SUMMARY
Molecular factors that define stem cell identity have recently emerged as oncogenic drivers. For instance, brachyury, a key developmental transcriptional factor, is also implicated in carcinogenesis, most notably of chordoma, through mechanisms that remain elusive. Here, we show that brachyury is a crucial regulator of stemness in chordoma and in more common aggressive cancers. Furthermore, this effect of brachyury is mediated by control of synthesis and stability of Yes-associated protein (YAP), a key regulator of tissue growth and homeostasis, providing an unexpected mechanism of control of YAP expression. We further demonstrate that the Brachyury-YAP regulatory pathway is associated with tumor aggressiveness. These results elucidate a mechanism of controlling both tumor stemness and aggressiveness through regulatory coupling of two developmental factors.
eTOC BLURB
Malignant neoplasms exhibit uninhibited and dysregulated growth coupled with acquisition of stem-like properties that are integral to the development and progression of disease. Shah et al demonstrate a critical role of brachyury in regulating stemness and growth by activating YAP through direct transcriptional and post-transcriptional mechanisms in various cancers.
INTRODUCTION
Accumulating evidence suggests malignant neoplasms contain a distinct subset of cells that hijack stem cell transcriptional programs to attain tumor initiating and propagating capability (Ben-Porath et al., 2008; Kreso and Dick, 2014; Mani et al., 2008; Reya et al., 2001). These tumor initiating cells (TIC) have the capacity to self-renew, develop into all tumor cell subtypes, and seed new lesions. In addition, numerous studies have shown that these TICs are particularly chemo- and radio-resistant, providing a source for tumor recurrence after therapy (Frank et al., 2010; Holohan et al., 2013; Hsu et al., 2012). Together, these findings suggest that TICs are necessary and sufficient to sustain prolonged oncogenic growth and feed the progression of tumor malignancy (Beck and Blanpain, 2013). However, the regulatory pathways that confer the TIC phenotype remain poorly understood.
Given that TIC exhibit properties similar to normal stem cells, transcriptional programs coordinating early embryogenesis have recently emerged as drivers of oncogenesis and potential therapeutic targets (Ben-Porath et al., 2008; Kim and Orkin, 2011; Wong et al., 2008). Brachyury, a core T-box transcription factor, plays a vital role during development in early embryonic gastrulation events and notochord formation (Edwards et al., 1996; Herrmann et al., 1990; Herrmann and Lehrach, 1988; Kavka and Green, 1997; Kispert and Herrmann, 1993; Morrison et al., 1996; Showell et al., 2004; Smith, 1997; Smith et al., 1997; Wilkinson et al., 1990). Post-developmentally, brachyury is expressed in the testes and some thyroid tissues, but is undetectable in all other non-neoplastic adult tissues (Edwards et al., 1996; Hamilton et al., 2015). Interestingly, recent studies have reported the expression of brachyury in several epithelial cancers where it promotes growth, confers resistance to chemo- and radiotherapy, and drives epithelial-to-mesenchymal transition (EMT) (Cho et al., 2010; Fernando et al., 2010; Haro et al., 2013; Huang et al., 2013; Imajyo et al., 2012; Jezkova et al., 2016; Kobayashi et al., 2014; Larocca et al., 2013; Li et al., 2016; Miettinen et al., 2015; Palena et al., 2014; Park et al., 2008; Pinto et al., 2015; Pinto et al., 2014; Pires and Aaronson, 2014; Roselli et al., 2012; Sarkar et al., 2012; Shao et al., 2015; Shimoda et al., 2012; Vujovic et al., 2006; Xu et al., 2015; Yoshihama et al., 2016); however, the mechanistic details of how brachyury mediates these features of tumor progression have not been fully elucidated. Furthermore, the lack of brachyury expression in most adult non-neoplastic tissues and exclusive tumor-specific expression underscores its value as a potential diagnostic and therapeutic target in cancer. These observations provide a strong impetus to better understand the transcriptional network driven by brachyury in cancer.
Chordomas are rare tumors of the osseous spine and skull base that may serve as an ideal model system to understand brachyury-driven networks in cancer (Sarabia-Estrada et al., 2017). These tumors are believed to arise from remnants of the notochord, a mesoderm-derived embryonic structure that is critical for neurulation and embryonic tissue organization (Chugh et al., 2007). Interestingly, familial cases of these neoplasms contain a genomic amplification of the locus harboring brachyury, and it is nearly ubiquitously expressed in both familial and sporadic chordomas (Barresi et al., 2014; Hsu et al., 2011; Hu et al., 2014; Jambhekar et al., 2010; Mathios et al., 2015; Miettinen et al., 2015; Nelson et al., 2012; Oakley et al., 2008; Presneau et al., 2011; Yang et al., 2009). However, our understanding of the role played by brachyury in this neoplasm is limited.
Three lines of evidence suggest that chordomas may harbor a cancer stem cell population that drives their progression. First, cancer stem cells are known to exhibit radio-resistance due to their enhanced DNA-repair capacity and reactive oxygen species (ROS) defenses, and their self-renewal potential (Bao et al., 2006; Rycaj and Tang, 2014), and chordomas demonstrate a remarkable amount of radio- and chemo-therapy resistance clinically (Walcott et al., 2012). Second, chordomas exhibit a predilection to seed new tumors in patients via iatrogenic spread of cells along the surgical route particularly when the tumor capsule is violated, a hallmark of TICs (Arnautovic and Al-Mefty, 2001; Dieter et al., 2011). Lastly, although not a prerequisite, it has been proposed that cancer stem cells may arise from transformed stem cell niches (Kreso and Dick, 2014; Sanai et al., 2005) and chordomas arise from transformed primordial remnants of the notochord (Salisbury et al., 1993). Furthermore, brachyury is predominantly a transcriptional regulator of early mesoderm development (Kispert et al., 1995a) and has been shown to be highly expressed in chordoma due to a gene duplication event (Kelley et al., 2014) and more recently has been implicated in orchestrating EMT in various metastatic carcinoma cell lines (Roselli et al., 2012). EMT is crucial for the acquisition of stem-like properties and malignant transformation (Caixeiro et al., 2014; Mani et al., 2008; Roselli et al., 2012). In summary, these considerations led us to explore the hypothesis that chordomas harbor a stem-like population of cells which is governed by brachyury signaling.
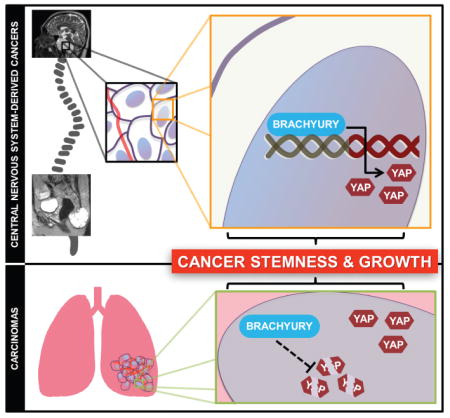
In this report, we demonstrate that brachyury drives cell cycle progression, stemness, and tumor growth in chordoma through direct transcriptional activation of Yes-associated protein (YAP), an effector of the Hippo pathway and a master regulator of organ development (Dong et al., 2007; Pan, 2010). By binding to the proximal region of the YAP promoter, brachyury was found to transactivate YAP-dependent signaling in chordoma. Furthermore, we find that brachyury expression is elevated in glioblastomas (GBM), the most common and aggressive type of primary brain cancer (Chaichana et al., 2013a; Chaichana et al., 2013b; Smith et al., 2015; Stupp et al., 2005), and in a majority of brain metastases derived from various carcinomas. Surprisingly, we also observed that brachyury enhanced YAP signaling in lung cancer cells by increasing protein stability instead of transcriptional activation, suggesting a dual role of brachyury in YAP expression. In addition, we find that this regulatory axis correlates with differentiation status of lung carcinomas, an indicator of tumor pluripotency and aggressiveness. Overall, our work provides molecular insights into brachyury-driven gene regulation in cancer and identifies it as a positive regulator of YAP. Moreover, our data provides important insights into how developmental programs are co-opted by cancer.
RESULTS
Brachyury regulates cancer stemness and tumor initiating capacity in chordoma
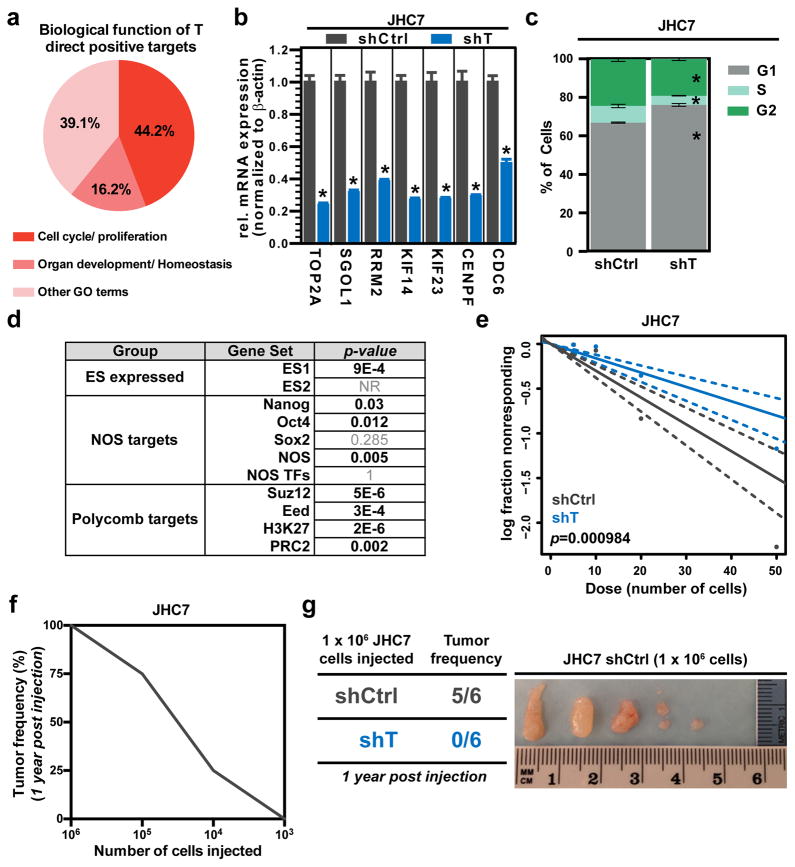
To delineate brachyury-driven mechanisms of cancer stemness and growth, we began our studies by utilizing patient-derived primary and recurrent chordoma cells due to their high and stable endogenous expression of brachyury. Using our previously established patient-derived primary sacral chordoma cell line JHC7 (Hsu et al., 2011), we sought to uncover the transcriptional network under the control of brachyury in cancer. To this end, we first generated lentiviral-based shRNA constructs that specifically target brachyury expression (shT) as well as a control non-targeting vector (shCtrl) (Fig. S1a). Next, using a genome-wide expression microarray analysis, we profiled RNA samples from JHC7 cells transduced with shT and shCtrl, in triplicate. Genes with greater than 2-fold decrease in RNA abundance relative to shCtrl were considered to be potential targets of brachyury and together defined the “T gene signature” (Table S1). We then performed functional annotation of this gene set using Gene Ontology (GO) to identify biological processes regulated by brachyury; significant enrichment of cell cycle/proliferation (44.2%) and organ development/homeostasis (16.2%) categorized biological functions was associated with brachyury positive gene targets (Fig. 1a). Consistent with this data, we observed a significant decrease in mRNA levels of cell cycle regulator genes following brachyury knockdown in two independent chordoma lines (JHC7 and UCH1, a recurrent chordoma cell line (Scheil et al., 2001)) (Fig. 1b and Fig. S1b). Also in agreement with these findings, we observed a decrease in proliferation in JHC7- and UCH2-shT cells (Fig. S1c, d) and augmented G1 phase accumulation in JHC7-shT cells (Fig. 1c). These results were consistent with a previous report (Presneau et al., 2011) demonstrating reduced proliferation of UCH1 cells silenced for brachyury expression. Taken together, these results demonstrate that brachyury regulates cell cycle progression and proliferation in chordoma.
Figure 1. Brachyury regulates cancer stemness and tumor-initiating capacity in chordoma.
a, Distribution of biological functions associated to brachyury (T) direct positive targets, identified by GO terms. b, mRNA expression of cell cycle regulator genes in shCtrl or shT JHC7 cells. c, Average percentage of cells in each cell cycle phase in shCtrl or shT JHC7 cells. d, Gene set enrichment analysis (GSEA) of T signature for signatures specific for embryonic stem cells (ES expressed), Nanog, Oct4, and Sox2 (NOS) targets, and polycomb targets. e, Representative in vitro extreme limiting dilution assay plating decreasing numbers of shCtrl or shT JHC7 cells. Solid lines: mean; dotted lines: 95% confidence interval; circles: values obtained in each cell dilution. f, In vivo limiting dilution assay showing relationship between number of JHC7 cells injected subcutaneously in mice and tumor formation capacity after a year (as assessed by tumor frequency in %). g, Left: Table showing tumor frequency in mice injected subcutaneously with the 1×106 shCtrl or shT JHC7 cells after 1 year. Right: Images of isolated JHC7 tumor mass from mice injected subcutaneously with 1×106 shCtrl cells after 1 year. All error bars are s.e.m. * = P<0.05. See also Figures S1, S2, and S3.
To investigate the possibility that chordomas may harbor a cancer stem cell population that drives their progression, JHC7 cells were evaluated for expression of stem cell markers frequently exhibited by multipotent mesenchymal stem cells. JHC7 cells and xenografts derived from this line exhibited strong expression of vimentin, CD90, CD105, Oct4 and nestin (Fig. S1e, f, g). In vitro, these cells demonstrated transdifferentiation capacity by readily differentiating into oil red-positive adipocytes and GFAP- and Tuj1-positive neuroglial cells (Fig. S1h, i), a quality that often distinguishes multipotent mesenchymal stem cells. Furthermore, a subpopulation of chordoma cells exhibited increased aldehyde dehydrogenase activity (ALDH), a functional marker of cancer stem cells, and this population was significantly reduced when the cells were grown in adherent conditions that promote differentiation (Fig. S2a.) Chordoma xenografts also exhibited the ability to undergo serial tumor formation in mice, which is a hallmark of cancer stem cells (Fig. S2b). Serial passaging of chordoma xenografts enriched for the cancer stem cell population as determined by side population analysis (Fig. S2c). Finally, brachyury knockdown resulted in a significant drop in the enriched cancer stem cell population, with a decrease in side population frequency (28.95% vs. 4.31%; Fig. S2d). Collectively, these findings suggest that chordomas harbor a putative stem-like cell population.
Given the role of brachyury in stem cell identity and fate determination during development, we explored whether brachyury modulates any of the core stem-cell regulatory networks. Using the T signature for gene set enrichment analysis (Irizarry et al., 2009), we found a significant enrichment of signatures specific for embryonic stem cells (ES1, ES2), Nanog, Oct4, Sox2, NOS (Nanog, Oct4, Sox2) targets, and the polycomb repressive complex-2 (PRC2) (Fig. 1d). Moreover, we observed a significant decrease in the mRNA levels of stemness-related genes including ABCG2, OCT4, SOX2, and ABCB1 following brachyury knockdown in JHC7 cells (Fig. S2e, f). Based on this data, we investigated the functional relevance of brachyury in self-renewal capacity through an extreme limiting dilution assay (ELDA) using previously described methods (Hu and Smyth, 2009). Interestingly, we found a significant reduction in self-renewing capacity and stem cell frequency in shT cells as compared to the shCtrl JHC7 cells (Fig. 1e). To determine whether brachyury, through its control of stemness, also affects tumor-initiating capacity in vivo, we used a murine subcutaneous xenograft model of chordoma. We subcutaneously injected JHC7 cells at limiting dilutions (103–106 cells) into immunocompromised mice and quantified tumor-initiation frequency 12 months following tumor implantation. Consistent with brachyury’s role in modulation of stemness properties in vitro, we observed a significant decrease in tumor-initiating capacity with decreasing cell dilutions implanted, demonstrating that implantation of 106 JHC7 cells is necessary to successfully form tumors with 100% frequency (Fig. 1f). Based on this data, we investigated whether brachyury regulates tumor-initiating capacity in vivo by injecting 106 shCtrl or shT JHC7 cells and monitoring tumor formation using the same protocol. While none of the JHC7 shT-injected mice developed tumors (0/6), nearly all (5/6) of the JHC7 shCtrl-injected mice formed tumors (Fig. 1g). It is important to point out that there was significant size variability among the tumors in the JHC7 shCtrl group; it remains to be investigated whether these differences correlated with the level of brachyury expression in the various xenografts. Overall, these results demonstrate that chordomas harbor a putative cancer stem cell population, and that brachyury regulates cancer stemness and tumor-initiating capacity.
Brachyury regulates proliferation and stemness in glioblastoma
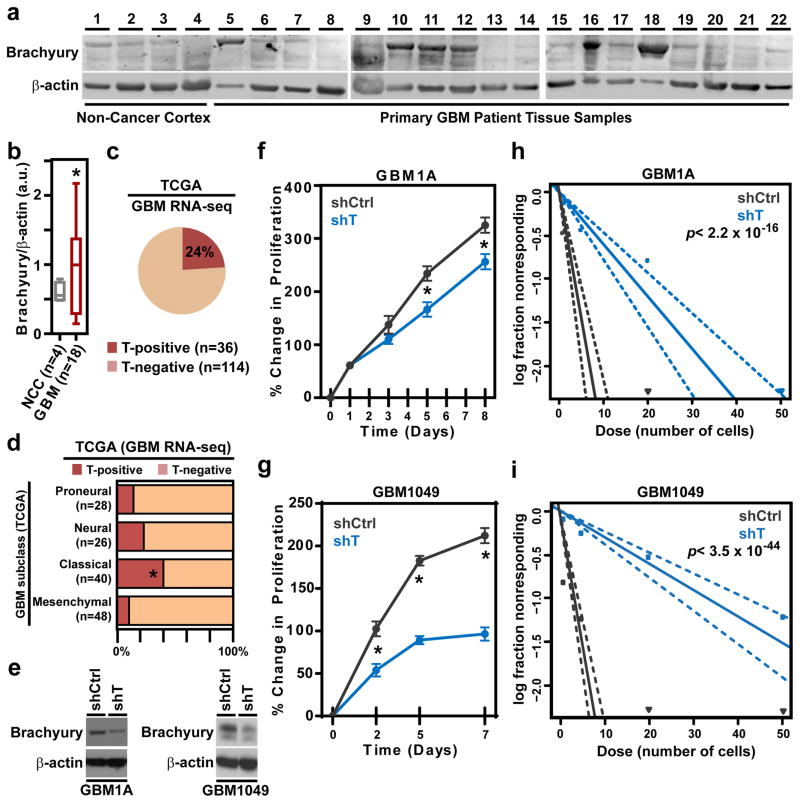
Given our results showing brachyury as a potent regulator of chordoma cell proliferation, stemness, and tumor-initiating capacity, we inquired whether brachyury plays a role in other central nervous system (CNS) derived cancers. While chordomas are characteristically slow-growing tumors with a median patient survival of 7 years (McMaster et al., 2001), GBM are the most common and aggressive type of primary brain cancer, and have a dismal patient prognosis with an overall median survival of only 14.6 months (Chaichana et al., 2013a; Chaichana et al., 2013b; Smith et al., 2015; Stupp et al., 2005). Thus, it was of interest to investigate the potential role of brachyury in driving stemness and growth in this highly proliferative and aggressive CNS-derived cancer. We found that in a fraction of intraoperatively obtained primary GBM tissues from patients, the brachyury protein expression was significantly elevated compared to non-cancer cortex tissue (Fig. 2a, b). Next, we surveyed RNA-sequencing profiles of patient-derived GBM tissues from The Cancer Genome Atlas (TCGA) to determine whether our observations using intraoperatively-obtained patient tissues were consistent with a larger cohort of clinical samples. As shown in Fig. 2c, 24% of GBM patient tissues exhibited detectable brachyury mRNA (T) in the TCGA dataset, suggesting that brachyury may play a role in a subset of GBM. Given these observations and the recent stratification of GBM to account for its molecular heterogeneity (Brennan et al., 2013; Verhaak et al., 2010), we examined the prognostic value of T mRNA and whether its expression is associated with one or more genetic subclasses of GBM. While T-transcript expression does not predict progression-free survival (Fig. S3a), we found a significant over-representation of T-positive patients among the Classical GBM compared with the other subclasses (Fig. 2d). These observations warrant further studies to evaluate the potential clinical value of T expression in GBM.
Figure 2. Brachyury regulates proliferation and stemness in glioblastoma.
a, Immunoblots of brachyury expression in non-cancer cortex and primary glioblastoma patient tissues. b, Densitometric quantification of brachyury expression (normalized to β-actin) from the immunoblots shown in a. a.u. = arbitrary units. c, Percentage of T-positive or T-negative expression in patient-derived glioblastoma tissues from the RNA-seq dataset of TCGA. d, Percentage of T-positive or T-negative GBM patients in each TCGA subtype. Fisher’s exact test. e, Representative immunoblot of brachyury expression in shCtrl or shT GBM1A and GBM1049 cells. f, g, Representative long-term MTT proliferation assay of shCtrl or shT GBM1A and GBM1049 cells. h, i, Representative in vitro extreme limiting dilution assay plating decreasing numbers of shCtrl or shT GBM1A and GBM1049 cells. Solid lines: mean; dotted lines: 95% confidence interval; circles: values obtained in each cell dilution. All error bars are s.e.m. * = P<0.05. See also Figure S4.
The expression of brachyury was then evaluated in a panel of patient-derived primary cultures of GBM cells (Fig. S3b), where it was found to be upregulated in 2/4 cell lines. Using the brachyury-positive cell lines, GBM1A and GBM1049, we then sought to determine whether brachyury plays a role in glioblastoma proliferation and stemness. With both cell lines, we found a significant decrease in proliferation in shT- compared to shCtrl-GBM cells (Fig. 2e–g). Next, we inquired whether brachyury expression was enhanced in stem cell-enriched GBM spheroids compared to adherent cultures. Our results demonstrated that brachyury was significantly higher in GBM1A spheres than in adherent cultures, which was accompanied by elevated nestin expression, a marker of GBM stemness, and reduced expression of the differentiation markers GFAP and TUJ1 in spheroid vs. adherent cultures (Fig. S3c). Congruent with these results, nestin expression was significantly decreased after brachyury-knockdown in GBM1A and GBM1049 cells (Fig. S3d, e). Moreover, a significant reduction in self-renewing capacity and in stem cell frequency was observed in both cell lines following knockdown of brachyury (shT) versus shCtrl cells using ELDA (Fig. 2h, i). In addition, shT GBM1A cells formed significantly smaller spheroids than the corresponding shCtrl GBM1A cells (Fig. S3f). Taken together, our studies demonstrate that a subset of GBMs express brachyury and that it promotes tumor cell proliferation and stemness in this malignancy.
Brachyury regulates YAP in CNS-derived cancers
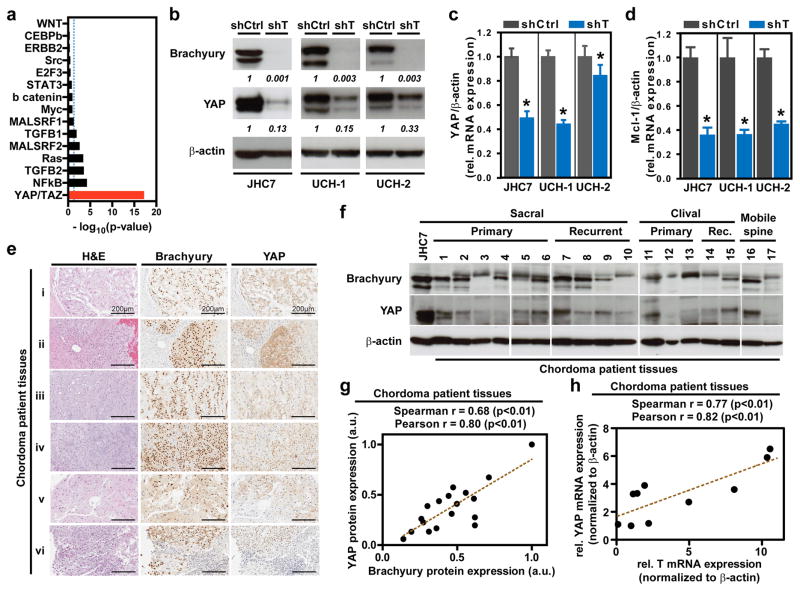
In cancer, transcription factors interact with oncogenic signaling pathways to synergistically drive disease progression. Thus, we sought to identify effectors of brachyury-driven regulatory networks in cancer. Using the T gene signature, we performed gene set enrichment analysis to check for higher-than-randomly-expected representation of individual signal transduction pathways. Notably, only a small subset of pro-oncogenic and pro-growth signatures were significantly associated with the T gene signature, of which the YAP/TAZ signature (canonically associated with activity of the Hippo pathway) registered as the strongest association (Fig. 3a). YAP, a transcriptional co-activator, is implicated in the regulation of the organ size during development, and has recently been reported to regulate cancer stemness and growth in numerous studies. However, the transcriptional regulators and drivers of YAP expression are poorly defined. Thus, we sought to identify a potential regulatory connection between YAP and brachyury. Using JHC7, UCH1, and UCH2 chordoma cell lines (Bruderlein et al., 2010), we observed a dramatic decrease in the expression of YAP protein and mRNA following brachyury silencing (Fig. 3b, c and Fig. S4a, b). Furthermore, the expression of MCL-1, a well-known YAP target, was also reduced in shT- compared to shCtrl-cells (Fig. 3d). Likewise, expression of Cyclin D1, CTGF, CYR61, ANKRD1 and c-MYC, indicators of YAP’s co-transcriptional activity, was significantly decreased in shT JHC7 cells while no significant change in AXL-1 was observed (Fig. S4c). Given these findings, we then evaluated the expression of brachyury and YAP in intraoperatively obtained primary chordoma tissues. As shown in Fig. 3e, f, the majority of chordoma tissues expressed high levels of both brachyury and YAP proteins, and their expression was restricted to tumor cells (Fig. 3e and Fig. S5). Moreover, a positive correlation was found between brachyury and YAP protein levels in these chordoma tissues (Fig. 3g), consistent with our findings in the chordoma cell lines (Fig. S4d). The expression of T and YAP mRNA levels also positively correlated in a subset of chordoma tissues analyzed (Fig. 3h). In addition, both T and YAP mRNA levels positively correlated with the expression of downstream targets of YAP such as MCL-1, Cyclin D1, and CTGF (Fig S6a–c, e–g). However, neither T nor YAP mRNA levels correlated with AXL-1 expression (Fig S6d, h). Altogether, these results demonstrate that brachyury regulates the expression and activity of YAP in chordoma.
Figure 3. Brachyury regulates YAP in CNS-derived cancers.
a, Statistical enrichment of the T signature genes among other pathway-specific signatures (Fisher’s exact test). b, Representative immunoblot of brachyury and YAP expression in shCtrl or shT JHC7, UCH1, and UCH2 cells. c, YAP mRNA expression in shCtrl vs. shT JHC7, UCH1, and UCH2 cells. d, Mcl-1 mRNA expression in shCtrl vs. shT JHC7, UCH1, and UCH2 cells. e, Representative images of immunohistochemical staining of Brachyury, YAP, and H&E in patient-derived chordoma tissues. Scale bar = 200μm. f, Immunoblots of brachyury and YAP expression in primary and recurrent sacral, clival, or mobile spine chordoma patient tissues. g, Correlation plot of relative brachyury and YAP protein expression (normalized to β-actin) based on densitometric quantification of immunoblots in e; a.u. = arbitrary units. h, Correlation plot of relative brachyury and YAP mRNA expression (normalized to β-actin) in a subset of patient-derived chordoma tissues (n=10). All error bars are s.e.m. * = P<0.05. See also Figure S4, S5, S6, and S7.
Since our previous work and that of others have demonstrated that GBMs have an elevated expression of YAP (Artinian et al., 2015; Orr et al., 2011; Xu et al., 2010), we also investigated whether a brachyury-driven mechanism of YAP regulation might be co-opted in the primary cultures of GBM cells. A significant decrease in YAP protein and mRNA expression were observed in primary GBM cells silenced for brachyury expression (Fig. S3g–k). Collectively, our data suggest that brachyury may regulate YAP expression and activity in CNS-derived tumors.
Brachyury directly activates the transcription of YAP
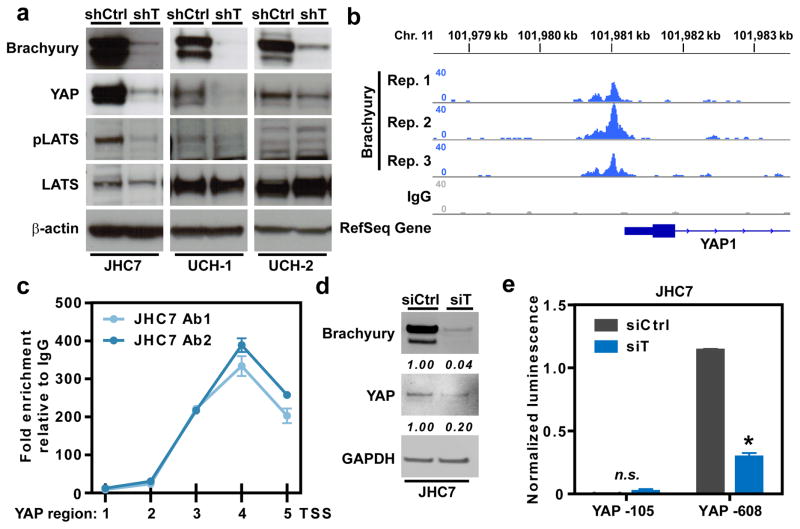
Next, we sought to determine the molecular mechanism of the brachyury-YAP regulatory network. Analysis of chordoma cells silenced for the expression of brachyury demonstrated reduced levels of pLATS1 expression compared to the corresponding shCtrl cells, thus ruling out the role of canonical Hippo signaling pathway in the control of YAP (Fig. 4a). Given brachyury’s function as a transcription factor, and our findings that brachyury regulates YAP transcript levels, we posited that brachyury may modulate YAP through direct transcriptional regulation. For this reason, a publicly available chromatin immunoprecipitation (ChIP) sequencing dataset of brachyury in UCH1 cells was analyzed. Interestingly, brachyury was found to be associated with the proximal promoter region of YAP, corresponding to a region spanning −1 kb directly upstream of the transcriptional start site (TSS) (Fig. 4b). This binding pattern was then validated by performing a ChIP-PCR querying across the −1.5 kb region upstream from the TSS of the YAP promoter in JHC7 cells. Utilizing two independent antibodies specific for epitopes on the C- and N-terminus of brachyury, a significant enrichment of brachyury binding was observed in the −1 kb region upstream of the TTS of the YAP promoter, suggesting a conserved chromatin occupancy pattern in JHC7 cells (Fig. 4c). To evaluate the functional consequence of this binding, we conducted a YAP gene promoter luciferase assay using the −105bp and −608bp regions upstream of the TSS in JHC7 cells +/− siRNA-mediated knockdown of brachyury. As expected, a significantly enhanced signal was observed with the luciferase reporter vector containing the longer segment of the YAP promoter region (−608bp to TSS), compared to the shorter promoter sequence (−105bp to TSS) (Fig. 4d, e). Furthermore, the luminescence signal of the −608bp YAP-luciferase reporter was significantly decreased after brachyury knockdown (siT, Fig. 4e). These findings led us to investigate whether YAP’s paralog, TAZ, is similarly regulated by brachyury. While TAZ expression was lower in JHC7 cells silenced for brachyury expression (Fig. S4e), we observed no significant enrichment of brachyury in the promoter region of TAZ (Fig. S4f). Taken together, these results demonstrate that brachyury directly activates the transcription of the YAP gene in chordoma cells.
Figure 4. Brachyury directly activates transcription of YAP.
a, Brachyury binding to the proximal promoter region of YAP as observed in a publicly available ChIP-sequencing dataset using UCH1 cells. b, ChIP-PCR querying across the −1.5 kb region upstream of the transcriptional start site (TSS) of the YAP promoter in JHC7 cells using two independent antibodies specific for epitopes on the C- (Ab1) and N-terminus (Ab2) of brachyury. c, Representative immunoblot of brachyury and YAP expression in siCtrl or siT JHC7 cells. d, YAP gene promoter luciferase assay using the −105bp and −608bp regions upstream of the TSS in siCtrl or siT JHC7 cells. e, Schematic of transcriptional regulation of YAP by brachyury. All error bars are s.e.m. * = P<0.05. See also Figure S5.
Brachyury regulates stemness and proliferation through YAP
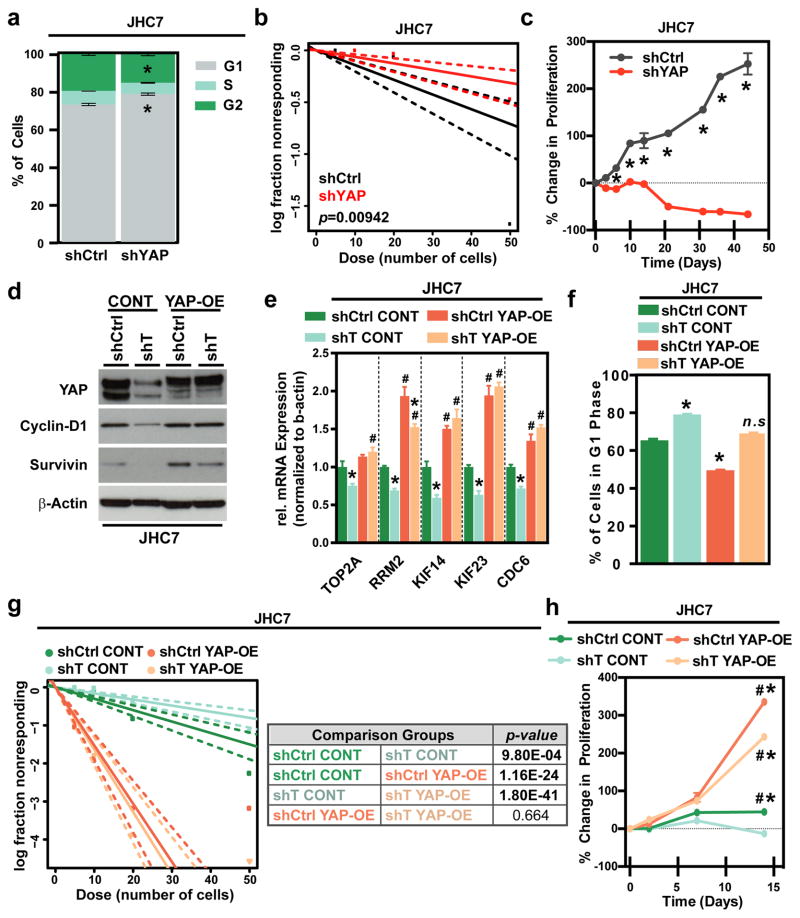
To further investigate whether brachyury potentiates cancer stemness and proliferation in chordoma through the activation of YAP, lentiviral-based shRNA constructs that specifically target YAP expression were used (Fig. S6i) with JHC7 cells. A G1 phase accumulation was observed in shYAP JHC7 versus shCtrl cells (Fig. 5a), which phenocopied the effect of brachyury knockdown (Fig. 1c). Similarly, YAP knockdown significantly reduced the self-renewing capacity and stem cell frequency based on ELDA (Fig. 5b), and decreased the side population and expression of the stemness marker ABCG2 (Fig. S6j,k) in JHC7 cells. In addition, the proliferation of shYAP JHC7 cells was markedly decreased compared to shCtrl cells (Fig. 5c, Fig. S6i), indicating that YAP is a critical regulator of stemness and growth in chordoma.
Figure 5. Brachyury regulates stemness and proliferation through YAP.
a, Average percentage of cells in each cell cycle phase in shCtrl or shYAP JHC7 cells. b, Representative in vitro extreme limiting dilution assay plating decreasing numbers of shCtrl or shYAP JHC7 cells. c, Representative long-term MTT proliferation assay of shCtrl or shYAP JHC7 cells. d, Representative immunoblot of YAP, Cyclin-D1, and Survivin expression in shCtrl or shT JHC7 cells transfected with empty vector (CONT) or YAP-overexpression (YAP-OE) plasmids. e, mRNA expression of cell cycle regulator genes in shCtrl or shT JHC7 cells with CONT or YAP-OE overexpression. f, Average percentage of cells in G1 phase in shCtrl or shYAP JHC7 cells with CONT or YAP-OE overexpression. g, Representative in vitro extreme limiting dilution assay plating decreasing numbers of shCtrl or shYAP JHC7 cells with CONT or YAP-OE overexpression. Solid lines: mean; dotted lines: 95% confidence interval; circles: values obtained in each cell dilution. h, Representative long-term MTT proliferation assay of shCtrl or shYAP JHC7 cells with CONT or YAP-OE overexpression. In h, * = significant versus Day 0, # = significant versus shCtrl CONT group for the same day. All error bars are s.e.m. *, # = P<0.05. See also Figure S6.
Next, we inquired whether YAP is sufficient to rescue the self-renewal and proliferative capacity of chordoma cells silenced for brachyury expression. To this end, we stably overexpressed a constitutively hyperactive form of YAP, bearing mutations at S127/128/131/381A (YAP-OE), or an empty control vector (CONT), in shT and shCtrl JHC7 cells. YAP overexpression rescued the expression of various critical cell cycle regulators in shT JHC7 cells (Fig. 5d, e, and Fig. S6m). Moreover, reintroduction of YAP reversed the cell cycle arrest observed in shT cells (shT YAP-OE cells), restoring the G1 phase levels to levels similar to those of shCtrl CONT cells (Fig. 5f). More importantly, overexpression of YAP was also sufficient to rescue self-renewal capacity in shT JHC7 cells based on ELDA (Fig. 5g). We also observed higher proliferation in shCtrl YAP-OE cells compared to all of the other groups tested relative percent decrease in proliferative capacity in shT YAP-OE versus shCtrl YAP-OE cells was not as pronounced as that seen in shT CONT than shCtrl CONT cells (Fig. 5h). Thus, these experiments demonstrated that YAP is an effector of brachyury-mediated growth and stemness in cancer.
Brachyury-YAP regulatory axis is evident in non-CNS-derived carcinomas
Brachyury has recently been implicated in potentiating tumor stemness and growth in epithelial cancers (Cho et al., 2010; Fernando et al., 2010; Haro et al., 2013; Hung et al., 2013; Imajyo et al., 2012; Jezkova et al., 2016; Kobayashi et al., 2014; Larocca et al., 2013; Li et al., 2016; Miettinen et al., 2015; Palena et al., 2014; Park et al., 2008; Pinto et al., 2015; Pinto et al., 2014; Pires and Aaronson, 2014; Roselli et al., 2012; Sarkar et al., 2012; Shao et al., 2015; Shimoda et al., 2012; Vujovic et al., 2006; Xu et al., 2015; Yoshihama et al., 2016); however, the molecular details are poorly defined and it is not known whether the brachyury-YAP signaling is also co-opted by non-CNS-derived carcinomas. Hence, we first examined the expression of brachyury and YAP in a panel of patient-derived primary metastases to the brain from different types of carcinomas. As shown in Fig. S7a, elevated expression of brachyury and YAP were observed in multiple cases of metastases to the brain, as compared to non-cancer cortex tissues (Fig. 2a). The analysis of expression levels in these clinical specimens also revealed a significant positive association between brachyury and YAP proteins (Fig. S8a). In particular, we found a significant positive correlation between these two proteins in brain metastatic lesions originating from lung carcinoma (Fig. S8b, c), which is one of the most common sources of brain metastases encountered clinically. Congruent with the results at the protein level, elevated expression of the T gene signature was also found to be associated with higher expression of genes regulated by YAP (termed YAP gene signature) in the TCGA lung cancer database (Fig. S7b), although a direct correlation between T and YAP mRNA was not observed in these samples.
Given the above findings, we next explored whether brachyury regulates YAP in lung carcinoma cells by establishing single clonal populations of H460 cells with different levels of brachyury (high, intermediate, and low T-expressing clones) generated via the CRISPR/Cas9 methodology, and subsequently evaluating YAP expression. As shown in Fig. S7c, the levels of YAP protein directly correlated with those of brachyury in the H460 cells, and the expression of TAZ protein (Fig. S8d) and various downstream target genes of YAP (Fig. S8f) were all decreased in the brachyury low clones. Like the results with chordoma cell lines, T-low H460 cells exhibited lower levels of pLATS1 and pMST1/2 expression than T-high cells, thus ruling out the role of canonical Hippo signaling pathway in the modulation of YAP in this system (Fig. S8e). Interestingly, no significant changes were observed in YAP mRNA levels among these clones (Fig. S7d), suggesting that unlike CNS-derived cancers, brachyury might regulate YAP through a non-transcriptional mechanism in lung carcinoma cells. In agreement with this idea, the analysis of YAP protein stability in the high and intermediate T-expressing H460 clones treated with cycloheximide for blockade of protein translation demonstrated a pronounced reduction in the apparent half-life of YAP protein in the intermediate clone (~43min) versus the high clone (>4hr) (Fig. S7e). To confirm this result, the high and intermediate T-expressing H460 clones were treated with the proteasomal inhibitor MG132, which increased YAP protein levels in the intermediate but not the high T-expressing clone (Fig. S7f). A similar experiment conducted with JHC7 chordoma cells treated with MG132 (Fig. S4g), however, showed no changes in YAP expression, thus ruling out protein stabilization as a mechanism of YAP control in chordoma cells.
To ascertain the role of brachyury in the control of YAP expression in H460 cells, a rescue experiment was conducted where the T-low expressing clone was transfected with a vector encoding for brachyury (pT) vs. an empty control (pCMV). The results are shown in Fig. S8g, h, demonstrating that re-introduction of brachyury in the low-T H460 clone is sufficient to reconstitute the expression of YAP and TAZ. Collectively, these experiments demonstrate that brachyury is necessary and sufficient to regulate YAP expression in these cancer cells.
Given the above results, the lung adenocarcinoma TCGA database was interrogated to elucidate a potential association between brachyury and stemness-related gene signatures (ES1, ES2, NOS) in primary lung cancer tissues. Elevated expression of the T gene signature in lung tumor samples was associated with higher expression of gene signatures registering the presence of embryonic stem cells, e.g., Nanog, Oct4, and Sox2 (Fig. S7g). Validating these correlations, we observed a significant decrease in in vitro anchorage-independent colony formation, a measure of cancer stemness, in shT H460 cells versus shCtrl cells (Fig. S7h) as well as in shYAP vs. shCtrl H460 cells (Fig. S8i). In agreement with these results, H460 clones with various levels of brachyury also demonstrated a positive association between brachyury levels and the expression of the stemness markers, Oct4 and ABCB1 (Fig. S8j).
Since lung adenocarcinomas are classified by differentiation status as a correlate of aggressiveness (Sun et al., 2006), we hypothesized that the transcriptional activity of brachyury would be higher in the poorly differentiated subtype. As shown in Fig. S7i, patient samples corresponding to poorly differentiated lung cancer had a significantly higher T signature expression than those with a well-differentiated subtype. Since tumor-initiating cells are thought to be enriched in the poorly differentiated and more aggressive subtypes of cancer, this clinical data lends support to the role of brachyury in governing cancer stemness. Thus, the brachyury-YAP regulatory network in lung cancer may serve as a potent driver of aggressive behavior.
To expand our observations to other non-CNS carcinoma models, lung H1299 cells were transfected to overexpress brachyury. In this model system where basal YAP expression is endogenously high, however, the addition of brachyury had no effect on the levels of YAP (Fig. S8k). This result suggested that expression of YAP expression may be independent of brachyury in certain brachyury-negative cells. However, other model systems, including pancreatic PANC-1, prostate ONYCAP23, Colon SW480 and SW620 cells, also demonstrated a positive correlation between brachyury and YAP protein levels when brachyury was either overexpressed or silenced (Fig. S9a–c). In agreement with a non-transcriptional mechanism of control, all these cell lines also showed modulation of YAP protein without an effect on the expression of YAP mRNA levels (Fig. S9d–g) in response to manipulations of brachyury expression. Collectively, our study demonstrates that brachyury-based regulation of YAP can occur through a transcriptional and/or post-transcriptional mechanism in different tumors, leading to enhanced YAP-dependent oncogenic activity.
DISCUSSION
Our work suggests an important role for the embryonic transcription factor brachyury in driving cancer stemness and growth. During development, brachyury maintains stem and progenitor cells during vertebrate neuro-axis formation (Kispert et al., 1995b), suggestive of a role for brachyury in the transformation of notochordal remnants and subsequent progression of chordoma. Our studies demonstrate that brachyury controls cell cycle progression, proliferation, and cancer stemness to regulate tumor initiating capacity in chordoma and other, more common cancers. Furthermore, surprisingly, we find that this function of brachyury is achieved through control of expression of another developmental factor, the transcriptional co-activator YAP (Dong et al., 2007). Given that YAP is overexpressed and hyperactive in numerous cancers (Zanconato et al., 2016), it is of vital interest to identify critical regulators of YAP. Thus, our study identifies a transcriptional regulator of this proto-oncogene YAP (Wang et al., 2013; Wu et al., 2013; Zhu et al., 2015). Furthermore, we demonstrate the conservation of this brachyury-YAP regulatory mechanism in another type of aggressive CNS-derived cancer, glioblastoma, suggesting a common presence of this regulatory linkage across different tissues of origin, and making this regulatory interaction a potential new biomarker of cancer stemness and aggressiveness.
Unlike with chordomas, the present study also demonstrates that brachyury-driven YAP expression can also be controlled by enhancing protein stability in lung carcinomas, highlighting different regulatory scenarios leading to equivalent functional outcomes. It is possible that both transcriptional and translational regulation of YAP expression by brachyury co-exist in any tissue of origin, but to different relative degrees, subject to additional regulatory constraints.
This analysis also highlights the importance of co-opting developmental factors in carcinogenesis. Both brachyury and YAP are key controllers of developmental patterning and growth and homeostasis of normal tissues. As such, their roles in controlling stemness of both normal and cancerous tissues is perhaps not surprising. However, the regulatory linkage of these factors is not completely understood, making it of interest to examine its role in both normal development and across a larger set of different cancer types. Rosenbluh J. et al. reported that YAP binds to TBX5, a T-box family transcription factor, along with other components to form a transcriptional complex to regulate synthesis of antiapoptotic genes (Rosenbluh et al., 2012). More recently, Mohamed et al showed that overexpression of constitutively active YAP increased brachyury-promoter reporter activity, suggesting that YAP positively regulates expression of brachyury (Mohamed et al., 2016). However, it remained unclear whether brachyury’s pro-oncogeneic function was mediated by YAP signaling. Thus, the unexpected role of brachyury as a regulator of YAP expression uncovered by our study can reveal further details of regulation of YAP both in normal and pathological conditions, suggesting a putative mechanism for the common observation of YAP overexpression across multiple cancers.
Given that our findings indicate that brachyury-YAP signaling is co-opted by a variety of cancer types, from some of the rarest and slow-growing but lethal tumors to the most prevalent and aggressive types, this brachyury-YAP regulatory cascade may serve as potential potent therapeutic target. Clinically, with the advent of vaccines and small molecule inhibitors against brachyury and YAP (Hamilton et al., 2013), respectively, targeting this hyperactive regulatory network may offer prognostic benefit to cancer patients. Hence, our study underscores the clinical value of identifying and delineating aberrant regulatory networks in cancer.
EXPERIMENTAL PROCEDURE
Cell culture
The patient-derived primary chordoma cell line, JHC7, was established from tissue obtained intraoperatively and processed for cell culture as described in Hsu, Mohyeldin, Shah et al., 2011. UCH1 and UCH2, were obtained from The Chordoma Foundation. Primary patient-derived glioblastoma tissue samples were obtained at the Johns Hopkins Hospital under the approval of the Institutional Review Board (IRB). All primary cell lines were established from excess tumor tissue from patients undergoing surgical resection for glioblastoma.
Lentiviral transduction
Cells were transduced with equal titers of virus in growth media supplemented with polybrene (Sigma) for 24 hours. After transduction, cells were cultured in normal media for 24 hours prior to selection.
In vivo experiments: subcutaneous xenografts
Animal protocols were approved by the Johns Hopkins School of Medicine Animal Care and Use Committee. Subcutaneous tumor cell implantation into mice was conducted according to the protocol as described previously (Hsu et al., 2011).
Supplementary Material
HIGHLIGHTS.
Chordomas harbor a putative cancer stem cell population which is driven by brachyury
Brachyury-YAP regulatory axis: Identification of a direct transcriptional regulator of YAP
Brachyury can enhance YAP activity through a post-transcriptional mechanism in carcinomas
Brachyury-YAP activity is tied to tumor aggressiveness in several types of cancers
Acknowledgments
We are thankful to the Chordoma Foundation for their continued support of our work and for access to the chordoma lines, UCH1 and UCH2. We deeply appreciate the technical assistance and guidance of Dr. Hao Zhang of Johns Hopkins Bloomberg School of Public Health (for all flow-cytometry based experiments), Dr. Colette Ap Rhys (for generation of lentiviral expression vectors and particles), Mr. William Ruff of Yale University (for assistance with murine xenograft experiments), and Ms. Brandy Edenfield of the Mayo Clinic Cancer Basic Science Histology Resource (for immunohistochemistry experiments). We are grateful to Ms. Vanessa C. Borotz for her artistic contributions to the Graphical Abstract and other manuscript-related artwork. This work was supported by the following grants: National Science Foundation Graduate Research Fellowship to S.R.S., NIH NCI U54-CA209992 to A.L., NIH R01 NS070024 to A.Q-H., and CP, JMD, and DHH were supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute (NCI), National Institutes of Health.
Footnotes
AUTHOR CONTRIBUTIONS
All authors contributed extensively to the work presented in this paper. SRS and AM conceived the initial study. SRS designed the experiments and overall study direction. SRS, JMD, NDT, AM, JCM, SG, and DHH conducted the experiments and performed data analyses. SRS wrote the manuscript. All authors edited the manuscript. CP, AL, and AQH supervised the work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnautovic KI, Al-Mefty O. Surgical seeding of chordomas. Journal of neurosurgery. 2001;95:798–803. doi: 10.3171/jns.2001.95.5.0798. [DOI] [PubMed] [Google Scholar]
- Artinian N, Cloninger C, Holmes B, Benavides-Serrato A, Bashir T, Gera J. Phosphorylation of the Hippo Pathway Component AMOTL2 by the mTORC2 Kinase Promotes YAP Signaling, Resulting in Enhanced Glioblastoma Growth and Invasiveness. J Biol Chem. 2015;290:19387–19401. doi: 10.1074/jbc.M115.656587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Barresi V, Ieni A, Branca G, Tuccari G. Brachyury: a diagnostic marker for the differential diagnosis of chordoma and hemangioblastoma versus neoplastic histological mimickers. Dis Markers. 2014;2014:514753. doi: 10.1155/2014/514753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck B, Blanpain C. Unravelling cancer stem cell potential. Nature reviews Cancer. 2013;13:727–738. doi: 10.1038/nrc3597. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nature genetics. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruderlein S, Sommer JB, Meltzer PS, Li S, Osada T, Ng D, Moller P, Alcorta DA, Kelley MJ. Molecular characterization of putative chordoma cell lines. Sarcoma. 2010;2010:630129. doi: 10.1155/2010/630129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caixeiro NJ, Kienzle N, Lim SH, Spring KJ, Tognela A, Scott KF, de Souza P, Becker TM. Circulating tumour cells--a bona fide cause of metastatic cancer. Cancer Metastasis Rev. 2014;33:747–756. doi: 10.1007/s10555-014-9502-8. [DOI] [PubMed] [Google Scholar]
- Chaichana KL, Martinez-Gutierrez JC, De la Garza-Ramos R, Weingart JD, Olivi A, Gallia GL, Lim M, Brem H, Quinones-Hinojosa A. Factors associated with survival for patients with glioblastoma with poor pre-operative functional status. J Clin Neurosci. 2013a;20:818–823. doi: 10.1016/j.jocn.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaichana KL, Pendleton C, Chambless L, Camara-Quintana J, Nathan JK, Hassam-Malani L, Li G, Harsh GRt, Thompson RC, Lim M, Quinones-Hinojosa A. Multi-institutional validation of a preoperative scoring system which predicts survival for patients with glioblastoma. J Clin Neurosci. 2013b;20:1422–1426. doi: 10.1016/j.jocn.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MS, Chan IL, Flores ER. DeltaNp63 transcriptionally regulates brachyury, a gene with diverse roles in limb development, tumorigenesis and metastasis. Cell Cycle. 2010;9:2434–2441. doi: 10.4161/cc.9.12.12051. [DOI] [PubMed] [Google Scholar]
- Chugh R, Tawbi H, Lucas DR, Biermann JS, Schuetze SM, Baker LH. Chordoma: the nonsarcoma primary bone tumor. Oncologist. 2007;12:1344–1350. doi: 10.1634/theoncologist.12-11-1344. [DOI] [PubMed] [Google Scholar]
- Dieter SM, Ball CR, Hoffmann CM, Nowrouzi A, Herbst F, Zavidij O, Abel U, Arens A, Weichert W, Brand K, et al. Distinct types of tumor-initiating cells form human colon cancer tumors and metastases. Cell stem cell. 2011;9:357–365. doi: 10.1016/j.stem.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards YH, Putt W, Lekoape KM, Stott D, Fox M, Hopkinson DA, Sowden J. The human homolog T of the mouse T(Brachyury) gene; gene structure, cDNA sequence, and assignment to chromosome 6q27. Genome Res. 1996;6:226–233. doi: 10.1101/gr.6.3.226. [DOI] [PubMed] [Google Scholar]
- Fernando RI, Litzinger M, Trono P, Hamilton DH, Schlom J, Palena C. The T-box transcription factor Brachyury promotes epithelial-mesenchymal transition in human tumor cells. J Clin Invest. 2010;120:533–544. doi: 10.1172/JCI38379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank NY, Schatton T, Frank MH. The therapeutic promise of the cancer stem cell concept. J Clin Invest. 2010;120:41–50. doi: 10.1172/JCI41004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton DH, Fernando RI, Schlom J, Palena C. Aberrant expression of the embryonic transcription factor brachyury in human tumors detected with a novel rabbit monoclonal antibody. Oncotarget. 2015;6:4853–4862. doi: 10.18632/oncotarget.3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton DH, Litzinger MT, Jales A, Huang B, Fernando RI, Hodge JW, Ardiani A, Apelian D, Schlom J, Palena C. Immunological targeting of tumor cells undergoing an epithelial-mesenchymal transition via a recombinant brachyury-yeast vaccine. Oncotarget. 2013;4:1777–1790. doi: 10.18632/oncotarget.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haro A, Yano T, Kohno M, Yoshida T, Koga T, Okamoto T, Takenoyama M, Maehara Y. Expression of Brachyury gene is a significant prognostic factor for primary lung carcinoma. Ann Surg Oncol. 2013;20(Suppl 3):S509–516. doi: 10.1245/s10434-013-2914-9. [DOI] [PubMed] [Google Scholar]
- Herrmann BG, Labeit S, Poustka A, King TR, Lehrach H. Cloning of the T gene required in mesoderm formation in the mouse. Nature. 1990;343:617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- Herrmann BG, Lehrach H. From phenotype to gene: molecular cloning in the Brachyury (T) locus region. Curr Top Microbiol Immunol. 1988;137:77–81. doi: 10.1007/978-3-642-50059-6_12. [DOI] [PubMed] [Google Scholar]
- Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nature reviews Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- Hsu W, Mohyeldin A, Shah SR, ap Rhys CM, Johnson LF, Sedora-Roman NI, Kosztowski TA, Awad OA, McCarthy EF, Loeb DM, et al. Generation of chordoma cell line JHC7 and the identification of Brachyury as a novel molecular target. Journal of neurosurgery. 2011;115:760–769. doi: 10.3171/2011.5.JNS11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu W, Mohyeldin A, Shah SR, Gokaslan ZL, Quinones-Hinojosa A. Role of cancer stem cells in spine tumors: review of current literature. Neurosurgery. 2012;71:117–125. doi: 10.1227/NEU.0b013e3182532e71. [DOI] [PubMed] [Google Scholar]
- Hu Y, Mintz A, Shah SR, Quinones-Hinojosa A, Hsu W. The FGFR/MEK/ERK/brachyury pathway is critical for chordoma cell growth and survival. Carcinogenesis. 2014;35:1491–1499. doi: 10.1093/carcin/bgu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Huang B, Cohen JR, Fernando RI, Hamilton DH, Litzinger MT, Hodge JW, Palena C. The embryonic transcription factor Brachyury blocks cell cycle progression and mediates tumor resistance to conventional antitumor therapies. Cell Death Dis. 2013;4:e682. doi: 10.1038/cddis.2013.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung IC, Cherng BW, Hsu WM, Lee SJ. Calnexin is required for zebrafish posterior lateral line development. Int J Dev Biol. 2013;57:427–438. doi: 10.1387/ijdb.120166sl. [DOI] [PubMed] [Google Scholar]
- Imajyo I, Sugiura T, Kobayashi Y, Shimoda M, Ishii K, Akimoto N, Yoshihama N, Kobayashi I, Mori Y. T-box transcription factor Brachyury expression is correlated with epithelial-mesenchymal transition and lymph node metastasis in oral squamous cell carcinoma. Int J Oncol. 2012;41:1985–1995. doi: 10.3892/ijo.2012.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Wang C, Zhou Y, Speed TP. Gene set enrichment analysis made simple. Stat Methods Med Res. 2009;18:565–575. doi: 10.1177/0962280209351908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar NA, Rekhi B, Thorat K, Dikshit R, Agrawal M, Puri A. Revisiting chordoma with brachyury, a “new age” marker: analysis of a validation study on 51 cases. Arch Pathol Lab Med. 2010;134:1181–1187. doi: 10.5858/2009-0476-OA.1. [DOI] [PubMed] [Google Scholar]
- Jezkova J, Williams JS, Pinto F, Sammut SJ, Williams GT, Gollins S, McFarlane RJ, Reis RM, Wakeman JA. Brachyury identifies a class of enteroendocrine cells in normal human intestinal crypts and colorectal cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavka AI, Green JB. Tales of tails: Brachyury and the T-box genes. Biochim Biophys Acta. 1997;1333:F73–84. doi: 10.1016/s0304-419x(97)00016-4. [DOI] [PubMed] [Google Scholar]
- Kelley MJ, Shi J, Ballew B, Hyland PL, Li WQ, Rotunno M, Alcorta DA, Liebsch NJ, Mitchell J, Bass S, et al. Characterization of T gene sequence variants and germline duplications in familial and sporadic chordoma. Hum Genet. 2014;133:1289–1297. doi: 10.1007/s00439-014-1463-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Orkin SH. Embryonic stem cell-specific signatures in cancer: insights into genomic regulatory networks and implications for medicine. Genome Med. 2011;3:75. doi: 10.1186/gm291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispert A, Herrmann BG. The Brachyury gene encodes a novel DNA binding protein. EMBO J. 1993;12:3211–3220. doi: 10.1002/j.1460-2075.1993.tb05990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispert A, Koschorz B, Herrmann BG. The T protein encoded by Brachyury is a tissue-specific transcription factor. EMBO J. 1995a;14:4763–4772. doi: 10.1002/j.1460-2075.1995.tb00158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispert A, Ortner H, Cooke J, Herrmann BG. The chick Brachyury gene: developmental expression pattern and response to axial induction by localized activin. Dev Biol. 1995b;168:406–415. doi: 10.1006/dbio.1995.1090. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Sugiura T, Imajyo I, Shimoda M, Ishii K, Akimoto N, Yoshihama N, Mori Y. Knockdown of the T-box transcription factor Brachyury increases sensitivity of adenoid cystic carcinoma cells to chemotherapy and radiation in vitro: implications for a new therapeutic principle. Int J Oncol. 2014;44:1107–1117. doi: 10.3892/ijo.2014.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell stem cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Larocca C, Cohen JR, Fernando RI, Huang B, Hamilton DH, Palena C. An autocrine loop between TGF-beta1 and the transcription factor brachyury controls the transition of human carcinoma cells into a mesenchymal phenotype. Mol Cancer Ther. 2013;12:1805–1815. doi: 10.1158/1535-7163.MCT-12-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Ying M, Feng D, Chen Y, Wang J, Wang Y. SMC1 promotes epithelial-mesenchymal transition in triple-negative breast cancer through upregulating Brachyury. Oncol Rep. 2016 doi: 10.3892/or.2016.4564. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathios D, Ruzevick J, Jackson CM, Xu H, Shah S, Taube JM, Burger PC, McCarthy EF, Quinones-Hinojosa A, Pardoll DM, Lim M. PD-1, PD-L1, PD-L2 expression in the chordoma microenvironment. J Neurooncol. 2015;121:251–259. doi: 10.1007/s11060-014-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster ML, Goldstein AM, Bromley CM, Ishibe N, Parry DM. Chordoma: incidence and survival patterns in the United States, 1973–1995. Cancer Causes Control. 2001;12:1–11. doi: 10.1023/a:1008947301735. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Wang Z, Lasota J, Heery C, Schlom J, Palena C. Nuclear Brachyury Expression Is Consistent in Chordoma, Common in Germ Cell Tumors and Small Cell Carcinomas, and Rare in Other Carcinomas and Sarcomas: An Immunohistochemical Study of 5229 Cases. Am J Surg Pathol. 2015;39:1305–1312. doi: 10.1097/PAS.0000000000000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed A, Sun C, De Mello V, Selfe J, Missiaglia E, Shipley J, Murray GI, Zammit PS, Wackerhage H. The Hippo effector TAZ (WWTR1) transforms myoblasts and TAZ abundance is associated with reduced survival in embryonal rhabdomyosarcoma. J Pathol. 2016;240:3–14. doi: 10.1002/path.4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison K, Papapetrou C, Attwood J, Hol F, Lynch SA, Sampath A, Hamel B, Burn J, Sowden J, Stott D, et al. Genetic mapping of the human homologue (T) of mouse T(Brachyury) and a search for allele association between human T and spina bifida. Hum Mol Genet. 1996;5:669–674. doi: 10.1093/hmg/5.5.669. [DOI] [PubMed] [Google Scholar]
- Nelson AC, Pillay N, Henderson S, Presneau N, Tirabosco R, Halai D, Berisha F, Flicek P, Stemple DL, Stern CD, et al. An integrated functional genomics approach identifies the regulatory network directed by brachyury (T) in chordoma. J Pathol. 2012;228:274–285. doi: 10.1002/path.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley GJ, Fuhrer K, Seethala RR. Brachyury, SOX-9, and podoplanin, new markers in the skull base chordoma vs chondrosarcoma differential: a tissue microarray-based comparative analysis. Mod Pathol. 2008;21:1461–1469. doi: 10.1038/modpathol.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr BA, Bai H, Odia Y, Jain D, Anders RA, Eberhart CG. Yes-associated protein 1 is widely expressed in human brain tumors and promotes glioblastoma growth. Journal of neuropathology and experimental neurology. 2011;70:568–577. doi: 10.1097/NEN.0b013e31821ff8d8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palena C, Roselli M, Litzinger MT, Ferroni P, Costarelli L, Spila A, Cavaliere F, Huang B, Fernando RI, Hamilton DH, et al. Overexpression of the EMT driver brachyury in breast carcinomas: association with poor prognosis. J Natl Cancer Inst. 2014:106. doi: 10.1093/jnci/dju054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Developmental cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JC, Chae YK, Son CH, Kim MS, Lee J, Ostrow K, Sidransky D, Hoque MO, Moon C. Epigenetic silencing of human T (brachyury homologue) gene in non-small-cell lung cancer. Biochem Biophys Res Commun. 2008;365:221–226. doi: 10.1016/j.bbrc.2007.10.144. [DOI] [PubMed] [Google Scholar]
- Pinto F, Campanella NC, Abrahao-Machado LF, Scapulatempo-Neto C, de Oliveira AT, Brito MJ, Andrade RP, Guimaraes DP, Reis RM. The embryonic Brachyury transcription factor is a novel biomarker of GIST aggressiveness and poor survival. Gastric Cancer. 2015 doi: 10.1007/s10120-015-0505-0. [DOI] [PubMed] [Google Scholar]
- Pinto F, Pertega-Gomes N, Pereira MS, Vizcaino JR, Monteiro P, Henrique RM, Baltazar F, Andrade RP, Reis RM. T-box transcription factor brachyury is associated with prostate cancer progression and aggressiveness. Clin Cancer Res. 2014;20:4949–4961. doi: 10.1158/1078-0432.CCR-14-0421. [DOI] [PubMed] [Google Scholar]
- Pires MM, Aaronson SA. Brachyury: a new player in promoting breast cancer aggressiveness. J Natl Cancer Inst. 2014:106. doi: 10.1093/jnci/dju094. [DOI] [PubMed] [Google Scholar]
- Presneau N, Shalaby A, Ye H, Pillay N, Halai D, Idowu B, Tirabosco R, Whitwell D, Jacques TS, Kindblom LG, et al. Role of the transcription factor T (brachyury) in the pathogenesis of sporadic chordoma: a genetic and functional-based study. J Pathol. 2011;223:327–335. doi: 10.1002/path.2816. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Roselli M, Fernando RI, Guadagni F, Spila A, Alessandroni J, Palmirotta R, Costarelli L, Litzinger M, Hamilton D, Huang B, et al. Brachyury, a driver of the epithelial-mesenchymal transition, is overexpressed in human lung tumors: an opportunity for novel interventions against lung cancer. Clin Cancer Res. 2012;18:3868–3879. doi: 10.1158/1078-0432.CCR-11-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ, Zack TI, Wang X, Tsherniak A, Schinzel AC, et al. beta-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell. 2012;151:1457–1473. doi: 10.1016/j.cell.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rycaj K, Tang DG. Cancer stem cells and radioresistance. Int J Radiat Biol. 2014;90:615–621. doi: 10.3109/09553002.2014.892227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury JR, Deverell MH, Cookson MJ, Whimster WF. Three-dimensional reconstruction of human embryonic notochords: clue to the pathogenesis of chordoma. J Pathol. 1993;171:59–62. doi: 10.1002/path.1711710112. [DOI] [PubMed] [Google Scholar]
- Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. The New England journal of medicine. 2005;353:811–822. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- Sarabia-Estrada R, Ruiz-Valls A, Shah SR, Ahmed AK, Ordonez AA, Rodriguez FJ, Guerrero-Cazares H, Jimenez-Estrada I, Velarde E, Tyler B, et al. Effects of primary and recurrent sacral chordoma on the motor and nociceptive function of hindlimbs in rats: an orthotopic spine model. J Neurosurg Spine. 2017:1–12. doi: 10.3171/2016.12.SPINE16917. [DOI] [PubMed] [Google Scholar]
- Sarkar D, Shields B, Davies ML, Muller J, Wakeman JA. BRACHYURY confers cancer stem cell characteristics on colorectal cancer cells. Int J Cancer. 2012;130:328–337. doi: 10.1002/ijc.26029. [DOI] [PubMed] [Google Scholar]
- Scheil S, Bruderlein S, Liehr T, Starke H, Herms J, Schulte M, Moller P. Genome-wide analysis of sixteen chordomas by comparative genomic hybridization and cytogenetics of the first human chordoma cell line, U-CH1. Genes Chromosomes Cancer. 2001;32:203–211. doi: 10.1002/gcc.1184. [DOI] [PubMed] [Google Scholar]
- Shao C, Zhang J, Fu J, Ling F. The potential role of Brachyury in inducing epithelial-to-mesenchymal transition (EMT) and HIF-1alpha expression in breast cancer cells. Biochem Biophys Res Commun. 2015;467:1083–1089. doi: 10.1016/j.bbrc.2015.09.076. [DOI] [PubMed] [Google Scholar]
- Shimoda M, Sugiura T, Imajyo I, Ishii K, Chigita S, Seki K, Kobayashi Y, Shirasuna K. The T-box transcription factor Brachyury regulates epithelial-mesenchymal transition in association with cancer stem-like cells in adenoid cystic carcinoma cells. BMC Cancer. 2012;12:377. doi: 10.1186/1471-2407-12-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showell C, Binder O, Conlon FL. T-box genes in early embryogenesis. Dev Dyn. 2004;229:201–218. doi: 10.1002/dvdy.10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Chaichana KL, Lee YM, Lin B, Stanko KM, O’Donnell T, Gupta S, Shah SR, Wang J, Wijesekera O, et al. Pre-exposure of human adipose mesenchymal stem cells to soluble factors enhances their homing to brain cancer. Stem Cells Transl Med. 2015;4:239–251. doi: 10.5966/sctm.2014-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. Brachyury and the T-box genes. Curr Opin Genet Dev. 1997;7:474–480. doi: 10.1016/s0959-437x(97)80073-1. [DOI] [PubMed] [Google Scholar]
- Smith JC, Armes NA, Conlon FL, Tada M, Umbhauer M, Weston KM. Upstream and downstream from Brachyury, a gene required for vertebrate mesoderm formation. Cold Spring Harb Symp Quant Biol. 1997;62:337–346. [PubMed] [Google Scholar]
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. The New England journal of medicine. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- Sun Z, Aubry MC, Deschamps C, Marks RS, Okuno SH, Williams BA, Sugimura H, Pankratz VS, Yang P. Histologic grade is an independent prognostic factor for survival in non-small cell lung cancer: an analysis of 5018 hospital- and 712 population-based cases. J Thorac Cardiovasc Surg. 2006;131:1014–1020. doi: 10.1016/j.jtcvs.2005.12.057. [DOI] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujovic S, Henderson S, Presneau N, Odell E, Jacques TS, Tirabosco R, Boshoff C, Flanagan AM. Brachyury, a crucial regulator of notochordal development, is a novel biomarker for chordomas. J Pathol. 2006;209:157–165. doi: 10.1002/path.1969. [DOI] [PubMed] [Google Scholar]
- Walcott BP, Nahed BV, Mohyeldin A, Coumans JV, Kahle KT, Ferreira MJ. Chordoma: current concepts, management, and future directions. Lancet Oncol. 2012;13:e69–76. doi: 10.1016/S1470-2045(11)70337-0. [DOI] [PubMed] [Google Scholar]
- Wang J, Ma L, Weng W, Qiao Y, Zhang Y, He J, Wang H, Xiao W, Li L, Chu Q, et al. Mutual interaction between YAP and CREB promotes tumorigenesis in liver cancer. Hepatology. 2013;58:1011–1020. doi: 10.1002/hep.26420. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, Herrmann BG. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990;343:657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell stem cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Xiao Y, Zhang S, Ji S, Wei L, Fan F, Geng J, Tian J, Sun X, Qin F, et al. The Ets transcription factor GABP is a component of the hippo pathway essential for growth and antioxidant defense. Cell Rep. 2013;3:1663–1677. doi: 10.1016/j.celrep.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Liu B, Liu Y. Impact of Brachyury on epithelial-mesenchymal transitions and chemosensitivity in non-small cell lung cancer. Mol Med Rep. 2015;12:995–1001. doi: 10.3892/mmr.2015.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Stamenkovic I, Yu Q. CD44 attenuates activation of the hippo signaling pathway and is a prime therapeutic target for glioblastoma. Cancer research. 2010;70:2455–2464. doi: 10.1158/0008-5472.CAN-09-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XR, Ng D, Alcorta DA, Liebsch NJ, Sheridan E, Li S, Goldstein AM, Parry DM, Kelley MJ. T (brachyury) gene duplication confers major susceptibility to familial chordoma. Nature genetics. 2009;41:1176–1178. doi: 10.1038/ng.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihama R, Yamaguchi K, Imajyo I, Mine M, Hiyake N, Akimoto N, Kobayashi Y, Chigita S, Kumamaru W, Kiyoshima T, et al. Expression levels of SOX2, KLF4 and brachyury transcription factors are associated with metastasis and poor prognosis in oral squamous cell carcinoma. Oncol Lett. 2016;11:1435–1446. doi: 10.3892/ol.2015.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the Roots of Cancer. Cancer cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Li L, Zhao B. The regulation and function of YAP transcription co-activator. Acta Biochim Biophys Sin (Shanghai) 2015;47:16–28. doi: 10.1093/abbs/gmu110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.