Significance
R-loops form when transcribed RNA remains bound to its DNA template to form a stable RNA:DNA hybrid. Stable R-loops at a CAG/CTG repeat tract, a sequence that can expand to cause human disease, cause DNA breaks as well as repeat instability. We found that R-loop–induced deamination of cytosines followed by base excision repair is responsible for causing CAG repeat breaks and contractions. Intriguingly, R-loop–dependent double-strand breaks were caused by the MutL gamma endonuclease, which is known to recognize structured DNA and cause nicks, defining a new mechanism for how R-loops can generate DNA breaks. Our results have implications for human repeat expansion diseases and provide a paradigm for how RNA:DNA hybrids can cause genome instability at structure-forming DNA sequences.
Keywords: R-loop, CAG repeat instability, chromosome fragility, Fcy1 cytosine deaminase, Mlh1/Mlh3
Abstract
CAG/CTG repeats are structure-forming repetitive DNA sequences, and expansion beyond a threshold of ∼35 CAG repeats is the cause of several human diseases. Expanded CAG repeats are prone to breakage, and repair of the breaks can cause repeat contractions and expansions. In this study, we found that cotranscriptional R-loops formed at a CAG-70 repeat inserted into a yeast chromosome. R-loops were further elevated upon deletion of yeast RNaseH genes and caused repeat fragility. A significant increase in CAG repeat contractions was also observed, consistent with previous human cell studies. Deletion of yeast cytosine deaminase Fcy1 significantly decreased the rate of CAG repeat fragility and contractions in the rnh1Δrnh201Δ background, indicating that Fcy1-mediated deamination is one cause of breakage and contractions in the presence of R-loops. Furthermore, base excision repair (BER) is responsible for causing CAG repeat contractions downstream of Fcy1, but not fragility. The Rad1/XPF and Rad2/XPG nucleases were also important in protecting against contractions, but through BER rather than nucleotide excision repair. Surprisingly, the MutLγ (Mlh1/Mlh3) endonuclease caused R-loop–dependent CAG fragility, defining an alternative function for this complex. These findings provide evidence that breakage at expanded CAG repeats occurs due to R-loop formation and reveal two mechanisms for CAG repeat instability: one mediated by cytosine deamination of DNA engaged in R-loops and the other by MutLγ cleavage. Since disease-causing CAG repeats occur in transcribed regions, our results suggest that R-loop–mediated fragility is a mechanism that could cause DNA damage and repeat-length changes in human cells.
An R-loop is a transcription-associated extended RNA:DNA hybrid structure with an exposed tract of single-stranded DNA. The RNA:DNA hybrid within an R-loop can be longer than the transient RNA:DNA hybrid that forms in the ∼8-nt transcription bubble or the ∼11-nt RNA primer region of an Okazaki fragment (1). Genome-wide studies have shown that R-loops exist frequently throughout genomes in many organisms, from yeast to humans (2–5). The single-stranded DNA (ssDNA) displaced by an R-loop is prone to DNA breaks, which can have positive or negative biological functions. During class-switch recombination (CSR), stable R-loops form on the G-rich transcription template in the Sμ region of the human immunoglobulin locus (Ig locus). This process exposes cytosine on the displaced strand, which is deaminated to uracil by activation-induced deaminase (AID) (6). Uracil DNA glycosylase (Ung1)–dependent base excision repair (BER) then creates single-stranded breaks (SSBs) by removal of uracil, followed by APE1- and APE2-dependent cleavage of abasic sites (7–9). These SSBs can be converted to double-strand breaks (DSBs) when close to each other on opposite strands, and religation of these DSBs achieves CSR. Mismatch repair (MMR) was reported to be required for efficient CSR to occur (10). AID can also cause off-target effects to create DSBs outside of the Ig locus (11). In addition to AID, there are also other cytosine deaminases, such as the APOBEC family, which can cause DNA damage (12).
Despite their prevalence and physiological functions, R-loops can be detrimental to cells due to increased R-loop–associated DNA damage and genome instability (13–15). In human cells, transcription-coupled nucleotide excision repair (TC-NER) nucleases XPG and XPF have been shown to process R-loops into DSBs (16). Accumulation of unscheduled and deleterious R-loops is found in cells defective in RNA-processing factors, RNA:DNA helicases, and mutants defective in RNA biogenesis (17–19). RNaseH family proteins are important in controlling R-loop levels as they directly degrade the RNA moiety in a RNA:DNA hybrid, a mechanism that is highly conserved from bacteria to humans (20). Budding yeast, Saccharomyces cerevisiae, has two RNaseH proteins: RNaseH1 and RNaseH2, which have distinct but also overlapping functions. A double deletion of RNH1 and RNH201 genes causes a global increase in R-loop levels (2, 5, 19).
R-loops have been shown to form in vivo and in vitro upon transcription through a CAG/CTG (CAG) repeat region, with the level of R-loop formation increasing with CAG tract length (21–23). R-loop formation destabilizes expanded CAG repeats, as depletion of RNaseH stimulates repeat contractions in a human cell line (21). Both CAG and CTG single strands can form intramolecular hairpin structures, which could act to stabilize R-loops and also mediate repeat contractions. Convergent transcription through a CAG/CTG tract further destabilizes the repeat, and high levels are toxic to human cells (22, 24). Interestingly, all known expandable CAG repeats are located within transcribed regions in either the coding or noncoding regions of genes (25). Thus, mechanisms causing R-loop–induced repeat instability could be highly relevant for understanding CAG repeat expansions, which are the cause of numerous human neurological and neuromuscular disorders, including Huntington’s disease, myotonic dystrophy, and multiple types of spinocerebellar ataxia. At most loci, expansion above a threshold of ∼35 CAG repeats causes the repeat to become highly unstable, and expansions above a length of ∼30–60 repeats (depending on the locus) cause disease (25).
In addition to instability (contractions and expansions), expanded CAG tracts are fragile sites that break in a length-dependent manner (25–28). Mutants defective in DNA break repair or the checkpoint response exhibit increased CAG repeat fragility rates and also elevated instability levels. Some contractions and expansions were shown to be dependent on DSB repair pathways, including both end-joining and homologous recombination (HR) pathways (29); for review, see ref. 30. However, the initial cause of the breaks at expanded CAG tracts is not known. Additionally, it is not known whether R-loop–associated CAG instability is mostly due to R-loop processing without induction of DSBs, for example, through excision repair, or whether R-loop–induced DNA breakage and repair events contribute.
To elucidate these important questions, we used a highly sensitive yeast artificial chromosome (YAC)-based system that carries a CAG-70 repeat tract to investigate CAG repeat breakage (fragility) and instability in conditions of increased R-loops. We observed a dramatic increase of CAG-70 repeat breakage when R-loops are present and identified an endogenous yeast cytosine deaminase, Fcy1, as being partially responsible for R-loop–induced CAG repeat fragility. Fcy1-mediated deamination followed by Ung1-dependent BER is a primary pathway resulting in repeat contractions in conditions of increased R-loops. Furthermore, the MutLγ complex (Mlh1/Mlh3), a dual-function nuclease complex involved in both meiotic recombination and MMR, also causes both repeat fragility and contractions when R-loops are present. Rad52-mediated repair is important in healing the breaks to reduce chromosome end loss. Altogether, our study reveals that CAG repeat breakage occurs when R-loops accumulate and that cytosine deamination followed by BER and MutLγ cleavage are two repair pathways that control R-loop–induced repeat fragility and instability.
Results
Transcription-Coupled R-Loop Formation Causes Fragility and Instability of Expanded CAG Repeats.
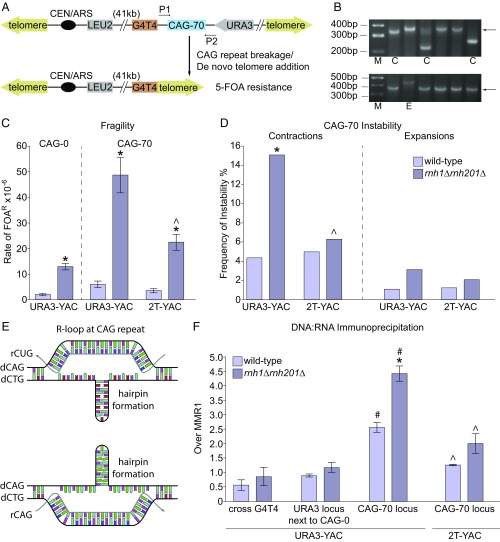
To investigate if in vivo CAG repeat breakage and instability occur when R-loop levels are elevated, fragility and instability of an expanded (CAG)70 repeat were tested in rnh1rnh201 double-deletion strains, using two previously established assays (27) (Fig. 1 A and B). CAG fragility was tested by measuring the rate of loss of a URA3 marker gene adjacent to the repeat tract on YAC CF1 by plating cells on media containing 5-fluoro-orotic acid (5-FOA); recovery of the broken chromosome is facilitated by the presence of a (G4T4)13 telomere seed proximal to the repeat tract (Fig. 1A). DNA isolated from a subset of 5-FOA–resistant (FOAR) colonies was analyzed by PCR and/or Southern blot to confirm the fraction that exhibited YAC end loss (27) (SI Appendix, Table S5). Using this assay, the CAG-70 repeat increases the rate of fragility by approximately threefold over a no-tract control (CAG-0). In the rnh1rnh201 deletion background, an eightfold increase in the rate of FOAR was observed compared with wild-type strains containing a CAG-70 repeat (Fig. 1C and SI Appendix, Table S1). The CAG-0 control showed a sixfold increase of FOAR in the rnh1rnh201 mutants compared with wild type, indicating that the effect is not specific to the CAG tract, as expected from previous YAC end loss data (19) (Fig. 1C and SI Appendix, Table S2). PCR analysis confirmed that the increase of FOA resistance is not due to mutation of the URA3 marker gene, with 100% end loss events for the CAG-70 YAC in FOAR colonies from both wild-type and rnh1rnh201 strains (SI Appendix, Table S5), therefore justifying the correlation of the rate of FOAR as an indicator of chromosome fragility. Single deletion of either rnh1 or rnh201 results in a much milder two- to threefold increase in CAG repeat fragility (SI Appendix, Fig. S1 and Table S1), indicating that RNase H1 and H2 can partially compensate for each other at CAG repeats to reduce fragility and suggesting that R-loops are the cause of the fragility since R-loop processing is a redundant activity.
Fig. 1.
CAG fragility and instability are increased in strains deleted for RNase H1 and RNase H2 in a manner dependent on transcription and R-loop formation. Assay system for (A) CAG fragility and (B) instability; P1 and P2 indicate site-specific primers used for PCR amplification of CAG repeats for sizing. The arrows in B indicate size of the CAG-70 repeat PCR product; C, contraction; E, expansion; M, marker. Two typical pictures from rnh1Δrnh201Δ deletion strains are shown. (C) Rate of FOAR × 10−6 in indicated strains; *P < 0.05 compared with wild type; average ± SEM of at least three experiments is shown (SI Appendix, Tables S1–S3). The 2 terminator (2T)-YAC contains transcription terminators flanking the CAG tract (SI Appendix, Fig. S2A). (D) CAG-70 expansion and contraction frequencies. *P < 0.05 compared with wild type of same tract length; ^P < 0.05 compared with same mutant with URA3-YAC by Fisher’s exact test (SI Appendix, Table S4). (E) Predicted R-loops due to transcription of either CAG or CTG strand. Purines are depicted in green/aqua, pyrimidines in red/pink, and rUTP in blue. Mismatches in the hairpins are depicted as conflicting/nonpaired bases. (F) DRIP in wild-type and rnh1Δrnh201Δ strains. YAC structures and qPCR primer locations are in SI Appendix, Fig. S2A. Each bar represents the mean ± SEM of at least two experiments from two biological replicates; *P ≤ 0.05, compared with wild type for the same locus, #P ≤ 0.05, compared with URA3 or G4T4 flanking loci of the same strain, ^P ≤ 0.05, 2T-YAC compared with URA3-YAC for the same strain background, all by t test. The MMR1 locus IP/input was similar and low in R-loop signal in both wild-type and rnh1Δrnh201Δ strains, as in ref. 2.
RNase H deletion also significantly increased CAG contractions 3.5-fold over wild-type (Fig. 1D and SI Appendix, Table S4), as has been observed in human cells (21). Because we analyzed instability by PCR without any selection (Fig. 1B), we were also able to monitor repeat expansion frequency changes in our assay, which were not evaluated in the human cell system. We observed a 2.8-fold increase in CAG-70 expansions in the rnh1rnh201 mutants compared with wild type (P = 0.07). We note that for contractions, deletion of RNase H1 or H2 alone also increased the observed frequency (SI Appendix, Fig. S1B). Since RNaseH1 preferentially targets long RNA:DNA hybrids (20), this supports a role for R-loops in causing contractions. RNase H2 targets both multiple contiguous ribonucleotide monophosphates (rNMPs) and single rNMPs, so the contractions occurring in this mutant could be due to either increased R-loops or increased rNMPs in DNA (31). In conclusion, deletion of RNase H1 and H2 causes an increase in both fragility and instability of an expanded CAG tract, with elevated contractions as the more prominent instability phenotype.
To determine whether the CAG repeat phenotypes were dependent on transcription, a YAC with two different terminators flanking the CAG repeats was constructed, oriented to block transcription initiating from either direction outside of the repeats (SI Appendix, Fig. S2A; 2T YAC). In the parent YAC without terminators, the main transcript detected was rCUG (Fig. 1E and SI Appendix, Fig. S2B). Addition of two terminators decreases these read-through transcripts from the URA3 gene (rCUG) by >70% (SI Appendix, Fig. S2B). Significant levels of rCAG transcript were detected only when a single bidirectional terminator between URA3 and the CAG tract was added, which is presumably due to stabilization of a cryptic transcript. This cryptic transcript was reduced by 40% upon addition of the second terminator, indicating that some cryptic transcription may initiate from within the repeat (SI Appendix, Fig. S2B). CAG fragility was significantly reduced by 54%, and contractions were reduced to wild-type levels on the 2T YAC in the rnh1rnh201 strain, indicating that the main source of fragility and instability in this background was dependent on transcription through the repeat tract (Fig. 1 C and D and SI Appendix, Tables S1, S3, and S4).
To establish if cotranscriptional R-loops were formed at the CAG repeats and if R-loop formation relates to repeat fragility, we performed a DNA:RNA immunoprecipitation using primers flanking or on either side of the CAG tract. We found that CAG repeats accumulated significantly more DNA:RNA hybrids than the adjacent URA3 gene or (G4T4)13 tract and that the level of hybrids at the CAG tract was further elevated in the rnh1rnh201 background (Fig. 1F). Addition of terminators flanking the CAG repeat significantly decreased the level of RNA:DNA hybrids at the CAG tract in both the wild-type and rnh1rnh201 strains (Fig. 1F). We conclude that transcription through the CAG tract promotes R-loop formation and that depletion of RNase H enzymes results in a further increase in R-loop formation at the CAG repeats. Taken together, our results indicate that the increase in CAG-70 repeat fragility in the rnh1rnh201 strains is most likely due to cotranscriptional R-loop accumulation.
A Yeast Endogenous Cytosine Deaminase, Fcy1, Mediates R-Loop–Induced CAG Repeat Fragility and Contractions.
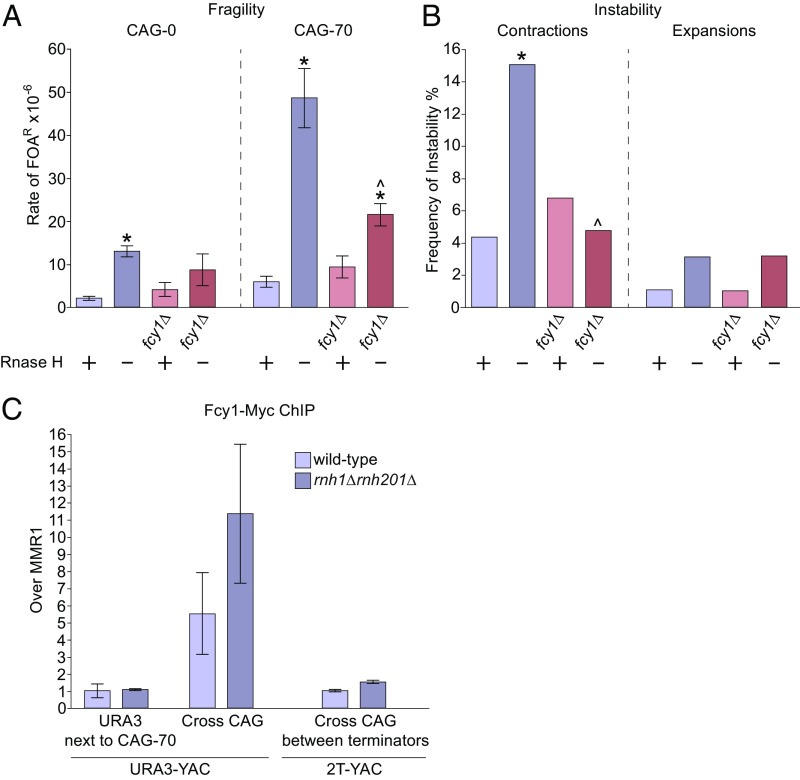
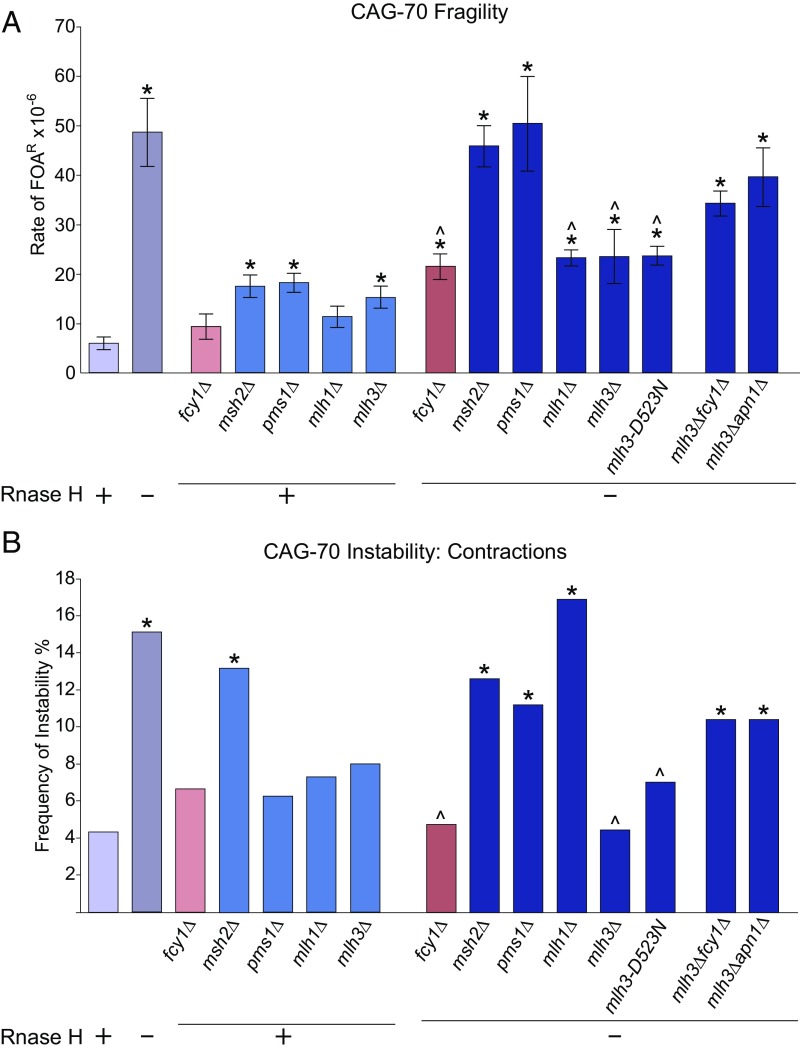
Ectopic expression of human AID was shown to generate transcription-associated and R-loop–induced site-specific recombination mutations and DSBs in yeast (32–34). We questioned whether an endogenous cytosine deamination mechanism could be mediating CAG repeat fragility and instability detected in the rnh1rnh201 mutants. To investigate this question, the FCY1 gene was deleted in the wild-type and rnh1rnh201 strains. FCY1 encodes a yeast cytosine deaminase, which exhibits in vitro biochemical activity to deaminate cytosine to uracil (35). Interestingly, deletion of Fcy1 suppressed the rate of CAG-70 fragility in the rnh1rnh201 background by 56% compared with the level of rnh1rnh201 (Fig. 2A and SI Appendix, Table S1). CAG-70 repeat contractions in rnh1rnh201 mutants were also significantly decreased by deletion of Fcy1: the rnh1rnh201fcy1 triple mutant showed complete suppression of contractions to the wild-type or fcy1 single-mutation level (Fig. 2B). However, the expansion frequency in rnh1rnh201 cells was not affected by fcy1 deletion.
Fig. 2.
Deletion of the yeast cytosine deaminase Fcy1 decreases R-loop–dependent CAG repeat fragility and contractions and binds to the CAG tract. (A) Rate of FOAR × 10−6 in indicated mutants; on the x axis, “+” indicates the presence of RNH1 and RNH201 genes, and “−” indicates the rnh1Δrnh201Δ condition. *P < 0.05 compared with wild type and ^P < 0.05 compared with rnh1Δrnh201Δ of same tract length by t test. Average ± SEM of at least three experiments is shown (see also SI Appendix, Tables S1 and S2). (B) CAG-70 expansion and contraction frequencies, *P < 0.05 compared with wild type and ^P < 0.05 compared with rnh1Δrnh201Δ by Fisher’s exact test (see also SI Appendix, Table S4). (C) Detection of Fcy1-Myc interaction with different loci on URA3-YAC and 2T-YAC by ChIP. Each bar represents the mean ± SEM of at least two experiments from two biological replicates.
To obtain evidence that the Fcy1 effect was direct, chromatin immunoprecipitation (ChIP) of Myc-tagged Fcy1 was performed. Fcy1 binding to the CAG tract was enriched compared with flanking DNA, with an additional twofold increase in the rnh1rnh201 strain (Fig. 2C), similar to the increase of R-loop levels shown by DNA:RNA immunoprecipitation (DRIP) in Fig. 1F. In addition, detection of the tagged Fcy1-myc by immunofluorescence showed increased nuclear accumulation in the rnh1rnh201 strain (SI Appendix, Fig. S3). The enriched level of Fcy1-Myc protein at the CAG-70 repeat in both wild-type and rnh1rnh201 mutants supports the conclusion that Fcy1 could be causing deamination at the CAG repeats when R-loops are present. Although an fcy1 deletion does not cause a growth defect (36), we tested whether the suppression of fragility and contractions could be due to an indirect effect of lowering transcription levels through the CAG repeat in the rnh1rnh201 background; however, no difference from the rnh1rnh201 strain was observed (SI Appendix, Fig. S2C) Note, however, that transcript levels were somewhat decreased in the rnh1rnh201 strain compared with wild type, perhaps due to the presence of persistent R-loops. As a comparison, we also examined the role of yeast cytidine deaminase, Cdd1, which functions in the pyrimidine salvage pathway (37), in repeat fragility and instability in rnh1rnh201 mutants. As expected for an RNA-specific enzyme, no significant decrease in either CAG-70 repeat fragility or contractions was detected in the rnh1rnh201cdd1 strains (SI Appendix, Tables S1 and S4). In conclusion, we identified that R-loop–induced CAG-70 repeat fragility is partially dependent on yeast Fcy1 cytosine deaminase and that Fcy1 also mediates CAG repeat contractions when R-loops are present. We conclude that Fcy1 could deaminate cytosine in the CTG (or CAG) strand when ssDNA is exposed by R-loop formation.
Ung1-Dependent BER Contributes to R-Loop–Induced CAG Repeat Contractions.
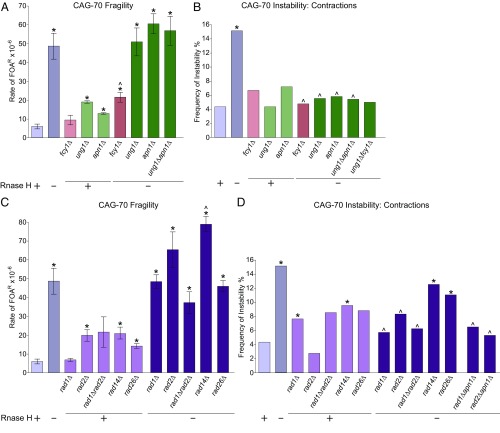
Since cytosine deamination was identified as a mediator of R-loop–induced repeat fragility and contractions, we wondered if the presence of uracil in DNA triggers excision by Ung1, which can remove uracil from ssDNA or double-stranded DNA to generate an abasic site, a substrate for BER (38). Surprisingly, deletion of the UNG1 gene in the rnh1rnh201 background did not decrease CAG repeat fragility (Fig. 3A), which is different from the fragility decrease in the rnh1rnh201fcy1 mutants. We further examined the role of BER by deletion of the yeast Apn1 endonuclease, which cleaves abasic sites to cause nicks in the DNA (39). Again, deletion of APN1 in the rnh1rnh201 background did not decrease R-loop–induced CAG-70 fragility (Fig. 3A). Moreover, the rnh1rnh201ung1apn1 quadruple mutation strains showed a similar level of fragility, compared with either the rnh1rnh201 double or each of the triple mutation strains (Fig. 3A). The rnh1rnh201ung1fcy1 strain is not viable on 5-FOA synthetic media and therefore could not be tested. Altogether, the lack of suppression of the rnh1rnh201 CAG fragility phenotype by deletion of BER pathway members indicates that BER is not causing R-loop–dependent CAG repeat fragility.
Fig. 3.
R-loop–induced CAG repeat contractions, but not fragility, are caused by Ung1-dependent BER, together with Apn1, Rad1, and Rad2 nucleases. (A and C) Fragility analysis analyzed as in Fig. 2A; *P < 0.05 compared with wild type and ^P < 0.05 compared with rnh1Δrnh201Δ of same tract length by t test. Average ± SEM of at least three experiments is shown (see also SI Appendix, Table S1). (B and D) CAG-70 contraction frequencies analyzed as in Fig. 2B; *P < 0.05 compared with wild type and ^P < 0.05 compared with rnh1Δrnh201Δ by Fisher’s exact test (see also SI Appendix, Table S4).
We next tested whether the BER pathway was involved in R-loop–induced CAG-70 repeat instability. Deletion of ung1 or apn1 in the rnh1rnh201 background decreases the contraction frequency to the wild-type level, as does the rnh1rnh201ung1apn1 quadruple mutant (Fig. 3B). Importantly, the contraction frequency for the rnh1rnh201ung1fcy1 strain decreased to the same level as in either the deamination-defective or BER-defective backgrounds (Fig. 3B). These data support the conclusion that Fcy1-mediated deamination followed by Ung1-dependent BER leads to CAG repeat contractions. This pathway could lead to contractions by exonuclease processing of Apn1-generated nicks, followed by fill-in synthesis of the gap across from a template hairpin (Fig. 6). To test this model, the effect of deleting Exo1, one of the exonucleases that could create the gap, was determined. Consistent with previous findings (29), deletion of Exo1 on its own greatly elevates CAG repeat fragility and contractions due to its important function in DNA damage repair (SI Appendix, Fig. S4 and Table S4). The rnh1rnh201exo1 mutants showed an additive increase in fragility, compared with the exo1 single and rnh1rnh201 double mutants (SI Appendix, Fig. S4A), indicating that Exo1 does not play a role in causing or preventing R-loop–induced CAG fragility. However, the rnh1rnh201exo1 triple mutants exhibited a significant decrease in CAG repeat contraction frequency compared with the rnh1rnh201 mutants, and the rnh1rnh201ung1exo1 quadruple mutants also had a decreased level of contractions compared with the level expected for additive pathways (SI Appendix, Fig. S4B). These data lend support to the conclusion that R-loop–induced CAG repeat contractions are caused by BER-dependent processing of deaminated cytosines followed by exonuclease-dependent gap formation.
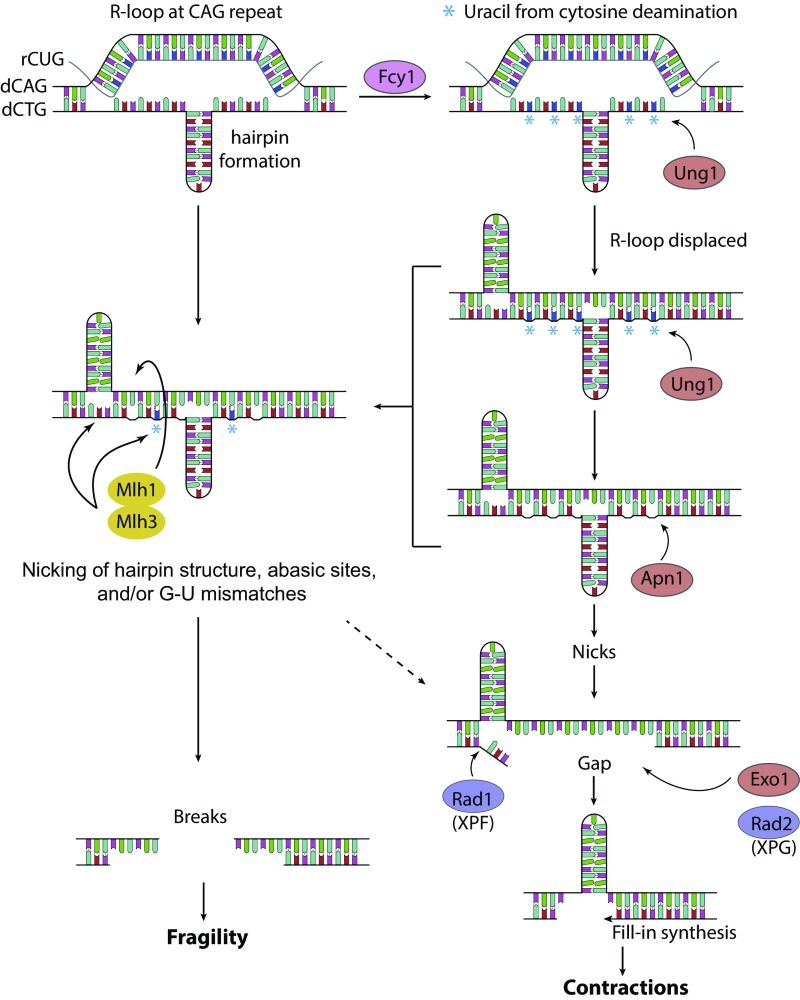
Fig. 6.
Model for how cytosine deamination, base excision repair, and Mlh1-Mlh3 cleavage cooperate to cause R-loop–induced CAG repeat fragility and contractions. Transcription through the CAG repeat promotes the formation of R-loops, and this is exacerbated in the rnh1Δrnh201Δ mutants. Formation of R-loops can promote hairpin formation on the nontemplate strand. Cytosine DNA on the exposed ssDNA is deaminated by Fcy1. Ung1 incises the uracil before or after R-loops are displaced by incoming transcription, leaving abasic sites. Due to hairpin formation at the repeats, slipped-stranded structures can form after the R-loop is displaced. Apn1 cleaves abasic sites and creates DNA nicks, which can be further processed into single-stranded gaps by Exo1, Rad2, and Rad1. Fill-in synthesis can create a shorter tract of repeats by skipping the template hairpin, resulting in a CAG contraction (Right). The R-loop or slipped-stranded structure may also be directly recognized and cleaved by MutLγ to cause R-loop–dependent fragility (Left). In addition, some MutLγ-dependent events occur after action by Fcy1 and Apn1 (arrow from right pathway). For example, dU:dG mismatches could be cleaved by the Mlh1/Mlh3 (MutLγ) nuclease, predicted to act on both strands to create a DSB, or nicks created by BER action at dUTPs may direct MutLγ-dependent nicking on the opposite strand. Rad52-dependent DNA repair is crucial to heal the breaks and reduce YAC end loss.
Transcription-Coupled NER Does Not Cause R-Loop–Induced CAG Fragility or Contractions, Although Rad1/XPF and Rad2/XPG Nucleases May Play a Role in the BER-Mediated Contraction Pathway.
In a human cell line, transcription-induced CAG repeat contractions are dependent on CSB, XPA, and XPG (40, 41). To investigate if CAG repeat fragility induced by cotranscriptional R-loop formation is dependent on nucleotide excision repair, yeast NER factors Rad14 (homolog of human XPA), Rad26 (hCSB), and the endonucleases Rad1 (hXPF) and Rad2 (hXPG) were deleted in the rnh1rnh201 background. A double deletion of both rad1 and rad2 in the rnh1rnh201 deletion background slightly decreased CAG fragility by 23%, although this decrease was not significant or observed in either of the rnh1rnh201rad1 or rnh1rnh201rad2 triple mutants (Fig. 3C and SI Appendix, Table S1). Deletion of Rad14 or Rad26 in the rnh1rnh201 strains resulted in an additive or no increase in fragility compared with single mutant levels (Fig. 3C). Taken together, these data suggest that nuclease activity of Rad1 and Rad2 together may cause a small portion of R-loop–induced CAG repeat breakage, but this role is likely independent of TC-NER.
Interestingly, depletion of either Rad1 or Rad2 significantly decreased CAG repeat contractions in the rnh1rnh201 background to the same level as the BER mutants; however, this was not the case for deletion of Rad14 or Rad26 (Fig. 3D and SI Appendix, Table S4). To test whether these nucleases could be acting in the BER pathway, we determined contraction frequencies in mutants missing Apn1 and either Rad1 or Rad2 in the rnh1rnh201 background. Indeed, the level of suppression indicates that Rad1, Rad2, and Apn1 are working in the same pathway to cause CAG contractions (Fig. 3D and SI Appendix, Table S4). This suggests that Rad1 and Rad2 nuclease activity contribute to R-loop–induced CAG repeat contractions through the BER rather than the TC-NER pathway. Based on their functions, we speculate that Rad1 and Rad2 assist Exo1 in the generation of gaps, which are necessary for contractions (Fig. 6).
MutLγ Directly Causes CAG Repeat Fragility and Instability Through Its Nuclease Cleavage Activity.
Since BER and NER nucleases were not responsible for the R-loop–dependent CAG fragility, we tested the MMR pathway nucleases. We hypothesized that there were two potential R-loop–dependent sources of mismatches that could be recognized by MMR: in CAG or CTG hairpins formed due to R-loops or G-U mismatches that occur due to cytosine deamination. Deletion of any one of the MMR components (msh2, pms1, mlh1, or mlh3) caused a two- to threefold increase in the rate of FOAR (Fig. 4A and SI Appendix, Table S1). FOAR rates adjusted to only reflect end loss slightly decreased this increase to 1.5- to 2.5-fold (SI Appendix, Fig. S5). Surprisingly, deletion of either component of MutLγ, Mlh1, or Mlh3 in the rnh1rnh201 background significantly decreased R-loop–induced fragility, whereas deletion of canonical MMR components Msh2 or Pms1 had no effect (Fig. 4A). This decrease is similar to the reduction of fragility in the rnh1rnh201fcy1 deletion strain. Furthermore, an endonuclease-dead mutation in Mlh3 [mlh3-D523N (42, 43)] also decreased fragility to the same level (Fig. 4A). We conclude that Mlh3 nuclease activity is responsible for creating CAG repeat breaks when R-loops are present, and that MutLγ may recognize the CAG repeat or R-loop independent of a MutS complex. A similar pattern was observed for CAG contractions, with an msh2 or pms1 deletion showing no decrease compared with rnh1rnh201, but with mlh3Δ or mlh3-D523N showing a suppression of R-loop–mediated contractions (Fig. 4B). Deletion of Mlh1 did not suppress contractions, but this could be because it functions both in the MutLα complex (Mlh1/Pms1), which is recruited by the MutS complex, and in the MutLγ complex. Since absence of MutS (msh2Δ) increases contractions, this phenotype could mask a suppression of contractions in the mlh1Δ strain. We note that CAG repeat instability was affected by msh2 and pms1 single-deletion mutations as has been observed in other systems: msh2Δ significantly increased CAG-70 contractions and expansions, and pms1Δ significantly elevated expansions (Fig. 4B and SI Appendix, Table S4).
Fig. 4.
The Mlh1/Mlh3 (MutLγ) complex causes R-loop–induced CAG repeat fragility and contractions. (A) Fragility analysis analyzed as in Fig. 2A; *P < 0.05 compared with wild type and ^P < 0.05 compared with rnh1Δrnh201Δ of same tract length by t test. Average ± SEM of at least three experiments is shown (see also SI Appendix, Table S1). (B) CAG-70 contraction frequencies analyzed as in Fig. 2B; *P < 0.05 compared with wild type and ^P < 0.05 compared with rnh1Δrnh201Δ by Fisher’s exact test (see also SI Appendix, Table S4).
To determine whether MutLγ causes R-loop–induced CAG repeat fragility and contractions downstream of cytosine deamination, rnh1rnh201mlh3fcy1 or rnh1rnh201mlh3apn1 quadruple mutants were tested. Although both mutants showed a reduction in fragility and instability, neither reduction was significant (Fig. 4). Therefore, MutLγ-induced events are dependent on R-loop formation, but not necessarily on cytosine deamination. One caveat is that these quadruple mutants were fairly sick, so it is possible that a general increase in chromosome instability masked a decrease in the CAG fragility and contractions to the rnh1rnh201fcy1 levels. Also, the rnh1rnh201mlh3apn1 fragility level is significantly decreased compared with rnh1rnh201apn1, suggesting that Mlh3 functions upstream of Apn1. Taken together, our data are most consistent with MutLγ acting on structures at the CAG/CTG tract independently of MMR but partially dependent on cytosine deamination, presumably to cause nicks on both strands since either contractions or fragility can result from MutLγ nuclease action.
R-Loop–Induced Breaks at the CAG Repeats Are Healed by a Rad52-Dependent Process.
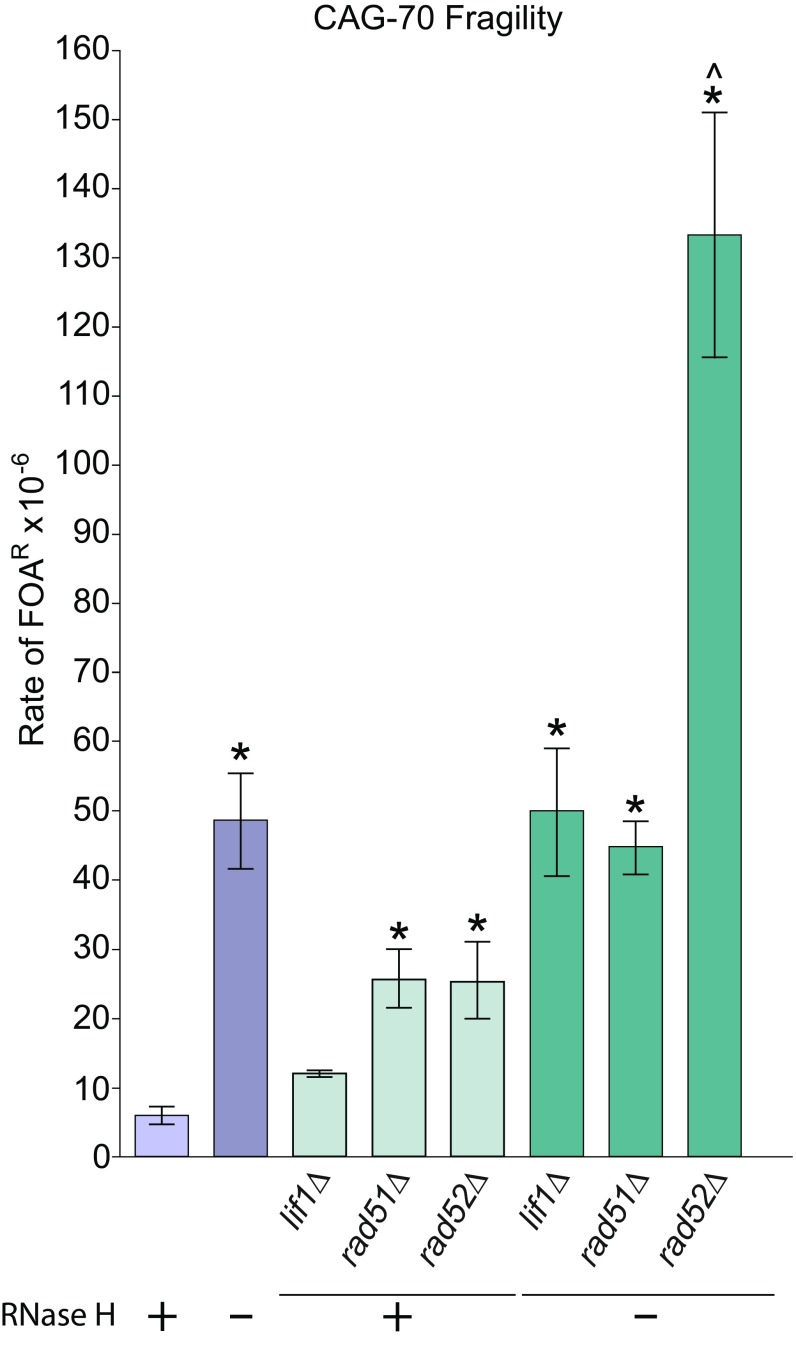
To investigate which repair mechanisms are responsible for healing the R-loop–induced CAG repeat breaks, we tested the role of the two major DSB repair pathways, HR and nonhomologous end-joining (NHEJ). As previously published, depletion of HR proteins Rad51 and Rad52 significantly increased CAG repeat fragility, with a lesser role for NHEJ Ligase IV cofactor Lif1 (Fig. 5) (29, 44). Deletion of rad51 or lif1 in the rnh1rnh201 background did not affect the level of R-loop–induced CAG repeat fragility (Fig. 5), indicating that neither Rad51-dependent HR nor NHEJ is a primary pathway to repair R-loop–induced repeat fragility. Since deletion of Rad51 on its own increased fragility, and the rnh1rnh201rad51 did not show a further additive increase, Rad51-dependent HR may be involved in repair of some R-loop–induced CAG repeat DSBs. However, deletion of Rad52 in the rnh1rnh201 background synergistically increased CAG repeat fragility compared with the rnh1rnh201 mutants (Fig. 5). These data indicate that the majority of R-loop–induced CAG repeat breaks are repaired through a Rad52-mediated repair event not requiring Rad51. One possibility is single-strand annealing repair, which is Rad51-independent but mediated by Rad52 (45, 46). CAG repeat contractions in the rnh1rnh201 background were not affected by deleting Lif1, Rad51, or Rad52 (SI Appendix, Fig. S6). Therefore, most R-loop–induced CAG repeat contractions do not arise from NHEJ or HR repair, consistent with our data that cytosine deamination followed by BER is the primary pathway involved.
Fig. 5.
Rad52-mediated events repair R-loop–induced CAG breaks. Fragility analysis analyzed as in Fig. 2A; *P < 0.05 compared with wild type and ^P < 0.05 compared with rnh1Δrnh201Δ of same tract length by t test. Average ± SEM of at least three experiments is shown (see also SI Appendix, Table S1).
Discussion
In this study, we show that cotranscriptional R-loop formation causes breakage of an expanded CAG repeat tract. The increased R-loop–associated CAG fragility has a strong correlation with repeat instability with increased contractions as the prominent phenotype. Occurrence of DNA breaks at CAG R-loops is highly dependent on a newly identified Fcy1 cytosine deamination pathway and the MutLγ complex and only mildly dependent on the NER nucleases Rad1 and Rad2. However, MutLγ nuclease activity, Fcy1-instigated Ung1/Apn1-dependent BER, and Rad1/Rad2-dependent processing were all shown to generate R-loop–induced repeat contractions. Since all nuclease pathways tested were involved in contractions, our results suggest that nicks, gaps, or breaks can all lead to repeat contractions. However, DSBs are rarer, and only deamination and MutLγ nuclease action were shown to be efficient in causing breaks. These results suggest that only pathways that act on both strands were efficient at causing double-strand breaks that lead to chromosome end loss.
R-loop–induced DNA breaks at the G-C rich Sµ regions during class-switch recombination are caused by both BER and MMR, but not by TC-NER (reviewed in ref. 6). In contrast, R-loops formed in the absence of RNA/DNA helicases Aquarius or Senataxin were processed into DSBs by TC-NER nucleases (16). Thus, it appears that different R-loops, caused by different means or at different sequences, may be processed into DSBs in multiple ways. Similar to our findings, RnaseH1 + H2 double knockdown increased CAG contractions in a selectable mammalian cell culture system (24). In this same system but without RnaseH knockdown, transcription through CAG repeats was shown to induce contractions through MMR (MutSβ) and TC-NER but did not require the human BER nuclease APEX1 (yeast homolog of Apn1) (40, 41). In contrast, the contractions in our system were primarily mediated through BER. Thus, the pathways that cause contractions in the presence of stable R-loops may be different from those operating under conditions of high transcription, which could actually displace R-loops.
In this study, we identified Fcy1-mediated cytosine deamination as a cause of CAG repeat fragility and repeat contractions upon R-loop induction. Previous biochemical studies showed that Fcy1 deaminates deoxynucleotides (47, 48), but a function on chromosomal DNA had not been characterized. Our results revealed Fcy1 binding to the CAG repeat by ChIP and chromosome breaks dependent on both the presence of R-loops at the CAG tract and Fcy1, suggesting that this deaminase acts directly upon chromosomal DNA. Human cells also have many deaminases that could act in a similar fashion at expanded CAG repeats and other sequences prone to forming stable R-loops.
Our genetic assay for CAG fragility detects only those DSB events that result in YAC end loss and thus is an underestimate of the level of DSBs that likely occur in the region. Nonetheless, it is likely that DSBs are rare in comparison with the level of nicks and gaps, given the high percentage of CAG contractions observed in the rnh1rnh201 background (15%). Thus, the majority of deaminated cytosines are likely processed by Ung1-dependent BER and Apn1 into nicks and gaps. The formation of R-loops would create an exposed single strand, providing a preferred substrate for Fcy1 to deaminate the exposed cytosines and explaining the complete suppression of contractions in the fcy1Δ background. R-loop displacement could increase the likelihood of hairpin formation on the template strand, resulting in contractions during the fill-in step (Fig. 6). However, we cannot rule out that excess ribonucleotides in the DNA provide another source of nicks that contribute to some contractions in the RNase H-deficient backgrounds. We speculate that, in addition to Exo1, Rad1/XPF and Rad2/XPG may work at the gap-processing step of BER since they operate in the same pathway as Ung1/Apn1, but not NER- or TC-NER–specific factors, to create contractions. Rad1 has a 5′ flap endonuclease activity that could process a displaced strand, and Rad2 exhibits 5′-3′ exonuclease activity (similar to Exo1) and can complement Rad27 (hFEN1) during BER (49, 50).
Directional RT-PCR data indicated that most R-loops form on the top strand (rCUG) of the CAG repeat tested, which would result in clustering of deaminated cytosines and the resulting nicks on the bottom strand, which can explain how BER mostly causes contractions and not fragility (Fig. 6). This finding suggests that one of the most common types of R-loop–mediated damage is strand-specific nicking, rather than DSBs. However, this model does not account for why deletion of Fcy1 but not BER pathway members suppressed fragility. Since we also detected a low level of cryptic transcript on the bottom strand, it is likely that sometimes the top strand is in a single-stranded state, allowing cytosine deamination on both strands. One possibility is that other pathways in addition to BER process some of the uracils resulting from cytosine deamination to cause nicks on both strands that lead to breaks. One candidate is Top1, which has been shown to cleave ribonucleotides in DNA to cause breaks (51, 52). Another possibility is that the U-G mismatches created by deamination attract the MMR pathway and/or MutLγ, which could cleave in a nonstrand-specific manner to create DSBs (Fig. 6). This type of nondirected cleavage has been shown to occur at hairpin loops in an in vitro assay (53). We also note that deletion of Fcy1 does not completely suppress R-loop–induced CAG fragility. There could be other mechanisms responsible for these remaining breaks such as ribonucleotide incorporation because of lack of RNase H2 in the rnh1rnh201 mutants (54, 55) or transcription-replication collisions (56, 57).
MutLγ nicking contributes to about 50% of R-loop–induced CAG repeat breakage. This is another important finding from our study. A portion of the MutLγ-induced CAG repeat fragility may be due to MMR recognition and excision of dU:dG mismatches because rnh1rnh201mlh3 (or –mlh1) fcy1 quadruple mutants slightly decreased R-loop–induced fragility, although not to the same level of rnh1rnh201mlh3 (or –mlh1) triple mutants, suggesting that MutLγ is partially epistatic to Fcy1 in causing repeat fragility. This model predicts recognition by a MutS complex, and we did not observe a decrease in fragility in the rnh1rnh201msh2 strain background. However, msh2 deletion on its own causes a significant increase in CAG repeat fragility and instability, indicating an important role in maintaining repeat stability under normal conditions without the presence of R-loops. Thus, we may not have been able to detect the predicted reduction in the rate of FOAR.
However, a second possibility is that MutLγ functions primarily independently of the Msh2-MMR machinery. MutLα (Mlh1-Pms1) is the primary endonuclease complex functioning in the MMR pathway, and MutLγ (Mlh1-Mlh3) plays only a minor role (58, 59). In contrast, MutLγ has a major role in meiotic recombination during Holiday junction (HJ) resolution (60). Indeed, in vitro studies showed that MutLγ can bind to HJs and other heteroduplex structures independently of Msh2, although loading of the Msh2/Msh3 complex enhanced MutLγ nicking of supercoiled DNA (43, 61). R-loops can promote secondary structure formation on the nontemplate strand of CAG repeats, and once the R-loop is displaced, the excess unpaired DNA on the template strand folds into a hairpin to form a slipped-stranded structure (Fig. 6) (22). MutLγ could bind to these hairpin junctions, which are similar to HJs. Based on its inherent endonuclease activity in supercoiled DNA, MutLγ could create nicks at the hairpin junctions that are open and accessible; nicks on both strands could be converted into a DSB (Fig. 6). Alternatively, recent results show that the MutLγ endonuclease activity can be directed by a nick to cleave the opposite strand in a concerted mechanism to create a DSB (62). This mechanism could account for the partial dependence on Fcy1 and Apn1, that is, an initial nick created by processing of a deaminated cytosine could direct Mlh1-Mlh3 cleavage to the opposite strand to create a DSB. This same concerted mechanism can also occur without a preexisting nick, so another possibility is that recruitment to the DNA structure initiates MutLγ nondirectional cleavage in the vicinity (62). Our data have potentially significant implications for the Huntington’s disease (HD) process because MutLγ was reported to be important in driving somatic CAG repeat instability in a HD mouse model (63). In addition, MutLγ action could be a general mechanism for causing R-loop–dependent breaks at other structure-forming DNA sequences.
Materials and Methods
Yeast Strains.
Yeast strains used in this work are listed in SI Appendix, Table S7. Gene deletions were created by one-step gene replacement with KANMX6, TRP1, HIS3MX6 (64), or natNT2 (65). PCR was used to confirm the successful replacement of a gene by confirming the presence of the selectable marker and the absence of the gene at the targeted locus. Strains with mlh3-D523N and MLH3-WT genes were obtained from Dr. E. Alani and made as in ref. 42. Transformants were confirmed by PCR and sequencing. Fcy1-Myc was tagged with one Myc at its endogenous locus and verified by PCR and Western blot analysis. Strains containing terminator YACs were constructed by integrating plasmids pVS20-CAG70-Tcyc1-URA3 and pVS20-Ttef1-CAG70-Tcyc1-URA3 into a truncated YAC (strain CFY 1690). The successful clones were confirmed by PCR, Southern blot, and sequencing. CAG tract length was verified in all strains using colony PCR as described below.
Analysis of CAG Repeat Fragility and Instability.
The CAG tract was amplified from yeast colonies using primers listed in SI Appendix, Table S6, and Taq polymerase as described in ref. 29. Product sizes were analyzed by gel electrophoresis and Adobe Illustrator. The repeat size prominent in each individual colony was determined; any less intense bands representing instability occurring during colony growth were not counted. A total of 100–200 daughter colonies were sized from at least two transformants in at least three independent assays. To assay fragility, colonies growing on FOA-Leu and YC-Leu were counted, and the rate of 5-FOA resistance was calculated using the MSS-Maximum Likelihood Estimator Method with the online web tool Fluctuation AnaLysis CalculatOR (66).
Chromatin and DNA:RNA Immunoprecipitation.
ChIP of Myc-tagged proteins was done using anti-Myc antibody 9E10 in unsynchronized yeast cells as in ref. 44, except formaldehyde (1%) cross-linking was for 20 min at room temperature. qPCR amplification reactions were run in duplicate (primers, SI Appendix, Table S6), and at least two biological replicates were done for each condition. The DRIP procedure was performed in a similar manner except 4 μg of S9.6 antibody (Kerafast) was used to coat 40 μL of Protein G Dynabeads (Invitrogen), and cross-link reversal was done in the presence of RNase H and RNase A.
Analysis of Directional CAG Repeat Transcription by RT-PCR.
Yeast strains were grown to five to six divisions to OD600 0.7–1 before lysis. Total cell lysis and RNA extraction were carried out using the RNAspin Mini kit (GE Healthcare). RT-PCR was done with the SuperScript First-Strand Synthesis System RT-PCR kit (Invitrogen). Locus-specific primers (SI Appendix, Table S6) were used for the first-strand synthesis of transcripts. qPCR was done as in ref. 44.
Supplementary Material
Acknowledgments
We thank Eric Alani for sharing reagents, Alina Chan for sharing technical expertise on the DRIP protocol, and Assya Abourabia and Meaghan McGoldrick for help generating and testing rnh1 single mutants. Funding for this work was provided by National Science Foundation Award MCB1330743 (to C.H.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711283114/-/DCSupplemental.
References
- 1.Aguilera A, García-Muse T. R loops: From transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 2.Chan YA, et al. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet. 2014;10:e1004288. doi: 10.1371/journal.pgen.1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ginno PA, Lott PL, Christensen HC, Korf I, Chédin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El Hage A, Webb S, Kerr A, Tollervey D. Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genet. 2014;10:e1004716. doi: 10.1371/journal.pgen.1004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wahba L, Costantino L, Tan FJ, Zimmer A, Koshland D. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 2016;30:1327–1338. doi: 10.1101/gad.280834.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schrader CE, Guikema JE, Wu X, Stavnezer J. The roles of APE1, APE2, DNA polymerase beta and mismatch repair in creating S region DNA breaks during antibody class switch. Philos Trans R Soc Lond B Biol Sci. 2009;364:645–652. doi: 10.1098/rstb.2008.0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bregenhorn S, Kallenberger L, Artola-Borán M, Peña-Diaz J, Jiricny J. Non-canonical uracil processing in DNA gives rise to double-strand breaks and deletions: Relevance to class switch recombination. Nucleic Acids Res. 2016;44:2691–2705. doi: 10.1093/nar/gkv1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guikema JE, et al. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J Exp Med. 2007;204:3017–3026. doi: 10.1084/jem.20071289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rada C, Di Noia JM, Neuberger MS. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell. 2004;16:163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 11.Khair L, Baker RE, Linehan EK, Schrader CE, Stavnezer J. Nbs1 ChIP-seq identifies off-target DNA double-strand breaks induced by AID in activated splenic B cells. PLoS Genet. 2015;11:e1005438. doi: 10.1371/journal.pgen.1005438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knisbacher BA, Gerber D, Levanon EY. DNA editing by APOBECs: A genomic preserver and transformer. Trends Genet. 2016;32:16–28. doi: 10.1016/j.tig.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Sollier J, Cimprich KA. Breaking bad: R-loops and genome integrity. Trends Cell Biol. 2015;25:514–522. doi: 10.1016/j.tcb.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamperl S, Cimprich KA. The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Repair (Amst) 2014;19:84–94. doi: 10.1016/j.dnarep.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costantino L, Koshland D. The Yin and Yang of R-loop biology. Curr Opin Cell Biol. 2015;34:39–45. doi: 10.1016/j.ceb.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sollier J, et al. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell. 2014;56:777–785. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Domínguez-Sánchez MS, Barroso S, Gómez-González B, Luna R, Aguilera A. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet. 2011;7:e1002386. doi: 10.1371/journal.pgen.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wahba L, Amon JD, Koshland D, Vuica-Ross M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell. 2011;44:978–988. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerritelli SM, Crouch RJ. Ribonuclease H: The enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin Y, Dent SY, Wilson JH, Wells RD, Napierala M. R loops stimulate genetic instability of CTG.CAG repeats. Proc Natl Acad Sci USA. 2010;107:692–697. doi: 10.1073/pnas.0909740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reddy K, et al. Processing of double-R-loops in (CAG)·(CTG) and C9orf72 (GGGGCC)·(GGCCCC) repeats causes instability. Nucleic Acids Res. 2014;42:10473–10487. doi: 10.1093/nar/gku658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddy K, et al. Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res. 2011;39:1749–1762. doi: 10.1093/nar/gkq935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin Y, Wilson JH. Nucleotide excision repair, mismatch repair, and R-loops modulate convergent transcription-induced cell death and repeat instability. PLoS One. 2012;7:e46807. doi: 10.1371/journal.pone.0046807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Usdin K, House NC, Freudenreich CH. Repeat instability during DNA repair: Insights from model systems. Crit Rev Biochem Mol Biol. 2015;50:142–167. doi: 10.3109/10409238.2014.999192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freudenreich CH, Kantrow SM, Zakian VA. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 27.Callahan JL, Andrews KJ, Zakian VA, Freudenreich CH. Mutations in yeast replication proteins that increase CAG/CTG expansions also increase repeat fragility. Mol Cell Biol. 2003;23:7849–7860. doi: 10.1128/MCB.23.21.7849-7860.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casella M, et al. Spontaneous chromosome loss and colcemid resistance in lymphocytes from patients with myotonic dystrophy type 1. Cytogenet Genome Res. 2003;100:224–229. doi: 10.1159/000072858. [DOI] [PubMed] [Google Scholar]
- 29.Sundararajan R, Gellon L, Zunder RM, Freudenreich CH. Double-strand break repair pathways protect against CAG/CTG repeat expansions, contractions and repeat-mediated chromosomal fragility in Saccharomyces cerevisiae. Genetics. 2010;184:65–77. doi: 10.1534/genetics.109.111039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polleys EJ, House NCM, Freudenreich CH. Role of recombination and replication fork restart in repeat instability. DNA Repair (Amst) 2017;56:156–165. doi: 10.1016/j.dnarep.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein HL. Genome instabilities arising from ribonucleotides in DNA. DNA Repair (Amst) 2017;56:26–32. doi: 10.1016/j.dnarep.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz JF, Gómez-González B, Aguilera A. AID induces double-strand breaks at immunoglobulin switch regions and c-MYC causing chromosomal translocations in yeast THO mutants. PLoS Genet. 2011;7:e1002009. doi: 10.1371/journal.pgen.1002009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim N, Jinks-Robertson S. Guanine repeat-containing sequences confer transcription-dependent instability in an orientation-specific manner in yeast. DNA Repair (Amst) 2011;10:953–960. doi: 10.1016/j.dnarep.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gómez-González B, Aguilera A. Activation-induced cytidine deaminase action is strongly stimulated by mutations of the THO complex. Proc Natl Acad Sci USA. 2007;104:8409–8414. doi: 10.1073/pnas.0702836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erbs P, Exinger F, Jund R. Characterization of the Saccharomyces cerevisiae FCY1 gene encoding cytosine deaminase and its homologue FCA1 of Candida albicans. Curr Genet. 1997;31:1–6. doi: 10.1007/s002940050169. [DOI] [PubMed] [Google Scholar]
- 36.Hartzog PE, Nicholson BP, McCusker JH. Cytosine deaminase MX cassettes as positive/negative selectable markers in Saccharomyces cerevisiae. Yeast. 2005;22:789–798. doi: 10.1002/yea.1245. [DOI] [PubMed] [Google Scholar]
- 37.Kurtz JE, Exinger F, Erbs P, Jund R. New insights into the pyrimidine salvage pathway of Saccharomyces cerevisiae: Requirement of six genes for cytidine metabolism. Curr Genet. 1999;36:130–136. doi: 10.1007/s002940050482. [DOI] [PubMed] [Google Scholar]
- 38.Percival KJ, Klein MB, Burgers PM. Molecular cloning and primary structure of the uracil-DNA-glycosylase gene from Saccharomyces cerevisiae. J Biol Chem. 1989;264:2593–2598. [PubMed] [Google Scholar]
- 39.Popoff SC, Spira AI, Johnson AW, Demple B. Yeast structural gene (APN1) for the major apurinic endonuclease: Homology to Escherichia coli endonuclease IV. Proc Natl Acad Sci USA. 1990;87:4193–4197. doi: 10.1073/pnas.87.11.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol Cell Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin Y, Dion V, Wilson JH. Transcription promotes contraction of CAG repeat tracts in human cells. Nat Struct Mol Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 42.Nishant KT, Plys AJ, Alani E. A mutation in the putative MLH3 endonuclease domain confers a defect in both mismatch repair and meiosis in Saccharomyces cerevisiae. Genetics. 2008;179:747–755. doi: 10.1534/genetics.108.086645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rogacheva MV, et al. Mlh1-Mlh3, a meiotic crossover and DNA mismatch repair factor, is a Msh2-Msh3-stimulated endonuclease. J Biol Chem. 2014;289:5664–5673. doi: 10.1074/jbc.M113.534644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su XA, Dion V, Gasser SM, Freudenreich CH. Regulation of recombination at yeast nuclear pores controls repair and triplet repeat stability. Genes Dev. 2015;29:1006–1017. doi: 10.1101/gad.256404.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 46.Krejci L, Altmannova V, Spirek M, Zhao X. Homologous recombination and its regulation. Nucleic Acids Res. 2012;40:5795–5818. doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ireton GC, Black ME, Stoddard BL. The 1.14 A crystal structure of yeast cytosine deaminase: Evolution of nucleotide salvage enzymes and implications for genetic chemotherapy. Structure. 2003;11:961–972. doi: 10.1016/s0969-2126(03)00153-9. [DOI] [PubMed] [Google Scholar]
- 48.Ko TP, et al. Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution. J Biol Chem. 2003;278:19111–19117. doi: 10.1074/jbc.M300874200. [DOI] [PubMed] [Google Scholar]
- 49.Sun X, et al. Complementary functions of the Saccharomyces cerevisiae Rad2 family nucleases in Okazaki fragment maturation, mutation avoidance, and chromosome stability. DNA Repair (Amst) 2003;2:925–940. doi: 10.1016/s1568-7864(03)00093-4. [DOI] [PubMed] [Google Scholar]
- 50.Habraken Y, Sung P, Prakash L, Prakash S. A conserved 5′ to 3′ exonuclease activity in the yeast and human nucleotide excision repair proteins RAD2 and XPG. J Biol Chem. 1994;269:31342–31345. [PubMed] [Google Scholar]
- 51.Pourquier P, et al. Effects of uracil incorporation, DNA mismatches, and abasic sites on cleavage and religation activities of mammalian topoisomerase I. J Biol Chem. 1997;272:7792–7796. doi: 10.1074/jbc.272.12.7792. [DOI] [PubMed] [Google Scholar]
- 52.Huang SN, Williams JS, Arana ME, Kunkel TA, Pommier Y. Topoisomerase I-mediated cleavage at unrepaired ribonucleotides generates DNA double-strand breaks. EMBO J. 2017;36:361–373. doi: 10.15252/embj.201592426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pluciennik A, et al. Extrahelical (CAG)/(CTG) triplet repeat elements support proliferating cell nuclear antigen loading and MutLα endonuclease activation. Proc Natl Acad Sci USA. 2013;110:12277–12282. doi: 10.1073/pnas.1311325110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim N, et al. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jinks-Robertson S, Klein HL. Ribonucleotides in DNA: Hidden in plain sight. Nat Struct Mol Biol. 2015;22:176–178. doi: 10.1038/nsmb.2981. [DOI] [PubMed] [Google Scholar]
- 56.Hamperl S, Cimprich KA. Conflict resolution in the genome: How transcription and replication make it work. Cell. 2016;167:1455–1467. doi: 10.1016/j.cell.2016.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.García-Muse T, Aguilera A. Transcription-replication conflicts: How they occur and how they are resolved. Nat Rev Mol Cell Biol. 2016;17:553–563. doi: 10.1038/nrm.2016.88. [DOI] [PubMed] [Google Scholar]
- 58.Romanova NV, Crouse GF. Different roles of eukaryotic MutS and MutL complexes in repair of small insertion and deletion loops in yeast. PLoS Genet. 2013;9:e1003920. doi: 10.1371/journal.pgen.1003920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flores-Rozas H, Kolodner RD. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc Natl Acad Sci USA. 1998;95:12404–12409. doi: 10.1073/pnas.95.21.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manhart CM, Alani E. Roles for mismatch repair family proteins in promoting meiotic crossing over. DNA Repair (Amst) 2016;38:84–93. doi: 10.1016/j.dnarep.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ranjha L, Anand R, Cejka P. The Saccharomyces cerevisiae Mlh1-Mlh3 heterodimer is an endonuclease that preferentially binds to Holliday junctions. J Biol Chem. 2014;289:5674–5686. doi: 10.1074/jbc.M113.533810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manhart CM, et al. The mismatch repair and meiotic recombination endonuclease Mlh1-Mlh3 is activated by polymer formation and can cleave DNA substrates in trans. PLoS Biol. 2017;15:e2001164. doi: 10.1371/journal.pbio.2001164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pinto RM, et al. Mismatch repair genes Mlh1 and Mlh3 modify CAG instability in Huntington’s disease mice: Genome-wide and candidate approaches. PLoS Genet. 2013;9:e1003930. doi: 10.1371/journal.pgen.1003930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 65.Janke C, et al. A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 66.Hall BM, Ma CX, Liang P, Singh KK. Fluctuation analysis CalculatOR: A web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics. 2009;25:1564–1565. doi: 10.1093/bioinformatics/btp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.