Abstract
We have extended our studies of Trp/Trp to other Aryl/Aryl through-space interactions that stabilize hairpins and other small polypeptide folds. Herein we detail the NMR and CD spectroscopic features of these types of interactions. NMR data remains the best diagnostic for characterizing the common T-shape orientation. Designated as an edge-to-face (EtF or FtE) interaction, large ring current shifts are produced at the edge aryl ring hydrogens and, in most cases, large exciton couplets appear in the far UV circular dichroic (CD) spectrum. The preference for the face aryl in FtE clusters is W≫Y≥F (there are some exceptions in the Y/F order); this sequence corresponds to the order of fold stability enhancement and always predicts the amplitude of the lower energy feature of the exciton couplet in the CD spectrum. The CD spectra for FtE W/W, W/Y, Y/W, and Y/Y pairs all include an intense feature at 225–232 nm. An additional couplet feature seen for W/Y, W/F, Y/Y and F/Y clusters, is a negative feature at 197–200 nm. Tyr/Tyr (as well as F/Y and F/F) interactions produce much smaller exciton couplet amplitudes. The Trp-cage fold was employed to search for the CD effects of other Trp/Trp and Trp/Tyr cluster geometries: several were identified. In this account, we provide additional examples of the application of cross-strand aryl/aryl clusters for the design of stable β-sheet models and a scale of fold stability increments associated with all possible FtE Ar/Ar clusters in several structural contexts.
Introduction
The contributions of chiral aryl/aryl interactions, particularly the indole/indole interactions of Trp pairs has been recognized, initially as a complication in deriving protein secondary structure content from the deconvolution of circular dichroic (CD) spectra, since at least 19941,2. Aryl/aryl clusters can also provide favorable enthalpic terms; of these, cross-strand Trp/Trp interactions have been particularly important as a means for creating stable hairpin structures3–5. Although rare in protein β-sheets (likely the result of the rarity of Trp residues in nature), this motif has appeared in numerous β-hairpin designs.3–19
The first utilization of aromatic clusters in hairpin design was attributed to Robinson and co-workers3, in a model of the binding loop of anti-influenza hemagglutinin antibody HC19. The cyclic hairpin that was studied, cyclo-(NHWF-VpPL-WEYS) p = D-Pro, displayed an exciton couplet, with the low energy band at 225 nm, and Hε3 of the Trp designated as WE was nearly 1.6 ppm upfield of a normal position in the NMR spectrum (IUPAC naming of aromatic side chains is shown in Figure 1). The WE designation, which will be used throughout this report, refers to an edge aromatic in a T-shaped, edge-to-face (EtF) aromatic cluster. This is the most common arrangement that produces strong CD exciton couplets and that stabilizes hairpins when flanking a turn. The two Trp’s are in this arrangement in the antibody as well –WE-YSNH-WF–, with the “face” at the C-terminus of the native turn sequence. In the case of hairpins with classic 4-residue β-turns that are aryl-cluster flanked, Ar-turn-Ar, there are examples in which the edge-ring can be either at the N- or C-terminus4,6,8,9 of the turn. Either arrangement can produce the classic spectroscopic indicators of a chiral EtF indole/indole interaction, given in Fig. 2. The factors governing EtF location selectivity, “Why is one arrangement, the face aryl unit at the N- or C-terminus of a turn, preferred?” is dealt with in the present account. The NMR diagnostics listed, upfield ring current shifts at the edge-Trp, have now been observed in numerous peptides with cross-strand Trp/Trp interactions. In addition, it has been established that this feature provides notable stabilization of hairpin folds (5 to >12 kJ/mol) when the Trp/Trp pair is turn flanking5,7,9–11 (S±2 using the nomenclature shown in Fig. 3) or at the terminal non-H-bonded sites in the hairpin β-strands. The latter motif has been extensively explored as a β-capping unit that serves to stabilize β-sheet models and reduce end-fraying in β-hairpins8,12,17,20, and more recently to cap the ends of protein domains21 and prepare highly structured oligomeric peptide assemblies22. Stabilizing EtF W/W interactions have also been observed, but do not always form at other non-H-bonded sites (S±4, S±6, etc.) more remote from the turn. To date, this interaction has not been observed for H-bonding site placements12–15,17. The absence of fixed-geometry Trp/Trp clusters at H-bonded β-strand sites is confirmed in the present study.
Figure 1.
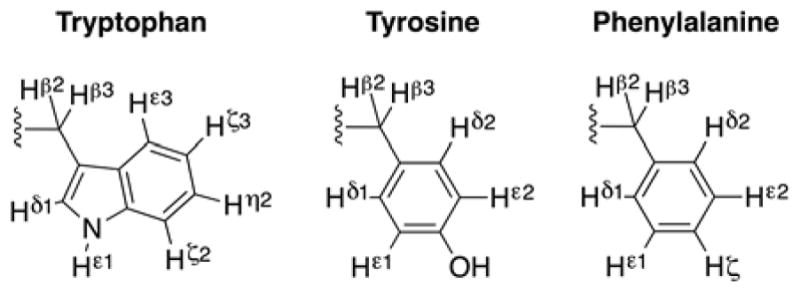
Proton IUPAC naming of aromatic residues.
Fig. 2.
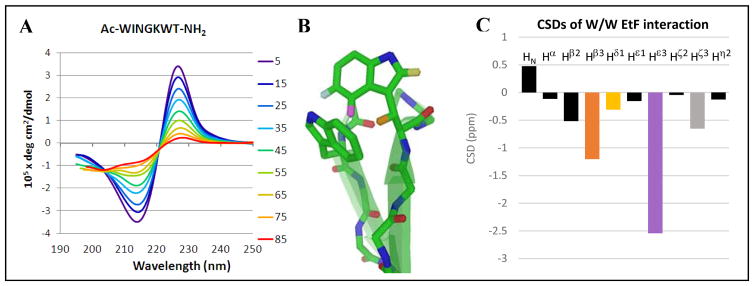
A) The CD melt for a minimized turn-flanking W/W interaction; in this sequence (Ac-WINGKWT-NH2), the bisignate exciton couplet is essentially the only feature evident in the CD spectrum; the fraction folded derived for this peptide motif was χF = 0.76 at the lowest temperature examined. Adjusting to 100% folded, [θ]228 = +460,000, a typical value for this interaction. B) The EtF indole/indole cluster geometry, which gives rise to a 227–229 nm ellipticity maximum in the CD spectrum, also produces upfield ring current shifts at the edge-Trp. Diagnostic protons are color coded and labeled here and in panel C. C) The structuring shifts of the edge-Trp, given as chemical shift deviations (CSDs) from statistical coil norms, typical values (ppm) are: Hβ3 (orange, −1.4 ± 0.45), Hε3 (lavender, −2.25 ± 0.40), Hζ3 (light gray, −0.65 ± 0.15), and Hδ (yellow, −0.45 ± 0.2).
Figure 3.
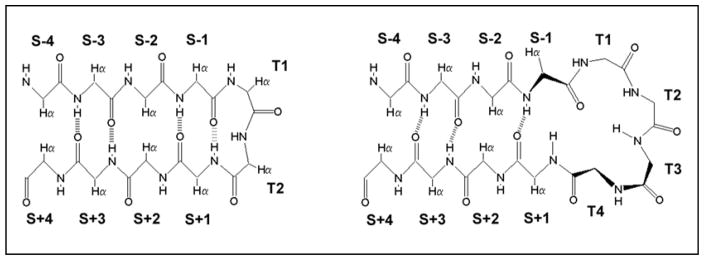
The nomenclature for positions in [2:2]/[2:4] and [4:4]/[4:6] (Thornton23 classification) β hairpins; T indicates turn or loop positions, S indicates strand positions. The [2:2]/[2:4]-hairpins have type I′ or II′ β-turns. The S±even positions have their Hα atoms directed inward and display short Hα/Hα distances; they are designated as “non-H-bonded”. The S± odd positions have their Hα’s outwardly directed and are designated as “H-bonded pairs”.
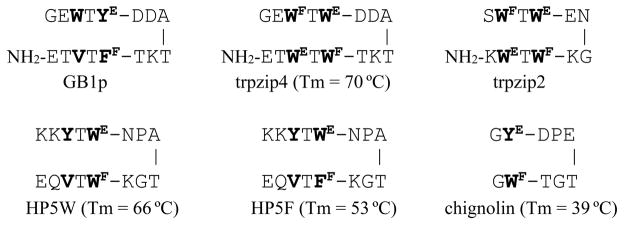
Multiple EtF aryl interactions have also appeared in hairpin designs. The trpzip (TZ) peptides, reported by Cochran4, and other analogs of the key hairpin in the B1 domain of protein G, display two EtF cross-strand pairings as shown below (Fig. 4).
Fig. 4.
Previously reported examples of hairpins with Aryl/Aryl-flanked turns.
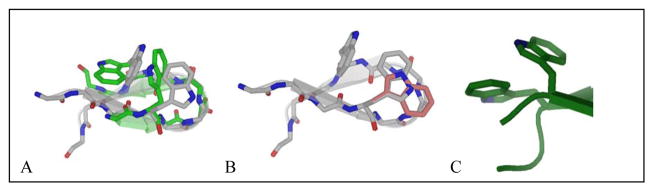
The comparison of the chemical shifts of the edge-Trp in HP5W and HP5F provide a basis for assigning expectation CSDs for an EtF FF/WE interaction. The upfield shift at W5Hβ3 is significantly reduced in HP5F but the Hε3 shift is only slightly decreased: −1.50 ppm in HP5F (versus −2.06 ppm in HP5W).
Peptides that can be viewed as truncated versions of trpzip4 with an optimized [4:6]-hairpin turn5 (e.g. NPATGK), such as HP7 (KTWE-NPATGK-WFTE)7 and even Ac-WE-NPATGK-WF-NH28 retained the edge-to-face W/W interaction with the indole ring at the C-terminus of the turn as the face-species and display the same CD exciton couplet. Peptides HP5W and HP5F5 provide additional examples, and in all cases NMR data indicates that the C-terminal aryl ring is the ‘face’ in an EtF cluster. Subsequent studies have shown that there is a strong preference for the more electron-rich aromatic (W>Y>F) to be the face residue9,12; nonetheless, the opposite situation prevailed in the case of GB1p and HP5F. The N-term–ArE-turn-ArF–C-term feature appears to be a strict requirement in [4:6]-hairpins with the turn flanked by aryl residues. However, when we prepared peptides that can be viewed as a highly truncated version of trpzip2, such as AWF-SNGK-WET, Ac-WF-INGK-WET-NH2 and PWF-IpGK-WE-NH27–9,17, the N-terminal Trp became the face-residue. NMR spectroscopy always provides a clear distinction for the edge-residue: the upfield shifts at an aryl-H (WHε3 or YHδ) and one of the Hβ resonances. The exchange broadening that appears at these sites when the EtF folded state is in equilibrium with the unfolded state that lacks the aryl cluster has emerged as a method for determining the hairpin folding and unfolding rates16. The addition of another residue in front of the N-terminal Trp of one of these peptides, ATW-INGK-WTG, led to a case of EtF confusion: both Trp indole-Hε3 signals were significantly upfield suggesting that both were the edge ring in sampled conformations. Even though an EtF to FtE conformation switch represents only changes in Trp chi angles, the extent of exchange broadening observed indicated a ≥3 μs exchange time, suggesting that a significant degree of hairpin unfolding is required to effect the Trp/Trp geometry flip9.
The other EtF aryl interaction that has figured heavily in the design of stable hairpins is the YE/WF interaction. This was first reported in chignolin (GYDPETGTWG), a very small hairpin peptide reported by Honda et al.24. Chignolin employed a somewhat different optimized loop sequence, but the turn geometry and EtF pairing are completely analogous to that of the other [4:6]-hairpins in Fig. 4. The edge-Tyr ring was readily recognized by the upfield shifts at Hδ (−0.98 ppm) and one of the Hβ’s (−1.26 ppm, based on our coil values and a prochirality re-assignment); we have observed similar, often somewhat larger, ring-current shifts in other hairpin systems by replacing an edge-W in an EtF pair with a tyrosine9,11. Chignolin also displays a CD exciton couplet with a positive low energy band at 228–230 nm (Fig. 4, vide infra).
Chignolin and the trpzips have been the subject of numerous additional studies and the exciton couplets have been a key spectroscopic tool throughout. Trpzip4 and trpzip2 display a very intense couplet with a molar [θ]228 value a bit greater than +106 °. Peptide HP5W (Fig. 4), with only the turn flanking W/W pair, displayed a value almost exactly one-half of that observed for trpzip45. Prior to the present study, we had observed molar [θ]228 values, when corrected to fully-folded state, between +310,000 and +560,000° for hairpin peptides with a single EtF W/W interaction. In the initial trpzip (TZ) study4, TZ4 (=WFWEWFWE) was compared to WFYEFFWE and WWEWFV and the lesser stability of the latter has been noted25 as evidence for a larger hairpin stabilization effect by the W/W interaction near the chain termini. Our studies5,7 imply a greater stabilizing effect of the turn-flanking W/W pair in trpzips although some confusion remains on this point25,26. The Keiderling laboratory has reported the effects of W→V and W→Y mutations in both the TZ2 and TZ1 (SW2TW4EGNKW9TW11K) systems25–27. In their examination of W→V mutations, it was noted that W→V mutations at W4 and W11 (the two “face” rings) resulted in the complete disappearance of the 228 nm maximum and that this peptide did not display the downfield backbone NMR shifts expected for hairpin formation. The two other double W→V mutations, each of which preserve a single cross-strand W/W pairing, displayed a CD exciton couplet that was one-third (rather than one-half) the magnitude observed for TZ2. A computational collaboration28 provided a rationale, predicting a positive CD contribution from the “stack-shifted face-to-face” interaction of the diagonal W2/W9 pairing in TZ species. As expected, an unfavorable turn-replacement can also eliminate CD exciton couplets29.
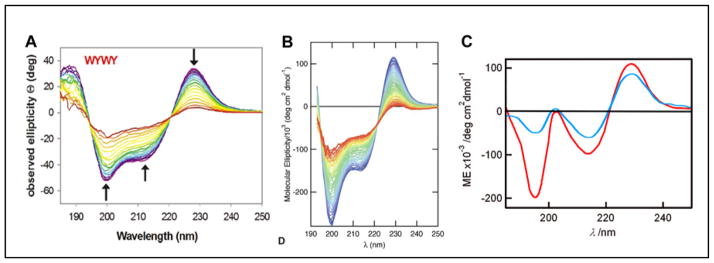
The W→Y mutations25,27 by Keiderling and co-workers provide CD data for comparison with chignolin24 and its more stable analog, CLN025 (YYE-DPETGT-WFY)29. Turning to the W→Y mutations of TZ2, the NMR data reported by Keiderling indicates that all of the TZ edge versus face preferences remain ( -ArFTArE-ENGK-ArFTArE-); however, W/Y pairs were noted as less structure-stabilizing and a cross-strand Y/Y interaction was viewed as even less effective than a V/V interaction with regard to hairpin stabilization. All of the species with at least one WF/WE or WF/YE interaction displayed CD exciton couplets with a low energy maximum at 228 – 230 nm. Fig. 5 provides a comparison of the CD spectra reported for -WFTYE-ENGK-WFTYE-, chignolin and CLN025. In the case of CLN025, quite different CD spectra were reported by the Sandor Lovas group30–32 in an extensive investigation of the effects of organic solvents and chemical denaturants which was supported by MD simulations.
Fig. 5.
A) Keiderling’s WFYE--WFYE-TZ2 -- when converted to molar units rather than residue-molar units, the [θ]229 maximum is circa 260,000°. B) chignolin, C) two CD spectra reported for peptide CLN025 spectra (red trace, the original report29, blue trace that reported by Lovas32).
There is a clear analogy between the chignolin spectrum and that observed by Keiderling for the TZ2 analog with two WF/YE interactions including the appearance of a ~199 nm minimum. This extends to quantitative correlations for [θ]229; the molar ellipticity at this exciton couplet maximum is about twice as large for Keiderling’s WFYE--WFYE construct with two cross-strand WF/YE interactions as that in chignolin. However, the chromophore contributions to these spectra suggested in the original reports are quite different. Since there are only two aryl groups in chignolin, it would seem that all of the features, including the new minimum at 199 nm, are associated with the WF/YE paring. CLN025 is a more stable analog of chignolin with an additional tyrosine added at each end, a YY-turn-WY motif. The CD changes associated with the added tyrosines are exclusively at λ < 210 nm, most notably a maximum at 203 nm. In his discussion of CLN025 CD spectra, Lovas30,32 viewed the extrema at 197 and 203 nm as another exciton couplet, but did not attribute it to a specific interaction. Keiderling noted27 that WF/YE interactions were expected28 to produce a −[θ]195/+[θ]200 exciton couplet feature and indicated that the dominant −[θ]212/+[θ]228 feature of WFYE--WFYE-TZ2 was a W/W exciton couplet, presumably associated with the diagonal stack-shifted face-to-face interaction of the W2 and W9.
The NMR data reported for both chignolin, CLN025 and WFYE--WFYE-TZ2 suggest that the same WF/YE interaction is present in all species. This conclusion is supported by ring current shifts in the case of chignolin24. In the case of CLN025 the NMR assignment does not appear to have been published, but a rich web of NOE between Y2 and W9 is shown and there were no Y1/W9- or Y1/Y10-sidechain NOEs listed29. Lovas has suggested that there are other aryl/aryl interactions besides the Y2/W9 cluster contributing to CD spectral changes. We have suggested that the enhanced stability of CLN025 reflects a generic hydrophobic interaction between the added tyrosines (Y1,10) with no fixed geometry preferred16. Herein, we rely on ring current shifts as the primary indicator of fixed geometry aryl cluster formation.
Clearly a number of questions remain, particularly regarding the CD signatures expected for different Ar/Ar clusters. In the present study we have taken a strictly empirical approach to: 1) elucidate the spectral signatures of W/W, Y/W, F/W and Y/Y clusters in classic EtF geometries as well as other geometric arrangements, 2) define the features that lead to the preference of one EtF pairing versus the alternative geometry, and 3) quantitate the fold stabilizations that result from different EtF aryl/aryl cluster placement in hairpins and other β-sheet models.
Materials and Peptide Synthesis
All of the peptides prepared or examined in this study (Table S1) were constructed using FMOC chemistry on an ABI 433A or Liberty Blue peptide synthesizer and purified by reverse-phase HPLC (C18 column) with a water (0.1% TFA)/acetonitrile (0.085% TFA) gradient. All peptides displayed the expected (M+2H)+2 and/or (M+3H)+3 mass spectral ions on a Bruker Esquire ion trap mass spectrometer. In some cases, an additional HPLC purification using a C8 column was required to obtain peptide samples meeting our purity criteria by NMR analysis. All of the sequences were confirmed by observing the expected vicinal residue connectivities in 2D NOESY or ROESY spectra.
Buffer components (ACS reagent grade, Fisher Scientific) were dissolved in deionized, 0.2 μm-filtered water (NANOpure Diamond water system, Barnstead). D2O (99.95%-D) for NMR spectroscopy was obtained from Cambridge Isotope Laboratories, Inc.
CD Spectroscopy
CD stock solutions were prepared by dissolving weighed amounts of peptides in 20 mM aqueous pH 6 potassium phosphate buffer to make a 100 – 500 μM stock solutions. The concentration of stock solutions was measured by the combined expectation UV absorption of Trp (ε = 5690 M−1 cm−1 per Trp) and Tyr (ε = 1280 M−1 cm−1 per Tyr) at 280 nm. CD samples were diluted into buffers to obtain 15 – 30 μM peptide solutions. Spectra were recorded on a Jasco J720 spectropolarimeter using 0.10 cm pathlength cells. The calibration of the wavelength and degree ellipticity scales and spectral accumulation parameters as well as the method for CD melting studies have been described previously.5,7,9,12 Although peptide CD data is typically reported in residue-molar ellipticity units (deg cm2 residue–dmol−1), we employ molar units (deg cm2 dmol−1): the dominant feature in most of the CD spectra is due to a single chromophore pair in the peptide rather than the backbone amide units. This dominant feature is an exciton couplet appearing as a maximum at 227–229 nm and minima at circa 213 nm and, in some instances also near 198 nm.
The variable overlap of the 214–216 nm minimum due to the alignment of β strands with the circa 213 minima of the exciton couplet makes it difficult to evaluate the amplitude of the exciton couplet. This, together with the near absence of secondary-structure CD contributions at 227–229 nm, suggested the use of the couplet maximum as the fold measure.
NMR Spectroscopy
NMR spectra were recorded on 500, 700 and 750 MHz NMR instruments with Bruker consoles. All 2D spectral experiments employed a WATERGATE33 pulse train for solvent suppression. TOCSY spectra employed a 60 ms MLEV-17 spinlock34 and NOESY spectra had a 150 ms mixing time. Samples contained 1 – 2 mM peptide in buffered H2O with 10% D2O. Sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) was used as an internal chemical shift reference and was set to 0 ppm for all temperature and pH conditions. The buffers employed were as follows: pH 6 (20 mM sodium phosphate for the HP5W4 structure ensemble calculation), otherwise pH 6 (50 mM sodium phosphate) and pH 3 (20mM formate/formic acid).
With the exception of resonances that display CSDs in excess of 1 ppm, all 1H-C resonances were sharp singlets or multiplets implying linewidths (Δ0 < 2.4 Hz) consistent with the rapid tumbling times expected for monomeric species in the 1000 – 2100 amu molecular weight range. Sites with large folded-state CSDs display differential exchange broadening that scales as (CSD)2 consistent with folded/unfolded state relaxation phenomena typically on the 0.5 – 10 μsec timescale but there were some slower folding systems approaching a 70 μsec timescale.16,21,35
Chemical Shift Deviation (CSD) and Fraction fold (χF) calculations
Chemical shift deviations (CSDs) were calculated by subtracting the random coil value from the observed chemical shift. Using these, fraction folded (χF) values were calculated by dividing the observed CSD by the known CSD at 100% folded for multiple diagnostic 1H probes in the peptide. The probes used for each calculation can be found in the cited reference where the system was introduced or in the supporting information for completely new systems (S2–S4). Fraction folded estimates made from CD data was based on the fraction of the projected maxima of the [θ]228 at 100% folded.
A full list of the peptides synthesized and referenced can be found in the supporting information, along with a unique identifying name (“CSDb Identifyer”) that can be used to search for the complete NMR chemical shift data within an in-house open access database. (Available online at: http://andersenlab.chem.washington.edu/CSDb/) Peptides being described for the first time are marked as “new”, peptides reported previously are referenced in the right column. The data for each peptide includes a complete assignment as well as how the chemical shifts change with temperature in the 280 – 320 K range. The graphical features allow these to be plotted as sequence-position/CSD histograms.
Results & Discussion
We have previously reported the systematic replacement of S±2, S±3 and S±4 cross-strand pairs in hairpins with the KKLTVS-IXGK-KITVSA sequence (X= Asn and/or D-Pro) by Trp and Tyr residues9,13,17; however, complete structural characterizations of the resulting species were not given at that time. Hairpin stabilization was modest with a S±3 Y/Y13,15 or W/W17 pairing; with notable increases in the Tm and extent of fold formation at 280K with an S±4 (ΔΔGU ≈ 4 kJ/mol) and particularly an S±2 (ΔΔGU ≥ 6 kJ/mol) W/W pair. The CD diagnostics of EtF cluster formation were observed only with the S±2 W/W pairing in which the Trp flanking the C-terminus the turn was the face residue. The absence of these diagnostics for S±3 W/W pairs has been confirmed in another modestly stable hairpin, RWV-NPATGK-IWE (vide infra). The S±4 W/W pairing, although stabilizing, does not always produce a specific EtF cluster; this has now been confirmed in a stable hairpin with a RAVWTV-NPATGK-ITWVIE sequence20. Additional data on this and related systems appears in a later section.
With regard to S±2 W/W pairings, even these do not always result in a complete set of EtF cluster formation spectral diagnostics. For example, in the case of a hairpin stabilizing N → D-Pro mutation in the turn of the peptide with a KKLTVW-INGK-WITVSA sequence, while the hairpin backbone CSDs indicated a higher hairpin fold population, a comparable increase was not observed for the exciton couplet magnitude. A loss of CD exciton couplet amplitude has also been reported25 for an N → D-Pro mutation in TZ2 and was also evident in another early trpzip analog with a VpGK turn6.
Upon adjusting the medium to 20 vol-% hexafluoro-isopropanol (HFIP), the CD exciton couplet of KKLTVW-IpGK-WITVSA completely disappeared even though the hairpin fold was more stable based on the CSDs observed for HN and Hα sites in the β strands. The NMR spectral features revealed a new EtF geometry. This geometry is compared to the typical one for a turn flanking W/W pair in Figure 6 (panel B). The differences results from a change in χ2 at the edge-Trp: Hδ has a 1.8 ppm upfield shift (versus 0.4 ppm at Hε3) and the turn Gly amide hydrogen appears 3.5 ppm upfield of its random coil value.
Fig. 6.
Edge-to-face indole/indole clusters in designed hairpins: A) turn flanking W/W units showing examples in which the indole at the N-terminus takes on, alternatively, edge and face conformation in the edge-to-face cluster8,9, B) The W/W cluster of KKLTVW-IpGK-WITVSA in aqueous HFIP medium with a circa 180° flip in chi29,11 of the edge-Trp in the gray cluster of panel A, and C) the β-cap arrangement of hairpins with a C-terminal WTG unit in which this Trp bears the edge indole of the EtF cluster.
The dramatic upfield shift of the amide HN in the turn reflects an H-bond with the indole π-system serving as the Lewis base, the upfield (rather than downfield H-bonding shift) results due to the shielding ring current effect of the facing aromatic ring. As it turns out, this is an important auxiliary Trp interaction that can serve to enhance its propensity to become the edge-indole in an EtF pairing. It is also a key feature of the W-strand-loop-strand-WTG β-capping motif12 which routinely display > 3 ppm upfield shifts at the Gly-NH. It was first observed8,9 in Ac-WINGKWT-NH2, in which the terminal amide shifts are 6.84 and 4.42 ppm, that of far upfield entgegen-NH corresponding to CSD ≈ −2.7 ppm, reflecting the indole ring-current effect. Auxiliary interactions can also enhance face preferences. Of these cation-π interactions are the most notable, with the sidechain of an Arg or Lys as the cationic site.7,16,20,36
Aryl clusters are not limited to hairpins. Indeed most of the ones found in proteins in the early CD studies1,2 were not within β-sheet or β–hairpin structures. Such alternative placements have, however, never been examined in any detail.
Aryl clusters within Trp-cage miniproteins
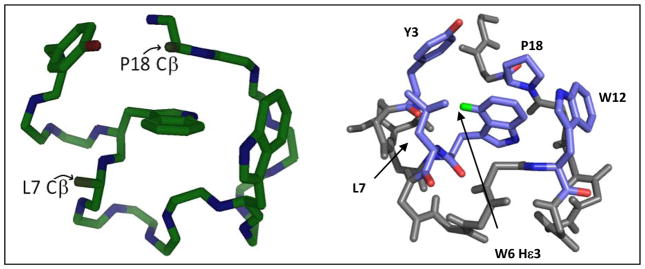
We turned to the Trp-cage miniprotein fold37–40 in our first search for additional W/W and Y/W clusters and their CD spectral contributions. The common, rigid fold of sequence-optimized, e.g. DAYAQ WLADX GPASX RPPPS, X = Gly (TC13b) or D-Ala (TC16b), Trp-cage species has been established by numerous NMR structures and two crystal structures41 and is stable enough to allow substitutions at L7, P12 and P18, as well as swapping Y3 for a Phe or Trp residue. This allowed us to examine the effects of potential W/W as well as Y/W cluster geometries upon the CD spectrum of the Trp-cage. Of the mutations examined, only one, the P12W mutation38,42,43, provides added fold stability. The structural features of one such mutant (the X = D-Ala species) are shown in Figure 7.
Fig. 7.
The [P12W]-TC16b structure determined by NMR40 showing the backbone and the three aryl residues. The Cβ locations at P18 and L7 are shown to indicate the position that would be taken up by added indole rings in P18W and L7W mutants. The right panel clearly shows the hydrophobic core, with W6Hε3 labeled in green. The distance between Y3 and W6 is ~7 Å.
The P12W mutation introduces an indole ring that serves as the face of EtF indole/indole interaction of a geometry that hadn’t been observed in hairpin constructs. Whether this feature, or just an added degree of apolar surface burial, is the basis for the circa 3 kJ/mol fold stability increase associated P12W mutations cannot be determined with certainty, but the interaction is clearly evidenced by additional ring-current shifts. The Hζ2 and Hη2 sites of W6 move 1.7 and 1.2 ppm upfield, respectively.
The structure in Fig. 7 shows the Y3/W6 cluster of the Trp-cage. This Y3/W6 geometry produces upfield shifts, notably a 0.5 ppm shift at W6Hε3, the same site that experiences large shifts in hairpins with cross-strand W/W pairs. The Y/W cluster in the Trp-cage is intermediate between a displaced-stacked and EtF geometry with the Tyr as the face. The upfield shift of W6Hε3 shift is retained with both Y3F and Y3W mutations indicating that the Ar/Ar cluster geometry remains the same throughout.
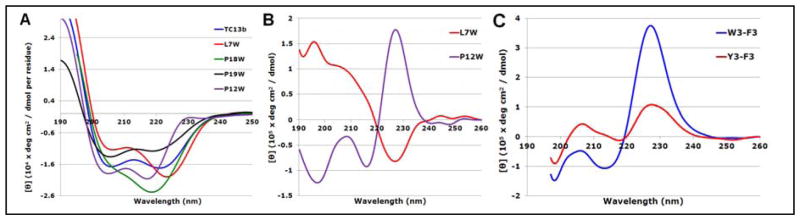
The fixed geometry of Trp-cages makes this an ideal system for exploring the CD spectral contributions of Ar/Ar clusters. Trp-cage species display a helical CD signature (panel A, Fig. 8), with the specific Ar/Ar cluster contributions best visualized in difference spectra (panels B and C). The spectra in panel A were recorded for the X = Gly species (TC13b), with Trp units introduced in place of L7, P12, P18 and P19. Each Trp introduction led to significant changes in the CD spectrum. The P19W mutation was particularly fold-destabilizing (Fig. 8, panel A) and no useful data could be gained regarding possible interactions between W19 and the other aryl units. The P18W mutation also failed to show a defined exciton couplet by CD differencing. As can be seen by examining the CD traces in panel A, the major effect of introducing a Trp at site 18 was an increased negative ellipticity throughout the 208–227 nm range (with a 217 nm extremum) with no evidence of a 228 – 230 nm feature that would indicate either a W/W or W/Y exciton couplet. This feature likely represents an “indole ring in a chiral environment” rather than a Trp contribution that is due to a specific Ar/Ar chromophore interaction; The P12W- and L7W-mutations resulted in exciton couplet difference spectra (Fig. 8, panel B).
Fig. 8.
A) The CD traces for the TC13b and its L7W, P12W, P18W and P19W mutants at 280 K, B) difference CDs generated from panel A, C) the TC16b minus [Y3F]-TC16b and [Y3W]-TC minus [Y3F]-TC spectra.
The difference spectrum for [P12W]-TC13b revealed a typical W/W exciton couplet with an amplitude that was about one-third of that observed in hairpins and β-capping units. As previously noted, the effect of the facing W12 unit on W6 was evidenced by upfield shifts at W6Hζ2 and Hη2. The [P12Y]-mutant of TC13b was examined. This mutant also displayed upfield shifts at W6Hζ2 and Hη2 (1.26 and 1.06 ppm, respectively). These were similar to those of the [P12W]-mutant, but difference CDs failed to show a significant exciton couplet (data not shown), suggesting that this EtF W/Y geometry results in much smaller CD values than the corresponding W/W cluster. The L7W-mutant provided our first observation of an exciton couplet with negative ellipticities at the 228–9 nm extremum.
The contributions of the Y3/W6 interaction in Trp-cage species to the CD spectrum was examined by CD difference as well, using mutations at Y3 of TC16b. The [Y3F]-mutants of TC16b confirmed the contribution of an edge-Tyr to face-Trp interaction to net CD spectrum of Trp-cages which had previously been postulated39. The value of the [θ]229 component (circa +100,000°) evident in the (TC16b minus Y3F-mutant)-difference spectrum (panel C) was similar to that seen for the face-Trp/edge-Tyr exciton couplets shown in Fig. 5. In the case of the W/W interaction in the [Y3W]-mutant, differencing versus the [Y3F]-mutant reveals a larger exciton couplet comparable to those we have observed for W/W interactions in hairpins. In both panels B and C of Figure 8, the difference spectra reveal a 197–199 nm minimum which could be coupled to the ca. 206 nm maximum or to a feature at smaller wavelengths. This is also observed in hairpin systems with W/Y and Y/Y exciton couplet CD spectra (vide infra).
W/W pairs in a turnless β-sheet model
In this section, we highlight key observations from studies in which we have added W/W pairs to β-sheet mimics. Here, we begin with studies aimed at finding improved alternatives to the Ac-W//WTG β-cap which inform many of the studies to follow, some of these were published recently20. In our announcement12 of the β-cap, we included the disulfide form of CH3CH2CO-WTTVCIRKWTGPK-NH2 as an example of a “turnless” β-sheet model (Figure 9) in which the N-terminal alkanoyl unit was essential for fold stability. This model had a χF = 0.985 at 280K based on amide NH exchange protection factors, which was reduced to χF = 0.72 upon N-terminal de-acylation. The N-propanoyl species displayed the usual diagnostics for an EtF W/W interaction with the N-terminal Trp as the face species: W9Hε3 was 2.01 ppm upfield. Since it would be difficult to quantitate fold stability improvements in this system, we turned to a somewhat less stable disulfide system, (XWTTHCHRKWZ)2, to examine alternatives, X and Z, to the acyl and TG-NH2 units, respectively20. The formation of EtF W/W clusters at the end of each strand results in a single orientation of the strands in the disulfide dimer and attractive Coulombic interactions between the chain terminal –NH3+ and –CO2− groups. In the absence of Z=TG-NH2, the best choices for X were residues with a positively charged sidechain, Lys and Arg. The retention of diagnostic structuring shifts (CSDs) at moderate temperatures was the best measure of relative fold stability. Some of the data collected for (XWTTHCHRKWZ)2 with (X = Ac, Z = WTG-NH2), (X = K, Z = A, E), and (X = R, Z = E) are gathered in Table 1. The T3HN, R8Hα and K9HN CSDs report on the extent of hairpin formation17,44, the W10Hε3 shift is included since it reports the extent to which this residue’s aryl ring is held rigidly in the edge-position of an EtF cluster.
Figure 9.
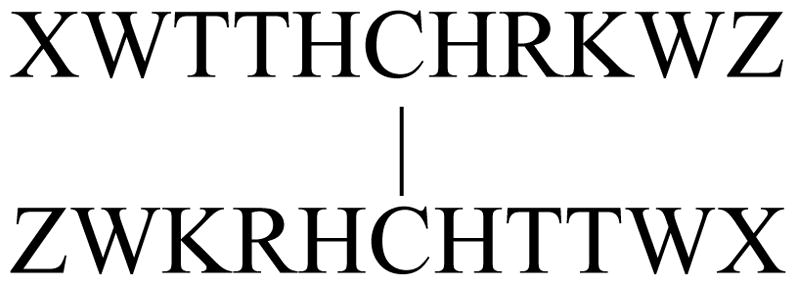
“Turnless” b-Sheet model
Table 1.
CSDs observed for (Ac-WTTHCHRKWTG)2 and (XWTTHCHRKWZ)2 at pH 8.
| χF | CSDs (ppm) | ||||||
|---|---|---|---|---|---|---|---|
| Sequence | T (K) | pH 2 | pH 8 | T3HN | R8Hα | K9HN | W10Hε3 |
| (Ac-WTTHCHRKWTG-NH2)2 | 280 | 0.39 | 0.83 | 1.229 | 0.981 | 0.986 | −1.677 |
| 310 | -- | 0.56 | 0.992 | 0.545 | 0.810 | −0.969 | |
| (KWTTHCHRKWA)2 | 280 | 0.60 | 0.93 | 1.240 | 1.123 | 1.014 | −2.329 |
| 310 | 0.20 | 0.69 | 0.89 | 0.81 | 0.71 | −1.521 | |
| (KWTTHCHRKWE)2 | 280 | 0.61 | 0.98 | 1.446 | 1.15 | 1.057 | −2.445 |
| 310 | 0.36 | 0.88 | 1.26 | 0.945 | 0.89 | −1.993 | |
| (RWTTHCHRKWE)2 | 280 | 0.73 | 0.99 | 1.463 | 1.15 | 1.052 | −2.462 |
| 310 | 0.46 | 0.92 | 1.393 | 1.013 | 0.95 | −2.158 | |
Inspection of Table 1 shows that the fold population estimates for the Ac-W/WTG-NH2 and the KW/WA species range from 0.61 – 0.93 at 280K depending on the probe selected for comparison. In contrast, the KW/WE and RW/WE species, with an additional Coulombic attraction in the cap, have fold populations χF ≥ 0.98 at 280 K. The RW/WE cap displays greatly enhanced thermal stability; the extent of additional fold stability associated with the K→R mutation is also evident when the peptides are examined at pH 2, KW/WE χF = 0.61 versus RW/WE χF = 0.73 at 280 K, with both versions being far superior to the Ac-W/WTG-NH2 cap (χF ≤ 0.39) at this acidic pH. At pH 2, all of the Coulombic attractive interactions between the termini are abolished; there are clearly other interactions that reinforce the W/W interactions in these β-caps. Based on this study, we have employed RW/WE (as well as KW/WE) caps in the remaining studies reported herein; we have also used RW/WD21 and HW/WE caps in other successful β-sheet construct designs. Henceforth, we refer to these as ‘Coulombic β-caps’ or ‘hydrophilic β-caps’ with the X-W/WTG- units called ‘hydrophobic β-caps’ to distinguish them.
Studies inspired by hairpins in WW domains provide additional examples of fold stabilization by RW/WD β-caps. While the N-terminal hairpin of the Pin1 WW domain is known to be the fold-nucleating feature of this protein motif45, we recently established21, that this peptide sequence (GWEKRM-SRSSGR-VYYFNH) does not form a stable hairpin fold outside of the protein context. (Peptide R-WEKRM-SRSSGR-VYYFN-S, with termini that should provide more support for fold stabilization displays a fold fraction less than 0.2 at 280K21.) As another example of β-capping effects, we prepared RWEKRM-SRSSGR-VYYFWD; this modification brought the hairpin population from < 0.28 to 0.90 (ΔΔGU ≥ 7 kJ/mol at 280K)20. We were also able21 to make a stable version of this hairpin by including tryptophans flanking a somewhat improved turn sequence, RWEKRW-DRGSGR-WFYFND (Tm = 60 °C, 1/kF = 67 μs at 300K). These changes set the stage for a circular permutation of the hairpin sequence; the permuted sequence, RW-FYFN-DRGSGR-WEKR-WD which has an RW/WD β-cap, proved to be faster folding (1/kF = 39 μs) and nearly as stable as the Trp-flanked-turn version. The RW/WD β-cap species displayed the expected EtF interaction (W17Hε3 upfield by 2.23 ppm) and exciton couplet ([θ]228=+340,000, Tm = 37 °C).
W/W pairs in a three stranded β-sheet model
We have also added cross-strand W/W interactions to other β-sheet models; the Schenk-Gellman43 double hairpin (Ac-VFIT-SpGK-TYTE-VpGO-KILQ-NH2), a three-stranded sheet provides a number of cases. Our experimental studies of this model, which included D-Pro → Asn and L-Pro mutations at each and both of the turn loci44,46 as well as computational studies47 have revealed that the second hairpin is more stable than the first and that folding is a multistate equilibrium that includes single hairpin species. Strand site CSDs report on the “net folding” in each strand and as such can be converted to a “net χF” for each hairpin. We chose the less stable N-terminal hairpin (HP1) as the site for cross-strand introduction of Trp pairs: [F2W,T11W]- and [T4W,T9W]-mutations corresponding to S±4 and S±2 pairs, respectively. Both placements produced systems in which HP1 became the more stable of the two hairpins and resulted in well-defined EtF clusters. The NMR data clearly showed 2WF/11WE and 4WE/9WF clusters, analogous to those observed in TZ peptides (see Table 2). The hairpin-stabilization associated with the added cross-strand Trp/Trp interactions in HP1 (Table 3) reduced the extent of cooperativity in N-terminal hairpin formation associated with turn locus mutations - L-Pro-Gly to NG to D-Pro-Gly (pG) – at the second turn locus. Indeed, the first hairpin populations were still quite high even with an L-Pro-Gly replacing the second turn precluding double hairpin formation. In this series, the stabilization afforded by these cross-strand Trp/Trp placements was 6.4 and 8.3 kJ/mol (for the [T4W,T9W]- and [F2W,T11W]-changes, respectively).
Table 2.
Trp/Trp mutants of the Schenk-Gellman double hairpin: ring current shifts, CD exciton couplet amplitudes, and fold populations (including those for unmodified reference compounds).
| Turn1/2 | HP1 | HP2 | S±4 W’s | S±2 W’s | ||||
|---|---|---|---|---|---|---|---|---|
| χF(1) | χF(2) | χF (1) | W11Hε3b | [θ]228 c | χF (1) | W4Hε3b | [θ]228 c | |
| pG/pG | 0.718 | 0.811 | 0.92 | −2.03 | +360 | ≥0.96 | −1.55 | +309 |
| pG/NG | 0.668 | 0.563 | ||||||
| pG/PG | 0.557 | -- | ||||||
| NG/pG | 0.365 | 0.682 | 0.84 | −1.94 | +330 | 0.90 | −1.70 | +285 |
| NG/NG | 0.247 | 0.461 | 0.85 | −1.68 | n. d. | |||
| NG/PG | 0.104 | -- | 0.80 | −2.05 | +370 | 0.73 | −1.73 | +120 |
| T4A | ||||||||
| pG/pG | n.d. | 0.82 | −1.83 | +440 | ||||
| pG/PG | n.d. | 0.65 | −1.91 | +470 | ||||
Fold population are given for each hairpin that can form, χF(1) and χF(2). The data for the species lacking Trp substitution are taken from Hudson46.
The CSD of Hε3 of the edge indole at 280K is given in ppm. These were not corrected so as to represent the values at 100% folded. The largest upfield shift observed for any proton sites on the other indole ring was 0.27 ppm.
Ellipticity values are reported in 10−3 molar ellipticity units.
Table 3.
Effects of Moving the W/W Cap to Different Distances from the Turn.
| Spectroscopic diagnostics at 280K (corrected to χF = 1) |
||||||
|---|---|---|---|---|---|---|
| W/W position | Turn Type | Sequence | χF (300K) | Hε3 CSD | [θ]228 CD max. | |
| 1 | S±2 | 2:4 | KKLTVW-INGK-WITVSA | 0.85 | − 2.08 | +264,000 |
| 2 | S±4 | 2:4 | KKLWVS-INGK-KIWVSA | 0.49 | − 0.55 & − 0.53 | +88,000 |
| 3 | S±4 | 2:4 | KWVS-INGK-KIWE | 0.91 | − 2.46 | +596,000 |
| 4 | S±6 | 2:4 | KWLTVS-INGK-KITVWE | 0.95 | − 2.66 | +342,000 |
| 5 | -- | 2:4 | KKLTVS-INGK-KITVSA | 0.30 | n.a. | n.a. |
| 6 | S±4 | 4:6 | RWTV-NPATGK-ITWE | 0.83 | − 2.40 | +387,000 |
| 7 | S±4 | 4:6 | RAVWTV-NPATGK-ITWVIE | 0.34 | − 0.30 & − 0.62 | +395,000 |
| 8 | S±3 | 4:6 | RWV-NPATGK-IWE | < 0.61 | − 0.52 at W11 | +35,000 |
| 9 | S±2 | 4:6 | RVW-NPATGK-WIE | 0.97 | − 2.40 | +297,000 |
| 10 | S±2 | 3:5 | KKLTVW-SADGR-WITVSA | 0.78 | − 2.07 | + 282,000 |
Turning to the cluster geometries observed in this series, the 4WE/9WF geometry observed with the S±2 W/W placement accords with prior observations for all turn-flanking W/W pairs in hairpins with at least 3 strand residues. The well-defined 2WF/11WE geometry and greater stabilization associated with the S±4 W/W pairing was, however, surprising. As previously noted, and recently confirmed20, strand-internal S±4 W/W pairings are much less stabilizing and do not necessarily display a specific EtF cluster preference. The observed 2WF/11WE geometry is that expected for a β-capping interaction. Indeed, there is no evidence for two cross-strand H-bonds between E12 and V1 in the Ac-VWIT-SXGK-TYWE-VXGO- sequence. The proximate VXGO turn appears to preclude a fully “beta” conformation in the E-V unit; W11 is effectively the C-terminal NHB strand site.
A more critical analysis of the spectroscopic diagnostics for the EtF indole/indole clusters in these three-stranded sheet analogs provides further insights and also raises some questions. The characteristic upfield shift of the edge WHε3 and [θ]228 CD maximum are more evident for the S±2 W/W placement when the second turn locus, VXGO, is rigid (X = D-Pro). At first this suggested a cooperative role for 3rd strand association. However, this trend was still observed in species with C-terminal truncations that completely removed the 3rd strand (data not shown). Indeed, the truncated species showed larger W4Hε3 shifts suggesting that there are steric features of the fully-formed double hairpin motif that are not optimal for, and thus can interfere with, the formation of the preferred EtF W/W geometry.
Both the ring current shifts and the magnitude of the [θ]228 exciton couplet maximum were larger in the S±4 series. The extent of hairpin stabilization was also greater, particularly when the second turn was disabled by mutating it to a PG locus that does not allow three-stranded sheet formation. The S±4 W/W series was stabilized to such an extent that a [T4A]-mutation was tolerated with only a modest loss in hairpin stability. In this series, particularly with the second hairpin disabled by an L-Pro insertion in its turn, both the ring current shift for W11Hε3 and the exciton couplet amplitude increased, with [θ]228-values as large as any observed for prototypic β-caps.
Optimization of Cross-strand Aryl/Aryl Interactions in Hairpins
The effects of moving a cross-strand W/W interaction to different locations relative to the turn locus and strand ends was studied in a new series of hairpin models (Table 3). It includes examples of adding, and moving the location of, a cross-strand W/W pair and another instance of a β-cap at an S±6 location (Table 3, entry 4). It is clear that the S±6 location provides as much, if not more, hairpin stabilization than the turn-flanking position (entry 1). The peptide with an S±4 placement in the 6-residue strands (entry 2) fails to impart significant fold stabilization and does not afford an EtF indole/indole interaction. A strand truncation (entry 3), makes this S±4 location a terminal β-cap and increases the fold population even though there are fewer cross-strand H-bonds.
These results were, with the examples in entries 6 through 9, extended to hairpins with a [4:6]-turn. The comparison of the peptides of entries 6 and 7 illustrates yet another failure to form a stabilizing EtF W/W pair with an S±4 pairing in longer strands. The entry 6 to 7 change corresponds to adding two residues to each β-strand, RWV→RAVWTV and IWE→IWVIE, even though the added strands have high β-propensity, actually decreases the hairpin fold population from 0.83 to 0.34 at 27°C. This series also provided a dramatic example of the total failure of S±3 pairings (H-bonded sites) to provide either hairpin stabilization or an EtF cluster (see entry 8). Entry 10 establishes that the same EtF preference applies to Trp’s flanking a [3:5]-turn.
Further Exploration of GB1p and Trpzip analogs with a [4:6]-hairpin configuration
In 2004, we reported5 a [θ]228-value for KKWTWE-NPATGK-WFTWQE (HP5W4) that was only 60% of that observed for trpzip4 and attributed that to the lack of well-formed terminal (W3/W14) indole/indole cluster. We have now prepared analogs of this system in order to: a) prepare a construct with two EtF W/W interactions but lacking the diagonal face-to-face W/W interaction which Keiderling viewed27,28 as an important contributor to the −[θ]212/+[θ]228 exciton couplet amplitude, b) examine the effect of introducing a better β-cap, and c) compare the fold-stabilizing effects of turn-flanking versus β-capping W/W interactions.
When we prepared RWTW-NPATGK-WTWE ([θ]228 = +972,000°) the full expectation spectrum for two EtF interactions appeared. We attribute this increase in the CD exciton couplet to the superior capping associated with the RW/WE capping unit rather than a contribution from a W2/W11 (WF/WF) interaction which would be a displaced stacked face/face interaction (DSFF). A definitive answer to question of whether a DSFF interaction can yield CD couplets required an examination of three additional peptides. The key one is RWATVW-NPATGK-WITVWE, which we view as a trpzip/HP5W4 analog without the possibility of a DSFF contribution; it displays a large exciton couplet: [θ]228 = +888,000°. This is nearly as large as that of RWTW-NPATGK-WTWE and trpzip4. The individual contributions of the two clusters was gauged from RWVTVW-NPATGK-AITVWE ([θ]228 +453,000°) and RAVTVW-NPATGK-WITVWE ([θ]228 +362,000°). The Trp to Ala mutations in peptide RWATVW-NPATGK-WITVWE were more deleterious when the mutation abolishes the β-capping Trp/Trp pair. This data, and the variability in spectral diagnostics for turn-flanking W/W cluster, leads us to suggest that aryl clusters in the end-capping position are generally preferred over turn-flanking motifs in β-hairpin design.
Returning to the question of possible CD contributions of DSFF W/W interactions, the specific couplet amplitudes observed for the full series of analogs, particularly the large amplitude observed for RWATVW-NPATGK-WITVWE, leads us to conclude that DSFF W/W interactions do not result in a significant [θ]228 feature. As a result, we view the CD spectrum of WFYE--WFYE-TZ2 in Fig. 5A as exclusively the result of the cross-strand WF/YE interactions. Additional examples of single WF/YE interactions which result in spectra equivalent to those in Fig. 4A appear in the Discussion section.
At this point, it is clear that W/W indole clusters at non-H-bonded positions when placed adjacent to the turn or near the strand termini provide significant hairpin stabilization. The one apparent literature exception to this, a W/W insertion in the first hairpin of the Pin1 WW domain, which resulted in only a 2.3 kJ/mol folding increment due to the W/W interaction48, now stands in contrast to numerous studies of the hairpins outside of the protein context (vide supra). In the isolated hairpins, such W/W interactions afford at least 5.6 kJ/mol of fold stabilization. A lesser contribution to β-sheet stability may apply in larger β sheets and when the W/W pair is at a non-H-bonded position more centrally-located in the β-strands. In some contrast, we have already suggested8,12,20,21 that W/W interactions in β-caps can provide as much as 11 kJ/mol of fold stabilization.
Hairpin stabilization effects of different Ar groups within EtF Ar/Ar clusters
We have previously reported some data on hairpin structures with W/Y, W/F, and Y/F cross-strand pairs.9,12 For Ar/Ar placements without a strong inherent preference for the edge-Ar ring, changes in an EtF⇔FtE equilibrium position have been observed based on another intrinsic preference: the superiority of Trp over Tyr as the electron-rich face ring in an Ar/Ar cluster. For example, peptides KAVWE-INGK-WFTVE and KAVYE-INGK-WFTVE show exclusively the cluster geometry indicated; however, there is a 1.5 kJ/mole drop in stability for WE/WF versus YE/WF.9 But KAVW-INGK-YTVE, which is even less stable (an additional 3.2 kJ/mol stability loss), displays upfield shifts at both YHδ and WHε3. We interpret this as a Trp-face preference overwhelming the EtF preference associated with a turn-flanking aryl pair.
Makwana and Mahalakshimi49,50 have reported on a Ac-L-ArF-VpGL-ArE-V-NH2 peptide series with all possible combinations of F, Y and W at the Ar sites. In the initial 2014 report, the edge-to-face geometry preference shown in the previous sentence was said to be universal throughout the series and the relative stabilities were reported to be surprising in several respects, most notably the position of WF/WE in the ranking. Independent of the specific chemical shift criterion employed, WF/FE and WF/YE both appeared to provide greater fold stabilization than WF/WE. By one measure, WF/WE ranked 7th in stability over the nine possible Ar/Ar permutations. Our examination of the data corroborates the authors’ conclusion that there is some contribution of the altered χ2 conformation at the edge Trp (similar to that shown Fig. 6B, vide supra). We first observed9 this altered edge conformation for a hairpin with a -VWE-IpGK-WFI- sequence and cluster geometry about the turn region. With the latest publication50, which appeared after this work was completed, it is clear that for the Ac-L-Ar-VpGL-W-V-NH2 series there are also, in some cases, alternative folded species present due to an EtF⇔FtE equilibrium. Steric problems associated with the placement of the Trp indole as the edge species are the likely cause. There is, perhaps, some concern about extrapolating these results to proteins since all of the NMR data (and most of the CD data) is for neat CH3OH solutions rather than aqueous medium.
Our prior studies9,11,12, with a bit of further analysis, provide additional estimates of both W/Y and W/F cluster contributions to stability. In a system where W/W affords a 7.6 kJ stabilization (Ac-WIpGKWTG-NH2, shown in Table S5A of original β-cap report: the stabilization for WF/YE is 6 kJ and only 2 kJ/mol for the YF/WE cluster analog.12 In another β-capping system (Table S5B, same article) where W/W provides 9.4 kJ of stabilization: WF/FE is 4.7 kJ while FF/WE is only 1 kJ/mol. In all of our prior studies, the preference for the face position is (W > Y > F) is clear. Over all available comparisons the following stability sequence was suggested: WF/WE > WF/YE > YF/WE ≥ WF/FE > FF/WE. On this basis, we examined strategies for improving the stability of some previously reported hairpins. One of our first efforts of this type, was a Tyr/Phe residue position exchange in second hairpin of protein-G, GEWTYE-DDATKT-FFTVTE (GB1p, Fig. 4). An increase in fold stability was expected. Although both peptides are rather poorly folded, the data indicates that YE/FF is more stabilizing that FE/YF (Fig. S1). We then moved on to WE/FF and FE/WF pairings as the GB1p analogs. Significant fold stabilization was observed with superiority of indole as an electron-releasing “face unit” in EtF clusters shown by a 6.3 kJ/mol ΔΔGU280 for the FE/YF→FE/WF mutation. A stabilizing effect was also observed at the edge site: the YE/FF→WE/FF mutation is stabilizing by 3 kJ/mol at 280K. The CSD comparisons appear in Fig. S1a–c.
The absence of data for Y/Y as well as the sparsity of Y/F and W/F observations, as well as difference between our stability sequences and those reported by Makwana and Mahalakshimi prompted a more focused study. Also, we needed to address the possibility that prior structure-stabilizing rankings apply only to Ar/Ar pairings that are either in Ac-Ar/Ar-TG β-cap and/or flanking 4-residue IXGK turns. To obtain quantitative comparisons of other Ar/Ar clusters to W/W clusters, we turned to specific motifs that have the strongest inherent EtF positional preferences. In both the Ac-Ar/Ar-TG and (R/K)-ArF/ArE-E caps, we expect the C-terminal unit to contain, as shown, the edge-Ar based on prior examples.12,20–22 This proved to be the case in all species prepared, even in cases where an FFWE cluster results. In turn-flanking Ar/Ar units of the form (X)n-Ar-turn-Ar-(X)n with n≥3, the edge-Ar needs to be at the N-terminus of the turn. This is a particularly strong preference in the case of [4:6]-turns such as that formed by the NPATGK sequence. We selected HP7 (KTWE-NPATGK-WFTE)7,16, HP5F (KKYTWE-NPATGK-FFTVQE)5, and KWF-LTVS-INGK-KITV-WEE for specific mutational studies at the aryl units.
Of these systems, the last was the easiest to analyze. The aryl mutations are within a distinct β-cap connected to the turn by rather long β-strands that contain numerous backbone proton sites with structuring-specific chemical shift changes. The sites within the turn combined with one adjacent site give a range of 300K χF-values from 0.93 (WFWE) to 0.58 (FFFE). Moving the probes to more distant locations from the common turn, results in a wider range (χF = 0.93 to 0.29) which we view as better estimates of the incremental effects of the aryl clusters. The aryl cluster series is ranked, from the most stabilizing to least stabilizing interaction, below with some of the χF-values at 300K shown. A more complete presentation of the data appears in Table S2. KK-LTVS-INGK-KITV-AE (χF = 0.26) served as a control for converting χF-values to ΔG increments for the various clusters examined. This system also demonstrated the extent of structuring about the favorable9,51,52 INGK turn.
NMR studies indicated that the KArF-LT----TV-ArEE cluster geometry was strictly conserved throughout the series even for the YFFE and FFFE systems that provide less than 1 kJ/mol of fold stabilization and systems with the less favorable aryl unit forced into the face position (e.g. YFWE). All clusters containing a Trp unit provided at least 4.6 kJ/mol of stabilization with the cluster from W/W producing a 9 kJ/mol stabilization at 300K.
In the case of the HP7 hairpin system, all possible probe sites (folding dependent chemical shifts) are either vicinal to, or interacting in a cross-strand fashion with, an aryl unit. As a result, the 100%-folded CSDs are expected to change upon essentially any aryl to alternate aryl ring mutation. This prevents us from calculating quantitative ΔΔG-values, but the relative ranking of fold stabilities could be discerned: WEWF > YEWF > WEYF ≥ FEWF ≥ YEYF. The FEWF species still displayed the full set of diagnostic CSDs at the edge aryl site; on this basis alone, its position in the stability series would involve a WEYF/FEWF switch. However, the FEWF displayed the largest loss of structure on warming from 280 to 320K in the entire series. Some of the key CSD comparisons are collected in Fig. S3
In the case of HP5F analogs, the greater length of the β-strands beyond the turn–flanking aryl groups provides, unlike HP7, a number of reliable chemical shift probes for obtained χF-values. Some specifics regarding the probe choices and results appear in the supporting material (Fig S5a–b). The aryl groups of HP5F (KKYTWE-NPATGK-FFTVQE, Fig. 4, Tm = 53ºC) appeared to be reversed relative to our expectations for optimum fold stabilization. As expected, reversing the aryl functions produced a much more stable hairpin with the FEWF cluster geometry (Tm = 55 ºC). Fold measures at 300K and a ranking of fold stability is given below. In instances where stability measures are similar at 300K, the ranking takes into account data at 320 and 330K; the system which displays greater resistance to structure melting is ranked as “more stable”. In the rank listing below, the parenthetic values are the χF-values at 300K.
The W/W cluster was the most stabilizing at ambient and lower temperatures. But, somewhat of a surprise, at temperatures above 310K, the FEWF hairpin was even more stable than the WEWF hairpin.
Peptide WA (KKYTWNPATGKATVQE) serves as the control for the absence of a turn-flanking aryl cluster. In this system, even the least favorable aryl clusters provide at least 4.5 kJ/mol of stabilization, much more than in the KArF-LT----TV-ArEE cluster system. The optimal WEWF and FEWF turn-flanking clusters provided a 10 kJ/mol folding increment. These were also the most favorable clusters for both the Coulombic and hydrophobic β-cap systems providing the same 10 kJ/mol fold-enhancement at 300K. Tryptophan, the most electron rich of the series, is preferred as the face of an aryl cluster.
As has been noted previously16, a turn-flanking aryl cluster has a particularly large stabilizing effect on [4:6]-hairpin systems. The new data from this study confirms this, but the even larger effect (11 – 14 kJ/mol) for NAAAGX loops has not been fully rationalized. The stabilizing effects of a turn-flanking aryl cluster are smaller for –INGK- turns, maxing out at 5 kJ/mol, and much smaller for –IpXX- turns. We have now extended this to [3:5]-Gly-bulge turns such as –SADGR- and –NPDGT-. Flanking Trp residues are stabilizing, but the fold stability increment is only on the order of 3–4 kJ/mol (see Supporting Material). The differences in fold-enhancement with turn type are likely due to contacts between the edge-aryl ring and the turn region and are discussed later. NMR studies provide insights into cluster geometries in both turn-flanking and capping motifs.
NMR signatures of EtF Ar/Ar clusters
With the examples from our effort to determine the relatively stabilization increments of aryl clusters, we also have additional data on the ring current shifts within these clusters. Since there do appear to be some context effects, we have divided these into three categories: 1) turn-flanking Ar/Ar systems, predominantly ArE-NPATGK-ArF and ArE-I(N/p)GK-ArF, 2) mutants of the original (hydrophobic) β-capping motif, AcArE-----ArFTG-NH2, and 3) the newer Coulombic β-cap20, (K/R)ArF-------ArEE.
Throughout, we have followed the CSDs observed for the edge aryl group of the EtF cluster. The CSDs observed for different aryl clusters are collected in Table 4. Table 4 lists only ring current shifts associated with the face aryl rings in these clusters; these are observed at the edge ring. There are also quite large and diagnostic shifts due to the edge-ring. The most sizeable of these is the >3ppm upfield i+2 Gly HN of the (alkanoyl)-W/WTG cap.
Table 4.
The range of structuring CSDs at 280K within FtE and EtF aryl/aryl clusters in a variety of contexts. Throughout, values (in ppm) are extrapolated to 100%-folded. A few categories are represented by only a single observation. Single entries that represent distant extrapolations are in parentheses. All values in the table are negative (upfield) CSDs.
| Turn-flanking series: (G/E)-X1,3-ArE-turn-ArF-X1,3-E | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| WEW | WEY | WEF | YEW | YEY | YEF | FEW | FEY | FEF | |
| ArE Hβ3 | 1.3–2.05 | 0.28–0.36 | 0.39 | 2.3–2.7 | 0.48–0.63 | 0.52 | 2.29 | 0.46 | 0.49 |
| WE Hε3 | 2.2–2.7 | 1.95–2.1 | 2.22 | ||||||
| WE Hζ3 | 0.64–0.82 | 0.57–0.63 | 0.62 | ||||||
| ArE Hδ | 0.35–0.63 | 0.44–0.52 | 0.54 | 1.5–1.74 | 1.0–1.56 | 1.23 | 1.58 | 1.34 | 1.38 |
| YE/FE Hε | 0.5–0.59 | ~0.55 | 0.53 | 0.47 | 0.47 | 0.49 | |||
| Classic β-cap series: alkanoyl-ArF-X2,4-turn-X2,4-ArE-TG | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| WWE | YWE | FWE | WYE | YYE | FYE | WFE | YFE | FFE | |
| ArE Hβ3 | 1.3–1.6 | 0.49 | 0.25 | 2.12 | (0.50) | 2.14 | |||
| WE Hε3 | 1.9–2.25 | 1.69 | 1.69 | ||||||
| WE Hζ3 | 0.35–0.45 | 0.26 | 0.28 | ||||||
| ArE Hδ | 0.46–0.61 | 0.4 | 0.25 | 1.22 | (0.54) | 1.33 | |||
| YE/FE Hε | 0.32 | (0.23) | 0.28 | ||||||
| Coulombic-cap series: (R/K)-ArF-X2,4-turn-X2,4-ArE-(E/D) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| WWE | YWE | FWE | WYE | YYE | FYE | WFE | YFE | FFE | |
| ArE Hβ3 | 0.9–1.2 | 0.37 | 0.38 | 1.17 | 0.3–0.5 | 0.47 | 1.65 | 0.47 | (0.86) |
| WE Hε3 | 2.4–2.7 | 1.72 | 2.06 | ||||||
| WE Hζ3 | 0.57–0.84 | 0.44 | 0.5 | ||||||
| ArE Hδ | 0.3–0.4 | 0.23 | 0.26 | 0.99 | ~0.46 | 0.96 | 1.54 | 0.69 | (1.32) |
| YE/FE Hε | 0.4 | 0.12–0.22 | 0.28 | 0.6 | 0.49 | −0.7 | |||
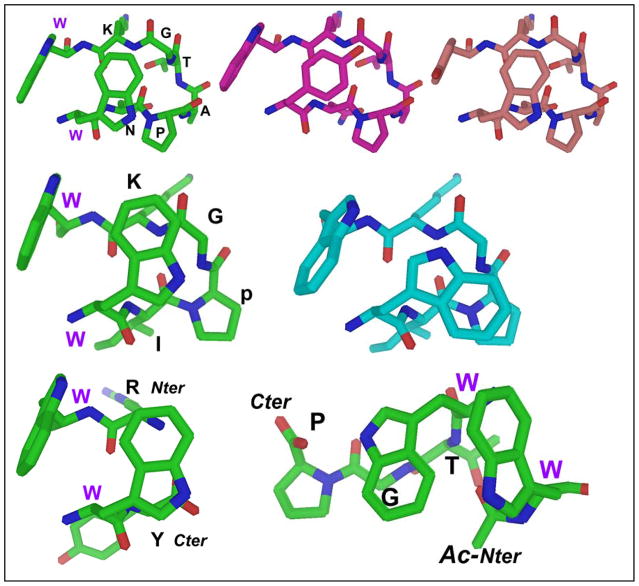
Other key diagnostics include upfield shifts for the Hα (>0.5 ppm) and Hβ protons of residue 1 of the (R/K)W/W(E/D) cap, and >1 ppm upfield CSDs for Gly Hα and Lys HN protons in the Trp-flanked NPATGK and I(p/N)GK turns. In the latter case, minor changes to solvent (e.g., 20% HFIP) induce an inversion in the χ2 rotamer of the edge Trp, increasing the negative CSD on Gly HN from −1.5 to −3.5, but shrinking the Lys HN from −1.8 to −1.0. Other changes are observed when the edge Trp is replaced with Tyr or Phe; chemical shift deviations become smaller and/or fewer. Figure 10 provides structural rationales for these changes.
Figure 10.
Top panel -- views of the Trp/Trp interaction in an NPATGK-turn-flanking context. From left to right: Trp/Trp, TrpF/TyrE, TyrF/TrpE. The rationale for more highly shifted Hβ3 ArylE protons for face = Trp is apparent: for phenyl rings the Hβ3 position on the cross-strand partner is not as close and not in the middle of the shielding cone. Middle panel -- two different Trp/Trp conformers available to WIpGKW-hairpins: the standard and inverted χ2 rotamers. The alternate conformer found flanking 2:4 turns (especially XpGX turns, or in the presence of fluoroalcohol cosolvent) is on the right. For the typical conformer, TrpE presents its Hε3 proton to TrpF, but in the alternate conformer Hδ1 is presented to TrpF. Major CSD changes are observed for the turn (e.g., the Gly HN CSD shifts from −1.0 ppm to −3.5 ppm) as its position changes relative to the TrpE indole. Bottom – the Coulombic +XW/WX- (left) versus the hydrophobic β-cap (Ac-Ar/ArTG). In all cases, extrapolated CSD variations (Table 4) for 100% folded constructs are consistent with the aromatic ring orientations shown.
After controlling for fold population, chemical shift deviations can be used to infer details regarding ring orientations within the aryl cluster. Nuances like the lower Hε3/Hβ3 shift ratio for the W/WTG versus Coulombic (RW/WE, etc.) β-caps suggest a context dependence for the precise edge-to-face geometry, even when the aryl residues remain constant. With changes in the aryl residues other features are evident; e.g., the near-complete loss of edge-aryl Hβ3 shifts when the “face” residue is Phe or Tyr rather than Trp even as the large edge Hδ/Hε3 shifts remain. This establishes that the shielding cone for these sidechains is smaller than the indole of Trp, and is specifically centered on the edge of the cross-strand aromatic ring (and not the sidechain in general). Hβ3 shifts are at their largest when the face is Trp but the edge is Phe or Tyr; these smaller rings allow the beta methylene to make a closer approach. See figure 10 for some examples.
The CD spectroscopic signatures of EtF Ar/Ar clusters
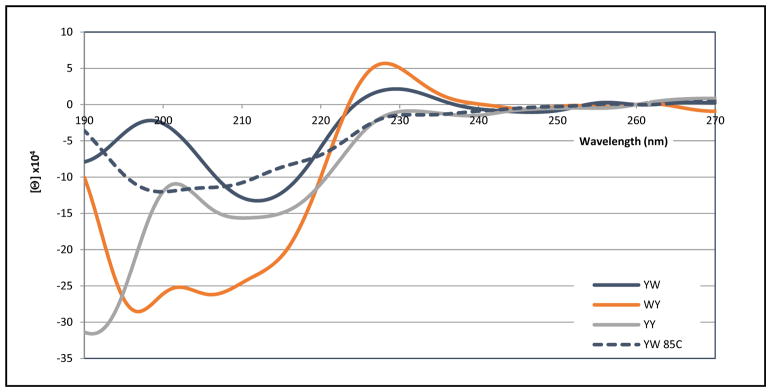
Examples in the previous sections indicate that amplitude of the [θ]228 band of EtF indole/indole clusters can vary significantly and that the upfield shifts of aryl-H signals (particularly Hε3 of an indole) on the edge aryl ring are a more dependable criterion for the formation of an EtF cluster. There does not appear to be any strict correlation between the ring current shift observed at the edge WHε3 and the amplitude of the exciton couplet even after correcting these values to their extrapolated 100%-folded values. Restricting our analysis to CD exciton couplets due to a single W/W pair in which the edge-W has negative Hε3 CSDs in the 1.8 – 2.7 ppm range, we have now observed a wide range of 100%-folded [θ]228 values (positive ellipticities in all but one non-hairpin case), 160,000 – 655,000°. The magnitude of the other portion of this couplet (−[θ]212), generally correlates with [θ]228, but is often obscured by other CD features (including the β-sheet backbone minimum at 215 – 218 nm51–54). Other features, the amplitude and even the appearance of extrema at 204 and 198 nm, are even more variable. In an indole/indole EtF cluster the position of Hε3 relative to the face indole ring appears to be rather rigidly fixed, but small changes in the angle between the two indole ring transition moments can have large effects on the chirality of the interactions between the two chromophores.
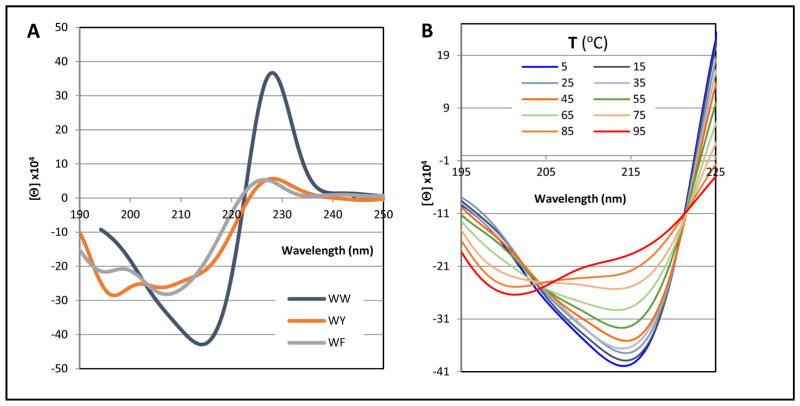
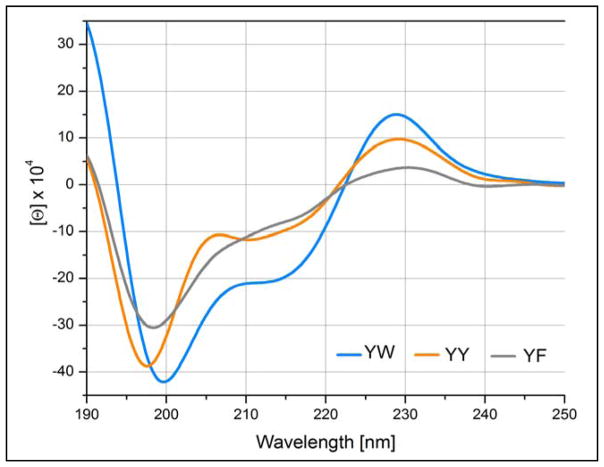
To ascertain the corresponding spectral diagnostics of W/Y (with either aryl as the edge) and Y/Y pairings, we employed specific motifs that have the strongest positional EtF preferences. In some cases, these could be extended to W/F interactions as well. We have collected CD spectra for a variety of aryl clusters in Figures 11 – 13. A complete set CD spectra including the melts appears in the supporting material (Figures S6–11). CD melts can be particularly useful since they reveal the wavelengths of features and provide a Δ[θ] that can suggest the nature of the effects being observed. A few CD melts are included here as well, panel B of Fig. 11 and Figs. 14 and 15 (vide infra).
Figure 11.
CD features of three KArF-LTVS-INGK-KITV-ArEE hairpin peptides at 280K, listed as ArFArE on the graph. All spectra have been normalized to reflect the expectation for the fully-folded state. The right-hand panel shows the melt (5–95° C) for the WF/WE species.
Fig. 13.
CD spectra of KKYTArE-NPATGK-ArFTVQE analogs with a constant ArE = tyrosine and the face residue varied through the complete series.
Fig. 14.
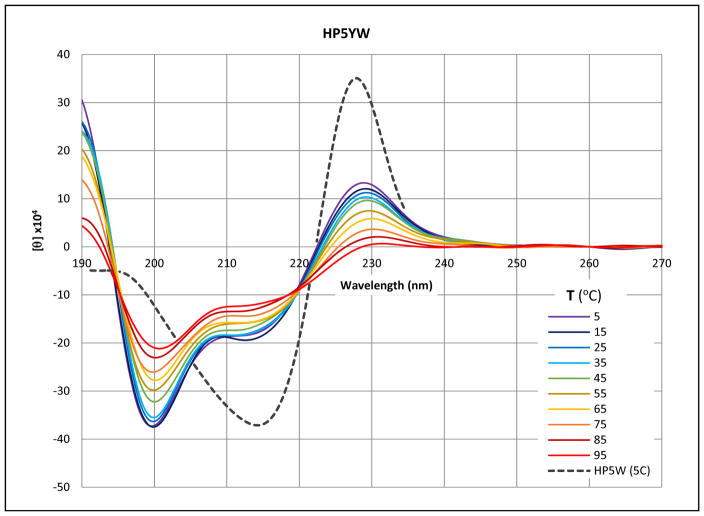
The CD melt (5–95 °C) of melt of KKYTYE-NPATGK-WFTVQE is overlaid on the low temperature spectrum (broken line) of the corresponding WEWF analog.
Fig. 15.
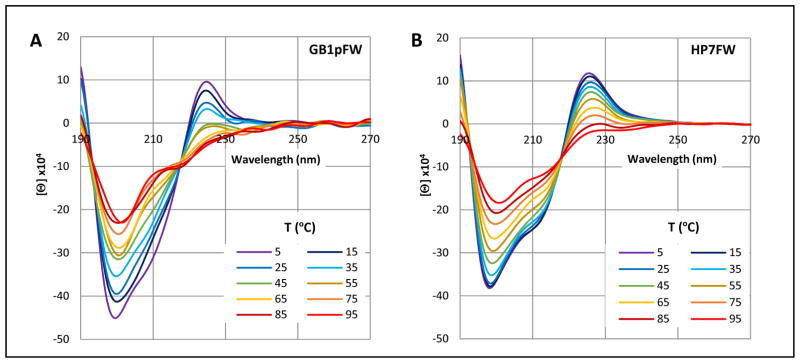
CD melts (5–95 °C) of GB1pFW (GEWTFE-DDATKT-WFTVTE) and HP7FW (KTFE-NPATGK-WFTE); panels A and B, respectively. Another example, HP5FW, appears in the Supporting Material.
With regard to the ellipticity at the 225 – 232 nm extremum (a maximum in all hairpins examined), an indole face unit in the cluster appears to be the prime determinant for unusually large exciton couplets.
Figure 11 shows the CD spectra of aryl clusters in a Coulombic β-cap with a common indole face ring and variation in the edge aryl. All three species show a positive ellipticity feature at 225 – 229 nm, appearing to be the low-energy portion of an exciton couplet. The amplitude is greatly reduced when either phenyl ring replaces the edge indole. Further reductions in amplitude for this band occur when the face-indole is swapped for another aryl ring (Fig. 12 and Supporting Material). There is some evidence for a 225 – 232 nm ellipticity maximum in all the KArF-LTVS-INGK-KITV-ArEE species examined so long as ArF ≠ F. Figure 11 illustrates the three CD spectra with the largest ~228nm band. Figure 11 show the further loss in amplitude of the 228 nm band as Tyr provides the face ring in the cluster. While all three spectra show evidence of a circa 212 nm minimum, it is difficult to view this as only the other half of the exciton couplet with the 228 nm maximum. Of interest here is the observation that the FFWE species has nearly the same +196/−212nm feature as YFWE is, but the 228nm feature disappears. The aryl cluster species of Figure 12 presents quite different spectra in the 190 – 210 nm region.
Figure 12.
The CD spectra of three additional KArF-LTVS-INGK-KITV-ArEE hairpins at 280K. The YFWE spectrum at 358K is shown as a broken line.
In examining CD spectra of aryl-cluster-bearing hairpins it should be kept in mind that the antiparallel association of β-strands also results in a CD signature: a minimum at 216–219 nm and a maximum at 196–199 nm.52,53,55 At the hairpin lengths examined herein, the latter results in negative Δmelt[θ]198-values of 105 to 2×105° in Molar units. A comparison of the two YFWE traces in Fig. 11 provides an example of expected melting behavior for the 198 nm maximum. Another example appears in the right-hand panel of Fig. 11. The expectation of a positive hairpin feature near 198 nm requires us to carefully consider systems that display strong negative features at this wavelength. This was the case for the WFYE trace in Fig. 12, and the even more distinct features in Fig. 5A and 13. In Fig. 13 (and Fig. 14, vide infra) the negative 198–200 nm feature becomes the largest band in the spectra.
With figures 13 – 15, we move on to turn-flanking clusters, in all cases the first listed Ar group, at the N-terminus of a [4:6]-hairpin turn provides the edge ring in an edge-to-face cluster: these are listed as ArE/ArF. In the analogs shown in figure 13, the face ring varied through the complete series: W→Y→F, as the edge ring was kept constant as tyrosine. Through this series, the amplitude of the exciton couplet centered about 220 – 222 nm decreases. However, unlike the β-cap series, there is still evidence of this feature even in the case where ArF = F. There is a red shift, 228 – 231 nm, at the exciton couplet maximum for the series W→Y→F. In addition, an intense minimum at 198 – 200 nm is the largest feature of all three spectra. Insights into the nature of this feature follow from the CD melts shown in figures 14 and 15. In these melts there are a large positive changes in [θ] near 198 nm upon warming; these must have a much larger amplitude than the negative Δ[θ] expected from the loss of β backbone structure.
Both the initial spectrum and the melting behavior of KKYTYE-NPATGK-WFTVQE precisely mirror that shown in panels A and B of Fig. 5 for more complicated YE/WF-containing peptides. The melts show distinct isodichroics at 195 and 219.5 nm. The lower energy isodichroic reflects an exciton couplet melt that is very similar to the 221 nm feature of the WEWF analog and that shown in panel B of figure 11. The λ < 250nm absorption bands of the three aryl amino acids and their designations (λ, 10−3ε) are: Trp - 1Bb (197nm, 20), 1La (220nm, 35); Tyr - 1Bb (193nm, 47), 1La (224nm, 8.3); and Phe - 1Bb (188nm, 60), 1La (206nm, 9). Thus, the 195 nm isodichroic appears to be an exciton coupling of the two 1Bb bands. As will be apparent from fig. 15, the same feature and rationale applies to an FE/WF cluster. However, we have seen little or no evidence for couplets arising from the 1Bb bands of two Trp indoles in an EtF cluster. The 219.5nm isodichroic reflects the melt-out of the 1La exciton couplet between the Tyr and Trp rings.
Upon examining the spectrum in panel A, it is tempting to view the 225 nm maximum as the result of a WW interaction; there is a Trp in each strand of this hairpin. However, this clearly is not the case. The spectrum of the much simpler HP7FW analog, with only the turn-flanking FE/WF cluster, is essentially the same. If we assign the 225 nm maximum as the low energy band of a couplet between the 1La bands of Phe and Trp, even though they have quite different transition energies, this does fit with the blue shift to 225 nm (versus 228 for a WW interaction) and the smaller red shift observed for the comparable WY interaction. The 198 minimum and 193 isodichroic represent 1Bb band interactions, also with a slight blue shift for the Y → F change.
Conclusions
With the additional designs prepared in the present study, we have determined the structural preferences of all permutations of both beta-sheet-terminating and turn-flanking aryl/aryl clusters in hairpins, establishing specific NMR and circular dichroism signatures for each. It is now apparent that EtF aryl clusters can, for all possible combinations of aryl rings in proteins, produce CD signals in the 190 – 230 nm span that are of comparable or larger amplitude than the backbone-derived CD signals used for secondary structure analysis. Consequently, CD data for any peptide with the possibility of forming aryl clusters (or for that matter having more than one aryl group per ten residues) should not be analyzed for secondary structure content.
The specific finding regarding the CD spectral contributions of EtF aryl clusters can be summarized as follows. Two exciton couplets, which can be characterized by their melting isodichroics of 193–197 and 218–222nm are produced with most aryl pairs. The 195nm couplet appears to be either absent or of greatly diminished amplitude for W/W clusters, but has been observed for all other combinations. For many of the clusters the 195nm exciton couplet is the dominant CD feature. We found no evidence of the −[θ]196/+[θ]202 exciton couplet predicted by Roy and Keiderling27,28 for the YE/WF cluster and attributed to a Y/Y interaction by Lovas30,32. We also have seen no evidence for a significant CD contribution from diagonal face-to-face interaction W/W interactions in hairpins. Turning to the circa 220nm couplet, the amplitude is maximal for W/W producing the now classic 228 nm maximum. All of the other WF/Ar interactions, even WF/FF, produce quite large positive bands in the 224 – 230nm span. Despite characterizing diverse aryl/aryl interactions with varied geometries, we have yet to observe an “inverted” or “negative” couplet at ~220 nm.
While we observed large variability in the amplitudes of CD exciton couplets due to aryl clusters of what appear to be rather similar geometry, the NMR characteristics (predominantly the upfield ring current shifts in the edge aryl ring) are remarkably constant (Table 4). This reflects both the orientation of the two rings and the relative rigidity of the peptide backbone within and adjacent to a cross-strand Ar/Ar interaction in a hairpin. Aryl/backbone interactions are essential to the stability and rigidity of the cluster, such that the backbone could be considered part of the extended cluster. Strand terminal β caps are not just energetically stabilizing; they also inhibit fraying. Removal of the strand-capping aryl cluster (or to a lesser extent, replacing a Trp with Tyr or Phe) increases local disorder, even when the hairpin has a similar global fold population. The edge residues of turn-flanking aryl/aryl clusters can interact strongly with the turn, and appear to populate mostly turn-interacting rotamers even in the absence of a cross-strand aromatic partner. In fact, the face residue of the EtF cluster could be thought of as a way to anchor the edge residue in a favorable geometry for an interaction with turn residues, as well as the key component of the aryl cluster.
The relative hairpin-fold stabilizing effects of aryl/aryl pairs differ depending on context due to changing ArE/backbone interactions. For NPATGK- (and other [4:6]-) turns, an edge Phe is nearly as stabilizing as Trp (and imparts greater resistance to melting) because Phe makes the best contacts with the turn. In the case of Phe, it’s not immediately obvious from model structures why it might be superior to Trp, but the model does suggest a reason for its superiority to Tyr: the tyrosine hydroxyl may have either a steric clash or repulsive Coulombic interaction with the turn glycine carbonyl oxygen.
The Coulombic (hydrophilic) β-cap also has a key ancillary stabilization feature, a π-cation interaction between the edge aryl ring and the N-terminal amine. Pi-cation interactions are stronger with indole versus phenyl rings, thus the significant decrease in stability on swapping edge Trp for a different aryl residue. The effect is not as extreme for the hydrophobic cap, where the analogous i-i+2 aryl/amide interaction is less polar.
Aryl/aryl clusters have a special place in β-sheet model design. In the absence of particularly favorable (and often un-natural) turn units strand-terminating aryl/aryl clusters (one or another of the β-cap designs) are near-essential for the design of stable systems. When properly inserted into the structure they provide greater rigidity and reduce fraying to a greater extent than cyclizing disulfide linkages. They have the additional advantage of presenting NMR and CD signatures that indicate their presence and effectiveness and can serve as probes for dynamics studies. In summary, we have determined that, in addition to their strong preference for non-H-bonding strand-terminal positions (flanking either a turn or the chain termini) aryl clusters prefer indoles as the “face” in edge-to-face pairings. They also have a particularly large stabilizing effect when flanking classic [4:6]-turns. These take the form, ArE-(N/D)1-P2X-(T/S)4-G5X6-ArF, with Trp or Phe as ArE making particularly favorable interactions with the backbone at P2, G5, X6. These are among the most stable EtF clusters we observed. In contrast to this, it was found that end-capping aryl clusters always form with a FtE geometry, regardless of aromatic makeup the aryl unit at the N-terminus serves as the face. The only exception to this is when a cluster is both turn-flanking and end-capping (i.e. Ac-WINGKWTG-NH2), where evidence of both EtF and FtE are present, with a larger chance of FtE. Steric interactions with bulkier turn locus residues can offset some of the stabilizing effects of turn-flanking Aryl clusters and even set up an EtF to FtE equilibrium.
Strand-terminating aryl/aryl clusters are near-essential for the design of fold-stable β-sheet models. Thus it behooves protein designers and bioengineers to become familiar with their application and limitations, as well as the NMR and CD signatures that signify their presence and effectiveness. In addition to their strong preference for non-H-bonding strand-terminal positions, (flanking either a turn or the chain termini) aryl clusters prefer indoles as the “face” in edge-to-face pairings. They also have a special preference for flanking [4:6]-turns.
Supplementary Material
References
- 1.Grishina IB, Woody RW. Faraday Discuss. 1994;(99):245. doi: 10.1039/fd9949900245. [DOI] [PubMed] [Google Scholar]
- 2.Woody RW, Dunker AK. Circular Dichroism and the Conformational Analysis of Biomolecules. Plenum Press; New York: 1996. [Google Scholar]
- 3.Favre M, Moehle K, Jiang L, Pfeiffer B, Robinson Ja. J Am Chem Soc. 1999;121(12):2679. [Google Scholar]
- 4.Cochran AG, Skelton NJ, Starovasnik MA. Proc Natl Acad Sci U S A. 2001;98(10):5578. doi: 10.1073/pnas.091100898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fesinmeyer RM, Hudson FM, Andersen NH. J Am Chem Soc. 2004;126(13):7238. doi: 10.1021/ja0379520. [DOI] [PubMed] [Google Scholar]
- 6.Dhanasekaran M, Prakash O, Gong YX, Baures PW. Org Biomol Chem. 2004;2(14):2071. doi: 10.1039/b315228f. [DOI] [PubMed] [Google Scholar]
- 7.Andersen NH, Olsen Ka, Fesinmeyer RM, Tan X, Hudson FM, Eidenschink La, Farazi SR. J Am Chem Soc. 2006;128(18):6101. doi: 10.1021/ja054971w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kier BL, Andersen NH. J Am Chem Soc. 2008;130(44):14675. doi: 10.1021/ja804656h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eidenschink L, Kier BL, Huggins KNL, Andersen NH. Proteins Struct Funct Bioinforma. 2009;75(2):308. doi: 10.1002/prot.22240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mirassou Y, Santiveri CM, de Vega MJP, González-Muñiz R, Jiménez MA. ChemBioChem. 2009;10(5):902. doi: 10.1002/cbic.200800834. [DOI] [PubMed] [Google Scholar]
- 11.Eidenschink L, Crabbe E, Andersen NH. Biopolymers. 2009;91(7):557. doi: 10.1002/bip.21177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kier BL, Shu I, Eidenschink La, Andersen NH. Proc Natl Acad Sci U S A. 2010;107(23):10466. doi: 10.1073/pnas.0913534107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersen NH, Huggins KNL, Bisaglia M, Bubacco L. 31st EPS Symp. 2010:22–23. [Google Scholar]
- 14.Santiveri CM, Pérez de Vega MJ, González-Muñiz R, Jiménez MA. Org Biomol Chem. 2011;9(15):5487. doi: 10.1039/c1ob05353a. [DOI] [PubMed] [Google Scholar]
- 15.Huggins KNL, Bisaglia M, Bubacco L, Tatarek-Nossol M, Kapurniotu A, Andersen NH. Biochemistry. 2011;50(38):8202. doi: 10.1021/bi200760h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scian M, Shu I, Olsen Ka, Hassam K, Andersen NH. Biochemistry. 2013;52(15):2556. doi: 10.1021/bi400146c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shu I, Scian M, Stewart JM, Kier BL, Andersen NH. J Biomol NMR. 2013;56(4):313. doi: 10.1007/s10858-013-9749-3. [DOI] [PubMed] [Google Scholar]
- 18.Wu L, Mcelheny D, Setnicka V, Hilario J, Keiderling Ta. Proteins Struct Funct Bioinforma. 2012;80(1):44. doi: 10.1002/prot.23140. [DOI] [PubMed] [Google Scholar]
- 19.Santiveri CM, Jiménez MA. Biopolymers. 2010;94(6):779. doi: 10.1002/bip.21436. [DOI] [PubMed] [Google Scholar]
- 20.Anderson JM, Kier BL, Shcherbakov Aa, Andersen NH. FEBS Lett. 2014;588(24):4749. doi: 10.1016/j.febslet.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kier BL, Anderson JM, Andersen NH. J Am Chem Soc. 2014;136(2):741. doi: 10.1021/ja410824x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kier BL, Andersen NH. J Pept Sci. 2014;20(9):704. doi: 10.1002/psc.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sibanda BL, Blundell TL, Thornton JM. J Mol Biol. 1989;206(4):759. doi: 10.1016/0022-2836(89)90583-4. [DOI] [PubMed] [Google Scholar]
- 24.Honda S, Yamasaki K, Sawada Y, Morii H. Structure. 2004;12(8):1507. doi: 10.1016/j.str.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 25.Takekiyo T, Wu L, Yoshimura Y, Shimizu A, Keiderling TA. Biochemistry. 2009;48(7):1543. doi: 10.1021/bi8019838. [DOI] [PubMed] [Google Scholar]
- 26.Wu L, McElheny D, Huang R, Keiderling Ta. Biochemistry. 2009;48(43):10362. doi: 10.1021/bi901249d. [DOI] [PubMed] [Google Scholar]
- 27.Wu L, McElheny D, Takekiyo T, Keiderling Ta. Biochemistry. 2010;49(22):4705. doi: 10.1021/bi100491s. [DOI] [PubMed] [Google Scholar]
- 28.Keiderling TA, Roy A, Bour P. Chirality. 2009;171(November):163. doi: 10.1002/chir.20792. [DOI] [PubMed] [Google Scholar]
- 29.Honda S, Akiba T, Kato YS, Sawada Y, Sekijima M, Ishimura M, Ooishi A, Watanabe H, Odahara T, Harata K. J Am Chem Soc. 2008;130(46):15327. doi: 10.1021/ja8030533. [DOI] [PubMed] [Google Scholar]
- 30.Hatfield MPD, Murphy RF, Lovas S. Biopolymers. 2010;93(5):442. doi: 10.1002/bip.21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatfield MPD, Murphy RF, Lovas S. J Phys Chem B. 2010;114(8):3028. doi: 10.1021/jp910465e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hatfield MPD, Murphy RF, Lovas S. J Phys Chem B. 2011;115(17):4971. doi: 10.1021/jp111076j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piotto M, Saudek V, Sklenar V. J Biomol NMR. 1992;2(6):661. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 34.Bax A, Davis DG. J Magn Reson. 1985;65(2):355. [Google Scholar]
- 35.Olsen Ka, Fesinmeyer RM, Stewart JM, Andersen NH. Proc Natl Acad Sci U S A. 2005;102(43):15483. doi: 10.1073/pnas.0504392102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riemen AJ, Waters ML. Biochemistry. 2009;48(7):1525. doi: 10.1021/bi801706k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neidigh JW, Fesinmeyer RM, Andersen NH. Nat Struct Biol. 2002;9(6):425. doi: 10.1038/nsb798. [DOI] [PubMed] [Google Scholar]
- 38.Bunagan MR, Yang X, Saven JG, Gai F. J Phys Chem B. 2006;110(8):3759. doi: 10.1021/jp055288z. [DOI] [PubMed] [Google Scholar]
- 39.Barua B, Lin JC, Williams VD, Kummler P, Neidigh JW, Andersen NH. Protein Eng Des Sel. 2008;21(3):171. doi: 10.1093/protein/gzm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams DV, Barua B, Andersen NH. Org Biomol Chem. 2008;6(23):4287. doi: 10.1039/b814314e. [DOI] [PubMed] [Google Scholar]
- 41.Scian M, Lin JC, Le Trong I, Makhatadze GI, Stenkamp RE, Andersen NH. Proc Natl Acad Sci. 2012;109(31):12521. doi: 10.1073/pnas.1121421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Byrne A, Kier BL, Williams DV, Scian M, Andersen NH. RSC Adv. 2013;3(43):19824. doi: 10.1039/C3RA43674H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Probe T, Schenck HL, Gellman SH. J Am Chem Soc. 1998;120(11):4869. [Google Scholar]
- 44.Fesinmeyer RM, Hudson FM, Olsen Ka, White GWN, Euser A, Andersen NH. J Biomol NMR. 2005;33(4):213. doi: 10.1007/s10858-005-3731-7. [DOI] [PubMed] [Google Scholar]
- 45.Jäger M, Zhang Y, Bieschke J, Nguyen H, Dendle M, Bowman ME, Noel JP, Gruebele M, Kelly JW. Proc Natl Acad Sci U S A. 2006;103(28):10648. doi: 10.1073/pnas.0600511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hudson FM, Andersen NH. Biopolymers. 2006;83(4):424. doi: 10.1002/bip.20575. [DOI] [PubMed] [Google Scholar]
- 47.Roe DR, Hornak V, Simmerling C. J Mol Biol. 2005;352(2):370. doi: 10.1016/j.jmb.2005.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jäger M, Dendle M, Fuller Aa, Kelly JW. Protein Sci. 2007;16:2306. doi: 10.1110/ps.072904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Makwana KM, Mahalakshmi R. ChemBioChem. 2014;15(16):2357. doi: 10.1002/cbic.201402340. [DOI] [PubMed] [Google Scholar]
- 50.Makwana KM, Mahalakshmi R. J Phys Chem B. 2015;119(17):5376. doi: 10.1021/acs.jpcb.5b00554. [DOI] [PubMed] [Google Scholar]
- 51.Andersen NH, Dyer RB, Fesinmeyer RM, Gai F, Liu Z, Neidigh JW, Tong H. J Am Chem Soc. 1999;121(42):9879. [Google Scholar]
- 52.Dyer RB, Maness SJ, Franzen S, Fesinmeyer RM, Olsen Ka, Andersen NH. Biochemistry. 2005;44(30):10406. doi: 10.1021/bi050698z. [DOI] [PubMed] [Google Scholar]
- 53.Cort J, Liu Z, Lee G, Harris SM, Prickett KS, Gaeta LSL, Andersen NH. Biochem Biophys Res Commun. 1994;204(3):1088. doi: 10.1006/bbrc.1994.2574. [DOI] [PubMed] [Google Scholar]
- 54.Sivanesam K, Byrne a, Bisaglia M, Bubacco L, Andersen N. RSC Adv. 2015;5(15):11577. doi: 10.1039/C5RA00325C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.López M, Lacroix E, Ramírez-Alvarado M, Serrano L. J Mol Biol. 2001;312(1):229. doi: 10.1006/jmbi.2001.4918. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.