Transcription profiles generated from maize gametes and zygotes at different stages reveal a highly dynamic zygotic genome activation pattern, providing insights into early embryo development.
Abstract
The formation of a zygote via the fusion of an egg and sperm cell and its subsequent asymmetric division herald the start of the plant’s life cycle. Zygotic genome activation (ZGA) is thought to occur gradually, with the initial steps of zygote and embryo development being primarily maternally controlled, and subsequent steps being governed by the zygotic genome. Here, using maize (Zea mays) as a model plant system, we determined the timing of zygote development and generated RNA-seq transcriptome profiles of gametes, zygotes, and apical and basal daughter cells. ZGA occurs shortly after fertilization and involves ∼10% of the genome being activated in a highly dynamic pattern. In particular, genes encoding transcriptional regulators of various families are activated shortly after fertilization. Further analyses suggested that chromatin assembly is strongly modified after fertilization, that the egg cell is primed to activate the translational machinery, and that hormones likely play a minor role in the initial steps of early embryo development in maize. Our findings provide important insights into gamete and zygote activity in plants, and our RNA-seq transcriptome profiles represent a comprehensive, unique RNA-seq data set that can be used by the research community.
INTRODUCTION
The life cycles of animals and plants begin with the formation of a zygote, containing the cytoplasm from two gametes, a large egg cell and a small sperm cell. This single cell then develops via embryogenesis into an entire organism consisting of hundreds of different cell types. In contrast to most animal systems, the flowering plant zygote divides asymmetrically into daughter cells of completely different cell fates. While the small, cytoplasm-rich apical daughter cell further develops into the embryo proper, the highly vacuolated basal cell gives rise to the suspensor, which delivers nutrients to the embryo and positions the embryo proper in the surrounding endosperm tissue of the developing seed. Little is known about the establishment of zygote polarity and the gene regulatory network that leads to asymmetric cell division and cell fate determination in both daughter cells (Zhao et al., 2017). Due to the unequal distribution of their cytoplasm, it is generally thought that maternal factors contributed by the egg regulate zygote and early embryo development. The maternal-to-zygotic transition (MZT) depends on both zygotic genome activation (ZGA) and the degradation of maternal components. In animals, ZGA occurs after the first cell cycle in mammals and as late as the sixth to eighth round of the cell cycle in insects, fish, and amphibians (Schier, 2007; Lee et al., 2014). The time point at which ZGA occurs in plants has long been debated. Currently, the zygote is thought to be in a relatively quiescent transcriptional state, and ZGA is thought to occur gradually rather than as an all-or-none process initiated before the first division (Baroux and Grossniklaus, 2015; Zhao and Sun, 2015). However, analyses of a few candidate genes have indicated that ZGA in flowering plants occurs before zygotic division (Zhao et al., 2017).
To characterize the onset of ZGA at the whole-genome level, it is necessary to determine the transcriptome profiles of both gametes and to identify de novo-generated transcripts from the zygotic genome. In this study, we established methods to manually isolate living male and female gametes, zygotes at different stages, and their daughter cells using maize (Zea mays) as a model flowering plant. We then generated RNA-seq data from these cells and investigated the transcriptome dynamics after fertilization. We compared the transcriptomes of maize and rice (Oryza sativa) gametes and explored how the cell cycle, chromatin, and auxin pathways are regulated after fertilization. Finally, we identified transcription factor (TF) and receptor-like kinase genes associated with the various cell types and zygotic stages and found that ZGA in maize occurs shortly after fertilization, displaying a highly dynamic pattern.
RESULTS AND DISCUSSION
Manual Cell Isolation, RNA-Seq, Validation, and Data Quality
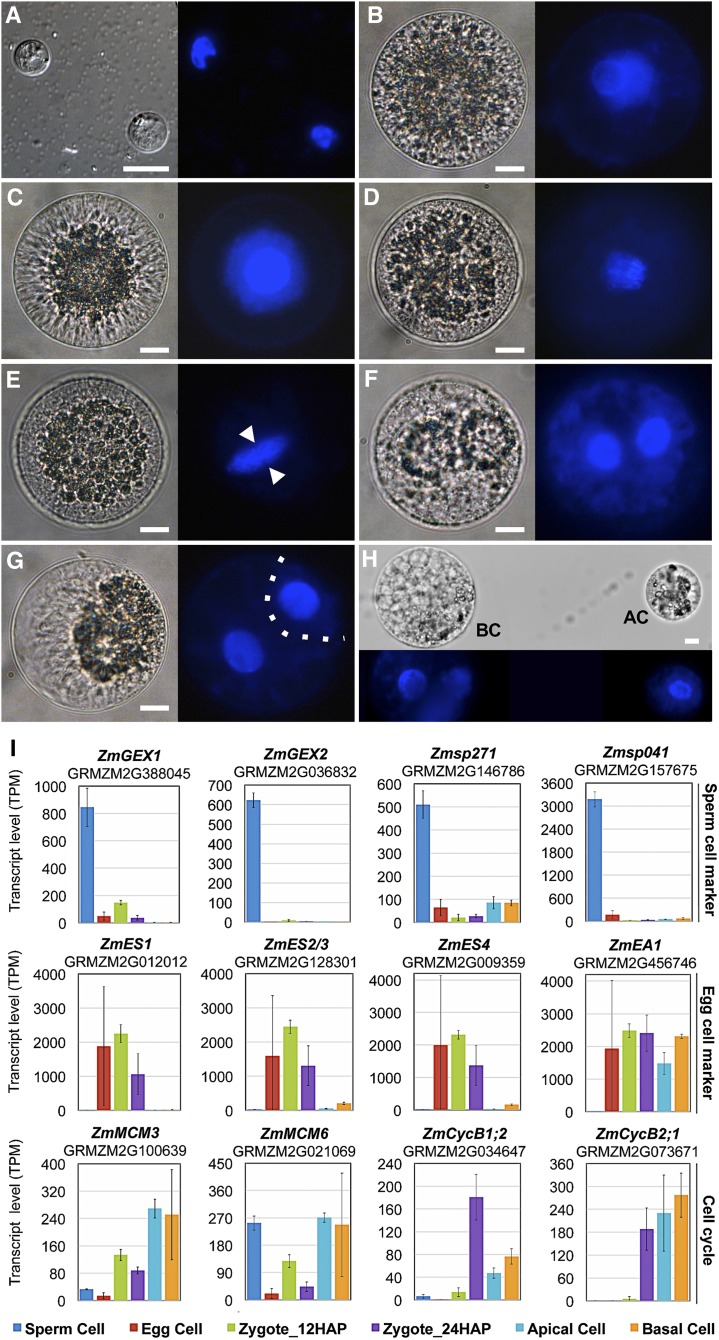
Maize plants were grown in indoor walk-in growth rooms to reduce seasonal variability in zygote development. We manually isolated cells of the inbred line B73 at 2-h intervals over a 2.5-d period as described in Methods. We used 4′,6-diamidino-2-phenylindole (DAPI) staining as a gross indicator to investigate cell cycle stages. As shown in Figure 1A, sperm cells consist mainly of the nucleus, containing highly condensed chromatin. Egg cell chromatin appeared less condensed than sperm cell chromatin and stained very weakly with DAPI. The strongest DAPI staining was detected in zygotes at 24 h after pollination (HAP), indicating that S-phase was complete. Early anaphase occurred at ∼30 HAP, and telophase began at 35 HAP. Cytokinesis was visible at 43 HAP, and asymmetric cell division (ACD) was completed by 48 HAP. The small cytoplasm-rich apical and large vacuolated basal cells could be manually separated at ∼52 HAP.
Figure 1.
Time Course of Zygote Development and Validation of RNA-Seq Data.
(A) Sperm cells.
(B) Egg cell.
(C) Zygote at 24 HAP.
(D) Zygote at 30 HAP at anaphase.
(E) Zygote at 35 HAP at telophase. Arrowheads indicate phragmoplast between daughter chromosome sets.
(F) Zygote at 43 HAP during ACD.
(G) Zygote at 48 HAP. Cytokinesis is completed. Dotted line indicates the cell division plane.
(H) Apical (AC) and basal cell (BC) have been separated at 54 HAP. Bright-field microscopy images are shown on the left and UV images of DAPI-stained cells on the right. Bars = 10 μm.
(I) Validation of RNA-seq data with known genes, as indicated. Top row: Genes preferentially expressed in sperm cells. Middle row: Genes highly expressed in egg cells and downregulated after fertilization. Bottom row: Expression of cell cycle regulators. Transcript levels are shown as TPM values (means ± sd) of three biological replicates.
The protocol used to generate RNA-seq data from only a few plant cells is described in Methods. Three biological replicates were prepared from ∼1000 sperm cells each and 13 to 20 cells each for the other cell types (Table 1; see also Supplemental Table 1 and Supplemental Figures 1 and 2 for samples, sequencing details, and library quality). A gene expression master list, containing the median expression values of each cell type for the 39,469 annotated maize genes, protein annotations, and homologs from Arabidopsis thaliana and rice is provided in Supplemental Data Set 1.
Table 1. Summary of Samples, NGS Runs, Alignment to the Ensembl Genome (AGPv3, Version 82.6), and Annotation to Ensembl Genebuild (AGPv3_5b, Version 82.6).
| Sample | Cell No. per Replicate | All Reads after Trimming | Aligned Pairs Mapped Unique | % Mapped Reads per Trimmed Reads | Expressed Genes (TPM > 1) |
|---|---|---|---|---|---|
| Sperm Cell1 | ∼1,000 | 68,988,450 | 26,514,492 | 77 | 11,819 |
| Sperm Cell2 | ∼1,000 | 102,929,736 | 38,689,716 | 75 | |
| Sperm Cell3 | ∼1,000 | 86,448,904 | 33,271,851 | 77 | |
| Egg Cell1 | 20 | 57,338,078 | 13,901,359 | 48 | 16,026 |
| Egg Cell2 | 20 | 63,488,722 | 22,978,462 | 72 | |
| Egg Cell3 | 20 | 69,199,124 | 25,147,141 | 73 | |
| Zygote_12HAP1 | 14 | 70,632,412 | 15,761,484 | 45 | 19,865 |
| Zygote_12HAP2 | 15 | 76,840,694 | 19,297,245 | 50 | |
| Zygote_12HAP3 | 15 | 55,130,854 | 11,323,005 | 41 | |
| Zygote_24HAP1 | 16 | 82,825,752 | 20,424,191 | 49 | 19,171 |
| Zygote_24HAP2 | 16 | 80,911,418 | 20,173,448 | 50 | |
| Zygote_24HAP3 | 17 | 87,134,202 | 19,443,455 | 45 | |
| Apical Cell1 | 16 | 94,779,064 | 29,713,124 | 63 | 17,747 |
| Apical Cell2 | 16 | 68,636,668 | 20,927,946 | 61 | |
| Apical Cell3 | 16 | 74,074,566 | 23,422,549 | 63 | |
| Basal Cell1 | 13 | 71,456,410 | 22,440,462 | 63 | 18,069 |
| Basal Cell2 | 13 | 68,963,988 | 21,238,495 | 62 | |
| Basal Cell3 | 14 | 67,950,552 | 20,886,111 | 61 |
The number of genes expressed in at least two of three replicates are given. See Supplemental Table 1 for details.
For validation and initial analysis of the dynamics of gene expression patterns obtained from the RNA-seq data, we focused on the transcript levels of 12 genes that were previously shown (by single-cell RT-PCR) to be highly or differentially expressed during fertilization and zygote development in maize (Engel et al., 2003). In agreement with previous results (Engel et al., 2003), the gamete-expressed membrane protein genes ZmGEX1 (SP versus all: log2FC > 4.0*; asterisk indicates an adjusted P value below 0.05) and ZmGEX2 (SP versus all: log2FC > 7.4*) were highly and specifically expressed in sperm cells, as were Zmsp271 (SP versus all: log2FC > 3.8*) and Zmsp041 (SP versus all: log2FC > 5.2*), which were identified in the same screen (Figure 1I; Supplemental Data Sets 1 to 3). ZmEA1 (EC versus SP: log2FC = 8.7*) and ZmES1-4 (EC versus AC/BC: log2FC > 2.9 to 9.7*, Zy24 versus AC/BC: log2FC > 2.4 to 8.7*), encoding secreted peptides required for micropylar pollen tube guidance and pollen tube burst, respectively, were highly expressed in egg cells and synergids and were significantly downregulated after fertilization (Cordts et al., 2001; Márton et al., 2005; Amien et al., 2010) (Supplemental Data Set 3). The cell cycle genes ZmMCM3, ZmMCM6, ZmCycB1;2, and ZmCycB2;1 were previously shown to be induced after fertilization (Sauter et al., 1998; Dresselhaus et al., 1999b, 2006). Expression of ZmMCM3 (Zy12 versus EC: = 2.7*, AC versus Zy24: log2FC = 1.8*) and ZmMCM6 (Zy12 versus EC: log2FC = 2.0*, AC versus Zy24: log2FC = 2.7*), marking the onset of DNA replication during S-phase (Maiorano et al., 2006), peaked in the zygote at 12 HAP, as well as after the first asymmetric zygote division in the apical cell, which divides more rapidly than the basal cell. The cell cycle regulatory genes ZmCycB2;1 (Zy24 versus Zy12 log2FC = 3.6*) and ZmCycB1;2 (Zy24 versus Zy12 log2FC = 5.0*), which mark the G2/M-transition (Maiorano et al., 2006), were strongly induced at 24 HAP. In contrast to ZmCycB1;2 (AC/BC versus Zy12 log2FC > 1.9*), the expression levels of ZmCycB2;1 (AC/BC versus Zy12 log2FC > 5.5*) were also high in apical and basal cells after zygote division (Sauter et al., 1998). In summary, these dynamic changes in gene expression (Figure 1B) are in perfect agreement with previous reports, which together with strong correlation between biological replicates (Supplemental Figure 2) assures the high quality and reliability of our data.
Contamination of transcriptomes by RNA from maternal tissues has recently been discussed as a serious issue that can result in poor reproducibility and misinterpretation of data sets (Schon and Nodine, 2017). We therefore investigated the presence of transcripts derived from genes expressed in maternal nucellus tissue surrounding embryo sacs (Chettoor et al., 2014) to evaluate the possibility of contamination. None of the nucellus-expressed genes, including GRMZM2G570791 (α-subunit of DNA-directed RNA polymerase), GRMZM2G125823 (heparanase-like protein), GRMZM2G099420 (cinnamoyl CoA reductase), and GRMZM5G803276 and GRMZM2G336859 (encoding unknown proteins), were detected in any of our data sets. These results indicate that our data sets are free of maternal RNA contamination and that the two washing steps were sufficient for removing maternal RNA from the burst maternal nucellus cells.
Comparison of Transcriptomic Data from Maize and Rice Gametes
A comprehensive comparison of gene expression activity after fertilization has not been reported yet for any plant species, and this study thus represents the first report of global gene expression patterns in gametes, zygotes, and daughter cells. Therefore, we restricted our comparisons to the transcriptomes of maize and rice gametes (egg and sperm cells). It was not possible to include the transcriptomes of Arabidopsis gametes in the comparison, as RNA-seq data were not available, and the available microarray data (Borges et al., 2008; Wuest et al., 2010) could not be accurately normalized to allow us to draw conclusions and lacked information for thousands of genes. In addition, each gamete in the data set was measured in a different experiment.
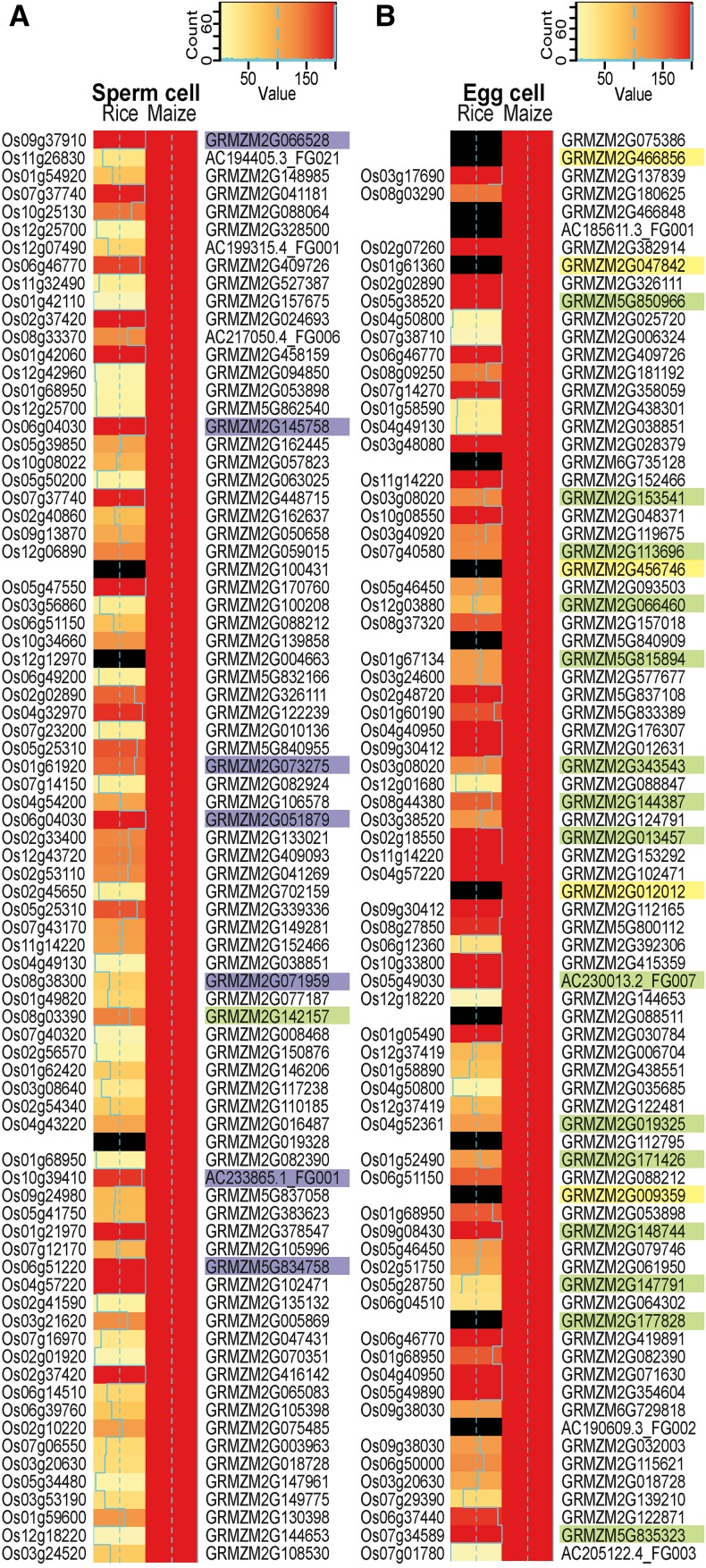
We used published RNA-seq data from rice sperm and egg cells (Anderson et al., 2013) and initially identified the rice homologs using public databases, i.e., EnsemblPlants and RiceAnnotationGenomeProject, which combine data from many species to identify putative orthologs. If the identity of the homologs/orthologs was unclear or unknown due to a lack of sequence information, we did not include them in the comparison. To compare transcription patterns in rice versus maize gametes, the gene expression values were binned into 200 expression level categories using the 99th percentile per species as the highest category (see also Supplemental Data Set 4). We selected the 80 most strongly expressed genes (TOP80 genes) in maize sperm and egg cells and compared their expression levels with those of the respective genes in rice (Figure 2). A summary of the TOP30 genes of all maize cell types, including their annotation, is provided in Supplemental Data Set 5.
Figure 2.
Heat Map Showing a Comparison of the 80 Most Highly Expressed Genes in Maize Gametes and Their Predicted Orthologs in the Corresponding Rice Gametes.
TOP80 most highly expressed genes in maize sperm cells (A) and TOP80 genes in maize egg cells (B). Rice and maize gene expression values (TPM) were square root transformed and classified into 200 expression bins using the 99th percentile of the data as the maximum value. Note that all maize genes shown are in the bin with maximum expression and are therefore represented by red bars. Plastid genomes were excluded, as they showed overshooting of expression in the rice data. Nonexpressed genes in the rice data and genes lacking a clear homolog are marked by black bars. Orthologous gene information is based on the EnsemblPlants Compara database, The Rice Genome Annotation Project (RGAP), and orthologs from RGAP based on OrthoMCL. Maize genes encoding histones and high-mobility group genes are shaded in purple, proteins involved in translation are in green, and EA1-box proteins and predicted secreted CRPs are in yellow. Proteins were classified according to InterPro.
Many of the predicted rice homologs/orthologs displayed similar, strong expression patterns. The proportion of genes with high expression levels was greater in egg cells than in sperm cells (Figure 2). The observation that many predicted rice homologs/orthologs displayed weaker expression patterns in rice than in maize might be due to our methods (as we summarized rice ortholog data using the median), the difficulty in predicting true orthologs within larger gene families, and/or the lack of common controls to normalize these two studies. Tightly controlled parallel RNA extraction from cells of different species and the identification of an appropriate control cell type common to both plant species may improve interspecies comparisons. However, our comparison pointed to some similarities and general findings regarding genome activity. Among the TOP80 genes expressed in maize sperm cells, 10% encode histones and high-mobility group (HMG) proteins (Figure 2A). This finding might explain the strongly condensed, compact sperm cell chromatin (see also Figure 1A). Indeed, a similar observation was reported for rice sperm cells (Russell et al., 2012; Anderson et al., 2013). Notably, no chromatin gene is among the TOP80 genes in maize egg cells. This finding correlates well with the less condensed chromatin in these cells and the difficulty in staining egg cells with DAPI (Figure 1A). However, in strong contrast to sperm cells, 20% of the TOP80 genes in maize egg cells encode proteins of the translational machinery, most of which also displayed similar expression patterns in rice egg cells. These include two genes encoding the translation initiation factor IF5A (Dresselhaus et al., 1999a), a gene encoding the translation initiation factor SUI1 (Cui et al., 1998), two genes encoding the translation elongation factor EF1A (Budkevich et al., 2002), and many ribosomal protein genes. These observations indicate that sperm cells are translationally inactive, whereas egg cells are either highly active or well prepared to strongly enhance translation after being activated during the fertilization process.
Another interesting observation relates to transcripts encoding polymorphic EA1-box proteins and small secreted cysteine-rich proteins, which we found in maize egg cells, but not in sperm cells. The corresponding proteins play key roles in female gamete cell identity, as well as pollen tube guidance and burst in maize (reviewed in Dresselhaus et al., 2011, 2016). However, due to their polymorphic nature, no unambiguous homologs of the individual members were identified in the databases from Ensembl Compara or the Rice Genome Annotation Project. Through manual searches, we identified similar genes in rice (Márton et al., 2005; Uebler et al., 2015), but true orthologs could not be predicted based on protein comparisons, and none of the candidate genes have been functionally tested in rice (Figure 2B). However, this finding indicates that genes involved in processes directly associated with fertilization appear to be polymorphic and species-specific, thus representing prime candidate genes involved in speciation mechanisms (Rieseberg and Willis, 2007).
Histone Variants and Chromatin-Based Gene Regulation
Histones are the major protein components of chromatin and, more importantly, dynamic regulators of transcription. To begin to uncover the molecular basis of chromatin remodeling and epigenetic reprogramming in plant gametes and the onset of embryo development, we investigated the expression patterns of histone variants and chromatin assembly factor genes in more detail (Figure 3; Supplemental Data Set 3G; important gene families).
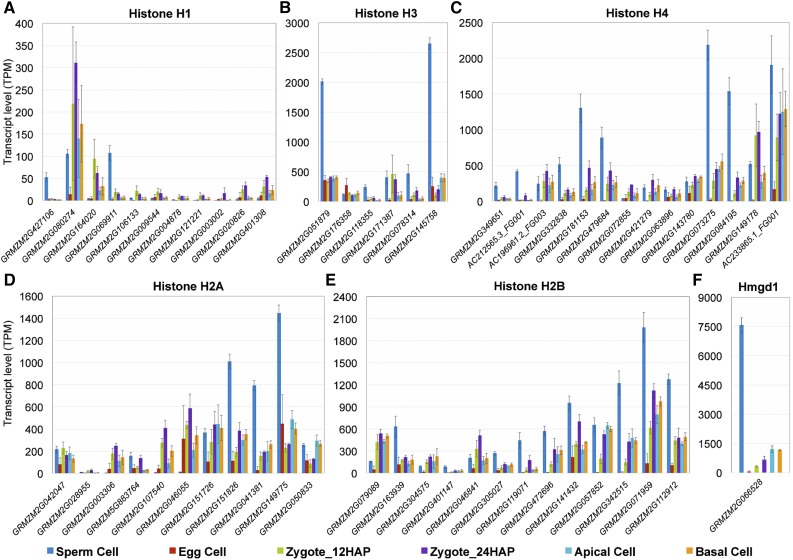
Figure 3.
Expression of Major Chromatin Structure Protein Genes in Maize Gametes, Zygotes, and Daughter Cells.
Histone H1 (A), histone H3 (B), histone H4 (C), histone H2A (D), histone H2B (E), and HMG protein Hmgd1 (F). All genes of the various families differentially expressed in at least one comparison are presented. See Supplemental Figure 3 for phylogenetic analysis of male-specific and active chromatin-specific variants of histones H3 and H2A. Gene identifiers are listed in Supplemental Data Set 7 (important gene families). Transcript levels in the cells indicated are given as TPM values (means ± sd).
As mentioned above, some canonical core histones, including two H3 (7 of 17 genes: SP versus all: log2FC > 1.5*) genes and at least three H4 (9 of 15 genes: SP versus all: log2FC > 1.4*) genes were predominantly and highly expressed in sperm cells (Figures 3B and 3C) and may contribute to the compactness of the chromatin and suggesting that sperm cells are already prepared for S-phase. Moreover, the most highly expressed gene in sperm, ZmHmgd1 (SP versus all: log2FC > 3.9*), was expressed at much higher levels in sperm cells than in any other cell type examined (Figure 3F). ZmHmgd1 encodes an HMG box protein that plays a role in chromosome condensation (Thomas and Travers, 2001) and is thought to possess a role similar to that of histone H1 in chromatin assembly, as both proteins bend linker DNA at the entry and exit points of the nucleosome. This hypothesis may explain why linker histone H1 genes are expressed at rather low levels in sperm cells, although their DNA is densely packed to ensure chromatin stability during sperm delivery. After fertilization, Hmgd1 appeared to be partially replaced by histone H1, as the expression of the H1 gene was strongly activated and peaked in 24 HAP zygotes (9 of 11 genes: Zy24 versus EC: log2FC > 1.8*) (Figure 3A), whereas ZmHmgd1 (Zy24 versus SP: log2FC = −5.0*) was strongly repressed.
The most abundant H3 gene in maize sperm (GRMZM2G145758, SP versus all: log2FC > 4.0*) encodes an unusual replication-independent (RI) H3.3-like variant that was also predominantly expressed in anthers and was previously designated as ZmAPH3. Phylogenetic analysis showed that ZmAPH3 is most similar to the Arabidopsis male gamete-specific histone H3 variant gene AtMGH3 (Okada et al., 2005) (Supplemental Figure 3A). However, the horizontal distance and the low number of histone variants and species included in the phylogenetic analysis suggest that they share a rather distant relationship. All other sperm cell-enriched H3 variants belong to the RI H3.3 group (Supplemental Figure 3A) (four orthologous genes, RI H3.3: SP versus Zy24: log2FC > 1.6*). In Arabidopsis, a limited subset of H3.3 variants referred to as the main HTR (HISTONE THREE-RELATED) protein genes are predominately expressed in the male gamete (Ingouff et al., 2010). Three AtMGH3 homologs and two other RI H3.3 variants are also highly expressed in rice sperm cells, while other core H3 genes exhibit only limited expression in these cells (Russell et al., 2012). These findings suggest that RI H3.3 variants encoded by only a few genes act as major histone H3s in the sperm cells of flowering plants.
Two replication-coupled canonical H2As (GRMZM2G151826 and GRMZM2G041381) exhibited high transcript levels in maize sperm cells, but the most highly expressed H2A (GRMZM2G149775, SP log2FC all: log2FC > 2.7) belongs to the RI H2A.Z class (Supplemental Figure 3B), which is associated with nucleosomes at transcription start sites, especially those also containing H3.3 (Deal and Henikoff, 2011). Therefore, together with histone H3.3 and H2A.Z, which mark active chromatin via RI nucleosome assembly (Ahmad and Henikoff, 2002; Deal and Henikoff, 2011), HMGD1 (encoded by ZmHmgd1), which likely represents the most abundant chromatin architectural protein in sperm that binds to highly accessible regulatory chromatin and active promoters, probably keeps the highly condensed sperm cell chromatin at least partially accessible. This hypothesis is supported by our RNA-seq data set, which includes transcripts of ∼11,000 differentially upregulated genes in sperm cells versus zygotes at 12 HAP (Figure 4F), including a subset of TF genes, despite the compact chromatin in sperm cells.
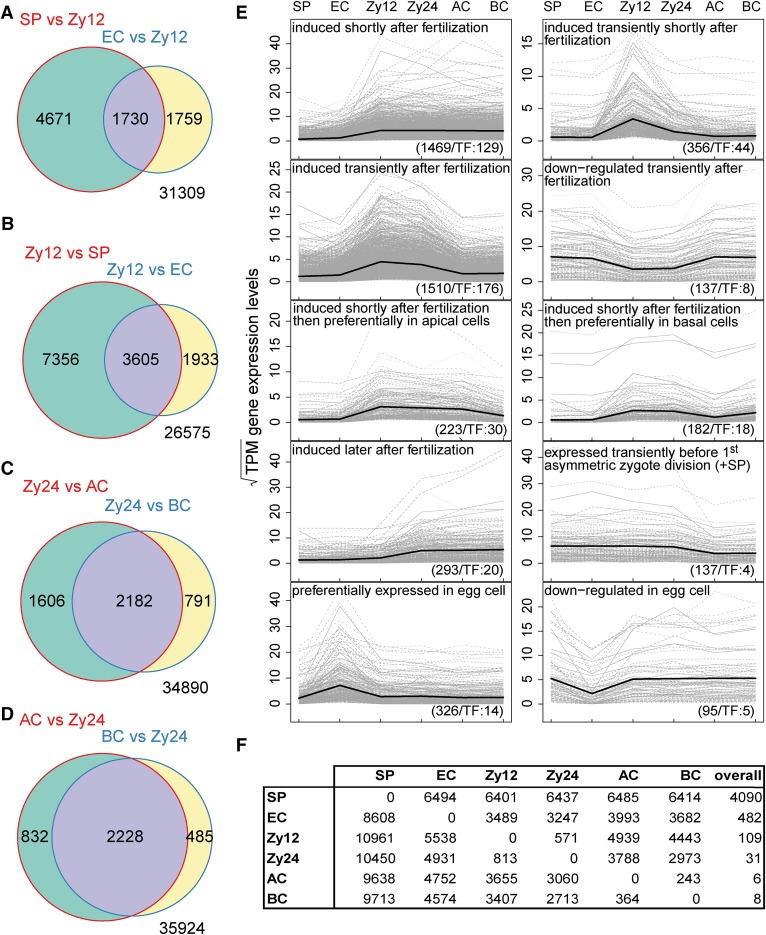
Figure 4.
Gene Expression Dynamics in Maize Gametes, Zygotes, and Two-Celled Pro-Embryo Cells.
Genes with an absolute log2FC > 1 and P-adjusted < 0.05 were considered to be biologically meaningful.
(A) Comparison of the number of genes induced in sperm cells (SP) and egg cells (EC) versus zygotes at 12 HAP (Zy12).
(B) Comparisons of genes upregulated in zygotes at 12 HAP versus sperm and egg cells.
(C) Number of genes induced in zygotes at 24 HAP compared with apical (AC) and basal (BC) cells.
(D) Comparison of genes upregulated in apical and basal cells versus zygotes at 24 HAP (Zy24).
(E) Selected gene expression profiles across the specific cell types analyzed (Supplemental Data Set 6). The Pearson correlation (>0.9) of square root transformed TPM values per gene and binary profile vectors were used to identify genes belonging to a specific profile. The total number of genes including TFs contained in each profile is shown at the bottom. The mean expression level of all genes per specific cell type is plotted in each profile (black line).
(F) Table of differentially upregulated genes (log2FC > 1, P-adjusted<0.05) of row cell type versus column cell type. The last column shows the number of differentially upregulated genes in a row cell type versus all other cell types.
Upon fertilization, H3 variants from male and female gametes are actively removed in Arabidopsis (Ingouff et al., 2010), a scenario that likely also occurs in maize, as the expression of the canonical core histone repertoire was activated in conjunction with a dramatic reduction in H2A.Z, H3.3, and Hmgd1 expression and a strong increase in H1 expression (Figure 3A). Thus, it appears that the paternal chromatin was reprogrammed by newly synthesized and entirely different sets of histones in the zygote. This theme may even extend to apical and basal cells, since the list of expressed orthologs of H3 and H4 again shifted to genes different from those expressed in zygotes.
Finally, while the canonical core histones were mostly expressed at very low levels in egg cells (Figure 3), they were induced in zygotes at 12 HAP, indicating the G1-phase of the cell cycle. Expression peaked in zygotes at 24 HAP, suggesting that DNA replication was almost complete (Figure 3). These observations support the hypothesis that the egg cell is arrested and requires activation after gamete fusion. Thus, chromatin-based transcriptional reprogramming in the zygote may represent a key step in MZT and the initiation of the sporophytic generation.
ZGA Occurs Shortly after Fertilization in Maize
To identify the global onset of ZGA, it is important to examine the transcriptomes of both gametes, i.e., egg and sperm cells, as well as zygotes at different stages. We therefore investigated zygotes collected at 12 and 24 HAP. At 12 HAP, i.e., only ∼4 h after fertilization, all egg cells were fertilized, as indicated by the presence of degenerated receptive synergid cells. These egg cells were considered to represent early zygotes. We chose to investigate zygotes at 24 HAP, as they appeared to be activated at this point, as indicated by the duplication of DNA content (Figure 1A). At later time points after pollination, the zygotes underwent mitosis, but the stages appeared less synchronous than those at earlier time points. We chose genes with an abs(log2FC) >1 and adjusted P value < 0.05 in the respective comparison as genes with a potential biological function and determined whether they were induced or repressed. Based on these criteria, sperm cells formed the most distinct group, expressing 4090 differentially upregulated genes in all comparisons (Figure 4F). As all of the other cell types are similar, fewer genes were induced in these cells compared with all other cell types; 482 differentially upregulated genes were detected in egg cells in all comparisons, 109 were detected in zygotes at 12 HAP, 31 were detected in zygotes at 24 HAP, and 6 and 8 were detected in apical and basal cells, respectively (Figure 4F; Supplemental Data Set 6).
We compared zygotes at 12 HAP to both sperm and egg cells and identified 3605 induced genes (9.1% of the maize transcriptome of 39,469 annotated genes; Schnable et al., 2009) over all chromosomes shortly after fertilization (Figure 4B). This high number of activated genes indicates that global ZGA in maize already occurs in the early zygote and not several days after fertilization, as previously reported (Grimanelli et al., 2005). Although de novo transcription has also been observed in Arabidopsis zygotes (Autran et al., 2011; Nodine and Bartel, 2012; Del Toro-De León et al., 2014), the timing of global ZGA in this species is unclear, and some reported results are currently under debate due to contamination from maternal tissues (Schon and Nodine, 2017).
We compared zygotes at 12 HAP with sperm and egg cells and identified 7356 and 1933 induced genes, respectively (Figure 4B). Furthermore, we identified 1730 differentially upregulated genes in sperm and egg cells versus zygotes at 12 HAP (Figure 4A), which can also be viewed as genes downregulated in zygotes at 12 HAP compared with gametes. These findings also suggest that major rearrangements in the transcriptome occur following fertilization.
To obtain a global overview of transcriptome dynamics during zygote development, we defined various gene expression profiles as capturing not only on and off states, but also the induction or repression of genes during the transition from gametes to zygotes and their descendant cells (Figure 4E; Supplemental Data Set 6). Very specific gene expression profiles were found for egg cells: 326 genes were upregulated in this cell type, whereas 95 genes were downregulated. Shortly after fertilization, 356 and 1510 genes were transiently induced only in zygotes at 12 HAP and in zygotes at both stages, respectively. Approximately 10% of the genes in each group encode transcriptional regulators. Of the 3808 genes induced after fertilization, 223 were predominantly expressed in apical cells, whereas 182 were predominantly expressed in basal cells. These results indicate that the expression levels of many genes are higher in the zygote than in its progenitor cells, supporting the notion of an early onset for ZGA. Few genes were transiently repressed after fertilization (137 genes). Together, these findings reveal a highly dynamic transcriptional landscape after fertilization in maize and demonstrate that ZGA occurs shortly after fertilization in this plant. Studies involving pollination with other inbred lines are now needed to distinguish between maternal and paternal transcripts and thus to determine whether both genomes contribute equally to ZGA in this species.
Transcription Factor Activation Schemes in Gametes and Zygotes
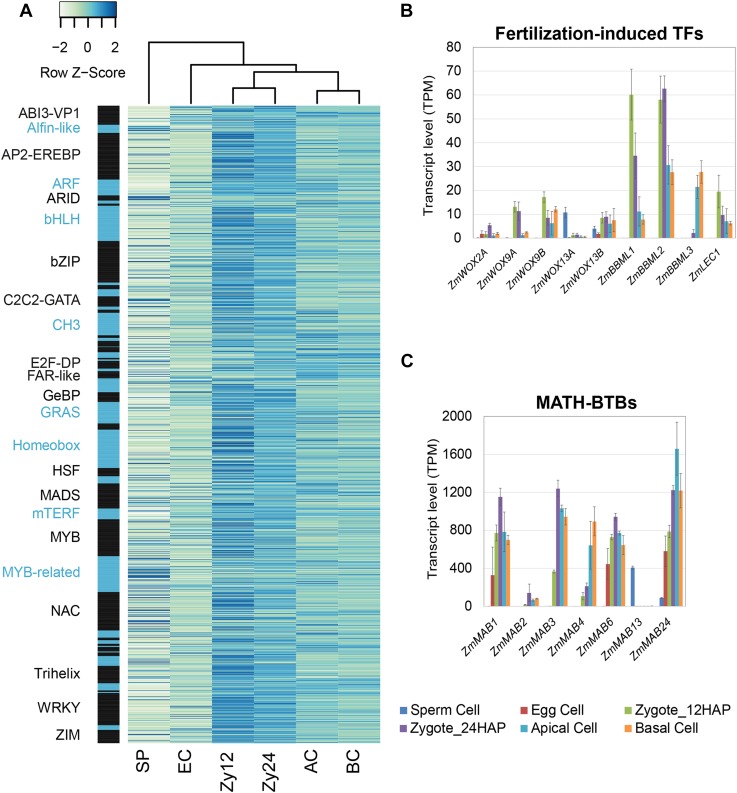
As TFs are major regulators of gene expression, we next examined TF gene expression levels in gametes and early zygotic embryos based on the maize TF database Grassius (www.grassius.org). Across all cell types, 1478 of 2630 maize TF genes were expressed in gametes and zygotes (log2FC > 1 and adjusted P value < 0.05 in at least one comparison). Comparing their transcription levels during early development (Figure 5A; Supplemental Data Set 3) showed that zygotes formed a group distinct from apical and basal cells and that both groups differed from gametes. We identified 428 TF genes that were induced in zygotes (12 and 24 HAP), 189 of which were strongly activated shortly after fertilization (Zy12 versus SP and EC, log2FC > 3). We detected 25 TF genes specifically expressed in pro-embryonic cells. Only 23 paternal and 103 maternal TF genes were expressed at similar levels in gametes and zygotes (Supplemental Data Set 3). While we could not distinguish between paternal and maternal mRNAs in our assays and were thus unable to identify the MZT (Baroux and Grossniklaus, 2015), the results suggest that ∼8.6% (25 + 103/1478) of expressed TF genes are parentally transmitted, while 29% (428/1478) are newly or more intensely transcribed in maize zygotes. These data support the notion that TF genes are activated early in maize zygotes (at 12 HAP; 403 of 428 TF genes were induced in zygotes versus 25 of 428 at 24 HAP) (Figure 5A).
Figure 5.
Expression Levels of TF and MAB Genes in Gametes, Zygotes, and Early Two-Celled Pro-Embryo Cells in Maize.
(A) Expression levels of TF genes in gametes and at early developmental stages after fertilization. Genes with abs(log2FC) > 1 and P-adjusted < 0.05 in at least one cell type comparison were considered. To make the expression levels comparable across multiple cell types, z-scores were calculated from the square root transformed TPM values corresponding to the number of standard deviations between the cell type-specific expression value and the mean expression value of the respective gene. Genes were ordered by TF classes (black/blue bars). Black and blue fonts were used alternatively to distinguish the various classes. See Supplemental Data Sets 2 and 7 for details.
(B) and (C) Expression analysis of selected maize genes encoding homologs of Arabidopsis early embryogenesis-related TFs (B) and MATH-BTB (MAB) family genes (C) involved in ACD.
We then identified important TF classes from the various expression profiles (Supplemental Data Set 6; profiles of TF classes are summarized in Supplemental Data Set 7). In sperm cells, many genes belong to the TF class AT-rich interactive domain (ARID 5/10) proteins. Members of this class have been implicated in sperm cell development in plants and mammals. In Arabidopsis, ARID1 is necessary for the appropriate expression of DUO1, a major TF required for sperm cell formation (Zheng et al., 2014). In mice, the loss of ARID4A combined with ARID4B haploinsufficiency leads to spermatogenic arrest (Wu et al., 2013). In egg cells, we detected high proportions of TFs from the classes FAR1-like (3/15), mTERF (8/30), Sigma70-like (2/9), S1Fa-like (2/2), and GeBP (5/21) compared with the other cell types analyzed. The first four classes are related to plastid development (Zhou et al., 1995; Ouyang et al., 2011; Kleine, 2012; Wei et al., 2012). This finding suggests that a tightly controlled regulatory network controls plastid development during the first steps in the plant life cycle. Members of the GL1 enhancer binding protein (GeBP) class of leucine-zipper TFs have been linked to the cytokinin response in Arabidopsis and are thought to be mainly expressed in vegetative meristems and in the primordia of young leaves (Chevalier et al., 2008). Our data suggest that this class of TFs also plays a role in the transition from egg cells to early zygotes.
As discussed above, at the first zygotic time point (12 HAP), genes for many TF classes were induced (Supplemental Data Set 7). The auxin-responsive ARF TFs (7/38) and the ethylene-responsive AP2-EREBP TFs (34/212) might be important at this stage, as crosstalk between these pathways is thought to be essential during zygote and pro-embryo development (see below). In addition, many genes from the TF classes C3H/CCCH (18/54), Trihelix (11/41), ZIM (10/36), MADS (15/77), NAC (26/134), bZIP (24/128), and Homeobox (26/133) were induced at this time point. MADS box TFs are associated with reproductive organ development and play a role in gametes and in zygotic embryogenesis (Schreiber et al., 2004; Lehti-Shiu et al., 2005).
Activation of Embryo Patterning
After ACD, WOX genes encoding homeodomain TFs mark apical and basal cell fate upon zygote division in Arabidopsis (Breuninger et al., 2008). In maize, ZmWOX9A and ZmWOX9B likely represent the homologs of AtWOX8 and AtWOX9, respectively (Salvo et al., 2014). Both ZmWOX9A and ZmWOX9B were induced shortly after fertilization (Zy12 versus SP/EC:log2FC > 5.9* and 7.0*, respectively) and were expressed at higher levels in basal cells than in apical cells, like their counterparts in Arabidopsis (Figure 5B; Supplemental Data Set 3B), indicating that they might play a similar role in early embryonic patterning. However, unlike AtWOX2, which marks apical descendants of the zygote, ZmWOX2A was expressed at very low levels in basal cells, with almost no expression in apical cells. Instead, this gene was expressed much later during seed development in the endosperm (Maize eFP Browser at bar.utoronto.ca). This finding indicates that pattern formation regulated by ZmWOX2A and other genes likely occurs later in maize embryogenesis than in Arabidopsis embryogenesis (Zhao et al., 2017). Notably, ZmWOX13A is the only WOX gene that was transcribed at high levels in sperm cells (SP versus all: log2FC > 4.3*); whether it marks cell identity of the male gamete or represents a zygote activator remains to be investigated. In Arabidopsis, BBM and LEC TF genes are key players in embryogenesis, and the presence of either gene is sufficient to induce competence for embryogenesis (Lotan et al., 1998; Boutilier et al., 2002). Overexpressing a combination of the maize homologs WUS2 and BBM was recently shown to significantly increase embryogenic potential in tissue culture and thus represents a key mechanism to increase the transformation efficiency in maize (Lowe et al., 2016). Notably, the maize homologs ZmBBML1 (Zy12 versus SP/EC: log2FC > 10.5*), ZmBBML2 (Zy12 versus SP/EC: log2FC > 10.2*), and ZmLEC1 (Zy12 versus SP/EC: log2FC > 7.9*) were already induced at 12 HAP (Figure 5B), suggesting that the egg cell quickly acquires embryogenic competence and that the characteristic embryogenic transcription program is activated shortly after fertilization. ZmBBML3 was induced in zygotes at 24 HAP (Zy24 versus Zy12: log2FC = 6.5*) and after zygote division (AP/BC versus Zy24: log2FC = 3.5/3.9*), suggesting step-by-step activation of the embryonic program. A comparison of apical and basal cells with zygotes at 24 HAP revealed 2228 genes induced in both cell types, 832 induced only in apical cells, and 485 induced only in basal cells (Figure 4D). This induction was accompanied by the downregulation of 2182 genes in apical and basal cells versus zygotes at 24 HAP (Figure 4C). These data suggest that global rearrangements also occur in the transcriptomes of apical and basal cells compared with their predecessor.
The development of multicellular organisms often involves ACDs to generate daughter cells with different cell fates. Spindle positioning is particularly associated with the generation of symmetric or asymmetric cell fates (Siller and Doe, 2009). The MATH-BTB domain protein ZmMAB1, a component of a CUL3-E3 ubiquitin ligase complex, regulates spindle length during the development of the male and female germline in maize (Juranič et al., 2012). ZmMAB1 may also function like its animal homolog, the key ACD regulator MEL-26, a factor required for embryogenic morphogenesis that regulates the formation of mitotic spindles in the early embryo (Pintard et al., 2003). As shown in Figure 5C, MAB family genes were strongly upregulated (ZmMAB1-3, ZmMAB6, and ZmMAB24, Zy12 versus SP: log2FC > 1.5 to 10.7*) after fertilization in maize. The highest expression levels of these genes were detected in zygotes at 24 HAP (Supplemental Data Set 3A; important gene families), suggesting that they play roles in processes such as spindle positioning during the first asymmetric zygote division (ZmMAB2-3, Zy24 versus Zy12 log2FC = 3.1* and 1.8*, respectively). Functional studies of ZmMABs are now needed to investigate this hypothesis.
Cell Cycle Regulation during Zygote Development
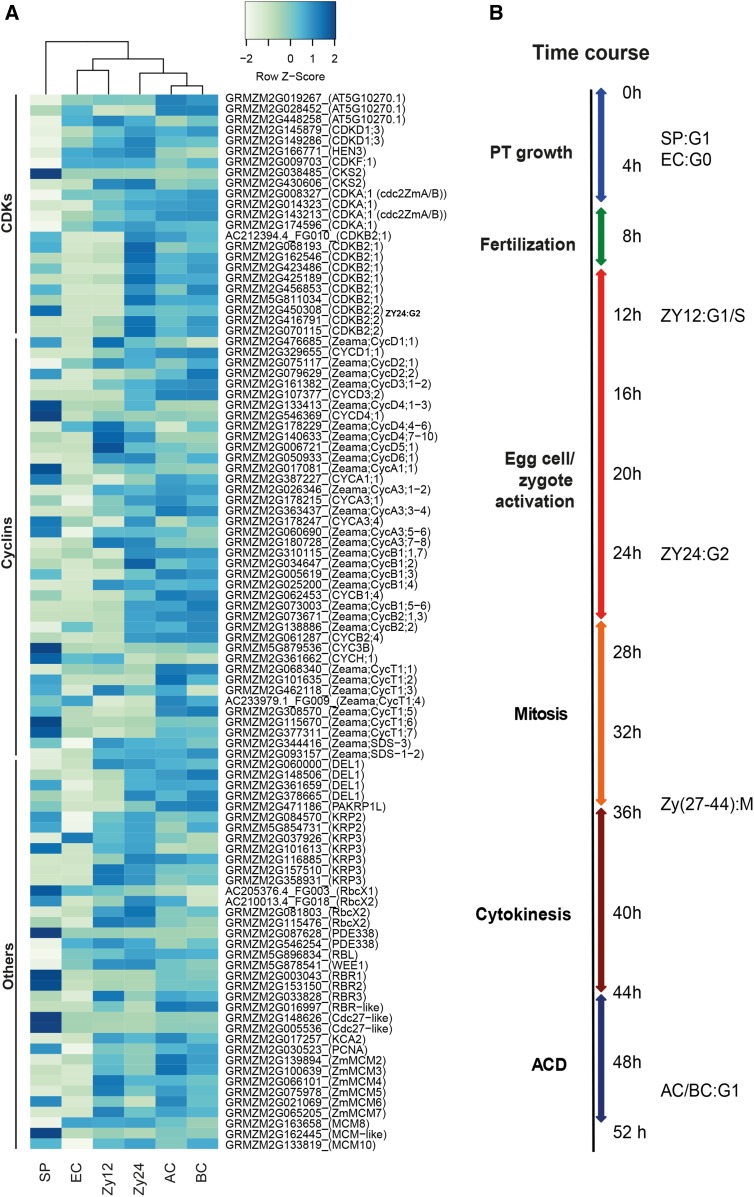
Since previous reports provide only a glimpse of cell cycle regulation in plant gametes and during zygote development, we investigated the expression patterns of important cell cycle regulator genes. First, we searched for orthologs of Arabidopsis cell cycle genes (Vandepoele et al., 2002) and then included cyclins and other cell cycle-related factors described previously for maize (see Methods), resulting in a list of 89 cell cycle genes (Supplemental Data Set 3). As shown in Figure 6A, hierarchical clustering of these genes from different cell types clearly separated sperm cell genes from a group of genes from egg cells and zygotes at 12 HAP. The expression patterns of the genes from zygotes at 24 HAP were more similar to those of apical and basal cells.
Figure 6.
Gene Expression Analysis of Cell Cycle Regulators.
(A) Major regulators of the maize cell cycle (Supplemental Data Set 2). Transformation of gene expression values as described above. Genes were ordered into protein classes based on data from the literature (see Methods for details).
(B) Summary of the time course of pollen tube (PT) growth, fertilization, and zygote development. Cell cycle stages of the selected cells are indicated based on the currently reported expression data.
The G1-phase of sperm cells is characterized by high expression levels of Cdc27-like genes (GRMZM2G148626 and GRMZM2G005536, SP versus all: log2FC > 5.7*) encoding subunits of the APC complex, which controls CDK degradation in M- and G1-phase, and relatively low levels of CDK gene expression. Furthermore, retinoblastoma-related protein genes RBR1 and RBR2 were highly expressed in sperm cells (Figure 6A, SP versus all: log2FC > 2.9*). RBR1 and RBR2 mediate G1-phase arrest by inhibiting E2F TFs, which in turn promote DNA replication (Sabelli et al., 2013). Compared with the other cell types, egg cells showed by far the lowest expression levels of cell cycle genes and lacked a typical cell cycle phase-specific gene expression pattern. This finding, together with the results of cell cycle gene expression analysis and the microscopy evidence reported above (Figure 1), indicate that the egg cell is in a resting G0 stage rather than in G1 (Figure 6B). Thus, the egg cell must be activated and its cell cycle synchronized with the sperm cell cycle stage before karyogamy (fusion of both nuclei) is executed. A typical G1-phase expression pattern was observed in zygotes at 12 HAP (Figure 6A), with slightly upregulated expression of E2F TF genes (GRMZM2G060000, GRMZM2G361659, and GRMZM2G378665, Zy12 versus EC, log2FC > 2.5*) and (especially) their cell cycle target genes (Supplemental Data Set 3). The latter include minichromosome maintenance genes (ZmMCM3-6, Zy12 versus EC: log2FC > 2.0*), encoding DNA replication licensing factors required for replication initiation, and the gene encoding proliferating cell nuclear antigen, which acts as a scaffold to recruit proteins involved in DNA replication and repair (Sabelli et al., 2009; Tuteja et al., 2011). Notably, RBR3, encoding an activating factor of MCM2-7 transcription, was specifically induced in zygotes at 12 HAP (Zy12 versus EC: log2FC = 2.2*), while RBR1 and RBR2, two genes encoding repressors of E2F TFs, were repressed in these zygotes compared with sperm cells (Zy12 versus SP: log2FC< −3.8*). This expression pattern coincided with the induction of CDK A genes (Zy12 versus SP: log2FC > 1.1 to 7.0*) and low levels of expression of CDK B genes, indicating that fertilized egg cells had been activated and zygotes at 12 HAP were in G1-phase. MCM2-7 expression levels were lower in zygotes at 24 HAP versus 12 HAP, whereas CDK B2 genes (9 of 14 genes, Zy24 versus Zy12: log2FC > 2.8*) and B-type cyclin genes were induced (8 of 9 genes, Zy24 versus Zy12: log2FC > 1.9*), especially ZmCycB1;2, suggesting that S-phase was completed and the zygotes were prepared for the G2/M-phase transition (Meijer and Murray, 2001). M-phase took place at 27 to 35 HAP (Figure 1A). Notably, G1-phase markers such as MCM2-7 were expressed at slightly higher levels in apical versus basal cells, hinting at more rapid cell cycle progression in the apical cell after ACD.
Taken together, our cell cycle analysis by microscopy of DNA staining patterns, transcription data from selected gene sets, and global cell expression analysis of cell cycle genes allowed us to determine the timing of zygote development in maize (Figure 6B). On average, fertilization occurs at ∼8 HAP. Sperm cells appear in G1-phase, and egg cells are in a resting G0-phase of the cell cycle. Activated zygotes are in G1 phase at 12 HAP (∼4 h after fertilization) and at G1/S-phase at 24 HAP. Mitosis typically occurs at 26 to 36 HAP, and cytokinesis lasts until 44 HAP. ACD is completed between 44 and 50 HAP, generating both apical and basal cells in G1-phase.
The Role of Auxin in Early Embryogenesis in Maize
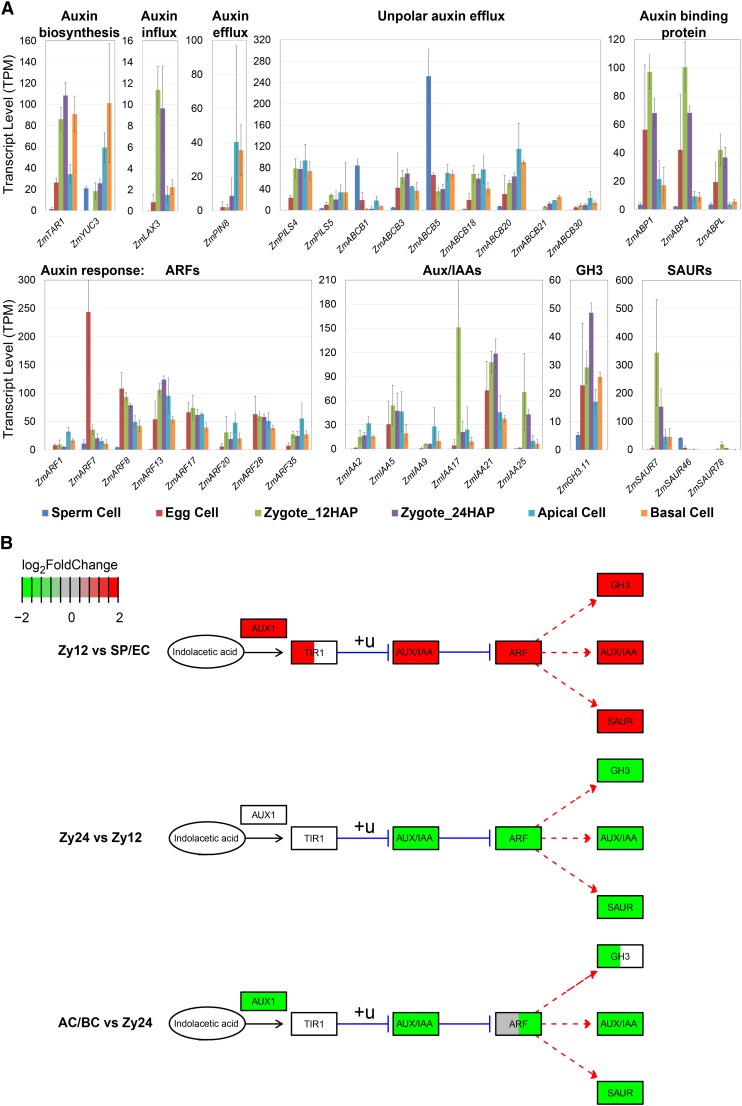
To demonstrate the utility of our data sets, we chose the auxin pathway, which plays a key role in early embryo patterning in Arabidopsis, as an example for analysis. Auxin gradients generated by PIN efflux carrier proteins establish the apical-basal axis upon the first ACD in the Arabidopsis zygote (Friml et al., 2003). The molecular mechanisms that determine axis formation during early embryogenesis in monocots are largely unknown. We therefore analyzed the expression of auxin biosynthesis, transport, and response genes, as shown in Figure 7 (Supplemental Data Set 3D; important gene families). In maize, the earliest ZmPIN1a localization was observed at 6 DAP at the adaxial side of the embryo proper (Chen et al., 2014). To identify potential PIN proteins that function at the earliest stage of embryo patterning, we analyzed the expression of all PIN family genes. Among the 15 PIN genes in maize (Yue et al., 2015), only ZmPIN8 was weakly expressed in the zygote and upregulated after ACD in both apical and basal cells (AC/BC versus Zy12: log2FC = 3.8/3.7*). A number of ZmABCBs, representing potential auxin transporter genes (Yue et al., 2015), were also active in maize gametes and/or zygotes. Auxin biosynthesis genes ZmTAR1 (Zy12 versus SP/EC: log2FC > 1.2*) and ZmYUC3 (Zy12 versus EC: log2FC = 9.4*) were significantly induced after fertilization, with higher expression levels in basal cells versus apical cells (ZmTAR1 BC versus AC: log2FC = 1.4*). Auxin receptor genes ZmABP1, ZmABP4, and ZmABPL were also expressed in egg cells (EC versus SP: log2FC > 1.1*) but not in sperm cells and became transiently activated after fertilization (Zy12 versus SP: log2FC > 1.9*, AC/BC versus Zy24: log2FC < −1.4*). Of the auxin-responsive factor (ARF) genes examined, ZmARF7 had the highest expression level in the egg cell and was almost completely switched off after fertilization (EC versus SP/Zy12/Zy24: log2FC > 3.1*, AC/BC versus EC: log2FC< −4.1*). Other ARF genes, such as ZmARF8, 13, 17, and 28, were expressed at similar levels in all cells except sperm cells. In general, ARF transcript levels were higher in the apical daughter cell of the zygote than in the other daughter cell (Figure 7B, AC/BC versus Zy24). A few AUX/IAA repressor genes encoding proteins that interact with ARF regulators were activated after fertilization (Figure 7B, Zy12 versus SP/EC). In particular, ZmIAA17 (Zy12 versus SP/EC log2FC > 4.3) was transiently expressed only shortly after fertilization; ZmIAA17 might be involved in the inactivation of ZmARF17-regulated gene expression patterns. Another highly upregulated gene, ZmSAUR7 (Zy12 versus SP/EC: log2FC > 5.2*, AC/BC versus Zy12: log2FC < −2.6*), is one of 79 SAUR (SMALL AUXIN UP RNAs) genes in maize, representing the largest family of auxin response genes (Ren and Gray, 2015). Globally, we found that auxin pathway genes were highly induced in the early zygote at 12 HAP and expressed at decreasing levels from 24 HAP zygotes to apical and basal cells (Figure 7B). By contrast, in Arabidopsis, these genes continue to show a strong auxin response, especially in the apical cell after ACD (reviewed in Petrásek and Friml, 2009). Moreover, transcripts encoding homologs of key players in auxin-regulated early embryo patterning in Arabidopsis, such as ARF5 (MP), IAA12 (BDL), PIN1, and PIN7, were absent in zygotes and their daughter cells in maize.
Figure 7.
Analysis of Gene Expression Changes in the Auxin Pathway in Gametes, Zygotes, and during Early Embryogenesis.
(A) Expression analysis of the most highly expressed auxin biosynthesis, transport, and auxin response-related genes.
(B) Auxin pathway analysis using three developmental transitions during zygote development. The median values over the log2FCs of all differentially expressed genes (P-adjusted <0.05) from the respective comparison are color-coded in green (downregulated) and red (upregulated). All log2FCs above 2 or below –2 were set to 2/–2 to improve visualization. White boxes, no significant log2FC; +u, ubiquitination; blue line, inhibition; red dashed line, expression. Pathway based on KEGG database. See Supplemental Data Set 2 for genes.
The observation that gametes and early zygotes are equipped with transcripts encoding proteins for auxin biosynthesis, transport, and perception, as well as the identification of strongly regulated auxin response genes (Supplemental Data Set 8), indicates that auxin-regulated early embryo patterning is likely different in maize and other grasses compared with Arabidopsis, providing an entry point for investigating the role of this important hormone during early embryogenesis in grasses. In addition to auxin signaling, we also obtained hints about brassinosteroid and ethylene signaling during early embryo development in maize, which will be investigated in the future.
Cell Signaling during Fertilization and Early Embryogenesis
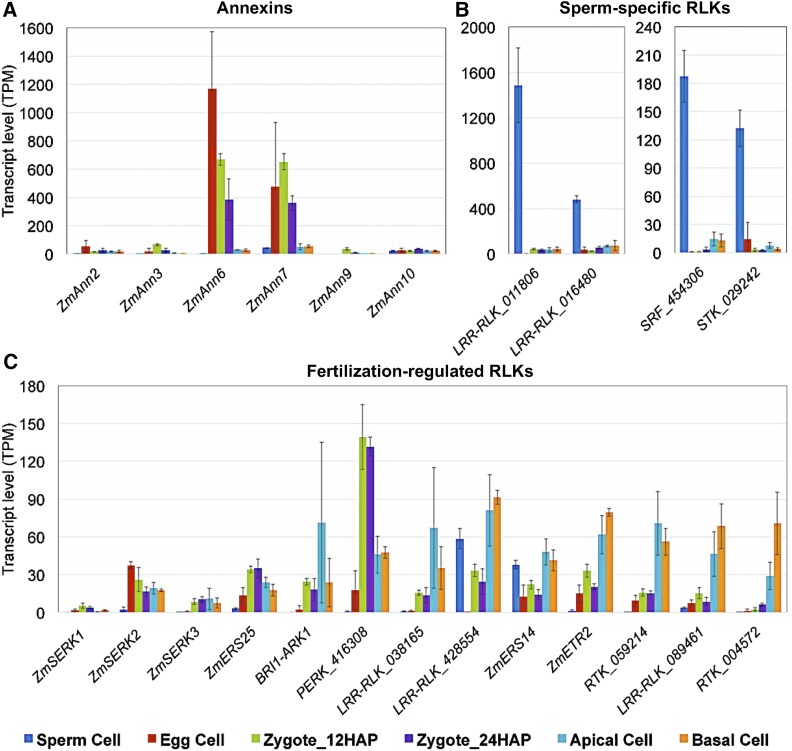
Ca2+ signaling is thought to play a pivotal role in fertilization by regulating a plethora of cellular processes (Chen et al., 2015). Annexins are a class of Ca2+-regulated proteins that link Ca2+ signaling to membrane and cytoskeleton dynamics (Gerke et al., 2005). Of the 12 genes encoding annexins in maize (Zhang et al., 2015), ZmAnn6 and ZmAnn7 exhibited very high, transient expression in egg cells (EC versus SP: log2FC = 8.5/2.1*, EC versus Zy12: log2FC = 1.1/−*) and early zygotes (AC/BC versus Zy12: log2FC < −4.1/−3.2*), respectively (Figure 8A; Supplemental Data Set 3C; important gene families). Thus, these proteins might function as key players in intra- and intercellular Ca2+ signaling during fertilization and early embryo development.
Figure 8.
Expression Analysis of Selected Maize Genes with Putative Roles in Signaling during Gamete Interaction and Early Embryogenesis.
Ca2+-dependent phospholipid binding annexin family proteins (A), sperm-specific receptor-like kinases (B), and fertilization-regulated receptor-like kinases (C). Gene identifiers are listed in Supplemental Data Set 2 (important gene families). Transcript levels in the cells indicated are given as TPM values (means ± sd).
Signal perception and transduction through cell surface receptor-like kinases (RLKs) likely play also roles in gamete interaction, fertilization, and early seed development in plants. We detected at least three RLK genes that were preferentially expressed in sperm cells (GRMZM2G011806, GRMZM2G016480, and GRMZM2G428554, SP versus EC/Zy12/Zy24: log2FC > 2.3*) (Figure 8B; Supplemental Data Set 3E; important gene families), representing potential players in gamete recognition and/or sperm activation. We did not identify RLK genes that were preferentially expressed in egg cells. In Arabidopsis, SOMATIC EMBRYOGENESIS RECEPTOR KINASE1 (AtSERK1) is expressed in developing ovules and early embryos, and enhances embryonic competence in cell culture (Hecht et al., 2001). While its maize ortholog ZmSERK1 (Salvo et al., 2014) was expressed at very low levels in gametes and zygotes, ZmSERK2 and ZmSERK3 were expressed in zygotes (e.g., ZmSERK3 Zy12 versus EC: log2FC = 3.7*) and daughter cells (Figure 8C), indicating the involvement of similar signaling pathways in embryonic initiation. Moreover, several RLK genes (GRMZM2G038165, GRMZM2G428554, and GRMZM2G089461) were upregulated and differentially expressed in apical and basal cells (AC/BC versus Zy24: log2FC > 1.5*), thus representing exciting candidates for future functional studies investigating cellular communication during early embryo development in grasses.
Conclusions
Detailed analysis of global gene expression patterns in plant gametes, zygotes, and manually separated apical and basal cells has allowed the onset of global ZGA in maize to be determined. The observation that ZGA occurs soon after fertilization, displaying a highly dynamic and partially transient pattern, is surprising and contradicts previous studies using a limited number of genes. These studies indicated that the zygote is in a relatively quiescent transcriptional state, that only a few genes are de novo activated in the zygote, and that ZGA occurs gradually rather than all at once (Baroux and Grossniklaus, 2015; Zhao and Sun, 2015). The striking differences in the expression patterns of cell cycle regulators between sperm and egg cells coincide with a distinct chromatin state in sperm cells and define a quiescent cell cycle state in egg cells, although egg cells appear to be translationally highly active or well prepared to quickly activate the translational machinery after fertilization. The chromatin state in sperm appears to depend on replication-independent histone assembly and the HMG protein ZmHmgd1, which likely keeps the highly condensed sperm cell chromatin at least partially accessible, as demonstrated by the numerous transcribed genes. In addition, our data allowed us to differentiate between the stages during G1-phase that occur in zygotes at 12 HAP, apical cells, and basal cells, and they suggest a preference for certain CDKs and cyclins during the first two cell cycles in plants. Analysis of the expression levels of TFs, structural regulators, and signaling pathway genes allowed us to identify relevant genes homologous to key, well-known Arabidopsis regulators as well as novel candidate genes, which will serve as a starting point for many future studies.
In summary, our analyses of the genes described above represent only a few examples of how our comprehensive data set can be used. This gene expression atlas should further accelerate the identification of key players involved in many biological processes, including fertilization, early embryogenesis, and the cell cycle, as well as the translational machinery. In addition, our data set could be used to uncover genes (and their corresponding promoters) for use in future efforts aimed at increasing seed yield and quality in maize and other crops.
METHODS
Plant Materials and Growth Conditions
Maize (Zea mays) inbred line B73 was cultivated in a walk-in plant growth room at 26°C under illumination of 24,000 lux using alternating SON-T Agro and HPI-T Plus bulbs with a 16-h-light/8-h-dark cycle and a relative humidity of 60%. Flowers at anthesis and pollinated cobs were used to isolate cells for RNA-seq.
Isolation of Cells from Male and Female Gametophytes
Hundreds of maize plants were grown to collect sufficient numbers of manually isolated cells. Each biological replicate consisted of pooled cells from different plants. Only the middle part of the cob was used for cell isolation from excised ovules. A whole cob was used to isolate cells at a defined developmental stage. Sperm cells were released from maize pollen grains (male gametophytes) by osmotic shock and separated using density gradient centrifugation on a discontinuous Percoll gradient. The detailed protocol is given below. Egg cells were isolated from embryo sacs (female gametophytes) of unpollinated ovules as described (Kranz et al., 1991). Early and late developmental stage zygotes were isolated from ovules at 12 and 24 HAP, respectively, as previously described (Cordts et al., 2001). Apical and basal cells were dissected from two-celled pro-embryos isolated from ovules at between 48 and 52 HAP following the procedure used for zygotes with some modifications (described in detail below). All cells isolated from ovules were individually collected using a microcapillary and washed twice in mannitol solution (480 mOsmol⋅kg−1 H2O). Cells showing cytoplasmic streaming were individually transferred to 0.5 mL Eppendorf RNA/DNA LoBind microcentrifuge tubes, immediately frozen in liquid nitrogen, and stored at −80°C for mRNA extraction. Three biological replicates (each representing an independent pool of cells) were performed, each with ∼1000 sperm cells, 20 egg cells, 14 to 15 zygotes at 12 HAP, 16 to 17 zygotes at 24 HAP, 16 apical cells, and 13 to 14 basal cells (Table 1). All three biological replicates of each cell type were used for RNA-seq and subsequent transcriptome analyses.
Sperm Cell Isolation
Sperm cells were isolated as described (Dupuis et al., 1987) with some modifications. Pollen grains were collected upon shedding, immersed in 550 mOsmol∙kg−1 H2O mannitol solution (100 mg pollen/mL solution), and incubated on a platform shaker with slow agitation (80 rpm) for 1 h. The resulting lysate was filtered through a 40-μm cell strainer to remove exines and unruptured pollen grains, resulting in a yellowish filtrate containing sperm cells and starch granules. A Percoll gradient was prepared in a 30-mL Corex tube, consisting of 5 mL 30% (v/v) Percoll in 550 mOsmol∙kg−1 H2O mannitol solution at the bottom, 6 mL 20% (v/v) Percoll in 550 mOsmol∙kg−1 H2O mannitol solution in the middle, and 6 mL 15% (v/v) Percoll in 550 mOsmol∙kg−1 H2O mannitol solution at the top. Sperm-containing filtrate (10 mL) was layered on top of the Percoll gradient and centrifuged in a swing-out rotor at 12,000g for 1 h at 4°C. After centrifugation, distinct white layers were visible in the 15/20% Percoll interphase and the 20/30% Percoll interphase. The 20/30% interphase, which was enriched in sperm cells, was carefully aspirated using a Pasteur pipette and transferred to 15- or 50-mL Falcon tubes. At least 10 volumes of fresh 550 mOsmol∙kg−1 H2O mannitol solution were added to the sperm cell-enriched fraction, and the cells were washed by carefully inverting the tube several times. The sperm cells were pelleted by centrifugation at 2500g for 15 min at 4°C and the supernatant was removed without disturbing the pellet, leaving a volume of ∼50 to 100 μL. The pellet was resuspended in the remaining supernatant by careful pipetting, resulting in a solution highly enriched in sperm cells. Cell counting was performed using a Neubauer counting chamber. Isolated sperm cells were used immediately or shock-frozen in liquid nitrogen and stored at −80°C.
Isolation of Apical and Basal Cells
To identify the time point of ACD of the zygote, several cobs were pollinated and analyzed at different intervals after pollination. The first zygotes were analyzed at 24 HAP. Subsequent examinations were performed at 1-h intervals; on average, zygote ACD was observed at ∼48 HAP. Apical and basal cells were subsequently separated using cell wall-degrading enzyme solution containing 1.5% Driselase (Sigma-Aldrich), 1.5% pectinase (Fluka), 0.5% pectolyase Y23 (Karlan), 1.0% hemicellulase (Sigma-Aldrich), 1.0% cellulase “Onozuka R10” (Serva), and 1.5% maceroenzyme (Karlan) in mannitol solution (480 mOsmol∙kg−1 H2O). The enzyme solution (100 μL) was combined with 1 mL mannitol solution (480 mOsmol∙kg−1 H2O), and ovary sections containing embryo sacs at 48 HAP were incubated in the diluted enzyme solution for 30 min at room temperature, followed by manual dissection of two-celled pro-embryos. The attachment between the apical and basal cell protoplasts was gently touched with a very thin glass needle to separate both cells. The cells were washed twice in mannitol solution (480 mOsmol∙kg−1 H2O), collected in 0.5-mL Eppendorf RNA/DNA LoBind microcentrifuge tubes, immediately frozen in liquid nitrogen, and stored at −80°C for mRNA isolation.
RNA Extraction, cDNA Preparation, and Purification
The mRNA was extracted from cell samples using a Dynabeads mRNA DIRECT Micro Kit (Life Technologies). A SMARTer Ultra Low RNA Kit for Illumina Sequencing (Clontech Laboratories) was used to generate first-strand cDNA. Double-stranded cDNA was amplified by LD PCR (15 cycles) and purified via magnetic bead cleanup using an Agencourt AMPure PCR purification kit (Beckman Coulter). The quality of the purified cDNA was measured using an Agilent 2100 Bioanalyzer with an Agilent High Sensitivity DNA kit (Agilent Technologies), frozen in liquid nitrogen, and stored at −80°C.
Library Construction and Illumina Sequencing
Library preparation was performed as described in the Adapted Nextera Sample Preparation protocol (Clontech Laboratories) for use with the SMARTer Ultra Low RNA Kit for Illumina Sequencing. Input cDNA (5 ng) was tagmented (tagged and fragmented) via the Nextera transposome. The products were purified and amplified via a limited-cycle PCR program to generate multiplexed sequencing libraries. The libraries were quantified using a KAPA SYBR FAST ABI Prism Library Quantification Kit (Kapa Biosystems). Equimolar amounts of each library were pooled and used for cluster generation on the cBot system with Illumina TruSeq PE Cluster v3 and Illumina TruSeq Dual Index Sequencing Primer paired-end kits. Sequencing runs were performed on a HiSeq 1000 instrument using the dual indexed 2 × 100 cycles paired-end protocol and TruSeq SBS v3 reagents according to the Illumina HiSeq 1000 System User Guide. Image analysis and base calling resulted in .bcl files, which were converted into .fastq files with CASAVA1.8.2 software. Library preparation and Illumina sequencing runs were performed at the Genomics Core Facility “KFB Center for Fluorescent Bioanalytics” (www.kfb-regensburg.de). The raw data (.fastq) plus supplemental tables, including count and TPM (transcripts per million) data for all replicates, were uploaded to Gene Expression Omnibus and are available under accession number GSE98379.
Bioinformatic and Statistical Analyses
Quality Control and Alignment
The quality of sequencing data from the RNA-seq libraries was assessed with FASTQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Primer contamination from the SMARTer Ultra Low RNA Kit was removed with cutadapt (version 1.13; Martin, 2011). Size and quality trimming were then performed using Trimmomatic (ver. 0.32 [Bolger et al., 2014], ILLUMINACLIP: NexteraPE-PE_SMARTer.fa:2:30:10:2:true TRAILING:26 LEADING:26 MINLEN:25). The results from trimming are summarized in Supplemental Table 1 (mapping information). Trimmed reads were aligned to the maize genome (AGPv3, INSEC Assembly GCA_000005005.5, release 23, ftp://ftp.ensemblgenomes.org/pub/plants/). STAR (Dobin et al., 2013) was used to align the mRNA reads. Alignment statistics are summarized in Supplemental Table 1. Duplicate reads were identified with picard MarkDuplicates (http://broadinstitute.github.io/picard). The remaining reads were then assessed on the gene level using featureCounts from the Rsubread R-library (Liao et al., 2014) using the annotation information supplied by Gramene, release 5b+ (http://ftp.gramene.org/maizesequence.org/release-5b+/zea_mays.protein_coding.gff). Gene annotation and ortholog information was retrieved from EnsemblPlants (www.plants.ensembl.org) via Biomart (BiomaRt7, release 29), for maize genes [AGPv3 (5b)], Arabidopsis thaliana genes (2010-09-TAIR10), and Oryza sativa ssp japonica genes (IRGSP-1.0), restricting the gene model to AGPv3 (5b) from EnsemblPlants.
Measurement of mRNA Transcript Abundance
TPM was used as a measure of mRNA abundance, which takes into account the length of the RNA transcripts and the sequencing depth (Wagner et al., 2012). Gene length was approximated by determining the sum of the exon lengths of the gene model. The TPMs were calculated for each gene in each sample. The median TPM value was calculated from three biological replicates for each cell type. An annotated list of genes with their expression levels at each developmental stage (cell type) and orthologs (based on Ensembl Compara from Arabidopsis and rice) was assembled (Supplemental Data Set 1).
Differential Expression Analysis and Venn Diagrams
Count data at the gene level were analyzed with DESeq2 (Love et al., 2014). All 15 cell-type-to-cell-type comparisons were performed and corrected for multiple testing over all genes and cell type comparisons using false discovery rate (FDR) (Benjamini and Hochberg, 1995) (Supplemental Data Set 2). Genes with a log2FC > 1 and FDR < 0.05 (“padj” in supplemental tables) were considered significant differentially expressed. Log2FCs with an adjusted P value < 0.05 are indicated by an asterisk in the text. Venn diagrams (R-library: vennerable; https://r-forge.r-project.org/projects/vennerable) were generated for certain comparisons (Figure 4).
Expression Profiles and Pathway Analysis
Genes showing differential expression (log2FC > 1 and FDR < 0.05) in at least one comparison were subjected to further analyses. To visualize gene expression values in heat maps and to compare gene expression profiles during different stages of very early plant development, square root transformed TPM values were used. The median expression values were then transformed to standard units to follow the expression values of one gene across different developmental stages in the heat maps. The dendrogram of the samples is based on Euclidean distances combined with hierarchical clustering with complete linkage. To classify genes into specific expression profiles, Pearson correlation analysis of the gene expression vector (square root transformed median TPM values as above) versus a binary vector encoding the different expression profiles, i.e., expressed only in egg cells and during fertilization (0,1,1,1,0,0)/(SP,EC,ZY12,ZY24,AC,BC) was performed (Supplemental Data Set 1). A stringent cutoff value of >0.9 (Pearson correlation coefficient) was used to define a positive correlation with the respective profile.
Comparison of Gene Expression Levels in Maize and Rice Gametes
To compare the expression of the 80 most highly expressed genes from maize sperm and egg cells with their orthologs in rice (O. sativa ssp japonica), data from Ensembl Compara (http://plants.ensembl.org, based on protein information for 53 species) and the Rice Genome Annotation project (RGAP) (http://rapdb.dna.affrc.go.jp/download/archive/RAP-MSU_2016-08-05.txt.gz) and orthologs from RGAP based on OrthoMCL were used (Li et al., 2003; Ouyang et al., 2007; Vilella et al., 2009). While OrthoMCL is based only on a comparison of protein sequences, Ensembl Compara also uses information from phylogenetic trees and therefore interprets sequence similarities based on the evolutionary development of a gene. Rice gene expression data (TPM) were obtained from Anderson et al. (2013). Maize and rice TPMs were square root transformed, and plastid transcripts were removed. Then, for each organism, the data were binned into 200 equally spaced expression categories, with the 99th percentile representing the maximum expression level. The color scale for the heat maps shown in Figure 2 represents the expression bin, indicating the relative expression level in the organism shown. For each maize gene, the respective rice orthologs in both resources were identified and compared. If there were common orthologs predicted by both resources, these were chosen, and if not, all predictions were used. From this selected set of orthologs, the most highly expressed ortholog in rice was used for plotting. All orthologs are listed in Supplemental Data Set 4. In the heat maps (Figure 2), the maximum of the color scale represents the 99th percentile, resulting in the same color for all maize genes shown (because all of the top 80 genes are within the most highly expressed 1% of the genes). Dark-red bars in the rice column indicate that the gene was also in the top 1% most highly expressed genes in the rice data, and the lighter color indicates lower expression. Cyan lines represent expression levels as bar charts, with the dotted line indicating the median value of the column.
Transcription Factor Gene List
A list of potential TFs in maize based on conserved domains at the protein level was retrieved from Grassius (http://grassius.org/tfomecollection.html, Maize_TFome_Bulk_data.txt). A list of median expression values (TPM > 1) of all TF genes, including their gene names and TF families, is shown in Supplemental Data Set 7. Important TF classes were identified by analyzing the fraction of expressed genes (TPM >1 in percent). Whenever the fraction of expressed genes of one cell type in one TF class exceeded the fractions in the other cell types, this class was discussed in the main text.
Cell Cycle Gene List
A list of cell cycle regulators identified in Arabidopsis (Vandepoele et al., 2002) was used to retrieve maize orthologs from the Rice Genome Annotation Project Database (University of Michigan), also comprising a full data set for maize (Kawahara et al., 2013) (http://rice.plantbiology.msu.edu/annotation_pseudo_apk.shtml). Predicted cyclins (Hu et al., 2010) were added to the list, as well as genes reported to be involved in cell cycle regulation in previous publications (Sauter et al., 1998; Dresselhaus et al., 1999b, 2006; Buendía-Monreal et al., 2011; Dante et al., 2014). Finally, the DNA replication-indicating genes encoding proliferating cell nuclear antigen and MCM homologous proteins were added to the list manually based on Ensemble plants (www.plants.ensemble.org) (Sabelli et al., 2009). The genes were sorted into different classes: CDKs and cyclins, followed by additional cell cycle-related factors (Supplemental Data Set 3).
Auxin Pathway Analysis
Information about auxin pathways was downloaded from (http://www.genome.jp/dbget-bin/www_bget?ath04075). Maize genes in the different categories were obtained from the literature based on the gene list Supplemental Data Set 3D (important gene families). For each category (i.e., SAURs), all significantly (P-adjusted < 0.05) differentially expressed genes from the respective comparisons were selected. The median of the log2FCs of these genes was calculated and represented by a color scale ranging from green (−2) to red (2). All log2FCs above or below these values were set to 2 (−2).
Accession Numbers
The raw data (.fastq) plus supplemental tables, including count and TPM data for all replicates, were uploaded to the Gene Expression Omnibus and are available under accession number GSE98379. Gene identifiers are listed in Supplemental Data Set 1.
Supplemental Data
Supplemental Figure 1. cDNA prepared from maize gametes/zygotes and initial validation based on the presence of selected transcripts.
Supplemental Figure 2. RNA-seq correlation plot and hierarchical clustering of maize gamete and zygote samples.
Supplemental Figure 3. Phylogenetic analysis of protein sequences of histone H3 and H2A variants.
Supplemental Table 1. Summary of NGS runs, alignment process to Ensembl genome (AGPv3, version 82.6), and annotation to Ensembl genebuild (AGPv3_5b, version 82.6).
Supplemental Data Set 1. Master table: binary identifier for venn diagram category membership.
Supplemental Data Set 2. List of differentially expressed genes for all 15 cell type comparisons, with abs(log2FC) > 1 and P-adjusted < 0.05.
Supplemental Data Set 3. Lists of transcript levels of known genes required for fertilization and zygote development (see Figure 1B), expression profiles of transcription factors, cell cycle genes, and important gene families (see Figures 5 to 8).
Supplemental Data Set 4. Lists of TOP80 maize sperm and egg cell genes compared with their predicted homologs/orthologs in rice gametes (see also Figure 2).
Supplemental Data Set 5. Lists of 30 most highly expressed genes in maize sperm cell, egg cell, zygote 12 HAP, zygote 24 HAP, apical cell, and basal cell, respectively (for detailed annotation see Supplemental Data Set 1).
Supplemental Data Set 6. Lists of gene expression profiles selected for Figure 4E.
Supplemental Data Set 7. List of expressed transcription factor classes (see also Figures 5A and 5B).
Supplemental Data Set 8. List of genes in the auxin pathway (see also Figure 7).
Acknowledgments
We thank Thomas Stempfl and Christoph Möhle from the Kompetenzzentrum Fluoreszente Bioanalytik (KFB), a genomics core facility based at the University of Regensburg, for their support in optimizing the procedure used to generate libraries for Illumina sequencing from only a few plant cells. This work was funded by the German Research Foundation (DFG) Collaborate Research Center (SFB960) to T.D., J.C.E., and S.S.
AUTHOR CONTRIBUTIONS
T.D. initiated and designed the project. J.C., N.G.K., and P.C. performed wet lab experiments and interpreted results. J.C., N.S., S.S., and J.C.E. performed bioinformatics analyses. J.C., N.S., and T.D. wrote the manuscript with input from all authors.
Glossary
- MZT
maternal-to-zygotic transition
- ZGA
zygotic genome activation
- TF
transcription factor
- HAP
hours after pollination
- ACD
asymmetric cell division
- DAPI
4′,6-diamidino-2-phenylindole
- HMG
high-mobility group
- RLK
receptor-like kinase
- TPM
transcripts per million
- FDR
false discovery rate
Footnotes
Articles can be viewed without a subscription.
References
- Ahmad K., Henikoff S. (2002). The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9: 1191–1200. [DOI] [PubMed] [Google Scholar]
- Amien S., Kliwer I., Márton M.L., Debener T., Geiger D., Becker D., Dresselhaus T. (2010). Defensin-like ZmES4 mediates pollen tube burst in maize via opening of the potassium channel KZM1. PLoS Biol. 8: e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S.N., Johnson C.S., Jones D.S., Conrad L.J., Gou X., Russell S.D., Sundaresan V. (2013). Transcriptomes of isolated Oryza sativa gametes characterized by deep sequencing: evidence for distinct sex-dependent chromatin and epigenetic states before fertilization. Plant J. 76: 729–741. [DOI] [PubMed] [Google Scholar]
- Autran D., et al. (2011). Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell 145: 707–719. [DOI] [PubMed] [Google Scholar]
- Baroux C., Grossniklaus U. (2015). The maternal-to-zygotic transition in flowering plants: Evidence, mechanisms, and plasticity. Curr. Top. Dev. Biol. 113: 351–371. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F., Gomes G., Gardner R., Moreno N., McCormick S., Feijó J.A., Becker J.D. (2008). Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol. 148: 1168–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutilier K., Offringa R., Sharma V.K., Kieft H., Ouellet T., Zhang L., Hattori J., Liu C.-M., van Lammeren A.A.M., Miki B.L.A., Custers J.B.M., van Lookeren Campagne M.M. (2002). Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14: 1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuninger H., Rikirsch E., Hermann M., Ueda M., Laux T. (2008). Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell 14: 867–876. [DOI] [PubMed] [Google Scholar]
- Budkevich T.V., Timchenko A.A., Tiktopulo E.I., Negrutskii B.S., Shalak V.F., Petrushenko Z.M., Aksenov V.L., Willumeit R., Kohlbrecher J., Serdyuk I.N., El’skaya A.V. (2002). Extended conformation of mammalian translation elongation factor 1A in solution. Biochemistry 41: 15342–15349. [DOI] [PubMed] [Google Scholar]
- Buendía-Monreal M., Rentería-Canett I., Guerrero-Andrade O., Bravo-Alberto C.E., Martínez-Castilla L.P., García E., Vázquez-Ramos J.M. (2011). The family of maize D-type cyclins: genomic organization, phylogeny and expression patterns. Physiol. Plant. 143: 297–308. [DOI] [PubMed] [Google Scholar]
- Chen J., Lausser A., Dresselhaus T. (2014). Hormonal responses during early embryogenesis in maize. Biochem. Soc. Trans. 42: 325–331. [DOI] [PubMed] [Google Scholar]
- Chen J., Gutjahr C., Bleckmann A., Dresselhaus T. (2015). Calcium signaling during reproduction and biotrophic fungal interactions in plants. Mol. Plant 8: 595–611. [DOI] [PubMed] [Google Scholar]
- Chettoor A.M., Givan S.A., Cole R.A., Coker C.T., Unger-Wallace E., Vejlupkova Z., Vollbrecht E., Fowler J.E., Evans M.M. (2014). Discovery of novel transcripts and gametophytic functions via RNA-seq analysis of maize gametophytic transcriptomes. Genome Biol. 15: 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier F., Perazza D., Laporte F., Le Hénanff G., Hornitschek P., Bonneville J.-M., Herzog M., Vachon G. (2008). GeBP and GeBP-like proteins are noncanonical leucine-zipper transcription factors that regulate cytokinin response in Arabidopsis. Plant Physiol. 146: 1142–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordts S., Bantin J., Wittich P.E., Kranz E., Lörz H., Dresselhaus T. (2001). ZmES genes encode peptides with structural homology to defensins and are specifically expressed in the female gametophyte of maize. Plant J. 25: 103–114. [DOI] [PubMed] [Google Scholar]
- Cui Y., Dinman J.D., Kinzy T.G., Peltz S.W. (1998). The Mof2/Sui1 protein is a general monitor of translational accuracy. Mol. Cell. Biol. 18: 1506–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dante R.A., Larkins B.A., Sabelli P.A. (2014). Cell cycle control and seed development. Front. Plant Sci. 5: 493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal R.B., Henikoff S. (2011). Histone variants and modifications in plant gene regulation. Curr. Opin. Plant Biol. 14: 116–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Toro-De León G., García-Aguilar M., Gillmor C.S. (2014). Non-equivalent contributions of maternal and paternal genomes to early plant embryogenesis. Nature 514: 624–627. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresselhaus T., Cordts S., Lörz H. (1999a). A transcript encoding translation initiation factor eIF-5A is stored in unfertilized egg cells of maize. Plant Mol. Biol. 39: 1063–1071. [DOI] [PubMed] [Google Scholar]
- Dresselhaus T., Lausser A., Márton M.L. (2011). Using maize as a model to study pollen tube growth and guidance, cross-incompatibility and sperm delivery in grasses. Ann. Bot. (Lond.) 108: 727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresselhaus T., Sprunck S., Wessel G.M. (2016). Fertilization mechanisms in flowering plants. Curr. Biol. 26: R125–R139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresselhaus T., Srilunchang K.-O., Leljak-Levanic D., Schreiber D.N., Garg P. (2006). The fertilization-induced DNA replication factor MCM6 of maize shuttles between cytoplasm and nucleus, and is essential for plant growth and development. Plant Physiol. 140: 512–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresselhaus T., Cordts S., Heuer S., Sauter M., Lörz H., Kranz E. (1999b). Novel ribosomal genes from maize are differentially expressed in the zygotic and somatic cell cycles. Mol. Gen. Genet. 261: 416–427. [DOI] [PubMed] [Google Scholar]
- Dupuis I., Roeckel P., Matthys-Rochon E., Dumas C. (1987). Procedure to isolate viable sperm cells from corn (Zea mays L.) pollen grains. Plant Physiol. 85: 876–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel M.L., Chaboud A., Dumas C., McCormick S. (2003). Sperm cells of Zea mays have a complex complement of mRNAs. Plant J. 34: 697–707. [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153. [DOI] [PubMed] [Google Scholar]
- Gerke V., Creutz C.E., Moss S.E. (2005). Annexins: linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 6: 449–461. [DOI] [PubMed] [Google Scholar]
- Grimanelli D., Perotti E., Ramirez J., Leblanc O. (2005). Timing of the maternal-to-zygotic transition during early seed development in maize. Plant Cell 17: 1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V., Vielle-Calzada J.-P., Hartog M.V., Schmidt E.D.L., Boutilier K., Grossniklaus U., de Vries S.C. (2001). The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 127: 803–816. [PMC free article] [PubMed] [Google Scholar]
- Hu X., Cheng X., Jiang H., Zhu S., Cheng B., Xiang Y. (2010). Genome-wide analysis of cyclins in maize (Zea mays). Genet. Mol. Res. 9: 1490–1503. [DOI] [PubMed] [Google Scholar]
- Ingouff M., Rademacher S., Holec S., Soljić L., Xin N., Readshaw A., Foo S.H., Lahouze B., Sprunck S., Berger F. (2010). Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr. Biol. 20: 2137–2143. [DOI] [PubMed] [Google Scholar]
- Juranič M., Srilunchang K.O., Krohn N.G., Leljak-Levanić D., Sprunck S., Dresselhaus T. (2012). Germline-specific MATH-BTB substrate adaptor MAB1 regulates spindle length and nuclei identity in maize. Plant Cell 24: 4974–4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N. Y.) 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T. (2012). Arabidopsis thaliana mTERF proteins: evolution and functional classification. Front. Plant Sci. 3: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz E., Bautor J., Lorz H. (1991). In vitro fertilization of single, isolated gametes of maize mediated by electrofusion. Sex. Plant Reprod. 4: 12–16. [Google Scholar]
- Lee M.T., Bonneau A.R., Giraldez A.J. (2014). Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 30: 581–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti-Shiu M.D., Adamczyk B.J., Fernandez D.E. (2005). Expression of MADS-box genes during the embryonic phase in Arabidopsis. Plant Mol. Biol. 58: 89–107. [DOI] [PubMed] [Google Scholar]
- Li L., Stoeckert C.J. Jr., Roos D.S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. [DOI] [PubMed] [Google Scholar]
- Lotan T., Ohto M., Yee K.M., West M.A., Lo R., Kwong R.W., Yamagishi K., Fischer R.L., Goldberg R.B., Harada J.J. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe K., et al. (2016). Morphogenic regulators Baby Boom and Wuschel improve monocot transformation. Plant Cell 28: 1998–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorano D., Lutzmann M., Méchali M. (2006). MCM proteins and DNA replication. Curr. Opin. Cell Biol. 18: 130–136. [DOI] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17: 10–12. [Google Scholar]
- Márton M.L., Cordts S., Broadhvest J., Dresselhaus T. (2005). Micropylar pollen tube guidance by egg apparatus 1 of maize. Science 307: 573–576. [DOI] [PubMed] [Google Scholar]
- Meijer M., Murray J.A. (2001). Cell cycle controls and the development of plant form. Curr. Opin. Plant Biol. 4: 44–49. [DOI] [PubMed] [Google Scholar]
- Nodine M.D., Bartel D.P. (2012). Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos. Nature 482: 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T., Endo M., Singh M.B., Bhalla P.L. (2005). Analysis of the histone H3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3. Plant J. 44: 557–568. [DOI] [PubMed] [Google Scholar]
- Ouyang S., et al. (2007). The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 35: D883–D887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang X., et al. (2011). Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell 23: 2514–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrásek J., Friml J. (2009). Auxin transport routes in plant development. Development 136: 2675–2688. [DOI] [PubMed] [Google Scholar]
- Pintard L., Willis J.H., Willems A., Johnson J.-L.F., Srayko M., Kurz T., Glaser S., Mains P.E., Tyers M., Bowerman B., Peter M. (2003). The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425: 311–316. [DOI] [PubMed] [Google Scholar]
- Ren H., Gray W.M. (2015). SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol. Plant 8: 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L.H., Willis J.H. (2007). Plant speciation. Science 317: 910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S.D., Gou X., Wong C.E., Wang X., Yuan T., Wei X., Bhalla P.L., Singh M.B. (2012). Genomic profiling of rice sperm cell transcripts reveals conserved and distinct elements in the flowering plant male germ lineage. New Phytol. 195: 560–573. [DOI] [PubMed] [Google Scholar]
- Sabelli P.A., Hoerster G., Lizarraga L.E., Brown S.W., Gordon-Kamm W.J., Larkins B.A. (2009). Positive regulation of minichromosome maintenance gene expression, DNA replication, and cell transformation by a plant retinoblastoma gene. Proc. Natl. Acad. Sci. USA 106: 4042–4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli P.A., Liu Y., Dante R.A., Lizarraga L.E., Nguyen H.N., Brown S.W., Klingler J.P., Yu J., LaBrant E., Layton T.M., Feldman M., Larkins B.A. (2013). Control of cell proliferation, endoreduplication, cell size, and cell death by the retinoblastoma-related pathway in maize endosperm. Proc. Natl. Acad. Sci. USA 110: E1827–E1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvo S.A.G.D., Hirsch C.N., Buell C.R., Kaeppler S.M., Kaeppler H.F. (2014). Whole transcriptome profiling of maize during early somatic embryogenesis reveals altered expression of stress factors and embryogenesis-related genes. PLoS One 9: e111407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter M., von Wiegen P., Lörz H., Kranz E. (1998). Cell cycle regulatory genes from maize are differentially controlled during fertilization and first embryonic cell division. Sex. Plant Reprod. 11: 41–48. [Google Scholar]
- Schier A.F. (2007). The maternal-zygotic transition: death and birth of RNAs. Science 316: 406–407. [DOI] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Schon M.A., Nodine M.D. (2017). Widespread contamination of Arabidopsis embryo and endosperm transcriptome data sets. Plant Cell 29: 608–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber D.N., Bantin J., Dresselhaus T. (2004). The MADS box transcription factor ZmMADS2 is required for anther and pollen maturation in maize and accumulates in apoptotic bodies during anther dehiscence. Plant Physiol. 134: 1069–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller K.H., Doe C.Q. (2009). Spindle orientation during asymmetric cell division. Nat. Cell Biol. 11: 365–374. [DOI] [PubMed] [Google Scholar]
- Thomas J.O., Travers A.A. (2001). HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 26: 167–174. [DOI] [PubMed] [Google Scholar]
- Tuteja N., Tran N.Q., Dang H.Q., Tuteja R. (2011). Plant MCM proteins: role in DNA replication and beyond. Plant Mol. Biol. 77: 537–545. [DOI] [PubMed] [Google Scholar]
- Uebler S., Márton M.L., Dresselhaus T. (2015). Classification of EA1-box proteins and new insights into their role during reproduction in grasses. Plant Reprod. 28: 183–197. [DOI] [PubMed] [Google Scholar]
- Vandepoele K., Raes J., De Veylder L., Rouzé P., Rombauts S., Inzé D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella A.J., Severin J., Ureta-Vidal A., Heng L., Durbin R., Birney E. (2009). EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 19: 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G.P., Kin K., Lynch V.J. (2012). Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 131: 281–285. [DOI] [PubMed] [Google Scholar]
- Wei K., Chen J., Wang Y., Chen Y., Chen S., Lin Y., Pan S., Zhong X., Xie D. (2012). Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 19: 463–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R.-C., Jiang M., Beaudet A.L., Wu M.-Y. (2013). ARID4A and ARID4B regulate male fertility, a functional link to the AR and RB pathways. Proc. Natl. Acad. Sci. USA 110: 4616–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest S.E., Vijverberg K., Schmidt A., Weiss M., Gheyselinck J., Lohr M., Wellmer F., Rahnenführer J., von Mering C., Grossniklaus U. (2010). Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr. Biol. 20: 506–512. [DOI] [PubMed] [Google Scholar]
- Yue R., Tie S., Sun T., Zhang L., Yang Y., Qi J., Yan S., Han X., Wang H., Shen C. (2015). Genome-wide identification and expression profiling analysis of ZmPIN, ZmPILS, ZmLAX and ZmABCB auxin transporter gene families in maize (Zea mays L.) under various abiotic stresses. PLoS One 10: e0118751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Li X., Han M., Wu Z. (2015). Genome-wide analysis and functional identification of the annexin gene family in maize (Zea mays L.). Plant Omics 8: 420–428. [Google Scholar]
- Zhao P., Sun M.X. (2015). The maternal-to-zygotic transition in higher plants: Available approaches, critical limitations, and technical requirements. Curr. Top. Dev. Biol. 113: 373–398. [DOI] [PubMed] [Google Scholar]
- Zhao P., Begcy K., Dresselhaus T., Sun M.X. (2017). Does early embryogenesis in eudicots and monocots involve the same mechanism and molecular players? Plant Physiol. 173: 130–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., He H., Zheng Y., Wu W., McCormick S. (2014). An ARID domain-containing protein within nuclear bodies is required for sperm cell formation in Arabidopsis thaliana. PLoS Genet. 10: e1004421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D.X., Bisanz-Seyer C., Mache R. (1995). Molecular cloning of a small DNA binding protein with specificity for a tissue-specific negative element within the rps1 promoter. Nucleic Acids Res. 23: 1165–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]