Significance
The tumor suppressor protein p53 acts as a transcription factor by binding as a tetramer to response elements made of two DNA half-sites separated by a variable spacer, leading to diverse cellular outcomes. Fifty percent of all p53 response elements contain spacers, but their significance is unclear at present. Here, we show that p53 binds to response elements containing long spacers in two different modes: fully specific and hemispecific. In the latter, only one p53 dimer is specifically bound to a DNA half-site, whereas the other dimer is bound to the spacer DNA. Nonetheless, the two modes have comparable binding affinity and specificity. Our findings expand and diversify the p53 regulatory network.
Keywords: p53, protein/DNA interaction, hemispecific interaction, cyclization kinetics, binding affinity
Abstract
The tumor suppressor protein p53 acts as a transcription factor, binding sequence-specifically to defined DNA sites, thereby activating the expression of genes leading to diverse cellular outcomes. Canonical p53 response elements (REs) are made of two decameric half-sites separated by a variable number of base pairs (spacers). Fifty percent of all validated p53 REs contain spacers between 1 and 18 bp; however, their functional significance is unclear at present. Here, we show that p53 forms two different tetrameric complexes with consensus or natural REs, both with long spacers: a fully specific complex where two p53 dimers bind to two specific half-sites, and a hemispecific complex where one dimer binds to a specific half-site and the second binds to an adjacent spacer sequence. The two types of complexes have comparable binding affinity and specificity, as judged from binding competition against bulk genomic DNA. Structural analysis of the p53 REs in solution shows that these sites are not bent in both their free and p53-bound states when the two half-sites are either abutting or separated by spacers. Cell-based assay supports the physiological relevance of our findings. We propose that p53 REs with long spacers comprise separate specific half-sites that can lead to several different tetrameric complexes. This finding expands the universe of p53 binding sites and demonstrates that even isolated p53 half-sites can form tetrameric complexes. Moreover, it explains the manner in which p53 binds to clusters of more than one canonical binding site, common in many natural REs.
The tumor suppressor protein p53 acts as a transcription factor that binds sequence-specifically to more than 200 validated p53 response elements (REs) (1–3). The p53 protein is one of the most central hubs in human cells activating or repressing target genes involved in various aspects of cellular response to stress, such as cell cycle arrest, DNA repair, senescence, and apoptosis (4, 5). p53 molecules consist of three major functional domains (6, 7). The N terminus contains a transactivation domain, the core domain contains the sequence-specific DNA binding domain (DBD), and the C-terminal domain (CTD) includes a tetramerization domain and a regulatory domain with a sequence-nonspecific DNA binding activity. In ∼50% of all cancers, p53 is inactivated by point mutations where ∼95% of them are in the DBD (8), thus compromising sequence-specific binding of p53 to its REs (9). p53 REs consist of two decameric half-site repeats of the general form RRRCWWGYYY (R = A, G; W = A, T; Y = C, T). Initial determination of the consensus sequence, by selection methods, showed that the two half-sites can be separated by spacers of up to 13 bp (10). On the other hand, studies using chromatin immunoprecipitation (ChIP) recovered mainly sequences that contain 0- to 1-bp spacers (11–14). It should be noted that ∼50% of all validated p53 REs contain spacers now estimated to be from 1 to 18 bp between half-sites (1, 2).
The functional unit of p53 is a tetramer made of a dimer of dimers (15, 16). The ability to dimerize is supported not only by the tetramerization domain but also by the core domains (17–19). Tetramerization occurs on the DNA target during the binding event (20). Cooperative binding of p53 tetramers to DNA (16, 20) is stabilized by both intradimer and interdimer interactions mediated by the core domains (18, 19). Moreover, the level of this cooperativity is encoded by the DNA base sequence (21). Spacer sequences inserted between DNA half-sites are expected to decrease interdimer stabilization of p53–DNA complexes, and hence an open question is how spacer sequences and especially long ones accommodate cooperative binding of p53 tetramers to its REs. It has been previously suggested that p53 bends significantly its REs (e.g., refs. 22 and 23). Here, we show that p53 does not confer significant bending upon its consensus DNA sites, when the sites are abutting or when they contain spacer sequences up to 20-bp long. REs with long spacers (equal or greater than 10 bp) reveal a unique binding mode, termed “hemispecific,” in addition to the conventional specific mode. In the regular binding mode, two p53 dimers are bound to two specific half-sites, whereas in the unexpected hemispecific mode, one dimer binds to a specific half-site and the other to an adjacent nonspecific spacer DNA. These two binding modes are also observed for two natural p53 binding sites incorporating long spacers: CHMP4c (24) and PTEN (25). The physiological relevance of the hemispecific binding mode is further supported by cell-based assays.
Results and Discussion
Binding Patterns of p53 Bound to REs with Spacers.
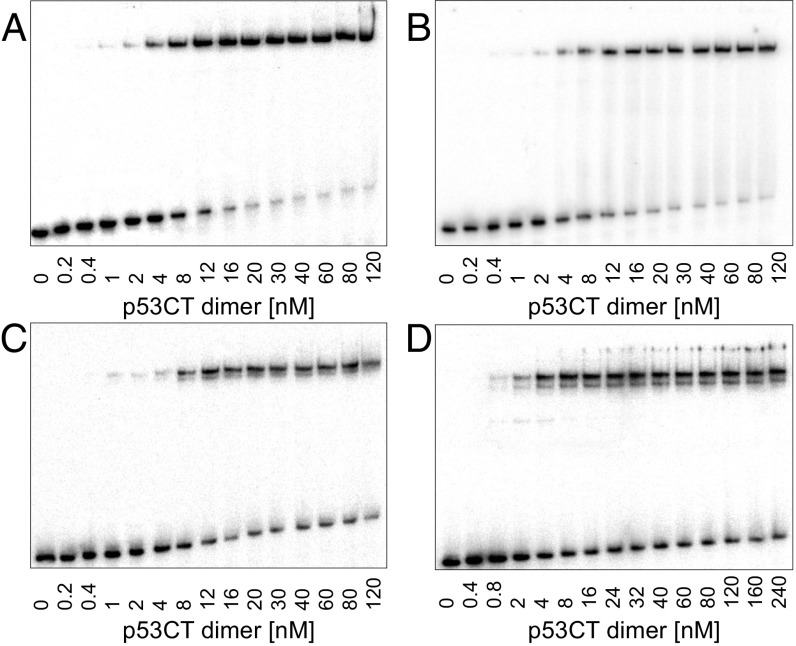
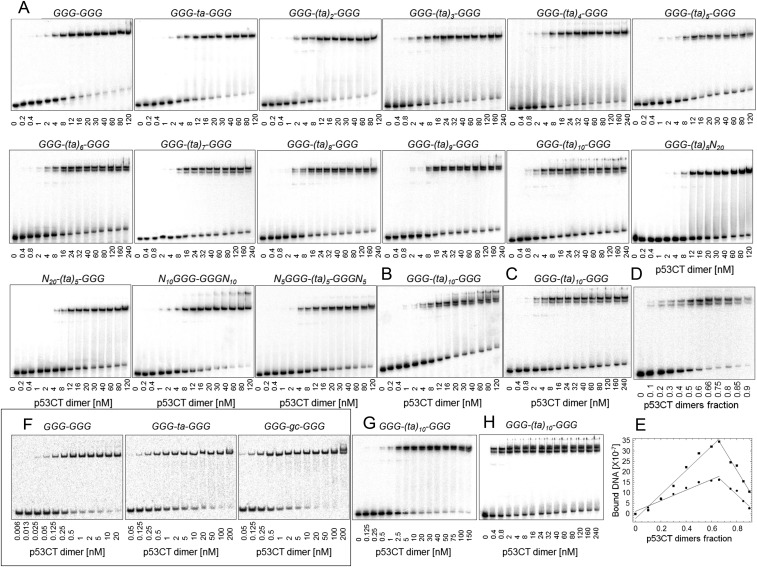
To investigate the role of spacer sequences in p53/DNA interaction, we determined the DNA binding of human p53CT, which incorporates the core (C) and the tetramerization (T) domains (residues 94–360). We used the consensus GGG half-site (GGGCATGCCC) previously investigated by us (18, 19, 21), and repeats of TA dinucleotides as the spacer elements, (ta)n (n = 1–10). p53CT binding to the GGG-(ta)n-GGG series (Table 1 and Table S1) was determined by the electrophoretic mobility shift assay (EMSA) (Fig. 1 and Fig. S1). Gel patterns for sequences with up to 8-bp spacers show the expected arrangement, of a major upper band and a minor lower band, corresponding to tetramer and dimer complexes shown previously (21). Surprisingly, from spacer length of 10 bp, (ta)5, onward, two closely spaced bands appear at the position of the upper tetramer complex (Fig. 1 C and D and Fig. S1). This pattern of two closely spaced bands was insensitive to changes in incubation and running temperature (Fig. S1B), as well as to an increase in the total ionic strength beyond 320 mM, used for sequence-specific binding conditions (Fig. S1C).
Table 1.
Macroscopic binding constants of p53CT/DNA complexes
| DNA sequence* | Specific tetramer KD (×1015 M2)†,‡ | Hemispecific tetramer KD (×1015 M2)† |
| GGG-GGG | 0.2 (0.1) | |
| GGG-ta-GGG | 0.3 (0.1) | |
| GGG-(ta)2-GGG | 0.5 (0.2) | |
| GGG-(ta)3-GGG | 1.3 (0.3) | |
| GGG-(ta)4-GGG | 4.0 (0.3) | |
| GGG-(ta)5-GGG | 1.6 (0.1) | 6.5 (0.5) |
| GGG-(ta)6-GGG | 1.6 (0.2) | 6.4 (1.2) |
| GGG-(ta)7-GGG | 3.0 (0.6) | 10.2 (2.2) |
| GGG-(ta)8-GGG | 6.5 (1.8) | 13.9 (3.8) |
| GGG-(ta)9-GGG | 5.8 (1.6) | 13.1 (3.4) |
| GGG-(ta)10-GGG | 0.9 (0.1) | 3.0 (0.3) |
| N20(ta)5-GGG | 1.3 (0.3) | |
| GGG-(ta)5N20 | 1.3 (0.1) | |
| N10GGG-GGGN10 | 0.3 (0.1) | |
| N5GGG-(ta)5-GGGN5 | 2.0 (0.1) | 5.4 (0.5) |
| CHMP4c | 0.6 (0.1)§ | |
| CHMP4c RHS¶ | 1.4 (0.3) | |
| CHMP4c LHS¶ | 1.6 (0.3) | |
| PTEN | 0.4 (0.1)§ | |
| PTEN RHS¶ | 1.0 (0.1) | |
| PTEN LHS¶ | 1.2 (0.2) |
GGG refers to the half-site motif: GGGCATGCCC; N5, N10, and N20 are random sequences; see Table S1 for further details and for the DNA sequence of the natural REs.
The values are averages of four to eight independent experiments conducted with each sequence; the values in parentheses are the SEMs.
These are macroscopic constants. See Table S1 for dimer-equivalent specific tetramer KD.
The EMSA patterns of CHMP4c/p53 and PTEN/p53 show a single p53–DNA band, and hence the derived value for each system refers to a weighted average of all species: specific and hemispecific complexes.
Right half-site (RHS) incorporates the specific right-handed half-site decamer and the DNA spacer; left half-site (LHS) incorporates the specific left-handed half-site decamer and the spacer DNA.
Table S1.
Sequences of p53 REs studied by EMSA experiments
| Name* | Sequence* | Dimer KD (nM)†,‡ | Dimer equivalent specific tetramer KD (nM)‡,§ | Cooperativity of the specific tetramer‡,¶ |
| GGG-GGG | cGGGCATGCCCGGGCATGCCCtg | 151 (30) | 6 (1) | 543 (74) |
| GGG-ta-GGG | cGGGCATGCCCtaGGGCATGCCCt | 172 (48) | 7 (1) | 535 (177) |
| GGG-(ta)2-GGG | cGGGCATGCCCtataGGGCATGCCCt | 216 (36) | 9 (2) | 542 (81) |
| GGG-(ta)3-GGG | cGGGCATGCCCtatataGGGCATGCCCt | 350 (46) | 16 (2) | 486 (67) |
| GGG-(ta)4-GGG | cGGGCATGCCCtatatataGGGCATGCCCt | 533 (60) | 24 (1) | 478 (48) |
| GGG-(ta)5-GGG | cGGGCATGCCCtatatatataGGGCATGCCCt | 450 (27) | 18 (1) | 656 (68) |
| GGG-(ta)6-GGG | cGGGCATGCCCtatatatatataGGGCATGCCCt | 409 (56) | 18 (1) | 538 (89) |
| GGG-(ta)7-GGG | cGGGCATGCCCtatatatatatataGGGCATGCCCt | 530 (68) | 28 (1) | 368 (62) |
| GGG-(ta)8-GGG | cGGGCATGCCCtatatatatatatataGGGCATGCCCt | 643 (116) | 37 (4) | 298 (56) |
| GGG-(ta)9-GGG | cGGGCATGCCCtatatatatatatatataGGGCATGCCCt | 627 (116) | 36 (3) | 292 (61) |
| GGG-(ta)10-GGG | cGGGCATGCCCtatatatatatatatatataGGGCATGCCCt | 311 (49) | 16 (1) | 449 (120) |
| N20(ta)5-GGG | cgcgcagtagggaggattcgatatatatataGGGCATGCCCt | 214 (34) | ||
| GGG-(ta)5N20 | cGGGCATGCCCtatatatatagaggattcgacgcgcagtagg | 220 (17) | ||
| N10GGG-GGGN10 | ccagagacaccGGGCATGCCCGGGCATGCCCgaagaggaaat | 230 (40) | 8 (1) | 945 (202) |
| N5GGG-(ta)5-GGGN5 | gacaccGGGCATGCCCtatatatataGGGCATGCCCgaagag | 481 (56) | 20 (1) | 596 (111) |
| GGG HS# | tggttgcGGGCATGCCCtgggta | |||
| CHMP4c | gAAACAAGCCCagtagcagcagctgctccGAGCTTGCCCa | |||
| CHMP4c RHS‖ | gcggaggcgatagtagcagcagctgctccGAGCTTGCCCa | 201 (26) | ||
| CHMP4c LHS‖ | gAAACAAGCCCagtagcagcagctgctccgcggaggcgta | 289 (24) | ||
| PTEN | gGAGCAAGCCCcaggcagctacactGGGCATGCTCg | |||
| PTEN RHS‖ | gcggaggcgtgcaggcagctacactGGGCATGCTCg | 151 (9) | ||
| PTEN LHS‖ | gGAGCAAGCCCcaggcagctacactgcggaggcgtg | 174 (10) |
The sequences are embedded in the stems of hairpin constructs with a 5-bp loop (21). The specific sites are shown in uppercase bold letters; spacer sequences are in lowercase underlined; nonspecific sites flanking the specific and spacer sites are in lowercase italics.
KD values of the p53 dimers bound to the specific sites.
The values are averages of four to eight independent experiments conducted with each sequence; the values in parentheses are the SEMs.
Dimer-equivalent tetramer KD is derived from the square root of Ka3, that is, the macroscopic specific tetramer value in Table 1. This KD and cooperativity cannot be derived for the hemispecific complex because Ka2 is composed of two different dimers, with binding constants k1 and k2. They cannot be computed for the natural REs either because the gel pattern of these REs was composed of a single bound tetrameric complex, and therefore the binding constant is a weighted average of specific and hemispecific values.
See definitions in SI Materials and Methods.
HS is half site, that is, a DNA binding site with a single specific half-site. This sequence is from Jordan et al. (31).
Right half-site (RHS) and left half-site (LHS) incorporate the specific right-handed and left-handed half-site decamer, respectively (uppercase bold), and the corresponding DNA spacer (lowercase underlined); nonspecific sequences were added to make the total length identical to that of the full site.
Fig. 1.
Patterns of gel migration of four sequences from the GGG-(ta)n-GGG series bound to p53CT. DNA targets concentration is <0.1 nM. The gels were run in a buffer containing 320 mM total ionic strength at 21 °C. The gels are representative examples of four to eight independent experiments conducted with each sequence. (A) GGG-GGG, (B) GGG-ta-GGG, (C) GGG-(ta)5-GGG, and (D) GGG-(ta)10-GGG.
Fig. S1.
Patterns of gel migration of p53CT dimers binding to the GGG-(ta)n-GGG RE series. DNA targets were embedded in hairpin constructs. The number below each gel is the concentration of p53CT dimers (nanomolar), active for DNA binding. DNA concentration for A–C is <0.1 nM. (A) These gels were run in a buffer containing 320 mM total ionic strength at 21 °C. (B and C) Effect of elevated temperature (30 °C) and elevated total ionic strength (420 mM at 21 °C), respectively, on p53 interactions with the GGG-(ta)10-GGG site. (D) p53CT dimers binding to GGG-(ta)10-GGG RE at the conditions used to determine the stoichiometry of interaction of the bound complexes using the continuous variation (Job plot) assay. Total macromolecular concentration (of p53CT dimers and DNA) was fixed at either 100 or 200 nM. Increasing molar fraction of the p53CT dimers is shown below the gel. (E) Analysis by a Job plot of the data from D. The y-axis units are photostimulated luminescence, as a measure of radioactivity from the storage phosphor plates. The lines are the least-square fitting results to the rising and falling subsets of the bound DNA. The values for the upper and lower bands are shown by squares and circles, respectively. The gels were running in a buffer containing 320 mM total ionic strength and 0.05% Nonidet P-40. The values shown in the main text are an average of nine experiments. (F) Gel migration pattern of p53CT bound to DNA sites containing GGG (GGGCATGCCC) half-sites with 0- or 2-bp spacers between them run at the cyclization kinetics conditions (low total ionic strength of 96 mM and 0.05% Nonidet P-40). DNA concentration is <0.001 nM. KD values for the specific tetramer are 0.6 (0.1), 0.4 (0.1), and 0.2 (0.1) nM, respectively. (G) Effect of Nonidet P-40 and low (96 mM) total ionic strength on p53 binding to GGG-(ta)10-GGG site. DNA concentration is <0.01 nM. KD value for the specific tetramer is 1.6 (0.1) nM. (H) Effect of Nonidet P-40 and high (320 mM) total ionic strength on p53 binding to GGG-(ta)10-GGG site. KD value for the specific tetramer is 1.7 (0.1) nM. All gels are representative examples of four to eight independent experiments conducted with each sequence.
We determined the stoichiometry of the binding interaction of the two bands by using the continuous variation method (Job plot) (26), as in our previous study (21). A representative gel and its analysis presented in Fig. S1 D and E, shows that the maximum amount of complex is achieved when the protein fraction of the upper complex is 0.65 (±0.02) and that of the lower complex is 0.66 (±0.02). The derived stoichiometry is thus two p53 dimers or a p53 tetramer bound to one DNA duplex for both complexes.
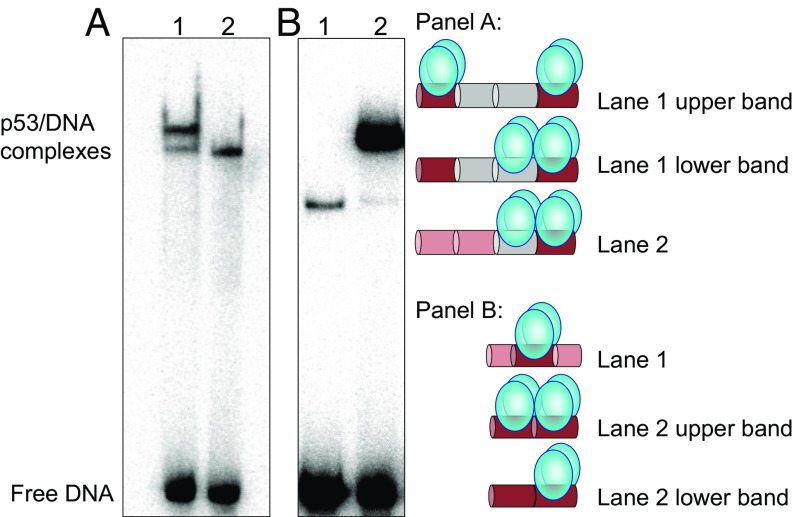
What is the difference between the two tetrameric complexes formed with p53 binding sites containing long spacers? We propose that one species is the regular sequence-specific complex referred to as “fully specific” or “specific,” whereas the second one is formed when a p53 tetramer is bound specifically to a single 10-bp half-site and nonspecifically to a 10-bp spacer sequence, referred to as “hemispecific complex,” also proposed previously on the basis of p53 sliding kinetics (27). To verify this conjecture, two DNA sequences which could form only hemispecific complexes were designed, by incorporating a single specific half-site adjacent to a (ta)5 motif, N20(ta)5-GGG and GGG-(ta)5N20 (Table S1). Fig. 2A compares the migration of the two complexes: p53 bound to the specific site versus one of the hemispecific sites, both being embedded in DNA sequences of equal length. The similar migration of the lower band in lane 1 to that of the single band in lane 2, suggests that the lower band corresponds to the hemispecific complex of p53 tetramer bound to GGG-(ta)10-GGG, whereas the upper band corresponds to the fully specific complex. The electrophoretic friction of the specific complex is likely larger than that of the hemispecific complex, due to the arrangement of the tetrameric complexes in the two species, and the presence of a free DNA leading segment in the hemispecific complex, but not in the specific complex (see schematic models in Fig. 2). Such factors could explain the slower migration of the specific complex through the polyacrylamide pores relative to the hemispecific complex (28). As described below, the hemispecific complex appears to be supported by interactions between adjacent p53 core dimers, facilitated by the linearity of the DNA binding sites.
Fig. 2.
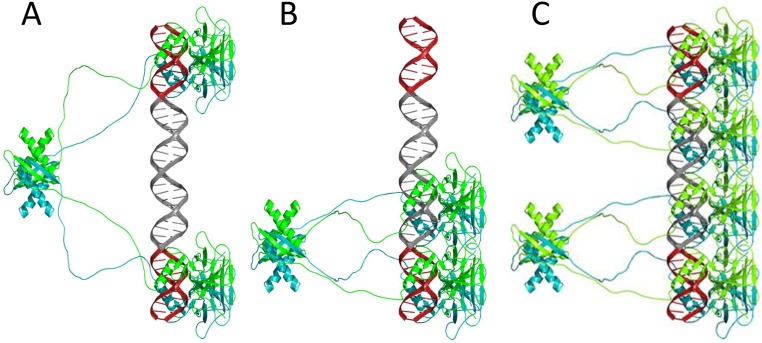
Comparison between the gel migration patterns of DNA sites bound to p53CT. (A) Lane 1, GGG-(ta)10-GGG; lane 2, N20(ta)5-GGG. p53 dimer concentration is 50 nM and DNA concentration is <0.1 nM. (B) Lane 1, GGG HS; lane 2, GGG-GGG. p53 dimer concentration is 24 nM, and DNA concentration is <0.1 nM. Other reaction conditions are those of Fig. 1. The proposed arrangements of p53 tetramers on the DNA are illustrated by the cartoons: p53 dimers are shown as two cyan bulbs. Specific half-sites are shown in dark red, (ta)5 tracts are shown in gray, and random DNA is in light red. Each colored cylinder corresponds to either 5 or 10 bp. The drawing is not to scale. The samples were loaded on the gel without an applied electric current.
Recently, it has been proposed that two p53 tetramers can interact sequence-specifically with a single DNA RE (29). This is in contrast to the present findings showing a binding ratio of two p53 dimers to a DNA site made of two specific half-sites in the fully specific complex, or to a DNA site made of a single specific half-site and a nonspecific decamer in the hemispecific complex. To prove that a single DNA half-site interacts only with a single p53 dimer, the binding modes of p53 to a GGG half-site (GGG HS in Table S1) and to a full site (GGG-GGG in Table S1) were compared. Fig. 2B demonstrates that p53 binds GGG HS as a dimer (lane 1), but mainly as a tetramer to GGG-GGG (lane 2), as determined previously by a Job plot for similar sequences (21). This is because GGG HS is too short to accommodate either a hemispecific or a fully specific tetramer. Such observations are in agreement with crystal structures (e.g., ref. 18) and most recently with a cryo-EM structure (30).
Structural Analysis of p53 Response Elements.
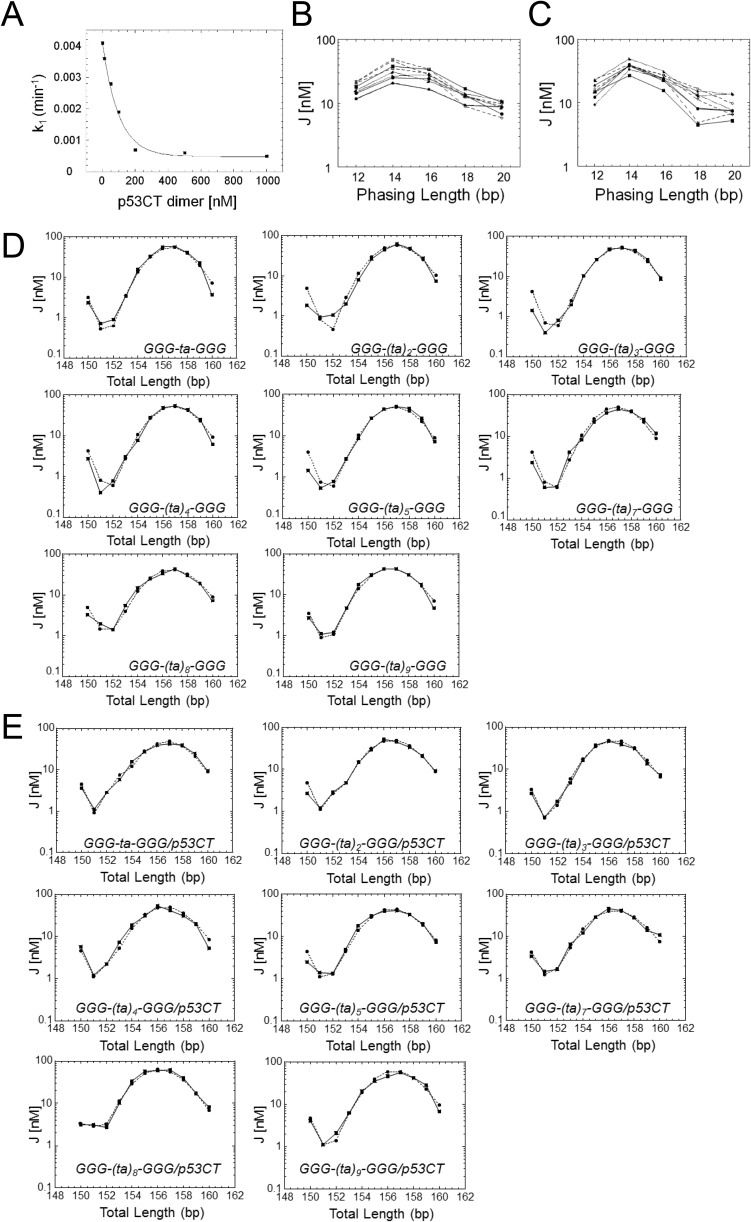
DNA bending was previously suggested to accompany p53 binding to its REs (e.g., refs. 22 and 23). Here, we determined the global conformations of p53 REs made of consensus half-sites previously investigated by us (18, 19, 21, 31). These REs were studied in both their free and p53-bound states in solution, using cyclization kinetics of DNA minicircles (32). As in our previous study of E2 protein–DNA complexes (33) we used a single p53 binding site (made of two half-sites), corresponding to the functional unit of p53 (Table S2). The 30-bp test sequence is composed of 20- to 22-bp binding sites and five or four nonspecific base pairs flanking them on either side (for sequence details, see Table S2). The individual overall bend angles of the various sequences in the free state are small (ranging from –9.9° to 0.3°; Table S3) and accessible within thermal energy (kBT) at 21° (34). The only parameter that changes significantly, depending on the DNA sequence, is the twisting (torsional) flexibility (Table S3), as also observed in our previous study of p53 half-sites (21). The observed changes in torsional flexibility are small (force constants are 1.5-fold different at most), but they are significantly different from each other because the extent of variation in the torsional and bending force constants, as a function of DNA sequence, is limited to about twofold and fourfold, respectively, as previously observed (35, 36).
Table S2.
Bottom template sequences for cyclization kinetics studies of p53 REs
 |
In red is the TAMRA primer, t in bold is the labeled residue, boxed cg is the sticky ends generated by ClaI digestion, and in square bracket is the test sequences. The specific half-sites are shown in capital letters. Underlined is the 3′-end of the hybridization region to the top template strand. The A-tract region starts right after this hybridization region, such that the test sequence is always 32 bp from the middle of the last A tract (or the first T tract on the test sequence strand) to the middle of the test sequence.
Table S3.
Best-fit parameters for p53 REs and their complexes obtained from cyclization kinetics data
| Name* | Bend angle per HS (degrees)†,‡ | Overall bend angle (degrees)§,‡ | Twist angle (degrees)‡ | Bending flexibility (degrees)‡ | Twisting flexibility (degrees)‡ | Torsional force constant(×1019 erg·cm) |
| GGG-GGG | −0.1 (0.3) | 0.3 (0.6) | 34.0 (0.3) | 4.3 (0.3) | 5.1 (0.3) | 1.713 |
| GGA-GGA | 0.1 (0.3) | −0.1 (0.6) | 34.1 (0.3) | 4.3 (0.3) | 5.2 (0.3) | 1.681 |
| AGG-AGG | 0.9 (0.4) | −1.7 (0.8) | 34.5 (0.4) | 4.3 (0.4) | 5.8 (0.4) | 1.335 |
| GGG-ta-GGG | 1.3 (0.1) | −2.6 (0.2) | 34.1 (0.2) | 4.4 (0.2) | 5.1 (0.2) | 1.729 |
| GGA-ta-GGA | 0.5 (0.1) | −1.0 (0.2) | 34.1 (0.1) | 4.3 (0.1) | 5.5 (0.2) | 1.525 |
| AGG-ta-AGG | 2.2 (0.1) | −4.4 (0.2) | 34.0 (0.2) | 4.2 (0.2) | 5.7 (0.2) | 1.385 |
| GGG-gc-GGG | 2.6 (0.2) | −5.1 (0.4) | 33.8 (0.2) | 4.1 (0.3) | 5.6 (0.3) | 1.445 |
| GGA-gc-GGA | −0.1 (0.1) | 0.3 (0.2) | 34.0 (0.2) | 4.1 (0.2) | 5.2 (0.2) | 1.676 |
| AGG-gc-AGG | 5.0 (0.4) | −9.9 (0.8) | 34.5 (0.4) | 4.3 (0.4) | 6.3 (0.4) | 1.128 |
| GGG-GGG/P | 2.9 (0.5) | −5.7 (1.0) | 33.8 (0.3) | 3.9 (0.3) | 5.6 (0.3) | 1.445 |
| GGA-GGA/P | 0.6 (0.3) | −1.2 (0.6) | 34.8 (0.3) | 4.1 (0.4) | 5.3 (0.4) | 1.614 |
| AGG-AGG/P | 2.5 (0.6) | −4.9 (1.2) | 33.2 (0.4) | 4.2 (0.4) | 6.1 (0.4) | 1.218 |
| GGG-ta-GGG/P | 2.5 (0.2) | −5.0 (0.4) | 34.0 (0.2) | 4.4 (0.3) | 6.3 (0.2) | 1.142 |
| GGA-ta-GGA/P | 3.1 (0.2) | −6.1 (0.4) | 34.0 (0.2) | 4.4 (0.2) | 6.4 (0.2) | 1.107 |
| AGG-ta-AGG/P | −0.3 (0.2) | 0.5 (0.4) | 34.3 (0.4) | 4.4 (0.3) | 6.4 (0.2) | 1.107 |
| GGG-gc-GGG/P | 1.4 (0.2) | −2.8 (0.4) | 35.0 (0.1) | 4.2 (0.3) | 5.7 (0.2) | 1.395 |
| GGA-gc-GGA/P | 3.3 (0.2) | −6.6 (0.4) | 34.7 (0.2) | 4.3 (0.2) | 4.8 (0.2) | 1.967 |
| AGG-gc-AGG/P | 0.5 (0.3) | −0.9 (0.6) | 34.3 (0.3) | 4.4 (0.3) | 6.5 (0.2) | 1.073 |
The measured REs contain two identical half-sites. GGG, GGA, and AGG refer to the sequences GGGCATGCCC, GGACATGTCC, and AGGCATGCCT, respectively. See Table S2 for further details on the constructs. P refers to p53CT protein.
The simulations of the cyclization kinetics data were performed with one bend angle at the center of each half-site, that is, 5 and 15 or 5 and 17 bases from the 5′-end for sites with zero or 2-bp spacers, respectively.
Numbers in parentheses are the simulation errors, calculated as described in ref. 50.
The overall bend angles were calculated as vectorial sums from the half-site values. The bend position is at the center of the full site, that is, 10 or 11 bases from the 5′-end for sites with 0- or 2-bp spacers, respectively. By convention, the sign of the bend angles are positive or negative when bending is toward the major or minor groove, respectively. Thus, the change in sign from the half-site bend to the overall bend reflects the change in the bend position between the two bends.
The protein concentration needed to achieve full binding of p53 to the tested sequences was established from gel shift experiments run under the conditions of the cyclization kinetics experiments (Fig. S1F). The structural parameters of the studied DNA sequences bound to p53 (Table S3) show no appreciable DNA bending upon binding to the protein (ranging from –6.6° to 0.5°), consistent with crystal structures of similar consensus DNA sites (18, 19, 37, 38), and contrary to the static bending by p53 observed previously (e.g., refs. 22 and 23). Moreover, the complexes with p53 show no change in DNA bending flexibility from that of naked DNA, but they do show significant increase in twisting flexibility for three REs that incorporate ta spacers (Table S3). This finding can be rationalized on the basis of our previous studies on high-resolution crystal structures of p53DBD tetramers bound to similar consensus REs (18, 19), revealing a noncanonical Hoogsteen geometry (HG) of A/T base pairs, instead of the regular Watson–Crick geometry (WC). The HG geometry was observed at the CATG motifs of two REs studied here: GGG-GGG (19), and more recently in GGA-GGA (39). In addition, this geometry was observed at a ta spacer between two AGG motifs (18), similar to AGG-ta-AGG studied here. The unique base pair geometry was shown to affect the local DNA shape contributing to the stabilization of the tetrameric complex (19). As the transition from WC to HG base pairs involves unwinding and rewinding of the double helix, it can lead to enhanced torsional flexibility of these noncontacted DNA regions. Protein binding usually leads to a stiffening of the double helix (e.g., ref. 33). However, both the central AT step, of the CATG motif, and the ta spacer regions are not contacted directly by the protein (18, 19). Thus, it could be that the bound segments of the DNA become stiffer upon binding, but the substantial increase in apparent flexibility of the unbound DNA regions obscure this phenomenon.
To determine the structural characteristics of p53 REs, as a function of spacer length, the GGG-(ta)n-GGG series was used (Tables S2 and S4). As a result of inhibition of the ligation reaction at high p53CT concentration (Fig. S2A), these simulations (Fig. S2 D and E) were conducted without phasing data (Materials and Methods). However, comparing the values for GGG-ta-GGG, measured with and without phasing data (Tables S3 and S4, respectively), shows that the parameters derived from the two experiments are similar within the simulation errors. The overall bend angles shown by the DNA sequences of the GGG-(ta)n-GGG series are small, for both the free and p53-bound DNA, ranging from –8.5° to 8.2°, and –3.1° to 9.6°, respectively (Table S4). Unbent DNA helices with long spacers between specific half-sites can be easily accommodated in complexes with p53, as shown by modeling of a complex between p53CT and a binding site incorporating a 20-bp spacer (Fig. S3). Here, both the axial (bending) as well as the torsional (twisting) flexibility of the free DNA increase as the spacer is lengthened (especially for the two longest spacers). This finding is expected, due to the flexible nature of the T-A dinucleotide steps (40). Less expected is the finding that the flexibility in the series (ta)n is systematically larger for DNA sites in complex with p53, except for the two longest spacers that are already highly flexible. This pattern could result from conformational transitions between WC and HG geometry at A/T base pairs, as described above.
Table S4.
Best-fit parameters for GGG-(ta)n-GGG series (n = 1–9) and their complexes obtained from cyclization kinetics data
| Name* | Overall bend angle (degrees)†‡ | Twist angle (degrees)‡ | Bending flexibility (degrees)‡ | Twisting flexibility (degrees)‡ |
| GGG-ta-GGG | −6.2 (0.7) | 34.5 (0.2) | 4.0 (0.2) | 5.0 (0.2) |
| GGG-(ta)2-GGG | −6.9 (0.7) | 34.1 (0.2) | 4.1 (0.2) | 5.1 (0.2) |
| GGG-(ta)3-GGG | −8.5 (0.9) | 34.3 (0.3) | 4.1 (0.3) | 5.1 (0.2) |
| GGG-(ta)4-GGG | −8.0 (0.7) | 34.1 (0.3) | 4.2 (0.3) | 5.1 (0.3) |
| GGG-(ta)5-GGG | −6.7 (1.4) | 34.1 (0.3) | 4.2 (0.3) | 5.2 (0.3) |
| GGG-(ta)7-GGG | −8.1 (0.4) | 34.1 (0.4) | 4.2 (0.4) | 5.3 (0.4) |
| GGG-(ta)8-GGG | 8.2 (0.2) | 33.7 (0.3) | 4.9 (0.3) | 6.2 (0.3) |
| GGG-(ta)9-GGG | 4.0 (0.2) | 34.1 (0.3) | 4.7 (0.3) | 6.1 (0.2) |
| GGG-ta-GGG/P | 0.3 (0.8) | 34.0 (0.2) | 4.7 (0.2) | 6.2 (0.2) |
| GGG-(ta)2-GGG/P | 2.3 (0.2) | 34.0 (0.2) | 4.8 (0.2) | 6.1 (0.2) |
| GGG-(ta)3-GGG/P | 3.3 (0.2) | 34.5 (0.2) | 4.7 (0.1) | 6.0 (0.1) |
| GGG-(ta)4-GGG/P | 6.5 (1.2) | 34.0 (0.2) | 5.0 (0.3) | 6.3 (0.2) |
| GGG-(ta)5-GGG/P | 4.2 (0.2) | 34.0 (0.2) | 4.8 (0.2) | 6.2 (0.2) |
| GGG-(ta)7-GGG/P | 9.6 (0.2) | 33.9 (0.2) | 5.0 (0.3) | 6.3 (0.2) |
| GGG-(ta)8-GGG/P | −3.1 (0.9) | 34.7 (0.2) | 4.6 (0.3) | 6.2 (0.2) |
| GGG-(ta)9-GGG/P | −2.8 (0.4) | 34.1 (0.3) | 4.6 (0.3) | 6.1 (0.2) |
The measured sequences contain two half-sites of GGG (GGGCATGCCC), separated by a variable number of base pairs. See Table S2 for further details on the constructs. The sequences were measured without phasing data (see text for details). P refers to p53CT protein.
The overall bend angle is positioned at the center of the full site.
Numbers in parentheses are the simulation errors, calculated as described in ref. 50.
Fig. S2.
Cyclization kinetics of consensus p53 binding sites and their complexes with p53. (A) Ligation rate of DNA minicircles versus protein concentration. The DNA construct contains the GGG-GGG RE with a phasing length of 14 and total length of 157 bp. k1 is the first-order rate constant for covalent closure of circles. Note that, at p53CT dimer concentrations used for sequences with 2-bp spacers (50 nM), there is already a ∼30% reduction in ligation rate. This may stem from electrostatic interaction between p53CT dimers and the ligase enzyme, when both are present at high concentrations, because of the low salt conditions used in the cyclization kinetics assay (96 mM total ionic strength). This is especially true for the out-of-phase constructs that require higher ligase concentrations (SI Materials and Methods). For test sequences with longer spacers that require larger amounts of p53CT for complete protein binding (Fig. 1 and Fig. S1), the inhibition of ligation does not allow to determine ligation rates. (B and C) The experimental J factors as a function of the phasing length for the free consensus DNA constructs and their complexes with p53CT, respectively (Table S2). Filled squares, GGG-GGG; filled circles, GGA-GGA; up filled circles, AGG-AGG; down triangles, GGG-ta-GGG; up triangles, GGA-ta-GGA; diamonds, AGG-ta-AGG; right-sided filled triangle, GGG-gc-GGG; left-sided filled triangle, GGA-gc-GGA; filled diamond, AGG-gc-AGG. (D and E) Cyclization kinetics of p53 consensus binding sites and their complexes with p53, respectively, as a function of spacer length. Shown are the experimental and simulated J factors for the series GGG-(ta)n-GGG as a function of the total DNA length for the free DNA constructs and their complexes with p53CT, respectively. The solid lines are the experimental curves and the dashed lines are the curves from the data simulation.
Fig. S3.
Models of p53CT tetramers (A and B) and a p53 octamer (C) bound to DNA. The DNA is made of two sequence-specific half-sites of 10 bp each (GGGCATGCCC, shown in red) and separated by a 20-bp spacer of (TA)10 (shown in gray). The monomers of each p53 dimer are shown in cyan and green. The p53CT tetramer shown in A presents a model for the fully specific complex, and the tetramer shown in B presents a model for the hemispecific complexes described in the text. Two adjacent hemispecific tetramers can bind on sequences harboring a 20-bp spacer, forming an octamer (C). The models are based on the published crystal structure coordinates of the p53 core-domain tetramer bound to DNA (PDB ID code 3XZ8), and the coordinates of the tetramerization domain (PDB ID code 1AIE). The unstructured linkers between the core domains and the tetramerization domains were modeled for human p53 residues 294–326. The figures were drawn by PyMol (52).
The Effect of Spacers on p53–DNA Binding Affinity.
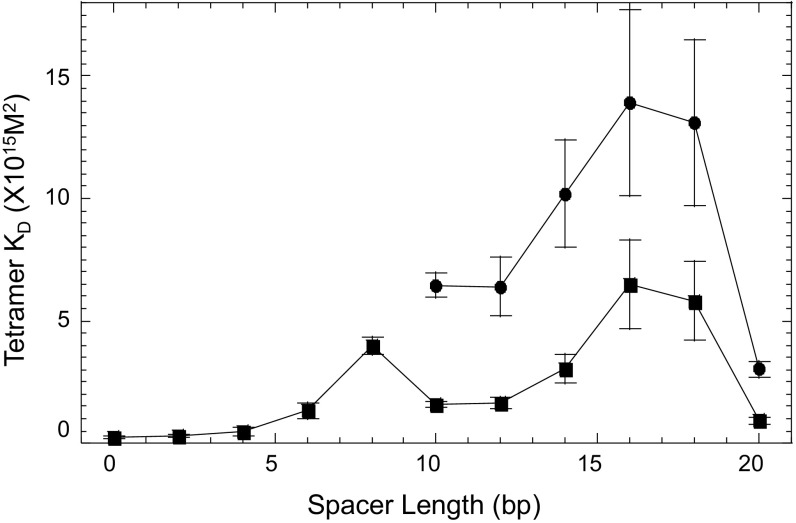
Binding analysis of the complexes varying in the length of their DNA spacers (SI Materials and Methods), shows that the binding affinity of the specific complex is rotationally modulated depending on the relative orientation between the two p53 dimers along the DNA helix (Fig. 3 and Table 1 and Table S1). The binding affinity of the specific p53–DNA tetramer (Table 1) goes down (i.e., KD goes up) from GGG-GGG to GGG-(ta)4-GGG, as well as the cooperativity. Then, the affinity goes up for GGG-(ta)5-GGG and GGG-(ta)6-GGG, and then goes down again until GGG-(ta)9-GGG, followed by a large KD drop (affinity goes up) for GGG-(ta)10-GGG. This means that the rotational positioning of the two DNA half-sites relative to each other modulates the binding affinity of the specific complex. This is a consequence of structural constraints imposed on the 3D architecture of the tetrameric complex, resulting from the coupling between the two core-domain dimers and the C-terminal tetramer via the four p53 linkers. When the two half-sites are rotated nearly in phase with the helical repeat (0, 10, or 20 bp apart), the two core dimers on the linear DNA helix are nearly parallel to each other. In such arrangement, the tetramer is likely to adopt a time-averaged symmetrical structure, shown by the model of a complex between p53CT and a binding site incorporating a 20-bp spacer (Fig. S3). In this configuration, the linkers between the four core domains and the tetramerization domains are minimally constrained, and hence the binding affinity (Table 1) and cooperativity (Table S1) are high relative to the other species in the GGG-(ta)n-GGG series.
Fig. 3.
Macroscopic tetramer KD values of complexes between p53CT and the GGG-(ta)n-GGG sequences as a function of spacer length. Squares represent fully specific tetramer complexes; circles represent hemispecific tetramers.
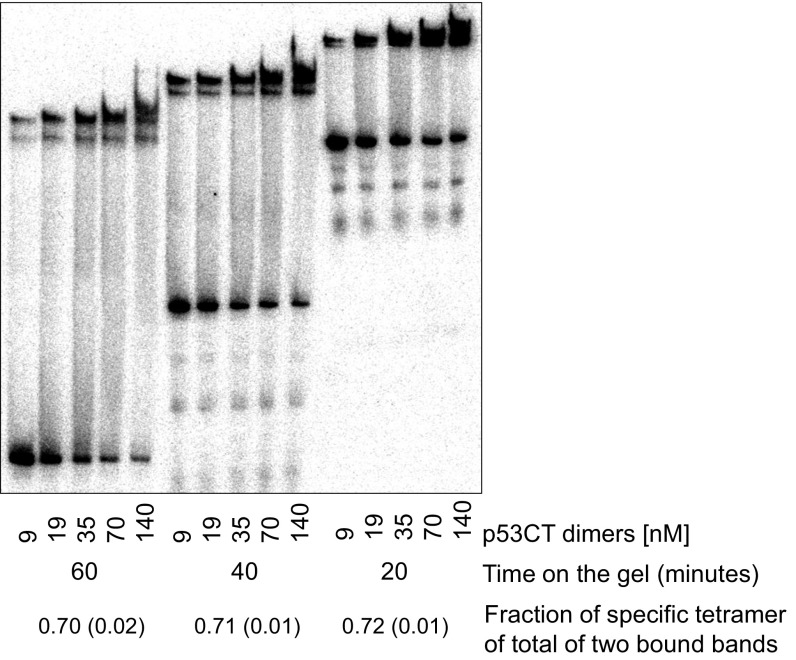
Surprisingly, the binding affinity of the hemispecific complex follows a similar spacer-dependent pattern as that of the specific complex and is only about twofold to fourfold lower than that of the specific one. The variations in the binding affinity of dimeric complexes in this series exhibit a similar spacer-dependent trend to that of the tetrameric species (Table S1), as also observed previously for p53DBD/DNA complexes (21). The hemispecific binding mode is not expected to be spacer-length dependent, since the two core dimers are adjacent and parallel to each other as for a zero-spacer DNA complex (19), as displayed in the model of Fig. S3. We currently do not understand these observations. However, this phenomenon is not due to leakage of the specific tetramer onto the lower hemispecific band, since a time course analysis indicates that the percentage of the specific complex on the gel is constant with time (Fig. S4). Interestingly, the binding affinity of p53 tetramers bound to the isolated hemispecific sites is higher than that of the hemispecific sites within GGG-(ta)10-GGG (Table 1) and is identical within experimental error to that of the specific tetramer. It is noteworthy that large variability in the quaternary structures of p53 tetramers complexed to a 20-bp RE flanked by random long sequences was observed by EM studies (41). The authors proposed that this finding might be related to specific and nonspecific recognition of the DNA helix, and thus provide snapshots of dynamic p53 tetramers supporting a two-binding site model for p53 (41).
Fig. S4.
Time course analysis of p53CT binding to GGG-(ta)10-GGG. Reaction conditions are those of Fig. 1 of the main text. Equal aliquots of a reaction mixture containing DNA (<0.1 nM) and protein at five different concentrations were loaded on the gel at 20-min intervals after 2-h incubation at 21 °C. The values of the fraction of complexes of GGG-(ta)10-GGG and specific p53CT tetramers that remained bound at the different time points (shown at the Bottom) show that there is no depletion of the fully specific tetramer species with time.
We have verified that the DNA binding affinity for the specific complexes is not affected by the length of the DNA used in the various RE constructs by designing elongated forms of GGG-GGG and GGG-(ta)5-GGG (Table S1). The measured KD values for these sequences are comparable to the KD values for sequences lacking the flanking nucleotides (Table 1; Fig. S1 and Table S1). For REs with 20-bp spacer [GGG-(ta)10-GGG], or two 10-bp flanking sequences (N10GGG-GGGN10, Fig. S1), we observe lower mobility bands of octameric complexes, which can be formed by binding of two adjacent hemispecific tetramers [modeled for GGG-(ta)10-GGG in Fig. S3C].
Binding Competition Experiment with Calf Thymus DNA.
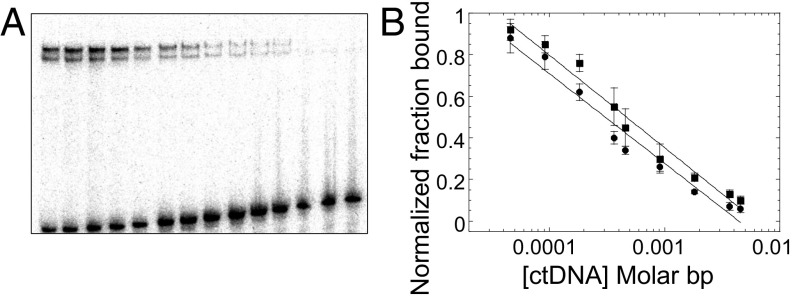
To assess the binding specificity [defined as the difference in binding affinity between the specific sites and all other possible binding sites in the genome (42)] of the two types of tetrameric complexes, fully specific and hemispecific, p53CT was incubated with the sequence GGG-(ta)10-GGG and then various concentrations of genomic calf thymus DNA (ctDNA) were added as a nonspecific competitor (Fig. 4A). Fig. 4B displays the bound fraction of the specific and hemispecific complexes of p53 (each normalized to their first lane) as a function of ctDNA concentration (in molar base pair). It shows two parallel lines; the specific complex is the upper line, and the lower line is the hemispecific complex. This means that both kinds of complexes are similarly thermodynamically stable with respect to competition by ctDNA. Hence, they have similar binding specificity relative to bulk genomic DNA binding sites. Thus, protein–protein interactions between adjacent core-domain dimers appear to compensate for the lack of base-specific contacts in the hemispecific complex. The stable hemispecific tetrameric complexes observed here demonstrate that, even in cases of a single specific DNA half-site, p53 can bind as a functional tetramer with high affinity (Table 1). This finding extends the range of possible p53 REs, as observed previously (43, 44), but emphasizes the notion that, even in cases of isolated DNA half-sites, the interacting protein species is a p53 tetramer.
Fig. 4.
Binding competition experiment. (A) p53CT and GGG-(ta)10-GGG were incubated together for 2 h, followed by adding increasing concentrations of ctDNA. Reaction conditions are those described in Fig. 1. DNA concentration is 3.5 nM. p53CT concentration is 200 mM dimers. ctDNA concentration is increased from 0 to 1.64 10−2 in M bp. (B) Concentration of intact specific complexes (squares) or hemispecific complexes (circles) as a function of ctDNA concentration.
Binding of p53 to Natural Response Elements Containing Long Spacers.
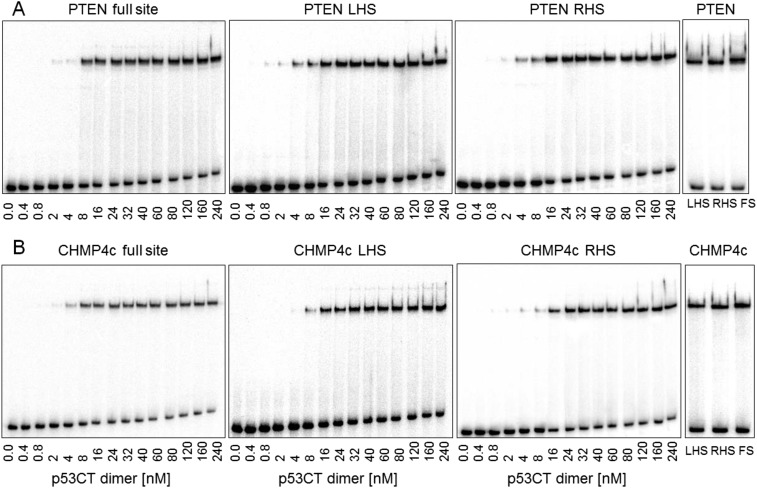
To confirm the biological relevance of the hemispecific binding mode, we probed the binding of p53 to two natural p53 REs: chromatin-modifying protein 4C (CHMP4c) (24) and phosphatase and tensin homolog (PTEN) (25), with 18- and 14-bp spacers, respectively. The binding pattern of p53 to CHMP4c shows a single shifted band (Fig. S5B), whereas that of PTEN shows two very close bands that are barely distinguishable (Fig. S5A). This finding suggests that the bands corresponding to the hemispecific p53 complexes comigrate with that of the specific complex, as shown by comparing the p53 binding patterns of the natural full sites (FS) to those of the designed isolated half-sites [left half-site (LHS), right half-site (RHS)] in Fig. S5. Hence, the single p53–DNA band has to be treated as consisting of a specific complex and two hemispecific complexes, and the derived KD values (Table 1) as a weighted average of specific and hemispecific values. However, the individual values of the two hemispecific complexes can be derived from designed hemispecific binding sites (RHS and LHS in Table 1 and Table S1). We find that the binding affinities of p53 tetramers with the isolated hemispecific sites of the natural and consensus REs are similar (Table 1). Moreover, the binding affinities of the designed hemispecific complexes of CHMP4c and PTEN are at most threefold lower than those of the composite (specific and hemispecific) complex. Hence, the hemispecific binding mode could play a role in sequence-specific transcriptional activities of p53.
Fig. S5.
Patterns of gel migration of p53CT dimers binding to natural p53 REs. (A) p53CT dimers bound to the PTEN RE. (B) p53CT dimers bound to the CHMP4c RE. DNA targets were embedded in hairpin constructs (concentration, <0.1 nM). LHS, RHS, and FS refer to left-handed site, right-handed site, and full site, respectively (defined in Table S1). The number below each gel is the concentration of p53CT dimers active for DNA binding. Gels were run in a buffer containing 320 mM total ionic strength at 21 °C. The gels at the right-handed side show a comparison between the migration patterns of the three variants of each sequence.
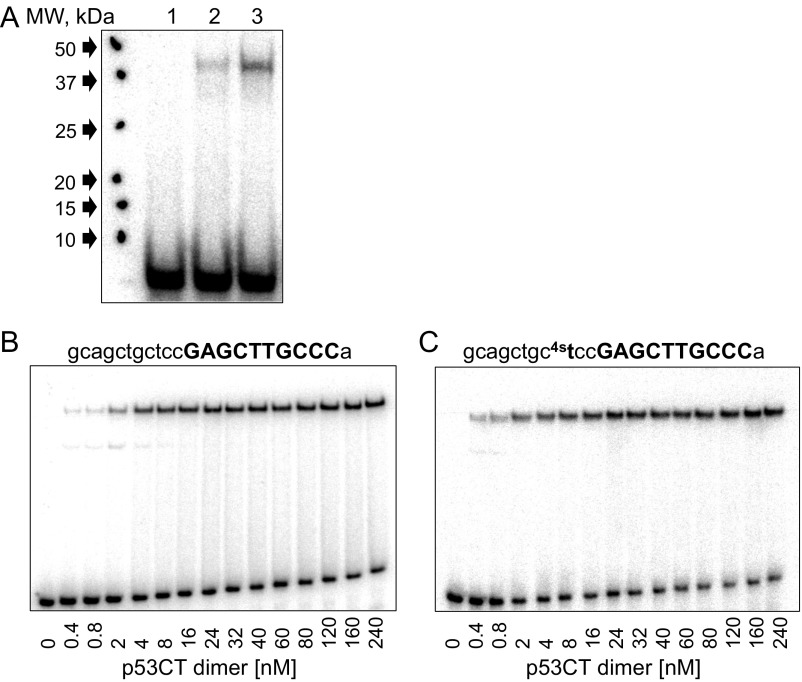
To support our observations that p53 dimers interact with the natural spacer DNA in hemispecific complexes, we used the UV-induced cross-linking reaction between p53 and the hemispecific binding site RHS of CHMP4c (Fig. S6), using a procedure described previously (45). This is the only RE, in the current study, where a T base is in a position potentially close to a cysteine residue at the p53/DNA interface (18). Hence, the T nucleotide at the third position from the 3′-end of the spacer sequence (gcagctgctcc) was replaced by a 4-thiothymidine (Fig. S6). Fig. S6A shows that a minute amount of cross-linked complex forms without UV exposure (lane 2) but is significantly enhanced upon exposure to UV (lane 3). We have measured the binding affinity of p53CT to this binding site to verify that the thio-T moiety does not interfere or enhance the binding of p53CT to this site. Fig. S6 B and C show that the binding patterns of a shortened version of CHMP4c RHS without or with 4-thiothymidine, are similar.
Fig. S6.
Results of cross-linking experiments of the hemispecific complex between p53CT dimers and CHMP4c RHS. (A) The 120 nM p53CT dimers were incubated with 6 nM of the hemispecific binding site made of a specific right-handed half-site of CHMP4c and 11 bp of the adjacent spacer sequence containing the UV-reactive nucleotide analog, 4-thiothymidine. Lane 1, DNA only; lane 2, untreated complex; lane 3, complex cross-linked at 312 nm for 10 min. Note that there is a minute amount of cross-linked complex also without applying UV light (lane 2) and that the molecular weight of the complex corresponds to a p53CT monomer cross-linked to DNA. (B and C) EMSA experiments to determine the binding affinity of the short version of CHMP4c RHS and its 4-thiothymidine analog, respectively. Other details are as in Fig. 1 and Fig. S1.
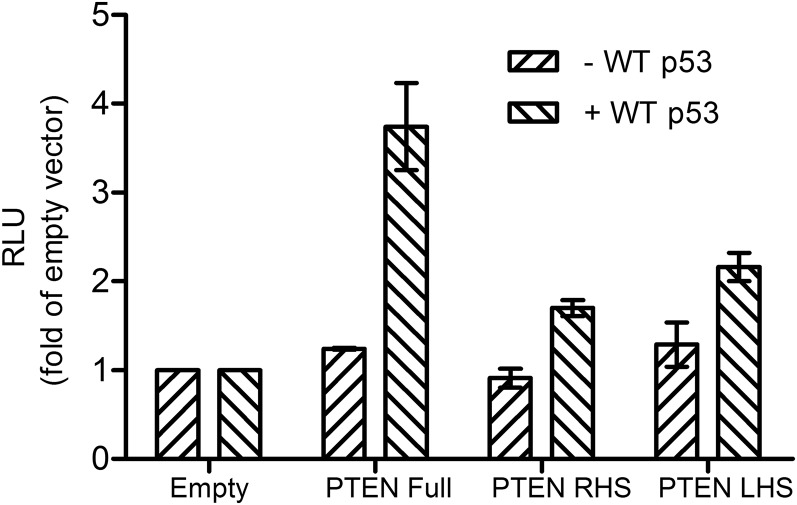
To examine the potential consequence of p53 binding to such hemispecific binding sites in living cells, we measured the transactivation of PTEN RHS and LHS, in comparison with PTEN full-site, which was previously shown to be transactivated by wild-type (WT) p53 (25). Fig. 5 shows that p53 transactivation from PTEN hemispecific sites (RHS, LHS) is about 50% of that of PTEN full site. These data demonstrate that the hemispecific binding modes can lead to transactivation of downstream genes in living cells.
Fig. 5.
p53 transactivation from PTEN full-site and PTEN hemispecific sites. p53-null H1299 cells were transiently cotransfected with different luciferase constructs harboring PTEN full site, PTEN hemispecific sites (RHS and LHS) or the original luciferase vector (empty cLuc), together with a WT p53-expressing plasmid or an empty vector. Luminescence was measured 24 h after transfection. Luminescence values were normalized to the transfection efficiency of cotransfected, constitutively expressed Gaussia luciferase. Results are presented as relative light units of the REs over that of the empty cLuc vector. Error bars represent mean ± SD of four experiments.
General Discussion and Biological Implications.
The current studies show that p53 can bind its DNA targets as a tetramer in two different modes: fully specific binding and hemispecific binding. In the latter mode, only one dimer is specifically bound to a p53 half-site. Of all (∼280 REs) currently validated p53 sites (1–3), ∼50% have spacer sequences between their two half-sites, of which ∼110 REs have spacers longer than 1 bp, and ∼30 REs have spacers which are 10 bp or longer (1–3). We demonstrate that both consensus and natural REs containing spacers of 10 bp or longer can form both fully specific and hemispecific tetrameric complexes. Hemispecific binding allows isolated p53 half-sites reported previously (13, 44) to form functional p53 complexes. Moreover, because many p53 REs have additional half-sites, or full sites that are 1–20 bp away from the main binding site (1), the hemispecific binding mode could bridge between the main and the secondary binding sites or between two main binding sites, shown for example, in the promoter of MDM2 (46).
The hemispecific binding mode shows similar specificity to that of the fully specific mode relative to bulk genomic DNA, which is probably due to protein–protein interactions between adjacent core-domain dimers of p53 bound to a straight DNA helix. Such interactions, together with those that support the C-terminal tetramer, are likely to play a major role in p53 functionality. Recently, it has been shown that the positively charged extreme CTD of p53 influences the stability of p53/DNA complexes, by facilitating cooperative contacts between the four core domains in the functional p53 tetramer (45), and that the extent of this influence is correlated with the deviation of the DNA REs from the consensus. Here, we use the p53CT construct to demonstrate that binding affinity and cooperativity of sequence-specific p53/DNA complexes depend on the length of the nonspecific DNA spacer between the DNA half-sites. p53CT is the minimal construct to study such effects for REs with long spacers that are unaffected by an additional cooperativity enabled by the CTD. All of the specific DNA half-sites studied here are entirely within the consensus. However, it is plausible that the CTD affects the hemispecific interactions, where one dimer of the p53 tetramer interacts with nonspecific DNA, and this remains to be explored. Hemispecific complexes were identified in other transcription factors such as bZip proteins (47) and steroid receptors (48), suggesting that asymmetry of binding could offer a mechanism of varying the affinity of protein–DNA interactions over a wide range of values, and that protein–protein interactions play an important stabilizing role in such complexes.
Studies using ChIP usually recover only REs containing 0- to 1-bp spacers (11–14). However, such studies may be biased toward relatively stable and high-affinity p53 REs (43). In contrast, reporter gene assays show mixed observations related to the effect of spacers on the transactivation potential of p53 REs. Artificial insertion of spacer sequences into abutting natural binding sites, or those containing short spacers (<10 bp), show decreased transactivation from reporter genes (43). However, transactivation level from the natural MDM2 RE, which contains two complete sites, RE1 and RE2, that are separated by 17 bp, is threefold higher relative to RE1 alone, and sixfold higher relative to RE2 alone (43). Interestingly, MDM2 transactivation level goes further up upon decreasing the spacer between RE1 and RE2 from 17 to 10 bp (43), consistent with the cyclical modulation of the binding affinity as a function of the DNA spacer shown here. The Luciferase reporter gene assays performed here shows that p53 interactions with PTEN hemispecific sites result in transactivation at 50% level of that with PTEN full site. Previously, comparable levels of p53 transactivation were observed from the PTEN and the p21 promoter (25), the latter known to be a highly responsive promoter (2) and a primary mediator of cell cycle arrest (5). Thus, transactivation from hemispecific sites, such as observed here for the PTEN sites, substantiate their relevance in the cellular context.
SI Materials and Methods
DNA.
All DNA sequences for the electrophoretic mobility shift assay (EMSA) were synthesized by Sigma Genosys (Israel) and purified by a reverse-phase cartridge. The sequences for the EMSA experiments were designed as intramolecular hairpin constructs, with 23–42 bp in the stem, and five cytosines in the loop as previously described (21, 31). The right-handed half-site decamer of CHMP4c with the UV–cross-linkable nucleotide analog 4-thio-dTMP was synthesized by Sigma Genosys and purified by HPLC. The DNA test sequences for cyclization experiments (bottom PCR templates), the library DNA sequences (top PCR templates), and the fluorescein- and tetramethylrhodamine (TAMRA)-labeled oligonucleotide primers were synthesized by the Keck Foundation Laboratory at Yale University, and purified as previously described (21, 32).
DNA Constructs for Cyclization Kinetics.
In previous studies of p53 REs by this method (21), the test sequences were three repetitions of the half-site decameric sequence. Here, the studied DNA site is 20-bp long consisting of two half-sites and flanked by 5 bp on either side. When spacer sequences were inserted between specific half-sites, these flanking sequences were shortened or deleted to conserve the total length of the DNA construct (Table S2). For test sequences longer than 30 bp, the hybridization region was reduced from 10 to 6 bp at the 3′-end, as well as deleting up to 4 bp from the constant flanking region of the bottom template at the 5′-end of the test sequence. We could not design a test sequence with a 20-bp spacer since this would have invaded the bottom primer sequence containing the TAMRA dye. Regardless of spacer length, the phasing of the A-tracts relative to the test sequence is always 32 bp from the middle of the last A-tract to the middle of the test sequence (32).
Cyclization Kinetics Measurements and Simulation of Cyclization Data.
Ligase concentration was varied as a function of phasing length (0.8 kU/μL for the in-phase 156 L14 construct and 1.6 kU/μL for all other constructs) and total length (from 0.8 kU/μL for the 156–158 constructs, to 1.6 kU/μL for 155 and 159; 3.2 kU/μL for 150, 153–154, and 160; and 6.4 kU/μL for the 151–152 constructs). The 5 nM p53CT was used for constructs without spacers and 50 nM p53CT for all constructs with spacers, which are the concentrations at which full binding of p53CT to the studied DNA sequences is observed, at the conditions of the cyclization kinetics assay (Fig. S1F).
For sequences with spacer length longer than 2 bp, we could not measure variation in phasing length, because of p53 inhibition of ligase activity (Fig. S2A). Hence, for the study of the GGG-(ta)n-GGG series (Table S2), the simulations were conducted using only 11 total-length J factors instead of 16 J factors (11 total-length and 5 phasing length), as for sequences without spacers and in ref. 21. This necessitated carrying out the simulations many times with various initial values for the analyzed parameters, which were automated and run at the Technion High Energy Physics Grid Computing cluster. Sequences with 0- or 2-bp spacer sequences were simulated using one kink (roll) position per half-site, as in our previous study (21). For the simulation of the GGG-(ta)n-GGG series that lack phasing data, we used a single kink (roll) position at the center of the full site, deriving an overall bending.
Binding Affinity Measurements.
Binding affinity measurements were conducted as previously described (21, 31). In short, radiolabeled and gel-purified DNA hairpin duplexes (concentration, <0.1 nM) and increasing amounts of p53CT were incubated at 21 °C for 2 h in a buffer containing 50 mM Tris⋅HCl (pH 7.5), 10 mM MgCl2, 1 mM ATP, 25 U/mL BSA, 10% glycerol, 10 mM DTT, and 100 mM KCl. By taking into account the protein dilution buffer of 20 mM Tris⋅HCl (pH 7.5) and 120 mM NaCl, the total ionic strength of the binding buffer was 320 mM. To check the effect of ionic strength, additional 100 mM KCl was used for a total ionic strength of 420 mM. We also ran experiments at the conditions used in the cyclization kinetics assays: 50 mM Tris⋅HCl (pH 7.5), 10 mM MgCl2, 1 mM ATP, 25 U/mL BSA, 10% glycerol, 10 mM DTT, and 0.05% of the nonionic detergent Nonidet P-40 (Thermo Scientific). The protein was diluted in this buffer by 1:5 and added to the reaction mix. The total ionic strength of this buffer is 96 mM. Complexes were resolved from free DNA by electrophoresis on native gels (6%, 37.5:1 acrylamide/bisacrylamide ratio). Samples were loaded while the gels were running, in all experiments except of those described in Fig. 2. The gels were run at 550 V and at temperatures of 21 or 30 °C, in a running buffer containing 1× TG [25 mM Tris⋅HCl (pH 8.3), 190 mM glycine], until the bromophenol blue dye migrated 10 cm. Gels were dried and quantified as described previously (21, 31) using a GE Typhoon FLA7000 phosphoimager. We have analyzed the system separately for each band using TotalLab (Nonlinear Dynamics). For p53CT/DNA complexes with 0- to 9-bp spacers, we used a regular two binding site model (51), where we assumed zero values, as initial values, when the dimer bands are unobserved (21, 31). Thus, the following equations were used:
| [S1] |
| [S2] |
| [S3] |
Θi, the fraction of DNA molecules with i protein molecules bound, was calculated from the equation: Θi = (PSL − bg)i/Σi(PSL − bg)i, where PSL is the photostimulated luminescence and bg is the background, and the summation is over all of the bands in a given lane. [P] is the protein concentration, and Ka1 and Ka2 are the macroscopic association binding constants for the dimeric and the tetrameric species, respectively. For the GGG-(ta)n-GGG series, microscopic (intrinsic) constants can be calculated using the following:
| [S4] |
| [S5] |
k1 is the intrinsic microscopic binding constant for each specific half-site, and k11 is the cooperativity constant. k11 measures the increase (or decrease) in the binding affinity of the second dimer relative to that of the first dimer due to the cooperativity of the binding interaction.
For p53CT/DNA complexes with 10- to 18-bp spacers, we used a two binding site model, in which two different tetrameric species were included, one for the specific complex and the other for two hemispecific tetrameric complexes:
| [S6] |
| [S7] |
| [S8] |
| [S9] |
For the GGG-(ta)n-GGG series, the microscopic (intrinsic) equations corresponding to the above system are as follows:
| [S10] |
| [S11] |
| [S12] |
k1 and k2 measure the intrinsic microscopic binding constants of p53CT dimers on the specific and nonspecific spacer sequence, respectively. We can estimate the cooperativity of interaction of p53CT on the specific complex, k11, from these equations by substituting the value of k1 from Eq. S10 into Eq. S12. However, we cannot estimate k12, the cooperativity constant for the hemispecific complex, since we do not know the values for k2, the intrinsic microscopic binding constants of p53CT dimers on the hemispecific sites. For the 20-bp spacer construct, an equation for an octamer complex was also included. For sequences that contain only one specific half-site sequence and one nonspecific site [such as N20(ta)5-GGG], we used a regular two binding site model composed of dimers and tetramers, because dimers were also observed. In cases where the two specific half-sites are identical and by assuming equipartition of free energies of binding, we can calculate an apparent dimer-equivalent tetramer association constant by taking the square root of the macroscopic constant (21). Here, we can calculate such values only for the specific tetramer (that is only for Ka3), because Ka2 is composed of two different dimers, with very different binding constants, k1 and k2. The reported values (Table 1 and Table S1) are the dissociation binding constants (KD), which are the reciprocal of the association binding constants. The values in parentheses in all tables, figure legends, and text are the SEMs. The error bars in the graphs are SDs.
Binding Stoichiometry Measurements.
The experiments were carried out by the continuous-variation (Job plot) method (21, 26). In this method, the ratio of two reacting species is varied but the total molar concentration is kept constant at a value that is significantly above the KD of their interaction. The maximum amount of complex is formed when the two species are present in their stoichiometric molar ratio. Here, we use this method for the complex with the GGG-(ta)10-GGG sequence. We used either 100 or 200 nM as the total molar concentration (protein dimers and DNA) with the inclusion of 0.05% Nonidet P-40 in the regular high ionic strength buffer. Under these conditions, 100 nM concentration is significantly above the KD of this sequence (Fig. S1H and Table S1). We varied the total molar concentration (using both 100 and 200 nM) since the maximum in the Job plot for a cooperative binding event is independent of the total protein and DNA concentration (26). The protein molar fraction was varied regularly from 0 to 0.9 (concentration of 0–90 or 0–180 nM). Other reaction conditions are as described above for the binding affinity measurements.
UV Cross-Linking Experiments of p53CT with 4-Thiothymidine–Containing DNA.
The hemispecific binding site made of the right-handed half-site decamer of CHMP4c and 11 bp from the adjacent spacer sequence, designed as intramolecular hairpin construct, was synthesized with the UV-reactive nucleotide analog, 4-thio-dTMP, replacing the T nucleotide at the tcc triplet of the spacer sequence (gcagctgc4stccGAGCTTGCCCa). The EMSA conditions are those described in ref. 45. The 6 nM radiolabeled and gel-purified oligonucleotide was incubated with 120 nM p53CT in a 50-μL reaction mixture containing 20 mM Hepes (pH 7.8 at 21 °C), 25 mM KCl, 10% glycerol, 2 mM MgCl2, 0.1 mM EDTA, 0.02% Nonidet P-40, 0.25 mM DTT, 0.4 mM spermidine, 0.5 mM CaCl2, and 100 ng of BSA, for 2 h at 21 °C in 96-well plates. The reactions were carried out under very dim-light conditions. The lid of a 96-well plate was removed, and the plate was then inverted onto a UVB/Vis transilluminator (Spectroline TVD-1000F), propped up by supports, such that the sample was at a distance of about 4 cm from the UV source. The bottom of the plate, which faces upward, was kept cold by placing a chilled metal block on it. The samples were then irradiated with UV light (312 nm) for 10 min at 100% intensity setting. The samples were then mixed with SDS containing sample buffer and electrophoresed on 8% Tris-glycine SDS gels, at 150 V for 2 h without heat inactivation. Gels were fixed, dried, and visualized using a GE Typhoon FLA7000 phosphoimager.
Cell Culture.
H1299 non–small-cell lung carcinoma cells were obtained from the ATCC and cultured in RPMI-1640 supplemented with 10% FCS, 2 mM l-glutamine, and 1% Pen-Strep solution (Biological Industries). The cells were maintained in a humidified incubator at 37 °C and 5% CO2.
Reporter Gene Constructs.
The reporter gene vectors pCLuc Mini-TK 2, encoding Cypridina secreted luciferase under the control of minimal promoter, and pCMV-GLuc, encoding constitutively expressed Gaussia secreted luciferase, were purchased from New England Biolabs. The pC53-SN3 construct that was used for wild-type p53 expression in cells was kindly provided by Prof. Bert Vogelstein (Johns Hopkins University School of Medicine, Baltimore, MD). The PTEN full responsive element, PTEN RHS, and PTEN LHS were cloned upstream to the CLuc gene (encoding the Cypridina luciferase) in the pCLuc Mini-TK 2 plasmid, by inverse PCR, using Kapa HiFi DNA polymerase (Kapa Biosystems) according to manufacturer’s recommendations. The correct insertion of the various responsive elements was validated by sequencing.
The primer sequences that were used for cloning using inverse PCR are as follows: PTEN full RE, CTACACTGGGCATGCTCGGGATCCTTCGCATATTAAGGT (forward) and CTGCCTGGGGCTTGCTCCGAGCTCGGTACCAAGCTTGGG (reverse); PTEN LHS, GGCAGCTACACTGGGATCCTTCGCATATTAAGGTGAC (forward) and TGGGGCTTGCTCCGAATTCCAAGATCTCCCGATCCG (reverse); PTEN RHS, CTGGGCATGCTCGGGATCCTTCGCATATTAAGGTGAC (forward) and TGTAGCTGCCTGCGAATTCCAAGATCTCCCGATCCG (reverse).
Materials and Methods
A detailed description of materials and methods is available in SI Materials and Methods.
Protein Expression and Purification.
The expression plasmid and production of human p53CT (residues 94–360) were as reported previously (20). Final protein buffer consisted of 50 mM Tris, pH 7.2, 300 mM NaCl, 10 mM DTT, and 10% glycerol. The protein was concentrated to 0.3–0.6 mg/mL, aliquoted, and stored at –80 °C. Before the binding studies, the fraction of p53CT dimers active for DNA binding was determined as described previously (49).
DNA Constructs for Cyclization Kinetics.
Constructs were synthesized using the PCR scheme and top template previously described (21, 32). Here, each bottom template contained a single p53 binding site, which varied from 20 bp, with no spacer sequences between the two half-sites, to 38 bp, with an 18-bp spacer sequence (Table S2).
Cyclization Kinetics Measurements and Structural Simulations.
Cyclization kinetics measurements were carried out as previously described (21, 32). The data were analyzed based on the theory developed by Zhang and Crothers (50), and modified here to account for variations in test-sequence length due to spacer sequences. For sequences with spacer length longer than 2 bp, we could not measure variation in phasing length, because of p53 inhibition of ligase activity (Fig. S2). However, since the phasing length did not change with sequence identity (Fig. S2B), or upon protein binding (Fig. S2C), for REs with 0- or 2-bp spacers, we measured the total-length dependence of all spacer-containing sequences in the in-phase position, 14, of the 0- to 2-bp spacer constructs (Fig. S2 D and E). Hence, for the study of the GGG-(ta)n-GGG series (Tables S2 and S4), the simulations were conducted using 11 total-length J factors instead of 16 J factors (11 total length and 5 phasing length), done for sequences without spacers (21). For measuring the global conformation of p53 REs bound to p53CT, we used 5 and 50 nM p53CT protein, for sequences without and with spacers, respectively, estimated from gel shift experiments conducted at the conditions of the cyclization kinetics assay (Fig. S1F).
Binding Experiments.
Binding affinity measurements were conducted as previously described (21, 31), at 320 mM total ionic strength, except that the incubation temperature was 21 °C. We also ran binding experiments at the conditions used for the cyclization kinetics assays, where the total ionic strength of the buffer was 96 mM, and included 0.05% of the nonionic detergent Nonidet P-40. The experiments to determine the binding stoichiometry were carried out by the continuous-variation (Job plot) method (21, 26).
Competition Experiments.
Competition experiments were performed with ctDNA using a mixture of radiolabeled and cold GGG-(ta)10-GGG sequence (total of 3.5 nM), and 200 nM p53CT dimers. The protein/DNA complexes were incubated at 21 °C for 2 h, and then the mixture was titrated with increasing concentration (0–1.64 × 10−2 in M bp) of double-stranded ctDNA (Sigma; D4764), and further incubated at 21 °C for 2 h. Other reaction conditions were as described for the binding affinity measurements. Based on the total DNA concentration in each gel lane, we determined the concentration of the specific and hemispecific complexes at each competitor concentration from the observed fractional concentration of the free and p53-bound DNA bands in each gel lane.
UV Cross-Linking with 4-Thiothymidine–Containing DNA.
A sequence composed of the specific right-handed half-site of CHMP4c and 11 bp from the adjacent spacer sequence was synthesized by incorporating the UV-reactive nucleotide analog 4-thio-dTMP instead of the T nucleotide at the third position from the 3′-end of the spacer sequence. EMSA was performed as described in ref. 45, except that the UV source was a UVB transilluminator (312 nm; Spectroline TVD-1000F), and the samples were contained in an inverted 96-well plate and kept cold by a chilled metal block.
Reporter Gene Assays.
H1299 cells were seeded in 24-well plates (40,000 cells per well) and 24 h later were transiently cotransfected with 910 ng of pCLuc Mini-TK 2 harboring PTEN REs, 50 ng of pCMV-GLuc, and 40 ng of either pC53-SN3 or a corresponding empty vector, using jetPEI (Polyplus-transfection), according to the manufacturer’s protocol. Luminescence was measured from 10 μL of cells’ media 24 h following transfection using BioLux Gaussia Luciferase Assay Kit and BioLux Cypridina Luciferase Assay Kit (both from New England Biolabs) according to the manufacturer’s protocol, and the signal was detected by Modulus Microplate Reader (Turner BioSystems). To correct for transfection variabilities, the Cypridina signal was normalized to the corresponding Gaussia signal in each sample.
Acknowledgments
We thank Mike Fried, Yongli Zhang, Udayan Mohanty, and Alberto Inga for many helpful discussions. We thank Malka Kitayner for help with the structural models and Aviva Kapitkovsky for help with protein production. We thank Yoav Peleg for help with DNA cloning. We thank the Technion High-Energy Physics Grid Computing Cluster for enabling the simulation of the data in this work. Support for this work was provided by Israel Science Foundation Grants 1517/14 (to T.E.H.) and 349/12 (to Z.S.) and the Kimmelman Center for Biomolecular Structure and Assembly (Z.S.). This article is dedicated to the memory of Professor Donald M. Crothers of Yale University. Professor Crothers helped our work by noticing an interesting observation based on preliminary data, which he encouraged us to further explore. This observation is related to the unique pattern of p53 binding affinity to DNA as a function of DNA spacer and the structure of DNA in such complex systems. However, more important are Crothers' contributions to the field of nucleic acids, by inventing the EMSA method (1981) and being one of the developers of the method of cyclization kinetics of DNA minicircles (1986), without which studies such as ours would have not been possible.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1618005114/-/DCSupplemental.
References
- 1.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 2.Menendez D, Inga A, Resnick MA. The expanding universe of p53 targets. Nat Rev Cancer. 2009;9:724–737. doi: 10.1038/nrc2730. [DOI] [PubMed] [Google Scholar]
- 3.Fischer M, Steiner L, Engeland K. The transcription factor p53: Not a repressor, solely an activator. Cell Cycle. 2014;13:3037–3058. doi: 10.4161/15384101.2014.949083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine AJ, Oren M. The first 30 years of p53: Growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 6.Laptenko O, Prives C. Transcriptional regulation by p53: One protein, many possibilities. Cell Death Differ. 2006;13:951–961. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- 7.Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53. Annu Rev Biochem. 2008;77:557–582. doi: 10.1146/annurev.biochem.77.060806.091238. [DOI] [PubMed] [Google Scholar]
- 8.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weisz L, Oren M, Rotter V. Transcription regulation by mutant p53. Oncogene. 2007;26:2202–2211. doi: 10.1038/sj.onc.1210294. [DOI] [PubMed] [Google Scholar]
- 10.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 11.Wei CL, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 12.Smeenk L, et al. Characterization of genome-wide p53-binding sites upon stress response. Nucleic Acids Res. 2008;36:3639–3654. doi: 10.1093/nar/gkn232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menendez D, et al. Diverse stresses dramatically alter genome-wide p53 binding and transactivation landscape in human cancer cells. Nucleic Acids Res. 2013;41:7286–7301. doi: 10.1093/nar/gkt504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang GS, et al. A comprehensive and high-resolution genome-wide response of p53 to stress. Cell Rep. 2014;8:514–527. doi: 10.1016/j.celrep.2014.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman PN, Chen X, Bargonetti J, Prives C. The p53 protein is an unusually shaped tetramer that binds directly to DNA. Proc Natl Acad Sci USA. 1993;90:3319–3323. doi: 10.1073/pnas.90.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLure KG, Lee PWK. How p53 binds DNA as a tetramer. EMBO J. 1998;17:3342–3350. doi: 10.1093/emboj/17.12.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veprintsev DB, et al. Core domain interactions in full-length p53 in solution. Proc Natl Acad Sci USA. 2006;103:2115–2119. doi: 10.1073/pnas.0511130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitayner M, et al. Structural basis of DNA recognition by p53 tetramers. Mol Cell. 2006;22:741–753. doi: 10.1016/j.molcel.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 19.Kitayner M, et al. Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nat Struct Mol Biol. 2010;17:423–429. doi: 10.1038/nsmb.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weinberg RL, Veprintsev DB, Fersht AR. Cooperative binding of tetrameric p53 to DNA. J Mol Biol. 2004;341:1145–1159. doi: 10.1016/j.jmb.2004.06.071. [DOI] [PubMed] [Google Scholar]
- 21.Beno I, Rosenthal K, Levitine M, Shaulov L, Haran TE. Sequence-dependent cooperative binding of p53 to DNA targets and its relationship to the structural properties of the DNA targets. Nucleic Acids Res. 2011;39:1919–1932. doi: 10.1093/nar/gkq1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balagurumoorthy P, et al. Four p53 DNA-binding domain peptides bind natural p53-response elements and bend the DNA. Proc Natl Acad Sci USA. 1995;92:8591–8595. doi: 10.1073/pnas.92.19.8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagaich AK, et al. p53-induced DNA bending and twisting: p53 tetramer binds on the outer side of a DNA loop and increases DNA twisting. Proc Natl Acad Sci USA. 1999;96:1875–1880. doi: 10.1073/pnas.96.5.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu X, Riley T, Levine AJ. The regulation of the endosomal compartment by p53 the tumor suppressor gene. FEBS J. 2009;276:2201–2212. doi: 10.1111/j.1742-4658.2009.06949.x. [DOI] [PubMed] [Google Scholar]
- 25.Stambolic V, et al. Regulation of PTEN transcription by p53. Mol Cell. 2001;8:317–325. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 26.Huang CY. Determination of binding stoichiometry by the continuous variation method: The Job plot. Methods Enzymol. 1982;87:509–525. doi: 10.1016/s0076-6879(82)87029-8. [DOI] [PubMed] [Google Scholar]
- 27.Leith JS, et al. Sequence-dependent sliding kinetics of p53. Proc Natl Acad Sci USA. 2012;109:16552–16557. doi: 10.1073/pnas.1120452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lumpkin OJ, Déjardin P, Zimm BH. Theory of gel electrophoresis of DNA. Biopolymers. 1985;24:1573–1593. doi: 10.1002/bip.360240812. [DOI] [PubMed] [Google Scholar]
- 29.Kearns S, Lurz R, Orlova EV, Okorokov AL. Two p53 tetramers bind one consensus DNA response element. Nucleic Acids Res. 2016;44:6185–6199. doi: 10.1093/nar/gkw215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin TG, et al. Design of a molecular support for cryo-EM structure determination. Proc Natl Acad Sci USA. 2016;113:E7456–E7463. doi: 10.1073/pnas.1612720113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jordan JJ, et al. Low-level p53 expression changes transactivation rules and reveals superactivating sequences. Proc Natl Acad Sci USA. 2012;109:14387–14392. doi: 10.1073/pnas.1205971109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Crothers DM. High-throughput approach for detection of DNA bending and flexibility based on cyclization. Proc Natl Acad Sci USA. 2003;100:3161–3166. doi: 10.1073/pnas.0530189100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Xi Z, Hegde RS, Shakked Z, Crothers DM. Predicting indirect readout effects in protein-DNA interactions. Proc Natl Acad Sci USA. 2004;101:8337–8341. doi: 10.1073/pnas.0402319101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bloomfield VA, Crothers DM, Tinoco IJ. Nucleic Acids: Structures, Properties, and Functions. University Science Books; Sausalito, CA: 2000. [Google Scholar]
- 35.Chen HH, Rau DC, Charney E. The flexibility of alternating dA-dT sequences. J Biomol Struct Dyn. 1985;2:709–719. doi: 10.1080/07391102.1985.10506318. [DOI] [PubMed] [Google Scholar]
- 36.Okonogi TM, Alley SC, Reese AW, Hopkins PB, Robinson BH. Sequence-dependent dynamics in duplex DNA. Biophys J. 2000;78:2560–2571. doi: 10.1016/S0006-3495(00)76800-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malecka KA, Ho WC, Marmorstein R. Crystal structure of a p53 core tetramer bound to DNA. Oncogene. 2009;28:325–333. doi: 10.1038/onc.2008.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y, Dey R, Chen L. Crystal structure of the p53 core domain bound to a full consensus site as a self-assembled tetramer. Structure. 2010;18:246–256. doi: 10.1016/j.str.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vainer R, Cohen S, Shahar A, Zarivach R, Arbely E. Structural basis for p53 Lys120-acetylation-dependent DNA-binding mode. J Mol Biol. 2016;428:3013–3025. doi: 10.1016/j.jmb.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 40.Rohs R, et al. Origins of specificity in protein-DNA recognition. Annu Rev Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melero R, et al. Electron microscopy studies on the quaternary structure of p53 reveal different binding modes for p53 tetramers in complex with DNA. Proc Natl Acad Sci USA. 2011;108:557–562. doi: 10.1073/pnas.1015520107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stormo GD. Introduction to Protein-DNA Interactions. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 2013. [Google Scholar]
- 43.Jordan JJ, et al. Noncanonical DNA motifs as transactivation targets by wild type and mutant p53. PLoS Genet. 2008;4:e1000104, and erratum (2008) 4, 10.1371/annotation/13bc83be-2345-401d-b953-f1886e9fbdff. doi: 10.1371/journal.pgen.1000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Menendez D, Inga A, Resnick MA. Estrogen receptor acting in cis enhances WT and mutant p53 transactivation at canonical and noncanonical p53 target sequences. Proc Natl Acad Sci USA. 2010;107:1500–1505. doi: 10.1073/pnas.0909129107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laptenko O, et al. The p53 C terminus controls site-specific DNA binding and promotes structural changes within the central DNA binding domain. Mol Cell. 2015;57:1034–1046. doi: 10.1016/j.molcel.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zauberman A, Flusberg D, Haupt Y, Barak Y, Oren M. A functional p53-responsive intronic promoter is contained within the human mdm2 gene. Nucleic Acids Res. 1995;23:2584–2592. doi: 10.1093/nar/23.14.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hollenbeck JJ, Oakley MG. GCN4 binds with high affinity to DNA sequences containing a single consensus half-site. Biochemistry. 2000;39:6380–6389. doi: 10.1021/bi992705n. [DOI] [PubMed] [Google Scholar]
- 48.Luisi BF, et al. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 49.Bareket-Samish A, Cohen I, Haran TE. Signals for TBP/TATA box recognition. J Mol Biol. 2000;299:965–977. doi: 10.1006/jmbi.2000.3797. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y, Crothers DM. Statistical mechanics of sequence-dependent circular DNA and its application for DNA cyclization. Biophys J. 2003;84:136–153. doi: 10.1016/S0006-3495(03)74838-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Senear DF, Brenowitz M. Determination of binding constants for cooperative site-specific protein-DNA interactions using the gel mobility-shift assay. J Biol Chem. 1991;266:13661–13671. [PubMed] [Google Scholar]
- 52.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]