Abstract
The NALCN/NCA ion channel is a cation channel related to voltage-gated sodium and calcium channels. NALCN has been reported to be a sodium leak channel with a conserved role in establishing neuronal resting membrane potential, but its precise cellular role and regulation are unclear. The Caenorhabditis elegans orthologs of NALCN, NCA-1 and NCA-2, act in premotor interneurons to regulate motor circuit activity that sustains locomotion. Recently we found that NCA-1 and NCA-2 are activated by a signal transduction pathway acting downstream of the heterotrimeric G protein Gq and the small GTPase Rho. Through a forward genetic screen, here we identify the GPCR kinase GRK-2 as a new player affecting signaling through the Gq-Rho-NCA pathway. Using structure-function analysis, we find that the GPCR phosphorylation and membrane association domains of GRK-2 are required for its function. Genetic epistasis experiments suggest that GRK-2 acts on the D2-like dopamine receptor DOP-3 to inhibit Go signaling and positively modulate NCA-1 and NCA-2 activity. Through cell-specific rescuing experiments, we find that GRK-2 and DOP-3 act in premotor interneurons to modulate NCA channel function. Finally, we demonstrate that dopamine, through DOP-3, negatively regulates NCA activity. Thus, this study identifies a pathway by which dopamine modulates the activity of the NCA channels.
Author summary
Dopamine is a neurotransmitter that acts in the brain by binding seven transmembrane receptors that are coupled to heterotrimeric GTP-binding proteins (G proteins). Neuronal G proteins often function by modulating ion channels that control membrane excitability. Here we identify a molecular cascade downstream of dopamine in the nematode C. elegans that involves activation of the dopamine receptor DOP-3, activation of the G protein GOA-1, and inactivation of the NCA-1 and NCA-2 ion channels. We also identify a G protein-coupled receptor kinase (GRK-2) that inactivates the dopamine receptor DOP-3, thus leading to inactivation of GOA-1 and activation of the NCA channels. Thus, this study connects dopamine signaling to activity of the NCA channels through G protein signaling pathways.
Introduction
Heterotrimeric G proteins modulate neuronal activity in response to experience or environmental changes. Gq is one of the four types of heterotrimeric G protein alpha subunits [1] and is a positive regulator of neuronal activity and synaptic transmission [2–4]. In the canonical Gq pathway, Gq activates phospholipase Cβ (PLCβ) to cleave the lipid phosphatidylinositol 4,5-bisphosphate (PIP2) into diacylglycerol (DAG) and inositol trisphosphate (IP3), which act as second messengers. In a second major Gq signal transduction pathway, Gq directly binds and activates Rho guanine nucleotide exchange factors (GEFs), activators of the small GTPase Rho [5–8]. Rho regulates many biological functions including actin cytoskeleton dynamics and neuronal development, but less is known about Rho function in mature neurons. In C. elegans, Rho has been reported to stimulate synaptic transmission through multiple pathways [9–11]. We recently identified the C. elegans orthologs of the NALCN ion channel, NCA-1 and NCA-2, as downstream targets of a Gq-Rho signaling pathway [12]. We aim to understand the mechanism of activation of this pathway.
The NALCN/NCA ion channel is a nonselective cation channel that is a member of the voltage-gated sodium and calcium channel family [13–15]. The NALCN channel was proposed to be the major contributor to the sodium leak current that helps set the resting membrane potential of neurons [16], though there is controversy whether NALCN is indeed a sodium leak channel [17–19]. In humans, mutations in NALCN or its accessory subunit UNC80 have been associated with a number of neurological symptoms, including cognitive and developmental delay [20–33]. In other organisms, mutations in NALCN/NCA or its accessory subunits lead to defects in rhythmic behaviors [16,34–42]. Specifically in C. elegans, the NCA channels act in premotor interneurons where they regulate persistent motor circuit activity that sustains locomotion [43]. In addition to the Gq-Rho pathway described above, two other mechanisms have been reported to regulate the activity of the NALCN channel: a G protein-independent activation of NALCN by G protein-coupled receptors [44,45] and a G protein-dependent regulation by extracellular Ca2+ [46]. Here we identify a molecular cascade downstream of dopamine in the nematode C. elegans that involves the D2-like dopamine receptor DOP-3 and the G protein-coupled receptor kinase GRK-2 to modulate activity of the NCA-1 and NCA-2 ion channels.
G protein-coupled receptor kinases (GRKs) are protein kinases that phosphorylate and desensitize G protein-coupled receptors (GPCRs). Mammalian GRKs have been divided into three groups based on their sequences and function: 1) GRK1 and GRK7, 2) GRK2 and GRK3, and 3) GRK4, GRK5 and GRK6 [47]. C. elegans has two GRKs: GRK-1 and GRK-2, orthologs of the GRK4/5/6 and GRK2/3 families respectively [48]. Mammalian GRK2 is ubiquitously expressed [49,50] and GRK2 knock-out mice die as embryos [51]. In C. elegans, grk-2 is expressed in the nervous system and required for normal chemosensation [52] and egg-laying [53]. In this study, we find that C. elegans grk-2 mutants have locomotion defects due to decreased Gq signaling. We identify the D2-like dopamine receptor DOP-3 as the putative GRK-2 target and find that GRK-2 acts through DOP-3 to inhibit Go signaling. This in turn leads to activation of the NCA channels through the Gq-Rho signaling pathway. We also find that GRK-2 and DOP-3 exert their effect by acting in the premotor interneurons, where the NCA channels also act to regulate persistent motor neuron activity [43].
The D2-like receptors are GPCRs that couple to members of the inhibitory Gi/o family [54]. In mammals, GRK2 has been connected to the regulation of D2-type dopamine receptors, but the reported results are based mainly on effects of GRK2 overexpression in heterologous expression systems [55–59]. The results reported here provide a direct connection between GRK-2 and D2-type receptor signaling in a behaviorally relevant in vivo system. In C. elegans, dopamine, through dop-3, causes the slowing of the worm’s locomotion rate on food [60]; DOP-3 signals through Go to inhibit locomotion [61]. Here we find that dopamine, through activation of DOP-3, negatively modulates the activity of the NCA channels. This suggests a model in which dopamine signaling negatively regulates NCA channel activity and sustained locomotion through G protein signaling acting in premotor interneurons.
Results
The G protein-coupled receptor kinase GRK-2 promotes Gq signaling
To identify regulators of Gq signaling, we performed a forward genetic screen in the nematode C. elegans for suppressors of the activated Gq mutant egl-30(tg26) [62,63]. The egl-30(tg26) mutant is hyperactive and has a tightly coiled “loopy” posture (Fig 1A and 1B). These phenotypes were suppressed by the yak18 mutation isolated in our screen (Fig 1A). When outcrossed away from the egl-30(tg26) mutation, yak18 mutant animals are shorter than wild-type animals, have slow locomotion (Fig 1C, Right), and are egg-laying defective.
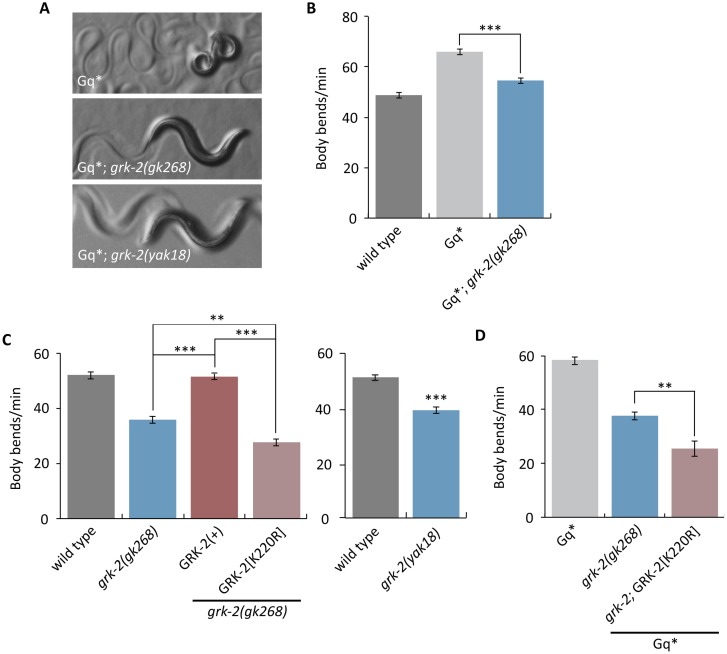
Fig 1. The GRK-2 kinase regulates locomotion and Gq signaling.
(A,B) grk-2 mutations suppress activated Gq. The activated Gq mutant egl-30(tg26) (Gq*) has hyperactive locomotion and a tightly coiled loopy posture. (A) The grk-2(gk268) and grk-2(yak18) mutations suppress the loopy posture of activated Gq. (B) The grk-2(gk268) mutation suppresses the hyperactive locomotion of activated Gq. (***, P<0.001. Error bars = SEM; n = 10). (C) The kinase activity of GRK-2 is required for proper locomotion. The grk-2(gk268) and grk-2(yak18) mutants have slow locomotion. The grk-2(gk268) slow locomotion is rescued by expression of the wild-type grk-2 cDNA under the control of its own promoter (GRK-2(+)), but is not rescued by expression of the kinase-dead GRK-2[K220R]. (**, P<0.01; ***, P<0.001. Error bars = SEM; n = 10–15). (D) The kinase-dead GRK-2 does not reverse the grk-2 suppression of activated Gq. A grk-2(gk268) mutation suppresses the hyperactive locomotion of egl-30(tg26) (Gq*). Expression of the kinase-dead GRK-2[K220R] does not rescue the grk-2 mutant for this phenotype. (**, P<0.01. Error bars = SEM; n = 10).
We mapped yak18 to the left arm of Chromosome III and cloned it by whole-genome sequencing and a complementation test with the deletion allele grk-2(gk268) (see Methods). yak18 is a G to A transition mutation in the W02B3.2 (grk-2) ORF that leads to the missense mutation G379E in the kinase domain of GRK-2. GRK-2 is a serine/threonine protein kinase orthologous to the human GPCR kinases GRK2 and GRK3 [52]. The deletion allele grk-2(gk268) also suppresses the loopy posture and hyperactive locomotion of activated Gq (Fig 1A and 1B) and causes defects in locomotion, egg-laying, and body-size similar to grk-2(yak18) (Fig 1C Left, S1A and S1B Fig). We also found that grk-2 mutant animals are defective in swimming (S2 Fig), a locomotion behavior that has distinct kinematics to crawling [64]. Additionally, grk-2 mutants restrict their movements to a limited region of a bacterial lawn, whereas wild-type animals explore the entire lawn (S1C Fig).
Our data suggest that GRK-2 regulates locomotion and is a positive regulator of Gq signaling. The standard model of GRK action is that GPCR phosphorylation by GRK triggers GPCR binding to the inhibitory protein beta-arrestin; binding of arrestin blocks GPCR signaling and mediates receptor internalization [65]. We tested whether loss of arrestin causes defects similar to loss of grk-2 by using a deletion allele of arr-1, the only C. elegans beta-arrestin homolog. We found that arr-1(ok401) mutant animals do not have slow locomotion (S3A Fig). To test whether an arr-1 mutation suppresses activated Gq, we constructed an egl-30(tg26) mutant strain carrying an arr-1 mutation in trans to a closely linked RFP marker (that is, an egl-30(tg26); arr-1/RFP strain). Surprisingly, this strain segregated few viable non-red animals, suggesting that egl-30(tg26); arr-1 double mutants are subviable. The few egl-30(tg26); arr-1 viable animals looked similar to the egl-30(tg26) single mutant (S3B Fig), but died as young adults. These results suggest that GRK-2 acts independently of arrestin to regulate locomotion rate and Gq signaling.
In addition to phosphorylation of GPCRs, mammalian GRK2 can also regulate signaling in a phosphorylation-independent manner [66,67]. Thus, we tested whether the kinase activity of GRK-2 is required for proper locomotion and Gq signaling by assaying whether a kinase-dead GRK-2[K220R] mutant [48,68] is capable of rescuing the grk-2(gk268) and egl-30(tg26); grk-2(gk268) mutants. Wild-type GRK-2 rescued the locomotion defect of grk-2(gk268) mutants (Fig 1C, Left), but the kinase-dead GRK-2[K220R], although it was properly expressed (Fig 2G), did not rescue either the locomotion defect or the suppression of activated Gq (Fig 1C and 1D). We conclude that GRK-2 acts as a kinase to regulate locomotion rate and Gq signaling.
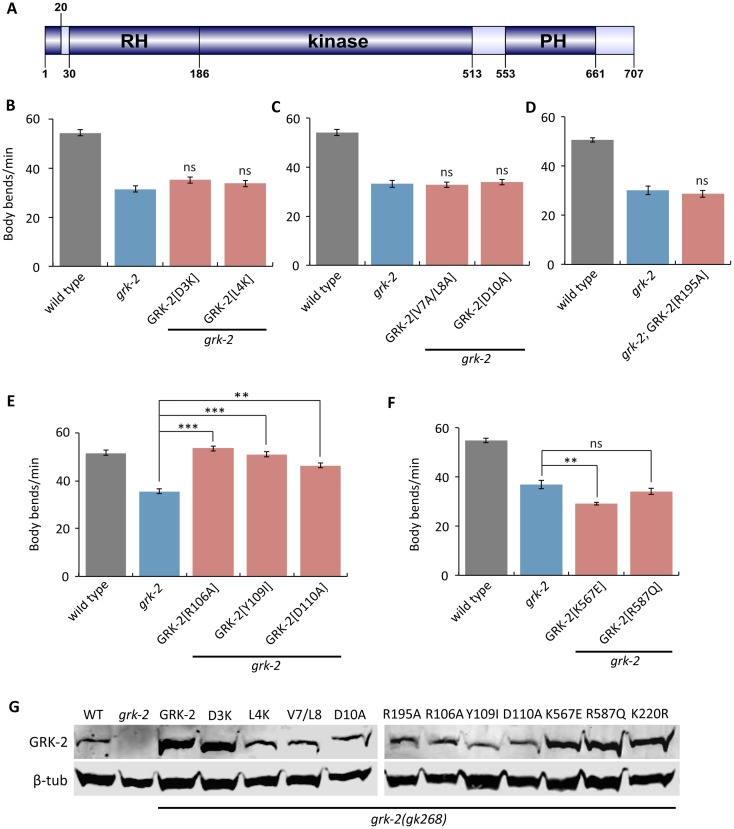
Fig 2. GRK-2 regulation of locomotion requires GPCR-phosphorylation and membrane association.
(A) Domain structure of GRK-2. GRK-2 is a 707 amino acid protein with three well-characterized domains: the RGS homology (RH) domain, the kinase domain, and the pleckstrin homology (PH) domain. The protein structure was drawn using DOG 1.0. (B-D) Residues required for GPCR phosphorylation are required for GRK-2 function in locomotion. The D3K (transgene yakEx77), L4K (transgene yakEx78), V7A/L8A (transgene yakEx79), and D10A (transgene yakEx80) mutations are predicted to block GPCR phosphorylation. The R195A mutation (transgene yakEx95) disrupts predicted intramolecular stabilizing interactions that are required for effective phosphorylation. In each case, expression of the mutant grk-2 cDNA under the control of its own promoter did not rescue the slow locomotion of the grk-2(gk268) mutant (ns, P>0.05, each strain compared to grk-2. Error bars = SEM; n = 10–20). (E) Residues in the RH domain predicted to disrupt Gq binding are not required for GRK-2 function in locomotion. The R106A (transgene yakEx57), Y109I (transgene yakEx55), and D110A (transgene yakEx56) mutations are predicted to disrupt Gq binding. In each case, expression of the mutant grk-2 cDNA under the control of the grk-2 promoter significantly rescued the slow locomotion of the grk-2(gk268) mutant (**, P<0.01; ***, P<0.001. Error bars = SEM; n = 10). (F) Residues in the PH domain predicted to disrupt GRK-2 phospholipid binding or binding to Gβγ are required for GRK-2 function in locomotion. Mutation K567E (transgene yakEx87) is predicted to disrupt GRK-2 phospholipid binding, and mutation R587Q (transgene yakEx88) is predicted to disrupt binding to Gβγ. In both cases, expression of the mutant grk-2 cDNA under the control of the grk-2 promoter did not rescue the slow locomotion of the grk-2(gk268) mutant. (**, P<0.01. ns, P>0.05. Error bars = SEM; n = 10). (G) Verification of the expression of the mutant grk-2 cDNAs used for the experiments shown in Figs 1D and 2B–2F. Western blot analysis of whole worm extracts from grk-2(gk268) mutants expressing the indicated grk-2 mutant cDNAs as extrachromosomal arrays.
GPCR-phosphorylation and membrane association domains of GRK-2 are required for its function in locomotion
To examine whether GRK-2 acts as a GPCR kinase to control locomotion, we took a structure-function approach (Fig 2A). We took advantage of previously-described mutations that disrupt specific activities of GRK-2, but do not disrupt GRK-2 protein expression or stability [48]. These mutations all affect conserved residues in well-characterized domains of GRK-2 [48].
Although GRKs act as kinases for activated GPCRs, mammalian GRKs have been shown to interact with and phosphorylate other molecules as well [66,67]. Therefore, although the kinase activity of GRK-2 is required for locomotion, it is possible that the relevant targets are proteins other than GPCRs. To examine whether phosphorylation of GPCRs is required for GRK-2 function in locomotion, we expressed GRK-2 with mutations (D3K, L4K, V7A/L8A, and D10A) that have been shown to reduce mammalian GRK2 phosphorylation of GPCRs, but that do not affect phosphorylation of other targets [69]. These N-terminal residues of mammalian GRKs form an amphipathic α-helix that contributes specifically to GPCR phosphorylation [70–74]. grk-2(gk268) mutants expressing any of these mutant GRK-2 constructs had slow locomotion like grk-2(gk268) (Fig 2B, 2C and 2G), indicating that GPCR phosphorylation is required for GRK-2 function in locomotion in vivo.
In mammalian GRKs, interaction of the N-terminal region with the kinase domain stabilizes a closed and more active conformation of the enzyme, important for phosphorylation of GPCRs and other substrates [70–72]. Specifically, mutation of mammalian GRK1 Arg191 disrupted phosphorylation of target substrates in addition to GPCRs, suggesting that this residue is critical for conformational changes important for GRK function as a kinase [71]. To determine whether the analogous residue in GRK-2 is required for its function in locomotion, we expressed GRK-2[R195A] in grk-2(gk268) mutants. GRK-2[R195A] did not rescue the grk-2(gk268) locomotion phenotype (Fig 2D and 2G), further supporting the model that GRK-2 acts as a GPCR kinase to regulate locomotion.
The RH (Regulator of G protein Signaling Homology) domain of mammalian GRK2 (Fig 2A) does not act like other RGS domains as an accelerator of the intrinsic GTPase activity of the Gq subunit, but instead interacts with Gq and participates in the uncoupling of GPCRs linked to Gq via a phosphorylation-independent mechanism [67,74]. To examine whether the Gq-binding residues of the RH domain are needed for GRK-2 function in locomotion, we expressed GRK-2[R106A], Y109I, and D110A that correspond to mutations previously shown to disrupt mammalian GRK2 binding to Gq/11 [75]. All three mutant GRK-2 constructs rescued the slow locomotion defect of grk-2(gk268) (Fig 2E and 2G). These results suggest that GRK-2 binding to Gq and phosphorylation-independent desensitization of GPCR signaling are not required for GRK-2 function in locomotion.
The pleckstrin homology (PH) domain of mammalian GRK2 (Fig 2A) mediates interactions of GRK2 with membrane phospholipids and Gβγ subunits [67,76–78]. To examine whether these activities are required for GRK-2 function in locomotion, we expressed GRK-2[K567E] that disrupts phospholipid binding [79] and GRK-2[R587Q] that disrupts binding to Gβγ [79]. Neither of these GRK-2 mutants rescued the locomotion defect of the grk-2(gk268) mutant (Fig 2F and 2G), suggesting that both phospholipid and Gβγ binding through the PH domain of GRK-2 are required for GRK-2 function in locomotion.
GRK-2 acts in head acetylcholine neurons to control locomotion and Gq signaling
GRK-2 is broadly expressed in body and head neurons [52]. To determine where GRK-2 acts to control locomotion, we expressed the grk-2 cDNA under the control of neuron-specific promoters. Expression of grk-2 under the pan-neuronal (Prab-3) or acetylcholine neuron (Punc-17) promoters fully rescued grk-2(gk268) mutant locomotion (Fig 3A). Interestingly, expression in ventral cord acetylcholine motor neurons (Pacr-2) did not rescue the locomotion phenotype, but expression driven by an unc-17 promoter derivative that is expressed mainly in the head acetylcholine neurons (Punc-17H [80,81]) rescued the locomotion phenotype (Fig 3A). Additionally, expression driven in a number of interneurons and head motorneurons by the glr-1 promoter did not rescue (Fig 3A). To exclude the possibility that the described role of GRK-2 in chemosensation [52] contributes to the slow locomotion phenotype of grk-2 mutants, we expressed grk-2 under ciliated sensory neuron promoters (Pxbx-1 and Posm-6). Expression of grk-2 in ciliated sensory neurons did not rescue the slow locomotion of grk-2 mutants (Fig 3A). We conclude that grk-2 acts in head acetylcholine neurons to regulate locomotion.
Fig 3. GRK-2 acts in head acetylcholine neurons.
(A) grk-2 acts in head acetylcholine neurons to control locomotion. The grk-2 cDNA was expressed in grk-2(gk268) mutants under a pan-neuronal promoter (Prab-3, transgene yakEx44), acetylcholine neuron promoter (Punc-17, transgene yakEx45), ventral cord acetylcholine motor neuron promoter (Pacr-2, transgene yakEx47), head acetylcholine neuron promoter (Punc-17H, transgene yakEx51), glutamate receptor promoter (Pglr-1, transgene yakEx52), and ciliated sensory neuron promoter (Pxbx-1, transgene yakEx71). Expression driven by the pan-neuronal, acetylcholine neuron, and head acetylcholine neuron promoters rescued the slow locomotion of grk-2 mutants. (***, P<0.001. Error bars = SEM; n = 10–25). (B,C) GRK-2 acts in head acetylcholine neurons to positively regulate Gq signaling. A grk-2(gk268) mutant suppresses the loopy posture and hyperactive locomotion of the activated Gq mutant egl-30(tg26) (Gq*). Expression of the grk-2 cDNA under a head acetylcholine neuron promoter (Punc-17H, transgene yakEx51) reverses the grk-2 suppression of the loopy posture (B) and hyperactive locomotion (C) of activated Gq. (***, P<0.001. Error bars = SEM; n = 10). (D) grk-2 is expressed in head acetylcholine neurons. Representative images of a Z-stack projection of the area around the nerve ring in the head of an animal coexpressing tagRFP fused to the GRK-2 ORF driven by the grk-2 promoter (grk-2::tagRFP, integration yakIs19) and GFP under a head acetylcholine neuron promoter (Punc-17H::eGFP, transgene yakEx94). Anterior to the left. Because Punc-17H::GFP is highly expressed and diffuse throughout the cell but grk-2::tagRFP is dimmer and localized only in the cytoplasm, their coexpression is hard to see in the merged image. For this reason, we have circled the cells where there is coexpression. Scale bar: 10 μm.
To determine if grk-2 also acts in head acetylcholine neurons to regulate Gq signaling, we expressed the grk-2 cDNA in the head acetylcholine neurons of egl-30(tg26); grk-2 double mutants. Expression in head acetylcholine neurons reversed the grk-2 suppression of the loopy posture and hyperactive locomotion of activated Gq−that is, the egl-30(tg26); grk-2 double mutants expressing grk-2 cDNA in the head acetylcholine neurons resemble the activated Gq single mutant (Fig 3B and 3C). These results suggest that grk-2 acts in head acetylcholine neurons to positively regulate Gq signaling.
To confirm that grk-2 is expressed in the head acetylcholine neurons, we coexpressed tagRFP fused to GRK-2 driven by the grk-2 promoter (grk-2::tagRFP) and GFP driven by the head acetylcholine neuron promoter (Punc-17H::GFP). We observed that grk-2::tagRFP is expressed broadly in head neurons and colocalizes with GFP in several head acetylcholine neurons (Fig 3D). We conclude that GRK-2 is expressed in head acetylcholine neurons to regulate locomotion and Gq signaling.
GRK-2 acts upstream of Go to regulate locomotion
Our results suggest that GRK-2 acts as a GPCR kinase to regulate locomotion. If GRK-2 were a kinase for a GPCR coupled to Gq (EGL-30 in C. elegans) then we would expect GRK-2 to negatively regulate Gq, which does not agree with our data. Alternatively, GRK-2 could be a kinase for a GPCR coupled to Go (GOA-1 in C. elegans). The C. elegans Gq and Go pathways act in opposite ways to regulate locomotion by controlling acetylcholine release [82]. EGL-30 is a positive regulator of acetylcholine release whereas GOA-1 negatively regulates the EGL-30 pathway through activation of the RGS protein EAT-16 and the diacylglycerol kinase DGK-1. egl-30 loss-of-function mutants are immobile whereas egl-30 gain-of-function mutants are hyperactive and have a loopy posture [83,84]. goa-1 and eat-16 mutants have locomotion phenotypes opposite those of egl-30; they are hyperactive and have a loopy posture [85–87]. dgk-1 loss-of-function mutants are hyperactive but do not have a loopy posture [88]. To test whether GRK-2 acts on a Go-coupled GPCR, we examined whether goa-1 mutations suppress grk-2 mutants. We found that the goa-1; grk-2 double mutant is hyperactive and has a loopy posture like the goa-1 single mutant (S4A, S4C and S4D Fig), indicating that GRK-2 acts upstream of goa-1. This result suggests that GRK-2 could be acting on GPCR(s) coupled to GOA-1.
To further dissect the GRK-2 pathway, we examined whether grk-2 mutations suppress the hyperactive phenotypes of eat-16 and dgk-1 mutants. The eat-16; grk-2 double mutant is hyperactive and has a loopy posture like the eat-16 single mutant (S4A, S4C and S4D Fig) indicating that eat-16, like goa-1, acts downstream of GRK-2. By contrast, the grk-2; dgk-1 double mutant is similar to grk-2 (S4B Fig). Expression of the kinase-dead GRK-2[K220R] in grk-2; dgk-1 mutants does not restore dgk-1 hyperactive locomotion (S4E Fig). In addition, expression of GRK-2 under a head acetylcholine neuron promoter in grk-2; dgk-1 mutants restores dgk-1 hyperactive locomotion (S4F Fig). Thus, GRK-2 regulation of the locomotion rate, Gq signaling, and DAG signaling all depend on the GRK-2 kinase activity and a function of GRK-2 in head acetylcholine neurons.
GRK-2 regulates the DOP-3 dopamine receptor
In a search for potential Go-coupled GPCR targets for GRK-2, we considered the Go-coupled D2-like dopamine receptor DOP-3. In C. elegans, dopamine is required for the “basal slowing response”, a behavior in which wild-type animals slow down when on a bacterial lawn [89]. This behavior is mediated by the mechanosensory activation of dopamine neurons caused by physical contact of the worm body with bacteria. cat-2 mutants that are deficient in dopamine biosynthesis [90] or dop-3 mutants that lack the D2-like dopamine receptor DOP-3, are defective in basal slowing [61,89]. DOP-3 has been proposed to act through Go in ventral cord acetylcholine motor neurons to decrease acetylcholine release and promote the basal slowing response [61].
If grk-2 acts in the dopamine pathway to mediate proper locomotion, possibly by phosphorylating and inactivating DOP-3, then mutations in dop-3 and cat-2 should suppress the grk-2 locomotion phenotype. Indeed, the grk-2 mutant slow locomotion phenotype was suppressed by mutations in dop-3 and cat-2 (Fig 4A). A dop-3 mutation also suppressed the swimming defect of the grk-2 mutant (S2 Fig). In addition, the dop-3 and cat-2 mutations reversed the grk-2 suppression of the loopy posture and hyperactive locomotion of activated Gq−that is, the triple mutants resemble the activated Gq single mutant (Fig 4B–4E and S5 Fig). These results suggest that GRK-2 acts in the dopamine pathway to regulate locomotion and Gq signaling by negatively regulating the D2-like dopamine receptor DOP-3.
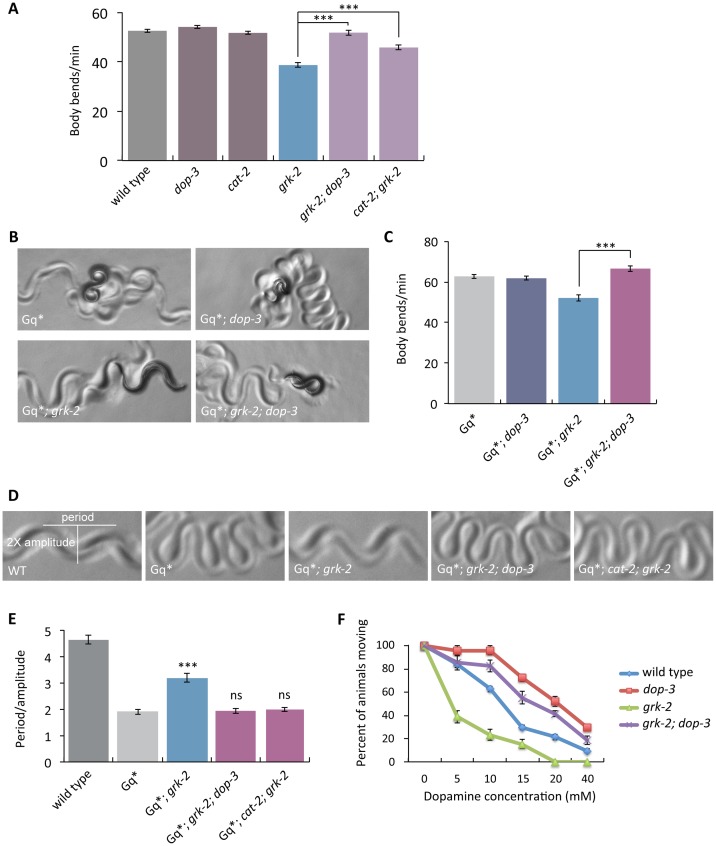
Fig 4. Mutations in dop-3 and cat-2 suppress grk-2.
(A) Mutations in dop-3 and cat-2 suppress the slow locomotion of grk-2 mutants. The grk-2(gk268) mutant has a slow locomotion phenotype. The dop-3(vs106) mutation fully suppresses and the cat-2(e1112) mutation partially suppresses the slow locomotion of the grk-2(gk268) mutant (***, P<0.001. Error bars = SEM; n = 32–72). (B,C) A dop-3 mutation reverses the grk-2 mutant suppression of activated Gq. The grk-2(gk268) mutation suppresses the loopy posture and hyperactive locomotion of the activated Gq mutant egl-30(tg26) (Gq*). The dop-3(vs106) mutation reverses the grk-2 suppression of the loopy posture (B) and hyperactive locomotion (C) of Gq*. (***, P<0.001. Error bars = SEM; n = 15–20). (D,E) dop-3 and cat-2 mutations reverse the grk-2 mutant suppression of the loopy posture of activated Gq. The grk-2(gk268) mutation suppresses the loopy posture of the activated Gq mutant egl-30(tg26) (Gq*). The dop-3 (vs106) and cat-2(e1112) mutations reverse the grk-2 suppression of the loopy posture of Gq*. (***, P<0.001. ns, P>0.05. Error bars = SEM; n = 5). (F) grk-2 mutants are hypersensitive to dopamine in a dop-3-dependent manner. Shown is the percentage of wild-type, dop-3(vs106), grk-2(gk268), or grk-2(gk268); dop-3(vs106) animals that moved ten body bends after a 20 min exposure to the indicated concentrations of dopamine. Every data point represents the mean +/- SEM of three trials (15–20 animals per experiment and strain).
Our results suggest that GRK-2 acts in head acetylcholine neurons to regulate locomotion. To test if DOP-3 acts in the same neurons as GRK-2, we expressed the dop-3 cDNA under a pan-neuronal promoter (Prab-3), an acetylcholine neuron promoter (Punc-17), a head acetylcholine neuron promoter (Punc-17H), and an acetylcholine motor neuron promoter (Pacr-2) in the grk-2; dop-3 double mutant. Expression driven by the pan-neuronal, acetylcholine neuron, and head acetylcholine neuron promoters reversed the dop-3 suppression of the slow locomotion of grk-2(gk268) mutant animals—that is, grk-2; dop-3 mutants expressing dop-3 cDNA by these three promoters resemble the grk-2 mutant (S6A Fig). By contrast, expression of the dop-3 cDNA by an acetylcholine ventral cord motor neuron promoter did not reverse the grk-2; dop-3 locomotion phenotype (S6A Fig) or the hyperactive locomotion and loopy posture of egl-30(tg26); grk-2; dop-3 mutant animals (S6C–S6E Fig). We conclude that dop-3, like grk-2, acts in head acetylcholine neurons, consistent with the model that GRK-2 acts directly on DOP-3. Moreover, dop-3 expression under the grk-2 promoter reversed the dop-3 suppression of the slow locomotion of grk-2 mutants (S7A Fig), supporting the idea that GRK-2 and DOP-3 act in the same neurons.
We observed that grk-2; dop-3 and cat-2; grk-2 double mutant animals still retain some of the characteristic grk-2 phenotypes: the animals have shorter bodies and are egg-laying defective. In addition, grk-2 mutants do not fully explore a bacterial lawn and this behavior remains in the grk-2; dop-3 double mutant (S1C Fig). Thus, GRK-2 has additional neuronal functions that do not depend on dop-3.
The D1-like dopamine receptor DOP-1 has been shown to act antagonistically to DOP-3 to regulate the basal slowing response: dop-1 mutations suppress the dop-3 basal slowing phenotype [61]. By contrast, we found that DOP-1 is not involved in the GRK-2 and DOP-3 pathway that regulates locomotion rate because dop-1 mutations do not affect the locomotion rate of the grk-2; dop-3 double mutant (S6B Fig). Thus, the role of DOP-3 in GRK-2-regulated locomotion is independent of its role in the basal slowing response.
Exposure of C. elegans to exogenous dopamine causes DOP-3-dependent paralysis—dop-3 mutants are significantly resistant to the paralytic effects of exogenous dopamine [61]. If GRK-2 negatively regulates DOP-3, then grk-2 mutants might be hypersensitive to dopamine due to increased DOP-3 activity. Indeed, we found that grk-2 mutants are hypersensitive to dopamine and this hypersensitivity depends on dop-3 (Fig 4F).
In an effort to dissect the molecular mechanism by which grk-2 regulates DOP-3 activity, we expressed GFP-tagged DOP-3 under the grk-2 promoter in dop-3 and grk-2; dop-3 mutant animals and examined the levels of expression of DOP-3::GFP both by Western and by fluorescence microscopy (S7 Fig). Although Pgrk-2::DOP-3::GFP fully reversed the dop-3 suppression of the slow locomotion of grk-2 mutants (S7A Fig), we did not observe any difference in the level of DOP-3 expression in a grk-2 mutant (S7B–S7D Fig) nor did we observe any obvious change in the subcellular localization of DOP-3::GFP in a grk-2 mutant (S7C Fig). However, one caveat is that we do not have the resolution to distinguish between DOP-3 localization on the plasma membrane or in an intracellular compartment.
GRK-2 is a positive modulator of NCA-1 and NCA-2 channel activity
In addition to genes within the canonical Gq-PLCβ pathway, our screen for suppressors of activated Gq also identified the Trio RhoGEF (UNC-73 in C. elegans) as a new direct Gq effector [8]. Recently, we identified the cation channels NCA-1 and NCA-2 as downstream targets of this Gq-Rho pathway. Specifically, we found that mutations in genes that encode accessory subunits of the NCA channels (unc-79, unc-80) or in the NCA channels per se (nca-1; nca-2) suppress the neuronal phenotypes of activated Gq and activated Rho [12]. Moreover, mutations in the Rho-NCA pathway suppress the loopy posture of the activated Gq mutant more strongly than do mutations in the canonical PLCβ pathway [12]. Like mutations in the Rho-NCA pathway, grk-2 mutants also strongly suppress the loopy posture of an activated Gq mutant (Figs 1A, 4D and 4E), suggesting that grk-2 may affect signaling through the Rho-NCA pathway.
To further examine whether grk-2 affects Rho-NCA signaling, we built double mutants of grk-2 with an activated Rho mutant (G14V), referred to here as Rho*, expressed in acetylcholine neurons. Rho* has a loopy posture and slow locomotion and a grk-2 mutation partially suppresses these phenotypes (Fig 5A and 5B), consistent with grk-2 affecting signaling through the Rho-NCA pathway.
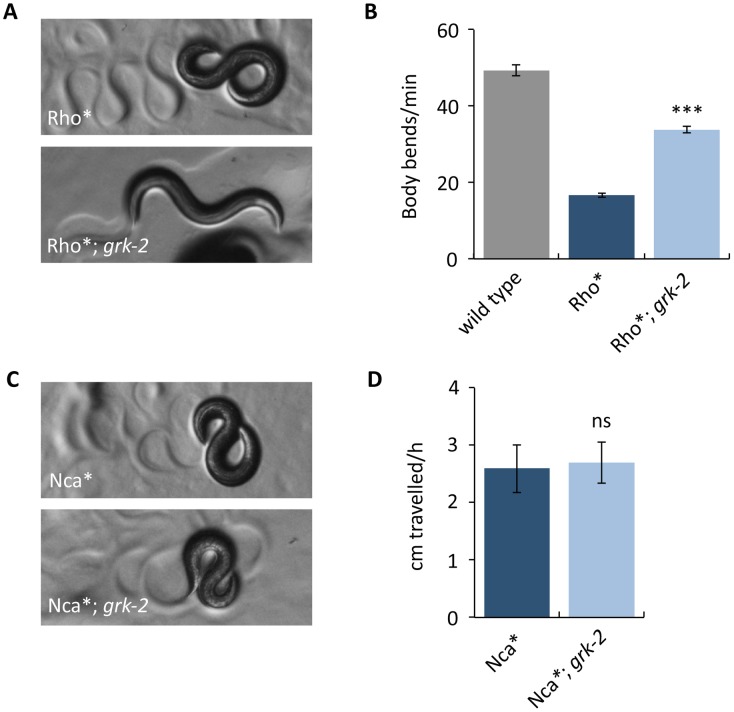
Fig 5. A grk-2 mutation partially suppresses activated Rho but does not suppress activated NCA-1.
(A,B) A grk-2 mutation partially suppresses activated Rho. Animals expressing activated RHO-1 [RHO-1[G14V]) under an acetylcholine promoter (Rho*, transgene nzIs29) have slow locomotion and loopy posture. The grk-2(gk268) mutation partially suppresses both the loopy posture (A) and slow locomotion (B) of the Rho* animals. (***, P<0.001. Error bars = SEM; n = 10). (C,D) A grk-2 mutation does not suppress activated NCA-1. The activated NCA-1 mutant (Nca*, nca-1(ox352)) has slow locomotion and loopy posture. The grk-2(gk268) mutation does not suppress the loopy posture (C) or the slow locomotion (D) of Nca*. To measure the locomotion of the slow moving Nca* animals, we used a radial locomotion assay in which we placed animals in the center of a 10 cm plate and measured how far the animals had moved in one hour. (ns, P>0.05. Error bars = SEM; n = 10).
We also built double mutants of grk-2 and a dominant activating mutation in the NCA-1 channel gene, nca-1(ox352), referred to here as Nca* [12,24]. Like Rho*, Nca* mutants have a loopy posture and slow locomotion. However, grk-2 mutants do not suppress either of these phenotypes because Nca*; grk-2 double mutants behave identically to Nca* mutants (Fig 5C and 5D). This suggests that grk-2 acts upstream of NCA.
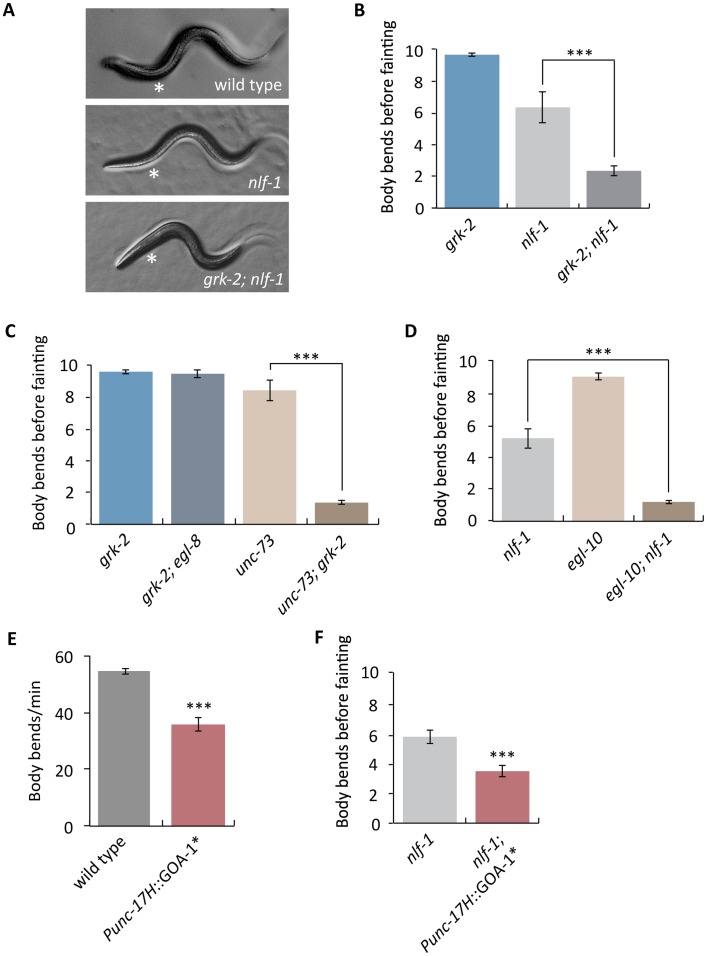
C. elegans has two proteins that encode pore-forming subunits of NCA channels, NCA-1 and NCA-2. Mutations that disrupt both NCA-1 and NCA-2 channel activity cause a characteristic “fainter” phenotype in which worms suddenly arrest their locomotion and acquire a straightened posture [35]. Our genetic experiments indicate that GRK-2 affects Rho-NCA signaling, but grk-2 mutants are not fainters. Given that grk-2 partially suppresses Rho*, we hypothesized that GRK-2 is not absolutely required for Rho-NCA signaling, but provides modulatory input. To test this hypothesis, we built double mutants between grk-2 and nlf-1, which is partially required for localization of the NCA-1 and NCA-2 channels and has a weak fainter mutant phenotype [12,91]. A grk-2 mutation strongly enhanced the weak fainter phenotype of an nlf-1 mutant so that the double mutants resembled the stronger fainter mutants that completely abolish NCA-1 and NCA-2 channel activity (Fig 6A and 6B). Moreover, double mutants between grk-2 and the RhoGEF Trio unc-73 were also strong fainters, supporting the hypothesis that GRK-2 modulates the Rho-NCA pathway (Fig 6C). By contrast, double mutants between grk-2 and the egl-8 PLCβ do not have a fainter phenotype (Fig 6C). These results suggest that GRK-2 is a positive modulator of NCA-1 and NCA-2 channel activity.
Fig 6. GRK-2 is a positive modulator of NCA-1 and NCA-2 channel activity.
(A) A grk-2 mutation enhances the weak forward fainting phenotype of an nlf-1 mutant. Representative images of wild-type, nlf-1(tm3631), and grk-2(gk268); nlf-1(tm3631) mutant animals. The asterisk shows the anterior part of the worm that becomes straight when an animal faints. (B) A grk-2 mutation enhances the weak forward fainting phenotype of an nlf-1 mutant. The nlf-1(tm3631) mutant is a weak fainter. The grk-2(gk268) mutation enhances the nlf-1 mutant so that the double is a strong fainter. (***, P<0.001. Error bars = SEM; n = 10–20). The number shown is the number of body bends before the animal faints. If the animal made ten body bends without fainting, the assay was stopped and we recorded ten as the number (see Methods). (C) The grk-2(gk268) mutation enhances the unc-73(ox317) mutant so that the double mutant is a strong fainter. The grk-2(gk268) mutation has no effect on an egl-8(sa47) mutant. (***, P<0.001. Error bars = SEM; n = 15). (D) The egl-10(md176) mutation enhances the nlf-1(tm3631) mutant so that the double mutant is a strong fainter. (***, P<0.001. Error bars = SEM; n = 25). (E) Expression of activated Go in head acetylcholine neurons inhibits locomotion. Animals expressing an activated Go mutant (GOA-1[Q205L]) under a head acetylcholine neuron promoter (Punc-17H::GOA-1*, transgene yakEx103) move more slowly than wild-type animals. (***, P<0.001. Error bars = SEM; n = 17). (F) Expression of activated Go in head acetylcholine neurons enhances the weak forward fainting phenotype of an nlf-1 mutant. The nlf-1(tm3631) mutant is a weak fainter in forward movement. The nlf-1(tm3631) mutant expressing an activated Go mutant (GOA-1[Q205L]) under a head acetylcholine neuron promoter (Punc-17H::GOA-1*, transgene yakEx103) is a stronger fainter than the nlf-1(tm3631) mutant. (***, P<0.001. Error bars = SEM; n = 54).
If GRK-2 modulates the NCA channels by acting as a negative regulator of Go then we would expect that mutations in other proteins that act as negative regulators of Go might enhance the fainter phenotype of nlf-1 mutants. Indeed, a mutation in egl-10, encoding the RGS that negatively regulates Go [92], strongly enhances the nlf-1 fainter phenotype (Fig 6D). As controls, mutations in genes involved in dense-core vesicle biogenesis (eipr-1 and cccp-1), that cause locomotion defects comparable to grk-2 or egl-10 [63,80], did not enhance the nlf-1 fainter phenotype, indicating that the interactions of grk-2 and egl-10 with nlf-1 are specific.
grk-2 acts in head acetylcholine neurons to mediate locomotion. We recently used the same Punc-17H promoter construct to show that nlf-1 also acts in head acetylcholine neurons and not in ventral cord motor neurons to regulate locomotion [12]. Therefore, we predicted that expression of an activated Go mutant under a head acetylcholine neuron promoter would enhance the fainter phenotype of nlf-1 mutants. Indeed, we found that expression of the activated Go mutant GOA-1[Q205L] in head acetylcholine neurons makes the animals slow (Fig 6E) and significantly enhances the fainter phenotype of nlf-1 mutants (Fig 6F). These results support the model that GRK-2 negatively regulates Go, and that Go negatively regulates NCA-1 and NCA-2 channel activity.
Dopamine acts through DOP-3 to negatively modulate NCA-1 and NCA-2 channel activity
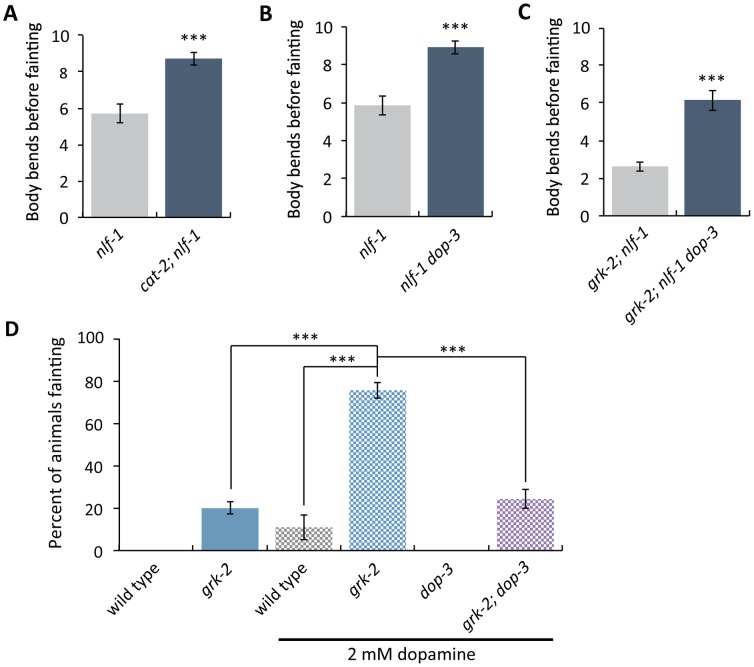
Our results are consistent with the model that GRK-2 acts in locomotion by negatively regulating DOP-3 and that GRK-2 is a positive modulator of NCA-1 and NCA-2 activity. These data predict that DOP-3 would be a negative modulator of NCA-1 and NCA-2 channel activity. Consistent with this model, mutations in cat-2 and dop-3 almost fully suppress the nlf-1 fainter phenotype during forward movement (Fig 7A and 7B). Additionally, dop-3 mutants partially suppress the strong grk-2; nlf-1 fainter phenotype, consistent with the model that DOP-3 is a substrate for GRK-2 (Fig 7C). These results suggest that dopamine, through DOP-3, negatively modulates NCA-1 and NCA-2 channel activity.
Fig 7. Dopamine negatively modulates NCA-1 and NCA-2 channel activity.
(A) The cat-2(e1112) mutation suppresses the weak forward fainting phenotype of the nlf-1(tm3631) mutant. (***, P<0.001. Error bars = SEM; n = 40). (B) The dop-3(vs106) mutation suppresses the weak forward fainting phenotype of the nlf-1(tm3631) mutant. (***, P<0.001. Error bars = SEM; n = 40). (C) The dop-3(vs106) mutation partially suppresses the strong forward fainting phenotype of the grk-2(gk268); nlf-1(tm3631) double mutant. (***, P<0.001. Error bars = SEM; n = 40). (D) Exogenous dopamine causes the grk-2(gk268) mutant to faint in a dop-3 dependent manner. Shown is the percentage of animals that faint within a period of ten body bends when moving backwards after exposure to 2 mM dopamine for 20 min. (***, P<0.001. Error bars = SEM; n = 2–5 trials of 14–25 animals each).
To more directly test whether grk-2 and dop-3 modulate the NCA channel per se, we created double mutants between grk-2 and the pore-forming subunit gene nca-1. nca-1 mutants have a low penetrance, very weak backward-fainting phenotype that is strongly enhanced in a grk-2 mutant background (S3C and S6F Figs). arr-1 mutants, on the other hand, do not enhance the nca-1 phenotype, further supporting the conclusion that arrestin does not play a role in this pathway (S3C Fig). As expected, dop-3 suppresses the enhanced fainting phenotype of the grk-2; nca-1 double mutant (S6F Fig).
Our data suggest that GRK-2 and DOP-3 play modulatory and not essential roles in the regulation of NCA-1 (and possibly NCA-2) channel activity. By contrast, UNC-80 is necessary for the stability and function of NCA-1 and NCA-2, so unc-80 mutants are strong fainters [36,39]. As expected for a modulatory role in regulating NCA-1 and NCA-2 activity, mutations in dop-3 and cat-2 do not suppress the strong fainter phenotype of unc-80 mutants (S8A and S8B Fig).
We showed above that grk-2 mutants are hypersensitive to the paralytic effects of dopamine. We also found that low concentrations of dopamine do not paralyze grk-2 mutants but instead cause them to faint, and that this effect depends on dop-3 (Fig 7D). This is consistent with the model that dopamine acts through DOP-3 to negatively modulate NCA-1 and NCA-2.
GRK-2 and DOP-3 act in command interneurons to control grk-2 dependent locomotion
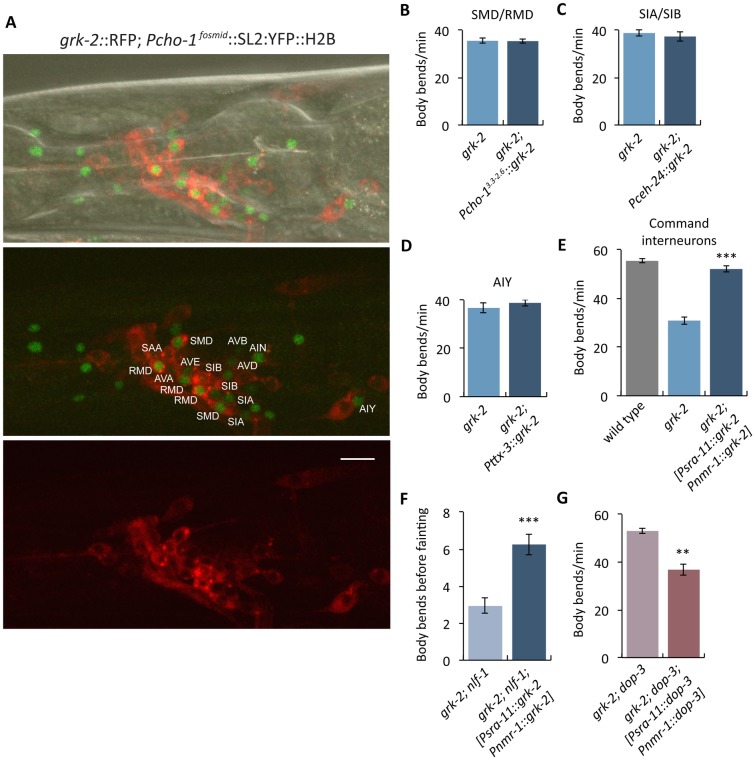
In C. elegans, the NCA channels act in premotor interneurons [12,43,91]. To determine whether grk-2 acts in a cell autonomous way to regulate NCA, we identified the head acetylcholine neurons where GRK-2 is expressed. We coexpressed GRK-2 fused to tagRFP driven by the grk-2 promoter (grk-2::RFP) and nuclear YFP driven by the choline transporter cho-1 promoter (Pcho-1fosmid::SL2::YFP::H2B), which is expressed in all acetylcholine neurons [93]. We found that grk-2 is expressed in the following head acetylcholine neurons: the AVA, AVB, AVD, and AVE premotor interneurons; SMD and RMD head motor neurons; and in the AIN, AIY, SIA, SIB, and SAA interneurons (Fig 8A).
Fig 8. GRK-2 is expressed and acts in command interneurons to regulate locomotion.
(A) grk-2 is expressed in command interneurons. Representative images of a Z-stack projection of the area around the nerve ring (head) of an animal coexpressing tagRFP fused to the GRK-2 cDNA driven by the grk-2 promoter (grk-2::RFP, transgene yakIs19) and a cho-1 fosmid YFP reporter (cho-1fosmid::SL2::YFP::H2B, transgene otIs534). For the cho-1 fosmid reporter, an SL2-spliced, nuclear-localized YFP::H2B sequence was engineered right after the stop codon of the gene [93,110]. As indicated in the figure, grk-2::RFP is expressed in the AVA, AVB, AVD, and AVE command interneurons; SMD and RMD head motor neurons; and in the AIN, AIY, SIA, SIB, and SAA interneurons. Scale bar: 10 μm. (B-D) grk-2 cDNA expression in (B) SMD/RMD (Pcho-1, 3.3 to 2.6 kb upstream of the ATG, transgene yakEx135), (C) SIA/SIB (Pceh-24, transgene yakEx149), or (D) AIY (Pttx-3, transgene yakEx138) neurons does not rescue the slow locomotion of grk-2(gk268) mutants. (Error bars = SEM; n = 15). (E) grk-2 cDNA expression in command interneurons (Psra-11 + Pnmr-1, transgene yakEx141) is sufficient to rescue the slow locomotion of grk-2(gk268) mutants. (***, P<0.001. Error bars = SEM; n = 25). (F) grk-2 cDNA expression in command interneurons (Psra-11 + Pnmr-1, transgene yakEx141) is sufficient to rescue the strong fainting phenotype of grk-2(gk268); nlf-1(tm3631) mutants. (***, P<0.001. Error bars = SEM; n = 40). (G) dop-3 cDNA expression in command interneurons (Psra-11 + Pnmr-1, transgene yakEx148) is sufficient to reverse the dop-3(vs106) mutant suppression of the slow locomotion of grk-2(gk268) mutant animals. (**, P<0.01. Error bars = SEM; n = 23).
To further determine where GRK-2 acts to control locomotion, we expressed the grk-2 cDNA under additional neuron-specific promoters in grk-2 mutants. We used a cho-1 promoter fragment for expression in the SMD and RMD head motor neurons [94], the ceh-24 promoter for expression in the SIA and SIB interneurons, and a ttx-3 promoter fragment for AIY-specific expression [95]. For expression in premotor command interneurons we used a combination of the nmr-1 promoter for AVA, AVD and AVE (also PVC and RIM) expression together with the sra-11 promoter for AVB (also AIY and AIA) expression, as previously described [91]. We found that grk-2 expression in command interneurons fully rescued the slow locomotion of grk-2 mutants, but expression in the other neuron types failed to rescue (Fig 8B–8E). However, expression of grk-2 in only sra-11 or only nmr-1 expressing neurons did not rescue the slow locomotion defect (S9 Fig). Additionally, grk-2 expression in the command interneurons was sufficient to rescue the enhanced fainting phenotype of grk-2; nlf-1 mutants (Fig 8F). Similarly, dop-3 expression in the command interneurons was sufficient to reverse the dop-3 suppression of the slow locomotion of grk-2 mutants (Fig 8G). Given that the fainting phenotypes of nlf-1 mutants and nca mutants were also rescued by expression in command interneurons [43,91], our results suggest that GRK-2, DOP-3, and the NCA channels act in the same neurons. However, we did not see rescue of the grk-2 locomotion phenotype using the glr-1 promoter (Fig 3A), which in principle is also expressed in the command interneurons, and similarly we did not observe statistically significant rescue of the nlf-1 fainting phenotype using the same glr-1 promoter [12]. The different results seen between the glr-1 and the nmr-1 + sra-11 promoters may be due to different levels of expression or because of expression in some different neuron types.
Discussion
In this study we identified a pathway that modulates the activity of the NCA-1 and NCA-2 channels through dopamine and Gq signaling (Fig 9). We found that dopamine acts through the D2-like receptor DOP-3 to negatively modulate NCA-1 and NCA-2. Furthermore, we identified the GPCR kinase GRK-2 as a positive (indirect) regulator of Gq and negative regulator of NCA-1 and NCA-2. Our results suggest that GRK-2 mediates its regulatory effects by inhibiting DOP-3.
Fig 9. Model for GRK-2 and dopamine action in modulating activity of the NCA channels.
Schematic representation of the dopamine, Gq and Go signaling pathways [61,82]. Solid arrows indicate direct actions or direct physical interactions. Dashed arrows indicate interactions that may be indirect. Our results suggest that dopamine decreases activity of the NCA-1 and NCA-2 channels (shown here collectively as “NCA”) by binding to DOP-3 and activating Go signaling. GRK-2 acts as a kinase for the D2-like dopamine receptor DOP-3 to inhibit DOP-3, and thereby inhibit Go, activate Gq, and positively regulate NCA-1 and NCA-2 channel activity.
In C. elegans, GRK-2 was previously found to act in sensory neurons to regulate chemosensation [52]. Here we found that GRK-2 acts in command interneurons to regulate locomotion and Gq signaling. Using a structure-function approach, we found that GPCR phosphorylation, Gβγ-binding, and membrane-binding are required for GRK-2 function in locomotion, but binding to Gq is not required. Similar results were reported for the function of GRK-2 in chemosensation [48], suggesting that in both cases GRK-2 acts as a GPCR kinase and that membrane localization is critical for its function. Additionally, GRK-2 seems to act independently of arrestin to regulate both locomotion and chemosensation [52]. Because cat-2 and dop-3 mutants are hypersensitive to the aversive odorant octanol [96–98] and grk-2 mutants are insensitive to octanol [52], GRK-2 might act as a GPCR kinase for DOP-3 in chemosensory neurons as well.
GRK-induced phosphorylation of GPCRs induces endocytosis, which leads to their sorting to either lysosomes for degradation or to recycling endosomes. GRK-dependent recruitment of arrestins to the phosphorylated receptor is typically required for endocytosis, but GRK2 was also reported to utilize arrestin-independent mechanisms to mediate receptor internalization [66]. GRK2 associates with a large number of proteins with known roles in receptor internalization and signaling. For example, the C-terminus of GRK2 directly binds clathrin and this interaction has been proposed to be involved in arrestin-independent internalization [99]. Our data suggest an arrestin-independent role for C. elegans GRK-2 in GPCR regulation, supporting the idea that the role of GRK-2 extends beyond just the recruitment of arrestin.
The D2-type dopamine receptors, like DOP-3, are GPCRs that couple to members of the inhibitory Gi/o family. Mammalian GRK2 and GRK3 (the orthologs of GRK-2) have been connected to the desensitization, internalization, and recycling of D2-type dopamine receptors [55–59,100]. Interestingly, some of the effects of GRK2 on D2 receptor function may be independent of receptor phosphorylation [57,58,100], though one caveat of these studies is that they involve GRK2 overexpression in heterologous cells. Our structure-function approach indicates that GPCR phosphorylation is important for GRK-2 function in locomotion and Gq signaling in C. elegans, although we cannot exclude the possibility that phosphorylation of additional substrates may also be required. In vivo studies of the role of mammalian GRKs in the regulation of dopamine receptors have focused on the analysis of behaviors that are induced by psychostimulatory drugs such as cocaine that elevate the extracellular concentration of dopamine [59]. Mice with a cell-specific knockout of GRK2 in D2 receptor-expressing neurons have altered spontaneous locomotion and sensitivity to cocaine [101], though the cellular mechanisms underlying these behavioral effects are not known. Our findings provide evidence of a direct association between GRK-2 and D2-type receptor signaling that regulates locomotion in an in vivo system.
In C. elegans, the Gq and Go pathways act in opposite ways to regulate locomotion by controlling synaptic vesicle release [82]. Gq acts as a positive regulator of acetylcholine release while Go negatively regulates Gq signaling, through activation of the Gq RGS EAT-16 and the diacylglycerol kinase DGK-1. DGK-1 phosphorylates the second messenger DAG and thus inhibits its action. Using genetic epistasis, we demonstrated that GRK-2 acts upstream of GOA-1/Go and EAT-16 to positively regulate locomotion and body posture. Given this result, our cell-specific rescue data, and our data indicating that GRK-2 acts as a GPCR kinase for a locomotion-related GPCR, we propose that GRK-2 acts as a kinase for the Go-coupled GPCR DOP-3 in premotor interneurons. In this model, GRK-2 driven phosphorylation of DOP-3 reduces Go signaling and thereby promotes Gq signaling (Fig 9). Inhibition of Go by GRK-2 could promote Gq-Rho signaling by two mechanisms: (1) by inhibiting the Gq RGS EAT-16 and thus activating Gq itself, and (2) by inhibiting DGK-1 which acts in parallel to Gq-Rho to regulate DAG levels (Fig 9).
Interestingly, a grk-2 mutant is suppressed by mutations in goa-1 and eat-16, but not by dgk-1. This finding supports other literature that suggests that goa-1 and eat-16 have similar interactions with Gq signaling, but that dgk-1 is distinct [87,102]. GOA-1 and EAT-16 act upstream of Gq to inhibit Gq signaling. DGK-1, on the other hand, acts downstream of Gq to reduce the pool of the Gq-generated second messenger DAG. Adding to the complexity, DAG levels may be controlled by both the PLCβ and Rho branches of the Gq pathway (Fig 9). Previously, it has been shown that mutations in dgk-1 partially suppress the strong locomotion defect of egl-30/Gq loss-of-function mutations [102]. Surprisingly, we found that a grk-2 mutation fully suppresses a dgk-1 mutant. This suggests that the effect of GRK-2 on locomotion is more complex and may be partially independent of Gq signaling and of Gq-generated DAG. This agrees with our data showing that GRK-2 has additional neuronal functions that do not depend on DOP-3.
Gq signaling regulates several genetically separable aspects of locomotion behavior including locomotion rate and waveform. The Gq-PLCβ signaling pathway has been reported to act in ventral cord motor neurons to regulate acetylcholine release and locomotion rate [103], whereas the Gq-Rho pathway has been reported to act in at least two different classes of neurons including head acetylcholine neurons to regulate locomotion rate, waveform, and fainting behavior [12]. Our data here further suggest that DOP-3, GRK-2, and the Gq-Rho pathway all act together in the premotor command interneurons to regulate activity of the NCA channels. The command interneurons have been previously shown to regulate several aspects of locomotion behavior including the propensity to go forward or reverse [104,105] and the tendency of the worm to sustain persistent locomotion [43]. Our data here suggest that the command interneurons also regulate the locomotion rate and the posture of the animals. As we reported previously, mutations in the Rho-NCA pathway suppress both the locomotion rate and loopy posture of activated Gq mutants whereas mutations in the PLCβ pathway suppress mainly the locomotion rate [12]. Thus, Gq acts through both the PLCβ pathway and the Rho-NCA pathway to regulate locomotion rate, probably by acting in different neurons. By contrast, Gq acts primarily through the Rho-NCA pathway to regulate the posture of the worms. This agrees with our data showing that grk-2 mutations, which affect signaling through the Rho-NCA pathway, strongly suppress the loopy posture of activated Gq.
The identification of GRK-2 as a putative DOP-3 kinase and positive modulator of Gq-Rho signaling connects dopamine signaling to modulation of the NCA channels (Fig 9). NCA channels have been shown in recent years to be important for neuronal excitability and a number of rhythmic behaviors [16,34–42]. In humans, mutations affecting the NCA channel NALCN cause neurological diseases [20–33]. However, despite the relevance of this channel to neuronal function it is unclear how it is gated and activated. Two studies have shown that NALCN-dependent currents can be activated by G protein-coupled receptors in a G protein independent way [44,45] whereas another study showed that NALCN can be activated by low extracellular calcium via a G protein-dependent pathway [46], but the specific mechanisms remain unknown. Our results suggest that dopamine acts through the DOP-3 G protein-coupled receptor and downstream G protein signaling pathways to modulate activity of the NCA channels in a physiologically relevant setting. This is the first study connecting dopamine to the activation of these important channels.
Methods
Strains
Strains were maintained at room temperature or 20° on the OP50 strain of E. coli [106]. The Supplementary Information contains full genotypes of all the strains we used (S1 Table; List of strains).
Isolation and identification of the grk-2(yak18) mutation
The grk-2(yak18) mutant was isolated in an ENU screen as a suppressor of the hyperactive locomotion and loopy posture of the activated Gq mutant egl-30(tg26) [63]. We mapped the yak18 mutation to the left arm of Chromosome III (between -27 and -21.8 m.u.) using a PCR mapping strategy that takes advantage of PCR length polymorphisms due to indels in the Hawaiian strain CB4856 (Jihong Bai, personal communication). Using whole-genome sequencing (see below), we found that yak18 is a G to A transition mutation in the W02B3.2 (grk-2) ORF that creates a G379E missense mutation in the kinase domain of GRK-2. We confirmed the gene identification by performing a complementation test between grk-2(yak18) and the grk-2(gk268) deletion mutant, finding that they fail to complement for the slow locomotion phenotype.
Whole-genome sequencing
Genomic DNA from grk-2(yak18) animals was isolated and purified according to the Worm Genomic DNA prep protocol from the Hobert lab website (http://hobertlab.org/wp-content/uploads/2013/02/Worm_Genomic_DNA_Prep.pdf). The sample was sequenced using Ion Torrent sequencing (DNA Sequencing Core Facility, University of Utah). The sequencing data were uploaded to the Galaxy web platform and were analyzed as described [107].
Constructs and transgenes
The Supplemental Information contains a complete list of constructs used (S2 Table; List of plasmids). All constructs made in this study were constructed using the multisite Gateway system (Invitrogen). Specifically, a promoter region, a gene region (cDNA), and an N- or C-terminal 3’UTR or fluorescent tag (GFP or tagRFP) fused to a 3’UTR were cloned into the destination vector pCFJ150. For the cell-specific rescuing experiments, an operon GFP was included in the expression constructs downstream of the 3’UTR [108]. This resulted in expression of untagged grk-2, dop-3, or goa-1, but allowed for confirmation of proper promoter expression by monitoring GFP expression. The cho-1 fosmid reporter construct otIs534 carries an SL2-spliced nuclear localized YFP::H2B immediately after the stop codon of the cho-1 gene [93].
Extrachromosomal arrays were made by standard injection and transformation methods [109]. In all cases we injected 5–10 ng/ul of the expression vector and isolated multiple independent lines. At least two lines were tested that behaved similarly.
Expression of grk-2
We made a construct driving expression of the grk-2 cDNA fused to tagRFP under the grk-2 promoter and generated worms with extrachromosomal arrays. For the grk-2 promoter region, we PCR amplified 2892 bp upstream of the start codon using genomic DNA as a template and the following set of primers: forward primer 5’cacgacagtttccatagtgattgg3’ and reverse primer 5’tttttgttctgcaaaatcgaattg3’. grk-2 was expressed in neurons in the head, ventral cord, and tail, consistent with the published expression pattern [52]. Neurons were identified by the stereotypical positions of cells expressing the acetylcholine neuron reporter cho-1fosmid::SL2::YFP::H2B [93,110] that colocalized with grk-2::tagRFP.
Locomotion and egg-laying assays
For most experiments, we measured locomotion rate using the body bend assay. Specifically, first-day adults were picked to a three-day-old lawn of OP50 and stimulated by poking the tail of the animal with a worm pick. Body bends were then immediately counted for one minute. A body bend was defined as the movement of the worm from maximum to minimum amplitude of the sine wave [102]. Specifically for the experiment described in Fig 5D we used a radial locomotion assay. Animals were placed in the center of 10 cm plates with thin one to two-day-old lawns of OP50 and left for one hour. The position of each worm was marked and the radial distance from the center of the plate was measured (cm travelled/h).
Egg-laying assays were performed as described [80]. L4 larvae were placed on plates with OP50 at 25°C overnight. The next day, five animals were moved to a fresh plate and allowed to lay eggs at 25°C for two hours. The number of eggs present on the plate was counted.
Waveform quantification
First-day adult animals were placed on an OP50 plate and allowed to move forward until when they had completed five to ten tracks. Each animal's tracks were imaged at 40X magnification using a Nikon SMZ18 microscope with the DS-L3 camera control system. Period and 2X amplitude were measured using the line tool in Image J. For each worm, five period/ amplitude ratios were averaged and five worms were used per experiment.
Fainting assays
The fainting phenotype is characterized by frequent arrest of locomotion, accompanied by a straightening of the anterior part of the body. We scored fainting as a sudden halt in movement accompanied by a straightened posture.
First-day adults were transferred to plates with two to three-day-old lawns of OP50 and left undisturbed for one minute. Animals were then poked either on the head (for backward movement) or on the tail (for forward movement), and we counted the number of body bends before the animal faints. If the animal made ten body bends, the assay was stopped and we recorded ten as the number. Thus, animals that never faint (for example, wild-type) are scored as 10 in these experiments. Specifically for the experiment described in Fig 7D the number reported was the percentage of animals that fainted before making 10 body bends.
Swimming assays
Single, first-day adults were transferred to a 25 ul drop of M9 buffer at the center of an empty NGM plate and video recorded for 30 sec. The swimming behavior was analyzed as described [38,64].
Body length measurements
First-day adults were mounted on 2% agarose pads and anesthetized in M9 buffer containing 50 mM sodium azide for ten minutes. The image of each animal was obtained using a Nikon 80i wide-field compound microscope. Body size was measured using ImageJ software.
Dopamine resistance assays
We used a method similar to the one described [61]. Specifically, first-day adults were transferred to plates containing dopamine (5 mM, 10 mM, 15 mM, 20 mM, 40 mM) and incubated for 20 min at room temperature. Animals were then poked using a worm-pick and the number of body bends was counted, stopping the assay at 10 body bends. We report the percent of animals that moved 10 body bends without stopping (Percent of animals moving). A body bend was defined as the movement of the worm from maximum to minimum amplitude of the sine wave. Dopamine plates were prepared fresh just before use, as described [61].
Immunoblotting
For the Western analysis shown in Fig 2G, worm lysates were prepared as follows. Ten transgenic animals from each strain were transferred to a 6 cm OP50 plate and grown until most of their progeny had reached adult stage. Animals from five such plates were washed off with M9, collected in a 15 ml Falcon tube, and spun down at 2000 rpm for 3 min. Animals were washed twice with M9. The pelleted worms were then resuspended in 2X SDS loading dye and lysed by incubation at 95°C for 20 min. For the Western analysis shown in S7B Fig, worm lysates were prepared as follows. Two hundred transgenic worms were individually picked and transferred in a microfuge tube in 10 ul M9. An equal volume of 2X SDS loading dye was added to the tube and the animals were lysed by incubation at 95°C for 20 min.
Samples were resolved on 10% SDS-polyacrylamide gels and blotted onto PVDF membranes. To detect the desired proteins, we added the following primary antibodies: monoclonal anti-GRK2/3, clone C5/1.1 (1:1000, EMD Millipore #05–465), monoclonal anti-beta-tubulin antibody (1:1000, ThermoFisher, BT7R, #MA5-16308), rabbit polyclonal anti-GFP (1:1000, Santa Cruz #sc-8334), and monoclonal anti-mCherry (1:50, a gift from Jihong Bai and the Fred Hutchinson Cancer Research Center antibody development shared resource center). The secondary antibodies were an Alexa Fluor 680-conjugated goat anti-mouse antibody (1:20,000, Jackson Laboratory #115-625-166) and an Alexa Fluor 680-conjugated goat anti-rabbit antibody (1:20,000, Jackson Laboratory #111-625-144). A LI-COR processor was used to develop images.
Imaging
For fluorescence imaging, first-day adult animals were mounted on 2% agarose pads and anesthetized with 50 mM sodium azide for ten minutes before placing the cover slip. The images shown in Fig 3D and S7C Fig were obtained using an Olympus FLUOVIEW FV1200 confocal microscope. The images shown in Fig 8A were acquired using a Zeiss confocal microscope (LSM880) with Z-stack analysis and reconstruction performed using the ZEN software tool.
For pictures of worms, first-day adult animals were placed on an assay plate and photographed at 50 or 60X using a Nikon SMZ18 dissecting microscope with a DS-L3 camera control system. The images were processed using ImageJ.
Statistics
P values were determined using GraphPad Prism 5.0d (GraphPad Software). Normally distributed data sets requiring multiple comparisons were analyzed by a one-way ANOVA followed by a Bonferroni or Dunnett test. Normally distributed pairwise data comparisons were analyzed by two-tailed unpaired t tests. Non-normally distributed data sets with multiple comparisons were analyzed by a Kruskal-Wallis nonparametric ANOVA followed by Dunn’s test to examine selected comparisons. Non-normally distributed pairwise data comparisons were analyzed by a Mann-Whitney test. For the experiments shown in S3C and S6F Figs a chi-square test for multiple comparisons was used.
Supporting information
(A) The grk-2(gk268) mutant has an egg-laying defect. The graph shows the number of eggs laid by 5 animals in a 2 h period. (**, P<0.01. Error bars = SEM; n = 2 plates of 5 animals each). (B) The grk-2(gk268) mutant animals have short bodies. (***, P<0.001. Error bars = SEM; n = 10).(C) A dop-3 mutation does not suppress the restricted exploration behavior of grk-2 mutants. Shown are images of tracks of five wild-type, grk-2(gk268), and grk-2(gk268); dop-3(vs106) mutant animals that were allowed to explore a bacterial lawn for 2 hours at room temperature.
(TIF)
Shown are plots of bending angle (midpoint) versus time for representative individual animals. The two plots of grk-2(gk268) mutant animals show individuals with strong and weak swimming defects. The dop-3(vs106) mutation suppresses the swimming defects of the grk-2(gk268) mutant.
(TIF)
(A) The arr-1(ok401) mutant has no locomotion defect. (ns, P>0.05. Error bars = SEM; n = 10). (B) The arr-1(ok401) mutation does not suppress the loopy posture of the egl-30(tg26) mutant. (C) The arr-1(ok401) mutation, in contrast to a grk-2(gk268) mutation, does not cause a fainting phenotype in an nca-1(gk9) mutant background. Shown is the percentage of animals that faint when moving backwards. The wild-type, nca-1, grk-2, and grk-2; nca-1 data are the same data shown in S6F Fig. The graph shows the combined data from two independent experiments, each with n = 20–40. (**, P<0.01; ***, P<0.001; ns, P>0.05).
(TIF)
(A) The grk-2(gk268) mutation does not suppress the hyperactive locomotion of the eat-16(tm775) and goa-1(sa734) mutants. (ns, P>0.05. Error bars = SEM; n = 10–20). (B) The grk-2(gk268) mutation suppresses the hyperactive locomotion phenotype of the dgk-1(sy428) mutant. (***, P<0.001. ns, P>0.05. Error bars = SEM; n = 10–20). (C-D) The grk-2(gk268) mutation does not suppress the loopy posture of the eat-16(tm775) and goa-1(sa734) mutants. (ns, P>0.05. Error bars = SEM; n = 5). (E) The kinase-dead GRK-2 does not reverse the grk-2 suppression of the dgk-1 hyperactive locomotion phenotype. Expression of the kinase-dead GRK-2[K220R] mutant under its own promoter (transgene yakEx48) does not reverse the grk-2 suppression of dgk-1 hyperactivity. (ns, P>0.05. Error bars = SEM; n = 10–20). (F) Expression of the grk-2 cDNA under a head acetylcholine neuron promoter (transgene yakEx51) reverses the grk-2 suppression of the hyperactive locomotion of the dgk-1(sy428) mutant. (***, P<0.001. Error bars = SEM; n = 10–20).
(TIF)
The grk-2(gk268) mutation suppresses the loopy posture and hyperactive locomotion of the activated Gq mutant egl-30(tg26) (Gq*). The cat-2(e1112) mutation reverses the grk-2 suppression of the loopy posture (A) and hyperactive locomotion (B) of Gq*. (***, P<0.001. Error bars = SEM; n = 15–20).
(TIF)
(A) The dop-3 suppression of grk-2 is reversed by dop-3 expression in head acetylcholine neurons. The dop-3 cDNA was expressed in the grk-2(gk268); dop-3(vs106) double mutant under a pan-neuronal promoter (Prab-3, transgene yakEx112), acetylcholine neuron promoter (Punc-17, transgene yakEx111), head acetylcholine neuron promoter (Punc-17H, transgene yakEx110) and ventral cord acetylcholine motor neuron promoter (Pacr-2, transgene yakEx109). Expression of dop-3 driven by the pan-neuronal, acetylcholine neuron, and head acetylcholine neuron promoters reversed the dop-3(vs106) mutant suppression of the slow locomotion of grk-2(gk268) mutant animals. (*, P<0.05; **, P<0.01; ***, P<0.001; ns, P>0.05. Error bars = SEM; n = 10–33). (B) A dop-1 mutation does not affect the dop-3 suppression of the grk-2 slow locomotion phenotype. grk-2; dop-3 mutants move more rapidly than the grk-2 mutant. The dop-1(vs100) mutation does not affect grk-2(gk268); dop-3(vs106) locomotion. (ns, P>0.05. Error bars = SEM; n = 23–34). (C-E) Expression of dop-3 in ventral cord motor neurons is not sufficient to reverse the hyperactive locomotion and loopy posture of egl-30(tg26); grk-2; dop-3 mutant animals. (C) Expression of dop-3 driven by the ventral cord neuron promoter (Pacr-2, transgene yakEx109) does not reduce the hyperactivity of egl-30(tg26); grk-2(gk268); dop-3(vs106) mutant animals. (***, P<0.001; ns, P>0.05. Error bars = SEM; n = 10). (D-E) Expression of dop-3 driven by the ventral cord neuron promoter (Pacr-2, transgene yakEx109) does not reverse the loopy waveform of egl-30(tg26); grk-2(gk268); dop-3(vs106) mutant animals. (**, P<0.01; ***, P<0.001; ns, P>0.05. Error bars = SEM; n = 10). (F) A dop-3 mutation suppresses the fainting phenotype of grk-2; nca-1 mutants. Shown is the percentage of animals that faint when moving backwards. The wild-type, nca-1, grk-2, and grk-2; nca-1 data are the same data shown in S3C Fig. The graph shows the combined data from two independent experiments, each with n = 20–40. (**, P<0.01; ***, P<0.001; ns, P>0.05).
(TIF)
(A) DOP-3::GFP expression driven by the grk-2 promoter (transgene yakEx130) reverses the dop-3 mutant suppression of the slow locomotion phenotype of grk-2 mutants. (**, P<0.01; ***, P<0.001. Error bars = SEM; n = 10). (B) DOP-3::GFP levels remain unaffected in grk-2 mutants. Immunoblot of extracts derived from dop-3 or grk-2; dop-3 animals expressing Pgrk-2::DOP-3::GFP and Pmyo-2::mCherry from an extrachromosomal array (transgene yakEx130). The experiment was repeated twice with similar results. (C,D) DOP-3::GFP subcellular localization and level of expression remain unaffected in grk-2 mutants. (C) Representative images of a Z-stack projection of the area around the nerve ring in the head of dop-3 or grk-2; dop-3 mutant animals expressing Pgrk-2::DOP-3::GFP (transgene yakEx130). (D) Quantification of the ratio of DOP-3::GFP to mCherry in the region around the nerve ring of dop-3 or grk-2; dop-3 animals expressing Pgrk-2::DOP-3::GFP and Pmyo-2::mCherry (transgene yakEx130). (ns, P>0.05. Error bars = SEM; n = 10).
(TIF)
(A) The dop-3(vs106) mutation does not suppress the strong forward fainting phenotype of the unc-80(ox330) mutant. (ns, P>0.05. Error bars = SEM; n = 20). (B) The cat-2(e1112) mutation does not suppress the strong forward fainting phenotype of the unc-80(ox330) mutant. (ns, P>0.05. Error bars = SEM; n = 36–38).
(TIF)
(A), (B) grk-2 cDNA expression driven by the (A) sra-11 (Psra-11, transgene yakEx147) or (B) nmr-1 (Pnmr-1, transgene yakEx85) promoter does not rescue the slow locomotion of the grk-2(gk268) mutant. (ns, P>0.05. Error bars = SEM; n = 10–20).
(TIF)
(DOCX)
(DOCX)
Acknowledgments
Special thanks to Denise Ferkey and Jordan Wood for generously providing the grk-2 mutant constructs, Jihong Bai for discussions and sharing unpublished methods and equipment, Yongming Dong for help with the swimming assay, Ithai Rabinowitch for the sra-11 promoter plasmid, Jill Hoyt for making the grk-2; nlf-1 double mutant, Jill Hoyt and Jordan Hoyt for help with the analysis of the whole-genome sequencing data, Jérôme Cattin-Ortolá for help with confocal microscopy, Dana Miller for sharing equipment, Oliver Hobert for providing materials and methodology, and Brian Kraemer and Rebecca Kow for insightful ideas. Some strains were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by US National Institutes of Health (NIH, http://www.nih.gov/) grant R00 MH082109 and by an Ellison Medical Foundation (http://www.ellisonfoundation.org/) New Scholar Award (AG-NS-1035-13) to MA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wilkie TM, Gilbert DJ, Olsen AS, Chen XN, Amatruda TT, Korenberg JR, et al. Evolution of the mammalian G protein alpha subunit multigene family. Nat Genet. 1992;1: 85–91. doi: 10.1038/ng0592-85 [DOI] [PubMed] [Google Scholar]
- 2.Coulon P, Kanyshkova T, Broicher T, Munsch T, Wettschureck N, Seidenbecher T, et al. Activity Modes in Thalamocortical Relay Neurons are Modulated by G(q)/G(11) Family G-proteins—Serotonergic and Glutamatergic Signaling. Front Cell Neurosci. 2010;4: 132 doi: 10.3389/fncel.2010.00132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gamper N, Reznikov V, Yamada Y, Yang J, Shapiro MS. Phosphatidylinositol 4,5-bisphosphate signals underlie receptor-specific Gq/11-mediated modulation of N-type Ca2+ channels. J Neurosci. 2004;24: 10980–10992. doi: 10.1523/JNEUROSCI.3869-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krause M, Offermanns S, Stocker M, Pedarzani P. Functional specificity of G alpha q and G alpha 11 in the cholinergic and glutamatergic modulation of potassium currents and excitability in hippocampal neurons. J Neurosci. 2002;22: 666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lutz S, Freichel-Blomquist A, Yang Y, Rümenapp U, Jakobs KH, Schmidt M, et al. The guanine nucleotide exchange factor p63RhoGEF, a specific link between Gq/11-coupled receptor signaling and RhoA. J Biol Chem. 2005;280: 11134–11139. doi: 10.1074/jbc.M411322200 [DOI] [PubMed] [Google Scholar]
- 6.Lutz S, Shankaranarayanan A, Coco C, Ridilla M, Nance MR, Vettel C, et al. Structure of Galphaq-p63RhoGEF-RhoA complex reveals a pathway for the activation of RhoA by GPCRs. Science. 2007;318: 1923–1927. doi: 10.1126/science.1147554 [DOI] [PubMed] [Google Scholar]
- 7.Rojas RJ, Yohe ME, Gershburg S, Kawano T, Kozasa T, Sondek J. Galphaq directly activates p63RhoGEF and Trio via a conserved extension of the Dbl homology-associated pleckstrin homology domain. J Biol Chem. 2007;282: 29201–29210. doi: 10.1074/jbc.M703458200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams SL, Lutz S, Charlie NK, Vettel C, Ailion M, Coco C, et al. Trio’s Rho-specific GEF domain is the missing Galpha q effector in C. elegans. Genes Dev. 2007;21: 2731–2746. doi: 10.1101/gad.1592007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan JP, Hu Z, Sieburth D. Recruitment of sphingosine kinase to presynaptic terminals by a conserved muscarinic signaling pathway promotes neurotransmitter release. Genes Dev. 2012;26: 1070–1085. doi: 10.1101/gad.188003.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiley E, McMullan R, Nurrish SJ. The Galpha12-RGS RhoGEF-RhoA signalling pathway regulates neurotransmitter release in C. elegans. EMBO J. 2006;25: 5884–5895. doi: 10.1038/sj.emboj.7601458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McMullan R, Hiley E, Morrison P, Nurrish SJ. Rho is a presynaptic activator of neurotransmitter release at pre-existing synapses in C. elegans. Genes Dev. 2006;20: 65–76. doi: 10.1101/gad.359706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Topalidou I, Chen P-A, Cooper K, Watanabe S, Jorgensen EM, Ailion M. The NCA-1 and NCA-2 ion channels function downstream of Gq and Rho to regulate locomotion in Caenorhabditis elegans. Genetics. 2017;206: 265–282. doi: 10.1534/genetics.116.198820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren D. Sodium leak channels in neuronal excitability and rhythmic behaviors. Neuron. 2011;72: 899–911. doi: 10.1016/j.neuron.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liebeskind BJ, Hillis DM, Zakon HH. Phylogeny unites animal sodium leak channels with fungal calcium channels in an ancient, voltage-insensitive clade. Mol Biol Evol. 2012;29: 3613–3616. doi: 10.1093/molbev/mss182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JH, Cribbs LL, Perez-Reyes E. Cloning of a novel four repeat protein related to voltage-gated sodium and calcium channels. FEBS Lett. 1999;445: 231–236. [DOI] [PubMed] [Google Scholar]
- 16.Lu B, Su Y, Das S, Liu J, Xia J, Ren D. The neuronal channel NALCN contributes resting sodium permeability and is required for normal respiratory rhythm. Cell. 2007;129: 371–383. doi: 10.1016/j.cell.2007.02.041 [DOI] [PubMed] [Google Scholar]
- 17.Boone AN, Senatore A, Chemin J, Monteil A, Spafford JD. Gd3+ and calcium sensitive, sodium leak currents are features of weak membrane-glass seals in patch clamp recordings. PloS One. 2014;9: e98808 doi: 10.1371/journal.pone.0098808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Senatore A, Spafford JD. A uniquely adaptable pore is consistent with NALCN being an ion sensor. Channels. 2013;7: 60–68. doi: 10.4161/chan.23981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Senatore A, Monteil A, van Minnen J, Smit AB, Spafford JD. NALCN ion channels have alternative selectivity filters resembling calcium channels or sodium channels. PloS One. 2013;8: e55088 doi: 10.1371/journal.pone.0055088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Sayed MD, Al-Zaidan H, Albakheet A, Hakami H, Kenana R, Al-Yafee Y, et al. Mutations in NALCN cause an autosomal-recessive syndrome with severe hypotonia, speech impairment, and cognitive delay. Am J Hum Genet. 2013;93: 721–726. doi: 10.1016/j.ajhg.2013.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chong JX, McMillin MJ, Shively KM, Beck AE, Marvin CT, Armenteros JR, et al. De novo mutations in NALCN cause a syndrome characterized by congenital contractures of the limbs and face, hypotonia, and developmental delay. Am J Hum Genet. 2015;96: 462–473. doi: 10.1016/j.ajhg.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fukai R, Saitsu H, Okamoto N, Sakai Y, Fattal-Valevski A, Masaaki S, et al. De novo missense mutations in NALCN cause developmental and intellectual impairment with hypotonia. J Hum Genet. 2016;61: 451–455. doi: 10.1038/jhg.2015.163 [DOI] [PubMed] [Google Scholar]
- 23.Aoyagi K, Rossignol E, Hamdan FF, Mulcahy B, Xie L, Nagamatsu S, et al. A Gain-of-Function Mutation in NALCN in a Child with Intellectual Disability, Ataxia, and Arthrogryposis. Hum Mutat. 2015;36: 753–757. doi: 10.1002/humu.22797 [DOI] [PubMed] [Google Scholar]
- 24.Bend EG, Si Y, Stevenson DA, Bayrak-Toydemir P, Newcomb TM, Jorgensen EM, et al. NALCN channelopathies: Distinguishing gain-of-function and loss-of-function mutations. Neurology. 2016;87: 1131–1139. doi: 10.1212/WNL.0000000000003095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gal M, Magen D, Zahran Y, Ravid S, Eran A, Khayat M, et al. A novel homozygous splice site mutation in NALCN identified in siblings with cachexia, strabismus, severe intellectual disability, epilepsy and abnormal respiratory rhythm. Eur J Med Genet. 2016;59: 204–209. doi: 10.1016/j.ejmg.2016.02.007 [DOI] [PubMed] [Google Scholar]
- 26.Karakaya M, Heller R, Kunde V, Zimmer K-P, Chao C-M, Nürnberg P, et al. Novel Mutations in the Nonselective Sodium Leak Channel (NALCN) Lead to Distal Arthrogryposis with Increased Muscle Tone. Neuropediatrics. 2016;47: 273–277. doi: 10.1055/s-0036-1584084 [DOI] [PubMed] [Google Scholar]
- 27.Köroğlu Ç, Seven M, Tolun A. Recessive truncating NALCN mutation in infantile neuroaxonal dystrophy with facial dysmorphism. J Med Genet. 2013;50: 515–520. doi: 10.1136/jmedgenet-2013-101634 [DOI] [PubMed] [Google Scholar]
- 28.Perez Y, Kadir R, Volodarsky M, Noyman I, Flusser H, Shorer Z, et al. UNC80 mutation causes a syndrome of hypotonia, severe intellectual disability, dyskinesia and dysmorphism, similar to that caused by mutations in its interacting cation channel NALCN. J Med Genet. 2016;53: 397–402. doi: 10.1136/jmedgenet-2015-103352 [DOI] [PubMed] [Google Scholar]
- 29.Valkanas E, Schaffer K, Dunham C, Maduro V, du Souich C, Rupps R, et al. Phenotypic evolution of UNC80 loss of function. Am J Med Genet A. 2016;170: 3106–3114. doi: 10.1002/ajmg.a.37929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shamseldin HE, Faqeih E, Alasmari A, Zaki MS, Gleeson JG, Alkuraya FS. Mutations in UNC80, Encoding Part of the UNC79-UNC80-NALCN Channel Complex, Cause Autosomal-Recessive Severe Infantile Encephalopathy. Am J Hum Genet. 2016;98: 210–215. doi: 10.1016/j.ajhg.2015.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stray-Pedersen A, Cobben J-M, Prescott TE, Lee S, Cang C, Aranda K, et al. Biallelic Mutations in UNC80 Cause Persistent Hypotonia, Encephalopathy, Growth Retardation, and Severe Intellectual Disability. Am J Hum Genet. 2016;98: 202–209. doi: 10.1016/j.ajhg.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Koh K, Ichinose Y, Yasumura M, Ohtsuka T, Takiyama Y. A de novo mutation in the NALCN gene in an adult patient with cerebellar ataxia associated with intellectual disability and arthrogryposis. Clin Genet. 2016;90: 556–557. doi: 10.1111/cge.12851 [DOI] [PubMed] [Google Scholar]
- 33.Lozic B, Johansson S, Lovric Kojundzic S, Markic J, Knappskog PM, Hahn AF, et al. Novel NALCN variant: altered respiratory and circadian rhythm, anesthetic sensitivity. Ann Clin Transl Neurol. 2016;3: 876–883. doi: 10.1002/acn3.362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Funato H, Miyoshi C, Fujiyama T, Kanda T, Sato M, Wang Z, et al. Forward-genetics analysis of sleep in randomly mutagenized mice. Nature. 2016;539: 378–383. doi: 10.1038/nature20142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphrey JA, Hamming KS, Thacker CM, Scott RL, Sedensky MM, Snutch TP, et al. A putative cation channel and its novel regulator: cross-species conservation of effects on general anesthesia. Curr Biol. 2007;17: 624–629. doi: 10.1016/j.cub.2007.02.037 [DOI] [PubMed] [Google Scholar]
- 36.Jospin M, Watanabe S, Joshi D, Young S, Hamming K, Thacker C, et al. UNC-80 and the NCA ion channels contribute to endocytosis defects in synaptojanin mutants. Curr Biol. 2007;17: 1595–1600. doi: 10.1016/j.cub.2007.08.036 [DOI] [PubMed] [Google Scholar]
- 37.Nash HA, Scott RL, Lear BC, Allada R. An unusual cation channel mediates photic control of locomotion in Drosophila. Curr Biol. 2002;12: 2152–2158. [DOI] [PubMed] [Google Scholar]
- 38.Pierce-Shimomura JT, Chen BL, Mun JJ, Ho R, Sarkis R, McIntire SL. Genetic analysis of crawling and swimming locomotory patterns in C. elegans. Proc Natl Acad Sci U S A. 2008;105: 20982–20987. doi: 10.1073/pnas.0810359105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeh E, Ng S, Zhang M, Bouhours M, Wang Y, Wang M, et al. A putative cation channel, NCA-1, and a novel protein, UNC-80, transmit neuronal activity in C. elegans. PLoS Biol. 2008;6: e55 doi: 10.1371/journal.pbio.0060055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lear BC, Darrah EJ, Aldrich BT, Gebre S, Scott RL, Nash HA, et al. UNC79 and UNC80, Putative Auxiliary Subunits of the NARROW ABDOMEN Ion Channel, Are Indispensable for Robust Circadian Locomotor Rhythms in Drosophila. PloS One. 2013;8: e78147 doi: 10.1371/journal.pone.0078147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lear BC, Lin J-M, Keath JR, McGill JJ, Raman IM, Allada R. The ion channel narrow abdomen is critical for neural output of the Drosophila circadian pacemaker. Neuron. 2005;48: 965–976. doi: 10.1016/j.neuron.2005.10.030 [DOI] [PubMed] [Google Scholar]
- 42.Yeh S-Y, Huang W-H, Wang W, Ward CS, Chao ES, Wu Z, et al. Respiratory Network Stability and Modulatory Response to Substance P Require Nalcn. Neuron. 2017;94: 294–303. doi: 10.1016/j.neuron.2017.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao S, Xie L, Kawano T, Po MD, Pirri JK, Guan S, et al. The NCA sodium leak channel is required for persistent motor circuit activity that sustains locomotion. Nat Commun. 2015;6: 6323 doi: 10.1038/ncomms7323 [DOI] [PubMed] [Google Scholar]
- 44.Lu B, Su Y, Das S, Wang H, Wang Y, Liu J, et al. Peptide neurotransmitters activate a cation channel complex of NALCN and UNC-80. Nature. 2009;457: 741–744. doi: 10.1038/nature07579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swayne LA, Mezghrani A, Varrault A, Chemin J, Bertrand G, Dalle S, et al. The NALCN ion channel is activated by M3 muscarinic receptors in a pancreatic beta-cell line. EMBO Rep. 2009;10: 873–880. doi: 10.1038/embor.2009.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu B, Zhang Q, Wang H, Wang Y, Nakayama M, Ren D. Extracellular calcium controls background current and neuronal excitability via an UNC79-UNC80-NALCN cation channel complex. Neuron. 2010;68: 488–499. doi: 10.1016/j.neuron.2010.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferguson SS. Evolving concepts in G protein-coupled receptor endocytosis: the role in receptor desensitization and signaling. Pharmacol Rev. 2001;53: 1–24. [PubMed] [Google Scholar]
- 48.Wood JF, Wang J, Benovic JL, Ferkey DM. Structural domains required for Caenorhabditis elegans G protein-coupled receptor kinase 2 (GRK-2) function in vivo. J Biol Chem. 2012;287: 12634–12644. doi: 10.1074/jbc.M111.336818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benovic JL, Onorato JJ, Arriza JL, Stone WC, Lohse M, Jenkins NA, et al. Cloning, expression, and chromosomal localization of beta-adrenergic receptor kinase 2. A new member of the receptor kinase family. J Biol Chem. 1991;266: 14939–14946. [PubMed] [Google Scholar]
- 50.Schleicher S, Boekhoff I, Arriza J, Lefkowitz RJ, Breer H. A beta-adrenergic receptor kinase-like enzyme is involved in olfactory signal termination. Proc Natl Acad Sci U S A. 1993;90: 1420–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jaber M, Koch WJ, Rockman H, Smith B, Bond RA, Sulik KK, et al. Essential role of beta-adrenergic receptor kinase 1 in cardiac development and function. Proc Natl Acad Sci U S A. 1996;93: 12974–12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fukuto HS, Ferkey DM, Apicella AJ, Lans H, Sharmeen T, Chen W, et al. G protein-coupled receptor kinase function is essential for chemosensation in C. elegans. Neuron. 2004;42: 581–593. [DOI] [PubMed] [Google Scholar]
- 53.Wang J, Luo J, Aryal DK, Wetsel WC, Nass R, Benovic JL. G protein-coupled receptor kinase-2 (GRK-2) regulates serotonin metabolism through the monoamine oxidase AMX-2 in Caenorhabditis elegans. J Biol Chem. 2017;292: 5943–5956. doi: 10.1074/jbc.M116.760850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3: 639–650. doi: 10.1038/nrm908 [DOI] [PubMed] [Google Scholar]
- 55.Ito K, Haga T, Lameh J, Sadée W. Sequestration of dopamine D2 receptors depends on coexpression of G-protein-coupled receptor kinases 2 or 5. Eur J Biochem. 1999;260: 112–119. doi: 10.1046/j.1432-1327.1999.00125.x [DOI] [PubMed] [Google Scholar]
- 56.Kim KM, Valenzano KJ, Robinson SR, Yao WD, Barak LS, Caron MG. Differential regulation of the dopamine D2 and D3 receptors by G protein-coupled receptor kinases and beta-arrestins. J Biol Chem. 2001;276: 37409–37414. doi: 10.1074/jbc.M106728200 [DOI] [PubMed] [Google Scholar]
- 57.Cho D, Zheng M, Min C, Ma L, Kurose H, Park JH, et al. Agonist-induced endocytosis and receptor phosphorylation mediate resensitization of dopamine D(2) receptors. Mol Endocrinol. 2010;24: 574–586. doi: 10.1210/me.2009-0369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Namkung Y, Dipace C, Javitch JA, Sibley DR. G protein-coupled receptor kinase-mediated phosphorylation regulates post-endocytic trafficking of the D2 dopamine receptor. J Biol Chem. 2009;284: 15038–15051. doi: 10.1074/jbc.M900388200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gurevich EV, Gainetdinov RR, Gurevich VV. G protein-coupled receptor kinases as regulators of dopamine receptor functions. Pharmacol Res. 2016;111: 1–16. doi: 10.1016/j.phrs.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sawin ER, Ranganathan R, Horvitz HR. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron. 2000;26: 619–631. [DOI] [PubMed] [Google Scholar]
- 61.Chase DL, Pepper JS, Koelle MR. Mechanism of extrasynaptic dopamine signaling in Caenorhabditis elegans. Nat Neurosci. 2004;7: 1096–1103. doi: 10.1038/nn1316 [DOI] [PubMed] [Google Scholar]
- 62.Doi M, Iwasaki K. Regulation of retrograde signaling at neuromuscular junctions by the novel C2 domain protein AEX-1. Neuron. 2002;33: 249–259. [DOI] [PubMed] [Google Scholar]
- 63.Ailion M, Hannemann M, Dalton S, Pappas A, Watanabe S, Hegermann J, et al. Two Rab2 interactors regulate dense-core vesicle maturation. Neuron. 2014;82: 167–180. doi: 10.1016/j.neuron.2014.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vidal-Gadea A, Topper S, Young L, Crisp A, Kressin L, Elbel E, et al. Caenorhabditis elegans selects distinct crawling and swimming gaits via dopamine and serotonin. Proc Natl Acad Sci U S A. 2011;108: 17504–17509. doi: 10.1073/pnas.1108673108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pitcher JA, Freedman NJ, Lefkowitz RJ. G protein-coupled receptor kinases. Annu Rev Biochem. 1998;67: 653–692. doi: 10.1146/annurev.biochem.67.1.653 [DOI] [PubMed] [Google Scholar]
- 66.Evron T, Daigle TL, Caron MG. GRK2: multiple roles beyond G protein-coupled receptor desensitization. Trends Pharmacol Sci. 2012;33: 154–164. doi: 10.1016/j.tips.2011.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ribas C, Penela P, Murga C, Salcedo A, García-Hoz C, Jurado-Pueyo M, et al. The G protein-coupled receptor kinase (GRK) interactome: role of GRKs in GPCR regulation and signaling. Biochim Biophys Acta. 2007;1768: 913–922. [DOI] [PubMed] [Google Scholar]
- 68.Kong G, Penn R, Benovic JL. A beta-adrenergic receptor kinase dominant negative mutant attenuates desensitization of the beta 2-adrenergic receptor. J Biol Chem. 1994;269: 13084–13087. [PubMed] [Google Scholar]
- 69.Pao CS, Barker BL, Benovic JL. Role of the amino terminus of G protein-coupled receptor kinase 2 in receptor phosphorylation. Biochemistry. 2009;48: 7325–7333. doi: 10.1021/bi900408g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boguth CA, Singh P, Huang C, Tesmer JJG. Molecular basis for activation of G protein-coupled receptor kinases. EMBO J. 2010;29: 3249–3259. doi: 10.1038/emboj.2010.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang C, Yoshino-Koh K, Tesmer JJG. A surface of the kinase domain critical for the allosteric activation of G protein-coupled receptor kinases. J Biol Chem. 2009;284: 17206–17215. doi: 10.1074/jbc.M809544200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang C-C, Orban T, Jastrzebska B, Palczewski K, Tesmer JJG. Activation of G protein-coupled receptor kinase 1 involves interactions between its N-terminal region and its kinase domain. Biochemistry. 2011;50: 1940–1949. doi: 10.1021/bi101606e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Noble B, Kallal LA, Pausch MH, Benovic JL. Development of a yeast bioassay to characterize G protein-coupled receptor kinases. Identification of an NH2-terminal region essential for receptor phosphorylation. J Biol Chem. 2003;278: 47466–47476. doi: 10.1074/jbc.M308257200 [DOI] [PubMed] [Google Scholar]
- 74.Pao CS, Benovic JL. Phosphorylation-independent desensitization of G protein-coupled receptors? Sci STKE. 2002;2002: pe42 doi: 10.1126/stke.2002.153.pe42 [DOI] [PubMed] [Google Scholar]
- 75.Sterne-Marr R, Tesmer JJG, Day PW, Stracquatanio RP, Cilente J-AE, O’Connor KE, et al. G protein-coupled receptor Kinase 2/G alpha q/11 interaction. A novel surface on a regulator of G protein signaling homology domain for binding G alpha subunits. J Biol Chem. 2003;278: 6050–6058. doi: 10.1074/jbc.M208787200 [DOI] [PubMed] [Google Scholar]
- 76.Boekhoff I, Inglese J, Schleicher S, Koch WJ, Lefkowitz RJ, Breer H. Olfactory desensitization requires membrane targeting of receptor kinase mediated by beta gamma-subunits of heterotrimeric G proteins. J Biol Chem. 1994;269: 37–40. [PubMed] [Google Scholar]
- 77.Koch WJ, Inglese J, Stone WC, Lefkowitz RJ. The binding site for the beta gamma subunits of heterotrimeric G proteins on the beta-adrenergic receptor kinase. J Biol Chem. 1993;268: 8256–8260. [PubMed] [Google Scholar]
- 78.Touhara K, Koch WJ, Hawes BE, Lefkowitz RJ. Mutational analysis of the pleckstrin homology domain of the beta-adrenergic receptor kinase. Differential effects on G beta gamma and phosphatidylinositol 4,5-bisphosphate binding. J Biol Chem. 1995;270: 17000–17005. [DOI] [PubMed] [Google Scholar]
- 79.Carman CV, Barak LS, Chen C, Liu-Chen LY, Onorato JJ, Kennedy SP, et al. Mutational analysis of Gbetagamma and phospholipid interaction with G protein-coupled receptor kinase 2. J Biol Chem. 2000;275: 10443–10452. [DOI] [PubMed] [Google Scholar]
- 80.Topalidou I, Cattin-Ortolá J, Pappas AL, Cooper K, Merrihew GE, MacCoss MJ, et al. The EARP Complex and Its Interactor EIPR-1 Are Required for Cargo Sorting to Dense-Core Vesicles. PLoS Genet. 2016;12: e1006074 doi: 10.1371/journal.pgen.1006074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hammarlund M, Palfreyman MT, Watanabe S, Olsen S, Jorgensen EM. Open syntaxin docks synaptic vesicles. PLoS Biol. 2007;5: e198 doi: 10.1371/journal.pbio.0050198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Koelle MR. Neurotransmitter signaling through heterotrimeric G proteins: insights from studies in C. elegans. WormBook Online Rev C Elegans Biol. 2016; 1–78. doi: 10.1895/wormbook.1.75.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bastiani CA, Gharib S, Simon MI, Sternberg PW. Caenorhabditis elegans Galphaq regulates egg-laying behavior via a PLCbeta-independent and serotonin-dependent signaling pathway and likely functions both in the nervous system and in muscle. Genetics. 2003;165: 1805–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brundage L, Avery L, Katz A, Kim UJ, Mendel JE, Sternberg PW, et al. Mutations in a C. elegans Gqalpha gene disrupt movement, egg laying, and viability. Neuron. 1996;16: 999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mendel JE, Korswagen HC, Liu KS, Hajdu-Cronin YM, Simon MI, Plasterk RH, et al. Participation of the protein Go in multiple aspects of behavior in C. elegans. Science. 1995;267: 1652–1655. [DOI] [PubMed] [Google Scholar]
- 86.Ségalat L, Elkes DA, Kaplan JM. Modulation of serotonin-controlled behaviors by Go in Caenorhabditis elegans. Science. 1995;267: 1648–1651. [DOI] [PubMed] [Google Scholar]
- 87.Hajdu-Cronin YM, Chen WJ, Patikoglou G, Koelle MR, Sternberg PW. Antagonism between G(o)alpha and G(q)alpha in Caenorhabditis elegans: the RGS protein EAT-16 is necessary for G(o)alpha signaling and regulates G(q)alpha activity. Genes Dev. 1999;13: 1780–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nurrish S, Ségalat L, Kaplan JM. Serotonin inhibition of synaptic transmission: Galpha(0) decreases the abundance of UNC-13 at release sites. Neuron. 1999;24: 231–242. [DOI] [PubMed] [Google Scholar]
- 89.Sawin ER, Ranganathan R, Horvitz HR. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron. 2000;26: 619–631. [DOI] [PubMed] [Google Scholar]
- 90.Lints R, Emmons SW. Patterning of dopaminergic neurotransmitter identity among Caenorhabditis elegans ray sensory neurons by a TGFbeta family signaling pathway and a Hox gene. Development. 1999;126: 5819–5831. [DOI] [PubMed] [Google Scholar]
- 91.Xie L, Gao S, Alcaire SM, Aoyagi K, Wang Y, Griffin JK, et al. NLF-1 delivers a sodium leak channel to regulate neuronal excitability and modulate rhythmic locomotion. Neuron. 2013;77: 1069–1082. doi: 10.1016/j.neuron.2013.01.018 [DOI] [PubMed] [Google Scholar]
- 92.Koelle MR, Horvitz HR. EGL-10 regulates G protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins. Cell. 1996;84: 115–125. [DOI] [PubMed] [Google Scholar]
- 93.Stefanakis N, Carrera I, Hobert O. Regulatory Logic of Pan-Neuronal Gene Expression in C. elegans. Neuron. 2015;87: 733–750. doi: 10.1016/j.neuron.2015.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang F, Bhattacharya A, Nelson JC, Abe N, Gordon P, Lloret-Fernandez C, et al. The LIM and POU homeobox genes ttx-3 and unc-86 act as terminal selectors in distinct cholinergic and serotonergic neuron types. Development. 2014;141: 422–435. doi: 10.1242/dev.099721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wenick AS, Hobert O. Genomic cis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C. elegans. Dev Cell. 2004;6: 757–770. doi: 10.1016/j.devcel.2004.05.004 [DOI] [PubMed] [Google Scholar]
- 96.Ezak MJ, Ferkey DM. The C. elegans D2-like dopamine receptor DOP-3 decreases behavioral sensitivity to the olfactory stimulus 1-octanol. PloS One. 2010;5: e9487 doi: 10.1371/journal.pone.0009487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ferkey DM, Hyde R, Haspel G, Dionne HM, Hess HA, Suzuki H, et al. C. elegans G protein regulator RGS-3 controls sensitivity to sensory stimuli. Neuron. 2007;53: 39–52. doi: 10.1016/j.neuron.2006.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wragg RT, Hapiak V, Miller SB, Harris GP, Gray J, Komuniecki PR, et al. Tyramine and octopamine independently inhibit serotonin-stimulated aversive behaviors in Caenorhabditis elegans through two novel amine receptors. J Neurosci. 2007;27: 13402–13412. doi: 10.1523/JNEUROSCI.3495-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shiina T, Arai K, Tanabe S, Yoshida N, Haga T, Nagao T, et al. Clathrin box in G protein-coupled receptor kinase 2. J Biol Chem. 2001;276: 33019–33026. doi: 10.1074/jbc.M100140200 [DOI] [PubMed] [Google Scholar]
- 100.Namkung Y, Dipace C, Urizar E, Javitch JA, Sibley DR. G protein-coupled receptor kinase-2 constitutively regulates D2 dopamine receptor expression and signaling independently of receptor phosphorylation. J Biol Chem. 2009;284: 34103–34115. doi: 10.1074/jbc.M109.055707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Daigle TL, Ferris MJ, Gainetdinov RR, Sotnikova TD, Urs NM, Jones SR, et al. Selective deletion of GRK2 alters psychostimulant-induced behaviors and dopamine neurotransmission. Neuropsychopharmacology. 2014;39: 2450–2462. doi: 10.1038/npp.2014.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Miller KG, Emerson MD, Rand JB. Goalpha and diacylglycerol kinase negatively regulate the Gqalpha pathway in C. elegans. Neuron. 1999;24: 323–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lackner MR, Nurrish SJ, Kaplan JM. Facilitation of synaptic transmission by EGL-30 Gqalpha and EGL-8 PLCbeta: DAG binding to UNC-13 is required to stimulate acetylcholine release. Neuron. 1999;24: 335–346. [DOI] [PubMed] [Google Scholar]
- 104.Chalfie M, Sulston JE, White JG, Southgate E, Thomson JN, Brenner S. The neural circuit for touch sensitivity in Caenorhabditis elegans. J Neurosci. 1985;5: 956–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zheng Y, Brockie PJ, Mellem JE, Madsen DM, Maricq AV. Neuronal control of locomotion in C. elegans is modified by a dominant mutation in the GLR-1 ionotropic glutamate receptor. Neuron. 1999;24: 347–361. [DOI] [PubMed] [Google Scholar]
- 106.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hoyt JM, Wilson SK, Kasa M, Rise JS, Topalidou I, Ailion M. The SEK-1 p38 MAP Kinase Pathway Modulates Gq Signaling in Caenorhabditis elegans. G3. 2017;7:2979–2989. doi: 10.1534/g3.117.043273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Frøkjær-Jensen C, Davis MW, Ailion M, Jorgensen EM. Improved Mos1-mediated transgenesis in C. elegans. Nat Methods. 2012;9: 117–118. doi: 10.1038/nmeth.1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pereira L, Kratsios P, Serrano-Saiz E, Sheftel H, Mayo AE, Hall DH, et al. A cellular and regulatory map of the cholinergic nervous system of C. elegans. eLife. 2015;4: e12432 doi: 10.7554/eLife.12432 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) The grk-2(gk268) mutant has an egg-laying defect. The graph shows the number of eggs laid by 5 animals in a 2 h period. (**, P<0.01. Error bars = SEM; n = 2 plates of 5 animals each). (B) The grk-2(gk268) mutant animals have short bodies. (***, P<0.001. Error bars = SEM; n = 10).(C) A dop-3 mutation does not suppress the restricted exploration behavior of grk-2 mutants. Shown are images of tracks of five wild-type, grk-2(gk268), and grk-2(gk268); dop-3(vs106) mutant animals that were allowed to explore a bacterial lawn for 2 hours at room temperature.
(TIF)
Shown are plots of bending angle (midpoint) versus time for representative individual animals. The two plots of grk-2(gk268) mutant animals show individuals with strong and weak swimming defects. The dop-3(vs106) mutation suppresses the swimming defects of the grk-2(gk268) mutant.
(TIF)
(A) The arr-1(ok401) mutant has no locomotion defect. (ns, P>0.05. Error bars = SEM; n = 10). (B) The arr-1(ok401) mutation does not suppress the loopy posture of the egl-30(tg26) mutant. (C) The arr-1(ok401) mutation, in contrast to a grk-2(gk268) mutation, does not cause a fainting phenotype in an nca-1(gk9) mutant background. Shown is the percentage of animals that faint when moving backwards. The wild-type, nca-1, grk-2, and grk-2; nca-1 data are the same data shown in S6F Fig. The graph shows the combined data from two independent experiments, each with n = 20–40. (**, P<0.01; ***, P<0.001; ns, P>0.05).
(TIF)
(A) The grk-2(gk268) mutation does not suppress the hyperactive locomotion of the eat-16(tm775) and goa-1(sa734) mutants. (ns, P>0.05. Error bars = SEM; n = 10–20). (B) The grk-2(gk268) mutation suppresses the hyperactive locomotion phenotype of the dgk-1(sy428) mutant. (***, P<0.001. ns, P>0.05. Error bars = SEM; n = 10–20). (C-D) The grk-2(gk268) mutation does not suppress the loopy posture of the eat-16(tm775) and goa-1(sa734) mutants. (ns, P>0.05. Error bars = SEM; n = 5). (E) The kinase-dead GRK-2 does not reverse the grk-2 suppression of the dgk-1 hyperactive locomotion phenotype. Expression of the kinase-dead GRK-2[K220R] mutant under its own promoter (transgene yakEx48) does not reverse the grk-2 suppression of dgk-1 hyperactivity. (ns, P>0.05. Error bars = SEM; n = 10–20). (F) Expression of the grk-2 cDNA under a head acetylcholine neuron promoter (transgene yakEx51) reverses the grk-2 suppression of the hyperactive locomotion of the dgk-1(sy428) mutant. (***, P<0.001. Error bars = SEM; n = 10–20).
(TIF)
The grk-2(gk268) mutation suppresses the loopy posture and hyperactive locomotion of the activated Gq mutant egl-30(tg26) (Gq*). The cat-2(e1112) mutation reverses the grk-2 suppression of the loopy posture (A) and hyperactive locomotion (B) of Gq*. (***, P<0.001. Error bars = SEM; n = 15–20).
(TIF)
(A) The dop-3 suppression of grk-2 is reversed by dop-3 expression in head acetylcholine neurons. The dop-3 cDNA was expressed in the grk-2(gk268); dop-3(vs106) double mutant under a pan-neuronal promoter (Prab-3, transgene yakEx112), acetylcholine neuron promoter (Punc-17, transgene yakEx111), head acetylcholine neuron promoter (Punc-17H, transgene yakEx110) and ventral cord acetylcholine motor neuron promoter (Pacr-2, transgene yakEx109). Expression of dop-3 driven by the pan-neuronal, acetylcholine neuron, and head acetylcholine neuron promoters reversed the dop-3(vs106) mutant suppression of the slow locomotion of grk-2(gk268) mutant animals. (*, P<0.05; **, P<0.01; ***, P<0.001; ns, P>0.05. Error bars = SEM; n = 10–33). (B) A dop-1 mutation does not affect the dop-3 suppression of the grk-2 slow locomotion phenotype. grk-2; dop-3 mutants move more rapidly than the grk-2 mutant. The dop-1(vs100) mutation does not affect grk-2(gk268); dop-3(vs106) locomotion. (ns, P>0.05. Error bars = SEM; n = 23–34). (C-E) Expression of dop-3 in ventral cord motor neurons is not sufficient to reverse the hyperactive locomotion and loopy posture of egl-30(tg26); grk-2; dop-3 mutant animals. (C) Expression of dop-3 driven by the ventral cord neuron promoter (Pacr-2, transgene yakEx109) does not reduce the hyperactivity of egl-30(tg26); grk-2(gk268); dop-3(vs106) mutant animals. (***, P<0.001; ns, P>0.05. Error bars = SEM; n = 10). (D-E) Expression of dop-3 driven by the ventral cord neuron promoter (Pacr-2, transgene yakEx109) does not reverse the loopy waveform of egl-30(tg26); grk-2(gk268); dop-3(vs106) mutant animals. (**, P<0.01; ***, P<0.001; ns, P>0.05. Error bars = SEM; n = 10). (F) A dop-3 mutation suppresses the fainting phenotype of grk-2; nca-1 mutants. Shown is the percentage of animals that faint when moving backwards. The wild-type, nca-1, grk-2, and grk-2; nca-1 data are the same data shown in S3C Fig. The graph shows the combined data from two independent experiments, each with n = 20–40. (**, P<0.01; ***, P<0.001; ns, P>0.05).
(TIF)
(A) DOP-3::GFP expression driven by the grk-2 promoter (transgene yakEx130) reverses the dop-3 mutant suppression of the slow locomotion phenotype of grk-2 mutants. (**, P<0.01; ***, P<0.001. Error bars = SEM; n = 10). (B) DOP-3::GFP levels remain unaffected in grk-2 mutants. Immunoblot of extracts derived from dop-3 or grk-2; dop-3 animals expressing Pgrk-2::DOP-3::GFP and Pmyo-2::mCherry from an extrachromosomal array (transgene yakEx130). The experiment was repeated twice with similar results. (C,D) DOP-3::GFP subcellular localization and level of expression remain unaffected in grk-2 mutants. (C) Representative images of a Z-stack projection of the area around the nerve ring in the head of dop-3 or grk-2; dop-3 mutant animals expressing Pgrk-2::DOP-3::GFP (transgene yakEx130). (D) Quantification of the ratio of DOP-3::GFP to mCherry in the region around the nerve ring of dop-3 or grk-2; dop-3 animals expressing Pgrk-2::DOP-3::GFP and Pmyo-2::mCherry (transgene yakEx130). (ns, P>0.05. Error bars = SEM; n = 10).
(TIF)
(A) The dop-3(vs106) mutation does not suppress the strong forward fainting phenotype of the unc-80(ox330) mutant. (ns, P>0.05. Error bars = SEM; n = 20). (B) The cat-2(e1112) mutation does not suppress the strong forward fainting phenotype of the unc-80(ox330) mutant. (ns, P>0.05. Error bars = SEM; n = 36–38).
(TIF)
(A), (B) grk-2 cDNA expression driven by the (A) sra-11 (Psra-11, transgene yakEx147) or (B) nmr-1 (Pnmr-1, transgene yakEx85) promoter does not rescue the slow locomotion of the grk-2(gk268) mutant. (ns, P>0.05. Error bars = SEM; n = 10–20).
(TIF)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.