Abstract
Plants tolerate large variations in the intensity of the light environment by controlling the efficiency of solar to chemical energy conversion. To do this, plants have a mechanism to detect the intensity, duration, and change in light as they experience moving shadows, flickering light, and cloud cover. Sugars are the primary products of CO2 fixation, a metabolic pathway that is rate limited by this solar energy conversion. We propose that sugar is a signal encoding information about the intensity, duration and change in the light environment. We previously showed that the Arabidopsis heterotrimeric G protein complex including its receptor-like Regulator of G signaling protein, AtRGS1, detects both the concentration and the exposure time of sugars [Fu, et al 2014 Cell 156: 1084–1095]. This unique property, designated dose-duration reciprocity, is a behavior that emerges from the system architecture / system motif. Here, we show that another property of the signaling system is to detect large changes in light while at the same time, filtering types of fluctuation in light that do not affect photosynthesis efficiency. When AtRGS1 is genetically ablated, photosynthesis efficiency is reduced in a changing-but not a constant-light environment. Mathematical modeling revealed that information about changes in the light environment is encoded in the amount of free AtRGS1 that becomes compartmentalized following stimulation. We propose that this property determines when to adjust photosynthetic efficiency in an environment where light intensity changes abruptly caused by moving shadows on top of a background of light changing gradually from sun rise to sun set and fluctuating light such as that caused by fluttering leaves.
Keywords: Photosynthesis efficiency, Regulator of G Signaling, WNK kinase, Heterotrimeric G protein, Endocytosis, shadow detector
1. Introduction
Cells detect different chemical stimuli through binding the corresponding signaling molecule to specific receptors that consequently trigger an appropriate response. In many situations, the concentration of these signaling molecules is not static but changes over time. Therefore, cells not only need to act as threshold detectors, but must also be able to interpret or filter temporal variations in the signal. Understanding the control mechanisms that allow cells to respond to changing environmental conditions is a fundamental problem in cell biology.
Two examples demonstrate the importance of time-dependent signaling. The first example pertains to killer T-cell activation by antigen presentation. T-cells must tolerate slowly-changing levels of antigens while still remaining competent to become activated in respond to acute changes in antigen presentation. This is a critical decision because commitment to activate is both energetically costly and precarious to healthy cells. To accomplish this, a change detector for T-cell activation was proposed by Kim and Lee (1) by which T cells compare instantaneous changes in signals to long-term steady-state levels. The second example pertains to plant cells deciding how to allocate newly-fixed carbon, mostly as sugars. Sugar levels change dramatically between the day and night (2) and sources of and sinks for sugars also change over time independently of the changes over the diel cycle. Recently, we proposed a mechanism called Dose-Duration Reciprocity to explain how plant cells are able to generate a maximal response to both low-sustained and high-transient glucose levels (3).
Here, we present a third example that demonstrates how plants are able to respond to rapid changes in both signal frequency and amplitude on top of slow variations in baseline levels. Plants are sessile and unable to escape their environment, therefore they evolved cellular and chemical control mechanisms to cope with the variability of sunlight impinging on the plant. The time scales of this variability can range from sub-seconds to hours. Moving clouds reduce light to different levels and for different times, shadows produce large changes in light over minutes, and moving leaves produce flickering light in a time scale of sub-seconds to seconds. These types of fluctuations occur in the presence of large changes in light caused by the Earth’s daily rotation. In other words, the light signal fluctuates on top of a periodically changing baseline. Therefore, a simple threshold detector is not sufficient for plants to efficiently utilize sunlight; rather additional control mechanisms that allow plants to interpret changing environmental conditions are required.
Without the ability to respond to temporal changes in light, unquenched solar energy would destroy the photosynthetic antenna and potentially kill the entire leaf. On the other hand, not maximizing photosynthesis efficiency reduces competitiveness with potential lethal consequences when neighbors consume the limited light resource. The key question then is how photosynthetic cells detect light change and maximize photosynthetic efficiency accordingly while at the same time guard against photobleaching in a light environment that is unpredictable in duration and intensity.
The identity of the control signals is not known but it is logical to assume that they are products of the light-dependent processes. The immediate products of the light reaction are NADPH, ATP, O2, and protons, which are pumped across the thylakoid membrane of the chloroplast to create a steep pH gradient between the stroma and thylakoid space (lumen). CO2 fixation occurs in the stromal space and is strictly dependent on pH and ATP (4). The immediate product of CO2 fixation is glycerate-3-phosphate which requires ATP and NADPH for final conversion to hexoses, including glucose, thus glucose is rate-limited by the light reactions.
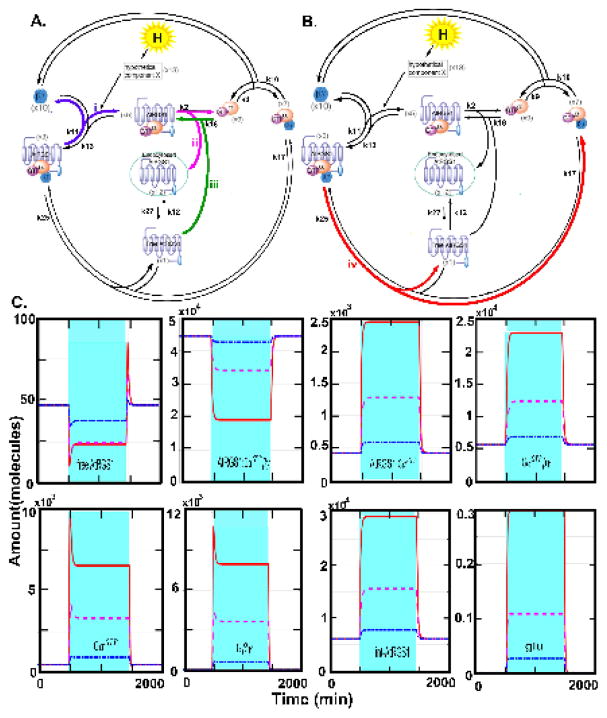
The AtRGS1/heterotrimeric G protein complex is one of three well-studied detectors of glucose in plants (5–9). AtRGS1 is the prototype 7-transmembrane (7TM) Regulator of G Signaling (RGS) family of GTPase-accelerating proteins (5). We constructed a mathematical model of the G protein signaling pathway in plant cells that enables both the concentration and duration of glucose to control cell behavior. The model is shown in Fig. 1A. In this model, the AtRGS1/G protein cycle is not described by a simple two-state system, rather the model describes frequent oscillation between an inactive set of intermediates (left cycle in Fig. 1A) and an active set of intermediates (right cycle in Fig. 1A). In contrast to animal G proteins, the Gα subunit of the plant heterotrimeric G protein complex spontaneously exchanges bound GDP for GTP (10, 11). GTP is not limiting in cells, therefore control occurs by holding the G protein complex in its GDP-bound, inactive state. The Gα binding is in equilibrium between the RGS and Gβγ sites, but D-glucose shifts the equilibrium toward the RGS site and releases Gβγ. AtRGS1 accelerates GTP hydrolysis and interacts with different G protein complex intermediates at the plasma membrane. Thus, removal of AtRGS1 from the plasma membrane allows the G protein complex to move from the inactive cycle to the active cycle. This occurs by AtRGS1 endocytosis (the central part of Fig. 1A). Previously, we found that to appropriately respond to glucose signals of varying strength and duration required two kinases with distinct biochemical properties (3). The first kinase has fast activation kinetics used for pulses of high concentrations of glucose. The second kinase has slow kinetics used for long periods of low concentrations of glucose. Both kinases are members of the WITH NO LYSINE (WNK) kinase family (11); kinase 1 is encoded by the WNK8 and WNK10 genes (x15) and kinase 2 is encoded by the WNK1 gene (x14)
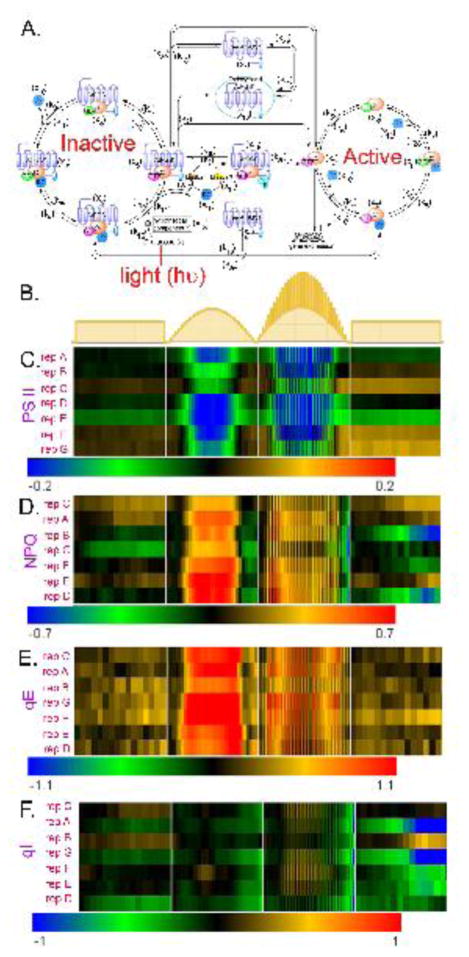
Figure 1. The light-glucose-sensitizing network of the model and relative photosynthesis efficiency and nonphotochemical quenching of rgs1 mutants under 3 different light regimes, and the illumination flicker.
(A) This network has three cycles: (1) the inactivated cycle (left), (2) endocytosis (center), and (3) the active cycle (right). The input of this network is light intensity (hν) which drives photosynthesis to produce glucose (L). When the presumed D-glucose co-receptor x13 receives the glucose stimulus, x13 promotes the AtRGS1/heterotrimer complex (x5) to enter the endocytosis cycle. There are two entry points for endocytosed AtRGS1: (1) phosphorylation by kinase 1 and/or kinase 2 and (2) a phosphorylation-independent pathway controlled by rate k2. This system architecture confers the ability to use both the concentration and the timing of glucose to activate G signaling, a property coined “dose-duration reciprocity” by Yu and coworkers (10). Kinase 1 null mutations abrogate the ability to activate G signaling at high concentrations of glucose for short pulses, but have no effect on the ability to activate G signaling when presented with low concentrations of glucose over a longer time. In contrast, kinase 2 null mutations abrogate the ability to activate G signaling at low concentrations of glucose presented over a long period, but have no effect on the ability to activate G signaling at high concentrations of glucose for short pulses. The components circled in red are “change detectors”: free AtRGS1 (x1) and Gβγ (x10) in the endocytosis, AtRGS1:GαGDP (x4) and AtRGS1:GαGTP (x5) in the inactivated cycle, and GαGDP (x8) and GαGTP (x9) in the activated cycle. These detect the light-on and light-off transitions (this work). (B.) Schematics of the light presentations to 5-week-old plants in chambers under 16:8 light:dark cycles. Details of the day length, light intensity, change in intensity, and fluctuations in light are described in Dutta, et al 2015 (33). The light presentations are labeled as “flat day” followed by “sinusoidal day”, followed by a “fluctuating sinusoidal day” and ending on a flat day. They are not drawn to the same time scale, rather to sampling cycle. (C–F). Heat maps showing the differences in the indicated photosynthesis parameter between an rgs1 null mutant and wild type with the color bar indicating the numerical values. Black on the heat map means that the rgs1 mutant has the wild type value at the indicated time of sampling. Seven replicates of plants were obtained on different days. The replicates are clustered according to similarity. The photosynthesis parameters quantitated are: (C.) PS II Efficiency. (D.) NPQ, nonphotochemical quenching. (E.) qE, (F.) qI.
Here, we show that under conditions of fluctuating light the photosynthesis efficiency of Arabidopsis depends on AtRGS1. Therefore, in addition to dose and duration, the AtRGS1/G protein system detects other temporal properties of natural sunlight. In particular, we use mathematical modeling to demonstrate that the AtRGS1 system has the ability to detect the temporal boundaries between changes in sunlight intensity through free AtRGS1, GαGTP, and Gβγ while filtering noise and remembering the immediate past signaling history. We propose a system motif that encodes the requisite properties of change detector, filter, and memory.
In the following sections, we show that detection of light fluctuations is a property of the AtRGS1/G signaling system and is a form of adaptation behavior. Disruption of this system abrogates the ability of plant cells to regulate the efficiency of photosynthesis in leaf cells. By assuming a direct positive correlation between light intensity/duration and glucose, the signaling system has the emergent property of detecting changes in light intensity and duration such as occurs with shadows passing across the leaf. Interestingly, this property does not require the WNK kinases previously shown to be important for dose-duration reciprocity whereas system memory does. The primary mechanism for this emergent property is the formation and dissociation of AtRGS1:GαGTP βγ and AtRGS1:GαGTP. The amounts of free AtRGS1, GαGTP, and Gβγ are the major constituent of the change detector, and the amount of free AtRGS1 is also the major constituent of the filter. Finally, we speculate that this change detector informs the plant of fluctuations in sunlight intensity used to make the appropriate adjustments in photosynthesis efficiency. This is important in locations such as canopy understory where neighboring plants temporarily shade each other.
2. The emergent property: change detection
Considerable evidence supports AtRGS1 as a component of a glucose-sensing pathway (12–15), however, because the grasses lack homologous 7TM-RGS proteins yet retain a functional heterotrimeric G protein complex (11, 16), the role of a 7TM-RGS protein in glucose sensing must be peripheral such as an add-on component that provides a unique function. In this sense, AtRGS1 is a modulator. Therefore, we hypothesized that AtRGS1 imparts some glucose sensing property needed for non-grass plants, specifically the ability to compete in fluctuating light such as within a canopy. Grasses are unable to compete well with other plant species for access to light and are not successful on forest floors. Biomes where grasses are found are open, typified by the savannahs of the pampas fields in Argentina, the grassland prairies of the Midwest US, and wild rice marshes throughout the world. These biomes do not have forest canopies and thus are not heavily subjected to fluctuation by shadows. Plants that contain 7TM RGS proteins grow in communities where they must compete with other species for light access with maximum efficiency. This suggests that AtRGS1 improves plant fitness in fluctuating light environments. We expect that RGS proteins extracts important features and information from the light pattern that provides plants using RGS proteins with a property that affords a competitive advantage over the grasses which lack RGS proteins.
We assumed that this property involved glucose produced by photosynthesis therefore we compared photosynthesis parameters in plants having or lacking a functional AtRGS1 protein. We tested this idea by subjecting 5-week-old plants to stable and changing light conditions (Fig. 1B) while simultaneously determining photosystem II (PS II) efficiency, nonphotochemical quenching (NPQ, total ability to dissipate energy), the reversible component of NPQ (qE), and the irreversible component of NPQ or photodamage (qI). As shown in Fig. 1C–F, rgs1 null mutants behaved similarly to wild type (WT) under flat day conditions (lights on → lights off, Fig. 1B). However, under varying light, the rgs1 mutants were less efficient in photosystem II. In the first condition, the light environment approximated a sinusoidal pattern using small steps in order to mimic the natural environment of light slowly increased from the experimental dawn time point up to its maximum then decreased to the experimental dusk time point. The second changing light environment was like the first except with pulses of light added. The difference in photosynthesis efficiency between wild type and rgs1 mutants was observed in all 7 biological replicates and to similar degree in most of the replicates. rgs1 mutants had a decreased PS II efficiency (Fig. 1C) due to increased NPQ (Fig. 1D). The regulated and reversible qE component of NPQ and not photochemical damage (qI) accounted for the increased NPQ in the rgs1 mutant (cf. Fig. 1E and F). Under a sinusoidal light environment with fluctuations in light intensity added, the rgs1 mutant more profoundly deviated from wild type behavior in adjusting the NPQ suggesting the component in the light environment that is of most importance is the change in light intensity. Importantly, the behavior does not appear to involve a signal threshold.
3. Identification of the change detectors within the system motif
We used our mathematical model to investigate which components of the G protein signaling pathway respond to the temporal light profiles shown in Fig. 1B. Our previous model used glucose as input whereas here we input light, which will generate glucose from photosynthesis. In order to connect light-intensity to glucose production we made use of the findings of Mäkelä, et al (17) showing a non-linear relationship between light and photosynthesis efficiency. In this case, photosynthesis efficiency saturates as the light intensity increases. Based on these findings, we used the following equation for the time-dependent glucose concentration, L, as a function of the temporal varying light intensity hν(t):
| (3.1) |
where the bi’s are parameters whose values are chosen such that the glucose-input model and light-input model have approximately the same proportion of internalized AtGRS1. In section 5 (Fig. 4), we demonstrated the validity of this equation by comparing the proportion of internalized AtRGS1 between simulation output and the observed experimental data. The light-input model simulated an approximate 15% increment in internalized AtRGS1 at 430 μEm−2s−1 illumination for 10 min. This is what was observed experimentally, therefore, Equation 3.1 properly describes the light-induced (photosynthesis) change in glucose level.
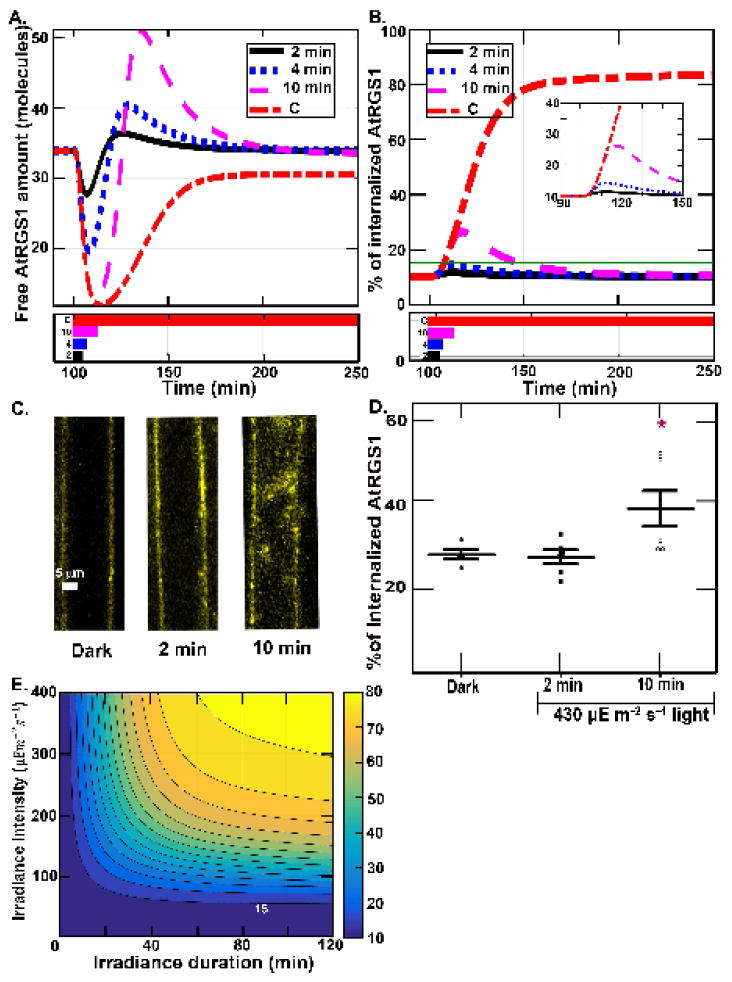
Figure 4. Dose-duration requirement for shadow detector.
(A) Profiles of the amount of free AtRGS1 (x1) after a single illumination for different lengths of time. All simulations start with 100 minutes in darkness and then are illuminated at 430μEm−2s−1 for 2 min. (black solid black), 4 min. (blue dotted blue), 10 min. (magenta dashed) or constantly (red dashed-dotted), as shown by the illumination profiles in the lower panel. The spikes associated with the change in light are dose dependent with the full amplitude reached by a 10-min pulse. (B) The proportion of internalized AtRGS1 from the illumination conditions shown for panel A: 2 min. (black solid black), 4 min. (blue dotted blue), 10 min. (magenta dashed) and constant (red dashed-dotted) illumination. (C–D). Predictions tested. Hypocotyl epidermal cells expressing AtRGS1-YFP (yellow) with chloroplast autofluroescence shown (C) were irradiated at 430 μEm−2s−1 intensity for 2 and 10 minutes. (D) AtRGS1 endocytosis. The proportion of internalized AtRGS1-YFP for the three cases in Fig. 4C. Each dot represents the result for one seedling. As predicted based on the dose and duration properties revealed in panels A and B, a 10-min, but not a 2-min irradiation at 430 μEm−2s−1 produced a significant increase in AtRGS1 endocytosis. (E.) Simulation of dose-duration requirement. All simulations start with 100 minutes in darkness and then are illuminated with sustained irradiance intensity. For all cases, the irradiance intensity is varying from 0 to 400 μEm−2s−1 with an increment of 10 μEm−2s−1 (vertical axis) and the irradiance pulse duration varying from 0 to 2 hours with an increment of 2 minutes ( horizontal axis). The heat map shows the maximum of the proportion of internalized AtRGS1 for each case, where blue and yellow represent 10% and 80% AtRGS1 endocytosis, respectively. All cases have a baseline at 10% maximum of AtRGS1 endocytosis. The contours represent the value of the maximum of AtRGS1 endocytosis from 15% to 80% with increments of 5%. For the highest intensity (i.e., 400μEm−2s−1), the proportion of AtRGS1 endocytosis is significantly increased after 4 minutes stimulation. At the lowest intensity (i.e., 42μEm−2s−1), the proportion of AtRGS1 endocytosis is not detectable, specifically the change in the internal pool of AtRGS1 is less than the detection limits of our technique. For the longest duration (i.e., 2 hours), the proportion of AtRGS1 endocytosis is significantly increased when the intensity is higher than 50μEm−2s−1.
We used our model to investigate the responses of each system component for wild type under periodic light presentation (16 h on-8 h off) as well as constant light (430 μEm−2s−1, 100 μEm−2s−1, and 42 μEm−2s−1) and darkness (Fig. 2). We used the dark steady-state concentrations as initial conditions for the equations (SS1). Fig. 2A shows that AtRGS1:GαGTP (x5), GαGTP (x9), and Gβγ (x10) spiked at lights-on, and free AtRGS1 (x1), AtRGS1:GαGDP (x4), and GαGDP (x8) spiked both at lights-on and lights-off. The other nine model components did not show this behavior. Free AtRGS1 (x1) and GαGDP (x8) had similar decay profiles and the respective amounts at steady state were similar to the steady state for constant irradiation and darkness (Fig. 2A, open and closed circles, respectively). During the light-on period, the amounts of these species returned to pre-illumination levels, indicating a form of adaptation behavior (18). Among these six components, free AtRGS1 and GαGDP were the only ones that showed the spikes at both lights-on and lights-off. Because plant cells need to detect rapid change in light, the response system must contain components that respond on appropriate time scales. This suggests that the G protein system uses free AtRGS1 and GαGDP to detect rapid changes in light.
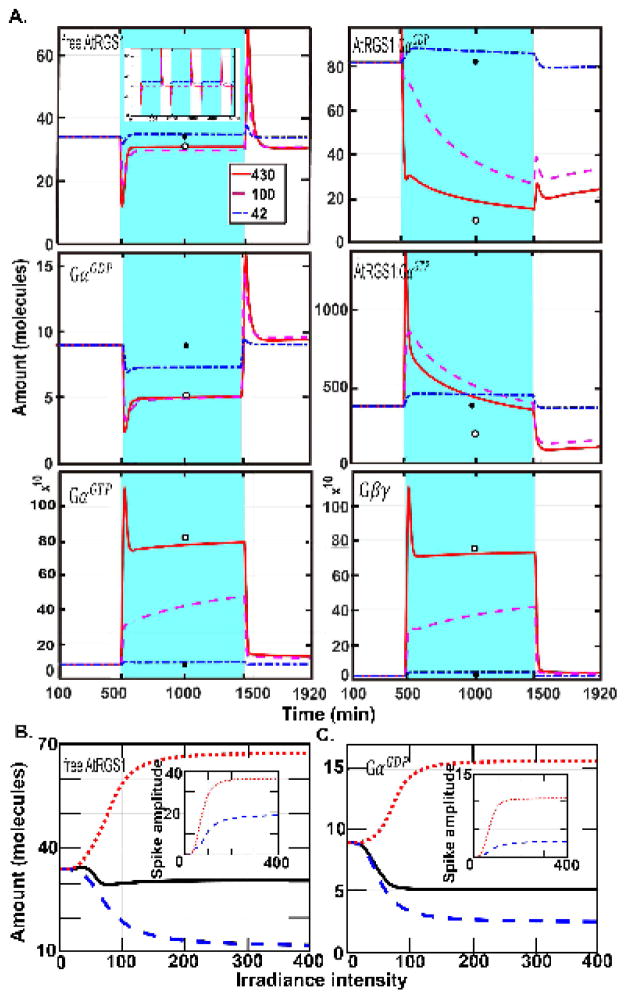
Figure 2. The dynamics of all change detectors for under single illumination pulse and the dependence on intensity.
(A.) The abundance of the six components, free AtRGS1 (x1), AtRGS1:GαGDP (x4), AtRGS1:GαGTP (x5), GαGDP (x8), GαGTP (x9), and Gβγ (x10), highlighted in Fig. 1A show transients over time (spikes). The time and duration of the light is indicated by blue shading. The length of the day is 16 hours. The response of the system to variable illumination from high intensity to low intensity is displayed by the red solid, magenta dashed, and blue dashed-dotted curves. These curves are initialized to the steady state values for darkness, designated SS_dark. The white (open) circle is the steady state for continuous illumination under 430 μEm−2s−1 intensity with no variation and is designated SS_light. When the irradiance intensity is high, these six components spike at the time of illumination or darkness. Hence, free AtRGS1, AtRGS1:GαGDP, AtRGS1:GαGTP, GαGDP, GαGTP, and Gβγ are “change detectors” of illumination. More precisely, AtRGS1:GαGTP, GαGTP, and Gβγ detect the time the irradiance is on, but free AtRGS1, AtRGS1:GαGDP, and GαGDP detect both light on and light off transitions. In addition, free AtRGS1 and GαGDP have a similar profile; and GαGDP and GαGTP also have a similar profile during illumination and darkness. For free AtRGS1, the light pulse mainly changes the value of free AtRGS1 at the time of illumination. At the light on and off transitions, the spike is maintained, but its amplitude decreases when the irradiance level decreases. During a low-level irradiance free AtRGS1 returns to the darkness steady state while a during a higher irradiance level, free AtRGS1 returns to the SS_light, even the value of free AtRGS1 is only slightly changed. Similar behavior occurs at GαGDP. However, for the other change detectors components, AtRGS1:GαGDP (x4), AtRGS1:GαGTP (x5), GαGTP (x9), and Gβγ (x10), their spike behavior vanishes when the irradiance level is reduced and the curves of these components significantly changed between SS light (i.e., the steady state for the continuous high illumination case) and SS_dark (i.e., the steady state for the constant darkness case). Therefore, one of the major functions of free AtRGS1 and GαGDP is light-dark transitions, since they instantly react to the change and return to the pre-stimuli level quickly. (B–C) The relationship between light intensity and the spike amplitude. The solid black curve is the SS dark value for the indicated intensity. The irradiance intensity varied from 0 μEm−2s−1 to 400 μEm−2s−1 with increment 2 μEm−2s−1. The red dotted line represents the amplitude of the spike of free AtRGS1 (left panel) and GαGDP (right panel) induced from light to darkness and the blue dashed lines are the inverse amplitude induced upon darkness to light. The inserted panels account for the amplitude of these spikes shown for free AtRGS1 and GαGDP, where the blue dashed and red dotted curves represent the amplitudes of the first and second spikes, respectively. The maximum, minimum, and the steady state for free AtRGS1 and GαGDP have a similar profile. The distance between the minimum and steady state and the distance between the maximum and steady state are increasing as the irradiance intensity is larger than 50 μEm−2s−1 suggesting that free AtRGS1 and GαGDP spikes when the irradiance intensity is larger than 50 μEm−2s−1.
To further characterize the system’s ability to detect changes in light, we generated stimulus-response curves in which the maximum change and steady state values of free AtRGS1 and GαGDP are computed as a function of light intensity (Fig. 2B–C). The panels in Fig. 2B–C show the relationship between the amplitude of the response and the change in light intensity. Free AtRGS1 and GαGDP spiked when the irradiance intensity was larger than 50 μEm−2s−1, suggesting that there is a threshold for the stimuli intensity needed to trigger a response. Above this threshold, the system only responded in the narrow range of 50 μEm−2s−1 to 150 μEm−2s−1, suggesting that the system behaves in a quasi-switch-like manner.
The amount of free AtRGS1 and GαGDP at steady state under constant light changed only slightly as intensity increased (black line, Fig. 2B), indicating that these components display adaptation behavior. This was not true for the other 4 change detectors, AtRGS1:GαGDP (x4), AtRGS1:GαGTP (x5), GαGTP (x9), and Gβγ (x10) (Supplementary S1).
Because kinases 1 (x15) and 2 (x14) are important in the dose-duration reciprocity, we examined their importance in detecting changes in light. The spike of free AtRGS1, AtRGS1:GαGDP, GαGDP, GαGTP, and Gβγ still occurred in the absence of the fast kinase 1 (x15) and the slow kinase 2 (x14), but only AtRGS1:GαGTP (x5) lost the spike feature in the absence of Wnk8/10 (data not shown). Hence, the model predicts that the kinases do not play a major role in detecting changes in light intensity. Taken together, the change detectors consisted of a sensitizing subsystem that included the components: AtRGS1 complexed with either GPAGDP (x4) or GPAGTP (x5), GPAGTP (x9), GPAGDP (x8), Gβγ (x10), and free AtRGS1 (x1).
4. Change detectors under natural conditions
In order to understand the change detection property under light conditions found in nature, we gradually changed the illumination to mimic a cloudless long (16 h) day from sunrise to sunset. As shown in Fig. 3A, under this condition, free AtRGS1 (x1), and to a lesser degree, GαGDP (x8) displayed the change detection property, but only at the beginning and end of the day. This suggests that small incremental increases in light as occurs from the rotation of the Earth are blind to the change detector.
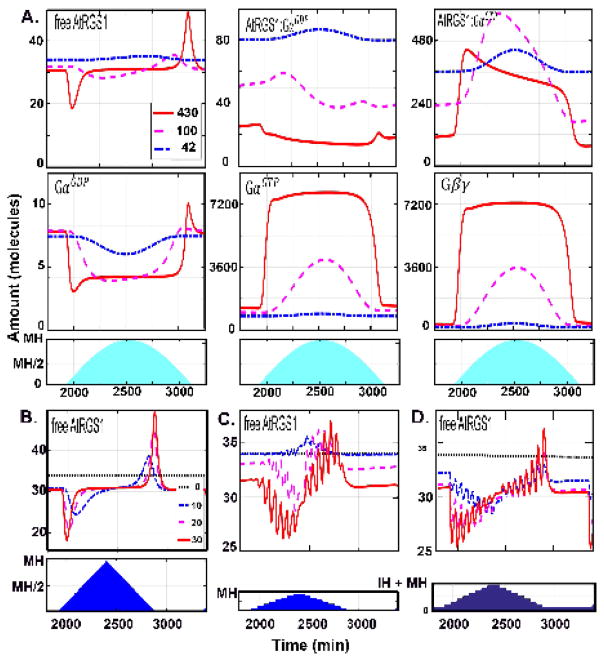
Figure 3. The dynamics of the change detectors for WT under natural settings.
(A.) he change detection property for the six components was analyzed for light presentations mimicking irradiance intensity that over the time of day (namely, a sine function) with different levels of constant cloud cover. The blue shading indicates the irradiance duration and each line represents a day with different intensity maxima. During this day, the intensity increases the first 8 hours, and decreases the next 8 hours with maximal irradiance intensity (MH) at the 8th hour (i.e., noon). Three MH were tested to mimic different levels of constant cloud cover: red solid curve, MH= 430 μEm−2s−1; magenta dashed lines, MH = 100 μEm−2s−1, and blue dashed-dotted lines, MH = 42 μEm−2s−1 lines in the first and second rows. Under these natural light profiles, free AtRGS1 and GαGDP have the most notable spike behavior. (B–D) The frequency of a change in irradiance and the intensity of that change affects the dynamics of free AtRGS1. The irradiance is for 16 hours followed by darkness another 8 hours. The blue shading in the second row indicates how the illumination intensity changed. During the first 8 hours of irradiance period, the intensity increased 10 μEm−2s−1 (thick black dotted curve), 20 μEm−2s−1 (blue dashed-dotted curve), and 30 μEm−2s−1 (red sold curve) every five minutes in (B.), every one hour in (C.) and (D), and the intensity is decreased with the same frequency for the last 8 hours. During darkness, the light intensity = 0μEm−2s−1 in (B. and C.) and 50 μEm−2s−1 in (D). For frequent changes in light intensity (B.), free AtRGS1 only spikes at beginning and end of the day (i.e., irradiance duration). However, at less frequent light intensity changes (C. and D.), free AtRGS1 spikes at each change throughout the day. Moreover, the spike amplitude of free AtRGS1 is larger at the beginning and ending of the day. The light intensity during a night with a full moon does not change the spike feature.
This implies that either large changes in amplitude and/or a sustained period of light are needed before free AtRGS1 (x1) levels change significantly. To distinguish between these two possibilities, we simulated conditions with varying steps in amplitude, and duration, and maximum intensity of the day (cloudy to sunny days) (Fig. 3B–D). The irradiance changed every 10 (Fig. 3B) or 60 minutes (Fig. 3C–D) with the same increment (0 μEm−2s−1, 10 μEm−2s−1, 20 μEm−2s−1, and 30 μEm−2s−1) over the linear light period. The maximum light level (MH) at noon changed accordingly. The amount of light at night was also increased to mimic a full moon (Fig. 3D). When the irradiance was 10 minutes (Fig. 3B), then free AtRGS1 (x1) displayed spikes only at the beginning and end of the day as simulated above using gradual changes in light (Fig. 3A). However, if the irradiance intensity was sustained for 60 minutes, the amount of free AtRGS1 (x1) (Fig. 3C–D) and GαGDP (data not shown) spiked at every change in light intensity. These results indicate that the duration of the change is more important for change detection than the increment of the change itself.
5. Shadow detector
Our hypothesis is that the G protein system uses free AtRGS1 (x1) to detect light/dark changes caused by shadows that meet intensity and duration thresholds. For example, a shadow slowly passing over a leaf may be distinguished from rapid flickering light caused by a leaf in the wind, from light changes caused by the Earth’s rotation. To test this hypothesis, we used the model to compute the amount of internalized AtRGS1 and then performed experiments to test the model’s prediction.
The profiles of the free AtRGS1 under transient illumination and high light intensity (Fig. 4A) showed that the spike amplitude decreased when the duration decreased. The proportion of internalized AtGRS1 (Fig. 4B) indicates that the proportion of internalized AtRGS1 is only significantly increased for the 10 minutes case. Note that there are seven model components that contain AtRGS1, free AtRGS1 (x1), AtRGS1:GαGDP βγ (x2), AtRGS1:GαGTP βγ (x3), AtRGS1:GαGDP (x4), AtRGS1:GαGTP (x5), phosphorylated AtRGS1 (x11), and internalized AtRGS1 (x12), therefore the proportion of internalized AtRGS1 in Fig. 4B is the amount of x12/C1, where C1=x1+x2+x3+x4+x5+x11+x12 is the total amount of AtRGS1. Figs. 4C–D show experimental results verifying the Equation (3.1) and model predictions in Figs. 4A–B. In the experiment, the simulated Arabidopsis seedlings were under high intensity illumination (i.e., 430 μEm−2s−1) for short duration (i.e., 2 and 10 minutes) with the control case being darkness. The proportion of internalized AtRGS1 was only significantly increased in the 10 minute treatment by ~ 15%, which approximated the prediction in Fig. 4B that the proportion of internalized AtRGS1 increased from 10% to ~30%. Hence, the experimental data in Fig. 4C and 4D validates the light-input model (Equation 3.1) quantitatively capturing the proportion of internalized AtGRS1. Moreover, the experimental data in Fig. 4C and 4D supports our major assumption of the light-input model, namely, that there is a correlation between glucose level and light intensity. While it is not possible to measure the glucose level in single cells, the simulated proportion of internalized AtRGS1 correlated with the illumination treatment greater than 2 min duration indicating that the time scale of changing glucose levels match the light intensity change. This means that cells distal to the source of glucose production react roughly with nearly real-time kinetics.
The results shown in Fig. 4E are the model predictions for the maximum proportion of internalized AtRGS1 under varying irradiance duration and intensity. This suggests that most passing clouds that cause short changes in light duration or small changes in intensity do not generate a response by the system.
6. Amplifiers
The above dynamics indicates that the free AtRGS1 (x1) is a critical component in the G protein system, even though its total amount is much smaller than other components in the system. This fact suggests two possibilities: 1) The parameter rate used in simulation is not optimized. For example, a small change in k18, k25, and k27 enlarged the range of free AtRRGS1 (data not shown). 2) There exists an amplifier outside the proposed system motif that detects and respond to changes in free AtRGS1. Because this amplifier should mimic the changes of AtRGS1 (x1) by having both the adaptation behavior and change by a large amount, none of the remaining components in the original light-input model are candidates. Therefore, the candidates must be some proteins or mechanism outside the system. For example, downstream of GαGTP (x9) includes critical gene expression that affect the photon reaction or photosynthesis efficiency which could also feedback on the inactivated cycle (glucose reaction).
7. Robustness of the shadow detector using free AtRGS1, GαGTP, and Gβγ
Our results were based on the final parameter set used in Fu et al. (3). However, they showed that another 50 parameter sets also approximated the wild type experimental data. Therefore, we tested these other 50 parameter sets at different light intensities and found that all 50 sets of parameters approximated the observed data at different irradiance (data not shown).
The shadow detector property of components AtRGS1:GαGDP (x4), AtRGS1:GαGTP (x5), and GαGDP (x8) were sensitive to the parameter values and light intensity. Some depended on kinase 1 (data not shown). However, we found that free AtRGS1 (x1), GαGTP (x9), and Gβγ (x10) operated under all the tested parameter sets and therefore are robust.
8. System memory
We sought to determine how kinase 1 (x15) and 2 (x14) contribute to AtRGS1 internalization in the presence of a fluctuating light input. Toward this end, we tested two versions of our model to simulate and predict AtRGS1 internalization in wild type, kinase 1 minus, and kinase 2 minus cells. Fu et al. (3) determined that the kinases operate on different time scales, with kinase 2 having slower activation kinetics than kinase 1. To investigate how these differential kinetics influence long-term AtRGS1 dynamics, we simulated each model in the presence of constant 100 μEm−2s−1 light intensity until each model achieved steady state (Figs. 5A). While the steady state amount of internalized AtRGS1 in the absence of kinase 1 was only slightly reduced, in the absence of kinase 2 minus, the amount of internalized AtRGS1 reached ~50% of wild type. At 430μEm−2s−1 light intensity, both kinase 1 and kinase 2 minus-models reached steady-levels similar to models containing both kinases (data not shown), suggesting that the differential kinetic contributions of each kinase to AtRGS1 internalization are dose-dependent.
Figure 5. System memory. The system motif has memory encoded by kinase 1.
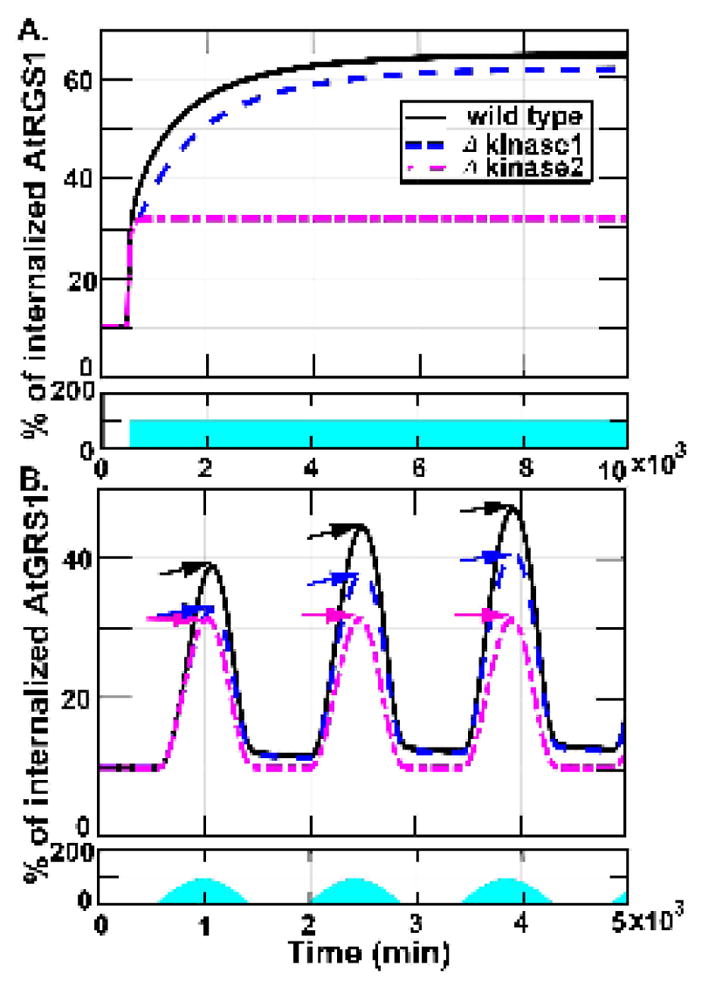
These figures show the proportion of AtRGS1 internalization in wild type, kinase 1, and kinase 2-minus strains under 100μEm−2s−1 light input. The first row in each figure shows the light input that the blue region accounts for light intensity and duration. The second row in each figure shows the proportion of AtRGS1 internalization in wild type (sold curve), kinase 1 minus (dashed curve) strains, and kinase 2-minus (dashed-dotted curve). (A). The AtRGS1 internalization under sustained 100 μEm−2s−1 light intensity. The steady state amount of internalized AtRGS1 in the kinase 1-minus strain is slightly lower. The kinase 2-minus model reaches an amount of internalized AtRGS1 that is approximately 50% of wild type. (B). The AtRGS1 internalization under light treatment as a sinusoidal input function that reached peak amplitude for 100μEm−2s−1. The wild type, kinase 1-minus, and kinase 2-minus models exhibited discreet increases and decreases in AtRGS1 internalization during light intensity increased period and decreased period, respectively. In the wild type and kinase 1-minus models, the internalized AtRGS1 levels are increased following subsequent sinusoidal illumination pulses. In contrast, the kinase 2-minus mutant reaches significantly reduced AtRGS1 internalization levels. Additionally, only the kinase 2-minus model returned to pre-stimulus levels following removal of the illumination. AtRGS1 internalization peaked at approximately 30% at 100 μEm−2s−1 illumination for the kinase 2-minus model (magenta arrows), and increased from approximately 32% (blue arrows) for the kinase 1-minus model and 40% for wild type (black arrows). Hence, the memory is genetically encoded by WNK1 (kinase 2). The return of AtRGS1 to the membrane was slightly delayed with respect to the discreet decreases in the light input in the wild type and kinase 1-minus models, while return of AtRGS1 to the membrane in the kinase 2minus model perfectly aligned with the decreases in the illumination.
Next, we investigated how each theoretical mutant responds to light inputs that change slowly in time mimicking a diel cycle (Figs. 5B). We simulated AtRGS1 internalization for each model using a sinusoidal input function constructed from small step changes in concentration that reached peak amplitude for 100 μEm−2s−1 over a 1000 minute time period. The control, kinase 1 minus, and kinase 2 minus models exhibited discreet increases in AtRGS1 internalization that were coincident with discreet increases in the light input. However, AtRGS1 returning to the membrane was slightly delayed with respect to the discreet decreases in the light input and the level did not return to its pre-stimulus levels in the wild type and kinase 1-minus models. In contrast, AtRGS1 return to the membrane in the kinase 2-minus model perfectly aligned with decreases in the light input. Thus, while the kinase 2-minus model system exhibited a squared sinusoidal AtRGS1 internalization profile, the wild type and kinase 1-minus model systems became more asymmetric with each subsequent squared-sinuosidal pulse of light (note positions of amplitude arrows in succession). Moreover, during irradiation, AtRGS1 internalization peaked at approximately from 40% to 48% for control (i.e., solid line in Fig. 5B), and peaked at approximately from 32% to 40% for the kinase 1-minus model (i.e., dashed line in Fig. 5B). However, AtRGS1 internalization peaked always at approximately 30% for the kinase 2-minus model (i.e., dashed-dotted line in Fig. 5B). On the other hand, only the amount of AtRGS1 internalized in the kinase 2-minus model returned to pre-stimulus levels following removal of the light stimulus. Taken together, these results suggest that the memory of stimulus is genetically encoded by kinase 2 (x14), not kinase 1 (x15). In genetic terms, kinase 1 is encoded by WNKs8/10 and kinase 2 is encoded by WNK1 (3).
9. The system motif underlying the shadow detector
In our system, free AtRGS1 has adaptation behavior such that it returns to its pre-stimulus levels during illumination. Examples of such adaptation behavior are found in many biological systems, such as bacteria (19, 20), yeast (20), and human stem cells (21). Many physiological responses also possess adaptation behavior whereby the system components return to their pre-stimulus levels in the presence of a sustained signal. Examples of such processes include bacterial chemotaxis (22, 23) and MAP kinase signaling in yeast (24, 25).
To explore the system motif responsible for the adaptation behavior, the model was simplified to its critical elements that preserved the dynamics of the complete model (designated toy model). The amount and profile of all components in the toy model was similar to the amount and profile of the corresponding components in the unabridged light-input model (Fig. 1A.). The toy model was formulated as the following process: Gα was more stable when bound with GTP than when bound with GDP, so the components related to GDP, namely, AtRGS1:GαGDP βγ (x2), AtRGS1:GαGDP (x4), GαGDP βγ (x6), and GαGDP (x8), were effectively removed from the system. Moreover, in order to test whether phosphorylation was the critical process to generate the adaptation behavior, the components related to phosphorylation, namely, phosphorylated AtRGS1 (x11), kinase1 (x15), and kinase2 (x14), were also removed from the system. The toy model lacked GαGDP (x8), indicating that GαGDP is not critical components for the shadow detector, even though GαGDP has a profile similar to free AtRGS1 (x1).
Based on the toy model, we propose the mechanism for the dynamic behavior underlying the shadow detector. As illustrated in Fig. 6, the analysis revealed four predominate processes: (i) dissociation of AtRGS1:GαGTP βγ, (ii) dissociation of AtRGS1:GαGTP, (iii) formation of AtRGS1:GαGTP, and (iv) dissociation of AtRGS1:GαGTP βγ to generate the spike and adaptation behavior of free AtRGS1 (x1). The loop contained AtRGS1:GαGTP βγ (x3), AtRGS1:GαGTP (x5), internalized AtRGS1 (x12), free AtRGS1 (x1) generated a feedback loop for free AtRGS1 (x1). A substrate depletion of AtRGS1:GαGTP βγ with a delayed return of free AtRGS1 (x1) generated the adaptation behavior of free AtRGS1 during illumination. Substrate restoration of AtRGS1:GαGTP βγ caused a rapid increment of free AtRGS1 onset of a shadow. Formation of AtRGS1:GαGTP through process (iii) generates the spike of GαGTP (x9) and Gβγ (x10).
Figure 6. System architecture for change detection property.
(A–B). The full model shown in Fig. 1A is condensed to the key elements that impart the change detection property. The minimal structure of the light-input model that generates the shadow detector property are free AtRGS1 (x1), GαGTP (x9), and Gβγ (x10) and the adaptation of free AtRGS1. Components AtRGS1:GαGTP βγ (x3) and AtRGS1:GαGTP (x5) are in the inactive cycle (left), GαGTP βγ (x7) and GαGTP (x9) are in the active cycle (right). Note that the kinases (kinase 1, x15, and kinase 2, x14 5) and the phosphorylated AtRGS1 (x11) in Fig. 1 are removed. The marked processes (i)–(iv) represent: (i) the dissociation of AtRGS1:GαGTP βγ (x3) to AtRGS1:GαGTP (x5) and Gβγ (x10), (ii) the dissociation of AtRGS1:GαGTP (x5) to GαGTP (x9) and internalized AtRGS1 (x12), (iii) the formation of AtRGS1:GαGTP (x5) from the association of GαGTP (x9) and free AtRGS1 (x1), and (iv) the dissociation of AtRGS1:GαGTP βγ (x3) to free AtRGS1 (x1) and GαGTP βγ (x7). (A). The major processes that affect the system behavior during passing of the shadow are indicated by (i)–(iii). (B). The major process that affects the system behavior during onset of a shadow is indicated by (iv). (C) The dynamics of this reduced system under sustained irradiance presentation recapitulates the full model of Fig. 1. The blue region indicates the period when the lights are on although at different irradiance intensities where the red solid curve = 430 μEm−2s−1, the magenta dashed curve = 100 μEm−2s−1, and the blue dotted curve = 42 μEm−2s−1. When the irradiance intensity is high, free AtRGS1 (x1) spikes at both the dark to light and the light to dark transitions while GαGTP (x9) and Gβγ (x10) spike at the lights on transition. When the irradiance intensity is reduced, the spikes of GαGTP and Gβγ are lost; only free AtRGS1 spikes albeit with a smaller amplitude. The combination of four processes (i)–(iv) generates the spike and adaptation behavior of free AtRGS1. The spike feature of GαGTP and Gβγ is generated by the amount of free AtRGS1 and the process (i).
Passing of the shadow
At the light-off->light-on transition (Fig. 6A), the G protein equilibrium shifts from AtRGS1:GαGTP βγ (x3) to AtRGS1:GαGTP (x5) through process (i) which reduces the free AtRGS1 instantly by depleting the AtRGS1:GαGTP βγ (x3) pool. After time, the generated AtRGS1:GαGTP (x5) dissociates into internalized AtRGS1 (x12) and GαGTP (x9) through process (ii), and the free AtRGS1 (x1) is replenished from the internalized AtRGS1 (x12). This process generates the spike of free AtRGS1 at the light off->on transition. Free AtRGS1 associated with GαGTP (x9) forms AtRGS1:GαGTP (x5) through process (iii), followed by process (ii) to replenish the free AtRGS1 pool. These two processes keep free AtRGS1 as a sustained level (between the minimum and the pre-stimulus level) after the first spike, and thus underlie the mechanism for the adaptation behavior of free AtRGS1. Moreover, the association rate k18 is much larger than k9, which means that GαGTP (x9) preferentially associates with free AtRGS1 (x1) over Gβγ (x10). Hence, process (iii) is the predominant consumer of GαGTP and the generator of the spike of GαGTP. Since Gβγ (x10) is downstream of GαGTP, Gβγ has a similar profile to GαGTP.
Onset of a shadow
At the light-on-> light-off transition (Fig. 6B), the G protein equilibrium shifts back from AtRGS1:GαGTP (x5) to AtRGS1:GαGTP βγ (x3), which increases free AtRGS1 because it increases the AtRGS1:GαGTP βγ (x3) pool through process (iv), and hence generates the spike of free AtRGS1 at the light-on-> light-off transition. Therefore, G protein shifting between AtRGS1:GαGTP βγ (x3) and AtRGS1:GαGTP (x5) generates the spike of free AtRGS1 and the two processes (ii)–(iii) are the reason for adaptation of free AtRGS1.
10. Synopsis
The architecture of heterotrimeric G signaling in plant cells embodies not only the dose-duration reciprocity property previously described but, as shown here, the property to detect light-dark transitions such as occur at the beginning and ending of the day and during the movement of shadows. Hence, the architecture contains a shadow detector. Such a property is useful in maximizing photosynthesis efficiency in a fluctuating light environment.
Integral control is a commonly-used process to control the output error. Adaptation behavior can be generated by integral control such in bacterial chemotaxis (26) and calcium homeostasis (27). However, adaptation behavior can be generated by processes other than integral control (28). A negative feedback loop or an incoherent feedforward loop in models describing the dynamics of some enzymes (e.g. three-node enzyme network) can also generate adaptation behavior (28). In addition, Hao, et al and Ni, et al showed that integral control, feed-forward motifs, and negative feedback loops also generate adaptation behavior (29, 30). In fact, integral control and feed-forward motifs achieve better adaptation behavior than negative-feedback motif (31). Our light-input model showed that the feedback loop (i.e. i. in Fig 6, substrate depletion of AtRGS1:GαGTP βγ with a delayed return of free AtRGS1) can also generate adaptation behavior.
Specifically, our mathematical modeling demonstrated that the AtRGS1 system has the ability to detect the temporal boundaries between changes in sunlight intensity through free AtRGS1, GαGTP, and Gβγ while filtering noise and remembering the immediate past signaling history. Hence, the amounts of free AtRGS1, GαGTP, and Gβγ were the major constituent of the change detector, and the amount of free AtRGS1 was also the major constituent of the filter. Detection of light fluctuations was a property of the AtRGS1/G signaling system and was a form of adaptation behavior. For this adaptation behavior, our reduced system showed that G protein shifting between AtRGS1:GαGTP βγ and AtRGS1:GαGTP generated the spike of free AtRGS1 and the two processes, dissociation of AtRGS1:GαGTP and formation of AtRGS1:GαGTP, are the reason for adaptation of free AtRGS1. Therefore, we hypothesized that this change detector informed the plant of fluctuations in sunlight intensity and was used to make the appropriate adjustments in photosynthesis efficiency.
11. Materials and Methods
Photosynthesis efficiency and nonphotochemical quenching of rgs1 mutants
Experiments measuring photosynthesis efficiency and the calculation of NPQ, qE, and qI are described in Dutta, et al 2015 (32).
AtRGS1 endocytosis
Arabidopsis seeds expressing AtRGS1-YFP were sterilized with ethanol (first with 70%, 10 min, 95% 10 min). Twenty to thirty seeds were then sown on 1-mL liquid 1/4 X Murashige and Skoog (MS) medium without sucrose in 24-well plates and stratified at 5°C for 2 days, followed by 2 hours light, then grown in darkness at 27 °C for 4 days. For best results, the plates kept in darkness should be moved to the microscope room on the third day. Seedlings were moved to a well with 2-mL sterile water for 5–10 minutes to wash out MS. After the wash out, the seedling was irradiated in the well with 430μEm−2s−1 or 100 μEm−2s−1 for 10 minute then moved to the slide for imaging. The hypocotyl epidermal cells imaged for this study were located 2–4 mm below the cotyledon hook. A Zeiss LSM710 confocal laser scanning microscope with C-Apochromat 40X 1.2N.A. water immersion objective was used. The YFP fluorescence was excited by a 514-nm argon laser and the photomultiplier detector was set between 526 nm and 569 nm for quantification. The proportion of internalized AtRGS1 was analyzed by Image J. Illumination was as short as possible to avoid heating or photodamage.
Simulation setting for light-input model
For the final parameter set, the initial steady state denoted as SS0, namely,
used in Fu et al (3) when L= 0 (no glucose) was used as the initial value in the current simulations. This steady state represents plant cells at the end of their dark phase in a diel cycle, a time when the cellular glucose level is extremely low. For the light-input model, the initial value was changed to SS1, by adding the initial value of glucose, L=0, into the 16th component of SS0, namely,
The ODEs described by Fu and coworkers (3) were used except in simulations of the kinase mutants. In these simulations, kinase 1 and 2 mutants were tested by setting x15 (Kinase 1) and x14 (Kinase 2) component values to zero, respectively, while other setting was the same as wild type. The light-input model was performed by the MatLab solver ODE 15S and the Euler implicit method when the illumination was sustained and non-sustained, respectively.
Simulation setting for robustness of the shadow detector
Fu et al. used an evolutionary algorithm to estimate the parameter sets that fit the observed endocytosis data. The uncertainty measurement for each parameter set was measured by the score: squared difference between the model output and experimental data at each time course in the experiment. The method was as described in detail in (3). The simulation output of the top 50 scored parameter sets (k1, …, k30) fitted the experimental data (3) well, namely, the scores were less than 30. Hence, ODEs with the top 50 parameter sets captured the endocytosis behavior in wild type. Moreover, these ODEs also captured the relaxation experimental data (Figure 4 in (3)). The final parameter set in (9) was taken from the median values of these 50 parameter sets, which represented the mean of these parameter values. In order to examine the robustness of the shadow detector, we tested the simulation output of the ODEs based on these 50 parameter sets. We randomly chose 1000 parameter sets within a range of 1/2-2-fold of these 50 parameter sets. The initial value was chosen from the steady state of the light-input model with hv=0 (no light) and initial value at (0, 1000, 50000, 100, 1000, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0). We then used the same simulation setting as in Fig. 2 to test the shadow detector profiles for the ODEs with the 1000 parameter sets.
Highlights.
Plants regulate photosynthesis efficiency to avoid photodamage and to maximize sugar
Trafficking of RGS1 protein is the key mechanism detecting changes in light and dark
System motifs encode a noise filter, memory, and a shadow detector
Acknowledgments
This work was supported by grants from the NIGMS (R01GM065989) and NSF (MCB-0718202) to A.M.J, NSF (MCB-1021363) and NIGMS (R01GM079271) to T.C.E. The Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy through the grant DE-FG02-05er15671 to A.M.J. funded the plant-based experimental part of this project.
Appendix
1. Equations
Glucose-input model
The parameter values are chosen from (11).
Light-input model
where b1 = 0.0426, b2 = 143.6, and b3 = 0.129.
Reduced Light-input model
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kim PS, Lee PP. T cell state transition produces an emergent change detector. J Theor Biol. 2011;275:59–69. doi: 10.1016/j.jtbi.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deuschle K, Chaudhuri B, Okumoto S, Lager I, Lalonde S, Frommer WB. Rapid metabolism of glucose detected with FRET glucose nanosensors in epidermal cells and intact roots of Arabidopsis RNA-Silencing mutants. Plant Cell. 2006;18(9):2314–25. doi: 10.1105/tpc.106.044073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu Y, Lim S, Urano D, Tunc-Ozdemir M, Phan N, Elston T, et al. Reciprocal encoding of signal intensity and duration in a glucose-sensing circuit. Cell. 2014;156(5):1084–96. doi: 10.1016/j.cell.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Werdan K, Heldt HW, Milovancev M. The role of pH in the regulation of carbon fixation in the chloroplast stroma. Studies on CO2 fixation in the light and dark. Biochimica et Biophysica Acta. 1975;396:276–92. doi: 10.1016/0005-2728(75)90041-9. [DOI] [PubMed] [Google Scholar]
- 5.Chen J-G, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, et al. A seven-transmembrane RGS protein that modulates plant cell proliferation. Science. 2003;301:1728–31. doi: 10.1126/science.1087790. [DOI] [PubMed] [Google Scholar]
- 6.Booker K, Schwarz J, Garrett M, Jones A. Glucose attenuation of auxin-mediated bimodality in lateral root formation is partly coupled by the heterotrimeric G protein complex. PLoS ONE. 2010;5:e12833. doi: 10.1371/journal.pone.0012833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J-G, Jones AM. Methods in Enzymology. Academic Press; 2004. AtRGS1 function in Arabidopsis thaliana; pp. 338–50. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Ji F, Xie H, Liang J, Zhang J. The regulator of G-protein signaling proteins involved in sugar and abscisic acid signaling in Arabidopsis seed germination. Plant Physiol. 2006;140:302–10. doi: 10.1104/pp.105.069872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Urano D, Phan N, Jones JC, Yang J, Huang J, Grigston J, et al. Endocytosis of the seven-transmembrane RGS1 protein activates G-protein-coupled signalling in Arabidopsis. Nat Cell Biol. 2012;14(10):1079–88. doi: 10.1038/ncb2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnston CA, Taylor JP, Gao Y, Kimple AJ, Grigston JC, Chen J-G, et al. GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proceedings of the National Academy of Sciences. 2007;104(44):17317–22. doi: 10.1073/pnas.0704751104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urano D, Jones JC, Wang H, Matthews M, Bradford W, Bennetzen JL, Jones AM. G protein activation without a GEF in the plant kingdom. PLoS Genetics. 2012;8:e1002756. doi: 10.1371/journal.pgen.1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ullah H, Chen JG, Young J, Im KH, Sussman MR, Jones AM. Modulation of cell proliferation by heterotrimeric G-protein in Arabidopsis. Science. 2001;292:2066–2069. doi: 10.1126/science.1059040. [DOI] [PubMed] [Google Scholar]
- 13.Grigston JC, Osuna D, Scheible WR, Stitt M, Jones AM. Structural requisites for acute vs. chronic D-glucose sensing mediated by AtRGS1 and AtGPA1. FEBS Lett. 2008;582:3577–3584. doi: 10.1016/j.febslet.2008.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Booker KS, Schwarz J, Jones AM. Auxin and glucose signaling mediate a novel G protein regulated bimodality in lateral roots. PLoS One published. 2010:17. doi: 10.1371/journal.pone.0012833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urano D, Phan N, Jones JC, Yang J, Huang J, Grigston J, Taylor JP, Jones AM. Endocytosis of seven-transmembrane RGS protein activates G-coupled signaling in Arabidopsis. Nature Cell Biology. 2012;14:1079–1088. doi: 10.1038/ncb2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urano D, Dong T, Bennetzen JL, Jones AM. Adaptive evolution of signaling partners. Mol Biol Evol advance access. 2015 Jan 6; doi: 10.1093/molbev/msu404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mäkelä A, Pulkkinen M, Kolari P, Lagergren F, Berbigier P, Lindroth A, et al. Developing an empirical model of stand GPP with the LUE approach: analysis of eddy covariance data at five contrasting conifer sites in Europe. Global Change Biology. 2008;14:92–108. [Google Scholar]
- 18.Tyson JJ, Chen KC, Novak B. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr Opin Cell Biol. 2003;15:221–31. doi: 10.1016/s0955-0674(03)00017-6. [DOI] [PubMed] [Google Scholar]
- 19.Eichenberger P, Fujita M, Jensen ST, Conlon EM, Rudner DZ, Wang ST, Ferguson C, Haga K, Sato T, Liu JS, Losick R. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2004;2:e328. doi: 10.1371/journal.pbio.0020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 21.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi T-M, Huang Y, Simon MI, Doyle J. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. PNAS. 2000;97:4649–4653. doi: 10.1073/pnas.97.9.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barkai N, Leibler S. Robustness in simple biochemical networks. Nature. 1997;387:913–917. doi: 10.1038/43199. [DOI] [PubMed] [Google Scholar]
- 24.Behar M, Hao N, Dohlman HG, Elston TC. Mathematical and Computational Analysis of Adaptation via Feedback Inhibition in Signal Transduction Pathways. Biophys J. 2007;93(3):806–821. doi: 10.1529/biophysj.107.107516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hao N, Behar M, Elston TC, Dohlman HG. Systems biology analysis of G protein and MAP kinase signaling in yeast. Oncogene. 2007;26(22):3254–66. doi: 10.1038/sj.onc.1210416. [DOI] [PubMed] [Google Scholar]
- 26.Yi TM, Huang Y, Simon MI, Doyle J. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. PNAS. 2000;97(9):4649–4653. doi: 10.1073/pnas.97.9.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Samad H, Goff JP, Khammash M. Calcium homeostasis and parturient hypocalcemia: An integral feedback perspective. Journal of Theoretical Biology. 2002;214:17–29. doi: 10.1006/jtbi.2001.2422. [DOI] [PubMed] [Google Scholar]
- 28.Ma W, Trusina A, El-Samad H, Lim WA, Tang C. Defining network topologies that can achieve biochemical adaptation. Cell. 2009;138:760–773. doi: 10.1016/j.cell.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hao N, Behar M, Elston TC, Dohlman HG. System biology analysis of G protein and MAP kinase signaling in yeast. Oncogene. 2007;14(26):3254–3266. doi: 10.1038/sj.onc.1210416. [DOI] [PubMed] [Google Scholar]
- 30.Ni XY, Drengstig T, Ruoff P. The control of the controller: molecular mechanisms for robust perfect adaptation and temperature compensation. Biophys J. 2009;97(5):1244–1253. doi: 10.1016/j.bpj.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bleris L, Xie Z, Glass D, Adadey A, Sontag E, Benenson Y. Synthetic incoherent feedforward circuits show adaptation to the amount of theor genetic template. Mol Syst Biol. 2011;7:519. doi: 10.1038/msb.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deuschle K, Chaudhuri B, Okumoto S, Lager I, Lalonde S, Frommer WB. Rapid metabolism of glucose detected with FRET glucose nanosensors in epidermal cells and intact roots of Arabidopsis RNA-Silencing mutants. Plant Cell. 2006;18(9):2314–25. doi: 10.1105/tpc.106.044073. [DOI] [PMC free article] [PubMed] [Google Scholar]