Abstract
Chitin, a polymer of N-acetyl-D-glucosamine (GlcNAc), is a major structural component in chitin-containing organism including crustaceans, insects and fungi. Mammals express two chitinases, chitotriosidase (Chit1) and acidic mammalian chitinase (AMCase). Here, we report that pig AMCase is stable in the presence of other digestive proteases and functions as chitinolytic enzyme under the gastrointestinal conditions. Quantification of chitinases expression in pig tissues using quantitative real-time PCR showed that Chit1 mRNA was highly expressed in eyes, whereas the AMCase mRNA was predominantly expressed in stomach at even higher levels than the housekeeping genes. AMCase purified from pig stomach has highest activity at pH of around 2–4 and remains active at up to pH 7.0. It was resistant to robust proteolytic activities of pepsin at pH 2.0 and trypsin and chymotrypsin at pH 7.6. AMCase degraded polymeric chitin substrates including mealworm shells to GlcNAc dimers. Furthermore, we visualized chitin digestion of fly wings by endogenous AMCase and pepsin in stomach extract. Thus, pig AMCase can function as a protease resistant chitin digestive enzyme at broad pH range present in stomach as well as in the intestine. These results indicate that chitin-containing organisms may be a sustainable feed ingredient in pig diet.
Introduction
Pigs are an ideal biomedical model for human diseases and conditions bridging the gaps between mouse models and humans1. Pigs are more similar to humans in terms of genetics, digestive physiology2,3 and metabolism4 than mouse, and potentially, they could be used as a source of organs for human transplantations in the future5,6. Besides the biomedical purpose, pork is also an important dietary source for humans, accounting for more than half of the world’s meat consumption. Recently, its demands are increasing due to growing human population7,8.
Chitin, a linear polymer of β-1, 4-linked N-acetyl-D-glucosamine (GlcNAc), is the second most abundant natural polysaccharide in nature and functions as a major structural component in fungi, crustaceans, and insects9,10. Although mammals do not produce chitin, humans and mice express two active chitinases which belong to the family 18 of glycoside hydrolases10,11. Firstly, chitotriosidase (Chit1) was identified in macrophages of Gaucher disease patients12–14. AMCase was discovered later and was named for its acidic isoelectric point15. These mammalian chitinases have been considered as a protection against chitin-containing pathogens10,16.
Since AMCase expression is significantly altered under several pathological conditions such as asthma, allergic inflammation, ocular allergy, dry eye syndrome and gastric cancer17–23, it has attracted considerable attention. Some polymorphisms and haplotypes in the AMCase gene are associated with bronchial asthma in humans24–26. Recently, AMCase was shown to be a constitutively produced enzyme essential for chitin degradation in the airways to maintain lung functions27,28. In addition, AMCase plays role in the protective immune response to gastrointestinal nematodes in the host gastrointestinal tract (GIT)29.
Murine AMCase is most active at pH of around 2.015,30,31. Mouse stomach produces enormous quantities of AMCase mRNA and protein32,33. Thus, AMCase seems to function as a digestive enzyme that breaks down chitin-containing ingested material15,30–32. Since chitin has long been thought to be not degraded in the mammalian digestive system, it is sometimes included in animal feeds as dietary fiber34. Recently we showed that mouse AMCase and chicken Chia, a homologue of AMCase, can function as a protease-resistant major glycosidase under stomach and intestine conditions while degrading chitin substrates to GlcNAc dimer [(GlcNAc)2], a source of carbon, nitrogen and energy35,36. However, the physiological roles of the AMCase in other mammals remain unknown.
Here, we quantified expression levels of the chitinases in pig tissues by quantitative real-time PCR (qPCR). Also, we purified AMCase from pig stomach tissue and characterized its optimal condition and protease-resistance. We provide evidence that chitin-containing organisms can be digested under pig GIT condition which is supported by degradation products analysis and morphological analysis.
Results
Gene expression analysis of Chit1 and AMCase mRNAs in pig tissues
To study the in vivo regulation of pig Chit1 and AMCase genes expression, total RNA from various normal pig tissues was analyzed using a qPCR assay with a specifically designed standard DNA (Supplementary Fig. S1) as described in Methods.
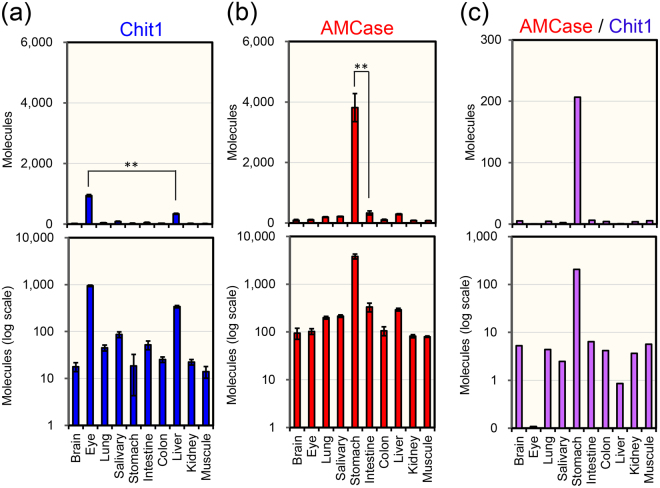
Clear tissue-specific pattern was observed in both chitinases mRNAs expression (Fig. 1a and b, upper panels). High levels of Chit1 mRNA were detected in the eye (Fig. 1a, upper panel), followed by liver, salivary gland, intestine and lung (Fig. 1a, lower panel). AMCase mRNA was predominantly detected in the stomach (Fig. 1b, upper panel), followed by intestine, liver, salivary gland and lung (Fig. 1b, lower panel). The levels of AMCase in all tissues, and particularly in stomach were markedly higher than those of Chit1 (Fig. 1c, upper panel) except for the eye (Fig. 1c, lower panel).
Figure 1.
Expression of Chit1 and AMCase mRNAs in pig tissues. Quantification of (a) Chit1 and (b) AMCase mRNAs in ten major pig tissues. Both chitinases were quantified by qPCR using the standard DNA. All values obtained were expressed as molecules per 10 ng of total RNA. (c) Ratios of AMCase to Chit1. All mRNA copy numbers were derived based on the same standard dilutions. The upper panels indicate the actual number, whereas the lower panel shows each value on logarithmic scale. Each reaction was performed in triplicate. **p < 0.01. P-values were determined using Student’s t-test.
AMCase mRNA levels in pig stomach
Next, we evaluated the AMCase mRNA levels in pig stomach in detail. The mRNAs of pepsinogen A and C, H+/K+-ATPase and AMCase were expressed at much higher levels than those of the housekeeping genes (Fig. 2). The expression of pepsinogen A and C mRNAs were ~300 times and ~15 times higher than that of AMCase, respectively (Fig. 2, upper panel). AMCase mRNA level was comparable to H+/K+-ATPase and 26 times higher than that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5 times higher than β-actin, and hundreds-to-thousands of times higher than other four tested gastric mucosa genes (Fig. 2, lower panel). These results indicate that AMCase is one of the major transcript in the pig stomach.
Figure 2.
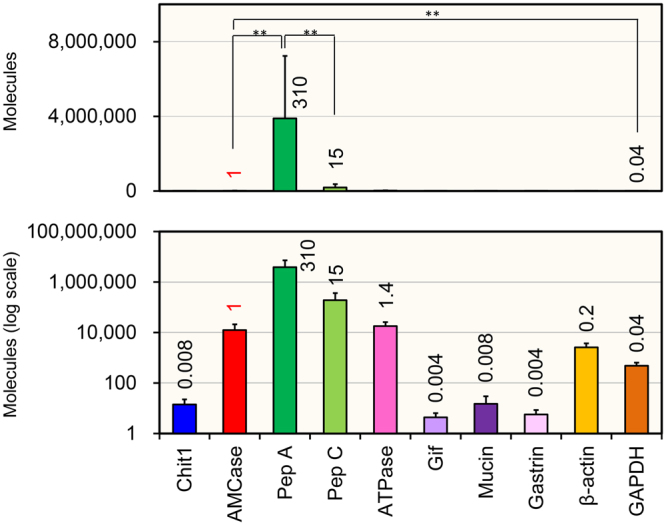
Quantification of mRNA expressions in pig stomach by real-time PCR. Evaluation of AMCase mRNA levels in pig stomach tissues using standard DNA containing 10 genes fragments: Chit1, AMCase, pepsinogen A (Pep A), pepsinogen C (Pep C), H + /K + -ATPase, Gastric intrinsic factor (Gif), mucin, Gastrin, β-actin and GAPDH. The expression levels of the 10 genes determined using the cDNAs prepared from stomach tissues from 6-month-old pig (n = 3) were quantified by qPCR. Expression level of the AMCase gene was set to 1 (written in red); the values on the bars indicate the relative expression levels compared to the expression level of AMCase. The upper panels indicate the actual number, whereas the lower panel shows each value on logarithmic scale. Each reaction was performed in triplicate. **p < 0.01. P-values were determined using Student’s t-test.
Endogenous pepsins degrade soluble proteins in the stomach extract
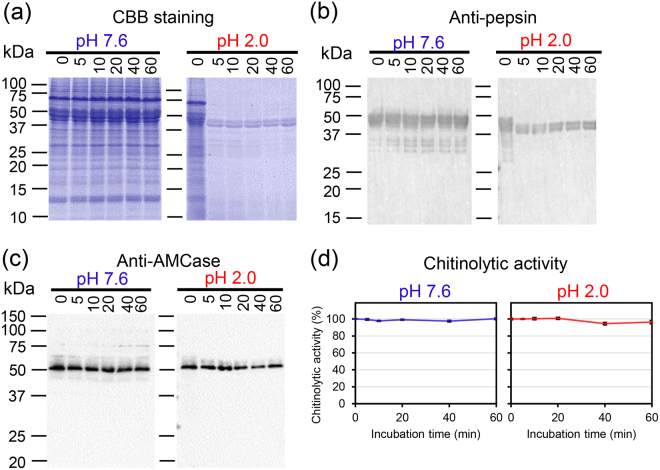
Next, we investigated the protease activity of endogenous pepsins in artificially created pig stomach environment at pH 2.0 and 37 °C. Soluble protein fraction was prepared from the pig stomach in the absence of protease inhibitor and incubated at pH 7.6 or pH 2.0 for up to 60 min. The protein fractions were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), followed by Coomassie Brilliant Blue (CBB) staining (Fig. 3a). At pH 7.6, no changes in the band pattern and intensities were noticed within 60 min incubation (Fig. 3a). In contrast, we observed time-dependent decrease of the soluble proteins with a marked reduction after as early as 5 min of incubation at pH 2.0, although several bands remained unchanged after 60 min of incubation (Fig. 3a). Western blot analysis using anti-pepsin antibody showed a shift of the respective bands within 5 min of incubation at pH 2.0, indicating pepsinogen-to-pepsin conversion (Fig. 3b).
Figure 3.
Pepsinogens were converted into active forms and degraded soluble proteins in stomach. Analysis of the endogenous pepsins protease activity in artificially created pig stomach environment (pH 2.0, 37 °C). Soluble protein fractions were prepared from pig stomach tissue in the absence of protease inhibitor and incubated at pH 7.6 or pH 2.0 for up to 60 min and analysed by (a) SDS-PAGE and CBB staining. Full-length gel image is shown in Supplementary Fig. S2. (b) Western blotting using anti-pepsin or (c) AMCase protein using anti-AMCase in the soluble proteins from pig tissue incubated and conducted as described in (a). Full-length blots are shown in Supplementary Fig. S3. (d) AMCase chitinolytic activity in the extract incubated at pH 7.6 or pH 2.0.
Western blot analysis using anti-mouse AMCase showed that this enzyme was stable during the 60-min incubation at both pH 7.6 and 2.0 (Fig. 3c). Moreover, the chitinolytic activity measured using 4-nitrophenyl N,N′-diacetyl-β-D-chitobioside (4-NP-chitobioside) also remained virtually unchanged (Fig. 3d). These results indicated that AMCase has chitinolytic activity even in the presence of large amount of endogenous pepsins in the pig stomach extract.
Purification and characterization of pig AMCase from stomach tissue
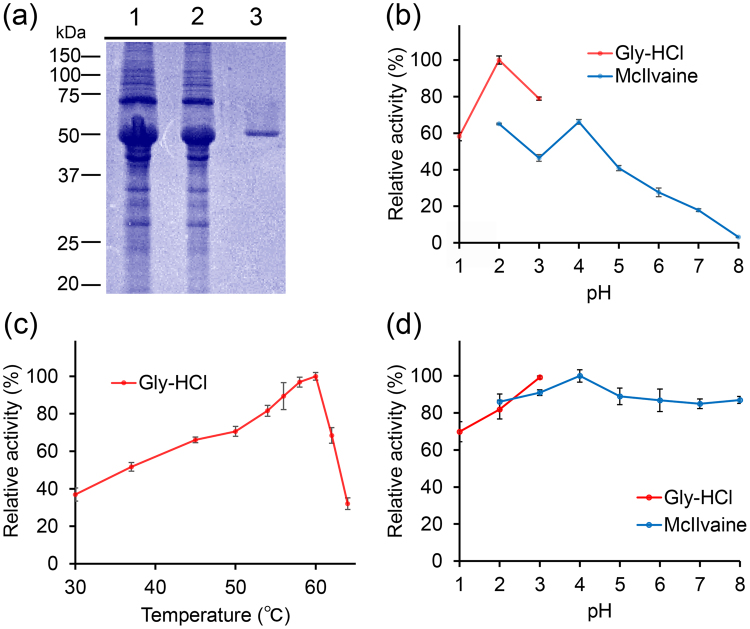
For further characterization, we purified AMCase by application of the stomach extract onto the chitin beads column. Bound chitinase was eluted from the column with 8 M urea, subsequently removed from the sample as described in Methods. SDS-PAGE analysis of the protein fractions showed one major band at 54 kDa (Fig. 4a). Thus, we obtained purified AMCase usable for in vitro enzymatic assays. Purification of the enzyme is summarized in Table 1.
Figure 4.
Purification and characterization of AMCase from pig stomach. (a) AMCase was purified from pig stomach tissues using chitin beads chromatography as described in the Methods and analyzed by SDS-PAGE and visualized by Coomassie Brilliant Blue (CBB) staining. 1, extract; 2, flow-through; 3, purified enzyme. Full-length gel is shown in Supplementary Fig. S4. (b) Optimal pH, (c) Optimal temperature and (d) pH stability for AMCase activity. Values in (b), (c,d) represent mean ± SD conducted in triplicate.
Table 1.
Purification of AMCase from pig stomach.
| Purification step | Total activity (mU) | Total Protein (mg) | Specific activity (mU/mg) | Yield (%) |
|---|---|---|---|---|
| Total soluble fraction | 542.9 ± 3.3 | 20.2 ± 1.5 | 26.8 ± 0.2 | 100 |
| Chitin beads Flow through | 61.5 ± 2.4 | 12.2 ± 2.1 | 5.0 ± 0.2 | 11.3 |
| Purified enzyme | 49.6 ± 2.1 | 0.023 ± 0.006 | 2174.0 ± 90.4 | 10.3 |
The purified protein was prepared from 1 G of stomach tissue as described in the Methods.
The pH optima were determined by monitoring enzyme activity at different pH in 0.1 M Gly-HCl (pH 1.0–3.0) or McIlvaine’s (pH 2.0–8.0) buffers using 4-NP-chitobioside as a substrate for 30 min at 37 °C. Highest activity was detected at pH 2.0 in 0.1 M Gly-HCl buffer. Using McIlvaine’s buffer, higher enzymatic activity was observed at pH 2.0–5.0 with peaks at pH 2.0 and pH 4.0 with gradual decrease in less acidic environments (pH 6.0–8.0) (Fig. 4b). Thus, the chitinolytic activity of AMCase has a slightly different pH-related pattern depending on used buffer.
The effect of temperature on enzyme activity was determined in 0.1 M Gly-HCl buffer at pH 2.0 at temperatures ranging from 30 to 64 °C using same substrate for 30 min. As shown in Fig. 3c, the rate of the AMCase-catalyzed reaction was gradually enhanced with increasing temperature and reached the maximum level at 60 °C, then abruptly declined.
We next determined the pH stability of the pig AMCase. The enzyme was pre-incubated on ice for 60 min at various pH in Gly-HCl or McIlvaine’s buffers. The enzyme activity was then analyzed at 37 °C and pH 2.0. As shown in Fig. 4d, the pig AMCase has a remarkable acid stability as the pre-incubation at pH 2.0 caused no measurable decrease in chitinase activity.
Pig AMCase degrades polymeric chitin substrates under the stomach condition
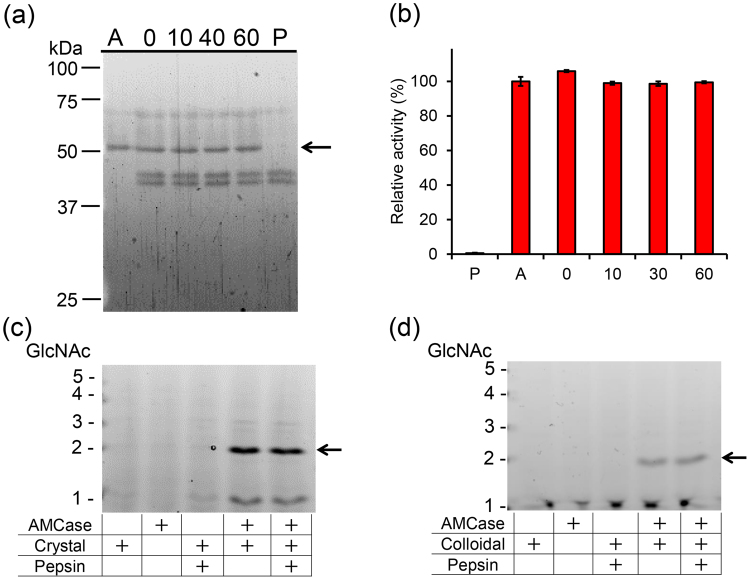
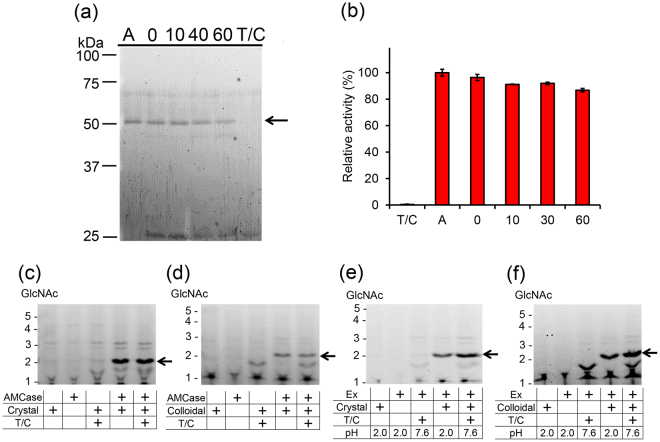
We incubated the purified protein with equal amount of pepsin (0.4 µg) at pH 2.0 for 1 hour and we confirmed the above-observed stability of AMCase at pH 2.0 (Fig. 5a) as well as the maintenance of its chitinolytic activity (Fig. 5b).
Figure 5.
Functional stability of AMCase against pepsin. Purified AMCase was incubated at 37 °C for 0, 10, 40, and 60 min in stomach-like environment in the presence of pepsin. (a) Samples were analyzed by SDS-PAGE followed by SYPRO Ruby staining. Full-length gel is shown in Supplementary Fig. S5. (b) Determination of the chitinolytic activity. A, AMCase only; P, pepsin only; numbers, incubation time of AMCase and pepsin in minutes. Values in (b) represent mean ± SD conducted in triplicate. Degradation products generated by incubation of (c) crystalline or (d) colloidal chitin with purified enzyme were analyzed by FACE. Full-length gels are shown in Supplementary Fig. S5.
Next, we tested whether AMCase can degrade chitin substrates under the pig stomach condition. We incubated colloidal or crystalline chitin with purified AMCase and analyzed the products by an improved fluorophore-assisted carbohydrate electrophoresis (FACE)37,38. Purified enzyme degraded both substrates at pH 2.0 as early as after 1 hour incubation and produced primarily (GlcNAc)2 fragments in the presence of pepsin (Fig. 5c and d).
Pig AMCase is resistant to trypsin and chymotrypsin and degrades chitin substrates under intestinal condition
Next, we investigated whether pig AMCase is also stable under intestinal condition. We incubated the purified protein with equal amount of trypsin/chymotrypsin (0.4 µg) at pH 7.6 and found that AMCase remained stable and active throughout the incubation (Fig. 6a and b).
Figure 6.
Chitin substrates are degraded by AMCase under gastrointestinal condition. Purified AMCase was incubated at 37 °C for 0, 10, 40, and 60 min under intestine-like environment in the presence of trypsin/chymotrypsin. (a) The samples were analyzed by SDS-PAGE followed by SYPRO Ruby staining. Full-length gel is shown in Supplementary Fig. S6. (b) Determination of the chitinolytic activity. A, AMCase only; T/C, trypsin/chymotrypsin only; numbers, incubation time of AMCase and trypsin/chymotrypsin in minutes. Values in (b) represent mean ± SD conducted in triplicate. Degradation products generated by incubation of (c,e) crystalline or (d,f) colloidal chitin with (c,d) purified enzyme under the intestine condition or with (e,f) the stomach extract mimicking GIT conditions were analyzed by FACE. Full-length gels are shown in Supplementary Fig. S6.
We incubated crystalline and colloidal chitin with purified AMCase and equal amount of trypsin/chymotrypsin (0.4 µg) at pH 7.6 for 1 hour and the degradation products were analyzed as described above (Fig. 6c and d). Similarly to stomach condition, (GlcNAc)2 was produced under the intestine-like condition by AMCase activity. Then, we mimicked the GIT physiology regarding the movement of stomach contents to intestine by pre-incubation of the stomach extract at pH 2.0 for 1 hour, followed by neutralization to pH 7.6 and addition of trypsin/chymotrypsin and further 1-hour incubation (Fig. 6e and f). We observed more (GlcNAc)2 degradation products from both colloidal and crystalline chitin after incubation in intestinal environment as compared to single stomach conditions (Fig. 6e and f). Thus, pig AMCase functions as a protease-resistant glycoside hydrolase and can degrade polymeric chitin substrates in both stomach and intestine.
Chitin in mealworm shells and fruit fly wings is degraded by AMCase and pepsin in the stomach extract
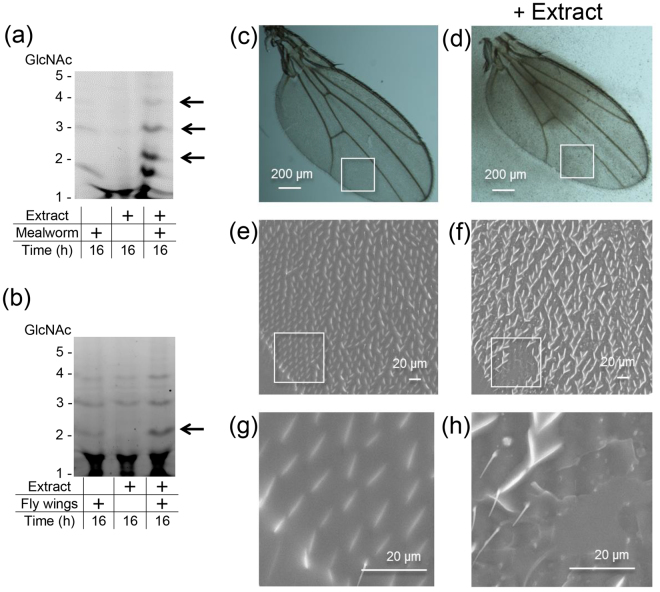
Next, we tested whether AMCase and pepsin can degrade chitin-protein substrates present in chitin-containing organism under pig stomach condition. We incubated mealworm (Tenebrio molitor) larvae shells with stomach extract at pH 2.0 followed by FACE analysis as described above. AMCase in the stomach extract degraded mealworm shells at pH 2.0 after 16 hours incubation and produced different GlcNAc fragments with the dimer being most abundant (Fig. 7a).
Figure 7.
Degradation of mealworm shells and fly wings by stomach extract. Degradation products generated by incubation of (a) mealworm shells or (b) fruit fly wings with stomach extract were analyzed by FACE as described in the Methods. Full-length gels are shown in Supplementary Fig. S7. Microscopic observations of fruit fly wings incubated with (d,f,h) or without (c,e,g) stomach extract in 0.1 M Gly-HCl (pH 2.0). (c,d) steromicroscopic photographs. (e–h) SEM photographs.
To gain further insights into the digestibility of chitin-containing organisms, we next tested fruit fly (Drosophila melanogaster Oregon-R) wings. We first homogenized wings and incubated with the stomach extract and analyzed the products by FACE. We detected (GlcNAc)2 fragments (Fig. 7b).
We also visualized the fruit fly wings surface using stereo microscope (Fig. 7c and d) and scanning electron microscope (SEM) (Fig. 7e–h) after 16 hours treatment with (Fig. 7d,f and h) or without (Fig. 7c,e and g) stomach extract in 0.1 M Gly-HCl (pH 2.0). The extract-treated fruit fly wings and the solution became hazy containing some particles (Fig. 7d) and we observed partially damaged regions (Fig. 7f and h). We detected no such damage in wings incubated only with the buffer (Fig. 7c,e and g). These results indicate that AMCase in the stomach extract can deteriorate chitinous fruit fly wing integrity.
Discussion
Mammalian chitinases have extensively been studied mainly in mice and humans, however relatively little is known about the enzymes in other mammals. In this report, we showed that AMCase mRNA was predominantly expressed in pig stomach tissue and it was much higher than Chit1, housekeeping genes and gastric mucosal genes. We purified AMCase from pig stomach and determined its optimal activity at pH 2.0–4.0 and 60 °C. This enzyme degraded polymeric chitin substrates including mealworm shells and fruit fly wings in the environment mimicking pig GIT conditions. These results support our hypothesis on AMCase functioning as a protease-resistant glycoside hydrolase in the pig digestive system.
AMCase mRNA level was comparable with of H+/K+-ATPase (role in maintaining the stomach acidity)39, and it was substantially higher than mRNA of two housekeeping genes and other gastric mucosa proteins in the pig stomach. This is in agreement with our previous report on high AMCase expression in mouse stomach32,35. These data suggest that AMCase in these animals might be able to digest chitin in those animal bodies.
It has been reported that AMCase expression is elevated under several pathological conditions including dry eye syndrome20,21. Our qPCR analysis showed that AMCase mRNA expression was ten times lower than that of Chit1 in healthy pig eye. These data imply that AMCase may be a key mediator of innate immune responses in certain ocular pathologies. Chit1 mRNA, on the other hand, is constitutively expressed in healthy mouse eye32, human lacrimal gland40 and pig eye. Lysozyme is thought to have anti-bacterial effects, whereas Chit1 protects against fungi. Thus, Chit1 probably protects mammalian eyes from chitin-containing pathogens such as fungi, whose cell wall contains high levels of chitin.
Pig pepsin A and C have been purified, crystallized and extensively studied, in the past41–45. However, to our knowledge, multiple comparisons of pepsin with other mucosal genes and reference genes mRNA levels have not been performed. Here, we show that pepsinogen A mRNA level was substantially higher than housekeeping genes and gastric mucosa genes in the stomach while exceeding 20 times that the level of pepsinogen C. Although the functional difference between pepsin A and C is still unclear, our results suggest that pepsin A is a major protease and pepsin C acts as a co-protease in the pig stomach.
Pig AMCase was more active in Gly-HCl buffer than in McIlvaine at pH 2.0 (Fig. 4b). Thus, the chitinolytic activity of AMCase has a slightly different pH-related pattern depending on used buffer. Similar results were also obtained in mouse AMCase35 and chicken Chia36, although with less significant difference in chicken Chia between the two buffers. The reason is not well understood, but some lessons could be learned from human pancreatic α-amylase, whose activation has been shown to be catalyzed by chloride ion46. Some members of the amylase protein family require chloride for maximal activity47–49. Hydrochloric acid is secreted in the stomach, creating acidic conditions (pH ~2), which may induce similar activation of AMCase. This assumption warrants further scrutiny.
Murine AMCase has been well studied and its optimal activity has been reported at pH 2.0 with a decrease at less acidic conditions (pH 3.0–7.0)15,35,38. Also, chicken Chia (AMCase homologue) was most active at pH 2.0 and it retained at less acidic condition36. We analyzed effect of pH on the pig AMCase chitinolytic activity and showed that the activity was highest in pH 2.0–4.0 and remained active at up to pH 7.0. Although the pig AMCase shares 81 to 89% primary sequence homology with mouse and chicken counterparts, those specificities to pH and buffer are different. These species particularities can be attributed to differences during the molecular evolution based on changes in feeding habitat.
In our study, we showed that pig AMCase degraded chitin substrates including shells of mealworm larvae and fruit fly wings as well as crystalline and colloidal chitins in the presence of digestive proteases and produced (GlcNAc)2. Accordingly, mouse and pig AMCase as well as chicken Chia mainly produced (GlcNAc)2 31,35,36. In addition, we found that mealworm shells digestion by pig AMCase also resulted in (GlcNAc)3 and (GlcNAc)4 fragments. The product patterns were slightly different from those resulting from colloidal and crystalline chitin degradation. Similarly, we confirmed formation of longer chitooligosaccharides from mealworm larvae shells by chicken Chia36. Importantly, we have recently shown that mouse AMCase catalyzes transglycosylation as well as hydrolysis50. Thus, it is feasible to assume that distinct chitooligosaccharides can be produced from partially deacetylated chitin in mealworm shells or transglycosylation by pig AMCase.
Pig is one of the major protein resource for humans worldwide. Increasing demand of meat protein requires more feed resources for the livestock. There have been published several studies reporting on application of insects as a sustainable high protein feed ingredient for pig. For example, Jin et al.51 showed that supplementation of dried mealworm in weaning pigs’ diet improves their growth performance and nutrient digestibility without any detrimental effect on immune responses51. Furthermore, chitin derivatives can enhance the immune response and act as an antibiotic/probiotic in pregnant pig52. Various biological activities, and especially anti-cancer and anti-inflammatory action of chitin oligosaccharide and chitosan oligosaccharide, have been well studied9,53–55. Distinct chitooligosaccharides from chitin-containing organisms may improve immune systems or act as probiotics providing benefits for animal health. Therefore, further evaluation of nutrient value, digestibility and potential side-effects of chitin-containing organisms used as feed ingredient on pig growth performance is needed.
In this report, we showed that AMCase mRNA was highly expressed in pig stomach, having a remarkable protease resistance and degraded chitin or chitin-containing organisms into (GlcNAc)2 and several chitooligosaccharides under the GIT condition. We previously reported that similar properties of AMCase and Chia were found in mouse35 and chicken36, respectively. These animals primarily feed on chitin-containing organisms such as insects and fungi. It is plausible to suggest that their food habitat leads to high expression levels of these enzymes in the stomach and their intense chitinolytic activity in the GIT. According to recent knowledge, chitin-containing organisms can be used as good energy source in pig, chicken and mouse organisms because proteolytic enzyme accessibility is improved by degrading chitinous cuticles of insects and crustaceans by respective AMCase enzymes. From practical point of view, we need further research in other animal species including those with limited ingestion of chitin-containing organisms to reveal whether chitinous diets could be implemented in such species including cattle, sheep, horse, dog, etc.
Methods
Pig stomach tissues
Six months-old male pig stomach tissues (Landrace F1) were purchased from Funakoshi Co., Ltd (Tokyo, Japan), which were dissected from the animals, quickly frozen on dry ice and kept at −80 °C.
RNA and cDNA preparation
Pig Total RNA Panel (Zyagen, San Diego, CA, USA) was used to examine the distribution of transcripts in various tissues. In addition, total RNA was isolated from the pig stomach tissues using TRIzol Reagent (Thermo Fisher Scientific, Waltham, MA, USA) per manufacturer’s instructions and reverse transcribed into cDNA essentially as described previously32,35,36.
Selection of primer pairs for qPCR
Primers for qPCR were designed using PrimerQuest Input (Integrated DNA Technologies, Coralville, IA, USA) and their suitability was evaluated based on a single product generation, as reflected by a single melting temperature as describe previously32,33,36. The primers’ sequences are listed in Supplementary Table S1.
Construction of the DNA standard and qPCR
Construction of the 10 genes standard DNA coding sequences of AMCase, pepsinogen A, pepsinogen C, H+/K+-ATPase, gastrin, gastric intrinsic factor and mucin were commercially synthesized and inserted into pTAKN-2 vector (Eurofins Genomics, Tokyo, Japan). The standard DNA (956 bases; see Supplementary Fig. S1) was prepared by PCR reamplification from the plasmid DNA using the forward primer 5′-TTGCCGTCCGTGCATATT-3′ and the reverse primer 5′-CAAGGTCAAGGCCATCAAA-3′ and was thereafter used as the standard DNA for qPCR. Each reaction was performed in triplicate.
Pig stomach extract preparation
Soluble fraction was prepared from pig stomach tissues (0.2 g) by homogenization followed by centrifugation at 15,000 g for 10 min at 4 °C35,36. The supernatants were used as the stomach extract and pre-incubated at 37 °C for 0, 10, 20, 40 or 60 min at pH 7.6 or pH 2.0. After incubation at pH 2.0 and 37 °C, the solutions were neutralized.
SDS-polyacrylamide gel electrophoresis and Western blot
The obtained protein fractions were analyzed using standard SDS-PAGE, followed by Coomassie Brilliant Blue R-250 (CBB, Sigma-Aldrich, St. Louis, MO, USA) or Western blot using polyclonal anti-mouse C-terminal AMCase33 or polyclonal pig anti-pepsin antibody (GeneTex, Irvine, CA, USA), followed by peroxidase-conjugated AffiniPure F (ab’)2 Fragment Donkey Anti-Rabbit IgG (H + L) (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) or AffiniPure Donkey Anti-Goat IgG-HRP (Jackson ImmunoResearch laboratories). The immunoblots were analyzed and quantified by Luminescent Image Analyzer (ImageQuant LAS 4000, GE Healthcare, Piscataway, NJ, USA) according to the manufacturer’s instructions.
Chitinase enzymatic assays
Chitinolytic activity was determined using a synthetic chromogenic substrate, 4-nitrophenyl N,N′-diacetyl-β-D-chitobioside (4-NP-chitobioside, Sigma-Aldrich) essentially as described previously31. All enzymatic reactions for optimum pH and temperature determination of were conducted in a volume of 50 μL as described previously31,56.
Purification of pig AMCase
AMCase was purified from pig stomach tissue (1 g) using chitin beads column and eluted with 8 M urea as performed previously36. Protein concentrations were determined by the Bradford Protein Assay (Bio-Rad Laboratories, Hercules, CA, USA) using the BioPhotometer Plus UV/Vis photometer (Eppendorf, Hamburg, Germany), with bovine serum albumin as a standard. AMCase unit definition was also reported previously31.
The effects of pH and temperature on chitinase activity
For determination of the optimal pH, the chitinase activity was investigated by incubating the enzyme with 4-NP-chitobioside as a substrate in 0.1 M Gly-HCl buffer (pH 1.0–3.0) or McIlvaine’s buffer (0.1 M citric acid and 0.2 M Na2HPO4; pH 2.0–8.0) at 37 °C for 30 min. To measure the optimal temperature, chitinase activity was assayed between 30 °C and 64 °C in 0.1 M Gly-HCl buffer (pH 2.0).
For the determination of the pH stability, samples were incubated for 1 hour on ice in 0.1 M Gly-HCl buffer (pH 1.0–3.0) or McIlvaine’s buffer (pH 2.0–8.0). After the pre-incubation at the indicated pH, the residual activity was analyzed at pH 2.0 in 0.1 M Gly-HCl buffer, as described above.
Mealworms
Jumbo mealworm (Tenebrio molitor) larvae were purchased from local commercial supplier (Lumberjack Co., Ltd., Tokyo, Japan). We used the shells containing connective tissue as chitin-protein polymer substrates as described previously36.
Degradation of colloidal and crystalline chitin substrates and mealworm larvae shell
Colloidal and crystalline chitin were incubated in a volume of 50 µL containing purified AMCase (4 mU) or soluble protein (4 mU) from pig stomach as described previously35,36. Mealworm larvae shells were also incubated with soluble protein fraction (20 mU) in an analogous way. Generated chitin fragments were analyzed by fluorophore-assisted carbohydrate electrophoresis (FACE) as originally described by Jackson37 and recently improved by us38.
Chitin degradation of fruit fly wings by stomach extract
Fruit flies (D. melanogaster Oregon-R) were bred at the facility in Okayama University. Flies were immersed once in ethanol. Fifty wings were homogenized using TaKaRa BioMasher Standard (TaKaRa Bio, Shiga, Japan) and then treated with stomach extract as described in mealworm. After incubation at 37 °C for 16 hours, degradation products were analyzed by FACE as described above. For morphological examination, wing was treated with stomach extract containing 9 mU AMCase activity in 0.1 M Gly-HCl (pH 2.0) at final volume of 10 µL using a glass slide printed with water-repellent mark (TF1205, Matsunami Glass Ind., Ltd., Osaka, Japan). The morphological changes of the wings were assessed using a stereo microscope (M205 C, Leica Microsystems, Wetzlar, Germany). Observations of fly wings by scanning electron microscope (SEM, JCM-6000, acceleration voltage: 15 kV, JEOL Ltd., Tokyo, Japan) were achieved by ionic liquids coating method without accumulation of electron charges, indicating that the liquid behaves as an electronically conducting material57. Observed sample was immersed into mixture of ionic liquid, 10% 1-hexyl-3-methylimidazolium bis (fluorosulfonyl) imide (Mitsubishi Materials Electronic Chemicals Co., Ltd., Akita, Japan) in ethanol. Immersed fly wing samples were dried at room temperature over 1 hour prior to SEM observations for removing ethanol. Thin and uniform ionic liquid coating layer enabled clear SEM observation without charge up of surface of measurement samples.
Statistical analysis
Biochemical data were compared by Student’s t-test. We carried out experiments in triplicate for the statistical analysis
Electronic supplementary material
Acknowledgements
We are grateful to Haruko Miyazaki and Nobuyuki Nukina for their encouragement, to Kazuaki Okawa, Shotaro Honda, Masahiro Kimura, Natsumi Yamashita and Rieko Oyama for valuable suggestions. This work was supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS) (grant numbers 15J10960, 16K07699 to M.O. and F.O., respectively); by the Project Research Grant from the Research Institute of Science and Technology, Kogakuin University (to F.O.); by a Grant from the Science Research Promotion Fund of the Promotion and Mutual Aid Corporation for Private Schools of Japan (to F.O.); and a grant of the Strategic Research Foundation Grant-aided Project for Private Universities (S1411005 to M.S. Y.I. and F.O.) from the Ministry of Education, Culture, Sport, Science and Technology, Japan.
Author Contributions
Conceived and designed the experiments: E.T., A.K., S.W., M.O., M.S., Y.I., S.S., H.U., Y.S., V.M., P.O.B., F.O. Performed research: E.T., A.K., S.W., M.O., Y.I., S.S., H.U., F.O. Analyzed data: E.T., A.K., S.W., M.O., Y.I., S.S., H.U., F.O. Wrote the paper: E.T., V.M., P.O.B., F.O. Contributed to the critical appraisal of the paper and approved the final version: E.T., A.K., S.W., M.O., M.S., Y.S., H.U., V.M., P.O.B., F.O.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-13526-6.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cui D, et al. Generation of a miniature pig disease model for human Laron syndrome. Sci. Rep. 2015;5:15603. doi: 10.1038/srep15603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pattengale PK, et al. Animal models of human disease. Pathology and molecular biology of spontaneous neoplasms occurring in transgenic mice carrying and expressing activated cellular oncogenes. Am. J. Pathol. 1989;135:39–61. [PMC free article] [PubMed] [Google Scholar]
- 3.Gotz J, et al. Transgenic animal models of Alzheimer’s disease and related disorders: histopathology, behavior and therapy. Mol. Psychiatry. 2004;9:664–683. doi: 10.1038/sj.mp.4001508. [DOI] [PubMed] [Google Scholar]
- 4.Imaizumi T, Lankford KL, Burton WV, Fodor WL, Kocsis JD. Xenotransplantation of transgenic pig olfactory ensheathing cells promotes axonal regeneration in rat spinal cord. Nat. Biotechnol. 2000;18:949–953. doi: 10.1038/79432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Litten-Brown JC, Corson AM, Clarke L. Porcine models for the metabolic syndrome, digestive and bone disorders: a general overview. Animal. 2010;4:899–920. doi: 10.1017/S1751731110000200. [DOI] [PubMed] [Google Scholar]
- 6.Qian L, et al. Targeted mutations in myostatin by zinc-finger nucleases result in double-muscled phenotype in Meishan pigs. Sci. Rep. 2015;5:14435. doi: 10.1038/srep14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Huis A, et al. Edible insects: future prospects for food and feed security. FAO Forestry Paper. 2013;171:1–201. [Google Scholar]
- 8.Kupferschmidt K. Buzz food. Science. 2015;350:267–269. doi: 10.1126/science.350.6258.267. [DOI] [PubMed] [Google Scholar]
- 9.Khoushab F, Yamabhai M. Chitin research revisited. Mar. Drugs. 2010;8:1988–2012. doi: 10.3390/md8071988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bueter CL, Specht CA, Levitz SM. Innate sensing of chitin and chitosan. PLoS Pathog. 2013;9:e1003080. doi: 10.1371/journal.ppat.1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bussink AP, Speijer D, Aerts JM, Boot RG. Evolution of mammalian chitinase(-like) members of family 18 glycosyl hydrolases. Genetics. 2007;177:959–970. doi: 10.1534/genetics.107.075846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hollak CE, van Weely S, van Oers MH, Aerts JM. Marked elevation of plasma chitotriosidase activity. A novel hallmark of Gaucher disease. J. Clin. Invest. 1994;93:1288–1292. doi: 10.1172/JCI117084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Renkema GH, Boot RG, Muijsers AO, Donker-Koopman WE, Aerts JM. Purification and characterization of human chitotriosidase, a novel member of the chitinase family of proteins. J. Biol. Chem. 1995;270:2198–2202. doi: 10.1074/jbc.270.5.2198. [DOI] [PubMed] [Google Scholar]
- 14.Boot RG, Renkema GH, Strijland A, van Zonneveld AJ, Aerts JM. Cloning of a cDNA encoding chitotriosidase, a human chitinase produced by macrophages. J. Biol. Chem. 1995;270:26252–26256. doi: 10.1074/jbc.270.44.26252. [DOI] [PubMed] [Google Scholar]
- 15.Boot RG, et al. Identification of a novel acidic mammalian chitinase distinct from chitotriosidase. J. Biol. Chem. 2001;276:6770–6778. doi: 10.1074/jbc.M009886200. [DOI] [PubMed] [Google Scholar]
- 16.Koch BE, Stougaard J, Spaink HP. Keeping track of the growing number of biological functions of chitin and its interaction partners in biomedical research. Glycobiology. 2015;25:469–482. doi: 10.1093/glycob/cwv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Z, et al. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science. 2004;304:1678–1682. doi: 10.1126/science.1095336. [DOI] [PubMed] [Google Scholar]
- 18.Reese TA, et al. Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature. 2007;447:92–96. doi: 10.1038/nature05746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bucolo C, Musumeci M, Maltese A, Drago F, Musumeci S. Effect of chitinase inhibitors on endotoxin-induced uveitis (EIU) in rabbits. Pharmacol. Res. 2008;57:247–252. doi: 10.1016/j.phrs.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Musumeci M, et al. Acidic mammalian chitinase in dry eye conditions. Cornea. 2009;28:667–672. doi: 10.1097/ICO.0b013e31819bc308. [DOI] [PubMed] [Google Scholar]
- 21.Bucolo C, Musumeci M, Musumeci S, Drago F. Acidic mammalian chitinase and the eye: implications for ocular inflammatory diseases. Front. Pharmacol. 2011;2:43. doi: 10.3389/fphar.2011.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cozzarini E, et al. CHIT1 and AMCase expression in human gastric mucosa: correlation with inflammation and Helicobacter pylori infection. Eur. J. Gastroenterol. Hepatol. 2009;21:1119–1126. doi: 10.1097/MEG.0b013e328329742a. [DOI] [PubMed] [Google Scholar]
- 23.Nookaew I, et al. Transcriptome signatures in Helicobacter pylori-infected mucosa identifies acidic mammalian chitinase loss as a corpus atrophy marker. BMC Med. Genomics. 2013;6:41. doi: 10.1186/1755-8794-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bierbaum S, et al. Polymorphisms and haplotypes of acid mammalian chitinase are associated with bronchial asthma. Am. J. Respir. Crit. Care Med. 2005;172:1505–1509. doi: 10.1164/rccm.200506-890OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seibold MA, et al. Differential enzymatic activity of common haplotypic versions of the human acidic mammalian chitinase protein. J. Biol. Chem. 2009;284:19650–19658. doi: 10.1074/jbc.M109.012443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okawa K, et al. Loss and gain of human acidic mammalian chitinase activity by nonsynonymous SNPs. Mol. Biol. Evol. 2016;33:3183–3193. doi: 10.1093/molbev/msw198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fitz LJ, et al. Acidic mammalian chitinase is not a critical target for allergic airway disease. Am. J. Respir. Cell Mol. Biol. 2012;46:71–79. doi: 10.1165/rcmb.2011-0095OC. [DOI] [PubMed] [Google Scholar]
- 28.Van Dyken SJ, et al. Spontaneous chitin accumulation in airways and age-related fibrotic lung disease. Cell. 2017;169:497–509 e413. doi: 10.1016/j.cell.2017.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vannella KM, et al. Acidic chitinase primes the protective immune response to gastrointestinal nematodes. Nat. Immunol. 2016;17:538–544. doi: 10.1038/ni.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boot RG, et al. Marked differences in tissue-specific expression of chitinases in mouse and man. J. Histochem. Cytochem. 2005;53:1283–1292. doi: 10.1369/jhc.4A6547.2005. [DOI] [PubMed] [Google Scholar]
- 31.Kashimura A, et al. Protein A-mouse acidic mammalian chitinase-V5-His expressed in periplasmic space of Escherichia coli possesses chitinase functions comparable to CHO-expressed protein. PLoS One. 2013;8:e78669. doi: 10.1371/journal.pone.0078669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohno M, Tsuda K, Sakaguchi M, Sugahara Y, Oyama F. Chitinase mRNA levels by quantitative PCR using the single standard DNA: acidic mammalian chitinase is a major transcript in the mouse stomach. PLoS One. 2012;7:e50381. doi: 10.1371/journal.pone.0050381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohno M, et al. Quantification of chitinase mRNA levels in human and mouse tissues by real-time PCR: species-specific expression of acidic mammalian chitinase in stomach tissues. PLoS One. 2013;8:e67399. doi: 10.1371/journal.pone.0067399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bays HE, et al. Chitin-glucan fiber effects on oxidized low-density lipoprotein: a randomized controlled trial. Eur. J. Clin. Nutr. 2013;67:2–7. doi: 10.1038/ejcn.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohno M, et al. Acidic mammalian chitinase is a proteases-resistant glycosidase in mouse digestive system. Sci. Rep. 2016;6:37756. doi: 10.1038/srep37756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tabata E, et al. Gastric and intestinal proteases resistance of chicken acidic chitinase nominates chitin-containing organisms for alternative whole edible diets for poultry. Sci. Rep. 2017;7:6662. doi: 10.1038/s41598-017-07146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackson P. The use of polyacrylamide-gel electrophoresis for the high-resolution separation of reducing saccharides labelled with the fluorophore 8-aminonaphthalene-1,3,6-trisulphonic acid. Detection of picomolar quantities by an imaging system based on a cooled charge-coupled device. Biochem. J. 1990;270:705–713. doi: 10.1042/bj2700705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wakita S, et al. Improved fluorescent labeling of chitin oligomers: Chitinolytic properties of acidic mammalian chitinase under somatic tissue pH conditions. Carbohydr Polym. 2017;164:145–153. doi: 10.1016/j.carbpol.2017.01.095. [DOI] [PubMed] [Google Scholar]
- 39.Canfield VA, Levenson R. Structural organization and transcription of the mouse gastric H+, K(+)-ATPase beta subunit gene. Proc. Natl. Acad. Sci. USA. 1991;88:8247–8251. doi: 10.1073/pnas.88.18.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall AJ, Morroll S, Tighe P, Gotz F, Falcone FH. Human chitotriosidase is expressed in the eye and lacrimal gland and has an antimicrobial spectrum different from lysozyme. Microbes Infect. 2008;10:69–78. doi: 10.1016/j.micinf.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 41.Ong EB, Perlmann GE. The amino-terminal sequence of porcine pepsinogen. J. Biol. Chem. 1968;243:6104–6109. [PubMed] [Google Scholar]
- 42.Tang J, et al. Amino-acid sequence of porcine pepsin. Proc. Natl. Acad. Sci. USA. 1973;70:3437–3439. doi: 10.1073/pnas.70.12.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen KC, Tao N, Tang J. Primary structure of porcine pepsin. I. Purification and placement of cyanogen bromide fragments and the amino acid sequence of fragment CB5. J. Biol. Chem. 1975;250:5068–5075. [PubMed] [Google Scholar]
- 44.Bank RA, et al. Identification of a Glu greater than Lys substitution in the activation segment of human pepsinogen A-3 and -5 isozymogens by peptide mapping using endoproteinase Lys-C. FEBS Lett. 1988;238:105–108. doi: 10.1016/0014-5793(88)80235-7. [DOI] [PubMed] [Google Scholar]
- 45.Lin XL, Wong RN, Tang J. Synthesis, purification, and active site mutagenesis of recombinant porcine pepsinogen. J. Biol. Chem. 1989;264:4482–4489. [PubMed] [Google Scholar]
- 46.Numao S, et al. Probing the role of the chloride ion in the mechanism of human pancreatic alpha-amylase. Biochemistry. 2002;41:215–225. doi: 10.1021/bi0115636. [DOI] [PubMed] [Google Scholar]
- 47.Maurus R, et al. Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity. Biochemistry. 2008;47:3332–3344. doi: 10.1021/bi701652t. [DOI] [PubMed] [Google Scholar]
- 48.Feller G, Bussy O, Houssier C, Gerday C. Structural and functional aspects of chloride binding to Alteromonas haloplanctis alpha-amylase. J. Biol. Chem. 1996;271:23836–23841. doi: 10.1074/jbc.271.39.23836. [DOI] [PubMed] [Google Scholar]
- 49.Aghajari N, Feller G, Gerday C, Haser R. Structural basis of alpha-amylase activation by chloride. Protein Sci. 2002;11:1435–1441. doi: 10.1110/ps.0202602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wakita, S. et al. Mouse acidic mammalian chitinase exhibits transglycosylation activity at somatic tissue pH. FEBS Lett., in press (2017). [DOI] [PubMed]
- 51.Jin XH, Heo PS, Hong JS, Kim NJ, Kim YY. Supplementation of dried mealworm (Tenebrio molitor larva) on growth performance, nutrient digestibility and blood profiles in weaning pigs. Asian-Australas J Anim Sci. 2016;29:979–986. doi: 10.5713/ajas.15.0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie C, et al. Chitosan oligosaccharide affects antioxidant defense capacity and placental amino acids transport of sows. BMC Vet. Res. 2016;12:243. doi: 10.1186/s12917-016-0872-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Masuda S, et al. Anti-tumor properties of orally administered glucosamine and N-acetyl-D-glucosamine oligomers in a mouse model. Carbohydr Polym. 2014;111:783–787. doi: 10.1016/j.carbpol.2014.04.102. [DOI] [PubMed] [Google Scholar]
- 54.Azuma K, Osaki T, Minami S, Okamoto Y. Anticancer and anti-inflammatory properties of chitin and chitosan oligosaccharides. J Funct Biomater. 2015;6:33–49. doi: 10.3390/jfb6010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aam BB, et al. Production of chitooligosaccharides and their potential applications in medicine. Mar. Drugs. 2010;8:1482–1517. doi: 10.3390/md8051482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kashimura A, et al. Functional properties of the catalytic domain of mouse acidic mammalian chitinase expressed in Escherichia coli. Int. J. Mol. Sci. 2015;16:4028–4042. doi: 10.3390/ijms16024028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuwabata S, Kongkanand A, Oyamatsu D, Torimoto T. Observation of ionic liquid by scanning electron microscope. Chem. Lett. 2006;35:600–601. doi: 10.1246/cl.2006.600. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.