternoon of long days. To test whether CDF1-dependent recruitment of TPL to the TSF locus occurs, we performed a ChIP assay looking at the TSF promoter in WT, TPL:TPL-3HA and TPL:TPL-3HA/35S:CDF1 plants at the ZT12 time point of long days. We found that TPL strongly associated with a region of the TSF promoter upstream of the transcription start site in TPL:TPL-3HA/35S:CDF1 plants (Figure 7e), suggesting that TPL recruitment to the TSF promoter can directly or indirectly occur through CDF1. Taken together, these data suggest that the long-day specific early flowering phenotype of SUC2:HA-tpl-1/ft-101 and SUC2:HA-tpl-1/co-101 plants may be in part due to the upregulation of TSF, and that CDF1 recruitment is partially responsible for TPL-dependent regulation of TSF.
Figure 7. Introduction of SUC2:HA-tpl-1 into co and ft mutants caused earlier flowering, and TPL directly regulates TSF expression in long days.
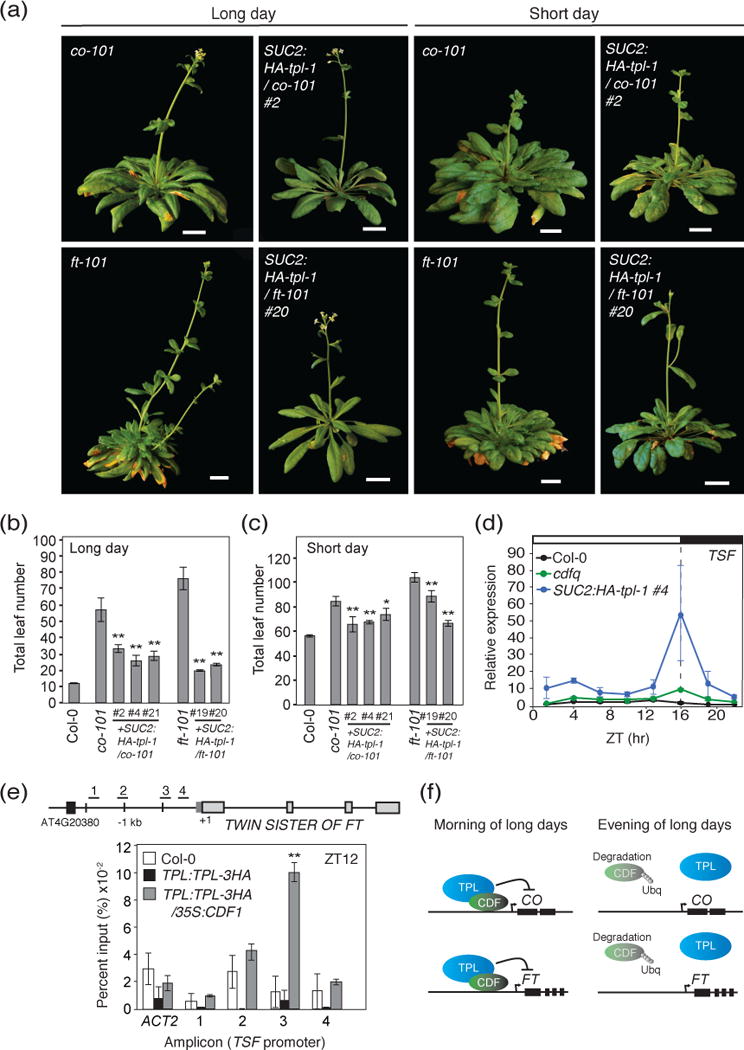
(a) Representative images of SUC2:HA-tpl-1/co-101 and SUC2:HA-tpl-1/ft-101 plants and their parental genetic backgrounds (co-101 and ft-101) under long-day and short-day photoperiods at flowering. Scale bars=2 cm. (b and c) Quantification of flowering time by total leaf number at bolting from (a) under long days (b) and short days (c). Means +/− SEM were calculated from N=16 individuals. *P < 0.05,**P < 0.01 (one-tailed t test). (d) Diurnal gene expression analysis of TSF in Col-0, cdfq, and SUC2:HA-tpl-1 plants in long days. Experiments were performed on 14-day-old seedlings grown in long days, with 3-hour resolution. Means +/− SEM were calculated from four independent experiments. (e) ChIP experiment for TPL binding to the TSF genomic locus at ZT12, in Col-0, TPL:TPL:3HA, and TPL:TPL:3HA/35S:CDF1. Means and +/− SEM were calculated from four independent experiments. **P < 0.01 (one-tailed t test). Plants were 2 weeks old and grown under long-day photoperiods. (f) Model for CDF1-TPL dependent regulation of CO and FT in the morning and the evening.
Discussion
Here we show that the mechanism by which CDF1 represses transcription of photoperiodic flowering target genes is through interactions with a co-repressor protein TPL. CDF proteins contain a conserved motif responsible for interaction with TPL (Figure 1), and eliminating this motif attenuates the function of CDF1 as a transcriptional repressor (Figure 2). TPL protein binds to the CO and FT promoters during the morning, which occurs at the same time as CDF1 binding to CO and FT (Figure 3). Increasing the expression of CDF1 brings additional TPL to target loci (Figure 3), suggesting CDF dependent recruitment of TPL. Removing TPL dependent repression exclusively within leaf phloem causes early flowering and photoperiodic insensitivity, similar to cdfq loss of function (Figure 4,6,7). SUC2:HA-tpl-1 plants are largely insensitive to changes in CDF1 expression, highlighting that loss of co-repressor perturbs CDF function (Figure 4). These data together demonstrate that a CDF-TPL transcriptional complex regulates CO and FT during the morning to limit their expression to the afternoon (Figure 7f).
TPL-dependent repression mechanism and its similarity to auxin signaling
TPL-dependent repression is typified by potential interactions such as mediator blocking and chromatin remodeling, though the exact mechanisms of TPL activity remain unclear (Long et al., 2006; Liu and Karmarkar, 2008; Wang et al., 2013; Ito et al., 2016). TPL interacts with several HDACs, and it has been postulated that TPL is able to recruit HDAC enzymes to target promoters to bring about transcriptional silencing through chromatin remodeling. In addition, TPL and LEUNIG (LUG), have both been shown to interact with HEN3/CDK8 (Gonzalez et al., 2007; Ito et al., 2016). This HEN3/CDK8 module of mediator has been implicated in a repressive role by preventing mediator holoenzyme formation (Tsai et al., 2013). A recent study of D. melanogaster Gro, a general co-repressor that shares similar functional domains with TPL, points to discrete Gro binding rather than spreading of hypoacetylated histones and heterochromatin for long-range silencing (Chambers et al., 2017). The same study found that bona fide Gro targets were enriched at loci having paused or stalled RNA polymerase II. This suggests that Gro plays a similar role to TPL-CDK8 in potentially preventing maturation of Mediator complex to initiate transcription. The TPL ChIP data presented here also implicates discrete binding rather than a spreading mechanism at CO and FT (Figure 3). How TPL-dependent histone deacetylation versus mediator interaction might function is unknown. Although recent work has progressed the understanding of histone modifications at the FT promoter, the temporal and cell specific nature of these changes still poses many questions (Turck et al., 2007; Jiang et al., 2008; Adrian et al., 2010; Pazhouhandeh et al., 2011; Cao et al., 2014).
This CDF-TPL-dependent transcriptional regulatory module bears many similarities to the molecular architecture of the IAA-TPL auxin circuit; the specific transcriptional repressor brings in TPL to solidify the repressive status of target loci. Further, the removal of the transcription factor through ubiquitin dependent proteasomal degradation (in this case through FKF1 E3 ubiquitin ligase) relieves repression and enables activation in the afternoon of long days (Pierre-Jerome et al., 2013). Due to the fact that several other FT regulators appear to bind to TPL, this positions TPL as a key mechanistic player in CO and FT regulation, and may play a more general role in mediating the accessibility of transcription factor binding sites over the relatively large space of the FT promoter (Causier et al., 2012; Graeff et al., 2016).
CDF and TPL function in the context of temporal and spatial regulation of transcription
Daily temporal regulation of photoperiodic regulators is critical for the timing and amplitude of FT expression, and thus the quantitative flowering time phenotype in Arabidopsis (Song et al., 2015). Due to the transcriptional complexity of the CO and FT gene regulatory network, being able to predict the combinatorial effects of many regulators can be challenging, and knowing more about the interactions between multiple transcription factors has the potential to better characterize the system (Andrés and Coupland, 2012). CDF transcription factors are key regulators of the timing of CO and FT expression, and their position as direct outputs of the circadian clock makes them a primary means through which to shut down CO and FT expression during the morning. In general, morning dependent repression at CO and FT cannot be overcome by the presence of activators during this time period. Overexpression of various CO activators, such as FLOWERING BHLH (FBH) and class II TEOSINTE BRANCHED 1/CYCLOIDEA/PROLIFERATING CELL FACTOR 1 (TCP) transcription factors, is insufficient to strongly upregulate CO during the morning (Ito et al., 2012; Kubota et al., 2017; Liu et al., 2017). Similarly at FT, overexpression of some activators, such as CRYPTOCHROME-INTERACTING BASIC-HELIX-LOOP-HELIX 1 (CIB1), are unable to increase FT during the morning (Liu et al., 2008). Potentially, CDF-TPL dependent repression can restrict the access of activators during the morning period. Either through physical blocking of mediator or the changing of chromatin structure at either the CO or FT locus (Wang et al., 2013; Ito et al., 2016), a CDF-TPL morning complex can prevent activation of transcription until the afternoon of long days, after FKF1-GI dependent degradation of CDFs.
Although proper temporal regulation of CO and FT transcription is crucial for inducing the photoperiodic flowering response, spatial regulation of their expression also plays an important role (Song et al., 2013). To more precisely investigate the function of the TPL general repressor in the photoperiodic flowering pathway, reducing TPL/TPR function in leaf phloem companion cells was an effective method for understanding the tissue-specific roles of TPL/TPR. Generally speaking, as TPL is involved in many different pathways in various cell types and interacts with wide varieties of transcriptional regulators (Liu and Karmarkar, 2008), the combination of functional analysis of specific TPL interactors and temporal, spatial, and developmental modification of TPL function using ectopic expression of tpl-1 will likely provide us with insight into the more specific roles of TPL in different regulatory networks.
Insights into evolution of the CDF-TPL module
Many of the components of the photoperiodic flowering pathway are highly conserved among flowering plants, suggesting a common module for regulation of target pathways in response to seasonal change (Song et al., 2015). This is evident from both the conservation of factors in the photoperiodic flowering pathway, as well as its role in multiple different kinds of seasonal organ development. These include the regulation of bud burst in tree species such as Populus triocharpa as well as for tuberization in potato (Hsu et al., 2011; Kloosterman et al., 2013). CDF1 homologs in in P. sativum and S. tuberosum are repressors of photoperiodic flowering and tuberization, respectively (Kloosterman et al., 2013; Ridge et al., 2016). Recently, it has been shown that the FKF1 and GI components of the photoperiodic time sensing module are also present in the basal linage of the land plants, in the bryophyte Marchantia polymorpha (Kubota et al., 2014). In addition, one DOF transcription factor in Marchantia is similar to CDFs and possesses the TPL binding site (Figure S1), although its function is unknown. Moreover, the moss Physcomitrella patens, the fern Selaginella moellendorffii, as well as several gymnosperms possess CDF-related DOF transcription factors that contain the TPL binding sites (Figure S1) (Sugiyama et al., 2012). These findings suggest that an ancient photoperiod mechanism including the DOF factors may have emerged and remained conserved with the evolution of land plants. There are also additional indications that CDF like proteins are part of this seasonal regulatory circuit. A recent study showed that a CDF-like protein in Chlamydomonas reinhardtii regulated growth in response to seasonal change (Lucas-Reina et al., 2015). This algae DOF protein bears the most sequence homology to the DOF domain of the CDF clade, although this specific DOF protein lacks the TPL binding sequences described in this manuscript (Lucas-Reina et al., 2015). The presence of this TPL binding domain is not universal amongst the DOF transcription factor family but is conserved within the CDF-like subclade of the DOFs through long periods of evolutionary time (Figure S1) (Cai et al., 2013; Lucas-Reina et al., 2015; Ma et al., 2015; Kang et al., 2016). The presence or absence of this domain likely determines other transcriptional interactions; indeed several DOF factors that were originally characterized function as activators, and these DOF factors lack the TPL binding motif (Cavalar et al., 2003). Based on the similar function as repressors amongst the CDF clade in Arabidopsis as well as their orthologs in other plant species, the N-terminal TPL binding motif is likely a conserved mechanism through which DOF factors function as transcriptional repressors (Kloosterman et al., 2013; Corrales et al., 2014; Ridge et al., 2016). While the conserved TPL binding domain is less common than the EAR-domain in TPL clients, its presence in transcription factors other than flowering regulators suggests that this domain is general rather than specific to flowering targets (Causier et al.,2012). Due to their conservation amongst many basal lineages of green algae and plants, it will be interesting to see if there is a functional CDF/TPL-FKF1/GI module that is part of a core conserved photoperiod circuit.
Experimental procedures
Plant materials
All genetic resources in this work are the Columbia-0 (Col-0) background, and Col-0 plants are used as wild type in all experiments. TPL:TPL:3HA (Wang et al., 2013), cdfq (Fornara et al., 2009), 35S:CDF1 (Imaizumi et al., 2005), fkf1-2 (Imaizumi et al., 2003), gi-2 (Fowler et al., 1999), co-101 and ft-101 (Takada and Goto, 2003) were previously described. To generate 35S:CDF1-3F6H, 35S:CDF1-ΔN-3F6H, and 35S:CDF1-mut-3F6H constructs, full length, truncated, and PCR mutagenized CDF1 cDNAs were cloned using the following forward primers 5′-TCCCCATGGGACTGGAAACTAAAGATCCTGCGATAAAGC-3′ (for CDF1-3F6H), 5′- TCCCCATGGGAACGGTTTTAGAGGTTGCTGATGAAGA-3′ (for CDF1-ΔN-3F6H), 5′- TCCCCATGGGACTGGAAACTAAAGATCCTGCGATAAAGGCCGCTGCTATGAAAATTCCTTTCCCGAC-3′ (for CDF1-mut-3F6H, the mutated sequences that encode three alanines are underlined) and the same reverse primer 5′-GGAGGATCCCCATCTGCTCATGGAAATTGATTGATCTTG-3′. These fragments were then inserted using restriction enzyme sites (NcoI, BamHI) into pENTR-3F6H (Ito et al., 2012; Song et al., 2014), which is modified pENTR D-TOPO vector (Invitrogen) containing sequences encoding a 3xFLAG 6xHis (3F6H) peptides to generate the constructs that contains CDF1 variant genes translationally fused to 3xFLAG 6xHis peptides. After confirming the sequences, these cDNAs were transferred into pH7WG2 (Karimi et al., 2002) destination vector, which harbors a 35S expression cassette, using the LR clonase II enzyme mix (Invitrogen), and subsequently transformed into plants using the conventional Agrobacterium mediated transformation method.
To generate SUC2:HA-tpl-1, we first cloned the wild-type version of TPL CDS into pENTR D-TOPO using cDNA as a template, forward primer 5′- CACCATGTCTTCTCTTAGTAGAGAGCTCGTTTTC-3′ and 5′- TCATCTCTGAGGCTGATCAGATGCAG as the reverse primer. We named this construct pENTR-TPL. The tpl-1 allele contains two mutations in the 5′ part of TPL CDS (Long et al., 2006). To generate a vector containing the tpl-1 mutation cDNA, we planned to replace about a 500-bp fragment of 5′ part of TPL CDS, which can be cut by NotI and NcoI, with that of tpl-1 CDS, which contains these mutations. We first amplified the 5′ fragment of the tpl-1 gene using tpl-1(Ler) cDNA as a template, Forward primer 5′- CACCATGTATCCATATGATGTTCCAGATTATGCTATGTCTTCTCTTAGTAGAGAGCTCGTT-3′ (the underlined sequences encode HA-tag) and 5′-GTACCAACTAGAAGCAGGGTCTGTTTAAT-3′ as the reverse primer. This fragment also contains a NotI site plus an N-terminal single HA tag. This fragment was cloned into pCR-Blunt II-TOPO (Invitrogen). We named this vector pCR-HA-tpl-1. As one of two tpl-1 mutations created one extra NcoI site, to replace the ca. 500 bp fragment of TPL with that of tpl-1, which contains two NcoI sites, instead of one, we decided to clone the 500 bp fragments in two steps. First, we digested pCR-HA-tpl-1 using NotI and NcoI (5′ half of ca. 500 bp fragment, size 303 bp), and ligated the fragment into the NotI-NcoI sites of pENTR-TPL vector. We named this vector pENTR-HA-tpl-1′. To place the remaining the NcoI-NcoI tpl-1 fragment (size 252 bp) into the vector, the NcoI-NcoI tpl-1 fragment was ligated into NcoI cut pENTR-HA-tpl-1′. After confirming the sequence and orientation of the NcoI-NcoI fragment, we named this vector pENTR HA-tpl-1. The finished pENTR HA-tpl-1 cassette was transferred into an pH7WG2 vector containing a SUC2 promoter and 5′-UTR (Truernit and Sauer, 1995; Sawa and Kay, 2011) using LR clonase II enzyme mix (Invitrogen), and transformed into plants using the conventional Agrobacterium mediated transformation method. The transgenic plants used in this work were selected based on having similar expression levels of CDF1 (variants), tpl-1 genes, or TPL-3HA protein.
Flowering time analysis
For flowering time analysis, seeds were directly sown on the soil (Sunshine Mix #4; Sun Gro Horticulture) and stratified for 2–3 days at 4°C in darkness to synchronize the timing of germination. Plants were grown at 22°C under long days (16 h light/8 h dark) or short days (8 h light/16 h dark). Light was provided by full-spectrum white fluorescent light bulbs (F017/950/48′′ Octron; Osram Sylvania) with a fluence rate of 60–90 μmol photons m−2 sec−1 in long days and 75–115 μmol photons m−2 sec−1 in short days. Flowering time was measured by counting the number of rosette and cauline leaves on the main stem when they bolted and the inflorescence was between 3–5 cm. Plant lines were sown in rows in horticultural 32-cell flats, with 16 individual plants per line split into two flats.
Yeast 2-hybrid analysis
To generate full-length products (without epitope tags) of CDF1, CDF1-ΔN, and CDF1-mut for Y2H analysis, the respective forward primers 5′- CACCATGCTGGAAACTAAAGATCC TGCGATAAAGC -3′ (for CDF1), 5′- CACCATGACGGTTTTAGAGGTTGCTGATGAAGA-3′ (for CDF1-ΔN), and 5′ CACCATGCTGGAAACTAAAGATCCTGCGATAAAGGCCGCTGCTATGAAAATTCCTTTCCCGAC-3′ (for CDF1-mut) were used and the same reverse primer 5′-TCACATCTGCTCATGGAA ATTGATTGATC-3′, using an cDNA clone as template. These PCR products were then cloned into pENTR D-TOPO (Invitrogen) using the standard TOPO reaction. The pENTR-TPL clone described above was used for this analysis. Plasmid cassettes were then transferred to pACT2-GW and pAS-GW, two gateway compatible prey and bait vectors (Song et al., 2014) using LR clonase II (Invitrogen). Yeast strains Y187 and AH109 were transformed with prey and bait vectors, respectively using the standard yeast PEG based plasmid transformation (Clontech). After colonies formed on –W or –L containing media, three independent colonies were grown together, then mated against their corresponding pairs for 3 days on YPDA media. After mating, yeast colonies were transferred onto –WL media. After checking for mating confirmation, yeast sectors were retransferred at the same time onto –WL and –WLH media. The GI clone contains the N-terminus of the GI protein used in this analysis was described previously (Sawa et al., 2007).
Bimolecular fluorescence complementation assays
BiFC experiments were performed on 3-week-old N. benthamiana plants grown at 22°C under long days (16 h light/8 h dark) on soil (Sunshine Mix #3; Sun Gro Horticulture) according to (Martin et al. 2009). Light was provided by full-spectrum white fluorescent light bulbs (F017/950/48′′ Octron; Osram Sylvania) with a fluence rate of 60–90 μmol photons m−2 sec−1. pSITE vectors were used to generate BiFC constructs for CDF1, CDF1-ΔN, CDF1-mut, and TPL proteins (Martin et al., 2009). In all cases the combinations are N-terminal fusions of either the nEYFP or cEYFP to the cDNA of CDF1 or TPL (Martin et al., 2009). RFP fused Histone H2B was used as a nucleus marker (Goodin et al., 2002). Injection of agrobacterium strains into tobacco leaves was performed as in (Goodin et al., 2002), but the OD600 of the Agrobacterium culture used was adjusted to 0.1, and the ABI Agrobacterium strain was used. 3 days after transfection, plant leaves were imaged using a confocal microscope (TCS SP5; Leica Microsystems).
RNA isolation and gene expression analysis
For gene expression analyses, 2-week-old seedlings were grown on LS agar plates containing 3% sucrose in long days and short days. Plates were grown in a self-contained growth chamber (Percival), light was provided by full-spectrum white fluorescent light bulbs (F017/950/24″ Octron; Osram Sylvania) with a fluence rate of 60–90 μmol photons m−2 sec−1. Tissue was harvested at every 3 hours during a 24-h period, and was used for RNA extraction using illustra RNAspin Mini kit (GE Healthcare). To synthesize cDNA, 2 μg of total RNA was reverse-transcribed using iScript cDNA synthesis kit (Bio- Rad). The cDNA was diluted to 100 μl of water (1:9 ratio), and 4 μl of diluted cDNA was used for quantitative polymerase chain reaction (Q-PCR) using Bio-Rad real-time thermal cycler (MyiQ). Primers and PCR conditions used for IPP2, CDF1, CO, FT, SOC1, AP1, LFY, and TSF amplification were described previously; IPP2, CDF1, CO and FT (Song et al., 2012), SOC1 and AP1 (Han et al., 2008), LFY (Kotake et al., 2003), and TSF (Yan et al., 2014). TPL and FUL Q-PCR primers are listed in supplementary table S1. The PCR conditions for detecting TPL transcripts were 45 total cycles using the protocol of 95°C for 10 sec, 55°C for 20 sec, and 72°C for 20 sec. For FUL, 45 cycles were run using the protocol of 95°C for 10 sec, 65.5°C for 15 sec, and 72°C for 15 sec. With the exception of the developmental time courses in Figure 5e,f and S6, IPP2 expression was used as an internal control to normalize cDNA amount. For Figure 5e,f and S6, IPP2 and PP2A were used as internal controls to minimize variation over the experimental period. All expression data were calculated from at least three independent biological experiments.
Immunoblot analysis
Total crude protein was extracted from frozen-ground seedlings in the extraction buffer [50 mM sodium phosphate (pH 7.0), 100 mM NaCl, 5 mM EDTA, 0.1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate]. The supernatant was collected after centrifugation at 21,000 × g for 5 min. Then protein samples were separated by 8 or 12% SDS-PAGE gels (8% for TPL, and 12% for CDF1) and transferred to nitrocellulose membranes (for each sample, 5–10 μg of total protein was used). The 3×FLAG and 6xHis epitope-tagged CDF1, CDF1-ΔN, CDF1-mut and HA-tagged TPL, HA-tagged tpl-1 fusion proteins were detected using HRP conjugated anti-FLAG (Sigma) and anti-HA (Roche) antibodies. Super Signal West Pico and Femto Chemiluminescent substrate kits (Thermo Fisher Scientific) were used to detect signals. All experiments were performed at least three times with independent biological replicates.
Chromatin immunoprecipitation
ChIP experiments were performed on 2-week-old seedlings, which were grown on the same conditions described in RNA isolation and gene expression analysis section. 600 mg of tissue was harvested and frozen in liquid nitrogen in small sachets and kept at −80°C until the ChIP procedure was started. ChIP experiments were performed as illustrated in (Yamaguchi et al., 2014) with minor modifications. The initial extraction buffer used contains 0.4 M sucrose, 10 mM HEPES pH 8.0, 2 mM EDTA, 5 mM β-mercaptoethanol, EDTA-free protease inhibitor tablet (Pierce), 1% Formaldehyde. Plants were harvested, ground in liquid nitrogen into a fine powder, then the extraction buffer containing formaldehyde was added and incubated for 10 min at 4°C, then quenched with glycine to a total concentration of 200 mM glycine for 5 min, then filtered twice through a miracloth filter. For the qPCR analysis of DNA, the reaction sizes were scaled down to a 15 μl reaction size, and 1.5 μl of DNA was used for input into the reaction for both purified input and immunopurified samples. For the CO promoter, PCR reactions were run for 60 cycles using the protocol of 95°C for 10 sec, 57°C for 20 sec, and 72°C for 20 sec. For the FT promoter, PCR reactions were run for 65 cycles using the protocol of 95°C for 10 sec, 52°C for 20 sec, 72°C for 20 sec. For the TSF promoter, PCR reactions were run for 60 cycles using the protocol of 95°C for 10 sec, 57°C for 20 sec, 72°C for 20 sec. Primer sequences for each amplicon of CO and FT loci are previously listed (Ito et al. 2012, Song et al. 2012). Primer sequences for each amplicon of TSF locus are listed in supplementary table S1.
Supplementary Material
Figure S1. Amino acid sequence alignment of CDF related proteins.
Figure S2. BiFC negative controls and subcellular localization of TPL, CDF1, CDF1-ΔN, and CDF1-mut.
Figure S3. Flowering phenotype of Col-0, 35S:CDF1-3F6H, 35S:CDF1-ΔN-3F6H, and 35S:CDF1-mut-3F6H transgenic lines in short days.
Figure S4. Protein expression level of TPL in TPL:TPL-3HA/35S:CDF1 transgenic line, CDF1 expression in TPL:TPL-3HA/35S:CDF1 transgenic line, and TPL expression in SUC2:HA-tpl-1 transgenic lines.
Figure S5. Gene expression profiles of TPL, LFY, AP1, FUL, and SOC1 in WT, cdfq mutants, and SUC2:HA-tpl-1 transgenic lines.
Figure S6. Gene expression levels of TPL, SOC1, LFY, and AP1 over developmental time in WT, cdfq mutants, and SUC2:HA-tpl-1 transgenic lines.
Table S1. ChIP and Q-PCR primers used in this study.
Significance Statement.
Temporal control of expression profiles of photoperiodic flowering genes is important for regulating timing of flowering. The mechanism of CDF-dependent repression on the CO and FT promoters is currently unknown. Here we demonstrate that CDFs recruit the co-repressor TPL to repress transcription during the morning. Understanding the time-dependent abundance of various transcriptional complexes that regulate CO and FT expression is key to understanding flowering time regulation.
Acknowledgments
The authors thank Drs. D.E. Somers and J.A. Long for sharing seeds and constructs, Dr. M. Goodin for providing H2B-RFP expressing construct, Drs. A. Kubota and J.S. Shim for critical reading of the manuscript and scientific advice. We thank T. Mitsuhiko, T. Chandran, R. Lillinas, E. Kow, M. Khan, M. Kwon, U. Okorafor, and R. Wang for experimental assistance. This work was supported by NIH grants (GM079712) and Next-Generation BioGreen 21 Program (SSAC, PJ011175, Rural Development Administration, Republic of Korea) to T.I. G.S.G. was supported from the NIH Developmental Biology Training Grant (5T32HD007183).
Footnotes
The authors declare that no conflict of interest exists.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- Adrian J, Farrona S, Reimer JJ, Albani MC, Coupland G, Turck F. cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell. 2010;22:1425–1440. doi: 10.1105/tpc.110.074682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13:627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- Cai X, Zhang Y, Zhang C, Zhang T, Hu T, Ye J, Zhang J, Wang T, Li H, Ye Z. Genome-wide analysis of plant-specific Dof transcription factor family in tomato. J Integr Plant Biol. 2013;55:552–566. doi: 10.1111/jipb.12043. [DOI] [PubMed] [Google Scholar]
- Cao S, Kumimoto RW, Gnesutta N, Calogero AM, Mantovani R, Holt BF., 3rd A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell. 2014;26:1009–1017. doi: 10.1105/tpc.113.120352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C, Pelaz S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol. 2008;18:1338–1343. doi: 10.1016/j.cub.2008.07.075. [DOI] [PubMed] [Google Scholar]
- Causier B, Ashworth M, Guo W, Davies B. The TOPLESS Interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 2012;158:423–438. doi: 10.1104/pp.111.186999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalar M, Moller C, Offermann S, Krohn NM, Grasser KD, Peterhansel C. The interaction of DOF transcription factors with nucleosomes depends on the positioning of the binding site and is facilitated by maize HMGB5. Biochemistry. 2003;42:2149–2157. doi: 10.1021/bi026761r. [DOI] [PubMed] [Google Scholar]
- Chambers M, Turki-Judeh W, Kim MW, Chen K, Gallaher SD, Courey AJ. Mechanisms of Groucho-mediated repression revealed by genome-wide analysis of Groucho binding and activity. BMC Genomics. 2017;18:215. doi: 10.1186/s12864-017-3589-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Corrales AR, Nebauer SG, Carrillo L, Fernández-Nohales P, Marqués J, Renau-Morata B, Granell A, Pollmann S, Vicente-Carbajosa J, Molina RV, Medina J. Characterization of tomato Cycling Dof Factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J Exp Bot. 2014;65:995–1012. doi: 10.1093/jxb/ert451. [DOI] [PubMed] [Google Scholar]
- Espinosa-Ruiz A, Martínez C, de Lucas M, Fàbregas N, Bosch N, Caño-Delgado AI, Prat S. TOPLESS mediates brassinosteroid control of shoot boundaries and root meristem development in Arabidopsis thaliana. Development. 2017;144:1619–1628. doi: 10.1242/dev.143214. [DOI] [PubMed] [Google Scholar]
- Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, Coupland G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell. 2009;17:75–86. doi: 10.1016/j.devcel.2009.06.015. [DOI] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999;18:4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez D, Bowen AJ, Carroll TS, Conlan RS. The Transcription Corepressor LEUNIG Interacts with the Histone Deacetylase HDA19 and Mediator Components MED14 (SWP) and CDK8 (HEN3) To Repress Transcription. Mol Cell Biol. 2007;27:5306–5315. doi: 10.1128/MCB.01912-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 2002;31:375–383. doi: 10.1046/j.1365-313x.2002.01360.x. [DOI] [PubMed] [Google Scholar]
- Graeff M, Straub D, Eguen T, Dolde U, Rodrigues V, Brandt R, Wenkel S. MicroProtein-mediated recruitment of CONSTANS into a TOPLESS trimeric complex represses flowering in Arabidopsis. PLoS Genet. 2016;12:e1005959. doi: 10.1371/journal.pgen.1005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF TIR1-dependent degradation of AUX/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- Han P, García-Ponce B, Fonseca-Salazar G, Alvarez-Buylla ER, Yu H. AGAMOUS-LIKE 17, a novel flowering promoter, acts in a FT-independent photoperiod pathway. Plant J. 2008;55:253–265. doi: 10.1111/j.1365-313X.2008.03499.x. [DOI] [PubMed] [Google Scholar]
- Hayama R, Sarid‐Krebs L, Richter R, Fernández V, Jang S, Coupland G. PSEUDO RESPONSE REGULATORs stabilize CONSTANS protein to promote flowering in response to day length. EMBO J. 2017;36:904–918. doi: 10.15252/embj.201693907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando-Amado S, González-Calle V, Carbonero P, Barrero-Sicilia C. The family of DOF transcription factors in Brachypodium distachyon: phylogenetic comparison with rice and barley DOFs and expression profiling. BMC Plant Biol. 2012;12:202. doi: 10.1186/1471-2229-12-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CY, Adams JP, Kim H, No K, Ma C, Strauss SH, Drnevich J, Vandervelde L, Ellis JD, Rice BM, Wickett N, Gunter LE, Tuskan GA, Brunner AM, Page GP, Barakat A, Carlson JE, dePamphilis CW, Luthe DS, Yuceer C. FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc Natl Acad Sci U S A. 2011;108:10756–10761. doi: 10.1073/pnas.1104713108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005;309:293–297. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature. 2003;426:302–306. doi: 10.1038/nature02090. [DOI] [PubMed] [Google Scholar]
- Ito J, Fukaki H, Onoda M, Li L, Li C, Tasaka M, Furutani M. Auxin-dependent compositional change in Mediator in ARF7- and ARF19-mediated transcription. Proc Natl Acad SCI U S A. 2016;113:6562–6567. doi: 10.1073/pnas.1600739113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Song YH, Josephson-Day AR, Miller RJ, Breton G, Olmstead RG, Imaizumi T. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109:3582–3587. doi: 10.1073/pnas.1118876109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Pullen N, Lamzin S, Morris RJ, Wigge PA. Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell. 2013;25:820–833. doi: 10.1105/tpc.113.109355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA. FT Protein acts as a long-range signal in Arabidopsis. Curr Biol. 2007;17:1050–1054. doi: 10.1016/j.cub.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Jiang D, Wang Y, Wang Y, He Y. Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis polycomb repressive complex 2 components. PLoS One. 2008;3:e3404. doi: 10.1371/journal.pone.0003404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Staiger D. Time to flower: interplay between photoperiod and the circadian clock. J Exp Bot. 2015;66:719–730. doi: 10.1093/jxb/eru441. [DOI] [PubMed] [Google Scholar]
- Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua NH, Park CM. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell. 2007;19:2736–2748. doi: 10.1105/tpc.107.054528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang WH, Kim S, Lee HA, Choi D, Yeom SI. Genome-wide analysis of Dof transcription factors reveals functional characteristics during development and response to biotic stresses in pepper. Sci Rep. 2016;6:33332. doi: 10.1038/srep33332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Ke J, Ma H, Gu X, Thelen A, Brunzelle JS, Li J, Xu HE, Melcher K. Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors. Sci Adv. 2015;1:e1500107. doi: 10.1126/sciadv.1500107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman B, Abelenda JA, Gomez Mdel M, Oortwijn M, de Boer JM, Kowitwanich K, Horvath BM, van Eck HJ, Smaczniak C, Prat S, Visser RG, Bachem CW. Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature. 2013;495:246–250. doi: 10.1038/nature11912. [DOI] [PubMed] [Google Scholar]
- Kotake T, Takada S, Nakahigashi K, Ohto M, Goto K. Arabidopsis TERMINAL FLOWER 2 gene encodes a heterochromatin protein 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes. Plant Cell Physiol. 2003;44:555–564. doi: 10.1093/pcp/pcg091. [DOI] [PubMed] [Google Scholar]
- Kubota A, Ito S, Shim JS, Johnson RS, Song YH, Breton G, Goralogia GS, Kwon MS, Laboy Cintrón D, Koyama T, Ohme-Takagi M, Pruneda-Paz JL, Kay SA, MacCoss MJ, Imaizumi T. TCP4-dependent induction of CONSTANS transcription requires GIGANTEA in photoperiodic flowering in Arabidopsis. PLoS Genet. 2017;13:e1006856. doi: 10.1371/journal.pgen.1006856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota A, Kita S, Ishizaki K, Nishihama R, Yamato KT, Kohchi T. Co-option of a photoperiodic growth-phase transition system during land plant evolution. Nat Commun. 2014;5:3668. doi: 10.1038/ncomms4668. [DOI] [PubMed] [Google Scholar]
- Lijavetzky D, Carbonero P, Vicente-Carbajosa J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol Biol. 2003;3:17. doi: 10.1186/1471-2148-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Cheng X, Liu P, Li D, Chen T, Gu X, Sun J. MicroRNA319-regulated TCPs interact with FBHs and PFT1 to activate CO transcription and control flowering time in Arabidopsis. PLoS Genet. 2017;13:e1006833. doi: 10.1371/journal.pgen.1006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008;322:1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- Liu L, Liu C, Hou X, Xi W, Shen L, Tao Z, Wang Y, Yu H. FTIP1 is an essential regulator required for florigen transport. PLoS Biol. 2012;10:e1001313. doi: 10.1371/journal.pbio.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Karmarkar V. Groucho/Tup1 family co-repressors in plant development. Trends Plant Sci. 2008;13:137–144. doi: 10.1016/j.tplants.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM. TOPLESS regulates apical embryonic fate in Arabidopsis. Science. 2006;312:1520–1523. doi: 10.1126/science.1123841. [DOI] [PubMed] [Google Scholar]
- Lucas-Reina E, Romero-Campero FJ, Romero JM, Valverde F. An evolutionarily conserved DOF-CONSTANS module controls plant photoperiodic signaling. Plant Physiol. 2015;168:561–574. doi: 10.1104/pp.15.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Li MY, Wang F, Tang J, Xiong AS. Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genomics. 2015;16:33. doi: 10.1186/s12864-015-1242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM. Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 2009;59:150–162. doi: 10.1111/j.1365-313X.2009.03850.x. [DOI] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Kuttner F, Schmid M. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol. 2007;17:1055–1060. doi: 10.1016/j.cub.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Niimura K, Ito S, Yamashino T, Mizoguchi T, Mizuno T. Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol. 2007;48:822–832. doi: 10.1093/pcp/pcm056. [DOI] [PubMed] [Google Scholar]
- Niwa Y, Ito S, Nakamichi N, Mizoguchi T, Niinuma K, Yamashino T, Mizuno T. Genetic linkages of the circadian clock-associated genes, TOC1, CCA1 and LHY, in the photoperiodic control of flowering time in Arabidopsis thaliana. Plant Cell Physiol. 2007;48:925–937. doi: 10.1093/pcp/pcm067. [DOI] [PubMed] [Google Scholar]
- Notaguchi M, Abe M, Kimura T, Daimon Y, Kobayashi T, Yamaguchi A, Tomita Y, Dohi K, Mori M, Araki T. Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol. 2008;49:1645–1658. doi: 10.1093/pcp/pcn154. [DOI] [PubMed] [Google Scholar]
- Osnato M, Castillejo C, Matias-Hernandez L, Pelaz S. TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat Commun. 2012;3:808. doi: 10.1038/ncomms1810. [DOI] [PubMed] [Google Scholar]
- Pazhouhandeh M, Molinier J, Berr A, Genschik P. MSI4/FVE interacts with CUL4-DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108:3430–3435. doi: 10.1073/pnas.1018242108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre-Jerome E, Moss BL, Nemhauser JL. Tuning the auxin transcriptional response. J Exp Bot. 2013;64:2557–2563. doi: 10.1093/jxb/ert100. [DOI] [PubMed] [Google Scholar]
- Ridge S, Sussmilch FC, Hecht V, Vander Schoor JK, Lee R, Aubert G, Burstin J, Macknight RC, Weller JL. Identification of LATE BLOOMER2 as a CYCLING DOF FACTOR homolog reveals conserved and divergent features of the flowering response to photoperiod in pea. Plant Cell. 2016;28:2545–2559. doi: 10.1105/tpc.15.01011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas U, Mei Y, Xie Q, Banta JA, Zhou RW, Seufferheld G, Gerard S, Chou L, Bhambhra N, Parks JD, Flowers JM, McClung CR, Hanzawa Y, Purugganan MD. Variation in Arabidopsis flowering time associated with cis-regulatory variation in CONSTANS. Nat Commun. 2014;5:3651. doi: 10.1038/ncomms4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M, Kay SA. GIGANTEA directly activates FLOWERING LOCUS T in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2011;108:11698–11703. doi: 10.1073/pnas.1106771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318:261–265. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU. Dissection of floral induction pathways using global expression analysis. Development. 2003;130:6001–6012. doi: 10.1242/dev.00842. [DOI] [PubMed] [Google Scholar]
- Simon S, Rühl M, de Montaigu A, Wötzel S, Coupland G. Evolution of CONSTANS Regulation and Function after Gene Duplication Produced a Photoperiodic Flowering Switch in the Brassicaceae. Mol Biol Evol. 2015;32:2284–2301. doi: 10.1093/molbev/msv110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Estrada DA, Johnson RS, Kim SK, Lee SY, MacCoss MJ, Imaizumi T. Distinct roles of FKF1, GIGANTEA, and ZEITLUPE proteins in the regulation of CONSTANS stability in Arabidopsis photoperiodic flowering. Proc Natl Acad Sci U S A. 2014;111:17672–17677. doi: 10.1073/pnas.1415375111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Ito S, Imaizumi T. Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 2013;18:575–583. doi: 10.1016/j.tplants.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T. Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol. 2015;66:441–464. doi: 10.1146/annurev-arplant-043014-115555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science. 2012;336:1045–1049. doi: 10.1126/science.1219644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Ishida T, Tabei N, Shigyo M, Konishi M, Yoneyama T, Yanagisawa S. Involvement of PpDof1 transcriptional repressor in the nutrient condition-dependent growth control of protonemal filaments in Physcomitrella patens. J Exp Bot. 2012;63:3185–3197. doi: 10.1093/jxb/ers042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Goto K. TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell. 2003;15:2856–2865. doi: 10.1105/tpc.016345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, Ogaki Y, Shimada C, Nakagawa A, Kojima C, Shimamoto K. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476:332–335. doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- Thomas B, Vince-Prue D. Photoperiodism in plants. Academic Press; 1996. [Google Scholar]
- Truernit E, Sauer N. The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of β-glucuronidase to the phloem: evidence for phloem loading and unloading by SUC2. Planta. 1995;196:564–570. doi: 10.1007/BF00203657. [DOI] [PubMed] [Google Scholar]
- Tsai KL, Sato S, Tomomori-Sato C, Conaway RC, Conaway JW, Asturias FJ. A conserved Mediator–CDK8 kinase module association regulates Mediator–RNA polymerase II interaction. Nat Struct Mol Biol. 2013;20:611–619. doi: 10.1038/nsmb.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007;3:e86. doi: 10.1371/journal.pgen.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Kim J, Somers DE. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci U S A. 2013;110:761–766. doi: 10.1073/pnas.1215010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B, Zhang J, Pang C, Yu H, Guo D, Jiang H, Ding M, Chen Z, Tao Q, Gu H, Qu LJ, Qin G. The molecular mechanism of SPOROCYTELESS/NOZZLE in controlling Arabidopsis ovule development. Cell Res. 2015;25:121–134. doi: 10.1038/cr.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- Wu MF, Yamaguchi N, Xiao J, Bargmann B, Estelle M, Sang Y, Wagner D. Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. eLife. 2015;4:e09269. doi: 10.7554/eLife.09269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 2005;46:1175–1189. doi: 10.1093/pcp/pci151. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Winter CM, Wu MF, Kwon CS, William DA, Wagner D. PROTOCOL: Chromatin Immunoprecipitation from Arabidopsis Tissues. Arabidopsis Book. 2014;12:e0170. doi: 10.1199/tab.0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Shen L, Chen Y, Bao S, Thong Z, Yu H. A MYB-domain protein EFM mediates flowering responses to environmental cues in Arabidopsis. Dev Cell. 2014;30:437–448. doi: 10.1016/j.devcel.2014.07.004. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. The Dof family of plant transcription factors. Trends Plant Sci. 2002;7:555–560. doi: 10.1016/s1360-1385(02)02362-2. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol. 2004;45:386–391. doi: 10.1093/pcp/pch055. [DOI] [PubMed] [Google Scholar]
- Yang J, Yang MF, Wang D, Chen F, Shen SH. JcDof1, a Dof transcription factor gene, is associated with the light-mediated circadian clock in Jatropha curcas. Physiol Plant. 2010;139:324–334. doi: 10.1111/j.1399-3054.2010.01363.x. [DOI] [PubMed] [Google Scholar]
- Zhang B, Wang L, Zeng L, Zhang C, Ma H. Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev. 2015;29:975–987. doi: 10.1101/gad.251520.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Amino acid sequence alignment of CDF related proteins.
Figure S2. BiFC negative controls and subcellular localization of TPL, CDF1, CDF1-ΔN, and CDF1-mut.
Figure S3. Flowering phenotype of Col-0, 35S:CDF1-3F6H, 35S:CDF1-ΔN-3F6H, and 35S:CDF1-mut-3F6H transgenic lines in short days.
Figure S4. Protein expression level of TPL in TPL:TPL-3HA/35S:CDF1 transgenic line, CDF1 expression in TPL:TPL-3HA/35S:CDF1 transgenic line, and TPL expression in SUC2:HA-tpl-1 transgenic lines.
Figure S5. Gene expression profiles of TPL, LFY, AP1, FUL, and SOC1 in WT, cdfq mutants, and SUC2:HA-tpl-1 transgenic lines.
Figure S6. Gene expression levels of TPL, SOC1, LFY, and AP1 over developmental time in WT, cdfq mutants, and SUC2:HA-tpl-1 transgenic lines.
Table S1. ChIP and Q-PCR primers used in this study.


