Abstract
Mutations, serving as the raw materials of evolution, have been extensively utilized to increase the chances of engineering molecules or microbes with tailor-made functions. Global and targeted mutagenesis are two main methods of obtaining various mutations, distinguished by the range of action they can cover. While the former one stresses the mining of novel genetic loci within the whole genomic background, targeted mutagenesis performs in a more straightforward manner, bringing evolutionary escape and error catastrophe under control. In this review, we classify the existing techniques of targeted mutagenesis into two categories in terms of whether the diversity is generated in vitro or in vivo, and briefly introduce the mechanisms and applications of them separately. The inherent connections and development trends of the two classes are also discussed to provide an insight into the next generation evolution research.
Keywords: Mutations, Targeted mutagenesis, Evolution, Synthetic biology
Abbreviations used: non-GMO, non-genetically modified organism; error-prone PCR, error-prone polymerase chain reaction; MAGE, multiplex automated genome engineering; TaGTEAM, targeting glycosylase to embedded arrays for mutagenesis; dNTP, deoxy-ribonucleoside triphosphate; ssDNA, single-stranded DNA; YOGE, yeast oligo-mediated genome engineering; CRISPR/Cas9, clustered regularly interspaced short palindromic repeats and associated protein 9; dsDNA, double-stranded DNA; ZFN, zinc-finger nuclease; TALEN, transcription activator-like effector nuclease; DSB, double strand break; NHEJ, error-prone non-homologous end-joining; MMEJ, microhomology-mediated end-joining; HDR, homology-directed repair; TALE, transcription activator-like effector; RVD, repeat variable di-residue; ZF, zinc-finger protein; FLASH, fast ligation-based automatable solid-phase high-throughput; LIC, ligation-independent cloning; PAM, protospacer-adjacent motif; pre-crRNA, pre-CRISPR RNA; tracrRNA, trans-encoded RNA; sgRNA, single-guide RNA; DNA PolI, DNA polymerase I; DNA Pol III, DNA polymerase III; TP, terminal protein; TP-DNAP, TP-DNA polymerase fusion; sctetR, single chain tetR; ICE, in vivo continuous evolution; HIV, human immunodeficiency virus; ORF, open reading frame; 3′-LTR, 3’-long terminal repeat; 5-FOA, 5-fluoro-orotic acid; dCas9, catalytically dead Cas9
1. Introduction
Though it has been chronically disputed by geneticists to what extent mutation affects the course of evolution [1], mutation per se can provide a direct source of genetic diversity [2], supporting the rapid engineering of phenotypes demanded. Generally, the naturally occurring mutations of most microbes is rare, approximating one in a billion base pairs per generation [3], [4], far from satisfying the requirements in engineering applications.
To this end, various mutagenesis methods have been developed ever since the year 1927, when X-ray was first found to be capable of triggering phenotype changes in fruit flies [5]. A number of global mutagenesis techniques flourished thereafter, and are assorted into physical [6], [7], chemical [8], [9], and biological mutagen-guided mutagenesis [10], [11], [12], [13]. These methods have been extensively applied in breeding microbes with superior phenotypes, including enhanced carbon source uptake rate and environmental tolerance [14], [15]. Furthermore, when coupled with whole genome sequencing, global mutagenesis can efficiently mine essential genes and mutations rendering microbes with improved phenotypes [16], [17] or specialized functions [18], [19], [20], [21].
As another genre of mutagenesis method, targeted mutagenesis lagged behind at one time, mainly due to underdeveloped DNA manipulation techniques and the requirement for prior knowledge of the target region. With molecular biology and bioinformatics ascending, targeted mutagenesis has made its way into protein and metabolic engineering fields, largely expanding the repertoire of mutagenesis by bringing the two major problems encountered by global mutagenesis under control, namely error catastrophe and evolutionary escape.
Orgel [22] first used the term error catastrophe to describe the situation where the mutation rate reached a threshold only to impair the cellular processes vital to cell viability. On one hand, we would estimate an exponentially increasing error frequency if the genes involved in the processing of genetic information were mutated. On the other, the DNA repair system would soon be saturated by the avalanche-like accumulated errors and crashing down [23], [24], falling into a vicious circle between repair dysfunction and error accumulation. Obviously, single cell microorganisms are more susceptible to the crisis, which is more likely to occur when global mutation rate is raised. As for evolutionary escape, it is almost an insurmountable problem encountered in adaptive laboratory evolution when biosensor is used to couple target molecule productivity and host cell fitness. Once the global mutation rate was upregulated, escapees would rapidly emerge, survive and predominate in the population without any target molecule production either by mutating and crippling the selection device [25], or by activating physical efflux pumps and oxidative-stress protective mechanisms [26], [27].
Thus, it can be seen from above that global mutagenesis is more applicable for gene mining, especially within an unclear genome, or to meet non-GMO (non-genetically modified organism) standards. When it comes to more straightforward and robust product-oriented evolution processes, the advantages of targeted mutagenesis are highlighted. By simply improving the mutation rate of the target region while maintaining a relatively stable genetic background, targeted mutagenesis can partially sidestep the risk of error catastrophe and evolutionary escape at the cost of comprehensive genomic loci discovery, which can be compensated through bioinformatics analysis.
In this review, brief introductions and comments will be given to existing targeted mutagenesis methods, including error-prone PCR (error-prone polymerase chain reaction), MAGE (multiplex automated genome engineering), PFunkel, programmable nucleases, orthogonal DNA polymerase-plasmid pairs, TaGTEAM (targeting glycosylase to embedded arrays for mutagenesis) and retrotransposon-based targeted mutagenesis. Some of these techniques are not formerly regarded as or confined to targeted mutagenesis, but they inherently have the ability or have been used to generate diversity of a local area. Here we discuss them together to unveil the internal logic among them for a more in-depth comprehension.
2. Existing targeted mutagenesis methods
The development and application of targeted mutagenesis started with error-prone PCR, which is not commonly defined as a targeted mutagenesis method but a random mutagenesis tool in directed evolution. In vitro manipulation endues it with the ability to target a specific DNA region of interest. It is the same with many other conventional in vitro mutagenesis methods. Recent years, developing synthetic biology has enabled the targeted diversification to be carried out in vivo, saving a large amount of labor, time and cost. Besides, in vivo targeted mutagenesis in conjunction with selection, can set up an in vivo targeted continuous evolution platform for protein and metabolic engineering with high efficiency. Here we roughly classified the existing methods into two categories based on whether the diversity is generated in vitro or in vivo. Typical methods are given below respectively.
2.1. In vitro targeted mutagenesis methods
2.1.1. Error-prone PCR
Error-prone Polymerase Chain Reaction is carried out under highly mutagenic conditions compared with the conventional PCR process. In 1989, Leung first established the error-prone PCR method by using the low-fidelity Thermus aquaticus DNA polymerase [28]. A well-developed protocol for the method was described by Cadwell where the reaction conditions were modified, including concentrations of MgCl2, MnCl2, Taq polymerase and the four dNTPs (deoxy-ribonucleoside triphosphates) [29]. Under optimal conditions, the overall mutation rate approximates to 0.007 per base per reaction in a relatively unbiased manner [29].
Because of its easy manipulation and adjustable mutation rate, error-prone PCR remains to be the most widely used targeted mutagenesis method in directed evolution, especially for protein engineering. Shortly after invented, it was applied in a number of research work to assist the engineering of ribozymes with novel catalytic [30], [31], [32] or allosteric [33] functions. Since ribozymes are short and their comprehensive mutant libraries are easy to synthesize, error-prone PCR was merely the icing on the cake. It was not used until several rounds of selective amplification had been carried out. The time came when it was introduced into more complex protein engineering, where a considerable mutant library is impossible to synthesize. Error-prone PCR functions as a propeller to drive the rapid engineering of enzymes with excellent specificities [34], [35], stabilities [36], [37], [38] and catalytic activities [39], [40], [41], [42], [43], [44]. In addition, some other types of problems have also been subject to it for identification and optimization, such as core residues analysis [45], [46], protein-protein interactions [47], [48], protein-nucleotide interactions [49] and pathway evolution [50].
Many efforts have been devoted to optimize the method itself [51], [52], [53], [54], as the constrained low mutation frequencies and inherently biased mutational spectra are somewhat insufficient to generate rich diversity [53], [54]. These adapted methods give new insights into mutagenic PCR, though some of them are sophisticated and demand high technical operations.
2.1.2. MAGE
MAGE, abbreviated from multiplex automated genome engineering, opened a new direction for targeted mutagenesis two decades after the establishment of mutagenic PCR. Church described the whole landscape of MAGE in 2009 [55], where different loci on the chromosome can be targeted simultaneously, generating a multiplex genomic mutant library. This is achieved by single-stranded oligonucleotide homologous recombineering. In the method, the artificially designed ssDNA (single-stranded DNA) with flanking homology arms to the target genomic region is imported into the host Escherichia coli strain, then hybridizes with the exposed complementary lagging strand at the replication fork during replication, leading to a defined diversity of the target site [56].
MAGE is actually derived from the λ-Red homologous recombineering system, which employs the three proteins Exo, Beta and Gamma from λ-bacteriophage to reinforce the recombination process [57]. The phage proteins are also applied in MAGE. Besides, several conditions have been optimized to further increase the recombination rate and mutation rate, including genomic mismatch repair gene mutS knock-out, ssDNA 5’-terminus phosphorothioated modification and ssDNA orientation/length/structure optimization [56], [58]. Far more than these conventional optimizations, a prototype device has been constructed to mechanize MAGE. In the proof-of-concept experiment, the target locus diversity was generated at an overwhelming rate of more than 4.3 billion base pairs of variation a day [55], greatly expanding our perceptions about mutagenesis.
The power of MAGE lies in its high efficiency in multiple genomic loci editing, thus it is preferred in genome-level scientific research, typified by artificial genomic recoding [59], [60], [61], [62], [63]. As for bioengineering applications, the practicability of MAGE is its automation. Just like other targeted mutagenesis methods, MAGE has been applied to identify the genetic bases of cell tolerance to organic solvent like isobutanol [64], and evolve strains with improved productivities of target compounds such as lycopene [55], naringenin and glucaric acid [25].
There are some transparent drawbacks within MAGE [56]. Firstly, the specific E. coli host strain with defined genome modification is required. Secondly, the 90-nt oligo with 4 phosphorothioated bases on its 5’-terminus largely increases the cost. Finally, the frequency of obtaining multi-site mutants is low. To deal with these disadvantages, modified MAGE and some accessories have been developed. In 2013, a MAGE-like genome editing method emerged in Corynebacterium glutamicum [65]. The same year, Church transplanted MAGE to yeast to form YOGE [66], short for yeast oligo-mediated genome engineering, though with very low efficiency. To cut down the expense, high-fidelity DNA microarray technology was recommended to synthesize large oligo libraries [67]. As for the multi-site recombination frequency, coselection-MAGE was proposed [60], where simultaneous editing of target regions with a genomic selectable marker gene would improve the frequency of attaining multi-site mutants. Fine regulation of MAGE is also concerned. Nyerges introduced temperature-sensitive mismatch repair proteins into the E. coli host [68]. They are active to sustain a steady genomic background when MAGE is not performed. Once temperature is raised to induce λ-Red proteins, they become dead to ensure an improved recombination frequency. Recently, the cooperation of CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats and associated protein 9) and MAGE has been described [69], where the nuclease was used to reduce the false positive rate after recombination.
2.1.3. PFunkel
Unlike MAGE, PFunkel demonstrates a radically different way to utilize single-stranded DNA to carry out targeted mutagenesis. This method is adapted from Kunkel mutagenesis, which is named after its founder. Kunkel was originally applied in introducing single-base substitution mutations. In the method [70], the primer harboring the defined mutation hybridizes onto the single-stranded uracil-containing circular DNA template prepared from M13mp2 phage, and is extended along it and ligated to form a covalently closed circular heteroduplex. Uracil bases are then removed by uracil glycosylase, leaving many abasic sites on the template strand. After imported into E. coli, the intact complementary strand is regarded as the template to guide the repair of base deletions, thus the mutation on the complementary strand is preserved. Kunkel relies on accurate mismatch repair processes, and glycosylase treatment significantly impairs transformation efficiency, leading to a high wildtype proportion of 48% when performing site-directed mutagenesis.
Three major adaptations are made in PFunkel [71]. Firstly, a thermostable DNA polymerase and ligase are used, enabling a high temperature reaction, therefore non-specific annealing and extension are restrained. Secondly, an optional PCR-form thermal cycling is provided with mutagenic primers added at the beginning of each cycle, which makes the mutation frequency controllable. Thirdly, the uracil-containing viral strand is degraded by uracil glycosylase and exonuclease III, and replaced by a newly synthesized strand using the mutated strand as the template. In this way, the percentage of the wildtype declined drastically to zero in site-directed mutagenesis.
The well-developed PFunkel can be used to perform site-directed or multi-site mutagenesis, and has particular expertise in one-pot comprehensive codon mutagenesis with no need to do site-saturated mutagenesis separately on each codon. By keeping a low primer-template ratio of 5%, a comprehensive codon mutation library of the TEM-1 gene was constructed covering more than 96% of the 18,081 designed single codon substitutions [71], after which, a highly resolved sequence-fitness landscape of the gene was unraveled assisted by deep sequencing [72]. Enlightened by this, a large proportion of the following protein engineering studies were dedicated to the coupling of PFunkel and next generation sequencing to comprehensively elucidate the genotype-phenotype relationship of a protein, making it a reality to have a God's eye view. In these studies, breakthroughs were made in illuminating protein-protein interactions [73], [74], [75], tracing protein evolution dynamics [76] and evolving synthetic metabolic pathways [77]. As a universal protocol, deep mutational scanning based on comprehensive codon mutagenesis and deep sequencing has been described [78], [79].
The requirement for phage-derived uracil-containing circular ssDNA template hampered the extensive application of PFunkel to some extent. The founders themselves proposed to use uracil-containing circular dsDNA (double-stranded DNA) as the template, however the multi-site mutagenesis efficiency dropped significantly [71]. In other modified protocols, linear templates [80], [81] or paired nickases [82] are used.
2.1.4. Programmable nucleases
Programmable nucleases, as opposed to the site-specific nucleases like I-SceI, can be easily tailored to cleave DNA at almost any desired site. Usually they refer to the nuclease triad, namely ZFNs (zinc-finger nucleases), TALENs (transcription activator-like effector nucleases) and CRISPR/Cas (CRISPR-associated protein) [83]. Actually, programmable nucleases are commonly considered as a practical genome editing tool for precise gene knock-out, knock-in and substitution. They can be guided to a designated genomic site and induce DSBs (double strand breaks) within the target. Then the DSBs are repaired through the NHEJ (error-prone non-homologous end-joining) or MMEJ (microhomology-mediated end-joining) pathway when no homologous template is available, or through the HDR (homology-directed repair) pathway when provided with a designed homologous donor DNA [84]. NHEJ and MMEJ are mainly responsible for gene knock-out, while HDR is versatile in all the editing types. In the targeted mutagenesis method, these programmable nucleases act in a similar way, in which a synthesized pool of mutagenic DNA donors flanked by homology arms serves as the template for repair. Therefore, mutations encoded in the donors can be delivered to the target loci, creating a site-directed mutant library on the genome.
A ZFN is an editable FokI type IIS restriction enzyme. The feature that the binding and cleavage activities of the wildtype enzyme are separable [85] makes it possible to replace its binding domain with a zinc-finger protein [86], enabling a programmable recognition pattern. Since a homodimer of FokI is required to cleave DNA, ZFNs are always working in pairs [87], doubling the recognition sites with a 5-7bp interspace between them [88]. Typically, a ZFN employs 3–6 Cys2His2 zinc-fingers with each one recognizing a triplet [83]. In this way, one can expect to shape a ZFN to recognize any 9-18bp sequence simply by modular assembly. While the fact is that the binding ability of a zinc-finger is susceptible to its neighbors [89], rearranged ZFNs often endure dramatic loss of binding affinity or specificity [90], [91]. It is toxic to the host cell. Though intensively optimized [92], [93], up to now, no comprehensive coding table between ZFNs and the 64 triplets is available [89]. At a time, ZFNs was mainly used in higher eukaryotes for targeted editing [94], [95], and seldom seen in microbes probably owing to its prohibitive cost in getting functional tailor-made ZFNs.
TALENs differ from ZFNs only in the DNA-binding domain. Instead of zinc-finger proteins, TALEs (transcription activator-like effectors) from the Xanthomonas plant pathogens are fused to FokI to give the guidance [83]. A TALE contains 33–35 highly conserved amino acid residues [96], except for the two residues at position 12 and 13 specifically recognize one single base, which are termed RVDs (repeat variable di-residues) [97]. Commonly used RVDs are Asn-Ile for A, Asn-Asn for G, His-Asp for C and Asn-Gly for T [97]. TALEs' much larger size compared with ZFs (zinc-finger proteins) [89] and conserved composition may account for their functional insulation from each other, surpassing ZFNs in high-performing modular assembly [98]. However, TALENs generally consist of almost 20 tandem repeats of TALEs [83], the conserved sequences among them are prone to rearrange through recombination, producing random truncations, which is a main cause for construction in and delivery failure [99]. Attempts have been made to pave the way for easy manipulation, including Golden Gate Assembly [100], FLASH (fast ligation-based automatable solid-phase high-throughput) Assembly [101] and hierarchical LIC (ligation-independent cloning) Assembly [102]. Most studies involving TALENs are carried out in higher eukaryotes [103], [104]. The unstable repetitive sequence structure and still the high pricing may be responsible for their diminishing applications in prokaryotes like E. coli.
An air of faded prosperity soon pervaded the research fields of ZFNs and TALENs, when CRISPR/Cas, abbreviated from clustered regularly interspaced short palindromic repeats and associated protein, rose from the horizon. Hailing from 47% of the bacteria and archaea sequenced [105], CRISPR/Cas systems are a Red Queen strategy [106] adopted by the host to withstand foreign genetic element invasion [107]. Among all the CRISPR/Cas systems uncovered, Class 2 Type II systems possess the simplest constitution [105]. Cas9, the core Cas protein in this system, has proved to be a preeminent programmable nuclease in targeted genome editing. It works in an RNA-guided recognition and cleavage fashion. In the application of targeted mutagenesis, a specific DNA sequence of the target region, which is typically 20 bp in length and followed by a short PAM (protospacer-adjacent motif), is embedded in the artificial CRISPR operon, and transcribed into a pre-crRNA (pre-CRISPR RNA). After partially hybridized to a conserved tracrRNA (trans-encoded RNA) and cleaved by the ribonuclease RNase III [108], the pre-crRNA-tracrRNA heteroduplex is processed into a mature targeting complex which then bind and activate Cas9 [109], [110]. For simplification, a single chimeric RNA, which is later on called sgRNA (single-guide RNA), was successfully engineered to mimic and substitute the natural duplex and dispense with the use of RNase III [111]. The dualRNA-Cas9 or sgRNA-Cas9 nuclease can survey the whole genome for PAM sites via Cas9 itself and unwind the double strand in the vicinity of a PAM to enable a base-pairing inspection, performed by the 20-bp seed in the crRNA or sgRNA, of the target sequence. Upon an identical sequence is detected near a PAM, the nuclease will induce a DSB within the target site [109]. Diversity of the site can be created following the HDR pathway, if a library of mutation-containing DNA donors, either ssDNA or dsDNA, with flanking homology to the target is introduced. CRISPR/Cas9 can also be modified into a multiplex genome editing tool by the simultaneous expression of an array of crRNAs [112] or sgRNAs [113]. Generally, the λ-Red system is incorporated to facilitate the homologous recombination [114].
Salute must be extended to the CRISPR/Cas9 system, for the RNA-DNA pairing rather than the protein-DNA interaction significantly simplifies the design and engineering processes before the programmable nuclease is ready for use, rendering it an affordable technology for various studies, especially for high-throughput genome research. Recent year, studies on microbial metabolic engineering using CRISPR/Cas9 has sprung up. Researchers endeavor to make the full use of the method to engineer microbes with enhanced carbon source uptake rates [115], [116] and target molecule productivities [113], [117], [118]. In a study carried out by Ryan in 2014 [119], a mutant library of the cellodextrin transporter gene cdt-1 was generated through error-prone PCR, and integrated into the URA3 locus of a yeast strain using CRISPR/Cas9 for the directed evolution of the transporter protein. After one round of mutagenesis and selection, a mutant cdt-1 resulting in a 2.6-fold improved cellobiose utilization ability of the host strain was obtained. Analysis of the beneficial mutations was made to instruct the further engineering of the protein.
The off-target effect is universally observed in any process involving recognition. So it is with the three programmable nucleases [120], [121], [122], [123], [124], [125]. Researchers conjectured that their off-target effects might be indispensable to the original systems' tolerance to evolving genetic elements [89]. To defense against the effect, methods predicting and quantifying the non-canonical cleavage have been delivered [126]. In the light of these theoretical studies, optimizations of the recognition and cleavage modules proceed with progressive results. For ZFNs and TALENs, an engineered FokI with more stringent dimerization was applied to eliminate undesired homodimers, which effectively improved the specificity of cleavage [127], [128]. For CRISPR/Cas9, the structure and target sequence of the sgRNA proved to be central to its activity and specificity. Various schemes of designing high-performed sgRNA are available now [129], [130], [131], [132]. Recently, a modified Cas9 with mutations reducing the non-specific DNA contacts was described based on energy analysis, significantly lessening the off-target effect [133]. Meanwhile, some researchers are paying attention to the repair process. They have demonstrated that the use of nickases instead of nucleases are beneficial to specific recombination, since the single-strand breaks they create are not favored by the error-prone NHEJ repair pathway [134], [135], [136].
2.2. In vivo targeted mutagenesis methods
2.2.1. Orthogonal DNA polymerase-plasmid pairs
In 2000, Fabret and coworkers described an in vivo targeted random mutagenesis method in E. coli based on the deepening understanding of plasmid replication processes. To the best of our knowledge, it was the first time that targeted mutagenesis had been implemented in vivo. The method takes the advantage of ColE1 and its derivative plasmids, the replication of which demands the participation of the host's DNA PolI (DNA polymerase I) [137]. It is clear that PolI is essential in lagging strand synthesis [138], DNA excision repair [139] and ColE1-type plasmid replication initiation [140]. In the third situation, DNA PolI unidirectionally initiates the synthesis of the leading strand by extending the RNA primer annealed at the start point. The conformation of the primer is anticipated to sterically block the entry of DNA Pol III (DNA polymerase III) responsible for bulk replication. The DNA PolI-initiated synthesis ends at the formation of a primosome, which marks the beginning of lagging strand synthesis and the switch of DNA PolI to the more efficient Pol III [140]. Though the molecular process is explicit, the precise initial synthesis length covered by DNA PolI still remains indefinite, ranging from 400 to 2000 nucleotides claimed by different studies [137], [140], [141]. Anyway, the research work has testified to the possibility of generating a random mutation library of several hundred base pairs downstream of the ColE1 ori by using a proofreading-deficient error-prone DNA PolI encoded on the genome.
In the proof-of-concept case of LacI targeted mutagenesis [137], where LacI negative mutant would interrupt the binding of wild-type LacI to lacO by competition or forming dead heterotetramers, the mutation frequency was about 57 per million cells after 30 generations, representing a more than 5000-fold improvement against the genome background indicated by the rifampicin resistance, which is a critical index in targeted mutagenesis in vivo. A paralyzed mismatch repair system gave another raise in absolute targeted mutation frequency of 20–40-fold, while obscured the contrast between targeted and global mutation frequencies. The researchers themselves evaluated an actual targeted mutation frequency of more than 1%, according to an underestimate by the incomprehensive LacI mutation assay. To be detailed, the area close to the origin mutated 6–20-fold more frequently compared with the remote area.
Modification to the method was made by Loeb lab. They adopted a conditional inviable E. coli strain of JS200 as the host cell to enable a plug-and-play form of the error-prone DNA PolI [142]. In this strain, the wildtype DNA PolI is replaced by a temperature-sensitive allele, which is lethal in rich media at elevated temperature, and can be complemented by a variety of polymerases [143]. They first constructed novel error-prone DNA PolI mutants with different performances based on their previous study [144]. By co-transforming the two plasmids respectively carrying the error-prone DNA PolI and the target gene into JS200, an in vivo targeted mutagenesis platform is ready to use. In the β-lactamase reversion assay, under optimal conditions, the targeted mutation frequency reached strikingly 2.1 per thousand cells after one round of cultivation with a targeting effect of 400-fold. A per-base mutation frequency of 0.81 per kb was deduced and a 3-kb mutable region following the ColE1 ori was observed in this study.
An analogous orthogonal replication system was reported in yeast exploiting the cytoplasmic plasmid pair originated from Kluveromyces lactis [145]. The two double-stranded linear plasmids replicate independently of each other as well as the host's replication system. Either of them is covalently tethered to TPs (terminal proteins) at the 5’-terminus of both strands. The hydroxyl group of a specific serine in the protein functions as the replication primer, which cannot be recognized by the host's polymerase. A self-encoded TP-DNAP (TP-DNA polymerase fusion) is the leading role in the autonomous replication process [146]. The strict orthogonality of the system ensured the targeted mutagenesis to be carried out by Ravikumar and coworkers. In the study [145], they engineered an error-prone TP-DNAP to specially mutate the corresponding TP-plasmid containing the target gene. One of the plasmid carried the target gene only, with its TP-DNAP encoded separately in a yeast nuclear plasmid for better performance. The other plasmid harbored all the components for replication and transcription. In the LEU2 reversion assay, the per-base mutation frequency approximated 40 per billion bp after 10-day passaging, which was 400-fold higher than that of the genome.
Owing to its high host specificity, the application of this method is rare. In the E. coli version, the founders themselves presented an enzyme engineering work, where they evolved the TEM-1 β-lactamase to confer a 160-fold improvement of aztreonam resistance on the host cell. Subsequent sequencing analysis revealed prevailing beneficial mutations afore identified in clinical research [142]. Besides, two other studies focused as well on protein engineering, respectively for a better catalytic activity [147] and a stronger binding ability [148]. Interestingly, coupled with β-lactamase assay, the platform was also reversely employed to guide the directed evolution of active polymerases with reduced fidelity [149], [150]. As for the yeast version, it is more like an orthogonal module in synthetic biology, and was proposed by many to help build synthetic circuits [151], [152] and sustain biological containment [153], [154].
As is mentioned above, DNA PolI also functions in gap-filling process between any two adjacent Okazaki fragments in E. coli. Thus, the use of the error-prone PolI leaves hidden dangers of error catastrophe scattering over the genome, and greatly reduces the orthogonality between the plasmid and the genome. This may account for the abnormally high global mutation frequency of up to one per million bp in the JS200 strain. Things could be better for yeast, since the replication of the TP-plasmids is strictly autonomous by adopting a special protein-primed mechanism. However, it is cumbersome to edit the TP-plasmids, for they are protein-linked and can only be effectively handled through in vivo homologous recombination. Besides, to transfer the plasmid to another type of yeast, a protoplast fusion process is needed [155]. Future work is demanded to improve the practicability of the method.
2.2.2. TaGTEAM
TaGTEAM, standing for targeting glycosylase to embedded arrays for mutagenesis, came into sight one year before the orthogonal replication system in yeast was established, and was reckoned to be the first case of in vivo targeted mutagenesis in yeast. In the method [156], the yeast 3-methyladenine DNA glycosylase Mag1p acts as a mutagenic agent, and is fused to the tet repressor protein (tetR). The tetR protein can specifically recognize and bind to an array of tet operator sequences (tetO), anchoring the mutator Mag1p in the site where the tetO array is embedded. In this way, the mutator protein can be enclosed in a user-defined area on the genome or a plasmid, ensuring a form of in vivo targeted mutagenesis.
Since a homodimer tetR is active in DNA binding, to avoid steric hindrance between two Mag1p proteins upon tetR dimerization, a sctetR (single chain tetR) having two normal tetR proteins fused together is applied in this method. The Mag1p enzyme, which is primarily involved in DNA base excision repair by removing the alkylated bases, is reported to have a much broader substrate spectrum [157] and be able to induce an approximate 600-fold increase of spontaneous mutation frequency in yeast if overexpressed compared with the apurinic/apyrimidinic endonuclease [158]. These all lay an experimental base for the method.
In the proof-of-concept assay, a 240-copy tetO array of target region was embedded in the yeast chromosome I. The auxotroph marker URA3 was integrated at different sites flanking the array to profile the distance-dependent feature of the mutation rate. The global mutation rate was indicated by a CAN1 marker on chromosome V. According to the study, the relationship between distances and mutation rates was bell-shaped. It peaked at the URA3 marker 0.3-kb next to the target array with a mutation rate of 31 per billion cell per generation, which was more than 800-fold higher than that without the protein mutator. Significant increase in URA3 mutation rates covered a 20-kb region centered on the target array, and still existed with a moderate increase of 40-fold when there was no array for some unknown mechanism. The global mutation rate in CAN1 marker remained relatively unchanged throughout the assay. A comparable target mutation rate was seen with the case of sctetR-FokI, where the nuclease was anchored in certain area for repair mutagenesis, albeit with a negligible increase of URA3 markers in the absence of the target array. Elaborate analyses of the two systems revealed different repair pathways they underwent after double strand ends or breaks. For the glycosylase, a high proportion of HDR-mediated point mutagenesis was preferred, while an NHEJ repair was dominant to the nuclease [156].
No direct utilization of the method has been reported. It is quite laborious to integrate a highly repetitive target array into the genome. Whether the insertion is detrimental to the target gene and genes nearby needs evaluation. In addition, the 20-kb mutagenic length is unadjustable and inappropriate for fine modification. As for the off-target effect, since the function of Mag1p does not rely on the binding of tetR to tetO, it has the potential to import a considerable number of non-specific mutations across the genome. Though remains to be optimized, this method has raised a novel thinking of in vivo targeting. A more strict and convenient way of target binding needs to be introduced in future work.
2.2.3. Retrotransposon-based targeted mutagenesis
Last year, Crook and coworkers depicted an original yeast ICE (in vivo continuous evolution) approach, and released the first demonstration of ICE-mediated gene and pathway optimization in yeast [159]. The yeast native retrotransposon Ty1 is the core element in the method. It behaves in a quite similar way to the class of ssRNA retroviruses like the notorious lentivirus HIV (human immunodeficiency virus). In fact, they have a shared replication strategy. The genome of the virus-like particle Ty1 contains two open reading frames, and they are the Gag ORF (open reading frame) encoding a single protein that serves as the capsid and chaperone, and the Pol ORF encoding three enzymes, namely the protease, integrase and reverse transcriptase with RNase H activity. Briefly, Ty1 on the genome is first transcribed into a ssRNA, which is then reverse transcribed into a Ty1 dsDNA by the Ty1 reverse transcriptase using the host initiator methionine tRNA as its primer. The newly synthesized Ty1 dsDNA is finally reintegrated into the genome with the help of the integrase [160]. Since the replication process is in the charge of the inherently error-prone Ty1 reverse transcriptase, which presents an error rate of around 10 per billion bp per replication [161], it allows the continuous mutating of the Ty1 cassette, forming a prototype of retrotransposon-based in vivo targeted mutagenesis. The evidence that heterologous genes can be expressed when inserted between the Pol ORF and the 3′-LTR (3’-long terminal repeat) further pushes it to reality [162]. Based on these knowledge of Ty1, Crook established this method.
After several rounds of optimization to the transposition efficiency, the retrotransposon-based method was able to bring about a mutation rate of 0.15 per kb at the URA3 site according to next generation sequencing analysis. The mutation spectrum of the method resembles that of other error-prone polymerases, with a uniform distribution of mutations. In addition, the method can include as long as 5-kb heterologous insert with no significant drop in transposition rate, making it feasible to manipulate large DNA fragments such as a pathway [159].
Crook made three demonstrations of applying the method [159]. In the first case, the substrate specificity of the orothidine-5’-phosphate decarboxylase Ura3p was the evolution target. After two rounds of targeted mutagenesis, a Ura3p mutant with a 2.5-fold decrease in 5-FOA (5-fluoro-orotic acid) utilization and an unchanged uracil synthesis activity was obtained, conferring 5-FOA resistance and uracil prototroph on a new host cell. The global transcriptional regulator Spt15p was also involved. In this case, two rounds of mutagenesis gave rise to the emergence of a Spt15p variant rendering the host 3.5% butanol resistance nearly twice that of the wildtype in the early stage of cultivation. The xylose catabolism pathway consisting of a xylose isomerase and a xylulokinase was set as the pathway evolution case. After one week of real sense of ICE, several variants with distinct mutations were isolated with increased exponential growth rates and shortened lag phases, which indicated the practicability of the method in long cassette engineering.
Though proved feasible, the retrotransposon-based method still poses a problem that requires attention. The native Ty1 is a mobile genetic element. Its insertion into the genome relies on the integrase rather than the sequence-similarity-based homologous recombination. Therefore, the wildtype Ty1 is not to be replaced by the newly mutated ones. Generally, Ty1 tends to integrate into the upstream regions of all RNA polymerase III transcribed genes, including tRNA genes and 5S rRNA gene [163]. They are all ‘housekeeping’ genes essential to cell viability. Although the native Ty1 proves not lethal to yeast, it remains to be confirmed that the multi-site integration of the engineered Ty1 with heterologous gene expression is always harmless and globally non-mutagenic. Even so, the relatively high copy number of Ty1 on the genome, about 32 full-length Ty1 out of 200–300 in total [164], will bring trouble to the selection process, for a pool of mutated Ty1 is spread in one genome, and only when the optimal genotype predominates the population may it alter the phenotype and be isolated, which is also encountered by many other multi-copy mutagenesis systems.
Actually, early in 2014, a study by Farzadfard adopted a similar reverse transcription design [165]. The study was carried out in E. coli, and the E. coli native retron Ec86 was the core functional module. Unlike Ty1, after transcription and reverse transcription, Ec86 only produces ssDNA corresponding to its encoded context. A ssDNA homologous recombination mechanism was employed in their work to rewrite the information of the target area on the genome. The single-copy and site specific features of the system may be enlightening to this type of targeted mutagenesis.
3. Internal logic among existing targeted mutagenesis methods
In most cases, mutations originate from replication and repair errors. Three factors are directly involved in an error-prone replication process. The first one is error-prone polymerases. Once the active center of a polymerase is modified, mis-incorporation of dNTPs may occur, and they can hardly be removed by a polymerase lacking the proofreading ability. The second one is substrate analogues. Chemicals similar to dNTPs in structure may confuse the replication machine, giving rise to mis-insertion. The last comes from environment. Altered catalytic conditions are likely to confer altered polymerase fidelity. As for repair errors, DNA damage followed by a low-fidelity or human-intervened repair process is often the case. Common damages include base modifications, mismatches and double strand breaks. For base errors, an impaired or interfered repair system would be unable to make corrections or make wrong decisions. For DSBs, they can be repaired either through a user-defined HDR pathway when homologous donors are provided, or through an error-prone NHEJ pathway in the absence of the donors. These understandings about mutations are always concerned before a novel mutagenesis method is born.
However, when it comes to proposing a targeted mutagenesis method, more things should be taken into consideration. The major challenge is how to make it targeted, that is how to increase the mutation rate of a target region while maintaining a stable genetic background.
A basic thinking of it was provided by in vitro targeted mutagenesis methods, which represent the early-to-middle stage of targeted mutagenesis development. Generally, in vitro targeted mutagenesis methods follow the principle of isolation. To retain an undisturbed host genome, the target gene is always isolated for in vitro diversification, and then reintroduced into the host cell for subsequent characterization. Error-prone PCR is the earliest demonstration of targeted mutagenesis. It takes the advantage of the low-fidelity Taq polymerase and a mutagenic reaction condition to make replication errors to an isolated gene of interest. Owing to its relatively low mutation rate and biased spectrum, error-prone PCR acts more like a perturbation probe, telling us which nucleotides or amino acids probably matter, thus narrowing down the optimization space to the more focused single-site or multi-site analysis, which is later on fulfilled by MAGE and PFunkel. These two methods have many features in common. They both create diversities through in vitro random synthesis of degenerate primers partially complementary to the target gene, followed by the annealing of the primer pool to the target site to form mismatches. After which they both rely on repair errors to fix these mutations. In MAGE, a mutS-deficient strain is used to endure mismatched base pairs. While in PFunkel, the host repair system is guided by design to modify the uracil-containing template strand rather than the mutated one. The main difference lies in their positioning, for MAGE is aimed at genome editing, and PFunkel is plasmid manipulation. Though proved effective in genome editing, an optimal primer design and continuous passaging are necessary to sustain a moderate recombination rate of 30% per 2–2.5 h in MAGE [55], which can be resource-consuming in the case with a huge library size. Efforts have always been made to compensate for the low recombination efficiency. Early in 2003, a remarkable 2-5 orders-of-magnitude increase in recombination level after the introduction of a DSB was reported in yeast [166], which is also seen in other organisms [167]. It is reasonable to single cell organisms since a DSB within the genome can stimulate the HDR pathway when a homologous donor exists. On the other hand, if the host cannot repair the DSB using the tailor-made donor, a broken genome will usually lead to cell death, efficiently bringing down the wildtype ratio. This is what the programmable nuclease method is founded on. In this method, repair errors are generated by favoring a user-defined HDR pathway at the target cleavage site. The diversity is created by in vitro random synthesis of homologous fragments of the target gene. Replication errors can also be included if these fragments are produced by error-prone PCR.
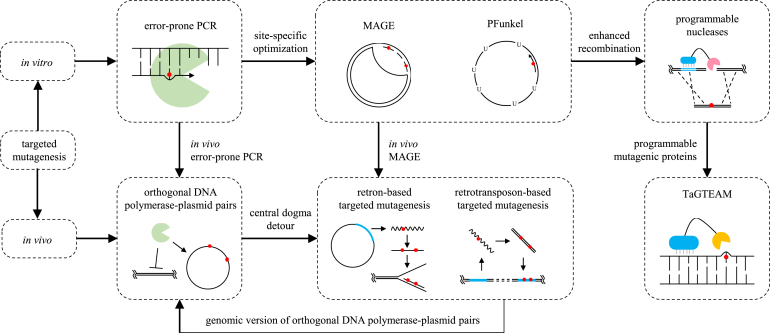
In response to the ascending in vivo continuous evolution, or so-called next-generation evolution [159], research on developing in vivo targeted mutagenesis methods booms. Existing in vivo methods all established on specialized genetic elements, some of which can be highly species-specific, but inherently, they have taken lessons from these in vitro methods. The principle of in vivo targeted mutagenesis methods is orthogonality. Without any human intervention, it is almost impossible to isolate a gene and individually modify it inside a cell. However, we can exploit orthogonal genetic tools to make it targeted. By restrict the target gene under the control of orthogonal tools, one can expect a separate manipulation to the gene with little impact on the genome. The orthogonal DNA polymerase-plasmid pairs imitate the in vitro error-prone PCR, albeit with a specialized error-prone polymerase which solely responsible for the replication of an area or whole sequence of a specialized plasmid. They create mutations through replication errors by using error-prone polymerases. A weakened repair system can also help by allowing repair errors. Nevertheless, there are too few naturally occurring orthogonal replication pairs to satisfy our need. According to the central dogma, instead of the DNA replication process, the cooperation of transcription and reverse transcription can also pass messages from DNA to DNA. If low-fidelity polymerases are involved, both routes can lead to DNA errors, which opened a new direction for proposing targeted mutagenesis methods. Therefore, in addition to the orthogonal pairs, we have the retro-element-based method. This method, either performed in E. coli or yeast, generates mutations through replication errors. The utilization of a reverse transcriptase turns the originally non-heritable transcription errors into heritable mutations in DNA. The low-fidelity reverse transcription process also contributes to the mutation rate. The orthogonality of the method is ensured by the exclusive reverse transcription mechanisms. Though closely related, the E. coli version, which is still in proposal, and the yeast version differs in mutation fixation. The E. coli native retron produces ssDNAs, which then undergo the ssDNA recombination pathway similar to MAGE. Thus, we can consider it as in vivo MAGE. While the yeast retrotransposon generates dsDNAs, which is then inserted into a genome site by the self-encoded integrase. In a sense, the yeast version is just like a genomic orthogonal polymerase-plasmid pair, since the target gene is always within itself, and no replacement is happened on the genome. As for TaGTEAM, it may be more straightforward to call it programmable mutagenic proteins. Repair errors account for the mutations. The protein mutators are guided to the neighborhood of the target gene through their DNA binding domains to sustain the orthogonality to the non-target area. An inspiration from the programmable nucleases is that the DNA recognition module of TaGTEAM can be substituted by CRISPR/dCas9 (catalytically dead Cas9) system, which may remarkably increase the target accuracy and feasibility. A similar application has proved workable where a cytidine deaminase-dCas9 fusion is used to make precise modifications to the target cytidine [168]. An illustration of the internal logic is shown in Fig. 1.
Fig. 1.
Internal logic among existing targeted mutagenesis methods. Targeted mutagenesis methods are classified into the in vitro type and in vivo type. The in vitro type includes error-prone PCR, MAGE, PFunkel and programmable nucleases. Error-prone PCR is adept at discovering key residues within a target gene. Then these residues can be subject to MAGE or PFunkel for further optimization. To compensate for the insufficient recombination level in microbes, programmable nucleases are developed to induce DSBs in the target sites, stimulating the HDR pathway. The in vivo methods take lessons from the in vitro ones. They are mainly orthogonal DNA polymerase-plasmid pairs, retro-element-based targeted mutagenesis and TaGTEAM. Orthogonal DNA polymerase-plasmid pairs are responsible for in vivo error-prone PCR, but there are too few of them to satisfy our need. DNA polymerases directly transfer messages from DNA to DNA, which can also be achieved through the coupling of transcription and reverse transcription processes according to the central dogma. This is what the retro-element-based targeted mutagenesis method is founded on. The E. coli retron-based version of this method is just like an in vivo form of MAGE, while the yeast retrotransposon-based version resembles orthogonal DNA polymerase-plasmid pairs albeit on the genome. As for TaGTEAM, it is actually programmable mutagenic proteins.
With these tools in hand, we need think it over which one is best suited in a certain condition. Each method has its own positioning. As for MAGE, programmable nucleases, retron-based targeted mutagenesis and TaGTEAM, they all aims at genome modification. If the mutagenesis target is an endogenous gene located on the genome, it is recommended to use these four methods. While for error-prone PCR, PFunkel, orthogonal DNA polymerase-plasmid pairs and retrotransposon-based targeted mutagenesis, their target genes are always harbored on mobile genetic elements like plasmids and retrotransposons. So, they are preferred when we intend to optimize exogenous genes encoded in mobile elements. These are merely suggestions. Things may be different in practice.
4. Prospects
A method with extraordinary performance is always demanded, while the road to it can be quite arduous. Instead of establishing a brand-new method which generally depends on novel findings, the development of existing methods is usually more practical. Traditional targeted mutagenesis methods have received extensive modifications individually as mentioned above. Nowadays, combinatorial optimization is pursued for synergetic effects. Typical cases are CRISPR-MAGE [69], CRISPR-error-prone PCR [119] and CRISPR-base editing enzyme [168]. Additional combinations will emerge to adapt to different situations. Technologies and concepts from other fields can also be infused, especially those in molecular biology and synthetic biology. Novel DNA editing tools can be applied in mutation fixation. Synthetic circuits may provide new thoughts of ‘how to make it targeted’. To conventional in vitro targeted mutagenesis methods, the isolated manipulation of mutant library makes it universally applicable. While for other host-reliant methods, a more portable version is urgently needed in future studies, which means that all the functional components in these methods should be independent of the genome, and act in a ‘plug-and-play’ manner or have their counterparts across different species.
Targeted mutagenesis primarily deals with protein engineering, and is now extended to non-coding region and short pathway optimization. Choosing target sites requires basic knowledge about the research system. Actually, vague information is enough, since targeted mutagenesis also answers yes-or-no questions. In the next step, it tells us which are the key points within the target site and their optimal alternatives. Therefore, a general engineering scheme of global mutagenesis mining whole genome sites followed by targeted mutagenesis handling them in detail comes into sight. In this way, an intricate global optimization issue can be narrowed down to studies on a few hotspots. Aside from these engineering applications, we can imagine that when coupled with in vivo selection, targeted mutagenesis will show its matchless power in tracing the evolutionary trajectory of a user-defined region, which is a vital long-term topic in scientific research. In addition to its conventional role, proposals of innovative targeted mutagenesis applications have arisen. The highly orthogonal in vivo targeted mutagenesis methods provide basic genetic elements for synthetic biology to construct hierarchical circuits. Orthogonality can also erect a firewall between artificiality and nature in biocontainment design as is aforementioned.
Targeted mutagenesis helps to elucidate the unknown biological field, and is conversely benefit from its new findings through novel method establishment. This iterative relationship will expedite the refinement of the methods, making opportunities for accessing and better exploiting the micro-world.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (NSFC 21627812 & 21676156).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.RC W., JN T.J. vol. 7. Springer Netherlands; Dordrecht: 1998. (Mutation and evolution). [Google Scholar]
- 2.Bassalo M.C., Liu R., Gill R.T. Directed evolution and synthetic biology applications to microbial systems. Curr Opin Biotechnol. 2016;39:126–133. doi: 10.1016/j.copbio.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 3.Lynch M. Evolution of the mutation rate. Trends Genet. 2010;26:345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tenaillon O., Barrick J.E., Ribeck N., Deatherage D.E., Blanchard J.L., Dasgupta A. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature. 2016;536:165–170. doi: 10.1038/nature18959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller H.J. Artificial transmutation of the gene. Science (80- ) 1927;66:84–87. doi: 10.1126/science.66.1699.84. [DOI] [PubMed] [Google Scholar]
- 6.Stadler L.J., Sprague G.F. Genetic effects of ultra-violet radiation in maize: I. Unfiltered radiation. Proc Natl Acad Sci. 1936;22:572–578. doi: 10.1073/pnas.22.10.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X., Zhang X.F., Li H.P., Wang L.Y., Zhang C., Xing X.H. Atmospheric and room temperature plasma (ARTP) as a new powerful mutagenesis tool. Appl Microbiol Biotechnol. 2014;98:5387–5396. doi: 10.1007/s00253-014-5755-y. [DOI] [PubMed] [Google Scholar]
- 8.Auerbach C., Robson J.M. Chemical production of mutations. Nature. 1947;105:243–247. doi: 10.1126/science.105.2723.243. [DOI] [PubMed] [Google Scholar]
- 9.Kim Y., Schumaker K.S., Zhu J.-K. vol. 323. Humana Press; New Jersey: 2006. EMS mutagenesis of arabidopsis; pp. 101–103. (Arab. Protoc.). [Google Scholar]
- 10.Greener A., Callahan M., Jerpseth B. vol. 57. Humana Press; New Jersey: 1996. An efficient random mutagenesis technique using an E. coli mutator strain; pp. 375–385. (Vitr. Mutagen. Protoc.). [DOI] [PubMed] [Google Scholar]
- 11.Esvelt K.M., Carlson J.C., Liu D.R. A system for the continuous directed evolution of biomolecules. Nature. 2011;472:499–503. doi: 10.1038/nature09929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Badran A.H., Liu D.R. Development of potent in vivo mutagenesis plasmids with broad mutational spectra. Nat Commun. 2015;6:8425. doi: 10.1038/ncomms9425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kazazian H.H. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 14.Hughes S.R., Gibbons W.R., Bang S.S., Pinkelman R., BischoV K.M., Slininger P.J. Random UV-C mutagenesis of ScheVersomyces (formerly Pichia) stipitis NRRL Y-7124 to improve anaerobic growth on lignocellulosic sugars. J Ind Microbiol Biotechnol. 2012;39:163–173. doi: 10.1007/s10295-011-1012-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luan G., Cai Z., Li Y., Ma Y. Genome replication engineering assisted continuous evolution (GREACE) to improve microbial tolerance for biofuels production. Biotechnol Biofuels. 2013;6:137. doi: 10.1186/1754-6834-6-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luan G., Bao G., Lin Z., Li Y., Chen Z., Li Y. Comparative genome analysis of a thermotolerant Escherichia coli obtained by Genome Replication Engineering Assisted Continuous Evolution (GREACE) and its parent strain provides new understanding of microbial heat tolerance. N Biotechnol. 2015;32:732–738. doi: 10.1016/j.nbt.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 17.Huang M., Bai Y., Sjostrom S.L., Hallström B.M., Liu Z., Petranovic D. Microfluidic screening and whole-genome sequencing identifies mutations associated with improved protein secretion by yeast. Proc Natl Acad Sci U. S. A. 2015;112:E4689–E4696. doi: 10.1073/pnas.1506460112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robins W.P., Faruque S.M., Mekalanos J.J. Coupling mutagenesis and parallel deep sequencing to probe essential residues in a genome or gene. Pnas. 2013;110:E848–E857. doi: 10.1073/pnas.1222538110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kokes M., Dunn J.D., Granek J.A., Nguyen B.D., Barker J.R., Valdivia R.H. Integrating chemical mutagenesis and whole-genome sequencing as a platform for forward and reverse genetic analysis of Chlamydia. Cell Host Microbe. 2015;17:716–725. doi: 10.1016/j.chom.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brune W., Ménard C., Hobom U., Odenbreit S., Messerle M., Koszinowski U.H. Rapid identification of essential and nonessential herpesvirus genes by direct transposon mutagenesis. Nat Biotechnol. 1999;17:360–364. doi: 10.1038/7914. [DOI] [PubMed] [Google Scholar]
- 21.Camacho L.R., Ensergueix D., Perez E., Gicquel B., Guilhot C. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol Microbiol. 1999;34:257–267. doi: 10.1046/j.1365-2958.1999.01593.x. [DOI] [PubMed] [Google Scholar]
- 22.ORGEL L.E. The maintenance of the accuracy of protein synthesis and its relevance to ageing. Proc Natl Acad Sci U. S. A. 1963;49:517–521. doi: 10.1073/pnas.49.4.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaaper R.M. Mechanisms of mutagenesis in the Escherichia coli mutator mutD5: role of DNA mismatch repair. Proc Natl Acad Sci U. S. A. 1988;85:8126–8130. doi: 10.1073/pnas.85.21.8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fijalkowska I.J., Schaaper R.M. Mutants in the Exo I motif of Escherichia coli dnaQ: defective proofreading and inviability due to error catastrophe. Proc Natl Acad Sci U. S. A. 1996;93:2856–2861. doi: 10.1073/pnas.93.7.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raman S., Rogers J.K., Taylor N.D., Church G.M. Evolution-guided optimization of biosynthetic pathways. Proc Natl Acad Sci. 2014;111:201409523. doi: 10.1073/pnas.1409523111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee H.H., Molla M.N., Cantor C.R., Collins J.J. Bacterial charity work leads to population-wide resistance. Nature. 2010;467:82–85. doi: 10.1038/nature09354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toprak E., Veres A., Michel J.-B., Chait R., Hartl D.L., Kishony R. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet. 2012;44:101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leung D.W., Chen E., Goeddel D.V. A Method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique. 1989;1:11–15. [Google Scholar]
- 29.Cadwell R.C., Joyce G.F. Randomization of genes by PCR mutagenesis. Genome Res. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 30.Beaudry A.A., Joyce G.F. Directed evolution of an RNA enzyme. Science. 1992;257:635–641. doi: 10.1126/science.1496376. [DOI] [PubMed] [Google Scholar]
- 31.Bartel D.P., Szostak J.W. Isolation of new ribozymes from a large pool of random sequences. Science (80- ) 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- 32.Unrau P.J., Bartel D.P. RNA-catalysed nucleotide synthesis. Nature. 1998;395:260–263. doi: 10.1038/26193. [DOI] [PubMed] [Google Scholar]
- 33.Piganeau N., Jenne A., Thuillier V., Famulok M. An allosteric ribozyme regulated by doxycyline. Angew Chem Int Ed. 2000;39:4369–4373. doi: 10.1002/1521-3773(20001201)39:23<4369::AID-ANIE4369>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 34.Reetz M.T., Zonta A., Schimossek K., Jaeger K.-E., Liebeton K., Jaeger K.-E. Creation of enantioselective biocatalysts for organic chemistry by in vitro evolution. Angew Chem Int Ed Engl. 1997;36:2830–2832. [Google Scholar]
- 35.Chahar S., Elsawy H., Ragozin S., Jeltsch A. Changing the DNA recognition specificity of the EcoDam DNA-(adenine-N6)-methyltransferase by directed evolution. J Mol Biol. 2010;395:79–88. doi: 10.1016/j.jmb.2009.09.027. [DOI] [PubMed] [Google Scholar]
- 36.Song J.K., Rhee J.S., Song J. a EK. Simultaneous enhancement of thermostability and catalytic activity of phospholipase a 1 by evolutionary molecular engineering simultaneous enhancement of thermostability and catalytic activity of phospholipase a 1 by evolutionary molecular engineering. Appl Environ Microbiol. 2000;66:890–894. doi: 10.1128/aem.66.3.890-894.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogino H., Uchiho T., Doukyu N., Yasuda M., Ishimi K., Ishikawa H. Effect of exchange of amino acid residues of the surface region of the PST-01 protease on its organic solvent-stability. Biochem Biophys Res Commun. 2007;358:1028–1033. doi: 10.1016/j.bbrc.2007.05.047. [DOI] [PubMed] [Google Scholar]
- 38.Bordes F., Tarquis L., Nicaud J.M., Marty A. Isolation of a thermostable variant of Lip2 lipase from Yarrowia lipolytica by directed evolution and deeper insight into the denaturation mechanisms involved. J Biotechnol. 2011;156:117–124. doi: 10.1016/j.jbiotec.2011.06.035. [DOI] [PubMed] [Google Scholar]
- 39.Seelig B., Szostak J.W. Selection and evolution of enzymes from a partially randomized non-catalytic scaffold. Nat (London, U K) 2007;448:828–831. doi: 10.1038/nature06032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bosma T., Damborský J. Biodegradation of 1, 2, 3-trichloropropane through directed evolution and heterologous expression of a haloalkane dehalogenase gene. Appl Environ Microbiol. 2002;68:3582–3587. doi: 10.1128/AEM.68.7.3582-3587.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lingen B., Grötzinger J., Kolter D., Kula M.R., Pohl M. Improving the carboligase activity of benzoylformate decarboxylase from Pseudomonas putida by a combination of directed evolution and site-directed mutagenesis. Protein Eng. 2002;15:585–593. doi: 10.1093/protein/15.7.585. [DOI] [PubMed] [Google Scholar]
- 42.Sheppard T.L., Ordoukhanian P., Joyce G.F. A DNA enzyme with N-glycosylase activity. Proc Natl Acad Sci. 2000;97:7802–7807. doi: 10.1073/pnas.97.14.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mijts B.N., Lee P.C., Schmidt-Dannert C. Identification of a carotenoid oxygenase synthesizing acyclic xanthophylls: combinatorial biosynthesis and directed evolution. Chem Biol. 2005;12:453–460. doi: 10.1016/j.chembiol.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 44.Parikh M.R., Greene D.N., Woods K.K., Matsumura I. Directed evolution of RuBisCO hypermorphs through genetic selection in engineered E. coli. Protein Eng Des Sel. 2006;19:113–119. doi: 10.1093/protein/gzj010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z., Clawson a, Realini C., Jensen C.C., Knowlton J.R., Hill C.P. Identification of an activation region in the proteasome activator REGalpha. Proc Natl Acad Sci U. S. A. 1998;95:2807–2811. doi: 10.1073/pnas.95.6.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parnot C., Bardin S., Miserey-Lenkei S., Guedin D., Corvol P., Clauser E. Systematic identification of mutations that constitutively activate the angiotensin II type 1A receptor by screening a randomly mutated cDNA library with an original pharmacological bioassay. Proc Natl Acad Sci U. S. A. 2000;97:7615–7620. doi: 10.1073/pnas.110142297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haldimann A., Prahalad M.K., Fisher S.L., Kim S.-K., Walsh C.T., Wanner B.L. Altered recognition mutants of the response regulator PhoB: a new genetic strategy for studying protein-protein interactions. Proc Natl Acad Sci. 1996;93:14361–14366. doi: 10.1073/pnas.93.25.14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colas P., Cohen B., Ko Ferrigno P., Silver P.A., Brent R. Targeted modification and transportation of cellular proteins. Proc Natl Acad Sci U. S. A. 2000;97:13720–13725. doi: 10.1073/pnas.97.25.13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qin X., Qian J., Yao G., Zhuang Y., Zhang S., Chu J. GAP promoter library for fine-tuning of gene expression in Pichia pastoris. Appl Environ Microbiol. 2011;77:3600–3608. doi: 10.1128/AEM.02843-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yokobayashi Y., Weiss R., Arnold F.H. Directed evolution of a genetic circuit. Proc Natl Acad Sci U. S. A. 2002;99:16587–16591. doi: 10.1073/pnas.252535999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vartanian J.P., Henry M., Wain-Hobson S. Hypermutagenic PCR involving all four transitions and a sizeable proportion of transversions. Nucleic Acids Res. 1996;24:2627–2631. doi: 10.1093/nar/24.14.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao H., Giver L., Shao Z., Affholter J.A., Arnold F.H. Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat Biotechnol. 1998;16:258–261. doi: 10.1038/nbt0398-258. [DOI] [PubMed] [Google Scholar]
- 53.Wong T.S., Tee K.L., Hauer B., Schwaneberg U. Sequence saturation mutagenesis (SeSaM): a novel method for directed evolution. Nucleic Acids Res. 2004;32 doi: 10.1093/nar/gnh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang V.Q., Hogrefe H.H. Easy two-step method for randomizing and cloning gene fragments. Methods Mol Biol. 2010;634:399–407. doi: 10.1007/978-1-60761-652-8_28. [DOI] [PubMed] [Google Scholar]
- 55.Wang H.H., Isaacs F.J., Carr P.A., Sun Z.Z., Xu G., Forest C.R. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gallagher R.R., Li Z., Lewis A.O., Isaacs F.J. Rapid editing and evolution of bacterial genomes using libraries of synthetic DNA. Nat Protoc. 2014;9:2301–2316. doi: 10.1038/nprot.2014.082. [DOI] [PubMed] [Google Scholar]
- 57.Yu D., Ellis H.M., Lee E.C., Jenkins N.A., Copeland N.G., Court D.L. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U. S. A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang H.H., Church G.M. first ed. vol. 498. Elsevier Inc.; 2011. (Multiplexed genome engineering and genotyping methods: applications for synthetic biology and metabolic engineering). [DOI] [PubMed] [Google Scholar]
- 59.Isaacs F.J., Carr P.A., Wang H.H., Lajoie M.J., Sterling B., Kraal L. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science (80- ) 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang H.H., Kim H., Cong L., Jeong J., Bang D., Church G.M. Genome-scale promoter engineering by coselection MAGE. Nat Methods. 2012;9:591–593. doi: 10.1038/nmeth.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang H.H., Huang P.Y., Xu G., Haas W., Marblestone A., Li J. Multiplexed in vivo his-tagging of enzyme pathways for in vitro single-pot multienzyme catalysis. ACS Synth Biol. 2012;1:43–52. doi: 10.1021/sb3000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lajoie M.J., Rovner A.J., Goodman D.B., Aerni H.-R., Haimovich A.D., Kuznetsov G. Genomically recoded organisms expand biological functions. Science (80- ) 2013;342:357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rovner A.J., Haimovich A.D., Katz S.R., Li Z., Grome M.W., Gassaway B.M. Recoded organisms engineered to depend on synthetic amino acids. Nature. 2015;518:89–93. doi: 10.1038/nature14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Minty J.J., Lesnefsky A a, Lin F., Chen Y., Zaroff T a, Veloso A.B. Evolution combined with genomic study elucidates genetic bases of isobutanol tolerance in Escherichia coli. Microb Cell Fact. 2011;10:18. doi: 10.1186/1475-2859-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Binder S., Siedler S., Marienhagen J., Bott M., Eggeling L. Recombineering in Corynebacterium glutamicum combined with optical nanosensors: a general strategy for fast producer strain generation. Nucleic Acids Res. 2013;41:6360–6369. doi: 10.1093/nar/gkt312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dicarlo J.E., Conley A.J., Penttilä M., Jä J., Wang H.H., Church G.M. 2013. Yeast oligo-mediated genome engineering (YOGE) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bonde M.T., Kosuri S., Genee H.J., Sarup-Lytzen K., Church G.M., Sommer MO a. Direct mutagenesis of thousands of genomic targets using microarray-derived oligonucleotides. ACS Synth Biol. 2014 doi: 10.1021/sb5001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nyerges Á., Csörgo B., Nagy I., Latinovics D., Szamecz B., Pósfai G. Conditional DNA repair mutants enable highly precise genome engineering. Nucleic Acids Res. 2014:42. doi: 10.1093/nar/gku105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ronda C., Pedersen L.E., Sommer M.O.A., Nielsen A.T. CRMAGE: CRISPR optimized MAGE recombineering. Sci Rep. 2016;6:19452. doi: 10.1038/srep19452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kunkel T.A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U. S. A. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Firnberg E., Ostermeier M. PFunkel: efficient, expansive, user-defined mutagenesis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0052031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Firnberg E., Labonte J.W., Gray J.J., Ostermeier M. A comprehensive, high-resolution map of a Gene's fitness landscape. Mol Biol Evol. 2014;31:1581–1592. doi: 10.1093/molbev/msu081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kowalsky C.A., Faber M.S., Nath A., Dann H.E., Kelly V.W., Liu L. Rapid fine conformational epitope mapping using comprehensive mutagenesis and deep sequencing. J Biol Chem. 2015;290:26457–26470. doi: 10.1074/jbc.M115.676635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guntas G., Hallett R. a., Zimmerman S.P., Williams T., Yumerefendi H., Bear J.E. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc Natl Acad Sci. 2015;112:112–117. doi: 10.1073/pnas.1417910112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kowalsky C.A., Whitehead T.A. Determination of binding affinity upon mutation for type I dockerin-cohesin complexes from C lostridium thermocellum and C lostridium cellulolyticum using deep sequencing. Proteins Struct Funct Bioinforma. 2016;84:1914–1928. doi: 10.1002/prot.25175. [DOI] [PubMed] [Google Scholar]
- 76.Steinberg B., Ostermeier M. Shifting fitness and epistatic landscapes reflect trade-offs along an evolutionary pathway. J Mol Biol. 2016;428:2730–2743. doi: 10.1016/j.jmb.2016.04.033. [DOI] [PubMed] [Google Scholar]
- 77.Klesmith J.R., Bacik J., Michalczyk R., Whitehead T.A. Comprehensive sequence-flux mapping of a levoglucosan utilization pathway in E. coli. ACS Synth Biol. 2015;4:1235–1243. doi: 10.1021/acssynbio.5b00131. [DOI] [PubMed] [Google Scholar]
- 78.Fowler D.M., Fields S. Deep mutational scanning: a new style of protein science. Nat Methods. 2014;11:801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fowler D.M., Stephany J.J., Fields S. Measuring the activity of protein variants on a large scale using deep mutational scanning. Nat Protoc. 2014;9:2267–2284. doi: 10.1038/nprot.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kitzman J.O., Starita L.M., Lo R.S., Fields S., Shendure J. Massively parallel single-amino-acid mutagenesis. Nat Methods. 2015;12:203–206. doi: 10.1038/nmeth.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jin P., Kang Z., Zhang J., Zhang L., Du G., Chen J. Combinatorial evolution of enzymes and synthetic pathways using one-step PCR. ACS Synth Biol. 2016;5:259–268. doi: 10.1021/acssynbio.5b00240. [DOI] [PubMed] [Google Scholar]
- 82.Wrenbeck E.E., Klesmith J.R., Stapleton J.A., Adeniran A., Tyo K.E.J., Whitehead T.A. Plasmid-based one-pot saturation mutagenesis. Nat Methods. 2016;13:928–930. doi: 10.1038/nmeth.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim H., Kim J.S. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 84.McVey M., Lee S.E. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li L., Wu L.P., Chandrasegaran S. Functional domains in Fok I restriction endonuclease. Proc Natl Acad Sci U. S. A. 1992;89:4275–4279. doi: 10.1073/pnas.89.10.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim Y.G., Cha J., Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U. S. A. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bitinaite J., Wah D.A., Aggarwal A.K., Schildkraut I. FokI dimerization is required for DNA cleavage. Proc Natl Acad Sci. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Carroll D. Origins of programmable nucleases for genome engineering. J Mol Biol. 2016;428:963–989. doi: 10.1016/j.jmb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ramirez C.L., Foley J.E., Wright D a, Müller-Lerch F., Rahman S.H., Cornu T.I. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cornu T.I., Thibodeau-Beganny S., Guhl E., Alwin S., Eichtinger M., Joung J.K. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol Ther. 2008;16:352–358. doi: 10.1038/sj.mt.6300357. [DOI] [PubMed] [Google Scholar]
- 92.Isalan M., Klug A., Choo Y. A rapid, generally applicable method to engineer zinc fingers illustrated by targeting the HIV-1 promoter. Nat Biotechnol. 2001;19:656–660. doi: 10.1038/90264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Greisman H.A., Pabo C.O. A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- 94.Meyer M., de Angelis M.H., Wurst W., Kühn R. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc Natl Acad Sci U. S. A. 2010;107:15022–15026. doi: 10.1073/pnas.1009424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shukla V.K., Doyon Y., Miller J.C., DeKelver R.C., Moehle E a, Worden S.E. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 2009;459:437–441. doi: 10.1038/nature07992. [DOI] [PubMed] [Google Scholar]
- 96.Boch J., Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol. 2010;48:419–436. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- 97.Miller J.C., Tan S., Qiao G., Barlow K.A., Wang J., Xia D.F. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 98.Morbitzer R., Römer P., Boch J., Lahaye T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc Natl Acad Sci. 2010;107:21617–21622. doi: 10.1073/pnas.1013133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Holkers M., Maggio I., Liu J., Janssen J.M., Miselli F., Mussolino C. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2013:41. doi: 10.1093/nar/gks1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cermak T., Doyle E.L., Christian M., Wang L., Zhang Y., Schmidt C. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:7879. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reyon D., Tsai S.Q., Khayter C., Foden J.A., Sander J.D., Joung J.K. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schmid-Burgk J.L., Schmidt T., Kaiser V., Höning K., Hornung V. A ligation-independent cloning technique for high-throughput assembly of transcription activator–like effector genes. Nat Biotechnol. 2013;31:76–81. doi: 10.1038/nbt.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mussolino C., Morbitzer R., Lütge F., Dannemann N., Lahaye T., Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bedell V.M., Wang Y., Campbell J.M., Poshusta T.L., Starker C.G., Krug R.G. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Benton M.J. The red queen and the court jester: species diversity and the role of biotic and abiotic factors through time. Science (80- ) 2009;323:728–732. doi: 10.1126/science.1157719. [DOI] [PubMed] [Google Scholar]
- 107.Jackson S.A., McKenzie R.E., Fagerlund R.D., Kieper S.N., Fineran P.C., Brouns S.J.J. CRISPR-Cas: adapting to change. Science (80- ) 2017:356. doi: 10.1126/science.aal5056. [DOI] [PubMed] [Google Scholar]
- 108.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang H., La Russa M., Qi L.S. CRISPR/Cas9 in genome editing and beyond. Annu Rev Biochem. 2016;85:227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- 110.Barrangou R., van Pijkeren J.P. Exploiting CRISPR-Cas immune systems for genome editing in bacteria. Curr Opin Biotechnol. 2016;37:61–68. doi: 10.1016/j.copbio.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 111.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA – guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–822. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N. Multiplex genome engineering using CRISPR/cas systems. Science (80- ) 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jakočinas T., Bonde I., Herrgård M., Harrison S.J., Kristensen M., Pedersen L.E. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng. 2015;28:213–222. doi: 10.1016/j.ymben.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 114.Jiang W., Bikard D., Cox D., Zhang F., Marraffini La. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tsai C.S., Kong, Lesmana A., Million G., Zhang G.C., Kim S.R. Rapid and marker-free refactoring of xylose-fermenting yeast strains with Cas9/CRISPR. Biotechnol Bioeng. 2015;112:2406–2411. doi: 10.1002/bit.25632. [DOI] [PubMed] [Google Scholar]
- 116.Shi S., Liang Y., Zhang M.M., Ang E.L., Zhao H. A highly efficient single-step, markerless strategy for multi-copy chromosomal integration of large biochemical pathways in Saccharomyces cerevisiae. Metab Eng. 2016;33:19–27. doi: 10.1016/j.ymben.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 117.Jakočinas T., Rajkumar A.S., Zhang J., Arsovska D., Rodriguez A., ille Jendresen CB. CasEMBLR: cas9-facilitated multiloci genomic integration of in vivo assembled DNA parts in Saccharomyces cerevisiae. ACS Synth Biol. 2015;4:1226–1234. doi: 10.1021/acssynbio.5b00007. [DOI] [PubMed] [Google Scholar]
- 118.Li Y., Lin Z., Huang C., Zhang Y., Wang Z., Tang Y Jie. Metabolic engineering of Escherichia coli using CRISPR-Cas9 meditated genome editing. Metab Eng. 2015;31:13–21. doi: 10.1016/j.ymben.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 119.Ryan O.W., Skerker J.M., Maurer M.J., Li X., Tsai J.C., Poddar S. Selection of chromosomal DNA libraries using a multiplex CRISPR system. Elife. 2014;3:1–15. doi: 10.7554/eLife.03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gupta A., Meng X., Zhu L.J., Lawson N.D., Wolfe S.A. Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Res. 2011;39:381–392. doi: 10.1093/nar/gkq787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gabriel R., Lombardo A., Arens A., Miller J.C., Genovese P., Kaeppel C. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 122.Osborn M.J., Starker C.G., McElroy A.N., Webber B.R., Riddle M.J., Xia L. TALEN-based gene correction for epidermolysis bullosa. Mol Ther. 2013;21:1151–1159. doi: 10.1038/mt.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hockemeyer D., Wang H., Kiani S., Lai C.S., Gao Q., Cassady J.P. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pattanayak V., Lin S., Guilinger J.P., Ma E., Doudna J a, Liu D.R. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fu Y., Foden J a, Khayter C., Maeder M.L., Reyon D., Joung J.K. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fine E.J., Cradick T.J., Bao G. Strategies to determine off-target effects of engineered nucleases. In: Cathomen T., Hirsch M., Porteus M., editors. Genome Ed. Next step gene ther. Springer New York; New York, NY: 2016. pp. 187–222. [Google Scholar]
- 127.Szczepek M., Brondani V., Büchel J., Serrano L., Segal D.J., Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 128.Doyon Y., Vo T.D., Mendel M.C., Greenberg S.G., Wang J., Xia D.F. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Meth. 2011;8:74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 129.Dang Y., Jia G., Choi J., Ma H., Anaya E., Ye C. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol. 2015;16:280. doi: 10.1186/s13059-015-0846-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:1–12. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chuai G Hui, Wang Q.L., Liu Q. In silico meets in vivo: towards computational CRISPR-based sgRNA design. Trends Biotechnol. 2016;35:12–21. doi: 10.1016/j.tibtech.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 132.Chari R., Yeo N.C., Chavez A., Church G.M. sgRNA Scorer 2.0 – a species independent model to predict CRISPR/Cas9 activity. ACS Synth Biol. 2017 doi: 10.1021/acssynbio.6b00343. acssynbio.6b00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ran F.A., Hsu P.D., Lin C.-Y., Gootenberg J.S., Konermann S., Trevino A.E. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gabsalilow L., Schierling B., Friedhoff P., Pingoud A., Wende W. Site- and strand-specific nicking of DNA by fusion proteins derived from MutH and I-SceI or TALE repeats. Nucleic Acids Res. 2013;41:e83. doi: 10.1093/nar/gkt080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ramirez C.L., Certo M.T., Mussolino C., Goodwin M.J., Cradick T.J., McCaffrey A.P. Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res. 2012;40:5560–5568. doi: 10.1093/nar/gks179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fabret C., Poncet S., Danielsen S., Borchert T.V., Ehrlich S.D., Jannière L. Efficient gene targeted random mutagenesis in genetically stable Escherichia coli strains. Nucleic Acids Res. 2000;28:E95. doi: 10.1093/nar/28.21.e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Okazaki R., Arisawa M., Sugino A. Slow joining of newly replicated DNA chains in DNA polymerase I-Deficient Escherichia coli mutants. Proc Natl Acad Sci. 1971;68:2954–2957. doi: 10.1073/pnas.68.12.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cooper P.K., Hanawalt P.C. Role of DNA polymerase I and the rec system in excision-repair in Escherichia coli. Proc Natl Acad Sci. 1972;69:1156–1160. doi: 10.1073/pnas.69.5.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.del Solar G., Giraldo R., Ruiz-Echevarría M.J., Espinosa M., Díaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. doi:1092-2172/98/$04.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Allen J.M., Simcha D.M., Ericson N.G., Alexander D.L., Marquette J.T., Van Biber B.P. Roles of DNA polymerase I in leading and lagging-strand replication defined by a high-resolution mutation footprint of ColE1 plasmid replication. Nucleic Acids Res. 2011;39:7020–7033. doi: 10.1093/nar/gkr157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Camps M., Naukkarinen J., Johnson B.P., Loeb L.A. Targeted gene evolution in Escherichia coli using a highly error-prone DNA polymerase I. Proc Natl Acad Sci. 2003;100:9727–9732. doi: 10.1073/pnas.1333928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Camps M., Loeb L.A. Use of Pol I-Deficient E. coli for functional complementation of DNA polymerase. In: Arnold F.H., Georgiou G., editors. Dir. Enzym. Evol. Screen. Sel. Methods. Humana Press; Totowa, NJ: 2003. pp. 11–18. [DOI] [PubMed] [Google Scholar]
- 144.Shinkai A., Loeb L.A. In vivo mutagenesis by Escherichia coliDNA polymerase I: ILE709 in MOTIF a functions in base selection. J Biol Chem. 2001;276:46759–46764. doi: 10.1074/jbc.M104780200. [DOI] [PubMed] [Google Scholar]
- 145.Ravikumar A., Arrieta A., Liu C.C. An orthogonal DNA replication system in yeast. Nat Chem Biol. 2014;10:175–177. doi: 10.1038/nchembio.1439. [DOI] [PubMed] [Google Scholar]
- 146.Salas M. Springer US; Boston, MA: 1999. Mechanisms of initiation of linear DNA replication in prokaryotes. Genet. Eng. (N. Y) pp. 159–171. [DOI] [PubMed] [Google Scholar]
- 147.Koch D.J., Chen M.M., Van Beilen J.B., Arnold F.H. In vivo evolution of butane oxidation by terminal alkane hydroxylases AlkB and CYP153A6. Appl Environ Microbiol. 2009;75:337–344. doi: 10.1128/AEM.01758-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Na D., Lee S., Yi G.S., Lee D. Synthetic inter-species cooperation of host and virus for targeted genetic evolution. J Biotechnol. 2011;153:35–41. doi: 10.1016/j.jbiotec.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 149.Kennedy S.R., Chen C.Y., Schmitt M.W., Bower C.N., Loeb L.A. The biochemistry and fidelity of synthesis by the apicoplast genome replication DNA polymerase Pfprex from the malaria parasite Plasmodium falciparum. J Mol Biol. 2011;410:27–38. doi: 10.1016/j.jmb.2011.04.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Söte S., Kleine S., Schlicke M., Brakmann S. Directed evolution of an error-prone T7 DNA polymerase that attenuates viral replication. ChemBioChem. 2011;12:1551–1558. doi: 10.1002/cbic.201000799. [DOI] [PubMed] [Google Scholar]
- 151.Segall-Shapiro T.H., Meyer A.J., Ellington A.D., Sontag E.D., Voigt Ca. A “resource allocator” for transcription based on a highly fragmented T7 RNA polymerase. Mol Syst Biol. 2014;10:742. doi: 10.15252/msb.20145299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Myhrvold C. Silver P a. Using synthetic RNAs as scaffolds and regulators. Nat Struct Mol Biol. 2015;22:8–10. doi: 10.1038/nsmb.2944. [DOI] [PubMed] [Google Scholar]
- 153.Ravikumar A., Liu C.C. Biocontainment through reengineered genetic codes. ChemBioChem. 2015;16:1149–1151. doi: 10.1002/cbic.201500157. [DOI] [PubMed] [Google Scholar]
- 154.Torres L., Kruger A., Csibra E., Gianni E., Pinheiro V.B. Synthetic biology approaches to biological containment: pre-emptively tackling potential risks. Essays Biochem. 2016;60:393–410. doi: 10.1042/EBC20160013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gunge N., Sakaguchi K. Intergeneric transfer of deoxyribonucleic acid killer plasmids, pGKll and pGK12, from kluyveromyces lactis into Saccharomyces cerevisiae by cell fusion. J Bacteriol. 1981;147:155–160. doi: 10.1128/jb.147.1.155-160.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Finney-Manchester S.P., Maheshri N. Harnessing mutagenic homologous recombination for targeted mutagenesis in vivo by TaGTEAM. Nucleic Acids Res. 2013;41:e99. doi: 10.1093/nar/gkt150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.O'Brien P.J., Ellenberger T. Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. J Biol Chem. 2004;279:9750–9757. doi: 10.1074/jbc.M312232200. [DOI] [PubMed] [Google Scholar]
- 158.Glassner B.J., Rasmussen L.J., Najarian M.T., Posnick L.M., Samson L.D. Generation of a strong mutator phenotype in yeast by imbalanced base excision repair. Proc Natl Acad Sci U. S. A. 1998;95:9997–10002. doi: 10.1073/pnas.95.17.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Crook N., Abatemarco J., Sun J., Wagner J.M., Schmitz A., Alper H.S. In vivo continuous evolution of genes and pathways in yeast. Nat Commun. 2016;7:13051. doi: 10.1038/ncomms13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Curcio M.J., Lutz S., Lesage P. The Ty1 LTR-retrotransposon of budding yeast, Saccharomyces cerevisiae. Microbiol Spectr. 2015;3:1–35. doi: 10.1128/microbiolspec.MDNA3-0053-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Boutabout M. DNA synthesis fidelity by the reverse transcriptase of the yeast retrotransposon Ty1. Nucleic Acids Res. 2001;29:2217–2222. doi: 10.1093/nar/29.11.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Curcio M.J., Garfinkel D.J. Single-step selection for Ty1 element retrotransposition. Proc Natl Acad Sci. 1991;88:936–940. doi: 10.1073/pnas.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Devine S.E., Boeke J.D. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 1996;10:620–633. doi: 10.1101/gad.10.5.620. [DOI] [PubMed] [Google Scholar]
- 164.Carr M., Bensasson D., Bergman C.M. Evolutionary genomics of transposable elements in Saccharomyces cerevisiae. PLoS One. 2012:7. doi: 10.1371/journal.pone.0050978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Farzadfard F., Lu T.K. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science (80- ) 2014;346:1256272. doi: 10.1126/science.1256272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cho S.W., Kim S., Kim Y., Kweon J., Kim H.S., Bae S. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Porteus M.H., Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 168.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;61:5985–5991. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]