Abstract
In utero exposure to diesel exhaust air pollution has been associated with increased adult susceptibility to heart failure in mice, but the mechanisms by which this exposure promotes susceptibility to heart failure are poorly understood. To identify the potential transcriptional effects that mediate this susceptibility, we have performed RNA sequencing analysis on adult hearts from mice that were exposed to diesel exhaust in utero and that have subsequently undergone transverse aortic constriction. We identified 3 target genes, Mir133a-2, Ptprf, and Pamr1, which demonstrate dysregulation after exposure and aortic constriction. Examination of expression patterns in human heart tissues indicates a correlation between expression and heart failure. We subsequently assessed DNA methylation modifications at these candidate loci in neonatal cultured cardiac myocytes after in utero exposure to diesel exhaust and found that the promoter for Mir133a-2 is differentially methylated. These target genes in the heart are the first genes to be identified that likely play an important role in mediating adult sensitivity to heart failure. We have also shown a change in DNA methylation within cardiomyocytes as a result of in utero exposure to diesel exhaust.—Goodson, J. M., Weldy, C. S., MacDonald, J. W., Liu, Y., Bammler, T. K., Chien, W.-M., Chin, M. T. In utero exposure to diesel exhaust particulates is associated with an altered cardiac transcriptional response to transverse aortic constriction and altered DNA methylation.
Keywords: PM2.5, heart failure, miR133a-2, Ptprf, Pamr1
Exposure to particulate matter air pollution has been associated with respiratory (1, 2), cardiovascular (3–5), and cerebrovascular (6) disease. Exposure to fine particulate matter—particulate matter ≤2.5 μm (PM2.5)—has been linked to increased cardiovascular morbidity and mortality (3). Early life exposure to PM2.5 has been increasingly recognized for its impact on newborn health. Exposure to PM10 during pregnancy has been linked with decreased birth weight, reduced placental weight (7), and placental oxidative and nitrosative stress (8), which further suggests that particulate matter air pollution contributes to placental insufficiency and intrauterine growth restriction. Placental insufficiency and intrauterine growth restriction has been implicated as an important determinant of adult cardiovascular health via the fetal origins of adult disease or Barker hypothesis pathway (9). These in utero PM exposures that induce placental insufficiency may predispose offspring to increased risk of disease, and, indeed, exposure to PM2.5 during the last trimester of pregnancy has been reported to be associated with increased newborn systolic blood pressure (10). Whereas prospective studies have not yet been carried out to determine whether the effects of maternal exposure affect the health of the offspring into adulthood, experimental studies that have used animal models raise the concern that in utero exposure to air pollution may be a strong determinant of adult health.
We have previously shown that in utero and early life exposure to diesel exhaust (DE) air pollution increases adult susceptibility to heart failure in mice, for which—after transverse aortic constriction (TAC) surgery—we observed decreased fractional shortening and increased left ventricular hypertrophy compared with filtered air (FA)–exposed control animals (11, 12). We have also reported that this increased adult susceptibility to heart failure is conferred with in utero exposure only, which highlights the importance of the gestational window in mediating this long-lasting effect. Although we have identified placental vascular oxidative stress and inflammation as possible physiologic mechanisms, the molecular basis of increased heart failure susceptibility that persists from gestation to adulthood has not yet been elucidated.
One potential molecular mechanism with which to explain our observation of increased susceptibility to heart failure after in utero DE exposure involves epigenetic modification and altered transcription of key mediators. Air pollution has been demonstrated to alter DNA methylation both globally and at specific target genes. Human workplace exposure to PM10 has been found to be associated with global hypomethylation at long interspersed nuclear element-1 and Alu repetitive DNA elements in circulating leukocytes, as well as decreased methylation at the iNOS promoter (13). A study of exposure to PM2.5 demonstrated a similar hypomethylation of DNA in peripheral blood leukocyte DNA (14). Exposure to PM2.5 during pregnancy has been associated with altered global DNA methylation profiles within placental tissue, where increased PM2.5 exposure was associated with lower global DNA methylation (15). This observation supports the hypothesis that developing fetal tissues in utero may be subject to particulate matter–induced alterations in DNA methylation. Altered DNA methylation in the developing myocardium as a result of exposure could persist into adulthood and influence the transcription of genes that are related to heart failure in our mouse model. To identify DE-responsive target genes that may be relevant to heart failure, we performed RNA sequencing of adult heart mRNA after in utero exposure to either DE or FA, followed by either TAC or sham surgery as adults, and have identified 3 potential target genes: Mir133a-2, Ptprf, and Pamr1. To determine whether alterations in DNA methylation play a role in the differential expression of these genes, we performed targeted bisulfite sequencing on CpG islands and promoters associated with these genes. We discuss the associated changes in DNA methylation and the potential role of these genes in mediating the increased susceptibility to heart failure that has been observed in this model.
MATERIALS AND METHODS
DE exposure and mice
C57BL/6 male and female mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA) and maintained as an inbred line. All mice were housed in specific pathogen–free conditions on a 12-h light/dark cycle. Female and male mice between ages of 12 and 14 wk were transferred to our Northlake Diesel Exposure Facility located near the University of Washington and housed under specific pathogen–free conditions in Allentown caging systems (Allentown, NJ, USA) as previously described (11, 12, 16–19). All animal experiments were approved by the University of Washington Institutional Animal Care and Use Committee (4134-01). DE was generated from a single cylinder Yanmar diesel engine (Model YDG5500EV-6EI; Osaka, Japan) operating on 82% load. A detailed analysis of DE particulate components in this system has been previously reported (17). DE exposures were conducted for 6 h/d (9 Am–3 pm) 5 d/wk (Monday–Friday), and DE concentrations were regulated to ∼300 μg/m3 of PM2.5. A 300-μg/m3 concentration of PM2.5 6 h/d, 5 d/wk equates to a time-weighted hourly average of 53 μg/m3. Exposure characteristics that detailed gas, particle-bound polycyclic aromatic hydrocarbons (PAHs), and particle diameter were recently measured and reported (11, 19).
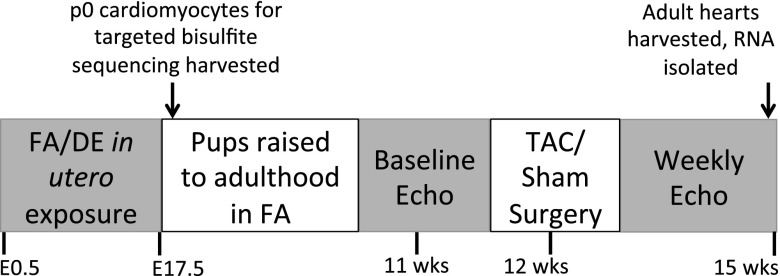
Female mice were paired with males for timed mating in FA. After observation of a vaginal plug, pregnant mice were put into FA or DE, with exposures beginning at embryonic day (E)0.5 and lasting until E17.5. Neonates were used for neonatal cardiomyocyte preparation (described below) or were saved for adult surgery. Offspring were raised in FA until 11 wk, at which time they underwent baseline echocardiography and TAC at 12 wk using a 27-gauge needle. Serial echocardiographic measurement, gravimetry, and histology for these mice have been previously published (12). Hearts from these mice were harvested at 15 wk, rapidly frozen in liquid nitrogen, and stored at −80°C before RNA isolation. The in utero exposure and sample collection scheme is displayed in Fig. 1.
Figure 1.
Exposure and sample collection scheme.
RNA isolation, sequencing, and bioinformatics analysis
RNA was isolated from fresh-frozen hearts of 3 mice per group by using an miRNeasy Kit (Qiagen, Germantown, MD, USA) according to manufacturer protocol. RNA sequencing was performed using the Ion Torrent Proton (Thermo Fisher Scientific, Waltham, MA, USA). In brief, RNA samples were depleted of ribosomal RNA by using the Low Input RiboMinus Eukaryotic System kit (v.2; Thermo Fisher Scientific). cDNA library was then created with the ribosomal-depleted RNA by using the Ion Total RNA sequencing kit (v.2; Thermo Fisher Scientific). This included fragmentation via RNase III, hybridizing and ligating the RNA, performing reverse transcription, and amplification. Samples were labeled with the Ion Xpress RNA sequencing barcode 1–16 kit (Thermo Fisher Scientific) to enable the multiplexing of 4 samples per run. Templating, loading, and sequencing were carried out by using the Ion PI Template OT2 200 Kit (v.2), the Ion PI Chip Kit (v.2), and the Ion PI Sequencing 200 Kit (v.2), respectively (all from Thermo Fisher Scientific). Data were generated by using the included Ion Reporter software (Thermo Fisher Scientific). Sequences were aligned to the mm10 mouse genome by using the subread aligner (20), and counts were generated by using the Bioconductor Rsubread package (University of California, Santa Cruz, CA, USA). Comparisons were performed by using the Bioconductor edgeR package (Roswell Park Cancer Institute, Buffalo, NY, USA; bioconductor.org). Genes were identified as significantly different between groups if they had both a false discovery rate (FDR)–adjusted value of P ≤ 0.05 and an absolute fold change of ≥1.5. The full data set has been deposited in the National Center for Biotechnology Information (NCBI) GEO Database (Accession No. GSE91398; Bethesda, MD, USA: https://www.ncbi.nlm.nih.gov/genbank/).
Quantitative RT-PCR analysis
cDNA was synthesized with the iScript Reverse Transcription Supermix kit (Bio-Rad, Hercules, CA, USA) using RNA from 4 FA sham-treated, 7 FA TAC, 4 DE sham-treated, and 6 DE TAC hearts, each group including the 3 samples used in RNA sequencing, and differential gene expression was validated by quantitative RT-PCR using the iTaq Universal Sybr Green Supermix (Bio-Rad). The following primers were used for validation: miR133a-2: (forward) 5′-GCCAAATGCTTTGCTGAAGCTG-3′, (reverse) 5′-GCTGGTTGAAGGGGACCAAA-3′; Ptprf: (forward) 5′-CAACGATGGGCTCAAGTTCT-3′, (reverse) 5′-TTCTTGGGCTTGTTCACCTC-3′; Pamr1: (forward) 5′-CCCAGGAAAGAAGGAAGTGG-3′, (reverse) 5′-GGCAGCTCTTGCAGTTTTCA-3′; and 18S: (forward) 5′- GGACAGGATTGACAGATTGATAG-3′, (reverse) 5′-ATCGCTCCACCAACTAAGAA-3′. Samples were cycled at 95°C for 15 s, 60°C for 1 min for 40 cycles. Samples were run on the 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Expression of Ptprf, miR133a-2, and Pamr1 was normalized to ribosomal RNA 18S. Statistical analysis was carried out by using 2-way ANOVA for comparison of expression values and with linear regression for comparison of expression value to ventricle weight.
Assessment of candidate gene expression in human heart failure samples
To determine whether the identified candidate genes demonstrated differential expression in human heart failure, we examined their expression patterns in a published human nondiabetic heart failure and control myocardial biopsy data set (21) that is available via the NCBI GEO database (Accession No. GSE26887). Statistically significant differences in gene expression were determined by using 1-way ANOVA with Bonferroni correction.
Isolation of neonatal cardiomyocytes
Upon birth, neonatal hearts were harvested, trimmed of surrounding vascular and atrial tissue, and dissociated as previously described (22, 23). Dissociation was performed by using 1 mg/ml Liberase TH (Roche, Pleasanton, CA, USA) in 1× HBSS by incubating hearts at 37°C for 5 min in solution, with pipetting to release cells after incubation. Medium that contained released neonatal cardiomyocytes was adjusted to 20% FBS-DMEM, and cellular dissociation was continued until the majority of cells were released. Cells were then filtered by using a 70-μm sieve, re-eluted in 20% FBS-DMEM with 20 μm Ara-C, and incubated at 37° for 1 h to allow fibroblasts to attach to the plate. After incubation, media with suspended cardiomyocytes was carefully removed and spun, and purified cardiomyocyte pellets were collected and frozen at −80°C.
DNA isolation and targeted bisulfite sequencing
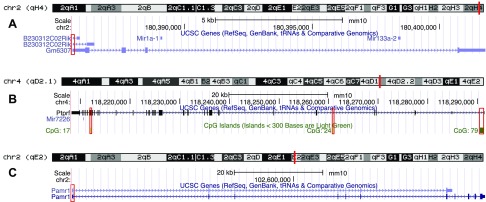
DNA was isolated from purified neonatal cardiomyocytes of 8 DE- and 6 FA-exposed p0 mice by using the DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer’s protocol. DNA was bisulfite-converted by using EpiTect Fast Bisulfite Conversion kit (Qiagen) according to the manufacturer’s protocol. Converted DNA was amplified by using HotStarTaq Master Mix (Qiagen), activated at 95°C for 15 min, cycled at 94°C for 30 s, 63°C for 45 s (−1°C/cycle for the first 10 cycles), 68°C for 1 min for 10 cycles, for 40 cycles. The following primers were used, targeting the regions boxed in red shown in Fig. 2:
Figure 2.
Genomic regions for bisulfite sequencing. Genomic regions highlighted in red boxes are areas that were targeted for bisulfite sequencing. Targets correspond to GM6307 promoter and first exon (A); Ptprf CpG islands 17, 24, and 79 (B); and Pamr1 promoter and first exon (C).
GM6307 promoter: (forward) 5′-GTATTGGGTTTTGTTAAAAATAGTAGG-3′, (reverse) 3′-CCTCACCTTAATAATTCTTATATACCC-5′; GM6307 exon 1.1: (forward) 5′-GGGTATATAAGAATTATTAAGGTGAGG-3′, (reverse) 5′-CAAAACCCAACTACACACCTTAC-3′; GM6307 exon 1.2: (forward) 5′-GTAGGAGGGTTGGGGGAA-3′, reverse: 5′-CAACCAACTAAAAAAAACTCACC-3′; Ptprf CpG17: (forward) 5′-GTATTTGGTTTAGTTTTTGGATGTG-3′, (reverse) 5′-CAAACTCTACTCTTCTCCTCAATAC-3′; Ptprf CpG24: (forward) 5′-TTTTTAGGTGTTGTAAAGGAAGTTTAAT-3′, (reverse) 5′-TACACTATTCTAAACCCTAACAAAC-3′; Ptprf CpG79.1: (forward) 5′-ATTGGAAAGGGAGAGAAATTTA-3′, (reverse) 5′-CCCCCRTACAAAATTTCCC-3′; Ptprf CpG79.2: (forward) 5′-GGGAAATTTTGTAYGGGGG-3′, (reverse) 5′-CCTACAACTCCACTCCTCTACAAAC-3′; Pamr1 promoter: (forward) 5′-GGTTTTAGTTAGTGTTGTTTGTTATA-3′, (reverse) 5′-AACTCTCCCACCCTCAACTAAAA-3′; and Pamr1 exon 1: (forward) 5′-TTTTAGTTGAGGGTGGGAGAGTT-3′, (reverse) 5′-CAAAAAACTCCTAATCACTAACAATAC-3′.
Bisulfite-converted DNA was sequenced in the University of Washington Northwest Genomics Center by using an Illumina MiSeq (Illumina, San Diego, CA, USA) in paired-end mode, 250-bp read length. Samples were sequenced to a mean depth of approximately 0.5 million reads. Adapter sequences and low-quality bases were removed from the raw reads by using Trimmomatic (v.0.32) (24). In brief, we removed Illumina TruSeq3 adaptors, then excluded any bases on either end of the read with a quality score <3, then used a moving window approach to trim off any portion of the read in which the average quality score over a 4-base moving window dropped <15. Any reads that were <36 bp in length after trimming were excluded. We then aligned trimmed reads against the mouse mm10 genome by using the Bismark aligner (v.0.15.0) (25) in conjunction with the bowtie aligner (v.0.12.8) (26). The trimming process resulted in a set of reads that remained paired as well as sets of reads that were unpaired—because the matching pair is missing or was excluded. We aligned the paired and unpaired reads separately, then combined the resulting BAM files before extracting methylation calls by using Bismark’s methylation_extractor function.
We read the per-base methylation calls from Bismark into R by using the Bioconductor bsseq package (27). To minimize low-quality methylation calls, we excluded any CpG with a read coverage of <5 in more than one half of samples. Individual CpG methylation estimates are known to be highly variable, so we smoothed the methylation estimates over adjacent bases by computing a moving average of all CpG sites within a 100-base moving window. We then made comparisons between groups at each CpG site by using the Bioconductor DSS package (28). In brief, we modeled the smoothed methylation estimates (at each CpG) on the basis of the β-binomial distribution, estimating the dispersion parameter by using a bayesian hierarchical model. We then computed a Wald statistic for each CpG site. Our goal was to find genomic regions that seem to be consistently differentially methylated between DE- and FA-exposed mice. We defined a differentially methylated genomic region on the basis of several criteria: the region had to be at least 50 bp long and contain at least 3 CpGs, and at least 50% of the CpGs had to have a Wald value of P < 0.01.
RESULTS
Transcriptional profiling by RNA sequencing reveals candidate target genes in the heart induced by in utero DE exposure
Table 1 shows the various comparison groups, the number of candidates that were identified by unadjusted values of P < 0.05, and those that were identified after multiple hypothesis correction controlling for FDR. The number of genes that were differentially expressed (FDR < 0.05 and absolute fold change > 1.5) after TAC was 880 in DE-exposed groups and 432 in FA-exposed groups. In contrast, the number of genes that were differentially expressed after DE exposure was 5 in the sham-treated group and 2 in the TAC group. These findings indicate that the magnitude of the TAC effect is much greater than that of the DE effect. Because gene expression after TAC has been studied extensively, we have focused primarily on genes that were affected by DE and do not discuss genes induced by TAC any further.
TABLE 1.
Number of genes that were altered in expression as a result of exposure, surgery type, or interaction
| Comparison | Unadjusted P < 0.05 | FDR < 0.05 |
|---|---|---|
| DE TAC vs. DE sham | 1375 | 880 |
| DE sham vs. FA sham | 316 | 5 |
| DE TAC vs. FA TAC | 153 | 2 |
| FA TAC vs. FA sham | 931 | 432 |
| Interaction | 321 | 3 |
Table 2 shows the 5 genes that were differentially expressed in DE and FA groups after sham surgery. Three of these genes, Gm8841, Pcdh1, and Bbip1, demonstrate a reduction in expression after DE exposure, whereas the remaining 2 genes, Mir133a-2 and Gck, show increased expression. Table 3 shows the 2 genes that were differentially expressed in DE and FA groups after TAC surgery. Ptprf demonstrates increased expression, whereas Pdk4 shows reduced expression. Table 4 lists genes that were identified in the interaction comparison, which identifies genes that respond differently to TAC, depending on exposure. These genes included Ptprf, Mir133a-2, and Pamr1.
TABLE 2.
Top 5 specific genes induced by DE in sham-treated animals (i.e., DE sham-treated vs. FA sham-treated)
| Gene symbol | LogFC | P | FDR |
|---|---|---|---|
| Gm8841 | −1.140 | 3.33e–08 | 0.000344 |
| Pcdh1 | −0.895 | 6.97e–07 | 0.002490 |
| Bbip1 | −1.130 | 7.24e–07 | 0.002490 |
| Mir133a-2 | 1.970 | 1.21e–05 | 0.027200 |
| Gck | 1.180 | 1.32e–05 | 0.027200 |
TABLE 3.
Specific genes induced by DE in TAC-treated animals (i.e., DE TAC vs. FA TAC)
| Gene | LogFC | P | FDR |
|---|---|---|---|
| Ptprf | 1.580 | 6.09e–08 | 0.000628 |
| Pdk4 | −1.100 | 1.25e–06 | 0.006440 |
FDR, false discovery rate.
TABLE 4.
Specific genes showing a differential response to TAC, depending on exposure (i.e., interaction)
| Gene | logFC | P | FDR |
|---|---|---|---|
| Ptprf | −2.380 | 2.29e–08 | 0.000236 |
| Mir133a-2 | 3.120 | 6.64e–07 | 0.003430 |
| Pamr1 | −2.630 | 4.03e–06 | 0.013900 |
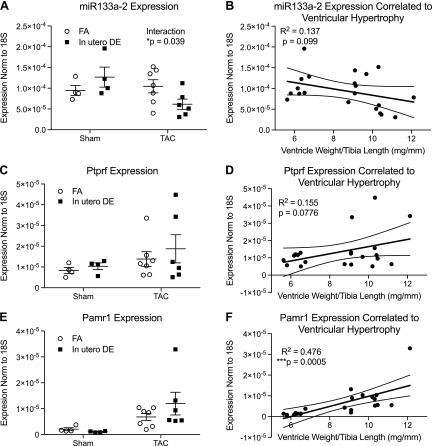
Additional validation of individual candidate gene expression from the interaction group by quantitative RT-PCR revealed interesting patterns of gene expression. On the basis of their statistically significant differences in expression across all 4 surgery and exposure conditions, we concluded that these candidates are likely to be important in the mediation of DE-induced susceptibility to heart failure. Expression of miR133a-2 demonstrated divergent regulation in adult hearts after exposure to DE in utero. DE-exposed mice that underwent sham surgery had elevated expression compared with FA controls, but TAC surgery induced a marked suppression of expression, whereas FA-exposed animals showed an induction of expression (Fig. 3A). Reduced expression of miR133a-2 after TAC is associated with worse heart failure and fibrosis. These findings are consistent with the previously reported role of miR133a-2 in functioning to suppress pathologic cardiac hypertrophy and fibrosis (29, 30).
Figure 3.
Expression of miR133a-2, PTPRF, and Pamr1 after in utero DE exposure and TAC surgery, correlated with ventricular weight/tibia length measurements. A, C, E) ΔΔCt values for miR133a-2 (A), Ptprf (C), and Pamr1 (E) in the 4 categories: FA sham-treated, DE sham-treated, FA TAC, and DE TAC. B, D, F) Corresponding linear regression values for miR311a-2 (B), Ptprf (D), and Pamr1 (F), plotting expression as a function of ventricle weight normalized to tibia length. *P < 0.05.
Expression of Ptprf, which encodes the protein, tyrosine phosphatase, receptor type, F (also known as leukocyte common antigen-related phosphatase), was not significantly changed by DE in mice that underwent sham surgery. In mice that underwent TAC, those that were exposed to DE in utero showed an increase in expression compared with those that were exposed to FA (Fig. 3C). This elevated expression is associated with worse heart failure and cardiac fibrosis, and is consistent with a causal role for PTPRF in promoting pathologic hypertrophy and heart failure.
Expression of Pamr1, which encodes peptidase domain-containing associated with muscle regeneration 1, was also increased as a result of exposure. Pamr1 expression is increased in both FA- and DE-exposed mice after TAC, although the increase is larger in the DE cohort. Expression of Pamr1 also demonstrated a significant correlation with the ventricular weight-to-tibia length ratio (Fig. 3E). This elevated Pamr1 expression, like that of Ptprf, is associated with worse heart failure and cardiac fibrosis, and is also consistent with a causal role for Pamr1 in the promotion of pathologic hypertrophy and heart failure.
Ptprf, miR133a-2, and Pamr1 gene expression are altered in a human heart failure gene expression data set
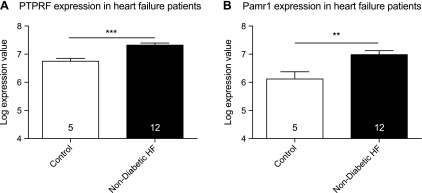
To determine whether the identified target genes demonstrated altered expression in human heart failure, we examined their expression in a publicly available Affymetrix data set (NCBI, Accession No. GSE26887) that was derived from human myocardial biopsies that were taken from patients with nondiabetic heart failure or control hearts (21). Mir133a expression is reduced in both forms of heart failure, which is consistent with a role in protecting against heart failure; however, examination of Ptprf and Pamr1 expression indicates that both genes show significant increases in expression in nondiabetic heart failure compared with controls (Fig. 4). These expression patterns are consistent with a role for these genes in contributing to human heart failure.
Figure 4.
Expression of Ptprf (A) and Pamr1 (B) in human heart failure samples. Significance was calculated by using 1-way ANOVA with post hoc Bonferroni. Data were derived from publicly available Affymetrix data set (NCBI GEO Accession No. GSE26887). **P < 0.005, ***P < 0.0005.
DNA methylation patterns at the Ptprf, Pamr1, and miR133a-2 loci
In our model of DE-related heart failure, exposure to DE occurred only during gestation, with no exposure after birth, yet these mice developed significantly worsened heart failure as adults after TAC. This suggests that long-lasting epigenetic modifications, such as DNA methylation, are responsible for this susceptibility. To test this, we performed targeted bisulfite sequencing at CpG islands and promoter regions that were associated with Ptprf, miR133a-2, and Pamr1 from neonatal cardiomyocytes of mice that were exposed to FA or DE in utero. We found that whereas in utero DE exposure did not induce significant differences in DNA methylation in either the Pamr1 or Ptprf loci (data not shown), the first exon of GM6307—the gene in which miR133a-2 resides—had decreased methylation on the basis of the criteria discussed above and in the legend for Fig. 5. This finding is consistent with the elevated expression of miR133a-2 in hearts from mice that were exposed to DE in the sham-surgery group (Fig. 3A).
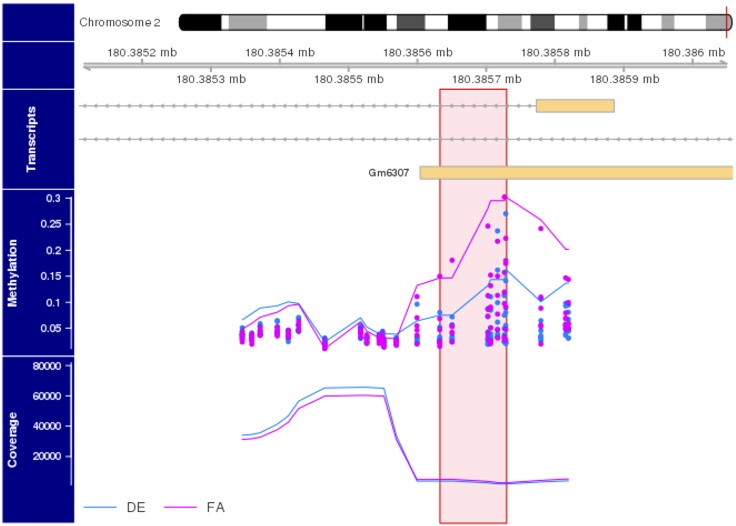
Figure 5.
Targeted bisulfite sequencing of GM6307 promoter and exon 1. The upper portion of the plot contains a karyogram of chromosome2, with a vertical red bar showing the region of interest; the horizontal bar just below the karyogram shows the zoomed-in region of the chromosome being inspected. The transcripts portion of the plot presents Universiy of California, Santa Cruz (UCSC) genome browser-style transcripts, where the orange bars represent exons and the gray lines represent introns. Arrows in the intronic region represent the direction of transcription. Below that, we present the observed (individual points) and smoothed (lines) percent methylation estimates for CpGs in this genomic region. Pink points and lines represent FA samples, whereas blue represent DE samples. In the lowest (coverage) portion of the plot we present the sequencing read depth (e.g., the number of sequencing reads that aligned over each CpG). The pink vertical bar indicates the genomic region that seems to be differentially methylated. This region spans 96 bp and contains 7 CpGs, all of which indicate a decrease in methylation in DE-exposed animals at a Wald value of P < 0.01.
DISCUSSION
We have demonstrated that gene transcription is significantly altered in mice as result of in utero DE exposure and adult TAC surgery. We identified the genes Ptprf, miR133a-2, and Pamr1 as being differentially regulated and verified these findings by quantitative RT-PCR. To the best of our knowledge, these genes have not been previously identified as being responsive to DE exposure in the heart. Whereas the exposure level used in this study (300 μg/m3) is well above the 2015 national average for the United States (8 μg/m3), such countries as Egypt and Saudi Arabia have averages of approximately 100 μg/m3 (31), and in 2013, Beijing reached a record of almost 1000 μg/m3. In addition, occupational exposure is of great concern. One study demonstrated that workers in high-exposure settings, such as tunnel construction, can receive exposures of up to 400 μg/m3 PM2.5 (32), and those who work in steel plants can reach exposure levels >1200 μg/m3 PM10 (13). Our exposure levels in this study thus reflect real-world human exposure.
It has been postulated that PAMR1 plays a role in the regeneration of skeletal muscle on the basis of its expression pattern (33), to play a role in the acute insulin response to glucose (34), and to be suppressed by promoter methylation in breast cancer (35). To the best of our knowledge, no functional studies have been published, and a role for PAMR1 in cardiac hypertrophy and heart failure has not been reported. It is unclear whether these changes in gene expression have individual roles in the development of DE-induced heart failure, or if the combination of these gene changes is required. The expression pattern indicates that increased expression is associated with increased cardiac hypertrophy and fibrosis.
PTPRF and other tyrosine phosphatases play critical roles in the mediation of signal transduction by limiting intracellular phosphorylation cascades that occur after autophosphorylation of receptor tyrosine kinases (36, 37). Overexpression of Ptprf has been shown to limit insulin sensitivity and, thus, promote insulin resistance in mouse models, whereas knockdown enhances insulin sensitivity in cultured cells (38–40). Common single-nucleotide polymorphisms in Ptprf are associated with the development of coronary artery disease in patients with type 2 diabetes (41). Previous work using in vitro models has shown that Ptprf expression is decreased after treatment with TNF-α, and that Ptprf expression is readily modified to regulate cell signaling (42). To the best of our knowledge, PTPRF has not previously been associated with the development of heart failure. On the basis of its expression pattern, we hypothesize that exaggerated expression in hearts after in utero DE exposure likely either potentiates pathologic hypertrophic signaling or limits antihypertrophic signaling.
A role for miR133a in cardiac hypertrophy is better established. miR133a-2 encodes a premature form of miR133a, which controls cardiac development (43). Reduced expression of miR133a leads to the enhancement of cardiac hypertrophy in vitro by increased expression of its target, inositol 1,4,5-trisphosphate type II (44). Overexpression of miR133a in the heart has been reported to suppress cardiac fibrosis, but not cardiac hypertrophy, after TAC (30), which is consistent with our results. MiR133a loss of function has previously been associated with myocyte apoptosis after ischemia-reperfusion via increased expression of caspase 9 (45) and skeletal muscle myopathy via an effect on its target, dynamin 2 (46). DNA hypermethylation of a CpG island that was associated with the miR133a-2 cluster and reduced miR133a-2 expression have been associated with colorectal cancer (47). In comparison, our results demonstrate that in utero DE resulted in a ∼2-fold increase in miR133a-2 expression at baseline in our sham-treated mice (Fig. 3A and Table 2). Our previously reported baseline phenotypic assessment in sham-treated mice did not reveal in utero DE exposure to have any effect on cardiac function or myocyte hypertrophy (12). We observed this increased miR133a-2 expression to be down-regulated after TAC in in utero DE-exposed mice (Fig. 3A and Table 1), with a simultaneous increase in left ventricle wall thickness, fibrosis, and decreased fractional shortening, as previously reported (12). This down-regulation of miR133a-2 with concomitant exacerbation of heart failure with reduced ejection fraction may suggest that the increased miR133a-2 expression at baseline played a protective role in preserving cardiac function, and that loss of this increased expression results in the exacerbation of heart failure. Our DNA methylation studies in neonatal cardiomyocytes reveal hypomethylation within the first exon of GM6307, which is consistent with our observation of increased miR133a-2 expression in baseline sham-treated mice. The loss of increased miR133a-2 expression in in utero DE-exposed mice in heart failure implies that there may be dynamic modifications of DNA methylation or multifactorial changes in chromatin assembly and transcription factor accessibility modified by in utero DE exposure. These observations suggest that baseline hypomethylation and increased miR133a-2 expression at baseline are advantageous adaptations, but that the dysregulation of its expression in heart failure is uniquely affected by in utero DE exposure. Loss of miR133a-2 overexpression in DE-treated hearts after TAC may facilitate pathologic hypertrophy by enhanced IP(3)RII signaling and increased apoptosis mediated by caspase 9 (44).
The effect of environmental toxicant exposure on cardiac hypertrophy via regulation of miR133a expression and DNA methylation has been previously reported (48). Exposure to the PAH, phenanthrene, causes cardiac and myocyte hypertrophy in both in vivo and in vitro models. In male Sprague-Dawley rats and the H9C2 rat cardiomyoblast cell line, phenanthrene exposure was associated with a decrease in miR133a expression and a simultaneous increase in protein levels of important miR133a cardiac hypertrophy targets, Cdc42 and RhoA. This decrease in miR133a expression was associated with an increase in DNA methylation at 5 CpG sites that were located near the putative transcription start site of miR133a in H9C2 cells. Overexpression of miR133a via transfection of miR133a mimetics, or inhibition of DNA methylation with a DNA methylation inhibitor (5-aza-2′-deoxycytodine), largely eliminated the hypertrophic effect of phenanthrene. PAHs are a significant component of DE particulate mass, and we have previously reported our DE exposure system to generate DE with a mass fraction of particle-bound PAH at 20 ng/μg (11). Phenanthrene is a major constituent of environmental PAHs and has been reported to be a significant component of DE (49), which suggests that PAHs, including phenanthrene, may possibly contribute to our observed effects on heart failure and DNA methylation. Although we observed in utero DE exposure to result in altered miR133a-2 expression in a manner that was similar to that reported previously (48), our observed DNA methylation results of hypomethylation of the first exon are in contrast and perhaps suggests a unique epigenetic mechanism of action in in utero DE-induced adult susceptibility to heart failure.
One surprising finding of our work is that relatively few genes are significantly affected in the heart after in utero DE exposure, but we believe that this finding is likely a result of dilutional effects from using whole-heart RNA. Future studies using adult cardiomyocyte RNA sequencing would likely generate additional candidate genes. Another unaddressed question is the effect of in utero DE exposure on the global DNA methylation pattern in cardiomyocytes. We have attempted to perform genome-wide bisulfine sequencing in whole adult heart DNA, but to date no differentially methylated regions have been consistently identified, likely because of the dilutional effect of noncardiomyocytes. Previous studies of methylation in adult cardiomyocytes have relied on the purification of adult cardiomyocytes (50). A future study that analyzes DNA methylation patterns in isolated adult cardiomyocytes will address this question.
Another potential caveat of studying whole-heart RNA is that gene expression changes may reflect a shift in cell population rather than a change in expression within a given cell type. We do not favor this hypothesis, as both miR133a-2 and Pamr1 are expressed predominantly in muscle tissue (30, 33), and the relative contribution of additional cells, such as inflammatory cells, is still likely to be modest compared with cardiomyocytes and fibroblasts.
Although it has been demonstrated in multiple studies that exposure to PM2.5 is associated with changes in DNA methylation (13–15), the mechanism is unclear. In our study, we initially assessed whether the DNA methylases, DNMT1, DNMT2, or DNMT3, were differentially expressed by using quantitative RT-PCR, but did not observe any significant difference, which is consistent with our RNA sequencing data (data not shown). It has been shown that exposure to DE increases reactive oxygen species production (51, 52), which, in turn, can interact with DNA to oxidize methylcytosine to hydroxymethylcytosine (hmC). hmC has been shown to prevent methyl-binding proteins (MBPs) from binding to methylated cytosines (53). This can prevent the normal chromatin silencing that would occur at these sites, which allows for transcriptional expression. Furthermore, oxidation to hmC has been shown itself to be an intermediary to the potential loss of DNA methylation (54, 55). In addition to the role of hmC as both a blockade of chromatin silencing and a stepping stone to loss of methylation, the guanine oxidative lesion, 8-oxoguanine, also can inhibit the binding of MBPs, which prevents the silencing of chromatin regions (53). Taken together, these processes could explain the observation of changes in DNA methylation associated with exposure to DE.
The findings of decreased DNA methylation at the first exon of GM6307 are intriguing, as it gives credence to the idea that in utero exposure to DE affects the developing heart via altered epigenetic patterning. DNA methylation of the first exon has been shown to control gene regulation (56), and decreased DNA methylation in DE-exposed mice correlates with the higher expression observed in adults that have undergone sham surgery. However, the DE-exposed, TAC surgery group demonstrated a reduction in expression, which is not explained by our DNA methylation findings and perhaps is because of other epigenetic modifications or altered signaling as a result of the surgery.
A limitation of our work is that the changes in gene expression represent associations and do not demonstrate causality. Our initial attempts to investigate effects of in utero DE exposure on TAC-induced heart failure in mice that overexpress miR133a in the heart were inconclusive because the wild-type FVB strain that has been used to generate these mice is not susceptible to TAC-induced heart failure after in utero DE exposure (data not shown), in contrast to the C57BL/6 strain that was used in this study and previously reported studies (12). We also isolated neonatal cardiomyocytes from this strain after in utero DE exposure to assess whether cardiomyocytes from exposed animals were more susceptible to hydrogen peroxide–induced apoptosis but did not observe increased susceptibility (unpublished data). Furthermore, we overexpressed both miR133a and PTPRF in H9c2 myoblasts by lentiviral transduction to assess whether stable changes in the expression of these genes affected the susceptibility of these cells to hydrogen peroxide–induced apoptosis. Unfortunately, in our studies, these cells rapidly undergo necrosis and, thus, an effect of candidate gene overexpression on apoptosis could not be measured (data not shown). Future studies on genetically modified mice that were generated on a C57BL/6 background are needed to truly assess the contribution of these genes to DE-induced susceptibility to heart failure, but are beyond the scope of the current study.
CONCLUSIONS
We have reported significant transcriptional and DNA methylation changes associated with in utero DE exposure and adult TAC surgery. Additional analysis of how these genes impact adult susceptibility to heart failure is in progress. Both gain- and loss-of-function models will be informative for the development of air pollution–associated heart failure treatments. miR133a has already been identified as a biomarker of patient outcome after valve replacement (57) and is currently being studied as a treatment postinfarction to improve patient outcomes (58). Determining whether miR133a can play a similarly protective role in our model of DE-induced heart failure—alongside the potential benefits of decreasing Ptprf and Pamr1 expression—will pave the way for further drug development for patients with air pollution–associated heart failure.
ACKNOWLEDGMENTS
The authors thank James Stewart (University of Washington) for expert technical assistance with diesel exhaust exposures. J.M.G. was supported by U.S. National Institutes of Health (NIH) National Institute of Environmental Health Sciences (NIEHS) Training Grant T32 ES007032. C.S.W. was supported by NIH National Heart, Lung, and Blood Institute Training Grant T32 HL007312. J.W.M. and T.K.B. were supported by NIH NIEHS Grant P30ES07033 to the University of Washington Center for Exposures, Diseases, Genomics and Environment (EDGE). M.T.C. was supported by NIH NIEHS Grant ES023015 and American Heart Association Grant 15GRNT25390012.
Glossary
- DE
diesel exhaust
- FA
filtered air
- FDR
false discovery rate
- hmC
hydroxymethylcytosine
- MBP
methyl-binding protein
- NCBI
National Center for Biotechnology Information
- PAH
polycyclic aromatic hydrocarbon
- PM2.5
particulate matter ≤2.5 μm
- TAC
transverse aortic constriction
AUTHOR CONTRIBUTIONS
J. M. Goodson contributed to hypotheses generation and experimental design, conducted primary cell harvests, prepared samples for bisulfite sequencing, interpreted data, and is the primary author of the manuscript; C. S. Weldy prepared RNA, performed quantitative RT-PCR and echocardiography, interpreted data, and contributed to the preparation of the manuscript; J. W. MacDonald performed key bioinformatics analyses for RNA sequencing and targeted bisulfite sequencing and contributed to the preparation of the manuscript; Y. Liu performed animal surgeries; T. K. Bammler contributed to bioinformatics analyses and to the preparation of the manuscript; W.-M. Chien assisted in molecular biology experiments and contributed to the preparation of the manuscript; M. T. Chin contributed to hypotheses generation and experimental design, contributed to the preparation of the manuscript, and was responsible for overall coordination of the project; and all authors read and approved of the final copy of the manuscript.
REFERENCES
- 1.Pope C. A. III, Burnett R. T., Thun M. J., Calle E. E., Krewski D., Ito K., Thurston G. D. (2002) Lung cancer, cardiopulmonary mortality, and long-term exposure to fine particulate air pollution. JAMA. 287, 1132–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ni L., Chuang C. C., Zuo L. (2015) Fine particulate matter in acute exacerbation of COPD. Front. Physiol. 6, 294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brook R. D., Rajagopalan S., Pope C. A. III, Brook J. R., Bhatnagar A., Diez-Roux A. V., Holguin F., Hong Y., Luepker R. V., Mittleman M. A., Peters A., Siscovick D., Smith S. C. Jr., Whitsel L., Kaufman J. D.; American Heart Association Council on Epidemiology and Prevention, Council on the Kidney in Cardiovascular Disease, and Council on Nutrition, Physical Activity and Metabolism (2010) Particulate matter air pollution and cardiovascular disease: an update to the scientific statement from the American Heart Association. Circulation 121, 2331–2378 [DOI] [PubMed] [Google Scholar]
- 4.Krishnan R. M., Adar S. D., Szpiro A. A., Jorgensen N. W., Van Hee V. C., Barr R. G., O’Neill M. S., Herrington D. M., Polak J. F., Kaufman J. D. (2012) Vascular responses to long- and short-term exposure to fine particulate matter: MESA air (multi-ethnic study of atherosclerosis and air pollution). J. Am. Coll. Cardiol. 60, 2158–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pope C. A. III, Muhlestein J. B., May H. T., Renlund D. G., Anderson J. L., Horne B. D. (2006) Ischemic heart disease events triggered by short-term exposure to fine particulate air pollution. Circulation 114, 2443–2448 [DOI] [PubMed] [Google Scholar]
- 6.Wang Y., Eliot M. N., Wellenius G. A. (2014) Short-term changes in ambient particulate matter and risk of stroke: a systematic review and meta-analysis. J. Am. Heart Assoc. 3, e000983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van den Hooven E. H., Pierik F. H., de Kluizenaar Y., Willemsen S. P., Hofman A., van Ratingen S. W., Zandveld P. Y., Mackenbach J. P., Steegers E. A., Miedema H. M., Jaddoe V. W. (2012) Air pollution exposure during pregnancy, ultrasound measures of fetal growth, and adverse birth outcomes: a prospective cohort study. Environ. Health Perspect. 120, 150–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saenen N. D., Vrijens K., Janssen B. G., Madhloum N., Peusens M., Gyselaers W., Vanpoucke C., Lefebvre W., Roels H. A., Nawrot T. S. (2016) Placental nitrosative stress and exposure to ambient air pollution during gestation: a population study. Am. J. Epidemiol. 184, 442–449 [DOI] [PubMed] [Google Scholar]
- 9.Barker D. J., Osmond C., Golding J., Kuh D., Wadsworth M. E. (1989) Growth in utero, blood pressure in childhood and adult life, and mortality from cardiovascular disease. BMJ 298, 564–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Rossem L., Rifas-Shiman S. L., Melly S. J., Kloog I., Luttmann-Gibson H., Zanobetti A., Coull B. A., Schwartz J. D., Mittleman M. A., Oken E., Gillman M. W., Koutrakis P., Gold D. R. (2015) Prenatal air pollution exposure and newborn blood pressure. Environ. Health Perspect. 123, 353–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weldy C. S., Liu Y., Chang Y. C., Medvedev I. O., Fox J. R., Larson T. V., Chien W. M., Chin M. T. (2013) In utero and early life exposure to diesel exhaust air pollution increases adult susceptibility to heart failure in mice. Part. Fibre Toxicol. 10, 59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weldy C. S., Liu Y., Liggitt H. D., Chin M. T. (2014) In utero exposure to diesel exhaust air pollution promotes adverse intrauterine conditions, resulting in weight gain, altered blood pressure, and increased susceptibility to heart failure in adult mice. PLoS One 9, e88582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tarantini L., Bonzini M., Apostoli P., Pegoraro V., Bollati V., Marinelli B., Cantone L., Rizzo G., Hou L., Schwartz J., Bertazzi P. A., Baccarelli A. (2009) Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Environ. Health Perspect. 117, 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baccarelli A., Wright R. O., Bollati V., Tarantini L., Litonjua A. A., Suh H. H., Zanobetti A., Sparrow D., Vokonas P. S., Schwartz J. (2009) Rapid DNA methylation changes after exposure to traffic particles. Am. J. Respir. Crit. Care Med. 179, 572–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janssen B. G., Godderis L., Pieters N., Poels K., Kiciński M., Cuypers A., Fierens F., Penders J., Plusquin M., Gyselaers W., Nawrot T. S. (2013) Placental DNA hypomethylation in association with particulate air pollution in early life. Part. Fibre Toxicol. 10, 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weldy C. S., Luttrell I. P., White C. C., Morgan-Stevenson V., Cox D. P., Carosino C. M., Larson T. V., Stewart J. A., Kaufman J. D., Kim F., Chitaley K., Kavanagh T. J. (2013) Glutathione (GSH) and the GSH synthesis gene Gclm modulate plasma redox and vascular responses to acute diesel exhaust inhalation in mice. Inhal. Toxicol. 25, 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gould T., Larson T., Stewart J., Kaufman J. D., Slater D., McEwen N. (2008) A controlled inhalation diesel exhaust exposure facility with dynamic feedback control of PM concentration. Inhal. Toxicol. 20, 49–52 [DOI] [PubMed] [Google Scholar]
- 18.Yin F., Lawal A., Ricks J., Fox J. R., Larson T., Navab M., Fogelman A. M., Rosenfeld M. E., Araujo J. A. (2013) Diesel exhaust induces systemic lipid peroxidation and development of dysfunctional pro-oxidant and pro-inflammatory high-density lipoprotein. Arterioscler. Thromb. Vasc. Biol. 33, 1153–1161 [DOI] [PubMed] [Google Scholar]
- 19.Liu Y., Chien W. M., Medvedev I. O., Weldy C. S., Luchtel D. L., Rosenfeld M. E., Chin M. T. (2013) Inhalation of diesel exhaust does not exacerbate cardiac hypertrophy or heart failure in two mouse models of cardiac hypertrophy. Part. Fibre Toxicol. 10, 49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao Y., Smyth G. K., Shi W. (2013) The subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greco S., Fasanaro P., Castelvecchio S., D’Alessandra Y., Arcelli D., Di Donato M., Malavazos A., Capogrossi M. C., Menicanti L., Martelli F. (2012) MicroRNA dysregulation in diabetic ischemic heart failure patients. Diabetes 61, 1633–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y., Yu M., Wu L., Chin M. T. (2010) The bHLH transcription factor CHF1/Hey2 regulates susceptibility to apoptosis and heart failure after pressure overload. Am. J. Physiol. Heart Circ. Physiol. 298, H2082–H2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiang F., Sakata Y., Cui L., Youngblood J. M., Nakagami H., Liao J. K., Liao R., Chin M. T. (2006) Transcription factor CHF1/Hey2 suppresses cardiac hypertrophy through an inhibitory interaction with GATA4. Am. J. Physiol. Heart Circ. Physiol. 290, H1997–H2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolger A. M., Lohse M., Usadel B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krueger F., Andrews S. R. (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen K. D., Langmead B., Irizarry R. A. (2012) BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 13, R83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng H., Conneely K. N., Wu H. (2014) A bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res. 42, e69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carè A., Catalucci D., Felicetti F., Bonci D., Addario A., Gallo P., Bang M. L., Segnalini P., Gu Y., Dalton N. D., Elia L., Latronico M. V., Høydal M., Autore C., Russo M. A., Dorn G. W. II, Ellingsen O., Ruiz-Lozano P., Peterson K. L., Croce C. M., Peschle C., Condorelli G. (2007) MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 13, 613–618 [DOI] [PubMed] [Google Scholar]
- 30.Matkovich S. J., Wang W., Tu Y., Eschenbacher W. H., Dorn L. E., Condorelli G., Diwan A., Nerbonne J. M., Dorn G. W. II (2010) MicroRNA-133a protects against myocardial fibrosis and modulates electrical repolarization without affecting hypertrophy in pressure-overloaded adult hearts. Circ. Res. 106, 166–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brauer M., Freedman G., Frostad J., van Donkelaar A., Martin R. V., Dentener F., van Dingenen R., Estep K., Amini H., Apte J. S., Balakrishnan K., Barregard L., Broday D., Feigin V., Ghosh S., Hopke P. K., Knibbs L. D., Kokubo Y., Liu Y., Ma S., Morawska L., Sangrador J. L., Shaddick G., Anderson H. R., Vos T., Forouzanfar M. H., Burnett R. T., Cohen A. (2016) Ambient air pollution exposure estimation for the Global Burden of Disease 2013. Environ. Sci. Technol. 50, 79–88 [DOI] [PubMed] [Google Scholar]
- 32.Lewné M., Plato N., Gustavsson P. (2007) Exposure to particles, elemental carbon and nitrogen dioxide in workers exposed to motor exhaust. Ann. Occup. Hyg. 51, 693–701 [DOI] [PubMed] [Google Scholar]
- 33.Nakayama Y., Nara N., Kawakita Y., Takeshima Y., Arakawa M., Katoh M., Morita S., Iwatsuki K., Tanaka K., Okamoto S., Kitamura T., Seki N., Matsuda R., Matsuo M., Saito K., Hara T. (2004) Cloning of cDNA encoding a regeneration-associated muscle protease whose expression is attenuated in cell lines derived from Duchenne muscular dystrophy patients. Am. J. Pathol. 164, 1773–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma P. R., Mackey A. J., Dejene E. A., Ramadan J. W., Langefeld C. D., Palmer N. D., Taylor K. D., Wagenknecht L. E., Watanabe R. M., Rich S. S., Nunemaker C. S. (2015) An islet-targeted genome-wide association scan identifies novel genes implicated in cytokine-mediated islet stress in type 2 diabetes. Endocrinology 156, 3147–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lo P. H., Tanikawa C., Katagiri T., Nakamura Y., Matsuda K. (2015) Identification of novel epigenetically inactivated gene PAMR1 in breast carcinoma. Oncol. Rep. 33, 267–273 [DOI] [PubMed] [Google Scholar]
- 36.Tonks N. K. (2013) Protein tyrosine phosphatases--from housekeeping enzymes to master regulators of signal transduction. FEBS J. 280, 346–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chagnon M. J., Uetani N., Tremblay M. L. (2004) Functional significance of the LAR receptor protein tyrosine phosphatase family in development and diseases. Biochem. Cell Biol. 82, 664–675 [DOI] [PubMed] [Google Scholar]
- 38.Zhang W. R., Li P. M., Oswald M. A., Goldstein B. J. (1996) Modulation of insulin signal transduction by eutopic overexpression of the receptor-type protein-tyrosine phosphatase LAR. Mol. Endocrinol. 10, 575–584 [DOI] [PubMed] [Google Scholar]
- 39.Zabolotny J. M., Kim Y. B., Peroni O. D., Kim J. K., Pani M. A., Boss O., Klaman L. D., Kamatkar S., Shulman G. I., Kahn B. B., Neel B. G. (2001) Overexpression of the LAR (leukocyte antigen-related) protein-tyrosine phosphatase in muscle causes insulin resistance. Proc. Natl. Acad. Sci. USA 98, 5187–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kulas D. T., Zhang W. R., Goldstein B. J., Furlanetto R. W., Mooney R. A. (1995) Insulin receptor signaling is augmented by antisense inhibition of the protein tyrosine phosphatase LAR. J. Biol. Chem. 270, 2435–2438 [DOI] [PubMed] [Google Scholar]
- 41.Menzaghi C., Paroni G., De Bonis C., Coco A., Vigna C., Miscio G., Lanna P., Tassi V., Bacci S., Trischitta V. (2008) The protein tyrosine phosphatase receptor type f (PTPRF) locus is associated with coronary artery disease in type 2 diabetes. J. Intern. Med. 263, 653–654 [DOI] [PubMed] [Google Scholar]
- 42.Ahmad F., Goldstein B. J. (1997) Functional association between the insulin receptor and the transmembrane protein-tyrosine phosphatase LAR in intact cells. J. Biol. Chem. 272, 448–457 [PubMed] [Google Scholar]
- 43.Liu N., Bezprozvannaya S., Williams A. H., Qi X., Richardson J. A., Bassel-Duby R., Olson E. N. (2008) microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 22, 3242–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drawnel F. M., Wachten D., Molkentin J. D., Maillet M., Aronsen J. M., Swift F., Sjaastad I., Liu N., Catalucci D., Mikoshiba K., Hisatsune C., Okkenhaug H., Andrews S. R., Bootman M. D., Roderick H. L. (2012) Mutual antagonism between IP(3)RII and miRNA-133a regulates calcium signals and cardiac hypertrophy. J. Cell Biol. 199, 783–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He B., Xiao J., Ren A. J., Zhang Y. F., Zhang H., Chen M., Xie B., Gao X. G., Wang Y. W. (2011) Role of miR-1 and miR-133a in myocardial ischemic postconditioning. J. Biomed. Sci. 18, 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu N., Bezprozvannaya S., Shelton J. M., Frisard M. I., Hulver M. W., McMillan R. P., Wu Y., Voelker K. A., Grange R. W., Richardson J. A., Bassel-Duby R., Olson E. N. (2011) Mice lacking microRNA 133a develop dynamin 2–dependent centronuclear myopathy. J. Clin. Invest. 121, 3258–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen W. S., Leung C. M., Pan H. W., Hu L. Y., Li S. C., Ho M. R., Tsai K. W. (2012) Silencing of miR-1-1 and miR-133a-2 cluster expression by DNA hypermethylation in colorectal cancer. Oncol. Rep. 28, 1069–1076 [DOI] [PubMed] [Google Scholar]
- 48.Huang L., Xi Z., Wang C., Zhang Y., Yang Z., Zhang S., Chen Y., Zuo Z. (2016) Phenanthrene exposure induces cardiac hypertrophy via reducing miR-133a expression by DNA methylation. Sci. Rep. 6, 20105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Westerholm R., Christensen A., Törnqvist M., Ehrenberg L., Rannug U., Sjögren M., Rafter J., Soontjens C., Almén J., Gränn K. (2001) Comparison of exhaust emissions from Swedish Environmental classified diesel fuel (MK1) and European Program on Emissions, Fuels and Engine Technologies (EPEFE) reference fuel: a chemical and biological characterization, with viewpoints on cancer risk. Environ. Sci. Technol. 35, 1748–1754 [DOI] [PubMed] [Google Scholar]
- 50.Gilsbach R., Preissl S., Grüning B. A., Schnick T., Burger L., Benes V., Würch A., Bönisch U., Günther S., Backofen R., Fleischmann B. K., Schübeler D., Hein L. (2014) Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nat. Commun. 5, 5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wauters A., Dreyfuss C., Pochet S., Hendrick P., Berkenboom G., van de Borne P., Argacha J. F. (2013) Acute exposure to diesel exhaust impairs nitric oxide-mediated endothelial vasomotor function by increasing endothelial oxidative stress. Hypertension 62, 352–358 [DOI] [PubMed] [Google Scholar]
- 52.Zuo L., Youtz D. J., Wold L. E. (2011) Particulate matter exposure exacerbates high glucose-induced cardiomyocyte dysfunction through ROS generation. PLoS One 6, e23116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valinluck V., Tsai H. H., Rogstad D. K., Burdzy A., Bird A., Sowers L. C. (2004) Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2). Nucleic Acids Res. 32, 4100–4108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hackett J. A., Sengupta R., Zylicz J. J., Murakami K., Lee C., Down T. A., Surani M. A. (2013) Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339, 448–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo J. U., Su Y., Zhong C., Ming G. L., Song H. (2011) Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145, 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brenet F., Moh M., Funk P., Feierstein E., Viale A. J., Socci N. D., Scandura J. M. (2011) DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One 6, e14524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.García R., Villar A. V., Cobo M., Llano M., Martín-Durán R., Hurlé M. A., Nistal J. F. (2013) Circulating levels of miR-133a predict the regression potential of left ventricular hypertrophy after valve replacement surgery in patients with aortic stenosis. J. Am. Heart Assoc. 2, e000211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dakhlallah D., Zhang J., Yu L., Marsh C. B., Angelos M. G., Khan M. (2015) MicroRNA-133a engineered mesenchymal stem cells augment cardiac function and cell survival in the infarct heart. J. Cardiovasc. Pharmacol. 65, 241–251 [DOI] [PMC free article] [PubMed] [Google Scholar]