Abstract
The intestinal microbiota is a plastic ecosystem that is shaped by environmental and genetic factors, interacting with virtually all tissues of the host. Many signals result from the interplay between the microbiota with its mammalian symbiont that can lead to altered metabolism. Disruptions in the microbial composition are associated with a number of comorbidities linked to the metabolic syndrome. Promoting the niche expansion of beneficial bacteria through diet and supplements can improve metabolic disorders. Reintroducing bacteria through probiotic treatment or fecal transplant is a strategy under active investigation for multiple pathological conditions. Here, we review the recent knowledge of microbiota’s contribution to host pathology, the modulation of the microbiota by dietary habits, and the potential therapeutic benefits of reshaping the gut bacterial landscape in context of metabolic disorders such as obesity.
Keywords: microbiota, obesity, metabolism, dysbiosis, diet, probiotics, fecal transplant, co-metabolism
Introduction
The intestinal microbiota is a highly dynamic ecosystem in which hundreds of bacterial species and other microorganisms coexist along with their neighboring mammalian cells. Estimated numbers vary across studies, but it is believed that there are at least as many bacteria as host cells in humans, if not drastically more (1). The most abundant phyla in humans and rodent models are Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes, sharing functional structure among hosts species despite having low taxonomic identity (2). Environmental factors such as early microbial exposure and lifestyle, as well as host genetics, shape its composition and function (3, 4). The gut microbiota in turn affects the host metabolic phenotype, contributes to food and drug metabolism, and helps the immune system to develop (5). From the first observation that obese individuals have a distinct gut microflora compared to lean people (6), and the following efforts to elucidate the function of this altered microbiota (7), the past 10 years have seen a growing body of evidence on the impact of the gut microflora on the host. By transplanting the gut microbiota to germ-free (GF) animals, it has become possible to directly assess the causality of microbiota composition with diseases. In this review, we focus on the relationship between gut microbiota composition and host pathophysiology, and on how shaping the microbiota can be beneficial to promote host health and combat metabolic disorders.
Deregulation of the Gut Microbiota and the Metabolic Syndrome
Microbiota Compositional Changes in Metabolic Disorders
The gut microbiota is sensitive to external cues that can reshape it to new stable compositions, resulting sometimes in a deranged, or dysbiotic gut flora. Dysbiotic states are often associated with metabolic alterations in both humans and rodent models such as obesity (6), type 2 diabetes (8), non-alcoholic fatty liver disease (NAFLD) spectrum (9), and dyslipidemia (10). These metabolic traits are often clustered in metabolic syndrome patients (11); it is, therefore, relevant, but challenging, to distinguish the implication of the gut microbiota with each of these pathologies separately.
Germ-free mice are extensively used as a model for studying the importance of the dysbiotic gut flora. Interestingly, GF mice are resistant to obesity following high-fat diets (HFD), and their colonization leads to an increased adiposity along with decreased insulin sensitivity (12) and altered lipid metabolism (13). Nevertheless, when colonization occurs from an obese dysbiotic donor, recipient mice gain even more adiposity and increase their systemic inflammation (14). Considering that HFD challenges elicit heterogeneous responses in terms of weight gain and glucose homeostasis, the differences in the pre-HFD gut microbiota–host interactions in mice can be predictive of the diet outcome (15), where final HFD-driven microbial differences are determinant to transfer these acquired phenotypes following microbiota transplantation and diet challenge (16). Specifically, Le Roy et al. show that GF mice populated with microbiota from two donors similarly obese but discordant in glycemia phenocopy their response to HFD, with a similar increase in the body weight but hyperglycemia and steatosis only in one group. This suggests that components of the gut microbiota can influence liver steatosis and hyperglycemia independently from their effect on adiposity and systemic inflammation.
Dysbiotic microbial composition can be, at least in the case of the obesogenic microbiota, resilient over time. Whereas dieting rapidly reverses the metabolic defects associated with HFD, the dysbiosis provoked in mice after a 4-week HFD persists up to 21 weeks after returning to normal chow diet (17). Importantly, this persistent post-HFD dysbiotic microbiota is not sufficient to drive obesity by itself, but can induce weight gain and glucose intolerance upon exposure to a second HFD stimulus. This two-step obesogenic mechanism relies on a reduced bioavailability of flavonoids (dietary compounds that can promote brown adipose tissue activation and increase energy expenditure) due to the combination of their scarcity in high-fat food and increased flavonoid-degrading ability of the obese microbiota (17). Weight gain upon second exposure to calorie-rich food is a common problem in dieting individuals. Human data are, therefore, needed to assess the plasticity of the microbiota in obese individuals and determine an ideal diet length and composition that would be accompanied with complete and lasting microbial reshaping (17). Another evidence that dysbiosis by itself may not be sufficient to drive metabolic defects comes from the observation that transplant of dysbiotic microbiota to healthy conventional mice neither causes metabolic dysfunctions nor alters the hepatic metabolism (18).
Microbiome analysis on two independent human cohorts described an intestinal microbial signature predicting the glycemic status (19, 20). The stratification of the microbiota analysis for metformin medication highlighted a commonly deregulated pathway in untreated type-2 diabetes (T2D) patients, characterized by decreased abundance of bacteria such as Roseburia spp. and Subdoligranulum spp., which produce butyrate, a known regulator of hepatic function through intestinal gluconeogenesis (8). Indeed, the metformin in part improves T2D by rescuing the decreased butyrate production through reshaping the microbiota, since microbial transplant from metformin-treated patients was sufficient to improve glucose control in GF mice (21, 22). Of note, other studies using meta-analyses, however, called for the need of large human cohorts to further generalize the predictive power of the microbiota (23–25).
Microbiota-Driven Regulation of Metabolism
Absence of microbiota in GF mice or through antibiotic treatment improves glucose and lipid metabolism (12, 13, 26, 27), protecting against diet-induced metabolic diseases. These improvements can, at least in part, be explained by increased activity of the thermogenic fat depots (26, 27), and can be reversed by microbial recolonization of the microbiota-depleted animals (27). Cold exposure, the most potent environmental trigger for brown and beige fat development and activation (28), drastically reshapes microbiota composition. Transplantation of this cold-adapted microbiota to GF mice is sufficient to induce tolerance to cold, improve insulin sensitivity, increase energy expenditure, and lower their fat content, largely due to increased brown and beige fat activity in the cold-microbiota transplanted mice (26, 29).
The complexity of the gut microbiota is reflected in its interplay with the host, with a great variety of signaling cues and relay organs (summarized in Table 1). Bile acids (BAs) are released after a meal directly in the proximal intestinal lumen and help lipid absorption by enterocytes. Since around 95% of BAs are reabsorbed in the distal intestine, the total BA pool is relatively stable across the enterohepatic circulation. The gut microbiota metabolizes primary BAs produced by the liver giving rise to secondary BAs, and this microbiota–liver cross talk is responsible of the BA pool (30). BAs act as signaling molecules through intracellular farnesoid X receptor with effect on the overall metabolism (31–35) and membrane-bound G-coupled bile acid receptor (TGR5). TGR5 stimulates intestinal glucagon-like peptide 1 (GLP1) production, brown fat activity, and improves hepatic metabolism in obese animals (36, 37). Interestingly, BAs signaling on intestinal cells can trigger their antimicrobial action (38), suggesting a negative feedback loop. In addition, it was suggested that the brown adipose tissue can also intervene into the gut microbiota–liver regulation of BA pool, since changes in cholesterol metabolism due to the brown adipose tissue activity during cold exposure can increase BAs biosynthesis and drive compositional changes in the gut microbiota (39).
Table 1.
List of microbe-derived signals that can impact host metabolism.
| Signal | Target organ | Effect | Reference |
|---|---|---|---|
| Bile acids (BAs) | Adipose tissue, intestine, liver | Hepatic metabolism, bacterial regulation, lipid metabolism | (36, 38) |
| Short chain fatty acids (SCFAs) | Adipose tissue, brain, intestine, liver, muscle | Lipid metabolism, regulation of appetite | (40–42) |
| Neuroactive molecules [g-aminobutyric acid (GABA), serotonine] | Central and peripheral nervous system | Regulation of appetite | (43, 44) |
| Lipopolysaccharide (LPS) | Adipose tissue, liver, brain | Systemic inflammation, hepatic glucose metabolism, adipose tissue fibrosis | (45–47) |
| Trimethylamine N-oxide | Adipose tissue, liver, kidney | Higher atherosclerosis risk, reduced beige fat | (48, 49) |
| Branched-chain amino acids (BCAAs) | Adipose tissue, endothelium, skeletal muscle | Adipogenesis, lipid trafficking, lipogenesis, and insulin resistance | (12, 13, 26, 27, 50, 51) |
LPS, BAs, SCFAs, BCAAs, trimethylamine N-oxide, and neuroactive molecules are major known signals of microbial origin that can affect different metabolic organs listed together with the proposed model of action.
Short chain fatty acids (SCFAs) derive from bacterial fermentation of dietary fibers. They can enter circulation and signal through their cognate receptors to many organs (52, 53) including the central nervous system, which in turn regulates other tissues (40). The SCFA acetate can act in the gut–brain communication, by directly suppressing appetite through hypothalamic activation (41). Conversely, evidence suggested that increased acetate levels in HFD microbiota relay into the parasympathetic nervous system activation driving ghrelin secretion and glucose-stimulated insulin secretion, leading to hyperphagia and metabolic syndrome (42). Other SCFAs are also involved in energy regulation through the gut–brain axis after being sensed in the portal vein and signaling to the autonomous nervous system (54). The gut microbiota also produces or controls the synthesis of other neuroactive signals that can affect the enteric and central nervous system, like g-aminobutyric acid (43) and serotonin (44), both of which could influence appetite and energy balance (55, 56). The contribution of the microbiota-produced neuropeptides to these mechanisms is under active investigation (57).
A group of receptors that senses bacteria-derived metabolites and has been implicated in metabolism is the toll-like receptor family, with TLR2 and TLR4 being particularly important (58). Lipopolysacharide (LPS), a component of the bacterial wall of Gram-negative species, plays a major role in metabolism pathophysiology. Metabolic endotoxemia, in part caused by increased LPS production, is a common consequence of high caloric diets and can affect host metabolism by inducing systemic inflammation and adipose tissue fibrosis, as well as decreasing thermogenesis and hepatic glucose metabolism (45–47). Accordingly, genetic inactivation of TLR4 in hematopoietic cells protects from NAFLD occurrence in mice housed at thermoneutrality (59).
An example of microbial–host interaction is trimethylamine N-oxide (TMAO), a product of the co-metabolism of commensal bacteria, producing trimethylamine from dietary precursors, and the liver, which metabolizes it into TMAO through the flavin monooxigenase proteins family (FMOs). Thus, TMAO levels depend on diet, commensal bacteria, and genetics of the host (48). Plasma levels of TMAO have been associated to atherosclerotic plaques and stroke risk in the past (60). Surprisingly, while the knock-out of Fmo3, the main enzyme for TMAO production, protects against HFD in mice by promoting beige fat development (49), chronic TMAO infusion improves glucose control and increases insulin secretion in vivo and in vitro by reducing endoplasmic reticulum stress potentially through a chaperon property (15).
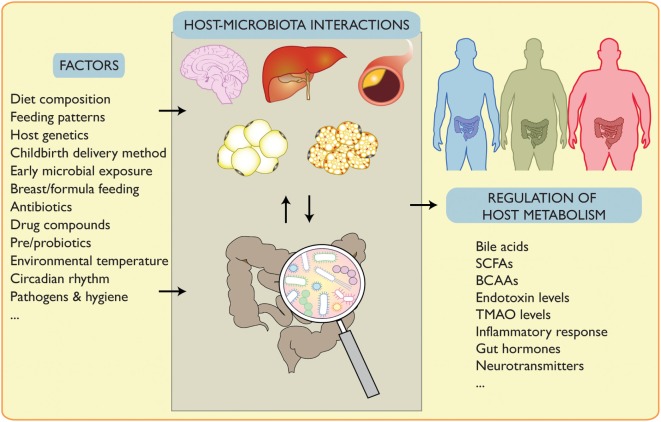
Using integrative metabolomics–metagenomics approaches, Pedersen and colleagues identified Prevotella copri and Bacteroides vulgatus as main species positively correlating their branched-chain amino acids (BCAAs) biosynthesis capacity with insulin resistance in humans. When supplementing P. copri to mice during HFD, BCAAs circulating levels increased, inducing insulin resistance and glucose intolerance (61). This is in line with several studies showing that elevated BCAAs can lead to metabolic disorders (62, 63) and provide correlation between their levels and diabetic status (64, 65) (Figure 1).
Figure 1.
Signals affecting host–microbiota interplay and its regulation of metabolism. Gut microbiota composition is affected by endogenous and exogenous factors such as lifestyle interventions. Changes in the microbiota affect its interplay with several organs and can regulate pathophysiological conditions. This can be mediated by altered bile acids, short chain fatty acids (SCFAs), branched-chain amino acids (BCAAs), endotoxin, trimethylamine N-oxide (TMAO), inflammation, gut hormones and neurotransmitters, and potentially other factors.
Dietary Interventions and Therapeutic Potential of Microbiota Reshaping
Feeding Patterns and Microbiota Compositional Fluctuations
Different lifestyles are associated with changes in microbiota composition, which can result in different efficacy in energy extraction from food and, therefore, impact host metabolism (7, 12). The general microbial composition, as well as the abundance of multiple taxa, undergoes circadian oscillations (66–68). This rhythmicity is dictated by the feeding pattern of the host controlled by its own circadian clock, as genetic depletion of the clock machinery, or its disruption due to jet lag induces dysbiosis and loss of diurnal cycling. In turn, the microbiota too can influence the circadian fluctuation of intestinal epithelial cells (69) and affect intestinal and hepatic metabolism through rhythmic patterns of attachment to the mucosa and metabolomic changes (70). The timing of meals, therefore, influences acute compositional fluctuations in the gut microbiota and modulate microbial-dependent effects on the host. For instance, time-restricted feeding (limiting food access to 10–11 h/day) reduces body weight and improves well-being in overweight individuals (71). While the authors did not explore the subsequent changes in the microbiota of the people involved in their study, in HFD-fed mice, time restriction is associated with a decrease of obesogenic taxa and an increase in beneficial bacteria, thus improving host metabolism (67).
Dietary Fibers and Prebiotics
Diet composition is one of the most important factors that shape the gut microflora. Diets rich in saturated fatty acids are associated with insulin resistance and adipose inflammation (72), whereas polyunsaturated fats have an insulin-sensitizing role (73). Both kinds of dietary lipids affect metabolism through compositional changes in the gut microbiota and signaling of microbial byproducts to the host (74). Protein intake and protein–carbohydrate ratio also impact the production of multiple bacterial metabolites (75). Carbohydrates constitute an important source of energy for the microbiota and, as mentioned above, their byproducts—the SCFAs influence host metabolism. The reduced fiber intake in western diets is associated with reduced bacterial richness and metabolic disorders (76), both of which can be rescued in overweight individuals by dieting (77, 78). Increased fiber consumption leads to improved postprandial glucose metabolism in response to whole grain-based meals (79) and is associated with an increase in Prevotella abundance (80) and a higher ratio of Prevotella over Bacteroides, the two main genera of the Bacteroidetes phylum. During fiber-rich diet, Prevotella appears to positively interact with species from the Actinobacteria, Firmicutes, Proteobacteria, and Archaea phyla to form a niche of bacteria involved in carbohydrate fermentation (81). This contributes to an improved glucose metabolism through increased hepatic glycogen storage (81).
Administration of oligofructose in obese mice regulates appetite, reduces obesity, and the related metabolic disturbances. These improvements are associated with 100-fold increase in the abundance of Akkermansia muciniphila, increased growth of Bifidobacteria, and Lactobacilli, and expression of antimicrobial peptides by the host (82, 83). Studies on healthy and obese individuals demonstrate expansion in Bifidobacterium species and Faecalibacterium prausnitzii during prebiotic treatment. Prebiotics can also induce satiety by regulating the SCFAs (84) and increasing Peptide YY and GLP1 production by the L cells in the ileon and the colon (85–87). In turn, these enteroendocrine hormones inhibit the hypothalamic orexigenic (hunger-inducing) regions and stimulate the anorexigenic (satiety-inducing) neurons (85–88). Fiber-rich diets also impact the interaction between the microbiota and the intestinal mucosal layer, a barrier that separates the epithelium from direct contact with bacteria, constituting a first level of defense against pathogen infection (89). Prebiotic treatment promotes production of the glucagon-like peptide-2, which increases mucosal barrier function and reduces endotoxin-driven inflammation in obese mice (90). Conversely, disruption or ablation of the mucose layer leads to intestinal inflammation, colitis, and even cancer (91–93). In absence of dietary fibers, the mucus layer is dramatically reduced due to expansion of a mucin-degrading bacterial niche. This causes susceptibility to enteric pathogens (94) and increases the predisposition toward metabolic disorders. Indeed, monocolonization with Bacteroides thetaiotamicron, a mucin degrader in absence of other available sources of energy, causes impaired glucose tolerance through decreased hepatic glycogen storage (81). Conversely, fiber-rich diets in humans promote the presence of species from the Xylanibacter, Prevotella, Butyrivibrio, and Treponema genera, preventing the colonization of intestinal pathogens like Enterobacteriaceae (95, 96). A vertical study in mice addressed the long-term effects on microbial changes in response to a low fiber diet. Whereas reverting to a fiber-rich diet within a single generation mostly restores the microbial composition, the loss of microbial taxa under fiber-low diets is not reversible after several generations (97). These results suggest that in addition to the dietary changes, it may be necessary to reintroduce beneficial taxa that are currently lost in the Western microbiota in order to prevent the diseases associated with it.
Probiotics
Probiotics are live bacteria, usually present in fermented foods, whose intake improves metabolic health. Their supplementation in diet has been associated with protective effects against irritable bowel syndrome (IBS), ulcerative colitis, allergic diseases, and obesity in both rodents and humans. They are mostly Gram-positive bacteria belonging either to the Lactobacillus or Bifidobacterium genera, although a Gram-negative, non-pathogenic, Escherichia coli strain has also a probiotic effect (98). The mechanism of action of probiotics is quite heterogeneous and depends on the specific strain used. The anti-obesity effects include reducing metabolic endotoxemia (99–101), improving endothelial dysfunction in obese mice (102, 103), improving hepatic steatosis (104), and limiting free fatty acids available to the host (105). This wide range of effects is mediated by multiple, mutually linked mechanisms like increased intestinal adhesion and colonization that limit the colonization of less beneficial bacteria, production of metabolites such as SFCA and poly-unsaturated fatty acids (106, 107), release of antibacterial molecules called bacterocins (108), and strengthening of the intestinal epithelial integrity and the intestinal mucus layer (109). Recently, in addition to “traditional” probiotic species, A. muciniphila has gained a lot of interest. Abundance of this species is inversely correlated with body weight and insulin resistance, and its increase is another effect of metformin treatment (110). Daily supplementation of A. muciniphila in mice ameliorates HFD-induced metabolic dysfunctions (111), and prevents the increased intestinal absorptive surface and caloric uptake during cold exposure (26). Even pasteurized, A. muciniphila potently reduces body weight gain and insulin resistance in obese mice, due to an outer membrane protein called Amuc_1100, which activates TLR2 and restores intestinal gut barrier function (112). Since A. muciniphila is a strict anaerobic species, the discovery that it can exert its protective function against metabolic disorders after pasteurization makes it a more manageable and therapeutically interesting tool.
Fecal Transplants
Another way to restore a dysbiotic state and reintroduce beneficial taxa is through fecal microbiota transplant (FMT) from a healthy donor. It is currently mainly used to restore intestinal balance in patients affected by recurrent Clostridium difficile infections, with a success rate up to 94% and without adverse effects (113). Since dysbiotic states are clinically similar regardless of the origin, this therapy is currently being tried also for non-infectious intestinal pathologies, like intestinal bowel disease and IBS (114, 115), with first few randomized trials that suggest, at least for IBS, a recovery in bacterial richness after transplantation and an attenuation of the symptoms (114, 116). In the context of the metabolic syndrome, a first human trial on obese Caucasian male subjects showed an increase in peripheral insulin sensitivity in patients receiving allogenic gut microbiota, as well as a tendency to increased hepatic insulin sensitivity (117). Subsequent analyses on this and other cohorts of human patients undergoing FMT (118) have suggested that the stimulation of the recipient microbiota with the donor one has an important impact on the efficiency of the microbial transfer and its persistence in the host and that, therefore, the outcome of FMT depends on the composition of both microbiota (119, 120). With FMT being suggested also for a plethora of other pathologies including anxiety, depression, and even autism (116, 121), increasing our knowledge on the function and the interaction of the gut microflora within itself and with the host will, therefore, be paramount in order to design microbiota-based therapies.
Perspectives
Dissecting how bacterial cues are sensed and act on host physiology is essential to either modulate the microbiota or mimic its signals in a therapeutic perspective. Nevertheless, the known variability of microbial ecosystems in humans is currently a constraint for standard treatments. We can envision an approach where the advances in gut microbiota profiling applied to personalized medicine could allow the definition of pipelines for treatments aiming at re-establishing a healthy microflora. These considerations can potentially overcome the current obstacles in single taxa reintroduction or fecal microbiota transfer and could rely on sequential treatments to reopen ecological niches for beneficial bacteria.
Author Contributions
The authors reviewed literature, conceived, and wrote the manuscript and artwork.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the Trajkovski lab members for meaningful discussions and apologize to the many researchers whose contribution could not be mentioned here.
Footnotes
Funding. SF is supported by a postdoctoral fellowship of the Peter und Traudl Engelhorn Stiftung. This work was funded by the European Research Council under the European Union’s Seventh Framework Programme (FP/2007 2013)/ERC Grant Agreement n. 336607 (ERC-2013-StG-336607) and the Swiss National Science Foundation Professorship grants (PP00P3_144886 and PP00P3_172906) to MT.
References
- 1.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol (2016) 14:e1002533. 10.1371/journal.pbio.1002533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao L, Feng Q, Liang S, Sonne SB, Xia Z, Qiu X, et al. A catalog of the mouse gut metagenome. Nat Biotechnol (2015) 33:1103–8. 10.1038/nbt.3353 [DOI] [PubMed] [Google Scholar]
- 3.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A (2010) 107:11971–5. 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, et al. Human genetics shape the gut microbiome. Cell (2014) 159:789–99. 10.1016/j.cell.2014.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macpherson AJ, de Aguero MG, Ganal-Vonarburg SC. How nutrition and the maternal microbiota shape the neonatal immune system. Nat Rev Immunol (2017) 17:508–17. 10.1038/nri.2017.58 [DOI] [PubMed] [Google Scholar]
- 6.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A (2005) 102:11070–5. 10.1073/pnas.0504978102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature (2006) 444:1027–31. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- 8.Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature (2015) 528:262–6. 10.1038/nature15766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dumas ME, Barton RH, Toye A, Cloarec O, Blancher C, Rothwell A, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci U S A (2006) 103:12511–6. 10.1073/pnas.0601056103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karlsson FH, Fak F, Nookaew I, Tremaroli V, Fagerberg B, Petranovic D, et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun (2012) 3:1245. 10.1038/ncomms2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckel RH, Grundy SM, Zimmet PZ. The metabolic syndrome. Lancet (2005) 365:1415–28. 10.1016/S0140-6736(05)66378-7 [DOI] [PubMed] [Google Scholar]
- 12.Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A (2004) 101:15718–23. 10.1073/pnas.0407076101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mestdagh R, Dumas ME, Rezzi S, Kochhar S, Holmes E, Claus SP, et al. Gut microbiota modulate the metabolism of brown adipose tissue in mice. J Proteome Res (2012) 11:620–30. 10.1021/pr200938v [DOI] [PubMed] [Google Scholar]
- 14.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science (2013) 341:1241214. 10.1126/science.1241214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dumas ME, Rothwell AR, Hoyles L, Aranias T, Chilloux J, Calderari S, et al. Microbial-host co-metabolites are prodromal markers predicting phenotypic heterogeneity in behavior, obesity, and impaired glucose tolerance. Cell Rep (2017) 20:136–48. 10.1016/j.celrep.2017.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Roy T, Llopis M, Lepage P, Bruneau A, Rabot S, Bevilacqua C, et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut (2013) 62:1787–94. 10.1136/gutjnl-2012-303816 [DOI] [PubMed] [Google Scholar]
- 17.Thaiss CA, Itav S, Rothschild D, Meijer M, Levy M, Moresi C, et al. Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature (2016) 540:544–51. 10.1038/nature20796 [DOI] [PubMed] [Google Scholar]
- 18.Nicolas S, Blasco-Baque V, Fournel A, Gilleron J, Klopp P, Waget A, et al. Transfer of dysbiotic gut microbiota has beneficial effects on host liver metabolism. Mol Syst Biol (2017) 13:921. 10.15252/msb.20167356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature (2012) 490:55–60. 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- 20.Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature (2013) 498:99–103. 10.1038/nature12198 [DOI] [PubMed] [Google Scholar]
- 21.Lee H, Ko G. Effect of metformin on metabolic improvement and gut microbiota. Appl Environ Microbiol (2014) 80:5935–43. 10.1128/AEM.01357-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Manneras-Holm L, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med (2017) 23:850–8. 10.1038/nm.4345 [DOI] [PubMed] [Google Scholar]
- 23.Walters WA, Xu Z, Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett (2014) 588:4223–33. 10.1016/j.febslet.2014.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sze MA, Schloss PD. Looking for a signal in the noise: revisiting obesity and the microbiome. MBio (2016) 7(4):e01018-16. 10.1128/mBio.01018-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finucane MM, Sharpton TJ, Laurent TJ, Pollard KS. A taxonomic signature of obesity in the microbiome? Getting to the guts of the matter. PLoS One (2014) 9:e84689. 10.1371/journal.pone.0084689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chevalier C, Stojanovic O, Colin DJ, Suarez-Zamorano N, Tarallo V, Veyrat-Durebex C, et al. Gut microbiota orchestrates energy homeostasis during cold. Cell (2015) 163:1360–74. 10.1016/j.cell.2015.11.004 [DOI] [PubMed] [Google Scholar]
- 27.Suarez-Zamorano N, Fabbiano S, Chevalier C, Stojanovic O, Colin DJ, Stevanovic A, et al. Microbiota depletion promotes browning of white adipose tissue and reduces obesity. Nat Med (2015) 21:1497–501. 10.1038/nm.3994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev (2004) 84:277–359. 10.1152/physrev.00015.2003 [DOI] [PubMed] [Google Scholar]
- 29.Zietak M, Kovatcheva-Datchary P, Markiewicz LH, Stahlman M, Kozak LP, Backhed F. Altered microbiota contributes to reduced diet-induced obesity upon cold exposure. Cell Metab (2016) 23:1216–23. 10.1016/j.cmet.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, et al. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci U S A (2011) 108(Suppl 1):4523–30. 10.1073/pnas.1006734107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flynn CR, Albaugh VL, Cai S, Cheung-Flynn J, Williams PE, Brucker RM, et al. Bile diversion to the distal small intestine has comparable metabolic benefits to bariatric surgery. Nat Commun (2015) 6:7715. 10.1038/ncomms8715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parseus A, Sommer N, Sommer F, Caesar R, Molinaro A, Stahlman M, et al. Microbiota-induced obesity requires farnesoid X receptor. Gut (2017) 66:429–37. 10.1136/gutjnl-2015-310283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang S, Suh JM, Reilly SM, Yu E, Osborn O, Lackey D, et al. Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance. Nat Med (2015) 21:159–65. 10.1038/nm.3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang C, Xie C, Lv Y, Li J, Krausz KW, Shi J, et al. Intestine-selective farnesoid X receptor inhibition improves obesity-related metabolic dysfunction. Nat Commun (2015) 6:10166. 10.1038/ncomms10166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie C, Jiang C, Shi J, Gao X, Sun D, Sun L, et al. An intestinal farnesoid X receptor-ceramide signaling axis modulates hepatic gluconeogenesis in mice. Diabetes (2017) 66:613–26. 10.2337/db16-0663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab (2009) 10:167–77. 10.1016/j.cmet.2009.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Broeders EP, Nascimento EB, Havekes B, Brans B, Roumans KH, Tailleux A, et al. The bile acid chenodeoxycholic acid increases human brown adipose tissue activity. Cell Metab (2015) 22:418–26. 10.1016/j.cmet.2015.07.002 [DOI] [PubMed] [Google Scholar]
- 38.Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A (2006) 103:3920–5. 10.1073/pnas.0509592103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Worthmann A, John C, Ruhlemann MC, Baguhl M, Heinsen FA, Schaltenberg N, et al. Cold-induced conversion of cholesterol to bile acids in mice shapes the gut microbiome and promotes adaptive thermogenesis. Nat Med (2017) 23:839–49. 10.1038/nm.4357 [DOI] [PubMed] [Google Scholar]
- 40.Inoue D, Kimura I, Wakabayashi M, Tsumoto H, Ozawa K, Hara T, et al. Short-chain fatty acid receptor GPR41-mediated activation of sympathetic neurons involves synapsin 2b phosphorylation. FEBS Lett (2012) 586:1547–54. 10.1016/j.febslet.2012.04.021 [DOI] [PubMed] [Google Scholar]
- 41.Frost G, Sleeth ML, Sahuri-Arisoylu M, Lizarbe B, Cerdan S, Brody L, et al. The short-chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat Commun (2014) 5:3611. 10.1038/ncomms4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perry RJ, Peng L, Barry NA, Cline GW, Zhang D, Cardone RL, et al. Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature (2016) 534:213–7. 10.1038/nature18309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yunes RA, Poluektova EU, Dyachkova MS, Klimina KM, Kovtun AS, Averina OV, et al. GABA production and structure of gadB/gadC genes in Lactobacillus and Bifidobacterium strains from human microbiota. Anaerobe (2016) 42:197–204. 10.1016/j.anaerobe.2016.10.011 [DOI] [PubMed] [Google Scholar]
- 44.O’Mahony SM, Clarke G, Borre YE, Dinan TG, Cryan JF. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav Brain Res (2015) 277:32–48. 10.1016/j.bbr.2014.07.027 [DOI] [PubMed] [Google Scholar]
- 45.Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes (2007) 56:1761–72. 10.2337/db06-1491 [DOI] [PubMed] [Google Scholar]
- 46.Okla M, Wang W, Kang I, Pashaj A, Carr T, Chung S. Activation of toll-like receptor 4 (TLR4) attenuates adaptive thermogenesis via endoplasmic reticulum stress. J Biol Chem (2015) 290:26476–90. 10.1074/jbc.M115.677724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vila IK, Badin PM, Marques MA, Monbrun L, Lefort C, Mir L, et al. Immune cell Toll-like receptor 4 mediates the development of obesity- and endotoxemia-associated adipose tissue fibrosis. Cell Rep (2014) 7:1116–29. 10.1016/j.celrep.2014.03.062 [DOI] [PubMed] [Google Scholar]
- 48.Bennett BJ, de Aguiar Vallim TQ, Wang Z, Shih DM, Meng Y, Gregory J, et al. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab (2013) 17:49–60. 10.1016/j.cmet.2012.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schugar RC, Shih DM, Warrier M, Helsley RN, Burrows A, Ferguson D, et al. The TMAO-producing enzyme flavin-containing monooxygenase 3 regulates obesity and the beiging of white adipose tissue. Cell Rep (2017) 19:2451–61. 10.1016/j.celrep.2017.05.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Green CR, Wallace M, Divakaruni AS, Phillips SA, Murphy AN, Ciaraldi TP, et al. Branched-chain amino acid catabolism fuels adipocyte differentiation and lipogenesis. Nat Chem Biol (2016) 12:15–21. 10.1038/nchembio.1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herman MA, She P, Peroni OD, Lynch CJ, Kahn BB. Adipose tissue branched chain amino acid (BCAA) metabolism modulates circulating BCAA levels. J Biol Chem (2010) 285:11348–56. 10.1074/jbc.M109.075184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cornall LM, Mathai ML, Hryciw DH, McAinch AJ. Diet-induced obesity up-regulates the abundance of GPR43 and GPR120 in a tissue specific manner. Cell Physiol Biochem (2011) 28:949–58. 10.1159/000335820 [DOI] [PubMed] [Google Scholar]
- 53.Kimura I, Ozawa K, Inoue D, Imamura T, Kimura K, Maeda T, et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun (2013) 4:1829. 10.1038/ncomms2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell (2014) 156:84–96. 10.1016/j.cell.2013.12.016 [DOI] [PubMed] [Google Scholar]
- 55.Heisler LK, Jobst EE, Sutton GM, Zhou L, Borok E, Thornton-Jones Z, et al. Serotonin reciprocally regulates melanocortin neurons to modulate food intake. Neuron (2006) 51:239–49. 10.1016/j.neuron.2006.06.004 [DOI] [PubMed] [Google Scholar]
- 56.Delgado TC. Glutamate and GABA in appetite regulation. Front Endocrinol (2013) 4:103. 10.3389/fendo.2013.00103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mazzoli R, Pessione E. The neuro-endocrinological role of microbial glutamate and GABA signaling. Front Microbiol (2016) 7:1934. 10.3389/fmicb.2016.01934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jialal I, Kaur H, Devaraj S. Toll-like receptor status in obesity and metabolic syndrome: a translational perspective. J Clin Endocrinol Metab (2014) 99:39–48. 10.1210/jc.2013-3092 [DOI] [PubMed] [Google Scholar]
- 59.Giles DA, Moreno-Fernandez ME, Stankiewicz TE, Graspeuntner S, Cappelletti M, Wu D, et al. Thermoneutral housing exacerbates nonalcoholic fatty liver disease in mice and allows for sex-independent disease modeling. Nat Med (2017) 23:829–38. 10.1038/nm.4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med (2013) 19:576–85. 10.1038/nm.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BA, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature (2016) 535:376–81. 10.1038/nature18646 [DOI] [PubMed] [Google Scholar]
- 62.Jang C, Oh SF, Wada S, Rowe GC, Liu L, Chan MC, et al. A branched-chain amino acid metabolite drives vascular fatty acid transport and causes insulin resistance. Nat Med (2016) 22:421–6. 10.1038/nm.4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab (2009) 9:311–26. 10.1016/j.cmet.2009.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Laferrere B, Reilly D, Arias S, Swerdlow N, Gorroochurn P, Bawa B, et al. Differential metabolic impact of gastric bypass surgery versus dietary intervention in obese diabetic subjects despite identical weight loss. Sci Transl Med (2011) 3:80re2. 10.1126/scitranslmed.3002043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, et al. Metabolite profiles and the risk of developing diabetes. Nat Med (2011) 17:448–53. 10.1038/nm.2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell (2014) 159:514–29. 10.1016/j.cell.2014.09.048 [DOI] [PubMed] [Google Scholar]
- 67.Zarrinpar A, Chaix A, Yooseph S, Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab (2014) 20:1006–17. 10.1016/j.cmet.2014.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe (2015) 17:681–9. 10.1016/j.chom.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell (2013) 153:812–27. 10.1016/j.cell.2013.04.020 [DOI] [PubMed] [Google Scholar]
- 70.Thaiss CA, Levy M, Korem T, Dohnalova L, Shapiro H, Jaitin DA, et al. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell (2016) 167:1495–510.e12. 10.1016/j.cell.2016.11.003 [DOI] [PubMed] [Google Scholar]
- 71.Gill S, Panda S, Smartphone App A. Reveals erratic diurnal eating patterns in humans that can be modulated for health benefits. Cell Metab (2015) 22:789–98. 10.1016/j.cmet.2015.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kennedy A, Martinez K, Chuang CC, LaPoint K, McIntosh M. Saturated fatty acid-mediated inflammation and insulin resistance in adipose tissue: mechanisms of action and implications. J Nutr (2009) 139:1–4. 10.3945/jn.108.098269 [DOI] [PubMed] [Google Scholar]
- 73.Oh DY, Talukdar S, Bae EJ, Imamura T, Morinaga H, Fan W, et al. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell (2010) 142:687–98. 10.1016/j.cell.2010.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Caesar R, Tremaroli V, Kovatcheva-Datchary P, Cani PD, Backhed F. Crosstalk between gut microbiota and dietary lipids aggravates wat inflammation through TLR signaling. Cell Metab (2015) 22:658–68. 10.1016/j.cmet.2015.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scott KP, Gratz SW, Sheridan PO, Flint HJ, Duncan SH. The influence of diet on the gut microbiota. Pharmacol Res (2013) 69:52–60. 10.1016/j.phrs.2012.10.020 [DOI] [PubMed] [Google Scholar]
- 76.Hou JK, Abraham B, El-Serag H. Dietary intake and risk of developing inflammatory bowel disease: a systematic review of the literature. Am J Gastroenterol (2011) 106:563–73. 10.1038/ajg.2011.44 [DOI] [PubMed] [Google Scholar]
- 77.Griffin NW, Ahern PP, Cheng J, Heath AC, Ilkayeva O, Newgard CB, et al. Prior dietary practices and connections to a human gut microbial metacommunity alter responses to diet interventions. Cell Host Microbe (2017) 21:84–96. 10.1016/j.chom.2016.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, et al. Dietary intervention impact on gut microbial gene richness. Nature (2013) 500:585–8. 10.1038/nature12480 [DOI] [PubMed] [Google Scholar]
- 79.Nilsson A, Granfeldt Y, Ostman E, Preston T, Bjorck I. Effects of GI and content of indigestible carbohydrates of cereal-based evening meals on glucose tolerance at a subsequent standardised breakfast. Eur J Clin Nutr (2006) 60:1092–9. 10.1038/sj.ejcn.1602423 [DOI] [PubMed] [Google Scholar]
- 80.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature (2014) 505:559–63. 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab (2015) 22:971–82. 10.1016/j.cmet.2015.10.001 [DOI] [PubMed] [Google Scholar]
- 82.Cani PD, Dewever C, Delzenne NM. Inulin-type fructans modulate gastrointestinal peptides involved in appetite regulation (glucagon-like peptide-1 and ghrelin) in rats. Br J Nutr (2004) 92:521–6. 10.1079/BJN20041225 [DOI] [PubMed] [Google Scholar]
- 83.Everard A, Lazarevic V, Derrien M, Girard M, Muccioli GG, Neyrinck AM, et al. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes (2011) 60:2775–86. 10.2337/db11-0227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Salazar N, Dewulf EM, Neyrinck AM, Bindels LB, Cani PD, Mahillon J, et al. Inulin-type fructans modulate intestinal Bifidobacterium species populations and decrease fecal short-chain fatty acids in obese women. Clin Nutr (2015) 34:501–7. 10.1016/j.clnu.2014.06.001 [DOI] [PubMed] [Google Scholar]
- 85.Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr (2009) 101:541–50. 10.1017/S0007114508019880 [DOI] [PubMed] [Google Scholar]
- 86.Parnell JA, Reimer RA. Weight loss during oligofructose supplementation is associated with decreased ghrelin and increased peptide YY in overweight and obese adults. Am J Clin Nutr (2009) 89:1751–9. 10.3945/ajcn.2009.27465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cani PD, Lecourt E, Dewulf EM, Sohet FM, Pachikian BD, Naslain D, et al. Gut microbiota fermentation of prebiotics increases satietogenic and incretin gut peptide production with consequences for appetite sensation and glucose response after a meal. Am J Clin Nutr (2009) 90:1236–43. 10.3945/ajcn.2009.28095 [DOI] [PubMed] [Google Scholar]
- 88.Kaviani S, Cooper JA. Appetite responses to high-fat meals or diets of varying fatty acid composition: a comprehensive review. Eur J Clin Nutr (2017). 10.1038/ejcn.2016.250 [DOI] [PubMed] [Google Scholar]
- 89.McGuckin MA, Linden SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nat Rev Microbiol (2011) 9:265–78. 10.1038/nrmicro2538 [DOI] [PubMed] [Google Scholar]
- 90.Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut (2009) 58:1091–103. 10.1136/gut.2008.165886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Johansson ME, Gustafsson JK, Holmen-Larsson J, Jabbar KS, Xia L, Xu H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut (2014) 63:281–91. 10.1136/gutjnl-2012-303207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fu J, Wei B, Wen T, Johansson ME, Liu X, Bradford E, et al. Loss of intestinal core 1-derived O-glycans causes spontaneous colitis in mice. J Clin Invest (2011) 121:1657–66. 10.1172/JCI45538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Van der Sluis M, De Koning BA, De Bruijn AC, Velcich A, Meijerink JP, Van Goudoever JB, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology (2006) 131:117–29. 10.1053/j.gastro.2006.04.020 [DOI] [PubMed] [Google Scholar]
- 94.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell (2016) 167:1339–53.e21. 10.1016/j.cell.2016.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A (2010) 107:14691–6. 10.1073/pnas.1005963107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gomez A, Petrzelkova KJ, Burns MB, Yeoman CJ, Amato KR, Vlckova K, et al. Gut microbiome of coexisting BaAka pygmies and bantu reflects gradients of traditional subsistence patterns. Cell Rep (2016) 14:2142–53. 10.1016/j.celrep.2016.02.013 [DOI] [PubMed] [Google Scholar]
- 97.Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature (2016) 529:212–5. 10.1038/nature16504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sonnenborn U, Schulze J. The non-pathogenic Escherichia coli strain Nissle 1917 – features of a versatile probiotic. Microb Ecol Health Dis (2009) 21:122–58. 10.3109/08910600903444267 [DOI] [Google Scholar]
- 99.Lim SM, Jeong JJ, Woo KH, Han MJ, Kim DH. Lactobacillus sakei OK67 ameliorates high-fat diet-induced blood glucose intolerance and obesity in mice by inhibiting gut microbiota lipopolysaccharide production and inducing colon tight junction protein expression. Nutr Res (2016) 36:337–48. 10.1016/j.nutres.2015.12.001 [DOI] [PubMed] [Google Scholar]
- 100.Lim SM, Kim DH. Bifidobacterium adolescentis IM38 ameliorates high-fat diet-induced colitis in mice by inhibiting NF-kappaB activation and lipopolysaccharide production by gut microbiota. Nutr Res (2017) 41:86–96. 10.1016/j.nutres.2017.04.003 [DOI] [PubMed] [Google Scholar]
- 101.Moya-Perez A, Neef A, Sanz Y. Bifidobacterium pseudocatenulatum CECT 7765 reduces obesity-associated inflammation by restoring the lymphocyte-macrophage balance and gut microbiota structure in high-fat diet-fed mice. PLoS One (2015) 10:e0126976. 10.1371/journal.pone.0126976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mauricio MD, Serna E, Fernandez-Murga ML, Portero J, Aldasoro M, Valles SL, et al. Bifidobacterium pseudocatenulatum CECT 7765 supplementation restores altered vascular function in an experimental model of obese mice. Int J Med Sci (2017) 14:444–51. 10.7150/ijms.18354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Toral M, Gomez-Guzman M, Jimenez R, Romero M, Sanchez M, Utrilla MP, et al. The probiotic Lactobacillus coryniformis CECT5711 reduces the vascular pro-oxidant and pro-inflammatory status in obese mice. Clin Sci (Lond) (2014) 127:33–45. 10.1042/CS20130339 [DOI] [PubMed] [Google Scholar]
- 104.Li Z, Jin H, Oh SY, Ji GE. Anti-obese effects of two Lactobacilli and two Bifidobacteria on ICR mice fed on a high fat diet. Biochem Biophys Res Commun (2016) 480:222–7. 10.1016/j.bbrc.2016.10.031 [DOI] [PubMed] [Google Scholar]
- 105.Chung HJ, Yu JG, Lee IA, Liu MJ, Shen YF, Sharma SP, et al. Intestinal removal of free fatty acids from hosts by Lactobacilli for the treatment of obesity. FEBS Open Bio (2016) 6:64–76. 10.1002/2211-5463.12024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Matsuki T, Pedron T, Regnault B, Mulet C, Hara T, Sansonetti PJ. Epithelial cell proliferation arrest induced by lactate and acetate from Lactobacillus casei and Bifidobacterium breve. PLoS One (2013) 8:e63053. 10.1371/journal.pone.0063053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Aoki R, Kamikado K, Suda W, Takii H, Mikami Y, Suganuma N, et al. A proliferative probiotic Bifidobacterium strain in the gut ameliorates progression of metabolic disorders via microbiota modulation and acetate elevation. Sci Rep (2017) 7:43522. 10.1038/srep43522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Martinez FA, Balciunas EM, Converti A, Cotter PD, de Souza Oliveira RP. Bacteriocin production by Bifidobacterium spp. A review. Biotechnol Adv (2013) 31:482–8. 10.1016/j.biotechadv.2013.01.010 [DOI] [PubMed] [Google Scholar]
- 109.Hsieh CY, Osaka T, Moriyama E, Date Y, Kikuchi J, Tsuneda S. Strengthening of the intestinal epithelial tight junction by Bifidobacterium bifidum. Physiol Rep (2015) 3:e12327. 10.14814/phy2.12327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shin NR, Lee JC, Lee HY, Kim MS, Whon TW, Lee MS, et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut (2014) 63:727–35. 10.1136/gutjnl-2012-303839 [DOI] [PubMed] [Google Scholar]
- 111.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A (2013) 110:9066–71. 10.1073/pnas.1219451110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med (2017) 23:107–13. 10.1038/nm.4236 [DOI] [PubMed] [Google Scholar]
- 113.Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis (2011) 53:994–1002. 10.1093/cid/cir632 [DOI] [PubMed] [Google Scholar]
- 114.Halkjaer SI, Boolsen AW, Gunther S, Christensen AH, Petersen AM. Can fecal microbiota transplantation cure irritable bowel syndrome? World J Gastroenterol (2017) 23:4112–20. 10.3748/wjg.v23.i22.4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lane ER, Zisman TL, Suskind DL. The microbiota in inflammatory bowel disease: current and therapeutic insights. J Inflamm Res (2017) 10:63–73. 10.2147/JIR.S116088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mizuno S, Masaoka T, Naganuma M, Kishimoto T, Kitazawa M, Kurokawa S, et al. Bifidobacterium-rich fecal donor may be a positive predictor for successful fecal microbiota transplantation in patients with irritable bowel syndrome. Digestion (2017) 96:29–38. 10.1159/000471919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology (2012) 143:913–6.e7. 10.1053/j.gastro.2012.06.031 [DOI] [PubMed] [Google Scholar]
- 118.Schloissnig S, Arumugam M, Sunagawa S, Mitreva M, Tap J, Zhu A, et al. Genomic variation landscape of the human gut microbiome. Nature (2013) 493:45–50. 10.1038/nature11711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li SS, Zhu A, Benes V, Costea PI, Hercog R, Hildebrand F, et al. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science (2016) 352:586–9. 10.1126/science.aad8852 [DOI] [PubMed] [Google Scholar]
- 120.de Groot PF, Frissen MN, de Clercq NC, Nieuwdorp M. Fecal microbiota transplantation in metabolic syndrome: history, present and future. Gut Microbes (2017) 8:253–67. 10.1080/19490976.2017.1293224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kang DW, Adams JB, Gregory AC, Borody T, Chittick L, Fasano A, et al. Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome (2017) 5:10. 10.1186/s40168-016-0225-7 [DOI] [PMC free article] [PubMed] [Google Scholar]